A concerted function of multiple sequence motifs in Arabidopsis KORRIGAN1 protein determines its destination in the secretory pathway and physiological function.
Abstract
Among many glycoproteins within the plant secretory system, KORRIGAN1 (KOR1), a membrane-anchored endo-β-1,4-glucanase involved in cellulose biosynthesis, provides a link between N-glycosylation, cell wall biosynthesis, and abiotic stress tolerance. After insertion into the endoplasmic reticulum, KOR1 cycles between the trans-Golgi network (TGN) and the plasma membrane (PM). From the TGN, the protein is targeted to growing cell plates during cell division. These processes are governed by multiple sequence motifs and also host genotypes. Here, we investigated the interaction and hierarchy of known and newly identified sorting signals in KOR1 and how they affect KOR1 transport at various stages in the secretory pathway. Conventional steady-state localization showed that structurally compromised KOR1 variants were directed to tonoplasts. In addition, a tandem fluorescent timer technology allowed for differential visualization of young versus aged KOR1 proteins, enabling the analysis of single-pass transport through the secretory pathway. Observations suggest the presence of multiple checkpoints/branches during KOR1 trafficking, where the destination is determined based on KOR1’s sequence motifs and folding status. Moreover, growth analyses of dominant PM-confined KOR1-L48L49→A48A49 variants revealed the importance of active removal of KOR1 from the PM during salt stress, which otherwise interfered with stress acclimation.
INTRODUCTION
Cell wall biosynthesis is a complex process that involves numerous proteins/enzymes responsible for the production of fibrous materials and matrix components, as well as for regulating substrate delivery and organization of coordinated assembly/catalysis (Somerville, 2006; McFarlane et al., 2014). Cellulose is the major constituent of the cell wall and produced by the cellulose synthase complex (CSC), consisting of multiple isoforms of cellulose synthase subunits (CESA; Turner and Kumar, 2018). Several other proteins required for cellulose biosynthesis include CESA binding proteins that tether the CSC complex to microtubules (MTs), apoplastic glucanases, and several cell wall/plasma membrane (PM) proteins (Nicol et al., 1998; Roudier et al., 2005; Li et al., 2012; Sánchez-Rodríguez et al., 2012, 2018; Bashline et al., 2013; Endler et al., 2015; Liu et al., 2016; Zhang et al., 2016; Polko and Kieber, 2019). KORRIGAN1/RADIALLY SWOLLEN2 (KOR1/RSW2) is an essential endo-β-1,4-glucanase of Arabidopsis (Arabidopsis thaliana) that is involved in cellulose biosynthesis of primary and secondary cell walls and belongs to the Glycoside Hydrolase9 (GH9) family (Robert et al., 2005; Takahashi et al., 2009; Mansoori et al., 2014; Rips et al., 2014; Vain et al., 2014; von Schaewen et al., 2015). Unlike many other GH9 family proteins, which are soluble secreted proteins with an N-terminal signal peptide, KOR1 is a type II integral membrane protein with an N-terminal cytosolic domain followed by a transmembrane anchor, a stalk region, and a catalytic N-glycosylated ectodomain.
While mutant kor1 alleles result in cellulose deficiency and growth retardation (Nicol et al., 1998; Peng et al., 2000; Lane et al., 2001; Szyjanowicz et al., 2004), the exact function of KOR1 in cellulose biosynthesis is unclear. However, the physical interaction of KOR1 and the CSC has been reported previously by Mansoori et al. (2014) and Vain et al. (2014). An early study suggested that KOR1 is involved in the cleavage of sterol-linked primers during cellulose elongation (Peng et al., 2002). Later, transgenic overexpression studies using the aspen homolog of KOR1 showed that one of its catalytic functions might be to reduce the crystallinity of cellulose fibrils (Takahashi et al., 2009). These observations led to a few hypotheses, with KOR1 cutting off β-glucoside primers in the apoplast during initiation, cleaving stalled cellulose chains to reduce fibril size, or resolving macrofibril aggregates of nascent cellulose polymers (Ding and Himmel, 2006). Interestingly, compared to secreted bacterial GH9 enzymes, plant KOR1 homologs showed more than two orders of magnitude lower catalytic rates (Master et al., 2004). Furthermore, the catalytically inactive jia1 allele of KOR1 was partially functional in vivo, also suggesting a noncatalytic physiological role for KOR1 (Lei et al., 2014).
The subcellular distribution of KOR1 is intrinsic to its in vivo function. In the endoplasmic reticulum (ER), the ectodomain of KOR1 becomes heavily N-glycosylated. In the Golgi apparatus (GA), subsequent N-glycan modification reactions convert at least some of the KOR1 N-glycans to complex N-glycans (Kang et al., 2008; Liebminger et al., 2013). Maturation of plant complex N-glycans involves multiple enzymes in the GA, including Golgi α-mannosidases I and II, N-acetylglucosaminyltransferases I and II, β-1,3-fucosyltransferases, and α-1,2-xylosyltransferase (Strasser, 2016; Nagashima et al., 2018). Defects in many of the N-glycosylation and maturation enzymes compromise the function of KOR1 and likely also of other cell wall biosynthetic proteins, as demonstrated by extensive genetic analyses (Kang et al., 2008; Liebminger et al., 2009; Farid et al., 2013; Rips et al., 2014; Nagashima et al., 2018). Interestingly, the catalytic activity of KOR1 was mostly unaffected by lack of any of its eight N-glycans; however, loss of multiple N-glycans affected subcellular localization of the protein and root growth, thus emphasizing their importance for ectodomain folding and enzymatic capacity of KOR1 (Liebminger et al., 2013; Rips et al., 2014). Normally, KOR1 proteins are distributed among the trans-Golgi network (TGN) and the PM and were also found at the newly forming cell plate of dividing cells (Zuo et al., 2000b; Robert et al., 2005; Rips et al., 2014). In the underglycosylation mutant stt3a, however, KOR1 accumulated at the tonoplast (TP), likely for subsequent degradation. Similarly, KOR1 variants missing several N-glycosylation sites, or temperature-sensitive KOR1G429R (carrying the G429→R mutation encoded by the rsw2-1 allele; Lane et al., 2001), accumulated in TPs at high levels as well (Rips et al., 2014). Currently, the distribution and retention mechanisms of KOR1 at each subcellular location are not well established. Zuo et al. (2000b) found that KOR1 variants mutated at two hydrophobic residues (L48L49) within the cytoplasmic domain were no longer targeted to the cell plate of tobacco (Nicotiana tabacum) BY-2 cells but distributed uniformly across the PM. This suggested a role for polarized targeting, directing KOR1 to specific delivery routes in dividing cells.
The cell wall is the first physical barrier toward the outside and reacts to various environmental perturbations. Previous studies, including ours, found that the growth and root swelling phenotype of kor1/rsw2 or cesA1/rsw1 was similar to the salt stress–induced phenotype of the N-glycosylation–related mutants, leading to the identification of KOR1/RSW2 as a link between N-glycosylation, cellulose biosynthesis, and salt-stress tolerance (Koiwa et al., 2003; Kang et al., 2008; Koiwa, 2009; Liebminger et al., 2009; Farid et al., 2013; Rips et al., 2014; Liu et al., 2018; Nagashima et al., 2018). Cellulose synthase subunits such as CESA1/RSW1, on the other hand, are not N glycosylated (Gillmor et al., 2002). The precise mode of action of N-glycans in salt stress and cellulose biosynthesis is unknown, but salt stress induces reorganization of the microtubule network as well as subcellular distribution of cellulose synthase complexes in plant cells (Shoji et al., 2006; Wang et al., 2007, 2011; Endler et al., 2015). Microtubules regulate intracellular and cell surface movements of CSC and of KOR1 (Vain et al., 2014) and determine the direction of polymerization of the cellulose fibrils in the apoplast. In fact, directional deposition of cellulose microfibrils and organization of the microtubule network are mutually dependent (Paredez et al., 2008; Gutierrez et al., 2009; Lei et al., 2014). Despite accumulating knowledge on the regulation of CSC transport within the cellular vesicle trafficking system (McFarlane et al., 2014; Kesten et al., 2017; Turner and Kumar, 2018; Polko and Kieber, 2019), little is known about the factors that govern the dynamic behavior of KOR1 along the secretory pathway. Recent studies showed the physical association of CSC and KOR1 (Mansoori et al., 2014; Vain et al., 2014); yet, only partial colocalization of CSC and KOR1 was observed in planta, suggesting that their interaction may be transient. Instead, the intracellular distribution of KOR1 extensively overlapped with SYP61, a TGN marker (Rips et al., 2014). Therefore, the paradigm established for CSC transport may not be entirely applicable to KOR1 transport; hence, understanding KOR1 transport requires empirical studies. Protein sorting and transport at the TGN are complex processes, in which distinct TGN domains are responsible for targeting a unique suite of proteins to the cell surface or vacuole (Robinson and Pimpl, 2014).
Here, we expanded our previous studies and investigated multiple determinants for subcellular localization of KOR1 and their physiological significance. We identified a sequence motif essential for stably retaining KOR1 in the PM/TGN and preventing TP accumulation of KOR1. The routes directing compromised KOR1 variants to the TP were followed by time-resolved transport analysis using fluorescent timer assays. Our data revealed that various sequence motifs support active surveillance of KOR1 transport between different subcellular locations, which is vital for KOR1 to execute its roles in plant growth and stress tolerance.
RESULTS
The C-Terminal Pro-Rich Motif of KOR1 Is Required to Retain KOR1 in the TGN/PM Cycle
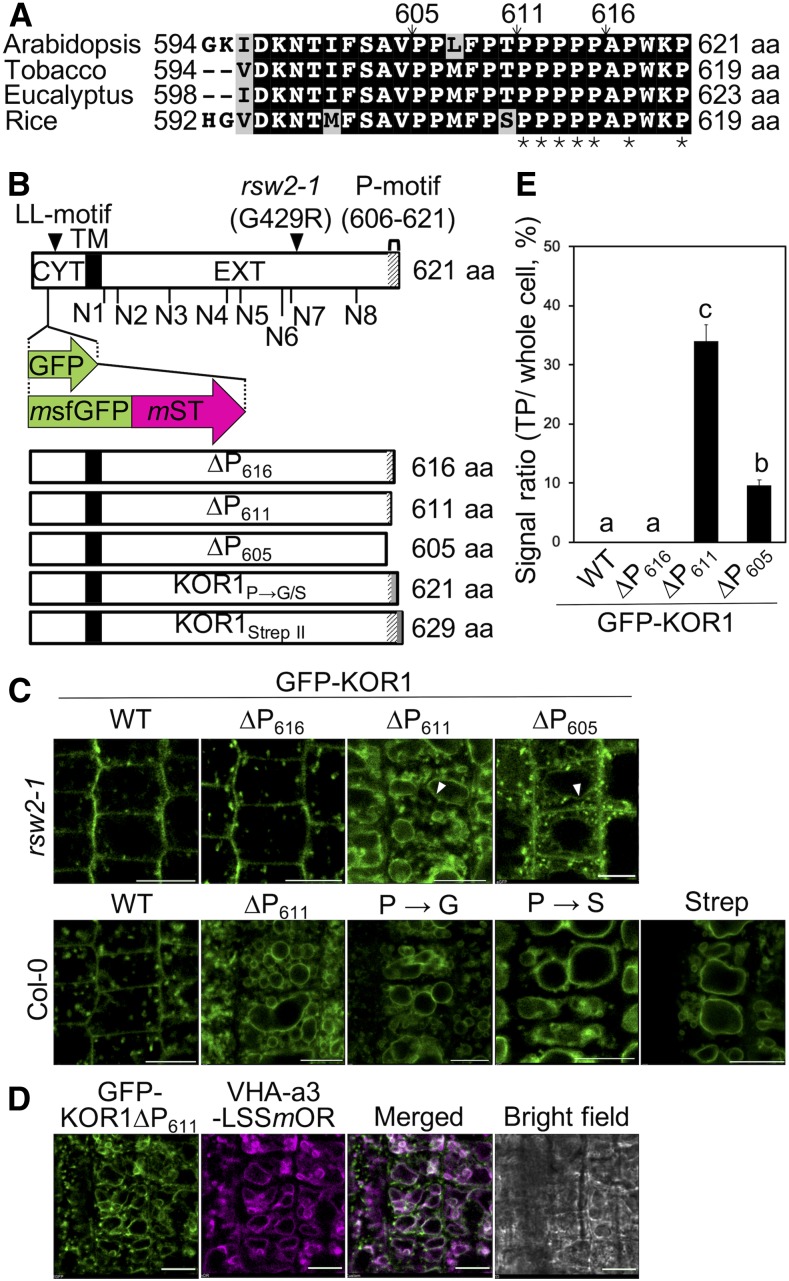
Arabidopsis KOR1 and other plant KOR1-family members adopt a type II membrane protein structure with N-terminal cytoplasmic domain, transmembrane anchor, stalk region, and an extracellular catalytic domain (Figure 1). Under normal conditions, KOR1 is present in multiple subcellular compartments, that is, the TGN, PM, and cell plate. Because mutations in the catalytic extracellular domain induce accumulation of KOR1 at the TP (Rips et al., 2014), we hypothesized that this domain might contain a signal important for retaining KOR1 in the TGN/PM cycle. Upon inspection of multiple GH9 family members, we noticed that plant KOR proteins do not contain carbohydrate binding motifs, which often follow the catalytic domain in many other GH9 glycohydrolases, such as Oryza sativa Cel9A (Supplemental Figure 1; Yoshida et al., 2006; Yoshida and Komae, 2006). Instead, KOR proteins display a different C-terminal ending with a highly conserved Pro-rich sequence (Figure 1A; Supplemental Figure 1). In our survey, all 52 KOR proteins from 22 plant species inspected contained the Pro-rich sequence. In Arabidopsis KOR1, 10 of 16 C-terminal amino acids are prolines. This region was termed the P-motif.
