Highlights
-
•
We present an approach for autism classification based on neuroimaging MRI.
-
•
The pipeline relies on connectivity matrices and machine learning techniques.
-
•
Accuracy is 85.06 ± 3.52% evaluated in more than 800 cases of the ABIDE I dataset.
-
•
The most important correlations for autism classification are highlighted.
-
•
Merging functional and structural information outperforms the monomodal pipelines.
Keywords: Autism, Functional MRI, Structural MRI, ABIDE, Classification
Abstract
Autism Spectrum Disorder (ASD) is a brain disorder that is typically characterized by deficits in social communication and interaction, as well as restrictive and repetitive behaviors and interests. During the last years, there has been an increase in the use of magnetic resonance imaging (MRI) to help in the detection of common patterns in autism subjects versus typical controls for classification purposes. In this work, we propose a method for the classification of ASD patients versus control subjects using both functional and structural MRI information. Functional connectivity patterns among brain regions, together with volumetric correspondences of gray matter volumes among cortical parcels are used as features for functional and structural processing pipelines, respectively. The classification network is a combination of stacked autoencoders trained in an unsupervised manner and multilayer perceptrons trained in a supervised manner. Quantitative analysis is performed on 817 cases from the multisite international Autism Brain Imaging Data Exchange I (ABIDE I) dataset, consisting of 368 ASD patients and 449 control subjects and obtaining a classification accuracy of 85.06 ± 3.52% when using an ensemble of classifiers. Merging information from functional and structural sources significantly outperforms the implemented individual pipelines.
1. Introduction
Autism Spectrum Disorder (ASD) is a neurological disorder characterized by persistent deficits in social communication (APA, 2013) that tends to evolve in severity over time (Gotham, Pickles, Lord, 2012, Szatmari, Georgiades, Duku, Bennett, Bryson, Fombonne, Mirenda, Roberts, Smith, Vaillancourt, et al., 2015). The symptoms generally appear in the first two years of life and include difficulty with communication and interaction, restricted interests and repetitive behaviors, as well as the degraded ability to function properly in various areas of life. The estimated prevalence of ASD in the United States is 1.47%, with average lifetime costs exceeding one million dollars per patient (Buescher et al., 2014).
The cause of the disorder, as suggested by the research, is a result of a combination of factors that include genetics, brain structure and function, as well as environmental influences (APA, 2013, Ha, Sohn, Kim, Sim, Cheon, 2015). Current diagnoses are interview-based and are most commonly performed by conducting the Autism Diagnostic Observation Schedule (Lord et al., 1989) or the Autism Diagnostic Interview - Revised (Lord et al., 1994). Although these methods are quite accurate, they are unable to point out the biological basis behind behavioral symptoms since the neuroanatomy is unclear (Riddle, Cascio, Woodward, 2017, Subbaraju, Suresh, Sundaram, Narasimhan, 2017). Nevertheless, in the last few decades there has been an increase in works focusing on magnetic resonance imaging (MRI) structural and functional brain abnormalities that would be symptomatic for the autism spectrum. However, the findings typically do not hold over the entire set of ASD subjects, although MRI studies have provided many implications of neurodevelopmental characteristics underlying ASD (Ecker et al., 2015). Structural MRI studies usually focus on volumetric and morphometric analyses to examine abnormal brain anatomy, while functional MRI studies have tried to investigate connectivity patterns in the brain, both locally and globally.
The vast majority of methods in structural MRI, instead of solving the inherent classification problem, try to point out common patterns among ASD patients versus the control group. For instance, voxel-based morphometry analysis (Riddle et al., 2017) showed an increase in total brain volume in children aged 2 to 4 with ASD, as well as an enlargement of the left anterior superior temporal gyrus. However, the picture is not so clear at a later age. While Aylward et al. (2002) observed no volumetric differences between ASD and control adult subjects, other studies concluded that the increase in total brain volume was still observable (Herbert, Ziegler, Deutsch, Oâ brien, Lange, Bakardjiev, Hodgson, Adrien, Steele, Makris, et al., 2003, Palmen, Pol, Kemner, Schnack, Durston, Lahuis, Kahn, Van Engeland, 2005). Other works investigated volumetric changes in particular regions of interest (ROIs) in the brain, but failed to reach strong conclusions. Palmen et al. (2005) reported an increase in gray matter in all lobes of the brain, whereas Courchesne et al. (2007) observed an increase in gray matter volume particularly in temporal lobes. In contrast, Herbert et al. (2003) reported findings on increased white matter, while Palmen et al. (2005) noted no difference in ASD versus control subjects regarding white matter volume. Conversely, Jou et al. (2011) reported a decrease in white matter volume in ASD patients. These inconsistent findings are likely due to the small sample sizes or because data were collected at a single site in each case (Riddle et al., 2017), and the choice of acquisition site has significant effects on the basic image properties (Nielsen et al., 2013). A promising study to solve the classification problem based on structural information was performed by Kong et al. (2019) based on the construction of an individual brain network for each subject in order to extract connectivity features between each pair of regions of interest. These features were then ranked and used to perform ASD versus control classification via a deep neural network classifier.
Functional connectivity in resting-state functional MRI is widely used to describe remote relationships in studies of the cerebral cortex parcellation and brain disorders (Du, Fu, Calhoun, 2018, Jiang, Zuo, 2016). The principal idea is to detect brain networks among functionally interconnected regions, which is done by using connectivity matrices that contain the correlations from different regions of the brain (Heinsfeld, Franco, Craddock, Buchweitz, Meneguzzi, 2018, Subbaraju, Suresh, Sundaram, Narasimhan, 2017). Recently, deep learning techniques have emerged as the main trend in ASD classification (Calhoun, Sui, 2016, Iidaka, 2015, Ju, Hu, Zhou, Li, 2019, Plis, Hjelm, Salakhutdinov, Allen, Bockholt, Long, Johnson, Paulsen, Turner, Calhoun, 2014). Popular approaches include simple multi-layer perceptron (MLP) networks combined with the unsupervised training of stacked autoencoders (Guo, Dominick, Minai, Li, Erickson, Lu, 2017, Kim, Calhoun, Shim, Lee, 2016). Even though several methods managed to obtain relatively high classification accuracy, there are some drawbacks of the proposed strategies that need to be addressed. First, most of the studies used a small number of subjects to perform the classification. This tends to lead to unreliable results, because of poor generalization. The real challenge is to replicate findings across large datasets. An accuracy above 0.9 is obtained only when using dozens of cases (Arbabshirani et al., 2017), and accuracy drops significantly when a larger dataset is introduced (Heinsfeld et al., 2018). Second, most of the studies used data acquired on a single site. This procedure also does not generalize the problem efficiently since the image properties highly depend on the imaging protocol conducted at each institution. In summary, few studies used multisite data with a number of subjects higher than 800. Moreover, these studies considered an approach in which they focused only on functional findings while neglecting structural information. In our knowledge, only one study conducted a fusion approach, combining fMRI and diffusion tensor imaging (DTI) information for ASD identification, although it only used a sample size of 30 subjects in total (Deshpande et al., 2013).
