Abstract
Communication between the gametophytes is vital for angiosperm fertilisation. Multiple Cr RLK1L‐type receptor kinases prevent premature pollen tube burst, while another Cr RLK1L protein, FERONIA (FER), is required for pollen tube reception in the female gametophyte. We report here the identification of two additional Cr RLK1L homologues, HERCULES RECEPTOR KINASE 1 (HERK1) and ANJEA (ANJ), which act redundantly to promote pollen tube growth arrest at the synergid cells. HERK1 and ANJ localise to the filiform apparatus of the synergid cells in unfertilised ovules, and in herk1 anj mutants, a majority of ovules remain unfertilised due to pollen tube overgrowth, together indicating that HERK1 and ANJ act as female determinants for fertilisation. As in fer mutants, the synergid cell‐specific, endomembrane protein NORTIA (NTA) is not relocalised after pollen tube reception; however, unlike fer mutants, reactive oxygen species levels are unaffected in herk1 anj double mutants. Both ANJ and HERK1 associate with FER and its proposed co‐receptor LORELEI (LRE) in planta. Together, our data indicate that HERK1 and ANJ act with FER to mediate female–male gametophyte interactions during plant fertilisation.
Keywords: angiosperm, CrRLK1L, fertilisation, receptor kinase, synergid cells
Subject Categories: Development & Differentiation, Plant Biology, Signal Transduction
The Cr RLK1L receptor kinases HERK1 and ANJ are genetically redundant during pollen tube reception in Arabidopsis. Both proteins interact with the Cr RLK1L receptor FER and its putative co‐receptor LRE and contribute to NTA relocalisation but not to ROS production.

Introduction
Fertilisation is a critical point in the life cycle of any sexually reproducing organism. In flowering plants, gametes are enclosed in gametophytes, multicellular structures that develop in the reproductive organs of the flower. The pollen grain constitutes the male gametophyte, with each grain generating a pollen tube in the form of a rapidly growing cellular protrusion that delivers the male gametes, or sperm cells, through the style tissues into the ovule. Female gametophytes develop inside the ovule and contain the female gametes within an embryo sac; the egg cell and central cell. The process of double fertilisation in angiosperms consists of the fusion of a sperm cell with each of the female gametes. If fertilisation is successful, the embryo and endosperm develop from the egg cell and central cell fertilisations, respectively. For double fertilisation to occur, the male and female gametophytes must engage in a molecular dialog that controls pollen tube attraction towards the ovule entrance, or micropyle, the arrest of pollen tube growth and the release of the sperm cells in the correct location within the ovule (see Refs 1, 2 for a detailed review).
The synergid cells occupy the micropylar portion of the female gametophyte and aid communication between the gametophytes. As such, their cytoplasm is densely occupied by endomembrane compartments, reflective of a highly active secretion system generating messenger molecules 3. The filiform apparatus appears at the outermost pole, a thickened and intricate cell wall structure that represents the first contact point between female and male gametophytes prior to fertilisation 4. Synergid cells secrete small cysteine‐rich LURE and XIUQIU peptides to guide pollen tubes towards the embryo sac 5, 6. AtLURE1 peptides are sensed by two pairs of pollen‐specific receptor‐like kinases (RLKs), MALE DISCOVERER 1 (MDIS1) and MDIS1‐INTERACTING RLK 1 (MIK1), and POLLEN‐SPECIFIC RECEPTOR KINASE 6 (PRK6) and PRK3 in Arabidopsis 7, 8. These RLKs bind AtLURE1 peptides through their extracellular domains at the growing tip of the pollen tubes, promoting their exit from the transmitting tract in a species‐specific manner 6, 7, 8, 9. XIUQIU peptides, on the other hand, attract pollen tubes towards the synergid cells regardless of the species, and signal through a pollen tube receptor that is yet to be described 6.
Within the expanded family of RLKs in Arabidopsis, the Catharanthus roseus RLK1‐like (CrRLK1L) subfamily has been demonstrated to play several roles during fertilisation (see Ref. 10 for a detailed review). Two pairs of functionally redundant CrRLK1Ls are integral in controlling pollen tip growth; ANXUR1 and 2 (ANX1/2), and BUDDHA'S PAPER SEAL 1 and 2 (BUPS1/2), heterodimerise and ensure pollen tube growth by sensing of two autocrine secreted peptides belonging to the RAPID ALKALINIZATION FACTOR (RALF) family, RALF4 and RALF19 11, 12, 13, 14. A fifth CrRLK1L protein, ERULUS (ERU), has also been implicated in male‐determined pollen tube growth via regulation of Ca2+ oscillations 15. The CrRLK1L protein FERONIA (FER) accumulates in the filiform apparatus of the synergids where it functions as a female determinant of pollen tube reception and subsequent sperm cell release 16, 17. Although no extracellular ligand has been identified for FER in a reproductive context, there is evidence for FER activation of a synergid‐specific signalling cascade upon pollen tube arrival. This signalling pathway involves the glycosyl‐phosphatidylinositol (GPI)‐anchored protein LORELEI (LRE) 18, 19, activation of NADPH oxidases to generate reactive oxygen species (ROS) in the micropyle 20, generation of specific Ca2+ signatures in the synergid cytoplasm 21, and relocalisation of the Mildew resistance locus O (MLO)‐like NORTIA (NTA), an endomembrane compartment protein that affects pollen tube‐induced Ca2+ signatures in the synergids 21, 22, 23.
CrRLK1L receptor kinases have also been assigned a number of other functions beyond fertilisation. For example, cell elongation during vegetative growth requires several members of the CrRLK1L family; HERCULES RECEPTOR KINASE 1 and 2 (HERK1 and 2), THESEUS1 (THE1) and FER 24, 25. FER has also been linked to pathogen responses 26, while THE1 and other CrRLK1L receptors detect cell wall integrity 27.
Many questions remain about the nature of the communication between gametophytes that controls sperm cell release, and CrRLK1Ls FER, ANX1/2 and BUPS1/2 are potential receptor candidates to mediate this dialog. Here, we report the characterisation of CrRLK1Ls HERK1 and ANJEA (AT5G59700; ANJ) as female determinants of pollen tube reception in Arabidopsis. We show that HERK1 and ANJ act redundantly at the filiform apparatus of the synergids to control pollen tube growth arrest, representing two new mediators of gametophytic communication and therefore expanding the female‐specific toolbox required for fertilisation.
Results
HERK1 and ANJ function redundantly in seed set
To test whether additional Arabidopsis CrRLK1L proteins are involved in reproduction, we obtained T‐DNA insertion lines for all 17 family members. Presence of a homozygous insertion was verified for ten CrRLK1L genes. These verified lines were crossed and double homozygous plants selected in the F2 generation by PCR genotyping (Fig EV1A and B for T‐DNA lines used further in this study). Stable double homozygous lines were qualitatively examined for fertility. Through this screen, we identified that double mutants in HERCULES RECEPTOR KINASE 1 (HERK1) and AT5G59700 (hereafter referred to as ANJEA/ANJ) have high rates of unfertilised ovules or seeds that abort very early in development, and shorter siliques (Fig 1A). The qualitative nature of our preliminary screen for fertility defects in CrRLK1L mutants does not preclude the involvement of additional CrRLK1Ls in reproduction as quantitative investigation may uncover more subtle fertility defects among the mutants of this family of receptors. HERK1 and ANJEA are close homologues within the CrRLK1L family 28, with 75% identity and 86% similarity at the amino acid level. Loss of ANJ gene expression in the double homozygous herk1‐1 anj‐1 T‐DNA line (hereafter referred to as herk1 anj) was confirmed by RT–qPCR (Fig EV1C). Although the herk1‐1 T‐DNA insertion has previously been reported to knockout gene expression 24, our RT–qPCR results indicate that transcripts are present at wild‐type levels 5′ of the T‐DNA insertion, and at ~ 20% of wild‐type levels 3′ of the T‐DNA insertion. Whether these transcripts are translated into truncated proteins would require generation of αHERK1 antibodies. However, as the herk1‐1 anj‐1 phenotype can be complemented by the expression of HERK1 and the herk1‐1 anj‐1 phenotype is equivalent to other mutants in the pathway (see below), we conclude that herk1‐1 likely does not act as a dominant negative or hypermorphic allele within a reproductive context.
Figure EV1. Confirmation of ANJEA gene expression knock out and genotyping of T‐DNA lines used in this study.
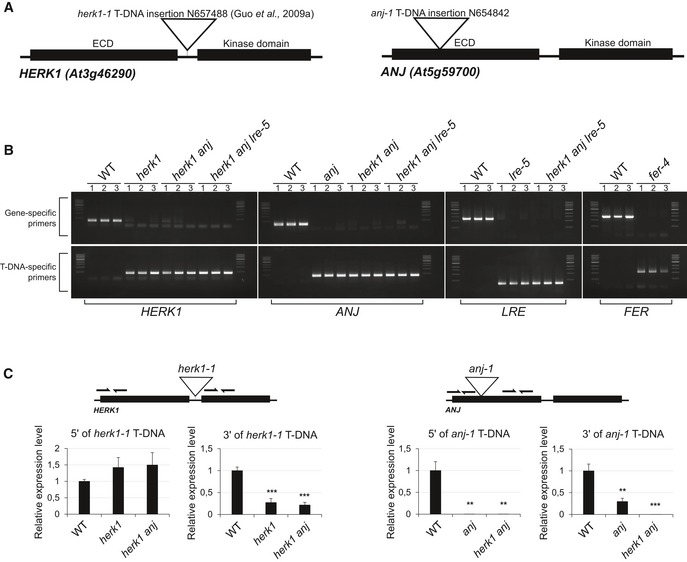
- Domain organisation of HERK1 and ANJEA and T‐DNA insertion sites in the lines used in this study, herk1‐1 and anj‐1.
- Genotyping PCRs to verify homozygosity in the lines used in this study. DNA from three independent seedlings per line was analysed.
- RT–qPCR analysis of HERK1 gene expression in wild‐type, herk1 and herk1 anj plants, and ANJ gene expression in wild‐type, anj and herk1 anj plants. RNA was extracted from multiple inflorescences from three plants per line. Data presented are means ± SD. **P < 0.01; ***P < 0.001 (Student's t‐test).
Source data are available online for this figure.
Figure 1. The herk1 anj fertility defect is caused by maternally mediated pollen tube overgrowth.
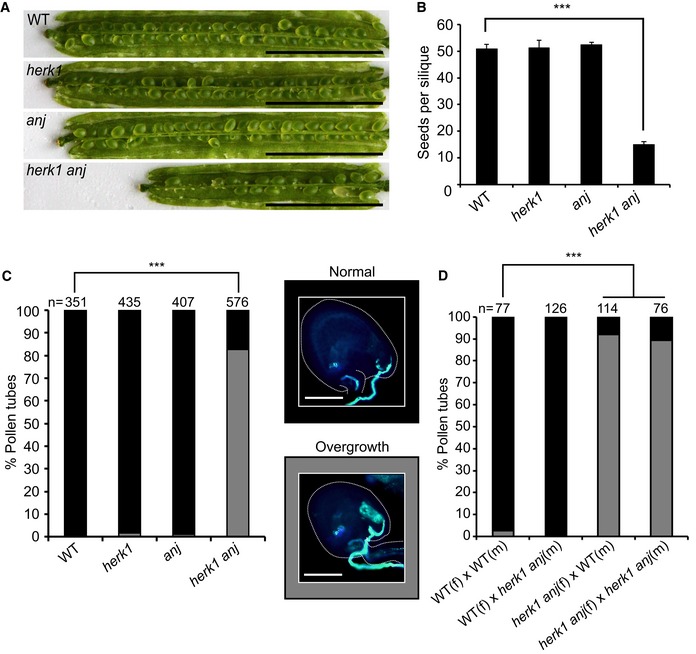
- Representative siliques from wild‐type (WT; Col‐0), herk1, anj and herk1 anj plants prior to dehiscence. Siliques were placed on double‐sided sticky tape and carpel walls separated from the replum to expose the developing seeds. Scale bar = 5 mm.
- Developing seeds per silique in wild‐type, herk1, anj and herk1 anj plants. Fully expanded siliques were dissected and photographed under a stereomicroscope. n = 15 (four independent experiments with at least three plants per line and five siliques per plant). Data presented are means ± SEM. ***P < 0.001 (Student's t‐test).
- Percentage of pollen tubes with normal reception at the female gametophyte (black bars; representative image middle centre of figure) and with overgrowth (grey bars; representative image lower centre) as assessed by aniline blue staining. Fifteen self‐pollinated stage 16 flowers from wild‐type, herk1, anj and herk1 anj were analysed. Legend scale bars = 50 μm. ***P < 0.001 (χ2 tests).
