Abstract
Over 50% of colorectal cancer (CRC) patients develop resistance after a transient response to therapy. Understanding tumor resistance from an evolutionary perspective leads to better predictions of treatment outcomes. The objectives of this study were to develop a computational framework to analyze tumor longitudinal measurements and recapitulate the individual evolutionary dynamics in metastatic CRC (mCRC) patients. A stochastic modeling framework was developed to depict the whole spectrum of tumor evolution prior to diagnosis and during and after therapy. The evolutionary model was optimized using a non-linear mixed effect (NLME) method based on the longitudinal measurements of liver metastatic lesions from 599 mCRC patients. The deterministic limits in the NLME model were applied to optimize the stochastic model for each patient. Cox proportional hazards models coupled with the least absolute shrinkage and selection operator (LASSO) algorithm were applied to predict patients’ progression-free survival (PFS) and overall survival (OS). The stochastic evolutionary model well described the longitudinal profiles of tumor sizes. The evolutionary parameters optimized for each patient indicated substantial inter-patient variability. The number of resistant subclones at diagnosis was found to be a significant predictor to survival, and the hazard ratios with 95% CI were 1.09 [0.79–1.49] and 1.54 [1.01–2.34] for patients with three or more resistant subclones. Coupled with several patient characteristics, evolutionary parameters strongly predict patients’ PFS and OS. A stochastic computational framework was successfully developed to recapitulate individual patient evolutionary dynamics, which could predict clinical survival outcomes in mCRC patients.
Keywords: evolutionary dynamic, survival prediction
Introduction
As the third-leading cause of cancer mortality in the United States, colorectal cancer (CRC) is expected to cause approximately 51,020 deaths during 2019 (8.4% of all cancer deaths). (1) More than 35% CRC patients already have liver metastases at diagnosis, and the five-year survival rate is only 11% for metastatic CRC (mCRC) patients. (2) Targeted therapies (cetuximab and panitumumab for wild-type KRAS patients, bevacizumab) combined with the chemotherapy FOLFIRI (fluorouracil, leucovorin, and irinotecan) were approved as the first- and second- line treatment for unresectable mCRC patients. (3) Despite the broad use of combination therapy, the development of resistance to treatment in mCRC patients continues to grow, which drastically limits patients’ long-term survival. (4,5) Most patients only show a transient response to therapy; the residual cancer cells can quickly adapt and gain resistant phenotypes through either genetic or epigenetic mechanisms, leading to relapse, disease progression, and eventually patient death. (6)
CRC harbors extensive intratumor heterogeneity with numerous subclones. (7) Many pre-existing small CRC subclones that are below the detection limit at diagnosis could rapidly accumulate during therapy and build resistance to treatment. (8) The selective pressure imposed by therapeutics typically expedites the accumulations of certain small pre-existing subclones that would cause tumor resistance and treatment failure. (9,10) Therefore, it is crucial to elucidate the evolutionary dynamics of CRC prior to, during, and after the therapy and identify the drivers to the evolution process. This will enable treatment to be proactively designed to delay or possibly thwart tumor evolution and further prolong patient survival. As shown in many studies, tumors from mCRC patients are comprised of genetically and epigenetically distinct cell populations, and the therapy could change the composition of these cell populations throughout the treatment process, making it quite challenging to predict evolutionary trajectories at the individual level. (7,11–13)
Multiple genetic biomarkers, including microsatellite instability, mutant KRAS, mutant BRAF, and mutant PIK3CA, have been implemented to select molecularly targeted therapies for CRC. (14,15) These genetic biomarkers have proven to be useful to predict the initial response to targeted treatments, but their predictability on patients’ long-term survival benefit remains unclear without considering clonal evolution. Existing tumor evolutionary studies on clonal dynamics have mainly relied on the sequencing data of biopsy samples from either pre- or post-treatment time points to speculate the evolutionary relationships among subclones (phylogenetics). (12,16,17) These analyses could yield many insights into tumor evolution. However, such analyses based on genomic profiling and computational/statistical examination remain limited to predict treatment efficacy. Tumor clone selection is made not only on genotypes but also phenotypes. (18,19) The prediction of tumor evolution solely based upon genetic compositions is often biased and lacks clinical validation, particularly when predicting patients’ long-term survival benefit.
Bulk tumor measurements, recorded in almost all clinical visits, are generally thought to have limited information on tumor evolution. In this study, we developed a parsimonious evolutionary modeling framework to analyze longitudinally measured bulk tumors and depict the evolutionary dynamics for each individual patient. These evolutionary constants were found to be closely associated with patients’ long-term survival outcomes.
Materials and Methods
Data source and mCRC patient characteristics
A phase III clinical trial data was acquired from Project Data Sphere (ClinicalTrials.gov ID ). It was an open-label, randomized, multicenter trial including two treatment arms—a FOLFIRI alone group and a panitumumab with FOLFIRI treatment group. (20) Patients’ clinical characteristics, progression-free survival (PFS), overall survival (OS), patients’ RECIST responses (21), and the longest diameter of liver metastatic lesion segmented from all CT scans were extracted for the following analysis.
The volume of liver metastatic lesions were calculated based on the longest lesion diameter assuming the ellipsoidal shape of tumor lesions and the average ratio of the tumor long versus short axis as 1.31. (22) The volume was converted to the tumor cell number assuming that 1 cm3 in lesion volume was comprised of approximately 109 tumor cells. (23) The equation is shown in equation 1. The detection limit of tumor cells was assumed to be 107.
| (equation 1) |
Patients with one of the following conditions were excluded from the analysis: no liver metastasis, with prior resection surgery on a liver metastasis, or with prior radiation therapy on a liver metastasis. In total, 599 patients were selected for the following analysis. The clinical demographics of all patients are summarized in Table 1.
Table 1.