In this paper, we propose an approach for the classification of ASD versus control group that merges information from functional and structural MRI evaluated on a large multisite dataset. Functional data connectivity matrices contain information about correlation coefficients of mean blood-oxygen level dependent (BOLD) signals from pairs of regions of interest. Structural data connectivity matrices contain information about cortical gray matter volumes. The principal hypothesis is that using multisite data and combining both structural and functional information could potentially unveil patterns that have not been exploited so far, while at the same time improving generalization in terms of classification due to the lack of reliance on a specific protocol. Our proposed method consists of several steps that include structural and functional data preprocessing, extraction of the features represented by the connectivity matrices, utilization of the Fisher score as a feature dimensionality reduction technique and, ultimately, classification of the data by means of an ensemble of autoencoders and multilayer perceptrons. Specifically, our proposal was inspired by the works of Heinsfeld et al. (2018) and Kong et al. (2019), which dealt with the classification of ASD based on either functional or structural data, respectively. The classification task itself was performed in a similar manner in both of these papers, even though the approach of Heinsfeld et al. (2018) did not include any dimensionality reduction technique, and the approach of Kong et al. (2019) was only based on a subset of images coming from a single screening site. Our approach is evaluated using the large and international multisite Autism Brain Imaging Data Exchange I (ABIDE I) dataset (Di Martino et al., 2014). Moreover, we show the generalization of the proposed approach including a leave-one-site-out cross-validation experiment.
2. Material and methods
2.1. Dataset
The ABIDE I dataset was used to conduct our study (Di Martino et al., 2014). It was released in August 2012 as a result of a collaboration involving 17 international sites and it consists of 1112 cases, including 539 from individuals with ASD and 573 from typical controls. The subjects ages ranges from with a median age of 14.7 years across the groups. The cases include structural MRI images and resting-state fMRI series of images, along with clinical data not utilized in this work.
Structural data and phenotypic information about the subjects were obtained directly from the ABIDE I initiative. However, the preprocessed dataset that includes functional information was acquired from the Preprocessed Connectomes Project (PCP) (Craddock et al., 2013). All rs-fMRI series were subjected to the processing pipeline CPAC (Configurable Pipeline for the Analysis of Connectomes), which includes slice time correction, motion correction, and intensity normalization, bandpass filtering (0.01 Hz - 0.1 Hz) and spatial registration to the MNI152 template space. Various derivatives of the functional data are available at PCP, however, the one of interest for our classification pipeline is the time series of BOLD signals in different areas of the brain. Two different, common used atlases are tested: the AAL (Automated Anatomical Labeling) atlas (Tzourio-Mazoyer et al., 2002) and the CC200 (Cameron Craddock’s 200 ROI) parcellation atlas (Craddock et al., 2012). Notice, however, that functional data is rendered useless in some cases, due to the motion artifacts, and it is therefore not available from PCP. Motion artefacts were computed for each individual case using mean framewise displacement, and if it surpassed the value of 0.2, the corresponding subject was discarded. Mean framewise displacement is a measure of head motion, which compares the motion between the current and previous volumes. This left us with a dataset of 884 rs-fMRI subjects, including 408 ASD patients and 476 control cases.
Regarding the structural information, cortical parcellation using the Destrieux atlas (Destrieux et al., 2010) was performed for each MRI volume. To achieve this, we employed the well-known Freesurfer software (Fischl et al., 2002). The pipeline that was used to extract useful information involves multiple stages, with the most notable ones being intensity normalization, skull stripping, registration of the volumes to a common space, segmentation and, ultimately, cortical parcellation. Apart from the division of cortex, a series of statistical measures, such as gray matter volume, cortical thickness and curvature, were computed for each of the parcels. However, due to artifacts that are present in some of the structural MRI volumes, this processing was not always possible. A total of 1014 cases were successfully processed, including 475 ASD patients and 539 control subjects.
Since our goal is to propose a classification strategy dealing with both structural and functional information, it is important to note that cross-referencing the remaining cases after using the preprocessing pipelines yields a total of 817 cases (368 ASD + 449 control) that are present in both subsets of the original dataset. Table 1 summarizes the details of the dataset used, including separate listings for every acquisition site involved in the project.
Table 1.
Summary of the number of subjects used from every screening site in the ABIDE I dataset in each of the strategies conducted: functional, structural and combined classification pipelines. For each center, we removed the cases that failed the preprocessing step. ASD refers to Autism Spectrum Disorder patients and TC to Typical Control.
| Site | ABIDE I |
Functional |
Structural |
Combined |
||||
|---|---|---|---|---|---|---|---|---|
| ASD | TC | ASD | TC | ASD | TC | ASD | TC | |
| Caltech | 19 | 19 | 19 | 18 | 17 | 18 | 17 | 17 |
| CMU | 14 | 13 | 3 | 2 | 14 | 12 | 3 | 2 |
| KKI | 22 | 33 | 12 | 27 | 20 | 32 | 11 | 26 |
| Leuven | 29 | 35 | 27 | 34 | 26 | 35 | 24 | 34 |
| MAX MUN | 24 | 33 | 18 | 24 | 21 | 31 | 17 | 22 |
| NYU | 79 | 105 | 73 | 98 | 74 | 103 | 69 | 96 |
| OHSU | 13 | 15 | 12 | 11 | 9 | 14 | 8 | 11 |
| OLIN | 20 | 16 | 14 | 11 | 17 | 16 | 11 | 11 |
| PITT | 30 | 27 | 22 | 23 | 27 | 24 | 20 | 20 |
| SBL | 15 | 15 | 14 | 12 | 14 | 14 | 13 | 11 |
| SDSU | 14 | 22 | 12 | 21 | 13 | 18 | 12 | 17 |
| Stanford | 20 | 20 | 17 | 19 | 19 | 16 | 16 | 15 |
| Trinity | 24 | 25 | 21 | 23 | 19 | 24 | 17 | 22 |
| UCLA | 62 | 47 | 36 | 39 | 50 | 43 | 34 | 37 |
| UM | 68 | 77 | 48 | 65 | 52 | 72 | 38 | 62 |
| USM | 58 | 43 | 38 | 23 | 56 | 42 | 37 | 23 |
| Yale | 28 | 28 | 22 | 26 | 27 | 25 | 21 | 23 |
| Total | 539 | 573 | 408 | 476 | 475 | 539 | 368 | 449 |
| Total Cases | 1112 | 884 | 1014 | 817 | ||||
2.2. Functional data classification pipeline
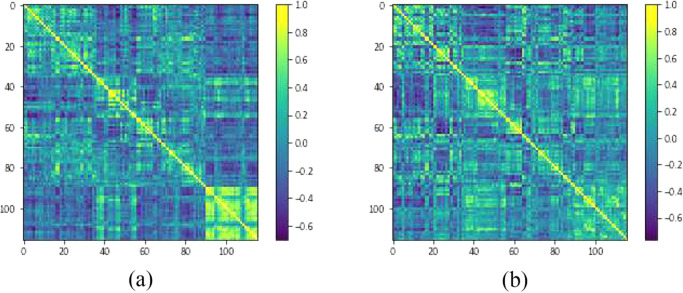
Once the data has been preprocessed with the CPAC pipeline and the time series of mean BOLD signals from different brain regions are extracted, the next step is to build a connectivity matrix. Such a matrix is constructed for each case individually and contains information about the correlation of the BOLD series between each pair of the regions defined by an atlas (the AAL atlas consists of 116 regions, whereas the CC200 atlas consists of 200). Therefore, the dimensions of a connectivity matrix are 116 × 116 or 200 × 200, depending on the atlas used, and each element ij inside a matrix is the Pearson correlation coefficient computed for the mean BOLD series from regions i and j. By definition of the Pearson correlation coefficient, elements of the matrix range from to 1, and all elements on the main diagonal are equal to 1 since they correspond to the correlation of a signal with itself. Additionally, such a matrix is symmetrical because of the commutative property of the correlation coefficient computation. Examples of connectivity matrices are shown in Fig. 1. We are, therefore, only interested in the upper triangle of the connectivity matrix (also excluding the main diagonal), as the remainder of the matrix is redundant. The part of interest is then flattened into a 1-dimensional vector for further manipulation. In the case of the AAL atlas, such a vector contains 6670 elements, whereas in case of using the CC200 atlas it contains 19, 900 elements.
