Abstract
A mutant form of the ataxin-1 protein with an expanded polyglutamine (polyQ) tract is the underlying cause of the inherited neurodegenerative disease spinocerebellar ataxia 1 (SCA1). In probing the biophysical features of the nuclear bodies (NBs) formed by polyQ-ataxin-1, we defined ataxin-1 NBs as spherical liquid protein/RNA droplets capable of rapid fusion. We observed dynamic exchange of the ataxin-1 protein into these NBs; notably, cell exposure to a pro-oxidant stress could trigger a transition to slower ataxin-1 exchange, typical of a hydrogel state, which no longer showed the same dependence on RNA or sensitivity to 1,6-hexanediol. Furthermore, we could alter ataxin-1 exchange dynamics either through modulating intracellular ATP levels, RNA helicase inhibition, or siRNA-mediated depletion of select RNA helicases. Collectively, these findings reveal the tunable dynamics of the liquid RNA/protein droplets formed by polyQ-ataxin-1.
Subject terms: Imaging, Nuclear organization
Introduction
Within eukaryotic cells, complex biochemical reactions are facilitated by the concentration and restriction of key enzymes, substrates and regulators within well-defined membrane-bound organelles. In addition, numerous membrane-less intracellular compartments are important for a range of essential activities in both the cytoplasm (e.g. stress granules storing RNA molecules from stalled translation in response to environmental stresses1) and the nucleus (e.g. nucleoli functioning in the biogenesis of ribosome subunits2, Cajal bodies in the processing and modification of short non-coding RNAs3, or promyelocytic leukemia (PML) bodies acting as sumoylation factories in response to interferon and oxidative stress4). The formation of many of these membrane-less organelles is now understood to proceed via a phase separation process of specific constituent proteins, RNA and/or DNA molecules5. Thus, after a certain critical concentration threshold is exceeded, molecular assemblies of these constituents are formed with liquid-like behaviors that include fusing ability, viscous fluid dynamics, and high exchange rates with their surroundings in the nucleoplasm or cytoplasm6–10. This process of protein phase separation is now viewed as an essential mechanism for efficient compartmentalization that can be rapidly responsive to environmental challenges or intracellular changes11,12.
Proteins that can undergo phase separation usually contain sequences conforming to either a low complexity region (LCR) or prion-like domain (PrLD)8,11,13; these are protein domains typically with low amino acid diversity and little conformational heterogeneity5,11. These disordered structural characteristics can also contribute to an additional change known as protein phase transition, in which liquid-like condensates continue to become less dynamic and so form a more viscoelastic hydrogel or solid-like fibrous aggregates12,14. Many factors can promote the phase transition process such as time, pH, and altered amino acid sequences arising from gene mutations8,9,15,16. Thus, sequences typically conforming to LCR or PrLD motifs have been proposed as indicators of a protein’s propensity to undergo protein phase separation and phase transition17,18.
In addition to this importance of membrane-less organelle formation as part of normal cellular physiology, many mutant proteins linked particularly to neurodegenerative diseases have also been identified with LCR or PrLD motifs. Notable examples include the liquid-like condensates and further phase transition into hydrogel by amyotrophic lateral sclerosis (ALS) mutants of FUS or hnRNPA19,19,20. In assessing the processes of phase separation and phase transition for other neurodegenerative diseases, it is notable that trinucleotide repeat expansions can be considered as a type of LCR or PrLD17,21,22. In spinocerebellar ataxia 1 (SCA1) patients, the mutant ATXN1 gene contains CAG repeats that encode an expanded polyglutamine (polyQ) region, and the formation of distinct nuclear “inclusions” of these polyQ-ataxin-1 proteins in SCA1 patients and transgenic mice23–25. Initially these inclusions may lack the fibrillar structure typical of disease-causing amyloids22,26 but instead show highly dynamic exchange27. The main aim of our study is to provide the first comprehensive evaluation of the physical properties of these NBs, to allow us to define a relationship between NB dynamic exchange and toxicity. Here, we implement a suite of microscopy and biochemical approaches to define the nuclear bodies (NBs) formed by polyQ-ataxin-1 as dynamic liquid protein/RNA droplets. These NBs exhibit ready-to-fuse ability and high dynamics revealed by fluorescence fluctuation spectroscopy (FFS) and fluorescence recovery after photobleaching (FRAP). More importantly, we have observed the tunable dynamics of these ataxin-1 NBs, with their high dynamic liquid phase maintained by ATP and RNA helicases, and their low dynamic hydrogel phase triggered by environmental stress. Thus, models that explain the protein aggregation process and pathogenesis mechanism in SCA1 neurodegeneration should now be extended to include polyQ-ataxin-1 protein phase separation and transition.
Results
PolyQ-ataxin-1 phase separates into liquid droplets in cells
PolyQ proteins can form larger protein structures that have been implicated as part of their toxicity mechanisms leading to neurodegeneration; this is clearly documented for the polyQ-huntingtin protein that forms heterogeneously-shaped nuclear aggregates28. In exploring the physical nature of the larger protein structures formed by polyQ-ataxin-1, we note that ataxin-1 NBs have been observed in SCA1 patients29 and that GFP-ataxin-1 forms distinctive NBs within the nucleoplasm of different cell lines30,31. Importantly, the remarkably spherical appearance of the ataxin-1[85Q] NBs (Fig. 1A, upper panel) raises the possibility that these NBs arise from phase separation of the ataxin-1[85Q] protein. Phase separation is a phenomenon that gives rise to membrane-less liquid-like compartments that are dependent on protein concentration11,32, are dynamic in composition9, and that display increased coordinated movement at the domain boundary due to the free energy cost to leave the compartment phase33. Thus, we exploited live cell imaging to explore these properties of the ataxin-1 NBs.
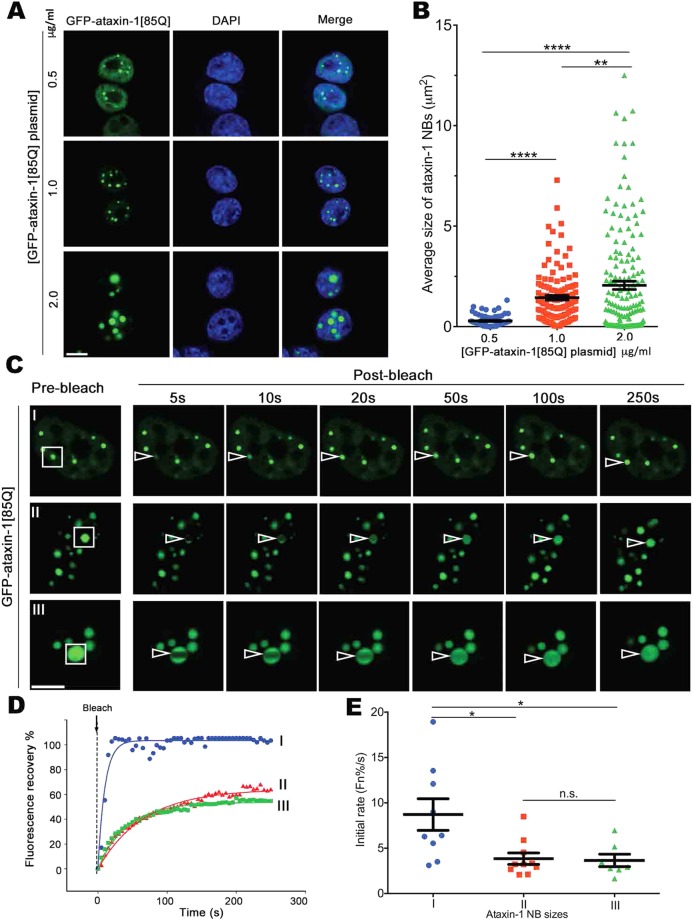
Figure 1.
Ataxin-1 forms concentration-dependent nuclear bodies (NBs) that are highly dynamic. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. (A) At 24 h post-transfection with different plasmid concentrations (0.5, 1.0, or 2.0 μg/ml), cells were fixed and stained with DAPI before CLSM imaging. Representative images are shown from 3 independent experiments. (B) Average sizes of ataxin-1 NBs corresponding to the conditions as per (A) were measured using CellProfiler. Results represent the mean ± SEM (n > 70). Significance values calculated by ANOVA, **p < 0.01, ****p < 0.0001. (C–E) At 24 h post-transfection, cells were incubated in an imaging chamber equilibrated with 5% CO2 at 37 °C prior to FRAP using CLSM. (C) Representative images are shown from 3 independent experiments with FRAP assessments of exchange dynamics of GFP-ataxin-1[85Q] for different size NBs (denoted as I with diameter ≤0.75 μm, II with diameter 0.75–2 μm, and III diameter >2 μm). White rectangles indicate the ataxin-1 NBs analyzed; white open arrowhead indicates photobleached area. All scale bars = 10 μm. (D) Plot of the percentage recovery of fluorescence from experiments as shown in (C I-III) Each symbol represents fluorescence measured at the indicated time for the indicated ROI. (E) Recovery initial rates (average percentage recovery of fluorescence in the first 15 s) (Fn%/s) were calculated and shown by the pooled data. Each symbol represents a single data point obtained from one ROI across 3 independent experiments. Results represent mean ± SEM (n > 7 measured ROIs). Significance values were calculated by ANOVA, *p < 0.05.
First, we expressed GFP-ataxin-1[85Q] in Neuro-2a cells at different levels by varying the concentrations of the transfected expression plasmid (0.5, 1.0 and 2.0 μg/ml plasmid), noting an increase in ataxin-1 NB size with increased transfected plasmid (Fig. 1A,B). To examine exchange dynamics for these ataxin-1 GFP-ataxin-1[85Q] NBs across these transfection conditions (Fig. 1C, rectangles, where I corresponds to NBs with a diameter ≤0.75 μm, II with a 0.75–2 μm diameter, III with a >2 μm diameter), we monitored fluorescence recovery after photobleaching (FRAP) (Fig. 1D). A rapid recovery of fluorescence for all NBs indicated a high exchange rate of GFP-ataxin-1[85Q] (Fig. 1C–E). Notably, the smaller NBs (group I) showed the fastest initial recovery rates (Fig. 1E) achieving near complete recovery within 20 s (Fig. 1D). Fluorescence recovery was demonstrated from either within the NB (i.e. non-bleached NB area) or the nucleoplasm surrounding the NB (Supp Fig. 1). These observations support our hypothesis that ataxin-1 NBs have liquid-like properties. Furthermore, during our FRAP analyses we frequently observed fusion events in which the movement of GFP-ataxin-1[85Q] NBs through the nucleoplasm brought two spherical NBs into close proximity, followed by their rapid fusion and re-shaping into a single, larger spherical NB (Supp. Fig. 2 and Supp. Movie 1). This observation suggests an ataxin-1 NB domain boundary that spatially confines diffusion, and upon fusion with another NB, undergoes a change in interfacial tension that permits mixing before re-establishment of a larger phase-separated compartment.
