Abstract
Many transition metals are essential trace nutrients for living organisms, but they are also cytotoxic in high concentrations. Bacteria maintain the delicate balance between metal starvation and toxicity through a complex network of metal homeostasis pathways. These systems are coordinated by the activities of metal-responsive transcription factors—also known as metal-sensor proteins or metalloregulators—that are tuned to sense the bioavailability of specific metals in the cell in order to regulate the expression of genes encoding proteins that contribute to metal homeostasis. Metal binding to a metalloregulator allosterically influences its ability to bind specific DNA sequences through a variety of intricate mechanisms that lie on a continuum between large conformational changes and subtle changes in internal dynamics. This review summarizes recent advances in our understanding of how metal sensor proteins respond to intracellular metal concentrations. In particular, we highlight the allosteric mechanisms used for metal-responsive regulation of several prokaryotic single-component metalloregulators, and we briefly discuss current open questions of how metalloregulators function in bacterial cells. Understanding the regulation and function of metal-responsive transcription factors is a fundamental aspect of metallobiochemistry and is important for gaining insights into bacterial growth and virulence.
Keywords: allosteric regulation, metal homeostasis, metal ion–protein interaction, metalloprotein, transcription factor, DNA-binding protein, bacterial transcription
Introduction
Many metals play vital roles for living organisms, often serving as structural or catalytic components of biomolecules that are critical to a variety of fundamental biological processes. Included in the list of essential metals are the first-row transition metals from manganese (Mn) to zinc (Zn), which supply a rich chemical versatility that is not available with purely organic compounds (1–3). A few “textbook” examples of metalloproteins include cytochrome c oxidase in respiration, superoxide dismutase in cellular protection, nitrogenase in nitrogen fixation, photosystem II in photosynthesis, and aconitase in central metabolism. Furthermore, we are still learning about the breadth of influence of these biologically relevant metals. For example, one recent area of discovery is the gut microbiota, specifically the unique metabolic transformations performed by the metalloenzymes that support these microorganisms that have such a pervasive impact on human health and disease (4).
Due to the essential nature of the trace nutrient metals, organisms devote significant resources to ensure a sufficient supply, often in the face of limited environmental availability (5). However, despite being indispensable, metal ion uptake must be restricted for several reasons (6–8). Metal transport across a membrane is often an energy-intensive process (9), so import in excess of the nutritional needs would be a waste of resources. In addition, surplus accumulation must be avoided because the chemical properties of the metal ions render them potentially toxic; excess metal ions can catalyze unwanted reactions and biomolecular damage or lead to the wrong metal binding to metalloproteins or to adventitious sites of other proteins, resulting in inactivation or inappropriate allosteric effects (6, 8).
Maintaining the delicate balance between metal starvation and toxicity is a particular challenge for bacterial pathogens because they face host defense mechanisms that take advantage of the dual nature of the nutrient metal ions to either 1) restrict metal ion access through nutritional immunity strategies, or 2) attack the pathogens with excessive amounts of metal ions (7, 10–12). To maintain the desirable levels of each essential metal, bacteria employ extensive networks of metal acquisition, storage, delivery, and efflux pathways, with each network typically dedicated to controlling the availability and distribution of a single type of metal ion (13–15). These systems are coordinated by metal-responsive transcription factors—also known as metal-sensor proteins or metalloregulators—that detect the level of bioavailability of a specific type of metal (or a subset of metals) in the cell and subsequently regulate the transcription of genes encoding the proteins involved in the networks corresponding to that particular metal (16–19). These metal-responsive transcription factors can also regulate genes encoding metal-independent substitute factors in order to reduce the demand for a limited nutrient (17). Altogether, these critical factors sustain the healthy levels of each transition metal nutrient and enable adaptation to shifting environmental conditions and/or metabolic demands.
Each metal-responsive transcription factor is allosterically regulated, typically by the type of metal for which it is responsible, resulting in positive or negative feedback to the production of the corresponding metal homeostasis systems. In this context, allosteric regulation is the process by which metal binding to specific sites on the protein influences DNA binding by a distant region. Metal either activates or inhibits binding to specific DNA sequences in the promoters of target genes to—depending on the system—activate, repress, or de-repress transcription (Fig. 1). How metal binding to the different types of metalloregulators controls DNA binding is a fundamental question in bioinorganic chemistry. In addition, given the nutritional requirement of microorganisms for various transition metals, as well as the potential for toxicity if these elements are not handled correctly, uncovering the mechanisms of metal homeostasis in pathogens may provide clues about how to combat virulence (17, 20, 21). These metal-responsive proteins also have applications in the design of biosensors, including microorganisms engineered to be sensitive and specific monitoring systems of metal pollution in the environment, as well as for bioremediation of these contaminated environments (22–24). Finally, the bacterial systems are the most extensively studied, perhaps because of the conceptual and technical tractability of these single-cell organisms, but eukaryotes also employ metal-responsive transcription factors to maintain metal homeostasis (for a few recent reviews see Refs. 25–31). These latter systems are more complex, and it is not yet clear to what extent they follow the same principles as the prokaryotic systems.
Figure 1.
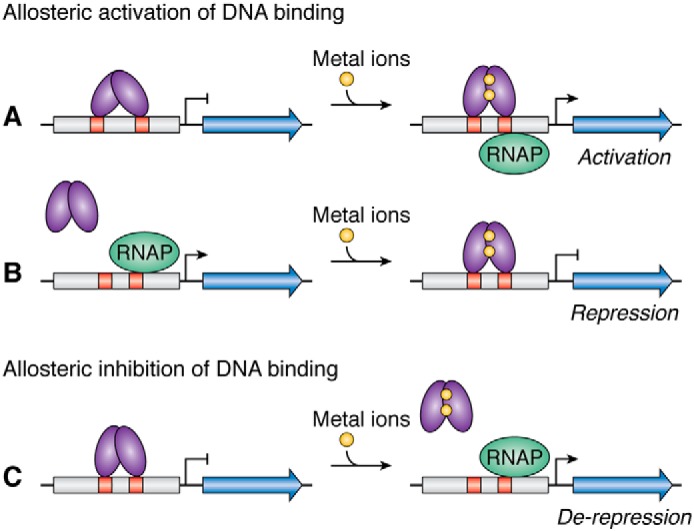
Schematic of metalloregulator responses to metal binding. Metalloregulators (purple) bind to specific recognition sequences (orange) in the promoters (gray) of genes to block access to RNA polymerase (RNAP) or to enhance its activity. Metal binding allosterically stimulates or inhibits DNA binding by the metalloregulator to activate (A), repress (B), or de-repress (C) transcription. For members of the MerR family, the allosteric response to metal binding is similar to the situation in A, and metal binding causes the proteins to distort the DNA structure in a manner that promotes RNAP activity.
This review will highlight recent advances in our understanding of how bacterial metalloregulatory proteins sense and react to metal ions. Given that these sensors must operate in the context of multiple metal ion pools, we start with a brief discussion of the factors that impact the metal selectivity and sensitivity of the sensors. Following an overview of some of the common traits of these proteins, we then focus on the allosteric mechanisms employed by members of specific families. Finally, we briefly touch upon outstanding questions about the mechanistic features of these fascinating proteins.
Set point and selectivity
One of the chemical challenges of employing many metal ions as nutrients is that an adequate supply of each essential metal must be maintained under conditions that minimize mismetallation of metalloproteins. The relative affinities of metal–protein complexes are dictated by the properties of the metal ions and typically follow the Irving-Williams (IW)2 series (Mg(II) < Ca(II) < Mn(II) < Fe(II) < Co(II) < Ni(II) < Cu(II) > Zn(II)) (18, 32). Consequently, there is a risk of mismetallation by metals that are higher in this series than the correct metal (referred to as the cognate metal), resulting in loss of function and/or gain of undesirable reactions (18, 33). For instance, it is estimated that ∼5% of all Escherichia coli proteins utilize iron–sulfur clusters, and mismetallation in these clusters by zinc, copper, or cobalt is known to cause toxicity in E. coli (33–37). To deal with this inherent property of the metals, intracellular metal ions are buffered by cytosolic components at levels inversely related to the IW series, such that the metals at the top of this series, Ni(II)/Cu(II)/Zn(II), are the most strictly limited (18, 38). The cytosolic components that buffer each metal are largely undefined, but along with along with the dedicated storage and delivery proteins as well as adventitious sites on the surface of macromolecules, they likely include a variety of small molecules such as glutathione, amino acids and other organic acids, and inorganic ligands (such as phosphates and sulfates) (18, 39–42). For example, in Bacillus subtilis, bacillithiol was identified as a major component of the Zn(II)-buffering system (43), whereas histidine functions as a Zn(II) buffer in Acinetobacter baumannii (44).
As key players in metal homeostasis, the metalloregulators themselves are tuned to respond to buffered metal concentrations that are inversely related to the IW series (45, 46). This point was highlighted by a recent study that brought together the metal- and DNA-binding affinities—as well as target DNA and protein concentrations—of a collection of Salmonella metalloregulators to calculate the bioavailable concentrations of the cognate metals that elicit a response from each regulator, revealing a spread of a dozen orders of magnitude between the metal ion concentrations that activated the copper regulator versus the Mn(II) sensor (45). Furthermore, the window of acceptable intracellular metal concentrations can be quite narrow, as revealed by analysis of pairs of regulators that control uptake and efflux (40, 47). Although the metal-responsive activities of metalloregulators indicate the healthy levels of each nutrient metal, they may not set these levels (45, 46). For instance, mutant versions of the cyanobacteria Ni(II) regulator InrS with weakened Ni(II) affinity did not cause a corresponding shift in cellular Ni(II) levels (46). Similarly, the loss of function upon moving metalloregulators into new organisms demonstrates that the set points for a given metal depends on the cellular context (48, 49). Altogether, it appears that the set points are defined by cytosolic components that buffer the metals at concentrations inversely related to the IW series, and thus the metalloregulators are tuned to these concentrations to avoid saturating or depleting the buffered systems (46).
