Abstract
Background
Dysregulation of splicing variants (SVs) expression has recently emerged as a novel cancer hallmark. Although the generation of aberrant SVs (e.g. AR-v7/sst5TMD4/etc.) is associated to prostate-cancer (PCa) aggressiveness and/or castration-resistant PCa (CRPC) development, whether the molecular reason behind such phenomena might be linked to a dysregulation of the cellular machinery responsible for the splicing process [spliceosome-components (SCs) and splicing-factors (SFs)] has not been yet explored.
Methods
Expression levels of 43 key SCs and SFs were measured in two cohorts of PCa-samples: 1) Clinically-localized formalin-fixed paraffin-embedded PCa-samples (n = 84), and 2) highly-aggressive freshly-obtained PCa-samples (n = 42).
Findings
A profound dysregulation in the expression of multiple components of the splicing machinery (i.e. 7 SCs/19 SFs) were found in PCa compared to their non-tumor adjacent-regions. Notably, overexpression of SNRNP200, SRSF3 and SRRM1 (mRNA and/or protein) were associated with relevant clinical (e.g. Gleason score, T-Stage, metastasis, biochemical recurrence, etc.) and molecular (e.g. AR-v7 expression) parameters of aggressiveness in PCa-samples. Functional (cell-proliferation/migration) and mechanistic [gene-expression (qPCR) and protein-levels (western-blot)] assays were performed in normal prostate cells (PNT2) and PCa-cells (LNCaP/22Rv1/PC-3/DU145 cell-lines) in response to SNRNP200, SRSF3 and/or SRRM1 silencing (using specific siRNAs) revealed an overall decrease in proliferation/migration-rate in PCa-cells through the modulation of key oncogenic SVs expression levels (e.g. AR-v7/PKM2/XBP1s) and alteration of oncogenic signaling pathways (e.g. p-AKT/p-JNK).
Interpretation
These results demonstrate that the spliceosome is drastically altered in PCa wherein SNRNP200, SRSF3 and SRRM1 could represent attractive novel diagnostic/prognostic and therapeutic targets for PCa and CRPC.
Keywords: Prostate cancer, Splicing, Spliceosome, SNRNP200, SRSF3, SRRM1, Therapeutic target
Research in context.
Evidence before this study
Early studies have demonstrated that the dysregulation of the expression of several splicing variants [e.g. Androgen receptor variant-7 (AR-v7), truncated somatostatin receptor SST5TMD4, In1-ghrelin, etc.) increases prostate cancer (PCa) aggressiveness and/or promotes resistance to the available drugs currently used in clinical practice (e.g. antiandrogens), hampering the treatment and the management of PCa patients. In this context, it is well known that this process is catalyzed by a macromolecular machinery (i.e. spliceosome) and regulated by specific proteins (i.e. splicing factors; SFs). However, the putative dysregulation of spliceosome components (SCs) and splicing factors in PCa as well as the potential of these elements as diagnostic, prognostic and/or therapeutic tools for this pathology remain poorly known.
Added value of this study
Our study demonstrates that the expression of several SCs and SFs are drastically dysregulated in PCa tissues compared with control (non-tumoral) tissues. Of particular importance is the demonstration that the changes in the expression of SNRNP200, SRSF3 and SRRM1 (at the mRNA and protein level) are associated to key clinical features of aggressiveness (e.g. Gleason score, biochemical recurrence, presence of metastasis, vascular and perineural invasion) in PCa patients as well as to important molecular phenotypes (e.g. AR-v7 expression) of PCa. Notably, this study also shows that the silencing of the expression of SNRNP200, SRRM1and SRSF3: 1) evoked clear antitumor actions in PCa cells (e.g. inhibition of proliferation, migration) through the modulation of key oncogenic signaling pathways (e.g. PI3K/AKT, ERK, JNK) and splicing variants (e.g. AR-v7, PKM2, XBP1s), and, 2) was able to re-sensitizes PCa cells to enzalutamide treatment, pointing out the potential therapeutic utility of these elements for the most aggressive phenotype of PCa, castration resistant PCa, which remains lethal nowadays.
Implications of all the available evidence
Altered expression of splicing machinery elements (spliceosome components and splicing factors) might be associated with the development, progression and aggressiveness of PCa. Our data demonstrate the existence of a splicing machinery-associated molecular dysregulation that could be potentially considered as a source of novel diagnostic and prognostic biomarkers as well as therapeutic targets for PCa. Specifically, our results reveal that three components of the splicing machinery (SNRNP200, SRSF3 and SRRM1) could represent potential, global and effective therapeutic targets to tackle this devastating pathology.
Alt-text: Unlabelled box
1. Introduction
Dysregulation of the alternative splicing process is considered a key hallmark of cancer and could represent a novel source for the identification of diagnostic, prognostic and therapeutic targets in highly prevalent tumor pathologies [1]. Specifically, alternative splicing represents an essential biological process by which the introns of an immature pre-mRNA are excised, and the exons are fused to generate mature mRNAs capable to be translated into functional proteins [2]. Indeed, most eukaryotic genes undergo alternative splicing, leading to a higher molecular flexibility through the increase of transcripts generated from a given gene [2]. Unfortunately, dysregulations of this process are associated to the appearance of diverse protein isoforms that could exhibit strong pathological potential [1]. In eukaryotes, the control of the appropriate splicing process is orchestrated by the spliceosome, a complex cellular machinery comprised by different small nuclear ribonucleoproteins (RNU1, RNU2, etc.) and core spliceosome-associated proteins (PRPF8, PRPF40A, U2AF2, SF3B1, etc.). The spliceosome dynamically interacts with additional proteins (splicing factors; SFs) to finely recognize the introns and exons to be processed and to catalyze the splicing process of the pre-RNAs [1]. The appropriate functioning of this cellular machinery is essential to maintain the cellular, tissue and body homeostasis and, therefore, the dysregulation of this process has been associated to different diseases, including endocrine-metabolic and tumor pathologies [2].
In this scenario, it has been shown that alternative splicing contributes to the heterogeneity, aggressiveness and resistance to medical treatment of prostate cancer (PCa), one of the most important public health problems worldwide with numbers of cases increasing significantly every year [3]. In this context, several studies have demonstrated that the androgen receptor (AR) gene is a source of splicing variants (SVs), especially AR-v7, which is a key driver for PCa progression (e.g. increased risk of biochemical relapse and inferior overall survival outcomes). In addition, a number of cancer-specific SVs has been identified in PCa, including SST5TMD4 [4], PKM2 [5], REST4 [6], XBP1s [7], In1-Ghrelin [8] etc., which also play an oncogenic role in this tumor pathology. In fact, it has been proposed that the “splicing signature” represents a more accurate parameter to stratify patients than the “transcriptome signature”, which is typically analysed by conventional microarray analyses [9]. Thus, the understanding of the regulation of splicing in normal and pathological prostate cells may help to identify novel biomarkers and therapeutic targets for this devastating tumor pathology. However, to the best of our knowledge, to date no studies have been focused on the mechanisms driving the generation of the SVs (i.e. the regulation of the splicing machinery), which might provide new contexts for the development of novel strategies to tackle PCa, since targeting the activity of selected spliceosome components that may be dysregulated in PCa could serve to inhibit the generation of oncogenic SVs (e.g. AR-v7), representing an attractive therapeutic strategy for PCa. Therefore, in this study we aimed to determine for the first time the expression levels of a representative set of spliceosome components (SCs) and SFs and their relationship with clinical and molecular features of PCa-aggressiveness, as well as its pathological role in this disease.
2. Material and methods
2.1. Patients and samples
This study was approved by the Reina Sofia University Hospital Ethics Committee and was conducted in accordance with the principles of the Declaration of Helsinki. The regional Biobank coordinated the collection, processing, management and assignment of the biological samples used in the present study according to the standard procedures established for this purpose. Written informed consent was obtained from all patients. Two different cohorts of prostate samples were included in this study:
-
-
Cohort 1) formalin-fixed, paraffin-embedded (FFPE) PCa tissues (n = 84) and their non-tumor adjacent region (N-TAR; used as control tissues; n = 84), taken from radical prostatectomies from patients diagnosed with clinically localized PCa (Table 1).
-
-
Cohort 2) fresh PCa samples (n = 42) that were obtained by core needle biopsies from patients with suspect of presenting significant PCa [defined as Gleason score (GS) ≥ 7; highly aggressive PCa], which was further confirmed histologically by uro-pathologists (Table 2).
Table 1.
Demographic, biochemical and clinical parameters of the patients with clinically localized PCa (Cohort 1). PSA: Prostate specific antigen; SigPCa: Significant PCa, defined as Gleason score ≥ 7; pT: Pathological primary tumor staging; PI: Perineural invasion; VI: Vascular invasion.
| Parameter | |
|---|---|
| Patients [n] | 84 |
| Age, years [median (IQR)] | 61 (57–66) |
| PSA levels, ng/mL [median (IQR)] | 5.2 (4.2–8.0) |
| Sig PCa [n (%)] | 76 (90.5%) |
| pT ≥ 3a [n (%)] | 59 (70.2%) |
| PI [n (%)] | 72 (85.7%) |
| VI [n (%)] | 8 (9.52%) |
| Recurrence [n (%)] | 35 (41.7%) |
| Metastasis [n (%)] | 0 (0%) |
Table 2.