- Aniline blue staining of pollen tube reception in reciprocal crosses between wild‐type and herk1 anj plants with at least two siliques per cross. Legend as per (C). ***P < 0.001 (χ2 tests).
Source data are available online for this figure.
To verify that the low rate of seed set results from functional redundancy between HERK1 and ANJ, we examined seed development in dissected siliques of wild‐type, herk1, anj and herk1 anj plants grown in parallel. While single mutants herk1 and anj did not have elevated numbers of unfertilised/aborted seeds compared to wild type, a high proportion of ovules in herk1 anj siliques had not developed into mature seeds, leading to a reduced number of seeds per silique (Fig 1B). We therefore concluded that there is functional redundancy between the HERK1 and ANJ proteins during fertilisation or early seed development.
HERK1 has previously been described to influence cell elongation in vegetative tissues with THE1 and HERK2, with the herk1 the1‐4 and herk1 herk2 the1‐4 mutants displaying a short petiole phenotype, similarly to fer mutants 24, 25. We further examined the herk1 anj mutants for developmental defects in vegetative and reproductive growth, finding no other developmental aberrations (Appendix Fig S1). Thus, HERK1 and ANJ do not act redundantly during vegetative growth.
HERK1 and ANJ are female determinants of pollen tube reception
Previous studies of CrRLK1L proteins where mutation results in low or absent seed set have identified functions in pollen tube growth (ANX1, ANX2, BUPS1, BUPS2 and ERU 11, 12, 13, 14, 15) and female‐mediated pollen tube growth arrest at the synergids (FER 17). To test which step in fertilisation is impaired in the herk1 anj mutant, we tracked pollen tube growth through the style and ovary in single and double mutants. In all plant lines, aniline blue staining revealed that the pollen tubes targeted the female gametophytes correctly (Appendix Fig S2). However, closer examination of the ovules revealed pollen tube overgrowth at high frequency in herk1 anj mutants. While pollen tube overgrowth is rare in wild‐type and single mutants, 83% of pollen tubes failed to burst upon entering ovules in the double mutant (Fig 1C). The 83% of ovules exhibiting pollen tube overgrowth is notably higher than the 71% of ovules that fail to develop into seeds (Fig 1B and C), indicating that in some cases fertilisation occurs in the presence of pollen tube overgrowth.
In fer mutants, pollen tube overgrowth occurs due to maternal defects in male–female gametophyte communications 16, 17, 20. To confirm that HERK1 and ANJ are female determinants of pollen tube reception, we performed reciprocal crosses between the herk1 anj mutant and wild‐type plants, as well as control crosses within each plant line. While wild‐type Col‐0 (female; f) × herk1 anj (male; m) crosses resulted in 1% of ovules with pollen tube overgrowth, over 90% of pollen tubes exhibited overgrowth in herk1 anj (f) × wild‐type (m) crosses, indicating that pollen tube overgrowth is a maternally derived phenotype in herk1 anj mutants (Fig 1D). As expected, pollen tube overgrowth was observed in only 3% of the ovules in the control wild‐type (f) × wild‐type (m) crosses, while 89% of ovules had overgrowth of the pollen tube in herk1 anj (f) × herk1 anj (m) crosses.
To verify that the reproductive defect is due to the disruption of the HERK1 and ANJ genes and does not arise from additional T‐DNA insertions, we reintroduced the HERK1 and ANJ genes into the herk1 anj background to test for complementation of the pollen tube overgrowth phenotype. We generated pHERK1::HERK1 and pANJ::ANJ‐GFP constructs and found that while pHERK1::HERK1 could be generated, pHERK1::HERK1‐GFP could not be cloned due to toxicity in several bacterial strains. This could explain why a pBRI1::HERK1‐GFP construct has previously been used to complement the herk1 mutant 24. FERONIA's promoter presents a broad expression pattern in ovules 29, and given the maternal origin of the reproductive defect in herk1 anj plants, we decided to use pFER::HERK1‐GFP to test for complementation. In the developing ovules of five independent T1 plants where a hemizygous insertion would segregate 50:50, expression of pFER::HERK1‐GFP or pANJ::ANJ‐GFP constructs in the herk1 anj background reduced pollen tube overgrowth by ~ 50%, as did a pHERK1::HERK1 construct (Appendix Fig S3). Complementation indicates that these reporter constructs produce functional proteins and confirms that the T‐DNA insertions in the HERK1 and ANJ genes are responsible for pollen tube overgrowth. We conclude that HERK1 and ANJ are female determinants of pollen tube reception and therefore named AT5G59700 after a fertility goddess in Australian aboriginal mythology, Anjea.
The kinase activity of FER is not required for its control of pollen tube reception in ovules 29. We therefore tested for complementation of the herk1 anj reproductive defect with kinase‐dead (KD) versions of HERK1 and ANJ. HERK1‐KD and ANJ‐KD were generated by targeted mutagenesis of key residues within the kinase activation loop (D609N/K611R for HERK1 and D606N/K608R for ANJ 30) that render the kinase domains inactive, as demonstrated by in vitro phosphorylation assays using recombinant HERK1(D609N/K611R) and ANJ(D606N/K608R) kinase domains (Fig EV2A). pHERK1::HERK1‐KD and pANJ::ANJ‐KD‐GFP were also able to complement the pollen overgrowth phenotype, indicating that the kinase activity of these RLKs is not required for their function in fertilisation (Fig EV2B). As kinase activity was not required for complementation of the herk1 anj phenotype, we also made a pHERK1::HERK1‐KD‐GFP construct to test for complementation by HERK1 when expressed under its native promoter. Seed set was confirmed to be complemented to the expected extent in T1 plants (Appendix Fig S4A). The similarity in the mutant phenotypes and the dispensable kinase activity in HERK1/ANJ and FER suggests they may act in the same signalling pathway as co‐receptors or as parallel receptor systems.
Figure EV2. Kinase activity of HERK1 and ANJ is not required for complementation of the pollen tube overgrowth phenotype.
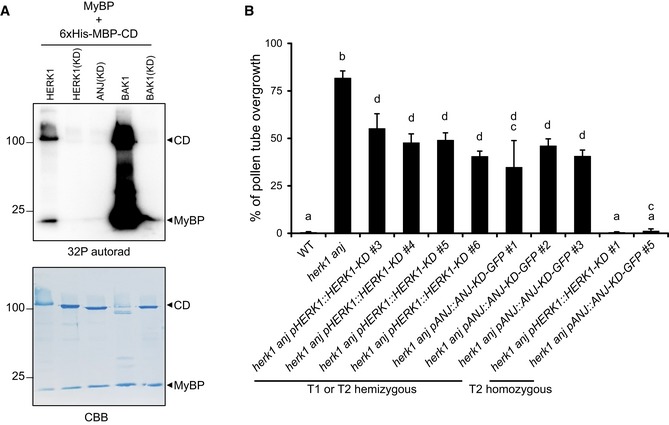
- Kinase activity was assayed for wild‐type HERK1, wild‐type BAK1 (positive control) and kinase‐dead versions (KD) of HERK1, ANJ and BAK1 using 32P incorporation into myelin basic protein (MyBP; trans‐phosphorylation) and the cytosolic domains of the receptor kinases (CD; auto‐phosphorylation). Coomassie Brilliant Blue (CBB) staining of the membrane is shown below as a loading control.
- Percentage of pollen tubes displaying overgrowth at the female gametophyte in WT, herk1 anj plants and at least four independent lines of herk1 anj transformed with pHERK1::HERK1‐KD or pANJ::ANJ‐KD‐GFP from generations T1 or T2. Pollen tube reception was scored for ovules in at least three siliques per line (n ≥ 3). Data presented are means ± SD (one‐way ANOVA followed by Bonferroni's post hoc test; letters mark statistically significant differences between samples, P < 0.05). pANJ::ANJ‐KD‐GFP T1 line 4 was excluded from the figure as it likely had multiple T‐DNA insertions.
Source data are available online for this figure.
HERK1 and ANJ are localised to the filiform apparatus
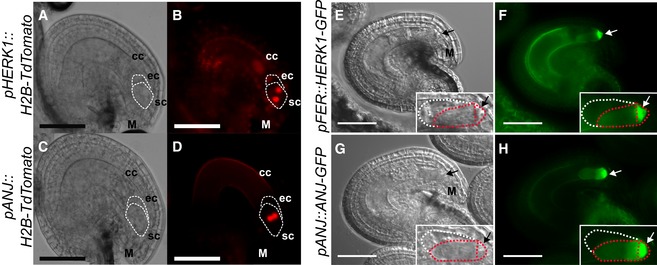
To explore the localisation of HERK1 and ANJ in the female gametophyte and hence gain insight into the possible function of HERK1/ANJ in fertilisation, we made promoter::H2B‐TdTomato transcriptional fusions where expression of either the HERK1 or ANJ promoter should direct nuclear localisation of an RFP signal. Both HERK1 and ANJ were strongly expressed in unfertilised embryo sacs, with expression of HERK1 in the two synergid cells, egg cell and central cell of 4‐cell stage female gametophytes and ANJ expression restricted to the two synergid cells (Fig 2A–D). As HERK1 and ANJ must be expressed in the same cells for a genetic interaction to occur, this restricts their potential function in the female gametophyte during fertilisation to the synergid cells.
Figure 2. HERK1 and ANJ are expressed in the female gametophyte and localise to the filiform apparatus of the synergid cells.
-
A, BExpression of pHERK1::H2B‐TdTomato in mature ovules. White dotted lines delineate the egg cell and synergid cells.
-
C, DExpression of pANJ::H2B‐TdTomato in mature ovules. White dotted lines delineate the egg cell and synergid cells.
-
E, FLocalisation of HERK1‐GFP in the synergid cell from the pFER::HERK1‐GFP construct in (F) and corresponding differential interference contrast (DIC) image in (E). White and red dotted lines delineate the egg cell and synergid cells, respectively.
-
G, HLocalisation of ANJ‐GFP in the synergid cell from the pANJ::ANJ‐GFP construct in (H) and corresponding DIC image in (G). White and red dotted lines delineate the egg cell and synergid cells, respectively.
We next generated promoter::GUS (β‐glucuronidase) transcriptional fusions to gain insight into the expression of these genes at a tissue level. pHERK1::GUS is expressed in the style, ovary walls and stamens (Appendix Fig S5A–E), whereas pANJ::GUS expression is detected in stigmas and stamen filaments (Appendix Fig S5F–J). No expression was detected in pollen grains within mature anthers, although HERK1 was expressed in some developing pollen grains (Appendix Fig S5B, D and I). Within the siliques, HERK1 was most highly expressed close to the stigma, while ANJ appears to be expressed in the funiculus (Appendix Fig S5E and J). Thus, HERK1 and ANJ are expressed in multiple reproductive tissues, with the pattern of expression suggesting the fertilisation defect may arise through a biological function in the junction of the stigma and style, or in the female gametophyte where HERK1 and ANJ gene expression overlaps in the synergid cells.
To further examine HERK1 and ANJ expression and subcellular localisation in ovules, we used the pANJ::ANJ‐GFP, pFER::HERK1‐GFP and pHERK1::HERK1‐KD‐GFP constructs that complement the fertilisation phenotype. Examination of fluorescent signals from HERK1‐GFP and ANJ‐GFP fusion proteins in the female gametophyte showed that they were strongly localised to the filiform apparatus of the synergid cells (Fig 2E–H, Appendix Fig S4B and C). The filiform apparatus is a structure formed by dense folds in the plasma membrane and cell wall where the regulators of fertilisation FER and LRE also localise 17, 19, 31. This specific cellular localisation supports the hypothesis that HERK1 and ANJ could function in the same pathway as FER and LRE. While loss of FER or LRE alone leads to a reproductive defect caused by pollen tube overgrowth in the ovule 17, 19, HERK1 and ANJ are functionally redundant, such that HERK1 and ANJ could act as alternative co‐receptors for FER and/or LRE during male–female interactions.
