Abstract
Mouse has emerged as the most common model organism in biomedicine. Here, we analyzed the tolerance to the loss-of-function (LoF) of selenoprotein genes, estimated from mouse knockouts and the frequency of LoF variants in humans. We found not only a general correspondence in tolerance (e.g., GPX1, GPX2) and intolerance (TXNRD1, SELENOT) to gene LoF between humans and mice but also important differences. Notably, humans are intolerant to the loss of iodothyronine deiodinases, whereas their deletion in mice leads to mild phenotypes, and this is consistent with phenotype differences in selenocysteine machinery loss between these species. In contrast, loss of TXNRD2 and GPX4 is lethal in mice but may be tolerated in humans. We further identified the first human SELENOP variants coding for proteins varying in selenocysteine content. Finally, our analyses suggested that premature termination codons in selenoprotein genes trigger nonsense-mediated decay, but do this inefficiently when UGA codon is gained. Overall, our study highlights differences in the physiological importance of selenoproteins between human and mouse.
Keywords: selenocysteine, selenoprotein, human, mouse, knockout, mutation
Introduction
Selenoproteins are a unique class of proteins that use selenium in the form of selenocysteine (Sec), the 21st amino acid. Sec is normally present at active sites of enzymes to perform catalytic redox reactions, such as reduction of thioredoxin (thioredoxin reductases), removal of hydroperoxides (glutathione peroxidases), repair of oxidized methionines in proteins (methionine-R-sulfoxide reductases), and activation and inactivation of thyroid hormones (iodothyronine deiodinases). The synthesis of selenoproteins involves recoding, whereby specific UGA codons are redefined to specify Sec insertion. UGA recoding is dependent on a cis-acting stem loop in the 3′UTR of the mRNA (SECIS element) and a dedicated set of trans-acting factors (Labunskyy et al. 2014).
The human genome encodes 25 selenoprotein genes (Kryukov et al. 2003), most of which are conserved across mammals (Mariotti et al. 2012). Selenium is an essential trace element in mammals, whose biological effects are mediated by selenoproteins encoded by these 25 genes. Many of these proteins evolved in early eukaryotes but were either lost or replaced Sec with cysteine (Cys) in different lineages. Nonmammal model organisms typically have fewer selenoprotein genes than humans, for example, three in Drosophila melanogaster (Castellano et al. 2001; Martin-Romero et al. 2001), one in Caenorhabditis elegans (Taskov et al. 2005), and none in Saccharomyces cerevisiae (Lobanov et al. 2009).
Mice have 24 selenoprotein genes, with the only difference with the human selenoproteome being Gpx6, which inserts Cys in place of the human Sec (Gladyshev et al. 2016). Studies in mice offered invaluable insights into selenoprotein functions, expression patterns, and responses to dietary selenium. The mouse models of selenoprotein deficiency have been particularly useful. Knockout (KO) studies showed that selenoproteins Txnrd1 (cytosolic thioredoxin reductase) (Jakupoglu et al. 2005), Txnrd2 (mitochondrial thioredoxin reductase) (Conrad et al. 2004), Gpx4 (phospholipid hydroperoxide glutathione peroxidase) (Imai et al. 2003; Yant et al. 2003), and Selenot (oxidoreductase of unknown function) (Boukhzar et al. 2016) are essential for embryonic development, further supporting the essential role of selenium.
Consistent with the indispensability of the four aforementioned selenoproteins, factors required for their synthesis are also essential in mice, including Sec tRNA (Trsp) (Bösl et al. 1997; Kumaraswamy et al. 2003) and SECIS-binding protein 2 (Secisbp2) (Seeher et al. 2014). The known set of factors needed for Sec biosynthesis and insertion in eukaryotes, here referred as Sec machinery, consists of Trsp, SECISBP2, phosphoseryl-tRNA kinase (PSTK), Sec synthase (SECS or SEPSECS), selenophosphate synthetase 2, and Sec-specific elongation factor (EEFSEC). Additional factors have been found to be involved in Sec insertion, such as SECP43 (TRNAU1AP), whose inactivation is embryonic lethal (Mahdi et al. 2015), but it may not have a direct role in Sec insertion or to be specific to Sec. We also included in our analysis two genes with homology to Sec machinery genes: selenophosphate synthetase 1 (SPS1 or SEPHS1) and SECIS-binding protein 2-like (SBP2L or SECISBP2L); these genes, however, have unclear functions and are not known to be essential for selenoprotein biosynthesis (Persson et al. 1997; Donovan and Copeland 2012).
Mouse is currently the most common model organism to assess the role of selenium in human health. Yet, mouse studies cannot always be extrapolated to humans, for example, drugs developed in mouse models of human disease often do not work in humans. Differences between humans and mice encompass many physiological and molecular aspects; mice are among the smallest and shortest-lived mammals, whereas humans are large and long-lived. Considering that many insights into human selenium biology are inferred from studies in mice, it is important to determine the functional correspondence between mouse and human selenoproteins and establish the relative tolerance of these organisms to selenoprotein deficiency or loss.
The dispensability of individual genes in humans can be assessed by quantifying the natural occurrence of loss-of-function (LoF) variants in populations. LoF variants that cause disruption of essential genes are strongly selected against, and their frequency in the general population is very low. In essence, abundant human genome sequence data allow to estimate the gene-specific constraints against LoF variants, by counting occurrences of natural human KOs. The large-scale joint-genotyping database gnomAD is sufficiently powered for this purpose (Karczewski et al. 2019). gnomAD provides with gene constraint metrics based on the observed and expected LoF variants. In contrast, the loss of mouse selenoproteins is typically engineered in the lab through various KO models. However, large-scale projects such as IMPC (Dickinson et al. 2016) aim to assess the consequences of loss of every mouse gene. Here, we use human LoF variant metrics from gnomAD, mouse KO data from IMPC and other sources, to assess the relative importance of human and mouse selenoproteins.
Results
Tolerance to Inactivation of Selenoprotein Genes
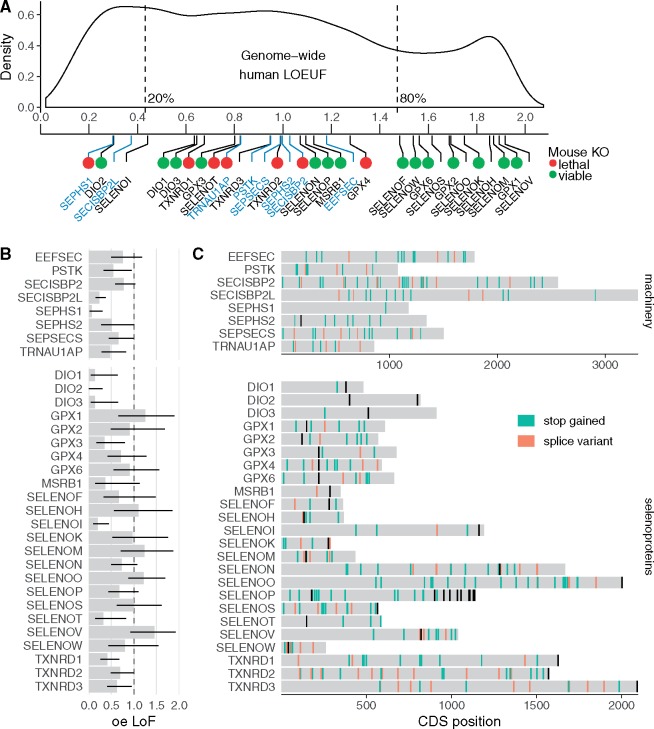
To assess the tolerance to inactivation of selenoproteins and Sec machinery genes, we first used the human constraint metrics provided by gnomAD v2.1 (Karczewski et al. 2019). gnomAD considers LoF variants leading to truncated gene products (stop gains and splice variants), and in the current study we use the same definition of LoF variants (i.e., do not consider missense mutations). We employed the oe LoF score (ratio of observed to expected occurrence of LoF variants; the lower the ratio, the stronger the selective constraints) and LoF observed/expected upper-bound fraction (LOEUF), the oe LoF 90% confidence interval upper limit, and the probability of being LoF intolerant (pLI). We then compared those estimates with phenotypes in mouse models of selenoprotein deficiency. It has been established that the LOEUF score correlates with measures of genic sensitivity to disruption (Karczewski et al. 2019): The lower extreme of the LOEUF distribution is enriched with haploinsufficient genes, whereas unconstrained genes and nonessential genes are found at the upper extreme. Autosomal recessive genes are found in the middle of the distribution, where selection against heterozygous LoF is present but weak. We classified selenoproteins and Sec machinery genes based on their LOEUF score (fig. 1A).
Fig. 1.
Loss-of-function (LoF) variants in human selenoproteins and tolerance to gene inactivation. (A) Genome-wide LOEUF score distribution of human genes and the corresponding values for selenoproteins (black labels) and Sec machinery genes (blue labels). A lower LOUEF indicates stronger selective constraints against LoF variants. Green and red dots indicate whether global homozygous deletion of the ortholog in mice is viable or lethal, respectively. IMPC reports contradictory phenotype for GPX3 and SECISBP2 (supplementary table S2, Supplementary Material online). (B) oe (observed/expected) LoF score for machinery genes and selenoproteins. Thin black lines correspond to the 90% confidence interval (its upper limit, LOEUF, is shown in panel A). (C) Observed nonsense and splice variants in human Sec machinery genes and selenoproteins (allele frequency <= 0.1 and median coverage >= 1) in the CDS of the representative transcripts (supplementary table S5, Supplementary Material online). Black sites correspond to annotated UGA-Sec. Variants and gene constraint scores from gnomAD v2.1. LOEUF, LoF oe upper-bound fraction (oe LoF 90% CI upper limit); oe LoF, observed/expected loss-of-function ratio; CDS, coding sequence.
