Significance
Living cells possess dynamical cytoskeletons capable of remodeling themselves to all shapes. In this work, we use the bacterium actin analog, MreB, to dynamically remodel the shape of spherical synthetic cells, thus breaking the spherical symmetry, an essential step toward the development of complex biological functions such as liposome division. Using an effective cell-free expression system, the synthesis of MreB in the presence of a large concentration of the PEG polymer at the lipid membrane deforms the cell bilayer by two-dimensional crowding. This membrane crowding has broad applications. This mechanical force acting at the membrane is a new physical phenomenon for synthetic cells.
Keywords: cell-free transcription−translation, MreB cytoskeleton, synthetic cell, molecular crowding, symmetry breaking
Abstract
Executing gene circuits by cell-free transcription−translation into cell-sized compartments, such as liposomes, is one of the major bottom-up approaches to building minimal cells. The dynamic synthesis and proper self-assembly of macromolecular structures inside liposomes, the cytoskeleton in particular, stands as a central limitation to the development of cell analogs genetically programmed. In this work, we express the Escherichia coli gene mreB inside vesicles with bilayers made of lipid-polyethylene glycol (PEG). We demonstrate that two-dimensional molecular crowding, emulated by the PEG molecules at the lipid bilayer, is enough to promote the polymerization of the protein MreB at the inner membrane into a sturdy cytoskeleton capable of transforming spherical liposomes into elongated shapes, such as rod-like compartments. We quantitatively describe this mechanism with respect to the size of liposomes, lipid composition of the membrane, crowding at the membrane, and strength of MreB synthesis. So far unexplored, molecular crowding at the surface of synthetic cells emerges as an additional development with potential broad applications. The symmetry breaking observed could be an important step toward compartment self-reproduction.
The construction of synthetic cells by molecular assembly is growing as a major multidisciplinary research area (1, 2). On the one hand, making cell analogs carries promises as a means for delivering basic information about unicellular life (3). On the other hand, building synthetic cells from molecules offers a convenient and rather safe environment for carrying out bioengineering at the scale of living cells (4). The primary motivation and the ultimate goal of the bottom-up assembly of synthetic cells are the creation of microscopic compartments capable of self-reproduction based on a genetic program. One of the objectives toward this ambitious goal is to develop a quantitative understanding of the cooperative links between the parts that are used, or, in other words, how the integrated parts interact to deliver more than their individual sum. The major experimental challenge toward making synthetic cells from molecular components is captured by the well-known quote from Virchow (5), “onnis cellula e cellula,” living cells arise from other living cells. Synthetic cells have to be assembled from scratch, which requires the integration of many molecular components characterized by a broad variety of biochemical properties.
Cell-free transcription−translation (TXTL) has emerged as a convenient technology for the synthesis of cell analogs because it reproduces the process of gene expression outside of living organisms. Encapsulated into liposomes, for instance, TXTL can be used to program cell-sized vesicles in isolation with specific gene sets. This approach incorporates the three molecular blocks, information−metabolism−compartment (3, 4), necessary to look far toward making synthetic cells programmed for sophisticated biological functions, including self-reproduction. Many experimental efforts are currently performed to boot up cell analogs, besides gene circuits only, using TXTL in liposomes (6–9). Among the numerous challenges encountered in this research area, the development of biological functions at the lipid membrane remains difficult.
In our previous studies of cell-sized biological reactors programmed with gene circuits, we were able to express reporter genes for several days in lipid vesicles by synthesizing the α-hemolysin channel to establish exchanges of building blocks (e.g., adenosine 5′-triphosphate [ATP], adenosine 5′-diphosphate, and amino acids) through the membrane (9). Later, we showed that the actin-like protein MreB assembles into filaments at the inner membrane of liposomes (10), but no mechanical deformations were observed. The spherical symmetry of the vesicles could not be broken by the assembly of MreB filaments. Similarly, when the FtsZ protein was synthesized in liposomes, it failed to assemble into contractile rings (11), despite evidence of such a pattern in an experimental setting using purified proteins (12). The difficulty of achieving the synthesis and self-assembly of a cytoskeleton at the inner membrane of liposomes and obtaining the proper phenotype remains a true limitation to the development of genetically programmed synthetic cells. Here we set out to understand how a cytoskeleton, dynamically synthesized inside phospholipid vesicles, can give rise to relevant morphologies. We focused on MreB based on our previous work (10).
The MreB protein is a bacterial analog of actin found in eukaryotic cells (13). MreB polymerizes at the inner membrane of living cells into a cortex-like cytoskeleton responsible for the rod-like shape of bacteria. When the mreB gene is knocked out, the default shape taken by bacterial cells is a sphere (14, 15). MreB interacts with the transmembrane protein RodZ to coordinate the synthesis of the cell wall and thus determine the shape of bacteria (16–18). The whole enzymatic Rod complex rotates to maintain robust morphogenesis (19–21). The Escherichia coli MreB protein is 37 kDa and has a critical concentration of polymerization, measured in vitro, on the order of 1.5 µM (22), similar to MreB proteins from other bacteria (23). The number of MreB proteins per cell has been estimated to be around 8,000 (15), which corresponds to about 5 to 10 µM (taking that 1 molecule per µm3 is about 1 nM), or 0.2 to 0.4 mg/mL. In vitro, it has been demonstrated that MreB alone, when added externally to a vesicle solution, interacts with the phospholipid bilayer, and polymerizes into filament bundles that mechanically distort the membranes (24). These observations, together with our prior work (10), motivated us to explore, in a synthetic cell setting, what could drive the assembly of MreB into a cytoskeleton capable of shaping liposomes into rod-like compartments resembling E. coli cells. The synthetic cell system consists of a TXTL reaction encapsulated in cell-sized liposomes, ranging from 1 to 50 μm in diameter. Genes are cloned separately into plasmids under silent promoters that are turned on through a transcriptional activation cascade. In this manner, the strength of expression can be adjusted precisely by choosing the concentration of each gene added to a TXTL reaction. The types of phospholipids can also be changed relatively easily. Such an experimental setting is highly convenient for isolating a specific mechanism. The TXTL system used in this work is strong enough (>50 µM) to synthesize MreB far above its critical concentration of polymerization (1.5 µM) and its concentration measured in vivo (<10 µM).
