The human gut microbiome is made of trillions of microbial cells, most of which are Bacteria, with a subset of Archaea. The bacterial family Christensenellaceae and the archaeal family Methanobacteriaceae are widespread in human guts. They correlate with each other and with a lean body type. Whether species of these two families interact and how they affect the body type are unanswered questions. Here, we show that species within these families correlate with each other across people. We also demonstrate that particular species of these two families grow together in dense flocs, wherein the bacteria provide hydrogen gas to the archaea, which then make methane. When the archaea are present, the ratio of bacterial products (which are nutrients for humans) is changed. These observations indicate that when these species grow together, their products have the potential to affect the physiology of their human host.
KEYWORDS: human gut, methanogens, microbiome, syntrophy
ABSTRACT
Across human populations, 16S rRNA gene-based surveys of gut microbiomes have revealed that the bacterial family Christensenellaceae and the archaeal family Methanobacteriaceae cooccur and are enriched in individuals with a lean, compared to an obese, body mass index (BMI). Whether these association patterns reflect interactions between metabolic partners, as well as whether these associations play a role in the lean host phenotype with which they associate, remains to be ascertained. Here, we validated previously reported cooccurrence patterns of the two families and their association with a lean BMI with a meta-analysis of 1,821 metagenomes derived from 10 independent studies. Furthermore, we report positive associations at the genus and species levels between Christensenella spp. and Methanobrevibacter smithii, the most abundant methanogen of the human gut. By coculturing three Christensenella spp. with M. smithii, we show that Christensenella spp. efficiently support the metabolism of M. smithii via H2 production far better than Bacteroides thetaiotaomicron does. Christensenella minuta forms flocs colonized by M. smithii even when H2 is in excess. In culture with C. minuta, H2 consumption by M. smithii shifts the metabolic output of C. minuta’s fermentation toward acetate rather than butyrate. Together, these results indicate that the widespread cooccurrence of these microorganisms is underpinned by both physical and metabolic interactions. Their combined metabolic activity may provide insights into their association with a lean host BMI.
INTRODUCTION
Obesity was the first human disease phenotype to be associated with an altered microbial ecology of the gut (1, 2). The link between the relative abundance in the gut of the bacterial family Christensenellaceae and a low host body mass index (BMI) now stands as one of the most robust associations described between the human gut microbiome and host BMI (3–15). Compared to other families of bacteria that comprise the human gut microbiome, the family Christensenellaceae was described relatively recently, when the type strain, Christensenella minuta, was reported in 2012 (16). Prior to the description of C. minuta, 16S rRNA sequences from this genus escaped notice in the gut microbiome, though these sequences accumulated steadily in small-subunit (SSU) rRNA gene databases. A positive association between a lean host BMI and the relative abundance in the gut of Christensenellaceae 16S rRNA genes was first reported in 2014 (4). The association was shown to have existed in earlier data sets (4) but was likely undetected, as this family had not yet been named. Goodrich et al. showed a causal link between the Christensenellaceae and host BMI in gnotobiotic mice: the addition of C. minuta to the gut microbiome of an obese human donor prior to transplantation reduced adiposity gains in the recipient mice compared to those of controls receiving the unsupplemented microbiome (4). The mechanism underlying this host response remains to be elucidated. One step toward this goal is a better understanding of how the members of the Christensenellaceae interact ecologically with other members of the gut microbiome.
Across human populations, the gut microbiota often forms patterns of cooccurrence (e.g., when these consortia exist in a subset of human subjects, they are termed enterotypes [17]). Such cooccurrences of taxa across subjects reflect shared environmental preferences, but to determine if they represent metabolic or physical interactions requires further study. The family Christensenellaceae consistently forms the hub of cooccurrence networks with other taxa (6, 8, 9, 18, 19). Notably, gut methanogens (specifically, of the archaeal family Methanobacteriaceae) are often reported as part of the Christensenellaceae cooccurrence consortium (4, 20–22). The most widespread and abundant of the gut methanogens, Methanobrevibacter smithii, produces CH4 from H2 and CO2, the products of bacterial fermentation of dietary fibers. Such cross-feeding likely explains why the relative abundances of M. smithii and fermenting bacteria are often positively correlated (21, 23, 24). Several studies have shown that in the laboratory, M. smithii can grow from the H2 provided by Bacteroides thetaiotaomicron, a common gut commensal bacterium (25–27). Given that the cultured representatives of the Christensenellaceae ferment simple sugars (16, 28) and that their genomes contain hydrogenases (29), we predicted that members of the Christensenellaceae produce H2 used by M. smithii as a substrate in methanogenesis.
Here, we explored the association between the Christensenellaceae and the Methanobacteriaceae in two ways. First, we analyzed metagenomes for statistical associations between the two families and their subtaxa. Compared to 16S rRNA gene surveys, metagenomes often can better resolve the taxonomic assignments of sequence reads below the genus level (30). Metagenome-based studies have so far been blind to the Christensenellaceae, however, because their genomes have been lacking from reference databases. Here, we customized a reference database to include Christensenellaceae genomes, which we used in a meta-analysis of >1,800 metagenomes from 10 studies. Second, to assess for metabolic interactions between members of the Christensenellaceae and Methanobacteriaceae, we measured methane production by M. smithii when grown in coculture with Christensenella spp. Our results show that (i) the positive association between the Christensenellaceae and the Methanobacteriaceae is robust to the genus/species level across multiple studies, (ii) these taxa associate with a lean host BMI, (iii) Christensenella spp. support the growth of M. smithii by interspecies H2 transfer far better than B. thetaiotaomicron does, and (iv) M. smithii directs the metabolic output of C. minuta toward less butyrate and more acetate and H2, which is consistent with reduced energy availability to the host and consistent with the association with a low BMI.
RESULTS
Christensenella relative abundance is significantly correlated with leanness across populations.
Both the Christensenellaceae family and the genus Christensenella had very high prevalences, as they were present in more than 99% of the 1,821 samples; both the family and the genus have a mean abundance of 0.07% ± 0.05% (Fig. 1b and d and see Fig. S1 in the supplemental material). To correct for the influence of environmental factors on the relative abundances of members of the Christensenellaceae family and of the Christensenella genus, we first constructed null models in which we selected covariates (see Appendix 1 in Text S1) that explained a significant proportion of the variance of the transformed relative abundance of the family Christensenellaceae and in the same manner as that of the Christensenella genus. BMI and age were significantly correlated with the transformed relative abundances of members of the Christensenellaceae family and of the Christensenella genus (Cf-tra and Cg-tra, respectively, where the suffix “-tra” indicates transformed abundances) and were retained in the null models (Cf-null and Cg-null).
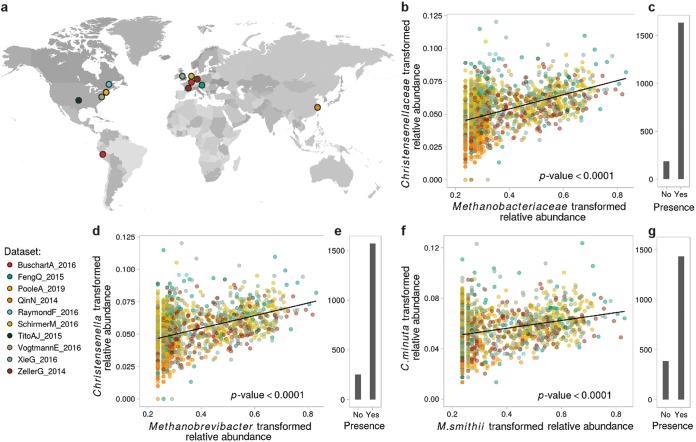
FIG 1.
Abundances of the Methanobacteriaceae and Christensenellaceae families across populations. (a) Countries where the human gut metagenomes used in our meta-analysis (n = 1,821 samples) were recruited by 10 independent studies (summarized in Table S3); (b) association between the transformed relative abundances of Christensenellaceae and Methanobacteriaceae in samples where the a member of the Methanobacteriaceae was detected; (c) numbers of samples in which the Methanobacteriaceae were detected; (d and e and f and g) same as panels b and c at the genus and species levels, respectively. The correlation between the transformed relative abundances of both taxa at each taxonomic level was evaluated using linear mixed models to correct for covariates (ANOVA, P values < 0.0001).
