Abstract
The preparation of hydrogels and stable emulsions is important in the formulation of many functional nanostructured soft materials. We investigate the multifunctional self-assembly and bioactivity properties of a novel surfactant-like peptide (SLP) that shows antimicrobial activity, is able to form hydrogels without pH adjustment, and is able to stabilize oil-in-water emulsions. Furthermore, we demonstrate on-demand de-emulsification in response to the protease enzyme elastase. We show that SLP (Ala)9-Arg (A9R) forms β-sheet fibers above a critical aggregation concentration and that water-in-oil emulsions are stabilized by a coating of β-sheet fibers around the emulsion droplets. Furthermore, we demonstrate enzyme-responsive de-emulsification, which has potential in the development of responsive release systems. The peptide shows selective antimicrobial activity against Gram-negative pathogens including Pseudomonas aeruginosa, which causes serious infections. Our results highlight the utility of SLPs in the stabilization of oil/water emulsions and the potential for these to be used to formulate antimicrobial peptide emulsions which are additionally responsive to protease. The peptide A9R has pronounced antibacterial activity against clinically challenging pathogens, and its ability to form β-sheet fibers plays a key role in its diverse structural properties, ranging from hydrogel formation to emulsion stabilization.
Keywords: peptides, emulsions, enzyme-reponsive biomaterials, hydrogels, fibers
Introduction
The global healthcare challenge of emerging antimicrobial resistance is stimulating intense research activity into the development of new antimicrobial agents. Peptides (and biomaterials incorporating peptide motifs) are attracting considerable attention in several respects. First, powerful antimicrobial peptides have already been evolved in nature, and if these show antimicrobial activity whilst having minimal toxicity to human cells, such peptides can be used directly as antimicrobial agents. Alternatively, they may be used as a basis to design and screen related compounds. Smaller de novo-designed peptides also show promise and are simple to synthesize in high purity. These peptides typically contain cationic residues such as arginine or lysine or the aromatic residue tryptophan.1−10 Surfactant-like peptides (SLPs) are a class of peptide with sequences of uncharged residues capped with charged residues; examples include peptides with alanine repeats capped with charged residues such as lysine, arginine, aspartic acid, or glutamic acid.11−14 The amphiphilic properties of SLPs lead to self-assembly into distinct nanostructures in aqueous solution depending on the SLP sequence and the solution conditions.2,11−29 SLPs are a potentially valuable class of peptide antibacterial agents because the self-assembled structure can lead to high-density presentation of the active motif (i.e., the sequence of charged residues in the case of antimicrobial SLPs).
Our group has been studying the self-assembly of SLPs and has recently begun to investigate their potentially valuable bioactivities. SLP NH2-A6R-OH (A6R) self-assembles into 3 nm thick nanotapes at low concentration but at high concentration, these wrap into helical ribbon and nanotube structures.23 The interaction of this peptide with model zwitterionic lipid membranes (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) was examined. The interaction leads to loss of peptide β-sheet conformation and restructuring of the lipid membrane (from multilamellar to unilamellar) but not membrane lysis.30 In the same paper, a preliminary study of antimicrobial activity showed activity against Gram-positive Staphylococcus aureus.30 More recently, the self-assembly and antimicrobial properties of A6R with free termini and capped termini has been investigated.31 The capped variant, CH3CONH-A6R-NH2, forms nanofibrils, whereas the uncapped peptide NH2-A6R-OH assembles into nanotapes. From measurements of lamellar spacings, small-angle X-ray scattering (SAXS) showed the selective interaction of the capped peptide with the anionic lipid POPG [2-oleoyl-1-palmitoyl-sn-glycero-3-phospho-rac-(1-glycerol)] in mixed POPG/POPE [POPE: 2-oleoyl-1-palmitoyl-sn-glycero-3-phosphoethanolamine] anionic/zwitterionic vesicles. The capped peptide also showed selective activity against Gram-positive Listeria monocytogenes, and the uncapped version showed greater antimicrobial activity against S. aureus, L. monocytogenes, and Escherichia coli.31
An alternative design of SLP has charged residues at both termini (“triblock polymer”-like architecture or bola-amphiphile structure). The self-assembly of SLPs including I2K2I2 and KI4K were investigated, and the sequence change was found to profoundly influence aggregation because the former shows no defined nanostructure (and there is no evidence for β-sheet formation), whereas the latter forms nanotubes based on β-sheet hydrogen bonding.25,32 Bolaamphiphile RFL4FR self-assembles into nanosheets and nanotubes, and its compatibility with fibroblast cells was demonstrated.33 This peptide forms a shear-aligned nematic phase of nanotubes.34 SLP RA3R adopts a polyproline II conformation in water.35 The peptide interacts strongly with POPG in mixed POPG/POPE vesicles leading to enhanced correlation of lipid bilayers in liposomes, but in contrast, the interaction of RA3R with multilamellar POPC/DOPC [POPC: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, DOPC: 1,2-dioleoyl-sn-glycero-3-phosphocholine] vesicles leads to the decorrelation of the lipid bilayers. RA3R was found to be particularly active against L. monocytogenes through a proposed mechanism of membrane reorganization.35
Building on our recent work on alanine/arginine SLPs, in the present paper, we first investigate the self-assembly and antimicrobial activity of SLP A9R before investigating whether such an SLP can be used to stabilize an oil/water emulsion. We hypothesized that increasing the length of the alanine sequence would favorably alter the amphiphilicity of the molecule compared to A6R. To the best of our knowledge, emulsion formation by SLPs has not previously been considered, and we also examine the mechanism behind the observed stabilization. Peptide A9R has a longer alanine sequence and is thus more hydrophobic than both A6R and RA3R. However, remarkably even just the single cationic arginine is sufficient to enable solubility in aqueous solution and self-assembly behavior. The conformation of the peptide is probed using circular dichroism (CD) spectroscopy. The presence of a critical aggregation concentration (cac) is assayed through fluorescence probe measurements. The nature and structure of the self-assembled nanostructures are then investigated using SAXS, cryogenic transmission electron microscopy (cryo-TEM), and X-ray diffraction (XRD). Unexpectedly, the peptide is observed to form a self-supporting hydrogel at sufficiently high concentration without pH adjustment, a feature not previously reported for this type of SLP, and the rheological properties of the gel are measured. Because long alanine repeats are known substrates for the enzyme elastase (which plays an important role in remodeling the extracellular matrix), we also investigate the elastase-induced degradation of the peptide, inspired by our work on related SLP KA6E.36
It has been suggested that the multivalent display of the active group (here arginine) in self-assembled structures can enhance bioactivity, as exemplified, for example, by studies on lipopeptides containing cationic residues.37 Futhermore, the antibacterial activity of peptides is generally due to physical disruption of bacterial membranes which may reduce the development of antimicrobial resistance.38 The disruption of membranes may be enhanced by peptide self-assembly. Related to peptide A9R, it has been shown that peptide A9K, which forms fibrils, has marked antimicrobial activity against E. coli and S. aureus, and in fact, this is stronger than for the homologues A3K and A6K at a given concentration.17 It has recently been shown that peptide A9K2 forms fibrillar hydrogels and these have antimicrobial activity.38 Here, we assayed the antimicrobial activity and cytocompatibility (using model human fibroblast cells) of A9R. The combination of antimicrobial activity (and cytocompatibility with human cells) along with the degradability of the peptide in the presence of an essential extracellular matrix enzyme points to potential utility of SLP A9R in the development of a wound healing biomaterial.
Emulsions offer a potentially powerful route to stabilize formulations of active peptides or to create emulsions stabilized by biocompatible, biofunctional, or bioresponsive peptides. These systems are less well understood than protein-stabilized emulsions where proteins serve as Pickering stabilizers, acting as particles that reduce interfacial tension at the oil/water interface.39−41 Recently, Ulijn’s group has shown that Fmoc-dipeptides can stabilize chloroform-in-water emulsions due to the formation of a peptide layer at the droplet surface stabilized by π-stacking and H-bonding of peptides.42 In addition, this group has shown that tripeptides (with a propensity to aggregate assessed from a screen of all 203 = 8000 tripeptides containing native residues) stabilize oil/water emulsions, in some cases by forming nanofibrous networks.43 In addition, the self-assembly of the peptide can be triggered by biocatalysis using an enzyme to dephosphorylate a precursor to produce a hydrogelating peptide.44 Here, we show that a distinct class of peptide, that is, an SLP such as A9R can be formulated into an emulsion where it acts as a stabilizer at the oil/water interface due to the formation of a coating of β-sheet fibrils. Finally, we demonstrate on-demand de-emulsification induced by the catalytic activity of elastase on A9R emulsions.
Experimental Section
Materials
Peptide A9R was supplied by Biomatik (Cambridge, Ontario, Canada). The purity was 98.93% by high-performance liquid chromatography using an Inertsil ODS-SP column with acetonitrile [0.1% trifluoroacetate (TFA)]/water (0.1% TFA) gradient. The molar mass by electrospray ionization mass spectroscopy (ESI-MS) was 814.90 g mol–1 (M + H+, 813.92 g mol–1 expected). Scheme S1 shows the chemical structure of A9R. Elastase from porcine pancreas (MW = 25.9 kDa) and Trizma base were purchased from Sigma-Aldrich. 1-bromohexadecane was purchased from Sigma-Aldrich. Lipids DOPC, POPC, POPG, and POPE were obtained from Sigma-Aldrich.
