Significance
Infection by the bacterium Helicobacter pylori is the main risk factor for stomach cancer. Strains of H. pylori that produce the virulence factor CagA substantially increase stomach cancer risk compared with strains without CagA. CagA disrupts cell polarity—the specialized asymmetric organization within cells—but the mechanism it uses and its influence on colonization are not fully understood. Using a three-dimensional culture of stomach cells (human gastric organoids) and cell lines, we find that the tumor suppressor ASPP2 is crucial in disruption of cell polarity by CagA during H. pylori infection. Interfering with the CagA–ASPP2 interaction, using small molecules or a specific peptide, blocks loss of cell polarity and reduces bacterial colonization in organoids.
Keywords: microbial pathogenesis, cell polarity, tumor suppressors
Abstract
The main risk factor for stomach cancer, the third most common cause of cancer death worldwide, is infection with Helicobacter pylori bacterial strains that inject cytotoxin-associated gene A (CagA). As the first described bacterial oncoprotein, CagA causes gastric epithelial cell transformation by promoting an epithelial-to-mesenchymal transition (EMT)-like phenotype that disrupts junctions and enhances motility and invasiveness of the infected cells. However, the mechanism by which CagA disrupts gastric epithelial cell polarity to achieve its oncogenicity is not fully understood. Here we found that the apoptosis-stimulating protein of p53 2 (ASPP2), a host tumor suppressor and an important CagA target, contributes to the survival of cagA-positive H. pylori in the lumen of infected gastric organoids. Mechanistically, the CagA–ASPP2 interaction is a key event that promotes remodeling of the partitioning-defective (PAR) polarity complex and leads to loss of cell polarity of infected cells. Blockade of cagA-positive H. pylori ASPP2 signaling by inhibitors of the EGFR (epidermal growth factor receptor) signaling pathway—identified by a high-content imaging screen—or by a CagA-binding ASPP2 peptide, prevents the loss of cell polarity and decreases the survival of H. pylori in infected organoids. These findings suggest that maintaining the host cell-polarity barrier would reduce the detrimental consequences of infection by pathogenic bacteria, such as H. pylori, that exploit the epithelial mucosal surface to colonize the host environment.
Half of the world’s population is infected by the gram-negative bacterium Helicobacter pylori, a pathogen known to cause gastritis, peptic ulcers, and gastric cancer (1, 2). The two major known types of H. pylori are type I strains that express cytotoxin-associated gene A (CagA) with the type IV secretion system (TFSS), a secretion apparatus used to inject CagA inside the host cell, and type II strains that are cagA-negative or TFSS-defective (3–7). Infection with cagA-positive H. pylori increases the risk of stomach cancer by 5- to 10-fold compared with infection with cagA-negative strains (8). Consistently, Mongolian gerbils infected with an H. pylori TFSS- and CagA-proficient strain develop gastric carcinoma within 12 wk of infection in a CagA-dependent manner (9), supporting the importance of CagA in oncogenesis. The World Health Organization has identified H. pylori as a type I carcinogen (10) and recently ranked H. pylori as a high-priority pathogen for which new antibiotics are urgently needed (11). How to eliminate the pathogenic impact of cagA-positive H. pylori remains an important challenge due to the rise in antibiotic-resistant H. pylori strains (12). Some epidemiological studies have observed an inverse correlation between reduced incidence of H. pylori infection and increased incidence of Barrett’s esophagus and esophageal adenocarcinoma in Western countries, suggesting a potential protective impact of H. pylori (13). Therefore, strategies to reduce cancer incidence need specifically to consider the mechanisms by which CagA transforms gastric epithelial cells. It is important to note that cagA-positive and -negative H. pylori, and mutant cagA-positive strains that are unable to inject CagA, can colonize human cells in vivo. It has also been reported that some injection-defective mutants colonize in model systems more efficiently than wild-type strains (14, 15). This raises the interesting questions of whether injection of CagA might elicit host cell responses that limit infection and whether H. pylori has strategies to overcome these responses.
In the stomach, the mucosal barrier is formed by highly polarized gastric epithelial cells and H. pylori is often found at the mucous barrier or directly attached to epithelial cells where it uses the apical surface of the cells as a replicative niche (16, 17). Polarization of epithelial cells provides a barrier function and allows compartmentalization of molecules to either the apical side, which interfaces with the external environment, or to the basolateral side, where connections with neighboring cells and the extracellular matrix are established (18). The three major polarity complexes are partitioning-defective (PAR) (19), Crumbs (CRB) (20), and Scribble (SCR) (21). CRB and SCR localize at the apical and basolateral membranes, respectively, whereas different components of PAR are at the basolateral side (Par1 kinases), apical side (aPKC, Par6, cdc42), and tight junctions (Par3). Polarity complex localization is tightly regulated by kinases, including the Par1 kinases that normally localize at the basolateral membrane (22). The importance of cell polarity as the first line of defense against infection is reflected by many pathogenic bacteria—including Pseudomonas aeruginosa and Neisseria meningitidis as well as H. pylori—targeting host polarity complexes. P. aeruginosa and N. meningitidis target the apical components of the PAR complex (23), whereas H. pylori is known to target the basolateral Par1b kinase (24). Perturbation of host cell polarity is required for pathogens to colonize the host environment effectively (17, 25). However, the mechanism for the H. pylori-induced loss of cell polarity is only partially understood. Furthermore, an intriguing question is whether interfering with bacterial mechanisms that disrupt host cell polarity might provide new approaches to target bacteria. Doing so would require the identification of a targetable mechanistic step in the process of pathogen-mediated disruption of polarity without any adverse effects on host cell polarity.
Injection of CagA by H. pylori triggers a plethora of effects on epithelial cells, with loss of cell polarity, adhesion, and increased cell motility as prominent phenotypes (26, 27). Thus, CagA induces an epithelial-to-mesenchymal transition (EMT)-like phenotype. In a previous unbiased interactome study, we identified apoptosis-stimulating protein of p53 2 (ASPP2) as a prominent cellular target of CagA (28) that binds to the N terminus of CagA (residues 19 to 235) (29). ASPP2 is a haploinsufficient tumor suppressor (30) that functions as a shuttling transcriptional regulator that is non–DNA-binding (31). ASPP2 shuttles from the tight junctions to the nucleus where it binds p53 and regulates p53 transcriptional target selectivity to enhance p53-induced apoptosis (32). Binding to CagA subverts the proapoptotic function of ASPP2 and induces p53 degradation (28, 29). ASPP2 is also a regulator of the PAR3 complex at tight junctions (33, 34). Reduced ASPP2 expression induces EMT whereas increased ASPP2 expression induces the reverse phenomenon, namely a mesenchymal-to-epithelial transition (35). We therefore reasoned that ASPP2 might be a central hub through which CagA deregulates not only p53 activity but also cell polarity.
To investigate this, we studied the role of ASPP2 in mediating CagA-induced loss of cell polarity using the gastric cancer cell line AGS, the polarized epithelial cell line Madin–Darby canine kidney (MDCK), and an ex vivo primary human gastric organoid model. We show the importance of the CagA–ASPP2 interaction in CagA-mediated disruption of polarity using peptide-mediated inhibition of the interaction. Notably, kinase inhibitors—identified in a high-content imaging screen—that interfere with CagA–ASPP2 complex formation also block loss of cell polarity, revealing the importance of receptor tyrosine kinase (RTK)/phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) signaling in the recruitment of ASPP2 by CagA. We validate these findings in primary human gastric organoids and demonstrate the importance of the CagA–ASPP2 complex in colonization by cagA-positive H. pylori.
Results
ASPP2 Is the Key Node in CagA-Induced Remodeling of the PAR Complex.