Abundances of the Methanobacteriaceae and Christensenellaceae families across studies. (a to j) Transformed relative abundances of Christensenellaceae (Cf-tra) and Methanobacteriaceae (Mf-tra) across 1,821 samples from 10 countries and generated from 10 independent studies. The data generated for this study are grouped with the first time series published in the work of Poole et al. (53). The gap between 0 and ∼0.2 is due to the detection limit of the sequencing method; the minimal relative abundance is 10−3%. Hence, 0 indicates that the microorganism was not detected, which introduces a gap after transformation of the data. Download FIG S1, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental appendices. Download Text S1, DOCX file, 0.01 MB (15.8KB, docx) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data sets used for the statistical analysis. The BMI and age values for each data set are reported as average values, with minimum and maximum values in parentheses. Download Table S3, PDF file, 0.1 MB (118.1KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
BMI was negatively correlated with both Cf-tra (type II analysis of variance [ANOVA], P value = 0.0002 and F value [339] = 14.46) and Cg-tra (type II ANOVA, P value = 0.0002 and F value [339] = 14.29), indicating that leaner individuals harbor higher relative abundances of Christensenellaceae and Christensenella. Age was negatively correlated with Cf-tra (type II ANOVA, P value = 0.01 and F value [1,468] = 6.56) and with Cg-tra (type II ANOVA, P value = 0.01 and F value [1,468] = 6.53), indicating that younger subjects carry greater relative abundances of Christensenellaceae and Christensenella. However, the interaction term between BMI and age was not significantly correlated with the transformed relative abundances (type II ANOVA, P values > 0.1), indicating that their effects are additive. These results show that regardless of their BMIs, younger subjects have higher levels of Christensenellaceae and Christensenella and that the lower a subject’s BMI, the more of these microbes they harbor, regardless of their age.
Methanobrevibacter relative abundance is significantly correlated with leanness across populations.
The Methanobacteriaceae family and Methanobrevibacter genus also had high prevalences, with 92% and 89% of people harboring them, respectively, and with mean abundances of 0.48% ± 1.55% and 0.49% ± 1.54%, respectively. As described above, we evaluated the association between the Methanobacteriaceae family and of the Methanobrevibacter genus by using models for BMI and age (models Mf-null and Mg-null). The transformed relative abundances of Methanobacteriaceae, Mf-tra, and of Methanobrevibacter, Mg-tra, were also negatively correlated with BMI (type II ANOVA, respective P values = 0.01 and 0.02, F values [341, 341] = 6.66 and 5.11, respectively). In contrast to the Christensenellaceae, both Methanobacteriaceae and Methanobrevibacter were positively correlated with age (type II ANOVA, respective P values = 0.001 and 4.27 × 10−4 and F values [1,468, 1,468] = 10.35 and 12.47), indicating that older people carry a greater proportion of methanogens. Moreover, M. smithii, the most abundant and prevalent methanogen species within the human gut, was also positively correlated with age and negatively with BMI regardless of age; i.e., the interaction term between age and BMI was not significantly correlated (see Appendix 2 in Text S1 for additional statistics).
The relative abundances of the Christensenella and Methanobrevibacter genera are significantly correlated across populations.
Next, we looked into how the Christensenellaceae and the Methanobacteriaceae correlated with each other across subjects while controlling for BMI and age. We constructed a model where Mf-tra was included in addition to BMI and age (model Cf-Mf). This allowed us to test whether adding Mf-tra to the model improved its fit and, if so, how much of the variance of Cf-tra not explained by age and BMI could be explained by Mf-tra. We also evaluated the interaction terms between Mf-tra and BMI and between Mf-tra and age to assess whether the correlation between Cf-tra and Mf-tra was dependent on age and BMI. The interaction term for BMI and Mf-tra was not significant and was removed from the model; the interaction term for age and Mf-tra was significant and was retained (type I ANOVA, F value [339] = 8.30 and P value = 0.0042). We compared the log likelihoods of the null and full models (Cf-null and Cf-Mf) to confirm that the relative abundances of the Methanobacteriaceae and Christensenellaceae families were significantly correlated (χ2 test, P value = 1.78 × 10−59). Furthermore, the model Cf-Mf showed that Mf-tra was significantly positively correlated with Cf-tra (Fig. 1b) (type I ANOVA, F value [339] = 287.03, P value < 0.0001) and that the interaction term between Mf-tra and age was positively correlated with Cf-tra as well. These results indicate that the relative abundances of the Christensenellaceae and Methanobacteriaceae families are positively correlated across multiple populations/studies. In addition, although both families are enriched in low-BMI people, they are correlated regardless of a subject’s BMI. Moreover, their association is stronger in older people, suggesting that although elders are less likely to carry as much Christensenellaceae as youths, the more Methanobacteriaceae they have, the more Christensenellaceae they have.
We performed a similar analysis using the abundances of the two most prominent genera belonging to these families (Christensenella and Methanobrevibacter, models Cg-null and Cg-Mg) and obtained equivalent results. First, the interaction term between Mg-tra and age was positively correlated with Cg-tra (Fig. 1c) (type I ANOVA, F value [339] = 10.19, P value = 0.0015). Then, by comparing model Cg-null with our full model Cg-Mg, we showed that the relative abundances of the two genera were also correlated (χ2 test, P value = 1.50 × 10−57). Our full model Cg-Mg showed that Mg-tra was significant for predicting Cg-tra while controlling for BMI and age (type I ANOVA, F value [339] = 274.35, P value < 0.0001), with the interaction term between Mg-tra and age also positively correlating with Cg-tra. These results indicate that the correlations between the relative abundances of the two families, explained above, hold true at the level of two representative genera. The association between Methanobrevibacter and Christensenella is stronger in older people regardless of BMI.
A similar analysis at the species level indicated that C. minuta and M. smithii were the most abundant species of each of their genera, and similarly to the family and genus ranks, their relative abundances across samples were significantly correlated (Fig. 1d and Appendix 2 in Text S1). The less abundant Christensenella gut species C. massiliensis and C. timonensis also correlated with Methanobrevibacter smithii across the 1,821 metagenomes (see Appendix 2 in Text S1). C. minuta and C. timonensis’s transformed relative abundances were significantly negatively correlated with both BMI and age, while C. massiliensis’s transformed relative abundance was significantly correlated with BMI but not with age. Leaner people are thus enriched in members of the Christensenellaceae family, and C. minuta and C. timonensis are more abundant in young people than in older people.
C. minuta forms flocs alone and in coculture with M. smithii.
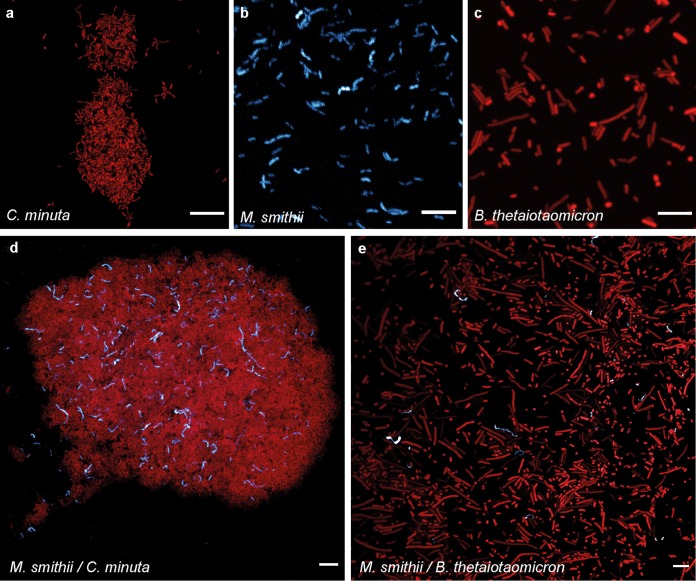
To assess the physical and metabolic interactions of two representative species, we used C. minuta DSM-22607, previously shown to reduce adiposity in germfree mouse fecal transplant experiments (4), and M. smithii DSM-861, which is the most abundant and prevalent methanogen in the human gut (31). Confocal and scanning electron imaging of 2- to 7-day-old cultures revealed that C. minuta organisms flocculate in mono- and cocultures (Fig. 2a and b and Fig. 3a to c and g to j). M. smithii is present within the C. minuta flocs (Fig. 2d and Fig. 3g to j) but does not aggregate in monoculture before 7 to 10 days of culture (data not shown). In contrast, B. thetaiotaomicron, used here as a positive control based on previous reports that it supports the growth of M. smithii via H2 production (25, 26), did not flocculate when grown alone (Fig. 2c) and, when cocultured with M. smithii, displayed very limited aggregation (Fig. 2e, Fig. 3k to n, and Fig. S2).
FIG 2.
