Abstract
Background
Floral transition initiates reproductive development of plants and occurs in response to environmental and endogenous signals. In Arabidopsis thaliana, this process is accelerated by several environmental cues, including exposure to long days. The photoperiod-dependent promotion of flowering involves the transcriptional induction of FLOWERING LOCUS T (FT) in the phloem of the leaf. FT encodes a mobile protein that is transported from the leaves to the shoot apical meristem, where it forms part of a regulatory complex that induces flowering. Whether FT also has biological functions in leaves of wild-type plants remains unclear.
Results
In order to address this issue, we first studied the leaf transcriptomic changes associated with FT overexpression in the companion cells of the phloem. We found that FT induces the transcription of SWEET10, which encodes a bidirectional sucrose transporter, specifically in the leaf veins. Moreover, SWEET10 is transcriptionally activated by long photoperiods, and this activation depends on FT and one of its earliest target genes SUPPRESSOR OF CONSTANS OVEREXPRESSION 1 (SOC1). The ectopic expression of SWEET10 causes early flowering and leads to higher levels of transcription of flowering-time related genes in the shoot apex.
Conclusions
Collectively, our results suggest that the FT-signaling pathway activates the transcription of a sucrose uptake/efflux carrier during floral transition, indicating that it alters the metabolism of flowering plants as well as reprogramming the transcription of floral regulators in the shoot meristem.
Keywords: Flowering time, FLOWERING LOCUS T, Photoperiod, Sugar transporter
Background
In plants, the transition from vegetative growth to flowering is regulated by several environmental and endogenous stimuli. This complexity is conferred by a network of genetic pathways that has been characterized in most detail in the model species A. thaliana. This network includes the vernalisation, gibberellin, thermosensory, age, sugar and photoperiod-dependent pathways [1–3]. FLOWERING LOCUS T (FT) is a positive regulator of flowering whose expression leads to rapid transcriptional reprogramming of the meristem associated with inflorescence and flower development, and is often described as a floral integrator because its transcription is activated by several genetic pathways that promote flowering [1, 4, 5]. Sugars such as sucrose and trehalose-6-phosphate also promote flowering, and there is evidence that these act both upstream and downstream of FT in the flowering process [6–9]. However, our understanding of the relationship between FT and sugar metabolism is fragmentary. Here, we demonstrate that FT is required for the transcriptional activation of a gene encoding a sugar uptake/efflux carrier in the vasculature of the leaf and at the shoot apex during floral transition, providing a specific link between FT function and sugar transport.
FT was first placed within the photoperiodic flowering pathway of A. thaliana based on physiological and genetic analyses [10]. Furthermore, simultaneous loss-of-function of FT and its closest relative TWIN SISTER OF FT (TSF) leads to late flowering plants under long days (LDs) that are almost insensitive to photoperiod [11, 12]. Transcription of FT is induced by exposure to LDs downstream of the GIGANTEA (GI) and CONSTANS (CO) genes in specialized companion cells of the phloem [13–16]. FT encodes a small globular protein that shares high homology with mammalian phosphatidylethanolamine-binding proteins (PEBP) [4, 5], and is a major component of a systemic signal that induces flowering in response to photoperiod (a “florigen”) [17–20]. FT protein moves through the phloem to the shoot apical meristem (SAM) by an active mechanism [21, 22] and binds lipids in vitro [23]. An endoplasmic reticulum-membrane protein, FT-INTERACTING PROTEIN 1 (FTIP1), interacts with FT in companion cells of the phloem and mediates its export into sieve elements [21]. In the sieve elements, FT interacts with a heavy metal-associated isoprenylated plant protein called SODIUM POTASSIUM ROOT DEFECTIVE 1 (NaKR1), which regulates the long-distance transport of FT to the SAM [22]. In the SAM, FT is proposed to interact with two bZIP transcription factors (FD and FD PARALOG [FDP]) [24–26]. The transcriptional complex that is formed between FT, these bZIPs and 14–3-3 proteins is proposed to trigger transcriptional activation of genes that promote flowering, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), FRUITFULL (FUL) and APETALA1 (AP1), which encode MADS-box transcription factors, and several members of the SQUAMOSA PROMOTER BINDING LIKE (SPL) gene family [24, 25, 27–30]. Transcriptomic and in situ hybridization studies identified SOC1 mRNA as the earliest activated transcript detected in the SAM during FT-mediated photoperiodic induction of flowering [28, 29, 31–33], while genome-wide experiments showed that SOC1 binds to the promoters of numerous genes involved in the floral transition and floral meristem identity [34, 35]. Therefore, SOC1 acts as an intermediate component in the FT-signaling pathway during the activation of flowering of A. thaliana. Consistently, the flowering response to FT overexpression is attenuated in the soc1 single mutant [36], and this effect is even more pronounced in the soc1 ful double mutant [29, 36].
Ectopic expression of FT from heterologous promoters leads to early flowering [4, 5]. For example, overexpression of FT from constitutive promoters such as the Cauliflower mosaic virus CaMV 35S promoter [p35S] [4, 5] or phloem-specific promoters such as those of the GALACTINOL SYNTHASE1 [GAS1] and SUCROSE TRANSPORTER2 [SUC2] genes [17, 19, 20, 37] induces early flowering of A. thaliana. This effect is highly conserved among Angiosperms, so that overexpression of FT or its homologues causes early flowering in a wide range of species [18, 38, 39]. Overexpression of FT also induces the transcription of FUL and SEPALLATA3 (SEP3) in leaves of A. thaliana, conferring changes in leaf morphology that are suppressed by ful and sep3 mutations [40]. Thus, at least when overexpressed, FT can influence the development of leaves by affecting the expression of regulatory genes, and upon transport from the leaves it promotes the floral transition at the SAM.
Here, we have further studied the regulatory role of FT. We analyzed global transcriptomic changes in leaves associated with the specific expression of FT in the phloem companion cells. Our results indicate that FT promotes the expression of SWEET10, a gene encoding a sucrose bidirectional transporter, in the leaf veins and at the shoot apex. This effect is also mediated by photoperiod and by SOC1. Moreover, the overexpression of SWEET10 slightly accelerates flowering, leading us to discuss possible roles for this gene during floral transition mediated by the FT-signaling pathway in A. thaliana.
Results
FT induces the expression of SWEET10
The global effects on gene expression of FT overexpression in the phloem companion cells of the leaves were examined. To this end, transgenic A. thaliana plants that overexpress FT from the pGAS1 promoter in a ft − 10 tsf − 1 double mutant background were employed (pGAS1:FT ft − 10 tsf-1). In these transgenic plants, the use of the pGAS1 promoter ensures that the FT transgene is expressed in phloem companion cells of the minor veins, recreating the spatial pattern of expression described for the native gene [15]. Indeed, the overexpression of FT from the pGAS1 promoter complements the late-flowering phenotype of ft-10 tsf-1 double mutants [11, 17]. The transcriptome of leaves of pGAS1:FT ft-10 tsf-1 transgenic plants was compared to that of ft-10 tsf-1 and Col-0 plants using Tiling Arrays. Bioinformatic analysis showed that 699 genes (p-Value ≤0.01) were differentially expressed between pGAS1:FT ft-10 tsf-1 and ft-10 tsf-1 (Additional file 1). A final list of 14 genes (Table 1) was selected by applying more restrictive statistical criteria (adj. P. Value ≤0.05). The majority of these genes were well-known flowering-related regulators that act downstream of FT [28, 29], such as SOC1, FUL, SEPALLATA1 (SEP1) and SEP3, which were up-regulated in pGAS1:FT ft-10 tsf-1. In addition, SWEET10 and SWEET13, two members of Clade III of the SWEET family that encode sucrose transporters [41], were in the list of genes induced by FT (Table 1). In particular, the mRNA levels of SWEET10 were strongly up-regulated in the pGAS1:FT ft-10 tsf-1 and other genetic backgrounds overexpressing FT (Fig. 1a). The expression of SWEET10 mRNA was clearly induced in plants overexpressing FT from companion cell specific promoters pGAS1 and pSUC2 [42, 43] (Fig. 1a). This experiment demonstrated that in all cases tested, FT overexpression increased transcription of SWEET10 mRNA. Additionally, in silico gene co-expression analyses indicated that FT is highly co-regulated with SWEET10 during Arabidopsis development (Additional file 2: Figure S1A). These analyses also showed that SWEET10 is co-expressed with many other genes that are regulated by FT (Additional file 2: Figure S1B), such as SEP3 and APETALA1 (AP1) [40].
