Abstract
Multiple clinical trials employing recombinant adeno-associated viral (rAAV) vectors have been initiated for neuromuscular disorders, including Duchenne and limb-girdle muscular dystrophies, spinal muscular atrophy, and recently X-linked myotubular myopathy (XLMTM). Our previous work on a canine model of XLMTM showed that a single rAAV8-cMTM1 systemic infusion corrected structural abnormalities within the muscle and restored contractile function, with affected dogs surviving more than 4 years post injection. This remarkable therapeutic efficacy presents a unique opportunity to identify the downstream molecular drivers of XLMTM pathology and to what extent the whole muscle transcriptome is restored to normal after gene transfer. Herein, RNA-sequencing was used to examine the transcriptomes of the Biceps femoris and Vastus lateralis in a previously described canine cohort that showed dose-dependent clinical improvements after rAAV8-cMTM1 gene transfer. Our analysis confirmed several dysregulated genes previously observed in XLMTM mice but also identified transcripts linked to XLMTM pathology. We demonstrated XLMTM transcriptome remodeling and dose-dependent normalization of gene expression after gene transfer and created metrics to pinpoint potential biomarkers of disease progression and correction.
Keywords: XLMTM, myotubularin, AAV, transcriptome, biomarkers, gene therapy
Mack and colleagues created a novel array of visualization metrics from RNA-sequencing data to quantify disease correction after AAV8-mediated gene transfer. In dogs suffering from X-linked myotubular myopathy and previously treated with increasing doses of recombinant AAV, they showed that transcriptome remodeling correlated with clinical observations, and they identified biomarkers of disease correction.
Introduction
The last 30 years have seen the field of neuromuscular disease (NMD) genetics grow from one known gene for Duchenne muscular dystrophy (DMD) in 1987 to hundreds of disease genes identified today. NMDs are complex diseases whose pathological processes are often not entirely understood. Gene therapy offers a unique treatment approach for single-gene NMDs, because fixing the root cause of the defect does not necessarily require a comprehensive understanding of the pathophysiology. However, successful gene therapy provides an extraordinary opportunity to delve more deeply into the complex molecular underpinnings of disease initiation, progression, and correction.
In the past several years, treatment breakthroughs have been made in small and large animal models of NMDs,1, 2, 3, 4, 5, 6, 7 while clinical benefits have been reported after a single injection of recombinant adeno-associated virus (rAAV) in patients with spinal muscular atrophy.8 In most of these studies, results consisted of correlations between transgene expression levels and clinical outcome measures. However, this level of insight may prove insufficient when sub-therapeutic effects are the outcome or, conversely, when adverse events occur in vivo,9, 10, 11 a limitation that strongly argues for a deeper understanding of disease mechanisms. Therefore, the identification of disease-associated gene dysregulation and signaling pathways that are not—or only partially—corrected after gene transfer is of considerable interest. Also, as more gene therapies are approved for NMDs, it will become increasingly important to track their long-term durability using non-invasive biomarkers that appear prior to clinical deterioration.
Recent advances in -omics technologies have provided valuable tools for the diagnosis of NMDs and for the understanding of disease mechanisms at the molecular level.12, 13, 14, 15, 16 In this study, we investigated the underlying mechanisms of rAAV-mediated correction of muscle pathology utilizing the canine model of X-linked myotubular myopathy (XLMTM). XLMTM is a centronuclear myopathy caused by mutations in the MTM1 gene, which encodes the myotubularin protein.17 Patients exhibit severe muscle weakness, breathing difficulties, and a high mortality rate in the first years of life.18,19 XLMTM muscle cells have smaller diameters, an abnormal architecture, and poor excitation-contraction coupling (ECC).20, 21, 22, 23 At the molecular level, myotubularin is involved in phospholipid metabolism, and its absence leads to structural and functional defects in key membrane compartments such as the sarcoplasmic reticulum, the T-tubule network, and the neuromuscular junction.20,24, 25, 26, 27 In addition, the insulin-like growth factor 1 (IGF1) and downstream signaling pathways (Akt, mTOR, autophagy, ubiquitin-proteasome) are dysregulated, which correlates with marked muscle hypotrophy.28, 29, 30, 31 During the past decade, the use of rAAV vectors in XLMTM mice and dogs demonstrated remarkable therapeutic efficacy, paving the way for a recently initiated clinical trial,3,5,24,32 (ClinicalTrials.gov: NCT03199469). Treated XLMTM dogs showed dose-dependent correction of multiple disease phenotypes, resulting in improved muscle function and survival up to 4 years of age.5,33 Here, we explored the transcriptional changes driving this correction of muscle morphology and function in treated XLMTM dogs using an unbiased, comprehensive RNA-sequencing (RNA-seq) approach. Untreated XLMTM dogs exhibited transcriptional changes consistent with well-described disease phenotypes, and several genes emerged as potential candidates to explain disease development and progression. Importantly, rAAV gene therapy leads to a dose-dependent restoration of the muscle transcriptome toward a more normal profile. We also present an array of metrics based on RNA-seq data to quantify gene therapy efficacy and distinguish the transcripts corrected to normal after treatment from those escaping correction. Finally, we highlight potential RNA biomarkers to monitor and predict the efficacy of current and future gene therapy protocols.
Results
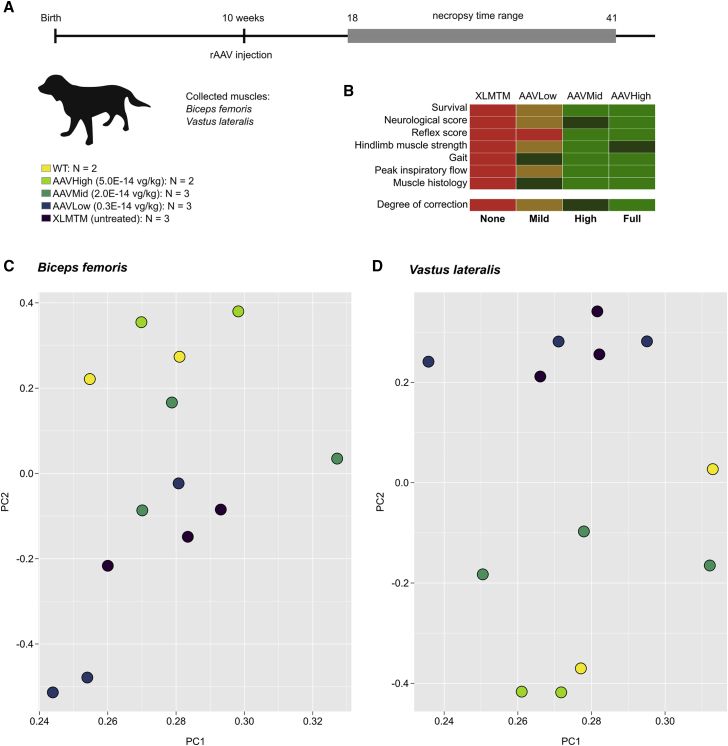
Unsupervised Analysis Discriminates XLMTM Dogs According to Treatment Groups
Muscle samples for the RNA-seq analysis herein were obtained from a previous study5 in which XLMTM dogs were randomized to receive one of three increasing doses of rAAV8-cMTM1 or saline as a negative control. Groups were named AAVLow (0.3E+14 vector genomes [vg]/kg), AAVMid (2.0E+14 vg/kg), AAVHigh (5.0E+14 vg/kg), and XLMTM, respectively, and were compared to wild-type (WT) controls (Figure 1A). The AAVMid and AAVHigh doses were characterized as therapeutic with full correction of disease phenotypes such as survival, neurological scores, and muscular and respiratory functions (Figure 1B).5 Conversely, the AAVLow dose resulted in sub-therapeutic clinical outcomes, including significant weakness, significant muscle pathology, and death of two animals. In this a posteriori study, the transcriptome of these dogs was analyzed using RNA-seq on two muscles of the hindlimb: Biceps femoris and Vastus lateralis. Due to premature death encountered in the XLMTM and AAVLow groups, dogs were not age matched, but samples were all collected at necropsy to reflect the final extent of disease progression or correction (Table S1). In each muscle, the top 500 most informative genes were selected based on their expression variance across samples and were used for principal component analysis (PCA). It showed segregation of the 13 dogs in 2 groups (Figures 1C and 1D): AAVLow and XLMTM dogs in one group; WT and AAVHigh dogs in the other. The AAVMid group also clustered with WT and AAVHigh dogs in the Vastus and was spread between the two groups in the Biceps. Gene ontology analysis was performed on the top 500 genes in each muscle and identified multiple terms related to muscle biology, particularly in the Vastus (Figure S1).
