PP2A-B′γ interacts with CALCIUM-DEPENDENT PROTEIN KINASE1 and age-dependently controls salicylic acid-related signaling in plant immunity and developmental leaf senescence.
Abstract
Plants optimize their growth and survival through highly integrated regulatory networks that coordinate defensive measures and developmental transitions in response to environmental cues. Protein phosphatase 2A (PP2A) is a key signaling component that controls stress reactions and growth at different stages of plant development, and the PP2A regulatory subunit PP2A‐B′γ is required for negative regulation of pathogenesis responses and for maintenance of cell homeostasis in short-day conditions. Here, we report molecular mechanisms by which PP2A‐B′γ regulates Botrytis cinerea resistance and leaf senescence in Arabidopsis (Arabidopsis thaliana). We extend the molecular functionality of PP2A‐B′γ to a protein kinase–phosphatase interaction with the defense-associated calcium-dependent protein kinase CPK1 and present indications this interaction may function to control CPK1 activity. In presenescent leaf tissues, PP2A-B′γ is also required to negatively control the expression of salicylic acid-related defense genes, which have recently proven vital in plant resistance to necrotrophic fungal pathogens. In addition, we find the premature leaf yellowing of pp2a-b′γ depends on salicylic acid biosynthesis via SALICYLIC ACID INDUCTION DEFICIENT2 and bears the hallmarks of developmental leaf senescence. We propose PP2A-B′γ age-dependently controls salicylic acid-related signaling in plant immunity and developmental leaf senescence.
Plants optimize their growth and survival through highly integrated signaling networks that modulate cellular functions upon environmental changes. During the life span of a plant, negative regulators can significantly affect plant fitness and productivity by preventing unnecessary defense activation and premature aging, which may severely penalize reproductive success. To date, transcript profiling has elucidated the dynamic nature of gene expression occurring in the time-course of age-dependent senescence or infection by phytopathogens (Breeze et al., 2011; Windram et al., 2012; Woo et al., 2016). A multitude of transcription factors and signaling components underlying immunity reactions and senescence have been identified and functionally characterized, but posttranslational control of the elicited responses remains poorly understood.
Reversible protein phosphorylation is one of the key mechanisms that control the perception and relay of internal and external signals. In plant immunity, mitogen-activated protein kinases and calcium-dependent protein kinases (CDPKs; termed “CPKs” in Arabidopsis [Arabidopsis thaliana]) are central in eliciting appropriate defense reactions against a variety of biotic stress agents (Suarez-Rodriguez et al., 2010; Boudsocq and Sheen, 2013; Meng and Zhang, 2013; Romeis and Herde, 2014). Controlled protein dephosphorylation by protein phosphatases, in turn, is vital in containing the extent of defensive measures. Well-characterized examples are MAP KINASE PHOSPHATASE1 (MKP1) and MKP2, which are monomeric dual-specificity phosphatases that control the activation state and signaling through the key signaling kinases MPK3 and MPK6 (Bartels et al., 2009; Lumbreras et al., 2010; Jiang et al., 2017). Upstream protein kinases and phosphatases responsible for posttranslational regulation of CPKs have so far remained unidentified. In developmental leaf senescence, a few protein phosphatases that positively or negatively influence the induction or progression of this process have been identified. These include e.g. SENESCENCE-ASSOCIATED GENE113 (SAG113)/HAI1, a protein phosphatase 2C (PP2C) that acts in prosenescence signaling (Zhang et al., 2012; Zhang and Gan, 2012); and SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE (SSPP), which in turn retains leaf longevity by negatively regulating prosenescence signals (Xiao et al., 2015).
An increasing number of studies indicate type 2A protein phosphatases regulate central processes in plant stress resistance, immunity, and growth (He et al., 2004; Trotta et al., 2011; Heidari et al., 2013; Li et al., 2014; Segonzac et al., 2014; reviewed by Durian et al., 2016). PP2A forms a heterotrimeric holoenzyme that consists of a catalytic C subunit, a scaffold A subunit, and a variable regulatory B subunit. The Arabidopsis genome encodes five PP2A-C subunits, three A subunits, and 17 B subunits, which are divided into evolutionarily conserved B, B′, and B″ families (Booker and DeLong, 2017). The specificity of PP2As is conferred by the variable PP2A-B subunits, which direct the holoenzymes toward specific target phosphoproteins (Farkas et al., 2007; Moorhead et al., 2009; Sents et al., 2013; Lillo et al., 2014).
Segonzac et al. (2014) revealed a mechanistic interaction where PP2A negatively regulated plasma membrane sensory systems. An Arabidopsis PP2A holoenzyme, composed of a catalytic subunit PP2A-C4, scaffold subunit A1, and either B′η or B′ζ, limited the activity of BRI1-ASSOCIATED KINASE1 (BAK1), an essential coreceptor involved in early defense signaling upon perception of conserved microbial-associated molecular patterns (Segonzac et al., 2014). Within intracellular networks, PP2A-B′γ is required for negative regulation of pathogen responses and maintenance of cellular integrity. This was first evidenced by phenotypic analysis of the knock-down pp2a‐b′γ mutant, which showed resistance against the necrotrophic fungal pathogen Botrytis cinerea and green peach aphid (Myzus persicae; Trotta et al., 2011; Rasool et al., 2014). Healthy pp2a‐b′γ mutants in turn undergo premature yellowing conditionally when grown under moderate light intensity in short-day conditions (Trotta et al., 2011). Studies on a pp2a‐b′γ cat2 double mutant deficient in PP2A-B′γ and the main peroxisomal antioxidant enzyme CATALASE2 elaborated the functional importance of PP2A‐B′γ in controlling intracellular oxidative stress responses and associated metabolite signatures (Li et al., 2014). Mechanistic insights into secondary metabolism were provided by Rahikainen et al. (2017), who found PP2A‐B′γ interacted with components of the activated methyl cycle and negatively regulated the formation of a deterring 4-methoxy-indol-3-yl-methyl glucosinolate in Arabidopsis leaves.
Recently, Zhu et al. (2018) demonstrated the importance of PP2A in host-necrotroph interactions in crop species. Silencing of a wheat (Triticum aestivum) PP2A catalytic subunit conferred resistance against the necrotrophic fungus Rhizoctonia cerealis, the causative agent of wheat sharp eyespot disease. The resistance phenotype was accompanied by increased transcript abundance for the wheat pathogenesis-related (PR) protein PR2 and antioxidant enzymes (Zhu et al., 2018). However, the exact molecular mechanisms by which PP2A contributes to plant resistance against necrotrophic fungal pathogens remain unresolved.
Here we report molecular mechanisms by which PP2A‐B′γ regulates B. cinerea resistance and prevents premature yellowing and cell death in Arabidopsis leaves. We provide evidence indicating 4-week–old pp2a-b′γ mutants are primed for salicylic acid (SA)-related immune reactions, the importance of which is increasingly recognized in plant resistance to necrotrophic fungal pathogens. Furthermore, we present a protein kinase–phosphatase interaction between PP2A-B′γ and Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE 1 (CPK1), which is involved in the resistance to necrotrophic fungi. By biochemical characterization and transcriptome analysis of 7-week–old pp2a-b´γ, we further demonstrate the premature yellowing is SA-dependent and bears the hallmarks of age-dependent leaf senescence. We propose PP2A-B′γ negatively controls the SA-component in the onset of developmental leaf senescence.
RESULTS
PP2A-B′γ Interacts with CPK1 and SAG12
To elucidate PP2A-B′γ dependent signaling interactions, full-length PP2A-B′γ was used as bait in a yeast two-hybrid screen against a complementary DNA (cDNA) library enriched for stress-related components (Jaspers et al., 2009). This led to identification of CPK1 (AT5G04870) and the Cys protease SAG12 (AT5G45890) as candidate PP2A-B′γ-interacting proteins (Supplemental Table S1).
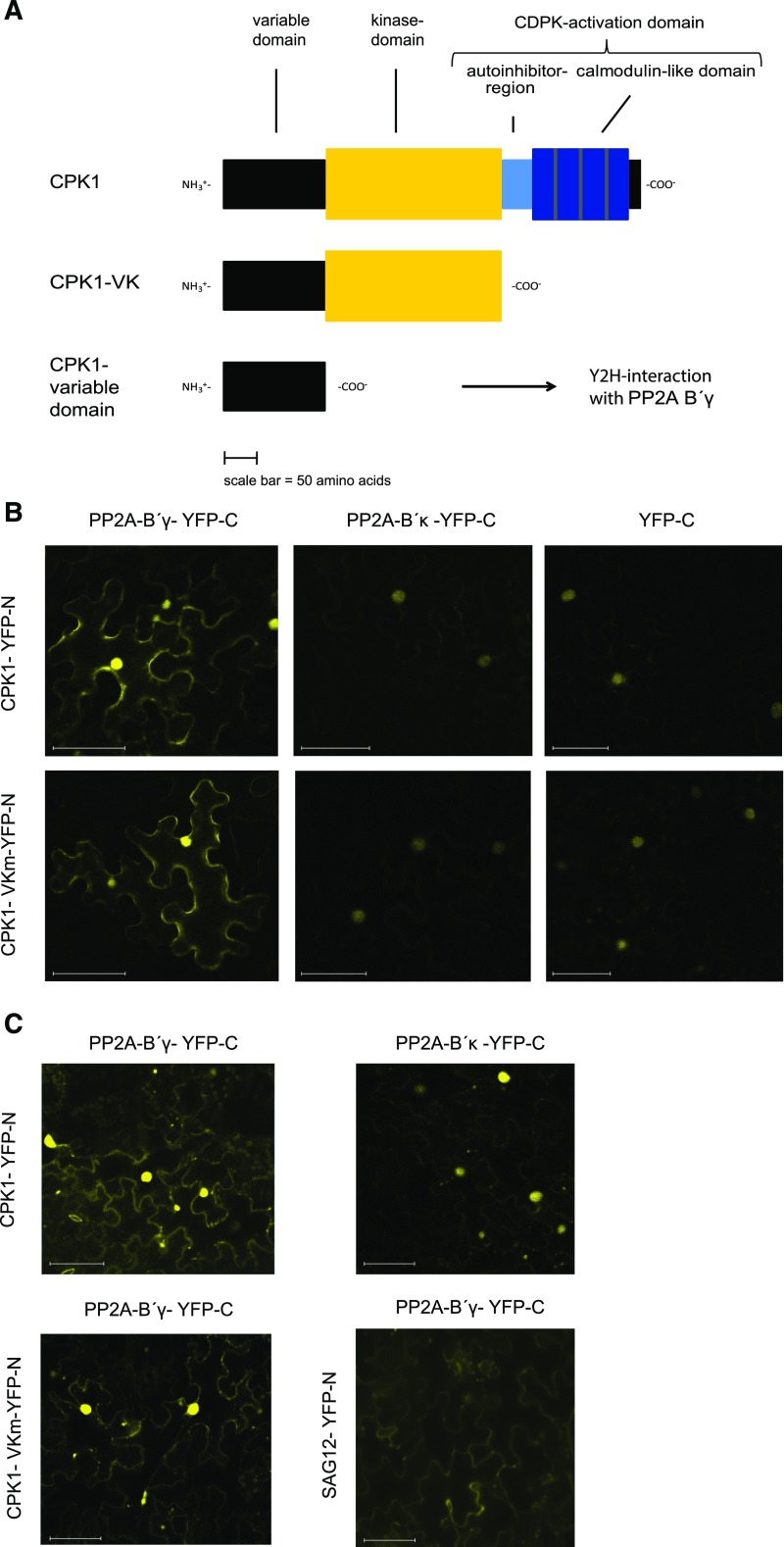
CDPKs are composed of a variable N-terminal domain, a Ser/Thr kinase domain, and a Ca2+-binding CDPK-activation domain (Fig. 1A; Liese and Romeis, 2013). The variable N terminus, which is the most distinctive feature between different CDPK isoforms, carries nine experimentally verified in vivo phosphorylation sites in CPK1, while its C terminus has been found phosphorylated on one site (Supplemental Table S1; PhosPhAt-database, Durek et al., 2010). It was therefore of interest that the yeast two-hybrid screen trapped a polypeptide comprising the first 149 amino acids corresponding to the N terminus of CPK1 (Fig. 1A).
Figure 1.
Analysis of protein interactions among PP2A-B′γ, CPK1, and SAG12. A, Schematic representation of full-length CPK1, a truncated VKm form that consists of the variable (V) N terminus and the kinase (K) domain, and the V N terminus that was found to interact with PP2A-B′γ in the yeast two-hybrid screen. The schemes and their domains are drawn to scale. B, BiFC analysis of protein interactions in N. benthamiana leaves transiently expressing the fusion protein pairs CPK1-YN/PP2A-B′γ-YC, CPK1-YN/PP2A-B′κ-YC, CPK1-YN/YFP-YC, CPK1-VKm-YN/PP2A-B′γ-YC, CPK1-VKm-YN/PP2A-B′κ-YC, and CPK1-VKm-YN/YFP-YC. C, BiFC analysis of protein interactions in Arabidopsis leaves. Transgenic Arabidopsis plants expressing the bacterial effector AvrPto under the control of a dexamethasone-inducible promoter (Hauck et al., 2003) transiently expressing the fusion protein pairs CPK1-YN/PP2A-B′γ-YC, CPK1-YN/PP2A-B′κ-YC, CPK1-VKm-YN/PP2A-B′γ-YC, and SAG12-YN/PP2A-B′γ-YC. Scale bars = 50 µm.
Despite intensive efforts, pp2a-b′γ cpk1 double mutants could not be identified even when screening the progeny of a PP2A-B′γ −/− CPK1+/− parental line, suggesting the two signaling components may be functionally connected. To verify the interaction between PP2A-B′γ and CPK1, we used a bimolecular fluorescence complementation (BiFC) system, which allows visualization of interactions that are highly transient in nature, such as those between phosphatases and kinases and their protein substrates. To assess whether the protein interaction with PP2A-B′γ requires the C-terminal CDPK-activation domain, we employed both a full-length CPK1 and a truncated form consisting of the variable (V) N terminus and the kinase (K) domain, denoted as CPK1-VK (Fig. 1A). This form is commonly used to mimic the conformation of the calcium-activated full-length CDPK (Harper et al., 1994). To avoid influences potentially caused by the kinase activity of a constitutively active CPK1, especially in the heterologous Nicotiana benthamiana system, we employed a form of CPK1-VK that is kinase-inactive due to a point-mutation in the ATP-binding site, denoted CPK1-VKm.
First we tested the interaction between PP2A-B′γ and the full-length and VKm forms of CPK1 using the well-established N. benthamiana system for BiFC (Bracha-Drori et al., 2004; Walter et al., 2004). Both the full-length CPK1 and the truncated CPK1-VKm interacted with PP2A-B′γ in the cytosol (Fig. 1B). This interaction was not visible when the related PP2A-B subunit isoform PP2A-B′κ was coexpressed with CPK1 or CPK1-VKm, or when the C-terminal part of yellow fluorescent protein (YFP) was coexpressed with either of the CPK1 constructs (Fig. 1B). The fluorescence detected in the nucleus, in contrast, was also visible when PP2A-B′κ or just the C-terminal part of YFP was coexpressed with either CPK1 or CPK1-VKm (Fig. 1B) and was hence a false positive, likely arising from split-off of the YFP-halves from the coexpressed proteins and a consequent formation of free YFP. Fluorescence signals indicative of a PP2A-B′γ-SAG12 interaction could not be detected using the N. benthamiana system.
