Abstract
Background
Phenylketonuria (PKU) is the most common inborn error of amino acid metabolism in Europe. The reasons underlying the high prevalence of heterozygous carriers are not clearly understood. We aimed to look for pathogenic PAH variant enrichment according to geographical areas and patients’ ethnicity using a multiethnic nationwide cohort of patients with PKU in France. We subsequently appraised the population differentiation, balancing selection and the molecular evolutionary history of the PAH locus.
Methods
The French nationwide PKU study included patients who have been referred at the national level to the University Hospital of Nancy, and for whom a molecular diagnosis of phenylketonuria was made by Sanger sequencing. We performed enrichment analyses by comparing alternative allele frequencies using Fisher's exact test with Bonferroni adjustment. We estimated the amount of genetic differentiation among populations using Wright's fixation index (Fst). To estimate the molecular evolutionary history of the PAH gene, we performed phylogenetic and evolutionary analyses using whole-genome and exome-sequencing data from healthy individuals and non-PKU patients, respectively. Finally, we used exome-wide association study to decipher potential genetic loci associated with population divergence on PAH.
Findings
The study included 696 patients and revealed 132 pathogenic PAH variants. Three geographical areas showed significant enrichment for a pathogenic PAH variant: North of France (p.Arg243Leu), North-West of France (p.Leu348Val), and Mediterranean coast (p.Ala403Val). One PAH variant (p.Glu280Gln) was significantly enriched among North-Africans (OR = 23·23; 95% CI: 9·75–55·38). PAH variants exhibiting a strong genetic differentiation were significantly enriched in the ‘Biopterin_H’ domain (OR = 6·45; 95% CI: 1·99–20·84), suggesting a balancing selection pressure on the biopterin function of PAH. Phylogenetic and timetree analyses were consistent with population differentiation events on European-, African-, and Asian-ancestry populations. The five PAH variants most strongly associated with a high selection pressure were phylogenetically close and were located within the biopterin domain coding region of PAH or in its vicinity. Among the non-PAH loci potentially associated with population divergence, two reached exome-wide significance: SSPO (SCO-spondin) and DBH (dopamine beta-hydroxylase), involved in neuroprotection and metabolic adaptation, respectively.
Interpretation
Our data provide evidence on the combination of evolutionary and adaptive events in populations with distinct ancestries, which may explain the overdominance of some genetic variants on PAH.
Funding
French National Institute of Health and Medical Research (INSERM) UMR_S 1256.
Keywords: Phenylketonuria, Balancing selection, Population divergence, Overdominance, Metabolic adaptation
Research in context.
Evidence before this study
Phenylketonuria (PKU) is the most common inborn error of amino acid metabolism in Europe, with an estimated prevalence of heterozygous carriers of PAH pathogenic variants around 2–3%. The reasons underlying the high prevalence of heterozygous carriers are not clearly understood. Numerous factors, including geographic isolation and adaptation, led to a divergence between human populations with regards to their genetic heritage, a concept known as population stratification. In the present study, we combined Sanger sequencing and exome-wide association study on a multiethnic nationwide cohort of patients with PKU in France to assess the effects of population divergence and balancing selection on the PAH locus. To better appraise the sequence of phylogenetic events on the PAH locus, we also performed phylogenetic and evolutionary analyses using whole-genome and exome-sequencing data from healthy individuals and non-PKU patients, respectively.
Added value of this study
Our study confirmed the high mutation burden on the PAH gene by reporting more than 130 pathogenic variants. Interestingly, within the PAH locus, the variants that were subject to a high population divergence were significantly enriched in the C-terminal catalytic ‘Biopterin_H’ domain by comparison to the N-terminal regulatory ‘ACT domain’, suggesting a potential role of balancing selection pressures in the shaping of the PAH locus. Phylogenetic and timetree analyses on the PAH locus were consistent with population differentiation events on European-, African-, and Asian-ancestry populations. The five PAH variants most strongly associated with a high selection pressure were phylogenetically close and were located within the biopterin domain coding region of PAH or in its vicinity. Among the loci potentially associated with population divergence, two reached exome-wide significance and were positioned on the SSPO (SCO-spondin) and DBH (dopamine beta-hydroxylase) genes. SSPO and DBH are involved in neuroprotection and metabolic adaptation, respectively. Interestingly, the balancing selection pressure on the PAH locus, especially on the biopterin domain, may reflect a mechanism of adaptation to environmental factors but also to other enzymatic reactions that may involve the common BH4 cofactor in the PAH and NOS pathways.
Implications of all the available evidence
Our findings suggest a contributing role of population divergence and balanced selection to uncover heterozygote advantage and overdominance mechanisms on the PAH gene. Furthermore, our data provide a new paradigm regarding the combination of evolutionary and adaptive events in various populations, which may explain the overdominance of some genetic variants on the PAH gene. Future studies are needed to decipher the adaptive mechanisms that underlie the influence of PAH, SSPO, and DBH genes, including those related to overdominance and heterozygote advantage, in response to the selection pressures exerted on the PAH gene.
Alt-text: Unlabelled box
1. Introduction
Phenylketonuria (PKU) is an inherited autosomal recessive disorder of phenylalanine metabolism caused by a deficiency in the enzyme phenylalanine hydroxylase (PAH). The PAH gene (HGNC: 8582) located in 12q23.2 codes for the PAH enzyme that converts phenylalanine into tyrosine in the presence of molecular oxygen and catalytic amounts of tetrahydrobiopterin (BH4) [1,2]. Clinical manifestations of PKU are related to the toxic accumulation of phenylalanine in the blood and brain and, if left untreated, could lead to irreversible severe physical, neurological, and cognitive abnormalities.
During human peopling of the world and the different processes of human migration over time, several subgroups have progressively differentiated and experienced specific stresses and environmental conditions [3], [4], [5]. Numerous factors, including geographic isolation and adaptation, led to a divergence between human populations with regards to their genetic heritage, a concept known as population stratification [3], [4], [5]. The study of population stratification helped to better evaluate the influence of genetic determinants on human phenotypes, including disease phenotypes [6]. Fine-scale population stratification has been described in Europe and France with rare genetic variants that tend to be more geographically localised than common variants [7,8]. The geographical borders that surround the French territory and the several waves of population migration (e.g., Celts, Barbarians, Roman empire) have impacted the genetic structure of the modern French population [7,8].
PKU is the most common inborn error of amino acid metabolism in Europe, with an estimated prevalence of heterozygous carriers of PAH pathogenic variants around 2–3% [9,10]. From an evolutionary perspective and population genetics point of view, Krawczak & Zschocke nicely demonstrated that the high prevalence burden of heterozygous carriers for pathogenic PAH variants could not have resulted from a high mutation rate in the PAH locus or a high initial frequency of PAH gene mutations [10]. In this context, the use of the PKU model as a study framework for population genetics seems relevant, mainly because of the large variability of mutations in the PAH gene, an aggregated alternative allele frequency (AAF) of pathogenic variants on the PAH gene ~ 1% in Caucasian and Asian Oriental populations, and the ethnic diversity of populations affected by the PKU.
Several studies have listed the PAH gene variants observed among patients with PKU [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. These studies assessed the genotype-phenotype correlation between the PAH gene variants and the PKU phenotype. None of these studies was explicitly devoted to the evaluation of population divergence on the PAH locus. In the present study, we combined Sanger sequencing and exome-wide association study on a multiethnic nationwide cohort of patients with PKU in France to assess the effects of population divergence and balancing selection on the PAH locus. We also performed phylogenetic and evolutionary analyses using whole-genome and exome-sequencing data on healthy individuals and non-PKU patients to better appraise the sequence of phylogenetic events on the PAH locus.