Confocal micrographs of the cultures after 3 days of growth. (a) C. minuta alone; (b) M. smithii alone; (c) B. thetaiotaomicron alone; (d) M. smithii and C. minuta together; (e) M. smithii and B. thetaiotaomicron together. SYBR green I fluorescence (DNA staining) is shown in red, and M. smithii’s coenzyme F420 autofluorescence is shown in blue. Scale bars represent 10 μm. Based on gas production, at 3 days of growth, B. thetaiotaomicron was already at stationary phase (explaining the elongated cells) (see Fig. S2 for confocal micrographs of B. thetaiotaomicron and M. smithii at 2 days of growth), C. minuta was at the end of the exponential phase, and M. smithii was still in exponential phase.
FIG 3.
Scanning electron micrographs of the cultures at 3 to 7 days of growth. (a, d, g, and k) Representative Balch tubes of cultures of C. minuta (Cm), M. smithii (Ms), C. minuta and M. smithii (Cm/Ms), and B. thetaiotaomicron and M. smithii (Bt/Ms) after 7 days of growth; (b and c) scanning electron micrographs (SEMs) of monocultures of C. minuta at 5 days of growth; (e and f) SEMs of monocultures of M. smithii at 5 days of growth; (h to j) SEMs of cocultures of C. minuta and M. smithii at 7, 5, and 2 days of growth, respectively; (l to n) SEMs of cocultures of B. thetaiotaomicron and M. smithii at 7 days of growth. The floc formed by C. minuta and M. smithii is indicated with a white arrow in panel g; other arrows indicate M. smithii cells. Metal bars in panels a, d, and g are from the tube rack.
Confocal imaging of cocultures of B. thetaiotaomicron and M. smithii at different time points. (a and b) Cells at day 2, when B. thetaiotaomicron enters stationary phase (Fig. 4d); (c and d) cells at day 7, the end of the experiment, when maximal CH4 concentrations were observed both in monocultures of M. smithii and in cocultures with B. thetaiotaomicron (Fig. 4b and e). In exponential phase, B. thetaiotaomicron cells are rod shaped (a), while during stationary phase, they suffer stress, leading to elongated cells (c). The bright fields (a and c) and M. smithii’s coenzyme F420 (b and d) channels are displayed. Scale bars represent 10 μm. Download FIG S2, JPG file, 0.1 MB (123.2KB, jpg) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
H2 and CH4 production.
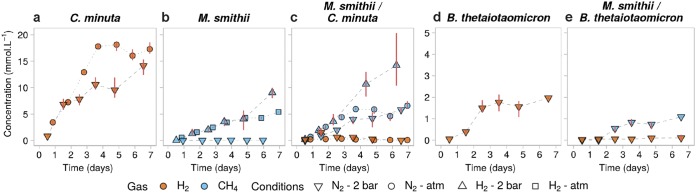
After 6 days in monoculture, C. minuta had produced 7 times more H2 than B. thetaiotaomicron (14.2 ± 1.6 mmol · liter−1 versus 2.0 ± 0.0 mmol · liter−1) (Fig. 4a and d and Fig. 5a; Wilcoxon rank sum test, P value = 0.1). As expected, M. smithii did not grow in monoculture when H2 was not supplied (80:20, vol/vol, N2-CO2 headspace) (Fig. 4b). After 6 days, M. smithii had produced 9.0 ± 1.0 mmol · liter−1 of CH4 when H2 was provided in excess (i.e., 80:20, vol/vol, H2-CO2 atmosphere at 2 × 105 Pa) (Fig. 4b and Fig. 5b).
FIG 4.
Gas concentrations over time in mono- and cocultures of C. minuta, B. thetaiotaomicron, and M. smithii grown under different conditions. (a to e) H2 (orange) and CH4 (blue) concentrations in the headspace over time in cultures from batches 1 to 3 (Table S1). Points represent the averages of results from 3 biological replicates for each condition, and red bars join the minimal and maximal values. In conditions where H2 was provided in excess (H2, 2 × 105 Pa, and atmospheric (atm) H2, with the headspace initially composed of 80:20, vol/vol, H2-CO2), its concentrations are not shown for scale reasons. Initial concentrations of H2 under conditions where it was not provided in the headspace were undetectable (N2, 2 × 105 Pa, and atmospheric N2, with the headspace initially composed of 80:20, vol/vol, N2-CO2) and stayed null in the monocultures of M. smithii (not shown). CH4 concentrations in the bacterial monocultures were undetectable and are not shown as well. (a to c) Same y scale as in panels d and e.
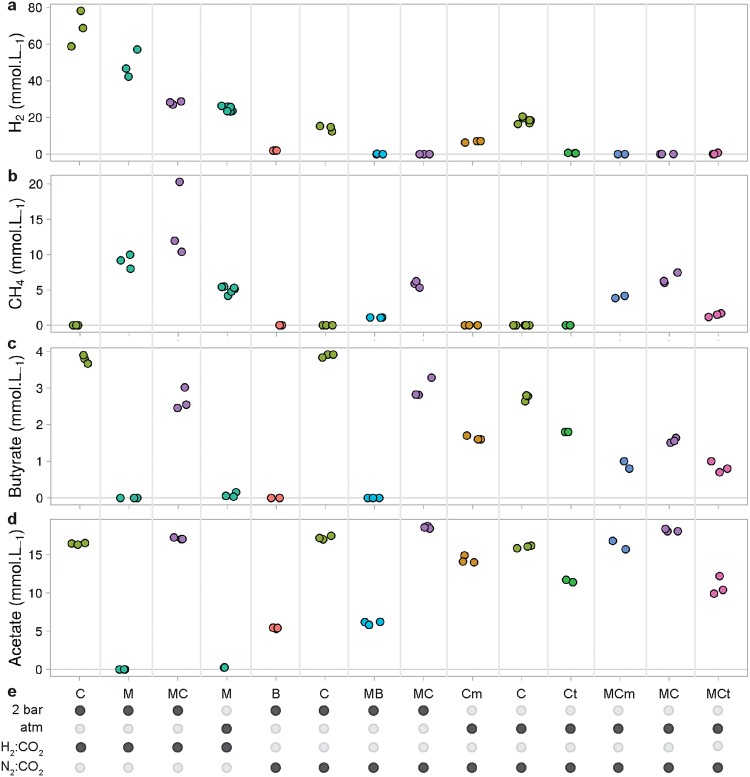
FIG 5.
Summary of gases and SCFAs produced in mono- and cocultures of C. minuta, C. timonensis, C. massiliensis, B. thetaiotaomicron, and M. smithii after 6 days of growth. (a to d) H2, CH4, butyrate, and acetate production after 6 days of growth in all mono- and cocultures presented in this study (batches 1 to 4) (Table S1). Points represent the concentration of each biological replicate. (e) Table summarizing the conditions for each culture. The conditions include the gas mixture (H2-CO2 or N2-CO2 at 80:20, vol/vol), the initial pressure (2 × 105 Pa or atmospheric), and the microorganisms inoculated. C, C. minuta; Ct, C. timonensis; Cm, C. massiliensis; B, B. thetaiotaomicron; M, M. smithii; MC, M. smithii and C. minuta; MB, M. smithii and B. thetaiotaomicron; MCm, M. smithii and C. massiliensis; MCt, M. smithii and C. timonensis. Samples inoculated with the same microorganisms are the same color.
Total pressure, headspace composition, and culture inocula for each batch of experiments described in the main text. Download Table S1, PDF file, 0.02 MB (16.2KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In accordance with the higher levels of H2 produced by C. minuta than by B. thetaiotaomicron, day 6 CH4 concentrations were higher for M. smithii cocultured with C. minuta than with B. thetaiotaomicron (respectively, 5.8 ± 0.5 mmol · liter−1 and 1.1 ± 0.0 mmol · liter−1; Wilcoxon rank sum test, P value = 0.1) (Fig. 4c and e and Fig. 5b). For both coculture conditions, H2 concentrations were very low (on average, across time points and replicates, H2 concentrations were 0.5 ± 0.6 mmol · liter−1 in cocultures with C. minuta and 0.1 ± 0.1 mmol · liter−1 in cocultures with B. thetaiotaomicron), indicating that almost all the H2 that had been produced was also consumed (Fig. 4c and e and Fig. 5a).
Pressure effects on gas production and aggregation.
Gas-consuming microbes, including hydrogenotrophic methanogens, grow better in a pressurized environment (32–34) due to a higher gas solubility at higher pressure, as described by Henry’s law. We compared levels of CH4 production by M. smithii in monoculture and in coculture with C. minuta under 2 different pressures (i.e., 2 × 105 Pa and atmospheric pressure). As with their flocculation at 2 × 105 Pa (Fig. 2d), C. minuta and M. smithii aggregated at atmospheric pressure (Fig. S3a and b). Accordingly, C. minuta supported CH4 production by M. smithii to similar extents under both pressure conditions (ANOVA followed by Tukey’s post hoc test, adjusted P value = 1.0) (Fig. 4c and 5b), even though the putative H2 produced by C. minuta (estimated based on the monocultures) was much lower than the amount of H2 provided in the headspace for M. smithii (Fig. 5a).
