Abstract
The retinoblastoma (RB) tumour suppressor gene is functionally inactivated in a broad range of paediatric and adult cancers, and a plethora of cellular functions and partners have been identified for the RB protein. Data from human tumours and studies from mouse models indicate that loss of RB function contributes to both cancer initiation and progression. However, we still do not know the identity of the cell types in which RB normally prevents cancer initiation in vivo, and the specific functions of RB that suppress distinct aspects of the tumorigenic process are poorly understood.
The initial functional characterization of the retinoblastoma protein (RB) following the seminal discovery of the RB gene as the first tumour suppressor focused on its role as a central regulator of cell cycle progression. RB tumour suppressor function was originally thought to be largely due to its capacity to arrest cells in G1 by inhibiting the activity of E2F transcription factors1,2. It is now believed that RB has many cellular roles in addition to serving as a G1 checkpoint, including control of cellular differentiation during embryogenesis and in adult tissues, regulation of apoptotic cell death, maintenance of permanent cell cycle arrest and preservation of chromosomal stability3,4. Recent studies have also demonstrated that control of the stability of the p27 cell cycle inhibitor (which is encoded by CDKN1B) by RB, through the interaction of RB with the anaphase-promoting complex/cyclosome (APC/C), is an important part of the capacity of RB to arrest cells in G1; therefore, E2Fs are not the sole mediators of the capacity of RB to control the G1–S transition5,6. In mammalian cells, RB belongs to a family of three proteins that also includes p107 and p130, which are structurally and functionally related to RB and belong to the same cellular pathway, but display distinct functions from RB in specific contexts4,7–10 (BOX 1). RB is now viewed as a transcriptional co-factor that can bind to and either antagonize or potentiate the function of numerous transcription factors11,12. Furthermore, RB is also an adaptor protein that recruits chromatin remodelling enzymes to control the expression of specific target genes and to modify chromatin structure at a chromosome-wide level13,14 (FIG. 1).
Box 1 |. The Rb gene family and the RB pathway.
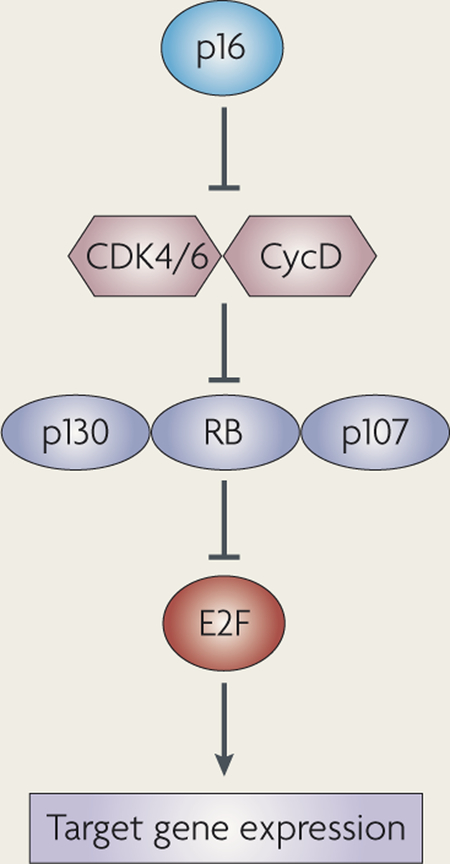
Three members of the ‘pocket’ protein family exist in mammalian cells — RB (retinoblastoma), p107 and p130. The three family members bind specific subsets of E2F transcription factors168, and their activity is thought to be largely controlled by phosphorylation. They all contain an LXCXE binding domain to which a number of common cellular partners can bind. In most cell types, overexpressing any Rb family member results in cell cycle arrest in G1.
RB is expressed in both cycling and non-cycling cells, and it seems to be regulated both transcriptionally and post-translationally. Although all three family members bind E2F transcription factors, RB has a unique domain in its C-terminal region that specifically binds E2F1. p107 expression is mostly controlled at the transcriptional level and is expressed in cycling cells. Interestingly, p107 expression often increases after loss of RB. p107 and p130 share a cyclin-binding domain and Cdk (cyclin-dependent kinase)-inhibitor activity. p130 is thought to be transcribed in all cells but p130 stability is increased in non-cycling cells169,170.
One major difference between RB, p107 and p130 is that RB is commonly mutated in human cancers while p107 and p130 are rarely directly inactivated. Instead, when RB is not mutated in human cancers, upstream regulators of all three pocket proteins are altered. These upstream regulators, such as p16 and cyclin D, are mutated in a largely mutually exclusive pattern with RB mutations, suggesting that these molecules function in a linear pathway to suppress human tumour formation.
Mouse models have demonstrated that E2F is a crucial downstream mediator of the tumour suppressor function of RB; concurrent mutation of E2F1 significantly inhibits the formation of the pituitary tumours and reduces the development of the thyroid tumours that both characterize Rb-mutant mice171. Surprisingly, activating mutations of the E2F transcription factors themselves are rare in human cancers, suggesting that the tumour suppressor function of RB extends beyond E2F repression172,173.
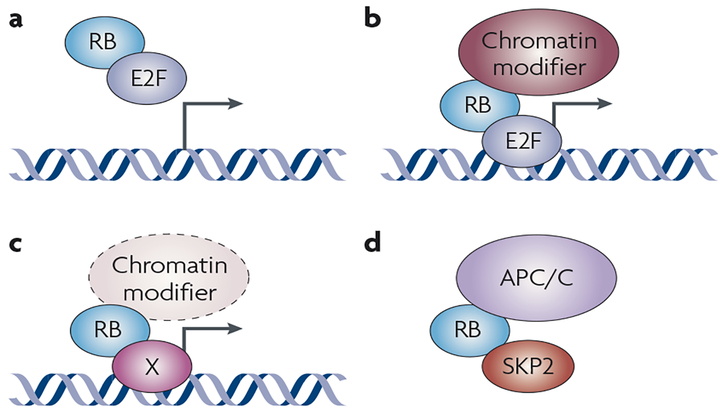
Figure 1 |. RB is a transcriptional co-factor and an adaptor protein that can function through at least four different types of protein interaction.
a | Classically, RB (retinoblastoma) binds to E2F transcription factors and recruits them away from their target genes. b | Alternatively, RB is recruited to the promoter of target genes by E2F and inhibits their transactivation activity and further recruits chromatin remodelling complexes (including HDAC (histone deactylase), DNMT1 (DNA methyltransferase 1), HP1A (heterochromatin protein 1A) and SUV39H1) to repress transcription. c | RB is a transcriptional co-factor for non-E2F transcription factors or other co-factors, such as the HIF1α (hypoxia-induced factor 1α), MYOD and SP1 transcription factors. d | RB serves as a non-chromatin-associated protein adaptor: illustrated is one example of RB acting to recruit APC/C (anaphase promoting complex/cyclosome) and SKP2 (S-phase kinase-associated protein 2) to the same complex, promoting SKP2 degradation.
Overall, although many cellular functions and binding partner proteins have been identified for RB, it remains unclear which of these functions are necessary or crucial for suppressing cancer, and several outstanding gaps remain in our understanding of the mode of action of RB in cells. The goal of this Review is to highlight the potential tumour suppressor role of the known cellular functions of RB, including and beyond its role at the G1–S transition. These functions might each be of particular importance at distinct steps of tumorigenesis — initiation, progression and invasion. Moreover, different functions of RB might be needed to suppress tumour formation in different cell types, such as stem cells and differentiated cells. Understanding unique cell type-specific and tumour stage-specific functions of RB might explain why it is such a potent tumour suppressor in humans, and will potentially provide the knowledge necessary to design novel targeted therapeutics against RB-deficient cancers.
Loss of RB function and cancer initiation
RB was originally identified through the pedigrees of families whose children developed retinoblastoma. Although tumour penetrance and progression can be under the control of several other factors and genetic events, it is well-accepted that loss of RB is the initiating event in these familial retinal tumours, as well as in sporadic cases15–17. Loss of RB also increases the risk of osteosarcoma development in children and teenagers18–20. In adults, human papillomavirus (HPV) is thought to initiate cervical carcinoma and squamous cell carcinoma of the head and neck in part by inactivating RB through expression of the E7 oncoprotein21,22, and similar mechanisms are possibly involved in virus-induced liver cancers23. RB is inactivated in more than 90% of human small-cell lung carcinomas (SCLC), and mouse genetic studies have confirmed that RB is crucial in preventing the initiation of this lung cancer sub-type24 (TABLE 1). Finally, upstream regulators of RB have been involved in cancer initiation in patients, both in familial and sporadic cases, and in several organs and tissues25–27. These observations in humans are corroborated by experiments with genetically modified mice: the high penetrance of pituitary and thyroid tumours in Rb+/− mice and the development of other tumours in mice that have been subjected to tissue-specific Rb deletion using the Cre–lox system demonstrate that loss of Rb is a causal event in many cancer types7.
Table 1 |.
Alterations of the RB gene in common human cancer types
| Tumour type | Frequency of RB inactivation (genetic or epigenetic) | Presumed consequence of RB inactivation | Refs |
|---|---|---|---|
| Lung cancer | Germline RB mutations predispose to small cell lung carcinoma (SCLC), and RB is inactivated in >90% of sporadic SCLC cases. In contrast, RB is mutated in only 15–30% of non-SCLC cases. | SCLC initiation; progression to invasive forms of non-SCLC | 182–184 |
| Melanoma | RB inactivation is rare in sporadic cases, but inherited mutation predisposes to melanoma | Initiating event in familial cases | 185,186 |
| Prostate cancer | ~20% | Progression to invasive carcinoma‡ | 187–189 |
| Breast cancer | ~20% | Progression | 190,191 |
| Bladder cancer | 20–50% | Progression to invasive tumours | 184,192–194 |
| Leukaemia | Reduced levels of expression are frequent, but mutations in RB are rare in leukaemias, except in 20% of chronic myeloid leukaemia (CML) cases | Progression (CML blast crisis) | 195–197 |
| Brain cancer | Rb-mutant mice develop pituitary tumours, but RB mutations are rare in human cases. 15–30% of advanced gliomas have RB mutations | Progression | 198–201 |
| Oesophageal cancer | RB deletion are found in 15–50% of adenocarcinomas or squamous cell carcinomas | Early progression | 202,203 |
| Liver cancer | Mutations in RB are found in 15–30% of the advanced hepatocellular carcinomas§ | Progression | 137,204–206 |
Inactivation of Rb in the lung epithelial of mice is sufficient to initiate neuroendocrine hyperplasia216 but the additional loss of Trp53 is required for SCLC development24.