2. Patients and methods
2.1. Patients population
The French nationwide PKU study included patients who have been referred at the national level to the Division of Biochemistry and Molecular Biology of the University Hospital of Nancy, and for whom a molecular diagnosis of phenylketonuria was made by Sanger sequencing (Figure S1). Parental and patient consent was obtained after oral and written information. All eligible patients were included in the study, and no patient declined to participate.
2.2. Study aims
The aims of the study were: i) to describe the pathogenic PAH variants and to look for potential pathogenic variant enrichment according to main geographical areas in France; ii) to perform ancestry marker analysis and look for potential pathogenic PAH variant enrichment according to patients’ ethnicity; and iii) to assess the potential involvement of balancing selection on the PAH locus using population divergence analysis.
2.3. Sanger sequencing of the PAHlocus
Genomic DNA from EDTA-treated peripheral blood samples was isolated using the Nucleon™ BACC3 Kit (GE Healthcare, Aulnay-sous-Bois, France). The 13 exons of the PAH gene were sequenced by bi-directional sequencing using the BigDye® Terminator v.1.1 cycle sequencing (Applied Biosystems, Foster, Ca, USA) and capillary electrophoresis (Applied Biosystems 3130 Genetic Analyzer). The PCR primer sequences are shown in Supplemental Table S1.
2.4. Exome-wide genotyping
High-throughput genotyping of DNA samples from patients included in the study was performed at the INSERM Unit UMR_S 1256 (NGERE) (UMS2008 IBSLor) using the Illumina Infinium HumanExome BeadChip v1.2 (Illumina Inc., San Diego, CA, USA) according to the manufacturer's recommendations. The Illumina HumanExome BeadChip includes 247,870 markers focused on protein-altering variants selected from >12,000 exome and genome sequences representing multiple ethnicities and complex traits [21]. Additional array content includes variants associated with complex traits in previous GWAS, HLA tags, ancestry-informative markers (n = 4,388), markers for identity-by-descent estimation, and random synonymous SNPs [21]. Bead intensity data were processed and normalised for each sample Illumina's GenomeStudio Genotyping Module. The Genome Reference Consortium GRCh37 (hg19; Feb. 2009) map was used (Illumina manifest file Immuno_BeadChip_11419691_B.bpm), and normalised probe intensities were extracted for all samples that passed standard laboratory quality-control thresholds. Genotype calling was performed with the GenomeStudio GenTrain 2.0 algorithm (Illumina's GenomeStudio data analysis software). Sample quality control measures included: sample call rate (>90%), overall heterozygosity, and relatedness testing. Duplicated samples and those showing cryptic relatedness were assessed by calculating identity by descent (IBD). We estimated IBD sharing and carried out PCA on an LD-pruned set of autosomal variants obtained by removing large-scale high-LD regions [22,23], variants with a call rate <0·99, variants with a minor allele frequency (MAF) <5% or variants with Hardy-Weinberg equilibrium (HWE) P-value <10–4. By estimating the probability of sharing zero, one, or two alleles for any two individuals as P(Z = 0), P(Z = 1), and P(Z = 2), respectively, a proportion of IBD was calculated using the following formula: PI-HAT = PI = P(Z = 1)/2 + P(Z = 2). A PI-HAT threshold >0·2 was used to suggest cryptic relatedness. Genetic variants were removed from the primary analysis if they had a call rate <95%, a significant departure from Hardy-Weinberg equilibrium (exact HWE-P < 10−4), or a MAF <0·1%.
2.5. Next-generation sequencing (Nancy Reference Centre Population)
We used the next-generation sequencing (NGS) data that was generated from the molecular diagnostics activity conducted on non-PKU patients in the Department of Molecular Medicine at the University Hospital of Nancy, Reference Centre for Inborn Errors of Metabolism (Nancy, France). NGS analysis was carried out on an Illumina MiSeq sequencer using the TruSight One sequencing panel, targeting 4813 genes, including the PAH gene (Illumina, San Diego, USA). Among the genetic variants with validated quality metrics (read depth >20, alt-read ratio >0·2, and genotype quality of 99), only those on the PAH gene were considered for the analysis in the present study. Variants were aligned to GRCh37. All the variants retrieved on the PAH locus were visually inspected by checking the BAM files alignment.
2.6. Extraction of whole-genome sequencing data of ancestral populations (1 kG population)
We extracted whole-genome sequencing data from the 1 kG project phase 3 which provides a database of genomic variants of 2504 individuals across five ancestral populations: Africans (AFR, n = 661), Europeans (EUR, n = 503), Admixed Americans (AMR, n = 347), East Asians (EAS, n = 504), and South Asians (SAS, n = 489) [24]. Genomic and phenotypic data were downloaded at http://www.internationalgenome.org/data/. Variants were aligned to GRCh37. The total number of genetic variants available in chromosome 12 was 3,985,172, including 2387 markers in the PAH gene.
2.7. Principal component analysis and ancestry inference
Principal-component analysis (PCA) was performed on the samples merged with 1092 subjects from the 1 kG phase 1 super populations as reference populations (Africans (AFR), n = 246; Americans (AMR), n = 181; Asians (ASN), n = 286; and Europeans (EUR), n = 379) (data were retrieved at: http://www.internationalgenome.org/data/). PCA analysis was performed under the additive model on 116,880 markers to find up to 10 components (Eigenvalues). PCA analysis results were reported using two-dimensional plots based on the two top Eigenvalues. PKU patients were plotted against all the 1092 subjects of the 1 kG phase 1 project. We used the same approach for PCA analysis on NGS data from patients included in the Department of Molecular Medicine at the University Hospital of Nancy.
2.8. Statistical analysis
We performed the enrichment analysis by comparing AAFs using Fisher's exact test with Bonferroni adjustment. Enrichment odds ratios and their 95% CI were reported. We estimated the amount of genetic differentiation among populations using the Wright's fixation index Fst (co-ancestry Coefficient θ) [25], [26], [27]. We calculated the 95% CI around the Fst using a percentile-t bootstrapping technique [28]. We estimated the fixation index Fst between pairs of subpopulations and the overall Fst index across all the studied populations. We used the following thresholds for Fst interpretation according to Hartl and Clark [26]: Fst <0·05: little genetic differentiation; Fst [0·05–0·15]: moderate genetic differentiation; Fst [0·15–0·25]: great genetic differentiation; Fst >0·25: very great genetic differentiation. To better visualise Fst indexes between pairs of subpopulations, we used dendrogram with heat map representation according to the following criteria: distance metric for top dendrogram: Euclidean; linkage algorithm for top dendrogram: weighted; distance metric for side dendrogram: Euclidean; and linkage algorithm for side dendrogram: weighted. To assess the haplotype structure of the PAH locus, we performed haplotype block analysis using the following criteria: upper confidence bound: 0·98; lower confidence bound: 0·7; reject criteria threshold: 0·9; MAF threshold: 0·05; Maximum number of markers in a block: 30; maximum length of a block: 160; display threshold: 0·01, maximum EM iterations: 50; and EM convergence tolerance: 0·0001. Linkage disequilibrium (LD)-pairwise analysis was performed on all adjacent pairs of genetic variants in a chromosome or within a haplotype block using a matrix output for both the expectation-maximization algorithm (EM) and the composite-haplotype method (CHM) [29,30]. R2 and D’ values were used in the Linkage disequilibrium plots. We performed the exome wide-association study through haplotype trend regression analysis to assess potential genetic loci associated with population differentiation and balancing selection pressure. For the haplotype association test, we used a moving window with a dynamic width of 10 kb and calculated the association per haplotype on 37,933 haplotype blocks. We estimated haplotype frequencies using the expectation/maximization (EM) algorithm with maximum EM iterations of 50 and an EM convergence tolerance of 0·0001 [31]. We compared haplotype frequencies using the Chi-squared test with Bonferroni and false discovery rate (FDR) adjustment. In a second step, we performed per-variant analysis on top significant loci obtained from exome-wide haplotype regression analysis by using linear regression with Bonferroni and FDR adjustment for assessing the same outcome. All quality controls and statistical analyses were performed using the SNP & Variation Suite (v8.8.3; Golden Helix, Inc., Bozeman, MT, USA).