Demographic, biochemical and clinical parameters of the patients with highly aggressive PCa (Cohort 2). PSA: Prostate specific antigen; SigPCa: Significant PCa defined as Gleason score ≥ 7.
| Parameter | |
|---|---|
| Patients [n] | 42 |
| Age, years [median (IQR)] | 75 (69–81) |
| PSA levels, ng/mL [median (IQR)] | 62.0 (36.2–254.5) |
| SigPCa [n (%)] | 42 (100%) |
| Metastasis [n (%)] | 28 (66.7%) |
The clinical parameters collected from each patient were GS (analysed by uro-pathologists following the modified 2005, 2010 and 2014 ISUP criteria, based on the sample collection date), T-Stage, perineural invasion, lymphovascular invasion, presence of metastases at diagnose (determined by computed tomography and bone scan) and biochemical recurrence (defined by two consecutive PSA values > 0.2 ng/mL and rising, after radical prostatectomy).
2.2. Cell cultures
PCa cell lines (LNCaP, 22Rv1, PC-3 and DU145) were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA) while normal prostate cell line PNT2 was a kind gift from Dr. J. De Bono. These cell lines were cultured according to manufacturer instructions as previously described [4,8,10], validated by analysis of short tandem repeats (STRs) sequences using GenePrint 10 System (Promega, Barcelona, Spain) and checked for mycoplasma contamination by PCR as previously reported [4]. For functional assays, LNCaP, 22Rv1 and DU145 cell lines were used. For mechanistic assays, 22Rv1 cells were used since this cell line represents a PCa model with AR and AR-v7 expression.
2.3. Transfection with specific siRNAs
For silencing assays, LNCaP, 22Rv1 and DU145 cell lines were used. Specifically, 200,000 cells were seeded in 6-well plates and grown until 70% of confluence was reached. Then, cells were transfected with specific siRNAs against SNRNP200(#124735; Thermo Fisher Scientific, Madrid, Spain), SRRM1(s20018; Thermo Fisher Scientific) and SRSF3(s12733; Thermo Fisher Scientific) at 100 nM, using Lipofectamine-RNAiMAX (Thermo Fisher Scientific) as previously reported [22]. After 24h, cells were collected for quantitative-PCR (qPCR), western blot, cell proliferation and/or cell migration assays.
2.4. Cell proliferation
In order to determine the effect of the silencing of SNRNP200, SRRM1 and SRSF3 on cell proliferation, Alamar-Blue assay (Bio-Source International, Camarillo, CA, USA) was performed in LNCaP, 22Rv1 and DU145 cell lines, as previously reported [4]. Briefly, cells were seeded in 96-well plates at a density of 3,000–5,000 cells/well and serum-starved for 24h, then cell proliferation was evaluated using the FlexStation III system (Molecular Devices, Sunnyvale, CA, USA) until 72 h.
2.5. Enzalutamide-sensitization assay
To test the role of SNRNP200, SRRM1 and SRSF3 on the response to enzalutamide treatment (#1613, Axon Medchem, Groningen, The Netherlands), cell proliferation was evaluated. Specifically, LNCaP and 22Rv1 cells were acclimated during 24h to RPMI 1640 without phenol-red supplemented with charcoal-stripped serum (#A3382101; Thermo Fisher Scientific). Then, scrambled- or siRNA-transfected cells were treated with enzalutamide at 1 µM. All cells were treated with 5α-dihydrotestosterone (DHT; # d-073; Merck, Madrid, Spain) at 10 nM. Cell proliferation was calculated, after 24h of treatment, as described above. Results were expressed as percentage referred to scramble treated with vehicle (DMSO) plus DHT treatment.
2.6. Cell migration
Cell migration was evaluated by wound-healing assay in DU145 cell line in response to SNRNP200, SRRM1, SRSF3 silencing, due to the inability of LNCaP and 22Rv1 cells to migrate. Specifically, images of the scratch were taken at 0 and 12 h and wound healing was calculated as the area observed 12 h after the wound was made vs. the area observed just after wounding, as previously described [4]. Results were expressed as percentage referred to scramble.
2.7. Western blot
Protein levels of several PCa-related genes were analysed in 22Rv1 cells as previously reported [4]. Briefly, 200,000 cells were seeded in 12-well plates and after two days, proteins were extracted using pre-warmed (65 °C) SDS-DTT buffer (62.5 mM Tris–HCl, 2% SDS, 20% glycerol, 100 mM DTT, and 0.005% bromophenol blue). Then, proteins were sonicated for 10 s and boiled for 5 min at 95 °C. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline/0.05% Tween 20 and incubated overnight with the specific antibodies for phospho-AKT (#4060S; Cell Signaling, Leiden, NLD), phospho-ERK (#4370S; Cell Signaling), phospho-JNK (AF1205; R&D-Systems, Minneapolis, MN, USA), AKT (#9272S; Cell Signaling), ERK (sc-154; Santa Cruz Biotechnology, Dallas, TX, USA), JNK (AF1387; R&D Systems), SNRNP200 (ab241589; Abcam, Camdridge, UK), SRRM1 (PA5-69086; Thermo Fisher Scientific), SRSF3 (ab198291; Abcam) and the secondary antibody HRP-conjugated goat anti-rabbit IgG (#7074 s; Cell Signaling). Specifically, the specificity of SNRNP200 and SRSF3 antibodies was validated in our laboratory by western blot and ICC (only 1 band was recognized by western blot and depletion of protein quantity was observed in response to specific siRNAs using western blot and ICC approaches). SRRM1 specificity was not completely validated since more than 1 band was recognized in western blot (however, depletion of protein quantity was observed in response to a specific siRNAs using western blot and ICC approaches). Proteins were detected using an enhanced chemiluminescence detection system (GEHealthcare, Madrid, Spain) with dyed molecular weight markers (Bio-Rad, Madrid, Spain). A densitometry analysis of the bands obtained was carried out with ImageJ software, using total AKT, ERK and JNK protein levels as normalizing factor for phospho-AKT, phospho-ERK and phospho-JNK, respectively.
2.8. RNA extraction
Total RNA from FFPE samples was isolated and DNase-treated using the Maxwell 16 LEVRNA FFPE Kit (Promega, Madison, WI, USA) according to manufacturer instructions in the Maxwell MDx 16 Instrument (Promega, Madrid, Spain). Additionally, total RNA was extracted from fresh samples using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen) and from PCa cell lines using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), followed, in both cases, by DNase treatment using RNase-Free DNase Kit (Qiagen, Hilden, DEU). Total RNA concentration and purity was assessed using Nanodrop One Spectrophotometer (Thermo Fisher Scientific). Total RNA was retrotranscribed using random hexamer primers and the cDNA First Strand Synthesis kit (Thermo Fisher Scientific).
2.9. qPCR dynamic array based on microfluidic technology
A qPCR dynamic array based on microfluidic technology that allows the determination of the expression of 45 transcripts in 48 samples, simultaneously, (Fluidigm, San Francisco, CA, USA) was implemented. Specific primers for human transcripts including components of the major spliceosome (n = 10), minor spliceosome (n = 4), associated SFs (n = 25) and three housekeeping genes (ACTB, GAPDHand HPRT) were specifically designed with the Primer3 software and StepOne™ Real-Time PCR System software v2.3 (Applied Biosystems, Foster City, CA, USA) (Supplemental Table 1). Preamplification, exonuclease treatment and qPCR dynamic array based on microfluidic technology were implemented as recently reported [11,12], following manufacturer's instructions using the Biomark System and the Real-Time PCR Analysis Software (Fluidigm). To control for variations in the efficiency of the retro-transcription reaction, mRNA copy numbers of the different transcripts analysed were adjusted by normalization factor, calculated with the expression levels of ACTBand GAPDHusing GeNorm 3.3 [13].
2.10. RNA retrotranscription and quantitative real-time PCR
Details regarding the development, validation, and application of the quantitative real-time PCR to measure expression levels of the transcripts of interest have been previously reported by our laboratory [14], [15], [16], [17]. Specific primers set used to measure the expression levels of the genes of interest in this study are described in Supplemental Table 2. Specifically, primers for KLF6-SV1, KLF6, PKM2, PKM1, REST4 and REST transcripts were obtained from previous studies [18], [19], [20]. To control for variations in the efficiency of the retro-transcription reaction, mRNA copy numbers of the different transcripts analysed were adjusted by a normalization factor which was calculated with the expression levels of ACTBand GAPDHusing GeNorm 3.3 [13].
2.11. Immunohistochemistry (IHC) analysis
IHC analysis was performed on FFPE prostate samples that were obtained by core needle biopsies from patients of cohort 2, since they represent the most aggressive cohort of our study (GS ≥ 7). Samples of benign prostatic hyperplasia (BPH), prostatic intraepithelial neoplasia (PIN) and non-significant PCa with GS =6 (n = 7, 6 and 5, respectively) also taken by core needle biopsies were included in this analysis as control samples and low aggressive PCa, respectively. Briefly, deparaffinized sections were incubated overnight (4 °C) with the primary antibodies against the proteins of interest [i.e. SNRNP200 (ab241589; Abcam), SRRM1 (PA5-69086; Thermo Fisher Scientific) or SRSF3 (ab198291; Abcam)] at 1:250 dilution, followed by incubation with an anti-rabbit horseradish peroxidase–conjugated secondary antibody (#7074; Cell Signaling). Finally, sections were developed with 3,39-diaminobenzidine (Envision system 2-Kit Solution DAB) and contrasted with haematoxylin. Two independent pathologists performed histopathologic analyses indicating low, moderate, and high intensities of staining, following a blinded protocol.
2.12. Statistical and bioinformatic analyses
Statistical differences between two groups were calculated by unpaired parametric t-test or nonparametric Mann Whitney U test, according to normality, assessed by Kolmogorov-Smirnov test. For differences among three groups, One-Way ANOVA analysis was performed. Spearman's or Pearson´s bivariate correlations were performed for quantitative variables according to normality. Significant relation between categorized mRNA expression and biochemical PCa recurrence was studied using the long-rank-p-value method. Predictive models were constructed by Random Forest algorithm (with R language) as classifier as previously reported [11,12]. The rest of the statistical analyses were assessed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA) or SPSS version 17.0. All the experiments were performed in, at least, 3 independent times (n ≥ 3), and with at least 2 technical replicates. Statistical significance was considered when p < 0.05. A trend for significance was indicated when p-values ranged between > 0.05 and < 0.1.