NORTIA relocalisation after fertilisation is impaired in herk1 anj mutants
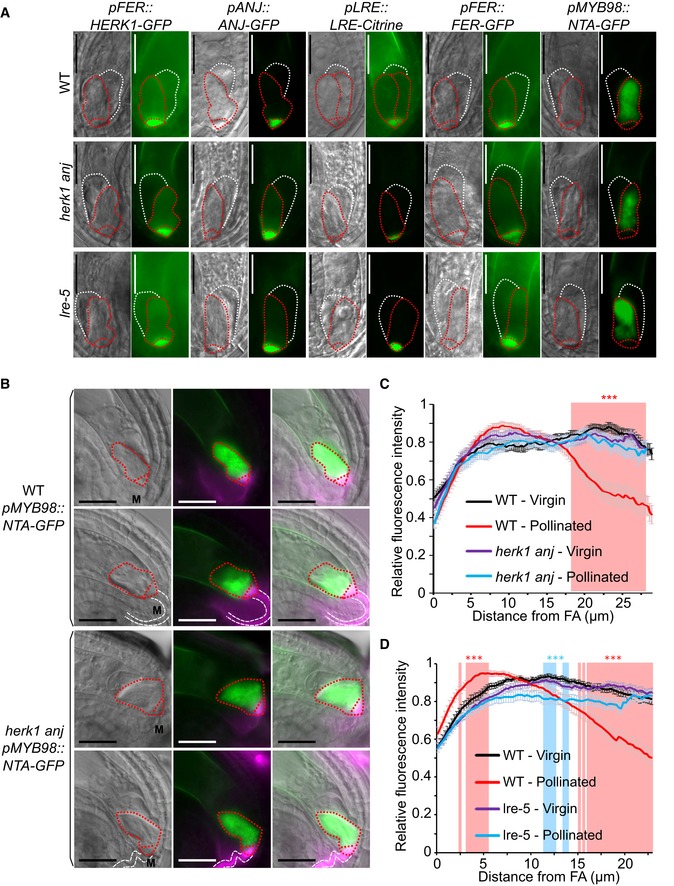
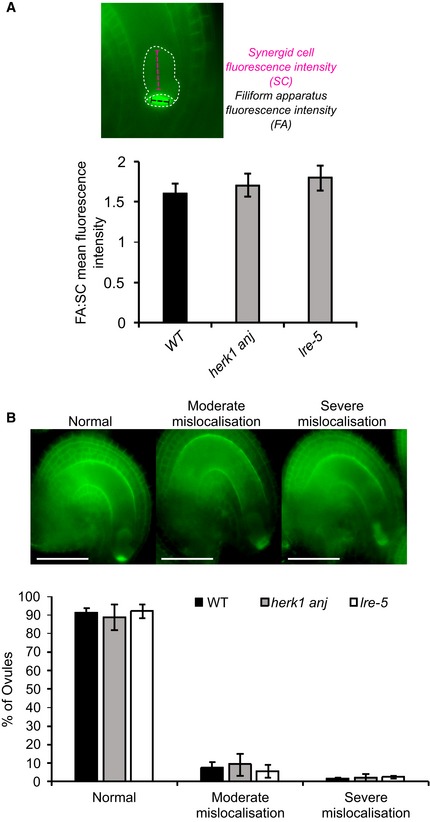
Previous reports point to an interdependence between FER, LRE and NTA in their respective cellular localisations 18, 22. FER only accumulates in the filiform apparatus if functional LRE is present, and NTA relocalisation towards the filiform apparatus upon pollen tube arrival is dependent on FER 18, 22. As HERK1 and ANJ may act in the same signalling pathway as FER, we tested the localisation of fluorescence‐tagged HERK1, ANJ, FER, LRE and NTA in the herk1 anj and lre‐5 backgrounds (Fig 3A). Localisation within the synergids of FER‐GFP, LRE‐Citrine and NTA‐GFP was not affected by herk1 anj mutations. Similarly, HERK1‐GFP and ANJ‐GFP localised to the filiform apparatus in the lre‐5 background. Contrary to previous findings 18, under our conditions FER‐GFP accumulation in the filiform apparatus was not impaired in lre‐5 plants (n > 25; FER‐GFP was found at the filiform apparatus in all ovules checked). To verify that FER subcellular localisation was not affected in lre‐5 under our growth conditions, we quantified the mean fluorescence intensity across the filiform apparatus (FA) and synergid cytoplasm (SC) to calculate the ratio of FA:SC fluorescence intensity (Fig EV3A). When compared across the wild‐type, herk1 anj and lre‐5 genotypes, the mean FA:SC fluorescence intensity ratios were not significantly different, indicating no effect on FER‐GFP localisation to the FA in plants lacking LRE or HERK1/ANJ. Furthermore, we found no differences in the percentage of ovules presenting moderate or severe mislocalisation of FER‐GFP in the synergid cells in wild‐type, herk1 anj or lre‐5 plants (Student's t‐tests, P > 0.05; Fig EV3B). Therefore, we found no dependency on HERK1/ANJ or LRE for localisation of FER, LRE, HERK1, ANJ or NTA within the synergids of unfertilised ovules.
Figure 3. Normal synergid localisation of HERK1, ANJ, LRE, FER and NTA pre‐fertilisation and impaired relocalisation of NTA after pollen tube reception in herk1 anj and lre‐5 .
-
ALocalisation of HERK1, ANJ, LRE, FER and NTA in the synergid cell of wild type (Col‐0; WT), herk1 anj and lre‐5 in unfertilised ovules, as shown by pFER::HERK1‐GFP, pANJ::ANJ‐GFP, pLRE::LRE‐Citrine, pFER::FER‐GFP and pMYB98::NTA‐GFP. DIC and fluorescence images are shown, left to right, respectively. White and red dotted lines delineate the egg cell and synergid cells, respectively. Scale bars = 25 μm.
-
BLocalisation of NTA in the synergid cell of wild‐type and herk1 anj plants before (upper panels) and after (lower panels) pollen tube arrival. In green, NTA localisation as shown by pMYB98::NTA‐GFP fluorescence. In magenta, callose of the filiform apparatus and pollen tube stained with SR2200. From left to right, images shown are DIC, merged fluorescence images, and merged images of DIC and fluorescence. White and red dotted lines delineate the pollen tube and synergid cells, respectively. Scale bars = 25 μm. M, micropyle.
-
C, DProfile of relative fluorescence intensity of NTA‐GFP along the synergid cells of wild‐type and herk1 anj ovules (C); and wild‐type and lre‐5 ovules (D) before (virgin) and after (pollinated) pollen arrival. Data shown are means ± SEM, n = 25. ***P < 0.001 (Student's t‐test). FA, filiform apparatus.
Source data are available online for this figure.
Figure EV3. Quantification of FER‐GFP mislocalisation in the synergid cells of herk1 anj and lre‐5 ovules.
-
ARatio between fluorescence intensities at the filiform apparatus (FA) and the synergid cell cytoplasmic region (SC) in mature ovules from wild‐type (Col‐0), herk1 anj and lre‐5 emasculated flowers expressing pFER::FER‐GFP. Fluorescence profiles for each region of the synergid cells were recorded as exemplified in the upper panel and averaged prior to the ratio calculation. At least 23 ovules obtained from three siliques per plant were scored for three plants per line, with means per plant (n = 3) used for the Student's t‐tests (P > 0.05). Data presented are means ± SD.
-
BQuantification of moderate and severe mislocalisation defects in the accumulation of FER‐GFP at the filiform apparatus in mature ovules from wild‐type (Col‐0), herk1 anj and lre‐5 emasculated flowers expressing pFER::FER‐GFP. Ovules with clear FER‐GFP expression were assigned to one of the three categories presented in the upper panel, as per Ref. 76. No statistically significant differences were detected in Student's t‐test comparisons with wild type. At least 23 ovules obtained from three siliques per plant were scored for three plants per line, with means per plant (n = 3) used for the Student's t‐tests (P > 0.05). Data presented are means ± SD.
Source data are available online for this figure.
To determine whether NTA relocalisation in synergid cells upon pollen tube arrival depends on functional HERK1 and ANJ, we transformed pMYB98::NTA‐GFP into the herk1 anj background. Using SR2200‐based callose staining to visualise the filiform apparatus and pollen tube, we observed NTA‐GFP fluorescence intensity across the length of the synergid cell. In unfertilised ovules, NTA‐GFP fluorescence is evenly distributed across the length of the synergid cell in wild‐type and herk1 anj plants (Fig 3B). Wild‐type fertilised ovules have a shift in the fluorescence intensity pattern, with NTA accumulation towards the micropylar end of the synergid cytoplasm and a decrease in relative fluorescence intensity towards the chalazal end (Fig 3B and C). This response is absent in herk1 anj‐fertilised ovules in which the relative fluorescence intensity pattern is indistinguishable from that of unfertilised ovules, indicating a requirement for HERK1/ANJ in NTA relocalisation upon pollen tube perception.
Whether LRE is dispensable for NTA relocalisation upon pollen tube arrival has not previously been tested. We therefore transformed the pMYB98::NTA‐GFP construct into the lre‐5 genetic background and repeated the assay above to examine whether LRE is required for NTA relocalisation as are HERK1, ANJ and FER 22. While a region of statistically lower signal intensity was present around the middle of the synergids in pollinated lre‐5 ovules compared to wild‐type virgin ovules (Fig 3D), there was no significant shift in signal towards the filiform apparatus upon fertilisation as observed for wild‐type pollinated ovules. Therefore, under our growth conditions, NTA relocalisation at pollen tube arrival is also affected by a loss of LRE.
As reported by Ngo et al 21, the journey of the pollen tube does not conclude upon contact with the filiform apparatus of the synergid cells. Pollen tubes transiently arrest growth upon contact with the synergid; they then grow rapidly along the receptive synergid and towards the chalazal end, before burst and release of the sperm cells 21. To observe this process in detail, we used TdTomato‐tagged pollen and monitored NTA‐GFP localisation at different stages of pollen tube growth within the ovule. The shift in NTA‐GFP localisation was noted in ovules in which the pollen tube had grown past the filiform apparatus and ruptured, rather than upon pollen tube arrival at the filiform apparatus (Appendix Fig S6A). Interestingly, in rare cases when pollen tube burst occurred normally in the herk1 anj background, the fluorescence shift towards the micropyle had also taken place (Appendix Fig S6A). In both cases, NTA‐GFP did not appear to accumulate in the filiform apparatus (Appendix Fig S6B). Our results differ from the interpretation of previous reports that NTA is polarly relocalised from endomembrane compartments to the plasma membrane in the filiform apparatus, instead supporting a more generalised relocalisation within the synergid cytoplasm towards the micropylar end, at least under our growth conditions. We propose that HERK1, ANJ and LRE, similarly to FER, act upstream of NTA relocalisation in the signalling pathway.
ROS production is not affected in mature herk1 anj ovules
ROS levels in fer‐4 and lre‐5 ovules have been reported to be significantly lower than in wild type with the implication that, as hydroxyl free radicals can induce pollen tube burst 20, reduced ROS levels could be responsible for pollen tube overgrowth. To assess whether HERK1 and ANJ also act upstream of ROS accumulation in the ovules, we used H2DCF‐DA to measure ROS levels on a categorical scale in herk1 anj, lre‐5 and fer‐4 ovules (Appendix Fig S7A and B). To ensure that all ovules were fully developed prior to ROS measurement, we emasculated stage 14 flowers and allowed them to develop for a further 20 h. At 20 h after emasculation (HAE), all ovules had reached the mature 7‐celled or 4‐celled pollen‐receptive stages and presented callose accumulation at the filiform apparatus in all backgrounds tested (Fig 4A, Appendix Figs S7C and S8 32, 33). Across three independent experiments, we confirmed that ROS levels are significantly lower in fer‐4 ovules compared to wild type (Fig 4B), indicating that the ROS assay is functional in our hands and able to distinguish changes in ROS levels. However, we found that ROS levels are consistently comparable to wild type in mature ovules of herk1 anj and lre‐5 (Fig 4B). To verify that the fertilisation defect is not rescued in the herk1 anj and lre‐5 genotypes at 20 HAE, we confirmed that pollen tube overgrowth still occurs when ovules are fertilised at this stage (Fig 4C). Taken together, these results suggest that FER acts upstream of ROS accumulation in ovules prior to pollen tube arrival while, under our experimental conditions, HERK1, ANJ and LRE are not required for this process. As these results conflict with a previous study showing lower ROS levels in lre‐5 ovules 20, this suggests that the function of LRE in ROS production may be environmentally sensitive. Our results do not preclude that pollen tube arrival‐induced ROS signalling in the synergid cells is affected in herk1 anj and lre‐5, however differences in transient synergid‐specific ROS burst cannot be quantified in our in vitro system.
Figure 4. herk1 anj mature female gametophytes are morphologically normal and unaffected in ROS production at the micropyle.
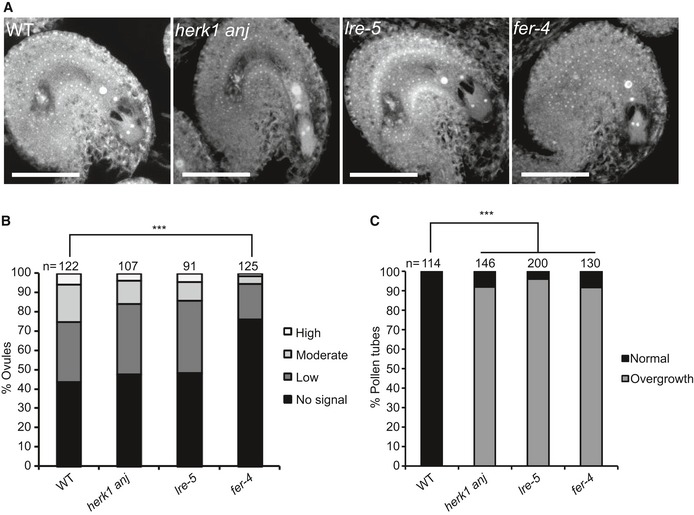
- Representative images of ovules from wild type (Col‐0), herk1 anj, lre‐5 and fer‐4 20 h after emasculation (HAE) displaying the mature female gametophyte structure. Images presented here are maximum intensity projections from confocal microscopy images across several z‐planes of ovules stained as per Ref. 69. Scale bars = 50 μm.
