Abstract
Immune checkpoint blockade therapies (ICBs) are a prominent breakthrough in cancer immunotherapy in recent years (named the 2013 “Breakthrough of the Year” by the Science magazine). Thus far, FDA-approved ICBs primarily target immune checkpoints CTLA-4, PD-1, and PD-L1. Notwithstanding their impressive long-term therapeutic benefits, their efficacy is limited to a small subset of cancer patients. In addition, ICBs induce inadvertent immune-related adverse events (irAEs) and can be costly for long-term use. To overcome these limitations, two strategies are actively being pursued: identification of predictive biomarkers for clinical response to ICBs and multi-pronged combination therapies. Biomarkers will allow clinicians to practice a precision medicine approach in ICBs (biomarker-based patient selection) such as treating triple-negative breast cancer patients that exhibit PD-L1 staining of tumor-infiltrating immune cells in ≥1% of the tumor area with nanoparticle albumin-bound (nab)–paclitaxel plus anti-PD-L1 and treating patients of MSI-H or MMR deficient unresectable or metastatic solid tumors with pembrolizumab (anti-PD-1). Importantly, the insights gained from these biomarker studies can guide rational combinatorial strategies such as CDK4/6 inhibitor/fractionated radiotherapy/HDACi in conjunction with ICBs to maximize therapeutic benefits. Further, with the rapid technological advents (e.g., ATCT-Seq), we predict more reliable biomarkers will be identified, which in turn will inspire more promising combination therapies.
Keywords: IFN-γ, Immune checkpoint, Microbiota, Microsatellite instability, Neoantigen, PD-L1, Radiotherapy
Introduction
Harnessing the patients' own immune system to attack tumor cells (cancer immunotherapy) is not recent. In 1891, William B. Coley injected live streptococcal organisms into a patient with inoperable cancer to stimulate the patient's own immune system and successfully cured the patient, which was the first documented case of cancer immunotherapy.1 However, due to the lack of understanding of the immune system back then and the rapid development of chemotherapy and radiotherapy in 1950s, immunotherapy has not become a mainstay in cancer care until recently. With the increased understanding of the cancer-immunity cycle,2 it has become evident that immune cells play a key role in constraining malignant cell growth. This dynamic “seek-and-hide game” between immune cells and cancer cells was best described by Schreiber and colleagues in 2002,3 termed “the cancer immuno-editing”, consisting of three stages: elimination, equilibrium, and escape. While detailed mechanisms underlying each stage are not fully understood, a clear realization is that elegantly-coordinated cooperation between the innate and adaptive immunity is indispensable for successful tumor eradication.4 Dysregulation of this process results in tumor escape from immunosurveillance. Among the multiple mechanisms proposed for tumor immune evasion, hijacking the “immune checkpoints” by tumor cells has taken the central stage in recent years, owing to the impressive clinical responses of therapeutic agents targeting “immune checkpoints”. In 2013, the Breakthrough Prize of the Year in Life Sciences was awarded to Jim Allison, in recognition of his pioneering work in this field.
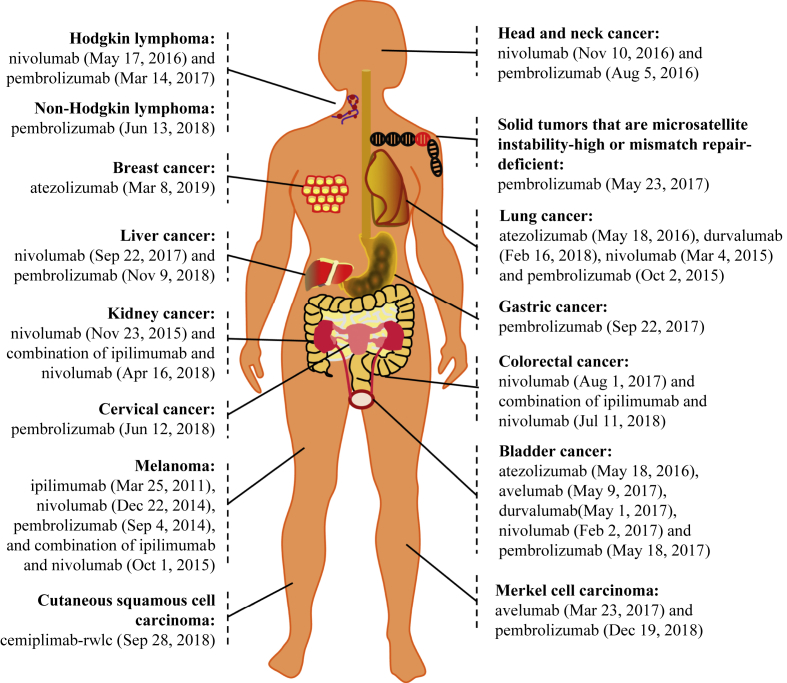
Immune checkpoints are a group of inhibitory molecules that are induced during an active immune response, serving as a negative feedback mechanism to limit collateral tissue damage. CTLA-4 and PD-1 were the first characterized immune checkpoints.5 As aforementioned, tumor cells can coopt CTLA-4 and PD-1 negative signals to evade immunosurveillance. Thus, blocking CTLA-4 and PD-1 with antibodies (collectively known as ICBs) significantly suppresses tumor growth and promotes long-term survival of tumor-bearing mice,6, 7 associated with rejuvenated effector functions of tumor-infiltrating T cells (TILs) including IFN-γ signaling, as demonstrated by us and others.8, 9 In 2011, based on substantially-improved survival of patients in phase III clinical trials,10, 11 the FDA approved the very first ICB, ipilimumab, which is an anti-human CTLA-4 monoclonal antibody, to treat patients with advanced melanoma. This opened a new era in cancer immunotherapy, and since then, anti-PD-1 (pembrolizumab and nivolumab) and anti-PD-L1 (atezolizumab, avelumab, and durvalumab) immunotherapies have also gained FDA approvals to treat patients with various types of cancer (https://www.fda.gov/Drugs), culminating in the awarding of the 2018 Nobel Prize to Jim Allison and Tasaku Honjo. This ever-growing list of ICB approvals is depicted in Fig. 1 with their respective FDA approval times shown.
Figure 1.
The FDA approvals of ICBs for cancer treatment (as of 5/6/2019).
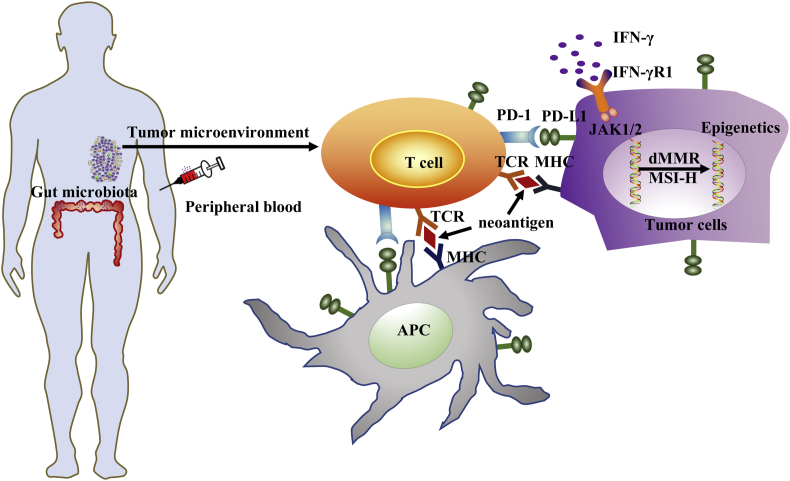
In spite of the impressive clinical response of ICBs, their therapeutic efficacy is generally limited to a minority of cancer patients (∼20–40%),12 except for the Hodgkin's lymphoma that shows ∼80% response rate (discussed below). In addition, ICBs induce inadvertent life-threatening irAEs in some patients and the cost for long-term treatment is high. Therefore, intensive research efforts have been focused on identifying predictive biomarkers for clinical response to ICBs and designing rational combination therapies of ICBs with other therapeutic agents that can synergize with ICBs to augment overall therapeutic efficacy. Here, we provide a focused review on the most-investigated biomarkers (Fig. 2) and discuss combination strategies that are underway or under development.
Figure 2.
The most-investigated predictive biomarkers for clinical response of immune checkpoint blockade therapies (ICBs):1. PD-L1 expression; 2. Tumor-infiltrating immune cells (TIICs); 3. IFN-γ signaling; 4. Neoantigens and tumor mutational burden; 5. Microsatellite instability-high (MSI-H) or mismatch repair (MMR) deficiency; 6. Epigenetics; 7. Peripheral blood biomarkers; 8. Microbiota.
Predictive biomarkers
PD-L1 expression
The PD-1-PD-L1 signaling pathway is an integral component of peripheral tolerance, serving as a rheostat in maintaining immune homeostasis. PD-L1 was successfully cloned in 1996.13 Ligation of PD-L1 to PD-1 inhibits T cell receptor-mediated T cell proliferation and cytokine production14 and prevents exuberant immune responses. PD-L1 is abundantly expressed on tumor cells as well as tumor-infiltrating immune cells (TIICs) including dendritic cells, macrophage and T cells. Since anti-PD-1/L1 therapies primarily counteract PD-L1-mediated negative signals, understandably, PD-L1 expression in the tumor microenvironment (TME) has been the most-studied biomarkers, particularly for anti-PD-1/L1, which are summarized in Table 1.
