Abstract
Nuclear receptors, such as liver X receptors (LXRs) and sterol regulatory element-binding proteins (SREBPs), are key regulators of lipogenic genes, including fatty acid synthase (FASN). It has been reported that several oxycholesterols (OCs) act as LXR ligands; however, it is unclear whether all OC molecular species act as ligands. We previously demonstrated that the absorption rate of dietary 6-ketocholestanol (6-keto), an oxycholesterol, is the highest of all the OCs using thoracic lymph duct-cannulated rats. However, limited information is available about the physiological significance of 6-keto. In this study, we investigated whether treatment with 6-keto increases intracellular triacylglycerol (TAG) levels through up-regulation of lipogenic gene expression in HepG2 cells. 6-Keto treatment significantly reduced intracellular TAG levels through down-regulation of lipogenic genes including FASN. Although 6-keto significantly suppressed FASN gene promoter activities, the action was completely diminished when mutations were present in the SREBP promoter site. TO901317 (TO) significantly increased FASN gene promoter activities, whereas simultaneous treatment with TO and 6-keto significantly reduced this activity. We also compared the effects of several OCs that are oxidized at the carbon-6 and -7 in the B-ring of cholesterol on FASN gene promoter activities. Similar to 6-keto, 6α-OH and 6β-OH significantly reduced the activity of the FASN gene promoter, which suggests that oxidation of carbon-6 in the B-ring may play an important role in the reduction of FASN expression. Our results indicate that 6-keto suppresses lipid accumulation by decreasing FASN gene expression through SREBP-dependent regulation in HepG2 cells.
Electronic supplementary material
The online version of this article (10.1007/s10616-019-00368-5) contains supplementary material, which is available to authorized users.
Keywords: 6-Ketocholestanol, Fatty acid synthase, SREBP, Lipid accumulation, HepG2 cell
Introduction
Fatty acid synthase (FASN), a central enzyme in the de novo lipogenesis pathway, catalyzes all the steps in the conversion of malonyl-CoA to palmitate (Sul and Wang 1998). Expression of the FASN gene is regulated at the transcription level and is responsive to both hormonal and nutritional signals (Sul and Wang 1998). Among the sterol regulatory element-binding proteins (SREBPs), SREBP-1c plays a critical role in the transcriptional regulation of lipogenic genes, including both FASN and stearoyl-CoA desaturase (SCD) (Brown and Goldstein 1997). It has been shown that SREBPs regulate FASN expression through direct interaction with the FASN promoter (Magaña and Osborne 1996; Latasa et al. 2000). The nuclear receptor liver X receptor alpha (LXRα), which is predominantly expressed in the central organs of lipid metabolism such as the liver and white adipose tissue, binds to DNA and regulates transcription of target genes, including FASN, in a heterodimeric complex with retinoid X receptor (RXR) (Willy et al. 1995). Most nuclear receptors are activated by endogenous lipophilic ligands, while LXRα is activated by several cholesterol oxidation products (oxycholesterols; OCs) (Janowski et al. 1996; Schroepfer 2000).
OCs are produced from cholesterol by spontaneous and/or enzymatic oxidation of the steroidal backbone and side chains. It has been previously suggested that OCs are associated with the development and progression of several diseases, such as osteoporosis, atherosclerosis, Alzheimer’s disease, cataracts, diabetes, dyslipidemia, and fatty liver (Sottero et al. 2009; Vejux and Lizard 2009; Sato and Shirouchi 2012; Zarrouk et al. 2014; Kulig et al. 2016). OCs have been found in several foods, especially those derived from animal sources (van de Bovenkamp et al. 1988; Paniangvait et al. 1995; Ichi et al. 2005). Although circulating OC levels increase with an increase in the intake of OCs (Emanuel et al. 1991), the absorption rates (5–25%) of these OCs vary depending on the oxidized position of the cholesterol (Shirouchi et al. 2019). Carbons-5, -6, and -7 in the B-ring of cholesterol are easily oxidized (Smith et al. 1978). The absorption rates of α- and β-cholestanetriols, with two hydroxyl groups at the carbon-5 and -6 positions of cholesterol, are less than 10% (Shirouchi et al. 2019). The absorption rate of 6-ketocholestanol (6-keto) (~ 25%), having a keto group at the carbon-6 position, was reported to be the highest of 10 different dietary OCs produced by heat (Shirouchi et al. 2019). Furthermore, 6-keto inhibits the effect of uncouplers of oxidative phosphorylation in mitochondria (Starkov et al. 1994, 1997; Cuéllar et al. 1997), suggesting that it can prevent, or reverse, decreased ATP production caused by mitochondrial dysfunction. However, the physiological significance of 6-keto, except for the above-mentioned, remains unclear.
To gain insight into the physiological significance of 6-keto, we investigated whether treatment with 6-keto, similar to several OCs previously reported as LXR ligands, increases intracellular triacylglycerol (TAG) levels through lipogenic gene expression in HepG2 cells.
Materials and methods
Materials
5α-Cholestan-3β,6α-diol (6α-hydroxycholestanol, 6α-OH), 5α-cholestan-3β,6β-diol (6β-hydroxycholestanol, 6β-OH), 5-cholesten-3β,7α-diol (7α-hydroxycholesterol, 7α-OH), 5-cholesten-3β,7β-diol (7β-hydroxycholesterol, 7β-OH), 5α-cholestan-3β-ol-6-one (6-ketocholestanol, 6-keto) were purchased from Steraloids Inc. (Newport, RI, USA) (Fig. 1). TO901317 (TO) was purchased from Cayman Chemical Company (East Ellsworth Road Ann Arbor, MI, USA). Dulbecco’s modified Eagle’s media (DMEM) with high glucose (4500 mg/L) or with low glucose (1000 mg/L) were purchased from Gibco-BRL (Grand Island, NY, USA). Fetal bovine serum (FBS) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Penicillin and streptomycin were purchased from Meiji Seika Pharma Co., Ltd. (Tokyo, Japan). Lipoprotein deficient FBS (LPDS) was prepared from FBS by ultracentrifugation as described previously (Kinoshita et al. 2000).