Figure 1.
The C-Terminal Pro-Rich P-Motif Prevents Mislocalization of KOR1 to the TP.
(A) Alignment of the C-terminal peptide sequences of KOR1 homologs from different plant species. Arrows indicate positions of C-terminal truncations and asterisks (*) of prolines mutated in P→G/S variants. aa, amino acid.
(B) Schematic diagram of the GFP/tdFT-KOR1 fusion proteins. LL motif, di-Leu motif in the cytoplasmic region (CYT, white boxes). N1 to N8, N-glycosylation sites of the extracellular domain (EXT, white boxes). Transmembrane domain (TM; black boxes). Position of the GFP/tdFT insertion, rsw2-1 mutation, and P-motif deletions/mutations (bottom, gray shading) are also indicated. aa, amino acid.
(C) Subcellular localization of the GFP-KOR1∆P variants in root tip cells. The white arrowheads indicate GFP signals in TP-like endomembrane structures of 3-d-old seedlings. Experiments were repeated with three independent lines per construct, which gave similar results. Bar = 10 µm.
(D) Colocalization analysis of GFP-∆P611 and VHA-a3-LSSmOrange (mOR), a TP marker. Bar = 10 µm. WT, the wild type.
(E) Quantitative data of (C), n = 15. The signal at the PM was divided by the total signal of each cell. Error bars represent the se of the mean. Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test). WT, the wild type.
To determine the functional significance of the P-motif for protein localization, we prepared variants of GFP-KOR1 with progressive deletions into the P-motif (∆P series; Figure 1B). This reporter construct is based on the one used previously (Rips et al., 2014) and contains the entire genomic fragment of KOR1. For each of the reporter constructs, we evaluated ∼20 transformants and analyzed three to four lines with a representative GFP localization pattern further. Fluorescent signals from the wild-type GFP-KOR1 were detected in the TGN and PM, as reported previously (Rips et al., 2014). Removal of 16 (∆P605) or 10 (∆P611) residues from the KOR1 C terminus, but not the five last residues (∆P616), promoted accumulation of GFP-KOR1 at an endomembrane compartment, likely the TP, in addition to the TGN and PM (Figure 1C). GFP-KOR1 signals at the altered location overlapped with Vacuolar H+-ATPase (VHA)-a3-LssmOrange (Brüx et al., 2008), confirming TP localization of the C-terminally truncated proteins (Figure 1D). GFP signals at the TP were strongest with ∆P611, reaching 34.1% of the total GFP signal in the cells, but weaker with further truncation (∆P605; 9.7% of the total cellular GFP signal; Figures 1C and 1E; Supplemental Figure 2A). This may indicate that the 16 amino-acid truncation destabilizes the protein and promotes degradation upon arrival at the TP; however, we cannot exclude other possibilities. Therefore, we used ∆P611 for further analysis. Importantly, the genotype used (the Columbia [Col]-0 wild type or rsw2-1 as background) did not affect the localization pattern of GFP-KOR1 (Figure 1C). TP targeting of truncated KOR1∆P611 protein was likely not caused by misfolding because subtler changes, such as amino acid replacements (Pro to Gly or Pro to Ser) or addition of an eight amino-acid-long Strep-tag II to the C terminus, similarly caused TP targeting (Figure 1C). These results indicated that the P-motif at the C terminus is vital for retaining KOR1 in the TGN/PM cycle. Hereafter, all GFP-KOR1 reporter constructs were expressed in the rsw2-1 mutant host to test localization profiles and functional complementation of growth phenotypes, unless stated otherwise.
Time-Course Analysis of tdFT-KOR1 Trafficking Reveals Two Routes for KOR1 to the TP
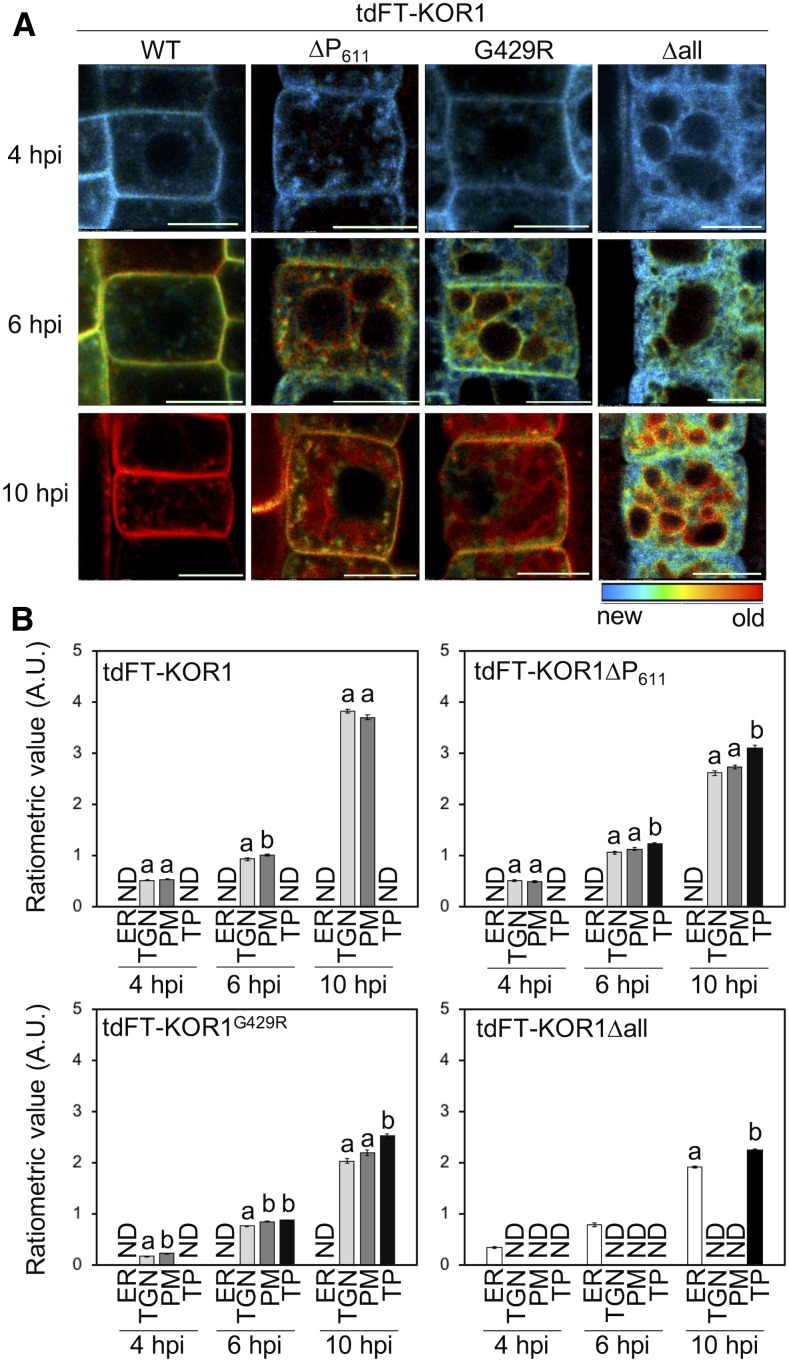
ER quality control (ERQC) is the initial protein quality control system in the secretory pathway, preventing misfolded proteins from being transported to the GA by sending them to the cytoplasm or vacuole for degradation (Shin et al., 2018). Even though the folding of KOR1∆P611 is likely unaffected, substantial accumulation of KOR1∆P611 at the TP was observed, together with at the TGN/PM. We hypothesized that KOR1∆P611 could exit the ER normally and that TP accumulation of KOR1∆P611 results from quality control at the TGN to remove expired KOR1 from the TGN/PM cycle. Because the conventional GFP-fusion approach only shows endpoint but not pre-equilibrium localization(s), we designed an estradiol-inducible KOR1 reporter construct to track KOR1 upon induced de novo synthesis and monitor its traffic en route to the final destination(s). To reliably distinguish between newly synthesized KOR1 and aged KOR1, a tandem fluorescent timer (tdFT) protein was designed to replace GFP within the cytosolic N terminus of KOR1 (Figure 1B). The new tdFT reporter consists of a tandem fusion of monomeric superfolder GFP (msfGFP) and monomeric Strawberry (mST; Figure 1B), with a t0.5 value (half-maximal fluorescence maturation at 37°C) of <1 min and 50 min, respectively (Khmelinskii et al., 2012; Barry et al., 2016). Upon induced biosynthesis, newly synthesized tdFT-KOR1 will initially produce only green fluorescence. Red fluorescence will develop more slowly, specifically marking aged tdFT-KOR1 molecules. Of note, the t0.5 values were obtained for the maturation of presynthesized proteins at 37°C. Upon triggered induction, plant cells take longer to synthesize mRNA and polypeptide for the accumulation of detectable levels of tdFT-KOR1 at the ambient growth temperature.
The tdFT-fusions were prepared for four types of KOR1. These included the wild type, ΔP611 from Figure 1, G429R containing the same point mutation (Gly429 to Arg) as the temperature-sensitive rsw2-1 allele, and Δall, in which all N-glycosylation sites were disrupted by amino acid replacement (Rips et al., 2014), so that we could compare the transport routes for different variants. Coding sequences of tdFT-KOR1 variants (genomic fragments) were placed under an estradiol-inducible promoter (Zuo et al., 2000a) and introduced into Arabidopsis. To start the expression, 5-d-old seedlings were incubated with estradiol solution. With tdFT-KOR1 in the Col-0 wild-type host, expression patterns were patchy and inconsistent as reported previously (Supplemental Figure 3; Zuo et al., 2000a). However, rdr6-11 mutant plants lacking posttranscriptional gene silencing showed more uniform and reproducible expression, as reported previously for a dimeric Förster resonance energy transfer (FRET) sensor (Deuschle et al., 2006). Therefore, rdr6-11 was used as host for all tdFT experiments. In preliminary analyses, we observed similar patterns in Col-0 and rdr6-11, as long as there was no loss of expression (Supplemental Figure 3).
