Summary
Bacterial type 4a pili are dynamic surface filaments that promote bacterial adherence, motility and macromolecular transport. Their genes are highly conserved amongst enterobacteria and their expression in Enterohemorrhagic Escherichia coli (EHEC) promotes adhesion to intestinal epithelia and pro-inflammatory signaling. To define the molecular basis of EHEC pilus assembly, we determined the structure of the periplasmic domain of its major subunit PpdD (PpdDp), a prototype of enterobacterial pilin subfamily containing two disulfide bonds. The structure of PpdDp, determined by NMR, was then docked into the electron density envelope of purified EHEC pili obtained by cryo-electron microscopy (cryo-EM). Cryo-EM reconstruction of EHEC pili at ~8 Å resolution revealed extremely high pilus flexibility correlating with a large extended region of the pilin stem. Systematic mutagenesis and functional analyses identified charged residues essential for pilus assembly. Structural information on exposed regions and interfaces between EHEC pilins is relevant for vaccine and drug discovery.
Keywords: type 4 pili, EHEC, NMR, cryo-EM, pilin structure, pilus assembly, molecular modeling, hemorrhagic coli pilus (HCP)
Introduction
Type 4 pili (T4P) are thin filamentous polymers that bacteria use to bind to their substrates, move across surfaces by twitching or gliding, and transport macromolecules in and out of the cell (Berry and Pelicic, 2015). T4P also promote formation of bacterial aggregates and biofilms and signaling to host cells during infection. The Enterobacteriaceae family of gamma proteobacteria comprises many commensal and environmental species, but also important human, animal and plant pathogens from the genus Klebsiella, Yersinia or Dickeya. Most enterobacteria contain gene clusters encoding pili of the T4a subclass in conserved chromosomal loci (Pelicic, 2008) and share high sequence conservation of major pilin subunits, designated PpdD in E. coli (Luna Rico et al., 2018b). However, their expression and function have been described only in few species. In the plant pathogen Erwinia amylovora, PpdD pili promote biofilm formation and mutants deleted for the ppdD operon show reduced virulence (Koczan et al., 2011). In specific nutrient-limiting conditions, PpdD pili are also assembled by enterohemorrhagic E. coli (EHEC) (Xicohtencatl-Cortes et al., 2007). EHEC is an important human pathogen causing outbreaks of severe intestinal infections, with hemolytic uremic syndrome (HUS) as a life-threatening complication of the disease. Sera of patients recovering from HUS contain antibodies against PpdD, suggesting that T4P represent one of the virulence factors in the arsenal of this pathogen (Monteiro et al., 2016).
The EHEC pili, also named HCP for hemorrhagic coli pili, promote adhesion to intestinal epithelia in bovines, the main reservoir of this pathogen (Xicohtencatl-Cortes et al., 2007) and induce pro-inflammatory signaling (Ledesma et al., 2010). Expression of pilin genes in a laboratory E. coli strain HB101 promotes biofilm formation and twitching motility (Xicohtencatl-Cortes et al., 2009), phenotypes typically associated with T4P dynamics. Transcriptional studies in E. coli K-12 revealed that PpdD pili assembly genes are co-regulated with DNA uptake genes via Sxy (TfoX) competence activator (Sinha et al., 2009), suggesting a role of these pili in natural transformation and horizontal gene transfer. PpdD pilus assembly was achieved in a heterologous type 2 secretion system from Klebsiella oxytoca (Sauvonnet et al., 2000) (Cisneros et al., 2012), and more recently in the reconstituted EHEC pilus assembly system in E. coli K-12 (Luna Rico et al., 2018b). The highly similar symmetry parameters of T4aP assembled in these different systems indicate the key role of major pilins as determinants of pilus structure (Luna Rico et al., 2018b).
EHEC pili are polymers of the major subunit PpdD, which is a prototype of the enterobacterial class of pilins, present in many important human, animal and plant pathogens (Luna Rico et al., 2018b). In order to understand the molecular and structural basis of assembly of these major subunits into a pilus, we determined the solution NMR structure of the periplasmic domain of the EHEC PpdD (PpdDp). We then combined this structure with the 3-D cryo-EM reconstruction of EHEC pilus to build a pseudoatomic model of the assembled fiber at sub-nanometer resolution. In addition, site-directed mutagenesis and functional assays identified PpdD residues crucial for pilus assembly and stability.
Results
Solution structure of the periplasmic PpdD domain
In addition to the well-conserved N-terminal hydrophobic segments (α 1 helix), PpdD homologues share similar periplasmic domains stabilized by two disulfide bonds at conserved positions (de Amorim et al., 2014; Luna Rico et al., 2018b). To gain molecular insight into this group of pilins, we solved the structure of the soluble periplasmic domain of EHEC PpdD (hereafter designated PpdDp) comprising residues 26 to 140 of the mature protein. The structure is very well defined from 2219 NOE-derived distance, 216 dihedral angle and 45 RDC (residual dipolar coupling) restraints as well as 30 hydrogen bonds. The details of restraints and structural statistics of the 20 lowest energy conformers representing the solution structure of PpdDp are summarized in Supplementary Table 1. The structures show high convergence with a mean pairwise root mean square deviation (RMSD) of 0.55 and 1.06 Å for the backbone and the heavy atoms of ordered regions, respectively. The 3D structure of PpdDp displays a canonical alpha-beta pilin fold with two disulphide bonds (Figure 1A,B). It is composed of two α-helices at both extremities (α 1C as the C-terminal part of α1: N26-H54 and α3: D123-F134) that are separated by a long α/β loop and a tilted 4-stranded antiparallel beta-sheet (strands β1-β2-β3-β4) facing the α1C helix. The structure is highly stabilized by two disulphide bridges (Figure 1A,D); one is conserved in type IV pilins (C118-C130) and connects α 3 with β4. The second (C50-C60) is only present in enterobacterial pilins and connects α1C to the beginning of the α/β loop leading into a short 310 helix (α 2: G56-C60). This loop is enriched in His, Gly and Asp residues conserved in the PpdD family.
Figure 1. Solution NMR structure of PpdDp.