Fig. 1.
Examples of connectivity matrices from two subjects in the Caltech subset of the ABIDE I dataset constructed using the AAL atlas. (a) is from an ASD patient while (b) is from a control.
Due to the high dimensionality of the feature vectors, we included a dimensionality reduction technique in the pipeline. The advantage of this step is that we avoid overfitting and make the model much more generalizable. The technique used to achieve this goal is the Fisher score computation, which ranks the features in order of distinctiveness and, consequently, decides which of them are of lesser importance (Chen and Lin, 2006). It measures the discrimination of two sets of real numbers, the greater the score value is, the higher the rank of a certain feature. Given training vectors xk, if the number of positive instances is and the number of negative instances is (where positive and negative instances indicate belonging to one class or the other), the Fisher score of the ith feature is:
| (1) |
where and are the mean of the ith feature of the whole, positive and negative sets, respectively, is the ith feature of the kth positive instance and is the ith feature of the kth negative instance (Kong et al., 2019). The numerator indicates interset discrimination, whereas the denominator indicates between-set discrimination.
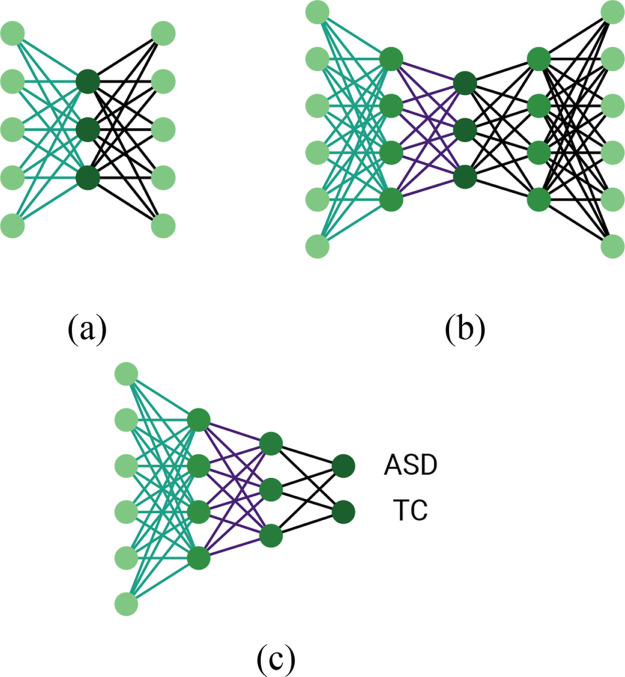
What is left after application of the Fisher score is a reduced feature vector that becomes the input vector for the classifier. The classification step itself is performed in two stages. The first one consists of an unsupervised training of stacked autoencoders. An autoencoder is a simple network that tries to reconstruct the input as precisely as possible. Given the input vector, it tries to learn a lower-dimensional representation of it, from which it can then reconstruct the original vector. These two steps are referred to as encoding and decoding. Simply, it has an input layer, a hidden fully connected layer that encodes the input and a fully connected output layer that decodes the encoded representation. Parameters of the model are adjusted by back-propagation until the difference between input and output has been minimized. A stacked autoencoder basically consists of two or more autoencoders and has a better learning ability than a single one. When the stack consists of two autoencoders, the output of the first encoding stage is given as input to the second autoencoder. Then, the decoding stage is done in a two-fold manner again, the second autoencoder decodes its input, and then the first autoencoder decodes the original input vector. An illustration of such a structure is shown in Fig. 2(b).
Fig. 2.
Graphical representation of (a) the simple and (b) stacked autoencoder structures and (c) the multilayer perceptron (MLP). The colored weights of the encoding part of stacked autoencoder (b) are used as initializing weights of the MLP (c).
The end result of the autoencoder training is apparent in the second stage of the classification step, which is a supervised training of a multilayer perceptron (MLP) with two hidden layers and a binary output layer. The number of nodes in the hidden layers corresponds to the number of nodes in the encoding layers of the stacked autoencoder. This ensures that the weights of the MLP can be initialized using the weights from the trained autoencoder. Hence, the MLP is able to learn hidden features from input vectors in the hidden layers and then classify the subjects accordingly in the ultimate layer with softmax activation. This is illustrated in Fig. 2(c), where the corresponding weights initialized from the autoencoder are color-coded in the same manner as in Fig. 2(b). To prevent overfitting, dropout is introduced in the hidden layers, as well as additional regularization terms and batch normalization that improves convergence.
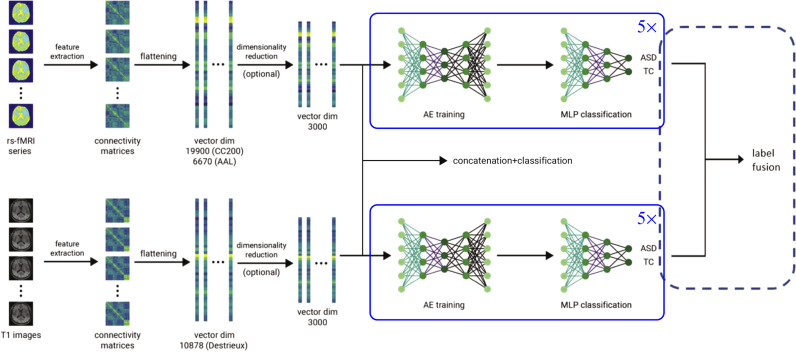
To get more robust results, we use an ensemble of classifiers in the last steps, so that each one can learn different feature representations. Specifically, we trained an ensemble of a total of 5 classifiers obtained by changing the number of nodes in the hidden layers of the autoencoders and MLPs. In this case, each subject’s feature vectors are given as an input to all classifiers, and the label assignment is conducted by averaging the softmax activation probabilities. The underlying hypothesis is that additional classifiers with different ways of learning feature representations can add a certain margin of improvement in terms of classification accuracy and make the decision-making more robust (Kamnitsas et al., 2017). As a summary, the complete functional pipeline is illustrated in Fig. 3 (top), which shows all the notable steps, including the flattening of connectivity matrices into vectors, dimensionality reduction, training of the stacked autoencoder and the MLP classification.
Fig. 3.
Graphical representation of the functional (top) and structural (bottom) data classification pipelines, together with combined strategies, including concatenation branch and label fusion after separate pipelines.