A major difference between cell membranes and liposomes is the 2-dimensional (2D) molecular crowding. The membrane of synthetic cells is typically deprived of proteins, as opposed to natural membranes hosting hundreds of different proteins, either into or attached to the bilayer. While the effect of bulk molecular crowding on protein self-assembly is well established (25–27), 2D crowding at the membrane has been mostly ignored in synthetic cells so far. This prompts us to emulate this biophysical property at the surface of TXTL-based minimal cells and quantify its effect on the polymerization of MreB, which is known to interact with the surface of lipid bilayers (24). Surprisingly, we show that molecular crowding at the bilayer, emulated by lipid-polyethylene glycols (PEGs), is sufficient to drive the assembly of MreB into a cytoskeleton capable of deforming spherical liposomes into elongated shapes, such as rods (Fig. 1). The physical process by which the vesicle’s spherical symmetry is broken is induced by the presence of PEG polymers that act as molecular crowders attached to the bilayer. We measure that the smaller the radius the larger the ratio of long axis over small axis. Deformations are observed for liposomes of radius smaller than 10 µm. We show that changing lipid head types, as opposed to the PEG moiety, has no effect on the process. Our study demonstrates that the self-assembly of MreB into a mechanically dynamic cytoskeleton is driven by molecular crowding in two dimensions, independent of the complexity of the interactions of MreB with other proteins. We anticipate that achieving symmetry breaking could lead to the development of other functions such as division by FtsZ fission ring.
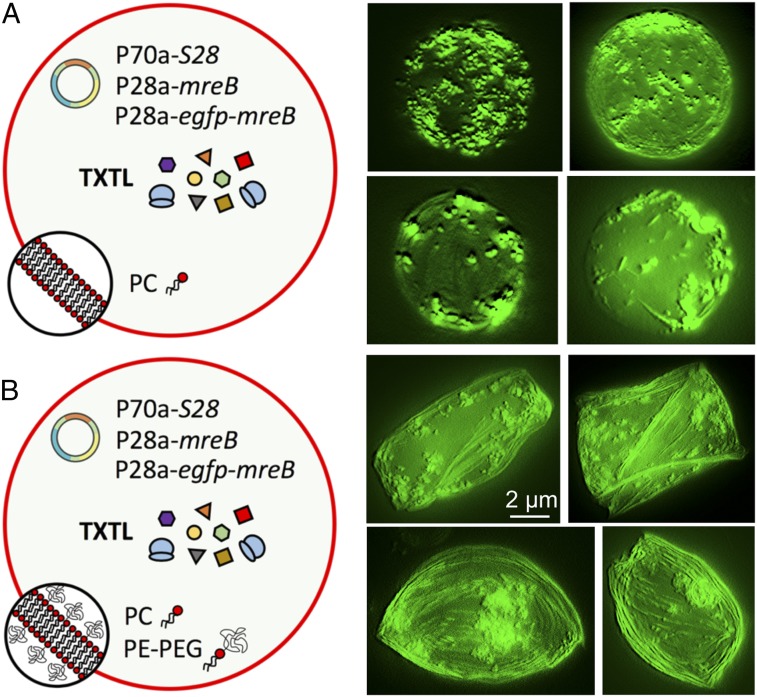
Fig. 1.
Image reconstruction from z scan in fluorescence microscopy. (A) Cell-free expression of mreB inside synthetic cells with a membrane composed of 100% PC. (B) Cell-free expression of mreB inside synthetic cells with a membrane composed of 99.33% PC and 0.66% PE-PEG5000. The cell-free reactions encapsulated inside the synthetic cells were programmed with the plasmids P70a-S28 (0.5 nM), P28a-mreB (2 nM), and P28a-egfp-mreB (0.2 nM). The fluorescence images of several liposomes (taken after 12 h of incubation) show MreB filament accumulating at the membrane in both cases. Deformations are observed when a lipid−PEG is used, as opposed to the case with no PEG at the membrane. These images were obtained from z-stack images reconstructed by the software MetaMorph. The scale bar is the same for all the images.
Results and Discussion
TXTL and Minimal Cell Systems.
The TXTL system employed in this work, known under the name of myTXTL, has been extensively described in several recent publications (28, 29). The lysate that provides the transcription and translation components does not contain any living cells (SI Appendix, Fig. S1). It is convenient to clone the genes of interest under a silent promoter, such as T7p14 or P28a (29), and synthetize the respective RNA polymerase or transcriptional factor for expression in TXTL (SI Appendix, Fig. S2). We used the S28 transcription cascade based on the endogenous E. coli transcription components. The strength of expression using this circuit is large enough to synthesize MreB at concentrations greater than the concentration of MreB in E. coli, estimated to be around 5 to 10 µM (15) (SI Appendix, Fig. S3). The TXTL system recapitulates physiological conditions and contains all of the nutrients necessary for MreB stability and polymerization, magnesium (∼10 mM) and ATP (∼2 mM) in particular. None of the proteins expressed in this work are present in the lysate, as evidenced recently by mass spectrometry analysis of this cell-free system (30). All of the performed reactions contained the same ingredients, except for the plasmids that were changed based on the proteins synthesized.