3. Results
3.1. Expression levels of spliceosome components and splicing factors are dysregulated in PCa and are associated with clinical and molecular aggressiveness features
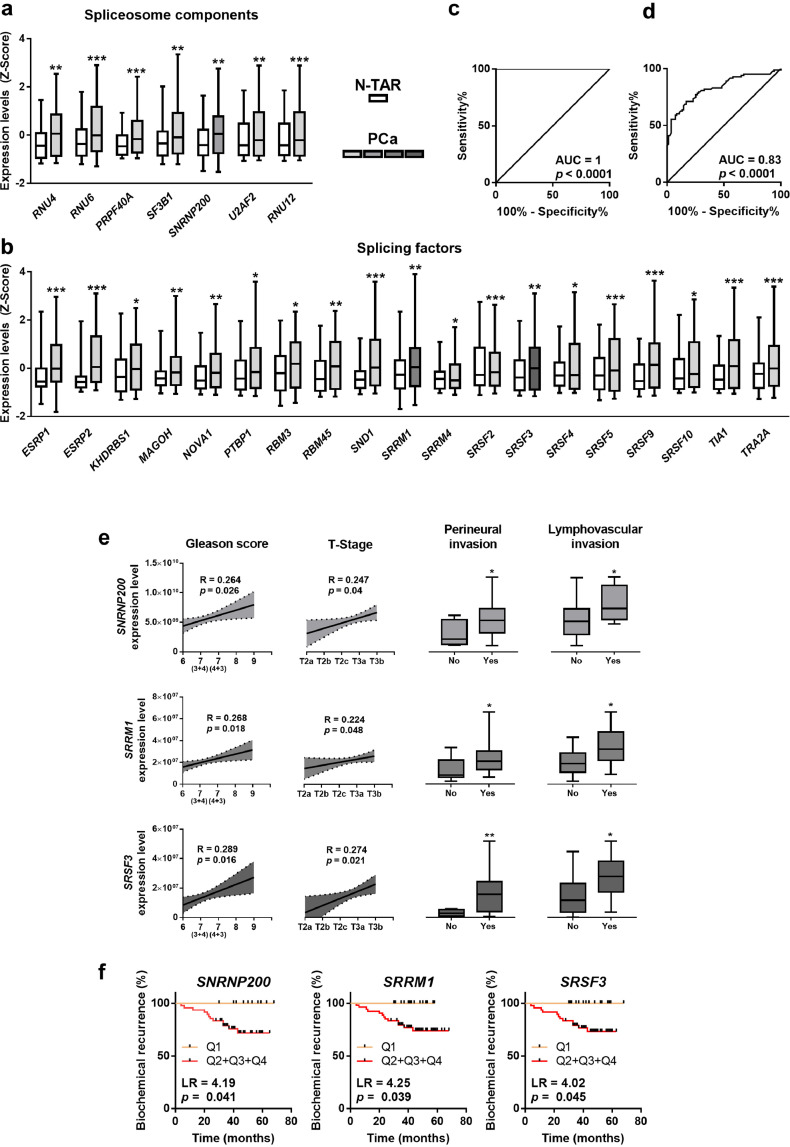
The expression levels of 26 out of 45 (58%) genes involved in the pre-RNA splicing process were found to be significantly dysregulated in PCa-tissues compared to their respective control-tissues (N-TAR) from a cohort of patients with clinically-localized PCa (n = 84; Table 1; Fig. 1a/b). Specifically, the SCs RNU4, RNU6, PRPF40A, SF3B1, SNRNP200, U2AF2 and RNU12 were overexpressed in PCa-tissues (Fig. 1a). Additionally, the SFs ESRP1, ESRP2, KHDRBS1, MAGOH, NOVA1, PTBP1, RBM3, RBM45, SND1, SRSF2, SRSF3, SRSF4, SRSF5, SRSF9, SRSF10, SRRM1, TIA1 and TRA2A were also overexpressed while the SF SRRM4 was down-regulated in PCa-tissues (Fig. 1b). No statistically significant differences were observed in the expression levels of the rest of SCs and SFs analysed (Supplemental Figure 1). Non-supervised clustering bioinformatic approaches (Random Forest algorithm) revealed that the molecular fingerprint comprised by the combination of the expression of SNRNP200, SRRM1, SRSF3, TIA1, SRSF10, U2AF2, SRSF9, SRRM4, SND1, ESRP1, and KHDRBS1 perfectly discriminated between PCa and N-TARs samples with an AUC =1 (Fig. 1c). This was further supported by cross-validation analysis of this molecular fingerprint (AUC =0.83, p < 0.0001; Fig. 1d).
Fig. 1.
Expression of spliceosome components and splicing factors in prostate cancer (PCa) samples. (a–b) Comparison of mRNA levels of spliceosome components (a) and splicing factors (b) between formalin-fixed paraffin embedded (FFPE) samples from PCa samples and non-tumor adjacent regions (N-TAR) (n = 84) determined by a microfluidic-based qPCR array. Data represent the mean ± SEM of mRNA expression levels adjusted by normalization factor (calculated from ACTB and GAPDH expression levels) and standardized by Z-score. c-d) ROC curves of a subset of spliceosome components and splicing factors generated by Random Forest computational algorithm (c) followed by cross validation analysis (d) to distinguish between tumor and N-TAR samples. e) Association between the expression levels of selected spliceosome components and splicing factors (SNRNP200, SRRM1 and SRSF3) and clinical parameters (Gleason score, T-Stage, perineural and lymphovascular invasion) in the same cohort of FFPE samples (n = 84). Correlations are represented by mean (connecting line) and error bands (pointed line) of expression levels. Data of associations represent the mean ± SEM of mRNA expression levels adjusted by normalization factor (calculated from ACTB and GAPDH expression levels). f) Association between SNRNP200, SRRM1 and SRSF3 expression levels and biochemical PCa recurrence in 67 samples from FFPE cohort (samples from patients who underwent adjuvant radiotherapy were not included), calculated by Log Rank analysis (LR). mRNA levels were determined by a microfluidic-based qPCR array and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.
Notably, the expression levels of the 85% (22 out of 26) SCs and SFs found dysregulated herein were significantly associated and/or correlated with at least one clinical parameter of PCa-aggressiveness (Tables 3 and 4). Remarkably, SNRNP200, SRRM1 and SRSF3 were the only genes whose expression was directly associated/correlated with all the clinical parameters of aggressiveness (Gleason score, T-Stage, perineural- and lymphovascular-invasion; Table 3; Fig. 1e) and progression (biochemical recurrence; Table 4; Fig. 1f) available in this cohort of patients. In addition, in silico data showed the overexpression in PCa samples of SNRNP200 in Singh, Wallace and Welsh datasets. Moreover, SRRM1 was found to be overexpressed in PCa samples in Welsh and Tomlins datasets, while a trend was observed in Wallace dataset (p = 0.089). On the other hand, an overexpression of SRSF3 in PCa samples was found in Singh and Tomlins datasets (Supplemental Figure 2).
Table 3.
Correlation and association of SNRNP200, SRRM1 and SRSF3 expression levels with clinical features of prostate cancer aggressiveness. Data of correlations represent the coefficient "r". Data of associations represent the difference between means of each group ± standard deviation. Significant prostate cancer (SigPCa) is defined as Gleason score ≥ 7. Asterisks (* p < 0.05; ** p < 0.01; ** p < 0.001) indicate statistically significant differences between groups.
