Abstract
Combinations of BRAF inhibitors and MEK inhibitors (BRAFi + MEKi) are FDA-approved to treat BRAF V600E/K mutant melanoma. Efficacy of BRAFi + MEKi associates with cancer cell death and alterations in the tumor immune microenvironment; however, the links are poorly understood. We show that BRAFi + MEKi caused durable melanoma regression in an immune-mediated manner. BRAFi + MEKi treatment promoted cleavage of gasdermin E (GSDME) and release of high mobility group protein B1 (HMGB1), markers of pyroptotic cell death. GSDME-deficient melanoma showed defective HMGB1 release, reduced tumor-associated T cell and activated dendritic cell infiltrates in response to BRAFi + MEKi, and more frequent tumor regrowth after drug removal. Importantly, BRAFi + MEKi-resistant disease lacked pyroptosis markers, showed decreased intra-tumoral T cell infiltration but was sensitive to pyroptosis-inducing chemotherapy. These data implicate BRAFi + MEKi-induced pyroptosis in anti-tumor immune responses and highlight new therapeutic strategies for resistant melanoma.
Keywords: Melanoma, MEK, resistance, BRAF, pyroptosis, gasdermin E
INTRODUCTION
Melanoma represents a small fraction of cutaneous malignancies yet accounts for the majority of skin cancer-related mortalities (1). Agents targeting the MEK-ERK1/2 pathway or immune checkpoints have emerged as effective treatment modalities that significantly improve progression-free survival and overall survival for stage III and stage IV melanoma patients (2–4). Targeted therapy elicits high response rates with the majority of BRAF V600E/K mutant melanoma patients exhibiting tumor shrinkage in response to the combination of BRAF and MEK inhibitors (BRAFi + MEKi). A limitation of targeted therapies is that tumors frequently recur within 13 months (5). Acquired resistance is often due to re-activation of the MEK-ERK1/2 pathway caused by mechanisms including NRAS mutation, increased BRAF copy number and aberrant BRAF splicing (5–7). Immune checkpoint inhibitors have come to the forefront of melanoma treatment, as they reverse dysfunctional anti-tumor T cell states and induce durable anti-tumor responses in ~50% of patients (8).
Given the clinical momentum in combining these two classes of therapies, it is important to understand the actions of targeted therapies on the tumor immune microenvironment. BRAFi and/or MEKi are known to induce anti-tumor immune responses. BRAFi increase MHC expression and induce CD4+ and CD8+ T cell-dependent anti-tumor immunity (9–19). Furthermore, MEKi improve anti-cancer T cell responses by impairing T-cell receptor (TCR)-mediated apoptosis of tumor antigen-specific T cells (19–23). Generally, BRAFi and/or MEKi efficacy correlates with T cell infiltration of tumors, while the loss of intra-tumoral CD8+ T cells and influx of tumor-associated macrophages are associated with acquired resistance in metastatic melanoma (10,17,19,24). Despite this knowledge, the mechanisms by which targeted inhibitors affect the phenotype and function of tumor-associated T cells are incompletely understood. Furthermore, the functional relationship between BRAFi + MEKi-mediated tumor cell death and alterations in the tumor immune environment remains to be elucidated.
It is well established that BRAFi and/or MEKi cause programmed cell death of BRAF V600E mutant melanoma cells. Mechanistically, inhibition of MEK-ERK1/2 signaling induces BIM-EL and BMF-mediated mitochondrial depolarization, leading to cytochrome C release and activation of caspase-3 (16,25–27). It has recently been shown that the intrinsic apoptotic pathway intersects with a distinct form of cell death termed pyroptosis that is gasdermin-mediated and involves pore-based release of immune stimulatory factors (28–31). We and others have demonstrated that caspase-3 cleavage leads to pyroptosis by inducing gasdermin E (GSDME or DFNA5) cleavage and subsequent pore formation within the plasma membrane (31–34). This pore formation causes the release of immune stimulants including HMGB1, which are able to induce dendritic cell (DC) activation and, in turn, propagate anti-tumor T cell activity (32,33,35). Cleaved gasdermin E also permeates the mitochondria to positively feedback to the intrinsic apoptotic pathway (32,34). Recent evidence shows MEKi-induced GSDME cleavage in lung cancer cell lines (36); however, how these effects contributed to anti-tumor immune responses remained unclear. We hypothesized that targeted inhibitor-mediated pyroptosis leads to activation of anti-tumor immune responses in BRAF mutant melanoma.
In this study, we used human and syngeneic mouse melanoma models to analyze GSDME-associated pyroptosis as it relates to efficacy of BRAFi + MEKi treatment and modulation of the tumor immune microenvironment. We demonstrated that therapeutic efficacy of BRAFi + MEKi is modulated by a functional immune system, specifically CD4+ and CD8+ T cells. Treatment-induced HMGB1 release, tumor-associated T cell alterations and tumor eradication were dependent on GSDME. Conversely, BRAFi + MEKi-resistant tumors did not undergo pyroptosis and lacked robust T cell responses. Finally, restoring GSDME cleavage and HMGB1 release delayed the growth of BRAFi + MEKi-resistant tumors. These data define a novel mechanism connecting BRAFi + MEKi-induced pyroptosis to immune responses and present new salvage options for targeted therapy-resistant melanoma.
RESULTS
Therapeutic efficacy of BRAFi + MEKi combination treatment in vivo depends on an intact immune system
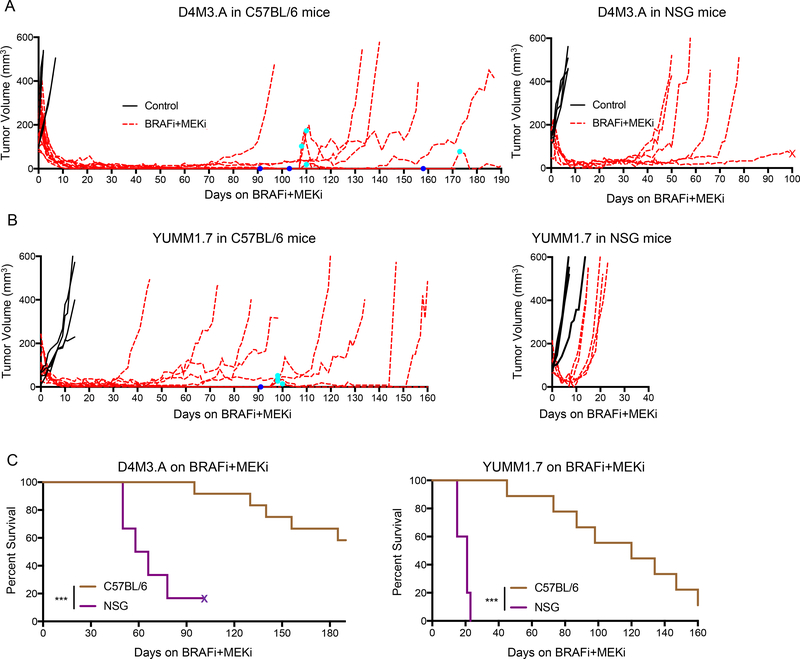
Acquired resistance to BRAFi + MEKi treatment is accompanied by reduced intra-tumoral infiltration of T cells (17). To ascertain the functional contribution of the immune system in BRAFi + MEKi therapeutic efficacy, we compared tumor responses in syngeneic Braf V600E mouse melanoma allografts of D4M3.A and YUMM1.7 cells (37,38). Intradermal tumors were established in either immunocompetent (C57BL/6 mice) or immune-deficient (NOD scid gamma, NSG) mice and mice treated with/without BRAFi + MEKi. D4M3.A tumors in either immunocompetent C57BL/6 mice or immune-deficient NSG mice showed a robust tumor regression following BRAFi + MEKi treatment (Fig. 1A). However, BRAFi + MEKi induced prolonged tumor regressions in C57BL/6 mice with tumors taking an average of 138 days to re-grow to 200 mm3 compared to short-term regressions averaging 57.4 days in NSG mice (Fig. 1A). In a second model, YUMM1.7 tumors took an average of 104.2 days to regrow to 200 mm3 following initial regressions in C57BL/6 mice compared to 16.8 days in NSG mice (Fig. 1A, B). Immunocompetent mice lacking palpable lesions after ≥ 90 days of treatment, regrew tumors when taken off BRAFi + MEKi, indicating the presence of residual disease (dark blue dots, Fig. 1A, B and Supp. Fig. 1A). These tumors regressed upon re-administration of BRAFi + MEKi (cyan dots). The differences in tumor growth kinetics in different mouse strains was not attributed to altered baseline tumor growth rates (Supp. Fig. 1B). Importantly, we observed extended overall survival of BRAFi + MEKi-treated C57BL/6 mice compared to NSG mice when utilizing both the D4M3.A and YUMM1.7 models (Fig. 1C). Together, these data suggest that an intact immune system significantly contributes to the therapeutic efficacy of BRAFi + MEKi.
Figure 1: Time to acquired resistance of BRAFi + MEKi is immune-mediated.
A) Male C57BL/6 or NSG mice were intradermally implanted with D4M3.A (3×105) mouse Braf V600E melanoma cells. Tumors were grown to ~50–250 mm3 after which animals were given either control (AIN-76A) or PLX4720 and PD0325901 (200 ppm PLX4720 and 7 ppm PD0325901 AIN-76A) laced chow (BRAFi + MEKi). Tumor growth, represented as the change in volume (mm3) over time, is shown from the start of treatment. Dark blue dots indicate removal of combination chow due to lack of visible tumors and cyan dots indicate when combination chow was restarted due to recurrent, visible tumors. B) Same as A, except that YUMM1.7 (2.5×105) cells were injected. C) Survival curves of D4M3.A or YUMM1.7 tumor-bearing, C57BL/6 or NSG mice treated with BRAFi + MEKi. Significance was determined by a log-rank test, ***p<0.001. X’s indicate if mice died for a non-experiment related reason.