C. minuta and M. smithii aggregate at atmospheric pressure and even when there is excess H2 in the medium. Confocal imaging of C. minuta and M. smithii at 3 days of growth. (a and b) Coculture grown at atmospheric pressure; (c and d) coculture grown under a pressurized H2-CO2 atmosphere. The bright fields (a and c) and M. smithii‘s coenzyme F420 (b and d) channels are displayed. Scale bars represent 10 μm. Download FIG S3, JPG file, 0.2 MB (246KB, jpg) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next sought to assess whether the mixed aggregation of M. smithii with C. minuta could be disrupted if H2 was pressurized in the medium, reducing M. smithii’s reliance on C. minuta as a H2 source. We observed that M. smithii aggregated with C. minuta (Fig. S3c and d) even though H2 was abundant. Total CH4 production was higher than in monocultures under the same headspace, reaching 14.2 ± 5.3 mmol · liter−1 in coculture versus 9.0 ± 1.0 mmol · liter−1 in monoculture after 6 days (ANOVA followed by Tukey’s post hoc test, adjusted P value = 0.1) (Fig. 4b and c). This indicates that interspecies H2 transfer occurs even when H2 is added to the headspace and leads to greater methanogenesis.
The SCFA production of C. minuta is influenced by the presence of M. smithii.
Regardless of headspace composition and pressure conditions, the only short-chain fatty acids (SCFAs) detected as produced by C. minuta in monoculture were acetate and butyrate (among 10 short and medium-chain fatty acids analyzed) (see Appendix 1 in Text S1 and Fig. 5). To investigate if the consumption of H2 by M. smithii influenced the SCFA production profile of C. minuta, we compared acetate and butyrate concentrations between the cocultures and C. minuta’s monocultures under all conditions tested (i.e., cultures at 2 × 105 Pa or atmospheric pressure with an 80:20, vol/vol, N2-CO2 or H2-CO2 headspace (Table S1).
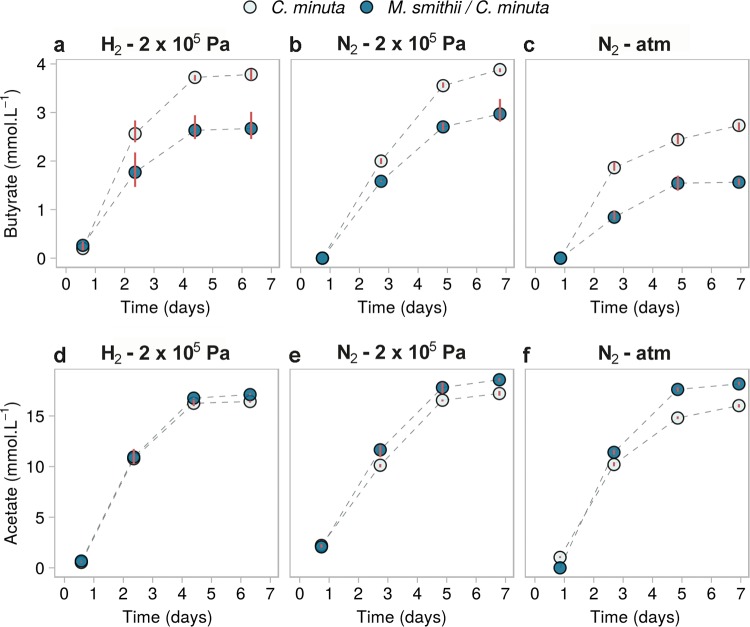
We consistently observed lower butyrate concentrations in all cocultures than in monocultures (Fig. 6a to c and 5c) (ANOVA, F value [1] = 161.461 and adjusted P value = 7.7 × 10−8). For all conditions, butyrate concentrations in coculture after 6 days were 1.1 ± 0.24 mmol · liter−1 lower than in monocultures (Fig. 6a to c and Table A3 in Text S1). The interaction factor between the mono/coculture conditions and the growth condition was not significantly correlated with butyrate concentrations (ANOVA, F value [2] = 0.862, adjusted P value = 0.4). The observation that butyrate concentrations in cocultures were lower than in monocultures regardless of pressure and headspace composition suggest that the methanogen’s presence shapes the metabolite output of C. minuta.
FIG 6.
SCFA concentrations over time in mono- and cocultures of C. minuta and M. smithii grown under different conditions and in cultures from batches 1 to 3 (Table S1). (a to c) Butyrate concentrations; (d to f) acetate concentrations. Only these SCFAs were detected among the fatty acids tested (fatty acids from C1 to C8, iso-valerate, and iso-butyrate). Points represent the averages of results from 3 biological replicates for each condition, and red bars join the minimal and maximal values. Monocultures of M. smithii are not shown, as they did not differ from the blanks (negative controls).
We observed, along with the reduced butyrate production, slightly but significantly higher acetate production in cocultures than in monocultures (Fig. 6d to f and 5d) (ANOVA, F value [1] = 317.41 and adjusted P value = 3.2 × 10−9). This difference was also observed in three additional batches performed at 2 × 105 Pa (Fig. S4). The differences in acetate production between mono- and coculture conditions significantly varied with the headspace and pressure conditions (the interaction term between the mono- or coculture and the growth condition was significantly correlated with acetate production; ANOVA, F value [2] = 29.09 and adjusted P value = 3.0 × 10−5). The differences in final acetate production (after 6 days) ranged from +0.7 mmol · liter−1 at 2 × 105 Pa under a H2-CO2 (80:20, vol/vol) atmosphere to +2.2 mmol · liter−1 at atmospheric pressure under a N2-CO2 (80:20, vol/vol) atmosphere. Furthermore, we observed in coculture more CH4 than what M. smithii could have produced based on the H2 production in C. minuta’s monocultures (see Appendix 3 in Text S1). This observation implies that C. minuta likely produced a greater amount of H2 in the cocultures, along with greater acetate production, than in monocultures.
Additional batches. H2, CH4, acetate, and butyrate concentrations in mono- and cocultures of M. smithii and C. minuta grown at 2 × 105 Pa, as described in the main text. The SCFAs of batch S1 were measured by gas chromatography instead of high-performance liquid chromatography. The points represent the averages of results from 2 to 3 biological cultures, and red bars join the minimal and maximal values. Download FIG S4, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. massiliensis and C. timonensis also support the metabolism of M. smithii.
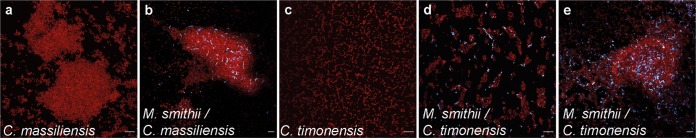
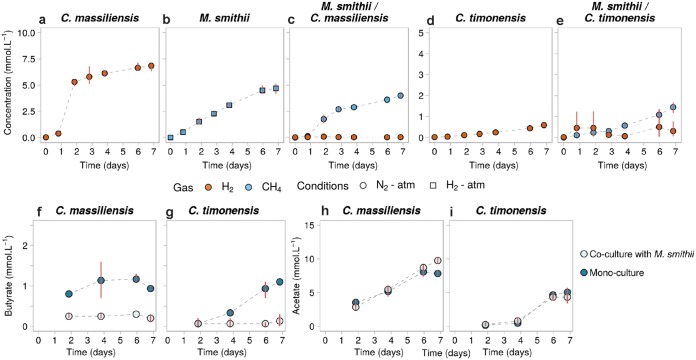
We performed similar coculture experiments of M. smithii with C. massiliensis and C. timonensis at atmospheric pressure. C. massiliensis and C. timonensis aggregated in monoculture, and M. smithii grew within its flocs in coculture (Fig. 7). The H2 produced by the bacteria in monoculture after 6 days of growth (6.9 ± 0.5 mmol · liter−1 for C. massiliensis and 0.6 ± 0.1 mmol · liter−1 for C. timonensis) (Fig. 8a and d) was lower than the levels produced by C. minuta (Fig. 4a). CH4 production in the cocultures reached 4.0 ± 0.2 mmol · liter−1 with C. massiliensis and 1.5 ± 0.3 mmol · liter−1 with C. timonensis. These amounts of methane are significantly lower than what we observed for M. smithii with C. minuta (6.6 ± 0.8 mmol · liter−1; ANOVA followed by a Tukey’s post hoc test, adjusted P values = 6.8 × 10−2 and 1.7 × 10−3 for cocultures, respectively, with C. massiliensis and C. timonensis against C. minuta) (Fig. 4c and 8c and e).