Patient characteristics in model estimation.
| Demographics | Total = 599 |
|---|---|
| Male – N (%) | 360 (60.1) |
| Age, years – mean (SD) | 60.4 (10.4) |
| White race – N (%) | 595 (99.3) |
| Panitumumab arm- N (%) | 308 (51.4) |
| ECOG performance status – N (%) | |
| 0–1 | 571 (95.3) |
| ≥2 | 28 (4.7) |
| Primary tumor type – N (%) | |
| Colon | 383 (63.9) |
| Rectal | 216 (36.1) |
| No. of metastatic sites – N (%) | |
| 1 | 118 (19.7) |
| 2 | 195 (32.6) |
| ≥3 | 282 (47.1) |
| LDH baseline≥1.5UL – N (%) | 205 (34.2) |
| KRAS state – N (%) | |
| Wild-type | 296 (49.4) |
| Mutant | 242 (40.4) |
| Unknown | 61 (10.2) |
| BMI, kg/m2 – mean (sd) | 26.3 (4.8) |
| Response – N (%) | |
| Complete response | 0 (0) |
| Partial response | 137 (22.9) |
| Stable disease | 290 (48.4) |
| Progressive disease | 154 (25.7) |
| Unable to evaluate | 12 (2.0) |
| Not done | 6 (1.0) |
Tumor evolutionary dynamics
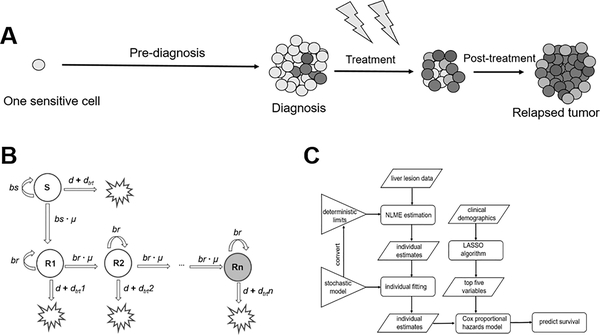
To recapitulate the tumor evolutionary dynamics at the individual patient level, a parsimonious stochastic branching model (24) was constructed and optimized using longitudinal tumor measurements. The whole spectrum of tumor evolution was considered in the model starting from one malignant cell to a bulk tumor lesion at diagnosis, followed by a transient response to treatment, until the point of resistance and final relapse. (Figure 1A)
Figure 1. The stochastic model structure and methods flow chart.
a. The model starts from one sensitive cell and simulates the tumor evolution trajectory including the pre-diagnosis, treatment, and follow-up (post-treatment) phases. During the follow-up phase, patients are off targeted therapies. b. Schematic of the stochastic tumor growth model. S represents the sensitive cell subclone, and R represents the resistant cell subclones. c. The workflow of our methods. The stochastic model was converted to its deterministic limits for non-linear mixed effect (NLME) estimation. Individual estimates from NLME estimation were used in later stochastic individual fitting as the initials.
As shown in Figure 1B, the sensitive cell population divides at a rate bs and dies at a rate d. Cells acquire resistance at a transition rate μ per cell division independently of therapy. Resistant cell populations emerge in a cascade manner, and resistant cells divide at the same rate br and die at the same rate d. To reflect the resistant phenotypes, response to treatment dtrt decreases to 1% of the previous population for each following resistant population. More details regarding model structure and the underlying model assumptions were given in the Supplementary Text S1.
As shown in Figure 1C, a workflow was constructed to optimize the evolutionary parameters at individual patient level using longitudinal tumor measurements. The degrees at which tumor cells proliferate (bs and br), die (d), transit to resistance (μ), and respond to treatments (dtrt) were all estimated for each patient using the measured liver metastatic lesions.
To enable model optimization at the patient-specific level, a non-linear mixed effect (NLME) method was first applied to search the global optimum considering many individuals having sparse or missing tumor measurements in real-world settings. Directly optimizing the stochastic model using the NLME method is not feasible because the likelihood function used in solving NLME equations is defined as a multiple integral over a high-dimension event space. Currently no statistical method is available to support such a high computational space. (25) In order to estimate parameters for stochastic NLME models, several assumptions have been made, such as the stochastic model could be approximated as deterministic model when the species number is large. We therefore estimated the population as well as individual parameters through its deterministic limits on MonolixSuite 2018R2 using the stochastic approximation expectation-maximization (SAEM) algorithm. (26) The stochastic model was first simplified to one sensitive subclone (S) and three resistant subclones (R1, R2, R3), and the ODEs are shown in equation 2–5.
| (equation 2) |
| (equation 3) |
| (equation 4) |
| (equation 5) |
In the equations above, the next higher index of dtrt is 1% of that with the last index one (dtrt n+1 = 0.01 × dtrt n).
Using the NLME method, the model parameters are described in equation 6–9.
| (equation 6) |
| (equation 7) |
| (equation 8) |
| (equation 9) |
where θx is the typical value of x in the patient population, and ηxj is the random effect describing the difference between the typical and individual values of parameter x for subject j. The random effects and ηnet growth were assumed to have a log-normal distribution with a mean of zero and estimated variances reflecting inter-individual variability. The random effect of malignant cell natural death rate (d) was assumed as a logit-normal distribution to limit d in a reasonable range (from 0 to 1).The difference between model predictions and measurements were assumed to be additive with log-normally distributed error. Notably here, we estimated the resistant subclones regrowth rate (θnet growth = br - d). The sensitive cell net growth rate was optimized by selecting a lower model Akaike information criterion (AIC) and fixed in NLME estimation for all subjects. More details regarding model structure and the underlying assumptions were given in the Supplementary Text S1.
The original stochastic model (Figure 1B), with the “pattern search” algorithm (27) to find the minimum of the objective function (equation 10) in MATLAB R2017b, was applied to search individual parameters for all 599 patients. In this stochastic estimation, the NLME-derived individual estimates were used as initials. We also set the parameter searching range within the 50% to 150% of the NLME-derived individual estimates to reduce the computational workload. We averaged the estimation on 1000 replicates each time to diminish the deviations caused by the stochastic process. The estimated parameters were further used to simulate the tumor growth trajectory and the sizes of each subclone at diagnosis for each single patient.
| (equation 10) |
where the Mobs is the observed tumor cell number and Mpred is the model-predicted cell number. The details of the stochastic model estimation were provided in the Supplementary Text S1.