Overexpression of Gankyrin in liver tumours might functionally inactivate RB, potentially bypassing the need to mutate RB in many cases219, similar to the expression of E7 by HPV in cervical cancer. Note that cells from common cancers such as colorectal carcinoma207, pancreatic carcinoma208, renal cell carcinoma209, endometrial carcinoma, ovarian carcinoma210, non-Hodgkin lymphoma211,212, myeloma213, thyroid carcinoma214, HPV-negative head and neck carcinoma22 and gastric carcinoma215 only rarely carry mutations in RB. However, in many cases, RB is hyperphosphorylated or shows decreased levels in these tumours, and mutations in other members of the RB pathway are usually present.
Although strong genetic evidence indicates that the RB pathway prevents cancer initiation in multiple cell lineages in humans and mice, it is still not clear how and in what cell types cancer initiation occurs on loss of RB function. Generally, adult tissues are composed of stem cells, progenitor cells and differentiated cells (FIG. 2). Stem cells are often quiescent but have a strong regenerative potential and the ability to self-renew. When needed, these stem cells give rise to transit amplifying progenitors, which are actively cycling but lack a significant self-renewing capacity. These progenitors eventually cease cycling and undergo a maturation process that results in differentiated, non-cycling cells.
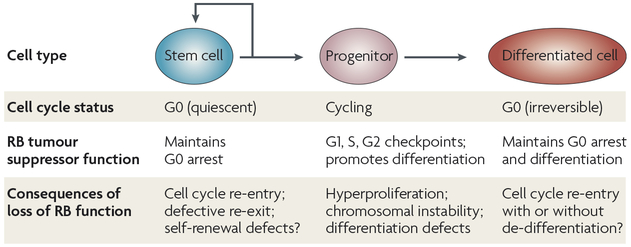
Figure 2 |. Functions of RB that are potentially involved in the prevention of cancer initiation in adult cells.
Adult tissues consist of stem cells that are able to produce cycling progenitor cells; these progenitors produce cells that eventually undergo terminal differentiation. In each of these cell types, RB (retinoblastoma) controls distinct cellular processes, which might be partly E2F-dependent and partly E2F-independent, and might involve extensive chromatin remodelling. Loss of RB will have different effects in each cell type. Although the control of the G1–S checkpoint is the most studied function of RB, the abrogation of this function might only be crucial under specific conditions, such as during tumour initiation from cycling progenitor cells.
Loss of RB function and cell cycle re-entry in quiescent stem cells.
Stem cells share many characteristics with tumour cells, including their strong regenerative potential. However, although stem cells have the ability to proliferate and renew, they are more often held in a quiescent state and, unlike cancer cells, only cycle rarely. Maintenance of quiescence in stem cells is necessary to maintain tissue homeostasis, and the observed role of RB in maintaining quiescence in cell culture models suggests that in vivo control of quiescence in stem cells might be a crucial tumour suppressing function of Rb.
The role of RB in the maintenance of quiescence in stem cells in vivo is largely unknown. Whereas p130, rather than RB, binds to the promoters of genes in G0 cells28–30, acute loss of Rb results in cell cycle re-entry from quiescence in mouse embryonic fibroblasts in culture31. In vivo, data from several groups indicate that loss of Rb in haematopoietic stem cells (HSCs) does not increase their proliferation, although Rb-mutant HSCs that are induced to enter the cell cycle under conditions that induce cellular stress might not be able to properly re-enter a quiescent state and can undergo enforced differentiation32–34. Deletion of Rb in the skin results in a decrease in the number of label-retaining, slowly cycling cells in the stem cell compartment35. These data raise two different possibilities: first, RB-mutant skin stem cells might die or differentiate more than controls. Second, these mutant epidermal stem cells might abnormally enter the cell cycle, diluting the BrdU (bromodeoxyuridine) signal; this potential hyperproliferative phenotype might or might not be accompanied by a loss of self-renewal. In addition, the cell autonomy of this phenotype with a decreased number of label-retaining cells remains unknown. Surprisingly, the clearest evidence that RB functions to maintain quiescence in stem cells comes from observations in plants. In Arabidopsis roots, loss of the RB homologue RBR leads to expansion of stem cells in the stem cell pool, seemingly without affecting their self-renewal potential and their proliferative status but by preventing their differentiation36. It is interesting to note that if loss of RB function leads to exit from quiescence and an increased number of stem cells without loss of self-renewal capacity, controlled and transient inactivation of RB function in adult stem cells could become a tool to exploit the regenerative potential of these cells in patients.
To date, not enough evidence is available in mammalian cells to conclusively show that loss of RB is sufficient to force stem cells to exit quiescence permanently and whether this cell cycle re-entry phenotype is related to cancer initiation. However, RB is a strong candidate to regulate quiescence in stem cells, and even a slight increase in the proliferation of largely quiescent stem cell pools on loss of RB might be sufficient to increase the probability of cancer development as an organism ages — a model that will need to be thoroughly tested.
Cell cycle re-entry in post-mitotic differentiated cells mutant for RB function.
The self-renewal properties and proliferation potential of stem cells and some progenitor cells resemble those of cancer cells. By contrast, fully mature, differentiated cells are thought to have little or no proliferative potential (FIG. 2). Therefore, the probability that a fully differentiated cell re-enters the cell cycle and becomes cancerous might seem low compared with the malignant potential of stem cells. However, post-mitotic differentiated cells largely outnumber stem cells in adult organs and tissues, and evidence indicates that loss of RB might initiate cancer by allowing fully differentiated cells to re-enter the cell cycle. Given the analogy between ‘permanently’ arrested differentiated cells and senescent cells37,38, the reversal of cell cycle arrest upon acute loss of RB in senescent cells31,39 offers initial evidence for this model. RB has also been shown to participate in and potentially modulate the structure and location of heterochromatin formation in senescent cells40, and loss of RB function might result in chromatin remodelling in differentiated cells, allowing the expression of cell cycle genes and resulting in the de-differentiation of the mutant cells41,42. Further support for this model comes from the observation that loss of Rb results in cell cycle re-entry in mature hepatocytes43. In neurons, however, loss of RB, or even inactivation of the entire Rb gene family, does not allow these cells to re-enter the cell cycle44. In muscle cells, loss of Rb function can result in no cell cycle re-entry45,46 or in cell cycle re-entry followed by a block in G2 (REF. 47), depending on the culture conditions. In the cochlea, Rb deletion allows increased proliferation of precursor cells, and although mature mechanosensory hair cells are capable of re-entering the cell cycle, they fail to complete cellular division48.
Therefore, cell cycle re-entry in terminally differentiated cells owing to loss of RB function might be context-dependent and most of the time insufficient to initiate cancer. Nevertheless, cells that lose RB function before cell cycle exit and retain the ability to differentiate might also exhibit an increased capacity to re-enter the cell cycle and initiate cancer in the presence of certain stimuli (see below). This idea was suggested by several previous studies49–51 and was recently illustrated in the retina of mice genetically engineered to develop retinoblastoma52. Because it is difficult to imagine how both alleles of the RB gene can become mutated in non-cycling cells in vivo, this model in which cells lose RB function before they differentiate and are induced to re-enter the cell cycle after they differentiate might be more representative of what is happening during cancer initiation in differentiated cells.
Loss of RB function in cycling progenitor cells and differentiating cells.
Although cancer initiation from quiescent stem cells and post-mitotic differentiated cells upon loss of RB can take place, loss of RB function is more likely to initiate cancer from cycling progenitor cells. In these cells, loss of RB might accelerate proliferation and prevent normal cell cycle exit in G1, which is associated with differentiation (FIG. 2). In support of this model, in HPV-induced cervical cancers, the virus initiates cancer in dividing cells in the basal layer of the epithelium, preventing their normal cell cycle exit and maturation21. In the brain, RB levels increase as progenitor cells normally undergo cell cycle exit and loss of Rb results in delayed cell cycle exit in these cells44,49,53,54. Similarly, Rb inactivation in mouse retinal progenitors results in cell-autonomous defects in cell cycle exit, specific differentiation defects, proliferation in differentiating cells and increased cell death. Although loss of Rb alone is not sufficient for brain cancer or retinoblastoma development in mice, similar phenotypes are observed in Rb;p107 double-deficient animals that develop retinoblastoma, indicating that failure to exit the cell cycle properly as they differentiate is a mechanism by which mutant Rb cells can begin to become tumour cells50,51,55–57.
In this model, loss of RB might also decrease the ability of cells to differentiate. The role of RB in promoting cellular differentiation during embryonic development and in adult lineages has been well characterized in mutant mice and in vitro systems20,42,58–61. Mechanistically, RB promotes the differentiation of multiple lineages by binding and regulating tissue-specific transcription factors11,42 and inhibitors of differentiation such as ID2 and EID1 (REFS 62–65), as well as general transcription factors and chromatin modifiers66,67. Whereas E2F transcription factors were previously thought to mediate only the action of RB on cell cycle progression, recent evidence suggests that the activity of these factors also controls cellular differentiation in RB-mutant cells68,69. A seminal finding that this function of RB might be crucial in tumour development came from the observation that some mutant forms of RB from families with low penetrance retinoblastoma were deficient in E2F binding but retained the ability to induce the differentiation of tumour cells. This suggests that retention of the ability to induce differentiation might be sufficient to prevent fully penetrant retinoblastoma, and similar observations have been made for osteosarcoma70–72. The existence of these mutant forms of RB that lack the cell cycle inhibitory function but retain pro-differentiation activity suggests that different regions or modifications of the RB protein mediate its control over cell cycle progression and differentiation (BOX 2).