Table 1.
Top 14 differentially expressed genes between pGAS1:FT ft-10 tsf-1 and ft-10 tsf-1
| ID | logFC | P. Value | adj.P.Val | Gene Name |
|---|---|---|---|---|
| AT1G65480 | 6,03 | 4E-10 | 7,8E-06 | FT (FLOWERING LOCUS T) |
| AT5G60910 | 2,83 | 2E-09 | 2,5E-05 | FUL (FRUITFULL) |
| AT1G24260 | 2,66 | 8E-09 | 6E-05 | SEP3 (SEPALLATA3) |
| AT5G50790 | 2,28 | 2E-07 | 0,00144 | SWEET10 |
| AT2G45660 | 2,25 | 5E-10 | 7,8E-06 | SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO 1) |
| AT5G15800 | 1,92 | 4E-06 | 0,01516 | SEP1 (SEPALLATA1) |
| AT1G80130 | 1,24 | 2E-05 | 0,03677 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| AT5G50800 | 1,09 | 8E-06 | 0,02541 | SWEET13 |
| AT3G58200 | 0,82 | 1E-05 | 0,02772 | TRAF-like family protein |
| AT3G56080 | 0,78 | 2E-05 | 0,04354 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| AT1G62290 | -1,18 | 8E-06 | 0,02541 | PASPA2 (PUTATIVE ASPARTIC PROTEINASE A2) |
| AT5G44400 | -1,31 | 1E-05 | 0,02772 | ATBBE26 (BERBERINE BRIDGE ENZYME-LIKE 26) |
| AT5G23020 | -1,52 | 3E-06 | 0,01516 | IMS2/MAM-L/MAM3 (METHYLTHIOALKYMALATE SYNTHASE-LIKE) |
| AT2G42540 | -3,21 | 9E-06 | 0,02541 | COR15A (COLD-REGULATED 15A) |
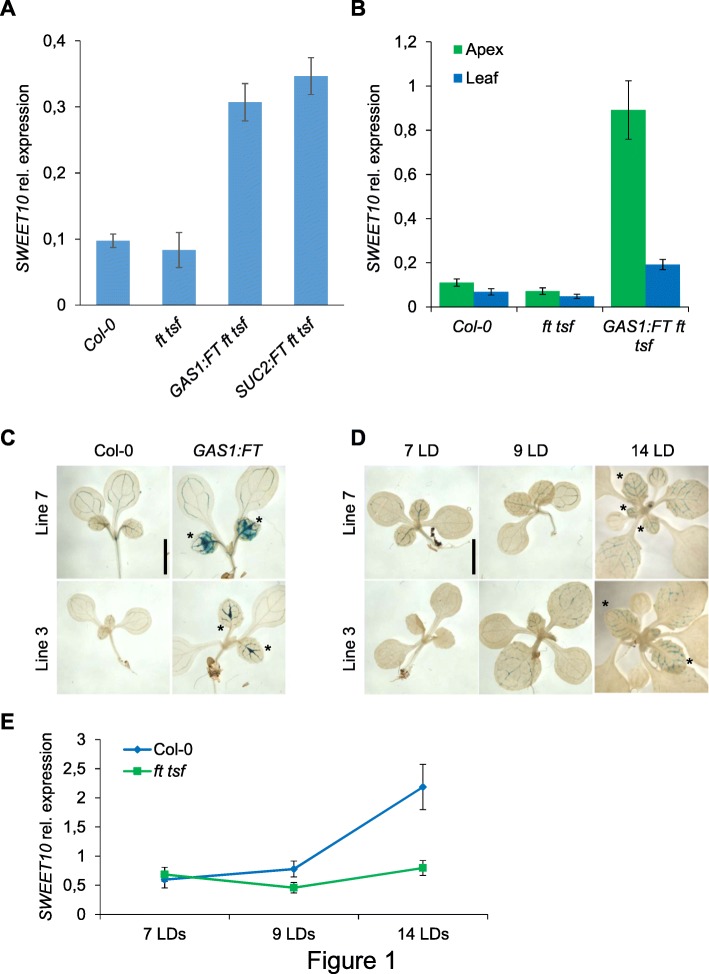
Fig. 1.
FT activates the transcription of SWEET10 mRNA. a Quantification of SWEET10 mRNA levels in leaves of different FT-overexpressing plants (9 LDs at ZT16). b Comparison of SWEET10 mRNA levels in leaves and shoot apices of different FT-overexpressing plants (9 LDs at ZT16). c GUS expression levels in T3 pSWEET10:GUS (Col-0) independent transgenic lines #3 and #7 (left) and in T1 plants from the cross between pGAS1:FT and pSWEET10:GUS lines #3 and #7 (right) at 7 LDs. d GUS staining of pSWEET10:GUS transgenic lines #3 and #7 in Col-0 background (T3 generation) during a time-course in LDs. Pictures of (c) and (d) were taken at ZT8. Scale bar = 5 mm. Asterisks indicate young leaves. e Quantification of SWEET10 expression levels in shoot apices of Col-0 and ft tsf double mutants in a time-course under LDs. Shoot apices were sampled at ZT8. Errors bars in A, B and E indicate Standard Deviation
FT promotes the expression of SWEET10 mRNA in the leaf veins
Next, FT-mediated spatial and temporal regulation of SWEET10 mRNA expression was characterized. The levels of SWEET10 mRNA in leaves and hand-dissected shoot apices (containing SAM, a segment of the apical stem and young leaves) were quantified by RT-qPCR. As shown in Fig. 1b, SWEET10 expression levels were higher in shoot apices compared to mature leaves and this difference was more pronounced in pGAS1:FT ft-10 tsf-1 plants. To better characterize the spatial pattern of expression of SWEET10, we fused the 3 Kb region 5′ of the translational start codon to a GUS reporter gene to create pSWEET10:GUS. In transgenic plants harboring this reporter, GUS signal was restricted to the leaf veins and notably enhanced in young leaves of pGAS1:FT plants (Fig. 1c). Changes in pSWEET10:GUS expression were also studied during the floral transition under LDs. To this end, we monitored GUS signal in pSWEET10:GUS transgenic plants grown under LDs for 7, 9 and 14 days, the time window during which the floral transition occurs in our growth conditions. As observed in the previous experiment, the GUS signal was restricted to the leaf vasculature and it was stronger in young leaves (Fig. 1d). Furthermore, the GUS signal was more evident in plants undergoing the transition to reproductive phase (i.e. 9 to 14 days) compared to those at vegetative stage (i.e. 7 days) (Fig. 1d). This result was confirmed by a RT-qPCR experiment performed during the same time-course, in which an increase of SWEET10 mRNA expression was observed in shoot apices containing young leaves of Col-0 plants from day 9 (Fig. 1e). The increase of SWEET10 expression was largely suppressed in the ft-10 tsf-1 mutant (Fig. 1 e). This indicates that the up-regulation of SWEET10 during the floral transition of A. thaliana partially depends on the presence of a functional FT allele.