To investigate the biophysical properties of the ataxin-1 NB domain boundary and whether it imparts coordinated movement that is disrupted upon fusion with another ataxin-1 NB, we employed fluorescence fluctuation spectroscopy (FFS). Traditionally developed for the detection of protein oligomerisation34, a moment-based analysis of the fluctuation in fluorescence intensity recorded in each pixel of a confocal image can detect the localisation of coordinated protein movement in the context of phase separation33. Thus, from rapid acquisition of a time series of GFP-ataxin-1[85Q] fluorescence intensity images during a NB fusion event (1000 frames, 120 ms/frame), we calculated the ratio of the variance (2nd moment) to the mean (1st moment) of GFP-ataxin-1[85Q] fluorescence fluctuations recorded in each pixel of 80 frame segments (~10 s) (Fig. 2A,B). This analysis enables us to derive a spatial map of where GFP-ataxin-1[85Q] undergoes coordinated movement within a NB (i.e. high variance to mean ratio, Fig. 2C), and to trace this feature as a function of time during NB fusion (Fig. 2D).
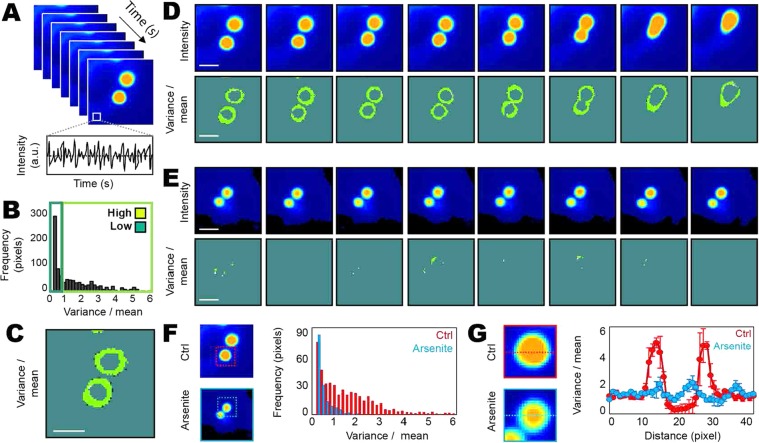
Figure 2.
Fluorescence Fluctuation Spectroscopy (FFS) analysis of ataxin-1 NB fusion reveals liquid droplet-like behavior. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q] for live cell imaging. (A) A time series acquisition (1000 frames) of ataxin-1 NBs enables the fluctuation in GFP-ataxin-1[85Q] fluorescence intensity to be acquired in each pixel of an image. (B,C) Estimate of coordinated GFP-ataxin-1[85Q] mobility in each pixel location. (B) Calculation of the ratio of the first and second moment (variance/mean) of the fluorescence fluctuation recorded in each pixel, directly corresponding to the image presented in (C). (C) The spatial map of a low versus high ratio of the variance to the mean demonstrates coordinated GFP-ataxin-1[85Q] mobility is at the NB boundary. (D) Time series acquisition (1000 frames) of two ataxin-1 NBs under control (Ctrl) conditions undergoing fusion, segmented into 10 s intervals (80 frames): the top row presents the intensity maps of GFP-ataxin-1[85Q] localization during each time segment and the bottom row presents the maps of GFP-ataxin-1[85Q] coordinated movement during each time segment. (E) Time series acquisition (1000 frames) of two ataxin-1 NBs not undergoing fusion in arsenite-treated cells, segmented into 10 s intervals (80 frames): the top row presents intensity maps of GFP-ataxin-1[85Q] localization during each time segment and the bottom row presents maps of GFP-ataxin-1[85Q] coordinated movement during each time segment. (F,G) A comparison of the degree of coordinated GFP-ataxin-1[85Q] movement (variance/mean) within (F) and across the axis (G) of GFP-ataxin-1[85Q] NBs undergoing fusion in Ctrl cells versus NBs not undergoing fusion in arsenite-treated cells. All scale bars in (C–E) = 2 μm. Images presented in (D–G) are representative of n = 3 cells under each condition, with average data presented in histograms in (F) and (G). Results in (G) represent mean ± SEM (n = 3 cells).
As seen in a comparison of the fluorescent intensity images of GFP-ataxin-1[85Q] localization (top row, Fig. 2D) with the maps of coordinated GFP-ataxin-1[85Q] mobility (bottom row, Fig. 2D), we observed a high variance to mean ratio only at the NB boundaries. Then, as the two NBs become physically closer and start to fuse, there is a loss of coordinated GFP-ataxin-1[85Q] movement at the point of contact, which results in the high variance to mean ratio localization surrounding a single, larger spherical NB. Interestingly, upon exposure of the cells to an environmental pro-oxidant stress agent such as arsenite35,36, that is well-established for driving formation of the biomolecular condensates known as stress granules37–39, we observe a significant reduction in coordinated GFP-ataxin-1[85Q] mobility at the edge of the NB boundary (Fig. 2E). From quantitation of the frequency at which pixels are detected with a high variance to mean ratio in untreated control versus arsenite-treated cells (Fig. 2F) and then plotting this parameter as a function of position along a NB axis (Fig. 2G), we demonstrate that arsenite treatment results in a five-fold reduction in protein mobility at the NB boundary. Thus, in agreement with the protein concentration (Fig. 1A,B) and FRAP (Fig. 1C,D) analyses, fluorescence fluctuation spectroscopy (FFS) demonstrates a critical phase-separation hallmark for ataxin-1 NBs.
In addition, arsenite treatment not only decreased ataxin-1 NB dynamic exchange, but also increased ataxin-1[85Q] NB size (Supp. Fig. 3). Specifically, many larger ataxin-1 NBs were visible (Supp. Fig. 3A) and quantitative analyses demonstrated a statistically significant increase in average NB size following arsenite treatment (Supp. Fig. 3B). As the total size of the population of ataxin-1[85Q] NBs per nucleus did not also significantly increase upon arsenite exposure (Supp. Fig. 3C), these results are consistent with increased fusion of the ataxin-1 NBs in the presence of arsenite. To explore these relationships further, we also examined the behavior of an ataxin-1[85Q] mutant with a deletion in its self-association domain (SAD)40 (amino acids 495–605, herein termed ataxin-1[85Q]∆SAD) (Supp. Fig. 4). In transgenic mice models, polyQ-ataxin-1[77Q]∆SAD was reported as toxic (also tested in Neuro-2a cells as shown in Supp. Fig. 4C), but without visible NBs41. In our cultured Neuro-2a cell system, we observed that ataxin-1[85Q]∆SAD retained the ability to NBs potentially due to the presence of other motifs (such as CT2AXH) outside of the SAD42, but these differed from those formed by wild-type ataxin-1[85Q] being significantly smaller in size and not being further impacted by arsenite treatment (Supp. Fig. 4). Thus, the formation of ataxin-1[85Q] NBs can be observed in the absence of the reported self-association domain, and when integrated with the previous in vivo studies41, these results suggest that these smaller sized NBs may retain the potential for neurotoxicity.
In addition to evaluating the NBs formed by ataxin-1[85Q], we examined the properties of NBs formed by the wild-type ataxin-1[30Q] protein. Expression of GFP-ataxin-1[30Q] protein in Neuro-2a cells showed the formation of spherical NBs indistinguishable morphologically from those observed for GFP-ataxin-1[85Q] (Supp. Fig. 5A), and consistent with previous studies43,44, thus defining higher ataxin-1 protein concentration as a major contributor to NB formation. Furthermore, ataxin-1[30Q] NBs showed high dynamics (Supp Fig. 5A–C) and fusion (Supp Fig. 5D) revealed by FRAP and live cell imaging, respectively. Taken together, these results demonstrate ataxin-1 protein phase separation as liquid droplets with high dynamics and fusion.
RNA/transcription is required for polyQ-ataxin-1 phase separation
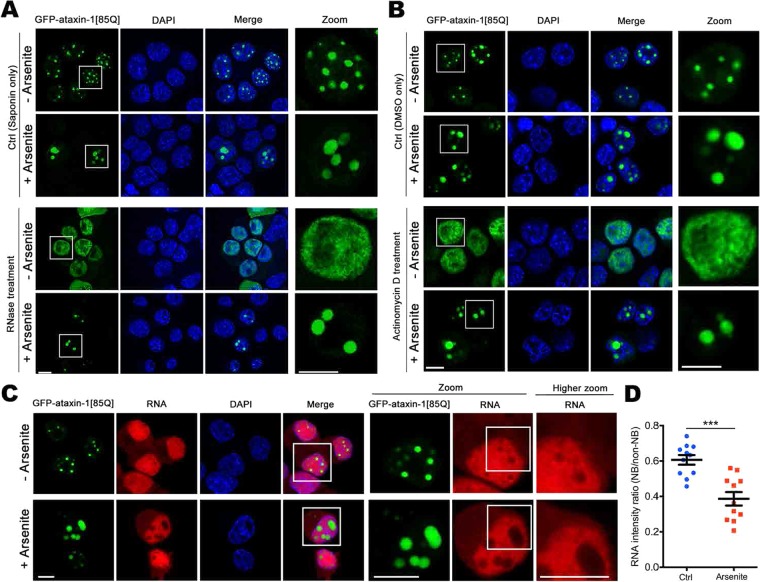
We next explored the dependence of the formation of ataxin-1 NBs on the presence of RNA, building on previous studies demonstrating an interaction of ataxin-1 with RNA in vitro45 and also that RNA can contribute to protein phase separation32,46. We addressed this requirement for RNA/transcription in two independent protocols, and we further included an oxidant stress treatment by incubation of the cells in the presence of arsenite35,36. First, we assessed the impact of RNase-mediated degradation on ataxin-1 NBs. Pretreatment with the mild detergent saponin to permeabilise cell and nuclear membranes allowed RNase A access to hydrolyse RNA. Importantly, saponin treatment alone did not impact the ataxin-1 NBs, and so GFP-ataxin-1[85Q] was visualized within distinct NBs (Fig. 3A, upper two rows). In contrast, saponin followed by RNase A treatment resulted in a diffuse nuclear distribution of GFP-ataxin-1[85Q] for control cells; however, for cells previously treated with arsenite, we noted that the ataxin-1 NBs remained following RNase A treatment (Fig. 3A). We next considered the impact of the transcriptional inhibitor actinomycin D47. In control cells treated with actinomycin D for 3 h, we again noted a diffuse distribution of GFP-ataxin-1[85Q] in the nucleus; but for cells pre-treated with arsenite for 1 h, followed by washout and actinomycin D treatment for 3 h, we continued to observe distinct ataxin-1 NBs (Fig. 3B). We further examined ataxin-1 NBs for the presence of newly-synthesized RNA48. We noted intense staining for RNA throughout the nucleus; this included regions occupied by the GFP-ataxin-1[85Q] nuclear bodies, albeit at levels lower than in the surrounding nucleoplasm; under arsenite-treated conditions, we continued to detect RNA throughout the nucleus, without distinctive accumulation within ataxin-1 NBs (Fig. 3C). Indeed, quantitative analysis showed that arsenite treatment resulted in lower RNA levels in areas corresponding to the ataxin-1 NBs (Fig. 3D). These observations support a model in which low RNA levels are critical in initiating liquid droplet formation whereas higher RNA levels trigger droplet disassembly49,50. Thus, ataxin-1 NBs are less reliant on RNA levels when in the presence of arsenite stress.