A9R Dissolved as a Substrate for Elastase
Elastase solubility and enzymatic activity increases at pH 8. Weighed amounts of A9R or elastase were dissolved in a Trizma base solution (titrated with 1 M HCl to pH 8) to evaluate the enzymatic activity of elastase on A9R. The molar ratio of A9R to elastase is indicated as [A9R]/[elastase] = Mre ([ ]: molar concentration).
Vesicle Preparation
Vesicles were prepared by the thin-layer hydration method, as reported in the literature for hybrid POPE/POPG vesicles.45 Measured quantities of lipids were dissolved in chloroform, dried under a stream of nitrogen and placed in a vacuum chamber for 2 h in order to remove traces of organic solvent. The lipid film was then resuspended in a weighed quantity of water to obtain 0.5 wt % lipid, vortexed at 1800 rpm at 55 °C for 5 min, and left to equilibrate before experiments. Anionic POPG/POPE vesicles were made to provide a POPG molar fraction ϕPOPG = [POPG]/([POPG] + [POPE]) = 0.2 (here [ ] refers to the molar concentration), which corresponds to a POPG/POPE content of 27/73 wt %. Zwitterionic vesicles were prepared by mixing POPC and DOPC lipids at a fixed molar fraction of DOPC, ϕDOPC = [DOPC]/([POPC] + [DOPC]) = 0.2 which corresponds to a POPC/DOPC content of 80/20 wt %. The ratios 27/73 and 80/20 are calculated from the weight of each lipid used to prepare the vesicles. The vesicles, containing ϕPOPG or ϕDOPC, were always prepared with a fixed total 0.5 wt % lipid concentration in the aqueous dispersion. For mixed peptide–lipid samples, a weighed quantity of A9R powder was added to solutions containing the lipid vesicles described above, to obtain 0.08 wt % A9R. The mixtures were then vortexed at 1800 rpm and 55 °C for 5 min and left to equilibrate before experiments.
A9R Emulsion Preparation
Emulsions were prepared by mixing measured volumes of 1-bromohexadecane (density 1 g mL–1) with aqueous solutions of A9R. This oil was chosen to match the density of both phases in order to reduce the possible spontaneous de-emulsification process.46 Preliminary tests were performed to investigate the time stability of emulsions containing a range of different volume ratios of water/1-bromohexadecane emulsions stabilized by 0.05 wt % A9R. Visual inspection revealed emulsions containing 30/70 v/v water/1-bromohexadecane were the most stable. Such emulsions were made by vigorously stirring 300 μL of 0.16 wt % A9R in water with 700 μL of 1-bromohexadecane, to obtain a final 0.05 wt % A9R concentration calculated with respect to the total mass of liquid. In the following, we will refer to 0.05 wt % A9R in water/1-bromohexadecane 30/70 v/v emulsion simply as a 0.05 wt % A9R emulsion.
Fluorescence Assays
Samples were placed inside a quartz cell with 10.0 × 5.0 mm2 internal cross section, and the fluorescence was measured using a Varian Cary Eclipse spectrofluorimeter. The cac was determined via thioflavin T (ThT) or pyrene (Pyr) fluorescence experiments. The ThT fluorescence assay was performed to detect amyloid formation because it is well known that ThT fluorescence depends on the formation of amyloid-like structures (β-sheet fibrils).47,48 For the ThT assay, emission spectra were recorded from 460 to 600 nm using an excitation wavelength λex = 440 nm, for peptide solutions containing 5.0 × 10–3 wt % ThT.
The Pyr assay was performed to identify the formation of a hydrophobic environment by the peptide, to provide complementary information to the ThT assay. A solution containing 1.3 × 10–5 M Pyr in water was used to dilute peptide solutions within the ranges (1.3 × 10–3 to 0.13) wt % A9R. The fluorescence of Pyr was excited at 335 nm at room temperature, and the emission spectra were recorded from 360 to 460 nm.
Fluorescamine (FLC) reacts with primary amines to form fluorescent products. Therefore, FLC assay was used to detect the formation of primary amines due to the cleavage of A9R by elastase. Solutions containing 1 × 10–3 to 5 × 10–2 wt % A9R, at Mre = [A9R]/[elastase] ≈ 3–158, were prepared using 0.07 wt % FLC as a solvent in Trizma base solution (pH 8). The samples were excited at λex = 380 nm, and the fluorescence emission was measured for λ = (410–600) nm.
CD Spectroscopy
CD spectra were recorded using a Chirascan spectropolarimeter (Applied Photophysics, UK). Solutions were placed in a quartz coverslip cuvette (0.01 mm thick). Spectra are presented for absorbance A < 2 at any measured point with a 0.5 nm step, 1 nm bandwidth, and 1 s collection time per step. The CD signal from the water background was subtracted from the CD data of the sample solutions.
Fourier Transform Infrared Spectroscopy
Spectra were recorded using a Nexus Fourier transform infrared (FTIR) spectrometer equipped with a DTGS detector. Samples were measured using an ATR configuration with a PEARL liquid cell. Spectra were scanned 128 times over the range of 900–4000 cm–1.
X-ray Diffraction
Measurements were performed on stalks prepared by drying a drop of solution suspended between the ends of wax-coated capillaries. The stalks were mounted onto a four axis goniometer of an Oxford Diffraction Gemini Ultra instrument. The sample-detector distance was 44 mm. The X-ray wavelength λ = 1.54 Å was used to calculate the scattering vector q = 4π sin θ/λ (2θ: scattering angle). The detector was a Sapphire CCD.
Transmission Electron Microscopy
TEM imaging was performed using a JEOL 2100Plus TEM microscope operated at 200 kV. Droplets of A9R emulsion was placed on Cu grids coated with a carbon film (Agar Scientific, UK), stained with uranyl acetate (1 wt %) (Sigma-Aldrich, UK), and dried.
Cryogenic Transmission Electron Microscopy
Imaging was carried out using a field emission cryoelectron microscope (JEOL JEM-3200FSC), operating at 200 kV. Images were taken in the bright-field mode and using zero loss energy filtering (omega type) with a slit width of 20 eV. Micrographs were recorded using a Gatan UltraScan 4000 CCD camera. The specimen temperature was maintained at −187 °C during the imaging. Vitrified specimens were prepared using an automated FEI Vitrobot device using Quantifoil 3.5/1 holey carbon copper grids with a hole size of 3.5 μm. Just prior to use, grids were plasma-cleaned using a Gatan Solarus 9500 plasma cleaner and then transferred into the environmental chamber of a FEI Vitrobot at room temperature and 100% humidity. Thereafter, 3 μL of sample solution was applied on the grid, and it was blotted twice for 5 s and then vitrified in a 1/1 mixture of liquid ethane and propane at a temperature of −180 °C. The grids with vitrified sample solution were maintained at the liquid nitrogen temperature and then cryotransferred to the microscope.
Cryogenic Scanning Electron Microscopy
Imaging was performed using an FEI Quanta 600F instrument in the high vacuum mode. Samples were mounted onto aluminum stubs and frozen in liquid nitrogen slush at approximately −210 °C. Once frozen, samples were transferred under high vacuum to a sample preparation chamber and allowed to equilibrate to the appropriate temperature prior to fracturing. Solutions were fractured at −185 °C and allowed to sublime at −90 °C for approximately 20 min. The temperature was then reset to −135 °C in the preparation chamber, and the sample was coated with a thin layer of platinum for 80 s, prior to imaging at 12.5 kV.
Small-Angle X-ray Scattering
Synchrotron SAXS experiments on solutions were performed using BioSAXS robots on beamline BM29 (ESRF, France) and on beamline B21 (Diamond Light Source Ltd., UK). On beamlines BM29 and B21, solutions were loaded into the 96-well plate of an EMBL BioSAXS robot and then injected via an automated sample exchanger into a quartz capillary (1.8 mm internal diameter) in the X-ray beam. The quartz capillary was enclosed in a vacuum chamber, in order to avoid air scattering. After the sample was injected in the capillary and reached the X-ray beam, the flow was stopped during the SAXS data acquisition. B21 was operated with a fixed camera length (3.9 m) and fixed wavelength λ = 1 Å. The images were captured using a PILATUS 2M detector. Data processing (background subtraction, radial averaging) was performed using the dedicated beamline software ScÅtter. BM29 was operated with an X-ray wavelength of λ = 1.03 Å. The images were captured using a PILATUS 1M detector, while data processing was performed using dedicated beamline software ISPyB.
Synchrotron SAXS and wide-angle X-ray scattering (WAXS) on gels were performed on BM26B (ESRF, France). The peptide gels were introduced in differential scanning calorimetry pans with mica windows to enable transmission of the X-ray beam. The distance to the 2D multiwire SAXS detector was ca. 5 m using a wavelength of 1.45 Å. WAXS images were recorded using a CCD-based X-ray digital camera (photonic science).