The photoperiodic flowering pathway of A. thaliana regulates SWEET10 expression
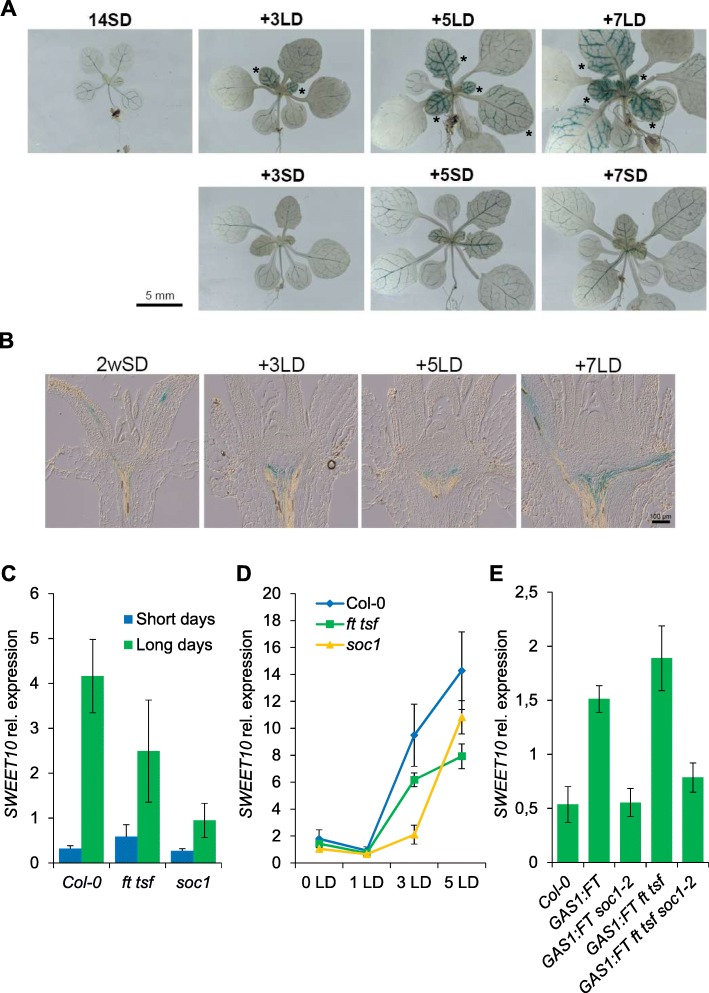
FT is a major component of the photoperiodic flowering pathway that promotes floral induction of A. thaliana in response to LDs. As the above experiments suggest that FT regulates SWEET10 mRNA expression levels during floral transition, we tested whether the photoperiodic pathway activates SWEET10 transcription. The expression of GUS in pSWEET10:GUS was monitored in plants grown under SDs and then shifted to LDs for 3, 5 and 7 days. An increase in intensity of the GUS signal was observed in the vascular tissue of leaves shifted to LDs compared to those grown under SDs (Fig. 2a), indicating that the SWEET10 promoter responds to LDs. Furthermore, in cross-sections of the shoot apex of pSWEET10:GUS plants, GUS expression increased in the mature vascular tissue at the apex of plants shifted to LDs (Fig. 2b). In agreement with these observations, RT-qPCR analysis demonstrated that the levels of SWEET10 mRNA were higher in plants grown under LDs compared to SDs (Fig. 2c) or after the shift of SD-grown plants to LDs (Fig. 2d). Interestingly, the GUS expression disappeared from leaves of pSWEET10:GUS plants at the end of the flowering phase. Instead, GUS expression was observed in reproductive organs, including anthers and siliques (Additional file 2: Figure S2). The photoperiod-dependent up-regulation of SWEET10 mRNA also involves FT, because it is reduced in ft-10 tsf-1 mutant plants (Fig. 2c and d). SOC1 is a key component of the photoperiod signaling pathway that acts immediately downstream of FT [31, 44]. Therefore, whether SWEET10 transcriptional regulation requires SOC1 downstream of FT was investigated. Remarkably, SWEET10 mRNA levels were strongly reduced in a null mutant allele of SOC1 (soc1–2) (Fig. 2d and e). Moreover, the introduction of the soc1–2 mutation into transgenic plants overexpressing FT from the GAS1 promoter was sufficient to largely suppress the enhanced transcriptional induction of SWEET10 mediated by higher levels of FT (Fig. 2e). Collectively, these results indicate that SWEET10 is transcriptionally regulated by the photoperiodic flowering pathway and this regulation involves the activities of FT and SOC1.
Fig. 2.
Photoperiod signaling pathway contributes to SWEET10 mRNA induction. a and b GUS expression in plants expressing pSWEET10:GUS (line #7, T4 generation) grown under SDs for 2 weeks, shifted to LDs and collected for GUS staining at ZT8 after exposure to different numbers of long days. a Whole seedlings. Scale bar = 5 mm. Asterisks indicate young leaves. b Shoot apices were dissected and visualized under an optical microscope (× 20). Bar scale: 100 μm. c, d and e shows the expression levels of SWEET10 mRNA in shoot apices of different genotypes at ZT8. In (c), plants were grown under LDs and SDs. In (d), plants were grown under SDs for 2 weeks, shifted to LDs shoot apices were harvested at ZT8 in different days. In (e), plants were grown under LDs and shoot apices sampled at ZT8. Errors bars in c, d and e indicate Standard Deviation
Overexpression of SWEET10 causes early flowering and affects the expression levels of genes that promote floral induction
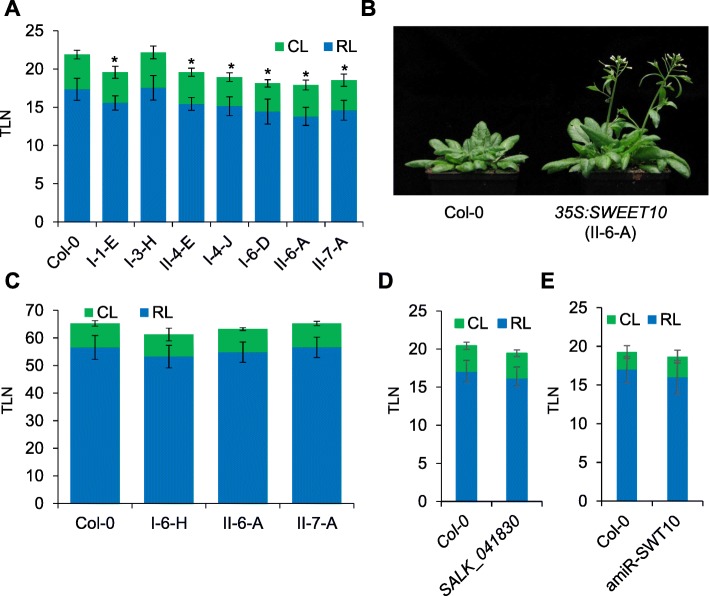
The results presented so far suggest that SWEET10 transcription is induced by FT-signaling pathway via SOC1. In order to explore this possibility, we overexpressed SWEET10 in A. thaliana plants and evaluated its effect on flowering time. We obtained several T1 transgenic lines that ectopically expressed SWEET10 from the 35S promoter (p35S:SWEET10). A higher level of SWEET10 mRNA expression was observed for several of these lines compared to the control Col-0 lines (Additional file 2: Figure S3). We scored the flowering-time of homozygous single copy T3 transgenic lines. Seven out of 8 tested independent transgenic lines displayed a significant acceleration of flowering compared to the control plants under LDs (Fig. 3a and b). We also overexpressed SWEET10 in the companion cells of the phloem from the SUC2 promoter. However, most of the pSUC2:SWEET10 transgenic plants did not flower earlier than the controls (Additional file 2: Figure S4). To address whether the overexpression of SWEET10 could accelerate flowering independently of the photoperiodic pathway, we scored the flowering time of p35S:SWEET10 plants under SD conditions. Under these conditions, p35S:SWEET10 transformants flowered at similar times to the controls (Fig. 3c). This result suggests that the acceleration of flowering mediated by increased SWEET10 mRNA levels requires LDs. Therefore, the flowering function of SWEET10 could also depend on FT function. To further characterize the function of SWEET10, a T-DNA insertion line and transgenic plants expressing an artificial microRNA (amiR) that targets SWEET10 mRNA were employed. None of these genetic backgrounds displayed a significant change in flowering time compared to the wild-type plants (Fig. 3d and e; and Additional file 2: Figure S5).
Fig. 3.
