Significance
Electron transfer flavoproteins (ETFs: ETFA and ETFB) shuttle electrons between multiple flavoprotein dehydrogenases involved in fatty acid β-oxidation and one-carbon metabolism and the membrane-bound electron transfer flavoprotein-ubiquinone oxidoreductase. Defects in this system have been implicated in glutaric aciduria type II (GA-II), a rare autosomal recessive inherited metabolic disorder. The mechanism by which the protein levels of ETFA and ETFB are regulated at the posttranscriptional level remains unknown. This study demonstrates that neddylation, a ubiquitination-like pathway, targets and stabilizes ETFs by preventing their ubiquitination and degradation in hepatocytes. Consequently, hepatic neddylation prevents GA-II–like abnormalities in neonatal mice and delays fasting-induced mortality in adults. Our study adds insight into GA-II and establishes substrates for neddylation.
Keywords: neddylation, GA-II, ETFs, ubiquitination, steatosis
Abstract
Neddylation is a ubiquitination-like pathway that controls cell survival and proliferation by covalently conjugating NEDD8 to lysines in specific substrate proteins. However, the physiological role of neddylation in mammalian metabolism remains elusive, and no mitochondrial targets have been identified. Here, we report that mouse models with liver-specific deficiency of NEDD8 or ubiquitin-like modifier activating enzyme 3 (UBA3), the catalytic subunit of the NEDD8-activating enzyme, exhibit neonatal death with spontaneous fatty liver as well as hepatic cellular senescence. In particular, liver-specific UBA3 deficiency leads to systemic abnormalities similar to glutaric aciduria type II (GA-II), a rare autosomal recessive inherited fatty acid oxidation disorder resulting from defects in mitochondrial electron transfer flavoproteins (ETFs: ETFA and ETFB) or the corresponding ubiquinone oxidoreductase. Neddylation inhibition by various strategies results in decreased protein levels of ETFs in neonatal livers and embryonic hepatocytes. Hepatic neddylation also enhances ETF expression in adult mice and prevents fasting-induced steatosis and mortality. Interestingly, neddylation is active in hepatic mitochondria. ETFs are neddylation substrates, and neddylation stabilizes ETFs by inhibiting their ubiquitination and degradation. Moreover, certain mutations of ETFs found in GA-II patients hinder the neddylation of these substrates. Taken together, our results reveal substrates for neddylation and add insight into GA-II.
Electron transfer flavoproteins (ETFs: ETFA and ETFB) and electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO) are components of the electron transport chain in mitochondria. Together, ETFA and ETFB transfer electrons from dehydrogenation reactions of multiple flavoprotein dehydrogenases involved in fatty acid β-oxidation (FAO) and one-carbon metabolism to ETF-QO to reduce ubiquinone, which is essential for oxidative phosphorylation (1). Defects in this system leave the body unable to metabolize lipids for energy within the liver and muscles and lead to the accumulation of various intramitochondrial acyl-CoA esters. Secondarily, free acids and other conjugation products, including large amounts of the lysine metabolic intermediate glutaric acid, accumulate in the blood and urine, leading to a disease called glutaric aciduria type II (GA-II, Online Mendelian Inheritance in Man 231680), also known as multiple acyl-CoA dehydrogenation deficiency (2, 3). Clinical symptoms of GA-II include varied degrees of hypoketotic hypoglycemia, hyperammonemia, acidosis, fatty changes in the liver, accumulation of acylcarnitines of various chain lengths in the blood, and a characteristic urinary organic acid profile (2, 3). In most cases, Etfa/Etfb/Etf-qo mutations affecting the stability of the corresponding messenger RNAs (mRNAs) or proteins are the molecular basis of GA-II. However, in some patients, no mutation in the Etfa/Etfb/Etf-qo genes can be found (3, 4), and the underlying mechanism(s) remain elusive.
Ubiquitin-like protein NEDD8 is covalently attached to certain lysine(s) in a substrate protein, very similarly to ubiquitination (5, 6), and this process is termed neddylation. Neddylation is triggered by the successive action of NEDD8-activating enzyme E1 (NAE), NEDD8-conjugating enzyme E2 (usually Ubc12), and certain NEDD8-E3 ligase(s) (7–10). NAE is the heterodimer of the regulatory subunit amyloid precursor protein binding protein-1 (APPBP1) and the catalytic subunit ubiquitin-like modifier activating enzyme 3 (UBA3) (7, 8). The best-characterized substrates of neddylation are Cullins, which are essential components of Cullin-RING E3 ubiquitin-ligase complexes (CRLs) (5, 6). The neddylation of Cullins augments the activity of CRLs and thereby contributes to cell-cycle progression and cellular survival (7, 8). Therefore, neddylation inhibition has been proposed to be a potential therapeutic strategy for various malignancies (8–10).
The discovery of non-Cullin neddylation targets indicates that neddylation may occur in various cellular organelles and have diverse biological functions (9–14). For example, neddylation targets p53 to inhibit its transcriptional activity (11), targets membrane receptor TGF-βRII to promote its stability and endocytosis (12), and targets a subset of ribosomal proteins to promote their stability and correct localization (13, 14). Neddylation is also active within synapses and regulates the maturation, stability, and function of dendritic spines, at least partially by targeting the scaffolding protein PSD-95 (15). The characterization of neddylation substrates and cellular organelles involved remains the focus of this field.
Recently, neddylation has been implicated in metabolism. For example, NEDD8 is induced in preadipocytes undergoing differentiation (16), and a selective inhibitor of NAE, MLN4924, inhibits adipogenesis and lipid droplet formation (16, 17). MLN4924 also depletes intracellular nucleotide pools in acute myeloid leukemia cells (18). The role of neddylation in mitochondrial function seems to be cell-type dependent. For example, MLN4924 triggers oxidative stress and consequently promotes autophagy in various malignant cells (19–21), whereas it antagonizes hydrogen-peroxide–induced reactive oxygen species production in primary cerebellar granule neurons (22). For oxidative phosphorylation, MLN4924 induces mitochondrial fission-to-fusion conversion in breast cancer cells, which inhibits the tricarboxyilic acid cycle but promotes both basal and maximal oxidative phosphorylation (23). In contrast, MLN4924 suppresses basal but not maximal oxidative phosphorylation in protumoral hepatocytes (24). Although the role of neddylation in metabolism has been attributed to certain Cullin-dependent mechanism(s) in most cases, the neddylation of PPARγ has been demonstrated to be essential for adipogenesis (16). In addition, the neddylation of LKB1 and Akt has been detected under the conditions of co-overexpression with NEDD8 in hepatocytes (24). However, the physiological role of neddylation in mammalian metabolism remains elusive, and no mitochondrial targets have been reported.
As the liver is an essential metabolic organ, we aimed to explore the above issues with the strategy of liver-specific blockade of neddylation. Our data suggest that hepatic neddylation prevents GA-II–like abnormalities in neonatal mice and delays fasting-induced mortality in adults, at least partially by targeting and stabilizing the mitochondrial substrates ETFA and ETFB.
Results
Hepatic Neddylation Facilitates FAO in Neonatal Mice.