The liver metastatic lesion cell number predictions, obtained with the stochastic model, were evaluated. Relative standard error (RSE) and diagnostic plots (i.e., plots of observations vs predictions and of prediction errors) were used as major criteria. The observed and simulated lesion cell number were also visually compared for each individual to ensure the model captures tumor growth profiles. (28)
Predictability of clinical outcomes by evolutionary parameters
The individually optimized evolutionary parameters were summarized under different clinical characteristics. ANOVA, multiple paired Student’s t-tests, Cohen’s d effect size comparisons and Pearson correlation tests were performed. The parameter distributions were tested with different distribution functions and were selected based on the lowest AIC. We compared Weibull, Gamma, and log-normal distributions for the response to treatment, and Cauchy and Laplace distributions for the transition rate to resistance. All the analyses were performed in R 3.5.1 and RStudio (Version 1.1.456).
Patients were grouped based on their evolutionary parameters to compare their survival outcomes. PFS and OS were plotted to compare patients with a high response to treatment (response to treatment ≥ median) vs a low response (< median). PFS and OS were also compared based on other model parameters using median values as cut-off and the number of resistant subclones at diagnosis (3, 4, or ≥5 subclones). The statistical analysis was done in R3.5.1 and RStudio.
Cox proportional hazards models were built to integrate the available baseline clinical characteristics into the evolutionary parameters to predict patients’ long-term survival. Patients with complete clinical characteristic profiles were included in the following analysis. In total, 526 patients were selected for this analysis. To identify potential survival-related covariates, a machine-learning algorithm LASSO (29) was applied on all available baseline clinical demographics. Non-baseline covariates including treatment duration and post-treatment factors were excluded. The most prognostic covariates were selected by the lambda value within one standard error from the minimum (lambda.1se) to predict PFS and OS. The estimated hazard ratios (HR) and two-sided 95% CIs (confidence intervals) of the evolutionary parameters after adjustment by the clinical demographics selected by LASSO were calculated. All variables were standardized to a mean of zero and variance of one for consistency. Forest plots were depicted to compare the contributions of each selected covariate. Cross-validation was performed by splitting the dataset half-half randomly and measuring the C-index of the split test datasets on the Cox proportional model as an internal validation. The statistical analysis was done in R.3.5.1 and RStudio.
Results
Evolutionary dynamics of individual patients
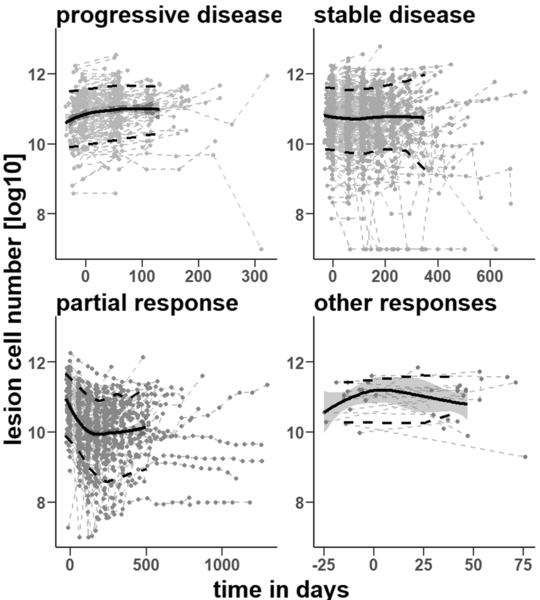
A total of 599 mCRC patients were included in our analysis. All patients had longitudinal measurements of the liver metastatic lesion size. The longitudinal tumor profiles are depicted in Figure 2. According to the RECIST criteria (21), the dataset included 137 patients with a partial response (PR), 290 with a stable disease (SD), and 154 with a progressive disease (PD). The remaining 18 patients were either “unable to evaluate” (UE) or “not done” (ND).
Figure 2. Liver lesion cell number data.
The liver metastatic lesion cell numbers change along with time for patients with a progressive disease (N=154), a stable disease (N=290), partial response (N=137), and other responses including “unable to detect” and “not done” (N=18). The black solid line with the blue shadow is the Loess regression line with a 95% confidence interval. The two broken black lines are the 10% and 90% percentiles of the data measurements, respectively.