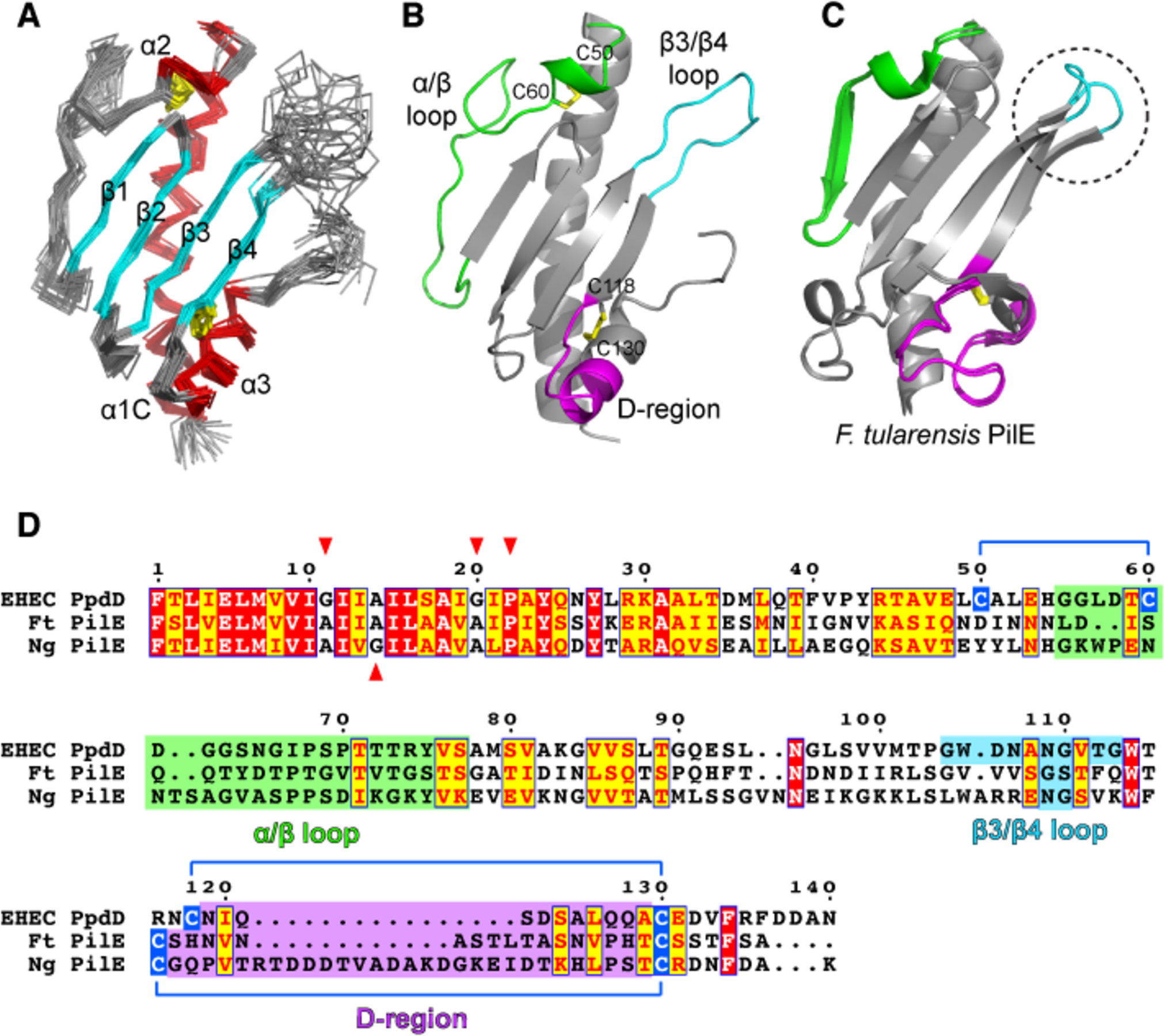
(A) Backbone representation of the NMR ensemble of PpdDp colored by secondary structure elements. Helices α1C, α2 and α3 are shown in red and the β1-β2-β3-β4 sheet is shown in cyan. Disulfide bridges are colored in yellow. (B) Ribbon representation of the lowest energy model of the PpdDp NMR ensemble. Characteristic hypervariable regions in T4a major pilins are colored as follows: α/β loop in green, β3/β4 loop in cyan and D-region in magenta. Disulfide bridges C50-C60 and C118-C130 are shown as yellow sticks. (C) Superposition of the two molecules in the asymmetric unit of the Francisella tularensis PilE crystallographic structure (PDB 3SOJ). Hypervariable regions are colored as in (B). The conformationally variable β3/β4 loop is highlighted by a dotted circle. (D) Sequence alignment of full-length PpdD from EHEC, PilE pilins from Francisella tularensis (Ft) and Neisseria gonorrhoeae (Ng). Positions of helix breaking residues (G, P) in the N-terminal region are highlighted with red arrows. Cysteine residues involved in disulfide bridges are connected by blue lines.
The 1H-15N heteronuclear NOE data for the backbone confirm an overall well-ordered structure (Supplementary Figure 1A). Besides the C-terminal tag, which is highly flexible and without any stable interaction with the rest of the protein, two regions with low 1H-15N NOE value are present, including the β3/β4 (G104-G113) and β4/a3 (I120-S122) loops. The former includes the two residues whose signals were not observed in the NMR spectra (D106 and V111) indicating the presence of intermediate exchange on the chemical shift time scale (μs-ms) between different conformations. Consequently, very few long-range NOE could be assigned in the β3/β4 loop, consistent with the high variability of this region in the PpdDp structure ensemble (Supplementary Figure 1B, C). Surprisingly, residues of the long α/β loop display 1H-15N NOE values representative of a well-structured region with low flexibility and slow internal motions.
Structure of the PpdD pilus filament
We determined that T4PS PpdD pili and T2SS PpdD pili (Luna Rico et al., 2018b) have the same morphology and indistinguishable helical parameters. We therefore combined the segments extracted from cryo-EM images of PpdD pili assembled by T4PS and T2SS for helical reconstruction. The variability in the helical parameters was evident (Supplementary Figure 2) from a reference-based classification for sorting the 99,678 segments in terms of variable twist and rise. The mode of this distribution corresponded to an axial rise of 10.8 Å and twist of 96°, accounting for ~25% of the whole dataset, and this group was used for the helical reconstruction. After multiple cycles of IHRSR (Egelman, 2000), the symmetry of this reconstruction converged to a rise of 11.2 Å and a rotation of ~96° per subunit. The overall resolution of the reconstruction was ~8 Å according to the half-map FSC criterion (Supplementary Figure 3), allowing us to identify the location of the well separated globular pilin domains forming the outer shell of the filament (Figure 2A). However, the inner core of the pilus has less density. At high contour level (~5σ), small rod-shaped densities are observed along the filament axis, but remain disconnected from the main densities of the globular pilin heads (Figure 2B). At lower contour level, weaker densities appear, connecting the outer globular heads to the inner rods (Figure 2C). Since the PpdDp NMR structure fitted very nicely into the cryo-EM density at the expected location but did not explain the inner rod-shape densities, we inferred that they actually correspond to the N-terminal transmembrane segments (TMS). We thus built an atomic model of the PpdD pilus by docking a homology model of the helical TMS, connected by an extended linker to the fitted PpdDp structure. After symmetrization and refinement in real-space, the structure of the PpdDp was conserved with a Cα RMSD of 2.3 Å between the NMR structure and the structure in the pilus. In the context of the pilus, the N-terminal part of α1 (α1N) stops at G11 and is followed by an unstructured region until around P22, where the α1C helix begins (Figure 2D).
Figure 2. Cryo-EM reconstruction of the EHEC T4P filament at ~8 Å resolution and PpdD pilin structure fitting.
