In this study, Cornelissen et al. set out to elucidate the role of the GATA-type zinc finger transcription factor TRPS1 in breast cancer. Using in vitro and in vivo loss-of-function approaches, the authors demonstrate that TRPS1 can function as a context-dependent tumor suppressor in breast cancer, while being essential for growth and differentiation of normal mammary epithelial cells.
Keywords: TRPS1, breast cancer, mammary gland development, E-cadherin, context-dependent regulator, ILC
Abstract
The GATA-type zinc finger transcription factor TRPS1 has been implicated in breast cancer. However, its precise role remains unclear, as both amplifications and inactivating mutations in TRPS1 have been reported. Here, we used in vitro and in vivo loss-of-function approaches to dissect the role of TRPS1 in mammary gland development and invasive lobular breast carcinoma, which is hallmarked by functional loss of E-cadherin. We show that TRPS1 is essential in mammary epithelial cells, since TRPS1-mediated suppression of interferon signaling promotes in vitro proliferation and lactogenic differentiation. Similarly, TRPS1 expression is indispensable for proliferation of mammary organoids and in vivo survival of luminal epithelial cells during mammary gland development. However, the consequences of TRPS1 loss are dependent on E-cadherin status, as combined inactivation of E-cadherin and TRPS1 causes persistent proliferation of mammary organoids and accelerated mammary tumor formation in mice. Together, our results demonstrate that TRPS1 can function as a context-dependent tumor suppressor in breast cancer, while being essential for growth and differentiation of normal mammary epithelial cells.
Breast cancer is a leading cause of cancer death among women worldwide (Bray et al. 2018). The majority of breast cancers are histologically classified as invasive ductal carcinoma (IDC; 60%–80%) or invasive lobular carcinoma (ILC; 10%–15%) (Li et al. 2003; Ciriello et al. 2015). While IDCs are represented among all PAM50 molecular subtypes, the majority of ILCs are classified as luminal A tumors (Parker et al. 2009; Ciriello et al. 2015). A hallmark of ILCs is the loss of E-cadherin (CDH1) expression, which drives the growth of noncohesive tumor cells invading the stroma in single files without forming dense masses (Vos et al. 1997; Bharat et al. 2009). Using Sleeping Beauty (SB) transposon-based in vivo insertional mutagenesis screens in mice, we previously identified recurrent SB insertions in Trps1, a member of the GATA-type zinc finger transcription factor family, in a mouse model for E-cadherin-deficient ILC (Kas et al. 2017). Although these data identify TRPS1 as a potential breast cancer driver, its functions in the context of breast cancer remain largely understudied and the potential role of TRPS1 in ILC formation remains fully elusive.
The transcription factor TRPS1 contains nine putative zinc finger domains; one GATA-type zinc finger domain required for DNA-binding, two IKAROS-type zinc finger domains associated with transcriptional repression, and six domains with unknown functions (Georgopoulos et al. 1997; Momeni et al. 2000; Malik et al. 2001). In contrast to other GATA-type transcription factors, which function as transcriptional activators, TRPS1 acts as a transcriptional repressor (Malik et al. 2001). Mutations and deletions identified in the TRPS1 locus are implicated in the autosomal dominant genetic disorder tricho–rhino–phalangeal syndrome (TRPS), a developmental disorder characterized by craniofacial and skeletal malformations (Momeni et al. 2000; Maas et al. 2015). In addition, TRPS1 is essential in the regulation of bone, kidney, and hair development (Malik et al. 2002; Suemoto et al. 2007; Gai et al. 2009; Wuelling et al. 2009; Fantauzzo et al. 2012). In summary, TRPS1 is crucial in the development of multiple tissues, and disrupted TRPS1 expression is associated with severe developmental malformations.
The relevance of TRPS1 in breast cancer is still rather unclear. On the one hand, TRPS1 amplifications are frequently observed in breast cancers with poor survival (Radvanyi et al. 2005; Chen et al. 2010; Serandour et al. 2018), but this observation might in part be confounded by the fact that TRPS1 is located near MYC, resulting in frequent coamplification of TRPS1 with MYC, which is associated with poor survival (Berns et al. 1992; Ciriello et al. 2015; Pereira et al. 2016). On the other hand, expression of TRPS1 correlates with a favorable outcome in breast cancer patients (Lin et al. 2017), but this might in part be explained by the fact that TRPS1 mRNA expression correlates with expression of GATA3, which is a marker of luminal breast cancer with good prognosis (Mehra et al. 2005; Chen et al. 2011).
By integrating DNA copy number alteration data and genome-wide pooled RNAi screens, Sanchez-Garcia et al. (2014) identified TRPS1 as a potential breast cancer driver gene and showed that overexpression of TRPS1 in nontumorigenic mammary epithelial MCF10A cells increased in vitro colony formation. However, conflicting results have been obtained in in vivo mouse models. Trps1 was identified as a potential tumor suppressor gene in an insertional mutagenesis screen in a triple-negative breast cancer (TNBC) mouse model and reduced expression of TRPS1 was reported to increase in vivo growth of multiple TNBC cell lines (Rangel et al. 2016). However, for other breast cancer cell lines in vivo cell growth is reported to decrease upon reduced TRPS1 expression (Elster et al. 2018; Wang et al. 2018b; Witwicki et al. 2018). In vitro, TRPS1 was reported to be involved in regulation and restriction of ERα DNA binding and histone acetylation at enhancers (Serandour et al. 2018) and required for maintenance of epithelial differentiation by suppression of ZEB2 (Stinson et al. 2011; Huang et al. 2016). Furthermore, TRPS1 was found to attenuate YAP activity by regulating genome-wide YAP-dependent gene transcription (Elster et al. 2018). Together, these reports indicate that both (over)expression and loss of TRPS1 are associated with breast cancer.
Here, we set out to evaluate TRPS1 function both in mammary gland development and tumor formation. We identified TRPS1 expression to be essential for the lactogenic differentiation capacity of nontransformed mammary cells in vitro by suppression of interferon signaling. Furthermore, by generating a conditional Trps1 mouse model, we found TRPS1 expression to be essential for proliferation and survival of luminal epithelial cells in mammary organoids and the mouse mammary gland. In contrast, combined loss of TRPS1 and E-cadherin is tolerated, resulting in persistent proliferation of mammary organoids and induction of mammary tumors in mice.
Results
Sleeping Beauty-induced mouse ILCs contain recurrent insertions in Trps1 that result in a functionally impaired truncated protein
Even though ILCs are characterized by functional loss of E-cadherin, mammary-specific inactivation of E-cadherin by itself does not induce ILC formation in female mice, indicating that additional mutations are required (Boussadia et al. 2002; Derksen et al. 2006, 2011). To identify cancer genes that collaborate with E-cadherin loss in ILC formation, we previously performed an in vivo Sleeping Beauty (SB) insertional mutagenesis screen in mice with mammary gland-specific loss of E-cadherin, which yielded Trps1 as one of the top hits (Kas et al. 2017). SB insertions at the Trps1 locus were present in 51 out of 99 SB-induced mammary tumors and distributed throughout the intronic regions of the gene (Fig. 1A; Kas et al. 2017). In addition, about two-thirds of all tumors with SB insertions in Trps1 contained more than one insertion in this gene (Fig. 1B; Kas et al. 2017). Together, these results indicate that SB insertions at the Trps1 locus result in inactivation of the gene.
Figure 1.
Recurrent Sleeping Beauty (SB) insertions in Trps1 results in a truncated protein that accumulates in the cytoplasm and does not interact with the DNA. (A) Schematic overview of SB insertions identified in intronic regions of the Trps1 gene in all tumors analyzed (Kas et al. 2017). Each arrow represents an unique SB insertion. Schematic overviews of the Trps1 locus and TRPS1 protein are indicated below the SB insertions. Numbered boxes represent exons of the canonical gene transcript. (B) Distribution of the number of insertions per tumor in Trps1. Tumors without insertions in the intronic region between exons 4 and 5 are indicated in black, and tumors with insertions in this intronic region are indicated in gray. (C) Western blot analysis of TRPS1 expression in tumors with the indicated number of SB insertions in Trps1. β-Actin was used as a loading control. (Black arrowhead) Full-length TRPS1 (FL); (gray arrowhead) truncated TRPS1 (TR). (D) Representative microscopic images of TRPS1 expression by immunohistochemistry in a tumor without insertions in Trps1 (left) and with insertions in Trps1 (right). Scale bars, 100 μm (top), 25 μm (bottom). (E) Representative microscopic images of TRPS1 expression detected by immunofluorescence in tumor-derived cell lines without (left) and with (right) insertions in Trps1. Scale bars, 20 μm. (F) Heat map illustrating raw peak intensity of TRPS1 ChIP-seq in tumor-derived cell lines without (red) and with (blue) insertions in Trps1. A window of 5 kb around the peak is shown. (G) Average read count profiles of the total TRPS1 peak signal.
