Significance
Genome integrity is required for clinical applications of pluripotent stem cells. Two major strategies, maintaining low levels of reactive oxygen species (ROS) and high DNA damage repair (DDR) activities, are applied by embryonic stem cells (ESCs) to secure their genomic stability. Here, we demonstrate the pivotal role of Cops5 in safeguarding genome integrity of ESCs. Through regulating the autophagic degradation of Mtch2, Cops5 suppresses oxidative phosphorylation and ROS production, thereby reducing endogenous DNA damage. Meanwhile, Cops5 is also involved in maintaining high DDR activities, including nucleotide excision repair, nonhomologous end joining, and homologous recombination, in ESCs.
Keywords: Cops5, genomic stability, cellular metabolism, DNA repair, embryonic stem cells
Abstract
The highly conserved COP9 signalosome (CSN), composed of 8 subunits (Cops1 to Cops8), has been implicated in pluripotency maintenance of human embryonic stem cells (ESCs). Yet, the mechanism for the CSN to regulate pluripotency remains elusive. We previously showed that Cops2, independent of the CSN, is essential for the pluripotency maintenance of mouse ESCs. In this study, we set out to investigate how Cops5 and Cops8 regulate ESC differentiation and tried to establish Cops5 and Cops8 knockout (KO) ESC lines by CRISPR/Cas9. To our surprise, no Cops5 KO ESC clones were identified out of 127 clones, while three Cops8 KO ESC lines were established out of 70 clones. We then constructed an inducible Cops5 KO ESC line. Cops5 KO leads to decreased expression of the pluripotency marker Nanog, proliferation defect, G2/M cell-cycle arrest, and apoptosis of ESCs. Further analysis revealed dual roles of Cops5 in maintaining genomic stability of ESCs. On one hand, Cops5 suppresses the autophagic degradation of Mtch2 to direct cellular metabolism toward glycolysis and minimize reactive oxygen species (ROS) production, thereby reducing endogenous DNA damage. On the other hand, Cops5 is required for high DNA damage repair (DDR) activities in ESCs. Without Cops5, elevated ROS and reduced DDR activities lead to DNA damage accumulation in ESCs. Subsequently, p53 is activated to trigger G2/M arrest and apoptosis. Altogether, our studies reveal an essential role of Cops5 in maintaining genome integrity and self-renewal of ESCs by regulating cellular metabolism and DDR pathways.
The unlimited self-renewing capacity and differentiation potential into all types of cells in the body, which is called pluripotency, renders embryonic stem cells (ESCs) a promising donor cell source for regenerative medicine. However, genomic stability and tumorigenicity of ESCs raise safety issues for their clinical applications.
To maintain genome stability, endogenous DNA lesions caused by transcription, replication, and oxidative stresses need to be repaired by various DNA damage repair (DDR) pathways, including base excision repair, mismatch repair, nucleotide excision repair (NER), homologous recombination (HR), and nonhomologous end-joining (NHEJ) (1, 2). Compared with differentiated cells, ESCs have a higher risk to acquire more DNA lesions due to their fast proliferation rate and hyperactive global transcription (3, 4). Yet, mutation frequency in ESCs is lower than that in mouse embryonic fibroblasts (5). At least two strategies, high DDR activities and low levels of reactive oxygen species (ROS), are applied by ESCs to secure the genome integrity (6, 7). To maintain high DDR activities, genes involved in DDR are expressed at higher levels in ESCs than in differentiated cells (8, 9). And ESCs preferentially use HR, rather than NHEJ, to repair DNA double-stranded breaks (DSBs) with high fidelity (10). Moreover, some ESC-specific factors also contribute to efficient DDR. For example, Zscan4, which is transiently expressed in about 5% of ESCs at a given time, promotes rapid telomere elongation by telomere recombination and regulates genomic stability (11). Induced by genotoxic stress, Filia stimulates the PARP1 activity and relocates from centrosomes to DNA damage sites and mitochondria to regulate DDR and apoptosis (12). Sall4, a pluripotency transcription factor, facilitates the ataxia telangiectasia-mutated activation in response to DSBs (13). To minimize the ROS-induced genomic DNA damage, ESCs produce lower levels of mitochondrial ROS and express higher levels of antioxidants than differentiated cells (14, 15). ESCs predominantly produce ATP through glycolysis, rather than through oxidative phosphorylation (OXPHOS), even though glycolysis is less efficient in energy production (15). The so-called Warburg effect allows sufficient supply of anabolic intermediates for proliferation, as well as minimizing the production of ROS (16). It has been reported that restricting the entry of pyruvate into mitochondria by uncoupling protein 2, together with high levels of hexokinase II and inactive pyruvate dehydrogenase, might rewire the cellular metabolism favoring glycolysis over OXPHOS (17, 18).
The highly conserved COP9 signalosome (CSN) is composed of eight subunits (Cops1 to Cops8). Its most studied function is to regulate protein degradation through suppressing the activity of the cullin-RING-E3 ligases by deneddylation of cullins (19–21). In addition, the CSN is associated with damage specific DNA binding protein 2 (DDB2) and Cockayne syndrome type A protein (CSA) complexes involved in two NER pathways, global genome repair (GGR) and transcription coupled repair (TCR), respectively. Knockdown of COPS5 leads to NER defect (22). A whole-genome RNA interference screening revealed that COPS1, COPS2, and COPS4 are required for maintaining the expression of the OCT4-GFP reporter in human ESCs, implicating a role of the CSN in pluripotency maintenance (23). However, by knocking down individual CSN subunits, we found that only Cops2, but not any other CSN subunits, is essential for the self-renewal and G2/M transition of mouse ESCs (24, 25). Moreover, Cops5 and Cops8 null embryos die after embryonic day 7.5, while no Cops2 null mice survive to embryonic day 7.5 (26–28). These data implicate that Cops5 and Cops8 might be involved in late differentiation events, while Cops2 is essential for the establishment of pluripotency in the inner cell mass.