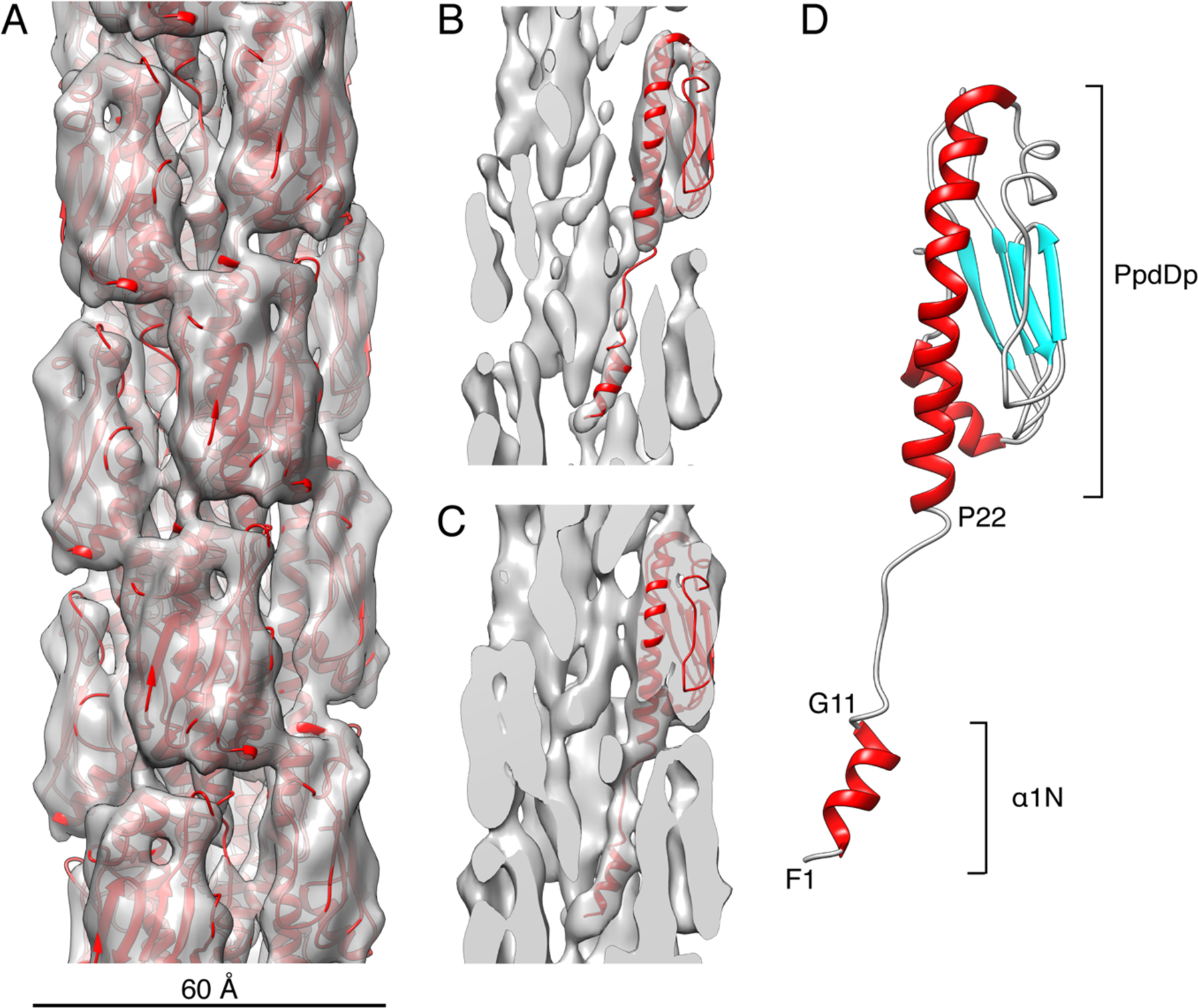
(A) Surface view of the EHEC T4P cryo-EM reconstruction at ~4.2σ contour level with the refined atomic model shown in ribbons (red). The diameter of the pilus is ~60Å. (B-C) Cross-sections of the cryo-EM reconstruction at ~5σ (B) and ~3.8σ (C) contour levels and with a single PpdD subunit shown in ribbons. (D) Structure of a PpdD pilin subunit in the refined EHEC T4P structure colored by secondary structures and showing the extended region between the short α1N helix and the PpdDp domain. The positions of helix breaking between residues G11 and P22 are indicated.
The PpdD pilus has a diameter of ~60 Å with somewhat loosely packed pilin subunits forming a right-handed 1-start helix (Figure 3A). Each PpdD subunit P has contacts with six neighbors, forming three different interfaces with subunits P±1, P±3 and P±4 (Figure 3), with respective buried surface areas of 569 Å2, 943 Å2 and 1529 Å2. Potential salt-bridges are present in the P-P+1 and P-P+3 interfaces (Figure 3C), while the P-P+4 interface mostly involves hydrophobic contacts. The β3/β4 loop, well visible and ordered in the cryo-EM density, folds as an elongated extension of the β-sheet, making inter-subunit contacts with the D-region and with the N-terminal region at the P-P+4 interface (Figure 3C).
Figure 3. Structure of the EHEC PpdD pilus.
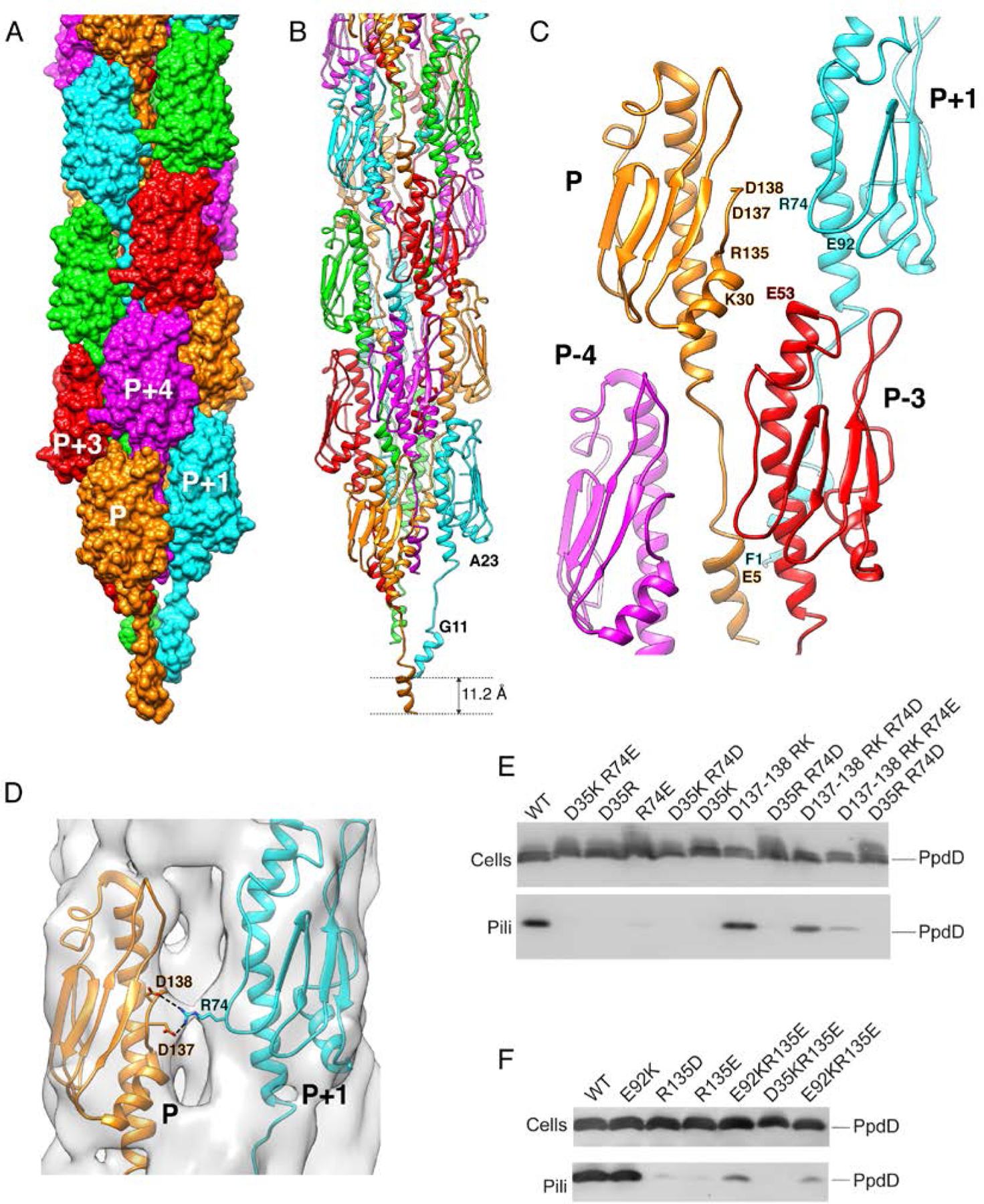
(A) Surface representation of the PpdD pilus structure with subunits P, P+1, P+2, P+3 and P+4 (along the 1-start helix) colored in orange, cyan, green, red and magenta, respectively. (B) Structure of PpdD pilus shown in ribbons where subunits are colored as in (A). The helical rise of the 1-start helix is 11.2Å. (C) Topological arrangement of neighboring subunits in the PpdD pilus structure, colored as in (A). The positions of charged residues involved in inter-subunit interactions are labeled. (D) Close-up view of the interface between protomer P and P+1 showing the side-chains of D137/D138 and R74 making potential salt-bridges in the PpdD pilus structure. The cryo-EM reconstruction is shown as surface at ~3.2σ contour level. (E-F) Piliation assay with single charge inversion PpdD variants. Cells (top) and sheared pilus fractions (Pili, bottom) corresponding to 0.05 OD600nm of bacteria were separated on SDS-PAGE and analyzed by immunoblot using anti-PpdD antibodies. PpdD residue substitutions are indicated above each lane. Migration of PpdD is indicated on the right.