T cells are required for sustained tumor growth inhibition by BRAFi + MEKi
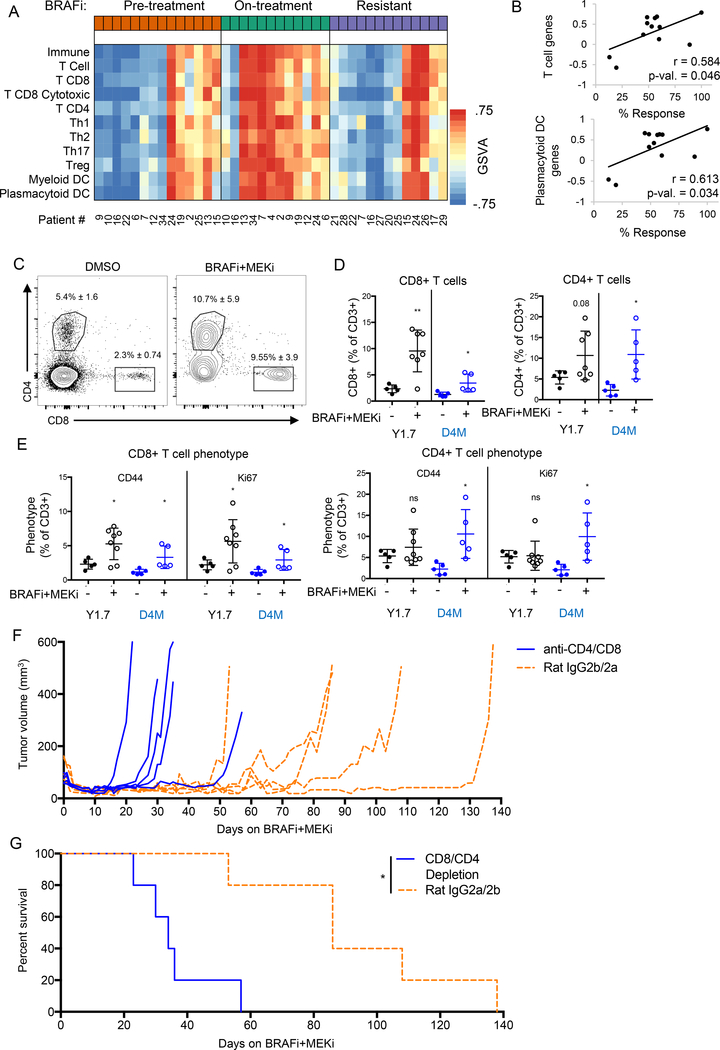
Next, we determined how ERK1/2 pathway inhibition affected the immune cell composition in patient tumors. RNA-seq analysis of BRAFi-treated patient tumors (European Genome-phenome Archive, EGAS000010000992 (39)) highlighted gene signatures consistent with increased expression of T cell and dendritic cell (DC) infiltration in on-treatment samples (Fig. 2A). These findings were contrasted by lower intra-tumoral immune transcript abundance upon tumor relapse. Expression levels of genes associated with T cells and plasmacytoid DCs positively correlated with % tumor response in this patient population (Fig. 2B and Supp. Fig. 1C). These data suggest that tumor-associated T cells and DCs associate with the efficacy of ERK1/2 pathway inhibition in BRAF mutant melanoma patients.
Figure 2: BRAFi + MEKi is T cell-mediated.
A) Heat map of GSVA scores for immune cell gene sets from patient tumors before and during BRAFi, and after the onset of resistance to BRAFi. Patient indicator numbers are included below. B) Scatter plots of GSVA scores and percent tumor regression data for on-treatment samples. Pearson correlation coefficient (r) and p-values are displayed. C) YUMM1.7 (Y1.7, black) or D4M3.A (D4M, blue) tumor-bearing mice were treated PLX4720 (1 μM) and PD0325901 (35 nM). Cohorts of mice were sacrificed pre-treatment or after four days of BRAFi + MEKi treatment, and intra-tumoral T cells were assessed by FACS. Representative FACS plots of tumor-associated CD8+ and CD4+ T cells (of CD3+ cells). D) Quantification of FACS plots of YUMM1.7 (Y1.7, black) or D4M3.A (D4M, blue) tumors. E) Phenotype of tumor-associated T cells, CD44 = activated, Ki67 = proliferating. F) Tumor growth during 4BRAFi + MEKi of CD4 and CD8-depleted mice compared to their appropriate isotype controls. G) Kaplan-Meier survival plot of mice from F. Significance was determined by a log-rank test, *p<0.05.
To further characterize alterations, we analyzed the immune cell infiltrates of syngeneic tumors harvested either pre-treatment or after four days of BRAFi + MEKi treatment. Compared to untreated controls, the proportion of CD8+ and CD4+ cells among CD3+ T cells was increased in YUMM1.7 and D4M3.A tumors treated with BRAFi + MEKi (Fig. 2C–D). The overall amount of CD3+ cells slightly decreased within the tumor and the percentage of CD8+ and CD4+ T cells of total cells were not affected (Supp. Fig. 2 and Supp. Fig. 3A). Furthermore, we observed higher levels of activated (CD44+) and proliferating (Ki67+) T cells in tumors of BRAFi + MEKi-treated mice compared to tumors from untreated mice (Fig. 2E and Supp. Fig. 2). Treatment-associated changes in T cell abundance were selective to the tumor in that they were not observed in the spleen (Supp. Fig. 3B–C). Markers of T cell function, IFNγ and IL-2, were unchanged following BRAFi + MEKi treatment and, by contrast, the production of TNFα from intra-tumoral and splenic CD4+ and CD8+ T cells was decreased (Supp. Fig. 3D–E).
To assess the functional contribution of T cells to tumor regression and acquired resistance following BRAFi + MEKi, we depleted CD4+ and CD8+ T cells in YUMM1.7 tumor-bearing mice (Supp. Fig. 3F–G). Concurrent depletion of CD4+ and CD8 + T cells significantly shortened the time to tumor growth (Fig. 2F) and reduced survival of mice following BRAFi + MEKi (Fig. 2G). These data suggest that T cells contribute to maintaining tumor regressions caused by BRAFi + MEKi therapy.
To characterize other effects of BRAFi + MEKi on the tumor immune microenvironment, we analyzed myeloid-derived cells and a panel of markers for immune activity (MHC-I, MHC-II; PD-L1, IDO-1, FasL, GalS9, and OX40L). Both tumor-associated macrophages (TAM) and myeloid-derived suppressor cells (MDSCs) were decreased intra-tumorally during BRAFi + MEKi treatment (Supp. Fig. 3H–I). We also observed a consistent decrease in the expression of IDO-1, FasL, LGasS9, and OX40L on CD45.2-negative and CD45.2-positive cells within tumors (Supp. Fig. 3J–M). Together, these data suggest that BRAFi + MEKi treatment reduce the immune-suppressive cells in the microenvironment of Braf V600E melanomas.
BRAFi + MEKi induces markers of immune stimulatory cell death in Braf V600E melanomas
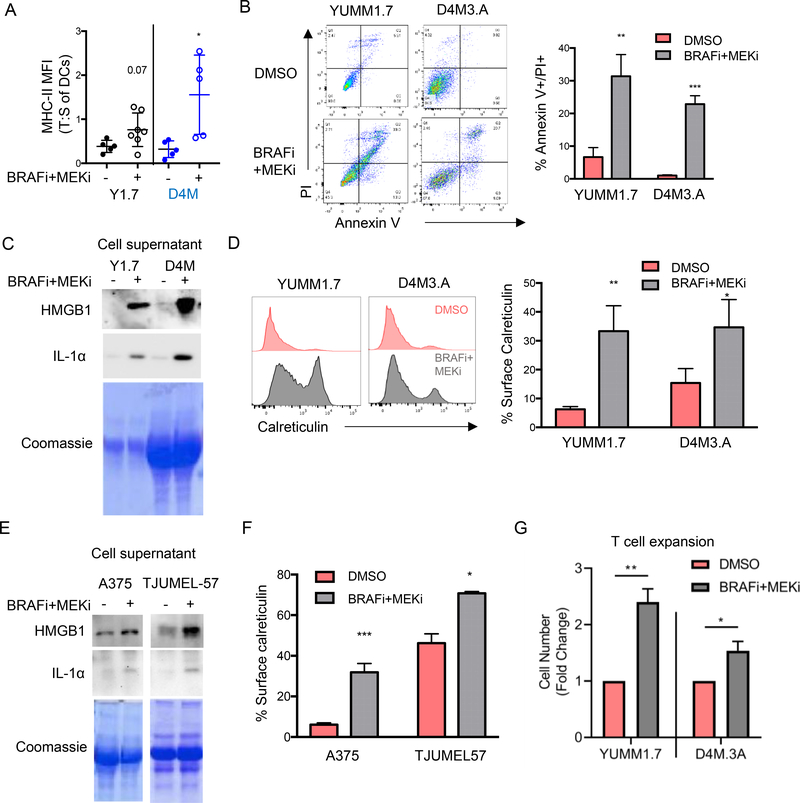
The basis of BRAFi + MEKi-mediated T cell activation is unclear. We detected increased expression of the activation marker MHC-II on tumor-infiltrating DCs following BRAFi + MEKi treatment from both YUMM1.7 and D4M3.A tumors (Fig. 3A and Supp. Fig. 4A). These data are consistent with the possibility that activated DCs contribute to the T cell expansion observed during BRAFi + MEKi treatment. Next, we analyzed the release and cell surface expression of immune stimulants from dying cells that could potentiate anti-tumor immune responses in part via effects on DCs, specifically HMGB1 and calreticulin (24, 25). As expected treatment of YUMM1.7 and D4M3.A mono-cultures with BRAFi + MEKi in vitro increased cell death as determined by annexin V staining and propidium iodide (PI) uptake compared to DMSO-treated cells (Fig. 3B). BRAFi + MEKi-mediated tumor cell death correlated with release of HMGB1 from cells (Fig. 3C and Supp. Fig. 4B). The release of additional inflammatory factors has been associated with of pyroptotic cell death (40,41) and we did detect BRAFi + MEKi-dependent release of another inflammatory mediator, IL-1α, from melanoma cells (Fig. 3C). In addition, BRAFi + MEKi treatment increased cell surface expression of calreticulin (Fig. 3D). Similar results were obtained in human BRAF V600E melanoma cells including A375, a frequently used cell line and TJUMEL-57, a short-term ex vivo culture of a patient melanoma tumor (Fig. 3E–F and Supp. Fig. 4C). These data suggest that combination BRAFi + MEKi treatment induces immune stimulatory forms of cell death.
