Abstract
Filament-forming proteins in bacteria function in stabilization and localization of proteinaceous complexes and replicons; hence they are instrumental for myriad cellular processes such as cell division and growth. Here we present two novel filament-forming proteins in cyanobacteria. Surveying cyanobacterial genomes for coiled-coil-rich proteins (CCRPs) that are predicted as putative filament-forming proteins, we observed a higher proportion of CCRPs in filamentous cyanobacteria in comparison to unicellular cyanobacteria. Using our predictions, we identified nine protein families with putative intermediate filament (IF) properties. Polymerization assays revealed four proteins that formed polymers in vitro and three proteins that formed polymers in vivo. Fm7001 from Fischerella muscicola PCC 7414 polymerized in vitro and formed filaments in vivo in several organisms. Additionally, we identified a tetratricopeptide repeat protein - All4981 - in Anabaena sp. PCC 7120 that polymerized into filaments in vitro and in vivo. All4981 interacts with known cytoskeletal proteins and is indispensable for Anabaena viability. Although it did not form filaments in vitro, Syc2039 from Synechococcus elongatus PCC 7942 assembled into filaments in vivo and a Δsyc2039 mutant was characterized by an impaired cytokinesis. Our results expand the repertoire of known prokaryotic filament-forming CCRPs and demonstrate that cyanobacterial CCRPs are involved in cell morphology, motility, cytokinesis and colony integrity.
Subject terms: Cellular microbiology, Bacterial genetics
Introduction
Species in the phylum Cyanobacteria present a wide morphological diversity, ranging from unicellular to multicellular organisms. Unicellular cyanobacteria of the Synechocystis and Synechococcus genera are characterized by a round or rod-shaped morphology, respectively, and many strains are motile. Species of the Nostocales order are multicellular and differentiate three types of specialized cells including heterocysts, which fix atmospheric nitrogen under aerobic conditions, hormogonia that are reproductive motile filaments and akinetes, which are dormant cells that are resistant to desiccation. Within the Nostocales, species of the Nostocaceae (e.g., Anabaena, Nostoc) form linear trichomes, while cells in the Hapalosiphonaceae and Chlorogloepsidaceae divide in more than one plane to form true-branching trichomes as in Fischerella or multiseriate trichomes (more than one filament in a row) as in Chlorogloeopsis1. Notably, cells within a single trichome of a multicellular cyanobacterium can differ in size, form or cell wall composition, which may be attributed to different stages of cell differentiation (or phenotypic heterogeneity) and varying environmental cues2,3. Cells in the Anabaena sp. PCC 7120 (hereafter Anabaena) trichome are linked by a shared peptidoglycan sheet and an outer membrane4. Anabaena cells communicate and exchange nutrients through intercellular cell-cell connections, called septal junctions, which are thought to comprise the septal junction proteins SepJ, FraC and FraD5,6. SepJ is essential for the multicellular phenotype in Anabaena7,8.
Studies of the molecular basis of cyanobacterial morphogenesis have so far focused on the function of FtsZ and MreB, the prokaryotic homologs of tubulin and actin, respectively9. FtsZ functions in a multi-protein complex called the divisome, and is known as a key regulator of cell division and septal peptidoglycan (PG) biogenesis9,10. FtsZ has been shown to be an essential cellular protein in Anabaena and in the coccoid cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis)11. The FtsZ cellular concentration in Anabaena is tightly controlled by a so far undescribed protease12. Apart from its function in cell division, the FtsZ-driven divisome also mediates the localization of SepJ13. MreB functions in a multi-protein complex called the elongasome, where it is a key mediator of longitudinal PG biogenesis that controls the cell shape9,14. In cyanobacteria, MreB plays a role in cell shape determination in Anabaena, nonetheless, it is not essential for cell viability15. In contrast, in Synechococcus sp. PCC 7942 (hereafter Synechococcus) MreB is essential, where partially segregated mutants display a coccoid morphology resembling the morphology of E. coli mreB deletion strains16,17. In Nostoc punctiforme ATCC 29113, the MreBCD operon was shown to be regulated by the hormogonium-specific sigma factor SigJ and is likely involved in the transition of coccoid vegetative cells to the more rod-shaped cells that are characteristic to hormogonia18.
Proteins resembling the eukaryotic intermediate filaments (IFs) have been discovered in several bacterial species and were shown to form filaments in vitro and in vivo and to impact essential cellular processes19. IF proteins exhibit an intrinsic nucleotide-independent in vitro polymerization capability that is mediated by the high frequency of coiled-coil-rich regions in their amino acid sequence9,20–22. Eukaryotic IF proteins are generally characterized by a conserved domain buildup consisting of discontinuous coiled-coil segments that form a central rod domain. This rod domain is N- and C-terminally flanked by globular head and tail domains of variable length22–24. Crescentin is a bacterial IF-like CCRP from Caulobacter crescentus, which exhibits a striking domain similarity to eukaryotic IF proteins. Crescentin filaments that align at the inner cell curvature are essential for the typical crescent-like cell shape of C. crescentus; possibly, by locally exuding a constriction force which coordinates the MreB-driven peptidoglycan (PG) synthesis machinery25–27. Reminiscent of eukaryotic IF proteins, Crescentin was found to assemble into filamentous structures in vitro in a nucleotide-independent manner25. However, so far no Crescentin homologs have been found in other bacteria, indicating that non-spherical or rod-shaped prokaryotic morphologies are putatively controlled by other polymerizing proteins28,29. Apart from Crescentin, many other coiled-coil-rich proteins (CCRPs) with IF-like functions have been identified to polymerize into filamentous structures and to perform cytoskeletal-like roles; however, none of them resembled the eukaryotic IF domain architecture (reviewed by Lin & Thanbichler (2013)19). Examples are two proteins from Streptomyces coelicolor whose function has been studied in more detail: FilP and Scy29–31. Gradients of FilP localize at the tip of a growing hyphae and contribute to cellular stiffness29. Scy forms patchy clusters at the sites of novel tip-formation and, together with the scaffolding CCRP DivIVA, orchestrates the polar hyphal growth30. Together with FilP and a cellulose-synthase, these proteins form the polarisome, which guides peptidoglycan biogenesis and hyphal tip growth in S. coelicolor30,32,33. Another example are four CCRPs in the human pathogen Helicobacter pylori, which were found to assemble into filaments in vitro and in vivo, with a function in determination of the helical cell shape as well as cell motility34,35. Consequently, filament-forming CCRPs with essential cellular functions have been found in numerous prokaryotes having various cellular morphologies. The presence of filament-forming CCRPs in cyanobacteria is so far understudied. Here we search for CCRPs with presumed IF-like functions in cyanobacteria using a computational prediction of CCRPs. Putative filament-forming proteins were further investigated experimentally by structural analyses and in vitro and in vivo localization assays in morphologically diverse cyanobacteria.
Results
Coiled-coil-rich proteins are widespread in cyanobacteria
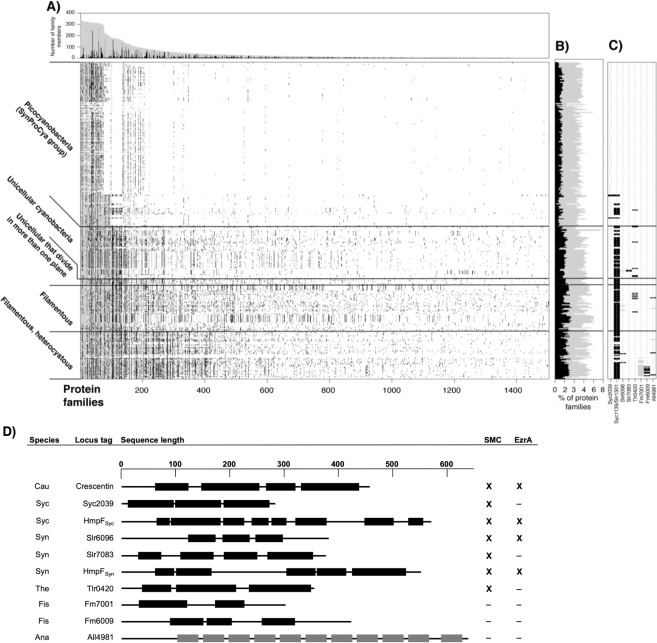
For the computational prediction of putative filament-forming proteins, we surveyed 364 cyanobacterial genomes including 1,225,314 protein-coding sequences (CDSs) for CCRPs. All CDSs in the cyanobacterial genomes where clustered by sequence similarity into families of homologous proteins (see Methods). The frequency of CCRPs in each CDS was calculated using the COILS algorithm36. The algorithm yielded a list of 28,737 CDSs with high coiled-coil content (≥80 amino acids in coiled-coil conformation; Supplementary File 1). CCRPs were predicted in 158,466 protein families covering all cyanobacterial species. To examine the overall distribution of CCRPs in cyanobacterial genomes, we investigated 1,504 families of homologous proteins that include at least three CCRP members (Fig. 1). Notably, most protein families (1,142; 76%) include CCRP and non-CCRP members, indicating that coiled-coil content may differ among homologous proteins. The presence/absence pattern of families including CCRPs further shows that those are less abundant in picocyanobacterial genomes (SynProCya group) in comparison to the remaining species in the phylum. Furthermore, the proportion of CCRPs in the genome is significantly higher in multicellular cyanobacteria in comparison to unicellular cyanobacteria (P = 2.65 × 10−46 using Kruskal-Wallis test and Tukey test with α = 0.05). This indicates that a high frequency of CCRPs is one characteristic of multicellular cyanobacteria.
Figure 1.
Distribution of CCRP protein families within cyanobacteria. (A) Lines in the presence/absence matrix designate cyanobacterial genomes; each column shows a protein family. Gray dots designate any homologous protein in the same protein family and black dots represent CCRP members. Protein families are sorted according to the number of members. Protein family size and the number of CCRP members are presented in a bar graph above. (B) The proportion of protein families containing CCRPs (gray) and CCRP proteins (black) in each genome. (C) Presence/absence pattern of CCRP candidate protein families. Only protein families with at least three members predicted to be CCRPs are shown. (D) Domain prediction of CCRP candidates. Scale on top is given in amino acid residues. Amino acid sequences in coiled-coil conformation are depicted by black bars with non-coiled-coil sequences represented by black lines. Tetratricopeptide repeats (TPR), also predicted by the COILS algorithm, are shown as grey bars. Proteins are given as cyanobase locus tags. Fm7001 and Fm6009 correspond to NCBI accession numbers WP_016868005.1 and WP_020476706, respectively. Abbreviations: Cau: C. crescentus; Syc: Synechococcus, Syn: Synechocystis; Ana: Anabaena; The: Thermosynechococcus elongatus BP-1; Fis: Fischerella. Cyanobacterial CCRPs had conserved domains present in prokaryotic IF-like CCRPs and eukaryotic IF proteins (Supplementary Table 1). Presence of a structural maintenance of chromosomes (SMC) domain or structural similarities to the cell division protein EzrA are marked with “X”, absence is indicated with “−”. Full list is given in Supplementary Table 1.