The classical BiFC system employs heterologous coexpression of Arabidopsis proteins in leaf cells of N. benthamiana, which, as a member of the superasterids, is only distantly related to the superrosid Arabidopsis (APG IV, 2016). Furthermore, the subcellular localization of PP2A-B′γ, as well as almost all other Arabidopsis PP2A-B′-subunits, can vary depending on the species it is expressed in and the expression system used in protein localization assays (Durian et al., 2016). Hence, to assess the protein interactions in a more natural environment, we developed a BiFC-system for Arabidopsis leaves. For this purpose, we employed a transgenic Arabidopsis line that expresses the bacterial effector protein AvrPto under the control of a dexamethasone-inducible promoter (Hauck et al., 2003). This ectopic expression of the bacterial effector weakens the innate immune system of the host plant and allows efficient Agrobacterium tumefaciens transfection of leaf cells and subsequent transgenic protein expression (Tsuda et al., 2012). In our Arabidopsis BiFC-system, we were able to reproduce the PP2A-B′γ-CPK1 and PP2A-B′γ-CPK1-VKm interactions (Fig. 1C). In addition, the Arabidopsis system revealed fluorescence signals indicative of PP2A-B′γ-SAG12 interaction in the cytosol (Fig. 1C).
Nonstressed pp2a-b′γ Mutants Display Increased In-Gel Kinase Activity of CPK1
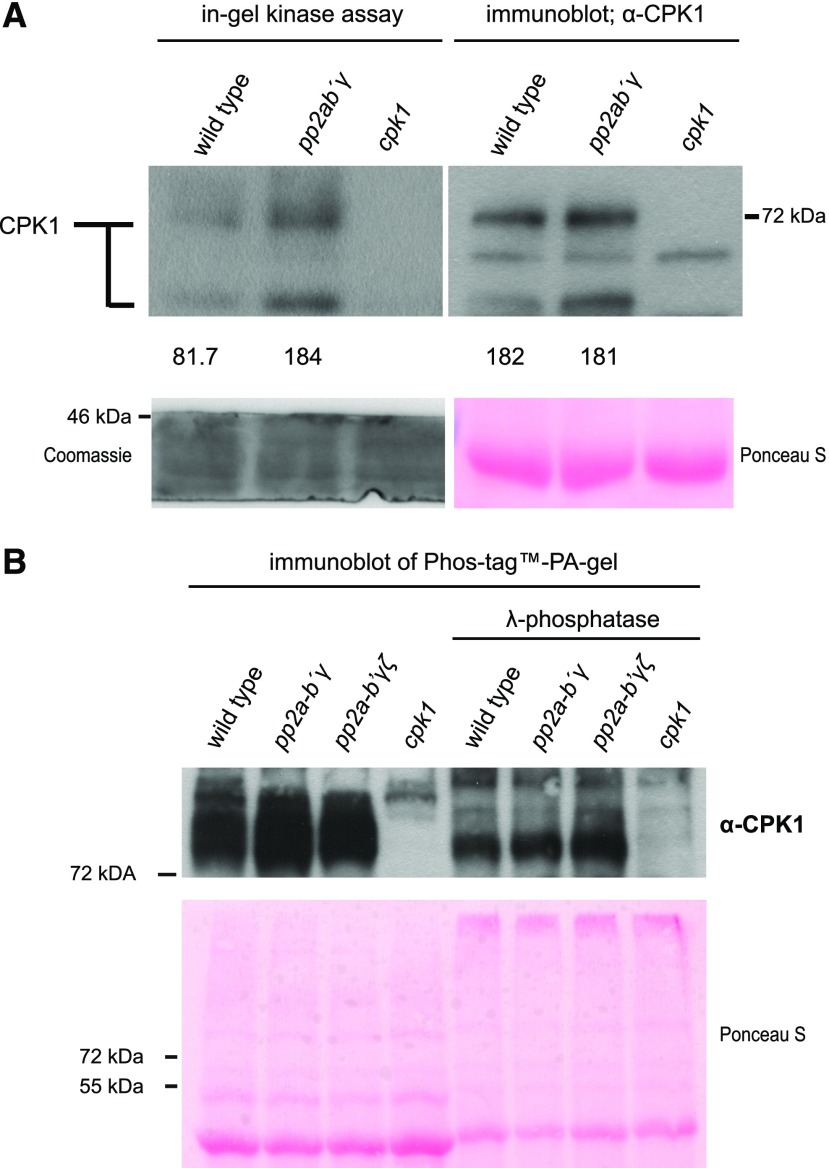
CPK1 is a phosphoprotein that acts as a positive component in immunity signaling (Coca and San Segundo, 2010; PhosPhAt database, Durek et al., 2010). Therefore, the observed interaction between PP2A-B′γ and CPK1 raised the question whether PP2A-B′γ exerts a control over CPK1 kinase activity. To answer this question, we first analyzed the abundance of CPK1 using an antibody raised against a CPK1-specific peptide that resides within an area of the N-terminal domain devoid of known in vivo phosphorylation sites. We chose an 18-amino acid CPK1-epitope for this antibody that is unique for the CPK-isoform CPK1 (Supplemental Fig. S1B). The generated CPK1 antibody bound the 18-amino acid CPK1-epitope (Supplemental Fig. S2) and reacted with polypeptides that were not detected in two independent cpk1 knock-out lines (Supplemental Fig. S1). Two major bands of CPK1 were observed in wild type and pp2a-b′γ, but not in cpk1, when leaf extracts were isolated from visually healthy plants grown in standard short-day conditions (Fig. 2A, right). When the same samples were subjected to CPK-specific in-gel kinase assays, in which calcium is added to activate the CPKs, a higher activity of the upper band of CPK1 was observed in pp2a-b′γ as compared to wild-type plants (Fig. 2A, left), while both genotypes displayed similar abundance of this protein band (Fig. 2A, right; full-length immunoblot for Fig. 2 in Supplemental Fig. S3).
Figure 2.
Biochemical characterization of the PP2A-B′γ-CPK1 interaction. A, CPK1 in-gel kinase activity and protein abundance in total leaf extracts isolated from Arabidopsis wild type, pp2a-b′γ, and cpk1. CPK1 in-gel kinase activity (left) was assessed in the presence of 0.2 mm CaCl2 and histone type IIIS. Subsequently, the same samples were separated on SDS-gel electrophoresis and subjected to immunoblot analysis with a CPK1-specific antibody (right). Numbers (× 1,000) under the panels indicate intensity measurements for the uppermost band (Image Studio Lite; Li-Cor Biosciences). Loading controls are shown in the panels below the numbers; left: Coomassie Brilliant Blue R250-staining of the lower rim of the in-gel kinase SDS-PA gel; right: Ponceau S-staining of the large subunit of Rubisco on the PVDF-membrane. B, CPK1 immunoblot after separation of total leaf extracts from wild type, pp2a-b′γ, pp2a-b′γζ, and cpk1 on PhosTag PA gel. To assess the presence of phosphorylated residues among the high-molecular weight CPK1 bands, the samples were prepared in the presence and absence of lambda-phosphatase. Lower: Ponceau S-staining.
PP2A-B′γ-mediated dephosphorylation of CPK1 could affect the pattern of phosphorylation among the multiple experimentally verified in vivo phosphorylation sites (Supplemental Table S1; PhosPhAt-database, Durek et al., 2010), which could specifically modulate the activity of the affected CPK1-species. Phosphorylated CPK-species leading to multiple bands in normal SDS-polyacrylamide (PA) gels are already known for Arabidopsis CPK5 (Dubiella et al., 2013) and tobacco (Nicotiana tabacum) CDPK2, which is the ortholog of Arabidopsis CPK1 (Witte et al., 2010). To assess whether CPK1 bands migrating at higher apparent Mr values on SDS gels represent phosphorylated CPK1 species, a subset of the samples was treated with lambda-phosphatase and thereafter separated on PhosTag gels. After the lambda-phosphatase treatment, CPK1 bands with a higher apparent Mr were not present in the PhosTag gels, demonstrating that they were phosphorylated forms of CPK1 (Fig. 2B).
PP2A-B′γ Controls the Abundance of CPK1 in B. cinerea-Infected Leaves
CPK1 is a positive regulator of defense reactions against the necrotrophic fungal pathogen B. cinerea (Coca and San Segundo, 2010). PP2A-B′γ in turn acts as a negative regulator, as demonstrated by reduced lesion size in fully expanded pp2a-b′γ leaves upon drop-inoculation with B. cinerea (Trotta et al., 2011). The regulatory interaction between PP2A-B′γ and CPK1 (Figs. 1 and 2) therefore prompted us to test whether PP2A-B′γ controls CPK1 function in B. cinerea-infected tissues.
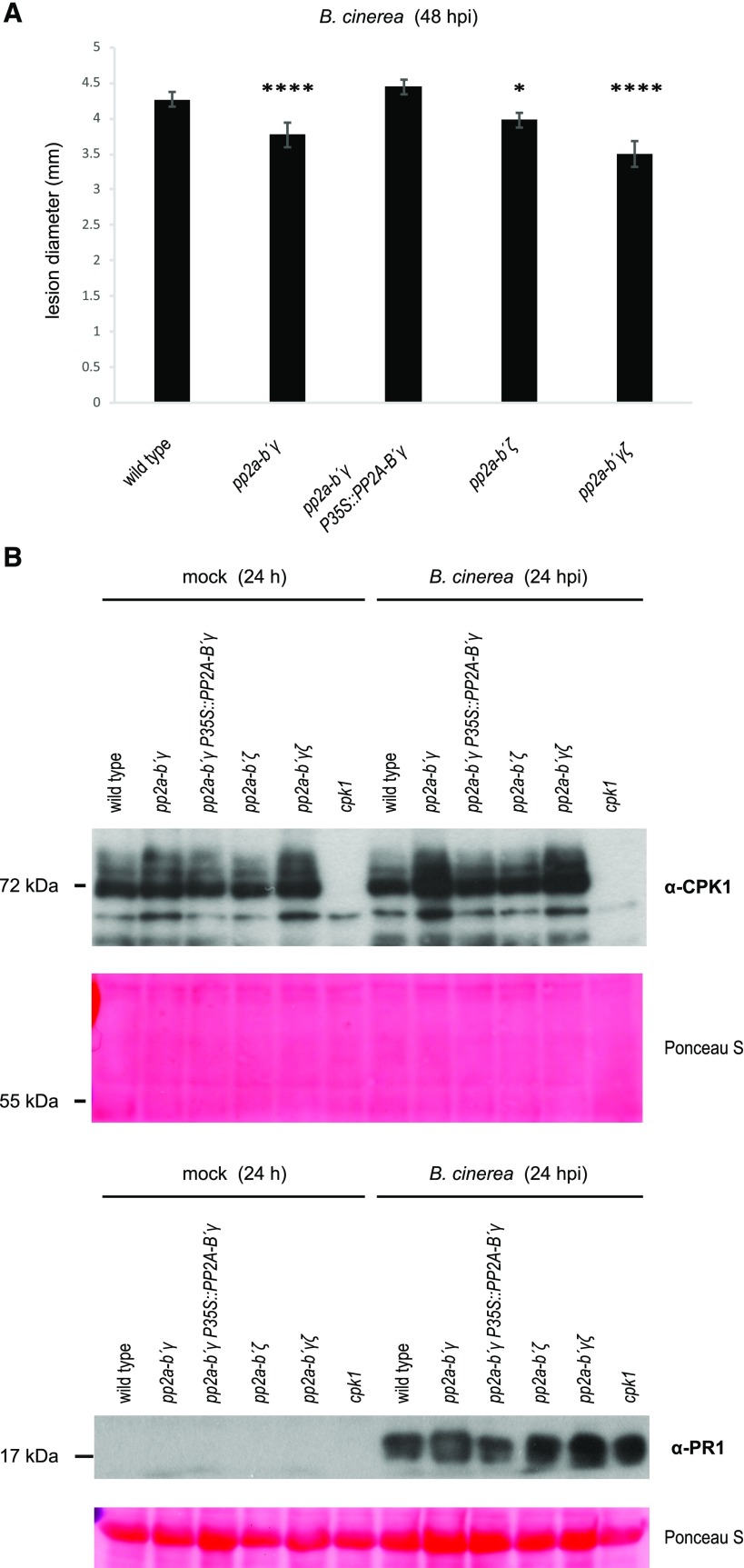
First, we tested PP2A-B subunit specificity on the resistance phenotype and subjected pp2a-b′γ, pp2a-b′ζ, deficient in the closest homolog of PP2A-B′γ, a respective pp2a-b′γζ double mutant (Rasool et al., 2014) and a complementation line expressing PP2A-B′γ under the 35S-promoter in a genetic pp2a-b′γ background (Trotta et al., 2011) to lesion size assays with B. cinerea. To quantify the resistance in these genotypes, 4-week–old short day-grown plants were selected for drop inoculation with B. cinerea spores, and the lesion size on infected leaves was measured 48-h post infection (hpi). The pp2a-b′γ and pp2a-b′ζ single mutants and the double mutant pp2a-b′γζ displayed significantly reduced lesion size compared to wild type and thereby proved more resistant to infection by B. cinerea (Fig. 3A; photographs in Supplemental Fig. S4). The complementation line expressing 35S:PP2A-B′γ in pp2a-b′γ showed no alterations in disease resistance when compared to wild type (Fig. 3A).
Figure 3.
Analysis of disease resistance and the abundance of CPK1 and PR1 in 4-week–old B. cinerea-infected pp2a mutant plants. A, Quantification of disease resistance in B. cinerea-infected wild type, pp2a-b′γ, a complementation line expressing PP2A-B′γ under the control of the 35S-promoter in pp2a-b′γ (pp2a-b′γ P35S:PP2A-B′γ), pp2a-b′ζ and the double mutant pp2a-b´γζ. Four-week–old short day-grown plants were drop-inoculated with B. cinerea spores, and the lesion size on infected leaves was measured 48 hpi. Means ± se, n ≥ 170; statistically significant differences compared to wild type are indicated by asterisks (linear mixed effect model, Tukey-Post hoc test: ****P < 0.0001; *P < 0.05). B, Representative immunoblots depicting CPK1 (upper) and the SA-signaling marker PR1 (lower) abundance in mock-treated and B. cinerea-infected pp2a mutant rosettes. Total leaf extracts isolated from wild type, pp2a-b′γ, pp2a-b′γ P35S:PP2A-B′γ, pp2a-b′ζ, pp2a-b′γζ, and cpk1 were separated by SDS-gel electrophoresis and subjected for immunoblot analysis with antibodies specific to CPK1 or PR1.
To assess whether PP2A-B′γ controls CPK1 abundance in B. cinerea-infected tissues, 4-week–old short day-grown wild-type plants and pp2a mutants were spray-infected with B. cinerea and the level of CPK1 was explored by immunoblotting from SDS gels. The mock treatment, i.e. spraying with half-strength potato dextrose broth (PDB) and keeping the plants under the fungal growth atmosphere of close to 100% humidity, showed slightly increased CPK1 abundance in pp2a-b′γ and the pp2a-b′γζ plants (Fig. 3B; full-length immunoblot for Fig. 3B in Supplemental Fig. S3). All genotypes showed increased CPK1 abundance 24 h after spraying with B. cinerea spores. This pathogen-induced upregulation of CPK1 was particularly strong in pp2a-b′γ and the pp2a-b′γζ double mutant (Fig. 3B). Hence, the B. cinerea-induced increase in CPK1 abundance is controlled by PP2A-B′γ.
