Abstract
Advances in Next Generation Sequencing (NGS) have made available a wealth of information that had previously been inaccessible to researchers and clinicians. NGS has been applied to understand genomic, transcriptomic, and epigenomic changes and gained traction as a significant tool capable of accelerating diagnosis, prognosis, and biomarker discovery. However, these NGS assays have yet to be practical methods for patient stratification or diagnosis because of the gap between the tiny quantities of biomaterials provided by a clinical sample and the large DNA input required by most of these assays. Current library preparation methodologies typically require large input amounts of DNA and a long and complicated manual process. Here, we present a microfluidic droplet-based system for NGS library preparation, capable of reducing the number of pipetting steps significantly, reducing reagent consumption by 10x, automating much of the process, while supporting extremely low DNA input requirement (10 pg per library). This semiautomated technology will allow for low-input preparations of 8 libraries simultaneously while reducing batch-to-batch variation and operator hands-on time.
Introduction
The human genome project was completed in 2003 and cost approximately $3 billion. Since that time the field of sequencing has advanced drastically, moving from Sanger Sequencing1 to Next Generation Sequencing (NGS)2,3. NGS differs from previous methods of DNA sequencing by being characterized as highly scalable methods. This typically is accomplished by fragmenting the genome into small pieces, and randomly sampling these pieces to construct a full genomic library. These fragments can be sequenced in a number of different ways, using one of the many commercially available systems such as: Illumina HiSeq4, ThermoFisher IonTorrent5,6, PacBio SMRT7, Oxford Nanopore8–11, and others. All of these systems have different advantages and disadvantages in terms of cost, speed, performance with long reads, and error rate.
The Illumina platform is currently the most widely-used NGS method12. It works by ligating specific Illumina adapters to both ends of purified DNA fragments. These Illumina adapter ligated DNA sequences are known as a prepared sequencing library. The sequencing library is flowed into a surface modified flow chamber where single stranded library fragments bind to a primer lawn on the flow cell. The single stranded fragments are amplified through bridge amplification, denatured back into single stranded fragments, and amplified more until dense clusters are generated. The exact sequence of these clusters is determined by fluorescently tagging and imaging each individual base in the fragment in order. This method is capable of doing reads up to 150 bases from one or both ends of the fragment. The sequencing data can be deconvoluted based on the ligated adapters and aligned to reference genomes for further analysis.
With the advent of NGS and the reduction of sequencing cost, NGS has become the foundation for a number of genome-wide analyses other than examining genetic changes13,14. Such sequencing technologies include transcriptome profiling (RNA-seq)15, protein-DNA interaction profiling (ChIP-seq)16–18, and DNA methylation profiling (bisulfite sequencing and MeDIP-seq)19,20. These assays give researchers access to information that helps identify biomarkers for diagnostic purposes, understand disease progression or pathways, and stratify patients for targeted precision medical treatment12,21,22.
For various NGS methods, there are slight differences between their respective library preparation protocols. However, in general, library preparation can be broken up into 3 major steps: template preparation, adapter ligation, and amplification23–27. The template preparation and adapter ligation can include a wide variety of steps carried out using different principles and enzymes, which may include random priming, reverse transcription, A tailing and others24,25,27–30. When working with low quantities of genetic material, amplification is a critical step for meeting minimum input requirements for optimal sequencing24. Library preparation protocols, in the interest of producing high quality datasets with low bias amplification, have become increasingly complicated to work with for low-input samples (< 1 ng starting DNA), involving 5 or more reaction steps24,27,30,31 with a number of purifications. The purification steps tend to be an important source of DNA loss32,33.
A small number of methods have been developed to simplify and improve the existing library preparation processes. Digital microfluidics was used to manipulate droplets and prepare the libraries using a bead-based assay34. Others used conventional flow microfluidics performing library preparation in high throughput using either purification columns or a filter using as little as 50 pg gDNA from bacteria35,36. Additionally there was a system utilizing the principle of purification with magnetic beads, capable of library preparation using 660 pg of human gDNA37. In this report, we demonstrate a droplet-based open microfluidic system based on the principle of phase separation purification of DNA38. This platform is capable of producing 8 sequencing libraries simultaneously using as little as 10 pg DNA per assay, following a protocol involving multiple steps for DNA end repair and ligation. We demonstrate its use for preparing low-input ChIP-seq libraries of high quality. Our system offers automation that drastically reduces manual labor and assay time, significant reduction in reagent consumption (by 10x), and a medium to high throughput that is suitable for rapid processing in an individual lab.
