Abstract
Group 3 innate lymphoid cells (ILC3) critically orchestrate host-microbe interactions in the healthy mammalian intestine and become substantially impaired in the context of inflammatory bowel disease (IBD). However, the molecular pathways controlling the homeostasis of ILC3 remain incompletely defined. Here, we identify that intestinal ILC3 are highly enriched in expression of genes involved in the circadian clock and exhibit diurnal oscillations of these pathways in response to light cues. Classical ILC3 effector functions also exhibited diurnal oscillations and lineage-specific deletion of BMAL1, a master regulator of the circadian clock, resulted in dramatically reduced ILC3s selectively in the intestine. BMAL1-deficient ILC3 exhibit impaired expression of Nr1d1 and Per3, hyper-activation of RORγt-dependent target genes, and elevated pro-apoptotic pathways. Depletion of the microbiota with antibiotics partially reduced the hyper-activation of BMAL1-deficient ILC3 and restored cellular homeostasis in the intestine. Finally, ILC3 isolated from the inflamed intestine of patients with IBD exhibit substantial alterations in expression of several circadian-related genes. Our results collectively define that circadian regulation is essential for the homeostasis of ILC3 in the presence of a complex intestinal microbiota, and further that this pathway is disrupted in the context of IBD.
One Sentence Summary:
Circadian regulation is important for intestinal ILC3 homeostasis.
INTRODUCTION
The mammalian gastrointestinal tract is continuously exposed to both pathogenic and commensal microbes, and the immune system must carefully regulate immunity, inflammation and tolerance at this site in order to maintain tissue homeostasis (1–3). Innate lymphoid cells (ILCs) have emerged as critical regulators of these processes due to their enrichment and tissue residency in the intestine, ability to rapidly respond to microbial stimuli, and multiple protective, anti-microbial or immune-modulatory functions (4–8). In particular, group 3 ILCs (ILC3) play numerous roles in regulating intestinal health through production of IL-22, as well as their ability to control adaptive immune responses to dietary antigens or the intestinal microbiota (9–16). Consistent with this, experimental manipulation of ILC3 in mice frequently results in spontaneous inflammation, microbial dysbiosis, or increased susceptibility to tissue damage and infection in the intestine (13, 14, 17, 18). Furthermore, humans with chronic intestinal inflammation or infections exhibit substantially impaired ILC3 responses (14, 15, 19–22), implicating a critical role for these cells in regulating human health.
Although many advances have shed light on how ILC3 maintain intestinal health and become impaired in human diseases, there is a limited understanding of the cellular and molecular pathways that are essential for ILC3 homeostasis in the gut. One identified factor is the microbiota, which can support the development of T-bet+ ILC3 responses in the intestine (23–25). Intriguingly, recent studies have demonstrated that the intestinal microbiota exhibits diurnal oscillation in luminal abundance, epithelial attachment and metabolism (26–29). This circadian cycle of the microbiota also influences the transcriptomic and epigenomic patterns of host cells both local and distal to the gut (27, 30). Mammalian hosts also exhibit diurnal oscillations or a circadian clock. These pathways are regulated by, or consist of, complex neuronal signaling from the synchronization of environmental light cues by the suprachiasmatic nuclei (SCN) in the hypothalamus and release of hormones or metabolites to the periphery (27, 31–33). This can in turn influence multiple immune responses and non-hematopoietic cells through entrainment and a cell autonomous circadian clock (34, 35). In the intestine, it was recently identified that intestinal epithelial cells (IEC) exhibit diurnal oscillation in transcription patterns that are required to orchestrate normal lipid metabolism, and this was in part regulated by the microbiota, IL-22 and IEC-intrinsic expression of NFIL3 (36). However, it remains unclear whether ILC3 exhibit circadian regulation and whether this is important for their cellular homeostasis. Here we provide the first report that ILC3 exhibit diurnal oscillations in expression of genes related to the circadian clock and classical effector functions, and further, that ILC3-specific BMAL1 is required for cellular homeostasis in the presence of a diverse microbiota. Importantly we identify that ILC3 in the inflamed intestine of IBD patients exhibit alterations in circadian gene expression. These findings reveal that circadian regulation critically orchestrates ILC3 homeostasis in the gut, and suggest that disruption in this pathway drives the impaired ILC3 responses in the inflamed intestine of IBD patients.
RESULTS
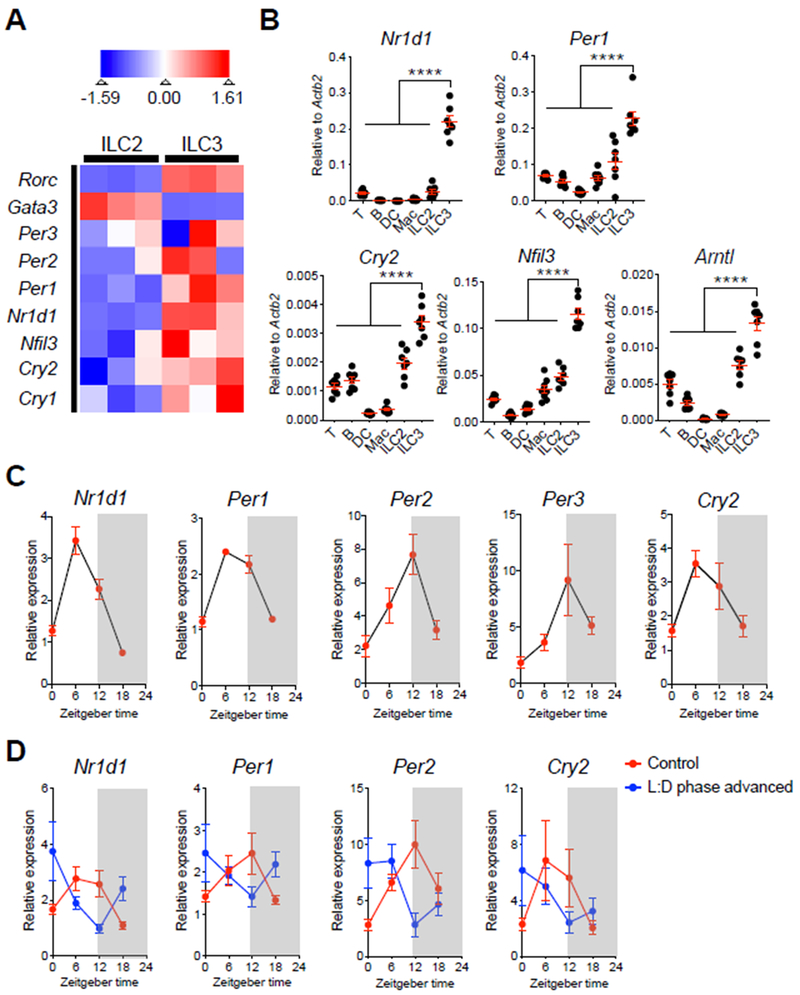
To begin to examine whether ILC3 exhibit circadian regulation, we sort-purified ILC3 from the small intestinal lamina propria (SI-LP) of healthy mice and profiled them with RNA sequencing. Strikingly, in comparison to the related GATA3+ group 2 ILC (ILC2) population, RORγt+ ILC3 exhibited enrichment in multiple transcripts associated with a cell-intrinsic circadian clock, including Per1, Per2, Per3, Cry1, Cry2, Nr1d1 and Nfil3 (Fig. 1A). To verify these findings, we sort-purified several immune cell subsets from the SI-LP of C57BL/6 mice at Zeitgeber time (ZT) 6 (Fig. S1). Quantitative PCR (qPCR) analyses confirmed that among major immune cell subsets in the intestine, ILC3 exhibited significantly higher expression of key circadian genes, including Nr1d1, Per1, Cry2, Nfil3 and Arntl (Fig. 1B). We next examined whether ILC3 express these genes in a diurnal manner by sort-purifying SI-LP ILC3 at 4 time points during a 24-hour cycle. We observed by qPCR analyses that ILC3 express Nr1d1, Per1, Per2, Per3 and Cry2 in a diurnal oscillation pattern (Fig. 1C). Diurnal oscillations of peripheral cell types can be entrained by the suprachiasmatic nuclei (SCN) in the hypothalamus in response to light cues (27, 31–33). Consistent with this, a 9-hour phase shift in the light-dark cycle resulted in a comparable shift in the diurnal expression patterns of Nr1d1, Per1, Per2 and Cry2 (Fig. 1D). In contrast, depletion of a diverse microbiota with antibiotics (Abx) modestly impacted the diurnal oscillations of these circadian genes in ILC3 (Fig. S2A). Thus, our data indicate that intestinal ILC3 exhibit a cell-intrinsic circadian clock that is regulated in part by light cues.