2.3. Structural data classification pipeline
The approach for structural data classification is very similar to the functional classification pipeline by design in order to obtain a subsequent smooth integration of the two. Hence, again, a connectivity matrix is built for each subject, and the upper triangular part is extracted and flattened to become a feature vector. The main difference between the functional and structural pipelines is the way connectivity matrices are built. Instead of computing the Pearson correlation coefficient, we are interested in the relations between gray matter volumes in each pair of the cortical parcels defined by the Destrieux atlas (148 regions, 74 in each hemisphere). Even though the features computed after the Freesurfer pipeline are readily usable, the underlying idea was to construct such a matrix so that the features reflect the volumetric correspondences among regions for a particular subject. That way, the networks represented by matrices and built for each subject become comparable with each other, even though the dataset is highly-varying and encompasses large age span among the subjects. Thus, each element ij of the matrix is the volumetric correspondence between two parcels i and j, which is defined by:
| (2) |
where gm(i) and gm(j) are the gray matter volumes of ROIs i and j (Kong et al., 2019).
As a result, the flattened vector extracted from the connectivity matrix has 10878 features. Similar to the functional pipeline, the Fisher score is used to reduce the dimensionality of the feature vectors, and the new obtained ones were fed to the ensemble of 5 stacked autoencoders and the MLP for classification task. The structural pipeline is illustrated in Fig. 3 (bottom).
2.4. Combined data classification pipeline
One of the main contributions proposed in this work involves combining the two previously described pipelines into one, with the goal of improving the classification results by accounting for different types of information. Since the functional and structural pipelines learn independent features, merging them together could mitigate errors to some extent. Notice that only cases that successfully underwent both preprocessing pipelines, functional and structural, can be considered as part of the dataset for the combined classification pipeline. In the ABIDE I dataset, this turns out to be a set of 817 cases.
The merging of functional and structural information was performed using two different strategies. The first strategy involves concatenating the structural and the functional feature vectors after the dimensionality reduction stage. This way, we merge the functional and structural data and then perform the autoencoder training, so that the autocander can decide which patterns are more informative. We opted to do the merging after the dimensionality reduction step since the sizes of the original feature vectors are dependent on the choice of atlas and therefore different for structural and functional data, which introduces a potential bias towards one side or the other. Subsequently, the classification was done using either the newly obtained vector (of length 6000) or reducing its dimensionality again with the Fisher score to get another vector to be used as an input to the network. The classification stage remained unchanged, consisting of the ensemble composed of the unsupervised stacked autoencoder training followed by the supervised training of an MLP (Fig. 3, concatenation branch). The second strategy consists of using separate classification pipelines, as previously described, followed by a decision-making method for choosing the final labels. In other words, we independently trained the functional and structural ensembles of autoencoders and multi-layer perceptrons using the corresponding functional and structural training and validation data, resulting in 10 softmax activations. The final output is obtained by either averaging the 10 softmax activation probabilities or by a majority voting strategy. This approach is illustrated in Fig. 3 (label fusion strategy).
2.5. Validation
Every model was validated by performing 10-fold cross-validation. In each fold, 10% of the corresponding dataset was used to test the classifier, while the remaining 90% of cases were used for training and validation, where training encompassed 70% and validation 30% of the available set. This strategy allowed for the evaluation of the model’s robustness and behavioral effects when training and testing with different subsets of data. It is important to note that in each fold, the set was split in such a way that the subsets retained class balance and contained cases coming from all or almost all screening sites. This approach was utilized to keep the model as generalizable as possible. Naturally, in the case of training 2 classifiers or an ensemble of classifiers in the combined approach, the split into training, validation and testing sets was the same for all the classifiers in a certain fold.
The models were evaluated using accuracy as a metric, which is the most common measurement in the state-of-the-art works. This allowed for quantitative comparison of our results with previous models, although there is no guarantee that the dataset used in our method is identical to the ones of previous works. Additionally, due to the class imbalance, we quantified sensitivity and specificity values for each of the experiments. Ultimately, we also used the best model to perform a leave-one-site-out cross-validation, in order to analyze the accuracy, sensitivity and specificity obtained for each of the 17 screening sites individually. This allowed for some qualitative and quantitative comparison of our results with other state-of-the-art works, especially since some of the previous research was conducted using only the data collected at one of the sites.
To statistically analyze the results obtained through the presented models, we performed a one-way ANOVA (ANalysis Of VAriance) test, together with a post hoc Tukey HSD (honestly significant difference) test, with the goal of indicating which models were significantly different. ANOVA and Tukey HSD tests were conducted separately on each of the three groups of models, functional data-based, structural data-based and combined data-based. A 95% confidence interval was chosen, meaning that a p-value less than 0.05 indicates a high statistical significance of a certain result when compared to another. Notice that the t-test estimation has inherent errors when evaluating the results of a 10-fold cross-validation procedure. However, it has a high statistical discrimination power when analyzing the Type II errors (i.e. the failure to detect a real difference between algorithms).
2.6. Qualitative analysis
According to Ha et al. (2015), the default mode network (DMN), which is one of the most commonly analyzed functional brain networks, shows a difference in brain activity between ASD and control subjects. DMN generally tends to be hypo-connected in adults with the disorder and hyperconnected in children with the same pathology. It comprises several parts of the brain and includes the posterior cingulate gyrus, retrosplenial cortex, lateral parietal cortex, medial prefrontal cortex, superior frontal gyrus and temporal lobe. It has shown greater activity during resting-state functional MRI than during task-based screenings (Greicius et al., 2003), which is why it is of a particular interest for the ABIDE I dataset.
To compare our method with common clinical findings, we propose a qualitative test of feature ranking. Because the Fisher score ranks all features by their distinctiveness, the idea is to analyze whether the top ranked features correspond or do not correspond to discoveries observed in previous clinical and scientific research and to check for the presence of potential common patterns.
3. Results
Table 2 summarizes the conducted experiments and accuracies obtained, providing additional details on standard deviations over folds in cross-validation, as well as sensitivity and specificity values. We tested the accuracy of the functional and structural pipelines alone and in combination. The latter is obtained by either concatenating the former two before the classification (i.e. training the neural network with functional and structural information together) or optimizing functional and structural pipelines independently and then combining the results (separate pipelines).
Table 2.
Summary of the conducted experiments and the obtained results. Vec. dim. - dimensionality of the input feature vectors and indicative of whether dimensionality reduction was used or not; Strategy - indication of whether the functional and structural feature vectors were concatenated prior to classification or classified separately; Ensemble - indication of whether an ensemble of classifiers was or was not used, and if yes, how many classifiers were considered; * - label fusion was performed using the average of softmax probabilities; ** - label fusion was performed by majority voting, using average softmax probabilities only in the case of a tie). Values of mean accuracy and its standard deviation are shown in percentages. Sensitivity and specificity values are also shown. The best result is highlighted in bold.
