Summary
Potato virus Y (PVY) is a major potato (Solanum tuberosum L.) pathogen that causes severe annual crop losses worth billions of dollars worldwide. PVY is transmitted by aphids, and successful control of virus transmission requires the extensive use of environmentally damaging insecticides to reduce vector populations. Ry sto , from the wild relative S. stoloniferum, confers extreme resistance (ER) to PVY and related viruses and is a valuable trait that is widely employed in potato resistance breeding programmes. Ry sto was previously mapped to a region of potato chromosome XII, but the specific gene has not been identified to date. In this study, we isolated Ry sto using resistance gene enrichment sequencing (RenSeq) and PacBio SMRT (Pacific Biosciences single‐molecule real‐time sequencing). Ry sto was found to encode a nucleotide‐binding leucine‐rich repeat (NLR) protein with an N‐terminal TIR domain and was sufficient for PVY perception and ER in transgenic potato plants. Ry sto ‐dependent extreme resistance was temperature‐independent and requires EDS1 and NRG1 proteins. Ry sto may prove valuable for creating PVY‐resistant cultivars of potato and other Solanaceae crops.
Keywords: potato, PVY, extreme resistance, RenSeq, Ry sto , TIR‐NLR immune receptor
Introduction
Potato virus Y (PVY), a member of the Potyvirus genus (Potyviridae family), is the most economically important virus of potato crops, affecting tuber yield and quality and resulting in losses of 10–90% depending on the year, cultivar and location (De Bokx and der van Want, 1987; Valkonen, 2007). PVY can also infect other Solanaceae species such as bell pepper, tomato and tobacco (Aramburu et al., 2006). PVY is mechanically transmitted by more than 40 aphid species in a non‐persistent manner (Brunt, 2001). Intracellular pathogens such as viruses are difficult to control chemically, and PVY outbreaks are managed by the destruction of infected plants or by pesticide treatment to reduce insect vector populations (Kopp and Bánfalvi, 2015). Selective breeding of plants with resistance to viral pathogens remains the best method to limit the impact of viral infections. PVY variants are highly variable at the biological, serological and molecular levels (Scholthof et al., 2011) and can be classified into different strain groups according to the host response. Virus strains PVYN and PVYNW produce barely noticeable foliar symptoms, whereas PVY0 and PVYC usually induce mild symptoms of infection such as leaf mosaic lesions, crinkling, leaf drop and dwarfing. By contrast, as well as inducing severe foliar symptoms, PVYNTN infection leads to potato tuber necrotic ringspot disease, which severely affects tuber marketability (Schubert et al., 2007).
Plant defence responses to viruses are multifaceted and depend on several factors, including plant age and tissue type. For PVY, resistance genes provide the most effective and durable control. Two main types of resistance against PVY are present in potato: extreme resistance (ER), conferred by the Ry genes, and hypersensitive resistance involving programmed cell death (HR), conferred by the Ny genes (Valkonen, 2015). However, HR is not effective in restricting PVY in plants under some conditions (Vidal et al., 2002). By contrast, ER genes are broad‐spectrum and confer strong and durable resistance, characterized by lack of visible symptoms after inoculation (Flis et al., 2005).
Resistance genes against PVY infection were previously introduced into potato cultivars from wild or domesticated Solanum species. Ten genes for resistance to PVY and one gene for resistance to PVA (Potato Virus A, a potyvirus related to PVY) were mapped to four potato genome segments on chromosomes IV, IX, XI and XII (Valkonen et al., 2017; Van Eck et al., 2017). Some of these genes, for example Ry sto and Ry‐f sto from S. stoloniferum, were introduced into multiple potato cultivars worldwide and shown to confer durable resistance against multiple PVY strains. However, no genes conferring effective HR‐ or ER‐type response have been cloned and characterized to date (Valkonen et al., 2017).
Previous estimates suggested that controlling major diseases through the informed deployment of resistance genes could increase crop yields by more than 30% while reducing the application of chemical pesticides (Gebhardt and Valkonen, 2001). This might be achieved by using new breeding tools to select resistant genotypes that can then be deployed in new crop varieties (Armstrong et al., 2019).
Most plant resistance genes encode intracellular nucleotide‐binding leucine‐rich repeat (NLR) receptors. Some of these genes carry an N‐terminal TIR domain and are termed TIR‐NLRs, whereas others carry an N‐terminal coiled coil domain and are termed CC‐NLRs (Jones et al., 2016). Pacific Biosciences single‐molecule real‐time sequencing combined with resistance gene enrichment (SMRT RenSeq) (Witek et al., 2016) is a useful tool for the identification and assembly of full‐length NLR‐encoding genes that cosegregate with resistance phenotypes. SMRT RenSeq methodology is more robust and cost‐effective for monitoring NLR sequences than whole‐genome sequencing. All the currently known NLRs effective against viruses, nematodes and the late blight pathogen Phytophthora infestans can be tracked with SMRT RenSeq in potato, and their polymorphisms have been defined (Armstrong et al., 2019).
Demand is high for strong, durable resistance genes to combat PVY infection, and R genes conferring ER to PVY fulfil these requirements. The mechanisms underlying the ER response to viruses have not been fully elucidated, and identifying the network of factors contributing to effective pathogen resistance is of key importance.
The aim of this study was to use SMRT RenSeq to isolate the Ry sto gene that governs the ER trait. Multiple TIR‐NLR‐encoding paralogues were identified that cosegregated with resistance. Paralogue Ry sto conferred resistance to PVY and the related potyvirus PVA in different Solanaceae backgrounds through recognition of PVY and PVA coat proteins (CPs) as avr factors for Ry sto ‐triggered immunity. Finally, investigation of the functional relationship between ER and HR showed that ER was EDS1‐ and NRG1‐dependent but was not dependent on salicylic acid (SA) level or temperature.
Results
Ry sto originates from S. stoloniferum
ER to PVY was introgressed into commercial potato cultivars from three different wild Solanum spp. In European cultivars, resistance originates mostly from S. stoloniferum (Ry sto and Ry‐f sto ) and has not yet been overcome (Valkonen et al., 2017). Therefore, we set out to clone Ry sto using dihaploid clone dH Alicja, which has PVY‐resistant S. stoloniferum in its ancestry. A diploid mapping population consisting of 391 F1 individuals was generated and evaluated for resistance to PVY by mechanical and graft inoculations as described by Flis et al. (2005). The segregation ratio of resistant versus susceptible progeny in the mapping population deviated from the 1:1 ratio expected for the segregation of a single dominant gene and was distorted towards resistance (149 susceptible and 242 PVY‐resistant F1 individuals).
RenSeq combined with SMRT sequencing and bulked segregant analysis led to successful cloning of Ry sto
Previous research mapped Ry sto to the distal end of the long arm of potato chromosome XII (Flis et al., 2005; Song et al., 2005; Van Eck et al., 2017), corresponding to the region downstream of 58 Mb in the DM reference potato genome (Potato Genome Sequencing Consortium (95 authors), 2011). Jupe et al. (2013) showed that this region contained sequences encoding 18 complete or partial NLR immune receptors from both the TIR‐NLR and CC‐NLR classes, organized in four clusters (Figure 1a). We therefore hypothesized that Ry sto likely encoded a NLR and used RenSeq to predict candidate genes. SMRT RenSeq was used to assemble the NLR‐encoding genes of the resistant parent with high confidence. Assembly of Pacific Biosciences (PacBio) reads of interest (ROIs) resulted in 1555 contigs, 1254 of which were annotated as NLRs using NLR‐parser software (Steuernagel et al., 2006). Mapping of short‐read RenSeq data from resistant (R), susceptible (S) and bulked susceptible (BS) plants, and subsequent polymorphism calling, resulted in 10 linked contigs with presence/absence polymorphisms. Expression of candidate genes was confirmed using cDNA‐RenSeq data obtained from the resistant parent. Eleven candidate NLRs were identified from these analyses, four of which belonged to the CC‐NLR class and seven of which belonged to the TIR‐NLR class. A phylogenetic tree constructed with full‐length predicted amino acid sequences showed that the candidate genes belonged to three different clades and were only distantly related to cloned functional Solanaceae genes. All TIR‐NLRs belonged to the same clade and exhibited amino acid identities of 75–90%. The CC‐NLRs split into two distinct clades, consistent with their physical separation in the potato genome (Figure 1 and Figure S1).
Figure 1.
