 This review summarizes the recent development of IDO1 inhibitors, focusing on structures, enzymatic inhibitory activity, selectivity and other biological activities.
This review summarizes the recent development of IDO1 inhibitors, focusing on structures, enzymatic inhibitory activity, selectivity and other biological activities.
Abstract
Indoleamine 2,3-dioxygenase 1 (IDO1), an important immunoregulatory enzyme ubiquitously expressed in various tissues and cells, plays a key role in tryptophan metabolism via the kynurenine pathway and has emerged as an attractive therapeutic target for the treatment of cancer and other diseases, such as Alzheimer's disease and arthritis. IDO1 has diverse biological roles in immune suppression and tumor progression by tryptophan catabolism. In addition, IDO1-mediated immune tolerance assists tumor cells in escaping the immune surveillance. Recently, extensive and enormous investigations have been made in the discovery of IDO1 inhibitors in both academia and pharmaceutical companies. In this review, IDO1 inhibitors are grouped as tryptophan derivatives, inhibitors with an imidazole, 1,2,3-triazole or tetrazole scaffold, inhibitors with quinone or iminoquinone, N-hydroxyamidines and other derivatives, and their enzymatic inhibitory activity, selectivity and other biological activities are also introduced and summarized.
1. Introduction
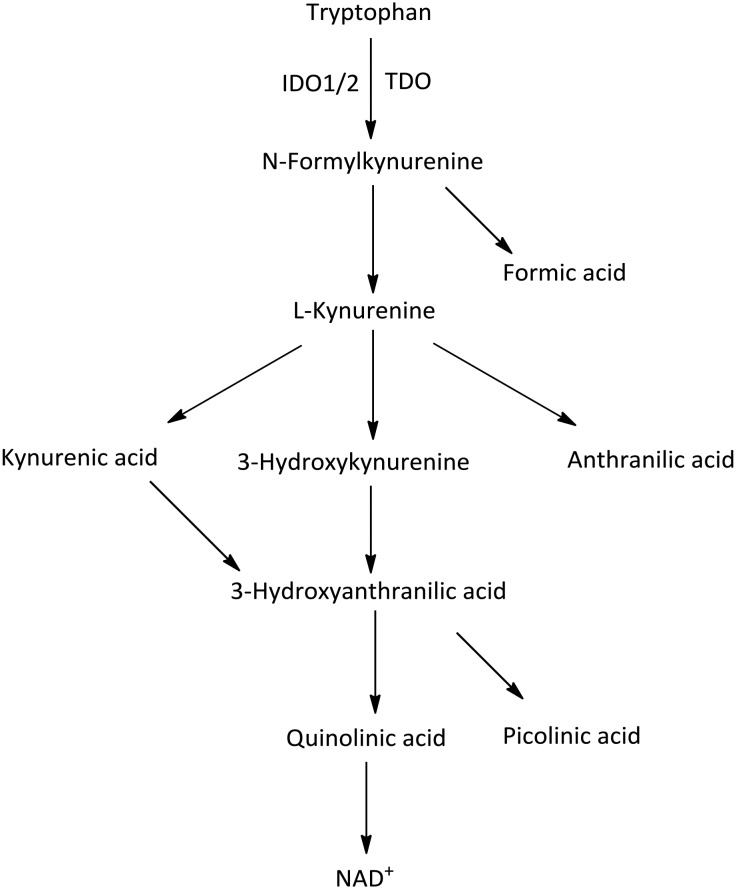
Tryptophan (Trp) is an essential amino acid for protein synthesis and is used in a variety of catabolic processes which is metabolized into serotonin, melatonin, niacin, auxins, and kynurenine (Kyn).1 The first, rate-limiting step of the kynurenine pathway is the oxidative cleavage of the 2,3-double bond of the indole ring in tryptophan to produce N-formylkynurenine by three distinct heme-containing enzymes, indoleamine 2,3-dioxygenase 1 (IDO1), IDO2, and tryptophan 2,3-dioxygenase (TDO).2 Then the generated N-formylkynurenine is further metabolized to bioactive metabolites, including kynurenine, kynurenic acid, 3-hydroxykynurenine and quinolinic acid (Fig. 1).3 However, the three enzymes show obvious differences in tissue distribution and substrate specificity.4 IDO1 is a monomeric enzyme mainly expressed in various tissues and cells throughout the body, such as the small intestine, epididymis, placenta, central nervous system (CNS), thymus, spleen, pancreas, kidney, macrophages, dendritic cells and microglial cells. IDO1 has broad substrate specificity for tryptophan compounds, such as d- and l-tryptophan, tryptamine, 5-hydroxy-l-tryptophan, serotonin, and melatonin.5 IDO2, with 42% structural similarity to IDO1 at the amino acid level, is constitutively expressed in the kidney, liver, spermatozoa, and dendritic cells, but it is oxidatively capable of cleaving l-tryptophan with a low turnover rate.6,7 TDO is predominantly expressed in the liver and is highly specific for l-tryptophan.8
Fig. 1. Kynurenine pathway of tryptophan metabolism induced by IDO1/IDO2/TDO.
IDO1 activity is low and exerts limited physiological effects in healthy humans, but IDO1 is overexpressed in response to inflammatory challenges under pathophysiological conditions (e.g., cancer, allergic inflammation, infection).9 IDO1-mediated immune tolerance plays a key role in helping tumor cells to escape the immune surveillance in the tumor microenvironment by depleting Trp and accumulating toxic Trp metabolites, resulting in the suppression of T-cell responses and the enhancement of immunosuppression mediated by regulatory T cells (Tregs).10–12 It has been demonstrated that IDO1 is constitutively expressed in a large number of human cancers13 and high IDO1 expression is associated with poor prognosis.14 IDO1 expression can be induced by interferon-γ (IFN-γ), tumor necrosis factor α (TNF-α) and other inflammatory mediators.15–17 IDO1 inhibition suppresses tumor proliferation and improves response to cancer chemotherapy, radiotherapy and immunotherapy.18–21 Therefore, IDO1 has emerged as a promising drug target for cancer immunotherapy.3,19 Recently, TDO and IDO2 have been elucidated in immunosuppression, but IDO2 is less well studied due to its very low Trp degradation activity.22,23
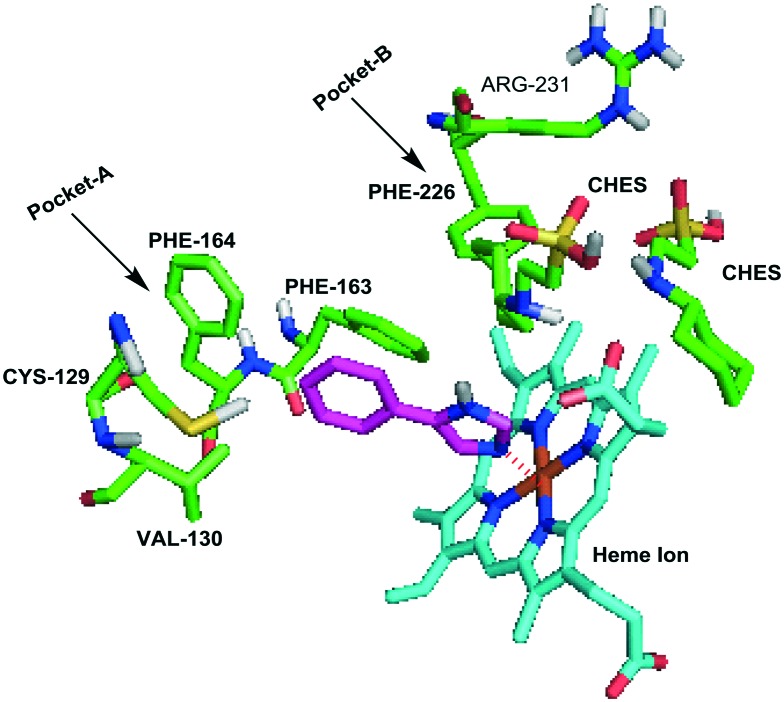
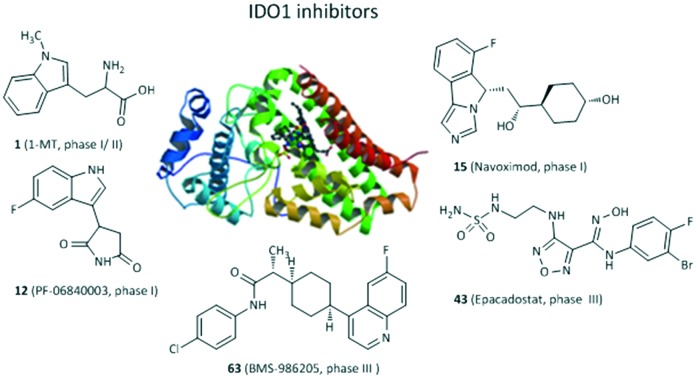
Since 2006, the X-ray crystal structures of human IDO1 in a complex with ligands 4-phenylimidazole (PDB ID: 2D0T, Fig. 2) and cyanide (PDB ID: ; 2D0U) have opened the door for in silico design of new IDO1 inhibitors,24 accelerating the development of new IDO1 inhibitor scaffolds and the discovery of a number of co-crystals of IDO1 with novel structures except the tryptophan-based scaffolds, such as PF-06840003,25 NLG919 analogues,26 and imidazothiazole derivatives.27 The active site of IDO1 is divided into three regions: pocket A, pocket B, and a heme iron. There are several reviews of IDO1 inhibitors published in recent years.19,28–32 They mainly focused on the function of IDO1 and its role in cancer as well as the latest clinical results of potential candidates in clinical trials. In this review, we provide the discovery and basic biological activities of IDO1 inhibitors, which is the first step in the development of IDO1 inhibitors, and hope to help researchers who are designing IDO1 inhibitors. These IDO1 inhibitors are grouped into structurally different classes including tryptophan derivatives, inhibitors with an imidazole, 1,2,3-triazole or tetrazole scaffold, inhibitors with quinone or iminoquinone, N-hydroxyamidines and other derivatives, and introduced in the following sections.
Fig. 2. The co-crystallized structure of 4-PI with IDO1 (PDB ID: 2D0T).
2. Tryptophan or indole analogues
IDO1 was first isolated in 1967 and at the same time, its natural substrate l-tryptophan could inhibit the activity of IDO1 at high concentration, while d-tryptophan had almost no inhibitory effect on IDO1.33 Although tryptophan analogues are the first discovered IDO1 inhibitors and studies of tryptophan derivatives have been carried out for many years, the IDO1 activity of most inhibitors still remains at the level of micromolar activity, which requires further study.