Given that metalloregulators themselves bind metals, they are also vulnerable to mismetallation. Furthermore, these proteins typically control the expression of homeostasis factors for one type of metal, so their metal selectivity is of critical importance. Although the details are still being uncovered, it appears that the selectivity of metalloregulators is achieved through a combination of mechanisms. Analysis of individual metalloregulators in vitro suggests that the cognate metal may bind with a unique coordination, signaling that the correct metal is bound and allosterically stimulating the subsequent DNA-binding response (50, 51). However, it is clear that this is not a complete explanation, as the difference between the responses to the cognate versus noncognate metals is often not very large, and there are examples of regulators responding in cells to the wrong metal under conditions when homeostasis is disrupted (52, 53). It is likely that regulators benefit from functioning in the metal-buffered environment of the cytoplasm, limiting exposure to the more competitive metals. Furthermore, it appears that these proteins are designed to function in the company of other metalloregulators that are each fine-tuned to respond to the lowest concentration of their cognate metals (54). Altogether, each metalloregulator is optimized to provide metal-selective genetic control in the host organism, highlighting the exquisite tuning of allosteric regulation for each system.
Metalloregulator families and mechanisms of allosteric control
Metalloregulators are typically homo-oligomeric proteins (dimeric or tetrameric) bearing two DNA-binding domains. The metal-binding sites (i.e. allosteric sites) are at distinct locations from the DNA-binding regions, often at the interface between the two protomers. A common organization is with the DNA-binding domain (DBD) at the N terminus and the metal-binding domain (MBD) at the C terminus. There are several distinct families of metalloregulators, usually named after the founding member(s). Members of a given family generally have similar DNA-binding motifs and protein architecture, but have variations within the signal-sensing motifs, such that they respond to different types of metals, or even nonmetal stimuli (16, 19, 50, 55). Those that respond to nonmetal stimuli are generally not involved in maintaining metal homeostasis, with a few notable exceptions linked to oxidative stress (16, 50, 55).
There are representatives of different metalloregulator families that control the same type of response to the same type of metal, but they are generally found in different organisms, keeping functional overlap to a minimum. For instance, both of the Ni(II)-responsive metalloregulators Nur and NikR repress nickel uptake, but Nur (a member of the Fur family) is found in Streptomyces coelicolor (56), whereas NikR is found in other bacteria such as E. coli and Helicobacter pylori (57). In the cases where organisms express two different regulators that respond to the same metal, they typically regulate metal homeostasis factors with distinct functions, such as import versus export, and are therefore tuned to coordinate metal uptake and efflux in order to keep the amounts of available metals at optimal levels for that organism. For example, B. subtilis employs two Zn(II)-responsive metalloregulators, Zur (from the Fur family) to regulate Zn(II) uptake, and CzrA (from the ArsR/SmtB family) to regulate Zn(II) efflux (58). As an exception, Mycobacterium tuberculosis has two Ni(II)/Co(II) sensors—NmtR and KmtR—from the ArsR/SmtB regulator family that both regulate expression of efflux transporters, albeit with different metal sensitivities (59). Why M. tuberculosis has two similar regulators is not clear, but perhaps it is advantageous for an organism that moves through a variety of physiological conditions to have the ability to cover a wider range of metal concentrations or to potentially coordinate responses to other environmental factors such as pH (60). However, the scarcity of examples of such a strategy suggests that it is not widespread.
Depending on the family, metal binding can either activate or inhibit DNA binding so allosteric regulation is considered in the context of the DNA-bound and DNA-free states, although the MerR family discussed below provides an exception to this model. Cognate metal binding allosterically influences the protein structure, either inhibiting or promoting the conformation in which the relative orientations of the DBDs are amenable to binding to a DNA recognition sequence composed of two-half sites (61). The thermodynamics of the metal- and DNA-binding activities of metalloregulators can be described by the coupling free energy ΔGc, which is calculated using the ratio of KDNA values of the protein when metal-bound and metal-free (ΔGc = −RT ln(KDNA, metal-bound/KDNA, apo) (50, 51). The value of ΔGc is negative when metal binding activates DNA binding (e.g. AdcR from the MarR family, NikR, and the Fur and DtxR families) and positive when metal binding inhibits DNA binding (e.g. CsoR/RcnR, MarR, and ArsR/SmtB families). The magnitude of ΔGc indicates the extent to which binding a specific metal and DNA are linked, and can therefore quantify the relative impact of cognate versus noncognate metals on allosteric control of a metalloregulator (50).
Our current knowledge of the allosteric mechanisms of metal-regulated DNA binding is not yet comprehensive. As ongoing research uncovers more details it is becoming clear that multiple strategies are employed and that they lie on a continuum between the imposition of large changes in three-dimensional structure to more subtle changes in protein dynamics (Fig. 2). As such, these systems can be considered within the broader paradigm of allosteric modulation of protein ensembles (62, 63). The following sections will highlight recent advances in our understanding of metal-responsive regulation of several well-known prokaryotic single-component metalloregulator families, starting with families that exhibit clear structural changes upon metal binding to those that are controlled by changes in internal dynamics, although these mechanisms are not mutually exclusive. This review is not exhaustive, and it focuses on families with well-studied allosteric mechanisms that are representative of the various mechanisms that can be employed and will therefore not discuss all of the known metal sensor families, including those of TetR and CopY (64, 65).
Figure 2.
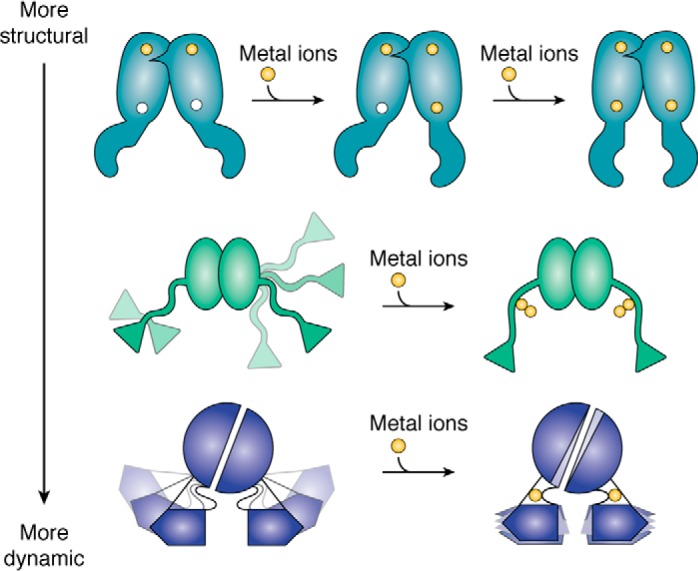
Continuum of the allosteric responses to metal binding that influence DNA binding by metalloregulators. Top to bottom: schematics of the metal ion-induced changes in Zur from the Fur family (teal; adapted from Ref. 79), MntR from the DtxR family (green; adapted from Ref. 98), and AdcR from the MarR family (blue (135)). In each of these examples the apo-protein is in a DNA-binding–incompetent state, and DNA binding is activated upon binding metal (yellow circles). The lightly-colored features of MntR and AdcR represent movement of those regions. For instance, MntR exhibits interdomain flexibility, allowing the DBDs to adopt different orientations relative to the MBD before metal binding (97, 98, 102).
MerR family
Members of the MerR family of transcriptional activators respond to various environmental stimuli—such as drugs, heavy metals (MerR itself responds to Hg(II)), and other chemical species that cause cellular damage—to regulate genes involved in efflux and detoxification (66, 67). This family also includes several proteins that respond to essential metals, such as the Cu(I)-responsive CueR and Zn(II)-responsive ZntR (66, 67). Compared with other metalloregulators, this family exhibits a unique response to metal binding; rather than binding or releasing DNA, the protein remains bound to the DNA and alters the DNA architecture to activate transcription (67–69).
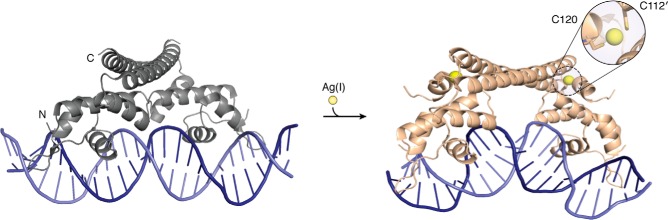
MerR proteins are homodimers with a winged helix–turn–helix DBD of one monomer connected to a long dimerization helix that forms an antiparallel coiled coil with the opposing monomer (Fig. 3) (67, 68). Metals are generally coordinated by a minimum of two residues at the metal-binding loop at the end of the dimerization helix (e.g. CueR), but some members also use a third residue supplied by the N-terminal region of the other dimerization helix (70). In the absence of metal, transcription is repressed due to the suboptimal spacing of the promoter elements, and metal binding allosterically induces a structural change that allows the protein to kink and undertwist the DNA to promote binding by RNA polymerase (67, 68, 70, 71). Recent crystallographic and double electron–electron resonance studies on CueR indicated that the protein–DNA contacts remain the same regardless of metal binding, but metal binding orders the structure of the metal-binding loop, displacing key residues in the hinge between the DBD and dimerization helix, which form new hydrogen bonds to the DBD (68, 71). This rearrangement, supported by a change in hydrophobic packing, pulls the rigid-body DBDs closer together via a scissor-like movement in the dimerization helix, forcing a distortion in the DNA structure (68, 71). Nanometer-precision single-molecule tracking of CueR and ZntR in living E. coli cells revealed distinct concentration-dependent dissociation kinetics from the promoter DNA for the metal-bound and metal-free proteins, adding another layer of complexity to transcriptional regulation by members of this family (72).