Figure 1.
Visualization of Gene Therapy Efficacy by Transcriptome Profiling in XLMTM Dogs
(A) Experimental timeline in weeks and details of the 5 groups of dogs. The rAAV vector was injected at 10 weeks of age, and dogs were monitored until the age of 39 to 41 weeks, unless euthanasia criteria were reached prematurely, as encountered in untreated XLMTM and AAVLow dogs. Additional details are provided in Table 1. (B) Heatmap representing the degree of corrections of key XLMTM phenotypes achieved in the three groups of dogs, as measured in our previous study.5 (C) Principal component analysis (PCA) on RNA-seq data in the Biceps femoris after selection of the top 500 most informative genes across samples. (D) PCA on RNA-seq data in the Vastus lateralis after selection of the top 500 most informative genes across samples. For (C) and (D), PC1 and PC2 were projected in a two-dimensional plot, with each dog represented as a color-coded dot.
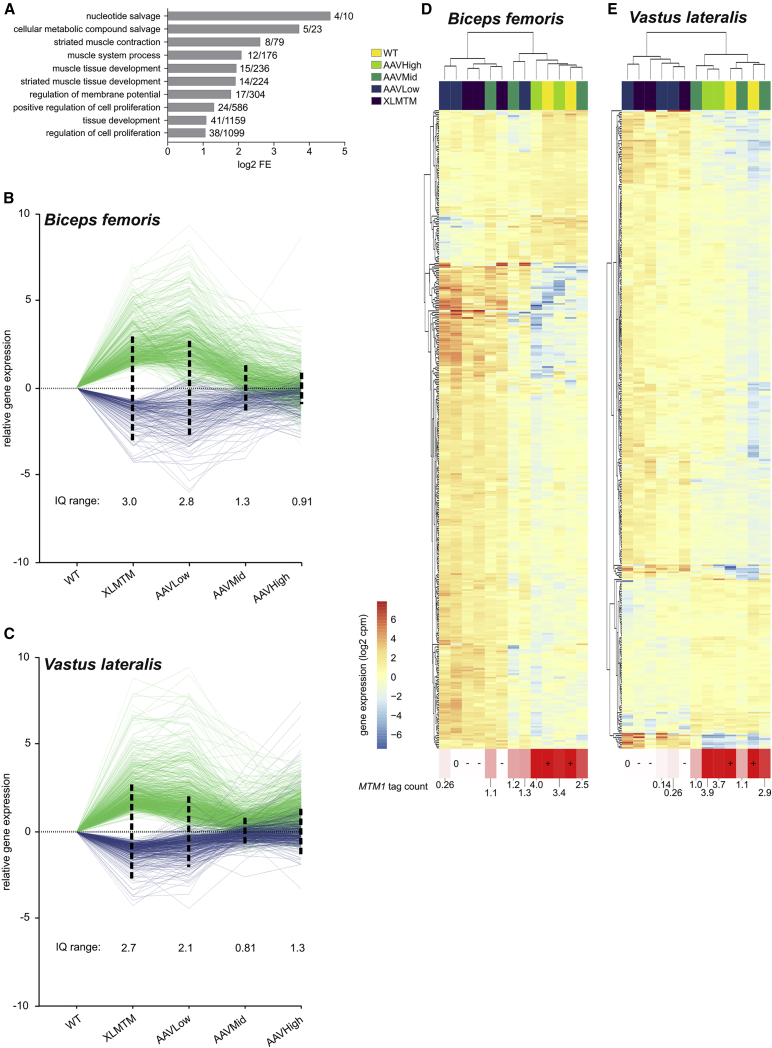
rAAV Gene Therapy Remodels the XLMTM Transcriptome in a Dose-Dependent Manner
To gain further insight into the development of XLMTM disease phenotypes in these two muscles, and their correction by gene therapy, we determined the differentially expressed (DE) genes between XLMTM dogs and WT controls using the edgeR package.34 In the Biceps and the Vastus, 824 and 1,122 DE genes were obtained, respectively. Among them, 400 were identified in both muscles, suggesting overlap in XLMTM disease processes between different muscles. Gene ontology analysis of the 400 common DE genes pointed to muscle development, muscle contraction, and various metabolic responses (Figure 2A). Of note, individual gene deregulations previously identified in XLMTM mice or in human patients were confirmed in the dog model. For instance, three acetylcholine receptor subunits were overexpressed in both muscles of XLMTM dogs: CHRNA1, CHRND, and CHRNG26,35,36 (Figure S2). Ryanodine receptor 1 (RYR1) and dihydropyridine receptor (CACNA1S) transcript levels were also both upregulated in XLMTM patient cells in vitro,37 and this result was confirmed in our datasets, although not significantly. Finally, the growth differentiation factor 8 (myostatin, MSTN) and its antagonist follistatin (FST) were also respectively down- and upregulated, similar to what was recently observed in Mtm1-knockout (KO) mice36,38 (Figure S2). Together, these data suggest an overlap between the transcriptome dysregulation induced by XLMTM in different species. The lists of DE genes in both Biceps and Vastus are available in Tables S2 and S3. Importantly, the expression profiles of the three CHRN genes, MSTN, and FST, were verified by qPCR, and comparison between the different groups of dogs showed a trend almost identical to the RNA-seq data (Figure S3). To visualize the extent of XLMTM transcriptome remodeling achieved by the three injected rAAV doses, we plotted the entire set of DE transcripts from both muscles of XLMTM untreated dogs and all three treatment groups relative to WT control dogs (log fold-change values, log FC) (Figures 2B and 2C). This plot showed that both upregulated and downregulated DE genes (in green and blue, respectively) were corrected, with log FC values progressively returning to zero. The log FC interquartile (IQ) range was then calculated to quantify transcriptome remodeling. This number represents the length of the interval comprising the central half (25%–75%) of ranked log FC values. Therefore, it estimates how the expression values of the DE gene set diverge from WT controls. In the Biceps, log FC IQ range values varied from 3.0 to 2.8, 1.3, and 0.91 in untreated XLMTM, AAVLow, AAVMid, and AAVHigh dogs, respectively (Figure 2B). In the Vastus, AAVMid dogs had a lower IQ range than AAVHigh dogs, reflecting a higher transcriptome remodeling effect with the intermediate rAAV dose (XLMTM, 2.7; AAVLow, 2.1; AAVMid, 0.81; and AAVHigh, 1.3; Figure 2C). Hierarchical clustering using the common DE genes distinguished therapeutic (AAVMid and AAVHigh) and sub-therapeutic (AAVLow) doses (Figure 2D). The only exception was observed between dog 04 (AAVMid) and dog 13 (AAVLow) Biceps samples, which clustered with the sub-therapeutic and therapeutic groups, respectively. Interestingly, disease correction in dog 13 (SSAN_18) was more pronounced than in the two other AAVLow dogs, and it did not reach euthanasia criteria before the end of the study period (Table S1).5 MTM1 transgene expression was then measured directly from RNA-seq data using a custom-made workflow for rAAV-derived mRNA tag counting and showed that transgene expression strongly correlated with the extent of transcriptome correction (Figure 2D). Transgene expression level in dog 13 Biceps was comparable to that of AAVMid dogs (MTM1 tag count = 1.3 versus a median of 1.2 in AAVMid dogs), suggesting that evaluation of transgene expression directly from RNA-seq data is a better predictor of transcriptome remodeling than the injected dose of gene therapy vector and that it reflects itself on overall treatment outcome. Therefore, injected Biceps samples were re-ordered in two distinct groups: the Therapeutic group with the two AAVHigh dogs, two AAVMid dogs, and dog 13; and the Sub-therapeutic group, with the two remaining AAVLow dogs, and dog 04. In the Vastus, all AAVMid and AAVHigh clustered together with WT controls and formed the Therapeutic group, while AAVLow dogs formed the Sub-therapeutic group.