2.9. Phylogenetic analyses of the PAHlocus
We used two datasets for phylogenetic analyses: i) the 1 kG dataset that included all the PAH variants that exhibit the highest population divergence (Fst > 0·50 for AFR- vs EAS-ancestry differentiation) (n = 64) and ii) the ‘Nancy Reference Centre Population’ dataset that included all the PAH variants that were reported through NGS analysis (n = 35). We extracted all the potential DNA sequences on the PAH locus taking into account the PAH variants in their reference and alternative forms to produce DNA FASTA files using the SeqTailor tool [32]. Phylogenetic trees were developed using the maximum likelihood-based method on aligned sequences. The evolutionary history was inferred by using the maximum likelihood method and the Tamura-Nei model [33]. Initial trees for the heuristic search were obtained automatically by applying Neighbour-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. Bootstraps with 500 replicates were applied. The phylogenetic tree was drawn to scale, with branch lengths measured in the number of substitutions per site. We conducted a timetree analysis using the RelTime method [34]. Divergence times for all branching points in the topology were calculated using the maximum likelihood method and Tamura-Nei model [33]. The timetree was drawn to scale, with branch lengths measured in the relative number of substitutions per site. The final timetree, showing relative node times, was normalised such that the maximum relative node time is 1. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version X [35].
3. Results
3.1. Description of the pathogenic PAHvariants found in the French Nationwide PKU Study
Between January 2004 and December 2012, 696 patients had a molecular diagnosis of PKU at the Division of Biochemistry and Molecular Biology of the University Hospital of Nancy using Sanger sequencing. Patients were recruited from seven geographical areas: North, North-East, North-West, Paris area, South-East, South-West, and Mediterranean coast (Figure S2). The descriptive statistics regarding the geographical origin and parent's ethnicity are available in Table 1. A total of 132 pathogenic PAH variants were found (Supplemental Table S2). The top three pathogenic PAH variants were: rs5030855 (7·97%; intronic variant, c.1066–11G>A), rs5030858 (6·11%; Missense, c.1222C>T, p.Arg408Trp), and rs5030861 (4·60%; splice donor variant, c.1315+1G>A). All the details related to the genomic description and the AAFs of the 132 PAH variants are reported in Supplemental Tables S2 and S3, respectively.
Table 1.
Characteristics of the 696 patients included in the French Nationwide PKU Study.
| Gender | Number of patients | Percentage |
| Female | 364 | 52·3 |
| Male | 332 | 47·7 |
| Geographical area / Main recruitment centre | Number of patients | Percentage |
| Mediterranean coast | 158 | 22·7 |
| North-West of France | 151 | 21·7 |
| North-East of France | 115 | 16·5 |
| North of France | 96 | 13·8 |
| South-East of France | 87 | 12·5 |
| Paris area | 55 | 7·9 |
| South-West of France | 34 | 4·9 |
| Ethnicity of the Mother* | Number of patients | Percentage |
| European Caucasian | 395 | 71·3 |
| North African | 82 | 14·8 |
| South European | 37 | 6·7 |
| Turkish | 17 | 3·1 |
| East European | 10 | 1·8 |
| Other | 6 | 1·1 |
| African sub-Saharan | 3 | 0·5 |
| Admixed: Caucasian/South European | 3 | 0·5 |
| Admixed: Caucasian/ North Africans | 1 | 0·2 |
| Ethnicity of the Father† | Number of patients | Percentage |
| European Caucasian | 405 | 73·6 |
| North African | 78 | 14·2 |
| South European | 25 | 4·5 |
| Turkish | 17 | 3·1 |
| East European | 14 | 2·5 |
| Other | 8 | 1·5 |
| Admixed: Caucasian/South European | 3 | 0·5 |
data available in 554 cases.
data available in 550 cases.
3.2. Enrichment analysis according to the geographical area
We performed an enrichment analysis to assess whether some pathogenic PAH variants were significantly enriched in a prespecified geographical area. Three geographical areas showed significant enrichment for a pathogenic PAH variant: rs62508588 (c.728G>T, p.Arg243Leu) in the North of France (OR = 22·41; 95% CI: 4·31–116·59; P = 1·96 × 10–4), rs62516092 (c.1042C>G, p.Leu348Val) in the North-West of France (OR = 4·29; 95% CI: 2·08–8·85; P = 8·16 × 10–5), and rs5030857 (c.1208C>T, p.Ala403Val) in the Mediterranean coast (OR = 4·88; 95% CI: 2·73–8·73; P = 1·14 × 10–7) (Supplemental Table S4, Fig. 1 and Supplemental Figure S2). The AAFs of all the PAH variants are reported in Supplemental Table S3. Enrichment analyses for all geographical areas are reported in Supplemental Tables S5-S11.
Fig. 1.
(A) Genomic position and (B) alternative allele frequencies of the three pathogenic PAH gene variants enriched in the North (rs62508588), North-West (rs62516092) and Mediterranean coastal (rs5030857) areas of France and the variant enriched among patients with North-African ancestry (rs62508698).
3.3. Enrichment analysis according to ethnicity
Among the 696 patients included in the study, 576 were studied using the Illumina Infinium HumanExome BeadChip v1.2. We performed PCA analysis on the 576 samples merged with 1092 subjects from the 1 kG phase 1 super populations (AFR, AMR, EAS, EUR, and SAS) as reference populations (Fig. 2). The PCA analysis demonstrated that the study population was represented by two main categories: a subpopulation with a European-background and a subpopulation with a genomic background that lies between European-ancestry subjects and those with African-ancestry subjects (Fig. 2). In sensitivity analyses, the geographical areas of the recruiting centre were not associated with a specific clustering pattern on the PCA plot (Figure S3). When the ethnicity of PKU patients was considered as a classification variable, the PCA plot exhibited a typical clustering pattern with Caucasian-ancestry PKU patients that clustered around the EUR-ancestry HapMap populations and the North-African PKU patients that clustered between the EUR- and the AFR-ancestry HapMap populations (Fig. 3A).
Fig. 2.
(A) Principal component analysis reporting patients included in the French nationwide PKU study. (B) Zoomed view of the Fig. part defined by the inset in panel A. According to PCA-defined ancestry analysis, European-, North-African, and Turkish-ancestry patients represent the three main patients’ ethnicities in the study.
Fig. 3.
(A) Principal component analysis according to patients’ ethnicity. (B) The PAH gene variant rs62508698 was significantly enriched among North-African ancestry patients and significantly under-represented among patients with European-ancestry.