Rheology
Rheological properties were determined using controlled stress TA Instruments AR-2000 rheometer (TA Instruments). The experiments to characterize A9R gels were performed using a cone-and-plate geometry (cone radius = 20 cm; cone angle = 1°). The linear regime was first determined performing stress sweep experiments in the range (0.08–1000) Pa at a constant angular frequency of 6.28 rad/s. Frequency sweep experiments were performed at a constant stress within the linear regime and angular frequencies between 0.1 and 627 rad/s.
The viscosity of the A9R emulsions was measured performing controlled stress experiments with a cone-and-plate geometry (cone radius = 40 cm; cone angle = 1°) and shear stress between 0.05 and 23 Pa.
Electrospray Ionization Mass Spectroscopy
Electrospray ionization mass spectra were recorded using a Thermofisher Obitrap XL instrument. Samples, which were presented as 1 mg/mL, were diluted 33-fold (30 μL sample + 970 μL MeOH). The two LCMS mobile phase buffers (water and acetonitrile) included 0.1% formic acid to assist the reverse phase separation and to aid protonation.
Polarized Optical Microscopy
Images were obtained with an Olympus BX41 polarized microscope by placing the sample between crossed polarizers. Samples were placed between a glass slide and a coverslip before capturing the images with a Canon G2 digital camera.
Laser Scanning Confocal Microscopy
Experiments were performed on a Nikon A1-R plus confocal microscope, using objective Plan Apo VC DIC N2 with a 20× magnification and a numerical aperture 0.75. For laser scanning confocal microscopy (LSCM), A9R emulsion was prepared by vigorously stirring 300 μL of 0.16 wt % A9R, dissolved in 5 × 10–3 wt % ThT, with 700 μL of 1-bromohexadecane. The excitation wavelength generated by an argon laser was 488 nm, while the emission detection was in the range of 525 nm. Data were collected with a pinhole radius of 17.88 μm. Samples were placed between a glass slide and a coverslip.
Cytotoxicity Assays
The cytotoxicity of A9R was examined using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. In vitro cell culture was conducted using the human skin fibroblast cell line, 161Br (European Collection of Authenticated Cell Cultures, ECACC) cells. Cells were cultured in EMEM, with 2 mM glutamine, enriched with 15% fetal bovine serum, 1% nonessential amino acids, and 1% antimycotic/antibiotic. Cells were maintained in a humidified atmosphere at 37 °C and 5% CO2.
For the MTT assay, cells were seeded into a 96-well plate at a concentration of 4 × 104 cells/mL and were allowed to adhere for 24 h in 100 μL of complete medium. The peptide was dissolved in complete medium and added to the cells to give a final volume of 200 μL at concentrations between 0.005 and 0.5 wt % A9R. One well, containing 200 μL of complete medium with no peptide, was used as a control. Then, the fibroblasts were incubated for 67 h. After this, 20 μL MTT (5 mg/mL, in PBS) was added to each well plate and allowed to incubate for 5 h (72 h total). Following the incubation time, the solution was removed from the wells and replaced with 100 μL dimethyl sulfoxide per well to dissolve the formazan crystals. Plates were incubated for 30 min, and then were analyzed using a UV microplate reader (λ = 570 nm). Results are reported as % cell viability compared to control (untreated) values. ANOVA and Bonferroni post hoc tests were used to assess statistical significance.
Antimicrobial Assays
The antimicrobial assays were performed with three strains of bacteria, S. aureus SH1000, E. coli K12 MG1655, and Pseudomonas aeruginosa PA01. Stock cultures were stored in 20% glycerol at −80 °C. Prior to experiments, all three strains were streaked out onto LB (Lysogeny Broth) agar and grown overnight at 37 °C. From these plates, one colony was then transferred into 3 mL sterile tryptone soy broth supplemented with a 0.3% (w/v) yeast extract and grown at 37 °C under agitation at 150 rev/min on an orbital shaker overnight, and these cultures were used for ongoing experiments. Cultures were then transferred into a 15 mL falcon tube, and cells were harvested by centrifugation at 9000 rpm and 4 °C for 10 min. The supernatant was discarded, and the pellet was resuspended in 1.5 mL ice-chilled PBS (phosphate-buffered saline). After this, 30 μL of this solution was transferred into 300 μL of 0.05 wt % A9R in sterile water or control solutions of 300 μL of sterile water. Solutions were then vortexed for 5 s, and 3 × 20 μL aliquots were taken at times 0, 30, 60, 120, and 1440 min. These samples were then serially diluted in PBS, and 10 μL of each dilution was plated onto LB agar and incubated at 37 °C overnight before colony counting.
Results and Discussion
We first examined the self-assembly of A9R in water, including potential hydrogel formation, together with the interaction of A9R with model membranes simulating mammalian or microbial cell walls. We then investigate the degradation and defibrillization of A9R by cleavage of the oligoalanine sequence through elastase proteolysis. The cytotoxicity and antimicrobial activity of A9R is then investigated, and the results are understood in terms of the interaction of the peptide with model lipid membranes mimicking the membranes of bacterial and mammalian cells. Finally, we report a method to prepare peptide emulsions as a novel method to deliver A9R at a nontoxic and antimicrobially active concentration. In addition, we show that the catalytic activity of elastase on A9R can be used as a route for de-emulsification.
Self-Assembly in Water and Interaction with Model Biological Membranes
We investigated the self-assembly of A9R in water, performing fluorescence spectroscopy assays and CD, FTIR, XRD, SAXS, WAXS, cryo-TEM, and rheology experiments. The results are summarized in Figures 1 and 2. The pKa of A9R is estimated to be approximately 11,49 and the charge is +1 in aqueous solution.
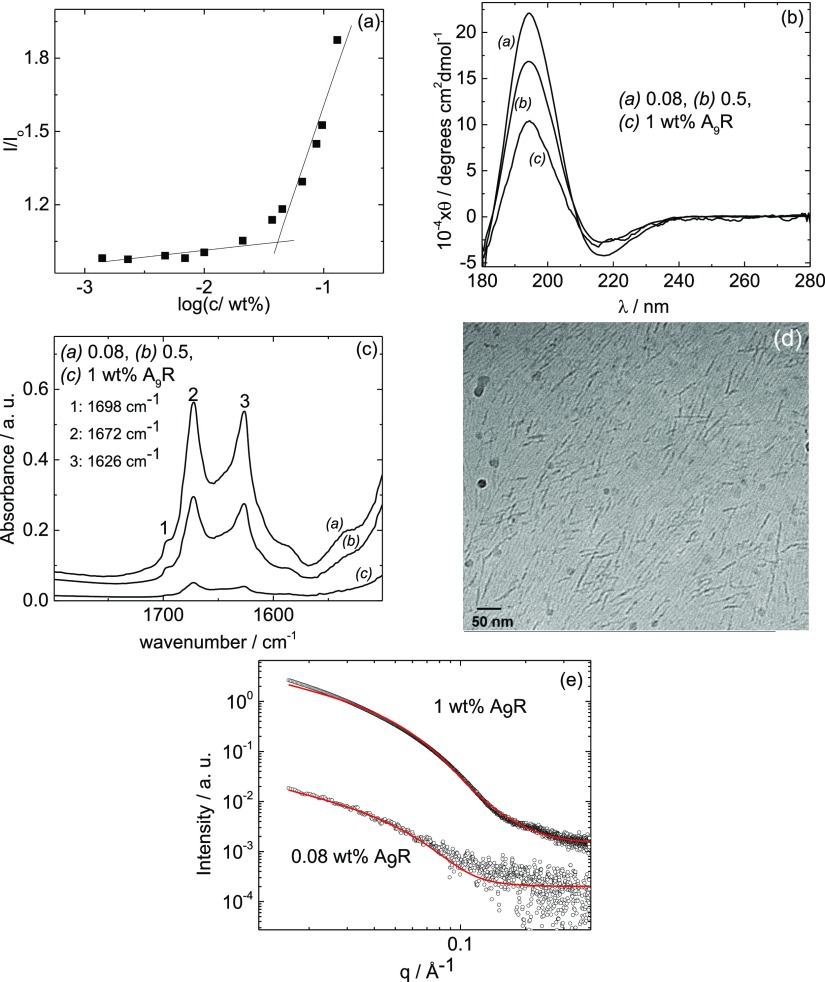
Figure 1.
(a) ThT assay for A9R. 1 wt % A9R solution: (b) CD data, (c) FTIR spectra, (d) cryo-TEM image, and (e) SAXS data along with fitting of the data.
