Abstract
Fusobacterium nucleatum has been increasingly implicated as a causative agent of various diseases, such as inflammatory bowel disease. Moreover, the gastrointestinal tracts of patients with colorectal cancer (CRC) also have been shown to be colonized by this bacterium. We aimed to determine the prevalence of F. nucleatum among CRC and non-CRC Iranian patients and to investigate potential associations between fadA-positive F. nucleatum and diagnosed CRC cases. Eighty patients admitted to two main hospitals in Tehran, Iran, were enrolled. The patients were aged between 20 and 75 and were diagnosed by a gastroenterologist. A trained surgeon used standard surgical protocols to collect two CRC biopsy samples per patient. One of the samples was used for pathologic examination, and the other was subjected to DNA extraction and PCR. Lesion colonization by F. nucleatum and expression of its major virulence factor, fadA, were investigated. The fadA-positive F. nucleatum strain was absent in all the lesions obtained from non-CRC patients. All patients with lesions that were colonized with fadA-positive F. nucleatum were diagnosed as CRC (p < 0.05); selected patients were sent for further intensive treatment. We found a significant association between the presence of F. nucleatum colonization and lesions from CRC patients (p 0.0001; odds ratio, 6.74; 95% confidence interval, 2.5–18.07). Our study confirmed colonization of the fadA-positive F. nucleatum on lesions from 80 Iranian CRC patients. New therapeutic strategies to achieve eradication of F. nucleatum are necessary for clinical management of patients suspected of having or prone to developing CRC.
Keywords: colorectal cancer, fadA, fusobacterium nucleatum, iranian patients, PCR
Introduction
Fusobacterium nucleatum is an anaerobic, nonsporulating, Gram-negative bacterium which is commensal in the healthy human oral cavity [1,2]. F. nucleatum is, however, implicated in different types of periodontal disease under abnormal conditions. F. nucleatum has also been associated with adverse pregnancy outcomes, gastrointestinal disorders and speculatively with other diseases, such as Alzheimer disease and atherosclerosis [2,3]. A significant correlation between severity of clinical prognoses of patients with colorectal cancer (CRC) and the extent of colonization by this bacterium has been reported [[4], [5], [6]]. Generally, the microbiologic cause of CRC is unknown, but various factors have been implicated [7,8]. Because complex populations of both commensal and pathogenic microorganisms colonize the human gastrointestinal tract, dysbiosis, an abnormal imbalance in the populating microbial communities—a dysbiotic gut microbiome—is presumed to contribute to colorectal adenomas and CRC [[7], [8], [9], [10]]. Accordingly, prevalence of F. nucleatum is reportedly high among patients with CRC [11,12], and, as expected, severe clinical symptoms manifest in subjects colonized by virulent strains of fusobacteria.
FadA is a major adhesion/invasion factor that exerts a significant role in the pathogenic mechanisms orchestrated by F. nucleatum [13,14]. Accordingly, molecular interactions between FadA and VE-cadherin weakens the endothelial cell–cell junctions, thus increasing the permeability of the endothelial layer, consequently facilitating the transendothelial transmission of F. nucleatum and other bacteria [14]. FadA interactions with E-cadherin have been postulated to trigger inflammatory and oncogenic responses through activation of β-catenin signaling [12], resulting in development of CRC [4,12,14,15]. Such a positive association between colonization with F. nucleatum and CRC has been reported by many different studies (reviewed in [16]).
Because F. nucleatum is a fastidious microorganism, its culture and maintenance are restricted to specialized research laboratories; however, developing easy and routine methods for detecting F. nucleatum and phenotyping it for its major virulence factor—fadA—in biopsy samples abrogates the need for specialized and costly culturing and microbiologic setups. CRC colonization with virulent F. nucleatum strains has not been documented in clinical samples—at least, not in the Middle East.