| Exp. no. | Pipeline | Atlas | Cases | Vec. dim. | Acc. mean | Acc. std | Sens. | Spec. |
|---|---|---|---|---|---|---|---|---|
| Functional data classification | ||||||||
| 1 | functional | AAL | 884 | 6670 | 64.46 | 4.73 | 0.53 | 0.72 |
| 2 | functional | AAL | 884 | 3000 | 67.96 | 3.50 | 0.60 | 0.73 |
| 3 | functional | CC200 | 884 | 3000 | 70.37 | 4.88 | 0.64 | 0.75 |
| 4 | ensemble of 5 | CC200 | 884 | 3000 | 74.90 | 3.48 | 0.74 | 0.76 |
| Structural data classification | ||||||||
| 5 | structural | Destrieux | 1014 | 10,878 | 52.27 | 3.51 | 0.47 | 0.55 |
| 6 | structural | Destrieux | 1014 | 3000 | 77.41 | 3.99 | 0.71 | 0.76 |
| 7 | ensemble of 5 | Destrieux | 1014 | 3000 | 78.69 | 2.86 | 0.78 | 0.79 |
| Combined data classification | ||||||||
| Exp. no. | Strategy | Ensemble | Cases | Vec. dim. | Acc. mean | Acc. std | Sens. | Spec. |
| 8 | concatenate | no | 817 | 6000 | 70.38 | 2.17 | 0.62 | 0.76 |
| 9 | concatenate | no | 817 | 3000 | 70.37 | 4.01 | 0.65 | 0.77 |
| 10 | concatenate | yes (5) | 817 | 6000 | 73.44 | 4.84 | 0.69 | 0.77 |
| 11 | concatenate | yes (5) | 817 | 3000 | 73.31 | 3.86 | 0.71 | 0.75 |
| 12 | separate | no | 817 | 3000 | 82.97 | 3.72 | 0.82 | 0.84 |
| 13 | separate* | yes (5+5) | 817 | 3000 | 85.06 | 3.52 | 0.81 | 0.89 |
| 14 | separate** | yes (5+5) | 817 | 3000 | 84.45 | 2.90 | 0.80 | 0.88 |
To test the pipeline for functional data classification, four experiments were proposed. Two of those considered data preprocessed using the AAL atlas, whereas the other two used data obtained with the CC200 atlas. In both approaches, we considered two sizes for the input feature vectors, both without and with the application of the dimensionality reduction method. When using the AAL atlas, accuracies of 64.46% and 67.96% were reached without and with dimensionality reduction, respectively. The accuracy was higher overall when using the CC200 atlas. We report an accuracy of 70.37% when using the Fisher score to reduce the size of the input vector. The last model, using the CC200 atlas with dimensionality reduction, was further explored by conducting the classification using the ensemble of classifiers. An improvement was reached, resulting in an accuracy of 74.90%. When comparing the results among these strategies, all pairwise p-values were lower than 0.05.
For structural information we also tested two scenarios, when using the original feature vector and when using the reduced one. The two obtained accuracies differ greatly. Using the entire feature vector we only obtained 52.27% accuracy, while including the dimensionality reduction, the accuracy was 77.41%. Furthermore, using the ensemble classification on the latter approach, the accuracy increased, yielding in an average accuracy of 78.69% over the 10 folds of the cross-validation. All three pairwise p-values were statistically significant (p < 0.05)
The combined approach was split into two major strategies. The first utilizes concatenation of the reduced vectors obtained via structural and functional data preprocessing. The second considers separate classification of the functional and structural data followed by the fusion of the obtained labels, either by averaging the softmax outputs or by majority voting. Several models were proposed for both strategies, including additional dimensionality reduction of the concatenated vector or classification using an ensemble of classifiers. The best result was obtained when considering the separate classification strategy, using an ensemble of 5 functional and 5 structural data classification models, followed by averaging of all 10 softmax probabilities. A mean accuracy of 85.06% was achieved with this approach. All pairwise comparisons were significant (p < 0.05) except for Experiments 13 and 14, which differ only in the way the labels were fused together.
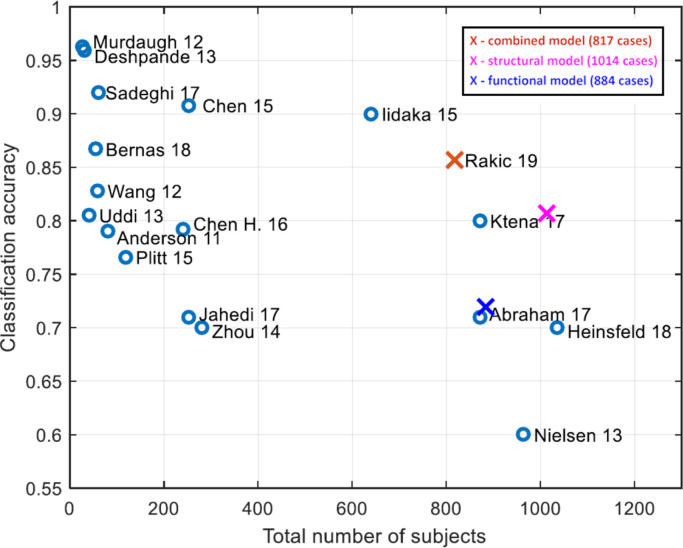
Fig. 4 shows how our best model for each functional, structural and combined pipelines compared to previous state-of-the-art works. The works reported in this figure were based on functional data only, since the work by Kong et al. (2019) is the only one, as far as we are aware of, that dealt with structural data-based classification task. Even though the accuracy we obtained is not the highest, every other result that is quantitatively better was based on a smaller dataset and often included data only originating from a single screening site. Furthermore, the works based on a larger dataset than ours yielded in a significantly lower accuracy. However, the behavior of our model when tested on the entirety of the ABIDE I dataset remains unknown since a portion of the full subject set was discarded due to various artifacts. Notice also in Fig. 4 the advantage of combining both functional and structural information, obtaining a significant improvement (p < 0.05) against the individual pipelines.
Fig. 4.
Comparison of the results obtained using the functional, the structural, and the merged classification pipelines with the ones reached in previous works that dealt only with functional data-based classification task. Figure modified from Du et al. (2018). The references are Anderson et al. (2011), Murdaugh et al. (2012), Wang et al. (2012), Deshpande et al. (2013), Nielsen et al. (2013), Uddin et al. (2013), Zhou et al. (2014), Chen et al. (2015), Iidaka (2015), Plitt et al. (2015), Chen et al. (2016), Abraham et al. (2017), Jahedi et al. (2017), Ktena et al. (2018), Sadeghi et al. (2017), Bernas et al. (2018) and Heinsfeld et al. (2018)
Finally, with the best model obtained, we did a quantitative analysis of the accuracy reached for each of the 17 imaging sites, by performing a leave-one-site-out cross-validation. Table 3 summarizes the obtained values, including sensitivity and specificity values reached. We obtained a consistent accuracy over 70% for all but the CMU center, where the accuracy was 60%. However, notice the small number of cases used at this center, which potentially biases the classification accuracy. This shows that the approach is able to generalize well to unseen data coming from different screening sites, machines and/or acquisition protocols.
Table 3.