Figure 1. Intestinal ILC3 are enriched for, and exhibit diurnal oscillations in, circadian-related gene expression.
A. Heatmap showing expression Z-scores of the indicated genes in ILC2 (CD45+Lin−CD90.2+CD127+CD27−KLRG1+) and ILC3 (CD45+CD3ε−CD127+RORγtGFP+) from SI-LPs of naïve Rorc(γt)-GfpTG mice, as measured by RNA-sequencing (Lineage markers: CD3ε, CD5, CD8α, NK1.1, CD11c, CD11b, B220). B. qPCR analyses of indicated circadian genes from sort-purified T cells, B cells, dendritic cells (DC), macrophages (Mac), ILC2 and ILC3 from SI-LP of C57BL/6 mice sacrificed at ZT6. The expression of each target gene was normalized to Actb2 (n=7, pooled from 2 independent assays). C. qPCR analyses of diurnal expression patterns in indicated circadian genes on sort-purified ILC3 from SI-LP of C57BL/6 mice sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 (n=4, representative of 2 independent assays). D. qPCR analyses of diurnal expression patterns in indicated circadian genes on sort-purified ILC3 from SI-LP of C57BL/6 mice housed in standard light:dark cycle (control) or mice housed in 9-hour advanced light:dark cycle (L:D phase advanced). Both groups of mice were sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 in the control group (n=6/group, pooled from 2 independent assays). Results are shown as the mean ± s.e.m. Statistics are calculated by one-way ANOVA.
A role for the circadian clock has been implicated in the function of multiple other immune cell types (35). Therefore, we next examined whether the classical effector functions of ILC3 exhibit circadian regulation. ILC3 are characterized by expression of the master regulator transcription factor RORγt and high expression of the effector cytokines IL-17 and IL-22 (10, 37). We observed that both Rorc, Il17a, and Il22 transcripts exhibited diurnal oscillations within a 24-hour cycle (Fig. 2A). We were also able to isolate differential numbers of ILC3s from the intestine in a diurnal manner, observed oscillations of Csf2 expression in ILC3, and found a diurnal expression of Reg3b in intestinal epithelial cells (Fig. S2B–D). We verified diurnal oscillation patterns in intestinal ILC3s at the protein level by the staining intensity of RORγt (Fig. 2B). To verify oscillations in IL-22, we developed a novel IL-22 reporter mouse to permit the analyses of IL-22 production without the need for ex vivo re-stimulation (Fig. S3A) and validated that the GFP signal could robustly represent IL-22 protein production by ILC3 (Fig. S3B, C). Consistent with the oscillations of Il22 transcript, we observed significant changes in IL-22-eGFP levels between ZT6 and ZT18 in intestinal ILC3 from the reporter mice (Fig. 2C). The diurnal oscillations of Rorc and Il22 transcripts were regulated by both light cues and microbiota-derived signals (Fig. 2D, E and S2E, F). Taken together, these data demonstrate that major ILC3 regulators and effector functions also exhibit diurnal oscillations across a 24-hour cycle.
Figure 2. ILC3-specific effector and regulatory pathways exhibit diurnal oscillations.
A. qPCR analyses of Rorc, Il17a and Il22 expression on sort-purified ILC3 from SI-LP of C57BL/6 mice sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 (n=4, representative of 2 independent assays). B. MFI of RORγt diurnal expression in ILC3 from large intestine lamina propria (LI-LP) of mice from Fig. 2A, (n=4, representative of 2 independent assays). C. Representative flow cytometry plots and bar graphs of frequency of IL-22-eGFP expressing ILC3 in SI-LP of IL-22-eGFP mice. Mice were sacrificed at ZT6 and ZT18, cells were gated on live CD45dimLin−CD90.2+CD127+CD27−KLRG1− (n=10-11, pooled from 3 independent assays). D. qPCR analysis of Il22 expression on sort-purified ILC3 from SI-LP of C57BL/6 mice housed in standard light:dark cycle or mice housed in 9-hour advanced light:dark cycle. Both groups of mice were sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The relative gene expression was normalized as Fig. 1D (n=6/group, pooled from 2 independent assays). E. qPCR analysis of Il22 expression on sort-purified ILC3 from SI-LP of conventionally housed C57BL/6 mice or mice treated with Abx for 2 weeks. Both groups of mice were sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The gene expression at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 in the control group (n=6/group, pooled from 2 independent assays). Results are shown as the mean ± s.e.m. Statistic is calculated by unpaired two-tailed Student’s t-test.