Methods
Microfluidic device fabrication
The microfluidic master molds were made by the Virginia Tech Chemical Engineering Machine Shop using a Computer Numerical Control (CNC) milling machine. The reaction channels had a cross sectional width of 1000 μm and height of 500 μm (Figure 1). The reaction channels were ~ 5 cm in length. The molds were machined from a single block of stainless steel 303. PDMS (Sylgard 184) was prepared in a 20:1 ratio of prepolymer to crosslinking agent to improve the hydrophobicity of the cured surface. Prepolymer PDMS was poured onto the master mold, making a 5 mm layer. The PDMS covered mold was degassed for 1 h under vacuum. Degassed prepolymer PDMS was used to coat cleaned 75 mm × 50 mm glass slides with a thin layer of PDMS using a spin coater at 500 rpm for 10 s followed by 1500 rpm for 30 s. For the 8 channel system, 2 coated glass slides were needed. The coated glass slide(s) and the PDMS covered mold were baked for 2 h at 75 °C. The PDMS was then removed from the mold with one set of ends of the channels cut perpendicular to the channel length and 1.5 mm holes punched at the opposite set of ends. The PDMS molded device was bound to the PDMS covered glass slide(s) using plasma bonding (1 min, Harrick plasma cleaner PDC-32G) After plasma bonding, the entire microfluidic chip was baked for 1 h at 75 °C. The channels had all their interior walls in PDMS to provide needed hydrophobicity. These microfluidic devices are made of low-cost PDMS and are disposable after a single use. The stainless-steel master mold used in this project was robust and led to very little chip-to-chip variation (<2% channel failure rate).
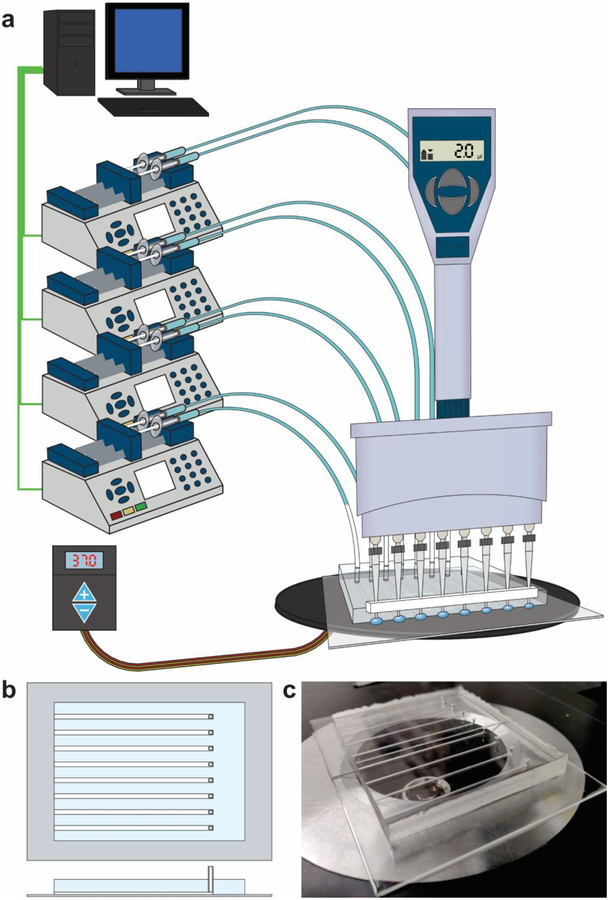
Figure 1.
Overview of the microfluidic library preparation system. (a) The setup including computer-controlled syringe pumps connected to a microfluidic chip on a stage heater. Reagents are delivered to the microfluidic channels using an 8-channel multichannel pipette using a Teflon spacer to align pipette tips with the open ends of microfluidic channels. (b) A schematic of the microfluidic device with 8 channels. Each channel has an open end and a connected end that is with a syringe pump. (c) a picture of the PDMS/glass device (sitting on a microscope stage).
Microfluidic device setup
The microfluidic system was set up as shown in Figure 1 and connected to 4 computer controlled infuse/withdraw syringe pumps, each loaded with two syringes (Chemyx). The syringe pumps were controlled using a custom LabVIEW program. The LabVIEW program was designed to allow for rapid switching between infuse and withdraw modes. The syringes were loaded with water, ensuring that there were no bubbles in the syringes or the connected PFA tubing. The syringe pumps were set to withdraw mode, and a 100 μl air plug was generated in the PFA tubing. The tubing with air plug was then connected to the channels via the 1.5 mm holes punched at one end of each channel.
ChIP DNA preparation
The ChIP DNA was prepared using the MOWChIP system previously reported18,39. Stock ChIP DNA sample was prepared from 10,000 gm12878 cells or the same number of nuclei from mouse prefrontal cortex39. ChIP samples were purified using phenol:chloroform extraction and ethanol precipitation. Samples were resuspended in 10 μl of low EDTA TE buffer. After samples were purified and resuspended the ChIP sample quality was confirmed using qPCR. The quantity of ChIP DNA was determined using Nanodrop. Mouse prefrontal cortex was dissected from 10-week old male CD-1 mice (Charles River Laboratories) and the production of nuclei was detailed in our recent work39.
Library preparation
Libraries were prepared using the microfluidic system or manually following the instructions of the kit manufacturer (ACCEL-NGS 2s plus DNA library kit, Swift Biosciences, Ann Arbor, Michigan, USA) (Figure 2). When using the 8-channel system, an 8-channel pipette was used to deliver the reagents simultaneously, using a Teflon spacer placed on the pipette tips to properly align the reagent dispensing to the open ends of the channels. Library preparation mixes were prepared immediately prior to use. For each reaction, 1/10 of the reaction volume suggested by the kit manufacturer was used in every step. Multiple reagents were thoroughly mixed (in some cases with the contents that were already in the channel) by using a flow rate of 150 μl/min and switching between infuse and withdraw modes in 8 s intervals for a total of 128 s (i.e. 8 cycles of infuse/withdraw). The droplet moved back and forth but stayed within the channel during infusion or withdrawal. This allowed for the reactants (including beads in some cases) to be fully mixed and homogenized.
