Significance
We show that kinetic proofreading of activator–promoter interactions solves a fundamental problem in gene regulation: that the specificity of eukaryotic activators cannot be explained by activator–DNA recognition alone. In light of our theory, multiple observations point to a role of nucleosomes in the kinetic proofreading of activator-DNA recognition. Our theory explains, among other findings, why transcription may occur in stochastic bursts.
Keywords: transcriptional regulation, nucleosome, kinetic proofreading, irreversibility, entropy production
Abstract
Specificity in transcriptional regulation is imparted by transcriptional activators that bind to specific DNA sequences from which they stimulate transcription. Specificity may be increased by slowing down the kinetics of regulation: by increasing the energy for dissociation of the activator–DNA complex or decreasing activator concentration. In general, higher dissociation energies imply longer DNA dwell times of the activator; the activator-bound gene may not readily turn off again. Lower activator concentrations entail longer pauses between binding events; the activator-unbound gene is not easily turned on again and activated transcription occurs in stochastic bursts. We show that kinetic proofreading of activator–DNA recognition—insertion of an energy-dissipating delay step into the activation pathway for transcription—reconciles high specificity of transcriptional regulation with fast regulatory kinetics. We show that kinetic proofreading results from the stochastic removal and reformation of promoter nucleosomes, at a distance from equilibrium.
Specificity in transcriptional regulation is imparted by the binding of gene-specific transcriptional activators to specific DNA sequences (“enhancer sequences”). Mutation of a single activator-binding site may abolish activated transcription (1). Once bound, activators stimulate transcription from enhancers in a tenuous, indirect fashion: by recruitment of other, gene-nonspecific, factors such as chromatin remodelers, histone-modifying enzymes, Mediator and SAGA (2–5).
Activators find their DNA-binding sites by trial and error (6). Consistently, the affinity for nonspecific DNA is high—equilibrium dissociation constants fall into the micromolar range (7). Enhancers and core promoters can, by and large, be freely mixed and matched (8), indicating that activators may stimulate transcription from both cognate enhancers, which bear specific binding sites for the activator, and noncognate enhancers, which do not. Thus, the question arises of how binding at correct and not incorrect enhancers triggers transcription.
High regulatory specificities may be attained either by activator on-rates close to zero, or large affinity differences between correct and incorrect sequence binding. While small activator on-rates result in long search times for target sequences, higher affinities generally imply extended DNA dwell times, for rate constants of the on-reaction are closely similar for different DNA sequences of the same activator (7, 9). Gene regulation, however, requires both high specificity and openness to change—i.e., finite activator search times and finite DNA residence times. Accordingly, measured affinity differences between correct and incorrect sequences are notably small: −3 kcal/mol—about three times the average kinetic energy of a molecule at 25 °C—or less (9). The specificity problem, no doubt, is further exacerbated by the fact that noncognate enhancers, in general, greatly outnumber cognate enhancers.
Molecular biological enzymes (DNA and RNA polymerases, aminoacyl-transfer RNA synthetases, the spliceosome and ribosome) face the same specificity problem, as they must discriminate between correct and incorrect substrates on the basis of small differences in binding energy, ∆∆G°. Remarkably, molecular biological enzymes exhibit error frequencies well below the lower limit imposed by the energetics of substrate–enzyme binding of , which furthermore is attainable only in the asymptotic limit of infinitely slow catalysis (10). This astounding feat is made possible by insertion of an additional reaction step—the “proofreading reaction”—into the Michaelis–Menten pathway, which delays product formation. Provided the enzyme dynamics are maintained away from equilibrium, differences in dwell time at the catalytic center between correct and incorrect substrate may be exploited twice, before and after the proofreading reaction, reducing the minimal error frequency from to (11).
The results of chromatin-structure analysis at the level of single-gene molecules (12–14) proffer a similar solution to the activator specificity problem. The nuclear DNA of eukaryotic cells is spooled onto octamers of histone proteins (15, 16). These spools, or “nucleosomes,” the basic structural unit of chromatin, impede access to DNA and thus are universal repressors of transcription (17).
Promoter nucleosomes have been viewed as an impediment to transcription that is overcome once, during the transition from transcriptionally repressed to active chromatin (18). Analysis of the PHO5 gene of yeast led to a different conclusion: promoter nucleosomes are continually removed and reformed as the promoter stochastically transitions between alternative nucleosome configurations, including the fully nucleosomal and nucleosome-free promoter (12, 19). Activator binding increases the transition probability from configurations with more to those with fewer nucleosomes, increasing the structural heterogeneity of promoter chromatin (12).