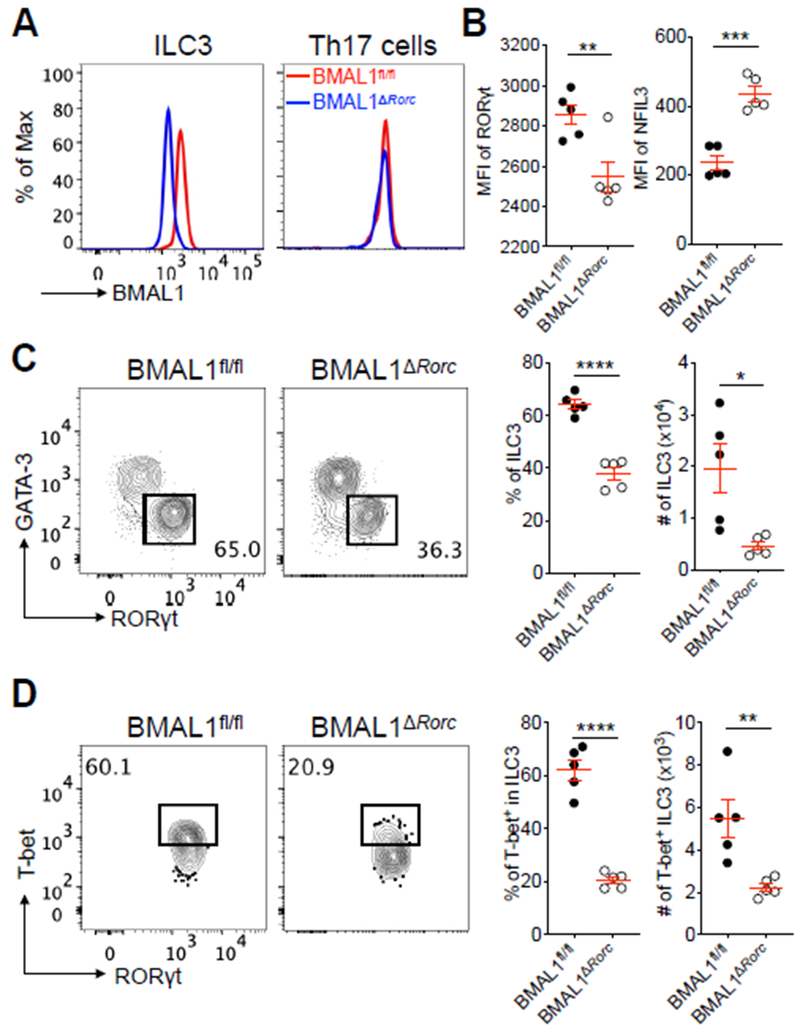
We next examined the functional significance of these diurnal oscillation patterns by generating mice with an ILC3-specific deletion in BMAL1, a master regulator of the circadian cycle encoded by Arntl (38). This was accomplished by crossing Rorccre mice (37) with BMAL1fl/fl mice (39), to generate BMAL1ΔRorc mice that exhibit an efficient loss of BMAL1 protein in intestinal ILC3 (Fig. 3A). Although RORγt is also expressed in double positive thymocytes and maintained in Th17 cells, we did not observe a loss of BMAL1 protein in intestinal Th17 cells in these mice (Fig. 3A), suggesting a selective targeting of ILC3. We found ILC3 from BMAL1ΔRorc mice exhibited significantly altered expression of RORγt and NFIL3 protein (Fig. 3B), indicating a functional impairment in this population. The lack of ILC3-intrinsic BMAL1 also led to significant reductions in both the frequency and cell number of ILC3 in the small intestine (Fig. 3C). This is in contrast to a recent study demonstrating that intestinal Th17 cells, an adaptive counterpart of ILC3, are modestly impacted by BMAL1 deletion (40). Interestingly, we found a preferential requirement of BMAL1 in the T-bet+ subset of ILC3 (Fig. 3D), a population that requires the microbiota for development (23–25). This was selective to small intestinal ILC3, as the cellularity of intestinal ILC2 and Th17 cells, along with ILC3 in lymphoid tissues, remained comparable to littermate controls (Fig. S4A–C). There were also comparable Peyer’s patches (PPs) and myeloid cell-derived cytokines in both groups of mice, and there was no impact on the homeostasis of ILC3 following selective deletion of BMAL1 in T cells (Fig. S4D–F). The frequency and total numbers of ILC3 in the large intestine were comparable in the context of cell-intrinsic BMAL1 deletion, but significant reductions were observed in the T-bet+ ILC3 subset (Fig. S5A, B). Littermate control and BMAL1ΔRorc mice exhibited comparable susceptibility to dextran sodium-induced intestinal damage and inflammation (Fig. S5C, D), which is consistent with the potential redundancy of T-bet+ ILC3 in this model (41). Collectively, these data critically demonstrate that the master regulator of circadian cycles, BMAL1, is essential for the homeostasis of ILC3 selectively within the intestine.
Figure 3. Cell-intrinsic BMAL1 is required for ILC3 homeostasis in the intestine.
A. Representative histogram overlay of BMAL1 expression in SI-LP ILC3 and Th17 cells from BMAL1fl/fl mice and BMAL1ΔRorc mice. ILC3 were gated as live CD45+Lin−CD90.2+CD127+GATA-3−RORγt+, Th17 cells were gated as live CD45+CD3ε+CD4+RORγt+IL-17A+. B. Bar graphs of MFI of RORγt and NFIL3 in SI-LP ILC3 from BMAL1fl/fl mice and BMAL1ΔRorc mice (representative of 3 independent assays). C. Representative flow cytometry plots and bar graphs of frequency and cell numbers of SI-LP ILC3 from BMAL1fl/fl mice and BMAL1ΔRorc mice. Cells were gated on live CD45+Lin−CD90.2+CD127+ (representative of 4 independent assays). D. Representative flow cytometry plots and bar graphs of frequency and cell numbers of T-bet+ ILC3 from SI-LP of BMAL1fl/fl mice and BMAL1ΔRorc mice. Cells were gated on live CD45+Lin−CD90.2+CD127+GATA-3−RORγt+ (representative of 4 independent assays). Results are shown as the mean ± s.e.m. Statistics are calculated by unpaired two-tailed Student’s t-test.
