Significance
Sustainable utilization of cellulosic biomasses to produce valuable compounds is an ideal approach but hydrolysis of recalcitrant cellulose is complex and time-consuming. Several cellulolytic bacteria produced multienzyme complexes called “cellulosomes” that efficiently degrade the cellulose. Hence, we engineered the yeast Kluyveromyces marxianus to express the “largest cellulolytic complex,” which can accommodate up to 63 enzymes, on its cell surface. Due to the synergistic effects of cellulase in cellulosomes, our engineered yeast exhibited greater degradation efficiency and released significantly higher quantities of reducing sugars and ethanol from cellulosic substrates than any previous constructs. In future, this superb cellulosome complex may also be used for the synthesis of various biopharmaceutical products (e.g., astaxanthin and morphine) which involve multiple enzymatic conversion steps.
Keywords: scaffoldin protein, anchoring protein, cellulose-binding module, cellulosome
Abstract
Cellulosomes, which are multienzyme complexes from anaerobic bacteria, are considered nature’s finest cellulolytic machinery. Thus, constructing a cellulosome in an industrial yeast has long been a goal pursued by scientists. However, it remains highly challenging due to the size and complexity of cellulosomal genes. Here, we overcame the difficulties by synthesizing the Clostridium thermocellum scaffoldin gene (CipA) and the anchoring protein gene (OlpB) using advanced synthetic biology techniques. The engineered Kluyveromyces marxianus, a probiotic yeast, secreted a mixture of dockerin-fused fungal cellulases, including an endoglucanase (TrEgIII), exoglucanase (CBHII), β-glucosidase (NpaBGS), and cellulase boosters (TaLPMO and MtCDH). The confocal microscopy results confirmed the cell-surface display of OlpB-ScGPI and fluorescence-activated cell sorting analysis results revealed that almost 81% of yeast cells displayed OlpB-ScGPI. We have also demonstrated the cellulosome complex formation using purified and crude cellulosomal proteins. Native polyacrylamide gel electrophoresis and mass spectrometric analysis further confirmed the cellulosome complex formation. Our engineered cellulosome can accommodate up to 63 enzymes, whereas the largest engineered cellulosome reported thus far could accommodate only 12 enzymes and was expressed by a plasmid instead of chromosomal integration. Interestingly, CipA 2B9C (with two cellulose binding modules, CBM) released significantly higher quantities of reducing sugars compared with other CipA variants, thus confirming the importance of cohesin numbers and CBM domain on cellulosome complex. The engineered yeast host efficiently degraded cellulosic substrates and released 3.09 g/L and 8.61 g/L of ethanol from avicel and phosphoric acid-swollen cellulose, respectively, which is higher than any previously constructed yeast cellulosome.
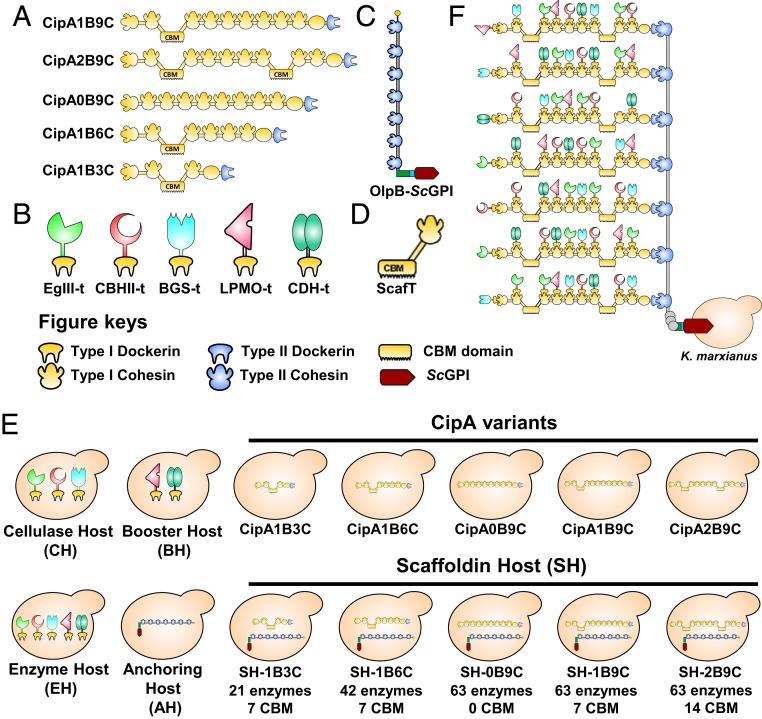
A cellulosome is a highly efficient supramolecular enzyme complex produced by Clostridium thermocellum and other anaerobic cellulolytic bacteria (1, 2). A typical C. thermocellum cellulosome is composed of a central nonenzymatic scaffoldin subunit known as “cellulosome integrating protein A” (CipA) with nine type I cohesins (3) (Fig. 1A). CipA contains an integral cellulose-binding module (CBM), which binds cellulosic substrates (4, 5). A cellulosomal enzyme contains a type I dockerin, which interacts with the type I cohesin of CipA (6) (Fig. 1B). The entire scaffoldin and enzymatic subunits are attached to the bacterial cell surface through the interaction between type II dockerins of CipA and type II cohesins of one of three surface anchoring proteins, SdbA, Orf2p, or OlpB (7, 8). The “outer layer protein B” (OlpB) typically includes seven type II cohesins (Fig. 1C), so a cellulosome can accommodate up to 63 cellulases to form one of the largest known cellulolytic enzyme complexes (8). The cohesin–dockerin interaction of a cellulosome facilitates spatial proximity among various cellulases (proximity effect) and the cellulose-binding capacity of CBM enhances the cellulose utilization (targeting effect) (9, 10). These advantages of cellulosomes provide clues for developing an efficient way to elevate the saccharification of cellulosic substrates (11).
Fig. 1.
Schematic representation of cellulosomal proteins. (A) Biomimetic scaffoldins (CipA) with varying numbers of cohesins and CBMs. In a scaffoldin name, “B” represents the number of CBMs and “C” represents the number of cohesins. For example, CipA1B9C means one CBM and nine cohesins are present in a CipA. (B) Fungal cellulases fused with dockerin T. (C) Outer layer scaffoldin protein B (OlpB) with seven type II cohesins and the ScGPI cell surface anchor. (D) ScafT containing a CBM and a cohesin T. (E) Overview of the engineered yeast hosts used in this study. (F) Schematic diagram of the engineered cellulosome complex with 63 enzymes on the cell surface of K. marxianus.
In recent years, several groups of researchers successfully expressed miniaturized versions of cellulosomes, called “minicellulosomes,” on the Saccharomyces cerevisiae cell surface and demonstrated their cellulolytic and ethanol-producing abilities using microcrystalline cellulose (12–18). Enzymes in minicellulosomes displayed enhanced activity compared to free or immobilized enzymes (19). However, a minicellulosome contains only a few cohesins and hence can accommodate only a few enzymes (12 so far), thus limiting the enzyme synergism. Engineering a large cellulosomal complex into the yeast genome remains highly challenging due to the massive TRs in the cohesins of CipA and OlpB genes, lack of stable chromosome integration strategy, low protein expression, and secretion capability of the host (20). The existing strategies use episomal plasmids to express minicellulosomes for higher protein yields. However, the episomal expression requires induction and constant selection, thus increasing the production cost and raising the stability concern (21).
The purpose of this study was to engineer Kluyveromyces marxianus with the largest cellulosome complex that can accommodate up to 63 enzymes on the cell surface. To do so, we synthesized the CipA gene (with nine type I cohesin repeats) and the largest OlpB gene (with seven type II cohesin repeats). The multiple repeats in the CipA and OlpB genes make it extremely difficult for their cloning or even DNA synthesis. We overcame this problem by randomizing the codons in the repeats and synthesized CipA and OlpB genes with advanced DNA synthesis techniques. These two genes were then integrated into the K. marxianus genome. Moreover, to understand the importance of cohesin numbers and CBMs of a CipA in avicel degradation, we constructed CipA variants with different numbers of cohesins and CBMs.
The conversion of cellulose into simple sugars requires at least three types of enzymes: endoglucanases (EGs), exoglucanases (CBHs) and beta-glucosidases (BGSs) (21). Moreover, a new class of oxidative enzymes, called lytic polysaccharide monooxygenases (LPMO), has been reported (22). LPMO can efficiently degrade crystalline cellulose and increase the soluble sugar release by 2.6-fold, so it was named a “cellulase booster” (22). LPMO requires electrons for its activity, so an electron donor called cellobiose dehydrogenase (CDH) was used together with LPMO. We therefore selected three types of fungal cellulases, namely an EG from Trichoderma reesei (TrEgIII) (23), a synthetic CBH (CBHII) (24), a β-glucosidase from Neocallimastix patriciarum (NpaBGS) (25), and the cellulase booster from Thermoascus aurantiacus (TaLPMO) (26) and an electron donor from Myceliophthora thermophila (Thermothelomyces thermophila) (MtCDH) (27), which were then fused with type I dockerin of C. thermocellum to facilitate the cellulosomal integration. The engineered cellulosomal yeast strains efficiently converted the microcrystalline cellulose into reducing sugars or/and ethanol and are suitable for consolidated bioprocessing (CBP).