Figure 3.
Ataxin-1 NBs are sensitive to RNase/transcription inhibition in control but not in arsenite stress conditions. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. At 24 h post-transfection, cells were treated with arsenite as indicated (A–C), followed by treatment with (A) Saponin only (0.005%, 15 s) for membrane permeabilization, or Saponin + RNase A (100 μg/ml, 1 h, on ice) for RNA degradation, or (B) DMSO only, or Actinomycin D in DMSO (2 μg/ml, 3 h 37 °C). Cells were then fixed and stained with DAPI before CLSM imaging. Zoom images (right panels) correspond to the boxed regions. Representative images are shown from 3 independent experiments. (C) RNA synthesis was detected using imaging including the Click-iT RNA Alexa Fluor 594. 5-ethynyl uridine was added for 24 h before arsenite treatment, then incorporated 5-ethynyl uridine was detected (red). Zoom images (right panels) correspond to the boxed regions. (D) Quantitative analysis of RNA staining intensity ratio (NB area/non-NB area in the nucleoplasm). Each symbol represents a single data point. Results represent the mean ± SEM (n > 10). Significance values were calculated by unpaired two-tailed student’s t-test, ***p < 0.001. All scale bars = 10 μm.
Different forms of stresses trigger phase transition of liquid ataxin-1 NBs to a reversible or an irreversible hydrogel
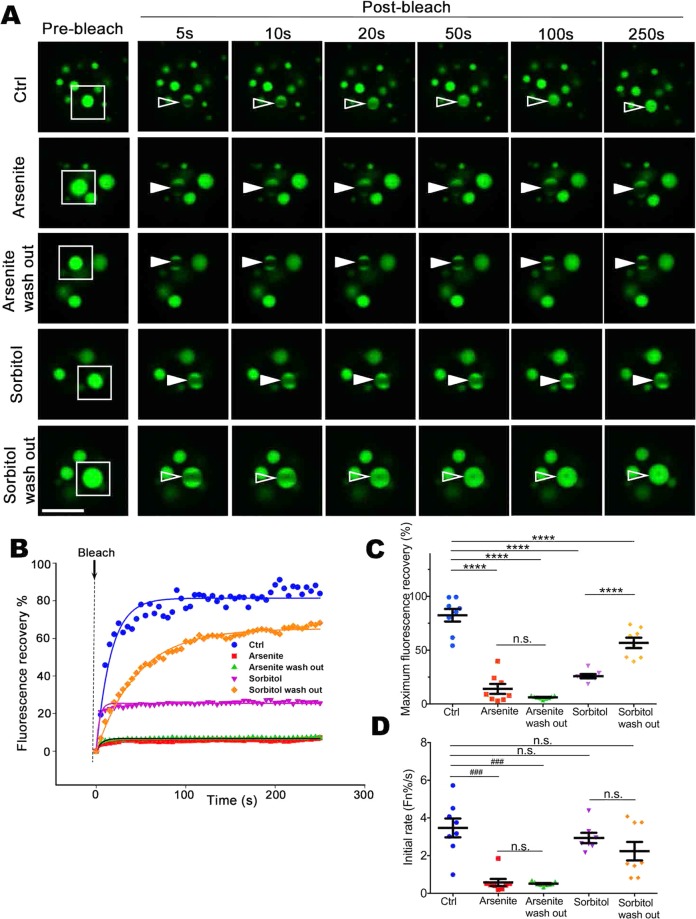
The liquid droplets formed by proteins with low complexity region (LCR) or prion-like domain (PrLD) motifs can show a time-dependent transition through an aging/maturation process20,51. Furthermore, exposure of cells to a range of environmental stress agents, such as arsenite, can enhance stress granule formation37–39. Therefore, we next assessed the impact of arsenite on the maturation/phase transition of ataxin-1 NBs. Our FRAP analyses revealed a significant slowing of GFP-ataxin-1[85Q] maximum recovery and initial rate of recovery, an impact not reversed upon arsenite washout (Fig. 4).
Figure 4.
Different stresses can inhibit ataxin-1 NB exchange dynamics either irreversibly or reversibly. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. At 24 h post-transfection, cells were left untreated (Ctrl) or treated with arsenite (0.3 mM, 1 h) (Arsenite) or sorbitol (0.5 M, 1 h) (Sorbitol). Following wash out (three washes with PBS), cells were left to recover from the stress treatment for 3 h (Arsenite wash out or Sorbitol wash out). (A) Representative images are shown from 3 independent experiments for quantitative assessments of GFP-ataxin-1[85Q] exchange dynamics. White rectangle indicates ataxin-1 NB imaged analyzed. A small ROI (indicated by white open arrowhead for dynamic NB or white solid arrowhead for non-dynamic NB) was photobleached and the fluorescence recovery was subsequently monitored as post-bleach at 5 s intervals for 250 s. (B) Plot of the percentage recovery of fluorescence from experiments as shown in (A). Each symbol represents fluorescence measured at the indicated time. (C,D) Results for pooled data for FRAP experiments. The recovery of fluorescence for GFP-ataxin-1[85Q] over time was analyzed. Each symbol represents a single data point. Results represent the mean ± SEM for (C) fluorescence maximum recovery percentage in the bleached area (where 100% represents full recovery) and (D) the initial rate of fluorescence recovery percentage in the bleached area for first three time points (0–15 s of post-bleach). Significance values were calculated by (C) ANOVA or (D) Mann-Whitney and Kruskal-Wallis non-parametric test, n > 7, ****p < 0.0001, ###p < 0.001, n.s. = not significant. All scale bars = 10 μm.
We extended our interrogations to include a second environmental insult in the form of hyperosmotic stress induced by addition to the cell culture medium of the non-metabolisable sugar, sorbitol; this change in osmolarity can also promote the formation of cytoplasmic stress granules52,53. We again observed significant slowing of GFP-ataxin-1[85Q] maximum recovery and initial rate of recovery, but these changes could be reversed upon the return of the cells to normal osmotic conditions (Fig. 4). Thus, the impact of stress on the transition to form non-dynamic ataxin-1 NBs is dependent on the nature of the initiating stress. This altered physical state of the ataxin-1 NBs does not appear to be caused by altered levels of ataxin-1 expression under these stress conditions, with no significant differences in total fluorescence intensity observed across the control, arsenite, or sorbitol treatments (Supp. Fig. 6). To further test the state of the ataxin-1 NBs under different conditions, 1,6-hexanediol, a chemical probe to differentiate liquid-like assemblies from solid-like assemblies8,54, was added to the cells expressing GFP-ataxin-1[85Q] under control or stress conditions (arsenite or sorbitol). Whilst ataxin-1 NBs exhibit a liquid-like state sensitive to 1,6-hexanediol, NBs pre-treated with stress (arsenite but not sorbitol) exhibit a solid-like state as they are 1,6-hexanediol resistant (Fig. 5). Thus, ataxin-1 protein exchange and the properties of ataxin-1 NBs are rapidly responsive to environmental changes.
Figure 5.
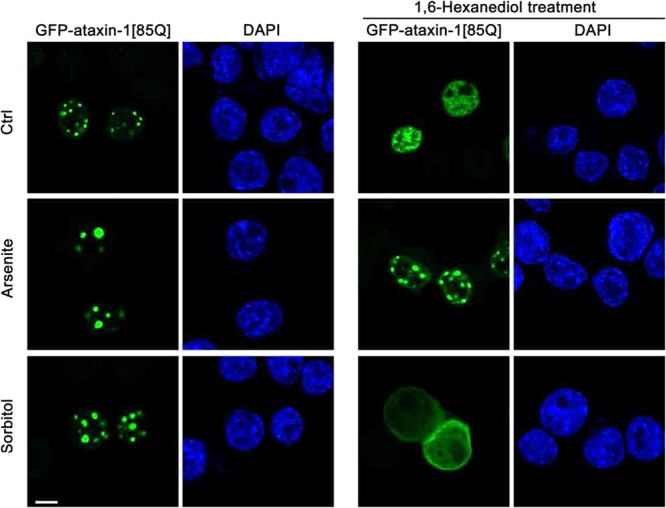
Liquid-like or solid-like properties of ataxin-1 NBs under control or stress conditions. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. At 24 h post-transfection, cells were left untreated (Ctrl) or treated with arsenite (0.3 mM, 1 h) (Arsenite) or sorbitol (0.5 M, 1 h) (Sorbitol). Cells were further treated with 1,6-hexanediol (5%, 5 min) to discriminate between liquid-like assemblies and solid-like assemblies. Cells were then fixed and stained with DAPI before CLSM imaging. Each symbol represents a single data point. Representative images are shown from 3 independent experiments. All scale bars = 10 μm.