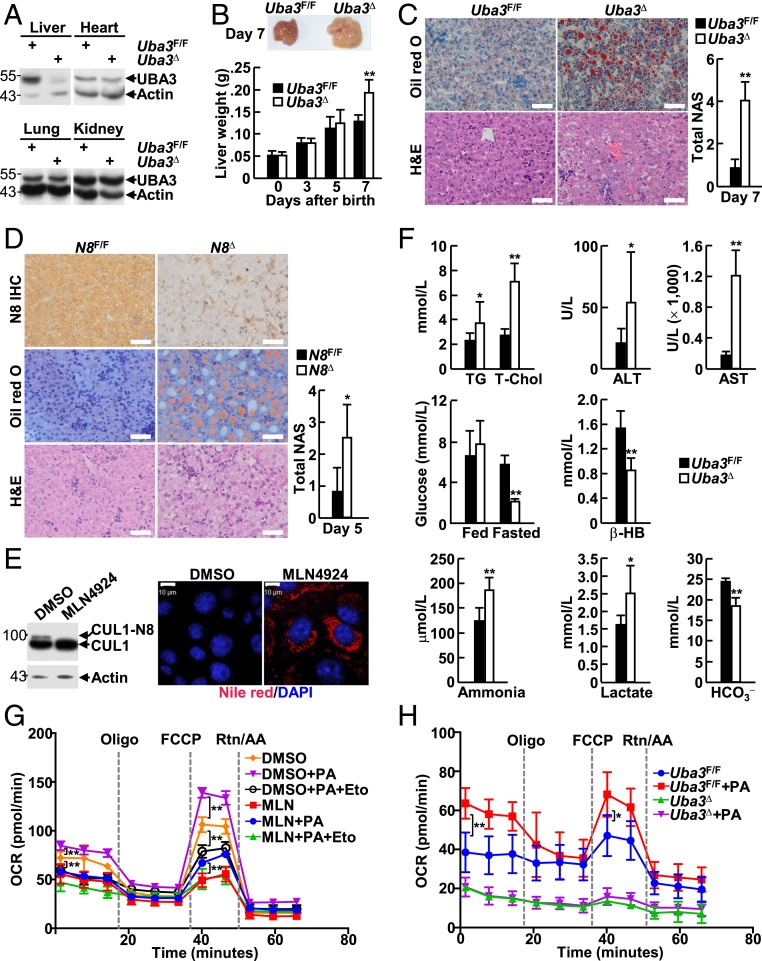
Crossing mice carrying the floxed Uba3 allele (25, 26) with mice expressing Cre recombinase under the control of the albumin promoter and enhancer (27) allowed us to generate a liver-specific UBA3 knockout mouse model (Alb-Cre:Uba3F/F, hereafter referred to as Uba3Δ). Immunoblotting (IB) analysis confirmed liver-specific UBA3 deficiency in Uba3Δ mice (Fig. 1A). Uba3Δ mice were born at Mendelian frequency but died within postnatal days 13 to 18 with hepatic cellular senescence (SI Appendix, Fig. S1), echoing the reported role of neddylation in cell-cycle progression (7, 8). A more careful examination revealed that Uba3Δ mice began to exhibit enlarged and discolored livers, which weighed more than those from their littermate controls (Uba3F/F) on postnatal day 7 (Fig. 1B), suggesting lipid accumulation. Indeed, Uba3Δ livers stained positive with the neutral lipid dye oil red O (Fig. 1C). Furthermore, ultrastructural analysis revealed an increased number of cytosolic lipid droplets and a lack of glycogen deposition in Uba3Δ livers (SI Appendix, Fig. S2). Histopathological evaluation using the nonalcoholic fatty liver disease activity score (NAS) (28) system suggested borderline or even definitive nonalcoholic steatohepatitis (Fig. 1C).
Fig. 1.
Hepatic neddylation facilitates FAO in neonatal mice. (A) IB analysis of UBA3 expression in the indicated tissues of postnatal-day-7 Uba3F/F and Uba3Δ mice. (B) The liver weights of neonatal Uba3F/F and Uba3Δ mice (n = 8 per group) were monitored from day 1 to day 7 (Bottom). A representative image of postnatal-day-7 livers is shown (Top). (C and D) Liver sections from postnatal-day-7 Uba3Δ mice (C) or postnatal-day-5 Nedd8Δ mice (D) and their littermates were subjected to IHC analysis (D), oil red O staining (C and D), and hematoxylin-eosin (H&E) staining (C and D), as indicated (Left). N8, NEDD8. The NAS was assessed (Right, n = 6 per group). (Scale bars, 50 μm.) (E) BNL CL.2 cells treated with or without 0.5 μM MLN4924 for 7 d were subjected to IB analysis with antibodies against CUL1 and β-actin (Left) or Nile red staining (Right). (Scale bar, 10 μm.) DAPI, 4′,6-diamidine-2-phenylindole. (F) The blood parameters of Uba3Δ mice and their littermates were measured. TG, triglycerides; T-Chol, total cholesterol; ALT, alanine aminotransferase; AST, aspartate transaminase; β-HB, β-hydroxybutyrate. n = 15 per group for ALT/AST and n = 9 per group for the other assays. (G and H) BNL CL.2 cells treated with or without 0.5 μM MLN4924 for 24 h (G) as well as UBA3-sufficient and UBA3-deficient neonatal primary hepatocytes (H) were subjected to the FAO assay. Eto, etomoxir; FCCP, fluorocarbonyl cyanide phenylhydrazone; Rtn, rotenone; AA, antimycin A; PA, palmitic acid. *P < 0.05; **P < 0.01.
To examine whether UBA3 has such effects via neddylation, we also generated a liver-specific conditional knockout mouse model for NEDD8 (Alb-Cre:Nedd8F/F, hereafter named Nedd8Δ). Immunohistochemistry (IHC) analysis confirmed NEDD8 deficiency in liver parenchymal cells (Fig. 1D). Nedd8Δ mice were born at Mendelian frequency but died within postnatal days 1 to 7 with hepatic cellular senescence (SI Appendix, Fig. S3). Nedd8Δ livers also stained positive with oil red O and showed higher NAS values (Fig. 1D). Consistent with these data, staining with the neutral lipid dye Nile red revealed that neddylation inhibition with MLN4924 induced lipid accumulation in BNL CL.2 murine embryonic hepatocytes (Fig. 1E).
Uba3Δ mice and their littermates were then subjected to clinical characterization for possible basic biochemical changes. Uba3Δ mice showed higher levels of serum triglycerides and total cholesterol (Fig. 1F). They also exhibited elevated levels of serum alanine aminotransferase and aspartate transaminase, markers of hepatocyte damage (Fig. 1F). On the other hand, Uba3Δ mice showed normal levels of blood glucose under the fed condition (Fig. 1F), suggesting that the mortality of Uba3Δ mice was not due to weaning. However, hypoglycemia was detected in Uba3Δ mice upon fasting for 4 h (Fig. 1F). Fasting hypoglycemia was associated with reduced levels of the serum ketone body β-hydroxybutyrate (Fig. 1F), suggesting impaired FAO (29). Consistently, hyperammonemia and accumulation of lactate were observed in fasted Uba3Δ mice (Fig. 1F). Furthermore, Uba3Δ mice exhibited mildly reduced levels of plasma bicarbonate (HCO3−) (Fig. 1F), an indicator of acidosis (30).
In this context, determination of the cellular oxygen consumption rate (OCR) was employed to evaluate FAO in real time with the use of etomoxir, an inhibitor of the FAO rate-limiting enzyme carnitine palmitoyltransferase-1 (31). As expected, neddylation inhibition by MLN4924 blocked the utilization of both endogenous and exogenous fatty acids by BNL CL.2 cells, impairing basal and maximal respiration (Fig. 1G). Consistent with this result, UBA3-deficient neonatal primary hepatocytes showed reduced basal and maximal respiration and lost the ability to utilize exogenous fatty acids (Fig. 1H).
Hepatic Neddylation Promotes FAO by Maintaining ETF Protein Levels in Neonatal Mice.