In this study we engineered a cellulosome complex in K. marxianus, a nontraditional yeast. K. marxianus has several advantages over S. cerevisiae and other yeast strains (23, 24, 28). It is Crabtree-negative, thermotolerant (up to 52 °C), and capable of fermenting various sugars, including inulin and other pentose sugars (e.g., xylose, and arabinose) (28, 29). It grows faster than S. cerevisiae and other yeasts (30–32). Moreover, its secretory capacity is higher than that of S. cerevisiae, due to its efficient signal peptides (28). Finally, it is a generally regarded as safe and qualified presumption of safety certified organism (33). These attributes make K. marxianus an excellent host for industrial applications (34).
In this study we have generated several cellulosomal hosts including a cellulase host (CH: expressing TrEgIII-t, CBHII-t, and NpaBGS-t), a booster host (BH: expressing TaLPMO-t and MtCDH-t), an enzyme host (EH: expressing TrEgIII-t, CBHII-t, NpaBGS-t, TaLPMO-t, and MtCDH-t), and an AH (expressing OlpB-ScGPI). Similarly, five different types of scaffoldin host (SH) were developed by integrating the CipA variants into an AH expressing OlpB-ScGPI. Finally, an EH and an AH were cocultured together to form the largest cellulosomes and then microcrystalline cellulose (avicel) and phosphoric acid-swollen cellulose (PASC) was separately used to demonstrate the ethanol production.
Results
Design and Synthesis of Cellulosomal Scaffoldins.
Several components were designed and synthesized to engineer an entire cellulosome system into the yeast genome. A synthetic scaffoldin was designed according to the sequence of the CipA gene (5.6 kb) in C. thermocellum ATCC 27405 (CP000568). To evaluate the effect of cohesin number, we designed three synthetic scaffoldins containing three, six, and nine cohesins with a single CBM (denoted 1B3C, 1B6C, and 1B9C) (Fig. 1A). Similarly, to examine the effect of CBM number on cellulose binding, two more synthetic scaffoldins were designed with no or two CBMs (0B9C and 2B9C).
An anchoring protein plays a critical role in the attachment of the cellulosome complex to the yeast cell surface. However, the anchoring domain of C. thermocellum is not suitable for a eukaryotic host. We therefore tested the anchoring efficiency of glycosylphosphatidylinositol (ScGPI), a cell-surface protein from S. cerevisiae, on K. marxianus (35). The pKlac2 plasmid containing ScGPI fused with a green fluorescent protein (GFP) and a 12x His was expressed in K. marxianus and its anchoring efficiency was confirmed by fluorescence microscopic analysis (SI Appendix, Fig. S1). The imaging data showed a successful display of ScGPI on the K. marxianus cell surface. Then, the largest cell surface scaffoldin was designed using OlpB as the backbone. The seven type II cohesins of OlpB were selected and the original anchoring domain (SLH) was replaced with the ScGPI domain to facilitate surface display. OlpB contains long tandem repeats (TRs) with highly repetitive TPSDEP amino acid sequences (36). In view of the repetitiveness of the OlpB DNA sequence, the TR length was reduced and seven cohesins were directly fused with the trimmed TRs and with the ScGPI domain (Fig. 1C). For efficient translation in K. marxianus, both CipA and OlpB gene sequences were codon-optimized for K. marxianus and the repeats in the cohesins were randomized to avoid DNA synthesis constraints.
Conversion of Free Cellulases into Cellulosomal Mode.
As fungal cellulases have no dockerins, two kinds of dockerin fusion plasmids were designed based on the type I dockerin (DocT) of the celS from C. thermocellum (SI Appendix, Fig. S2 A and B). The dockerin module was fused at either the N or C terminal, depending on the domain organization of each enzyme (SI Appendix, Fig. S2). A 36-bp glycine-rich linker was used between the dockerin module and the catalytic domain to reduce the conformational constraint and steric hindrance between cellulase enzymes (22, 37) and an 8xHis tag was fused for purification and detection purposes. For efficient secretion of each cellulase into the medium, the native secretory signal peptide (α-mating type) of K. lactis was fused at the N terminal. In our previous studies, we successfully expressed several fungal cellulases in K. marxianus, including TrEgIII, CBHII, and NpaBGS (24, 38). In this study, in addition to cellulases we also included the cellulase booster LPMO (from T. aurantiacus, TaLPMO) and an electron donor CDH (from M. thermophila, MtCDH). To convert the free fungal cellulases into the cellulosomal mode, DocT was fused at either the N or C terminal of TrEgIII, CBHII, and NpaBGS. Interestingly, CBHII and TrEgIII retained their activity after C-terminal dockerin fusion but not after N-terminal dockerin fusion (SI Appendix, Fig. S3 A and B). However, the enzyme activity was recovered after adding a scaffoldin T (ScafT) that contains a CBM and a cohesin T (CohT) (SI Appendix, Fig. S3). On the other hand, NpaBGS displayed similar amounts of enzyme activity in either N- or C-terminal dockerin fusion (SI Appendix, Fig. S3C). Based on a previous study (22), a dockerin was fused at the C termini of TaLPMO and MtCDH, since the N-terminal histidine residue has been evolutionarily conserved in LPMO and is essential for its function. The suffix “t” after the enzyme name (i.e., TrEgIII-t) denotes the type I dockerin fusion. Three-dimensional (3D) protein structures of dockerin-fused chimeric enzymes were predicted using homology modeling and the results showed that the dockerin fusion did not alter their substrate-binding pockets (SI Appendix, Fig. S4).
Chromosomal Integration of Cellulosomal Genes.
The cellulosomal genes were integrated into the LAC4 promoter region of the K. marxianus genome using yeast integration plasmid pKLac2 (SI Appendix, Fig. S5 A and B). To study the effects of the numbers of cohesins and CBMs in CipA on cellulose degradation, five different CipA variants—CipA1B3C, 1B6C, 1B9C, 2B9C, and 0B9C—were transformed individually into K. marxianus (Fig. 1E). The chromosomal integration was confirmed by PCR verification (SI Appendix, Fig. S5C) and their expression was confirmed by Western blot (WB) analysis using anti-His antibody. All of the CipA constructs were secreted out to the medium because they have an α-matting-type signal peptide. The WB results of CipA1B3C showed a distinctive band at ∼89 kDa (SI Appendix, Fig. S5C). Similarly, transformants expressing 1B6C, 1B9C, 0B9C, and 2B9C displayed bands at ∼142 kDa, ∼194 kDa, ∼172 kDa, and ∼217 kDa, respectively (SI Appendix, Fig. S5C). The AH was developed by transforming OlpB-ScGPI into the K. marxianus genome and their expression was confirmed by WB analysis, showing a band at ∼211 kDa (SI Appendix, Fig. S6). Finally, an SH was constructed by integrating the CipA variants into the AH (Fig. 1E). Based on the types of CipA integrated into AH five types of SH were constructed (SH-1B3C, SH-1B6C, SH-1B9C, SH-2B9C, and SH-0B9C).
Demonstration of Cell-Surface Display Using Immunofluorescence Analysis.
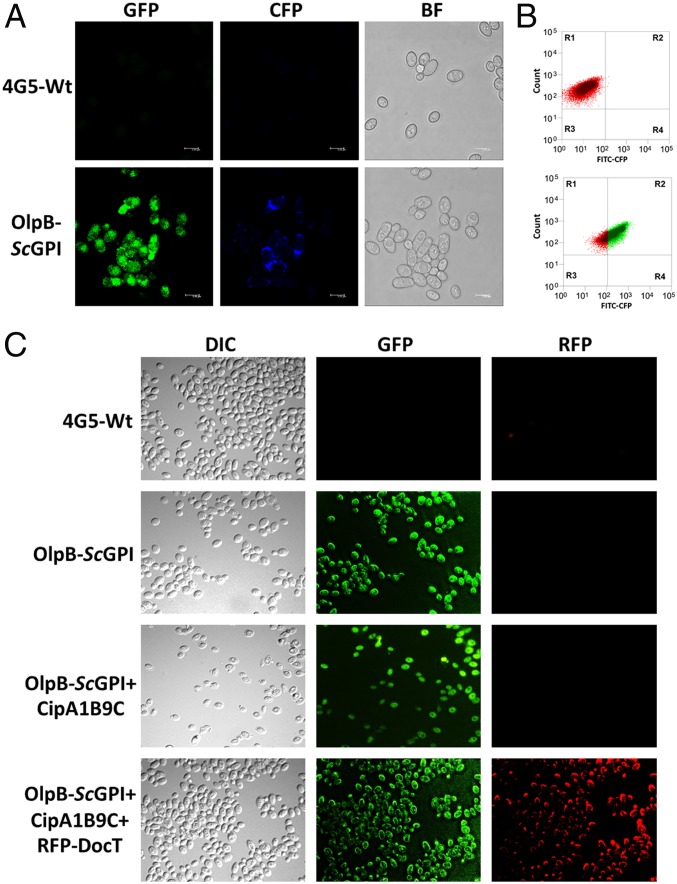
Anchoring of the entire OlpB-ScGPI on the cell surface was confirmed by immunofluorescence microscopy and fluorescence-activated cell sorting (FACS) analysis. The OlpB-ScGPI contained a FLAG tag and a GFP protein at the C terminal. The yeast cells expressing OlpB exhibited a strong green fluorescence signal throughout the cell, making it difficult to distinguish the protein anchored on the cell surface from the internal OlpB. Since the monoclonal antibodies did not penetrate the yeast cell wall, the yeast cells expressing OlpB were immunostained with DyeLight 405 secondary antibody conjugated with the cyan fluorescent protein (CFP) that specifically binds to the anti-FLAG tag. A clear blue fluorescence was observed around the AH, thus confirming the display of the OlpB protein on the K. marxianus cell surface, whereas no fluorescence was observed on wild-type strains (Fig. 2A and SI Appendix, Fig. S7). Similarly, the cells immunostained with DyeLight 405 were analyzed using FACS, and among ∼50,000 cells examined ∼81.47% showed a fluorescence signal, confirming the anchoring of OlpB-ScGPI on the yeast cell surface (Fig. 2B).