Figure 3: BRAFi + MEKi-induces immune stimulatory cell death.
A) Activation status (ratio of MHC-II MFI in tumors compared to spleens) of F4/80 -ve, CD11c +ve, CD11b -ve DCs in tumors during BRAFi + MEKi treatment. B) YUMM1.7 or D4M3.A cells were treated with PLX4720 (1 μM) and PD0325901 (35 nM) in vitro (n=3) for 72 hours (B) or 24 hours (C-D). Cell death as indicated by % annexin V+ and PI+ cells after 72 hours of treatment (representative gating on left, quantitation on right). C) Levels of HMGB1 and IL-1α in supernatant from YUMM1.7 or D4M3.A cells from B. Coomassie stained gel as loading control. D) Calreticulin surface expression of YUMM1.7 and D4M3.A mouse melanoma cell lines (left shows representative plots and right shows percentages). E) Same as C for A375 cells or short-term patient tumor cells (TJUMEL57) treated with BRAFi + MEKi for 48 hours (n=3). F) Same as D for human cells treated with BRAFi + MEKi for 48 hours. G) Pmel-I T cells were expanded ex vivo for 5 days in the presence of supernatant from YUMM1.7 or D4M3.A cells treated with DMSO or PLX4720 (1 μM) and PD0325901 (35 nM) for 48 hours. Shown is the cell number represented as a ratio of BRAFi + MEKi-treated:control supernatants (n=3). Statistical analysis was completed by student’s T-test, *p<0.05, **p<0.01.
Next, we determined whether tumor cell death was immune stimulatory in terms of inducing T cell expansion. To this end, we cultured splenocytes from Pmel-I mice (a transgenic mouse model with T cells specific for the melanoma antigen gp100) with conditioned medium from BRAFi + MEKi-treated tumor cells (42). This system incubates antigen presenting cells, like DCs, with the immune stimulants present in the supernatant of BRAFi + MEKi-treated melanoma cells, allowing us to test for promotion of T cell expansion. The addition of conditioned medium from BRAFi + MEKi-treated YUMM1.7 or D4M3.A cells increased T cell proliferation compared to medium from DMSO-treated cells (Fig. 3G). These data suggest that during BRAFi + MEKi treatment, dying melanoma cells release factors that promote T cell expansion.
BRAFi + MEKi mediates GSDME-dependent pyroptosis
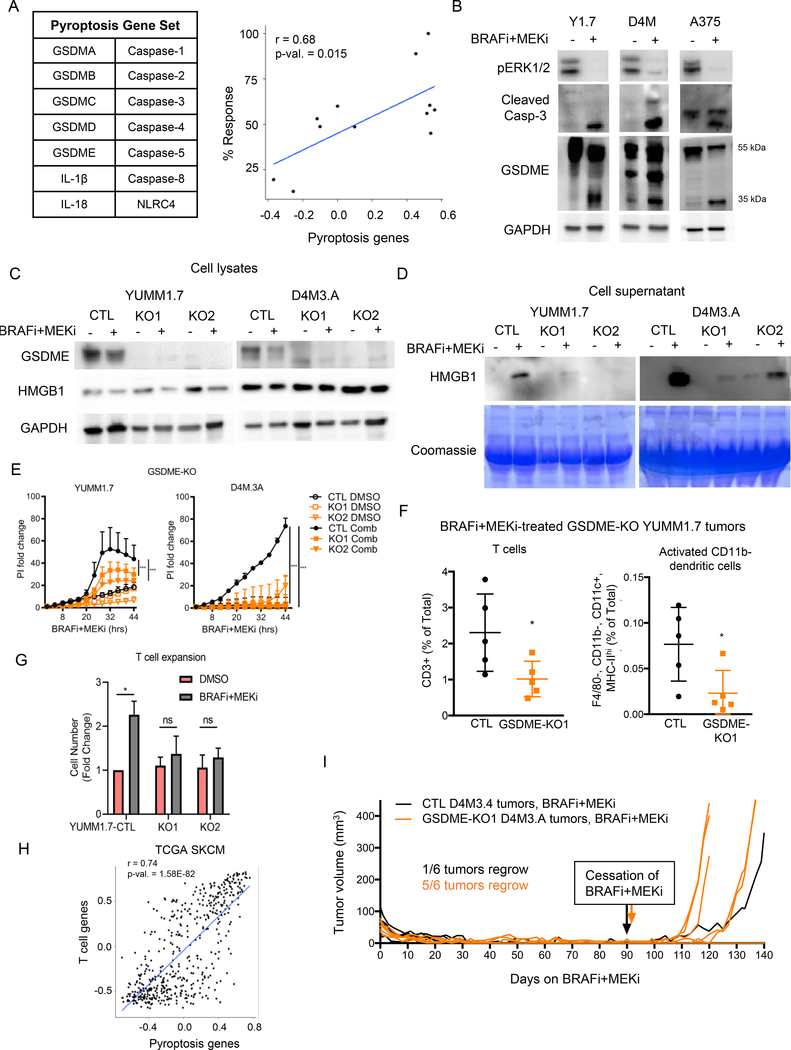
Given our data indicating that BRAFi + MEKi-induced release of factors promotes T cell expansion, we hypothesized that BRAFi + MEKi cause pyroptotic cell death. From RNA-seq datasets from BRAFi +/− MEKi-treated melanoma patient samples (EGAS000010000992 (39)), we determined that a pyroptosis gene set positively correlated with percent tumor response to targeted therapy (Fig. 4A), further suggesting BRAFi + MEKi induces pyroptotic cell death. Expression of pyroptosis genes was increased on-treatment in partial and complete responders (Supp. Fig. 4D). We examined a second dataset (GSE99898; (43)), containing patient-matched pre-treatment, on-treatment and progression samples but lacking initial response data. This analysis also showed up-regulation of the pyroptosis signature on-treatment and that was decreased following onset of resistance (Supp. Fig. 4E). We have recently shown that pyroptosis can be initiated by caspase-3-mediated GSDME cleavage in response to various apoptotic stimuli (32,34). Since BRAFi + MEKi treatment induces caspase-3 activation (44), we tested for GSDME processing in melanoma cell lines. In addition to decreased phospho-ERK1/2, and increased cleavage of caspase-3, BRAFi + MEKi treatment caused the production of the 35 kDa GSDME cleavage fragment in mouse and human melanoma cells (Fig. 4B and Supp. Fig. 5A). Cleavage of caspase 3 and GSDME by the combination was dose-dependent (Supp. Fig. 5B) and BRAFi and MEKi were individually able to induce GSDME cleavage but to a lesser extent than the combination (Supp. Fig. 5C).
Figure 4: BRAFi + MEKi-induces GSDME-dependent pyroptosis.
A) Pyroptosis genes (left) and scatter plot of pyroptosis genes GSVA scores and percent tumor regression data for BRAFi and BRAFi + MEKi on-treatment samples from patient tumors. Pearson correlation coefficient (r) and p-values are displayed (right). B) Levels of pERK1/2, cleaved caspase-3 and GSDME in the cell lysates from mouse YUMM1.7 and D4M3.A treated for 24 hours and human A375 cells treated for 48 hours with PLX4720 (1 μM) and PD0325901 (35 nM). HMGB1 and GAPDH as loading controls. Full-length GSDME runs at 55 kDa and cleaved GSDME runs at 35 kDa. C) Full-length GSDME and cellular HMGB1 levels in empty vector controls (CTL) and GSDME CRISPR knockout (KO1 and KO2) YUMM1.7 or D4M3.A cells were analyzed by Western blot. GAPDH as loading control, n=3–4. D) Level of secreted HMGB1 in supernatants from CTL, KO1 or KO2 YUMM1.7 or D4M3.A cells treated with PLX4720 (1 μM) or PD0325901 (35 nM) for 16 hours. Coomassie stained gel shows protein loading, n=3. E) PI incorporation over time, normalized to 0 hours for CTL, KO1 or KO2 cell lines treated with either DMSO or BRAFi + MEKi (n=3, data are representative of two independent experiments). Significance determined by area under the curve, ***p<0.001. F) T Cells (CD3+) and activated CD11b- DCs (F4/80-, CD11b-, CD11c+, MHC-IIhi) in CTL or KO1 YUMM1.7 tumors four days after beginning BRAFi + MEKi treatment. Statistical analysis was completed by student’s t-test, *p<0.05. G) T cell expansion stimulated by diafiltered supernatants from control and GSDME-KO treated cells. Fold change of PmeI-I T cell numbers after 5 days is shown (n=3). Statistical analysis by student’s T-test *p<0.05. H) Scatter plot comparing GSVA signature scores using RNA-seq data from TCGA cutaneous melanoma patient samples. Pearson correlation coefficient (r) and p-values are displayed. I) Tumor growth of CTL or KO1 D4M3.A treated with BRAFi + MEKi in C57BL/6 mice. Mice were removed from BRAFi + MEKi at day 91 (CTL) and day 92 (sg1GSDME) when tumors were undetectable.