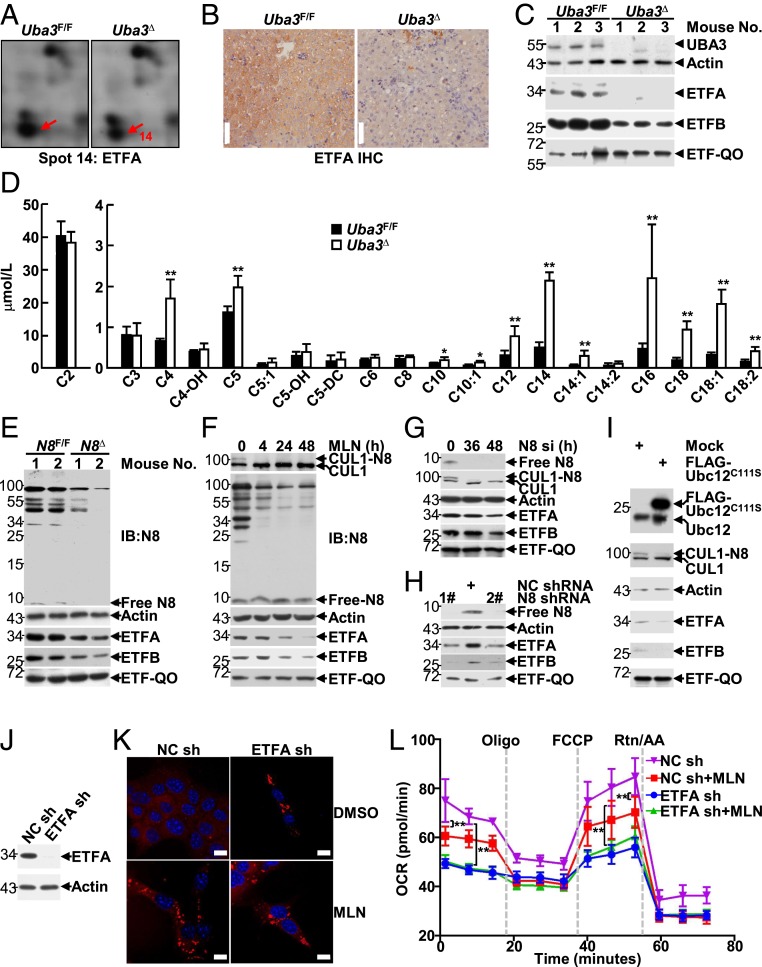
Our aforementioned data suggest that hepatic neddylation is essential for FAO in neonatal mice. To identify the key mitochondrial protein(s) involved, we compared the total liver proteomes of postnatal-day-7 Uba3Δ mice and their littermates (SI Appendix, Fig. S4). Among the 25 changed spots identified, ETFA expression was observed in spot 14, which was decreased in UBA3-deficient conditions in two independent experiments (Fig. 2A and SI Appendix, Table S1). IHC and IB analyses confirmed the reduction in ETFA expression in Uba3Δ livers (Fig. 2 B and C). Furthermore, the protein level of ETFB, but not that of ETF-QO, also decreased in Uba3Δ livers (Fig. 2C). Consistent with those findings, tandem mass spectrometry analysis revealed higher levels of serum acylcarnitines of various chain lengths in Uba3Δ mice than in their littermates (Fig. 2D and SI Appendix, Fig. S5). The metabolic aberrance in Uba3Δ mice, including the fatty changes in the liver, hypoketotic hypoglycemia, hyperammonemia, reduced plasma bicarbonate, and most importantly, the characteristic acylcarnitine profile, resembled GA-II (2, 3). Thus, the reduction in ETF expression should be a key factor contributing to metabolic aberrance in Uba3Δ mice.
Fig. 2.
Hepatic neddylation promotes FAO by maintaining ETF protein levels in neonatal mice. (A–C) Postnatal-day-7 Uba3F/F and Uba3Δ livers were subjected to two-dimensional (2D) electrophoresis mapping. Close-up sections of spot 14 on silver-stained 2D gels are shown (A). The same liver tissues were then subjected to IHC (B) and IB analyses (C) to examine ETF and ETF-QO expression. (D) The serum acylcarnitine levels in Uba3Δ mice and their littermates were measured by tandem mass spectrometry analysis. The blood of three mice was combined, and the experiment was repeated three times. (E) IB analysis of ETF and ETF-QO expression in liver tissues of postnatal-day-3 Nedd8F/F and Nedd8Δ mice. (F–I) At 0, 4, 24, or 48 h after treatment with 0.5 μM MLN4924 (F), at 0, 36, or 48 h after transfection with NEDD8 siRNA (G), at 96 h after infection with lentiviral vectors carrying the indicated shRNAs (H), or at 24 h after transfection with the indicated mammalian expression vectors (I), BNL CL.2 cells were subjected to IB analysis to examine ETF and ETF-QO expression. NC, nontargeting control. (J–L) At 96 h after infection with the indicated lentiviral vectors, a small portion of BNL CL.2 cells were subjected to IB analysis to confirm ETFA knockdown (J). Then the other cells were treated with or without 0.5 μM MLN4924 for 7 d (K) or 24 h (L), followed by Nile red staining (K) or the FAO assay (L). *P < 0.05; **P < 0.01. (Scale bars in K, 10 μm.)
The maintenance of ETFA and ETFB expression by neddylation was further confirmed in Nedd8Δ livers (Fig. 2E) and in BNL CL.2 cells with the following neddylation inhibition strategies: MLN4924 treatment (Fig. 2F), silencing endogenous NEDD8 by small interfering RNA (siRNA) (Fig. 2G) or short hairpin RNAs (shRNAs) (Fig. 2H), or overexpression of a dominant-negative mutant of Ubc12 (Ubc12C111S) (32) (Fig. 2I). Intriguingly, both silencing NEDD8 and MLN4924 treatment gradually decreased the protein levels of ETFA and ETFB in BNL CL.2 cells, but the decrease occurred much more slowly than that of Cullin1 (CUL1) neddylation (Fig. 2 F and G).
We then analyzed whether reduced ETF expression contributes to impaired FAO upon neddylation blockade. As expected, Nile red staining revealed that ETFA knockdown in BNL CL.2 cells (Fig. 2J) substantially increased the accumulation of lipid droplets, as did MLN4924 treatment (Fig. 2K). Furthermore, measurement of the OCR revealed that ETFA knockdown led to even lower basal and maximal respiration than MLN4924 treatment (Fig. 2L). The deleterious roles of MLN4924 treatment diminished upon ETFA knockdown (Fig. 2 K and L), suggesting that hepatic neddylation facilitates FAO in neonatal mice, at least partially through the maintenance of ETF protein levels.
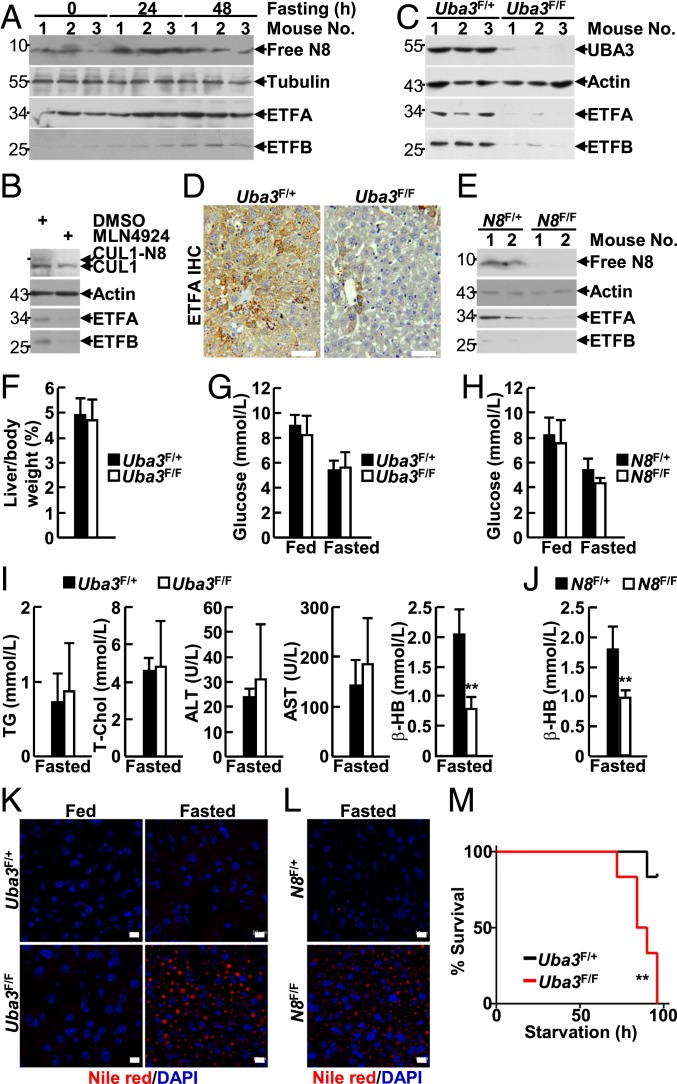
Hepatic Neddylation also Enhances ETF Protein Levels in Adult Mice and Prevents Fasting-Induced Steatosis and Mortality.