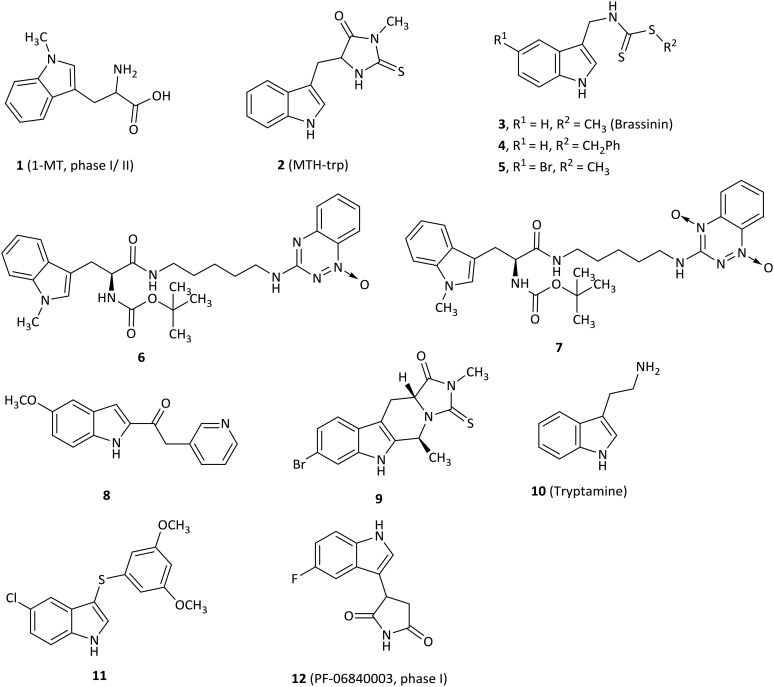
The most frequently used IDO1 inhibitor in preclinical studies, 1-methyl-d,l-tryptophan (1, 1MT, Fig. 3), had a reported Ki of 34 μM by NewLink Genetics.34,35 Further research studies revealed that the l isomer (L-1MT) was actually the more potent IDO1 inhibitor, but the d isomer (D-1MT, also known as indoximod) was more active for IDO2.36–39 Preclinical findings supported that D-1MT was selected as the lead compound for human trials because D-1MT was significantly effective in reversing the suppression of T cells and enhancing anticancer activity.36,40 Strikingly, indoximod was involved in a novel IDO1 effector mechanism in which indoximod reversed mTORC1 inhibition substantially under tryptophan-depleted conditions.41 A phase I trial of indoximod combined with docetaxel suggested that in a dose escalation study of indoximod, 22 of 27 patients were found to have good tolerance to the maximum dose of 1200 mg orally twice per day with no increase in expected toxicities or pharmacokinetic interactions in patients with metastatic solid tumors. In addition, there were 4 partial responses (2 breast, 1 NSCLC, 1 thymic tumor) and no drug–drug interactions were noted. The most common adverse events were fatigue, anemia, hyperglycemia, infection and nausea (NCT01191216).42 Another phase I study of indoximod showed that indoximod was safe at doses up to 2000 mg orally twice per day in 48 advanced cancer patients. The plasma AUC and Cmax of indoximod plateaued above 1200 mg. Cmax occurred at 2.9 hours, and the half-life was 10.5 hours. Notably, 5 patients showed stable disease at >6 months and 3 patients previously treated with the checkpoint inhibitor ipilimumab developed hypophysitis (an autoimmune signature of this inhibitor) at 200 mg once per day (NCT00567931).40 Currently, 3 phase I or II trials of indoximod in combination with chemotherapeutic agents or checkpoint inhibitors in patients with a variety of solid tumors are ongoing (NCT01560923, NCT02073123, NCT02052648). 4 phase I or II trials are recruiting, combining indoximod with different chemotherapeutic agents or checkpoint inhibitors in patients with a variety of malignancies (NCT02835729, NCT02502708, NCT03301636, NCT02913430).
Fig. 3. Tryptophan or indole analogues.
An indole derivative methyl-thiohydantoin-tryptophan (2, MTH-trp, also called necrostatin 1), as a necroptosis inhibitor, was screened among commercially available compounds and was determined as a competitive inhibitor of IDO1 (Ki = 11.6 μM).43 MTH-trp was about 20-fold more potent than 1MT in a cell-based assay (EC50 = 12.85 μM) and the combination therapy of MTH-trp with paclitaxel also produced better tumor regression than with 1MT.
SAR studies of brassinin-derived analogues revealed that substitution of the S-methyl group on the brassinin core with large aromatic groups enabled the inhibitors to be more potent for IDO1 activity than 3 (brassinin, Ki = 97.7 μM), such as compound 4 (Ki = 13.22 μM).44 Compound 5 (5-Br-brassinin) behaved as an IDO1 inhibitor (Ki = 24.5 μM), which inhibited both human and mouse COS-1 cell lines with EC50 values of 24.0 μM and 26.1 μM, respectively.45 Notably, 5 suppressed B16-F10 tumor outgrowth substantially in wild-type mice, but not in IDO-null mice and athymic nude mice, indicating that the mechanism of anticancer action of brassinin-based compounds might require IDO1 activity which is essential for T cell immunity.
1-Methyltryptophan (1MT)–tirapazamine hybrids, which combined the scaffolds of 1MT and the hypoxic cytotoxin tirapazamine (TPZ) moiety, were expected to have dual roles as antineoplastic agents.46 TPZ-monoxide 6 was the most potent IDO1 inhibitor among these hybrids with a Ki value of 76.3 μM, but with low hypoxic cytotoxicity (IC50 = 33 μM). In marked contrast, the corresponding parent dioxide hybrids 7 (Ki = 197 μM) first acted as a hypoxic cytotoxin and then was metabolized to TPZ-monoxide 6 as an IDO1 inhibitor.
The keto-indole derivative 8 was described as an IDO1 inhibitor with an IC50 value of 13.1 μM by a virtual screening and exhibited an uncompetitive inhibition mode by detailed kinetics.47,48 Importantly, SAR studies showed that the ketone moiety was crucial for IDO1 inhibitory activity, consistent with the docking results that the oxygen atom of ketone was coordinated with the heme iron of the IDO1 active site. The tryptoline analogue 9 with a bromo substituent at the C6′ position on the phenyl ring exhibited IDO1 activity with an IC50 value of 46.1 μM, better than that of MTH-trp (2, IC50 = 76.9 μM),43 indicating that the substitution at the C6′ position was preferred for potent biological activity.49
A natural endogenous compound, tryptamine (10), exhibited noncompetitive inhibitory activity against IDO1 (Ki = 156 μM) and suppressed kynurenine production without affecting A172 cell viability.50 Furthermore, tryptamine improved the antitumor activity of peripheral blood mononuclear cells (PBMCs) in co-culture assays.
The most effective compound (11) of this class of indole-based derivatives was obtained by a structure-based virtual screening study and had an IC50 value of 7 μM for IDO1.51 The SAR data highlighted that the single sulfur atom bridge contributed to the more remarkable potency on IDO1 compared to its oxidation to sulphone and substitution with ketone or a methylene group. Furthermore, 11 significantly induced a dose-dependent growth inhibition of HTC116 and HT29 cancer cell lines, both of which expressed IDO1,52 while it exhibited a reversible cell cycle arrest in both cells.
PF-06840003 (12) was identified as a novel potent and selective IDO1 inhibitor with an IC50 of 0.41 μM by a new and sensitive high-throughput mass spectrometry assay.25 Actually, only the R-enantiomer was responsible for the IDO1 inhibitory activity (IC50 = 0.2 μM), and the IC50 value of the S-enantiomer was 38 μM. Surprisingly, the rapid conversion of the R-enantiomer to the S-enantiomer was found in plasma of all preclinical species, indicating that the racemate was more valuable in the following preclinical and clinical trials than the active pure enantiomer. In a cellular assay, PF-0684003 presented excellent activity in both HeLa cells and THP1 cells (IC50 = 1.8 and 1.7 μM, respectively). X-ray and spectroscopy results showed that PF-06840003 was a noncompetitive, non-heme binding IDO1 inhibitor. It had good potency in a human whole blood assay (IC50 = 4.7 μM). Based on the excellent pharmacokinetic characteristics in preclinical species, PF-06840003 was predicted to possess favorable PK profiles in humans with a long half-life of 19 hours and bioavailability of 64%. It could also permeate CNS well, which ensured that it has the potential to treat brain metastases. The first clinical study to evaluate the safety and tolerability of increasing doses of PF-06840003 in patients with malignant gliomas had been completed by Pfizer Pharmaceuticals and initiated in 2016 (NCT02764151).
3. Inhibitors with imidazole, 1,2,3-triazole or tetrazole
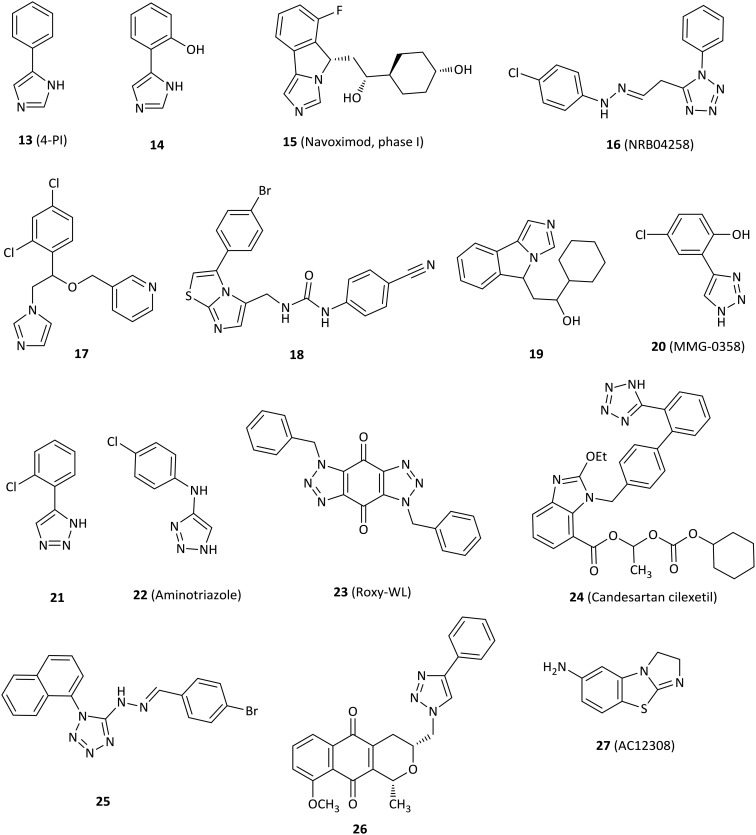
The first crystal structure of IDO1 in complex with 4-phenylimidazole (13, 4-PI, Fig. 4) that was a weak noncompetitive IDO1 inhibitor (IC50 = 48 μM) gained much attention in 2006.24,53 4-PI interacts directly with heme iron at the imidazole nitrogen and its phenyl group occupies a hydrophobic domain (pocket A). Two molecules of the buffer N-cyclohexyl-2-aminoethanesulfonic acid (CHES) occupy the adjacent hydrophobic domain (pocket B). In pocket A, Phe163 interacts with the phenyl group of 4-PI in the π–π stacking. At the back of the pocket, the amino acids Phe164, Val130 and Cys129 contribute to the wall but are far from the iron. In pocket B, both residues of Phe226 and Arg231 form part of the entrance of pocket B and are directly involved in substrate recognition by hydrophobic interactions (Fig. 2). This binding mode of IDO1 with 4-PI provides a basis for the design and development of 4-PI-derived IDO1 inhibitors. Structural modification of 4-PI or substitution of imidazole with 1,2,3-triazole and tetrazole gave a series of active compounds.