Genes with Strong Selective Constraints
We found that the lowest 20% of human genes in the LOEUF distribution includes two selenoproteins, DIO2 and SELENOI, and two Sec machinery paralogs of unknown function, SEPHS1 and SECISBP2L. These four genes appeared to be strongly depleted of LoF variants suggesting that they do not tolerate the loss of even one of the two alleles in humans.
Unconstrained Genes
At the higher end of the LOEUF distribution (ninth to tenth deciles genome-wide), we found 11 selenoproteins (fig. 1A), representing almost half of the human selenoproteome. Seven of them are not essential for development in mice: GPX1, GPX2, GPX6 (Cys homolog in mice), SELENOF, SELENOK, SELENOM, and SELENOW. No mouse models have been reported for the remaining four selenoproteins: SELENOH, SELENOO, SELENOS, and SELENOV. Interestingly, SELENOV was lost in gorilla (Ventura et al. 2011). High LOEUF scores indicate a relatively high tolerance to inactivation. Typically, many such genes harbor homozygous LoF variants (Karczewski et al. 2019). Indeed, we detected three homozygous LoF variants in human selenoprotein genes among 141,565 individuals. These were three frameshift-causing variants in SELENOH, SELENOO, and SELENOV (supplementary table S1, Supplementary Material online).
Weak Selective Constraints against Heterozygous LoF
The rest of the genes fell in the middle of the LOEUF distribution (fig. 1A), including the factors for selenoprotein synthesis PSTK, SEPSECS, SEPHS2, SECISBP2, and EEFSEC. Genes with similar LOEUF scores are enriched for autosomal recessive genes (Karczewski et al. 2019). Consistently, heterozygous deletion of Secisbp2 (Seeher et al. 2014) and Trsp (Kumaraswamy et al. 2003; Carlson et al. 2004) in mice is tolerated, whereas homozygous deletion is lethal. We also found the remaining 11 human selenoproteins in this category: DIO1, DIO3, GPX3, GPX4, MSRB1, SELENON, SELENOT, SELENOP, TXNRD1, TXNRD2, and TXNRD3. Consistent with this classification, homozygous KO mouse models of Txnrd1, Txnrd2, Selenot, and Gpx4 are embryonic lethal, but the corresponding heterozygous mutants are viable and mostly indistinguishable from wild type (reviewed in [Conrad and Schweizer 2016]).
Differences in Tolerance to Selenoprotein Loss between Human and Mouse
Iodothyronine Deiodinases Differ in Tolerance to LoF between Human and Mouse
The three deiodinases (DIO1, DIO2, and DIO3) appeared to be among the selenoproteins with strongest intolerance against LoF variants in human (fig. 1B). Their oe LoF scores ranged between 0 and 0.18, thus, at most, only 18% of the expected LoF variance was observed. No human LoF variants were found in DIO2 (ENST00000438257), and with the pLI > 0.9 (table 1), this gene was predicted to be intolerant to LoF variants even in heterozygous state. The three DIO enzymes are involved in the regulation of thyroid hormone by reductive deiodination. Thyroid hormone regulates a variety of processes, including growth, development, and metabolic rate. Thyroxine (T4) is the main product of the thyroid gland, but it has low biological activity, whereas the active triiodothyronine (T3) has a higher ∼10-fold affinity for thyroid hormone receptors. DIO1 and DIO2 catalyze the activation of T4 to T3. Conversely, DIO3, and in some conditions DIO1, can inactivate T3 by producing the inactive metabolites T2 and reverse T3 (rT3), respectively. Studies in deiodinase-deficient mice have confirmed the function of deiodinases for T3 formation (Schneider et al. 2001, 2006; Fonseca et al. 2014; Schmitt et al. 2014; Martinez et al. 2016). However, Dio1 and Dio2 KO mice showed only a mild phenotype, with normal serum T3 level and unimpaired growth, reproductive capacity, and general health (Ng et al. 2004, 2009; Schneider et al. 2006; Hernandez et al. 2007; Williams and Bassett 2011). Even double Dio1/Dio2 KO mice were viable and showed roughly additive effects of the two phenotypes (Galton et al. 2009). Thus, our analyses revealed major differences in the physiological importance of deiodinases between human and mouse, wherein these genes are essentially intolerant to the loss of even one allele in humans, but tolerate complete KO in mice.
Table 1.
Gene Constraint Metrics for Human Selenoproteins from gnomAD v2.1.
| Gene | LOEUF | Decile | oe lof | pLI | LoF |
|---|---|---|---|---|---|
| Selenoproteins | |||||
| DIO1 | 0.63 | 3 | 0.13 | 0.63 | 1 |
| DIO2 | 0.30 | 1 | 0.00 | 0.96 | 0 |
| DIO3 | 0.65 | 3 | 0.14 | 0.62 | 1 |
| GPX1 | 1.89 | 9 | 1.25 | 0.00 | 6 |
| GPX2 | 1.69 | 8 | 0.90 | 0.00 | 6 |
| GPX3 | 0.80 | 4 | 0.35 | 0.05 | 4 |
| GPX4 | 1.28 | 6 | 0.71 | 0.00 | 8 |
| GPX6 | 1.56 | 8 | 0.91 | 0.00 | 9 |
| MSRB1 | 1.13 | 6 | 0.36 | 0.13 | 2 |
| SELENOF | 1.48 | 7 | 0.66 | 0.01 | 4 |
| SELENOH | 1.85 | 9 | 1.10 | 0.00 | 5 |
| SELENOI | 0.44 | 1 | 0.19 | 0.64 | 4 |
| SELENOK | 1.75 | 8 | 0.97 | 0.00 | 6 |
| SELENOM | 1.87 | 9 | 1.23 | 0.00 | 8 |
| SELENON | 1.07 | 5 | 0.72 | 0.00 | 18 |
| SELENOO | 1.69 | 8 | 1.21 | 0.00 | 24 |
| SELENOP | 1.09 | 5 | 0.68 | 0.00 | 12 |
| SELENOS | 1.62 | 8 | 0.99 | 0.00 | 11 |
| SELENOT | 0.82 | 4 | 0.32 | 0.12 | 3 |
| SELENOV | 1.92 | 9 | 1.45 | 0.00 | 13 |
| SELENOW | 1.54 | 7 | 0.80 | 0.00 | 6 |
| SEPHS2 | 1.00 | 5 | 0.51 | 0.00 | 6 |
| TXNRD1 | 0.68 | 3 | 0.41 | 0.00 | 11 |
| TXNRD2 | 0.99 | 5 | 0.69 | 0.00 | 21 |
| TXNRD3 | 0.95 | 4 | 0.62 | 0.00 | 15 |
| Machinery | |||||
| EEFSEC | 1.18 | 6 | 0.76 | 0.00 | 14 |
| PSTK | 0.95 | 5 | 0.55 | 0.00 | 9 |
| SECISBP2 | 1.03 | 5 | 0.77 | 0.00 | 33 |
| SECISBP2L | 0.37 | 1 | 0.23 | 0.29 | 12 |
| SEPHS1 | 0.30 | 1 | 0.06 | 0.98 | 1 |
| SEPSECS | 0.99 | 5 | 0.65 | 0.00 | 16 |
| TRNAU1AP | 0.83 | 4 | 0.47 | 0.00 | 9 |
Weak Selection against LoF Variants in Human GPX4
GPX4 is the only member of the glutathione peroxidase family with the capacity to reduce phospholipid hydroperoxides. Due to its role in clearing this form of oxidative damage, it is considered the main player in the recently discovered form of nonapoptotic cell-death ferroptosis (Friedmann Angeli et al. 2014; Ingold et al. 2018). Consistently, deletion of Gpx4 in mice causes early embryonic lethality (Imai et al. 2003; Yant et al. 2003). Recently, it was shown that mouse embryonic fibroblasts could tolerate Trsp (Sec tRNA) loss if selenocysteine in Gpx4 is replaced by cysteine, but not otherwise, leading to the conclusion that the essentiality of selenocysteine could be mainly ascribed to the function of Gpx4 (Ingold et al. 2018). We thus expected human GPX4 to present the highest intolerance to LoF in humans. Instead, its oe LoF score was not particularly low (0.65; 64th percentile genome-wide) and its LOEUF (1.28) estimates a relatively weak selection against LoF variants. In fact, a patient with a homozygous nonsense variant was reported to be born (Smith et al. 2014). Nonetheless, all variants observed in gnomAD were heterozygous, which is consistent with the fact that heterozygous Gpx4 +/− mice showed no phenotype and lived normally (Imai et al. 2003; Yant et al. 2003).
SEPHS1 Is Intolerant to LoF Variants in Humans
Humans and mice have two genes in the selenophosphate synthetase family: SEPHS2 and SEPHS1. SEPHS2 is a selenoprotein that synthesizes selenophosphate using ATP and selenide as substrates (Veres et al. 1994) and is a member of the Sec machinery. SEPHS1 is a SEPHS2 paralog found in vertebrates, insects, and other animals (Mariotti et al. 2015), but it is not a selenoprotein and has no catalytic activity for selenophosphate synthesis (Persson et al. 1997). Deletion of Sephs1 in mice is embryonic lethal (Tobe et al. 2016). SEPHS1 had the second lowest oe LoF score in our analysis (0.06; tenth percentile genome-wide), and a pLI > 0.9 (table 1), indicating intolerance to loss of even one allele in humans. Although humans seem to not tolerate the loss of one copy of SEPHS1, Sephs1 heterozygous KO mice are viable and show no obvious phenotypes (Tobe et al. 2016).