Phase I clinical trials have revealed that the safety profile of MLN4924 in adult patients with various malignancies is generally tolerable (33–38), suggesting that neddylation in adults is not as essential as it is in infants. Because hepatic FAO is critical for liver physiology during starvation, which causes the mobilization of lipids from peripheral depots into the liver (39, 40), we first examined the relationship between starvation and the neddylation/ETF axis in the liver. Adult male mice were used for this experiment because male mice are more sensitive to starvation than females (41). IB analysis revealed that a 24- to 48-h fast induced NEDD8, ETFA, and ETFB expression in the liver of 8-wk-old male mice (Fig. 3A). In this scenario, we tried to explore whether hepatic neddylation also contributes to ETF expression during adulthood. Indeed, treatment of adult primary hepatocytes with MLN4924 led to reduced protein levels of ETFA and ETFB (Fig. 3B). To confirm this result in vivo, recombinant adeno-associated virus DJ (AAV-DJ) expressing Cre recombinase driven by a strong synthetic CAG promoter was injected into the tail vein of 8-wk-old male Uba3F/F and littermate Uba3F/+ mice. AAV-DJ transduction is highly enriched in the liver, and transgene expression persists for at least 120 d (42–44). Six weeks after the injection, IB analysis confirmed UBA3 deficiency in the liver, but not in the heart or kidneys, of Uba3F/F mice (Fig. 3C and SI Appendix, Fig. S6). As expected, the protein levels of ETFA and ETFB significantly decreased in the absence of UBA3 (Fig. 3 C and D). Reduced ETF expression was also observed in AAV-DJ-Cre–transduced adult Nedd8F/F livers (Fig. 3E). Because AAV-DJ-Cre–transduced Uba3F/F mice looked normal and showed body and liver weights comparable to those of their littermate controls during our 6-mo observation period (Fig. 3F), AAV-DJ-Cre–transduced mice were subjected to a 48-h fast. Surprisingly, AAV-DJ-Cre–transduced Uba3F/F and Nedd8F/F mice exhibited blood glucose levels comparable to those of their littermate controls before and after 48 h of starvation (Fig. 3 G and H). AAV-DJ-Cre–transduced Uba3F/F mice and their littermate controls also showed comparable serum triglycerides, total cholesterol, alanine aminotransferase, and aspartate transaminase levels after 48 h of starvation (Fig. 3I). Nevertheless, AAV-DJ-Cre–transduced Uba3F/F and Nedd8F/F mice exhibited reduced levels of the serum ketone body β-hydroxybutyrate (Fig. 3 I and J) and significantly increased lipid storage in the liver after 48 h of starvation (Fig. 3 K and L). Thus, hepatic neddylation also contributes to FAO in adult mice. Consequently, all AAV-DJ-Cre–transduced Uba3F/F mice succumbed to a 96-h fast, whereas most littermate controls survived under the same conditions (Fig. 3M). Moreover, AAV-DJ-Cre–transduced Uba3F/F mice showed undetectable blood glucose just before their death. Together, our data suggest that the neddylation/ETF axis in adult murine livers prevents fasting-induced steatosis and mortality.
Fig. 3.
Hepatic neddylation also enhances ETF protein levels in adult mice and prevents fasting-induced steatosis and mortality. (A) IB analysis of NEDD8 and ETF expression in liver tissues of 8-wk-old male mice fasted for 0, 24, or 48 h. (B) IB analysis of ETF expression in adult primary hepatocytes treated with or without 2 μM MLN4924 for 24 h. (C–M) Recombinant AAV-DJ expressing Cre recombinase driven by a strong synthetic CAG promoter was injected into the tail vein of 8-wk-old male Uba3F/F or Nedd8F/F mice and their heterozygous littermates. Six weeks later, the mice were subjected to the following assays: IB (C and E) and IHC (D) analysis of ETF expression in liver tissues of the fed mice; liver weights relative to body weights of the fed mice (F, n = 6 per group); blood parameters before and after a 48-h fast (G–J, n = 6 per group); Nile red staining of liver tissues before and after a 48-h fast (K and L); and survival curves during a 96-h fast (M, n = 6 per group). **P < 0.01. (Scale bars, 50 μm in D; 10 μm in K and L.)
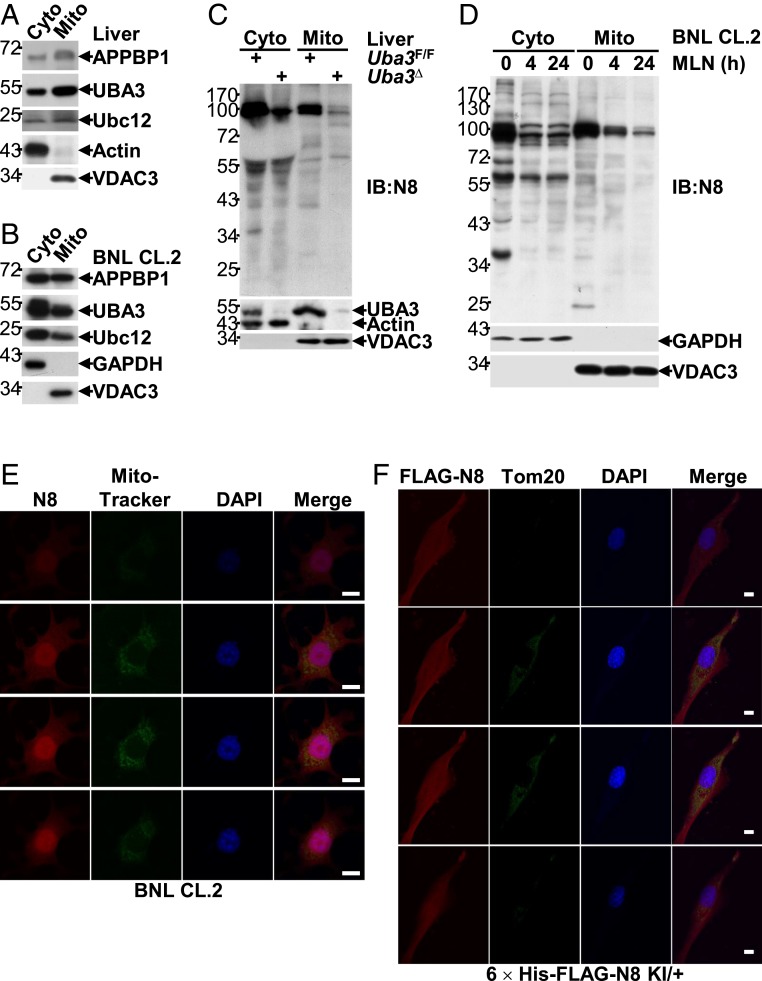
Neddylation Is Active in Hepatic Mitochondria.
Our aforementioned data suggest that the maintenance of ETF protein levels by neddylation in the liver plays pivotal roles in both neonatal and adult mice. Therefore, it was important to investigate the underlying mechanism(s). After verifying the Etfa/Etfb primers used for quantitative real-time PCR (qRT-PCR) analysis (SI Appendix, Fig. S7), we first analyzed whether neddylation maintains the mRNA levels of Etfa and Etfb. As shown in SI Appendix, Fig. S8 A and B, Uba3Δ and Nedd8Δ livers exhibited comparable mRNA levels of Etfa and Etfb to those of their littermates. Because the transcriptional landscape in Uba3Δ and Nedd8Δ livers might adapt to a knockout condition over time, it was absolutely necessary to also assess the time course of these transcripts upon neddylation blockade. For this purpose, BNL CL.2 cells were transfected with NEDD8 siRNA or treated with MLN4924 for various periods of time. qRT-PCR analysis revealed no significant reduction in the mRNA levels of Etfa and Etfb (SI Appendix, Fig. S8 C and D) throughout the entire time course observed.
In this scenario, we tried to explore whether neddylation is active in hepatic mitochondria. Purification of mitochondria from neonatal liver tissues followed by IB analysis demonstrated that the key components of the neddylation system, APPBP1, UBA3, and Ubc12, were present in both mitochondria and the cytosol (Fig. 4A). Similarly, APPBP1, UBA3, and Ubc12 were also present in mitochondrial proteins purified from BNL CL.2 cells (Fig. 4B). Because the anti-NEDD8 antibody that we used in IB analysis showed good specificity for recognizing neddylated proteins as well as free NEDD8 (Fig. 2 E and F), we further employed this antibody to probe possible neddylated proteins in mitochondria. Indeed, the anti-NEDD8 antibody recognized both cytosolic and mitochondrial proteins of neonatal liver tissues, as multiple bands were observed, and most of the band intensities decreased in UBA3-deficient conditions (Fig. 4C). Similarly, the anti-NEDD8 antibody recognized both cytosolic and mitochondrial proteins of BNL CL.2 cells, as multiple bands were once again observed (Fig. 4D). Intriguingly, the intensities of most cytosolic bands rapidly decreased within 4 h of MLN4924 treatment, whereas those of many mitochondrial bands became diminished only upon prolonged treatment (Fig. 4D). The above data suggest that mitochondrial proteins also undergo neddylation.