Fig. 4. Inhibitors with imidazole, 1,2,3-triazole or tetrazole.
Modification of 4-PI led to derivative 14 designed by utilizing the first reported crystal structure of 4-PI bound to IDO1, which had an IC50 value of 4.8 μM and showed a 10-fold improvement on IDO1 potency relative to that of 4-PI.24,54
The imidazoleisoindole derivative navoximod (15, NLG919) was developed as a noncompetitive IDO1 inhibitor by a rational structural design based on the X-ray crystal structure of IDO1 in complex with 4-PI,24 displaying an EC50 value of 75 nM in a cell-based assay with 10- to 20-fold selectivity against TDO.55 The in vivo study revealed that navoximod had good pharmacokinetic profiles as an orally active agent and significantly reduced the concentration of kynurenine in mice plasma by approximately 50%. Navoximod greatly enhanced vaccine responses against B16 melanoma, thus reducing the tumor size by approximately 95% within 4 days of vaccination.56 Combination treatment of navoximod with anti-PD-L1 blockade showed stronger efficacy in activating intratumoral CD8+ T cells and inhibiting tumor growth than either of the two compounds alone.57 Despite having good preclinical profiles, only one phase Ia study of navoximod in monotherapy had been completed (NCT02048709) and one study is being evaluated in combination with atezolizumab for the treatment of advanced or metastatic solid tumors (NCT02471846). Navoximod alone was well tolerated at doses of up to 800 mg twice per day on a 21/28 day cycle, but the majority of patients showed slight clinical anticancer effects. The best response was limited to stable disease observed in 7 of 17 patients.57
A series of imidazole derivatives and drug-like molecules were screened by the Prestwick Chemical Library and the Maybridge HitFinder Collection.58 Regrettably, most of the compounds failed structural filters. Compound NRB04258 (16) displayed micromolar activity for IDO1 (IC50 = 8.8 μM), but it demonstrated stronger potency at the cellular level (IC50 = 0.34 μM). The imidazole antifungal agents showed micromolar IC50 values against IDO1, such as miconazole (IC50 = 6.7 μM) and econazole (IC50 = 8.1 μM). Structure-based lead optimization of the imidazole scaffold provided more soluble but less active compounds compared to imidazole antifungal drugs in an enzymatic assay, such as compound 17 (IC50 = 84 μM).
The imidazothiazole derivative 18 with a carbamido linker showed the best enzymatic IC50 value of 77 nM among the designed compounds.27 The highlights of the research were that IDO1 inhibitors interacted directly with both pocket A and pocket B and formed an interaction with Phe226 and Arg231 of pocket B, which were essential for potent inhibitory activity against IDO1.
A navoximod analogue 19 was a potent inhibitor with an IC50 value of 38 nM against the IDO1 enzyme and an EC50 value of 61 nM against HeLa cells.26 Structural biology studies of 19 and its analogues indicated that 19 occupied both pocket A and pocket B. In detail, the imidazoleisoindole core was located in pocket A and formed extensive hydrophobic interactions. The nitrogen atom of imidazole coordinated with the heme iron. The 1-cyclohexyl ethanol moiety extended to pocket B and formed hydrophobic interactions with the surrounding residues. Additionally, 19 formed an extensive hydrogen bond network with IDO1, which contributed to the great potency of imidazoleisoindole derivatives.
Rational optimization based on a previous study59 resulted in the 4-aryl-1,2,3-triazole scaffolds as IDO1 inhibitors by the in silico strategy.60 The most active compound 20 (MMG-0358) showed an IC50 value of 330 nM against IDO1 in an enzymatic assay without activity against TDO. It also showed IC50 values of 2 nM and 80 nM on mIDO1 and hIDO1 in a cellular assay without cellular toxicity.
Compound 21, identified as a reversible and uncompetitive IDO1 inhibitor (Ki = 14.5 μM), showed enzymatic and cellular IC50 values of 86 μM and 19.3 μM, respectively.61 Strikingly, 21 also improved T cell proliferation in the presence of LLC cells. The SAR results and molecular docking studies indicated that an electron-withdrawing group with small steric hindrance near the NH group of triazole was essential for the IDO1 inhibition.
1,2,3-Triazole derivative 22 (aminotriazole) was a remarkable noncompetitive IDO1 inhibitor discovered and optimized by high-throughput screening in both HeLa cells and HEK293 cells (IC50 = 23 nM and IC50 = 67 nM, respectively), despite poor inhibition in an enzymatic IDO1 assay (IC50 = 11.3 μM) compared to cell potency.62 Comprehensive studies using biochemical, spectroscopic and crystallographic methods showed that 22 could form a tight complex with ferrous IDO1 with slow association and dissociation kinetics, which partially accounted for the potent cellular activity.
Compound 23 (Roxy-WL), a very potent IDO1 inhibitor (IC50 = 1 nM), was discovered by molecular docking and pharmacophore modeling, with outstanding selectivity over 337 kinases.63 Roxy-WL not only reduced the conversion of native CD4+ T cells to Treg cells but also effectively suppressed tumor growth (inhibition of 91.5%), inhibited IDO1 expression, reduced the number of Foxp3+ Tregs and decreased the Kyn/Trp ratio.
The antihypertensive agent candesartan cilexetil (24) demonstrated IDO1 inhibitory activity with a noncompetitive inhibition mode and had both micromolar enzymatic and cellular potency with IC50 values of 12 μM and 2.6 μM, respectively.64 SAR and docking studies suggested that candesartan derivatives occupied the entrance at the active site of IDO1, but not the heme region.
Compound 25 with a tetrazole motif displayed low micromolar inhibitory potency (IC50 = 8.8 μM) by a novel and effective high-throughput virtual screening combining both pharmacophore modeling and molecular docking, which might be used to find potential IDO1 inhibitors in the future.65
The pyranonaphthoquinone derivative 26 displayed micromolar potency against IDO1 (IC50 = 6 μM) with little cytotoxicity and loss of cell viability, but it still remarkably decreased kynurenine production by over 60% in drug-treated cells at low concentrations.66
Small molecule AC12308 (27) displayed slight micromolar IDO1 enzymatic activity (IC50 = 50 μM) and possessed high ligand efficiency by multiple pharmacophores in conjunction with docking techniques to perform in silico screening, which could be a good starting point for further optimization studies.67
4. Inhibitors with quinone or iminoquinone
As early as 1961, it was found that compounds with redox activity such as benzoquinone and hydroquinone potently inhibited TDO.68 In 2006, annulin and other hydroxynaphthoquinones were first found to have high inhibitory activity against IDO1 and quinine oxygen interacted with heme iron.69 Since then, the study of quinine derivatives became a hot spot.
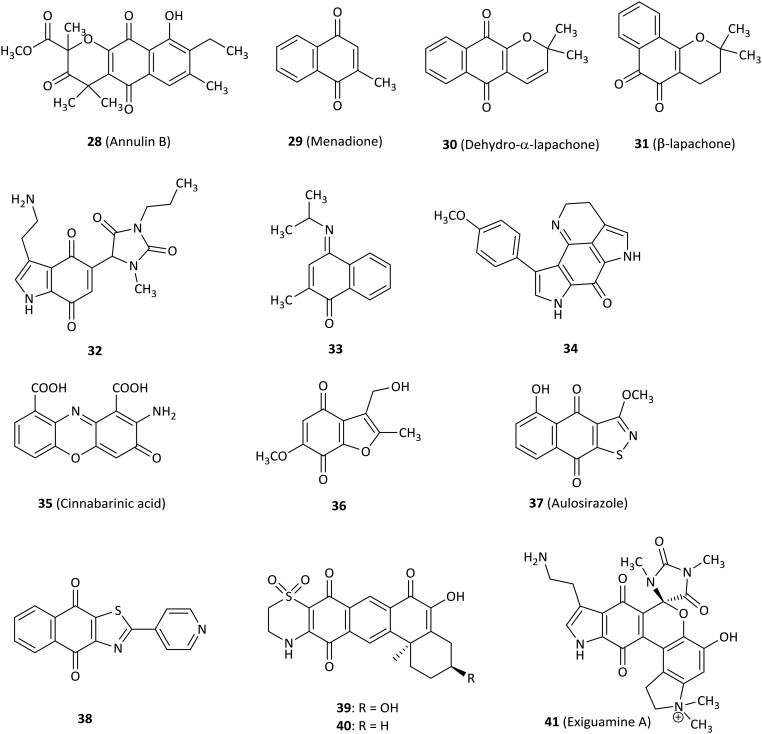
Compound 28 (annulin B, Fig. 5) was extracted from the marine hydroid Garveia annulata and showed submicromolar inhibitory activity for IDO1 (Ki = 0.12 μM), which was a known compound with naphthoquinone as the key pharmacophore identified by comparing the spectroscopic data with the literature values.69 Even though 28 was unexpectedly inactive in a yeast cell-based IDO1 inhibition assay, it remained to determine whether 28 was active in human cells.70 A novel series of 1,4-naphthoquinone-based derivatives were synthesized and evaluated as IDO1 inhibitors.71 Compound 29 (menadione, known as vitamin K3)72 was active against IDO1 (IC50 = 1.1 μM) and substantially showed reduced tumor growth in a mouse B16F10 melanoma tumor graft model, demonstrating both functional T cell immunity and IDO1 inhibition involved in antitumor activity. Docking studies of tricyclic pyranonaphthoquinone 30 (dehydro-α-lapachone) with an IC50 value of 0.21 μM indicated that naphthoquinone structures displayed a noncompetitive mode of inhibition for IDO1, even though molecular docking predicted direct binding at the IDO1 active site. However, the cell-based IDO1 activity of several pyranonaphthoquinones tested was attenuated. To improve cell-based potency, a 1,2-naphthoquinone-based derivative 31 (β-lapachone) was identified as an IDO1 inhibitor with low potency (IC50 = 0.44 μM) relative to 30, but it exhibited superior intracellular IDO1 inhibitory activity with an IC50 of 1.0 μM in IFN-γ-induced HeLa cells.73 Nonlinear regression analysis also confirmed the uncompetitive mode of inhibition. The results were consistent with the conclusion obtained from compound 30 (ref. 71) and IDO1 expression was unaffected by exposing HeLa cells to 5 μM β-lapachone by western blot analysis. These data showed that these naphthoquinones are excellent lead compounds for further study as potential IDO1 inhibitors.74
Fig. 5. Inhibitors with quinone or iminoquinone.
