Significance
Copper is an essential metal ion facilitating many biological functions. To avoid the toxicity of free copper, there are protein-based transport systems that deliver the metal to copper-dependent enzymes. Tumors have high demands for copper, but it is unclear why. Thus it is of importance to reveal copper-binding proteins involved in cancer progression. This work investigated the putative role of a copper-binding protein, Antioxidant 1 copper chaperone (Atox1), in breast cancer cell migration, which is a key step in cancer metastasis. We demonstrated that Atox1 is needed for efficient cell migration and identify partner proteins. Since primary breast tumors with high levels of Atox1 correlate with poorer patient survival, Atox1 may be a biomarker of metastatic breast cancer.
Keywords: Atox1, breast cancer, cell migration, live imaging, ATP7A
Abstract
Copper ions are needed for several hallmarks of cancer. However, the involved pathways, mechanisms, and copper-binding proteins are mostly unknown. We recently found that cytoplasmic Antioxidant 1 copper chaperone (Atox1), which is up-regulated in breast cancer, is localized at the lamellipodia edges of aggressive breast cancer cells. To reveal molecular insights into a putative role in cell migration, we here investigated breast cancer cell (MDA-MB-231) migration by video microscopy as a function of Atox1. Tracking of hundreds of individual cells (per condition) over a 9-h time series revealed that cell migration velocity and directionality are significantly reduced upon Atox1 silencing in the cells. Because silencing of the copper transporter ATP7A also reduced cell migration, these proteins appear to be on the same pathway, suggesting that their well-known copper transport activity is involved. In-cell proximity ligation assays demonstrated that Atox1, ATP7A, and the proenzyme of lysyl oxidase (LOX; copper-loaded via ATP7A) are all in close proximity and that LOX activity is reduced upon Atox1 silencing in the cells. Since LOX is an established player in cancer cell migration, our results imply that Atox1 mediates breast cancer cell migration via coordinated copper transport in the ATP7A-LOX axis. Because individual cell migration is an early step in breast cancer metastasis, Atox1 levels in tumor cells may be a predictive measure of metastasis potential and serve as a biomarker for copper depletion therapy.
Many proteins coordinate copper (Cu) ions to facilitate biological processes such as cellular respiration, protection against oxidative stress, biosynthesis of chemical messengers, modulation of connective tissue, and pigment construction (1–3). The concentrations and locations of Cu in cells are regulated via a set of so-called Cu transport proteins that handle Cu uptake and export as well as selective delivery to Cu-dependent enzymes (4–6). Soluble, cytoplasmic Antioxidant 1 copper chaperone (Atox1) (7) collects Cu that has entered the cell and delivers the metal to two multidomain P1B-type ATPases (adenosine triphosphatases) in the trans-Golgi network called ATP7A and ATP7B. Several Cu-dependent enzymes found in humans acquire Cu from ATP7A (e.g., lysyl oxidase) and ATP7B (e.g., ceruloplasmin) before further transport to specific locations for functioning (4–6). Whereas Atox1 is expressed in all cells, ATP7A and ATP7B expression depends on cell type: ATP7B is found mostly in hepatic cells; ATP7A, in most other cells (http://www.proteinatlas.org/).
Because Cu is important as a cofactor in many biological processes (8–10), it is no big surprise that Cu is required in several distinguishing cancer phenomena (such as metastasis) and that cancer patients’ serum and tumors have increased Cu levels (11). Metastasis begins with migration of individual cancer cells that detach from the primary tumor. Such cancer cells first invade local tissue and then migrate through the tumor stroma and blood vessels to eventually reach distant metastatic sites where they start to grow (12). Local cell migration is accomplished by the creation of actin-rich plasma membrane protrusions (e.g., lamellipodia) at the leading edge of the cell. Cu can promote the capacity of cancer cells to metastasize through activation of enzymes involved in cell growth and metabolism, such as the Cu-dependent proteins lysyl oxidase (LOX) (13, 14), SPARC (secreted protein acidic and rich in cysteine or osteonectin) (15), MEK1 (mitogen-activated protein kinase kinase 1) (16), and MEMO1 (mediator of cell motility 1) (17). Whereas it is not known how SPARC, MEK1, and MEMO1 are loaded with Cu for LOX, a Cu-dependent amine oxidase, there is information on both Cu loading and functions. ATP7A loads Cu into the secretory compartment for eventual loading onto LOX before secretion and proteolytic cleavage to a propeptide (LOXPP) and mature LOX (18–20). The Cu-dependent activity of LOX includes cross-linking of collagen and elastin in the extracellular matrix (ECM) (21). Cancer cells secrete LOX to create premetastatic niches by stimulating collagen cross-linking and fibronectin synthesis (14, 22) that in turn promote tumor cell migration and adhesion (23–25). In addition to extracellular activities, LOX may also regulate cancer cell migration via (less-known) intracellular activities, such as modulation of actin polymerization (26, 27).