Interestingly, ∼90% of Trps1 mutated tumors contained at least one insertion in the intronic region between exons 4 and 5 (Fig. 1A,B). Protein analysis showed expression of a smaller TRPS1 protein variant in all Trps1 mutated tumors analyzed (Fig. 1C), even though the pattern of insertions differed between tumors (Supplemental Fig. S1A). This smaller protein appeared to be a truncated form of TRPS1, since an shRNA targeting exon 3 of Trps1 resulted in reduced expression levels of this smaller protein in tumor-derived cell lines, whereas an shRNA directed at the 3′UTR of Trps1 did not (Supplemental Fig. S1B,C). In tumors with SB insertions, mRNA transcripts of exons 1–4 of Trps1 were much more abundant than transcripts of exons 5 and 6, indicating that the SB insertions of all analyzed tumors resulted in a truncated form of TRPS1 that is encoded by exons 1–4 (Supplemental Fig. S1D,E). In addition, expression levels of truncated TRPS1 were greatly increased when more than one insertion was identified in Trps1 (Fig. 1C; Supplemental Fig. S1A,D,E). Truncation of Trps1 by SB insertions in intron 4 results in loss of part of the nuclear localization signal (Fig. 1A). In line with this, SB-induced mouse ILCs with insertions in Trps1 showed diffuse expression of truncated TRPS1 throughout the cells compared with nuclear staining of wild-type (WT) TRPS1 in tumors without Trps1 insertions (Fig. 1D). Cytoplasmic localization of truncated TRPS1 was confirmed using cell lines derived from SB-induced tumors with and without Trps1 insertions (Fig. 1E; Supplemental Fig. S1F). Furthermore, ChIP sequencing experiments revealed impaired chromatin binding of truncated TRPS1 (Fig. 1F,G). Together, these results show that insertions in Trps1 in SB-induced mouse ILCs result in expression of a truncated nonfunctional protein.
TRPS1 interacts with NuRD members and mediates chromatin binding of the NuRD complex
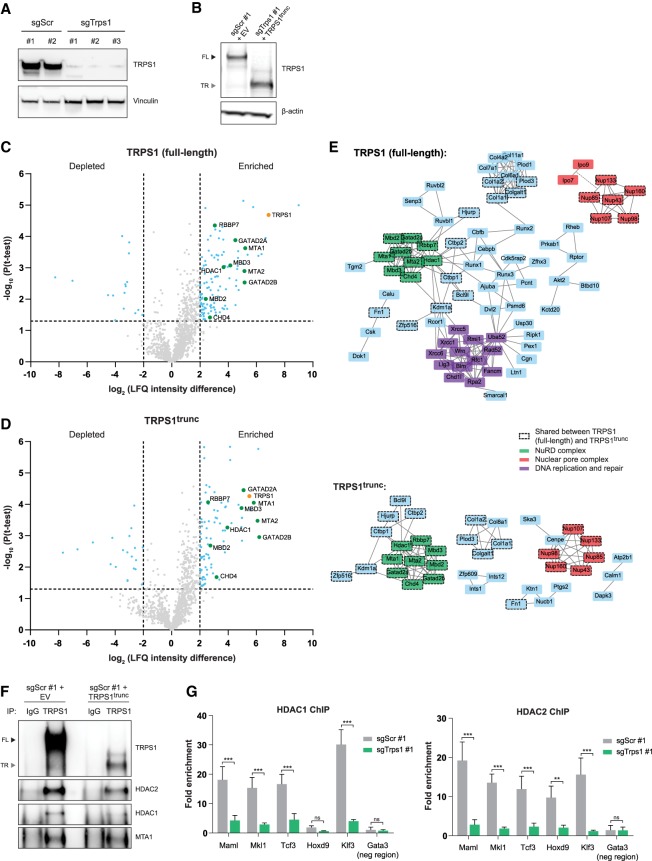
TRPS1 regulates gene expression by acting as a transcriptional repressor (Malik et al. 2001). To analyze the protein interactome of full-length and truncated TRPS1, we compared mouse mammary epithelial HC11 cells expressing full-length TRPS1 with CRISPR-mediated TRPS1 knockout HC11 cells with or without expression of truncated TRPS1 (TRPS1trunc, comprising AA1-887 encoded by exons 2–4) (Fig. 2A,B; Supplemental Fig. S2A,B). Using an antibody that recognizes both WT and mutant TRPS1, we performed immunoprecipitation followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Fig. 2C,D; Supplemental Table S1). Significant interactors compared with the IgG control were analyzed for functional protein–protein associations. Both full-length and truncated TRPS1 showed interactions with nuclear pore complex proteins (Fig. 2E). However, TRPS1trunc did not interact with importin proteins (IPO7 and IPO9) required for nuclear import (Sorokin et al. 2007), in line with the lack of nuclear localization of truncated TRPS1 (Fig. 2E). In addition, full-length TRPS1 interacted with DNA replication and repair proteins that were not identified as interactors for truncated TRPS1 (Fig. 2E), suggesting that upon truncation of TRPS1 this interaction is disrupted or that this interaction only takes place in the nucleus. Consistent with previous reports (Serandour et al. 2018; Wang et al. 2018a; Witwicki et al. 2018), we observed interactions of full-length TRPS1 with members of the nucleosome remodeling and deacetylation (NuRD) corepressor complex (Fig. 2C,E,F) that is involved in transcriptional regulation of basic cellular processes (Basta and Rauchman 2015). Interestingly, this interaction was maintained in the cytoplasm upon truncation of TRPS1, even though NuRD complex members were lowly expressed in the cytoplasm and localization of these proteins was not affected (Supplemental Fig. S2C).
Figure 2.
TRPS1 interacts with the NuRD complex and loss of TRPS1 expression reduces DNA binding of HDAC1 and HDAC2. (A) Western blot analysis of TRPS1 expression in CRISPR-mediated TRPS1 knockout (sgTrps1) HC11 clones compared with scrambled controls (sgScr). Vinculin was used as a loading control. (B) Western blot analysis of TRPS1 expression in control cells transduced with empty vector and TRPS1 knockout cells transduced with TRPS1trunc. β-Actin was used as a loading control. (Black arrowhead) Full-length TRPS1 (FL); (gray arrowhead) truncated TRPS1 (TR). (C,D) Volcano plots showing protein interactors of full-length (C) and truncated (D) TRPS1 compared with IgG control in HC11 cells in four independent experiments. TRPS1 (orange) and members of the NuRD complex (green) are indicated. LFQ, label-free quantification. (E) Projection of all significant interactors identified in C and D onto the STRING protein–protein interaction network (version 11). Only proteins that interact with two or more proteins are shown based on known interactions from curated databases and experimentally determined interactions. (F) Coimmunoprecipitation of full-length TRPS1 and TRPS1trunc with members of the NuRD complex in HC11 cells. (Black arrowhead) Full-length TRPS1 (FL); (gray arrowhead) truncated TRPS1 (TR). (G) HDAC1 and HDAC2 ChIP-qPCR analysis in TRPS1-proficient and -deficient HC11 cells. Five target regions and one negative control region of a representative experiment are shown. Data represent mean + standard deviation (SD), n = 3. Two-way ANOVA: (***) P < 0.001; (**) P < 0.01; (ns) P > 0.05.
To further explore the interaction between TRPS1 and the NuRD complex, we evaluated the DNA-binding potential of HDAC1 and HDAC2, core components that drive histone deacetylation activity of the NuRD complex (Lai and Wade 2011) in TRPS1-proficient and CRISPR-mediated TRPS1 knockout HC11 cells (Fig. 2A; Supplemental Fig. S2A). Both HDAC1 and HDAC2 DNA binding was greatly impaired upon loss of TRPS1 expression (Fig. 2G), while protein expression of both HDAC1 and HDAC2 was not affected (Supplemental Fig. S2D), indicating that TRPS1 recruits the NuRD complex to the DNA. Expression of truncated TRPS1 did not affect DNA binding of HDAC1 or HDAC2 in both TRPS1-proficient and TRPS1-deficient cells (Supplemental Fig. S2E). Lactogenic differentiation induced by dexamethasone, insulin, and prolactin (DIP mix) in HC11 cells results in phosphorylation of STAT5 and production of milk proteins such as β-casein (Ball et al. 1988; Gouilleux et al. 1994). The necessity of the NuRD complex for lactogenic differentiation potential was tested by treating HC11 cells with the HDAC inhibitor trichostatin A (TSA) (Yoshida et al. 1990), resulting in decreased phosphorylation of STAT5 and diminished β-casein (Csn2) production following stimulation with DIP (Supplemental Fig. S2D,E). Together, these results show that TRPS1 loss in HC11 cells impairs DNA-binding potential of HDAC1 and HDAC2, while inhibition of HDAC activity in these cells interferes with lactogenic differentiation potential.