The minimal cell system was prepared by encapsulating TXTL reactions into cell-sized liposomes, based on the emulsion transfer method (9, 31), following a procedure reported thoroughly by our group recently (32). The difference in osmolarity between the TXTL reactions encapsulated inside the liposomes and the external solution was measured to be between 25 and 75 mOsm, a magnitude that we found adequate for our study. At larger osmotic pressure, it is known that liposomes burst or that micrometer-sized pores spontaneously form in the membrane (33), a configuration that we wanted to avoid so as to keep the reaction components inside the vesicles. Furthermore, with such an imbalance of osmolarity, the liposomes under tension have a spherical shape, with a membrane that does not fluctuate when observed under the microscope. To measure liposome deformation, we decided to characterize the ratio L/S. This ratio is calculated by measuring the long axis (L) and the short axis (S) of each liposomes after 12 h of incubation. The L/S ratio is determined with respect to the spherical nondeformed liposomes just after they are prepared. By definition, L/S is equal to one for a spherical object. The liposomes were binned as a function of their radius, by 2-µm increments (SI Appendix, Fig. S4). In such a manner, the statistics are converted into histograms that give a much better overview of deformations for each 2-µm segment. Besides, the segment 0 to 2 µm, for which the deformations are the largest, is relevant to the size of E. coli.
Lipid Membrane Composition and Molecular Crowding.
We chose PC (egg phosphatidylcholine) as the default phospholipid, given that liposome production is the greatest with this lipid and with respect to the method employed in this work (32). Making hundreds of vesicles per preparation is practical in a context of deformation measurement. To emulate crowding at the membrane, PE-PEG (phosphatidylethanolamine−polyethylene glycol) was added to PC at different molar concentrations. PEG is the molecular crowder of choice, due to its low nonspecific interactions with biomolecules and its high compatibility with biological solutions. Besides this, PE-PEG is the only type of commercially available lipid−polymer molecule that we found, and at different PEG sizes in the ideal range. To begin our study, we looked for a PEG size comparable to the MreB monomer size and filament diameter, determined to be 5.1 and 3.9 nm, respectively (13). The persistence length of PEG, on the order of 3.8 Å (34, 35), is comparable to the monomer length (made of three single covalent bonds). Considering an ideal chain polymer conformation, a PEG of molar mass of 5,000 g/mol containing 114 monomers has a root mean square end-to-end distance of 5.7 nm (effective segment length: 7.6 Å; number of effective segments: 57) (SI Appendix, Table S1), an ideal size to mimic crowding at the membrane. The surface area of a phospholipid is on the order of 40 to 50 Å2. Thus, a PEG polymer of molar mass 5,000 g/mol covers about 65 to 80 phospholipids. Consequently, we estimated that, at a molar concentration of PE-PEG5000 greater than a few percent (∼1 to 2%), the polymer covers the whole membrane, potentially limiting the interaction of MreB with the lipid bilayer. This estimation is valid when the PE-PEG5000 molecules are only found in the membrane and the PEG8000 remains in solution. This assumption is supported by our control experiments, as we shall see later. The TXTL reactions contain PEG8000 in solution to increase cell-free expression at a concentration of 2 mM, which corresponds to an optimum. The presence of PEG8000, a standard component in TXTL, was kept fixed in all of the reactions, except for a control experiment, so as to not bias our observations with the lipid−PEG at the membrane.
MreB Polymerization Is Enhanced in the Presence of 2D Molecular Crowding.
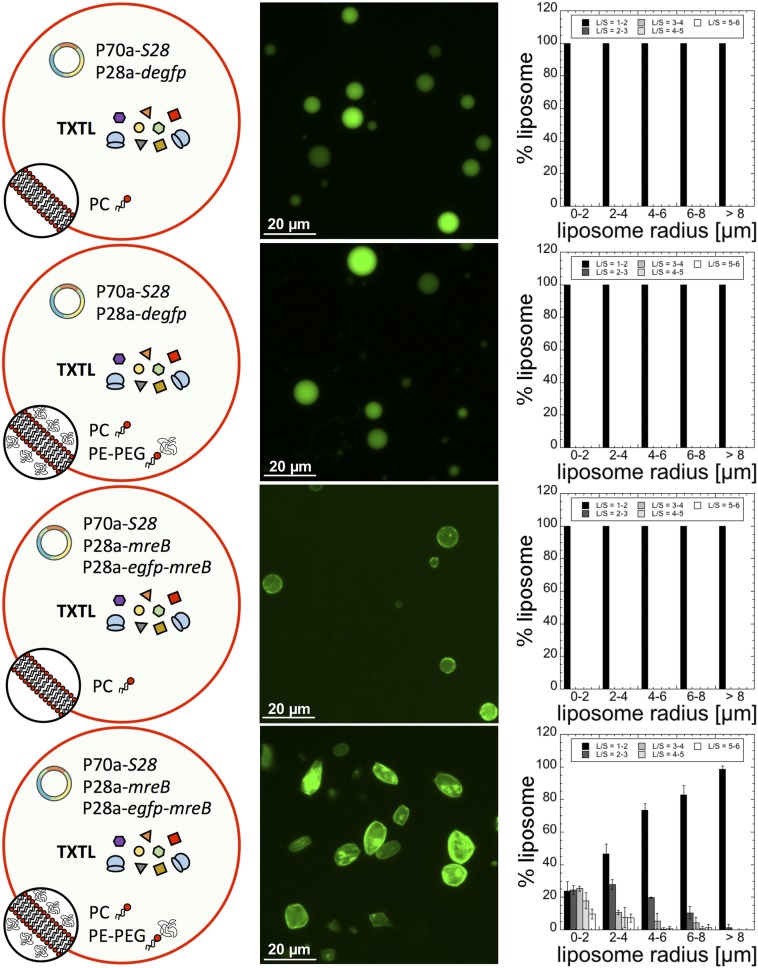
Our first experiment consisted of expressing either degfp or mreB inside liposomes made of 100% PC or a mixture of 99.33% PC and 0.66% PE-PEG5000. Strikingly, after just a few hours, a large fraction of the liposome population appeared as rod-like shapes when MreB was synthesized in the presence of PE-PEG5000 in the membrane (Fig. 2). A net accumulation of fluorescently labeled MreB was observed at the membrane. No deformations were noticed when egfp was expressed or when MreB was synthesized in the absence of PE-PEG5000, despite a net interaction with the membrane. The statistics of the ratio L/S revealed that the smaller the liposome the larger the L/S ratio. About 75% of the liposomes of radii between 0 and 2 µm had an L/S larger than two. This result is relevant to the size of