Table 1.
Clinical trials assessing PD-L1 expression as a predictive biomarker for clinical response to ICBs.
| Therapies | Tumor type | Target cells | Detection Ab | Cut-off | PD-L1+ | PD-L1− | P value | Reference |
|---|---|---|---|---|---|---|---|---|
| Nivolumab | Melanoma, NSCLC, CRC, RCC, prostate cancer | Tumor | 5H1 | 5% | 36% (9/25) | 0% (0/17) | P = 0.006 | 15 |
| Ipilimumab + Nivolumab (concurrent) | Melanoma | Tumor | 28–8 | 5% | 46% (6/13) | 41% (9/22) | P > 0.99 | 39 |
| Ipilimumab + Nivolumab (sequenced) | 50% (4/8) | 8% (1/13) | Unknown | |||||
| Atezolizumab | NSCLC | TILs | SP142 | 1% | 31% (8/26) | 20% (4/20) | P = 0.015 | 34 |
| 5% | 40% (6/13) | 18% (6/33) | ||||||
| 10% | 83% (5/6) | 18% (7/40) | ||||||
| All tumors studied (melanoma, NSCLC and others) | TILs | 1% | 29% (26/90) | 13% (8/60) | P = 0.007 | |||
| 5% | 34% (19/56) | 16% (15/94) | ||||||
| 10% | 46% (15/33) | 16% (19/117) | ||||||
| NSCLC | Tumor | 1% | 25% (3/12) | 26% (9/34) | P = 0.920 | |||
| 5% | 33% (3/9) | 24% (9/37) | ||||||
| 10% | 38% (3/8) | 24% (9/38) | ||||||
| All tumors studied (melanoma, NSCLC and others) | Tumor | 1% | 31% (9/29) | 18% (22/121) | P = 0.079 | |||
| 5% | 39% (7/18) | 18% (24/132) | ||||||
| 10% | 47% (7/15) | 18% (24/135) | ||||||
| Nivolumab | Nonsquamous NSCLC | Tumor | 28–8 | 1% | 31% (28/123) | 9% (10/108) | P = 0.002 | 21 |
| 5% | 36% (34/95) | 10% (14/136) | P = 0.002 | |||||
| 10% | 37% (32/86) | 11% (16/145) | P = 0.002 | |||||
| Nivolumab | Squamous NSCLC | Tumor | 28–8 | 1% | 17% (11/63) | 17% (9/54) | P = 0.9364 | 22 |
| 5% | 21% (9/42) | 15% (11/75) | P = 0.2908 | |||||
| 10% | 19% (7/36) | 16% (13/81) | P = 0.6411 | |||||
| Pembrolizumab | NSCLC | Tumor | 22C3 | 50% | 45% (33/73) | 15% (20/131) | p < 0.01 | 18 |
| 1% | 28% (50/176) | 11% (3/28) | ||||||
| Ipilimumab + Nivolumab | Melanoma | Tumor | 28–8 | 5% | 72% (49/68) | 55% (115/210) | Unknown | 12 |
| Nivolumab | Melanoma | 58% (46/80) | 41% (86/208) | |||||
| Ipilimumab | Melanoma | 21% (16/75) | 18% (36/202) | |||||
| Ipilimumab + Nivolumab | Melanoma | Tumor | SP142 | 5% | 58% (14/24) | 55% (31/56) | Unknown | 25 |
| Ipilimumab | Melanoma | 18% (2/11) | 4% (1/27) | |||||
| Nivolumab | Untreated metastatic melanoma | Tumor | 28–8 | 5% | 53% (39/74) | 33% (45/136) | Unknown | 19 |
| Pembrolizumab | NSCLC | Tumor | 50% | 45% (69/154) | Unknown | Unknown | 24 | |
| Atezolizumab | Urothelial carcinoma | TILs | SP142 | 1% | 22% (45/207) | 13% (13/103) | Unknown | 35 |
| 5% | 27% (27/100) | 15% (31/210) | ||||||
| Nivolumab | NSCLC | Tumor | 28–8 | 5% | 26 (55/211) | Unknown | Unknown | 26 |
| Atezolizumab + Paclitaxel | TNBC | TIICs | 1% | 59 (109/185) | 54 (143/265) | 38 |
Note: Ipilimumab: anti-CTLA-4; Nivolumab, Pembrolizumab: anti-PD-1; Atezolizumab: anti-PD-L1; NSCLC: non-small cell lung cancer; CRC: colorectal cancer; RCC: renal cell carcinoma; TNBC: triple-negative breast cancer; TILs: tumor-infiltration T cells; TIICs: tumor-infiltrating immune cells; percentages (%) indicate rates of objective response.
There is a vast body of work assessing PD-L1 expression in tumor cells as a predictive biomarker to ICBs. However, the results have been splitting with both positive and negative correlations reported. In an initial clinical trial with limited number of patients, Topalian, et al found that while 9 out of 25 patients with PD-L1+ advanced melanoma (36%) showed objective response to anti-PD-1 treatment, none of 17 patients with PD-L1– tumors exhibited objective response, suggesting that PD-L1 expression on tumor cells is a promising predictive biomarker for clinical responses to anti-PD-1 therapy.15 In support of this, it was demonstrated preclinically that PD-L1 expression on tumor cells indicates an immune-active TME, a prerequisite for effective antitumor responses induced by anti-PD-1 treatment.16 Subsequently, favorable clinical response to ICBs was reported in patients with PD-L1+ Hodgkin's lymphoma (reaching 87%),17 non-small-cell lung cancer (NSCLC),18 and melanoma,12, 19 which was further confirmed in expanded large scale analyses of patients with NSCLC.20, 21, 22, 23 Not only were PD-L1+ NSCLCs more responsive to ICBs, more importantly, milder side effects were observed, leading to the approval of pembrolizumab (anti-PD-1) to treat patients with metastatic PD-L1-expressing NSCLC by the FDA.24 It is noteworthy to mention that in most of these studies PD-L1– tumors still responded to ICBs, albeit at lower rate, suggesting that PD-L1 expression in tumor cells is not a definitive biomarker. To this end, another study reported that patients with previously-untreated PD-L1+ melanoma did not show better clinical responses to combined anti-CTLA-4 and anti-PD-1 therapy than those with PD-L1– tumors.25 This was corroborated by a more recent clinical trial of patients with previously-untreated stage IV or recurrent NSCLC after chemotherapy where distinguishable clinical responses of anti-PD-1 in PD-L1+ vs PD-L1– tumors were not observed.26 To reconcile these discrepant results, four factors can be considered: 1. The heterogeneity of PD-L1 expression in the tumor (in clusters) imposing a challenge to obtain a “representative” biopsy that accurately reflects the overall expression of PD-L1.27, 28 2. Detection methods of PD-L1 expression: currently, PD-L1 expression is primarily evaluated by immunohistochemistry (IHC) of formalin-fixed paraffin-embedded (FFPE) tissues, which is highly dependent on the reliability and sensitivity of the detection antibodies29 (Table 1). 3. Different cut-offs of PD-L1 expression (spanning from 1% to 50%) were used; additionally, the intensity of PD-L1 expression per cell need to be assessed, given the important role of tumor cell-intrinsic PD-L1 signaling in tumor cell proliferation30 and metabolism.31 4. Different mechanisms for PD-L1 upregulation: intrinsically, PD-L1 expression can be modulated by key transcriptional factors such as Myc and HIF1α and by epigenetic factors such as EZH2 and DNMT132; extrinsically, IFN-γ induces PD-L1 expression on the tumor cells.13 Considering all these contributing factors and the inconsistent results, we argue that PD-L1 expression on tumor cells is more of a dynamic and inducible biomarker, reflecting a greater likelihood of response rather than a definitive predictor of clinical response to ICBs.
TIICs are essential cellular components of the TME. Many of these cells express PD-L1 and govern anti-tumor response elicited by ICBs.33 It is therefore pertinent to assess if PD-L1 expression on these cells predicts clinical response to ICBs with greater certainty than that of tumor cells. In support of this notion, PD-L1 expression on TIICs, but not tumor cells, was associated with better response to anti-PD-L1 therapy across multiple cancer types.34 This was further confirmed in a separate clinical trial in patients with metastatic urothelial carcinoma treated with anti-PD-L1.35 PD-L1 expression on peripheral T cells was also positively correlated with progression-free and overall survival of patients with metastatic melanoma treated with anti-CTLA-4.36 Importantly, PD-L1 expression in TIICs of any intensity covering ≥1% of the tumor area, detected by the designated staining assay (the VENTANA PD-L1 (SP142) Assay),37 was included in a recent approval by the FDA to treat TNBC patients with nanoparticle albumin-bound (nab)–paclitaxel plus PD-L1 blocking antibody.38 Taken together, these results support that PD-L1 expression on TIICs represents a more predictive biomarker to clinical benefit of ICBs compared to that of tumor cells.