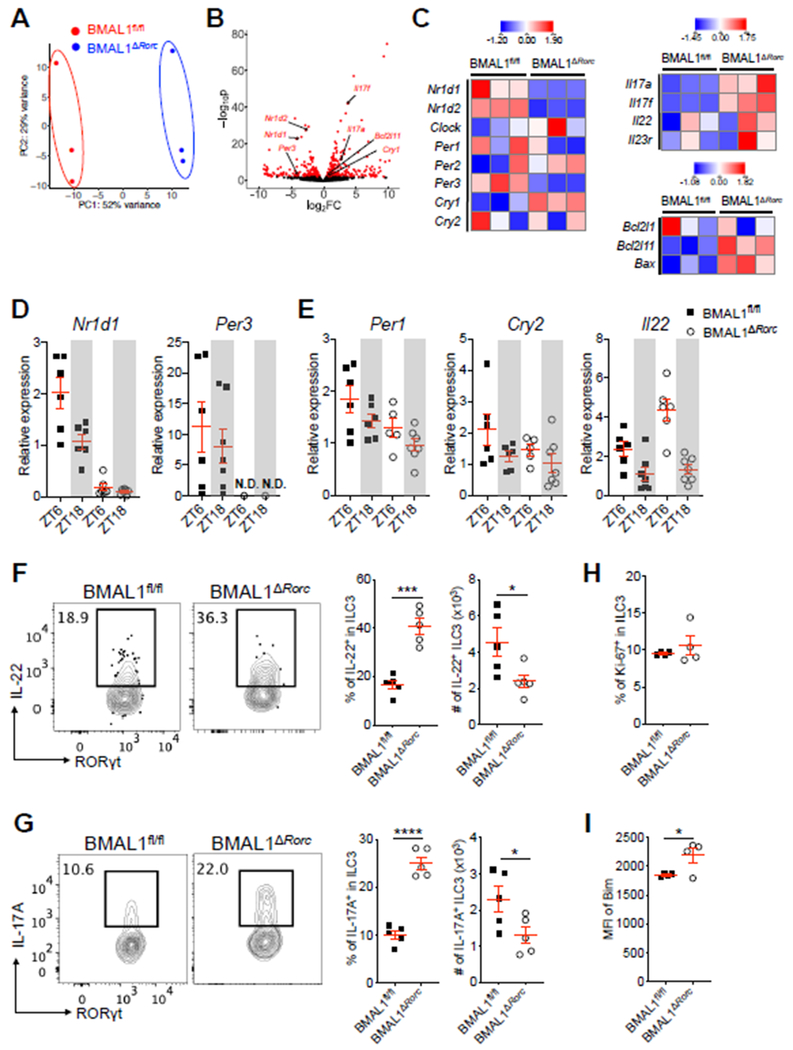
To define how BMAL1 regulates ILC3 cellular homeostasis, we performed RNA-sequencing on sort-purified populations from the small intestine of littermate control and BMAL1ΔRorc mice. Unbiased analyses revealed significant differences in transcriptional signatures, which were driven in part by altered expression of genes associated with circadian-regulation, RORγt-mediated transcription, and apoptosis pathways (Fig. 4A, B). Specifically, BMAL1-deficiency in ILC3 resulted in significantly reduced expression on key circadian-associated genes Nr1d1, Nr1d2, and Per3, while Cry1 was significantly increased and other circadian-related genes remained unchanged (Fig. 4C). NR1D1 (also known as Rev-erbα) is a transcriptional repressor that competes for binding to RORγt DNA consensus sequences (42, 43). Consistent with this, BMAL1-deificient ILC3 exhibited hyper-activation of RORγt-dependent target genes Il17a, Il17f and Il22, as well as the pro-apoptotic pathways Bcl2l11 (encoding Bim) and Bax (Fig. 4C). We verified that BMAL1-deficient ILC3 exhibited significantly impaired expression of Nr1d1 and Per3 at both ZT6 and ZT18, while Per1 and Cry2 were comparable at these time points, and Il22 was significantly increased at ZT6, relative to littermate controls (Fig. 4D, E). Expression of Reg3b in the intestinal epithelium was comparable between both groups of mice, which is likely the result of significantly elevated frequencies of IL-22- and IL-17A-producing ILC3 but reduced total numbers (Fig. 4F, G and S6A). Mice lacking ILC3-specific BMAL1 also exhibited comparable frequencies of proliferating Ki-67+ ILC3 but significantly increased levels of Bim+ ILC3 relative to littermate controls (Fig 4.H, I). The reduced frequencies of ILC3 and increased levels of Bim+ ILC3s were observed as early as 4 weeks of age (Fig. S6B-D). These results collectively demonstrate that ILC3-specific BMAL1 critically promotes expression of several circadian-associated genes (Nr1d1 and Per3), limits hyper-activation of RORγt-dependent target genes (Il17a, Il17f and Il22), and prevents elevated expression of pro-apoptotic pathways (Bim and Bax).
Figure 4. BMAL1-deficiency results in circadian dysregulation, cytokine hyper-production and increased cell death in intestinal ILC3.
A. Principle component analysis (PCA) of genome-wide transcriptional profiles of sort-purified ILC3 from SI-LP of BMAL1fl/fl mice and BMAL1ΔRorc mice, as measured by RNA-sequencing. n=3/group. B. Volcano plot of differential expression between BMAL1fl/fl (positive log2(fold change (FC))) and BMAL1ΔRorc (negative log2(FC)) groups. Differentially expressed genes (defined as False Discovery Rate (FDR) < 0.1) are shown in red. C. Heatmap showing expression Z-scores of the indicated genes in ILC3 from SI-LP of BMAL1fl/fl mice and BMAL1ΔRorc mice. D and E. qPCR analyses of indicated genes on sort-purified ILC3 from SI-LP of BMAL1fl/fl mice and BMAL1ΔRorc mice. Both groups of mice were sacrificed at ZT6 and ZT18. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT6 in the BMAL1fl/fl group (n=6–7/group, pooled from 2 independent assays), N.D., non-detectable. F and G. Representative flow cytometry plots and bar graphs of frequency and cell numbers of IL-22+ ILC3 (F) and IL-17A+ ILC3 (G) from SI-LP of BMAL1fl/fl mice and BMAL1ΔRorc mice. Cells were gated on live CD45+Lin−CD90.2+CD127+GATA-3−RORγt+ (representative of 4 independent assays). H and I. Bar graphs of frequency of Ki-67+ ILC3 (H) and MFI of Bim expression in ILC3 (I) from SI-LP of 8 weeks old BMAL1fl/fl mice and BMAL1ΔRorc mice. (representative of 4 independent assays). Results are shown as the mean ± s.e.m. Statistics are calculated by unpaired two-tailed Student’s t-test.