E. coli (1 × 3 µm) and its L/S ratio, which is around 2.5 to 3 in most of the growing conditions (36). Deformations were negligible for liposomes of radius greater than 10 µm. This result is consistent with the persistence length of the MreB filament, estimated to be similar to actin on the order of 15 µm (37). For the same lipid composition (99.33% PC and 0.66% PE-PEG5000), we determined how the statistics of deformations change when the concentration of mreB plasmids is varied (SI Appendix, Fig. S5). The concentration of P70a-S28 was fixed to 0.5 nM because it corresponds to an optimum for this transcription cascade (29). No major differences in the statistics of liposome deformation were found at a plasmid concentration larger than 1 nM (1 nM P28a-mreB + 0.1 nM P28a-egfp-mreB). The transition from undeformed to deformed liposomes occurs when the plasmid concentration is set between 0.5 and 1 nM. Our quantitative measurements of protein synthesis (SI Appendix, Fig. S3) roughly agree with the estimation of the endogenous concentration of MreB in E. coli (5 to 10 µM). In a TXTL reaction, a concentration of about 10 µM MreB is attained for plasmid concentration between 0.2 and 0.5 nM (SI Appendix, Fig. S5). Protein synthesis is linear up to 1 nM plasmid, and saturates slowly toward a maximum of 50 µM between 1 and 5 nM plasmid.
Fig. 2.
Statistics of liposome deformation (ratio L/S) as a function of the presence of PE-PEG5000 in the membrane. The bilayers of the synthetic cells are either composed of 100% PC or of 99.33% PC + 0.66% PE-PEG5000 (% indicates molarity %). No deformation is observed when degfp is expressed with or without PE-PEG5000 in the membrane (0.5 nM P70a-S28, 5 nM P28a-degfp). When mreB is expressed (0.5 nM P70a-S28, 5 nM P28a-mreB, 0.5 nM P28a-egfp-mreB), negligible deformations are observed in the absence of PE-PEG5000 at the membrane. In the presence of PE-PEG5000, strong deformations are observed. The liposome radii are the radii measured before deformation when liposomes are all spherical. Fluorescence images were acquired after 12 h of incubation.
No major differences in the statistics of the L/S ratio were found when the experiment was performed with a different extract preparation (SI Appendix, Fig. S6). To determine whether the liposome deformations could be due to a stronger gene expression in the presence of PEG at the membrane, we compared the level of deGFP synthesis through the S28 cascade inside liposomes made of PC only or of PC and PE-PEG5000. Our measurements show that deGFP synthesis is similar in both cases (SI Appendix, Fig. S7).
To clarify whether the soluble PEG8000 added to TXTL reactions at a concentration of 2 mM participates in the mechanism of liposome deformation as a molecular crowder, we performed an experiment without PEG8000, with a lipid composition of 99.33% PC and 0.66% PE-PEG5000. Our statistics of liposome deformation show that there is no difference, whether PEG8000 is present or not in the TXTL reaction (SI Appendix, Fig. S8). Protein synthesis in the liposomes without PEG8000 is as strong as in the presence of PEG8000 (SI Appendix, Fig. S9). With a critical micelle concentration of about 1 µM (38), the lipid−PEG is at negligible concentration in the solution, far below the PEG8000 concentration. These experiments indicate that only the PEG5000 located at the membrane is responsible for boosting the polymerization of MreB into filaments and bundles. A consequence of this result is the unique optimal concentration of PE-PEG5000 at the membrane, estimated to be around 0.5 to 1% molar.
Liposome Deformations by MreB Depend on the Size of the Crowding Polymer.
It is well established that molecular crowding results in an excluded volume that enhances the thermodynamic activity of proteins in phase solutions (25, 27). Although less studied in two dimensions, similar effects of excluded volume produced by molecular crowding have been discussed or evidenced on the activity of proteins on lipid membranes (39–41). One of the major consequences of excluded volume in three or two dimensions is to affect macromolecular interactions. In particular, in crowded media, the self-assembly of proteins that self-associate is enhanced, as demonstrated for cytoskeleton proteins in phase solutions (26, 42). This mechanism relies on an increase of entropy in crowding molecules when reactants associate (SI Appendix, Fig. S10). Although not exclusive, it is our main assumption in this work: The PEG polymer at the membrane results in 2D excluded volume that promotes and enhances self-assembly of MreB at the membrane. Polymerization occurs predominantly at the membrane because MreB interacts with lipids via its N terminus end (24).