Fig. 2.
Confirmation of cellulosome assembly on the K. marxianus cell surface. (A) Anchoring of OlpB-ScGPI on the cell surface confirmed by immunostaining and confocal microscopy using GFP or CFP. (B) FACS analysis of immunostained cells expressing OlpB-ScGPI. (C) Confirmation of entire cellulosome complex assembly on yeast cell surface using fluorescence microscopy. To form a cellulosome complex cells expressing OlpB-ScGPI were mixed with the concentrated supernatant of CipA1B9C or RFP-DocT or both. The complex formation was performed at 37 °C and cells were washed to remove the unbound CipA1B9C and RFP-DocT. Cells expressing OlpB-ScGPI have green fluorescence around the cells. The complex formation of RFP-DocT with CipA1B9C and OlpB-ScGPI exhibited both red and green florescence, thus confirming the complex formation on the cell surface.
Similarly, the anchoring of the entire cellulosome complex on the cell surface was demonstrated using fluorescence microscopy. The cells expressing OlpB-ScGPI and the concentrated culture supernatant of CipA1B9C and red fluorescent protein (RFP)-DocT were allowed to form a complex, which was observed under an epifluorescence microscope. Green color fluorescence was observed on the cells expressing OlpB-ScGPI (Fig. 2C). Since CipA1B9C does not contain any fluorescence tag, the complex of OlpB-ScGPI and CipA1B9C also showed green fluorescence. The cellulosome complex composed of OlpB-ScGPI, CipA1B9C, and RFP-DocT exhibited both green and red color signals (Fig. 2C). The red color fluorescence was observed only on the cell surface, due to the presence of RFP in the RFP-DocT protein. Thus, DocT indeed binds to CipA and CipA binds to the cells expressing OlpB-ScGPI.
Confirmation of Cellulosome Complex Assembly.
As the molecular weight of the entire cellulosome complex (with CipA, OlpB, and DocT) is expected to be ∼2.6 MDa, purifying this large cellulosome complex is challenging. Therefore, the complex formation study was divided into three steps (Fig. 3A). The CipA1B9C, OlpB, and DocT proteins were purified (SI Appendix, Fig. S8). First, DocT was used for complex formation study with CipA1B9C and the CipA:DocT complex was purified by size-exclusion chromatography (SEC). The elution volume of the SEC CipA:DocT complex peak was equal to the theoretical molecular weight (∼303 kDa) of the CipA:DocT complex (Fig. 3B). As expected, DocT (type I) binds randomly to available type I cohesins of CipA. Fractions of the corresponding peaks were pooled together and observed on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Two clear bands were seen (Fig. 3 B, Inset), which represented exactly the CipA and DocT molecular weights, and identified as CipA and DocT by mass spectrometry (SI Appendix, Fig. S9). Similarly, formation of the OlpB:DocT complex was tried and analyzed using SEC, but no peak that matched the molecular weight of the OlpB:DocT complex was found (Fig. 3C). Thus, as anticipated, DocT (type I) did not interact with OlpB, which does not contain the type I cohesins. Second, the complex formation between CipA and OlpB was found by native PAGE (Fig. 3D). The data showed five different protein bands on the gel, each of which was analyzed to identify its protein by mass spectrometry. The data revealed that the top three bands represented the CipA:OlpB complex, while the fourth and fifth bands represented CipA and OlpB, respectively (SI Appendix, Fig. S10). When mixing all three proteins in vitro they might not always form a full complex, so there were differences in the number of CipAs binding to OlpB and different lengths of cellulosome complexes were observed. Therefore, our study showed that CipA and DocT interacted and CipA could form a complex with OlpB. Finally, complex formation between CipA and DocT fused enzymes was examined using the concentrated supernatants of the CipA1B9C host and the EH (EH-P1-44) expressing five different dockerin fused enzymes. After the complex formation cellulose was added and allowed to interact with the CipA:enzyme complex. Then, the protein complex was eluted. Enzymatic assays confirmed the complex formation of CipA with DocT fused enzymes using the purified cellulosome complex. The data showed activity in all tested enzymatic assays (Fig. 3 E–I), thus confirming the presence of all enzymes in the purified complex. Taken together, these multifaceted data confirmed that our cellulosome proteins indeed formed an active complex.
Fig. 3.
Cellulosome complex assembly demonstration. (A) Flow diagram of cellulosome complex formation. (B) The SEC chromatogram represents the CipA:DocT complex colored in cyan and the corresponding complex verified on 4 to 15% SDS–PAGE. (C) The chromatogram in pink denotes the unbound OlpB and DocT complex. (D) Native PAGE analysis revealed the complex formation of CipA:OlpB. Each protein band was identified using mass spectrometry. (E–I) Enzymatic assays of purified cellulosome complex contain CipA and five types of enzymes. The CipA alone and cellulosome complex were purified using cellulose. All enzyme assays were performed at 40 °C at 1,200 rpm. (E) EG activity assay using dye CMC as substrate. (F) CBH activity assay using PASC as substrate. (G) β-Glucosidase assay using pNPG as substrate. (H and I) The H2O2 release was measured using the Amplex Red/HRP assay kit and excitation/emission was measured at 530/590 nm using a fluorescence spectrophotometer. The assay was performed using cellobiose (H), and ascorbate (I) as an electron donor.
Apart from this, the cellulosome complex assembly was demonstrated using the concentrated supernatants of CipA1B9C and BGS-t and the cell extract of OlpB-ScGPI. They were allowed to form complexes in vitro at 37 °C and were analyzed using native PAGE followed by WB analysis. In native PAGE WB a shift in the band was observed, confirming that the cellulosome complex had been formed (SI Appendix, Fig. S11A). A smear of the band in the BGS-t lane was observed, which might be due to the N-glycosylation of the DocT (SI Appendix, Fig. S11B) domain or improper folding or degradation during the electrophoresis study. As expected, the type I dockerin of BGS-t interacted randomly to available type I cohesins of the CipA scaffoldin, causing the band to shift upward. Similarly, the type II dockerins of CipA interacted with the type II cohesins of OlpB-ScGPI to form a complex. When mixing the three components together, a clear band was observed at the top of the lane, thus confirming the assembly of all three components to form cellulosomes. Thus, the cellulosome complex formed even using the culture supernatant of each component protein.
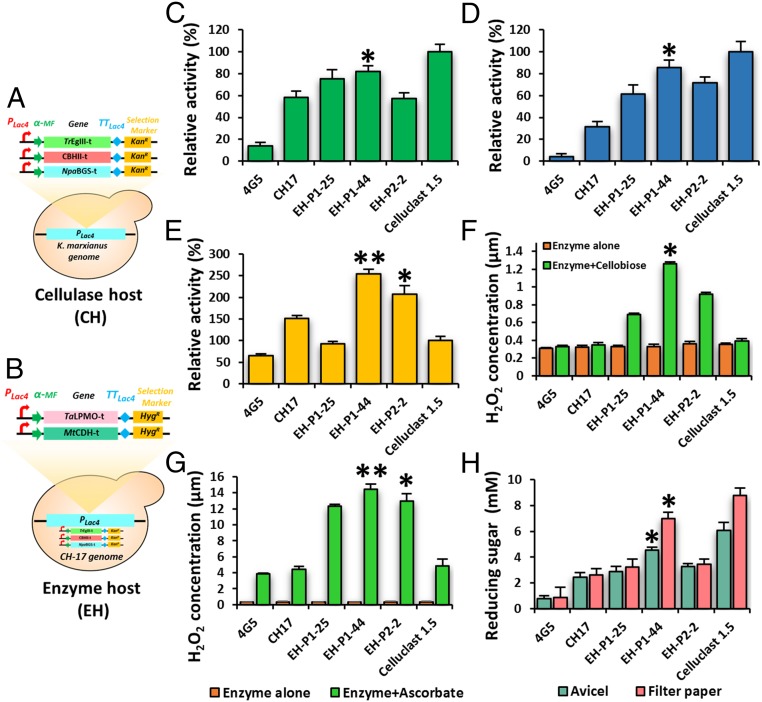
Constructing EHs.