Classification accuracy obtained for each of the 17 screening sites by performing leave-one-site-out cross-validation using the best model, which considers a combined data classification approach with an ensemble of classifiers. NoS - Number of subjects, Sens. - Sensitivity, Spec. - Specificity
| Site | NoS | Accuracy [%] | Sens. | Spec. |
|---|---|---|---|---|
| Caltech | 34 | 73.53 | 0.76 | 0.71 |
| CMU | 5 | 60.00 | 0.67 | 0.50 |
| KKI | 37 | 75.68 | 0.82 | 0.73 |
| Leuven | 58 | 79.31 | 0.58 | 0.94 |
| MAX MUN | 39 | 82.05 | 0.88 | 0.77 |
| NYU | 165 | 86.67 | 0.96 | 0.80 |
| OHSU | 19 | 73.68 | 0.62 | 0.82 |
| OLIN | 22 | 86.36 | 0.73 | 1.00 |
| PITT | 40 | 85.00 | 0.75 | 0.95 |
| SBL | 24 | 66.67 | 0.46 | 0.91 |
| SDSU | 29 | 79.31 | 0.75 | 0.82 |
| Stanford | 31 | 80.64 | 0.88 | 0.73 |
| Trinity | 39 | 82.05 | 0.82 | 0.82 |
| UCLA | 71 | 90.14 | 0.85 | 0.95 |
| UM | 100 | 87.00 | 0.84 | 0.89 |
| USM | 60 | 86.67 | 0.97 | 0.70 |
| Yale | 44 | 93.18 | 0.95 | 0.91 |
3.1. Qualitative feature analysis
Since the Fisher score ranks features according to how discriminant they are, we wanted to analyze whether or not the top features (i.e. top functional connectivity patterns between pairs of regions) correspond to common findings in clinical research based on functional connectivity. Table 4 lists the top 15 pairs of regions whose connectivity patterns are of the most interest in regard to the classification task.
Table 4.
Top 15 most discriminant pairs of regions for the classification task, corresponding to the top 15 functional connectivity features. The labels are defined by the AAL atlas.
| Feature rank | Pair of brain regions |
|---|---|
| 1 | Right Superior Frontal Gyrus |
| Right Middle Temporal Gyrus | |
| 2 | Left Superior Medial Gyrus |
| Left Superior Frontal Gyrus | |
| 3 | Right Middle Frontal Gyrus |
| Right Insula Lobe | |
| 4 | Right Inferior Frontal Gyrus |
| Left Middle Occipital Gyrus | |
| 5 | Right Caudate Nucleus |
| Right Insula Lobe | |
| 6 | Right Inferior Parietal Lobule |
| Right Middle Frontal Gyrus | |
| 7 | Right Anterior Cingulate Cortex |
| Right Calcarine Gyrus | |
| 8 | Left Inferior Frontal Gyrus |
| Left Cerebellum | |
| 9 | Right Fusiform Gyrus |
| Left Insula Lobe | |
| 10 | Right Middle Frontal Gyrus |
| Left Subcallosal Gyrus | |
| 11 | Right Inferior Parietal Lobule |
| Left Cuneus | |
| 12 | Right Thalamus |
| Left Inferior Frontal Gyrus | |
| 13 | Right Angular Gyrus |
| Left Superior Frontal Gyrus | |
| 14 | Left Thalamus |
| Right Calcarine Gyrus | |
| 15 | Left Precuneus |
| Left Angular Gyrus |
According to the table, the most interesting correlation is between the Right Superior Frontal Gyrus and the Right Middle Temporal Gyrus. On the other hand, the Right Middle Frontal Gyrus appears in 3 of the 15 correlations, while the Left Inferior Frontal Gyrus, the Left Superior Frontal Gyrus, the Right Calcarine Gyrus, the Right Inferior Parietal Lobule, and the Right Insula Lobe each appear in two of the correlations.
We conducted an analogous analysis to examine top features in the case of structural data. There are some observations and recurring patterns of note. For instance, in the top 15 features (i.e. top 15 most discriminant correlations between pairs of cortical regions), left transverse temporal sulcus appears in 5 of them. This can signify importance of that particular cortical parcel in regard to ASD classification, since it is one of 148 possible parcels and appear in total in five of the top 15 features. Other recurring regions on the list are the left intraparietal sulcus and the transverse parietal sulci, appearing four times, as well as the right subcallosal gyrus, which appears in three features.
4. Discussion
In this work, we aimed to improve ASD detection by means of combining structural and functional MRI information. In particular, we implemented and combined two baseline implementations based on the approaches developed by Heinsfeld et al. (2018) for functional data classification and Kong et al. (2019) for structural data classification, obtaining comparable results as the ones presented in these two works. Indeed, discrepancies between our results and the reported ones were expected since there is no guarantee that the cases used for classification were the same, the split between training, validation and testing was randomly generated and, finally, some details of the original implementations were not available. When designing our baselines implementations, it is important to note that we kept the best parameters analyzed in those works. Therefore, the number of nodes in the hidden layers of the default autoencoder and the MLP (1000 and 600 in two layers) were the ones used in the work of Heinsfeld et al. (2018). The same architecture was preserved for the structural data classification pipeline. On the other hand, the choice to lower the input vectors’ dimensionality to 3000 was shown to be the best by Kong et al. (2019) (different models were tested by varying dimensionality of the input vectors from 2000 to 5000), and, consequently, we applied the same reduction constraint when conducting the functional data classification strategy. Additionally, an analogous test was performed on functional data (by varying dimensionality of input vectors from 2000 to 5000 with a step of 200) and it was shown that 3000 elements was an optimal choice indeed.
Using the Fisher score as a dimensionality reduction technique improved the results, both when considering the functional and structural pipelines separately as well as in the combined approach. It prevents overfitting the classifier by selecting the most discriminant features from the defined set of features. Consequently, it minimizes redundancy. Dimensionality reduction is particularly beneficial in regard to structural data classification. From Table 2, we observe a significant increase in the classification accuracy by removing the redundant features (p < 0.05). This may be due to the fact that the shallow architecture of the proposed stacked the autoencoder is not able to handle such long original feature vectors, thus Fisher score served as a pre-processing step to reduce its size. This accuracy difference is much lower in regard to the functional data classification, but that pipeline also shows variation in the results as a consequence of the atlas choice. As shown in the functional pipeline, we used both the AAL and CC200 atlases to preprocess the functional data. When comparing the obtained results, CC200 outperformed AAL in all of the conducted experiments (p < 0.05). This may be because the CC200 atlas has 200 defined regions, whereas AAL only has 116. We think that the additional regions unveil more information and connectivity patterns, which may not have been present or distinctive when using the AAL atlas alone.
If we consider separate approaches for the structural and functional data classification, we can conclude that the structural pipeline significantly outperforms the functional one in terms of the obtained accuracy (p < 0.05). This could be because there were more cases available for classification after the structural preprocessing pipeline. Furthermore, even though the features are defined independently in the two pipelines, they come from separate modalities, thus, it is shown that the dimensionality reduction technique has a greater effect on the structural data classification than on the functional, which is another justification for the difference in the quantitative results. In other words, the improvement in terms of classification accuracy is higher in the structural pipeline than in the functional pipeline when the Fisher score is applied. In regard to the implementation using an ensemble of classifiers, there is a statistically significant improvement in classification accuracy in both the functional and structural pipelines (p < 0.05). Each classifier in the ensemble is able to learn different representations of the input feature vectors, and by fusing the output labels, errors can be mitigated up to a certain extent. This compensation of errors is much more significant in the case of the combined classification pipeline because the input vectors encompass more information from different modalities. Regarding label fusion, we tried both majority voting and averaging the softmax activations. As it turns out, the latter approach slightly outperforms the former. When conducting majority voting, all the labels are given the same weight, whereas when considering softmax probabilities, the classifiers that output probabilities with higher certainty are given more weight in the decision making than the ones with lower certainty. However, this improvement was not statistically significant.