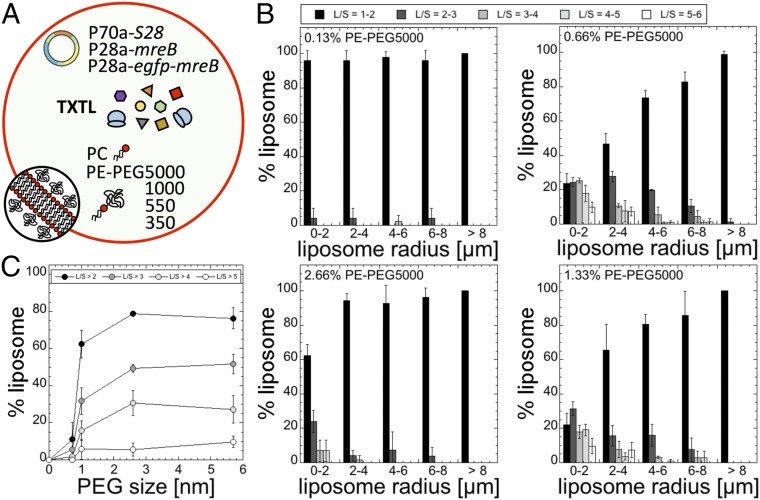
The magnitude of the effect produced by molecular crowding depends on the volume fraction (or surface fraction) and on the size of crowders (43). We first varied the concentration of PE-PEG5000 added to the PC membrane in a synthetic cell programmed to express mreB (Fig. 3A). Two expected trends were observed (Fig. 3B): 1) The fraction of deformed liposomes increases as the concentration of PE-PEG5000 is increased, and 2) the effect of molecular crowding vanishes as the concentration of PE-PEG5000 exceeds a few percent, likely because the PEG polymers cover the whole bilayer surface area, limiting the interaction of MreB with the membrane. Experimentally, we measured a net decrease of the number of deformed liposomes starting at about 2% PE-PEG5000. It is in agreement with our estimation of full membrane coverage for a PEG5000 polymer (estimated to be between 1% and 2%), taking as a first approximation an ideal polymer chain for which the characteristics are well known in the case of PEG (effective segment length) (SI Appendix, Table S1). We performed the same experiments for PE-PEG of PEG molar masses 350, 550, and 1,000 g/mol. We determined that the number of deformed liposomes was the greatest for molar concentrations of 7% (PEG350), 6% (PEG550), 2.3% (PEG1000), and 0.66% (PEG5000). We focused our analysis on liposomes of radii between 0 and 2 µm and plotted the fraction of liposomes in that size that have an L/S larger than two with respect to the root-mean-square end-to-end distance of the PEG polymer (Fig. 3C). We observe a sharp transition for a PEG molar mass of 550 g/mol, which corresponds to a root-mean-square end-to-end distance of 1 nm. A PE-PEG350 has little effect on MreB self-assembly at any concentration in the membrane, while the number of liposomes with large deformations starts being significant for a PE-PEG550. No major differences were noticed for PE-PEG1000 and PE-PEG5000.
Fig. 3.
Statistics of liposome deformation (ratio L/S) as a function of the concentration of PE-PEG in the membrane (added to pure PC; % indicates molarity) and size of the PEG polymer attached to PE (350, 550, 1,000, 5,000 g/mol). (A) Schematic of the synthetic cell system. The PE-PEGs of different sizes were used separately. (B) Statistics of liposome deformation as a function of PE-PEG5000 added to PC for four concentrations: 0.13%, 0.66%, 1.33%, and 2.66%. The same experiment was done for PEG350, PEG550, and PEG1000 to make the plot in C. (C) Percentage of liposomes of radius comprised between 0 and 2 µm that have an L/S larger than either 2, 3, 4, or 5 as a function of the PEG polymer physical size (350: 0.72 nm; 550: 1 nm; 1,000: 2.6 nm; 5000: 5.7 nm). The optimum PE-PEG concentrations (for which the greatest number of liposomes were deformed) were the following (% indicates molarity): 7% (350), 6% (550), 2.3% (1,000), and 0.66% (5,000).
MreB Structures and Morphology of Liposomes.
Our experiments support, although not exclusively, the hypothesis that two physical mechanisms are responsible for liposome deformation. The excluded volume produced by 2D molecular crowding enhances the polymerization of MreB at the inner membrane. The mechanical deformation of the liposomes is due to the polymerization of MreB into bundles of filaments with persistence length on the order of 10 to 20 µm (37). This mechanical stiffness is consistent with the fact that 1) deformations are only observed for liposomes of radii small than 10 µm, and, 2) on average, the most deformed liposomes are the smallest.
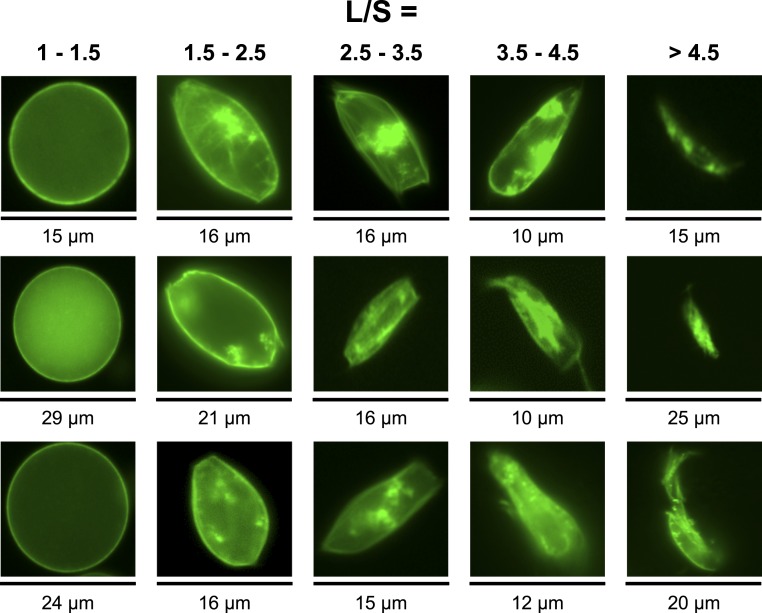
The two most abundant morphologies observed were ovals and rectangles, and combinations of both independent of the original size of the liposomes (Fig. 4). The z-stack reconstruction of several liposomes shows a net accumulation of the MreB filaments at the inner membrane of the liposomes (Fig. 1). The formation of such MreB filament bundles has been observed in vitro using purified MreB (24). In the oval liposomes, the filament bundles are wrapped around the inner membrane (Fig. 1). In the rectangular liposomes, it appears that independent filament bundles stretch the membrane into parallelepipedal shapes, creating endpoints. In the absence of 2D crowding, we observed the formation of MreB spots located at the membrane and the formation of a few filaments in some cases (Fig. 1).