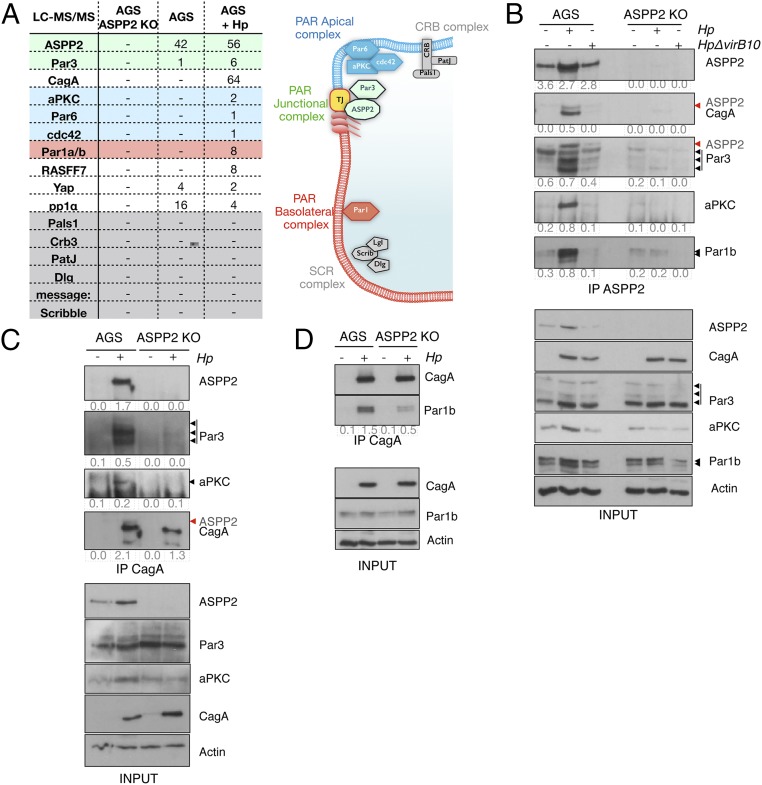
ASPP2 has previously been implicated in regulating epithelial cell polarity (33, 34). We therefore hypothesized that ASPP2 is essential for CagA-induced loss of host cell polarity. To test this, we examined the ASPP2 interactome by analyzing affinity-purified ASPP2 from cagA-positive H. pylori-infected or -uninfected stomach gastric adenocarcinoma (AGS) cells by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Several known ASPP2-interacting proteins, including PP1α, Yap1, and Par3, were identified in uninfected cells (Fig. 1A). In infected cells, there was increased association between ASPP2 and Par3, and the entire PAR complex copurified with ASPP2: namely, the apical PAR polarity complex (aPKC, Par6, and cdc42), the junctional Par3, and the basolateral proteins Par1a and Par1b (Fig. 1 A, Right). Specificity was confirmed using a cell line in which ASPP2 has been genetically ablated through CRISPR/Cas9-mediated genome engineering (Fig. 1A and SI Appendix, Fig. S1A).
Fig. 1.
ASPP2 mediates CagA–PAR complex formation. (A, Left) Number of unique peptides identified by LC-MS/MS after immunoprecipitation of endogenous ASPP2 from parental AGS cells, ASPP2 KO AGS (clone 2) cells, or AGS cells infected with wt H. pylori. Colors correspond to the adjacent diagram. (A, Right) Diagram of polarity complex distribution in epithelial cells (apical membrane, blue; basolateral membrane, red; TJ, tight junction). (B) Parental or ASPP2 KO AGS cells were left uninfected or infected with the wt or ΔvirB10 mutant H. pylori strains. After 7 h, cells were lysed and subjected to ASPP2 immunoprecipitation (IP) followed by SDS/PAGE and immunoblotting using the indicated antibodies. Black arrowheads indicate the different isoforms of Par3 and Par1b. Red arrowheads indicate residual ASPP2 signal, as the membrane was first immunoblotted for ASPP2 and subsequently for CagA and Par3. Actin serves as a loading control. (C and D) AGS cells were infected as described in B. CagA immunoprecipitates were immunoblotted using the indicated antibodies. In C, fourth panel, the membrane was immunoblotted for ASPP2 (red arrowhead) and subsequently immunoblotted for CagA. Actin serves as a loading control. In B–D, the immunoprecipitated levels were calculated by densitometry and the enrichment has been stated as a fold change compared with the input. See also SI Appendix, Fig. S1.
We confirmed these findings by testing the interaction of ASPP2 with a representative member of each of the PAR subcomplexes (aPKC for apical, Par3 for junctional, and Par1b for basolateral) from cells that were uninfected, or infected with wild-type (wt) H. pylori or the ΔvirB10 H. pylori strain. ΔvirB10 carries a mutation in CagY, meaning that the mutant bacteria have a defective TFSS that cannot inject CagA; we use ΔvirB10 to compare CagA-positive versus CagA-negative host cells after infection. Note that immunostaining of cells infected with wt or ΔvirB10 H. pylori strains readily detects the bacterium, as well as the CagA–ASPP2 association in cells infected with the wt H. pylori strain (SI Appendix, Fig. S1B). Infection with wt, but not ΔvirB10, H. pylori increases the association between ASPP2 and each of the tested PAR components (Fig. 1B). Next, we tested whether CagA is part of the H. pylori-induced ASPP2–PAR component complex. Immunoprecipitation of CagA from wt H. pylori-infected control cells retrieved ASPP2, Par3, aPKC, and Par1b (Fig. 1 C and D). Importantly, in H. pylori-infected ASPP2-deficient cells, immunoprecipitation of CagA does not retrieve aPKC or Par3 (Fig. 1C) and the interaction with Par1b is reduced (Fig. 1D).
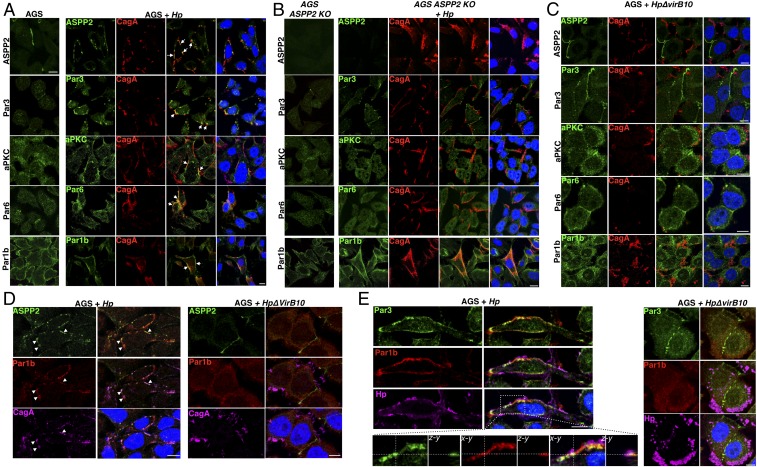
To further corroborate our results, we assessed by immunofluorescence the distribution of the PAR components in cells that were uninfected or infected with wt or ΔvirB10 H. pylori strains. In line with the immunoprecipitation results, infection of AGS cells with wt H. pylori results in colocalization of CagA and all of the apical PAR components tested, including Par3, Par6, aPKC, and Par1b, at the sites where the bacterium is attached to the cells (Fig. 2A). However, in H. pylori-infected ASPP2-deficient cells, PAR3 complex components rarely colocalize with CagA, so their distribution is not affected by infection (Fig. 2B and SI Appendix, Fig. S1C); a similar result was observed in AGS cells infected with the ΔvirB10 H. pylori strain (Fig. 2C). In addition, we observed a strong reduction in the redistribution at the plasma membrane of Par1b in infected ASPP2 knockout (KO) cells compared with the infected wt AGS cells (Fig. 2B and SI Appendix, Fig. S1 C and D). To assess the specificity of these findings, we tested changes in the distribution of two unrelated polarity complexes, CRB and SCR. Both members of the apical CRB complex (Cbr3 and Pals1), as well as a member of the SCR complex (Scribble), were not affected by infection with H. pylori (SI Appendix, Fig. S2A), confirming that the PAR complex is the main polarity target of the H. pylori infection.
Fig. 2.
ASPP2 mediates colocalization of the CagA–PAR complex. (A) Confocal images of AGS cells that were uninfected (Left) or infected with wt H. pylori (Hp) (Right). Seven hours postinfection, cells were fixed and stained for markers of the PAR complex (ASPP2, Par3, aPKC, Par6, and Par1b, green), CagA (red), and nuclei (DAPI, blue). White arrows point to the colocalization between CagA and PAR members. (B) ASPP2 KO AGS cells were infected with wt H. pylori and treated as described in A. (C) AGS cells were infected with the ΔvirB10 mutant H. pylori strain and treated as described in A. (D and E) AGS cells were infected with the wt or ΔvirB10 mutant H. pylori strains. After 7 h, cells were fixed and stained for (D) ASPP2 (green), Par1b (red), CagA (magenta), and nuclei (DAPI, blue) and (E) Par1b (green), Par3 (red), H. pylori (magenta), and nuclei (DAPI, blue). (E, Insets) Z sections of the colocalization between Par1b and Par3. White arrowheads in D point to the colocalization between ASPP2, Par1b, and CagA. (Scale bars, 10 μm.) See also SI Appendix, Fig. S2.