Box 2 |. RB structure and post-translational modifications.
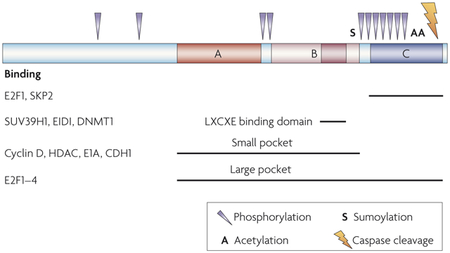
Human RB (retinoblastoma) consists of 928 amino acids and does not contain any commonly recognized DNA-binding or protein-interacting domains. Deletion and mutagenesis analysis as well as structural studies174–176 have uncovered several regions that mediate RB binding to individual protein binding partners. Most protein binding partners seem to bind to the pocket region, but some proteins bind unique residues (see figure), making it possible to separate some functions by structure. Furthermore, specific post-translational modifications, which are further described below, might regulate the role of RB in individual cellular functions78. The RB C terminus binds specifically to E2F1 and inhibits apoptosis177. E1A binding disrupts E2F-dependent proliferation but not apoptosis92. Low penetrance retinoblastoma mutations deregulate proliferation but not differentiation71. Mutation of the LXCXE binding domain disrupts the ability of RB to mediate H4K20 (histone 4 lysine 20) trimethylation but not proliferation109.
Phosphorylation
RB can be phosphorylated by several kinases, including cyclin-D–CDK4 (cyclin-dependent kinase 4), cyclin-D–CDK6, cyclin-A–CDK2, cyclin-E–CDK2, CHK2 (checkpoint homologue 2) and RAF1. Phosphorylation of RB enables cell cycle progression, which is thought to occur through hyperphosphorylation-induced release of the E2F transcription factors from the large pocket. Although the binding of many of the partners of RB seems to be disrupted by phosphorylation of RB, it is possible that individual phosphorylation sites might selectively inhibit RB functions; for example, potentially inactivating cell cycle inhibition while allowing apoptosis protection.
Acetylation
Acetylation of RB has been demonstrated to occur at lysines 873 and 874, and this acetylation might prevent RB inactivation by phosphorylation178. Acetylated RB might therefore make up a pool of RB that remains active despite the presence of Cdks. Furthermore, double-stranded DNA breaks induce acetylation of RB and acetylation77 might have a role in differentiation179.
Sumoylation
RB can be sumoylated at lysine 720, in the small pocket near the LXCXE binding domain, but the function of this sumoylation is unknown180.
Caspase-cleavage
RB can be cleaved by caspase 8 at the C terminus. Mice lacking the caspase recognition sequence in RB are resistant to apoptosis induced by TNFα (tumour necrosis factor-α) and are more cancer-prone181.
CDH1, cadherin 1; DNMT1, DNA methylatransferase 1; HDAC, histone deacetylase.
RB retention and early cancer progression
Despite the clear evidence that RB normally prevents the initiation of retinoblastoma, osteosarcoma and SCLC, it is striking to note that the vast majority of tumour types only display RB alterations later in tumour progression (TABLE 1) and that patients with familial retinoblastoma are not strongly predisposed to a wide variety of other tumours73. These observations suggest that there might be a disadvantage to losing RB too early during tumour development in some contexts. Indeed, RB has pro-survival functions that might seem paradoxical to its role as a tumour suppressor.
Loss of RB function increases cell death and DNA repair.
E2F1 activity is an important mediator of p53-dependent apoptosis in response to loss of RB74,75, and recent evidence suggests that this cell death might reflect a normal function of E2F1 during the DNA damage response. Double-strand breaks activate the kinases ATM (ataxia telangiectasia mutated) and CHK2 (checkpoint homologue 2), which phosphorylates RB at serine 612, resulting in the formation of a RB–E2F1 complex that can inhibit E2F1-specific target genes that are necessary for apoptosis, such as Apaf1 and Trp73 (REF. 76). Similarly, DNA damage increases the acetylation of human RB at lysines 873 and 874, which decreases RB phosphorylation by cyclin–Cdk (cyclin-dependent kinase) complexes and therefore increases its repression over E2F1 (REF. 77). Interestingly, among the E2F family, induction of apoptosis is a function largely mediated by E2F1 and its repression is mediated by specific RB and E2F1 protein domains that are distinct from those through which RB interacts with E2F1, E2F2, E2F3 and E2F4 to control cell cycle progression78. Nevertheless, E2F3 can also induce cell death in Rb-deficient cells in vivo79,80. Through these mechanisms, the presence of RB in tumour cells might prevent E2F-mediated apoptosis in response to DNA damage, and might prevent the elimination of tumour cells.
RB seems to have a separate function in the ATR (ATM and RAD3-related) pathway, which responds to single-stranded DNA resulting from stalled replication forks and induces DNA repair. In Rb-deficient cells, UV-induced lesions fail to induce cell cycle arrest, unlike in Rb wild-type cells; however, DNA repair mechanisms are rapidly engaged81. In primary Rb-null mouse hepatocytes, cyclob-utane pyrimidine dimers are removed more quickly than in wild-type cells following UV irradiation82. In these hepatocytes, E2F activity has been shown to activate Ddb2 (damage-specific DNA binding 2), a gene that is crucial for DNA repair, and loss of RB might therefore increase the DNA repair activity of mutant cells82. However, UV treatment of RB-deficient cancer cells results in apoptosis, similar to that seen with other types of DNA damage83, and an increase in double-strand breaks is observed in Rb–/– mouse adult fibroblasts treated with DNA-damaging chemotherapeutic drugs84. These diverse results suggest that the role of RB in the response to DNA damage might depend significantly on cellular context.
In both of these cases, presence of RB seems to be beneficial to tumour progression, which is counterintuitive given the role of RB as a potent tumour suppressor; these observations might be highly relevant to cancer initiation, following the model that DNA damage signals are induced by oncogenic activation in early human tumours85. The idea that RB could promote cancer under certain conditions by increasing survival is supported by experiments in transgenic mice expressing a phosphorylation-resistant, constitutively active allele of Rb in the mammary gland epithelium. Strikingly, these transgenic mice develop focal hyperplastic lesions and mammary tumours, probably because the survival of differentiated mammary epithelial cells is extended after pregnancy86.
A related point is that abnormal proliferation that is induced by loss of RB is often accompanied by increased cell death, through p53-dependent and independent mechanisms49,57,87–91. Interestingly, it is possible that functional inactivation of RB by phosphorylation or by viral oncoproteins does not induce the same amount of cell death as deletion of RB, potentially explaining why mutations in upstream members of the pathway are selected in some cancers92,93. For example, loss of INK4A (which is encoded by CDKN2A and is also known as p16) or expression of E1A is insufficient to disrupt RB inhibition of E2F1-dependent apoptosis92,94. Both decreased INK4A and presence of E1A result in inhibition of protein interactions that are mediated through the RB pocket region, suggesting the structure and/or specific modifications of RB might distinctly regulate individual functions (BOX 2). However, if cancer cells carry a mutation that protects them from apoptosis, such as inactivation of p53, before losing RB, then the tumour will benefit from the loss of the other tumour suppressive functions of RB (described below) and cancer will progress.
Loss of RB function increases autophagy in response to hypoxia.
During tumour progression, solid tumours eventually achieve a size at which the hypoxic environment limits further growth. Autophagy, which is induced in hypoxic conditions, can initially allow survival of a hypoxic cell by enabling metabolism to continue; however, extended autophagy can eventually result in cell death95. The role of autophagy in cancer is currently unclear: one hypothesis is that the absence of both apoptosis and autophagy pathways simultaneously might be beneficial to tumour development by promoting the survival of abnormal cells that accumulate mutations, enabling faster progression to a more malignant phenotype96. Recent work has demonstrated that the presence of functional RB might prevent autophagy in response to hypoxia through its inhibition of E2F target genes97,98. So, similar to apoptosis, loss of RB might increase autophagy-mediated cell death, and presence of RB might be beneficial to early tumours under some conditions, although more studies are required to better understand the role of RB in autophagy.
Loss of RB function and cancer progression
Expression and loss of heterozygosity studies indicate that loss of RB function is associated with the progression of human cancers, in addition to its role in cancer initiation (TABLE 1). However, one has to keep in mind that loss of the chromosomal region in which RB resides in unstable human tumour cells could be coincidental and not causal. To our knowledge, there is no mouse model yet to specifically test the consequences of loss of Rb function during tumour progression, as most current mouse models are based on the simultaneous rather than sequential mutation of cancer genes. Nevertheless, loss of several of the cellular functions attributed to RB might directly participate in tumour progression.
Loss of RB might decrease the differentiation potential of mutant cells.
The early stages of human cancer, such as neoplasia, clearly display proliferation phenotypes while retaining at least some markers of differentiated cells. The degree of differentiation of human cancers is often used to designate the pathological grade of an individual tumour, with the most differentiated being the lowest grade and the least differentiated being the highest grade. Altered RB expression has been observed to be correlated with higher grade, poorly differentiated gastrointestinal cancers99. Similar to its role in progenitor cells undergoing differentiation (see above), the presence of RB in tumour cells could in theory promote their differentiation and thereby restrict their proliferative potential, which might explain why RB must be lost during the progression from a well differentiated to a poorly differentiated tumour. Because some anticancer strategies force some tumour cells to undergo differentiation100, this aspect of RB function might have important therapeutic implications.