Combining the hydantoin and tryptaminequinone structures produced the uncharged compound 32, which was a potent and uncompetitive IDO1 inhibitor with a Ki of 200 nM based on the lead structure of exiguamine A, expecting the structure to be stable and to easily cross the cell membrane in the following research.75
Various structural compounds were developed as IDO1 inhibitors at the enzymatic or the cellular level, including the 4-aryl-1,2,3-triazole scaffold.59 For example, compound 33 showed an IC50 of 200 nM for IDO1 enzyme, but the result of cells transfected with human IDO1 displayed disappointing data compared to enzyme activity (IC50 > 50 μM). Nevertheless, in silico screening combining both pharmacophore-based lead design and fragment-based virtual lead design was a precedent to search for novel IDO1 inhibitors.
The tsitsikammamine A analogue 34 exhibited good IC50 activity in the submicromolar range in an enzyme test (IC50 = 0.9 μM) rationalized by molecular modeling studies, but it showed a drop in potency at the cellular level, which probably resulted from the fact that 34 had low cell membrane permeability due to its high polarity.76 Cinnabarinic acid (35), a member of 2-aminophenoxazin-3-ones, showed submicromolar inhibitory activity against IDO1 with Ki and IC50 values of 362 nM and 460 nM, respectively.77 It was also noted that containing an additional electron-withdrawing group was beneficial for IDO1 activity.
Compound 36 was the most potent IDO1 inhibitor (IC50 = 0.24 μM) among a range of benzofuranquinones.78 Importantly, 36 not only did not generate visible damage to microtubules or cytoskeleton but also did not produce significant levels of oxidative stress and cytotoxicity at concentrations that inhibited IDO1.
A natural quinine derivative aulosirazole (37, isolated from blue-green alga) and its analogues were described as IDO1 inhibitors.79 Aulosirazole showed potent inhibitory activity against IDO1 with an IC50 value of 80.3 nM and was also involved in NQO1-mediated reduction to produce unstable hydroquinones with the generation of reactive oxygen species, which might prompt quinones for the development as antitumor agents.
Out of a series of novel naphthoquinone derivatives as IDO1 inhibitors, several compounds displayed excellent nanomolar IDO1 inhibitory activity (18–61 nM) without cytotoxicity tested in several types of cells.80 Compound 38 was one of the most potent IDO1 inhibitors with an IC50 value of 26 nM and had a high selectivity index of 61.5 against TDO. 38 further decreased the kynurenine level in rat plasma by 50.1%.
In 2014, compound 39, a natural product isolated from the sponge Xestospongia vansoesti, exhibited inhibitory activity for the IDO1 enzyme (IC50 = 4 μM).81 Compound 40 only missing the C-3 hydroxyl substituent compared with natural product 39, was synthesized by the photochemical coupling reaction which was firstly used in a natural product synthesis. 40 represented IC50 of 0.11 μM for IDO1 with 40 times more potent than 39.
Among a library of marine invertebrate extracts, exiguamine A (41, Fig. 6), a natural product isolated from the marine sponge Neopetrosia exigua, exhibited a Ki of 210 nM for purified recombinant human IDO1 in vitro. The data presented that it should be a valuable guide for determining other novel natural products as a new structural class of IDO1 inhibitors.82
Fig. 6. N-Hydroxyamidine inhibitors.
5. N-Hydroxyamidines
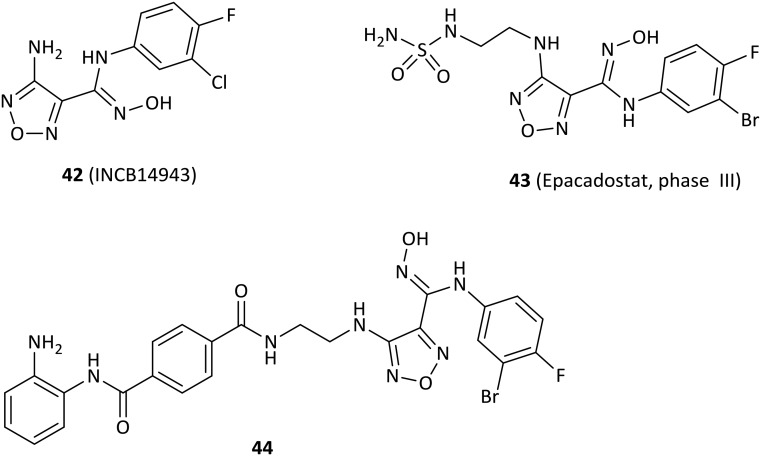
Over the past 10 years, high-throughput screening has played an important role in the discovery of new IDO1 inhibitor scaffolds. High-throughput screening of Incyte's corporate collection led to the discovery of N-hydroxyamidines as potent IDO1 inhibitors.83
Compound 42 (INCB14943, Fig. 6), the most potent compound of a novel series of hydroxyamidines, was found to be a competitive inhibitor of IDO1 with enzymatic and cellular IC50 values of 67 nM and 19 nM, respectively, which were identified and initiated by high-throughput screening of Incyte's corporate collection.83 Importantly, 42 decreased kynurenine generation by >50% in plasma and inhibited B16-GM-CSF tumor growth in a dose-dependent manner, but it had poor oral bioavailability in rodent pharmacokinetics. The hydroxyamidine moiety was responsible for binding IDO1, of which the oxygen coordinated with the heme iron at the active site by the docking model, in agreement with the results of enzyme kinetics and SAR results. Further modification of the lead compound 42 yielded the clinical lead agent epacadostat (43, INCB024360) developed by Incyte Corporation, which was a highly potent and selective IDO1 inhibitor in both HeLa cells (IC50 = 7.4 nM) and enzymatic assays (IC50 = 73 nM) with great selectivity over TDO, IDO2 (>1000-fold) and a panel of 50 other proteins.84 Although epacadostat failed the calculated “drug-like” filters, the in vitro ADME data were consistent with good cell permeability and oral bioavailability observed in all species tested (rat, dog, monkey), which were attributed to the two intramolecular hydrogen bonds observed in the crystal structure. Additionally, epacadostat was well tolerated in preclinical IND toxicology studies, enhanced the immunogenicity of dendritic cells and lytic ability of tumor antigen-specifc T cells,85 promoted the growth of effector T cells and NK cells, increased IFN-γ production, and reduced conversion to Treg cells in vitro.86In vivo, it expectedly suppressed tryptophan catabolism and impeded the growth of IDO1-expressing tumors.87 Furthermore, epacadostat was found to enhance the antitumor effect of anti-CTLA4 or anti-PD-L1 antibodies in the B16 melanoma mode.88 However, recent data showed that the phase III clinical trial of epacadostat in combination with pembrolizumab for the treatment of patients with unresectable or metastatic melanoma did not meet the primary endpoint of improving progression-free survival in the overall population compared to pembrolizumab alone (ECHO-301, NCT02752074). The negative outcome resulted in the termination or withdrawal of other clinical trials of epacadostat. Currently, 12 phase I or II trials are recruiting and 19 phase I, II or III clinical studies are evaluating epacadostat alone or combining epacadostat with checkpoint agents, chemotherapy or radiotherapy in patients with a variety of tumors.
The first generation of dual IDO1 and HDAC inhibitors were designed by pharmacophore fusion strategy.89 Particularly, the highly active dual inhibitor, compound 44 exerted excellent activity against both IDO1 (IC50 = 69.0 nM) and HDAC1 (IC50 = 66.5 nM), and showed excellent anticancer activity in the HCT-116 cell line (IC50 = 5.12 μM) without cellular toxicity. Importantly, 44 drastically reduced the kynurenine level in plasma and showed good in vivo antitumor efficacy in the murine LLC tumor model.
6. Others
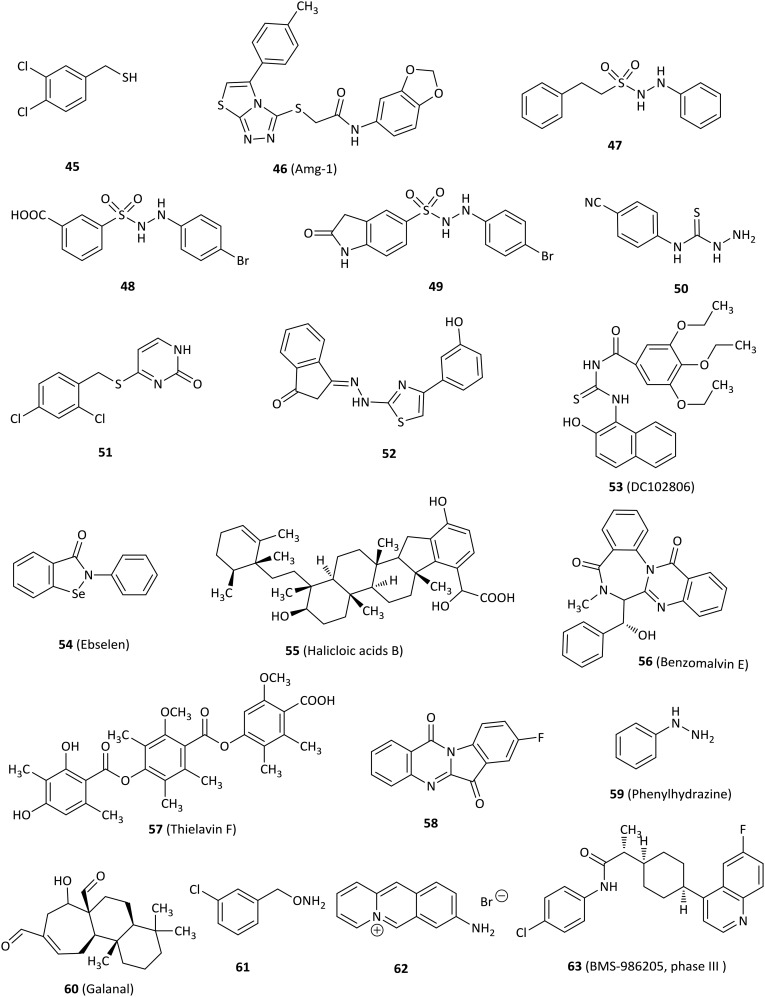
In addition to the above IDO1 inhibitors, there are many other kinds of IDO1 inhibitors, such as benzenesulfonyl hydrazides and thioureas. Fig. 7 shows some other types of IDO1 inhibitors.
Fig. 7. Other IDO1 inhibitors.
3,4-Dichlorophenylmethanethiol (45) significantly inhibited IDO1 (IC50 = 0.1 μM) and displayed inhibition of kynurenine production with an IC50 of 1.1 μM in human epithelial carcinoma A431 cells.90 The binding mode of 45 at the active site of IDO1 might be that the sulfhydryl group of benzylthiol directly chelated with the heme iron, because sulfhydryl group has potent metal coordinating property.