Commonalities in Tolerance to Selenoprotein Loss between Mouse and Human
Essential Selenoproteins in Mice
To date, KO mice were developed for 18 of the 24 selenoproteins. Global homozygous deletion resulted in embryonic lethality for Txnrd1, Txnrd2, Selenot, and GPx4. The gnomAD gene constraints indicate that GPX4 has weaker selection against inactivation in humans than in mice (see above), whereas TXNRD1, TXNRD2, and SELENOT had rather low oe LoF scores (fig. 1B). The LOEUF score in all four genes suggests tolerance to heterozygous LoF variants but not to homozygous, which is consistent with observations in mice. The cytosolic thioredoxin reductase Txnrd1 keeps thioredoxin 1 (Txn1) in the reduced state and is important for cell cycle progression. The mitochondrial thioredoxin reductase Txnrd2 catalyzes the reduction of mitochondrial thioredoxin and is involved in hematopoiesis and heart function. Deletion of either Txnrd1 (Jakupoglu et al. 2005) or Txnrd2 (Conrad et al. 2004) is embryonic lethal in mice, but heterozygous mutants are viable and fertile, and show no obvious phenotype (Conrad et al. 2004; Jakupoglu et al. 2005). Similarly, deletion of Selenot, a thioredoxin-like selenoprotein localized in the endoplasmic reticulum, is lethal during embryogenesis, but heterozygous deletion is viable. However, the percentage of collected animals was lower than expected and had smaller size compared with wild type (Boukhzar et al. 2016). SELENOT in humans exhibited one of the lowest oe LoF scores among selenoproteins (only the three DIOs and SELENOI scored lower). Consistently, it is the selenoprotein with the highest degree of sequence conservation among mammals (Mariotti et al. 2012).
Nonessential Selenoproteins in Mice
Studies in mice revealed a number of selenoprotein genes whose deletion resulted in viable animals, indicating they are not essential during development. Deletion of Gpx1 (Ho et al. 1997), Gpx2 (Esworthy et al. 2001), and Gpx3 (Olson et al. 2010; Jin et al. 2011) produced viable animals with no gross phenotypes (although IMPC reports Gpx3 as lethal [supplementary table S2, Supplementary Material online], we consider more reliable the significant body of experimental data suggesting that Gpx3 is not essential); Selenom KO mice were also viable and their phenotype was, apart from weight gain, indistinguishable from wild-type littermates (Pitts et al. 2013); Selenok gene loss in mice led to Ca2+ flux defects during immune cell activation and an impaired immune response, but the animals were viable (Verma et al. 2011); Selenow KO mice are reported as viable in IMPC, but with reduced leukocyte count (supplementary table S2, Supplementary Material online); Selenof KO mice are viable and showed only a mild phenotype of congenital cataract (Kasaikina et al. 2011). Consistent with disposable functions in humans too, the LOEUF scores for GPX1, GPX2, SELENOM, SELENOK, SELENOF, and SELENOW were among the highest for selenoproteins (fig. 1A).
Selenoprotein Machinery Genes
The Sec machinery consists of Trsp, PSTK, SEPSECS, SEPHS2, SECISBP2, and EEFSEC. Our analysis supports that selenoprotein machinery genes are intolerant to homozygous LoF variants both in humans and mice. The selenoprotein machinery factors had a LOEUF score roughly centered at 1 (fig. 1A), indicating weak selection against heterozygous LoF variants, but no homozygous high-impact LoF variants, as defined by gnomAD (i.e., stop codon gain and splice variants), were observed. Likewise, the missense mutations in machinery genes SECISBP2 and SEPSECS described in patients (Schoenmakers, Schoenmakers, et al. 2016) were not observed in homozygous state.
Humans and mice have two paralogous SECIS-binding proteins, SECISBP2 and SECISBP2L. The former is thought to be the obligate factor for selenoprotein synthesis and is embryonic lethal in mice (Seeher et al. 2014), although recent data led to question its exclusive requirement for Sec insertion (Fradejas-Villar et al. 2017). Despite conservation of the Sec insertion domain and the RNA binding domain (Donovan and Copeland 2009), SECISBP2L does not support Sec insertion and was found to bind only weakly to GPX4 SECIS (Copeland and Driscoll 2001). SECISBP2L appeared to have a particularly low oe LoF score in humans (0.19; 24th percentile genome-wide), and pLI > 0.9, suggesting intolerance to LoF variants, even in one of the two alleles. Yet, its role in selenoprotein synthesis remains unclear. In contrast, SECISBP2 appeared to be more tolerant to heterozygous LoF (oe = 0.77; 75th percentile genome-wide). Several mutations in SECISBP2 have been identified in patients with abnormal thyroid function tests (due to deficiency of Sec-dependent deiodinase activity) and low Se plasma levels, among other signs (reviewed in Schoenmakers, Schoenmakers, et al. [2016] and Fradejas-Villar [2018]).
The Sec tRNA is a key factor in selenoprotein synthesis. Deletion of Trsp, the Sec tRNA gene, is embryonic lethal in mice (Bösl et al. 1997; Kumaraswamy et al. 2003). A human homozygous mutation G65C in Trsp was detected in a patient with impaired deiodinase activity and low selenium plasma levels. Expression profiles of fibroblasts and plasma from the patient showed selective reduction of stress-related selenoproteins, for example, GPX1 and GPX3, but preservation of housekeeping selenoproteins, for example, TXNRDs and GPX4. The study concluded that the homozygous variant G65C impairs maturation of tRNA[Ser]Sec with the reduction of tRNA[Ser]Sec levels (Schoenmakers, Carlson, et al. 2016). Maturation of tRNA[Ser]Sec involves base modifications yielding two major isoforms containing either 5-methoxycarbonylmethyluridine (mcm5U) or 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) at position 34. Each subtype has different roles in selenoprotein synthesis, with housekeeping selenoproteins being dependent on mcm5U, whereas stress-related selenoproteins depend on mcm5Um. The heterozygous variant G65C yielded lower levels of tRNA[Ser]Sec (40% reduction), but selenoprotein synthesis was not affected, suggesting that the moderate reduction in tRNA[Ser]Sec levels is not rate limiting for selenoprotein synthesis (Schoenmakers, Carlson, et al. 2016). We explored the genetic variance in Trsp in human populations. The functional constraint scores in gnomAD only exist for protein coding genes. Instead, we explored the occurrence of variants affecting the Trsp locus using the 15,708 whole genomes (exomes could not be considered because they do not include tRNAs). A total of 35 variants were observed in the Trsp locus, all of them heterozygous (supplementary table S3, Supplementary Material online). It is not clear to what extent the observed variants would affect tRNA maturation and functionality, but we expect all those individuals to have at least one fully functional allele, which may be sufficient for normal selenoprotein synthesis levels, as observed in heterozygous G65C (Schoenmakers, Carlson, et al. 2016). This agrees with the fact that heterozygous Trsp KO in mice can be well tolerated (Kumaraswamy et al. 2003; Carlson et al. 2004).
Selenoproteins with No Mouse KO Model
To our knowledge, no published mouse models were produced for seven selenoproteins: SELENOH, SELENOI, SELENOO, SELENOS, SELENOV, SEPHS2, and TXNRD3. We explored their tolerance to LoF in humans through the gnomAD database. SELENOI (or EPT1) had the second lowest LOEUF score among selenoproteins (fig. 1A), indicating strong selection against LoF, thus, potentially, it is one of the most important selenoproteins in humans. SELENOI was proposed to be involved in the synthesis of ethanolamine glycerophospholipids (Horibata and Hirabayashi 2007), but further studies are needed to establish its function. The protein has a conserved CDP-alcohol phosphatidyltransferase domain, which is also found in choline phosphotransferases (CHPT1) and choline/ethanolamine phosphotransferases (CEPT1). The gene is a selenoprotein only in vertebrates (Mariotti et al. 2012), but the role of Sec in its C-terminal extension is currently unknown. On the other hand, SELENOH, SELENOO, SELENOS, SELENOV, and GPX6 had high LOUEF scores, all found among the top 20% of the genome-wide distribution (fig. 1A). Deletion of selenoproteins with similar LOEUF in mice produced viable animals (e.g., GPX1, GPX2, GPX6, SELENOM, and SELENOK). We considered those genes to be unconstrained in terms of selection against LoF. Finally, SEPHS2 and TXNRD3 were found in the middle of the LOEUF distribution consistent with autosomal recessive inheritance mechanism.
Selenoproteins with Frequent UGA Gain Variants
Stop gain is a class of LoF variant that introduce a premature termination codon (PTC) in the coding sequence. Translation of such mRNAs would normally result in a truncated protein product. The most notable feature of selenoprotein transcripts is the ability to redefine UGA codons during translation to specify Sec insertion, mediated by the presence of a SECIS stem loop in the 3′UTR. Previously, it was shown that Sec could be inserted at any tested position in TXNRD2 (Turanov et al. 2013) and in GPX1 (Wen et al. 1998), but only at its natural position and its vicinity in the C-terminal region of TXNRD1 (Turanov et al. 2013). We analyzed the occurrence of stop gain variants in human selenoprotein genes. In this analysis, we discriminated TGA variants from TAG and TAA, to account for the fact that UGA could potentially be translated as Sec, therefore resulting in functional proteins with additional Sec residues.