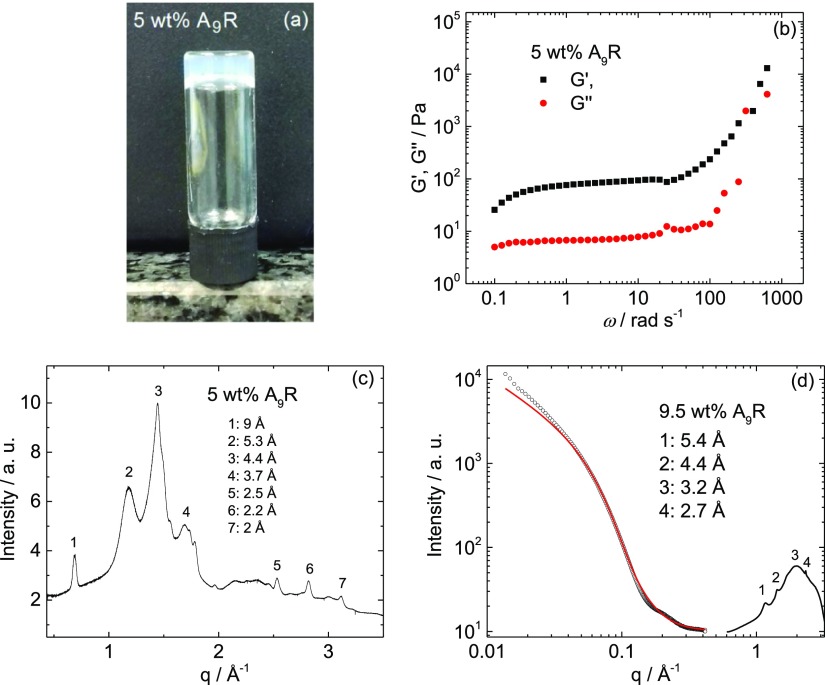
Figure 2.
A9R hydrogels: (a) tube inversion test, (b) storage and shear modulus, (c) XRD spectra on a dry gel stalk, and (d) SAXS and WAXS profiles. The full line in (d) is the fitting to the experimental data.
Fluorescence assays using dye ThT were performed to determine the cac for amyloid formation by A9R. Figure 1a shows the fluorescence emission intensity of ThT at 486 nm for solutions containing A9R (I), normalized by the fluorescence emission intensity of ThT at 486 nm for solutions without A9R (Io). A discontinuity in I/Io as a function of A9R concentration shows that the peptide forms amyloid fibrils at concentrations equal or higher than cac = (0.05 ± 0.01) wt % A9R (Figure 1a), which may be compared with a cac = 1.4 wt % for the related peptide A6R previously reported by us.23 The lower cac for A9R is consistent with the increase in the hydrophobicity of the peptide as a result of a longer alanine sequence.
Fluorescence assays using dye Pyr were performed to determine the concentration for the formation of a hydrophobic environment by the A9R peptide and therefore provide complementary information to the ThT assay. Figure S1 shows the fluorescence emission intensity of Pyr at 373 nm for solutions containing A9R (I1), normalized by the fluorescence emission intensity of Pyr at 373 nm for solutions without A9R (I0,1). According to the data in Figure S1, A9R self-assembles into a structure with a hydrophobic core for concentrations equal or higher than (0.07 ± 0.01) wt % A9R (Figure 1a). This is the same as the cac determined from ThT fluorescence within respective uncertainties and indicates that the formation of β-sheet fibrils occurs at the same concentration as sequestration of the hydrophobic alanine residues into the fibril cores.
The CD spectra for A9R (Figure 1b) show a maximum at 194 nm and a minimum at 217 nm corresponding to a β-sheet structure over a concentration range of 0.08–1 wt % A9R.50 The FTIR spectra for A9R in Figure 1c (for the same concentration range) show FTIR bands centered at 1626 and 1698 cm–1, which confirms the β-sheet structure of the self-assembled peptide.51 The peak at 1672 cm–1 is due to bound TFA counterions.52−54 These results confirm the retention of the β-sheet structure over a wide concentration range above the cac.
Figure 1d shows a cryo-TEM image of a sample containing 1 wt % A9R. The image shows the formation of long fibers, ∼5 nm in diameter. Figure 1e shows the SAXS data measured for 0.08 and 1 wt % A9R (data for samples with 0.5 and 9.5 wt % A9R are shown in Figures S4 and 2c, respectively), along with model form factor fits using a long cylindrical shell model, performed with the software SASfit.55 The parameters of the model are the core radius R (with polydispersity ΔR), the shell thickness Dr, and the scattering length density of the core, shell, and solvent ηcore, ηshell, and ηsolv. The parameters extracted from the fitting are listed in Table S1. The parameters obtained from the SAXS fittings for 0.08–1 wt % sample are consistent. In particular, at 1 wt % A9R, R ± ΔR = (20.3 ± 10.8) Å and Dr = 3 Å indicate that the cylinder core radius is smaller than the length of the oligoalanine A9 sequence = 28.8 Å (spacing per residue in a parallel β-sheet is 3.2 Å56), while the external shell has a thickness very similar to the length of one arginine residue. This result shows that there is an overlapping of the alanine chains in the core of the fibers, with the arginine residue exposed at the surface of the fibers. The fiber radius from SAXS is in good agreement with the dimensions obtained from cryo-TEM images.
We unexpectedly found that, even without pH adjustment, A9R forms a self-standing hydrogel at a higher peptide concentration in water (Figure 2a). Rheology experiments were performed to evaluate the viscoelastic properties of the hydrogel. Stress sweeps show approximately linear viscoelastic behavior up to a stress of σ = 20 Pa (Figure S2). Frequency sweeps at a fixed shear stress within the linear regime (σ = 4.78 Pa) reveal a low frequency plateau in the moduli with G′ > G″, consistent with a gel-like response (Figure 2b). The XRD spectra for a peptide stalk dried from a 5 wt % A9R gel has peaks at 9, 5.3, 4.4, 3.7, 2.5, 2.2, and 2 Å (Figure 2c). Figure 2d shows the SAXS data measured for a 9.5 wt % A9R hydrogel, along with the fitting of the experimental data using a long cylindrical shell form factor model. The parameters extracted from the fitting are listed in Table S1. The values, R ± ΔR = (15.0 ± 10) Å and Dr = 2 Å, indicate that the hydrogel contains peptide fibers with similar dimensions to those in solution. The WAXS data for 9.5 wt % A9R hydrogel (Figure 2d) shows peaks at 5.4, 4.4, 3.2, and 2.7 Å. Both XRD (Figure 2c) and WAXS (Figure 2d) data indicate a β-sheet structure and are very similar to the XRD patterns already reported by us for 17 wt % A6R23 and 1 wt % capA6R.31 In analogy with our previous XRD analysis of oligoalanine SLPs,23,31 we assign reflections in Figure 2d to the packing of the oligoalanine β-sheets (5.3 Å), and intrasheet spacings (4.4, 3.7, 2.5, 2.2, and 2 Å). The β-sheet spacing 5.3 Å is very small compared to typical amyloid structures because oligoalanine sequences facilitate close packing of the peptide chains. In summary, we demonstrated that A9R forms β-sheet fibrils above a cac in water, and that the peptide is able to form self-supporting hydrogels.
A9R Degradation in Response to Elastase Enzymatic Activity
Having established that A9R self-assembles into fibrils in solution, and because alanine is a substrate for elastase catalytic activity, we investigated the enzyme-induced degradation and defibrillization of the peptide.
Figure S3a shows the CD spectra for 0.5 wt % A9R compared to the CD spectra for 0.5 wt % A9R (Mre = 158) after 3 and 68 h incubation. Similar results are displayed for 0.1 wt % A9R (Mre = 31) in Figure S3b. Samples studied at 68 h incubation were diluted and used for the FLC assay. The results are displayed in Figure S3c. The CD spectrum for 0.5 and 0.1 wt % A9R shows β-sheet features; however, these are eliminated upon incubation with elastase for 68 h, and a spectrum of a disordered peptide conformation is observed (Figure S3a,b). FLC fluorescence assay was performed to confirm the existence of free amine groups in the solution. The emission fluorescence spectrum of FLC was characterized by a broad maximum centered at 475 nm (results not shown). Figure S3c shows that the fluorescence emission at 475 nm increases with the Mre ratio. The results in Figure S3 are consistent with the degradation of the A9R fibrils by elastase.
SAXS was used to investigate the structure of A9R fibers with and without elastase. Figure S4 compares the SAXS data for a 0.5 wt % A9R solution and a solution containing elastase and the same concentration of A9R, after incubation at Mre = 158 for 68 h. The SAXS data for 0.5 wt % A9R can be fitted using the form factor for a cylindrical shell (fitting parameters listed in Table S1). However, the SAXS data show the loss of the cylinder form factor shape due to the fiber fragmentation after exposure to the elastase.
Finally, the effect of elastase on the peptide (0.1 wt % A9R, Mre = 31, 68 h incubation) was studied by ESI-MS. The mass spectra are displayed in Figure S5. The identification of different fragments of A9R in the ESI-MS spectra, together with the absence of a peak corresponding to the expected molecular weight of A9R, confirms the fragmentation of the peptide in response to elastase. We hypothesized that this could be exploited in enzyme-responsive de-emulsification (vide infra).