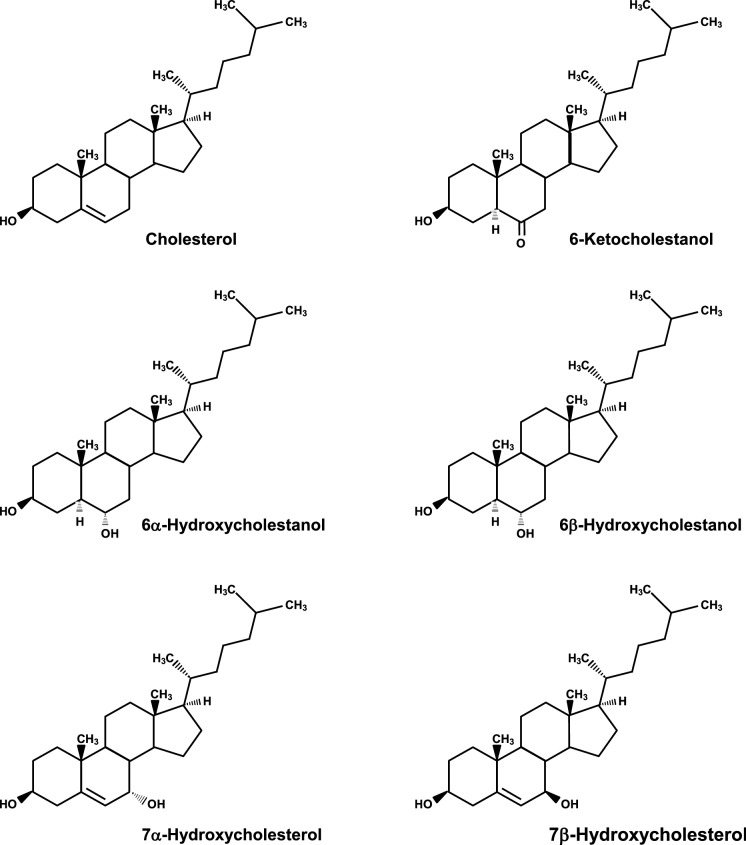
Fig. 1.
Chemical structures of cholesterol, 6-ketocholestanol, 6α-hydroxycholestanol, 6β-hydroxycholestanol, 7α-hydroxycholesterol, and 7β-hydroxycholesterol
Cell culture and treatment
HepG2 cells (JCRB1054) were maintained in culture media (DMEM containing high glucose supplemented with 10% FBS, 1.0 × 105 IU/L penicillin, and 100 mg/L streptomycin) at 37 °C in 5% CO2. 6-Keto and TO were dissolved in ethanol. Cells were treated with ethanol (as a vehicle control), 1–9 μM 6-keto, or 1 μM TO. The final concentration of ethanol was adjusted to 1% so as not to affect cell growth.
Measurement of intracellular triacylglycerol contents in HepG2 cells
HepG2 cells (1 × 106 cells/well) were seeded into 6-well plates and maintained in the culture media (DMEM containing high glucose supplemented with 10% LPDS, 1.0 × 105 IU/L penicillin, and 100 mg/L streptomycin) at 37 °C in 5% CO2. After 24 h incubation, the cells were treated with ethanol (as a vehicle control), 7 μM 6-keto, or 1 μM TO for 96 h with the medium changed every 48 h. After the treatment, the cells were washed twice with ice-cold PBS buffer containing 0.02% EDTA, harvested in ice-cold RIPA buffer, and lysed by sonication. Intracellular TAG contents in the lysates were measured using a commercial enzyme assay kit (Triglyceride E-Test from Wako Pure Chemicals). Protein contents in the lysates were measured using a commercial kit (DC Protein Assay kit from Bio-Rad Laboratories, CA, USA). Intracellular TAG contents were expressed as a relative value after being normalized to protein content.
Oil Red O staining
HepG2 cells (1 × 106 cells/well) were seeded into 6-well plates and maintained in the culture media (DMEM containing high glucose supplemented with 10% LPDS, 1.0 × 105 IU/L penicillin, and 100 mg/L streptomycin) at 37 °C in 5% CO2. After 24 h incubation, the cells were treated with ethanol (as a vehicle control), 7 μM 6-keto, or 1 μM TO for 96 h with the medium changed every 48 h. After the treatment, the cells were washed twice with PBS buffer, fixed with 10% formalin for 1 h, washed twice with distilled water, and then stained with Oil Red O dye (Lipid assay kit from Cosmo Bio Co., Ltd., Tokyo, Japan) for 15 min. Stained cells were washed three times with distilled water, and then photographed.
Analysis of mRNA expression
HepG2 cells (1 × 106 cells/well) were seeded into 6-well plates and maintained in the culture media (DMEM containing low glucose supplemented with 10% LPDS, 1.0 × 105 IU/L penicillin, and 100 mg/L streptomycin) at 37 °C in 5% CO2. After 48 h incubation, the cells were treated with ethanol (as a vehicle control) or 7 μM 6-keto for 6 h. Total RNA from the HepG2 cells was isolated using a guanidinium thiocyanate/cesium chloride ultracentrifugation method (Chirgwin et al. 1979), and the total RNA was reverse transcribed with a Transcriptor First Strand cDNA Synthesis kit (Roche, Berlin, Germany) to obtain cDNA. Quantitative real-time RT-PCR analysis was performed with the SYBR Premix EX Taq II kit (Takara, Shiga, Japan) and a real-time PCR system (Mx3000P qPCR system; Agilent Technologies, Santa Clara, CA, USA). The primer sequences that were used are as follows: nuclear receptor subfamily 1 group H member 3 (NR1H3) [encoding liver X receptor α (LXRα)] (forward primer, 5′-TCAGCATCTTCTCTGCAGACCGG-3′; reverse primer, 5′-TCATTAGCATCCGTGGGAACA-3′), ATP binding cassette subfamily A member 1 (ABCA1) (forward primer, 5′-CGAACAGTACACATTTGTCAGC-3′; reverse primer, 5′-GTGTCTGGGATTGGGTTTCCTTC-3′), sterol regulatory element binding transcription factor 1 (SREBF1) [encoding sterol regulatory element-binding protein-1c (SREBP-1c)] (forward primer, 5′-ATGCCATGGGCAAGTACACA-3′; reverse primer, 5′-ACGTGTCAAGAAGTGCAAGG-3′), fatty acid synthase (FASN) (forward primer, 5′-ATGCACACAGTGCTCAAAGG-3′; reverse primer, 5′-GTATCCTCCACAGGCAGGAA-3′), stearoyl-CoA desaturase (SCD) [encoding SCD1] (forward primer, 5′-CCCCACCTACAAGGATAAGGAAG-3′; reverse primer, 5′-GTAGAATACCCCCCAAAGCCAGG-3′), tubulin beta 2A class IIa (TUBB2A) [encoding β-tubulin] (forward primer, 5′-CAATGAGGCTGCTGGTAAC-3′; reverse primer, 5′-GAACACGAAGTTGTCTGGTCTG-3′). Results were expressed as a relative value after normalization to TUBB2A expression.