Cu transport proteins may play key roles in cancer through loading of Cu-dependent enzymes with Cu (28, 29), but they may also have additional functions related to cancer. For example, Atox1 was shown to act as a Cu-dependent transcription factor that promotes expression of cyclin D1 (30), a key protein involved in cell proliferation (31), and it promotes inflammatory neovascularization by acting as both a transcription factor and a cytoplasmic Cu chaperone (32). In addition, Atox1 was described to be essential for platelet-derived growth factor (PDGF-) –induced cell migration (33) through a mechanism that involves translocation of ATP7A and the small G protein known as Rac family small GTPase 1 (Rac1) to the leading edge of cells for lamellipodia formation (34).
To identify overexpressed Cu transport proteins in cancer, we previously analyzed the RNA transcript level changes of the whole Cu proteome (i.e., for 54 identified Cu-binding proteins) in different cancer tissues using information from the Cancer Genome Atlas (TCGA) database (35). Our analysis revealed that, with respect to Atox1, it is up-regulated in breast, colorectal, uterus, and liver tumors (35), and the breast cancer data were confirmed by us in tissue microarray experiments. Next, using two breast cancer cell lines, MDA-MB-231 (i.e., MDA-231) and MCF7, we found that Atox1 accumulates at lamellipodia borders of the cells. Atox1 silencing in MDA-231 cells resulted in apparent cell migration defects as evidenced by reduced wound closure in a wound-healing assay (36), although this assay not only probes cell migration but also reflects cell proliferation and cell-cell interactions (37). To specifically probe the role of Atox1 in the migration of individual breast cancer cells, we here applied live-cell video microscopy in combination with Atox1 silencing. Notably, this methodology allows for quantitative measurements of individual cell migration parameters; in addition, morphological changes in cells during migration, as well as cell death and proliferation, can be identified. The results we present demonstrate that Atox1 is needed for two-dimensional (2D) MDA-231 single-cell migration with respect to both velocity and directionality. Building on a recent report that showed the ATP7A-LOX pathway to be required for breast cancer metastasis (20), we used in-cell proximity ligation and LOX activity assays to demonstrate that the same pathway, with Atox1 added as an upstream partner, is responsible for single-cell migration in MDA-231 breast cancer cells.
Results
Atox1 Silencing Reduces Single-Cell Migration Velocity and Directionality.
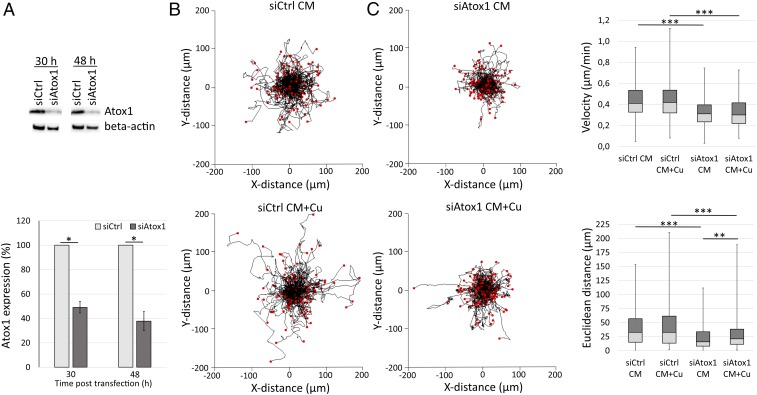
Boyden chamber migration assays demonstrated that knockdown of Atox1 expression with silencing RNA (siRNA) reduces MDA-231 cell migration (SI Appendix, Fig. S1). This observation supports our earlier finding of reduced wound healing for MDA-231 cells upon Atox1 silencing (36). To study breast cancer cell migration more quantitatively and at the single-cell level, we turned to 2D video microscopy of low-density cell cultures followed by single-cell tracking. In most experiments presented in this study, we analyzed MDA-231 breast cancer cells with control (control nontargeting siRNA) and silenced Atox1 expression levels in both standard (0.26 µM Cu according to inductively coupled plasma mass spectrometry [ICP-MS]) and 5 µM Cu-supplemented culture medium (CM). Cell transfection with 28 nM Atox1-targeting siRNA resulted in 50 and 60% reduced Atox1 expression levels at 30 and 48 h posttransfection, respectively (Fig. 1A). Thus, silencing reduced the Atox1 level but did not abolish all of the protein in the cells. This was important as we did not want the cells to be dramatically perturbed. Tracking analysis of hundreds of MDA-231 cells per condition for the 9-h time series (Materials and Methods; examples of tracking data are given in Movies S1–S4 with explanatory legends at the end of the SI Appendix) demonstrated that cells with reduced Atox1 levels have less distributed tracks (Fig. 1B). Specifically, the quantitative analysis showed that the Atox1-silenced cells migrated with reduced velocity (median value of 0.41 versus 0.31 µm/min [standard CM] and 0.42 versus 0.30 µm/min [Cu-supplemented CM] for control versus Atox1-silenced cells) and shorter Euclidean distance (median value of 32.6 versus 15.7 µm [standard CM] and 32.6 versus 21.1 µm [Cu-supplemented CM] for control versus Atox1-silenced cells) in both CM conditions (Fig. 1C and SI Appendix, Fig. S2).
