Significance
The burgeoning intestinal microbiota pose a unique challenge to the developing immune system of the newborn. Extensive crosstalk between mucosal immune cells and microbes is necessary to establish healthy microbial communities and promote the development of productive mucosal immunity. However, it is yet unknown whether intestinal microbes also influence the development of lymphocytes themselves in the thymus. Here, we report that thymic distribution of transcription factor PLZF-expressing innate lymphocytes is regulated by intestinal microbes in early life. Migratory plasmacytoid dendritic cells are one conveyor of microbial information to developing thymic cells and perturbation of this entero-thymic communication in early life impacts disease susceptibility in adulthood. Our data highlight a novel regulation of thymic lymphocyte development by early-life microbiota.
Keywords: early-life immunity, mucosal immunity, thymic lymphocyte development
Abstract
The thymus generates cells of the T cell lineage that seed the lymphatic and blood systems. Transcription factor regulatory networks control the lineage programming and maturation of thymic precursor cells. Whether extrathymic antigenic events, such as the microbial colonization of the mucosal tract also shape the thymic T cell repertoire is unclear. We show here that intestinal microbes influence the thymic homeostasis of PLZF-expressing cells in early life. Impaired thymic development of PLZF+ innate lymphocytes in germ-free (GF) neonatal mice is restored by colonization with a human commensal, Bacteroides fragilis, but not with a polysaccharide A (PSA) deficient isogenic strain. Plasmacytoid dendritic cells influenced by microbes migrate from the colon to the thymus in early life to regulate PLZF+ cell homeostasis. Importantly, perturbations in thymic PLZF+ cells brought about by alterations in early gut microbiota persist into adulthood and are associated with increased susceptibility to experimental colitis. Our studies identify a pathway of communication between intestinal microbes and thymic lymphocytes in the neonatal period that can modulate host susceptibility to immune-mediated diseases later in life.
Mammalian intestinal microbial communities harbor both an enormous source of commensals as well as potential pathogens. In the postnatal period, the immune cells must rapidly discriminate enteric friend from foe. Lymphocytes of the T cell lineage that participate in this mucosal process develop in the thymus. Whether gut bacteria influence their development in early life is not well understood. In humans, early-life fluctuations in intestinal microbial communities have been well documented and may be influenced by the mode of delivery, maternal and infant diet, as well as the introduction of solid weaning foods (1–3). Importantly, changes in microbial diversity introduced as a result of antibiotic use, for example, may have negative long-term health consequences including increased susceptibility to autoimmune diseases and allergies (4, 5). Studies in GF mice that lack indigenous microbiota have greatly advanced the concept of the microbial influence of the immune function, especially in early life (6, 7). While the role of the microbiota in regulating mucosal immune responses has been well studied, extraintestinal effects of the gut bacteria on developing T cells in the thymus are less understood.
Thymus specific progenitors undergo an ordered process of differentiation to become conventional αβ—or γδ—T cell receptor (TCR) expressing T cells (8). However, multiple unconventional lineage choices are also available to progenitor cells that lead to the development of innate-like T cells, such as Vα14-Jα28 TCR expressing invariant natural killer T (NKT) (iNKT) (9) cells and innate lymphoid cells (ILCs) that do not express antigen specific receptors but closely mirror T cells in their development and function (10–12). ILCs and other innate-like T cells mediate crosstalk with the microbiota and provide immune protection at mucosal sites (13).
The BTB-zinc finger protein PLZF (encoded by Zbtb16) is a signature transcription factor (TF) that is expressed by a common multilineage progenitor that gives rise to ILC subsets (14) and is also expressed in innate-like T cells (15) and iNKT cells (16). In this paper, we investigated the influence of intestinal microbes on PLZF-expressing cells in the thymus in early life. We report that intestinal bacteria regulate the thymic distribution of thymic PLZF+ cells and identify a role for host Toll-like Receptor 2 (TLR2) and migratory plasmacytoid dendritic cells (pDCs) in the process.
Results
Intestinal Commensal B. fragilis Promotes Early-Life Thymic PLZF+ Cell Development.
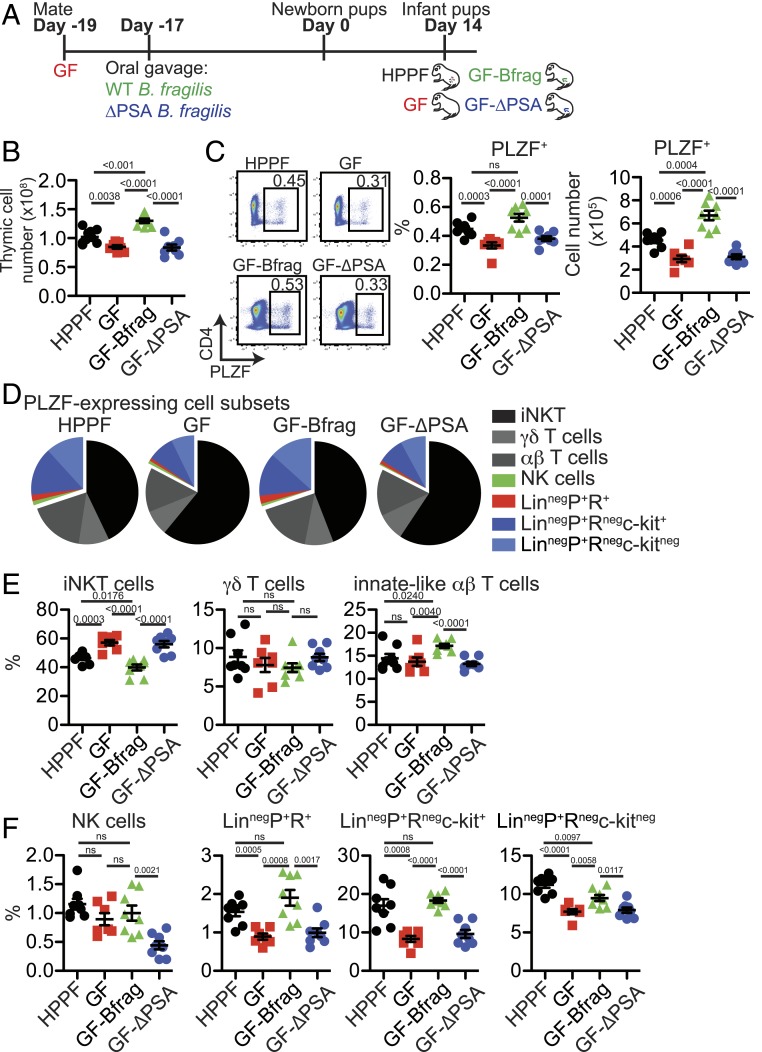
Intestinal microbes regulate multiple aspects of the immune function at mucosal and nonmucosal sites in early life (17, 18). To determine whether they also impact thymic T cell development, we evaluated the effect of monocolonization of GF mice with the human commensal B. fragilis (GF-Bfrag) and a PSA deficient isogenic mutant (GF-ΔPSA) (19–21) (Fig. 1A). B. fragilis and its capsular polysaccharide, PSA, have potent immunomodulatory effects on several mucosal cell types including DCs, T cells, and enterocytes (20–23). Monocolonization of GF mice with B. fragilis restored thymic and splenic cellularity of d14 GF pups to levels similar to conventionally housed Helicobacter and Pasteurella pneumotropica free (HPPF) mice while ΔPSA B. fragilis did not (Fig. 1B and SI Appendix, Fig. S1B). No differences in the distribution of major thymic and splenic cell subsets were noted after intestinal microbial colonization (SI Appendix, Fig. S1 A and B). However, there was a decrease in CD4+FOXP3+ cells in the thymus but not in the spleen of GF-Bfrag pups compared to GF pups (SI Appendix, Fig. S1 A and B).