Plasmid constructs
Genomic DNA was isolated from HepG2 cells and the livers of Sprague–Dawley rats. Human FASN and rat Fasn promoter regions were generated by PCR amplification. Human FASN and rat Fasn promoter regions and each primer sequence are shown in Supplemental Figures 1 and 2. For human FASN promoter amplification, the primer sequences used were as follows: forward primer, 5′-ACCCGCGAGGAAAACCGGGGATGCGCTGCG-3′; reverse primer, 5′-AGCGGGAGGCTGAAGCGCGGCGGAGAGGGAG-3′. For rat Fasn promoter amplification, the primer sequences used were as follows: 5′-GCCACAGAAAGGGTGGGTGTCTGAGAAAGC-3′; reverse primer, 5′-TCTAGGCCGCGCCGGCGCTATTTAAACCGCGGCCATCCC-3′. The amplified insert DNA fragments were subcloned into the pGEM®-T Easy Vector using the pGEM®-T Easy Vector System (Promega, Madison, WI, USA) and JM109 Escherichia coli competent cells (Toyobo, Tokyo, Japan). The subcloned plasmids were digested with EcoRI (Toyobo, Tokyo, Japan) for 4 h at 37 °C. The digest products were separated on 2% agarose gel and purified using the Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Spin Columns (Bio-Rad, Tokyo, Japan). The pGL4.10 [luc2] Vector (Promega, Madison, WI, USA) encodes the luciferase reporter gene luc2 (Photinus pyralis). The pGL4.10 [luc2] Vector was digested with EcoRV (Toyobo, Tokyo, Japan) for 4 h at 37 °C. The digested vector was incubated with calf intestinal alkaline phosphatase (Takara, Tokyo, Japan) for 24 h at 37 °C to remove 5′ phosphate groups, which prevents the vector from reclosing on itself without each insert. Subsequently, the vector was separated on 0.8% agarose gel and purified using the Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction Spin Columns. Each insert was ligated with the vector using 2× Rapid Ligation Buffer and T4 DNA ligase (Promega) to form the plasmids. Each ligation plasmid was transformed into JM109 Escherichia coli competent cells (Toyobo) and was then purified using Viogene® Maxi Plus Ultrapure Plasmid Extraction System according to the manufacturer’s protocol.
The mutant constructs at the LXRE and/or SREBP binding site of the rat Fasn promoter were generated by site-directed mutagenesis using the KOD Plus mutagenesis kit (Toyobo) according to the manufacturer’s protocol. LXRE mutation and SREBP mutation elements of the rat Fasn promoter region (insert DNA) are shown in Supplemental Figure 2. The LXRE mutation and SREBP mutation elements of the rat Fasn promoter region (insert DNA) were also generated by PCR amplification. For the rat Fasn-LXRE mutation promoter amplification, the primer sequences used were as follows (underline indicates mutation bases): forward primer, 5′-CGGTAGTAACCCCGCCTGAGGCGCCCTC-3′; reverse primer, 5′-AAAATCGTGGCCCAGCTTTCTCAGACACCCACC-3′. For the rat Fasn-SERBP mutation promoter amplification, the primer sequences used were as follows (underline indicates mutation bases): 5′-TTCGGCGTGGCCGCGCGGGGATGGCCGCGGTTT-3′; reverse primer, 5′-TTCGCTGACAGCTTGGCTGCGCCGCCCAGGCC-3′.
Luciferase assay
HepG2 cells (2.5 × 105 cells/well) were seeded into 24-well plates. After 24 h incubation, the cells were transfected with the reporter plasmid (180 ng/well) and pGL4.74TK plasmid (Tk promoter-driven Renilla luciferase) (20 ng/well) using the FuGENE® 6 Transfection Reagent (Roche, Indianapolis, IN) according to the manufacturer’s protocol. The cells were treated with ethanol (as a vehicle control) or 1–9 μM 6-keto for 24 h. The cells were harvested 24 h post-transfection and assessed using the luciferase assay. The luciferase assays were carried out using the Dual-Luciferase Assay System (Promega) according to the manufacturer’s protocol on the Lumat LB 9507 tube luminometer (Berthold Japan Co. Ltd., Tokyo, Japan). The relative luciferase activity was calculated by the ratio of firefly luciferase activity to Renilla luciferase activity (internal transfection control) and relating it to each pGL4.10[luc2] vector to reflect each FAS promoter activity.
Statistical analysis
All values are expressed as the mean ± standard error of mean (SEM). The data were analyzed using the Student’s t-test (Figs. 2, 3, 5a, b), one-way ANOVA followed by the Dunnett’s multiple comparison post hoc test (Figs. 4 and 8) or the Tukey–Kramer multiple comparison post hoc test (Fig. 7), and two-way ANOVA (6-keto treatment and glucose level) (Fig. 6). Differences were considered significant at P < 0.05. Statistical analysis was performed using Excel 2011 (Microsoft, USA) with the add-in software Statcel 4 (Yanai 2015).