Fig. 1.
Silencing of Atox1 expression in MDA-231 breast cancer cells results in reduced cell migration velocity and Euclidean distance. Cell migration evaluation by 2D phase contrast video microscopy and single-cell tracking analysis (9-h video recorded between 30 and 48 h posttransfection, 1-min intervals) of MDA-231 cells with control or reduced Atox1 expression levels in standard CM or 5 µM Cu-supplemented CM (CM Cu). (A) Western blot analysis of Atox1 expression at 30 and 48 h post transfection with control (siCtrl) or Atox1-targeting siRNA (siAtox1). Beta-actin expression was determined as loading control. Error bars indicate SD of the mean (nExp = 3; statistics using Student’s t test). (B) Wind-rose plots show cell tracks (nExp = 3, nROI/Exp = 2, ntracked cells = 166, 170, 219, and 214 for siCtrl CM, siCtrl CM + Cu, siAtox1 CM, and siAtox1 CM + Cu, respectively). (C) Box-whisker plots present migration velocity and Euclidean distance of the tracked cells (nExp = 3, nROI/Exp = 2; Mann–Whitney U statistical test). The error bars indicate the minimum and maximum values for the data sets (thus, the entire range of data is shown in each case). *P < 0.05, **P < 0.01, ***P < 0.001.
ICP-MS analysis of cell-associated Cu content showed that control and Atox1-silenced cells had similar amounts of Cu. Moreover, the 5 µM Cu treatment of the cells increased the cell-associated Cu concentration four times in control cells and five times in Atox1-silenced cells (SI Appendix, Table S1). We note that in all presented experiments, regardless of Cu supplementation or not, the cells had access to Cu and thus Atox1 could be Cu-loaded.
As a control for the tracking experiments, we counted the average number of cells per region of interest (ROI; 632.5 µm2). The analysis revealed that the determined cell density, 84 cells (±9 SD) per ROI, was comparable for control and Atox1-silenced cells in both standard and Cu-supplemented CM (SI Appendix, Fig. S3). This parameter is important to probe since variation in cell density may influence the cell tracks.
ATP7A Silencing Reduces Single Breast Cancer Cell Migration Velocity and Directionality.