Figure 3.
CueR from MerR family. Crystal structures of apo (gray; PDB code 4WLS) and Ag(I)-bound (light orange; PDB code 4WLW) CueR show clear rearrangements in the protein and DNA upon metal binding. The Ag(I) ions are shown as yellow spheres.
Fur family
The Fur family of transcriptional repressors—including Fe(II)-responsive Fur, Zn(II)-responsive Zur, Mn(II)-responsive Mur, and Ni(II)-responsive Nur—are widespread in bacteria and act as global regulators of many operons (Fig. 1B) (73–75). Metal binding activates DNA binding by the proteins in this family, which play important and diverse biological roles. For instance, Mur and Zur regulate the expression of the corresponding metal uptake systems, and Zur is known to regulate additional genes involved in Zn(II) homeostasis as well as protection from oxidative stress, whereas Fur is considered to be a global transcriptional regulator because it regulates the expression of various genes encoding proteins involved in Fe(II) homeostasis, DNA synthesis, energy metabolism, and many more (73, 75).
Members of this family are typically homodimers with two winged-helix DBDs and metal-binding sites at the dimerization domain (73, 75). These proteins generally bind two to three metal ions per monomer, including in many cases a Zn(II) ion required for folding and dimerization that is often referred to as the “structural” metal (73, 76). A key metal-sensing site is located near the hinge region that links the two domains and includes a conserved coordinating histidine (73, 75, 77, 78). Metal binding to this site bridges both domains, drawing them in closer together, along with re-orientation of the N-terminal domains. This rearrangement is thought to stabilize the DBDs in a “caliper-like” conformation that is compatible with DNA binding (73, 77, 78). Furthermore, the Zur dimer can form several metallated species as it binds Zn(II) sequentially at two regulatory sites with negative cooperativity, producing a graded response to intracellular metal availability in B. subtilis that prioritizes scavenging of Zn(II) from cytosolic sources over activation of Zn(II) uptake and emergency back-up systems (76, 79).
Initially classed as a metal-dependent repressor, Fur can also modulate transcriptional activation, and in some organisms there is evidence of direct genetic regulation by apo-protein (73, 74). These complex modes of action may be modulated by the ability of Fur to compact the DNA structure (80), as well as variations between the structures of the different homologs (75). An additional means of regulation may arise through the quaternary structure of the proteins, given that a subclass of Fur homologs are isolated as tetramers in both the apo- and metal-bound states (81, 82). The organization of the proteins within the tetramer prevents DNA binding, but dissociation into the active dimers is promoted by the presence of DNA.
CsoR/RcnR family
The CsoR/RcnR family of transcriptional repressors (Fig. 1C) are tetramers that form all α-helical disk-shaped structures (83). Members of this family have a WXYZ amino acid fingerprint for coordinating metal ions, which inhibits DNA binding (83–85). Proteins in this family have a novel DNA-binding motif, and a structure of a complex with DNA has not yet been solved, but it is thought that two proteins bind per operator in an interaction that is mediated by several surface-exposed clusters of basic residues (83, 86). In the case of the Cu(I)-responsive CsoR—which regulates expression of a Cu-efflux pump—Cu(I) binding is believed to influence a hydrogen-bonding network that is anchored by a coordinating His and spans two subunits, generating a more compact structure with fewer exposed positively-charged residues in order to allosterically inhibit DNA binding (61, 87, 88). Furthermore, analysis of CsoR from Geobacillus thermodenitrificans revealed that Cu(I) binding quenches the mean residue mobility, and an N-terminal region packs against the Cu(I)-binding pocket to stabilize the metal-bound state (84, 89), although these changes may vary between copper-binding homologs of this family.
The other founding member of this family is the Ni(II)/Co(II)-responsive RcnR that regulates the expression of a Ni(II) and Co(II) efflux pump (60). The response to cognate metal binding includes residues right at the N terminus of the protein (83, 85), unlike CsoR, suggesting a variation on the mechanism of allosteric regulation. This was highlighted by a recent study that investigated the structural flexibility of the protein using hydrogen–deuterium exchange coupled with mass spectrometry (90). The biggest impact of cognate metal binding was a decrease in flexibility at the N terminus and mutation of two highly-conserved basic residues in this region greatly abrogated the transcriptional inhibition by apo-protein. Altogether, the results suggest that re-organization upon metal binding would impact key nonspecific electrostatic interactions with the DNA. Another recent study demonstrated that the molecular mechanisms for responding to Co(II) and Ni(II) are distinct, because coordination of Co(II)—but not Ni(II)—forms an inter-subunit linkage to drive allostery in a manner similar to that of formaldehyde-responsive FrmR from the same family (91, 92). The response to Ni(II) is still being investigated but does not appear to depend on its coordination geometry (91).
DtxR family
Members of the Fe(II)-responsive DtxR family and the Mn(II)-dependent subfamily generally repress transcription of metal uptake systems upon metal binding (Fig. 1B) (93, 94). These proteins are homodimers composed of a helix–turn–helix DNA-binding motif at each N terminus, a dimerization domain, and usually an additional C-terminal domain referred to as the FeoA domain because its fold is similar to that of the bacterial Fe(II)-transport protein FeoA (95–97). Members of this family bind metals at the interface between the N- and C-terminal domains in two conserved and structurally distinct metal-binding sites per monomer, which appear to have differing roles depending on the system (95–99). For instance, DxtR requires stepwise binding to both sites for full allosteric activation, but the Mn(II)-responsive PsaR uses only one site as the primary regulatory site (96, 99). Recent studies on the Mn(II)-sensing MtsR from Streptococcus pyogenes revealed a second site that is structurally unique compared with its paralogs, and a single Ala substitution of a metal ligand only caused a partial loss of repression, suggesting that metal sensing by the two sites under varying levels of manganese limitation may fine-tune the degree of repression of the Mn(II) acquisition system (100).
The common mechanism of allosteric regulation of the DtxR family involves a metal-induced disorder–to–order transition in the N terminus that reorients the DBDs to promote DNA binding (98, 101). The Mn(II)-responsive MntR exhibits interdomain flexibility such that the DBDs can adopt different orientations relative to the dimerization domain, until Mn(II) or Cd(II) binding causes rigidification of an α-helix connecting the dimerization domain and DBDs to restrict the conformation of the protein and promote DNA binding (97, 98, 102). Oligomerization on target promoters is also necessary for gene regulation by some family members (100, 103, 104), which may involve the FeoA domain (100). The residues involved in the FeoA domain interactions and protein oligomerization are highly conserved among the Mn(II)-responsive subfamily, suggesting that FeoA-dependent oligomerization on the target promoter may be part of their mechanism of regulation (100).
NikR
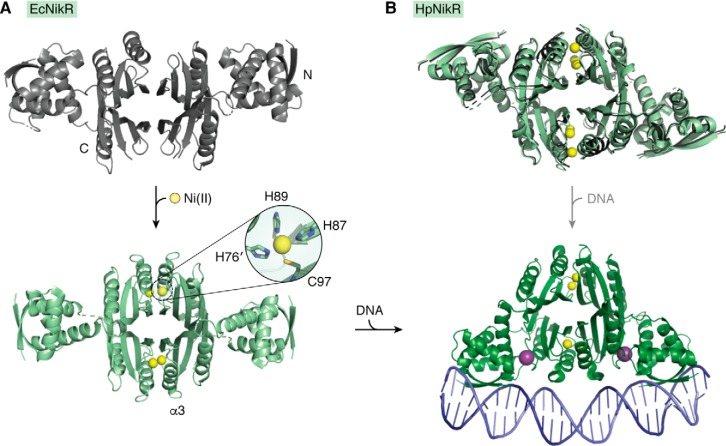
The Ni(II)-responsive NikR is a homotetramer, with a central MBD connected by flexible linkers to two ribbon–helix–helix DBDs (105–107). NikR is believed to exist in a conformational equilibrium between different states characterized by the positions of the DBDs relative to the MBD regardless of Ni(II) binding, which is uncommon for metalloregulators (105–108). In the DNA-bound complex, the DBDs face the same side of the protein in the “cis” conformation (109), but nickel binding does not activate the large change in tertiary structure needed to achieve this conformation (Fig. 4) (105–108), so metal-dependent activation of DNA binding must employ alternative strategies that appear to differ between NikR homologs (57, 110). In the case of E. coli NikR (EcNikR), which is a transcriptional repressor of nickel import (Fig. 1B) (111), Ni(II) binding induces short-range effects, causing a disorder–to–order transition of α-helix 3 in the MBD and its proceeding loop that contains a Ni(II)-binding ligand (Fig. 4A) (107, 112, 113). Ordering of this region is thought to localize EcNikR to DNA through nonspecific electrostatic contacts with the phosphodiester backbone, allowing the protein to initiate a one-dimensional search along the DNA (109, 114). As it moves along the DNA, the DBDs continuously make transient contacts, only forming specific contacts and adopting the “cis” conformation upon finding the recognition sequence, which is then bolstered by potassium binding at the inter-domain interface (109, 114).