Loss of RB results in chromosomal instability.
Human cancers demonstrate a high degree of genomic instability; mutations in RB might participate in this phenotype by inducing defects during DNA replication and abnormal chromosomal segregation in mitosis101–103, which could result in abnormal expression of other cancer genes, enabling tumour progression. Wild-type RB activity might normally slow down the proliferation of tumour cells by acting as a checkpoint not only at the G1–S transition but also during S phase and at the G2–M transition, and loss of this checkpoint could contribute to the ability of a tumour cell to proliferate in the presence of genomic abnormalities. In mouse embryonic stem cells, in which there is no active G1 checkpoint, Rb deficiency leads to increased chromosomal alterations104,105, and in adult mouse fibroblasts loss of Rb deregulates S phase, resulting in polyploidy106. In normal adult hepatocytes, which are often tetraploid, acute deletion of Rb results in cell cycle re-entry and increased aneuploidy43. It has been suggested that this type of chromosomal instability is linked to the progression of benign eye lesions to malignant retinoblastoma17. Although the mechanisms underlying these aneuploid phenotypes are still only partly understood, RB–E2F complexes are thought normally to restrict the expression of MAD2 (REF. 107), a spindle checkpoint component. MAD2 levels are increased in Rb-deficient cells, correlating with increased levels of activating E2F, and overexpression of MAD2 is sufficient to induce aneuploidy and tumour formation in transgenic mice108. In addition, RB might help maintain genomic stability by regulating the expression of, and associating with, chromatin remodelling complexes, thereby regulating chromatin structure, in particular at centromeres and telomeres109–112. Many chromatin modifiers bind to RB through the LXCXE binding domain (BOX 2), and recent evidence has demonstrated that an LXCXE mutant that retains E2F-binding capability, and therefore has no effect on MAD2 levels, still contributes to chromosome missegregation and genomic instability through its failure to regulate appropriate pericentric heterochromatin formation109. Furthermore, loss of Rb can lead to chromosome segregation defects through the misregulation of genes that are important for processes such as centrosome duplication113,114 and DNA replication106. Recent experiments in Drosophila melanogaster and human cells also indicate that RB might normally promote chromosomal condensation and preserve chromosomal stability through a direct interaction with a condensin protein115. Finally, as mentioned above, Rb-deficient cells fail to properly arrest in G1 upon DNA damage and might replicate mutated DNA, leading to the accumulation of mutations102,105,111,116.
Beyond inducing gross chromosomal changes, loss of RB function might also alter the epigenetic definition of the genome through misregulation of chromatin remodelling enzymes117 and DNA modifiers. DNA methyltransferase DNMT1, which is both an E2F target112 and a RB–E2F protein binding partner118, has been shown to be upregulated in human cancers and in response to loss of RB. Both chromatin remodelling and DNA methylation have been linked to tumorigenesis119,120. These epigenetic changes might result in abnormal inactivation of tumour suppressors and activation of oncogenes without the physical mutation of these genes.
Although still poorly understood, the role of RB in maintaining chromosomal stability might be crucial to the prevention of cancer progression.
Loss of RB prevents induction of cellular senescence.
Cellular senescence acts to suppress tumours in vivo in response to oncogenic stress, and strong evidence places RB–E2F activity and associated chromatin-regulating complexes, such as SUV39H1, as a key regulators of senescence in cells in culture121–124. This is mediated, at least in part, through the formation of senescence-specific heterochromatin at cell cycle gene loci40. Furthermore, the RB family controls the length of telomeres and mediates the cellular signals to induce senescence in cells with shortened telomeres125–127. Therefore, loss of RB function allows a tumour cell to bypass the cell cycle arrest that is associated with both replication and oncogene-induced senescence during cancer progression. One recent example supports this model: loss of function of the Von Hippel Lindau (VHL) tumour suppressor creates an oncogenic stress that triggers a senescence response both in vitro and in vivo. This senescent cell cycle arrest depends on the presence of functional RB128 and is in part mediated by the RB–SKP2 (S-phase kinase-associated protein 2)–p27 pathway. Through this pathway, the presence of RB leads to downregulation of SKP2 and therefore stabilization of p27, which inhibits cyclin-E-associated kinase activity129.
Loss of RB promotes angiogenesis.
In order to over-come the growth-limiting effects of hypoxia, tumours select for mutations that increase their ability to recruit endothelial cells to form novel blood vessels in a process termed angiogenesis. Emerging evidence shows that vascular endothelial growth factor (VEGF) and other angiogenic factors that are secreted by tumour cells to recruit endothelial cells are transcriptional targets of the RB–E2F pathway130. For example, pituitary tumours arising in Rb-mutant mice have high levels of VEGF131. Another mechanism by which loss of RB might promote angiogenesis in these pituitary tumours is through the loss of inhibition of ID2 (REF. 132). ID2 is one member of a family of regulators that prevent activity of basic helix-loop-helix (bHLH) transcription factors. Expression of several of the Id proteins has been associated with tumour progression and several genetic studies suggest that the presence of Id factors is necessary for tumour vascularization through regulation of pro-angiogenic factors133,134. Additionally, RB might relay receptor-mediated mitogenic signals to develop an angiogenic response135. Moreover, deletion of Rb in a mouse model of Trp53-deficient squamous cell carcinoma demonstrates increased vascularization when compared to similar Trp53–/– tumours136. So, although loss of RB might not be sufficient to trigger a full angiogenic response, it might participate in this crucial stage of tumour progression in vivo.
Loss of RB function is associated with increased metastatic potential.
Altered RB expression patterns have been observed in poorly differentiated, metastatic hepatocellular carcinomas137, and correlative analyses have suggested that changes in RB expression are significantly associated with invasion and metastasis of oesophageal cancers99. A recent report indicates that loss of RB function is associated with increased levels of cyclooxygenase 2 (COX2) and an increased chance of recurring invasive basal-like breast cancer138. COX2 overexpression has been observed in various epithelial cancers and has been demonstrated to increase motility of breast cancer cells in culture139, as well as invasiveness of colon cancer cells140. The correlation between loss of RB and an increase in COX2 expression might reflect a potential mechanism through which RB can prevent metastasis in a tissue-specific manner. Loss of RB has been shown to produce inappropriate migration of neurons in the developing mouse cortex141, a phenotype that is dependent on E2F3 activity69. A similar increase in E2F3 activity in metastatic tumour cells could be indirectly related to the capacity of these cells to be more motile. However, E2F3 inactivation has also been found to enhance the metastatic potential of RB-deficient thyroid carcinomas in vivo142. More experiments are required to clarify the specific role of E2F3 activity in the metastatic process, but a general role for E2F factors in metastasis is underscored by cell culture experiments, which have identified several E2F target genes with a potential role in invasion and metastasis143,144. Additionally, E2F1 over-expression has been shown to increase the invasiveness of human tumour cell lines145. Overall, however, our understanding of the molecular mechanisms mediating the potential role of RB in preventing tumour metastasis remains extremely limited.
Conclusions and discussion
In conclusion, a great deal of time and effort has gone into understanding the numerous cellular functions of RB that are mediated by over a hundred known protein binding partners and numerous transcriptional targets (FIG. 3). Some of these functions seem difficult to reconcile with others: RB promotes terminal differentiation and cell cycle arrest in progenitor cells undergoing maturation, but it is also important for maintaining reversible cell cycle arrest in quiescent stem cells that must not permanently withdraw from the cell cycle and differentiate. In addition, many consequences of RB inactivation are easily linked to an increase in the tumorigenic potential of the mutant cells, such as loss of a G1 checkpoint or increased genomic instability. But, loss of RB might also increase apoptotic and autophagic cell death and the ability to repair DNA lesions81,84, which might not be beneficial to tumour cells. Although cases have been made for the importance of several of these functions in specific cell types and cancer models, there is still much to be learned about the relative importance of each of these functions in the role of RB as a tumour suppressor in vivo. The cellular response to the presence or absence of RB might depend on complex regulatory networks that are poorly understood146.
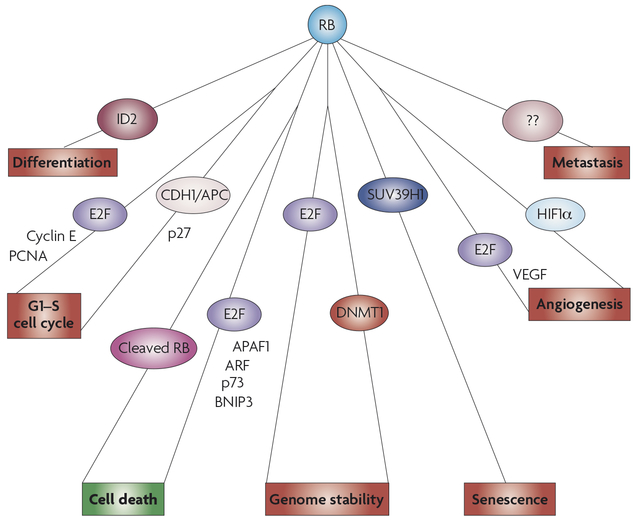
Figure 3 |. Overview of the numerous RB binding partners and transcriptional targets that might mediate its tumour suppressor ability.
Presence of RB (retinoblastoma) might prevent tumour formation by inducing differentiation, controlling cell-cycle arrest, maintaining genomic stability and inducing senescence in response to oncogenic stresses. Furthermore, the absence of RB has been associated with increased angiogenesis and metastasis, although the mediators of these functions are less well understood. Surprisingly, presence of RB has a pro-survival function because of its inhibition of cell death through apoptosis and, potentially, autophagy. This function, shown in green, might be necessary for early tumour cell survival in some contexts. This figure depicts a simplified representation of the potential role of RB in tumour suppression. Each function is illustrated with some of the key protein binding partners and transcriptional targets that might be necessary for the function, but they are not meant to be comprehensive. APAF1, apoptotic peptidase activating factor 1; BNIP3, BCL2-interacting protein 3; CDH1, cadherin 1; DNMT1, DNA methylatransferase 1; HIF1α, hypoxia-induced factor 1α; PCNA, proliferating cell nuclear antigen; VEGF, vascular endothelial growth factor.
