Abstract
Despite tremendous efforts to fight cancer, it remains a major public health problem and a leading cause of death worldwide. With increased knowledge of cancer pathways and improved technological platforms, precision therapeutics that specifically target aberrant cancer pathways have improved patient outcomes. Nevertheless, a primary cause of unsuccessful cancer therapy remains cancer drug resistance. In this review, we summarize the broad classes of resistance to cancer therapy, particularly pharmacokinetics, the tumor microenvironment, and drug resistance mechanisms. Furthermore, we describe how bacterial-mediated cancer therapy, a bygone mode of treatment, has been revitalized by synthetic biology and is uniquely suited to address the primary resistance mechanisms that confound traditional therapies. Through genetic engineering, we discuss how bacteria can be potent anticancer agents given their tumor targeting potential, anti-tumor activity, safety, and coordinated delivery of anti-cancer drugs.
Keywords: Cancer therapy, Synthetic biology, Drug resistance, Bacterial-mediated therapy
In 1971, the US government passed the National Cancer Act, which provided more funding and support for the nation's effort in what colloquially became known as the “war on cancer.” Despite the tremendous progress made in screening, detection, and treatment, the number of cancer deaths in the United States has nearly doubled from 335,000 in 1971 to 600,920 in 2017.1 Chemotherapy is a widely used treatment for cancers that have spread from the primary tumor location. However, chemotherapeutic drug resistance is a major impediment to patient survival and is the primary cause of patient death in most advanced stage cancers.2 In fact, unsuccessful chemotherapeutic treatment is often a result of multifactorial issues dependent on pharmacokinetics, the tumor microenvironment (TME), and drug resistance.3,4 As a targeted alternative to systemic chemotherapy, bacterial-mediated therapy could deliver tumor clearance in diverse and metastatic cancers. While it has been known for at least 200 years that infections with microbes could result in cancer remission, this remained a dormant field until recently, where advancements in synthetic biology now enable controlled targeting and delivery of therapeutic agents. In this review, we discuss the challenges of traditional chemotherapeutic treatment and advancements in bacterial-mediated therapy that overcome these obstacles.
Pharmacokinetic failure
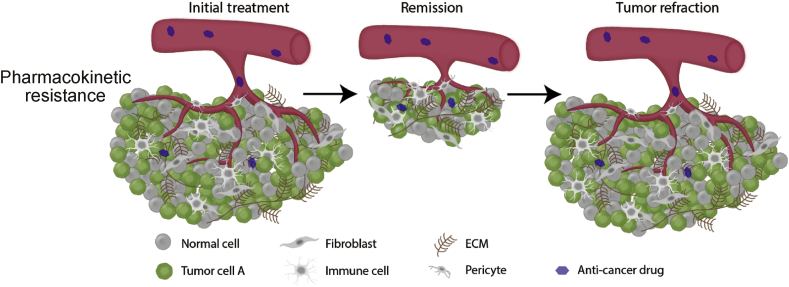
Therapeutic dose levels that can successfully treat a tumor site require a satisfactory ADME (Absorption, Distribution, Metabolism, Excretion) profile. Pharmacokinetic failure that results in insufficient dosing at the tumor site can lead to incomplete antitumor therapy resulting in residual disease (Fig. 1).5 Major sources of pharmacokinetic failure can be due to issues with drug solubility, distribution, and dose-limiting toxicity that incompletely suppress the targeted tumor pathway.6 Also, tumor cells themselves can actively reduce the intracellular drug concentration through efflux pumps that transport anticancer drugs, thereby providing another level of resistance.7 While mathematical models have been developed to predict drug delivery, drug concentration, and tumor clearance,8,9 the multifactorial nature of the problem makes successful targeted therapy a challenge.
Fig. 1.
Drug formulations with poor ADME profiles can result in incomplete tumor remission leading to a refractory response.
As such, there has been much effort in developing therapies that are target ligand specific. While these efforts have resulted in powerful advances in therapeutics, both targeted and untargeted therapies have very low levels of accumulation of the injected dose at the target site. Delivery of the injected dose to the targeted cancer site can range from below 0.1% for drugs without a targeting ligand (e.g. small molecule inhibitors) to over 1% of the injected dose for targeted drugs (e.g. antibody drug conjugates).10 However, while the low percentage of the total dose can be discouraging, the critical factor for successful treatment is the ratio between on-site and off-site accumulation. Increases in target site activation without an increase in toxicity from accumulation in normal tissue could thus greatly improve patient outcomes.