In this study, we used PCR to investigate the presence of F. nucleatum in tissue samples of CRC patients and to determine their positivity for fadA to investigate a potential correlation between the presence or absence of virulent fadA-positive F. nucleatum with CRC in Iranian patients.
Methods
Patients and sampling protocol
This cross-sectional study started in February 2017 and concluded in June 2019. Patients (n = 80; aged between 20 and 75 years) with signs and symptoms of colorectal disorders or with related digestive complaints with no record of abdominal surgery over the last 2 years were enrolled. Each participant provided written informed consent before enrolment; participation was voluntary. The study and relevant protocols were reviewed and approved by the ethical committee of Tarbiat Modares University, Tehran, Iran (ethics approval IR.MODARES.REC.1397.240), and complied with the principles of the Declaration of Helsinki [17]. Colonoscopic surgery was recommended and was performed by expert gastroenterologists after thorough diagnosis. All CRC samples were obtained at the Mehrad Hospital and Labafi-Nejad Hospital, Tehran, Iran, following standard surgical protocols.
For each case, two biopsy samples were taken adjacent to the tumor or from suspected tumorous tissues by colonoscopy performed by an expert gastroenterologist. One sample was transported, in normal saline solution (0.9% w/v, pH 7), to the pathology department, and the second sample was transported in the same solution to the microbiology laboratory, Tarbiat Modares University, for further testing. Possible associations between demographic data (gender, age) and presence of F. nucleatum and fadA positivity were examined.
Genomic DNA extraction and PCR
Genomic DNA was extracted from the biopsy samples by using a commercially available kit (Roje Technologies, Yazd, Iran) and purified according to the manufacturer's instructions. Concentrations of purified DNA was measured by using a NanoDrop system at 260 nm. 16S ribosomal RNA–based PCR was performed using the fus primers for confirming the presence of F. nucleatum and by using fadA primers for confirming its major virulence factor. Primer sequences for detecting F. nucleatum and fadA are listed in Table 1. We used the genomic DNA from F. nucleatum subsp. nucleatum Knorr strain VPI 4355 (ATCC 25586) as the positive control to check the quality of our experiments. Distilled water instead of genomic DNA served as the negative control in our PCR experiments. PCR reactions were set up in 20 μL, containing 0.75 μL of bacterial chromosomal DNA (≤180 ng), 0.5 μM of each primer, 1 mM of each dNTP, 1.5 mM MgCl2 and 0.10 U/μL Taq DNA polymerase. PCR was performed in a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). Both genes were amplified by 37 cycles of initial denaturation at 95 °C for 55 seconds, annealing at 61°C for 1 minute and polymerase extension at 72°C for 1 minute.
Table 1.
Primers used in this study
| Primer | Oligonucleotide sequence (5′–3′) | Temperature (°C) | Product (bp) | Reference |
|---|---|---|---|---|
| fus-F | CTTTAACAGAAGATGAAGCTG | 61 | 540 | This study |
| fus-R | CTAAGGCAGATATTCTTCTC | 61 | 540 | This study |
| fadA-F | CAC AAG CTG ACG CTG CTA GA | 61 | 232 | [18] |
| fadA-R | TTACCAGCTCTTAAAGCTTG | 61 | 232 | [18] |
F and R represent forward and reverse primers.
Gel electrophoresis
PCR products were electrophoresed in 1.5% agarose (Cinna Gen, Tehran, Iran) gels in single-strength TAE buffer (40 mM Tris, 20 mM acetic acid and 1 mM EDTA, pH 8.0) and visualized under an ultraviolet transilluminator (Biometra, Göttingen, Germany).
Statistical analyses
Statistical analyses including associations between presence of virulence gene (fadA) and reported cases of CRC were performed by SPSS 16.0 (IBM, Armonk, NY, USA). The Fisher exact test was used to assess the association of the fadA-positive samples with CRC subjects; p < 0.05 was deemed statistically significant.