We performed an enrichment analysis to assess whether some variants were significantly enriched among Caucasian- and North-African ancestry PKU patients. Among the 132 PAH variants, only one (rs62508698 [c.838G>C, p.Glu280Gln]) was significantly enriched among North-African PKU patients (OR = 23·23; 95% CI: 9·75–55·38; P = 8·47 × 10–13). The same variant was significantly under-represented among Caucasian-ancestry PKU patients (OR = 0·11; 95% CI: 0·05–0·26; P = 3·55 × 10–8) (Table 2). Enrichment analyses, according to the patient's ethnicity, are reported in Supplemental Tables S12-S14.
Table 2.
Enrichment analysis for pathogenic PAH gene variants according to according to patients’ ethnicity.
| Marker | Fisher's P-value | Fisher's Bonf. P-value | OR (95% CI), Alternative allele | AAF, PKU Nationwide | AAF, Studied ethnicity | AAF, Other ethnicities | gnomAD Exomes AAF |
|---|---|---|---|---|---|---|---|
| Caucasian-ancestry* | |||||||
| rs62508698 | 3·55 × 10–8 | 4·19 × 10–6 | 0·11 (0·05–0·26) | 3·08 × 10–2 | 1·04 × 10–2 | 8·58 × 10–2 | 5·57 × 10–5 |
| North-African-ancestry* | |||||||
| rs62508698 | 7·18 × 10–15 | 8·47 × 10–13 | 23·23 (9·75–55·38) | 3·08 × 10–2 | 1·69 × 10–1 | 8·70 × 10–3 | 5·57 × 10–5 |
AAF: alternative allele frequency; Bonf: Bonferroni; OR: odds ratio.
No significant variant enrichment was found in patients’ subgroups of other ethnicities.
3.4. Assessment of the population divergence on the PAHlocus
Given the highly discriminant pattern of the PAH variant rs62508698 between Caucasian-ancestry PKU and North-African PKU patients, we performed fixation index analysis by calculating the Fst index —which quantifies genetic differentiation between populations— on the 132 pathogenic PAH variants found in the study. The fixation index analysis confirmed that the rs62508698 variant exhibited the top Fst value (overall Fst = 0·249) across all ethnicities, which suggests a strong genetic differentiation [26] (Fig. 3B). The Fst value was even higher when the Caucasian- and North-African ancestries were considered (Fst = 0·280) (Supplemental Table S15). Considering the 132 variants in the PAH locus, the highest Fst values were observed in the ‘Biopterin_H’ domain (Figure S4A).
We used WGS data from the 2504 subjects included in the 1 kG project phase 3 and confirmed that the density of genetic variants with an Fst value ≥0·25 (strong genetic differentiation [26]) was significantly higher in the ‘Biopterin_H’ domain (1 variant per 0·936 kb) when compared to that observed in the ‘ACT’ domain (1 variant per 6.033 kb) with an OR of 6·45 (95% CI: 1·99–20·84; P = 1·27 × 10–4). The relatively high density of variants exhibiting an Fst value ≥0·25 in the ‘Biopterin_H’ domain is suggestive of a balancing selection pressure while the relatively low Fst values on the ACT domain was rather suggestive of a neutral selection across populations (Figure S4B).
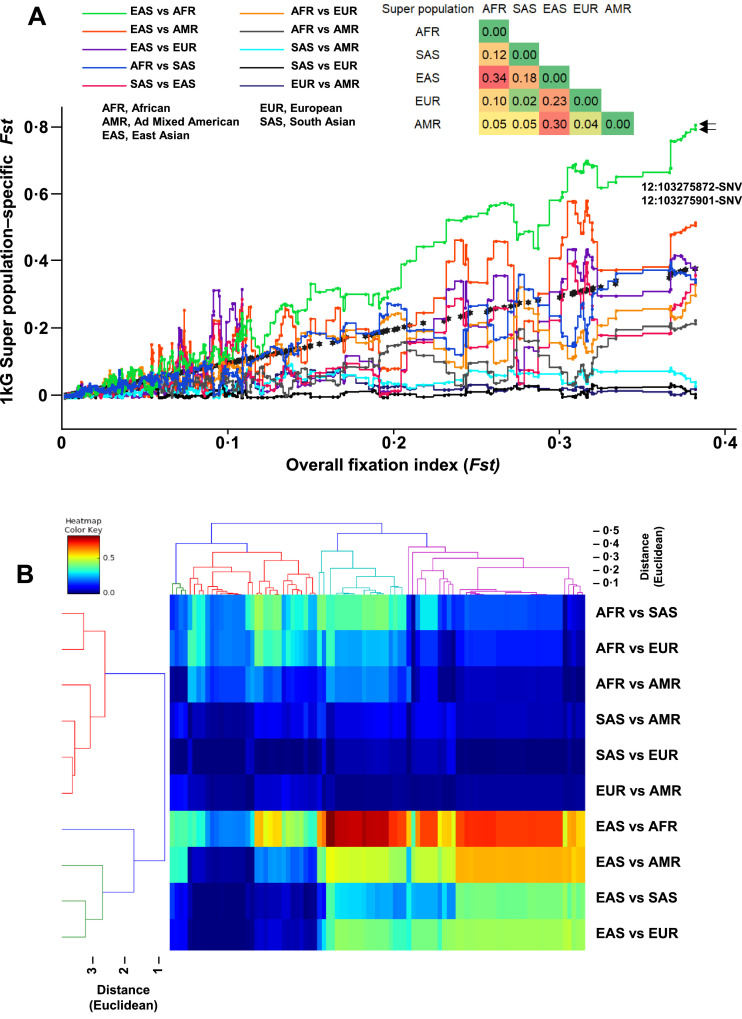
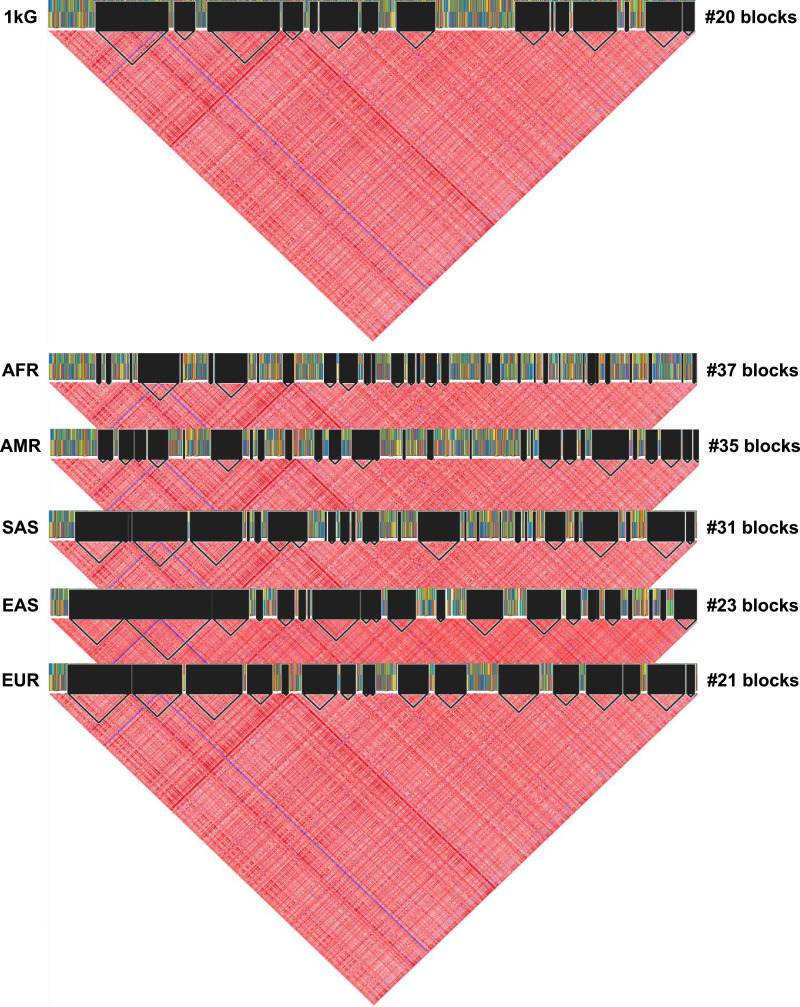
We used WGS data from the 1KG project to calculate the overall Fst values using the 2387 genetic variants on the PAH locus by comparing the five 1 kG super populations: AFR, AMR, EAS, EUR, and SAS. The PAH locus exhibited a population differentiation pattern, which was consistent with a significant balancing selection pressure with an overall Fst value of 0·16, indicating a great genetic differentiation (Supplemental Table S16). The top Fst values were observed between the AFR–EAS (0·34, 95% CI: 0·31–0·38), EAS–AMR (0·30, 95% CI: 0·27–0·33), and EUR–EAS (0·23, 95% CI: 0·20–0·25) indicating great to very great genetic differentiation (Fig. 4). We assessed the haplotype structure of the PAH locus in the whole 1 kG population and each super population using the same dataset of WGS. The haplotypic structure of the PAH locus significantly differed according to the 1 kG super population with a more complex structure in the AFR population (37 haplotype blocks) when compared to the less complex haplotype structure in the European population (21 haplotype blocks) (Fig. 5).