Figure 2.
Remodeling of the XLMTM Transcriptome by rAAV Gene Therapy
(A) Gene ontology analysis on the differentially expressed genes common to XLMTM Biceps and Vastus muscles. For each represented term, the log2 fold enrichment (FE) values are graphically represented, together with the x/y ratio, defined as the number of genes participating to this enrichment (x) over the total number of genes affiliated with the corresponding ontology (y). (B) Relative expression of the differentially expressed genes in the Biceps, expressed in log fold change (log FC) after normalization with healthy controls. The interquartile ranges were determined from positive log FC values (negative log FC were converted into absolute values) and represented as dashed vertical lines pointing in both directions around the y = 0 line. (C) Similar representation for the differentially expressed genes detected in the Vastus muscle. (D and E) Hierarchical clustering generated from log cpm values of the 400 common differentially expressed genes for the Biceps (D) and the Vastus (E). Each column represents one muscle sample, color-coded for treatment group; each row represents one individual gene, color coded for its expression level (log cpm). RNA-seq-based MTM1 mRNA tag counts are indicated under each column.
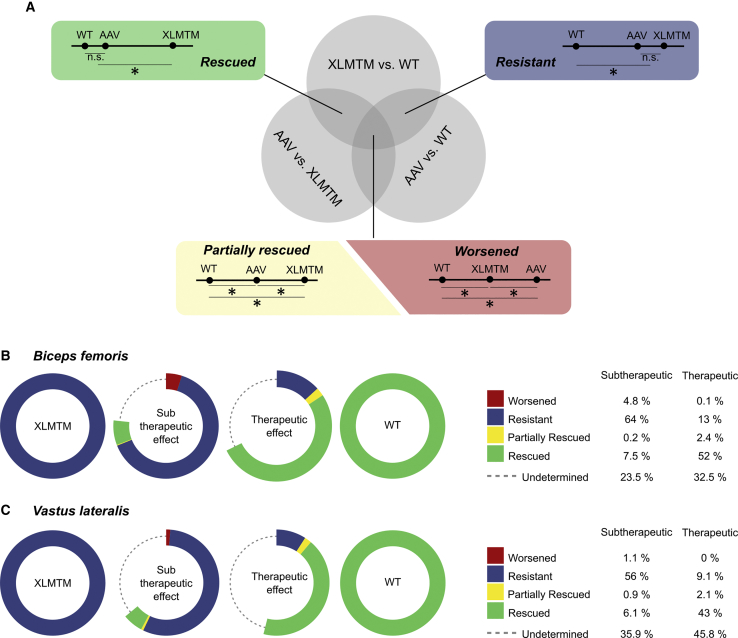
RNA-Seq Allows the Creation of Molecular Metrics of Gene Therapy Efficiency
Because distinct sub-populations of transcripts may respond differently to gene therapy, an array of metrics was created, based on the type of transcriptome remodeling effect. Rescued genes were defined as the XLMTM-associated DE genes that were no longer significantly different from WT controls in rAAV-treated dogs (Figure 3A). Conversely, Resistant genes were still differentially expressed between rAAV-treated dogs and WT controls. Two additional sub-populations of Partially Rescued and Worsened genes were also defined but were both marginally represented irrespective of the vector dose. These metrics were determined for the Sub-therapeutic and Therapeutic groups of dogs, in the Biceps and the Vastus muscles (Figures 3B and 3C). As anticipated, a sub-therapeutic effect was associated with only 6.1% and 7.5% of genes with a Rescued expression profile in the Biceps and Vastus muscles, respectively, while Resistant genes accounted for 64% and 57%. In sharp contrast, the proportion of Rescued genes reached 52.1% and 42.7% in the Biceps and Vastus of the dogs showing therapeutic benefits (Figures 3B and 3C). This indicates that an increase in MTM1 transgene expression drives a more profound restoration of the muscle transcriptome, providing for the first time a comprehensive transcriptomic explanation of physiological and histological outcome measures observed previously.5
Figure 3.
Quantifiers of Transcriptome Remodeling and Gene Therapy Efficacy in XLMTM Dogs
(A) Venn diagram representing the intersections of differentially expressed gene populations obtained after comparison of untreated XLMTM, rAAV-treated XLMTM, and healthy dogs. Focus is made on the Rescued and the Resistant two-circle domains in specific insets and on the central domain containing Partially Rescued and Worsened genes. Exemplar gene-expression levels are plotted on a line for the three groups: healthy, XLMTM, and rAAV-treated (AAV) dogs. n.s., non-significant gene expression difference; *, significant difference. (B and C) Evolution of the described metrics in the Sub-therapeutic and the Therapeutic groups of dogs—as defined in the text—in the Biceps (B) and the Vastus (C) muscles. The initial DE gene set in XLMTM dogs is represented as a blue ring, and its counterpart in healthy controls as a green ring. In rAAV-treated dogs, the areas corresponding to the above-described metrics have an arc length directly proportional to the proportion of initial DE genes presenting the corresponding expression profile.