Barriers of the tumor microenvironment
While once thought of as a detached spectator to cancer progression, the complex interplay between the tumor and stroma is a fundamental hallmark of cancer and is known to influence tumor progression, metastasis, and importantly, therapeutic resistance.11 The TME consists of a variety of malignant cells, stromal cells, immune cells, and soluble growth factors and cytokines that can secrete into an extracellular matrix (ECM).12 In various cancer types, the TME has been shown to reduce drug penetration, provide proliferative and anti-apoptotic advantages to the tumor cells, and modify the immune response.13
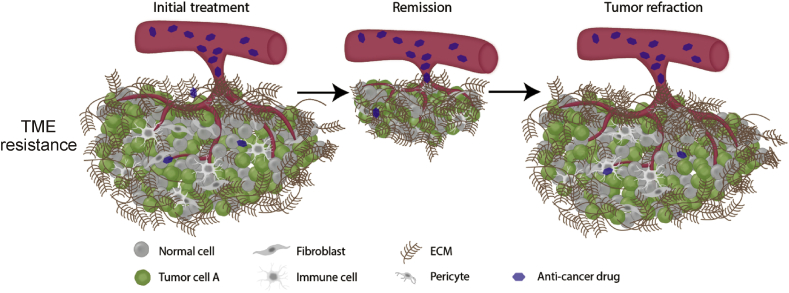
A major cause of drug resistance from the tumor microenvironment is the ECM physical barrier against cytotoxic therapeutics (Fig. 2). The ECM is mainly composed of glycoproteins, proteoglycans, elastin, collagen, and hyaluronan. Increasing amounts of ECM has been shown to have a direct impact on the intratumoral drug concentration, due to reduced penetration and distribution.14 A study on pancreatic ductal carcinoma has shown that excessive stroma can cause increased tumor stiffness and compressed tumor vessels resulting in decreased drug accumulation in the tumor.15 Moreover, increased stiffness in the ECM of hepatic carcinoma has also been shown to promote drug resistance.16 Originating from the ECM, exosomes are another TME component that reduces drug penetration by trapping therapeutic antibodies such as rituximab and trastuzumab, thereby limiting therapeutic efficacy.17,18
Fig. 2.
Tumors with an extensive extracellular matrix can result in incomplete penetration of anticancer drugs leading to incomplete anti-tumor therapy and tumor refraction.
Pharmaceuticals have been developed to specifically degrade the ECM to improve therapeutic delivery. Increased levels of hyaluronan, a linear polysaccharide found in the extracellular space of most tissues, results in increased interstitial pressure and reduced drug penetration.19 Collagen is also overexpressed in many tumors leading to proliferative oncogenic environment through structural and signaling interactions.20 As such, ECM remodeling enzymes such as hyaluronidase and collagenase have been used in conjunction with anticancer drugs to increase drug penetration.21,22 However, the timing and control of ECM degradation must be carefully controlled, as enhanced metastasis of cancer cells due to loss of ECM integrity can be a dangerous side effect.23
Drug resistance
While conventional cytotoxic drugs such as 5-fluorouracil have been widely used as cancer therapeutics, increased knowledge of molecular cancer mechanisms has allowed the development of precision medications (e.g. targeted therapies such as kinase inhibitors). These targeted therapies disrupt the function of oncogenic driver proteins and have revolutionized cancer therapy. A few examples include kinase inhibitors of epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), and BRAF. Unlike conventional cytotoxic drugs that simply target rapidly proliferative cells, these precision therapies specifically target molecular aberrations common in cancerous tissues, with a relatively lesser effect on normal tissue.24 A canonical example is the targeting of the kinase BRAF, a member of the mitogen-activated protein (MAP) kinase signal transduction pathway responsible for growth and cell differentiation by kinase inhibitors such as vemurafenib.25 The basis of these precision therapies could only be realized by the identification of the primary genetic drivers of cancer progression through rigorous mechanistic laboratory studies coupled with biomarker-driven clinical trials.26
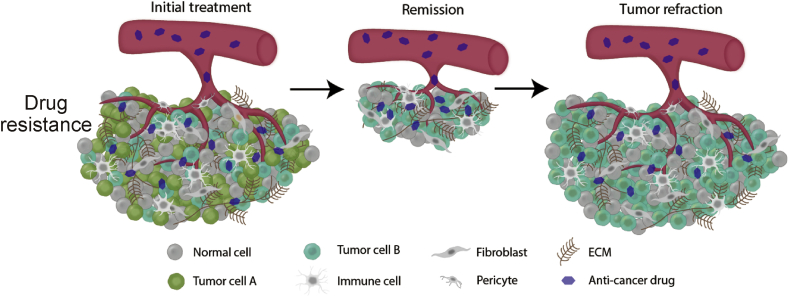
Despite the success of these treatments, many cancers eventually adapt to both conventional and precision pharmaceuticals, and this resistance is a primary cause of patient death in most advanced stage cancers.2 Resistance may be present at the time of initial therapy (intrinsic) or may develop during the course of the therapy (acquired). Drug resistance is multifactorial, and is further complicated by the genetic and epigenetic heterogeneity between and within tumor populations that can result in subpopulations with different drug sensitivities.4 The administration of an anticancer drug to a mixture of drug-sensitive and drug-resistant subpopulations in a tumor can be a significant cause of drug resistance (Fig. 3).27 A convergence-based classification of resistance mechanisms can illuminate polytherapy pathways that target parallel cancer dependencies. By simultaneously targeting these resistance pathways, polytherapies are less likely to result in resistance to a multi-drug mixture. Importantly, in some cases a polytherapy is better tolerated than each individual agent due to off-setting toxicities (e.g. BRAF + MEK inhibitor treatment),28 while increased off-target toxicity in others can result in greater toxicity with polytherapies.29
Fig. 3.