Fig. 4.
Neddylation is active in hepatic mitochondria. (A–D) IB analysis of the neddylation system in cytosolic and mitochondrial proteins isolated from liver tissues of postnatal-day-7 mice (A and C) or BNL CL.2 cells treated with 0.5 μM MLN4924 for 0, 4, or 24 h (B and D). VDAC is a mitochondrial marker, whereas β-actin or GAPDH is a cytosolic marker. (E) After incubation with MitoTracker Green (0.5 μg/mL) for 30 min, BNL CL.2 cells were subjected to indirect IF analysis with an anti-NEDD8 antibody. A Z-stack analysis is shown. (Scale bars, 10 μm.) (F) Primary hepatocytes from 8-wk-old 6 × His-FLAG-NEDD8 heterozygous KI mice were subjected to IF analysis with antibodies against FLAG-tag and Tom20. A Z-stack analysis is shown. (Scale bars, 10 μm.)
Because purified mitochondria may be contaminated with other subcellular fractions, we also employed an alternative strategy. Indirect immunofluorescence (IF) analysis with an anti-NEDD8 antibody and MitoTracker Green demonstrated significant colocalization of NEDD8 with mitochondria in BNL CL.2 cells (Fig. 4E). This anti-NEDD8 antibody was specific because the signal diminished upon NEDD8 knockdown (SI Appendix, Fig. S9). To further verify the colocalization of neddylation with mitochondria, a 6 × His-FLAG-NEDD8 constitutive knock-in (KI) mouse model was generated (SI Appendix, Fig. S10A). PCR analysis confirmed the constitutive presence of the KI allele in genomic DNA (SI Appendix, Fig. S10B). IB and IF analyses revealed that primary hepatocytes isolated from 8-wk-old heterozygous KI mice exhibited similar global neddylation levels but reduced free NEDD8 levels compared to those of littermate wild-type (WT) controls (SI Appendix, Fig. S10 C and D). Histidine pulldown confirmed the presence of His-tagged NEDD8, and IF analysis with an anti-FLAG antibody confirmed the presence of FLAG-tagged NEDD8, in primary hepatocytes from heterozygous KI mice. Thus, 6 × His-FLAG-NEDD8 should behave like endogenous NEDD8. Importantly, IF analysis with antibodies against FLAG-tag and Tom20 demonstrated good colocalization of 6 × His-FLAG-NEDD8 with hepatic mitochondria (Fig. 4F). These data further confirm the notion that mitochondrial proteins also undergo neddylation.
ETFA and ETFB Are Neddylation Substrates.
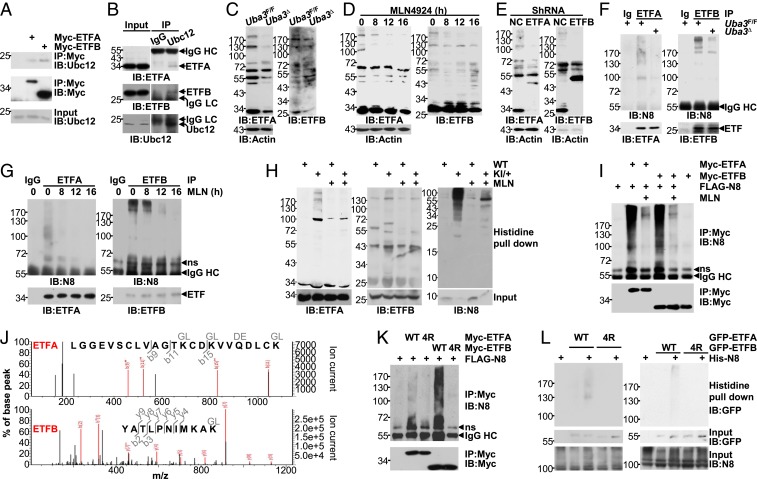
To explore whether ETF proteins are novel neddylation substrates, their possible interaction with Ubc12 was first confirmed by coimmunoprecipitation (Fig. 5 A and B). Then, we attempted to analyze the possible ETF neddylation by blotting an entire molecular-weight spectrum with protein lysates from neonatal liver tissues and BNL CL.2 cells. As expected, some high-molecular-weight smear bands were detected by antibodies against ETFA and ETFB after a long exposure, and their intensities diminished upon UBA3 deficiency (Fig. 5C), MLN4924 treatment (Fig. 5D), or ETF knockdown (Fig. 5E). Intriguingly, the reduction in the intensities of the smear bands preceded the reduction in free ETF protein expression despite that the smear bands were much less intense than the free ETF protein bands (Fig. 5D). Immunoprecipitation (IP) under partially denaturing conditions demonstrated that both anti-ETFA and anti-ETFB precipitates were detectable as smear bands by the anti-NEDD8 antibody, and the smear band intensities decreased upon UBA3 deficiency (Fig. 5F) or MLN4924 treatment (Fig. 5G). As shown in Fig. 5 D and G, the kinetics of the reduction in smear band intensities detected with the two methods was well correlated with each other. To confirm the covalent NEDD8 modification of endogenous ETFs, we employed adult primary hepatocytes from 6 × His-FLAG-NEDD8 heterozygous KI mice. As expected, histidine pulldown clearly demonstrated that ETFA and ETFB were neddylated and MLN4924 treatment abrogated the covalent modification (Fig. 5H). Thus, ETFA and ETFB are neddylation targets.
Fig. 5.
ETFA and ETFB are neddylation substrates. (A) The interaction between Myc-tagged ETFs and endogenous Ubc12 was analyzed by IB after IP with an anti-Myc antibody. (B) The interaction between endogenous ETFs and endogenous Ubc12 in postnatal-day-7 liver tissues was analyzed by IB after IP with an anti-Ubc12 antibody or a control IgG antibody. HC, heavy chain; LC, light chain. (C–E) IB analysis of an entire molecular weight spectrum of ETF expression with whole-cell lysates harvested from postnatal-day-7 liver tissues (C) or BNL CL.2 cells at 0, 8, 12, or 16 h after treatment with 0.5 μM MLN4924 (D) or 96 h after infection with the indicated lentiviral vectors (E). ShRNA, short hairpin RNA. (F and G) The neddylation of endogenous ETF proteins in postnatal-day-7 liver tissues (F) or BNL CL.2 cells treated with 0.5 μM MLN4924 for 0, 8, 12, or 16 h (G) was examined by IB after IP under partially denaturing conditions. ns, nonspecific. (H) Primary hepatocytes from 8-wk-old 6 × His-FLAG-NEDD8 heterozygous KI and littermate WT mice were treated with or without 2 μM MLN4924 for 24 h, and the neddylation of endogenous ETF proteins was then analyzed by histidine pulldown. Please note that in F–H, the amount of lysate in each sample was adjusted to ensure comparable protein levels of ETFA and ETFB. (I) Twenty-four hours after transfection with 2 μg FLAG-NEDD8 and varying amounts of Myc-ETF expression constructs, cells of hepatic origin in 60-mm dishes were treated with or without 0.5 μM MLN4924 for another 24 h. The neddylation of exogenous ETF proteins was then examined by IB after IP under partially denaturing conditions. (J) The smear bands obtained in SI Appendix, Fig. S11, were subjected to mass spectrometry analysis. Possibly modified peptides are shown. (K) Possibly neddylated lysines identified by mass spectrometry analysis and adjacent evolutionarily conserved lysines were mutated to arginines. The neddylation of Myc-tagged ETFs and the corresponding 4R mutants was then examined as described in I. (L) Forty-eight hours after BNL CL.2 cells were transfected with 0.5 μg His-NEDD8 and varying amounts of GFP-ETF expression constructs in 60-mm dishes, the neddylation of exogenous ETF proteins with or without the 4R mutation was then examined by histidine pulldown. Please note that in I–L the amounts of tagged ETFA or ETFB plasmid in each sample were adjusted to ensure comparable protein levels.