It is possible that novel functions for RB remain to be discovered, but it is clear that the crucial importance of the known functions in tumorigenesis need to be further investigated. One method will be to improve mouse models of human cancers that are associated with loss of RB7. Because loss of Rb in mouse models is not sufficient to initiate retinoblastoma, osteosarcoma or SCLC, we do not know if the phenotypes of Rb-deficient cells in these organs truly reflect the mechanisms of cancer initiation in the corresponding human cells. Furthermore, it is striking that the mechanisms of cancer initiation in the pituitary, thyroid and adrenal glands, in which loss of Rb is sufficient for tumorigenesis, are still largely unknown. When loss of Trp53 and Rb are combined, more tumours develop in mutant mice24,147–149 but it becomes difficult to dissect the respective roles of these two potent tumour suppressors.
Targeted alterations in upstream components of the RB pathway can result in the inactivation of all of the Rb family genes, but because the RB pathway is not strictly linear, these experiments might not directly inform us about the mechanisms of RB tumour suppressive action4,7,150–153. Alternatively, combined deletions in Rb family genes might eliminate the functional overlap within this gene family, but, so far, few models of human cancers have been analysed using this approach. The Van Dyke group has published a series of elegant studies in mice expressing a truncated form of the SV40 large T oncoprotein, T121, which is thought to specifically inactivate the three Rb family members87,154–160. These transgenic mice provide powerful models to investigate how loss of Rb family function initiates cancer in multiple cell types, including the brain, the mammary epithelium and the prostate epithelium. These experiments illustrate the point that different cell types respond differently to loss of Rb family members, which could dictate the requirement for specific mutations during tumour progression161. In addition, the analysis of mice expressing T121 in the prostate indicates that loss of Rb family function in epithelial cells triggers signals to surrounding stromal cells, underscoring that not all the mechanisms of cancer initiation are cell autonomous159. In conclusion, however, although RB inactivation is clearly an initiating event in many cancers, much work remains to be done and new systems need to be developed to better understand how loss of RB function can promote cancer initiation in vivo.
The generation of novel conditional alleles of Rb and other cancer genes, using a combination of Cre–lox, Flp–frt and tetracycline-inducible systems162, should enable the production of a new generation of mouse models that can test the consequences of Rb loss during tumour progression. These models will also be useful in understanding the continued dependency of a tumour on RB loss, a poorly studied question163,164. Similarly, because ectopic expression of RB in cells in culture can produce nonspecific effects, a detailed structure–function analysis of RB in vivo will be key to our understanding of RB tumour suppressor functions78. In particular, comparing the structure of RB with those of p107 and p130, which are rarely mutated in human cancers and yet are similar to RB in many ways, might help to identify mutant forms of RB that can distinguish specific cellular functions of RB.
Because RB and its upstream regulators belong to a pathway that is often referred to as the linear RB pathway, it is tempting to equate RB mutation with p16 inactivation, activating mutations in Cdk4 or overexpression of cyclin D1. However, alterations in upstream members of the RB pathway also impinge upon the activities of p107 and p130, as well as other cellular proteins165,166. In addition, even high levels of cyclin–Cdk complexes might be insufficient to fully phosphorylate — and functionally inactivate — RB. A clear example that RB and p16 mutations are not equivalent comes from the observations that RB is mutated in most, if not all cases of SCLC, whereas p16 mutations are never found in this lung cancer type but are prevalent in non-SCLC, a different type of lung cancer167.
A greater understanding of the interplay between Rb family members, regulators of the RB pathway and other binding partners is crucial for the design of therapeutics targeting RB-deficient human tumours. Identification of the tumour suppressing functions of RB that are most important within a particular tissue or cell type is necessary to identify compounds that can induce arrest or death of the tumour cells.
DATABASES
Entrez Gene:
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene Apaf1 | ATM | ATR | CDKN1B | CDKN2A | CHK2 | COX2 | Ddb2 | DNMT1 | E2F1 | E2F2 | E2F3 | E2F4 | EID1 | ID2 | MAD2 | p53 | RB | SKP2 | SUV39H1 | Trp73 | VHL
National Cancer Institute: http://www.cancer.gov brain cancer | cervical cancer | head and neck cancer | liver cancer | oesophageal cancer | osteosarcoma | retinoblastoma | small-cell lung carcinoma
FURTHER INFORMATION
J. Sage’s homepage: http://www.stanford.edu/group/sage
Cancer Genetics: http://www.cancerindex.org/geneweb/RB1.htm
Eye Cancer Network: http://www.eyecancer.com
Gene Cards: http://www.genecards.org/cgi-bin/carddisp.pl?gene=Rb1
Gene Clinics: http://www.geneclinics.org/profiles/retinoblastoma/details.html
NCBI Genes and Disease:
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View.ShowSection&rid=gnd.section.129
OMIM: http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=180200
Retinoblastoma.com:
http://retinoblastoma.com/retinoblastoma
Retinoblastoma genetics: http://www.verandi.de/joomla
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
At a glance.
RB, the retinoblastoma protein, has been identified as a crucial tumour suppressor. It is believed to be directly or indirectly inactivated in nearly all human cancers.
RB has been demonstrated to bind to over one hundred protein partners and has been shown to mediate transcriptional regulation of hundreds of target genes. These protein partners and transcriptional targets are thought to mediate the numerous cellular functions of RB, including temporary and permanent cell cycle arrest, genomic stability, apoptosis and differentiation.
The cellular functions of RB, as well as a potential role in angiogenesis and metastasis, might contribute to its role as a tumour suppressor, but it is currently unknown which function is most critical. Distinct cellular functions of RB might contribute to its role in preventing tumour initiation versus its role in preventing tumour progression.
The function of RB that is crucial for tumour suppression might also depend on in which type of cell RB is lost — stem cell, progenitor or differentiated cell — as well as in which tissue.
In some contexts, presence of RB during earlier stages might be beneficial to tumour progression. Effects of post-translational modifications of RB on individual cellular functions might contribute to preference for a tumour to mutate RB or an upstream regulator.
E2F transcription factors.
Five members (E2F1–5) of a family of eight mammalian transcription factors that are transcriptional regulators, function as heterodimers with DP1–3 and have been shown to be regulated by direct binding to the pocket proteins.
Transcriptional co-factor.
Protein that is recruited to promoters or enhancers of gene expression through binding to other proteins rather than to the DNA itself. Co-factors affect the transcriptional activity of transcription factors.
Cre–lox:
Cre is a recombinase that specifically deletes DNA sequences flanked by lox sites.
Autophagy:
A cellular stress response in which cellular proteins and organelles are digested and recycled by lysosomes in order to maintain active metabolism.
Aneuploidy:
The occurrence of extra or missing chromosomes.
Pericentric heterochromatin:
DNA regions around the centromeres of chromosomes that contain hypoacetylated and methylated histones, resulting in transcriptional silencing.
Senescence:
Permanent cell cycle arrest, induced by cellular stresses and telomere shortening. Epigenetic changes prevent a mitogenic growth response, induce a distinct cellular morphology and promote expression of senescence-associated markers. Cells remain metabolically active.
References
- 1.Riley DJ, Lee EY & Lee WH The retinoblastoma protein: more than a tumor suppressor. Annu. Rev. Cell Biol 10, 1–29 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Weinberg RA The retinoblastoma protein and cell cycle control. Cell 81, 323–330 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Zheng L & Lee WH The retinoblastoma gene: a prototypic and multifunctional tumor suppressor. Exp. Cell Res 264, 2–18 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Dannenberg JH & te Riele HP The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesi. Results Probl. Cell Differ 42, 183–225 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ji P et al. An Rb–Skp2–p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol. Cell 16, 47–58 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Binne UK et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nature Cell Biol. 9, 225–232 (2007). [DOI] [PubMed] [Google Scholar]; References 5 and 6 demonstrate that the role of RB in cell cycle control is not solely mediated through its interactions with E2F transcription factors.
- 7.Wikenheiser-Brokamp KA Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol. Life Sci 63, 767–780 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobrinik D Pocket proteins and cell cycle control. Oncogene 24, 2796–2809 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Claudio PP, Tonini T & Giordano A The retinoblastoma family: twins or distant cousins? Genome Biol. 3, reviews3012 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Classon M & Dyson N p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res 264, 135–147. (2001). [DOI] [PubMed] [Google Scholar]
- 11.Morris EJ & Dyson NJ Retinoblastoma protein partners. Adv. Cancer Res 82, 1–54 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Macaluso M, Montanari M & Giordano A Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene 25, 5263–5267 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Gonzalo S & Blasco MA Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 4, 752–755 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Brehm A & Kouzarides T Retinoblastoma protein meets chromatin. Trends Biochem. Sci 24, 142–145 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Knudson AG Jr. Genetic predisposition to cancer. Cancer Detect Prev. 7, 1–8 (1984). [PubMed] [Google Scholar]
- 16.Corson TW & Gallie BL One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer 46, 617–634 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Dimaras H et al. Loss of RB1 induces non-proliferative retinoma; increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet (2008). [DOI] [PubMed] [Google Scholar]
- 18.Friend SH et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323, 643–646 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Bookstein R & Lee WH Molecular genetics of the retinoblastoma suppressor gene. Crit. Rev. Oncog 2, 211–227 (1991). [PubMed] [Google Scholar]
- 20.Deshpande A & Hinds PW The retinoblastoma protein in osteoblast differentiation and osteosarcoma. Curr. Mol. Med 6, 809–817 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Doorbar J Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci 110, 525–541 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Perez-Ordonez B, Beauchemin M & Jordan RC Molecular biology of squamous cell carcinoma of the head and neck. J. Clin. Pathol 59, 445–453 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munakata T et al. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 3, 1335–1347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meuwissen R et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4, 181–189 (2003). [DOI] [PubMed] [Google Scholar]; Describes a mouse model of human SCLC, the only adult tumour type in which loss of RB function occurs in most, if not all cases.