Results
Eighty patients were included in the study; 33 (41%) were male and 47 (58%) female, averaging 54 years of age (range, 22–69 years). No statistically significant correlations were found between F. nucleatum colonization and demographic parameters, including age, diet and gender. Of 80 patients, 35 (43%) were positive for CRC and the rest were non-CRC cases according to pathologic and colonoscopy observations by a pathologist and a gastroenterologist, respectively.
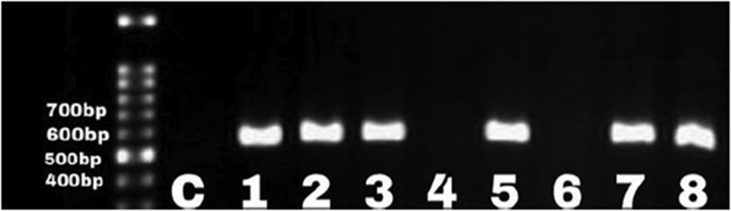
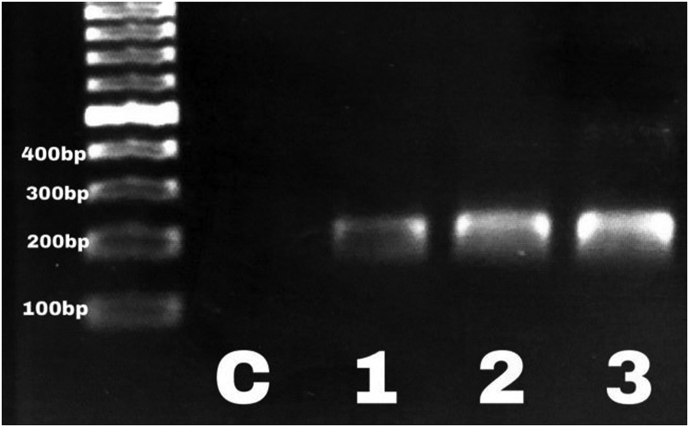
Incidence of F. nucleatum infection was 68% (24/35) and 24% (11/45) among the CRC and non-CRC patients, respectively. Eight (33%) of 24 CRC patients had fadA-positive F. nucleatum (p 0.03). All patients with fadA-positive F. nucleatum were symptomatic (p < 0.05) and were candidates for further treatment. We found a significant association between the presence of F. nucleatum infection among CRC patients (p 0.0001; odds ratio, 6.74; 95% confidence interval, 2.5–18.07). However, none of the 11 F. nucleatum samples isolated from the non-CRC patients was carrying the fadA gene (Fig. 1, Fig. 2).
Fig. 1.
Three representative cases positive for Fusobacterium nucleatum gene confirmed by PCR amplification of fus. Lane 1, molecular weight marker (bp); lane 2, distilled water as negative control (C).
Fig. 2.
PCR amplification of fadA in three representative clinical samples confirming presence of fadA-positive, virulent Fusobacterium nucleatum. Lane 1, molecular weight marker; lane 2, distilled water as negative control.