Amg-1 (46), a potent and reversible IDO1 inhibitor (IC50 = 3 μM), was discovered by high-throughput screening from Amgen's compound library and displayed at least 80-fold selectivity over IDO2 and at least 30-fold selectivity over TDO.91 The selectivity of Amg-1 to IDO1 might be rationally explained through molecular modeling results of Amg-1 binding to the structure of IDO1 and a homology model of IDO2. Amg-1 penetrated well into the active site of IDO1 and was predicted to interact with at least thirteen IDO1 residues including hydrogen bonding interactions between S167 of the side chain and two oxygens of the benzodioxole moiety, π–π stacking interaction between the benzodioxole group and F163, and hydrophobic interactions. By contrast, Amg-1 did not deeply penetrate into the IDO2 active site and was predicted to interact with nine IDO2 residues, not including hydrogen bonding interaction and π–π interaction.
A phenyl benzenesulfonylhydrazide derivative 48 was identified as a potent IDO1 inhibitor with an IC50 of 61 nM in an enzymatic assay and an EC50 of 171 nM in a HeLa cell-based assay based on the modification of 47, which acted as an IDO1 inhibitor (IC50 = 167 nM), via a high-throughput screening. Molecular docking studies of 48 with the IDO1 structure (PDB ID: ; 2D0T) suggested that the key interactions were the coordination of sulphone with heme iron, the hydrogen bond and hydrophobic interaction.92 Regrettably, 48 was degraded by 80% within 5 min in rat plasma. Further optimization to improve in vivo efficacy profiles yielded 49 which was a more potent and selective IDO1 inhibitor (hIDO, IC50 = 36 nM; HeLa cells, EC50 = 68 nM), with 59% oral bioavailability and no affinity to 67 tested proteins including receptors, transporters, and channels. 49 not only demonstrated tumor growth delay of 63% and 73% in a murine CT26 allograft model without body weight loss after oral administration with 200 mg kg–1 and 400 mg kg–1, respectively, but also showed 42% and 83% reduction of the ratios of Kyn/Trp in the plasma and tumor tissues of rats, respectively, after oral administration with 50 mg kg–1 at 5 h and 100 mg kg–1 at 6 h, respectively. 49 was also observed to reduce the final tumor weight by 64% relative to the vehicle in rats. Taken together, 49 might have potential for further research as an immunotherapeutic anticancer agent.93
Compound 50 bearing a cyano group at the 4-position on the aryl ring showed the best inhibitory activity for IDO1 among 32 phenylthiosemicarbazide, and the SAR results indicated that substitution at the 3- and 4-positions on the aromatic ring are more potent than in the 2-position.94 Compound 51, discovered by screening the compound library, was a moderate potent IDO1 inhibitor (IC50 = 7.5 μM). Moreover, 51 possessed increased activity in a cell-based assay (IC50 = 4.3 μM) and passed three structural filters.95 The SAR studies showed that Ser167 at the IDO1 active site was important for the potency of IDO1 inhibitors, in agreement with the molecular docking results.
Compound 52 containing a 1-indanone scaffold was found to be an IDO1 inhibitor by structure-based virtual screening, displaying an enzymatic IC50 value of 2.78 μM and a cellular EC50 value of 9.17 μM.96 SAR analysis revealed that 52 occupied both pocket A and pocket B of the IDO1, and the hydroxyl group at the 3-position was critical for activity, which formed a hydrogen bond with Phe226 validated by molecular docking.
Compound 53 (DC102806) acted as an IDO1 inhibitor with an IC50 of 18 μM in both enzymatic and HeLa cellular assays based on structure-based virtual screening.97 Molecular docking studies showed that the oxygen of 53 might bind to the heme of the IDO1 enzyme.
The antioxidant ebselen (54) represented a potent IDO1 inhibitor with a Ki value of 94 nM and inhibited IDO1 activity effectively in IFN-γ-stimulated human macrophages.98 Of note, ebselen inhibited IDO1 activity by reacting with the enzyme's cysteine residues, which altered the protein conformation and heme environment at the active site, resulting in disruption of the substrate binding pocket and increasing the level of nonproductive l-Trp binding.
Halicloic acid B (55) was discovered as an IDO1 inhibitor with an IC50 of 11 μM isolated from the marine sponge Haliclona sp. in an IDO1 inhibition assay in 2012.99
Benzomalvin E (56), a new benzodiazepine alkaloid isolated from fungal metabolites, showed activity in an IDO1 enzymatic assay and its IC50 value was determined as 21.4 μM, but no further research was done.100 In 2014, the same research group identified thielavin derivatives isolated from soil fungus as IDO1 inhibitors, among which thielavin F (57) was the most potent compound with an IC50 value of 14.5 μM.101
A series of tryptanthrin derivatives were synthesized and evaluated as novel potent IDO1 inhibitors and SAR studies indicated that an electron-withdrawing group at the 8-position of tryptanthrin was necessary for IDO1 inhibition.102 For example, compound 58 exhibited a Ki of 161 nM in an uncompetitive inhibition mode and excellent activity in both enzymatic and HEK 293 cell assays with IC50 values of 534 nM and 230 nM, respectively. In addition, 58 significantly enhanced T cell proliferations stimulated with Lewis lung cancer (LLC) cells, inhibited IDO1 activity, suppressed tumor growth and reduced the numbers of Foxp3+ Tregs when administered to LLC tumor bearing mice.
Phenylhydrazine (59) was identified as an IDO1 inhibitor by fragment screening and inhibited the IDO1 enzyme greatly (IC50 = 0.25 μM). It also inhibited mIDO1 and hIDO1 in transfected Lewis Lung carcinoma cells at noncytotoxic concentrations (IC50 = 0.2 μM and 1.3 μM, respectively).103 Nevertheless, its selectivity for IDO1 over other heme-containing proteins remains to be investigated to avoid off-target effects.
The phytochemical galanal (60) was discovered as an active IDO1 inhibitor in a competitive manner with an IC50 value of 7.7 μM against IDO1 and an IC50 value of 45 nM against LPS-stimulated THP-1 cells.104 Furthermore, galanal also inhibited expression of IDO1 mRNA induced by both the NFkB-dependent pathway and the IFN-γ-dependent pathway.
O-Benzylhydroxylamine derivatives that mimicked alkylperoxy species were designed based on the oxidative metabolism of Trp by IDO1. The coordination of oxygen to a ferrous heme iron led to formation of the alkylperoxy transition or intermediate state.105–107 One of the most potent derivatives, compound 61, had equivalent activity in both IDO1 enzymatic and cell-based assays (IC50 values of 0.3 μM and 0.14 μM, respectively), with good selectivity over other heme-containing enzymes (e.g. catalase, CYP3A4) and low cytotoxicity.
The 8-aminobenzo[b]quinolizinium bromide (62), derived from a screening of a natural compound library, was found to inhibit mIDO1 with an IC50 of 164 nM and showed the lowest cytotoxicity with GI50 values of 62 μM and 86 μM for Jurkat and A549 cells, respectively.10861 also inhibited parasite growth effectively in vitro (IC50 = 109 nM). However, 61 did not show an effect on parasite clearance or the life span of infected mice in vivo when it is administered orally.
BMS-986205 (63) was a novel and potent IDO1 inhibitor developed by Bristol-Myers Squibb for clinical research with an IC50 of 1.1 μM in HEK293 cells overexpressing human IDO1.109 In 2017, BMS-986205 alone and in combination with nivolumab in a phase 1/2a trial for the treatment of advanced cancer patients was reported at a meeting. 42 evaluable patients with advanced cancers were orally treated at 25–200 mg qd for 2 weeks, followed by a combination of BMS-986205 and nivolumab 240 mg every 2 weeks, presenting favorable preclinical profiles. All treatment-related adverse events were grade 1/2 except three grade 3 toxicities (dose-limiting autoimmune hepatitis, rash, and asymptomatic hypophosphatemia). Serum kynurenine reduction was observed at all doses including the lowest dose of 25 mg. Serum kynurenine was substantially reduced with >45% mean reduction at all doses and >60% mean reduction at the 100 and 200 mg qd doses. Importantly, BMS-986205 showed a significant reduction in intratumoral kynurenine in evaluable paired pre- and on-treatment samples. Due to the negative outcome of the combination of epacadostat with pembrolizumab in melanoma, 2 phase III clinical trials of BMS-986205 in combination with nivolumab in head and neck cancer and 1 in non-small cell lung cancer were terminated (NCT03386838, NCT03417037).
As an overview, Table 1 lists representative IDO1 inhibitors with their enzymatic and cellular activities, but we can see that most of the reported compounds are weak inhibitors against IDO1 with potency at the micromolar range. Among them, hydroxyamidine derivatives, imidazoleisoindole compounds and imidazothiazole derivatives exhibited superior inhibitory potency against IDO1 at the nanomolar range. Analysis and comparison with these crystal structures of IDO1 with 4-PI (13, IC50 = 48 μM),24 epacadostat (43, IC50 = 73 nM),8418 (IC50 = 77 nM)27 and 19 (IC50 = 38 nM)26 demonstrated that direct coordination to the heme iron is vital to the IDO1 inhibitory activity, such as chelating groups imidazole nitrogen and hydroxyl of hydroxyamidine. In the IDO1/4-PI complex structure, 4-PI only occupied the pocket A. In the IDO1/18 and IDO1/19 complex structures, the two compounds occupied both pocket A and pocket B to interact with the surrounding residues. Therefore, it may be beneficial to improve the activity against IDO1 by introducing aromatic groups that form π–π interactions with residues and introducing nonpolar functional groups, such as phenyl and cyclohexyl, that form hydrophobic interactions in pocket B. In the IDO1/19 and IDO1/epacadostat complex structures, there are extensive intramolecular and intermolecular hydrogen bond networks, which may contribute to the great potency of the two compounds. Briefly, an IDO1 inhibitor should be well situated in both pocket A and pocket B, interact with protein residues, bond with the heme iron atom, and contain a hydrogen bond acceptor and a hydrogen bond donor, all of which need to be considered in the future design of potent IDO1 inhibitors.