- 25.Ranade K et al. Mutations associated with familial melanoma impair p16INK4 function. Nature Genet. 10, 114–116 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Zuo L et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nature Genet. 12, 97–99 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Berman H et al. Genetic and epigenetic changes in mammary epithelial cells identify a subpopulation of cells involved in early carcinogenesis. Cold Spring Harb. Symp. Quant. Biol 70, 317–327 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Smith EJ, Leone G, DeGregori J, Jakoi L & Nevins JR The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol. Cell. Biol 16, 6965–6976 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balciunaite E et al. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol 25, 8166–8178 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y, Rayman JB & Dynlacht BD Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14, 804–816 (2000). [PMC free article] [PubMed] [Google Scholar]
- 31.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM & Jacks T Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424, 223–228 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Daria D et al. The retinoblastoma tumor suppressor is a critical intrinsic regulator for hematopoietic stem and progenitor cells under stress. Blood 111, 1894–1902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walkley CR & Orkin SH Rb is dispensable for self-renewal and multilineage differentiation of adult hematopoietic stem cells. Proc. Natl Acad. Sci. USA 103, 9057–9062 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walkley CR, Shea JM, Sims NA, Purton LE & Orkin SH Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell 129, 1081–1095 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz S et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development 131, 2737–2748 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Wildwater M et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349 (2005). [DOI] [PubMed] [Google Scholar]; Provides the first description of a role for RB in maintaining a stem cell pool in vivo.
- 37.Wier ML & Scott RE Regulation of the terminal event in cellular differentiation: biological mechanisms of the loss of proliferative potential. J. Cell Biol 102, 1955–1964 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peacocke M & Campisi J Cellular senescence: a reflection of normal growth control, differentiation, or aging? J. Cell Biochem 45, 147–155 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Beausejour CM et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 22, 4212–4222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narita M et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Ferreira R, Naguibneva I, Pritchard LL, Ait-Si-Ali S & Harel-Bellan A The Rb/chromatin connection and epigenetic control: opinion. Oncogene 20, 3128–3133. (2001). [DOI] [PubMed] [Google Scholar]
- 42.Skapek SX, Pan YR & Lee EY Regulation of cell lineage specification by the retinoblastoma tumor suppressor. Oncogene 25, 5268–5276 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Mayhew CN et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 65, 4568–4577 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Slack RS, El-Bizri H, Wong J, Belliveau DJ & Miller FD A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J. Cell Biol 140, 1497–1509 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camarda G et al. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J. Cell Biol 167, 417–423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh MS, Parker MH, Scime A, Parks R & Rudnicki MA Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol 166, 865–876 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D & Dynlacht BD Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol 179, 1399–1412 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber T et al. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc. Natl Acad. Sci. USA 105, 781–785 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferguson KL et al. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21, 3337–3346. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5, 539–551 (2004). [DOI] [PubMed] [Google Scholar]
- 51.MacPherson D et al. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 18, 1681–1694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ajioka I et al. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 131, 378–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes the first example of a fully differentiated cell (a neuron) that re-enters the cell cycle to initiate cancer.
- 53.Callaghan DA et al. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev. Biol 207, 257–270 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Ferguson KL & Slack RS The Rb pathway in neurogenesis. Neuroreport 12, A55–A62 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Zhang J et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nature Genet. 36, 351–360 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Dyer MA & Bremner R The search for the retinoblastoma cell of origin. Nature Rev. Cancer 5, 91–101 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Pacal M & Bremner R Insights from animal models on the origins and progression of retinoblastoma. Curr. Mol. Med 6, 759–781 (2006). [DOI] [PubMed] [Google Scholar]; References 50, 51 and 55, and the two reviews 56 and 57 describe the first breedable mouse models of retinoblastoma and investigate the mechanisms of tumorigenesis in the retina.
- 58.Khidr L & Chen PL RB, the conductor that orchestrates life, death and differentiation. Oncogene 25, 5210–5219 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Nguyen DX & McCance DJ Role of the retinoblastoma tumor suppressor protein in cellular differentiation. J. Cell Biochem 94, 870–879 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Thomas DM, Yang HS, Alexander K & Hinds PW Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol. Ther 2, 124–130 (2003). [PubMed] [Google Scholar]
- 61.Classon M & Harlow E The retinoblastoma tumour suppressor in development and cancer. Nature Rev. Cancer 2, 910–917 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Iavarone A et al. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature 432, 1040–1045 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Lasorella A, Noseda M, Beyna M, Yokota Y & Iavarone A Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407, 592–598 (2000). [DOI] [PubMed] [Google Scholar]; Describes a crucial link between MYC signalling and RB function, and provides mechanistic evidence that the interactions between ID2 and RB might regulate the proliferation and differentiation of cells.
- 64.Miyake S et al. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol 20, 8889–8902 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacLellan WR, Xiao G, Abdellatif M & Schneider MD A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol 20, 8903–8915 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benevolenskaya EV, Murray HL, Branton P, Young RA & Kaelin WG Jr. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18, 623–635 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Krutzfeldt M et al. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol. Cell 18, 213–224 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Chen D et al. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 5, e179 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McClellan KA et al. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol 27, 4825–4843 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 68 and 69 describe a novel function of RB in controlling cell migration, distinct from its role in the cell cycle, and show that E2F transcription factors might mediate this novel function of RB in vivo.
- 70.Sidle A et al. Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation. Crit. Rev. Biochem. Mol. Biol 31, 237–271 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Sellers WR et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12, 95–106 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]; A classical example dissecting the mechanisms of retinoblastoma development by studying various mutants of RB; these observations highlight the fact that a pro-differentiation activity of RB that is independent of E2F acts as a tumour suppressor mechanism.
- 72.Thomas DM et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8, 303–316 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Kleinerman RA et al. Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: an extended follow-up. J. Clin. Oncol 23, 2272–2279 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Tsai KY et al. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2, 293–304 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Macleod KF, Hu Y & Jacks T Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 15, 6178–6188 (1996). [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue Y, Kitagawa M & Taya Y Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 26, 2083–2093 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Markham D, Munro S, Soloway J, O’Connor DP & La Thangue NB DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 7, 192–198 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dick FA Structure-function analysis of the retinoblastoma tumor suppressor protein — is the whole a sum of its parts? Cell Div. 2, 26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziebold U, Reza T, Caron A & Lees JA E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15, 386–391 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saavedra HI et al. Specificity of E2F1, E2F2, and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 13, 215–225 (2002). [PubMed] [Google Scholar]
- 81.Bosco EE & Knudsen ES Differential role of RB in response to UV and IR damage. Nucleic Acids Res. 33, 1581–1592 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prost S, Lu P, Caldwell H & Harrison D E2F regulates DDB2: consequences for DNA repair in Rb-deficient cells. Oncogene 26, 3572–3581 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Billecke CA et al. Lack of functional pRb results in attenuated recovery of mRNA synthesis and increased apoptosis following UV radiation in human breast cancer cells. Oncogene 21, 4481–4489 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Bosco EE et al. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 32, 25–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halazonetis TD, Gorgoulis VG & Bartek J An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Jiang Z & Zacksenhaus E Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J. Cell Biol 156, 185–198 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Symonds H et al. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78, 703–711 (1994). [DOI] [PubMed] [Google Scholar]
- 88.Chau BN & Wang JY Coordinated regulation of life and death by RB.. Nature Rev. Cancer 3, 130–138 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Borges HL, Hunton IC & Wang JY Reduction of apoptosis in Rb-deficient embryos via Abl knockout. Oncogene 26, 3868–3877 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Moroni MC et al. Apaf-1 is a transcriptional target for E2F and p53. Nature Cell Biol. 3, 552–558 (2001). [DOI] [PubMed] [Google Scholar]
- 91.Simpson MT et al. Caspase 3 deficiency rescues peripheral nervous system defect in retinoblastoma nullizygous mice. J. Neurosci 21, 7089–7098 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seifried LA et al. pRB-E2F1 complexes are resistant to adenovirus E1A-mediated disruption. J. Virol 82, 4511–4520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weinstein IB Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis 21, 857–864 (2000). [DOI] [PubMed] [Google Scholar]
- 94.Pickering MT & Kowalik TF Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 25, 746–755 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Mathew R, Karantza-Wadsworth V & White E Role of autophagy in cancer. Nature Rev. Cancer 7, 961–967 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin S & White E Tumor suppression by autophagy through the management of metabolic stress. Autophagy 4, 563–566 (2008). [PMC free article] [PubMed] [Google Scholar]
- 97.Tracy K et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol 27, 6229–6242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Polager S, Ofir M & Ginsberg D E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 14 April 2008 (doi: 10.1038/onc.2008.117). [DOI] [PubMed] [Google Scholar]
- 99.Sanseverino F et al. pRb2/p130 and VEGF expression in endometrial carcinoma in relation to angiogenesis and histopathologic tumor grade. Cancer Biol. Ther 5, 84–88 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Sell S Leukemia: stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev. 1, 197–205 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Foijer F, Wolthuis RM, Doodeman V, Medema RH & te Riele H Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell 8, 455–466 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Eguchi T, Takaki T, Itadani H & Kotani H RB silencing compromises the DNA damage-induced G2/M checkpoint and causes deregulated expression of the ECT2 oncogene. Oncogene 26, 509–520 (2007). [DOI] [PubMed] [Google Scholar]
- 103.Kennedy BK, Barbie DA, Classon M, Dyson N & Harlow E Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14, 2855–2868 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zheng L, Flesken-Nikitin A, Chen PL & Lee WH Deficiency of Retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res. 62, 2498–2502 (2002). [PubMed] [Google Scholar]
- 105.Zheng L & Lee WH Retinoblastoma tumor suppressor and genome stability. Adv. Cancer Res 85, 13–50 (2002). [DOI] [PubMed] [Google Scholar]
- 106.Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W & Knudsen ES RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J. Biol. Chem 282, 23867–23877 (2007). [DOI] [PubMed] [Google Scholar]
- 107.Hernando E et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430, 797–802 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Sotillo R et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11, 9–23 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Isaac CE et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol 26, 3659–3671 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes one of the first in vivo models to use a knock-in mouse to understand the cellular function of individual protein residues of RB.