The PAR complex is asymmetrically distributed in epithelial cells and posttranslational modification, mainly by the aPKC and Par1 kinases, regulates its cellular distribution (36). We tested whether infection with H. pylori promotes the formation of an aberrant PAR complex by uniting components that are not normally complexed together (37). Indeed, H. pylori-infected cells showed a strong colocalization between CagA, ASPP2, and Par1b (Fig. 2D and SI Appendix, Fig. S2B), and Par3 and Par1b (Fig. 2E and SI Appendix, Fig. S2C), supporting the idea that these PAR members are simultaneously recruited by cagA-positive H. pylori. Together, these data indicate that ASPP2 is a key target via which CagA redistributes the PAR polarity complex.
ASPP2 Is Required for CagA to Disrupt Cell Polarity.
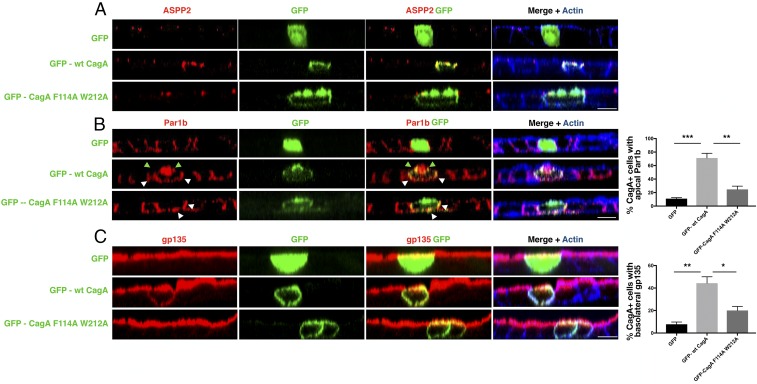
The ASPP2–CagA cocrystal structure shows that Phe114 and Thr212 of CagA are essential for the interaction of CagA with ASPP2 (29). Hence, to test whether the CagA–ASPP2 interaction is a prerequisite for CagA-induced cell-polarity loss, we generated a GFP-tagged CagA mutant that is defective in ASPP2 binding (GFP-CagA F114A/W212A) and we expressed this mutant, wt CagA, or GFP in a polarized epithelial cell line (MDCK). In polarized MDCK cells, GFP-CagA F114A/W212A localizes to the plasma membrane and promotes the same elongated phenotype as seen with GFP-tagged wt CagA (GFP-wt CagA) (SI Appendix, Fig. S3A). As expected, the CagA F114A/W212A mutant does not bind ASPP2 (SI Appendix, Fig. S3B) and its expression has minimal impact on the normal cellular localization of ASPP2 (Fig. 3A). Notably, this mutant is not able to mislocalize the basolateral marker Par1b (Fig. 3B) and the apical marker gp135 to the apical and basolateral membranes, respectively (Fig. 3C). These results indicate that the ASPP2 binding-defective CagA mutant is unable to disrupt cell polarity, in contrast to wt CagA (Fig. 3 B and C). Thus, full-length mutant CagA that is unable to bind ASPP2 is still able to induce cell elongation but is unable to break the junctional complex and promote the loss of cell polarity, similar to the phenotype induced by expression of the isolated CagA C terminus (26). Together, these results indicate that binding to ASPP2 is required for CagA to achieve full disruption of cell polarity.
Fig. 3.
Binding to ASPP2 is required for CagA to disrupt cell polarity. (A) Confocal images of polarized MDCK monolayers transfected to express either GFP, GFP-wt CagA, or mutant GFP-CagA F114A W212A (green). Monolayers were stained for ASPP2 (red) and actin (phalloidin, blue). (B) As in A, except monolayers were stained for Par1b (red) and actin (phalloidin, blue). Green arrowheads point to the colocalization between CagA and Par1b at both the apical and basolateral sides. White arrowheads point to the colocalization between CagA and Par1b at the basolateral side only. (C) As in A, except monolayers were stained for the apical marker gp135 (red) and actin (phalloidin, blue). (Scale bars, 10 μm.) Percentages of apical Par1b (B) and basolateral gp135 (C) were determined for cells expressing GFP (total n = 30, from three experiments; black bars), GFP-wt CagA (total n = 63, from three experiments; light gray bars), and GFP-CagA F114A W212A (total n = 69, from three experiments; dark gray bars). Significance was tested using a one-way ANOVA multiple-comparison test. Error bars, ±SEM. ***P < 0.001, **P < 0.01, *P < 0.05. See also SI Appendix, Fig. S3.
The apical redistribution of Par1b is thought to be important for the CagA-induced EMT-like phenotype (24). Although we still detected colocalization between Par1b and the mutant CagA F114A/W212A, this only occurred at the basolateral side of the cell (Fig. 3 B, Bottom, white arrowheads) and CagA was unable to redistribute Par1b to the apical membrane. Phosphorylation of Par1b Thr595 normally results in the dissociation of Par1b from the plasma membrane. We observed a small but detectable reduction in Par1b phosphorylation upon infection with wt H. pylori (SI Appendix, Fig. S3C) (24) and Par1b remains associated with CagA at the plasma membrane (SI Appendix, Fig. S1 D, Top). Importantly, in ASPP2-deficient cells, we did not observe a reduction in Par1b phosphorylation upon H. pylori infection (SI Appendix, Fig. S3C), and Par1b had an increased cytoplasmic distribution (SI Appendix, Figs. S1 D, Bottom and S3 D and E). Thus, when CagA is unable to bind ASPP2, it also fails to recruit Par1b efficiently and to promote cell-polarity loss. Combined with the above results, our findings show that CagA serves as a scaffold protein that, via ASPP2, brings together the tight-junctional, apical Par complex ASPP2–Par3–aPKC (Fig. 2 A and B and SI Appendix, Fig. S3D) and the basolateral complex Par1b (Figs. 2 A and B and 3B and SI Appendix, Fig. S3 D and E), thereby forming an abnormal complex that leads to the loss of cell polarity in epithelial cells infected with H. pylori.
CagA Colocalizes with ASPP2 and the PAR Complex at the Apical Side in Infected Gastric Organoid Cells.
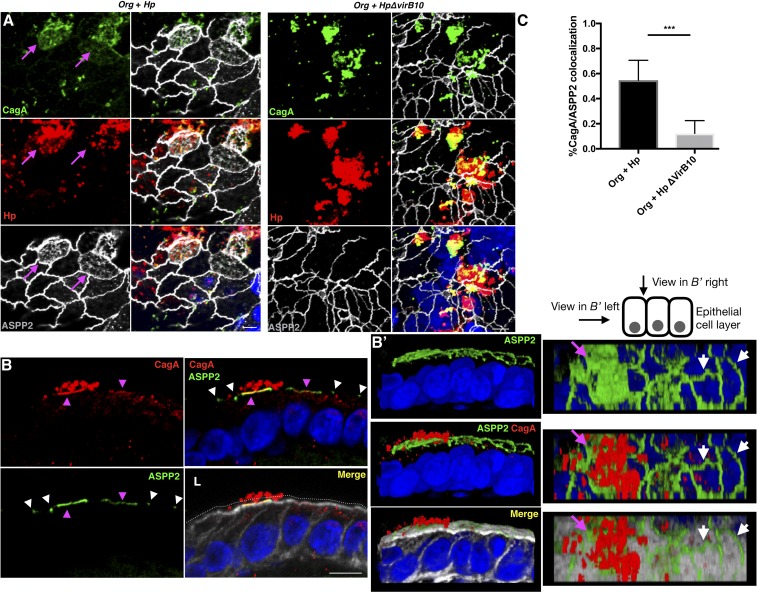
To place these findings in a physiological context, we used an ex vivo human gastric organoid (gastroid) system that has previously been used as a model to study H. pylori infection (38). Human gastric organoids closely mimic the gastric glands and recapitulate important features of the gastric compartment, including a well-polarized epithelium. Gastric antrum tissues from gastrectomy samples from two morbidly obese donors without known gastric disease were used to establish the organoid lines. In the organoids, ASPP2 has a distinct apical localization at the tight junctions that matches its cellular distribution in the tissue of origin (SI Appendix, Fig. S4A), and colocalizes with Par3 (SI Appendix, Fig. S4B). Mechanically disrupted gastroids were incubated with H. pylori and allowed to close the lumen with the internalized bacterium before reembedding in Matrigel. Upon infection with wt H. pylori, CagA is delivered into the epithelial cells of the organoids as phosphorylation of CagA is detected 24 h postinfection (SI Appendix, Fig. S4C). In the presence of H. pylori, ASPP2 is mislocalized from the apical junctions to the apical side of the epithelial cells in close proximity to the attached bacteria (Fig. 4, SI Appendix, Fig. S4D, and Movies S1 and S2). These effects are strictly dependent on the delivery of CagA, as the distribution of ASPP2 is unchanged in organoids infected with the H. pylori ΔvirB10 mutant (Fig. 4A and SI Appendix, Fig. S4D).