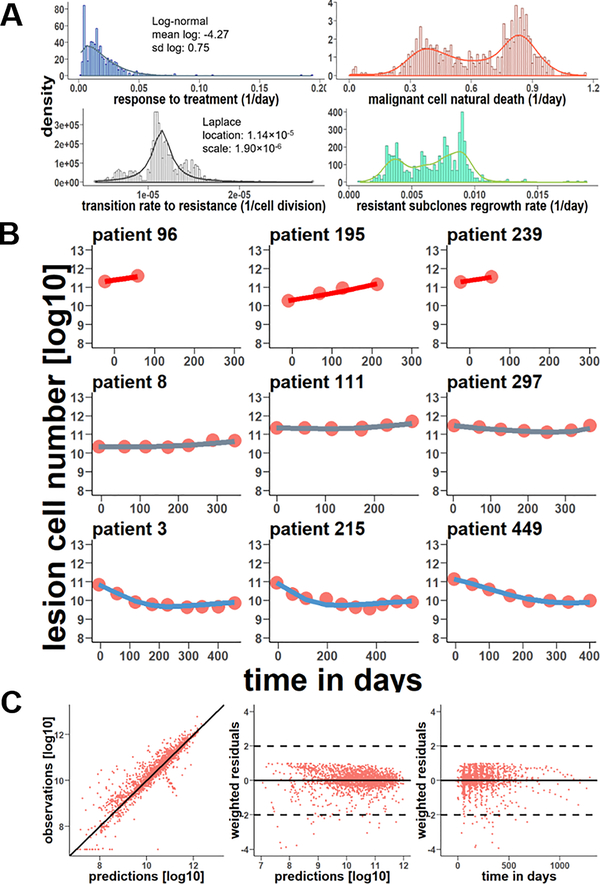
In the dataset, 149 patients (24.9%) had two lesion measurements, 114 patients (19.0%) had three measurements, and 336 patients (56.1%) had four or more. It is technically challenging to fit the evolutionary model for patients with limited sampling measurements. The NLME model was therefore applied to accommodate data sparsity and data missing issues by borrowing information from each other. The population average and inter-patient variabilities of model parameters are summarized in Table 2. The RSE of model parameters are all below 30%, indicating robust parameter estimation. The net growth rate of the sensitive cell population was estimated as 0.01 per day, predicting an approximate evolution time of 5.68 years (2072 days) from a single metastatic cell expanding into a 109 cell bulk (~ 1 gram tumor). The evolution time was approximated as the reciprocal of the sensitive cell net growth rate divided by the natural logarithm of 109. Notably, these estimations were based on deterministic ordinary differential equations and could not reflect the random genetic drift caused by fluctuations in cell proliferation. (30) With these deterministic model-derived parameters as initial values, we optimized the stochastic model for each patient. The distributions of stochastic model-derived parameters are shown in Figure 3A. Large inter-patient variabilities were observed in the model parameters. The individual patient response to treatment exhibits log-normal distribution. The transition rate to resistance shows Laplace distribution, with a median of approximately 1.14×10−5 per cell division. Both malignant cell natural death rate and resistant subclones regrowth rate follow bimodal distribution, suggesting two distinct subpopulations in the analyzed patient population. The broad distribution of these parameters indicated high heterogeneity in tumor evolution and variabilities in response to therapy.
Table 2.
Stochastic model deterministic limit estimates.
| Parameter | Unit | Description | Estimate | RSE%1 | IIV2 | RSE%3 |
|---|---|---|---|---|---|---|
| Response to treatment | 1/day | Treatment killing rate for sensitive cells | 0.012 | 3.8 | 0.693 | 4.82 |
| Malignant cell natural death rate | 1/day | Cell death rate for each individual | 0.696 | 10.4 | 2.41 | 11.1 |
| Transition rate to resistance | 1/cell division | Mutation rate of the cell-gaining resistant mechanism | 1.03×10−5 | 7.23 | 0.444 | 13.6 |
| Resistant subclones regrowth rate | 1/day | Resistant cell net growth rate | 0.00574 | 3.45 | 0.367 | 5.58 |
The relative standard error of parameter estimates in all 599 patients
The standard deviation of the inter-individual variability distribution
The relative standard error of the standard deviation estimates of inter-individual variability distribution
Figure 3. Stochastic model parameter distributions and diagnostics.
a. Histograms of the response to treatment, the malignant cell natural death rate, the transition rate to resistance, and resistant subclones regrowth rate with their density functions. b. Nine representative patients with a progressive disease (above row), stable disease (middle row), or partial response (bottom row) and their stochastic individual fitting results. The red circles are liver lesion observations along with time and the line is the stochastic model predictions. c. Diagnostic plots of the stochastic model. The left plot is the stochastic predictions vs observations in the logarithm, and the black line is y=x. The middle plot is the stochastic predictions vs weighted residuals ([observations-predictions]/observations), and the right plot is the time vs weighted residuals.
The evolutionary model adequately captured the individual tumor growth trajectory and effectively predicted the transient response phase quickly turning into a relapse phase. Nine individual profiles are provided in Figure 3B, representing cases with PD (upper row), SD (middle row), and PR (bottom row). Individual profiles of all 599 patients were provided in the Supplementary Figure 1. Figure 3C shows the diagnostic plots of stochastic model predictions vs observations. No apparent biases or deviations in the diagnostic plots were detected, indicating good model performance.
Evolutionary parameters categorized by treatments and KRAS status
The evolutionary parameters were evaluated in relation to clinical characteristics and treatment types. Patients in the panitumumab plus FOLFIRI group had a significantly higher response to treatment than those under FOLFIRI only (0.022 vs 0.015 per day, p < 0.001), suggesting the efficacy of panitumumab in combination with FOLFIRI in the treatment of mCRC. Compared to patients with mutant KRAS, those with wild-type KRAS showed a significantly lower transition rate to resistance (1.19×10−5 vs 1.14×10−5 per cell division, p = 0.045) and a significantly slower resistant subclones regrowth rate (0.006 vs 0.007 per day, p = 0.01). These observations are consistent with clinical observations that mCRC patients with mutant KRAS, compared to wild-types, usually have a more durable response to EGFR targeted therapy and also have better survival outcomes.
Predictability of patient survival by evolutionary parameters
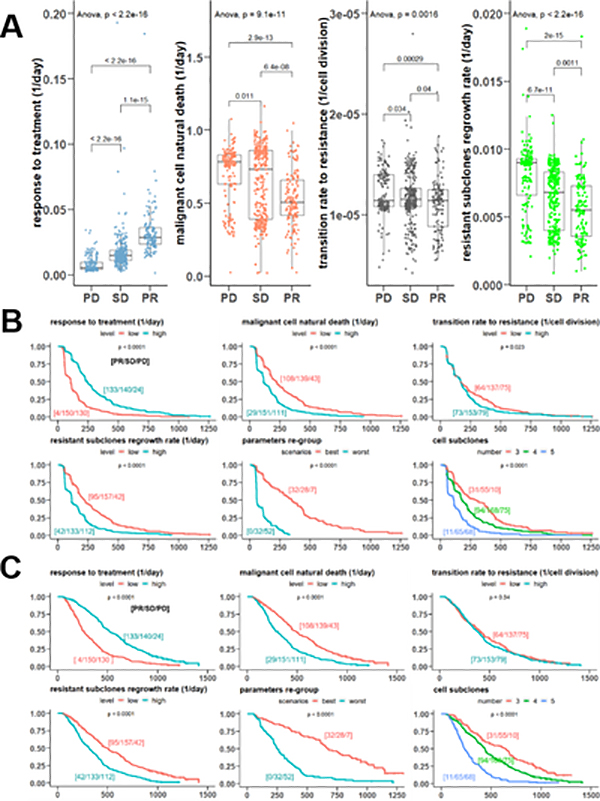
To assess the evolutionary parameters in the prediction of patients’ treatment responses, we first stratified patients based on their RECIST response (PR, SD, and PD) and compared the parameters among these groups. Patients in the ND and UE categories were excluded from this analysis. As shown in Figure 4A, compared to patients with an SD and PR, patients with a PD had a drastically weaker response to treatment, a higher malignant cell natural death rate, a faster transition rate to resistant cell populations, and a greater regrowth rate of resistant subclones (p < 0.005 in all ANOVA tests). Cohen’s d effect size tests also indicated significant differences in these four evolutionary parameters among patients with PR, SD or PD responses (see Supplementary Table 1).