| Correlations |
Associations |
||||
|---|---|---|---|---|---|
| Gleason score | T- Stage | Perineural Invasion | Lymphovascular Invasion | SigPca | |
| RNU4 | 0.122 | 0.239* | 9,664,718,823 ± 3,162,530,673 ** | 7,730,140,948 ± 3,078,119,236 * | 7,526,218,231 ± 3,298,861,131 * |
| RNU6 | 0.235* | 0.075 | 230,034,826 ± 72,616,606 ** | 119,362,920 ± 102,513,311 | 163,738,961 ± 75,084,561 * |
| PRPF40A | 0.184 | 0.214 | 1,801,231 ± 878,336 * | 1,669,010 ± 855,998 | 1,789,300 ± 832,618 * |
| SF3B1 | 0.312** | 0.121 | 9,003,875 ± 3,487,752 * | 10,853,954 ± 4,783,529 ** | 5,808,930 ± 3,941,225 |
| U2AF2 | 0.09 | 0.083 | 714,287 ± 348,778 * | 807,889 ± 481,128 | 347,329 ± 370,880 |
| SNRNP200 | 0.264* | 0.247* | 2,681,185,105 ± 1,175,955,224 * | 2,812,777,484 ± 1,275,566,115 * | 2,125,357,393 ± 1,042,911,480 * |
| RNU12 | 0.321** | 0.178 | 395,441 ± 132,486 ** | 133,816 ± 149,579 | 208,236 ± 139,628 |
| ESRP1 | 0.319** | 0.166 | 1,658,118 ± 916,550 | 2,151,271 ± 1,084,569 | 1,874,015 ± 936,328 * |
| ESRP2 | 0.281* | 0.254* | 1,953,298 ± 1,179,570 | 3,964,985 ± 1,266,549 ** | 2,941,102 ± 1,300,974 * |
| KHDRBS1 | 0.128 | 0.215 | 8,816,079 ± 4,355,945 * | 7,047,264 ± 5,942,380 | 10,326,770 ± 4,988,242 * |
| MAGOH | 0.331** | 0.173 | 572,836 ± 376,052 | 1,036,475 ± 489,737 * | 772,768 ± 395,132 |
| NOVA1 | 0.131 | 0.211 | 1,375,554 ± 1,240,422 | 2,891,594 ± 1,397,232 * | 1,968,158 ± 1,417,671 |
| PTB | 0.189 | 0.197 | 462,842 ± 504,081 | 707,443 ± 685,520 | 923,496 ± 547,502 |
| RBM3 | 0.141 | 0.139 | 1,831,860 ± 1,133,513 | 2,948,642 ± 1,333,148 * | 2,051,801 ± 1,189,264 |
| RBM45 | 0.145 | 0.159 | 2,144,099 ± 1,086,705 | 2,444,946 ± 1,430,495 | 1,613,514 ± 1,237,865 |
| SND1 | 0.236* | 0.167 | 6,829,797 ± 3,127,408 * | 8,116,316 ± 3,698,131 * | 7,483,290 ± 3,461,207 * |
| SRSF2 | 0.284* | 0.161 | 903,780 ± 409,990 * | 1,099,682 ± 533,886 * | 914,182 ± 460,892 |
| SRSF3 | 0.289* | 0.274* | 13,698,858 ± 5,050,576 ** | 12,653,631 ± 5,712,504 * | 10,070,038 ± 5,031,330 * |
| SRSF4 | 0.139 | 0.169 | 2,131,612 ± 857,251 * | 3,422,072 ± 1,068,338 ** | 941,268 ± 902,489 |
| SRSF5 | 0.055 | 0.129 | 105,348,741 ± 37,924,790 ** | 141,185,003 ± 48,026,387 ** | 35,477,260 ± 37,643,655 |
| SRSF9 | 0.088 | 0.211 | 33,397,890 ± 12,917,648 * | 28,501,453 ± 15,538,733 | 26,539,564 ± 14,468,693 |
| SRSF10 | 0.057 | 0.218 | 10,135,257 ± 4,077,596 * | 10,622,878 ± 5,604,567 | 4,248,356 ± 4,293,783 |
| SRRM1 | 0.211* | 0.224* | 9,522,292 ± 4,314,271 * | 9,444,197 ± 4,007,860 * | 9,478,531 ± 4,760,093 * |
| SRRM4 | −0.154 | 0.107 | 53,981 ± 33,971 | -2375 ± 34,689 | 47,458 ± 36,610 |
| TIA1 | 0.114 | 0.246* | 5,965,156 ± 2,459,924 * | 6,334,929 ± 2,989,097 | 5,493,989 ± 2,630,409* |
| TRA2A | 0.141 | 0.191 | 27,361,791 ± 12,693,441 * | 39,100,412 ± 15,368,002 | 29,010,940 ± 15,801,444 |
Table 4.
Association of expression levels of spliceosome components and splicing factors with the development of biochemical recurrence of prostate cancer.
| Factor | LongRank | p value |
|---|---|---|
| RNU4 | 1.822 | 0.177 |
| RNU6 | 5.046 | 0.025 |
| PRPF40A | 4.938 | 0.026 |
| SF3B1 | 4.012 | 0.045 |
| U2AF2 | 4.222 | 0.040 |
| RNU12 | 2.844 | 0.092 |
| ESRP1 | 3.201 | 0.074 |
| ESRP2 | 5.049 | 0.025 |
| KHDRBS1 | 4.455 | 0.035 |
| MAGOH | 2.540 | 0.111 |
| NOVA1 | 4.758 | 0.029 |
| PTBP1 | 4.407 | 0.036 |
| RBM3 | 1.631 | 0.202 |
| RBM45 | 5.075 | 0.024 |
| SND1 | 4.992 | 0.025 |
| SRSF2 | 4.632 | 0.031 |
| SRSF4 | 3.722 | 0.054 |
| SRSF5 | 3.944 | 0.047 |
| SRSF9 | 4.349 | 0.037 |
| SRSF10 | 3.971 | 0.046 |
| SRRM4 | 1.703 | 0.192 |
| TIA1 | 4.551 | 0.033 |
| TRA2A | 4.702 | 0.030 |
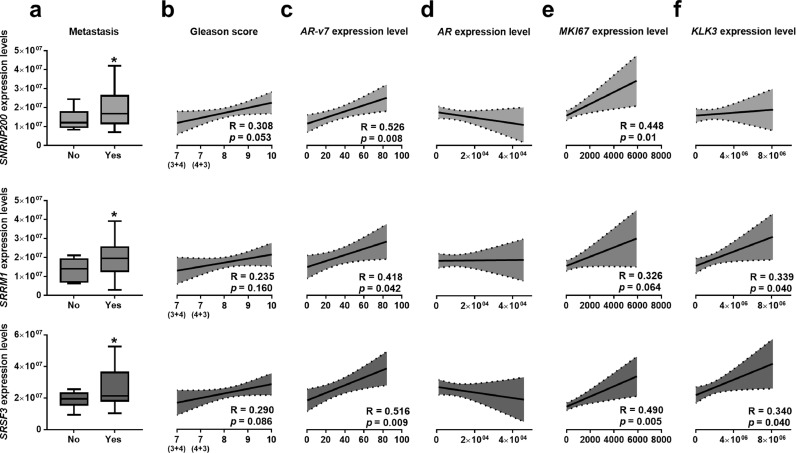
Based on these results, we further analysed the expression levels of SNRNP200, SRRM1 and SRSF3 in an independent and more aggressive cohort of PCa-samples (Table 2). These analyses revealed an association between the elevated expression levels of SNRNP200, SRRM1 and SRSF3 in PCa-samples and the presence of metastases at the moment of diagnosis (Fig. 2a). SNRNP200 and SRSF3 expression tended to be positively correlated with Gleason score (p = 0.053, R = 0.308; p = 0.086, R = 0.290; respectively; Fig. 2b). Furthermore, SNRNP200, SRRM1 and/or SRSF3 expression directly correlated with the expression levels of the oncogenic SV AR-v7 [(Fig. 2c); but not with ARexpression (Fig. 2d)], with MKI67 (Fig. 2e) and KLK3 (i.e. SRSF3 and SRRM1, but not SNRNP200; Fig. 2f).
Fig. 2.
Expression of SNRNP200, SRRM1 and SRSF3 in a highly aggressive cohort of prostate cancer (PCa) samples. (a) SNRNP200, SRRM1 and SRSF3 expression levels in a battery of highly-aggressive PCa samples, with or without the presence of metastasis (n = 42). represent the mean ± SEM of mRNA expression levels. b-f) Correlation between SNRNP200, SRRM1 and SRSF3 expression levels and Gleason score (b), expression levels of AR-v7 (c), AR (d) MKI67 (e) and KLK3 (f). mRNA levels were determined by a microfluidic-based qPCR array and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Asterisk (* p < 0.05) indicates statistically significant differences between groups.
3.2. Protein levels of SNRNP200, SRRM1 and SRSF3 are elevated in PCa samples and associated with clinical aggressiveness features
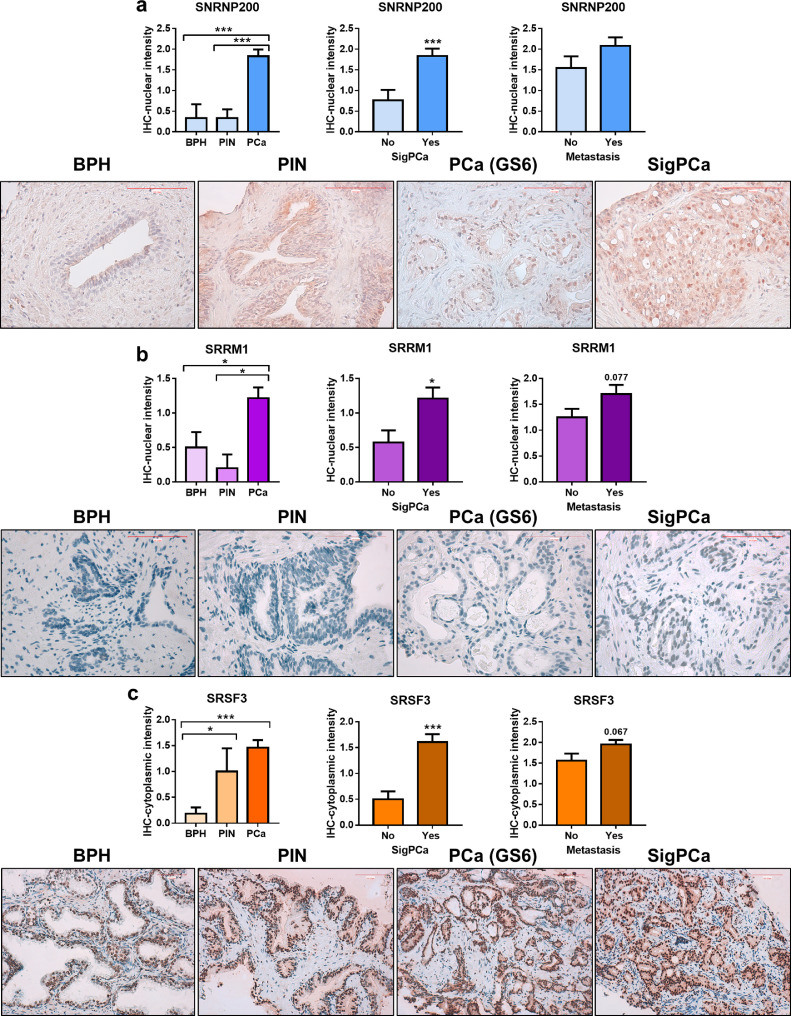
IHC analyses using FFPE samples from the cohort of patients with highly-aggressive PCa (Table 2) revealed that SNRNP200, SRSF3 and SRRM1 protein levels were significantly higher in PCa-samples compared to controls samples (FFPE samples of patients with PIN and/or BPH). Specifically, nuclear staining of SNRNP200 was higher in PCa samples compared to PIN and BPH samples. Low levels of cytoplasmic SNRNP200 protein were detected, while no changes were observed among groups (data not shown). Furthermore, although cytoplasmic protein level of SRRM1 was not detected, SRRM1-nuclear levels were significantly higher in PCa-samples compared to BPH and PIN samples (Fig. 3b). In contrast, no differences among SRSF3-nuclear staining were found between PCa, BPH and PIN samples (data not shown); however, SRSF3-cytoplasmic staining was significantly higher in PCa and PIN samples compared to BPH samples (Fig. 3c). Finally, SNRNP200, SRRM1 and SRSF3 protein levels were associated with clinically significant PCa (SigPCa; Gleason score ≥ 7) and, in the case of SRRM1 and SRSF3, tended to be associated with the presence of metastasis (p = =0.077, p = =0.067, respectively).