Fig. 1.
Intestinal commensal B. fragilis promotes early-life thymic PLZF+ cell development. (A) Thymuses from 14 d old pups born of C57BL/6 conventionally housed (HPPF) or GF breeders monocolonized with B. fragilis NCTC 9343 (GF-Bfrag) and ∆PSA B. fragilis (GF-∆PSA) were analyzed by flow cytometry. (B) Total thymic cellularity (HPPF n = 8; GF n = 7; GF-Bfrag n = 8; GF-∆PSA n = 8). (C, Left) Representative flow cytometry plots showing PLZF-expressing cells, (Right) frequency (HPPF n = 8; GF n = 7; GF-Bfrag n = 8; GF-∆PSA n = 8), and total numbers of PLZF+ cells (HPPF n = 8; GF n = 7; GF-Bfrag n = 8; GF-∆PSA n = 8). Effect size: HPPF vs. GF: 1.86; GF vs. GF-Bfrag: −2.2; GF-Bfrag vs. GF-PSA: 2.02. (D) Pie graphs showing distribution of indicated PLZF-expressing cell subsets. (E) Frequency of PLZF+mCD1d-PBS57-tetramer+β-TCR+ iNKT cells, PLZF+δ-TCR-expressing γδ-T cells, and PLZF+β-TCR-expressing (CD1d-PBS57neg) innate-like αβ-T cells in the thymus (HPPF n = 8; GF n = 7; GF-Bfrag n = 8; GF-∆PSA n = 8). (F) Frequency of PLZF+NK1.1+CD122+ NK cells, LinnegPLZF+RORγt+ cells, LinnegPLZF+RORγtnegc-kit+ cells, and LinnegPLZF+RORγtnegc-kitneg cells (Lin: δ-TCR, β-TCR, NK1.1, CD11c, CD11b, Ter119, CD19, GR-1, and CD8a) in the thymus (HPPF n = 8; GF n = 7; GF-Bfrag n = 8; GF-∆PSA n = 8). Data are from two independent experiments for each group. Bars are mean ± SEM.
Of interest, the frequency and distribution of TF PLZF expressing thymocytes changed with microbial reconstitution of pups (Fig. 1 C–F). There were more PLZF+ thymocytes in GF-Bfrag pups compared to GF and GF-ΔPSA pups (Fig. 1C). Major PLZF-expressing thymic cell subsets identified by flow cytometry were mCD1d-PBS57-tetramer+β-TCR+ iNKT cells, δ-TCR-expressing γδ T cells, and β-TCR-expressing (CD1d-PBS57neg) innate-like αβ-T cells (SI Appendix, Fig. S2A). While microbial colonization did not affect the distribution of PLZF+ γδ T cells, the frequency of PLZF+ iNKT cells in GF-Bfrag pups was lower, and that of PLZF+ innate-like αβ-T cells was higher compared to GF and HPPF pups (Fig. 1 D and E and SI Appendix, Fig. S3A). A significant proportion of PLZF+ thymocytes did not express the δ- or β-TCR (SI Appendix, Fig. S2A), and a minor fraction of these were NK cells (NK1.1+CD122+) cells (SI Appendix, Fig. S2B). TCRnegNK1.1negPLZF+ cells did not express lineage markers including CD11c, CD11b, Ter119, CD19, GR-1, and CD8α (henceforth referred to as LinnegPLZF+ cells). A rare subset of LinnegPLZF+ cells also expressed the TF RORγt (SI Appendix, Fig. S2C) and were identified as IL7Rα+GATA3+c-kitnegCD44hiCD25neg, potentially an ILC3 subset (SI Appendix, Fig. S2C). The remainder of the LinnegPLZF+ (RORγtneg) cells could be subdivided into c-kit+ and c-kitneg subsets (SI Appendix, Fig. S2C). LinnegPLZF+c-kit+ cells were IL7Rα+GATA3+CD44hiCD25+, while the LinnegPLZF+c-kitneg cells appeared to be a heterogeneous mix of cells that were IL7Rα+/−GATA3+CD44hi/loCD25neg cells (SI Appendix, Fig. S2C). The frequency and numbers of these minor PLZF-expressing cell subsets was significantly increased in the thymus of GF-Bfrag but not GF-ΔPSA pups (Fig. 1 D and F and SI Appendix, Fig. S3B). This thymic phenotype extended to the spleen but not the colon of GF, GF-Bfrag, and GF-∆PSA mice (SI Appendix, Fig. S4 A–D). Thus, in these proof-of-concept studies, intestinal bacteria, and their components appeared to impact thymic distribution of PLZF+ innate and innate-like cells in infant mice.
Host TLR2 Regulates Thymic PLZF+ Cell Homeostasis in Early Life.
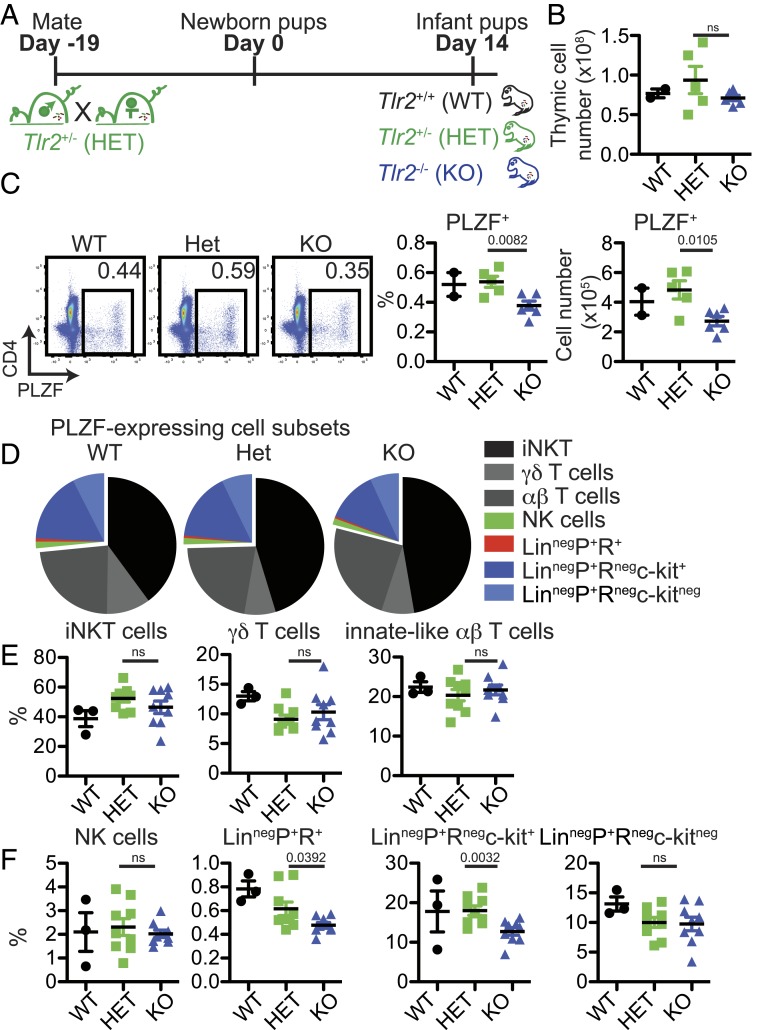
B. fragilis PSA appeared to play a role in the thymic response to intestinal colonization in early life (Fig. 1). Various B. fragilis components including PSA signal through TLR2 (20, 24–26). Therefore, to address the impact of early-life TLR2-mediated microbial interactions on thymic PLZF+ cell homeostasis, 14 d old Tlr2−/− pups were studied (Fig. 2A). There were no significant differences in thymic cellularity between Tlr2−/− and littermate wild-type (WT) and Tlr2+/− pups (Fig. 2B). Distribution of major thymic subsets as well as CD4+FOXP3+ cells was also comparable (SI Appendix, Fig. S5A). However, similar to GF-ΔPSA mice, d14 Tlr2−/− pups had fewer PLZF+ in the thymus, spleen, and colon compared to littermate controls (Ctrls) (Fig. 2C and SI Appendix, Fig. S7 A and C). The frequency of PLZF+ thymic iNKT cells, γδ T cells, and innate-like αβ-T cells were not significantly different (Fig. 2 D and E), but their numbers were reduced in Tlr2−/− pups (SI Appendix, Fig. S6A). Minor LinnegPLZF+ cell subsets including LinnegPLZF+RORγt+ and LinnegPLZF+c-kit+ were also decreased in d14 Tlr2−/− pups in the thymus and spleen (Fig. 2 D and F and SI Appendix, Figs. S6B and S7B). These results suggested that TLR2 signals might play a role in conveying microbial information to developing thymic cells in early life.