To assess the frequency of stop gain in a given transcript sequence, we developed a metric to normalize the number of observed stop gains to the number of codons that could become a stop signal by a single substitution in the transcript sequence (supplementary table S4, Supplementary Material online). We called this metric dSTOP, and its principle is similar to Ka (the number of nonsynonymous substitutions per nonsynonymous site) in the Ka/Ks ratio. The metric considers only single nucleotide variants (SNVs) and can be computed for each transcript and each stop codon independently. We used the stop gain variants present in gnomAD v2.1.
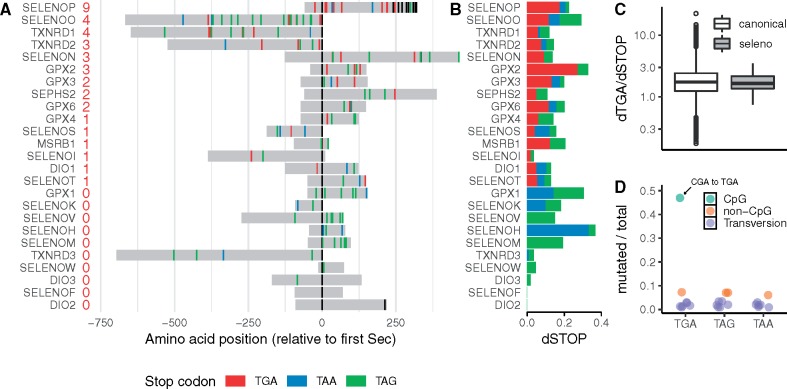
SELENOP was the selenoprotein with the highest number of TGAs gained, 9 in total (fig. 2A). Its dSTOP for TGA (dTGA = 0.18) was much higher than dTAA (0.03) and dTAG (0.02), thus TGA was the most frequently gained stop. Considering that SELENOP has the ability to incorporate multiple Sec, due to its two SECIS elements, it seems plausible that additional UGA codons could be translated as Sec residues, especially those in the C-terminal region (fig. 2A). On the other hand, there were selenoproteins in which TGA gain was not observed whereas the other two stops were (fig. 2B). A particularly interesting example is GPX1, the most abundant mammalian selenoprotein that accounts for ∼50% of Se in mouse liver (Hill et al. 2012). GPX1 had the 2nd highest LOEUF among selenoproteins, indicating that it tolerates LoF variants, including stop gain. Consistently, its global dSTOP score (0.11) was among the highest for selenoproteins (table 2). Its sequence (ENST00000419783) has eight codons that could potentially mutate to TGA, but no TGA gain was observed. We speculate that additional Sec residues in a highly abundant selenoprotein could sequester the available pool of Sec tRNA, affecting Sec insertion into other selenoproteins, and therefore TGA gain might be strongly selected against. However, we cannot exclude the possibility that the lack of TGA gain in GPX1 was by chance, due to the lack of CGA codons in its sequence (CGA to TGA is the most common stop gain mutation, as discussed below). Nonetheless, TGA gain was observed in other selenoproteins without CGA codons, that is, GPX6 and SELENOM (supplementary table S7, Supplementary Material online).
Fig. 2.
Stop gain variants in human selenoprotein genes. (A) Position of observed stop gain variants found in the analysis of more than 140,000 individuals. Selenoproteins (shown in gray) are aligned based on their Sec residues (shown in black). The numbers in red indicate observed TGA codons. (B) dTGA, dTAA, and dTAG metrics for each of the 25 selenoproteins. (C) Distribution of dTGA/dSTOP index (Materials and Methods) in canonical genes and selenoproteins (log scale). The ten selenoproteins with no TGA gain (see panel A) are not included. (D) Proportion of mutated codons for each of the potential stop gain codons genome-wide. Each dot corresponds to 1 codon among the 18 codons that are 1 mutation away from a stop (supplementary table S4, Supplementary Material online). The color indicates the type of mutation: CpG transition (green); non-CpG transition (orange); and transversion (purple). Stop gain variants obtained from gnomAD v2.1.
Table 2.
Stop Gain Metrics for Human Selenoproteins.
| Gene | dTGA/dSTOP | dSTOP | dTGA | dTAA | dTAG | oSTOP | oTGA | oTAA | oTAG | pSTOP | pTGA | pTAA | pTAG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DIO1 | 3.31 | 0.02 | 0.06 | 0.00 | 0.00 | 1 | 1 | 0 | 0 | 53 | 16 | 11 | 26 |
| DIO2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 | 0 | 0 | 0 | 96 | 26 | 23 | 47 |
| DIO3 | 0.00 | 0.01 | 0.00 | 0.00 | 0.02 | 1 | 0 | 0 | 1 | 86 | 12 | 17 | 57 |
| GPX1 | 0.00 | 0.11 | 0.00 | 0.00 | 0.16 | 5 | 0 | 0 | 5 | 46 | 8 | 7 | 31 |
| GPX2 | 3.22 | 0.08 | 0.27 | 0.00 | 0.06 | 5 | 3 | 0 | 2 | 59 | 11 | 13 | 35 |
| GPX3 | 2.80 | 0.02 | 0.07 | 0.00 | 0.02 | 2 | 1 | 0 | 1 | 84 | 15 | 22 | 47 |
| GPX4 | 1.35 | 0.09 | 0.13 | 0.00 | 0.11 | 6 | 2 | 0 | 4 | 65 | 16 | 11 | 38 |
| GPX6 | 1.48 | 0.08 | 0.12 | 0.08 | 0.07 | 7 | 2 | 2 | 3 | 88 | 17 | 25 | 46 |
| MSRB1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0 | 0 | 0 | 0 | 41 | 8 | 8 | 25 |
| SELENOF | 1.14 | 0.04 | 0.04 | 0.00 | 0.06 | 2 | 1 | 0 | 1 | 57 | 25 | 15 | 17 |
| SELENOH | 0.00 | 0.08 | 0.00 | 0.33 | 0.03 | 3 | 0 | 2 | 1 | 40 | 4 | 6 | 30 |
| SELENOI | 2.13 | 0.02 | 0.04 | 0.00 | 0.02 | 3 | 2 | 0 | 1 | 163 | 51 | 51 | 61 |
| SELENOK | 0.00 | 0.07 | 0.00 | 0.10 | 0.17 | 3 | 0 | 1 | 2 | 41 | 19 | 10 | 12 |
| SELENOM | 2.50 | 0.10 | 0.25 | 0.10 | 0.08 | 4 | 1 | 1 | 2 | 40 | 4 | 10 | 26 |
| SELENON | 2.21 | 0.07 | 0.15 | 0.00 | 0.07 | 11 | 4 | 0 | 7 | 158 | 26 | 29 | 103 |
| SELENOO | 1.17 | 0.10 | 0.12 | 0.06 | 0.11 | 19 | 4 | 2 | 13 | 189 | 34 | 35 | 120 |
| SELENOP | 2.13 | 0.08 | 0.18 | 0.02 | 0.07 | 14 | 9 | 1 | 4 | 169 | 51 | 64 | 54 |
| SELENOS | 1.50 | 0.11 | 0.17 | 0.14 | 0.03 | 10 | 4 | 5 | 1 | 90 | 24 | 37 | 29 |
| SELENOT | 1.37 | 0.04 | 0.06 | 0.04 | 0.03 | 3 | 1 | 1 | 1 | 74 | 18 | 26 | 30 |
| SELENOV | 0.00 | 0.11 | 0.00 | 0.00 | 0.18 | 7 | 0 | 0 | 7 | 65 | 10 | 15 | 40 |
| SELENOW | 2.47 | 0.08 | 0.20 | 0.00 | 0.09 | 3 | 1 | 0 | 2 | 37 | 5 | 10 | 22 |
| SEPHS2 | 0.84 | 0.06 | 0.05 | 0.00 | 0.12 | 8 | 2 | 0 | 6 | 135 | 40 | 44 | 51 |
| TXNRD1 | 1.53 | 0.04 | 0.07 | 0.01 | 0.06 | 10 | 4 | 1 | 5 | 226 | 59 | 82 | 85 |
| TXNRD2 | 1.82 | 0.08 | 0.15 | 0.03 | 0.08 | 13 | 6 | 1 | 6 | 154 | 39 | 37 | 78 |
| TXNRD3 | 0.00 | 0.02 | 0.00 | 0.02 | 0.03 | 5 | 0 | 2 | 3 | 278 | 65 | 100 | 113 |
Note.—Shown are dTGA/dSTOP index; frequency of stop gain considering all stops (dSTOP) and each stop codon separately (dTGA, dTAA, and dTAG); number of observed (o) and potential (p) variants in the transcript sequence for all stops (STOP) and each stop codon separately.
Next, we investigated whether selenoproteins had, in general, a higher frequency of TGA gain compared with canonical genes. We assessed the frequency of gain of each stop codon in selenoproteins and compared it to canonical genes. We computed the proportion of dTGA over the total dSTOP (considering all three stops). The index is designated as dTGA/dSTOP. We computed dTGA/dSTOP for all genes, including selenoproteins (table 2). The dTGA/dSTOP of selenoprotein genes did not significantly deviate from the rest of the genome as a whole (Kruskal–Wallis test; P value = 0.13) (fig. 2C). The null expectation for canonical genes, if mutations were completely random, would be an index of 1, where the frequency of TGA is not different from all stops. Instead, the median of dTGA/dSTOP in canonical genes was 1.53, thus, TGA is the most frequently gained stop codon genome-wide. Such enrichment in TGA gain is readily explained by the mutational bias in the human genome. Mutations are not randomly distributed, for example, CpG sites (a dinucleotide consisting in CG) are hotspots for mutations. The most common mutations are CpG transitions, for example, CG to TG (Lek et al. 2016). This mutation bias has a strong effect in stop gain variants because there is only one possible CpG transition SNV that can introduce a stop codon: CGA to TGA. This particular variant was by far the most frequently observed in the gnomAD data set (fig. 2D). Because of mutational bias in the human genome, TGA gain is particularly abundant and dilutes the statistical power to detect a difference in terms of tolerance to TGA gain, between selenoproteins and canonical genes.