PP2A-B′γ Is Required To Negatively Control the Abundance of Transcripts Involved in Defense-Related SA Signaling
CPK1 was previously assigned a role in SA signaling, as CPK1-overexpressing plants displayed enhanced induction of SA signaling marker genes after infection with Fusarium oxysporum, while mutants deficient in CPK1 showed reduced induction (Coca and San Segundo, 2010). To assess whether the increased B. cinerea resistance of pp2a-b′γ, pp2a-b′ζ, and pp2a-b′γζ is associated with constitutive transcriptional priming of SA responses, we performed microarray transcript profiling of unchallenged 4-week–old short day-grown wild type, pp2a-b′γ, pp2a-b′ζ, and pp2a-b′γζ.
The pp2a-b′γ mutant showed significantly altered transcript levels for 407 genes compared to wild-type plants, while the number of differentially expressed genes in the pp2a-b′γζ double mutant was 218. In contrast, the pp2a-b′ζ mutant had statistically significant changes in gene expression for only nine genes (Supplemental Table S2). Increasing evidence indicates PR1, which binds sterols and sequesters them from pathogens (Gamir et al., 2017), is an important player in plant resistance to necrotrophic pathogens. Among the 30 genes, which are most strongly coexpressed with PR1 according to the ATTED database (ATTEDII; http://atted.jp), 19 were upregulated more than 3-fold in pp2a-b′γ (listed in Table 1). The abundance of PR1 transcripts was upregulated 20-fold, PR2 was upregulated 6-fold, and PR5 was upregulated 4-fold in the noninfected pp2a-b′γ plants (Table 1). Transcriptional induction of PR1, PR2, and PR5 is generally considered a hallmark for defense-induced SA signaling and for systemic acquired resistance (SAR), a defense-primed state of the plant (Fu and Dong, 2013). RECEPTOR LIKE PROTEIN23 (RLP23), 2-OXOGLUTARATE AND Fe(II)-DEPENDENT OXYGENASE SUPERFAMILY PROTEIN (2OG), CYS-RICH RECEPTOR-LIKE PROTEIN KINASE45 (CRK45), and CHITINASE (CHI), which are well known for their roles in defense responses against pathogenic fungi, also exhibited severalfold increased transcript abundance in pp2a-b′γ. In addition, the key SAR-regulatory genes AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1) and FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1) were strongly transcriptionally induced. For PR1 and the strongly PR1-coexpressed genes PR2, PR5, NUCLEOSIDE DIPHOSPHATE LINKED TO SOME MOIETY X6 (NUDX6), and CRK45, which are all involved in SA-signaling (Table 1), we additionally tested the mRNA-abundance in 4-week–old short day-grown wild-type and pp2a-b′γ plants as well as plants of the pp2a-b′γ P35S:PP2A-B′γ complementation line by reverse transcription quantitative real-time PCR (RT-qPCR; Supplemental Fig. S5). With this method, the transcripts of these five genes showed in all cases a significantly higher abundance in pp2a-b′γ than in wild-type plants, with fold changes (FCs) in the relative gene expression compared to the wild type ranging from 4-fold (CRK45) to 20-fold (PR1). For the complementation line pp2a-b′γ P35S:PP2A-B′γ, however, no significant changes in mRNA amount of the five genes compared to the wild type could be detected (Supplemental Fig. S5). Taken together, PP2A-B′γ is required to suppress accumulation of SA-signaling related transcripts under nonstressed conditions.
Table 1. Abundance of PR1-coexpressed transcripts and key SA signaling-related transcripts in 4-week–old pp2a-b′γ and pp2a-b′ζ single mutants and pp2a-b′γζ double mutants relative to wild-type plants in microarray analysis.
Values are log2 (FC) and are the means of four independent biological replicates. Statistically significant values of log2 (FC) > 1 and q value of q < 0.05 are indicated in bold. Coexpression rank to PR1 (AT2G14610) is based on the Mutual Rank (MR, data platform Ath-m.c7-1; Obayashi et al., 2018), according to the Arabidopsis thaliana trans-factor and cis-element prediction database (ATTED-II) v9.2 (http://atted.jp). AGI, Arabidopsis genome initiative locus code.
| Rank ATTED-II | MR ATTED-II | AGI | Annotation and Involvement in Defense Responsesa | FC to Wild Type (Log2) | ||
|---|---|---|---|---|---|---|
| pp2a- b′γ | pp2a-b′γζ | pp2a-b′ζ | ||||
| 0 | 0.0 | At2g14610 | PR1; SA-signaling, defense response, SAR | 4.3 | 2.5 | 1.3 |
| 1 | 2.0 | At3g57260 | PR2; SA-signaling, SAR | 2.6 | 1.4 | 0.5 |
| 2 | 3.7 | At2g18660 | PNP-A; SAR | 2.7 | 1.7 | 0.0 |
| 3 | 4.4 | At1g75040 | PR5; SA-signaling, SAR | 2.1 | 1.4 | 0.2 |
| 4 | 4.4 | At1g33960 | AIG1; ETI | 2.6 | 1.5 | 0.4 |
| 5 | 4.8 | At5g10760 | AED1; SAR | 2.1 | 0.9 | 0.3 |
| 7 | 7.0 | At2g04450 | NUDX6; SA signaling | 2.3 | 1.1 | 0.3 |
| 8 | 8.9 | At3g57240 | BG3; response to bacterium | 2.4 | 1.6 | 0.6 |
| 9 | 9.9 | At4g23150 | CRK7; defense response to bacterium | 3.0 | 1.7 | 0.4 |
| 10 | 10.5 | At3g25010 | RLP41; defense response | 2.7 | 1.6 | 0.4 |
| 11 | 10.9 | At2g32680 | RLP23; defense response to fungus/bacterium | 2.4 | 1.3 | 0.2 |
| 16 | 17.4 | At3g47480 | CALMODULIN-LIKE47 | 1.6 | 0.7 | 0.3 |
| 18 | 19.2 | At5g55450 | ATLTP4.4; SAR | 1.6 | 0.7 | 0.1 |
| 20 | 20.9 | At1g21240 | WAK3 | 2.5 | 1.6 | 0.1 |
| 21 | 23.0 | At4g04500 | CRK37 | 2.2 | 1.1 | 0.3 |
| 22 | 23.3 | At2g26400 | ARD3 | 3.0 | 1.5 | 0.7 |
| 23 | 24.2 | At3g24900 | RLP39; defense response | 2.7 | 1.3 | 0.3 |
| 26 | 25.9 | At2g43570 | CHI; chitin catabolic process, SAR | 2.5 | 1.4 | −0.1 |
| 28 | 26.8 | At4g10500 | 2OG; response to SA, defense response to fungus | 2.9 | 1.4 | 0.5 |
| 29 | 28.3 | At4g11890 | CRK45; defense response to fungus, response to SA/SA-signalingb | 2.0 | 1.1 | 0.1 |
| 68 | 78.4 | At1g74710 | SID2/ICS1; defense response to fungus/bacterium, SAR | 0.8 | 0.4 | 0.1 |
| 106 | 124.3 | At2g13810 | ALD1; SA-signaling, SAR | 3.8 | 2.0 | 0.7 |
| 129 | 145.7 | At1g19250 | FMO1; SAR, defense resp. to fungus/bacterium | 3.5 | 2.3 | 0.4 |
| no | — | At5g26170 | WRKY50; PR1-transcriptional regulation,c defense response to fungus | 1.8 | 0.8 | 0.2 |
| no | — | At1g64280 | NPR1; SA-signaling, defense response to fungus/bacterium, SAR | 0.5 | 0.3 | 0.1 |
SA-signaling/defense involvement according to TAIR (www.arabidopsis.org).
Arabidopsis PR1 protein accumulates 12 h after infection with B. cinerea and significantly higher PR1 abundance is observed in plants in which induced systemic resistance has been triggered by exposure to nonpathogenic bacteria, leading to a higher resistance upon subsequent B. cinerea infection (Nie et al., 2017). In a leaf lesion assay very similar to ours (Fig. 3), Arabidopsis plants overexpressing PR1 in turn proved more resistant against another closely related necrotrophic fungus, Sclerotinia sclerotiorum (Yang et al., 2018). Furthermore, the tomato (Solanum lycopersicum) protein P14c, closely related to Arabidopsis PR1, reduces B. cinerea growth in vitro (Gamir et al., 2017). Hence, PR1 function appears to hinder the growth of B. cinerea and S. sclerotiorum, both of which belong to the family of Sclerotiniaceae. These findings prompted us to explore the abundance of PR1 in the B. cinerea-infected leaves of pp2a mutants and wild-type plants.
Despite the increased PR1 transcript abundance in unchallenged 4-week–old pp2a-b′γ (Table 1), immunoblot analysis did not detect PR1 protein in any of the plants sprayed with mock solution (Fig. 3B, lower). However, 24 h after infection with B. cinerea spores, PR1 was strongly induced at the protein level in all genotypes (Fig. 3B). The resistance phenotype of pp2a-b′γ and pp2a-b′γζ was not accompanied by a significantly stronger increase in PR1 protein abundance when compared to the other lines (Fig. 3B). Moreover, even though CPK1 was implicated in SA-related gene expression in F. oxysporum-infected leaves (Coca and San Segundo, 2010), CPK1 function was not a prerequisite for accumulation of PR1 protein after B. cinerea infection, as evidenced by accumulation of PR1 in cpk1 plants (Fig. 3B).
Finally, we also measured the contents of SA, jasmonic acid (JA), and 1-aminocyclopropane-1-carboxylic acid (ACC), the direct precursor of the gaseous plant hormone ethylene, in control plants and 24 h after spray-infection with B. cinerea spores. Even though SA, JA, and ethylene are all positively involved in plant defense against B. cinerea (Han et al., 2010; Smirnova et al., 2017), no significant differences in the levels of SA, JA, or ACC were detected between mock-treated versus spore-infected plants at 24 hpi in any of the genotypes tested (Supplemental Fig. S6).
The Yellowing Phenotype in the Apex of Aging pp2a-b′γ Leaves Coincides with Protein Expression of PR1 and SAG12
While pp2a-b′γ develops smaller lesions when infected by B. cinerea (Fig. 3A), noninfected pp2a-b′γ leaves are characterized by age-dependent formation of chlorotic regions and cell death spots, which are clearly observed on apical leaf areas after 7 weeks of growth in short-day conditions (Fig. 4A). Hence, the following experiments were designed to clarify which physiological process is responsible for the premature yellowing phenotype of pp2a-b′γ. The spatial occurrence of the chlorosis and cell death in pp2a-b′γ on the leaf apices resembled that of developmental leaf senescence in Arabidopsis. In contrast, no symptoms of premature yellowing were observed on 7-week–old pp2a-b′ζ, the pp2a-b′γζ double mutant, or pp2a-b′γ complemented with 35S::PP2A-B′γ (Fig. 4A).
Figure 4.
Analysis of SA-related responses in pp2a mutant leaves displaying age-dependent yellowing. A, Phenotypic characteristics of 7-week–old, short day-grown wild type, pp2a-b´γ, pp2a-b´γ P35S:PP2A-B′γ, pp2a-b′ζ, pp2a-b′γζ, and the pp2a-b′γ sid2 double mutant. Scale bars = 1 cm. B, Representative immunoblots depicting accumulation of the SA-signaling marker PR1 and the senescence marker SAG12 in the yellowing apical halves of pp2a-b′γ leaves. Leaves of 7-week–old wild type, pp2a-b′γ, pp2a-b′γ P35S:PP2A-B′γ, pp2a-b′ζ, pp2a-b′γζ, cpk1, sid2, and the pp2a-b′γ sid2 double mutant were harvested and split into apical and basal halves. Total extracts were isolated and separated by SDS-gel electrophoresis and subjected to immunoblot analysis with antibodies specific to PR1 or SAG12.
The macroscopic similarity between the pp2a-b′γ phenotype and leaf senescence, together with the identification of the well-known leaf senescence marker SAG12 as an interactor for PP2A-B′γ (Supplemental Table S1; Fig. 1C), suggested the chlorosis phenotype could constitute premature developmental leaf senescence in pp2a-b′γ. To elucidate this, we harvested fully expanded leaves from 7-week–old plants and split them into apical and basal halves. For pp2a-b′γ, this approach yielded pools of apical leaf halves that displayed symptoms of yellowing and cell death and basal leaf halves that were visually asymptomatic.
Strongly increased abundance of SAG12 transcript is routinely used as a marker for ongoing leaf senescence in Arabidopsis. We detected SAG12 protein specifically only in the apical, symptomatic halves of pp2a-b′γ mutant leaves (Fig. 4B, lower representation; full-length immunoblot for Fig. 4B in Supplemental Fig. S3), which strongly indicated senescence was progressing only in the apical parts of pp2a-b′γ leaves. We also assayed whether this Cys-endopeptidase is required to execute senescence-related processes in pp2a-b′γ. Phenotypic analysis revealed no differences between pp2a-b′γ and pp2a-b′γ sag12 double mutants with respect to leaf apex chlorosis or cell death (Supplemental Fig. S7). The lack of phenotypic differences between the pp2a-b′γ and pp2a-b′γ sag12 mutants mirrors the findings of Otegui et al., (2005) and James et al., (2018), who reported knock-out mutants lacking SAG12 function displayed no discernible change in the progression of developmental leaf senescence.