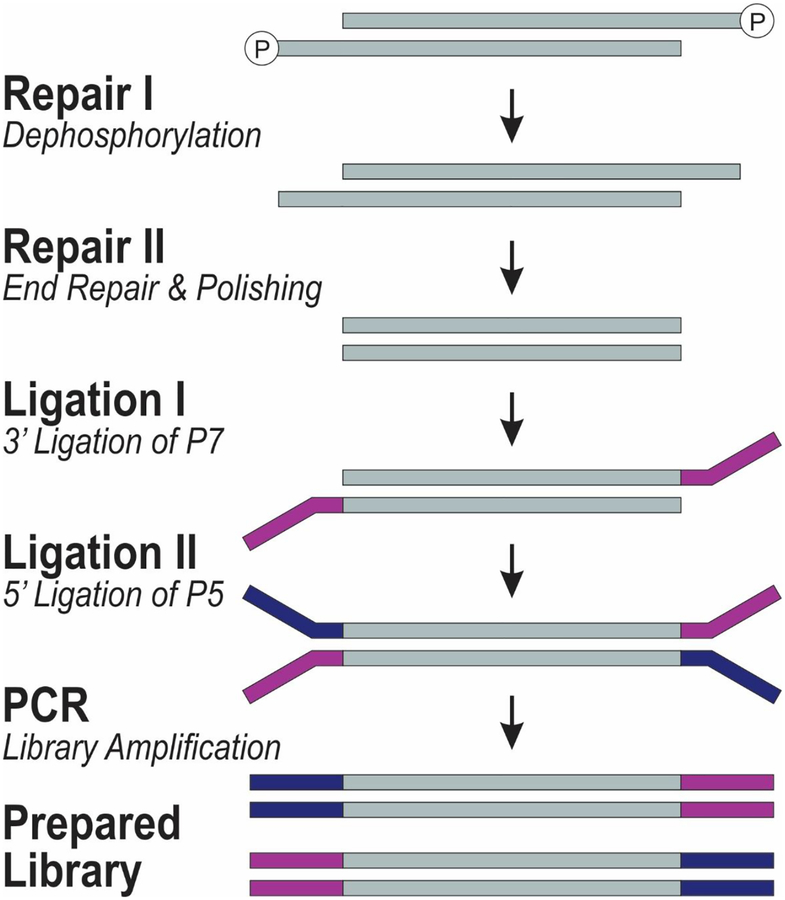
Figure 2.
The molecular process involved in the library preparation used in this work. The process is used in Swift Biosciences Accel 2S DNA library kit.
The library preparation protocol utilizes a series of repeating steps: Initial Reagent Addition, SPRI Cleanup, Reagent Removal, Ethanol Wash, Reagent Addition, and Product Elution, shown in Fig. 3. Initial Reagent Addition: 2 reagents are both pipetted onto the PDMS-covered glass slide at the channel’s open inlet. The reactants were withdrawn into the reaction channel at 15 μl /min, and mixed as described above using infuse/withdraw cycles (Video S1). The mixed reactants were incubated at 37 °C for 10 min. SPRI Cleanup: Next, the liquid droplet plug was withdrawn to the end of the channel to collect any liquid lost from the droplet due to evaporation. The droplet plug was moved to the end of the reaction channel and a droplet of SPRI Select beads (whose volume was dependent on reaction step, see Table 1) were pipetted onto the channel inlet, forming a single droplet plug. The droplet was then withdrawn into the reaction channel at 15 μl/min and mixed (Video S2). The channel contents were incubated for 5 min at room temperature. Reagent Removal: A neodymium magnet (K&J Magnetics, BZ088) was placed under the middle of the reaction chamber, and the droplet plug was flowed out of the reaction channels at 15 μl/min past the magnet, with the beads separated from the droplet plug (Video S3). Ethanol Wash: The channels were washed by rinsing them with 80% ethanol being withdrawn and infused at 15 μl/min, rinsing the channel and the SPRI beads (collected into a pellet using the magnet during this step) a single time (Video S4). Reagent Addition: After the reaction channel was dried, reagents for the next step were added, and the beads were resuspended by the mixing protocol (Video S5). The mixed reactants were incubated in the microfluidic system at temperatures up to 40 °C for between 10 and 20 min (see Table 1 for different conditions). Product Elution: After the final ethanol rinse, the prepared library was eluted into 5 μl of low EDTA TE buffer by mixing, incubating for 5 min, and collected using the multichannel pipette after the liquid droplet was separated from beads using the magnet. This step was similar to reagent removal except that the removed solution contained the product (i.e. libraries before amplification).