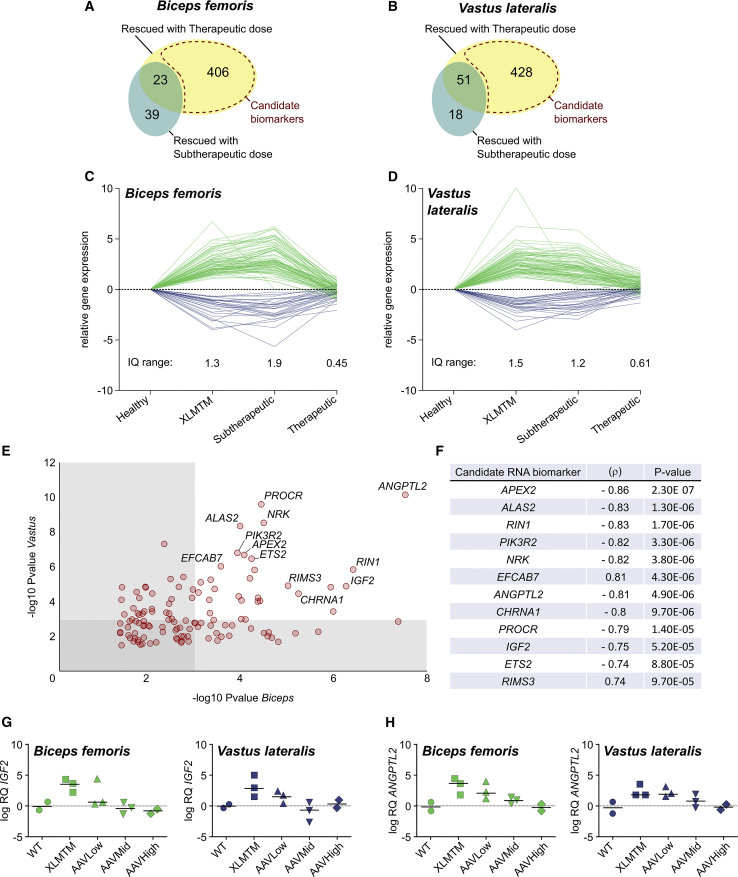
Differential Treatment Outcome Leads to Identification of Disease Biomarkers
Next, populations of Candidate Biomarkers of XLMTM correction were defined as the genes presenting a Rescued expression profile in the Therapeutic, but not in the Sub-therapeutic group of dogs (Figures 4A and 4B). This resulted in more than 400 genes in each muscle, 120 of which were detected in both Biceps and Vastus. As expected, the expression pattern of these 120 genes followed a similar trend, with sustained deregulation in the XLMTM and Sub-therapeutic groups, and a marked rescue in the Therapeutic group (Figures 4C and 4D). In the latter, IQ range values were only 0.45 and 0.61 in the Biceps and the Vastus, respectively. Finally, a list of 12 candidate biomarkers was isolated among the genes with the most significant expression changes in the two XLMTM muscles (p value < 0.001 in each muscle; Figure 4E) and their expression level was correlated with MTM1 tag counts in injected dogs and untreated controls (tag counts = 0, by definition). The most significant correlation was obtained for APEX2, an endonuclease involved in base-excision repair and progression through the cell cycle (correlation coefficient ρ = −0.86, p value = 2.3E−7). Interestingly, this list also contained ALAS2 (r = −0.83, p value = 1.3E−6) and PIK3R2 (r = −0.82, p value = 3.3E−6), two genes that were recently associated with muscle strength,39 and NRK (r = −0.82, p value = 3.8E−6), known to be specifically expressed during myotome formation and early muscle development in mice, but not in the adult.40 The expression of these 4 genes in injected dogs was inversely correlated with MTM1 transgene expression. Candidate Biomarkers also included RIN1, EFCAB7, ANGPTL2, CHRNA1, PROCR, IGF2, ETS2, and RIMS3 (Figure 4F). Interestingly, two of these genes are known to be secreted in the serum: IGF2 and ANGPTL2. Their expression was validated on independent biopsies by qPCR, which resulted in typical “biomarker” expression profiles (Figures 4G and 4H). The full lists of Rescued genes in both muscles and of Candidate Biomarkers are available in Tables S4 and S5.
Figure 4.
Candidate RNA Biomarkers of AAV Therapeutic Effect in XLMTM Dogs
(A and B) Venn diagrams showing the number of genes with a Rescued profile and their overlap between the Sub-therapeutic and the Therapeutic groups. The Candidate Biomarker gene subsets are delineated with a dashed red area, respectively in the Biceps (A) and the Vastus (B). (C) Relative expression of the candidate biomarkers in the Biceps (C) and the Vastus (D), expressed in log FC after normalization with healthy controls. The interquartile ranges were determined from positive log FC values (negative log FC were converted into absolute values). (E) Two-dimensional dot-plot of the 120 candidate biomarkers common to the Biceps and Vastus muscles, representing the significance of the differential expression between XLMTM dogs and healthy controls (p values). Each dot represents one gene. (F) Selection of twelve RNA biomarkers of gene therapy efficacy in XLMTM dogs, ranked according to their correlation coefficient (ρ) when compared to MTM1 mRNA tag counts. The significance of this correlation is indicated (p-value). (G and H) IGF2 (G) and ANGPTL2 (H) expression levels in both muscles assessed by qRT-PCR normalized with the housekeeping gene RPL32 and represented relative to WT dogs; RQ, relative quantity = 2−ΔΔCt; log, base 2 logarithm.
Discussion
With the recent success of preclinical and clinical trials using rAAV vectors, understanding the exact processes by which disease phenotypes are impacted—and eventually rescued—by gene therapy is of considerable interest for the field of molecular medicine. Here, we take advantage of the unprecedented therapeutic efficacy obtained in XLMTM dogs to investigate the molecular mechanisms of NMD correction, with a focus on pathological muscle transcriptome. Overall, our study demonstrates that corrective rAAV gene therapy induces a dose-dependent transcriptome remodeling, which was highly correlated with RNA-seq-based transgene expression measurements. These measures were comparable with our previous study despite being obtained by different techniques on distinct muscle biopsies5 and revealed high levels of recombinant MTM1 transcript in the dogs presenting with full disease correction.
For logistical and ethical considerations, it was decided to include both hemizygous MTM1-/y male and homozygous MTM1−/− female dogs in the study, since they both present comparable XLMTM phenotypes. Dog randomization into the different treatment groups led to uneven distribution of females in the untreated XLMTM and AAVLow groups, and therefore, sex-related differences cannot be entirely ruled out in this study.5 Nonetheless, unsupervised analyses have clearly demonstrated that the overall variability was driven by transgene expression, which can distinguish dogs showing therapeutic versus subtherapeutic responses to rAAV treatment.
To our knowledge, this study is one of the first to report a comprehensive transcriptome profile from myotubularin-deficient muscles, and the first in a large animal model. Previously, individual genes were queried in Mtm1-deficient mice as an attempt to explain clinical observations.26,29,38 In addition, muscle biopsies from 8 XLMTM patients have been analyzed for the expression of 4,200 genes using microarray and pointed to the upregulation of genes coding for cytoskeletal and extracellular matrix proteins.35 More recently, a publication used RNA-seq in Mtm1δ4 mice treated or not with tamoxifen, and highlighted a set of 849 differentially expressed genes between untreated Mtm1δ4 mice and WT controls.36 In our study, we gathered data on more than 32,000 dog genes, some of them not yet characterized. Importantly, several genes previously identified as differentially expressed in XLMTM patients and mice were also detected in dogs. In comparison with the mouse transcriptome,36 we found an overlap of 87 deregulated ortholog genes in our XLMTM dogs, and ranking of log FC values showed a high correlation between the two species (ρ = 0.6, p = 1.1E−9) (Table S6).