As neddylation occurs on specific lysines, we next tried to identify the potential neddylation sites on ETFA and ETFB by co-overexpressing Myc-tagged ETFA or ETFB with NEDD8. IP under partially denaturing conditions demonstrated that anti-Myc precipitates were detected as strong smear bands by the anti-NEDD8 antibody (Fig. 5I). Artificial conjugation of overexpressed NEDD8 reportedly depends on the ubiquitin E1 enzyme but not on NAE (45). Because the smear bands were eliminated by MLN4924, the modification of exogenous ETFA and ETFB was not artificial (Fig. 5I). Mass spectrometry analysis of the smear bands obtained with this strategy (SI Appendix, Fig. S11) revealed Lys59, Lys62, and Lys69 of ETFA and Lys202 of ETFB as potential modification sites (Fig. 5J). However, mutation of these lysines to arginines had only partial effects on ETF neddylation (SI Appendix, Fig. S12). Lysines that are near each other might play redundant roles as neddylation sites for certain NEDD8 substrates (9–11). Indeed, the evolutionarily conserved Lys75 is adjacent to Lys59, Lys62, and Lys69 of ETFA, and three evolutionarily conserved lysines, Lys200, Lys203, and Lys205, are adjacent to Lys202 of ETFB. The four lysines of ETFA are localized in the surface-exposed ETF domain, and the four lysines of ETFB are close to the surface-exposed recognition loop, which is responsible for the interaction with dehydrogenases (46). Mutation of these four lysines together (named 4R) significantly dampened the modification of Myc-tagged ETF proteins under the conditions of co-overexpression with NEDD8, as revealed by IB after IP under partially denaturing conditions (Fig. 5K). To confirm the neddylation sites, we coexpressed GFP-tagged ETFA or ETFB with His-NEDD8 at near-endogenous NEDD8 levels. Histidine pulldown clearly demonstrated that GFP-tagged ETF proteins were neddylated, and the 4R mutation abolished the covalent modification (Fig. 5L).
Neddylation of ETF Proteins Antagonizes Their Ubiquitination and Degradation.
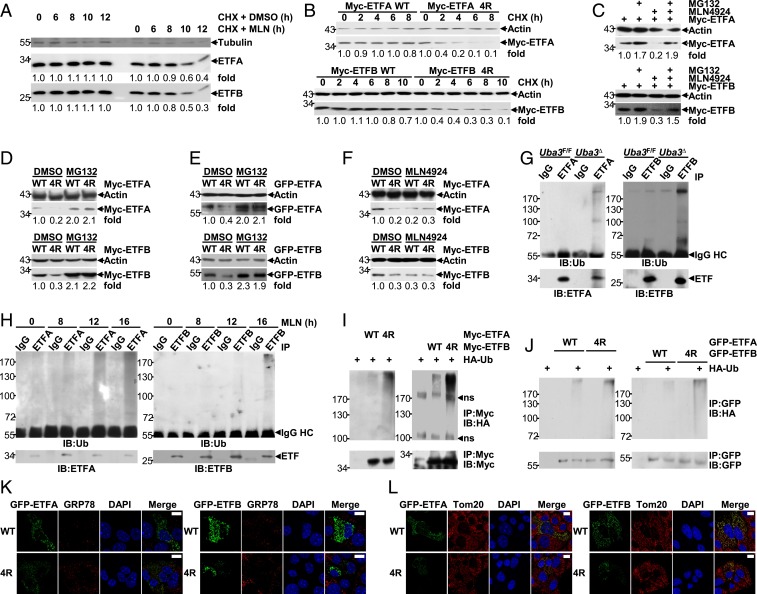
We next investigated whether neddylation could affect the stability of ETF proteins. For this purpose, we employed cycloheximide (CHX), which blocks de novo protein synthesis (47). As shown in Fig. 6A, a chase assay with CHX and MLN4924 shortened the half-lives of endogenous ETF proteins in BNL CL.2 cells. Consistently, ETF protein mutants defective in neddylation had a faster turnover than their WT counterparts (Fig. 6B). Because neddylation usually affects the stability of substrate proteins through the ubiquitin-proteasome system (13–15, 24), we tested whether neddylation inhibition promotes the degradation of ETF proteins in a proteasome-dependent manner. The proteasome inhibitor MG132 prevented the down-regulation of ETF proteins upon MLN4924 treatment (Fig. 6C). The protein levels of ETFA 4R and ETFB 4R were always lower than those of their WT counterparts when the same amount of plasmid was used (Fig. 6 D–F). However, the differences disappeared upon MG132 treatment (Fig. 6 D and E) or MLN4924 treatment (Fig. 6F). Together, these data suggest that ETF neddylation prevents their ubiquitin-proteasome–dependent degradation.
Fig. 6.
Neddylation of ETF proteins antagonizes their ubiquitination and degradation. (A) BNL CL.2 cells were treated with 10 μg/mL CHX in the presence or absence of 0.5 μM MLN4924 for various periods of time, as indicated. Then, the half-lives of endogenous ETF proteins were analyzed by IB. (B–F) Twenty-four hours after transfection with the same (C–F) or adjusted (B) amounts of the indicated plasmids, cells of hepatic origin were treated with 10 μg/mL CHX for various periods of time, as indicated (B) or treated with MG132 (20 μM, 6 h) and/or MLN4924 (0.5 μM, 24 h). (C–F). The expression of Myc-tagged (B–D and F) or GFP-tagged ETF proteins (E) was analyzed by IB. Please note that in A–F quantification is shown as numbers under each image, which were determined using ImageJ. (G and H) The ubiquitination of endogenous ETF proteins in postnatal-day-7 liver tissues (G) or BNL CL.2 cells treated with 0.5 μM MLN4924 for 0, 8, 12, or 16 h (H) was examined by IB after IP under partially denaturing conditions. Ub, ubiquitin. BNL CL.2 cells were treated with 20 μM MG132 for 6 h before harvest. (I and J) Twenty-four hours after transfection with 2 μg HA-ubiquitin and varying amounts of tagged ETF expression constructs, cells of hepatic origin in 60-mm dishes were treated with 20 μM MG132 for 6 h. The ubiquitination of exogenous ETF proteins with or without the 4R mutation was then examined by IB after IP under partially denaturing conditions. (K and L) Twenty-four hours after BNL CL.2 cells were transfected with GFP-tagged ETF expression constructs with or without the 4R mutation, the possible impact on ER stress (K) and the possible colocalization of these exogenous proteins with mitochondria (L) were examined by IF analysis with an antibody against GRP78 (K) or Tom20 (L). (Scale bars, 10 μm.)
In this scenario, we aimed to explore whether ETFA and ETFB undergo ubiquitination and how neddylation might affect this modification by performing IP under partially denaturing conditions. We found that the ubiquitination of endogenous ETFA and ETFB in neonatal liver tissues was augmented in UBA3-deficient conditions (Fig. 6G). Furthermore, MLN4924 treatment gradually enhanced the ubiquitination of endogenous ETF proteins in BNL CL.2 cells (Fig. 6H), which was inversely correlated with the reduction in neddylation (Fig. 5G) and preceded the reduction in free ETF protein expression (Figs. 2F and 5D). Consistent with these data, ubiquitination of either Myc-tagged or GFP-tagged ETF proteins occurred upon co-overexpression with ubiquitin (Fig. 6 I and J). The ubiquitination levels of neddylation-defective ETF protein mutants were higher than those of their WT counterparts (Fig. 6 I and J). It is possible that the 4R mutants were only improperly folded and degraded through a quality control pathway. However, the expression of GFP-ETFA 4R or GFP-ETFB 4R in BNL CL.2 cells did not result in up-regulation of GRP78 (Fig. 6K), a key indicator of the unfolded protein response (48), under the conditions that GRP78 was significantly induced by the endoplasmic reticulum (ER) stress-inducer Brefeldin A (49) (SI Appendix, Fig. S13). Moreover, these mutants demonstrated good colocalization with mitochondria, like their WT counterparts (Fig. 6L). Therefore, these data indicate that the neddylation of ETFA proteins could antagonize their ubiquitination and degradation.