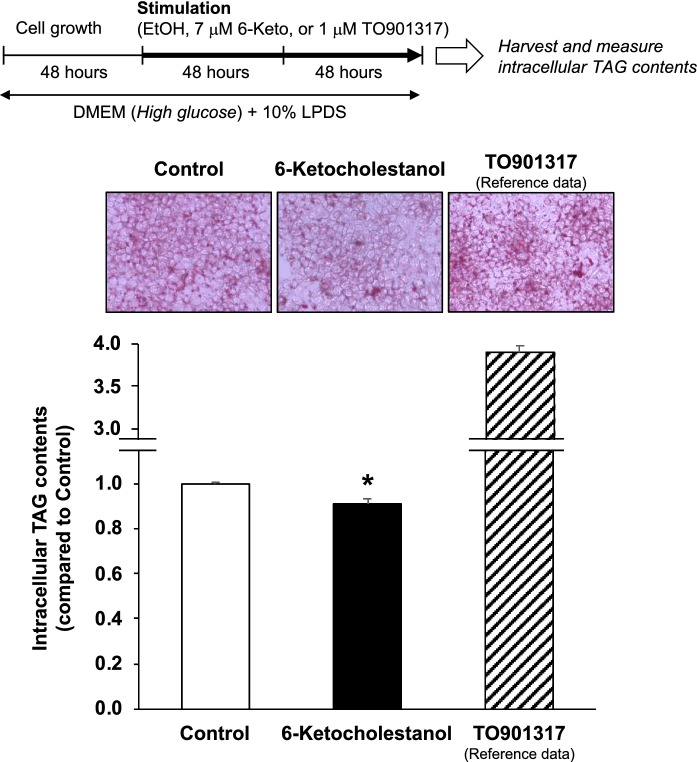
Fig. 2.
6-Ketocholestanol reduces intracellular TAG accumulation in HepG2 cells. HepG2 cells were treated with ethanol (vehicle control), 7 μM 6-keto, or 1 μM TO (as reference) for 4 days. TAG and protein levels in the cells were measured. TAG content of the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. The column marked with an asterisk is significantly different from the control (*P < 0.05), analyzed using the Student’s t-test. TAG triacylglycerol, TO TO901317
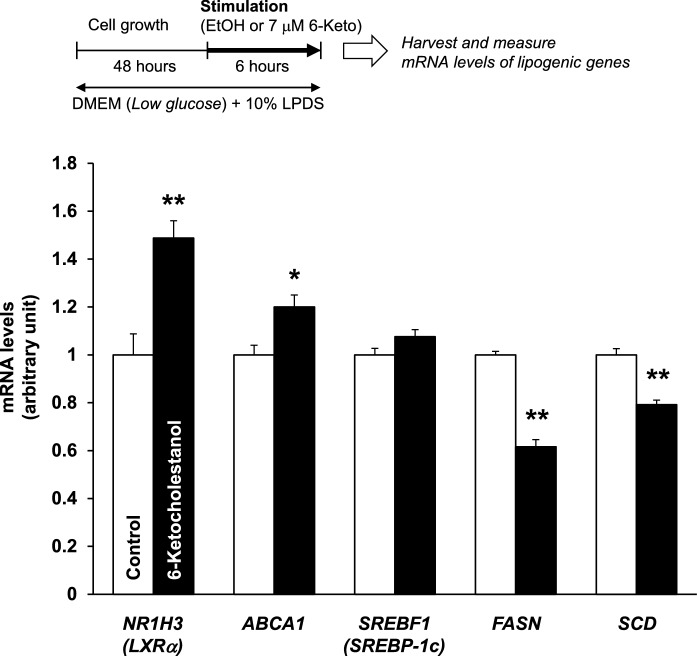
Fig. 3.
6-Ketocholestanol alters lipogenic gene expressions in HepG2 cells. HepG2 cells were treated with ethanol (vehicle control) or 7 μM 6-keto for 6 h. The expression levels of each gene (NR1H3, ABCA1, SREBF1, FASN, and SCD) were normalized to the TUBB2A expression level. The expression level of each gene in the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. Columns marked with asterisk(s) are significantly different from the control (*P < 0.05, **P < 0.01), analyzed using the Student’s t-test. ABCA1 ATP binding cassette subfamily A member 1, FASN fatty acid synthase, NR1H3 nuclear receptor subfamily 1 group H member 3 (encoding liver X receptor α), SCD stearoyl-CoA desaturase (encoding SCD1), SREBF1 sterol regulatory element binding transcription factor 1 (encoding sterol regulatory element-binding protein-1c)
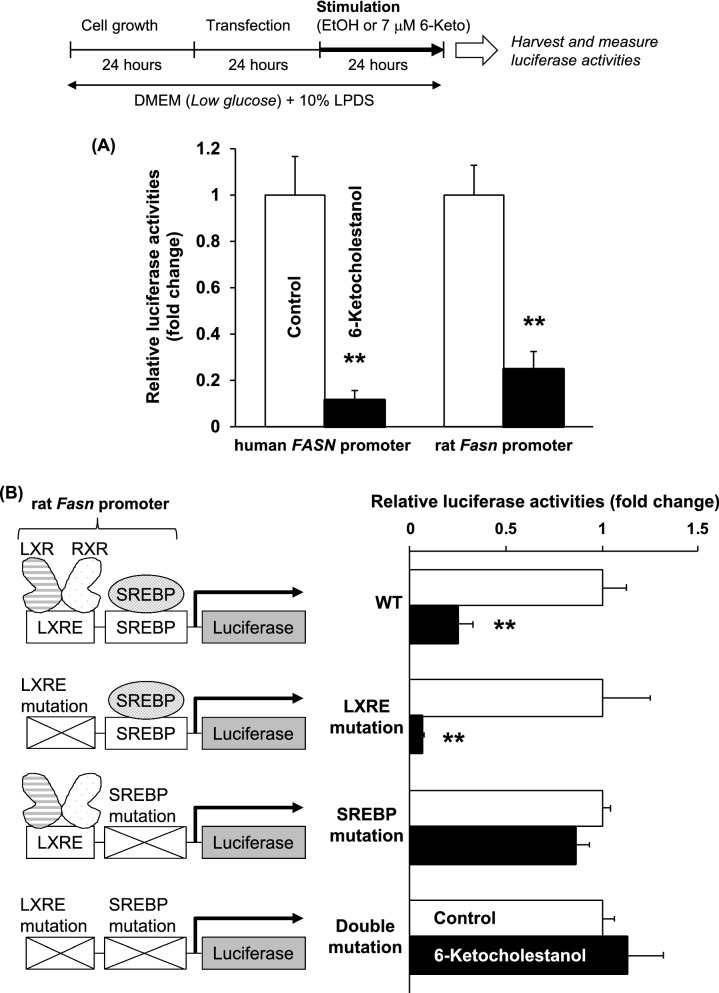
Fig. 5.
