Summary
The gastrointestinal tract is covered by a single layer of epithelial cells that, together with the mucus layers, protect the underlying tissue from microbial invasion. The epithelium has one of the highest turnover rates in the body. Using stable isotope labeling, high-resolution mass spectrometry, and computational analysis, we report a comprehensive dataset of the turnover of more than 3,000 and the expression of more than 5,000 intestinal epithelial cell proteins, analyzed under conventional and germ-free conditions across five different segments in mouse intestine. The median protein half-life is shorter in the small intestine than in the colon. Differences in protein turnover rates along the intestinal tract can be explained by distinct physiological and immune-related functions between the small and large intestine. An absence of microbiota results in an approximately 1 day longer protein half-life in germ-free animals.
Keywords: proteomics, mass spectrometry, protein turnover, bacteria, colonization, commensals, germ-free, colon, small intestine
Graphical Abstract
Highlights
-
•
Dataset of protein turnover rate and expression along the mice intestinal tract
-
•
Protein turnover rate is slower in colon than in small intestine
-
•
Median protein half-life is 1 day longer in germ-free mice
Arike et al. report a turnover rate of >3,000 and expression of >5,000 proteins in epithelial cells and mucus along the mice gastrointestinal tract. Protein turnover rate is faster in the small intestine than in the colon and slower in germ-free animals.
Introduction
The gastrointestinal tract is responsible for the digestion and absorption of nutrients. It is covered by a single layer of epithelial cells that together with the mucus layers protect the organism from bacterial invasion (Johansson et al., 2013, Muniz et al., 2012). All of the epithelial cells originate from the stem cells in the crypt bottom. The enterocytes, enteroendocrine, and goblet cells differentiate and migrate over 3–5 days from the crypt to the villus tip, where they eventually are sloughed off; Paneth cells in the small intestine remain at the base of the crypt (Barker, 2014, Clevers, 2013, van der Flier and Clevers, 2009). The gastrointestinal tract performs region-specific tasks. The small intestine continues digestion that started in the mouth and stomach, with the help of pancreatic enzymes and bile acids. Released nutrients are taken up mainly by the enterocytes of the duodenum and jejunum, whereas the bile acids are absorbed in the ileum (Dawson and Karpen, 2015). In the colon, water and salts are reabsorbed and commensal bacteria ferment complex indigestible carbohydrates from the food and mucus (Salyers et al., 1977) to produce short-chain fatty acids (SCFAs) that serve as an energy source for colonocytes (Bergman, 1990).
The gastrointestinal tract harbors a high number of bacteria. The small intestine is more exposed to the intestinal bacteria, as the mucus layer is unattached and permeable (Ermund et al., 2013). However, fewer microbes reside in the small intestine (O’Hara and Shanahan, 2006) due to the fast passage time (0.5–5 h) and a high concentration of antimicrobial peptides originating largely from the Paneth cells (Donaldson et al., 2016). The colon has no Paneth cells and instead has a thicker two-layered mucus, where the inner layer is attached to the epithelium and normally impenetrable to bacteria (Johansson et al., 2008). The outer mucus layer is non-attached and more expanded, allowing it to become the habitat for the commensal bacteria. The mucus in both the small and large intestine is composed of a limited set of proteins, where the MUC2 mucin is forming the structural skeleton of the water-rich mucus (Johansson et al., 2008, Rodríguez-Piñeiro et al., 2013).
The role and effect of the microbiota on the host has been extensively studied using germ-free (GF) animals. This has shown that the microbiota has a high impact on host metabolism by increasing energy uptake from food, modifying bile acids, activating G-coupled-receptors, and affecting lipid metabolism (Koh et al., 2016, Sommer and Bäckhed, 2013, Wahlström et al., 2016). The microbiota also modulates the immune system and affects intestinal epithelial cell proliferation, differentiation, and apoptosis (Belkaid and Hand, 2014, Louis et al., 2014, Peck et al., 2017). The gastrointestinal epithelium has one of the highest renewal rates in the body (Creamer et al., 1961), whereas the turnover time of epithelial cells in the duodenum and ileum of GF mice is twice as long as for conventionally raised (CR) mice (Abrams et al., 1963, Lesher et al., 1964). This is reflected in a faster general protein turnover in CR than in GF animals (Muramatsu et al., 1983). Such studies are based on radioactive pulse labeling, which is unable to provide detailed information on the turnover rate of individual proteins.
Protein turnover rate is defined as a balance between protein synthesis and degradation that provide information on overall cellular responses (Larance and Lamond, 2015). Proteins with a high turnover rate can change their abundance rapidly and, hence, are considered regulatory important (Claydon and Beynon, 2012). Identification of long-lived proteins suggests candidates that might have a higher risk for accumulating spontaneous chemical damages (Toyama et al., 2013). With the use of mass-spectrometry-based techniques, stable-isotope-labeled amino acids have become the preferred approach, allowing studies of individual protein turnover (Claydon et al., 2012). Stable isotope labeling by amino acids in cell culture (SILAC) (Ong et al., 2002) has successfully been used to study protein turnover of cells in culture (Boisvert et al., 2012, Cohen et al., 2013) and animals (Ruhs et al., 2012, Westman-Brinkmalm et al., 2011). Such studies have revealed that the protein turnover depends on organ (Claydon et al., 2012) or subcellular location (Boisvert et al., 2012, Johnson et al., 1999). The protein turnover has also been shown to been reduced during starvation (Price et al., 2012) and by aging (Dai et al., 2014). However, no studies have addressed the individual protein turnover in epithelial cells or mucus along the gastrointestinal tract and the influence of microbiota.
To obtain a detailed understanding of the turnover and the abundance of individual proteins in intestinal epithelial cells and mucus, we have fed mice with food containing stable-isotope-labeled lysine and followed its incorporation by mass-spectrometry-based proteomics. We analyzed different regions of the small and large intestine of CR and GF mice. We have, thus, generated a comprehensive database for general use that demonstrates that the individual protein turnover rate varies along the gastrointestinal tract and is affected by the microbiota.
Results
Protein Turnover Rates in CR and GF Mice
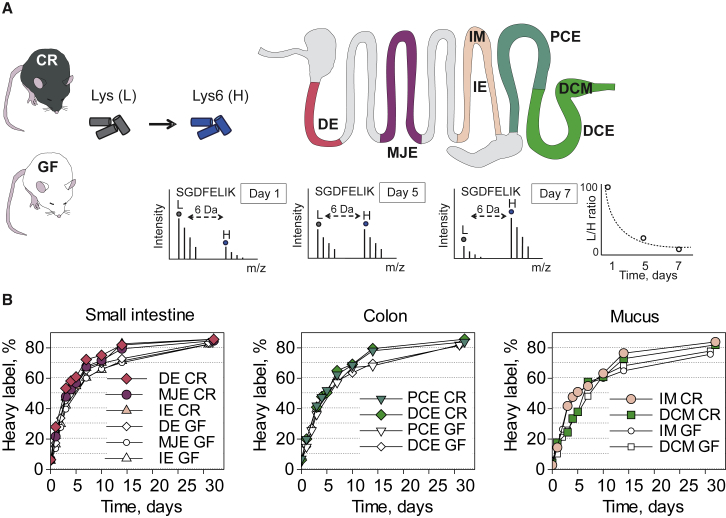
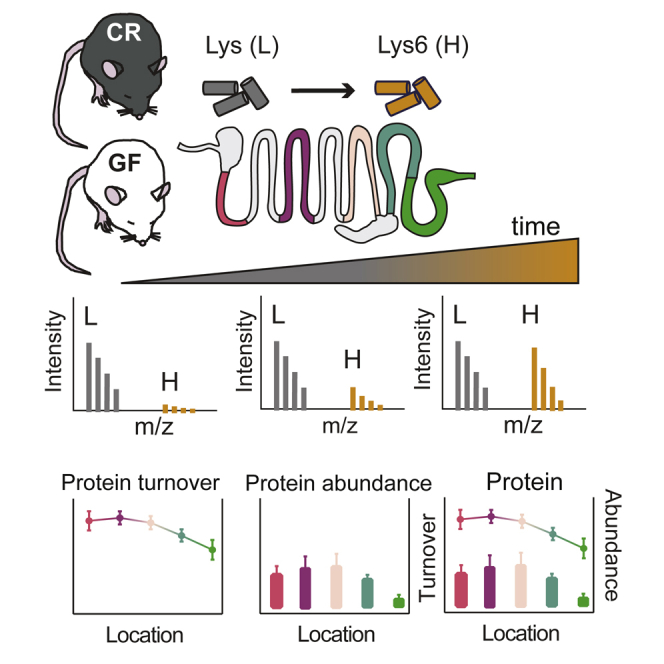
CR and GF mice were kept for 1 week on the amino-acid-defined diet containing unlabeled lysine, after which the diet was replaced with the 13C-lysine diet. Animals were sacrificed at 0, 1, 2, 3, 5, 7, 10, 14, and 32 days, and the incorporation of the heavy-labeled lysine into the epithelial cells and the mucus was analyzed by nanoscale liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS), as illustrated in Figure 1A. The incorporation of heavy label into the epithelial cell proteins reached 85% in the CR and 82% in GF mice after 32 days (Figure 1B). The incorporation of 13C-lysine was slower in the mucus and reached 83% in CR and 77% in GF mice after 32 days of labeling (Figure 1B).