Suppression of interferon signaling by TRPS1 is essential for proliferation and lactogenic differentiation capacity of HC11 cells
Next, we determined the functional consequences of TRPS1 loss in E-cadherin-proficient HC11 cells. TRPS1 knockout HC11 clones showed slower proliferation compared with control cells (Fig. 3A). In addition, the lactogenic differentiation potential of the TRPS1 knockout cells was greatly impaired, as determined by low phospho-STAT5 and β-casein levels upon treatment with DIP mix (Fig. 3B; Supplemental Fig. S2H). Similar effects were observed upon shRNA-mediated knockdown of TRPS1 in WT HC11 cells (Supplemental Fig. S2I–K). Expression of truncated TRPS1 did not affect proliferation or lactogenic differentiation potential (Supplemental Fig. S2L–N). Therefore, truncation of TRPS1 results in a nonfunctional protein, whereas loss of TRPS1 has pronounced effects.
Figure 3.
TRPS1 loss impairs cell proliferation and lactogenic differentiation, mediated by increased interferon signaling. (A) Cell proliferation of control and TRPS1 knockout HC11 clones, as quantified using IncuCyte imaging for 140 h. Data represent mean ± standard error of the mean (SEM). (B) RT-qPCR analysis of β-casein (Csn2) expression levels upon stimulation with DIP mix (dexamethasone, insulin, and prolactin) in control and TRPS1 knockout HC11 clones. Data represent mean + SD, n = 2. One-way ANOVA: (***) P < 0.001. (C) Pathway enrichment analysis performed on RNA-seq of HC11 control and TRPS1 knockout cells, using the MSigDB hallmark gene set collection (Liberzon et al. 2015). FDR < 0.05 was considered significant (blue bars). (D) Heat map reflecting gene expression changes at the genes listed in the interferon α and interferon γ gene sets indicated in C between HC11 control and TRPS1 knockout cells. Key factors in these signaling pathways are indicated. (E) Western blot analysis of STAT1 and STAT2 expression levels in HC11 control and TRPS1 knockout cells transduced with a nontargeting shRNA or a pool of shRNAs targeting Stat1. β-actin was used as a loading control. (F) RT-qPCR analysis of β-casein (Csn2) expression levels upon stimulation with DIP mix in control and TRPS1 knockout HC11 clones transduced with a nontargeting shRNA or a pool of shRNAs targeting Stat1. Data represent mean + SD, n = 3. One-way ANOVA: (***) P < 0.001.
To determine the effect of TRPS1 loss on the transcriptome of HC11 cells, RNA sequencing was performed on TRPS1 knockout and control cells (log2FC >2/−2; 1477 up/932 down). Pathway enrichment analysis using the MSigDB hallmark gene set collection (Liberzon et al. 2015) revealed several significantly enriched pathways, including epithelial–mesenchymal transition (EMT), in line with the role of TRPS1 in suppression of EMT (Stinson et al. 2011) and the interferon α and γ response pathways (Fig. 3C,D; Supplemental Fig. S3A). Key players in the interferon α and γ response pathways are STAT signaling proteins that allow the cell to respond to surrounding stimuli by induction of transcriptional programs (Haricharan and Li 2014). Increased protein expression of both STAT1 and STAT2 was observed in TRPS1 knockout clones compared with controls (Supplemental Fig. S3B). Stat1 targeting shRNAs resulted in efficient down-regulation of STAT1 expression, and to some extent STAT2 levels, thereby restoring the production of β-casein in DIP-treated TRPS1 knockout cells and increasing the β-casein production in DIP-treated control cells, without affecting phosphorylated STAT5 levels (Fig. 3E,F; Supplemental Fig. S3C). In addition, stimulation of HC11 cells with IFNγ increased Stat1 mRNA levels and severely impaired STAT5 phosphorylation and β-casein production (Supplemental Fig. S3D–F). However, stimulation with IFNγ increases total and phosphorylated STAT1 levels to a much higher extent than the induction upon TRPS1 loss (Supplemental Fig. S3G). In addition, interferon signaling is not only stimulated by reduced recruitment of the NuRD complex to the DNA caused by loss of TRPS1, but also by treatment with the HDAC inhibitor TSA (Supplemental Fig. S3H,I). Together, these results indicate that the increased interferon signaling, caused by loss of TRPS1, hampers lactogenic differentiation in HC11 cells.
TRPS1 is essential for survival of luminal epithelial cells in the developing and differentiating mammary gland
The transcription factor GATA3 is also a member of the GATA-type zinc finger transcription factor family, similar to TRPS1. GATA3 is required for mammary gland morphogenesis and for maintenance of the luminal epithelium in the adult mammary gland (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007). Since both TRPS1 and GATA3 are highly expressed in luminal epithelial cells in the adult mammary gland (Supplemental Fig. S4A,B), we sought to study the role of TRPS1 in mammary gland development and differentiation. To this end, we generated Trps1 “floxed” (Trps1F) mice carrying two loxP sites flanking Trps1 exons 2–6 (Fig. 4A). Cre-mediated recombination of these LoxP sites will result in loss of all coding exons of Trps1. We next generated MMTV-cre;Trps1F/F;mT/mG mice, in which mammary gland-specific expression of Cre results in combined loss of TRPS1 and tdTomato (mT) expression and concomitant induction GFP (mG) expression (Fig. 4A).
Figure 4.
Loss of TRPS1 expression during development of the mammary gland is not tolerated in E-cadherin-proficient luminal cells, but results in the formation of lesions in combination with E-cadherin loss. (A) Schematic overview of the engineered alleles in MMTV-cre;Trps1+/+;mT/mG (Trps1+/+) and MMTV-cre;Trps1F/F;mT/mG (Trps1F/F) mice. (B) Representative images of GFP and TRPS1 expression in mammary glands of Trps1+/+ (left) and Trps1F/F (right) animals at 6.5 and 10 wk of age and at day 15 of pregnancy, detected by immunofluorescence. Nuclei were counterstained with Hoechst. Scale bars, 50 μm. (C) Quantification of percentage GFP-positive cells per field. Data represent measurements of five fields in three animals per condition. Two-way ANOVA: (***) P < 0.001. (D) Schematic overview of the engineered alleles in MMTV-cre;Cdh1+/F;Trps1+/F;mT/mG (Cdh1+/F/Trps1+/F), MMTV-cre;Cdh1+/F;Trps1F/F;mT/mG (Cdh1+/F/Trps1F/F), MMTV-cre;Cdh1F/F;Trps1+/F;mT/mG (Cdh1F/F/Trps1+/F), and MMTV-cre;Cdh1F/F;Trps1F/F;mT/mG (Cdh1F/F/Trps1F/F) animals. (E) Representative images of GFP, TRPS1, and E-cadherin expression in mammary glands of the indicated genotypes at 10 wk of age, detected by immunofluorescence. Nuclei were counterstained with Hoechst. Scale bars, 50 μm. (F) Quantification of percentage of GFP-positive cells per field. Data represent measurements of five fields in three animals per condition. One-way ANOVA: (***) P < 0.001; (**) P < 0.01; (ns) P > 0.05.
To determine the effect of TRPS1 loss on development and differentiation of the gland, we analyzed carmine-stained mammary glands from MMTV-cre;Trps1+/+;mT/mG and MMTV-cre;Trps1F/F;mT/mG mice at 6.5 and 10wk of age and at day 15 of pregnancy. In contrast to what has been reported for MMTV-cre;Gata3F/F mice (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007), MMTV-cre;Trps1F/F;mT/mG mice did not show any consistent impairment in outgrowth and differentiation of the ductal-tree (Supplemental Fig. S4C–F). However, the number of GFP-positive cells that mark cells in which Cre-induced recombination has taken place was significantly decreased in MMTV-cre;Trps1F/F;mT/mG mammary glands compared with MMTV-cre;Trps1+/+;mT/mG glands (Fig. 4B,C). This indicates that TRPS1-deficient mammary epithelial cells in MMTV-cre;Trps1F/F;mT/mG mice did not survive, resulting in selective depletion of these cells and formation of the ductal structure by TRPS1-proficient epithelial cells. GFP-positive cells in MMTV-cre;Trps1+/F;mT/mG mice showed half-dose expression levels of TRPS1, while the percentage of GFP-positive cells was comparable with MMTV-cre;Trps1+/+;mT/mG mice (Supplemental Fig. S4G–I). Together, this indicates that complete loss of TRPS1 expression is not tolerated in luminal epithelial cells.