- Quantification of H2DCF‐DA staining of ROS in ovules from wild‐type, herk1 anj, lre‐5 and fer‐4 plants at 20 HAE. Categories are listed in the legend (see also Appendix Fig S7A). Ovules dissected from at least five siliques per line. ***P < 0.001 (χ2 tests).
- Percentage of pollen tubes with normal reception at the female gametophyte (black bars) and displaying overgrowth (grey bars) in wild‐type, herk1 anj, lre‐5 and fer‐4 plants, manually selfed at 20 HAE. Fertilisation events counted from at least three siliques per line. ***P < 0.001 (Student's t‐test).
Source data are available online for this figure.
HERK1 and ANJ interact with LRE and FER
LRE and its homolog LORELEI‐LIKE GPI‐ANCHORED PROTEIN 1 (LLG1) physically interact with RLKs FER, FLAGELLIN SENSING 2 (FLS2) and EF‐TU RECEPTOR (EFR) 18, 34. Mutations in these GPI‐anchored proteins and their associated RLKs result in similar phenotypes, with LRE and LLG1 regarded as co‐receptors and/or stabilisers of RLK function 18, 34, 35. HERK1, ANJ and FER are closely related RLKs and, given the similarities in reproduction defects and subcellular localisation in synergid cells (Fig 3A), we hypothesised that HERK1 and ANJ may act in complex with LRE and/or FER at the filiform apparatus. To examine this hypothesis, we used yeast two‐hybrid assays to test for direct interactions between the extracellular juxtamembrane domains of HERK1, ANJ (HERK1exJM, ANJexJM) and LRE, as well as the complete extracellular domains of HERK1, ANJ and FER (HERK1‐ECD, ANJ‐ECD and FER‐ECD). Interactions between HERK1exJM and ANJexJM with LRE were detected, as were interactions of FER‐ECD and HERK1‐ECD with FER‐ECD, HERK1‐ECD and ANJ‐ECD, and of ANJ‐ECD with FER‐ECD and HERK1‐ECD, indicative of a possible direct interaction between these four proteins (Fig 5A and B). Weaker interactions of HERK1‐ECD and FER‐ECD with ANJ‐ECD, and the lack of interaction of ANJ‐ECD with itself could indicate that they do not form complexes in vivo or could be the result of a lower expression in yeast of the activation domain (AD) version of ANJ‐ECD (ANJ‐ECD‐AD) in comparison with its HERK1‐ECD and FER‐ECD counterparts (Appendix Fig S9). Interactions were also tested by yeast two‐hybrid assays between the kinase domains of HERK1, ANJ and FER (HERK1‐KIN, ANJ‐KIN and FER‐KIN), but interaction between these domains was much weaker (Appendix Fig S9).
Figure 5. HERK1 and ANJ interact with LRE and FER .
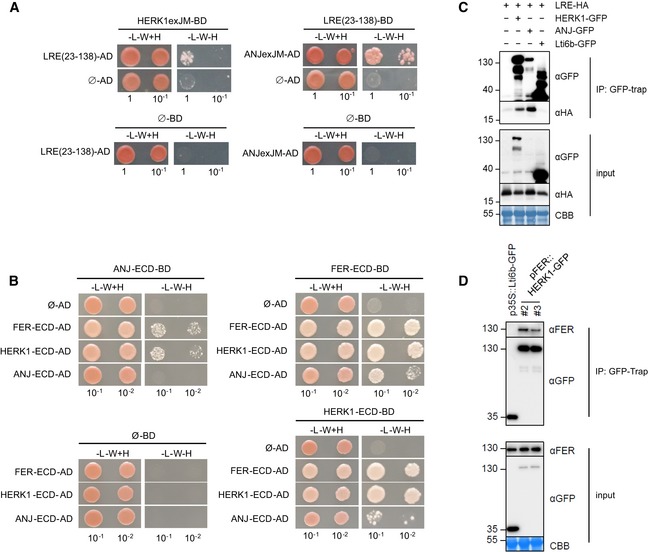
- Yeast two‐hybrid (Y2H) assays of the extracellular juxtamembrane domains of HERK1 and ANJ (HERK1exJM and ANJexJM, respectively) with LRE (residues 23–138; signal peptide and C‐terminal domains excluded).
- Y2H assays with the extracellular domains of HERK1, ANJ and FER (HERK1‐ECD, ANJ‐ECD and FER‐ECD, respectively). Ø represents negative controls where no sequence was cloned into the activating domain (AD) or DNA binding domain (BD) constructs. ‐L‐W‐H, growth medium depleted of leucine (‐L), tryptophan (‐W) and histidine (‐H).
- Co‐immunoprecipitation of HA‐LRE with HERK1‐GFP or ANJ‐GFP following 2 days of transient expression in Nicotiana benthamiana leaves.
- Co‐immunoprecipitation of FER with HERK1‐GFP in Arabidopsis seedlings expressing pFER::HERK1‐GFP. Numbers indicate molecular weight marker sizes in kDa. Assays were performed twice with similar results. CBB refers to Coomassie Brilliant Blue staining of total proteins.
Source data are available online for this figure.
To corroborate interactions of HERK1, ANJ, FER and LRE in planta, co‐immunoprecipitation assays were performed. In a heterologous system using Agrobacterium‐mediated transient expression of pFER::HERK1‐GFP, pFER::ANJ‐GFP and p35S::HA‐LRE in Nicotiana benthamiana leaves, HA‐LRE co‐immunoprecipitated with HERK1‐GFP and ANJ‐GFP (Fig 5C), confirming that these proteins form complexes in planta. Furthermore, herk1 anj lines complemented with pFER::HERK1‐GFP were used to assay the association of HERK1 with endogenous FER using an α‐FER antibody 35. FER co‐immunoprecipitated with both HERK1‐GFP independent transformants in several independent experiments (Fig 5D), again confirming that these complexes form in planta. In an additional genetic approach, we introduced the lre‐5 mutation into the herk1 anj background and characterised fertility impairment in triple homozygous herk1 anj lre‐5 plants. No additive effect was observed in the seed set defect in herk1 anj lre‐5 plants compared to herk1 anj and lre‐5 mutants (Fig EV4A).
Figure EV4. HERK1, ANJ and LRE do not act additively in seed set or ROS production, but mutants attract multiple pollen tubes.
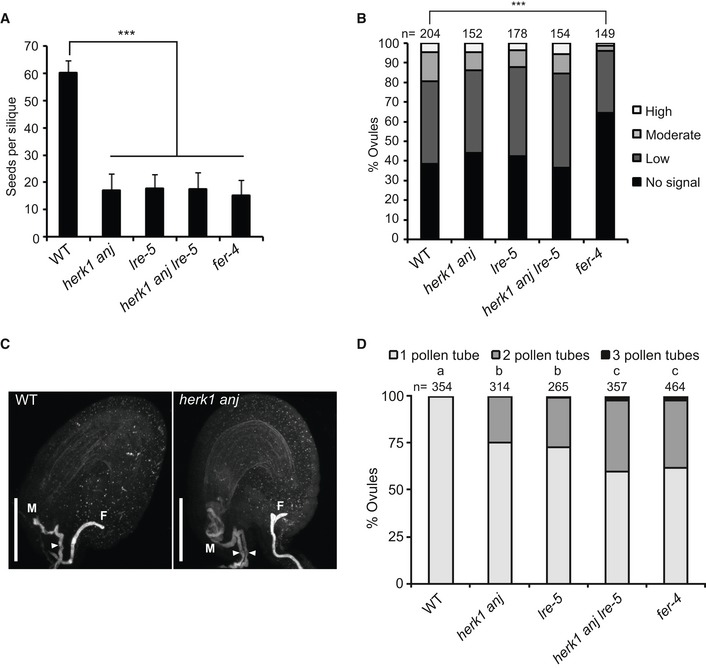
- Quantification of developing seeds per silique in wild‐type, herk1 anj, lre‐5, herk1 anj lre‐5 and fer‐4 plants. Fully expanded siliques were dissected and photographed under a stereomicroscope. n = 25. Data presented are means ± SD. ***P < 0.001 (Student's t‐test).
- Quantification of the H2DCF‐DA staining of ROS in ovules from wild‐type, herk1 anj, lre‐5, herk1 anj lre‐5 and fer‐4 plants at 20 HAE. Categories are listed in the legend (see also Appendix Fig S7A). Ovules dissected from at least five siliques per line. ***P < 0.001 (χ2 tests).
- Representative image of a normal pollen tube reception event in a wild‐type ovule by confocal microscopy on the left and a herk1 anj ovule displaying pollen tube overgrowth and multiple pollen tubes in the micropyle on the right. Images are maximum intensity projections from confocal microscopy images across several z‐planes of ovules stained with aniline blue. M, micropyle. F, funiculus. White arrowhead, pollen tube. Scale bars = 50 μm.
- Polytubey quantification in wild‐type (Col‐0), herk1 anj, lre‐5, herk1 anj lre‐5 and fer‐4 ovules by epifluorescence microscopy following hand pollination at 24 h after emasculation. Ovules from 10 to 13 siliques per line were scored for the number of pollen tubes present at the micropyle if fertilised (total fertilised ovules analysed per line > 265). Letters (a, b, c) mark statistically significant differences between samples in multiple Fisher's exact test pairwise comparisons (P < 0.001).
Source data are available online for this figure.
ROS production in ovules of the triple herk1 anj lre‐5 mutant was measured using H2DCF‐DA at 20 HAE. In agreement with the seed set phenotype, ROS levels were unaffected in the triple homozygous line (Fig EV4B). These results reinforce the hypothesis that HERK1, ANJ and LRE act in the same signalling pathway and, given their cellular localisation and our protein–protein interaction results, we propose that HERK1‐LRE‐FER and ANJ‐LRE‐FER form part of a receptor complex in the filiform apparatus of synergid cells, which mediates pollen tube reception.
To test for any additional additive interaction between HERK1, ANJ, FER and LRE at the level of seed set, CRISPR‐Cas9 was used with two guide RNAs to generate deletions in FER in wild‐type, herk1 anj and herk1 anj lre‐5 genetic backgrounds. Plants were selected based on the fer rosette phenotype. PCR genotyping was used to check each line for deletions, however only two of the eight lines showed smaller PCR bands (Fig EV5A). No PCR products could be amplified for lines 5 or 27 in the herk1 anj background, even when primers at least 1.7 kb upstream and 1.1 kb downstream of the two target sites were used (Fig EV5B), which is interpreted as these lines containing larger deletions or inversions than expected. Amplified PCR products were sequenced in the other lines to characterise each of the CRISPR‐Cas9 lines, and ranged from single nucleotide insertions which caused a frame shift, to an inversion and deletions (Fig EV5C). Seed set was analysed in T2 plants grown in parallel with wild‐type, herk1 anj, lre‐5, fer‐4 and herk1 anj lre‐5 mutants, with further analysis of pollen tube overgrowth in selected lines. No statistically significant difference was found between single, double, triple or quadruple mutants, while all mutants produced significantly fewer seeds and higher levels of pollen tube overgrowth than wild type (Fig EV5D and E).
Figure EV5. Quantification of seed set in CRISPR‐Cas9 fer mutants.
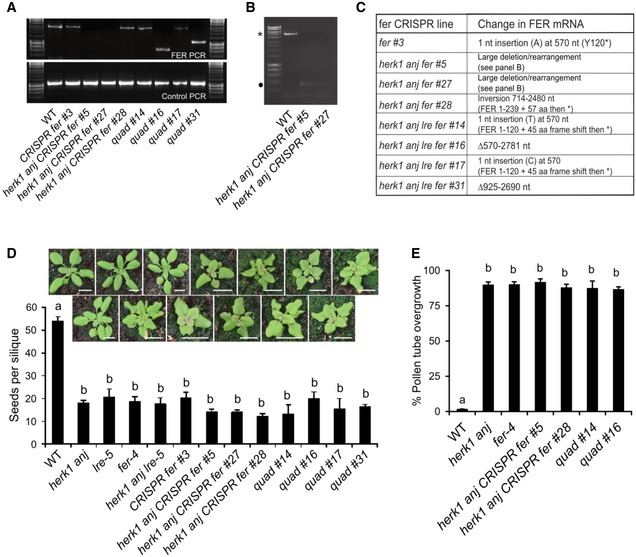
-
APCR amplification of FER and control genomic DNA from wild‐type and CRISPR‐Cas9 fer mutants.