A9R Cytotoxicity and Antimicrobial Activity
To examine the cytocompatibility of A9R, the viability of the peptide was tested using 161Br skin fibroblast cells via MTT assays (Figure 3a). Peptide A9R was tolerated by the cells up to a concentration of 0.05 wt % (∼75% viability), which corresponds to the minimal concentration for amyloid fibril formation (cac; Figure 1a). A lower cell viability <70% was observed from 0.1 to 0.5 wt % A9R. The antimicrobial activity of the A9R peptide was studied using two model Gram-negative microorganisms, E. coli (K12) and P. aeruginosa and one Gram-positive microorganism, S. aureus. Microorganisms were treated with 0.05 wt % A9R, the highest concentration for peptide viability. Preliminary tests were performed using 0.01 wt % A9R, but at that concentration, no significant antimicrobial activity (results not shown) was observed.
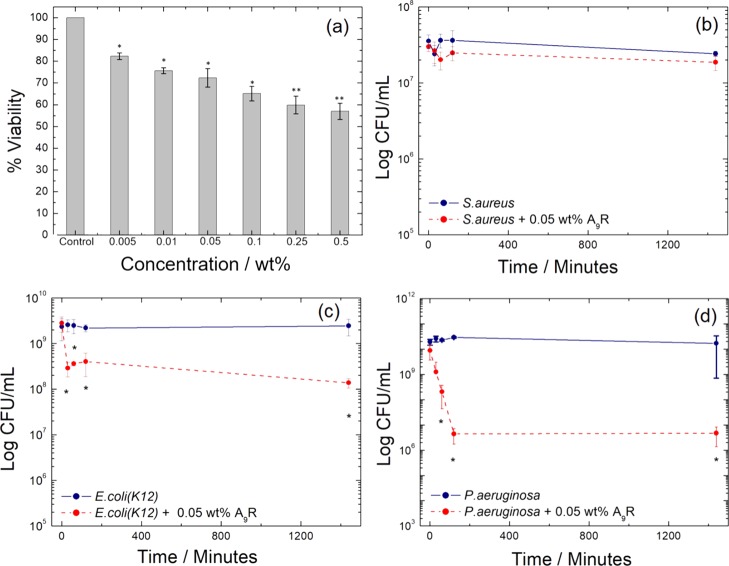
Figure 3.
(a) Cell viability profiles of A9R solutions. Error bars are SEM (n = 3). * = p < 0.05, ** = p < 0.01. Survival of three bacteria strains without (blue line) and in the presence of 0.05 wt % A9R (red dashed line) for (b) S. aureus, (c) E. coli, and (d) P. aeruginosa, where CFU = colony forming units, and the error bars represent the standard error of the mean. * = p < 0.05 as assessed by the t-test between treated and untreated strains.
The antimicrobial assays of the activity of A9R against S. Aureus, E. coli, and P. aeruginosa are displayed in Figure 3b–d. The effect of A9R on S. Aureus was not statistically significant (Figure 3b). However, a statistically significant antimicrobial effect was found with E. coli and P. aeruginosa (Figure 3c,d). In particular, a major effect of four orders of magnitude reduction in CFU/mL was found with P. aeruginosa, while a less pronounced effect was seen with E. coli. Overall, the data in Figure 3c,d show a dramatic effect of A9R on the numbers of bacteria within the first 1–2 h. Subsequently, the numbers of E. coli and P. aeruginosa remained stable up to 24 h.
The results in Figure 3 suggest that A9R shows selective antimicrobial activity against Gram-negative microorganisms, which cause infections that are particularly challenging to treat. In contrast, we previously reported that closely related peptides RA3R, A6R, and capA6R have a major antimicrobial activity against the Gram-positive bacteria L. monocytogenes.31,35
To examine the potential mode of antibacterial activity, we studied the interaction of A9R with vesicles comprising lipid mixtures representing model bacterial membranes (POPG/POPE) or eukaryotic membranes (DOPC/POPC), using SAXS to probe the lipid bilayer stacking. These results provide insights into the mechanism of selective antimicrobial activity of A9R. We studied samples containing 0.08 wt % A9R because, although a relatively low concentration, that was the highest concentration of the peptide which did not induce sample precipitation in the presence of the lipid vesicles. The lipid mixture composition was fixed with a mole fraction of ϕDOPC = 0.2 in the DOPC/POPC mixture or ϕPOPG = 0.2 in the POPG/POPE mixture. The peptide powder was dissolved in the solution of vesicles. The corresponding SAXS data are displayed in Figure S6. The SAXS data for the liposomes without the peptide in Figure S6 were already reported by us,31,35 and it is presented here only as a reference. The SAXS data for ϕDOPC = 0.2 display two peaks in a positional ratio 1:2 corresponding to a lamellar spacing of 64.4 Å. The intensity of the lamellar peaks is reduced by the addition of 0.08 wt % A9R (Figure S6a).
The SAXS data for the lipid vesicles with ϕPOPG = 0.2 (Figure S6b) show a broad peak centered at ∼0.1 Å arising from a single bilayer form factor.31,35,45 Addition of 0.08 wt % A9R induces the formation of multilamellar ordering with a bilayer spacing of d = 88.4 Å.
The SAXS data in Figure S6 were further analyzed using models for the form factor and structure factor of a lamellar structure. Details of the model57 are provided in the Supporting Information. Fits to the SAXS data are displayed in Figure S6, while the parameters extracted from the fitting are listed in Table S2.
Addition of 0.08 wt % A9R to ϕDOPC = 0.2 vesicles changes the bilayer structure by decreasing the distance between headgroups (lH), increasing the thickness of the water layer (lw) and reducing the correlation between bilayers (N) (Table S2). Accordingly, A9R is not inserted in the DOPC/POPC bilayers but decorates the surface of those bilayers as previously reported by us for the study of the interaction of A6R with zwitterionic multilamellar vesicles.31 Addition of 0.08 wt % A9R to ϕPOPG = 0.2 vesicles induces the correlation of lipid bilayers (N) and increases the distance between headgroups (lH) suggesting the insertion of A9R in the lipid bilayers (Table S2).
These findings are significant in terms of the possible mode of antimicrobial activity of A9R. Several mechanisms have been proposed by which bacterial cell membranes are restructured, involving different modes of incorporation of peptides into the lipid membrane (to form pores as in the barrel-stave model) or adsorption onto the surface of the membrane (carpet model) with subsequent membrane breakup or the formation of toroidal pores due to peptide-induced lipid curvature.2,3,5,8 A9R appears to restructure bacterial cell membrane-mimicking ϕPOPG = 0.2 vesicles by inserting into the lipid membrane due to electrostatic attraction, which induces a correlation between bilayers. In contrast, A9R does not insert in the zwitterionic membrane-mimicking ϕDOPC = 0.2 vesicles, which may underlie the low observed cytotoxicity.
A9R Emulsification
We investigated the ability of A9R to stabilize emulsions containing 0.05 wt % A9R, a dosage of peptide that allows for cell viability but is simultaneously effective against E. coli and P. aeruginosa. The stability of 0.05 wt % A9R emulsion was checked through its physical appearance a few minutes and 3 days after emulsification (Figure S7). After one day of mixing, phase separation occurs in the mixture without the peptide, which becomes more visible after 3 days of mixing (Figure S7b,d). In contrast, the peptide stabilizes the emulsion, and no phase separation is observed even after 3 days of mixing (Figure S7a,c). The emulsion shows shear-thinning behavior (Figure S8) with a limiting viscosity of ∼19 mPa·s, significantly higher than the viscosity of water (0.89 mPa·s at 25 °C).59
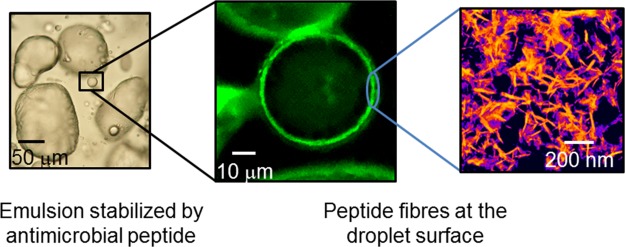
The structure of the 0.05 wt % A9R emulsion was studied by a range of microscopy techniques. Results obtained using polarized optical microscopy (POM), LSCM, and TEM are displayed in Figure 4. POM images (Figure 4a,b) show that the peptide stabilizes the emulsion through the formation of micrometer-sized droplets with irregular shape. LSCM was used to assess the presence of peptide fibrils within a 0.05 wt % A9R emulsion prepared with A9R fibers stained with ThT, because ThT is known to bind to amyloid fibers.47 LSCM images (Figure 4c,d) show that the fluorescent peptide fibers are adsorbed at the surface of the droplets making a ∼1 μm thick layer. TEM was used to confirm that the peptide fibers stabilized at the surface of the droplets in the emulsion are ∼25 nm in diameter (Figure 4e). The CD spectra in Figure 4f show that the population of β-sheets is higher for A9R fibers in the emulsion than for a peptide solution with the same peptide concentration in the aqueous phase. The combination of images and CD data in Figure 4 clearly shows that the emulsion is stabilized by A9R fibers adsorbed at the surface of the droplets. These fibers form a micron thick layer on the surface of the droplets. This appears to indicate an unexpected inverse Pickering-type emulsion stabilization mechanism by peptide fibers, rather than simple interfacial adsorption of the SLP molecules. The fibers are assembled from peptides with an amphiphilic character, and it is expected that the hydrophilic arginine residues will be exposed to the aqueous phase.