FIG 7.
Confocal imaging of C. massiliensis and C. timonensis in mono- and cocultures with M. smithii. Confocal micrographs after 5 days of growth of C. massiliensis (a), M. smithii and C. massiliensis in coculture (b), C. timonensis (c), and M. smithii and C. timonensis in coculture (d and e). SYBR green I fluorescence (DNA staining) is shown in red, and M. smithii’s coenzyme F420 autofluorescence is shown in blue. Scale bars represent 10 μm.
FIG 8.
Gas and SCFA concentrations in mono- and cocultures of C. massiliensis and C. timonensis with M. smithii. (a to e) H2 (orange) and CH4 (blue) concentrations in the headspace of cultures from batch 4 (Table S1); (f to i) butyrate (f and g) and acetate (h and i) concentrations in these cultures. Points represent the averages of results of 3 biological replicates, and red bars join the minimal and maximal values. (b) In the monocultures of M. smithii where H2 was provided in excess (condition, H2 atmosphere, with the headspace initially composed of 80:20, vol/vol, H2-CO2), its concentrations are not shown for scale reasons.
We observed less butyrate production in the cocultures than in the monocultures (Wilcoxon rank sum test, P values = 0.33 for C. massiliensis and 0.5 for C. timonensis) (Fig. 8f and g), with butyrate measured barely above the detection limit in cocultures. While in monocultures, C. massiliensis and C. timonensis produced 0.93 ± 0.06 mmol · liter−1 and 1.10 ± 0.00 mmol · liter−1 of butyrate, respectively; in coculture with M. smithii, they produced 0.20 ± 0.14 mmol · liter−1 and 0.13 ± 0.15 mmol · liter−1, respectively. Acetate production by C. massiliensis was higher in coculture than in monoculture (7.83 ± 0.49 mmol · liter−1 of acetate produced in monoculture by day 6 and 9.75 ± 0.78 mmol · liter−1 produced in coculture with M. smithii), although this difference was not significant (Wilcoxon rank sum test, P value = 0.2). And in contrast with the cocultures of C. minuta with M. smithii, acetate production by C. timonensis was not higher in the cocultures than in monocultures: C. timonensis produced 5.05 ± 0.21 mmol · liter−1 in monoculture and 4.33 ± 1.21 mmol · liter−1 in coculture (Wilcoxon rank sum test, P value = 0.8) (Fig. 8h and i).
DISCUSSION
The link between the relative abundance of the Christensenellaceae and host BMI now stands as one of the most reproducible associations described between the gut microbiome and obesity (4–15). Here, we confirm in a meta-analysis of metagenomes across 10 populations the previously observed association between leanness and the Christensenellaceae family (4, 20–22). We could also show that the Christensenella genus and Christensenella spp. also correlated with leanness. Similarly, we observed correlations between leanness and the Methanobacteriaceae family, the Methanobrevibacter genus, and M. smithii. These methanogens were positively correlated with members of the Christensenellaceae family. The relative abundances of the Christensenellaceae were higher in young people, whereas conversely, Methanobacteriaceae were enriched in older people. Despite these opposite patterns, the two families correlate with each other regardless of age and BMI.
We selected the two most prominent members of the two families, C. minuta and M. smithii, to ask if physical and metabolic interactions may underlie these positive associations. C. minuta produced copious amounts of H2 during fermentation. In coculture with C. minuta, M. smithii produced amounts of CH4 comparable to those in monoculture with an excess of H2, indicating that C. minuta can efficiently support the growth of M. smithii via interspecies H2 transfer. C. minuta formed flocs visible by eye, and M. smithii grew within these flocs.
M. smithii would likely benefit by associating with the flocs formed by C. minuta through better access to H2. Interspecies metabolite transfer corresponds to the diffusion of a metabolite (e.g., H2) from the producer (e.g., C. minuta) to the consumer (e.g., M. smithii). As described by Fick’s law of diffusion, the flux of a metabolite between two microorganisms is directly proportional to the concentration gradient and inversely proportional to the distance, such that the closer the microorganisms are, the better the H2 transfer (35, 36). Thus, within the flocs, the H2 interspecies transfer would be more efficient, to the benefit of M. smithii. In accord, we observed greater methane production under excess H2 when C. minuta was present.
When grown in coculture, M. smithii influenced the metabolism of C. minuta. The presence of the methanogen inhibited the production of butyrate while enhancing acetate production by C. minuta under all growth conditions, on average among all experimental batches. This observation suggests that H2 consumption by M. smithii decreased the PH2 within the floc enough to favor acetate production (37). The consumption of H2 causes the cell to produce more oxidized fermentation products, such as acetate (38–41), and the interspecies H2 transfer leads to greater CH4 production.
Both the methane production and the coflocculation were far more pronounced when M. smithii was grown with C. minuta than with B. thetaiotaomicron. B. thetaiotaomicron has previously been shown to support the growth of M. smithii in coculture (25, 26). B. thetaiotaomicron barely aggregated, in contrast to C. minuta’s very large (visible to the naked eye) flocs. When grown together, B. thetaiotaomicron and M. smithii showed very poor aggregation. Moreover, acetate was the only SCFA detected in monocultures of B. thetaiotaomicron, and its production was less affected by the methanogen than in C. minuta. Methane produced by M. smithii in coculture with B. thetaiotaomicron was one-fifth of that produced with C. minuta, possibly as a result of the smaller amount of H2 produced and the reduced contact between cells. Given that M. smithii does not cooccur with B. thetaiotaomicron in human microbiome data sets, this is another indication that cooccurrence patterns may point to metabolic interactions.
C. massiliensis and C. timonensis also produced H2, acetate, and butyrate and also flocculated in monoculture. C. massiliensis and C. timonensis supported methane production by M. smithii, which grew within the bacterial flocs. However, M. smithii also grew outside the flocs when cocultured with these two species, which we did not observe in the cocultures with C. minuta, and although M. smithii also influenced the fermentation of C. massiliensis and C. timonensis, the overall changes in SCFA production in coculture were different from what we observed with C. minuta: butyrate production was almost undetectable, while acetate production was not significantly affected.
These results suggest that the interaction between M. smithii and C. minuta leads to higher methane production than with B. thetaiotaomicron and other species of the Christensenellaceae, possibly due to the higher levels of interspecies H2 transfer. Nevertheless, C. massiliensis and C. timonensis supported CH4 production better than B. thetaiotaomicron did. The higher H2 production of C. massiliensis than of B. thetaiotaomicron might explain this. In the case of C. timonensis, although it produced half the H2 produced by B. thetaiotaomicron in monoculture, M. smithii produced more CH4 in coculture with C. timonensis than with B. thetaiotaomicron. This suggests that, as with its effect on C. minuta, M. smithii triggered the production of H2 by C. timonensis.
Altogether, our work demonstrates that members of the Christensenellaceae act as a H2 source to methanogens, and this process is enhanced via close physical proximity. Such interactions also likely underlie the cooccurrence patterns of the Christensenellaceae with other members of the microbiome. Many of these families lack cultured representatives, such as Firmicutes unclassified SHA-98, Tenericutes unclassified RF39, and unclassified ML615J-28 (4). Based on our results, cultivation of these elusive members of the microbiome may require H2 (or the provision of another metabolite that C. minuta produces when H2 is being consumed). Despite their very low abundance in the human gut, members of the Christensenellaceae may shape the composition of the gut microbiome by favoring the colonization and persistence of certain hydrogenotrophs and by supplying other butyrate producers with acetate (42).
Here, we confirmed an association of M. smithii and leanness based on metagenomes from 10 studies. In contrast, some studies have reported an association between M. smithii and obesity (2, 43). In this scenario, H2 uptake by M. smithii would promote the breakdown of nondigestible carbon sources by fermenters, such as acetogens, thereby increasing the amount of acetate or other SCFAs that can be absorbed and utilized by the host and promoting fat storage (2, 44). In contrast, and consistently with our results, M. smithii has also been repeatedly associated with anorexia and leanness (4, 45–48). In this case, the production of CH4 would decrease the amount of energy available for the host via carbon loss, as has been observed in livestock (49–52). Thus, our observation that the presence of M. smithii directs the metabolic output of the C. minuta toward greater H2 availability for methanogenesis via increased acetate production is consistent with their association with a lean phenotype. To assess quantitatively how the presence and activity of these microbes impact host physiology will require careful modeling of energy flow in vivo.