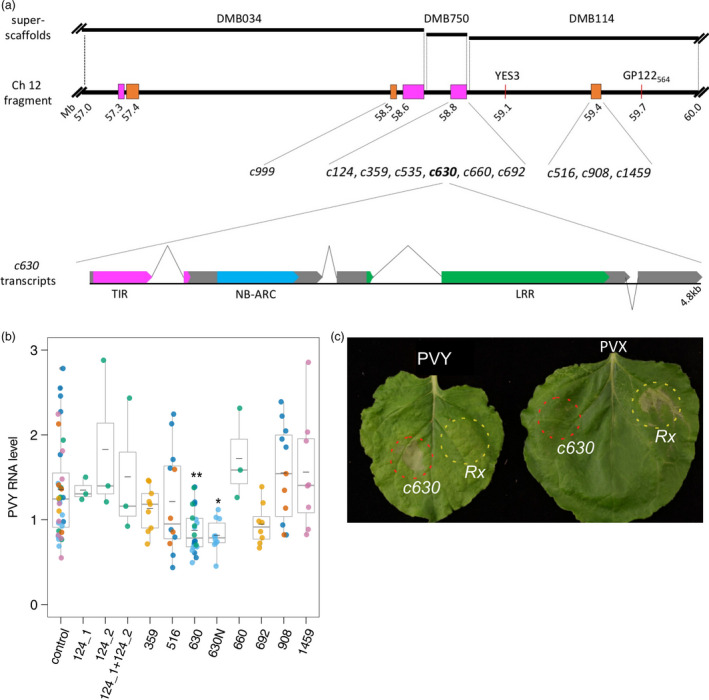
Functional analysis of candidate genes using transient expression assays in Nicotiana benthamiana. (a) Schematic representation of candidate genes and a fragment of chromosome XII linked to the Ry sto gene. Potato superscaffolds (DMB034, DMB750 and DMB114; top panel) were aligned to the distal end of the longer arm of potato chromosome XII (57–60 Mb fragment shown; middle panel), which is linked to the Ry sto gene. Annotated NLRs/NLR clusters are depicted in orange (CNL) and magenta (TNL). Red lines indicate known markers linked to Ry sto . Distances are given in Mb according to the DM potato reference genome (Potato Genome Sequencing Consortium (95 authors), 2011), with numbers indicating proximal positions of marked sequences. Candidate contigs for Ry sto derived from SMRT RenSeq were aligned to the DM potato genome, and best hit positions are shown on the map. Map drawn to scale. Schematic structure of c630 transcripts (lower panel). Approximately 80% of cDNA‐RenSeq reads supported a version of c630 with four exons. An additional intron (depicted with inverted lines) positioned 24 nt upstream of the initial stop codon results in a fifth exon at the 3′ end. Exons are drawn in grey; colours indicate canonical NLR domains: TIR (magenta), NB‐ARC (blue) and LRR (green). Schematic drawn to scale. (b) Relative levels of PVY RNA after infection of Nicotiana benthamiana leaves transiently expressing candidate contigs. Three N. benthamiana leaves were infiltrated with vector pICSLUS0003:35S overexpressing c124_1, c124_2, c359, c516, c630, c660, c692, c908 c1459 and pICSLUS0001 vector overexpressing c630 under the control of native regulatory elements or an empty vector. After 48 h, leaves were inoculated with PVYNTN . Seven days after PVY inoculation, mRNA was isolated from upper, non‐inoculated leaves and PVY RNA levels were quantified with qPCR. EF1 and L23 were used as standardization references. Error bars represent the standard deviation of the means, and medians are also presented. One‐way ANOVA with Dunnett's test was used for statistical analysis. Experiments were performed on 3–10 plants for each construct and were repeated three times. (c) HR response of N. benthamiana plants expressing c630 or Rx genes. Two fully developed leaves of N. benthamiana plants were infiltrated with a suspension of A. tumefaciens carrying pICSLUS0003:c630 (area marked with red) or pGBT‐Rx‐GFP (area marked with yellow) 14 days after PVYNTN (left) or PVX (right) infection. Seventy‐two hours after infiltration, expression of c630 in PVY‐infected plants resulted in HR symptoms similar to those observed when Rx was delivered into plants infected with PVX virus. Experiments were repeated three times.
Homology searches of the reference potato genome positioned the candidate genes in three clusters located at the distal end of the long arm of chromosome XII (58–59 Mb of the DM reference genome, Figure 1a), consistent with marker locations linked to Ry sto (GP122564, Witek et al., 2006, and YES3, Nie et al., 2016). This region of chromosome XII consists of three neighbouring superscaffolds, DMB034, DMB750 and DMB114, one of which (DMB114) was proposed as a likely location for Ry sto (Van Eck et al., 2017). All the selected candidate NLR genes from SMRT RenSeq data therefore localized to the distal end of chromosome XII in a region previously identified with Ry sto .
Functional analysis of candidate contigs by transient expression in Nicotiana benthamiana
To determine whether candidate genes were functional in planta, the open reading frames (ORFs) of 11 cosegregating NLRs were PCR‐amplified and placed under the control of a 35S promoter in the pICSLUS0003 binary vector, as described previously (Witek et al., 2016).
Constructs were transiently expressed in N. benthamiana, followed by PVYNTN inoculation. At 7 dpi, leaf samples were collected and levels of viral RNA were measured using qPCR. One candidate gene (c630) reduced virus multiplication as compared to control N. benthamiana plants (Figure 1b). The c630 ORF with its native 5′ and 3′ regulatory sequences (2263 nt upstream from 5′ and 1013 nt downstream from 3′ end, respectively) was cloned into the pICSLUS0001 binary vector as described (Witek et al., 2016). Transient assays in N. benthamiana also showed a statistically significant (P = 0.011475) inhibition of virus multiplication (Figure 1b). Candidate gene c999 showed autoactivity, even when infiltrated with low OD (0.1), and another candidate gene, c535, could not be cloned.
Next, N. benthamiana plants were systemically infected with PVYNTN or Potato virus X (PVX) prior to transient expression of c630. A HR response was observed upon expression of c630 in plants carrying PVYNTN (Figure 1c), similar to the response seen when the Rx gene was delivered into plants infected with PVX (Bendahmane et al., 1999).
Expression of c630 triggers HR in response to a range of viruses
To test whether c630 elicited an immune response against different strains of PVY, N. benthamiana plants were infected with the PVY0, PVYN, PVYN‐Wi and PVYNTN isolates or cross related Potato Virus A (PVA). Leaves showing symptoms of viral infection at 7 dpi were infiltrated with Agrobacterium strains carrying c630 or an empty vector. Transient expression of c630 resulted in a strong HR in plants infected with all tested PVY isolates and related PVA, but not Potato virus X (PVX) or Tobacco Mosaic Virus (TMV) (Table S1). No HR was observed in control plants without c630 expression. These experiments provided further support that c630 was the functional Ry sto gene.
Ry sto expression restricts systemic spread of PVY and PVA
To further investigate c630/Ry sto function, stable transgenic Solanaceae plants (potato and tobacco) carrying Ry sto or two other non‐functional homologues under the control of either a 35S promoter or native regulatory elements were created using Agrobacterium‐mediated transformation.
Two PVY‐susceptible potato cultivars (Maris Piper [MP] and Russet Burbank [RB]) were then used to test Ry sto functionality. Transgenic plants of both cultivars carrying Ry sto under the control of either a 35S promoter or a native promoter (MP only) were inoculated with PVYNTN or mock treated with water. Systemic virus spread was monitored in upper, non‐inoculated leaves using enzyme‐linked immunosorbent assay (ELISA) 3 weeks after inoculation. In the MP background expressing Ry sto under control of 35S, no PVY was detected in upper plant parts in 10 of the 12 tested transgenic lines (Table S2). To confirm these ELISA results, three randomly chosen transgenic lines (two resistant and one susceptible) were tested using qPCR (Figure 2b). The inhibition of virus multiplication and spread in the two resistant transgenic plants correlated with elevated levels of Ry sto gene expression. In the susceptible transgenic line, no expression of the Ry sto transgene was detected. No PVY was detected in RB in any of the four independently isolated transgenic lines, either by ELISA or by qPCR (Figure 2c and Table S3). Furthermore, all five lines in which Ry sto was expressed under the control of its native promoter were PVY‐free (Figure 2d). No macroscopic symptoms of HR were observed in all resistant lines regardless of the genotype of potato used (Figure 2a).
Figure 2.
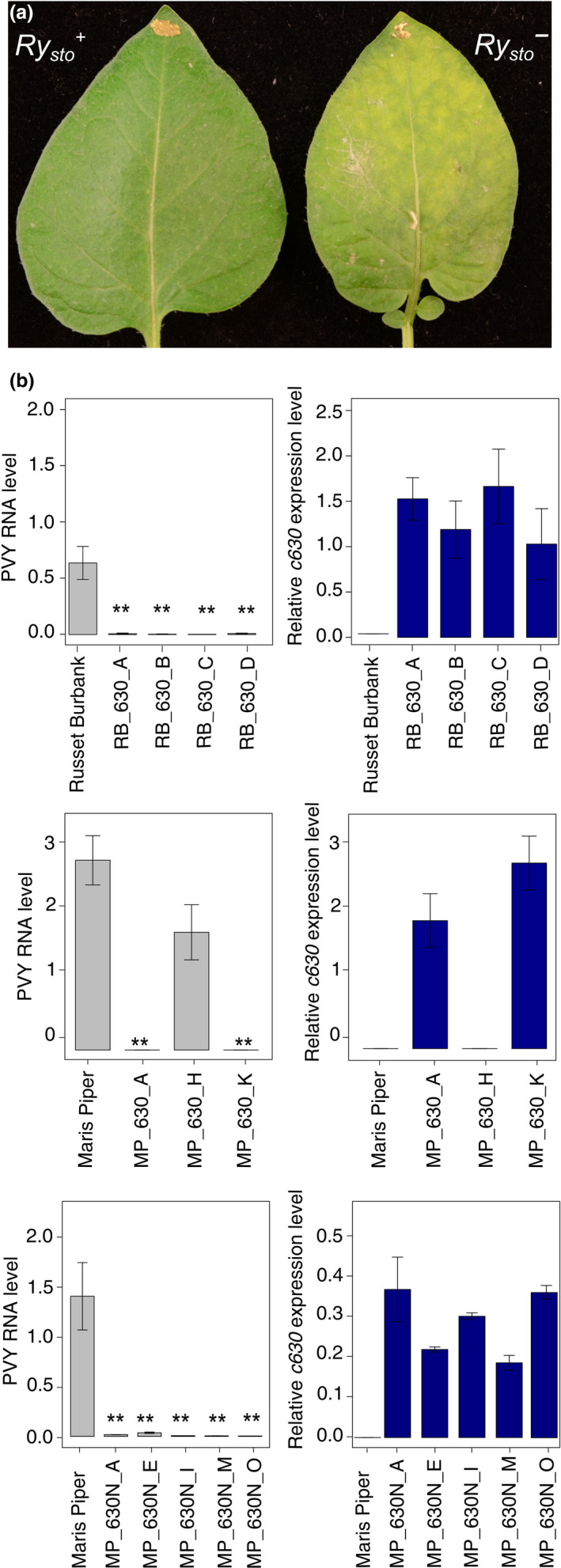
Expression of Ry sto leads to immunity of transgenic plants to PVY infection. (a) Illustration of ER ‐type response of PVY ‐inoculated leaves of Solanum tuberosum cv Russet Burbank plants expressing Ry sto . Four‐week‐old transgenic potato plants cv. Russet Burbank carrying construct 35S:Ry sto , and non‐transformed plants, were inoculated with PVYNTN . Two weeks after infection, chlorosis was observed on inoculated leaves of non‐transformed plants (right), whereas Ry sto transgenic plants (left) remained symptomless. (b) Ry sto expression completely blocks PVY spreading in transgenic potato plants. Four‐week‐old transgenic potato plants cv. Russet Burbank (upper graph) or Maris Piper (middle graph) carrying construct 35S:Ry sto , Maris Piper (lower graph) plants carrying Ry sto construct under the control of native 5′ and 3′ regulatory elements and non‐transformed plants were inoculated with PVYNTN . Three weeks after viral inoculation, mRNA was isolated from upper, non‐inoculated leaves. PVY RNA levels and expression of Ry sto were quantified using qPCR , relative to EF 1 and Sec3 reference genes, and expressed as means ± SD calculated from three biological replicates per plant line. One‐way ANOVA with Dunnett′s test was used for statistical analysis.
