Mycobacterial pathogens use the ESX-1 system to transport protein substrates that mediate essential interactions with the host during infection. We previously demonstrated that in addition to transporting proteins, the ESX-1 secretion system regulates gene expression. Here, we identify a conserved transcription factor that regulates gene expression in response to the ESX-1 system. We demonstrate that this transcription factor is functionally conserved in M. marinum, a pathogen of ectothermic animals; M. tuberculosis, the human-pathogenic species that causes tuberculosis; and M. smegmatis, a nonpathogenic mycobacterial species. These findings provide the first mechanistic insight into how the ESX-1 system elicits a transcriptional response, a function of this protein transport system that was previously unknown.
KEYWORDS: ESAT-6, ESX-1, Mycobacterium, protein secretion, regulation, feedback control
ABSTRACT
Pathogenic mycobacteria encounter multiple environments during macrophage infection. Temporally, the bacteria are engulfed into the phagosome, lyse the phagosomal membrane, and interact with the cytosol before spreading to another cell. Virulence factors secreted by the mycobacterial ESX-1 (ESAT-6-system-1) secretion system mediate the essential transition from the phagosome to the cytosol. It was recently discovered that the ESX-1 system also regulates mycobacterial gene expression in Mycobacterium marinum (R. E. Bosserman, T. T. Nguyen, K. G. Sanchez, A. E. Chirakos, et al., Proc Natl Acad Sci U S A 114:E10772–E10781, 2017, https://doi.org/10.1073/pnas.1710167114), a nontuberculous mycobacterial pathogen, and in the human-pathogenic species M. tuberculosis (A. M. Abdallah, E. M. Weerdenburg, Q. Guan, R. Ummels, et al., PLoS One 14:e0211003, 2019, https://doi.org/10.1371/journal.pone.0211003). It is not known how the ESX-1 system regulates gene expression. Here, we identify the first transcription factor required for the ESX-1-dependent transcriptional response in pathogenic mycobacteria. We demonstrate that the gene divergently transcribed from the whiB6 gene and adjacent to the ESX-1 locus in mycobacterial pathogens encodes a conserved transcription factor (MMAR_5438, Rv3863, now espM). We prove that EspM from both M. marinum and M. tuberculosis directly and specifically binds the whiB6-espM intergenic region. We show that EspM is required for ESX-1-dependent repression of whiB6 expression and for the regulation of ESX-1-associated gene expression. Finally, we demonstrate that EspM functions to fine-tune ESX-1 activity in M. marinum. Taking the data together, this report extends the esx-1 locus, defines a conserved regulator of the ESX-1 virulence pathway, and begins to elucidate how the ESX-1 system regulates gene expression.
INTRODUCTION
Following infection, pathogenic mycobacteria, including Mycobacterium tuberculosis, are engulfed by macrophages and reside in the phagosome (1–3). Survival in the phagosome requires regulated changes in bacterial gene expression (1, 4). Pathogenic mycobacteria use the ESX-1 secretion system (SS) to lyse the phagosome and mediate bacterial access the cytoplasm (5–13). The ESX-1 system is functionally conserved between M. tuberculosis, the cause of human tuberculosis, and Mycobacterium marinum, a pathogen of poikilothermic fish and an established model for the ESX-1 system (14–18). Phagosomal lysis releases secreted bacterial factors and triggers the host response to infection (7, 8, 19–27). In the absence of an ESX-1 system, both mycobacterial pathogens remain in the phagosome and are attenuated (7–9, 22).
Several ESX-1 conserved components (Ecc’s) form a complex in the cytoplasmic membrane (CM). The ESX-1 membrane complex recognizes ESX-1 substrates and provides the energy and the pore for the export of ESX-1 substrates across the CM (28, 29). The protein substrates are then translocated across the periplasm and mycolate outer membrane via an unknown process (30). ESX-1 substrates can be localized to the cell surface and/or secreted from the bacterial cell into the extracellular environment (31–34). We recently demonstrated that, in addition to transporting proteins, the presence or absence of the ESX-1 membrane complex in the CM elicits a widespread transcriptional response, a previously unrecognized function of the ESX-1 system (35). ESX-1-dependent gene expression has since been confirmed in M. marinum and reported in M. tuberculosis (36, 37).
The ESX-1-dependent transcriptional response includes a negative-feedback mechanism linking the levels of ESX-1 substrates to the presence or absence of the ESX-1 membrane complex (35). WhiB6 is a stress-responsive transcription factor (38, 39) that directly activates ESX-1 substrate gene expression in M. marinum and in M. tuberculosis (38, 39). The ESX-1 system regulates whiB6 gene expression both in M. marinum and in M. tuberculosis (35–37). In the presence of the ESX-1 membrane complex, the whiB6 gene is expressed, and there is WhiB6-dependent expression of the genes encoding ESX-1 substrates. In the absence of the ESX-1 membrane complex, whiB6 gene expression, as well as the expression of ESX-1 substrate genes, is significantly reduced (35, 36). How the ESX-1 membrane complex regulates whiB6 gene expression is unknown.
On the basis of our published data and of those published previously by independent groups, ESX-1-dependent changes in gene expression cannot be explained by the loss of the WhiB6 transcription factor alone (35–37). Therefore, we hypothesized that additional transcription factors regulate genes in response to the presence of the ESX-1 membrane complex.
RESULTS
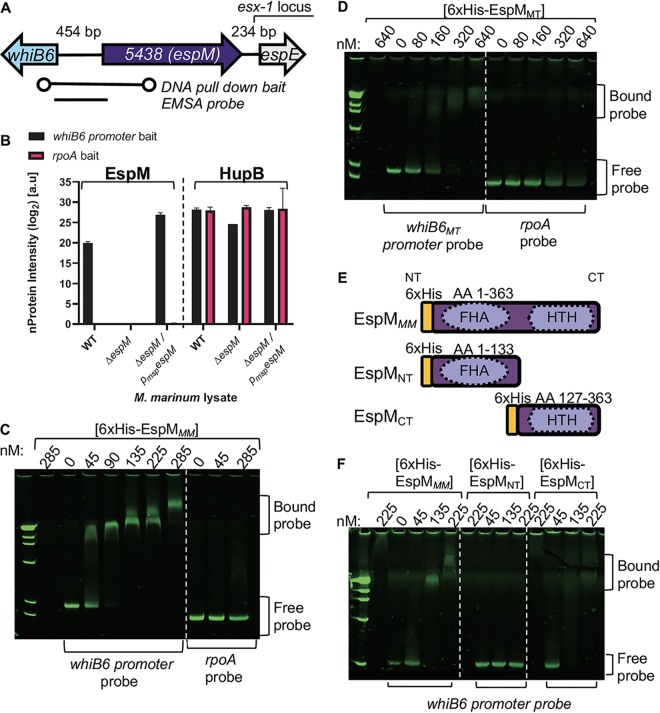
The EspM protein binds upstream of the whiB6 gene.
To identify transcription factors that regulate genes in response to the ESX-1 membrane complex, we focused on the regulation of the whiB6 gene. The 1 kb of DNA upstream of the whiB6 gene is sufficient for regulation of whiB6 gene expression by the ESX-1 membrane complex (35). We used a DNA pulldown to enrich proteins from M. marinum lysate that specifically bind the 1 kb of DNA upstream of the whiB6 gene (“whiB6 promoter bait,” Fig. 1A; bp 6577326 to 6578305 in the M. marinum genome). Using liquid chromatograph-tandem mass spectrometry (LC-MS/MS)-based quantitative proteomics on the proteins eluted from the DNA, we identified several proteins that were specifically and reproducibly enriched for binding the whiB6 promoter bait relative to binding nonspecific DNA (rpoA bait; see Table S1 in the supplemental material). MMAR_5438 was enriched for binding the whiB6 promoter bait ≥64.0-fold ± 0.4-fold relative to the rpoA bait (Fig. 1B). We propose renaming the MMAR_5438 gene “espM,” consistent with current ESX-1 nomenclature (40). We generated an M. marinum strain with an unmarked deletion of the espM gene (ΔespM; Fig. S1) and a complementation strain with an integrated constitutive espM expression plasmid (ΔespM/pmspespM). The EspM protein was not identified in the DNA pulldown performed with lysate from the ΔespM strain and was further enriched for whiB6 promoter bait binding compared to the rpoA bait in lysates from the complemented strain (Fig. 1B). We also identified several M. marinum proteins with known DNA binding activity that were not significantly or reproducibly enriched for binding the whiB6 promoter bait relative to the rpoA bait (Table S1). For example, the M. marinum DNA-binding protein Hu homolog HupB (MMAR_1728) bound the two baits comparably following incubation with any M. marinum lysate (Fig. 1B).
FIG 1.
Identification of MMAR_5438 (EspM) as a DNA-binding protein in M. marinum. (A) The whiB6 gene is separated from the esx-1 locus by the MMAR_5438 gene. The biotinylated 1-kb probe (circles) for the DNA pulldown is indicated. The 500-bp probe for the EMSA analysis is indicated in panel C. (B) MS analysis of the DNA pulldown showing the enrichment of the EspM and HupB proteins. The scale represents normalized MS peak area intensity levels. a.u, arbitrary units. (C and D) EMSAs performed with increasing concentrations of the 6×His-EspM protein from M. marinum (EspMMM) (C) or the 6×His-EspM protein from M. tuberculosis (EspMMT) (D). The control probe used as indicated in both panels was 500 bp of the rpoA open reading frame (ORF) (bp 1309999 to 1310499) from M. marinum. (E) Schematic of the 6×His-EspMMM proteins affinity purified from E. coli used in the EMSAs. (F) EMSA performed with the whiB6-espM probe with increasing amounts of EspMMM, EspMNT, and EspMCT from M. marinum.