Maintenance of the ataxin-1 NB liquid phase requires ATP
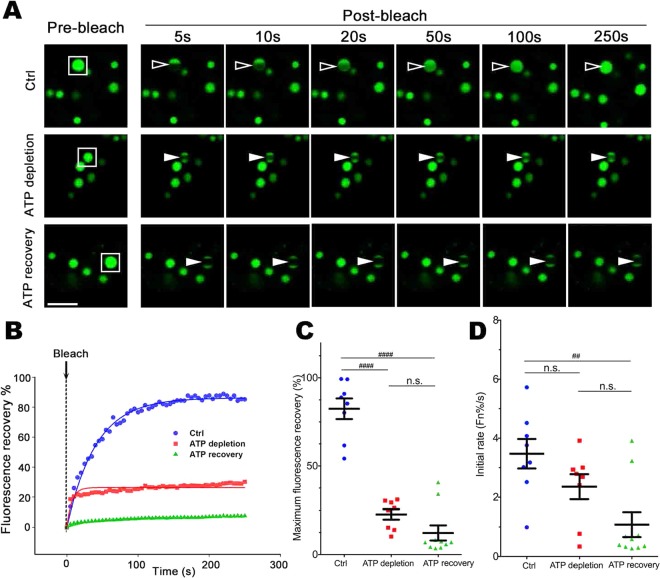
The maintenance of liquid droplet high dynamics, for membrane-less organelles such as nucleoli, is ATP-dependent7. Therefore, we next addressed the ATP requirement in the dynamic exchange of GFP-ataxin-1[85Q] in ataxin-1 NBs. We lowered ATP levels with 2-Deoxy-D-glucose (2DG) and Carbonyl cyanide m-chlorophenyl hydrazone (CCCP)55,56 and confirmed ATP recovery upon inhibitor washout (Supp. Fig. 7). Strikingly, maximum and initial rates of fluorescence recovery were significantly and irreversibly inhibited by ATP depletion (Fig. 6A–D), indicating the irreversibility of the phase transition process following ATP depletion.
Figure 6.
Dynamic ataxin-1 NBs transit to irreversible non-dynamic hydrogel following intracellular ATP depletion. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. At 24 h post-transfection, cells were left untreated (Ctrl) or treated with 2DG + CCCP (ATP depletion). Inhibitors were washed out (3 washes with PBS) and cells were left to recover for 3 h (ATP recovery). (A) Representative images are shown from 3 independent experiments for quantitative assessments of GFP-ataxin-1[85Q] exchange dynamics. White rectangle indicates ataxin-1 NB analyzed. A small ROI (indicated by white open arrowhead for dynamic NB or white solid arrowhead for non-dynamic NB) was photobleached and the fluorescence recovery was subsequently monitored as post-bleach at 5 s intervals for 250 s. (B) Plot of the percentage recovery of fluorescence from experiments as shown in (A). Each symbol represents fluorescence measured at the indicated time. (C,D) Results for pooled data for FRAP experiments. The recovery of fluorescence for GFP-ataxin-1[85Q] over time was analyzed. Each symbol represents a single data point. Results represent the mean ± SEM for (C) fluorescence maximum recovery percentage in the bleached area (where 100% represents full recovery) and (D) the initial rate of fluorescence recovery percentage in the bleached area for first three time points (0–15 s of post-bleach). Significance values were calculated by Mann-Whitney and Kruskal-Wallis non-parametric test, n > 7, ##p < 0.01, ####p < 0.0001, n.s. = not significant. All scale bars = 10 μm.
Down regulation of DEAD/H-box RNA helicases decreases ataxin-1 NB dynamic exchange
Whilst ATP can directly influence the phase transition process57,58, intracellular ATP levels are also critical for the actions of multiple ATP-dependent enzymes and chaperones59,60. We previously identified 13 RNA helicases in the ataxin-1 interactome (see ProteomeXchange, PXD010352)61. RNA helicases have ubiquitous roles in regulating RNA structures as well as RNA-RNA and RNA-protein interactions in multiple different aspects of RNA metabolism, including transcription, mRNA splicing, RNA export, translation and RNA degradation62,63. The two largest groups within the RNA helicase family are the DEAD-box (DDX) and DEAH-box (DHX) helicases63. That these RNA helicases may contribute to protein phase transition of membrane-less NBs is seen with siRNA-driven decrease in RNA helicase CGH-1/DDX6 levels resulting in non-dynamic ribonucleoprotein (RNP) granules64.
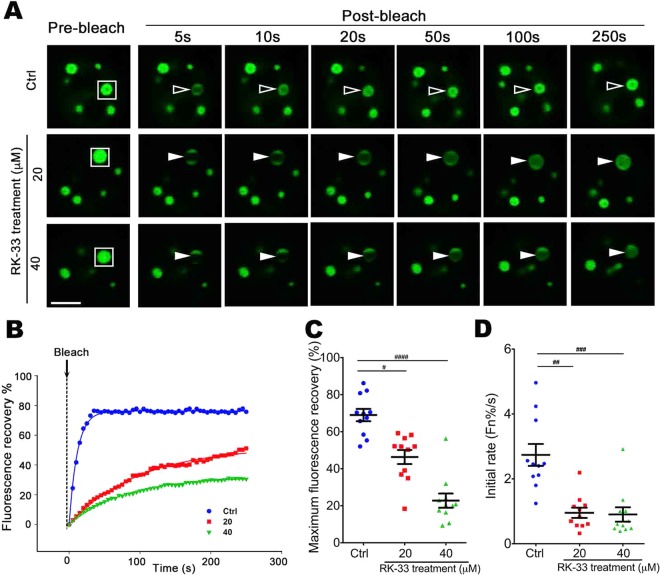
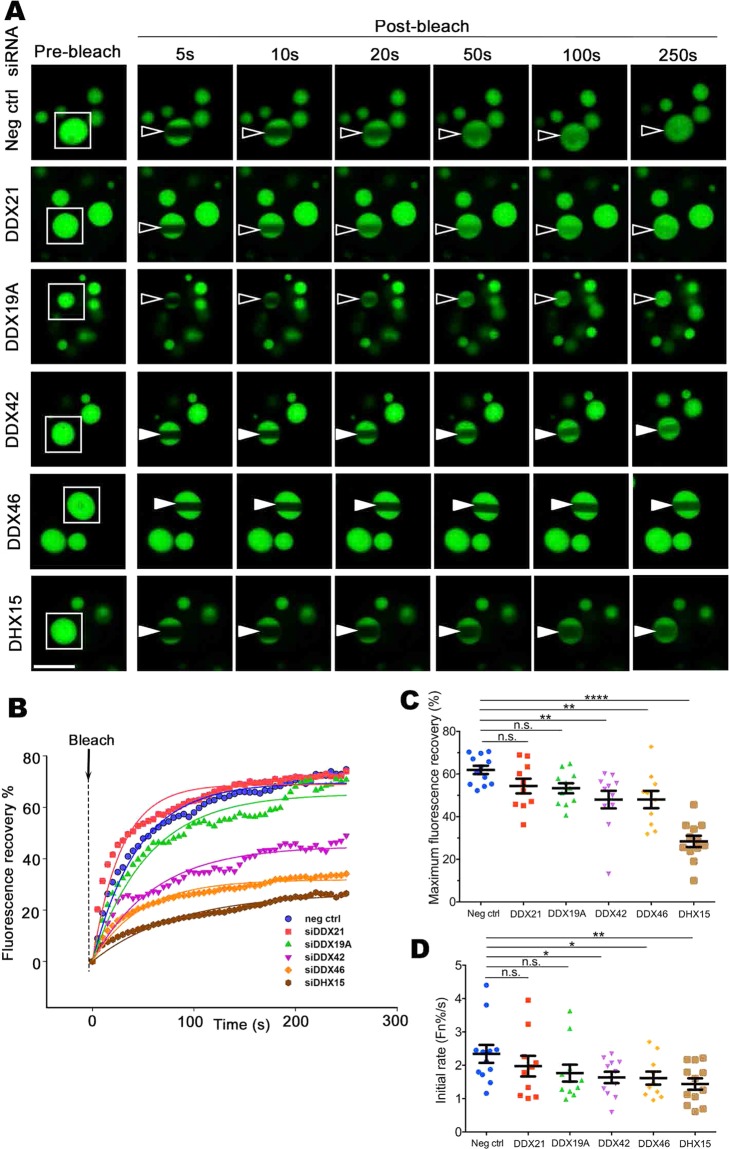
We initially showed that RK-33, an inhibitor that binds the ATP-binding cleft of RNA helicase(s)65,66, could decrease the exchange dynamics of ataxin-1 NBs (Fig. 7). We next incubated cells in the presence of siRNAs targeting each of the 13 RNA helicases of the ataxin-1 interactome. Our initial FRAP assessments directed our focus to siRNA targeting of the helicases DDX21, DDX19A, DDX42, DDX46, and DHX15. We ensured that the levels of each, as assessed by Western blot, were decreased by siRNA treatment (Supp. Fig. 8). Using FRAP protocols, we assessed how these helicase-targeted siRNA treatments, alongside control siRNA treatment, altered the dynamics of recovery. Our quantitative analysis of the pooled data confirmed that DDX42, DDX46, or DDX15 siRNA significantly decreased the dynamic exchange of the ataxin-1 NBs (Fig. 8) by decreasing both the maximum fluorescence recovery (Fig. 8C) and the initial recovery rate (Fig. 8D). When we assessed the intracellular localization of each helicase, we further noted that DDX42, DDX46, and DHX15 were more nuclear than cytoplasmic (Supp. Fig. 9A,B). Taken together, these results reinforce the observation that DDX42, DDX46 and DHX15 are predominantly nuclear RNA helicases that contribute to the regulation of the dynamic exchange of ataxin-1 NBs.
Figure 7.