To test for requirement, we reduced GSDME expression by knockdown (siRNA) and GSDME-knockout (KO) using two different GSDME CRISPR/Cas9 guide sequences. GSDME knockdown/KO decreased the release of HMGB1 from melanoma cells into the supernatant, indicating inhibition of pyroptosis (Fig. 4C–D and Supp. Fig. 5D–G). GSDME knockout did not alter the level of cellular HMGB1 expression (Fig. 4C). To further test if GSDME was required for pyroptosis, we measured PI uptake in BRAFi + MEKi-treated GSDME-KO D4M3.A and YUMM1.7 cell lines (32,34). GSDME-KO cells internalized less PI than control cells, suggesting a reduction of pyroptosis levels (Fig. 4E). Together, these data imply that BRAFi + MEKi induced GSDME-mediated pyroptotic cell death.
To determine if the anti-tumor immune responses observed during BRAFi + MEKi were dependent on mediators of pyroptosis, we compared tumor-infiltrating lymphocytes (TIL) populations from CTL or GSDME-KO tumors treated for four days with BRAFi + MEKi. Higher levels of T cells and activated DCs (MHC-II+; CD11b-) were detected in CTL tumors in comparison with the GSDME-KO counterparts (Fig. 4F and Supp. Fig. 5H). Furthermore, diafiltered medium from BRAFi + MEKi-treated GSDME-KO cells was ineffective at promoting T cell expansion (Fig. 4G). These data provide evidence that BRAFi + MEKi-induced anti-tumor immune responses are dependent on pyroptotic cell death. In support of this notion, RNA-seq analysis of cutaneous melanoma samples from The Cancer Genome Atlas (TCGA) displayed a positive correlation between T cell genes and pyroptosis genes (Fig. 4H). To confirm whether GSDME impacts on tumor growth during treatment, we measured the size of BRAFi + MEKi-treated CTL and GSDME-KO D4M3.A tumors (Supp Fig. 5I). There was no noticeable difference in the initial regression of CTL or GSDME-KO tumors treated with BRAFi + MEKi (Fig. 4I). However, when testing for the re-growth of residual disease by removing drug from mice lacking palpable lesions, only 1 of 6 (16.7%) of CTL tumors regrew whereas 5 of 6 (83.3%) of GSDME-KO tumors regrew (Fig. 4I). These data suggest that GSDME-dependent pyroptosis is associated with lower levels of residual disease after BRAFi + MEKi treatment.
BRAFi + MEKi combination resistant cells do not undergo pyroptosis
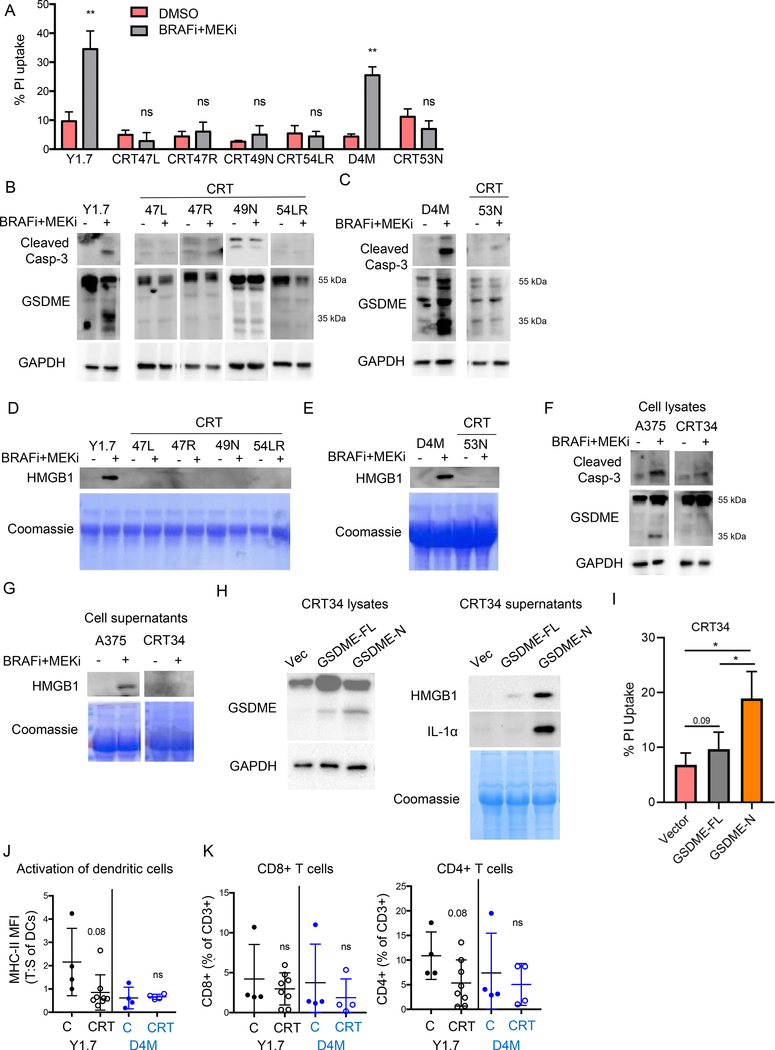
Acquired resistance to combination BRAFi + MEKi in patient samples is associated with T cell exclusion (5,17), findings we corroborated using available data sets (Fig. 2A–B). To assess pyroptosis in the setting of BRAFi + MEKi resistance, we treated established YUMM1.7 and D4M3.A tumors in C57BL/6 mice with BRAFi + MEKi until they became resistant. Cell lines generated from combination resistant tumors (CRTs) did not uptake PI or cleave caspase-3 when treated with BRAFi + MEKi (Fig. 5A–C). These effects were associated with weak induction of the pro-apoptotic BH3 proteins, Bim-EL and Bmf, in CTRs compared to parental cells (Supp. Fig. 6A). BRAFi + MEKi also failed to induce GSDME cleavage, HMGB1 release, or increase calreticulin surface expression in CRT cells (Fig. 5B–E and Supp. Fig. 6B). Similar findings were obtained in human melanoma A375-derived CRT cells upon treatment with the combination therapy (Fig. 5F–G). Importantly, expression of the GSDME N-terminal sequence (amino acids 1–270) and, to a lesser extent full-length GSDME, in BRAFi + MEKi-resistant CRT34 cells was sufficient to induce both the release of HMGB1 and IL-1α as well as PI uptake (Fig. 5H–I). These data show that expression of pore-forming GSDME in BRAFi + MEKi-resistant cells is sufficient to cause release of pro-inflammatory mediators and melanoma cell death.
Figure 5: BRAFi + MEKi-resistant cell lines (CRT cells) do not undergo pyroptosis.
YUMM1.7 CRT cells (CRT47L, 47R, 49N, and 54LR), D4M3.A CRT cells (CRT 53L), or A375 CRT cells (CRT34) were treated with PLX4720 (1 μM) and PD0325901 (35 nM) for 72 (A) or 24 hours (B-G). A) Cell death as indicated by PI uptake of cells after BRAFi + MEKi treatment. B) Levels of cleaved caspase-3 and GSDME in YUMM1.7 CRT cells after treatment. GAPDH as loading control. Full-length GSDME runs at 55 kDa and cleaved GSDME runs at 35 kDa. C) Same as B for parental D4M3.A cells and D4M3.A CRT cells. D) Levels of HMGB1 in supernatants from parental YUMM1.7 and CRT cells. Coomassie stained gel showing protein loading. E) Same as D for D4M3.A CRT cells. F) Same as B for parental A375 and derived CRT cell lysates. G) Same as D for human cell lines. H) A375 CRT34 cells were transfected with pLenti3-hygro vector (EV), full-length human GSDME (FL) or amino acids 1–270 of GSDME (N) for 24 hours. Cell lysates (left) and supernatant (right) were analyzed by Western blotting with indicated antibodies. GAPDH or Coomassie stained gel served as loading control. I) Cell death, as measured by PI uptake (n=3). Statistical analysis for this figure was completed by student’s T-test *p<0.05. J) Activation status of dendritic cells in control treated (C) and CRT tumors. Ratio of MHC-II MFI in tumors compared to spleens. K) Tumor-associated CD8+ and CD4+ T cells as % of CD3+ cells.
As GSDME cleavage was associated with intra-tumoral immune responses (Fig. 4G), we compared TILs from YUMM1.7 CRTs and D4M3.A CRTs to TILs from treatment-naive tumors taken at similar sizes. Whereas responsive tumors had increased DC activation and T cell counts during BRAFi + MEKi (Fig. 2 and 3), there was no difference in these populations between control and CRT tumors (Fig. 5J–K, and Supp. Fig. 6C). In addition, we observed lower TNFα production from intra-tumoral and splenic CD4+ and CD8+ T cells from CRTs (Supp. Fig. 3D–E), consistent with previous publications suggesting that MEKi inhibits T cell function (21,23). Intra-tumoral TAM and MDSC populations were increased in resistant tumors compared to on-treatment tumors but were lower than in control progressing tumors, at least in the case of YUMM1.7 (Supp. Fig. 3H–I). Lastly, expression of IDO-1 and OX40L was consistently decreased in CD45.2-negative cells within resistant tumors compared to control progressing tumors (Supp. Fig. 3J). MHC-I, PD-L1, FasL, and OX40L expression were all consistently decreased in CD45.2+ cells in BRAFi + MEKi-resistant tumors compared to control progressing tumors (Supp. Fig. 3K). Together, these data suggest that the loss of BRAFi + MEKi-induced pyroptosis and GSDME cleavage in BRAFi + MEKi-resistant tumors is associated with reduced anti-tumor immune responses.