Figure 4.
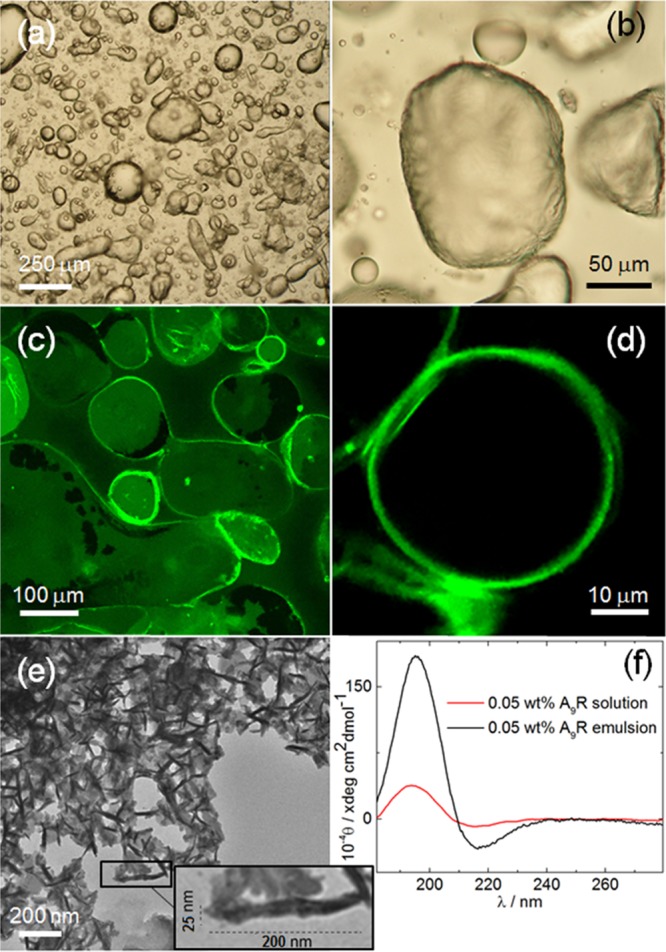
0.05 wt % A9R emulsion: (a,b) POM, (c,d) LCSM, and (e) TEM images. (f) CD data measured for a 0.05 wt % A9R emulsion and for 0 0.05 wt % A9R sol.
Figure 5 shows cryo-SEM images obtained for 0.05 wt % A9R emulsion. The images show droplets on the micrometer length scale, wrapped by a network of fibers. This result is in good agreement with LSCM images in Figure 4c,d showing the absorption of A9R peptide fibers at the surface of the droplets in the emulsion.
Figure 5.
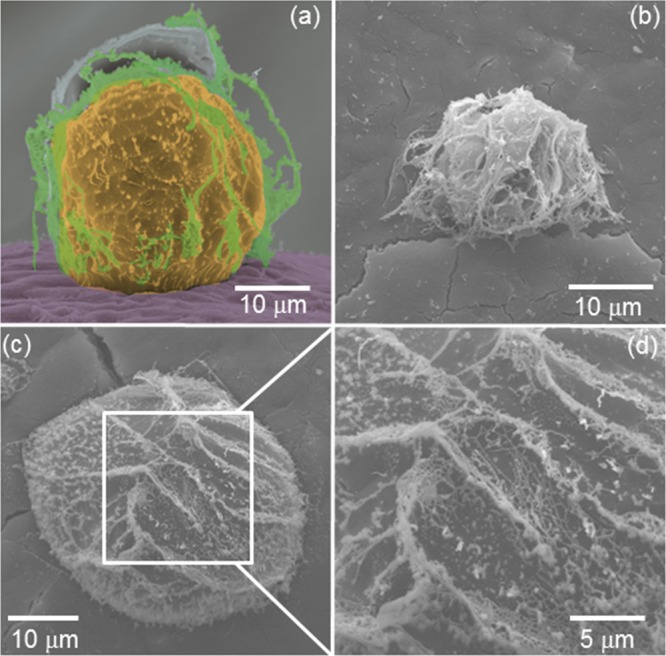
(a–d) cryo-SEM images obtained for a 0.05 wt % A9R emulsion. (a) Colorization assists the visualization of the fibril coating. The image in (d) is an enlargement of the region shown in (c).
As a further development toward a multifunctional responsive peptide biomaterial, we examined de-emulsification driven by the elastase-induced degradation of the A9R fibers.
The 0.05 wt % A9R emulsion, stained with ThT and used for LSCM experiments (Figure 4c,d), was treated with elastase at Mre = 8.3. The ThT fluorescence emission of this emulsion was characterized by a broad peak centered at 490 nm (results not shown). Figure S9a plots the time dependence of the fluorescence emission intensity maximum for 0.05 wt % A9R emulsion, stained with ThT (I), normalized by the fluorescence emission maximum for a sample containing only ThT (Io). The spectra show a drop in I/Io after 3 h due to the breakup of A9R fibers induced by the elastase. Simultaneously, the initial homogeneous emulsion (a; Figure S9a) undergoes phase separation (b; Figure S9a).
Figure S9b shows the time dependence of the CD signal measured for a 0.05 wt % A9R emulsion treated with elastase at Mre = 8.3, that is, the same sample studied in Figure S9a but without ThT dye. The CD spectra in Figure S9b show that there is an evolution from a β-sheet conformation into a disordered secondary structure after 2 days incubation of the emulsion with the elastase.
Conclusions
We first established that SLP A9R self-assembles into β-sheet fibrils above cac. XRD shows that the β-sheets are unusually closely spaced due to the tight packing facilitated by the small alanine sequence. Remarkably, the peptide forms a self-supporting hydrogel structure without pH adjustment, simply by increasing the concentration sufficiently in water. This is not typical for peptide hydrogelation which typically necessitates pH control.60 We believe this is due to the presence of only a single-charged residue in A9R. Although the termini are uncapped, the location of the arginine residue at the C terminus gives rise to a type of “zwitterionic” structure. We furthermore established that A9R can be degraded into smaller peptide fragments by the enzyme elastase, this being used as the basis to subsequently demonstrate an enzyme-triggered de-emulsification process. We next showed that A9R has activity against Gram-negative bacteria, in particular P. aeruginosa. Gram-negative bacterial infections are particularly challenging, and P. aeruginosa is the source of serious hospital infections, and strains have evolved antimicrobial resistance.61−63 Our results suggest that peptide A9R has great potential in the development of future antibacterial therapeutics based on our initial assays.
We furthermore hypothesized that A9R, being a SLP, would potentially be able to stabilize water/oil emulsions. This indeed proved to be the case, and stable emulsions in the model system water in 1-bromohexadecane were observed. A combination of CD spectroscopy and fluorescent dye staining revealed that the emulsion is stabilized by a coating of β-sheet fibrils around emulsion droplets. These findings provide evidence for the previously unreported formation of stable emulsions by an antimicrobial SLP. The emulsions are distinct from conventional surfactant-stabilized emulsions because the peptides form β-sheet fibril coatings of the emulsion droplets (Scheme 1), rather than simply forming a surfactant monolayer at the interface. They can also be compared with protein-stabilized emulsions which are a type of Pickering (particle-stabilized) emulsion.39,40 As a further demonstration of the utility of SLPs in the development of novel responsive nanomaterials, we showed that it is possible to de-emulsify the A9R water-in-oil emulsions using the enzyme elastase. This enzyme has a key role in the remodeling of the extracellular matrix (it degrades elastin), and so our findings point to the possible future development of wound-healing materials with antimicrobial activity. On-demand enzyme-triggered de-emulsification may find other applications in the creation of responsive nanomaterials, for example, for drug delivery.
Scheme 1. A9R Peptide Fibers Stabilize the 0.05 wt % A9R Emulsion by Forming a Fibrillar Mesh at the Interface of the Water Droplets.
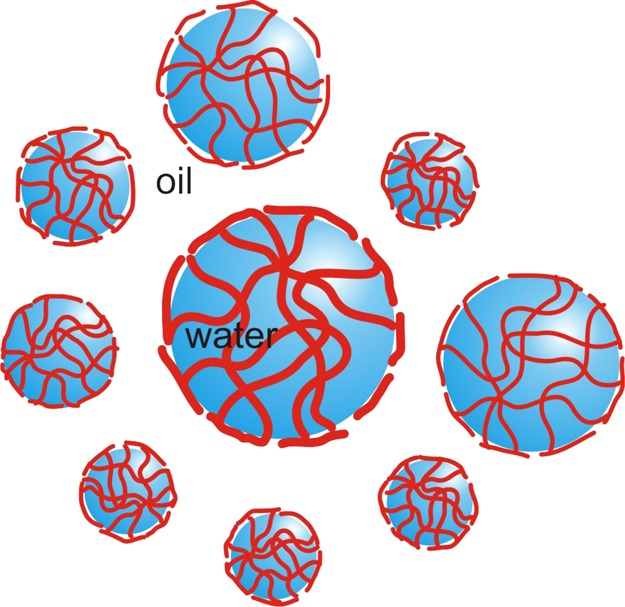
Considering the unique properties of peptides in terms of biofunctionality and biodegradability, emulsions and gels of antimicrobial SLPs have great promise in the future production of formulated personal care and food products. Taken together, our results show that A9R has an unprecedented diversity of bionanomaterial properties relevant to such applications.