Initially, this probabilistic theory of promoter chromatin dynamics was conceived to reconcile apparently contradictory experimental findings that suggested both loss and presence of nucleosomes at transcriptionally active promoter sequences (20, 21). The theory could subsequently be employed to explain the statistical distribution of promoter nucleosome configurations observed by electron microscopy (12). However, the biological question remained of why cells allow for, or perhaps prefer, structurally heterogeneous over homogeneous promoter chromatin, given that random transitioning between nucleosome configurations, some conducive to transcription and others not, should increase noise and thus reduce the signal-to-noise ratio in gene expression (22).
Here, we show that, to the contrary, stochastic structural dynamics in the promoter chromatin, when maintained at a distance from equilibrium, may both attenuate transcription noise and, at least in part, solve the specificity problem by kinetically proofreading activator–promoter interactions.
Results
To model activator specificity, we consider two genes that are identical, except that one copy bears the binding site for a specific activator (represented by a triangle in Fig. 1), whereas the other does not. As a measure of the activator’s ability to distinguish between both copies, between correct and incorrect genes, we define “regulatory specificity” or “activator fidelity,” f, as the ratio of
where and are the (average) steady-state rates of transcription for correct and incorrect promoter binding, respectively. Thus, when the activator promotes transcription indiscriminately, or transcription is activator-independent, .
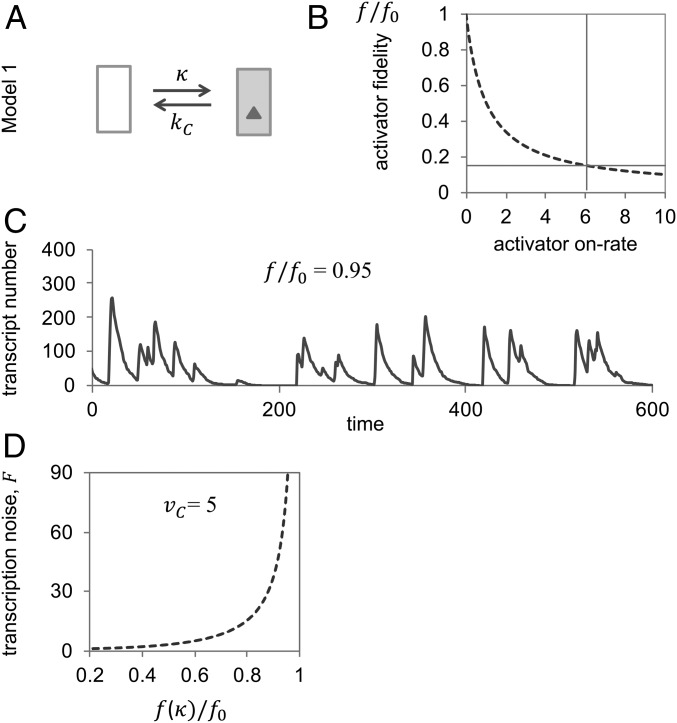
Fig. 1.
Standard two-state promoter model (Model 1): activator fidelity is bounded by the Hopfield barrier. (A) Transition graph of Model 1. (B) Activator fidelity approaches its upper limit or Hopfield barrier, , as the activator on rate, , tends to zero. To calculate the graph, we assumed , . Actual fidelities must be markedly lower than : for instance, measured off-rates for Pho4 of yeast (the activator of PHO5) for specific and nonspecific sequences are ∼0.01 and 1 s−1, respectively (7, 9). From Pho4’s equilibrium dissociation constant for correct binding of nM (9), and nuclear concentration of nM (47) (assuming a nuclear volume of 4 femtoliters), both the on-rate, (indicated by a vertical line), and relative fidelity (indicated by horizontal line) may be calculated; the unit on the abscissa, then, is . (C) Representative “sample path” (single cell trajectory of mRNA abundance) at relative activator fidelity of 0.95; the sample path was obtained with the Gillespie stochastic simulation algorithm (48) with , , (rate constant for mRNA degradation), and average rate of transcription, . (D) The Fano factor tends to infinity as activator fidelity, , approaches the Hopfield barrier . Calculations were based on the assumption of , , and average rate of transcription . Both Fano factor and fidelity were calculated as functions of the activator on-rate, κ (SI Appendix).
Free-energy differences for activator–DNA binding reactions, ∆∆G°, are generally determined by differences in the activator’s DNA-residence time (7, 9). Thus, , where and are the dissociation rate constants at nonspecific (incorrect) and specific (correct) DNA-binding sites, respectively, and .