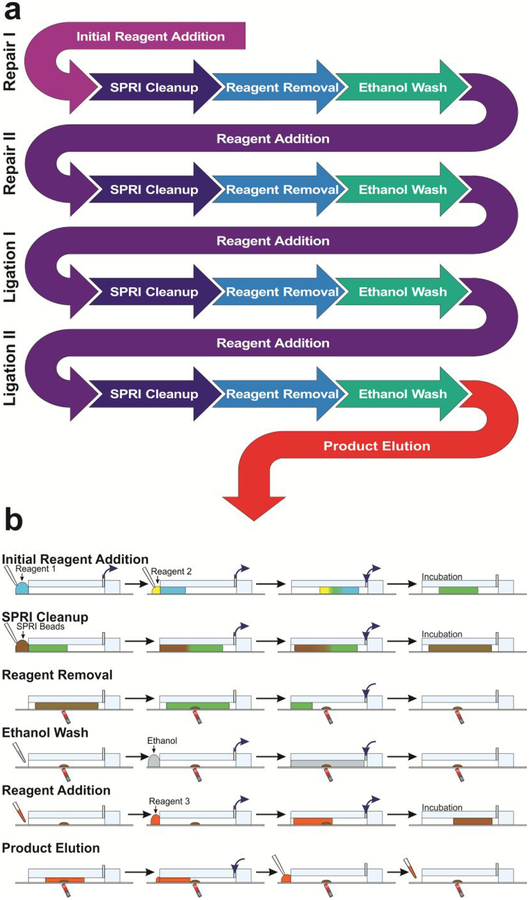
Figure 3.
Overview of microfluidic library preparation process. (a) The flowchart of the library preparation protocol (before PCR) in terms of steps involved. Each process in the library preparation involves one reaction mix (prepared based on manufacturer instructions) and is a repeating cycle of reagent addition, SPRI Cleanup, Reagent Removal, and Ethanol Washing. (b) Various steps involved and their implementation on our microfluidic platform. Reagents are delivered simultaneously using a multichannel pipette. Fluidic manipulation is controlled using the computer-controlled pumps connected to the outlets. The infuse/withdraw motion is denoted using blue arrows at the outlets. Multidirectional arrows denote an oscillatory movement of the droplet.
Table 1.
Summary of library preparation reagent amounts and incubation times
| Step | Reaction Mix Volume (μl) | Incubation Time (min) | Incubation Temperature (°C) | SPRI Bead Amount (μl) |
|---|---|---|---|---|
| Repair I | 1 (Mix) and 2 (DNA) | 10 | 37 | 4.2 |
| Repair II | 2.5 | 20 | 25 | 3.0 |
| Ligation I | 2.4 | 15 | 25 | 2.0 |
| Ligation II | 2.5 | 10 | 40 | 2.1 |
The library preparation protocol was carried out using these repeating steps to complete Repair I, Repair II, Ligation I, and Ligation II. The reaction mix volumes, reaction temperatures, incubation times, and SPRI bead amounts are described in Table 1.
Library amplification, quantification, and pooling
The libraries were amplified using the manufacturer-recommended protocol with the exception of supplementing the reaction mixture with one part of 20x EvaGreen, which allowed for precise control of the library amplification. Libraries were amplified using a BioRad CFX Connect qPCR, with amplification being terminated after a >3000 RFU fluorescence increase was observed. This ensured that the libraries were not over-amplified. This helped to reduce amplification bias and improve library quality. After the libraries were prepared and amplified, the amount of DNA in the library was quantified using KAPA Biosystems Library Quantification Kit according to the manufacturer’s instructions. Libraries were then pooled in equal parts to a total concentration of 10 nM, with few enough samples to get at least 10 million reads per sample (typically <24). The samples were then sequenced using the Illumina HiSeq 4000 sequencer with single-end 50 nt read.
Bioinformatics
The sequencing data was analyzed using methods previously published16,17,39. The raw sequencing data was first trimmed using TrimGalore! before being aligned to the proper genome hg19 and mm10 for the gm12878 and mouse samples, respectively. The samples are analyzed using MACS40 and SICER41 to determine the number of peaks and the redundancy rate. The samples are normalized to allow for comparison between samples with different sequencing read numbers. The normalized signal is converted to bigwig format and visualized using Integrative Genomics Viewer. The normalized signal is averaged over each of the promotor regions, which are defined as 2000 bp up and downstream of the transcription start site. The averaged normalized signal is used to compute Pearson correlation between samples.
Results and Discussion
Here we demonstrate the use of a droplet-based microfluidic system for semi-automated and high-throughput library preparation. We adopt a low-input protocol used in ACCEL-NGS 2s plus DNA library kit (Swift Biosciences) on our microfluidic platform. In addition to significant reduction in manual steps such as pipetting, the microfluidic system is capable of reducing the quantities of required reagents by a factor of 10 without compromising the library quality.
The microfluidic system is setup as shown in Figure 1a. The PDMS microfluidic chip is placed on a microscope stage heater, which can be heated to ensure proper reaction temperatures in the reaction channels. Syringes with attached PFA tubing are each loaded with 500 μl of water and placed on the 4 computer-controlled infuse/withdraw pumps and a 100 μl air gap is generated in each tubing by withdrawing (Fig. 1a). Each tubing is then connected to one end of an individual reaction channel with all inside walls in PDMS and cross-sectional dimensions of 500 μm × 1000 μm (Fig. 1b and 1c). Each reaction channel has a cut-end opening for delivery and removal of reagents on the other end. The delivery of reagents can be conducted simultaneously at all 8 channels using a multichannel pipette.