Overexpression of SWEET10 promotes flowering under LDs. Flowering time of transgenic plants overexpressing SWEET10 from the 35S promoter under LDs (a) and (b), and under SDs (c). d Flowering time of the SALK_041830 T-DNA line compared to Col-0 under LDs. e Flowering time of a T3 transgenic line (#17–3) silencing SWEET10 gene expression compared to Col-0 under LDs. At least 10 plants were used for each experiment. Asterisk indicates a significant difference compared to Col-0 (T-test, p-Value ≤0.05). TLN: Total Leaf Number; RL: Rosette Leaf number; CL: Cauline Leaf number. Errors bars in A, C, D and E indicate Standard Deviation
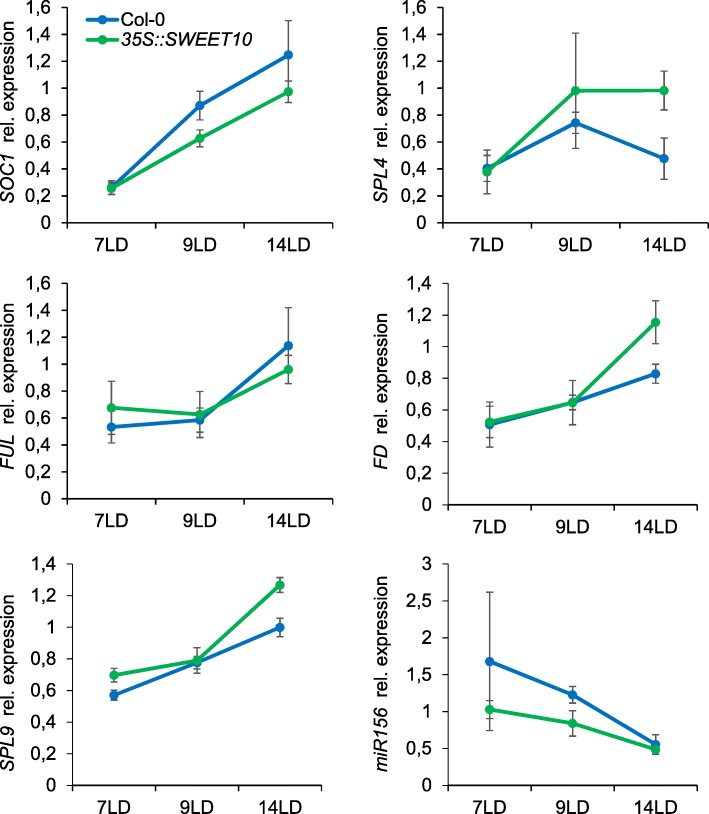
In order to clarify the nature of the effect of SWEET10 overexpression on flowering time, the expression levels of key regulators of flowering in A. thaliana were quantified in plants overexpressing SWEET10 (Fig. 4). In this analysis, the mRNA levels of FD and some SPL genes (SPL4 and 9) were higher in shoot apices of p35S:SWEET10 during reproductive development (14 LDs). This pattern of expression correlates with a possible role of SWEET10 in promoting flowering. However, SOC1 expression was slightly lower in p35S:SWEET10 transgenics compared wild-type plants, whereas FUL mRNA levels were not differentially expressed at this developmental stage. Notably, the expression level of one precursor of miR156 (MIRNA156C), which targets several mRNAs encoding SPLs, was reduced after 7 and 9 LDs in the p35S:SWEET10 compared to wild type plants.
Fig. 4.
Expression levels of flowering-time related genes in SWEET10 overexpressing plants. The expression levels of flowering-time related genes was quantified in Col-0 and p35S:SWEET10 transgenic plants (Line II-6-A, T4 generation) under LDs. Shoot apices were collected at ZT8. Errors bars indicate Standard Deviation
SWEET10 might be the only member of the SWEET family involved in the FT-flowering pathway in A. thaliana
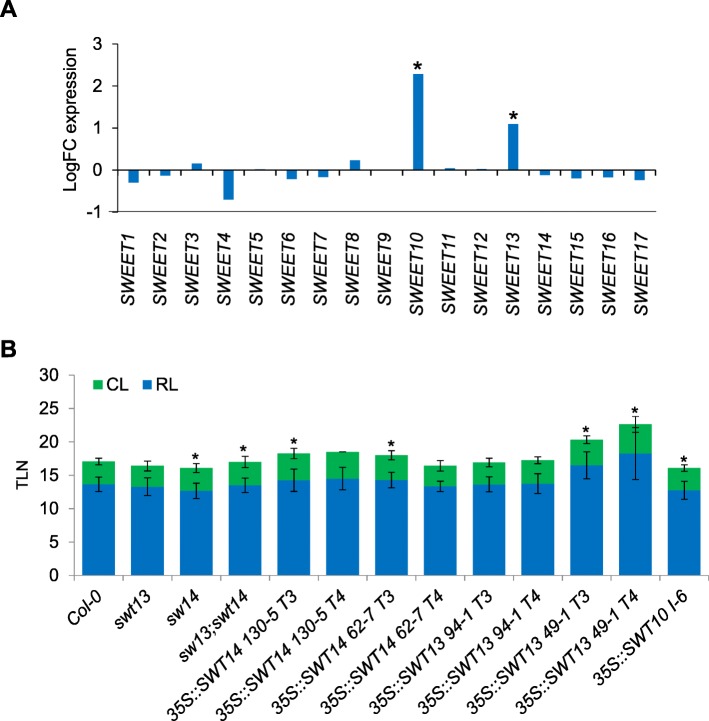
SWEET10 belongs to a large family of genes composed by 17 members in A. thaliana [45]. At least two of them, SWEET10 and 13, were deregulated in the microarray experiment that we performed (Table 1). We extracted the expression data of all 17 members of the family from the microarray analysis (Additional file 1). As shown in Fig. 5a, only the mRNA levels of SWEET10 and 13 were significantly affected in pGAS1:FT ft-10 tsf-1 plants compared to ft-10 tsf-1 double mutants. Furthermore, we made use of promoter:GUS fusions to monitor the spatial and temporal expression of some other Clade III SWEET genes (SWEET11, 12, 13 and 14). In all the tested transgenic plants the GUS signal was detected in the vasculature under SDs, but did not increase after exposure to LDs, as was observed for pSWEET10:GUS (Fig. 2a and Additional file 2: Figure S6). Moreover, plants overexpressing SWEET13 and SWEET14 did not show acceleration of flowering under LDs. Instead, some of the tested lines displayed late flowering compared to the wild-type plants (Fig. 5b).
Fig. 5.
Involvement of SWEET family members the FT-flowering pathway. a Expression of SWEET family members in GAS1:FT ft tsf plants. The logFC between GAS1:FT ft tsf and Col-0 is represented. Significant differences are indicated with an asterisk (adj. P. Value ≤0.05). b Effect of the overexpression of SWEET13 and 14 in flowering-time under LDs. At least 10 plants were used for each experiment. Asterisk indicates a significant difference compared to Col-0 (T-test, p-Value ≤0.05). Scale bar = 5 mm. Errors bars in B indicate Standard Deviation
Measurement of concentrations of fructose, glucose and sucrose at shoot apices during floral transition
The increased expression of SWEET10 at the shoot apex during floral transition (Fig. 2), suggested that sugar levels might increase in this tissue during the transition. Therefore, concentrations of sucrose, fructose and glucose were tested in shoot apices harvested from plants grown for 2 weeks under short days and then transferred to LDs for 7 days to induce the transition. Although SWEET10 expression increases under these conditions (Fig. 2d), no significant change in concentration of any of the sugars was detected across the time course (Additional file 2: Figure S7). Also, there was no significant difference in levels of these sugars between Col-0 and ft tsf genotypes (Additional file 2: Figure S7). Thus, based on this analytical method, no changes in sugar levels that correlated with the floral transition could be detected in apical samples.
Discussion
Here, we demonstrate that FT and SOC1 activate the transcription of the SWEET10 gene. The pattern of transcription of SWEET10 and the effect of its overexpression suggests that the sugar transporter it encodes contribute to the floral transition in A. thaliana under LDs. SWEET10 represents a previously undescribed link between sugar transport and the photoperiod-dependent control of flowering time.