For all following calculations, we set (i.e., we normalized all rate constants to ) and assumed , which corresponds to an energy difference between correct and incorrect promoter binding of kcal/mol (9). For stochastic simulations and noise calculations, we assumed an average expression level of 50 transcripts per cell, corresponding to a strongly transcribed gene of yeast, e.g., the fully induced PHO5 gene (1). While the numerical results of our calculations depend on the choice of specific parameter values, our principle conclusions do not (SI Appendix).
In the simplest case, the standard model of transcriptional regulation (Model 1; Fig. 1A), the promoter transitions between two states: activator-bound and unbound, where only the activator-bound state is transcriptionally active (23). We assume that and linearly depend on the steady-state probability of finding the promoter in its transcriptionally active state. Activator fidelity for Model 1, thus, is given by
(SI Appendix). Maximal fidelity, , is attained as the activator on-rate, , which linearly depends on the concentration of the activator, tends to zero (Fig. 1B). We will refer to (the upper limit to regulatory specificity imposed by the energetics of activator–DNA binding) as the “Hopfield barrier” to activator fidelity (24).
As tends to zero, the activator’s search time for its binding sequence tends to infinity. Periods of transcriptional activity with short pauses between initiation events in the activator-bound state are interrupted, then, by longer pauses of inactivity in the unbound state. To maintain a constant average rate of transcription, , the rate of transcription in the activator-bound state, , must increase as decreases, which further exacerbates the discrepancy between short pauses (average length: ) and long pauses (average length: ). As long pauses grow longer and short pauses shorter, transcription in stochastic bursts becomes increasingly manifest (Fig. 1C). The strength of bursting or magnitude of “transcription noise” may be expressed in terms of a population statistic, the Fano factor—i.e., variance of mRNA abundance normalized by its mean (SI Appendix). As activator fidelity approaches the Hopfield barrier, the noise of transcription tends to infinity (Fig. 1D). This limiting case, of course, is unrealistic: cannot and does not come arbitrarily close to zero (Fig. 1B), nor may tend to infinity. The biological problem to be solved is how to reconcile the requirements for finite activator on-rates and finite off-rates with the need for high regulatory specificity.
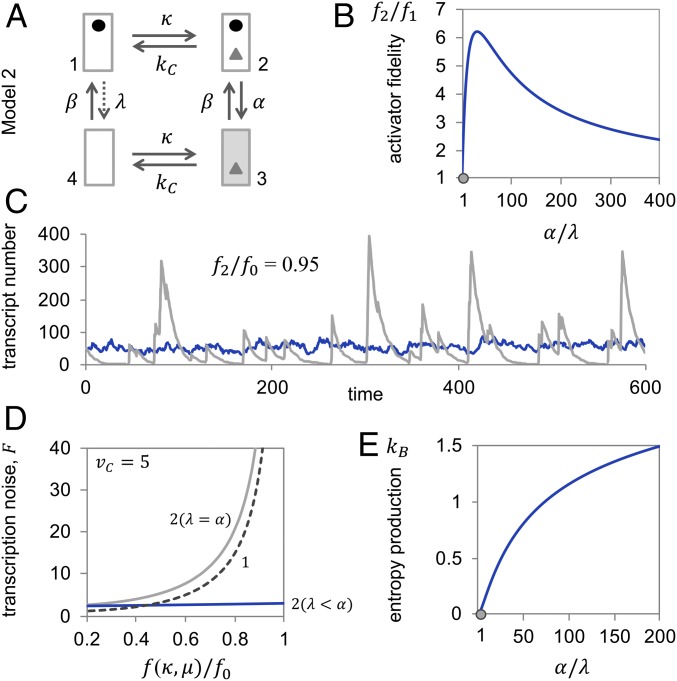
We now lay out such a solution. For simplicity, we consider a promoter (enhancer plus core promoter) with a single nucleosome position and single activator-binding site (Model 2; Fig. 2A). The nucleosome is subject to stochastic removal and reformation (12). Removal may occur by nucleosome sliding away from promoter sequences (25) or disassembly (26)—although, for theoretical reasons, we favor disassembly as the mechanism of removal (see below). Our promoter model encompasses four states: with nucleosome but without activator (state 1), with nucleosome and activator (state 2), with activator but without nucleosome (state 3), and without both (state 4).
Fig. 2.