KOR1 trafficking was analyzed using stably transformed homozygous lines for each construct in a 10-h time course. Induction of tdFT-KOR1 was triggered by treating seedlings with 2 µM β-estradiol for 2 h, and KOR1 levels/localizations were monitored for an additional 8 h on media without estradiol. In Supplemental Figure 4, the green (GFP) channel shows total KOR1 protein and the red (mST) channel aged KOR1. Early after estradiol induction (1 h postinduction, hpi), the GFP signal of the wild-type tdFT-KOR1 became detectable in intracellular compartments, likely the TGN, and then also at the PM (2 hpi). Up to 6 hpi, GFP signals at the PM became stronger and exceeded those at the TGN. The mST signal of aged KOR1 was first detected at the PM (6 hpi), increasing further, with intracellular signals corresponding to the TGN also becoming visible (10 hpi). Somewhat weaker signals in the green channel at 10 hpi are likely the result of FRET because the emission spectrum of msfGFP significantly overlaps with the excitation spectrum of mST. Previous study indicated that FRET between GFP and red fluorescent protein enhances timer signal and is a desirable feature for designing timers (Barry et al., 2016). These results suggested that newly synthesized KOR1 proteins quickly move along the secretory pathway, that is, from the ER via Golgi stacks to the TGN and then to the PM. After PM delivery, some of the aged KOR1 proteins returned, that is, started to accumulate at the TGN.
Next, we tested three KOR1 variants with enhanced TP accumulation. The time-course profile of tdFT-KOR1∆P611 was similar to that of the wild-type tdFT-KOR1, consistent with the model that newly synthesized KOR1∆P611 is first delivered to the PM (Supplemental Figure 4). However, at 10 hpi, red fluorescence of aged KOR1∆P611 was detected at the TGN again, and also at TPs, suggesting KOR1∆P611 was retrieved from the PM and transported via the TGN to the TP (Supplemental Figure 4). By contrast, both tdFT-KOR1G429R and tdFT-KOR1∆all dwelled in the ER at 1 and 2 hpi. At 4 hpi, KOR1G429R was detected at the TGN and the PM, whereas KOR1∆all remained in the ER (Supplemental Figure 4). At 6 hpi, aged KOR1G429R produced strong red signals at the PM and TP, which continued to develop until 10 hpi. This indicated that the G429R mutation slows down the ER exit of tdFT-KOR1G429R, but this variant reaches the PM prior to accumulation at the TP. By contrast, KOR1∆all did not produce clear signals at the PM and continued to label the ER, even at 6 and 10 hpi. Aged KOR1∆all started to label the TP at 6 hpi, with increased signal intensity at 10 hpi. Together, this suggested that KOR1∆all uses a unique route from the ER to the TP without passing through the PM.
The relative age of tdFT-KOR1 was visualized and quantified by ratiometric analysis of GFP and mST signals (Figure 2; Supplemental Figure 2B). Over the time course, progressive aging of KOR1 was evident for all variants (Figure 2A). The wild-type KOR1 showed slight enrichment of more aged KOR1 in the PM at 6 hpi, but at 10 hpi, the age difference compared with the intracellular (TGN) KOR1 populations became insignificant (Figure 2B). For KOR1∆P611 and KOR1G429R, KOR1 proteins at the TP at 10 hpi were significantly older than at the PM or at the TGN (intracellular), supporting the idea that KOR1 reaches the TP via the TGN/PM. By contrast, KOR1∆all continued to age in the ER before appearing at the TP without detectable accumulation at the TGN or PM (Figures 2A and 2B). Overall, results obtained from the time-course induction and tdFT analysis suggested that direction of KOR1 variants to the TP could be either determined at the ER (KOR1∆all) or the TGN/PM (KOR1∆P611 and KOR1G429R).
Figure 2.
Age Dependency of Subcellular tdFT-KOR1 Localization.
(A) Time-course ratiometric images of tdFT-KOR1 variants revealing KOR1 distribution of different age. Ratiometric (mST/msfGFP) images are color coded as indicated. Details of experimental conditions are described in the Methods. Bar = 10 µm. WT, the wild type.
(B) Quantitative data of (A). Error bars represent the se of the mean (n = 15). Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test). Experiments were performed with two independent lines per construct with similar results. A.U., arbitrary units; ND, not detected.
Blocking Endocytosis of KOR1 Inhibits TP Targeting via the PM, but Not via ERQC
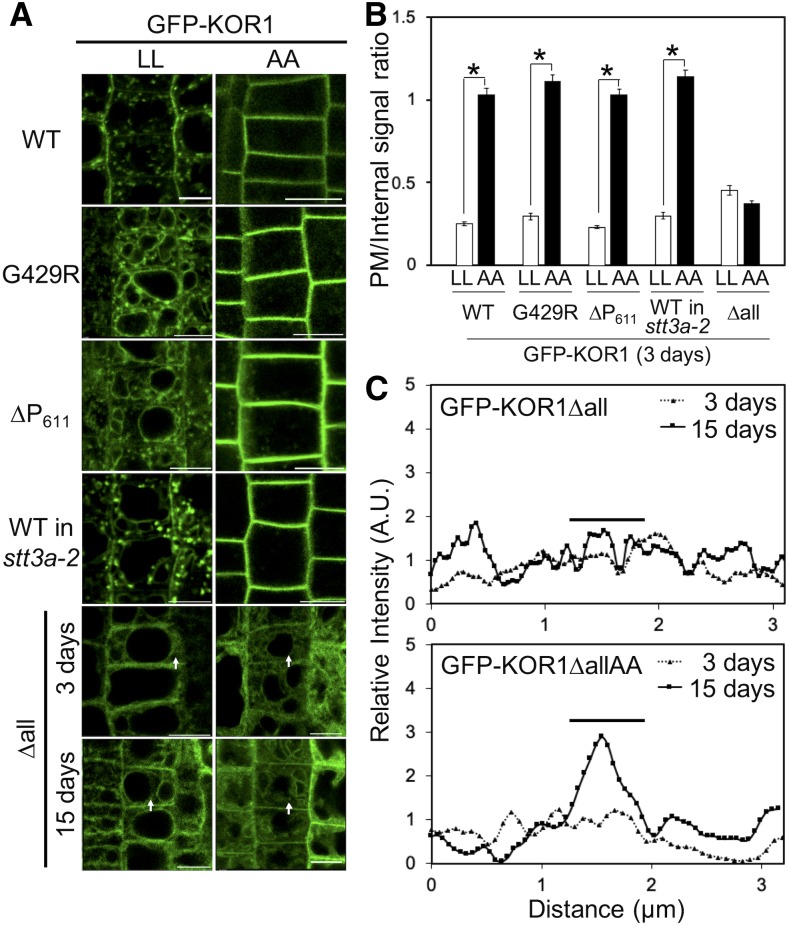
To firmly establish the TP targeting routes/mechanism for each KOR1 variant, we analyzed the behavior of a GFP-KOR1 variant whose transport is blocked at the PM. The cytosolic domain of KOR1 contains L48L49 followed by a Y59xxΦ motif, both known to be bound by ADAPTOR PROTEIN COMPLEX-2 (AP-2), which is involved in clathrin-mediated endocytosis (CME) from the PM (Kelly et al., 2008; Di Rubbo et al., 2013). If KOR1 is delivered to the PM before being recycled to the TGN and sorted to the TP, mutation of the di-Leu (LL) motif should trap KOR1 at the PM and prevent its accumulation at the TGN and the TP. We introduced A48A49 mutations (AA) into the wild-type KOR1 and its variants (using KOR1 genomic fragments) to produce GFP-KOR1AA, GFP-KOR1∆P611AA, GFP-KOR1G429RAA, and GFP-KOR1ΔallAA. Also, KOR1AA was expressed in stt3a-2 (underglycosylation background; Koiwa et al., 2003). Surprisingly, all KOR1AA variants, except for KOR1ΔallAA, almost exclusively accumulated at the PM, producing on average three to four times higher GFP signal levels at the PM compared with the corresponding variants with intact LL motif (Figures 3A and 3B; Supplemental Figure 2C). By contrast, neither KOR1Δall nor KOR1ΔallAA showed clear PM signals in 3-d-old seedlings. However, some weak but distinct signals were observed in 15-d-old KOR1ΔallAA plants at PM/cell wall regions, suggesting that minute amounts of KOR1Δall/KOR1ΔallAA can escape ERQC and reach the PM (Figures 3A and 3C).
Figure 3.
Subcellular Localization of GFP-KOR1 Variants with Intact (LL) or Mutated (AA) Cytoplasmic LL Motif.
(A) Three-day-old plants of all lines and 15-d-old plants of KOR1∆all were used for observation. Bar =10 µm. WT, the wild-type GFP-KOR1.
(B) Quantitative data of (A), n = 30. The signal at the PM was divided by the internal signal of cells. Error bars represent the se of the mean (P < 0.01, Welch’s t test). WT, the wild type.
(C) Signal intensity of GFP-KOR1∆all and ∆allAA fluorescence along the arrows in (A). For relative intensity, each signal intensity was normalized by the average signal intensity of each genotype. The bars indicate the position of PM/cell wall. A.U. = arbitrary units.
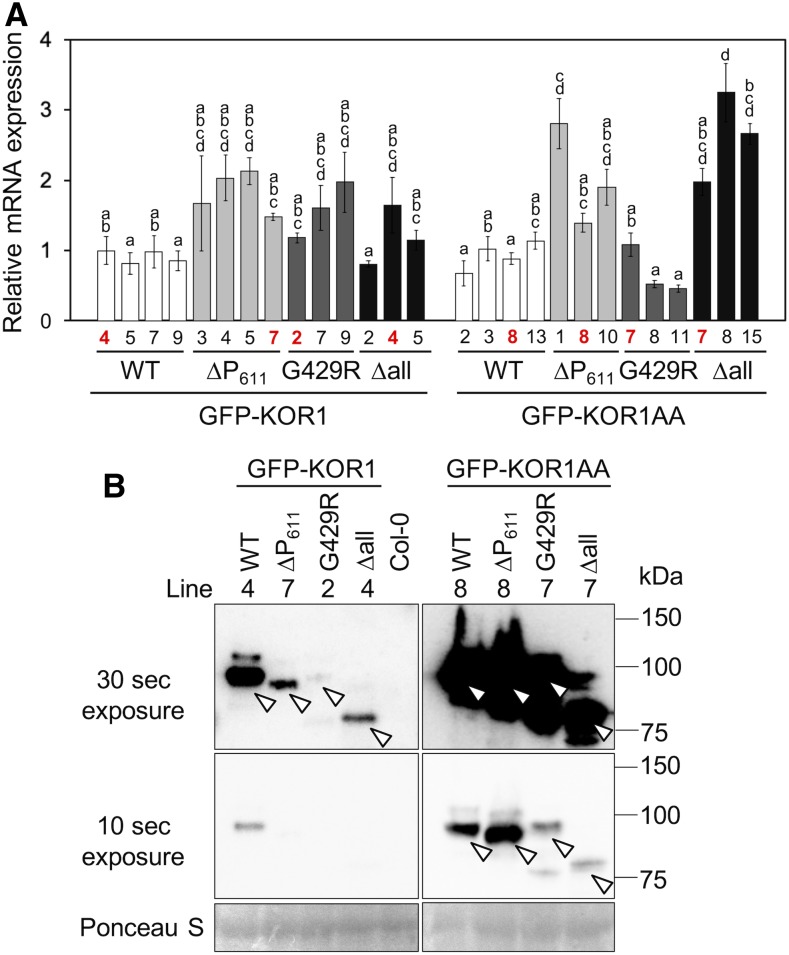
ER-to-TP sorting of KOR1Δall is likely the result of ERQC, which specifically marks misfolded proteins for degradation. By analogy, diverting KOR1∆P611 and KOR1G429R from the TGN/PM cycle to the TP likely functions as a quality control mechanism to sort dysfunctional/expired proteins for degradation. If so, preventing TP targeting should lead to stabilization of the proteins. To test whether the AA mutation increases the amount of detectable KOR1, KOR1 mRNA and protein levels were determined in the transgenic lines, using RT-qPCR and anti-GFP immunoblot analyses (Figure 4). Transcript levels of the various GFP-KOR1 transgenes were within a threefold range compared with the wild-type GFP-KOR1 (Figure 4A). Relatively high transcript levels found for the ΔallAA lines are likely due to the selection of high expression lines during the T1 transformant screen, because overall GFP signals obtained for ΔallAA were weaker than those for the other variants. Lines with similar transcript levels were subjected to immunoblot analysis (Figure 4B). When the LL motif was left intact, protein levels of GFP-KOR1∆P611, GFP-KOR1G429R, and GFP-KOR1Δall were substantially less abundant than of the wild-type GFP-KOR1, suggesting that these variants are prone to degradation. By contrast, the AA mutation substantially increased protein detection for all GFP-KOR1 variants tested, albeit to a lesser extent for GFP-KOR1G429R and GFP-KOR1ΔallAA (with little PM accumulation). This result indicated that the LL motif functions in retrieval of KOR1 from the PM to promote cycling and turnover of KOR1 proteins, and KOR1 is likely degraded upon delivery to the TP.