Figure 1.
Experimental Setup for Protein Turnover Rate Studies in Mice Intestinal Epithelial Cells and Mucus
(A) Conventionally raised (CR) and germ-free (GF) mice were kept for 1 week on an amino-acid-defined diet (Lys [L]). Thereafter, the food was replaced by the same diet where lysine was substituted with 13C-lysine (Lys6 [H]). Animals were sacrificed at time points 0, 1, 2, 3, 4, 5, 7, 10, 14, and 32 days after the start of the heavy-labeled diet. Small intestine was divided into five parts and colon into two; caecum was excluded. For the proteomics analysis we used first (duodenum [DE]), third (middle jejunum [MJE]), and fifth (ileum [IE]) part of small intestine and both proximal (PCE) and distal colon (DCE) epithelial cells. Mucus was collected from the ileum and distal colon (IM and DCM, respectively). Mass-spectrometry-based proteomic analyses were used to follow the 13C-lysine incorporation into the freshly synthesized proteins, as indicated by the tryptic peptide SGDFELIK from the Muc2 mucin.
(B) Heavy label incorporation into proteins in epithelial cells and mucus. Data points represent the average value of heavy label in at each time point for the different segments and mucus.
See also Figures S1 and S2.
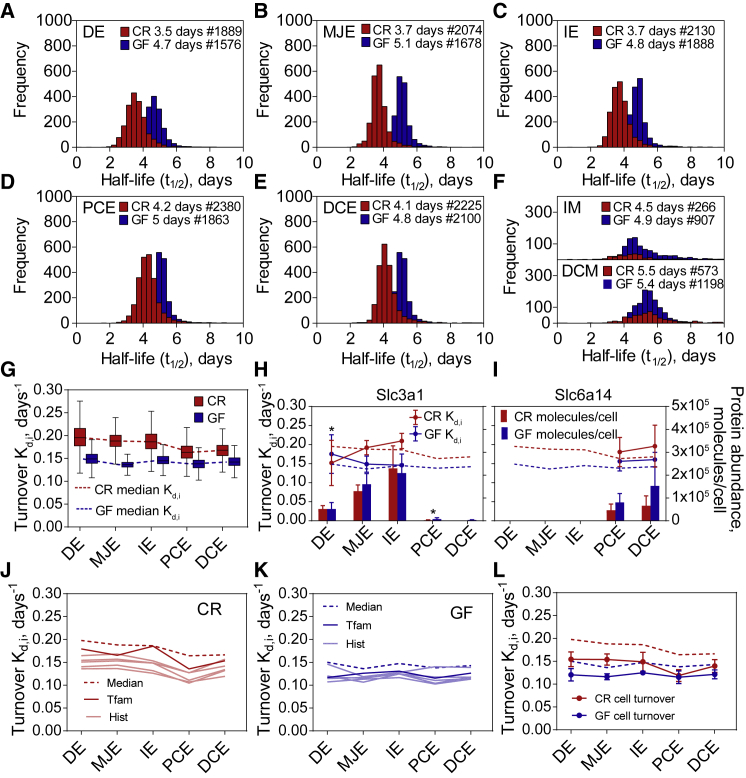
The half-life of individual proteins was calculated when at least three time points were present from the same location, something that was possible for 3,041 proteins. Details for the calculation are presented in STAR Methods, and the complete dataset is available in Table S1. The median half-life of CR mice proteins ranged from 3.5–4.2 days in intestinal epithelial cells (Figures 2A–2E, red histograms) to 4.5–5.5 days in the mucus (Figure 2F). The median protein half-life in intestinal epithelial cells of GF mice (Figures 2A–2E, blue histograms) was approximately 1 day longer than in CR animals (red histograms).
Figure 2.
Protein Turnover Rates in CR and GF Mice
(A–F) Histograms of protein half-lives along the gastrointestinal tract divided into 50 bins. The median half-life and the number of protein half-lives for each of the locations are stated on top of the corresponding histogram. DE, duodenum epithelial cells; MJE, middle jejunum epithelial cells; IE, ileum epithelial cells; PCE, proximal colon epithelial cells; DCE, distal colon epithelial cells; IM, ileum mucus; DCM, distal colon mucus as explained in Figure 1A. See also Figure S3.
(G) Boxplots represent the distribution of turnover rates for all of the proteins, 25th and 75th percentiles, and the whiskers indicate the values within 1.5 times the interquartile range [IQR]. The dotted line represents the median of protein turnover rates.
(H and I) Protein abundances and turnover rates for lysine transporters Slc3a1 and Slc6a14. Dots connected with solid line represent the specific protein turnover rate; error bars represent coefficient of variance (CV). Dotted lines represent median protein turnover rate of all proteins. Bars represent median protein abundance with standard deviation (SD). Asterisk above turnover rate dots represents protein turnover rate difference GF versus CR, with p < 0.1 based on Perseus (version 1.5.0.0) Significance A test. Asterisk above bars represents protein abundance difference GF versus CR, with p < 0.05 based on Significance A test.
(J and K) Median protein turnover rate (dotted line) and turnover rates of proteins used for cell turnover estimation in CR (J) and GF mice (K). Tfam - Turnover rate of Tfam. Hist - turnover rates of histones (Hist1h1b, Hist1h1c, Hist1h1d, Hist2h2aa1, and Hist1h4a).
(L) Median protein turnover rates and estimated cell turnover rate for CR and GF mice.
For further analysis, we used turnover rate, which is the first-order rate constant for degradation and more directly illustrates protein dynamics than half-life (Claydon and Beynon, 2012). The median CR mice protein turnover rate was faster in the small intestine than in the colon, a difference not noticed in GF mice (Figure 2G). One explanation for a slower protein turnover rate in the colon of CR mice is that the colonocytes are using bacterium-produced light lysine (Neis et al., 2015). Lysine is an essential amino acid that can be synthesized by microbiota and is also a preferred amino acid substrate used by colonic bacteria (Dai et al., 2011).We analyzed the relative levels of heavy label in bacterial and mouse peptides present in feces (Figure S3). The incorporation of the label was similar until the 6th day, and from the 11th day, the bacterial proteins lagged behind (Figure S3), suggesting that the microbiota is using an alternative light lysine source. It has been suggested that 1%–20% of the lysine in the host is derived from the intestinal microbiota (Metges, 2000). Modeling of microbial community metabolism suggested that the human gut microbiota produces less than 1 mmol/L of lysine per day (Shoaie et al., 2015). As the diet used here contained approximately 40 mmol/L of lysine, the bacterially produced lysine should, in theory, contribute less than 2.5% to the lysine pool. Therefore, it can be suggested that the CR mice colon epithelial cells are not using any notable amounts of bacterial lysine.