Nucleosome dynamics away from, but not in, equilibrium allow for increased activator fidelity and attenuation of transcription noise. (A) Transition graph of Model 2. (B) Activator fidelity of Model 2 normalized by the fidelity of Model 1 , as a function of the rate of nucleosome removal in the activator-bound state, , normalized by the rate of removal in the unbound state, . For calculations, we assumed , , , , and . The gray dot indicates the equilibrium state. (C) Representative sample paths at relative activator fidelity and for nonequilibrium nucleosome dynamics (dark gray; , ), which required and ; and equilibrium dynamics (light gray; ), which required and . For both simulations, we assumed . (D) Transcription noise as a function of relative activator fidelity, , for Model 2 in equilibrium (light gray, 2 [α = λ]; α, λ = 2), away from equilibrium (blue, 2 [α > λ]; , ), and Model 1 (dashed line, 1; same as in Fig. 1D). For all calculations, we assumed, as above, , , and . Fano factor and activator fidelity were calculated as functions of the activator on-rate, κ, and the rate of transcription in the active state, (SI Appendix). (E) Entropy production (in units of , the Boltzmann constant) as a function of nucleosome removal rate in the activator-bound state, , relative to the rate in the unbound state, , for , , and . The gray dot indicates the equilibrium state.
We assume that transcription requires both activator binding and absence of the nucleosome (i.e., transcription occurs in state 3 alone), that the kinetics of activator binding are not altered by the nucleosome (i.e., the energetics of activator binding are the same for all models discussed here), and that nucleosome removal in the presence of the activator occurs at a faster rate than its absence (i.e., in Fig. 2A) because the activator recruits chromatin-remodeling activities to the promoter that catalyze removal of the nucleosome.
Our model, thus, entails two types of promoter states, activator-bound and unbound, with different nucleosome removal kinetics. This kinetic asymmetry engenders a closed loop of reactions where transition cycles in one direction (clockwise in Fig. 2A) are more probable than transition cycles in the reverse direction (for proof, see SI Appendix).
For every clockwise, but not counterclockwise, cycle of promoter-state transitions, transcription requires the bound activator twice: for removal of the nucleosome and initiation of transcription. This sequential twofold requirement allows for kinetic discrimination between correct and incorrect promoter binding twice (transitions and in Fig. 2A). As a consequence, the upper limit of activator fidelity increases from to (for proof, see SI Appendix), without changing the energetics of activator–DNA binding. Activator fidelity, thus, may significantly exceed the Hopfield barrier, and it always exceeds the fidelity afforded by Model 1 (Fig. 2B; for proof, see SI Appendix). An analogous result was obtained by Hopfield for enzyme kinetics (11). Following Hopfield, we call the mechanism that affords this increase in activator fidelity “kinetic proofreading” (24).
With kinetic proofreading, fidelities close to the Hopfield barrier may be attained with faster activator on-rates, , than without—because the nucleosome “filters out” many incorrect activator-binding events. Fast on-rates for the activator markedly dampen (temporal) fluctuations in transcript number (compare blue trace in Fig. 2C with trace in Fig. 1C). Thus, at sufficiently high fidelities, kinetic proofreading affords lower transcription noise (compare blue and dashed curve in Fig. 2D). At sufficiently low fidelities, the random dynamics of proofreading increase noise (Fig. 2D).
Activator-dependent nucleosome removal , which entails the preference of clockwise over counterclockwise transition cycles, is essential for nucleosome-mediated kinetic proofreading. In steady state with , the system is also in thermodynamic equilibrium or “detailed balance”: forward and reverse transition of all reactions are equally probable (SI Appendix). Clockwise and anticlockwise transition cycles, therefore, are equally probable and kinetic proofreading is lost: (Fig. 2B). Activator fidelity, again, is limited from above by the Hopfield barrier, (for proof, see SI Appendix).
In equilibrium, therefore, the transcript number wildly fluctuates for fidelities close to (Fig. 2C, compare blue and gray traces), because fidelities close to require activator on-rates, , close to zero (see equation for ). Random transitioning between transcriptionally conducive and inconducive states now exacerbates the noise in transcription, as expected (Fig. 2D); the slower the nucleosome dynamics the higher the noise.
Maintenance of nucleosome dynamics away from equilibrium requires entropy production (i.e., free-energy dissipation); the greater the distance from equilibrium, the more energy must be dissipated per unit time (Fig. 2E). Although kinetic proofreading in Model 2 requires nonequilibrium promoter dynamics , increasing free-energy expenditure to increase the rate of nucleosome removal does not monotonically improve activator fidelity (Fig. 2B). The reason is simple: kinetic proofreading requires removal kinetics that are slow relative to the dissociation kinetics of activators that bind the promoter nonspecifically.