Generation and confirmation of M. marinum ΔespM strain. (A) Schematic of parental, ΔMMAR_5438 (ΔespM), and complemented strains. (B) Confirmation of ΔespM genotype. Diluted lysates of resolved strains were checked by PCR performed with primers OMF619 and OMF620. Download FIG S1, PDF file, 0.4 MB (457.6KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A to D) Processed MS data from the DNA affinity chromatography assay performed as described in the Fig. 1 legend. (A) Unfiltered processed protein data. (B) Filtered data used for quantitation, (C) Normalized LFQ enrichment for MMAR_5438 and HupB DNA binding. (D) Proteins enriched for binding of the whiB6-espM probe (>2-fold enrichment over the rpoA probe level). (E and F) Relative quantification of the proteins described in the Fig. S4E legend. (E) Unfiltered processed protein group data. (F) Filtered data used for quantification. For panels E and F, two biological replicates each were performed in technical duplicate. The data shown are representative of results from one biological replicate, performed in technical duplicate. Download Table S1, XLSX file, 1.9 MB (2MB, xlsx) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To confirm the interaction of the EspM protein with the whiB6 promoter region, we expressed and purified an N-terminally 6×His-tagged EspMMM fusion protein from Escherichia coli (the MM subscript refers to the protein from M. marinum [40]) (see Fig. S2 in the supplemental material) and performed electrophoretic mobility shift assays (EMSAs). We observed a specific shift in mobility of the whiB6 promoter probe (550 bp) (Fig. 1A, “EMSA probe”) and a concomitant loss of free whiB6 promoter probe with increasing concentrations of the 6×His-EspMMM protein (Fig. 1C). We did not observe a mobility shift of the rpoA probe, confirming the specific binding of the EspMMM protein to the whiB6 promoter probe.
Heterologous expression of EspM proteins from M. marinum and M. tuberculosis. (A to C) Coomassie-stained gels representing purification of 6×His-EspMMM protein (A) and of denatured 6×His-EspMMT with refolding (B) and dilution series of purified 6×His-EspMNT (aa 1 to 133) and 6×His-EspMCT (aa 127 to 363) (C) from M. marinum. (D) Purification of WhiB6MM-6×His protein. Final concentrations and buffer conditions are listed in Text S1. Download FIG S2, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed Materials and Methods section regarding the approaches used in this article. Download Text S1, PDF file, 0.2 MB (217.3KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The espM gene is conserved in M. tuberculosis. The EspM proteins in M. marinum and M. tuberculosis Erdman (ERDMAN_4236, EspMMT) are 76.25% identical at the amino acid level (41, 42). To test if EspM binds the genomic region upstream of the whiB6MT gene, we expressed and purified 6×His-tagged EspMMT in E. coli (the MT subscript refers to the EspM protein from M. tuberculosis) (Fig. S2). We amplified the 500 bp upstream of the whiB6 gene from M. tuberculosis Erdman and tested if EspMMT specifically bound the whiB6MT promoter region using EMSAs. Increasing concentrations of 6×His-EspMMT protein led to a specific mobility shift of the whiB6MT promoter probe and to a corresponding loss of free probe (Fig. 1D). Although bound rpoA probe was not observed at the highest concentrations of 6×His-EspMMT protein, the free probe was reduced, indicating weak binding at the highest protein concentrations. Together, these data indicate that EspM, from both M. marinum and M. tuberculosis, directly and specifically bound the whiB6-espM intergenic region.
espM is divergently transcribed from the whiB6 gene and is immediately adjacent to the esx-1 locus (Fig. 1A). EspM is a predicted conserved regulatory protein (42), but the corresponding function has not been investigated. The EspMMM protein is predicted to have an N-terminal forkhead-associated (FHA) domain (amino acids [aa] 32 to 89) and a C-terminal helix-turn-helix domain (Fig. 1E). We hypothesized that the C-terminal half of the protein mediated DNA binding. We expressed and purified 6×His-tagged EspMNT (aa 1 to 133) and EspMCT (aa 127 to 363) M. marinum proteins from E. coli (Fig. S2). We tested the ability of each protein to bind the whiB6 promoter probe using EMSA. The 6×His-EspMNT protein did not shift the mobility of the whiB6 promoter probe (Fig. 1F). Incubation with increasing concentrations of the 6×His-EspMCT protein caused a shift in mobility of the whiB6 promoter probe and a loss of free probe. We conclude that the C-terminal half of the EspM protein is required for DNA binding.
EspM is a conserved regulator of whiB6 and esx-1 gene expression.
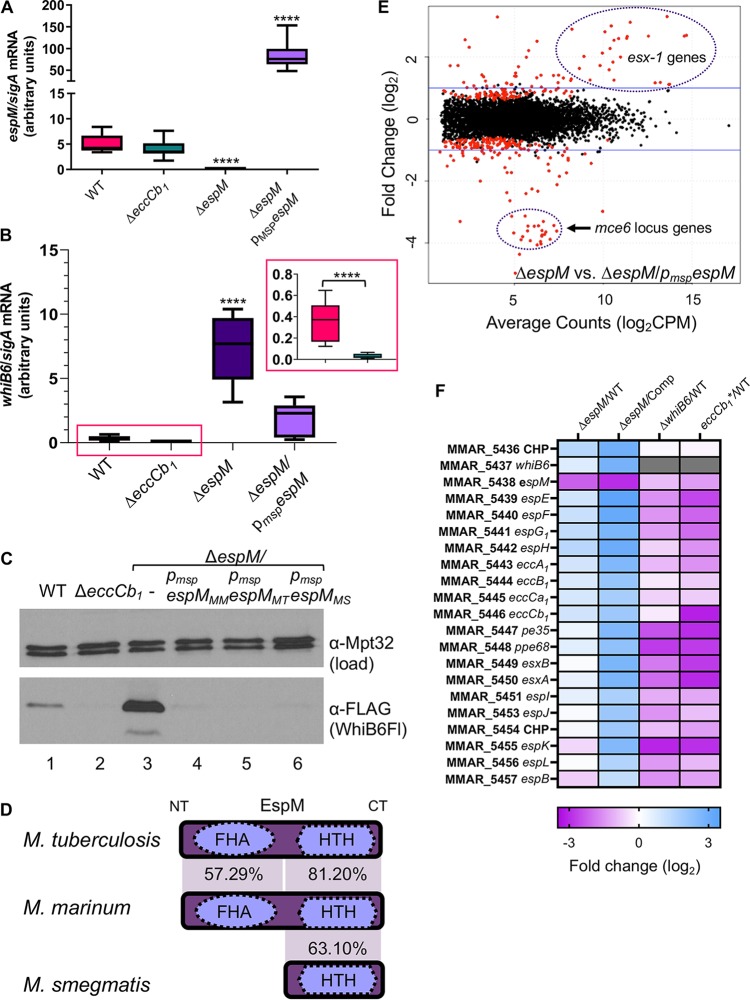
We confirmed that the espM transcript was absent in the ΔespM M. marinum strain using quantitative reverse transcription-PCR (qRT-PCR) (Fig. 2A). The espM expression level was significantly higher in the ΔespM/pmspespM complemented strain than in the wild-type (WT) strain (P < 0.0001). These data indicate that the complementation strain is an espM overexpression strain. We did not observe a significant reduction of espM gene expression in the ΔeccCb1 strain relative to the WT strain. These data confirm that espM expression is not regulated by the ESX-1 system in M. marinum, consistent with our previously published transcriptomic analysis (35).
FIG 2.
EspM is a conserved regulator of whiB6 and esx-1 gene expression. (A) qRT-PCR measuring the levels of espM expression relative to sigA expression. A one-way ordinary analysis of variance (ANOVA) (P < 0.0001), followed by a Dunnett’s multiple-comparison test relative to the WT strain, was performed. ****, P < 0.0001. (B) qRT-PCR measuring the levels of whiB6 expression relative to sigA expression. A one-way ordinary ANOVA (P < 0.0001), followed by a Sidak’s multiple-comparison test relative to the WT strain, was performed. ****, P < 0.0001. The inset shows just the comparison between the WT and ΔeccCb1 strains. A Student’s unpaired, two-tailed t test was used to define the significance of the results of the comparisons between the two strains. For panels A and B, the data represent averages of results from at least three biological replicates, each performed in technical triplicate. (C) Western blot analysis of 10 μg of protein per lane. Anti-Mpt32 was used as the loading control. All M. marinum strains indicated in this panel contained a C-terminal FLAG epitope tag on the whiB6 gene. Samples were resolved on an 18% Tris-glycine gel. The Western blot shown is representative of at least three independent biological replicates. All strains indicated in panel D contained a C-terminal epitope tag on the whiB6 gene. (D) Conservation of the EspM proteins (percent identity at the amino acid level) from M. tuberculosis, M. marinum, and M. smegmatis. (E) Scatterplot of genes differentially expressed in the ΔespM strain versus the ΔespM/pmspespM complemented strain. Genes indicated in red had a q value of <0.05. Regions enriched with the esx-1 locus or mce6 locus are highlighted. Full gene lists are available in Table S3. (F) Heat map of esx-1 locus genes that are significantly differentially regulated in the ΔespM strain versus the ΔespM/pmspespM complemented strain compared to genes expressed in the ΔespM, ΔwhiB6, or eccCb1 mutant strains relative to the WT strain.
Because EspM bound the region upstream of the whiB6 gene, we tested if EspM regulates whiB6 gene expression. We measured whiB6 gene expression in M. marinum using qRT-PCR. Consistent with our prior findings (35), whiB6 gene expression was significantly reduced in the ΔeccCb1 strain compared to the WT strain (Fig. 2B, inset, P < 0.0001). Deletion of the espM gene resulted in a significant increase in whiB6 expression relative to the WT strain (P < 0.0001). Overexpression of the espM gene resulted in espM expression that was not significantly different from that seen with the WT strain. We conclude that EspM is a repressor of whiB6 gene expression.
We measured the levels of WhiB6 protein in the presence and absence of the espM gene (Fig. 2C). The parental M. marinum strain for these strains includes a whiB6 gene with a C-terminal FLAG epitope tag (WhiB6Fl [35]). Consistent with our previously published data (35), the WhiB6Fl protein was absent from the lysate generated from the ΔeccCb1 strain (Fig. 2C, lane 2). Consistent with the expression data (Fig. 2B), deletion of the espM gene resulted in increased WhiB6Fl protein levels relative to those seen with the WT strain (Fig. 2C, compare lane 3 to lane 1). The WhiB6Fl protein levels in the ΔespM/pmspespMMM complemented strain (espM overexpression) were lower than those in the WT strain (Fig. 2C, lane 4 versus lane 1). Together, these data strongly support the conclusion that EspM represses whiB6 gene expression in M. marinum.