Another explanation for the difference in protein turnover between CR mice small intestine and colon could be that the labeled lysine is taken up faster in the proximal parts of the intestine. Absorption of amino acids is rapid in the duodenum and jejunum, slower in the ileum, and not significant in the large intestine, where amino acids are rather used by microbial metabolism (Barrett et al., 2010, Davila et al., 2013). We identified two amino acid transporters capable of transporting the cationic amino acid lysine: transporter Slc3a1, which was mainly present in the small intestine (Figure 2H), and the sodium- and chloride-dependent transporter Slc6a14 (Anderson et al., 2008, Bröer, 2008) present only in the colon (Figure 2I). Different transporters in the small intestine and colon could lead to differences in lysine uptake. Despite the low Slc3a1 abundance in the colon, the protein was three times higher in GF mice proximal colon and not detectable in the CR mice distal colon. The Slc6a4 levels were also increased in the GF mice colon. This might suggest an increased absorption of the labeled lysine in the colon of GF mice that could explain a more similar turnover rate between GF mice small intestine and colon than that observed for CR mice.
A third explanation for the observed differences between the small intestine and colon of CR and GF mice is that protein turnover is affected by the epithelial cell turnover rate, which is a combination of cell proliferation in the crypt, cell migration, and cell shedding. Long-lived proteins with slower turnover rate than the cell turnover are primarily removed by cell division (Zhang et al., 2017); therefore, to estimate cell turnover, we analyzed slow-turnover proteins dependent on replication, namely, the histone proteins: H1 (Hist1h1b, Hist1h1c, and Hist1h1d), H2A (Hist2h2aa1), and H4 (Hist1h4a) (Banday et al., 2014) and mitochondrial transcription factor Tfam (Figures 2J–2K). Tfam regulates mitochondrial DNA copy number in mammals (Ekstrand et al., 2004), and as mitochondria only divides at mitosis, the turnover of Tfam can be taken as an estimate of a cell turnover. The estimated epithelial cell turnover based on the mean of these six proteins was 0.154–0.149 days−1, corresponding to 4.5–4.7 days in CR mice small intestine, and 0.140–0.119 days−1, corresponding to 5–5.9 days in CR mice colon (Figure 2L). In GF mice, the cell turnover was 0.125–0.115 days−1, corresponding to 5.6–6.1 days (Figure 2L), and there was no difference between small and large intestine. Our estimated epithelial cell turnover rates follow the median protein turnover rate (Figure 2L) and are in line with previous estimations of 3–5 days for small intestine and 5–7 days in colon (Barker, 2014). In GF mice, epithelial cell turnover has been shown to be longer (Park et al., 2016).
Protein Turnover Rate Correlation with Function
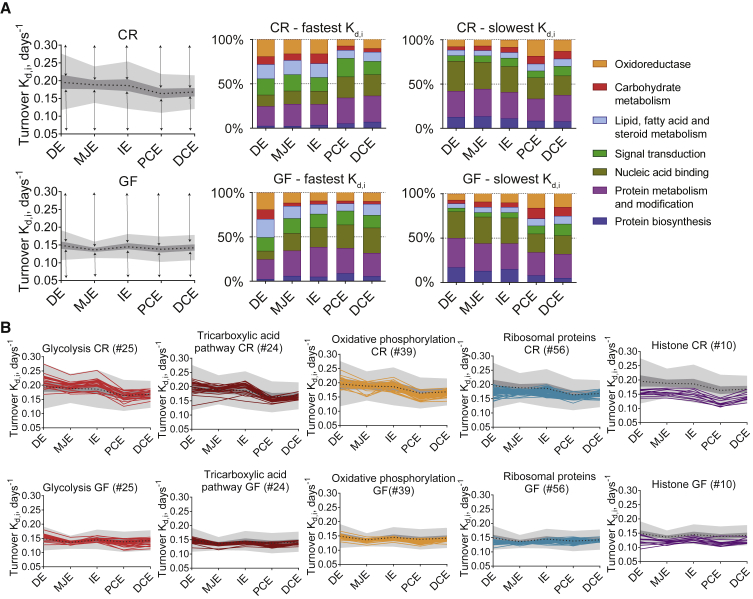
To determine functions associated with the proteins having the fastest or slowest turnover rates (top 25%), enrichment analysis was performed using the Gene Ontology (GO) terms “biological process” and “molecular function” (Figure 3A). Proteins belonging to the functional groups “oxidoreductase,” “carbohydrate metabolism,” and “lipid, fatty acid and steroid metabolism” had typically fast turnover in the small intestine and slow turnover in the colon (Figure 3A). For GF mice, this switch happened in the middle jejunum (Figure 3A).
Figure 3.
Functional Characterization and Dynamics of Protein Turnover Rate along the Intestine
(A) Enrichment analysis for proteins with fastest and slowest turnover rate in CR and GF mice. Black dotted line represents median of turnover rates, 50% of the protein turnover rates fall into dark gray area, and light gray area represents 1.5 times the IQR. The arrows show the area for top 25% of proteins with the fastest or slowest turnover rate, and the bar graphs show distribution of Panther GO “biological process” and “molecular function” terms associated with the proteins with the fastest and slowest turnover rate for each location. See also Figure S4.
(B) Proteins with turnover rate calculated in all epithelial cell locations and belonging into energy conversion pathways or protein complexes.
See also Figure S5.
Next, we were interested in how the protein abundance correlates with protein turnover rate. The abundance was calculated as molecules per cell for 5,012 proteins based on at least three replicates (Table S1) and compared to protein turnover (Figure S4). Although there was no correlation between protein turnover rate and abundance (Figure S4), we found that proteins with high abundance and fast turnover in small intestine were enriched in proteins involved in metabolism, whereas in colon, metabolic pathways had high abundance and slow turnover (Figure S4). High-abundance proteins with slow turnover in small intestine were involved in protein biosynthesis, which had high abundance and fast turnover in colon (Figure S4). Low-abundance proteins were mainly associated with translation, nucleic acid binding, and signal transduction. As mass-spectrometry-based proteomics has limitations in detecting low-abundance proteins, less turnover rate values were available for those proteins (Figure S4).
To study turnover rate dynamics along the intestine, we focused on proteins with turnover calculated in all of the epithelial cell locations (1,518 proteins in CR and 1,230 proteins in GF mice). This included approximately 60%–80% of the entire rates determined; as for the remaining proteins, turnover rates were not possible to calculate in every segment due to low abundance. Proteins belonging to glycolysis had a higher than median turnover along the CR mice small intestine; tricarboxylic acid and oxidative phosphorylation pathways followed the glycolytic protein patterns, with slightly slower turnover in small intestine and slower than median turnover in colon (Figure 3B). Proteins involved in energy production had a decreasing turnover trend toward the distal parts of intestine. In GF mice, glycolysis and tricarboxylic acid pathway proteins had higher turnover in duodenum and close to median in rest of the small intestine (Figure 3B). Additional examples of protein turnover and abundances for enzymes in those pathways are presented in Figure S5.
It is known that long-lived proteins often are components of large protein assemblies as histones, nuclear pore complex, or structural proteins like filaments, lamins, and myelin proteins (Toyama et al., 2013). We confirmed this, as histone and ribosomal proteins had slower than median turnover (Figure 3B). The longest living proteins in gastrointestinal epithelial cells of all locations were structural components Transgelin (Tagln), Filamin A (Flna), and Plastin-2 (Lcp1) that all had an exceptionally slow turnover (<0.13 days−1, corresponding to a half-life more than 5 days) (Figure S5).