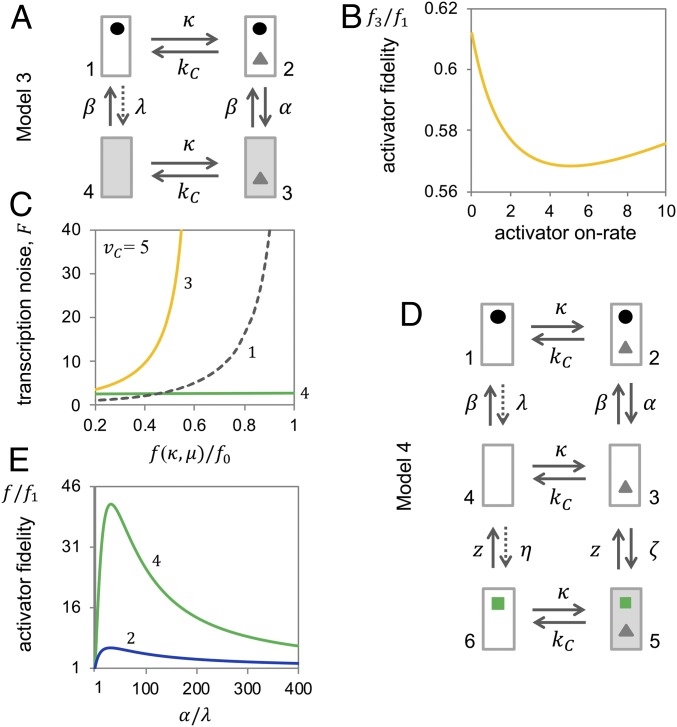
Effective kinetic proofreading requires that initiation of transcription is tied to activator binding. If transcription persists after dissociation of the activator (Fig. 3A; Model 3), kinetic discrimination between correct and incorrect activators occurs only once: before removal of the nucleosome (transition ). Since transcription is partially uncoupled from activator binding, activator fidelity remains well below the fidelity afforded by Model 1, despite nucleosome dynamics away from equilibrium (Fig. 3B; for proof, see SI Appendix); and because the upper limit of fidelity lies below , the noise in transcription rises faster in Model 3 than Model 1 as fidelity increases with decreasing activator on-rate (Fig. 3C).
Fig. 3.
Kinetic proofreading requires coupling of transcript initiation to activator binding; multiple proofreading steps improve fidelity. (A) Transition graph of Model 3. (B) Activator fidelity of Model 3 relative to the fidelity of Model 1 , as a function of activator on-rate, ; with , , α = , and (for , fidelities further decrease). (C) Transcription noise as a function of relative activator fidelity, , for Model 3 (yellow, 3), Model 4 (green, 4), and Model 1 (gray dashed line, 1). As in calculations for Fig. 2, we assumed and ; for Model 4 alone: and to reflect both active removal (by Mot1) and high concentration of TBP; all other parameters were as indicated above. Fano factor and fidelity were calculated as functions of the activator on-rate, κ, and the rate of transcription in the active state, (SI Appendix). (D) Transition graph of Model 4. (E) Activator fidelities of Model 4 (green, 4) and Model 2 (blue, 2) relative to fidelity for Model 1 as a function of . For Model 4, we assumed (thus, both parameters are varied equally) and . Other rate constants were , , , and . (For smaller , and z than assumed here, fidelity further increases; SI Appendix.)
Additional steps of kinetic proofreading may further increase activator fidelity. The central component for core promoter recognition of all genes, the TATA box binding protein (TBP) is thought to be recruited to promoters by activators as a subunit of either the SAGA or TFIID complex (27). The enzyme Mot1 couples adenosine triphosphate (ATP) hydrolysis to the removal of TBP from DNA (28, 29). Thus, Mot1 may drive the TBP–DNA binding reaction away from equilibrium, which affords a second step of activator proofreading: if transcriptional initiation requires continued activator binding for steps downstream of TBP binding (Fig. 3D, Model 4), the system can discriminate between correct and incorrect activator binding thrice—before nucleosome removal (transition ), after nucleosome removal , and after TBP binding —which may significantly improve activator fidelity (Fig. 3E). The second proofreading step increases the upper limit of fidelity to (for proof, see SI Appendix) and, as might be expected, affords further noise suppression at sufficiently high fidelities (SI Appendix, Fig. S1).
Discussion
In equilibrium, there is a fundamental limit, the “Hopfield barrier,” to how well any information processing task—e.g., transcription of specific genes in response to an environmental signal—can be undertaken (24). At the expense of free energy to maintain the system away from equilibrium, the Hopfield barrier may be breached. The system dynamics, then, are irreversible—i.e., for some sequence of events, forward and reverse direction are statistically distinguishable (SI Appendix); e.g., in Model 2 (Fig. 2A), clockwise cycles are more probable than counterclockwise cycles for .