Tumor-infiltrating immune cells (TIICs)
Responsive “hot” tumors to ICBs (e.g., melanoma and NSCLC) contain more TIICs than non-responsive “cold” tumors (e.g., prostate and pancreatic cancer), underscoring the importance of TIICs in predicting the efficaciousness of ICBs. However, absolute lymphocyte count alone was not correlative with clinical responses in cohorts of melanoma patients treated with either concurrent regimen or sequenced regimen of anti-CTLLA-4 and anti-PD-139; rather, the profile of TIICs appeared to be a strong predictor of patient survival in an early study using large cohorts of human colorectal cancer (CRC).40 In this regard, RNA expression of genes associated with T cell activation and function in the TME positively correlated with clinical response to anti-CTLA-4 in patients with metastatic melanoma41 and to anti-PD-L1 in patients with bladder tumors42 and other tumors.34 Besides T cell activation makers, the location of pre-existing CD8+ cells (that is, at the invasive tumor margin) directly correlated with clinical response to anti-PD-1, which was further validated in a separate cohort of 15 patients with metastatic melanoma in the same study.43 This result was confirmed from a different perspective in another study wherein constitutive activation of the WNT/β-catenin signaling in tumor cells mediates therapeutic resistance to ICBs by precluding T-cell infiltration into the tumor bed.44 Collectively, these studies indicate that the immune contexture including functional state and localization of TIICs may predict clinical response of ICBs.45
IFN-γ signaling
One of the essential effector molecules in antitumor immunity is IFN-γ. Pioneering work from the Schreiber group using either IFN-γ blocking antibody46 or mouse models lacking essential IFN-γ signaling molecules47 convincingly demonstrated a pivotal role of IFN-γ signaling in immunosurveillance. Binding of IFN-γ to its receptors (IFNGR1 and IFNGR2) recruits and activates the Janus kinases, JAK1 and JAK2, leading to subsequent phosphorylation, dimerization, and activation of STAT1 (signal transducer and activator of transcription 1). Dimerized STAT1 translocates to the nucleus and modulates transcription of IFN-γ-regulated genes,48 resulting in immune cell activation. As such, we contemplated that ICBs may act through this pathway to exert their therapeutic benefits. Using IFNGR1−/− mice and adoptive T-cell therapy (ACT) approach wherein T cells harvested from the IFNGR1−/− mice were injected into recipient mice bearing palpable tumors, we found that loss of IFNGR1 in T cells completely abrogates the efficaciousness of combined anti-CTLA-4 and anti-PD-1 therapy.9 Furthermore, melanoma patients that were not responsive to anti-CTLA-4 therapy harbor more copy number loss of IFN-γ signaling genes. Consistent with this, knock-down of IFNGR1 in B16 melanoma tumor cells attenuates therapeutic effects of anti-CTLA-4.49 In a separate study, Zaretsky, et al reported that loss-of-function mutations in JAK1 and JAK2 in melanoma patients causes therapeutic resistance to anti-PD-1.50 These results together point to an indispensable role of IFN-γ signaling in determining therapeutic efficacy of ICBs, suggesting that activation of IFN-γ signaling may serve as a predictive biomarker for clinical response to ICBs. To this end, a recent clinical trial of patients with melanoma, head and neck squamous carcinoma, and gastric cancer showed that gene expression profiles (GEPs) including IFN-γ signature genes successfully separate responders from non-responders to anti-PD-1 therapy.51 More recently, another study revealed that Ifng expression correlates with longer progression-free survival of patients with melanoma and NSCLC in response to anti-PD-1.52 Paradoxically, a separate study reported that chronic IFN-γ signaling in the tumor cells induced acquired resistance to ICBs via STAT1-related epigenomic changes.53 In sum, expression of IFN-γ signature genes may predict positive clinical response to ICBs in the early stage, but once resistance is developed as in recurrent tumors persistent IFN-γ signaling may attenuate therapeutic efficacy of ICBs.
Neoantigens and tumor mutational burden (TMB)
Tumor progression is accompanied with acquisition and accumulation of mutations. These mutations can lead to aberrant expression of self-antigens, known as tumor-associated antigens <TAAs>, or expression of sequence-modified proteins, known as tumor-specific neoantigens. Both TAAs and neoantigens can be recognized by the immune system, initiating anti-cancer immune responses. T cells recognizing TAAs and neoantigens have been utilized in ACT, a therapeutic modality utilizing patients' own T cells that are genetically or pharmacologically manipulated, expanded ex vivo, and reinfused into the patients to cure cancer. While ACT of T cells against TAAs has thus far only generated modest clinical effects,54 ACT of T cells recognizing mutant neoantigens such as ERBB2IP in metastatic cholangiocarcinoma55 and PPP1R3B in advanced melanoma56 has exerted potent tumor suppression, indicating a predominant role of immune responses against neoantigens in tumor control. Interestingly, clinical activity of ICBs also relies on immune responses against neoantigens.57 Combining genomics and bioinformatics approaches, Gubin, et al showed that anti-CTLA-4 and anti-PD-1 improve both the quality and magnitude of neoantigen-specific intratumoral T cell responses, which orchestrate tumor rejection.58 This was also observed in a patient with stage IV melanoma whose T cell response against ATR (ataxia telangiectasia and Rad3 related) mutation was greatly expanded by anti-CTLA-4, resulting in objective clinical response.59 Further, patients with advanced NSCLC and melanoma who have enriched clonal neoantigens are more sensitive to anti-PD-1 and anti-CTLA-4 therapies.60
In keeping with an important role of neoantigen in ICBs, tumor mutational burden (TMB) also exhibits positive correlation with clinical responses to ICBs. Melanoma patients with higher TMB showed improved long-term benefits after anti-CTLA-4 therapy.61 Likewise, NSCLC patients with higher TMB were more responsive to anti-PD-1 therapy and had enhanced neoantigen-specific CD8+ T cell reactivity.62 In an expanded analysis of accumulative data from 1638 patients with different types of tumor, high TMB was found to be an independent predictor of clinical response to anti-PD-1/PD-L1 immunotherapy; response rates of cancer patients with high and low TMB were 58% and 20%, respectively; and PFS of high vs low TMB was 12.8 months vs 3.3 months, respectively.63 To pinpoint the role of specific mutations, exploratory efforts revealed that cancer patients with concurrent mutations of TP53 and KRAS64 or BRCA-2 mutation65 had greater clinical benefit to anti-PD-1.
Nevertheless, not all mutations and neoantigens correlate positively with therapeutic benefits of ICBs. Some mutations are not translated into neoantigen; rather, they mediate acquired resistance to ICBs.50 For instances, loss-of-function mutation of PTEN66 and activation of PI3K-AKT67 render tumors resistant to ICBs. Given the essential role of T cells in governing therapeutic effects of ICBs, Heemskerk, et al proposed the concept of cancer antigenome, which states that only those mutations capable of producing T cell-recognizable neoantigens without inducing therapeutic resistance may predict clinical response.68 Using whole-exome cDNA sequencing, Robbins et al successfully identified mutated antigens that can be recognized by adoptively transferred tumor-reactive T cells.69 Subsequently, various other “readable” neoantigens have been reported and summarized as in Table 2.55, 56, 59, 61, 69, 70, 71, 72, 73, 74, 75 To name a few, neoantigen IDH1 (R132H) can be presented by MHC class II and subsequently activates CD4+ TH1 response, which has the potential to predict response to immunotherapy.74 Hydroxysteroid dehydrogenase-like protein 1 (HSDL1L25V) mutation can be recognized by CD8+ T cell initially, but this CD8+ T cell response was then suppressed by immunosuppressive TME,75 suggesting treatment with ICBs may revamp this CD8+ T cell response and eradicate tumors. There are other neoantigens ALMS1, C6ORF89 and FNDC3B identified in two chronic lymphocytic leukemia patients, but their T cell-reactivity awaits to be evaluated.72 Using a neoantigen fitness model that integrates antigen presentation by MHC and effector T cell recognition, Luksza, et al successfully predicted the survival of patients with melanoma treated with anti-CTLA-4 and patients with NSCLC treated with anti-PD-1.76 Moreover, newly-developed deep-sequencing technologies will enable systematic analysis of T cell reactivity in an individual tumor to provide a profile of patient-specific neoantigens. The personalized neoantigen profile may be THE predictive biomarker for ICBs. Given that not all mutations will be translated to neoantigen, TMB can be of some reference value, but will remain as an imperfect biomarker to clinical responses to ICBs.
Table 2.
Neoantigens in anti-tumor immune responses.
| Neoantigens | Functions | Tumor | Recognized by | Reference |
|---|---|---|---|---|
| PPP1R3B | Regulate glycogen synthesis in liver | Melanoma | CD4+ and CD8+ | 56 |
| PPP1R3B and PLEKHM2 | PLEKHM2 related to abnormal localization of lysosomes | Melanoma | CD4+ and CD8+ | 69 |
| ATR | DNA damage sensor | Melanoma | CD4+ and CD8+ | 59 |
| FND3CB,ALMS1, and C6ORF89 | Chronic lymphocytic leukemia | CD8+ | 72 | |
| IDH1 (R132H) | Catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate | Gliomas | CD4+ TH1 | 74 |
| Candidates for each patients | Unknown | Melanoma | CD8+ | 61 |
| ERBB2IP | Regulate ERBB2 function and localization | Metastatic cholangiocarcinoma | CD4+ TH1 | 55 |
| HSDL1 (L25V) | Unknown | Ovarian Cancer | CD8+ | 75 |
| B2M, HLA-A, -B and –C and CASP8 | B2M associated with MHC I heavy chain | CRC and others | CD8+ | 73 |
| MUC16 | Form a protective mucous barrier | Pancreatic cancer | CD8+ | 70 |
| PBRM1 | Related to transcriptional activation of nuclear hormone receptors | Clear cell renal cell carcinoma | Unknown | 71 |
Note: TH1: IFN-γ-producing CD4+ T cells.