The transcriptional activation of SWEET10 might be part of a genetic network controlled by the FT-pathway in leaves
FT protein is expressed in the vascular tissue of leaves and is transported to the shoot apex as a component of the florigenic signal that activates flowering [17, 19, 20]. However, whether FT has additional roles in the vascular tissue or in leaves remains unclear. Furthermore, overexpression of FT from the constitutive 35S promoter caused the transcriptional induction in leaves of FUL and SEP3, which in wild-type plants are activated by FT signaling at the shoot apex [40]. Thus, we reasoned that transcriptomic analysis of plants overexpressing FT from a promoter specific to companion cells of the phloem, the cell type in which FT is expressed [15, 37], could identify additional genes that respond to FT function in wild-type plants. The majority of genes identified by this approach as significantly regulated by FT were already known to act downstream of FT during the floral transition and flower development, such as SOC1, FUL, SEP1 and SEP3 (Table 1). In addition, SWEET10 was one of the most significantly differentially expressed genes in leaves of GAS1:FT plants and was co-regulated with these flowering genes (Additional file 2: Figure S1), suggesting that it could be activated in leaves by FT along with other known floral regulators.
FT is proposed to activate gene expression directly by interacting with the bZIP transcription factor FD that is expressed in the shoot meristem [24, 25]. However, it could also activate expression of genes such as SWEET10 indirectly through the action of downstream transcription factors. We have shown that SOC1 regulates the transcription of SWEET10 (Fig. 2d and e). This regulation would probably occur in the leaves, as SOC1 is also expressed in these organs [46]. Thus, SWEET10 is placed downstream of FT and SOC1, within a genetic network that operates in the leaves.
Spatial-temporal distribution of SWEET10 mRNA
Several SWEET genes belonging to Clade III (e.g. SWEET11, 12, 13 and 14) have been shown to be expressed in the vascular tissue of A. thaliana, most likely in the phloem [41, 47] (Chen et al., 2011; Kanno et al., 2016). In particular, SWEET11 and 12 are expressed in specialized cells that form files along the veins and probably correspond to phloem parenchyma cells [41]. SWEET10 was also expressed in the phloem (Fig. 1 c, d; Fig 3a and b), most likely in phloem parenchyma cells as was suggested for other members of the Clade III [41]. After floral transition, the GUS expression driven by pSWEET10:GUS was dramatically reduced in the leaves and highly detected in the reproductive organs (Additional file 2: Figure S2). This pattern of expression suggests diverse SWEET10 functions throughout plant development as proposed for some SWEET genes [48] In the presence of high levels of FT in the phloem such as in GAS1:FT transgenic plants, the expression of SWEET10 was remarkably increased and restricted to the vasculature (Fig. 1c). This poses the question of how FT activates the transcription of SWEET10 in the phloem parenchyma cells. One possibility is that FT is expressed in this cell type, as suggested for the rice FT-like gene Hd3a mRNA [49]. However, it was recently shown that in A. thaliana FT mRNA is synthesized in specific companion cells of the phloem [15] from where its protein is exported to the sieve elements. In this context, it would be more plausible that the movement of FT and/or SOC1 to the phloem parenchyma cells is responsible for the transcriptional activation of SWEET10. Detailed imaging studies of the spatial distribution of FT, SOC1 and SWEET10 using fluorescent markers would contribute to address specifically in which phloem cells they are present. The possibility that SOC1 acts as a mediator of FT-signaling to activate the transcription of SWEET10 is particularly interesting. Recently, the direct targets of SOC1 were identified using genome-wide approaches [34, 35]. In these studies, the binding of SOC1 to SWEET10 was not found, suggesting the existence of a third SWEET10 activating-factor downstream of SOC1 and FT. This factor could be encoded by one of the genes that are highly co-expressed with SWEET10 such as SEP3 and FTM5 (Additional file 2: Figure S1). The identification of transcription factors that bind to SWEET10 regulatory regions would help understand how the FT-signaling pathway induces the expression of this gene in the vasculature.
Potential functions of SWEET10 in regulating flowering
Sugars are believed to promote flowering in several species [50]. In many of these species, floral induction correlates with a rapid increase in the concentration of sucrose in the phloem, especially near the shoot apex. This phenomenon was shown for instance in Sinapis alba (white mustard) [51] and Xanthium strumarium (rough cocklebur) [52]. In A. thaliana, the inductive LD treatment triggers a transient increase of sucrose in the leaf exudate [53]. Moreover, transgenic plants of different species, such as tomato, potato and A. thaliana, which over-accumulated sucrose in leaves flowered earlier than the control wild-type plants [53–55]. These results suggest that sucrose acts as a signal during the photoperiod flowering induction. Whether the levels of sucrose or other sugars change during floral transition in the SAM remains unclear, as its quantification in this tissue is technically challenging. Indeed, we did not detect significant changes in the concentrations of sucrose, fructose or glucose in shoot apices of A. thaliana plants shifted from SDs to LDs (Additional file 2: Figure S7). However, these apical samples include young leaves, a segment of the apical stem and meristems, so we cannot exclude that local changes in sugar concentration occur. Furthermore, in a previous report, sucrose was found to increase in concentration during the floral transition in shoot apices of plants grown under continuous LDs [6]. Therefore, sugars, and sucrose in particular, could act in the SAM to induce or facilitate the floral transition in response to LDs. In this context, sugar transporters such as SWEET proteins might play an important role in this process. In agreement with this, the overexpression of SWEET10 in A. thaliana triggered a significant acceleration of flowering (Fig. 3a and b). Interestingly, other sugar transporters have also been related to flowering-time control. For example, A. thaliana mutants deficient in SUCROSE TRANSPORTER 9 (SUC9) were early flowering under SDs, probably by an increase in the phloem-loading of sucrose [56]. Therefore, the transport of sugars from leaves to the SAM mediated by specialized transporters could contribute to the floral transition in A. thaliana and other species. However, the precise role of SWEET10 in controlling flowering time is still unknown. One possible scenario is that SWEET10 is transcriptionally induced downstream of FT (and SOC1) in order to supply sugars to the SAM at the time that floral transition occurs. This would contribute to satisfying increased energy requirements of the shoot meristem in order to undergo the increased growth and cell division associated with the floral transition and the initiation of floral organogenesis. Remarkably, the transcription factor CO, which is part of the photoperiodic flowering pathway of Arabidopsis, is responsible for the mobilization of sugars from amylose during the floral transition [57]. Therefore, the photoperiod pathway could affect sugar transport at least at two distinct levels: through CO to mobilize sugars [57] and then through FT to facilitate sugar transport to the SAM. In an alternative scenario, sugars transported by SWEET10 would contribute to the movement of FT towards the SAM. However, so far there is no evidence that sugars are involved in FT transport, although it is proposed to move through the phloem in the photosynthate stream. In both situations, the effect of SWEET10 overexpression on flowering time would depend on FT activity. In agreement with this, the early-flowering phenotype of 35S:SWEET10 transgenics was suppressed under SD conditions (Fig. 3c). Moreover, the overexpression of SWEET10 resulted in the induction of genes in the shoot apex related to FT function (Fig. 4). Among them, SPL4 and SPL9 that are also known to be upregulated by gibberellin signaling under inductive LD conditions [58]. Interestingly, SWEET proteins were proposed to transport gibberellins as well as sucrose [47], suggesting that FT could regulate both sucrose and gibberellin levels at the apex during flowering by upregulating SWEET10. Also, in potato FT was proposed to regulate SWEET function at the post-translational level to prevent leakage of sugar into the apoplast [59].