Nonequilibrium dynamics, irreversibility, corresponds to entropy production (Fig. 2E). Nucleosomal proofreading of activator–DNA binding, therefore, calls for enzymes that couple the catalysis of nucleosome dynamics to exergonic (i.e., entropy-producing) reactions. That such enzymes indeed exist—ATP-dependent chromatin remodelers (30)—fulfills a critical demand of our theory.
Irreversibility is a necessary but by no means sufficient condition for kinetic proofreading. For instance, if nucleosome removal was much faster than the off-rate of the incorrect activator—i.e., if transition in Model 2 was effectively disallowed—the promoter dynamics would be irreversible. Yet, the expenditure of energy would afford no increase in activator fidelity, for the ability of distinguishing between correct and incorrect promoter binding in state 2 would no longer exist. This explains the monotonic decrease in activator fidelity beyond an optimal rate for activator-controlled nucleosome removal (Fig. 2B). To increase activator fidelity, activators must promote nucleosome removal but not too effectively. Therefore, induced promoters must never remain in a nucleosome-free state—in good agreement with experimental observation (12, 31)—and transcription cannot be devoid of noise due to the stochastic dynamics of nucleosome removal and reformation.
Our theory implies that ATP consumption by one or more chromatin remodelers recruited to the promoter is used not to speed up the approach to equilibrium but to maintain nucleosome dynamics away from equilibrium. This demand may be difficult to satisfy if removal of nucleosomes occurred by sliding alone, for the same remodeler may use ATP hydrolysis both to slide nucleosomes away from the promoter and back. Removal of nucleosomes by sliding, therefore, might be an ineffective use of ATP hydrolysis to drive nucleosome dynamics away from equilibrium. In contrast, nucleosome removal by ATP-dependent nucleosome disassembly couples ATP hydrolysis to removal alone—the reverse reaction, nucleosome reassembly, entails synthesis of ATP from adenosine diphosphate (ADP) and phosphate which, under physiological conditions, renders the reverse reaction highly improbable. This may explain why nucleosomes are removed from transcriptionally active promoters by disassembly rather than sliding (26).
Are the dynamics of activated promoter nucleosomes nonequilibrium dynamics? In our model, irreversibility is engendered by activator-stimulated nucleosome removal. Consistently, Pho4, the transcriptional activator of the PHO5 gene, promotes loss of PHO5 promoter nucleosomes (18, 21), not by occluding nucleosomes but recruitment of ATP-dependent chromatin remodelers (12, 32, 33). It may be no accident, therefore, that assumptions of irreversibility—unidirectional sliding and ordered removal of nucleosomes (12, 19)—helped to explain the observed statistical frequencies of PHO5 promoter nucleosome configurations. Accordingly, nucleosome occupancy at many promoter sequences in yeast is not explained by the thermodynamics of nucleosome formation (34).
Kinetic proofreading involves continual reactivation of transcription: to test for the continued presence of the (correct) activator, the promoter stochastically returns to nucleosome configurations that suppress transcription, despite environmental conditions that induce gene activity. The variation in promoter chromatin structure, therefore, must be intrinsic—i.e., independent of the environment. This prediction of our theory has been tested (22). If the nucleosomal variation was imposed by environmental variation, the nucleosome configuration of one promoter copy would be stochastically dependent on the configuration of another copy within the same cell. Contrary to this expectation, electron microscopic analyses of PHO5 promoter pairs in single cells showed that both copies were stochastically independent (22). The heterogeneity of promoter chromatin cannot be reduced to environmental variation and, therefore, must arise “intrinsically.” Thus, promoter nucleosome dynamics fulfill another critical demand of our theory.
Another implication of our theory is that activators must promote multiple steps toward transcription, which may explain why eukaryotic activators do not bear specific activities to stimulate transcription but promiscuously recruit other factors instead (2, 3): promiscuous recruitment easily affords the same activator the ability to promote multiple, biochemically distinct, steps toward transcription. In addition, recruitment delays the activator’s effect on transcription, a critical requirement for effective kinetic proofreading (Fig. 2B).
However, transcription may not strictly be limited to activator-bound promoter states because past activator-binding events are remembered in form of other factors (e.g., TBP) that are recruited by the activator but may remain at the promoter after dissociation of the activator. This uncoupling of transcription from activator binding diminishes the effectiveness of kinetic proofreading (Fig. 3B). Thus, our theory requires the existence of an enzyme that removes TBP from promoter DNA: Mot1 (29). TBP removal by Mot1 erases the memory of past activator-binding events and, thus, maintains a close relationship between transcription and activator binding; as a consequence, maintenance of transcription requires continual reactivation.