Fig. 4.
Apical redistribution of ASPP2 in gastric organoids infected with H. pylori. (A) Human gastroids (Org) were infected for 24 h with wt or ΔvirB10 mutant H. pylori strains. Infected organoids were fixed and stained for CagA (green), Hp (red), ASPP2 (gray), and nuclei (DAPI, blue). Magenta arrows point to the apical distribution of ASPP2 in cells infected with H. pylori. (Scale bars, 10 μm.) (B) Confocal images of human gastroids infected with wt H. pylori for 24 h and stained for ASPP2 (green), CagA (red), actin (phalloidin, gray), and nuclei (DAPI, blue). Magenta arrowheads point to the colocalization of ASPP2 with CagA and to the apical redistribution of ASPP2. White arrowheads point to the junctional localization of ASPP2. The white dotted line indicates the lumen (L) of the organoid. (Scale bar, 10 μm.) (B′) Three-dimensional reconstruction of the images shown in B. (B′, Left) View from the side of the epithelial layer. (B′, Right) View from the top. Magenta arrowheads point to the colocalization of ASPP2 with CagA and to the apical redistribution of ASPP2. White arrowheads point to the junctional localization of ASPP2. (C) Quantification of the percentage of colocalization between CagA and ASPP2. See method C in Quantification of Microscopy for further explanation of the analysis method. Significance was tested using a one-way ANOVA Tukey’s multiple-comparison test. Error bars, ±SEM. ***P < 0.001. Redistribution of ASPP2 was assessed for >100 H. pylori-infected cells from three or more independent experiments. See also SI Appendix, Figs. S4 and S5.
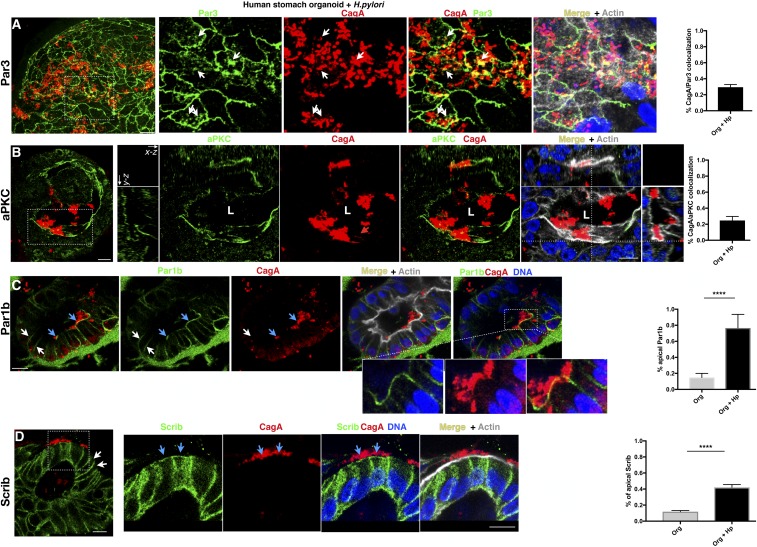
Similar to ASPP2, Par3 colocalizes with CagA on the apical side, at the site of bacterial attachment in H. pylori-infected organoids (Fig. 5A and SI Appendix, Fig. S5A). Although the localization of aPKC at the apical side of the epithelial cells (SI Appendix, Fig. S5B) is not visibly altered in H. pylori-infected organoids, we nonetheless detected CagA colocalization with aPKC (Fig. 5B).
Fig. 5.
Relocalization of the junctional Par3 complex and basolateral Par1b complex by H. pylori in gastric organoids. (A–C) Confocal images of human gastroids infected with H. pylori and stained for Par3 (green; A), aPKC (green; B), Par1b (green; C), CagA (red), nuclei (DAPI, blue), and actin (phalloidin, gray). In A, white arrows point to the colocalization between Par3 and CagA. (B, Insets) All three projections of the epithelial cells infected with H. pylori. L, lumen of the organoid. In C, blue arrows point to the apical localization of Par1b in epithelial cells infected with H. pylori; white arrows point to the normal basolateral distribution of Par1b. The enlarged insets show the tight association between CagA and Par1b. (D) Confocal images of human gastroids infected as described in A and stained for the basolateral marker Scribble (Scrib; green), CagA (red), actin (phalloidin, gray), and nuclei (DAPI, blue). Blue arrows point to the apical distribution of Scribble in the epithelial cells infected with H. pylori; white arrows point to the normal basolateral distribution of Scribble. (Scale bars, 10 μm.) Quantification of the percentage of colocalization between CagA and each polarity marker (Par3, aPKC, Par1b, and Scrib) is shown (Right). See method C in Quantification of Microscopy for further explanation of the analysis method. Quantification of the percentage of apical localized Par1b and Scrib was measured as a percentage of colocalization between these markers and cortical actin. Significance was tested using a one-way ANOVA Tukey’s multiple-comparison test. Error bars, ±SEM. ****P < 0.0001. Redistribution of the polarity markers was assessed for >75 H. pylori-infected cells from three or more independent experiments. See also SI Appendix, Fig. S5.
In the organoids, Par1b shows a symmetrical distribution to the basolateral domain of the epithelial cells (Fig. 5C, white arrows and SI Appendix, Fig. S5C). Upon infection with H. pylori, Par1b is relocalized just underneath the site of bacterial attachment at the apical side of the infected cells (Fig. 5C, light blue arrows, SI Appendix, Fig. S5D, and Movies S3 and S4). Delocalization of basolateral and tight-junctional proteins is one of the first steps that leads to the loss of cell polarity (39). Accordingly, epithelial cells of organoids infected with H. pylori show an increased cytoplasmic and apical distribution of the basolateral protein Scribble (Fig. 5D) compared with uninfected organoids in which Scribble is restricted to the basolateral side (SI Appendix, Fig. S5E). This suggests that the tight-junctional fence that regulates paracellular transit of molecules is at least temporarily compromised upon infection. The organoid model shows similar results to our in vitro findings and shows that H. pylori recruits basolateral (Par1b) and apical-junctional PAR proteins (ASPP2, Par3, aPKC) to the apical membrane of the host cells at the sites where H. pylori is attached to the cells. These effects perturb the host epithelial cell polarity and promote the mislocalization of other cell-polarity markers such as Scribble, which is not detected in a nonpolarized system such as AGS cells.
CagA–ASPP2 Complex Formation Is Blocked by RTK/PI3K/AKT Inhibitors Identified by a High-Content Imaging Screen.