Discussion
Understanding the gastrointestinal microbiome and its contribution to human physiology or pathology is a challenging but emerging field of research that is engaging microbiologists and gastroenterologists globally [19,20]. Recent reports on prevalence of CRC in different countries conclude that certain deleterious microorganisms likely contribute to the pathogenesis of CRC [11,16,21]. Accordingly, some studies have confirmed that prevalence of F. nucleatum has a major effect on unfavourable clinical prognoses of CRC patients, e.g. metastasis to lymph nodes [22]. Intriguingly, F. nucleatum was thought to be responsible only for chronic inflammatory diseases, mostly inflammatory bowel disease and subgingival plaques [11,23]. Historically, many microbiologists considered F. nucleatum to be a transient, commensal microbe in the human oral cavity and gastrointestinal tract; however, the current body of literature challenges the previous understanding about this mysterious pathogen, which likely plays a crucial role in the pathogenesis of CRC [11]. Association of fusobacteria with CRC paints a similar picture to that of Helicobacter pylori with downstream gastric cancer [4]. Our cross-sectional study was performed to confirm the association between the prevalence of virulent fadA-positive F. nucleatum and the clinical diagnosis of the patients with CRC. We found that 68% of CRC patients had lesions colonized by F. nucleatum, indicating a relatively high rate of prevalence among symptomatic CRC patients. In contrast, F. nucleatum reportedly was enriched in lesions from only 8.6% of CRC patients (n = 511) in Japan [24]. The two main discrepancies between our study and the report by Nosho et al. [24] are use of fresh biopsy samples by us instead of formalin-fixed, paraffin-embedded (FFPE) specimens and inclusion of geographically and racially different patient populations. Suehiro et al. [25] reported that more than 260 copies of F. nucleatum were detectable in 54% (85/158) of fecal samples from Japanese CRC patients, as detected and quantified by droplet digital PCR. Like the Japanese study in 2016, which used droplet digital PCR [24], Ito et al. [26] reported that 56% (286/511) of FFPE samples from the Japanese CRC patients were colonized by F. nucleatum as assessed by conventional PCR. A Chinese group reported that >87% of FFPE samples from CRC patients were positive for F. nucleatum, as detected by fluorescence in situ hybridization [11]; no other study reported such a high prevalence rate of F. nucleatum among CRC patients [11]. Additionally, Tahara et al. [27] reported a prevalence of 74% (111/149) among the Japanese population by real-time PCR detection of extracted genomic DNA from the CRC samples, indicating a 1.1-fold higher prevalence rate than the Iranian CRC patients (68%) (p 0.0001; odds ratio, 6.74; 95% confidence interval, 2.5–18.07) found in this study.
An important aspect of our study is population selection because epidemiologic studies have reported that almost half of the global death rate associated with CRC occurs in Asian and African countries [23]. In addition to a high prevalence of F. nucleatum among CRC patients in our study, we also found that fadA-positive F. nucleatum had colonized the biopsy samples from the patients with CRC (p 0.03). The association between the virulent F. nucleatum and CRC samples also positively correlated with worse clinical outcomes for CRC patients.
Iran has alarming rates of mortality and morbidity due to CRC [28,29], and our results can serve as a pilot study for testing colonization of F. nucleatum in CRC patients [28,29]. We did not seek to confirm the presence of F. nucleatum among CRC patients but to prove the existence of a high prevalence of fadA-positive F. nucleatum in samples from CRC patients; this observation is a novel one. Similar studies using larger population sizes are required to further support our findings. Our study corroborates the significance of the hypothesis that F. nucleatum may cause or promote CRC [6,30]. Therefore, new therapeutic strategies aiming to eradicate F. nucleatum could be promising for treating subjects suspected of developing CRC. Studies aiming to establish such an association between CRC development have great potential in improving the management and prevention of CRC.
In conclusion, following the discovery of H. pylori as an important culprit in the development of gastric cancer [31], proving a mechanistic causative role for F. nucleatum in the development of CRC is forthcoming.
Conflict of Interest
None declared.
Acknowledgements
Financially supported by the research deputy of Tarbiat Modares University, Tehran, Iran. We acknowledge N. Kamali for her technical assistance with preliminary experiments and collecting samples.