Table 1. Representative IDO1 inhibitors with their selected biological data.
| Compd | IDO1 a IC50 or Ki | Cell-based potency EC50 or IC50 | Ref. |
| 1 | 34 μM (Ki) | — b | 35 |
| 5 | 24.5 μM (Ki) | COS-1 cells; 24 μM | 45 |
| 12 | 0.41 μM (IC50) | HeLa cells; 1.8 μM; THP1 cells; 1.8 μM | 25 |
| 13 | 48 μM (IC50) | — | 53 |
| 15 | — | 75 nM | 55 |
| 16 | 54 μM (IC50) | P815 cells; 0.34 μM | 58 |
| 18 | 77 nM (IC50) | — | 27 |
| 19 | 38 nM (IC50) | HeLa cells; 61 nM | 26 |
| 20 | 330 nM (IC50) | P815 cells; 2 nM | 60 |
| 21 | 14.5 μM (Ki); 86 μM (IC50) | HEK293 cells; 19.3 μM | 61 |
| 22 | 11.3 μM (IC50) | HeLa cells; 23 nM; HEK293 cells; 67 nM | 62 |
| 23 | 1 nM (IC50) | — | 63 |
| 24 | 12 μM (IC50) | A431 cells; 2.6 μM | 64 |
| 31 | 0.44 μM (IC50) | HeLa cells; 1.0 μM | 73 |
| 32 | 200 nM (Ki) | — | 75 |
| 33 | 200 nM (IC50) | P815 cells; > 50 μM | 59 |
| 35 | 362 nM (Ki); 460 nM (IC50) | — | 77 |
| 37 | 80.3 nM (IC50) | — | 79 |
| 42 | 67 nM (IC50) | HeLa cells; 16 nM | 83 |
| 43 | 73 nM (IC50) | HeLa cells; 7.4 nM | 84 |
| 44 | 69 nM (IC50) | HCT116 cells; 5.12 μM | 89 |
| 45 | 0.1 μM (IC50) | A431 cells; 1.1 μM | 90 |
| 48 | 61 nM (IC50) | HeLa cells; 171 nM | 92 |
| 49 | 36 nM (IC50) | HeLa cells; 68 nM | 93 |
| 51 | 7.5 μM (IC50) | LLTC cells; 4.3 μM | 95 |
| 52 | 2.78 μM (IC50) | HeLa cells; 9.17 μM | 96 |
| 53 | 18 μM (IC50) | HeLa cells; 18 μM | 97 |
| 58 | 161 nM (Ki); 534 nM (IC50) | HEK293 cells; 230 nM | 102 |
| 59 | 0.25 μM (IC50) | LLTC cells; 0.2 μM | 103 |
| 60 | 7.7 μM (IC50) | THP1 cells; 45 nM | 104 |
| 61 | 0.3 μM (IC50) | HeLa cells; 0.14 μM; Treg cells; 0.077 μM | 105 |
| 62 | 164 nM (IC50) | — | 108 |
| 63 | — | HEK293 cells; 1.1 μM | 109 |
aValues are the average of n > 3 experiments without noting the standard deviation (SD).
b“—”: Not tested or unknown from the corresponding original reference.
7. Conclusion
IDO1 inhibitors are an emerging class of pharmaceuticals due to the involvement of IDO1 in the kynurenine pathway responsible for immune escape. In addition to the analogues of the substrate l-tryptophan, many novel IDO1 inhibitor scaffolds have been discovered based on virtual screening, structure-guided drug design approach and natural product screening, but the majority of newly reported IDO1 inhibitors are limited to in vitro enzyme activity and cell activity without further investigations. To date, several orally available IDO1 inhibitors including epacadostat, indoximod, and navoximod have entered clinical trials.
However, the latest results of the phase III trial combining epacadostat with pembrolizumab in melanoma did not show better responses compared to pembrolizumab alone, frustrating the enthusiasm for the discovery and development of IDO1 inhibitors. It is a lack of calmness to carry out large phase III trial quickly based on the results of phase I/II with a small amount of data. First, the dose chosen to test in ECHO-301 based on pharmacodynamic data for epacadostat is derived from serum, not a tumor (100 mg bid). Thus, it is uncertain whether the dose tested was sufficient to inhibit IDO1 activity in tumor cells. Second, it is not fully understood what kind of people are the best patient population suitable for IDO1 inhibitors. Therefore, the discovery of biomarker information is crucial to help address the dose tested in clinical trials and the best population suited to IDO1 inhibitors. In addition, exploring IDO/TDO pan-inhibitors may be an appealing research direction to attenuate tryptophan catabolism effectively in human cancers.
Given the critical immunoregulatory function of IDO1, therefore it is of great interest to find IDO1 inhibitors with novel scaffolds. We hope IDO1 inhibitors will contribute to the treatment of cancer and other disorders.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This paper has not been funded.
Biographies

Xiu-Xiu Wang
Xiu-Xiu Wang obtained her B.Sc. degree in pharmaceutical science from Anhui University of Traditional Chinese Medicine, China, in 2015, and obtained her M.Sc. degree from Shandong University in 2018. Now, she is a clinical pharmacist in the Second Affiliated Hospital of Bengbu Medical College. Her research interests involve the design and synthesis of IDO1 inhibitors with antitumor property.

Ya-Qun Xing
Ya-Qun Xing graduated from the Department of Pharmacy of Dalian Railway Health College in 1986. She studied at China Pharmaceutical University and Anhui University of Traditional Chinese Medicine. She has accumulated rich experience in drug supply, dispensing and management, and is good at various drug preparations, drug testing and clinical pharmacy.
References
- Munn D. H., Mellor A. L. Trends Immunol. 2013;4:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa O., Yoshida R., Kido R., Hayaishi O. J. Biol. Chem. 1986;261:3648–3653. [PubMed] [Google Scholar]
- Dounay A. B., Tuttle J. B., Verhoest P. R. J. Med. Chem. 2015;58:8762–8782. doi: 10.1021/acs.jmedchem.5b00461. [DOI] [PubMed] [Google Scholar]
- Austin C. J. D., Rendina L. M. Drug Discovery Today. 2015;20:609–617. doi: 10.1016/j.drudis.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Taylor M. W., Feng G. S. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- Aitken J. B., Austin C. J. D., Hunt N. H., Ball H. J., Lay P. A. Biochem. Biophys. Res. Commun. 2014;450:25–29. doi: 10.1016/j.bbrc.2014.05.054. [DOI] [PubMed] [Google Scholar]
- Austin C. J. D., Mailu B. M., Maghzal G. J., Sanchez-Perez A., Rahlfs S., Zocher K., Yuasa H. J., Arthur J. W., Becker K., Stocker R. J., Hunt N. H., Ball H. J. Amino Acids. 2010;39:565–578. doi: 10.1007/s00726-010-0475-9. [DOI] [PubMed] [Google Scholar]
- Rafice S. A., Chauhan N., Efimor I., Basran J., Raven E. L. Biochem. Soc. Trans. 2009;37:408–412. doi: 10.1042/BST0370408. [DOI] [PubMed] [Google Scholar]
- Ban N. V., Van den Eynde B. J. Front. Immunol. 2015;6:34. doi: 10.3389/fimmu.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D. H., Mellor A. L. J. Clin. Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski T. F., Meng Y., Blank C., Brown I., Kacha A., Kline J., Harlin H. Immunol. Rev. 2010;213:131–145. doi: 10.1111/j.1600-065X.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- Bauer T. M., Jiga L. P., Chuang J. J., Randazzo M., Opelz G., Terness P. Transplant Int. 2010;18:95–100. doi: 10.1111/j.1432-2277.2004.00031.x. [DOI] [PubMed] [Google Scholar]
- Uyttenhove C., Pilotte L., Theate I., Stroobant V., Colau D., Parmentier N., Boon T., Van den Eynde B. J. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- Godin-Ethier J., Hanafi L. A., Piccirillo C. A., Lapointe R. Clin. Cancer Res. 2011;17:6985–6991. doi: 10.1158/1078-0432.CCR-11-1331. [DOI] [PubMed] [Google Scholar]
- Muller A. J., Sharma M. D., Chandler P. R., Duhadaway J. B., Everhart M. E., Johnson B. A., Kahler D. J., Jeanene P., Alejandro Peralta S., Munn D. H. Proc. Natl. Acad. Sci. U. S. A. 2018;105:17073–17078. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Mondal A., Dey S., Laury-Kleintop L. D., Muller A. J. Trends Cancer. 2018;4:38–58. doi: 10.1016/j.trecan.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming L., Xu W., Lei W., Ma X., Gong Z., Zhang S., Yong L. J. Hematol. Oncol. 2018;11:100. doi: 10.1186/s13045-018-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Malachowski W. P., DuHadaway J. B., Muller A. J. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig U. F., Majjigapu S. R., Vogel P., Zoete V., Michielin O. J. Med. Chem. 2015;58:9421–9437. doi: 10.1021/acs.jmedchem.5b00326. [DOI] [PubMed] [Google Scholar]
- Ladomersky E., Zhai L., Lenzen A., Lauing K. L., Qian J., Scholtens D. M., Gritsina G., Sun X., Liu Y., Yu F., Gong W., Liu Y., Jiang B., Tang T., Patel R., Platanias L. C., James C. D., Stupp R., Lukas R. V., Binder D. C., Wainwright D. A. Clin. Cancer Res. 2018;24:2559–2573. doi: 10.1158/1078-0432.CCR-17-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Xia J., Wang L., Wang X., Ma X., Deng Q., Lu Y., Kumar M., Zhou Z., Li L., Zeng Z., Young K. H., Yi Q., Zhang M., Li Y. J. Hematol. Oncol. 2018;11:58. doi: 10.1186/s13045-018-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotte L., Larrieu P., Stroobant V., Colau D., Dolusic E., Frederick R., Plaen E. D., Uyttenhove C., Wouters J., Masereel B., Van den Eynde B. J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun A. A., Hunt N. H., Ball H. J. Amino Acids. 2013;45:1319–1329. doi: 10.1007/s00726-013-1602-1. [DOI] [PubMed] [Google Scholar]
- Sugimoto H., Oda S.-I., Otsuki T., Hino T., Yoshida T., Shiro Y. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosignani S., Bingham P., Bottemanne P., Cannelle H., Cauwenberghs S., Cordonnier M., Dalvie D., Deroose F., Feng J. L., Gomes B., Kaiser S. E., Kraus M., Négrerie M., Maegley K., Miller N., Murray B. W., Schneider M., Soloweij J., Stewart A. E., Tumang J., Torti V. R., Van Den Eynde B., Wythes M. J. Med. Chem. 2017;60:9617–9629. doi: 10.1021/acs.jmedchem.7b00974. [DOI] [PubMed] [Google Scholar]