A CH was constructed by electroporating equimolar ratios of CBHII-t, TrEgIII-t, and NpaBGS-t genes into K. marxianus (Fig. 4A). The transformants were selected on YPG (1% yeast extract, 2% peptone, and 2% galactose) plates supplemented with kanamycin (G418). The transformants integrated with all of the cellulase genes were selected by PCR screening (SI Appendix, Fig. S12) and their activity was confirmed by enzyme assays (SI Appendix, Fig. S13 A–C). Transformant CH-17 showed the highest enzyme activity and released 3.24 mM and 4.91 mM reducing sugars from avicel and filter paper, respectively (SI Appendix, Fig. S13D). Similarly, to demonstrate the effects of the cellulase booster, the TaLPMO-t and MtCDH-t genes were transformed into K. marxianus (BH) (SI Appendix, Fig. S14) and the booster functionality was confirmed by measuring the H2O2 production using the Amplex Red/HRP (horseradish peroxidase) assay kit. The production of H2O2 was directly proportional to the LPMO activity, which requires enzymatic (CDH together with cellobiose) or nonenzymatic (ascorbate) electron donors. Hence, the enzyme activity was assayed using two kinds of electron donors: cellobiose and ascorbate. The transformant BH-20 released 1.60 µM and 12.13 µM of H2O2 using the substrate cellobiose and ascorbate, respectively (SI Appendix, Fig. S15). The lowest H2O2 production was observed while using cellobiose as the substrate and its concentration was increased in a time-dependent manner. In this case, CDH needs to oxidize the cellobiose and transfers an electron through its heme domain to LPMO (27). In contrast, ascorbate can directly donate the electron to LMPO, thereby producing higher quantities of H2O2 in a short time.
Fig. 4.
Construction of cellulase and EH and their activity assay. (A) Schematic representation of CH expressing TrEGIII-t, CBHII-t, and NpaBGS-t. (B) Schematic representation of EH expressing TrEGIII-t, CBHII-t, NpaBGS-t, TaLPMO-t, and MtCDH-t. The numbers after CH or EH represent the transformant numbers. (C) CBH activity assay using PASC as the substrate. (D) EG activity assay using dye CMC as the substrate. (E) β-Glucosidase activity assay using pNPG as the substrate. (F and G) LPMO and CDH activity assay using cellobiose (G) or ascorbate (H) as the substrate for CDH, which donates the electrons to LPMO. The H2O2 release was measured using the Amplex Red/HRP assay kit and excitation/emission was measured at 530/590 nm using a fluorescence spectrophotometer. (H) Soluble sugar release assay using avicel or filter paper as the substrate. For all enzyme assays, concentrated culture supernatant of the CH or EH host was used as an enzyme source and equal protein concentrations were calculated using BSA as a standard and used for enzyme assay. All enzyme assays were performed at 40 °C at 1,200 rpm. The reducing sugar release was calculated using the DNS method. The results are expressed as mean (n = 3) ± SD. *P < 0.05, **P < 0.01; one-way ANOVA followed by Bonferroni post hoc test.
Boosting Effects of Cellulase Boosters.
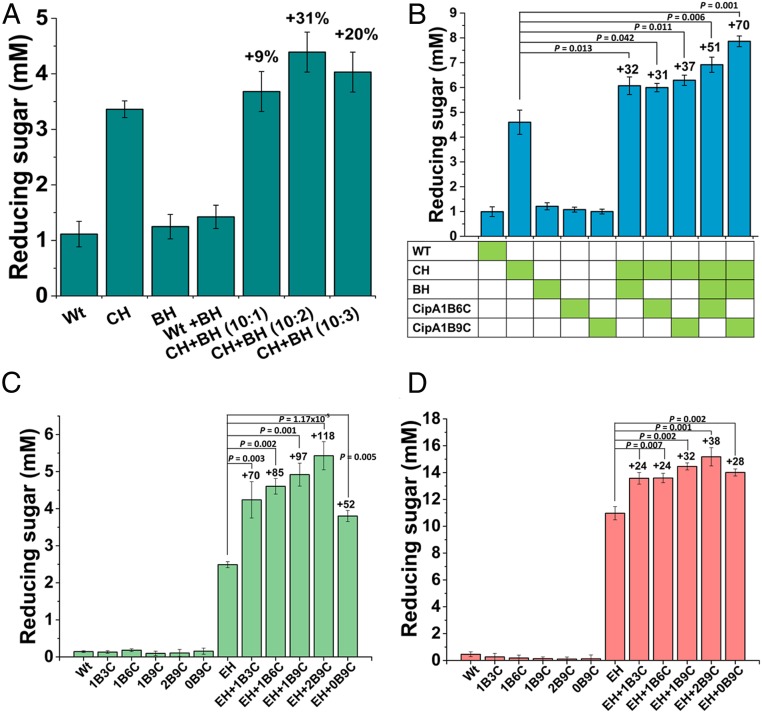
The selected BH, BH-20, which expresses TaLPMO-t and MtCDH-t, and the selected CH, CH-17, which expresses CBHII-t, TrEgIII-t, and NpaBGS-t, were cultured separately. Then, the concentrated supernatants of CH-17 and BH-20 were mixed at three different ratios (10:1, 10:2, and 10:3) and incubated along with avicel. The addition of CH-17 supernatant to the BH-20 supernatant at the ratio of 10:2 increased the soluble sugar release by 31%, confirming that LPMO has a proper function in K. marxianus (Fig. 5A). Similarly, the effects of cellulase boosters on the cellulosome complex were examined by mixing the supernatants of CH-17, BH-20, and CipA variants (including 1B3C, 1B6C, 1B9C, 0B9C, or 2B9C) in the molar ratio of 10:2:2 (CH:BH:CipA) and the enzyme mix was allowed to form the cellulosome complex at 37 °C. Avicel was added to the cellulosome complex and reducing sugar release was measured. The results showed that the activity of the CH improved by 32% (6.07 mM) with TaLPMO-t and MtCDH-t, and the addition of CipA 1B6C or CipA1B9C along with CH-17 increased the sugar release by 31% and 37%, respectively (Fig. 5B). Importantly, the addition of CH-17 and BH-20 supernatants together with either CipA1B6C or 1B9C increased the sugar release by 51% and 70%, respectively.
Fig. 5.
Effects of cellulase booster and CipA variants on avicel degradation. (A) Optimization of booster ratio. Wild type (Wt), CH-17, and BH-20 were cultured separately and concentrated supernatants were used for the experiment. The CH and BH supernatants were mixed at different ratio (10:1, 10:2 and 10:3) and reaction was performed at 40 °C using avicel. (B) Effects of cellulosome complex on avicel. The concentrated supernatants of Wt, CH-17, BH-20, and CipA (1B6C and 1B9C) were mixed in the ratio of 10:2:2 and allowed to form complexes at 37 °C, for 3 h, which were then mixed with avicel and incubated at 40 °C. (C and D) Effects of CipA variants on avicel (C) and PASC (D) degradation. Wt, EH (EH-P1-44), and CipA variants (1B3C, 1B6C, 1B9C, 0B9C, and 2B9C) were cultured separately and concentrated supernatants were used for the experiment. The EH and CipA were mixed in the ratio of 10:2 and allowed to form complexes at 37 °C for 3 h, which were then mixed with avicel or PASC and incubated at 40 °C. The reducing sugar release was measured using the DNS method. The results are expressed as mean (n = 3) ± SD. P values were determined using two-tailed Student t tests.
These results corroborate the effects of TaLPMO-t and MtCDH-t on the cellulosome complex. Therefore, we constructed another host, called EH, with three cellulases (TrEgIII-t, CBHII-t, and NpaBGS-t) and TaLPMO-t and MtCDH-t (Fig. 4B). Transformants with all of the integrated genes were selected (SI Appendix, Fig. S16A) and their expression was confirmed by qPCR analysis using actin as a reference gene (SI Appendix, Fig. S16B). Transformant EH-P1-44 demonstrated the highest enzyme activity on all of the tested substrates (Fig. 4 C–G) and released 4.55 mM and 6.97 mM of reducing sugar from avicel and filter paper, respectively (Fig. 4H).
Effects of Cohesin and CBM Numbers on Cellulosome Efficiency.
The effects of cohesin and CBM numbers on sugar release were assayed using avicel or PASC as the substrate. The concentrated supernatant of EH-P1-44 (expressing CBHII-t, TrEgIII-t, NpaBGS-t, TaLPMO-t, and MtCDH-t) and the hosts expressing CipA variants were mixed at different ratios (10:1, 10:2, and 10:3). After the complex formation at 37 °C, substrates were added to the mixture and sugar release was measured after 48 h incubation at 37 °C. The data exhibited a significant increase in enzyme activity when the EH-P1-44 supernatant was mixed with CipA variants at the ratio of 10:2. CipA2B9C exhibited the highest increase, 118%, in sugar release, followed by 97%, 85%, and 70% increases for CipA1B9C, CipA1B6C, and CipA1B3C, respectively (Fig. 5C and SI Appendix, Fig. S17A). These data indicate that the cohesin number has a strong influence on sugar release when using avicel as the substrate. However, when PASC was used as the substrate, the influence of cohesin number was less significant (Fig. 5D). The addition of CipA1B9C, CipA1B6C, or CipA1B3C only increased 32%, 24%, or 24% in the sugar release compared to the activity of EH-P1-44 alone (Fig. 5D and SI Appendix, Fig. S17B). These results substantiate the vital role of the cohesin number in enzyme synergism, which is important for digesting complex substrates (i.e., microcrystalline cellulose or plant biomass).
The number of CBMs also plays an important role in the efficiency of the cellulosome complex because they bring the entire complex toward the substrate. The data for sugar release from avicel showed that the construct with two CBMs (CipA2B9C) remarkably increased the sugar release by 118%, followed by 97% for CipA with one CBM. Although CipA without any CBM increased the sugar release by 52%, it was the lowest among all of the tested CipA variants (Fig. 5C). However, the number of CBMs did not significantly improve the sugar release from PASC. The CipA2B9C exhibited 38% increased sugar release whereas CipA1B9C and CipA0B9C increased the sugar release by 32% and 28%, respectively (Fig. 5D). These results confirm the significance of the CBM on synthetic scaffoldin for microcrystalline cellulose degradation.