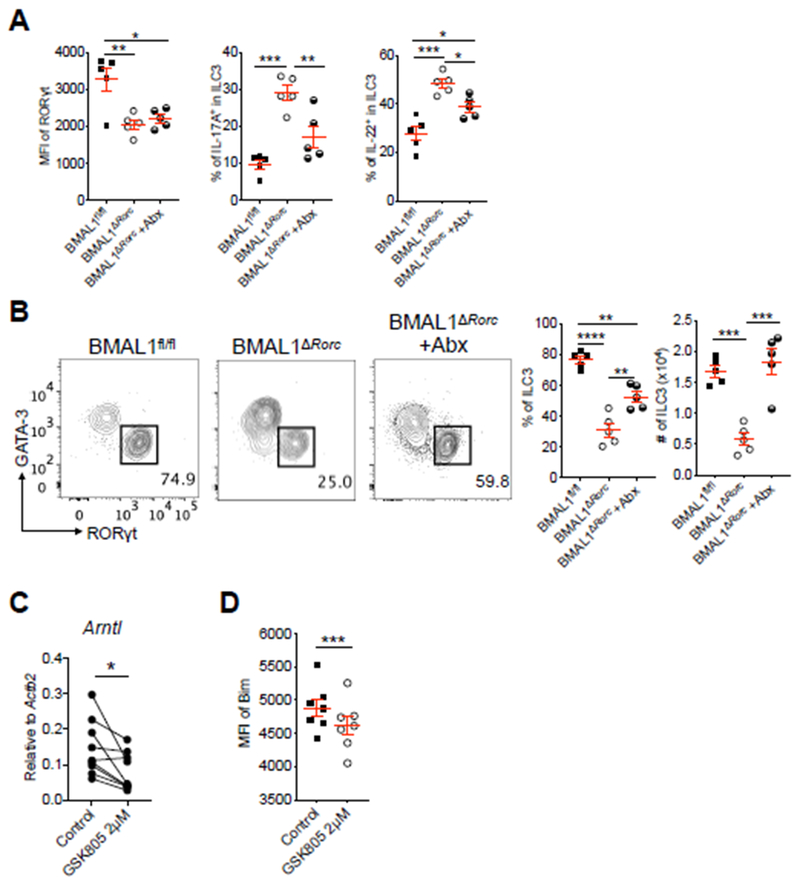
There is a dynamic and reciprocal regulation of ILC3 with intestinal microbiota, which promotes expression of RORγt and its associated target genes (18, 44–46). Given our new findings and previous reports that the microbiota exhibit diurnal oscillations (26–29), we next examined whether the microbiota is driving the observed alterations of ILC3 homeostasis following loss of BMAL1. This was accomplished by administering antibiotics to BMAL1ΔRorc mice. After two weeks of Abx administration, we observed comparable protein levels of RORγt, but significantly reduced levels of IL-17A and IL-22 in ILC3 from Abx treated BMAL1ΔRorc mice relative to non-treated BMAL1ΔRorc mice (Fig. 5A). Strikingly, Abx-treatment also resulted in a partial restoration of ILC3 frequencies and a complete restoration of ILC3 numbers in BMAL1ΔRorc mice (Fig. 5B). These results demonstrate that circadian regulation is essential for orchestrating homeostasis between ILC3 and the microbiota, and implicates that hyper-activation of RORγt is associated with the reduction of intestinal ILC3 in the context of BMAL1-deficiency. In support of this, we identified that small molecule inhibition of RORγt in purified ILC3 cultures was sufficient to reduce expression of BMAL1, as well as significantly reduce Bim+ ILC3 in the context of simulation with pro-inflammatory cytokines that are typically elicited by the microbiota (Fig. 5C, D). Thus, hyper-activation and loss of intestinal ILC3 in the context of cell-intrinsic BMAL deletion is driven in part by the microbiota.
Figure 5. Microbiota drives the dysfunction of ILC3 in the absence of BMAL1.
A. Bar graphs of MFI of RORγt in ILC3, frequency of IL-17+ and IL-22+ ILC3 from SI-LP of Abx treated or control BMAL1fl/fl mice and BMAL1ΔRorc mice (representative of 3 independent assays). B. Representative flow cytometry plots and bar graphs of frequency and cell numbers of SI-LP ILC3 from Abx treated or control BMAL1fl/fl mice and BMAL1ΔRorc mice. Cells were gated as Fig. 3C (representative of 3 independent assays). C. qPCR analysis of Arntl expression on sort-purified SI-LP ILC3 from Rorc(γt)-GfpTG mice treated with 2μM of the RORγt inhibitor GSK805 or vehicle control. The gene expression was normalized to Actb2 (n=9, pooled from 4 independent assays). D. Bar graph of MFI of Bim in SI-LP ILC3 from C57BL/6 mice after incubation with 2μM of the GSK805 or vehicle control, in the presence of recombinant mouse IL-1β, IL-6, IL-12, IL-23 (20ng/ml each) (representative of 2 independent assays). Results are shown as the mean ± s.e.m. Statistics are calculated by one-way ANOVA or paired two-tailed Student’s t-test.
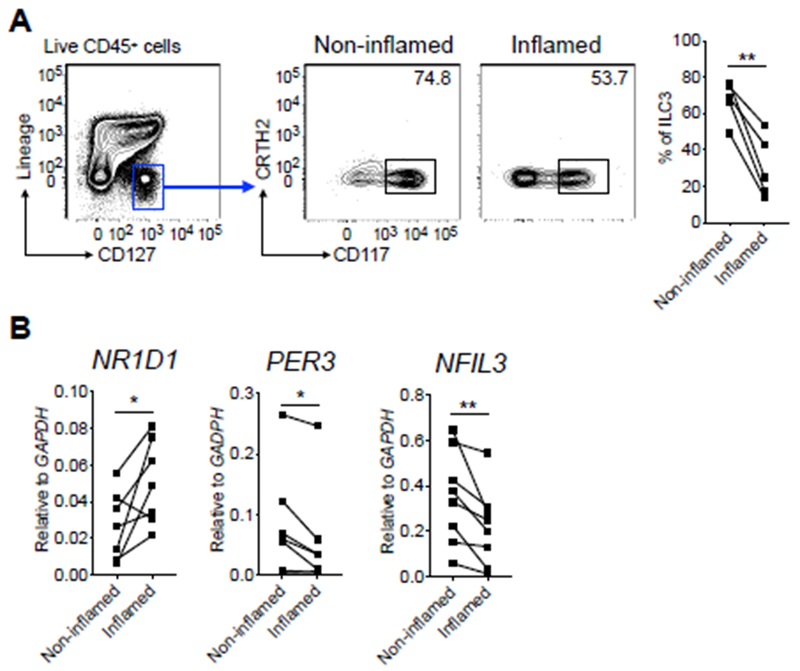
IBD is an inflammatory disease of humans that is accompanied with microbial dysbiosis and disruption of intestinal ILC3 responses (14, 15, 22). To investigate for a contribution of circadian regulation, we sort purified ILC3 from the inflamed or non-inflamed regions of intestinal resections from IBD patients. We observed significantly reduced frequencies of ILC3 from the inflamed versus non-inflamed intestine of IBD patients, and strikingly, this correlated with significantly altered expression of the circadian-related genes NR1D1, PER3 and NFIL3 (Fig. 6A, B), while expression of ARNTL and CRY1 were not substantially altered. These results suggest that in the context of chronic intestinal inflammation, human ILC3 exhibit alterations in circadian gene expression, which may contribute to the altered homeostasis of these protective cell types.
Figure 6. Circadian regulation of ILC3 is disrupted in IBD patients.