The library preparation process in ACCEL-NGS 2s plus kit involves 4 basic steps of Repair I, Repair II, Ligation I, and Ligation II, which corresponds with dephosphorylation, end repair, 3’ ligation of P7, and 5’ ligation of P5 (Figure 2). After library preparation, the library can then be amplified using PCR.
The microfluidic system is able to automate the mixing and washing steps repeatedly used in the protocol. Using the programmable infuse/withdraw pumps, we are able to move a liquid droplet in/out of the channel and also back and forth within the channel to create mixing. While the library preparation process involves 4 individual reactions, the reactions all involve the same basic steps of reagent addition, SPRI cleanup, reagent removal, and ethanol washing with the first (“initial reagent addition”) and last (“product elution”) steps of the overall process being slightly different (Fig. 3a). During “initial reagent addition”, DNA and the first reaction mix (for Repair I) are piecewise added to the reaction channel for mixing. During “SPRI cleanup”, SPRI beads are used to reversibly immobilize the library DNA on the bead surface in order to separate DNA from solution. During “reagent removal”, solution is pushed out the channel while the SPRI beads with adsorbed DNA are retained by a magnet. During “ethanol washing”, ethanol is used to remove all traces of reagent mixture in the channel. Then in the next cycle, during “reagent addition”, a reaction mix for Repair II is added, involving mixing the new solution with dried SPRI beads to elute the library DNA from the bead surface into the reaction solution. This is followed by SPRI cleanup, reagent removal and ethanol washing again. Another two cycles are then conducted for Ligation I and Ligation II with corresponding reaction mix in each case (Fig. 3a). Finally, during “product elution”, library DNA on the bead surface is eluted into low EDTA TE buffer for removal from the library preparation system. These basic steps can be readily carried out on our microfluidic droplet platform in a semi-automated fashion (Figure 3b). For reagent addition, solution is distributed to all of the reaction channels using a multichannel pipette. Using a Teflon spacer to ensure tip-to-channel alignment, solutions are pipetted onto the channel inlets and withdrawn into the channel using the syringe pumps. If multiple reagents are to be combined, the two solutions merge at the inlets (as in Fig. 3b “initial reagent addition” and “SPRI cleanup”) by withdrawing two droplets into the channel sequentially. The solution in the channel can be mixed using rapid repeating infuse and withdraw steps (as in Fig. 3b “initial reagent addition”, “SPRI cleanup”, and “reagent addition”). After 8 cycles of mixing the solution is fully homogeneous and mixed. The solution can then be incubated at elevated or room temperature to allow the reaction to take place (as in Fig. 3b “Initial reagent addition”), DNA to precipitate onto the SPRI bead surface (as in Fig. 3b “SPRI cleanup”), or DNA to release from SPRI bead surface (as in Fig. 3b “reagent addition”). After SPRI cleanup steps, the beads can be separated from the solution by using a magnet below the channels at an angle (Fig. 3b “reagent removal”). The magnetic field aggregates the beads to the bottom of the channel while the syringe pump infuses air to push the liquid segment out of the channel. The solution is subsequently removed with a pipette. Using the same above principles for droplet manipulation with the syringe pumps and magnetic bead concentration and retention, we can also conduct bead treatment during ethanol wash, reagent addition, and product elution (Fig. 3b). The entire microfluidic process took ~ 2 h. Because the PCR amplification readily allows parallel processing of a number of samples, we conduct PCR amplification of the 8 DNA samples yielded from the microfluidic device using a qPCR machine. For each mixing-washing cycle, the microfluidic system requires ~1 min hands-on time per 8 samples. In comparison, manual preparation requires careful pipetting for full liquid removal in addition to 3–5 min of pipetting for the same mixing-washing cycle, per sample.