Acknowledgments
The work of V.C. was supported by EPSRC Platform grant EP/L020599/1 “Nanostructured Polymeric Materials for Healthcare” to I.W.H. C.J.C.E.-G. was supported by a studentship co-funded by the University of Reading and Diamond Light Source. We are grateful to the ESRF for beamtime on BM29 (refs MX1880 and MX1869) and Diamond for beamtime on B21 (ref SM18523-1). We acknowledge Gabrielle Giachin (BM29) and Katsuaki Inoue (B21) for support during beamtime. We thank Daniel Hermida-Merino for the SAXS/WAXS measurements on BM26B at the ESRF. We are grateful to Nick Michael for assistance with MS and Nick Spencer for assistance with XRD experiments. We acknowledge access to the Chemical Analysis Facility Laboratory (University of Reading).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.9b00581.
Description of the method to fit SAXS data, tables of parameters obtained from SAXS data fittings, rheology, CD, fluorescence, ESI data, and images of emulsion stability and rheology (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Nicolas P.; Mor A. Peptides as Weapons Against Microorganisms in the Chemical Defense System of Vertebrates. Annu. Rev. Microbiol. 1995, 49, 277–304. 10.1146/annurev.micro.49.1.277. [DOI] [PubMed] [Google Scholar]
- Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta, Biomembr. 1999, 1462, 55–70. 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- Epand R. M.; Vogel H. J. Diversity of Antimicrobial Peptides And Their Mechanisms Of Action. Biochim. Biophys. Acta, Biomembr. 1999, 1462, 11–28. 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Chapple D. S. Peptide Antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Sahl H.-G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24, 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Jenssen H.; Hamill P.; Hancock R. E. W. Peptide Antimicrobial Agents. Clin. Microbiol. Rev. 2006, 19, 491–511. 10.1128/cmr.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. I.; Prenner E. J.; Vogel H. J. Tryptophan- and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. Biochim. Biophys. Acta, Biomembr. 2006, 1758, 1184–1202. 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Nawrot R.; Barylski J.; Nowicki G.; Broniarczyk J.; Buchwald W.; Goździcka-Józefiak A. Plant Antimicrobial Peptides. Folia Microbiol. 2014, 59, 181–196. 10.1007/s12223-013-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M.; Hakansson J.; Ringstad L.; Bjorn C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauthey S.; Santoso S.; Gong H.; Watson N.; Zhang S. Molecular Self-Assembly of Surfactant-Like Peptides to Form Nanotubes and Nanovesicles. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 5355–5360. 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso S. S.; Vauthey S.; Zhang S. Structures, Function and Applications of Amphiphilic Peptides. Curr. Opin. Colloid Interface Sci. 2002, 7, 262–266. 10.1016/s1359-0294(02)00072-9. [DOI] [Google Scholar]
- Von Maltzahn G.; Vauthey S.; Santoso S.; Zhang S. Positively Charged Surfactant-Like Peptides Self-Assemble into Nanostructures. Langmuir 2003, 19, 4332–4337. 10.1021/la026526+. [DOI] [Google Scholar]
- Hauser C. A. E.; Zhang S. Designer Self-Assembling Peptide Nanofiber Biological Materials. Chem. Soc. Rev. 2010, 39, 2780–2790. 10.1039/b921448h. [DOI] [PubMed] [Google Scholar]
- Xu H.; Wang J.; Han S.; Wang J.; Yu D.; Zhang H.; Xia D.; Zhao X.; Waigh T. A.; Lu J. R. Hydrophobic-Region-Induced Transitions in Self-Assembled Peptide Nanostructures. Langmuir 2009, 25, 4115–4123. 10.1021/la802499n. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Pan F.; Xu H.; Yaseen M.; Shan H.; Hauser C. A. E.; Zhang S.; Lu J. R. Molecular Self-Assembly and Applications of Designer Peptide Amphiphiles. Chem. Soc. Rev. 2010, 39, 3480–3498. 10.1039/b915923c. [DOI] [PubMed] [Google Scholar]
- Chen C.; Pan F.; Zhang S.; Hu J.; Cao M.; Wang J.; Xu H.; Zhao X.; Lu J. R. Antibacterial Activities of Short Designer Peptides: A Link Between Propensity for Nanostructuring and Capacity for Membrane Destabilization. Biomacromolecules 2010, 11, 402–411. 10.1021/bm901130u. [DOI] [PubMed] [Google Scholar]
- Castelletto V.; Nutt D. R.; Hamley I. W.; Bucak S.; Cenker Ç.; Olsson U. Structure of Single-Wall Peptide Nanotubes: In Situ Flow Aligning X-Ray Diffraction. Chem. Commun. 2010, 46, 6270–6272. 10.1039/c0cc00212g. [DOI] [PubMed] [Google Scholar]
- Han S.; Cao S.; Wang Y.; Wang J.; Xia D.; Xu H.; Zhao X.; Lu J. R. Self-Assembly of Short Peptide Amphiphiles: The Cooperative Effect of Hydrophobic Interaction and Hydrogen Bonding. Chem.—Eur. J. 2011, 17, 13095–13102. 10.1002/chem.201101970. [DOI] [PubMed] [Google Scholar]
- Hamley I. W. Self-Assembly Of Amphiphilic Peptides. Soft Matter 2011, 7, 4122–4138. 10.1039/c0sm01218a. [DOI] [Google Scholar]
- Cenker Ç. Ç.; Bucak S.; Olsson U. Nanotubes and Bilayers in a Model Peptide System. Soft Matter 2011, 7, 4868–4875. 10.1039/c0sm01186j. [DOI] [Google Scholar]
- Middleton D. A.; Castelletto V.; Hamley I. W.; Madine J. New Insights into the Molecular Architecture of a Peptide Nanotube using FTIR and Solid-State NMR Combined with Sample Alignment. Angew. Chem., Int. Ed. 2013, 52, 10537–10540. 10.1002/anie.201301960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamley I. W.; Dehsorkhi A.; Castelletto V. Self-Assembled Arginine-Coated Peptide Nanosheets in Water. Chem. Commun. 2013, 49, 1850–1852. 10.1039/c3cc39057h. [DOI] [PubMed] [Google Scholar]
- Castelletto V.; Hamley I. W.; Segarra-Maset M. D.; Gumbau C. B.; Miravet J. F.; Escuder B.; Seitsonen J.; Ruokolainen J. Tuning Chelation by the Surfactant-Like Peptide A6H Using Predetermined pH Values. Biomacromolecules 2014, 15, 591–598. 10.1021/bm401640j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Wang J.; Deng L.; Zhou P.; Wang S.; Wang Y.; Xu H.; Lu J. R. Tuning The Self-Assembly of Short Peptides via Sequence Variations. Langmuir 2013, 29, 13457–13464. 10.1021/la402441w. [DOI] [PubMed] [Google Scholar]
- Dehsorkhi A.; Castelletto V.; Hamley I. W. Self-Assembling Amphiphilic Peptides. J. Pept. Sci. 2014, 20, 453–467. 10.1002/psc.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletto V.; Gouveia R. M.; Connon C. J.; Hamley I. W.; Seitsonen J.; Nykänen A.; Ruokolainen J. Alanine-Rich Amphiphilic Peptide Containing The RGD Cell Adhesion Motif: a Coating Material for Human Fibroblast Attachment and Culture. Biomater. Sci. 2014, 2, 362–369. 10.1039/c3bm60232j. [DOI] [PubMed] [Google Scholar]
- Hamley I. W. Lipopeptides: From Self-Assembly to Bioactivity. Chem. Commun. 2015, 51, 8574–8583. 10.1039/c5cc01535a. [DOI] [PubMed] [Google Scholar]
- Hamley I. W.; Hutchinson J.; Kirkham S.; Castelletto V.; Kaur A.; Reza M.; Ruokolainen J. Nanosheet Formation by an Anionic Surfactant-Like Peptide and Modulation of Self-Assembly Through Ionic Complexation. Langmuir 2016, 32, 10387–10393. 10.1021/acs.langmuir.6b02180. [DOI] [PubMed] [Google Scholar]