Fig. 3.
Immunohistochemical analysis of SNRNP200,SRRM1 and SRSF3 in prostate cancer (PCa) samples. a) Comparison of SNRNP200,SRRM1 (b) and SRSF3 (c) protein levels by immunohistochemistry (IHC) between a representative set of PCa samples (n = 47), prostatic intraepithelial neoplasia (PIN; n = 6) and benign prostatic hyperplasia (BPH; n = 7). Association of protein levels with clinically significant PCa (SigPCa; defined as Gleason score higher than 7) and the presence of metastasis at diagnosis (central panel and right panel, respectively). Representative images of BPH, PIN, PCa with Gleason score = 6 and SigPCa stained with SNRNP200 (400× magnification), SRRM1 (400× magnification) and SRSF3 (200× magnification) antibodies are showed below a, b and c panels, respectively. Scale bar indicates 100 µm. Data are expressed as mean ± SEM of IHC staining scaled from low [1] to high [3] intensity. Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.
3.3. Silencing of SNRNP200, SRRM1 and SRSF3 expression decreases functional parameters of aggressiveness in PCa cells
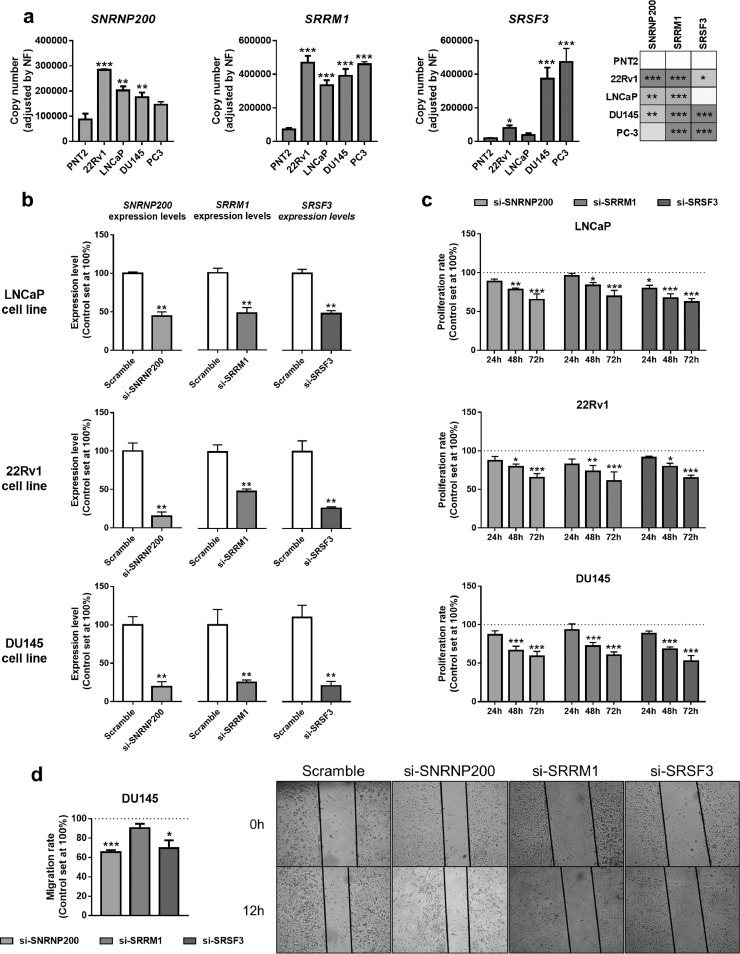
The analysis of the expression levels of SNRNP200, SRRM1 and SRSF3 in prostate-derived cell lines showed that: 1) SNRNP200 was overexpressed in the PCa-derived cell lines 22Rv1, LNCaP and DU145 compared to the normal-like prostate cell line PNT2 (Fig. 4a, left-panel); 2) SRRM1was significantly overexpressed in all the PCa cell lines compared to PNT2 (Fig. 4a, middle-panel); and, 3) that SRSF3was overexpressed in 22Rv1, DU145 and PC-3 compared to PNT2 (Fig. 4a, right-panel). Therefore, we selected an androgen-dependent AR+/AR-v7− model (LNCaP), an androgen-independent AR+/AR-v7+model (22Rv1) and an androgen independent AR−/AR-v7− model (DU145) to perform functional experiments in response to the silencing of these three genes. The silencing of SNRNP200, SRRM1 and SRSF3 in response to specific siRNAs was validated at mRNA levels in LNCaP, 22Rv1 and DU145 cells (Fig. 4b), as well as at protein levels in DU145 cells (Supplemental Figure 3). Remarkably, no effects in proliferation rate were observed in response to scramble-siRNA in LNCaP, 22Rv1 and DU145 as compared to non-transfected cells (Supplemental Figure 4). Specifically, proliferation rate of LNCaP cells significantly decreased at 48 and 72 h in response to SNRNP200- and SRRM1-silencing, as well as at 24, 48 and 72 h in response to SRSF3-silencing (Fig. 4c) compared to scramble-transfected cells. Moreover, the silencing of SNRNP200, SRRM1 and SRSF3 significantly decreased proliferation rate (at 48 and 72 h) in 22Rv1 and DU145 (Fig. 4c). In addition, silencing of SNRNP200, SRSF3, but not SRRM1, reduced migration rate in DU145 cells (Fig. 4d).
Fig. 4.
Functional consequences of SNRNP200, SRRM1 and SRSF3 silencing in prostate-derived cell lines. a) Comparison of SNRNP200, SRRM1 and SRSF3 expression levels between a non-tumor prostate cell line (PNT2) and PCa cell lines LNCaP, 22Rv1, DU145 and PC-3 (n = 5). mRNA levels were determined by qPCR and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. b) Validation by qPCR of SNRNP200, SRRM1 and SRSF3 silencing (si-SNRNP200, si-SRRM1 and si-SRSF3, respectively). mRNA levels were determined by qPCR and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Data were represented as percent of scramble cells (mean ± SEM). c) Proliferation rate of LNCaP (upper panel), 22Rv1 (middlepanel) and DU145 (bottom panel) cell lines after 24-, 48- and 72 h of SNRNP200-, SRRM1- and SRSF3-silencing (n = 4). d) Effect of SNRNP200-, SRRM1- and SRSF3-silencing on the migration rate of DU145 cell line was determined by wound-healing assay (12 h; n ≥ 3). Representative images are depicted in right panel. Data were represented as percent of scramble cells (mean ± SEM). Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.
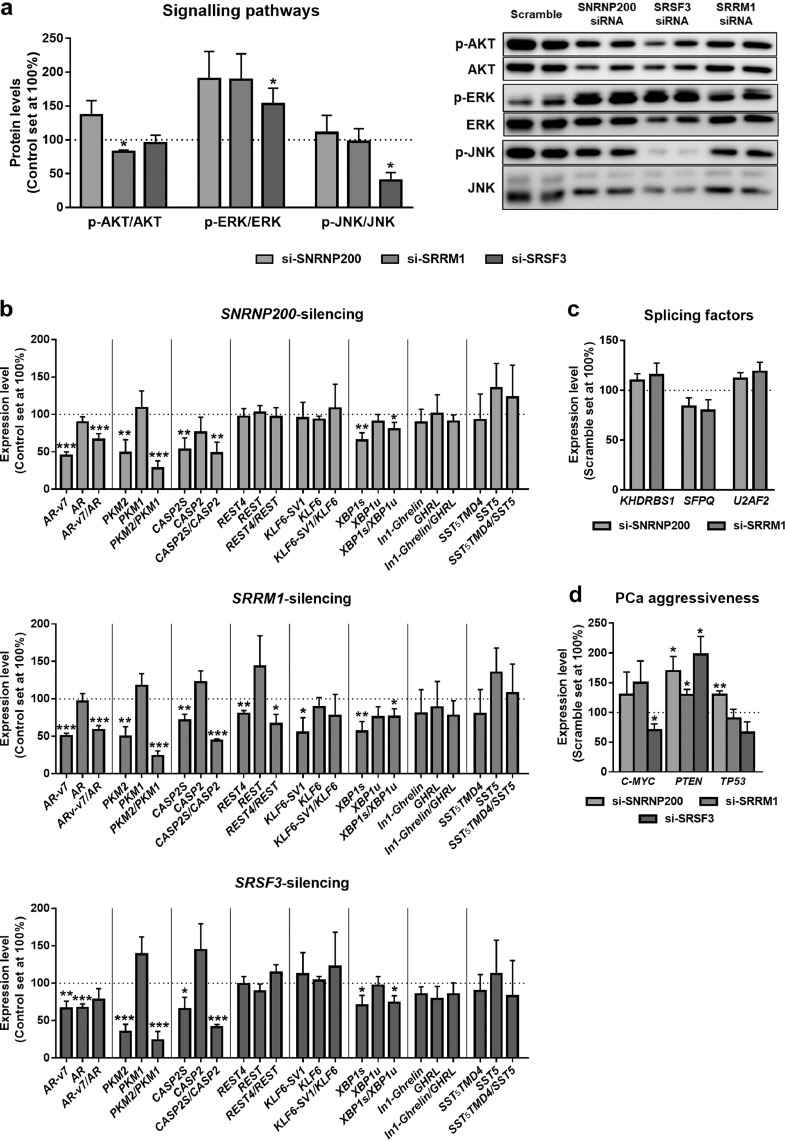
3.4. Silencing of SNRNP200, SRRM1 and SRSF3 modulates relevant signaling pathways and the expression of oncogenic splicing variants in PCa
The silencing of SRRM1 significantly decreased the phosphorylation levels of AKT, while silencing of SRSF3 increased phosphorylation levels of ERK and decreased that of JNK (Fig. 5a). In contrast, no statistically significant changes in the phosphorylation levels of AKT, ERK or JNK were observed in response to SNRNP200-silencing (Fig. 5a).