Demonstration of CBP.
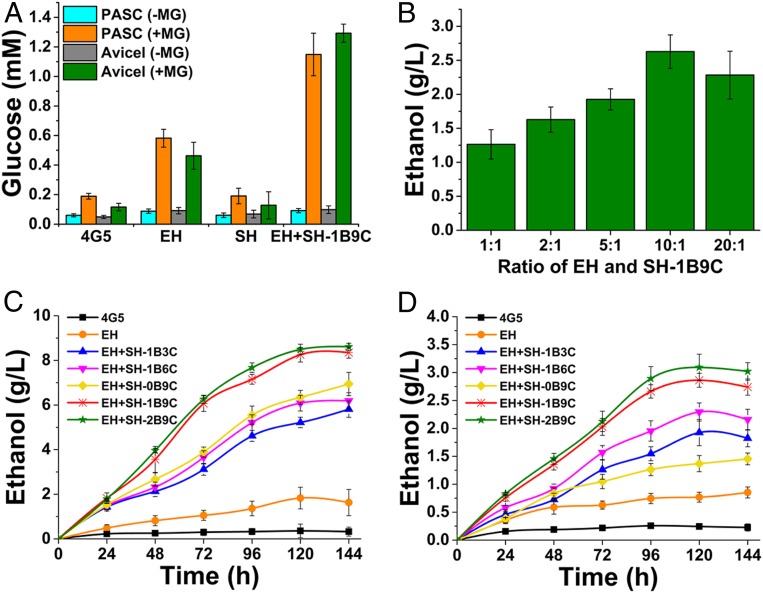
CBP is a single-step approach, including the production of cellulase enzymes, biomass saccharification, and fermentation and could be an ideal approach to cellulosic ethanol production. The hydrolytic activity of cellulosome on cellulosic substrates was demonstrated with or without methylglyoxal, a glucose metabolism inhibitor. Cells were cultured in YP (1% yeast extract, 2% peptone, and 10 mM CaCl2) containing 1% avicel or PASC supplemented with or without 100 mM methylglyoxal for 6 h. The lowest amount of glucose was observed in all conditions in the absence of methylglyoxal (Fig. 6A). The concentration of glucose increased significantly in the presence of 100 mM methylglyoxal. Notably, EH-P1-44 along with SH released a higher amount of glucose using avicel than PASC as the substrate. This confirms the above observation that our scaffoldin increased the cellulolytic activity on microcrystalline cellulose.
Fig. 6.
Hydrolysis of avicel or PASC and production of glucose or ethanol by the engineered cellulosome. (A) Analysis of glucose release from avicel or PASC. The cells were precultured and 10 OD cells were inoculated into YP medium supplemented with 1% avicel or PASC with or without 100 mM methylglyoxal (MG). In the presence of MG, the glucose metabolism of host cells was arrested, while the glucose produced during the hydrolysis remained intact. (B) Optimization of the ratio of EH and the SH (with OlpB-ScGPI and CipA1B9C). The cells were precultured in YPG medium and washed with PBS twice then inoculated at different ratio in YP media and supplemented with 0.042% Tween 80 and 1% avicel. Fermentation was carried out at 37 °C at 300 rpm for 4 d and ethanol concentration was analyzed using HPLC. (C and D) Time course of cellulosic ethanol production by yeast consortium composed of EH (EH-P1-44) and either SH-1B3C, SH-1B6C, SH-0B9C, SH-1B9C, or SH-2B9C at the optimized ratio of 10:1. Ethanol production from 10 g/L avicel (C) and 20 g/L PASC (D) was analyzed using HPLC.
For efficient secretion and assembly of the cellulosomal protein complex, the EH-P1-44 host and the SH-1B9C host expressing CipA1B9C and OlpB were cocultured in different ratios and their ethanol-producing ability was analyzed using avicel. The direct ethanol fermentation was conducted under a microaerobic condition to provide the oxygen required for LPMO activity. The microaerobic condition was achieved by culturing the cells with 5% headspace in 50-mL serum bottles. The maximal ethanol production (2.68 g/L) was achieved on avicel when the EH-P1-44 and SH-1B9C hosts were cocultured at the ratio of 10:1 (Fig. 6B). The cellulosic ethanol production rates of SH-CipA variants (1B3C, 1B6C, 0B9C, 1B9C, and 2B9C) along with EH-P1-44 were investigated to show the influences of CBM and cohesin numbers on enzyme–substrate–microbe complex synergism. As shown in Fig. 6C, when using avicel as the substrate, the maximal ethanol production (3.09 g/L) was achieved in the consortium that contained EH-P1-44 and SH-2B9C. In comparison, EH-P1-44+SH-1B9C and EH-P1-44+SH-0B9C produced 2.86 g/L and 1.37 g/L of ethanol, respectively. Cellulosome hosts expressing 1B3C and 1B6C produced 1.93 and 2.29 g/L of ethanol, which are higher than the full scaffoldin without the CBM (0B9C). These observations indicate a strong influence of the number of CBMs on ethanol production using microcrystalline cellulose. In contrast, while using PASC as the substrate, EH-P1-44+SH-1B9C and EH-P1-44+SH-2B9C produced similar quantities of ethanol, 8.25 g/L and 8.49 g/L (Fig. 6D). Interestingly, when PASC was used as the substrate, the ethanol titer of the consortium containing SH-0B9C cells (lacking CBM) was higher (6.95 g/L) than the cellulosomes containing SH-1B3C and SH-1B6C (5.81 g/L and 6. 91 g/L, respectively).
Discussion
In the past few decades, researchers have tried to engineer cellulosomes into various industrial hosts, including S. cerevisiae, Bacillus subtilis, Lactobacillus plantarum, and Lactococcus lactis (12–18, 39–43). Fan et al. (13) engineered S. cerevisiae with two scaffoldins, which can accommodate up to 12 cellulase enzymes, while Liang et al. (15) developed a S. cerevisiae host expressing 5 different dockerin-fused enzymes, including LPMO and CDH. Our study developed a K. marxianus host that can accommodate up to 63 enzymes in a single cellulosome complex, which is to date the largest engineered cellulosome in yeasts. Note that Fan et al. (13) and Liang et al. (15) used episomal plasmids to express the cellulosomal genes, which might be impractical for industrial applications. In contrast, we integrated our cellulosome genes into the yeast genome.
We have demonstrated the complex formation between CipA and DocT and between CipA and OlpB, confirming that our expressed cellulosomal proteins indeed formed a complex. The enzymatic activity of purified cellulosome confirmed the functionality of our cellulosome (Fig. 3). During our analysis of CipA and OlpB complex on native PAGE, we found three bands, and mass spectrometry data showed that all of the three bands contained mixtures of CipA and OlpB (Fig. 3D and SI Appendix, Fig. S10), but the ratios were different. OlpB has seven binding sites for CipA but sometimes fewer than seven CipAs might bind to OlpB, so that we found three different populations with different molecular weights. Apart from this, we have demonstrated the assembly of the entire cellulosome complex on the cell surface using epifluorescence microcopy (Fig. 2C).
In this study, we have used two kinds of cellulosic substrates: PASC, a pretreated cellulose, and avicel, a microcrystalline cellulose. Both of these substrates are widely used to demonstrate ethanol production. In terms of ethanol production, the minicellulosome engineered by Fan et al. (13), which could display up to 12 enzymes, produced 1.41 g/L and 1.09 g/L of ethanol from avicel and PASC, respectively. By including the LPMO and CDH genes into the cellulosome, Liang et al. (15) improved the ethanol titer to 1.5 g/L and 2.7 g/L for avicel and PASC, respectively. Compared to Liang et al.’s study, we achieved twofold higher ethanol production (3.09 g/L) using avicel and threefold higher ethanol production (8.61 g/L) using PASC as the sole carbon source (Fig. 6 C and D and SI Appendix, Table S5). The performance improvement of our construct over that of Liang et al. (15) was not dramatic, but we used genome integration, whereas Liang et al. used an episomal plasmid, which has the advantage of increasing the copy number in the yeast cell, but it is not stable in the absence of selection and thus not suitable for industrial applications. Since the copy numbers of our genes are low, we have used higher initial inoculum (20 optical density [OD]) to quicken the digestion. In the future, we will investigate whether increasing gene copy numbers or using new promoters (i.e., TEF, ADH3, or TDH3) in our hosts can improve the performance of our engineered hosts.
In addition to showing the advantage of increasing the number of cohesins on the scaffoldin (CipA), we also showed that a CipA with two CBMs produced a higher ethanol titer than the CipA containing only one CBM, which in turn produced a higher ethanol titer than a CipA with no CBM. Thus, both the scaffoldin length (number of cohesins) and the CBM number influence the cellulose degradation efficiency of the cellulosome.