Figure 4.
NikR from E. coli and H. pylori. A, crystal structures of EcNikR in the apo (gray; PDB code 1Q5V) and Ni(II)-bound (light green; PDB code 2HZA) forms reveal ordering of α-helix 3 in the metal-binding domain. B, the superimposed structures of HpNikR in the apo (gray; PDB code 2CA9) and Ni(II)-bound (light green; PDB code 2CAD) forms exhibit little structural differences. The Ni(II)–EcNikR–DNA structure (green; PDB code 2HZV) has the DBDs in the “cis” conformation. Although there is no DNA-bound structure of HpNikR, it is believed to bind DNA in the same conformation as EcNikR (depicted by gray arrow). Ni(II) ions are shown as yellow spheres and K+ are shown as purple spheres.
The H. pylori homolog of NikR (HpNikR) has a more complex activity than other known NikR proteins because it can function as either an activator or a repressor to regulate the transcription of a slate of genes in a temporally graded fashion (Fig. 1B, repression and activation of transcription) (60, 115, 116). HpNikR also binds with a range of affinities to the various target sequences (117), which have a weak consensus, and there is evidence that the DNA-bound complexes are in different conformations (118). How Ni(II) activates this DNA-binding response by HpNikR is not yet clear, and unlike EcNikR there is no evidence for a local structural change (Fig. 4B) (105, 119, 120). However, HpNikR has unique features compared with its homologs, including longer interdomain linkers and a nine-residue extension on its N terminus (105, 121), as well as several Ni(II) coordination sites observed under different conditions (120, 122). In addition, it exhibits a functional response to the acidic pH that H. pylori would experience in its colonization environment, the human stomach (123–125). It is likely that Ni(II) binding induces an allosteric effect propagated through either the linkers or the inter-domain interface to influence the motion of the DBDs and promote DNA binding (108, 120, 126). In addition, the protein is proposed to become stabilized in the “cis” conformation upon binding its target DNA sequence, so the DNA may play a significant role in tuning the activity of HpNikR.
MarR family
The ubiquitous MarR family of transcription factors includes proteins that respond to antibiotics, metabolic intermediates, and indicators of oxidative stress, among other inducers (127). Only a few metal-responsive members of this family have been identified, including the Zn(II)-responsive AdcR that represses the transcription of genes involved in Zn(II) uptake (128). MarR proteins share a homodimeric structure with two winged helix–turn–helix motifs at the base of a “pyramidal” structure. In many of these proteins, ligand binding or oxidation occurs in roughly the same relative location sandwiched between the dimerization and DNA-binding regions, which modulates DNA binding.
Distinct allosteric mechanisms have been proposed for different MarR proteins. For example, Cu(II) oxidizes a cysteine residue in the DNA-binding helices of E. coli MarR, creating disulfide bonds between two MarR dimers to induce tetramer formation and block DNA binding (Fig. 1C) (129). In contrast, DNA binding by AdcR is activated by Zn(II) binding (Fig. 1B) (128). Initial structural analysis of AdcR suggested that Zn(II) binding activates a conformational switch that rotates and re-orients the DNA-binding motifs with respect to the central core so that the protein is poised to bind DNA, possibly through a hydrogen-bond network connecting a Zn(II) ligand to the DNA recognition helix (130, 131). Analysis of the homologous ZitR from Lactococcus lactis revealed that stepwise binding of two Zn(II) ions to neighboring sites induced switching of the coordinating nitrogen of a His ligand from site 1 to site 2, which “locks” the protein into the optimal DNA-binding conformation (131). Further biophysical solution studies of AdcR revealed that Zn(II) binding results in an allosteric redistribution of protein dynamics, quenching the local and global motions to focus the conformational ensemble around the more compact structure required to bind DNA, while simultaneously activating fast timescale motions in the DNA-binding motifs, resulting in both entropic and enthalpic contributions to ΔGc (132). The proposed model, which remains to be tested, is that the dynamics of the DNA-binding competent structure are conserved; for cases of ligand-dependent activation, the effector would quench larger-scale dynamics to activate DNA binding (as with AdcR), whereas for ligand-responsive inhibition, effector binding would further narrow the conformational ensemble to a state incompatible with DNA binding (most of the rest of the MarR family), a scenario supported by studies of other members of this family (127).
ArsR/SmtB family
The ArsR/SmtB family of transcriptional repressors is widespread, with at least one version of ArsR/SmtB encoded in most of the sequenced bacterial genomes (55), and this family exhibits an unusual diversity in terms of the location and composition of the inducer-binding sites. Members adopt a homodimeric winged-helical structure (55, 133), and binding of the metal inducer, or in some cases metalloids or nonmetal signals, inhibits DNA binding and results in de-repression of the downstream genes (Fig. 1C). In the apo-form, the metal-responsive members of this family repress the expression of genes involved in metal efflux or metal ion sequestration (134).
One of the best-studied proteins in this family is Zn(II)-responsive CzrA from Staphylococcus aureus, which regulates the expression of a Zn(II) efflux pump and provides an example of an entropic model of allostery in a metalloregulator (135). Analysis of apo- and Zn(II)-CzrA revealed similar structures that do not resemble the DNA-bound protein, which exhibits global rearrangements of the wing domains into a more “closed” conformation that allow for concurrent contacts of the DNA major grooves by the two DNA-binding domains (Fig. 5) (134, 136). The lack of structural changes upon metal binding led to the model that Zn(II) binding impacts the internal dynamics of the protein, limiting the conformational ensemble and the ability to adopt the structure that is appropriate for DNA binding, and that the protein undergoes an induced fit to the more “closed” conformation upon binding DNA (136, 137). Furthermore, an H-bond network originating from the nonligating nitrogen of a His ligand was identified as key for allosteric coupling (133). Additional NMR studies demonstrated that the target DNA increases the flexibility of methyl-side chains throughout the apo-protein, consistent with an entropic driving force for DNA complex formation (135). Zn(II) binding was found to redistribute the fast dynamics, thereby quenching a network of conditional motions and causing inhibition of DNA binding (135). The functional dynamics of CzrA may also include surface water molecules, which should be considered a part of the conformational ensemble that responds to Zn(II) binding (138). This model of allosteric regulation bypasses the need for large-scale structural rearrangements or even a specific molecular path linking the metal-binding site to the DNA-binding motifs, and it may provide a common mechanism that explains the regulation of the many proteins in this family with distinct inducer binding sites.
Figure 5.
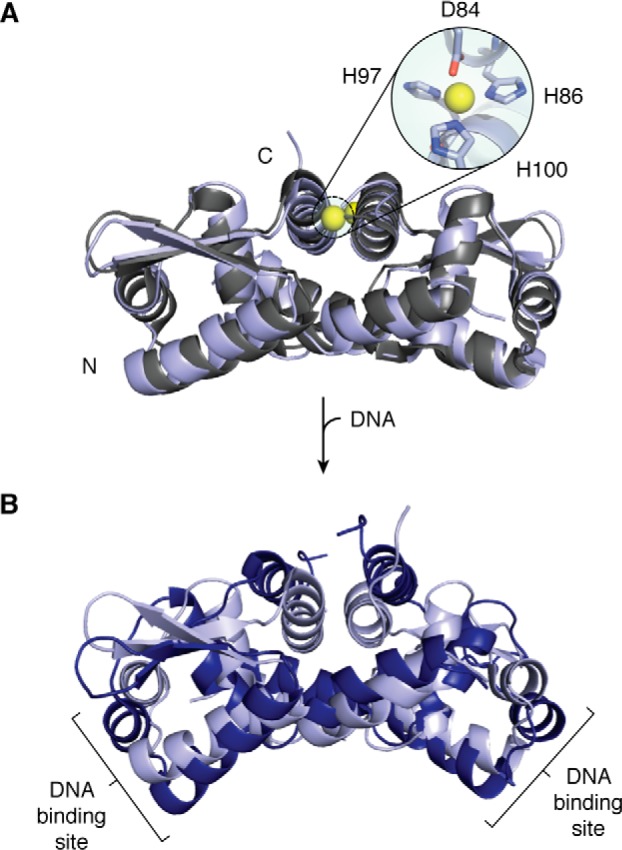
CzrA from ArsR/SmtB family. A, overlay of the crystal structures of apo- (gray; PDB code 1R1U) and Zn(II)-CzrA (light blue; PDB code 2M30) reveals very similar structures. The Zn(II) ions are shown as yellow spheres. B, crystal structure of the DNA-bound protein (blue; PDB code 2KJB, DNA not shown) superimposed on the Zn(II)-bound protein (light blue; PDB code 2M30) shows some structural changes, corresponding to an induced fit upon DNA binding.
Concluding remarks and perspectives
Metalloregulators are key players in maintaining homeostasis of essential transition metals; therefore, understanding how they sense and respond to the intracellular availability of their cognate metal is of fundamental importance. Recent research revealed that metalloregulators are tuned to sense and respond to changes within a narrow range of healthy intracellular metal concentrations and that this window differs substantially for each type of nutrient metal. In general, cognate metal binding elicits an allosteric change in the metalloregulator to activate or inhibit DNA binding. Research into various metalloregulators has made it clear that the allosteric response can occur through a variety of different mechanisms, falling on a continuum from large conformational changes in tertiary structure to more subtle adjustments in dynamics. In some cases, metal binding has a domino effect on the protein structure, resulting in a conformation that is activated for, or prevented from, binding the target DNA sequence. However, in other cases the metal-binding residues seem to be largely pre-organized in the absence of metals, so metal binding does not have a dramatic impact on tertiary structure, and the manner in which DNA binding is impacted involves changes in secondary structure and/or a redistribution of protein dynamics.