To corroborate PVY resistance of MP, T1 generation progeny tubers of selected lines were collected and subsequently planted following by graft inoculation with tobacco scions carrying PVYNTN. In agreement with previous results, any symptoms of PVY infection were detected using ELISA assay (Figure S2; Table S4).
N. tabacum 35S:Ry sto transgenic and non‐transformed control plants were inoculated with PVYNTN as previously. At 5 dpi, some necrotic lesions were visible on the inoculated leaves of Ry sto plants, whereas no macroscopic symptoms of HR were observed on the non‐transformed control plants (Figure S3). In addition, control susceptible plants developed typical systemic symptoms of PVY infection. qPCR analysis of upper, non‐inoculated leaves showed that systemic spread of the virus was fully restricted in Ry sto ‐expressing lines, whereas PVY RNA levels in non‐transformed plants were high. These results were confirmed in T0 (Figure S4a) and T1 (Figure S4b) generation plants. Transgenic tobacco lines expressing Ry sto under the control of its natural 5′ and 3′ regulatory elements exhibited a local HR response at 5 dpi similar to that seen in 35S::Ry sto plants. No viral RNA was detected with qPCR (Figure S5) at 7 and 14 dpi, and this corresponded with Ry sto transcript expression.
Introduction of Ry genes from S. stoloniferum to S. tuberosum may condition resistance not only to PVY but also to PVA, which is closely related (Cockerham, 1970). To test whether expression of Ry sto provided resistance to PVA, tobacco plants with and without the functional Ry sto transgene were inoculated with PVA. Western blotting analysis with anti‐PVA antibodies revealed inhibition of PVA systemic spread in Ry sto transgenic plants but not in control plants (Figure S6).
To compare Ry sto and Ry‐f sto alleles, a relevant region from PW363 breeding line was amplified using primers corresponding to the genomic sequence of Ry sto (see Experimental Procedures). An alignment of these two Ry alleles revealed 100% sequence identity in the coding and non‐coding regions of the 4.85 kb amplified product (Figure S7).
PVY and PVA coat proteins are elicitors of Ry sto ‐triggered immunity
A single ORF in the PVY genome encodes a polyprotein that is then cleaved by three viral proteases to form functional proteins (Jakab et al., 1997). To identify the elicitor of the Ry sto ‐mediated immune response, the ORFs encoding putative viral proteins Hc‐Pro, NIa, NIb and CP were cloned and transiently expressed in transgenic Ry sto and non‐transformed tobacco plants. Only the CP induced strong HR at 2–3 dpi in Ry sto plants, whereas no HR was observed in control plants (Figure 3a). Similarly, only the CP of PVA elicited HR when expressed in transgenic Ry sto plants (Figure 3b). The expression of specific viral proteins was detected by immunoblotting (Figure 3c), indicating that the absence of HR was not caused by a lack of protein expression. Additionally, cellular localization of each protein was determined (Figure 3d).
Figure 3.
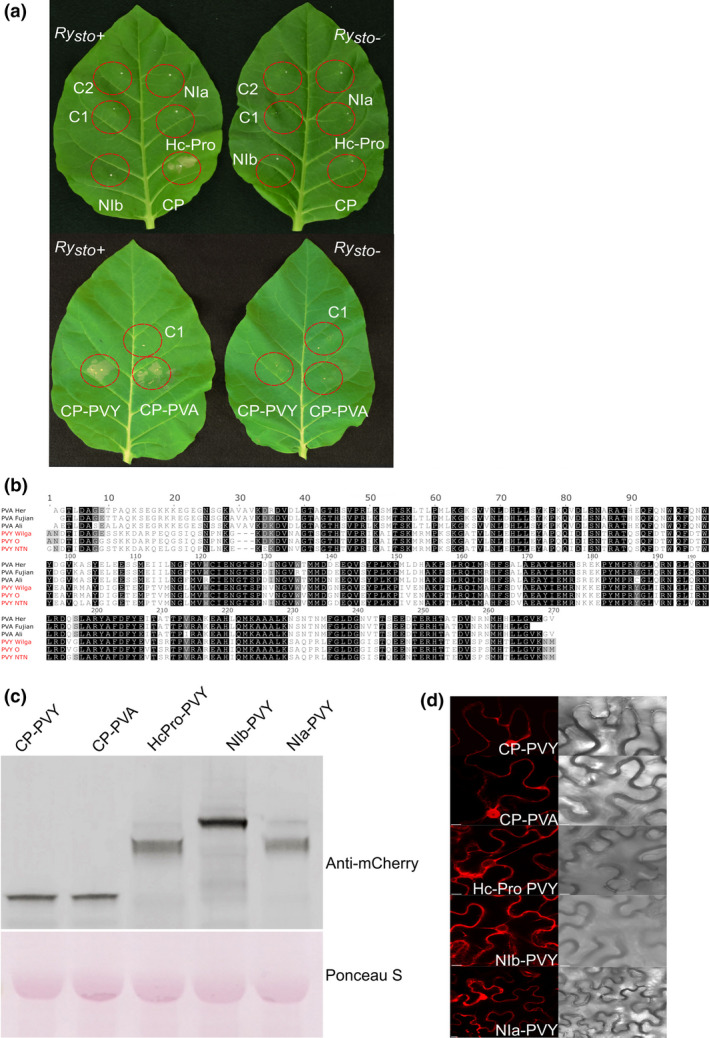
Coat proteins of two closely related potyviruses elicit Ry sto ‐mediated immunity. (a) Rysto recognizes PVY (upper panel) and PVA (lower panel) coat proteins as avirulence factors in transient expression assays. To identify the elicitor of the Ry sto ‐mediated resistance against PVY or PVA viruses, open reading frames encoding putative viral proteins (Hc‐Pro, Nia, Nib and CP) were cloned into the pBIN‐mCherry vector and transiently expressed in N. tabacum Ry sto transgenic and non‐transformed plants. As a control (C1, C2), Agrobacterium with empty vector or buffer infiltration was used. Three days after treatment, HR was observed only for PVY or PVA coat proteins (CPs). (b) Comparison of PVY and PVA coat protein amino acid sequences. PVY and PVA CP amino acid sequences were compared with PVYNTN CP. Identical residues are shaded in black. PVA CP shares >59% identity with the PVY sequence. The alignment was generated using MAFFT‐L‐INS‐I (Katoh and Toh, 2008) and visualized in Jalview 2,10,4b1 (Waterhouse et al., 2009). (c) Western blot analysis of viral protein expression. The indicated combinations of PVY‐CP‐mCherry, PVA‐CP‐mCherry, PVY‐Hc‐Pro‐mCherry, PVY‐NIb‐mCherry and PVY‐NIa‐mCherry were transiently expressed in leaves of N. tabacum. Total proteins were extracted and analysed by protein gel blotting with polyclonal anti‐mCherry antibody (Abcam). Staining of RuBisCO with Ponceau S was used as a loading control. (d) Cellular localization of viral proteins. Confocal images show representative N. benthamiana leaf epidermal cells transiently expressing indicated proteins. Images were taken 72 h after Agro‐infiltration. For each variant, approximately 50 transformed cells were examined. Bars = 10 μm.
ER mediated by Ry sto is epistatic to Ny‐1‐mediated HR
Next, the functional relationship between the two types of defence response to PVY infection, ER and HR, was investigated. Potato cultivar Rywal, carrying the Ny‐1 gene responsible for HR to PVY infection (Baebler et al., 2014; Szajko et al., 2008), was transformed with Ry sto . Plants of both genotypes (Ny‐1 or Ry sto /Ny‐1) were inoculated with PVYNTN, as described previously. Five days after infection, HR developed only in the parental genotype (Ny‐1), whereas no HR was observed in the Ry sto /Ny‐1 genotype (Figure 4b). This indicated that ER conferred by Ry sto was epistatic to Ny‐1‐dependent HR.
Figure 4.