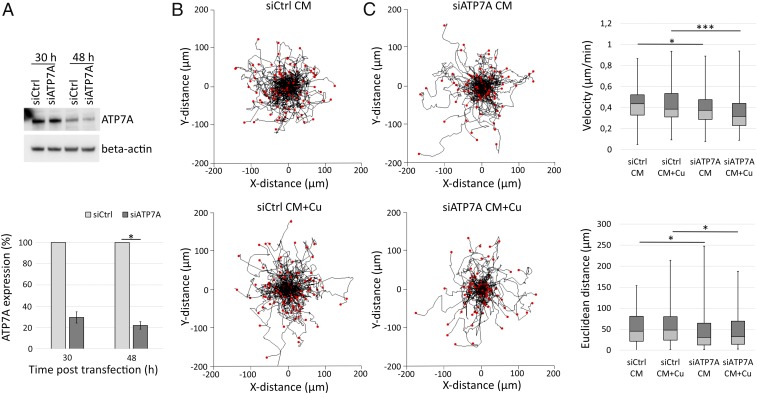
Since recent studies showed ATP7A to be important for metastasis in a different breast cancer system (20) and for stimulated migration of vascular smooth-muscle cells (38), we decided to test if ATP7A is also involved here, with Atox1, to promote MDA-231 breast cancer cell migration. To test involvement of ATP7A, we assessed cell migration in control versus ATP7A-silenced cells with the same methodology as for Atox1. Cell transfections with 28 nM ATP7A siRNA resulted in a 70 and 78% reduction in ATP7A expression at 30 and 48 h posttransfection, respectively (Fig. 2A). Live-cell video microscopy followed by cell-tracking analysis demonstrated significantly reduced migration velocity (median 0.44 versus 0.37 µm/min [standard CM] and 0.38 versus 0.32 µm/min [Cu-supplemented CM] for control versus ATP7A-silenced cells) and Euclidean distance (median 45.17 versus 30.66 µm [standard CM] and 47.90 versus 32.39 µm [Cu-supplemented CM] for control versus ATP7A-silenced cells) for cells with reduced ATP7A expression levels in standard and Cu-supplemented CM (Fig. 2 B and C and SI Appendix, Fig. S4). ATP7A silencing did not change cell-associated Cu levels, as compared to control cells, in either standard or Cu-supplemented CM as probed by ICP-MS (SI Appendix, Table S1). In comparison to Atox1 silencing, the decrease in cell migration velocity upon ATP7A silencing was not large. However, since the consequences on cell migration are similar upon Atox1 silencing and ATP7A silencing, the two proteins appear to be on the same pathway. As for the siAtox1 cell-tracking experiments, we analyzed average cell number per ROI in the ATP7A-related tracking experiments and found comparable cell density: 90 cells (±8 SD) per ROI for control and ATP7A-silenced cell conditions in both standard and Cu-supplemented CM (SI Appendix, Fig. S5).
Fig. 2.
Silencing of ATP7A expression results in reduced cell migration velocity and Euclidean distance. Cell migration evaluation by 2D phase contrast video microscopy and single-cell tracking analysis (9-h video recorded between 30 and 48 h posttransfection, 10-min intervals) of MDA-231 cells with control or reduced ATP7A expression levels in standard CM or 5 µM Cu-supplemented CM (CM + Cu). (A) Western blot analysis of Atox1 expression at 30 and 48 h posttransfection with control siRNA (siCtrl) or ATP7A-targeting siRNA (siATP7A). Beta-actin expression was determined as loading control. Error bars indicate SD of the mean (nExp = 2; statistics using Student’s t test). (B) Wind-rose plots with cell tracks for control (siCtrl) and ATP7A-silenced (siATP7A) cells in CM and CM + Cu (nExp = 2, nROI/Exp = 2, ntracked cells = 134, 130, 132, and 106 for siCtrl CM, siCtrl CM + Cu, siATP7A CM, and siATP7A CM + Cu, respectively). (C) Box-whisker plots presenting migration velocity and Euclidean distance of the tracked cells (nExp = 2, nROI/Exp = 2; Mann–Whitney U statistical test). The error bars indicate the minimum and maximum values for the data sets. *P < 0.05, ***P < 0.001.
In control experiments, we assessed cell viability and proliferation after treatment with siRNA and Cu. Cell viability was not affected by any of the treatments (tested by erythrosine B staining 24 h posttransfection). To evaluate cell proliferation, we visually identified dividing cells in the time-lapse microscopy videos. We observed no decrease in proliferation for Atox1-silenced cells in standard CM, but in Cu-supplemented CM, Atox1-silenced cells showed somewhat decreased proliferation compared to control cells [similar to what was reported in HEK cells (39)]. Also, there was a decrease in proliferation of ATP7A-silenced cells as compared to control cells for both standard and Cu-supplemented CM (SI Appendix, Fig. S6). Importantly, these variations in proliferation for the different culture conditions did not affect our cell-tracking results as only nonproliferative cells were tracked and analyzed.
Atox1, ATP7A, and LOX Are in Proximity with Interdependence in Breast Cancer Cells.