A. Representative flow cytometry plots and bar graph of frequency of ILC3 from distal non-inflamed versus matched inflamed surgical resection tissues of IBD patients. Cells were gated on live CD45+Lin−CD127+ (Lineage markers: CD3, CD5, CD11b, CD11c, CD19, FcεR1). B. qPCR analyses of indicated circadian genes on sort-purified ILC3 from distal non-inflamed versus matched inflamed surgical resection tissues of IBD patients. The expression of each target gene was normalized to GAPDH. Statistics are calculated by paired two-tailed Student’s t-test.
DISCUSSION
Our results define that cell-intrinsic circadian regulation is critically required for the homeostasis of ILC3 in the intestine. The mammalian circadian clock plays important roles in regulating multiple immune cell functions (34, 35, 47), recent studies have shed light on how circadian clock influences both innate and adaptive counterparts of immunity, as well as non-immune cells to properly respond to environmental stimuli and maintain homeostasis (36, 48, 49). However, very little is known about the circadian clock and ILC3. An important earlier study demonstrated that ILC2 exhibit circadian regulation and that this was important to support eosinophil homeostasis (50). Here we provide the first evidence that intestinal ILC3 require a circadian clock to better sense and interact with the microbiota, adding another layer of complexity in the dialogue between ILCs and microbes (Fig. S8). This may be the result of coordinating diurnal cycles to facilitate normal ILC3 responses and fine-tune the function of RORγt. In the context of IBD patients, we identified that ILC3 in the inflamed intestine exhibit altered circadian gene expression. This is consistent with prior reports that characterized altered circadian gene expression in whole intestinal biopsies from IBD patients (51). Given that our findings come from the same patient, it suggests that the altered expression is not being driven by body wide disruptions in circadian cycles or entrainment, but rather the result of chronic inflammation or microbial dysbiosis. Inflammatory disruption of circadian cycles has been reported in multiple other cell types (52–54). Moreover, it has become increasingly appreciated that there are certain signatures of immune responses and therapies that exert day and night differences (termed as chrono-immunotherapy), and our understanding of this is already being applied at different levels in diseases such a COPD and rheumatoid arthritis (55, 56). Therefore, our critical finding that ILC3 in the inflamed intestine of IBD patients exhibit altered circadian gene expression suggests that this is an important pathway in human health and disease, and may hold a key for developing novel strategies to boost ILC3 responses in the context of impaired intestinal homeostasis or microbial dysbiosis.
MATERIALS AND METHODS
Study design
The objective of this study was to interrogate the role of circadian regulation on ILC3 homeostasis in the gut. To do this, we employed a combination of ex vivo and in vivo assays with both mouse and human samples. We designed and performed the experiments mainly in the fields of cellular immunology and molecular biology. The number of replicates for each experiment is indicated in the figure legends.
Mice
Wild-type, BMAL1fl/fl and Cd4cre mice on a C57BL/6 background were purchased from the Jackson Laboratory. C57BL/6 Rorccre mice and Rorc(γt)-GfpTG mice were provided by G. Eberl. BMAL1ΔRorc mice were generated by crossing Rorccre mice with BMAL1fl/fl mice and BMAL1ΔCd4 mice were generated by crossing Cd4cre mice with BMAL1fl/fl mice. IL-22-eGFP mice were generated in collaboration with Cyagen US Inc by modifying a Bacterial Artificial Chromosome as noted in Figure S3A and injection into fertilized C57BL/6 embryos. All mice were bred and maintained in specific pathogen-free facilities under a standard 24 hour light:dark cycle (light on for 12 hours from 6am to 6pm as Zeitgeber times [ZT] 0-12) at Weill Cornell Medicine). In one noted experimental set mice were housed with a 9-hour advanced light:dark cycle, and the light was set on from 9pm to 9am. Mice were fed ad libitum. Sex- and age-matched littermates were used as controls in all experiments and mice were between 7 and 11 weeks of age where used for all experiments unless otherwise indicated. Mice were sacrificed at ZT5 to ZT6 unless otherwise indicated. All animal experiments were approved and are in accordance with the Institutional Animal Care and Use Committee guidelines at Weill Cornell Medicine.
Isolation of cells from the intestinal epithelium and lamina propria of mice and humans
Mouse intestines were removed, cleaned from remaining fat tissue and washed in ice-cold PBS (Corning). Peyer’s patches on the small intestine were identified and completely eliminated. Intestines were opened longitudinally and washed in ice-cold PBS. Afterwards, mucus was gently removed by forceps and intestines were cut into approximately 0.5 cm sections. Dissociation of epithelial cells was performed by incubation on a shaker in HBSS (Sigma-Aldrich) containing 5 mM EDTA (Thermo Fisher Scientific), 1mM DTT (Sigma-Aldrich) and 2% heat inactivated FBS two times for 20 min each at 37 °C. After each incubation, samples were vortexed, after the 2nd incubation, the epithelial fraction was washed by ice-cold PBS and proceeded with qPCR analysis. The remaining samples were then washed by cold PBS and enzymatic digestion was performed using dispase (0.4 U/ml; Thermo Fisher Scientific), collagenase III (1 mg/ml; Worthington) and DNase I (20 μg/ml; Sigma-Aldrich) in 10% FBS DMEM (Corning) on a shaker for 45 min at 37 °C. Leukocytes were further enriched by a 40/80% Percoll gradient centrifugation (GE Healthcare).
Surgical resection samples from IBD patients (see Table S1) were obtained through Institutional Review Board approved protocols from the Centers for Advanced Digestive Care at Weill Cornell Medicine following informed consent. Single cell suspensions from intestinal tissues were obtained by incubating tissues for 30 min at 37°C with shaking in stripping buffer (1 mM EDTA, 1 mM DTT and 5% FCS) to remove the epithelial layer. Supernatants were then discarded. Tissues were then mechanically dissociated with a sterile scalpel. The lamina propria fraction was obtained by incubating the dissociated tissues for 1 hour at 37°C with shaking in 2 mg/ml collagenase D (Roche), 0.1 mg/ml DNase I (Sigma) and 1 mg/ml of Trypsin Inhibitor (Gibco) digestion solution. Remaining tissues were then filtered through a 70 μm cell strainer. All cells were then viably cryopreserved in 90% FBS and 10% DMSO for side-by-side analysis at a later time point. Following thawing and filtered through a 70μm cell strainer, cells were stained with antibodies for flow cytometry acquisition.