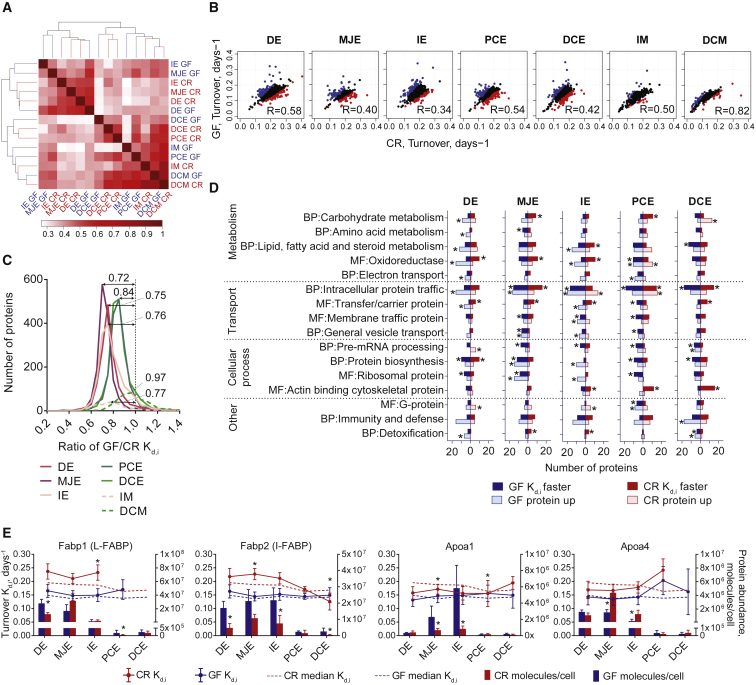
Protein Turnover Rate Comparison between GF and CR Mice
Protein turnover rates clustered according to their locations rather than to colonization status (Figure 4A). Segments closer to each other clustered together and mucus samples clustered together with colon epithelial cell samples (Figure 4A). This shows that the turnover rate is dependent on the protein location along the intestinal tract and is less affected by colonization. Although the turnover rate in GF mice is slower, the overall differences are much bigger, driven by the varying biological roles of each individual segment. Protein turnover rates correlate the best for GF and CR mice in duodenal epithelial cells (Figure 4B) and distal colon mucus samples, with correlations of 0.58 and 0.82, respectively (Figure 4B). The ileum showed the lowest correlation between GF and CR mice, with a correlation of 0.34 (Figure 4B).
Figure 4.
Comparison of Protein Turnover Rate between GF and CR Mice
(A) Heatmap representing the similarity in global protein turnover rates clustered based on Pearson correlation.
(B) Correlation of GF versus CR protein turnover rates between each sample analyzed. Blue and red dots represent the most significantly changed protein turnover rates (p < 0.1) in GF or CR mice, respectively. R, Pearson correlation.
(C) Distribution of protein turnover rate ratios (GF/CR) before normalizing to 1. Median values are indicated above curves.
(D) Most significantly changed protein turnover rates and abundances at each intestinal location annotated into Panther biological process (BP) and molecular function (MF) terms. The proteins with significantly faster turnover rate (p < 0.1) are given for GF in dark blue and CR in dark red. The proteins with higher abundance (p < 0.05) in GF or CR mice are presented in the light blue or light red boxes, respectively. Asterisk shows significantly enriched terms (p < 0.05). Extended data can be found in Table S2.
(E) Proteins involved in lipid transport. Fabp1, liver-type fatty acid binding protein; Fabp2, intestinal fatty acid binding protein; Apoa1, Apolipoprotein A-I; Apoa4, Apolipoprotein A-IV. Bars represent protein abundance calculated based on MS peak intensity with SD; dots with connecting lines represent specific protein turnover rate and error bars coefficient of variance; dotted lines represent median protein turnover rate of all proteins; asterisk above dots represents protein turnover rate difference of GF versus CR, with p < 0.1 based on Significance A test; asterisk above bars represents protein abundance difference of GF versus CR, with p < 0.05 based on Significance A test. GF, blue; CR, red.
The GF-to-CR ratio of the median turnover rate was 0.75 in duodenum, 0.72 in middle jejunum, 0.76 in ileum, 0.84 in colon, 0.77 in ileum mucus, and 0.97 in colon mucus (Figure 4C). Proteins with a turnover rate significantly different in GF mice compared to CR mice and, thus, affected by microbiota were largely found in the biological process and molecular function categories (all the data are presented in Table S2). Selected terms are presented as a bar graph in Figure 4D (relatively faster in GF mice, dark blue; relatively faster in CR mice, dark red). The same was done for proteins with a significantly different abundance (higher in CR mice, light red; higher in GF mice, light blue). The dynamics between protein turnover and alterations in abundance was different; for example lipid, fatty acid, and steroid metabolism proteins were increased in GF mice small intestine, whereas similar levels of proteins had higher turnover either in GF or CR mice small intestine (Figure 4D). Proteins involved in oxidoreduction had more proteins with faster turnover in CR mice small intestine, whereas more oxidoreductases had higher abundance in GF mice duodenum and ileum (Figure 4D). Protein biosynthesis members had mostly higher turnover in GF mice and higher abundance in jejunum and ileum (Figure 4D). Only a few proteins affected by microbiota were shared between the locations. Furthermore, microbiota-dependent differences in protein turnover were not related to the alterations in protein amounts. A complex relationship between protein turnover and abundance is demonstrated by fatty-acid-binding proteins Fabp1 and Fabp2 and apolipoproteins Apoa1 and Apoa4 (Figure 4E). Protein abundance (bars) and turnover (lines as compared to all proteins, dotted lines) are presented on the same graph showing that all of those lipid-binding proteins are strongly expressed along the small intestine, responding to microbiota with different protein turnover rate and abundance regulation (Figure 4E).
Turnover Rate of Mucus and Other Secreted Proteins
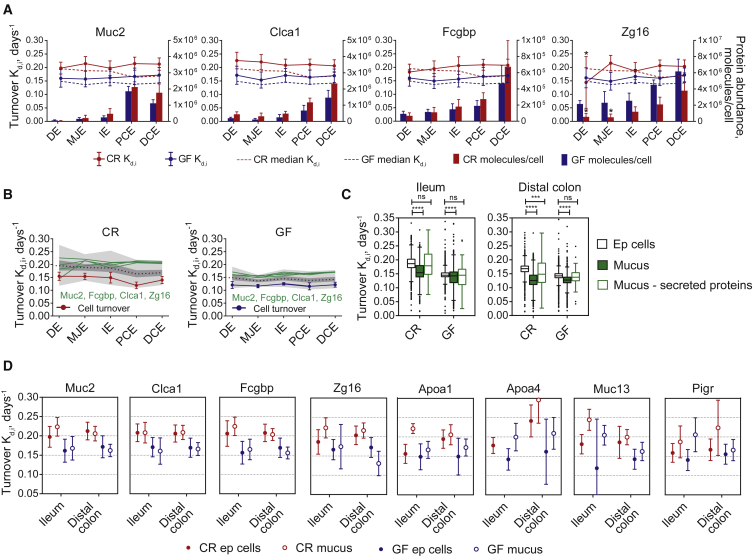
The heavily glycosylated Muc2 mucin is produced and secreted by the goblet cells and is the major component of the protective mucus layer together with the mucus related proteins Clca1, Fcgbp, and Zg16 (Johansson et al., 2008). The Muc2, Clca1, and Fcgbp epithelial cell protein abundances (bars on Figure 5A) were unchanged by bacteria, but interestingly, the Gram-positive bacteria binding protein Zg16 was decreased in small intestine (Figure 5A). Mucus proteins in both CR and GF mice had faster turnover (dots with connecting lines) than median (dotted lines) along the intestine (Figure 5A). The mucus main components had an almost two times faster turnover rate in the colon than the cell turnover rate (Figure 5B), suggesting that mucus is resynthesized almost two times in the colon during the average lifetime of epithelial cells. In GF mice, the difference between mucus components and estimated cell turnover was similar between small intestine and colon (Figure 5B).
Figure 5.
Turnover Rate and Protein Abundance for Mucus Components
(A) Turnover and abundance of Muc2, Clca1, Fcgbp, and Zg16. Bars represent protein abundance calculated based on MS peak intensity with SD; dots with connecting lines represent specific protein turnover rate and error bars coefficient of variance; dotted lines represent median protein turnover rate of all proteins; asterisk above dot represents protein turnover rate difference GF versus CR, with p < 0.1 based on Significance A test. GF, blue; CR, red.
(B) Turnover rate of mucus main components and estimated cell turnover (median of Tfam, Hist1h1b, Hist1h1c, Hist1h1d, Hist2h2aa1, and Hist1h4a) on the background of all of the turnover rates.
(C) Turnover of all proteins: black empty boxes represent all proteins in epithelial cells; green filled boxes represent all of mucus proteins; and empty green boxes represent all secreted proteins in mucus (mucus proteins with a signal sequence). Boxes represent the medians and 25th and 75th percentiles, and the whiskers indicate the values within 1.5 times the IQR. Median protein turnover rate for mucus and for secreted proteins in mucus has been compared to epithelial cells; ns, non-significant. ∗∗∗p ≤ 0.0002; ∗∗∗∗p ≤ 0.0001 as determined with one-way ANOVA followed by Dunnett’s multiple comparison test.