We set out to investigate how Cops5 and Cops8 regulate the differentiation of ESCs and tried to establish Cops5 and Cops8 knockout (KO) ESC lines by CRISPR/Cas9. To our surprise, no Cops5 KO ESC clones were identified out of 127 clones, while three Cops8 KO ESC lines were established out of 70 clones. We then constructed an inducible Cops5 KO (iC5; C5 KO) ESC line. Cops5 KO leads to decreased expression of the pluripotency marker Nanog, proliferation defect, G2/M cell-cycle arrest, and apoptosis of ESCs. Further analysis demonstrated that, without Cops5, DDR activities are reduced. In addition, loss of Cops5 accelerates the turnover of Mtch2 through autophagy, thus altering cellular metabolism toward OXPHOS and enhancing ROS level. Consequently, DNA damages are accumulated in ESCs, and p53 is activated to trigger apoptosis. Altogether, our studies reveal an essential role of Cops5 in maintaining genome integrity of ESCs by regulating cellular metabolism and DDR pathways.
Results
Cops8, but Not Cops5, Can Be Knocked Out in Mouse ESCs.
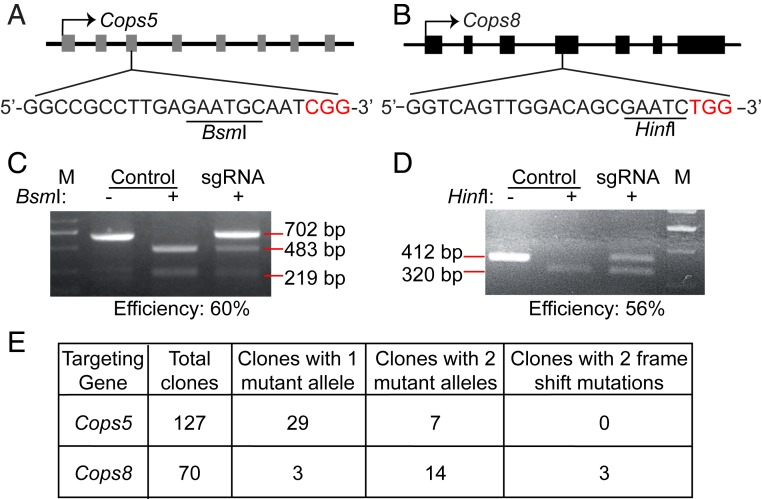
Aiming to study the roles of Cops5 and Cops8 in ESC differentiation, we designed single guide RNAs (sgRNAs) targeting Cops5 and Cops8 (Fig. 1 A and B). The cutting efficiencies of Cas9 at Cops5 and Cops8 loci were about 60 and 56%, as demonstrated by the disruption of the BsmI and HinfI sites, respectively (Fig. 1 C and D). Three Cops8 KO ESC lines were established out of 70 clones. In contrast, no Cops5 KO ESC line was obtained after screening 127 clones (Fig. 1E). Even though seven clones harboring two disrupted BsmI sites in the Cops5 alleles were identified, none of the seven clones had frameshift mutations in both Cops5 alleles. These data implied that Cops5 might be essential for mouse ESCs.
Fig. 1.
Failure in construction of Cops5 KO ESC line by CRISPR/Cas9. (A and B) Schematic illustration of sgRNA design for Cops5 (A) and Cops8 (B). Gray and black rectangles represent the exons of Cops5 and Cops8, respectively. The restriction endonuclease sites in the sgRNA recognition sequences are underlined, and the protospacer-adjacent motifs are shown in red. (C and D) Cutting efficiency of Cas9 at the Cops5 (C) and Cops8 (D) loci. ESCs were transfected with pX330 plasmids targeting Cops5 (C) and Cops8 (D) or with the pX330 plasmid without an sgRNA insert. Forty-eight hours after transfection, cells were harvested for genomic DNA purification. DNA fragments of 702- and 412-bp around the Cas9 target sites at the Cops5 and Cops8 loci were amplified by PCR, respectively. The DNA fragments were digested by BsmI or HinfI. The intensities of DNA bands were quantified using Image J software, and the cutting efficiency was calculated. (E) Genotypes of ESC clones after Cas9 treatment. ESCs were transfected as described in C and D. Forty-eight hours after transfection, ESCs were treated with trypsin and plated at low density. After 5 to 7 d, individual colonies were picked, expanded, and subjected to genotyping.
Cops5 KO Compromises the Self-Renewal and Differentiation of ESCs.
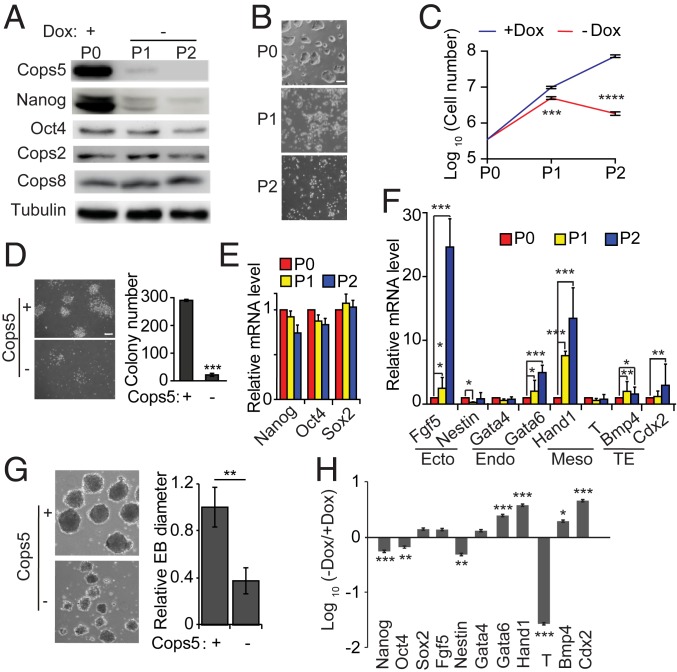
To directly demonstrate the pivotal role of Cops5 in pluripotency maintenance of ESCs, we constructed an inducible Cops5 KO ESC line, using a strategy that we had developed previously (29). A doxycycline-inducible Cops5 (iC5) ESC line was then generated from KH2 cells (30). The sgRNA recognition site in the exogenous inducible Cops5 gene was mutated to render it resistant to Cas9. Then the endogenous Cops5 alleles were disrupted in iC5 ESCs by CRISPR/Cas9 in the presence of doxycycline (Dox), resulting in iC5; C5 KO ESCs. Only a residual amount of Cops5 remains in iC5; C5 KO ESCs at 48 h after Dox withdrawal, and no Cops5 is detectable by Western blot at 96 h after Dox withdrawal (Fig. 2A). Notably, depletion of Cops5 compromises the self-renewal of ESCs, demonstrated by loss of ESC colony morphology, reduced proliferation rate, and colony-forming capacity (Fig. 2 B–D). Moreover, iC5; C5 KO ESCs cannot be cultured for more than two passages without Dox. The messenger RNA (mRNA) levels of the pluripotency genes Nanog, Oct4, and Sox2 are not altered by Cops5 KO (Fig. 2E). Yet, Nanog protein, but not Oct4, is decreased upon Cops5 KO (Fig. 2A), suggesting that Cops5 regulates Nanog expression posttranscriptionally. With Nanog truncation mutants and individual domains fused to luciferase, we found that both N-terminal and C-terminal domains of Nanog, but not the Homeobox domain of Nanog, mediate the degradation of Nanog in Cops5 KO ESCs (SI Appendix, Fig. S1). Cops5 KO also perturbs the expression of differentiation genes, such as Fgf5, Gata6, Hand1, Bmp4, and Cdx2 (Fig. 2F). All these data suggest an essential role of Cops5 in ESC self-renewal.