Overexpression of SWEET10 caused early flowering, but loss of function mutants were not affected in flowering time. Overexpression from the 35S promoter is widely used to address the function of genes, but loss-of-function genetics would provides more definitive evidence on the role of SWEET genes in flowering time-control. SWEET10 single mutants examined here did not show any striking phenotype related to flowering-time (Fig. 3d and e), which could be explained by functional redundancy between members of the SWEET family. SWEET13 is an obvious candidate to play a redundant function, as its expression was also upregulated in pGAS1:FT ft-10 tsf-1 plants compared to ft-10 tsf-1 double mutants (Table 1). However, SWEET13 overexpression did not result in early flowering (Fig. 5b), and higher order mutants might also show pleiotropic phenotypes. Thus, a systematic study of higher order loss-of-function mutants could be necessary to obtain a more complete picture of the SWEET genes function in flowering-time. Furthermore, induction of SWEET proteins during flowering might contribute to the altered metabolic state of the vasculature during floral transition without visibly altering leaf number or flowering time. Nevertheless, the reduced expression of SWEET10 in ft tsf double mutants and its increased expression after transfer to LDs, support a relationship between SWEET10 transcription and flowering.
Conclusions
The data shown here indicate that transcriptional activation of SWEET10 by FT and SOC1 occurs during the promotion of flowering mediated by inductive photoperiod and that overexpression of SWEET10 causes early flowering consistent with a functional role in this process. This emphasizes the likely significance of changing patterns in sugar transport during the floral transition. Moreover, it supports the idea that FT not only plays a role as a long-distance signaling molecule but that it can also function in leaves to bring about transcriptional changes that eventually contribute to flowering-time regulation in the SAM.
Methods
Plant materials
Arabidopsis thaliana Columbia-0 (Col) was used as wild-type in all experiments and for plant transformation. The transgenic plants pGAS1:FT ft-10 tsf-1, pGAS1:FT and pGAS1:FT soc1–2 were previously described [11, 60]. The mutant alleles used were soc1–2 [33] and ft-10 tsf-1 [11]. The SWEET10 CDS sequence was obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/) (clone U15254) and cloned in the pAlligator-2 [61] and pSUC2:GW [11] vectors to generate the p35S:SWEET10 and pSUC2:SWEET10 lines, respectively. To generate 35S:SW13 and 35S:SW14 transgenic plants, SWEET13 and SWEET14 cDNAs were amplified (primer combinations in Additional file 2: Table S1) and inserted into a cloning vector. The inserted sequences were then cloned into the binary vector pBE2113 [62] with XbaI and SmaI restriction sites. The pSWEET10:GUS lines were obtained by cloning a 3 Kb region upstream of the transcriptional starting site of the SWEET10 gene (primers in Additional file 2: Table S1) into the pGreen-GW-GUS vector [63]. For pSWEET11:GUS and pSWEET12:GUS constructs, promoter regions (approximately 2 kb) of SWEET11 and SWEET12 were amplified (primer combinations in Additional file 2: Table S1). The amplified fragments were cloned into pENTR/D-TOPO and then into pGWB3 [64]. pSWEET13:GUS and pSWEET14:GUS transgenic plants were described previously [47]. T-DNA line SALK_041830 was obtained from the Nottingham Arabidopsis Stock Center (NASC) (http://arabidopsis.info/). For the production of the SWEET10 silencing lines, a amiRNA targeting this gene was generated by using the online tool WMD3 (primers in Additional file 2: Table S1) and the artificial miRNA vector pRS300 [65]. The resulting amiRNA-SWEET10 construct was cloned in the vector pAlligator-2. Arabidopsis plants were transformed following the floral dip method [66].
Plant growth conditions
Seeds were stratified on soil for 3 day in the dark at 4 °C. Plants were grown under controlled environmental conditions at 22 °C and white fluorescent light (150 μmol/m2/s), either in LDs (16 h light/8 h dark) or in SDs (8 h light/16 h dark). Flowering time was scored by counting total leaves number (caulines and rosettes) of at least 10 plants per genotype. Each experiment was performed at least twice. For RT-qPCR experiments leaves and shoot apices (containing a segment of the apical stem, SAM and young leaves) were dissected manually.
Microarray experiment
Col-0, ft-10 tsf-1 and pGAS1:FT ft-10 tsf-1 plants were grown under LD conditions during 9 days. Leaves of each genotypes were harvested at the end of the light period (ZT16). RNA from three independent biological replicates was extracted using the RNA Plant Mini kit, QIAGEN (www1.qiagen.com/). The concentration of the total RNA was determined using a NanoDrop ND1000 spectrophotometer. The probe synthesis and the hybridization were performed as previously described in [67]. One microgram of total RNA was reverse transcribed into cDNA using an oligo(dT)-T7 primer, and was then converted into cRNA and linearly amplified by T7 in vitro transcription reaction using the standard Ambion protocol (MessageAmp aRNA Kit, Ambion). cRNA was then reverse transcribed with random primers to dUTP-containing ds cDNA (WT ds cDNA Synthesis Kit, catalog no. 900813; Affymetrix). Fragmentation and labeling was performed with the GeneChip WT double-stranded DNA Terminal Labeling Kit (catalog no. 900812, Affymetrix). After fragmentation, 7.5 μg of ds-cDNA was hybridized for 16 h at 45C on GeneChip Arabidopsis Tiling 1.0R Array. GeneChips were washed and stained with Fluidics Script FS450_0001 in the Affymetrix Fluidics Station 450. Then, the GeneChips were scanned using the GeneChip Scanner 3000 7G. Data was processed in R v2.8.1 using the probe annotation athtiling1.0rcdf as described in [68]. Probe-level data were pre-processed using the RMA algorithm implemented in the Bioconductor package Affy v1.24.2. Linear models and empirical Bayes methods from the Limma package v2.14 of Bioconductor were applied to derive a P value, false discovery rate (FDR; P adjusted), and mean of log2-based ratio across replicates. The data were deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (GEO accession number GSE125054).
RT-qPCR
RNA expression analyses were performed as described in [69]. The RNA was extracted from plant tissue (leaves or shoot apices) by using the RNeasy Plant Mini Kit (Qiagen) and treated with DNA-free DNase (Ambion). One microgram of total RNA (quantified in a Nanodrop ND-1000) was used for reverse transcription by using the Superscript III (Invitrogen). Levels of gene expression were quantified by qPCR in a LightCycler 480 instrument (Roche) using the PEX4 gene (AT5G25760) as a reference. Three biological replicates were performed for each qRT-PCR assay. The average of the three replicates is shown. The list of primers used for expression analyses can be found in the Additional file 2: Table S1.
Histochemical staining for GUS activity
Transgenic plants of pSWEET10:GUS, pSWEET11:GUS, pSWEET12:GUS, pSWEET13:GUS and pSWEET14:GUS were fixed with cold 90% (v/v) acetone for 30 min on ice, then washed with 50 mM sodium phosphate buffer twice. The samples were then immersed in the X-Gluc staining solution [50 mM NaPO4 buffer (pH 7.0), 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% (v/v) Triton X-100, 0.5 mg/ml 5-bromo-4-chloro− 3-indolyl-beta-D-glucuronide (X-Gluc) in H2O] under vacuum for 15 min, and then incubated at 37 °C in the dark for 40 h. After the reaction, the samples were washed with 50 mM sodium phosphate buffer, dehydrated through an ethanol series and observed under stereo microscope (Zeiss, Stemi 508).
For histological analysis, the samples were embedded in paraffin, and sliced with the microtome (Leica, RM2125 RTS) to make serial sections of 8-μm thickness. After deparaffinization and rehydration, the sections were observed with the differential interference contrast (DIC) microscope (Zeiss, Axio Imager M2).
Sugar measurements
For each sample, 30 apexes were harvested and frozen in liquid nitrogen. The samples were extracted in chloroform/methanol/water according to [70]. The aqueous phase was used for sugar measurement and the chloroform phase for protein determination. Sucrose, fructose and glucose were determined photospectrometrically using a 96-well plate reader (Synergy HT from BioTek, U.S.A.) based on the method described in [71] and adapted to the 96-well format by [72]. Protein content was measured according to [73] using the DC™ Protein Assay kit (Bio-Rad Laboratories, U.S.A.) and the values were used for normalization of the sugar data.