RNA helicase inhibitor RK-33 slows ataxin-1 NB exchange dynamics. Neuro-2a cells were transfected to express GFP-ataxin-1[85Q]. At 4 h post-transfection, cells were left untreated (Ctrl) or treated with RK-33 (20 μM or 40 μM) for 24 h. (A) Representative images are shown from 3 independent experiments for quantitative assessments of GFP-ataxin-1[85Q] exchange dynamics. White rectangle indicates ataxin-1 NB analyzed. A small ROI (indicated by white open arrowhead for dynamic NB or white solid arrowhead for non-dynamic NB) was photobleached and the fluorescence recovery was subsequently monitored as post-bleach at 5 s intervals for 250 s. (B) Plot of the percentage recovery of fluorescence from experiments as shown in (A). Each symbol represents fluorescence measured at the indicated time. (C,D) Results for pooled data for FRAP experiments. The recovery of fluorescence for GFP-ataxin-1[85Q] over time was analyzed. Each symbol represents a single data point. Results represent the mean ± SEM for (C) fluorescence maximum recovery percentage in the bleached area (where 100% represents full recovery) and (D) the initial rate of fluorescence recovery percentage in the bleached area for first three time points (0–15 s of post-bleach). Significance values were calculated by Mann-Whitney and Kruskal-Wallis non-parametric test, n > 10, #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001. All scale bars = 10 μm.
Figure 8.
ATP-dependent RNA helicases DDX42, DDX46 and DHX15 are required for ataxin-1 NB rapid exchange dynamics. Neuro-2a cells were co-transfected with plasmid encoding GFP-ataxin-1[85Q] and siRNAs: negative siRNA ctrl, DDX21, DDX19A, DDX42, DDX46, or DHX15, as indicated. At 24 h post-transfection, cells were incubated in an imaging chamber equilibrated with 5% CO2 at 37 °C prior to FRAP using CLSM. (A) Representative images are shown from 4 independent experiments for quantitative assessments of GFP-ataxin-1[85Q] exchange dynamics under different siRNA conditions. White rectangle indicates ataxin-1 NB analyzed. A small ROI (indicated by white open arrowhead for dynamic NB or white solid arrowhead for non-dynamic NB) was photobleached and the fluorescence recovery was subsequently monitored as post-bleach at 5 s intervals for 250 s. (B) Plot of the percentage recovery of fluorescence from experiments as shown in (A). Each symbol represents fluorescence measured at the indicated time. (C,D) Results for pooled data for FRAP experiments. The recovery of fluorescence for GFP-ataxin-1[85Q] over time was analyzed. Each symbol represents a single data point. Results represent the mean ± SEM for (C) fluorescence maximum recovery percentage in the bleached area (where 100% represents full recovery) and (D) the initial rate of fluorescence recovery percentage in the bleached area for first three time points (0–15 s of post-bleach). Significance values were calculated by ANOVA, n > 10, *p < 0.05, **p < 0.01, **** p < 0.0001, n.s. = not significant. All scale bars = 6 μm.
The processes of protein phase transition have been previously discussed as part of a time-dependent or “aging” process that can be linked to neurodegeneration. For example, in ALS, protein condensates formed by FUS or hnRNPA1 can transit to a more solid hydrogel or aggregate phase8,9,19. Initially this process can be reversed by ATP-dependent chaperones or disaggregases67, but the increasing spatial order of these protein assemblies then promotes a transition to a solid-like fibrous aggregate, that may be toxic for neurons and so be a driver of ALS neurodegeneration14. We therefore tested the impact of down regulation of RNA helicases on cell viability as an approach to defining whether RNA helicase sequestration/disruption might underpin ataxin-1[85Q]-driven cell toxicity. Treatment of the cells with siRNAs directed to the helicases DDX42, DDX46, or DHX15 that decrease ataxin-1 NBs dynamics (Fig. 8) did not significantly increase cell death percentage in GFP-expressing cells, and indeed there was a small but significant increase in viability noted for the DDX42 siRNA-treated GFP-expressing cells (Supp Fig. 10). This demonstrates that a disruption of RNA helicases, and in particular the single disruption of DDX42, DDX46 or DHX15, is not a toxic event per se, but it also further suggests a decreased dynamics of ataxin-1[85Q] NBs that is observed upon the down regulation of specific RNA helicases is not caused by reduced cell viability.
Arsenite increases ataxin-1[85Q]S776A toxicity and decreases ataxin-1[85Q]S776A NB dynamic exchange
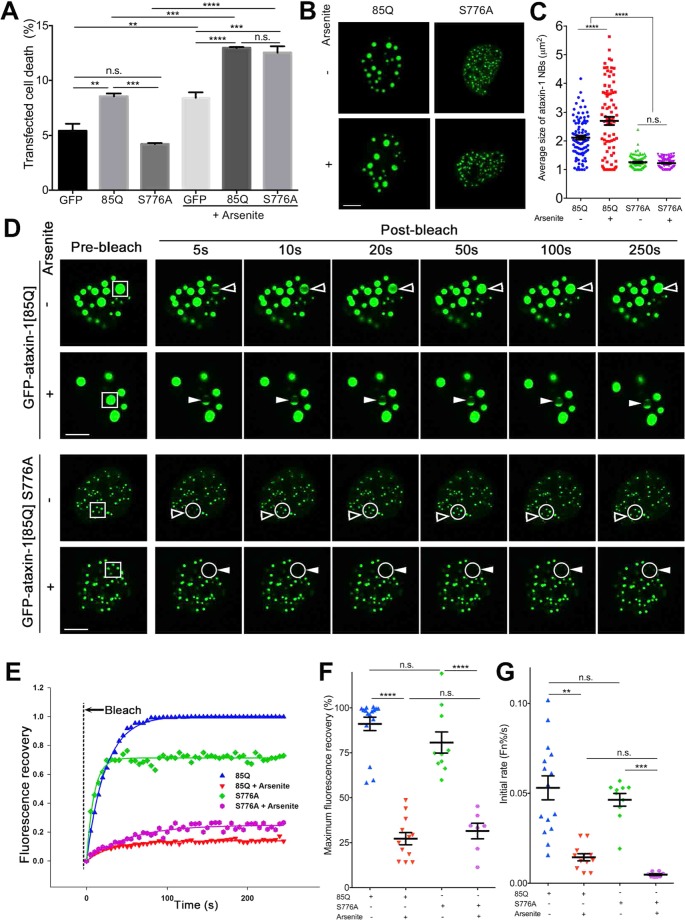
To further explore any correlation between NB size and dynamics and cell toxicity, an ataxin-1 S776A mutant (ataxin-1 serine 776 mutated to the non-phosphorylatable alanine)68,69 was examined (Fig. 9). Indeed, ataxin-1[85Q]S776A remained non-toxic under control conditions as reported68,69 with levels of cell death being not statistically different from those recorded for GFP-only transfected cells (Fig. 9A). However, upon arsenite treatment, we observed statistically significant increases in death of cells expressing GFP-only, GFP-ataxin-1[85Q] or GFP-ataxin-1[85Q]S776A such that toxicity for cells expressing either form of ataxin-1 was comparable (Fig. 9A). When we examined the morphology of ataxin-1 protein exchange for the NBs formed, we observed that ataxin-1[85Q]S776A formed smaller NBs that did not increase in size upon arsenite treatment (Fig. 9B). This suggests that size of the formed NB cannot itself be used as a predictor of toxicity, and thus we further considered the dynamics of these NBs, noting the similar level of high dynamics of ataxin-1 exchange for both the ataxin-1[85Q] and ataxin-1[85Q]S776A NBs, and with the dynamic exchange of both significantly slowing upon arsenite exposure (Fig. 9E–G). These results suggest that decreased NBs dynamics may be related to enhanced cytotoxicity, but this interpretation may be complicated by the broad spectrum of impacts of arsenite exposure35–39.
Figure 9.
Non-toxic ataxin-1 mutant S776A shows increased toxicity and decreased NB exchange dynamics similar to ataxin-1[85Q] level following arsenite stress. Neuro-2a cells were transfected with GFP, GFP-ataxin-1[85Q], or GFP-ataxin-1[85Q]S776A as indicated. At 24 h post-transfection, cells were left untreated or treated with arsenite (0.3 mM, 1 h). (A) Cell death percentage for the transfected cell population was assessed by staining with the SYTOX Red dead cell stain followed by flow cytometry analysis. (B,C) Cells were fixed and stained with DAPI before CLSM imaging. Representative images are shown from 3 independent experiments. (C) Average sizes of ataxin-1 NBs corresponding to the conditions as per (B) were measured using CellProfiler. Results represent the mean ± SEM (Significance values calculated by ANOVA, n > 70, ****p < 0.0001, n.s. = not significant). (D–G). cClls were incubated in an imaging chamber equilibrated with 5% CO2 at 37 °C prior to FRAP using CLSM. (D) Representative images are shown from 3 independent experiments for quantitative assessments of GFP-ataxin-1[85Q] or S776A exchange dynamics. A small ROI (indicated by white open arrowhead for dynamic NB or white solid arrowhead for non-dynamic NB; S776A ROI with multiple small NBs was indicated also by the white circle) was photobleached and the fluorescence recovery was subsequently monitored as post-bleach at 5 s intervals for 250 s. Scale bars = 10 μm. (E) Plot of the percentage recovery of fluorescence from experiments as shown in (D). Each symbol represents fluorescence measured at the indicated time. (D–E) Pooled data from FRAP experiments measuring recovery of fluorescence for GFP-ataxin-1[85Q] or GFP-ataxin-1[85Q] S776A over time. Each symbol represents a single data point. The results are also represented as the mean ± SEM for (F) fluorescence maximum recovery percentage in the bleached area (where 100% represents full recovery) and (F) initial rates (average percentage recovery of fluorescence in the first 15 s) (Fn%/s). Significance were values calculated by ANOVA, n > 7, ***p < 0.0001, ****p < 0.001, n.s. = not significant.
Discussion
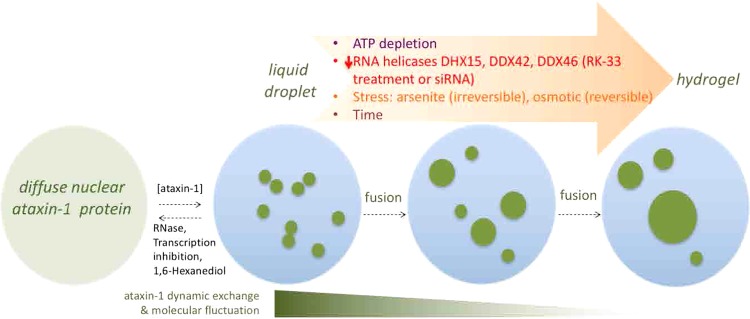
Proteins with LCR or PrLD have attracted much recent attention due to their ability to phase separate to liquid droplets and transition to disease-associated non-dynamic aggregates8,9,19. Here we interrogate polyQ-ataxin-1 NBs in cells to show, for the first time, their multiple liquid-like properties and phase transition to structures with lower dynamic exchange. Based on our findings of NB fusion and the altered protein exchange dynamics we propose a model, as depicted in Fig. 10, in which the ataxin-1 protein undergoes phase separation and then transitions via the liquid droplet state to ultimately form a less dynamic state typical of a hydrogel. In the following paragraphs, we discuss key features of this model and its integration with the current concepts of the formation of biomolecular condensates and the reported physical features and toxicity of polyQ proteins.