Microsatellite instability-high (MSI-H) or mismatch repair (MMR) deficiency
Normal cells rely on the DNA mismatch repair (MMR) system to rectify errors in association with DNA replication. In contrast, many types of cancer cells are defective in MMR, causing accumulation of erroneous genetic sequences that are typically repeated, known as microsatellites, presenting a microsatellite instability-high (MSI-H) phenotype. Mechanistically, epigenetic hyper-methylation of the promoter of MLH1 DNA mismatch repair gene can silence the MLH1 gene, significantly increasing microsatellite instability.77 Le, et al demonstrated for the first time that patients' MMR status faithfully predicts clinical response to anti-PD-1 immunotherapy; the objective response rates of patients with MMR-deficient vs MMR-proficient colorectal cancer (CRC) were 40% (4 of 10 patients) vs 0% (0 of 18 patients). The objective response rate was even higher in patients with other types of MMR-deficient cancer, reaching 78% (5 out of 7 patients).78 Subsequently, the same group conducted an extended clinical trial where they prospectively evaluated patients with different subtypes of MMR-deficient cancers treated with anti-PD-1.79 Remarkably, among these 12 different tumor types, they observed an overall objective radiographic response in 53% of patients, among whom 21% had complete responses. These responses were durable and median progression-free survival and overall survival had not yet been reached by the time that the study was published. Analysis of the peripheral blood T cells from a responding patient (subject 19) revealed a rapid in vivo expansion of neoantigen-specific T cell clones reactive to mutant neopeptides derived from the tumor.79 These data corroborated an early study showing that MSI-H colon cancer was infiltrated with activated CD8+ CTLs and TH1, but these cells were suppressed by multiple immune checkpoints including PD-1, PD-L1, CTLA-4, LAG-3 and IDO, which may explain why ICBs are highly efficient in MSI-H CRC.80 On the basis of these results, FDA approved pembrolizumab (anti-PD-1) to treat patients with unresectable or metastatic solid tumors that are either MSI-H or MMR-deficient, regardless of the tumor type. In addition, another anti-PD-1 antibody (nivolumab) and anti-CTLA-4 antibody (ipilimumab) were approved to treat patients with MSI-H or MMR-deficient CRC. Thus, MSI-H and MMR deficiency are arguably the most successful predictive biomarkers for clinical response to ICBs.
Epigenetic modifications
In addition to genetic alterations (mutations, MMR deficiency, loss of IFN-γ signaling genes, etc.) correlative with tumor sensitivity to ICBs, epigenetic modulations (mainly, DNA methylation and post-translational histone modifications) could also contribute to therapeutic effects of ICBs.66, 81, 82 In support, it was stated that ICB-induced functional rejuvenation of T cells largely depends on the DNA methylation states of exhausted T cells.83 Using assay for transposase-accessible chromatin with sequencing (ATAC-seq), Philip, et al found that tumor-infiltrating antigen-specific CD8+ T cells undergo two discrete chromatin states: a plastic dysfunctional state when loss of T cell functionality can be rejuvenated and a fixed dysfunctional state when T cell dysfunction cannot be rescued.84 Chromatin states are delicately controlled by “writers”, enzymes that introduce epigenetic changes such as DNA methyltransferases (DNMTs) and histone methyltransferase (EZH2), and “erasers”, enzymes that eliminate epigenetic alterations such as histone deacetylase (HDACs). In general, DNA methylation of the cytosine residues in the CpG islands is associated with the closed heterochromatin state and transcriptional repression/silencing; histone acetylation of lysine residues (H3K9, H3K14, H4K5, and H4K16) is associated with open euchromatin state and active gene transcription; with regard to histone methylation, the closed or open chromatin states depend on the extent of methylation, that is, mono-, di-, or tri-methylation: typically, monomethylation of H3K9, H3K2, and H3K79 histone proteins leads to active transcription, whereas trimethylation results in transcriptional repression.
Accordingly, DNMT inhibitors (DNMTi) demethylate the promoters of genes encoding cancer-testis antigens (CTAs), leading to increased expression of CTAs and immunogenicity.85 DNMTi can also induce interferon-stimulated genes (ISGs) and immune signaling in tumor cells by activating the transcription factor IRF7 and augmenting cytosolic dsRNAs derived from endogenous retroviral sequences (ERVs), a phenomenon known as “viral mimicry”.82, 86 Hence, DNMTi can expose hidden immunogenic signals in the tumor cells, which in turn enhance immune signaling in tumor cells. EZH2 is another epigenetic regulator that has well-known immunoregulatory functions in sustaining Treg stability,87 inhibiting TH1 cytokine production,66 driving T cell exhaustion,88 and suppressing NK cell activity.89 Expression of EZH2 in ovarian cancer cells epigenetically repressed production of chemokines CXCL9 and CXCL10 by mediating H3K27me3 (trimethylation of histone H3 lysine 27), which blocked T cell infiltration into the tumor bed.90 Similar effects were observed with expressing polycomb repressive complex 2 (PRC2) (EZH2 is the catalytic subunit of PRC2) in colon cancer.91 In Treg cells, Ezh2 sustains their immunosuppressive activity and prevents tumor-infiltration by CD4+ and CD8+ T cells.92 As such, genetic depletion of EZH2 in Treg led to robust antitumor immunity; pharmacological inhibition of EZH2 in human T cells elicited phenotypic and functional alterations of Treg cells and enhanced cytotoxic activity of Teff cells; and, more importantly, EZH2 inhibition worked in synergy with anti-CTLA-4 therapy to boost overall therapeutic efficacy.93 These findings corroborated a previous study where synergistic effects between EZH2 inhibition and anti-CTLA-4 and IL-2 immunotherapy in suppressing melanoma growth were described.94 The third group of epigenetic regulators are HDACs. Utilization of HDAC inhibitors has led to some clinical successes in the treatment of cutaneous T cell lymphomas.95 A recent study showed that a class I/IV HDAC inhibitor (mocetinostat) upregulated PD-L1 and class I/II human leukocyte antigen (HLA) antigen-presentation genes both in vitro and in vivo. In a syngeneic mouse tumor model, mocetinostat decreased intratumoral Treg cells and MDSCs and concurrently increased intratumoral CD8+ T cells. In ex vivo assays, patient-derived, mocetinostat-treated Treg showed significant downregulation of Foxp3. The combination of mocetinostat and anti-PD-L1 increased anti-tumor activity, as compared to monotherapies in two syngeneic HNSCC models.96 Similarly, a pan-HDAC inhibitor increased acetylation of the PD-L1 gene promoter and upregulated PD-L1 expression in both human and murine melanoma cells lines, and when combined with anti-PD-1, led to more pronounced tumor regression using the B16F10 syngeneic murine melanoma model.97 Given these immunomodulatory effects, epigenetic modulations (DNA methylation, histone methylation and acetylation) may represent putative biomarkers for ICB responsiveness. Targeting epigenetic alterations with inhibitors (DNMTi, EZH2i, and HDACi) may serve as an effective means to overcoming therapeutic resistance to ICBs, as they open up otherwise closed chromatin structures associated with therapeutic resistance.