TRPS1 loss is tolerated in E-cadherin-deficient mammary epithelial cells
Since the SB insertions in Trps1 were identified in E-cadherin-deficient mammary tumors, we investigated whether complete loss of TRPS1 expression is tolerated in E-cadherin-deficient mammary epithelium. MMTV-cre;Trps1F/F;mT/mG mice were crossed with Cdh1F/F mice (Derksen et al. 2006) to generate MMTV-cre;Cdh1F/F;Trps1F/F;mT/mG mice, in which Cre-mediated recombination induced combined loss of E-cadherin (encoded by the Cdh1 gene), TRPS1, and tdTomato, and concomitant expression of GFP (Fig. 4D). Similar to E-cadherin WT mice (Fig. 4B,C), the percentage of GFP-positive cells was high in ducts of MMTV-cre;Cdh1+/F;Trps1+/F;mT/mG glands, but almost absent in MMTV-cre;Cdh1+/F;Trps1F/F;mT/mG glands (Fig. 4E,F). In line with previous observations (Boelens et al. 2016), complete loss of E-cadherin expression in MMTV-cre;Cdh1F/F;TRPS1+/F;mT/mG glands resulted in migration of luminal cells out of the duct into the stroma, resulting in a similar percentage of GFP-positive cells, but at a different location (Fig. 4E,F; Supplemental Fig. S5A). These E-cadherin-deficient, TRPS1-proficient cells remained in the stromal compartment in organized structures. Also in MMTV-cre;Cdh1F/F;TRPS1F/F;mT/mG mice we observed migration of GFP-positive mammary epithelial cells into the stroma, resulting in extraductal clusters of GFP-positive cells that lost both E-cadherin and TRPS1 expression (Fig. 4E,F; Supplemental Fig. S5A). These clusters were less common but larger and more disorganized than the clusters observed in MMTV-cre;Cdh1F/F;TRPS1+/F;mT/mG glands (Supplemental Fig. S5A–C). Hence, while mammary-specific loss of E-cadherin alone results in organized but nonproliferating epithelial cell clusters in the stroma, combined loss of TRPS1 and E-cadherin results in more disorganized clusters with increased proliferation potential.
Combined loss of TRPS1 and E-cadherin induces continued proliferation of mammary organoids
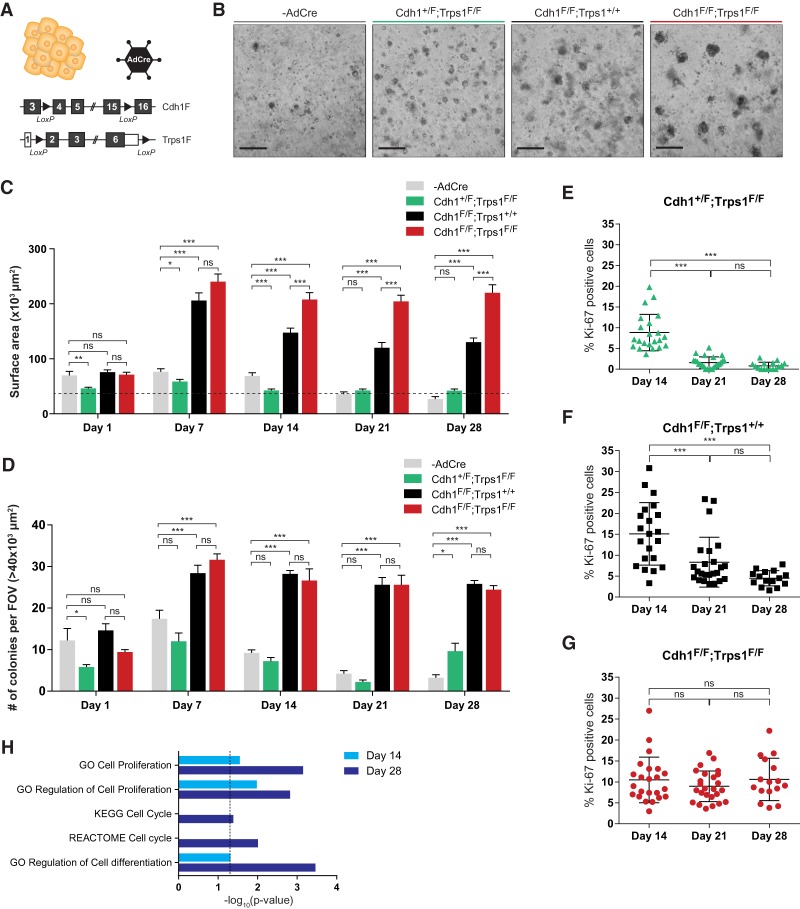
To further explore the effect of combined loss of TRPS1 and E-cadherin, primary mammary organoids were isolated from Cdh+/F;Trps1F/F, Cdh1F/F;Trps1+/+ and Cdh1F/F;Trps1F/F mice. These organoids were treated with Cre-encoding adenovirus (AdCre) to induce recombination of the floxed alleles and subsequently grown in Matrigel for 28 d (Fig. 5A; Supplemental Fig. S6A). The organoids contained both cytokeratin-8-positive luminal cells and cytokeratin-14-positive basal cells, whereas TRPS1 is expressed solely in the luminal cells (Supplemental Fig. S6B). AdCre-mediated loss of TRPS1 in Cdh1+/F;Trps1F/F cells resulted in organoids that were smaller than untransduced WT organoids, whereas E-cadherin loss in AdCre-infected Cdh1F/F;Trps1+/+ cells resulted in enlarged and noncohesive organoids (Fig. 5B–D; Boelens et al. 2016). Combined loss of E-cadherin and TRPS1 in AdCre-infected Cdh1F/F;Trps1F/F cells resulted in organoids with a similar noncohesive morphology as AdCre-treated Cdh1F/F;Trps1+/+ organoids, but these organoids grew larger over time (Fig. 5B–D). Cell proliferation (as determined by immunofluorescence staining for Ki-67) was strongly decreased in TRPS1-deficient organoids after 21 d of culture, in line with the small size of these organoids (Fig. 5E). Even though E-cadherin-deficient organoids grew initially, cell proliferation also decreased over time (Fig. 5F). In line with having the largest size after 28 d of culture, proliferation potential of E-cadherin/TRPS1-deficient organoids persisted over time (Fig. 5G). To identify global differences in gene expression, RNA sequencing was performed on Cdh1F/F;Trps1+/+ and Cdh1F/F;Trps1F/F organoids isolated after 14 and 28 d of culture (log2FC >2/−2; day 14, 101 up/124 down; day 28, 217 up/421 down). Pathway enrichment analysis using the MSigDB hallmark gene set collection (Liberzon et al. 2015) revealed several significantly enriched pathways, including G2M checkpoint and mitotic spindle, indicative for differences in proliferation (Supplemental Fig. S6C). Gene set enrichment analysis confirmed an enrichment of genes related to proliferation, cell cycle, and cell differentiation among the genes with a greater than fourfold difference in expression at day 14 and day 28 (Fig. 5H). Together, these results show that individual loss of TRPS1 or E-cadherin in mammary organoids results in decreased cell proliferation potential, whereas combined loss of TRPS1 and E-cadherin results in increased cell proliferation potential and larger organoids.
Figure 5.
Combined loss of E-cadherin and TRPS1 expression in primary mammary organoids results in prolonged proliferation. (A) Schematic overview of the engineered alleles in Cdh1+/F;Trps1F/F, Cdh1F/F;Trps1+/+ and Cdh1F/F;Trps1F/F primary organoids, in which recombination is induced in vitro by addition of adenovirus encoding Cre (AdCre). (B) Representative bright field images of primary untransduced and AdCre-transduced organoids 28 d after seeding. Scale bars, 500 μm. (C) Quantification of surface area of primary organoids as shown in B, at different time points. Data represent mean + SEM of measurements of organoids within five fields of view per condition. Two-way ANOVA: (***) P < 0.001; (**) P < 0.01; (*) P < 0.05; (ns) P > 0.05. Dotted line indicates the 40 × 103-μm2 size cutoff used to quantify the number of organoids in D. (D) Quantification of the number of organoids larger than 40 × 103 μm2 shown in C. Data represent mean + SD of the number of organoids within five fields of view per condition. Two-way ANOVA: (***) P < 0.001; (*) P < 0.05; (ns) P > 0.05. (FOV) Field of view. (E–G) Quantification of percentage of Ki-67-expressing cells in Cdh1+/F;Trps1F/F (E), Cdh1F/F;Trps1+/+ (F), and Cdh1F/F;Trps1F/F (G) primary organoids, detected by immunofluorescence. Data represent measurements from three independent organoid isolations. One-way ANOVA: (***) P < 0.001; (ns) P > 0.05. (H) Differentially expressed genes (log2FC >2/<–2) between Cdh1F/F;Trps1+/+ and Cdh1F/F;Trps1F/F primary organoids at days 14 and 28, as determined by RNA-seq, are enriched for cell cycle, cell proliferation, and cell differentiation related gene sets. Fisher exact test: P < 0.05 is considered significant.