Tumor heterogeneity can result in subpopulations of cells with distinct molecular signatures with varying drug sensitivities. Drug-sensitive cells can be eliminated while a drug-resistant subpopulation can cause tumor refraction.
New therapeutic platforms are needed to address the multifactorial challenges presented by drug delivery, the TME, and tumor heterogeneity. Synthetic biology has enabled the creation of “living therapeutics” that are biologically programmed to perform specific pre-designed therapeutic treatments. With the ability to actively move towards the nutrients at the cancer site via chemotaxis, modulate the TME, and deliver on-site therapies, genetically modified bacteria are a promising and relatively unexplored avenue in cancer therapeutics.
Bacterial-mediated therapy
In the late 19th century, Dr. William Coley began experimenting with treating his cancer patients with Streptococcus pyogenes.30 Now considered the father of cancer immunotherapy, Coley's toxins, as they came to be known, were largely set aside once radiotherapy was developed. In the past few decades, however, there has been renewed interest in both preclinical and clinical studies in using bacteria for cancer therapy.
Bacterial-mediated therapy may be used to treat nearly all cancer sites including blood cancer, sarcoma, melanoma, and solid carcinomas. Oral administration of genetically engineered probiotics is an exciting avenue to treat gastrointestinal cancer31 and enteral administration of heterologous bacteria, either as an isolated probiotic or a fecal transplant, is well-studied and has reduced safety concerns compared to parenteral administration.32 However, the vast majority of clinical and preclinical trials to treat cancer has been through direct tumor injection and intravenous systemic injection of tumor-targeting bacteria, which is the context we discuss in this review.
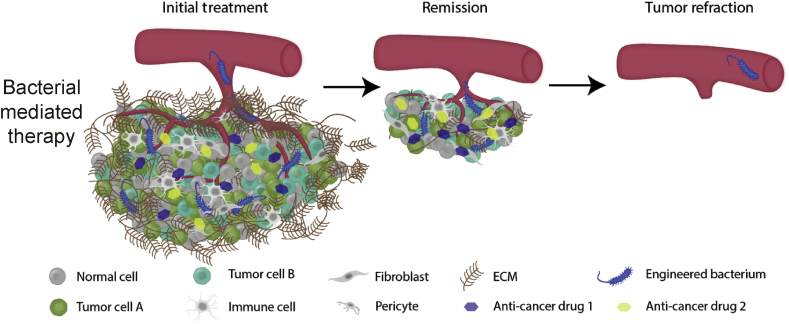
In principle, bacteria may be engineered to selectively target a tumor site, multiply within the tumor microenvironment, recruit the immune system, and release multiple drugs targeting parallel disease pathways resulting in complete elimination of the cancer cells (Fig. 4). With advancements in synthetic biology, bacterial mediated therapy can be programmed to address the shortcomings of conventional treatment in dealing with pharmacokinetics, the tumor microenvironment, and drug resistance. We discuss the progress made in addressing these challenges with bacterial-mediated therapy below as well as highlight safety and prospective directions and opportunities.
Fig. 4.
A genetically engineered bacterium can overcome the multifactorial challenges of drug resistance. The bacteria can actively target the tumor site without producing the anticancer compound, reducing off-site toxicity. After reaching the tumor, the bacteria can burrow past the extensive ECM into the hypoxic tumor core. Once the bacteria has colonized the tumor, genetic switches activated by quorum sensing or the tumor microenvironment produce multiple therapies that target parallel disease pathways resulting in complete tumor elimination.