MATERIALS AND METHODS
Metagenome data generation.
We generated 141 metagenomes from fecal samples obtained as part of a previous study (53) (see Table S2 in the supplemental material). Metagenomic libraries were prepared as described in Appendix 1 (additional methods) in Text S1.
Samples from the Poole et al. (53) study. Additional data were generated from time points that had not been sequenced for the Poole et al. study. For each individual and each time point, the color indicates whether the sample was prepared and sequenced as described previously (orange) or as described in Materials and Methods (blue). If the sample failed sequencing or was otherwise missing, the color is white. Download Table S2, PDF file, 0.04 MB (46.3KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data from public databases.
We constructed a metagenome sequence collection from (i) the newly generated data (above) to complement the 146 metagenomes previously reported by Poole et al. in 2019 (53) and (ii) publicly available shotgun-metagenome sequences from stool samples included in the curatedMetagenomicData package of Bioconductor (54) for which BMI information was provided. For the latter, we restricted our analyses to individuals for which the following information was available: gender, age, country of origin, and BMI. Individuals with Schistosoma (n = 4) or Wilson's disease (n = 2) were excluded from the analysis, as were samples from two pregnant women. In all, 1,534 samples from 9 studies were downloaded from the sequence read archive (SRA) and further processed (Table S3), for a total or 1,821 samples with at least 1 million sequence pairs per sample.
Data processing.
A detailed description of the processing of the raw sequences is given in Appendix 1 in Text S1. To obtain a taxonomic profile of the metagenome samples, we built a custom genome database (55) for Kraken v2.0.7 (56) and Bracken v2.2 (57) using the representative genomes from the proGenomes database (as available on 24 August 2018) (58), to which we added genome sequences of C. minuta (GenBank assembly accession number GCA_001652705.1), C. massiliensis (GCA_900155415.1), and C. timonensis (GCA_900087015.1). Reads were classified using Kraken2, and a Bayesian reestimation of the species-level abundance of each sample was then performed using Bracken2. We obtained complete taxonomic annotations from NCBI taxon IDs with TaxonKit v0.2.4 (https://bioinf.shenwei.me/taxonkit/). The detection limit for the relative abundances in samples was 10−3%; in consequence, all relative abundances below this threshold were equal to 0.
Meta-analysis of human gut metagenomes.
Linear mixed models (R package nlme) were used to evaluate the correlation between the relative abundances of taxa while correcting for the structure of the population; the study of origin was set as a random effect. In some data sets, individuals were sampled multiple times, in which case the individual effect was nested inside the data set effect. Relative abundances were transformed using Tukey's ladder of power transformation (59) and are designated with the suffix “-tra” (e.g., the transformed relative abundance of the family Christensenellaceae is Cf-tra). Covariates in null models were selected using a backward feature selection approach based on a type II ANOVA (i.e., by including all covariates and removing the nonsignificant ones step-by-step until all remaining variables were significant [see Appendix 2 in Text S1). We made 4 null models predicting the transformed relative abundances of members of the family Christensenellaceae (Cf-null), the genus Christensenella (Cg-null), the family Methanobacteriaceae (Mf-null), and the genus Methanobrevibacter (Mg-null). To evaluate the correlation between taxa, we made model Cf-Mf by adding Mf-tra and its interaction with age to the covariates of Cf-null. Reciprocally, we made model Cg-Mg by adding Mg-tra and its interaction with age to the covariates of Cg-null. The same approach was performed at the species level, and it is described in Appendix 2 in Text S1.
We used the likelihood ratio test to compare the nested models via the χ2 distribution (i.e., Cf-null versus Cf-Mf and Cg-null versus Cg-Mg). To characterize the correlation of Cf-tra with Mf-tra and Cg-tra with Mg-tra, after correcting for BMI and age, we used a type I ANOVA to evaluate the importance of the variables in the order in which they appear in Cf-Mf and Cg-Mg. The F value, degree of freedom, and P value are reported for each variable. All analyses were performed using R (60).
Culturing of methanogens and bacteria.
We obtained M. smithii DSM-861, C. minuta DSM-22607, C. massiliensis DSM 102344, C. timonensis DSM 102800, and B. thetaiotaomicron VPI-5482 from the German Collection of Microorganisms and Cell Cultures (DSMZ; Braunschweig, Germany). Each culture was thawed and inoculated into brain heart infusion (BHI) medium (Carl Roth, Karlsruhe, Germany) supplemented with yeast extract (5 g/liter), reduced with l-cysteine-HCl (0.5 g/liter) and Ti-NTA III (0.3 mM), and buffered with sodium bicarbonate (42 mM, pH 7, adjusted with HCl 6 M). Cultures (10 ml) were grown at 37°C without shaking in Balch tubes (total volume of 28 ml) under a headspace of N2-CO2 (80:20, vol/vol) in the case of the bacteria and H2-CO2 (80:20, vol/vol, with pressure adjusted to 2 × 105 Pa) for M. smithii. When initial cultures reached exponential growth and before floc formation, they were transferred into fresh medium, and these transfers were used as inocula for the experiments described below.
Coculture conditions.
M. smithii was cocultured with C. minuta, B. thetaiotaomicron, C. massiliensis, or C. timonensis, and in parallel, each microorganism was grown in monoculture (Table S1). Prior to inoculation, 1-day-old cultures of bacterial species or 4-day-old cultures of M. smithii were adjusted to an optical density at 600 nm (OD600) of 0.01 with sterile medium. For the cocultures, 0.5 ml of each adjusted culture was inoculated into 9 ml of fresh medium. For the monocultures, 0.5 ml of the adjusted culture and 0.5 ml of sterile medium were combined as an inoculum. For negative controls, sterile medium was transferred as a mock inoculum. Headspaces were exchanged with 80:20 (vol/vol) N2-CO2 or H2-CO2 and pressurized at 2 × 105 Pa or atmospheric pressure (Table S1). Each batch of experiments was carried out once with 3 biological replicates per culture condition (Table S1).
Imaging.
For confocal microscopy, SYBR green I staining was performed as previously described (61) with the modifications described in Appendix 1 in Text S1. Imaging by confocal microscopy (LSM 780 NLO; Zeiss) was used to detect the autofluorescence emission of coenzyme F420 of M. smithii and the emission of SYBR green I (Appendix 1 in Text S1). Images were acquired with the ZEN Black 2.3 SP1 software and processed with FIJI (62). Micrographs are representative of all replicate cultures within each experimental batch. The preparation of the samples for scanning electron microscopy is described in Appendix 1 in Text S1. Cells were examined with a field emission scanning electron microscope (Regulus 8230; Hitachi High Technologies, Tokyo, Japan) at an accelerating voltage of 10 kV.
Gas and SCFA measurements.
Headspace concentrations of H2, CO2, and CH4 were measured with a gas chromatograph (GC) (SRI 8610C; SRI Instruments, Torrance, USA) equipped with a packed column at 42°C (0.3-m HaySep-D packed Teflon; Restek, Bellefonte, PA, USA), a thermal conductivity detector (TCD) at 111°C, and a flame ionization detector (FID). The gas production and consumption were estimated from the total pressure in the vials (ECO2 manometer; Keller, Jestetten, Germany) and the gas concentrations in the headspace using the ideal gas equation. The concentrations are given in millimoles of gas in the headspace per liter of culture.
SCFA measurements were performed with liquid samples (0.5 ml) filtered through 0.2-μm-pore-size polyvinylidene fluoride filters (Carl Roth, GmbH, Karlsruhe, Germany). SCFA concentrations were measured with a CBM-20A high-performance liquid chromatography (HPLC) system equipped with an Aminex HPX-87P column (300 by 7.8 mm; Bio-Rad, CA, USA), maintained at 60°C, and a refractive index detector. A sulfuric acid solution (5 mM) was used as the eluent at a flow rate of 0.6 ml/min (∼40 × 105-Pa column pressure). Calibration curves for acetate and butyrate were prepared from 1.25 to 50 mM using acetic acid and butyric acid, respectively (Merck KGaA, Darmstadt, Germany). No other fatty acids were detected (see Appendix 1 in Text S1 [63–69]). The SCFA concentrations were estimated with the Shimadzu LabSolutions software.
Statistical analyses.