TRPS1 loss collaborates with E-cadherin loss in mammary tumorigenesis
To determine whether complete loss of TRPS1 is sufficient to accelerate mammary tumor formation induced by E-cadherin loss, Trps1F/F mice were crossed with WAP-cre;Cdh1F/F mice (Fig. 6A; Derksen et al. 2011). Both E-cadherin or TRPS1 status did not affect the number of pups in the first litter, indicative for normal lactation (Supplemental Fig. S7A). In line with previous results (Kas et al. 2017), a subset of WAP-cre;Cdh1F/F;Trps1+/+ mice developed mammary tumors with a long median latency of >843 d (Fig. 6B). This median latency was decreased to 610 d in WAP-cre;Cdh1F/F;Trps1F/F mice (Fig. 6B; P = 0.0127). In addition, we generated transgenic mice with Cre-inducible expression of TRPS1trunc in combination with firefly luciferase, by targeting the Frt-invCAG-mTrps1trunc-IRES-Luc allele into the Col1a1 locus of mouse embryonic stem cells (mESCs) derived from WAP-cre;Cdh1F/F mice (Supplemental Fig. S7B; Huijbers et al. 2015). Chimeric mice were generated through blastocyst injections of correctly targeted WAP-cre;Cdh1F/F mESCs and subsequently crossed back to the WAP-cre;Cdh1F/F model, resulting in WAP-cre;Cdh1F/F;Col1a1Frt-invCAG-mTrps1trunc-IRES-luc (WAP-cre;Cdh1F/F;Trps1trunc) mice, which showed a similar tumor latency compared with WAP-cre;Cdh1F/F;Trps1+/+ (Supplemental Fig. S7C). However, in full-blown tumors we did not observe luciferase activity, indicating no or low expression of the introduced transgene (Supplemental Fig. S7D), while TRPS1trunc was efficiently expressed in in vitro AdCre-transduced primary mouse mammary epithelial cells (MMECs) (Supplemental Fig. S7E). Together, this indicates that loss of TRPS1 expression is sufficient to accelerate tumor formation in WAP-cre;Cdh1F/F mice, while expression of TRPS1trunc is dispensable for tumor formation in WAP-cre;Cdh1F/F mice.
Figure 6.
Combined loss of E-cadherin and TRPS1 results in acceleration of tumor formation. (A) Schematic overview of the engineered alleles in WAP-cre;Cdh1+/F;Trps1+/+(Cdh1+/F;Trps1+/+), WAP-cre;Cdh1+/F;Trps1F/F(Cdh1+/F;Trps1F/F), WAP-cre;Cdh1F/F;Trps1+/+ (Cdh1F/F;Trps1+/+), and WAP-cre;Cdh1F/F;Trps1F/F (Cdh1F/F;Trps1F/F) mice. (B) Kaplan-Meier analysis of mammary tumor free survival of Cdh1+/F;Trps1+/+ (n = 6), Cdh1+/F;Trps1F/F (n = 22), Cdh1F/F;Trps1+/+ (n = 19), and Cdh1F/F;Trps1F/F (n = 23) mice. Mantel-Cox: (***) P < 0.001; (*) P < 0.05; (ns) P > 0.05. (C) Representative images of H&E staining, and TRPS1 and E-cadherin expression detected by immunohistochemistry in end-stage tumors derived from Cdh1+/F;TRPS1F/F, and Cdh1F/F;TRPS1F/F mice, and a healthy duct. Scale bars, 50 μm. (D) Distribution of tumor burden, determined in four mammary glands per mouse upon sacrifice. χ2: (***) P < 0.001; (*) P < 0.05. (E) Kaplan-Meier analysis of overall survival of breast cancer patients with high TRPS1 versus low TRPS1 expressing IDCs (left) or ILCs (right) from the METABRIC breast cancer data set (Curtis et al. 2012; Pereira et al. 2016). TRPS1 high: Z-score > 0; TRPS1 low: Z-score < 0. Mantel-Cox. (F) Kaplan-Meier analysis of overall survival of breast cancer patients with high TRPS1 versus low TRPS1 expressing ILCs from the METABRIC breast cancer data set (Curtis et al. 2012; Pereira et al. 2016), the RATHER project (Michaut et al. 2016), and Metzger-Filho et al. (2013) combined.
The decreased latency of WAP-cre;Cdh1F/F;Trps1F/F mice resulted in eight out of 23 mice that were sacrificed due to the tumor burden, compared with two out of 19 WAP-cre;Cdh1F/F;Trps1+/+ mice (Supplemental Fig. S7F). Similar to WAP-cre;Cdh1F/F;Trps1+/+ mice, about two-thirds of the full-blown mammary tumors that arose in WAP-cre;Cdh1F/F;Trps1F/F mice were classic or solid ILCs, while one-third of tumors were sarcomatoid tumors (Supplemental Fig. S7G). The mammary tumors that arose in WAP-cre;Cdh1F/F;Trps1F/F animals were negative for both E-cadherin and TRPS1 (Fig. 6C). In addition, overall tumor burden was determined in four glands per mouse. Interestingly, both the percentage of glands with a >10% tumor burden, and the percentage of glands with no lesions at all were higher in WAP-cre;Cdh1F/F;Trps1F/F mice compared with WAP-cre;Cdh1F/F;Trps1+/+ mice (Fig. 6D). The proliferative index of small lesions and full-blown tumors (as determined by immunofluorescence staining for Ki-67) was similar between WAP-cre;Cdh1F/F;Trps1+/+ and WAP-cre;Cdh1F/F;Trps1F/F mice (Supplemental Fig. S7H–J). Also lung metastases were observed to a similar extent in WAP-cre;Cdh1F/F;Trps1+/+ and WAP-cre;Cdh1F/F;Trps1F/F mice (Supplemental Fig. S7K). RNA sequencing was performed on full-blown mammary tumors derived from WAP-cre;Cdh1F/F;Trps1+/+ and WAP-cre;Cdh1F/F;Trps1F/F mice. Two WAP-cre;Cdh1F/F;Trps1F/F tumors showed gene expression patterns comparable with WAP-cre;Cdh1F/F;Trps1+/+ tumors, which showed both ILC and sarcomatoid morphologies (Supplemental Fig. S8A–C, mixed). WAP-cre;Cdh1F/F;Trps1F/F ILCs appeared to be most different from WAP-cre;Cdh1F/F;Trps1+/+ tumors (Supplemental Fig. S8A,B,D, ILC classic/solid). Also two WAP-cre;Cdh1F/F;Trps1F/F sarcomatoid tumors showed differential gene expression changes compared with WAP-cre;Cdh1F/F;Trps1+/+ tumors (Supplemental Fig. S8A,B,E, sarcomatoid). Interestingly, in patients with invasive ductal carcinoma (IDC), overall survival was not affected by TRPS1 expression status (Fig. 6E; Supplemental Table S4). In contrast, we observed a statistically significant association of low TRPS1 expression with poor overall survival of patients with ILC (Fig. 6F; Supplemental Table S4). Together, these results confirm that loss of TRPS1 accelerates tumor formation induced by E-cadherin loss. Hence, the oncogenic potential of TRPS1 loss depends on the E-cadherin status and TRPS1 represents a novel ILC subtype-specific tumor suppressor.
Discussion
In this study, we set out to identify the role of the transcription factor TRPS1 both in mammary gland development and tumor formation. We show that suppression of interferon signaling by TRPS1, mediated by interactions with the NuRD complex, is essential for lactogenic differentiation capacity of nontransformed mammary cells. TRPS1 expression is also indispensable for in vivo survival of luminal epithelial cells during mammary gland development, but TRPS1 loss is tolerated in mammary epithelial cells that have lost E-cadherin. Moreover, combined loss of TRPS1 and E-cadherin boosts persistent proliferation in mammary organoids and induces mammary tumors in mice.
In line with previous reports (Serandour et al. 2018; Wang et al. 2018a; Witwicki et al. 2018), we found TRPS1 to interact with members of the NuRD transcriptional repressor complex, which regulates gene transcription by histone deacetylation and ATP-dependent chromatin remodeling (Lai and Wade 2011). Activity of the NuRD complex is involved in gene regulation of numerous processes including pluripotency during embryonic development or transcriptional events involved in cancer formation or progression (Basta and Rauchman 2015). TRPS1 stabilizes HDAC2 expression, and reduced cell viability caused by low TRPS1 levels can be rescued by HDAC2 overexpression (Wang et al. 2018b), even though we did not observe any changes in HDAC1 or HDAC2 expression upon TRPS1 loss. In addition, we now show that DNA binding of the core enzymatic components of the NuRD complex (HDAC1 and HDAC2) is reduced upon TRPS1 loss. Together, this indicates that TRPS1 acts as a transcriptional regulator by recruiting the NuRD complex to the DNA.