References
- 1.Han Y.W. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tateishi F., Hasegawa-Nakamura K., Nakamura T., Oogai Y., Komatsuzawa H., Kawamata K. Detection of Fusobacterium nucleatum in chorionic tissues of high-risk pregnant women. J Clin Periodontol. 2012;39(5):417–424. doi: 10.1111/j.1600-051X.2012.01855.x. [DOI] [PubMed] [Google Scholar]
- 3.Hashemi Goradel N., Heidarzadeh S., Jahangiri S., Farhood B., Mortezaee K., Khanlarkhani N. Fusobacterium nucleatum and colorectal cancer: a mechanistic overview. J Cell Physiol. 2019;234(3):2337–2344. doi: 10.1002/jcp.27250. [DOI] [PubMed] [Google Scholar]
- 4.Mima K., Sukawa Y., Nishihara R., Qian Z.R., Yamauchi M., Inamura K. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan C.A., Garrett W.S. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flanagan L., Schmid J., Ebert M., Soucek P., Kunicka T., Liska V. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 7.Dulal S., Keku T.O. Gut microbiome and colorectal adenomas. Cancer J. 2014;20:225–231. doi: 10.1097/PPO.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mármol I., Sánchez-de-Diego C., Pradilla Dieste A., Cerrada E., Rodriguez Yoldi M.J. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1):197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vipperla K., O’Keefe S.J. Diet, microbiota, and dysbiosis: a ‘recipe’ for colorectal cancer. Food Funct. 2016;7:1731–1740. doi: 10.1039/c5fo01276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 12.Siegel R.L., Torre L.A., Soerjomataram I., Hayes R.B., Bray F., Weber T.K. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. doi: 10.1136/gutjnl-2019-319511. [DOI] [PubMed] [Google Scholar]
- 13.Coppenhagen-Glazer S., Sol A., Abed J., Naor R., Zhang X., Han Y.W. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83(3):1104–1113. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fardini Y., Wang X., Témoin S., Nithianantham S., Lee D., Shoham M. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82(6):1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bashir A., Miskeen A.Y., Bhat A., Fazili K.M., Ganai B.A. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev. 2015;24(5):373–385. doi: 10.1097/CEJ.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 16.Lee S.A., Liu F., Riordan S., Lee C.S., Zhang L. Global investigations of Fusobacterium nucleatum in human colorectal cancer. Front Oncol. 2019;9:566. doi: 10.3389/fonc.2019.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 18.Liu P., Liu Y., Wang J., Guo Y., Zhang Y., Xiao S. Detection of Fusobacterium nucleatum and fadA adhesin gene in patients with orthodontic gingivitis and non-orthodontic periodontal inflammation. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barko P.C., McMichael M.A., Swanson K.S., Williams D.A. The gastrointestinal microbiome: a review. J Vet Intern Med. 2018;32(1):9–25. doi: 10.1111/jvim.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cresci G.A., Bawden E. Gut microbiome: what we do and don’t know. Nutr Clin Pract. 2015;30:734–746. doi: 10.1177/0884533615609899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 22.Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haggar F.A., Boushey R.P. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosho K., Sukawa Y., Adachi Y., Ito M., Mitsuhashi K., Kurihara H. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22(2):557. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suehiro Y., Sakai K., Nishioka M., Hashimoto S., Takami T., Higaki S. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem. 2017;54(1):86–91. doi: 10.1177/0004563216643970. [DOI] [PubMed] [Google Scholar]
- 26.Ito M., Kanno S., Nosho K., Sukawa Y., Mitsuhashi K., Kurihara H. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137(6):1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 27.Tahara T., Yamamoto E., Suzuki H., Maruyama R., Chung W., Garriga J. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolah D.S., Sajadi A., Radmard A.R., Khademi H. Five common cancers in Iran. Arch Iran Med. 2010;13:143–146. [PubMed] [Google Scholar]
- 29.Pourhoseingholi M.A. Increased burden of colorectal cancer in Asia. World J Gastrointest Oncol. 2012;4:68–70. doi: 10.4251/wjgo.v4.i4.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keku T.O., McCoy A.N., Azcarate-Peril A.M. Fusobacterium spp. and colorectal cancer: cause or consequence? Trends Microbiol. 2013;21:506–508. doi: 10.1016/j.tim.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.IARC working group on the evaluation of carcinogenic risk to humans. Schistosomes, liver flukes and Helicobacter pylori. IARC; Lyon: 1994. [PMC free article] [PubMed] [Google Scholar]