PTCs in Selenoproteins Trigger Nonsense-Mediated Decay
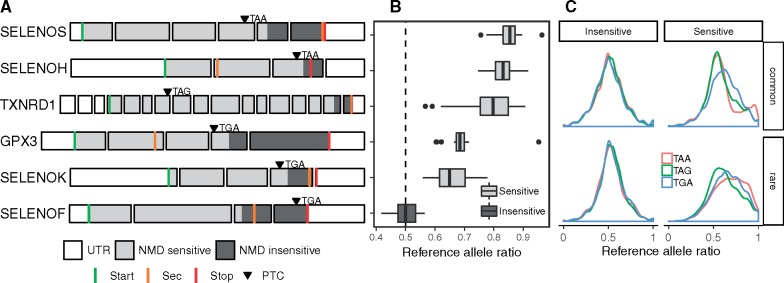
The presence of a PTC in mRNAs may trigger nonsense-mediated decay (NMD), the surveillance pathway that protects the cell against translation of aberrant mRNAs (Maquat 1995). PTCs are predicted to be sensitive to NMD if located more than ∼50 nucleotides upstream of the 3′-most exon–exon junction (Nagy and Maquat 1998). In case of heterozygous sensitive PTC, only one of the two alleles is degraded by NMD, resulting in allele-specific expression (ASE). We explored the transcriptomic effect of PTCs in selenoprotein mRNAs using expression data generated by the Genotype-Tissue Expression (GTEx) project (Rivas et al. 2015). We first searched for stop gain variants in the 652 subjects from GTEx v7 (Materials and Methods), which resulted in six heterozygous stop gain variants in selenoproteins (GPX3, SELENOF, SELENOH, SELENOK, SELENOS, and TXNRD1) and one in SECISBP2. We then analyzed ASE for those variants in GTEx expression data. Based on the 50 base-pair (bp) rule, all variants were expected to be sensitive to NMD, with the exception of SELENOF, whose PTC was located in the last exon (fig. 3A). Indeed, ASE was observed in SELENOS, SELENOH, TXNRD1, GPX3, and SELENOK (fig. 3B) and was not observed in SELENOF and SECISBP2. Interestingly, ASE appeared to be stronger in selenoprotein genes where the stop gained was TAA or TAG, compared with those with TGA (P value = 1.6e-09; fig. 3B). No difference in ASE between the three codons was observed in PTCs genome-wide across GTEx subjects and tissues (fig. 4C). In addition, indel variants were observed in GTEx subjects (supplementary table S6, Supplementary Material online), although no ASE information could be obtained for them in GTEx v7.
Fig. 3.
Presence of premature termination codon (PTC) in selenoprotein transcripts triggers their nonsense-mediated decay (NMD) in human tissues. (A) Exon structure of selenoprotein genes in which a PTC (shown by arrowheads) was identified in GTEx subjects. The corresponding stop codon is indicated. NMD sensitive and insensitive transcript regions (based on the 50-bp rule) are shaded in light and dark gray, respectively. (B) Allele-specific expression (ASE) of selenoprotein transcripts with PTC events. Distribution of the reference allele ratios (proportion of RNA-seq reads supporting the reference allele) observed across multiple tissues. Expression from both alleles results in 0.5 score (shown by the dashed vertical line). Imbalance toward 1 indicates depletion of the allele carrying the PTC. (C) Distribution of reference allele ratios in 2,456 genome-wide PTCs in GTEx subjects. PTCs are classified as insensitive or sensitive to NMD, and as common or rare based on allele frequency (threshold 0.01). UTR, untranslated region; Sec, selenocysteine UGA codon. Transcript length not at scale; only 100 nt of 3′UTR shown. PTC variants and ASE data obtained from GTEx v7.
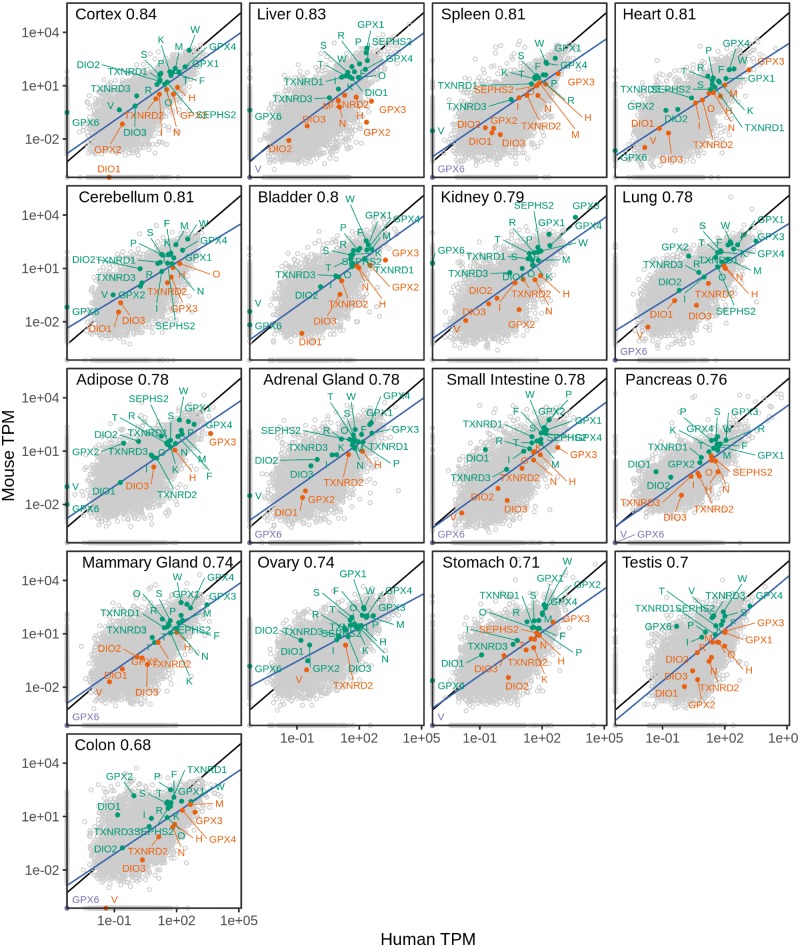
Fig. 4.
Correlation of selenoprotein gene expression between human and mouse across tissues. Each panel shows the tissue and the corresponding Spearman’s correlation coefficient (P value <1e-16 in all tissues). The blue line corresponds to the fitted linear model using 15,610 one-to-one orthologs (gray dots). Selenoproteins are labeled and colored based on the sign of the residuals, which indicates whether they are above or below the regression line. The prefix “SELENO” was removed from gene names, for example, V corresponds to SELENOV. Expression values in log scaled TPM (transcripts per million reads). RNA-seq quantification obtained from GTEx v7 (human) and ENCODE (mouse).
Correlation of Selenoprotein Gene Expression between Human and Mouse
We assessed the correspondence of selenoprotein gene expression between human and mouse using RNA-seq data from GTEx and mouse ENCODE. We compared the expression of 15,610 one-to-one orthologous genes, including selenoproteins and Sec machinery, in 17 different tissues (Materials and Methods). The highest correlation between the two species was found in frontal cortex (Spearman rho 0.84; P value <1e-16) (fig. 4). Expression was detected (TPM > 0) for almost all selenoproteins in all analyzed tissues, but GPX6 and SELENOV were consistently lowly expressed, or not detected, in all tissues on both species, except for testis. GPX6 and SELENOV are the most recently evolved selenoproteins in placental mammals (Mariotti et al. 2012), and recently evolved genes are known to be expressed at a lower level in mammals (Guschanski et al. 2017). DIO1 expression was not detected in the two available mouse cortex samples, whereas it was expressed in human cortex. Interestingly, GPX3 was the single most expressed gene (among all 15,610 one-to-one orthologs) in the kidney of both species. By fitting a regression linear model, we identified genes which were above or below the fitted expression value. Overall, we found that most selenoproteins had positive residuals (above the fitted regression line) across tissues, indicating that their expression level was higher in mouse than in human (supplementary fig. S2, Supplementary Material online). The Sec machinery genes, instead, showed residual values very close to 0, therefore their expression is more conserved between the two species compared with selenoproteins.
Discussion
The importance of selenoproteins in human health is manifested in health conditions associated with severe Se deficiency and by complex disorders caused by mutations in selenoprotein machinery genes, which impair the synthesis of multiple selenoproteins, as well as mutations in individual selenoproteins (Schoenmakers, Schoenmakers, et al. 2016; Fradejas-Villar 2018; Friedmann Angeli and Conrad 2018). Studies in mouse models provided mechanistic details of selenoprotein-dependent processes and had been particularly useful to study selenoproteins in organs that are not easily accessible in humans (Conrad and Schweizer 2016). It is commonly assumed that orthologs exhibit identical functions in related organisms. However, for most human and mouse orthologs, including selenoproteins, the correspondence of function and consequences of gene deficiency is not known. For example, SELENON-related myopathy, the first selenoprotein human genetic disorder identified (Moghadaszadeh et al. 2001), is not readily evident in mice lacking Selenon (Rederstorff et al. 2011; Moghadaszadeh et al. 2013).