Developing a cellulosic host capable of releasing an enzyme mixture comprising balanced amounts of cellulases from various cellulolytic organisms is an ideal approach to attain efficient hydrolysis of cellulose into fermentable sugars (44). In the present study, cellulases were selected from different biomass-degrading fungal species and incorporated into the cellulosome complex. Although TaLPMO is from thermophilic microbes, its expression and function have previously been demonstrated in S. cerevisiae (15). Our preliminary experiments demonstrated the function of TaLPMO and MtCDH in K. marxianus and their activity was measured using H2O2 release. To avoid steric hindrance and provide the flexibility to the enzyme, intermodular linkers were fused between an enzyme and its dockerin module (22, 45). The dockerin-fused cellulases retained their activity after the addition of the dockerin domain at the C termini but not at the N termini (SI Appendix, Fig. S3). This might be due to the conformational changes at the active site or steric hindrance caused by the dockerin fusion. We have simulated the 3D structures of dockerin-fused enzymes using homology modeling. Modeled protein structures revealed that dockerins fused at the C termini of the fungal cellulases were positioned far away from their active sites (SI Appendix, Fig. S4). EgIII and CBHII completely lost their activity in N-terminal dockerin fusion but retained their activity in C-terminal dockerin fusion. Based on the structural analysis, both enzymes have a CBM near their N terminal, so that dockerin fusion at the N terminal might affect the binding of CBM to the substrate. On the other hand, C-termini dockerin fusion did not affect the EgIII and CBHII activity because the CBMs are free to bind substrates (SI Appendix, Figs. S3 and S4). These results were further corroborated by CBM restoration experiments with ScafT, in which the enzyme activity of t-EgIII and t-CBHII was restored significantly. In this case, the CBM that was lost through dockerin fusion could be restored by the CBM in the ScafT. These results suggest that CBM plays an important role in substrate binding and enzyme activity. In the case of NpaBGS, dockerin fusion at both ends did not affect its enzyme activity as it contains two GH3 catalytic domains at both N and C terminals (SI Appendix, Fig. S3) (25). As K. marxianus has endogenous BGS enzyme (46), it showed significant BGS activity in all of the enzyme assays.
As expected, adding LPMO and CDH to the cellulase mixture greatly increased the soluble sugar release (22, 47). Interestingly, adding CipA2B9C to the enzyme mixture increased the sugar release from the avicel by 118% (Fig. 5C). However, the hydrolytic efficiency of CipA and enzyme complex on PASC was weaker than on avicel, which might be attributed to the poor binding of CBM to PASC (48). Generally, yeast cells utilize the glucose released from hydrolysis and the addition of methylglyoxal inhibits the host’s glucose metabolism (15). With this method, the amount of glucose released during the hydrolysis could be quantified readily. The glucose amount was low without the addition of methylglyoxal (Fig. 6A), due to the immediate uptake of glucose by the cells. In contrast, the coculture of the EH and the SH accumulated a higher amount of glucose in the presence of methylglyoxal. These results confirm that our cellulosome has stronger cellulolytic activity on higher order celluloses, that is, avicel. Since we only used the Lac4 region as an integration site we were unable to integrate both cellulosomal genes (i.e., CipA and OlpB) and dockerin-fused enzyme into a single host. Hence, to achieve efficient secretion and cellulosome assembly, we engineered the cellulosomal genes (SH) and dockerin-fused enzymes (EH) into two different hosts. We adopted the cell consortium approach and the ratio of each host was controlled according to the complexity of the cellulosic substrates to achieve a higher ethanol titer.
In conclusion, we have engineered the “largest enzyme complex” on K. marxianus cell surface and a cellulolytic host for cellulosic ethanol production. The assembly of up to 63 enzymes in a cellulosome complex facilitates enzyme synergism and exhibits much greater degradation potential due to its highly ordered structural organization and assembly, which enables enzyme proximity synergy and enzyme–substrate–microbe complex synergy. Our cellulosome provides a means for simple incorporation of other types of enzymes (i.e., hemicellulases and laccases) into the cellulosome complex. Moreover, this large cellulosome complex could be used for the synthesis of various biopharmaceutical products that involve multiple enzymatic conversion steps (e.g., astaxanthin and morphine).
Materials and Methods
Strains and Media.
Escherichia coli strain DH5α was used as the host for plasmid construction and propagation. E. coli strain JM110 was used to demethylate plasmid and to ligate large DNA fragments. E. coli strains were cultured in Luria–Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with 50 μg/mL ampicillin. Yeast K. marxianus 4G5 strain (30) was used to develop a CBP host, which was maintained in YPG medium (1% yeast extract, 2% peptone, and 2% galactose) and cultured at 30 °C at 300 rpm. Yeast transformants were cultured in YPG media supplemented with either G418 or hygromycin B at a concentration of 200 μg/mL.
Cellulosomal Construct Design and Synthesis.
Scaffoldins (CipA) were designed in a biomimetic manner using the C. thermocellum CipA gene as a backbone, which has nine type I cohesin domains with a CBM. The original CipA was named as CipA1B9C since it has one binding domain (B) and nine cohesins (C). Similarly, synthetic scaffoldins CipA1B6C and CipA1B3C were designed by removing the cohesins 6 through 9 and 4 through 9, respectively. The synthetic scaffoldin CipA2B9C was designed by adding one more CBM domain between cohesin 7 and 8, while CBM was removed to form CipA0B9C. Each CipA construct was fused with a K. marxianus alpha-factor signal peptide and a 12xHis tag at the N termini for secretion and purification purposes, respectively. The anchoring protein (OlpB) mimics the C. thermocellum OlpB, which contains seven type II cohesin domains with an SLH anchoring domain and a long TR. The SLH domain was replaced with the ScGPI domain and the TR domain was removed to avoid the synthesis constraints. A GFP and a FLAG tag were fused in between the cohesin and the ScGPI domain for fluorescence and detection purpose (Fig. 1C). All of the constructs were codon-optimized and difficult sequences were randomized using the Build Optimization Software Tools (BOOST) developed by DOE-Joint Genome Institute (JGI) (49). All of the constructs were synthesized and inserted into the yeast integration plasmid pKLac2-α-Kan using AvrII and NotI.
EgIII and NpaBGS genes were obtained from our previous studies (23, 25). The CBHII (KC311337), LPMO, and CDH genes were chemically synthesized by GeneScript Inc. and codon-optimized for K. marxianus expression (24) (SI Appendix, Table S2). To fuse the C-terminal dockerin each cellulase gene was amplified with gene-specific primers (SI Appendix, Table S3) using High-Fidelity DNA polymerases (New England Biolabs), digested with AvrII and SpeI and inserted into the yeast integration plasmid pKLac2-α-Kan-DocT, which carries a type I dockerin (DocT) of the celS from C. thermocellum. The pKLac2-α-Kan vector was derived from the K. lactis vector pKLac2 (50), and the alpha-factor of K. marxianus and kanamycin (G418) selection marker were cloned into the original vector. The pKLac2-α-Kan plasmids containing target gene was linearized by either enzyme digestion (i.e., SacII or BstXI) or PCR amplification. The gene cassettes with 5′ PLAC4 and 3′ PLAC4 homologous sequences were inserted into the LAC4 promoter region of the K. marxianus genome (50) (SI Appendix, Fig. S5 A and B). The LAC4 promoter was included in each gene integration, so that a copy of LAC4 promoter was available for further integration. The multiple genes were integrated simultaneously by cotransforming the gene cassettes. Previously, Read et al. (51) used this method to integrate four genes into the Lac4 region.
Yeast Transformation and Clone Screening.
K. marxianus cells were cultured in 5 mL YPG medium at 30 °C at 300 rpm for 16 h. Yeast competent cells were prepared as described in Chang et al. (23). SacII-digested gene cassettes (1 µg) were electroporated (1.0 kV, 400 Ω, and 25 μF capacitance) with 40 µL of competent cells in a Bio-Rad system (Gene Pulser Xcell; Bio-Rad) with an aluminum cuvette (2 mm). Then, electroporated cells were recovered at 30 °C for 3 h and plated onto YPG plates supplemented with G418 or hygromycin B (200 μg/mL). Transformed yeast colonies were mixed with QuickExtract DNA Extraction Solution (Epicentre) to remove the cell wall and used as a template for PCR verification. Chromosomal integration of each gene was confirmed by the PCR using gene-specific primers (SI Appendix, Table S3) and the stability was verified up to five generations.
Expression and Purification of Cellulosomal Proteins.
To purify the CipA and OlpB proteins, yeast cells were grown at 37 °C in YPG medium supplemented with hygromycin B/G418 for 48 h. Then, cells were harvested by centrifugation at 8,000 × g for 30 min and supernatant was collected, filtered by 0.2-µm Nalgene Rapid-Flow, and concentrated to 100 mL by an Amicon stirred cells ultrafiltration unit (Millipore). CipA was purified based on CBM, which has an inherent ability to bind cellulose, and 100 mL of concentrated supernatant was incubated with 1 g cellulose for 3 h with gentle stirring at 4 °C. Then, cellulose pellets were recovered by centrifugation at 4,000 × g for 30 min and pellets were washed thrice with Tris-buffered saline (TBS) with 1 M NaCl buffer and thrice with TBS buffer (pH 7.5). The CipA protein was eluted from the pellet by adding 1% triethylamine and the eluted fraction was neutralized using 1 M 2-(N-morpholino) ethanesulfonic acid (MES). The OlpB protein was purified by immobilized metal affinity chromatography (IMAC). Briefly, 100 mL of concentrated supernatant was incubated with 3 mL Ni-NTA Sepharose beads for 30 min at 4 °C with mild constant stirring. Then, beads were loaded onto the column and proteins eluted using TBS buffer, 7.5 pH, with different concentrations of imidazole. The fractions were analyzed using 4 to 15% SDS–PAGE.