Fig. 2.
Cops5 is required for ESC self-renewal and differentiation. (A) iC5; C5 KO ESCs cultured with Dox (Passage 0, P0) were switched into medium without Dox for two passages. The protein levels of the pluripotency factors Nanog and Oct4, as well as the CSN subunits, Cops2, Cops5, and Cops8, were measured by Western blot. (B) Colony morphology change upon Cops5 KO. Phase-contrast images of ESC colonies at each passage are shown. (Scale bar, 100 μm.) (C) Growth curves of Cops5 KO ESCs. iC5; C5 KO ESCs were cultured in ESC medium with or without Dox. The cell numbers were counted every passage, and an equal amount of ESCs was plated into tissue culture dishes. (D) Colony forming assay of iC5; C5 KO ESCs with or without Dox. (Scale bar, 200 μm.) (E) Quantitative RT-PCR was performed to measure the RNA levels of the pluripotency markers Nanog, Oct4, and Sox2. (F) Expression of differentiation genes after Cops5 KO. (G) iC5; C5 KO ESCs were cultured with Dox, and then the cells were used for EB differentiation with or without Dox. (Left) The images of day 4 EBs (40× magnification) with or without Dox. The relative diameters of day 4 EBs were measured and plotted (Right). (H) Quantitative RT-PCR analysis of pluripotency and differentiation genes in day 4 EBs, as described in G. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Next, we tested whether Cops5 is required for the differentiation of ESCs. iC5; C5 KO ESCs were cultured with Dox and then were allowed to form embryoid bodies (EBs) in hanging drops with or without Dox. EBs without Cops5 are smaller than EBs with Cops5 (Fig. 2G). Cops5 KO does not block the down-regulation of the pluripotency genes Nanog, Oct4, and Sox2. Rather, it leads to deregulation of differentiation genes, such as elevated expression of Gata6, Hand1, Bmp4, and Cdx2, and to reduced expression of Nestin and T (Fig. 2H). Thus, Cops5 is also required for EB differentiation of ESCs.
Cops5 KO Leads to G2/M Arrest and Apoptosis through p53 Activation.
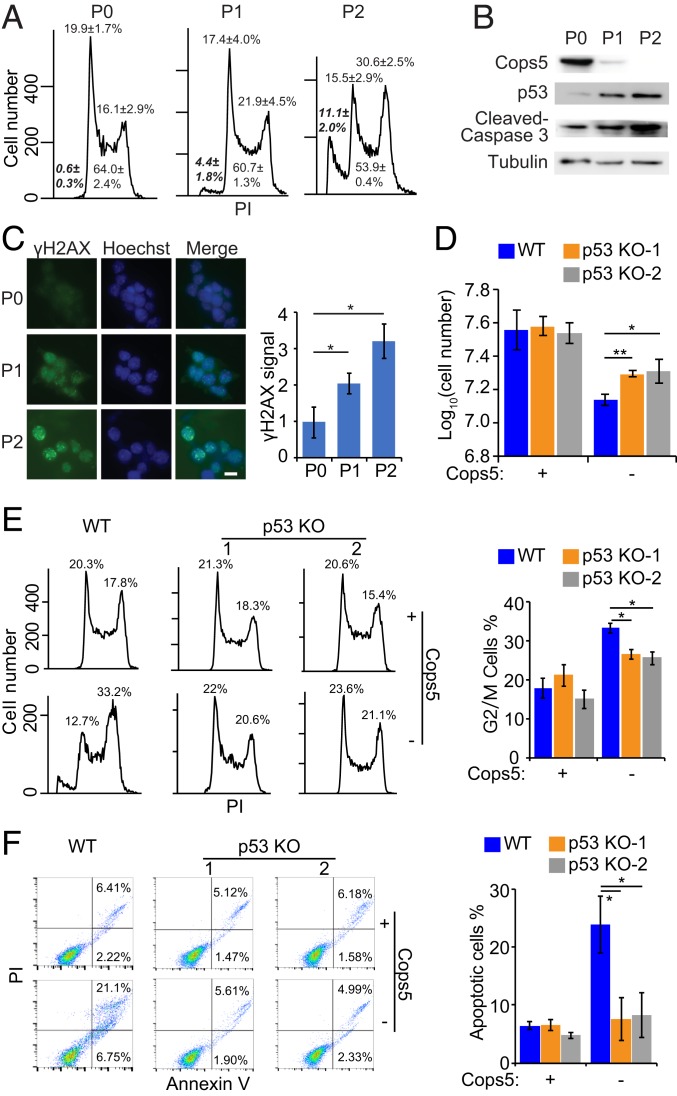
Given the growth defect of Cops5 KO ESCs, we analyzed the cell-cycle profile of iC5; C5 KO ESCs after Dox withdrawal by propidium iodide staining. G2/M cells, as well as the sub-G1 population (apoptotic cells), are increased upon Cops5 KO (Fig. 3A). Several key molecular markers for apoptosis, including cleaved Caspase 3, p53, and DNA damage (indicated by γH2AX and comet assay), are also activated after Cops5 KO (Fig. 3 B and C and SI Appendix, Fig. S2A). To prove that Cops5 KO activates p53 to induce G2/M arrest and apoptosis, we knocked out p53 in iC5; C5 KO ESCs (SI Appendix, Fig. S2B). p53 KO partially rescues the reduced proliferation rate and G2/M arrest caused by Cops5 KO, whereas Cops5 KO-induced apoptosis is completely rescued by p53 KO (Fig. 3 D–F). In contrast, the γH2AX signal, an indicator of DNA damage, is not affected by p53 KO (SI Appendix, Fig. S2C). These data suggest that Cops5 KO leads to DNA damage accumulation and subsequently to p53 activation, which in turn induces apoptosis, G2/M arrest, and reduced growth rate.