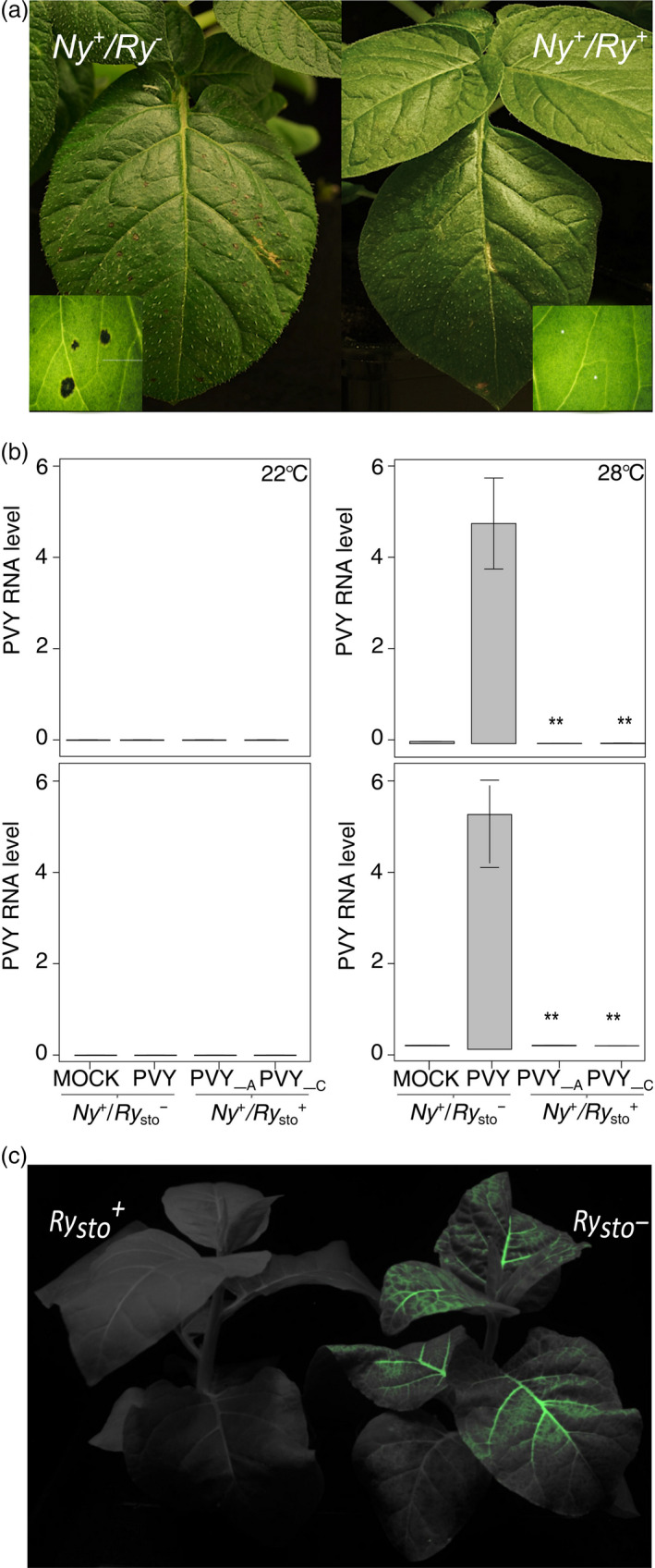
Ry sto ‐dependent ER is epistatic to HR and is temperature‐independent. (a) Determination of epistatic effect of ER to HR. Potato Ry sto /Ny‐1 and Ny‐1 plants were inoculated with PVYNTN . Five days after infection, symptoms of local cell death occurred only in Ny‐1 plants. The experiment was repeated three times. (b) Ry sto ‐dependent PVY resistance is not inhibited at elevated temperature. Potato Ry sto /Ny‐1 and Ny‐1 plants were inoculated with PVYNTN and divided into two groups, at 20 °C and 28 °C. Water‐treated plants were used as negative controls. Seven days after inoculation, samples of inoculated (lower graphs) and upper non‐inoculated leaves (upper graphs) were collected and PVY RNA levels were measured using qPCR. Values are expressed relative to EF1 and Sec3 reference genes and are expressed as means ± SD calculated from three biological replicates per plant line. A and C describe the names of transgenic Ry sto lines used. One‐way ANOVA with Tukey's test for statistical analysis was performed. (c) Stable transgenic N. tabacum plants carrying Ry sto under the control of a 35S promoter are resistant to PVY infection at elevated (32 °C) temperature. Seven‐week‐old N. tabacum 35S:Ry sto transgenic and non‐transformed plants were inoculated with a PVYN 205:GFP clone. Typical symptoms of PVY infection were observed 7 dpi in non‐transformed plants, whereas Ry sto lines remained symptomless. Image was taken at 14 dpi.
ER mediated by Ry sto is temperature‐independent
To investigate the role of temperature in Ry sto ‐mediated ER, the response to PVY infection was studied in a Ny‐1 or Ry sto /Ny‐1 genotype. PVY mRNA levels were measured 14 dpi in upper non‐inoculated leaves following mock and virus treatments at two temperatures (22 °C and 28 °C). In both genotypes, no PVY multiplication was observed when plants were kept at 22 °C. By contrast, at 28 °C, HR was inhibited in the Ny‐1 genotype and PVY RNA was detected systemically, whereas PVY spread remained fully inhibited in transgenic Ry sto /Ny‐1 lines (Figure 4a and b). To test whether Ry sto hindered systemic infection above 28 °C, transgenic and non‐transformed tobacco plants were infected with GFP‐tagged PVY (PVYN605‐GFP) at 32 °C. Expression of Ry sto at the higher 32 °C temperature still prevented systemic spread of the virus (Figure 4c).
These observations suggest that high (>28 °C) temperature does not compromise Ry sto immunity to PVY infection, unlike the tobacco N gene, which also encodes a TIR‐NLR (Samuel, 1931).
NRG1 and EDS1 mediate Ry sto ‐dependent HR
It has been recently shown that TIR‐NLR signalling pathway may involve, in addition to Enhanced Disease Susceptibility 1 (EDS1), a well studied component, other helper proteins, including N requirement gene 1 (NRG1) (Castel et al., 2019; Qi et al., 2018). To check whether EDS1 and/or NRG1 are involved the Ry sto ‐signalling, we assayed development of HR in response to PVY in EDS1 and NRG1 knockout background. To this aim, Ry sto was transiently expressed in eds1‐1 or nrg1‐1 CRISPR/Cas9‐mediated N. benthamiana mutant plants (Castel et al., 2019; Schultink et al.,2017) that had been pre‐inoculated with PVYNTN. While in the wild‐type control plants HR was observed 72 hpi, the eds1‐1 and nrg1‐1 plants remained symptomless (Figure 5a), suggesting that both EDS1 and NRG1 are required for Rysto‐mediated response. Having shown that Rysto‐mediated HR was abolished in eds1‐1 and nrg1‐1 lines, we then performed coexpression of Rysto either with EDS1 or NRG1 in the respective mutants to check whether HR phenotype will be restored. Consistently, expression of EDS1 and NRG1 restored the resistant phenotype in the mutants’ background (Figure 5b, c) implying that Rysto activates HR in EDS1‐/NRG1‐dependent manner. A similar analysis was performed with N. benthamiana plants expressing bacterial NahG, which encodes SA hydroxylase (Friedrich et al., 1995). Symptoms of HR were observed 72 hpi both in wild‐type and NahG plants (Figure 5a), indicating that elevated levels of SA were not essential for establishing Ry sto ‐dependent resistance.
Figure 5.
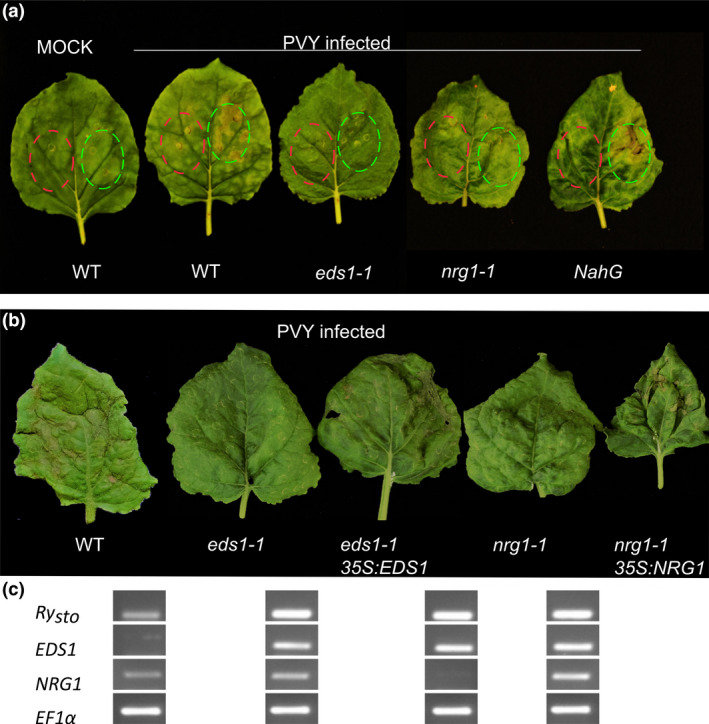
Downstream signalling components are crucial for Ry sto ‐mediated immunity. (a) Fully developed leaves of Nicotiana benthamiana eds1‐1 or nrg1‐1 knockout plants, NahG plants or non‐transformed plants were infected with PVYNTN or were mock treated with water. Two weeks later, leaves with symptoms of PVY infection were infiltrated with A. tumefaciens suspensions carrying pICSLUS 0001:Ry sto (highlighted in green) or pGBT : GFP (highlighted in red) as a control. Three days after infiltration, symptoms of cell death were observed in non‐transformed plants and NahG plants ( SA ‐free). Experiments were repeated three times with similar results. (b) Complementation tests for Ry sto ‐triggered HR in eds1‐1 or nrg1‐1 N. benthamiana mutants. PVY ‐infected leaves of N. benthamiana eds1‐1, nrg1‐1 knockouts or non‐transformed plants were infiltrated with Agrobacterium carrying Ry sto alone or Ry sto coexpressed with EDS 1 or NRG 1. Three days after infiltration, HR was observed only when Rysto was coexpressed with indicated proteins. Experiments were repeated three times with similar results. Transcripts expression was confirmed via semi‐quantitative RT ‐ PCR (c) Elongation factor‐1α ( EF 1α) was used as an internal control. Raw data are presented in the Figure S9.
Discussion
Genes conferring ER against PVY infection were previously introduced into potato cultivars from wild or domesticated species of the Solanaceae family, including S. chacoense, S. stoloniferum and S. tuberosum ssp. andigena. Two alleles of the Ry sto gene derived from S. stoloniferum (Ry‐f sto and Ry sto ) have been used in breeding programmes (Flis et al., 2005; Song et al., 2005). In this study, we report the isolation and characterization of a novel, broad‐spectrum resistance Ry sto /Ry‐f sto gene using RenSeq (Jupe et al., 2013; Witek et al., 2016).