Fig. 5.
Molecular consequences of SNRNP200, SRRM1 and SRSF3 silencing in 22Rv1 cell line. a) Basal phospho-AKT, phospho-ERK1/2 and phospho-JNK levels in SNRNP200-, SRRM1- and SRSF3 silenced 22Rv1 cells (si-SNRNP200, si-SRRM1 and si-SRSF3, respectively; 24 h; n ≥ 3). Protein levels were normalized by total AKT, ERK and JNK protein levels. Representative images are shown in right panel. Protein data were represented as percent of scramble cells. b) Expression levels of selected transcripts in response to SNRNP200 (upper panel), SRRM1 (central panel) and SRSF3 (bottom panel) silencing (24 h) in 22Rv1 cells. Ratio between the expression of splicing variants is shown in bars with dotted pattern. c) Expression levels of KHDRBS1, SFPQ and U2AF2 in response to SNRNP200- and SRRM1-silencing in 22Rv1 cells. d) Expression levels of C-MYC, PTEN and TP53 in response to SNRNP200-, SRRM1- and SRSF3-silencing in 22Rv1 cells. mRNA levels were determined by qPCR and adjusted by normalization factor calculated from ACTB and GAPDH expression levels. Data were represented as percent of scramble-treated control cells (mean ± SEM). Asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001) indicate statistically significant differences between groups.
The silencing of the expression of SNRNP200, SRRM1 and SRSF3 reduced the expression levels of the oncogenic splicing variants PKM2, CASP2S and XBP1s, without altering the expression of PKM1, CASP2 and XBP1u. Therefore, SNRNP200-, SRRM1- and SRSF3-silencing decreased PKM2/PKM1, CASP2S/CASP2 and XBP1s/XBP1u ratio (Fig. 5b). Moreover, only SRRM1-silencing resulted in a decrease in the REST4/REST expression ratio (by a reduction of REST4 without alteration of REST) and in a reduction of KLF6-SV1 expression (Fig. 5b). In the case of AR splicing process, silencing of SNRNP200 and SRRM1 (but not SRSF3) evoked a decrease in the ratio of the AR-v7/AR expression, by a reduction of AR-v7 without altering AR expression; while the silencing of SRSF3 decreased both AR-v7 and AR expression. Due to the importance of AR-v7 on PCa aggressiveness, we determined whether SNRNP200 and SRRM1 could control the expression levels of splicing factors previously associated to the modulation of AR gene splicing and with AR-v7 generation [i.e. KHDRBS1 [21], SFPQ [22] and U2AF2 [23]]. However, the expression levels of KHDRBS1, SFPQ and U2AF2 were not significantly altered in response to the silencing of SNRNP200 or SRRM1 (Fig. 5c). In addition, no statistically significant changes were observed in the expression of additional splicing variants, such as SST5TMD4 andIn1-ghrelin, in response to SNRNP200-, SRRM1- and SRSF3-silencing (Fig. 5b).
Finally, expression levels of classical markers of PCa aggressiveness were analysed in response to SNRNP200-, SRRM1- and SRSF3-silencing. Specifically, C-MYC expression levels were reduced in response to SRSF3-silencing, while those of TP53 were increased in response to SNRNP200-silencing (Fig. 5d). In addition, the silencing of SNRNP200, SRRM1 and SRSF3 resulted in a significantly increase of the expression levels of PTEN (Fig. 5d).
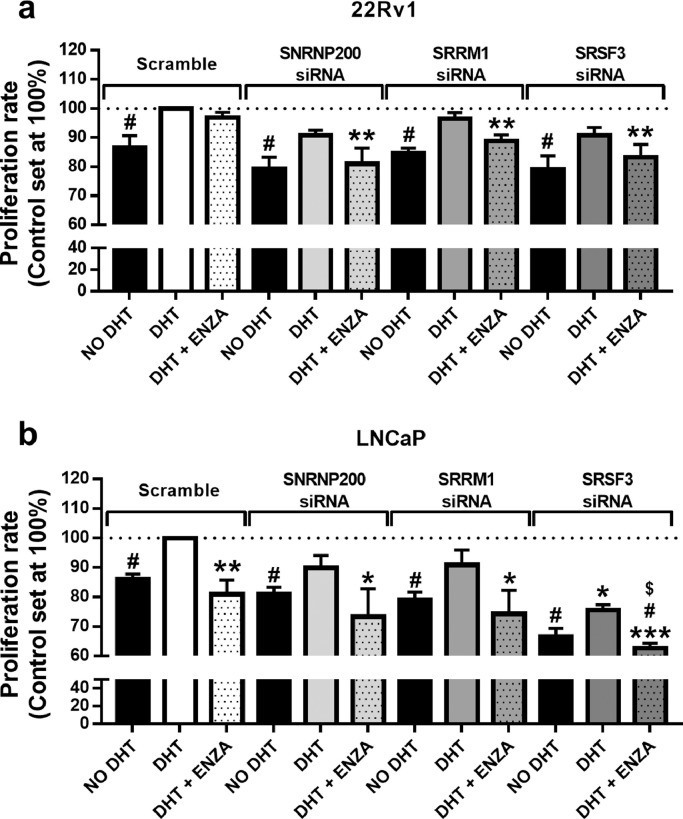
3.5. Silencing of SNRNP200, SRRM1 and SRSF3 enhanced the antitumor actions of enzalutamide in PCa cells
In order to test the effects of SNRNP200-, SRRM1- and SRSF3-silencing on androgens and enzalutamide responsiveness, we evaluated proliferation rate of 22Rv1 and LNCaP cells after 24h. We used this experimental paradigm in that cell proliferation was not significantly compromised after 24h of enzalutamide treatment nor by 24h of SNRNP200-, SRRM1- and SRSF3-silencing (Fig. 6a, open bars) in 22Rv1 cells. Firstly, we observed that the silencing of SNRNP200, SRRM1or SRSF3did not alter the normal (scramble-treated) response to DHT as a similar decrease of proliferation rate was found compared to DHT-treated cells, independently of the siRNA used (i.e. scramble, si-SNRNP200, si-SRRM1 or si-SRSF3) in 22Rv1 and LNCaP cell lines (Fig. 6a/b). However, the silencing of SNRNP200, SRRM1 and SRSF3 combined with enzalutamide treatment resulted in an additive, statistically significant antiproliferative effect in 22Rv1 cells at 24h of incubation, indicating that silencing of SNRNP200, SRRM1 or SRSF3 sensitized 22Rv1 cells to enzalutamide (Fig. 6a). Additionally, although no differences were observed when analysing 22Rv1 scramble-treated cells in response to enzalutamide at 24h, we found a significantly decrease in the proliferation rate of scramble-treated LNCaP cells in response to enzalutamide (Fig. 6a/b). Interestingly, the pattern of response of SNRNP200- and SRRM1-silenced LNCaP cells was similar to scramble-treated LNCaP cells (Fig. 6b), wherein the response to non-DHT and to enzalutamide was comparable in scramble-, si-SNRNP200- and si-SRRM1-treated LNCaP cells. In striking contrast, the response of SRSF3-silenced cells was markedly different. Specifically, the proliferation rate of SRSF3-silenced LNCaP cells in the presence of DHT was significantly lower compared to scramble-treated LNCaP cells in the presence of DHT. In addition, we observed that enzalutamide treatment and SRSF3-silencing exerted an additive antiproliferative effect in LNCaP cells, inasmuch as the proliferation rate of enzalutamide-treated SRSF3-silenced LNCaP cells was significantly lower than that of enzalutamide-treated scramble-treated cells (in presence of DHT) and than that of SRSF3-silenced cells (in presence of DHT) (Fig. 6b).
Fig. 6.
Cell proliferation assay in response to enzalutamide treatment combined with SNRNP200, SRRM1 and SRSF3 silencing. Proliferation rate of 22Rv1 (a) and LNCaP (b) cell line was measured after 24 h of SNRNP200-, SRRM1- and SRSF3-silencing in the presence (DHT) or absence (no DHT) of 5α-dihydrotestosterone with or without enzalutamide (ENZA; n = 4). Results were expressed as percentage referred to scramble vehicle-treated control with DHT (mean ± SEM). Asterisks (* p < 0.05; ** p < 0.01), dash (#p < 0.05) and dollar sign ($p < 0.05) indicate statistically significant differences compared to DHT of scramble, DHT of each condition and DHT+ENZA of scramble, respectively.