Targeting and on-site production
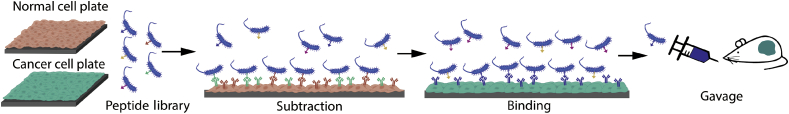
Bacteria have a unique ability to selectively target and colonize tumors compared to normal tissues. Small molecules produced by tumor cells can act as chemo-attractants to bacteria, and the suppressed immune response at the tumor site can prevent bacterial clearance. While an extensive ECM caused by cancer creates a hypoxic environment and reduces conventional therapeutic dose, both obligate anaerobes (e.g. Clostridium and Bifidobacterium) and facultative anaerobes (e.g. Escherichia and Salmonella) have been shown to colonize the necrotic and hypoxic conditions of the tumor.33 Moreover, bacteria have been genetically engineered to express binding peptides to selectively target cancer biomarkers and colonize tumors (Fig. 5).31,34 This selective colonization can be leveraged with other treatment modalities. Conventional chemotherapy and radiotherapy are much more effective in the well-perfused areas compared to the more dense, hypoxic core of a tumor where bacteria colonize. The synergy of these modalities has been shown in murine models by dosing Clostridium novyi-NT with radiotherapy or chemotherapeutic treatments.35,36
Fig. 5.
Workflow of process to identify tumor targeting peptides. A library of known peptides that bind specific cancer receptors is engineered to display on the bacterial cell surface and screened against normal cells and the target cancer cell line.
While precision medication can help reduce toxicity through the targeting of aberrant molecular signatures, its systematic delivery causes toxicity due to accumulation in healthy tissues. By encoding bacteria to target tumor sites and coordinating cellular actions through sensing of the TME, therapeutics can be released on-site, greatly reducing off-site toxicity. Through promoters that are activated by differential pH, nutrient, or oxygen availability, bacteria have been engineered to the TME thereby limiting off-site delivery.37,38 Leveraging the preferential accumulation of bacteria at the tumor site, genetic switches have been developed that respond to bacterial cell-density dependent quorum sensing (QS). As these bacteria accumulate at a site, the communication molecules they produce eventually reach a critical threshold activating the genetic switch and coordinating gene expression. This coupling of QS mechanisms to drug release enables coordinated therapeutic release and acts as a safety valve to prevent off-site accumulation and increase drug delivery.39
Tumor clearance through immune system activation and direct oncolysis
The intrinsic ability of bacterial cells to colonize the TME can result in remodeling of the environment, primarily through the activation of immune pathways. Differential expression of pathogen associated molecular patterns (PAMPs) such as flagella, pili, and lipopolysaccharide by bacteria elicit the immune system in a manner unique to each bacterial strain. This response includes repolarization of tumor associated macrophages, elimination of tumor associated myeloid derived suppressor cells, and promotion of dendritic cell maturation.40 A prominent example is the sensitization of cluster of differentiation (CD) 8+ T cells, a major component of the adaptive immune response, to tumor antigens by enhancing T-cell receptor signaling.41 Beyond the natural ability of some bacteria to elicit immune pathways, the immune-suppressive TME can be activated to become immune stimulating through the release of adjuvants, antigens, cytokines and checkpoint inhibitors.33 Salmonella enterica and C. novyi-NT have been engineered to release cytokines or tumor-specific antigens to convert the TME from immune-suppressive to immune-activated.42 Exciting new studies in Escherichia coli have shown that a lysis mechanism based on quorum sensing can be used to release nanobody fragments against receptors programmed death ligand-1 (PD-L1), cytotoxic T lymphocyte associated antigen-4 (CTLA-4) and CD47, thereby reducing or clearing tumor growth in syngeneic mouse models.43,44
Beyond bacterial recruitment of immune cells, genetically engineered bacteria can directly cause tumor regression by competing for nutrients, uncontrolled growth that causes tumor cells to lyse, or through secretion of exotoxins and pro-apoptotic molecules.45 In syngeneic mice models, the direct release of a clinical therapeutic along with an exotoxin haemolysin E, a pore-forming anti-tumor toxin, by genetically engineered E. coli, resulted in reduced tumor activity in a syngeneic mouse transplantation model with metastatic hepatic carcinoma.46
Systemic cytokines stimulate the immune system and directly cause preferential apoptosis of cancer cells compared to normal healthy cells. However, systemic cytokine injection cannot be used due to off-target toxicity, whereas localized release by bacteria could reduce tumor size without causing widespread toxicity. Such release of pro-apoptotic cytokines by genetically engineered bacteria has already begun to be explored, including the expression of FAS-ligand, tumor necrosis factor alpha (TNF-α), and TNF-related apoptosis-inducing ligand.47, 48, 49
A therapeutic platform that enables more selective on-target drug release of multiple therapies that act in parallel would represent a significant step in cancer therapeutics. Systemic administration of drug cocktails that target multiple disease pathways can have dangerous side effects. By genetically encoding the localized production of anti-tumor compounds, targeted bacteria could, in principle, selectively release multiple oncolytic therapeutics. Moreover, programmable design through genetic circuits could enable controllable timed release of therapeutics for maximum efficiency. While most bacteria have been engineered to release simple peptides, many chemotherapeutics are inspired or directly taken from natural products. Through heterologous natural product synthesis, bacteria could be engineered to release Food and Drug Administration (FDA)-approved chemotherapeutics.