Certain Mutations of ETFA and ETFB Found in GA-II Patients Hinder the Neddylation of These Substrates.
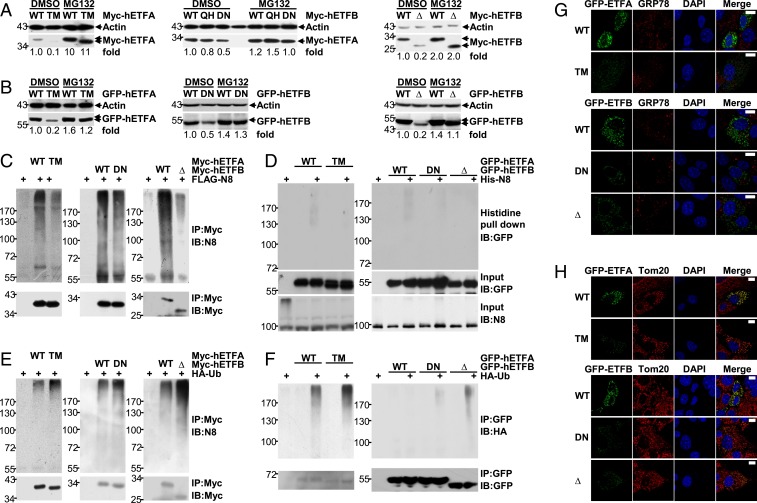
Several mutations of ETFA and ETFB, which either affect ETF activity or result in reduced ETF protein levels, have been found in GA-II patients (3, 4). It is possible that some mutations affect the neddylation of ETF proteins, thereby leading to augmented ubiquitin-proteasome–dependent degradation. To test this idea, we evaluated whether the reduced expression of ETF proteins with reported mutations could be reversed by MG132. As shown in Fig. 7 A and B, the reduced expression of either Myc-tagged or GFP-tagged ETFA T266M, ETFB Δ73–125, and ETFB D128N, reported mutations found in some GA-II patients (3, 4), was fully reversed upon MG132 treatment. Consistent with these findings, these mutations dampened the neddylation of Myc-tagged ETF proteins under the conditions of co-overexpression with NEDD8, as revealed by IB after IP under partially denaturing conditions (Fig. 7C). To confirm this effect, we coexpressed GFP-tagged ETFA or ETFB with His-NEDD8 at near-endogenous NEDD8 levels. Histidine pulldown clearly demonstrated that these mutations indeed hindered the covalent NEDD8 modification of GFP-tagged ETF proteins (Fig. 7D). Furthermore, these mutations augmented the ubiquitination of either Myc-tagged or GFP-tagged ETF proteins under the conditions of co-overexpression with ubiquitin, as revealed by IB after IP under partially denaturing conditions (Fig. 7 E and F). The expression of GFP-tagged ETF proteins with these mutations in BNL CL.2 cells did not result in up-regulation of GRP78 (Fig. 7G). Moreover, these mutants demonstrated good colocalization with mitochondria, like their WT counterparts (Fig. 7H). Thus, defective neddylation resulting from certain mutations in the Etf genes contributes to the pathogenesis of GA-II.
Fig. 7.
Certain mutations of ETFA and ETFB found in GA-II patients hinder the neddylation of these substrates. (A and B) IB analysis of the effects of MG132 on the expression of Myc-tagged (A) or GFP-tagged ETF proteins (B) with or without the indicated mutation found in certain GA-II patients. hETFA, human ETFA; hETFB, human ETFB; TM, T266M; QH, Q125H; DN, D128N; Δ, Δ73–125. Quantification is shown as numbers under each image, determined using ImageJ. (C and D) The neddylation of Myc-tagged (C) or GFP-tagged ETF proteins (D) with or without the indicated mutation was examined as described in Fig. 5I (C) or Fig. 5L (D), respectively. (E and F) The ubiquitination of Myc-tagged (E) or GFP-tagged ETF proteins (F) with or without the indicated mutation was examined as described in Fig. 6I. (G and H) Twenty-four hours after BNL CL.2 cells were transfected with GFP-tagged ETF expression constructs with or without the indicated mutation, the possible impact on ER stress (G) and the possible colocalization of these exogenous proteins with mitochondria (H) were examined by IF analysis with an antibody against GRP78 (G) or Tom20 (H). (Scale bars, 10 μm.)
Discussion
To date, little is known about the role of neddylation in postnatal mammalian physiology. In this work, we show that liver-specific blockade of neddylation leads to neonatal death with defective hepatic FAO. During the neonatal period, breast milk is the sole food, which underscores the importance of hepatic FAO. Our work indicates that neddylation facilitates the utilization of both endogenous and exogenous fatty acids by hepatocytes. Consequently, both Uba3Δ and Nedd8Δ mice showed spontaneous fatty liver. Moreover, Uba3Δ mice exhibited systemic abnormalities similar to GA-II, a rare autosomal recessive inherited metabolic disorder (2, 3). Thus, the liver is a central target organ for GA-II during the neonatal period. However, it should be noted that the blood parameters are not a very good explanation of neonatal death. Apparently, the impaired hepatic FAO is not the primary reason for the neonatal death even though it might accelerate its occurrence. As embryonic-day-17.5 Nedd8F/F and Nedd8Δ livers exhibited comparable percentages and numbers of hematopoietic stem cells and cells of different lineages (SI Appendix, Fig. S14), Nedd8Δ mice did not likely die from defective hematopoiesis (50) either. In contrast, hepatic cellular senescence was observed in both Uba3Δ and Nedd8Δ mice, which might be the major cause of neonatal death.
GA-II results from mitochondrial ETF/ETF-QO defects (1–4). Indeed, the protein levels of ETFA and ETFB were decreased upon neddylation blockade in neonatal livers and BNL CL.2 cells, while that of ETF-QO remained unchanged. Because the deleterious effects of MLN4924 on lipid accumulation and the OCR in BNL CL.2 cells diminished upon ETFA knockdown, it is reasonable to propose that hepatic neddylation facilitates FAO in neonatal mice, at least partially through the maintenance of ETF protein levels. The neddylation/ETF axis is also active in the adult murine liver. However, our data indicate that this axis is far less important in adult mice than in neonatal mice. In line with these observations, the use of MLN4924 in preclinical studies and phase I clinical trials for the treatment of various malignancies has been reported to be safe (8, 18–20, 33–38). We also found that adult mice with a liver neddylation blockade maintained their blood glucose levels even after a 48-h fast, consistent with a previous report that liver-specific knockout of carnitine palmitoyltransferase-2 (CPT-2), a key enzyme involved in long-chain FAO, does not affect blood glucose in adult mice during a 24-h fast (51). It has been speculated that loss of hepatic FAO during adulthood is compensated by the kidneys, muscles, and adipose tissues to maintain blood glucose (51). Nevertheless, reduced serum ketones and steatosis were observed in adult mice with a liver neddylation blockade as well as in adult mice with liver-specific CPT-2 deficiency (51). Moreover, a 96-h fast was lethal to adult mice with a liver neddylation blockade, and similarly, a ketogenic diet was shown to be lethal to adult mice with liver-specific CPT-2 deficiency (51). Therefore, patients using MLN4924 should avoid prolonged fasts or a ketogenic diet.
Interestingly, neddylation is active in hepatic mitochondria. Specifically, we identified the neddylation of ETFA and ETFB. Our study shows that these mitochondrial proteins function as neddylation substrates. Because coexpression of NEDD8 with lysine-containing target proteins may lead to artifactual conjugation with NEDD8 (45), we employed the 6 × His-FLAG-NEDD8 constitutive KI mouse model. We tried to obtain homozygous KI mice but failed (KI/+ × KI/+ yielded 55 KI/+, 22 WT, and 0 KI/KI offspring). Because free 6 × His-FLAG-NEDD8 was not detected in primary hepatocytes from homozygous KI mice by regular IB analysis under the same conditions that free untagged NEDD8 was detected in primary hepatocytes from their WT littermates, the expression efficiency of 6 × His-FLAG-NEDD8 was apparently lower than that of untagged NEDD8. Therefore, it is possible that homozygous KI embryos cannot maintain sufficient global neddylation and consequently die before birth despite the fact that heterozygous KI mice exhibit global neddylation levels comparable to those of their WT littermates. Nevertheless, we conducted histidine pulldown with heterozygous KI mice to confirm the neddylation of ETFA and ETFB.