-
BFor herk1 anj CRISPR fer lines 5 and 27, PCR of the FER locus using primers 1.7 kb upstream and 1.1 kb downstream of the CRISPR target sites (CRISPR‐Cas9 fer mutant genotyping outer primers) was also performed. The expected 5.1 kb band from the wild‐type Col‐0 plant is indicated by an asterisk. The band indicated by a black dot was cloned and sequenced but does not contain FER DNA and is therefore an artefact, leading to the conclusion that herk1 anj CRISPR fer lines #5 and #27 contain large deletions or rearrangements that extend beyond the targeted region.
-
CMolecular characterisation of the CRISPR lines.
-
D, EDeveloping seeds per silique (D) and pollen tube overgrowth (E) in wild‐type, single, double, triple and quadruple mutants as listed. Quad = herk1 anj lre‐5 CRISPR fer. Fully expanded siliques were dissected and photographed using an SLR camera. Three plants per line and five siliques per plant were analysed. Data presented are means per plant (n = 3) ± SD. Letters (a, b) mark statistically significant differences between samples in one‐way ANOVA analysis followed by Bonferroni's post hoc comparison of means (P < 0.05). Pictures above (D) are of plants at 21 days after sowing. Scale bars = 1 cm.
Source data are available online for this figure.
It has been reported for several mutations causing pollen tube overgrowth, including lre and fer, that pollen tube overgrowth is occasionally accompanied by polytubey, where more than one pollen tube enters the ovule (Fig EV4C 16, 19). This is indicative of uninterrupted secretion of attraction signals from the synergid cells, suggesting impaired degeneration of the receptive synergid cell upon pollen tube arrival 36, 37. Polytubey has been reported to occur at a rate of ~ 10% in the progeny of a heterozygous fer‐1 mutant 16. To assess whether polytubey occurs in the herk1 anj mutant at a similar rate, polytubey was quantified in herk1 anj mutants along with lre‐5 and fer‐4 mutants as controls (Fig EV4D). Under our growth conditions, polytubey was more frequent in fer‐4 mutants (38.6% of fertilised ovules) than previously reported for fer‐1. Compared to fer‐4, herk1 anj (24.8% of fertilised ovules) and lre‐5 mutants (27.2% of fertilised ovules) exhibited statistically lower rates of polytubey, whereas herk1 anj lre‐5 mutants presented similar rates to fer‐4 (40.3% of fertilised ovules), indicating that mutations in HERK1, ANJ and LRE may have an additive effect in the attraction of supernumerary pollen tubes.
Discussion
Successful reproduction in angiosperms relies on tightly controlled communication between gametophytes through the exchange of chemical and mechanical cues 1. Here, we describe the role of the RLKs HERK1 and ANJ in early stages of fertilisation in Arabidopsis. HERK1 and ANJ are widely expressed in female reproductive tissues including the synergid cells of ovules, where they are polarly localised to the filiform apparatus. herk1 anj plants fail to produce seeds from most ovules due to a maternally derived pollen tube overgrowth defect. As female gametophytes develop normally in herk1 anj mutants, pollen tube overgrowth is likely due to impaired signalling. To clarify the position of HERK1/ANJ in relation to the previously characterised signalling elements of the pollen tube reception pathway, we have shown that NTA relocalisation after pollen tube reception is impaired in herk1 anj as described for FER, whereas ROS production at the micropylar entrance of ovules prior to pollen arrival is not affected. Interactions between HERK1/ANJ, FER and LRE lead us to propose receptor complexes containing HERK1‐LRE‐FER and ANJ‐LRE‐FER at the filiform apparatus.
Associated with diverse hormonal, developmental and stress responses, FER is regarded as a connective hub of cellular responses through its interactions with multiple partners, including small secreted peptides, cell wall components, other RLKs, GPI‐anchored proteins and ROPGEFs 18, 38, 39, 40, 41, 42. As related members of the CrRLK1L family, HERK1 and ANJ have the potential to perform similar roles to FER, and as reported here control pollen tube rupture. Interestingly, control of tip growth in pollen tubes depends on two redundant pairs of CrRLK1Ls: ANX1 and ANX2; and BUPS1 and BUPS2 11, 12, 13, 14. ANX1/2 and BUPS1/2 form ANX‐BUPS heterodimers to control pollen tube growth by sensing autocrine RALF signals 12. In turn, ovular RALF34 efficiently induces pollen tube rupture at the pollen tip, likely through competition with autocrine RALF4/19 12. LEUCINE‐RICH REPEAT EXTENSINS (LRXs) constitute an additional layer of regulation during pollen tube growth 14. LRXs interact physically with RALF4/19 and are thought to facilitate RALF sensing during pollen tube growth [14, 43, preprint: 44]. Here, we propose that female control of pollen tube reception is executed by an analogous mechanism, where CrRLK1L heterocomplexes of FER with either HERK1 or ANJ potentially sense pollen tube‐derived cues to trigger the female gametophyte to induce pollen tube rupture. Given the multiple CrRLK1L‐RALF interactions identified to date 12, 14, 38, 45, pollen tube‐derived RALF signals constitute a potential candidate to induce synergid responses to pollen tube perception. RALF4/19 are continuously secreted at the growing tip of the pollen tube and, while their involvement in pollen growth has been thoroughly studied 12, 14, their possible dual role as synergid‐signalling activators remains unexplored. Disruption of synergid autocrine RALF signalling upon pollen arrival constitutes another possible model, comparable to that hypothesised for RALF34 and RALF4/19 during pollen growth 12. Additionally, LRXs could facilitate RALF perception at the synergid cell to control pollen tube reception.
A second category of putative pollen tube cues involves changes in cell wall properties of the filiform apparatus. As a polarised fast‐growing structure, pollen tubes present cell walls that differ from stationary cell types, with particular emphasis on the growing tip where active cell wall remodelling rapidly takes place 46. When the growing tip reaches the filiform apparatus, it temporarily arrests growth, subsequently growing along the receptive synergid cell prior to rupture 21. The prolonged direct physical contact between the growing tip and the filiform apparatus likely allows a direct exchange of signals which could result in modification of the filiform apparatus cell wall structure. CrRLK1L receptors present an extracellular malectin‐like domain 47, a tandem organisation of two malectin domains with structural similarity to the di‐glucose binding malectin protein 48. The malectin di‐glucose binding residues are not conserved in the malectin‐like domains of ANX1/2 according to structural data 49, 50. However, direct interactions of FER, ANX1/2 and BUPS1/2 malectin‐like domains with the pectin building block polygalacturonic acid have been recently reported [39, preprint: 51]. An extracellular domain anchored to cell wall components and a cytoplasmic kinase domain capable of inducing downstream signalling make FER and the other CrRLK1L proteins a putative link between cell wall status and cellular responses 52. Involvement of FER in root mechanosensing provides additional support for this hypothesis 53. Therefore, FER and the related receptors HERK1 and ANJ may be fulfilling a cell wall integrity surveillance function in the filiform apparatus, triggering cellular responses upon changes in the composition or mechanical forces registered at this specialised cell wall structure.
Receptor complexes are a common feature in signal transduction in multiple cellular processes 54, 55, 56. Our genetic and biochemical results support possible HERK1‐LRE‐FER/ANJ‐LRE‐FER heterocomplexes (Figs 5 and 6). LRE and related proteins form complexes with RLKs FER, FLS2 and EFR, making them versatile co‐receptors that mediate signal perception in multiple processes 18, 34. LRE functions in the maternal control of fertilisation and early seed development 31, 57, whereas its homolog LLG1 is restricted to vegetative growth and plant–pathogen interactions 34. Uncharacterised LLG2 and LLG3 show pollen‐specific expression in microarray data and therefore constitute likely candidates as ANX1/2 and BUPS1/2 receptor complex partners to control pollen tube growth. LRE proteins are thought to stabilise their receptor partners in the plasma membrane and act as direct co‐receptors for the extracellular cues sensed by the RLK 18, 35. As we found that FER localisation in the filiform apparatus is unaltered in lre‐5 plants, with HERK1/ANJ localisation also not affected, our results do not support the role previously reported for LRE as a chaperone for FER localisation in synergid cells 18. A strict requirement for LRE as a FER chaperone in the synergid cells has also been challenged by a previous report evidencing that the fertility defect in lre female gametophytes could be partially rescued by pollen‐expressed LRE 58. In the absence of synergid‐expressed LRE, the authors speculate that sufficient FER is still localised to the filiform apparatus to interact with LRE on the pollen tube plasma membrane, demonstrating a more minor role for LRE intracellular activity in the synergid cells to correctly localise FER 58. We hypothesise that LRE could act as co‐receptor for FER and HERK1 or ANJ at the filiform apparatus, forming tripartite HERK1‐LRE‐FER or ANJ‐LRE‐FER complexes that sense pollen‐derived ligands such as RALF peptides or cell wall components, in a mechanism analogous to that described for pollen tube growth through BUPS1/2‐ANX1/2‐LLG2/3‐RALF4/19 signalling. Further verification of the protein–protein interactions described here could be done via Förster resonance energy transfer (FRET) analysis, cryo‐electron microscopy 59, or super‐resolution microscopy techniques such as Stimulated Emission‐Depletion Measurements (STED 60).
Figure 6. Model of HERK1/ANJ involvement in pollen tube reception.
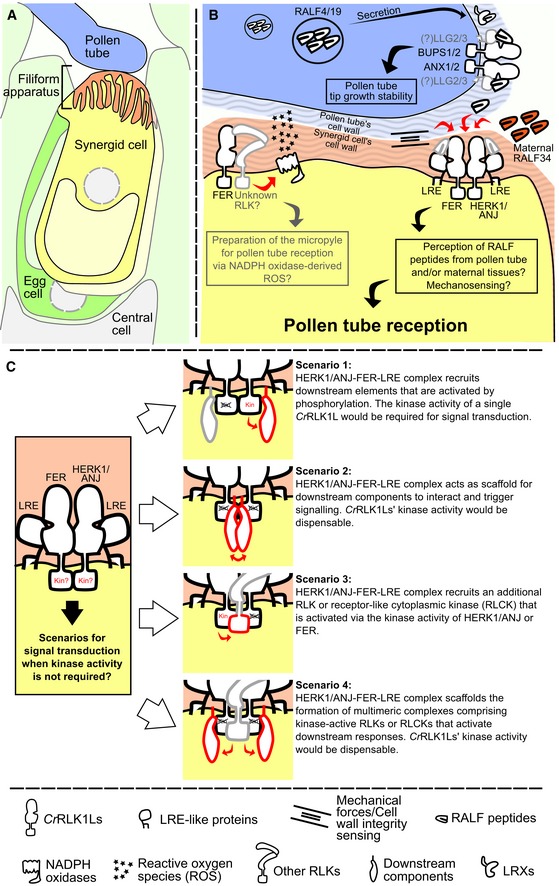
- Overview of the contact point between the male and female gametophytes at pollen tube reception.
- Proposed mechanism(s) of pollen tube reception at a molecular level where HERK1 and ANJ form alternative co‐receptors with FER and LRE. Unknown components or interactions are shown in grey. Maternally or paternally derived RALFs could act as ligands for the HERK1‐LRE‐FER/ANJ‐LRE‐FER heterocomplexes.
- Four possible scenarios where kinase activity of HERK1/ANJ are not required for signal transduction during pollen tube reception. Red components indicate proteins active in signal transduction, while black proteins act as scaffolds for complex assembly. Each scenario is discussed in more detail in the discussion section.
Confirmation of the role of CrRLK1Ls and LRE proteins as RALF peptide sensors has been recently obtained through an elegant combination of crystallographic and biochemical techniques 35. By solving the structure of a FER‐LLG1‐RALF23 complex, Xiao et al 35 have demonstrated that (i) LRE proteins play a central role in the recognition of RALFs; (ii) the N‐terminal region of a subgroup of RALFs is sufficient to induce the interaction between LRE proteins and FER; (iii) while LLG1‐3 proteins are capable of binding RALF23, interaction between LRE and RALF23 was not detected; and (iv) specific amino acid differences between LRE and LLG1‐3 proteins are responsible for such affinity differences. These findings reinforced the hypothesis that signalling specificity can be achieved by the combinatorial action of different CrRLK1Ls, LRE proteins, RALFs and their respective expression patterns and affinities towards each other. Pollen tube reception provides another layer of complexity to this scenario, as two independent cellular systems come into contact, with two putative RALF‐sensing complexes (BUPS1/2‐ANX1/2‐LLG2/3; HERK1/ANJ‐FER‐LRE) and RALF peptides secreted from both the pollen tube tip and the female gametophyte are brought together. The differences in affinity towards certain RALFs observed between LRE and LLG1‐3 allows us to speculate that the pollen‐derived RALF4/19 may not activate HERK1/ANJ‐FER‐LRE signalling, and rather, this activation may instead depend on maternally derived RALFs. Detailed dissection of the affinity of LRE towards pollen and ovule‐derived RALFs will shed light on how pollen tube reception is mediated. Additionally, while the molecular nature of the tripartite CrRLK1L‐LRE protein–RALF complex is now well understood, data presented in this report and previous studies point at CrRLK1L‐to‐CrRLK1L direct interactions 12, for which structural data remains elusive. It will be necessary to address how these higher order complexes are formed, whether there are tripartite complexes composed by two CrRLK1Ls and a single LRE protein, or whether two CrRLK1L‐LRE protein heterodimers dimerise to form a functional signalling unit.