Fig. 4.
(A) Two-dimension scatter plot reporting the pairwise population differentiation analysis using the Fst index against the overall Fst index (black stars) for PAH gene variants using whole-genome sequencing data from the 2504 subjects of the 1000 Genomes Project phase 3. The matrix at the top of the panel reports the pairwise population Fixation index. (B) Dendrogram with heat map reporting the pairwise population differentiation analysis for PAH gene variants using whole-genome sequencing data from the 1000 genomes project phase 3.
Fig. 5.
Haplotype block analysis of the PAH gene using whole-genome sequencing data from the 2504 subjects of the 1000 Genomes Project phase 3. The number of haplotype blocks is reported for each super population.
3.5. Phylogenetic analyses on the PAHlocus
The phylogenetic analysis of the 1 kG dataset highlighted five clades (Fig. 6). Among them, one exhibited clustering of five intronic PAH variants that were highly differentiated between African- and East Asian-ancestry populations with an Fst > 0·75. Three variants located in the ‘Biopterin_H’ domain coding region (12:103265899-G-A, 12:103238949-T-C, and 12:103260273-T-G) are underrepresented among Africans (AAF ≈ 0·05, gnomAD Genomes database) and over-enriched among East Asians (AAF ≈ 0·75) (Fig. 7 and Supplemental Table S17). The two remaining variants are located outside the ‘Biopterin_H’ domain coding region (12:103275872-T-C, 12:103275901-C-T) and are in high linkage disequilibrium (D’ = 1) (Fig. 7). To better estimate the relative ordering of population differentiation events, we conducted a timetree analysis using the two top differentiated PAH variants between African- and East Asian-ancestry populations as outgroup sequences (12:103275872-T-C, 12:103275901-C-T). The timetree analysis confirmed the anteriority of divergence events on the phylogeny of the two top differentiated PAH variants with a branch length of 0·64 (relative time) (Supplemental Figure 5).
Fig. 6.
Phylogenetic analysis of the PAH locus using 1 kG whole-genome sequencing data.
Fig. 7.
World map representation of the alternative allele frequencies of the top differentiated (African- vs East Asian-ancestry populations) and phylogenetically clustered PAH variants.
The ‘Nancy Reference Centre Population’ dataset included a total of 409 patients (European-ancestry, n = 319; North-African-ancestry, n = 49; and Asian-ancestry, n = 41). Two markers were associated with population differentiation between European- and North-African-ancestry populations and clustered under the same clade in phylogenetic analysis (Fig. 8A). The first variant corresponds to a splice acceptor variant (12:103234294-C-A, c.1200–1G>T), and the second is a synonymous variant (12:103234215-A-G, p.Asn426=). The splice acceptor variant (12:103234294-C-A) has an AAF of 1·59 × 10–5 (2/125,568) in the TOPMed (Trans-Omics for Precision Medicine) database [36] and is not reported in the gnomAD Genomes database [37]. The AAF of this variant was 7·18 × 10–4 (1/1392; enrichment ratio ≈ 45) among all PKU patients in the French Nationwide PKU Study, 1·27 × 10–3 (1/395; enrichment ratio ≈ 159) among Caucasian-ancestry PKU patients in the French Nationwide PKU Study, 2·44 × 10–3 (2/818; enrichment ratio ≈ 154) in the ‘Nancy Reference Centre Population’, and 2·04 × 10–2 (2/98; enrichment ratio ≈ 1281) among North-African-ancestry subjects of the ‘Nancy Reference Centre Population’.
Fig. 8.
(A) Two-dimension scatter plot reporting the pairwise population differentiation analysis using the Fst index against the overall Fst index for PAH gene variants evidenced in the ‘Nancy Reference center Population’. (B) Phylogenetic and timetree analyses of the PAH locus using the ‘Nancy Reference Centre Population’ exome-sequencing data on non-PKU patients.
3.6. Exome-wide association study: unveiling other potential loci associated with population differentiation
For the exome-wide high-throughput genotyping, among the 696 patients included in the study, DNA samples were available for 574 (83%). We performed an exome-wide association study to assess potential genetic loci associated with the population differentiation and balancing selection pressure between Caucasian-ancestry PKU and North-African PKU patients. We performed linear haplotype trend regression using the rs62508698 as the response variable and a moving window with a dynamic width of 10 kb. We adjusted the regression model for the three top principal components (EV = 15·8214, EV = 7·33095, EV = 2·12178) to account for potential confounding bias due to population stratification. Two loci reached statistical significance and were located in the SSPO (SCO-spondin, HGNC:21998) and DBH (dopamine beta-hydroxylase, HGNC:2689) genes (Fig. 9 and Supplemental Table S18). The genotype association study on the 37 variants located in the SSPO and DBH loci confirmed a significant association with four coding variants on the SSPO gene (rs73727633, p.Phe3209Leu; rs73727632, p.Cys3204Arg; rs56921891, p.Pro2393=; rs73727650, p.Arg5042Gly) and one coding variant on the DBH gene (rs4531, p.Ala318Thr) (Supplemental Table S19).
Fig. 9.
(A) Manhattan plot reporting the exome-wide association study for the association with population differentiation and balancing selection pressure (PAH gene variant rs62508698) in haplotype trend regression analysis. The two arrows indicate the top significant loci: SSPO (SCO-spondin, HGNC:21998) and DBH (dopamine beta-hydroxylase, HGNC:2689) genes. Panels (B) and (C) report the genomic context of the SSOP and SBH genes, respectively. The P-values were reported after a symmetric smoothing transformation method using a window radius value of 2, as previously described [64].