BRAFi + MEKi combination resistant tumors are susceptible to drugs that re-induce GSDME cleavage and pyroptosis
Since HMGB1 release and GSDME cleavage is impaired in BRAFi + MEKi-resistant cell lines, we determined whether alternative agents could induce pyroptosis and elicit anti-tumor effects. We tested the ability of several therapeutic modalities (chemotherapy, epigenetic inhibitors, targeted inhibitors, and radiation) to induce HMGB1 release from CRT-derived cell lines. Radiation therapy did not cause release of HMGB1, despite delaying tumor cell growth (Supp. Fig. 7A and B). In addition, neither BET inhibitor, CDK4/6 inhibitor, dacarbazine, tamoxifen, ERK1/2 inhibitor nor paradox breaking BRAFi induced release of HMGB1, despite the latter two agents inducing cell death (Supp. Fig. 7C and D). However, etoposide treatment induced both release of HMGB1 and cell death in CRT cells (Supp. Fig. 7C and D).
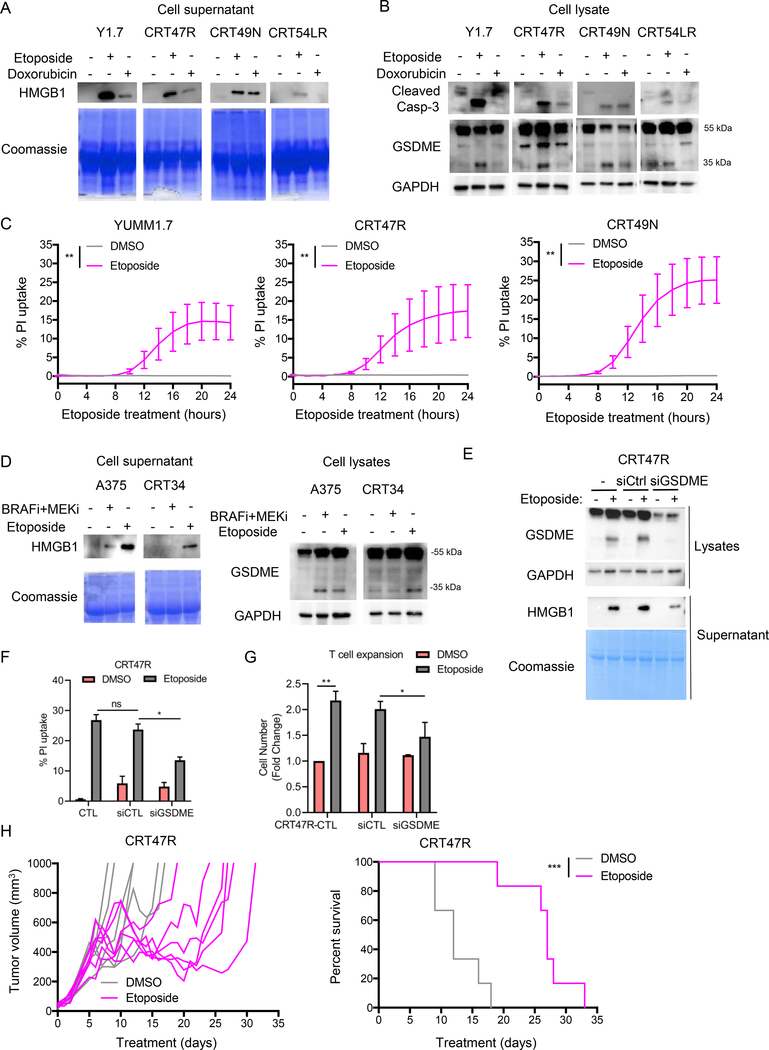
We confirmed the ability of etoposide and also tested doxorubicin, a known chemotherapeutic inducer of pyroptosis, for ability to cause GSDME cleavage in CRT cell lines (31,32,34). Etoposide treatment consistently induced HMGB1 release, caspase-3 cleavage, GSDME cleavage, and PI uptake in parental YUMM1.7 cells and YUMM1.7 CRT cells (Fig. 6A–C). Doxorubicin displayed more variable results but was able to induce HMGB1 release, caspase-3 cleavage, and GSDME cleavage in some lines (Fig. 6A–B). Etoposide treatment also induced GSDME cleavage and HMGB1 release into the supernatant from a human CRT cell line (Fig. 6D). Knockdown of GSDME in CRT47R cells reduced etoposide-induced HMGB1 release (Fig. 6E) and PI uptake (Fig. 6F), and supernatant from GSDME knockdown cells was significantly less effective at inducing T cell expansion compared to control cells (Fig. 6G). These findings implicate GSDME in etoposide-induced cell death.
Figure 6: BRAFi + MEKi-resistant cell lines are susceptible to pyroptosis.
YUMM1.7 CRT cells (CRT47R, 49N, and 54LR) or A375 CRT cells (CRT34) were treated with 37.5 μM etoposide or 1 μM doxorubicin for 24 hours. A) Levels of HMGB1 in supernatant from mouse cell lines. Coomassie stained gel as loading control, n=4. B) Cleavage of caspase-3 and GSDME in mouse cells after treatment. GAPDH as loading control. Full-length GSDME runs at 55 kDa and cleaved GSDME runs at 35 kDa. C) PI uptake overtime during etoposide treatment, n=3. Significance determined by area under the curve, **p<0.01. D) Same as A and B for human cells. E) YUMM1.7 CRT cells (CRT47R) were transiently transfected with either control or GSDME siRNA for 24 hours and then treated with etoposide (37.5 μM) for a further 24 hours. Cell lysates and supernatant were analyzed by Western blotting with indicated antibodies. GAPDH or Coomassie stained gel serve as loading controls. F) As above, except that cell death as indicated by PI uptake of cells after etoposide treatment (n=3). *p<0.05. G) As above, T cell expansion stimulated by diafiltered supernatants was analyzed. Fold change of PmeI-I T cell numbers after 5 days is shown (n=3). Statistical analysis by student’s T-test *p<0.05, **p<0.01. H) Mice were started on BRAFi + MEKi treatment one day before tumor cell implantation. YUMM1.7 CRT47R cells (2.5×105) were intradermally implanted on day 0. On day 5, mice were removed from BRAFi + MEKi chow and etoposide treatment was administered as described in Supp. Figure 7E–F. Shown are tumor growth (left) and percent survival (right) of treated mice.
In vivo, etoposide treatment slowed CRT47R tumor growth and significantly improved the survival of tumor-bearing mice (Fig 6H); however, mice lost weight during etoposide treatment necessitating scheduling that altered the frequency of the treatment during the experiment (Supp. Fig. 7E and 7F). Nonetheless, these data provide proof-of-principle that pharmacological re-induction of pyroptosis may be a potential salvage therapy for combination BRAFi + MEKi-resistant melanoma.
DISCUSSION
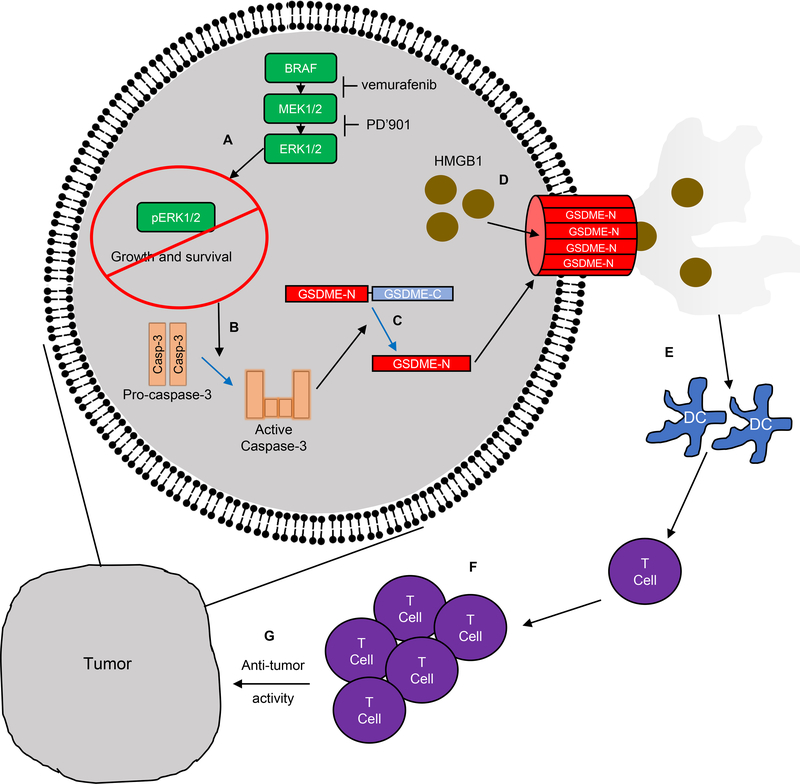
The salient findings of this study are that: i) an intact immune system is required for in vivo BRAFi + MEKi efficacy in BRAF mutant melanoma; ii) T lymphocytes are required for the sustained therapeutic effects of BRAFi + MEKi; iii) T cell activation and tumor regression are contingent on GSDME-dependent pyroptosis of tumor cells; iv) re-induction of pyroptosis may offer an effective salvage therapy for BRAFi + MEKi-resistant tumors. These findings provide a novel mechanistic link between BRAFi + MEKi-induced pyroptosis, regulation of the tumor immune microenvironment and anti-tumor immunity (Fig. 7). In addition, we provide proof-of principle evidence for a salvage therapy for BRAFi + MEKi-resistant BRAF mutant melanoma.