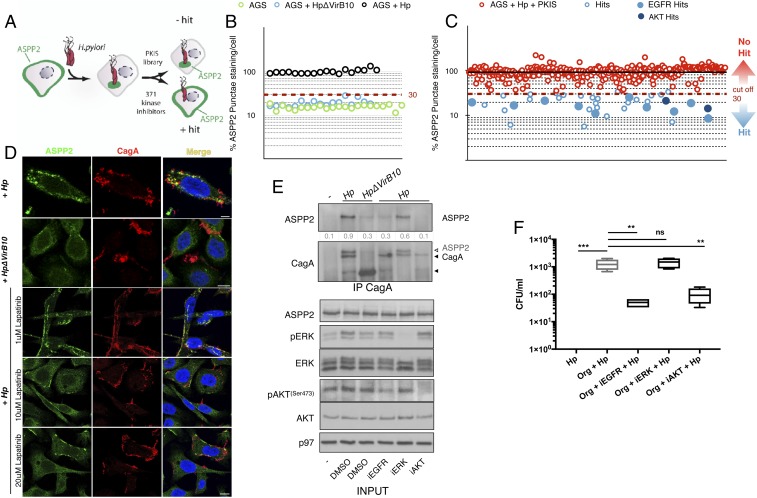
The observation that the association between CagA and ASPP2 is an upstream event involved in H. pylori-induced loss of cell polarity suggests that interference with this protein–protein interaction may provide a novel means by which to decrease the detrimental consequences of H. pylori infection. Previous studies have shown that the phosphorylation status of CagA influences its oncogenic functions and, importantly, H. pylori infection also has a profound impact on host cell signaling pathways (40–42). Therefore, to identify small-molecule inhibitors that prevent the CagA–ASPP2 interaction and to study how such inhibitors influence H. pylori fitness and colonization of the gastric environment, we performed a high-content fluorescence microscopy-based high-throughput screen using a GlaxoSmithKline (GSK) Published Kinase Inhibitor Set (PKIS) in AGS cells. This library comprises 371 small-molecule ATP-competitive kinase inhibitors with well-characterized bioactivity (43).
We analyzed the cellular distribution of ASPP2 in uninfected cells, cells infected with wt or ΔvirB10 mutant H. pylori strains, or cells infected with H. pylori in the presence of the small-molecule compounds (Fig. 6A and SI Appendix, Fig. S6 A and A′). ASPP2 distribution was quantified through automated image analysis (SI Appendix, Fig. S6 A and A′). Uninfected and H. pylori ΔvirB10 mutant-infected cells had ∼25% of the level of ASPP2 redistribution of cells infected with wt H. pylori (Fig. 6B and SI Appendix, Fig. S6B; where the average redistribution of ASPP2 in the cells infected with wt H. pylori was assigned as 100%). The screen yielded strong reproducibility between replicates, with a Pearson correlation of 0.88, and a good dynamic range between negative and positive controls, with the average Z factor for the screen being 0.78 and 0.74 when wt H. pylori-infected cells were compared with uninfected or mutant H. pylori-infected cells, respectively (SI Appendix, Fig. S6B). We set our “positive-hit” cutoff as below 30% (Fig. 6B) to identify robust inhibitors of the redistribution of ASPP2 by wt H. pylori, in line with the percent difference we observed with uninfected and H. pylori ΔvirB10 mutant-infected cells. This led to the identification of 39 compounds that prevented the recruitment of ASPP2 by H. pylori (Fig. 6C). Interestingly, around half of these inhibitors (21/39 compounds) target receptor tyrosine kinases (RTKs) (SI Appendix, Fig. S6 C and D). Of these RTK inhibitors, 15 targeted the epidermal growth factor receptor (EGFR) (Fig. 6C and SI Appendix, Fig. S6E), suggesting that EGFR signaling regulates the CagA–ASPP2 interaction. The remaining hits target the PKB/AKT pathway (11/39), regulators of the cytoskeleton (2/39), and regulators of the cell cycle (5/39) (SI Appendix, Fig. S6E).
Fig. 6.
High-content screen of gastric epithelial cells infected with H. pylori to identify inhibitors of the CagA–ASPP2 interaction. (A) Schematic of the high-content screen used to identify compounds blocking the CagA–ASPP2 interaction. PKIS, Protein Kinase Inhibitor Set library B. (B) ASPP2 relocalization was quantified as the number of ASPP2 punctae per cell in AGS cells that were uninfected (green circles), infected with wt H. pylori (black circles), or infected with the H. pylori ΔvirB10 mutant (blue circles). The dashed red line shows the cutoff (30%). See also method B in Quantification of Microscopy for further explanation of the analysis method. (C) Results of the high-content screening of a PKIS library of AGS cells infected with wt H. pylori and treated with the PKIS compound library (red circles). The solid black line indicates the median. The dashed red line represents the cutoff (30%). An average of n = 2 replicates is shown. (D) Confocal images of AGS cells incubated with DMSO or increasing concentrations of EGFR inhibitor (lapatinib) and infected with wt or ΔvirB10 mutant H. pylori. Seven hours after infection, cells were fixed and stained for ASPP2 (green), CagA (red), and nuclei (DAPI, blue). (Scale bar, 10 μm.) (E) AGS cells were treated with different kinase inhibitors (iEGFR, 20 μM lapatinib; iERK, 5 μM U126; iAKT, 10 μM AKTVIII) or DMSO and infected with wt or H. pylori ΔvirB10 mutant strains. Cell lysates were subjected to CagA immunoprecipitation followed by SDS/PAGE and immunoblotting using the indicated antibodies. p97 served as a loading control. In the second panel, the membrane was immunoblotted for ASPP2 and subsequently immunoblotted for CagA; residual ASPP2 signal is indicated by a white arrowhead. (F) Human gastroids were infected with wt H. pylori for 24 h and either treated with DMSO or EGFR inhibitor (lapatinib, 10 μM), ERK (U126, 5 μM), or AKT (AKTVIII, 10 μM). After 24 h, bacteria were replated to assess their viability in a colony-forming assay (CFUs, colony-forming units). Significance was tested using a one-way ANOVA Tukey’s multiple-comparison test. n = 3 (where n is a biological repeat and each biological repeat has two technical replicates that are averaged). Error bars, ±SEM. ***P < 0.001, **P < 0.01; ns, P > 0.05. See also SI Appendix, Figs. S6–S9.
We confirmed that H. pylori activates EGFR at 7 h postinfection (SI Appendix, Fig. S7A), which correlates with the recruitment and binding of ASPP2 by CagA (SI Appendix, Fig. S7A, second lane). We then analyzed ASPP2 colocalization and binding to CagA upon H. pylori infection of AGS cells pretreated with different concentrations of lapatinib, a well-known EGFR inhibitor (iEGFR) that targets the EGFR/Her2 pathways and is of the same chemotype as the EGFR inhibitors identified in the library. Under these conditions, we observed a dose-dependent inhibition of CagA–ASPP2 complex formation, as determined by immunoprecipitation (SI Appendix, Fig. S7A) and immunofluorescence staining of CagA (Fig. 6D and SI Appendix, Fig. S7B). These data show that inhibition of EGFR activity blocks the recruitment of ASPP2 by CagA. To further investigate how RTK signaling leads to the recruitment of ASPP2 by CagA, we inhibited the two major pathways downstream of EGFR, namely mitogen-activated protein kinase (MAPK) (also known as ERK) and PI3K/AKT. In accordance with our screening results, inhibition of AKT, but not MAPK (ERK), abrogates the binding of CagA to ASPP2 (Fig. 6E). To rule out the possibility that these inhibitors could affect the viability of H. pylori or the translocation of CagA into the host cell, we plotted a growth curve of the bacterium alone or in the presence of iEGFR or iAKT (SI Appendix, Fig. S7C). Neither inhibitor affects the growth of H. pylori (SI Appendix, Fig. S7C) or its ability to translocate CagA into the host cells (SI Appendix, Fig. S7D). These data suggest that inhibition of the RTK/PI3K/AKT pathway can block the recruitment of ASPP2 by cagA-positive H. pylori strains.
Inhibition of the CagA–ASPP2 Interaction Dampens H. pylori Colonization in Gastric Organoids.
Loss of cell polarity can favor the colonization of the host environment by bacterial pathogens (17, 25) and our data suggest that by blocking the recruitment of ASPP2, we can prevent the remodeling of the PAR complex and hence the loss of polarity upon infection. We therefore assessed whether interfering with the CagA–ASPP2 interaction reduces the fitness of H. pylori in the host environment, using the gastroid model. At 24 h postinfection with H. pylori, bacterial counts from gastroids show ∼103/mL actively replicating bacteria (as assessed by colony-forming units), whereas no bacteria survived when cultured in the absence of gastroids (Matrigel plus medium alone) (Fig. 6F, first column). Strikingly, we observed a nearly 2-log reduction in replicating bacteria in gastroids pretreated with either EGFR or AKT inhibitors (Fig. 6F), suggesting that this signaling pathway favors H. pylori colonization. In line with our interaction data, inhibition of MAPK (ERK) does not affect the ability of H. pylori to replicate in the gastroids (Fig. 6F).