Among the overlap, we notably found three subunits of the nicotinic acetylcholine receptor: CHRNA1, CHRNG, and CHRND, whose deregulation might be related to the compromised ability of XLMTM muscle cells to generate Ca2+ transients in response to electrical stimulation.20,21 In this regard, genes coding for RYR1 and CACNA1S are upregulated in XLMTM patients’ cells,37 and were also slightly overexpressed in XLMTM dogs, although no significant differences were measured (Figure S2). Interestingly, XLMTM dogs also showed deregulation of multiple additional genes coding for ion transporters that could also lead to ECC defects, including sodium (SCN4B, SCN5A, SCNN1A) and potassium (KCNK17, KCNQ5) channels, the sodium/potassium-transporting ATPase subunit alpha-3 (ATP1A3), and members of the solute carrier (SLC) family.
Another major consequence of myotubularin deficiency is an early muscle hypotrophy observed in patients and animal models.22,23,32,41, 42, 43 XLMTM myotubes have a small diameter and centrally positioned nuclei, and they are often considered abnormally developed or unable to maintain a fully differentiated state. In XLMTM mice, these characteristics correlate with dysregulated PI3K/Akt/mTOR signaling and abnormal autophagy.29,30,44,45 More recently, another study reported the downregulation of the myostatin pathway in Mtm1-KO mice, which could be a compensatory mechanism to counter muscle hypotrophy.38 Herein, the genes coding for myostatin (MSTN) and follistatin (FST) were down- and upregulated, respectively, in XLMTM dogs. These outcomes confirmed the involvement of this pathway downstream of myotubularin deficiency (Figure S2), and the potential of myostatin as a therapeutic target. Indeed, treatment with ActRIIB-mFc, a myostatin inhibitor, resulted in muscle hypertrophy and increased lifespan in XLMTM mice.46 More recently, these mice were co-injected with an rAAV vector coding for an inhibitory myostatin propeptide in addition to rAAV-Mtm1, and this led to a greater increase in muscle mass than mice treated with rAAV-Mtm1 only.38 This finding demonstrates that the tools created in our study have the potential to identify relevant targets for combinatorial gene therapy applications.
In light of our RNA-seq data, abnormal muscle development appears as an important component of XLMTM biology in dogs. The genes encoding the myogenic regulators MyoD (MYOD1) and myogenin (MYOG) are overexpressed in XLMTM dogs, together with PAX7, a marker of adult muscle stem (satellite) cells (Tables S2 and S3). In addition, several genes coding for isoforms of structural proteins transiently expressed during skeletal muscle embryonic development were found upregulated, including the acetylcholine receptor γ subunit (CHRNG) and myosin heavy chain 3 (MYH3) in the Biceps, or cardiac alpha-actin (ACTC1) and troponin T (TNNT2) in the Vastus. Of note, XLMTM dogs reached euthanasia criteria prematurely and their median age at necropsy was 22 weeks, as opposed to 39 weeks in WT dogs. At this age, muscles are expected to be in their terminally differentiated state, as seen by histological examination. However, subtle differences in their transcriptome cannot be ruled out and might confound the expression of developmental genes. Overall, transcriptome profiling of canine XLMTM muscles not only confirmed gene-expression data in mice and human patients, but also helped identify multiple genes not previously associated with XLMTM biology, and whose deregulation provides insight into disease phenotypes.
Gene therapy studies rarely investigate the molecular consequences of target cell transduction. With rAAV vectors injected in new patients every year, now including boys suffering from XLMTM (clinical trial NCT03199469), a better understanding of the molecular mechanisms by which NMDs are corrected will become crucial. Recently, one study reported the normalization of microarray gene expression data after dual rAAV gene therapy in a mouse model of oculopharyngeal muscular dystrophy (OPMD).6 Concerning XLMTM, tamoxifen-treated Mtm1δ4 mice only showed 29 differentially expressed genes compared to untreated controls,36 but these data were obtained at an earlier time after treatment compared to our study. In these two publications,6,36 the authors did not focus on specific genes or gene sub-populations. Herein, we provide for the first time a comprehensive picture of muscle transcriptome remodeling after rAAV gene therapy using RNA-seq. We used hierarchical clustering to group injected dogs based on the extent of transcriptome remodeling rather than on the dose of rAAV injected in vivo. In agreement with previous clinical observations,5 this clustering approach resulted in a Sub-therapeutic and a Therapeutic group of dogs. Strikingly, the Biceps sample collected from dog 13 was part of the Therapeutic group despite a treatment with an otherwise sub-therapeutic dose of vector. However, inclusion in the Therapeutic group correlated with the extended lifespan and phenotypic improvements observed in this specific dog.5 To illustrate different facets of transcriptome remodeling, we created several metrics and determined the proportion of genes whose expression was normalized or not after treatment (Figure 3A). We demonstrated that these metrics can be used to identify dogs exhibiting therapeutic benefits from those in which gene therapy efficacy was not durable. In specific cases, it may also be used to determine transcripts that have been induced by gene therapy treatment as a side effect independent of phenotypic correction. More precisely, this population corresponds to genes that are deregulated in treated dogs but not in their untreated counterparts, when compared to WT controls. Herein, we detected only marginal proportions of genes presenting this expression pattern, which correlates with the absence of adverse events, and of any humoral or cellular immune responses against the transgene product in rAAV-injected dogs.5 Overall, we obtained confirmation that treatment was well-tolerated at the level of the muscle transcriptome.
Finally, this analytical approach proved very effective to identify RNA biomarkers of XLMTM correction. Interestingly, several of the top 12 candidate genes not only align with XLMTM phenotypes but could also help deepen our understanding of disease development: ALAS2 and PIK3R2 have recently been negatively associated with muscle strength in a meta-analysis of more than 7,500 human subjects39 and were also found overexpressed in XLMTM dogs; Nik-related kinase (NRK) is a protein kinase involved in actin polymerization through phosphorylation and inhibition of cofilin, is only expressed during embryonic myogenesis,40 and is overexpressed in XLMTM dogs, which adds to the list of dysregulated developmental genes; APEX2 encodes an endonuclease physically associated with proliferating cell nuclear antigen (PCNA) and driving cell-cycle progression through base-excision repair on the replicative nuclear and mitochondrial DNA.47,48 APEX2 is also overexpressed in untreated XLMTM muscles and corrected in the Therapeutic group, suggesting that de-regulation/re-regulation of the cell cycle is a key component of disease development and rescue. Altogether our data provide a transcriptional characterization of the structural immaturity encountered in XLMTM muscles.
RNA biomarkers defined in this study could also help identify protein products known to be secreted in the blood and, therefore, would be easily quantifiable. In this regard, at least two genes highlighted in our list could be further investigated at the protein level: angiopoietin-like protein 2 (ANGPTL2), an inflammatory mediator participating in blood vessel formation, and insulin-like growth factor 2 (IGF2). Recently, increased ANGPTL2 expression has been reported in a mouse model of denervation-induced muscle atrophy, and when knocked out, improved muscle growth and satellite cell activity.49 Herein, ANGPTL2 is overexpressed in untreated XLMTM dogs, and reduced to basal levels in the Therapeutic group. Thus, ANGPTL2 itself might be an important mediator of rAAV-mediated therapeutic benefits. As a proinflammatory cytokine, it is accurately quantifiable in blood samples, and is already used in the context of colorectal cancer.50 This makes it an excellent candidate biomarker to monitor the evolution of XLMTM after clinical gene therapy.
In conclusion, this study brings evidence of XLMTM rescue by rAAV gene therapy at the molecular level, in the form of a dose-dependent transcriptome remodeling acting on specific aspects of disease biology. Furthermore, it introduces an array of RNA biomarkers in the form of individual genes or carefully defined gene populations, which brings unprecedented insight to the mechanisms of NMD rescue by gene therapy.