Effects of 6-ketocholestanol on human FASN and rat Fasn gene promoter activities. a 6-Ketocholestanol suppresses human FASN and rat Fasn gene promoter activities. b Suppression of rat Fasn promotor activity by 6-ketocholestanol is inhibited by specific mutations in the SREBP binding sites. After transfection, HepG2 cells were treated with ethanol (vehicle control) or 7 μM 6-keto for 24 h. Luciferase activity of the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. Columns marked with asterisk(s) are significantly different from the control (**P < 0.01) analyzed using the Student’s t-test
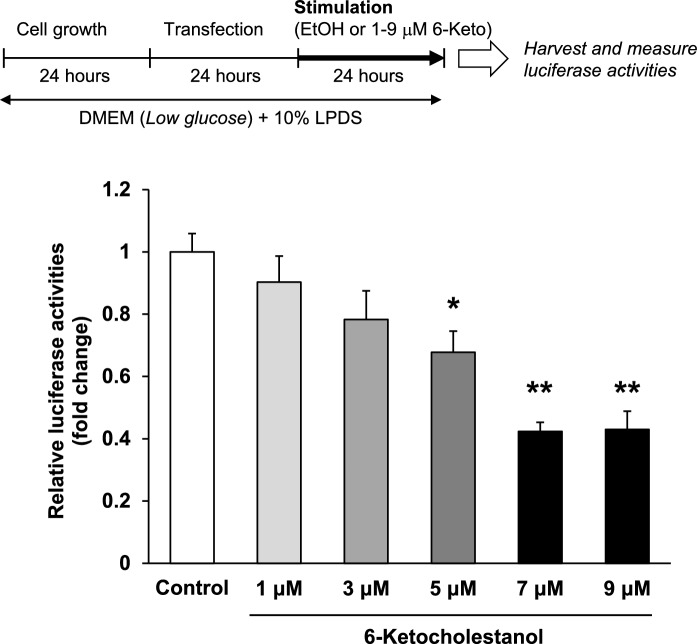
Fig. 4.
6-Ketocholestanol suppresses human FASN gene promoter activity in a dose-dependent manner. After transfection, HepG2 cells were treated with ethanol (vehicle control) or 1–9 μM 6-keto for 24 h. Luciferase activity of the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. Columns marked with asterisk(s) are significantly different from the control (*P < 0.05, **P < 0.01) analyzed using one-way ANOVA followed by the Dunnett’s multiple comparison post hoc test
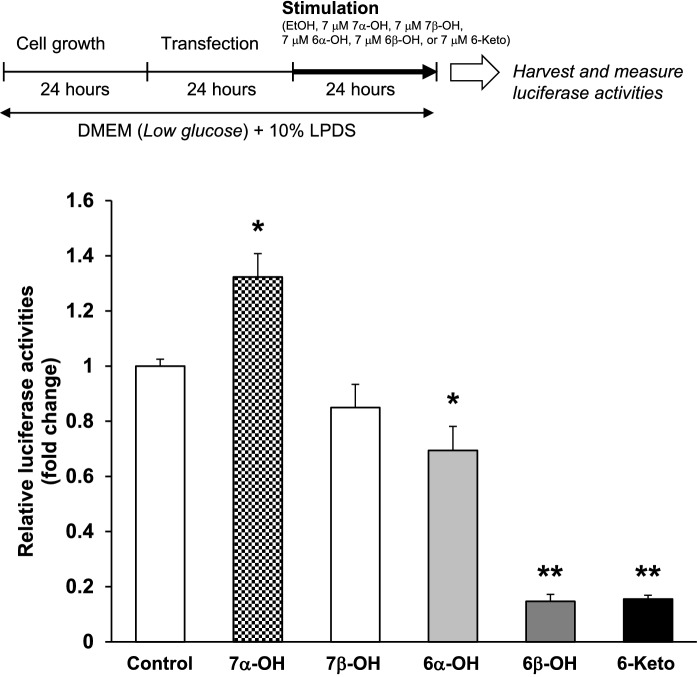
Fig. 8.
6-Ketocholestanol, 6α-hydroxycholestanol, and 6β-hydroxycholestanol, oxidized in the 6-carbon of B-ring, suppress human FASN gene promoter activities. After transfection, HepG2 cells were treated with ethanol (the vehicle control), 7 μM 7α-OH, 7 μM 7β-OH, 7 μM 6α-OH, 7 μM 6β-OH, or 7 μM 6-Keto for 24 h. Data are presented as the mean ± SEM of three independent experiments. Columns marked with asterisk(s) are significantly different from the control (*P < 0.05, **P < 0.01) by one-way ANOVA followed by the Dunnett’s multiple comparison post hoc test
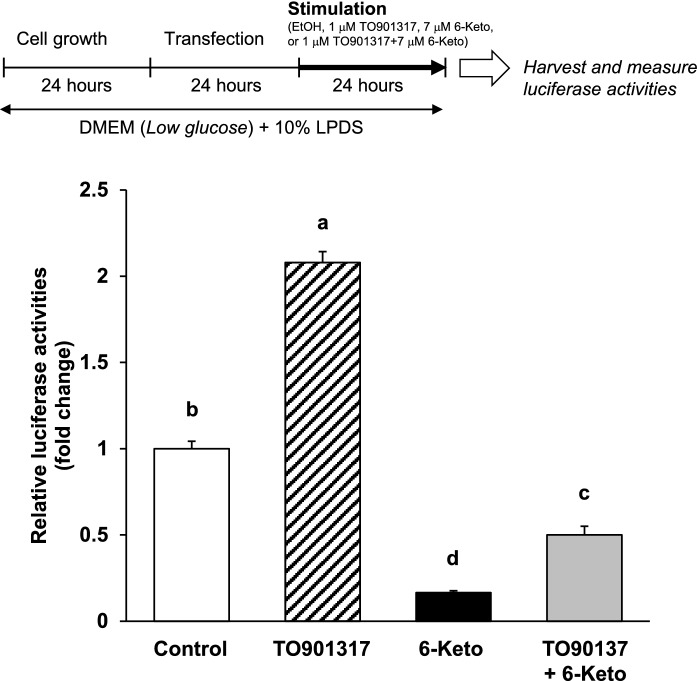
Fig. 7.