Supplementary information
Additional file 1. Results of the expression studies performed on the GeneChip Arabidopsis Tiling 1.0R Array.
Additional file 2: Table S1. Primers used in this study. Figure S1. In silico analyses of co-expressed gene networks around SWEET10. (A) Gene network representation and list of genes correlated to SWEET10 during development generated by the GENEVESTIGATOR software [74]. (B) The Arabidopsis thaliana trans-factor and cis-element prediction database ATTED-II [75], implemented in www.arabidopsis.org, was used to predict and visualize co-expressed genes around SWEET10. Figure S2. Expression of pSWEET10:GUS in adult Arabidopsis plants. (A) GUS expression in a whole plant expressing pSWEET10:GUS. The T4 transgenic plant shown in (A) was grown under LDs until siliques were produced. Detail of an inflorescence (B) and a silique (C) showing GUS expression. Figure S3. Expression levels of SWEET10 in T1 transgenic lines overexpressing SWEET10. The expression levels of SWEET10 was quantified in Col-0 and 35S:SWEET10 T1 lines under LDs. Leaves were collected at ZT8. Errors bars indicate Standard Deviation. Figure S4. Effect of the overexpression of SWEET10 from SUC2 promoter on flowering time under LDs. At least 10 plants were used for each experiment. Asterisk indicates a significant different compared to Col-0 (T-test, p-Value ≤0.05). Errors bars indicate Standard Deviation. Figure S5. Analysis of amiR-SWEET10 transgenic plants. (A) Flowering time of 44 amiR-SWEET10 T1 lines compared to Col-0 under LDs. TLN: Total Leaf Number. (B) SWEET10 expression levels in a subset of T3 amiR-SWEET10 lines. Figure S6. Photoperiod-dependent expression profile of SWEET11, 12, 13 and 14. GUS expression in plants expressing pSWEET10:GUS. Plants were grown under SDs for 2 weeks, shifted to LDs and collected for GUS staining at ZT8 in different days. Scale bar = 5 mm. Figure S7. Levels of sugar during the photoperiodic induction of flowering. Col-0 and ft tsf plants were grown under SDs for 2 weeks, shifted to LDs shoot apices were harvested at ZT8 in different days. Shoot apices were harvested and used to quantify the concentration of fructose (fru), glucose (glu) and sucrose (suc). Errors bars indicate Standard Deviation.
Acknowledgements
Not Applicable.
Abbreviations
- FDR
False discovery rate
- LDs
Long days
- RT-qPCR
Quantitative reverse transcription polymerase chain reaction
- SAM
Shoot apical meristem
- SDs
Short days
Authors’ contributions
FA performed experimental work and data analysis. FA and GC designed the study and contributed to writing and revising the manuscript. AK, MS, YC and TMA contributed to experimental work and revising the manuscript. NK, VF, VSF, TMDC, BH and SJ contributed to experimental work. All authors read and approved the final manuscript.
Authors’ information
All information is provided on the title page.
Funding
This work was financially supported by the Deutsche Forschung Gemeinschaft (DFG) through the Cluster of Excellence in Plant Science CEPLAS (EXC 1028) to T.M.A. and G.C., the European Union via a Marie Curie Intra-European Fellowship for Career Development (Project Intra-European Grant Agreement-2009-251839) (to F.A.), Alexander von Humboldt Fellowship for Postdoctoral Researchers and Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowships (to A.K.), and the Max Planck Society through a core grant (to G.C.). The Funding bodies were not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The transcriptomic datasets generated during the current study are available in the GEO NCBI repository under the accession number GSE125054 and in the supplementary information files. Arabidopsis thaliana Columbia, which was used throughout this work, is available from the Arabidopsis Stock Centre, Nottingham University, UK or the Arabidopsis Biological Resource Centre, Ohio State University or from the authors. No permissions were required to use Arabidopsis thaliana. All other plants materials are available from the corresponding authors.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fernando Andrés and Atsuko Kinoshita contributed equally to this work.
Contributor Information
Fernando Andrés, Email: fernando.andres-lalaguna@inrae.fr.
Atsuko Kinoshita, Email: kinoshita@mpipz.mpg.de.
Naveen Kalluri, Email: naveen.kalluri@cragenomica.es.
Virginia Fernández, Email: fernandez@mpipz.mpg.de.
Vítor S. Falavigna, Email: falavigna@mpipz.mpg.de
Tiago M. D. Cruz, Email: tiago.dias@biol.ethz.ch
Seonghoe Jang, Email: seonghoe.jang@worldveg.org.
Yasutaka Chiba, Email: s_per_10th@yahoo.co.jp.
Mitsunori Seo, Email: mitsunori.seo@riken.jp.
Tabea Mettler-Altmann, Email: tabea.mettler@hhu.de.
Bruno Huettel, Email: huettel@mpipz.mpg.de.
George Coupland, Email: coupland@mpipz.mpg.de.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-2266-0.
References
- 1.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 2.Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci. 2011;68(12):2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouche F, Lobet G, Tocquin P, Perilleux C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016;44(D1):D1167–D1171. doi: 10.1093/nar/gkv1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286(5446):1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 6.Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by Trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339(6120):704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- 7.Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in arabidopsis. Plant Physiol. 2001;127(1):252–261. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho LH, Pasriga R, Yoon J, Jeon JS, An G. Roles of sugars in controlling flowering time. J Plant Biol. 2018;61(3):121–130. doi: 10.1007/s12374-018-0081-z. [DOI] [Google Scholar]
- 9.Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 2010;64(1):1–13. doi: 10.1111/j.1365-313X.2010.04312.x. [DOI] [PubMed] [Google Scholar]
- 10.Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ. Genetic interactions among late-flowering mutants of Arabidopsis. Genetics. 1998;148(2):885–892. doi: 10.1093/genetics/148.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009;60(4):614–625. doi: 10.1111/j.1365-313X.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46(8):1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 13.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15(12):2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22(5):1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Payyavula RS, Chen L, Zhang J, Zhang C, Turgeon R. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland mammoth tobacco leaf veins. Proc Natl Acad Sci U S A. 2018;115(11):2830–2835. doi: 10.1073/pnas.1719455115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 17.Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 18.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17(12):1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Mathieu J, Warthmann N, Kuttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17(12):1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012;10(4):e1001313. doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Liu L, Shen L, Yu H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat Plants. 2016;2(6):16075. doi: 10.1038/nplants.2016.75. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y, Andres F, Kanehara K, Liu YC, Dormann P, Coupland G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat Commun. 2014;5:3553. doi: 10.1038/ncomms4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 25.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25(3):820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Searle I, He YH, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20(7):898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130(24):6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 29.Torti S, Fornara F, Vincent C, Andres F, Nordstrom K, Gobel U, Knoll D, Schoof H, Coupland G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell. 2012;24(2):444–462. doi: 10.1105/tpc.111.092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476(7360):332–U397. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 31.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 32.Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24(5):591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14(18):2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Immink RGH, Pose D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk ADJ, Schmid M, et al. Characterization of SOC1's central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012;160(1):433–449. doi: 10.1104/pp.112.202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao Z, Shen LS, Liu C, Liu L, Yan YY, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012;70(4):549–561. doi: 10.1111/j.1365-313X.2012.04919.x. [DOI] [PubMed] [Google Scholar]