(D) Protein turnover of mucus components in ileum and distal colon for epithelial cells (filled dots) and mucus (empty dots) in GF (blue) and CR (red) mice.
The median protein turnover rate in CR mice ileum and distal colon mucus was slower than in the corresponding epithelial cells (Figure 5C). In addition to the main secreted goblet cell components, mucus contained cellular proteins from shed cells that have longer half-life in mucus than in epithelial cells. Predicted secreted proteins based on the presence of a signal peptide had a median protein turnover in the mucus close to that of the epithelial cells (Figure 5C). Additional to known mucus proteins, apolipoproteins Apoa1 and Apoa4 had faster turnover in mucus (Figure 5D). Apoa1 is released as a free apolipoprotein from the apical side of enterocytes into the lumen in the fasting state (Danielsen et al., 2012, Danielsen et al., 1993). Also, the transmembrane mucin Muc13 and polymeric immunoglobulin receptor (Pigr) had faster turnover in the mucus samples (Figure 5D). Pigr translocates immunoglobulin A (IgA) over the epithelium and is secreted together with IgA into the mucus (Johansen and Kaetzel, 2011).
In GF mice, the difference between median protein turnover in mucus and epithelial cells was smaller, indicating an increased cell lifespan and a slower mucus turnover (Figure 5C). This finding is supported by the observation that GF mice have reduced cell shedding (Hughes et al., 2014), and the lack of bacterial products that stimulate mucus can be expected to cause a slower mucus turnover (Birchenough et al., 2016). Still, the secreted proteins have a relatively faster turnover also in GF mice mucus than that in cells (Figure 5D).
Discussion
The present results suggest a dynamic and variable protein turnover rate along the intestine that is affected by microbiota. Median protein turnover rate in CR mice epithelial cells was 0.195 day−1 in duodenum, 0.188 day−1 in jejunum, 0.186 day−1 in ileum, 0.163 day−1 in proximal colon, and 0.168 day−1 in distal colon. Median protein turnover rate values for different mouse tissues have been reported: for liver 0.162 day−1, for kidney 0.156 day−1, for cardiac muscle 0.059 day−1, and for skeletal muscle 0.023 day−1 (Claydon et al., 2012). Faster protein turnover rate in intestinal epithelial cells is in accordance with previous suggestions that the gastrointestinal epithelium is the organ with the highest turnover rate in the body (Creamer et al., 1961). We further observed that the protein turnover rate decreases in the same organ along the proximal-to-distal direction. A similar trend was found in the human small intestine after 13C-leucine administration (Nakshabendi et al., 1999). This might be due to the proximal small intestine being the site for the first contact with the ingested food and at a high demand for adaptation that is gradually decreasing distally. This also explains why proteins involved in energy metabolism, especially in glycolysis, have fast turnover in the small intestine.
In the colon, the metabolic pathways have a slower turnover rate. This can be explained by the observation that colonocytes use bacterium-produced SCFAs as their primary energy source, whereas small intestinal epithelial cells largely utilize glucose (den Besten et al., 2013a, Donohoe et al., 2011). To evaluate the effect of the microbiota, we analyzed protein turnover in GF mice and found that the protein turnover was slower and there was no decrease in the proximal-distal direction. It has previously been suggested that the small intestinal epithelial cells of GF mice have a slower turnover due to luminal bacterial signals controlling the epithelial cell apoptosis (Alam et al., 1994, Hausmann, 2010, Kim et al., 2010, Savage et al., 1981), induction of stem cell proliferation (Buchon et al., 2009, Jones et al., 2013), development of villus capillary networks (Reinhardt et al., 2012), and Paneth cell differentiation (Stappenbeck et al., 2002). Here, we also show that protein turnover rate is slower in GF mice intestinal epithelial cells.
Fast turnover proteins in the small intestine were involved in energy metabolism, and slow turnover proteins along the intestine were proteins like histones, ribosomes, and cell structural components. In GF mice, glycolysis and citric acid pathway proteins had higher turnover in only the duodenum and close to median in the rest of the small intestine; oxidative phosphorylation pathway enzymes had close or slower than median turnover along the whole intestine. Colonocytes of GF mice have been shown to be energy deprived and require autophagy-derived amino acids for their energy demand (Donohoe et al., 2011). This could be a reason for slower than median turnover of energy-generating pathways in GF mice. Interestingly, actin-binding cytoskeletal proteins had faster turnover in CR colon, something that could be related to microbiota-induced apoptosis in the CR colon, as actin-binding proteins are involved in apoptosis (Desouza et al., 2012, Gourlay and Ayscough, 2005). Ribosomal proteins and protein biosynthesis proteins had relatively faster turnover in GF mice, which may be a cellular compensation for slower turnover in metabolic pathways.
The bacterial influence on protein abundance and turnover rate was dependent on the intestinal segment, as very few proteins were affected in all epithelial cells samples. Microarray analyses have previously shown that the responses to microbiota are site specific along the intestine (Larsson et al., 2012) and along the crypt-tip axis (Sommer et al., 2015). One of the most affected pathways is lipid metabolism, as GF are leaner than CR mice and are protected against diet-induced obesity due to the increased fatty acid metabolism (Bäckhed et al., 2004, Bäckhed et al., 2007, Camp et al., 2014, Larsson et al., 2012, Sommer et al., 2015). Global transcriptome comparisons of GF and CR mice intestine have demonstrated that the gene expression of proteins involved in lipid metabolic processes and cholesterol biosynthesis is affected by microbiota (Camp et al., 2014, Larsson et al., 2012, Sommer et al., 2015). An important reason for this is likely that the bacterially produced SCFAs affect the epithelial cell metabolism (den Besten et al., 2013b). Current data demonstrate that microbiota affects the turnover of proteins involved in lipid, fatty acid, and steroid metabolism along the intestine, which do not correlate with alterations in protein abundance. Protein levels were increased in GF mice duodenum and ileum, and the corresponding turnover rate was faster in CR mice ileum and GF mice distal colon. Altered protein turnover rates in response to the luminal bacteria were sometimes accompanied with changes in protein abundance (Fabp2 in distal colon epithelial cells (DCE), Apoa1 in middle jejunum epithelial cells (MJE); Figure 4E). However, the change in protein turnover rate can also be only an indication of protein regulation (Fabp1 in ileum epithelial cells (IE), Apoa1 in proximal colon epithelial cells (PCE); Figure 4E).
The intestinal mucus is the first defense line against bacteria. In the small intestine, the mucus is unattached and penetrable to bacteria (Ermund et al., 2013). In the colon, on the other hand, the mucus is built by a two-layer system: an inner layer attached to the epithelium, impenetrable to and devoid of bacteria, and an outer non-attached mucus layer that hosts the commensal bacteria (Johansson et al., 2008). The turnover of mucus main components was faster in the colon than in small intestine compared to the cell turnover and accompanied with higher protein concentration. The colon relies more on a thick mucus layer for its protection, which demands an increased mucus synthesis capacity in the presence of bacteria, which is reflected in the present observations.
Extracellular proteins that are immediately secreted from the cell do not contribute to the intracellular pool of unlabeled proteins (Hammond et al., 2016). This also means that secreted proteins should be labeled more rapidly than the intracellular proteins. We show here that the secreted proteins have faster turnover in the mucus than in epithelial cells, something that could be utilized to distinguish between true mucus proteins and cellular contaminations due to the cell shedding.