The lack of effect from inhibition of the MAPK (ERK) pathway suggests that reduction in bacterial load in organoids treated with the EGFR and AKT inhibitors is unlikely to be a consequence of the toxicity of the compounds. To further confirm this, we tested the cell viability of organoids treated with iEGFR and iAKT (the two conditions in which there is a reduction of bacteria). Inhibition of EGFR has only a very mild toxicity on the epithelial cells of the organoids while AKT inhibition does not affect cell viability (SI Appendix, Fig. S8A). Next, we infected organoids either treated with iEGFR or dimethyl sulfoxide (DMSO) with wt or ΔvirB10 H. pylori strains and compared the reduction of viable bacteria in combination with the EGFR inhibitor. We note that H. pylori mutants lacking injected CagA can colonize host cells, particularly in the presence of nutrients and/or within 1 d of infection (17, 44, 45). In this experiment, we use ΔvirB10 H. pylori to check for nonspecific toxicity of iEGFR. Less than 10% of wt H. pylori survive upon iEGFR treatment compared with the untreated H. pylori-infected gastroids, whereas nearly 100% recovery was achieved when iEGFR-treated organoids were infected with the ΔvirB10 H. pylori strain (SI Appendix, Fig. S8B).
We then sought to understand whether the reduced ability of wt H. pylori to survive in the lumen of the organoids is linked to the loss of cell polarity. Gastroids treated with iEGFR (lapatinib) maintain a normal distribution of the polarity proteins (SI Appendix, Fig. S8 C and D). Upon infection, lapatinib-treated gastroids showed reduced ASPP2 apical localization to the sites of H. pylori attachment compared with mock-treated infected gastroids (SI Appendix, Fig. S9 A and E). Also, we observed a reduction in the apical distribution of both Par3 (SI Appendix, Fig. S9B) and Par1b (SI Appendix, Fig. S9C) and decreased redistribution of Scribble (SI Appendix, Fig. S9 D and E) in the organoids treated with the EGFR inhibitor and infected with H. pylori. These results suggest that inhibition of RTK/PI3K/AKT signaling interferes with the CagA–ASPP2 interaction, reduces the loss of polarity in H. pylori-infected gastroids, and lowers colonization by virulent H. pylori strains.
Direct Inhibition of the CagA–ASPP2 Interaction Affects H. pylori Colonization in Human Gastroids.
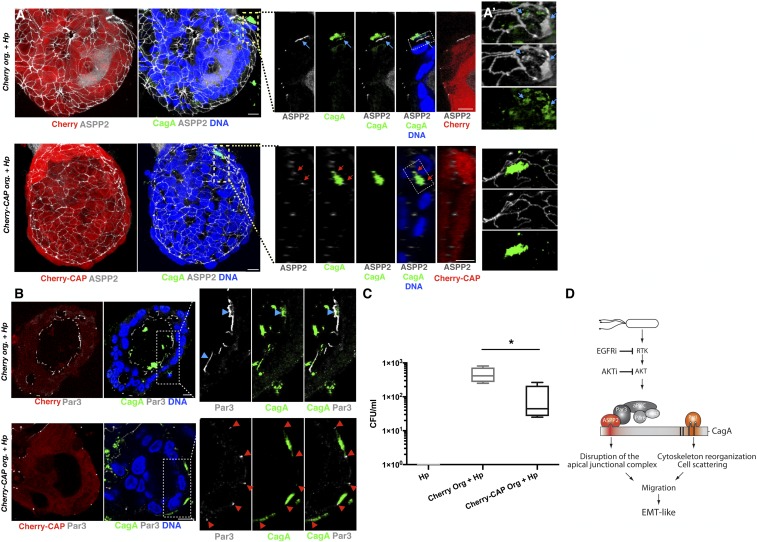
We have shown previously that expression of a Cherry-tagged 56-amino acid CagA-binding ASPP2 peptide (Cherry-CAP) functions in a dominant-negative manner in AGS cells and prevents the CagA–ASPP2 interaction, inhibiting the recruitment of p53 and restoring the apoptotic response in H. pylori-infected cells (29). In light of our findings, we examined whether expression of Cherry-CAP might also prevent the misregulation of the PAR complex by CagA. We stably expressed Cherry-CAP in gastroids (SI Appendix, Fig. S10A) and observed that Cherry-CAP precluded the relocalization of endogenous ASPP2 by CagA (Fig. 7 A and A′, Bottom and SI Appendix, Fig. S10 B, B′, and D). Importantly, the redistribution of Par3 was also reduced upon H. pylori infection of Cherry-CAP–expressing organoids (Fig. 7B and SI Appendix, Fig. S10D). In contrast, colocalization of ASPP2 and CagA or Par3 was unaffected in the control Cherry-expressing organoids (Fig. 7 A, Top and SI Appendix, Fig. S10D) and basolateral Scribble was not redistributed in H. pylori-infected organoids expressing Cherry-CAP compared with H. pylori-infected control Cherry-expressing organoids (SI Appendix, Fig. S10C). These data show that targeting the CagA–ASPP2 interaction prevents the deregulation of the PAR complex by H. pylori. To test whether this can affect the ability of H. pylori to colonize the lumen of the organoids, we repeated the colony-forming assay using the Cherry-CAP–expressing organoids (Fig. 7C). As expected, the number of H. pylori colony-forming units was indeed reduced in the Cherry-CAP but not in the Cherry control gastroids. These results show that blocking the CagA–ASPP2 interaction reduces H. pylori colonization of the host environment in an ex vivo organoid model, suggesting that disrupting the CagA–ASPP2 interaction is a viable option to reduce the pathogenic activity of cagA-positive H. pylori strains.
Fig. 7.
CagA–ASPP2–binding peptide prevents CagA from inducing apical redistribution of ASPP2 and reduces H. pylori colonization. (A) Confocal images of human gastroids expressing Cherry (Top) or Cherry-CAP (Bottom) and infected with wt H. pylori for 24 h. Infected organoids were stained with ASPP2 (gray), CagA (green), and nuclei (DAPI, blue). (A, Insets) Apical colocalization of CagA and ASPP2 in the H. pylori-infected organoids expressing Cherry (Top, blue arrows) versus the normal junctional distribution of ASPP2 in the H. pylori-infected organoids expressing Cherry-CAP (Bottom, red arrows). (A′) Three-dimensional reconstruction of the Insets in A. (B) Confocal images of human gastroids infected as described in A. Infected organoids were stained for Par3 (gray), CagA (green), and nuclei (DAPI, blue). (B, Insets) Apical colocalization of CagA and Par3 (Top, blue arrowheads) in the H. pylori-infected control Cherry organoids versus the normal junctional distribution of Par3 (Bottom, red arrowheads) in the H. pylori-infected organoids expressing Cherry-CAP. (Scale bars, 10 μm.) (C) Gastroids expressing Cherry or Cherry-CAP were infected with H. pylori and after 24 h, bacteria were replated for the CFU assay. n = 4 (where n is a biological repeat and each biological repeat has two technical replicates that are averaged). Error bars, ±SEM. *P < 0.05. (D) Model of PAR polarity complex remodeling by CagA and ASPP2 in epithelial cells. Infection with H. pylori activates an RTK pathway that is then sustained by the delivery of CagA. The apical-junctional PAR complex is redistributed by CagA via the direct interaction of CagA with ASPP2. This predisposes the cells receiving CagA to the loss of cell polarity. Through its C-terminal domain, CagA binds Par1b and this leads to cell scattering (hummingbird phenotype). The combination of these two events promotes the EMT-like phenotype induced by CagA. Inhibitors of the RTK/PI3K/AKT pathway prevent CagA’s association with ASPP2 and, under these conditions, the delivery of CagA no longer remodels the PAR complex and the cell does not lose cell polarity. See also SI Appendix, Fig. S10.