- 110.Gonzalo S et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nature Cell Biol. 7, 420–428 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Knudsen ES, Sexton CR & Mayhew CN Role of the retinoblastoma tumor suppressor in the maintenance of genome integrity. Curr. Mol. Med 6, 749–757 (2006). [DOI] [PubMed] [Google Scholar]
- 112.McCabe MT, Davis JN & Day ML Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 65, 3624–3632 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Iovino F, Lentini L, Amato A & Di Leonardo A RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol. Cancer 5, 38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meraldi P, Lukas J, Fry AM, Bartek J & Nigg EA Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nature Cell Biol. 1, 88–93 (1999). [DOI] [PubMed] [Google Scholar]
- 115.Longworth MS, Herr A, Ji JY & Dyson NJ RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 22, 1011–1024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harrington EA, Bruce JL, Harlow E & Dyson N pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl Acad. Sci. USA 95, 11945–11950 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Siddiqui H, Fox SR, Gunawardena RW & Knudsen ES Loss of RB compromises specific heterochromatin modifications and modulates HP1α dynamics. J. Cell Physiol 211, 131–137 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Robertson KD et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet. 25, 338–342 (2000). [DOI] [PubMed] [Google Scholar]
- 119.Lund AH & van Lohuizen M Epigenetics and cancer. Genes Dev. 18, 2315–2335 (2004). [DOI] [PubMed] [Google Scholar]
- 120.McCabe DC & Caudill MA DNA methylation, genomic silencing, and links to nutrition and cancer. Nutr. Rev. 63, 183–195 (2005). [DOI] [PubMed] [Google Scholar]
- 121.Sage J Making young tumors old: a new weapon against cancer? Sci. Aging Knowledge Environ. pe25 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Collado M, Blasco MA & Serrano M Cellular senescence in cancer and aging. Cell 130, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Courtois-Cox S, Jones SL & Cichowski K Many roads lead to oncogene-induced senescence. Oncogene 27, 2801–2089 (2008). [DOI] [PubMed] [Google Scholar]
- 124.Braig M et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005). [DOI] [PubMed] [Google Scholar]
- 125.Garcia-Cao M, Gonzalo S, Dean D & Blasco MA A role for the Rb family of proteins in controlling telomere length. Nature Genet. 32, 415–419 (2002). [DOI] [PubMed] [Google Scholar]
- 126.Kong LJ, Meloni AR & Nevins JR The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol. Cell 22, 63–71 (2006). [DOI] [PubMed] [Google Scholar]
- 127.Wei W, Herbig U, Wei S, Dutriaux A & Sedivy JM Loss of retinoblastoma but not p16 function allows bypass of replicative senescence in human fibroblasts. EMBO Rep. 4, 1061–1066 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Young AP et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nature Cell Biol. 10, 361–369 (2008). [DOI] [PubMed] [Google Scholar]
- 129.Alexander K & Hinds PW Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol. Cell. Biol 21, 3616–3631 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gabellini C, Del Bufalo D & Zupi G Involvement of RB gene family in tumor angiogenesis. Oncogene 25, 5326–5332 (2006). [DOI] [PubMed] [Google Scholar]
- 131.Chien WM et al. Differential gene expression of p27Kip1 and Rb knockout pituitary tumors associated with altered growth and angiogenesis. Cell Cycle 6, 750–757 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lasorella A, Rothschild G, Yokota Y, Russell RG & Iavarone A Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol. Cell. Biol 25, 3563–3574 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ruzinova MB & Benezra R Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13, 410–418 (2003). [DOI] [PubMed] [Google Scholar]
- 134.Benezra R, Rafii S & Lyden D The Id proteins and angiogenesis. Oncogene 20, 8334–8341 (2001). [DOI] [PubMed] [Google Scholar]
- 135.Dasgupta P et al. Disruption of the Rb–Raf-1 interaction inhibits tumor growth and angiogenesis. Mol. Cell. Biol 24, 9527–9541 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martinez-Cruz AB et al. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Res. 68, 683–692 (2008). [DOI] [PubMed] [Google Scholar]
- 137.Hui AM, Li X, Makuuchi M, Takayama T & Kubota K Over-expression and lack of retinoblastoma protein are associated with tumor progression and metastasis in hepatocellular carcinoma. Int. J. Cancer 84, 604–608 (1999). [DOI] [PubMed] [Google Scholar]
- 138.Gauthier ML et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 12, 479–491 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Singh B, Berry JA, Shoher A, Ramakrishnan V & Lucci A COX-2 overexpression increases motility and invasion of breast cancer cells. Int. J. Oncol 26, 1393–1399 (2005). [PubMed] [Google Scholar]
- 140.Tsujii M, Kawano S & DuBois RN Cyclooxygenase-2 expression in human colon cancer cells increases metastatic potential. Proc. Natl Acad. Sci. USA 94, 3336–3340 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ferguson KL et al. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 24, 4381–4391 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ziebold U, Lee EY, Bronson RT & Lees JA E2F3 loss has opposing effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol. Cell. Biol 23, 6542–6552 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stanelle J, Stiewe T, Theseling CC, Peter M & Putzer BM Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 30, 1859–1867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoon SO, Shin S & Mercurio AM Ras stimulation of E2F activity and a consequent E2F regulation of integrin alpha6beta4 promote the invasion of breast carcinoma cells. Cancer Res. 66, 6288–6295 (2006). [DOI] [PubMed] [Google Scholar]
- 145.Zhang SY, Liu SC, Johnson DG & Klein-Szanto AJ E2F-1 gene transfer enhances invasiveness of human head and neck carcinoma cell lines. Cancer Res. 60, 5972–5976 (2000). [PubMed] [Google Scholar]
- 146.Calzone L, Gelay A, Zinovyev A, Radvanyl F & Barillot E A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol. Syst. Biol 4, 173 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marino S, Vooijs M, van Der Gulden H, Jonkers J & Berns A Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004. (2000). [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou Z, Flesken-Nikitin A & Nikitin AY Prostate cancer associated with p53 and Rb deficiency arises from the stem/progenitor cell-enriched proximal region of prostatic ducts. Cancer Res. 67, 5683–5690 (2007). [DOI] [PubMed] [Google Scholar]
- 149.Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN & Nikitin AY Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 63, 3459–3463 (2003). [PubMed] [Google Scholar]
- 150.Dannenberg JH, Schuijff L, Dekker M, van der Valk M & te Riele H Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 18, 2952–2962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sotillo R et al. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 20, 6637–6647 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rane SG, Cosenza SC, Mettus RV & Reddy EP Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol. Cell. Biol 22, 644–656 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ortega S, Malumbres M & Barbacid M Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta 1602, 73–87 (2002). [DOI] [PubMed] [Google Scholar]
- 154.Yin C, Knudson CM, Korsmeyer SJ & Van Dyke T Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature 385, 637–640 (1997). [DOI] [PubMed] [Google Scholar]
- 155.Pan H et al. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol. Cell 2, 283–292 (1998). [DOI] [PubMed] [Google Scholar]
- 156.Lu X et al. Selective inactivation of p53 facilitates mouse epithelial tumor progression without chromosomal instability. Mol. Cell. Biol 21, 6017–6030 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xiao A, Wu H, Pandolfi PP, Louis DN & Van Dyke T Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell 1, 157–168 (2002). [DOI] [PubMed] [Google Scholar]
- 158.Simin K et al. pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2, E22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hill R, Song Y, Cardiff RD & Van Dyke T Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell 123, 1001–1011 (2005). [DOI] [PubMed] [Google Scholar]
- 160.Hill R, Song Y, Cardiff RD & Van Dyke T Heterogeneous tumor evolution initiated by loss of pRb function in a preclinical prostate cancer model. Cancer Res. 65, 10243–10254 (2005). [DOI] [PubMed] [Google Scholar]
- 161.Simin K et al. Deciphering cancer complexities in genetically engineered mice. Cold Spring Harb. Symp. Quant. Biol 70, 283–290 (2005). [DOI] [PubMed] [Google Scholar]; References 154–161 illustrate the efforts from the Van Dyke group to study the role of the entire Rb family in preventing cancer initiation in different cell types in mice.