The espM gene is syntenic in M. marinum, M. tuberculosis, and M. smegmatis (Fig. S3). M. smegmatis is a nonpathogenic, soil-dwelling mycobacterial species that uses the ESX-1 system to mediate conjugation (43, 44). The EspM orthologs in all three species are conserved at the protein level (Fig. 2D; see also Fig. S3). The M. smegmatis ortholog (MSMEG_0052; EspMMS) lacks the N-terminal FHA domain. Aligning the C-terminal halves of the EspMMM and EspMMT proteins with EspMMS revealed that the M. marinum and M. tuberculosis C-terminal halves are 81.20% identical at the amino acid level. EspMMS is 63.10% and 62.20% identical to the C-terminal half of EspMMM and EspMMT, respectively.
Conservation of the espM gene and protein in M. marinum, M. tuberculosis, and M. smegmatis. (A) Rv3863 and MSMEG_0052 are the espM orthologs in M. tuberculosis and M. smegmatis. The esx-1 locus begins with the espE gene in M. marinum and M. tuberculosis and with the espG gene in M. smegmatis. (B) The alignment was generated using Clustal Omega, followed by visualization with BoxShade. Black, identity; gray, similarity. The asterisk (*) indicates conservation across all three species. The predicted FHA domain in M. marinum and M. tuberculosis is indicated in pink. The sequence of EspMNT from M. marinum used as described for Fig. 1 ends at the red circle. EspMCT from M. marinum used as described for Fig. 1 starts at the green arrow. Download FIG S3, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Because EspM proteins are conserved across three mycobacterial species, we hypothesized that the repression of whiB6 expression by EspM would be functionally conserved. We generated integrating plasmids constitutively expressing the espM genes from M. tuberculosis Erdman (espMMT) and M. smegmatis mc2155 (espMMS) and introduced each plasmid into the ΔespM M. marinum strain. As shown in Fig. 2C, overexpression of the EspMMT protein or EspMMS protein reduced WhiB6Fl protein levels in the ΔespM M. marinum strain (Fig. 2C; compare lanes 5 and 6 with lane 3), similarly to the complemented strain overexpressing the espMMM gene (Fig. 2C, lane 4). These data demonstrate that repression of whiB6 expression is functionally conserved between the EspM orthologs in M. marinum, M. tuberculosis, and M. smegmatis.
We performed RNA-seq transcriptional profiling to determine if EspM regulates other genes in addition to whiB6 in M. marinum. Comparison of the WT strain to the ΔespM strain (both bearing the whiB6Fl allele) identified 134 genes that were upregulated and 300 genes that were downregulated (>2-fold; false-discovery rate [q value] of <0.05) (Fig. S4A; see also Table S3A). Genes controlled by EspM are also expected to be differentially regulated in the ΔespM strain compared to the complemented strain that overexpresses the repressor. We observed 44 genes that were upregulated and 55 genes that were downregulated in the ΔespM strain compared to the complemented strain (>2-fold; q value of <0.05) (Fig. 2E; see also Table S3B). Consistent with repression of whiB6 expression by EspM, we observed that whiB6 expression was induced 1.6-fold and 7.0-fold in the ΔespM strain compared to the WT strain and the complemented overexpression strain, respectively. Of the 44 genes that were induced in the ΔespM strain compared to the complemented strain, 21 genes from the esx-1 locus were identified (MMAR_5436 to MMAR_5457), including 8 genes that were also induced in the ΔespM strain compared to WT strain (Fig. 2F; see also Fig. S4B). Most of the other genes in the esx-1 locus were significantly induced in the ΔespM strain relative to the WT strain, but with induction levels below 2-fold.
EspM broadly regulates gene expression in M. marinum and fine-tunes ESX-1 protein levels. (A) Scatter plot of genes differentially expressed in the ΔespM strain versus the WT strain. Genes highlighted in red had a q value of <0.05. (B) Venn diagram of genes that were induced (>2×, q value of <0.05) in the ΔespM/WT or ΔespM/complemented strain from this study or that were repressed (>2×, q value of <0.05) in the ΔwhiB6 or eccCb1 ochre mutant (relative to the WT strain) from Bosserman et al. (35). (C) Heat map of genes in the mce6 locus and of surrounding genes that were significantly downregulated (>2×, q value of <0.05) in the ΔespM/WT or ΔespM/complemented comparison. These genes were induced in the ΔwhiB6 or eccCb1 ochre mutants relative to the WT strain, consistent with expression being whiB6 and ESX-1 dependent; however, the induction in gene expression did not achieve statistical significance for these strains. All strains in this figure contained the whiB6Fl allele at the whiB6 locus. (D) Western blot analysis of EsxA production in M. marinum lysates. The strains are the same as those shown in Fig. 5. A 10-μg or 5-μg volume of protein, as indicated, was loaded per lane and resolved on a 4% to 20% gel. Mpt-32 served as a loading control. The image shown is representative of results of three biological replicates. (E) Relative quantification of ESX-1-associated proteins using label-free proteomics. The proteins were derived from the strains described in the Fig. 5 legend. All protein levels are represented as log2-fold changes compared with those measured in the WT (whiB6Fl) strain or the ΔespM strain. Only the subset of ESX-1-associated proteins that were reproducibly quantified are shown in this heat map. The complete data are presented in Table S1E and F. The data shown are representative of results from one biological replicate, performed in technical duplicate. (F) Thin-layer chromatography of lipids extracted from the strains described in the Fig. 5 legend. PDIM purified from M. tuberculosis H37Rv was used as a positive control. TAG is triacylglycerols. The Rf values are as follows: for M. tuberculosis PDIM and the WT, espM, and eccCb1 strains, 3.17 cm; for the complemented strain, 3.07 cm, likely due to the slight curvature of the band. The results shown are representative of experiments performed in biological triplicate. Download FIG S4, PDF file, 0.8 MB (782.2KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also observed induction of unlinked esx-associated loci in the ΔespM strain compared to the complemented strain, including MMAR_0187-188 (esxB_1esxA_1), MMAR_3654 (esxP2), and the ESX-1 substrate locus MMAR_2894 (45) (Table S3B). Several of these genes were previously shown to be regulated by WhiB6 or ESX-1 (Fig. 2F; see also Fig. S4B).
Of the 55 genes downregulated in the ΔespM strain relative to the complemented strain, 39 were also downregulated in the ΔespM strain relative to WT strain (Table S3). These included 24 strongly downregulated genes between MMAR_0159 and MMAR_0182 (Fig. 2E; see also Table S3B), which includes the mce6 locus (Fig. S4C), and genes for amino acid metabolism and lipid anabolism. Prior studies with the ΔwhiB6 and eccCb1 mutant strains showed induction of the genes in the mce6 locus (Fig. S4C), supporting the idea of ESX-1-dependent regulation. Curiously, we also detected downregulation of an ESX-1-associated operon, MMAR_4166 to MMAR_4168 (espA, espC, and espD). Together, these data strongly support the conclusion that EspM is a regulator of genes broadly associated with the ESX-1 system in M. marinum.
EspM represses whiB6 expression in the absence of the ESX-1 membrane complex.
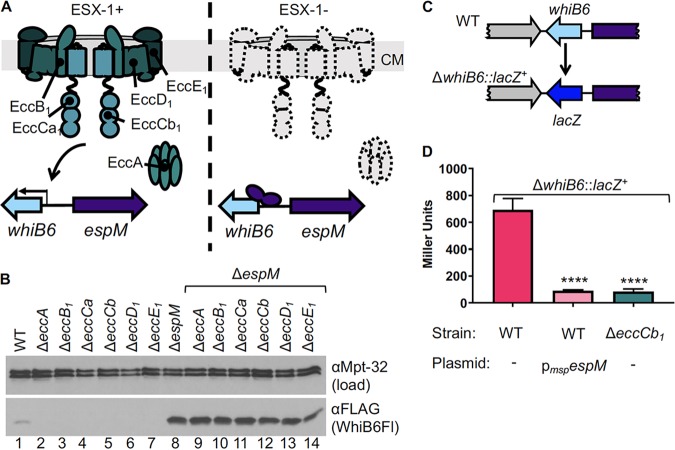
whiB6 gene expression levels are reduced in the absence of the ESX-1 membrane complex (35). We hypothesized that EspM represses whiB6 and esx-1 gene expression in the absence of the ESX-1 membrane complex (Fig. 3A). Four ESX-conserved components (Ecc’s) reside in the CM (Fig. 3A; EccB1, EccCa1, EccD1 and EccE1 [28, 29]), and two Ecc’s (EccCa1 and EccA) are cytoplasmic (10, 46–49).
FIG 3.
EspM is required for the repression of whiB6 expression in the absence of the ESX-1 membrane complex. (A) Schematic of the ESX-1 membrane complex and the proposed role of EspM in ESX-1-dependent gene expression. CM, cytoplasmic membrane. The depiction of the membrane complex was adapted from reference 28. (B) Western blot analysis of 10 μg per lane on an 18% gel. Mpt-32 was used as a loading control. The image is representative of three independent biological replicates. All strains indicated in panels B and C contained a C-terminal epitope tag on the whiB6 gene. (C) Schematic of the ΔwhiB6::lacZ+ reporter strain. (D) β-Galactosidase assay in WT M. marinum strains. The data in the figure represent averages of results from four independent biological replicates, each performed in technical triplicate. Significance was determined using a one-way ordinary ANOVA (P < 0.0001) followed by a Tukey’s multiple-comparison test. ****, P < 0.0001. The WT/pmspespM and ΔeccCb1 levels were not significantly different from each other (P = 0.9694). Error bars represent standard errors.