It is evident that TRPS1 expression is essential during embryonic development. Constitutive Trps1 mutant mice display severe developmental abnormalities mimicking the human tricho–rhino–phalangeal syndrome (TRPS), but die shortly after birth due to respiratory distress (Malik et al. 2002; Suemoto et al. 2007). The development of Cre-conditional Trps1F mice in this study allows for detailed in vivo analysis of the effects of TRPS1 loss on mouse mammary gland development and mammary tumor formation. We identified TRPS1 to be essential for survival of luminal epithelial cells during mammary gland development. Interestingly, loss of TRPS1 in mammary epithelial cells was associated with increased expression of STAT1 and STAT2, proteins that are differentially activated during different stages of mammary gland differentiation and involution during pregnancy. STAT1 activity is highest in virgin and late involuting glands and lower during gestation and lactation, while STAT5 is activated during gestation and lactation (Philp et al. 1996; Hughes and Watson 2012; Haricharan and Li 2014). STAT1 and STAT2 are also essential components of interferon signaling. Since this signaling cascade is of high importance in the tumor microenvironment, future research is required to determine whether TRPS1 might have a role in regulating interferon signaling in the tumor microenvironment and metastasis formation. Together, these findings identify TRPS1 as an essential regulator of STAT-signaling, required for mammary gland differentiation and involution.
Intriguingly, TRPS1 shares several features with the transcription factor GATA3. Both transcription factors are of the GATA-type and, similar to TRPS1, expression of GATA3 is required for survival of luminal epithelium in the adult mammary gland (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007). Similar to TRPS1, GATA3 is essential during embryonic development and reduced GATA3 expression by germline mutations and deletions in humans results in the HDR syndrome, characterized by Hypoparathyroidism, Sensorineural deafness, and Renal anomalies (Van Esch et al. 2000; Nesbit et al. 2004). Interestingly, TRPS1 and GATA3 form a complex with the IRX5 transcription factor. This tripartite complex regulates craniofacial development and germline mutations in either TRPS1, GATA3, or IRX5 cause craniofacial dysmorphisms (Bonnard et al. 2012). Together, these findings suggest that TRPS1 and GATA3 have similar or overlapping biological functions during embryonic development and postnatal mammary gland development.
Both TRPS1 and GATA3 have been implicated in breast cancer. GATA3 is one of the most frequently mutated genes in human breast cancer (Usary et al. 2004; The Cancer Genome Atlas Network 2012). Most mutations in GATA3 are heterozygous and predicted to result in a truncated GATA3 protein. Although the exact function of these truncating GATA3 mutations remains elusive, they are enriched in IDCs compared with ILCs (Stephens et al. 2012; Ciriello et al. 2015). In contrast to GATA3, the relevance of TRPS1 in breast cancer is still unclear. The most common aberrations reported in TRPS1 are amplifications that are associated with poor survival (Radvanyi et al. 2005; Chen et al. 2010; Serandour et al. 2018). It is however unclear whether TRPS1 is a bona fide driver of these amplifications, or whether it is simply coamplified with MYC, which is located close to TRPS1. TRPS1 is highly expressed in healthy mammary epithelial cells and expression is associated with differentiated tumors and a favorable clinical outcome in breast cancer patients (Lin et al. 2017), suggesting that TRPS1 is a marker for luminal epithelial cells.
We found TRPS1 expression to be essential for proliferation and differentiation of nontransformed epithelial cells in vitro and in mammary gland development in vivo. TRPS1 is also shown to be essential and required for growth of (mostly hormone receptor-positive) breast cancers (Witwicki et al. 2018). Indeed, several breast cancer cell lines show reduced growth both in vitro and in vivo upon loss of TRPS1 expression (Elster et al. 2018; Wang et al. 2018b; Witwicki et al. 2018). In other settings, loss of TRPS1 expression appears to be promoting mammary tumor growth. In vivo transposon mutagenesis screens identified TRPS1 as a candidate tumor suppressor gene in TNBC (Rangel et al. 2016) and ILC (Kas et al. 2017; this study). Hence, the effects of TRPS1 loss appear to be context dependent. Why TRPS1 loss is beneficial in ILC remains elusive and warrants further investigation in future studies. In this study, we provide direct evidence for the context-dependent tumor suppressor role of TRPS1 by demonstrating that the effect of TRPS1 loss in mammary epithelial cells is strongly dependent on their E-cadherin status. In an E-cadherin-proficient context, TRPS1 expression is essential for survival, while in an E-cadherin-deficient context, TRPS1 loss promotes persistent proliferation and mammary tumor formation.
Materials and methods
Plasmids and transfection/transduction methods
The truncated Trps1 sequence was isolated from mouse Trps1 (GE Dharmacon/Horizon Discovery MMM1013-202741213) cDNA using specific PCR primers (Supplemental Table S2A) with NheI and BamHI overhangs using Q5 high-fidelity DNA polymerase (New England Biolabs M0492) and subsequently cloned into a zero TOPO blunt vector (ThermoFisher Scientific 450245). The sequence-verified cDNA was inserted as a NheI–BamHI fragment into the lentiviral pCDH-CMV-MCS-EF1-copGFP (System Biosciences CD511B-1) vector. Broad Institute's Mission TRC-1 mouse library lentiviral shRNAs were used targeting Trps1 or Stat1 (Supplemental Table S2B). Lentiviral particles were produced by cotransfection of four plasmids in HEK293T cells as described previously (Follenzi et al. 2000). Conditioned media containing the lentiviral particles were collected 48 h after transfection and used for transduction. Stable cell lines were obtained after FACS sorting for GFP or puromycin (1.8 μg/mL; Sigma-Aldrich P7255) selection.
Nontargeting and sgRNAs targeting mouse Trps1 (Supplemental Table S2C) were cloned into a modified version of the pX330-U6-Chimeric_BB-CBh-SpCas9 (Addgene plasmid #42230) backbone, in which a puromycin resistance gene was introduced under the Pgk1 promoter (Harmsen et al. 2018). DNA oligos were phosphorylated, annealed, and subsequently ligated into the BbsI-digested pX330 backbone (Cong et al. 2013). All constructs were sequence verified by Sanger sequencing. Transfection was performed using Lipofectamine 2000 transfection reagent (ThermoFisher Scientific 11668-019).
Immunoprecipitation
Protein lysates were prepared using RIPA lysis buffer (10 mM Tris-HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% deoxycholate in milli-Q) supplemented with protease inhibitors. The whole-cell lysates were incubated with 100 μL of Protein A or G magnetic beads (ThermoFisher Scientific 10002D/10004D) prebound with anti-TRPS1 (R&D Systems), anti-GATA3, normal goat IgG control, or normal mouse IgG control (Supplemental Table S3). The immunoprecipitates were analyzed using Western blot after elution in sample buffer at 95°C or using LC/MS-MS analysis (Supplemental Material).
RNA expression analysis
Total RNA was extracted from cells using TRIzol reagent (ThermoFisher Scientific 15596026). For qPCR analysis, cDNA was synthesized from 1 μg of RNA using Tetro cDNA synthesis kit (Bioline BIO-65043) with random hexamer primers. qPCR was performed using SensiMix SYBR low-ROX kit (Bioline BIO-QT625) on a QuantStudio 6 system (Thermo Fisher Scientific). Primer sequences used are listed in Supplemental Table S2D. RNA sequencing details are in the Supplemental Material.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (Singh et al. 2019). In short, cells were cross-linked in 1% formaldehyde for 10 min. After addition of glycine (final concentration of 125 mM) to quench the cross-linking reaction and washing with PBS, cells were collected. The Bioruptor Pico (Diagenode SA) was used for sonication. The antibodies used are listed in Supplemental Table S3 together with Protein A or G magnetic beads. Isolated DNA was used for qPCR (primers in Supplemental Table S2E) and/or sequencing (Supplemental Material).