Fig. 2.
Altered thymic PLZF+ cell distribution in infant Tlr2−/− mice. (A) Thymuses from 14 d old pups born of Tlr2+/− (HET) crosses were analyzed by flow cytometry. (B) Total thymic cellularity (WT n = 2; HET n = 5; knockout [KO] n = 6). Data are from two experiments. (C, Left) Representative flow cytometry plots showing PLZF-expressing cells and (Right) frequency and total numbers of PLZF+ cells (WT n = 3; HET n = 9; KO n = 9). Effect size: Het vs. KO: 1.4. (D) Pie graphs showing distribution of indicated PLZF-expressing cell subsets. (E) Frequency of PLZF+mCD1d-PBS57-tetramer+β-TCR+ iNKT cells, PLZF+δ-TCR-expressing γδ-T cells, and PLZF+β-TCR-expressing (CD1d-PBS57neg) innate-like αβ-T cells in the thymus (WT n = 3; HET n = 9; KO n = 9). (F) Frequency of PLZF+NK1.1+CD122+ NK cells, LinnegPLZF+RORγt+ cells, LinnegPLZF+RORγtnegc-kit+ cells, and LinnegPLZF+RORγtnegc-kitneg cells (Lin: δ-TCR, β-TCR, NK1.1, CD11c, CD11b, Ter119, CD19, GR-1, and CD8a) cells in the thymus (WT n = 3; HET n = 9; KO n = 9). Data in C–F are from three experiments. Bars are mean ± SEM.
Cells from the Colon Migrate to the Thymus during the Neonatal Period.
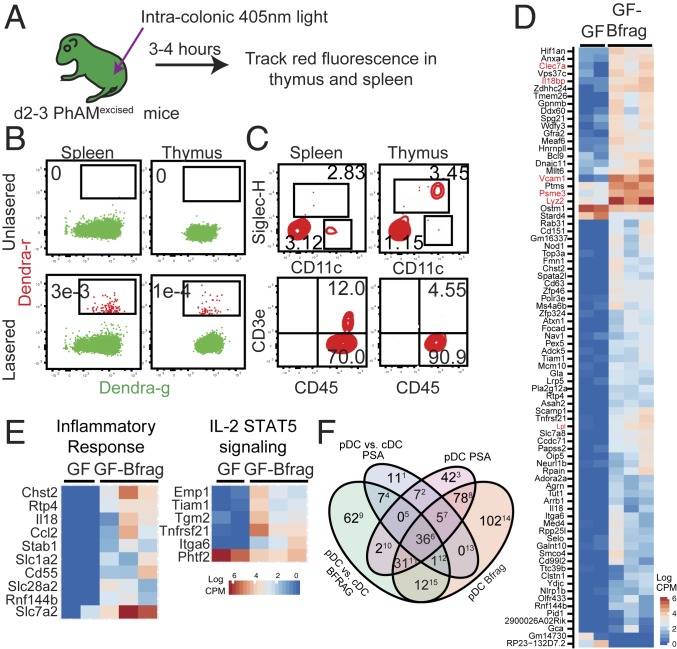
Among the formal possibilities for enterothymic communication are soluble mediators that are disseminated systemically or migratory cellular populations that convey microbial information to the thymus. Intestine-resident migratory cells carry bacteria and bacterial products to secondary lymphoid organs where they influence immunity (27–29). To determine whether colon-resident cells also migrate to the thymus, PhAMexcised mice expressing the photoconvertible Dendra protein were used (30, 31). Cells in the colon of newborn mice were photoconverted from Dendra-green (Dendra-g) to Dendra-red (Dendra-r) expression using a custom-made fiber-optic probe as described before (Fig. 3A) (31). We showed previously that Dendra-r+ cells were not detected at extraintestinal sites immediately after photoconversion (31). However, migratory Dendra-r+ cells of colonic origin could be detected in the thymus and spleen 3 to 4 h after photoconversion (Fig. 3B). While the identity of the majority of Dendra-r+ CD45+ cells remains unknown, a fraction was composed of CD11cintSiglecH+ pDCs and CD3+ T cells (Fig. 3C). We focused on pDCs as they have been shown previously to traffic from the periphery to the thymus to regulate central tolerance (32). pDCs could be found in total colonic lymphocyte preparations from neonatal d3, d5, and d7 mice (SI Appendix, Fig. S8A) as well as in the colonic lamina propria lymphocyte (LPL) and intraepithelial lymphocyte (IEL) fractions of d13 C57BL/6 mice (SI Appendix, Fig. S8B). In both IEL and LPL fractions, colonic pDCs expressed lower levels of MHC Class II compared to cDCs (CD11chi conventional DCs) (SI Appendix, Fig. S8C). pDCs also expressed lower levels of the αE-integrin CD103 but higher levels of CCR9 relative to cDCs (SI Appendix, Fig. S8C). Thus, colonic pDCs represented a population that was competent for thymus homing, migration, and potential antigen presentation.
Fig. 3.
pDCs from the colon migrate to the thymus in early life. (A) Newborn (d0–2) PhAMexcised mice were exposed to 405 nm light to photoconvert Dendra-g protein to Dendra-r protein by intracolonic exposure. Dendra-r+ cells were identified by flow cytometry 3 to 4 h after photoconversion. (B) Representative flow cytometry plots showing expression of Dendra-g and Dendra-r fluorescences in the thymus and spleen. Frequency of Dendra-r+ cells is shown. (C) Expression of (Top) CD11c (x axis) and SiglecH (y axis) and (Bottom) CD45 (x axis) and CD3e (y axis) on Dendra-r+ cells in the spleen and thymus of photoconverted pups. Frequency of CD11+SiglecH+ pDCs, CD11c+SiglecHneg conventional DCs (cDCs), and CD45+CD3e+ T cells is shown. Data in B and C are representative of a minimum of 10 independent experiments. (D) Thymic pDCs from d14 GF and GF-Bfrag pups were analyzed by RNA-sequencing (RNA-seq). Differentially expressed genes between thymic pDCs from GF and GF-Bfrag mice were filtered to include only those genes that were differentially expressed in a pDC^(GF vs. Bfrag) and cDC^(GF. vs. Bfrag) (cDC: CD11chi) comparison to exclude genes that changed in both pDCs and cDCs as a result of B. fragilis monocolonization. The heat map shows expression of these 80 genes in GF and GF-Bfrag thymic pDCs. (E) The heat map of differentially expressed genes between GF and GF-Bfrag thymic pDCs contained in selected significant (false discovery rate < 0.1) networks is shown. (F) Venn diagram showing distribution of genes and the extent of dissimilarity between pDCs from GF-Bfrag and GF-ΔPSA pups. Numbers in superscripts indicate column number in SI Appendix, Fig. S10, listing genes in each subset.