Peripheral blood biomarkers
While tumor biopsies may directly reflect the TME, they are difficult to obtain sometimes, especially for biopsies post-ICBs. Given this, some studies exploited peripheral blood samples for biomarker identification. Peripheral blood contains DNAs, RNAs and proteins released from tumor tissues, which to some extent reflect the dynamic changes in the TME. For example, the level of circulating tumor DNAs (ctDNAs) in the blood during the early stage of treatment revealed invaluable hints on therapeutic efficacy.98 Detection of hypermutated ctDNAs, as an indicator of the tumor mutational burden (TMB), positively correlated with better clinical responses to ICBs across various cancer types including melanoma, lung cancer, and head and neck cancer.99 Using blood-based cell-free DNA (cfDNA) assay to measure TMB in plasma, another group successfully identified patients with NSCLC that had significant improvement of PFS upon anti-PD-L1 treatment.100 Furthermore, Wang, et al constructed a cancer gene panel comprised of 150 genes (NCC-GP150), based on the plasma levels of ctDNAs. Their results indicated that NCC-GP150 matched well with tissue TMB measured by whole-exosome sequencing. More importantly, patients whose NCC-GP150 detected 6 or more blood-based-TMB exhibited superior response rates to anti-PD-1/L1 therapies.101 With respect to potential use of blood RNAs as predictive biomarkers, a previous report concluded that a four-gene signature including cathepsin D (CTSD), phospholipase A2 group VII (PLA2G7), thioredoxin reductase 1 (TXNRD1), and interleukin 1 receptor–associated kinase 3 (IRAK3) in the blood could predict survival of patients with melanoma treated with anti-CTLA-4 antibody.102 Serum level of protein VEGF was also found to be associated with clinical responses in melanoma patients treated with anti-CTLA.103
Blood cellular components including CD4+ and CD8+ T cells and myeloid cells (monocytes, neutrophils, etc.) have been explored as predictive biomarkers for clinical response to ICBs. Two groups found that increases of central memory CD4+ T cells (CD27+, FAS−, CD45RA−, and CCR7+)104 and IL-9-producing TH9 cells105 correlated with clinical responses of melanoma patients treated with anti-PD-1. Another report detected PD-1+ tumor antigen-specific CD8+ T cells in the peripheral blood of melanoma patients,106 which also existed in peripheral blood of patients responsive to anti-CTLA-4 and anti-PD-1.60 Proliferating Ki67+CD8+ effector-like T cells in the blood were another marker associated with better clinical outcomes of NSCLC patients treated with anti-PD-1-therapy.107 In addition, blood myeloid cells may predict clinical response to ICBs. In patients treated with anti-PD-1 antibody, lower baseline levels of MDSCs were seen in non-relapsing melanoma patients treated with anti-PD-1108; conversely, high numbers of MDSCs were associated with poor survival of melanoma patients treated with anti-PD-1.109 To track down the specific subtypes of MDSCs associated with clinical response of ICBs, Krieg, et al demonstrated that the frequency of CD14+CD16–HLA–DRHi monocytes in the pretreatment blood samples is a strong predictor for progression-free survival and overall survival of patients with stage IV melanoma treated with anti-PD-1 immunotherapy.110 With this key finding, blood CD14+CD16–HLA–DRHi monocytes as a predictive biomarker should be prospectively evaluated in clinical trials. Although peripheral blood analysis of ctDNAs, RNAs, and proteins can provide valuable insights into the clinical responses of cancer patients to ICBs, more investigations are needed before these biomarkers can be applied in the clinic, given the abundance of DNAs and proteins in peripheral blood is usually much lower than that in the TME. More sensitive methods need to be developed for accurate detection of ctDNAs, RNAs, and proteins.
Microbiota
An emerging concept in immunology is that commensal bacteria are not just a mere symbiotic partner living inside our body, but actively shape immune responses, contributing to antitumor immunosurveillance. Accumulating evidence indicates that microbiota affect DC function, T cell trafficking and infiltration into the tumor site, and Treg cell stability.111 The interconnection between microbiota and ICBs was initially suggested in a study where PD-1 deficiency can perturb microbial communities in the gut and mucosal immunity by acting through the axis of follicular helper T cells (TFH)- plasma cells-IgA secretion in the germinal center of Peyer's patches.112 Direct evidence on the importance of microbiota in therapeutic effects of ICBs came from a study showing that fecal transfer of commensal Bifidobacterium bacteria boosted therapeutic effects of anti-PD-L1 by enhancing DC function and subsequent CD8+ T cell priming and accumulation in the TME.113 Interestingly, unlike anti-PD-L1, anti-CTLA-4 required Bacteroides species, but not Bifodobacterium bacteria for its therapeutic efficacy; B. thetaiotaomicron or B. fragilis-specific T cell responses were associated with anti-CTLA-4 efficacy; adoptive transfer of B. fragilis–specific T cells and fecal microbial transplantation from humans to mice confirmed that treatment of melanoma patients with anti-CTLA-4 favored the outgrowth of B. fragilis with anticancer properties.114 Consistent with these preclinical results, the reliance of therapeutic effects of ICBs on specific commensal bacteria was also observed in cancer patients. In a clinical trial of melanoma patients treated with anti-PD-1, oral and fecal microbiome samples were prospectively collected for taxonomic profiling using 16S ribosomal RNA (rRNA) gene sequencing and metagenomic whole-genome shotgun (WGS) sequencing. Significantly higher alpha diversity and greater abundance of the Ruminococcaceae and Faecalibacterium were observed in responders; in contrast, low alpha diversity and high relative abundance of Bacteroidales were seen in non-responders.115 Perplexingly, a separate study revealed a different spectrum of commensal bacteria (Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium) abundantly present in the fecal samples of melanoma patients responding to anti-PD-1 therapy; and reconstitution of germ-free mice with fecal material from responding patients led to improved tumor control, augmented T cell responses, and greater efficacy of anti–PD-L1 therapy.116 The underlying reasons for this discrepancy are currently unknown, which may be due to the different methods of sampling and sequencing, different experimental designs, and different patient populations/diets in these clinical trials. In addition to melanoma, correlation of microbiome with clinical response to anti-PD-1 was observed in patients with advanced NSCLC, RCC, or urothelial carcinoma treated with anti-PD-1/PD-L1 antibodies117; metagenomic analyses of patient stool samples at diagnosis revealed correlations between clinical response to ICBs and the relative abundance of Akkermansia muciniphila. Consistent with previous reports, fecal microbiota transplantation (FMT) from responders into germ-free or antibiotic-treated mice restored the antitumor effects of PD-1 blockade. More importantly, supplementation of A. muciniphila to the FMT with non-responders’ feces can restore the efficaciousness of PD-1 blockade in an IL-12-dependent manner by enhancing the recruitment of CCR9–CXCR3+CD4+ T cells into the tumor bed.117 Taken together, the baseline profiles of commensal bacteria may predict clinical response to ICBs: Bacteroides thetaiotaomicron or B. fragilis in anti-CTLA-4; Bifidobacterium bacteria in anti-PD-L1; and Ruminococcaceae + Faecalibacterium, or Bifidobacterium longum + Collinsella aerofaciens + Enterococcus faecium, or Akkermansia muciniphila in anti-PD-1. Given this key information, two pertinent questions to address are how to nutritionally cultivate “good” endogenous commensal bacteria and how to utilize exogenously-expanded microorganisms as an immune-adjuvant to boost therapeutic efficacy of ICBs.
Combination strategies to boost therapeutic efficacy of ICBs
Although the “perfect” biomarkers await to be identified, the above-described seminal work provides promising leads for future explorations. Considering the complexity of the TME wherein the dynamic tug-of-war between host cells (including immune cells) and tumor cells takes place, we propose an interconnected network of multiple factors, rather than individual factors in isolation would be the eventual biomarker, following the philosophy “the whole is greater than the sum of its parts”. As such, we contemplate that therapies imparting multiple biomarkers would work in stronger synergy with ICBs to boost the overall therapeutic efficacy. One good example is cyclophosphamide, a commonly-used platinum-based chemotherapy. When combined with anti-PD-1, cyclophosphamide promotes tumor antigen-specific immunity by inducing immunogenic cell death and release of tumor antigens and DAMPs (danger-associated molecular patterns), decreasing immunosuppressive immune cells (Treg), and increasing tumor-infiltration of CTLs and non-Treg CD4+ T cells, leading to improved tumor-free survival of mice bearing cervical cancer.118 Several other chemotherapeutic agents with multifaceted immunogenic effects also synergize effectively with ICBs to drive tumor eradication. Readers interested in the combinatorial therapies of chemotherapy and ICBs are encouraged to read excellent reviews on this.119, 120 Here, we discuss the most recent developments of combinatorial therapies, including small molecule inhibitors, metabolic inhibitors, and radiotherapies, in conjunction with ICBs.
In this era of precision medicine, small molecule inhibitors have been developed to target specific pathways that are mutated in tumor cells, including BRAFi/MEKi, imatinib (an antagonistic agent against BCR-ABL tyrosine kinase), and CDK4/6 inhibitor (abemaciclib). Interestingly, in addition to inducing tumor cell death, these inhibitors also exerted potent multifaceted immunomodulatory impacts on both innate and adaptive immune cells. For example, administration of abemaciclib to mice bearing CRC resulted in endogenous retroviral gene/dsRNA response, anti-viral effector function, and significant downregulation of immunosuppressive mechanisms (Treg, expression of PD-1, TIM-3, CTLA-4, and LAG-3, etc.).121 Interestingly, it also increased PD-L1 expression, suggesting addition of anti-PD-2 to abemaciclib treatment would further enhance therapeutic efficacy. Indeed, combination therapy of abemaciclib and anti-PD-1 greatly enhanced tumor regression and improved OS rates in mice bearing breast cancers.122 Similarly, greater therapeutic benefits have been reported for combination therapies of BRAFi/MEKi + ICBs in patients with BRAFV600E mutated melanoma,123, 124 and imatinib + anti-CTLA-4 in patients with advanced malignancies.125
One unusual metabolic aspect of tumor cells is that they rewire their metabolic machinery to engage glycolysis (a rather inefficient energy-producing pathway compared to oxidative phosphorylation), even in the presence of ample oxygen, the well-known Warburg effect.126 This metabolic reprogramming of tumor cells produces substantial amount of immune-suppressive metabolites such as lactate, adenosine, and kynurenine that hinder immune cell infiltration into the tumor bed, interfering immune surveillance and leading to therapeutic resistance to ICBs. Multiple metabolic pathways have been shown to contribute to immunotherapeutic resistance, including PI3K-Akt-mTOR, CD73-CD39 adenosinergic pathway, IFN-γ-JAK/STAT, and Wnt/β-catenin. To overcome the therapeutic resistance, inhibitors against these metabolic regulators have been actively tested in conjunction with ICBs. However, provided the important roles of these metabolic pathways in mediating immune cells functions,127, 128 special considerations should be given to when (relative to the onset of ICBs) and how (locoregional or systemic) these inhibitors should be administered. Among the four PI3K isoforms (α, β, δ, and γ), PI3Kδ may represent an interesting target, in light of its indispensable role in Treg survival and proliferation but dispensable in non-Treg cells. Coadministration of a PI3Kδ-specific inhibitor (CAL101) with a tumor-specific vaccine to mice-bearing lung cancer significantly decreased Treg and increased vaccine-induced CD8+ T cells in the TME, inducing potent antitumor efficacy.129 In preclinical MC38-OVA (colon) and RM-1 (prostate) subcutaneous tumor models, concomitant blockade of CD73 with ICBs has also shown potent synergistic effects.130 As discussed above (the IFN-γ signaling section), the IFN-γ-JAK-STAT axis has dual functions in anti-tumor immunity: anti-tumorigenic during the early stage and pro-tumorigenic at the late stage when acquired resistance develops. Thus, prudent temporal administration of JAKi in relation to ICBs is of critical importance to achieve the optimal outcome.49, 50, 53 WNT inhibitors (PKF115-58, IWP-L6, and XAV939) have yielded promising immunogenic effects in mouse models of melanoma and lymphoma, justifying their use in combination with ICBs in cancer therapy.44, 131 Clinical testing of some these combinatorial therapies is underway and results will be available in the future (https://clinicaltrials.gov). On a further note, with the availability of advanced sequencing technologies, personalized combination therapy based on specific mutations in the patient may become feasible in the near future.