Mot1 may contribute to activator fidelity in yet another way. Energy expenditure by Mot1 affords a second kinetic proofreading step (Fig. 3D; Model 4). Mot1 must drive the TBP–DNA binding reaction away from equilibrium, which may significantly increase activator fidelity (Fig. 3E). In this context, it is of interest that both Mediator and TBP are recruited to promoters in association with repressing factors (35–37), which may provide additional proofreading steps. However, whether relief of this repression requires free-energy expenditure is not known. Multiple kinetic proofreading steps are likely required to solve the activator specificity problem in eukaryotes.
The free energy for regulatory specificity may be provided by activator–DNA binding alone, but higher binding affinities entail longer dwell times of the activator on its target sequence; activated genes are not easily turned off again. Activator fidelity may be increased by decreasing activator concentration. However, the price to be paid is long activator search times and, thus, erratic promoter activity, which blurs the correspondence between regulatory signal and transcriptional response (Figs. 1C and 2C). Kinetic proofreading of activator–promoter interactions resolves this dilemma. The free energy required to increase specificity and dampen transcription noise is not provided by increasing the binding energy for activator–promoter recognition—i.e., by increasing the average lifetime of the activator on the DNA—but ATP hydrolysis. Thus, kinetic proofreading reconciles seemingly competing demands of regulation: high specificity and fast promoter-state kinetics (i.e., openness to change).
Archaea possess a precursor of the nucleosome (38), but eubacteria lack nucleosome-like structures entirely. How do eubacteria solve the problem of regulatory specificity? Surprisingly, many regulators of transcription in eubacteria, e.g., the TetR and lac repressors, recognize their target sequences with greater specificity than most eukaryotic activators: equilibrium dissociation constants often fall into the picomolar range and energy differences between correct and incorrect promoter binding are two or three times larger compared to most eukaryotic activators (7, 9, 39, 40). The entailed problem of long DNA dwell times is solved by controlling the activity of transcriptional regulators via allosteric effectors—e.g., tetracycline and allolactose—that upon binding, induce large changes in the affinity of the transcription factor for its target sequence (41). The evolution of transcriptional activators in the presence of a ubiquitous repressor of transcription, the nucleosome (42), may have favored kinetic proofreading as a solution to the specificity problem instead.
Irreversibility is a probabilistic and not deterministic phenomenon (43). Thus, whether kinetic proofreading is employed to increase activator fidelity, as suggested here, sharpen the gene regulatory function (24), or increase the fidelity of substrate recognition by RNA polymerase (44) or promoter recognition by general transcription factors (45), transcription and its regulation may be fully understood only on the basis of probabilistic theories; random molecular motion may be a requirement for, rather than impediment of, biological function. Rigorous testing of theories that assert random molecular behavior requires novel methods for the analysis of gene expression at the level of single gene molecules rather than population averages (46).
Methods
Promoter-state dynamics and transcription were modeled as stationary Markov processes on strongly connected graphs (SI Appendix). Calculations where performed using Mathematica and Python.
Data Availability.
Mathematica notebooks and Python programs are available upon request.
Supplementary Material
Acknowledgments
We like to thank Drs. Joshua Deutsch, Namrita Dillon, John Field, Jeremy Gunawardena, Grant Hartzog, Rohinton Kamakaka, Roger Kornberg, and Michael Levitt, as well as Kevin Chen, for discussion, critical comments, encouragement, helpful suggestions, and proofreading. This work was supported by National Science Foundation Grant 1243957 (to H.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911188117/-/DCSupplemental.