There was a small but significant decrease in pDCs in the thymus of GF-∆PSA pups compared to GF-Bfrag (SI Appendix, Fig. S9 A–C). Furthermore, thymic pDCs had distinct gene expression patterns based on their intestinal microbial colonization state (Fig. 3 D–F). B. fragilis monocolonization induced the expression of ∼80 unique genes in thymic pDCs including Lyz2 [encoding Lysozyme C-2; antimicrobial function (33)], Clec7a [encoding C-type lectin domain family 7/Dectin-1; functions as a pattern-recognition receptor (34)], and Psme3 [proteasome activator subunit 3; functions in host bacterial defense pathways (35)] (Fig. 3D). A pathway analysis further revealed induction of genes involved in the inflammatory response and STAT5 signaling pathways in thymic pDCs from GF-Bfrag pups (Fig. 3E). Thymic pDCs from GF-ΔPSA pups were distinct with significantly fewer induced genes (∼60) compared to pDCs from GF-Bfrag mice (Fig. 3F and SI Appendix, Fig. S10). Thus, thymus-homing pDCs appeared to be a cell population that was responsive to intestinal microbes.
Correlation between Thymic pDCs and PLZF+ Lymphocytes.
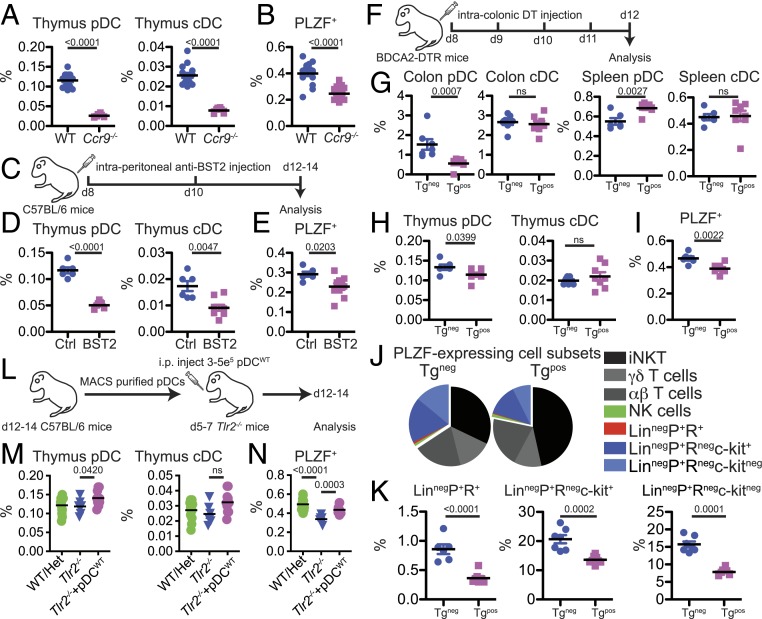
We next determined the impact of the perturbation of pDCs on thymic PLZF+ cell homeostasis. The chemokine receptor CCR9 regulates migration of pDCs to the intestine and thymus (32, 36). Thymuses from d14 Ccr9−/− mice had decreased frequency of pDCs (Fig. 4A), and this correlated with a significant decrease in the frequency of thymic PLZF+ cells (Fig. 4B). However, defective migration of bone marrow precursor cells as well as improper positioning of double-negative cells within the thymus of Ccr9−/− mice could potentially confound interpretation of these results (9, 37, 38). In an alternate approach, infant mice were administered intraperitoneal (i.p.) anti-BST2 antibody to reduce pDC numbers (Fig. 4C). Similar to Ccr9−/− mice, there was a significant decrease in the frequency of PLZF+ cells when there were fewer pDCs in the thymus (Fig. 4 D and E). However, in both Ccr9−/− mice and anti-BST2 treated mice, in addition to decreased pDCs frequency, we also observed a decrease in cDCs in the thymus (Fig. 4 A and D). To circumvent this issue and to target pDCs specifically in the colon, DT was administered by intracolonic injection to neonatal BDCA2-DTR-Tg mice (Fig. 4F) (39). DT administration markedly reduced pDC frequency in the colon but not in the spleen (Fig. 4G). Of interest, there was a small but significant decrease in the frequency of pDCs in the thymus upon DT administration (Fig. 4H). Most thymic pDCs are derived from the circulation (32, 40), and it is possible that pDCs transiting through the colon to the thymus might represent only a minor thymic pDC subset. However, they appeared to be an important population as the frequency of total thymic PLZF+ cells significantly decreased in colonic pDC-reduced pups (Fig. 4I). While most PLZF-expressing subsets were present at a similar frequency, all minor LinnegPLZF+ cell subsets were decreased in DT-treated mice (Fig. 4 J and K). Infant Tlr2−/− mice have lower thymic PLZF+ numbers (Fig. 2C), and we determined if the transfer of magnetic activated cell sorting (MACS) purified thymic pDCs from WT B6 mice (pDCWT) might restore their thymic homeostasis (Fig. 4L). Indeed, a small increase in thymic pDC upon adoptive transfer was accompanied by a significant increase in the frequency of PLZF+ cells in Tlr2−/− mice (Fig. 4 M and N). Thus, in multiple models of pDC manipulation, our experiments identified a correlation between these cells and thymic PLZF-expressing innate lymphocytes in early life.
Fig. 4.