Our results indicate that HERK1, ANJ and LRE are not required to generate the ROS‐enriched environment in the micropyle of mature ovules under our experimental conditions, while FER is involved in this process (Fig 4 20). The role of FER in ROS production has also been characterised in root hairs, where FER activates NADPH oxidase activity via ROPGEF and RAC/ROP GTPase signalling, ensuring root hair growth stability 40. Micropylar ROS accumulation prior to pollen tube arrival depends on NADPH oxidase activity and FER, suggesting a similar pathway to root hairs may take place in synergid cells 20. This evidence places FER upstream of ROS production, whereas FER, HERK1/ANJ and LRE would function upstream of pollen tube reception. One possible explanation is that FER is a dual regulator in synergid cells, promoting ROS production and regulating pollen tube reception, while HERK1/ANJ and LRE functions are restricted to the latter under our environmental conditions. Kinase‐inactive mutants of FER rescue the pollen tube overgrowth defect in fer mutants, but cannot restore the sensitivity to exogenous RALF1 in root elongation 61. These recent findings support multiple signal transduction mechanisms for FER in a context‐dependent manner 61. It would thus be informative to test whether the kinase‐inactive version of FER can restore the ovular ROS production defect in fer mutants. The use of genetic ROS reporters expressed in synergid cells and pollen tubes in live imaging experiments would allow us to observe specific changes in ROS production at the different stages of pollen tube perception in ovules, as performed with Ca2+ sensors 21, 62, 63. ROS production and Ca2+ pump activation in plant cells have been linked during plant–pathogen interactions and are thought to take place during gametophyte communication 64, 65. Thus, given the dynamic changes in Ca2+ during the different stages of pollen tube reception in synergids and pollen, it is likely that ROS production variations also take place in parallel. Studying ROS production profiles during pollen perception in the fer‐4, herk1 anj and lre‐5 backgrounds would provide the resolution required to link these receptors to dynamic ROS regulation during pollen reception. Induction of specific Ca2+ signatures in the synergids upon pollen tube arrival is dependent on FER, LRE and NTA 21. Given that NTA relocalisation after pollen reception depends on functional HERK1/ANJ and NTA is involved in modulating Ca2+ signatures in the synergids, it is possible that HERK1 and ANJ might also be required for Ca2+ signalling during pollen perception.
Downstream signalling after pollen tube reception in the synergid cells likely involves interactions of HERK1, ANJ and FER with cytoplasmic components through their kinase domain. Our results indicate that the kinase activity of HERK1/ANJ is not required for controlling pollen tube rupture (Appendix Fig S4B), as has been reported for FER 29. The fer‐1 pollen tube overgrowth defect could also be rescued with a chimeric protein comprising the FER extracellular domain and the HERK1 kinase domain 29. This implies that the FER and HERK1/ANJ kinase domains are likely redundant in controlling pollen tube reception and may transduce the signal in a similar manner. Testing whether FER‐dependent induction of ROS production in the micropyle is also independent of its kinase activity and whether the HERK1/ANJ kinase domains can also substitute for the FER kinase domain in this process would provide insight into how this signalling network is organised.
Our results suggest a model where FER and LRE form functionally redundant complexes with HERK1 and ANJ in the plasma membrane of synergid cells (Fig 6A and B). These complexes could sense maternally or paternally derived RALF peptides as has been characterised for analogous protein complexes involved in pollen tube growth. Alternatively, the HERK1/ANJ‐FER‐LRE complexes may sense changes in cell wall integrity through mechanosensing. As kinase‐inactive versions of FER can rescue the pollen tube reception phenotype in fer, and kinase‐inactive HERK1 or ANJ can likewise complement a herk1‐1 anj‐1 mutant, we envisage four possible signalling scenarios (Fig 6C). Firstly, the kinase activity of at least one CrRLK1L receptor may be required for activation of downstream signalling through phosphorylation. Secondly, the kinase activity of none of the CrRLK1L receptors may be required if they act as a scaffold to recruit cytoplasmic kinases. Thirdly, additional receptor kinases (either CrRLK1L or other families) may be present in the complex and be phosphorylated by either FER, or HERK1/ANJ to then activate downstream signalling. And lastly, if we combine scenarios two and three, additional receptor kinases along with the HERK1/ANJ‐FER‐LRE complex could recruit cytoplasmic kinases to trigger downstream events. Thus, a number of scenarios exist for the function of HERK and ANJ in pollen tube reception.
This study provides evidence for the involvement of multiple CrRLK1L detectors of pollen tube arrival at the female gametophyte, implicating HERK1 and ANJ as co‐receptors of FER. The action of multiple CrRLK1L proteins at the filiform apparatus highlights the relevance of the CrRLK1Ls in controlling reproduction in flowering plants. Future research in this field will undoubtedly provide new views on how these RLKs integrate pollen‐derived cues to ensure tight control of fertilisation.
Materials and Methods
Experimental model and subject details
Plant material
Arabidopsis thaliana T‐DNA insertion lines herk1 (At3g46290; N657488; herk1‐1; 24) and anj (At5g59700; N654842; anj‐1) were obtained from the Nottingham Arabidopsis Stock Centre (NASC 66, 67), along with cap1/eru (At5g61350; N666567), the1 (At5g54380; N829966), At2g23200 (N685400), cvy1 (At3g39360; N660329), herk2 (At1g30570; N663563), fer (At3g51550; N655026), anx1 (At3g04690; N659315) and anx2 (At5g28680; N656997). T‐DNA lines fer‐4 (At3g51550; N69044 20, 38) and lre‐5 (At4g26466; N66102 31) were kindly provided by Prof. Alice Cheung (University of Massachusetts) and Dr. Ravi Palanivelu (University of Arizona), respectively. Accession Col‐0 was used as a wild‐type control in all experiments. T‐DNA lines were confirmed as homozygous for the insertion by genotyping PCRs. The anj mutant line was characterised as a knockout of gene expression in this study by RT–qPCR. A full list of plant lines used in this study is given in Appendix Table S1.
Growth conditions
Seeds were stratified at 4°C for 3 days. Seeds were sown directly on soil and kept at high humidity for 4 days until seedlings emerged. The soil mix comprised a 4:1 (v:v) mixture of Levington M3 compost:sand. Plants were grown in walk‐in Conviron growth chambers with 22°C continuous temperature, 16 h/day of ~ 120/μmol/m2 light and 60% humidity. For selection of transformants, seeds were surface sterilised with chlorine gas, sown onto half‐strength Murashige and Skoog medium (MS 68), 0.8% (w/v) agar, pH 5.7 (adjusted with KOH), supplemented with the appropriate antibiotic (25 μg/ml of hygromycin B or 50 μg/ml of kanamycin). Seeds on plates were stratified for 3 days at 4°C and then transferred to a growth chamber (Snijders Scientific) at 22°C, 16 h/day of ~ 90 /μmol/m2 of light. Basta selection was carried out directly on soil soaked in a 1:1,000 dilution of Whippet (150 g/l glufosinate ammonium; AgChem Access Ltd).
Method details
Phenotyping
To quantify seed production, fully expanded green siliques were placed on double‐sided sticky tape, and valves were dissected along the replum with No. 5 forceps, exposing the developing seeds. Dissected siliques were kept in a high humidity chamber until photographed to avoid desiccation. Alternatively, mature siliques were collected prior to dehiscence and cleared in 0.4 M NaOH, 1% Triton X‐100 for at least 2 days before imaging with a Microtec dissection microscope. Seeds were counted from the micrographs.
Carpels from self‐pollinated or hand‐pollinated flowers at stage 16 were selected for aniline blue staining of pollen tubes. Carpels were fixed at least overnight in a 3:1 solution of ethanol:acetic acid, then softened overnight in 8 M NaOH, washed four times in water and incubated for 3 h in aniline blue staining solution [0.1% (w/v) aniline blue (Fisons Scientific) in 0.1 M K2PO4‐KOH buffer, pH 11]. Stained carpels were mounted in 50% glycerol, gently squashed onto the microscope slide and then visualised with epifluorescence or confocal microscopy. Aniline blue fluorescence was visualised on a Leica DM6 or Olympus BX51 epifluorescence microscope using a 400 nm LED light source and a filter set with 340–380 nm excitation, emission filter of 425 nm (long pass) and 400 nm dichroic mirror. Confocal images were acquired using a 403.5 nm laser line, 30.7 μm pinhole size and filter set with 405 nm dichroic mirror and 525/50 nm emission filter cube.
Quick callose staining was carried out by incubating freshly dissected tissue samples in a 1,000 × dilution of SR2200 (Renaissance Chemicals Ltd) in half‐strength MS, 5% (w/v) sucrose, pH 5.7. Samples were mounted in the staining solution directly and visualised under an epifluorescence microscope with the same settings as used for aniline blue staining. Callose‐enriched structures like pollen tubes and the filiform apparatus of ovules display a strong fluorescence within 10 min of incubation. Only structures directly exposed to the SR2200 solution are stained.
To observe the development of the female gametophyte, we used the confocal laser scanning microscopy method as described by Christensen 69. Ovules were dissected from unpollinated carpels, fixed for 2 h in a 4% (v/v) solution of glutaraldehyde, 12.5 mM sodium cacodylate buffer pH 6.9, dehydrated in an ethanol series (20–100%, 20% intervals, 30 min each) and cleared in a benzyl benzoate:benzyl alcohol 2:1 mixture for 2 h prior to visualisation. Samples were mounted in immersion oil, and coverslips sealed with clear nail varnish and visualised with an inverted Nikon A1 confocal microscope. Fluorescence was visualised with 35.8 μm pinhole size, 642.4 nm laser line and filter set of 640 nm dichroic mirror and 595/50 nm emission filter cube. Multiple z‐planes were taken and analysed with ImageJ.
Analyses of expression patterns of HERK1 and ANJ used promoter::reporter constructs. promoter::GUS reporters were analysed by testing β‐glucuronidase activity in Col‐0 plants from the T1 and T2 generations. Samples were fixed in ice‐cold 90% acetone for 20 min, then washed for 30 min in 50 mM NaPO4 buffer pH 7.2. Samples were transferred to X‐Gluc staining solution (2 mM X‐Gluc (Melford Laboratories Ltd), 50 mM NaPO4 buffer pH 7.2, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide and 0.2% (v/v) Triton X‐100), vacuum‐infiltrated for 30 min and incubated at 37°C for several hours or overnight. Samples were cleared in 75% ethanol and visualised under a light microscope or stereomicroscope. For the promoter::H2B‐TdTomato reporters, unpollinated ovules were dissected from the carpels and mounted in half‐strength MS, 5% (w/v) sucrose, pH 5.7. RFP signal was detected on a Leica DM6 epifluorescence microscope using a 535 nm LED light source and a filter set with 545/25 nm excitation filter, 605/70 nm emission filter and a 565 nm dichroic mirror. DIC images were taken in parallel.
H2DCF‐DA staining of ROS in ovules was carried out as per Ref. 20. Ovules from unpollinated carpels were dissected and incubated in staining solution (25 μM H2DCF‐DA (Thermo Scientific), 50 mM KCl, 10 mM MES buffer pH 6.15) for 15 min. Samples were subsequently washed three times in H2DCF‐DA‐free buffer for 5 min, mounted on slides and immediately visualised by epifluorescence microscopy. H2DCF‐DA fluorescence was visualised using a 470 nm LED light source and a filter set with 470/40 nm excitation filter, 460/50 nm emission filter and 495 nm dichroic mirror.
All steps were performed at room temperature unless otherwise specified. Ovules were dissected by placing carpels on double‐sided sticky tape, separating the ovary walls from the replum with a 0.3‐mm gauge needle, and by splitting the two halves of the ovary along the septum with No. 5 forceps. GFP was visualised by epifluorescence microscopy with the same settings used to visualise H2DCF‐DA fluorescence. TdTomato was visualised as described above.