Safety of bacterial-mediated therapy
Numerous preclinical pharmacological and toxicity studies have shown that select bacteria have satisfactory safety profiles in healthy and tumor bearing animals.50,51 The FDA has approved several clinical trials with tumor targeting bacteria.52, 53, 54 These studies showed acceptable safety profiles with promising results for anti-tumor activity. The most noteworthy clinical example is the FDA-approved treatment of bladder cancer with the Bacillus Calmette-Guerin vaccine, an attenuated Mycobacterium bovis.55
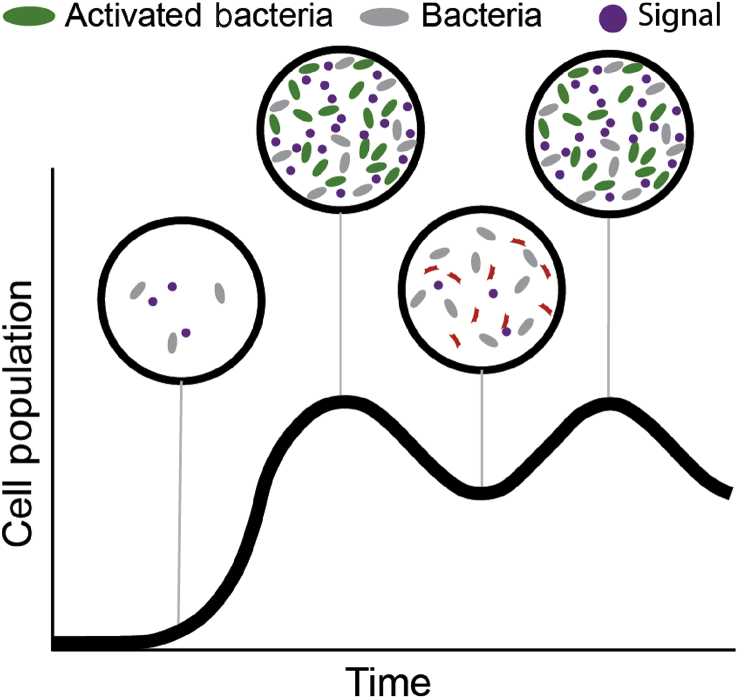
In the studies shown so far, substantial colonization has been required for clinical benefit, which highlights the uniqueness of the dose profile with a live therapeutic. As live bacteria generally multiply in the tumor site, the injected dose of bacteria plays less of a role than the type of tumor and bacterium used. Well-perfused tumors compared to necrotic tumors will have different colonization profiles depending on the type of bacterium employed. Genetic circuits programmed to maintain bacterial density at a certain threshold have been developed.46 If these systems were applied to a tumor site, a density-dependent signal would trigger a kill switch in a subset of the population, thereby maintain a bacterial density range (Fig. 6). With the maturation of synthetic biology enhancements allowing the precise delivery of oncolytic payloads, future Phase I clinical trials may reveal that lower bacterial loads are required for successful anti-tumor activity.
Fig. 6.
Control mechanisms to maintain bacterial population. Genetic circuits have been created to maintain bacterial population in microfluidic devices. Adapted to colonization of cancer sites, initial colonization would contain few cells and signaling molecules (left-most bubble). Cell growth reaches a critical threshold and the signaling molecule causes lysis in a subset of the population, which then undergoes growth again until the threshold is reached again.
Future directions
With its anti-tumor effects and preference to selectively colonize tumors, bacterial-mediated therapy has the potential to treat cancers that are resistant to current therapies, including refractory metastatic cancer and multi-drug resistant cancer. Depending on the payload and tissue target, bacterial-mediated therapy could be effective beyond direct treatment of manifested cancer. By targeting precancerous lesions, microbes could be engineered to prevent tumor occurrence or recurrence. While there is great potential in bacterial-mediated therapy, improved tools and knowledge are required for successful clinical translation.