For the experimental validation, the complete list of CCRPs was filtered to include candidates from freshwater unicellular and filamentous cyanobacteria that are genetically accessible, including Thermosynechococcus elongatus BP-1 (Thermosynechococccus), Synechocystis, Synechococcus, Anabaena and Fischerella muscicola PCC 7414 (Fischerella). In addition to cytoskeleton functions, coiled-coils are common motifs of proteins involved in other cellular processes such as transcription, the extracellular matrix, chemotaxis and host–pathogen interactions37. Consequently, the remaining CCPRs were further sorted to include proteins having similar properties to known prokaryotic IF-like CCRPs (e.g. Crescentin, FilP) and are annotated as hypothetical proteins with an unknown function. Additionally, proteins lacking an unstructured N-terminal head and C-terminal tail domain, which are characteristics of prokaryotic IF-like proteins29, were excluded. Furthermore, proteins with an assigned function or predicted to be involved in other cellular processes were excluded (using publicly available online bioinformatic tools: NCBI Blast, NCBI CD search, PSORTb, TMHMM, InterPro, PSIPRED and I-TASSER). In the screening for protein characteristics and annotation, Crescentin, FilP and other eukaryotic IF proteins (e.g., Vimentin and Desmin) were chosen as reference for our predictions, where proteins displaying similar results were favored. An additional Fischerella CDS, Fm7001, was added to the list as earlier analyses suggested that it has a cell shape-determining function. The preliminary filtration resulted in a list of nine candidates, which we investigated experimentally here (Fig. 1C,D and Supplementary Table 1).
Candidate coding sequences varied in size and ranged from ca. 280 amino acids (Synpcc7942_2039, abbreviated Syc2039) to ca. 650 amino acids (All4981). The coiled-coil domain distribution was variable among the candidates in both coiled-coil domain count and length (Fig. 1D). Only Slr7083 exhibited a somewhat characteristic domain architecture of eukaryotic IF proteins, whereas the coiled-coil domain distribution in the other candidates had major differences in coiled-coil domain number and lengths. None of the predicted CCRPs exhibited a stutter-like structure in the last coiled-coil segment. Besides coiled-coil domains, the COILS algorithm also predicted tetratricopeptide repeats (TPRs) as coiled-coils, thus we also included All4981 into our analysis, even though conserved domain searches reliably predicted these domains as TPRs and not coiled-coils. Many protein candidates contained conserved domains from eukaryotic IF proteins, found in Crescentin and FilP or from the bacterial cell division protein EzrA (Supplementary Table 1). The presence of these domains may be regarded as support for our classification. Additionally, structural maintenance of chromosomes (SMC) domains were predicted in almost all chosen candidates, all eukaryotic IF proteins as well as in Crescentin and FilP (Supplementary Table 1). The MscS_TM domain from Desmin was found in Slr7083 and Tlr0420 contains a Neuromodulin_N as well as a CCDC158 domain, both present in FilP or Crescentin, respectively.
The presence of homologs across all cyanobacterial morphotypes serves as a hint for universal protein function while a restricted distribution in specific subsections or morphotypes indicates a functional specialization within the respective taxon. An example for such species-specific candidate in our list is slr7083 that is encoded on the pSYSA toxin-antitoxin plasmid in Synechocystis, similarly to parM and tubZ, which mediate plasmid segregation38,39. Synpcc7942_1139 and Slr1301 are homologs to the previously characterized motility protein HmpF from N. punctiforme hence we term them here HmpFSyn and HmpFSyc, respectively, to highlight their relationship. In contrast to slr7083, the homologous proteins HmpFSyn and HmpFSyc are highly conserved and have homologs among all cyanobacterial groups but are absent from the picocyanobacterial (Fig. 1; Supplementary File 2). As most of the candidate CCRPs are annotated as hypothetical proteins, we initially verified the transcription of the respective genes by RT-PCR from cDNA (Supplementary Fig. 1A–D). Our results showed that slr7083 was only weakly transcribed during mid-exponential culture growth phase and all4981 was found to be transcribed in an operon with its upstream genes all4982 and all4983 (Supplementary Fig. 1B,C).
Cyanobacterial CCRPs assemble into diverse filament-like structures in vitro
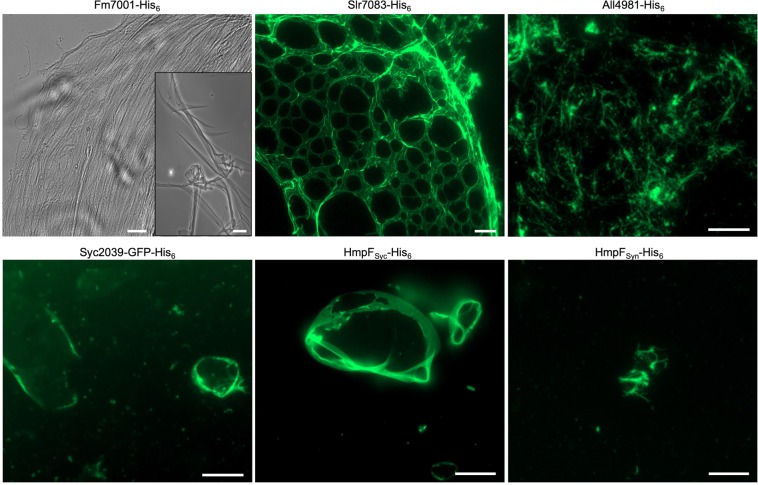
A major characteristic of filament-forming proteins is their ability to self-polymerize into filaments intra and extracellularly22,40. Unlike actin and tubulin, IFs are able to form filamentous structures in vitro in a nucleotide-independent manner without additional co-factors upon renaturation from a denaturing buffer40. To examine the self-polymerization property of the nine tested CCRPs, we purified His6-tagged CCRPs under denaturing conditions and subjected them to subsequent renaturation by dialysis. Here we used protein concentrations in a similar range (0.5–1 mg ml−1) to previously investigated proteins shown to form filaments in vitro (e.g., Crescentin25 and Scc41, the metabolic enzyme CtpS42 and the bactofilins BacA, BacB43 and BacM44). When applicable, the purified proteins were labeled with NHS-Fluorescein and the formation of in vitro filaments was assessed by epifluorescence or bright field microscopy. Several candidates did not form discernible structures in vitro and were consequently excluded from further investigation (including Slr6096, Tlr0420 and Fm6009; Supplementary Fig. 2A). The remaining CCRPs assembled into highly diverse structures in vitro (Fig. 2). Direct dialysis of Fm7001 from a high urea-containing buffer to a physiological buffer led to protein precipitation. However, upon slow stepwise renaturation (removing 0.5 M every 2 h), Fm7001 polymerized into a flat two-dimensional sheet floating on top of the dialysate in 4,5 M urea (Supplementary Fig. 2D). We addressed the eventuality that these structures could be the product of crystalized urea, but control experiments did not reveal filaments. Polymerized Fm7001 revealed two-dimensional filament-like sheets as well as single filament-like fibers (Fig. 2). Similar structures were observed for purified Fm7001-GFP and MBP-Fm7001-His6 (Supplementary Fig. 2E,F). A two-dimensional filament-like pattern was observed also for Slr7083, which formed single, long and straight strings that were interconnected by two-dimensional sheets, thereby producing an irregular net (Fig. 2). Similarly, All4981 assembled into an interconnected net of thin and single filament-like strings (Fig. 2). The heterologous expression of Syc2039-His6 in E. coli failed, but we successfully purified Syc2039-GFP-His6 from Synechococcus instead. The polymerization pattern of Syc2039-GFP-His6 revealed sphere or cell shape-like three-dimensional sheets (Fig. 2). However, we note that most of the protein precipitated upon renaturation, hence it is unlikely that Syc2039 has in vitro polymerizing properties. HmpFSyc polymerized into similar cell shape-like three-dimensional sheets but without any detectable aggregates (Fig. 2). The resemblance between Syc2039 and HmpFSyc sheets raised the possibility that the sheet-like structures observed in the Syc2039-GFP-His6 sample represented co-precipitated and polymerized HmpFSyc. In accordance with this suggestion, we identified direct interactions of HmpFSyc and Syc2039 using the bacterial adenylate cyclase two-hybrid (BACTH) assays (Supplementary Fig. 3A). For HmpFSyn, no clear in vitro structures were observed (Fig. 2). Notably, Crescentin, which we used as a positive control, polymerized into smooth and filigree filaments only in the presence of monovalent ions (i.e. NaCl; Supplementary Fig. 2B). This observation highlights the importance of suitable buffer conditions for the detection of filament-forming proteins. To further confirm our in vitro observations, we included the monomeric and highly soluble maltose binding protein (MBP) as well as the oligomeric proteins GroEL1.245 (from Chlorogloeopsis fritschii PCC 6912) and the UMP kinase (from Anabaena) as negative controls. While both, the MBP and the UMP kinase readily clumped into comparably small aggregates, GroEL1.2 formed large proteinaceous aggregates in vitro, likely as a result of uncoordinated multimerization (Supplementary Fig. 2C). Consequently, we conclude that the in vitro filament-like structures of the cyanobacterial CCRPs we observed here are unlikely to be oligomerization artifacts. We further validated the self-binding properties of the remaining six CCRPs using the BACTH assay and found that all proteins are able to self-interact (Supplementary Fig. 3A).
Figure 2.
Cyanobacterial CCRPs assemble into diverse filament-like structures in vitro. Bright field and epifluorescence micrographs of filament-like structures formed by purified and renatured Fm7001-His6 (0.7 mg ml−1), Slr7083-His6 (1 mg ml−1), All4981-His6 (0.5 mg ml−1), Syc2039-GFP-His6 (0.3 mg ml−1), HmpFSyc-His6 (0.5 mg ml−1) and HmpFSyn-His6 (0.5 mg ml−1). Proteins were dialyzed into 2 mM Tris-HCl, 4.5 M urea pH 7.5 (Fm7001), HLB (Slr7083), PLB (All4981, HmpFSyc, HmpFSyn) or BG11 (Syc2039). Renatured proteins were either directly analyzed by bright field microscopy (Fm7001) or stained with an excess of NHS-Fluorescein and analyzed by epifluorescence microscopy. The NHS-Fluorescein dye binds primary amines and is thus incompatible with urea, which is why Fm7001 filament-like structures were visualized by bright field microscopy. Scale bars: 10 µm or (Fm7001 inlay and Slr7083) 20 µm.
Putative filament-forming proteins form filament-like structures in vivo
To investigate whether the genetic background influences the polymerization properties of the candidate proteins, we expressed GFP or YFP translational fusion constructs of the putative filament-forming CCRPs in multiple hosts: (1) E. coli, (2) their native cyanobacterium and (3) in cyanobacteria of a different morphotype or subsection. Gene expression was driven by inducible or constitutive promoters commonly used in cyanobacteria. These included Pcpc560 (for Synechocystis)46, Ptrc (for E. coli, Synechocystis and Synechococcus)47 or PpetE (for Anabaena and Fischerella)48. As a positive control for in vivo polymerization, we expressed Crescentin-GFP in Anabaena, which formed round and helical filament-like structures in the cells, thereby showing that PpetE is suitable for studying filament-forming IF-like CCRPs in Anabaena. (Supplementary Fig. 4A).