Co-evolutionary analysis performed on the PpdD pilin sequence family provided intra- and inter-pilin contact predictions that support the atomic model of PpdD pilus (Supplementary Table 2; Supplementary Figure 4).
PpdD mutagenesis and effects on pilus assembly
The mature PpdD, N-terminally processed by the prepilin peptidase has seven positively and 13 negatively charged residues. The surface of the EHEC pilus is highly negatively charged (Figure 4A). To assess the role of electrostatic interactions in assembly and stabilization of PpdD pili, we introduced single charged residue substitutions that create electrostatic repulsion between PpdD subunits or between PpdD and its assembly components. We examined pilus assembly in an E. coli strain producing the K. oxytoca T2SS but lacking its cognate major pseudopilin PulG (Figure 4D,E). The mean piliation efficiency, expressed as a fraction of total PpdD detected on the bacterial surface, was determined for each of these variants from four independent experiments. About 50% of native PpdD (PpdDWT) was assembled into pili in this system, a fraction comparable to the assembly efficiency for the cognate T2SS substrate, the major pseudopilin PulG assembled into pili under the same conditions (Campos et al., 2010). As expected, piliation was abolished for the PpdDE5A variant lacking the conserved E5 residue, as in P. aeruginosa (Strom and Lory, 1991) and Neisseria gonorrhoeae (Aas et al., 2007). The E5 residue has been implicated in an early step of subunit recruitment to the assembly machinery (Nivaskumar et al., 2016), as well as in inter-subunit contacts in assembled pili (Kolappan et al., 2016; Wang et al., 2017). Similar to the behavior of PulGE5A, which is arrested in the membrane-embedded preassembly state, (Nivaskumar et al., 2014; Santos-Moreno et al., 2017) PpdDE5A levels were consistently higher compared to PpdDWT. Several PpdD variants showed reduced stability, including PpdDR29E and, more dramatically, PpdDR44D, consistent with the intra-protomer interaction of R44 with E48 identified by NMR. Charge inversion of residues mapping onto the fiber surface had little effect on pilus assembly (Figure 4B,C,D). With the exception of the surface-exposed residue K83, all positively charged residues were essential for piliation (Figure 4C,D,E). Several charge inversions localized at the interfaces with other subunits abolished pilus assembly, including K30E, E48K, D61R, R74D, R116D and R135E. Nearly all residues in the periplasmic part of the alpha helical PpdD stem appear to be very important for piliation (Figure 4D,E). Charge inversions at these positions affected interactions with other PpdD subunits in the membrane, prior to pilus assembly, as indicated by the bacterial two-hybrid (BACTH) analysis (Supplementary Figure 5).
Figure 4. Mapping of residues essential for pilus assembly and hypervariable regions on the PpdD pilus surface.
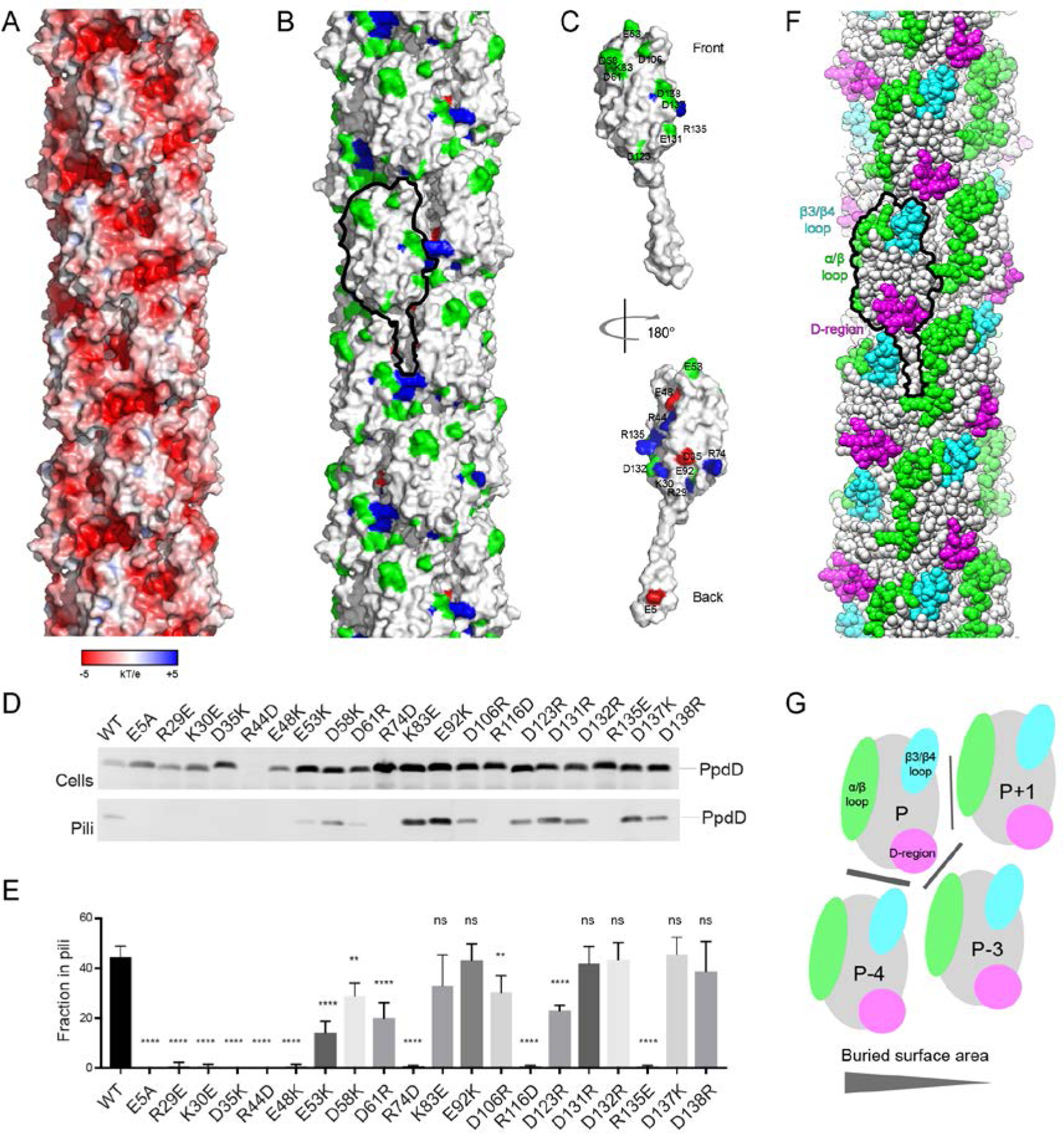
(A) Surface electrostatic potential of the PpdD pilus (B) Surface representation of the PpdD pilus structure showing the distribution of charged residues essential for pilus assembly (positively charged in blue and negatively charged in red). Non-essential charged residues are colored in green. Surface of a single PpdD subunit is outlined in black. (C) Surface representation of a PpdD pilin subunit in the pilus colored as in (A). The surface-exposed side of the subunit (front view) displays predominantly residues permissive for charge inversions. The back view (180° rotation) shows the clustering of charged residues essential for pilus assembly on the buried face of the pilin. (D) Piliation assay with single charge inversion PpdD variants. Cells (top) and sheared pilus fractions (Pili, bottom) corresponding to 0.05 OD600nm of bacteria were separated on SDS-PAGE and analyzed by immunoblot using anti-PpdD antibodies. (E) Quantification of the fraction of PpdD assembled into pili. The bars represent the mean values and error bars represent standard deviation from 4 independent experiments. Statistical analysis was performed by using ANOVA test to compare datasets from native PpdD and its variants: ns: non-significant difference; **: p<0.01; ****: p<0.0001. (F) Sphere representation of the PpdD pilus structure where the location of hypervariable regions is highlighted (α/β loop in green, β3/β4 loop in cyan and D-region in magenta). A single PpdD subunit is outlined in black. (G) Schematic representation of the topological arrangement of neighboring subunits in the PpdD pilus structure. The location of hypervariable regions is indicated and colored as in (F). The size of the buried surface area in each inter-protomer interface is proportional to the width of the grey line.