Figure 4.
Protein and Transcript Levels of GFP-KOR1 Variants.
(A) Expression levels of GFP-KOR1 transgenes determined by RT-qPCR using GFP-specific primers. Error bars represent the se of the biological triplicates. Lines marked with red were used for immunoblot analysis. Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test). WT, the wild type.
(B) Immunoblot of GFP-KOR1 proteins using an anti-GFP antibody. Positions of GFP-KOR1 main bands are marked by open arrowheads. Note that KOR1∆all and ∆allAA lack all eight N-glycans and migrate ∼16 kD smaller than the rest. Ponceau S staining is shown as the loading control. Two exposures (30 and 10 s) are shown to compare protein levels. WT, the wild type.
The LL Motif Increases Intracellular Availability of KOR1 for Cell Plate Targeting
The above-mentioned data showed that the LL motif of KOR1 functions in retrieving KOR1 from the PM. On the other hand, Zuo et al. (2000b) reported that the LL and YxxΦ motifs of KOR1 function as polarized targeting signals that direct KOR1 to the growing cell plate during cell division in telophase, using GFP-KOR1 expressed in liquid-cultured tobacco BY-2 cells. In light of our observations described above, in planta systems that can visualize the dynamics of KOR1 in differentiated tissues are necessary to clarify the role of the LL motif.
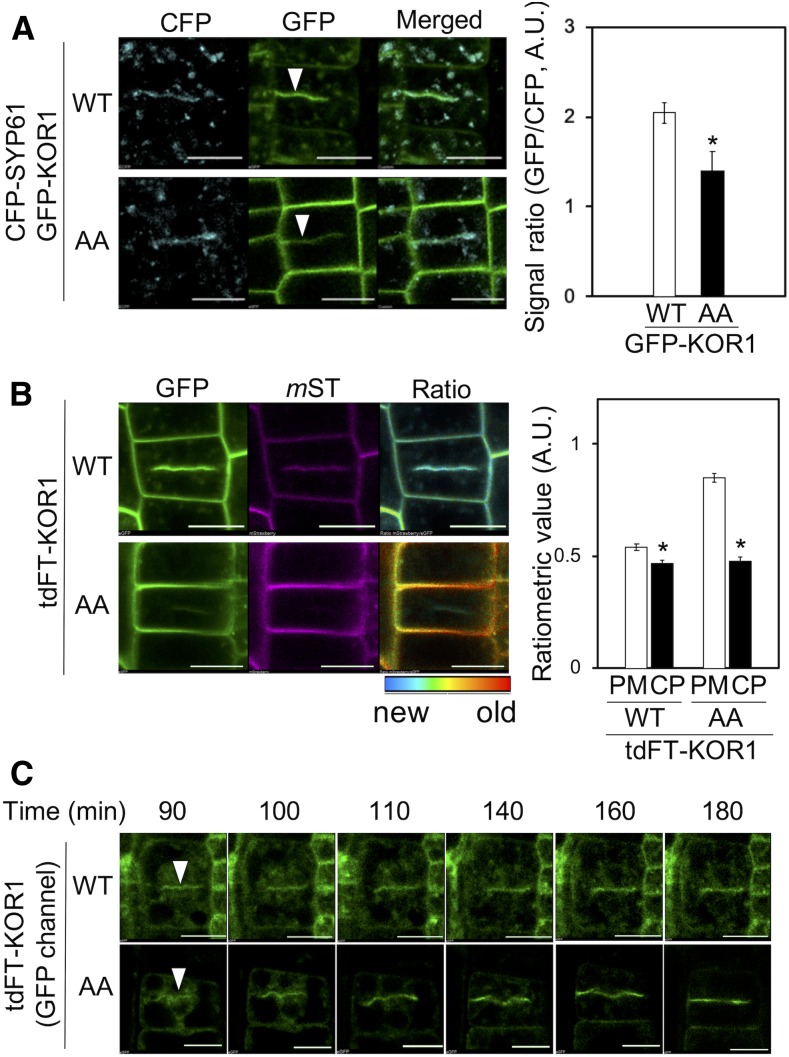
For this purpose, we crossed the GFP-KOR1 and GFP-KOR1AA lines with TGN-marker line cyan fluorescent protein (CFP)-SYP61 (Col-0 background), which also accumulates at cell plates (Steiner et al., 2016). Both the wild-type GFP-KOR1 and CFP-SYP61 were detected at developing cell plates, as well as at the PM and the TGN (Figure 5A, top left). In root tissues expressing GFP-KOR1AA, uniformly labeled cell outlines suggested that most GFP-KOR1AA is confined to the PM. However, we could also identify cells with GFP signals at the cell plate (Figure 5A, bottom left). At the cell plate, the GFP-KOR1/CFP-SYP61 signal ratio was consistently higher with the wild-type GFP-KOR1 than that of GFP-KOR1AA (Figure 5A, right; Supplemental Figure 2D). There could be two reasons for this observation. First, the KOR1-LL motif indeed functions as a polarized targeting signal in telophase. Second, the LL motif promotes retrieval of KOR1 from the PM, which increases the availability of intracellular KOR1 for targeting to the cell plate.
Figure 5.
Both GFP-KOR1 and GFP-KOR1AA Are Targeted to Cell Plates.
(A) GFP-KOR1 and GFP-KOR1AA colocalize with CFP-SYP61 at cell plates (white arrowheads). The graph shows the ratio of GFP and CFP at the cell plate (CP). A.U., arbitrary units; WT, the wild type.
(B) Ratiometric age analysis of de novo-synthesized tdFT-KOR1 and tdFT-KOR1AA at 6 hpi. For the quantification, values at the PM and CP were determined using the same method as described in Figure 2. Error bars represent the se of the mean (n = 10). The asterisks indicate significant differences to each value at the PM (P < 0.01, Welch’s t test). Bar = 10 µm. A.U., arbitrary units; WT, the wild type.
(C) De novo–synthesized tdFT-KOR1 and tdFT-KOR1AA are preferentially targeted to cell plates in dividing cells (white arrowheads). The time-course micrographs were taken starting from 90 min until 180 min after β-estradiol treatment. Only young KOR1 proteins (green fluorescence) were detected. WT, the wild type.
To assess the pre-equilibrium transport of KOR1 and KOR1AA, we analyzed tdFT-KOR1 transport to the cell plate (Figure 5B; Supplemental Figure 2B). Ratiometric analyses showed that young KOR1 is enriched at cell plates for both the wild-type KOR1 and KOR1AA at 6 hpi. The age difference of KOR1 between the PM and cell plate was more distinct with KOR1AA than with the wild-type KOR1. This may indicate that young KOR1 proteins are targeted to the cell plate in both the KOR1 and KOR1AA lines, but upon aging, the wild-type KOR1 is recycled and redistributed, making age distribution of KOR1 in the cell uniform, whereas KOR1AA is forced to age at the PM. On the other hand, we cannot exclude the possibility that the LL motif promotes targeting of KOR1 to the cell plate, because the wild-type KOR1 accumulated to a higher level at the cell plate than KOR1AA. This is due to the fact that the images for ratiometric analysis were taken at 6 hpi so that sufficient mST signals for analysis could be detected. At this time point, it is likely that some KOR1 proteins at the PM were already retrieved (via the function of their LL motifs) and redirected to the TGN, where they became a secondary source of KOR1 for targeting to the growing cell plate.
To avoid the contribution of recycled KOR1 at the PM, and test whether the wild-type KOR1 may be more preferred for targeting to the cell plate, we studied cell plate formation in tdFT-KOR1 at 90 to 180 min postinduction, the very early stage of the estradiol treatment (Figure 5C). At this stage, only young KOR1 molecules were present, since we did not detect mST signals. For the GFP signals, we could observe the delivery of KOR1 to the cell plate before substantial accumulation at the PM, suggesting only minimal contribution of PM-recycled KOR1 by this analysis. Both the wild-type KOR1 and KOR1AA accumulated at the cell plate in a similar fashion. Therefore, we concluded that the apparent enrichment of KOR1 with a functional LL motif at newly forming cell plates is not due to the polarized targeting by the LL motif as previously proposed but to the increased intracellular availability of KOR1 retrieved from the PM.
Proper Localization of KOR1 Is Essential for Root Growth under Abiotic Stress Conditions
The above-mentioned results showed that localization of KOR1 in the cell is determined by the hierarchical interaction of sequence motifs and protein structures. The functional significance of different KOR1 locations, on the other hand, is not fully understood. To quantify the functionality of KOR1 variants showing different localization patterns, we measured the growth of mutant plants (rsw2-1) complemented with the GFP-KOR1 variants. The suitability of rsw2-1 (kor1G429R) for complementation analyses was confirmed, since first, the localization patterns of GFP-KOR1, GFP-KOR1∆P611, and GFP-KOR1G429R were not affected by the host genotype (the Col-0 wild type or rsw2-1; Figure 1C; data not shown), and second, the growth phenotype of transgenic rsw2-1 plants expressing GFP-KOR1 was indistinguishable to that of the untransformed wild-type Col-0 (see below). These results are consistent with previous observations (Rips et al., 2014; Liu et al., 2018) and ensure that the recessive rsw2-1 mutant allele does not interfere with functions of the GFP-KOR1 transgenes.