Synthetic biology advancements have enabled bacteria to perform more coordinated and complex actions, which is a major advantage to their use as a “living therapeutic”. While most synthetic biology studies have centered on well-studied organisms, particularly E. coli, the recent genetic “domestication” of a wide variety of organisms could provide a better-suited chassis for applications such as anticancer therapy. The bacteria studied the most in this context have been E. coli, Salmonella typhimurium, and S. enterica because of their relative ease of genetic manipulation. Future efforts should develop genetic tools in other non-model organisms that naturally elicit the immune system, but were previously considered too difficult to engineer, such as Listeria monocytogenes and C. novyi.
An important therapeutic consideration is the level and timing of therapeutic dose. As tumor heterogeneity can contain a drug-insensitive population that can flourish if the drug-sensitive population is eliminated, intermittent dose programs to contain the tumor at a certain size could be the preferred course.56 Further, drug discontinuation can also result in the re-sensitization of tumor cells to the drug. Genetic circuits have been built in E. coli to oscillate cell population with varying growth dynamics within the TME.46 These forms of circuits could be used to not only maintain the bacterial population, but also oscillate therapeutic release without subsequent injected doses (Fig. 6).
Advancements in cancer treatment require an increased understanding of cancer pathogenesis, particularly as the cancer evolves. In fact, under-sampling of cancers has been noted as a critical knowledge gap in tumor progression.2 To help provide some of this information, tools such as liquid biopsies have been developed that monitor genetic, transcriptional and epigenetic changes by isolating circulating tumor cells from the blood.57 A promising method to further understand tumor development could be through colonized bacteria. For example, bacteria have been engineered to sense and record changes in their environment through targeted alterations in their own DNA.58,59 Applying these bacteria that record changes in tumor cells or tumor microenvironments in real time could provide vital information about cancer progression and regression.
Conflicts of interest
None.
Funding
This work was supported by a grant from National Institutes of Health Awards (F32GM125179).
Edited by Pei-Fang Wei and Yi Cui
Footnotes
Peer review under responsibility of Chinese Medical Association.
Contributor Information
Amin Zargar, Email: azargar@berkeley.edu.
Trever G. Bivona, Email: trever.bivona@ucsf.edu.
References
- 1.Lou E. The Art of War and oncology: applying the principles of strategy and tactics to greater effect in the era of targeted therapy. Ann Transl Med. 2018;6:168. doi: 10.21037/atm.2018.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konieczkowski D.J., Johannessen C.M., Garraway L.A. A convergence-based framework for cancer drug resistance. Cancer Cell. 2018;33:801–815. doi: 10.1016/j.ccell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee N., Bivona T.G. Polytherapy and targeted cancer drug resistance. Trends Cancer. 2019;5:170–182. doi: 10.1016/j.trecan.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabnis A.J., Bivona T.G. Principles of resistance to targeted cancer therapy: lessons from basic and translational cancer biology. Trends Mol Med. 2019;25:185–197. doi: 10.1016/j.molmed.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfarouk K.O., Stock C.M., Taylor S. Resistance to cancer chemotherapy: failure in drug response from ADME to P-gp. Cancer Cell Int. 2015;15:71. doi: 10.1186/s12935-015-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Undevia S.D., Gomez-Abuin G., Ratain M.J. Pharmacokinetic variability of anticancer agents. Nat Rev Cancer. 2005;5:447–458. doi: 10.1038/nrc1629. [DOI] [PubMed] [Google Scholar]
- 7.Nobili S., Landini I., Mazzei T., Mini E. Overcoming tumor multidrug resistance using drugs able to evade P-glycoprotein or to exploit its expression. Med Res Rev. 2012;32:1220–1262. doi: 10.1002/med.20239. [DOI] [PubMed] [Google Scholar]
- 8.Thurber G.M., Weissleder R. A systems approach for tumor pharmacokinetics. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizk M.L., Zou L., Savic R.M., Dooley K.E. Importance of drug pharmacokinetics at the site of action. Clin Transl Sci. 2017;10:133–142. doi: 10.1111/cts.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lammers T., Kiessling F., Ashford M., Hennink W., Crommelin D., Storm G. Cancer nanomedicine: is targeting our target? Nat Rev Mater. 2016;1:16069. doi: 10.1038/natrevmats.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouad Y.A., Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7:1016–1036. [PMC free article] [PubMed] [Google Scholar]