Mouse models
LoxP sites were targeted to the Trps1 locus in FVB zygotes using CRISPR/Cas9, as described previously (Pritchard et al. 2017). LoxP sites were inserted in intron 1 and downstream from exon 6, thereby flanking the complete coding region of Trps1. Sequences of the guideRNAs and homology-directed repair oligos are listed in Supplemental Table S2F. Mice with correct integration of both LoxP sites were intercrossed to produce Trps1F/F mice. The truncated Trps1 sequence was isolated from mouse Trps1 (GE Dharmacon/Horizon Discovery MMM1013-202741213) cDNA using specific PCR primers (Supplemental Table S2G) with FseI and PmeI overhangs using Q5 high-fidelity DNA polymerase (New England Biolabs M0492) and subsequently cloned into a zero TOPO blunt vector (Thermo Fisher Scientific 450245). Sequence-verified cDNA was inserted as FseI–PmeI fragment into the Frt-invCAG-IRES-Luc vector (Huijbers et al. 2015). Flp-mediated integration of the shuttle vector in WAP-cre;Cdh1F/F;Col1a1frt/+ embryonic stem cell (ESC) clones (FVB) and subsequent blastocyst injections of the modified ESCs were performed as described previously, resulting in chimeric mice carrying the Col1a1invCAG-mTrps1trunc-IRES-Luc (Trps1trunc) allele (Huijbers et al. 2015). Mice carrying the Trps1F or the Trps1trunc allele were crossed with mice carrying MMTV-cre, mT/mG reporter, WAP-cre or Cdh1F/F alleles (FVB) to generate cohorts with the required genotypes (Derksen et al. 2006, 2011; Muzumdar et al. 2007). The Trps1F, Trps1trunc, MMTV-cre, mT/mG, WAP-cre and Cdh1F alleles were confirmed by multiplex PCR using MyTaq HS Red Mix (Bioline BIO-25048) with an annealing temperature of 60°C (Supplemental Table S2H). Mice were monitored twice weekly for the development of palpable tumors. Mice were sacrificed when (total) mammary tumor burden reached a size of 1500 mm3 (0.5 × length × width2) or when the mice showed signs of pain or distress (e.g., weight loss and behavioral changes). All mouse experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute and performed in accordance with institutional, national, and European guidelines for animal care and use.
Immunofluorescence
Immunofluorescence stainings were performed as described previously on formalin-fixed tissue sections, cells, or organoids, with adaptations (Pasic et al. 2011; Boelens et al. 2016). The samples were permeabilized using 0.8% Triton X-100 and blocked using 2.5% BSA. All samples were incubated overnight with primary antibodies, followed by incubation with Alexa fluor-conjugated secondary antibodies and staining with Hoechst (Supplemental Table S3). The samples were mounted using Aqua-Poly/Mount (Polysciences, Inc. 18606-20 ). Images were acquired using a Leica TCS SP5 confocal and analyzed using LAS AF software (v2.6.3). Confocal images were analyzed automatically using an in-house-developed ImageJ macro that detects nuclei based on the DAPI signal and measures either the GFP signal in a band around each nucleus or the percentage of Ki-67-positive nuclei.
Statistics
Statistical analyses were performed with Graphpad Prism (v7.03). Statistical tests used were t-test, one-way ANOVA, two-way ANOVA, log-rank (Mantel-Cox) test, and χ2 test. P-values of <0.05 were considered to be significant.
Additional experimental details are described in the Supplemental Material. All sequencing data generated in this study are available on GEO repository (GSE133075).
Supplementary Material
Acknowledgments
We are grateful to Sjors Kas, Julian de Ruiter, Eva Schut, Ute Boon, Bram van den Broek, and Sjoerd Klarenbeek for their technical support with the experiments. We thank the Netherlands Cancer Institute Mass Spectrometry Facility, Genomics Core Facility, Mouse Clinic for Cancer and Aging, Animal Facility, and Animal Pathology Facility for their expert technical support. This work is part of the Oncode Institute, which is partly financed by the Dutch Cancer Society. This work was financially supported by the Netherlands Organization for Scientific Research (NWO), Cancer Genomics Netherlands (CGCNL), Zenith 93512009, Vici 91814643), the European Research Council (ERC Synergy project CombatCancer), and a National Roadmap grant for Large-Scale Research facilities from NWO.
Author contributions: L.M.C., W.Z., and J.J. designed the research. L.M.C., A.P.D., E.v.d.B., R.d.B., C.E.J.P., and I.J.H. performed the research. L.M.C., R.d.B., W.Z., and J.J. analyzed the data. L.M.C., W.Z., and J.J. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.331371.119.
References
- Asselin-Labat M-L, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209. 10.1038/ncb1530 [DOI] [PubMed] [Google Scholar]
- Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. 1988. Prolactin regulation of β-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J 7: 2089–2095. 10.1002/j.1460-2075.1988.tb03048.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta J, Rauchman M. 2015. The nucleosome remodeling and deacetylase (NuRD) complex in development and disease. Transl Res J Lab Clin Med 165: 36–47. 10.1016/j.trsl.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns EMJJ, Klijn JGM, van Putten WLJ, van Staveren IL, Portengen H, Foekens JA. 1992. c-myc amplification is a better prognostic factor than HER2/neu amplification in primary breast cancer. Cancer Res 52: 1107–1113. [PubMed] [Google Scholar]
- Bharat A, Gao F, Margenthaler JA. 2009. Tumor characteristics and patient outcomes are similar between invasive lobular and mixed invasive ductal/lobular breast cancers but differ from pure invasive ductal breast cancers. Am J Surg 198: 516–519. 10.1016/j.amjsurg.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Boelens MC, Nethe M, Klarenbeek S, de Ruiter JR, Schut E, Bonzanni N, Zeeman AL, Wientjens E, van der Burg E, Wessels L, et al. 2016. PTEN Loss in E-Cadherin-deficient mouse mammary epithelial cells rescues apoptosis and results in development of classical invasive lobular carcinoma. Cell Rep 16: 2087–2101. 10.1016/j.celrep.2016.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, Nelson SF, Ababneh OH, Uz E, Güran T, Kayserili H, et al. 2012. Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nat Genet 44: 709–713. 10.1038/ng.2259 [DOI] [PubMed] [Google Scholar]
- Boussadia O, Kutsch S, Hierholzer A, Delmas V, Kemler R. 2002. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech Dev 115: 53–62. 10.1016/S0925-4773(02)00090-4 [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68: 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chen JQ, Litton J, Xiao L, Zhang H-Z, Warneke CL, Wu Y, Shen X, Wu S, Sahin A, Katz R, et al. 2010. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer 1: 21–33. 10.1007/s12672-010-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Bao Y, Litton J, Xiao L, Zhang H-Z, Warneke CL, Wu Y, Shen X, Wu S, Katz RL, et al. 2011. Expression and relevance of TRPS-1: a new GATA transcription factor in breast cancer. Horm Cancer 2: 132–143. 10.1007/s12672-011-0067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. 2015. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163: 506–519. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen PWB, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. 2006. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 10: 437–449. 10.1016/j.ccr.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Derksen PWB, Braumuller TM, van der Burg E, Hornsveld M, Mesman E, Wesseling J, Krimpenfort P, Jonkers J. 2011. Mammary-specific inactivation of E-cadherin and p53 impairs functional gland development and leads to pleomorphic invasive lobular carcinoma in mice. Dis Model Mech 4: 347–358. 10.1242/dmm.006395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elster D, Tollot M, Schlegelmilch K, Ori A, Rosenwald A, Sahai E, von Eyss B. 2018. TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells. Nat Commun 9: 3115 10.1038/s41467-018-05370-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Kurban M, Levy B, Christiano AM. 2012. Trps1 and its target gene Sox9 regulate epithelial proliferation in the developing hair follicle and are associated with hypertrichosis. PLoS Genet 8: e1003002 10.1371/journal.pgen.1003002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet 25: 217–222. 10.1038/76095 [DOI] [PubMed] [Google Scholar]
- Gai Z, Zhou G, Itoh S, Morimoto Y, Tanishima H, Hatamura I, Uetani K, Ito M, Muragaki Y. 2009. Trps1 functions downstream of Bmp7 in kidney development. J Am Soc Nephrol 20: 2403–2411. 10.1681/ASN.2008091020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K, Winandy S, Avitahl N. 1997. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol 15: 155–176. 10.1146/annurev.immunol.15.1.155 [DOI] [PubMed] [Google Scholar]