Fm7001 forms filament-like structures in vivo independent of the host
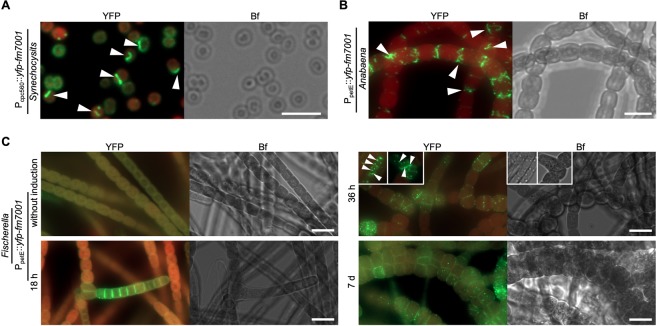
The in vivo localization of Fm7001 in Fischerella showed different results depending on the tag orientation. Only the expression of N-terminal YFP fusions of Fm7001 resulted in filament-like structures (Fig. 3 and Supplementary Fig. 4B). In Synechocystis, YFP-Fm7001 formed filament-like structures throughout the cell (Fig. 3A) while in Anabaena we observed septum-arising filament-like strings (Fig. 3B). In its host, Fischerella, YFP-Fm7001 only rarely assembled into short filament-like strings (Fig. 3C inlays). Despite of the low abundance of filament-like structures in Fischerella, induction of heterologous expression of YFP-Fm7001 induced an altered cell phenotype and trichomes seemingly divided in more than one plane resulting in a multiseriate (more than one trichome in a row) phenotype characteristic of C. fritschii. While under non-inducing conditions (i.e. in the absence of copper), Fischerella cells carrying a plasmid that expresses YFP-Fm7001 from PpetE had a WT phenotype, an altered morphotype and multiseriate growth was observed after around 4 rounds of replication (i.e. after 7 d) under inducing conditions (Fig. 3C). We also observed that, although expressed from a non-native promoter, YFP-Fm7001 was initially localized in the branching trichome, close to the branching points of putatively developing hormogonia (Fig. 3C, 18 h after induction). Those observations suggest that Fm7001 may be involved in cell shape control, branching phenotype or the development of hormogonia in Fischerella. Our attempts to generate a Fischerella ∆fm7001 mutant strain remained unsuccessful, hence the function of Fm7001 remains unknown.
Figure 3.
Host-independency for Fm7001 in vivo filamentation. Merged GFP fluorescence and chlorophyll autofluorescence (red) and bright field micrographs of (A) Synechocystis, (B) Anabaena or (C) Fischerella cells expressing YFP-Fm7001. Cells were either grown in (A,B) BG11 or (C) BG11 without copper and then induced with 0.5 µM CuSO4. (C) Micrographs were taken before induction of yfp-fm7001 expression (without induction) and 18 h, 36 h or 7 d post induction. White triangles point to selected YFP-Fm7001 filament-like strings within the cells. Notably, unlike in Anabaena and Fischerella, Fm7001-GFP induced a swollen morphotype in E. coli and a subpopulation of Synechocystis cells (Supplementary Fig. 1E). (B) Maximum intensity projection of a Z-stack. Scale bars: (A,B) 5 µm, (C) 10 µm.
Slr7083 and HmpFSyn are involved in twitching motility in Synechocystis
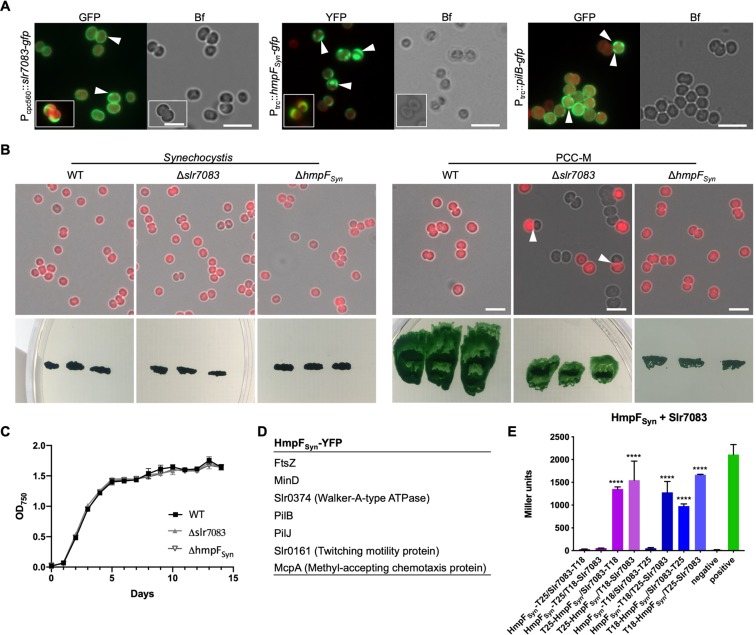
The in vivo localization of Slr7083-GFP in Synechocystis showed that it was localized to the cell periphery as well as rare focal spots spanning through the cell (Fig. 4A). We also attempted to localize Slr7083-GFP in the motile Synechocystis PCC-M substrain (hereafter PCC-M) but never obtained any successfully transformed clone, suggesting that overrepresentation of Slr7083 is deleterious for this strain. The localization of HmpFSyn-YFP in Synechocystis and PCC-M was at indistinct peripheral sites as assemblies of crescent-like shapes and rarely as focal spots spanning the cell (Fig. 4A and Supplementary Fig. 4C). Similar structures have been previously reported for the pilus ATPase PilB49. The localization of Slr7083-GFP and YFP-Slr7083 in Anabaena was at the cell periphery (Supplementary Fig. 4D). Furthermore, extended expression of YFP-Slr7083 in Anabaena altered the cellular morphology and disturbed the linear Anabaena trichome growth pattern (Supplementary Fig. 4E). In E. coli, Slr7083-GFP localized next to the cell poles (Supplementary Fig. 1E). When expressed in E. coli, HmpFSyn-GFP revealed a similar polar localization (Supplementary Fig. 1E). To further assess the role of HmpFSyn and Slr7083 in Synechocystis motility, we generated Synechocystis and PCC-M Δslr7083 and ΔhmpFSyn mutant strains. The Synechocystis Δslr7083 and ΔhmpFSyn mutants revealed no phenotypic defects compared to the WT (Fig. 4B,C). In contrast, the PCC-M Δslr7083 mutant is characterized by a decrease in twitching motility and a defect in cytokinesis (Fig. 4B). PCC-M Δslr7083 mutant cells often lacked internal chlorophyll signal entirely and failed to properly divide internal thylakoid membrane (assessed by the lack of chlorophyll autofluorescence) during cell division (Fig. 4B). Similarly, the PCC-M ΔhmpFSyn mutant lost its twitching motility (Fig. 4B) confirming previous results in Synechocystis50 and as also shown for N. punctiforme ΔhmpF mutants51. Attempts to complement the motility defect in the PCC-M ΔhmpFSyn mutant by expressing HmpFSyn-YFP from the conjugation plasmid pRL153 failed, possibly as a result of the comparably high expression of HmpFSyn-YFP from Ptrc (we note that Ptrc cannot be regulated in Synechocystis). Alternatively, the addition of a fluorescent protein to the C-terminus of HmpFSyn could have also rendered it non-functional. Notably, a C-terminal GFP fusion to HmpF from N. punctiforme was shown to be functional51, suggesting that HmpFSyn-YFP is likely also functional but complementation is prevented due to its overexpression. Additional attempts to complement the PCC-M Δslr7083 mutant never resulted in exconjugants. In order to further explore how HmpFSyn affects motility, we analyzed co-precipitated proteins of HmpFSyn-YFP expressed in Synechocystis by mass spectrometry. This revealed multiple putative interaction partners involved in motility, including a twitching motility protein (Slr0161), two methyl-accepting chemotaxis proteins (McpA and PilJ) and the type IV pilus assembly ATPase PilB (Fig. 4D). The interaction of HmpFSyn with PilB, together with their similar in vivo localization, prompted us to characterize the interaction of both proteins. For this purpose, we attempted to express PilB-GFP in Synechocystis WT, and in the ΔhmpFSyn and Δslr7083 mutants. In Synechocystis WT, PilB-GFP localized to the cell periphery and often formed crescent-like formations (reminiscent of HmpFSyn-YFP and Slr7083-GFP; Fig. 4A), confirming previous results49. However, we never observed any PilB-GFP expression in the Synechocystis or PCC-M Δslr7083 and ΔhmpFSyn mutants (Supplementary Fig. 5) and the few obtained exconjugants were never viable upon re-streaking on fresh selective plates or transfer to liquid growth medium. The similarity between our observations so far for HmpFSyn and Slr7083 led us to test for an interaction between these two proteins. Indeed, a bacterial two-hybrid assay confirmed a direct interaction between Slr7083 and HmpFSyn (Fig. 4E). Taken together, our investigation identified two Synechocystis CCRPs that are involved in cell motility and are localized to the cell periphery, often as crescent-like structures.
Figure 4.
Slr7083 and HmpFSyn are involved in twitching motility in Synechocystis. (A) Merged GFP fluorescence and chlorophyll autofluorescence (red) and bright field micrographs of Synechocystis cells expressing, Slr7083-GFP, HmpFSyn-YFP or PilB-GFP from Pcpc560 (Slr7083) or Ptrc (HmpFSyn, PilB). Expression of PilB-GFP in PCC-M resulted in the same localization pattern (data not shown). White triangles indicate focal spots and crescent-like formations. Scale bars: 5 μm. (B) Merged bright field and chlorophyll autofluorescence micrographs of motile and non-motile Synechocystis WT, Δslr7083 and ΔhmpFSyn mutant cells. Below, motility tests of three single colonies from indicated cells streaked on BG11 plates and illuminated from only one direction are shown. (C) Growth curve of Synechocystis WT, Δslr7083 and ΔhmpFSyn mutant strains grown in quadruples at standard growth conditions. OD750 values were recorded once a day for 15 d. Error bars show the standard deviation (n = 4). (D) Excerpt of interacting proteins of interest from mass spectrometry analysis of anti-GFP co-immunoprecipitations of Synechocystis cells expressing HmpFSyn-YFP from Ptrc. (E) Beta-galactosidase assays of E. coli cells co-expressing indicated translational fusion constructs of all possible pair-wise combinations of Slr7083 with HmpFSyn grown for 1 d at 30 °C. Quantity values are given in Miller Units per milligram LacZ of the mean results from three independent colonies. Error bars indicate standard deviations (n = 3). Neg: pKNT25 plasmid carrying hmpFSyn co-transformed with empty pUT18C. Pos: Zip/Zip control. Values indicated with * are significantly different from the negative control. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Dunnett’s multiple comparison test and one-way ANOVA).