Discussion
The establishment and maintenance of cell polarity in gastrointestinal epithelial cells is essential to form the mucosal barrier that is the first line of defense against invading microbes. Infection by P. aeruginosa or N. meningitidis targets the apical-junctional PAR complex, and our findings show that delivery of H. pylori CagA remodels the whole PAR complex, including the apical, junctional, and basolateral components. Therefore, CagA has more widespread effects on polarity complex proteins than previously anticipated. This study also provides a mechanistic demonstration that by binding to ASPP2, CagA mislocalizes the PAR members, thus predisposing the infected cells to the loss of cell polarity and EMT-like phenotype promoted by the interaction of CagA with Par1b. This is consistent with previous findings that the CagA C terminus, responsible for initiating signaling and for interaction with Par1b (24), is not able to recapitulate the phenotype caused by full-length CagA (26). Expression of the CagA N terminus alone did not cause EMT and cell migration either, but coexpression of the N and C termini of CagA is required to recapitulate the effects of the full-length protein (26). Now we show that the interaction of CagA with ASPP2, which occurs via the N-terminal domain of CagA (26, 27), remodels the PAR complex at the apical junctions of the cell, thus complementing the signal generated by the C terminus and enabling the infected cell to achieve complete loss of cell polarity (Fig. 7D). Notably, tight junctions and cell-polarity proteins are required for the maintenance of the epithelial barrier function of the gastrointestinal tract and in this regard they function as tumor suppressors. In line with this, it has been shown that Claudin 18, a tight-junctional protein, is down-regulated in mice infected with H. pylori. Claudin 18 loss increases cell proliferation and loss of nuclear polarity and accelerates neoplasia development, correlating with poor prognosis of patients with stomach cancer (46). Consistent with this finding, reduced expression of ASPP2 increases an invasive phenotype in patients with stomach cancer (47) and analysis of The Cancer Genome Atlas (TCGA) data reveals that five of six truncation mutations observed in gastric cancers occur at the N terminus of ASPP2, which is required to bind Par3 (33, 48). Loss of ASPP2 promotes an EMT phenotype, and enforced expression of ASPP2 reverts this phenotype (35). Thus, CagA reaches cell-polarity components via the interaction with ASPP2 to achieve its full oncogenic potential. In addition, the CagA–ASPP2 interaction inactivates the apical PAR complex by bringing together effectors (e.g., aPKC) and regulators (e.g., Par6, Par3) that normally are separated in a fully polarized epithelium (22, 49). Par1b, which is normally excluded from the apical membrane by aPKC-dependent phosphorylation (50), is no longer phosphorylated when complexed with CagA (24). However, without ASPP2, CagA associates less effectively with Par1b and phosphorylation is restored. This could be achieved through ASPP2’s ability to bind and regulate the activity of protein phosphatase 1 (PP1) (51), for example if ASPP2 brings PP1α to dephosphorylate Par1b via the CagA interaction.
The finding that EGFR inhibitors are the most prominent group of kinase inhibitors in our screen (15 hits out of 40 compounds; SI Appendix, Fig. S6E) is consistent with EGFR signaling being activated by infection with H. pylori (52). Compared with wt flies, those flies that carry a single copy of the egfr gene show an attenuated phenotype upon a transgenic expression of CagA (53). EGFR activation promotes the translocation of ASPP2 from the tight junctions to the cytosol, where ASPP2 binds RAS and potentiates the downstream MAPK (ERK) and AKT signaling pathways (54, 55). It is therefore conceivable that cagA-positive H. pylori strains might co-opt ASPP2 through the activation of the EGFR pathway. In agreement with this, the PI3K–AKT signaling axis is also activated by EGFR, and our results indicate that inhibitors of AKT also prevent the CagA–ASPP2 interaction. We have previously shown that the CagA–ASPP2 interaction is independent of CagA phosphorylation (28), therefore excluding activation of the MAPK (ERK) pathway by CagA as the mechanism for ASPP2 recruitment. In line with this, we did not identify any MAPK (ERK) inhibitors (0 hits out of 89 compounds; SI Appendix, Fig. S6E) in our screen.
Finally, the finding that a Cherry-tagged CagA-binding ASPP2 peptide reduces the CagA-induced phenotypes, including decreased loss of cell polarity and a reduction in H. pylori colonization, is of great interest. Given the role played by ASPP2 in connecting cell polarity to inflammation (56), blocking the CagA–ASPP2 interaction might evoke a stronger immune response from the infected epithelial cells. Alternatively, we have previously shown in vitro that blocking the CagA–ASPP2 interaction increases the apoptotic response specifically in wt H. pylori-infected cells (29). In organoids, blocking the CagA–ASPP2 interaction might specifically promote the apoptotic response of the infected cells, as seen in in vitro conditions, and therefore deplete the target cell population of the infection. Further studies will address how epithelial cells better control the infection with H. pylori when the bacterium is not able to reach ASPP2. Also, given that CagA is needed for colonization by H. pylori in nutrient-deprived conditions in vitro (17), in future studies it will be interesting to address whether the CagA–ASPP2 complex can influence H. pylori’s ability to access nutrients. The incomplete reduction in colonization observed in the organoid phenotypes may be due to partial degradation of the CagA-binding ASPP2 peptide, since a degradation fragment of the peptide was detected by immunoblot (SI Appendix, Fig. S10A). Heterogeneous expression of the peptide may also affect its efficacy in preventing the CagA–ASPP2 interaction and altering cellular phenotypes. Regardless, this study demonstrates that preventing bacterial pathogens from disrupting the host cell polarity could be a strategy for the development of new antimicrobial drugs that limit the damage during infection.
Materials and Methods
Human Tissue Materials.
Donor samples were obtained from the Oxford GI Biobank (Gastrointestinal Illness in Oxford: prospective cohort for outcomes, treatment, predictors and biobanking). Ethical approval was obtained from the Yorkshire & The Humber - Sheffield Research Ethics Committee (Reference no. 11/YH/0020) and informed written consent was given by all study patients. Isolation of the gastric glands was performed as previously described (38).
Immunofluorescence.
Samples for confocal immunofluorescence were processed as previously described (29) with minor modifications. Briefly, cells were grown on coverslips and organoids were grown on a µ-Slide 8 Well (Ibidi) and fixed in 4% paraformaldehyde (PFA). After permeabilization in 0.1% Triton X-100, samples were incubated with Image-iT FX signal enhancer (Thermo Scientific) and incubated overnight with the indicated antibodies. References for the antibodies used in this study and their concentration are shown in SI Appendix. Cells were incubated with Alexa Fluorochrome secondary antibodies, phalloidin-647 for actin, and DAPI for nuclei acids, and mounted with ProLong Gold Antifade Mountant (Thermo Scientific). A Zeiss LSM 710 confocal microscope was used to acquire the confocal images.
High-Content Screening for Regulators of the Recruitment of ASPP2 by H. pylori.
The screen was performed in duplicate. AGS cells were seeded in 96-well plates in DMEM supplemented with 5% FBS at 37 °C in 5% CO2. After 8 h, compounds or DMSO were transferred robotically from a library stock plate to a final concentration of 10 μM and cells were further incubated for 12 h. Wild-type or ΔvirB10 mutant H. pylori strains were added to the cells at a multiplicity of infection of 1:50 and allowed to infect the cells for 7 h. Cells were then fixed in 4% PFA, permeabilized in 2% BSA, 0.1% Triton-containing buffer, and stained for ASPP2, CagA, DAPI for nuclei, and phalloidin for actin. Assays were performed in black-walled, clear-bottom, 96-well plates (PerkinElmer) suitable for automated fluorescence imaging. Images were acquired using a high-content imaging system (InCell Analyzer 6000; GE Healthcare Life Sciences); a total of 18 different images were acquired per wavelength, well, and replicate.
Quantification of Microscopy.
Method A. Recruitment of the polarity markers in AGS cells.
A cell was scored as positive for redistribution if we saw at least some overlap between PARs and CagA in that cell (i.e., not the total % of the PAR members that colocalize with CagA).
Method B. High-content imaging screening.
The raw image files (.jpeg) were segmented and analyzed to extract changes in the cellular distribution of ASPP2 using ImageJ software (code used for the analysis is available at https://github.com/LButi/Code-CagA-ASPP2-image-analysis/releases/tag/v1.0). ASPP2 distribution was quantified as puncta area of staining per nucleus. The average punctate staining of ASPP2 in the positive control samples (infected with wt H. pylori) was assigned a maximum value of 100% of redistribution (Fig. 6 B and C and SI Appendix, Fig. S6B). ASPP2 punctate staining for all other samples (uninfected, ΔvirB10 H. pylori mutant-infected and wt H. pylori-infected cells + drugs) was represented as a percentage compared with the positive controls. Based on the Pearson correlation (0.88) and the average Z factors (0.74), our positive-hit cutoff was assigned a 30% value.