Figure 7: Proposed model of BRAFi + MEKi-induced pyroptosis.
A) BRAFi + MEKi treatment blocks ERK1/2 signaling, inhibiting growth and survival of BRAF V600E melanoma cells. B) ERK1/2 pathway blockade results in activation of caspase-3. C) leading to the cleavage of GSDME. The N-terminal cleaved region translocates to the plasma membrane leading to pore formation. D) After BRAFi + MEKi-induced GSDME pore formation, HMGB1 and other DAMPs are released from the cell. E-G) Extracellular DAMPs lead to the activation of dendritic cells, which induce T cell proliferation and contribute to anti-tumor effects during BRAFi + MEKi treatment.
Despite their clinical efficacy and evidence of inhibitor-induced T cell infiltration, it remains unclear how BRAFi + MEKi induce anti-tumor T cell responses. Furthermore, links between pyroptosis and tumor-associated immune infiltrates remain unclear. It is known that caspase-3 activation cleaves GSDME to induce pore formation and pyroptosis (32,33). Here, we show that BRAFi + MEKi induced caspase-3 activation and GSDME cleavage, and that GSDME is required for the release of the damage-associated molecular patterns (DAMP) molecule, HMGB1, from melanoma cells. Released HMGB1 is known to promote inflammation through its binding to toll-like receptor 4 on dendritic cells (45,46). While previous work demonstrated that the combination of BRAFi plus HDAC inhibitors induced release of HMGB1 from melanoma (47), our data underscore the mechanistic role of GSDME in HMGB1 release and effects on anti-melanoma immunity. Our findings are supported by a recent study that found that MEKi promote GSDME cleavage in lung cancer (36), although again anti-tumor immune responses were not assessed in that report. We demonstrate that GSDME cleavage is required not only for pyroptosis but also for anti-tumor T cell responses observed following BRAFi + MEKi administration. Thus, our data suggest that GSDME-dependent pyroptosis may be an indispensable mediator of immune-driven therapeutic response in BRAF mutant melanoma. Consistent with this notion, lack of GSDME and pyroptosis led to enhanced re-growth of residual disease after removal of BRAFi + MEKi. MEKi efficacy is known to be T cell-dependent via impaired TCR-driven apoptosis in CD8+ T cells (19–23); thus, GSDME-mediated pyroptosis during BRAFi + MEKi may be working in tandem with impaired T cell apoptosis to induce robust immune responses. Taken together, our data define a new functional intersection between BRAFi + MEKi-induced pyroptosis and T cell responses to melanoma.
The development of resistance to BRAFi + MEKi in metastatic melanoma remains a significant challenge in the clinic. While several melanoma cell-autonomous mechanisms of resistance to BRAFi and/or MEKi have been established (5,6,48), it has remained unclear how anti-melanoma immune responses in the tumor microenvironment can be leveraged to overcome treatment resistance. Resistance to BRAFi + MEKi is linked to loss of intra-tumoral T cell responses (17), data corroborated here using an independent human data set and mouse models. These findings are increasingly important as BRAFi + MEKi therapy in combination with immune checkpoint inhibition is being tested in patients with advanced melanoma with promising efficacy albeit toxicity challenges (49–51). Additionally, in pre-clinical models, CSF-1R inhibitors targeting macrophage accumulation improved efficacy of BRAFi (18), and the efficacy of MEKi can be improved by targeting the PD-1/PD-L1/L2 axis (19,22). The prevalence of resistance to targeted therapies and the relative lack of insight of immune system’s role in this process underscores the importance of our findings. We demonstrated that combination inhibitor-resistant tumor cells do not undergo pyroptosis with BRAFi + MEKi, resulting in a loss of anti-tumor immune responses. These data may help to explain the loss of intra-tumoral CD8+ T cells in patients who are no longer responsive to BRAFi + MEKi (17). Furthermore, re-induction of pyroptosis with etoposide in BRAFi + MEKi-resistant melanomas provides proof-of-concept that targeting this programmed cell death pathway represents a potential strategy for salvage therapy for melanoma patients who are resistant to BRAFi + MEKi.
In summary, this study establishes the requirement for T cells on the immune-mediated mechanisms of resistance to BRAFi + MEKi. Furthermore, we link ERK1/2 pathway inhibition to the induction of pyroptosis through cleavage of GSDME to produce a more productive anti-tumor immune response. Expanding on this knowledge may lead to new salvage therapies for metastatic melanoma patients with BRAFi + MEKi-resistant disease.
MATERIALS AND METHODS
Cell culture
D4M3.A cells (derived from Tyr::CreER; BrafV600E;Pten −/− mice; cells donated by Dr. Constance E. Brinckerhoff, Dartmouth University, 2016) were cultured in DMEM/F12 with 5% FBS, 1% PenStrep, and 1% L-Glutamine. YUMM1.7 cells (Braf V600E/wt, Pten−/−, Cdkn2−/−; donated by Dr. Marcus Bosenberg, Yale University, 2014) were cultured in DMEMF-12 50/50 with 10% FBS, 1% PenStrep, and 1% non-essential amino acids. A375 cells (purchased from ATCC in 2005) were cultured in DMEM with 10% FBS. Cell lines were STR analyzed, confirmed for BRAF/Braf V600E mutation, and IMPACT III PCR pathogen tested (IDEXX) to authenticate them and determine they were pathogen free. CRT cells were isolated from tumors, cultured in the same medium as parental cells with the addition of PLX4720 (1 μM) and PD0325901 (35 nM) and utilized within 5 passages for experiments. Drug concentrations utilized are close to published GI-50 values for PLX4720 and PD0325901 (52,53). Inhibitor levels maintain the same BRAFi to MEKi ratio as used for in vivo experiments. A375-derived CRT cells were previously published (54).
In vivo tumor growth studies
Animal experiments were approved by the IACUC and performed at Thomas Jefferson University in a facility accredited by the AAALAC. Male C57BL/6 mice (Jackson, 6–12 weeks) were used unless denoted. Tumors were implanted intradermally in 100 μL HBSS. 6-week-old male or female NSG mice were provided by Dr. Timothy Manser. Tumor volume was tracked with a caliper: volume = (length x width2)*0.52. When the volume reached ~50–250 mm3 animals were fed with either vehicle control chow or combination BRAFi + MEKi chow (200 ppm PLX4720 plus 7 ppm PD0325901). For etoposide experiments, mice were treated with intraperitoneal injections of 17 μM/animal of etoposide following the schedule outlined in Supplemental Figure 7E and F (55). PLX4720 and PD0325901 were generously provided by Plexxikon Inc. and chow was purchased from Research Diets Inc.
Western blot analysis and cell supernatant collection
Protein lysates were prepared in Laemmli sample buffer, separated by SDS-PAGE, and proteins were transferred to PVDF membranes. Immunoreactivity was detected using HRP-conjugated secondary antibodies (CalBioTech) and chemiluminescence substrate (Thermo Scientific) on a Versadoc Imaging System (BioRad). Primary antibodies, all from Cell Signaling Technologies unless otherwise stated, were as follows: anti-phospho-ERK1/2 (T202/Y204), anti-ERK2 (Santa Cruz Biotech.), anti-GSDME (Abcam), anti-cleaved caspase-3, anti-BIM/BOD (Enzo Life Sciences), anti-IL-1α (Santa Cruz Biotech), anti-HMGB1 and anti-GAPDH. Cell supernatants were harvested in absence of FBS in culture medium to avoid distortion of SDS-PAGE. After centrifugation to remove cell debris, cell supernatants were concentrated 10x using Amicon Ultra 10K (Sigma-Aldrich). Concentrates were mixed with Laemmli sample buffer (BioRad) and analyzed via Western blotting. For T cell culture, concentrates were further washed twice with PBS and once with RPMI1640 medium by spin, and sterilized by filtration through 0.2 μm filter. Protein gel staining was performed using Coomassie® Brilliant Blue R-250.
Calreticulin surface expression
Cells were treated with BRAFi + MEKi for the indicated times. Adherent cells were washed and stained with live/dead stain (Zombie UV, Biolegend) per company instructions. Cells were then primary surface stained with anti-calreticulin (Cell Signaling), then secondary stained with the appropriate anti-rabbit AF488 antibody (Invitrogen). Cells were analyzed on the BD Celeste flow cytometer and data quantified with FlowJo.
Annexin V/PI analysis
Cells were treated with BRAFi + MEKi for 72 hours. Adherent cells were washed and incubated with 5 μL annexin V-APC (BD Biosciences) in 100 μL of binding buffer and then incubated with 0.02 mg/mL propidium iodide for 15 minutes at room temperature. Cells were analyzed on the FACS Calibur or BD LSR II flow cytometers. Experiments were performed in triplicate, and statistical analysis was completed using a two-tailed t-test assuming equal variance with error bars representing SEM.