All4981 is an Anabaena TPR protein that forms septal-arising filament-like structures
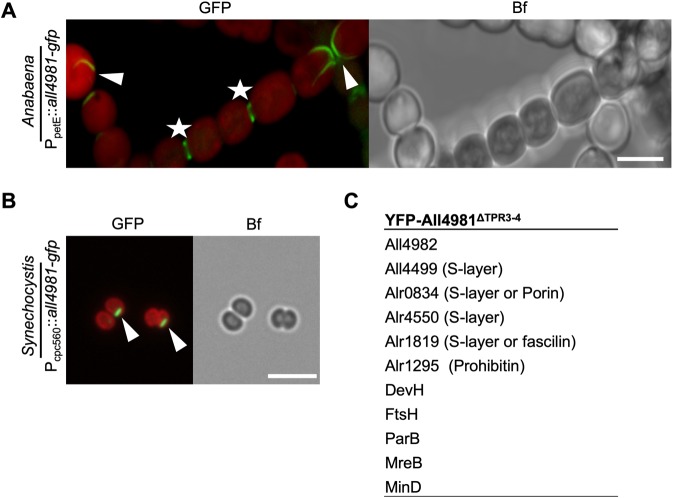
The expression of All4981-GFP in Anabaena revealed numerous filament-like structures that traversed the cell while in other cells All4981-GFP was associated with the cell septa (Fig. 5A). All4981-GFP filament-like structures also occasionally spread in a star-like pattern into the cytosol. Additionally, in freshly ruptured All4981-GFP-expressing cells, filament-like ex vivo structures assembled in the medium into an interconnected network (Supplementary Fig. 4F), resembling the in vitro polymerization pattern of All4981 (Fig. 2). We confirmed a host-independent in vivo polymerization capacity of All4981 by expressing All4981-GFP in Synechocystis, which lacks homologs to that protein (Fig. 5B). Intrigued by the septal localization, we tested for an interaction with SepJ, a septal junction protein in Anabaena7, and found weak, albeit significant physical interaction (Supplementary Fig. 3A). In addition, bacterial two-hybrid assays revealed that All4981 strongly interacted with the cell shape-determining protein MreB (Supplementary Fig. 3A). Notably, MreB has previously been shown to form similar filamentous structures in Anabaena. However, in contrast to genes in the mreBCD operon, whose overexpression induces abnormal cell morphologies15, no direct morphogenic influence was detected for All4981 in Anabaena. It is likely that All4981 is an essential protein in Anabaena as we were not able to generate an all4981 deletion strain. Initially, we accidently also created a YFP-All4981 fusion construct with a deletion of 240 bp between nt 735 and nt 975 of the all4981 CDS, resulting in a deletion of the third and fourth TPR (YFP-All4981ΔTPR3–4) leaving the remaining ORF intact. Remarkably, this fusion protein, like All4981-GFP, formed cell-traversing filament-like or spindle-like strings in Anabaena (Supplementary Fig. 4G). In contrast, full length YFP-All4981 localized to the septa between two neighboring cells but also revealed indistinct cytosolic localization (Supplementary Fig. 4G). Co-immunoprecipitation experiments following LC-MS/MS analytics from Anabaena WT expressing YFP-All4981ΔTPR3–4 revealed an association of YFP-All4981ΔTPR3–4 with ParB, MinD and MreB (Fig. 5C). Thus, All4981 might be involved in ParA/B/S-driven plasmid or chromosome segregation. The interaction with MreB agrees with the in vivo localization of YFP-All4981ΔTPR3–4 in Anabaena (Supplementary Fig. 4G) and the results from the bacterial two-hybrid assay (Supplementary Fig. 3A). Further interactions were found with a variety of putative S-layer and prohibitin-like proteins and with DevH, an essential protein for heterocyst glycolipid layer synthesis. Notably, we never observed All4981 expression in heterocysts, regardless of the fluorescence tag. All4981 also interacted with All4982, a protein encoded directly upstream of all4981, but not with All4983, which is encoded upstream of all4982 (Supplementary Fig. 1C). This observation, together with the common transcript of all4981 and all4982 (Supplementary Fig. 1C) suggests a common function of both proteins. Thus, we attempted to localize All4982 with an eCFP tag in Anabaena but could not observe a coherent localization pattern. Overall, our results demonstrate that All4981 is connected to the MreB cytoskeleton, the septal junctions and the protective S-layer. Additionally, All4981 polymerizes in vitro, in vivo and ex vivo, is likely essential for Anabaena and is thus accordingly classified as a novel cyanobacterial TPR-repeat protein with the capacity to polymerize.
Figure 5.
All4981 forms cell-traversing filament-like structures in cyanobacteria. (A,B) GFP fluorescence and merged GFP fluorescence and chlorophyll autofluorescence (red) and bright field micrographs of (A) Anabaena and (B) Synechocystis cells expressing All4981-GFP. Anabaena cells were grown in BG110 and Synechocystis cells were grown in BG11. (A) Maximum intensity projections of a Z-stack. White triangles indicate selected filament-like strings traversing through the cells. White arrows point to spindle-like All4981-GFP structures. White stars mark septal formations between two neighboring cells. Scale bars: 5 µm. (C) Excerpt of interacting proteins of interest from mass spectrometry analysis of anti-GFP co-immunoprecipitations of Anabaena cells expressing YFP-All4981ΔTPR3–4 from PpetE.
Synechococcus CCRPs are involved in cytokinesis and colony integrity
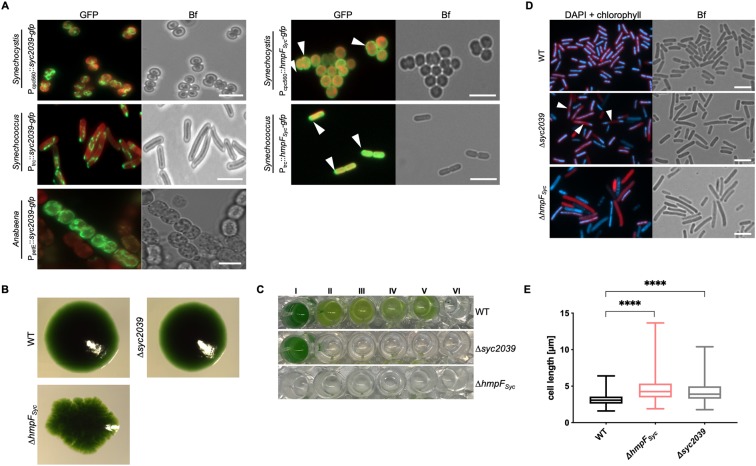
The results of the in vivo localization of a functional Syc2039-GFP fusion protein (Supplementary Fig. 6E,F) contrasted the ambiguous in vitro polymerization pattern (Fig. 2). Filament-like strings were readily observed in different cyanobacterial hosts, indicating that Syc2039 self-polymerization is independent of the host (Fig. 6A). However, Syc2039 formed different structures in each host. In Anabaena, the filament-like structures were long, curved and intertwined; in Synechocystis Syc2039-GFP appeared as spindle-like structures and in Synechococcus filament-like structures were long, sometimes helical and often aligned with or in close proximity to the cell envelope (Fig. 6A). A similar helical or cell periphery-aligned localization pattern was also observed in E. coli (Supplementary Fig. 1E). In Synechocystis and Synechococcus HmpFSyc-GFP localized as spots at the cell periphery, while in E. coli it seemingly coated the entire cell envelope (Fig. 6A, Supplementary Fig. 1E). HmpFSyc-GFP failed to be expressed in Anabaena, suggesting that (over-)expression of this protein has a negative impact on that organism. Using double homologous gene replacement, we generated a Synechococcus ∆syc2039 mutant strain and a non-segregated Synechococcus ∆hmpFSyc mutant strain (Supplementary Fig. 6A–C). The non-segregated nature of the ∆hmpFSyc mutant suggests that this gene performs an essential cellular function and cannot be fully deleted. Colony integrity of the ∆syc2039 mutant was unaltered while the ∆hmpFSyc mutant was characterized by apparent changes in colony morphology (Fig. 6B), which were lost upon growth on non-selective plates (Supplementary Fig. 6D). Additionally, both mutants presented an impairment in liquid culture growth: the ∆syc2039 mutant grew in standard BG11 medium but failed to grow upon addition of several osmotic stressors, whereas the ∆hmpFSyc mutant failed to grow in liquid culture entirely (Fig. 6C). Spot assays confirmed a decreased viability of the ∆hmpFSyc mutant and showed that it is highly sensitive to Proteinase K but unaffected by lysozyme (Supplementary Fig. 7A). These cell wall defects, together with the in vitro cell shape-like polymerization pattern suggest that HmpFSyc might form a protective and protease-resistant proteinaceous layer below the cytoplasmic membrane. This possibility would also be in concert with the distorted colony morphology of the non-segregated ∆hmpFSyc mutant strain. The Δsyc2039 mutant was unaffected by cell wall and membrane destabilizers (Supplementary Fig. 7B). To investigate the role of these proteins in cell division, we stained intracellular DNA with DAPI and localization of FtsZ was detected by immunofluorescence in Synechococcus WT and both mutant strains. A proportion of ∆syc2039 mutant cells exhibited a segregated DNA distribution either to both cell poles or to just one pole (Fig. 6D). Furthermore, some cells of both mutants lacked any discernible intracellular DNA or perceptible chlorophyll signal and were elongated compared to the WT (Fig. 6D,E). The WT phenotype of the ∆syc2039 mutant could be rescued by insertion of Ptrc::syc2039-gfp or Psyc2039::syc2039 into the neutral NS152 locus (Supplementary Fig. 6E,F). Although both mutant cells were elongated compared to WT cells (Fig. 6E), the intracellular localization of FtsZ was unaffected (Supplementary Fig. 7C). And despite the defect in cytokinesis, the Δsyc2039 mutant strain revealed similar liquid culture growth properties as the WT (Supplementary Fig. 7D). Taken together, Syc2039 forms abundant filament-like networks in vivo and is involved in cytokinesis or cell cycle control. We could further show that hmpFSyc is an essential gene important for cytokinesis, cellular integrity and colony formation, implicating structural functions.
Figure 6.
Synechococcus CCRPs affect cytokinesis and cellular integrity. (A) Merged GFP fluorescence and chlorophyll autofluorescence (red) and bright field micrographs of Synechocystis, Synechococcus and Anabaena cells expressing Syc2039-GFP or HmpFSyc-GFP from Pcpc560, PpetE or Ptrc. Synechocystis cells were grown in BG11, Anabaena cells were grown in BG110 supplemented with 0.25 µM CuSO4 for 1 day, and Synechococcus cells were grown on BG11 plates supplemented with 0.01 mM (Syc2039) or 1 mM (HmpFSyc) IPTG. Micrographs of Synechococcus and Anabaena cells expressing Syc2039-GFP are maximum intensity projections of a Z-stack. White triangles indicate HmpFSyc-GFP spots. Attempts to translationally fuse a YFP-tag to the N-terminus of Syc2039 were unsuccessful, possibly due to the transmembrane domain predicted to the Syc2039 N-terminus (Supplementary Table 1). (B) Colony formation of Synechococcus WT and mutant strains on BG11 plates. (C) Cell viability of Synechococcus WT and mutant strains grown in (I) BG11 or BG11 supplemented with (II) 5 mM glucose, (III) 200 mM glucose, (IV) 2 mM NH4Cl, (V) 200 mM maltose or (VI) 500 mM NaCl. (D) Merged DAPI fluorescence and chlorophyll autofluorescence (red) and bright field micrographs of Synechococcus WT and mutant strains grown on BG11 plates and stained with 10 μg ml−1 DAPI. White triangles indicate non-dividing cells revealing inhomogeneous DNA placement. (E) Cell length of Synechococcus WT (n = 648), non-segregated ΔhmpFSyc (n = 417) and Δsyc2039 (n = 711) mutant cells. Values indicated with * are significantly different from the WT. ****P < 0.0001 (one-way ANOVA, using Turkey’s multiple comparison test). Scale bars: 5 µm.