We reasoned that if EspM repressed whiB6 gene expression in the absence of the ESX-1 membrane complex, then deletion of the espM gene in strains lacking the ESX-1 membrane complex would restore whiB6 gene expression. We generated M. marinum strains bearing deletions of each ecc gene (eccA to eccE1) alone or in combination with deletion of the espM gene. Deletion of any ecc gene resulted in a loss of WhiB6Fl protein relative to the WT strain (Fig. 3B, lanes 2 to 7 versus lane 1). The deletion of the espM gene in combination with the ecc genes (ΔespM ΔeccA, ΔespM ΔeccB1, ΔespM ΔeccCa1, ΔespM ΔeccCb1, ΔespM ΔeccD1, and ΔespM ΔeccE1 mutant strains) resulted in levels of WhiB6Fl similar to those in the ΔespM strain (Fig. 3B, lanes 9 to 14 versus lane 8) and higher than those in the WT strain (Fig. 3B, lane 1). We further demonstrated that complementation with the eccA gene or the espM gene in the ΔespM ΔeccA strain resulted in levels of WhiB6Fl similar to those seen with the ΔespM deletion strain or the ΔeccA deletion strain, respectively (Fig. S5).
EspM is required for whiB6 repression in the ΔeccA M. marinum strain. Western blot analysis of 10 μg per lane on an 18% gel was performed. Mpt-32 was used as a loading control. The image is representative of results from three independent biological replicates. All strains contained a C-terminal epitope tag on the whiB6 gene. Download FIG S5, PDF file, 0.2 MB (260.4KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We reasoned that overexpression of the espM gene might be sufficient to repress whiB6 expression to levels similar to those seen with the ΔeccCb1 strain. We generated a strain bearing a whiB6 transcriptional reporter in M. marinum. We replaced the whiB6 gene with the lacZ gene, creating a strain lacking the whiB6 gene and with a reporter fusion to the whiB6 promoter (ΔwhiB6::lacZ+; Fig. 3C). We generated an isogenic ΔeccCb1 strain (no ESX-1 membrane complex [30]), as well as an isogenic WT strain overexpressing the espMMM gene. The level of β-galactosidase activity was significantly reduced in the ΔeccCb1 strain compared to the WT strain (Fig. 3D; P < 0.0001), confirming that the whiB6::lacZ+ reporter fusion was regulated by the ESX-1 membrane complex (35). Overexpression of the espM gene in the WT strain significantly reduced the levels of β-galactosidase activity compared to the WT strain levels (P < 0.0001). The levels of β-galactosidase activity in the ΔeccCb1 and espM overexpression strains were not significantly different from each other (P = 0.9915), demonstrating that overexpression of espM is sufficient to repress whiB6 gene expression in M. marinum. Collectively, our data demonstrate that EspM is required for repression of whiB6 gene expression in the absence of the ESX-1 membrane complex. Moreover, because the reporter strain lacks the whiB6 gene, these data indicate that EspM represses whiB6 expression in a WhiB6-independent manner.
The EspM and WhiB6 regulators coordinately control gene expression.
The espM and whiB6 genes are divergently organized in mycobacterial genomes (Fig. 4A). Because the whiB6 and espM genes share an intergenic region which likely controls the expression of both genes (Fig. 4A, pink), we sought to further define the relationship between the EspM and WhiB6 regulators.
FIG 4.
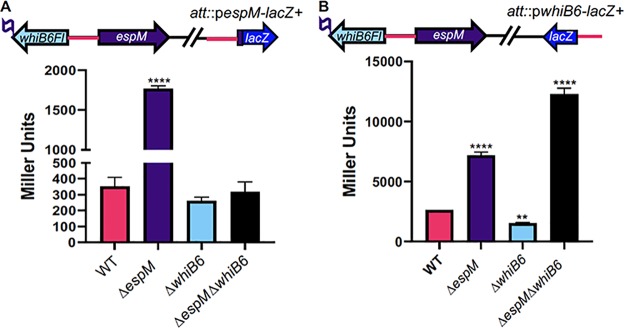
EspM and WhiB6 mutually regulate the expression of the espM and whiB6 genes. (A) The whiB6/espM locus and the espM-lacZ+ transcriptional fusion integrated at the attB site in M. marinum. The flag at the C terminus of the whiB6 gene indicates the presence of the whiB6Fl allele. Data represent β-galactosidase activity of the espM-lacZ+ transcriptional fusion. Error bars represent propagated errors. A one-way ordinary ANOVA (P < 0.0001) followed by a Sidak’s multiple-comparison test was performed. Significance is shown relative to the WT strain. ****, P < 0.0001. (B) The whiB6/espM locus and the whiB6-lacZ+ transcriptional fusion integrated at the attB site in M. marinum. Data represent β-galactosidase activity of the whiB6-lacZ+ transcriptional fusion. Error bars represent propagated errors. A one-way ordinary ANOVA (P < 0.0001) followed by a Tukey’s multiple-comparison test was performed. Significance data shown are relative to the WT strain. ****, P < 0.0001; **, P = 0.0078. For both panels, the data represent averages of results from at least three biological replicates, each performed in technical triplicate. All strains indicated in Fig. 4, with the exception of the ΔwhiB6 and ΔwhiB6 ΔespM strains, contained a C-terminal epitope tag on the whiB6 gene.
We generated espM-lacZ+ and whiB6-lacZ+ integrating transcriptional reporters (Fig. 4). The espM-lacZ+ reporter resulted in significantly increased β-galactosidase activity in the ΔespM strain compared to the WT strain (P < 0.0001; Fig. 4A). Loss of the whiB6 gene did not significantly impact β-galactosidase activity relative to the WT strain (P = 0.1195). Deletion of both the espM and whiB6 genes (ΔespM ΔwhiB6 mutant strain) resulted in β-galactosidase activity comparable to that seen with the WT M. marinum strain (P = 0.9305). We conclude from these data that espM expression is negatively autoregulated. Moreover, in the absence of EspM, WhiB6 is required for the observed increased espM gene expression. We confirmed that WhiB6 binds the whiB6-espM intergenic region by expressing and purifying a C-terminally 6×His-tagged WhiB6MM fusion protein from Escherichia coli (Fig. S2) and performing EMSAs (Fig. S6). We observed a specific shift in mobility of the whiB6 promoter probe (Fig. 1A, “EMSA probe”) and a concomitant loss of free whiB6 promoter probe with increasing concentrations of the WhiB6MM-6×His protein (Fig. S6). We did not observe a mobility shift of the rpoA probe, confirming the specific binding of the WhiB6MM protein to the whiB6 promoter probe.
WhiB6 binds the whiB6-espM intergenic region in vitro. EMSA was performed with increasing concentrations of WhiB6 protein from M. marinum. The whiB6 promoter probe was the same as that used as described in the Fig. 1 legend (bp 6577396 to 6577960 in the M. marinum M genome; 30 bp upstream of the whiB6 ORF to 141 bp into MMAR_5438). The control probe was 500 bp of the rpoA ORF (bp 1309999 to 1310499). Download FIG S6, PDF file, 0.7 MB (727.6KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The presence of the whiB6-lacZ+ reporter resulted in significantly increased β-galactosidase activity in the ΔespM strain compared to the WT strain (P < 0.0001; Fig. 4B). Loss of the whiB6 gene caused a significant reduction in β-galactosidase activity relative to the WT strain (P = 0.0078). Deletion of the espM and whiB6 genes together (ΔespM ΔwhiB6 mutant strain) resulted in significantly increased β-galactosidase activity relative to the WT and ΔespM M. marinum strains (P < 0.0001 for both comparisons). This further supports the idea that EspM represses whiB6 gene expression and confirms positive autoregulation of WhiB6, consistent with prior findings (38). Moreover, the significant increase in whiB6 expression in the absence of both EspM and WhiB6 suggests there is at least one more transcriptional activator of whiB6 expression in M. marinum.
EspM fine-tunes ESX-1 function in M. marinum.
Because EspM regulates whiB6 and esx-1 gene expression, we tested if EspM was required for ESX-1 activity. The WT strain produced the EsxA and EsxB substrates and secreted them into the culture supernatant during growth in vitro (Fig. 5A, lanes 1 and 2). Deletion of the eccCb1 gene, which is required for ESX-1 secretion (10, 12, 14), reduced production of EsxA and EsxB, and neither protein was secreted (Fig. 5A, lanes 3 and 4). The ΔespM strain exhibited at least WT levels of production and secretion of EsxA and EsxB (Fig. 5A, lanes 5 and 6). The espM complemented strains showed reduced levels of production of EsxA and EsxB (Fig. 5A, lanes 7 and 8, and Fig. S4D) but exhibited at least wild-type levels of secretion of EsxA and EsxB. To further confirm that the levels of ESX-1 proteins were altered, consistent with the observed EspM-dependent expression changes, we performed global proteomics on whole-cell lysates of the M. marinum strains represented in panel A (for the WT, ΔeccCb1, ΔespM, and complemented strains) (Table S1E and F; see also Fig. S4E). We identified 1,881 proteins at a 1% false-discovery rate. Protein quantification was performed by using label-free quantification (LFQ). We found that, similarly to the EsxA and EsxB proteins, the levels of several ESX-1 substrates (EspF, EspK, and EspB) and components (EccA) and other associated proteins (EspG, EspH, and EspL) were significantly reduced in the complemented strain, consistent with the expression data (Fig. 2). These data demonstrate that EspM is required for fine-tuning the levels of ESX-1-associated proteins, including the EsxA and EsxB substrates, in the mycobacterial cells but not for the secretory function of the ESX-1 system.
FIG 5.