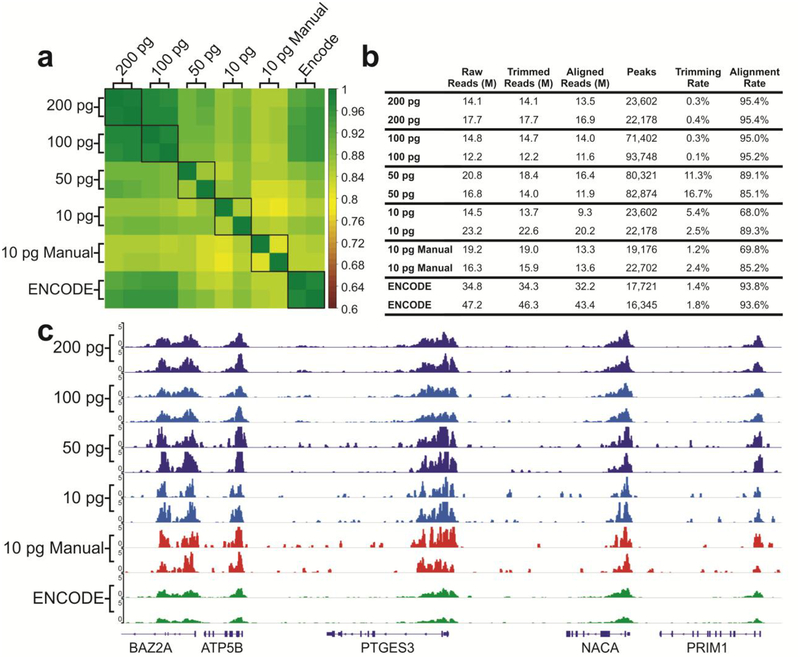
To validate the library preparation system and its compatibility with low-input samples, we prepared ChIP-seq libraries using ChIP DNA on H3K4me3 prepared in bulk from GM12878 cells using MOWChIP-seq developed in our lab18,39. ChIP-seq analysis allows us to examine the enrichment and peaks of ChIP DNA after library preparation to determine whether the library preparation process maintains the original distribution of ChIP DNA in the genome. We used 200, 100, 50, and 10 pg samples of ChIP DNA (anti-H3K4me3) for library preparation on our microfluidic droplet platform. Figure 4 demonstrates that the libraries prepared using 100 and 200 pg present very high quality (with Pearson correlation between replicates at 0.978 and 0.981, respectively, and alignment rates higher than 95%). These data with 100 pg or more ChIP DNA are highly correlated with (Pearson correlation ~ 0.975) and slightly superior in the alignment rate to the data published by ENCODE consortium42 which typically started with 1–10 ng DNA for library preparation. To challenge our system, we also used 10 pg ChIP DNA for library preparation using the microfluidic platform and compared the data to those by manually prepared libraries (“10 pg Manual”). The data quality is similar between the two approaches with the microfluidic system offering slightly higher consistency (Pearson correlation of 0.853 by the microfluidic system vs. 0.811 by manual process, as shown in Figure 4a). This consistency and data quality are further demonstrated in the consistent epigenomic profiles seen in Figure 4c. Overall the data produced by the microfluidic system show very low background outside the peak areas.
Figure 4.
Overview of the quality of sequencing libraries prepared using the microfluidic system compared to manually prepared samples (10 pg) and ENCODE data (prepared using > 1 ng ChIP DNA). The ChIP DNA was on H3K4me3. (a) Pearson correlation matrix of various samples. Signals in the promoter regions (±2 kb around TSSs) were used for computation of Pearson correlation coefficients. (b) Summary of library preparation sequencing metrics. (c) Genome browser tracks comparing various datasets with libraries prepared by the microfluidic system and manually and those published by ENCODE consortium.
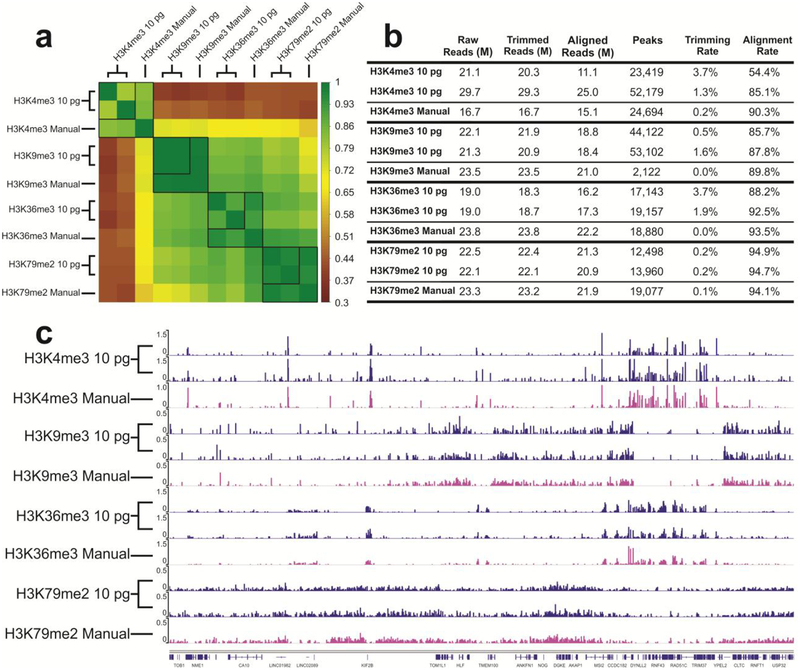
To further demonstrate the value of our microfluidic library preparation system for high-throughput and low-input processing starting with primary tissues, we prepared 8 libraries simultaneously using 4 types of ChIP DNA corresponding to 4 different histone modifications (H3K4me3, H3K9me3, H3K36me3, H3K79me2) with 10 pg DNA from mouse prefrontal cortex used as starting material for each library. All the 8 libraries were prepared in one run using one device. Figure S1 shows the DNA size distribution before and after library preparation using the microfluidic device. The fragment size roughly had a shift of 120 bp due to ligation of adapters. We additionally prepared samples using manual library preparation methods for comparison. ChIP DNA produced from different histone marks may cover various regions of the genome and have different peak patterns. As shown in Figure 5, the samples prepared by the microfluidic system have high correlation between technical replicates (0.796 for H3K4me3, 0.997 for H3K9me3, 0.897 for H3K36me3, 0.968 for H3K79me2), and high correlation with manually prepared samples (on average 0.836 for H3K4me3, 0.992 for H3K9me3, 0.934 for H3K36me3, 0.966 for H3K79me2) (Figure 5a). The profiles in Figure 5c shows that there is no crosstalk among different units and the library preparation is conducted independently without interference from other units.