Discussion
Earlier studies suggested that there is likely a broad spectrum of coiled-coil-rich and rod-domain containing proteins with IF-like function in prokaryotes29. And indeed, reports on such proteins followed with the discovery of Scy (in Streptomyces coelicolor) and several CCRPs from Helicobacter pylori30,31,34,35. Here we further investigated the presence and function of CCRPs with filament-forming IF-like properties in prokaryotes, by predicting and evaluating CCPRs in cyanobacteria. Our in vitro polymerization assay allowed for a rapid detection of proteins with the potential to form filament-like structures in vitro using fluorescence microscopy. However, we note that for conclusive proof of in vitro filaments, higher resolution microscopy like electron microscopy would be necessary and will be addressed in future studies. The observed protein filament-like lengths were in the range of previously described in vitro filaments of FtsZ53 and of the human prion protein in its amyloid form54 that were obtained by a similar experimental procedure.
Our results show that Fm7001 assembles into polymers in vitro upon renaturation from urea as well as in vivo, and that this protein has an impact on cellular and trichome morphology, thereby fulfilling major IF criteria40,55. Consequently, we propose that Fm7001 constitutes a novel filament-forming CCRP specific to multicellular, cell-differentiating and branching cyanobacteria. The floating Fm7001 polymer sheet in high molar urea (i.e. 4.5 M urea) indicates an exceptionally high self-association capacity of Fm7001. In comparison, the eukaryotic IF protein Vimentin exists only as tetramers in 5 M urea23. In vivo localization experiments revealed an essential role of the Fm7001 C-terminus for polymerization, which is a common observation for known prokaryotic filament-forming proteins, including MreB56, Crescentin25 as well as eukaryotic IF proteins23,57–60. Additionally, the assigned structural similarities of Fm7001 with the acetyl-CoA-carboxylase may provide further support for the theory that filament-forming proteins originated from metabolic enzymes that obtained polymerization features61. Notwithstanding, the metabolic activity of Fm7001 was not evaluated in our study hence its presumed enzymatic activity remains to be tested. Additionally, so far, no sufficient genome modification systems exist for Fischerella62,63, as such a precise analysis of the function of Fm7001 is currently not possible.
Several prokaryotic tubulin-like and actin-like cytoskeletal proteins, such as ParM and TubZ, are known to be encoded on plasmids or on bacteriophages9,64. In Synechocystis, slr7083 is encoded on the large toxin-antitoxin defense plasmid (pSYSA)65, thus it adds another protein to the list of those CCRPs with the property to assemble into filament-like structures carried by an autonomously replicating genetic element. Preliminarily, we suspected that Slr7083 has a role in plasmid-segregation similar to ParM. However, Slr7083 showed no indications of dynamic properties, which would be indispensable for a plasmid segregation mechanism. As Slr7083 was ectopically overexpressed, this observation has to be taken with a grain of salt as overexpression could interrupt the protein dynamics. Furthermore, unlike ParM66, Slr7083 did not localize in a spindle-like pattern in vivo and was only expressed at later growth phases, which is contradictory to a possible involvement in the cell cycle. In contrast, the polymers formed by Slr7083 in vitro and in vivo rather suggest that it could form a proteinaceous layer below the cytoplasmic membrane. Notably, Slr7083 in vitro structures resemble the nuclear lamina formed by nuclear lamins and FilP lace-like in vitro filaments29,67,68. It is thus conceivable that Slr7083 has a role in cellular stiffness as well as rigidity and mediates mechanical cell stabilization. However, restriction of transcription to only a comparably short period of the culture growth phase challenges the idea of a cell-stabilizing function for Slr7083. In contrast, cell motility in Synechocystis seems to be partially regulated by Slr7083, reminiscent of the role of the actin cytoskeleton in eukaryotes.
The role of Slr7083 in cell motility is possibly mediated by means of its interaction with HmpFSyn, which has been previously shown to be essential for twitching motility in Synechocystis50. So far it is unknown how photoreceptors transduce the perceived light stimuli to the motility apparatus in Synechocystis ultimately resulting in phototactic movements69. It is tenable to hypothesize that HmpFSyn might constitute the missing link between the two systems, possibly in combination with Slr7083. This hypothesis is supported by the physical interaction of HmpFSyn with PilB and the in vivo localization of HmpFSyn that is similar to that observed for PilB49. Direct interaction of HmpFSyn and PilB provides further support for the model from Cho et al.51 according to which, a direct interaction of HmpF with PilB could activate the pilus extension by the type IV system. In N. punctiforme, HmpF was found to dynamically localize to the leading or lagging pole of hormogonia depending on the light intensity and in a dependent manner with the type IV apparatus and the Ptx system51. In Synechocystis, localization of PilB was shown to correlate with the direction of movement49 and to be dependent on the direction of incoming light in a complex with other pili protein70. Given its similar localization with PilB, we hypothesize that like HmpF from N. punctiforme, HmpFSyn may be characterized by dynamic light-dependent properties in a complex with the type IV pilus system. A similar motility complex was observed in Pseudomonas aeruginosa, where FimL (a proposed scaffolding protein with a weakly predicted coiled-coil) was shown to connect the chemosensory receptor system to the type IV pili apparatus, regulating the chemotactic and virulence pathways71. In eukaryotes, cellular motility is strongly dependent on cytoskeletal proteins72, thus it is possible that filament-forming proteins are also key factors for cell locomotion in prokaryotes. Although IFs do not directly participate in cell motility in eukaryotes73, an adaptation of filament-forming CCRPs in prokaryotes for this task is conceivable. Bactofilins constitute a separate class of prokaryotic-specific polymerizing proteins and were proposed to be involved in coordinated motility in C. crescentus43. Additionally, the filament-forming CCRP AglZ from Myxococcus xanthus was previously shown to govern gliding motility together with a multi-protein complex that also involves the MreB cytoskeleton74,75. Notably, the interaction of HmpFSyn with FtsZ and MinD, which are essential cellular factors involved in cell division, cytokinesis and Z-ring placement76, might indicate that HmpFSyn function is not restricted to motility. Although thylakoid membranes pose no physical barrier for proper Min system oscillation in Synechococcus, suggesting no direct interaction, MinD is part of the thylakoid fractions in Synechocystis77. In concert with its localization to the cell envelope in Synechocystis, it would be intriguing to investigate a potential link between HmpFSyn and thylakoid development and cytokinesis during cell division. Despite their different cellular functions and in vitro polymerization properties, the homologous proteins HmpFSyn, HmpFSyc and HmpFAna retained the ability to cross-interact (Supplementary Fig. 3A). Further studies may focus on identifying the protein domains that mediate this interaction, likely residing within the highly conserved amino acid sequence region in this homologous protein family (Supplementary Fig. 3B). These regions are likely important for an interaction with species-specific proteins that lead to their species-specific cellular function.
TPR proteins are known to mediate protein-protein interactions and can assemble into multimers, but their ability to polymerize into filaments has not been described so far78. Nonetheless, All4981 polymerized in vitro and formed filament-like structures in vivo in all tested hosts. Additionally, it forms extracellular filament-like structures and is presumably an essential protein in Anabaena. These observations suggest that All4981 is a bona fide prokaryotic TPR protein with the property of filament-like assembly. The association of All4981 with MreB, FtsZ-regulators, the S-layer and SepJ indicates that it might function as a bridge that connects the shape-determinants outside of the cell wall and inside of the cytoplasmic membrane to the sites of cell-cell connections (i.e. septal junctions).
Considering the presence of an N-terminal transmembrane domain and the lack of clear in vitro filaments, it is unlikely that Syc2039 constitutes a genuine filament-forming protein. Nonetheless, the highly abundant filament-like network structures formed in all tested bacterial hosts suggests that Syc2039 is associated with cytoskeletal structures. Specifically, the elongated phenotype and the disturbed cytokinesis in the Synechococcus Δsyc2039 mutant and the non-segregated ΔhmpFSyc mutant suggest an association with the FtsZ-driven divisome. Direct interaction with FtsZ or MreB could not be shown, as such, future studies will likely attempt to unravel the presumed connection of the Synechococcus CCRPs to those two major cytoskeletal systems. Notably, besides its cytokinetic defect, the Δsyc2039 mutant showed growth characteristics like the WT, suggesting that feedback mechanisms between cytokinesis and cell division are disturbed in the Δsyc2039 mutant.
Our results reveal two novel filament-forming CCRPs - Fm7001 and All4981 - from different cyanobacterial subsections and morphotypes (Fig. 7). Our study thus extends the spectrum of known filament-forming CCRPs in prokaryotes and expands the set of functional properties associated with IF-like proteins in prokaryotes. As previously suggested29, we demonstrate that the sole observation of coiled-coil-rich regions within a protein sequence cannot be regarded as a sole predictor of protein polymerization, hence identification of novel filament-forming proteins requires additional in vitro and in vivo assays. The cyanobacterial CCRPs we report here, like other bacterial CCRPs25,29,30,34,35,79 and eukaryotic IFs80 are essential cellular components (All4981), are important for cell shape determination (Fm7001, HmpFSyc and Syc2039), mediate cellular motility (Slr7083 and HmpFSyn), DNA segregation (HmpFSyc and Syc2039) and colony integrity (HmpFSyc). Our study thus strengthens the perception that like eukaryotes, prokaryotes require organized internal complexes and even microcompartments to maintain cell shape, size and proper cell function and highlights the usefulness of polymerized proteinaceous structures for cellular processes. Remarkably, some of the identified CCRPs were highly conserved among all cyanobacterial morphotypes, suggesting that their function is conserved. Future studies are required in order to evaluate the functional conservation of homologous proteins in different cyanobacterial species and morphotypes. On the other hand, Syc2039 and Slr7083 are highly strain specific, possibly performing a function that is adapted to the very needs of their hosts. Similarly to the eukaryotic cytolinker proteins81,82, cyanobacterial CCRPs were often associated with other cytoskeletal systems (MreB, FtsZ and other filament-forming CCRPs) and sites of cell-cell connections (i.e. SepJ), which demonstrates the necessity for those structures to be in a constant interplay even in comparably small cells. The discovery of filament-forming CCRPs with different levels of conservation in various cyanobacterial morphotypes thus opens up new avenues of research on their contribution to cyanobacterial morphological diversity.
Figure 7.