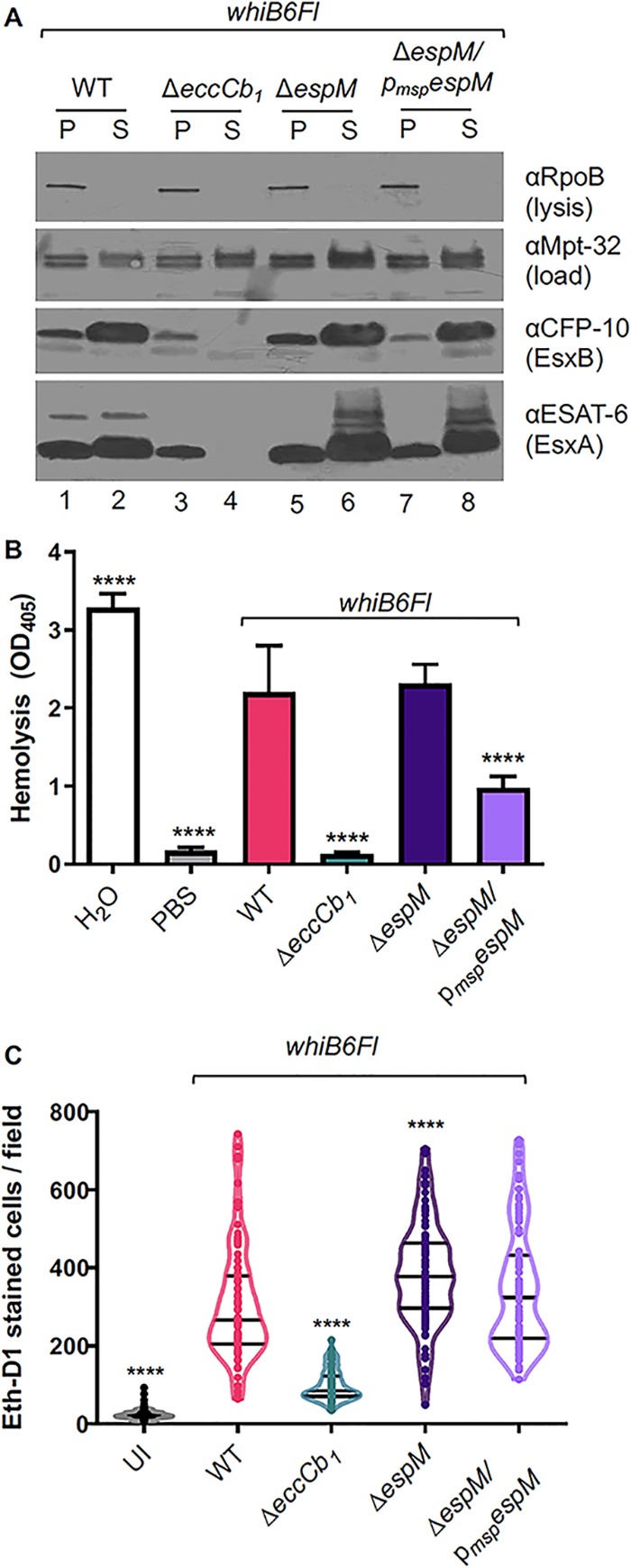
EspM fine-tunes ESX-1 function. (A) Western blot analysis of EsxA and EsxB secretion in vitro. 10 μg of protein was loaded per lane and resolved on a 4% to 20% gel. RpoB was used as the lysis control; Mpt-32 is a Sec-dependent secreted protein that served as a loading control. The image shown is representative of three biological replicates. (B) Hemolysis assay of M. marinum strains. The image shown represents at least three biological replicates, each performed in technical triplicate. Error bars represent the propagated errors. A one-way ordinary ANOVA (P < 0.0001) followed by a Tukey’s multiple-comparison test was performed. ****, P < 0.0001 (relative to the WT strain). OD405, optical density at 405 nm. (C) Cytolysis assay of RAW 264.7 cells following 24 h of infection with M. marinum at an MOI of 7. Black bars indicate median and quartiles. UI, uninfected. Statistical analysis was performed using a one-way ordinary ANOVA (P < 0.0001) followed by a Tukey’s multiple-comparison test. ****, P < 0.0001 (compared to the WT strain). Each dot represents the number of EthD-1-stained cells in a single field. A total of 10 fields were counted using ImageJ for each well. Processing of 3 wells was performed for each biological replicate. A total of 90 fields were counted for each strain.
The ESX-1 system promotes phagosomal lysis during macrophage infection (6, 9, 27). M. marinum lyses red blood cells (RBCs) in an ESX-1-dependent manner (14, 17, 50). Hemolysis analysis is a common way to measure the membranolytic activity of the ESX-1 system (14, 17, 50). The WT strain caused significantly increased hemolytic activity compared to the phosphate-buffered saline (PBS) control (no bacteria) (P < 0.0001; Fig. 5B). The ΔeccCb1 strain exhibited hemolytic activity that was not significantly different from that seen with the PBS control (P = 0.9996). The ΔespM strain exhibited hemolytic activity not significantly different from that seen with the WT strain (P = 0.9602). The complemented strain, which overexpresses espM relative to the WT strain (Fig. 2B), showed significantly reduced hemolytic activity relative to the WT and ΔespM strains (P < 0.0001).
ESX-1-deficient M. marinum strains fail to lyse the phagosome and fail to lyse macrophages (7, 51). We infected RAW 264.7 cells with M. marinum at a multiplicity of infection (MOI) of 7 and measured macrophage lysis by visualizing and quantifying the uptake of ethidium homodimer by permeabilized macrophages (52). Consistent with previous findings (51, 53), the WT strain caused macrophage lysis (Fig. 5C). Infection with the ΔeccCb1 strain resulted in a significant reduction in macrophage lysis compared to infection with the WT strain (P < 0.0001). In contrast, infection with the ΔespM strain resulted in significantly increased macrophage lysis compared to infection with the WT strain (P < 0.0001). Infection with the espM overexpression strain restored macrophage lysis to WT levels (P = 0.2138). These data show that in the absence of the espM gene, the ESX-1 system promoted higher levels of macrophage lysis. Moreover, combined with the hemolysis data, these findings indicate that the levels of EspM fine-tune the activity of the ESX-1 system in M. marinum.
DISCUSSION
Collectively, our findings identify EspM as a conserved transcription factor required for the ESX-1-dependent transcriptional response in pathogenic mycobacteria. Although the espM gene is adjacent to the esx-1 locus, EspM has not been previously characterized. The espM gene may not have been linked to the ESX-1 system previously because deletion of the espM gene in M. marinum had only a subtle impact on ESX-1 activity, likely because several transcription factors regulate whiB6 gene expression (39, 54–56). Moreover, the M. tuberculosis EspM ortholog Rv3863 was previously reported to be essential for growth in vitro in some genome-wide studies (57, 58) and nonessential in others (59), which may complicate study in M. tuberculosis.
The identification of EspM further expands our understanding of the feedback control mechanism that links the levels of ESX-1 substrates, and other genes, to the assembly of the secretory apparatus (35). We found that deletion of the espM gene resulted in levels of whiB6 expression that were higher than those seen with the WT strain (Fig. 2B and C). We propose that whiB6 expression is repressed by EspM in the WT strain and that whiB6 gene expression is further repressed by EspM in the absence of the ESX-1 system (Fig. 6). Therefore, regulation by EspM is relevant in WT bacteria and not simply when the ESX-1 system is absent, which may or may not be physiologically relevant.
FIG 6.
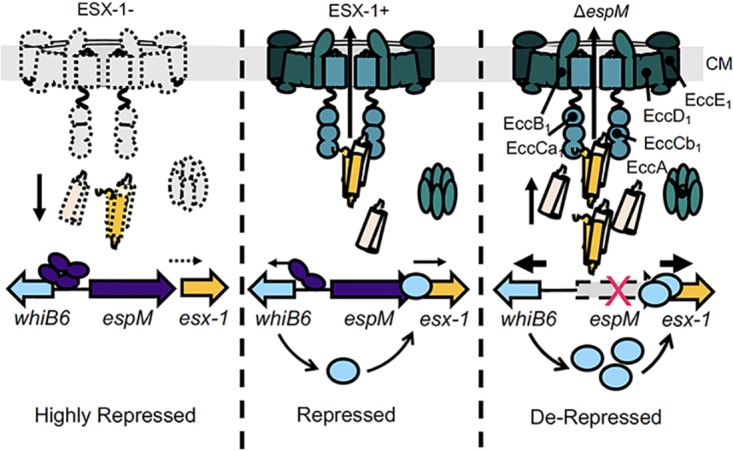
EspM regulates gene expression in response to the ESX-1 system. A model proposing a role for EspM in regulating whiB6 gene expression is shown. In the absence of the ESX-1 membrane complex, we propose that EspM represses whiB6 gene expression (highly repressed state). Reduced WhiB6 levels cause reduced activation of ESX-1 substrate gene expression, preventing substrate accumulation. In the presence of ESX-1, EspM still promotes repression of whiB6 gene expression, fine-tuning expression of the esx-1 substrate genes (repressed state). In the absence of espM, the levels of whiB6 and esx-1 substrate gene expression are derepressed.
We do not yet understand how the ESX-1 membrane complex controls the magnitude of whiB6 repression by EspM. We do not think that the ESX-1 system transcriptionally regulates the espM gene. We observed no ESX-1-dependent change in espM transcript levels either here (Fig. 2A) or in our prior work (35). These findings contrast those of Abdallah et al. (36), which indicated that the Rv3863 transcript (espMMT) is regulated by the ESX-1 system in M. tuberculosis, similarly to the whiB6 gene. This may be an example of differential regulation between M. marinum and M. tuberculosis. The presence or absence of an assembled ESX-1 membrane complex likely posttranscriptionally controls the levels of EspM in M. marinum. We recapitulated the levels of whiB6 gene expression in the ΔeccCb1 strain by overexpressing espM in the WT strain (Fig. 3D). However, our published proteomic data indicate that EspMMM protein levels are reduced 2-fold in the absence of EccCb1, when repression of whiB6 expression is strongest (35). Regulation of the EspM transcription factor may be similar to the control of gene expression by type III secretion systems (T3SS) in Gram-negative bacteria (60–63). The injectisome T3SS uses cytoplasmic substrates and/or chaperones to posttranscriptionally modulate the levels or activity of transcription factors that regulate secretion-associated genes (62, 64–69). ESX-1 substrates or chaperones may posttranscriptionally regulate the activity of EspM in response to the presence or absence of the ESX-1 membrane complex.
Posttranscriptional regulation of EspM could occur through the predicted N-terminal forkhead-associated (FHA) domain. FHA domain-containing proteins posttranscriptionally regulate Gram-negative type III and type VI protein secretion systems (70–72). Staphylococcus aureus has an Ess-type VII secretion system similar to the ESX-1 system (73, 74). The EccC-related protein EssCSA (75, 76) includes a twin-FHA domain that is essential for secretion (77). FHA domains also mediate oligomerization (78–80). We observed a second shift in mobility of the whiB6-espM probe by EMSA with increasing concentrations of EspMMM protein (Fig. 1C and F). We did not observe this supershifted product when using the C-terminal half of EspMMM alone (Fig. 1F), suggesting that the N-terminal half of the protein is important for this observation. The FHA domain may directly or indirectly control oligomerization of EspM in response to the ESX-1 membrane complex.