- Gouilleux F, Wakao H, Mundt M, Groner B. 1994. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 13: 4361–4369. 10.1002/j.1460-2075.1994.tb06756.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haricharan S, Li Y. 2014. STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol Cell Endocrinol 382: 560–569. 10.1016/j.mce.2013.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen T, Klaasen S, van de Vrugt H, te Riele H. 2018. DNA mismatch repair and oligonucleotide end-protection promote base-pair substitution distal from a CRISPR/Cas9-induced DNA break. Nucleic Acids Res 46: 2945–2955. 10.1093/nar/gky076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-Z, Chen M, Zeng M, Xu S-H, Zou F-Y, Chen D, Yan G-R. 2016. Down-regulation of TRPS1 stimulates epithelial–mesenchymal transition and metastasis through repression of FOXA1. J Pathol 239: 186–196. 10.1002/path.4716 [DOI] [PubMed] [Google Scholar]
- Hughes K, Watson CJ. 2012. The spectrum of STAT functions in mammary gland development. JAKSTAT 1: 151–158. 10.4161/jkst.19691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers IJ, Del Bravo J, Bin Ali R, Pritchard C, Braumuller TM, van Miltenburg MH, Henneman L, Michalak EM, Berns A, Jonkers J. 2015. Using the GEMM-ESC strategy to study gene function in mouse models. Nat Protoc 10: 1755–1785. 10.1038/nprot.2015.114 [DOI] [PubMed] [Google Scholar]
- Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S, Drenth AP, van der Burg E, Klijn C, ten Hoeve JJ, et al. 2017. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet 49: 1219–1230. 10.1038/ng.3905 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055. 10.1016/j.cell.2006.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. 2011. NuRD: a multi-faceted chromatin remodeling complex in regulating cancer biology. Nat Rev Cancer 11: 588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CI, Anderson BO, Daling JR, Moe RE. 2003. Trends in incidence rates of invasive lobular and ductal breast carcinoma. JAMA 289: 1421–1424. 10.1001/jama.289.11.1421 [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database hallmark gene set collection. Cell Syst 1: 417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-Y, Zeng D, Liang Y-K, Wei X-L, Chen C-F. 2017. GATA3 and TRPS1 are distinct biomarkers and prognostic factors in breast cancer: database mining for GATA family members in malignancies. Oncotarget 8: 34750–34761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SM, Shaw AC, Bikker H, Lüdecke H-J, van der Tuin K, Badura-Stronka M, Belligni E, Biamino E, Bonati MT, Carvalho DR, et al. 2015. Phenotype and genotype in 103 patients with tricho–rhino–phalangeal syndrome. Eur J Med Genet 58: 279–292. 10.1016/j.ejmg.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Malik TH, Shoichet SA, Latham P, Kroll TG, Peters LL, Shivdasani RA. 2001. Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J 20: 1715–1725. 10.1093/emboj/20.7.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik TH, von Stechow D, Bronson RT, Shivdasani RA. 2002. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho–rhino–phalangeal syndromes. Mol Cell Biol 22: 8592–8600. 10.1128/MCB.22.24.8592-8600.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG. 2005. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res 65: 11259–11264. 10.1158/0008-5472.CAN-05-2495 [DOI] [PubMed] [Google Scholar]
- Metzger-Filho O, Michiels S, Bertucci F, Catteau A, Salgado R, Galant C, Fumagalli D, Singhal SK, Desmedt C, Ignatiadis M, et al. 2013. Genomic grade adds prognostic value in invasive lobular carcinoma. Ann Oncol Off J Eur Soc Med Oncol 24: 377–384. 10.1093/annonc/mds280 [DOI] [PubMed] [Google Scholar]
- Michaut M, Chin S-F, Majewski I, Severson TM, Bismeijer T, de Koning L, Peeters JK, Schouten PC, Rueda OM, Bosma AJ, et al. 2016. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep 6: 18517 10.1038/srep18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni P, Glöckner G, Schmidt O, von Holtum D, Albrecht B, Gillessen-Kaesbach G, Hennekam R, Meinecke P, Zabel B, Rosenthal A, et al. 2000. Mutations in a new gene, encoding a zinc-finger protein, cause tricho–rhino–phalangeal syndrome type I. Nat Genet 24: 71–74. 10.1038/71717 [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis 45: 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nesbit MA, Bowl MR, Harding B, Ali A, Ayala A, Crowe C, Dobbie A, Hampson G, Holdaway I, Levine MA, et al. 2004. Characterization of GATA3 mutations in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. J Biol Chem 279: 22624–22634. 10.1074/jbc.M401797200 [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27: 1160–1167. 10.1200/JCO.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L, Eisinger-Mathason TSK, Velayudhan BT, Moskaluk CA, Brenin DR, Macara IG, Lannigan DA. 2011. Sustained activation of the HER1–ERK1/2–RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev 25: 1641–1653. 10.1101/gad.2025611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut S-J, et al. 2016. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 7: 11479 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp JAC, Burdon TG, Watson CJ. 1996. Differential activation of STATs 3 and 5 during mammary gland development. FEBS Lett 396: 77–80. 10.1016/0014-5793(96)01069-1 [DOI] [PubMed] [Google Scholar]
- Pritchard CEJ, Kroese LJ, Huijbers IJ. 2017. Direct generation of conditional alleles using CRISPR/Cas9 in mouse zygotes. Methods Mol Biol 1642: 21–35. 10.1007/978-1-4939-7169-5_2 [DOI] [PubMed] [Google Scholar]
- Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J, et al. 2005. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci 102: 11005–11010. 10.1073/pnas.0500904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel R, Lee S-C, Ban KH-K, Guzman-Rojas L, Mann MB, Newberg JY, Kodama T, McNoe LA, Selvanesan L, Ward JM, et al. 2016. Transposon mutagenesis identifies genes that cooperate with mutant Pten in breast cancer progression. Proc Natl Acad Sci 113: E7749–E7758. 10.1073/pnas.1613859113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia U-D, Chen B-J, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, et al. 2014. Integration of genomic data enables selective discovery of breast cancer drivers. Cell 159: 1461–1475. 10.1016/j.cell.2014.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Mohammed H, Miremadi A, Mulder KW, Carroll JS. 2018. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene 37: 5281–5291. 10.1038/s41388-018-0312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AA, Schuurman K, Nevedomskaya E, Stelloo S, Linder S, Droog M, Kim Y, Sanders J, van der Poel H, Bergman AM, et al. 2019. Optimized ChIP-seq method facilitates transcription factor profiling in human tumors. Life Sci Alliance 2: e201800115 10.26508/lsa.201800115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin AV, Kim ER, Ovchinnikov LP. 2007. Nucleocytoplasmic transport of proteins. Biochem Mosc 72: 1439–1457. 10.1134/S0006297907130032 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Loo PV, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. 2012. The landscape of cancer genes and mutational processes in breast cancer. Nature 486: 400–404. 10.1038/nature11017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson S, Lackner MR, Adai AT, Yu N, Kim H-J, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et al. 2011. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal 4: ra41 10.1126/scisignal.2001538 [DOI] [PubMed] [Google Scholar]
- Suemoto H, Muragaki Y, Nishioka K, Sato M, Ooshima A, Itoh S, Hatamura I, Ozaki M, Braun A, Gustafsson E, et al. 2007. Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev Biol 312: 572–581. 10.1016/j.ydbio.2007.10.001 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerød A, Kåresen R, Oh DS, Dressler LG, et al. 2004. Mutation of GATA3 in human breast tumors. Oncogene 23: 7669–7678. 10.1038/sj.onc.1207966 [DOI] [PubMed] [Google Scholar]
- Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, Beetz R, Bilous RW, Holdaway I, et al. 2000. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 406: 419–422. 10.1038/35019088 [DOI] [PubMed] [Google Scholar]
- Vos CB, Cleton-Jansen AM, Berx G, de Leeuw W, ter Haar N, van Roy F, Cornelisse CJ, Peterse JL, van de Vijver M. 1997. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br J Cancer 76: 1131–1133. 10.1038/bjc.1997.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lin X, Gong X, Wu L, Zhang J, Liu W, Li J, Chen L. 2018a. Atypical GATA transcription factor TRPS1 represses gene expression by recruiting CHD4/NuRD(MTA2) and suppresses cell migration and invasion by repressing TP63 expression. Oncogenesis 7: 96 10.1038/s41389-018-0108-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Wu L, Liu W, Wei G, Gong X, Liu Y, Ma Z, Ma F, Thiery JP, et al. 2018b. Tricho–rhino–phalangeal syndrome 1 protein functions as a scaffold required for ubiquitin-specific protease 4-directed histone deacetylase 2 de-ubiquitination and tumor growth. Breast Cancer Res BCR 20: 83 10.1186/s13058-018-1018-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwicki RM, Ekram MB, Qiu X, Janiszewska M, Shu S, Kwon M, Trinh A, Frias E, Ramadan N, Hoffman G, et al. 2018. TRPS1 is a lineage-specific transcriptional dependency in breast cancer. Cell Rep 25: 1255–1267.e5. 10.1016/j.celrep.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuelling M, Kaiser FJ, Buelens LA, Braunholz D, Shivdasani RA, Depping R, Vortkamp A. 2009. Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol 328: 40–53. 10.1016/j.ydbio.2009.01.012 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.