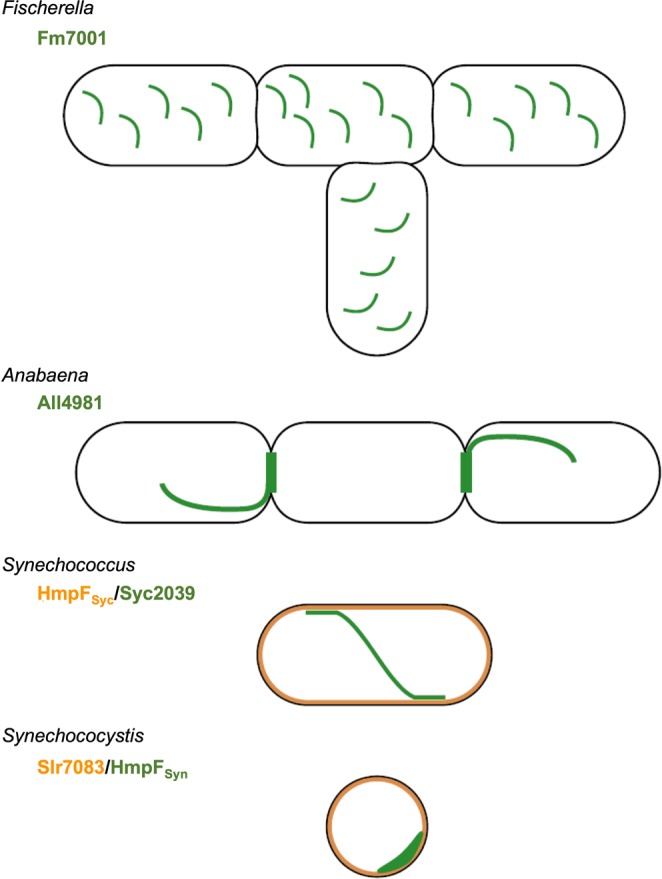
Cyanobacterial CCRP systems. Schematic models for the in vivo localization of cyanobacterial CCRPs in their respective hosts. Fm7001 forms filament-like strings in Fischerella. In Anabaena, All4981 assembles into pole-arising filament-like structures that traverse through the cell or forms septal-localized bridge-like formations. Syc2039, either independently of other Synechococcus proteins, or in direct cooperation with other putative filamentous proteins, forms long and sometimes helical strings that are often aligned with or in close proximity to the cell periphery. In Synechococcus, HmpFSyc likely forms a protective proteinaceous layer below the cytoplasmic membrane. In Synechocystis, HmpFSyn forms crescent-like structures while Slr7083 seemingly underlies the cytoplasmic membrane. Both localization types were also observed for PilB, suggesting a cooperative function.
Material and Methods
Data and CCRP prediction
The cyanobacterial protein families were constructed from completely sequenced genomes available in RefSeq database83 (V. May 2016; Supplementary File 3). For the construction of protein families, at the first stage, all protein sequences annotated in the genomes were blasted all-against-all using stand-alone BLAST84 (V. 2.2.26). Protein sequence pairs that were found as reciprocal best BLAST85 hits (rBBHs) with a threshold of E-value ≤ 1 × 10−5 were further compared by global alignment using needle86 (EMBOSS package, V. 6.6.0.0). Sequence pairs having ≥30% identical amino acids were clustered into protein families using the Markov clustering algorithm (MCL)87 (V. 12–135) with the default parameters. For the CCRPs prediction, 1,535 protein sequences containing non-standard amino acids were discarded. Coiled-coil regions in protein sequences were predicted using PEPCOIL86 (EMBOSS package, V. 6.6.0.0). The algorithm was executed with a window size of 21 and the threshold for amino acids in coiled-coil conformation was set to ≥80 amino acid residues similarly as described previously29. Statistical tests were performed with MatLab©. For the comparison of CCRPs proportion, the compared groups included: (1) SynProCya group, (2) unicellular cyanobacteria, (3) unicellular cyanobacteria that divide in more than one plane, and (4) multicellular cyanobacteria. Identification of conserved amino acid domains within cyanobacterial CCRP homologs (HmpFSyn (Slr1301) and HmpFSyc (Synpcc7942_1139)) was done using MULTALIGN88.
Protein candidates were further manually examined with online available bioinformatic tools (NCBI Conserved Domain (CD) Search89, TMHMM Server90 (V. 2.0), PSIPRED91, PSORTb92 (V. 3.0), I-TASSER93. CCRPs exhibiting similar predictions to known IF and IF-like proteins like CreS, FilP, Vimentin, Desmin or Keratin were selected, and proteins predicted to be involved in other cellular processes were excluded.
Bacterial strains and growth conditions
Fischerella, Anabaena and Synechocystis were obtained from the Pasteur Culture Collection (PCC) of cyanobacteria (France). Synechococcus was a gift from Martin Hagemann (University Rostock). Glucose-tolerant motile Synechocystis PCC-M substrain was a gift from Annegret Wilde (University Freiburg). Cells were grown photoautotropically in BG11 or without combined nitrogen (BG110) at a 16 h/8 h light/dark regime (Fischerella) or at constant light (Anabaena, Synechococcus and Synechocystis) with a light intensity of 20 µmol m−2 s−1. When appropriate, 50 µg ml−1 kanamycin (Km), 2.5 µg ml−1 spectinomycin (Sp), 2.5 µg ml−1 streptomycin (Sm) or 30 µg ml−1 neomycin (Nm) was added. Non-segregated ΔhmpFSyc cells were always grown in the presence of Km. E. coli strains DH5α, DH5αMCR, XL1-blue and HB101 were used for cloning and conjugation by triparental mating. BTH101 was used for BACTH assays and BL21 (DE3) was used for expression of His- and GFP-tagged proteins in E. coli. All E. coli strains (Supplementary Table 2) were grown in LB medium containing the appropriate antibiotics at standard concentrations.
Plasmid and strain construction
All plasmids employed in this study were either generated by using standard restriction enzyme-base cloning procedures or using Gibson assembly94. A detailed description of the cloning strategies for the respective plasmids is available upon request from the authors. All primers, plasmids and strains employed or generated in this study are listed in Supplementary Tables 2–5. GFP, YFP and eCFP protein tags were used as reporter proteins and His6 tag was used for protein affinity purification. For gene replacement mutants, homologous flanks for double homologous recombination comprised 1000 bp upstream and downstream of the gene of interest. Mutant strains harboring gene replacements with antibiotic resistance cassettes (nptII95 or CS.396) were verified by colony PCR testing for absence of gene of interest using primers #129/#130 for Δslr7083, primers #168/#169 for ΔhmpFSyn, primers #146/#147 for Δsyc2039 or primers #161/#162 for ΔhmpFSyc. We also attempted to generate gene replacement mutants for all4981 and fm7001 but remained unsuccessful.
Transformation of cyanobacteria
Transformation of Synechococcus was achieved by natural transformation as described previously97 and transformation of Synechocystis was accomplished by natural transformation or by conjugation as described previously98,99. Anabaena and Fischerella were transformed by conjugation as previously described62,99. Ex-conjugant colonies from Synechococcus and Synechocystis carrying gene replacements were re-streaked three to four times and absence of genes of interest was verified by colony PCR. Transformation of sonicated (fragmented) and NaCl-treated Fischerella cells followed by the conjugational method99 was also feasible for Fischerella, albeit with a lower transformation frequency.
Phenotypic characterization of the mutant strains
Defects in cell viability were evaluated by spot assays adapted from Dörrich et al. (2014)100. Wild type and mutant strains from liquid cultures or BG11 plates were adjusted to an OD750 of about 0.4 in liquid BG11 liquid. Next, 5 µl of cells were spotted in triplicates onto BG11 plates or BG11 plates supplemented with Proteinase K or lysozyme at indicated concentrations in 10-fold serial dilutions and incubated under standard growth conditions until no further colonies arose in the highest dilution.
Growth defects were assessed with growth curves. For this, cells were grown in liquid BG11 medium, washed three times by centrifugation (6500 × g, RT, 3 min) in BG11, adjusted to an OD750 of 0.1 and then grown in triplicates or quadruples at standard growth conditions in 15 ml culture volumes. OD750 values were recorded every 24 h.
Cell length of Synechococcus WT, mutant strains and mutant complementation strains was measured using the line tool from the imaging software Fiji and its plugin microbeJ.
Cell wall integrity defects were evaluated by testing the influence of osmotic factors on cell growth. Synechococcus WT and mutant strains were grown on BG11 agar plates, transferred to BG11 liquid medium and grown under standard growth conditions with or without 5 mM glucose, 200 mM glucose, 2 mM NH4Cl, 200 mM maltose or 500 mM NaCl.
To evaluate the motility of Synechocystis and PCC-M WT and mutant strains, three single colonies of the respective strain were streaked on a line on a BG11 growth plate. Growth plates were then placed into the standard culture incubator for 10 d with illumination limited from one direction.
Protein purification and in vitro filamentation assays
For protein purification, E. coli BL21 (DE3) cells carrying His6-tagged protein candidates were grown in overnight cultures at 37 °C and 250 rpm. The next day, overnight cultures were diluted 1:40 in the same medium and grown at 37 °C until they reached an OD600 of 0.5–0.6. Protein expression was induced with 0.5 mM IPTG for 3–4 h at 37 °C and 250 rpm. Afterwards, cell suspensions of 50 ml aliquots were harvested by centrifugation, washed once in PBS and stored at −80 °C until further use. For in vitro filamentation assays, cell pellets were resuspended in urea lysis buffer (ULB: 50 mM NaH2PO4, 300 mM NaCl, 25 mM imidazole, 6 M urea; pH 8.0) and lysed in a Precellys® 24 homogenizer (3 × 6500 rpm for 30 s) using the 2 ml microorganism lysis kit (VK01; Bertin) or self-packed Precellys tubes with 0.1 mm glass beads. The resulting cell debris was pelleted by centrifugation at 21,000 × g (30 min, 4 °C) and the supernatant was incubated with 1 ml HisPur™ Ni-NTA resin (Thermo Fischer Scientific) for 1 h at 4 °C in an overhead rotator. The resin was washed 5 times with 4x resin-bed volumes ULB and eluted in urea elution buffer (UEB: ULB supplemented with 225 mM imidazole). Total protein concentration was measured using the Qubit® 3.0 Fluorometer (Thermo Fischer Scientific) and generally adjusted to 0.5–1.0 mg ml−1 before dialysis. Purified proteins were dialyzed overnight against polymerization buffer (PLB: 50 mM PIPES, 100 mM KCl, pH 7.0; HLB: 25 mM HEPES, 150 mM NaCl, pH 7.4) at 18 °C and 180 rpm with three bath changes using a Slide-A-Lyzer™ MINI Dialysis Device (10 K MWCO, 0.5 ml or 2 ml; Thermo Fischer Scientific). Purified proteins were stained with 0.005 mg NHS-Fluorescein (Thermo Fischer Scientific) per 1 ml protein dialysate and in vitro filamentation was analyzed by epifluorescence microscopy.
For Fm7001-His6, proteins were slowly dialyzed against 2 mM Tris-HCl, 4.5 M urea, pH 7.5 (18 °C, 200 rpm) decreasing 0.5 M urea every 2 h (from 6 M to 4.5 M urea). The resulting floating filament-like web was then analyzed by bright field microscopy.