A cardinal feature of the TME (especially in solid tumors) is hypoxia, which is primarily regulated by HIF1α (hypoxia-inducible factor-1α). Hypoxia induces therapeutic resistance through downregulation of MHC class-I molecules,132 upregulation of immune checkpoints such as CTLA-4 and PD-L1 on tumor cells,133 and impairment of DNA damage response proteins (leading to more aggressive phenotype).134, 135 Intriguingly, many of these immunosuppressive effects can be reversed by radiotherapy (RI), particularly fractionated RT (leading to reoxygenation). Beyond direct cytotoxic effects, RT may enhance anti-tumor immune responses via releasing tumor antigens and DAMPs, increasing MHC-I expression on cancer cells,136 priming and mediating maturation of antigen-presenting cells,137 driving T cell infiltration to irradiated tumors,138 repolarizing tumor-associated macrophages to be inflammatory M1 macrophages,139 and activating natural killer cells.140 Of note, RT also leads to upregulation of PD-L1141 and accumulation of Treg142 in irradiated tumors, justifying combination therapy of fractionated RT, anti-CTLA-4 (efficiently depleting intratumoral Treg) and anti-PD-1/L1 (blocking increased PD-L1 signaling) to treat hypoxic tumors.143 This strategy was applied in a recent study with both mouse tumor models and patients with melanoma, resulting in optimal clinical response and immunity.144 Additional supporting evidence came from clinical trials of combination of RT (palliative or ablative) with single ICBs (anti-CTLA-4, anti-PD-1, and anti-PD-L1), which showed prolonged progression-free survival rates and increased overall survival.145, 146, 147, 148, 149, 150 In spite of these promising clinical outcomes, details on timing, dosage, and fractionation of RT awaits further elaboration to achieve the optimal efficacy when used in combination with ICBs. Moreover, specific mechanisms underlying the abscopal effects of these combination strategies need to be identified, which may manifest novel therapeutic targets. Considering the largely undistinguished killing of tumor cells by RT, combined RT and ICBs may represent an effective approach to overcome therapeutic resistance of some tumors.49, 50
Additionally, clinical successes with anti-CTLA-4 and anti-PD-1/L1 inspired research endeavors to characterize and target other co-inhibitory molecules including TIGIT (T cell immunoreceptor with Ig and ITIM domains),151 LAG-3 (Lymphocyte-activation gene 3),152 and TIM-3 (T-cell immunoglobulin and mucin domain-containing protein 3).153 Combination therapies by blocking multiple immune checkpoints are being tested in the clinic with promising results available from some of these clinical trials. Thus far, the best outcome of the combination therapies of ICBs is from dual blockade of CTLA-4 and PD-1, which significantly improves the survival of patients with advanced melanoma.12, 25, 39 One of the rationales to combine anti-CTLA-4 with anti-PD-1 is that anti-CTLA-4 upregulated PD-L19 and additional blockade with anti-PD-1 effectively annihilated this inhibitory signal, augmenting efficacy. HDACi or other epigenetic modulators can open closed chromatin structures and convert “cold” tumors to “hot” ones, which may also sensitize them to ICBs.154, 155 Inspired by the exciting findings on the role of microbiome in orchestrating therapeutic efficacy of ICBs, feces transplantation from responding patients and dietary supplementation boosting growth of beneficial bacterial genera may prove to be effective approaches to maximize clinical benefits of ICBs. With the identification of personalized neoantigen becoming feasible, ACT with T cells against these neoantigens in combination with ICBs is another promising approach.156 All in all, our improved understanding of how ICBs function and recent developments in precision medicine and RT open doors to new opportunities to develop combinatorial approaches with enhanced efficacy. Reliable biomarkers that responsively reflect the dynamics of the immune response, be it in isolation (for specific tumor types and specific therapies) or in combination, will provide guidance to select patients for those mechanistically-justified combination strategies.
Concluding remarks
With the burgeoning applications of ICBs in cancer treatment, a clinical concern would be “leaving some patients who may respond to ICBs behind”. In the meantime, inappropriate or overtreatment of patients with ICBs should be avoided, considering their limited efficacy to a minor subset of patients with cancer, inadvertent toxicity (some can be life-threatening) and high financial cost. This conundrum can be solved with the identification of reliable predictive biomarkers of clinical responses to ICBs and rational combination therapies. As far as the biomarker development is concerned, concerted efforts from onco-immunologists have already resulted in successes, evidenced by the incorporation of some biomarkers in the clinical decision-making (as discussed: PD-L1 staining of TIICs in ≥1% of the tumor area by the SP142 Assay as a pre-condition for anti-PD-1 therapy of TNBC patients37 and MSI-H/MMR deficiency in solid tumors (regardless of tumor types) for anti-PD-1 therapy). However, much more work needs to be done to improve the predictability of biomarkers. With the rapid advances in both immunology (e.g., immunometabolism, microbiota, etc.) and biotechnology (deep-sequencing, ATAC-seq, imaging tracer, etc.), “better” and more reliable biomarkers will become available. For example, utilization of radiolabeled high-affinity PD-1 as a PET imaging tracer would provide an accurate evaluation of otherwise highly heterogeneous PD-L1 expression within the tumor. Further, this is a noninvasive method and easier for cancer patients to accept.157 On the same token, with the neoantigen fitness model capable of predicting antigen presentation by MHC molecules and recognition by T cells and deep sequencing technology, “actual” immunogenic neoantigens can be discovered as more reliable biomarkers.76 Furthermore, the majority of work so far has been done with analysis of samples collected at a single static time point (mostly, pre-treatment baseline), missing essential information on the dynamic responses to ICBs, which can be gained by implementing longitudinal tumor sample collection and analysis. Doing this can detect molecular determinants after the first round of ICB treatment that better reflect the clinical response and irAEs.158 While it may be easier said than done, with the advancement of imaging technology using radio-labeled tracers, it is doable. In addition, most results have been generated through retrospective analyses and in some cases with limited number of patients, which should be confirmed prospectively in clinical trials with more patients.
With all the insights gained through the seminal work on predictive biomarkers, substantial efforts to combine agents that impact multiple biomarkers with ICBs are underway, in order to augment overall therapeutic efficacy. Of note, the majority of the therapeutic agents (small molecule inhibitors, metabolic inhibitors, radiotherapies, etc.) exerted therapeutic effects at the start, but eventually lost their efficaciousness. This may be ascribed to their immunostimulatory effects in the early stage, the “Yang” impact, and subsequent immunosuppressive effects when therapeutic resistance develops, the “Yin” counter-balance mechanism. Several stereotypical examples are MEK4/6 inhibitors, RT, and HDACi. Adding ICBs to these therapies will not only overcome the “Yin” counteractive responses (e.g. upregulation of PD-L1 and increase of Treg) but also further sustain and boost the short-lived “Yang” immunostimulatory effects, tipping the “Yin-Yang” balance to the benefit of tumor eradication. Deeper understanding of the underlying mechanisms of these different therapies will allow a more rational design of combination therapies. Considering the dynamic of induced immune responses, timing and dosing are the keys to achieving successful outcomes.
Conflict of interest
A consensus has been reached among authors on the content of this review. All authors declare no conflict of interest.