Figure 5.
Comparison of ChIP datasets of mouse prefrontal cortex samples against 4 different histone modifications (H3K4me3, H3K9me3, H3K36me3, H3K79me2). 10 pg ChIP DNA was used in each unit and 8 libraries were prepared in one run in one device. The “Manual” data on H3K9me3, H3K36me3, H3K79me2 were taken from our recently published work39. (a) Pearson correlation matrix. Signals in the promoter regions (±2 kb around TSSs) were used for computation of Pearson correlation coefficients. (b) Summary of ChIP-seq data metrics. (c) Genome browser tracks comparing microfluidic library preparation system to manual preparation for all 4 histone marks.
Conclusions
Library preparation is one of the biggest roadblocks in terms of cost and labor for performing NGS assays. To create a semi-automated and low-input platform for conducting library preparation using low-abundance samples (down to 10 pg starting DNA), we have developed a microfluidic system based on 8 parallel channels and droplet manipulation. Using our system, we are able to simultaneously reduce the cost of library preparation by using up to 10x less reagents, and the labor associated with pipetting by having programed operations. We demonstrate a scalable microfluidic technology capable of producing high-quality and reproducible sequencing libraries. Although the technology is demonstrated for preparing ChIP-seq libraries, the platform is universally applicable to all Illumina-based library preparation.
As the current microfluidic workflow consists of a number of common reaction steps across different library preparation kits (reaction, mixing, and bead-based purification), it could be utilized to improve the workflow for most other commercially available kits. This study chose to use the Swift Biosciences ACCEL-NGS 2s plus kit, as it requires as little as 10 pg input DNA.
Our system is ideal for handling operations of medium size, although the system can be easily scaled up by replicating the control and microfluidic devices. Sequencing library preparation in an individual academic or clinical lab typically requires a throughput of 10 samples or less per batch. This makes use of very high-throughput and expensive platform like liquid-handling robot excessive. Our system readily suits such need.
Supplementary Material
Acknowledgements
We thank Michael Vaught and Kevin Holshouser at Virginia Tech Chemical Engineering Machine Shop for technical support. This work was supported by US National Institutes of Health grants CA214176 (CL), EB017235 (CL), and seed grants from Center for Engineered Health of Virginia Tech Institute for Critical Technology and Applied Science.
Footnotes
Supporting Information
Fig. S1 on DNA size distribution before and after library preparation using the microfluidic device; Video S1 “Initial Reagent Addition.mp4”; Video S2 “SPRI Cleanup. mp4”; Video S3 “Reagent Removal.mp4”; Video S4 “Ethanol Wash.mp4”; Video S5 “Reagent Addition.mp4”
Accession codes
Gene Expression Omnibus: Our sequencing data are deposited under accession number GSE132786;
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132786
The authors declare the following competing financial interest(s): We purchased ACCEL-NGS 2s plus kits at a discount from Swift Biosciences.
References
- (1).Sanger F; Nicklen S; Coulson AR Proc. Natl. Acad. Sci 1977, 74, 5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Margulies M; Egholm M; Altman WE; Attiya S; Bader JS; Bemben LA; Berka J; Braverman MS; Chen Y-J; Chen Z; Dewell SB; Du L; Fierro JM; Gomes XV; Godwin BC; He W; Helgesen S; Ho CH; Irzyk GP; Jando SC, et al. Nature 2005, 437, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Shendure J; Porreca GJ; Reppas NB; Lin X; McCutcheon JP; Rosenbaum AM; Wang MD; Zhang K; Mitra RD; Church GM Science 2005, 309, 1728. [DOI] [PubMed] [Google Scholar]
- (4).Bentley DR; Balasubramanian S; Swerdlow HP; Smith GP; Milton J; Brown CG; Hall KP; Evers DJ; Barnes CL; Bignell HR; Boutell JM; Bryant J; Carter RJ; Keira Cheetham R; Cox AJ; Ellis DJ; Flatbush MR; Gormley NA; Humphray SJ; Irving LJ, et al. Nature 2008, 456, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Golan D; Medvedev P Bioinformatics 2013, 29, i344–i351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Merriman B; D Team IT; Rothberg JM Electrophoresis 2012, 33, 3397–3417. [DOI] [PubMed] [Google Scholar]
- (7).Chin C-S; Alexander DH; Marks P; Klammer AA; Drake J; Heiner C; Clum A; Copeland A; Huddleston J; Eichler EE; Turner SW; Korlach J Nat. Methods 2013, 10, 563. [DOI] [PubMed] [Google Scholar]
- (8).Ashkenasy N; Sánchez-Quesada J; Bayley H; Ghadiri MR Angewandte Chemie International Edition 2005, 44, 1401–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Clarke J; Wu H-C; Jayasinghe L; Patel A; Reid S; Bayley H Nat. Nanotechnol 2009, 4, 265. [DOI] [PubMed] [Google Scholar]