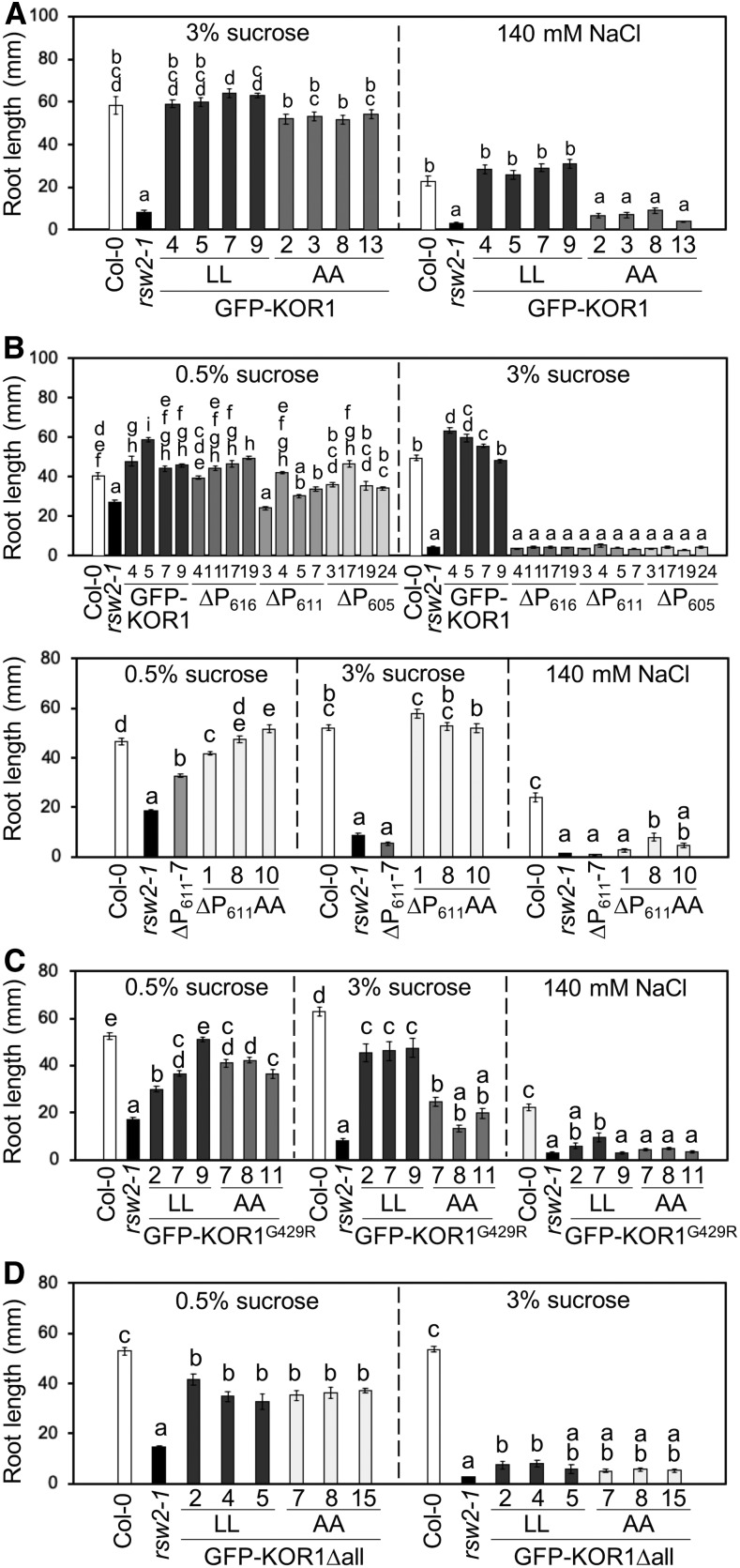
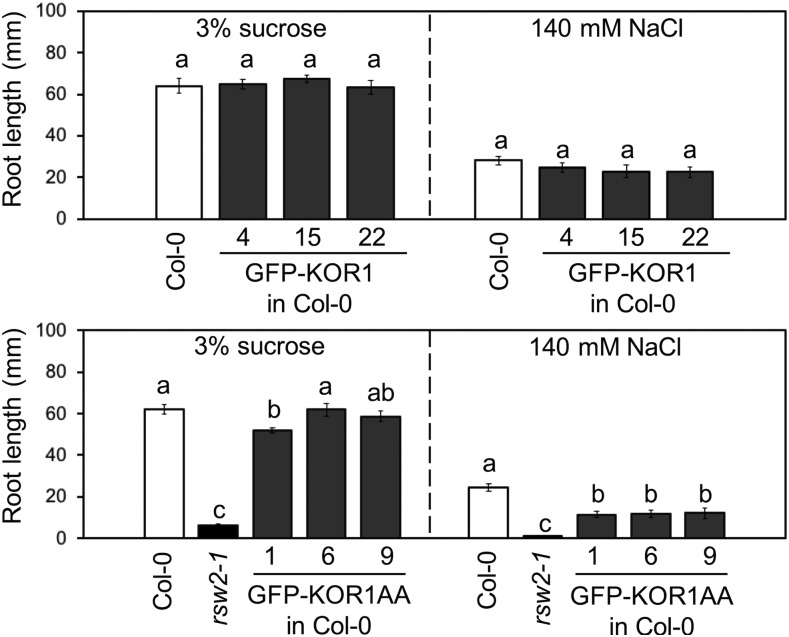
As shown in Figure 6A, the wild-type GFP-KOR1 fully complemented root growth of the rsw2-1 mutant, both under standard (1× Murashige and Skoog [MS] salts and 3% [w/v] Suc) and salt-stress conditions (140 mM NaCl). Growth of the PM-localized GFP-KOR1AA was comparable to the wild-type GFP-KOR1 under the standard condition; however, the ability to support growth under salt stress was lost, suggesting that locking KOR1 at the PM is detrimental under salt stress. For the GFP-KOR1∆P transformants (in the rsw2-1 host), the presence of 3% (w/v) Suc (standard condition) severely inhibited root growth, even though partial complementation was found at a lower Suc concentration (1× MS salts and 0.5% [w/v] Suc; Figure 6B, top). These results indicate that the C-terminal P-motif (amino acids 606 to 621; Figure 1) is necessary for full functionality of KOR1. The GFP-KOR1G429R lines (with extra copies of the KOR1G429R variant corresponding to the rsw2-1 host allele) showed up to 80% rescued root growth compared with the wild-type Col-0 under the standard condition, which was nearly abolished under salt stress (Figure 6C).
Figure 6.
GFP-KOR1 Transgene Variants Differentially Rescue Root Growth of the rsw2-1 Host.
(A) to (D) (A) GFP-KOR1 and GFP-KOR1AA transformants, (B) GFP-KOR1∆P and GFP-KOR1∆PAA series, (C) GFP-KOR1G429R and GFP-KOR1G429RAA transformants, and (D) GFP-KOR1Δall transformants. The growth condition for each experiment is listed above the graph. Three to four independent homozygous T3 lines were analyzed for each construct. Details of plant growth conditions are described in the Methods. Error bars represent the se of the mean with 13 to 35 plants per line. Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test).
Introduction of the A48A49 mutation (locking KOR1 at the PM) improved complementation for three GFP-KOR1 variants (∆P611, G429R, and ∆all) under low Suc (0.5% [w/v]), but growth under the standard condition was differentially affected: GFP-KOR1∆P611AA was comparable to the wild type (Figure 6B, bottom), suggesting that the P-motif is not necessary for KOR1 function at the PM. By contrast, that of GFP-KOR1G429RAA (Figure 6C) was less improved and that of GFP-KOR1∆allAA (Figure 6D) was not different from GFP-KOR1∆all. These results are consistent with the hypothesis that the overall function of KOR1G429R is compromised and that the majority of GFP-KOR1∆allAA does not reach the PM. Similar to GFP-KOR1AA, all AA variants failed to restore root growth under salt stress.
Internalization of KOR1 Is Essential for the Establishment of Salt Tolerance
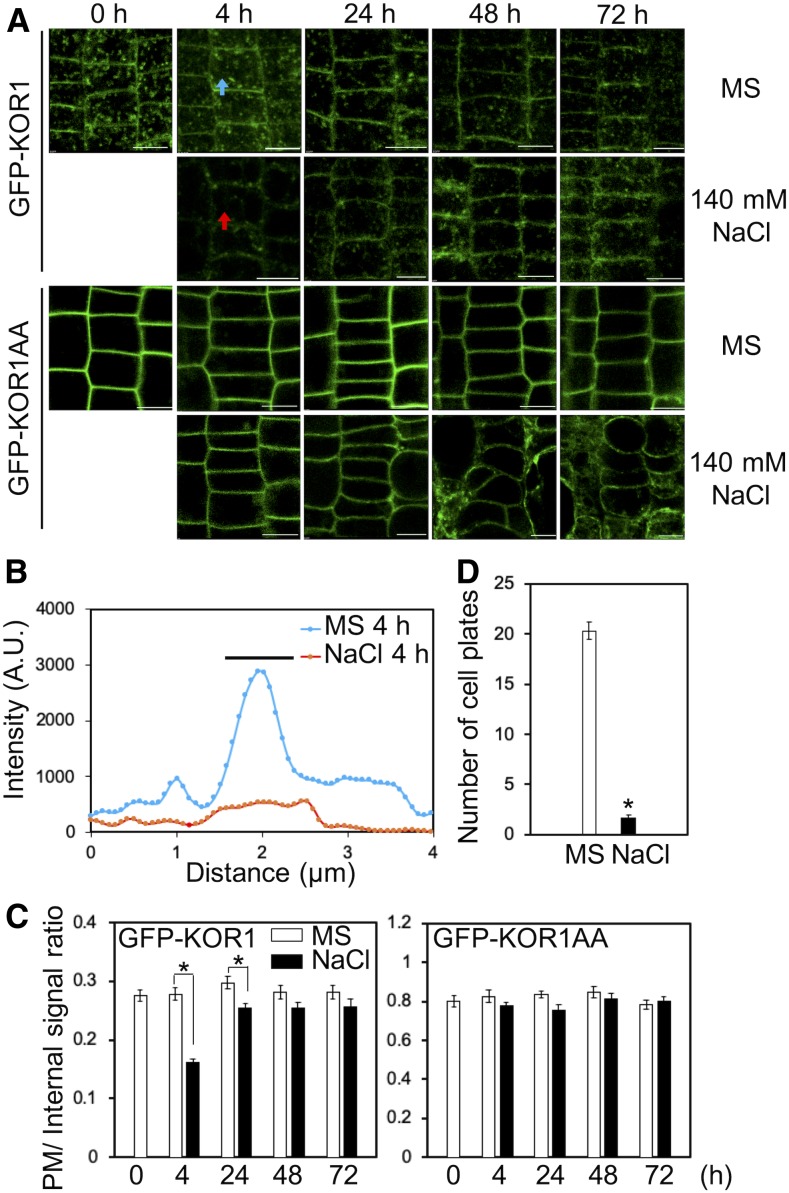
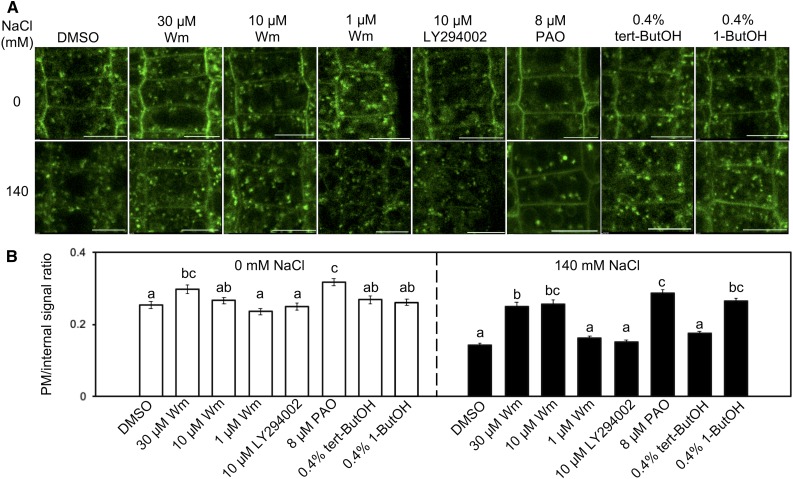
To determine why all PM-retained KOR1AA variants could not support root growth under salt stress, cellular KOR1 distribution patterns were analyzed during the onset of salt stress (Figure 7). Upon treatment with 140 mM NaCl, GFP-KOR1 signals at the PM decreased substantially after 4 h (Figures 7A to 7C), but recovered after 24 h. By contrast, GFP-KOR1AA signals at the PM did not change over the entire time course of the salt treatment. This strongly indicated that salt-induced internalization of KOR1 is essential for establishing stress-acclimated root growth. Salt stress also decreased the number of GFP-KOR1–positive cell plates (Figure 7D), consistent with reduced cell division activity under stress (Koiwa et al., 2003). To determine the nature of salt-induced GFP-KOR1 internalization, we tested the impact of vesicle transport inhibitors on GFP-KOR1 in rsw2-1 (Figure 8). Salt-induced internalization was blocked by 10 µM wortmannin (Wm), which inhibits phosphoinositide 3-kinase (PI3K) and phosphoinositide 4-kinase (PI4K; Jung et al., 2002; Aggarwal et al., 2013; Fujimoto et al., 2015). However, treatment with 10 µM LY294002 or only 1 µM Wm (both more specific for PI3K; Jung et al., 2002; Lee et al., 2008; Aggarwal et al., 2013; Fujimoto et al., 2015) did not inhibit salt-induced GFP-KOR1 internalization. On the other hand, 8 µM phenylarsine oxide (PAO, more specific for PI4K; Vermeer et al., 2009; Fujimoto et al., 2015) inhibited salt-induced internalization. Interestingly, PAO and high concentrations of Wm also increased the PM signal in the absence of salt stress, suggesting that PI4K function is part of the normal KOR1 cycling mechanism. This is consistent with a previous study, showing that PI4K is important for internalization of CesA3 (Fujimoto et al., 2015). Salt-induced KOR1 internalization was specifically inhibited by 0.4% (v/v) 1-butanol, which is a substrate of phospholipase D (PLD) and inhibits phosphatidic acid formation, but was not affected by tert-butanol, a poor substrate/inhibitor of PLD. Among the known processes involving PI4K and PLD are clathrin-mediated endocytosis and vesicle fission via induction of negative membrane curvature by phosphatidic acid (Li and Xue, 2007; Donaldson, 2009; Fujimoto et al., 2015), respectively. While it is impossible to exclude putative secondary effects of the inhibitors on other cellular targets, together with the result that salt-induced internalization was blocked in GFP-KOR1AA mutated in the AP-2 binding motif, the results indicate that salt-induced KOR1 internalization is indeed an endocytotic process (Donaldson, 2009; Seaman, 2012; McLoughlin et al., 2013; Fujimoto et al., 2015). Under normal conditions, KOR1 internalization was not affected by 1-butanol (Figure 8B). Thus, apparently a stress-specific, PLD-dependent mechanism controls KOR1 entrance to the endocytotic pathway.