IncuCyte® (Essen Bioscience, Ann Arbor, MI) imaging of PI uptake was used to measure cell death. Cells were treated with drugs indicated and PI (10 μg/ml). Red fluorescence was imaged and quantified with the IncuCyte®. Experiments were performed in triplicate, and statistical analysis performed by calculating the areas under the curve and a two-tailed t-test assuming equal variance with error bars representing SEM.
siRNA transfections
Cells were transfected for 24 hours with siRNAs at a final concentration of 25 nmol/L using Lipofectamine RNAiMAX (Invitrogen). Non-targeting control (5′-UGGUUUACAUGUCGACUAA-3′) and GSDME (D-041196-01-0005, Dharmacon). After transfection, cells were treated with the indicated drugs for another 24 hours.
Flow cytometry of tumor, spleen, and blood samples
Spleens were processed mechanically using a 70 μM nylon filter and a syringe plunger. For immunogenicity and TIL studies, tumors were removed and dissociated into single cell suspensions by placing tumors in digestion medium: HBSS (CellGro), 10% FBS, 0.3–0.5 mg/ml Collagenase 1A (Sigma), 60 U/mL DNase I (Sigma) and mincing using the gentleMACS Octo Dissociator using C Tubes (Miltenyi Biotec). Minced tumors were incubated at 37°C for 30 minutes while shaking, minced again, washed with T cell medium (TCM, RPMI 1640, with L-glutamine + 10% FBS + 1% PenStrep and 5×10−5 M 2-βME), and filtered through a 70 μM nylon filter. Blood was collected in heparin-coated tubes, and stained for flow as needed. Blood samples were lysed and fixed using the RBC Lysis/Fixation Solution (BioLegend). All samples were analyzed on the BD Fortessa and data quantified with FlowJo.
For TIL and blood studies, cells were first stained with a fixable live/dead stain (BioLegend) followed by surface antibody staining. Cells were surface stained with the following antibodies from BioLegend: CD45.2 (clone 104), CD3 (clone 17A2), CD8α (clone 53.6.7), CD8β (clone YTS156.7.7), CD4 (clone RM4.4 and GK1.5), CD44 (clone IM7), CD11c (clone N418), CD11b (clone M1/70), CD103 (clone 2E7), F4/80 (clone BM8), Gr-1 (clone RB6–8C5) and I-A/I-E (clone M5/114.15.1). For Ki67 staining, cells were fixed, and nuclear permeabilized using the eBioscience™ Foxp3/transcription factor buffer staining set and an antibody specific for Ki67 (clone 16A8) following company instructions. For immunogenicity studies, cells were stained with H-2 (clone M¼2), I-A/I-E (clone M5/114.15.1), PD-L1 (clone 10F.9G2), IDO-1 (clone 2E2/IDO1), FasL (clone MLF3), LGalS9 (clone 108A2), and Ox40L (clone 11C3.1). For T cell re-stimulation assays, cytokine production by T cells was assessed after ex vivo stimulation with the eBioscienceTM cell stimulation cocktail. Cells (0.5 – 4 × 106) were incubated in medium for 5 hours at 37°C in 5% CO2, with the stimulation cocktail and 1 μg/ml brefeldin A (GolgiPlug; BD Biosciences). Cells were washed with ice-cold FACS, stained with Zombie UV and antibodies specific for surface proteins, and then analyzed for intracellular cytokines. All cells were fixed using the BD Cytofix/cytoperm kit (BD Biosciences). Samples were run on the BD Fortessa and data analyzed on FlowJo software.
In vivo CD4 and CD8 cell depletion
To deplete CD4+ and CD8+ T cells, mice were treated with 300 μg of anti-CD8α (clone 53–6.72) and 250 μg of anti-CD4 (clone GK1.5), or IgG2a and IgG2b antibody controls every 3–7 days for the duration of the experiment. Depletions were initiated 2 days before tumor implantation.
In vitro T cell growth assay
Splenocytes were isolated from Pmel-17 mice (B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J, purchased from Jackson), and frozen in CryoStor C10 Freeze Media (BioLife Solutions). Pmel-I splenocytes were thawed into T cell media (RPMI 1640, 10% FBS, 1% PenStrep, 0.00038% β-mercaptoethanol) at 1×106 cells per 2 mL. On the day of thawing, gp100 peptide (0.5 μg/mL, EGSRNQDWL, GeneMed) and 50% diafiltrated supernatant from YUMM1.7 cells treated with DMSO or inhibitors for 48 hours were added to splenocytes. Two days after thawing, mouse rIL-2 30 U/mL (Biolegend) was added to the culture. Five days after thawing, cells were counted via a hemocytometer.
GSDME CRISPR-Cas9
The CRISPR design tools at MIT (http://crispr.mit.edu) or Benchling (https://benchling.com) were used to identify candidate sgRNA sequences. Sequences targeting murine GSDME exon 3 (5’-GTGTGAGAACCATAAGAGCG-3’ and 5’-GGGCTATTGGGACAGTCGTG-3’) were cloned into lentiCRISPRv2GFP vector (Addgene) containing Cas9 fused to EGFP. LentiCRISPRv2GFP sgRNA plasmids (5 μg) were cotransfected with 3.75 μg psPAX2, and 2.5 μg VSVg plasmids (Addgene) using 25 μL Lipofectamine in 293T cells. Seventy-two hours after transfection, media was collected and concentrated overnight using Lenti-X Concentrator (Takara) according to the manufacturer’s protocol. Infected 1×106 CEM-C7 or 2.5×104 YUMM1.7 or D4M3.A cells/ml were plated in a 12-well plate with concentrated lentivirus. Cells were then enriched for Cas9-EGFP expression by flow cytometry, and single cells were then isolated and screened by Western blot analysis for protein expression.
cDNA cloning
The human (BC099911) GSDME cDNA (Dharmacon Inc.) was cloned into pENTR-D TOPO vector by PCR amplification with primers: forward (CACCATGTTTGCCAAAGCAACCAGG) and either reverse with amino acid 496 stop (TCATGAATGTTCTCTGCCTAAAGC) for full-length GSDME or amino acid 270 stop (TCAATCTGGCATGTCTATGAATGC) for N terminal GSDME. Entry constructs were recombined into pLenti3-hygro by Gateway cloning (Thermo Fisher Scientific). For transient transfection, plasmids were transfected with Lipofectamine 2000 (Thermo Fisher Scientific).
RNA-seq data analysis
RNA-seq data were collected from European Genome-phenome Archive, EGAS000010000992 (39) and from the GSE99898 dataset (43). TCGA cutaneous melanoma RNA-seq V2 normalized gene expression and mutation call data were retrieved from the latest Broad GDAC Firehose data run (stddata__2016_01_28). The immune cell gene set analysis was performed as previously published (56). Gene set scores were calculated using the GSVA package (version 1.28.0) in R (version 3.5.1) (57).
Pyroptosis gene set
The pyroptosis gene set was determined using http://amigo.geneontology.org/amigo/term/GO:0070269 and the recent ‘Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018 (58). Genes used were GSDMA, B, C, D, and E, Caspase 1, 2, 3, 4, 5, and 8, NLRC4, IL-1β, and IL-18.
Radiation therapy
Ionizing radiation was administered at doses ranging from 5 to 20 Gy using a 250-kVp X-ray machine (PanTak) with 50 cm source-to-skin distance and a 2 mm aluminum filter. The dose rate was approximately 3.6 Gy/minute.
In vitro drug treatments
Cells were treated with BETi (2 μM PLX51107 provided by Plexxikon Inc.), CDK4/6i (1 μM PD0332991), etoposide (37.5 μM, Sigma), dacarbazine (20 μM, Sigma), tamoxifen (1 μM, Sigma), ERK1/2i (1 μM SCH772984, SelleckChem), or paradox-breaking BRAFi (500 nM PLX8394 provided by Plexxikon Inc.) for 24 hours. Etoposide (37.5 μM) and doxorubicin (1 μM) concentrations were based upon previous publications (59–61).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Targeted inhibitors and immune checkpoint agents have advanced the care of melanoma patients; however, a detailed knowledge of the intersection between these two research areas is lacking. We describe a molecular mechanism of targeted inhibitor regulation of an immune stimulatory form of cell death and provide a proof-of-principle salvage therapy concept for inhibitor-resistant melanoma.
ACKNOWLEDGMENTS
We acknowledge Dr. Gideon Bollag, Plexxikon Inc. (Berkeley, CA) for generously providing PLX4720, PD0325901, PLX51107, and PLX8394. We thank Dr. Timothy Manser (Thomas Jefferson University) for the NSG mice, Dr. Chris Snyder for comments on the manuscript and Ms. Carla Portocarrero for her assistance with the irradiation of cell lines.
Financial Support: This work is supported by grants from National Institutes of Health (R01 CA196278, R01 CA160495, R01 CA182635 to AEA, and AR055398, AR074564 to ESA), the Department of Defense (PC150650 to UR), and Dr. Ralph and Marian Falk Medical Research Catalyst Trust Award (to ESA). The Sidney Kimmel Cancer Center Flow Cytometry, Translational Pathology, and Meta-Omics core facilities are supported by National Cancer Center Support Grant (P30 CA056036). DAE is supported by an American Cancer Society CEO’s Against Cancer PA Chapter Postdoctoral Fellowship (PF-18-096-01-LIB). IMS was supported by a diversity supplement to R01 CA160495. EJH was supported by NIH K99 CA207855.