We analyzed the gene constraint metrics in gnomAD (Karczewski et al. 2019), which quantify tolerance to gene inactivation by comparing the number of observed and expected occurrences of LoF variants across 141,456 human exomes and genomes. We compared the selective constraints in human selenoproteins with phenotypes of mouse models of selenoprotein deficiency, to address the correspondence in tolerance to selenoprotein gene inactivation between human and mouse. Although insightful, our comparative analysis comes with caveats. Even though efforts were made to provide a confident set of high-impact LoF variants through a careful filtering (Karczewski et al. 2019), some of the variants may in fact not have a LoF effect (for example, due to alterations in splicing), or may be compensated by indirect genetic mechanisms (El-Brolosy and Stainier 2017). On the other hand, gnomAD only considers the LoF variants with the most obvious effect of removing functional protein products (such as stop gain). Missense variants that inactivate specific gene functions are missed (such the known missense variants in SECISBP2, SEPSECS [Schoenmakers, Schoenmakers, et al. 2016], and DIO2 [Jo et al. 2019]). Additionally, although gene KO in mice is introduced by gene engineering, such KOs in humans (or their lack in the population) are the consequence of spontaneous mutations and subsequent selection, therefore a phenotype which is seemingly mild may actually be deleterious in nature, and thus cleared out in natural populations by selection. For all these reasons, caution is advised in interpreting differences between the two species.
Except for a few striking cases discussed later, we observed a general consistency between human and mouse in terms of tolerance to LoF in selenoproteins. The selenoprotein machinery genes Secisbp2 and Trsp are essential in mice, but heterozygous KO mutants show no phenotype. Consistent with being autosomal recessive also in humans, the machinery genes reside in the middle of the LOEUF distribution, which is enriched with autosomal recessive genes (SECISBP2 and SEPSECS are known to have autosomal recessive inheritance in humans). Similarly, the selenoproteins Selenot, Txnrd1 and Txnrd2 are essential in mice, but the heterozygous deletion is well tolerated. The human orthologs revealed LOEUF scores consistent with having weak constraints against heterozygous LoF, but not in homozygous state. Moreover, consistent with a high LOEUF score (among the top 20% genome-wide) for GPX1, GPX2, GPX6, SELENOF, SELENOK, SELENOM, and SELENOW, homozygous global deletion of the corresponding homologs in mice produced viable animals with no gross phenotypes.
Notably, our results revealed some striking differences between human and mouse. The three deiodinases showed strong selection against LoF variants in humans, whereas their deletion in mice produced viable animals with mild phenotypes. The enzymes DIO1 and DIO2 catalyze the activation step (T4 to T3 conversion) for thyroid hormone activity. It is estimated that ∼80% of the daily circulating T3 in humans is produced from T4 deiodination, whereas the remaining ∼20% is derived from direct thyroidal secretion. In rats, those percentages are ∼60% and ∼40% (Bianco et al. 2002). Metabolic alterations in DIO2 KO mice include significant deficit in TSH regulation (Schneider et al. 2001), thermogenesis (de Jesus et al. 2001; Fonseca et al. 2014), and auditory function (Ng et al. 2004). In humans, patients with inactivating mutations in SECISBP2 or Trsp, essential genes for selenoprotein synthesis, showed a phenotype with a specific signature of abnormal thyroid hormone metabolism caused by an impairment of T4 to T3 conversion. Within the general framework of selenoprotein deficiency, the lack of deiodinases seemed to be the most severe consequence in those patients. In contrast, patients with mutations in SEPSECS did not show thyroid metabolism problems. Instead, they shared neuropathological disorders (Schoenmakers, Schoenmakers, et al. 2016), which is consistent with GPX4 deficiency. Due to the evident contrast in phenotype severity between humans and mice, we ascribe the difference of tolerance to LoF variants in deiodinases to physiological differences between species, rather than to the disparity of the type of data analyzed. Our analyses, however, did not include missense variants such as p.Thr92Ala (rs225014) in DIO2, which has an impact on thyroid function in humans (Jo et al. 2019). This variant has high allele frequency (∼0.4) and is commonly observed in homozygosis (17% of subjects in gnomAD), which indicates the lack of selective constraints. Possibly, its deleterious effects appear at advanced age, so that selection is blind to them. Nevertheless, altogether our results point to iodothyronine deiodinases as the selenoproteins with the highest physiological importance in humans, unlike in mice.
Among the investigated genes, SEPHS1 stood out with the lowest LOEUF score (0.3) and the highest pLI (0.98), indicating a strong selection in humans against LoF variants, even in the heterozygous form. SEPHS1 (or SPS1) is the paralog of the selenoprotein SEPHS2 (or SPS2). SEPHS2 synthesizes selenophosphate, the Se donor for the synthesis of selenoproteins, and is found in all organisms with selenoproteins across the tree of life. Unlike SEPHS2, SEPHS1 is not a selenoprotein, it does not catalyze selenophosphate synthesis (Persson et al. 1997), and has a much more restricted distribution, being only present in some metazoan lineages. SEPHS1 evolved by SEPHS2 gene duplication independently in different lineages including vertebrates, insects, and other animals (Mariotti et al. 2015). In addition, SEPHS1 is found in selenoproteinless insects (Lobanov et al. 2007; Chapple and Guigó 2008; Mariotti et al. 2015), suggesting a function unrelated to selenoprotein synthesis. Deletion of Sephs1 in mice and flies is embryonic lethal (Alsina et al. 1999; Tobe et al. 2016), but heterozygous Sephs1+/− mice are viable and have no apparent phenotype (Tobe et al. 2016). A notable difference between humans and mice is the tolerance to heterozygous deletion of Sephs1: LoF variants in SEPHS1 in humans are extremely rare, indicating that even heterozygous LoF variants are not tolerated. The function of SEPHS1 remains unknown.
The LOEUF metrics can be used to identify genes intolerant to loss, regardless of existing mouse models. SELENOI appeared to have strong constraints against LoF, but no mouse model has been reported. Two recent studies showed that SELENOI is associated with hereditary spastic paraplegia, a group of neurodegenerative disorders. Patients had a homozygous missense variant (chr2:26373391 G > C; p.Arg112Pro) in the highly conserved Arg residue in the CDP-alcohol phosphatidyltransferase domain, which was found in four affected siblings (Ahmed et al. 2017); and a homozygous splice site mutation that generates splice variants with truncation in exons 6 and 8 was described in a patient with a severe form of hereditary spastic paraplegia at a very young age (Horibata et al. 2018). SELENOI is required for myelination and neurodevelopment, and consistent with such an essential function, the LOEUF score was among the low 20% genome-wide, and second lowest for selenoproteins (behind DIO2). These observations prompt further research into the function of this essential selenoprotein.
It is also of interest that the two essential selenoproteins in mouse, Txnrd2 and Gpx4, showed high tolerance to LoF in humans. Both genes have been associated with human disease. Mutations in GPX4 have been found in two patients with the rare Sedaghatian-type spondylometaphyseal dysplasia (Smith et al. 2014). Sedaghatian-type spondylometaphyseal dysplasia is a neonatal lethal form of spondylometaphyseal dysplasia characterized by severe metaphyseal chondrodysplasia with mild limb shortening, platyspondyly, cardiac conduction defects, and central nervous system abnormalities. One of the patients was compound heterozygous of two variants affecting splicing (c.587 + 5G > A and c.588-8_588-4del) and the second patient had a homozygous nonsense mutation (p.Tyr127*). None of those variants were observed in gnomAD. Heterozygous missense mutations in human TXNRD2 have been associated with dilated cardiomyopathy (Sibbing et al. 2011), and a homozygous stop gain mutation had been associated with familial glucocorticoid deficiency (Prasad et al. 2014). The latter is caused by a nonsense variant (p.Y447X) that creates a TAG codon, which cannot be read as Sec, and truncates the protein before the TXNRD2 Sec residue, located in its penultimate position. This variant is observed in 143 individuals in gnomAD, all of them heterozygous. Heterozygous individuals are known to be unaffected for familial glucocorticoid deficiency (Prasad et al. 2014). Described consanguineal individuals homozygous for this mutation are from the Kashmir region (Prasad et al. 2014). Consistently, the p.Y447X variant in TXNRD2 is particularly frequent in South Asia populations according to gnomAD database. Nonetheless, the high oe scores in gnomAD suggest that both genes tolerate heterozygous LoF variants in human.
Through the ASE analysis from GTEx, we were able to analyze the transcriptomic effect of PTCs in selenoprotein transcripts. We observed that PTC in human selenoproteins follows the general 50-bp rule and triggers NMD if located in the NMD sensitive region of the transcript. The effect of NMD is observed as an allelic imbalance in RNA-seq samples from multiple tissues. In selenoproteins, the imbalance appeared to be to a lesser extent for PTC variants in which the gained stop codon is TGA, whereas this was not the case for other genes. Although based on few cases (six PTC variants in selenoproteins with ASE quantified in multiple tissues), our analysis is consistent with higher readthrough efficiency of TGA compared with other gained stop codons in selenoproteins, resulting in less efficient NMD.
To our knowledge, our comparative analysis of the consequences of gene loss between humans and mice is the first of its kind. Although our study focused on selenoproteins and related genes, it may serve as a test case for genome-wide analyses of orthologs between human and mouse.