4. Discussion
In 2007, Charles Scriver stated that: “The PKU story contains many messages, including what the human PAH gene tells us about human population genetics and evolution of modern humans” [1]. Our study suggests a contributing role of population divergence and balanced selection to uncover heterozygote advantage and overdominance mechanisms on the PAH gene, in a multiethnic nationwide sample population of patients with PKU in France.
Our study confirmed the high mutation burden on the PAH gene by reporting more than 130 pathogenic variants. These mutations are not distributed randomly across the studied geographical areas in France. Interestingly, within the PAH locus, the variants that were subject to a high population divergence were significantly enriched in the C-terminal catalytic Biopterin_H domain by comparison to the N-terminal regulatory ACT domain, suggesting a potential role of balancing selection pressures in the shaping of the PAH locus. One variant (rs62508698) was associated with a substantial population divergence with a significant enrichment among North-Africans and an under-representation among Europeans. The exome-wide association study that addressed potential loci related to population divergence emphasised two top significant loci, namely SSOP and DBH. These data could lead to a better understanding of the adaptive mechanisms in response to the selection pressures exerted on the PAH gene and pave the way for new hypotheses regarding the adaptive advantages conferred by PAH heterozygous mutations in the face of specific environmental factors, such as infectious diseases [38,39].
The reasons underlying the high prevalence of PAH heterozygous carriers are not clearly understood. One of the hypotheses is that the increased prevalence in heterozygotes is linked to an overdominant selection process [10,[40], [41], [42], [43], [44], [45]]. Overdominance could be described as a heterozygous advantage, wherein heterozygous individuals have higher fitness than either homozygous individuals [46]. Heterozygous advantage represents a balanced selection mechanism by which multiple alleles at a specific locus are actively maintained in the gene pool of a population longer than expected from genetic drift alone [39,47]. Woolf et al. reported that women who were heterozygous for PAH mutations were at a lower risk of miscarriage when compared to wild-type, homozygous women [40]. It is well known that host-pathogen interactions have shaped genetic variation in modern populations [39]. The development of high-throughput approaches, as well as analytical methods and expanding public data resources, has led to new insights on how human infectious disease influenced the natural selection and several adaptive mechanisms, including autoimmune and metabolic disorders [39]. Several examples of adaptive metabolic mechanisms providing benefit to defence against pathogens have been reported. An example regarding the overdominant selection in PKU could be the consequence of higher phenylalanine levels in heterozygous than in wild-type homozygous individuals on host-pathogen interactions [48]. The potential mechanism was that phenylalanine acts as a competitive inhibitor of ochratoxin A, which is a teratogenic mycotoxin produced by Aspergillus spp. in mouldy grains and lentils [10,38]. Ochratoxin A is an N-acyl derivative of phenylalanine that can cross the placenta and induce spontaneous miscarriage through the inhibition of phenylalanyl-tRNA synthetase-catalysed reaction and protein synthesis [38]. Other examples of inherited disorders suggesting that overdominance provides resistance to a specific infectious disease include sickle trait (HBB) and resistance to malaria, cystic fibrosis (CFTR) and resistance to cholera, Tay-Sachs disease (HEXA) and resistance to tuberculosis, congenital disorder of glycosylation type IIb (MOGS) and resistance to viral infections, and Niemann-Pick disease type C1 (NPC1) and resistance to filoviruses (e.g., Ebola, Marburg) [38].
Phylogenetic and timetree analyses on the PAH locus were consistent with population differentiation events on European-, African-, and Asian-ancestry populations. Few studies have assessed the potential relationship between population divergence, overdominance, and molecular phylogenetics in the setting of inherited metabolic disorders. Using three different populations, we have highlighted a selection pressure on the PAH gene with a substantial population divergence in association with specific variants. The assessment of the allelic frequencies of the most differentiated variants between Africans and East-Asians also revealed an enrichment gradient between Africans and Europeans but to a lesser extent. Phylogenetic analyses showed that the variants most strongly associated with a high selection pressure were close phylogenetically and were located within the ‘Biopterin_H’ domain coding region or in its vicinity. The ‘Biopterin_H’ domain of PAH contains a binding domain for the PAH cofactor BH4, which regulates the enzyme activity as well as being essential in catalysis [49]. Several experiments of coupled transcription-translation in vitro assay have shown that BH4 has a chaperone-like effect on PAH synthesis and/or is a protecting cofactor against enzyme auto-inactivation and degradation [49]. BH4 is also an essential oxidant-sensitive cofactor for the nitric oxide synthase that synthesises nitric oxide (NO) from arginine and oxygen [50]. Patients with severe malaria have BH4 depletion, which in turn compromises both NO synthesis and phenylalanine metabolism [50,51]. The decreased BH4 availability in severe malaria uncouples NO synthase and leads to impaired NO bioavailability and potentially increased oxidative stress [51]. These data may suggest the potential role of malaria as an environmental factor that may drive selection pressure. However, malaria could not explain the genetic differentiation between Latinos and East Asian populations since the distribution of the infectious agent is similar in both geographical regions [52]. On the other hand, several studies have shown a clear genetic differentiation between East-Asian and Latino populations in terms of hypoxic adaptation to altitude [53], [54], [55], [56], [57]. Indeed, balancing selection on NOS1 and NOS2 has been shown to participate in the adaptive mechanisms associated with higher oxygen delivery in Andean and Tibetan populations [56,57]. The balancing selection pressure on the PAH locus, especially on the biopterin domain, may reflect a mechanism of adaptation to environmental factors but also to other enzymatic reactions that may involve the common BH4 cofactor in the PAH and NOS pathways. Thus, a combination of evolutionary and adaptive events in various populations may potentially explain the overdominance of some genetic variants on the PAH gene.
In humans, few phylogenetic studies have been reported in the setting of inherited metabolic disorders. One example is an in silico study that identified significant evolutionary events on the NPC1 gene, which is associated with the Niemann-Pick disease type C disease [58]. This study did not evaluate the involvement of NPC1 genetic variants in population differentiation or their potential role in overdominance. The precise model that links the evolutionary aspects of a gene from a phylogenetic point of view with population differentiation and overdominance remains to be clarified. Future studies on genes in which polymorphisms are known to be maintained by selection will help better understanding the role of balancing selection in the setting of population differentiation and phylogenetic evolution [59].