Acknowledgements
Work in our lab has been funded by the V Foundation Scholar Award (V2018-023), ACS-IRG (91-022-19), and R21 (1R21CA230475-01A1) to L.S. All authors contributed to writing. L.S. was responsible for the overall construction and final editing of this manuscript.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Coley W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. Clin Orthop Relat Res. 1893;1991(262):3–11. [PubMed] [Google Scholar]
- 2.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 5.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Canc. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwai Y., Terawaki S., Honjo T. PD-1 blockade inhibits hematogenous spread of poorly immunogenic tumor cells by enhanced recruitment of effector T cells. Int Immunol. 2005;17(2):133–144. doi: 10.1093/intimm/dxh194. [DOI] [PubMed] [Google Scholar]
- 7.Leach D.R., Krummel M.F., Allison J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 8.Curran M.A., Montalvo W., Yagita H., Allison J.P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107(9):4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi L.Z., Fu T., Guan B. Interdependent IL-7 and IFN-gamma signalling in T-cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nat Commun. 2016;7:12335. doi: 10.1038/ncomms12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodi F.S., O'Day S.J., McDermott D.F. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robert C., Thomas L., Bondarenko I. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364(26):2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 12.Larkin J., Chiarion-Sileni V., Gonzalez R. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H., Strome S.E., Salomao D.R. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 14.Freeman G.J., Long A.J., Iwai Y. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian S.L., Hodi F.S., Brahmer J.R. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taube J.M., Klein A., Brahmer J.R. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansell S.M., Lesokhin A.M., Borrello I. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372(4):311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garon E.B., Rizvi N.A., Hui R. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 19.Robert C., Long G.V., Brady B. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 20.Aguiar P.N., Jr., De Mello R.A., Hall P., Tadokoro H., Lima Lopes G. PD-L1 expression as a predictive biomarker in advanced non-small-cell lung cancer: updated survival data. Immunotherapy. 2017;9(6):499–506. doi: 10.2217/imt-2016-0150. [DOI] [PubMed] [Google Scholar]
- 21.Borghaei H., Paz-Ares L., Horn L. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brahmer J., Reckamp K.L., Baas P. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passiglia F., Bronte G., Bazan V. PD-L1 expression as predictive biomarker in patients with NSCLC: a pooled analysis. Oncotarget. 2016;7(15):19738–19747. doi: 10.18632/oncotarget.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reck M., Rodriguez-Abreu D., Robinson A.G. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 25.Postow M.A., Chesney J., Pavlick A.C. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372(21):2006–2017. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbone D.P., Reck M., Paz-Ares L. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluger H.M., Zito C.R., Barr M.L. Characterization of PD-L1 expression and associated T-cell infiltrates in metastatic melanoma samples from variable anatomic sites. Clin Cancer Res. 2015;21(13):3052–3060. doi: 10.1158/1078-0432.CCR-14-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taube J.M., Anders R.A., Young G.D. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra137. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunshine J.C., Nguyen P.L., Kaunitz G.J. PD-L1 expression in melanoma: a quantitative immunohistochemical antibody comparison. Clin Cancer Res. 2017;23(16):4938–4944. doi: 10.1158/1078-0432.CCR-16-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark C.A., Gupta H.B., Sareddy G. Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 2016;76(23):6964–6974. doi: 10.1158/0008-5472.CAN-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C.H., Curtis J.D., Maggi L.B., Jr. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Jiang C.C., Jin L., Zhang X.D. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–416. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 33.Seidel J.A., Otsuka A., Kabashima K. Anti-PD-1 and anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and limitations. Front Oncol. 2018;8:86. doi: 10.3389/fonc.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbst R.S., Soria J.C., Kowanetz M. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg J.E., Hoffman-Censits J., Powles T. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacquelot N., Roberti M.P., Enot D.P. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun. 2017;8(1):592. doi: 10.1038/s41467-017-00608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rimm D.L., Han G., Taube J.M. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol. 2017;3(8):1051–1058. doi: 10.1001/jamaoncol.2017.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid P., Adams S., Rugo H.S. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 39.Wolchok J.D., Kluger H., Callahan M.K. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galon J., Costes A., Sanchez-Cabo F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 41.Van Allen E.M., Miao D., Schilling B. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powles T., Eder J.P., Fine G.D. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 43.Tumeh P.C., Harview C.L., Yearley J.H. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 45.Fridman W.H., Zitvogel L., Sautes-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 46.Dighe A.S., Richards E., Old L.J., Schreiber R.D. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity. 1994;1(6):447–456. doi: 10.1016/1074-7613(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 47.Kaplan D.H., Shankaran V., Dighe A.S. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95(13):7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda H., Old L.J., Schreiber R.D. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. doi: 10.1016/s1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 49.Gao J., Shi L.Z., Zhao H. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404. doi: 10.1016/j.cell.2016.08.069. e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaretsky J.M., Garcia-Diaz A., Shin D.S. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayers M., Lunceford J., Nebozhyn M. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Investig. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karachaliou N., Gonzalez-Cao M., Crespo G. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol. 2018;10 doi: 10.1177/1758834017749748. 1758834017749748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benci J.L., Xu B., Qiu Y. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554. doi: 10.1016/j.cell.2016.11.022. e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson L.A., Morgan R.A., Dudley M.E. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran E., Turcotte S., Gros A. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y.C., Yao X., Li Y.F. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol. 2013;190(12):6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 58.Gubin M.M., Zhang X., Schuster H. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Rooij N., van Buuren M.M., Philips D. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol – Off J Am Soc Clin Oncol. 2013;31(32):e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGranahan N., Furness A.J., Rosenthal R. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder A., Makarov V., Merghoub T. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rizvi N.A., Hellmann M.D., Snyder A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodman A.M., Kato S., Bazhenova L. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong Z.Y., Zhong W.Z., Zhang X.C. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 65.Hugo W., Zaretsky J.M., Sun L. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng W., Chen J.Q., Liu C. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.George S., Miao D., Demetri G.D. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46(2):197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heemskerk B., Kvistborg P., Schumacher T.N. The cancer antigenome. Embo j. 2013;32(2):194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robbins P.F., Lu Y.C., El-Gamil M. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balachandran V.P., Luksza M., Zhao J.N. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao D., Margolis C.A., Gao W. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajasagi M., Shukla S.A., Fritsch E.F. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schumacher T., Bunse L., Pusch S. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 75.Wick D.A., Webb J.R., Nielsen J.S. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20(5):1125–1134. doi: 10.1158/1078-0432.CCR-13-2147. [DOI] [PubMed] [Google Scholar]
- 76.Luksza M., Riaz N., Makarov V. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudley J.C., Lin M.T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22(4):813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 78.Le D.T., Uram J.N., Wang H. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le D.T., Durham J.N., Smith K.N. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Llosa N.J., Cruise M., Tam A. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghoneim H.E., Fan Y., Moustaki A. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1):142–157. doi: 10.1016/j.cell.2017.06.007. e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiappinelli K.B., Zahnow C.A., Ahuja N., Baylin S.B. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 2016;76(7):1683–1689. doi: 10.1158/0008-5472.CAN-15-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Philip M., Fairchild L., Sun L. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545(7655):452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.James S.R., Link P.A., Karpf A.R. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25(52):6975–6985. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 86.Roulois D., Loo Yau H., Singhania R. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DuPage M., Chopra G., Quiros J. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity. 2015;42(2):227–238. doi: 10.1016/j.immuni.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobenecker M.W., Park J.S., Marcello J. Signaling function of PRC2 is essential for TCR-driven T cell responses. J Exp Med. 2018;215(4):1101–1113. doi: 10.1084/jem.20170084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin J., Leavenworth J.W., Li Y. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci U S A. 2015;112(52):15988–15993. doi: 10.1073/pnas.1521740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peng D., Kryczek I., Nagarsheth N. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nagarsheth N., Peng D., Kryczek I. PRC2 epigenetically silences Th1-type chemokines to suppress effector T-cell trafficking in colon cancer. Cancer Res. 2016;76(2):275–282. doi: 10.1158/0008-5472.CAN-15-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang D., Quiros J., Mahuron K. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep. 2018;23(11):3262–3274. doi: 10.1016/j.celrep.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goswami S., Apostolou I., Zhang J. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J Clin Investig. 2018;128(9):3813–3818. doi: 10.1172/JCI99760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zingg D., Arenas-Ramirez N., Sahin D. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 2017;20(4):854–867. doi: 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 95.Whittaker S.J., Demierre M.F., Kim E.J. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol : Off J Am Soc Clin Oncol. 2010;28(29):4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 96.Briere D., Sudhakar N., Woods D.M. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol Immunother. 2018;67(3):381–392. doi: 10.1007/s00262-017-2091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woods D.M., Sodre A.L., Villagra A., Sarnaik A., Sotomayor E.M., Weber J. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Canc Immunol Res. 2015;3(12):1375–1385. doi: 10.1158/2326-6066.CIR-15-0077-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cabel L., Riva F., Servois V. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017;28(8):1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 99.Khagi Y., Goodman A.M., Daniels G.A. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res. 2017;23(19):5729–5736. doi: 10.1158/1078-0432.CCR-17-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gandara D.R., Paul S.M., Kowanetz M. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]