- 162.Frese KK & Tuveson DA Maximizing mouse cancer models. Nature Rev. Cancer 7, 645–658 (2007). [DOI] [PubMed] [Google Scholar]
- 163.Nikitin AY, Juarez-Perez MI, Li S, Huang L & Lee WH RB-mediated suppression of spontaneous multiple neuroendocrine neoplasia and lung metastases in Rb+/− mice. Proc. Natl Acad. Sci. USA 96, 3916–3921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou Z et al. Suppression of melanotroph carcinogenesis leads to accelerated progression of pituitary anterior lobe tumors and medullary thyroid carcinomas in Rb+/− mice. Cancer Res. 65, 787–796 (2005). [PubMed] [Google Scholar]
- 165.Zwijsen RM CDK-independent activation of estrogen receptor by cyclin D1. Cell 88, 405–415 (1997). [DOI] [PubMed] [Google Scholar]
- 166.Liu F Smad3 phosphorylation by cyclin-dependent kinases. Cytokine Growth Factor Rev. 17, 9–17 (2006). [DOI] [PubMed] [Google Scholar]
- 167.Kaye FJ RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene 21, 6908–6914 (2002). [DOI] [PubMed] [Google Scholar]
- 168.Trimarchi JM & Lees JA Sibling rivalry in the E2F family. Nature Rev. Mol. Cell Biol 3, 11–20 (2002). [DOI] [PubMed] [Google Scholar]
- 169.Smith EJ, Leone G & Nevins JR Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 9, 297–303 (1998). [PubMed] [Google Scholar]
- 170.Tedesco D, Lukas J & Reed SI The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16, 2946–2957 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yamasaki L et al. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/− mice. Nature Genet. 18, 360–364 (1998). [DOI] [PubMed] [Google Scholar]
- 172.Feber A et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 23, 1627–1630 (2004). [DOI] [PubMed] [Google Scholar]
- 173.Grasemann C et al. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene 24, 6441–6449 (2005). [DOI] [PubMed] [Google Scholar]
- 174.Lee C, Chang JH, Lee HS & Cho Y Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 16, 3199–3212 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim HY, Ahn BY & Cho Y Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20, 295–304 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee JO, Russo AA & Pavletich NP Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391, 859–865 (1998). [DOI] [PubMed] [Google Scholar]
- 177.Julian LM, Palander O, Seifried LA, Foster JE & Dick FA Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene 27, 1572–1579 (2008). [DOI] [PubMed] [Google Scholar]
- 178.Chan HM, Krstic-Demonacos M, Smith L, Demonacos C & La Thangue NB Acetylation control of the retinoblastoma tumour-suppressor protein. Nature Cell Biol. 3, 667–674 (2001). [DOI] [PubMed] [Google Scholar]
- 179.Nguyen DX, Baglia LA, Huang SM, Baker CM & McCance DJ Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. EMBO J. 23, 1609–1618 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ledl A, Schmidt D & Muller S Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24, 3810–3818 (2005). [DOI] [PubMed] [Google Scholar]
- 181.Borges HL et al. Tumor promotion by caspase-resistant retinoblastoma protein. Proc. Natl Acad. Sci. USA 102, 15587–15592 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wikenheiser-Brokamp KA Retinoblastoma regulatory pathway in lung cancer. Curr. Mol. Med 6, 783–793 (2006). [DOI] [PubMed] [Google Scholar]
- 183.Kaye FJ & Harbour JW For whom the bell tolls: susceptibility to common adult cancers in retinoblastoma survivors. J. Natl Cancer Inst 96, 342–343 (2004). [DOI] [PubMed] [Google Scholar]
- 184.Horowitz JM et al. Anti-oncogenes and the negative regulation of cell growth. Cold Spring Harb. Symp. Quant. Biol 53, 843–847 (1988). [DOI] [PubMed] [Google Scholar]
- 185.Delston RB & Harbour JW Rb at the interface between cell cycle and apoptotic decisions. Curr. Mol. Med 6, 713–718 (2006). [DOI] [PubMed] [Google Scholar]
- 186.Li W et al. The role of cell cycle regulatory proteins in the pathogenesis of melanoma. Pathology 38, 287–301 (2006). [DOI] [PubMed] [Google Scholar]
- 187.MacGrogan D & Bookstein R Tumour suppressor genes in prostate cancer. Semin. Cancer Biol 8, 11–19 (1997). [DOI] [PubMed] [Google Scholar]
- 188.Hugel A & Wernert N Loss of heterozygosity (LOH), malignancy grade and clonality in microdissected prostate cancer. Br. J. Cancer 79, 551–557 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Abate-Shen C & Shen MM Molecular genetics of prostate cancer. Genes Dev. 14, 2410–2434 (2000). [DOI] [PubMed] [Google Scholar]
- 190.Cox LA, Chen G & Lee EY Tumor suppressor genes and their roles in breast cancer. Breast Cancer Res. Treat 32, 19–38 (1994). [DOI] [PubMed] [Google Scholar]
- 191.Bosco EE & Knudsen ES RB in breast cancer: at the crossroads of tumorigenesis and treatment. Cell Cycle 6, 667–671 (2007). [DOI] [PubMed] [Google Scholar]
- 192.Miyamoto H, Shuin T, Ikeda I, Hosaka M & Kubota Y Loss of heterozygosity at the p53, RB, DCC and APC tumor suppressor gene loci in human bladder cancer. J. Urol 155, 1444–1447 (1996). [PubMed] [Google Scholar]
- 193.Cordon-Cardo C, Sheinfeld J & Dalbagni G Genetic studies and molecular markers of bladder cancer. Semin. Surg. Oncol 13, 319–327 (1997). [DOI] [PubMed] [Google Scholar]
- 194.Mitra AP, Datar RH & Cote RJ Molecular pathways in invasive bladder cancer: new insights into mechanisms, progression, and target identification. J. Clin. Oncol 24, 5552–5564 (2006). [DOI] [PubMed] [Google Scholar]
- 195.Krug U, Ganser A & Koeffler HP Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene 21, 3475–3495 (2002). [DOI] [PubMed] [Google Scholar]
- 196.Beck Z et al. Alterations of P53 and RB genes and the evolution of the accelerated phase of chronic myeloid leukemia. Leuk. Lymphoma 38, 587–597 (2000). [DOI] [PubMed] [Google Scholar]
- 197.Zhu YM, Haynes AP, Keith FJ & Russell NH Abnormalities of retinoblastoma gene expression in hematological malignancies. Leuk. Lymphoma 18, 61–67 (1995). [DOI] [PubMed] [Google Scholar]
- 198.Jacks T et al. Effects of an Rb mutation in the mouse. Nature 359, 295–300 (1992). [DOI] [PubMed] [Google Scholar]
- 199.Honda S et al. Human pituitary adenomas infrequently contain inactivation of retinoblastoma 1 gene and activation of cyclin dependent kinase 4 gene. Endocr. J 50, 309–318 (2003). [DOI] [PubMed] [Google Scholar]
- 200.Wechsler-Reya R & Scott MP The developmental biology of brain tumors. Annu. Rev. Neurosci 24, 385–428 (2001). [DOI] [PubMed] [Google Scholar]
- 201.Hulleman E & Helin K Molecular mechanisms in gliomagenesis. Adv. Cancer Res 94, 1–27 (2005). [DOI] [PubMed] [Google Scholar]
- 202.Jenkins GJ et al. Genetic pathways involved in the progression of Barrett’s metaplasia to adenocarcinoma. Br. J. Surg 89, 824–837 (2002). [DOI] [PubMed] [Google Scholar]
- 203.Lerut T & Decker G Esophageal cancer. Curr. Opin. Gastroenterol 15, 364–369 (1999). [DOI] [PubMed] [Google Scholar]
- 204.Zhang X et al. Deletions of chromosome 13q, mutations in Retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res. 54, 4177–4182 (1994). [PubMed] [Google Scholar]
- 205.Murakami Y, Hayashi K, Hirohashi S & Sekiya T Aberrations of the tumor suppressor p53 and retinoblastoma genes in human hepatocellular carcinomas. Cancer Res. 51, 5520–5525 (1991). [PubMed] [Google Scholar]
- 206.Villanueva A, Newell P, Chiang DY, Friedman SL & Llovet JM Genomics and signaling pathways in hepatocellular carcinoma. Semin. Liver Dis 27, 55–76 (2007). [DOI] [PubMed] [Google Scholar]
- 207.Kinzler KW & Vogelstein B Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996). [DOI] [PubMed] [Google Scholar]
- 208.Kowalski J et al. Chromosomal abnormalities of adenocarcinoma of the pancreas: identifying early and late changes. Cancer Genet. Cytogenet 178, 26–35 (2007). [DOI] [PubMed] [Google Scholar]
- 209.Decker HJ et al. Cytogenetic and molecular studies of a familial renal cell carcinoma. Cancer Genet. Cytogenet 63, 25–31 (1992). [DOI] [PubMed] [Google Scholar]
- 210.Sanseverino F, Torricelli M, Petraglia F & Giordano A Role of the retinoblastoma family in gynecological cancer. Cancer Biol. Ther 2, 636–641 (2003). [PubMed] [Google Scholar]
- 211.Moller MB et al. Frequent disruption of the RB1 pathway in diffuse large B cell lymphoma: prognostic significance of E2F-1 and p16INK4A. Leukemia 14, 898–904 (2000). [DOI] [PubMed] [Google Scholar]
- 212.Leoncini L, Bellan C & De Falco G Retinoblastoma gene family expression in lymphoid tissues. Oncogene 25, 5309–5314 (2006). [DOI] [PubMed] [Google Scholar]
- 213.Juge-Morineau N, Harousseau JL, Amiot M & Bataille R The retinoblastoma susceptibility gene RB-1 in multiple myeloma. Leuk. Lymphoma 24, 229–237 (1997). [DOI] [PubMed] [Google Scholar]
- 214.DeLellis RA Pathology and genetics of thyroid carcinoma. J. Surg. Oncol 94, 662–669 (2006). [DOI] [PubMed] [Google Scholar]
- 215.Zheng L, Wang L, Ajani J & Xie K Molecular basis of gastric cancer development and progression. Gastric Cancer 7, 61–77 (2004). [DOI] [PubMed] [Google Scholar]
- 216.Wikenheiser-Brokamp KA Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development 131, 4299–4310 (2004). [DOI] [PubMed] [Google Scholar]
- 217.Maddison LA, Sutherland BW, Barrios RJ & Greenberg NM Conditional deletion of Rb causes early stage prostate cancer. Cancer Res. 64, 6018–6025 (2004). [DOI] [PubMed] [Google Scholar]
- 218.Zhou Z et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 66, 7889–7898 (2006). [DOI] [PubMed] [Google Scholar]
- 219.Higashitsuji H et al. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nature Med. 6, 96–99 (2000). [DOI] [PubMed] [Google Scholar]