We used Wilcoxon rank sum tests to compare levels of gas production between cultures after 6 days of growth. We performed ANOVA tests when more than one culture condition (i.e., headspace composition and pressure) (Table S1) was included in the comparison. The conditions in the ANOVA tests (i.e., headspace composition and pressure in mono- or cocultures) were evaluated to explain the variance of CH4 production after 6 days of growth. A Tukey post hoc test was then performed to discriminate between the effects of the different conditions. SCFA concentrations were compared using a two-way ANOVA where the culture conditions (i.e., headspace composition and pressure) (Table S1) and the sample (mono- and cocultures) were evaluated to explain the variance of butyrate and acetate concentrations after 6 days of growth. P values were adjusted using the Benjamini-Hochberg method. Tukey’s post hoc test was performed to discriminate between the effects of the different conditions. All statistical analyses were done in R using the stats R package.
Data availability.
The metagenomic sequence data generated during this study have been deposited in the European Nucleotide Archive under accession number PRJEB34191 (http://www.ebi.ac.uk/ena/data/view/PRJEB34191). The jupyter notebooks and associated data are available at https://github.com/Albabune/Ruaud_EsquivelElizondo.
ACKNOWLEDGMENTS
We are grateful to Monika Temovska, Sophie Maisch, and Iris Holdermann for valuable help. We also thank Daren Heavens for the metagenome library preparation protocol.
This work was supported by the Max Planck Society and the Humboldt Foundation.
Footnotes
This article is a direct contribution from Ruth E. Ley, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Rob Knight, UCSD School of Medicine, and Daniel Buckley, Cornell University.
Citation Ruaud A, Esquivel-Elizondo S, de la Cuesta-Zuluaga J, Waters JL, Angenent LT, Youngblut ND, Ley RE. 2020. Syntrophy via interspecies H2 transfer between Christensenella and Methanobrevibacter underlies their global cooccurrence in the human gut. mBio 11:e03235-19. https://doi.org/10.1128/mBio.03235-19.
REFERENCES
- 1.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Waters JL, Ley RE. 2019. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol 17:83. doi: 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JAM, Brandsma E, Marczynska J, Imhann F, Weersma RK, Franke L, Poon TW, Xavier RJ, Gevers D, Hofker MH, Wijmenga C, Zhernakova A. 2015. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res 117:817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. 2016. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, Trøseid M, Marschall H-U, Schrumpf E, Moum B, Røsjø H, Aukrust P, Karlsen TH, Hov JR. 2017. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 66:611–619. doi: 10.1136/gutjnl-2015-310500. [DOI] [PubMed] [Google Scholar]
- 8.Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, Lee S, Song Y-M, Lee K, Sung J, Ko G. 2017. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66:1031–1038. doi: 10.1136/gutjnl-2015-311326. [DOI] [PubMed] [Google Scholar]
- 9.Oki K, Toyama M, Banno T, Chonan O, Benno Y, Watanabe K. 2016. Comprehensive analysis of the fecal microbiota of healthy Japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol 16:284. doi: 10.1186/s12866-016-0898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanislawski MA, Dabelea D, Wagner BD, Sontag MK, Lozupone CA, Eggesbø M. 2017. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome 5:113. doi: 10.1186/s40168-017-0332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun Y, Kim H-N, Kim SE, Heo SG, Chang Y, Ryu S, Shin H, Kim H-L. 2017. Comparative analysis of gut microbiota associated with body mass index in a large Korean cohort. BMC Microbiol 17:151. doi: 10.1186/s12866-017-1052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks AW, Priya S, Blekhman R, Bordenstein SR. 2018. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 16:e2006842. doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson MA, Bonder MJ, Kuncheva Z, Zierer J, Fu J, Kurilshikov A, Wijmenga C, Zhernakova A, Bell JT, Spector TD, Steves CJ. 2018. Detection of stable community structures within gut microbiota co-occurrence networks from different human populations. PeerJ 6:e4303. doi: 10.7717/peerj.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-Contreras BE, Morán-Ramos S, Villarruel-Vázquez R, Macías-Kauffer L, Villamil-Ramírez H, León-Mimila P, Vega-Badillo J, Sánchez-Muñoz F, Llanos-Moreno LE, Canizalez-Román A, Del Río-Navarro B, Ibarra-González I, Vela-Amieva M, Villarreal-Molina T, Ochoa-Leyva A, Aguilar-Salinas CA, Canizales-Quinteros S. 2018. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr Obes 13:381–388. doi: 10.1111/ijpo.12262. [DOI] [PubMed] [Google Scholar]
- 15.Peters BA, Shapiro JA, Church TR, Miller G, Trinh-Shevrin C, Yuen E, Friedlander C, Hayes RB, Ahn J. 2018. A taxonomic signature of obesity in a large study of American adults. Sci Rep 8:9749. doi: 10.1038/s41598-018-28126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morotomi M, Nagai F, Watanabe Y. 2012. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int J Syst Evol Microbiol 62:144–149. doi: 10.1099/ijs.0.026989-0. [DOI] [PubMed] [Google Scholar]
- 17.Costea PI, Hildebrand F, Arumugam M, Bäckhed F, Blaser MJ, Bushman FD, de Vos WM, Ehrlich SD, Fraser CM, Hattori M, Huttenhower C, Jeffery IB, Knights D, Lewis JD, Ley RE, Ochman H, O'Toole PW, Quince C, Relman DA, Shanahan F, Sunagawa S, Wang J, Weinstock GM, Wu GD, Zeller G, Zhao L, Raes J, Knight R, Bork P. 2018. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol 3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, Guttman DS, Griffiths A, Panaccione R, Otley A, Xu L, Shestopaloff K, Moreno-Hagelsieb G, GEM Project Research Consortium, Paterson AD, Croitoru K. 2016. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 19.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AAM, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. 2016. The effect of host genetics on the gut microbiome. Nat Genet 48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 20.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Upadhyaya B, McCormack L, Fardin-Kia AR, Juenemann R, Nichenametla S, Clapper J, Specker B, Dey M. 2016. Impact of dietary resistant starch type 4 on human gut microbiota and immunometabolic functions. Sci Rep 6:28797. doi: 10.1038/srep28797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimenko NS, Tyakht AV, Popenko AS, Vasiliev AS, Altukhov IA, Ischenko DS, Shashkova TI, Efimova DA, Nikogosov DA, Osipenko DA, Musienko SV, Selezneva KS, Baranova A, Kurilshikov AM, Toshchakov SM, Korzhenkov AA, Samarov NI, Shevchenko MA, Tepliuk AV, Alexeev DG. 2018. Microbiome responses to an uncontrolled short-term diet intervention in the frame of the citizen science project. Nutrients 10:576. doi: 10.3390/nu10050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanderhaeghen S, Lacroix C, Schwab C. 2015. Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol Lett 362:fnv092. doi: 10.1093/femsle/fnv092. [DOI] [PubMed] [Google Scholar]
- 24.Moore WEC, Johnson JL, Holdeman LV. 1976. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int J Syst Evol Microbiol 26:238–252. doi: 10.1099/00207713-26-2-238. [DOI] [Google Scholar]
- 25.Traore SI, Khelaifia S, Armstrong N, Lagier JC, Raoult D. 2019. Isolation and culture of Methanobrevibacter smithii by coculture with hydrogen-producing bacteria on agar plates. Clin Microbiol Infect 25:1561.e1–1561.e5. doi: 10.1016/j.cmi.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Khelaifia S, Lagier J-C, Nkamga VD, Guilhot E, Drancourt M, Raoult D. 2016. Aerobic culture of methanogenic archaea without an external source of hydrogen. Eur J Clin Microbiol Infect Dis 35:985–991. doi: 10.1007/s10096-016-2627-7. [DOI] [PubMed] [Google Scholar]
- 27.Nkamga VD, Lotte R, Roger P-M, Drancourt M, Ruimy R. 2016. Methanobrevibacter smithii and Bacteroides thetaiotaomicron cultivated from a chronic paravertebral muscle abscess. Clin Microbiol Infect 22:1008–1009. doi: 10.1016/j.cmi.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Lau SKP, McNabb A, Woo GKS, Hoang L, Fung AMY, Chung LMW, Woo PCY, Yuen K-Y. 2007. Catabacter hongkongensis gen. nov., sp. nov., isolated from blood cultures of patients from Hong Kong and Canada. J Clin Microbiol 45:395–401. doi: 10.1128/JCM.01831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosa BA, Hallsworth-Pepin K, Martin J, Wollam A, Mitreva M. 2017. Genome sequence of Christensenella minuta DSM 22607T. Genome Announc 5:e01451-16. doi: 10.1128/genomeA.01451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D, Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D. 2018. Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dridi B, Raoult D, Drancourt M. 2011. Archaea as emerging organisms in complex human microbiomes. Anaerobe 17:56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Edwards T, McBride BC. 1975. Biosynthesis and degradation of methylmercury in human faeces. Nature 253:463–464. doi: 10.1038/253462a0. [DOI] [PubMed] [Google Scholar]