An increase in human FASN gene promotor activity by TO901317 is inhibited by the addition of 6-ketocholestanol. After transfection, HepG2 cells were treated with ethanol (the vehicle control), 1 μM TO, 7 μM 6-keto, or 1 μM TO + 7 μM 6-Keto for 24 h. Each luciferase activity of the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. Columns marked with different letters are significantly different at P < 0.05 by one-way ANOVA followed by the Tukey–Kramer multiple comparison post hoc test
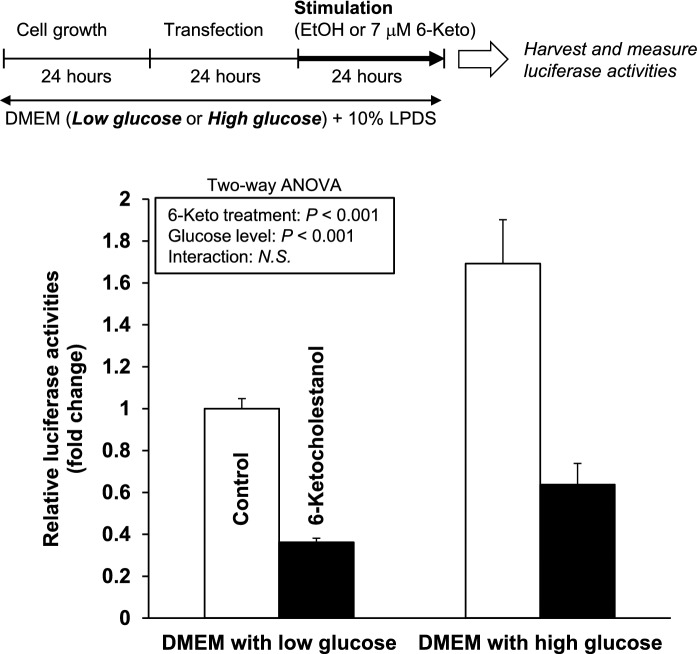
Fig. 6.
Effects of 6-ketocholestanol and glucose concentrations in the media on human FASN gene promoter activities. After transfection, HepG2 cells were treated with ethanol (vehicle control) or 7 μM 6-keto and DMEM containing high glucose or low glucose for 24 h. Luciferase activity of the vehicle control was arbitrarily set at 1. Data are presented as the mean ± SEM of three independent experiments. The data were analyzed using two-way ANOVA (6-keto treatment and glucose level)
Results and discussion
6-Ketocholestanol reduces intracellular TAG accumulation in HepG2 cells
Several OCs, such as 22(R)-hydroxycholesterol, 4β-hydroxycholesterol, and 7α-OH, act as ligands for liver X receptor alpha (LXRα) (Janowski et al. 1996; Schroepfer 2000). LXRα is highly expressed in the liver and white adipose tissues (Willy et al. 1995), and it upregulates expression of lipogenic genes (Joseph et al. 2002). However, it is unknown whether 6-keto acts similarly to LXRα agonists and upregulates lipogenesis. Therefore, in this study, we evaluated the effects of 6-keto on intracellular TAG accumulation in HepG2 cells. Treatment with TO, a potent and selective agonist for LXRα (Schultz et al. 2000), increased intracellular TAG levels, whereas treatment with 6-keto significantly reduced intracellular TAG levels (Fig. 2). Previous studies showed that 25-hydroxycholesterol-3-sulfate, a 25-hydroxycholesterol metabolite, acts as an LXRα antagonist and reduces cellular lipid levels in human THP-1-derived macrophages (Ma et al. 2008), human aortic endothelial cells (Bai et al. 2011), and HepG2 cells (Xu et al. 2010). Thus, we suggest that the suppression of TAG accumulation in hepatocytes by 6-keto may contribute to its antagonistic action on LXRα.
Effects of 6-ketocholestanol on lipogenic gene expression in HepG2 cells
To further examine the effects of 6-keto on lipogenesis in HepG2 cells, we analyzed the mRNA levels of lipogenic genes. As shown in Fig. 3, 6-keto treatment significantly increased NR1H3 (LXRα) and ABCA1 mRNA levels and decreased FASN and SCD mRNA levels. To explain the different mRNA levels of the two genes, ABCA1 and FASN, under LXRα regulation, we hypothesized that 6-keto may not only act as an LXRα agonist and antagonist, but it also may affect target genes through an LXRα-independent pathway. A previous study showed that TO, an LXRα ligand, triggers an autoregulatory loop, which leads to induction of LXRα gene expression and transcription of downstream target genes such as ABCA1 (Li et al. 2002). In concordance with the previous study, 6-keto acted as a LXRα ligand that led to increased LXRα and ABCA1 mRNA levels. Conversely, expression of FASN, a central enzyme in de novo lipogenesis and an established target of LXR, was also induced by the SREBP-1c pathway (Joseph et al. 2002). Therefore, suppression of FASN expression in hepatocytes by 6-keto may be occurring through the SREBP-1c pathway.
6-Ketocholestanol down-regulates human FASN gene promoter activities in an SREBP-dependent manner
To investigate the molecular mechanism by which 6-keto reduces FASN gene expression, we established human FASN (Figs. 4 and 5a) and rat Fasn (Fig. 5a and b) promoter luciferase reporter gene assays in HepG2 cells. As shown in Fig. 4, human FASN gene promoter activities were inhibited by 6-keto treatment in a dose-dependent manner. The inhibitory action by 6-keto reached a maximum inhibition at a concentration of 7 μM. Rat Fasn gene promoter activity was also significantly inhibited by 7 μM 6-keto treatment (Fig. 5a).
It is known that the LXRE and SREBP sites in the FASN promoter region are highly conserved between humans and rats (Joseph et al. 2002). Therefore, to clarify the effects of LXR and/or SREBP on the rat Fasn promoter after treatment with 6-keto, we constructed luciferase reporter vectors that have specific mutations in these sites. As shown in Fig. 5b, the inhibitory action of rat Fasn gene promoter activities by 6-keto occurred with mutations at the LXRE site. Conversely, the action of 6-keto was completely inhibited when mutations were present at the SREBP site. Thus, we conclude that 6-keto down-regulates FASN gene promoter activities in an SREBP-dependent manner but not via the LXR pathway.