Flow cytometry and cell sorting
Single cell suspensions were incubated on ice with conjugated antibodies in PBS containing 2% FBS and 1mM EDTA. Dead cells were excluded with Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific). The staining antibodies for flow cytometry were purchased from Thermo Fisher Scientific, Biolegend, BD Biosciences or Cell Signaling Technology. For mouse cell-surface staining: CD45 (30-F11), CD3ε (145-2C11), CD5 (53-7.3), CD8α (53-6.7), NK1.1 (PK136), CD11c (N418), CD11b (M1/70), B220 (RA3-6B2), CD19 (eBio1D3), TCRβ (H57-597), CD64 (X54-5/7.1), MHC-II (M5/114.15.2), CD90.2 (30-H12), CD127 (A7R34), CD27 (LG.7F9), CD4 (GK1.5 or RM4-5), KLRG1 (2F1). For mouse intracellular staining: T-bet (4B10), GATA3 (L50-823), RORγt (B2D or Q31-378), NFIL3 (S2M-E19), Ki-67 (SolA15), Bim (C34C5), IL-17A (eBio 17B7), IL-22 (IL22JOP). Intracellular staining of mouse BMAL1 was conducted by first staining polyclonal anti-BMAL1 antibody (Abcam) then further staining with fluorescent conjugated donkey anti-rabbit IgG antibody (BioLegend). Human samples were stained for CD3 (UCHT1), CD5 (UCHT2), CD11b (CBRM1/5), CD11c (3.9), CD19 (HIB19), CD45 (HI30), CD117 (104D2), CD127 (A019D5), FcεR1 (AER-37) and CRTH2 (BM16).
For intracellular transcription factor or cytokine staining, cells were stained for surface markers, followed by fixation and permeabilization before nuclear factor staining according to the manufacturer’s protocol (Foxp3 staining buffer set from Thermo Fisher Scientific). For intracellular cytokine staining, cells were incubated for 4 hours at 37 °C in complete medium (DMEM with 10% FBS, 10 mM HEPES, 1 mM sodium pyruvate, non-essential amino acids, 80 μM 2-mercaptoethanol, 100 U/ml penicillin and 100 μg/ml streptomycin, all from Gibco), supplied with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml), ionomycin (750 ng/ml) and brefeldin A (10 μg/ml) (all from Sigma-Aldrich). All flow cytometry experiments were performed using a Fortessa flow cytometer and the FACS Diva software (BD Biosciences) and analyzed with FlowJo V10 software (TreeStar) or sort-purified by using FACSAria II cell sorter (BD Biosciences).
RNA sequencing analysis
ILC2 and ILC3 were sort-purified from small intestine of Rorc(γt)-GfpTG mice or BMAL1fl/fl and BMAL1ΔRorc littermates. Sorted cells were used to prepare RNA-seq libraries by the Epigenomics Core at Weill Cornell Medicine using the Clontech SMARTer Ultra Low Input RNA Kit V4 (Clontech Laboratories). Sequencing was performed on an Illumina HiSeq 2500, yielding 50 bp single-end reads.
Raw sequencing reads were demultiplexed with Illumina CASAVA (v1.8.2). Adapters were trimmed from reads using FLEXBAR (v2.4) and reads were aligned to the NCBI GRCm38/mm10 mouse genome using the STAR aligner (v2.5.2b) with default settings. Reads per gene were counted using Rsubread. Genes with at least 50 or more counts in at least 2 samples were tested for differential expression. Differential expression was assessed using DESeq2 version 1.22.2 with default parameters and with a false discovery rate (FDR) of 0.1. Principal component analysis (PCA) was performed after applying the DESeq2 varianceStabilizingTransformation function, using the 500 genes with highest variance.
Quantitative PCR
Intestinal epithelial cells or sort-purified ILC3 were lysed in Buffer RLT (QIAGEN) or single-cell lysis buffer (Clontech-Takara). RNA was extracted via RNeasy mini kits (QIAGEN) as per the manufacturer’s instructions. Reverse transcription of RNA was performed using Maxima reverse transcription or SuperScript VILO according to the protocols provided by the manufacturer (Thermo Fisher Scientific). Real-time PCR was performed on cDNA using SYBR green chemistry (Applied Biosystems). Reactions were run on a real-time PCR system (ABI7500; Applied Biosystems). Samples were normalized to Actb2, Hprt1 or GAPDH and displayed as a fold change or relative values compared to controls (see Table S2 for detailed primer information).
In vivo administration of antibiotics
0.5 mg/mL of ampicillin (Santa Cruz) and 0.5 mg/mL gentamicin (Gemini BioProducts) were continuously administered via drinking water for 2 weeks. Mice were 7-8 weeks of age when antibiotic treatment started.
In vitro stimulation with GSK805
GSK805 (Small molecule RORγt inhibitor) was purchased from EMD Millipore at a purity ⩾ 95%. ILC3 were sort-purified from SI-LP of Rorc(γt)-GfpTG mice and incubated for 6 hours at 37 °C with GSK805 or control vehicle in complete medium, before proceeding with qPCR analysis. Bulk isolated SI-LP cells from C57BL/6 mice were incubated for 4 hours at 37 °C with GSK805 or control vehicle in complete medium, supplied with recombinant mouse IL-1β, IL-6, IL-12, IL-23 (20ng/ml each, all from Thermo Fisher Scientific) and analyzed by flow cytometry.
DSS induced colitis
3.5 grams DSS (MP Biomedicals) was dissolved in 100 ml drinking water. 6-7 weeks old BMAL1fl/fl and BMAL1ΔRorc littermates were given ad libitum access to DSS containing water for 7 consecutive days, the water was changed to normal drinking water from day 8. The mice were sacrificed on day 13 for measuring colon length and histology analysis.
For histology, distal colonic tissues were fixed with 4% paraformaldehyde (bioWORLD), embedded in paraffin, and 5 mm sections were stained with haematoxylin and eosin (H&E) by IDEXX BioResearch. Images were acquired using Nikon Eclipse Ti microscope (Nikon).
Statistical analysis
P values of mouse data sets were determined by one-way ANOVA, unpaired or paired two-tailed Student’s t-test with 95% confidence interval. For human data, significance was determined by a paired two-tailed Students’ t-test with 95% confidence interval. All statistical tests were performed with Graph Pad Prism V8 software. Data are shown as the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Fig. S1. Gating strategy for sorting immune cells from the small intestine of C57BL/6 mice.
Fig. S2. Circadian regulation of ILC3 by light dues and microbiota-derived signals.
Fig. S3. Construct design and experimental validation of BAC transgenic IL-22-eGFP reporter mice.
Figure S4. BMAL1ΔRorc mice have intact ILC2 and Th17 cell responses in the intestine, and similar cellularity of ILC3 in lymphoid tissues.
Fig. S5. BMAL1 deficiency impacts T-bet+ ILC3 in the large intestine.
Fig. S6. BMAL1 deficiency impairs intestinal ILC3 at weaning.
Fig. S7. Analysis of circadian genes in intestinal ILC3 from IBD patients.
Fig. S8. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut.
Table S1. Clinical metadata associated with the human samples.
Table S2. Primers used in the study.