Figure 10.
Model showing the multiple factors contributing to the tunable dynamics of ataxin-1 NBs. Ataxin-1 can transit through three stages: diffuse nuclear protein, liquid droplet and then hydrogel, with multiple factors controlling these transitions. Initially, perturbations in ataxin-1 protein level or RNA/transcription, or by 1,6-hexanediol, can influence the transition from diffuse protein to dynamic liquid droplets that are visualized as distinct spherical ataxin-1 NB; fusion can result in larger NBs, but additional factors of time, altered RNA helicases by inhibitor RK-33 or siRNA, ATP depletion, or environmental stresses impact these ataxin-1 NBs to establish a less exchange dynamic and molecular fluctuation hydrogel state.
Initially, the formation of NBs is favoured by an increased concentration of the ataxin-1 protein itself (Fig. 1). Whilst proteins at low concentrations show diffuse intracellular distributions, phase separation can be observed when a critical threshold concentration intrinsic to each protein is exceeded8,11–13,19. This concentration-dependent phase separation has been reported for multiple LCR/PrLD-containing proteins, including ALS disease-causing proteins FUS, hnRNPA1, TIA1, and polyQ protein WHI312; because the polyQ tract is a predicted PrLD21,22, polyQ-ataxin-1 is expected to behave in this way in cells. Consistent with previous reports70, here we have shown that both mutant polyQ-ataxin-1 (with an expanded polyQ tract) and wild-type ataxin-1 protein (with a shorter polyQ tract) can form NBs (Fig. 1 and Supp. Fig. 3). Importantly, although the toxic form of ataxin-1 was first identified with an expanded polyQ tract23,71, increased expression of even the wild-type ataxin-1 causes SCA1 phenotypic changes in flies and mice models43,44. Thus, increased ataxin-1 protein levels are important in the toxicity mechanisms in SCA1 and a significant consequence of an expanded polyQ tract is increased stability that favours the accumulation of higher levels of ataxin-172 and thus the formation of ataxin-1 NBs.
RNA also contributes to the processes of phase separation for ataxin-1 as demonstrated by our results showing the impact of RNase treatment or actinomycin D exposure to drive diffuse distribution of the ataxin-1 protein mainly in the nucleus (Fig. 3). Whilst RNA alone can undergo phase separation into liquid droplets10, RNA can also change the material properties of protein assemblies to influence phase separation and transition12,32,73. Ataxin-1’s ability to interact with RNA in vitro has been reported as showing strong binding to poly(rG) but weaker binding to poly(rU), poly(rA), and poly (rC)45. Our bioinformatics assessment of the ataxin-1 interactome61 highlights a link of ataxin-1 to the processes of RNA metabolism and specifically identified multiple RNA helicases. Our identification of the RNA helicases DDX42, DDX46, and DHX15 which function in pre-mRNA processing74–76 as regulators of ataxin-1 NBs dynamics (Figs. 7 and 8) reinforces a functional association of ataxin-1 with RNA metabolism68,77.
We also characterized the liquid droplet state of the ataxin-1 NBs (Figs. 1 and 2). Importantly, our FFS analyses revealed that the ataxin-1 NB boundary imparts coordinated movement and that upon fusion of two individual ataxin-1 NBs the interfacial tension established by this boundary is disrupted such that mixing between the two compartments is permitted and a single larger NB is formed. The application of FFS to quantify molecular mobility within intracellular compartments has contributed important new insights into accessibility of nuclear structures and the properties of biomolecular condensates, including the heterochromatin domain formed by phase separation33,78,79. By capitalizing on the capacity of FFS to detect the localisation of coordinated protein movement, we recover a map that reports the coordinated movement of ataxin-1 in and out of the NB, which can be tracked as a function of time and modified by arsenite stress (Fig. 2). Thus, FFS should be considered a critical inclusion in the approaches to study biomolecular condensates.
Importantly, our use of FRAP protocols revealed the tunable dynamics of ataxin-1 exchange for these NBs; exchange under control conditions was rapid, but multiple factors could slow this exchange. This reinforces the concepts of phase transition in which liquid-like condensates show rapid fusion and high dynamics, but then the processes of phase transition result in the formation of non-dynamic hydrogels, and ultimately solid-like fibrous aggregates, characterized by a higher spatial order of protein structures and decreased exchange dynamics with their environment14. For ataxin-1 NBs, we showed that this characteristic of the phase transition process could be promoted by the exposure of the cells to different forms of environmental stress (Figs. 4 and 5) or ATP depletion (Fig. 6). These changes echo the observations of previous studies that highlighted a maturation of dynamic ataxin-1 NBs over time to result in mixed populations with slow- or fast-exchanging components70, and support the notion that external factors such as temperature, pH, salinity, oxidative stress or osmolarity can promote phase separation and transition12,80. The application of arsenite or sorbitol stress is known to induce the formation of the biomolecular condensates known as stress granules37–39,52,53 by causing pro-oxidative or hyperosmotic stress responses, respectively, that ultimately stall protein translation37,53. Here we have noted a striking difference for ataxin-1 NBs: the transition to the non-dynamic hydrogel state is irreversible if initiated by arsenite, but reversible if initiated by osmotic stress. The exact mechanisms underlying these differences require further evaluation, but may reflect irreversible oxidative damage in the presence of arsenite, but reversible osmotic control via water efflux and influx81 when osmolarity is modulated by the presence of different concentrations of sorbitol.
Our study has provided the first comprehensive evaluation of the physical properties of these NBs (Figs. 1–6), evaluating a suite of manipulations that allow us to define a relationship between NB dynamic exchange and toxicity (Figs. 7, 8, Supp. Figs. 9 and 10). Thus, we propose that possible links between the regulation of ataxin-1 NB dynamics and the neurodegeneration observed in SCA1 patients should be an important area for future investigation. A dynamic exchange in the liquid droplet stage of the ataxin-1 NB formation would not restrict the transport of associated proteins into and/or exiting the NB27; thus ataxin-1 NBs are disassembled during mitosis and then ataxin-1 protein can be re-distributed into two cells82. With the phase transition to an irreversible hydrogel as that caused by ATP depletion or arsenite stress, it is inevitable that cellular function would be impacted if additional proteins essential for normal cellular functions are sequestered. Current SCA1 murine models (e.g. SCA1[82Q] transgenic mice83–85 or SCA1[154Q/2Q] knock-in mice86–88) rely on the assessment of downstream pathological impacts but, to date, have not included fluorescent protein tagging of the expressed polyQ-ataxin-1 protein. Thus, future studies in murine SCA1 models engineered to express fluorescent protein-tagged polyQ-ataxin-1 will enable the study of the dynamics of ataxin-1 NBs in Purkinje cells, for example by capitalizing on recent advances in lattice light sheet microscopy that offer advantages in spatial and temporal resolution as well as low phototoxicity89,90. The polyQ-ataxin-1 phase transition model that we propose (Fig. 10) will thus provide a framework for ongoing investigations to contribute new knowledge to a mechanistic understanding how polyQ-ataxin-1 can drive the pathological changes observed in the neurodegenerative disease SCA1.
Materials and Methods
Cell culture, transfection and treatments
Mouse neuroblastoma cells (Neuro-2a) were used in a majority of experiments and were cultured in growth medium: Opti-MEM (Gibco) supplemented with 2 mM L-glutamine (Gibco), 10% (v/v) fetal bovine serum (BOVOGEN) and 100 U/ml penicillin/streptomycin (Gibco). Where indicated in control experiments, Ad293 cells were cultured in Dulbecco’s Modified Eagle’s media (DMEM) supplemented with 10% [v/v] fetal bovine serum (FBS) (BOVOGEN) and 100 U/ml penicillin/streptomycin (Gibco). Cells were plated and cultured (16 h) for the following protocols: immunofluorescence and RNA staining (12-well plate, Corning), Western blot (6-well plate, Corning), live cell imaging (8-well μ-slides, Ibidi), and ATP measurements (96 well plate, Corning) then, transfected with the indicated GFP-ataxin-1[85Q] or GFP-ataxin-1[30Q] constructs using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions (24 h). Unless indicated otherwise, 1 µg plasmid DNA/mL culture medium was used per transfection.
Cells transfected to express GFP-ataxin-1 proteins were subjected to different treatments as follows. For environmental stress, cells were exposed to either the pro-oxidant stress of arsenite (0.3 mM, 1 h, 37 °C) (Sigma), whereas for hyperosmotic stress cells were exposed to the non-metabolisable sugar D-sorbitol (0.5 M, 1 h, 37 °C) (Ajax Finechem). For RNase treatment, cells were treated with saponin (0.005%, 15 s, room temperature) (Sigma) prior to the addition of RNase A (100 μg/ml, 1 h, ice) (Invitrogen); thus, saponin-only treatment was employed in parallel control studies. For transcription inhibition, cells were exposed to actinomycin D (2 μg/ml, 3 h, 37 °C) (Sigma), with an equivalent exposure to only the vehicle DMSO (0.2%, 3 h, 37 °C) (Merck), as a negative control. For 1,6-hexanediol treatment, cells were incubated with 5% 1,6-hexanediol (5 min, RT) (Sigma) prior to cell fixation. For RK-33 treatment, cells were treated with 20 μM or 40 μM RK-33 (Adooq, A16194) (24 h, 37 °C) 4 h after transfection. For ATP depletion, cells were exposed to 2-deoxyglucose (2DG) (20 mM, 1 h, 37 °C) (Adrich Chem), + Carbonyl cyanide m-chlorophenyl hydrazine (CCCP) (100 μM, 1 h, 37 °C) (Sigma).