The concept of allostery driven by changes in dynamics, in the absence of clear-cut conformational transformations, is becoming well-established, although it has only recently been applied to metalloregulators and has not yet been elucidated for any metalloregulator larger than a homodimer (132, 135, 139–144). As such, the classical models of allostery—the concerted (145) and sequential (146) models—describing rigid-body conformational changes cannot account for every case. In its place, due to advancements in solution NMR spectroscopy and molecular dynamics, the ensemble model of allostery has emerged in recent years. In this model, a protein exists as an ensemble of conformations sampled according to their energy landscape, and binding of an inducer remodels the landscape to promote the biological function (62, 147). As exemplified by CzrA (from the ArsR/SmtB family), a redistribution of fast internal dynamics can also contribute to allostery (135, 148). It is clear that our view of allostery is continuing to evolve, and because allosteric regulation is ubiquitous in biology, the mechanisms learned for metalloregulators can be applied to understand allosteric regulation of various other systems, such as signal transduction, metabolism, and enzymatic activity.
Beyond the mechanisms of allostery, there are a variety of other questions about metalloregulators that are becoming more tractable, building on our increasing understanding of these systems along with technical improvements. For example, as with other transcriptional regulators, one functional challenge to metalloregulators is finding and recognizing the specific target sequence within the haystack of genomic DNA. Given the small number of recognition sites and the strictly controlled regulation, metalloregulators afford excellent model systems to explore the strategies used by DNA-processing factors to locate their target sequences. Along the same lines, most of the studies to date have focused on thermodynamic analysis of the isolated DNA complexes, but it is likely that, at least in some cases, the activities of these factors will be impacted by the complex kinetic environment of the cell, and by cooperation with other protein factors, particularly in the cases of transcriptional activation.
In terms of understanding how each metalloregulator is able to properly function in cells and—in the context of each other—to regulate metal homeostasis, several outstanding questions remain. Some questions that we are just starting to unravel include the following. What is the source of the metal ions that the metalloregulators are sensing? Are the metalloregulators responding to slight changes in the balance of metals as buffered by the background of cytoplasmic metal chelators or are there specific factors that help tip the balance and load or unload the regulator? Beyond the initial response, is recycling needed to reset the systems? For example, in the case of regulators that are activated upon binding metals with very tight affinities, is there a mechanism to unload or degrade the proteins if the levels of available metal ions subside? As more research reveals the details of the different sensors, we gain a clearer picture of the regulation and function of metal-responsive transcription factors. This information is not only important for untangling the virulence of various bacterial pathogens but in the broader context of understanding how dysregulation of seemingly separate pools of labile metals contributes to various disease states in living systems (149, 150).
Acknowledgments
We thank Dr. M. D. Jones, Dr. M. J. Lacasse, and J. Augustine for critical comments on this manuscript.
This work was supported in part by funding from the Natural Science and Engineering Research Council (Canada). The authors declare that they have no conflicts of interest with the contents of this article.
- IW
- Irving-Williams
- DBD
- DNA-binding domain
- MBD
- metal-binding domain
- EcNikR
- E. coli NikR
- HpNikR
- H. pylori NikR
- PDB
- Protein Data Bank
- RNAP
- RNA polymerase.
References
- 1. Andreini C., Bertini I., Cavallaro G., Holliday G. L., and Thornton J. M. (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218 10.1007/s00775-008-0404-5 [DOI] [PubMed] [Google Scholar]
- 2. Zamble D. B., Rowińska-Żyrek M., and Kozlowski H. (eds) (2017) The Biological Chemistry of Nickel, Vol. 10, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 3. Bertini I., Gray H. B., Stiefel E. I., and Valentine J. S. (eds) (2007) Biological Inorganic Chemistry Structure and Reactivity, University Science Books, Sausalito, CA [Google Scholar]
- 4. Rajakovich L. J., and Balskus E. P. (2019) Metabolic functions of the human gut microbiota: the role of metalloenzymes. Nat. Prod. Rep. 36, 593–625 10.1039/C8NP00074C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Merchant S. S., and Helmann J. D. (2012) Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60, 91–210 10.1016/B978-0-12-398264-3.00002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lemire J. A., Harrison J. J., and Turner R. J. (2013) Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384 10.1038/nrmicro3028 [DOI] [PubMed] [Google Scholar]
- 7. Palmer L. D., and Skaar E. P. (2016) Transition metals and virulence in bacteria. Annu. Rev. Genet. 50, 67–91 10.1146/annurev-genet-120215-035146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macomber L., and Hausinger R. P. (2011) Mechanisms of nickel toxicity in microorganisms. Metallomics 3, 1153–1162 10.1039/c1mt00063b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klein J. S., and Lewinson O. (2011) Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics 3, 1098–1108 10.1039/c1mt00073j [DOI] [PubMed] [Google Scholar]
- 10. Becker K. W., and Skaar E. P. (2014) Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 38, 1235–1249 10.1111/1574-6976.12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hood M. I., and Skaar E. P. (2012) Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 10, 525–537 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheldon J. R., and Skaar E. P. (2019) Metals as phagocyte antimicrobial effectors. Curr. Opin. Immunol. 60, 1–9 10.1016/j.coi.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Outten F. W., and Twining B. S. (2008) in Wiley Encyclopedia of Chemical Biology (Begley T. P., ed) pp. 1–10, John Wiley & Sons, Inc., New York [Google Scholar]
- 14. Waldron K. J., and Robinson N. J. (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 10.1038/nrmicro2057 [DOI] [PubMed] [Google Scholar]
- 15. Sydor A. M., and Zamble D. B. (2013) Nickel metallomics: general themes guiding nickel homeostasis. Metal Ions Life Sci. 12, 375–416 10.1007/978-94-007-5561-1_11 [DOI] [PubMed] [Google Scholar]
- 16. Capdevila D. A., Edmonds K. A., and Giedroc D. P. (2017) Metallochaperones and metalloregulation in bacteria. Essays Biochem. 61, 177–200 10.1042/EBC20160076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandrangsu P., Rensing C., and Helmann J. D. (2017) Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350 10.1038/nrmicro.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foster A. W., Osman D., and Robinson N. J. (2014) Metal preferences and metallation. J. Biol. Chem. 289, 28095–28103 10.1074/jbc.R114.588145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Halloran T. V. (1993) Transition metals in control of gene expression. Science 261, 715–725 10.1126/science.8342038 [DOI] [PubMed] [Google Scholar]
- 20. de Reuse H., Vinella D., and Cavazza C. (2013) Common themes and unique proteins for the uptake and trafficking of nickel, a metal essential for the virulence of Helicobacter pylori. Front. Cell. Infect. Microbiol. 3, 94 10.3389/fcimb.2013.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rowinska-Zyrek M., Zakrzewska-Czerwinska J., Zawilak-Pawlik A., and Kozlowski H. (2014) Ni2+ chemistry in pathogens–a possible target for eradication. Dalton Trans. 43, 8976–8989 10.1039/C4DT00421C [DOI] [PubMed] [Google Scholar]
- 22. Bereza-Malcolm L. T., Mann G., and Franks A. E. (2015) Environmental sensing of heavy metals through whole cell microbial biosensors: a synthetic biology approach. ACS Synth. Biol. 4, 535–546 10.1021/sb500286r [DOI] [PubMed] [Google Scholar]
- 23. Fernandez-López R., Ruiz R., de la Cruz F., and Moncalián G. (2015) Transcription factor-based biosensors enlightened by the analyte. Front. Microbiol. 6, 648 10.3389/fmicb.2015.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jung J., and Lee S. J. (2019) Biochemical and biodiversity insights into heavy metal ion-responsive transcriptional regulators for synthetic biological heavy metal sensors. J. Microbiol. Biotechnol. 29, 1522–1542 10.4014/jmb.1908.08002 [DOI] [PubMed] [Google Scholar]
- 25. Andresen E., Peiter E., and Küpper H. (2018) Trace metal metabolism in plants. J. Exp. Bot. 69, 909–954 10.1093/jxb/erx465 [DOI] [PubMed] [Google Scholar]
- 26. Navarro J. A., and Schneuwly S. (2017) Copper and zinc homeostasis: lessons from Drosophila melanogaster. Front. Genet. 8, 223–223 10.3389/fgene.2017.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horianopoulos L. C., and Kronstad J. W. (2019) Connecting iron regulation and mitochondrial function in Cryptococcus neoformans. Curr. Opin. Microbiol. 52, 7–13 10.1016/j.mib.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raffa N., Osherov N., and Keller N. P. (2019) Copper utilization, regulation, and acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 20, E1980 10.3390/ijms20081980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ehrensberger K. M., and Bird A. J. (2011) Hammering out details: regulating metal levels in eukaryotes. Trends Biochem. Sci. 36, 524–531 10.1016/j.tibs.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 30. Martins T. S., Costa V., and Pereira C. (2018) Signaling pathways governing iron homeostasis in budding yeast. Mol. Microbiol. 109, 422–432 10.1111/mmi.14009 [DOI] [PubMed] [Google Scholar]