Correlation between thymic pDCs and PLZF+ cells in infant mice. (A and B) Analysis of d14 Ccr9−/− mice; pDC (CD11cintSiglecH+), cDC (CD11chiSiglecHneg). (A) Frequency of pDCs and cDCs in the thymuses of d14 Ccr9−/− and WT mice (WT n = 15; KO n = 8). Data are from two independent experiments. (B) Frequency of PLZF+ cells in the thymuses of d14 Ccr9−/− and WT mice (WT n = 16; KO n = 17). Data are from three independent experiments. Effect size: WT vs. Ccr9−/−: 2.44. (C–E) anti-BST2 treatment. (C) C57/BL6 pups were treated with saline control (Ctrl) or anti-BST2 antibodies at d8 and 10 after birth. (D) Frequency of pDCs and cDCs in the thymuses of d14 Ctrl and a-BST2 treated mice (Ctrl n = 6; a-BST2 n = 9). (E) Frequency of PLZF+ cells in the thymuses of d14 Ctrl and a-BST2 treated mice (Ctrl n = 6; a-BST2 n = 9). Effect size: Ctrl vs. a-BST2: 1.45. Data in D and E are from two independent experiments. F–K Diphtheria toxin (DT) treatment of infant BDCA2-DTRTg mice. (F) BDCA2-DTR Tg+ and Tgneg pups were given intracolonic injections of DT at d8–11 after birth. (G) Frequency of pDCs and cDCs in the colon and spleen of DT-treated 14 d old Tgneg and Tg+ mice (Tgneg n = 6; Tg+ n = 8). (H) Frequency of pDCs and cDCs in the thymuses of DT-treated d14 Tgneg and Tg+ mice (Tgneg n = 6; Tg+ n = 8). (I) Frequency of PLZF+ cells in the thymuses of DT-treated d14 Tgneg and Tg+ mice (Tgneg n = 6; Tg+ n = 8). Effect size: Tgneg vs. Tg+: 2.07. (J) Pie graphs showing distribution of indicated PLZF-expressing cell subsets. (K) Frequency of LinnegPLZF+RORγt+ cells, LinnegPLZF+RORγtnegc-kit+ cells, and LinnegPLZF+RORγtnegc-kitneg cells (Lin: δ-TCR, β-TCR, NK1.1, CD11c, CD11b, Ter119, CD19, GR-1, and CD8a) cells in the thymus (Tgneg n = 6–7; Tg+ n = 8). (L–N) Adoptive transfer of WT pDCs into Tlr2−/− pups. (L) Thymic pDCs were isolated from d12–14 C57BL/6 pups and i.p. injected into d5–7 Tlr2−/− pups. (M) Frequency of pDCs and cDCs in the thymuses of d12–14 TLR2 WT/Het, Tlr2−/−, and Tlr2−/− + pDCWT mice (WT/Het n = 13, Tlr2−/− n = 6, and Tlr2−/− + pDCWT n = 11). (N) Frequency of PLZF+ cells in the thymuses of d12–14 TLR2 WT/Het, Tlr2−/−, and Tlr2−/− + pDCWT mice (WT/Het n = 13, Tlr2−/− n = 6, and Tlr2−/− + pDCWT n = 11). Data are representative of two independent experiments. Bars are mean ± SEM.
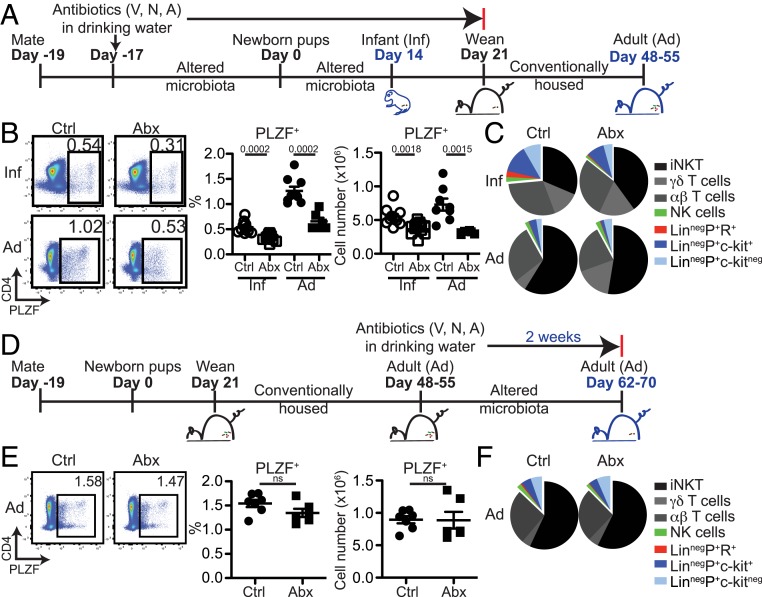
Antibiotics Treatment in Early Life Impacts PLZF+ Cell Distribution in Adulthood.
Experiments so far suggested that an early-life enterothymic circuit conveyed microbial information to influence the distribution of PLZF+ innate lymphocytes. Whether these early-life events persisted into adulthood or modulated disease susceptibility remained to be determined. We first analyzed 6 to 7 wk old littermate Tlr2+/− and Tlr2−/− mice and noted that similar to infant Tlr2−/− pups, adult Tlr2−/− mice also had significantly lower PLZF+ cell frequency in the thymus and spleen (SI Appendix, Fig. S11). Next, C57BL/6 breeders were treated with broad-spectrum antibiotics, and pups born of these microbiota-altered parents were either analyzed at 14 d after birth or weaned at 21 d and conventionally housed without antibiotics until analysis at 48–55 d of age (Fig. 5A). This strategy resulted in markedly reduced microbial diversity in d14 pups compared to Ctrls that persisted into adulthood after cessation of antibiotics at weaning (SI Appendix, Fig. S12 A and B). Thymic cellularity and distribution of major thymic subsets were unchanged in infant and adult antibiotics-treated mice (SI Appendix, Fig. S13 A and B). However, similar to GF pups (Fig. 1B), the frequency and numbers of PLZF+ cells were lower in antibiotic-treated d14 pups, and, importantly, this deficit persisted into adulthood (Fig. 5B). We also noted fewer PLZF+ cells in the spleen and colon of infant and adult mice that were treated with antibiotics in early life (SI Appendix, Fig. S14). The distribution of PLZF+ cell subsets changed significantly from infancy to adulthood, and early-life antibiotic treatment appeared to primarily impact the frequency and numbers of minor LinnegPLZF+ cell subsets (Fig. 5C and SI Appendix, Fig. S13 C and D). Importantly, antibiotic treatment in adulthood alone did not impact the homeostasis of thymic PLZF+ cells, suggesting that microbial regulation of these innate and innate-like immune subsets might occur specifically in an early-life period (Fig. 5 D–F).
Fig. 5.
Early-life antibiotics treatment impacts later-life thymic PLZF+ cell distribution. (A) C57BL/6 breeders were given antibiotics (vancomycin, neomycin, and ampicillin) in drinking water at the time of mating. Pups born of these “altered microbiota” breeders were analyzed at 14 d of age (Inf-Ctrl and Inf-Abx) or weaned at 21 d and housed in conventional isolators without antibiotics and analyzed at age 48–55 d (Ad-Ctrl and Ad-Abx). (B, Left) Representative flow cytometry plots showing PLZF-expressing cells and (Right) frequency (Inf-Ctrl n = 10; Inf-Abx n = 13; Ad-Ctrl n = 9; Ad-Abx n = 6) and total numbers of PLZF+ cells (Inf-Ctrl n = 10; Inf-Abx n = 13; Ad-Ctrl n = 9; Ad-Abx n = 6). Effect size: Inf-Ctrl vs. Inf-Abx: 1.90 and Ad-Ctrl vs. Ad-Abx: 2.82. (C) Pie graphs showing distribution of indicated PLZF-expressing cell subsets. (D) Conventionally housed adult (d48–55) C57BL/6 mice were given antibiotics (vancomycin, neomycin, and ampicillin) in drinking water for 2 wk. (E, Left) Representative flow cytometry plots showing PLZF-expressing cells, (Right) frequency (Ctrl n = 7; Abx n = 6), and total numbers of PLZF+ cells (Ctrl n = 7; Abx n = 6). (F) Pie graphs showing distribution of indicated PLZF-expressing cell subsets in adult mice treated with antibiotics. Data are representative of two independent experiments. Bars are mean ± SEM.