Fig. 4.
Sample fluorescence images of liposomes for different L/S ratios. The images were all shot with a 40× objective.
The macromolecular structures formed by MreB at the inner membrane of E. coli have been debated, in particular, due to the presence of the fluorescent reporter. It has been suggested that the helical patterns observed in E. coli cells are artifacts produced by YFP when it is fused to MreB (44). To confirm that the liposome deformations observed in our work are not the product of nonspecific interactions of the reporter protein with the membrane, we expressed mreB only together with degfp in minimal cells. The deGFP served as a fluorescent marker of the synthetic cell protoplasm. In such settings, the liposomes are as deformed as the ones when egfp-mreB is expressed (SI Appendix, Fig. S11). No interaction of the reporter deGFP was observed with the membrane.
The proteins MreC, MreD, and RodZ are known to form a complex with MreB at the membrane of E. coli for cell wall synthesis (45). We expressed mreB concurrently with mreC, mreD, and rodZ. Except for a reduction of the fraction of deformed liposomes, no other effect was noticed when these three proteins were expressed with mreB (SI Appendix, Fig. S12). In the context of a single membrane minimal cell, it is somewhat expected to not see any significant effect of those proteins.
An additional control experiment consisted of measuring the effect of 2D molecular crowding when membrane proteins tagged with eGFP are synthesized. We chose the membrane channels α-hemolysin and MscL because their activity in TXTL synthetic cells has been demonstrated in prior works (9, 29, 46). No deformation was noticed when these proteins are expressed in PC liposomes with PE-PEG lipids added to the membrane (SI Appendix, Fig. S13). In the presence of lipid−PEG at the membrane, we observed the formation of fluorescent aggregates when either MreB or membrane channels are synthesized (Figs. 1 and 5 and SI Appendix, Fig. S13). We assume that the formation of these aggregates is due to the strength of protein synthesis, which, in addition, is not regulated. Consequently, gene expression goes on for hours, and proteins are overexpressed, resulting in the formation of aggregates either attached to the membrane or in the cytoplasm.
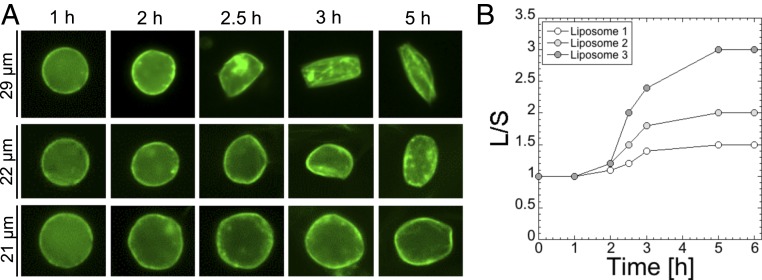
Fig. 5.
Kinetics of deformation for three liposomes. (A) Images for three liposomes for which the L/S is (Top to Bottom) 3, 2, and 1.5. (B) Plot showing the ratio L/S over time for the three liposomes shown in A.
Liposome Deformations Are Independent of the Lipid Composition.
The protein MreB interacts with the lipid membrane via its N-terminal end (24). We hypothesized that the lipid composition could alter MreB self-assembly at the membrane. It is known that the activity of membrane proteins or protein interacting with the membrane can depend, to some degree, on the lipid composition (47), either on the lipid flavor or on the lipid charge, for instance. We tested the effect of molecular crowding at the membrane when the lipid composition of the liposome is changed. We focused our study on the most abundant phospholipids found in the E. coli membrane, composed approximately of 60 to 70% PE (zwitterionic), 15% PG (phosphatidylglycerol, one negative charge), and 10% CA (cardiolipin, two negative charge), keeping PC (zwitterionic) as our default lipid. While liposomes composed of 15% PG and 10% CA were produced, we could not make liposomes with more than 30% PE. We fixed PE-PEG5000 at 0.66%, an optimum to generate deformations with this lipid PEG. Except for slight deformations observed for 10% CA, no effects were noticed in the absence of PE-PEG5000 (SI Appendix, Fig. S14) when PG and PE where present in the membrane. When PE-PEG5000 was present in the membrane, no difference was found, except for a decrease of the deformations in some cases. We concluded that, in the case of MreB, the lipid composition of the membrane has minor effects on the molecular crowding mechanism and self-assembly process.
Deformations Occur in the First 5 h.
In the last step of our study, we determined the kinetics of liposome deformation, fixing the settings in the optimum regime of observed deformations: 0.66% PE-PEG5000, 99.33% PC, plasmids P70a-S28 (0.5 nM), P28a-mreB (5 nM), and P28a-egfp-mreB (0.5 nM). The shape of more than 90% of the liposomes does not change after 5 h of incubation. The major change in the liposome morphology is found between 2 and 3 h, independent of the magnitude of the L/S ratio (Fig. 5). All of the liposomes were characterized first by a net accumulation of MreB on the membrane before being deformed.
Conclusions
Building synthetic cells from scratch, using the natural biological molecules, is a highly convenient environment for deconstructing and understanding mechanisms often entangled in living cells, or simply hard to approach in vivo. In this work, we provided the demonstration that 2D molecular crowding at the lipid membrane can literally change the self-assembly of a cytoskeleton protein, using a genetically programmed synthetic cell system that allowed us to isolate a particular biological function. This result is unexpected at several levels. First, the self-assembly of MreB alone enables shaping liposomes into the proper phenotype in the presence of 2D crowding. Second, our results strongly suggest that, despite the complexity of its interactions with other proteins in living cells, MreB’s self-assembly into a sturdy cytoskeleton is driven by the presence of a macromolecular background confined on a surface rather than being enzymatically guided. We expect 2D molecular crowding to be essential for other processes located at the membrane, especially the ones involved in cytoskeleton assembly. We anticipate that this result will contribute to the development of other active functions in synthetic cells engineered either for applications or for tackling basic questions. In particular, creating symmetry breaking based on a genetic program should facilitate moving toward more elaborate functions such as the self-reproduction of synthetic liposomes, the major goal of minimal cell engineering.