Among the inter-protomer contacts detected in the cryo-EM structure of the EHEC pilus, R74P+1 is in close proximity to both D137P and D138P (Figure 3D). Whereas the charge inversion of residue R74 abolished piliation, the double charge inversion D137R-D138K did not. However, piliation of PpdDR74E and PpdDR74D variants was restored in the presence of the D137R-D138K substitution, supporting the proximity of these residues and their importance at the P-P+1 interface (Figure 3E). Similarly, in a non-piliated PpdD variant R135E, pilus assembly could be partially rescued by an additional charge inversion of a non-essential residue E92P+1, facing the R135P (Figure 3F). This effect was specific since piliation was not restored in another double mutant variant, PpdD D35K-R135E and it is consistent with the relative proximity of R135 and E92 in the pilus structure (Figure 3F).
Dynamic behavior of different T4aP
We used normal mode analysis to obtain information on the dynamic behavior of EHEC, N. meningitides (Nm), N. gonorrhoeae (Ng) and P. aeruginosa strain PAK T4aP. For each T4aP structure, the first 3 non-trivial modes reveal similar dynamic behavior. Modes 1 and 2 correspond to the bending of the filaments in two orthogonal directions, while movements along mode 3 represent twisting of the filament around the helical axis (Supplementary Figure 6). For each of the 3 modes, the EHEC T4P structure displays the largest mean square fluctuations of the pilin head per subunit, while Nm, Ng and PAK T4P display similar behaviors of lower fluctuations (Supplementary Figure 6). As expected, the largest fluctuations are observed at the extremities of the filaments, and in the central region for the bending modes. In the twisting mode (mode 3), the fluctuation pattern along the pilus axis oscillates, with a periodicity of 3 subunits correlating with the left-handed 3-start helix present in all T4aP structures. This observation may indicate the importance of the P-P+3 interface in the plasticity of T4aP filaments. Interestingly, the EHEC T4P structure displays a significantly smaller P-P+3 interface compared to Nm, Ng and PAK (Supplementary Figure 7).
Discussion
The fold of the periplasmic domain of EHEC PpdD is very similar to known structures of T4a pilins with RMSD for Cα atoms ranging from 2.7 to 3.2 Å (Figure 1 and Supplementary Table 2). Major structural differences are restricted to the characteristic hypervariable regions in T4a pilins: the α α/β loop, the β3/β4 loop and the D-region, encompassed by the two cysteines forming the C-terminal disulfide bridge (Supplementary Figure 8). Among the pilins, the major pilin PilE from Francisella tularensis (PilEFt) is the closest structural homologue to PpdDp despite sharing the lowest sequence identity (only 16% for the periplasmic domain) (Supplementary Table 3), indicating the high sequence variability in the structurally conserved T4a pilins.
The PilEFt X-ray structure shows two molecules in the asymmetric unit (Hartung et al., 2011) with secondary structure features quite similar to that of PpdDp (Figure 1C). The most pronounced structural difference is an extra strand present in PilEFt in the α/β loop involved in the crystallographic dimer interface (Hartung et al., 2011). When the two molecules of PilE in the crystallographic dimer are superimposed (Figure 1C), the slightly different conformation in the β3/β4 loop between the two molecules suggests some flexibility. For PpdDp, the flexibility in this region is evidenced by the lower heteronuclear NOE values (Supplementary Figure 1). It was shown recently that a short segment in the α/β loop of truncated PilA pilin of P. aeruginosa PA14 is highly dynamic in solution while essential for piliation (Nguyen et al., 2018). Interestingly, increased backbone dynamics of both the α/β and β3/ β4 loops was also observed in monomeric P. aeruginosa K122–4 pilin (Suh et al., 2001). In PpdD, a charge inversion at position D106 within the dynamic β3/ β4 loop affected pilus assembly, presumably by altering contacts with another PpdD protomer (Supplementary Figure 5) or with an assembly factor.
So far, detailed cryo-EM reconstructions of bacterial T4aP have been determined for N. meningitidis (Kolappan et al., 2016), N. gonorrhoeae and P. aeruginosa (Wang et al., 2017). In all cases, pilin subunits show a similar axial rise of approximately 10 Å, whereas the twist angle is more variable and reflects specific packing of pilin globular domains. The new cryo-EM reconstruction of EHEC pili shows very similar subunit organization and helical parameters to that of other T4aP (Supplementary Figure 9). As observed for the other T4aP structures recently solved, the α1 helix of PpdD is interrupted by a melted and extended region. However, this extended region is longer in PpdD, going from G11 to P22, whereas in Ng, Nm and PAK pilin, it starts at G14, a residue which is not conserved in PpdD. We note that the interruption of the α1 helix correlates well with the T4a pilin-specific sequence-based secondary structure predictions (Supplementary Figure 10). This extended region at the α1 helix seems to be a structural feature of T4aP. It has been proposed that the interruption of the helical part may have a role in the ability of T4P to extend under force, and relax back to a native state when the force is removed (Wang et al., 2017). This flexibility, while crucial for pili functions, is an impediment for their structure determination at high resolution. The long, extended region of PpdD can explain the limited resolution of the pilus structure. The enhanced flexibility of EHEC pilus towards bending and twisting from normal mode analysis also indicates the crucial role of the P-P+3 interface in pilus rigidity. In EHEC, this reduced interface mainly involves the extended region of the pilin stem, thus supporting its implication in plasticity of pili.
Interestingly, when mapped onto the PpdD pilus structure, the location of hypervariable pilin regions forms a triangular junction between neighboring pilins at the filament surface (Figure 4F,G). Their exposed residues can define the specificity of interactions with different partners.
Overall, our mutagenesis and functional analyses show that the majority of PpdD residues required for pilus assembly mapped onto the side of the protein facing the fiber core, consistent with their involvement in inter-protomer interactions in the polymerized fiber. In addition, some of these residues might play a role during earlier assembly steps via binding to assembly factors in the plasma membrane (Luna Rico et al., 2018b). In the related Klebsiella T2SS, binding of the major pseudopilin PulG to PulM has been implicated in subunit targeting to the assembly machinery (Nivaskumar et al., 2016; Santos-Moreno et al., 2017). PpdD binding to PulM probably allows for its assembly via the T2SS, whereas the PulM equivalent HofN in the EHEC T4PS might play a similar role, as suggested by the BACTH-based interaction studies (Luna Rico et al., 2018b).
EHEC pili have been implicated in colonization of mammalian host epithelia specifically under starvation conditions and there is evidence for their synthesis in vivo (Xicohtencatl-Cortes et al., 2007). Antibodies against PpdD are produced in patients and they inhibit pro-inflammatory signaling in a dose-dependent manner (Ledesma et al., 2010). Therefore, targeting T4P is a possible therapeutic strategy for EHEC infections, which are life-threatening, and where antibiotic treatment is contra- indicated.