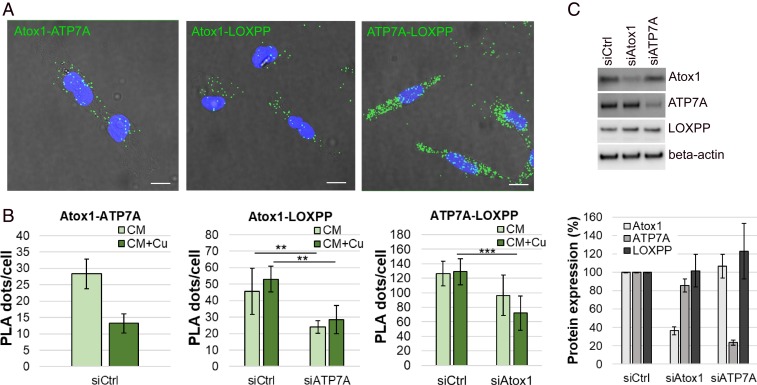
Because ATP7A-mediated activation of LOX was found to be important for breast cancer metastasis in another model system (20), we decided to test if LOX is downstream of Atox1-ATP7A in the pathway facilitating MDA-231 cell migration. Both ATP7A and LOX are overexpressed in MDA-231 cells (40). Here, we used an anti-LOXPP antibody that targets the intracellular LOX proenzyme. First, we applied immunofluorescent cell staining to locate Atox1, ATP7A, and LOXPP in the MDA-231 breast cancer cells. As reported before for Atox1 in these cells (36) and ATP7A in vascular smooth-muscle cells (33), we found all three proteins to, in part, localize at the lamellipodia borders (SI Appendix, Fig. S7). Then we tested possible interactions between Atox1, ATP7A, and LOXPP using in situ proximity ligation assays (PLAs) (41, 42). With this approach, protein-protein interactions could be probed with single-molecule resolution. The concept builds on attaching specific oligonucleotide probes to antibodies against two proteins of interest; if the antibodies are in close proximity (<40 nm), the oligonucleotides form circular DNA that can serve as a template for localized rolling-circle amplification, which is then visualized in situ as bright dots using fluorescence microscopy.
We first confirmed that we could detect the expected interaction of Atox1 and ATP7A in the MDA-231 cells using PLA (SI Appendix, Fig. S8A) and performed several technical and biological negative controls (one primary antibody only and Atox1-beta actin, respectively) (SI Appendix, Fig. S8B). Then we probed the proximity of Atox1 and ATP7A individually, with the LOX proenzyme (via LOXPP antibody) in the MDA-231 cells, in both standard and Cu-supplemented CM. As expected, since ATP7A delivers Cu to the LOX proenzyme, we detected ATP7A-LOXPP proximity in the MDA-231 cells (Fig. 3A). Surprisingly, we also detected Atox1-LOXPP proximity in the cells (Fig. 3B). This implies that Cu delivery from Atox1 to ATP7A, and then from ATP7A to the LOX proenzyme, is a synchronized chain of events.
Fig. 3.
PLAs reveal proximity and interdependence between Atox1, ATP7A, and LOXPP. PLA data for Atox1-ATP7A, Atox1-LOXPP, and ATP7A-LOXPP interactions in MDA-231 cells with control (siCtrl) or reduced expression levels of Atox1 (siAtox1) or ATP7A (siATP7A) in standard CM or 5 µM Cu-supplemented CM (CM + Cu). (A) Maximum projection confocal micrographs (blue = DAPI, indicating the nucleus; green = PLA dots) merged with phase contrast images. (Scale bars indicate 10 µm.) (B) Quantitative PLA results. Error bars indicate SD of the mean (nExp = 2 for Atox1-ATP7A, nExp = 3 for Atox1-LOXPP and ATP7A/LOXPP, nROI/Exp = 5; statistics using Student’s t test). (C) Western blot, representative blots (Top), and quantitative results (Bottom). Error bars indicate SD of the mean (nExp = 3). **P < 0.01, ***P < 0.001.
To further test the relationship between the three proteins in the MDA-231 cells, we evaluated Atox1-LOXPP and ATP7A-LOXPP proximities as a function of ATP7A and Atox1 expression levels, respectively. Notably, we found the number of Atox1-LOXPP interactions (fluorescent dots) per cell to decrease significantly upon ATP7A silencing (by 48 and 47% in standard and Cu-supplemented CM, respectively). This implies that the presence of ATP7A is required for Atox1-LOXPP proximity. Similarly, the number of ATP7A-LOXPP interactions (fluorescent dots) per cell decreased upon Atox1 silencing (by 25 and 44% in standard and Cu-supplemented CM, respectively). Thus, the presence of Atox1 appears necessary for ATP7A-LOXPP proximity (Fig. 3 A and B). We concluded that, in MDA-231 breast cancer cells, the three proteins (Atox1, ATP7A, and LOX) depend on each other for spatial proximity. As a control, we analyzed total cellular levels of the three proteins after silencing Atox1 and ATP7A. We found that neither Atox1 nor ATP7A silencing changed the cellular levels of the other two proteins (Fig. 3C). This supports that it is the spatial proximities of Atox1 and LOX proenzyme proteins, or ATP7A and LOX proenzyme proteins, that are disrupted upon ATP7A or Atox1 silencing, respectively.