Neddylation controls the stability, subcellular localization, or activity of its substrates (9–17, 23–26). For ETF proteins, neddylation prevents their ubiquitination and subsequent degradation. Theoretically, most EFTA and ETFB should be neddylated to truly prevent ubiquitination. However, substantially lower levels of neddylated ETF proteins were detected by IB analysis under physiological conditions, as demonstrated by the high-molecular-weight smear bands, compared with those of free ETF proteins. Similar phenomena were also observed for other reported substrates conjugated with poly-NEDD8 (11, 16, 24, 52) (SI Appendix, Fig. S15). Reasons for this discrepancy include that the conjugation with poly-NEDD8 might hinder the transfer efficiency or the binding of the antibody to the corresponding substrate, the neddylation might be removed during sample preparation, or conjugation with poly-NEDD8 might make the molecular weight beyond the detection scope. Future studies are required to address these issues. On the other hand, the deneddylation of ETFs is much slower than that of Cullins. In our hands, the neddylation of many mitochondrial proteins became diminished only upon prolonged neddylation inhibition. Thus, it is possible that some special deneddylation enzyme works in mitochondria. Nevertheless, the significant reduction in ETF protein expression due to defective neddylation might help explain the pathogenic mechanisms underlying some GA-II patients with no mutation in the Etf/Etf-qo genes. Furthermore, we have demonstrated that certain mutations of ETFA and ETFB found in GA-II patients hinder the neddylation of these substrates. Consequently, these mutants undergo enhanced ubiquitination and degradation. These findings suggest a possible therapeutic intervention.
The fact that the anti-NEDD8 antibody that we used detected numerous bands in mitochondrial lysates suggests that other mitochondrial neddylation targets might be involved in metabolic aberrance. In addition, potential extramitochondrial neddylation substrates, such as LKB1 and Akt (24), might also contribute to metabolic aberrance. Future studies are required to address these issues.
Materials and Methods
Animal experiments were approved by the ethics committee of the Institute of Basic Medical Sciences. A detailed description of the methodology of this study, including the generation of mouse models, in vivo neddylation assays, fatty acid oxidation assays, mass spectrometric analysis, and statistical analysis, is provided in SI Appendix, SI Materials and Methods. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the Proteomics Identifications Database (PRIDE) (53) partner repository with the dataset identifier PXD016111.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 81625010 and 31671482).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier PXD016111).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910765117/-/DCSupplemental.
References
- 1.Kim S. H., et al. , Multi-organ abnormalities and mTORC1 activation in zebrafish model of multiple acyl-CoA dehydrogenase deficiency. PLoS Genet. 9, e1003563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelius N., et al. , Molecular mechanisms of riboflavin responsiveness in patients with ETF-QO variations and multiple acyl-CoA dehydrogenation deficiency. Hum. Mol. Genet. 21, 3435–3448 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Olsen R. K., et al. , Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency. Hum. Mutat. 22, 12–23 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Schiff M., Froissart R., Olsen R. K., Acquaviva C., Vianey-Saban C., Electron transfer flavoprotein deficiency: Functional and molecular aspects. Mol. Genet. Metab. 88, 153–158 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Saha A., Deshaies R. J., Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang D. T., et al. , E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateishi K., Omata M., Tanaka K., Chiba T., The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Enchev R. I., Schulman B. A., Peter M., Protein neddylation: Beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xirodimas D. P., Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 36, 802–806 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Xirodimas D. P., Saville M. K., Bourdon J. C., Hay R. T., Lane D. P., Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118, 83–97 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Zuo W., et al. , c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol. Cell 49, 499–510 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Bai D., Ma X., Guan J., Zheng X., hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33, 246–254 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Mahata B., Sundqvist A., Xirodimas D. P., Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene 31, 3060–3071 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Vogl A. M., et al. , Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat. Neurosci. 18, 239–251 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Park H. S., et al. , PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 23, 1296–1311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubiel D., Bintig W., Kähne T., Dubiel W., Naumann M., Cul3 neddylation is crucial for gradual lipid droplet formation during adipogenesis. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1405–1412 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Nawrocki S. T., et al. , The NEDD8-activating enzyme inhibitor MLN4924 disrupts nucleotide metabolism and augments the efficacy of cytarabine. Clin. Cancer Res. 21, 439–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swords R. T., et al. , Inhibition of NEDD8-activating enzyme: A novel approach for the treatment of acute myeloid leukemia. Blood 115, 3796–3800 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Luo Z., et al. , The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 72, 3360–3371 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q., et al. , The novel protective role of P27 in MLN4924-treated gastric cancer cells. Cell Death Dis. 6, e1867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andérica-Romero A. C., et al. , The MLN4924 inhibitor exerts a neuroprotective effect against oxidative stress injury via Nrf2 protein accumulation. Redox Biol. 8, 341–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q., et al. , Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight 4, 121582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbier-Torres L., et al. , Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 6, 2509–2523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., et al. , Neddylation is required for herpes simplex virus type I (HSV-1)-induced early phase interferon-beta production. Cell. Mol. Immunol. 13, 578–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Q., et al. , Neddylation contributes to CD4+ T cell-mediated protective immunity against blood-stage Plasmodium infection. PLoS Pathog. 14, e1007440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postic C., et al. , Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Kleiner D. E., et al. ; Nonalcoholic Steatohepatitis Clinical Research Network , Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Houten S. M., et al. , Impaired amino acid metabolism contributes to fasting-induced hypoglycemia in fatty acid oxidation defects. Hum. Mol. Genet. 22, 5249–5261 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Christopher R., Sankaran B. P., An insight into the biochemistry of inborn errors of metabolism for a clinical neurologist. Ann. Indian Acad. Neurol. 11, 68–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers A. E., Characterization of hepatic mitochondrial injury induced by fatty acid oxidation inhibitors. Toxicol. Pathol. 37, 78–88 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Wada H., Yeh E. T., Kamitani T., A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. J. Biol. Chem. 275, 17008–17015 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Swords R. T., et al. , Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: A phase 1 study. Br. J. Haematol. 169, 534–543 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Shah J. J., et al. , Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin. Cancer Res. 22, 34–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarantopoulos J., et al. , Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin. Cancer Res. 22, 847–857 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Bhatia S., et al. , A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Invest. New Drugs 34, 439–449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swords R. T., et al. , Expanded safety analysis of pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukemia and myelodysplastic syndromes. Blood Cancer J. 7, e520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swords R. T., et al. , Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood 131, 1415–1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCue M. D., Starvation physiology: Reviewing the different strategies animals use to survive a common challenge. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 156, 1–18 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Zoncu R., Efeyan A., Sabatini D. M., mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jikumaru M., et al. , Effect of starvation on the survival of male and female mice. Physiol. Chem. Phys. Med. NMR 39, 247–257 (2007). [PubMed] [Google Scholar]
- 42.Grimm D., et al. , In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J., Moon Y. A., Simple purification of adeno-associated virus-DJ for liver-specific gene expression. Yonsei Med. J. 57, 790–794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y., et al. , Single point mutation in adeno-associated viral vectors -DJ capsid leads to improvement for gene delivery in vivo. BMC Biotechnol. 16, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hjerpe R., et al. , Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts D. L., Frerman F. E., Kim J. J., Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution. Proc. Natl. Acad. Sci. U.S.A. 93, 14355–14360 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider-Poetsch T., et al. , Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Samali A., Fitzgerald U., Deegan S., Gupta S., Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell Biol. 2010, 830307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang K. T., Sefc L., Psenák O., Vokurka M., Necas E., Early fetal liver readily repopulates B lymphopoiesis in adult bone marrow. Stem Cells 23, 230–239 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Lee J., Choi J., Scafidi S., Wolfgang M. J., Hepatic fatty acid oxidation restrains systemic catabolism during starvation. Cell Rep. 16, 201–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oved S., et al. , Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 281, 21640–21651 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., Comparison of the total liver proteomes of postnatal-day-7 mice with liver-specific UBA3 deficiency and their littermate controls. ProteomeXchange Consortium. http://www.ebi.ac.uk/pride/archive/projects/PXD016111. Deposited 1 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.