Syc2039-His6 failed to be expressed in E. coli BL21 (DE3). To bypass this, Syc2039-GFP-His, under the control of an IPTG-inducible Ptrc, was inserted into a neutral locus of Synechococcus. Cells were grown to an OD750 of 0.8 and protein expression was induced with 0.05 mM IPTG for 3 d. Induced cells were harvested and washed with PBS by centrifugation (4800 × g, 4 °C, 10 min) and stored at −80 °C. Protein purification, dialysis and labeling was then performed as described above with the exception that BG11 growth medium was used as dialysate.
Co-immunoprecipitation
For co-immunoprecipitations of fluorescently tagged CCRP candidates, cyanobacterial strains expressing YFP-All4981 or HmpFSyn-YFP were grown in BG11 or BG110 liquid medium. About 20–30 ml of the respective culture was pelleted by centrifugation (4800 × g, 10 min, RT), cells were washed twice by centrifugation (4800 × g, 10 min, RT) with 40 ml PBS and then resuspended in 1 ml lysis buffer (PBS-N: PBS supplemented with 1% NP-40) supplemented with protease inhibitor cocktail (PIC; cOmplete™, EDTA-free Protease Inhibitor Cocktail, Sigma-Aldrich). Cells were lysed using the VK05 lysis kit (Bertin) in a Precellys® 24 homogenizer (3 strokes for 30 seconds at 6500 rpm) and cell debris was pelleted by centrifugation (30 min, 21,100 × g, 4 °C). 50 µl μMACS anti-GFP MicroBeads (Miltenyi Biotec) was added to the resulting cell-free supernatant and incubated for 1 h at 4 °C with mild rotation. Afterwards, the sample was loaded onto µColumns (Miltenyl Biotec), washed two times with 1 ml lysis buffer and eluted in 50 µl elution Buffer (50 mM Tris HCl pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromphenol blue, 10% glycerol; Miltenyl Biotec). Until further use, samples were stored at −80 °C. Proteins were identified by mass spectrometry. A detailed protocol of the mass spectrometry analysis is available upon request from the authors.
Immunofluorescence
The localization of FtsZ in Synechococcus WT and mutant strains was evaluated by immunofluorescence using a modified protocol from Heinz et al.101. In contrast, cells were lysed in 50 mM Tris-HCl pH 7.4, 10 mM EDTA and 0.2 mg ml−1 lysozyme for 30 min at 37 °C and samples were blocked in 1x Roti®-ImmunoBlock (Carl Roth) in PBS supplemented with 0.05% Tween 20. Samples were incubated with rabbit anti-FtsZ primary antibody (Agrisera; raised against Anabaena FtsZ; 1:250 diluted) in blocking buffer followed by incubation with 7.5 µg ml−1 Alexa Fluor 488-conjugated goat anti-rabbit IgG (H + L) secondary antibody (Thermo Fischer Scientific) in blocking buffer. Before microscopy, cells were stained with 10 µg ml−1 DAPI (final concentration) in PBS.
Brightfield and fluorescence microscopy analysis
Bacterial strains grown in liquid culture were either directly applied to a microscope slide or previously immobilized on a 2% low-melting agarose in PBS agarose pad and air dried before microscopic analysis. Epifluorescence microscopy was performed using an Axio Imager.M2 light microscope (Carl Zeiss) equipped with Plan-Apochromat 63×/1.40 Oil M27 objective and the AxioCam MR R3 imaging device (Carl Zeiss). GFP, Alexa Fluor 488, eCFP and YFP fluorescence was visualized using filter set 38 (Carl Zeiss; excitation: 470/40 nm band pass (BP) filter; emission: 525/50 nm BP). Chlorophyll auto-fluorescence was recorded using filter set 15 (Carl Zeiss; excitation: 546/12 nm BP; emission: 590 nm long pass). When applicable, cells were previously incubated in the dark at RT for about 5 min with 10 µg ml−1 DAPI in PBS to stain intracellular DNA. For visualization of DAPI fluorescence filter set 49 (Carl Zeiss; excitation: G 365 nm; emission: 455/50 nm) was employed. E. coli BL21 (DE3) cells expressing C-terminally GFP-tagged protein candidates were grown over night in LB and then diluted 1:40 in the same medium the following day. Cells were grown for 2 h at 37 °C, briefly acclimated to 20 °C for 10 min and induced with 0.05 mM IPTG at 20 °C. Protein localization of GFP/YFP-tagged proteins was then observed after indicated time points of cells immobilized on an agarose pad.
Statistical analysis
Beta-galactosidase values were measured in triplicates from three independent colonies and significant differences compared to WT were determined by a one-way ANOVA using Dunnett’s multiple comparison test. For statistical evaluation of Synechococcus WT and mutant cell length, a one-way ANOVA using Turkey’s multiple comparison test was used. Significance levels are the same as for the beta-galactosidase assay. Statistical tests were performed with the GraphPad Prims 8.0.0 software. Significance levels are indicated by stars (*) and correspond to: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
RNA isolation and RT-PCR
Total RNA was isolated from 10 ml culture using either the Direct-zol™ RNA MiniPrep Kit (Zymo Research; Synechocystis, Synechococcus and Anabaena) according to the manufacturer’s instructions or the Plant RNA Reagent (Thermo Fischer Scientific; Anabaena, Fischerella and Synechocystis). For RNA isolation using the Plant RNA Reagent, a modified protocol was employed. To this end, cells were pelleted by centrifugation (4800 × g, 10 min, 4 °C) and the supernatant was discarded. The pellet was resuspended in 0.5 ml of Plant RNA Reagent und lysed in a Precellys® 24 homogenizer (Bertin) with 3 strokes at 6500 rpm for 30 s in 2 ml soil grinding (SK38) or tough microorganism (VK05) lysis tubes (Bertin). RNA was then isolated according to the manufacturer’s instructions. Isolated RNA was treated with DNA-free™ Kit (2 units rDNAs/reaction; Thermo Fischer Scientific) and 1 µg (Fischerella, Synechocystis and Synechococcus) or 200 ng (Anabaena) RNA was reverse transcribed using the Maxima™ H Minus cDNA Synthesis Master Mix (with dsDNase; Thermo Fischer Scientific, for Fischerella, Synechocystis and Synechococcus) or the qScript™ cDNA Synthesis Kit (Quanta Biosciences, for Anabaena). RT-PCR of cDNA samples for fm7001, ftsZ, slr7083, rnpB, hmpFSyn, syc2039, hmpFSyc, all4981, all4981 + all4982 and all4981 + all4983 was done using primer pairs #1/#2, #3/#4, #5/#6, #7/#8, #9/#10, #11/#12, #13/#14, #15/#16, #17/#15 and #18/#15, respectively.
Bacterial two hybrid assays
In this study, the BACTH system (Euromedex) was employed. Gene candidates were cloned into the expression vectors pKNT25, pKT25, pUT18 and pUT18C by GIBSON assembly, thereby generating C and N-terminal translational fusions to the T25 or T18 subunit. Chemically competent E. coli BTH101 (Δcya) cells were co-transformed with 5 ng of the indicated plasmids, plated onto LB plates supplemented with 200 µg ml−1X-gal, 0.5 mM IPTG, Amp, Km and grown at 30 °C for 24–36 h. Interactions were quantified by beta-galactosidase assays from three colonies for each combination according to the protocol described by Euromedex or in a 96 well format102. For this aim, cultures were either grown over night at 30 °C or for 2 d at 20 °C in LB Amp, Km, 0.5 mM IPTG and interaction strength of the investigated proteins was by quantified by beta-galactosidase-mediated hydrolyzation of ONPG (ortho-Nitrophenyl-β-galactoside), which is then recorded in Miller units103.
Supplementary information
Acknowledgements
We thank Katrin Schumann, Myriam Barz, Lisa Stuckenschneider, Lisa-Marie Philipp and Marius Lasse Theune for their assistance in the experimental work. Furthermore, we thank Martin Thanbichler and Daniela Kiekebusch (both from Philipps University, Marburg, Germany) for their support with mass spectrometry analysis. The study was supported by the German science foundation (DFG) (Grant No. STU513/2-1 awarded to K.S.).
Author contributions
B.L.S. and K.S. designed the study. B.L.S. established and performed the experimental work with contributions from J.W. C.W. and T.D. performed the comparative genomics analysis. A.O.H. analyzed protein samples by mass spectrometry. B.L.S., T.D. and K.S. drafted the manuscript with contributions from all coauthors.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Benjamin L. Springstein, Email: benjamin_springstein@hms.harvard.edu
Karina Stucken, Email: kstucken@userena.cl.
Supplementary information
is available for this paper at 10.1038/s41598-020-58726-9.