Protein turnover rate, as measured with heavy-isotope-coded amino acid incorporation in vivo, has to consider dilution of the protein pool by cell division and protein degradation. Although the effect of protein degradation that results in circulation of unlabeled amino acids from old proteins (as well as labeled peptides from newly synthesized proteins) has been considered by using miscleaved peptides to calculate maximum relative isotope abundance (see STAR Methods), cell division has been neglected. Comparisons between GF and CR mice protein turnover rate are further complicated by the differences in morphology of the villi, longer and thinner villi causes slower cell traveling time along the crypt-villus axis in GF (Park et al., 2016, Reinhardt et al., 2012). To evaluate differences in cell division between small intestine and colon in CR and GC mice, we have estimated cell turnover by using median turnover rate of five slow turnover histone proteins and replication-dependent Tfam. The estimated epithelial turnover was in line with the previous knowledge that cell turnover is slower in the colon than in the small intestine (Barker, 2014, Messier and Leblond, 1960), and it is slower in GF mice (Abrams et al., 1963, Lesher et al., 1964, Park et al., 2016). Despite the limitations, the presented results are in line with previous observations and extend these by providing individual protein turnover rates and protein amounts in epithelial cells and mucus along the gastrointestinal tract.
Conclusions
Using labeled-lysine-containing food combined with MS enabled us to estimate both individual protein turnover rates and abundances in isolated epithelial cells and mucus from different parts along the mouse intestine. We found slower turnover in colon than in small intestine, likely related to the slower metabolic activity of colonocytes. There was a general slower turnover in the GF mice, but this varied between spatial locations and individual proteins. The fastest turnover rates were observed for metabolic enzymes belonging to the glycolysis and citric acid cycle, whereas cell structural proteins had the slowest protein turnover. The present dataset (Table S1) provides a resource for mining proteins for turnover rate and abundance in the epithelial cells and mucus along the intestinal tract and their alterations upon bacterial conventionalization.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Amino Acid Defined Diet | Harlan Laboratories, Inc. | TD.99366 |
| Lys(6)-SILAC-Mouse Diet | Silantes GmbH, Munich, Germany | Product Number: 230924630 |
| GuHCl | Thermo Scientific | 24115 |
| DTT | Sigma-Aldrich | D9163 |
| Iodoacetamide | Sigma-Aldrich | I6125 |
| LysC | Wako | 125-05061 |
| AspN | Promega | V1621 |
| Deposited Data | ||
| Proteomics data raw data | This paper | ProteomeXchange: identifier PXD011457 |
| Experimental Models: Organisms/Strains | ||
| Male C57BL/6J mice | In-house breeding | N/A |
| Software and Algorithms | ||
| MaxQuant (Version 1.3.0.5) | Cox & Mann, 2008 | https://www.biochem.mpg.de/5111795/maxquant |
| Perseus (Version 1.5.0.0) | Tyanova et al., 2016 | https://www.biochem.mpg.de/5111810/perseus |
| MATLAB | MathWorks | https://se.mathworks.com/ |
| Prism (Version 7.02) | GraphPad | https://www.graphpad.com/ |
| Other | ||
| Mouse mucin sequence database | In-house database | http://www.medkem.gu.se/mucinbiology/databases/ |
| Nanosep 10K and 30K Omega | Pall, Life Sciences | OD010C35, OD030C35 |
| StageTip C18 columns | Rappsilber et al., 2007 | N/A |
| Reverse-phase column (150 × 0.075 mm inner diameter, C18-AQ 3 mm) | In-house packed | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gunnar C. Hansson (gunnar.hansson@medkem.gu.se).
This study did not generate new unique reagents.
Experimental Model and Subject Details
All animal procedures were approved by the local Laboratory Animal Ethics Committee, Gothenburg, Sweden. Male wild-type C57BL/6J mice were co-housed with 4-5 mice/cage at a 12 hour light/ dark cycle, had unlimited access to water and food and were bred in-house. Conventionally raised (CR) and germ-free (GF) 6-9 weeks old C57BL/6J male mice were kept first 1 week on an amino acid-based diet containing 12C6-lysine (light feed, Harlan Laboratories, Inc.). Thereafter feed was replaced by same diet but where 12C6-lysine was replaced by 13C6-lysine (heavy feed, Silantes GmbH, Munich, Germany) and animals were kept on this diet for 0, 1, 2, 3, 5, 7, 10, 14 and 32 days. Experimental setup is presented of Figure 1A. Bodyweight of the animals and the food consumption during the experiment is presented on Figure S1A.
Method Details
Collection of epithelial cells and mucus
Animals were sacrificed via cervical dislocation. Small intestine was divided into five parts and colon into two. For the proteomics analysis we used the first (duodenum), third (middle jejunum) and fifth (ileum) part of small intestine and both proximal and distal colon tissues (Figure 1A). Mesenteric tissue was removed and the intestinal tissues were cut open and incubated in PBS containing 3 mM EDTA and 1 mM DTT at 37°C for 60 min. The solution was replaced with fresh 37°C PBS and epithelial cells were dissociated from the tissue by vigorous shaking for 5x20 s on a vortex. Example of a colon tissue before and after removal of epithelial cells is presented on Figure S1B. Remaining tissue was removed and cells were pelleted by centrifugation at 1000 g for 5 min. Mucus was collected by mounting ileum and distal colon tissues in a horizontal perfusion chamber with a circular opening of 4.9 mm2, and the mucus was collected apically (Gustafsson et al., 2012). Pelleted cells and mucus were flash frozen in liquid nitrogen and stored at −80°C until further processing.
Protein extraction and digestion
Epithelial cells were solubilized in lysis buffer (4% SDS, 100 mM Tris-HCl pH 8, 100 mM DTT), heated 5 min at 95°C and cleared by centrifugation at 14 000 rpm 5 min. Thereafter samples were digested with LysC or AspN according to FASP (Filter Aided Sample Preparation) protocol (Wiśniewski et al., 2009b) with 30 kDa filters (PALL, Port Washington, NY). Overnight digestion was followed with fractionation according to the pipette tip SAX protocol (Wiśniewski et al., 2009a), and two fractions were eluted at pH of 11 and 3. Mucus samples were incubated overnight at 37°C in reduction buffer (6 M GuHCl, 0.1 M Tris/ HCl pH 8.5 (Merck), 5 mM EDTA, 0.1 M DTT (Merck)) followed by a FASP digestion protocol using 6 M guanidinium hydrochloride (GuHCl) instead of urea and 10kDa filters. Peptides were cleaned with StageTip C18 columns (Rappsilber et al., 2007) prior to mass-spectrometry analysis.
LC-MS/MS Analysis
Nano LC−MS/MS was performed on a Q-Exactive mass-spectrometer (Thermo Fischer Scientific) connected with an EASY-nLC 1000 system (Thermo Fischer Scientific) through a nanoelectrospray ion source. Peptides were loaded on a reverse-phase column (150 mm × 0.075 mm inner diameter, New Objective, New Objective, Woburn, MA) packed in-house with Reprosil-Pur C18-AQ 3 μm particles (Dr. Maisch, Ammerbuch, Germany). Peptides were separated with a 95-minute gradient from 5 to 30% B (A: 0.1% formic acid, B: 0.1% formic acid/80% acetonitrile) using a flow rate of 200 nl/min. In case of SAX fractionated samples two gradients were used: for pH 11 fraction in 80 min from 5 to 20% B and in next 15 min up to 30% B; for pH 3 fractions in 95 min from 10 to 30% B. Q-Exactive was operated at 200°C capillary temperature and 2.0 kV spray voltage. Full mass spectra were acquired in the Orbitrap mass analyzer over a mass range from m/z 350 to 1600 with resolution of 70 000 (m/z 200) after accumulation of ions to a 1 × 106 target value based on predictive AGC from the previous full scan. Twelve most intense peaks with a charge state ≥ 2 were fragmented in the HCD collision cell with normalized collision energy of 30%, and tandem mass spectrum was acquired in the Orbitrap mass analyzer with resolution of 35 000. Dynamic exclusion was set to 30 s. The maximum allowed ion accumulation times were 120 ms for full MS scans and 100 ms for tandem mass spectrum.
MS data analysis
MS raw files were processed with MaxQuant software version 1.3.0.5 (Cox and Mann, 2008), peak lists were identified by searching against the mouse UniProt protein database (release 2013.02) supplemented with an in-house database containing all the mouse mucin sequences (http://www.medkem.gu.se/mucinbiology/databases/). Searches were performed using either LysC or AspN as an enzyme, maximum 2 missed cleavages, precursor tolerance of 20 ppm in the first search used for recalibration, followed by 7 ppm for the main search and 0.5 Da for fragment ions. Carbamidomethylation of cysteine was set as a fixed modification and methionine oxidation and protein N-terminal acetylation were set as variable modifications. The required false discovery rate (FDR) was set to 1% both for peptide and protein levels and the minimum required peptide length was set to seven amino acids. Quantification of SILAC pairs was performed by MaxQuant with standard settings using a minimum ratio count of 2.