Fig. 3.
Loss of Cops5 leads to G2/M arrest and apoptosis through activating p53. (A) Cell-cycle analysis of iC5; C5 KO ESCs at different passages after Dox withdrawal. (B) Western blot to detect apoptotic markers, p53, and cleaved Caspase 3 upon Cops5 KO. (C) Immunofluorescence images of γH2AX upon Cops5 KO. (Scale bar, 10 μm.) Relative fluorescence intensities of γH2AX were quantified in around 100 nuclei for each condition. Quantification results were plotted (Right). (D) iC5; C5 KO and iC5; C5 KO; p53 KO ESCs were cultured in serum/LIF medium with or without Dox for two passages. Cell numbers were counted after two passages, and an equal amount of ESCs were plated into tissue culture dishes. (E and F) Cell-cycle (E) and apoptosis (F) analysis of iC5; C5 KO and iC5; C5 KO; p53 KO ESCs at different passages after Dox withdrawal. Quantification results are shown on the Right. *P < 0.05; **P < 0.01.
To address whether the essential role of Cops5 in ESC self-renewal is dependent on the CSN, we analyzed the effect of Cops8 KO on ESCs. Both Cops5 and Cops8 KO compromise the deneddylation activity of the CSN, indicating disruption of the CSN by Cops5 or Cops8 KO (SI Appendix, Fig. S3A). However, except for slightly reduced growth rate, Cops8 KO ESCs do not have defects in pluripotency marker expression, cell-cycle progression, or DNA damage accumulation (SI Appendix, Fig. S3 B–G). Moreover, upon differentiation, Cops8 KO ESCs fail to activate T and Cdx2, while Cdx2 is up-regulated in Cops5 KO EBs (Fig. 2H and SI Appendix, Fig. S3H). All these data suggest that the CSN-independent function of Cops5 contributes to pluripotency maintenance of ESCs.
Cops5 Depletion Impairs DDR Activities.
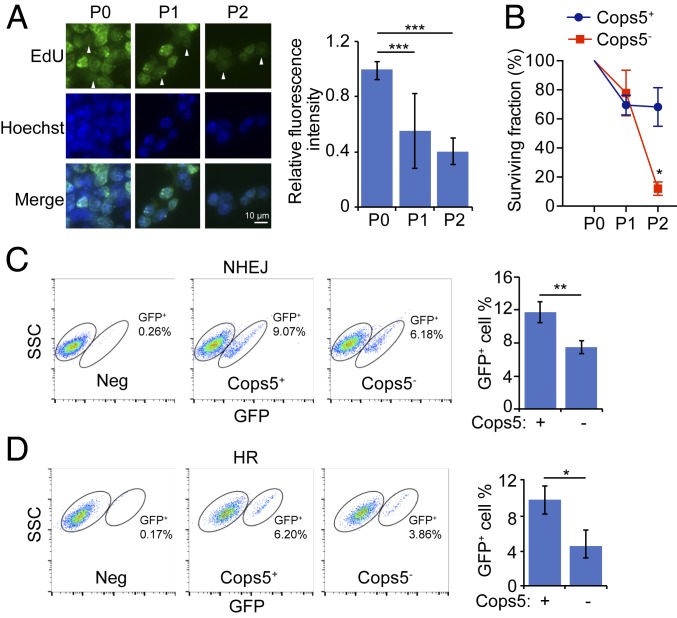
We then addressed the question how Cops5 KO induces DNA damage accumulation. It has been demonstrated that the CSN is associated with DDB2 and CSA complexes, which are involved in two NER pathways, GGR and TCR, respectively. Also, knockdown of COPS5 in human fibroblasts leads to defects in GGR and TCR (22). Moreover, osteosarcoma U2OS cells with COPS5 knockdown display HR repair defect (31). Thus, we measured the GGR, TCR, HR, and NHEJ activities in wild-type (WT) and Cops5 KO ESCs and found that the GGR, TCR, HR, and NHEJ activities are all reduced in Cops5 KO ESCs (Fig. 4). In contrast, Cops8 KO does not impair DDR activities (SI Appendix, Fig. S4 A–D), arguing that Cops5, rather than the CSN, is required for GGR and TCR activities.
Fig. 4.
Cops5 KO leads to DDR defects in ESCs. (A) iC5; C5 KO ESCs cultured with (P0) or without Dox for the indicated time (P1, P2) were subjected to GGR assay. Representative images are shown on the Left. Triangles indicate non–S-phase cells. (Scale bar, 10 μm.) Relative fluorescence intensities of 50 non–S-phase cells were quantified and plotted (Right). (B) iC5; C5 KO ESCs were cultured with or without Dox and exposed to 3 μg/mL illudin S for two passages. The surviving fraction was defined as the ratio of live cells with illudin S treatment to live cells without illudin S treatment. (C and D) iC5; C5 KO ESCs containing NHEJ (C) or HR (D) reporter were cultured with or without Dox for 24 h and then transfected with I-SceI–expressing plasmid. Forty-eight hours after transfection, the percentage of GFP+ cells was analyzed by flow cytometry. (Left three panels) The result of a representative experiment. (Right) The quantification result of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
It has been reported that, in mouse embryonic fibroblasts and osteosarcoma cells, Cops5 depletion activates p53, which subsequently suppresses the transcription of Rad51, a key gene for HR, and leads to reduced HR activity (31). The working mechanism for Cops5 to regulate HR in ESCs appears to be different. First, Rad51 protein, but not Rad51 mRNA, is reduced in Cops5 KO ESCs (SI Appendix, Fig. S4E). Second, decreased expression of Rad51 proteins in Cops5 KO ESCs are not completely abolished by p53 KO (SI Appendix, Fig. S4F). Third, even though p53 KO rescue the apoptosis induced by Cops5 KO, p53 KO does not affect DNA damage accumulation in Cops5 KO ESCs (SI Appendix, Fig. S2C), indicating that p53 is downstream of DNA damage accumulation. Thus, our data suggest that Cops5 KO impairs HR activity through down-regulating the Rad51 protein, independent of p53. Similarly, reduced NHEJ activity in Cops5 KO ESCs is likely due to decreased Ku70 protein, a key factor for NHEJ, again independent of p53 (SI Appendix, Fig. S4 E and F).