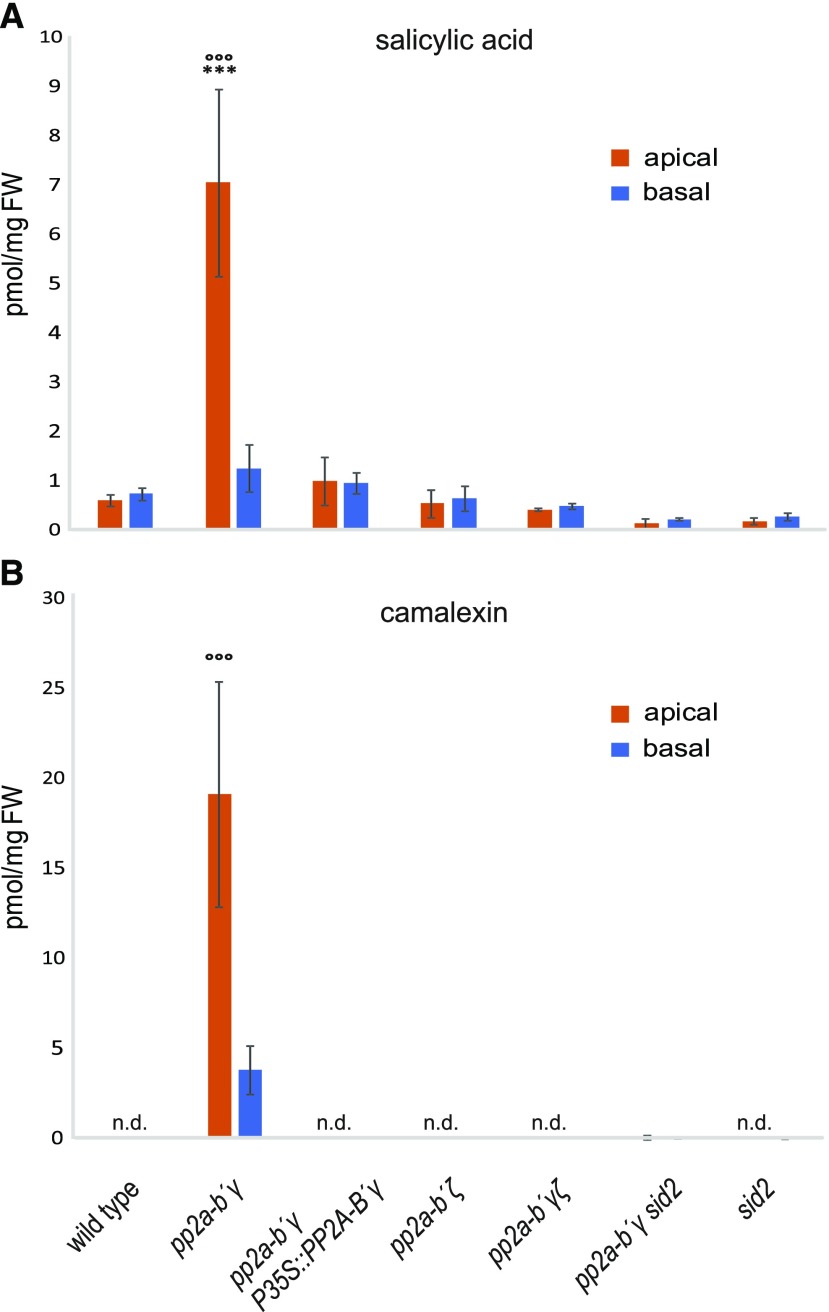
SA has long been known to play a positive role in Arabidopsis developmental leaf senescence, and more recently it has become clear that SA is a crucial factor in triggering the onset of this biological process (Zhang et al., 2013a, 2017; Guo et al., 2017; Kim et al., 2018a). Because PP2A-B′γ negatively regulates immunity-related SA signaling in 4-week–old unchallenged plants (Table 1), we hypothesized it could also repress SA signaling in developmental leaf senescence. To test this, we first assayed the presence of PR1 in 7-week–old wild type and pp2a mutant plants. Immunoblot analysis revealed the presence of PR1 in pp2a-b′γ leaves, the abundance of PR1 being significantly higher in the symptomatic apical leaf halves when compared to the nonsymptomatic basal halves (Fig. 4B, upper representation). This coincided with 7-fold higher SA levels in the apical leaf halves of pp2a-b′γ, compared to the corresponding basal halves, and ∼14-fold higher levels than the apical halves of wild-type leaves (Fig. 5A). In summary, SA accumulates to high levels in the apical leaf halves of pp2a-b′γ, and the strong expression of PR1 indicates the prevalence of active SA signaling.
Figure 5.
Contents of SA and camalexin in apical and basal leaf halves of 6-week–old plants. Leaves of 6-week–old wild type, pp2a-b′γ, pp2a-b′γ P35S:PP2A-B′γ, pp2a-b′ζ, pp2a-b′γζ, pp2a-b′γ sid2, and sid2 were harvested and split into apical and basal halves. Metabolites were extracted in 85% methanol and the contents of SA (A) and camalexin (B) were analyzed by liquid chromatography tandem mass spectrometry. Data are means ± sd, n = 4. Statistically significant differences between apical or basal leaf halves of a mutant to the corresponding leaf halves of the wild type are indicated by asterisks. Statistically significant differences between the apical and the basal leaf halves of the same genotype are indicated by circles. ANOVA: leaf half, P < 0.001 (F = 26.48), genotype, P < 0.001 (F = 48.53), leaf half*genotype, P < 0.001 (F = 26.57); Tukey posthoc test: ***P < 0.001, °°°P < 0.001.
The biosynthesis of camalexin, the major antimicrobial phytoalexin in Arabidopsis (Glawischnig, 2007; Frerigmann et al., 2015), is SA-dependent (Lenz et al., 2011). Camalexin accumulation was observed in the entire leaf in pp2a-b′γ plants but was ∼4-fold more abundant in the apical half (Fig. 5B). The other genotypes, which did not display leaf yellowing (Fig. 4), did not contain detectable levels of camalexin or a significant increase in SA (Fig. 5). These findings are in line with those of Mishina et al., (2007), who reported both SA and camalexin content increased upon developmental leaf senescence in short day-grown Arabidopsis accession Col-0, which was also the genetic background in our study.
The Phenotypes of pp2a-b′γ Leaves Require ICS1-Mediated SA Biosynthesis
To further analyze the SA-dependency of pp2a-b′γ phenotypes, we employed a pp2a-b′γ sid2 double mutant. SALICYLIC ACID INDUCTION DEFICIENT2/ISOCHORISMATE SYNTHASE1 (SID2/ICS1) is the enzyme required for the bulk of induced SA-biosynthesis in Arabidopsis. In the pp2a-b′γ sid2 double mutant, the macroscopic pp2a-b′γ phenotype with apical leaf chlorosis and cell death spots was not evident (Fig. 4A) and neither the accumulation of SA above wild-type levels, nor accumulation of PR1 and SAG12 proteins, could be detected (Figs. 4B and 5). In addition, the accumulation of camalexin in symptomatic pp2a-b′γ leaves proved to require functional SID2, and camalexin was not detected in the pp2a-b′γ sid2 double mutant (Fig. 5B). These findings demonstrate that the chlorosis, cell death, and SAG12 expression of pp2a-b′γ leaves, as well as the signaling leading to PR1 expression and accumulation of camalexin, depend on biosynthesis of SA via the SID2/ICS1 pathway.
To gain further insights into the role of PP2A-B′γ in SA signaling (Fig. 4B; Table 1), we assessed the genetic interaction between pp2a-b′γ and nonexpresser of pathogenesis related gene1 (npr1). NPR1 is an SA receptor, a transcriptional coactivator of the SA-inducible PR1, and a central hub in the transcriptional regulation of defense-related SA signaling (Saleh et al., 2015; Ding et al., 2018). In our short-day conditions, npr1 rosettes grew to a very small size and developed distinct lesion spots in the middle of the leaf rim (Supplemental Fig. S8). The pp2a-b′γ mutant grew considerably larger than npr1 plants. In the pp2a-b′γ npr1 double mutant, the dramatic growth effect and lesion formation phenotypes of npr1 were less evident (Supplemental Fig. S8). The npr1 plants accumulated camalexin at levels that were significantly higher than those of pp2a-b′γ. This biochemical characteristic displayed an intermediate phenotype in pp2a-b′γ npr1 (Supplemental Fig. S8B). Moreover, the early flowering of npr1 under short-day conditions did not occur in pp2a-b′γ npr1 (Supplemental Fig. S8C). These data corroborate PP2A-B′γ function interferes with SA-related signaling through NPR1.
The Transcriptome of Apical pp2a-b′γ Leaf Halves Suggests Induction of Major Transcription Factor Cascades Involved in Developmental Leaf Senescence
The dependence of leaf yellowing and SAG12 protein accumulation on functional SID2 in pp2a-b′γ leaves suggested PP2A-B′γ could regulate SA-dependent signaling in leaf senescence. To substantiate this, we performed a transcriptomics (RNA sequencing [RNA-seq]) experiment on the apical and basal leaf halves of 7-week–old wild type, pp2a-b′γ single mutant, pp2a-b′γζ, and pp2a-b′γ sid2 double mutants, and the pp2a-b′γ P35S:PP2A-B′γ complementation line in five independent biological replicates. Firstly, abundance of transcripts in apical leaf halves of pp2a-b′γ, pp2a-b′γ sid2, and pp2a-b′γζ double mutants and complementation line pp2a-b′γ P35S:PP2A-B′γ was compared with the respective abundance in apical leaf halves of wild-type plants (Supplemental Tables S3 and S4).
Genes with ≥2-fold differential expression in the apical leaf halves of pp2a-b′γ, compared to the apical leaf halves of wild-type plants, were analyzed for overrepresentation of gene ontology (GO) categories. GO terms related to innate immunity, SAR, response to hypoxia, phospholipid translocation, hormone catabolic process, positive regulation of leaf senescence, and negative regulation of cell death were significantly enriched in the apical leaf halves of pp2a-b′γ compared to wild type (Supplemental Table S5).
In the next approach, genes with ≥2-fold differential expression in the apical versus the basal sections in wild type and pp2a-b′γ were compared between these two genotypes. A total of 2,188 genes showed increased transcript abundance exclusively in the apical halves of pp2a-b′γ leaves versus pp2a-b′γ basal halves and not in the apical halves of wild-type leaves versus wild-type basal halves (Supplemental Table S6). These genes exhibited an enrichment in GO categories related to defense reactions and leaf senescence (Supplemental Table S6). Increased transcript abundance for 205 genes was evident in the apical parts of both wild type and pp2a-b′γ when compared to the respective basal parts. Intriguingly, these genes were enriched in GO categories of SA signaling and SAR (Supplemental Table S6).
When wild-type and pp2a-b′γ leaf apices were compared, 38 genes that are central in developmental leaf senescence in Arabidopsis were significantly upregulated in pp2a-b′γ (Table 2; color-coded depiction of the same result in Supplemental Table S7). Of these genes, 19 were transcription factors of the NAM, ATAF, and CUC (NAC) and WRKY families. For nine of these transcription factors, the direct downstream target genes, as reported in the literature, were also significantly upregulated in pp2a-b′γ leaf apices as compared to wild type (Table 2; these transcription factors with their targets are listed under subheadings in the table). Five of these target genes were transcription factors themselves, indicating transcriptional cascades that likely have a broad effect on the transcriptome in apical sections of pp2a-b′γ leaves.
Table 2. Abundance of senescence-related transcripts in apical leaf halves of 7-week–old pp2a-b′γ, pp2a-b′γ sid2, and pp2a-b′γζ double mutants and complementation line pp2a-b′γ P35S:PP2A-B′γ relative to those of wild-type plants in RNA-seq analysis.
Values are log2 (FC) and are the means of five independent biological replicates. Transcription factors (TFs) are denoted by x in the respective column. TFs, which have been experimentally shown to bind the promoter of, or directly transcriptionally activate, a down-stream target gene (reference given in the respective column for each target), are listed under subheadings together with their respective target gene transcripts. For the TF ORE1, its up-stream transcription factor PRR9 (Kim et al., 2018b) constitutes the uppermost level of a transcriptional cascade with three levels. AGI, Arabidopsis genome initiative locus code. False discovery rate (FDR) denotes the corrected P-values for differential expression across all genotypes.
| AGI | TF | Annotation | FC to Wild Type, Log2 | FDR | Reference | |||
|---|---|---|---|---|---|---|---|---|
| pp2a-b′γ | pp2a-b′γ sid2 | pp2ab′γζ | pp2a-b′γ P35S:PP2A-B′γ | |||||
| WRKY75 and target genes | ||||||||
| AT5G13080 | x | WRKY75 | 9.0 | 1.4 | 1.0 | 2.0 | 7.60e-16 | — |
| AT1G74710 | — | SID2/ICS1 | 3.2 | −1.0 | 0.6 | 0.3 | 4.70e-09 | Guo et al., 2017 |
| PRR9, ORE1, and Target Genes | ||||||||
| AT2G46790 | x | TL1/PRR9 | 0.1 | −2.8 | −1.4 | −1.5 | 1.58e-15 | — |
| AT5G39610 | x | ORE1/ANAC092 | 3.8 | −2.4 | −0.3 | 0.2 | 3.11e-14 | Kim et al., 2018b |
| AT1G11190 | — | BFN1 | 5.0 | 0.2 | −1.3 | −0.2 | 1.19e-06 | Matallana-Ramirez et al., 2013 |
| AT1G01480 | — | ACS2 | 9.1 | 0.1 | 0.1 | 1.1 | 8.05e-17 | Qiu et al., 2015 |
| AT4G22920 | — | NYE1/SGR1 | 1.8 | −0.2 | −0.1 | −0.2 | 2.04e-09 | Qiu et al., 2015 |
| AT5G01820 | — | CIPK14/SR1 | 2.3 | −0.4 | −0.5 | −0.4 | 1.73e-14 | Balazadeh et al., 2010 |
| AT5G14000 | x | ANAC084 | 4.0 | −0.5 | −0.7 | 0.3 | 1.41e-08 | Balazadeh et al., 2010 |
| AT5G13170 | — | SAG29 | 1.1 | −1.3 | −1.6 | −1.3 | 3.82e-03 | Balazadeh et al., 2010 |
| AT4G19810 | — | CHIC | 3.2 | 0.4 | 0.5 | 0.7 | 2.43e-10 | Balazadeh et al., 2010 |
| AT1G19200 | — | AT1G19200 | 1.9 | −0.5 | 0.4 | −0.1 | 1.94e-04 | Balazadeh et al., 2010 |
| WRKY53 and Target Genes | ||||||||
| AT4G23810 | x | WRKY53 | 1.2 | 0.5 | 1.7 | 0.2 | 4.27e-05 | — |
| AT3G01080 | x | WRKY58 | 1.8 | −0.3 | −0.2 | 0.1 | 1.37e-10 | Miao et al., 2004 |
| AT4G01250 | x | WRKY22 | 1.8 | 0.7 | 1.9 | 0.9 | 2.06e-05 | Miao et al., 2004 |