Immunostaining
Neuro-2a cells were cultured on coverslips (Proscitech) within 12-well plates, transfected and treated with 0.3 mM arsenite, as indicated. Subsequent processing was performed in phosphate-buffered saline at room temperature as follows: washing, permeabilization (0.2% (v/v) Triton X-100), fixation (4% (w/v) paraformaldehyde) and blocking (1% (w/v) bovine serum albumin (BSA)). Cells were incubated sequentially with primary antibodies then fluorophore-conjugated secondary antibodies as indicated, each in 1% (w/v) BSA in phosphate-buffered saline (PBS) for 1 h at room temperature: anti-DDX19A (Abcam, ab108462) (1:100), anti-DDX21 (Atlas antibodies, 10528-1-AP) (1:100), anti-DDX42 (Proteintech, HPA023571) (1:100), anti-DDX46 (Atlas antibodies, 16927-1-AP) (1:100), anti-DHX15 (Proteintech, HPA047047) (1:100). Subsequent detection used Alexa Fluor® 568-conjugated secondary antibodies (A-11004, Invitrogen) (1:400). Nuclei were visualized using 4’,6-diamidino-2-phenylindole (DAPI) (Sigma) (1:10,000 in PBS). Processed coverslips were then mounted in water-based Fluoro-Gel (Proscitech) onto glass slides for visualization.
RNA staining
For detection of RNA the Click-iT RNA Alexa Fluor 594 imaging kit (ThermoFisher, C10330) was used. Cells were co-incubated with 5-ethynyl uridine during the transfection. After 24 h, cells were fixed and washed; incorporated 5-ethynyl uridine was detected according to the manufacturer’s instructions.
Confocal laser scanning microscopy (CLSM) and image analysis
Fixed samples were visualized by confocal laser scanning microscopy (CLSM) (Leica TCS SP5 with 63X 1.4 Oil Objective). All imaging studies were performed on >3 independent occasions including preliminary optimization steps. The acquired images were quantitatively analyzed using CellProfiler cell image analysis software (version 2.1.1 for Mac) for ataxin-1 NB size (μm2; the size of individual NBs as well as the total size of NBs per nucleus calculated by summing the size of all individual NBs within a nucleus) and the fluorescence intensity of nucleus/cytoplasm ratio (Fn/c) for localization of RNA helicases. In the Fn/c analyses, after applying the “CorrectIllumination” function, cytoplasmic areas were defined by subtracting the area stained with DAPI. Where indicated, acquired images were quantitatively analyzed using Fiji for fluorescence intensity of RNA staining within NBs/non-NBs or total nuclear fluorescence intensity (pixels). Quantitation of the RNA fluorescence ratio of NB/non-NB was calculated using the formula: (FNB − Fbackground)/(Fnon-NB − Fbackground), in which FNB represents average RNA fluorescence intensity in the NB area, Fnon-NB represents average RNA fluorescence intensity in the non-NB area in the nucleoplasm and FBackground represents average RNA fluorescence intensity in the background of the image.
Live cell imaging, fluorescence recovery after photobleaching (FRAP) and fluorescence fluctuation spectroscopy (FFS)
At 24 h post-transfection, culture medium was replaced with pre-warmed Phenol-Red-Free DMEM growth medium (Gibco). Cells were kept in an environmental chamber (37 °C, 5% CO2) throughout live cell imaging, as well as the FRAP and FFS acquisitions. Image stacks acquired during GFP-ataxin-1 NB fusion events or FRAP analysis were collected using the CLSM Leica TCS SP5 (63X 1.4 Oil Objective). Images stacks acquired during GFP-ataxin-1 NB fusion events and subsequent fluorescence fluctuation analysis were collected using the Zeiss Elyra 780 (63X 1.2 Water Objective).
For FRAP, GFP was excited with the 488 nm emission line of an Argon laser with maximum laser power (50% for Leica TCS SP5) lasting one frame used for photobleaching. Three frames were recorded before photobleaching at a minimal interval (1.3 s). Fluorescence recovery was recorded at 5 s intervals for 250 s. Fluorescence intensity was measured using Fiji. Recovery curve fitting and maximum fluorescence recovery were calculated in SigmaPlot 12.0. Initial rate was calculated as the average fluorescence recovery rate for the first 15 s.
For FFS, 1000 frames (64 × 64 pixel) were acquired using a digital zoom of 10 (13.5 μm region of interest) and a 12.61 μs pixel dwell time which resulted in a line time of 1.89 ms and frame time of 121 ms. GFP was excited with the 488 nm emission line of an Argon laser and detected using the 510–560 nm emission range. The average and variance were calculated in each pixel of every 80 frames (approximately every 10 s per segment) within the 1000 frame acquisition using the Number and Brightness algorithms as executed by the SimFCS software developed at the University of California Irvine Laboratory for Fluorescence Dynamics (www.lfd.uci.edu).
ATP measurement
Cellular ATP levels were measured using a luminescent ATP detection assay kit (Abcam, ab113849) according to the manufacturer’s instructions.
Gene knockdown by siRNA
ON-TARGETplus SMARTpool siRNAs were purchased from Dharmacon: D1Pas1(L-059783-01-0005), DDX1(E-052098-00-0005), DDX18(L-063435-01-0005), DDX19A(L-045066-01-0005), DDX21(L-046395-01-0005), DDX24(L-042299-01-0005), DDX3Y(L-043317-01-0005), DDX41(L-052130-00-0005), DDX42(L-054910-01-0005), DDX46(L-066260-01-0005), DHX15(L-044833-01-0005), DHX9(L-056729-02-0005), UPF1(L-050929-01-0005). Each siRNA was co-transfected with GFP-ataxin-1[85Q] using DharmaFECT Duo Transfection Reagent (T-2010-03). Efficacy of knockdown by each siRNA was assessed by cell lysis and Western blot as described in the following section.
Cell lysis and western blot
Cell lysates were prepared in RIPA buffer [50 mM Tris-HCl, pH 7.3, 150 mM NaCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 1% (v/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% (w/v) NaF and 100 µM Na3VO4] supplemented with Complete protease inhibitor mix (Roche Diagnostic). Cell lysates were incubated on ice (20 min), cleared by centrifugation, and protein concentrations determined using the BioRad protein assay.
Protein samples were separated by SDS-PAGE (8% polyacrylamide gels, 1.5 h) and transferred to polyvinylidene difluoride (PVDF) membranes (Amersham Life Science; 2 h, room temperature). Subsequent steps were performed by blocking with 1% (w/v) BSA in phosphate-buffered saline (PBS; 0.5 h, room temperature), and incubating with primary antibodies (all anti-DDX antibodies as per those used in immunofluorescence analyses) and horseradish peroxidase-linked secondary antibodies (ThermoFisher Scientific) (1:1000) for visualization using an enhanced chemiluminescence detection system (ThermoFisher Scientific). Images were captured using ChemiDoc imager (Bio-Rad) operating in a single-channel protocol.
Cell death assessment
Neuro-2a cells (2 × 105 cells/well) with transfection as indicated in different conditions, were suspended in PBS and transferred to 5 ml glass round-bottom tubes (BD Bioscience). To determine the numbers of dead cells per sample, the SYTOX Red dead cell stain (ThermoFisher Scientific) was added to cell suspensions, and incubated at room temperature for 15 min before flow cytometry analysis according to the manufacturer’s instructions. All samples were prepared and measured in triplicate, with n = 3 independent experiments performed.
Fluorescence detection and recording was assessed by flow cytometry (BD LSRFortessa (BD Bioscience)) with the background fluorescence control estimated from using untransfected cells and negative controls determined by the GFP-only cells (GFP, Green channel) and SYTOX-only cells (SYTOX, Red channel). The voltage applied to each channel was adjusted according to the GFP-only or SYTOX-only controls to avoid the fluorescence count exceeding 100 K. For analysis, 50,000–75,000 cells were recorded by the cytometer for each sample. FlowJo software (Version vX.0.7) was used in the analysis. The gating of debris and intact Neuro-2a cells was based on forward scatter-height (FSC-H) and the dead cell stain (APC-A). Further gating of single cells was based on the side scatter-height (SSC-H) and side scatter-width (SSC-W) or FSC-H and forward scatter-width (FSC-W). The gating for transfected cells was based on fluorescence GFP-A (GFP, Green channel); the gate for dead cells was based on fluorescence APC-A (SYTOX, Red channel). Cell death percentage (%) in each fluorescence range was determined using the following formula: Number of 100 × (dead cells/number of total cells).
Statistical analysis
Graphpad Prism 6 (version 6.00 for Mac) was used for statistical analysis. Data are represented as mean ± standard error of the mean (SEM). Data sets were analyzed by D’Agostino-Pearson omnibus normality test before further comparison analysis. For datasets with normal distribution, an unpaired two-tailed student’s t-test (for 2 data sets) or analysis by ANOVA (for > 2 data sets) was applied to compare differences among different datasets, followed by Tukey’s multiple comparisons test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. For datasets with deviation from normal distribution, Mann-Whitney and Kruskal-Wallis non-parametric test was used for analysis; #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001.
Supplementary information
Acknowledgements
This work was supported by National Health and Medical Research Council (NHMRC) Project Grant APP1121907. S.Z. is a recipient of the Melbourne International Research Scholarship at the University of Melbourne, EH is an NHMRC Career Development Fellow (APP1124762) and D.A.J. is an NHMRC Senior Principal Research Fellow (APP1103050). The GFP-ataxin-1[85Q] and GFP-ataxin-1[30Q] plasmids were provided by D Hatters (University of Melbourne) and the GFP-ataxin-1[85Q] mutant plasmids (GFP-ataxin-1ΔSAD and GFP-ataxin-1S776A) were provided by W.-K. Chen (University of Melbourne). We acknowledge access to the Biological Optical Microscopy Platform at the University of Melbourne.
Author contributions
S.Z. and M.A.B. conceived the work and designed the experiments. S.Z. and E.H. and M.P.S. performed the experiments and all authors analyzed the data. M.A.B. and D.A.J. directed the work. S.Z. drafted the manuscript, and all authors contributed to discussions and reviewed the manuscript.
Data availability
Materials used and data analysed during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-57994-9.