Figure 7.
Salt Stress Leads to Internalization of GFP-KOR1, but Not PM-Locked GFP-KOR1AA.
(A) Time-course observation of GFP-KOR1 and GFP-KOR1AA under salt stress. Three-day-old seedlings growing on a cellophane membrane placed on medium containing 1× MS salts and 3% (w/v) Suc were transferred to the same medium supplemented with no or 140 mM NaCl for the indicated period before root cells were imaged using a confocal microscope. Bar = 10 µm.
(B) Signal intensity of GFP-KOR1 fluorescence along the arrows across the PM in (A). The bar indicates the position of the PM/cell wall. A.U., arbitrary units.
(C) Quantitative data of (A), n = 50. The signal at the PM was divided by the internal signal of cells.
(D) Number of GFP-KOR1–positive cell plates per plant without or with 140 mM NaCl treatment for 4 h. Error bars represent the se of the mean (n = 30). The asterisks indicate significant differences between treatments (P < 0.01, Welch’s t test).
Figure 8.
Salt Stress–Induced KOR1 Internalization from the PM Is Distinct from Normal PM/TGN Cycling.
(A) Effect of drug treatment on the internalization of GFP-KOR1 under salt stress: Wm, a PI3K/PI4K inhibitor; LY294002, a PI3K inhibitor; PAO, a PI4K inhibitor; 1-butanol (1-ButOH), a PLD inhibitor; tert-butanol (tert-ButOH), negative control for 1-butanol. Five-day-old plants growing on a cellophane membrane placed on medium containing 1× MS salts and 3% (w/v) Suc were transferred to the same medium supplemented with 140 mM NaCl and/or inhibitors as indicated for 4 h. Bar = 10 µm.
(B) Quantitative data of (A). For each cell, the signal intensity at the PM was divided by the intracellular signal intensity. Error bars represent the se of the mean (n = 50). Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test).
Two possible scenarios could explain how the PM-confined KOR1AA variants may confer salt sensitivity to the host. First, an intracellular function of KOR1 may be required for establishing salt tolerance, and second, KOR1 locked at the PM might prevent proper adaptive salt-stress responses. To assess these possibilities, we introduced GFP-KOR1AA into Col-0 (with functional endogenous KOR1). Interestingly, GFP-KOR1AA, but not GFP-KOR1, conferred salt hypersensitivity to the wild-type roots (Figure 9), strongly indicating that presence of KOR1AA at the PM during salt stress is inhibitory to establishing tolerance in the host plants.
Figure 9.
Root Growth of GFP-KOR1 and GFP-KOR1AA in the Col-0 Wild-Type Background.
Experiments were conducted as described in Figure 6. The growth condition for each experiment is listed above each graph. Error bars represent the se of the mean (10 to 21 plants per line). Different letters show significant differences between genotypes (P < 0.05, one-way ANOVA followed by Tukey’s honestly significant difference post hoc test).
DISCUSSION
In this study, we report on the regulation of subcellular dynamics of KOR1 that is targeted to multiple subcellular domains, such as the TGN, PM, and TP. TP accumulation of KOR1 was observed in several reporter-fusion studies but initially considered an artifact (Robert et al., 2005; Vain et al., 2014), because only low levels of endogenous KOR1 were detected in the TP fraction (Nicol et al., 1998). Also, reporter genes used in those earlier studies did not fully rescue the kor1 mutant phenotype (Robert et al., 2005; Vain et al., 2014). We previously developed a GFP-KOR1 reporter construct based on the entire genomic fragment with GFP embedded in the cytoplasmic domain (Szyjanowicz et al., 2004; Rips et al., 2014). The resulting GFP-KOR1 fusion fully complemented root and shoot growth of rsw2-1 and produced only low levels of TP signal (Rips et al., 2014), consistent with a previous biochemical study (Nicol et al., 1998). This allowed us to analyze which factors and modifications may divert KOR1 from the PM/TGN cycle to accumulate at the TP. We now implemented a tdFT approach to track pre-equilibrium transport of de novo–synthesized KOR1 in the secretory system. The key benefit of this new technology (compared to the simple inducible expression of reporter fusions) is that the aged protein population appears in a different color, informing about possible transport direction(s) of KOR1 in the cell. Data obtained by this analysis likely represent single-pass transport routes of KOR1 upon induced biogenesis, that is, before entering the steady state of cycling between the PM and TGN. To minimize reporter artifacts, the same strategy was used: tdFT was embedded in the cytosolic domain of a KOR1 genomic fragment.
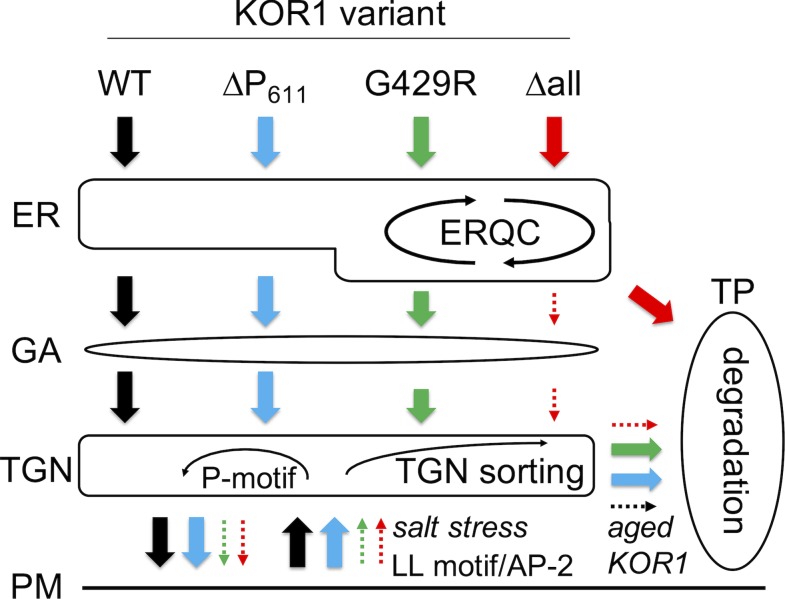
Using this carefully tuned system, we assessed transport patterns of KOR1 variants modified in various regulatory motifs or residues that likely affect protein structure. A model for KOR1 transport, as deduced from this study, is presented in Figure 10. By default, all newly synthesized KOR1 proteins except KOR1Δall are delivered to the PM using the conventional secretory route (ER→GA→TGN→PM). This is supported by the observations that nearly all tdFT-KOR1 proteins were quickly delivered to the PM with very short dwell time in the ER/GA/TGN, and all endocytosis-defective AA variants except for KOR1∆allAA accumulated at the PM almost exclusively. The presence of complex-type N-glycans on KOR1 also supports the transport of KOR1 through the GA (Liebminger et al., 2013; Rips et al., 2014). Moreover, although compromised KOR1 variants accumulated at the TP, the route to this final location could vary. The majority of KOR1∆all was segregated early. KOR1∆all had long dwell time at the ER after synthesis (Figure 2; Supplemental Figure 4, at 6 hpi) followed by accumulation at TPs. This likely reflects multiple rounds of folding trials in the lumen and sorting by ERQC mechanisms, through which membrane proteins that fail to fold correctly are sent to the vacuole for degradation. On the other hand, the transport of both KOR1G429R and KOR1∆P611 variants occurred after arriving at the PM. Supporting this model, the ratiometric analyses showed that tdFT-KOR1G429R and tdFT-KOR1∆P611 proteins at the TP were more aged than those at the TGN/PM, and endocytosis-defective AA variants abolished TP accumulation of KOR1G429R and KOR1∆P611. At the PM, KOR1 molecules destined for internalization by CME (aged, salt stress) are bound by the AP-2 complex (recognizing the LL and Y59xxΦ motifs) and possibly by the TPLATE complex that cooperates with the AP-2 complex (Van Damme et al., 2011; Gadeyne et al., 2014; Bashline et al., 2015; Zhang et al., 2015; Sánchez-Rodríguez et al., 2018) delivered to the TGN. During/after this retrieval process, the protein integrity status is likely monitored, and KOR1G429R and KOR1∆P611 variants are sorted to the TP. This probably reflects the operation of TGN sorting mechanisms to remove damaged/expired KOR1 proteins. The C-terminal P-motif may function in retaining KOR1 in TGN/PM cycles, ensuring that functional KOR1 is sent to the PM again. The degradative role of TGN sorting is also supported by the significant increase of protein levels for KOR1 variants that are blocked at the PM by the AA mutation.
Figure 10.
Model of KOR1 Transport Regulation.
Upon synthesis and insertion into the ER, KOR1 undergoes N-glycan–assisted luminal folding and ERQC; but in case KOR1 fails to fold correctly (such as KOR1∆all), the polypeptide is targeted to the TP for vacuolar degradation. After N-glycan maturation in the GA, folded KOR1 is transported to the TGN, and PM. Upon delivery to the PM, some KOR1 return to the TGN via an endocytotic pathway that is mediated by the LL motif and AP-2 complex. Internalized KOR1 is quality checked at the TGN (TGN sorting) and redirected to the PM, which is facilitated by the P-motif, or sorted to the TP for degradation. WT, the wild type.
Also assessed was the function of the LL motif of KOR1, which was previously studied as polarized sorting signal to the cell plate using tobacco BY-2 cells (Zuo et al., 2000b). Our data showed that the presence of the LL motif was not crucial for specific targeting of KOR1 to the cell plate. Instead, it was necessary for retrieving KOR1 from the PM and making it available for TGN-cell plate delivery. This was consistent with the fact that both the LL and Y59xxΦ motifs of KOR1 match the AP-2 recognition motif for CME at the PM (Kelly et al., 2008; Di Rubbo et al., 2013). Interestingly, LL motifs within acidic environments are also recognized by other AP complexes, such as AP-1, which sorts vacuolar iron/manganese transporter AtVIT1 (E16kqtLLD) to the TP (Wang et al., 2014). While the LL motif of KOR1 (D41EtqqswLL) slightly deviates from the consensus of acidic LL motifs found in proteins of mammalian cells ([DE]xxxL[LI]; Bonifacino and Traub, 2003), proteomic studies detected phosphorylation of T43/S46 upstream of the KOR1 L48L49 motif (Nühse et al., 2007). However, whether phosphorylation of these adjacent sites may function in conditional targeting of KOR1, for example, from the TGN to the TP, is not clear at this point.