To assess functional consequences of Atox1 silencing for LOX activity, we probed LOX activity in the conditioned CM of the cells using a LOX activity assay (fluorimetric) similar to what was used by Petris et al. (20). We used ATP7A silencing as a positive biological control, as Petris et al. showed that ATP7A knockout reduced LOX activity in another metastatic breast cancer cell model. In our experiments, silencing of ATP7A resulted in a 28% reduction in LOX activity and Atox1 silencing resulted in a 16% reduction in LOX activity (SI Appendix, Figs. S9 and S10 for positive and negative technical controls). Notably, in these experiments Atox1 and ATP7A expression levels were reduced by 54 and 80%, respectively (SI Appendix, Fig. S11). These findings demonstrate that Atox1 levels in the cells have direct effects on LOX activity.
Discussion
Atox1 is up-regulated in tissue from several types of cancers (35). In fact, if one analyzes patient data (e.g., https:/www.proteinatlas.org, but there are several data bases), it becomes evident that breast cancer patients with high Atox1 mRNA levels have poorer survival than those with low Atox1 levels (SI Appendix, Fig. S12). Thus, the level of Atox1 in cancer cells appears to be of direct clinical relevance. Here we used live-cell video microscopy for single-cell tracking, in combination with selective gene silencing, to demonstrate that Atox1 is needed for fast and directional breast cancer cell migration. This is an important result, as cell migration is directly related to metastasis potential and thus patient survival. We further showed that this effect appears mediated via the ATP7A-LOX axis. ATP7A silencing results in decreases in cell migration similar to those detected for Atox1 silencing, and the three proteins (Atox1, ATP7A, and LOX) are found in close proximity in breast cancer cells. Furthermore, we showed that silencing of Atox1 reduces LOX activity and, since LOX is a known mediator of cancer cell migration and metastasis (23–25) and LOX activity was found to be essential for migration of single MDA-231 cells (23), this provides a mechanistic explanation for the Atox1 effects. In other words, the promoting role of Atox1 for MDA-231 cell migration appears facilitated via coordinated Atox1-mediated Cu delivery to ATP7A and further to LOX, with Cu-loaded, activated LOX in turn promoting processes that stimulate cell migration.
Earlier work showed Atox1 to be important for PDGF-stimulated smooth-muscle cell migration, and it was suggested that in these cells Atox1 mediates migration via interactions with ATP7A and Rac1 (34). This work was followed by another study, using the same smooth-muscle cells, where it was shown that cell migration is promoted by ATP7A interactions with IQGAP1 (IQ motif containing GTPase activating protein 1) and Rac1, whereby all proteins translocate to the leading edge of the cells (38). IQGAP1 is a Rac1-binding scaffolding protein that plays a central role in the assembly of signaling complexes that regulate cellular motility, morphogenesis, and cell adhesion (43). Since we found ATP7A to be important for migration of MDA-231 breast cancer cells, we also tested if IQGAP1 plays a role. However, a pathway involving IQGAP1 appears not to dominate migration of MDA-231 breast cancer cells, as IQGAP1 silencing did not affect cell migration in live-cell–tracking experiments (SI Appendix, Fig. S13). Our presented results are instead similar to the findings described by Petris et al. (20). In this work, ATP7A-mediated activation of LOX was required for LOX-related mechanisms of metastasis of 4T1 mammary and Lewis lung carcinoma cells. In similarity to the Atox1 data in SI Appendix, Fig. S12, Petris et al. showed that breast cancer patients with high ATP7A expression levels in the primary tumor have reduced survival chances (20). Based on our current findings on Atox1 in MDA-231 cells, we predict that Atox1 is also a key upstream partner of ATP7A-LOX in 4T1 mammary and Lewis lung carcinoma cells that aids in promotion of metastasis.