DocT was cloned into pET9a vector as His-MBP fusion protein by the ligation-independent cloning method. The pET9a-His-MBP-DocT clone was transformed into BL21 (DE3) E. coli cells, which were grown at 37 °C in LB medium supplemented with 100 mg/mL ampicillin until the OD600 reached about 0.8. The proteins were overexpressed by addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.5 mM. After an additional incubation for 8 h at 16 °C, the culture was harvested by centrifugation at 8,000 × g for 20 min and stored at −80 °C. Cell pellets from a total of 6 L culture was resuspended in 100 mL buffer A (100 mM phosphate buffer, pH 7.5, 500 mM NaCl, 10% glycerol, and 10 mM β-mercaptoethanol). The suspension was lysed by sonication and centrifuged at 35,000 × g at 4 °C for 45 min. The supernatant was loaded onto a nickel affinity column (HisTrap 5 mL; GE Healthcare) preequilibrated with buffer A. The protein was eluted with buffer B (50 mM Tris⋅HCl, 7.5 pH, 500 mM NaCl, and 500 mM imidazole). The fractions containing His-MBP-DocT were pooled and treated with 1 mg of tobacco etch virus (TEV) protease per 50 mg of His-MBP-DocT and incubated at 16 °C for overnight. The digested proteins were then centrifuged at 6,000 rpm for 20 min to remove TEV protease/His-MBP aggregates. The cleaved DocT and His-MBP proteins were further separated by IMAC, and the flow-through was subsequently concentrated to 10 mg/mL. All three purified proteins were dialyzed against TBS buffer, 7.5 pH, and further purified in terms of molecular weight by SEC (Superdex 200 HR 10/30; GE Healthcare), which was preequilibrated with 20 mM Tris, pH 7.5, 150 mM NaCl, 5 mM dithiothreitol, and 5% glycerol. The proteins were further identified by protein mass spectrometry. The resulting proteins were utilized for complex preparation.
Cellulosome Complex Assembly and Purification.
Purification of the entire cellulosome complex is challenging, so we divided the complex into three portions to demonstrate cellulosome complex formation. All three proteins were buffer-exchanged to 50 mM citrate, pH 5.0, 2 mM EDTA, and 1 mM CaCl2 buffer. First, we pooled CipA: DocT (1:10 equimolar ratio) and incubated it for 3 h at 37 °C without shaking. The resulting complex was purified by SEC (Superdex 200 HR 10/30; GE Healthcare), which was preequilibrated with TBS buffer, 7.5 pH, and 1 mM CaCl2. The peak fractions were collected and subsequently analyzed on 4 to 15% SDS–PAGE. The protein bands were further identified by mass spectrometry. Second, we pooled OlpB:DocT (1:10 ratio) and incubated it for 3 h at 37 °C without shaking. The resultant was purified as in the case of CipA:DocT complex. Third, we pooled OlpB: CipA (1:10 ratio) and incubated it at 3 h at 37 °C without shaking. As it was difficulty to purify the large complex by SEC, we used native PAGE to analyze and identify the complex. The resultant CipA:OlpB complex bands were identified by mass spectrometry.
The complex formation of dockerin-fused cellulases and scaffoldins were confirmed by native PAGE. Briefly, culture supernatants of CipA1B9C and NpaBGS-t hosts were collected after 48-h cultivation at 37 °C at 300 rpm and concentrated 50-fold using Vivaspin 20 (10-kDa cutoff; Merck Millipore). Similarly, cells expressing OlpB-ScGPI were harvested and washed with phospate-buffered saline (PBS) and cell lysates were collected after sonication. The complex formation was examined by mixing the supernatants/cell lysates at different ratios in PBS containing 10 mM CaCl2 and the mixture was incubated for 3 h at 37 °C without shaking. After incubation, the enzyme mixture was loaded onto native PAGE (stacking gel 5%; separating gel 8%) with 2× sample loading dye (without SDS) (22, 52).
Immunofluorescence Microscopy and FACS.
Immunostaining was performed using the method described in Chang et al. (16). Yeast cells expressing anchoring protein was collected (A600 of 1.0) and washed three times with PBS (pH 7.4). Cells were resuspended in 4% paraformaldehyde (PFA) and incubated for 30 to 60 min at 4 °C. Then, cells were centrifuged and washed with PBS buffer to remove the excess PFA and resuspended in 500 µL of bovine serum albumin (BSA) (1 mg/mL in PBS) containing 0.5 µg of anti-FLAG primary antibody (Biotools) and incubated for 3 h. The cells were harvested by centrifugation and resuspended in 500 µL of PBS containing secondary antibody conjugated with DyLight 405 fluorophores (Jackson Immunoresearch) and incubated for 1 h. After incubation, the cells were washed three times and 2 µL of cell suspension were fixed on a slide and observed under Leica TCS SP5 II confocal microscopy (Wetzlar). Similarly, immunostained yeast cells were subjected to FACS analysis to further confirm the anchoring efficiency. Approximately 50,000 cells were used for the analysis using MoFlo XDP Flow Cytometer (Beckman Coulter, Inc.).
Cellulosome Assembly on the Cell Surface.
To confirm the assembly of the entire cellulosome complex on the cell surface the RFP was fused with dockerin T (RFP-DocT) and expressed in K. marxianus. The yeast cells expressing OlpB-ScGPI, CipA1B9C, and RFP-DocT were separately cultured for 48 h at 37 °C with 300 rpm. The supernatants of CipA1B9C and RFP-DocT were collected and concentrated as described earlier. The cells expressing OlpB-ScGPI were harvested by centrifugation at 3,000 rpm at 4 °C and suspended in 1 mL concentrated supernatants of CipA1B9C and RFP-DocT. To form cellulosome complex 10 mM CaCl2 was added to the cell mixture, which was then incubated for 3 h at 37 °C without shaking. The cells were harvested and washed thrice with PBS buffer to remove the unbound proteins. Finally, cells were fixed on slides as described earlier and observed using a Nikon Eclipse 90i epifluorescence microscope.
Real-Time Quantitative PCR.
Total RNA was isolated from the transformants using AccuPure Yeast RNA mini Kit in the iColumn automated system (iColumn12; AccuBioMed). Complementary DNA synthesis was conducted using a reverse transcription kit (SuperScript II kit; Invitrogen). The relative quantification of each gene was carried out using gene-specific Universal Probe Library probes (LightCycler W480 Probes Master; Roche) with a specific primer pair (amplicon size around 100 to 150 bp) on a LightCycler (LightCycler 480; Roche) (SI Appendix, Table S4). The actin gene was used as the reference gene and each analysis was repeated three times. The relative expression level was calculated using the 2−∆∆Ct method (53).
Enzyme Assays.
Enzyme activity was assayed as described in Chang et al. (38). The activity of dockerin-fused cellulases was determined by various assays using specific substrates and the commercial enzyme Celluclast 1.5L (cellulase from T. reesei; Novozymes) was used as a positive control. Endo-glucanase (EgIII-t) activity was measured using dye CMC (Megazyme) as a substrate. The 100-μL assay reaction contains 40 μL of concentrated supernatants with 60 μL of buffer solution (0.4% [wt/vol] dye CMC and 50 mM sodium acetate, pH 5), incubated at 40 °C for 6 h. After incubation, 250 μL precipitation solution (containing sodium acetate trihydrate, zinc acetate, and ethanol, pH 5.0) was added and centrifuged at 8,000 rpm for 10 min and absorption was measured at 590 nm. Similarly, exoglucanase (CBHII-t) activity was estimated using PASC. The assay was performed by adding 100 μL of supernatants with 900 μL buffer solution containing 50 mM sodium acetate (pH 5) and 0.4% PASC and incubated in a thermal shaker at 40 °C for 12 h. The released reducing sugars were measured by the dinitrosalicylic acid (DNS) method and sugar concentration was calculated using glucose standard (54). β-Glucosidase (NpaBGS-t) activity was determined by the formation of p-nitrophenol from the hydrolysis of substrate p-nitrophenyl-β-d-glucuronide (pNPG) and absorption was measured at 410 nm. Total cellulase activity was determined using filter paper as the substrate and the reducing sugar release was estimated by the DNS method.
Fermentation and Ethanol Production.
The yeast cells expressing cellulases, scaffoldin, and anchoring proteins were cultured and washed twice with YP medium (1% yeast extract, 2% peptone, and 10 mM CaCl2). Then, 20 OD cells (A600) were resuspended in YP medium, supplemented with 0.042% Tween 80, 1% avicel, or PASC. In the sugar release experiment, 100 mM methylglyoxal was added to the media to inhibit the glucose metabolism of the host. Fermentation was carried out in an anaerobic 50-mL serum tube with 5% volumetric headspace and incubated for several days at 37 °C with agitation at 300 rpm (24). The ethanol and glucose concentrations were analyzed using HPLCy (high-performance liquid chromatography, HPLC) (Jasco PU-2089 Quaternary HPLC pump; Jasco International Co., Ltd.) coupled with the ICSep ICE- COREGEL 87H3 column (Transgenomic) and Shodex RI-101 Refractive Index Detector (ECOM).
Homology Modeling of Dockerin-Fused Enzymes.