We hypothesized that the underlying PVY resistance gene encoded a NLR protein. Data obtained from SMRT and Illumina sequencing together with SNP calling and presence/absence polymorphism detection revealed 11 transcriptionally active NLR genes that were candidates for the Ry sto gene. These candidates mapped to the DM reference genome (Potato Genome Sequencing Consortium (95 authors), 2011) at the distal end of chromosome XII, in a region known to carry Ry sto ‐linked markers. Previous research using graphical genotyping with a panel of tetraploid potatoes suggested that Ry sto was located on superscaffold DMB114 (Van Eck et al., 2017). This was consistent with our data, as 3 of the 11 candidates were located on this superscaffold and the remaining candidates were located within 1 Mb, on neighbouring superscaffolds. All the linked NLRs showed presence/absence polymorphisms with the same allele ratios, indicating that the whole interval introgressed from S. stoloniferum and rarely undergoes recombination in cultivated potato.
Functional assays in N. benthamiana and cultivated potato plants confirmed that only one candidate, c630, prevented PVY multiplication and spread, hereafter termed Ry sto . The protein encoded by Ry sto had motifs and domains typical of TIR‐NLR‐type resistance proteins and shared <34% identity at the amino acid level with previously described TIR‐NLR‐type resistance proteins from Solanaceae, including N, Y‐1, Bs4 and Pvr4.
Sequence homologues of Ry sto were amplified from eight PVY‐resistant cultivars (Barbara, Pirola, Ute, Wega, Assia, Fanal, Heidrun and Esta [for details, see Świeżyński et al., 1997; www.europotato.org]) from the collection at the Max Planck Institute for Plant Breeding Research (MPI‐PZ). No SNPs were observed in either the coding or the non‐coding sequences of the 4.85 kb amplified regions.
Most potato cultivars lack broad‐spectrum resistance to circulating strains of PVY (Valkonen, 2015). Recently, two novel genes (Pvr4 and TPN1) were reported that conferred immunity to some PVY strains. Kim et al. (2017) isolated Pvr4, a dominant gene conferring a broad range of resistance to potyviruses, including some PVY isolates, from Capsicum annuum. However, Pvr4 was tested only in N. benthamiana, a heterologous system, and not in any Solanaceae crops. Therefore, there is no direct evidence that Pvr4 would be functional in a potato background. Similarly, TPN1, a single recessive gene, was isolated and tested only in tobacco where it causes venial HR‐like necroses (Michel et al., 2018). Our results suggest that Ry sto ‐mediated immunity is effective in potato and tobacco plants not only against different strains of PVY but also against the related virus PVA (Table S1, Figure 2a and S6). This is in contrast to Rx, Rsv3 and Sw‐5, which mediate ER to other viruses and to which immunity‐breaking isolates have emerged (Brommonschenkel et al., 2000; Moreira et al., 1980; Querci et al., 1995; Zhang et al., 2012).
Multiple PVY ORFs were tested, and viral CP was found to be the recognized in Rysto‐mediated immunity against PVY. Similarly, CP was the elicitor of Rysto‐mediated immunity for PVA (Figure 3a). The PVY and PVA CPs share ~59% identity (Figure 3b), and both display nucleocytoplasmic localization in infected cells (Figure 3d). The mechanism of CP recognition by Rysto remains unknown. As was previously shown for Rx, recognition may rely on a three‐dimensional structure containing a specific internal amino acid motif (Baurès et al., 2008; Candresse et al., 2010).
Rx induced HR when transiently expressed with PVX CP in a heterologous N. benthamiana system (Bendahmane et al., 1999). Similarly, PVY infection of N. benthamiana leaves transiently overexpressing Ry sto produced a strong HR reaction (Figure 1c). HR was also elicited when PVY CP was transiently produced under control of a constitutive 35S promoter in transgenic Ry sto potato plants. By contrast, CP produced during PVY infection led to ER, indicating that the level of CP determined the type of response. However, when Ry sto was introduced into N. tabacum, some necrotic lesions on inoculated leaves of resistant plants were observed upon PVY infection (Figure S3). This led us to the conclusion that the nature of the resistance response (ER or HR) is determined not only by the CP level but also by host factors differing between the plant models used. Modulation of the specificity of elicitor recognition was also observed for the Rx gene between PVX CP in Rx‐expressing potato and tobacco plants (Baurès et al., 2008).
ER or HR formation and control of viral spread might be determined by R gene expression level for different resistance genes. High levels of HRT transcript in susceptible Arabidopsis thaliana led to restriction of virus replication or movement without any visible symptoms of HR, whereas lower HRT transcript levels in resistant Arabidopsis lines led to micro‐HR or HR with systemic virus movement (Cooley et al., 2000). By contrast, our results suggest that ER is not affected by Ry sto expression levels. Transgenic potato lines with Ry sto under the control of native regulatory elements exhibited much lower transcript levels than lines with Ry sto under the control of 35S. ER phenotypes were observed throughout all the experiments regardless of relative Ry sto expression levels (Figure 2b and 2d).
Here, we showed that the Ry sto mode of action was, unusually for a TIR‐NLR‐type receptor, temperature‐independent (Figure 4b and 4c). Products of many R genes are non‐functional at or above 28 °C. Elevated temperatures reduced functionality for the N gene conferring resistance to TMV, the Mi‐1 gene responsible for resistance to root‐knot nematodes (Hwang et al., 2000; Jablonska et al., 2007), the Ny‐1 gene conferring resistance to PVY (Szajko et al., 2014), the suppressor of NPR1, constitutive 1 (SNC1) gene in Arabidopsis contributing to the resistance to the bacterial effector AvrRps4 (Yang and Hua, 2004) and many others. All these genes induced defence at permissive temperatures but experienced arrested HR development upon shifts to an elevated temperature. By contrast, Ry sto ‐mediated immunity remained effective in different Solanaceae species at or above 28 °C (Figure 4b and 4c). Temperature‐sensing characteristics have also been identified in Arabidopsis SNC1, a NB‐LRR‐type gene closely related to RPP4 and RPP5. Genetic screens of snc1 mutants identified a single mutation (Gly380 to Ser) in the encoded protein that reverted the heat‐stable resistance response (Zhu et al., 2010). This provides evidence that temperature sensitivity of R genes might be controlled internally rather than by other regulatory components. In the Rysto protein, glycine is present at position 380, immediately after the putative GLPL motif in the NB‐ARC domain. This motif was previously identified as important for nucleotide binding, and mutations in residues close to this motif might compromise activation of NB‐LRR proteins (Rairdan and Moffett, 2006). It remains to be determined whether elements of the Rysto structure are responsible for its temperature stability.
A detailed model of the functional relationship between HR and ER has not yet been proposed. In principle, the products of gene expression for ER act earlier and more efficiently than the genes for HR, as demonstrated in comparative studies of potato Rx and N genes (Bendahmane et al., 1999). Ry sto ‐mediated ER was epistatic to Ny‐1‐mediated HR against PVY in potato (Figure 4).
The epistatic Ry sto /Ny‐1 interaction was also observed in progeny of a cross between cv Rywal and clone PW 363 (Szajko et al., 2018). In contrast to Ny‐1 plants (Baebler et al., 2014; Szajko et al., 2008), Ry sto /Ny‐1 plants were PVY‐free at elevated temperature (28 °C). Collectively, Ry sto ‐mediated ER was epistatic to HR. Nevertheless, the ER and HR pathways might be interdependent. For instance, Rysto might compete with Ny‐1 for recruitment of some signalling components. Assuming that Rysto has higher affinity to selected components than Ny‐1, Ry sto ‐mediated immunity is stronger and progresses faster. These components might act as helper NLRs since many R proteins assemble into multi‐subunit complexes (Wu et al., 2018).
To activate immunity, most TIR‐NLRs require EDS1 protein (Aarts et al., 1998; Wiermer et al., 2005). Recently, it has been shown that also safeguarding proteins like activated disease resistance 1 (ADR1) and NRG1 may be involved in NLRs activation upon pathogen perception (Castel et al., 2019; Wu et al., 2018). In this study, we found that Rysto failed to initiate HR in eds1 and nrg1 knockout plants (Figure 5a, b). The HR phenotype was rescued by expression of EDS1 or NRG1 in the respective mutant background, consistent with the model, that Rysto‐mediated resistance depends on EDS1 and NRG1. Thus, our results provide corroborating evidence for a critical role of NRG1 in the TIR‐NLRs immune signalling pathway (Castel et al., 2019; Qi et al., 2018). In our studies, HR in response to PVY infection was established in SA‐depleted NahG plants transiently expressing Ry sto (Figure 5a). This indicates that, in contrast to EDS1, an increased SA level is not essential for Ry sto ‐HR development. These data support a model that EDS1 performs different functions during plant immunity and the role played in triggering local cell death is independent of SA (Bhandari et al., 2019; Rietz, 2011).
Taking our observations as a whole, we propose a model in which Ry sto ‐mediated immunity to ER against PVY results in Ry sto recognition of PVY CP and recruitment of downstream signalling components such as NRG1 and EDS1. Since ADR1 activity may also contribute to TIR‐NLR‐mediated immunity (Dong et al., 2016), we cannot exclude ADR1 involvement in the Ry sto mode of action. Conceivably, co‐chaperone complex components SGT1, RAR1 and HSP90 might also be involved in Ry sto ‐mediated immunity through their roles in supporting correct folding of resistance proteins (Peart et al., 2005). Ry sto might also be negatively regulated through miRNA activity through promotion of mRNA degradation, inhibition of translation or suppression of transcription by epigenetic modification (Križnik et al., 2017).
In summary, our results demonstrate that Ry sto plays an important role in defence and may prove valuable for breeding PVY‐resistant cultivars of potato and other Solanaceae crops. It remains unclear why some NLRs trigger ER and others trigger HR, but ER recognition appears to produce durable and efficient resistance.