References
- 1.Ivanov Pavel, Kedersha Nancy, Anderson Paul. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harbor Perspectives in Biology. 2018;11(5):a032813. doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahboubi H, Stochaj U. Nucleoli and stress granules: connecting distant relatives. Traffic. 2014;15:1179–1193. doi: 10.1111/tra.12191. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, et al. Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J. Cell Sci. 2014;127:2029–2039. doi: 10.1242/jcs.143453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin U, Lallemand-Breitenbach V, de The H. PML nuclear bodies: regulation, function and therapeutic perspectives. J. Pathol. 2014;234:289–291. doi: 10.1002/path.4426. [DOI] [PubMed] [Google Scholar]
- 5.Boeynaems S, et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell. Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Sci. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 7.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. U S Am. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel A, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 10.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nat. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat. reviews. Mol. Cell. Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbaum-Garfinkle S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. U S Am. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti S, Hyman AA. Are aberrant phase transitions a driver of cellular aging? BioEssays: N. Rev. molecular, Cell. developmental Biol. 2016;38:959–968. doi: 10.1002/bies.201600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munder, M. C. et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife5, 10.7554/eLife.09347 (2016). [DOI] [PMC free article] [PubMed]
- 16.Mackenzie IR, et al. TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron. 2017;95:808–816 e809. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami T, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasagna-Reeves, C. A. et al. A native interactor scaffolds and stabilizes toxic ATAXIN-1 oligomers in SCA1. Elife4, 10.7554/eLife.07558 (2015). [DOI] [PMC free article] [PubMed]
- 23.Cummings CJ, Orr HT, Zoghbi HY. Progress in pathogenesis studies of spinocerebellar ataxia type 1. Philos. Trans. R. Soc. London. Ser. B, Biol. Sci. 1999;354:1079–1081. doi: 10.1098/rstb.1999.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S, Hong S. Molecular pathogenesis of spinocerebellar ataxia type 1 disease. Molecules Cell. 2009;27:621–627. doi: 10.1007/s10059-009-0095-y. [DOI] [PubMed] [Google Scholar]
- 25.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiti F, Dobson CM. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 27.Chai Y, Shao J, Miller VM, Williams A, Paulson HL. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl Acad. Sci. U S Am. 2002;99:9310–9315. doi: 10.1073/pnas.152101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp. Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummings CJ, et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 30.Bolger TA, Zhao X, Cohen TJ, Tsai CC, Yao TP. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J. Biol. Chem. 2007;282:29186–29192. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 31.Hearst SM, Lopez ME, Shao Q, Liu Y, Vig PJ. Dopamine D2 receptor signaling modulates mutant ataxin-1 S776 phosphorylation and aggregation. J. neurochemistry. 2010;114:706–716. doi: 10.1111/j.1471-4159.2010.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, et al. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nat. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophysical J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu CW, Liao VH. Arsenite induces neurotoxic effects on AFD neurons via oxidative stress in Caenorhabditis elegans. Metallomics. 2014;6:1824–1831. doi: 10.1039/c4mt00160e. [DOI] [PubMed] [Google Scholar]
- 36.Prakash C, Soni M, Kumar V. Biochemical and Molecular Alterations Following Arsenic-Induced Oxidative Stress and Mitochondrial Dysfunction in Rat Brain. Biol. Trace Elem. Res. 2015;167:121–129. doi: 10.1007/s12011-015-0284-9. [DOI] [PubMed] [Google Scholar]
- 37.Jain S, et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilks N, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.e04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tourriere H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Burright EN, et al. Identification of a self-association region within the SCA1 gene product, ataxin-1. Hum. Mol. Genet. 1997;6:513–518. doi: 10.1093/hmg/6.4.513. [DOI] [PubMed] [Google Scholar]
- 41.Klement IA, et al. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 42.Menon RP, et al. Mapping the self-association domains of ataxin-1: identification of novel non overlapping motifs. PeerJ. 2014;2:e323. doi: 10.7717/peerj.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Funez P, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nat. 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 44.Gennarino VA, et al. Pumilio1 Haploinsufficiency Leads to SCA1-like Neurodegeneration by Increasing Wild-Type Ataxin1 Levels. Cell. 2015;160:1087–1098. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue S, Serra HG, Zoghbi HY, Orr HT. The spinocerebellar ataxia type 1 protein, ataxin-1, has RNA-binding activity that is inversely affected by the length of its polyglutamine tract. Hum. Mol. Genet. 2001;10:25–30. doi: 10.1093/hmg/10.1.25. [DOI] [PubMed] [Google Scholar]
- 46.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl Acad. Sci. U S Am. 2015;112:E5237–5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. U S Am. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA. Reentrant Phase Transition Drives Dynamic Substructure Formation in Ribonucleoprotein Droplets. Angew. Chem. Int. Ed. Engl. 2017;56:11354–11359. doi: 10.1002/anie.201703191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maharana S, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Sci. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904. doi: 10.1038/Nphys3532. [DOI] [Google Scholar]
- 52.Dewey CM, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kedersha N, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamazaki T, et al. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell. 2018;70:1038–1053 e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 55.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 56.JanssenDuijghuijsen LM, et al. Mitochondrial ATP Depletion Disrupts Caco-2 Monolayer Integrity and Internalizes Claudin 7. Front. Physiol. 2017;8:794. doi: 10.3389/fphys.2017.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel A, et al. ATP as a biological hydrotrope. Sci. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 58.Hayes, M. H., Peuchen, E. H., Dovichi, N. J. & Weeks, D. L. Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. Elife7, 10.7554/eLife.35224 (2018). [DOI] [PMC free article] [PubMed]
- 59.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeng Wilson, Lee Sukyeong, Sung Nuri, Lee Jungsoon, Tsai Francis T.F. Molecular chaperones: guardians of the proteome in normal and disease states. F1000Research. 2015;4:1448. doi: 10.12688/f1000research.7214.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S, Williamson NA, Bogoyevitch MA. Complementary proteomics strategies capture an ataxin-1 interactome in Neuro-2a cells. Sci. Data. 2018;5:180262. doi: 10.1038/sdata.2018.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jarmoskaite I, Russell R. RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 2014;83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. reviews. Mol. Cell Biol. 2016;17:426–438. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 64.Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev. Cell. 2013;27:161–173. doi: 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bol GM, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015;7:648–669. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tantravedi S, et al. Targeting DDX3 in Medulloblastoma Using the Small Molecule Inhibitor RK-33. Transl. Oncol. 2019;12:96–105. doi: 10.1016/j.tranon.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tariq, A. et al. Potentiating Hsp104 activity via phosphomimetic mutations in the middle domain. FEMS Yeast Res18, 10.1093/femsyr/foy042 (2018). [DOI] [PMC free article] [PubMed]
- 68.de Chiara C, Menon RP, Strom M, Gibson TJ, Pastore A. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS one. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stenoien DL, Mielke M, Mancini MA. Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 2002;4:806–810. doi: 10.1038/ncb859. [DOI] [PubMed] [Google Scholar]
- 71.Servadio A, et al. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat. Genet. 1995;10:94–98. doi: 10.1038/ng0595-94. [DOI] [PubMed] [Google Scholar]
- 72.Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saha S, Hyman AA. RNA gets in phase. J. Cell Biol. 2017;216:2235–2237. doi: 10.1083/jcb.201706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Will CL, et al. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wen X, Tannukit S, Paine ML. TFIP11 interacts with mDEAH9, an RNA helicase involved in spliceosome disassembly. Int. J. Mol. Sci. 2008;9:2105–2113. doi: 10.3390/ijms9112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hozumi S, et al. DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish. PLoS one. 2012;7:e33675. doi: 10.1371/journal.pone.0033675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nat. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hink MA. Quantifying intracellular dynamics using fluorescence fluctuation spectroscopy. Protoplasma. 2014;251:307–316. doi: 10.1007/s00709-013-0602-z. [DOI] [PubMed] [Google Scholar]
- 79.Hinde E, Cardarelli F, Gratton E. Spatiotemporal regulation of Heterochromatin Protein 1-alpha oligomerization and dynamics in live cells. Sci. Rep. 2015;5:12001. doi: 10.1038/srep12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holehouse AS, Pappu RV. Protein polymers: Encoding phase transitions. Nat. Mater. 2015;14:1083–1084. doi: 10.1038/nmat4459. [DOI] [PubMed] [Google Scholar]
- 81.Mager WH, de Boer AH, Siderius MH, Voss HP. Cellular responses to oxidative and osmotic stress. Cell Stress. Chaperones. 2000;5:73–75. doi: 10.1379/1466-1268(2000)005<0073:CRTOAO>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rich T, et al. Disassembly of nuclear inclusions in the dividing cell–a novel insight into neurodegeneration. Hum. Mol. Genet. 1999;8:2451–2459. doi: 10.1093/hmg/8.13.2451. [DOI] [PubMed] [Google Scholar]
- 83.Burright EN, et al. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 84.Xia H, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 85.Keiser MS, Kordasiewicz HB, McBride JL. Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington’s disease and spinocerebellar ataxia. Hum. Mol. Genet. 2016;25:R53–64. doi: 10.1093/hmg/ddv442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watase K, et al. Lithium therapy improves neurological function and hippocampal dendritic arborization in a spinocerebellar ataxia type 1 mouse model. PLoS Med. 2007;4:e182. doi: 10.1371/journal.pmed.0040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cvetanovic M, Patel JM, Marti HH, Kini AR, Opal P. Vascular endothelial growth factor ameliorates the ataxic phenotype in a mouse model of spinocerebellar ataxia type 1. Nat. Med. 2011;17:1445–1447. doi: 10.1038/nm.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taniguchi JB, et al. RpA1 ameliorates symptoms of mutant ataxin-1 knock-in mice and enhances DNA damage repair. Hum. Mol. Genet. 2016;25:4432–4447. doi: 10.1093/hmg/ddw272. [DOI] [PubMed] [Google Scholar]
- 89.Whitehead LW, McArthur K, Geoghegan ND, Rogers KL. The reinvention of twentieth century microscopy for three-dimensional imaging. Immunol. Cell Biol. 2017;95:520–524. doi: 10.1038/icb.2017.36. [DOI] [PubMed] [Google Scholar]
- 90.Ducros, M. et al. Lattice light sheet microscopy and photo-stimulation in brain slices. Neural Imaging and Sensing 2019, Proc. SPIE, 10865, 10.1117/12.2509467 (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials used and data analysed during the current study are available from the corresponding authors on reasonable request.