Figure 4. Model parameters predict patient outcomes.
a. Boxplots of the stochastic model parameters under different responses (PD is progressive disease, SD is stable disease, PR is partial response). The error bars show 1.5 times interquartile range. b. Kaplan-Meier curves of the progression-free survival according to the evolution parameters categorized by the median or cell subclone number at tumor diagnosis. The “low” level indicates the parameter < median, while the “high” level indicates the parameter ≥ median. The “best” scenario indicates patients with a response to treatment ≥ median and other parameters < medians, while the “worst” scenario indicates patients with a response to treatment < median and other parameters ≥ medians. The numbers in the brackets represent the number of patients under RECIST responses [PR/SD/PD]. 18 patients of other responses including “Not done” and “Unable to Evaluate” were not included in the summary. c. Kaplan-Meier curves of the overall survival according to the evolution parameters categorized by the median or cell subclone number at tumor diagnosis.
Patients’ PFS and OS under different evolutionary characteristics were also systematically assessed. The median values were used as cut-off points to stratify patients and compare their PFS and OS for all parameters. The median for the response to treatment is 0.0149 per day, for the malignant cell natural death rate is 0.6812 per day, for the transition rate to resistance is 1.14×10−5 per cell division, and for the resistant subclones regrowth rate is 0.0072 per day. The predicted resistant cell subclones at diagnosis were also applied to categorize patients for survival comparisons. As shown in Figure 4B, patients’ PFS curves are significantly different (p < 0.05) between groups stratified by the evolutionary parameters. As shown in Figure 4C, patients’ OS curves are also significantly correlated with the parameters, except for the cell transition rate, which does not seem to be correlated to OS. Patients’ PFS (median 300 days [95% CI, 198–394 days]) and OS (median 645 days [95% CI, 447–719 days]) were superior in patients with three resistant subclones at diagnosis than those with four cell subclones (PFS median 178 days [95% CI, 168–209 days] and OS median 437 days [95% CI, 401–455 days]). Patients with five or more subclones have the worst prognosis (PFS median 111 days [95% CI, 78–120 days] and OS median 233 days [95% CI, 213–277 days]). This result is consistent with a general perception that patients with higher intratumor heterogeneity, namely more diverse subpopulations, tend to have a shorter duration of survival. (31–33)
The best-case scenarios for patient survival were also defined (response to treatment ≥ median, other parameters < medians) and 69 patients were selected in this scenario, in comparison to 85 patients in the worst-case scenarios (response to treatment < median, other parameters ≥ medians). (Figure 4) The patients in the best-case scenarios have a significantly longer PFS (median 347 days [95% CI, 272–442 days]) and OS (median 674 days [95% CI, 613–872 days]) than in the worst-case scenarios (PFS median 111 days [95% CI, 57–104 days] and OS median 249 days [95% CI, 219–309 days]). Furthermore, the numbers of patients under different RECIST responses [PR/SD/PD] on each survival curve were specified. Notably, PR patients clustered in the groups with high responses to treatment, low malignant cell natural death rates, and low resistant subclones regrowth rates, and also in the best-case scenario group.
Integrated model parameters with clinical characteristics to predict survival
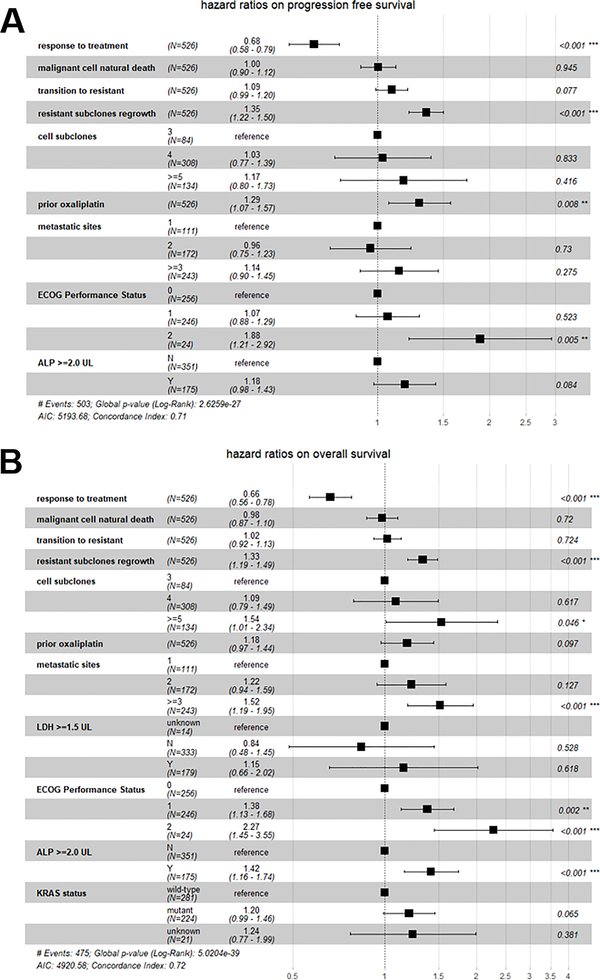
Of the 599 patients, 526 with a complete record of baseline clinical characteristics were analyzed to integrate model-derived evolutionary parameters with clinical characteristic factors into the predictions of patient survival. To adjust for potential confounders, the LASSO algorithm was applied to select the most survival-relevant variables from baseline clinical characteristics, which were then added into the Cox model. The PFS-relevant factors selected by lambda.1se in LASSO include prior oxaliplatin usage before the trial, the number of metastatic sites at diagnosis, ECOG performance status at diagnosis, and baseline alkaline phosphatase (ALP) values if greater than twice the upper limit. Similarly, the selected factors for OS are prior oxaliplatin usage, the number of metastatic sites at diagnosis, baseline lactic acid dehydrogenase (LDH) values if greater than 1.5 fold the upper limit, ECOG performance status at diagnosis, baseline ALP values if greater than twice the upper limit and KRAS status. We also compared the Cox proportional model results that only incorporated the selected clinical covariates with the full Cox model after integrating the evolutionary parameters. The C-index (0.707 vs 0.597 for PFS model, 0.72 vs 0.655 for OS model) and ANOVA tests (p < 0.001) indicated the model after incorporating evolutionary parameters is significantly better in survival prediction for both PFS and OS. A cross-validation was performed to further validate our Cox model. The average of C-index in the test datasets was 0.512 for PFS and 0.519 for OS, which indicated both fair predictions. The estimated HR and 95% CI for the model-derived evolutionary parameters in the Cox model accounting for the selected covariates are shown in Figure 5A and Figure 5B. The response degree to treatment is significantly correlated with both PFS (adjusted HR, 0.68 [95% CI, 0.58–0.79]) and OS (adjusted HR, 0.66 [95% CI, 0.56–0.78]). The resistant subclones regrowth rate was another significant predictor to both PFS (adjusted HR, 1.35 [95% CI, 1.22–1.5]) and OS (adjusted HR, 1.33 [95% CI, 1.19–1.49]). The number of resistant subclones was greatly correlated with OS, and the adjusted HR is 1.09 [95% CI, 0.79–1.49] for four subclones and 1.54 [95% CI, 1.01–2.34]) for five or more subclones, in comparison to three subclones. This observation further supports the view that tumor heterogeneity greatly accounts for treatment failure and high risks of patient death in mCRC patients.