Method C. Colocalization of CagA with polarity markers in the organoid model.
Confocal optical sections of organoids stained with the indicated antibodies were analyzed with Zen Zeiss (Carl Zeiss). Whole organoids or random fields of H. pylori-infected organoids were selected to measure the percentage of colocalization. The data were then transferred to an Excel worksheet and analyzed by Prism 7 (GraphPad).
All other experimental procedures, including bacterial infections and organoid culture, are detailed in SI Appendix, Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Mark Shipman, Richard Lisle, and Lennard Voortman for technical assistance with microscopy; Andrew Worth, Xiao Qin, and Carol Leung for assistance with fluorescence-activated cell sorting (FACS) acquisition and analysis; Felix Zhou for assistance with the development of the algorithms; Mary Muers and Françoise Howe for editorial assistance; and Annemarthe van der Veen, Mads Gyrd-Hansen, Jan Bornschein, and Colin Goding for critical reading of the manuscript. The kinase inhibitor set PKIS and PKIS2 were provided by the Structural Genomics Consortium at the University of North Carolina (SGC-UNC) and include kinase inhibitors donated by GlaxoSmithKline, Pfizer, and Takeda. This work was mainly funded by the Ludwig Institute for Cancer Research. R.P.O. was supported by the National Institute for Health Research Oxford Biomedical Research Centre, Oxford Health Services Research Committee, and Oxford University Clinical Academic Graduate School.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908787117/-/DCSupplemental.
References
- 1.Marshall B. J., Warren J. R., Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1, 1311–1315 (1984). [DOI] [PubMed] [Google Scholar]
- 2.Algood H. M. S., Cover T. L., Helicobacter pylori persistence: An overview of interactions between H. pylori and host immune defenses. Clin. Microbiol. Rev. 19, 597–613 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Censini S., et al. , cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U.S.A. 93, 14648–14653 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein M., Rappuoli R., Covacci A., Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. U.S.A. 97, 1263–1268 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahi M., et al. , Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191, 593–602 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odenbreit S., et al. , Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287, 1497–1500 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Hatakeyama M., Helicobacter pylori CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15, 306–316 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Ekström A. M., Held M., Hansson L.-E., Engstrand L., Nyrén O., Helicobacter pylori in gastric cancer established by CagA immunoblot as a marker of past infection. Gastroenterology 121, 784–791 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Franco A. T., et al. , Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 68, 379–387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Agency for Research on Cancer , Schistosomes, Liver Flukes and Helicobacter pylori, IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (IARC, Lyon, France, 1994), vol. 61. [PMC free article] [PubMed]
- 11.Tacconelli E., et al. ; WHO Pathogens Priority List Working Group , Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Malfertheiner P., et al. , Management of Helicobacter pylori infection—the Maastricht V/Florence Consensus Report. Gut 66, 6–30 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Gall A., et al. , Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS One 10, e0129055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold I. C., et al. , Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140, 199–209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigal M., et al. , R-spondin-3 induces secretory, antimicrobial Lgr5+ cells in the stomach. Nat. Cell Biol. 21, 812–823 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Amieva M. R., et al. , Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300, 1430–1434 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan S., Tompkins L. S., Amieva M. R., Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 5, e1000407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Belmonte F., Mostov K., Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20, 227–234 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Goehring N. W., PAR polarity: From complexity to design principles. Exp. Cell Res. 328, 258–266 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Román-Fernández A., Bryant D. M., Complex polarity: Building multicellular tissues through apical membrane traffic. Traffic 17, 1244–1261 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Su W.-H., Mruk D. D., Wong E. W. P., Lui W.-Y., Cheng C. Y., Polarity protein complex Scribble/Lgl/Dlg and epithelial cell barriers. Adv. Exp. Med. Biol. 763, 149–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A., Ohno S., The PAR-aPKC system: Lessons in polarity. J. Cell Sci. 119, 979–987 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Ruch T. R., Engel J. N., Targeting the mucosal barrier: How pathogens modulate the cellular polarity network. Cold Spring Harb. Perspect. Biol. 9, a027953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saadat I., et al. , Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447, 330–333 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Coureuil M., et al. , Meningococcal type IV pili recruit the polarity complex to cross the brain endothelium. Science 325, 83–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagnoli F., Buti L., Tompkins L., Covacci A., Amieva M. R., Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. U.S.A. 102, 16339–16344 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito Y., Murata-Kamiya N., Hirayama T., Ohba Y., Hatakeyama M., Conversion of Helicobacter pylori CagA from senescence inducer to oncogenic driver through polarity-dependent regulation of p21. J. Exp. Med. 207, 2157–2174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buti L., et al. , Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc. Natl. Acad. Sci. U.S.A. 108, 9238–9243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nešić D., Buti L., Lu X., Stebbins C. E., Structure of the Helicobacter pylori CagA oncoprotein bound to the human tumor suppressor ASPP2. Proc. Natl. Acad. Sci. U.S.A. 111, 1562–1567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vives V., et al. , ASPP2 is a haploinsufficient tumor suppressor that cooperates with p53 to suppress tumor growth. Genes Dev. 20, 1262–1267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu M., Muers M. R., Lu X., Introducing STRaNDs: Shuttling transcriptional regulators that are non-DNA binding. Nat. Rev. Mol. Cell Biol. 17, 523–532 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Samuels-Lev Y., et al. , ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8, 781–794 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Sottocornola R., et al. , ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev. Cell 19, 126–137 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Cong W., et al. , ASPP2 regulates epithelial cell polarity through the PAR complex. Curr. Biol. 20, 1408–1414 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., et al. , ASPP2 controls epithelial plasticity and inhibits metastasis through β-catenin-dependent regulation of ZEB1. Nat. Cell Biol. 16, 1092–1104 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Suzuki A., et al. , aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14, 1425–1435 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Yang Z., et al. , The signaling adaptor GAB1 regulates cell polarity by acting as a PAR protein scaffold. Mol. Cell 47, 469–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartfeld S., et al. , In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruch T. R., Bryant D. M., Mostov K. E., Engel J. N., Par3 integrates Tiam1 and phosphatidylinositol 3-kinase signaling to change apical membrane identity. Mol. Biol. Cell 28, 252–260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higashi H., et al. , SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295, 683–686 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Mimuro H., et al. , Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 10, 745–755 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Churin Y., et al. , Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161, 249–255 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drewry D. H., Willson T. M., Zuercher W. J., Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS). Curr. Top. Med. Chem. 14, 340–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertaux-Skeirik N., et al. , CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. 11, e1004663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schumacher M. A., et al. , Helicobacter pylori-induced Sonic Hedgehog expression is regulated by NFκB pathway activation: The use of a novel in vitro model to study epithelial response to infection. Helicobacter 20, 19–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagen S. J., et al. , Loss of tight junction protein Claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology 155, 1852–1867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gen Y., et al. , ASPP2 suppresses invasion and TGF-β1-induced epithelial-mesenchymal transition by inhibiting Smad7 degradation mediated by E3 ubiquitin ligase ITCH in gastric cancer. Cancer Lett. 398, 52–61 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Kakiuchi M., et al. , Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 46, 583–587 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Yamanaka T., et al. , PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells 6, 721–731 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Hurov J. B., Watkins J. L., Piwnica-Worms H., Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14, 736–741 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Royer C., et al. , ASPP2 links the apical lateral polarity complex to the regulation of YAP activity in epithelial cells. PLoS One 9, e111384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bauer B., et al. , The Helicobacter pylori virulence effector CagA abrogates human β-defensin 3 expression via inactivation of EGFR signaling. Cell Host Microbe 11, 576–586 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Reid D. W., et al. , Identification of genetic modifiers of CagA-induced epithelial disruption in Drosophila. Front. Cell. Infect. Microbiol. 2, 24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., et al. , ASPP1 and ASPP2 bind active RAS, potentiate RAS signalling and enhance p53 activity in cancer cells. Cell Death Differ. 20, 525–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z., et al. , N terminus of ASPP2 binds to Ras and enhances Ras/Raf/MEK/ERK activation to promote oncogene-induced senescence. Proc. Natl. Acad. Sci. U.S.A. 110, 312–317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnquist C., et al. , STAT1-induced ASPP2 transcription identifies a link between neuroinflammation, cell polarity, and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 111, 9834–9839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.