| AT2G23320 | x | WRKY15 | 1.8 | 0.2 | 1.0 | 0.2 | 8.50e-10 | Miao et al., 2004 |
| AT5G01900 | x | WRKY62 | 5.6 | 0.1 | 0.6 | 1.3 | 9.33e-10 | Miao et al., 2004 |
| AT5G45890 | — | SAG12 | 13.0 | 0.0 | 0.0 | 0.1 | 3.95e-26 | Miao et al., 2004 |
| AT5G14930 | — | SAG101 | 1.3 | 0.1 | 0.6 | 0.3 | 4.57e-07 | Miao et al., 2004 |
| WRKY45 and Target Genes | ||||||||
| AT3G01970 | x | WRKY45 | 4.7 | −0.1 | −0.1 | 0.5 | 1.89e-18 | — |
| AT5G45890 | — | SAG12 | 13.0 | 0.0 | 0.0 | 0.1 | 3.95e-26 | Chen et al., 2017 |
| AT2G29350 | — | SAG13 | 6.7 | 0.6 | 0.2 | 0.2 | 9.82e-15 | Chen et al., 2017 |
| WRKY6 and Target Gene | ||||||||
| AT1G62300 | x | WRKY6 | 3.1 | −0.1 | 0.5 | 0.2 | 5.25e-15 | — |
| AT2G19190 | — | FRK1/SIRK | 5.6 | −0.8 | −0.1 | 0.2 | 2.55e-19 | Robatzek and Somssich, 2002 |
| NAP/ANAC029 and Target Gene | ||||||||
| AT1G69490 | x | NAP/ANAC029 | 5.1 | −0.8 | −0.2 | −0.1 | 1.67e-16 | — |
| AT2G27150 | — | AAO3 | 1.3 | −0.1 | 0.0 | 0.1 | 3.21e-13 | Yang et al., 2014 |
| ANAC016 and Target Gene | ||||||||
| AT1G34180 | x | ANAC016 | 2.4 | 0.1 | 0.8 | 0.6 | 1.75e-15 | — |
| AT4G22920 | — | NYE1/SGR1 | 1.8 | −0.2 | −0.1 | −0.2 | 2.04e-09 | Sakuraba et al., 2016 |
| ANAC019 and Target Gene | ||||||||
| AT1G52890 | x | ANAC019 | 3.3 | 0.6 | 1.0 | 1.1 | 1.38e-06 | — |
| AT4G22920 | — | NYE1/SGR1 | 1.8 | −0.2 | −0.1 | −0.2 | 2.04e-09 | Zhu et al., 2015 |
| NAC3/ANAC055 and Target Gene | ||||||||
| AT3G15500 | x | NAC3/ANAC055 | 6.1 | −1.2 | 1.2 | 1.0 | 3.19e-11 | — |
| AT4G22920 | — | NYE1/SGR1 | 1.8 | −0.2 | −0.1 | −0.2 | 2.04e-09 | Zhu et al., 2015 |
| Further Genes Involved in Developmental Leaf Senescence | ||||||||
| AT3G04060 | x | ANAC046 | 1.3 | −2.3 | −0.9 | −0.7 | 4.46e-14 | — |
| AT3G29035 | x | ORS1/ANAC059 | 1.1 | −1.7 | −0.4 | −0.2 | 6.40e-11 | — |
| AT3G56400 | x | WRKY70 | 2.1 | 0.0 | −0.1 | −0.3 | 3.58e-07 | — |
| AT2G40750 | x | WRKY54 | 1.4 | −0.7 | −0.5 | −0.5 | 1.04e-12 | — |
| AT5G24110 | x | WRKY30 | 1.3 | 0.7 | 1.3 | 0.3 | 2.07e-02 | — |
| AT5G51070 | — | SAG15/ClpD | 1.0 | 0.2 | 0.3 | 0.4 | 1.32e-04 | — |
| AT4G02380 | — | SAG21 | 1.7 | −1.2 | 0.7 | 0.1 | 1.69e-08 | — |
| AT3G10985 | — | SAG20 | 1.5 | 0.0 | 0.7 | 0.2 | 3.73e-11 | — |
| AT2G45210 | — | SAG201/SAUR36 | 2.8 | −0.1 | −0.3 | 0.2 | 9.80e-12 | — |
| AT4G35770 | — | SEN1 | 1.8 | −1.5 | −1.0 | −0.8 | 4.53e-11 | — |
| AT2G13810 | — | ALD1 | 4.2 | −1.1 | −1.1 | −0.7 | 8.22e-08 | — |
| AT5G37600 | — | GLN1;1 | 1.2 | −0.9 | −0.4 | −0.1 | 5.90e-15 | — |
| Negative Transcriptional Markers of Developmental Leaf Senescence | ||||||||
| AT1G29930 | — | CAB1/LHCB1.3 | −1.9 | −0.6 | −0.6 | −1.1 | 1.30e-04 | — |
| AT3G54890 | — | LHCA1 | −1.6 | −0.8 | −0.6 | −0.9 | 2.79e-07 | — |
| AT5G38410 | — | RBCS3B | −1.3 | 0.9 | 0.2 | 0.3 | 5.68e-11 | — |
| AT1G03630 | — | PORC | −1.3 | −0.1 | −0.2 | −0.2 | 2.38e-10 | — |
The significantly increased abundance of senescence-related gene transcripts in the apical leaf halves of pp2a-b′γ was almost completely annulled in the SA-deficient pp2a-b′γ sid2 double mutant (Table 2, Supplemental Table S7). Transcripts for PSEUDO-RESPONSE REGULATOR9 (PRR9), ORESARA1/ANAC092 (ORE1), SAG29, ANAC055, ANAC046, SAG21, ORE1 SISTER1 (ORS1), SENESCENCE ASSOCIATED GENE1 (SEN1), and ALD1 were significantly less abundant in pp2a-b′γ sid2 when compared to wild type, while they were significantly more abundant in pp2a-b′γ (Table 2). Even though transcripts for WRKY75 were detected in elevated amounts in pp2a-b′γ sid2 compared to wild type (2.6-fold; Table 2), this was notably lower as compared to the 500-fold upregulation observed in pp2a-b′γ (Table 2). The circadian clock component PRR9 regulates the onset of age-dependent leaf senescence (Kim et al., 2018b) and acts as one of the upstream activating transcription factors for ORE1, which encodes the master regulator of cell death in Arabidopsis developmental leaf senescence (Kim et al., 2009). Transcripts for eight direct ORE1 target genes showed increased abundance in pp2a-b′γ, but seven of these were detected at wild-type levels in the pp2a-b′γ sid2 double mutant (Table 2). SAG29 and ORE1 itself showed even stronger downregulation with significantly diminished transcript abundance in pp2a-b′γ sid2 (Table 2).
The transcriptomic patterns of SAGs in the pp2a-b′γζ double mutant and the complementation line pp2a-b′γ P35S:PP2A-B′γ followed that of the pp2a-b′γ sid2 double mutant described in this article. This was evident with respect to diminished transcript levels for the key PRR9/ORE1 regulon as well as many other transcription factors involved in leaf senescence and SAGs (Table 2; Supplemental Table S7). These observations are in line with the finding that none of these genotypes showed macroscopic senescence-like phenotypes or SA-dependent expression of SAG12 or PR1 (Fig. 4).
The pp2a-b′γ mutant also showed upregulation of NAC DOMAIN CONTAINING PROTEIN29/ABSCISIC ALDEHYDE OXIDASE3 (NAP/AAO3), and ANAC016/STAY-GREEN1 (SGR1) regulons (Table 2), which are directly involved in chlorophyll breakdown in Arabidopsis developmental leaf senescence (Yang et al., 2014; Sakuraba et al., 2016). Chlorophyll-binding proteins (CABs) are commonly used as negative marker genes for developmental leaf senescence. Transcripts for two CABs, the photosynthetic light-harvesting antenna proteins LHCB1.3 (Besseau et al., 2012; Zhang et al., 2017) and LHCA1 (Mishina et al., 2007; Zhang et al., 2017), showed reduced abundance in the yellowing pp2a-b′γ leaf halves, as did Rubisco small subunit 3B (RBCS3B) and the chlorophyll biosynthesis gene PROTOCHLOROPHYLLIDE OXIDOREDUCTASE C (PORC), which represent additional negative marker genes for senescence (Table 2). In contrast, the transcript abundance for the ATP-dependent chloroplastic Clp-protease SAG15/ClpD (Zheng et al., 2002) was increased in pp2a-b′γ leaves (Table 2), as is known to occur during developmental leaf senescence (Weaver et al., 1999). Hence, the degradation processes in the chloroplasts also seem to follow the natural pattern of age-dependent leaf senescence in pp2a-b′γ.
It is worth noting almost all of the senescence-associated transcription factor regulons and the other senescence-related genes in Table 2 displayed significantly increased abundance in the biologically older, symptomatic apical leaf halves of pp2a-b′γ when compared to the basal leaf halves of pp2a-b′γ (Supplemental Table S8). Finally, to validate these results we performed RT-qPCR analysis using the apical leaf halves of wild type, pp2a-b′γ, pp2a-b′γ P35S:PP2A-B′γ, and the pp2a-b′γ sid2 double mutant (Supplemental Fig. S9). Transcripts for the senescence-induced genes WRKY75, FLG22-INDUCED RECEPTOR-LIKE KINASE1/SENESCENCE-INDUCED RECEPTOR-LIKE KINASE (FRK1/SIRK), SAG13, and SAG21 all showed high induction in apical leaf halves of pp2a-b′γ when compared to apical leaf halves of wild type, pp2a-b′γ P35S:PP2A-B′γ, or pp2a-b′γ sid2 (Supplemental Fig. S9).
The Premature Yellowing of pp2a-b′γ Involves Signaling through ORE1, the Master Regulator of Cell Death in Developmental Leaf Senescence
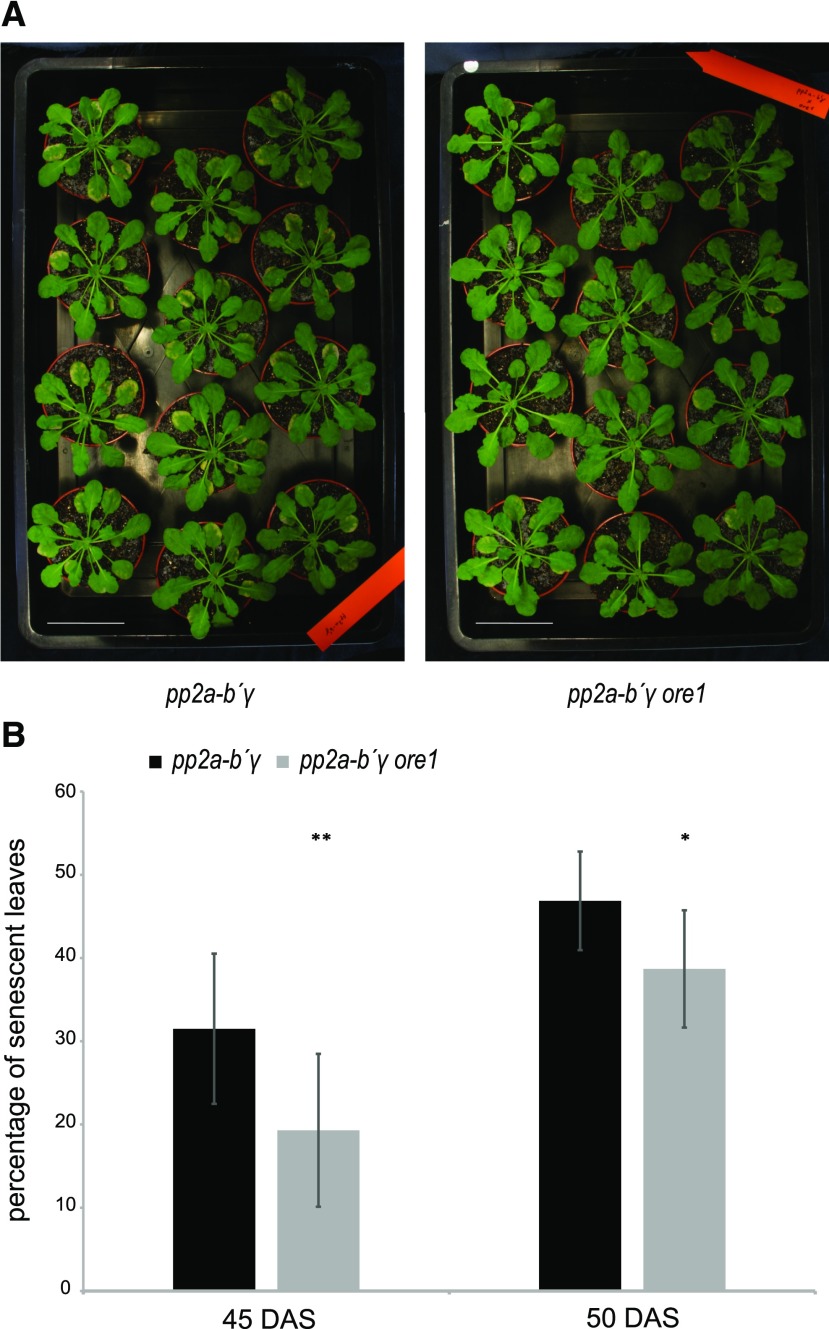
Results from the RNA-seq experiments suggested the yellowing of pp2a-b′γ leaves involves transcriptional upregulation of key components of developmental leaf senescence (Table 2). To assess whether processes related to developmental leaf senescence contribute to the phenotypic characteristics of pp2a-b′γ, we examined leaf yellowing and cell death in pp2a-b′γ ore1 double mutants. As shown in Figure 6, the yellowing and cell death phenotype of pp2a-b′γ was significantly delayed in pp2a-b′γ ore1 double mutants. At 45 d after sowing (DAS), 30% of pp2a-b′γ leaves showed symptoms of initial yellowing, while in pp2a-b′γ ore1, ∼20% of the leaves showed these symptoms (Fig. 6B, Supplemental Fig. S10). At 50 DAS, this phenotypical difference started to level off, with ∼45% of pp2a-b′γ and 38% of pp2a-b′γ ore1 showing symptoms of senescence (Fig. 6B). At this point, wild type and ore1 single mutants did not show any visual symptoms of leaf senescence (Supplemental Fig. S11).
Figure 6.
Senescence progression in pp2a-b′γ and pp2a-b′γ ore1 leaves. A, Photographs depicting leaf yellowing in pp2ab′γ and pp2ab′γ ore1 leaves, 45 DAS. Scale bars = 5 cm. B, Quantification of senescence progression in pp2ab′γ and pp2ab′γ ore1 leaves shown in (A), expressed as percentage of the number of leaves showing any visual symptoms of yellowing and cell death from all leaves of the plant. Data are means (nplants = 12) ± sd. Statistically significant differences to pp2ab′γ are indicated by asterisks. Student’s t test, **P < 0.005; *P < 0.01.
DISCUSSION
Age-dependent regulatory modules are critical in determining defensive measures and timing of developmental transitions in plants. Within regulatory networks, posttranslational control by reversible protein phosphorylation is one of the key mechanisms that regulate cellular functions. The PP2A regulatory subunit PP2A-B′γ is a versatile signaling protein that controls stress reactions and growth at different stages of plant development (Trotta et al., 2011; Heidari et al., 2013; Li et al., 2014; Konert et al., 2015a). In presenescent Arabidopsis leaves, PP2A-B′γ maintains chloroplast functionality and controls oxidative stress responses and the associated metabolic alterations in short-day conditions (Trotta et al., 2011; Li et al., 2014; Konert et al., 2015b). On a molecular level, PP2A-B′γ contributes to defensive metabolism through its interactions with INDOLE GLUCOSINOLATE METHYL TRANSFERASEs, forming a mechanism that negatively regulates the channeling of precursors for generation of deterring methoxylated indole glucosinolates (Rahikainen et al., 2017). With respect to cell death regulation pp2a‐b′γ develops smaller necrotic lesions when infected by B. cinerea, while unchallenged pp2a‐b′γ leaves undergo premature yellowing and cell death upon aging (Figs. 3 and 4; Trotta et al., 2011).
Here we have addressed the molecular mechanisms by which PP2A‐B′γ regulates B. cinerea interactions and leaf senescence in Arabidopsis under short-day conditions. We extend the regulatory function of PP2A‐B′γ to a protein kinase–phosphatase interaction, where PP2A-B′γ controls CPK1, which is a key player in Arabidopsis resistance against B. cinerea infection (Coca and San Segundo, 2010). In addition, our results support a dual age-dependent role for PP2A-B′γ in the regulation of SA signaling. In younger leaf tissues, PP2A-B′γ acts as a negative regulator of SA signaling-mediated defense gene expression, while at later stages of development it controls the SA component in the onset of developmental leaf senescence in short-day conditions.