Cloning and transformation of Arabidopsis
To study the cellular localisation and to complement the pollen overgrowth defect, we generated the constructs pANJ::ANJ‐GFP, pHERK1::HERK1, pFER::FER‐GFP, pANJ::ANJ‐KD‐GFP and pHERK1::HERK1‐KD. Genomic regions of interest (spanning 2 kb upstream of the start codon ATG and the full coding sequence excluding stop codon) were amplified by PCR with Phusion DNA polymerase (NEB). Promoter::CDS amplicons were cloned via KpnI/BamHI restriction sites into a pGreen‐IIS backbone (Basta resistance; from Detlef Weigel's group, Max Planck Institute for Developmental Biology 70), with or without an in‐frame C‐terminal GFP coding sequence. Kinase‐dead versions of HERK1 and ANJ were generated by site‐directed mutagenesis of the activation loop residues D606N/K608R of ANJ and D609N/K611R of HERK1 using pANJ::ANJ‐GFP and pHERK1::HERK1 constructs as template 71. To generate the GUS and H2B‐TdTomato reporter constructs, pHERK1 and pANJ (from 2 kb upstream of the ATG start codon) were cloned with a pENTR‐dTOPO system (Thermo Scientific) and then transferred to the GUS expression cassette in the pGWB433 destination vector or pAH/GW:H2B‐TdTomato via LR recombination (LR clonase II; Thermo Scientific 72). ASE Agrobacterium tumefaciens strain was used with pGreen vectors; GV3101pMP90 strain was used otherwise. Arabidopsis stable transformants were generated through the floral dip method. Primers used for cloning are listed in Appendix Table S2, and all plasmids used in this study are listed in Appendix Table S3.
To test interaction in vivo in co‐immunoprecipitation assays, we generated pFER::ANJ‐GFP via three‐way ligation cloning of KpnI‐pFER‐NotI and NotI‐ANJ‐BamHI fragments into a pGreen‐IIS backbone (Basta resistance; from Detlef Weigel's group, Max Planck Institute for Developmental Biology 70). To test direct interaction between HERK1exJM, ANJexJM and LRE in yeast, we cloned the extracellular juxtamembrane sequence corresponding to the 81 amino acids N‐terminal of the predicted transmembrane domain of HERK1 and ANJ, as well as the sequence corresponding to the amino acids 23–138 of LRE (as per Ref. 18). Interaction between HERK1, ANJ and FER was also assayed by Y2H and the extracellular domains excluding the signal peptide (HERK1‐ECD, amino acids 24–405; ANJ‐ECD, amino acids 25–405; FER‐ECD, amino acids 28–446) as well as the cytosolic kinase domains (HERK1‐KIN, amino acids 429–830; ANJ‐KIN, amino acids 429–830; FER‐KIN, amino acids 470–895). Amplicons of exJM and KIN domains were cloned into yeast two‐hybrid vectors pGADT7 and pGBKT7 via SmaI restriction digests, in frame with the activation or DNA binding domains (AD or BD, respectively). Amplicons of ECD domains were cloned into PCR8 entry vectors and subsequently recombined into pGADT7‐GW and pGBKT7‐GW via LR recombination. Col‐0 genomic DNA was used as the template for all cloning events unless otherwise specified.
To mutate FER in the Col‐0, herk1 anj and herk1 anj lre genotypes, CRISPR‐Cas9 with two guide RNAs was used to generate large deletions. The guide RNAs were designed with https://crispr.dbcls.jp 73 to target two regions of the FER gene 1.7–2.2 kb apart and were cloned into pBEE401E. T1 transformants were selected with BASTA and based on a fer‐4‐like phenotype. Seed set was assessed in the T2 generation and the lines genotyped at FER to verify either a large deletion in the gene or no amplification due to loss of the primer binding sites. Primers used for cloning are listed in Appendix Table S2.
For the kinase assays, the cytosolic domains (CDs) of WT or kinase‐dead (KD) variants of HERK1 (amino acids 429–830) or ANJ (amino acids 429–829) were cloned into the pOPINM expression vector in frame with an N‐terminal 6xHis‐maltose binding protein (‐MBP) tag using InFusion clonase (Takara) using the pOM primers listed in Appendix Table S2.
Genotyping PCRs and RT–qPCRs
Genomic DNA was extracted from leaves of 2‐week‐old seedlings by grinding fresh tissue in DNA extraction buffer (200 mM Tris–HCl pH 7.5, 250 mM NaCl, 25 mM EDTA and 0.5% SDS), precipitating DNA with isopropanol, washing pellets with 75% EtOH and resuspending DNA in water. Genotyping PCRs were performed with Taq polymerase and 35 cycles with 60°C annealing temperature and 1 min extension time. RNA was extracted a Spectrum Plant Total RNA extraction kit (Sigma) for qPCR, from 100 mg of floral tissue from three plants per line. RNA concentrations were normalised, and an aliquot was DNaseI‐treated and subsequently transcribed into first strand cDNA with the RevertAid cDNA synthesis kit (Thermo Scientific) using random hexamers. qPCRS were performed on a Qiagen Rotor‐Gene Q machine (40 cycles of 95°C for 10 s to denature and 60°C for 40 s to anneal and extend) using a Rotor‐Gene SYBR Green PCR kit (Qiagen). Expression was standardised to actin. Primers for genotyping and qPCR are listed in the Appendix Table S2.
Yeast two‐hybrid assays
Direct interaction assays in yeast were carried out following the Clontech small‐scale LiAc yeast transformation procedure. Yeast strain Y187 was transformed with pGADT7 constructs and yeast strain Y2HGold with pGBKT7 constructs (including empty vectors as controls). Yeast diploid cells carrying both plasmids were obtained by mating, and interaction tests were surveyed on selective media lacking leucine, tryptophan and histidine.
FER‐, HERK1‐ and ANJ‐ECD protein fusions to the Gal4‐BD and Gal4‐AD were detected by Western blots with antibodies anti‐Myc (1:1,000 dilution, clone 9E10; Roche) and anti‐HA (clone 3F10; Roche), respectively. For yeast protein extraction, cultures (OD600 0.7) were centrifuged and the pellets resuspended in sterile water. 0.2 M NaOH was used immediately to lyse the cells for 5 min at room temperature. After centrifugation, pellets were resuspended in Laemmli 1× buffer [0.034 M Tris–HCl pH 6.8, 1% SDS, 12.5% glycerol, 0.0075% bromophenol blue, 1 M 1,4‐dithiothreitol (DTT)] and heat to 95°C for 3 min. Extracts were centrifuged and the supernatants collected and stored at −80°C. 5 μg total protein of each simple was loaded on the gel.
Co‐immunoprecipitation and Western blots
For assays using transient expression, leaves of 4.5‐week‐old N. benthamiana were infiltrated with Agrobacterium tumefaciens strain GV3101 carrying constructs indicated in figure captions. In all cases, leaves were co‐infiltrated with A. tumefaciens carrying a P19 silencing suppressor. Leaves were harvested 2 days post‐infiltration and frozen in liquid nitrogen before extraction in buffer (20 mM MES pH 6.3, 100 mM NaCl, 10% glycerol, 2 mM EDTA, 5 mM DTT, supplemented with 1% IGEPAL and protease inhibitors). Immunoprecipitations were performed in the same buffer with 0.5% IGEPAL for 3–4 h at 4°C with GFP‐trap resin (Chromotek). Beads were washed three times with the same buffer, and bound proteins were eluted by addition of SDS loading dye and heating to 90°C for 10 min. Proteins were separated by SDS–PAGE and detected via Western blot following blocking (in TBS 0.1% Tween‐20 with 5% non‐fat milk powder) with the following antibody dilutions in the same blocking solution: α‐GFP‐HRP (B‐2, sc‐9996, Santa Cruz), 1:5,000; α‐HA‐HRP (3F10, Roche), 1:3,000.
To test whether HERK1 associates with FER in planta, T2 generation herk1 anj lines expressing pFER::HERK1‐GFP were germinated on selection for 5 days. Homozygous p35S::Lti6b‐GFP (Col‐0 background) was used a control membrane‐localised GFP‐tagged protein 74. Five‐day‐old seedlings were transferred to liquid MS culture and grown in 6‐well plates for an additional 7 days. Seedlings were harvested and ground in liquid nitrogen, and total protein was extracted in IP buffer (50 mM Tris–Cl pH 7.5, 150 mM NaCl, 2 mM EDTA, 10% glycerol, supplemented with 5 mM DTT, 0.5 mM PMSF, Sigma protease inhibitor cocktail P9599 and Sigma phosphatase inhibitor cocktails 2 and 3) + 1% IGEPAL. Extracts were clarified by centrifugation at 10,000 g, filtered through Miracloth (Millipore) and diluted with detergent‐free IP buffer to 0.5% IGEPAL (final concentration). Immunoprecipitations were performed with GFP‐trap resin (Chromotek) for 4 h at 4°C with mixing. Beads were collected by centrifugation at 500 g and washed three times with IP buffer + 0.5% IGEPAL. Bound proteins were eluted by heating to 80°C in 2 × SDS loading dye. FER was detected using anti‐FER (rabbit polyclonal, 1:1,000 35) and anti‐Rabbit IgG (whole molecule)–HRP (Sigma A0545, 1:5,000).
Recombinant protein expression, purification and kinase assays
6xHis‐MBP‐CD fusion proteins were expressed in BL21 Rosetta pLysS cells and purified via Ni2+‐affinity chromatography using Ni Sepharose High Performance resin (GE Healthcare). After purification, the proteins were concentrated into buffer (25 mM Tris–Cl pH 7.5, 100 mM NaCl, 2 mM DTT, 10% glycerol) using Amicon centrifugal concentrators (MWCO 10,000. Millipore) and stored at −80°C until use. For kinase assays, 1 μg of 6xHis‐MBP‐CD was mixed with 1 μg myelin basic protein (MyBP) in a 30‐μl reaction in kinase buffer (25 mM Tris–Cl pH 7.5, 3 mM MgCl2, 3 mM MnCl2, 1 mM DTT). Reactions were initiated with the addition of 10 μM ATP with 1 μCi 32P‐γ‐ATP and were carried out for 30 min at 25°C. Proteins were separated by SDS–PAGE, transferred to PVDF membrane, stained with Coomassie brilliant blue G‐250 and imaged using a Typhoon phosphorimager (GE Healthcare).
Microscopy and image building
Epifluorescence images were obtained with Leica DM6 or Olympus BX51 widefield microscopes equipped with HC PL Fluotar objectives or UPlanFl 4×, 10× and 20× objectives, respectively. A Nikon A1 inverted confocal laser scanning microscope fitted with Plan Fluor 40× oil and Plan Apo VC 60× oil objectives was used to obtain confocal micrographs. A Leica M165 FC stereomicroscope was used to visualise floral tissues from GUS stained samples. Leica LASX, NIS Elements Viewer and ImageJ software were used to analyse microscopy images. Inkscape was used to build all figures in this article.
Quantification and statistical analysis
Leica LASX software was used to obtain relative fluorescence intensity profiles from synergid cells by defining linear regions of interest across the synergid cytoplasm in a micropylar to chalazal orientation. Synergid cytoplasm area was defined between the filiform apparatus and the synergid‐egg cell chalazal limit using the corresponding DIC images.
Statistical significance in seed set averages and relative fluorescence averages (at equivalent distances from the filiform apparatus) were assessed with Student's t‐tests. χ2 tests were used to compare distributions obtained in pollen tube overgrowth assays and ROS measurements in ovules, using the distribution obtained in wild‐type plants as the expected distribution. In all tests, *P < 0.05, **P < 0.01, and ***P < 0.001. When more than five comparisons were required, one‐way ANOVA was performed using Origin Pro 2017 and 2018b, followed by Tukey's or Bonferroni's tests if differences were detected. Sample size n is indicated in the graphs or in figure legends.
Author contributions
Conceptualization, SG‐T and LMS; Methodology, SG‐T and LMS; Investigation, SG‐T, NB‐T, TAD, LMS, and ESW; Writing—Original Draft, SG‐T and LMS; Writing—Review and Editing, all authors; Supervision, CZ, JEG, and LMS.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank Andrew Fleming and his group at the University of Sheffield for early feedback and guidance on experiments; Alice Cheung and Qiaohong Duan from the University of Massachusetts for advice on the ROS assays and for sharing fer‐4 seeds with us; Chao Li from East China Normal University for the p35S::HA‐LRE construct; Ravi Palanivelu from the University of Arizona for lre‐5 seeds; Martin Bayer from the Max Planck Institute for Developmental Biology for the pLAT52::TdTomato line; Ueli Grossniklaus from the University of Zurich for the pFER::HERK1‐GFP and pLRE::LRE‐Citrine constructs; Sharon Kessler from Purdue University for sharing the pMYB98::NTA‐GFP construct; Daphne Goring from the University of Toronto for the pBEE401E CRISPR/Cas‐9 construct and Melinka Butenko at the University of Oslo for the pAH21\GW vector used to make promoter::H2B‐TdTomato reporters. S.G‐T. was supported by a Department of Animal and Plant Sciences postgraduate teaching fellowship. Research in J.E.G.'s laboratory is supported by RCUK grant BB/N004167/1. T.A.D. was supported by post‐doctoral fellowships from the European Molecular Biology Organisation (LTF 100‐2017) and the Natural Sciences and Engineering Research Council of Canada (PDF‐532561‐19). N.B‐T. was supported by a MINECO FPI Fellowship (BES‐2014‐068868), and we acknowledge David Alabadi for his supervision of N.B‐T. The Zipfel laboratory was supported by the Gatsby Charitable Foundation and European Research Council (PEPTALK). Confocal imaging work was performed at the Wolfson Light Microscope Facility using the Nikon A1 confocal microscope.