Conflict of interest: D. A. Erkes reports that he is now an employee of the journal publisher, the American Association for Cancer Research, and had no influence over any editorial decisions related to this article. A.E. Aplin reports receiving a commercial research grant from Pfizer Inc. (2013–2017), has ownership interest in patent number 9880150 and has consulted for SpringWorks Therapeutics and Fortress Biotech within the last 3 years. U. Rodeck has received research support from Bristol Myers Squibb, Imclone Inc., Merck Sharp & Dome, Advaxis Therapeutics and, Onconova Therapeutics Inc. He is a cofounder, serves on the SAB and holds equity in Akrevia Therapeutics. No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, et al. Melanoma. The Lancet 2018;392:971–84. [DOI] [PubMed] [Google Scholar]
- 2.Silva IP, Long GV. Systemic therapy in advanced melanoma: integrating targeted therapy and immunotherapy into clinical practice. Curr Opin Oncol 2017;29:484–92. [DOI] [PubMed] [Google Scholar]
- 3.Long GV, Hauschild A, Santinami M, Atkinson V, Mandala M, Chiarion-Sileni V, et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med 2017;377:1813–23. [DOI] [PubMed] [Google Scholar]
- 4.Amaria RN, Prieto PA, Tetzlaff MT, Reuben A, Andrews MC, Ross MI, et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol 2018;19:181–93. [DOI] [PubMed] [Google Scholar]
- 5.Hartsough E, Shao Y, Aplin AE. Resistance to RAF inhibitors revisited. J Invest Dermatol 2014;134:319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015;27:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 2014;4:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015;373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J Clin Invest 2013;123:1371–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Frederick DT, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res 2013;19:1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koya RC, Mok S, Otte N, Blacketor KJ, Comin-Anduix B, Tumeh PC, et al. BRAF inhibitor vemurafenib improves the antitumor activity of adoptive cell immunotherapy. Cancer Res 2012;72:3928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilmott JS, Long GV, Howle JR, Haydu LE, Sharma RN, Thompson JF, et al. Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin Cancer Res 2012;18:1386–94. [DOI] [PubMed] [Google Scholar]
- 13.Ho PC, Meeth KM, Tsui YC, Srivastava B, Bosenberg MW, Kaech SM. Immune-based antitumor effects of BRAF inhibitors rely on signaling by CD40L and IFNgamma. Cancer Res 2014;74:3205–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper ZA, Juneja VR, Sage PT, Frederick DT, Piris A, Mitra D, et al. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol Res 2014;2:643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilieva KM, Correa I, Josephs DH, Karagiannis P, Egbuniwe IU, Cafferkey MJ, et al. Effects of BRAF mutations and BRAF inhibition on immune responses to melanoma. Mol Cancer Ther 2014;13:2769–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper ZA, Reuben A, Austin-Breneman J, Wargo JA. Does It MEK a Difference? Understanding Immune Effects of Targeted Therapy. Clin Cancer Res 2015;21:3102–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015;162:1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mok S, Tsoi J, Koya RC, Hu-Lieskovan S, West BL, Bollag G, et al. Inhibition of colony stimulating factor-1 receptor improves antitumor efficacy of BRAF inhibition. BMC Cancer 2015;15:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X, et al. Recurrent Tumor Cell-Intrinsic and -Extrinsic Alterations during MAPKi-Induced Melanoma Regression and Early Adaptation. Cancer Discov 2017;7:1248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allegrezza MJ, Rutkowski MR, Stephen TL, Svoronos N, Tesone AJ, Perales-Puchalt A, et al. IL15 agonists overcome the immunosuppressive effects of MEK inhibitors. Cancer Res 2016;76:2561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allegrezza MJ, Rutkowski MR, Stephen TL, Svoronos N, Perales-Puchalt A, Nguyen JM, et al. Trametinib Drives T-cell-Dependent Control of KRAS-Mutated Tumors by Inhibiting Pathological Myelopoiesis. Cancer Res 2016;76:6253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert PJ, Cheung J, Yang Y, McNamara E, Hong R, Moskalenko M, et al. MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity 2016;44:609–21. [DOI] [PubMed] [Google Scholar]
- 23.Dushyanthen S, Teo ZL, Caramia F, Savas P, Mintoff CP, Virassamy B, et al. Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat Commun 2017;8:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu-Lieskovan S, Mok S, Moreno BH, Tsoi J, Robert L, Goedert L, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Sci Trans Med 2015;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger A, Quast SA, Plotz M, Kuhn NF, Trefzer U, Eberle J. RAF inhibition overcomes resistance to TRAIL-induced apoptosis in melanoma cells. J Invest Dermatol 2014;134:430–40. [DOI] [PubMed] [Google Scholar]
- 26.Geserick P, Herlyn M, Leverkus M. On the TRAIL to Overcome BRAF-Inhibitor Resistance. J Invest Dermatol 2014;134:315–8. [DOI] [PubMed] [Google Scholar]
- 27.Beck D, Niessner H, Smalley KS, Flaherty K, Paraiso KHT, Busch C, et al. Vemurafenib Potently Induces Endoplasmic Reticulum Stress–Mediated Apoptosis in BRAFV600E Melanoma Cells. Science Signaling 2013;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science 2016;352:aaf2154. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016;535:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J, Gao W, Shao F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem Sci 2017;42:245–54. [DOI] [PubMed] [Google Scholar]
- 31.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018;25:486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun 2017;8:14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017;547:99–103. [DOI] [PubMed] [Google Scholar]
- 34.Rogers C, Erkes DA, Nardone A, Aplin AE, Fernandes-Alnemri T, Alnemri ES. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun 2019;10:1689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. [DOI] [PubMed] [Google Scholar]
- 36.Lu H, Zhang S, Wu J, Chen M, Cai MC, Fu Y, et al. Molecular Targeted Therapies Elicit Concurrent Apoptotic and GSDME-Dependent Pyroptotic Tumor Cell Death. Clin Cancer Res 2018;24:6066–77. [DOI] [PubMed] [Google Scholar]
- 37.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res 2016;29:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins MH, Steinberg SM, Alexander MP, Fisher JL, Ernstoff MS, Turk MJ, et al. Multiple murine BRAF melanoma cell lines with sensitivity to PLX4032. Pigment cell & melanoma research 2014;27:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest 2015;125:1459–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eigenbrod T, Park JH, Harder J, Iwakura Y, Nunez G. Cutting edge: critical role for mesothelial cells in necrosis-induced inflammation through the recognition of IL-1 alpha released from dying cells. J Immunol 2008;181:8194–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J, et al. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis 2019;10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overwijk WW, Tsung A, Irvine KR, Parkhurst MR, Goletz TJ, Tsung K, et al. gp100/pmel 17 Is a Murine Tumor Rejection Antigen: Induction of “Self ”-reactive, Tumoricidal T Cells Using High-affinity, Altered Peptide Ligand. J Exp Med 1998;188:277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakavand H, Rawson RV, Pupo GM, Yang JYH, Menzies AM, Carlino MS, et al. PD-L1 Expression and Immune Escape in Melanoma Resistance to MAPK Inhibitors. Clin Cancer Res 2017;23:6054–61. [DOI] [PubMed] [Google Scholar]
- 44.Niessner H, Sinnberg T, Kosnopfel C, Smalley KSM, Beck D, Praetorius C, et al. BRAF Inhibitors Amplify the Proapoptotic Activity of MEK Inhibitors by Inducing ER Stress in NRAS-Mutant Melanoma. Clin Cancer Res 2017;23:6203–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002;418:191–5. [DOI] [PubMed] [Google Scholar]
- 46.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001;2:675–80. [DOI] [PubMed] [Google Scholar]
- 47.Lai F, Guo ST, Jin L, Jiang CC, Wang CY, Croft A, et al. Cotargeting histone deacetylases and oncogenic BRAF synergistically kills human melanoma cells by necrosis independently of RIPK1 and RIPK3. Cell Death Dis 2013;4:e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ascierto PA, Ferrucci PF, Fisher R, Del Vecchio M, Atkinson V, Schmidt H, et al. Dabrafenib, trametinib and pembrolizumab or placebo in BRAF-mutant melanoma. Nat Med 2019;25:941–6. [DOI] [PubMed] [Google Scholar]
- 50.Ribas A, Lawrence D, Atkinson V, Agarwal S, Miller WH Jr., Carlino MS, et al. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat Med 2019;25:936–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan RJ, Hamid O, Gonzalez R, Infante JR, Patel MR, Hodi FS, et al. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat Med 2019;25:929–35. [DOI] [PubMed] [Google Scholar]
- 52.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 2008;105:3041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henderson YC, Chen Y, Frederick MJ, Lai SY, Clayman GL. MEK inhibitor PD0325901 significantly reduces the growth of papillary thyroid carcinoma cells in vitro and in vivo. Mol Cancer Ther 2010;9:1968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez I, Purwin T, Chervoneva I, Erkes D, Nguyen M, Davies M, et al. In vivo ERK1/2 reporter predictively models response and resistance to combined BRAF and MEK inhibitors in melanoma. Mol Cancer Therap 2019;9:1637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo Prete AC, Maria DA, Rodrigues DG, Valduga CJ, Ibanez OC, Maranhao RC. Evaluation in melanoma-bearing mice of an etoposide derivative associated to a cholesterol-rich nano-emulsion. J Pharm Pharmacol 2006;58:801–8. [DOI] [PubMed] [Google Scholar]
- 56.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018;175:984–97 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 2013;14:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009;16:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tiago M, de Oliveira EM, Brohem CA, Pennacchi PC, Paes RD, Haga RB, et al. Fibroblasts protect melanoma cells from the cytotoxic effects of doxorubicin. Tissue Eng 2014;20:2412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calvani M, Bianchini F, Taddei ML, Becatti M, Giannoni E, Chiarugi P, et al. Etoposide-Bevacizumab a new strategy against human melanoma cells expressing stem-like traits. Oncotarget 2016;7:51138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lage H, Helmbach H, Grottke C, Dietel M, Schandendorf D. DFNA5 (ICERE-1) contributes to acquired etoposide resistance in melanoma cells. FEBS Lett 2001;494:54–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.