Materials and Methods
Animal Model
The XLMTM Labrador/Beagles used in this study have a p.N155K mutation in the MTM1 gene and were issued from a colony maintained at the University of Washington.3,41 More precisely, they were previously included in a gene therapy study, the methods and results of which were extensively described in a recent publication.5 Animal experimentation was reviewed and supervised by the University of Washington Institutional Review Board. In brief, XLMTM dogs were infused with three increasing doses of a rAAV vector expressing a canine MTM1 cDNA: 0.3E+14, 2.0E+14, and 5.0E+14 vg/kg at the age of 10 weeks, and sacrificed between 39 and 41 weeks old or when reaching humane euthanasia criteria. In the absence of treatment, XLMTM dogs, both hemizygous males and homozygous females, exhibit progressive clinical deterioration between about 12 and 26 weeks of age, resulting in an inability to stand, walk, and feed. Eight injected dogs were included in this study, together with 3 untreated XLMTM and 2 WT controls, as summarized in Table S1. Necropsy samples were collected from two muscles of the hindlimb to increase statistical power and not restrict the analysis to only one replicate per dog.
RNA Extraction and Sequencing
Muscle necropsy tissues were frozen on dry ice and stored at ≤−60°C. RNA extraction, library preparation, and sequencing on an Illumina HiSeq 4000 were conducted by Genewiz (NJ, USA). Specifically, total RNA was extracted with QIAGEN AllPrep mini kit. RNA samples were quantified using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked with an RNA Screen Tape on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing library preparation was prepared with a TruSeq Stranded mRNA Library Prep kit following the manufacturer’s protocol (Illumina, category number RS-122-2101). Sequencing libraries were validated using a DNA Analysis Screen Tape on the Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified by using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA), as well as by qPCR (Applied Biosystems, Carlsbad, CA, USA). Sequencing libraries were multiplexed and clustered on flow cell using the cBOT from Illumina. After clustering, the flow cell was loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 Pair-End (PE) High Output configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS) on the HiSeq instrument. Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina bcl2fastq program version 2.17. One mismatch was allowed for index sequence identification. The 13 Vastus-mRNA-derived libraries were sequenced to an average of 60 M paired-end reads/sample and the 13 Biceps-mRNA-derived libraries were sequenced to an average of 20 M paired-end reads/sample.
RNA Extraction and qRT-PCR
Dog tissues were homogenized using MagNa Pure LC RNA Isolation Tissue Lysis Buffer (Roche) and total RNA was isolated using automated nucleic acid extraction instruments, MagNA Pure 96 System (Cellular RNA Large Volume Kit [Roche]; 350 μL sample input, 50 μL sample output). RNA was subject to DNase treatment to remove genomic DNA contamination (Ambion, DNA-free DNA Removal Kit, Life Technologies) and reverse transcribed using random hexamers and RevertAid H minus Reverse Transcriptase (Fermentas) (250 ng of total RNA). qPCR was performed in a LightCycler 480 system (Roche) by using 4 μL of 1/10 diluted cDNA, SyberGreen mix (Thermo Fisher Scientific) (ANGPTL2), Taqman Gene Expression (Thermo Fisher Scientific) (CHRNA1, CHRND, CHRNG, IGF2, FST, MSTN) and primers (RPL32) listed below (Table 1).
Table 1.
Details of the Primers, Probe, and Gene-Expression Arrays Used in the qRT-PCR Validation Experiment
| RPL32 | Forward primer |
5′-TGGTTACGGGAGCAACAAGAAA-3′ |
|---|---|---|
| Reverse primer |
5′-GCACATCAGCAGCACTTCA-3′ |
|
| Probe | 5′-TGCTGCCCAGTGGCTTCTGG-3′ - VIC-QSY | |
| CHRNA1 | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02621957_m1 - FAM/MGB-NFQ | |
| CHRND | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02691404_m1 - FAM/MGB-NFQ | |
| CHRNG | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02704147_m1 - FAM/MGB-NFQ | |
| IGF2 | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02693069_m1 - FAM/MGB-NFQ | |
| FST | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02645950_m1 - FAM/MGB-NFQ | |
| MSTN | Thermo Fisher Scientific – TaqMan Gene Expression - Cf02704228_m1 - FAM/MGB-NFQ | |
| ANGPTL2 | QIAGEN - qRT-PCR2 primer assay for dog ANGPTL2 – product 330001 | |
Data Analysis
Initial data analysis, including alignment to reference genome and count table generation, was conducted on the DNAnexus platform (Palo Alto, CA, USA). Specifically, fastq files were first mapped onto the reference genome of Canis lupus familiaris (genome assembly: CanFam3.1 [GCA_000002285.2], downloaded from https://www.ensembl.org) using the Subread mapping package51,52 (settings: subread-align -t 0 -T 4 -d 50 -D 600 -i my_index -r read1 -S ff -R read2 -o $output_name). The resulting bam files were fed into featureCounts51 package to obtain the count-per-gene table (settings: featureCounts -p -P -d 50 -D 600 -t exon -g gene_id -a GTF_file -o $output_name bam_file1 bam_file2 bam_file3 bam_file4 bam_file5 bam_file6 bam_file7 bam_file8 bam_file9 bam_file10 bam_file11 bam_file12 bam_file13). Count tables were further analyzed with the open-source RStudio environment for R (https://www.r-project.org/) and the Bioconductor software (https://www.bioconductor.org/). The limma52 and edgeR34 packages were used to normalize, fit, and compare the data between groups following the analysis pipeline detailed here.53 Cutoff values for DE gene determination were as follows: p value < 0.05 and fold-change (FC) > 2.0. The code used to analyze count tables is comprehensively detailed in the Supplemental Information. Gene ontology analyses were performed on the GO consortium online platform (http://geneontology.org/), using the statistical overrepresentation test and the Bonferroni correction for multiple testing. Enrichments with a corrected p value lower than 0.05 were considered significant.
Data Representation
Heatmaps and PCA plots were automatically generated in R-Bioconductor. Other data representations were realized on Graphpad PRISM 7 and figures were assembled using the vector graphics Inkscape software. The color palette used in this manuscript to distinguish dog groups was extracted from the viridis package on R-Bioconductor and was carefully selected to be easily interpretable by individuals with colorblindness (https://cran.r-project.org/web/packages/viridis/vignettes/intro-to-viridis.html).
Data Availability
The RNA-sequencing data from this publication have been deposited to the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra), and assigned the Bioproject ID PRJNA49575. The read files are available at the following link: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP165150.
Author Contributions
J.-B.D. designed and performed the experiments, analyzed the data and wrote the manuscript. J.G. designed the experiments, analyzed the data, and assisted in drafting the manuscript. E.R.-G., K.P., and V.L. performed the experiments and analyzed the data. M.W.L. processed the canine muscle samples and assisted in drafting the manuscript. R.W.G. performed the assessments of muscle force and assisted in drafting the manuscript. J.T.G. designed the experiments and assisted in drafting the manuscript. A.B.-B. provided the gene therapy vector, designed the experiments, and assisted in drafting the manuscript. M.K.C. designed the experiments. D.L.M. designed the experiments, analyzed the data, and wrote the manuscript.
Conflicts of Interest
M.W.L. is a member of advisory boards for Audentes Therapeutics, Solid Biosciences, and Ichorion Therapeutics. He is also a consultant for Wave Life Sciences and Dynacure. A.B.-B. and M.K.C. are inventors of a patent on gene therapy for myotubular myopathy.