PP2A-B′γ Influences the Defense-Associated CPK1
Plant immunity is governed by converging regulatory pathways, which collectively determine the outcome of defensive reactions. Reversible protein phosphorylation is a central regulatory mechanism in these interactions, but connecting protein kinases to their counteracting protein phosphatases and understanding the physiological significance of kinase–phosphatase interactions are still matters of intensive investigation. PP2A-type protein phosphatases, which in Arabidopsis can theoretically form 255 different trimeric holoenzymes, and CDPKs, a 34-member family in Arabidopsis, are well known for their roles in plant defense signaling networks (Schulz et al., 2013; Romeis and Herde, 2014; Durian et al., 2016). In PP2A, the regulatory B-subunits function as substrate-binding modules (Farkas et al., 2007) and their in vivo interaction partners are therefore candidates for direct dephosphorylation by PP2A holoenzymes. Here, we present an in vivo interaction between PP2A-B′γ and CPK1 and describe a regulatory effect of PP2A-B′γ on the kinase activity of CPK1 (Figs. 1 and 2). As the upstream protein phosphatases regulating CPK1 have so far remained unidentified, this molecular interaction adds a potential layer of regulation in signaling through calcium-dependent protein kinases in plants.
The cytosolic interaction between CPK1 and PP2A-B′γ (Fig. 1) is consistent with previous reports, which have observed PP2A-B′γ-YFP or a fraction of CPK1 in this subcellular compartment (Trotta et al., 2011; Majeran et al., 2018). The localization of CPK1 depends on N-terminal myristoylation at Gly-2 and palmitoylation at Cys-5. CPK1 lacking myristoylation or both acylations is localized in the cytosol, as evidenced by transient overexpression of a mutated CPK1-GFP lacking one or both known acylation sites in Arabidopsis root tip cells or onion (Allium cepa) epithelial cells (Dammann et al., 2003; Coca and San Segundo, 2010). In its Gly-2-myristoylated form, CPK1 is readily detectable in the total cell extracts, enriched in plasma membrane and detergent-resistant membranes, and to a minor extent found in the microsomal fraction in Arabidopsis suspension cells (Majeran et al., 2018). Hence, CPK1 likely has compartment-specific roles in cellular signaling depending on its acylation status.
Our results support a model where PP2A-B′γ negatively regulates the activation state of CPK1 (Fig. 2). The knock-down pp2a-b′γ mutant showed increased in-gel kinase activity of the upper of the two major CPK1 band species (Fig. 2). The kinase activity observed from in-gel kinase assays for CPKs is always measured after the addition of calcium, which mimics the in planta activating Ca2+-spike required to overcome the intrinsic autoinhibition of CDPKs. Hence, CPK-kinase activity assays cannot directly reflect the in planta activity state of CPK1 in pp2a-b′γ. By directing the dephosphorylation of a subset of CPK1 in vivo phosphorylation sites, PP2A-B′γ may affect the priming of CPK1 kinase activity, which becomes evident upon perception of calcium and hence the activation of CPK1 in pp2a-b′γ in the experimental system. Tobacco CDPK2, the ortholog of Arabidopsis CPK1, exhibits several differentially phosphorylated bands in SDS-PAGE (Witte et al., 2010), and a correlation between in vivo phosphorylation and increased NtCDPK2 kinase activity has been demonstrated (Romeis et al., 2001). However, in addition to kinase activity, phosphorylation may also regulate the target specificity of CPKs (reviewed by Liese and Romeis, 2013). Differentially phosphorylated forms of CPK1 could therefore interact with different protein substrates, allowing specific CPK1 functions in different signaling pathways.
A role for CPK1 in immunity signaling was reported in resistance against different phytopathogens, including the necrotrophic fungi B. cinerea and F. oxysporum f.sp. matthiolae and the hemibiotrophic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst; Coca and San Segundo, 2010). In addition, CPK1 was also implicated in reactive oxygen species-production in effector-triggered immunity against the avirulent bacterial strains Pst avrRpm1 and Pst avrRpt2 in a cpk1 cpk2 double mutant (Gao et al., 2013). We found knocking-down PP2A-B′γ promoted constitutive immune responses and contributed to basal resistance against B. cinerea. Because this resistance coincided with increased protein expression of CPK1 (Fig. 3), one of the defense signaling functions of PP2A-B′γ appears to be to control the abundance of CPK1 in B. cinerea-infected leaf tissues (Fig. 3B).
PP2A-B′γ Regulates SA Signaling in Both Plant Immunity and Developmental Leaf Senescence
In presenescent leaves, PP2A-B′γ prevented extraneous SA-related gene expression and transcriptional defense priming, as evidenced by increased transcript abundance for a cluster of coexpressed genes with well-known roles in SA-signaling, SAR, and/or immune responses against fungi in 4-week–old pp2a-b′γ plants (Table 1). Notably, the constitutive transcriptional activation of SA-signaling genes in pp2a-b′γ (Table 1) was not concurrent with transcriptional activation of SID2 (Table 1). Similar to pp2a-b′γ and pp2a-b′γζ (Table 1; Fig. 3), plants overexpressing the fungal protein SsCP1 from S. sclerotiorum accumulated transcripts of PR1, PR2, and PR5 and were more resistant to B. cinerea in a leaf lesion assay (Yang et al., 2018). It is therefore intriguing that the increased PR1 transcript accumulation in 4-week–old pp2a-b′γ or pp2a-b′γζ was not reflected in a detectable PR1 protein level when assessed in mock-treated plants (Fig. 3B). At this developmental stage, translation of PR1 mRNA might be inhibited e.g. via posttranscriptional modifications or storage in mRNA-containing stress granules (Protter and Parker, 2016; Kosmacz et al., 2019) that could sequester PR1 mRNA from translation. In contrast, when exposed to B. cinerea, all the genotypes studied showed accumulation of PR1 protein (Fig. 3B), reflecting the important role of PR1 in plant resistance against necrotrophic fungi (Gamir et al., 2017; Nie et al., 2017; Yang et al., 2018). Evidently, the constitutive transcriptional activation of SA-related immune responses did not promote cell death when pp2a-b′γ was exposed to the necrotrophic pathogen B. cinerea (Table 1; Fig. 3).
The onset of developmental leaf senescence is highly responsive to internal and external cues and is tightly regulated on the levels of transcriptome (Breeze et al., 2011; Woo et al., 2016), proteome (Tamary et al., 2019) and metabolome (Watanabe et al., 2013; Chrobok et al., 2016; Jafari et al., 2017). By characterization of aging pp2a-b′γ leaves, we demonstrate the regulatory role of PP2A-B′γ in SA signaling extends to the SA component of developmental leaf senescence under short-day conditions (Figs. 4–6; Table 2; Supplemental Tables S7 and S8).
Posttranslational regulation of age-dependent leaf senescence has become an intensive field of research, but only a few protein phosphatases have been identified to date. In this study, we demonstrate PP2A exerts control over developmental leaf senescence. The occurrence of precocious senescence in pp2a-b′γ leaves manifested itself in the expression of the senescence marker protein SAG12 in the leaf apices of 7-week–old pp2a-b′γ plants, which also showed the degreening and cell death typical of this developmental stage in Arabidopsis leaves (Fig. 4). Concomitantly, there was a SID2-dependent accumulation of SA and camalexin in these leaves (Fig. 5), both of which are known to accumulate during age-dependent leaf senescence in short day-grown Arabidopsis wild-type plants (Mishina et al., 2007). In the yellowing leaf tips of pp2a-b′γ, active SA-signaling was reflected by strong accumulation of PR1-protein (Fig. 4B). Collectively, these data support a role for PP2A-B′γ as a regulator of SA-related signaling in leaf senescence.
PP2A-B′γ Controls the Expression of Regulatory Modules Involved in Developmental Leaf Senescence
Transcription factors of the WRKY and NAC families play key roles during developmental leaf senescence, and many of them are transcriptionally upregulated during this process (Breeze et al., 2011; Woo et al., 2016). We detected increased transcript abundance for several senescence-related transcription factors, including the key regulators WRKY75, WRKY53, and ORE1, in the yellowing apices of pp2a-b′γ leaves (Table 2; Supplemental Tables S7 and S8; Supplemental Fig. S9).
WRKY75 and WRKY53 act as important positive factors in the onset of leaf senescence (Miao et al., 2013; Guo et al., 2017; Ren et al., 2017; Zhang et al., 2017; Liu et al., 2019). Counter to the negative regulatory role of PP2A-B′γ in SA-related defense and senescence signaling (Tables 1 and 2; Figs. 3–5; Supplemental Tables S7 and S8), WRKY75 has a dual positive role in promoting defense reactions at earlier stages of plant development and mediating SA signaling and PR1 transcription in senescing tissues (Guo et al., 2017). WRKY75 binds to the promoter of SID2 and activates its transcription with consequent production of SA (Guo et al., 2017). Notably, WRKY75 itself is also transcriptionally induced by SA, creating a feed-forward loop that promotes leaf senescence (Guo et al., 2017; Zhang et al., 2017). In line with these findings, the early senescence phenotype of WRKY75-overexpressing plants was suppressed in the sid2 background, where biosynthesis of SA through the isochorismate-dependent pathway is not operational (Guo et al., 2017). We found the abundance of SID2 mRNA was strongly increased in pp2a-b′γ leaf apices (9-fold; Table 2) and SA produced by the SID2-pathway was a prerequisite for the formation of the precocious leaf senescence phenotype of pp2a-b′γ (Figs. 4 and 5). The strongly increased expression of WRKY75 and SID2 in the yellowing pp2a-b′γ leaf tips may therefore reflect activation of the positive feed-forward loop with a consequent fast progression of leaf senescence as observed in pp2a-b′γ. Interestingly, the apical leaf halves of pp2a-b′γ also showed a strong transcriptional upregulation of a gene encoding avrPphB SUSCEPTIBLE3 (Supplemental Table S4), which was only very recently shown to catalyze the subsequent step in SA-biosynthesis after the SID2-catalyzed production of the intermediate isochorismate (Rekhter et al., 2019).
In addition to WRKY75, WRKY53, and six of the known transcriptional target genes of WRKY53, among them SAG12, showed increased transcript abundance in pp2a-b′γ (Table 2). WRKY45 also directly activates the SAG12 expression (Chen et al., 2017) and was strongly transcriptionally induced in pp2a-b′γ (Table 2). The transcriptional pattern of age-dependent leaf senescence in pp2a-b′γ is further strengthened by elevated transcript abundance of negative regulators slowing down the process of senescence, notably WRKY70 and WRKY54 (Table 2), which are upregulated in this developmental stage in a partially SID2-dependent manner (Besseau et al., 2012).
Besides other NAC transcription factors, pp2a-b′γ showed enhanced expression of ORE1 and eight of its known target genes (Table 2). This is noteworthy, as ORE1 encodes the key transcriptional regulator of cell death in developmental leaf senescence and is additionally involved in regulating chlorophyll degradation in this process (Kim et al., 2009; Qiu et al., 2015). In contrast, the pp2a-b′γ sid2 and pp2a-b′γζ double mutants lacked transcriptional induction of the ORE1 cascade as well as the chlorosis and cell-death phenotypes (Fig. 4). One of the ORE1 targets upregulated in pp2a-b′γ, NON-YELLOWING1 (NYE1/SGR1), encodes a Mg-dechelatase, which is predominantly expressed during leaf senescence and catalyzes the first step in chlorophyll a degradation (Shimoda et al., 2016; Li et al., 2017). In addition to being an ORE1 target, NYE1 is transcriptionally activated by the senescence-associated NAC transcription factors ANAC016, ANAC019, and ANAC055, all of which were induced in the yellowing leaf apices of pp2a-b′γ (Table 2).
The involvement of the ORE1-dependent pathway in pp2a-b′γ leaf senescence was corroborated by a slower progression of apical leaf chlorosis in pp2a-b′γ ore1 as compared to pp2a-b′γ (Fig. 6). In addition, chlorosis and cell death in the leaf apices of pp2a-b′γ leaves require a functional PP2A-B′ζ gene, as shown by the absence of senescence-associated phenotypes in the pp2a-b′γζ double mutant (Fig. 4). This illustrates a nonredundant dependency of the two close homologs among PP2A-B-subunits, PP2A-B′γ and PP2A-B′ζ. There is no dominant effect of a transcriptional induction of PP2A-B′ζ in pp2a-b′γ, as evidenced by the slightly diminished transcript levels of PP2A-B′ζ in pp2a-b′γ when compared to the wild type (Supplemental Table S4). Because transcriptional induction of SID2 (Table 2), accumulation of SA and camalexin (Fig. 5), and protein expression of PR1 and SAG12 (Fig. 4B) were not detectable in pp2a-b′γζ, we conclude PP2A-B′ζ is indispensable for the SA-regulated premature senescence in pp2a-b′γ.
Summary
Taken together, we ascribe a dual regulatory role for PP2A-B′γ in the leaves of Arabidopsis. PP2A-B′γ prevents defense responses in unchallenged leaves of younger plants, while at later stages of development it prevents premature initiation and progression of developmental leaf senescence. In both cases, these regulatory functions of PP2A-B′γ involve SA-dependent signaling, and in the case of senescence, the concomitant accumulation of this phytohormone.
MATERIALS AND METHODS
Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accession Columbia (Col-0) wild type and mutants were grown under 130 μmol photons m−2 s−1, 22°C and 50% humidity at an 8-h light period. GVG-AvrPto transgenic plants (Hauck et al., 2003; Tsuda et al., 2012) were grown under 100 μmol photons m−2 s−1, 22°C, and 70% humidity at a 12-h light period. Homozygous pp2a-b′γ (SALK_039172), pp2a-b′ζ (SALK_107944C), pp2a-b′γζ, pp2a-b′γ sid2 (sid2-1 allele), and a pp2a-b′γ line complemented by 35S-driven expression of the PP2A-B′γ gene have been generated before (Trotta et al., 2011; Li et al., 2014; Rasool et al., 2014). Homozygous pp2a-b′γ ore1 (ore1: SALK_090154), pp2a-b′γ sag12 (sag12: SALK_203275C), and pp2a-b′γ npr1-1 (npr1-1; Cao et al., 1997) were identified with the primers listed in Supplemental Materials and Methods and in the case of the latter by subsequent restriction analysis of the CAPS marker of npr1-1 with NlaIII. The homozygous CPK1-KO lines used were SALK_096452 and SALK_080155c; the latter was used for the hormone measurements and as a second line in Supplemental Figure S1.
Botrytis cinerea strain B05.10 was grown on homemade potato dextrose agar plates. Potato dextrose agar (1 L) consisted of 300 g of potato (Solanum tuberosum), 50 g of tomato (Solanum lycopersicum), 25 g of carrot (Daucus carota), 15 g of agar, 10 g of dextrose, and 1.5 g of yeast extract. For infection, the plants were grown for 4 weeks at 23°C/18°C and 65%/75% humidity under an 8-h/16-h (day/night) photoperiod, and infected with a 1 × 106 B. cinerea spore suspension in 0.5× PDB (Difco) by 3-μL drop inoculations or by spraying, and covered to maintain 100% humidity. Mock-treated plants were sprayed with 0.5× PDB and treated in parallel to the infected plants. Lesion size on drop-inoculated leaves was determined after 48 h. Measurement of lesion diameters was performed with the software ImageJ (v1.47v; Schneider et al., 2012) and the data were analyzed with the software R (v3.4.0) using the nlme (Pinheiro et al., 2017) and multcomp (Hothorn et al., 2008) packages. Samples of spray-inoculated plants were collected 24-h postinfection and immediately frozen in liquid nitrogen.