Although WhiB6 directly binds the whiB6-espM promoter region, we did not identify WhiB6 in the DNA pulldown (Fig. 1; see also Table S1 in the supplemental material). We have not routinely identified WhiB6 from M. marinum lysates using mass spectrometry. We also did not identify the PhoP response regulator, which regulates whiB6 expression in M. tuberculosis. Under the conditions of our experiments, EspM may bind the intergenic region preferentially to other regulators, including WhiB6 and PhoP. This idea is supported by the finding that WhiB6 activates espM gene expression only in the absence of EspM (Fig. 4A). Also, it is possible that no single technique can identify all proteins that bind and regulate a specific region. For example, chromatin immunoprecipitation sequencing (ChIP-seq) experiments in strains overexpressing WhiB6 in M. tuberculosis did not identify direct binding of WhiB6 to the whiB6-espM intergenic region. And yet, overexpression of WhiB6 resulted in a significant upregulation of whiB6 gene expression in the same study (55, 56). Likewise, although PhoP bound the WhiB6 promoter directly in M. tuberculosis, overexpression of PhoP failed to significantly impact whiB6 gene expression (55, 56). Therefore, the absence of enrichment of regulators in our study does not preclude the possibility of a role for them in the regulation of whiB6 and espM expression. Finally, regulation of the whiB6 and espM genes may not be conserved between M. marinum and M. tuberculosis. In the case of whiB6 expression, it has already been established that there is variability in how PhoP regulates whiB6 in M. tuberculosis strains (39). It has not yet been established if whiB6 regulates esx-1 in M. marinum as part of the PhoPR regulon.
The back-to-back divergent arrangement of two regulators is a common theme in microorganisms (81), the best described of which are the cI and Cro regulators of bacteriophage λ (82). Divergence in organization allows tight coordination of the expression of both transcription factors and of their regulons from a single genetic locus. The intergenic region between the espM and whiB6 genes likely contains binding sites for both WhiB6 and EspM. Indeed, we demonstrated using EMSAs that both EspM and WhiB6 bind this region in vitro (Fig. 1; see also Fig. S6 in the supplemental material) and that both contribute to regulating the whiB6 and espM genes [Fig. 4]) (38, 39). Therefore, the genes regulated by WhiB6, the ESX-1 system, and EspM may be coordinated to fine-tune the ESX-1 secretion and for additional biological purposes. Moreover, whereas approximately half of the genes induced or repressed in the ΔespM strain versus the complemented strain are associated with the ESX-1 system, other EspM-regulated genes may have a currently unrecognized role in ESX-1-associated functions.
Our data clearly demonstrate that EspM impacts the expression of esx-1-associated genes and is associated with corresponding changes in ESX-1 protein levels, supporting the idea that EspM functions to fine-tune ESX-1 function. Consistent with these findings, we observed reduced hemolytic activity upon overexpression of espM and increased cytolytic activity in the absence of EspM. Although hemolysis and macrophage cytolysis are both measures of ESX-1 function, our prior work indicated that the results of the two assays do not always align, especially when using strains with intermediate ESX-1 production or secretion levels (45). It is possible that there are additional roles for EspM in ex vivo infection that differ from those seen in our studies in vitro. Alternatively, EspM could impact the expression of additional genes required for phagosomal lysis or macrophage cytolysis. For example, phthiocerol dimycocerosate (PDIM) has been implicated in both phagosomal lysis and macrophage cytolysis (83, 84). However, we did not see changes in the expression of genes required for PDIM synthesis and transport in our RNA sequencing analysis (Table S3) or in the production of PDIM (Fig. S4F).
Unlike most examples of T3SS-dependent gene expression, the genes regulated by EspM and the ESX-1 membrane complex are not restricted to the ESX-1-associated genes (35, 36). The C. trachomatis T3SS, which impacts global gene expression, may represent a temporal cue for regulating gene expression during infection (85). Likewise, the assembly of the ESX-1 system may serve as a temporal cue to regulate mycobacterial gene expression. While pathogenic mycobacteria elicit a transcriptional response essential for survival in the phagosome (1, 4), there has been no report of a transcriptional response to interaction with the macrophage cytosol. The cytoplasm is considered restrictive for bacterial survival and growth unless the pathogen adapts (86). Listeria monocytogenes, a pathogen that lyses the phagosomal membrane and accesses the cytoplasm (87), adapts by altering metabolism and inducing stress response pathways (86). We propose that the assembly of the ESX-1 membrane complex elicits gene expression pathways to link ESX-1-mediated phagosomal lysis and cytoplasmic adaptation. Indeed, several of the genes regulated by EspM and by the ESX-1 system are predicted to be associated with metabolism (Table S3). This is most notable in genes that are downregulated in the ΔespM strain or upregulated in the ΔwhiB6 and eccCb1 mutant strains. For example, the genes in the mce6 locus and surrounding genes were significantly downregulated in the ΔespM strain but were upregulated in the ΔwhiB6 and eccCb1 mutant strains (Fig. S4C), although, due to variability in the data, the results representing the gene induction in the ΔwhiB6 and eccCb1 mutant strains were not statistically significant. These data are supportive of the conclusion that the mce6 genes are repressed in a whiB6-dependent manner, although further characterization studies will be required to support this hypothesis. mce genes have been associated with carbon nutrient uptake, including mce1, promoting uptake of fatty acids (88), and mce4, promoting uptake of cholesterol (89). mce6 is absent in the M. tuberculosis genome but is present in the genomes of many nontuberculous mycobacterial species (90) and could play a role in controlling metabolite import to promote survival in the phagosome or cytosol. The mce6 locus may be important for the cytosolic lifestyle of M. marinum, which polymerizes host actin and exhibits cytosolic motility (5), which is not conserved in M. tuberculosis.
Finally, because EspM regulates a subset of genes controlled by the ESX-1 system, there are likely additional transcription factors that make up an ESX-1-dependent transcriptional network. We focused on proteins that specifically bound the whiB6/espM intergenic region. Studies aimed at identifying proteins that bind additional ESX-1-responsive promoters would identify additional transcription factors in the ESX-1-responsive network.
In conclusion, we have identified a conserved transcription factor, EspM, which is encoded by a gene adjacent to the esx-1 locus that is required for the repression of whiB6 gene expression in the absence of the ESX-1 system. Our study results begin to define a transcriptional network that links the assembly of the ESX-1 system to widespread changes in gene expression, including the regulation of the ESX-1 apparatus and substrates.
MATERIALS AND METHODS
A fully detailed explanation of the methods used in this study can be found in Text S1 in the supplemental material. All M. marinum strains were derived from M. marinum strain M (BAA-535). Where indicated, the parental strain included a FLAG epitope tag at the C terminus of the whiB6 gene (35). Maintenance of the M. marinum strains and E. coli strains is described in Text S1. Enriched proteins were analyzed using quantitative nano-high-performance liquid chromatography–tandem mass spectrometry (nano-UHPLC-MS/MS) proteomics. All mycobacterial strains were generated using the allelic exchange protocol developed by Parish and Stoker (91) as described previously (35, 45, 52, 92). All strains, constructs, and primers (IDT, Coralville, IA) used in this study are listed in Table S2 in the supplemental material. All plasmids and genetic deletions were confirmed by targeted DNA sequencing performed at the Notre Dame Genomics and Bioinformatics Facility. All proteins were expressed in E. coli with 6×His affinity tags and purified using metal chelation affinity chromatography as described in Text S1. EMSAs were performed as reported previously (93–95), with modifications listed in Text S1. β-Galactosidase assays on M. marinum strains bearing the whiB6::lacZ+, attB::pwhiB6-lacZ+, or attB::pespM-lacZ+ reporter were performed as described previously (52). Hemolysis assays were performed as described previously (35). ESX-1 secretion assays were performed as described previously (35), except that 10 μg of protein was analyzed for all protein fractions. Western blot analysis was performed as described previously (35). Macrophage (RAW 264.7 cells) infections were performed as described previously (45) at an estimated multiplicity of infection (MOI) of 7 (2.5 × 106 cells/ml). Cells were imaged and ethidium-homodimer uptake by perforated cells was quantified using ImageJ (35, 52). For transcriptional profiling, M. marinum strains were grown and RNA was extracted exactly as described previously (35). RNA sequencing was conducted as described previously (96), and the results were analyzed using SPARTA software (97). For analysis of differentially expressed genes (>2-fold; q value of <0.05), lists were filtered for genes with average counts greater than 4 (log2 CPM), with full unfiltered data sets available in Table S3.
M. marinum strains, plasmids, and primers used in this study. Download Table S2, PDF file, 1.1 MB (142.1KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression tables presenting results of the RNA sequencing analysis performed in this study. (A) Gene expression tables for the ΔespM strain versus the WT (whiB6Fl) strain. (B) Gene expression tables for the ΔespM strain versus the complemented strain. Download Table S3, XLSX file, 2.0 MB (2.1MB, xlsx) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
The transcriptional profiling data are available at the NCBI GEO database (accession number GSE135072). All statistical analysis was performed as described in each figure legend, using PRISM v8.1.
ACKNOWLEDGMENTS
We thank Brian Stevenson, Julia Van Kessel, and Christina Stallings for their helpful advice in establishing DNA pulldown experiments and EMSAs. We thank Josh Gillen, Kerry Hagedorn, Rachel Belans, Allison Huffman, and Su Jean Park for their technical support.
The research findings reported in this study were supported by the National Institute of Allergies and Infectious Diseases of the National Institutes of Health under award no. R01AI106872 to P.A.C. and R01AI116605 to R.B.A. K.G.S. is supported by a Graduate Student Fellowship from the Eck Institute for Global Health at the University of Notre Dame. M.J.F. is supported by the Center for Rare and Neglected Diseases at the University of Notre Dame. K.R.N. is supported in part by a Graduate Student Fellowship from the Arthur J. Schmitt Foundation.