Cops5 Regulates Cellular Metabolism to Maintain Genome Stability.
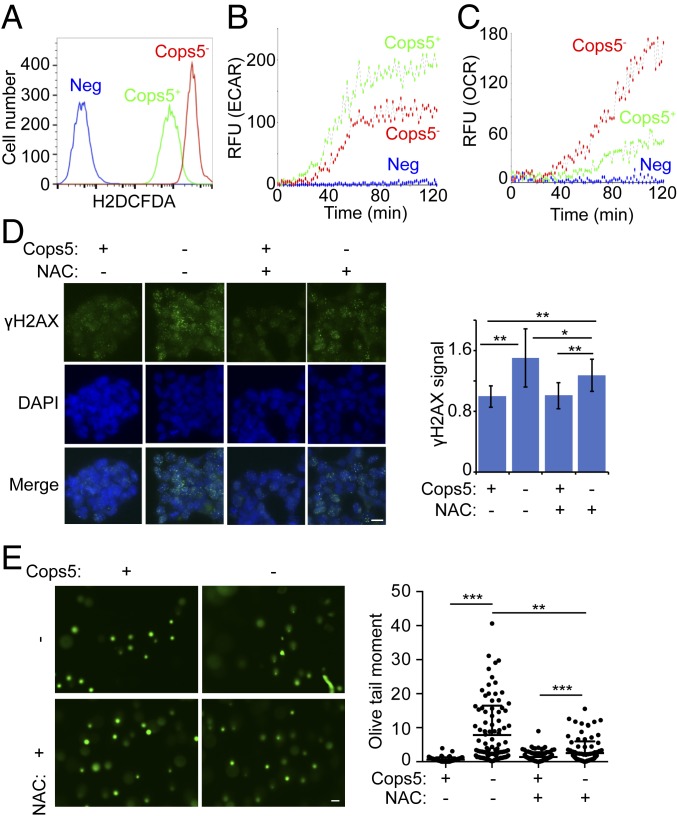
Xpa KO ESCs, defected in both GGR and TCR, are viable (32). No obvious DNA damage is accumulated in Xpa KO ESCs under normal culture conditions (SI Appendix, Fig. S4G). In addition, HR and NHEJ activities are only reduced, but not completely abolished, in Cops5 KO ESCs. These data imply that reduced DDR activities might not be the only reason for increased DNA damage in Cops5 KO ESCs under normal culture conditions. We suspected that DNA damage accumulation in Cops5 KO ESCs is due to enhanced endogenous DNA damage, combined with impaired DDR activities. To test this hypothesis, we examined the cellular level of ROS, which is an important oxidative molecule causing endogenous DNA damage. Indeed, Cops5 KO, but not Cops8 KO, enhances the level of ROS in ESCs (Fig. 5A and SI Appendix, Fig. S5A). Consistent with elevated ROS level, glycolysis activity is decreased and OXPHOS activity is enhanced in Cops5 KO ESCs, but not in Cops8 KO ESCs (Fig. 5 B and C and SI Appendix, Fig. S5 B and C). Moreover, Cops5 KO appears to have a negligible effect on ROS level, glycolysis, and OXPHOS activities in differentiated cells (SI Appendix, Fig. S5 D–F). To demonstrate the important role of ROS in elevated DNA damage induced by Cops5 KO, we treated ESCs with the reductant N-acetylcysteine (NAC). Both γH2AX immunostaining and a comet assay showed that NAC treatment suppresses DNA damage accumulation in Cops5 KO ESCs (Fig. 5 D and E).
Fig. 5.
Cops5 KO alters cellular metabolism and elevates ROS level. (A) Measurement of ROS in iC5; C5 KO ESCs cultured with or without Dox for 48 h. (B) Extracellular acidification rate (ECAR) of iC5; C5 KO ESCs with or without Dox for 48 h. (C) Oxygen consumption rate (OCR) of iC5; C5 KO ESCs with or without Dox for 48 h. (D) iC5; C5 KO ESCs were cultured with or without Dox for 48 h. For the NAC treatment group, 3 mM NAC was added during the 48 h. Representative immunofluorescence images of γH2AX (three independent experiments) are shown on the Left. (Scale bar, 10 μm.) Relative fluorescence intensities of γH2AX were quantified in ∼100 nuclei for each condition and plotted (Right). (E) Comet assay of iC5; C5 KO ESCs treated as D. (Left) The representative images. (Scale bar, 50 μm.) Comet tail lengths of 100 cells per sample are quantified (Right). *P < 0.05; **P < 0.01; ***P < 0.001.
Cops5 Regulates Cellular Metabolism through Mtch2.