- 34.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 36.Shen L, Zhao Q, Wu X, Li X, Li Q, Wang Y. 2016. Interspecies electron transfer in syntrophic methanogenic consortia: from cultures to bioreactors. Renew Sustain Energy Rev 54:1358–1367. doi: 10.1016/j.rser.2015.10.102. [DOI] [Google Scholar]
- 37.Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R. 2004. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol 22:477–485. doi: 10.1016/j.tibtech.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Macfarlane S, Macfarlane GT. 2003. Regulation of short-chain fatty acid production. Proc Nutr Soc 62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Whitman WB. 2008. Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea. Ann N Y Acad Sci 1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 40.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 42.Hoyles L, Swann J. 2019. Influence of the human gut microbiome on the metabolic phenotype, p 535–560. In Lindon JC, Nicholson JK, Holmes E (ed), The handbook of metabolic phenotyping. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 43.Mbakwa CA, Penders J, Savelkoul PH, Thijs C, Dagnelie PC, Mommers M, Arts I. 2015. Gut colonization with Methanobrevibacter smithii is associated with childhood weight development. Obesity (Silver Spring) 23:2508–2516. doi: 10.1002/oby.21266. [DOI] [PubMed] [Google Scholar]
- 44.Morrison DJ, Preston T. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mack I, Cuntz U, Grämer C, Niedermaier S, Pohl C, Schwiertz A, Zimmermann K, Zipfel S, Enck P, Penders J. 2016. Weight gain in anorexia nervosa does not ameliorate the faecal microbiota, branched chain fatty acid profiles, and gastrointestinal complaints. Sci Rep 6:26752. doi: 10.1038/srep26752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. 2009. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients. PLoS One 4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. 2012. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond) 36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 49.Blaxter KL, Clapperton JL. 1965. Prediction of the amount of methane produced by ruminants. Br J Nutr 19:511–522. doi: 10.1079/bjn19650046. [DOI] [PubMed] [Google Scholar]
- 50.Crutzen PJ, Aselmann I, Seiler W. 1986. Methane production by domestic animals, wild ruminants, other herbivorous fauna, and humans. Tellus B Chem Phys Meteorol 38B:271–284. doi: 10.1111/j.1600-0889.1986.tb00193.x. [DOI] [Google Scholar]
- 51.Callaway TR, Edrington TS, Rychlik JL, Genovese KJ, Poole TL, Jung YS, Bischoff KM, Anderson RC, Nisbet DJ. 2003. Ionophores: their use as ruminant growth promotants and impact on food safety. Curr Issues Intest Microbiol 4:43–51. [PubMed] [Google Scholar]
- 52.Kruger Ben Shabat S, Sasson G, Doron-Faigenboim A, Durman T, Yaacoby S, Berg Miller ME, White BA, Shterzer N, Mizrahi I. 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J 10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poole AC, Goodrich JK, Youngblut ND, Luque GG, Ruaud A, Sutter JL, Waters JL, Shi Q, El-Hadidi M, Johnson LM, Bar HY, Huson DH, Booth JG, Ley RE. 2019. Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 25:553–564.e7. doi: 10.1016/j.chom.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, Beghini F, Malik F, Ramos M, Dowd JB, Huttenhower C, Morgan M, Segata N, Waldron L. 2017. Accessible, curated metagenomic data through ExperimentHub. Nat Methods 14:1023–1024. doi: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de la Cuesta-Zuluaga J, Ley RE, Youngblut ND. 2019. Struo: a pipeline for building custom databases for common metagenome profilers. Bioinformatics 2019:btz899. doi: 10.1093/bioinformatics/btz899. [DOI] [PubMed] [Google Scholar]
- 56.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Sci 3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 58.Mende DR, Letunic I, Huerta-Cepas J, Li SS, Forslund K, Sunagawa S, Bork P. 2017. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res 45:D529–D534. doi: 10.1093/nar/gkw989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mangiafico SS. 2016. Summary and analysis of extension program evaluation in R, version 1.15.0. Rutgers Cooperative Extension, New Brunswick, NJ: https://rcompanion.org/handbook/. [Google Scholar]
- 60.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 61.Lambrecht J, Cichocki N, Hübschmann T, Koch C, Harms H, Müller S. 2017. Flow cytometric quantification, sorting and sequencing of methanogenic archaea based on F420 autofluorescence. Microb Cell Fact 16:180. doi: 10.1186/s12934-017-0793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karasov TL, Almario J, Friedemann C, Ding W, Giolai M, Heavens D, Kersten S, Lundberg DS, Neumann M, Regalado J, Neher RA, Kemen E, Weigel D. 2018. Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24:168–179.e4. doi: 10.1016/j.chom.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Droop AP. 2016. fqtools: an efficient software suite for modern FASTQ file manipulation. Bioinformatics 32:1883–1884. doi: 10.1093/bioinformatics/btw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang H, Lei R, Ding S-W, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Betancur JC, Yepes A, Schneider J, Lopez D. 2012. Single-cell analysis of Bacillus subtilis biofilms using fluorescence microscopy and flow cytometry. J Vis Exp 2012:3796. doi: 10.3791/3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. 2009. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abundances of the Methanobacteriaceae and Christensenellaceae families across studies. (a to j) Transformed relative abundances of Christensenellaceae (Cf-tra) and Methanobacteriaceae (Mf-tra) across 1,821 samples from 10 countries and generated from 10 independent studies. The data generated for this study are grouped with the first time series published in the work of Poole et al. (53). The gap between 0 and ∼0.2 is due to the detection limit of the sequencing method; the minimal relative abundance is 10−3%. Hence, 0 indicates that the microorganism was not detected, which introduces a gap after transformation of the data. Download FIG S1, PDF file, 2.6 MB (2.6MB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental appendices. Download Text S1, DOCX file, 0.01 MB (15.8KB, docx) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data sets used for the statistical analysis. The BMI and age values for each data set are reported as average values, with minimum and maximum values in parentheses. Download Table S3, PDF file, 0.1 MB (118.1KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confocal imaging of cocultures of B. thetaiotaomicron and M. smithii at different time points. (a and b) Cells at day 2, when B. thetaiotaomicron enters stationary phase (Fig. 4d); (c and d) cells at day 7, the end of the experiment, when maximal CH4 concentrations were observed both in monocultures of M. smithii and in cocultures with B. thetaiotaomicron (Fig. 4b and e). In exponential phase, B. thetaiotaomicron cells are rod shaped (a), while during stationary phase, they suffer stress, leading to elongated cells (c). The bright fields (a and c) and M. smithii’s coenzyme F420 (b and d) channels are displayed. Scale bars represent 10 μm. Download FIG S2, JPG file, 0.1 MB (123.2KB, jpg) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Total pressure, headspace composition, and culture inocula for each batch of experiments described in the main text. Download Table S1, PDF file, 0.02 MB (16.2KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. minuta and M. smithii aggregate at atmospheric pressure and even when there is excess H2 in the medium. Confocal imaging of C. minuta and M. smithii at 3 days of growth. (a and b) Coculture grown at atmospheric pressure; (c and d) coculture grown under a pressurized H2-CO2 atmosphere. The bright fields (a and c) and M. smithii‘s coenzyme F420 (b and d) channels are displayed. Scale bars represent 10 μm. Download FIG S3, JPG file, 0.2 MB (246KB, jpg) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional batches. H2, CH4, acetate, and butyrate concentrations in mono- and cocultures of M. smithii and C. minuta grown at 2 × 105 Pa, as described in the main text. The SCFAs of batch S1 were measured by gas chromatography instead of high-performance liquid chromatography. The points represent the averages of results from 2 to 3 biological cultures, and red bars join the minimal and maximal values. Download FIG S4, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Samples from the Poole et al. (53) study. Additional data were generated from time points that had not been sequenced for the Poole et al. study. For each individual and each time point, the color indicates whether the sample was prepared and sequenced as described previously (orange) or as described in Materials and Methods (blue). If the sample failed sequencing or was otherwise missing, the color is white. Download Table S2, PDF file, 0.04 MB (46.3KB, pdf) .
Copyright © 2020 Ruaud et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The metagenomic sequence data generated during this study have been deposited in the European Nucleotide Archive under accession number PRJEB34191 (http://www.ebi.ac.uk/ena/data/view/PRJEB34191). The jupyter notebooks and associated data are available at https://github.com/Albabune/Ruaud_EsquivelElizondo.