- Peng Y. H., Ueng S. H., Tseng C. T., Hung M. S., Song J. S., Wu J. S., Liao F. Y., Fan Y. S., Wu M. H., Hsiao W. C., Hsueh C. C., Lin S. Y., Cheng C. Y., Tu C. H., Lee L. C., Cheng M. F., Shia K. S., Shih C., Wu S. Y. J. Med. Chem. 2016;59:282–293. doi: 10.1021/acs.jmedchem.5b01390. [DOI] [PubMed] [Google Scholar]
- Tojo S., Kohno T., Tanaka T., Kamioka S., Ota Y., Ishii T., Kamimoto K., Asano S., Isobe Y. J. A. M. C. L. ACS Med. Chem. Lett. 2014;5:1119–1123. doi: 10.1021/ml500247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng T., Qiu X., Wang J., Li Z., Bian J. Eur. J. Med. Chem. 2018;143:656–669. doi: 10.1016/j.ejmech.2017.11.088. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Malachowski W. P., DuHadaway J. B., Muller A. J. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochez L., Chevolet I., Kruse V. Eur. J. Cancer. 2017;76:167–182. doi: 10.1016/j.ejca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Coletti A., Greco F. A., Dolciami D., Camaioni E., Sardella R., Pallotta M. T., Volpi C., Orabona C., Grohmann U., Macchiarulo A. MedChemComm. 2017;8:1378–1392. doi: 10.1039/c7md00109f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Huang C. H. Front. Oncol. 2018;8:423. doi: 10.3389/fonc.2018.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Hayaishi O. J. Biol. Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- Cady S. G., Sono M. Arch. Biochem. Biophys. 1991;291:326–333. doi: 10.1016/0003-9861(91)90142-6. [DOI] [PubMed] [Google Scholar]
- Peterson A. C., Martin M. J., Hamaker L. K., Czerwinski K. M., Zhang W. Med. Chem. Res. 1994;3:531–544. [Google Scholar]
- Hou D. Y., Muller A. J., Sharma M. D., DuHadaway J., Banerjee T., Johnson M., Mellor A. L., Prendergast G. C., Munn D. H. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Smith C., Thomas S., Mandik-Nayak L., Laury-Kleintop L., Metz R., Muller A. J. Cancer Immunol. Immunother. 2014;63:721–735. doi: 10.1007/s00262-014-1549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R., Duhadaway J. B., Kamasani U., Laury-Kleintop L., Muller A. J., Prendergast G. C. Cancer Res. 2007;67:7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- Merlo L. M. F., Pigott E., DuHadaway J. B., Grabler S., Metz R., Prendergast G. C., Mandik-Nayak L. J. Immunol. 2014;192:2082–2090. doi: 10.4049/jimmunol.1303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman H. H., Minton S. E., Han H. S., Ismail-Khan R., Neuger A., Khambati F., Noyes D., Lush R., Chiappori A. A., Roberts J. D., Link C., Vahanian N. N., Mautino M., Streicher H., Sullivan D. M., Antonia S. J. Oncotarget. 2016;7:22928–22938. doi: 10.18632/oncotarget.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R., Rust S., Duhadaway J. B., Mautino M. R., Munn D. H., Vahanian N. N., Link C. J., Prendergast G. C. Oncoimmunology. 2012;1:1460–1468. doi: 10.4161/onci.21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman H. H., Jackson E., Neuger T., Dees E. C., Harvey R. D., Han H., Ismail-Khan R., Minton S., Vahanian N. N., Link C., Sullivan D. M., Antonia S. Oncotarget. 2014;5:8136–8146. doi: 10.18632/oncotarget.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A. J., DuHadaway J. B., Donover P. S., Sutanto-Ward E., Prendergast G. C. Nat. Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- Paul G., Tinku B., Malachowski W. P., Muller A. J., Prendergast G. C., James D., Shauna B., Donovan A. M. J. Med. Chem. 2006;49:684–692. doi: 10.1021/jm0508888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T., Duhadaway J. B., Gaspari P., Sutanto-Ward E., Munn D. H., Mellor A. L., Malachowski W. P., Prendergast G. C., Muller A. J. Oncogene. 2008;27:2851–2857. doi: 10.1038/sj.onc.1210939. [DOI] [PubMed] [Google Scholar]
- Nakashima H., Uto Y., Nakata E., Nagasawa H., Ikkyu K., Hiraoka N., Nakashima K., Sasaki Y., Sugimoto H., Shiro Y., Hashimoto T., Okamoto Y., Asakawa Y., Hori H. Bioorg. Med. Chem. 2008;16:8661–8669. doi: 10.1016/j.bmc.2008.07.087. [DOI] [PubMed] [Google Scholar]
- Dolusic E., Larrieu P., Blanc S., Sapunaric F., Pouyez J., Moineaux L., Colette D., Stroobant V., Pilotte L., Colau D., Ferain T., Fraser G., Galleni M., Frere J. M., Masereel B., Van den Eynde B., Wouters J., Frederick R. Eur. J. Med. Chem. 2011;46:3058–3065. doi: 10.1016/j.ejmech.2011.02.049. [DOI] [PubMed] [Google Scholar]
- Dolusic E., Larrieu P., Blanc S., Sapunaric F., Norberg B., Moineaux L., Colette D., Stroobant V., Pilotte L., Colau D., Ferain T., Fraser G., Galleni M., Frere J. M., Masereel B., Van den Eynde B., Wouters J., Frederick R. Bioorg. Med. Chem. 2011;19:1550–1561. doi: 10.1016/j.bmc.2010.12.032. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Li X., Hikawa H., Suzuki T., Tsutsumi K., Sato M., Takikawa O., Suzuki H., Yokoyama Y. Bioorg. Med. Chem. 2013;21:1159–1165. doi: 10.1016/j.bmc.2012.12.028. [DOI] [PubMed] [Google Scholar]
- Tourino M. C., de Oliveira E. M., Belle L. P., Knebel F. H., Albuquerque R. C., Dorr F. A., Okada S. S., Migliorini S., Soares I. S., Campa A. Cell Biochem. Funct. 2013;31:361–364. doi: 10.1002/cbf.2980. [DOI] [PubMed] [Google Scholar]
- Coluccia A., Passacantilli S., Famiglini V., Sabatino M., Patsilinakos A., Ragno R., Mazzoccoli C., Sisinni L., Okuno A., Takikawa O., Silvestri R., La Regina G. J. Med. Chem. 2016;59:9760–9773. doi: 10.1021/acs.jmedchem.6b00718. [DOI] [PubMed] [Google Scholar]
- Thaker A. I., Rao M. S., Bishnupuri K. S., Kerr T. A., Foster L., Marinshaw J. M., Newberry R. D., Stenson W. F., Ciorba M. A. Gastroenterology. 2013;145:416–425. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sono M., Cady S. Biochemistry. 1989;28:5392–5399. doi: 10.1021/bi00439a012. [DOI] [PubMed] [Google Scholar]
- Kumar S., Jaller D., Patel B., LaLonde J. M., DuHadaway J. B., Malachowski W. P., Prendergast G. C., Muller A. J. J. Med. Chem. 2008;51:4968–4977. doi: 10.1021/jm800512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautino M. R., Jaipuri F. A., Waldo J., Kumar S., Adams J., Allen C. V., Marcinowicz-Flick A., Munn D., Vahanian N., Link C. Cancer Res. 2013;73:491–491. [Google Scholar]
- Spahn J., Peng J., Lorenzana E., Kan D., Hunsaker T., Segal E., Mautino M., Brincks E., Pirzkall A., Kelley S., Mahrus S., Liu L., Dale S., Quiason C., Jones E., Liu Y., Latham S., Salphati L., DeMent K., Merchant M., Hatzivassiliou G. J. Immunother. Cancer. 2015;3:303. [Google Scholar]
- Nayak-Kapoor A., Hao Z., Sadek R., Dobbins R., Marshall L., Vahanian N. N., Ramsey W. J., Kennedy E., Mautino M. R., Link C. J., Lin R. S., Royer-Joo S., Liang X., Salphati L., Morrissey K. M., Mahrus S., McCall B., Pirzkall A., Munn D. H., Janik J. E., Khleif S. N. J Immunother. Cancer. 2018;6:61. doi: 10.1186/s40425-018-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig U.U. F.F., Majjigapu S. R., Chambon M., Bron S., Pilotte L., Colau D., Van den Eynde B. J., Turcatti G., Vogel P., Zoete V., Michielin O. Eur. J. Med. Chem. 2014;84:284–301. doi: 10.1016/j.ejmech.2014.06.078. [DOI] [PubMed] [Google Scholar]
- Röhrig U. F., Awad L., Grosdidier A., Larrieu P., Stroobant V., Colau D., Cerundolo V., Simpson A. J., Vogel P., Van den Eynde B. J., Zoete V., Michielin O. J. Med. Chem. 2010;53:1172–1189. doi: 10.1021/jm9014718. [DOI] [PubMed] [Google Scholar]
- Rohrig U. F., Majjigapu S. R., Grosdidier A., Bron S., Stroobant V., Pilotte L., Colau D., Vogel P., Van den Eynde B. J., Zoete V., Michielin O. J. Med. Chem. 2012;55:5270–5290. doi: 10.1021/jm300260v. [DOI] [PubMed] [Google Scholar]
- Huang Q., Zheng M., Yang S., Kuang C., Yu C., Yang Q. Eur. J. Med. Chem. 2011;46:5680–5687. doi: 10.1016/j.ejmech.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Alexandre J. A. C., Swan M. K., Latchem M. J., Boyall D., Pollard J. R., Hughes S. W., Westcott J. ChemBioChem. 2018;19:552–561. doi: 10.1002/cbic.201700560. [DOI] [PubMed] [Google Scholar]
- Xu G., Wang T., Li Y., Huang Z., Wang X., Zheng J., Yang S., Fan Y., Xiang R. J. Enzyme Inhib. Med. Chem. 2018;33:1089–1094. doi: 10.1080/14756366.2018.1471688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K., Yamazaki H., Isaka Y., Takai K., Unno Y., Ogo N., Ishikawa Y., Fujii S., Takikawa O., Asai A. MedChemComm. 2012;3:475–479. [Google Scholar]
- Xu H., Song Y., Yang Q. Chem. Pharm. Bull. 2017;65:714–717. doi: 10.1248/cpb.c16-01010. [DOI] [PubMed] [Google Scholar]
- Bridewell D. J. A., Sperry J., Smith J. R., Kosim-Satyaputra P., Ching L.-M., Jamie J. F., Brimble M. A. Aust. J. Chem. 2013;66:40–49. [Google Scholar]
- Smith J. R., Evans K. J., Wright A., Willows R. D., Jamie J. F., Griffith R. Bioorg. Med. Chem. 2012;20:1354–1363. doi: 10.1016/j.bmc.2011.10.068. [DOI] [PubMed] [Google Scholar]