Materials and Methods
Materials.
DNA was purchased from Integrated DNA Technologies. Lipids were purchased from Avanti Polar Lipids Inc. Unless otherwise mentioned, all of the other reagents were purchased from Sigma Aldrich.
DNA Constructs.
The plasmids P70a-S28 and P28a-degfp have been described previously (29). P28a-mreB and P28a-egfp-mreB were obtained by cloning mreB and egfp-mreB into the P28a backbone. The fusion egfp-mreB was used in TXTL synthetic cells previously, under the name yfp-mreB in ref. 10. The genes mreC, mreD, rodZ, mscL, and alpha-hemolysin were amplified by PCR and cloned under a P28a promoter. All of the constructs have been sequenced. The sequences are reported in SI Appendix, Table S2. Both degfp (as single reporter) and egfp (used in the fusion proteins) were used. The gene degfp was used rather than egfp as single reporter gene because it is our reference reporter gene, used in many of our articles related to the cell-free system used in this work. The reporter protein deGFP was described earlier (28, 29). It is a slightly truncated version of eGFP, with the same fluorescent properties as eGFP. The plasmid P70a-degfp was described in ref. 29. Some of these constructions are available at Arbor Biosciences (Toolbox 2.0 Plasmid Collection). The other plasmids are available on demand.
TXTL Reactions.
The myTXTL kit (Arbor Biosciences) was used. TXTL reactions are composed of an E. coli lysate, an amino acid mixture, an energy buffer, and the desired DNA templates. The preparation of the lysate has been described in prior work (48). The lysate represents roughly one-third of the total reaction volume and contains the endogenous machinery required for both transcription and translation. The amino acid mixture is an equimolar blend of the 20 standard amino acids (49), with each amino acid concentration at a final reaction of 4 mM. The concentration of the plasmid P28a-egfp-mreB was systematically set to 10% of the concentration of P28a-mreB (e.g., 2 nM P28a-mreB, 0.2 nM P28a-egfp-mreB).
Batch Mode Expression.
Fluorescence from batch mode TXTL reactions was measured using the reporter protein deGFP (25.4 kDa, 1 mg/mL = 39.4 µM). Fluorescence was measured at 3-min intervals using monochromators (Ex/Em 488/525 nm) on Biotek Synergy H1 plate readers in Nunc polypropylene 96-well, V-bottom plates. Endpoint reactions were measured after 12 h of incubation. To measure protein concentration, a linear calibration curve of fluorescence intensity versus eGFP concentration was generated using purified recombinant eGFP obtained from Cell Biolabs, Inc. or purified in the laboratory (29). All reactions were incubated at 29 °C in either a bench-top incubator, for endpoint measurements, or the plate readers, for kinetic measurements.
Liposome Preparation.
The protocol to prepare the TXTL-loaded liposomes, based on the emulsion method, has been described, in detail, by our group in a recent publication (32). Briefly, the phospholipids were dissolved in chloroform at a concentration of around 20 mg/mL. The lipids were added to mineral oil at a concentration of 2 mg/mL and incubated for 1 h at 80 °C to evaporate the chloroform. Five microliters of TXTL reaction was added to 500 µL of mineral oil lipid mix, followed by gentle vortex for a few seconds to create an emulsion. Then, 200 µL of emulsion was placed on top of 50 µL of feeding solution, and this solution was centrifuged for 1 min at 5,000 × g. The liposome solution was recovered and placed on a microscope cover glass, sealed, and allowed to incubate at 29 °C. The osmolarity of TXTL reactions and of the external solution was estimated using an osmometer Advanced Instrument 3320. All of the lipid mixtures are given in percent molar. For instance, 98% PC + 2% PE-PEG means that, out of 100 phospholipids, 98 are PC and 2 are PE-PEG.
Imaging.
Fluorescence images of liposomes were taken on an inverted microscope Olympus IX-81 connected to an Andor iXon3 CCD camera. Images analysis and L/S ratio measurements were carried out using MetaMorph. Except for some control experiments (∼50 liposomes measured in the control experiments for which all of the liposomes were spherical), 200 to 300 liposomes were analyzed per case. All of the error bars indicate the SD of three independent repeats. An independent repeat consisted of preparing a synthetic cell solution from scratch, using new aliquots of materials.
Data Statement.
The datasets generated during and/or analyzed during this study are available from the public data repository for the University of Minnesota (DRUM) (50).
Supplementary Material
Acknowledgments
This work is supported by the Human Frontier Science Program Grant RGP0037/2015.
Footnotes
Competing interest statement: The V.N. laboratory receives research funds from Arbor Biosciences, a distributor of the myTXTL cell-free protein synthesis kit.
Data deposition: The datasets generated during and/or analyzed during this study are available from the public data repository for the University of Minnesota (DRUM), http://hdl.handle.net/11299/210183.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914656117/-/DCSupplemental.