Analysis of Protein Interactions by Yeast Two-Hybrid Screening and BiFC
Yeast two-hybrid screening was conducted with Hybrid Hunter (Invitrogen; http://www.invitrogen.com) using yeast (Saccharomyces cerevisiae) strain L40 and LexA DBD fusion of PP2A-B′γ in pHybLex as bait against a cDNA library that was enriched for stress-related factors and cloned into the LexA activating domain-containing pYESTrp2 plasmid as described in Jaspers et al. (2009) and Konert et al. (2015b). Twenty-five colony-forming units of Trp auxotrophic yeast were placed on –His selection supplemented with 10 mm of 3-aminotriazole (cat. no. 3AT; Sigma Aldrich; http://www.sigmaaldrich.com) to remove autoactivation. Colonies were picked after 4 d of growth at 28°C and tested for β-galactosidase activity according to the HybridHunter manual (Invitrogen). Putative interaction partners in the pYESTrp2 library plasmid were identified by sequencing.
For BiFC analyses, all constructs were introduced into pGPTVII-backbone BiFC vectors (Waadt and Kudla, 2008; Waadt et al., 2008) as described in Supplemental Materials and Methods. For the Nicotiana benthamiana system, Agrobacterium tumefaciens strain GV3101::pMP90 carrying the fluorescent fusion constructs was infiltrated into leaves according to Waadt and Kudla (2008). For the Arabidopsis system, leaves of 4.5-week–old GVG-AvrPto (Hauck et al., 2003) transgenic plants were sprayed with dexamethasone solution (2 μm of dexamethasone, 0.01% [v/v] Silwet l-77, and 0.1% ethanol; Tsuda et al., 2012) on both adaxial and abaxial leaf surfaces and infiltrated 24 h later with the two BiFC-carrying GV3101::pMP90 and A. tumefaciens strain C58C1 (pBin61-p19, pGV2260), expressing the silencing inhibitor p19, each at an OD600nm = 0.1 in infiltration buffer (10 mm of MES-KOH at pH 5.6, 10 mm of MgCl2, and 150 μm of acetosyringone). After 2.5 d under growth conditions, YFP-fluorescence in Arabidopsis leaf discs was analyzed.
YFP-fluorescence was imaged with a model no. LSM510 META confocal laser scanning microscope (Zeiss; http://www.zeiss.com) with excitation at 514 nm and detection at 535–590 nm and with a model no. LSM780 (Zeiss) with excitation at 514 nm and detection at 519–570 nm (Fig. 1C, SAG12). N. benthamiana leaf discs were incubated in water, and abaxial epidermal cells were visualized with a Plan-Neofluar 20×/0.5 objective (Zeiss). Arabidopsis leaf discs were preincubated for 7 min in Perfluorodecalin (cat. no. P9900; Sigma Aldrich), and abaxial epidermal cells were visualized with objectives Plan-Neofluar 40×/0.75 or C-Apochromat 40×/1.2 W Korr M27 (Zeiss; SAG12; Fig. 1C) of the respective microscopes. The images were exported with the software ZEN 2.1 (Zeiss).
Protein Extraction, PA Gels, and Lambda-Phosphatase Treatment
For protein extraction and subsequent immunoblotting, pools of three Arabidopsis rosettes per sample or pools of seven apical or basal leaf halves per sample were ground in liquid nitrogen and thawed under cooling in extraction buffer containing 100 mm of Tris at pH 8, 100 mm of NaCl, 10 mm of DTT, 0.5% (v/v) Triton X-100 protease inhibitor (cat. no. 88665, 1 tablet/10 mL; Pierce), and phosphatase inhibitor (PhosStop, 1 tablet/10 mL; Roche). Protein concentrations were analyzed with the Detergent-Compatible Protein Assay (Bio-Rad). Sample aliquots with equal total protein amounts (75 μg in the case of SAG12 gels, otherwise 30 μg) were mixed with Laemmli sample buffer (Laemmli, 1970) heated 10 min to 74°C and separated by discontinuous gel electrophoreses on 10% or 12% (w/v) separation PA gels. For immunodetection of CPK1, an isoform-specific CPK1-epitope of 18 amino acids was chosen to generate a custom-made antibody (product no. AS19 4315; Agrisera). The PR1- and SAG12-specific antibodies were acquired from Agrisera (AS10 687, AS14 2771). For treatment with lambda protein phosphatase (cat. no. P0753S; New England Biolabs), protein from rosette pool plant extract containing 82.5 μg of total protein was precipitated with 4-fold volume of 80% (v/v) acetone at 4°C under centrifugation for 15 min. The pellets were washed twice with 80% (v/v) acetone under centrifugation and dried in air. The pellets were incubated with 40 μL of lambda-phosphatase reaction mix for 5 min and then dissolved by pipetting. For the phosphatase reaction, samples were incubated for 30 min at room temperature and protein was precipitated with a 4-fold volume of 100% (v/v) acetone overnight. Pellets were heated for 10 min at 65°C in Laemmli sample buffer, and samples containing 55 μg of total protein of phosphatase-treated or control samples were loaded on PhosTag gels (WAKO) containing 7.5% (w/v) total acrylamide. PhosTag gel preparation and protein separation on the gels was performed according to manufacturer’s instructions (www.wako-chem.co.jp/english/labchem).
CPK In-Gel Kinase Assay
Protein extracts (three rosettes of 4-week–old plants per sample) were prepared with 25 mm of HEPES at pH 7.5, 10 mm of MgCl2, 10 mm of DTT, and 0.5% (v/v) Triton X-100, and samples containing 60 μg of total protein were heated at 65°C for 10 min in Laemmli sample buffer. Thereafter, 30 μg of total protein per sample was separated on 10% SDS-PA gels containing 0.25 mg/mL histone type III-S (cat. no. H5505; Sigma-Aldrich) as kinase substrate. For the in-gel kinase assay, the buffers and conditions for washing, renaturation, and stopping were as described in Gao et al. (2013). The kinase reaction was performed for 2 h in the reaction buffer (Gao et al., 2013) containing 10 μCi 32P-ATP. After stopping the reaction, the gel was dried and bands were visualized by autoradiography by exposure to Super RX film (product no. 47410-19236; Fujifilm). Aliquots of the same samples (containing 30 μg of total protein) were separated on PA gels for CPK1 immunoblotting.
Analysis of Gene Expression by Microarray and RT-qPCR
For microarray analysis of 4-week–old plants, rosettes of wild type, pp2a-b′γ, pp2a-b′ζ, and pp2a-b′γζ double mutant plants were collected 4 h after the onset of light period. RNA from four biological replicates was isolated using an Agilent Plant RNA Isolation Mini Kit (Agilent Technologies), and 200 ng of total RNA was amplified and Cy-3 labeled using a one-color Low Input Quick Amp Labeling Kit (product no. 5190-2331; Agilent Technologies) and processed with the RNA Spike-in Kit (product no. 5188-5282; Agilent Technologies). RNA/complementary RNA quality control was performed using a 2100 Bioanalyzer RNA 6000 Nano Kit (product no. 5067-1511; Agilent Technologies). Cy-3 labeled samples (1.65 μg) were hybridized to Arabidopsis (V4) Gene Expression Microarrays, 4×44K (Design ID 021169; Agilent Technologies) according to the manufacturer’s instructions, and finally scanned with a model no. G2565CA scanner with a profile AgilentHD_GX_1Color. Numeric data were produced with the program Feature Extraction, v0.7.3 (Agilent Technologies). Gene expression data were analyzed using scripts in the R software package (https://rstudio.com). First, the average of processed signal of probes targeting each gene was computed. Then, a linear model with genotype, tissue, and their interaction as fixed effects was estimated. The P values from the linear model were then subjected to false discovery rate (FDR) correction using the qvalues package. Genes with FDR-corrected P values < 0.05 were considered statistically significant. Model comparisons and their P values were estimated using the multcomp package in R (Storey, 2002; Hothorn et al., 2008).
For RT-qPCR analysis, RNA was isolated from rosettes of 4-week–old wild-type, pp2a-b′γ, and pp2a-b′γ P35S:PP2A-B′γ plants in three to four biological replicates, using the innuPREP Plant RNA Kit (cat. no. 845-KS-2060250; Analytik Jena). One μg of RNA was DNAseI-treated and used for cDNA synthesis with Maxima Reverse Transcriptase according to the manufacturer’s instructions (Thermo Fisher Scientific). The reverse transcription reaction was diluted to a final volume of 100 μL, and 1 μL was used per PCR reaction. qPCR was performed in triplicate with EvaGreen Supermix (Solis Biodyne) on a CFX384 Thermal Cycler 1000 (Bio-Rad), as described in Brosché et al. (2014). Normalization of the data were performed in the program qBase+ 2.3 (Biogazelle; https://www.qbaseplus.com/), with three reference genes (PP2AA3, TIP41, and YLS8), which were validated with the tool geNorm (https://genorm.cmgg.be) to have stable expression in the samples used in this study. Amplification efficiency of all primer pairs was calculated through amplification of serially diluted cDNA. Primer sequences and amplification efficiency are provided in Supplemental Materials and Methods. Data were tested with a one-way ANOVA followed by a Tukey posthoc test.
Analysis of Gene Expression by RNA-Seq
For RNA-seq, RNA was isolated from seven apical leaf halves and the corresponding seven basal halves in five biological replicates using the innuPREP Plant RNA Kit (cat. no. 845-KS-2060250; Analytik Jena). Libraries were prepared according to a TruSeq Stranded mRNA Sample Preparation Guide (part no. 15031047; Illumina). Total RNA and library quality was assessed with an Advanced Analytical Fragment Analyzer and Qubit Fluorometric Quantitation (Life Technologies). The libraries were sequenced in single-ended, 50-bp reads using HiSeq 3000 technology (Illumina) at the Turku Bioscience Centre. Gene expression was analyzed using the software Strand NGS 3.11 (Agilent Technologies). Sequence reads were aligned to the Arabidopsis TAIR10 (https://www.arabidopsis.org/download/index-auto.jsp?dir=/download_files/Genes) reference genome with Ensembl gene and transcript annotation. Aligned reads were normalized and quantified using DESeq Bioconductor package (R). Replicate analysis was performed by a two-way ANOVA and a Shapiro–Wilk test of normality (cutoff P < 0.1). The Benjamini–Hochberg procedure was applied to obtain corrected P values representing an FDR for each gene in each comparison (i.e. genotype or tissue).
GO term enrichment analyses were performed using the tool PANTHER (http://pantherdb.org; GO Ontology database release date: 2019-01-01). Statistically significant enrichment was calculated by a Fisher’s Exact Test, corrected according to the calculated FDR, with background reference set to all Arabidopsis genes in the database. Venn diagrams were drawn using the software Venny2.1 (http://bioinfogp.cnb.csic.es/tools/venny/).
Analysis of Metabolites
Frozen fresh material (<200 mg) was homogenized in a bead mill (3-mm chrome balls, 2 × 30 s at 30 Hz) and subsequently extracted with 1-mL ice-cold 85% (v/v) methanol (HPLC grade) containing 10 μM of p-hydroxybenzyl glucosinolate (pOHb, cat. No. 89793; PhytoLab), 0.215 μM of (2H6)-abscisic acid ([2H6]-ABA, cat. No. 0342722; Olchemim), and (2H5)-transzeatin ([2H5]-tZ, cat. No. 0010302; Olchemim) as internal standards. After centrifugation (20 min, 4700g, 4°C), 20 µL of the supernatant was diluted with 80 µL of MilliQ-grade water (1:5; Merck Millipore) and filtered (Durapore 0.22-µm PVDF filters; Merck Millipore) for the analysis of SA and camalexin (Sample 1). For the analysis of JA and ACC, 800 μL of the supernatant was dried in a vacuum concentrator, resuspended in 150-μL 20% (v/v) methanol, and filtered with 0.22-μm PVDF filters (Sample 2). Liquid chromatography tandem mass spectrometry analysis was carried out on an Advance UHPLC system (Bruker) equipped with a Kinetex XB-C18 column (100 × 2.1 mm, 1.7 μm, 100 Å; Phenomenex) coupled to an EVOQ Elite TripleQuad Mass Spectrometer (Bruker) equipped with an electrospray ionization source. The injection volume was 1 μL for Sample 1 and 5 μL for Sample 2. Separation was achieved with a gradient of water/0.05% (v/v) formic acid (solvent A) and acetonitrile (solvent B) at a flow rate of 0.4 mL/min at 40°C (formic acid, cat. no. F0507, reagent grade; acetonitrile, HPLC grade l; Sigma-Aldrich). The elution profile was: 0–0.5 min 2% (v/v) B; 0.5–1.2 min 2% to 30% (v/v) B; 1.2–2.0 min 30% to 100% (v/v) B; 2.0–2.5 min 100% (v/v) B; 2.5–2.6 min 100% to 2% (v/v) B; 2.6% to 4.0 min 2% (v/v) B. Instrument parameters were optimized by infusion experiments with pure standards. For Sample 1, the mass spectrometer parameters were as follows: ionspray voltage was maintained at −2,500 V, cone temperature was set to 300°C and heated probe temperature to 400°C, cone gas flow was set to 20 psi, probe gas flow to 40 psi, nebulizer gas to 60 psi, and collision gas to 1.5 mTorr. For Sample 2, the mass spectrometer parameters were as follows: ionspray voltage was maintained at 3,500 V and −3,300 V, cone temperature was set to 300°C and heated probe temperature to 120°C, cone gas flow was set to 20 psi, probe gas flow to 40 psi, nebulizer gas to 60 psi, and collision gas to 1.5 mTorr. Nitrogen was used as cone and nebulizer gas, and argon as collision gas. Multiple reaction monitoring was used to monitor analyte parent ion > product ion transitions [collision energy]: camalexin (negative) 199 > 141 [15 V], SA (negative) 137 > 93 [13 V], pOHb (negative) 425 > 97 [24 V], (2H5)-tZ (positive) 225 > 137 [15 V], (2H6)-ABA (negative) 269 > 159 [7 V], ACC (positive) 102 > 56 [15 V], JA (negative) 209 > 59 [11 V]. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Camalexin and SA were quantified relative pOHb, JA to (2H6)-ABA and ACC to (2H5)-tZ using experimentally determined response factors with commercially available standards in a representative plant matrix. Statistical analysis was performed using the software R (v3.3.2; http://www.R-project.org/). Data were controlled for statistical prerequisites such as homogeneity of variances and normality. Metabolite concentrations in apical and basal leaf parts of the genotype (Fig. 5) were analyzed with a two-way ANOVA using the linear model (general formula: metabolite ∼ genotype*leaf part) followed by a Tukey posthoc test. Metabolite concentrations of mock- and B. cinerea-treated genotypes (Supplemental Fig. S6) were analyzed with a Kruskal–Wallis one-way ANOVA against genotype and the combined genotype*treatment term, and the Mann–Whitney U test for the treatment term. Camalexin concentrations in rosettes (Supplemental Fig. S8B) were analyzed with a one-way ANOVA using the lm model (general formula: metabolite ∼ genotype) followed by a Tukey posthoc test.
Accession Numbers
The AGI locus numbers (www.arabidopsis.org) for the genes discussed in this article are as follows: At4g15415 (PP2A-B′γ), At3g21650 (PP2A-B′ζ), At5g04870 (CPK1), At1g74710 (SID2/ICS1), At5g45890 (SAG12), At1g64280 (NPR1), and At5g39610 (ORE1/ANAC092). The RNA-seq data discussed in this publication have been deposited in National Center for Biotechnology’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE136944 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136944).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Specificity of the affinity-purified anti-CPK1 antibody employed in this study.