Our data also suggest that other loci, potentially associated with population differentiation and balancing selection, may contribute to the adaptive mechanisms of overdominance and heterozygote advantage. The exome-wide association study that addressed potential loci in association with population divergence on the PAH locus unravelled two top significant loci on SSPO and DBH genes. The SSPO gene codes for the subcommissural organ spondin (SCO-spondin) protein which is involved in the modulation of neuronal aggregation and the formation of the central nervous system. In vitro, SCO-spondin has been shown to stimulate neuronal differentiation and neurite growth [60]. The thrombospondin type-1 (TSP1) repeat (TSR) domain supports the main neurogenic properties of the SCO-spondin protein [60]. In vitro and in vivo studies have shown that the NX210 oligopeptide —which was designed from SCO-spondin's specific TSR domain— was able to prevent neural cell death and to promote neurite outgrowth [60]. NX210 has been granted Orphan Drug designation by the European Medicine Agency for the treatment of spinal cord injury with an ongoing phase 1 study planned in patients with spinal cord injury. The association of a PAH gene variant serving as a marker of high balancing selection pressure with a gene locus involved in neuroprotection provides new insight into the potential adaptive mechanisms to PAH variants overdominance among populations. This hypothesis deserves further studies. The second locus associated with population divergence on PAH was on the DBH gene. The DBH gene codes for the dopamine beta-hydroxylase, which catalyses the conversion of dopamine to norepinephrine. The dopamine molecule is a down product of phenylalanine metabolism. It has been shown that the synthesis of dopamine is impaired in the human brain of patients with PKU [61]. The most relevant biochemical abnormalities among PKU patients are a decrease in the activity of tyrosine and tryptophan hydroxylases, which lead to deficiencies in dopamine and serotonin, which result in neurological abnormalities such as abnormal muscle tone, irritability and lethargy, seizures, and progressive developmental delay [62]. The treatment of PKU patients with a restricted phenylalanine diet induces a reduction of phenylalanine levels and an increase of dopamine and serotonin levels [61]. Consistently, the mice model of PKU showed that phenylalanine diet supplemented with tyrosine induce a normalisation of dopamine levels in the brain [62,63]. The association between a selection pressure on the PAH locus and a genomic signature on DBH allows considering the hypothesis of genome plasticity with regards to environmental factors, but also adaptive metabolic factors to hyperphenylalaninemia in the setting of PAH overdominance. The influence of genetic variants of DBH on the phenotypic variability and natural history of PKU patients deserves to be studied in large cohorts.
5. Conclusion
In conclusion, our study suggests a potential role of population divergence and balanced selection in the overdominance mechanisms of the PAH gene. Phylogenetic and timetree analyses confirm population differentiation as a potential driver of the PAH gene evolution among European-, African-, and Asian-ancestry populations, notably on the biopterin domain at the crossroad between PAH and NOS pathways. Future studies are needed to decipher the adaptive mechanisms that underlie the influence of PAH, SSPO, and DBH genes, including those related to overdominance and heterozygote advantage, in response to the selection pressures exerted on the PAH gene.
Declaration of Competing Interest
The authors who have taken part in this study declare that they do not have anything to disclose regarding conflicts of interest concerning this manuscript.
Acknowledgments
Acknowledgements
Dr. Abderrahim Oussalah and Prof. Jean-Louis Guéant had full access to all the data in the study. They took responsibility for the integrity of the data and the accuracy of the data analysis. The authors thank the patients and institutions involved in this study. Genomic analyses were performed on the Genomic Platform affiliated to UMS2008 IBSLor (Ingénierie, Biologie, Santé en Lorraine) and UMR_S 1256 NGERE (University of Lorraine, Nancy, France).
Funding sources
This work was supported by grants from the French National Institute of Health and Medical Research (INSERM) UMR_S 1256. The study is part of the “GEENAGE” project, Lorraine University of Excellence (LUE) i-Site ANR-15-IDEX-0004-LUE, funded by the French Ministry of Higher Education, Research and Innovation. The work was also supported by fundings from the University Hospital Federation (FHU) ARRIMAGE project, European Regional Development Fund (ERDF), and the Lorraine Region, France. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Author contributions
Study concept and design: AO, JLG, and FN; study supervision and guarantor of the article: AO, JLG, and FN; data collection: AO, EJT, CC, PP, PR, TJ, AC, MB, AF, KM, FL, JBA, FM, CL, CD, KW, DT, PB, LDP, CG, AK, AB, GB, DL, SO, AM, RMRG, FF, JLG, FN; Sanger sequencing: CC, PP, RMRG, FN; exome-wide genotyping: CC, PR, AO, JLG; next-generation clinical exome sequencing: CC, TJ, AO, JLG, FN; statistical analysis: AO; data analysis and interpretation: AO, JLG, and FN; report drafting, review, and approval: AO, JLG, and FN; drafting the first version of the manuscript: AO, JLG, and FN; critical revision of the manuscript for relevant intellectual content: AO, JLG, and FN; approval of the submitted final draft: AO, EJT, CC, PP, PR, TJ, AC, MB, AF, KM, FL, JBA, FM, CL, CD, KW, DT, PB, LDP, CG, AK, AB, GB, DL, SO, AM, RMRG, FF, JLG, FN.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102623.
Contributor Information
Abderrahim Oussalah, Email: abderrahim.oussalah@univ-lorraine.fr.
Jean-Louis Guéant, Email: jean-louis.gueant@univ-lorraine.fr.
Appendix. Supplementary materials
References
- 1.Scriver C.R. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat. 2007;28:831–845. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
- 2.van Wegberg A.M.J., MacDonald A., Ahring K. The complete European guidelines on phenylketonuria: diagnosis and treatment. Orphanet J Rare Dis. 2017;12:162. doi: 10.1186/s13023-017-0685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellwege J.N., Keaton J.M., Giri A., Gao X., Velez Edwards D.R., Edwards T.L. Population stratification in genetic association studies. Curr Protoc Hum Genet. 2017;95 doi: 10.1002/cphg.48. 1 22 1-1 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellenthal G., Busby G.B.J., Band G. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakachoff M., Duforet-Frebourg N., Simonet F. Fine-scale human genetic structure in Western France. Eur J Hum Genet. 2015;23:831–836. doi: 10.1038/ejhg.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Persyn E., Redon R., Bellanger L., Dina C. The impact of a fine-scale population stratification on rare variant association test results. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0207677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scriver C.R. Hyperphenylalaninemia: phenylalanine hydroxylase deficiency. Metabol Molecul Base Inherit Dis. 2002:1667–1724. [Google Scholar]
- 10.Krawczak M., Zschocke J. A role for overdominant selection in phenylketonuria? Evidence from molecular data. Hum Mutat. 2003;21:394–397. doi: 10.1002/humu.10205. [DOI] [PubMed] [Google Scholar]
- 11.Rozen R., Mascisch A., Lambert M., Laframboise R., Scriver C.R. Mutation profiles of phenylketonuria in Quebec populations: evidence of stratification and novel mutations. Am J Hum Genet. 1994;55:321–326. [PMC free article] [PubMed] [Google Scholar]
- 12.Guldberg P., Levy H.L., Hanley W.B. Phenylalanine hydroxylase gene mutations in the United States: report from the maternal PKU collaborative study. Am J Hum Genet. 1996;59:84–94. [PMC free article] [PubMed] [Google Scholar]
- 13.Guldberg P., Rey F., Zschocke J. A European multicenter study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am J Hum Genet. 1998;63:71–79. doi: 10.1086/301920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leuders S., Wolfgart E., Ott T. Influence of PAH genotype on sapropterin response in PKU: results of a single-center cohort study. JIMD Rep. 2014;13:101–109. doi: 10.1007/8904_2013_263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeannesson-Thivisol E., Feillet F., Chery C. Genotype-phenotype associations in French patients with phenylketonuria and importance of genotype for full assessment of tetrahydrobiopterin responsiveness. Orphanet J Rare Dis. 2015;10:158. doi: 10.1186/s13023-015-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N., Huang Q., Li Q. Spectrum of PAH gene variants among a population of Han Chinese patients with phenylketonuria from northern China. BMC Med Genet. 2017;18:108. doi: 10.1186/s12881-017-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohlsson A., Bruhn H., Nordenstrom A., Zetterstrom R.H., Wedell A., von Dobeln U. The spectrum of PAH mutations and increase of milder forms of phenylketonuria in Sweden during 1965-2014. JIMD Rep. 2017;34:19–26. doi: 10.1007/8904_2016_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton V., Santa Maria L., Fuenzalida K. Characterization of phenyalanine hydroxylase gene mutations in chilean PKU patients. JIMD Rep. 2018;42:71–77. doi: 10.1007/8904_2017_85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirzadeh T., Saeidian A.H., Bagherian H. Molecular genetics of a cohort of 635 cases of phenylketonuria in a consanguineous population. J Inherit Metab Dis. 2018;41:1159–1167. doi: 10.1007/s10545-018-0228-6. [DOI] [PubMed] [Google Scholar]
- 20.Li N., He C., Li J. Analysis of the genotype-phenotype correlation in patients with phenylketonuria in mainland China. Sci Rep. 2018;8:11251. doi: 10.1038/s41598-018-29640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huyghe J.R., Jackson A.U., Fogarty M.P. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price A.L., Weale M.E., Patterson N. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weale M.E. Quality control for genome-wide association studies. Methods Mol Biol. 2010;628:341–372. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 24.Genomes Project C., Auton A., Brooks L.D. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 26.Hartl DL, Clark AG, Clark AG. Principles of population genetics: Sinauer associates Sunderland, MA; 1997.