- Frieden E., Westmark G. W., Schor J. M. Arch. Biochem. Biophys. 1961;92:176–182. doi: 10.1016/0003-9861(61)90233-8. [DOI] [PubMed] [Google Scholar]
- Pereira A., Vottero E., Roberge M., Mauk A. G., Andersen R. J. J. Nat. Prod. 2006;69:1496–1499. doi: 10.1021/np060111x. [DOI] [PubMed] [Google Scholar]
- Vottero E., Balgi A., Woods K., Tugendreich S., Melese T., Andersen R. J., Mauk A. G., Roberge M. Biotechnol. J. 2010;1:282–288. doi: 10.1002/biot.200600001. [DOI] [PubMed] [Google Scholar]
- Kumar S., Malachowski W. P., Duhadaway J. B., Lalonde J. M., Carroll P. J., Jaller D., Metz R., Prendergast G. C., Muller A. J. J. Med. Chem. 2008;51:1706–1718. doi: 10.1021/jm7014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson D. W., Plaza S. M. Altern. Med. Rev. 2003;8:303–318. [PubMed] [Google Scholar]
- Flick H. E., Lalonde J. M., Malachowski W. P., Muller A. J. Int. J. Tryptophan Res. 2013;6:35–45. doi: 10.4137/IJTR.S12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C. J., Kahlert J., Issa F., Reed J. H., Smith J. R., Ioppolo J. A., Ong J. A., Jamie J. F., Hibbs D., Rendina L. M. Dalton Trans. 2014;43:10719–10724. doi: 10.1039/c4dt00444b. [DOI] [PubMed] [Google Scholar]
- Gavin C., Chung M. K. W., Grant M. A., Andersen R. J. J. Med. Chem. 2008;51:2634–2637. doi: 10.1021/jm800143h. [DOI] [PubMed] [Google Scholar]
- Dolusic E., Larrieu P., Meinguet C., Colette D., Rives A., Blanc S., Ferain T., Pilotte L., Stroobant V., Wouters J., Van den Eynde B., Masereel B., Delfourne E., Frederick R. Bioorg. Med. Chem. Lett. 2013;23:47–54. doi: 10.1016/j.bmcl.2012.11.036. [DOI] [PubMed] [Google Scholar]
- Raffaele P., David S., David R., Moody C. J. J. Med. Chem. 2013;56:3310–3317. doi: 10.1021/jm400049z. [DOI] [PubMed] [Google Scholar]
- Carvalho C., Siegel D., Inman M., Xiong R., Ross D., Moody C. J. Org. Biomol. Chem. 2014;12:2663–2674. doi: 10.1039/c3ob42258e. [DOI] [PubMed] [Google Scholar]
- Blunt C. E., Torcuk C., Liu Y., Lewis W., Siegel D., Ross D., Moody C. J. Angew. Chem., Int. Ed. 2015;54:8740–8745. doi: 10.1002/anie.201503323. [DOI] [PubMed] [Google Scholar]
- Pan L., Zheng Q., Chen Y., Yang R., Yang Y., Li Z., Meng X. Eur. J. Med. Chem. 2018;157:423–436. doi: 10.1016/j.ejmech.2018.08.013. [DOI] [PubMed] [Google Scholar]
- Centko R. M., Steino A., Rosell F. I., Patrick B. O., de Voogd N., Mauk A. G., Andersen R. J. Org. Lett. 2014;16:6480–6483. doi: 10.1021/ol503337f. [DOI] [PubMed] [Google Scholar]
- Brastianos H. C., Vottero E., Patrick B. O., Soest R. V., Matainaho T., Mauk A. G., Andersen R. J. J. Am. Chem. Soc. 2006;128:16046–16047. doi: 10.1021/ja067211+. [DOI] [PubMed] [Google Scholar]
- Yue E. W., Douty B., Wayland B., Bower M., Liu X., Leffet L., Wang Q., Bowman K. J., Hansbury M. J., Liu C., Wei M., Li Y., Wynn R., Burn T. C., Koblish H. K., Fridman J. S., Metcalf B., Scherle P. A., Combs A. P. J. Med. Chem. 2009;52:7364–7367. doi: 10.1021/jm900518f. [DOI] [PubMed] [Google Scholar]
- Yue E. W., Sparks R., Polam P., Modi D., Douty B., Wayland B., Glass B., Takvorian A., Glenn J., Zhu W., Bower M., Liu X., Leffet L., Wang Q., Bowman K. J., Hansbury M. J., Wei M., Li Y., Wynn R., Burn T. C., Koblish H. K., Fridman J. S., Emm T., Scherle P. A., Metcalf B., Combs A. P. ACS Med. Chem. Lett. 2017;8:486–491. doi: 10.1021/acsmedchemlett.6b00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochems C., Fantini M., Fernando R. I., Kwilas A. R., Donahue R. N., Lepone L. M., Grenga I., Kim Y. S., Brechbiel M. W., Gulley J. L., Madan R. A., Heery C. R., Hodge J. W., Newton R., Schlom J., Tsang K. Y. Oncotarget. 2016;7:37762–37772. doi: 10.18632/oncotarget.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Shin N., Koblish H. K., Yang G., Wang Q., Wang K., Leffet L., Hansbury M. J., Thomas B., Rupar M., Waeltz P., Bowman K. J., Polam P., Sparks R. B., Yue E. W., Li Y., Wynn R., Fridman J. S., Burn T. C., Combs A. P., Newton R. C., Scherle P. A. Blood. 2010;115:3520–3530. doi: 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- Koblish H. K., Hansbury M. J., Bowman K. J., Yang G., Neilan C. L., Haley P. J., Burn T. C., Waeltz P., Sparks R. B., Yue E. W., Combs A. P., Scherle P. A., Vaddi K., Fridman J. S. Mol. Cancer Ther. 2010;9:489–498. doi: 10.1158/1535-7163.MCT-09-0628. [DOI] [PubMed] [Google Scholar]
- Spranger S., Koblish H. K., Horton B., Scherle P. A., Newton R., Gajewski T. F. J. Immunother. Cancer. 2014;2:3. doi: 10.1186/2051-1426-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K., Dong G., Li Y., He S., Wu Y., Wu S., Wang W., Sheng C. ACS Med. Chem. Lett. 2018;9:312–317. doi: 10.1021/acsmedchemlett.7b00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K., Takai K., Isaka Y., Unno Y., Sato M., Takikawa O., Asai A. Bioorg. Med. Chem. Lett. 2010;20:5126–5129. doi: 10.1016/j.bmcl.2010.07.025. [DOI] [PubMed] [Google Scholar]
- Meininger D., Zalameda L., Liu Y., Stepan L. P., Borges L., McCarter J. D., Sutherland C. L. Biochim. Biophys. Acta. 2011;1814:1947–1954. doi: 10.1016/j.bbapap.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Cheng M. F., Hung M. S., Song J. S., Lin S. Y., Liao F. Y., Wu M. H., Hsiao W., Hsieh C. L., Wu J. S., Chao Y. S., Shih C., Wu S. Y., Ueng S. H. Bioorg. Med. Chem. Lett. 2014;24:3403–3406. doi: 10.1016/j.bmcl.2014.05.084. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Yeh T. K., Kuo C. C., Song J. S., Cheng M. F., Liao F. Y., Chao M. W., Huang H. L., Chen Y. L., Yang C. Y., Wu M. H., Hsieh C. L., Hsiao W., Peng Y. H., Wu J. S., Lin L. M., Sun M., Chao Y. S., Shih C., Wu S. Y., Pan S. L., Hung M. S., Ueng S. H. J. Med. Chem. 2016;59:419–430. doi: 10.1021/acs.jmedchem.5b01640. [DOI] [PubMed] [Google Scholar]
- Serra S., Moineaux L., Vancraeynest C., Masereel B., Wouters J., Pochet L., Frederick R. Eur. J. Med. Chem. 2014;82:96–105. doi: 10.1016/j.ejmech.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Tomek P., Palmer B. D., Flanagan J. U., Sun C., Raven E. L., Ching L. M. Eur. J. Med. Chem. 2017;126:983–996. doi: 10.1016/j.ejmech.2016.12.029. [DOI] [PubMed] [Google Scholar]
- Gao D., Li Y. Bioorg. Med. Chem. 2017;25:3780–3791. doi: 10.1016/j.bmc.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Zhang G., Xing J., Wang Y., Wang L., Ye Y., Lu D., Zhao J., Luo X., Zheng M., Yan S. Front. Pharmacol. 2018;9:277. doi: 10.3389/fphar.2018.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terentis A. C., Freewan M., Sempertegui Plaza T. S., Raftery M. J., Stocker R., Thomas S. R. Biochemistry. 2010;49:591–600. doi: 10.1021/bi901546e. [DOI] [PubMed] [Google Scholar]
- Williams D. E., Steino A., de Voogd N. J., Mauk A. G., Andersen R. J. J. Nat. Prod. 2012;75:1451–1458. doi: 10.1021/np300345j. [DOI] [PubMed] [Google Scholar]
- Jang J. P., Jang J. H., Soung N. K., Kim H. M., Jeong S. J., Asami Y., Shin K. S., Kim M. R., Oh H., Kim B. Y., Ahn J. S. J. Antibiot. 2012;65:215–217. doi: 10.1038/ja.2011.141. [DOI] [PubMed] [Google Scholar]
- Jang J. P., Jang J. H., Oh M., Son S., Kim S. M., Kim H. M., Shin K. S., Oh H., Soung N. K., Hong Y. S., Kim B. Y., Ahn J. S. J. Antibiot. 2014;67:331–333. doi: 10.1038/ja.2013.134. [DOI] [PubMed] [Google Scholar]
- Yang S., Li X., Hu F., Li Y., Yang Y., Yan J., Kuang C., Yang Q. J. Med. Chem. 2013;56:8321–8331. doi: 10.1021/jm401195n. [DOI] [PubMed] [Google Scholar]
- Fung S. P., Wang H., Tomek P., Squire C. J., Flanagan J. U., Palmer B. D., Bridewell D. J., Tijono S. M., Jamie J. F., Ching L. M. Bioorg. Med. Chem. 2013;21:7595–7603. doi: 10.1016/j.bmc.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Yamamoto Y., Imai S., Fukutomi R., Ozawa Y., Abe M., Matuo Y., Saito K. PLoS One. 2014;9:e88789. doi: 10.1371/journal.pone.0088789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malachowski W. P., Winters M., DuHadaway J. B., Lewis-Ballester A., Badir S., Wai J., Rahman M., Sheikh E., LaLonde J. M., Yeh S. R., Prendergast G. C., Muller A. J. Eur. J. Med. Chem. 2016;108:564–576. doi: 10.1016/j.ejmech.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Ballestera A., Batabyala D., Egawaa T., Lua C., Lina Y., Martib M. A., Capeceb L., Estrinb D. A., Yeh S.-R. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17371–17376. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capece L., Lewis-Ballester A., Yeh S. R., Estrin D. A., Marti M. A. J. Phys. Chem. B. 2012;116:1401–1413. doi: 10.1021/jp2082825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jortzik E., Zocher K., Isernhagen A., Mailu B. M., Rahlfs S., Viola G., Wittlin S., Hunt N. H., Ihmels H., Becker K. Antimicrob. Agents Chemother. 2016;60:115–125. doi: 10.1128/AAC.01066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu L. L., Gelmon K., Chu Q., Pachynski R., Alese O., Basciano P., Walker J., Mitra P., Zhu L., Phillips P., Hunt J., Desai J. Cancer Res. 2017;77:13. [Google Scholar]