Supplemental Figure S2. ELISA of the affinity-purified anti-CPK1 antibody.
Supplemental Figure S3. Loading controls and full-length immunoblots for Figures 2–4.
Supplemental Figure S4. Disease resistance in 4-week–old B. cinerea-infected pp2a mutant plants.
Supplemental Figure S5. RT-qPCR analysis of transcript abundance of PR1 and strongly PR1-coexpressed genes (genes chosen from microarray results; Table 1) in 4-week–old wild-type, pp2a-b′γ, and p2a-b′γ P35S:PP2A-B′γ plants.
Supplemental Figure S6. Contents of SA, JA, ACC, and jasmonoyl-Ile in 4-week–old mock-treated and B. cinerea-infected Arabidopsis plants.
Supplemental Figure S7. Phenotypic characteristics of 7-week–old short day-grown wild-type, pp2ab′γ, sag12 and pp2a-b′γ sag12 double mutant plants.
Supplemental Figure S8. Genetic interaction between pp2a-b′γ and the SA-signaling mutant npr1-1.
Supplemental Figure S9. RT-qPCR analysis of transcript abundance of WRKY75, FRK1/SIRK, SAG13, and SAG21 in apical leaf halves of 7-week–old wild type, pp2a-b′γ, pp2a-b′γ P35S:PP2A-B′γ, and pp2a-b′γ sid2.
Supplemental Figure S10. Phenotypic characteristics of short day-grown wild-type, pp2ab′γ, ore1, and pp2a-b′γ ore1 double mutant plants 45 DAS.
Supplemental Figure S11. Phenotypic characteristics of short day-grown wild-type, pp2ab′γ, ore1, and pp2a-b′γ ore1 double mutant plants 50 DAS.
Supplemental Table S1. Candidate Arabidopsis PP2A-B′γ interacting proteins related to SA signaling and senescence as identified by yeast two-hybrid screening.
Supplemental Table S2. Microarray analysis of gene expression in pp2a mutant plants after a 4-week growth period under 130-μmol photons m−2 s−1 in short-day conditions.
Supplemental Table S3. Abundance of transcripts in apical leaf halves of pp2a-b′γ, pp2a-b′γ sid2, and pp2a-b′γζ double mutants and complementation line pp2a-b′γ P35S:PP2A-B′γ relative to those of wild-type plants in RNA-seq analysis.
Supplemental Table S4. Abundance of transcripts of PP2A-B′γ, PP2A-B′ζ, SID2, and avrPphB SUSCEPTIBLE3 in apical leaf halves of pp2a-b′γ, pp2a-b′γ sid2, and pp2a-b′γζ double mutants and complementation line pp2a-b′γ P35S:PP2A-B′γ relative to those of wild-type plants in RNA-seq analysis.
Supplemental Table S5. GO terms overrepresented in transcripts significantly induced in apical leaf halves of pp2a-b′γ compared with apical leaf halves of wild type.
Supplemental Table S6. Comparison of gene expression between apical and basal halves of leaves in wild type and pp2a-b′γ.
Supplemental Table S7. Abundance of senescence-related transcripts in apical leaf halves of 7-week–old pp2a-b′γ, pp2a-b′γ sid2, and pp2a-b′γζ double mutants and complementation line pp2a-b′γ P35S:PP2A-B′γ relative to those of wild-type plants in RNA-seq analysis.
Supplemental Table S8. Abundance of senescence-related transcripts differentially expressed in apical leaf halves as compared to basal leaf halves of 7-week–old pp2a-b′γ in RNA-seq analysis.
Supplemental Materials and Methods. Cloning of BiFC constructs and oligonucleotides for qPCR.
Acknowledgments
We are grateful for Andrea Trotta and Grzegorz Konert, University of Turku, for excellent assistance. We thank Tina Romeis, Free University of Berlin, for the seeds of the cpk1-lines, Claus-Peter Witte, University of Hannover, for the GVG-AvrPto seeds, Teemu Teeri, University of Helsinki,for the bacterial strain C58C1, and Jörg Kudla, University of Münster, for BiFC vectors. The Salk Institute Genomic Analysis Laboratory is acknowledged for providing sequence-indexed T-DNA insertion mutants. Confocal imaging was performed with microscopes of the Cell Imaging and Cytometry Core at the Turku Bioscience Centre, University of Turku and at Åbo Akademi University.
Footnotes
This work was supported by the Academy of Finland (project 289687 to G.D., 307719 to S.K., 303757 to E.-M.A., and 317218 to J.K.); the Academy of Finland Center of Excellence in Primary Producers 2014–2019 (307335); the University of Turku Doctoral Programme in Molecular Life Sciences, the Turku University Foundation, and the Finnish Cultural Foundation Varsinais-Suomi Regional Fund (to M.R.); the Danish National Research Foundation (grant 99 to M.Bu.); Villumfonden (project no. 13169 to V.J.); the Scandinavian Society for Plant Physiology (travel funding to G.D.); a Deutsches Forschungsgemeinschaft Heisenberg Fellowship (GL346/5 to E.G.); the University of Helsinki Doctoral Program in Plant Science (to K.V., as a member of the doctoral program); The Ella and Georg Ehrnrooth Foundation (additional funding to K.V.); and the Finnish Functional Genomics Centre, University of Turku, and Åbo Akademi and Biocenter Finland.
Articles can be viewed without a subscription.
References
- APG IV (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181: 1–20 [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor M-I, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux J-P, Peck SC, Ulm R (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker MA, DeLong A (2017) Atypical Protein Phosphatase 2A gene families do not expand via paleopolyploidization. Plant Physiol 173: 1283–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosché M, Blomster T, Salojärvi J, Cui F, Sipari N, Leppälä J, Lamminmäki A, Tomai G, Narayanasamy S, Reddy RA, et al. (2014) Transcriptomics and functional genomics of ROS-induced cell death regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet 10: e1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D (2017) Arabidopsis WRKY45 interacts with the DELLA Protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. (2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol 172: 2132–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca M, San Segundo B (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63: 526–540 [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467.e15 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte C-P, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX (2010) PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durian G, Rahikainen M, Alegre S, Brosché M, Kangasjärvi S (2016) Protein Phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front Plant Sci 7: 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Glawischnig E, Gigolashvili T (2015) The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front Plant Sci 6: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Gamir J, Darwiche R, Van’t Hof P, Choudhary V, Stumpe M, Schneiter R, Mauch F (2017) The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J 89: 502–509 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E. (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A Tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Li G-J, Yang K-Y, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang JF, Lloyd SJ (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277 [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Anderson JC, del Pozo O, Gu Y-Q, Tang X, Martin GB (2004) Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J 38: 563–577 [DOI] [PubMed] [Google Scholar]
- Heidari B, Nemie-Feyissa D, Kangasjärvi S, Lillo C (2013) Antagonistic regulation of flowering time through distinct regulatory subunits of protein phosphatase 2A. PLoS One 8: e67987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Hussain RMF, Sheikh AH, Haider I, Quareshy M, Linthorst HJM (2018) Arabidopsis WRKY50 and TGA transcription factors synergistically activate expression of PR1. Front Plant Sci 9: 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari T, Durian G, Rahikainen M, Kortesniemi M, Kangasjärvi S, Sinkkonen J (2017) NMR study of age dependent metabolic adjustments in wild type and pp2a-b′γ mutant Arabidopsis thaliana. Phytochem Lett 22: 13–20 [Google Scholar]
- James M, Poret M, Masclaux-Daubresse C, Marmagne A, Coquet L, Jouenne T, Chan P, Trouverie J, Etienne P (2018) SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol 59: 2052–2063 [DOI] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jiang L, Wan Y, Anderson JC, Hou J, Islam SM, Cheng J, Peck SC (2017) Genetic dissection of Arabidopsis MAP kinase phosphatase 1-dependent PAMP-induced transcriptional responses. J Exp Bot 68: 5207–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park J-H, Kim J, Kim JJ, Hong S, Kim J, Kim JH, Woo HR, Hyeon C, Lim PO, et al. (2018a) Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115: E4930–E4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim HJ, Vu QT, Jung S, McClung CR, Hong S, Nam HG (2018b) Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA 115: 8448–8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Konert G, Rahikainen M, Trotta A, Durian G, Salojärvi J, Khorobrykh S, Tyystjärvi E, Kangasjärvi S (2015a) Subunits B′γ and B′ζ of protein phosphatase 2A regulate photo-oxidative stress responses and growth in Arabidopsis thaliana. Plant Cell Environ 38: 2641–2651 [DOI] [PubMed] [Google Scholar]
- Konert G, Trotta A, Kouvonen P, Rahikainen M, Durian G, Blokhina O, Fagerstedt K, Muth D, Corthals GL, Kangasjärvi S (2015b) Protein phosphatase 2A (PP2A) regulatory subunit B′γ interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol 205: 1250–1263 [DOI] [PubMed] [Google Scholar]
- Kosmacz M, Gorka M, Schmidt S, Luzarowski M, Moreno JC, Szlachetko J, Leniak E, Sokolowska EM, Sofroni K, Schnittger A, et al. (2019) Protein and metabolite composition of Arabidopsis stress granules. New Phytol 222: 1420–1433 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, et al. (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66: 818–830 [DOI] [PubMed] [Google Scholar]
- Li S, Mhamdi A, Trotta A, Kangasjärvi S, Noctor G (2014) The protein phosphatase subunit PP2A-B′γ is required to suppress day length-dependent pathogenesis responses triggered by intracellular oxidative stress. New Phytol 202: 145–160 [DOI] [PubMed] [Google Scholar]
- Li Z, Wu S, Chen J, Wang X, Gao J, Ren G, Kuai B (2017) NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis. Plant J 92: 650–661 [DOI] [PubMed] [Google Scholar]
- Liese A, Romeis T (2013) Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim Biophys Acta 1833: 1582–1589 [DOI] [PubMed] [Google Scholar]
- Lillo C, Kataya ARA, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z, Jonassen EM (2014) Protein phosphatases PP2A, PP4 and PP6: Mediators and regulators in development and responses to environmental cues. Plant Cell Environ 37: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang S, Zhou B, Luo X, Zhou XF, Cai B, Jin YH, Niu D, Lin J, Cao X, et al. (2019) The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell 31: 430–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Vilela B, Irar S, Solé M, Capellades M, Valls M, Coca M, Pagès M (2010) MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J 63: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Majeran W, Le Caer J-P, Ponnala L, Meinnel T, Giglione C (2018) Targeted profiling of Arabidopsis thaliana subproteomes illuminates co- and posttranslationally N-terminal myristoylated proteins. Plant Cell 30: 543–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue G-P, Dröge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant 6: 1438–1452 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang J, Ren Y, Zhao Z (2013) The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol 163: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30: 39–52 [DOI] [PubMed] [Google Scholar]
- Moorhead GBG, De Wever V, Templeton G, Kerk D (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417: 401–409 [DOI] [PubMed] [Google Scholar]
- Nie P, Li X, Wang S, Guo J, Zhao H, Niu D (2017) Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis. Front Plant Sci 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol 59: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Noh Y-S, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41: 831–844 [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D R Core Team (2017). nlme: Linear and nonlinear mixed effects models. R package version 3.1-131, https://cran.r-project.org/web/packages/nlme/index.html
- Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, Zhou X (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahikainen M, Trotta A, Alegre S, Pascual J, Vuorinen K, Overmyer K, Moffatt B, Ravanel S, Glawischnig E, Kangasjärvi S (2017) PP2A-B′γ modulates foliar trans-methylation capacity and the formation of 4-methoxy-indol-3-yl-methyl glucosinolate in Arabidopsis leaves. Plant J 89: 112–127 [DOI] [PubMed] [Google Scholar]
- Rasool B, Karpinska B, Konert G, Durian G, Denessiouk K, Kangasjärvi S, Foyer CH (2014) Effects of light and the regulatory B-subunit composition of protein phosphatase 2A on the susceptibility of Arabidopsis thaliana to aphid (Myzus persicae) infestation. Front Plant Sci 5: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I (2019) Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365: 498–502 [DOI] [PubMed] [Google Scholar]
- Ren Y, Li Y, Jiang Y, Wu B, Miao Y (2017) Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis. Mol Plant 10: 749–763 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Herde M (2014) From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr Opin Plant Biol 20: 1–10 [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JDG (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Han S-H, Lee S-H, Hörtensteiner S, Paek N-C (2016) Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep 35: 155–166 [DOI] [PubMed] [Google Scholar]
- Saleh A, Withers J, Mohan R, Marqués J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X (2015) Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P, Herde M, Romeis T (2013) Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol 163: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Macho AP, Sanmartín M, Ntoukakis V, Sánchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33: 2069–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V (2013) The biogenesis of active protein phosphatase 2A holoenzymes: A tightly regulated process creating phosphatase specificity. FEBS J 280: 644–661 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Marquis V, Poirier L, Aubert Y, Zumsteg J, Ménard R, Miesch L, Heitz T (2017) Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol Plant 10: 1159–1173 [DOI] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol 64: 479–498 [Google Scholar]
- Suarez-Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Tamary E, Nevo R, Naveh L, Levin-Zaidman S, Kiss V, Savidor A, Levin Y, Eyal Y, Reich Z, Adam Z (2019) Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct 3: e00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmström M, Hiltunen H-M, Rips S, Sipari N, et al. (2011) Regulatory subunit B′γ of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156: 1464–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Qi Y, Nguyen V, Bethke G, Tsuda Y, Glazebrook J, Katagiri F (2012) An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J 69: 713–719 [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Froehlich JE, Amasino RM (1999) Chloroplast-targeted ERD1 protein declines but its mRNA increases during senescence in Arabidopsis. Plant Physiol 119: 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins DJ, Penfold CA, Baxter L, Breeze E, et al. (2012) Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24: 3530–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C-P, Keinath N, Dubiella U, Demoulière R, Seal A, Romeis T (2010) Tobacco calcium-dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J Biol Chem 285: 9740–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Koo HJ, Kim J, Jeong H, Yang JO, Lee IH, Jun JH, Choi SH, Park SJ, Kang B, et al. (2016) Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol 171: 452–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Cui Y, Xu F, Xu X, Gao G, Wang Y, Guo Z, Wang D, Wang NN (2015) SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE directly interacts with the cytoplasmic domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and negatively regulates leaf senescence in Arabidopsis. Plant Physiol 169: 1275–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Tang L, Gong Y, Xie J, Fu Y, Jiang D, Li G, Collinge DB, Chen W, Cheng J (2018) A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol 217: 739–755 [DOI] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Gan S-S (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu C-J, Gan S-S (2013a) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan S-S (2012) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69: 667–678 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Han X, Shi R, Yang G, Qi L, Wang R, Li G (2013b) Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol Biochem 73: 383–391 [DOI] [PubMed] [Google Scholar]
- Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of Chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol Plant 114: 92–101 [DOI] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B (2015) Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84: 597–610 [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang Y, Su Z, Lv L, Zhang Z (2018) Silencing of the wheat protein phosphatase 2A catalytic subunit TaPP2Ac enhances host resistance to the necrotrophic pathogen Rhizoctonia cerealis. Front Plant Sci 9: 1437. [DOI] [PMC free article] [PubMed] [Google Scholar]