EMBO Reports (2020) 21: e48466
Data availability
The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through the IntAct Molecular Interaction Database 75, and assigned the identifier IM‐27345 (https://www.ebi.ac.uk/intact/search/do/search?searchString=pubid:unassigned2053).
References
- 1. Dresselhaus T, Sprunck S, Wessel GM (2016) Fertilization mechanisms in flowering plants. Curr Biol 26: R125–R139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson MA, Harper JF, Palanivelu R (2019) A fruitful journey: pollen tube navigation from germination to fertilization. Annu Rev Plant Biol 70: 809–837 [DOI] [PubMed] [Google Scholar]
- 3. Higashiyama T (2002) The synergid cell: attractor and acceptor of the pollen tube for double fertilization. J Plant Res 115: 149–160 [DOI] [PubMed] [Google Scholar]
- 4. Huang B‐Q, Russell SD (1992) Female germ unit: organization, isolation, and function. Int Rev Cytol 140: 233–293 [Google Scholar]
- 5. Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D et al (2009) Defensin‐like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- 6. Zhong S, Liu M, Wang Z, Huang Q, Hou S, Xu YC, Ge Z, Song Z, Huang J, Qiu X et al (2019) Cysteine‐rich peptides promote interspecific genetic isolation in Arabidopsis . Science 364: eaau9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang T, Liang L, Xue Y, Jia PF, Chen W, Zhang MX, Wang YC, Li HJ, Yang WC (2016) A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244 [DOI] [PubMed] [Google Scholar]
- 8. Takeuchi H, Higashiyama T (2016) Tip‐localized receptors control pollen tube growth and LURE sensing in Arabidopsis . Nature 531: 245–248 [DOI] [PubMed] [Google Scholar]
- 9. Zhang XX, Liu WJ, Nagae TT, Takeuchi H, Zhang HQ, Han ZF, Higashiyama T, Chai JJ (2017) Structural basis for receptor recognition of pollen tube attraction peptides. Nat Commun 8: 1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franck CM, Westermann J, Boisson‐Dernier A (2018) Plant malectin‐like receptor kinases: from cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69: 301–328 [DOI] [PubMed] [Google Scholar]
- 11. Boisson‐Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U (2009) Disruption of the pollen‐expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ge ZX, Bergonci T, Zhao YL, Zou YJ, Du S, Liu MC, Luo XJ, Ruan H, Garcia‐Valencia LE, Zhong S et al (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF‐mediated signaling. Science 358: 1596–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miyazaki S, Murata T, Sakurai‐Ozato N, Kubo M, Demura T, Fukuda H, Hasebe M (2009) ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol 19: 1327–1331 [DOI] [PubMed] [Google Scholar]
- 14. Mecchia MA, Santos‐Fernandez G, Duss NN, Somoza SC, Boisson‐Dernier A, Gagliardini V, Martinez‐Bernardini A, Fabrice TN, Ringli C, Muschietti JP et al (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis . Science 358: 1600–1603 [DOI] [PubMed] [Google Scholar]
- 15. Schoenaers S, Balcerowicz D, Costa A, Vissenberg K (2017) The kinase ERULUS controls pollen tube targeting and growth in Arabidopsis thaliana . Front Plant Sci 8: 1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huck N, Moore JM, Federer M, Grossniklaus U (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- 17. Escobar‐Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U (2007) The FERONIA receptor‐like kinase mediates male‐female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- 18. Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L et al (2015) Glycosylphosphatidylinositol‐anchored proteins as chaperones and co‐receptors for FERONIA receptor kinase signaling in Arabidopsis . Elife 4: e06587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Capron A, Gourgues M, Neiva LS, Faure J‐E, Berger F, Pagnussat G, Krishnan A, Alvarez‐Mejia C, Vielle‐Calzada J‐P, Lee Y‐R et al (2008) Maternal control of male‐gamete delivery in Arabidopsis involves a putative GPI‐anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, Cheung AY (2014) Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis . Nat Commun 5: 3129 [DOI] [PubMed] [Google Scholar]
- 21. Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U (2014) A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev Cell 29: 491–500 [DOI] [PubMed] [Google Scholar]
- 22. Kessler SA, Shimosato‐Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- 23. Jones DS, Kessler SA (2017) Cell type‐dependent localization of MLO proteins. Plant Signal Behav 12: e1393135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y (2009) Three related receptor‐like kinases are required for optimal cell elongation in Arabidopsis thaliana . Proc Natl Acad Sci USA 106: 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo H, Ye H, Li L, Yin Y (2009) A family of receptor‐like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana . Plant Signal Behav 4: 784–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato‐Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen‐associated molecular pattern)‐induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hematy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Hofte H (2007) A receptor‐like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17: 922–931 [DOI] [PubMed] [Google Scholar]
- 28. Galindo‐Trigo S, Gray JE, Smith LM (2016) Conserved roles of CrRLK1L receptor‐like kinases in cell expansion and reproduction from algae to angiosperms. Front Plant Sci 7: 1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kessler SA, Lindner H, Jones DS, Grossniklaus U (2015) Functional analysis of related CrRLK1L receptor‐like kinases in pollen tube reception. EMBO Rep 16: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knighton DR, Zheng JH, Teneyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM (1991) Crystal‐structure of the catalytic subunit of cyclic adenosine‐monophosphate dependent protein‐kinase. Science 253: 407–414 [DOI] [PubMed] [Google Scholar]
- 31. Tsukamoto T, Qin Y, Huang YD, Dunatunga D, Palanivelu R (2010) A role for LORELEI, a putative glycosylphosphatidylinositol‐anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J 62: 571–588 [DOI] [PubMed] [Google Scholar]
- 32. Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell 16: S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Christensen CA, Subramanian S, Drews GN (1998) Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis . Dev Biol 202: 136–151 [DOI] [PubMed] [Google Scholar]
- 34. Shen QJ, Bourdais G, Pan HR, Robatzek S, Tang DZ (2017) Arabidopsis glycosylphosphatidylinositol‐anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc Natl Acad Sci USA 114: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J (2019) Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572: 270–274 [DOI] [PubMed] [Google Scholar]
- 36. Beale KM, Leydon AR, Johnson MA (2012) Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr Biol 22: 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maruyama D, Volz R, Takeuchi H, Mori T, Igawa T, Kurihara D, Kawashima T, Ueda M, Ito M, Umeda M et al (2015) Rapid elimination of the persistent synergid through a cell fusion mechanism. Cell 161: 907–918 [DOI] [PubMed] [Google Scholar]
- 38. Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan QH, Liu MC, Maman J, Steinhorst L, Schmitz‐Thom I et al (2018) The FERONIA receptor kinase maintains cell‐wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor‐like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stegmann M, Monaghan J, Smakowska‐Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF‐regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- 42. Hou YN, Guo XY, Cyprys P, Zhang Y, Bleckmann A, Cai L, Huang QP, Luo Y, Gu HY, Dresselhaus T et al (2016) Maternal ENODLs are required for pollen tube reception in Arabidopsis . Curr Biol 26: 2343–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stegmann M, Zipfel C (2017) Complex regulation of plant sex by peptides. Science 358: 1544–1545 [DOI] [PubMed] [Google Scholar]
- 44. Moussu S, Broyart C, Santos‐Fernandez G, Augustin S, Wehrle S, Grossniklaus U, Santiago J (2019) Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. bioRxiv: 695874 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hématy K, Renou J, Landrein B et al (2018) Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis . Curr Biol 28: 2452–2458 [DOI] [PubMed] [Google Scholar]
- 46. Chebli Y, Kaneda M, Zerzour R, Geitmann A (2012) The cell wall of the Arabidopsis pollen tube‐spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boisson‐Dernier A, Kessler SA, Grossniklaus U (2011) The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot 62: 1581–1591 [DOI] [PubMed] [Google Scholar]
- 48. Schallus T, Jaeckh C, Feher K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T et al (2008) Malectin: a novel carbohydrate‐binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N‐glycosylation. Mol Biol Cell 19: 3404–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Du S, Qu LJ, Xiao J (2018) Crystal structures of the extracellular domains of the CrRLK1L receptor‐like kinases ANXUR1 and ANXUR2. Protein Sci 27: 886–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moussu S, Augustin S, Roman AO, Broyart C, Santiago J (2018) Crystal structures of two tandem malectin‐like receptor kinases involved in plant reproduction. Acta Crystallogr D Struct Biol 74: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin W, Tang W, Anderson C, Yang Z (2018) FERONIA's sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis . bioRxiv [PREPRINT] [Google Scholar]
- 52. Verger S, Hamant O (2018) Plant physiology: FERONIA defends the cell walls against corrosion. Curr Biol 28: R215–R217 [DOI] [PubMed] [Google Scholar]
- 53. Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB (2014) The receptor‐like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol 24: 1887–1892 [DOI] [PubMed] [Google Scholar]
- 54. Greeff C, Roux M, Mundy J, Petersen M (2012) Receptor‐like kinase complexes in plant innate immunity. Front Plant Sci 3: 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burkart RC, Stahl Y (2017) Dynamic complexity: plant receptor complexes at the plasma membrane. Curr Opin Plant Biol 40: 15–21 [DOI] [PubMed] [Google Scholar]
- 56. Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537 [DOI] [PubMed] [Google Scholar]
- 57. Wang YB, Tsukamoto T, Noble JA, Liu XL, Mosher RA, Palanivelu R (2017) Arabidopsis LORELEI, a maternally expressed imprinted gene, promotes early seed development. Plant Physiol 175: 758–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu X, Castro C, Wang Y, Noble J, Ponvert N, Bundy M, Hoel C, Shpak E, Palanivelu R (2016) The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight‐cysteine motif and the receptor‐like kinase FERONIA. Plant Cell 28: 1035–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miura K (2018) An overview of current methods to confirm protein‐protein interactions. Protein Pept Lett 25: 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D et al (2013) Arabidopsis nanodomain‐delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110: 8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haruta M, Gaddameedi V, Burch H, Fernandez D, Sussman MR (2018) Comparison of the effects of a kinase‐dead mutation of FERONIA on ovule fertilization and root growth of Arabidopsis . FEBS Lett 592: 2395–2402 [DOI] [PubMed] [Google Scholar]
- 62. Hamamura Y, Nishimaki M, Takeuchi H, Geitmann A, Kurihara D, Higashiyama T (2014) Live imaging of calcium spikes during double fertilization in Arabidopsis . Nat Commun 5: 4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Denninger P, Bleckmann A, Lausser A, Vogler F, Ott T, Ehrhardt DW, Frommer WB, Sprunck S, Dresselhaus T, Grossmann G (2014) Male‐female communication triggers calcium signatures during fertilization in Arabidopsis . Nat Commun 5: 4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bleckmann A, Alter S, Dresselhaus T (2014) The beginning of a seed: regulatory mechanisms of double fertilization. Front Plant Sci 5: 452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma W, Berkowitz GA (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- 66. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- 67. Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI‐Kat SimpleSearch: new features of the Arabidopsis thaliana T‐DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- 69. Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- 70. Mathieu J, Warthmann N, Kuttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis . Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- 71. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site‐directed mutagenesis by overlap extension using the polymerase chain‐reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- 72. Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y et al (2007) Improved gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- 73. Naito Y, Hino K, Bono H, Ui‐Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off‐target sites. Bioinformatics 31: 1120–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A et al (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- 75. Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes‐Carter F, Campbell NH, Chavali G, Chen C, del‐Toro N et al (2014) The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 42: D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L et al (2015) Glycosylphosphatidylinositol‐anchored proteins as chaperones and co‐receptors for FERONIA receptor kinase signaling in Arabidopsis . Elife 4: e06587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
The protein interactions from this publication have been submitted to the IMEx (http://www.imexconsortium.org) consortium through the IntAct Molecular Interaction Database 75, and assigned the identifier IM‐27345 (https://www.ebi.ac.uk/intact/search/do/search?searchString=pubid:unassigned2053).