Taken together, our results demonstrate a key role for Atox1 in individual cell migration of aggressive breast cancer cells. Individual cancer cell migration relates closely to what happens when a tumor begins to metastasize (44). With our methodology (i.e., live-cell time-lapse video microscopy followed by cell tracking), we specifically probed migration kinetics of individual cells and at the same time kept track of other cellular processes such as death and proliferation. In contrast, more common and easier cell migration assays (such as wound healing) often only report on endpoint outcomes and the results are affected by cell proliferation and cell-cell contacts. We further connected the Atox1 effect on cell migration to cellular activities of partner proteins ATP7A and LOX. Our work thus extends and links (in breast cancer) earlier work showing Atox1 being important for vascular smooth-muscle cell migration (33), and ATP7A and LOX being important for breast cancer metastasis (20). Although we did not prove a Cu dependence (in all experiments, cells had access to Cu; a four- to sixfold change in cell-associated Cu amount did not change the observed cell migration trends), Atox1 likely mediates breast cancer cell migration via Cu delivery to ATP7A, which delivers the metal to the LOX proenzyme. This in turn results in secreted, activated LOX. With all three proteins near each other in the cell and, in part, at the leading edge, the response to external cues may be at its most efficient, which is important when there is a high demand of Cu (such as in cancer). Due to this activity, Atox1 may act as a biomarker for breast cancer metastasis, such that patients with high Atox1 levels in primary tumor cells are at higher risk of metastasis than those with low Atox1 levels. Patients with high Atox1 would benefit the most from copper chelation therapy, which is an approach currently under investigation in clinical trials (45). In connection with clinical studies, quantification of migration properties of cancer cells using time-lapse microscopy may be a useful tool in the study of potential therapeutic anticancer drugs as well as in the discovery of molecular pathways of metastasis.
Materials and Methods
Video Microscopy and Cell Tracking.
Cells were seeded in plates with poly-L-lysine–coated glass bottoms. Upon cell adhesion, each plate was positioned into a stage top incubator on an Eclipse Ti 2 inverted microscope (Nikon). Cells were automatically imaged over time using phase contrast microscopy at multiple positions per well using a 10× air objective (Nikon). Cell tracking was performed manually for all selected cells. Further details are given in SI Appendix, Extended Materials and Methods.
In Situ Proximity Ligation Assay.
PLA was performed on fixed MDA-231 cells using the Duolink Detection Reagents Green Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Full details are given in the SI Appendix, Extended Materials and Methods.
LOX Activity.
LOX activity was measured in the conditioned CM of control Atox1- and ATP7A-silenced cells using a fluorescence-based LOX activity kit (Abcam) according to the manufacturer’s instructions. Full details are given in the SI Appendix, Extended Materials and Methods.
Other Methods.
Cell lines, siRNAs, and reagents used are described in the SI Appendix, Extended Materials and Methods. Details on the Boyden chamber assay, Western blotting, immunofluorescence microscopy, Cu treatment, cell-associated Cu analysis by ICP-MS, and clinical association analysis are given in the SI Appendix, Extended Materials and Methods.
Data Availability.
All data and procedures are provided here and in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the Knut and Alice Wallenberg foundation, the Swedish Research Council, and the Swedish Cancer Foundation for funding. Stellan Holgersson (Chalmers) is acknowledged for the ICP-MS measurements. High school students Anna Frisk and Alma Ahlberg helped with cell tracking as part of a school project.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910722117/-/DCSupplemental.
References
- 1.Huffman D. L., O’Halloran T. V., Function, structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 70, 677–701 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Puig S., Thiele D. J., Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 6, 171–180 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Harris E. D., Basic and clinical aspects of copper. Crit. Rev. Clin. Lab. Sci. 40, 547–586 (2003). [PubMed] [Google Scholar]
- 4.O’Halloran T. V., Culotta V. C., Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275, 25057–25060 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Festa R. A., Thiele D. J., Copper: An essential metal in biology. Curr. Biol. 21, R877–R883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson N. J., Winge D. R., Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamza I., Schaefer M., Klomp L. W., Gitlin J. D., Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc. Natl. Acad. Sci. U.S.A. 96, 13363–13368 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson Dzebo M., Ariöz C., Wittung-Stafshede P., Extended functional repertoire for human copper chaperones. Biomol. Concepts 7, 29–39 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Grubman A., White A. R., Copper as a key regulator of cell signalling pathways. Expert Rev. Mol. Med. 16, e11 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Turski M. L., Thiele D. J., New roles for copper metabolism in cell proliferation, signaling, and disease. J. Biol. Chem. 284, 717–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denoyer D., Masaldan S., La Fontaine S., Cater M. A., Targeting copper in cancer therapy: ‘Copper That Cancer.’ Metallomics 7, 1459–1476 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Erler J. T., et al. , Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 15, 35–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddikuzzaman V. M., Grace V. M., Guruvayoorappan C., Lysyl oxidase: A potential target for cancer therapy. Inflammopharmacology 19, 117–129 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Morrissey M. A., et al. , SPARC promotes cell invasion in vivo by decreasing type IV collagen levels in the basement membrane. PLoS Genet. 12, e1005905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M., Huang C. Z., Mitogen-activated protein kinase signaling pathway and invasion and metastasis of gastric cancer. World J. Gastroenterol. 21, 11673–11679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald G., et al. , Memo is a copper-dependent redox protein with an essential role in migration and metastasis. Sci. Signal. 7, ra56 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Tchaparian E. H., Uriu-Adams J. Y., Keen C. L., Mitchell A. E., Rucker R. B., Lysyl oxidase and P-ATPase-7A expression during embryonic development in the rat. Arch. Biochem. Biophys. 379, 71–77 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Kosonen T., Uriu-Hare J. Y., Clegg M. S., Keen C. L., Rucker R. B., Incorporation of copper into lysyl oxidase. Biochem. J. 327, 283–289 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanbhag V., et al. , ATP7A delivers copper to the lysyl oxidase family of enzymes and promotes tumorigenesis and metastasis. Proc. Natl. Acad. Sci. U.S.A. 116, 6836–6841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gacheru S. N., et al. , Structural and catalytic properties of copper in lysyl oxidase. J. Biol. Chem. 265, 19022–19027 (1990). [PubMed] [Google Scholar]
- 22.Xiao Q., Ge G., Lysyl oxidase, extracellular matrix remodeling and cancer metastasis. Cancer Microenviron. 5, 261–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne S. L., et al. , Lysyl oxidase regulates breast cancer cell migration and adhesion through a hydrogen peroxide-mediated mechanism. Cancer Res. 65, 11429–11436 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Erler J. T., et al. , Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Baker A. M., Bird D., Lang G., Cox T. R., Erler J. T., Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene 32, 1863–1868 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Payne S. L., Hendrix M. J., Kirschmann D. A., Lysyl oxidase regulates actin filament formation through the p130(Cas)/Crk/DOCK180 signaling complex. J. Cell. Biochem. 98, 827–837 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Cuevas E. P., et al. , LOXL2 drives epithelial-mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Sci. Rep. 7, 44988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brady D. C., et al. , Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509, 492–496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blockhuys S., Wittung-Stafshede P., Roles of copper-binding proteins in breast cancer. Int. J. Mol. Sci. 18, E871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh S., et al. , Novel role of antioxidant-1 (Atox1) as a copper-dependent transcription factor involved in cell proliferation. J. Biol. Chem. 283, 9157–9167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein E. A., Assoian R. K., Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell Sci. 121, 3853–3857 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G. F., et al. , Copper transport protein antioxidant-1 promotes inflammatory neovascularization via chaperone and transcription factor function. Sci. Rep. 5, 14780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashino T., et al. , Unexpected role of the copper transporter ATP7A in PDGF-induced vascular smooth muscle cell migration. Circ. Res. 107, 787–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohno T., et al. , Novel role of copper transport protein antioxidant-1 in neointimal formation after vascular injury. Arterioscler. Thromb. Vasc. Biol. 33, 805–813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blockhuys S., et al. , Defining the human copper proteome and analysis of its expression variation in cancers. Metallomics 9, 112–123 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Blockhuys S., Wittung-Stafshede P., Copper chaperone Atox1 plays role in breast cancer cell migration. Biochem. Biophys. Res. Commun. 483, 301–304 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Pijuan J., et al. , In vitro cell migration, invasion, and adhesion assays: From cell imaging to data analysis. Front. Cell Dev. Biol. 7, 107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashino T., et al. , Copper transporter ATP7A interacts with IQGAP1, a Rac1 binding scaffolding protein: Role in PDGF-induced VSMC migration and vascular remodeling. Am. J. Physiol. Cell Physiol. 315, C850–C862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson Dzebo M., et al. , Copper chaperone Atox1 interacts with cell cycle proteins. Comput. Struct. Biotechnol. J. 16, 443–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaraja G. M., et al. , Gene expression signatures and biomarkers of noninvasive and invasive breast cancer cells: Comprehensive profiles by representational difference analysis, microarrays and proteomics. Oncogene 25, 2328–2338 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Fredriksson S., et al. , Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Söderberg O., et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Brown M. D., Sacks D. B., IQGAP1 in cellular signaling: Bridging the GAP. Trends Cell Biol. 16, 242–249 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Giampieri S., et al. , Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 11, 1287–1296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez J., Ramchandani D., Vahdat L., Copper depletion as a therapeutic strategy in cancer. Met. Ions Life Sci. 19, 303–330 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and procedures are provided here and in the SI Appendix.