- 27.Weir B.S., Cockerham C.C. Estimating F-Statistics for the analysis of population structure. Evolution (N Y) 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 28.Leviyang S., Hamilton M.B. Properties of Weir and Cockerham's Fst estimators and associated bootstrap confidence intervals. Theor Popul Biol. 2011;79:39–52. doi: 10.1016/j.tpb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Remington D.L., Thornsberry J.M., Matsuoka Y. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewontin R.C. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriel S.B., Schaffner S.F., Nguyen H. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P., Boisson B., Stenson P.D. SeqTailor: a user-friendly webserver for the extraction of DNA or protein sequences from next-generation sequencing data. Nucleic Acids Res. 2019;47:W623–WW31. doi: 10.1093/nar/gkz326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K., Battistuzzi F.U., Billing-Ross P., Murillo O., Filipski A., Kumar S. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 2012;109:19333–19338. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taliun D., Harris D.N., Kessler M.D. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. BioRxiv. 2019 doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karczewski K.J., Francioli L.C., Tiao G. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019 [Google Scholar]
- 38.Withrock I.C., Anderson S.J., Jefferson M.A. Genetic diseases conferring resistance to infectious diseases. Genes Dis. 2015;2:247–254. doi: 10.1016/j.gendis.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson E.K., Kwiatkowski D.P., Sabeti P.C. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolf L.I., McBean M.S., Woolf F.M., Cahalane S.F. Phenylketonuria as a balanced polymorphism: the nature of the heterozygote advantage. Ann Hum Genet. 1975;38:461–469. doi: 10.1111/j.1469-1809.1975.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 41.Saugstad L.F. Anthropological significance of phenylketonuria and the importance of heterozygote advantage. Ir Med J. 1976;69:405–410. [PubMed] [Google Scholar]
- 42.Saugstad L.F. Heterozygote advantage for the phenylketonuria allele. J Med Genet. 1977;14:20–24. doi: 10.1136/jmg.14.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith I., Carter C.O., Wolff O.H. Heterozygote advantage for the phenylketonuria allele. J Med Genet. 1978;15:246–248. doi: 10.1136/jmg.15.3.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saugstad L.F. Heterozygote advantage for the phenylketonuria allele. J Med Genet. 1978;15:317–319. doi: 10.1136/jmg.15.4.317-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woolf L.I. The heterozygote advantage in phenylketonuria. Am J Hum Genet. 1986;38:773–775. [PMC free article] [PubMed] [Google Scholar]
- 46.DeGiorgio M., Lohmueller K.E., Nielsen R. A model-based approach for identifying signatures of ancient balancing selection in genetic data. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leffler E.M., Bullaughey K., Matute D.R. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolf L.I., Cranston W.I., Goodwin B.L. Genetics of phenylketonuria. Heterozygosity for phenylketonuria. Nature. 1967;213:882–883. doi: 10.1038/213882a0. [DOI] [PubMed] [Google Scholar]
- 49.Thony B., Ding Z., Martinez A. Tetrahydrobiopterin protects phenylalanine hydroxylase activity in vivo: implications for tetrahydrobiopterin-responsive hyperphenylalaninemia. FEBS Lett. 2004;577:507–511. doi: 10.1016/j.febslet.2004.10.056. [DOI] [PubMed] [Google Scholar]
- 50.Alkaitis M.S., Ackerman H.C. Tetrahydrobiopterin supplementation improves phenylalanine metabolism in a murine model of severe malaria. ACS Infect Dis. 2016;2:827–838. doi: 10.1021/acsinfecdis.6b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeo T.W., Lampah D.A., Kenangalem E. Impaired systemic tetrahydrobiopterin bioavailability and increased dihydrobiopterin in adult falciparum malaria: association with disease severity, impaired microvascular function and increased endothelial activation. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalrymple U., Mappin B., Gething P.W. Malaria mapping: understanding the global endemicity of falciparum and vivax malaria. BMC Med. 2015;13:140. doi: 10.1186/s12916-015-0372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beall C.M. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beall C.M., Laskowski D., Erzurum S.C. Nitric oxide in adaptation to altitude. Free Radic Biol Med. 2012;52:1123–1134. doi: 10.1016/j.freeradbiomed.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong C., Ozga A.T., Witonsky D.B. Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc Natl Acad Sci U S A. 2016;113:7485–7490. doi: 10.1073/pnas.1520844113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawford J.E., Amaru R., Song J. Natural selection on genes related to cardiovascular health in high-altitude adapted andeans. Am J Hum Genet. 2017;101:752–767. doi: 10.1016/j.ajhg.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhandari S., Cavalleri G.L. Population history and altitude-related adaptation in the Sherpa. Front Physiol. 2019;10:1116. doi: 10.3389/fphys.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adebali O., Reznik A.O., Ory D.S., Zhulin I.B. Establishing the precise evolutionary history of a gene improves prediction of disease-causing missense mutations. Genet Med. 2016;18:1029–1036. doi: 10.1038/gim.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakka L., Deletage N., Lalloue F. SCO-spondin derived peptide NX210 induces neuroprotection in vitro and promotes fiber regrowth and functional recovery after spinal cord injury. PLoS ONE. 2014;9:e93179. doi: 10.1371/journal.pone.0093179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guttler F., Lou H. Dietary problems of phenylketonuria: effect on CNS transmitters and their possible role in behaviour and neuropsychological function. J Inherit Metab Dis. 1986;9(Suppl 2):169–177. doi: 10.1007/BF01799701. [DOI] [PubMed] [Google Scholar]
- 62.Martynyuk A.E., van Spronsen F.J., Van der Zee E.A. Animal models of brain dysfunction in phenylketonuria. Mol Genet Metab. 2010;99(Suppl 1):S100–S105. doi: 10.1016/j.ymgme.2009.10.181. [DOI] [PubMed] [Google Scholar]
- 63.Joseph B., Dyer C.A. Relationship between myelin production and dopamine synthesis in the PKU mouse brain. J Neurochem. 2003;86:615–626. doi: 10.1046/j.1471-4159.2003.01887.x. [DOI] [PubMed] [Google Scholar]
- 64.Gueant J.L., Chery C., Oussalah A. APRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat Commun. 2018;9:67. doi: 10.1038/s41467-017-02306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.