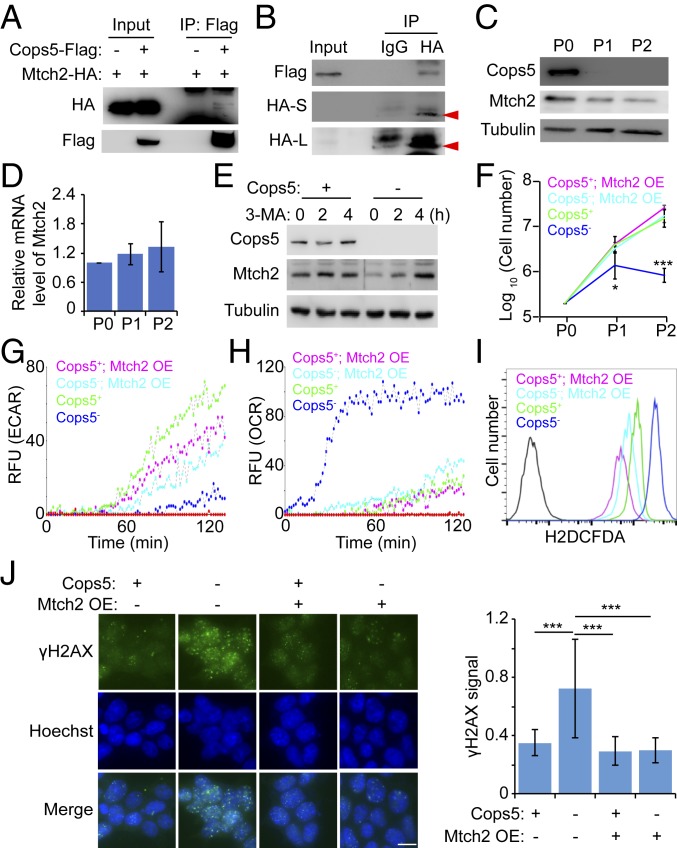
To understand the molecular mechanism of Cops5 in regulating cellular metabolism, we looked into the list of Cops5-interacting proteins identified by coimmunoprecipitation (co-IP) and mass spectrometric analysis (25). We focused on the 49 proteins interacting with Cops5, but not with Cops2 (Dataset S1), because the regulation of cellular metabolism by Cops5 is independent of the CSN. Among these 49 proteins, Mtch2, a transporter located in the mitochondrial inner membrane, drew our immediate attention. First, the interaction between Cops5 and Mtch2 was validated by co-IP and Western blot (Fig. 6 A and B). Next, we showed that Mtch2 protein, but not its mRNA, decreases upon Cops5 KO (Fig. 6 C and D). The down-regulation of Mtch2 protein upon Cops5 KO is due to autophagic degradation, as the autophagy inhibitor 3-methyladenine (3-MA) restored the level of Mtch2 protein in Cops5 KO ESCs (Fig. 6E). We then constructed an iC5; C5 KO ESC line stably overexpressing Mtch2 (SI Appendix, Fig. S6A). Mtch2 overexpression rescues the growth defect of Cops5 KO ESCs, as well as reduced glycolysis, enhanced OXPHOS, and elevated ROS level caused by Cops5 KO (Fig. 6 F–I). More importantly, Cops5 KO no longer induces DNA damage accumulation when Mtch2 is overexpressed (Fig. 6J and SI Appendix, Fig. S6B). These data indicate that Mtch2 is a key downstream target of Cops5 in regulating cellular metabolism.
Fig. 6.
Cops5 regulates cellular metabolism through Mtch2. (A and B) The co-IP experiment to detect the interaction between Cops5 and Mtch2. (A) ESCs with or without stable Cops5-Flag expression were transfected with Mtch2-HA–expressing plasmid. Forty-eight hours after transfection, cells were harvested for the co-IP experiment using anti-Flag M2 magnetic beads. (B) Cell extracts from ESCs stably expressing Cops5-Flag and Mtch2-HA were used for the co-IP experiment, using IgG or anti-HA antibody, together with protein G beads. (C and D) Mtch2 protein (C) and mRNA (D) levels in iC5; C5 KO ESCs at various time points after Dox withdrawal. (E) iC5; C5 KO ESCs were cultured with or without Dox for 48 h and then treated with 5 mM 3-MA for the indicated time. Mtch2 protein levels in these cells were detected by Western blot. (F) Mtch2 overexpression rescues the growth defect of Cops5 KO ESCs. iC5; C5 KO and iC5; C5 KO; Mtch2 OE ESCs were cultured with or without Dox for two passages. Cell numbers were counted for every passage, and an equal amount of ESCs was plated onto tissue culture dishes. (G) ECAR of iC5; C5 KO and iC5; C5 KO; Mtch2 OE ESCs with or without Dox for 48 h. (H) OCR of iC5; C5 KO and iC5; C5 KO; Mtch2 OE ESCs with or without Dox for 48 h. (I) Measurement of ROS in iC5; C5 KO and iC5; C5 KO; Mtch2 OE ESCs with or without Dox for 48 h. (J) Immunofluorescence images of γH2AX in iC5; C5 KO and iC5; C5 KO; Mtch2 OE ESCs with or without Dox for 48 h. (Left) Representative immunofluorescence images of three independent experiments. Relative fluorescence intensities of γH2AX were quantified in ∼100 nuclei for each condition and plotted (Right). (Scale bar, 10 μm.) *P < 0.05; ***P < 0.001.
Discussion
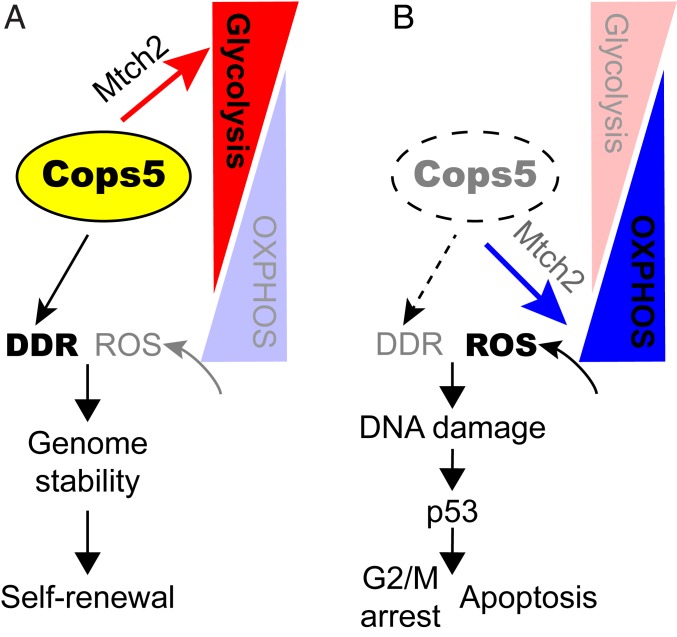
ESCs are able to proliferate infinitely under proper culture condition. For applications of ESCs and their derivatives, such as genetically modified animals and cell replacement therapy, it is necessary to maintain genomic stability of ESCs during the expansion phase. ESCs apply at least two strategies to secure exceptional genomic stability. First, the generation of endogenous DNA damage is minimized through preferentially utilizing glycolysis over OXPHOS to reduce the ROS level (15). Second, high DDR activities are maintained to remove DNA damage efficiently. Both high expression levels of DDR genes and ESC-specific factors, including Zscan4, Filia, and Sall4, contribute to high DDR activities in ESCs (8, 9, 11–13). In this study, we discovered that Cops5 is involved in both suppressing endogenous DNA damage by ROS and in maintaining high DDR activities to safeguard genome integrity of mouse ESCs. First, Cops5 ensures the biased energy production by the glycolysis pathway through maintaining the proper expression level of Mtch2. Consequently, the generation of ROS is suppressed, thus minimizing endogenous DNA damage caused by ROS. Second, Cops5 is required for high DDR activities, including NER, HR, and NHEJ, in ESCs. Thus, Cops5 contributes to genomic stability of ESCs through suppressing endogenous DNA damage and activating DDR activities simultaneously. Without Cops5, down-regulated Mtch2 rewires the cellular metabolism toward OXPHOS, resulting in elevated ROS and endogenous DNA damage. Enhanced endogenous DNA damage, together with impaired DDR activities, leads to accumulation of DNA damage in Cops5 KO ESCs, which in turn activates p53 and induces G2/M arrest, slow growth rate, and apoptosis (Fig. 7).