Materials and Methods
gnomAD Database Gene Constraint Scores
We used the Genome Aggregation Database (gnomAD) v2.1 (http://gnomad.broadinstitute.org; last accessed September 27, 2019) to assess the tolerance to LoF variants in human selenoproteins and Sec machinery genes. The database contains genetic variance across 141,456 individuals. The LoF variants considered for gene constraint metrics are nonsense and splice variants. The calculation of the expected number of variants for each gene takes into account sequence content and mutational bias (Lek et al. 2016). We used the oe (observed/expected) LoF score and the upper limit of its 90% confidence interval (LOEUF). A low oe score indicates strong selection against LoF variance. The number of variants in a particular gene, and importantly their functional impact, vary across the different transcript isoforms. For our genes of interest, we selected the representative transcript isoform based on the highest median expression from RNA-seq across multiple tissues and individuals from the GTEx project (https://gtexportal.org; last accessed September 27, 2019). We only considered annotated transcripts from Gencode v19 (Frankish et al. 2019) that include the known Sec-UGA positions. We also used the oe LoF scores for all genes in order to compute genome-wide percentiles.
dSTOP: Frequency of Stop Gain
Stop gain is a class of sequence variation wherein at least one base of a codon is changed resulting in a premature stop codon (TAA, TAG, or TGA), leading to a shortened coding sequence. We developed a metric designated dSTOP to assess the frequency of stop gain in a protein coding transcript, using the genotypes from gnomAD. The concept behind dSTOP is similar to Ka (or Ks) in the Ka/Ks ratio, exploiting the fact that, considering only single nucleotide changes, stops can only be acquired in a limited set of codons, here referred as potential stop codons. There is a total of 23 variants that can occur in 18 potential stop codons, whereby a stop codon is gained (5 codons are 1 mutation away from 2 different stops) (supplementary table S4, Supplementary Material online). dSTOP is calculated as the ratio of observed stop gain variants in the population to the number of possible stop gain variants in the sequence of our set of transcripts (supplementary table S5, Supplementary Material online). dSTOP can be computed for each stop codon separately (dTGA, dTAA, or dTAG). In order to compute the proportion of TGA gain among all three stops, we used the index dTGA/dSTOP based on the formula: (oTGA/pTGA)/[(oTGA + oTAA + oTAG)/(pTGA + pTAA + pTAG)]; where oTNN corresponds to the number of observed gains of a particular stop and pTNN corresponds to the number of potential stop codons in the coding sequence of a particular transcript. Since gnomAD v2.1 is based on GRCh37/hg19 and Gencode v19 (Frankish et al. 2019), we used those sources of transcript sequences.
RNA-seq Gene Expression Data
Human RNA-seq data used in this manuscript were obtained from The GTEx project v7, from dbGaP accession number phs000424.vN.pN on March 14, 2019. Mouse RNA-seq data were downloaded from the ENCODE portal (Yue et al. 2014) (https://www.encodeproject.org/; last accessed September 27, 2019). For ENCODE data, we downloaded gene expression values from the project web site using the following filters in the “Experiment matrix” page: Assay = polyA RNA-seq; Genome assembly = mm10; Biosample type = tissues; Life stage = adult; and Available data = tsv; which produced 49 results from 29 different tissues. The Biosample term names provided by ENCODE were matched with the Tissue Site Detail field (SMTSD) from GTEx, producing 17 pairs of matched tissue samples (fig. 4). Gene expression correlation between human and mouse was analyzed through 15,610 one-to-one orthologous protein coding genes (obtained from ensembl compara), by using gene expression TPM (Transcripts Per Million). For GTEx, the median TPM values across all samples for each tissue were computed. For ENCODE, we used the mean TPM across multiple samples, since the number of samples was much lower than in GTEx. A robust linear regression model was fitted for each tissue and the residuals were used to identify genes above or below the predicted value.
LoF Variants in GTEx Subjects and ASE
We searched for variants annotated as high impact in selenoproteins and Sec machinery genes, specifically in our set of transcripts, in the genomes of 652 subjects from GTEx v7. In all, 18 variants in 11 genes from 16 subjects were identified. All of them heterozygous and singletons. We used ASE data from GTEx (Rivas et al. 2015) to determine whether those variants showed ASE, and to obtain the proportion of reads supporting the reference allele in the different tissues available for each corresponding subject. ASE data for GTEx v7 are only available for SNV.
Statistical Analyses
The Kruskal–Wallis nonparametric statistical tests were performed using “kruskal.test” function from the stats package in R. Percentiles for genome-wide oe LoF score and dSTOP were computed using the function “ecdf” in R, which computes the cumulative distribution of a set of values. The gene expression correlation was computed using the nonparametric Spearman rho in the function cor.test in R. All conditions returned a P value <1e-16. Robust linear regression was performed using the function “rlm” from the MASS package in R.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health DK117149, AG021518, and CA080946.
Author Contributions
D.S. and M.M. designed the study, analyzed data, and wrote the manuscript; V.N.G. conceived and supervised the study, and wrote the manuscript.
References
- Ahmed MY, Al-Khayat A, Al-Murshedi F, Al-Futaisi A, Chioza BA, Fernandez-Murray JP, Self JE, Salter CG, Harlalka GV, Rawlins LE, et al. 2017. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain 140(3):547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsina B, Corominas M, Berry MJ, Baguñà J, Serras F.. 1999. Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J Cell Sci. 112 (Pt 17):2875–2884. [DOI] [PubMed] [Google Scholar]
- Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR.. 2002. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 23(1):38–89. [DOI] [PubMed] [Google Scholar]
- Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM.. 1997. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc Natl Acad Sci U S A. 94(11):5531–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhzar L, Hamieh A, Cartier D, Tanguy Y, Alsharif I, Castex M, Arabo A, El Hajji S, Bonnet J-J, Errami M, et al. 2016. Selenoprotein T exerts an essential oxidoreductase activity that protects dopaminergic neurons in mouse models of Parkinson’s disease. Antioxid Redox Signal. 24(11):557–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL.. 2004. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 279(9):8011–8017. [DOI] [PubMed] [Google Scholar]
- Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigó R.. 2001. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2(8):697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CE, Guigó R.. 2008. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS One 3(8):e2968.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, et al. 2004. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 24(21):9414–9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schweizer U. 2016. Mouse models that target individual selenoproteins. In: Dolph L. Hatfield, Ulrich Schweizer, Petra A. Tsuji, Vadim N. Gladyshev, editors. Selenium, Its Molecular Biology and Role in Human Health. Cham (Switzerland): Springer International Publishing. p. 567–578. [Google Scholar]
- Copeland PR, Driscoll DM.. 2001. RNA binding proteins and selenocysteine. Biofactors 14(1–4):11–16. [DOI] [PubMed] [Google Scholar]
- de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim S-W, Harney JW, Larsen PR, Bianco AC.. 2001. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 108(9):1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, et al. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537(7621):508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Copeland PR.. 2009. Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins. BMC Evol Biol. 9:229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Copeland PR.. 2012. Selenocysteine insertion sequence binding protein 2L is implicated as a novel post-transcriptional regulator of selenoprotein expression. PLoS One 7(4):e35581.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Stainier D.. 2017. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 13(7):e1006780.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy RS, Aranda R, Martín MG, Doroshow JH, Binder SW, Chu FF.. 2001. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 281(3):G848–G855. [DOI] [PubMed] [Google Scholar]
- Fonseca TL, Werneck-De-Castro JP, Castillo M, Bocco B, Fernandes GW, McAninch EA, Ignacio DL, Moises CCS, Ferreira AR, Ferreira A, et al. 2014. Tissue-specific inactivation of type 2 deiodinase reveals multilevel control of fatty acid oxidation by thyroid hormone in the mouse. Diabetes 63(5):1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradejas-Villar N. 2018. Consequences of mutations and inborn errors of selenoprotein biosynthesis and functions. Free Radic Biol Med. 127:206–214. [DOI] [PubMed] [Google Scholar]
- Fradejas-Villar N, Seeher S, Anderson CB, Doengi M, Carlson BA, Hatfield DL, Schweizer U, Howard MT.. 2017. The RNA-binding protein Secisbp2 differentially modulates UGA codon reassignment and RNA decay. Nucleic Acids Res. 45(7):4094–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47(D1):D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Conrad M.. 2018. Selenium and GPX4, a vital symbiosis. Free Radic Biol Med. 127:153–159. [DOI] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. 2014. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 16(12):1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galton VA, Schneider MJ, Clark AS, St. Germain DL.. 2009. Life without thyroxine to 3,5,3′-triiodothyronine conversion: studies in mice devoid of the 5′-deiodinases. Endocrinology 150(6):2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN, Arnér ES, Berry MJ, Brigelius-Flohé R, Bruford EA, Burk RF, Carlson BA, Castellano S, Chavatte L, Conrad M, et al. 2016. Selenoprotein gene nomenclature. J Biol Chem. 291(46):24036–24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschanski K, Warnefors M, Kaessmann H.. 2017. The evolution of duplicate gene expression in mammalian organs. Genome Res. 27(9):1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Garcia B, Obregon MJ.. 2007. Gene expression from the imprinted Dio3 locus is associated with cell proliferation of cultured brown adipocytes. Endocrinology 148(8):3968–3976. [DOI] [PubMed] [Google Scholar]
- Hill KE, Wu S, Motley AK, Stevenson TD, Winfrey VP, Capecchi MR, Atkins JF, Burk RF.. 2012. Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J Biol Chem. 287(48):40414–40424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YS, Magnenat JL, Bronson RT, Cao J, Gargano M, Sugawara M, Funk CD.. 1997. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 272(26):16644–16651. [DOI] [PubMed] [Google Scholar]
- Horibata Y, Elpeleg O, Eran A, Hirabayashi Y, Savitzki D, Tal G, Mandel H, Sugimoto H.. 2018. EPT1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in humans. J Lipid Res. 59(6):1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibata Y, Hirabayashi Y.. 2007. Identification and characterization of human ethanolaminephosphotransferase1. J Lipid Res. 48(3):503–508. [DOI] [PubMed] [Google Scholar]
- Imai H, Hirao F, Sakamoto T, Sekine K, Mizukura Y, Saito M, Kitamoto T, Hayasaka M, Hanaoka K, Nakagawa Y.. 2003. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 305(2):278–286. [DOI] [PubMed] [Google Scholar]
- Ingold I, Berndt C, Schmitt S, Doll S, Poschmann G, Buday K, Roveri A, Peng X, Porto Freitas F, Seibt T, et al. 2018. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 172(3):409–422.e21. [DOI] [PubMed] [Google Scholar]
- Jakupoglu C, Przemeck GKH, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, et al. 2005. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 25(5):1980–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin RC, Mahoney CE, (Coleman) Anderson L, Ottaviano F, Croce K, Leopold JA, Zhang Y-Y, Tang S-S, Handy DE, Loscalzo J.. 2011. Glutathione peroxidase-3 deficiency promotes platelet-dependent thrombosis in vivo. Circulation 123(18):1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Fonseca TL, Bocco B, Fernandes GW, McAninch EA, Bolin AP, Da Conceição RR, Werneck-de-Castro JP, Ignacio DL, Egri P, et al. 2019. Type 2 deiodinase polymorphism causes ER stress and hypothyroidism in the brain. J Clin Invest. 129(1):230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2019. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv: 531210.