References
- 1.Mao C., et al. , Quantitative analysis of the transcription control mechanism. Mol. Syst. Biol. 6, 431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ptashne M., Gann A., Transcriptional activation by recruitment. Nature 386, 569–577 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Bryant G. O., Ptashne M., Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11, 1301–1309 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Cosma M. P., Tanaka T., Nasmyth K., Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97, 299–311 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Kuras L., Borggrefe T., Kornberg R. D., Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. U.S.A. 100, 13887–13891 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halford S. E., Marko J. F., How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geertz M., Shore D., Maerkl S. J., Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc. Natl. Acad. Sci. U.S.A. 109, 16540–16545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen W. C., Green M. R., Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90, 615–624 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Maerkl S. J., Quake S. R., A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Pauling L., “The probability of errors in the process of synthesis of protein molecules” in Festschrift Arthur Stoll, Birkhaeuser A., Ed. (Birkaeuser, Basel, Switzerland, 1957), pp. 597–602. [Google Scholar]
- 11.Hopfield J. J., Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown C. R., Mao C., Falkovskaia E., Jurica M. S., Boeger H., Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 11, e1001621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jessen W. J., Hoose S. A., Kilgore J. A., Kladde M. P., Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 13, 256–263 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Small E. C., Xi L., Wang J. P., Widom J., Licht J. D., Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 111, E2462–E2471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornberg R. D., Chromatin structure: A repeating unit of histones and DNA. Science 184, 868–871 (1974). [DOI] [PubMed] [Google Scholar]
- 16.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Kornberg R. D., Lorch Y., Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Almer A., Rudolph H., Hinnen A., Hörz W., Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boeger H., Nucleosomes, transcription, and probability. Mol. Biol. Cell 25, 3451–3455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeger H., Griesenbeck J., Kornberg R. D., Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133, 716–726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D., Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11, 1587–1598 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Brown C. R., Boeger H., Nucleosomal promoter variation generates gene expression noise. Proc. Natl. Acad. Sci. U.S.A. 111, 17893–17898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kepler T. B., Elston T. C., Stochasticity in transcriptional regulation: Origins, consequences, and mathematical representations. Biophys. J. 81, 3116–3136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estrada J., Wong F., DePace A., Gunawardena J., Information integration and energy expenditure in gene regulation. Cell 166, 234–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C., ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97, 833–842 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D., Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14, 667–673 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Taatjes D. J., The continuing SAGA of TFIID and RNA polymerase II transcription. Mol. Cell 68, 1–2 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Adamkewicz J. I., Hansen K. E., Prud’homme W. A., Davis J. L., Thorner J., High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J. Biol. Chem. 276, 11883–11894 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Auble D. T., et al. , Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8, 1920–1934 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Zhou C. Y., Johnson S. L., Gamarra N. I., Narlikar G. J., Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 45, 153–181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griesenbeck J., Boeger H., Strattan J. S., Kornberg R. D., Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol. Cell. Biol. 23, 9275–9282 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao C., Brown C. R., Griesenbeck J., Boeger H., Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS One 6, e17521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown C. R., Mao C., Falkovskaia E., Law J. K., Boeger H., In vivo role for the chromatin-remodeling enzyme SWI/SNF in the removal of promoter nucleosomes by disassembly rather than sliding. J. Biol. Chem. 286, 40556–40565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorch Y., Maier-Davis B., Kornberg R. D., Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 28, 2492–2497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sermwittayawong D., Tan S., SAGA binds TBP via its Spt8 subunit in competition with DNA: Implications for TBP recruitment. EMBO J. 25, 3791–3800 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeronimo C., et al. , Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell 64, 455–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anandapadamanaban M., et al. , High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat. Struct. Mol. Biol. 20, 1008–1014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik H. S., Henikoff S., Phylogenomics of the nucleosome. Nat. Struct. Biol. 10, 882–891 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Hillen W., Gatz C., Altschmied L., Schollmeier K., Meier I., Control of expression of the Tn10-encoded tetracycline resistance genes. Equilibrium and kinetic investigation of the regulatory reactions. J. Mol. Biol. 169, 707–721 (1983). [DOI] [PubMed] [Google Scholar]
- 40.Forde G. M., et al. , LacO-LacI interaction in affinity adsorption of plasmid DNA. Biotechnol. Bioeng. 95, 67–75 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Schleif R. F., Modulation of DNA binding by gene-specific transcription factors. Biochemistry 52, 6755–6765 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Lorch Y., LaPointe J. W., Kornberg R. D., Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987). [DOI] [PubMed] [Google Scholar]
- 43.Boltzmann L., Ueber die Beziehung zwischen dem zweiten Hauptsatz der mechanischen Waermetheorie und der Wahrscheinlichkeitsrechnung, respective den Saetzen ueber das Waermegleichgewicht. Wiener Berichte 75, 373–435 (1877). [Google Scholar]
- 44.Mellenius H., Ehrenberg M., Transcriptional accuracy modeling suggests two-step proofreading by RNA polymerase. Nucleic Acids Res. 45, 11582–11593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Bushnell D. A., Silva D. A., Huang X., Kornberg R. D., Initiation complex structure and promoter proofreading. Science 333, 633–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H., Larson D. R., What have single-molecule studies taught us about gene expression? Genes Dev. 30, 1796–1810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Gillespie D. T., A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 (1976). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mathematica notebooks and Python programs are available upon request.