ACKNOWLEDGEMENTS:
We thank members of the Sonnenberg Laboratory for discussions and critical reading of the manuscript. We would also like to thank the Epigenomics Core of Weill Cornell Medicine, Gregory G. Putzel, and the Center for Advanced Digestive Care (CADC).
Funding: Research in the Sonnenberg Laboratory is supported by the National Institutes of Health (R01AI143842, R01AI123368, R01AI145989 and U01AI095608), the NIAID Mucosal Immunology Studies Team (MIST), the Crohn’s and Colitis Foundation of America, the Searle Scholars Program, the American Asthma Foundation Scholar Award, Pilot Project Funding from the Center for Advanced Digestive Care (CADC), an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, a Wade F.B. Thompson/Cancer Research Institute CLIP Investigator grant, the Meyer Cancer Center Collaborative Research Initiative, and the Jill Roberts Institute (JRI) for Research in IBD. Gregory F. Sonnenberg is a CRI Lloyd J. Old STAR. L.Z. and J.G. are supported by fellowships from the Crohn’s and Colitis Foundation (608975 and 519428).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and material availability: RNA sequencing data will be deposited and made publicly available in the Gene Expression Omnibus database upon acceptance of this manuscript.
REFERENCES AND NOTES
- 1.Turner JR, Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9, 799–809 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Belkaid Y, Harrison OJ, Homeostatic Immunity and the Microbiota. Immunity 46, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ, Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artis D, Spits H, The biology of innate lymphoid cells. Nature 517, 293–301 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Eberl G, Colonna M, Di Santo JP, McKenzie AN, Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose CS, Artis D, Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 17, 765–774 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Tait Wojno ED, Artis D, Emerging concepts and future challenges in innate lymphoid cell biology. J Exp Med 213, 2229–2248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vivier E et al. , Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Sonnenberg GF, Fouser LA, Artis D, Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12, 383–390 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D, CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer SP et al. , Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 343, 432–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Pavert SA et al. , Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hepworth MR et al. , Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature 498, 113–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hepworth MR et al. , Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science 348, 1031–1035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L et al. , Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 568, 405–409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenberg GF, Hepworth MR, Functional interactions between innate lymphoid cells and adaptive immunity. Nat Rev Immunol, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto Y et al. , Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenberg GF et al. , Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forkel M, Mjosberg J, Dysregulation of Group 3 Innate Lymphoid Cells in the Pathogenesis of Inflammatory Bowel Disease. Curr Allergy Asthma Rep 16, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geremia A, Arancibia-Carcamo CV, Innate Lymphoid Cells in Intestinal Inflammation. Front Immunol 8, 1296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloverpris HN et al. , Innate Lymphoid Cells Are Depleted Irreversibly during Acute HIV-1 Infection in the Absence of Viral Suppression. Immunity 44, 391–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernink JH et al. , Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol 14, 221–229 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Kiss EA et al. , Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Klose CS et al. , A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature 494, 261–265 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Vonarbourg C et al. , Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity 33, 736–751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaiss CA et al. , Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Thaiss CA et al. , Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 167, 1495–1510 e1412 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Leone V et al. , Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang X, Bushman FD, FitzGerald GA, Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A 112, 10479–10484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weger BD et al. , The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab 29, 362–382 e368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buijs FN et al. , The Circadian System: A Regulatory Feedback Network of Periphery and Brain. Physiology (Bethesda) 31, 170–181 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Dumbell R, Matveeva O, Oster H, Circadian Clocks, Stress, and Immunity. Front Endocrinol (Lausanne) 7, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheiermann C, Kunisaki Y, Frenette PS, Circadian control of the immune system. Nat Rev Immunol 13, 190–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheiermann C, Gibbs J, Ince L, Loudon A, Clocking in to immunity. Nat Rev Immunol 18, 423–437 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Wang Y et al. , The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawa S et al. , Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science 330, 665–669 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA, Circadian clock proteins and immunity. Immunity 40, 178–186 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Storch KF et al. , Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemmers S, Rudensky AY, The Cell-Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell Rep 11, 1339–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song C et al. , Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J Exp Med 212, 1869–1882 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kojetin DJ, Burris TP, REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 13, 197–216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amir M et al. , REV-ERBalpha Regulates TH17 Cell Development and Autoimmunity. Cell Rep 25, 3733–3749 e3738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mortha A et al. , Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343, 1249288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou L, Sonnenberg GF, Essential immunologic orchestrators of intestinal homeostasis. Sci Immunol 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnenberg GF, Artis D, Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med 21, 698–708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tognini P, Thaiss CA, Elinav E, Sassone-Corsi P, Circadian Coordination of Antimicrobial Responses. Cell Host Microbe 22, 185–192 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Yu X et al. , TH17 cell differentiation is regulated by the circadian clock. Science 342, 727–730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen NC et al. , Desynchronization of the molecular clock contributes to the heterogeneity of the inflammatory response. Sci Signal 12, (2019). [DOI] [PubMed] [Google Scholar]
- 50.Nussbaum JC et al. , Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Yu R, Zhu L, Hou X, Zou K, Bidirectional Regulation of Circadian Disturbance and Inflammation in Inflammatory Bowel Disease. Inflamm Bowel Dis 23, 1741–1751 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Leone MJ, Marpegan L, Duhart JM, Golombek DA, Role of proinflammatory cytokines on lipopolysaccharide-induced phase shifts in locomotor activity circadian rhythm. Chronobiol Int 29, 715–723 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Cavadini G et al. , TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci U S A 104, 12843–12848 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westfall S, Aguilar-Valles A, Mongrain V, Luheshi GN, Cermakian N, Time-dependent effects of localized inflammation on peripheral clock gene expression in rats. PLoS One 8, e59808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buttgereit F et al. , Efficacy of modified-release versus standard prednisone to reduce duration of morning stiffness of the joints in rheumatoid arthritis (CAPRA-1): a double-blind, randomised controlled trial. Lancet 371, 205–214 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Canonica GW et al. , Characterization of circadian COPD symptoms by phenotype: Methodology of the STORICO observational study. Eur J Intern Med 43, 62–68 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gating strategy for sorting immune cells from the small intestine of C57BL/6 mice.
Fig. S2. Circadian regulation of ILC3 by light dues and microbiota-derived signals.
Fig. S3. Construct design and experimental validation of BAC transgenic IL-22-eGFP reporter mice.
Figure S4. BMAL1ΔRorc mice have intact ILC2 and Th17 cell responses in the intestine, and similar cellularity of ILC3 in lymphoid tissues.
Fig. S5. BMAL1 deficiency impacts T-bet+ ILC3 in the large intestine.
Fig. S6. BMAL1 deficiency impairs intestinal ILC3 at weaning.
Fig. S7. Analysis of circadian genes in intestinal ILC3 from IBD patients.
Fig. S8. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut.
Table S1. Clinical metadata associated with the human samples.
Table S2. Primers used in the study.