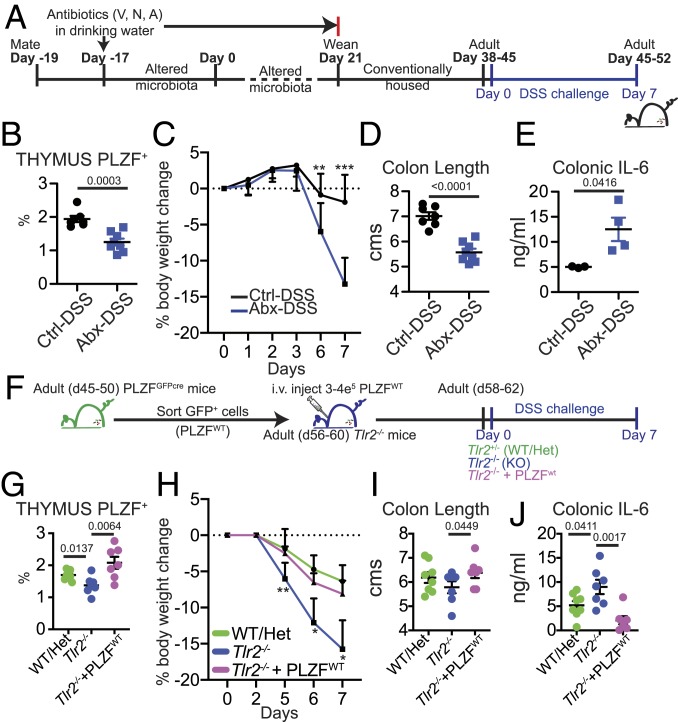
Perturbation of Early-Life Microbial-Thymic PLZF Axis Impacts Susceptibility to Dextran Sulfate Sodium Colitis in Later Life.
Early-life antibiotic use is associated with increased susceptibility to autoimmune diseases, such as celiac disease and inflammatory bowel disease in later life. In our studies, early-life antibiotics decreased the frequency of PLZF-expressing immune cells, a deficiency that persisted into adulthood (Fig. 5 A–C). To determine the consequences of this impaired immune development, we challenged adult mice treated with antibiotics in early life with dextran sulfate sodium (DSS) (Abx-DSS) (Fig. 6A). Flow cytometry analysis revealed fewer thymic PLZF+ cells in Abx-DSS mice (Fig. 6B). Compared to Ctrl-DSS mice, antibiotics-treated mice lost significantly more weight upon the DSS challenge (Fig. 6C). Colon shortening at day 7 of treatment in Abx-DSS mice was accompanied by increased production of IL-6 compared to Ctrl-DSS mice (Fig. 6 D and E). To determine if the increased susceptibility to DSS colitis was mediated by altered homeostasis and/or the function of PLZF+ cells, we performed adoptive transfer of sorted PLZF+ cells from the thymus and spleen of TLR2WT PLZFGFPcre reporter mice (referred to as PLZFWT) into adult Tlr2−/− mice (Fig. 6F). Tlr2−/− mice have previously been shown to be susceptible to DSS colitis (41). In our experiments, we showed that infant Tlr2−/− mice have fewer thymic PLZF+ cells, a deficiency that persisted into adulthood (Fig. 2 and SI Appendix, Fig. S11). Transfer of PLZFWT cells increased the frequency of total thymic PLZF+ cells in Tlr2−/− mice (Fig. 6G). Importantly, we noted moderated disease severity and reduced colonic inflammation in Tlr2−/− mice that had received PLZFWT cells (Fig. 6 H–J). Thus, our studies suggest that early-life microbial perturbation impacted PLZF+ cell subset homeostasis and increased susceptibility to DSS induced colitis in later life.
Fig. 6.
Early-life antibiotics and PLZF+ cells influence susceptibility to DSS. (A) C57BL/6 breeders were given antibiotics (vancomycin, neomycin, and ampicillin) in drinking water at the time of mating. Pups born of these altered microbiota breeders were weaned at 21 d and housed in conventional isolators without antibiotics. Adult (d38–45 d old) mice were administered DSS in drinking water and analyzed 7 d later. Ctrl-DSS: Mice that received no antibiotics in early life and were challenged with DSS. Abx-DSS: Mice that received antibiotics in early life and were challenged with DSS. (B) Frequency of PLZF+ cells in the thymus (Ctrl-DSS n = 7; Abx-DSS n = 8). Effect size: Ctrl-DSS vs. Abx-DSS: 2.56. (C) Percentage of body weight change in animals. (Ctrl-DSS n = 7; Abx-DSS n = 8; Day 6: P = 0.0344; Day 7: P = 0.0006). (D) Colon length at day 7 post-DSS treatment (Ctrl-DSS n = 7; Abx-DSS n = 8). (E) IL-6 ELISA on supernatants of colon sections cultured overnight at 37 °C (Ctrl-DSS n = 4; Abx-DSS n = 4). (F) PLZF-expressing cells were sorted from adult PLZFGFPcre reporter mice and i.v. injected into adult Tlr2−/− mice. Mice were then administered DSS in drinking water and analyzed 7 d later. WT/Het: Tlr2+/+ and Tlr2+/− mice that were challenged with DSS. Tlr2−/−: Tlr2−/− mice that were challenged with DSS. Tlr2−/− + PLZFWT: Tlr2−/− mice that received WT PLZF+ cells and were challenged with DSS. (G) Frequency of PLZF+ cells in the thymus (WT/Het n = 8; Tlr2−/− n = 7; Tlr2−/−+ PLZFWT n = 7). Effect size: Tlr2−/− vs. Tlr2−/−+ PLZFWT: 1.76. (H) Percentage of body weight change in animals. (WT/Het n = 8; Tlr2−/− n = 7; Tlr2−/−+ PLZFWT n = 7; Day 5 (KO vs. KO + PLZF): P = 0.0039, Day 6: P = 0.0453, Day 7: P = 0.0353). (I) Colon length at day 7 post-DSS treatment (WT/Het n = 8; Tlr2−/− n = 7; Tlr2−/−+ PLZFWT n = 7). (J) IL-6 ELISA on supernatants of colon sections cultured overnight at 37 °C (WT/Het n = 8; Tlr2−/− n = 7; Tlr2−/−+ PLZFWT n = 7). Data are from two independent experiments. Bars are mean ± SEM.
Discussion
The results presented here reveal a novel communication between intestinal microbes and developing cells in the thymus. The sterile intestine of a newborn receives its first microbial inoculum during and shortly after naissance. Local mucosal immune responses to intestinal bacteria are expected and have been widely reported (42), but gut-distal effects are also being recognized (43, 44). In our studies, commensal microbes influenced the distribution of thymic PLZF+ innate-like cells during the postnatal period. Migratory pDCs that traffic from the colon to the thymus during the postnatal period are one potential mediator of this effect. Missing bacterial species or bacterial products may perturb this regulatory circuit, reducing thymic PLZF+ cells and exacerbating intestinal inflammation in later life.
Intestinal microbes protect the host from inflammatory insults at mucosal sites as shown by the heightened susceptibility of adult mice treated with antibiotics to DSS induced experimental colitis (41). However, it has been reported that young-adult mice treated with antibiotics but, subsequently, allowed to recover for 6 wk are protected from DSS colitis via a mechanism that appeared independent of the adaptive immune system (45). In our experiments, we noted that adult mice treated with antibiotics in infancy were more susceptible to DSS colitis (Fig. 6 A–E), suggesting the existence of an early-life time window when thresholds of immune reactivity are set. Indeed, antibiotics treatment in later life had no impact on thymic PLZF+ cell homeostasis (Fig. 5 D–F). We further showed that adoptive transfer of WT PLZF+ cells could reduce severity of colitis in Tlr2−/− mice (Fig. 6 F–J). It remains to be determined which subset of PLZF-expressing cells mediates the protective effect observed in our studies. iNKT cells have previously been shown to protect mice from experimental colitis (46, 47). ILC2s (CD25+ckit+IL33R+) also expand in DSS colitis and are considered to have a tissue-protective function in the intestinal tract (48). ILC2s may arise in the thymus from T committed precursor cells (49, 50), and one possibility is that microbes regulate their development and functional imprinting in early life. Indeed, an altered thymic homeostasis of PLZF+c-kit+ cells was observed in GF monocolonization experiments as well as in Tlr2−/− mice (Figs. 1F and 2F). Furthermore, disruption of enterothymic communication by deletion of pDCs in the colon in BDCA2-DTR mice also reduced thymic PLZF+c-kit+ cells (Fig. 4K), indicating that this cell subset may be particularly responsive to intestinal microbial colonization state in early life.