The conserved luminal P-motif is important for KOR1 localization and intracellular function. Pro-rich C-terminal sequences are relatively rare in Arabidopsis, and besides KOR1 proteins, we did not find similar motifs in those previously reported for the TGN proteome (Drakakaki et al., 2012). Hence, it is unlikely that the P-motif has a universal function in retaining proteins at the TGN but instead seems related to KOR-specific processes. Interestingly, microbial xylanase A (isolated from the sheep ruminal microbiome) contains a C-terminal Pro-rich sequence that is absent from other homologous GH10-group β-1,4-xylanases (Li et al., 2014). This P-rich sequence broadened the pH and temperature optima of xylanase A, thereby improving catalytic activity and thermostability of the enzyme. By analogy, the P-motif may also improve enzymatic function and stability of KOR1 in plant cells. However, direct involvement of the P-motif in basal protein stability or catalysis is unlikely, because KOR1∆P611 was functional when targeted to the PM, and all KOR1∆P variants rescued growth of aerial plant parts. Moreover, OsCel9A, a secreted GH9 cellulase, produces active enzyme when posttranslationally cleaved at the C terminus further upstream of the truncations used in this study (Supplemental Figure 1; Yoshida et al., 2006; Yoshida and Komae, 2006). Hence, it is more plausible that the KOR1 P-motif functions as an interface with yet to be identified partners in the TGN. In that case, the intact P-motif may have functions for stabilizing KOR1 at the TGN, allowing re-entry into regions from which PM-targeted vesicles form over those destined for TP targeting (Sanderfoot et al., 2001). This would explain why steady-state cycling of KOR1∆P between the PM and TGN decreased, and TP accumulation increased. The localization-related role of the P-motif is also consistent with the observation that PM-confined KOR1∆P611AA showed similar functionality as KOR1AA with intact C terminus.
Overall, our findings demonstrate the existence of several layers of regulatory mechanisms that are required to retain KOR1 in TGN-to-PM cycles. The cytoplasmic LL motif, as well as the luminal P-motif, are necessary to maintain a sufficiently large pool of KOR1 at the TGN, from where the protein also reaches the cell plate during cell division. Although PM-localized KOR1 could fulfill the major part of KOR1 function, endocytotic internalization of KOR1 seems essential for salt adaptation of plants, because locking KOR1 at the PM during salt stress was detrimental. The mechanism of the negative effect of PM-localized KOR1 is not clear, but during the period of salt-induced KOR1 internalization, MT depolymerization also occurs, as well as MT reorientation and a reduction of cell expansion (West et al., 2004; Wang et al., 2011). Hence, KOR1 retention at the PM may interfere with the salt-induced reorganization of MTs, although other possibilities cannot be excluded. Interestingly, proteomic studies identified additional possible regulatory modifications of KOR1, such as phosphorylation and palmitoylation (Nühse et al., 2007; Hemsley et al., 2013; Jones et al., 2016). The described tdFT system, which is suitable for differentiating subtle changes in protein trafficking, will be useful for dissecting the roles of other KOR1 modifications further.
METHODS
Plant Materials
The Arabidopsis (Arabidopsis thaliana) Col-0 ecotype was used for this study. Mutant genotypes rsw2-1 and stt3a-2 were described previously (Kang et al., 2008), as well as the GFP-KOR1 CFP-SYP61 line (Rips et al., 2014). For the preparation of the tdFT-KOR1 CFP-SYP61 line, CFP-SYP61 (Robert et al., 2008) was introduced into tdFT-KOR1 by crossing.
Plant Growth Conditions
Unless otherwise stated, seeds were sown on germination media (1/4× MS salts; Murashige and Skoog, 1962) with 0.5% (w/v) Suc and 0.7% (w/v) agar before stratification at 4°C for 2 to 4 d. Media plates were moved to an incubator set to 25°C and 16-h-light (using 50% output of PHILLIPS F17T8TL741) versus 8-h-dark conditions. After 7 to 10 d, plants were transferred to soil (Sunshine Mix #1 Fafard-1P, Sun Gro Horticulture) and moved to a growth chamber set to 22°C and 16-h-light (using 50% output of PHILLIPS F96T12HL41) versus 8-h-dark conditions.
Preparation of tdFT-KOR1g (Genomic Fragment) Vectors
Primers used in this study are listed in Supplemental Table 1. The tdFT template was prepared by replacing mCherry of pEnEOimCherryF3SGThsp (GenBank accession number KF537341) with a PCR product (primers 1581/1605) of msfGFP (monomeric superfolder GFP) without stop codon and then inserting mST (primers 1583/1584) behind msfGFP. The resulting tdFT (msfGFP-mST) was amplified by PCR (primers 1590/1591) and ligated to the BamHI site of pEnKORg (Rips et al., 2014), and tdFT-KOR1g variants were derived as described below. Resulting tdFT-KOR1g series fragments were amplified by PCR (primers 1611/1612) and inserted between XhoI and SpeI sites of estradiol-inducible cassette vector pER8exFTf, a derivative of pER8 (Zuo et al., 2000a). pER8extdFT-KOR1g sequences are listed in Supplemental File 1.
Construction of KOR1 Expression Cassettes
pEnGFP-KOR1g was prepared by ligating the SacII-ApaLI fragment of pEnGFP-KORc (Rips et al., 2014), the ScaI-ApaL1 fragment of pEnKORg (Rips et al., 2014), and the SacII-ScaI digested PCR product prepared using pEnKORg as template and primers (765/603). pEnGFP-KOR1g∆all was prepared as follows. First, the SacII-ScaI fragment of pEnGFP-KOR1g was replaced by an overlap extension product of PCR fragments (1. template pEnGFP-KORc∆all, primers 1007/1014; 2. template pEnKORg, primers 602/649; and 3. template pEnGFP-KORc∆all, primers 1241/1036; Ho et al., 1989; Rips et al., 2014). Second, the PpuMI-BstXI fragment of the resulting pEnGFP-KOR1g∆123 was replaced by an overlap extension product of PCR fragments (1. template pEnGFP-KORc∆all, primers 1233/1234; 2. template pEnKORg, primers 1235/1242; and 3. template pEnGFP-KORc∆all, primers 988/997). Finally, the ScaI-PpuMI fragment of resulting pEnGFP-KOR1g∆123678 was replaced by an overlap extension product of PCR fragments (1. template pEnKORg, primers 1037/1013; 2. template pEnGFP-KORc∆all, primers 1237/1239; and 3. template pEnKORg, primers 1238/1236). pEnGFP-KOR1g∆P605, ∆P611, ∆P616, PtoG, PtoS, and EXT were prepared by replacing the PpuMI-BstXI fragment of pEnGFP-KOR1g by a PCR product using pEnGFP-KOR1g as template and primers (1233/1649, 1233/1229, 1233/1650, 1233/1238, 1233/1239, and 1233/1240, respectively). To produce pEnGFP-KOR1AA, the AA fragment was amplified using primers 1620/1621 and cloned into the BspEI-ScaI sites of pEntdFT-KOR1g (Rips et al., 2014). BspEI is located between msfGFP and mST; therefore, only the msfGFP domain of tdFT remained in the vector. pEnGFP-RSW2AA and pEnGFP-RSW2 were prepared by ligating the ScaI/BstXI fragment of pEnFTf-KOR1G429R (containing the rsw2-1 genomic fragment) with ScaI/BstXI-opened pEnGFP-KOR1 or pEnGFP-KOR1AA. For pEnGFP-KOR1∆allAA, the BspEI-BstXI fragment of pEnGFP-KOR1∆all was replaced by a PCR product (template pEnGFP-KOR1∆all, primers 1620/997). Similarly, for pEnGFP-KOR1∆P611AA, the BspEI-BstXI fragment of pEnGFP-KOR1g was replaced by a PCR product (template pEnGFP-KOR1g, primers 1620/1622). Sequences of the pEnGFP-KOR1g clones are listed in Supplemental File 1. These constructs were introduced into pMDC99 by Gateway LR reactions.
VHA-a3-LssmOR Expression Cassettes
VHA-a3-LssmOR (long stokes-shift mOrange) was prepared by recombineering (Rips et al., 2014). Briefly, the rescue vector fragment was prepared by PCR (primers 1549/1551) using pRSETUOiTIPLssmORThsp (Supplemental File 1) as a template. Bacterial artificial chromosome clone F19H22 containing the VHA-a3 gene was introduced into Escherichia coli SW102. The recombination reaction was performed as described by Rips et al. (2014). Resulting pRSETVHA-a3-LssmORThsp was subjected to Gateway BP and LR reactions using pDonr221 (Thermo Fisher Scientific) and pFAJ-GW (Rips et al., 2014) vectors to produce pFAJ-VHA-a3-LssmORThsp (Supplemental File 1).
Plant Transformation and Selection
GFP-KOR1∆P series, GFP-KOR1AA, GFP-KOR1G429R, GFP-KOR1G429RAA, and GFP-KOR1∆P611AA were introduced into Agrobacterium tumefaciens GV3101 and used to transform the Col-0 wild type or rsw2-1 mutant. The tdFT-KOR1 series and pFAJ-VHA-a3-mOR were introduced into A. tumefaciens ABI and used to transform rdr6-11 (CS24285) to avoid transgene silencing. All plant transformations were done by the floral dip method (Clough and Bent, 1998).
Microscopy Analysis
For the tdFT-KOR1 series, transgenic plants were grown on vertical plates with solid medium containing 1× MS salts, 3% (w/v) Suc, and 1.5% (w/v) agar for 5 d. To induce expression, plants were transferred to fresh agar plates and covered with 1 mL of induction solution (β-estradiol in 1× MS salts, and 3% [w/v] Suc), which was infiltrated by brief application of vacuum. Estradiol concentration was 2 µM for simple transport analysis (Figure 5C; Supplemental Figure 4) and 0.4 µM for ratiometric analysis (Figures 2 and 5B). Plants were incubated for 2 h for transgene induction and subsequently transferred to fresh medium without β-estradiol until observation, to avoid excess and delayed expression. Fluorescence images were acquired using a C1si confocal microscope equipped with NIS Elements confocal microscope software (NIS Elements, Nikon) and processed by the NIS Elements. To render images from different samples comparable, microscope settings were kept constant within the same series of experiments unless otherwise stated. Visualization and quantification of mST/msfGFP ratio were conducted using NIS Elements as described by Khmelinskii and Knop (2014). Segmentation of images were performed as described in Supplemental Figure 2.
Plant Growth Assay and Salt Treatment
To measure root growth, seeds were sown on plates containing 1× MS salts (Murashige and Skoog, 1962), with the indicated concentration of Suc and 1.5% (w/v) agar before stratification at 4°C for 2 to 4 d. Media plates were moved to an incubator set to 25°C and 16-h-light (using 50% output of PHILLIPS F17T8TL741) versus 8-h-dark conditions. Root positions were marked on the third day of culture, and growth was scored after 10 additional days. For salt treatment, seeds were germinated and grown on sterilized cellophane membrane, placed on medium containing 1× MS salts, 3% (w/v) Suc, and 1.5% (w/v) agar for 5 d and then membrane transferred to the same medium supplemented with 140 mM NaCl. Root growth was scored after 10 additional days.
Analysis of Salt Stress–Induced GFP-KOR1 Internalization
Five-day-old seedlings grown on medium containing 1× MS salts, 3% (w/v) Suc, and 1.5% (w/v) agar with the cellophane membrane were transferred to media containing 1× MS salts, 3% (w/v) Suc, 1.5% (w/v) agar, and an inhibitor and NaCl at the indicated concentration. GFP signals were observed at 4 h after the treatment. To obtain the signal ratio, GFP signal intensity at the PM is divided by intracellular GFP signal intensity for each cell.