Quantitative analysis per site was performed to test the robustness of the best model and its generalization capability. The idea was to collect misclassified samples over 10 folds of the cross-validation and, consequently, determine which screening site from where they were originated. Then, the classification accuracies were computed for each of the 17 sites. The lowest accuracy of 60% was obtained for the CMU site, however, the total number of cases considered from that particular site is only 5, which is much lower than the number of cases coming from the other sites. Having more available cases would arguably increase this site’s accuracy in an individual site analysis. In the rest of the centers, the classification was over 75%, and we even obtained an accuracy of more than 90% in 3 of them.
Another analysis we performed investigated whether the top discriminant features ranked by the Fisher score correspond to the common findings in functional brain connectivity, particularly in the research related to default mode network connectivity (Greicius, Krasnow, Reiss, Menon, 2003, Ha, Sohn, Kim, Sim, Cheon, 2015). By comparing the DMN regions with the regions corresponding to the top ranked features (shown in Table 4), some similarities can be observed. There is a presence of superior frontal gyri and temporal lobe in both, but there are also pairs of regions that the classification network is able to discriminate between, that are not present in DMN. The reasoning is that functional connectivity patterns are observed globally, not by focusing on one particular brain network. Our method quantifies the connectivity of all ROI pairs and, consequently, selects the most distinct connections in order to optimize classification accuracy. Instead, the DMN analyses only focus on the connectivity of regions that fall under the definition of DMN, disregarding the rest. This selection of the most discriminant features can justify the improvement in accuracy when conducting the combined approach; we consider two different modalities, with two separate types of features, and select the most distinguishable ones from both. When it comes to the analogous analysis we performed on the structural-based features, we presented some common findings among the top 15 features (i.e. pairs of regions with distinct volumetric correspondence). However, there is no standardized network or model like DMN that would provide grounds for comparison and deriving conclusions.
A total of 295 cases out of 1112 in the original dataset did not meet the required preprocessing criteria, either in the functional or structural datasets, or, in some cases, both. This is one of the main limitations of our work, because the reported results are not obtained using the entire ABIDE I dataset. Furthermore, this restriction obstructs the comparison with some of the other works on the same topic because the subset we used does not necessarily correspond to the ones used in other papers. However, we were able to obtain high classification results in comparison to the other works, even though the data originated from 17 different screening sites and was acquired using different protocols. This means that our method has a good generalization ability and does not rely on a specific protocol.
5. Conclusions
In this paper, we proposed a method for the classification of Autism Spectrum Disorder versus a control group. The proposed method, based on a network consisting of autoencoders and multilayer perceptrons, was tested on both functional and structural data (in both separate and combined manner) available from the ABIDE I dataset. We showcased the importance of the multimodal approach by analyzing the obtained results qualitatively and quantitatively. By encompassing different types of information in our classification algorithm, we were able to improve the results in a statistically significant manner. The highest classification accuracy obtained, 85.06%, was a result of a multimodal strategy that included an ensemble of classifiers for both structural and functional data classification.
Current diagnosis of ASD is based on two main criteria: impairments in social communication and interaction and a restrictive, repetitive range of interests, behaviors and activities (APA, 2013). An inexperienced clinician is likely to incorrectly apply the criteria for autism and related conditions, which is a major concern in diagnostics. Another significant problem in current clinical practice is delayed diagnosis since early initialization of treatment increases the probability for a favorable outcome. Taking this into consideration, our method may provide additional insight in regard to ASD diagnosis. Even though the neuroimaging studies in the field yielded inconsistent results and are still not considered robust enough for a diagnostic tool, our method may be used as a second opinion system for ASD detection, as it may potentially unveil some useful patterns and findings for discrimination of the disorder.
Acknowledgments
This work has been supported by Retos de Investigacin TIN2015-73563-JIN and DPI2017-86696-R from the Ministerio de Ciencia y Tecnología. Mladen Rakić holds an EACEA Erasmus+ grant for the master in Medical Imaging and Applications (MAIA), Kaisar Kushibar holds a FI-DGR2017 grant from the Catalan Government with reference number 2017FI_B00372, and Mariano Cabezas holds a Juan de la Cierva - Incorporación grant from the Spanish Government with reference number IJCI-2016-29240. The authors gratefully acknowledge the support of the NVIDIA Corporation with their donation of the TITAN-V GPU used in this research.
References
- Abraham A., Milham M.P., Di Martino A., Craddock R.C., Samaras D., Thirion B., Varoquaux G. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. NeuroImage. 2017;147:736–745. doi: 10.1016/j.neuroimage.2016.10.045. [DOI] [PubMed] [Google Scholar]
- Anderson J.S., Nielsen J.A., Froehlich A.L., DuBray M.B., Druzgal T.J., Cariello A.N., Cooperrider J.R., Zielinski B.A., Ravichandran C., Fletcher P.T. Functional connectivity magnetic resonance imaging classification of autism. Brain. 2011;134:3742–3754. doi: 10.1093/brain/awr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APA . American Psychiatric Pub; 2013. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) [Google Scholar]
- Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward E.H., Minshew N.J., Field K., Sparks B., Singh N. Effects of age on brain volume and head circumference in autism. Neurology. 2002;59:175–183. doi: 10.1212/wnl.59.2.175. [DOI] [PubMed] [Google Scholar]
- Bernas A., Aldenkamp A.P., Zinger S. Wavelet coherence-based classifier: a resting-state functional MRI study on neurodynamics in adolescents with high-functioning autism. Comput. Methods Programs Biomed. 2018;154:143–151. doi: 10.1016/j.cmpb.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Buescher A.V., Cidav Z., Knapp M., Mandell D.S. Costs of autism spectrum disorders in the united kingdom and the united states. J. Am. Med. Assoc. Pediatr. 2014;168:721–728. doi: 10.1001/jamapediatrics.2014.210. [DOI] [PubMed] [Google Scholar]
- Calhoun V.D., Sui J. Multimodal fusion of brain imaging data: a key to finding the missing link (s) in complex mental illness. Biolog. Psych. Cognit. Neurosci. Neuroimag. 2016;1:230–244. doi: 10.1016/j.bpsc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.P., Keown C.L., Jahedi A., Nair A., Pflieger M.E., Bailey B.A., Müller R.A. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage: Clinical. 2015;8:238–245. doi: 10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Duan X., Liu F., Lu F., Ma X., Zhang Y., Uddin L.Q., Chen H. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivityâ a multi-center study. Progr. Neuro-Psychopharmacol. Biolog. Psych. 2016;64:1–9. doi: 10.1016/j.pnpbp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Lin C.J. Feature Extraction. Springer; 2006. Combining SVMS with various feature selection strategies; pp. 315–324. [Google Scholar]
- Courchesne E., Pierce K., Schumann C.M., Redcay E., Buckwalter J.A., Kennedy D.P., Morgan J. Mapping early brain development in autism. Neuron. 2007;56:399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Craddock C., Benhajali Y., Chu C., Chouinard F., Evans A., Jakab A., Khundrakpam B.S., Lewis J.D., Li Q., Milham M. The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. Neuroinformatics. 2013;4 [Google Scholar]