Fig. 7.
A working model for Cops5 to regulate genomic stability of ESCs. (A) Cops5 promotes cellular metabolism toward glycolysis through maintaining Mtch2 expression, thereby suppressing ROS production. In addition, high DDR activities, which are dependent on Cops5, together with low ROS, ensure the genomic stability and self-renewal of ESCs. (B) Without Cops5, a reduced level of Mtch2 rewires cellular metabolism to OXPHOS, leading to elevated ROS. Enhanced ROS level, as well as compromised DDR activities, cause DNA damage accumulation in ESCs, which in turn activates p53, G2/M arrest, and apoptosis.
We previously reported that Cops2 is essential for pluripotency maintenance through stabilizing Nanog protein and as a transcriptional corepressor (24). We failed to demonstrate the requirement of Cops5 in ESC self-renewal by short hairpin RNA knockdown, most likely due to low efficiency of Cops5 knockdown. Nevertheless, both Cops2 and Cops5 contribute to pluripotency maintenance independent of the CSN. Nanog protein are down-regulated upon Cops2 knockdown or Cops5 KO. However, the Homeobox domain of Nanog is required for Cops2 to promote Nanog protein stability (24), while Cops5 stabilizes Nanog protein through the N-terminal and C-terminal domains (SI Appendix, Fig. S1), suggesting that Cops2 and Cops5 regulate Nanog stability through distinct mechanisms. Cops8 KO ESCs provide further supporting evidences. Both Cops5 and Cops8 KO compromise the deneddylation activity of the CSN (SI Appendix, Fig. S3A). However, except for a slightly reduced growth rate, Cops8 KO does not affect pluripotency marker expression, cell-cycle progression, or DNA damage accumulation in ESCs (SI Appendix, Fig. S3 B–G). Moreover, Cops8 KO does not affect DDR activities, cellular ROS level, glycolysis, and OXPHOS activities (SI Appendix, Fig. S4 A–D). All these data suggest that CSN-independent function of Cops5 contributes to pluripotency maintenance and genomic stability of ESCs.
Our data show that Cops5 KO reprograms the cellular metabolism toward OXPHOS through down-regulating Mtch2. However, there are conflicting results regarding Mtch2 in metabolism regulation. Mtch2 KO enhances glycolytic flux and suppresses OXPHOS in ESCs (33). In contrast, loss of Mtch2 in hematopoietic stem cells (HSCs) and skeleton muscle cells promotes the metabolic switch from glycolysis to mitochondrial OXPHOS (34, 35). Our data are consistent with the metabolic regulatory function of Mtch2 in HSCs and skeleton muscle cells, despite that we studied Mtch2 in ESCs. Down-regulation of Mtch2 in our study and Mtch2 KO in the work by Bahat et al. (33) might account for the opposite effects in regulating metabolic switch. Consistent with this note, knockdown of Mtch2 with small interfering RNA in ESCs leads to down-regulation of glycolysis and elevation of OXPHOS (SI Appendix, Fig. S6 C–F). Alternatively, the difference in ESC culture conditions might contribute to the seemingly conflicting observation. We cultured ESCs in the presence of leukemia inhibitory factor (LIF) and serum, while Mtch2 KO ESCs were cultured in 2i/LIF and serum-free condition.
Cops5 KO down-regulates Mtch2 through autophagic degradation. Yet, the detailed mechanism of how Cops5 regulates autophagic degradation of Mtch2 remains unclear. Cops5 is a metalloprotease with deneddylation and deubiquitination activities (22). In addition to its well-known role in proteasomal degradation, ubiquitin also serves as a key signal for selective autophagy (36, 37). Thus, it is possible that Cops5 regulates the autophagy of Mtch2 by deubiquitinating Mtch2. In addition, it has been shown that Cops5 regulates autophagy via the ERN1 and mTOR pathways in goat endometrial epithelial cells (38). Given that Cops5 interacts with the mitochondrial protein Mtch2, the possibility is raised that Cops5 regulates mitophagy or, even more specifically, the autophagy of Mtch2. Interestingly, mitophagy plays an important role in metabolic switch and cell-fate change (39, 40). Thus, further investigation of how Cops5 regulates the autophagic degradation of Mtch2 might shed light on metabolic regulation and cell-fate determination of ESCs.
Materials and Methods
Cell Culture and Transfection.
D3 and KH2 ESCs were cultured in ESC medium which consists of 15% fetal bovine serum (FBS) (HyClone), 85% Dulbecco’s Modified Eagle Media (DMEM) (high-glucose DMEM, Gibco), 2 mM L-glutamine, 5,000 units/mL penicillin and streptomycin, 0.1 mM nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), and 1,000 units/mL LIF (Millipore).
To establish the iC5 ESC line, KH2 ESCs were transfected with pBS31-Cops5m and pCAGGS FLPe using lipofectamine 3000 (Invitrogen) (30). After 10-d of hygromycin selection, individual colonies were picked up and validated by genotyping. To induce the expression of exogenous Cops5 gene, 1 μM Dox was added into medium. To knockout Cops5 or Cops8, pX330 plasmids targeting Cops5 or Cops8 were transfected into ESCs using lipofectamine 3000. Two days after transfection, ESCs were plated down at low density to allow the formation of colonies from single cells. Five to seven days later, individual colonies were picked up and subjected to further culture and analysis.
GGR Assay.