We contributed as follows. K.G.S., M.J.F., and P.A.C. conceptualized the study. K.G.S., M.J.F., M.M.C., R.B.A., and P.A.C. developed the methodology. All of us contributed to the investigation, validation, data visualization, formal data analysis, writing, and editing of the manuscript. P.A.C. and R.B.A. acquired funding. P.A.C., M.M.C., and R.B.A. provided resources. P.A.C. administered and supervised the project.
The content of this article is solely our responsibility and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation Sanchez KG, Ferrell MJ, Chirakos AE, Nicholson KR, Abramovitch RB, Champion MM, Champion PA. 2020. EspM is a conserved transcription factor that regulates gene expression in response to the ESX-1 system. mBio 11:e02807-19. https://doi.org/10.1128/mBio.02807-19.
REFERENCES
- 1.Russell DG. 2016. The ins and outs of the Mycobacterium tuberculosis-containing vacuole. Cell Microbiol 18:1065–1069. doi: 10.1111/cmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong JA, Hart PD. 1975. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med 142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong JA, Hart PD. 1971. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 7.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson RO, Manzanillo PS, Cox JS. 2012. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 10.Stanley SA, Raghavan S, Hwang WW, Cox JS. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR Jr.. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol 53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 15.Pozos TC, Ramakrishnan L, Ramakrishan L. 2004. New models for the study of Mycobacterium-host interactions. Curr Opin Immunol 16:499–505. doi: 10.1016/j.coi.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. 2004. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol 2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, Hopkinson-Woolley D, Brosch R, Ramakrishnan L. 2017. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci U S A 114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiloh MU, Champion PA. 2010. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol 13:86–92. doi: 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upadhyay S, Mittal E, Philips JA. 1 June 2018, posting date Tuberculosis and the art of macrophage manipulation. Pathog Dis doi: 10.1093/femspd/fty037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkowski EF, Zulauf KE, Weerakoon D, Hayden JD, Ioerger TR, Oreper D, Gomez SM, Sacchettini JC, Braunstein M. 2017. The EXIT strategy: an approach for identifying bacterial proteins exported during host infection. mBio 8:e00872-17. doi: 10.1128/mBio.00872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, Sherman DR, Gerner MY, Urdahl KB. 2018. Alveolar Macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 24:439–446.e4. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacGurn JA, Cox JS. 2007. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun 75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, Mehra A, Park HS, Ernst JD, Jacobs WR Jr, Philips JA. 2016. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4(+) T-cell activation. Nat Microbiol 2:16232. doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittal E, Skowyra ML, Uwase G, Tinaztepe E, Mehra A, Koster S, Hanson PI, Philips JA. 2018. Mycobacterium tuberculosis type VII secretion system effectors differentially impact the ESCRT endomembrane damage response. mBio 9:e01765-18. doi: 10.1128/mBio.01765-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley SA, Cox JS. 2013. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol 374:211–241. doi: 10.1007/82_2013_332. [DOI] [PubMed] [Google Scholar]
- 26.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol 178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 27.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN. 2017. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol 2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- 29.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol 86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 30.Bosserman RE, Champion PA. 2017. ESX systems and the mycobacterial cell envelope: what’s the connection? J Bacteriol 199:e00131-17. doi: 10.1128/JB.00131-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champion MM, Williams EA, Kennedy GM, Champion PA. 2012. Direct detection of bacterial protein secretion using whole colony proteomics. Mol Cell Proteomics 11:596–604. doi: 10.1074/mcp.M112.017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy GM, Hooley GC, Champion MM, Medie FM, Champion PA. 2014. A novel ESX-1 locus reveals that surface associated ESX-1 substrates mediate virulence in Mycobacterium marinum. J Bacteriol 196:1877–1888. doi: 10.1128/JB.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinhikar AG, Verma I, Chandra D, Singh KK, Weldingh K, Andersen P, Hsu T, Jacobs WR Jr, Laal S. 2010. Potential role for ESAT6 in dissemination of M. tuberculosis via human lung epithelial cells. Mol Microbiol 75:92–106. doi: 10.1111/j.1365-2958.2009.06959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlsson F, Joshi SA, Rangell L, Brown EJ. 2009. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog 5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosserman RE, Nguyen TT, Sanchez KG, Chirakos AE, Ferrell MJ, Thompson CR, Champion MM, Abramovitch RB, Champion PA. 27 November 2017, posting date WhiB6 regulation of ESX-1 gene expression is controlled by a negative feedback loop in Mycobacterium marinum. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1710167114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdallah AM, Weerdenburg EM, Guan Q, Ummels R, Borggreve S, Adroub SA, Malas TB, Naeem R, Zhang H, Otto TD, Bitter W, Pain A. 2019. Integrated transcriptomic and proteomic analysis of pathogenic mycobacteria and their esx-1 mutants reveal secretion-dependent regulation of ESX-1 substrates and WhiB6 as a transcriptional regulator. PLoS One 14:e0211003. doi: 10.1371/journal.pone.0211003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sala C, Odermatt NT, Soler-Arnedo P, Gulen MF, von Schultz S, Benjak A, Cole ST. 2018. EspL is essential for virulence and stabilizes EspE, EspF and EspH levels in Mycobacterium tuberculosis. PLoS Pathog 14:e1007491. doi: 10.1371/journal.ppat.1007491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Hu Y, Cumming BM, Lu P, Feng L, Deng J, Steyn AJ, Chen S. 2016. Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep 16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 39.Solans L, Aguilo N, Samper S, Pawlik A, Frigui W, Martin C, Brosch R, Gonzalo-Asensio J. 2014. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect Immun 82:3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol 69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. 2004. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci U S A 101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosserman RE, Nicholson KR, Champion MM, Champion PA. 21 June 2019, posting date A new ESX-1 substrate in Mycobacterium marinum that is required for hemolysis but not host cell lysis. J Bacteriol doi: 10.1128/JB.00760-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, Holton J, Cheng Y, Stroud RM, Cox JS. 2015. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII Secretion. Cell 161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Champion PA, Champion MM, Manzanillo P, Cox JS. 2009. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol 73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi SA, Ball DA, Sun MG, Carlsson F, Watkins BY, Aggarwal N, McCracken JM, Huynh KK, Brown EJ. 2012. EccA1, a component of the Mycobacterium marinum ESX-1 protein virulence factor secretion pathway, regulates mycolic acid lipid synthesis. Chem Biol 19:372–380. doi: 10.1016/j.chembiol.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Wagner JM, Evans TJ, Korotkov KV. 2014. Crystal structure of the N-terminal domain of EccA ATPase from the ESX-1 secretion system of Mycobacterium tuberculosis. Proteins 82:159–163. doi: 10.1002/prot.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King CH, Mundayoor S, Crawford JT, Shinnick TM. 1993. Expression of contact-dependent cytolytic activity by Mycobacterium tuberculosis and isolation of the genomic locus that encodes the activity. Infect Immun 61:2708–2712. doi: 10.1128/IAI.61.6.2708-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mba Medie F, Champion MM, Williams EA, Champion P. 2014. Homeostasis of N-alpha terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect Immun 82:4572–4586. doi: 10.1128/IAI.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams EA, Mba Medie F, Bosserman RE, Johnson BK, Reyna C, Ferrell MJ, Champion MM, Abramovitch RB, Champion PA. 26 January 2017, posting date A nonsense mutation in Mycobacterium marinum that is suppressible by a novel mechanism. Infect Immun doi: 10.1128/IAI.00653-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng H, Williams JT, Coulson GB, Haiderer ER, Abramovitch RB. 2018. HC2091 kills Mycobacterium tuberculosis by targeting the MmpL3 mycolic acid transporter. Antimicrob Agents Chemother 62:e02459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broset E, Martin C, Gonzalo-Asensio J. 2015. Evolutionary landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR: implications for virulence regulation and application to vaccine development. mBio 6:e01289-15. doi: 10.1128/mBio.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minch KJ, Rustad TR, Peterson EJ, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. 2015. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rustad TR, Minch KJ, Ma S, Winkler JK, Hobbs S, Hickey M, Brabant W, Turkarslan S, Price ND, Baliga NS, Sherman DR. 2014. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol 15:502. doi: 10.1186/PREACCEPT-1701638048134699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 60.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 61.Buttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brutinel ED, Yahr TL. 2008. Control of gene expression by type III secretory activity. Curr Opin Microbiol 11:128–133. doi: 10.1016/j.mib.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mou X, Souter S, Du J, Reeves AZ, Lesser CF. 4 June 2018, posting date Synthetic bottom-up approach reveals the complex interplay of Shigella effectors in regulation of epithelial cell death. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1801310115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulmeyer KH, Yahr TL. 2017. Post-transcriptional regulation of type III secretion in plant and animal pathogens. Curr Opin Microbiol 36:30–36. doi: 10.1016/j.mib.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. 2015. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 197:2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kopaskie KS, Ligtenberg KG, Schneewind O. 2013. Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J Biol Chem 288:35478–35488. doi: 10.1074/jbc.M113.504811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bohme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parsot C, Ageron E, Penno C, Mavris M, Jamoussi K, d'Hauteville H, Sansonetti P, Demers B. 2005. A secreted anti-activator, OspD1, and its chaperone, SpaXV, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol 56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- 69.Vakulskas CA, Brady KM, Yahr TL. 2009. Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J Bacteriol 191:6654–6664. doi: 10.1128/JB.00902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin JS, Pissaridou P, Wu HH, Tsai MD, Filloux A, Lai EM. 29 March 2018, posting date TagF-mediated repression of bacterial type VI secretion systems involves a direct interaction with the cytoplasmic protein Fha. J Biol Chem doi: 10.1074/jbc.RA117.001618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin JS, Wu HH, Hsu PH, Ma LS, Pang YY, Tsai MD, Lai EM. 2014. Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens. PLoS Pathog 10:e1003991. doi: 10.1371/journal.ppat.1003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 73.Garufi G, Butler E, Missiakas D. 2008. ESAT-6-like protein secretion in Bacillus anthracis. J Bacteriol 190:7004–7011. doi: 10.1128/JB.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burts ML, Williams WA, DeBord K, Missiakas DM. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A 102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jager F, Kneuper H, Palmer T. 2018. EssC is a specificity determinant for Staphylococcus aureus type VII secretion. Microbiology 164:816–820. doi: 10.1099/mic.0.000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jager F, Zoltner M, Kneuper H, Hunter WN, Palmer T. 2016. Membrane interactions and self-association of components of the Ess/type VII secretion system of Staphylococcus aureus. FEBS Lett 590:349–357. doi: 10.1002/1873-3468.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zoltner M, Ng WM, Money JJ, Fyfe PK, Kneuper H, Palmer T, Hunter WN. 2016. EssC: domain structures inform on the elusive translocation channel in the type VII secretion system. Biochem J 473:1941–1952. doi: 10.1042/BCJ20160257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almawi AW, Matthews LA, Guarne A. 2017. FHA domains: phosphopeptide binding and beyond. Prog Biophys Mol Biol 127:105–110. doi: 10.1016/j.pbiomolbio.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 79.Weng JH, Hsieh YC, Huang CC, Wei TY, Lim LH, Chen YH, Ho MR, Wang I, Huang KF, Chen CJ, Tsai MD. 2015. Uncovering the mechanism of forkhead-associated domain-mediated TIFA oligomerization that plays a central role in immune responses. Biochemistry 54:6219–6229. doi: 10.1021/acs.biochem.5b00500. [DOI] [PubMed] [Google Scholar]
- 80.Ahn JY, Li X, Davis HL, Canman CE. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 81.Beck CF, Warren RA. 1988. Divergent promoters, a common form of gene organization. Microbiol Rev 52:318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. 1981. lambda Repressor and cro–components of an efficient molecular switch. Nature 294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 83.Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. 2017. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. mBio 8:e00148-17. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, Hinman AE, Thompson M, Nietupski RM, Golas A, Montgomery P, Fitzgerald M, Smith RS, White DW, Tischler AD, Carpenter AE, Hung DT. 2017. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 13:e1006363. doi: 10.1371/journal.ppat.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanson BR, Slepenkin A, Peterson EM, Tan M. 2015. Chlamydia trachomatis type III secretion proteins regulate transcription. J Bacteriol 197:3238–3244. doi: 10.1128/JB.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen GY, Pensinger DA, Sauer JD. 21 Ju Ly 2017, posting date Listeria monocytogenes cytosolic metabolism promotes replication, survival, and evasion of innate immunity. Cell Microbiol doi: 10.1111/cmi.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nazarova EV, Montague CR, La T, Wilburn KM, Sukumar N, Lee W, Caldwell S, Russell DG, VanderVen BC. 2017. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 6:e26969. doi: 10.7554/eLife.26969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fedrizzi T, Meehan CJ, Grottola A, Giacobazzi E, Fregni Serpini G, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, De Sanctis V, Rumpianesi F, Pecorari M, Jousson O, Tortoli E, Segata N. 2017. Genomic characterization of nontuberculous mycobacteria. Sci Rep 7:45258. doi: 10.1038/srep45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 92.Bosserman RE, Thompson CR, Nicholson KR, Champion PA. 2018. Esx paralogs are functionally equivalent to ESX-1 proteins but are dispensable for virulence in Mycobacterium marinum. J Bacteriol 200:e00726-17. doi: 10.1128/JB.00726-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garner AL, Weiss LA, Manzano AR, Galburt EA, Stallings CL. 2014. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol Microbiol 93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alves-Rodrigues I, Ferreira PG, Moldón A, Vivancos AP, Hidalgo E, Guigó R, Ayté J. 2016. Spatiotemporal control of forkhead binding to DNA regulates the meiotic gene expression program. Cell Rep 14:885–895. doi: 10.1016/j.celrep.2015.12.074. [DOI] [PubMed] [Google Scholar]
- 95.Hellman LM, Fried MG. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc 2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baker JJ, Johnson BK, Abramovitch RB. 2014. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol Microbiol 94:56–69. doi: 10.1111/mmi.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson BK, Scholz MB, Teal TK, Abramovitch RB. 2016. SPARTA: Simple Program for Automated reference-based bacterial RNA-seq Transcriptome Analysis. BMC Bioinformatics 17:66. doi: 10.1186/s12859-016-0923-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation and confirmation of M. marinum ΔespM strain. (A) Schematic of parental, ΔMMAR_5438 (ΔespM), and complemented strains. (B) Confirmation of ΔespM genotype. Diluted lysates of resolved strains were checked by PCR performed with primers OMF619 and OMF620. Download FIG S1, PDF file, 0.4 MB (457.6KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A to D) Processed MS data from the DNA affinity chromatography assay performed as described in the Fig. 1 legend. (A) Unfiltered processed protein data. (B) Filtered data used for quantitation, (C) Normalized LFQ enrichment for MMAR_5438 and HupB DNA binding. (D) Proteins enriched for binding of the whiB6-espM probe (>2-fold enrichment over the rpoA probe level). (E and F) Relative quantification of the proteins described in the Fig. S4E legend. (E) Unfiltered processed protein group data. (F) Filtered data used for quantification. For panels E and F, two biological replicates each were performed in technical duplicate. The data shown are representative of results from one biological replicate, performed in technical duplicate. Download Table S1, XLSX file, 1.9 MB (2MB, xlsx) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heterologous expression of EspM proteins from M. marinum and M. tuberculosis. (A to C) Coomassie-stained gels representing purification of 6×His-EspMMM protein (A) and of denatured 6×His-EspMMT with refolding (B) and dilution series of purified 6×His-EspMNT (aa 1 to 133) and 6×His-EspMCT (aa 127 to 363) (C) from M. marinum. (D) Purification of WhiB6MM-6×His protein. Final concentrations and buffer conditions are listed in Text S1. Download FIG S2, PDF file, 1.3 MB (1.4MB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed Materials and Methods section regarding the approaches used in this article. Download Text S1, PDF file, 0.2 MB (217.3KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conservation of the espM gene and protein in M. marinum, M. tuberculosis, and M. smegmatis. (A) Rv3863 and MSMEG_0052 are the espM orthologs in M. tuberculosis and M. smegmatis. The esx-1 locus begins with the espE gene in M. marinum and M. tuberculosis and with the espG gene in M. smegmatis. (B) The alignment was generated using Clustal Omega, followed by visualization with BoxShade. Black, identity; gray, similarity. The asterisk (*) indicates conservation across all three species. The predicted FHA domain in M. marinum and M. tuberculosis is indicated in pink. The sequence of EspMNT from M. marinum used as described for Fig. 1 ends at the red circle. EspMCT from M. marinum used as described for Fig. 1 starts at the green arrow. Download FIG S3, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
EspM broadly regulates gene expression in M. marinum and fine-tunes ESX-1 protein levels. (A) Scatter plot of genes differentially expressed in the ΔespM strain versus the WT strain. Genes highlighted in red had a q value of <0.05. (B) Venn diagram of genes that were induced (>2×, q value of <0.05) in the ΔespM/WT or ΔespM/complemented strain from this study or that were repressed (>2×, q value of <0.05) in the ΔwhiB6 or eccCb1 ochre mutant (relative to the WT strain) from Bosserman et al. (35). (C) Heat map of genes in the mce6 locus and of surrounding genes that were significantly downregulated (>2×, q value of <0.05) in the ΔespM/WT or ΔespM/complemented comparison. These genes were induced in the ΔwhiB6 or eccCb1 ochre mutants relative to the WT strain, consistent with expression being whiB6 and ESX-1 dependent; however, the induction in gene expression did not achieve statistical significance for these strains. All strains in this figure contained the whiB6Fl allele at the whiB6 locus. (D) Western blot analysis of EsxA production in M. marinum lysates. The strains are the same as those shown in Fig. 5. A 10-μg or 5-μg volume of protein, as indicated, was loaded per lane and resolved on a 4% to 20% gel. Mpt-32 served as a loading control. The image shown is representative of results of three biological replicates. (E) Relative quantification of ESX-1-associated proteins using label-free proteomics. The proteins were derived from the strains described in the Fig. 5 legend. All protein levels are represented as log2-fold changes compared with those measured in the WT (whiB6Fl) strain or the ΔespM strain. Only the subset of ESX-1-associated proteins that were reproducibly quantified are shown in this heat map. The complete data are presented in Table S1E and F. The data shown are representative of results from one biological replicate, performed in technical duplicate. (F) Thin-layer chromatography of lipids extracted from the strains described in the Fig. 5 legend. PDIM purified from M. tuberculosis H37Rv was used as a positive control. TAG is triacylglycerols. The Rf values are as follows: for M. tuberculosis PDIM and the WT, espM, and eccCb1 strains, 3.17 cm; for the complemented strain, 3.07 cm, likely due to the slight curvature of the band. The results shown are representative of experiments performed in biological triplicate. Download FIG S4, PDF file, 0.8 MB (782.2KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
EspM is required for whiB6 repression in the ΔeccA M. marinum strain. Western blot analysis of 10 μg per lane on an 18% gel was performed. Mpt-32 was used as a loading control. The image is representative of results from three independent biological replicates. All strains contained a C-terminal epitope tag on the whiB6 gene. Download FIG S5, PDF file, 0.2 MB (260.4KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
WhiB6 binds the whiB6-espM intergenic region in vitro. EMSA was performed with increasing concentrations of WhiB6 protein from M. marinum. The whiB6 promoter probe was the same as that used as described in the Fig. 1 legend (bp 6577396 to 6577960 in the M. marinum M genome; 30 bp upstream of the whiB6 ORF to 141 bp into MMAR_5438). The control probe was 500 bp of the rpoA ORF (bp 1309999 to 1310499). Download FIG S6, PDF file, 0.7 MB (727.6KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M. marinum strains, plasmids, and primers used in this study. Download Table S2, PDF file, 1.1 MB (142.1KB, pdf) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression tables presenting results of the RNA sequencing analysis performed in this study. (A) Gene expression tables for the ΔespM strain versus the WT (whiB6Fl) strain. (B) Gene expression tables for the ΔespM strain versus the complemented strain. Download Table S3, XLSX file, 2.0 MB (2.1MB, xlsx) .
Copyright © 2020 Sanchez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The transcriptional profiling data are available at the NCBI GEO database (accession number GSE135072). All statistical analysis was performed as described in each figure legend, using PRISM v8.1.