- 36.Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet. 2008;40(12):1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- 37.An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131(15):3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 38.Navarro C, Abelenda JA, Cruz-Oro E, Cuellar CA, Tamaki S, Silva J, Shimamoto K, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Harry DE, Ma C, Yuceer C, Hsu CY, Vikram V, Shevchenko O, Etherington E, Strauss SH. Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. J Exp Bot. 2010;61(10):2549–2560. doi: 10.1093/jxb/erq092. [DOI] [PubMed] [Google Scholar]
- 40.Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17(10):2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335(6065):207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 42.Stadler R, Sauer N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta. 1996;109(4):299–306. doi: 10.1111/j.1438-8677.1996.tb00577.x. [DOI] [Google Scholar]
- 43.Haritatos E, Ayre BG, Turgeon R. Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 2000;123(3):929–937. doi: 10.1104/pp.123.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. CONSTANS activates SUPPRESSOR OFOVEREXPRESSION OFCONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139(2):770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002;21(16):4327–4337. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durand M, Mainson D, Porcheron B, Maurousset L, Lemoine R, Pourtau N. Carbon source-sink relationship in Arabidopsis thaliana: the role of sucrose transporters. Planta. 2018;247(3):587–611. doi: 10.1007/s00425-017-2807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasriga R, Cho LH, Yoon J, An G. Identification of the regulatory region responsible for vascular tissue-specific expression in the Rice Hd3a promoter. Mol Cells. 2018;41(4):342–350. doi: 10.14348/molcells.2018.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P. Physiological signals that induce flowering. Plant Cell. 1993;5(10):1147–1155. doi: 10.2307/3869768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pryke JA, Bernier G. Acid Invertase activity in apex of Sinapis-Alba during transition to flowering. Ann Bot-London. 1978;42(179):747–749. [Google Scholar]
- 52.Houssa P, Bernier G, Kinet JM. Qualitative and quantitative-analysis of carbohydrates in leaf exudate of the short-day plant, xanthium-Strumarium L during floral transition. J Plant Physiol. 1991;138(1):24–28. doi: 10.1016/S0176-1617(11)80724-8. [DOI] [Google Scholar]
- 53.Corbesier L, Lejeune P, Bernier G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta. 1998;206(1):131–137. doi: 10.1007/s004250050383. [DOI] [PubMed] [Google Scholar]
- 54.Micallef BJ, Haskins KA, Vanderveer PJ, Roh KS, Shewmaker CK, Sharkey TD. Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta. 1995;196(2):327–334. doi: 10.1007/BF00201392. [DOI] [Google Scholar]
- 55.Mullerrober B, Sonnewald U, Willmitzer L. Inhibition of the Adp-glucose Pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J. 1992;11(4):1229–1238. doi: 10.1002/j.1460-2075.1992.tb05167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sivitz AB, Reinders A, Johnson ME, Krentz AD, Grof CPL, Perroux JM, Ward JM. Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 2007;143(1):188–198. doi: 10.1104/pp.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ortiz-Marchena MI, Albi T, Lucas-Reina E, Said FE, Romero-Campero FJ, Cano B, Ruiz MT, Romero JM, Valverde F. Photoperiodic control of carbon distribution during the floral transition in Arabidopsis. Plant Cell. 2014;26(2):565–584. doi: 10.1105/tpc.114.122721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porri A, Torti S, Romera-Branchat M, Coupland G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012;139(12):2198–2209. doi: 10.1242/dev.077164. [DOI] [PubMed] [Google Scholar]
- 59.Abelenda JA, Bergonzi S, Oortwijn M, Sonnewald S, Du MR, Visser RGF, Sonnewald U, Bachem CWB. Source-sink regulation is mediated by interaction of an FT Homolog with a SWEET protein in potato. Curr Biol. 2019;29(7):1178−+. doi: 10.1016/j.cub.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 60.Andres F, Romera-Branchat M, Martinez-Gallegos R, Patel V, Schneeberger K, Jang S, Altmuller J, Nurnberg P, Coupland G. Floral induction in Arabidopsis by FLOWERING LOCUS T requires direct repression of BLADE-ON-PETIOLE genes by the Homeodomain protein PENNYWISE. Plant Physiol. 2015;169(3):2187–2199. doi: 10.1104/pp.15.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bensmihen S, To A. Lambert G, Kroj T, Giraudat J, Parcy F. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 2004;561(1–3):127–131. doi: 10.1016/S0014-5793(04)00148-6. [DOI] [PubMed] [Google Scholar]
- 62.Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37(1):49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y, Wang YJ, Krause K, Yang TT, Dongus JA, Zhang YJ, Turck F. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat Genet. 2018;50(5):638−+. doi: 10.1038/s41588-018-0109-9. [DOI] [PubMed] [Google Scholar]
- 64.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 65.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53(4):674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 67.Gregis V, Andres F, Sessa A, Guerra RF, Simonini S, Mateos JL, Torti S, Zambelli F, Prazzoli GM, Bjerkan KN, et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013;14(6):R56. doi: 10.1186/gb-2013-14-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Ratsch G, Inze D, Kuiper M, et al. Quantitative RNA expression analysis with Affymetrix tiling 1.0R arrays identifies new E2F target genes. Plant J. 2009;57(1):184–194. doi: 10.1111/j.1365-313X.2008.03662.x. [DOI] [PubMed] [Google Scholar]
- 69.Andres F, Porri A, Torti S, Mateos J, Romera-Branchat M, Garcia-Martinez JL, Fornara F, Gregis V, Kater MM, Coupland G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci U S A. 2014;111(26):E2760–E2769. doi: 10.1073/pnas.1409567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397:139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stitt M, Lilley RM, Gerhardt R, Heldt HW. Metabolite levels in specific cells and subcellular compartments of plant-leaves. Methods Enzymol. 1989;174:518–552. doi: 10.1016/0076-6879(89)74035-0. [DOI] [Google Scholar]
- 72.Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M. Variation of enzyme activities and metabolite levels in 24 arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006;142(4):1574–1588. doi: 10.1104/pp.106.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 74.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35(Database issue):D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Results of the expression studies performed on the GeneChip Arabidopsis Tiling 1.0R Array.
Additional file 2: Table S1. Primers used in this study. Figure S1. In silico analyses of co-expressed gene networks around SWEET10. (A) Gene network representation and list of genes correlated to SWEET10 during development generated by the GENEVESTIGATOR software [74]. (B) The Arabidopsis thaliana trans-factor and cis-element prediction database ATTED-II [75], implemented in www.arabidopsis.org, was used to predict and visualize co-expressed genes around SWEET10. Figure S2. Expression of pSWEET10:GUS in adult Arabidopsis plants. (A) GUS expression in a whole plant expressing pSWEET10:GUS. The T4 transgenic plant shown in (A) was grown under LDs until siliques were produced. Detail of an inflorescence (B) and a silique (C) showing GUS expression. Figure S3. Expression levels of SWEET10 in T1 transgenic lines overexpressing SWEET10. The expression levels of SWEET10 was quantified in Col-0 and 35S:SWEET10 T1 lines under LDs. Leaves were collected at ZT8. Errors bars indicate Standard Deviation. Figure S4. Effect of the overexpression of SWEET10 from SUC2 promoter on flowering time under LDs. At least 10 plants were used for each experiment. Asterisk indicates a significant different compared to Col-0 (T-test, p-Value ≤0.05). Errors bars indicate Standard Deviation. Figure S5. Analysis of amiR-SWEET10 transgenic plants. (A) Flowering time of 44 amiR-SWEET10 T1 lines compared to Col-0 under LDs. TLN: Total Leaf Number. (B) SWEET10 expression levels in a subset of T3 amiR-SWEET10 lines. Figure S6. Photoperiod-dependent expression profile of SWEET11, 12, 13 and 14. GUS expression in plants expressing pSWEET10:GUS. Plants were grown under SDs for 2 weeks, shifted to LDs and collected for GUS staining at ZT8 in different days. Scale bar = 5 mm. Figure S7. Levels of sugar during the photoperiodic induction of flowering. Col-0 and ft tsf plants were grown under SDs for 2 weeks, shifted to LDs shoot apices were harvested at ZT8 in different days. Shoot apices were harvested and used to quantify the concentration of fructose (fru), glucose (glu) and sucrose (suc). Errors bars indicate Standard Deviation.
Data Availability Statement
The transcriptomic datasets generated during the current study are available in the GEO NCBI repository under the accession number GSE125054 and in the supplementary information files. Arabidopsis thaliana Columbia, which was used throughout this work, is available from the Arabidopsis Stock Centre, Nottingham University, UK or the Arabidopsis Biological Resource Centre, Ohio State University or from the authors. No permissions were required to use Arabidopsis thaliana. All other plants materials are available from the corresponding authors.