Quantification and Statistical Analysis
Protein abundance determination
Protein abundances were calculated by dividing the protein MS intensity for each protein by the number of theoretically observable peptides (all fully LysC digested peptides between 700 and 2,500 Da while missed cleavages were neglected and only carbamidomethylation of cysteine was considered as fixed modification) and normalized to summed intensity of all identified proteins. In epithelial cells normalized values were multiplied with the number of protein copies in the mammalian cell - 109 (Milo, 2013). Protein abundances are available in Table S1.
Calculation of protein turnover rates
To estimate protein turnover rates data had to be fitted to a model describing the replacement of unlabeled proteins with labeled ones as a function of protein turnover rate. Model can be described with following differential equation:
| (1) |
Where kd,i describes the protein turnover rate, fp,i is heavy to light (H/L) ratio of labeled protein and R∞,i is a maximum relative isotope abundance of proteins in particular sample. Differential equation gives a following solution for incorporation of heavy labeled proteins:
| (2) |
Protein turnover rate kd,i is directly linked to average protein degradation time td,i. with the following formula:
| (3) |
Before fitting data to Equation 2, maximum relative isotope abundance had to be calculated. This was done using the data for samples collected after 31 days of labeling and digested with AspN in order to have information for miss-cleaved peptides. Resulting peptides contained a mixture of labeled and unlabeled lysine that can originate from degraded proteins or from gut microbiota metabolism. To calculate relative isotope abundance, first for each peptides that had data for fully labeled (heavy-heavy i.e., HH) and partly labeled (heavy-light i.e.HL) versions the ratio between heavy-heavy and heavy-light labeled peptides was calculated. Median value of HH/HL ratio of all peptides of particular sample was used for calculating relative isotope abundance (R∞) using following formula:
| (4) |
where M is median HH/HL value.
Calculated relative isotope abundance values for each sample are presented in the following table.
| DE | MJE | IE | IM | PCE | DCE | DCM | |
|---|---|---|---|---|---|---|---|
| CR | 0.8868 | 0.8634 | 0.8728 | 0.8728 | 0.8677 | 0.8689 | 0.8689 |
| GF | 0.8659 | 0.8535 | 0.8453 | 0.8453 | 0.8316 | 0.8263 | 0.8263 |
Calculation of protein turnover rates were done by fitting experimental data measured on the day when substrate feed was replaced with labeled feed and on 1st, 2nd, 3rd, 4rd, 5th, 7th, 10th, and 14th day after the feed was replaced into function (2).
Data fitting was done in Wolfram Mathematica using nonlinear fitting algorithm. This algorithm was preferred over linearization of exponential as the fitting results were better according to the comparison of the coefficient of variance (CV), coefficients of determination (R2), etc. Obtained results fit relatively well to the particular model, as median R2 and CV values were 0.88 and 14.0% for CR samples and 0.87 and 17.2% for GF samples (Figures S1C and S1D). Examples of the experimental data fit into the function are presented on Figure S2.
Resulting protein turnover rates were subjected to simple one-sided non-parametric outliers test, where protein turnover rates with CV above four times median were considered as outliers. Depending on the sample 3 to 53 protein turnover rates were eliminated. This formed relatively small part (0.2%–3.8%) of total number of estimated protein turnover rates. Protein half-lives and turnover rates are available in Table S1.
Statistical analysis
Perseus program (version 1.5.0.0) (Tyanova et al., 2016) function Significance A was used to calculate significant outliers in comparison of GF versus CR. Significance A values < 0.05 for log2 protein abundance ratios and < 0.1 for protein turnover rate ratios are marked with asterisk on the figures and are used in further data analysis. Comparison of turnover rate of proteins in epithelial cells versus mucus was performed with one-way ANOVA followed by Dunnett’s multiple comparison test (Figure 5C).
Enrichment analysis
Enrichment analysis was performed with DAVID (version 6.7) (Huang et al., 2009a, Huang et al., 2009b) using KEGG and PANTHER database biological process (BP) and molecular function (MF) annotations. Enrichment was performed for protein turnover rate ratios GF versus CR with Significance A value < 0.1 and for protein abundance ratios GF versus CR with Significance A value < 0.05. Full data of enrichment analysis is presented in Table S2.
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD011457.
Acknowledgments
This work was supported by the European Research Council (ERC) (694181); National Institute of Allergy and Infectious Diseases (U01AI095473; the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH); The Knut and Alice Wallenberg Foundation (2017.0028); Swedish Research Council (2017-00958); The Swedish Cancer Foundation, IngaBritt, and Arne Lundberg Foundation, Swedish State under the agreement between the Swedish Government and the County Council; the ALF agreement (236501); Sahlgren’s University Hospital (ALF); and Wilhelm and Martina Lundgren’s Foundation.
Author Contributions
Conceptualization, L.A., A. Seiman, S.v.d.P., and G.C.H.; Methodology, L.A., A. Seiman, S.v.d.P., and G.C.H.; Investigation, L.A., A.M.R.P., A.E., and A. Schütte; Resources, F.B.; Formal Analysis, L.A., A. Seiman, and S.v.d.P.; Data Curation, A. Seiman; Writing – Original Draft, L.A., S.v.d.P., and G.C.H.; Writing – Review & Editing, L.A., A. Seiman, S.v.d.P., A.M.R.P., M.E.V.J., F.B., and G.C.H.
Declaration of Interests
The authors declare no competing interests.
Published: January 28, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.068.
Supplemental Information
References
- Abrams G.D., Bauer H., Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab. Invest. 1963;12:355–364. [PubMed] [Google Scholar]
- Alam M., Midtvedt T., Uribe A. Differential cell kinetics in the ileum and colon of germfree rats. Scand. J. Gastroenterol. 1994;29:445–451. doi: 10.3109/00365529409096836. [DOI] [PubMed] [Google Scholar]
- Anderson C.M.H., Ganapathy V., Thwaites D.T. Human solute carrier SLC6A14 is the beta-alanine carrier. J. Physiol. 2008;586:4061–4067. doi: 10.1113/jphysiol.2008.154500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banday A.R., Baumgartner M., Al Seesi S., Karunakaran D.K.P., Venkatesh A., Congdon S., Lemoine C., Kilcollins A.M., Mandoiu I., Punzo C., Kanadia R.N. Replication-dependent histone genes are actively transcribed in differentiating and aging retinal neurons. Cell Cycle. 2014;13:2526–2541. doi: 10.4161/15384101.2015.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- Barrett K., Brooks H., Boitano S., Barman S. McGraw-Hill Medical; 2010. Ganong’s Review of Medical Physiology, Twenty-Third Edition. [Google Scholar]
- Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Birchenough G.M.H., Nyström E.E.L., Johansson M.E.V., Hansson G.C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–1542. doi: 10.1126/science.aaf7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.-M., Ahmad Y., Gierliński M., Charrière F., Lamont D., Scott M., Barton G., Lamond A.I. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.011429. M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 2008;88:249–286. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N.A., Poidevin M., Pradervand S., Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Camp J.G., Frank C.L., Lickwar C.R., Guturu H., Rube T., Wenger A.M., Chen J., Bejerano G., Crawford G.E., Rawls J.F. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014;24:1504–1516. doi: 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claydon A.J., Beynon R. Proteome dynamics: revisiting turnover with a global perspective. Mol. Cell. Proteomics. 2012;11:1551–1565. doi: 10.1074/mcp.O112.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claydon A.J., Thom M.D., Hurst J.L., Beynon R.J. Protein turnover: measurement of proteome dynamics by whole animal metabolic labelling with stable isotope labelled amino acids. Proteomics. 2012;12:1194–1206. doi: 10.1002/pmic.201100556. [DOI] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Cohen L.D., Zuchman R., Sorokina O., Müller A., Dieterich D.C., Armstrong J.D., Ziv T., Ziv N.E. Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS One. 2013;8:e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Creamer B., Shorter R.G., Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut. 1961;2:110–118. doi: 10.1136/gut.2.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z.-L., Wu G., Zhu W.-Y. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front. Biosci. 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- Dai D.F., Karunadharma P.P., Chiao Y.A., Basisty N., Crispin D., Hsieh E.J., Chen T., Gu H., Djukovic D., Raftery D. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen E.M., Hansen G.H., Poulsen M.D. Apical secretion of apolipoproteins from enterocytes. J. Cell Biol. 1993;120:1347–1356. doi: 10.1083/jcb.120.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen E.M., Hansen G.H., Rasmussen K., Niels-Christiansen L.-L., Frenzel F. Apolipoprotein A-1 (apoA-1) deposition in, and release from, the enterocyte brush border: a possible role in transintestinal cholesterol efflux (TICE)? Biochim. Biophys. Acta. 2012;1818:530–536. doi: 10.1016/j.bbamem.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Davila A.-M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.-H., Sanz Y., Tomé D. Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013;68:95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Dawson P.A., Karpen S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015;56:1085–1099. doi: 10.1194/jlr.R054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G., Lange K., Havinga R., van Dijk T.H., Gerding A., van Eunen K., Müller M., Groen A.K., Hooiveld G.J., Bakker B.M., Reijngoud D.-J. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G900–G910. doi: 10.1152/ajpgi.00265.2013. [DOI] [PubMed] [Google Scholar]