Acknowledgments
This work was funded by NIH/NIAMS R01-HL115001, the Will Cure Foundation, the Joshua Frase Foundation, the Association Française contre les Myopathies (AFM-Telethon), Myotubular Trust UK, and Audentes Therapeutics, Inc. (San Francisco, CA, USA).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.10.018.
Supplemental Information
References
- 1.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Corrigendum: Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:16007. doi: 10.1038/ncomms16007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Childers M.K., Joubert R., Poulard K., Moal C., Grange R.W., Doering J.A., Lawlor M.W., Rider B.E., Jamet T., Danièle N. Gene therapy prolongs survival and restores function in murine and canine models of myotubular myopathy. Sci. Transl. Med. 2014;6:220ra10. doi: 10.1126/scitranslmed.3007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Guiner C., Servais L., Montus M., Larcher T., Fraysse B., Moullec S., Allais M., François V., Dutilleul M., Malerba A. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun. 2017;8:16105. doi: 10.1038/ncomms16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mack D.L., Poulard K., Goddard M.A., Latournerie V., Snyder J.M., Grange R.W., Elverman M.R., Denard J., Veron P., Buscara L. Systemic AAV8-Mediated Gene Therapy Drives Whole-Body Correction of Myotubular Myopathy in Dogs. Mol. Ther. 2017;25:839–854. doi: 10.1016/j.ymthe.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malerba A., Klein P., Bachtarzi H., Jarmin S.A., Cordova G., Ferry A., Strings V., Espinoza M.P., Mamchaoui K., Blumen S.C. PABPN1 gene therapy for oculopharyngeal muscular dystrophy. Nat. Commun. 2017;8:14848. doi: 10.1038/ncomms14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozsgai E.R., Griffin D.A., Heller K.N., Mendell J.R., Rodino-Klapac L.R. Systemic AAV-Mediated β-Sarcoglycan Delivery Targeting Cardiac and Skeletal Muscle Ameliorates Histological and Functional Deficits in LGMD2E Mice. Mol. Ther. 2017;25:855–869. doi: 10.1016/j.ymthe.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 9.Gicquel E., Maizonnier N., Foltz S.J., Martin W.J., Bourg N., Svinartchouk F., Charton K., Beedle A.M., Richard I. AAV-mediated transfer of FKRP shows therapeutic efficacy in a murine model but requires control of gene expression. Hum. Mol. Genet. 2017;26:1952–1965. doi: 10.1093/hmg/ddx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell J.R., Campbell K., Rodino-Klapac L., Sahenk Z., Shilling C., Lewis S., Bowles D., Gray S., Li C., Galloway G. Dystrophin immunity in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomsen G.M., Alkaslasi M., Vit J.-P., Lawless G., Godoy M., Gowing G., Shelest O., Svendsen C.N. Systemic injection of AAV9-GDNF provides modest functional improvements in the SOD1G93A ALS rat but has adverse side effects. Gene Ther. 2017;24:245–252. doi: 10.1038/gt.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterfield R.J., Dunn D.M., Hu Y., Johnson K., Bönnemann C.G., Weiss R.B. Transcriptome profiling identifies regulators of pathogenesis in collagen VI related muscular dystrophy. PLoS ONE. 2017;12:e0189664. doi: 10.1371/journal.pone.0189664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., Genotype-Tissue Expression Consortium Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal5209. eaal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gama-Carvalho M., L Garcia-Vaquero M., R Pinto F., Besse F., Weis J., Voigt A., Schulz J.B., De Las Rivas J. Linking amyotrophic lateral sclerosis and spinal muscular atrophy through RNA-transcriptome homeostasis: a genomics perspective. J. Neurochem. 2017;141:12–30. doi: 10.1111/jnc.13945. [DOI] [PubMed] [Google Scholar]
- 15.Liang W.-C., Tian X., Yuo C.-Y., Chen W.-Z., Kan T.-M., Su Y.-N., Nishino I., Wong L.C., Jong Y.J. Comprehensive target capture/next-generation sequencing as a second-tier diagnostic approach for congenital muscular dystrophy in Taiwan. PLoS ONE. 2017;12:e0170517. doi: 10.1371/journal.pone.0170517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang W.-C., Tian X., Yuo C.-Y., Chen W.-Z., Kan T.-M., Su Y.-N., Nishino I., Wong L.C., Jong Y.J. Correction: Comprehensive target capture/next-generation sequencing as a second-tier diagnostic approach for congenital muscular dystrophy in Taiwan. PLoS ONE. 2017;12:e0183406. doi: 10.1371/journal.pone.0183406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laporte J., Hu L.J., Kretz C., Mandel J.L., Kioschis P., Coy J.F., Klauck S.M., Poustka A., Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 18.Amburgey K., Tsuchiya E., de Chastonay S., Glueck M., Alverez R., Nguyen C.-T., Rutkowski A., Hornyak J., Beggs A.H., Dowling J.J. A natural history study of X-linked myotubular myopathy. Neurology. 2017;89:1355–1364. doi: 10.1212/WNL.0000000000004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beggs A.H., Byrne B.J., De Chastonay S., Haselkorn T., Hughes I., James E.S., Kuntz N.L., Simon J., Swanson L.C., Yang M.L. A multicenter, retrospective medical record review of X-linked myotubular myopathy: The recensus study. Muscle Nerve. 2018;57:550–560. doi: 10.1002/mus.26018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A.H., Allard B., Mandel J.L. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. USA. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutchukian C., Lo Scrudato M., Tourneur Y., Poulard K., Vignaud A., Berthier C., Allard B., Lawlor M.W., Buj-Bello A., Jacquemond V. Phosphatidylinositol 3-kinase inhibition restores Ca2+ release defects and prolongs survival in myotubularin-deficient mice. Proc. Natl. Acad. Sci. USA. 2016;113:14432–14437. doi: 10.1073/pnas.1604099113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawlor M.W., Beggs A.H., Buj-Bello A., Childers M.K., Dowling J.J., James E.S., Meng H., Moore S.A., Prasad S., Schoser B., Sewry C.A. Skeletal Muscle Pathology in X-Linked Myotubular Myopathy: Review With Cross-Species Comparisons. J. Neuropathol. Exp. Neurol. 2016;75:102–110. doi: 10.1093/jnen/nlv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shichiji M., Biancalana V., Fardeau M., Hogrel J.-Y., Osawa M., Laporte J., Romero N.B. Extensive morphological and immunohistochemical characterization in myotubular myopathy. Brain Behav. 2013;3:476–486. doi: 10.1002/brb3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C.R., Durand M., Kretz C., Danos O., Douar A.M. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 2008;17:2132–2143. doi: 10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowling J.J., Joubert R., Low S.E., Durban A.N., Messaddeq N., Li X., Dulin-Smith A.N., Snyder A.D., Marshall M.L., Marshall J.T. Myotubular myopathy and the neuromuscular junction: a novel therapeutic approach from mouse models. Dis. Model. Mech. 2012;5:852–859. doi: 10.1242/dmm.009746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amoasii L., Hnia K., Chicanne G., Brech A., Cowling B.S., Müller M.M., Schwab Y., Koebel P., Ferry A., Payrastre B., Laporte J. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle in vivo. J. Cell Sci. 2013;126:1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 28.Razidlo G.L., Katafiasz D., Taylor G.S. Myotubularin regulates Akt-dependent survival signaling via phosphatidylinositol 3-phosphate. J. Biol. Chem. 2011;286:20005–20019. doi: 10.1074/jbc.M110.197749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Qusairi L., Prokic I., Amoasii L., Kretz C., Messaddeq N., Mandel J.-L., Laporte J. Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin-proteasome pathways. FASEB J. 2013;27:3384–3394. doi: 10.1096/fj.12-220947. [DOI] [PubMed] [Google Scholar]