In addition, the EHEC pili share common regulatory and structural features with the Vibrio T4aP that are involved in natural competence (Matthey and Blokesch, 2016). In a recent elegant study, Ellison and colleagues showed that Vibrio T4aP capture DNA via their tip and promote its uptake to the periplasm through pilus retraction (Ellison et al., 2018). Removing several positively charged residues of two minor pilin subunits, presumably localized at the tip of the pilus, significantly affected DNA binding and transformation efficiency (Ellison et al., 2018). Although DNA binding has been attributed to major pilins in certain bacteria like P. aeruginosa (van Schaik et al., 2005) or Neisseria spp. (Craig et al., 2006) (Imhaus and Duménil, 2014), direct biochemical evidence supports the role of specialized minor pilin ComP in this binding in N. meningitidis (Cehovin et al., 2013). Consistent with this, we did not observe any interaction of PpdDp with double stranded DNA by NMR. Moreover, the highly negatively charged surface of the EHEC pilus makes its direct interaction with DNA unlikely.
Specific nutritional or other signals so far unknown may activate DNA uptake in vivo, as suggested by the capacity of E. coli to take up exogenous DNA in vitro (Sinha and Redfield, 2012). Given the high conservation of PpdD and T4aP systems among enterobacteria, this capacity might be widespread among commensal strains, and among pathogens of the ESKAPE group like Klebsiella, Salmonella or Yersinia that a particular threat due to their antibiotic resistance. Further studies will be required to determine whether and how T4P in enterobacteria participate in DNA uptake and contribute to exchange of genetic material in the gut. Detailed structural information will be instrumental to gain a better understanding of the mechanisms underlying these processes in vivo, and is crucial for drug and vaccine discovery.
STAR Methods
Bacterial strains and plasmids
Escherichia coli strain DH5α was used for DNA cloning purposes and strain PAP7460 (Δ(lac-argF)U169 araD139 relA1 rpsL150 ΔmalE444 malG501 [F’ (lacIQ ΔlacZM15 pro+ Tn10)] was used for functional assays. Strain PAP5171 (PAP7460 degP::KmR) was used as a host for pilus purification. Bacteria were cultured in LB or M9 0.5% glycerol plates supplemented with antibiotics as required: Ap (100 μg.mL−1), Cm (25 μg.mL−1) or Km (25 μg.mL−1). Maltose (0.4 %) or isopropyl-thio-β-D galactoside (IPTG) was added to induce pul gene expression.
Purification of pili
Bacteria of strain PAP5171 containing plasmid pCHAP8565 encoding PpdD and one of the two plasmids encoding the pilus assembly machinery, pCHAP8184 (for the T2SS) or pMS41 (for T4PS), were densely inoculated on LB Cm Ap IPTG plates and cultured at 30°C for 5 days. Bacteria were harvested in LB and pili were sheared by extensive vortexing and ten passages through a 26-Gauge needle. Bacteria were pelleted for 10 min at 14000g and the supernatant was further pelleted in the same conditions in 1.5-ml eppendorf tubes. The collected cleared supernatant was subjected to ultracentrifugation for 1 hr in Ti60 Beckman rotor at 100 000g. Pellets were resuspended in 50 mM HEPES pH 7.2, 50 mM NaCl and stored on ice.
Plasmid constructions
Plasmid pCHAP8656 contains ppdD gene, amplified from the genomic DNA of strain EDL933 using the high-fidelity Pwo DNA polymerase and placed under the control of lacZ promoter (Supplementary Table 4). This plasmid was used as template for site-directed mutagenesis to create charged residue substitutions in PpdD listed in Supplementary Table 4. For mutagenesis, five cycles of PCR amplification were performed in parallel with reverse and forward complementary oligonucleotides (Eurofins, listed in Supplementary Table 5). The reaction mixtures were then combined and amplifications were continued for another 13 cycles (30 sec at 96°C, 30 sec at 50°C and 3 min at 72°C). After DpnI digestion, 10–20 μL of the reactions were introduced into ultra-competent DH5α cells and transformants were selected on LB Cm plates. The resulting plasmids were purified and verified by DNA sequencing (GATC and Eurofins).
Piliation assays
The piliation assay was performed as described previously (Luna Rico et al., 2018a). Bacteria of strain PAP7460 transformed with plasmid pCHAP8184 and either vector alone or pCHAP8565 derivatives carrying ppdD were grown 48 hrs at 30°C on LB agar containing Ap, Cm and 0.4% maltose. Bacteria were harvested and normalized to OD600nm of 1 in LB. Pili were detached by a 1-min vortex treatment and bacteria were spun for 5 min at 16000g. The bacterial pellet was resuspended in SDS sample buffer at 10 OD600nm. mL−1 and the supernatant was cleared from the remaining bacteria in a second 10-min centrifugation. The cleared supernatant was transferred to a new eppendorf tube and precipitated with 10% TCA for 30 min on ice. Pellets were collected by 30-min centrifugation at 16000g, washed twice with acetone, air-dried and taken up in SDS sample buffer at a concentration of 10 OD600nm equivalents per mL. Equivalent volumes of bacteria and pili fractions were analyzed by denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with a Tris-Tricin buffer system (Schagger and von Jagow, 1987). Proteins were transferred to a nitrocellulose membrane (ECL Amersham) and probed with antisera generated against MalE-PpdD fusion protein described previously (Sauvonnet et al., 2000).
NMR experiments
NMR data were acquired at 298 K on a Varian spectrometer operating at a proton frequency of 600 MHz and equipped with a cryogenically cooled triple resonance 1H{13C/15N} PFG probe. Proton chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as 0 ppm. 15N and 13C chemical shifts were referenced indirectly to DSS (Wishart et al., 1995). The pulse sequences were from VnmrJ Biopack. NMR data were processed with NMRPipe/NMRDraw (Delaglio et al., 1995) and analyzed with the CcpNmr Analysis software package (Vranken et al., 2005).
Residual dipolar couplings (RDCs) were determined by measuring the difference in JNH splitting between in-phase/antiphase (IPAP) NMR experiments under isotropic and anisotropic conditions (Ottiger et al., 1998). The protein sample for RDC determination was at 1 mM in 25 mM sodium phosphate, pH 7.5, 50 mM NaCl, 12% D2O. For protein alignment in the anisotropic sample, 10 mg/mL of the bacteriophage Pf1 (ASLA Biotech) was added and the sample was incubated 1 hour in the magnetic field prior to acquisition.
The backbone 15N dynamics experiments were performed with a 1 mM sample in 50 mM sodium phosphate, pH 7.5, 50 mM NaCl, 12% D2O. 1H-15N NOE values were determined as the ratio between the intensities of corresponding peaks in the spectra recorded with and without pre-saturation of 1H.
NMR structure determination
The structure determination strategy consisted in several rounds of automated iterative NOESY assignment and structure calculation with the ARIA 2.3 (Rieping et al., 2007); (Bardiaux et al., 2008) and CNS software packages (Brunger, 2007). Backbone dihedral angles were predicted with TALOS+ (Shen et al., 2009), and predictions classified as “good” or “dynamic” were converted into φ and ψ dihedral angle restraints. In addition, 45 DNH residual dipolar coupling (RDC) were introduced as restraints in the structure calculation. Axial and rhombic components of the alignment tensor were first estimated from an intermediate structure and further refined with the software PALES (Zweckstetter and Bax, 2000). A log-harmonic potential was employed (Nilges et al., 2008) during the cooling phase of the simulated annealing, for NOE derived and hydrogen bonds distances restraints. This potential was combined with an automated estimation of the optimal weighting of the distance restraints (Bernard et al., 2011; Nilges et al., 2008). For NOE restraints, the final average weight was 14.9 kcal/mol, and 11.9 kcal/mol for hydrogen bonds restraints. The disulphide bonds that had been confirmed by cysteine Cβ chemical shifts as reported previously (Amorim et al., 2014), were imposed in the structure calculation. In the final ARIA iteration, 100 conformers were calculated and the 20 lowest energy structures were refined in a shell of water molecules (Linge et al., 2003). Supplementary Table 1 gives a summary of NMR-derived restraints and statistics on the final ensemble of NMR structure.