- 12.Cui D., Huang Z., Liu Y., Ouyang G. The multifaceted role of periostin in priming the tumor microenvironments for tumor progression. Cell Mol Life Sci. 2017;74:4287–4291. doi: 10.1007/s00018-017-2646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y. Tumor microenvironment and cancer therapy resistance. Cancer Lett. 2016;380:205–215. doi: 10.1016/j.canlet.2015.07.044. [DOI] [PubMed] [Google Scholar]
- 14.Jain R.K. Vascular and interstitial barriers to delivery of therapeutic agents in tumors. Cancer Metastasis Rev. 1990;9:253–266. doi: 10.1007/BF00046364. [DOI] [PubMed] [Google Scholar]
- 15.Neesse A., Bauer C.A., Öhlund D. Stromal biology and therapy in pancreatic cancer: ready for clinical translation? Gut. 2019;68:159–171. doi: 10.1136/gutjnl-2018-316451. [DOI] [PubMed] [Google Scholar]
- 16.Schrader J., Gordon-Walker T.T., Aucott R.L. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53:1192–1205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aung T., Chapuy B., Vogel D. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci U S A. 2011;108:15336–15341. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciravolo V., Huber V., Ghedini G.C. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227:658–667. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 19.Whatcott C.J., Han H., Posner R.G., Hostetter G., Von Hoff D.D. Targeting the tumor microenvironment in cancer: why hyaluronidase deserves a second look. Cancer Discov. 2011;1:291–296. doi: 10.1158/2159-8290.CD-11-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khawar I.A., Kim J.H., Kuh H.J. Improving drug delivery to solid tumors: priming the tumor microenvironment. J Control Release. 2015;201:78–89. doi: 10.1016/j.jconrel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 21.St Croix B., Man S., Kerbel R.S. Reversal of intrinsic and acquired forms of drug resistance by hyaluronidase treatment of solid tumors. Cancer Lett. 1998;131:35–44. doi: 10.1016/s0304-3835(98)00199-2. [DOI] [PubMed] [Google Scholar]
- 22.Eikenes L., Tari M., Tufto I., Bruland O.S., de Lange Davies C. Hyaluronidase induces a transcapillary pressure gradient and improves the distribution and uptake of liposomal doxorubicin (Caelyx) in human osteosarcoma xenografts. Br J Cancer. 2005;93:81–88. doi: 10.1038/sj.bjc.6602626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovar J.L., Johnson M.A., Volcheck W.M., Chen J., Simpson M.A. Hyaluronidase expression induces prostate tumor metastasis in an orthotopic mouse model. Am J Pathol. 2006;169:1415–1426. doi: 10.2353/ajpath.2006.060324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garraway L.A., Jänne P.A. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 25.Zaman A., Bivona T.G. Emerging application of genomics-guided therapeutics in personalized lung cancer treatment. Ann Transl Med. 2018;6:160. doi: 10.21037/atm.2018.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bivona T.G., Doebele R.C. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med. 2016;22:472–478. doi: 10.1038/nm.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D., Wang D.C., Cheng Y. Roles of tumor heterogeneity in the development of drug resistance: a call for precision therapy. Semin Cancer Biol. 2017;42:13–19. doi: 10.1016/j.semcancer.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Mondaca S., Lacouture M., Hersch J., Yaeger R. Balancing RAF, MEK, and EGFR inhibitor doses to achieve clinical responses and modulate toxicity in BRAF V600E colorectal cancer. JCO Precis Oncol. 2018;2018 doi: 10.1200/PO.18.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S.R., Davis M., Doroshow J.H., Kummar S. Safety and feasibility of targeted agent combinations in solid tumours. Nat Rev Clin Oncol. 2013;10:154–168. doi: 10.1038/nrclinonc.2012.245. [DOI] [PubMed] [Google Scholar]
- 30.Richardson M.A., Ramirez T., Russell N.C., Moye L.A. Coley toxins immunotherapy: a retrospective review. Altern Ther Health Med. 1999;5:42–47. [PubMed] [Google Scholar]
- 31.Ho C.L., Tan H.Q., Chua K.J. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat Biomed Eng. 2018;2:27–37. doi: 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- 32.Bermúdez-Humarán L.G., Langella P. Live bacterial biotherapeutics in the clinic. Nat Biotechnol. 2018;36:816–818. doi: 10.1038/nbt.4248. [DOI] [PubMed] [Google Scholar]
- 33.Forbes N.S., Coffin R.S., Deng L. White paper on microbial anti-cancer therapy and prevention. J Immunother Cancer. 2018;6:78. doi: 10.1186/s40425-018-0381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasinskas R.W., Forbes N.S. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007;67:3201–3209. doi: 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- 35.Bettegowda C., Dang L.H., Abrams R. Overcoming the hypoxic barrier to radiation therapy with anaerobic bacteria. Proc Natl Acad Sci U S A. 2003;100:15083–15088. doi: 10.1073/pnas.2036598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang L.H., Bettegowda C., Agrawal N. Targeting vascular and avascular compartments of tumors with C. novyi-NT and anti-microtubule agents. Cancer Biol Ther. 2004;3:326–337. doi: 10.4161/cbt.3.3.704. [DOI] [PubMed] [Google Scholar]