References
- 1.Xu C., Hu S., Chen X., Artificial cells: From basic science to applications. Mater Today (Kidlington) 19, 516–532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia H., Heymann M., Bernhard F., Schwille P., Kai L., Cell-free protein synthesis in micro compartments: Building a minimal cell from biobricks. N. Biotechnol. 39, 199–205 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Noireaux V., Maeda Y. T., Libchaber A., Development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl. Acad. Sci. U.S.A. 108, 3473–3480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caschera F., Noireaux V., Integration of biological parts toward the synthesis of a minimal cell. Curr. Opin. Chem. Biol. 22, 85–91 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Virchow R. L. K., Die cellularpathologie in ihrer begründung auf physiologische und pathologische gewebelehre (Hirschwald, Berlin, Germany, 1858), vol. A. [Google Scholar]
- 6.van Nies P., et al. , Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat. Commun. 9, 1583 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berhanu S., Ueda T., Kuruma Y., Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 10, 1325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garamella J., Majumder S., Liu A. P., Noireaux V., An adaptive synthetic cell based on mechanosensing, biosensing, and inducible gene circuits. ACS Synth. Biol. 8, 1913–1920 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Noireaux V., Libchaber A., A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. U.S.A. 101, 17669–17674 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda Y. T., et al. , Assembly of MreB filaments on liposome membranes: A synthetic biology approach. ACS Synth. Biol. 1, 53–59 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Furusato T., et al. , De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth. Biol. 7, 953–961 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Osawa M., Anderson D. E., Erickson H. P., Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Ent F., Amos L. A., Löwe J., Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Doi M., et al. , Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J. Bacteriol. 170, 4619–4624 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones L. J. F., Carballido-López R., Errington J., Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001). [DOI] [PubMed] [Google Scholar]
- 16.van den Ent F., Johnson C. M., Persons L., de Boer P., Löwe J., Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29, 1081–1090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgenstein R. M., et al. , RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 12510–12515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bratton B. P., Shaevitz J. W., Gitai Z., Morgenstein R. M., MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. Nat. Commun. 9, 2797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Teeffelen S., et al. , The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 15822–15827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner E. C., et al. , Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domínguez-Escobar J., et al. , Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Nurse P., Marians K. J., Purification and characterization of Escherichia coli MreB protein. J. Biol. Chem. 288, 3469–3475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popp D., et al. , Filament structure, organization, and dynamics in MreB sheets. J. Biol. Chem. 285, 15858–15865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salje J., van den Ent F., de Boer P., Löwe J., Direct membrane binding by bacterial actin MreB. Mol. Cell 43, 478–487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minton A. P., How can biochemical reactions within cells differ from those in test tubes? J. Cell Sci. 119, 2863–2869 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Rivas G., Fernandez J. A., Minton A. P., Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: Indefinite linear self-association of bacterial cell division protein FtsZ. Proc. Natl. Acad. Sci. U.S.A. 98, 3150–3155 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minton A. P., The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 276, 10577–10580 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Shin J., Noireaux V., An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1, 29–41 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Garamella J., Marshall R., Rustad M., Noireaux V., The all E. coli TX-TL Toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol. 5, 344–355 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Garenne D., Beisel C. L., Noireaux V., Characterization of the all-E. coli transcription-translation system myTXTL by mass spectrometry. Rapid Commun. Mass Spectrom. 33, 1036–1048 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Pautot S., Frisken B. J., Weitz D. A., Production of unilamellar vesicles using an inverted emulsion. Langmuir 19, 2870–2879 (2003). [Google Scholar]
- 32.Garamella J., Garenne D., Noireaux V., TXTL-based approach to synthetic cells. Methods Enzymol. 617, 217–239 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ohno M., Hamada T., Takiguchi K., Homma M., Dynamic behavior of giant liposomes at desired osmotic pressures. Langmuir 25, 11680–11685 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kienberger F., et al. , Static and dynamical properties of single poly(Ethylene Glycol) molecules investigated by force spectroscopy. Single Mol. 1, 123–128 (2000). [Google Scholar]
- 35.Lee H., Venable R. M., Mackerell A. D. Jr, Pastor R. W., Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys. J. 95, 1590–1599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P. Y., et al. , Real-time measurement of single bacterium’s refractive index using optofluidic immersion refractometry. Procedia Eng. 87, 356–359 (2014). [Google Scholar]
- 37.Jiang H., Si F., Margolin W., Sun S. X., Mechanical control of bacterial cell shape. Biophys. J. 101, 327–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashok B., Arleth L., Hjelm R. P., Rubinstein I., Onyüksel H., In vitro characterization of PEGylated phospholipid micelles for improved drug solubilization: Effects of PEG chain length and PC incorporation. J. Pharm. Sci. 93, 2476–2487 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Ryan T. A., Myers J., Holowka D., Baird B., Webb W. W., Molecular crowding on the cell surface. Science 239, 61–64 (1988). [DOI] [PubMed] [Google Scholar]
- 40.Aisenbrey C., Bechinger B., Gröbner G., Macromolecular crowding at membrane interfaces: Adsorption and alignment of membrane peptides. J. Mol. Biol. 375, 376–385 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Zuckermann M. J., Heimburg T., Insertion and pore formation driven by adsorption of proteins onto lipid bilayer membrane-water interfaces. Biophys. J. 81, 2458–2472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindner R. A., Ralston G. B., Macromolecular crowding: Effects on actin polymerisation. Biophys. Chem. 66, 57–66 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Minton A. P., The effect of volume occupancy upon the thermodynamic activity of proteins: Some biochemical consequences. Mol. Cell. Biochem. 55, 119–140 (1983). [DOI] [PubMed] [Google Scholar]
- 44.Swulius M. T., Jensen G. J., The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J. Bacteriol. 194, 6382–6386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Divakaruni A. V., Loo R. R. O., Xie Y., Loo J. A., Gober J. W., The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 102, 18602–18607 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majumder S., et al. , Cell-sized mechanosensitive and biosensing compartment programmed with DNA. Chem. Commun. (Camb.) 53, 7349–7352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laganowsky A., et al. , Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z. Z., et al. , Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp., e50762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caschera F., Noireaux V., Preparation of amino acid mixtures for cell-free expression systems. Biotechniques 58, 40–43 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Garenne D., Libchaber A., Noireaux V., PNAS article Garenne et al, data used in the article, collected in 2019. Data repository for U of M. http://hdl.handle.net/11299/210183. Deposited 2 January 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.