- (10).Fologea D; Gershow M; Ledden B; McNabb DS; Golovchenko JA; Li J Nano Lett 2005, 5, 1905–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gribnau J; Hochedlinger K; Hata K; Li E; Jaenisch R Genes Dev 2003, 17, 759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Metzker ML Nat. Rev. Genet 2009, 11, 31. [DOI] [PubMed] [Google Scholar]
- (13).Ma S; Murphy TW; Lu C Biomicrofluidics 2017, 11, 021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Murphy TW; Zhang Q; Naler LB; Ma S; Lu C Analyst 2018, 143, 60–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sarma M; Lee J; Ma S; Li S; Lu C Lab Chip 2019, 19, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ma S; Hsieh Y-P; Ma J; Lu C Sci. Adv 2018, 4, eaar8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Murphy TW; Hsieh YP; Ma S; Zhu Y; Lu C Anal. Chem 2018, 90, 7666–7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cao Z; Chen C; He B; Tan K; Lu C Nat. Methods 2015, 12, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ma S; de la Fuente Revenga M; Sun Z; Sun C; Murphy TW; Xie H; González-Maeso J; Lu C Nat. Biomed. Eng 2018, 2, 183–194. [PMC free article] [PubMed] [Google Scholar]
- (20).Zhu Y; Cao Z; Lu C Analyst 2019, 144, 1904–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schuster SC Nat. Methods 2007, 5, 16. [DOI] [PubMed] [Google Scholar]
- (22).Shendure J; Ji H Nat. Biotechnol 2008, 26, 1135. [DOI] [PubMed] [Google Scholar]
- (23).Head SR; Komori HK; LaMere SA; Whisenant T; Van Nieuwerburgh F; Salomon DR; Ordoukhanian P BioTechniques 2014, 56, 61-passim. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sundaram AYM; Hughes T; Biondi S; Bolduc N; Bowman SK; Camilli A; Chew YC; Couture C; Farmer A; Jerome JP; Lazinski DW; McUsic A; Peng X; Shazand K; Xu F; Lyle R; Gilfillan GD BMC Genomics 2016, 17, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Quail MA; Kozarewa I; Smith F; Scally A; Stephens PJ; Durbin R; Swerdlow H; Turner DJ Nat. Methods 2008, 5, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tyler AD; Christianson S; Knox NC; Mabon P; Wolfe J; Van Domselaar G; Graham MR; Sharma MK PLOS ONE 2016, 11, e0148676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Aigrain L; Gu Y; Quail MA BMC Genomics 2016, 17, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bowman SK; Simon MD; Deaton AM; Tolstorukov M; Borowsky ML; Kingston RE BMC Genomics 2013, 14, 466–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lazinski DW; Camilli A BioTechniques 2013, 54, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Peng X; Wu J; Brunmeir R; Kim S-Y; Zhang Q; Ding C; Han W; Xie W; Xu F Nucleic Acids Res. 2015, 43, e35–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ding C; Zhang Q; Brunmeir R; Kim S-Y; Peng X; Xu F; Wu J; Xie W; Han W Nucleic Acids Res. 2014, 43, e35–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).DeAngelis MM; Wang DG; Hawkins TL Nucleic Acids Res. 1995, 23, 4742–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fisher S; Barry A; Abreu J; Minie B; Nolan J; Delorey TM; Young G; Fennell TJ; Allen A; Ambrogio L; Berlin AM; Blumenstiel B; Cibulskis K; Friedrich D; Johnson R; Juhn F; Reilly B; Shammas R; Stalker J; Sykes SM, et al. Genome Biol. 2011, 12, R1–R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kim H; Bartsch MS; Renzi RF; He J; Van de Vreugde JL; Claudnic MR; Patel KD JALA: Journal of the Association for Laboratory Automation 2011, 16, 405–414. [DOI] [PubMed] [Google Scholar]
- (35).Kim S; De Jonghe J; Kulesa AB; Feldman D; Vatanen T; Bhattacharyya RP; Berdy B; Gomez J; Nolan J; Epstein S; Blainey PC Nat. Commun 2017, 8, 13919–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tan SJ; Phan H; Gerry BM; Kuhn A; Hong LZ; Min Ong Y; Poon PSY; Unger MA; Jones RC; Quake SR; Burkholder WF PLOS ONE 2013, 8, e64084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Snider A; Nilsson M; Dupal M; Toloue M; Tripathi A SLAS TECHNOL.: Translating Life Sciences Innovation 2018, 24, 196–208. [DOI] [PubMed] [Google Scholar]
- (38).Berry SM; Alarid ET; Beebe DJ Lab Chip 2011, 11, 1747–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhu BH, Y. -P; Murphy TW; Zhang Q; Naler LB; Lu C Nat. Protoc 2019, 14, 3366–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang Y; Liu T; Meyer CA; Eeckhoute J; Johnson DS; Bernstein BE; Nusbaum C; Myers RM; Brown M; Li W; Liu XS Genome Biol. 2008, 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Xu S; Grullon S; Ge K; Peng W Methods Mol. Biol 2014, 1150, 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).The ENCODE project consortium. Nature 2012, 489, 57.22955616 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.