We identified pDCs as a key component of the enterothymic communication axis. Using in vivo photolabeling experiments, we could detect colonic Dendra-r+ pDCs in the thymus in early life (Fig. 3C). Furthermore, using the BDCA2-DTRtg model of depleting colonic pDCs, we could demonstrate a small but significant decrease in thymic pDC frequency upon DT administration (Fig. 4H). Flow cytometric analysis of colonic pDCs confirmed their expression of CCR9 (SI Appendix, Fig. S8), a receptor that has previously been shown to facilitate their migration from secondary lymphoid organs to the thymus (32). However, CCR9 also regulates cellular migration to the intestine itself and raises the issue of how CCR9+ pDCs might exit the colon to reach the thymus. Thus, while it appears that pDCs are competent to migrate from the colon to the thymus, the precise mechanism remains undetermined. pDCs are also notoriously poor at antigen presentation (51) and express low levels of MHC Class II (SI Appendix, Fig. S8). It is unlikely that pDCs transport microbial antigens to the thymus to regulate the development of immune cells. Instead, pDCs may respond to microbial molecules with changes in phenotype or gene expression that influence thymic PLZF+ cells (Fig. 3 D–F). Other gut related mechanisms, including soluble mediators, might operate in conjunction with migratory pDCs to regulate thymic innate PLZF+ cell distribution. In Dendra photoconversion experiments, the identity of the majority of cells that migrated from the colon to the thymus in early life is yet unknown. It is also a formal possibility that PLZF+ cells themselves may recirculate from the colon to the thymus. In humans, PLZF-expressing cells were enriched in fetal tissue especially the intestine where they contribute to both the protective and the pathologic arms of the early-life immune system (52). Thus, PLZF+ immune cells appear poised to regulate early-life immunity, and our studies offer a glimpse toward how their homeostasis may be regulated by intestinal microbes.
Recent studies have also highlighted the contribution of maternal microbes in guiding intestinal immune cell homeostasis in their offspring during gestation (17). In our studies, we did not distinguish gestational effects of maternal microbiota from postnatal acquisition of maternal microbiota by offspring on thymic development. Whether transplacental transfer of microbial antigens plays a role in offspring thymic cell development remains an open question. The present studies highlight a communication between the neonatal intestine and immune cells in the thymus that show developmental plasticity in response to microbial constitution. The gut is likely by far the largest contributor of novel antigens presented to the neonatal immune system. A graded and apposite response to this antigenic load is undoubtedly required to promote healthy immune system maturation. A deeper understanding of the mechanisms that control thymic-gut interactions in early life may lead to a better appreciation of the mechanisms by which intestinal flora sustain healthy immunity and influence the development of autoimmune disorders.
Materials and Methods
Animals and Care.
Mouse studies were conducted under protocols approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital (MGH). C57BL/6J mice (Stock No. 000664) were purchased from the Jackson Laboratory and housed in HPPF facilities. Tlr2−/− mice (Stock No. 004650) were purchased from the Jackson Laboratory and bred as heterozygotes. Ccr9−/− mice (Stock No. 027041), BDCA2-DTR Tg+ mice (Stock No. 01476), and PLZFGFPcre (Stock No. 024529) mice were also purchased from the Jackson Laboratory. For most experiments, entire litters of 14 d old mice were analyzed, and animals of both sexes were used as available.
GF Animal Studies.
GF animal experiments were performed at the Gnotobiotics, Microbiology and Metagenomics Core of the Harvard Digestive Diseases Center of the Brigham and Women’s Hospital, Boston. B. fragilis NCTC 9343 and B. fragilis ΔPSA (19) (from Dr. Laurie Comstock, Brigham and Women’s Hospital, Boston) were inoculated by oral gavage of adult GF B6 breeders and monocolonization confirmed by bacterial culture of fecal material. Monocolonized male and female mice were bred in dedicated isolators, and pups from these breeders were used for analysis at 14 d of age.
Preparation of Lymphocytes and Flow Cytometry.
Thymuses and spleens were prepared for flow cytometry as described before (31). Changes in antibody staining panels (e.g., antibody clones and reagent batches) across experiments were minimized. Strong discrepancies in staining from different experiments were noted, and the data in question were excluded from final analyses.
Photoconversion of Colon-Resident Cells.
Cells in the colon of newborn mice were photoconverted from Dendra-g to Dendra-r expression as described before (31). Briefly, a custom-made fiber-optic probe (0.9 cm length, 400 μm core diameter, and light diffuser at end, ∼3 mW) was coupled to a 405 nm laser source (Thor Labs, 1,400 mA and >20 mW) and inserted to a depth of 0.9 cm through the anuses of 0–5 d old PhAMexcised mice. Colonic tissues were exposed to 405 nm light for a total of 2 min (the fiber-optic cable was retracted by ∼0.25 cm every 30 s of exposure). Tissues were harvested at 3 to 4 h after light exposure for analysis.
RNA-Seq.
Libraries for RNA-seq. were prepared using the NuGEN Ovation RNA-seq v2 library preparation kit (NuGEN Technologies, Inc.) and sequenced on the Illumina platform at the MGH Bioinformatics Core. Sequenced reads were mapped to the mm9 reference genome using the STAR aligner (53). Reads were processed as described here (54). Briefly, reads were quality checked using FastQC, adapters trimmed using Cutadapt, reads mapped to the GRCm38.p4 mouse reference using STAR, and counts generated using htseq-count. Subsequent analyses and visualizations were performed using R, DE genes were called using edgeR, and networks tested for significance using the camera module (55) from the limma package (56). MSigDB v6.2 (57, 58) was used to generate the networks. Only the H, C2, and C7 datasets were tested.
Anti-BST2 Treatment.
A cohort of 8 and 10 d old C57BL/6 pups were given anti-BST2 antibody (Bio X Cell) by i.p. injection. Ctrl pups were given saline injections at the same time.
DT Treatment.
Transgene positive and negative littermate BDCA2-DTR-Tg pups were given four intracolonic injections of 50 ng DT (Sigma-Aldrich) at days 8–11 after birth.
Adoptive Transfer.
Thymic pDCs were isolated from d12–14 C57BL/6 mice using anti-PDCA1 MACS beads and 3–5e5 pDCs were injected i.p. into d5-7 Tlr2−/− mice. Transcription factor PLZF-expressing cells were sorted to >95% purity from the spleen and thymus of adult (d45–50) PLZFGFPcre mice on a FACS Aria. Cells (3–4e5/mouse) were injected i.v. into adult (d56–60) Tlr2−/− mice.
Antibiotics Treatment.
C57BL/6 breeding pairs were placed on antibiotic water (vancomycin [0.5 mg/mL], neomycin [1 mg/mL], and ampicillin [1 mg/mL]) at the time of mating as described before (59). Antibiotic water was continued until pups were weaned at 21 d of age.