Figure 5. Cox proportional hazards model integrating the evolution parameters and survival-relevant clinical characteristics.
a. HR estimates with a 95% confidence interval and p-values for each covariate on the progression-free survival. b. HR estimates with a 95% confidence interval and p-values for each covariate on the overall survival.
Discussion
The development of resistance to targeted therapy or cytotoxic chemotherapy remains the major hurdle in the treatment of mCRC. Patients with mCRC harbor enormous tumor heterogeneity at diagnosis and multiple drug-resistant mechanisms have been identified. (13) Typically, standard therapy can substantially eradicate sensitive subclones, which unfortunately often expedite the accumulation of resistant subclones. Enriched resistant subclones eventually lead to treatment failure and cancer relapse. (34–37) Therefore a deeper understanding of the process of cancer resistance and the underlying clonal dynamics would be of high clinical relevance to develop treatments for delaying resistance and likely prolonging patient survival. (6,22) In this study, we developed a mathematical framework to investigate tumor longitudinal measurements with which to depict the evolutionary dynamics in each individual patient. The optimized parameters revealed huge inter-patient variability in tumor evolution and were predictive of patients’ long-term survival.
The tumor lesion size is frequently monitored for mCRC patients during their clinical visits. However, bulk tumor measurements are often believed to hold limited information on cancer evolution and not reflect evolutionary dynamics. (38) With the developed framework, using longitudinal tumor measurements, patient evolutionary dynamics could be illustrated on both population and individual levels. Our results indicated that the obtained evolutionary parameters, as well as numbers of tumor-resistant subclones at diagnosis, could predict treatment efficacy and patients’ long-term survival. This is consistent in our results that panitumumab in combination with FOLFIRI exhibited better evolution control than FOLFIRI alone, and patients with wild-type KRAS showed significantly lower rates of resistant accumulation than those with mutant KRAS. In our simulation, all patients had no less than three tumor cell subclones at diagnosis with at least two resistant subclones. Approximately 22% of mCRC patients had already acquired more than three resistant subclones in liver metastatic lesions. The patients with more resistant subclones had a significantly shorter OS.
Real-world tumor quantification is often of high noise and is collected through opportunistic samples with very few data points for each patient. Many subjects had sparse or missing data in the real clinical setting. The rationale of NLME is to include the noise and account for inter-patient variability by collectively modeling all patient data, which could leverage the whole population to inform the parameters for each individuals, particularly individuals with missing or sparse data. (39) Due to the demanding computational space, we performed the model fittings in two steps, first by estimating the deterministic limits of the model parameters and then by optimizing a stochastic model for each patient. Such a two-stage method retained the accuracy of stochastic estimation at a very early stage and meanwhile prohibited local minima in the stochastic model solving process. The obtained parameters exhibited huge inter-patient variabilities in tumor evolution, which provide a basis to generate virtual mCRC patient populations and test many evolutionary theories in CRC patients.
Many studies have been done to find the prognostic predictors of CRC patient survival. (40,41) Our study is known to be the first to integrate tumor evolutionary parameters with clinical characteristics to predict patient survival. Prognostic clinical characteristics were selected by the LASSO algorithm to make survival predictions in mCRC patients. The tumor metastasis level, namely the number of metastatic sites at diagnosis, was found to be a positive prognostic factor to both PFS and OS in mCRC. This was consistent with observations in both non-small cell lung cancer and Hodgkin’s disease. (42,43) Notably the total tumor burden (i.e., the total tumor cell number) at diagnosis was not as relevant to patient survival as the tumor evolutionary parameters. In fact, there are mixed observations on whether the total tumor burden should be considered for cancer staging or for the prediction of long-term prognosis, particularly in the era of molecularly targeted cancer therapies and immunotherapies. (44,45) Tumor burden at diagnosis is mostly correlated with disease history, (46) but it does not necessarily indicate tumor heterogeneity and evolutionary plasticity, which is more predictive of patient survival in many cancer types.