- 31. Smith A. D., Logeman B. L., and Thiele D. J. (2017) Copper acquisition and utilization in fungi. Annu. Rev. Microbiol. 71, 597–623 10.1146/annurev-micro-030117-020444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Irving H., and Williams R. J. (1948) Order of stability of metal complexes. Nature 162, 746 10.1038/162746a0 [DOI] [Google Scholar]
- 33. Imlay J. A. (2014) The mismetallation of enzymes during oxidative stress. J. Biol. Chem. 289, 28121–28128 10.1074/jbc.R114.588814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fontecave M. (2006) Iron–sulfur clusters: ever-expanding roles. Nat. Chem. Biol. 2, 171–174 10.1038/nchembio0406-171 [DOI] [PubMed] [Google Scholar]
- 35. Macomber L., and Imlay J. A. (2009) The iron–sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ranquet C., Ollagnier-de-Choudens S., Loiseau L., Barras F., and Fontecave M. (2007) Cobalt stress in Escherichia coli: the effect on the iron–sulfur proteins. J. Biol. Chem. 282, 30442–30451 10.1074/jbc.M702519200 [DOI] [PubMed] [Google Scholar]
- 37. Li J., Ren X., Fan B., Huang Z., Wang W., Zhou H., Lou Z., Ding H., Lyu J., and Tan G. (2019) Zinc toxicity and iron–sulfur cluster biogenesis in Escherichia coli. Appl. Environ. Microbiol. 85, e01967 10.1128/AEM.01967-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carter K. P., Young A. M., and Palmer A. E. (2014) Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 114, 4564–4601 10.1021/cr400546e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hider R. C., and Kong X. L. (2011) Glutathione: a key component of the cytoplasmic labile iron pool. BioMetals 24, 1179–1187 10.1007/s10534-011-9476-8 [DOI] [PubMed] [Google Scholar]
- 40. Outten C. E., and O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 10.1126/science.1060331 [DOI] [PubMed] [Google Scholar]
- 41. Helbig K., Bleuel C., Krauss G. J., and Nies D. H. (2008) Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190, 5431–5438 10.1128/JB.00271-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Costello L. C., and Franklin R. B. (2016) A comprehensive review of the role of zinc in normal prostate function and metabolism; and its implications in prostate cancer. Arch. Biochem. Biophys. 611, 100–112 10.1016/j.abb.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma Z., Chandrangsu P., Helmann T. C., Romsang A., Gaballa A., and Helmann J. D. (2014) Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol. Microbiol. 94, 756–770 10.1111/mmi.12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nairn B. L., Lonergan Z. R., Wang J., Braymer J. J., Zhang Y., Calcutt M. W., Lisher J. P., Gilston B. A., Chazin W. J., de Crécy-Lagard V., Giedroc D. P., and Skaar E. P. (2016) The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19, 826–836 10.1016/j.chom.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osman D., Martini M. A., Foster A. W., Chen J., Scott A. J. P., Morton R. J., Steed J. W., Lurie-Luke E., Huggins T. G., Lawrence A. D., Deery E., Warren M. J., Chivers P. T., and Robinson N. J. (2019) Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 15, 241–249 10.1038/s41589-018-0211-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foster A. W., Pernil R., Patterson C. J., Scott A. J. P., Pålsson L.-O., Pal R., Cummins I., Chivers P. T., Pohl E., and Robinson N. J. (2017) A tight tunable range for Ni(II) sensing and buffering in cells. Nat. Chem. Biol. 13, 409–414 10.1038/nchembio.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iwig J. S., and Chivers P. T. (2010) Coordinating intracellular nickel-metal-site structure–function relationships and the NikR and RcnR repressors. Nat. Prod. Rep. 27, 658–667 10.1039/b906683g [DOI] [PubMed] [Google Scholar]
- 48. Cavet J. S., Meng W., Pennella M. A., Appelhoff R. J., Giedroc D. P., and Robinson N. J. (2002) A nickel-cobalt-sensing ArsR-SmtB family repressor. J. Biol. Chem. 277, 38441–38448 10.1074/jbc.M207677200 [DOI] [PubMed] [Google Scholar]
- 49. Guedon E., and Helmann J. D. (2003) Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48, 495–506 10.1046/j.1365-2958.2003.03445.x [DOI] [PubMed] [Google Scholar]
- 50. Giedroc D. P., and Arunkumar A. I. (2007) Metal sensor proteins: Nature's metalloregulated allosteric switches. Dalton Trans. 2007, 3107–3120 10.1039/b706769k [DOI] [PubMed] [Google Scholar]
- 51. Guerra A. J., and Giedroc D. P. (2012) Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch. Biochem. Biophys. 519, 210–222 10.1016/j.abb.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Begg S. L., Eijkelkamp B. A., Luo Z., Couñago R. M., Morey J. R., Maher M. J., Ong C. L., McEwan A. G., Kobe B., O'Mara M. L., Paton J. C., and McDevitt C. A. (2015) Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 6, 6418 10.1038/ncomms7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helmann J. D. (2014) Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J. Biol. Chem. 289, 28112–28120 10.1074/jbc.R114.587071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Osman D., Foster A. W., Chen J., Svedaite K., Steed J. W., Lurie-Luke E., Huggins T. G., and Robinson N. J. (2017) Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 8, 1884 10.1038/s41467-017-02085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Osman D., and Cavet J. S. (2010) Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27, 668–680 10.1039/b906682a [DOI] [PubMed] [Google Scholar]
- 56. Ahn B. E., Cha J., Lee E. J., Han A. R., Thompson C. J., and Roe J. H. (2006) Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59, 1848–1858 10.1111/j.1365-2958.2006.05065.x [DOI] [PubMed] [Google Scholar]
- 57. Jones M. D., Sydor A. M., and Zamble D. B. (2013) in Metals in Cells (Scott R. S., and Culotta V., eds) pp. 277–287, John Wiley & Sons, Inc., New York [Google Scholar]
- 58. Moore C. M., and Helmann J. D. (2005) Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8, 188–195 10.1016/j.mib.2005.02.007 [DOI] [PubMed] [Google Scholar]
- 59. Campbell D. R., Chapman K. E., Waldron K. J., Tottey S., Kendall S., Cavallaro G., Andreini C., Hinds J., Stoker N. G., Robinson N. J., and Cavet J. S. (2007) Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 282, 32298–32310 10.1074/jbc.M703451200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chivers P. T. (2017) in The Biological Chemisry of Nickel (Zamble D. B., Rowinska-Zyrek M., and Kozlowski H., eds) pp. 259–282, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 61. Ma Z., Jacobsen F. E., and Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 10.1021/cr900077w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Motlagh H. N., Wrabl J. O., Li J., and Hilser V. J. (2014) The ensemble nature of allostery. Nature 508, 331–339 10.1038/nature13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Smock R. G., and Gierasch L. M. (2009) Sending signals dynamically. Science 324, 198–203 10.1126/science.1169377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cuthbertson L., and Nodwell J. R. (2013) The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475 10.1128/MMBR.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Portmann R., Poulsen K. R., Wimmer R., and Solioz M. (2006) CopY-like copper inducible repressors are putative 'winged helix' proteins. BioMetals 19, 61–70 10.1007/s10534-005-5381-3 [DOI] [PubMed] [Google Scholar]
- 66. Brown N. L., Stoyanov J. V., Kidd S. P., and Hobman J. L. (2003) The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163 10.1016/S0168-6445(03)00051-2 [DOI] [PubMed] [Google Scholar]
- 67. Hobman J. L. (2007) MerR family transcription activators: similar designs, different specificities. Mol. Microbiol. 63, 1275–1278 10.1111/j.1365-2958.2007.05608.x [DOI] [PubMed] [Google Scholar]
- 68. Philips S. J., Canalizo-Hernandez M., Yildirim I., Schatz G. C., Mondragón A., and O'Halloran T. V. (2015) Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 349, 877–881 10.1126/science.aaa9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hobman J. L., Wilkie J., and Brown N. L. (2005) A design for life: prokaryotic metal-binding MerR family regulators. BioMetals 18, 429–436 10.1007/s10534-005-3717-7 [DOI] [PubMed] [Google Scholar]
- 70. Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., and Mondragón A. (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 10.1126/science.1085950 [DOI] [PubMed] [Google Scholar]
- 71. Sameach H., Narunsky A., Azoulay-Ginsburg S., Gevorkyan-Aiapetov L., Zehavi Y., Moskovitz Y., Juven-Gershon T., Ben-Tal N., and Ruthstein S. (2017) Structural and dynamics characterization of the MerR family metalloregulator CueR in its repression and activation states. Structure 25, 988–996.e3 10.1016/j.str.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 72. Chen T. Y., Santiago A. G., Jung W., Krzemiński L., Yang F., Martell D. J., Helmann J. D., and Chen P. (2015) Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat. Commun. 6, 7445 10.1038/ncomms8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fillat M. F. (2014) The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 74. Carpenter B. M., Whitmire J. M., and Merrell D. S. (2009) This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601 10.1128/IAI.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sarvan S., Butcher J., Stintzi A., and Couture J. F. (2018) Variation on a theme: investigating the structural repertoires used by ferric uptake regulators to control gene expression. BioMetals 31, 681–704 10.1007/s10534-018-0120-8 [DOI] [PubMed] [Google Scholar]