4. Discussion
PCa is one of the tumor pathologies whose development and, specially, progression is mostly influenced by the alteration of the normal gene expression pattern and the aberrant presence of oncogenic SVs [24]. Indeed, PCa is drastically influenced by the appearance of the AR splicing variant-7 (AR-v7), inasmuch as it has been strongly associated with PCa aggressiveness [25], as well as with the resistance to conventional therapies such as antiandrogens and chemotherapy [26,27]. Similarly, the altered expression pattern of additional SVs, such as SST5TMD4 [4], PKM2[5], REST4[6], XBP1s[7], In1-Ghrelin [8] among others, has also been found to be associated to PCa development and progression. In this sense, it is reasonable to think that a dysregulation of the cellular machinery involved in the control of splicing process would be responsible for the broad alteration of oncogenic SVs observed in PCa. However, although some specific SFs and SCs have been associated with PCa development and aggressiveness [22,23,[28], [29], [30]], to the best of our knowledge, no studies have comprehensively explored the global dysregulations of these elements in PCa.
In our study, we have demonstrated for the first time a profound and overt dysregulation of the expression levels of a representative set of SCs and associated SFs in PCa. In particular, more than 50% of the SCs and SFs analysed herein displayed an altered expression pattern, demonstrating that the components of the cellular machinery responsible for the processing of the splicing process are drastically dysregulated in PCa. Indeed, we have bioinformatically defined an expression-based molecular fingerprint (combining the mRNA expression levels of 11 SCs and SFs) able to perfectly discriminate between PCa and non-tumor adjacent regions, which further reinforces this contention and demonstrate that PCa curses with a global dysregulation of the splicing machinery. Even more important is the fact that the expression levels of many of the SCs and SFs determined in this study were associated or correlated with relevant clinical and molecular features of aggressiveness (e.g. Gleason score, presence of metastasis and AR-v7 expression), suggesting a causal link between the dysregulations of the splicing machinery and the aggressiveness of PCa. These results are consistent with and further expand previous observations indicating that high expression of specifics SFs, including RBM3, U2AF2, ESRP1, ESRP2and NOVA1among others, is associated to clinical and/or molecular PCa aggressiveness features [23,28,30,31].
Among all the SCs and SFs analysed herein, SNRNP200, SRRM1 and SRSF3 seemed to have special relevance in PCa pathophysiology in that their expression levels were significantly up-regulated and associated with all the relevant clinical features analysed in this study, including Gleason score, pathological stage, perineural invasion, lymphovascular invasion, biochemical recurrence, presence of metastasis at diagnosis or AR-v7 expression. Importantly, data available in silico further validated the overexpression of SNRNP200, SRRM1and SRSF3in PCa as compared to non-tumoral prostate tissues [i.e. Singh, Wallace, Tomlins and/or Welsh-dataset [32], [33], [34], [35]]. In addition, these splicing machinery components could represent novel biomarkers and/or therapeutic targets in PCa inasmuch as their dysregulation has not been previously described in this tumor pathology and their expression levels showed a potential utility as prognostic markers, since these levels were associated with biochemical recurrence. It should to be noted that, although SRSF3has been defined as an oncogene in different tumor pathologies [36], [37], [38], this is the first description of the overexpression of this SF in PCa. Specifically, it has been shown that SRSF3 can enhance aggressiveness features of several tumor types through the control of splicing process in the nucleus [37,39,40], but also through the alteration of translational efficiency of certain mRNAs in the cytoplasm [41]. This suggests that overexpression of SRSF3mRNA can exert oncogenic actions either by increasing SRSF3 protein levels in the nucleus or in the cytoplasm, as found herein in the case of PCa cells. On the other hand, the protein overexpression of SRRM1 and SNRNP200 was observed in the nucleus of PCa cells, suggesting an enhancement of their putative activity as splicing modulators in these cells.
Due to the increasing body of evidence pointing toward a strong dysregulation of the splicing process in cancer, many therapeutic strategies to prevent the expression of oncogenic SVs and/or to modulate the activity of the spliceosome have been reported hitherto [42]. In particular, during the last years, many spliceosome inhibitors (e.g. Pladienolide-B, spliceostatin-A) have been reported and suggested as therapeutic targets in different pathologies wherein the dysregulated splicing process has been shown to be relevant [43,44]. However, while blocking the activity of the spliceosome could be less specific, targeting specific SCs and/or SFs could represent a novel and more specific approach to tackle cancer diseases in that a more reduced number but better-defined splicing events may be altered. In this sense, we have demonstrated for the first time herein that the silencing of the expression of SNRNP200, SRRM1and SRSF3using specific siRNAs clearly decreased key functional parameters of aggressiveness, including proliferation and migration, in PCa-derived cell lines. These clinically relevant antitumor actions seemed to be associated to the modulation of key signaling pathways and the expression of certain oncogenic SVs. Indeed, SRRM1-and SRSF3-silencing evoked a decrease in phosphorylation levels of AKT and JNK proteins, respectively, probably leading to the inhibition of PI3K/AKT and JNK pathways, which have been broadly defined as oncogenic signaling pathways [45,46]. Intriguingly, the silencing of SRSF3increased ERK-phosphorylation, presumably leading to a higher activation of MAPK/ERK pathway, which has been reported to be highly susceptible to be dysregulated in response to splicing changes [47]. Remarkably, despite its well-known oncogenic role, MAPK/ERK-pathway has been postulated as a regulator of cell senescence, thus also exerting antitumor effects [48]. As expected, and providing a mechanistic explanation, SNRNP200-, SRRM1- and SRSF3-silencing dysregulated the splicing process of several genes involved in tumor aggressiveness, such as AR, PKM, CASP2or XBP1. Among them, especially relevant are the results obtained in ARsplicing, due to its well-known role in the development and aggressiveness of PCa [49]. Specifically, a decrease in the expression of AR-v7 (but not AR) was observed in response to SNRNP200-and SRRM1-silencing, thus altering the normal splicing process of AR. Interestingly, the modulation of AR splicing process exerted by SNRNP200 and SRRM1 may be not mediated by the regulation of splicing factors previously reported to be involved in AR-v7 generation such as KHDRBS1 [21], SFPQ [22] or U2AF2 [23], inasmuch as we found that the silencing of SNRNP200 and SRRM1 did not alter the expression levels of these AR-splicing modulators. On the other hand, the silencing of SRSF3decreased the expression levels of both ARand AR-v7. In any case, these results postulate these three factors as potential therapeutic target candidates to tackle castration resistant PCa (CRPC), since AR-v7 has been reported as a driver of CRPC-development [50]. Reinforcing the oncogenic role of SNRNP200, SRRM1and SRSF3in PCa, we found that the silencing of these genes resulted in the modulation of the expression of key genes involved in CRPC aggressiveness, including C-MYC, PTEN and/or TP53 [51], [52], [53]. Consistently, SNRNP200-, SRRM1- and SRSF3-silencing sensitized 22Rv1 cells to enzalutamide by showing additive effects in the inhibition of proliferation rate in these cells, possibly through AR-v7 downregulation. This hypothesis was reinforced by the fact that SNRNP200- or SRRM1-silencing did not alter the normal response to enzalutamide in LNCaP cell line (which express AR but lack AR-v7), while SRSF3-silencing (which is associated to a reduction in full-length ARexpression) in combination with enzalutamide treatment resulted in an additive antiproliferative effect in these cells.
Therefore, the data presented herein indicate that the cellular machinery responsible for the regulation of the splicing process is drastically altered in PCa and that certain SCs and SVs could represent novel candidates as potential diagnostic and prognostic biomarkers, as well as putative targets to develop novel therapeutic strategies against PCa. Specifically, our results demonstrated that SNRNP200, SRRM1 and SRSF3 could represent attractive novel diagnostic/prognostic and therapeutic targets for PCa and CRPC.
5. Role of the funding sources
This work was funded by Instituto de Salud Carlos III, co-funded by European Union (ERDF/ESF, “Investing in your future”) [PI16/00264, PI17-02287, CM16/00180, FI17/00282, CD16/00092], FEDER (CCB.030PM), MINECO/MECD (BFU2016-80360-R; FPU16/06190, FPU18/02485, FPU16/05059, FPU17/00263, FPU14/04290), Junta de Andalucía (BIO-0139), Spanish Association Against Cancer (AECC; INVES18057YUBE) and CIBERobn. CIBER is an initiative of Instituto de Salud Carlos III, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain. The biological samples repository node (Córdoba, Spain) is gratefully acknowledged for coordination tasks in the selection of samples.
All the funding sources were essential for data collection, analysis, interpretation and patient recruitment. The corresponding author declare that he had full access to all the data in the study and had the final responsibility for the decision to submit for publication.
Author's contributions
Juan Manuel Jiménez-Vacas: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft, Writing - review & editing. Vicente Herrero-Aguayo, Antonio Jesús Montero-Hidalgo: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - review & editing. Enrique Gómez-Gómez: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Resources, Writing - review & editing. Antonio C. Fuentes-Fayos, Antonio José León-González, Prudencio Sáez-Martínez, Emilia Alors-Pérez, Sergio Pedraza-Arévalo, Teresa González-Serrano, Oscar Reyes, Ana Martínez-López, Rafael Sánchez-Sánchez and Elena M Yubero-Serrano: Data curation, Formal analysis, Investigation, Writing - review and editing. Sebastián Ventura: Data curation, Formal analysis, Investigation, Software, Writing - review and editing. María J. Requena-Tapia, Justo P. Castaño and Manuel D. Gahete: Formal analysis, Methodology, Resources, Writing - original draft, Writing - original draft, Writing - review & editing. Raúl M. Luque: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.008.