Our analysis did not distinguish genotypic resistance from phenotypic resistance. To explicitly account for genetic or epigenetic attributes, DNA or RNA sequencing data are typically required to reveal the genetic or epigenetic traits and patterns in each subclone. However, these analyses are often based on a snapshot sample rather than longitudinally collected samples. (16,17) A series of biopsy is too invasive and often not feasible in clinical routine practice, let alone multiple biopsies to all metastatic lesions. In contrast, what we typically have for almost all cancer patients is their longitudinal measurements of tumor burdens and response to therapy. It has been broadly demonstrated in the literature that tumor growth trajectories potentially reveal tumor evolution processes, i.e., resistance accumulation process. (47) Although our analyses based on tumor growth trajectories would not reveal the fundamental genetic or epigenetic attributes about tumor evolution, these analyses can provide a general profile of resistance evolution on both individual and population levels and further apply this to study how tumor evolution would predict patient long-term survival.
More recent observations have highlighted the role of genetic driver (and passenger) mutations in clonal evolution and predictions of patient survival using whole genome or exome sequencing in acute myeloid leukemia (48) and non-small cell lung cancer (49). A fitness model on immunoediting also showed the predictability of patient survival under checkpoint blockages. (50) In theory, when data become available, our model could be expanded to include resistant mutations and immune interactions to improve its performance.
The prediction to patient long-term survival in our model was based on a few routinely available tumor measurements in CRC patients. These were not baseline predictions. It remains to be verified in a prospective study about how to use this approach for clinical decision making. In addition, the tumor evolutionary parameters obtained from our analysis should be considered along with other clinical factors to predict patient long-term benefits. However, there are two strengths of the model. First, the model can take advantage of a few routinely available measurements to predict the future growth. As shown in Supplementary Figure 1, among 599 patients, about 25% of patients only had two longitudinal measurements, and about 46% of patients had less than 4 measurements. Using the NLME modeling approach, the model could still predict each patient’s evolutionary parameters (through borrowing information from the whole population) to make the prediction. Secondly, even though the longitudinal measurements were not baselines, there were still “months or years” time gaps from “these measurements” to patient “long-term survival” (Supplementary Figure 2), which could be valuable for patients to know the future growth trajectories of their tumors and adjust the treatments accordingly. Furthermore, the modeling platform is an open system that could integrate both baseline and time dependent covariates to make more robust predictions. It could also be jointly modeled with time–to–event data to seek more quantitative relationships between tumor evolutionary parameters and patient long-term survivals. With the distributions of evolutionary parameter (Figure 3A), a Bayesian approach is also applicable to our model to make a priori predictions.
In conclusion, we developed a computational framework to analyze the longitudinal tumor measurements in mCRC patients and optimize a stochastic tumor growth model, which can predict individual tumor evolutionary dynamics at diagnosis, before, during, and after therapy. The model-derived parameters revealed high inter-patient variabilities in tumor evolution and disclosed many insights in predictions of PFS and OS in mCRC patients. This developed framework is translational to other tumor types, which can take advantage of the largely available tumor burden data in most cancer patients and generate insights into the evolutionary dynamics of each individual patient.
Supplementary Material
Statement of Significance.
A data analysis framework depicts the individual evolutionary dynamics of metastatic colorectal cancer patients, and can be generalized to project patient survival outcomes.
Acknowledgements
This work was supported by NIH grant GM119661.
Financial support: Y. Cao and J. Zhou were funded by National Institute of Health (GM119661).
Footnotes
Conflicts of interest: The authors declare that there is no conflict of interest regarding the publication of this article
Reference
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Valderrama-Treviño AI, Barrera-Mera B, Ceballos-Villalva JC, Montalvo-Javé EE. Hepatic metastasis from colorectal cancer. Euroasian J hepato-gastroenterology. 2017;7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards MS, Chadda SD, Zhao Z, Barber BL, Sykes DP. A systematic review of treatment guidelines for metastatic colorectal cancer. Color Dis. 2012;14:e31–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Annals of surgery, 1999, 230(3): 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lackner MR, Wilson TR, Settleman J. Mechanisms of acquired resistance to targeted cancer therapies. Futur Oncol. 2012;8:999–1014. [DOI] [PubMed] [Google Scholar]
- 6.Gatenby R, Brown J. The evolution and ecology of resistance in cancer therapy. Cold Spring Harb Perspect Med. 2018;8:a033415. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Uchi R, Takahashi Y, Niida A, Shimamura T, Hirata H, Sugimachi K, et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 2016;12:e1005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, et al. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin cancer Res. 2015;21:1087–97. [DOI] [PubMed] [Google Scholar]
- 10.Enriquez-Navas PM, Kam Y, Das T, Hassan S, Silva A, Foroutan P, et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med. 2016;8:327ra24 LP–327ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito T, Niida A, Uchi R, Hirata H, Komatsu H, Sakimura S, et al. A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer. Nat Commun. 2018;9:2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Heijden M, Miedema DM, Waclaw B, Veenstra VL, Lecca MC, Nijman LE, et al. Spatiotemporal regulation of clonogenicity in colorectal cancer xenografts. Proc Natl Acad Sci. 2019;116:6140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. 2008;1626:1634. [DOI] [PubMed] [Google Scholar]