- 37.Arrach N., Zhao M., Porwollik S., Hoffman R.M., McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68:4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 38.Mengesha A., Dubois L., Lambin P. Development of a flexible and potent hypoxia-inducible promoter for tumor-targeted gene expression in attenuated Salmonella. Cancer Biol Ther. 2006;5:1120–1128. doi: 10.4161/cbt.5.9.2951. [DOI] [PubMed] [Google Scholar]
- 39.Swofford C.A., Van Dessel N., Forbes N.S. Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc Natl Acad Sci U S A. 2015;112:3457–3462. doi: 10.1073/pnas.1414558112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mogensen T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [Table of Contents] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richer M.J., Nolz J.C., Harty J.T. Pathogen-specific inflammatory milieux tune the antigen sensitivity of CD8(+) T cells by enhancing T cell receptor signaling. Immunity. 2013;38:140–152. doi: 10.1016/j.immuni.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorenson B.S., Banton K.L., Frykman N.L., Leonard A.S., Saltzman D.A. Attenuated Salmonella typhimurium with interleukin 2 gene prevents the establishment of pulmonary metastases in a model of osteosarcoma. J Pediatr Surg. 2008;43:1153–1158. doi: 10.1016/j.jpedsurg.2008.02.048. [DOI] [PubMed] [Google Scholar]
- 43.Gurbatri C., Coker C., Hinchliffe T.E. biorxiv; 2019. Engineered Probiotics for Local Tumor Delivery of Checkpoint Blockade Nanobodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury S., Castro S., Coker C., Hinchliffe T.E., Arpaia N., Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. 2019;25:1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchugonova A., Zhang Y., Salz R. Imaging the different mechanisms of prostate cancer cell-killing by tumor-targeting Salmonella typhimurium A1-R. Anticancer Res. 2015;35:5225–5229. [PubMed] [Google Scholar]
- 46.Din M.O., Danino T., Prindle A. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganai S., Arenas R.B., Forbes N.S. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer. 2009;101:1683–1691. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theys J., Nuyts S., Landuyt W. Stable Escherichia coli-Clostridium acetobutylicum shuttle vector for secretion of murine tumor necrosis factor alpha. Appl Environ Microbiol. 1999;65:4295–4300. doi: 10.1128/aem.65.10.4295-4300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loeffler M., Le'Negrate G., Krajewska M., Reed J.C. Inhibition of tumor growth using salmonella expressing Fas ligand. J Natl Cancer Inst. 2008;100:1113–1116. doi: 10.1093/jnci/djn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clairmont C., Lee K.C., Pike J. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 51.Diaz L.A., Jr., Cheong I., Foss C.A. Pharmacologic and toxicologic evaluation of C. novyi-NT spores. Toxicol Sci. 2005;88:562–575. doi: 10.1093/toxsci/kfi316. [DOI] [PubMed] [Google Scholar]
- 52.Roberts N.J., Zhang L., Janku F. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci Transl Med. 2014;6:249ra111. doi: 10.1126/scitranslmed.3008982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham C., Nemunaitis J. A phase I trial of genetically modified Salmonella typhimurium expressing cytosine deaminase (TAPET-CD, VNP20029) administered by intratumoral injection in combination with 5-fluorocytosine for patients with advanced or metastatic cancer. Protocol no: CL-017. Version: April 9, 2001. Hum Gene Ther. 2001;12:1594–1596. [PubMed] [Google Scholar]
- 54.Toso J.F., Gill V.J., Hwu P. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koyama A. Re: History of bacillus Calmette-Guerin and bladder cancer: an immunotherapy success story: H. W. Herr and A. Morales. J Urol 2008; 179: 53-56. J Urol. 2008;180:2255. doi: 10.1016/j.juro.2007.08.122. author reply 2255. [DOI] [PubMed] [Google Scholar]
- 56.Salgia R., Kulkarni P. The genetic/non-genetic Duality of drug 'resistance' in cancer. Trends Cancer. 2018;4:110–118. doi: 10.1016/j.trecan.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murtaza M., Dawson S.J., Tsui D.W. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 58.Farzadfard F., Lu T.K. Emerging applications for DNA writers and molecular recorders. Science. 2018;361:870–875. doi: 10.1126/science.aat9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang W., Liu D.R. Rewritable multi-event analog recording in bacterial and mammalian cells. Science. 2018:360. doi: 10.1126/science.aap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]