Cryo-electron microscopy
Three microliters PpdD pili samples (assembled from either T4PS or T2SS) were applied to plasma-cleaned lacey carbon grids and vitrified using a Vitrobot Mark IV (FEI). The grids were imaged on a Titan Krios microscope operating at 300 kV equipped with a Falcon III camera with a sampling of 1.09 Å/px. Images were collected with a defocus range of 1.0μm to 3.0 μm. Motioncor2 was used for motion correcting all the images, followed by the CTFFIND3 program (Mindell and Grigorieff, 2003) for the defocus and astigmatism estimation. Long filament boxes were extracted from the cryo-EM images (after correction of phases and amplitudes through multiplying by the CTF) using the e2helixboxer program in EMAN2 (Tang, 2007). The SPIDER software package (Frank, 1996) was used for most other image processing. 384 px long overlapping boxes were cut from the long filament boxes (with a shift of 4% of the box size, which is ~ 1.5 times the axial rise), yielding 99,678 segments in total. A reference-based classification was used to sort the segments in terms of the axial rise and azimuthal rotation. After sorting, 25,669 segments from the dominant group, which accounted for ~25% of the whole dataset, were processed by IHRSR (Egelman, 2000) to produce the final reconstruction.
Model building and refinement
We first generated a full-length model of PpdD by adding the alpha-helical stem (F1-Q25) to the PpdDp structure by using the available structure of the full-length gonococcal PilE (PDB 1AY2) as template with the Modeller program version 9v8 (Sali and Blundell, 1993). Next, the full-length PpdD model was docked as a rigid-body in the cryo-EM density map with Situs version 2.7 (Wriggers, 2012). However, only the region A23-N140 (corresponding to PpdDp plus A23, Y24 and Q25) could be fitted inside the cryo-EM density. After symmetrization by making use of the helical symmetry parameters of the PpdD pilus cryoEM map, a theoretical density map was generated from the atomic coordinates of the fitted PpdD23–140 model. A difference map was created by subtracting the theoretical PpdD23–140 pilus map from the cryoEM density map. Next, we rigidly docked the modelled PpdD1–16 segment in the difference map with Situs (Wriggers, 2012). An initial real-space refinement was performed with 14 copies of the docked PpdD1–16 and PpdD23–140 segments with PHENIX version 1.11.1 (Adams et al., 2010). A full-length model of PpdD was then constructed with Modeller (Sali and Blundell, 1993) by connecting the refined PpdD1–16 and PpdD23–140 segments by an extended linker (S17-P22). Finally, full-atom real-space refinement, including morphing, minimization and simulated-annealing, was performed with PHENIX (Adams et al., 2010) to improve model geometry and the correlation with the experimental cryo-EM map. The final model of PpdD pilus was validated with Molprobity version 4.1 (Davis et al., 2007). Refinement statistics are given in Supplementary Table 6. Structure figures were generated with PyMOL (Schrödinger, 2015) and UCSF Chimera (Pettersen et al., 2004).
Analysis of residue co-evolution and secondary structure propensities
Long-range contact predictions from residue co-evolution were obtained with the Gremlin tool (Kamisetty et al., 2013). Using the EHEC PpdD sequence as query, we generated a Multiple Sequence Alignment of 1473 homologous sequences with HHblits (Remmert et al., 2011), for a final ratio of ~11 sequences per query position. Secondary structure propensities were derived from local residue co-evolutions predicted with CCMpred (Seemayer et al., 2014), following the strategy proposed recently by Toth-Petroczy et al. (Toth-Petroczy et al., 2016). Briefly, for each position in the query sequence, a propensity score for α helix or β strand (Sα or Sβ) is calculated based on the average prediction score of short-range contacts (Si,i+1 to Si,i+4 for helices and Si,i+1 to Si,i+2 for strands) involving the given position or its flanking residues (2 for helices and 1 for strands on both sides). Then, scores are normalized with regards to their correlation with Si,i+1 scores. The final scores are obtained by subtracting scores of non-specific contacts from scores specific to a particular secondary structure element, i.e. Sα≃Si+4+Si+3-Si+2-Si+1 and Sβ≃Si+2-Si+1.
Normal Mode Analysis of T4P structures
Normal modes were computed with Prody (Bakan et al., 2014) with an Anisotropic Network Model on Cα atoms of T4P filament structures and a cutoff of 15 Å for pairwise interactions in the elastic network. The different helical pitches of the EHEC, Nm, Ng and PAK T4P filaments lead to structures with significantly variable length for a fixed number of pilin subunits, that may influence the outcome of the normal mode analysis. Consequently, filament structures of equivalent length were generated using different number of subunits (30 for Nm and Ng, 29 for Pak and 27 for EHEC), the respective helical symmetry and the pilin structure. The following PDB entries were used: 5KUA for Nm, 5VXX for Ng and 5VXY for PAK pili. For each of the first 3 non-trivial lowest frequency mode, the average of the atomic mean square displacements was calculated for each subunit, excluding the first 24 residues.
Data availability
The PpdDp structure and NMR restraints have been deposited in the Protein Data Bank under the accession id 6GMS. The PpdD pilus structure and cryo-EM map are available from the Protein Data Bank (accession code 6GV9) and Electron Microscopy Data Bank (accession code EMD-0070), respectively.