Microbiome Analysis.
Colonic content was collected at the time of death, and bacterial DNA isolated using a Qiagen QIAamp PowerFecal Pro DNA kit. The 16S sequencing was performed by CosmosID and analyzed on their platform (https://app.cosmosid.com/login).
DSS Colitis.
Adult (d38–45) mice were given 3% (wt/vol) DSS (MP Biomedicals, colitis grade, mol wt 36,000–50,000) in drinking water for 7 d. Body weights were measured daily. At necropsy, the colon from the cecal junction to the rectum was removed, and its length recorded. Tissues were flushed with PBS to expel contents, and small sections were excised from the distal third for overnight culture in Dulbecco’s modified Eagle's medium with 10% heat-inactivated fetal calf serum and antibiotics. The supernatants of the cultured colon explants were used for measurement of IL-6.
IL-6 ELISA.
IL-6 levels in supernatants were measured using antibodies specific for the mouse cytokine (purified rat anti-mouse IL-6 (Cat No. 554400), and biotin rat anti-mouse IL-6 (Cat No. 554402), BD Pharmingen, Franklin Lakes, NJ) following manufacturer’s protocols.
Statistical Analysis.
For comparison between two cohorts of mice, a minimum of five mice per group was used, and the experiments were repeated, at least, two times. The D’Agostino and Pearson omnibus normality test was used to determine if values came from a Gaussian distribution. A two-tailed unpaired t test was used to calculate significance in the comparisons of two cohorts that were normally distributed. A Mann–Whitney test was performed for comparing groups that failed normality tests. Cohen’s d for effect size was calculated as the difference between the means of two groups divided by average SD. Graphs are vertical scatter plots showing the SE of mean. Animals of both sexes were used as available. No randomization was performed, and investigators were not blinded to group allocations.
Data Availability.
All data generated or analyzed in this paper are included in this published article and its supplementary information files.
Supplementary Material
Acknowledgments
We are thankful for the resources of the Center for Computational and Integrative Biology at MGH that supported this study. We thank Dr. Laurie Comstock for providing strain B. fragilis ΔPSA. L.B. and gnotobiotic studies were supported by NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant P30DK034854 (Harvard Digestive Disease Center) and the Massachusetts Life Science Center Grant. B.J.C. and S.V. were supported by Grant R01AI089700. S.I. received salary support from NIH NIDDK Grant R25DK103579. N.J. was supported by NIH National Institute of Allergy and Infectious Diseases (NIAID) Grants 1K22AI116661-01 and 1R21AI139735-01A1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915047117/-/DCSupplemental.
References
- 1.Zeissig S., Blumberg R. S., Life at the beginning: Perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 15, 307–310 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Gregory K. E., et al. , Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 4, 68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain N., Walker W. A., Diet and host-microbial crosstalk in postnatal intestinal immune homeostasis. Nat. Rev. Gastroenterol. Hepatol. 12, 14–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amenyogbe N., Kollmann T. R., Ben-Othman R., Early-life host-microbiome interphase: The key frontier for immune development. Front Pediatr. 5, 111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gensollen T., Blumberg R. S., Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 139, 1084–1091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y., Hand T. W., Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renz H., Brandtzaeg P., Hornef M., The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 12, 9–23 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Rothenberg E. V., Moore J. E., Yui M. A., Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendelac A., Savage P. B., Teyton L., The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Wong S. H., et al. , Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H. C., et al. , Downregulation of E protein activity augments an ILC2 differentiation program in the thymus. J. Immunol. 198, 3149–3156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrielli S., et al. , Murine thymic NK cells are distinct from ILC1s and have unique transcription factor requirements. Eur. J. Immunol. 47, 800–805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Constantinides M. G., Interactions between the microbiota and innate and innate-like lymphocytes. J. Leukoc. Biol. 103, 409–419 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A., A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prince A. L., et al. , Innate PLZF+CD4+ αβ T cells develop and expand in the absence of Itk. J. Immunol. 193, 673–687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage A. K., et al. , The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez de Aguero M., et al. , The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Pronovost G. N., Hsiao E. Y., Perinatal interactions between the microbiome, immunity, and neurodevelopment. Immunity 50, 18–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyne M. J., et al. , Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 69, 4342–4350 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Round J. L., et al. , The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L., An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 22.An D., et al. , Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang F., et al. , The symbiotic bacterial surface factor polysaccharide A on Bacteroides fragilis inhibits IL-1β-induced inflammation in human fetal enterocytes via toll receptors 2 and 4. PLoS One 12, e0172738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alhawi M., Stewart J., Erridge C., Patrick S., Poxton I. R., Bacteroides fragilis signals through Toll-like receptor (TLR) 2 and not through TLR4. J. Med. Microbiol. 58, 1015–1022 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Erridge C., Pridmore A., Eley A., Stewart J., Poxton I. R., Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2. J. Med. Microbiol. 53, 735–740 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Wang Q., et al. , A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 203, 2853–2863 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diehl G. E., et al. , Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 494, 116–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson A. J., Uhr T., Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Mowat A. M., Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 3, 331–341 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Pham A. H., McCaffery J. M., Chan D. C., Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter C., Ennamorati M., Jain N., In vivo photolabeling of cells in the colon to assess migratory potential of hematopoietic cells in neonatal mice. J. Vis. Exp. 138, e57929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadeiba H., et al. , Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity 36, 438–450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bevins C. L., Salzman N. H., Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Geijtenbeek T. B., Gringhuis S. I., C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 16, 433–448 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Sun J., et al. , The 11S proteasome subunit PSME3 is a positive feedforward regulator of NF-κB and important for host defense against bacterial pathogens. Cell Rep. 14, 737–749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendland M., et al. , CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc. Natl. Acad. Sci. U.S.A. 104, 6347–6352 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uehara S., Grinberg A., Farber J. M., Love P. E., A role for CCR9 in T lymphocyte development and migration. J. Immunol. 168, 2811–2819 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Zlotoff D. A., et al. , CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood 115, 1897–1905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M., Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Park J., Foss D., Goldschneider I., Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J. Exp. Med. 206, 607–622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R., Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Geva-Zatorsky N., et al. , Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H. J., et al. , Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao E. Y., et al. , Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward N. L., et al. , Antibiotic treatment induces long-lasting changes in the fecal microbiota that protect against colitis. Inflamm. Bowel Dis. 22, 2328–2340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saubermann L. J., et al. , Activation of natural killer T cells by alpha-galactosylceramide in the presence of CD1d provides protection against colitis in mice. Gastroenterology 119, 119–128 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Kim H. S., Chung D. H., IL-9-producing invariant NKT cells protect against DSS-induced colitis in an IL-4-dependent manner. Mucosal Immunol. 6, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Monticelli L. A., et al. , IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. U.S.A. 112, 10762–10767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian L., et al. , Suppression of ILC2 differentiation from committed T cell precursors by E protein transcription factors. J. Exp. Med. 216, 884–899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones R., et al. , Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur. J. Immunol. 48, 1481–1491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villadangos J. A., Young L., Antigen-presentation properties of plasmacytoid dendritic cells. Immunity 29, 352–361 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Halkias J., et al. , CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J. Clin. Invest. 130, 3562–3577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conesa A., et al. , A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D., Smyth G. K., Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liberzon A., et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olson C. A., et al. , The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173, 1728–1741.e13 (2018). Erratum in: Cell174, 497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this paper are included in this published article and its supplementary information files.