Experimental procedures
Plant material
All potato cultivars (S. tuberosum ssp. tuberosum) and viruses PVY0, PVYN, PVYN‐Wi, PVX and PVA were obtained from the Laboratory for Potato Gene Resources and In Vitro Cultures at the Institute of Plant Breeding and Acclimatization – National Research Institute, Bonin, Poland. The cultivar Alicja (the source of Ry sto ) was released from the breeding company HZ Zamarte, Poland, as a cross between clone OL‐21852 and cv. Ora (Figure S8). A S. stoloniferum accession from the collection of the Max Planck Institute for Plant Breeding Research (MPI‐PZ), Cologne, Germany, was in the ancestry of OL‐21852. PW363 (the source of Ry‐f sto ) was generated in the parental line breeding programme at the IHAR‐PIB, Młochów. The ancestry of PW363 contained a S. stoloniferum accession from the collection of the Vavilow Research Institute of Plant Industry (VIR), St. Petersburg, Russia. Cultivars, Barbara, Pirola, Ute, Wega, Assia, Fanal, Heidrun and Esta from MPI‐PZ collection, were obtained from the Laboratory for Potato Gene Resources and in Vitro Cultures at the IHAR‐PIB. Plants were grown for 4 weeks in soil under controlled environmental conditions (22 °C, 16‐h light; 18 °C, 8‐h dark) as described previously (Szajko et al., 2008). Tobacco plants Nicotiana tabacum cv. Xanthi‐nc and N. benthamiana were grown for 6 weeks in soil under controlled environmental conditions (22 °C; 16‐h light and 8‐h dark) as described previously (Hoser et al., 2013). Transgenic potato and tobacco plants were regenerated following Agrobacterium leaf disc transformation as described by Mac et al. (2004).
Mapping population
A diploid potato (2n = 2x = 24) mapping population was developed from a cross between PVY‐resistant heterozygous clone dH Alicja and susceptible clone DW 83‐3121. dH Alicja was obtained from potato cultivar Alicja (Świerzyński et al., 1997; www.europotato.org) via parthenogenesis. ER to PVY in dH Alicja was conferred by the Ry sto gene, which derived from clone MPI 55.957/54. The pedigree of MPI 55.957/54 included S. stoloniferum (Ross, 1958; Handbuch der Pflanzenzüchtung, 2nd, Vol. III:106‐125).
Short‐ and long‐read RenSeq and bioinformatic analysis of sequencing data
Construction and enrichment of short‐read RenSeq libraries was carried out on susceptible (S, clone DW 83‐3121), resistant (R, dHAlicja) and 149 bulked susceptible (BS) plants as described previously (Jupe et al., 2013; Witek et al., 2016). Enriched libraries were sequenced with an Illumina MiSeq platform using 500 cycles. RenSeq was also performed on cDNA from resistant parents as described previously (Witek et al., 2016) and sequenced using an Illumina HiSeq platform with 300 cycles. Long‐read PacBio RenSeq was performed as described in Witek et al. (2016) and sequenced using a single PacBio RSII SMRT cell, resulting in 69,737 ROIs with a minimum of three passes and accuracy >90% (available at accession number MN393235). Reads obtained from SMRT RenSeq were assembled using Geneious 8.1.2 software as described previously (Witek et al., 2016), resulting in 1555 contigs. R, S and BS reads from Illumina MiSeq 250PE sequencing were mapped to contigs derived from the SMRT RenSeq assembly using BWA (Li and Durbin, 2009), with default settings. SNP calling and candidate prediction were performed as described previously (Jupe et al., 2013; Witek et al., 2016). Contigs with >95% of susceptible alleles in mapped BS data were considered to be linked. Candidate NLRs showing presence/absence polymorphism between the R and S parents and linkage to resistance based on BS samples were also called. Briefly, numbers of pair‐end mapped reads relative to contigs derived from the SMRT RenSeq assembly were calculated for R, S and BS samples using TSL Galaxy built‐in scripts (Maclean and Kamoun, 2012). The resulting data were sorted and visualized using Microsoft Excel software. To define presence/absence polymorphism, contigs with at least 250 mapped reads from the R parent, and <10% and 18% of that number for S and BS samples, respectively, were considered. Expression of the candidate genes was determined using cDNA‐RenSeq data from the R parent as described previously (Andolfo et al., 2014; Witek et al., 2016).
DNA extraction
RenSeq and PCR were performed on freshly extracted genomic DNA obtained from young leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
RNA extraction and cDNA preparation for RenSeq
For cDNA‐RenSeq, RNA was extracted using TRI‐Reagent (Sigma‐Aldrich, Saint Louis, MO) and a Direct‐zol RNA MiniPrep Kit (Zymo Research, Tustin, CA) according to the manufacturers’ instructions. First‐strand cDNA was synthesized using a mix of oligo‐dT and random hexamer primers and the SuperScript II First‐Strand Synthesis System (Sigma‐Aldrich). Second‐strand cDNA was synthesized as described previously (Rallapalli, 2014).
Cloning of candidate ORFs
PCR primers flanking predicted ORFs were designed for each of 12 selected PacBio contigs. Specific 5′ and 3′ extensions were added to primers for compatibility with the custom USER expression vectors used in this study and described previously (Witek et al., 2016). Candidate genes were PCR‐amplified from R parent genomic DNA in 25 μL PCR reactions (35 cycles with annealing at 62 °C and 7 min extension at 72 °C) using Kappa HiFI HotStart Uracil+ Fidelity Polymerase (Manufacturing, R&D Cape Town, South Africa). Purified PCR product (30 ng) was hybridized with 30 ng pICSLUS0003 vector in the presence of 1 μL USER enzyme mix (New England Biolabs, Inc., Ipswich, MA) as described previously (Witek et al., 2016). All constructs were verified by DNA sequencing. Plasmids containing candidate genes were transformed into Agrobacterium strain GV3101 for transient complementation assays or strain LBA404 for stable potato transformation using the method described previously (Mac et al., 2004). To create a construct containing Ry sto (c630) under the control of its native regulatory elements, the whole contig (7.5 kB) assembled from PacBio reads was PCR‐amplified and introduced into USER‐vector pICSLUS0001 lacking the 35S promoter and OCS terminator. List of primers used in this study is enclosed as Table S5.
Transient expression assay
Agrobacterium GV3101 strains carrying plasmids containing the candidate genes or PVY‐derived sequences were used to infiltrate N. benthamiana. Bacteria harbouring plasmids were grown in YEB medium supplemented with gentamicin, rifampicin and kanamycin. Overnight cultures were collected by centrifugation at 3500 rpm, resuspended in infiltration buffer containing 10 mm MgCl2, 10 mM 2‐[N‐morpholino] ethanesulfonic acid (MES) (pH 6.5) and 100 μm acetosyringone to a final OD600 = 1 and infiltrated into leaves of 4‐week‐old plants using a 3 mL syringe. HR reactions were observed 2–3 days after infiltration. Each experiment was performed at least twice and included at least three independent biological replicates.
Resistance assay
PVY isolate NIB‐NTN (GenBank: AJ585342.1) was used in all experiments involving PVY infection monitoring. Seven‐week‐old N. tabacum plants or 4‐week‐old potato plants were infected under the previously described conditions (Baebler et al., 2014). Where different virus isolates were used, plants were infected under the same experimental conditions with PVY isolates 0 (AJ890349), N (FJ666337) or N‐Wilga (EF558545), or unrelated viruses PVA, PVX or TMV(U1).
Semi‐quantitative RT‐PCR
Total RNA was treated with DNase I (Thermo Fisher Scientific, Waltham, MA) and subjected to reverse transcription using a mix of random hexamers and oligo‐dT primers and a RevertAid First‐Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Semi‐quantitative RT‐PCR was performed using DreamTaq (Thermo Scientific) with 25 to 30 amplification cycles followed by electrophoresis with 2% agarose gel stained with Ethidium bromide. Primer pairs used in the PCR reaction are listed in Table S6.
Gene expression analysis‐ RT‐qPCR
Gene expression analysis via RT‐qPCR was performed using a LightCycler®480 instrument and LightCycler®480 SYBR Green I Master Kit reagents (Roche, Indiana, IN). Relative gene expression levels were determined using a standard curve method, and the value for each target gene was normalized against the mean of expression values of two reference genes, EF1 and L23 for N. tabacum and N. benthamiana or EF1 and Sec3 for potato cultivars, as described previously (Liu et al., 2012; Tang et al., 2017). Each sample was tested with four technical replicates and two dilutions. Primers for qPCR are provided in Table S6.
PVY ORFs cloning
A full‐length infectious PVY‐N605 clone (Jakab et al., 1997) was used as a template to create all PVY constructs. ORFs encoding each putative PVY protein were PCR‐amplified and cloned into the pBIN53‐mCherry vector with a CaMV 35S promoter, a polylinker separating the PVY 5′ and 3′ UTRs and a poly(A) tail as described previously (Mestre et al., 2000).
PVA CP cDNA isolation
PVA CP encoding cDNA was amplified by reverse transcription‐PCR from total RNA extracted from PVA‐infected tobacco leaf tissue. The PCR fragment was inserted into the pENTR/D‐TOPO vector (Invitrogen, Carlsbad, CA). The resulting entry clones were LR recombined with the Gateway pGWB 454 destination vector (Nakagawa et al., 2007).
Western blotting
Agrobacterium‐infiltrated leaves were collected, frozen and ground in liquid nitrogen. Total proteins were extracted by incubating ground leaf samples in extraction buffer containing 100 mm Tris HCl pH 8.0, 1 mm EDTA, 150 mm NaCl, 7.2 mm β‐mercaptoethanol, 0.5 mm 4‐(2‐aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) and 0.03 μm PMSF (phenylmethylsulfonyl fluoride) for 15 min. Homogenates were centrifuged at 21,000 g for 10 min, and the supernatants were collected. Samples were separated by 12.5% SDS‐PAGE and subjected to immunoblot analysis using polyclonal Anti‐mCherry antibody (Abcam, Cambridge, UK) or appropriate alkaline phosphatase‐conjugated anti‐PVA antibodies (Bioreba, Reinach, Switzerland). Immunoblots were developed using a NBT/BCIP colorimetric detection kit (BioShop Inc., Canada).