Supplementary Material
Acknowledgments
This work was funded by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS), the French Agence Nationale de la Recherche (ANR-14-CE09-0004), the Fondation pour la Recherche Médicale (Equipe FRM 2017M.DEQ20170839114), the European Union FP7-IDEAS-ERC 294809 (to M. Nilges) and the NIH R35GM122510 (to E.H.E.). ALR was funded by the Pasteur Paris University (PPU) international PhD program. We are grateful to Muriel Delepierre and Daniel Ladant for support and interest in this work.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aas FE, Winther-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JP, and Koomey M (2007). Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect Type IV pilus assembly, dynamics and associated functions. Mol. Microbiol 63, 69–85. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, and Echols N, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, Biological Crystallography, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim GC, Cisneros DA, Delepierre M, Francetic O, and Izadi-Pruneyre N (2014). ¹H, ¹⁵N and ¹³C resonance assignments of PpdD, a type IV pilin from enterohemorrhagic Escherichia coli. Biomol NMR Assign. 8, 43–6. [DOI] [PubMed] [Google Scholar]
- Bakan A, Dutta A, Mao W, Liu Y, Chennubhotla C, Lezon TR, and Bahar I (2014). Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics 30, 2681–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiaux B, Bernard A, Rieping W, Habeck M, Malliavin TE, and Nilges M (2008). Influence of different assignment conditions on the determination of symmetric homodimeric structures with ARIA. Proteins 75, 569–585. [DOI] [PubMed] [Google Scholar]
- Bernard A, Vranken WF, Bardiaux B, Nilges M, and Malliavin TE (2011). Bayesian estimation of NMR restraint potential and weight: a validation on a representative set of protein structures. Proteins 79, 1525–1537. [DOI] [PubMed] [Google Scholar]
- Berry JL, and Pelicic V (2015). Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39, 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007). Version 1.2 of the Crystallography and NMR system. Nat Protoc 2, 2728–2733. [DOI] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA, and Francetic O (2010). Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci U S A 107, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, and Pelicic V (2013). Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci U S A 110, 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros DA, Pehau-Arnaudet G, and Francetic O (2012). Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol Microbiol 86, 805–818. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, and Tainer JA (2006). Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell 23, 651–662. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. , (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl. Acids Res. 35, W375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Amorim GC, Cisneros DA, Delepierre M, Francetic O, and Izadi-Pruneyre N (2014). 1H, 15N and 13C resonance assignments of PpdD, a type IV pilin from enterohemorrhagic Escherichia coli. Biomol NMR Assign, 43–46. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Egelman EH (2000). A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Dalia TN, Ceballos AV, Wang JC, Biais N, Brun YV, and Dalia AB (2018). Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat Microbiol 3, 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J.e.a. (1996). SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, and Tainer JA (2011). Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J Biol Chem 286, 44254–44265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhaus A-F, and Duménil G (2014). The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J 33, 1767–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisetty H, Ovchinnikov S, and Baker D (2013). Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A 110, 15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczan JM, Lenneman BR, McGrath MJ, and Sundin GW (2011). Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol 77, 7031–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, and Craig L (2016). Structure of the Neisseria meningitidis Type IV pilus. Nat Commun 7, 13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma MA, Ochoa SA, Cruz A, Rocha-Ramírez LM, Mas-Oliva J, Eslava CA, Girón JA, and Xicohtencatl-Cortes J (2010). The Hemorrhagic Coli Pilus (HCP) of Escherichia coli O157:H7 is an inducer of proinflammatory cytokine secretion in intestinal epithelial cells. PLoS One 5, e12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, and Nilges M (2003). Refinement of protein structures in explicit solvent. Proteins 50, 496–506. [DOI] [PubMed] [Google Scholar]
- Luna Rico A, Thomassin J-L, and Francetic O (2018a). Analysis of bacterial pilus assembly by shearing and immunofluorescence microscopy. Methods Mol Biol 1764, 291–305. [DOI] [PubMed] [Google Scholar]
- Luna Rico A, Zheng W, Nadeau N, Egelman EH, and Francetic O (2018b). Functional reconstitution of enterohemorrhagic Escherichia coli O157:H7 type 4 pilus assembly Mol. Microbiol. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey N, and Blokesch M (2016). The DNA-uptake process of naturally competent Vibrio cholerae. Trends Microbiol. 24, 98–110. [DOI] [PubMed] [Google Scholar]
- Mindell JA, and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 142, 334–347. [DOI] [PubMed] [Google Scholar]
- Monteiro R, Ageorges V, Rojas-Lopez M, Schmidt H, Weiss A, Bertin Y, Forano E, Jubelin G, Henderson IR, Livrelli V, et al. (2016). A secretome view of colonisation factors in Shiga toxin-encoding Escherichia coli (STEC): from enterohaemorrhagic E. coli (EHEC) to related enteropathotypes. FEMS Microbiol Rev. 363, pii: fnw179. [DOI] [PubMed] [Google Scholar]
- Nguyen Y, Boulton S, McNicholl ET, Akimoto M, Harvey H, Aidoo F, Melacini G, and Burrows LL (2018). A highly dynamic loop of the Pseudomonas aeruginosa PA14 type IV pilin is essential for pilus assembly. ACS Infect Dis. 4, 936–943. [DOI] [PubMed] [Google Scholar]
- Nilges M, Bernard A, Bardiaux B, Malliavin T, Habeck M, and Rieping W (2008). Accurate NMR structures through minimization of an extended hybrid energy. Structure 16, 1305–1312. [DOI] [PubMed] [Google Scholar]
- Nivaskumar M, Bouvier G, Campos M, Nadeau N, Yu X, Egelman EH, Nilges M, and Francetic O (2014). Distinct docking and stabilization steps of the pseudopilus conformational transition path suggest rotational assembly of type IV pilus-like fibers. Structure 22, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M, Santos-Moreno J, Malosse C, Nadeau N, Chamot-Rooke J, Tran Van Nhieu G, and Francetic O (2016). Pseudopilin residue E5 is essential for recruitment by the type 2 secretion system assembly platform. Mol Microbiol 101, 924–941. [DOI] [PubMed] [Google Scholar]
- Ottiger M, Delaglio F, and Bax A (1998). Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson 131, 373–378. [DOI] [PubMed] [Google Scholar]
- Pelicic V (2008). Type IV pili: e pluribus unum? Mol Microbiol 68, 827–837. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, and Söding J (2011). HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9, 173–175. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, and Nilges M (2007). ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382. [DOI] [PubMed] [Google Scholar]
- Sali A, and Blundell TL (1993). Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Santos-Moreno J, East A, Bond PJ, Tran Van Nhieu G, and Francetic O (2017). Polar N-terminal residues conserved in type 2 secretion pseudopilins determine subunit targeting and membrane extraction during fibre assembly. J Mol Biol 429, 1746–1765. [DOI] [PubMed] [Google Scholar]
- Sauvonnet N, Gounon P, and Puglsey AP (2000). PpdD Type IV Pilin of Escherichia coli K-12 Can Be Assembled into Pili in Pseudomonas aeruginosa. J Bacteriol 182, 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, and von Jagow G (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schrödinger L (2015). The PyMOL Molecular Graphics System, Version 1.8.
- Seemayer S, Gruber M, and Söding J (2014). CCMpred-fast and precise prediction of protein residue-residue contacts from correlated mutations. Bioinformatics 30, 3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, and Bax A (2009). TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR 44, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Cameron ADS, and R.J. R (2009). Sxy induces a CRP-S regulon in Escherichia coli. J Bacteriol 191, 5180–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, and Redfield RJ (2012). Natural DNA uptake by Escherichia coli. PLoS One 7, e35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, and Lory S (1991). Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem 266, 1656–1664. [PubMed] [Google Scholar]
- Suh JY, Spyracopoulos L, Keizer DW, Irvin RT, and Sykes BD (2001). Backbone dynamics of receptor binding and antigenic regions of a Pseudomonas aeruginosa pilin monomer. Biochemistry 40, 3985–3995. [DOI] [PubMed] [Google Scholar]
- Tang G.e.a. (2007). EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Toth-Petroczy A, Palmedo P, Ingraham J, Hopf TA, Berger B, Sander C, and Marks DS (2016). Structured states of disordered proteins from genomic sequences. Cell 167, 158–170.e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik EJ, Giltner CL, Audette GF, Keizer DW, Bautista DL, Slupsky CM, Sykes BD, and Irvin RT (2005). DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol 187, 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH, and Craig L (2017). Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae Type IV Pili at Sub-nanometer Resolution. Structure. 25, 1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, and Sykes BD (1995). 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6, 135–140. [DOI] [PubMed] [Google Scholar]
- Wriggers W (2012). Conventions and workflows for using Situs. Acta Crystallographica Section D, Biological Crystallography, 68(Pt 4), 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Ledesma MA, Jordan D, Francetic O, Kaper JB, Puente JL, and Girón JA (2007). Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J Clin. Investig 117, 3519–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Saldana Z, Ledesma MA, Puente JL, and Giron JA (2009). The type 4 pili of Enterohemorrhagic Escherichia coli O157:H7 are multipurpose Structures with pathogenic attributes. J Bacteriol 191, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweckstetter M, and Bax A (2000). Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. J. Am. Chem. Soc 122, 3791–3792. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PpdDp structure and NMR restraints have been deposited in the Protein Data Bank under the accession id 6GMS. The PpdD pilus structure and cryo-EM map are available from the Protein Data Bank (accession code 6GV9) and Electron Microscopy Data Bank (accession code EMD-0070), respectively.