References
- 1.Rippka R, Stanier RY, Deruelles J, Herdman M, Waterbury JB. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 2.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2010;2:1–19. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SP, Montgomery BL. Determining cell shape: adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol. 2011;19:278–285. doi: 10.1016/j.tim.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Wilk L, et al. Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cell. Microbiol. 2011;13:1744–1754. doi: 10.1111/j.1462-5822.2011.01655.x. [DOI] [PubMed] [Google Scholar]
- 5.Herrero A, Stavans J, Flores E. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol. Rev. 2016;40:831–854. doi: 10.1093/femsre/fuw029. [DOI] [PubMed] [Google Scholar]
- 6.Weiss GL, Kieninger A-K, Maldener I, Forchhammer K, Pilhofer M. Structure and Function of a Bacterial Gap Junction Analog. Cell. 2019;178:374–384.e15. doi: 10.1016/j.cell.2019.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores E, et al. Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2007;189:3884–3890. doi: 10.1128/JB.00085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayar AS, Yamaura H, Rajagopalan R, Risser DD, Callahan SM. FraG is necessary for filament integrity and heterocyst maturation in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology. 2007;153:601–607. doi: 10.1099/mic.0.2006/002535-0. [DOI] [PubMed] [Google Scholar]
- 9.Wagstaff, J. & Löwe, J. Prokaryotic cytoskeletons: protein filaments organizing small cells. Nat. Rev. Microbiol. 10.1038/nrmicro.2017.153 (2018). [DOI] [PubMed]
- 10.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C-CC, Hugenin S, Friry A, Huguenin S, Friry A. Analysis of genes encoding the cell division protein FtsZ and a glutathione synthetase homologue in the cyanobacterium Anabaena sp. PCC 7120. Res. Microbiol. 1995;146:445–455. doi: 10.1016/0923-2508(96)80290-7. [DOI] [PubMed] [Google Scholar]
- 12.Lopes Pinto F, Erasmie S, Blikstad C, Lindblad P, Oliveira P. FtsZ degradation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Plant Physiol. 2011;168:1934–1942. doi: 10.1016/j.jplph.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 13.Ramos-León F, Mariscal V, Frías JE, Flores E, Herrero A. Divisome-dependent subcellular localization of cell-cell joining protein SepJ in the filamentous cyanobacterium Anabaena. Mol. Microbiol. 2015;96:566–580. doi: 10.1111/mmi.12956. [DOI] [PubMed] [Google Scholar]
- 14.Jones LJF, Carballido-López R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/S0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 15.Hu B, Yang G, Zhao W, Zhang Y, Zhao J. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 2007;63:1640–1652. doi: 10.1111/j.1365-2958.2007.05618.x. [DOI] [PubMed] [Google Scholar]
- 16.Jain IH, Vijayan V, O’Shea EK. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2012;109:13638–43. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez A, Riley KW, Harwood TV, Zuniga EG, Risser DD. A Tripartite, Hierarchical Sigma Factor Cascade Promotes Hormogonium Development in the Filamentous Cyanobacterium Nostoc punctiforme. mSphere. 2019;4:e00231–19. doi: 10.1128/mSphere.00231-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin L, Thanbichler M. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton. 2013;70:409–423. doi: 10.1002/cm.21126. [DOI] [PubMed] [Google Scholar]
- 20.Löwe J, Amos LA. Evolution of cytomotive filaments: The cytoskeleton from prokaryotes to eukaryotes. Int. J. Biochem. Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Shoeman RL, Traub P. Assembly of Intermediate Filaments. Bioessays. 1993;15:605–611. doi: 10.1002/bies.950150906. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs E, Weber K. INTERMEDIATE FILAMENTS: Structure, Dynamics, Function and Disease. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann H, et al. Structure and assembly properties of the intermediate filament protein vimentin: The role of its head, rod and tail domains. J. Mol. Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann H, Aebi U. Intermediate Filaments: Molecular Structure, Assembly Mechanism, and Integration Into Functionally Distinct Intracellular Scaffolds. Annu. Rev. Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 25.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/S0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 26.Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: In vivo assembly, organization, and dynamics of crescentin. Genes and Development. 2009;23:1131–1144. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabeen MT, et al. Bacterial cell curvature through mechanical control of cell growth. The EMBO Journal. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickstead B, Gull K. The evolution of the cytoskeleton. J. Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagchi S, Tomenius H, Belova LM, Ausmees N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 2008;70:1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes NA, et al. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proceedings of the National Academy of Sciences. 2013;110:E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walshaw J, Gillespie MD, Kelemen GH. A novel coiled-coil repeat variant in a class of bacterial cytoskeletal proteins. Journal of Structural Biology. 2010;170:202–215. doi: 10.1016/j.jsb.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Flärdh K, Richards DM, Hempel AM, Howard M, Buttner MJ. Regulation of apical growth and hyphal branching in Streptomyces. Curr. Opin. Microbiol. 2012;15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Hempel, A. M. et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. USA109, E2371–E2379 (2012). [DOI] [PMC free article] [PubMed]
- 34.Waidner, B. et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathogens5 (2009). [DOI] [PMC free article] [PubMed]
- 35.Specht M, Schätzle S, Graumann PL, Waidner B. Helicobacter pylori Possesses Four Coiled-Coil-Rich Proteins That Form Extended Filamentous Structures and Control Cell Shape and Motility. J. Bacteriol. 2011;193:4523–4530. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 37.Rackham OJL, et al. The Evolution and Structure Prediction of Coiled Coils across All Genomes. J. Mol. Biol. 2010;403:480–493. doi: 10.1016/j.jmb.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 38.Bharat TAM, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. Intermediate filament mechanics in vitro and in the cell: From coiled coils to filaments, fibers and networks. Curr. Opin. Cell Biol. 2015;32:82–91. doi: 10.1016/j.ceb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.England P, et al. The Scc Spirochetal Coiled-Coil Protein Forms Helix-Like Filaments and Binds to Nucleic Acids Generating Nucleoprotein Structures. J. Bacteriol. 2005;188:469–476. doi: 10.1128/JB.188.2.469-476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nature Cell Biology. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kühn J, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO Journal. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol. Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissenbach J, et al. Evolution of Chaperonin Gene Duplication in Stigonematalean Cyanobacteria (Subsection V) Genome Biol. Evol. 2017;9:241–252. doi: 10.1093/gbe/evw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, et al. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep. 2014;4:1–6. doi: 10.1038/srep04500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang H-H, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buikema WJ, Haselkorn R. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. 2001;98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuergers N, Nürnberg DJ, Wallner T, Mullineaux CW, Wilde A. PilB localization correlates with the direction of twitching motility in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2015;161:960–966. doi: 10.1099/mic.0.000064. [DOI] [PubMed] [Google Scholar]
- 50.Bhaya D, Takahashi A, Shahi P, Arthur R. Novel Motility Mutants of Synechocystis Strain PCC 6803 Generated by In Vitro Transposon Mutagenesis Novel Motility Mutants of Synechocystis Strain PCC 6803 Generated by In Vitro Transposon Mutagenesis †. J. Bacteriol. 2001;183:1–5. doi: 10.1128/JB.183.20.6140-6143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho YW, et al. Dynamic localization of HmpF regulates type IV pilus activity and directional motility in the filamentous cyanobacterium Nostoc punctiforme. Mol. Microbiol. 2017;106:252–265. doi: 10.1111/mmi.13761. [DOI] [PubMed] [Google Scholar]
- 52.Bustos SA, Golden SS. Light-regulated expression of the psbD gene family in Synecbococcus sp. strain PCC 7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol. Gen. Genet. MGG. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- 53.Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrPSc. J. Mol. Biol. 2005;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 55.Kelemen, G. H. Intermediate Filaments Supporting Cell Shape and Growth in Bacteria BT - Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells. In (eds. Löwe, J. & Amos, L. A.) 161–211, 10.1007/978-3-319-53047-5_6 (Springer International Publishing, 2017). [DOI] [PubMed]
- 56.Swulius MT, Jensen GJ. The helical mreb cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J. Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura Y, et al. Acceleration of bovine neurofilament L assembly by deprivation of acidic tail domain. Eur. J. Biochem. 1993;212:565–571. doi: 10.1111/j.1432-1033.1993.tb17694.x. [DOI] [PubMed] [Google Scholar]
- 58.Traub P, Vorgias CE. Involvement of the N-terminal polypeptide of vimentin in the formation of intermediate filaments. J. Cell Sci. 1983;63:43 LP–67. doi: 10.1242/jcs.63.1.43. [DOI] [PubMed] [Google Scholar]
- 59.Geisler N, Weber K. The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J. 1982;1:1649–1656. doi: 10.1002/j.1460-2075.1982.tb01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber K, Geisler N. The structural relation between intermediate filament proteins in living cells and the alpha-keratins of sheep wool. EMBO J. 1982;1:1155–1160. doi: 10.1002/j.1460-2075.1982.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingerson-Mahar M, Gitai Z. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 2012;36:256–266. doi: 10.1111/j.1574-6976.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF. Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis. Curr. Microbiol. 2012;65:552–560. doi: 10.1007/s00284-012-0193-5. [DOI] [PubMed] [Google Scholar]
- 63.Stucken K, Koch R, Dagan T. Cyanobacterial defense mechanisms against foreign DNA transfer and their impact on genetic engineering. Biol. Res. 2013;46:373–382. doi: 10.4067/S0716-97602013000400009. [DOI] [PubMed] [Google Scholar]
- 64.Hurme R, Namork E, Nurmiaho-Lassila EL, Rhen M. Intermediate filament-like network formed in vitro by a bacterial coiled coil protein. Journal of Biological Chemistry. 1994;269:10675–10682. [PubMed] [Google Scholar]
- 65.Kopfmann S, Hess WR. Toxin-antitoxin systems on the large defense plasmid pSYSA of Synechocystis sp. PCC 6803. J. Biol. Chem. 2013;288:7399–7409. doi: 10.1074/jbc.M112.434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carballido-Lopez R. The Bacterial Actin-Like Cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuurman N, Heins S, Aebi U. Nuclear lamins: Their structure, assembly, and interactions. J. Struct. Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 68.Fuchino K, et al. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proceedings of the National Academy of Sciences. 2013;110:E1889–E1897. doi: 10.1073/pnas.1305358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schuergers N, Mullineaux CW, Wilde A. Cyanobacteria in motion. Curr. Opin. Plant Biol. 2017;37:109–115. doi: 10.1016/j.pbi.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 70.Schuergers N, et al. Cyanobacteria use micro-optics to sense light direction. Elife. 2016;5:1–16. doi: 10.7554/eLife.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inclan YF, et al. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol. Microbiol. 2016;101:590–605. doi: 10.1111/mmi.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cappuccinelli, P. The movement of eukaryotic cells. in Motility of Living Cells 59–74, 10.1007/978-94-009-5812-8_4 (Springer Netherlands, 1980).
- 73.Lodish, H. et al. Molecular Cell Biology. (Freeman, W. H., 2000).
- 74.Nan B, Mauriello EMF, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang R, et al. AglZ Is a Filament-Forming Coiled-Coil Protein Required for Adventurous Gliding Motility of Myxococcus xanthus. J. Bacteriol. 2004;186:6168–6178. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 2004;52:1145–1158. doi: 10.1111/j.1365-2958.2004.04042.x. [DOI] [PubMed] [Google Scholar]
- 77.Agarwal R, Matros A, Melzer M, Mock H-P, Sainis JK. Heterogeneity in thylakoid membrane proteome of Synechocystis 6803. J. Proteomics. 2010;73:976–991. doi: 10.1016/j.jprot.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 78.Blatch GL, Lässle M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 79.Fiuza M, et al. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. Journal of Biological Chemistry. 2010;285:29387–29397. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alberts, B. et al. Molecular Biology of the Cell. (Garland Science, 2014).
- 81.Wiche G, Osmanagic-Myers S, Castañón MJ. Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 2015;32:21–29. doi: 10.1016/j.ceb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Leung CL, Green KJ, Liem RKH. Plakins: A family of versatile cytolinker proteins. Trends Cell Biol. 2002;12:37–45. doi: 10.1016/S0962-8924(01)02180-8. [DOI] [PubMed] [Google Scholar]
- 83.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 85.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science (80-.). 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 86.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 87.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marchler-Bauer A, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 91.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 92.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins Struct. Funct. Bioinforma. 2009;77:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 95.Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 96.Sandvang D. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 1999;43:3036–3038. doi: 10.1128/AAC.43.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vermaas WFJ, Kufryk GI, Schmetterer G, Sachet M. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: optimization of efficiency. FEMS Microbiol. Lett. 2002;206:215–219. doi: 10.1111/j.1574-6968.2002.tb11012.x. [DOI] [PubMed] [Google Scholar]
- 99.Ungerer J, Pakrasi HB. Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dörrich AK, Mitschke J, Siadat O, Wilde A. Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light-dark conditions. Microbiol. (United Kingdom) 2014;160:2538–2550. doi: 10.1099/mic.0.081695-0. [DOI] [PubMed] [Google Scholar]
- 101.Heinz S, et al. Thylakoid Membrane Architecture in Synechocystis Depends on CurT, a Homolog of the Granal CURVATURE THYLAKOID1 Proteins. Plant Cell. 2016;28:2238–2260. doi: 10.1105/tpc.16.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karimova G, Davi M, Ladant D. The β-lactam resistance protein Blr, a small membrane polypeptide, is a component of the Escherichia coli cell division machinery. J. Bacteriol. 2012;194:5576–5588. doi: 10.1128/JB.00774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miller, J. H. A Short Course in Bacterial Genetics – A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, 10.1002/jobm.3620330412 (Cold Spring Harbor, 1992).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.