Confocal laser scanning microscopy
Subcellular localization of the fusion proteins was evaluated using a Nikon C1 confocal system built on TE2000E and equipped with a 60×Plan‐Apochromat oil immersion objective (Nikon Instruments B.V. Europe, Amsterdam, The Netherlands). RFP fusion protein was excited by a 543 nm helium‐neon laser and detected using the 605/75 nm barrier filter. Confocal images were analysed using free viewer EZ‐C1 and ImageJ software.
Statistical analysis
Statistical analyses were conducted using R 3.2.2 within R Studio 0.99.483. Technical replicates consisted of replicate readings from the same plant in the same experiment, whereas biological replicates consisted of measurements obtained from independent plants. Data were analysed using the following pipeline: data were assessed for their suitability for parametric analysis by testing for the normal distribution of the residuals using a Shapiro–Wilk test. If the data were suitable for conducting parametric tests, then analysis of variance (ANOVA) was used. Dunnett's or Tukey's HSD (P < 0.001**) tests were used for post hoc analysis.
Author contributions
M.G‐B., K.W and J.H. performed most of the experimental work, data analyses and writing; A. W., I.W‐F., H.J., K.S. and K.M performed the research; J.J. and W.M. edited the article; J.H. supervised the work.
Competing interests
KW, MGB, JH, JDGJ, WM and KS have filed a US patent application 62/538.020 based on this work.
Supporting information
Figure S1 Phylogenetic analysis of Ry sto candidate genes and other functional Solanaceae NLRs.
Figure S2 Grafted transgenic Ry sto plants display resistance to PVY.
Figure S3 HR of transgenic N. tabacum expressing Ry sto .
Figure S4 Stable transgenic tobacco plants carrying Ry sto under the control of a 35S promoter display resistance to PVY. Seven‐week‐old N. tabacum 35S:Ry sto transgenic and non‐transformed plants were inoculated with PVYNTN or mock treated with water.
Figure S5 Stable transgenic tobacco plants carrying Ry sto under the control of native regulatory elements display resistance to PVY. Seven‐week‐old N. tabacum Ry sto (native) transgenic and non‐transformed plants were inoculated with PVYNTN.
Figure S6 Ry sto ‐mediated resistance to PVA infection.
Figure S7 Comparison of Ry sto and Ry‐f sto nucleotide sequences.
Figure S8 Pedigree chart of cultivar Alicja.
Figure S9 Semi‐quantitative RT‐PCR of Ry sto , NRG1 and EDS1 genes.
Table S1 PVY dependent HR in N. benthamiana plants expressing contig 630.
Table S2 ELISA titers of PVY in transgenic 35S::Rysto S. tuberosum cv. Maris Piper plants.
Table S3 ELISA titers of PVY in transgenic 35S::Rysto S. tuberosum cv. Russet Burbank plants.
Table S4 ELISA titers of PVY in grafted transgenic 35S::Rysto S. tuberosum cv. Maris Piper plants.
Table S5 Primers used to clone candidate Rysto genes.
Table S6 Primer sequences for quantitative RT and qRT‐PCR
ACKNOWLEDGMENTS
Research was supported by OPUS 8 (2014/15/B/NZ9/04384) and OPUS 14 (2017/27/B/NZ9/01803) grants from the National Science Centre, Poland, to Jacek Hennig. Kamil Witek was supported by BBSRC grants BB/L009293/1 and BB/P021646/1 to Jonathan Jones. The research at IHAR‐PIB Młochów was founded by statutory grant 1‐3‐00‐1‐01 from the Polish Ministry of Science and Higher Education. We would like to thank our co‐workers, in particular Dr. Magdalena Krzymowska, for critical comments on the manuscript, Prof Ewa Zimnoch‐Guzowska and Dr Bogdan Flis for helpful discussion. We would like to acknowledge Danuta Strzelczyk‐Żyta and Izabella Barymow for the technical support, Baptiste Castel for sharing seeds of nrg1 N. benthamiana knockout plants, Prof Brian Staskawicz for sharing seeds of eds1 N. benthamiana knockout plants and Prof Doil Choi for sharing seeds of NahG‐transgenic N. benthamiana plants.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis. Proc. Natl Acad. Sci. USA 95, 10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo, G. , Jupe, F. , Witek, K. , Etherington, G.J. , Ercolano, M.R. , et al. (2014) Defining the full tomato NB‐LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramburu, J. , Galipienso, L. and Matas, M. (2006) Characterization of potato virus Y isolates from tomato crops in northeast Spain. Eur. J. Plant Path. 115, 247–258. [Google Scholar]
- Armstrong, M.R. , Vossen, J. , Lim, T.Y. , Hutten, R.B.C. , Xu, J. et al. (2019) Tracking disease resistance deployment in potato breeding by enrichment sequencing. Plant Biotechnol. J., 17, 540–549. 10.1101/360644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler, Š. , Witek, K. , Petek, M. et al. (2014) Salicylic acid is an indispensable component of the Ny‐1 resistance‐gene‐mediated response against Potato virus Y infection in potato. J. Exp. Bot. 65, 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurès, I. , Candresse, T. , Leveau, A. , Bendahmane, A. and Sturbois, B. (2008) The Rx gene confers resistance to a range of potexviruses in transgenic Nicotiana plants. Mol Plant‐Microbe Interact. 21, 1154–1164. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari, D.D. , Lapin, D. , Kracher, B. , von Born, P. , Bautor, J. , Niefind, K. and Parker, J.E. (2019) An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Comm. 10, 772. 10.1038/s41467-019-08783-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommonschenkel, S.H. , Frary, A. and Tanksley, S.D. (2000) The broad–spectrum tospovirus resistance gene Sw–5 of tomato is a homolog of the root–knot nematode resistance gene Mi . Mol Plant–Microbe Interact 13, 1130–1138. [DOI] [PubMed] [Google Scholar]
- Brunt, A.A. (2001). Potyviruses. In: Virus and Virus‐Like Diseases of Potato and Production of Seed‐Potatoes ( Loebenstein, G. , Berger, P.H. , Brunt, A.A. and Lawson, R.H. eds), pp. 77–84. Drodrecht/Boston/London: Kluwier Academic Publishers. [Google Scholar]
- Candresse, T. , Marais, A. , Faure, C. , Dubrana, M.P. , Gombert, J. and Bendahmane, A. (2010) Multiple coat protein mutations abolish recognition of Pepino mosaic potexvirus (PepMV) by the potato Rx resistance gene in transgenic tomatoes. Mol. Plant‐Microbe Interact. 23, 376–383. [DOI] [PubMed] [Google Scholar]
- Castel, B. , Ngou, P. , Cevik, V. , Redkar, A. , Kim, D. , Yang, Y. , Ding, P. et al. (2019) Diverse NLR immune receptors activate defence via the RPW8‐NLR NRG1. New Phytol. 222, 966–980. [DOI] [PubMed] [Google Scholar]
- Cockerham, G. (1970) Genetical studies on resistance to potato viruses X and Y. Heredity 25, 309–348. [Google Scholar]
- Cooley, M.B. , Pathirana, S. , Wu, H.J. , Kachroo, P. and Klessig, D.F. (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bokx, J.A. and , der van Want, J.P.H. (1987). Viruses of Potatoes and Seed Production, 259 pp. Wageningen, The Netherlands: PUDOC. [Google Scholar]
- Dong, O.X. , Tong, M. , Bonardi, V. , El Kasmi, F. , Woloshen, V. et al. (2016) TNL‐mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol. 210, 960–973. [DOI] [PubMed] [Google Scholar]
- Flis, B. , Hennig, J. , Strzelczyk‐Żyta, D. , Gebhardt, C. and Marczewski, W. (2005) The Ry‐f sto gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122718 in PVY resistant potato cultivars. Mol. Breed. 15, 95–101. [Google Scholar]
- Friedrich, L. , Vernooij, B. , Gaffney, T. , Morse, A. and Ryals, J. (1995) Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 5, 959–968. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C. and Valkonen, J.P. (2001) Organization of genes controlling disease resistance in the potato genome. Annu. Rev. Phytopathol. 39, 79–102. [DOI] [PubMed] [Google Scholar]
- Hoser, R. , Żurczak, M. , Lichocka, M. , Zuzga, S. , Dadlez, M. , Samuel, M.A. , Ellis, B.E. et al. (2013) Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 200, 158–171. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F. , Bhakta, A.V. , Truesdell, G.M. , Pudlo, W.M. and Williamson, V.M. (2000) Evidence for a role of the N terminus and leucine‐rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell. 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska, B. , Ammiraju, J.S. , Bhattarai, K.K. , Mantelin, S. , Martinez de Ilarduya, O. et al. (2007) The Mi‐9 gene from Solanum arcanum conferring heat‐stable resistance to root‐knot nematodes is a homolog of Mi‐1 . Plant Physiol. 143, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab, G. , Droz, E. , Brigneti, G. , Baulcombe, D. and Malnoe, P. (1997) Infectious in vivo and in vitro transcripts from a full‐length cDNA clone of PVY‐N605, a Swiss necrotic isolate of Potato virus Y. J. Gen. Vir. 78, 3141–3145. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. , Vance, R.E. and Dangl, J.L. (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Jupe, F. , Witek, K. , Verweij, W. , Sliwka, J. , Pritchard, L. et al. (2013) Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB‐LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. and Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Kang, W. , Huy, H.N. , Yeom, S. , An, J. et al. (2017) Divergent evolution of multiple virus‐resistance genes from a progenitor in Capsicum spp. New Phytol. 213, 886–899. [DOI] [PubMed] [Google Scholar]
- Kopp, A. and Bánfalvi, Z. (2015) Molecular mechanisms of resistance to potato virus X and Y in potato. Acta Phytopathol. Entomol. Hung. 50(2), 151–160. [Google Scholar]