- 101.Wang Z., Duan J., Cai S. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5(5):696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saenger Y., Magidson J., Liaw B. Blood mRNA expression profiling predicts survival in patients treated with tremelimumab. Clin Cancer Res. 2014;20(12):3310–3318. doi: 10.1158/1078-0432.CCR-13-2906. [DOI] [PubMed] [Google Scholar]
- 103.Yuan J., Zhou J., Dong Z. Pretreatment serum VEGF is associated with clinical response and overall survival in advanced melanoma patients treated with ipilimumab. Canc Immunol Res. 2014;2(2):127–132. doi: 10.1158/2326-6066.CIR-13-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takeuchi Y., Tanemura A., Tada Y., Katayama I., Kumanogoh A., Nishikawa H. Clinical response to PD-1 blockade correlates with a sub-fraction of peripheral central memory CD4+ T cells in patients with malignant melanoma. Int Immunol. 2018;30(1):13–22. doi: 10.1093/intimm/dxx073. [DOI] [PubMed] [Google Scholar]
- 105.Nonomura Y., Otsuka A., Nakashima C. Peripheral blood Th9 cells are a possible pharmacodynamic biomarker of nivolumab treatment efficacy in metastatic melanoma patients. Oncoimmunology. 2016;5(12) doi: 10.1080/2162402X.2016.1248327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gros A., Parkhurst M.R., Tran E. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22(4):433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kamphorst A.O., Pillai R.N., Yang S. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A. 2017;114(19):4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gibney G.T., Kudchadkar R.R., DeConti R.C. Safety, correlative markers, and clinical results of adjuvant nivolumab in combination with vaccine in resected high-risk metastatic melanoma. Clin Cancer Res. 2015;21(4):712–720. doi: 10.1158/1078-0432.CCR-14-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weber J., Gibney G., Kudchadkar R. Phase I/II study of metastatic melanoma patients treated with nivolumab who had progressed after ipilimumab. Canc Immunol Res. 2016;4(4):345–353. doi: 10.1158/2326-6066.CIR-15-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krieg C., Nowicka M., Guglietta S. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24(2):144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 111.Routy B., Gopalakrishnan V., Daillere R., Zitvogel L., Wargo J.A., Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 112.Kawamoto S., Tran T.H., Maruya M. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336(6080):485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 113.Sivan A., Corrales L., Hubert N. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vetizou M., Pitt J.M., Daillere R. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gopalakrishnan V., Spencer C.N., Nezi L. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Matson V., Fessler J., Bao R. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Routy B., Le Chatelier E., Derosa L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 118.Mkrtichyan M., Najjar Y.G., Raulfs E.C. Anti-PD-1 synergizes with cyclophosphamide to induce potent anti-tumor vaccine effects through novel mechanisms. Eur J Immunol. 2011;41(10):2977–2986. doi: 10.1002/eji.201141639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Longo V., Brunetti O., Azzariti A. Strategies to improve cancer immune checkpoint inhibitors efficacy, other than abscopal effect: a systematic review. Cancers (Basel) 2019;11(4) doi: 10.3390/cancers11040539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Emens L.A., Middleton G. The interplay of immunotherapy and chemotherapy: harnessing potential synergies. Canc Immunol Res. 2015;3(5):436–443. doi: 10.1158/2326-6066.CIR-15-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goel S., DeCristo M.J., Watt A.C. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J., Bu X., Wang H. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Homet Moreno B., Mok S., Comin-Anduix B., Hu-Lieskovan S., Ribas A. Combined treatment with dabrafenib and trametinib with immune-stimulating antibodies for BRAF mutant melanoma. Oncoimmunology. 2016;5(7) doi: 10.1080/2162402X.2015.1052212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ribas A., Gonzalez R., Pavlick A. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol. 2014;15(9):954–965. doi: 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 125.Reilley M.J., Bailey A., Subbiah V. Phase I clinical trial of combination imatinib and ipilimumab in patients with advanced malignancies. J Immunother Cancer. 2017;5:35. doi: 10.1186/s40425-017-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 127.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12(5):325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen H., Shi L.Z. Metabolic regulation of TH17 cells. Mol Immunol. 2019;109:81–87. doi: 10.1016/j.molimm.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ahmad S., Abu-Eid R., Shrimali R. Differential PI3Kdelta signaling in CD4(+) T-cell subsets enables selective targeting of T regulatory cells to enhance cancer immunotherapy. Cancer Res. 2017;77(8):1892–1904. doi: 10.1158/0008-5472.CAN-16-1839. [DOI] [PubMed] [Google Scholar]
- 130.Allard B., Pommey S., Smyth M.J., Stagg J. Targeting CD73 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin Cancer Res. 2013;19(20):5626–5635. doi: 10.1158/1078-0432.CCR-13-0545. [DOI] [PubMed] [Google Scholar]
- 131.Hong Y., Manoharan I., Suryawanshi A. beta-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res. 2015;75(4):656–665. doi: 10.1158/0008-5472.CAN-14-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sethumadhavan S., Silva M., Philbrook P. Hypoxia and hypoxia-inducible factor (HIF) downregulate antigen-presenting MHC class I molecules limiting tumor cell recognition by T cells. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Noman M.Z., Desantis G., Janji B. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211(5):781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bindra R.S., Glazer P.M. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26(14):2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 135.Meng A.X., Jalali F., Cuddihy A. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol. 2005;76(2):168–176. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 136.Reits E.A., Hodge J.W., Herberts C.A. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J Exp Med. 2006;203(5):1259–1271. doi: 10.1084/jem.20052494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Demaria S., Coleman C.N., Formenti S.C. Radiotherapy: changing the game in immunotherapy. Trends Canc. 2016;2(6):286–294. doi: 10.1016/j.trecan.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dovedi S.J., Cheadle E.J., Popple A.L. Fractionated radiation therapy stimulates antitumor immunity mediated by both resident and infiltrating polyclonal T-cell populations when combined with PD-1 blockade. Clin Cancer Res. 2017;23(18):5514–5526. doi: 10.1158/1078-0432.CCR-16-1673. [DOI] [PubMed] [Google Scholar]
- 139.Klug F., Prakash H., Huber P.E. Low-dose irradiation programs macrophage differentiation to an iNOS(+)/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. 2013;24(5):589–602. doi: 10.1016/j.ccr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 140.Canter R.J., Grossenbacher S.K., Foltz J.A. Radiotherapy enhances natural killer cell cytotoxicity and localization in pre-clinical canine sarcomas and first-in-dog clinical trial. J Immunother Cancer. 2017;5(1):98. doi: 10.1186/s40425-017-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shen M.J., Xu L.J., Yang L. Radiation alters PD-L1/NKG2D ligand levels in lung cancer cells and leads to immune escape from NK cell cytotoxicity via IL-6-MEK/Erk signaling pathway. Oncotarget. 2017;8(46):80506–80520. doi: 10.18632/oncotarget.19193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Muroyama Y., Nirschl T.R., Kochel C.M. Stereotactic radiotherapy increases functionally suppressive regulatory T cells in the tumor microenvironment. Canc Immunol Res. 2017;5(11):992–1004. doi: 10.1158/2326-6066.CIR-17-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shahabi V., Postow M.A., Tuck D., Wolchok J.D. Immune-priming of the tumor microenvironment by radiotherapy: rationale for combination with immunotherapy to improve anticancer efficacy. Am J Clin Oncol. 2015;38(1):90–97. doi: 10.1097/COC.0b013e3182868ec8. [DOI] [PubMed] [Google Scholar]
- 144.Twyman-Saint Victor C., Rech A.J., Maity A. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tang C., Welsh J.W., de Groot P. Ipilimumab with stereotactic ablative radiation therapy: phase I results and immunologic correlates from peripheral T cells. Clin Cancer Res. 2017;23(6):1388–1396. doi: 10.1158/1078-0432.CCR-16-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Antonia S.J., Villegas A., Daniel D. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377(20):1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 147.Antonia S.J., Villegas A., Daniel D. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med. 2018;379(24):2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 148.Luke J.J., Lemons J.M., Karrison T.G. Safety and clinical activity of pembrolizumab and multisite stereotactic body radiotherapy in patients with advanced solid tumors. J Clin Oncol – Off J Am Soc Clin Oncol. 2018;36(16):1611–1618. doi: 10.1200/JCO.2017.76.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hiniker S.M., Reddy S.A., Maecker H.T. A prospective clinical trial combining radiation therapy with systemic immunotherapy in metastatic melanoma. Int J Radiat Oncol Biol Phys. 2016;96(3):578–588. doi: 10.1016/j.ijrobp.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Postow M.A., Callahan M.K., Barker C.A. Immunologic correlates of the abscopal effect in a patient with melanoma. N Engl J Med. 2012;366(10):925–931. doi: 10.1056/NEJMoa1112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu X., Harden K., Gonzalez L.C. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10(1):48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 152.Melero I., Berman D.M., Aznar M.A., Korman A.J., Perez Gracia J.L., Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Canc. 2015;15(8):457–472. doi: 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
- 153.Anderson A.C., Joller N., Kuchroo V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44(5):989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zamarin D., Holmgaard R.B., Ricca J. Intratumoral modulation of the inducible co-stimulator ICOS by recombinant oncolytic virus promotes systemic anti-tumour immunity. Nat Commun. 2017;8:14340. doi: 10.1038/ncomms14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ribas A., Dummer R., Puzanov I. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell. 2017;170(6):1109–1119 e1110. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zacharakis N., Chinnasamy H., Black M. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24(6):724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maute R.L., Gordon S.R., Mayer A.T. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A. 2015;112(47):E6506–E6514. doi: 10.1073/pnas.1519623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pedicord V.A., Montalvo W., Leiner I.M., Allison J.P. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proc Natl Acad Sci U S A. 2011;108(1):266–271. doi: 10.1073/pnas.1016791108. [DOI] [PMC free article] [PubMed] [Google Scholar]