6-Ketocholestanol down-regulates human FASN gene promoter activities regardless of glucose concentration in the culture medium
Glucose activates LXR at physiological concentrations in the liver and upregulates expression of the LXR target genes (Mitro et al. 2007). Using DMEM with high and low glucose, we investigated whether glucose concentrations in the culture media affect the reduction of human FASN gene promoter activities by 6-keto treatment. As shown in Fig. 6, 6-keto treatment significantly reduced human FASN gene promoter activities under both high and low glucose concentrations.
Human FASN gene promoter activity induced by TO901317, a potent and selective agonist for LXRα, is inhibited by the addition of 6-ketocholestanol
To further examine the effect of 6-keto on human FASN gene promoter activity in HepG2 cells, we investigated whether treatment with 6-keto would suppress human FASN gene promoter activities that are induced by TO treatment. TO treatment significantly increased human FASN gene promoter activities, whereas simultaneous treatment with TO and 6-keto significantly reduced the activities to less than half when compared with the control (Fig. 7).
Oxidation of carbon-6 in the B-ring of cholesterol is important for reduction of human FASN promoter activity
Carbons in the B-ring of cholesterol are easily oxidized (Smith et al. 1978). In this study, to gain additional insight into the molecular mechanism by which 6-keto reduces FASN gene expression, we compared the effects of several OCs oxidized at the positions of carbon-6 and -7 in the B-ring of cholesterol (Fig. 1) on human FASN gene promoter activities in HepG2 cells. As shown in Fig. 8, 7α-OH significantly increased the activities, suggesting that 7α-OH acts as a ligand for LXRα. This result was consistent with a previous report (Janowski et al. 1996). Conversely, 6α-OH, 6β-OH and 6-keto significantly reduced the activities of the human FASN gene promoter (Fig. 8). Oxidation of carbon-6 in the B-ring of cholesterol may play an important role in the SREBP-dependent reduction of FASN expression. SREBPs are transported from the endoplasmic reticulum to the Golgi apparatus through the action of SREBP cleavage-activating protein (Scap), an escort protein, which forms a complex with SREBPs (Goldstein et al. 2006). 25-Hydroxycholesterol (25-OH) acts by binding to Insig, which causes Insig to bind to Scap, and this then blocks the Scap-mediated movement of SREBPs to the Golgi apparatus (Radhakrishnan et al. 2007). Further studies are necessary to clarify whether OCs having carbon-6 oxidation as well as 25-OH block the movement of SREBPs to the Golgi apparatus by binding to Insig. Unlike the sterol regulation of SREBP-2, the gene expression and activity of SREBP-1c are dependent on the energy status (Shimano and Sato 2017). The mechanistic target of rapamycin (mTOR) is a serine-threonine protein kinase and regulates cell growth and metabolism with environmental inputs including in energy, nutrients, and growth factors (Saxton and Sabatini 2017). Recently, it has been revealed that the activation of SREBPs is regulated by mTOR signaling, especially mTOR complex 1 (mTORC1) (Porstmann et al. 2008; Yamauchi et al. 2011; Peterson et al. 2011). Therefore, whether the down-regulation of the SREBP pathway by 6-keto is due to the inhibition of the mTOR signaling would be of interest for future study.
In conclusion, this study is the first report that 6-keto down-regulates SREBP-1c regulated lipogenic enzyme expression to suppress intracellular TAG accumulation in HepG2 cells. Future studies are needed to determine how to increase circulating levels of 6-keto and thus prevent lipid accumulation in the liver and fatty liver disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. The sequence of the human FASN promoter region. Underlines indicate selected regulatory elements. LXRE, liver X receptor response element; SRE, sterol response element; SREBP, sterol regulatory element-binding protein. Supplemental Fig. 2. The sequences of wild type and mutant LXRE and SREBP elements from the rat Fasn promoter region. Underlines indicate selected regulatory elements. Dotted bases indicate mutations in the LXRE and SREBP elements. LXRE, liver X receptor response element; SRE, sterol response element; SREBP, sterol regulatory element-binding protein. (PPTX 52 kb)
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 21580144. The authors thank Editage (www.editage.jp) for English language editing.