To predict the 3D structures of dockerin-fused enzymes, the protein sequence was blasted against the Protein Data Bank database. The online software Protein Homology/analogY Recognition Engine V 2.04 (Phyre2) (55) and RaptorX (56) were used to predict the homology model of each enzyme. Bumps present in the predicted model were removed and missing side chains were added using the WHAT IF Web Interface. The predicted protein structure was validated using a protein structure validation tool. A Ramachandran plot was used to analyze the structure quality. The modeled protein structures were visualized and images were rendered using UCSF Chimera software (57).
Statistical Analysis.
All of the experiments were performed in triplicate and results were expressed as means ± SD. Statistical significance was analyzed using two-tailed Student t tests (with unequal variance) or one- or two-way ANOVA analyses followed by Bonferroni post hoc test. P < 0.05 was considered statistically significant throughout the study.
Data Availability Statement.
All of the data supporting the findings of this study are available within the paper and SI Appendix. The amino acid sequence of the cellulosomal constructs is provided in SI Appendix, Table S5. Additional data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We acknowledge the technical support provided by Institute of Plant and Microbial Biology Flow Cytometry Analysis and Sorting Services, Academia Sinica, Taiwan. This work was supported by Innovative Translational Agricultural Research, Academia Sinica, Taiwan, by the Joint Genome Institute, a Department of Energy (DOE) Office of Science User Facility, and by the Office of Science of the DOE under contract DE-AC02-05CH11231. This work was also supported by the Ministry of Science and Technology (MOST) in Taiwan (MOST 107-2311-B-039-002 and MOST 107-2621-M-039-001). Part of this work was also supported by US Army grant W911NF1910434 and Taiwan Protein Project grant AS-KPQ-105-TPP.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916529117/-/DCSupplemental.
References
- 1.Hirano K., et al. , Enzymatic diversity of the Clostridium thermocellum cellulosome is crucial for the degradation of crystalline cellulose and plant biomass. Sci. Rep. 6, 35709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer E. A., Belaich J.-P., Shoham Y., Lamed R., The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Kruus K., Lua A. C., Demain A. L., Wu J. H., The anchorage function of CipA (CelL), a scaffolding protein of the Clostridium thermocellum cellulosome. Proc. Natl. Acad. Sci. U.S.A. 92, 9254–9258 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J., Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poole D. M., et al. , Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol. Lett. 78, 181–186 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Bayer E. A., Setter E., Lamed R., Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163, 552–559 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demain A. L., Newcomb M., Wu J. H., Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69, 124–154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoav S., et al. , How does cellulosome composition influence deconstruction of lignocellulosic substrates in Clostridium (Ruminiclostridium) thermocellum DSM 1313? Biotechnol. Biofuels 10, 222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumer-Schuette S. E., et al. , Thermophilic lignocellulose deconstruction. FEMS Microbiol. Rev. 38, 393–448 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fierobe H.-P., et al. , Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277, 49621–49630 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Schwarz W. H., The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56, 634–649 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Wen F., Sun J., Zhao H., Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76, 1251–1260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan L. H., Zhang Z. J., Yu X. Y., Xue Y. X., Tan T. W., Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. U.S.A. 109, 13260–13265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai S. L., DaSilva N. A., Chen W., Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2, 14–21 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Liang Y., Si T., Ang E. L., Zhao H., Engineered pentafunctional minicellulosome for simultaneous saccharification and ethanol fermentation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 80, 6677–6684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang J. J., et al. , Biomimetic strategy for constructing Clostridium thermocellum cellulosomal operons in Bacillus subtilis. Biotechnol. Biofuels 11, 157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Currie D. H., et al. , Functional heterologous expression of an engineered full length CipA from Clostridium thermocellum in Thermoanaerobacterium saccharolyticum. Biotechnol. Biofuels 6, 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L. H., et al. , Engineering yeast with bifunctional minicellulosome and cellodextrin pathway for co-utilization of cellulose-mixed sugars. Biotechnol. Biofuels 9, 137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang G. L., Anderson T. D., Clubb R. T., Engineering microbial surfaces to degrade lignocellulosic biomass. Bioengineered 5, 96–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H., et al. , Efficient yeast surface-display of novel complex synthetic cellulosomes. Microb. Cell Fact. 17, 122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambertz C., et al. , Challenges and advances in the heterologous expression of cellulolytic enzymes: A review. Biotechnol. Biofuels 7, 135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arfi Y., Shamshoum M., Rogachev I., Peleg Y., Bayer E. A., Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc. Natl. Acad. Sci. U.S.A. 111, 9109–9114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang J.-J., et al. , PGASO: A synthetic biology tool for engineering a cellulolytic yeast. Biotechnol. Biofuels 5, 53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J.-J., et al. , Assembling a cellulase cocktail and a cellodextrin transporter into a yeast host for CBP ethanol production. Biotechnol. Biofuels 6, 19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H.-L., et al. , A highly efficient β-glucosidase from the buffalo rumen fungus Neocallimastix patriciarum W5. Biotechnol. Biofuels 5, 24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris P. V., et al. , Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Phillips C. M., Beeson W. T., Cate J. H., Marletta M. A., Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Fonseca G. G., Heinzle E., Wittmann C., Gombert A. K., The yeast Kluyveromyces marxianus and its biotechnological potential. Appl. Microbiol. Biotechnol. 79, 339–354 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Lertwattanasakul N., et al. , Genetic basis of the highly efficient yeast Kluyveromyces marxianus: Complete genome sequence and transcriptome analyses. Biotechnol. Biofuels 8, 47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee M. H., et al. , Genome-wide prediction of CRISPR/Cas9 targets in Kluyveromyces marxianus and its application to obtain a stable haploid strain. Sci. Rep. 8, 7305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banat I. M., Nigam P., Marchant R., Isolation of thermotolerant, fermentative yeasts growing at 52 degrees C and producing ethanol at 45 degrees C and 50 degrees C. World J. Microbiol. Biotechnol. 8, 259–263 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Ho C.-Y., et al. , Development of cellulosic ethanol production process via co-culturing of artificial cellulosomal Bacillus and kefir yeast. Appl. Energy 100, 27–32 (2012). [Google Scholar]
- 33.Spencer J. F., Ragout de Spencer A. L., Laluce C., Non-conventional yeasts. Appl. Microbiol. Biotechnol. 58, 147–156 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Chang J.-J., et al. , A thermo- and toxin-tolerant kefir yeast for biorefinery and biofuel production. Appl. Energy 132, 465–474 (2014). [Google Scholar]
- 35.Kondo A., Ueda M., Yeast cell-surface display–applications of molecular display. Appl. Microbiol. Biotechnol. 64, 28–40 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Lemaire M., Ohayon H., Gounon P., Fujino T., Béguin P., OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177, 2451–2459 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caspi J., et al. , Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl. Environ. Microbiol. 75, 7335–7342 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang J.-J., et al. , Constructing a cellulosic yeast host with an efficient cellulase cocktail. Biotechnol. Bioeng. 115, 751–761 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Wieczorek A. S., Martin V. J., Effects of synthetic cohesin-containing scaffold protein architecture on binding dockerin-enzyme fusions on the surface of Lactococcus lactis. Microb. Cell Fact. 11, 160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieczorek A. S., Martin V. J., Engineering the cell surface display of cohesins for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb. Cell Fact. 9, 69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moraïs S., et al. , Establishment of a simple Lactobacillus plantarum cell consortium for cellulase-xylanase synergistic interactions. Appl. Environ. Microbiol. 79, 5242–5249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraïs S., Shterzer N., Lamed R., Bayer E. A., Mizrahi I., A combined cell-consortium approach for lignocellulose degradation by specialized Lactobacillus plantarum cells. Biotechnol. Biofuels 7, 112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern J., et al. , Assembly of synthetic functional cellulosomal structures onto the cell surface of Lactobacillus plantarum, a potent member of the gut microbiome. Appl. Environ. Microbiol. 84, e00282-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sørensen A., Lübeck M., Lübeck P. S., Ahring B. K., Fungal beta-glucosidases: A bottleneck in industrial use of lignocellulosic materials. Biomolecules 3, 612–631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazana Y., et al. , A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnol. Biofuels 6, 182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida E., et al. , Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem. J. 431, 39–49 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Horn S. J., Vaaje-Kolstad G., Westereng B., Eijsink V. G., Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein M. A., et al. , Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175, 5762–5768 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberortner E., Cheng J.-F., Hillson N. J., Deutsch S., Streamlining the design-to-build transition with build-optimization software tools. ACS Synth. Biol. 6, 485–496 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Colussi P. A., Taron C. H., Kluyveromyces lactis LAC4 promoter variants that lack function in bacteria but retain full function in K. lactis. Appl. Environ. Microbiol. 71, 7092–7098 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read J. D., Colussi P. A., Ganatra M. B., Taron C. H., Acetamide selection of Kluyveromyces lactis cells transformed with an integrative vector leads to high-frequency formation of multicopy strains. Appl. Environ. Microbiol. 73, 5088–5096 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morais S., et al. , Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes. mBio 3, e00508-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Ghose T. K., Measurement of cellulase activities. Pure Appl. Chem. 59, 257–268 (1987). [Google Scholar]
- 55.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Källberg M., et al. , Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data supporting the findings of this study are available within the paper and SI Appendix. The amino acid sequence of the cellulosomal constructs is provided in SI Appendix, Table S5. Additional data that support the findings of this study are available from the corresponding authors upon request.