- Križnik, M. , Petek, M. , Dobnik, D. et al. (2017) Salicylic acid perturbs sRNA‐gibberellin regulatory network in immune response of potato to potato virus Y infection. Front Plant Sci. 8, 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Shi, L. , Han, C. , Yu, J. , Li, D. and Zhang, Y. (2012) Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS ONE, 7, e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac, A. , Krzymowska, M. , Barabasz, A. and Hennig, J. . (2004) Transcriptional regulation of the gluB promoter during plant response to infection. Cell. Mol. Biol. Lett. 9, 843–853. [PubMed] [Google Scholar]
- Maclean, D. and Kamoun, S. (2012) Big data in small places. Nat. Biotechnol. 30, 33–34. [DOI] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. and Baulcombe, D.C. (2000) An Ry‐mediated resistance response in potato requires the intact active site of the NIa proteinase from Potato virus Y. Plant J. 23, 653–661. [DOI] [PubMed] [Google Scholar]
- Michel, V. , Julio, E. , Candresse, T. , Cotucheau, J. , Decorps, C. , Volpatti, R. , Moury, B. et al. (2018) NtTPN1: a RPP8‐like R gene required for Potato virus Y‐induced veinal necrosis in tobacco. Plant J. 95, 700–714. [DOI] [PubMed] [Google Scholar]
- Moreira, A. , Jones, R.A.C. and Fribourg, C.E. (1980) Properties of a resistance breaking strain of potato virus X. Ann. App. Biol. 95, 93–103. [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Niwa, Y. , Toyooka, K. et al. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nie, X. , Lalany, F. , Dickison, V. , Wilson, D. , Singh, M. , Koyer, D. and Murphy, A. (2016) Detection of molecular markers linked to Ry genes in potato germplasm for marker‐assisted selection for extreme resistance to PVY in AAFC's potato breeding program. Canadian J. Plant Sci. 96, 737–742. [Google Scholar]
- Peart, J.R. , Mestre, P. , Lu, R. , Malcuit, I. and Baulcombe, D.C. (2005) NRG1, a CC‐NB‐LRR protein, together with N, a TIR‐NB‐LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15, 968–973. [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (95 authors) (2011) Genome sequence and analysis of the tuber crop potato. Nature. 475, 189–195. [DOI] [PubMed] [Google Scholar]
- Qi, T. , Seong, K. , Thomazella, D.P.T. , Kim, J.R. , Pham, J. , Seo, E. , Cho, M.J. et al. (2018) NRG1 functions downstream of EDS1 to regulate TIR‐NLR‐mediated plant immunity. PNAS, 115, E10979–E10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querci, M. , Baulcombe, D.C. , Goldbach, R.W. and Salazar, L.F. (1995) Analysis of the resistance‐breaking determinants of Potato virus X (PVX) strain Hb on different potato genotypes expressing extreme resistance to PVX. Phytopathology 85, 1003–1010. [Google Scholar]
- Rairdan, G. and Moffett, P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell. 18, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallapalli, G. (2014) EXPRSS: an Illumina based high‐throughput expression‐profiling method to reveal transcriptional dynamics. BMC Genom. 15, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz, S. , Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N. et al. (2011) Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Ross, H. (1958) Resistenzzüchtung gegen die Mosaic- und andere Viren der Kartoffel. In Handbuch der Pflanzenzüchtung, Vol. III ( Kappert, H. and Rudorf, W. , eds), 2nd edn, pp. 106–125. Berlin, Germany: Paul Parey. [Google Scholar]
- Samuel, G. (1931) Some experiments on inoculating methods with plant viruses, and on local lesions. Ann. Appl. Biol. 18, 494–507. [Google Scholar]
- Scholthof, K.B. , Adkins, S. , Czosnek, H. et al. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, J. , Fomitcheva, V. and Sztangret‐Wiśniewska, J. (2007) Differentiation of Potato virus Y strains using improved sets of diagnostic PCR‐primers. J Viroll Meth. 140, 66–74. [DOI] [PubMed] [Google Scholar]
- Schultink, A. , Tiancong, Q. , Arille, L. , Steinbrenner, A. and Staskawicz, B. (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 92, 787–795. [DOI] [PubMed] [Google Scholar]
- Song, Y.S. , Hepting, L. , Schweizer, G. , Hartl, L. , Wenzel, G. et al. (2005) Mapping of extreme resistance to PVY (Ry sto ) on chromosome XII using anther‐culture‐derived primary dihaploid potato lines. Theor. Appl. Genet. 111, 879–887. [DOI] [PubMed] [Google Scholar]
- Steuernagel, B. , Periyannan, S.K. , Hernández‐Pinzón, I. , Witek, K. , Rouse, M.N. , Yu, G. , Hatta, A. et al. (2006) Rapid cloning of disease‐resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652. [DOI] [PubMed] [Google Scholar]
- Świeżyński, K.M. , Haynes, K.G. , Hutten, R.C.B. , Sieczka, M.T. , Watts, P. and Zimnoch-Guzowska, E. . (1997) Pedigree of European and North-American potato varieties. Plant Breed Seed Sci. 41(1 supplement), 3–149. [Google Scholar]
- Szajko, K. , Chrzanowska, M. , Witek, K. , Strzelczyk‐Zyta, D. , Zagorska, H. et al. (2008) The novel gene Ny‐1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor. Appl. Genet. 116, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko, K. , Strzelczyk‐Zyta, D. and Marczewski, W. (2014) Ny‐1 and Ny‐2 genes conferring hypersensitive response to potato virus Y (PVY) in cultivated potatoes: mapping and marker‐assisted selection validation for PVY resistance in potato breeding. Mol. Breed. 34, 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko, K. and Strzelczyk‐Żyta, D. and Marczewski, W. (2018) Comparison of leaf proteomes of potato (Solanum tuberosum L.) genotypes with ER‐ and HR‐mediated resistance to PVY infection. Eur. J. Plant Pathol. 150, 375–385. [Google Scholar]
- Tang, X. , Zhang, N. , Si, H. and Calderón‐Urrea, A. (2017) Selection and validation of reference genes for RT‐qPCR analysis in potato under abiotic stress. Plant Meth. 13, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkonen, J.P.T. (2007) Potato viruses: economic losses and biotechnological potential. In Potato Biology and Biotechnology ( Vreugdenhil, D. , Bradshaw, J. , Gebhardt, C. and Glovers, F. et al. eds), pp. 619–641. The Netherlands: Elsvier. [Google Scholar]
- Valkonen, J.P.T. (2015) Elucidation of virus‐host interactions to enhance resistance breeding for control of virus diseases in potato. Breed. Sci. 65, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkonen, J.P.T. , Gebhardt, C.H. , Zimnoch‐Guzowska, E. and Watanabe, K.N. (2017). Resistance to potato virus Y in potato. Chapter in: Potato virus Y: biodiversity, pathogenicity, epidemiology and management.
- Van Eck, H.J. , Vos, P.G. , Valkonen, J.P.T. , Uitdewilligen, J. , Lensing, H. et al. (2017) Graphical genotyping as a method to map Ny (o, n)sto and Gpa5 using a reference panel of tetraploid potato cultivars. Theor. Appl. Genet. 130, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, S. , Cabrera, H. , Andersson, R.A. , Fredriksson, A. and Valkonen, J.P.T. (2002) Potato gene Y‐1 is an N gene homolog that confers cell death upon infection with Potato Virus Y. Am. Phytopathol. Soc. 15, 717–727. [DOI] [PubMed] [Google Scholar]
- Waterhouse, A.M. , Procter, J.B. , Martin, D.M. , Clamp, M. and Barton, G.J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Witek, K. , Strzelczyk‐Żyta, D. , Hennig, J. and Marczewski, W. (2006) A multiplex PCR approach to simultaneously genotype potato towards the resistance alleles Ry‐fsto and Ns. Mol. Breeding 18, 273–275. [Google Scholar]
- Witek, K. , Jupe, F. , Witek, A. , Baker, D. , Clark, M.D. et al. (2016) Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nat. Biotech. 34, 656–660. [DOI] [PubMed] [Google Scholar]
- Wu, C.‐H. , Derevnina, L. and Kamoun, S. (2018) Receptor networks underpin plant immunity. Science 360, 1300. [DOI] [PubMed] [Google Scholar]
- Yang, S. and Hua, J. (2004) A haplotype‐specific Resistance gene regulated by BONZAI1 mediates temperature‐dependent growth control in Arabidopsis . Plant Cell 16, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Grosic, S. , Whitham, S.A. and Hill, J.H. (2012) The requirement of multiple defense genes in soybean Rsv1–mediated extreme resistance to Soybean mosaic virus . Mol. Plant Microbe Interact. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Zhu, Z. , Xu, F. , Zhang, Y. , Cheng, Y.T. , Wiermer, M. , Li, X. and Zhang, Y. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl Acad. Sci. USA 107, 13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analysis of Ry sto candidate genes and other functional Solanaceae NLRs.
Figure S2 Grafted transgenic Ry sto plants display resistance to PVY.
Figure S3 HR of transgenic N. tabacum expressing Ry sto .
Figure S4 Stable transgenic tobacco plants carrying Ry sto under the control of a 35S promoter display resistance to PVY. Seven‐week‐old N. tabacum 35S:Ry sto transgenic and non‐transformed plants were inoculated with PVYNTN or mock treated with water.
Figure S5 Stable transgenic tobacco plants carrying Ry sto under the control of native regulatory elements display resistance to PVY. Seven‐week‐old N. tabacum Ry sto (native) transgenic and non‐transformed plants were inoculated with PVYNTN.
Figure S6 Ry sto ‐mediated resistance to PVA infection.
Figure S7 Comparison of Ry sto and Ry‐f sto nucleotide sequences.
Figure S8 Pedigree chart of cultivar Alicja.
Figure S9 Semi‐quantitative RT‐PCR of Ry sto , NRG1 and EDS1 genes.
Table S1 PVY dependent HR in N. benthamiana plants expressing contig 630.
Table S2 ELISA titers of PVY in transgenic 35S::Rysto S. tuberosum cv. Maris Piper plants.
Table S3 ELISA titers of PVY in transgenic 35S::Rysto S. tuberosum cv. Russet Burbank plants.
Table S4 ELISA titers of PVY in grafted transgenic 35S::Rysto S. tuberosum cv. Maris Piper plants.
Table S5 Primers used to clone candidate Rysto genes.
Table S6 Primer sequences for quantitative RT and qRT‐PCR