Author contributions
BS wrote the manuscript. SY, CO, and MK participated in the experimental work and collected and analyzed data. BS and MS contributed to the study design, supervised the study, and commented on the manuscript. All authors have read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bai Q, Xu L, Kakiyama G, Runge-Morris MA, Hylemon PB, Yin L, Pandak WM, Ren S. Sulfation of 25-hydroxycholesterol by SULT2B1b decreases cellular lipids via the LXR/SREBP-1c signaling pathway in human aortic endothelial cells. Atherosclerosis. 2011;214:350–356. doi: 10.1016/j.atherosclerosis.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przbyla AE, McDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cuéllar A, Ramirez J, Infante VM, Chavez E. Further studies on the recoupling effect of 6-ketocholestanol upon oxidative phosphorylation in uncoupled liver mitochondria. FEBS Lett. 1997;411:365–368. doi: 10.1016/S0014-5793(97)00741-2. [DOI] [PubMed] [Google Scholar]
- Emanuel HA, Hassel CA, Addis PB, Bergmann SD, Zavoral JH. Plasma cholesterol oxidation products in human subjects fed a meal rich in oxysterols. J Food Sci. 1991;56:843–847. doi: 10.1111/j.1365-2621.1991.tb05396.x. [DOI] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Ichi I, Iwamoto M, Tomoyori H, Sato M, Ikeda I, Imaizumi K. Intake of oxycholesterols and oxyphytosterols in Japanese meals. J Jpn Soc Nutr Food Sci. 2005;58:145–149. doi: 10.4327/jsnfs.58.145. [DOI] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem. 2002;277:11019–11025. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Kawamura M, Maeda T, Fujimaki Y, Fujita M, Kojima K, Teramoto T. Apolipoprotein E accelerates the efflux of cholesterol from macrophages: mechanism of xanthoma formation in apolipoprotein E deficiency. J Atheroscler Thromb. 2000;6:22–27. doi: 10.5551/jat1994.6.22. [DOI] [PubMed] [Google Scholar]
- Kulig W, Cwiklik L, Jurkiewicz P, Rog T, Vattulainen I. Cholesterol oxidation products and their biological importance. Chem Phys Lipids. 2016;199:144–160. doi: 10.1016/j.chemphyslip.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Latasa MJ, Moon YS, Kim KH, Sul HS. Nutritional regulation of the fatty acid synthase promoter in vivo: sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc Natl Acad Sci USA. 2000;97:10619–10624. doi: 10.1073/pnas.180306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bolten C, Bhat BG, Woodring-Dietz J, Li S, Prayaga SK, Xia C, Lala DS. Induction of human liver X receptor alpha gene expression via an autoregulatory loop mechanism. Mol Endocrinol. 2002;16:506–514. doi: 10.1210/mend.16.3.0789. [DOI] [PubMed] [Google Scholar]
- Ma Y, Xu L, Rodriguez-Agudo D, Li X, Heuman DM, Hylemon PB, Pandak WM, Ren S. 25-Hydroxycholesterol-3-sulfate regulates macrophage lipid metabolism via the LXR/SREBP-1 signaling pathway. Am J Physiol Endocrinol Metab. 2008;295:E1369–E1379. doi: 10.1152/ajpendo.90555.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magaña MM, Osborne TF. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V, Kreusch A, Saez E. The nuclear receptor LXR is a glucose sensor. Nature. 2007;445:219–223. doi: 10.1038/nature05449. [DOI] [PubMed] [Google Scholar]
- Paniangvait P, King AJ, Jones AD, German BG. Cholesterol oxides in foods of animal origin. J Food Sci. 1995;60:1159–1174. doi: 10.1111/j.1365-2621.1995.tb04548.x. [DOI] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Shirouchi B. Physiological significance of oxidized cholesterols. Oleoscience. 2012;12:115–123. doi: 10.5650/oleoscience.12.115. [DOI] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroepfer GJ., Jr Oxysterols: modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology—divergent pathophysiology. Nat Rev Endrocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- Shirouchi B, Furukawa Y, Nakamura Y, Kawauchi A, Imaizumi K, Oku H, Sato M. Inhibition of Niemann-Pick C1-like 1 by ezetimibe reduces dietary 5β,6β-epoxycholesterol absorption in rats. Cardiovasc Drugs Ther. 2019;33:35–44. doi: 10.1007/s10557-019-06854-4. [DOI] [PubMed] [Google Scholar]
- Smith LL, Kulig MJ, Miiller D, Ansari GAS. Sterol metabolism. 44. Oxidation of cholesterol by dioxygen species. J Am Chem Soc. 1978;100:6206–6211. doi: 10.1021/ja00487a042. [DOI] [Google Scholar]
- Sottero B, Gamba P, Gargiulo S, Leonarduzzi G, Poli G. Cholesterol oxidation products and disease: an emerging topic of interest in medicinal chemistry. Curr Med Chem. 2009;16:685–705. doi: 10.2174/092986709787458353. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Dedukhova VI, Skulachev VP. 6-Ketocholestanol abolishes the effect of the most potent uncouplers of oxidative phosphorylation in mitochondria. FEBS Lett. 1994;355:305–308. doi: 10.1016/0014-5793(94)01211-3. [DOI] [PubMed] [Google Scholar]
- Starkov AA, Bloch DA, Chernyak BV, Dedukhova VI, Mansurova SE, Severina II, Simonyan RA, Vygodina TV, Skulachev VP. 6-Ketocholestanol is a recoupler for mitochondria, chromatophores and cytochrome oxidase proteoliposomes. Biochim Biophys Acta. 1997;1318:159–172. doi: 10.1016/S0005-2728(96)00134-X. [DOI] [PubMed] [Google Scholar]
- Sul HS, Wang D. Nutritional and hormonal regulation of enzymes in fat synthesis: studies of fatty acid synthase and mitochondrial glycerol-3-phosphate acyltransferase gene transcription. Annu Rev Nutr. 1998;18:331–351. doi: 10.1146/annurev.nutr.18.1.331. [DOI] [PubMed] [Google Scholar]
- van de Bovenkamp P, Kosmeijer-Schuil TG, Katan MB. Quantification of oxysterols in Dutch foods: egg products and mixed diets. Lipids. 1988;23:1079–1085. doi: 10.1007/BF02535656. [DOI] [PubMed] [Google Scholar]
- Vejux A, Lizard G. Cytotoxic effects of oxysterols associated with human diseases: induction of cell death (apoptosis and/or oncosis), oxidative and inflammatory activities, and phospholipidosis. Mol Aspects Med. 2009;30:153–170. doi: 10.1016/j.mam.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Xu L, Bai Q, Rodriguez-Agudo D, Hylemon PB, Heuman DM, Pandak WM, Ren S. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45:821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Furukawa K, Hamamura K, Furukawa K. Positive feedback loop between PI3K-Akt-mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen. Cancer Res. 2011;71:4989–4997. doi: 10.1158/0008-5472.CAN-10-4108. [DOI] [PubMed] [Google Scholar]
- Yanai H. Statcel 4—the useful add-in software forms on Excel. 4. Tokyo: OMS; 2015. [Google Scholar]
- Zarrouk A, Vejux A, Mackrill J, O’Callaghan Y, Hammami M, O’Brien N, Lizard G. Involvement of oxysterols in age-related diseases and ageing processes. Ageing Res Rev. 2014;18:148–162. doi: 10.1016/j.arr.2014.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. The sequence of the human FASN promoter region. Underlines indicate selected regulatory elements. LXRE, liver X receptor response element; SRE, sterol response element; SREBP, sterol regulatory element-binding protein. Supplemental Fig. 2. The sequences of wild type and mutant LXRE and SREBP elements from the rat Fasn promoter region. Underlines indicate selected regulatory elements. Dotted bases indicate mutations in the LXRE and SREBP elements. LXRE, liver X receptor response element; SRE, sterol response element; SREBP, sterol regulatory element-binding protein. (PPTX 52 kb)