- 15.Bregni G, Sciallero S, Sobrero A. HER2 Amplification and Anti-EGFR Sensitivity in Advanced Colorectal Cancer. JAMA Oncol. 2019;605:606. [DOI] [PubMed] [Google Scholar]
- 16.Peeters M, Price T, Boedigheimer M, Kim TW, Ruff P, Gibbs P, et al. Evaluation of Emergent Mutations in Circulating Cell-Free DNA and Clinical Outcomes in Patients with Metastatic Colorectal Cancer Treated with Panitumumab in the ASPECCT Study. Clin Cancer Res. 2019;25:1216–25. [DOI] [PubMed] [Google Scholar]
- 17.Lin P-C, Yeh Y-M, Wu P-Y, Hsu K-F, Chang J-Y, Shen M-R. Germline susceptibility variants impact clinical outcome and therapeutic strategies for stage III colorectal cancer. Sci Rep. 2019;9:3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navin NE. Tumor evolution in response to chemotherapy: phenotype versus genotype. Cell Rep. 2014;6:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallaher J, Anderson ARA. Evolution of intratumoral phenotypic heterogeneity: the role of trait inheritance. Interface Focus. 2013;3:20130016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peeters M, Jay Price T. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;4706–13. [DOI] [PubMed] [Google Scholar]
- 21.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–16. [DOI] [PubMed] [Google Scholar]
- 22.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devita VT Jr, Young RC, Canellos GP. Combination versus single agent chemotherapy: a review of the basis for selection of drug treatment of cancer. Cancer. 1975;35:98–110. [DOI] [PubMed] [Google Scholar]
- 24.Kozłowska E, Färkkilä A, Vallius T, Carpén O, Kemppainen J, Grénman S, et al. Mathematical modeling predicts response to chemotherapy and drug combinations in ovarian cancer. Cancer Res. 2018;78:4036–44. [DOI] [PubMed] [Google Scholar]
- 25.Diabate M, Coquille L, Samson A. Parameter estimation and treatment optimization in a stochastic model for immunotherapy of cancer. arXiv Prepr arXiv180601915. 2018; [DOI] [PubMed] [Google Scholar]
- 26.Model T, Language C. Mlxtran The Model Coding Language for Monolix. 2013;1–34. [Google Scholar]
- 27.Sivanandam SN, Deepa SN. Genetic algorithm implementation using matlab. Introd to Genet algorithms. 2008. page 211–62. [Google Scholar]
- 28.Dartois C, Brendel K, Comets E, Laffont CM, Laveille C, Tranchand B, et al. Overview of model‐building strategies in population PK/PD analyses: 2002–2004 literature survey. Br J Clin Pharmacol. 2007;64:603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibshirani R Regression shrinkage and selection via the lasso. J R Stat Soc Ser B. 1996;58:267–88. [Google Scholar]
- 30.Lande R Natural selection and random genetic drift in phenotypic evolution. Evolution (N Y). 1976;30:314–34. [DOI] [PubMed] [Google Scholar]
- 31.Kidd EA, Grigsby PW. Intratumoral metabolic heterogeneity of cervical cancer. Clin cancer Res. 2008;14:5236–41. [DOI] [PubMed] [Google Scholar]
- 32.Kim PH, Cha EK, Sfakianos JP, Iyer G, Zabor EC, Scott SN, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr HA. Fitness and its role in evolutionary genetics. Nat Rev Genet. 2009;10:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staňková K, Brown JS, Dalton WS, Gatenby RA. Optimizing Cancer Treatment Using Game Theory: A ReviewOptimizing Cancer Treatment Using Game TheoryOptimizing Cancer Treatment Using Game Theory. JAMA Oncol. 2019;5:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You L, Brown JS, Thuijsman F, Cunningham JJ, Gatenby RA, Zhang J, et al. Spatial vs. non-spatial eco-evolutionary dynamics in a tumor growth model. J Theor Biol. 2017;435:78–97. [DOI] [PubMed] [Google Scholar]
- 37.Gallaher JA, Enriquez-Navas PM, Luddy KA, Gatenby RA, Anderson ARA. Spatial heterogeneity and evolutionary dynamics modulate time to recurrence in continuous and adaptive cancer therapies. Cancer Res. 2018;78:2127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson M, Janzén DLI, Durrieu L, Colman-Lerner A, Kjellsson MC, Cedersund G. Nonlinear mixed-effects modelling for single cell estimation: when, why, and how to use it. BMC Syst Biol. 2015;9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam R, Delvart V, Pascal G, Valeanu A, Castaing D, Azoulay D, et al. Rescue surgery for unresectable colorectal liver metastases downstaged by chemotherapy: a model to predict long-term survival. Ann Surg. 2004;240:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claret L, Girard P, Hoff PM, Van Cutsem E, Zuideveld KP, Jorga K, et al. Model-based prediction of phase III overall survival in colorectal cancer on the basis of phase II tumor dynamics. Clin Oncol. 2006; [DOI] [PubMed] [Google Scholar]
- 42.Gobbi PG, Ghirardelli ML, Solcia M, Di Giulio G, Merli F, Tavecchia L, et al. Image-aided estimate of tumor burden in Hodgkin’s disease: evidence of its primary prognostic importance. J Clin Oncol. 2001;19:1388–94. [DOI] [PubMed] [Google Scholar]
- 43.Park JH, Kim TM, Keam B, Jeon YK, Lee S-H, Kim D-W, et al. Tumor burden is predictive of survival in patients with non–small-cell lung cancer and with activating epidermal growth factor receptor mutations who receive gefitinib. Clin Lung Cancer. 2013;14:383–9. [DOI] [PubMed] [Google Scholar]
- 44.Saha S, Shaik M, Johnston G, Saha SK, Berbiglia L, Hicks M, et al. Tumor size predicts long-term survival in colon cancer: an analysis of the National Cancer Data Base. Am J Surg. 2015;209:570–4. [DOI] [PubMed] [Google Scholar]
- 45.Huang B, Feng Y, Mo S-B, Cai S-J, Huang L-Y. Smaller tumor size is associated with poor survival in T4b colon cancer. World J Gastroenterol. 2016;22:6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends in cancer. 2016;2:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkerson J, Abdallah K, Hugh-jones C, Curt G, Rothenberg M, Simantov R, et al. Estimation of tumour regression and growth rates during treatment in patients with advanced prostate cancer : a retrospective analysis. Lancet Oncol. 18:143–54. [DOI] [PubMed] [Google Scholar]
- 48.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 50.Luksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.