- Kasaikina MV, Kravtsova MA, Lee BC, Seravalli J, Peterson DA, Walter J, Legge R, Benson AK, Hatfield DL, Gladyshev VN.. 2011. Dietary selenium affects host selenoproteome expression by influencing the gut microbiota. FASEB J. 25(7):2492–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN.. 2003. Characterization of mammalian selenoproteomes. Science 300(5624):1439–1443. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, et al. 2003. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 23(5):1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunskyy VM, Hatfield DL, Gladyshev VN.. 2014. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 94(3):739–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, Gladyshev VN.. 2007. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 17(1):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, Gladyshev VN.. 2009. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta 1790(11):1424–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi Y, Xu X-M, Carlson BA, Fradejas N, Günter P, Braun D, Southon E, Tessarollo L, Hatfield DL, Schweizer U.. 2015. Expression of selenoproteins is maintained in mice carrying mutations in SECp43, the tRNA selenocysteine 1 associated protein (Trnau1ap). PLoS One 10(6):e0127349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. 1995. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1(5):453–465. [PMC free article] [PubMed] [Google Scholar]
- Mariotti M, Ridge PG, Zhang Y, Lobanov AV, Pringle TH, Guigo R, Hatfield DL, Gladyshev VN.. 2012. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS One 7(3):e33066.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M, Santesmasses D, Capella-Gutierrez S, Mateo A, Arnan C, Johnson R, D’Aniello S, Yim SH, Gladyshev VN, Serras F, et al. 2015. Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Res. 25(9):1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Romero FJ, Kryukov GV, Lobanov AV, Carlson BA, Lee BJ, Gladyshev VN, Hatfield DL.. 2001. Selenium metabolism in Drosophila. Selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem. 276(32):29798–29804. [DOI] [PubMed] [Google Scholar]
- Martinez ME, Karaczyn A, Stohn JP, Donnelly WT, Croteau W, Peeters RP, Galton VA, Forrest D, St. Germain D, Hernandez A.. 2016. The type 3 deiodinase is a critical determinant of appropriate thyroid hormone action in the developing testis. Endocrinology 157(3):1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Quijano Roy S, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, et al. 2001. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 29(1):17–18. [DOI] [PubMed] [Google Scholar]
- Moghadaszadeh B, Rider BE, Lawlor MW, Childers MK, Grange RW, Gupta K, Boukedes SS, Owen CA, Beggs AH.. 2013. Selenoprotein N deficiency in mice is associated with abnormal lung development. FASEB J. 27(4):1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE.. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 23(6):198–199. [DOI] [PubMed] [Google Scholar]
- Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, Richardson GP, Kelley MW, St. Germain DL, Galton VA, Forrest D.. 2004. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Natl Acad Sci U S A. 101(10):3474–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Hernandez A, He W, Ren T, Srinivas M, Michelle M, Galton VA, St. Germain DL, Forrest D.. 2009. A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and auditory function. Endocrinology 150(4):1952–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson GE, Whitin JC, Hill KE, Winfrey VP, Motley AK, Austin LM, Deal J, Cohen HJ, Burk RF.. 2010. Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am J Physiol Renal Physiol. 298(5):F1244–F1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BC, Böck A, Jäckle H, Vorbrüggen G.. 1997. SelD homolog from Drosophila lacking selenide-dependent monoselenophosphate synthetase activity. J Mol Biol. 274(2):174–180. [DOI] [PubMed] [Google Scholar]
- Pitts MW, Reeves MA, Hashimoto AC, Ogawa A, Kremer P, Seale LA, Berry MJ.. 2013. Deletion of selenoprotein M leads to obesity without cognitive deficits. J Biol Chem. 288(36):26121–26134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Chan LF, Hughes CR, Kaski JP, Kowalczyk JC, Savage MO, Peters CJ, Nathwani N, Clark AJL, Storr HL, et al. 2014. Thioredoxin reductase 2 (TXNRD2) mutation associated with familial glucocorticoid deficiency (FGD). J Clin Endocrinol Metab. 99(8):E1556–E1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rederstorff M, Castets P, Arbogast S, Lainé J, Vassilopoulos S, Beuvin M, Dubourg O, Vignaud A, Ferry A, Krol A, et al. 2011. Increased muscle stress-sensitivity induced by selenoprotein N inactivation in mouse: a mammalian model for SEPN1-related myopathy. PLoS One 6(8):e23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, et al. 2015. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348(6235):666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt BM, Rudolph KLM, Karagianni P, Fonseca NA, White RJ, Talianidis I, Odom DT, Marioni JC, Kutter C.. 2014. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res. 24(11):1797–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St. Germain DL, Galton VA.. 2001. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol. 15(12):2137–2148. [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Fiering SN, Thai B, Wu S, St. Germain E, Parlow AF, St. Germain DL, Galton VA.. 2006. Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology 147(1):580–589. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, et al. 2016. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J Clin Invest. 126(3):992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers E, Schoenmakers N, Chatterjee K. 2016. Mutations in humans that adversely affect the selenoprotein synthesis pathway. In: Dolph L. Hatfield, Ulrich Schweizer, Petra A. Tsuji, Vadim N. Gladyshev, editors. Selenium, Its Molecular Biology and Role in Human Health. Cham (Switzerland): Springer International Publishing. p. 523–538. [Google Scholar]
- Seeher S, Atassi T, Mahdi Y, Carlson BA, Braun D, Wirth EK, Klein MO, Reix N, Miniard AC, Schomburg L, et al. 2014. Secisbp2 is essential for embryonic development and enhances selenoprotein expression. Antioxid Redox Signal. 21(6):835–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbing D, Pfeufer A, Perisic T, Mannes AM, Fritz-Wolf K, Unwin S, Sinner MF, Gieger C, Gloeckner CJ, Wichmann H-E, et al. 2011. Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 cause dilated cardiomyopathy. Eur Heart J. 32(9):1121–1133. [DOI] [PubMed] [Google Scholar]
- Smith AC, Mears AJ, Bunker R, Ahmed A, MacKenzie M, Schwartzentruber JA, Beaulieu CL, Ferretti E, Majewski J, Bulman DE, et al. 2014. Mutations in the enzyme glutathione peroxidase 4 cause Sedaghatian-type spondylometaphyseal dysplasia. J Med Genet. 51(7):470–474. [DOI] [PubMed] [Google Scholar]
- Taskov K, Chapple C, Kryukov GV, Castellano S, Lobanov AV, Korotkov KV, Guigó R, Gladyshev VN.. 2005. Nematode selenoproteome: the use of the selenocysteine insertion system to decode one codon in an animal genome? Nucleic Acids Res. 33(7):2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe R, Carlson BA, Huh JH, Castro NP, Xu X-M, Tsuji PA, Lee S-G, Bang J, Na J-W, Kong Y-Y, et al. 2016. Selenophosphate synthetase 1 is an essential protein with roles in regulation of redox homoeostasis in mammals. Biochem J. 473(14):2141–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turanov AA, Lobanov AV, Hatfield DL, Gladyshev VN.. 2013. UGA codon position-dependent incorporation of selenocysteine into mammalian selenoproteins. Nucleic Acids Res. 41(14):6952–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. 2011. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 21(10):1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres Z, Kim IY, Scholz TD, Stadtman TC.. 1994. Selenophosphate synthetase. Enzyme properties and catalytic reaction. J Biol Chem. 269(14):10597–10603. [PubMed] [Google Scholar]
- Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, Hoffmann PR.. 2011. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 186(4):2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Weiss SL, Sunde RA.. 1998. UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J Biol Chem. 273(43):28533–28541. [DOI] [PubMed] [Google Scholar]
- Williams GR, Bassett J.. 2011. Local control of thyroid hormone action: role of type 2 deiodinase. J Endocrinol. 209(3):261–272. [DOI] [PubMed] [Google Scholar]
- Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA.. 2003. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 34(4):496–502. [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515(7527):355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.