- den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.-J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desouza M., Gunning P.W., Stehn J.R. The actin cytoskeleton as a sensor and mediator of apoptosis. Bioarchitecture. 2012;2:75–87. doi: 10.4161/bioa.20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G.P., Lee S.M., Mazmanian S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D.R., Garge N., Zhang X., Sun W., O’Connell T.M., Bunger M.K., Bultman S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.-G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Ermund A., Schütte A., Johansson M.E.V., Gustafsson J.K., Hansson G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C.W., Ayscough K.R. The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005;6:583–589. doi: 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- Gustafsson J.K., Ermund A., Johansson M.E.V., Schütte A., Hansson G.C., Sjövall H. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G430–G438. doi: 10.1152/ajpgi.00405.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond D.E., Claydon A.J., Simpson D.M., Edward D., Stockley P., Hurst J.L., Beynon R.J. Proteome dynamics: tissue variation in the kinetics of proteostasis in intact animals. Mol. Cell. Proteomics. 2016;15:1204–1219. doi: 10.1074/mcp.M115.053488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann M. How bacteria-induced apoptosis of intestinal epithelial cells contributes to mucosal inflammation. Int. J. Inflamm. 2010;2010:574568. doi: 10.4061/2010/574568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes K., Alcon-Giner C., Lawson M., McCoy K., Macpherson A., Hall L., Watson A. Absence of gut microbiota reduces lipopolysaccharide-induced epithelial cell shedding in the small intestine. Gut. 2014;63:A157. [Google Scholar]
- Johansen F.-E., Kaetzel C.S. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4:598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E.V., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E.V., Sjövall H., Hansson G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H.A., Baldwin R.L., France J., Calvert C.C. A model of whole-body protein turnover based on leucine kinetics in rodents. J. Nutr. 1999;129:728–739. doi: 10.1093/jn/129.3.728. [DOI] [PubMed] [Google Scholar]
- Jones R.M., Luo L., Ardita C.S., Richardson A.N., Kwon Y.M., Mercante J.W., Alam A., Gates C.L., Wu H., Swanson P.A. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Sasakawa C. Bacterial interactions with the host epithelium. Cell Host Microbe. 2010;8:20–35. doi: 10.1016/j.chom.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Larance M., Lamond A.I. Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 2015;16:269–280. doi: 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesher S., Walburg H.E., Jr., Sacher G.A., Jr. Generation cycle in the duodenal crypt cells of germ-free and conventional mice. Nature. 1964;202:884–886. doi: 10.1038/202884a0. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- Messier B., Leblond C.P. Cell proliferation and migration as revealed by radioautography after injection of thymidine-H3 into male rats and mice. Am. J. Anat. 1960;106:247–285. doi: 10.1002/aja.1001060305. [DOI] [PubMed] [Google Scholar]
- Metges C.C. Contribution of microbial amino acids to amino acid homeostasis of the host. J. Nutr. 2000;130:1857S–1864S. doi: 10.1093/jn/130.7.1857S. [DOI] [PubMed] [Google Scholar]
- Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays. 2013;35:1050–1055. doi: 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L.R., Knosp C., Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 2012;3:310. doi: 10.3389/fimmu.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T., Coates M.E., Hewitt D., Salter D.N., Garlick P.J. The influence of the gut microflora on protein synthesis in liver and jejunal mucosa in chicks. Br. J. Nutr. 1983;49:453–462. doi: 10.1079/bjn19830054. [DOI] [PubMed] [Google Scholar]
- Nakshabendi I.M., McKee R., Downie S., Russell R.I., Rennie M.J. Rates of small intestinal mucosal protein synthesis in human jejunum and ileum. Am. J. Physiol. 1999;277:E1028–E1031. doi: 10.1152/ajpendo.1999.277.6.E1028. [DOI] [PubMed] [Google Scholar]
- Neis E.P.J.G., Dejong C.H.C., Rensen S.S. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Park J.-H., Kotani T., Konno T., Setiawan J., Kitamura Y., Imada S., Usui Y., Hatano N., Shinohara M., Saito Y. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One. 2016;11:e0156334. doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B.C.E., Shanahan M.T., Singh A.P., Sethupathy P. Gut microbial influences on the mammalian intestinal stem cell niche. Stem Cells Int. 2017;2017:5604727. doi: 10.1155/2017/5604727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.C., Khambatta C.F., Li K.W., Bruss M.D., Shankaran M., Dalidd M., Floreani N.A., Roberts L.S., Turner S.M., Holmes W.E., Hellerstein M.K. The effect of long term calorie restriction on in vivo hepatic proteostatis: a novel combination of dynamic and quantitative proteomics. Mol. Cell. Proteomics. 2012;11:1801–1814. doi: 10.1074/mcp.M112.021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Reinhardt C., Bergentall M., Greiner T.U., Schaffner F., Ostergren-Lundén G., Petersen L.C., Ruf W., Bäckhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Piñeiro A.M., Bergström J.H., Ermund A., Gustafsson J.K., Johansson M.E.V., Hansson G.C., Schuette A. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G348–G356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhs A., Konzer A., Cemič F., Hemberger J., Braun T., Krueger M. Proteome wide determination of absolute turnover rates in mice. Pneumologie. 2012;66:A812. [Google Scholar]
- Salyers A.A., Vercellotti J.R., West S.E., Wilkins T.D. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D.C., Siegel J.E., Snellen J.E., Whitt D.D. Transit time of epithelial cells in the small intestines of germfree mice and ex-germfree mice associated with indigenous microorganisms. Appl. Environ. Microbiol. 1981;42:996–1001. doi: 10.1128/aem.42.6.996-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaie S., Ghaffari P., Kovatcheva-Datchary P., Mardinoglu A., Sen P., Pujos-Guillot E., de Wouters T., Juste C., Rizkalla S., Chilloux J., MICRO-Obes Consortium Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 2015;22:320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Sommer F., Bäckhed F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Sommer F., Nookaew I., Sommer N., Fogelstrand P., Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015;16:62. doi: 10.1186/s13059-015-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck T.S., Hooper L.V., Gordon J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R., III, Hetzer M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., Griss J., Alpi E., Birim M., Contell J. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A., Sayin S.I., Marschall H.-U., Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Westman-Brinkmalm A., Abramsson A., Pannee J., Gang C., Gustavsson M.K., von Otter M., Blennow K., Brinkmalm G., Heumann H., Zetterberg H. SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J. Proteomics. 2011;75:425–434. doi: 10.1016/j.jprot.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wolfe C., Pierle A., Welle K.A., Hryhorenko J.R., Ghaemmaghami S. Proteome-wide modulation of degradation dynamics in response to growth arrest. Proc. Natl. Acad. Sci. USA. 2017;114:E10329–E10338. doi: 10.1073/pnas.1710238114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the dataset identifier PXD011457.