- Craddock R.C., James G.A., HoltzheimerIII P.E., Hu X.P., Mayberg H.S. A whole brain FMRI atlas generated via spatially constrained spectral clustering. Human Brain Mapping. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G., Libero L., Sreenivasan K.R., Deshpande H., Kana R.K. Identification of neural connectivity signatures of autism using machine learning. Front. Human Neurosci. 2013;7:670. doi: 10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destrieux C., Fischl B., Dale A., Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., Yan C.G., Li Q., Denio E., Castellanos F.X., Alaerts K., Anderson J.S., Assaf M., Bookheimer S.Y., Dapretto M. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psych. 2014;19:659. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Fu Z., Calhoun V.D. Classification and prediction of brain disorders using functional connectivity: Promising but challenging. Frontiers Neurosci. 2018;12 doi: 10.3389/fnins.2018.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Bookheimer S.Y., Murphy D.G. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 2015;14:1121–1134. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Gotham K., Pickles A., Lord C. Trajectories of autism severity in children using standardized ADOS scores. Pediatrics. 2012;130:e1278–e1284. doi: 10.1542/peds.2011-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius M.D., Krasnow B., Reiss A.L., Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Dominick K.C., Minai A.A., Li H., Erickson C.A., Lu L.J. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front. Neurosci. 2017;11:460. doi: 10.3389/fnins.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S., Sohn I.J., Kim N., Sim H.J., Cheon K.A. Characteristics of brains in autism spectrum disorder: structure, function and connectivity across the lifespan. Experimental Neurobiol. 2015;24:273–284. doi: 10.5607/en.2015.24.4.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinsfeld A.S., Franco A.R., Craddock R.C., Buchweitz A., Meneguzzi F. Identification of autism spectrum disorder using deep learning and the abide dataset. NeuroImage: Clinical. 2018;17:16–23. doi: 10.1016/j.nicl.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert M., Ziegler D., Deutsch C., Oâ brien L., Lange N., Bakardjiev A., Hodgson J., Adrien K., Steele S., Makris N. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126:1182–1192. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- Iidaka T. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex. 2015;63:55–67. doi: 10.1016/j.cortex.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Jahedi A., Nasamran C.A., Faires B., Fan J., Müller R.A. Distributed intrinsic functional connectivity patterns predict diagnostic status in large autism cohort. Brain Connect. 2017;7:515–525. doi: 10.1089/brain.2017.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zuo X.N. Regional homogeneity: a multimodal, multiscale neuroimaging marker of the human connectome. The Neurosci. 2016;22:486–505. doi: 10.1177/1073858415595004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou R.J., Mateljevic N., Minshew N.J., Keshavan M.S., Hardan A.Y. Reduced central white matter volume in autism: Implications for long-range connectivity. Psych. Clinical Neurosci. 2011;65:98–101. doi: 10.1111/j.1440-1819.2010.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju R., Hu C., Zhou P., Li Q. Early diagnosis of Alzheimer’s disease based on resting-state brain networks and deep learning. IEEE/ACM Trans. Comput. Biol. Bioinf. (TCBB) 2019;16:244–257. doi: 10.1109/TCBB.2017.2776910. [DOI] [PubMed] [Google Scholar]
- Kamnitsas K., Bai W., Ferrante E., McDonagh S., Sinclair M., Pawlowski N., Rajchl M., Lee M., Kainz B., Rueckert D. Proceedings of the International MICCAI Brainlesion Workshop. Springer; 2017. Ensembles of multiple models and architectures for robust brain tumour segmentation. [Google Scholar]; 450–462
- Kim J., Calhoun V.D., Shim E., Lee J.H. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: Evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage. 2016;124:127–146. doi: 10.1016/j.neuroimage.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Gao J., Xu Y., Pan Y., Wang J., Liu J. Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing. 2019;324:63–68. [Google Scholar]
- Ktena S.I., Parisot S., Ferrante E., Rajchl M., Lee M., Glocker B., Rueckert D. Metric learning with spectral graph convolutions on brain connectivity networks. NeuroImage. 2018;169:431–442. doi: 10.1016/j.neuroimage.2017.12.052. [DOI] [PubMed] [Google Scholar]
- Lord C., Rutter M., Goode S., Heemsbergen J., Jordan H., Mawhood L., Schopler E. Austism diagnostic observation schedule: A standardized observation of communicative and social behavior. J. Autism Develop. Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- Lord C., Rutter M., Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Develop. Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Murdaugh D.L., Shinkareva S.V., Deshpande H.R., Wang J., Pennick M.R., Kana R.K. Differential deactivation during mentalizing and classification of autism based on default mode network connectivity. Publ. Lib. Sci. One. 2012;7:e50064. doi: 10.1371/journal.pone.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.A., Zielinski B.A., Fletcher P.T., Alexander A.L., Lange N., Bigler E.D., Lainhart J.E., Anderson J.S. Multisite functional connectivity mri classification of autism: Abide results. Front. Human Neurosci. 2013;7:599. doi: 10.3389/fnhum.2013.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmen S.J., Pol H.E.H., Kemner C., Schnack H.G., Durston S., Lahuis B.E., Kahn R.S., Van Engeland H. Increased gray-matter volume in medication-naive high-functioning children with autism spectrum disorder. Psychol. Med. 2005;35:561–570. doi: 10.1017/s0033291704003496. [DOI] [PubMed] [Google Scholar]
- Plis S.M., Hjelm D.R., Salakhutdinov R., Allen E.A., Bockholt H.J., Long J.D., Johnson H.J., Paulsen J.S., Turner J.A., Calhoun V.D. Deep learning for neuroimaging: a validation study. Front. Neurosci. 2014;8:229. doi: 10.3389/fnins.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitt M., Barnes K.A., Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical. 2015;7:359–366. doi: 10.1016/j.nicl.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle K., Cascio C.J., Woodward N.D. Brain structure in autism: a voxel-based morphometry analysis of the autism brain imaging database exchange (abide) Brain Imaging Behav. 2017;11:541–551. doi: 10.1007/s11682-016-9534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi M., Khosrowabadi R., Bakouie F., Mahdavi H., Eslahchi C., Pouretemad H. Screening of autism based on task-free fmri using graph theoretical approach. Psych. Res. Neuroimaging. 2017;263:48–56. doi: 10.1016/j.pscychresns.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Subbaraju V., Suresh M.B., Sundaram S., Narasimhan S. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Med. Image Anal. 2017;35:375–389. doi: 10.1016/j.media.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Szatmari P., Georgiades S., Duku E., Bennett T.A., Bryson S., Fombonne E., Mirenda P., Roberts W., Smith I.M., Vaillancourt T. Developmental trajectories of symptom severity and adaptive functioning in an inception cohort of preschool children with autism spectrum disorder. J. Am. Med. Assoc. Psych. 2015;72:276–283. doi: 10.1001/jamapsychiatry.2014.2463. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Uddin L.Q., Supekar K., Lynch C.J., Khouzam A., Phillips J., Feinstein C., Ryali S., Menon V. Salience network–based classification and prediction of symptom severity in children with autism. J. Am. Med. Assoc. Psych. 2013;70:869–879. doi: 10.1001/jamapsychiatry.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Chen C., Fushing H. Extracting multiscale pattern information of FMRI based functional brain connectivity with application on classification of autism spectrum disorders. Publ. Library Sci. One. 2012;7:e45502. doi: 10.1371/journal.pone.0045502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Yu F., Duong T. Multiparametric mri characterization and prediction in autism spectrum disorder using graph theory and machine learning. Publ. Library Sci. One. 2014;9:e90405. doi: 10.1371/journal.pone.0090405. [DOI] [PMC free article] [PubMed] [Google Scholar]