Appendix. Supplementary materials
References
- 1.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15(2):108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Hormaechea-Agulla D., Jimenez-Vacas J.M., Gomez-Gomez E., F L.L., Carrasco-Valiente J., Valero-Rosa J. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. Faseb J. 2017;31(11):4682–4696. doi: 10.1096/fj.201601264RRR. [DOI] [PubMed] [Google Scholar]
- 5.Wong N., Yan J., Ojo D., De Melo J., Cutz J.C., Tang D. Changes in PKM2 associate with prostate cancer progression. Cancer Invest. 2014;32(7):330–338. doi: 10.3109/07357907.2014.919306. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X., Coleman I.M., Brown L.G., True L.D., Kollath L., Lucas J.M. SRRM4 expression and the loss of rest activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(20):4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng X., Nenseth H.Z., Qu S., Kuzu O.F., Frahnow T., Simon L. IRE1α-XBP1s pathway promotes prostate cancer by activating c-MYC signaling. Nat Commun. 2019;10(1):323. doi: 10.1038/s41467-018-08152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hormaechea-Agulla D., Gahete M.D., Jimenez-Vacas J.M., Gomez-Gomez E., Ibanez-Costa A., F L.L. The oncogenic role of the In1-ghrelin splicing variant in prostate cancer aggressiveness. Mol Cancer. 2017;16(1):146. doi: 10.1186/s12943-017-0713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Li HR, Fan JB, Wang-Rodriguez J, Downs T, Fu XD. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformat. 2006;7:202. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedraza-Arevalo S., Hormaechea-Agulla D., Gomez-Gomez E., Requena M.J., Selth L.A., Gahete M.D. Somatostatin receptor subtype 1 as a potential diagnostic marker and therapeutic target in prostate cancer. Prostate. 2017;77(15):1499–1511. doi: 10.1002/pros.23426. [DOI] [PubMed] [Google Scholar]
- 11.Del Rio-Moreno M., Alors-Perez E., Gonzalez-Rubio S., Ferrin G., Reyes O., Rodriguez-Peralvarez M. Dysregulation of the splicing machinery is associated to the development of non-alcoholic fatty liver disease. J Clin Endocrinol Metab. 2019 doi: 10.1210/jc.2019-00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gahete MD, Del Rio-Moreno M, Camargo A, Alcala-Diaz JF, Alors-Perez E, Delgado-Lista J. Changes in splicing machinery components influence, precede, and early predict the development of type 2 diabetes: from the cordioprev study. EBioMedicine. 2018;37:356–365. doi: 10.1016/j.ebiom.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luque R.M., Ibanez-Costa A., Neto L.V., Taboada G.F., Hormaechea-Agulla D., Kasuki L. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett. 2015;359(2):299–306. doi: 10.1016/j.canlet.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibanez-Costa A., Gahete M.D., Rivero-Cortes E., Rincon-Fernandez D., Nelson R., Beltran M. In1-ghrelin splicing variant is overexpressed in pituitary adenomas and increases their aggressive features. Sci Rep. 2015;5:8714. doi: 10.1038/srep08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran-Prado M, Gahete MD, Hergueta-Redondo M, Martinez-Fuentes AJ, Cordoba-Chacon J, Palacios J. The new truncated somatostatin receptor variant sst5TMD4 is associated to poor prognosis in breast cancer and increases malignancy in MCF-7 cells. Oncogene. 2012;31(16):2049–2061. doi: 10.1038/onc.2011.389. [DOI] [PubMed] [Google Scholar]
- 17.Sampedro-Nunez M, Luque RM, Ramos-Levi AM, Gahete MD, Serrano-Somavilla A, Villa-Osaba A. Presence of sst5TMD4, a truncated splice variant of the somatostatin receptor subtype 5, is associated to features of increased aggressiveness in pancreatic neuroendocrine tumors. Oncotarget. 2016;7(6):6593–6608. doi: 10.18632/oncotarget.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatami R., Sieuwerts A.M., Izadmehr S., Yao Z., Qiao R.F., Papa L. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med. 2013;5(169) doi: 10.1126/scitranslmed.3004688. 169ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuranaga Y., Sugito N., Shinohara H., Tsujino T., Taniguchi K., Komura K. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Molecular Sci. 2018;19(10) doi: 10.3390/ijms19103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Donmez N., Sahinalp C., Xie N., Wang Y., Xue H. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2017;71(1):68–78. doi: 10.1016/j.eururo.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 21.Stockley J., Markert E., Zhou Y., Robson C.N., Elliott D.J., Lindberg J. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci Rep. 2015;5:13426. doi: 10.1038/srep13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takayama K.I., Suzuki T., Fujimura T., Yamada Y., Takahashi S., Homma Y. Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF. PNAS. 2017;114(39):10461–10466. doi: 10.1073/pnas.1706076114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33(24):3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paschalis A., Sharp A., Welti J.C., Neeb A., Raj G.V., Luo J. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15(11):663–675. doi: 10.1038/s41571-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 25.Kong D., Sethi S., Li Y., Chen W., Sakr W.A., Heath E. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. Prostate. 2015;75(2):161–174. doi: 10.1002/pros.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonarakis E.S., Lu C., Luber B., Wang H., Chen Y., Nakazawa M. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1(5):582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grupp K., Wilking J., Prien K., Hube-Magg C., Sirma H., Simon R. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50(4):852–861. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto R, Osawa T, Sasaki Y, Yamamoto S, Anai M, Izumi K. Overexpression of p54(nrb)/NONO induces differential EPHA6 splicing and contributes to castration-resistant prostate cancer growth. Oncotarget. 2018;9(12):10510–10524. doi: 10.18632/oncotarget.24063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell. 2018;34(6) doi: 10.1016/j.ccell.2018.10.016. 996-1011.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Z.X., Huang Q., Park J.W., Shen S., Lin L., Tokheim C.J. Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol Cancer Res. 2015;13(2):305–318. doi: 10.1158/1541-7786.MCR-14-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 33.Wallace T.A., Prueitt R.L., Yi M., Howe T.M., Gillespie J.W., Yfantis H.G. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68(3):927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.H., Dhanasekaran S.M., Mehra R., Tomlins S.A., Gu W., Yu J. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67(17):8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 35.Welsh J.B., Sapinoso L.M., Su A.I., Kern S.G., Wang-Rodriguez J., Moskaluk C.A. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61(16):5974–5978. [PubMed] [Google Scholar]
- 36.Peiqi L., Zhaozhong G., Yaotian Y., Jun J., Jihua G., Rong J. Expression of SRSF3 is correlated with carcinogenesis and progression of oral squamous cell carcinoma. Int J Med Sci. 2016;13(7):533–539. doi: 10.7150/ijms.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gautrey H., Jackson C., Dittrich A.L., Browell D., Lennard T., Tyson-Capper A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015;12(10):1139–1151. doi: 10.1080/15476286.2015.1076610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin JC, Lee YC, Tan TH, Liang YC, Chuang HC, Fann YC. RBM4-SRSF3-MAP4K4 splicing cascade modulates the metastatic signature of colorectal cancer cell. Biochimica et Biophysica Acta Molecular Cell Res. 2018;1865(2):259–272. doi: 10.1016/j.bbamcr.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Wang H., Zhang C.Z., Lu S.X., Zhang M.F., Liu L.L., Luo R.Z. A coiled-coil domain containing 50 splice variant is modulated by serine/arginine-rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the ras signaling pathway. Hepatology (Baltimore, Md) 2019;69(1):179–195. doi: 10.1002/hep.30147. [DOI] [PubMed] [Google Scholar]
- 40.Zhu S, Chen Z, Katsha A, Hong J, Belkhiri A, El-Rifai W. Regulation of CD44E by DARPP-32-dependent activation of SRp20 splicing factor in gastric tumorigenesis. Oncogene. 2016;35(14):1847–1856. doi: 10.1038/onc.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J., Park R.Y., Chen J.K., Kim J., Jeong S., Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell Death Differ. 2014;21(3):481–490. doi: 10.1038/cdd.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jyotsana N., Heuser M. Exploiting differential RNA splicing patterns: a potential new group of therapeutic targets in cancer. Expert Opin Ther Targets. 2018;22(2):107–121. doi: 10.1080/14728222.2018.1417390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato M., Muguruma N., Nakagawa T., Okamoto K., Kimura T., Kitamura S. High antitumor activity of pladienolide B and its derivative in gastric cancer. Cancer Sci. 2014;105(1):110–116. doi: 10.1111/cas.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larrayoz M, Blakemore SJ, Dobson RC, Blunt MD, Rose-Zerilli MJ, Walewska R. The SF3B1 inhibitor spliceostatin a (SSA) elicits apoptosis in chronic lymphocytic leukaemia cells through downregulation of Mcl-1. Leukemia. 2016;30(2):351–360. doi: 10.1038/leu.2015.286. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, Zhou L, Wu X, Li R, Wen J, Sha J. The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark Ed) 2016;21:1084–1091. doi: 10.2741/4443. [DOI] [PubMed] [Google Scholar]
- 46.Hubner A., Mulholland D.J., Standen C.L., Karasarides M., Cavanagh-Kyros J., Barrett T. JNK and pten cooperatively control the development of invasive adenocarcinoma of the prostate. PNAS. 2012;109(30):12046–12051. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegfried Z, Bonomi S, Ghigna C, Karni R. Regulation of the Ras-MAPK and PI3K-mTOR signalling pathways by alternative splicing in cancer. Int J Cell Biol. 2013;2013 doi: 10.1155/2013/568931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deschenes-Simard X, Gaumont-Leclerc MF, Bourdeau V, Lessard F, Moiseeva O, Forest V. Tumor suppressor activity of the erk/mapk pathway by promoting selective protein degradation. Genes Dev. 2013;27(8):900–915. doi: 10.1101/gad.203984.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan M.H., Li J., Xu H.E., Melcher K., Yong E.L. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36(1):3–23. doi: 10.1038/aps.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu Y., Dai B., Ye D., Kong Y., Chang K., Jia Z. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai S, Cao S, Jin L, Kobelski M, Schouest B, Wang X. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene. 2019;38(25):4977–4989. doi: 10.1038/s41388-019-0768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15(4):222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamid A.A., Gray K.P., Shaw G., MacConaill L.E., Evan C., Bernard B. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol. 2019;76(1):89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.