- 76. Ma Z., Gabriel S. E., and Helmann J. D. (2011) Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138 10.1093/nar/gkr625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jacquamet L., Traoré D. A., Ferrer J. L., Proux O., Testemale D., Hazemann J. L., Nazarenko E., El Ghazouani A., Caux-Thang C., Duarte V., and Latour J. M. (2009) Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31 10.1111/j.1365-2958.2009.06753.x [DOI] [PubMed] [Google Scholar]
- 78. Deng Z., Wang Q., Liu Z., Zhang M., Machado A. C., Chiu T. P., Feng C., Zhang Q., Yu L., Qi L., Zheng J., Wang X., Huo X., Qi X., Li X., Wu W., Rohs R., Li Y., and Chen Z. (2015) Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 6, 7642 10.1038/ncomms8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shin J. H., and Helmann J. D. (2016) Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis. Nat. Commun. 7, 12612 10.1038/ncomms12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roncarati D., Pelliciari S., Doniselli N., Maggi S., Vannini A., Valzania L., Mazzei L., Zambelli B., Rivetti C., and Danielli A. (2016) Metal-responsive promoter DNA compaction by the ferric uptake regulator. Nat. Commun. 7, 12593 10.1038/ncomms12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pérard J., Covès J., Castellan M., Solard C., Savard M., Miras R., Galop S., Signor L., Crouzy S., Michaud-Soret I., and de Rosny E. (2016) Quaternary structure of Fur proteins, a new subfamily of tetrameric proteins. Biochemistry 55, 1503–1515 10.1021/acs.biochem.5b01061 [DOI] [PubMed] [Google Scholar]
- 82. Pérard J., Nader S., Levert M., Arnaud L., Carpentier P., Siebert C., Blanquet F., Cavazza C., Renesto P., Schneider D., Maurin M., Coves J., Crouzy S., and Michaud-Soret I. (2018) Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role in Francisella tularensis virulence. Commun. Biol. 1, 93 10.1038/s42003-018-0095-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Higgins K. A., and Giedroc D. (2014) Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem. Lett. 43, 20–25 10.1246/cl.130965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chang F.-M., Martin J. E., and Giedroc D. P. (2015) Electrostatic occlusion and quaternary structural ion pairing are key determinants of Cu(I)-mediated allostery in the copper-sensing operon repressor (CsoR). Biochemistry 54, 2463–2472 10.1021/acs.biochem.5b00154 [DOI] [PubMed] [Google Scholar]
- 85. Iwig J. S., Leitch S., Herbst R. W., Maroney M. J., and Chivers P. T. (2008) Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 130, 7592–7606 10.1021/ja710067d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tan B. G., Vijgenboom E., and Worrall J. A. (2014) Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucleic Acids Res. 42, 1326–1340 10.1093/nar/gkt902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., and Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 10.1038/nchembio844 [DOI] [PubMed] [Google Scholar]
- 88. Chang F.-M., Lauber M. A., Running W. E., Reilly J. P., and Giedroc D. P. (2011) Ratiometric pulse–chase amidination mass spectrometry as a probe of biomolecular complex formation. Anal. Chem. 83, 9092–9099 10.1021/ac202154r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chang F. M., Coyne H. J., Cubillas C., Vinuesa P., Fang X., Ma Z., Ma D., Helmann J. D., García-de los Santos A., Wang Y. X., Dann C. E. 3rd., and Giedroc D. P. (2014) Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 289, 19204–19217 10.1074/jbc.M114.556704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huang H.-T., Bobst C. E., Iwig J. S., Chivers P. T., Kaltashov I. A., and Maroney M. J. (2018) Co(II) and Ni(II) binding of the Escherichia coli transcriptional repressor RcnR orders its N terminus, alters helix dynamics, and reduces DNA affinity. J. Biol. Chem. 293, 324–332 10.1074/jbc.RA117.000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang H.-T., and Maroney M. J. (2019) Ni(II) sensing by RcnR does not require an FrmR-like intersubunit linkage. Inorg. Chem. 58, 13639–13653 10.1021/acs.inorgchem.9b01096 [DOI] [PubMed] [Google Scholar]
- 92. Denby K. J., Iwig J., Bisson C., Westwood J., Rolfe M. D., Sedelnikova S. E., Higgins K., Maroney M. J., Baker P. J., Chivers P. T., and Green J. (2016) The mechanism of a formaldehyde-sensing transcriptional regulator. Sci. Rep. 6, 38879 10.1038/srep38879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Que Q., and Helmann J. D. (2000) Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468 10.1046/j.1365-2958.2000.01811.x [DOI] [PubMed] [Google Scholar]
- 94. Lieser S. A., Davis T. C., Helmann J. D., and Cohen S. M. (2003) DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry 42, 12634–12642 10.1021/bi0350248 [DOI] [PubMed] [Google Scholar]
- 95. McGuire A. M., Cuthbert B. J., Ma Z., Grauer-Gray K. D., Brunjes Brophy M., Spear K. A., Soonsanga S., Kliegman J. I., Griner S. L., Helmann J. D., and Glasfeld A. (2013) Roles of the A and C sites in the manganese-specific activation of MntR. Biochemistry 52, 701–713 10.1021/bi301550t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lisher J. P., Higgins K. A., Maroney M. J., and Giedroc D. P. (2013) Physical characterization of the manganese-sensing pneumococcal surface antigen repressor from Streptococcus pneumoniae. Biochemistry 52, 7689–7701 10.1021/bi401132w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cong X., Yuan Z., Wang Z., Wei B., Xu S., and Wang J. (2018) Crystal structures of manganese-dependent transcriptional repressor MntR (Rv2788) from Mycobacterium tuberculosis in apo and manganese bound forms. Biochem. Biophys. Res. Commun. 501, 423–427 10.1016/j.bbrc.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 98. Golynskiy M., Li S., Woods V. L. Jr., and Cohen S. M. (2007) Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. J. Biol. Inorg. Chem. 12, 699–709 10.1007/s00775-007-0216-z [DOI] [PubMed] [Google Scholar]
- 99. D'Aquino J. A., Tetenbaum-Novatt J., White A., Berkovitch F., and Ringe D. (2005) Mechanism of metal ion activation of the diphtheria toxin repressor DtxR. Proc. Natl. Acad. Sci. U.S.A. 102, 18408–18413 10.1073/pnas.0500908102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Do H., Makthal N., Chandrangsu P., Olsen R. J., Helmann J. D., Musser J. M., and Kumaraswami M. (2019) Metal sensing and regulation of adaptive responses to manganese limitation by MtsR is critical for group A streptococcus virulence. Nucleic Acids Res. 47, 7476–7493 10.1093/nar/gkz524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Twigg P. D., Parthasarathy G., Guerrero L., Logan T. M., and Caspar D. L. (2001) Disordered to ordered folding in the regulation of diphtheria toxin repressor activity. Proc. Natl. Acad. Sci. U.S.A. 98, 11259–11264 10.1073/pnas.191354798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. DeWitt M. A., Kliegman J. I., Helmann J. D., Brennan R. G., Farrens D. L., and Glasfeld A. (2007) The conformations of the manganese transport regulator of Bacillus subtilis in its metal-free state. J. Mol. Biol. 365, 1257–1265 10.1016/j.jmb.2006.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Spatafora G., Corbett J., Cornacchione L., Daly W., Galan D., Wysota M., Tivnan P., Collins J., Nye D., Levitz T., Breyer W. A., and Glasfeld A. (2015) Interactions of the metalloregulatory protein SloR from Streptococcus mutans with its metal ion effectors and DNA binding site. J. Bacteriol. 197, 3601–3615 10.1128/JB.00612-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jakubovics N. S., Smith A. W., and Jenkinson H. F. (2000) Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38, 140–153 10.1046/j.1365-2958.2000.02122.x [DOI] [PubMed] [Google Scholar]
- 105. Dian C., Schauer K., Kapp U., McSweeney S. M., Labigne A., and Terradot L. (2006) Structural basis of the nickel response in Helicobacter pylori: crystal structures of HpNikR in apo and nickel-bound states. J. Mol. Biol. 361, 715–730 10.1016/j.jmb.2006.06.058 [DOI] [PubMed] [Google Scholar]
- 106. Chivers P. T., and Tahirov T. H. (2005) Structure of Pyrococcus horikoshii NikR: nickel sensing and implications for the regulation of DNA recognition. J. Mol. Biol. 348, 597–607 10.1016/j.jmb.2005.03.017 [DOI] [PubMed] [Google Scholar]
- 107. Schreiter E. R., Sintchak M. D., Guo Y., Chivers P. T., Sauer R. T., and Drennan C. L. (2003) Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10, 794–799 10.1038/nsb985 [DOI] [PubMed] [Google Scholar]
- 108. Musiani F., Bertoša B., Magistrato A., Zambelli B., Turano P., Losasso V., Micheletti C., Ciurli S., and Carloni P. (2010) Computational study of the DNA-binding protein Helicobacter pylori NikR: the role of Ni2+. J. Chem. Theory Comput. 6, 3503–3515 10.1021/ct900635z [DOI] [PubMed] [Google Scholar]
- 109. Schreiter E. R., Wang S. C., Zamble D. B., and Drennan C. L. (2006) NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl. Acad. Sci. U.S.A. 103, 13676–13681 10.1073/pnas.0606247103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Phillips C. M., Stultz C. M., and Drennan C. L. (2010) Searching for the Nik operon: how a ligand-responsive transcription factor hunts for its DNA binding site. Biochemistry 49, 7757–7763 10.1021/bi100947k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chivers P. T., and Sauer R. T. (1999) NikR is a ribbon–helix–helix DNA-binding protein. Protein Sci. 8, 2494–2500 10.1110/ps.8.11.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang S. C., Dias A. V., Bloom S. L., and Zamble D. B. (2004) Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry 43, 10018–10028 10.1021/bi049405c [DOI] [PubMed] [Google Scholar]