GGR assay was performed as described elsewhere (41). Coverslips were placed into a 24-well plate and coated with gelatin in advance. ESCs (1 105) were plated into each well and cultured to 50% confluence. Cells were washed with phosphate-buffered saline (PBS) and irradiated with 5 J/m2 ultraviolet (UV) light for 5 s. Immediately after UV irradiation, cells were immediately incubated with 5-ethynyl-2′-deoxyuridine (final concentration 10 μM, Invitrogen, C10637) in serum-free DMEM for 4 h at 37 °C. After three washes with PBS, cells were fixed and permeabilized in buffer 1 (2% paraformaldehyde; 0.5% Triton-X; 0.3 M sucrose; diluted in PBS) on ice for 20 min, followed by three washes with 10% FBS in PBS for 5 min each. Cells were blocked with PBS containing 10% FBS for 30 min at room temperature. After aspirating PBS, 1 μL of 2.5 mM Alexa Fluor 488-azide and 100 μL buffer 2 (4 mM CuSO4; 10 mM sodium ascorbate; 50 mM Tris⋅HCl, pH 7.3) were mixed and added to the cells. After a 1-h incubation at room temperature, cells were washed three times with PBST (0.05% Tween 20) for 5 min each. Hoechst 33342 (5 μg/mL) diluted with PBS was added to the cells. After a 20-min incubation at room temperature, cells were fixed with 3.7% formaldehyde for 20 min. Images were captured by Zeiss Axio-Imager Z1 fluorescence microscope and analyzed by Image J software.
TCR Assay.
TCR assay was performed as described elsewhere (32). Cells (2.5 × 105) were plated into each well of a six-well plate, cultured with or without Dox. Simultaneously, 3 μg/mL Illudin S (Santa Cruz, SC-391575) was added to the medium for the experimental group. At passages 1 and 2, cells were harvested, and live cells were counted under a microscope after Trypan blue staining.
Measurement of NHEJ and HR.
The NHEJ and HR reporters were described elsewhere (42). The NHEJ or HR reporter plasmids were transfected into iC5; C5 KO ESCs, Cops8 KO, and WT ESCs, and stably integrated clones were established after puromycin selection. A 1-μg I-SceI–expressing plasmid was transfected into NHEJ or HR reporter ESC lines to induce a DSB. Forty-eight hours after transfection, cells were harvested for flow cytometry analysis.
Statistical Analysis.
All data were analyzed by Student’s t test. Statistically significant P values were indicated in figures as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Averages and SDs of at least three independent experiments are shown in figures when applicable.
Data Availability Statement.
All data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
L.C. was supported by the National Key R&D Program of China (Grants 2018YFC1313003 and 2018YFA0107002); the National Natural Science Foundation of China (Grants 31622038, 31671497, and 31871485); the Natural Science Foundation of Tianjin (Grant 18JCJQJC48400); the 111 Project Grant (Grant B08011); and the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915079117/-/DCSupplemental.
References
- 1.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeman M. K., Cimprich K. A., Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efroni S., et al. , Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdon T., Smith A., Savatier P., Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12, 432–438 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Cervantes R. B., Stringer J. R., Shao C., Tischfield J. A., Stambrook P. J., Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc. Natl. Acad. Sci. U.S.A. 99, 3586–3590 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X., Cui K., Yi Q., Yu L., Xu Y., DNA repair mechanisms in embryonic stem cells. Cell. Mol. Life Sci. 74, 487–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitale I., Manic G., De Maria R., Kroemer G., Galluzzi L., DNA damage in stem cells. Mol. Cell 66, 306–319 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Saretzki G., et al. , Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells 26, 455–464 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Maynard S., et al. , Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells 26, 2266–2274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tichy E. D., et al. , Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 19, 1699–1711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalzman M., et al. , Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B., et al. , Filia is an ESC-specific regulator of DNA damage response and safeguards genomic stability. Cell Stem Cell 16, 684–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J., et al. , Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J. Cell Biol. 208, 513–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saretzki G., Armstrong L., Leake A., Lako M., von Zglinicki T., Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells 22, 962–971 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Kondoh H., et al. , A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid. Redox Signal. 9, 293–299 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Shyh-Chang N., Daley G. Q., Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab. 21, 349–350 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Varum S., et al. , Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6, e20914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., et al. , UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30, 4860–4873 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyapina S., et al. , Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Wei N., Serino G., Deng X. W., The COP9 signalosome: More than a protease. Trends Biochem. Sci. 33, 592–600 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Chamovitz D. A., Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 10, 352–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman R., et al. , The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Chia N. Y., et al. , A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Zhang W., et al. , Cops2 promotes pluripotency maintenance by stabilizing Nanog protein and repressing transcription. Sci. Rep. 6, 26804 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P., Ding N., Zhang W., Chen L., COPS2 antagonizes OCT4 to accelerate the G2/M transition of mouse embryonic stem cells. Stem Cell Rep. 11, 317–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomoda K., Yoneda-Kato N., Fukumoto A., Yamanaka S., Kato J. Y., Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J. Biol. Chem. 279, 43013–43018 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Lykke-Andersen K., et al. , Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 23, 6790–6797 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menon S., et al. , COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat. Immunol. 8, 1236–1245 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Chen H., et al. , Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E5936–E5943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohozinski J., Agoulnik A. I., Boettger-Tong H. L., Bishop C. E., Successful targeting of mouse Y chromosome genes using a site-directed insertion vector. Genesis 32, 1–7 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Tian L., et al. , Essential roles of Jab1 in cell survival, spontaneous DNA damage and DNA repair. Oncogene 29, 6125–6137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Waard H., et al. , Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts. DNA Repair (Amst.) 7, 1659–1669 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Bahat A., et al. , MTCH2-mediated mitochondrial fusion drives exit from naïve pluripotency in embryonic stem cells. Nat. Commun. 9, 5132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzaglo-Azriel L., et al. , Loss of muscle MTCH2 increases whole-body energy utilization and protects from diet-induced obesity. Cell Rep. 14, 1602–1610 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Maryanovich M., et al. , An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat. Commun. 6, 7901 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Yamano K., Matsuda N., Tanaka K., The ubiquitin signal and autophagy: An orchestrated dance leading to mitochondrial degradation. EMBO Rep. 17, 300–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khaminets A., Behl C., Dikic I., Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26, 6–16 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Yang D., et al. , COPS5 negatively regulates goat endometrial function via the ERN1 and mTOR-autophagy pathways during early pregnancy. J. Cell. Physiol. 234, 18666–18678 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Naik P. P., Birbrair A., Bhutia S. K., Mitophagy-driven metabolic switch reprograms stem cell fate. Cell. Mol. Life Sci. 76, 27–43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez-Martin A., et al. , Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging (Albany N.Y.) 8, 1330–1352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limsirichaikul S., et al. , A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 37, e31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Z., Bozzella M., Seluanov A., Gorbunova V., Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and SI Appendix.