- Dehsorkhi A.; Castelletto V.; Hamley I. W.; Seitsonen J.; Ruokolainen J. Interaction Between a Cationic Surfactant-Like Peptide and Lipid Vesicles and its Relationship to Antimicrobial Activity. Langmuir 2013, 29, 14246–14253. 10.1021/la403447u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletto V.; Barnes R. H.; Karatzas K.-A.; Edwards-Gayle C. J. C.; Greco F.; Hamley I. W.; Rambo R.; Seitsonen J.; Ruokolainen J. Arginine-Containing Surfactant-Like Peptides: Interaction with Lipid Membranes and Antimicrobial Activity. Biomacromolecules 2018, 19, 2782–2794. 10.1021/acs.biomac.8b00391. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Deng L.; Yang W.; Wang D.; Pambou E.; Lu Z.; Li Z.; Wang J.; King S.; Rogers S.; Xu H.; Lu J. R. Tuning One-Dimensional Nanostructures of Bola-Like Peptide Amphiphiles by Varying the Hydrophilic Amino Acids. Chem.—Eur. J. 2016, 22, 11394–11404. 10.1002/chem.201601309. [DOI] [PubMed] [Google Scholar]
- da Silva E. R.; Walter M. N. M.; Reza M.; Castelletto V.; Ruokolainen J.; Connon C. J.; Alves W. A.; Hamley I. W. Self-Assembled Arginine-Capped Peptide Bolaamphiphile Nanosheets for Cell Culture and Controlled Wettability Surfaces. Biomacromolecules 2015, 16, 3180–3190. 10.1021/acs.biomac.5b00820. [DOI] [PubMed] [Google Scholar]
- Hamley I. W.; Burholt S.; Hutchinson J.; Castelletto V.; Da Silva E. R.; Alves W.; Gutfreund P.; Porcar L.; Dattani R.; Hermida-Merino D.; Newby G.; Reza M.; Ruokolainen J.; Stasiak J. Shear Alignment of Bola-Amphiphilic Arginine-Coated Peptide Nanotubes. Biomacromolecules 2017, 18, 141–149. 10.1021/acs.biomac.6b01425. [DOI] [PubMed] [Google Scholar]
- Castelletto V.; Barnes R. H.; Karatzas K.-A.; Edwards-Gayle C. J. C.; Greco F.; Hamley I. W.; Seitsonen J.; Ruokolainen J. Restructuring of Lipid Membranes by an Arginine-Capped Peptide Bolaamphiphile. Langmuir 2019, 35, 1302–1311. 10.1021/acs.langmuir.8b01014. [DOI] [PubMed] [Google Scholar]
- Castelletto V.; Gouveia R. J.; Connon C. J.; Hamley I. W.; Seitsonen J.; Ruokolainen J.; Longo E.; Siligardi G. Influence of Elastase on Alanine-Rich Peptide Hydrogels. Biomater. Sci. 2014, 2, 867–874. 10.1039/c4bm00001c. [DOI] [PubMed] [Google Scholar]
- Beter M.; Kara H. K.; Topal A. E.; Dana A.; Tekinay A. B.; Guler M. O. Multivalent Presentation of Cationic Peptides on Supramolecular Nanofibers for Antimicrobial Activity. Mol. Pharmaceutics 2017, 14, 3660–3668. 10.1021/acs.molpharmaceut.7b00434. [DOI] [PubMed] [Google Scholar]
- Bai J.; Chen C.; Wang J.; Zhang Y.; Cox H.; Zhang J.; Wang Y.; Penny J.; Waigh T.; Lu J. R.; Xu H. Enzymatic Regulation of Self-Assembling Peptide A9K2 Nanostructures and Hydrogelation with Highly Selective Antibacterial Activities. ACS Appl. Mater. Interfaces 2016, 8, 15093–15102. 10.1021/acsami.6b03770. [DOI] [PubMed] [Google Scholar]
- Dickinson E. Food Emulsions and Foams: Stabilization by Particles. Curr. Opin. Colloid Interface Sci. 2010, 15, 40–49. 10.1016/j.cocis.2009.11.001. [DOI] [Google Scholar]
- Tcholakova S.; Denkov N. D.; Lips A. Comparison of Solid Particles, Globular Proteins and Surfactants as Emulsifiers. Phys. Chem. Chem. Phys. 2008, 10, 1608–1627. 10.1039/b715933c. [DOI] [PubMed] [Google Scholar]
- Lam S.; Velikov K. P.; Velev O. D. Pickering Stabilization of Foams and Emulsions with Particles of Biological Origin. Curr. Opin. Colloid Interface Sci. 2014, 19, 490–500. 10.1016/j.cocis.2014.07.003. [DOI] [Google Scholar]
- Bai S.; Pappas C.; Debnath S.; Frederix P. W. J. M.; Leckie J.; Fleming S.; Ulijn R. V. Stable Emulsions Formed by Self-Assembly of Interfacial Networks Of Dipeptide Derivatives. ACS Nano 2014, 8, 7005–7013. 10.1021/nn501909j. [DOI] [PubMed] [Google Scholar]
- Scott G. G.; Mcknight P. J.; Tuttle T.; Ulijn R. V. Tripeptide Emulsifiers. Adv. Mater. 2016, 28, 1381–1386. 10.1002/adma.201504697. [DOI] [PubMed] [Google Scholar]
- Moreira I. P.; Piskorz T. K.; van Esch J. H.; Tuttle T.; Ulijn R. V. Biocatalytic Self-Assembly of Tripeptide Gels and Emulsions. Langmuir 2017, 33, 4986–4995. 10.1021/acs.langmuir.7b00428. [DOI] [PubMed] [Google Scholar]
- Pozo Navas B.; Lohner K.; Deutsch G.; Sevcsik E.; Riske K. A.; Dimova R.; Garidel P.; Pabst G. Composition Dependence of Vesicle Morphology and Mixing Properties in a Bacterial Model Membrane System. Biochim. Biophys. Acta, Biomembr. 2005, 1716, 40–48. 10.1016/j.bbamem.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Radford S. J.; Dickinson E.; Golding M. Stability And Rheology Of Emulsions Containing Sodium Caseinate: Combined Effects of Ionic Calcium and Alcohol. J. Colloid Interface Sci. 2004, 274, 673–686. 10.1016/j.jcis.2003.12.045. [DOI] [PubMed] [Google Scholar]
- Levine H. Thioflavine-T Interaction With Synthetic Alzheimers-Disease Beta-Amyloid Peptides- Detection of Amyloid Aggregation in Solution. Protein Sci. 1993, 2, 404–410. 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine H.Quantification of β-Sheet Amyloid Fibril Structures with Thioflavin T. In Methods in Enzymology; Wetzel R., Ed.; Academic Press: San Diego, 1999; Vol. 309, pp 274–284. [DOI] [PubMed] [Google Scholar]
- http://www.bachem.com/Service-Support/Peptide-Calculator/. Accessed in 2019.
- Haris P. I.; Chapman D. The Conformational Analysis of Peptides Using Fourier Transform IR Spectroscopy. Biopolymers 1995, 37, 251–263. 10.1002/bip.360370404. [DOI] [PubMed] [Google Scholar]
- Adochitei A.; Drochioiu G. Rapid Characterization of Peptide Secondary Structure by FT-IR Spectroscopy. Rev. Roum. Chim. 2011, 56, 783–791. [Google Scholar]
- Pelton J. T.; Mclean L. R. Spectroscopic Methods for Analysis of Protein Secondary Structure. Anal. Biochem. 2000, 277, 167–176. 10.1006/abio.1999.4320. [DOI] [PubMed] [Google Scholar]
- Gaussier H.; Morency H.; Lavoie M. C.; Subirade M. Replacement of Trifluoroacetic Acid with HCl in the Hydrophobic Purification Steps of Pediocin PA-1: a Structural Effect. Appl. Environ. Microbiol. 2002, 68, 4803–4808. 10.1128/aem.68.10.4803-4808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eker F.; Griebenow K.; Schweitzer-Stenner R. Aβ1-28Fragment of the Amyloid Peptide Predominantly Adopts a Polyproline II Conformation in an Acidic Solution. Biochemistry 2004, 43, 6893–6898. 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- Bressler I.; Kohlbrecher J.; Thünemann A. F. Sasfit: A Tool for Small-Angle Scattering Data Analysis Using a Library of Analytical Expressions. J. Appl. Crystallogr. 2015, 48, 1587–1598. 10.1107/s1600576715007347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E.Proteins. Structures and Molecular Properties; W.H.Freeman: New York, 1993. [Google Scholar]
- Pabst G.; Rappolt M.; Amenitsch H.; Laggner P. Structural information from multilamellar liposomes at full hydration: Fullq-range fitting with high quality x-ray data. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2000, 62, 4000–4009. 10.1103/physreve.62.4000. [DOI] [PubMed] [Google Scholar]
- The International Association For The Properties Of Water And Steam. http://www.iapws.org/. Accessed in 2019.
- De Leon Rodriguez L. M.; Hemar Y.; Cornish J.; Brimble M. A. Structure-Mechanical Property Correlations of Hydrogel Forming Beta-Sheet Peptides. Chem. Soc. Rev. 2016, 45, 4797–4824. 10.1039/c5cs00941c. [DOI] [PubMed] [Google Scholar]
- Costerton J. W.; Stewart P. S.; Greenberg E. P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Stewart P. S.; William Costerton J. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Lakemeyer M.; Zhao W.; Mandl F. A.; Hammann P.; Sieber S. A. Thinking Outside the Box-Novel Antibacterials to Tackle the Resistance Crisis. Angew. Chem., Int. Ed. 2018, 57, 14440–14475. 10.1002/anie.201804971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.