- 113. Dias A. V., and Zamble D. B. (2005) Protease digestion analysis of Escherichia coli NikR: evidence for conformational stabilization with Ni(II). J. Biol. Inorg. Chem. 10, 605–612 10.1007/s00775-005-0008-2 [DOI] [PubMed] [Google Scholar]
- 114. Krecisz S., Jones M. D., and Zamble D. B. (2012) Nonspecific interactions between Escherichia coli NikR and DNA are critical for nickel-activated DNA binding. Biochemistry 51, 7873–7879 10.1021/bi300510z [DOI] [PubMed] [Google Scholar]
- 115. Muller C., Bahlawane C., Aubert S., Delay C. M., Schauer K., Michaud-Soret I., and De Reuse H. (2011) Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 39, 7564–7575 10.1093/nar/gkr460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vannini A., Pinatel E., Costantini P. E., Pelliciari S., Roncarati D., Puccio S., De Bellis G., Peano C., and Danielli A. (2017) Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci. Rep. 7, 45458 10.1038/srep45458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dosanjh N. S., West A. L., and Michel S. L. (2009) Helicobacter pylori NikR's interaction with DNA: a two-tiered mode of recognition. Biochemistry 48, 527–536 10.1021/bi801481j [DOI] [PubMed] [Google Scholar]
- 118. Benanti E. L., and Chivers P. T. (2011) Helicobacter pylori NikR protein exhibits distinct conformations when bound to different promoters. J. Biol. Chem. 286, 15728–15737 10.1074/jbc.M110.196055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mazzei L., Dobrovolska O., Musiani F., Zambelli B., and Ciurli S. (2015) On the interaction of Helicobacter pylori NikR, a Ni(II)-responsive transcription factor, with the urease operator: in solution and in silico studies. J. Biol. Inorg. Chem. 20, 1021–1037 10.1007/s00775-015-1284-0 [DOI] [PubMed] [Google Scholar]
- 120. West A. L., Evans S. E., González J. M., Carter L. G., Tsuruta H., Pozharski E., and Michel S. L. (2012) Ni(II) coordination to mixed sites modulates DNA binding of HpNikR via a long-range effect. Proc. Natl. Acad. Sci. U.S.A. 109, 5633–5638 10.1073/pnas.1120283109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Benanti E. L., and Chivers P. T. (2007) The N-terminal arm of the Helicobacter pylori Ni2+-dependent transcription factor NikR is required for specific DNA binding. J. Biol. Chem. 282, 20365–20375 10.1074/jbc.M702982200 [DOI] [PubMed] [Google Scholar]
- 122. Bahlawane C., Dian C., Muller C., Round A., Fauquant C., Schauer K., de Reuse H., Terradot L., and Michaud-Soret I. (2010) Structural and mechanistic insights into Helicobacter pylori NikR activation. Nucleic Acids Res. 38, 3106–3118 10.1093/nar/gkp1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Jones M. D., Li Y., and Zamble D. B. (2018) Acid-responsive activity by the Helicobacter pylori metalloregulator NikR. Proc. Natl. Acad. Sci. U.S.A. 115, 8966–8971 10.1073/pnas.1808393115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Li Y., and Zamble D. B. (2009) The pH-responsive DNA-binding activity of Helicobacter pylori NikR. Biochemistry 48, 2486–2496 10.1021/bi801742r [DOI] [PubMed] [Google Scholar]
- 125. Ansari S., and Yamaoka Y. (2017) Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22, 10.1111/hel.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Musiani F., Zambelli B., Bazzani M., Mazzei L., and Ciurli S. (2015) Nickel-responsive transcriptional regulators. Metallomics 7, 1305–1318 10.1039/C5MT00072F [DOI] [PubMed] [Google Scholar]
- 127. Deochand D. K., and Grove A. (2017) MarR family transcription factors: dynamic variations on a common scaffold. Crit. Rev. Biochem. Mol. Biol. 52, 595–613 10.1080/10409238.2017.1344612 [DOI] [PubMed] [Google Scholar]
- 128. Reyes-Caballero H., Guerra A. J., Jacobsen F. E., Kazmierczak K. M., Cowart D., Koppolu U. M., Scott R. A., Winkler M. E., and Giedroc D. P. (2010) The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J. Mol. Biol. 403, 197–216 10.1016/j.jmb.2010.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hao Z., Lou H., Zhu R., Zhu J., Zhang D., Zhao B. S., Zeng S., Chen X., Chan J., He C., and Chen P. R. (2014) The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat. Chem. Biol. 10, 21–28 10.1038/nchembio.1380 [DOI] [PubMed] [Google Scholar]
- 130. Guerra A. J., Dann C. E. 3rd., and Giedroc D. P. (2011) Crystal structure of the zinc-dependent MarR family transcriptional regulator AdcR in the Zn(II)-bound state. J. Am. Chem. Soc. 133, 19614–19617 10.1021/ja2080532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhu R., Song Y., Liu H., Yang Y., Wang S., Yi C., and Chen P. R. (2017) Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation. Proc. Natl. Acad. Sci. U.S.A. 114, 13661–13666 10.1073/pnas.1708563115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Capdevila D. A., Huerta F., Edmonds K. A., Le M. T., Wu H., and Giedroc D. P. (2018) Tuning site-specific dynamics to drive allosteric activation in a pneumococcal zinc uptake regulator. Elife 7, e37268 10.7554/eLife.37268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Campanello G. C., Ma Z., Grossoehme N. E., Guerra A. J., Ward B. P., Dimarchi R. D., Ye Y., Dann C. E. 3rd., and Giedroc D. P. (2013) Allosteric inhibition of a zinc-sensing transcriptional repressor: insights into the arsenic repressor (ArsR) family. J. Mol. Biol. 425, 1143–1157 10.1016/j.jmb.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Eicken C., Pennella M. A., Chen X., Koshlap K. M., VanZile M. L., Sacchettini J. C., and Giedroc D. P. (2003) A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J. Mol. Biol. 333, 683–695 10.1016/j.jmb.2003.09.007 [DOI] [PubMed] [Google Scholar]
- 135. Capdevila D. A., Braymer J. J., Edmonds K. A., Wu H., and Giedroc D. P. (2017) Entropy redistribution controls allostery in a metalloregulatory protein. Proc. Natl. Acad. Sci. U.S.A. 114, 4424–4429 10.1073/pnas.1620665114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Arunkumar A. I., Campanello G. C., and Giedroc D. P. (2009) Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc. Natl. Acad. Sci. U.S.A. 106, 18177–18182 10.1073/pnas.0905558106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Grossoehme N. E., and Giedroc D. P. (2009) Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J. Am. Chem. Soc. 131, 17860–17870 10.1021/ja906131b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Capdevila D. A., Edmonds K. A., Campanello G. C., Wu H., Gonzalez-Gutierrez G., and Giedroc D. P. (2018) Functional role of solvent entropy and conformational entropy of metal binding in a dynamically driven allosteric system. J. Am. Chem. Soc. 140, 9108–9119 10.1021/jacs.8b02129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Nussinov R., and Tsai C. J. (2015) Allostery without a conformational change? Revisiting the paradigm. Curr. Opin. Struct. Biol. 30, 17–24 10.1016/j.sbi.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 140. Popovych N., Sun S., Ebright R. H., and Kalodimos C. G. (2006) Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 13, 831–838 10.1038/nsmb1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Tzeng S.-R., and Kalodimos C. G. (2012) Protein activity regulation by conformational entropy. Nature 488, 236–240 10.1038/nature11271 [DOI] [PubMed] [Google Scholar]
- 142. Tzeng S.-R., and Kalodimos C. G. (2009) Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 10.1038/nature08560 [DOI] [PubMed] [Google Scholar]
- 143. Tzeng S.-R., and Kalodimos C. G. (2013) Allosteric inhibition through suppression of transient conformational states. Nat. Chem. Biol. 9, 462–465 10.1038/nchembio.1250 [DOI] [PubMed] [Google Scholar]
- 144. Petit C. M., Zhang J., Sapienza P. J., Fuentes E. J., and Lee A. L. (2009) Hidden dynamic allostery in a PDZ domain. Proc. Natl. Acad. Sci. U.S.A. 106, 18249–18254 10.1073/pnas.0904492106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Monod J., Wyman J., and Changeux J.-P. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- 146. Koshland D. E. Jr., Némethy G., and Filmer D. (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5, 365–385 10.1021/bi00865a047 [DOI] [PubMed] [Google Scholar]
- 147. Wei G., Xi W., Nussinov R., and Ma B. (2016) Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem. Rev. 116, 6516–6551 10.1021/acs.chemrev.5b00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Law A. B., Sapienza P. J., Zhang J., Zuo X., and Petit C. M. (2017) Native state volume fluctuations in proteins as a mechanism for dynamic allostery. J. Am. Chem. Soc. 139, 3599–3602 10.1021/jacs.6b12058 [DOI] [PubMed] [Google Scholar]
- 149. Chung C. Y., Posimo J. M., Lee S., Tsang T., Davis J. M., Brady D. C., and Chang C. J. (2019) Activity-based ratiometric FRET probe reveals oncogene-driven changes in labile copper pools induced by altered glutathione metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 18285–18294 10.1073/pnas.1904610116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Aron A. T., Heffern M. C., Lonergan Z. R., Vander Wal M. N., Blank B. R., Spangler B., Zhang Y., Park H. M., Stahl A., Renslo A. R., Skaar E. P., and Chang C. J. (2017) In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. U.S.A. 114, 12669–12674 10.1073/pnas.1708747114 [DOI] [PMC free article] [PubMed] [Google Scholar]