- 30.Fetalvero K.M., Yu Y., Goetschkes M., Liang G., Valdez R.A., Gould T., Triantafellow E., Bergling S., Loureiro J., Eash J. Defective autophagy and mTORC1 signaling in myotubularin null mice. Mol. Cell. Biol. 2013;33:98–110. doi: 10.1128/MCB.01075-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavriilidis C., Laredj L., Solinhac R., Messaddeq N., Viaud J., Laporte J., Sumara I., Hnia K. The MTM1-UBQLN2-HSP complex mediates degradation of misfolded intermediate filaments in skeletal muscle. Nat. Cell Biol. 2018;20:198–210. doi: 10.1038/s41556-017-0024-9. [DOI] [PubMed] [Google Scholar]
- 32.Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J.-F., Mandel J.L. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. USA. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elverman M., Goddard M.A., Mack D., Snyder J.M., Lawlor M.W., Meng H., Beggs A.H., Buj-Bello A., Poulard K., Marsh A.P. Long-term effects of systemic gene therapy in a canine model of myotubular myopathy. Muscle Nerve. 2017;56:943–953. doi: 10.1002/mus.25658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noguchi S., Fujita M., Murayama K., Kurokawa R., Nishino I. Gene expression analyses in X-linked myotubular myopathy. Neurology. 2005;65:732–737. doi: 10.1212/01.wnl.0000174625.67484.4d. [DOI] [PubMed] [Google Scholar]
- 36.Maani N., Sabha N., Rezai K., Ramani A., Groom L., Eltayeb N., Mavandadnejad F., Pang A., Russo G., Brudno M. Tamoxifen therapy in a murine model of myotubular myopathy. Nat. Commun. 2018;9:4849. doi: 10.1038/s41467-018-07057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bachmann C., Jungbluth H., Muntoni F., Manzur A.Y., Zorzato F., Treves S. Cellular, biochemical and molecular changes in muscles from patients with X-linked myotubular myopathy due to MTM1 mutations. Hum. Mol. Genet. 2017;26:320–332. doi: 10.1093/hmg/ddw388. [DOI] [PubMed] [Google Scholar]
- 38.Mariot V., Joubert R., Hourdé C., Féasson L., Hanna M., Muntoni F., Maisonobe T., Servais L., Bogni C., Le Panse R. Downregulation of myostatin pathway in neuromuscular diseases may explain challenges of anti-myostatin therapeutic approaches. Nat. Commun. 2017;8:1859. doi: 10.1038/s41467-017-01486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilling L.C., Joehanes R., Kacprowski T., Peters M., Jansen R., Karasik D., Kiel D.P., Harries L.W., Teumer A., Powell J. Gene transcripts associated with muscle strength: a CHARGE meta-analysis of 7,781 persons. Physiol. Genomics. 2016;48:1–11. doi: 10.1152/physiolgenomics.00054.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanai-Azuma M., Kanai Y., Okamoto M., Hayashi Y., Yonekawa H., Yazaki K. Nrk: a murine X-linked NIK (Nck-interacting kinase)-related kinase gene expressed in skeletal muscle. Mech. Dev. 1999;89:155–159. doi: 10.1016/s0925-4773(99)00193-8. [DOI] [PubMed] [Google Scholar]
- 41.Beggs A.H., Böhm J., Snead E., Kozlowski M., Maurer M., Minor K., Childers M.K., Taylor S.M., Hitte C., Mickelson J.R. MTM1 mutation associated with X-linked myotubular myopathy in Labrador Retrievers. Proc. Natl. Acad. Sci. USA. 2010;107:14697–14702. doi: 10.1073/pnas.1003677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goddard M.A., Mack D.L., Czerniecki S.M., Kelly V.E., Snyder J.M., Grange R.W., Lawlor M.W., Smith B.K., Beggs A.H., Childers M.K. Muscle pathology, limb strength, walking gait, respiratory function and neurological impairment establish disease progression in the p.N155K canine model of X-linked myotubular myopathy. Ann. Transl. Med. 2015;3:262. doi: 10.3978/j.issn.2305-5839.2015.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierson C.R., Dulin-Smith A.N., Durban A.N., Marshall M.L., Marshall J.T., Snyder A.D., Naiyer N., Gladman J.T., Chandler D.S., Lawlor M.W. Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum. Mol. Genet. 2012;21:811–825. doi: 10.1093/hmg/ddr512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joubert R., Vignaud A., Le M., Moal C., Messaddeq N., Buj-Bello A. Site-specific Mtm1 mutagenesis by an AAV-Cre vector reveals that myotubularin is essential in adult muscle. Hum. Mol. Genet. 2013;22:1856–1866. doi: 10.1093/hmg/ddt038. [DOI] [PubMed] [Google Scholar]
- 45.Lawlor M.W., Viola M.G., Meng H., Edelstein R.V., Liu F., Yan K., Luna E.J., Lerch-Gaggl A., Hoffmann R.G., Pierson C.R. Differential muscle hypertrophy is associated with satellite cell numbers and Akt pathway activation following activin type IIB receptor inhibition in Mtm1 p.R69C mice. Am. J. Pathol. 2014;184:1831–1842. doi: 10.1016/j.ajpath.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawlor M.W., Read B.P., Edelstein R., Yang N., Pierson C.R., Stein M.J., Wermer-Colan A., Buj-Bello A., Lachey J.L., Seehra J.S., Beggs A.H. Inhibition of activin receptor type IIB increases strength and lifespan in myotubularin-deficient mice. Am. J. Pathol. 2011;178:784–793. doi: 10.1016/j.ajpath.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ide Y., Tsuchimoto D., Tominaga Y., Nakashima M., Watanabe T., Sakumi K., Ohno M., Nakabeppu Y. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104:4097–4103. doi: 10.1182/blood-2004-04-1476. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J., Tian Z., Kadomatsu T., Xie P., Miyata K., Sugizaki T., Endo M., Zhu S., Fan H., Horiguchi H. Age-dependent increase in angiopoietin-like protein 2 accelerates skeletal muscle loss in mice. J. Biol. Chem. 2018;293:1596–1609. doi: 10.1074/jbc.M117.814996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toiyama Y., Tanaka K., Kitajima T., Shimura T., Kawamura M., Kawamoto A., Okugawa Y., Saigusa S., Hiro J., Inoue Y. Elevated serum angiopoietin-like protein 2 correlates with the metastatic properties of colorectal cancer: a serum biomarker for early diagnosis and recurrence. Clin. Cancer Res. 2014;20:6175–6186. doi: 10.1158/1078-0432.CCR-14-0007. [DOI] [PubMed] [Google Scholar]
- 51.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 52.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law C.W., Alhamdoosh M., Su S., Dong X., Tian L., Smyth G.K., Ritchie M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res. 2016;5:1408. doi: 10.12688/f1000research.9005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data from this publication have been deposited to the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra), and assigned the Bioproject ID PRJNA49575. The read files are available at the following link: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP165150.