Immunoblot Analysis
Protein samples were extracted from 2-d-old seedlings, germinated and cultured on 1× MS salts, 3% (w/v) Suc, and 1.5% (w/v) agar. Thirty micrograms of total proteins each were prepared for SDS-PAGE, resolved on a 6% SDS gel, and transferred to nitrocellulose filters. Blots for the GFP-KOR1 series and GFP-KOR1AA series were processed in parallel, but separately. Immunoblotting was performed as described previously using anti-GFP primary antibody (1:4000 dilution; catalog no. A01704, Genscript) and goat anti-rabbit IgG horseradish peroxidase–conjugate (1:100,000 dilution; catalog no. 31460, Thermo Fisher Scientific; Rips et al., 2014).
RT-qPCR Analysis
Total RNA was extracted from 2-d-old seedlings, germinated and cultured on 1× MS salts, 3% (w/v) Suc, and 1.5% (w/v) agar, using TRIzol (Thermo Fisher), according to the manufacturer’s protocol. Reverse transcription using 1 µg of total RNA using Goscript (Promega) and qPCR using EvaGreen qPCR MasterMix (Bullseye) with primers N96/N97 for transgenes and E504/E505 for ACT2 were conducted as described by Fukudome et al. (2014).
Statistical Analysis
The data presented in Figures 3B, 5A, 5B, 7C, and 7D were evaluated by a Welch’s t test. One-way ANOVA was used to analyze the data presented in Figures 1E, 2B, 4A, 6, 8B, and 9 (Supplemental File 2).
Accession Numbers
Sequence data used for this article can be found in the EMBL/GenBank data libraries under accession numbers Arabidopsis KOR1 (At5g49720, NP_199783), tobacco KOR1 (XP_009770880), eucalyptus KOR1 (AFD33696), rice KOR1 (ABF95745), rice OsCel9A (BAF37260), Thermobifida fusca TfCel9A (WP_011292599), Arabidopsis VHA-a3 (At4g39080), Arabidopsis SYP61 (At1g28490).
Supplemental Data
Supplemental Figure 1. Alignment of the amino acid sequences of Glycosyl hydrolase 9 (GH9) family members from different plant species and one bacterium.
Supplemental Figure 2. rdr6-11 but not Col-0 host allowed uniform expression of estradiol-induced tdFT-KOR1.
Supplemental Figure 3. Time course of subcellular tdFT-KOR1 localization.
Supplemental Figure 4. Measurement methods of fluorescent signals in various subcellular locations.
Supplemental Table 1. Sequences of primers used in this study.
Supplemental File 1. FASTA file for vector sequences used in this study.
Supplemental File 2. Outputs of statistical analysis.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors thank Nam-Hai Chua for providing the pER8 vector, Georgia Drakakaki for CFP-SYP61 plants, and the Arabidopsis Biological Resource Center for mutant stocks. The authors thank Paul M. Hasegawa for critical reading of the article and for providing discussions and suggestions. This work was supported by the National Science Foundation (1547551 to H.K.) and by the German Research Foundation (SCHA541 to A.v.S.).
AUTHOR CONTRIBUTIONS
Y.N. and H.K. designed and performed the research; Y.N., Z.M., X.L., X.Q., X.Z., A.v.S., and H.K. analyzed data; and Y.N., A.v.S., and H.K. wrote the article.
References
- Aggarwal C., Labuz J., Gabryś H. (2013). Phosphoinositides play differential roles in regulating phototropin1- and phototropin2-mediated chloroplast movements in Arabidopsis. PLoS One 8: e55393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J.D., Donà E., Gilmour D., Huber W. (2016). TimerQuant: A modelling approach to tandem fluorescent timer design and data interpretation for measuring protein turnover in embryos. Development 143: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L., Li S., Anderson C.T., Lei L., Gu Y. (2013). The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 163: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L., Li S., Zhu X., Gu Y. (2015). The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 112: 12870–12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72: 395–447. [DOI] [PubMed] [Google Scholar]
- Brüx A., Liu T.Y., Krebs M., Stierhof Y.D., Lohmann J.U., Miersch O., Wasternack C., Schumacher K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth Inhibition in Arabidopsis. Plant Cell 20: 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Deuschle K., Chaudhuri B., Okumoto S., Lager I., Lalonde S., Frommer W.B. (2006). Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18: 2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S., et al. (2013). The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25: 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.Y., Himmel M.E. (2006). The maize primary cell wall microfibril: A new model derived from direct visualization. J. Agric. Food Chem. 54: 597–606. [DOI] [PubMed] [Google Scholar]
- Donaldson J.G. (2009). Phospholipase D in endocytosis and endosomal recycling pathways. Biochim. Biophys. Acta 1791: 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N. (2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A., Kesten C., Schneider R., Zhang Y., Ivakov A., Froehlich A., Funke N., Persson S. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162: 1353–1364. [DOI] [PubMed] [Google Scholar]
- Farid A., Malinovsky F.G., Veit C., Schoberer J., Zipfel C., Strasser R. (2013). Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 162: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Suda Y., Vernhettes S., Nakano A., Ueda T. (2015). Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol. 56: 287–298. [DOI] [PubMed] [Google Scholar]
- Fukudome A., Aksoy E., Wu X., Kumar K., Jeong I.S., May K., Russell W.K., Koiwa H. (2014). Arabidopsis CPL4 is an essential C-terminal domain phosphatase that suppresses xenobiotic stress responses. Plant J. 80: 27–39. [DOI] [PubMed] [Google Scholar]
- Gadeyne A., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704. [DOI] [PubMed] [Google Scholar]
- Gillmor C.S., Poindexter P., Lorieau J., Palcic M.M., Somerville C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Weimar T., Lilley K.S., Dupree P., Grierson C.S. (2013). A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197: 805–814. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Jones D.M., Murray C.M., Ketelaar K.J., Thomas J.J., Villalobos J.A., Wallace I.S. (2016). The emerging role of protein phosphorylation as a critical regulatory mechanism controlling cellulose biosynthesis. Front. Plant Sci. 7: 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.Y., Kim Y.W., Kwak J.M., Hwang J.U., Young J., Schroeder J.I., Hwang I., Lee Y. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14: 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.S., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105: 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B.T., McCoy A.J., Späte K., Miller S.E., Evans P.R., Höning S., Owen D.J. (2008). A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456: 976–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesten C., Menna A., Sánchez-Rodríguez C. (2017). Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 40: 106–113. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., et al. (2012). Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 30: 708–714. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Knop M. (2014). Analysis of protein dynamics with tandem fluorescent protein timers. Methods Mol. Biol. 1174: 195–210. [DOI] [PubMed] [Google Scholar]
- Koiwa H. (2009). Pathways and genetic determinants for cell wall–based osmotic stress tolerance in the Arabidopsis thaliana root system In Genes for Plant Abiotic Stress, Jenks M.A., and Woods A.J., eds (Ames: Wiley-Blackwell; ), pp. 35–53. [Google Scholar]
- Koiwa H., Li F., McCully M.G., Mendoza I., Koizumi N., Manabe Y., Nakagawa Y., Zhu J., Rus A., Pardo J.M., Bressan R.A., Hasegawa P.M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15: 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.R., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Bak G., Choi Y., Chuang W.I., Cho H.T., Lee Y. (2008). Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol. 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Zhang T., Strasser R., Lee C.M., Gonneau M., Mach L., Vernhettes S., Kim S.H., J Cosgrove D., Li S., Gu Y. (2014). The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in Arabidopsis. Plant Cell 26: 2601–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xue H.W. (2007). Arabidopsis PLDzeta2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lei L., Somerville C.R., Gu Y. (2012). Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. USA 109: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Xue X., Zhao H., Yang P., Luo H., Zhao J., Huang H., Yao B. (2014). A C-terminal proline-rich sequence simultaneously broadens the optimal temperature and pH ranges and improves the catalytic efficiency of glycosyl hydrolase family 10 ruminal xylanases. Appl. Environ. Microbiol. 80: 3426–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E., Grass J., Altmann F., Mach L., Strasser R. (2013). Characterizing the link between glycosylation state and enzymatic activity of the endo-β1,4-glucanase KORRIGAN1 from Arabidopsis thaliana. J. Biol. Chem. 288: 22270–22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E., Hüttner S., Vavra U., Fischl R., Schoberer J., Grass J., Blaukopf C., Seifert G.J., Altmann F., Mach L., Strasser R. (2009). Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Niu G., Zhang H., Sun Y., Sun S., Yu F., Lu S., Yang Y., Li J., Hong Z. (2018). Trimming of N-glycans by the Golgi-localized α-1,2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol. Plant 11: 678–690. [DOI] [PubMed] [Google Scholar]
- Liu Z., Schneider R., Kesten C., Zhang Y., Somssich M., Zhang Y., Fernie A.R., Persson S. (2016). Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev. Cell 38: 305–315. [DOI] [PubMed] [Google Scholar]
- Mansoori N., Timmers J., Desprez T., Alvim-Kamei C.L., Dees D.C., Vincken J.P., Visser R.G., Höfte H., Vernhettes S., Trindade L.M. (2014). KORRIGAN1 interacts specifically with integral components of the cellulose synthase machinery. PLoS One 9: e112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master E.R., Rudsander U.J., Zhou W., Henriksson H., Divne C., Denman S., Wilson D.B., Teeri T.T. (2004). Recombinant expression and enzymatic characterization of PttCel9A, a KOR homologue from Populus tremula × tremuloides. Biochemistry 43: 10080–10089. [DOI] [PubMed] [Google Scholar]
- McFarlane H.E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- McLoughlin F., Arisz S.A., Dekker H.L., Kramer G., de Koster C.G., Haring M.A., Munnik T., Testerink C. (2013). Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450: 573–581. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and biol. assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nagashima Y., von Schaewen A., Koiwa H. (2018). Function of N-glycosylation in plants. Plant Sci. 274: 70–79. [DOI] [PubMed] [Google Scholar]
- Nicol F., His I., Jauneau A., Vernhettes S., Canut H., Höfte H. (1998). A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17: 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse T.S., Bottrill A.R., Jones A.M., Peck S.C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Hocart C.H., Redmond J.W., Williamson R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211: 406–414. [DOI] [PubMed] [Google Scholar]
- Peng L., Kawagoe Y., Hogan P., Delmer D. (2002). Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150. [DOI] [PubMed] [Google Scholar]
- Polko J.K., Kieber J.J. (2019). The regulation of cellulose biosynthesis in plants. Plant Cell 31: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rips S., Bentley N., Jeong I.S., Welch J.L., von Schaewen A., Koiwa H. (2014). Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26: 3792–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Bichet A., Grandjean O., Kierzkowski D., Satiat-Jeunemaître B., Pelletier S., Hauser M.T., Höfte H., Vernhettes S. (2005). An Arabidopsis endo-1,4-beta-D-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17: 3378–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Pimpl P. (2014). Clathrin and post-Golgi trafficking: A very complicated issue. Trends Plant Sci. 19: 134–139. [DOI] [PubMed] [Google Scholar]
- Roudier F., Fernandez A.G., Fujita M., Himmelspach R., Borner G.H., Schindelman G., Song S., Baskin T.I., Dupree P., Wasteneys G.O., Benfey P.N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C., et al. (2012). Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24: 589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C., et al. (2018). The cellulose synthases are cargo of the TPLATE adaptor complex. Mol. Plant 11: 346–349. [DOI] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N. (2012). The retromer complex - Endosomal protein recycling and beyond. J. Cell Sci. 125: 4693–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.J., Vavra U., Veit C., Strasser R. (2018). The glycan-dependent ERAD machinery degrades topologically diverse misfolded proteins. Plant J. 94: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T., Suzuki K., Abe T., Kaneko Y., Shi H., Zhu J.K., Rus A., Hasegawa P.M., Hashimoto T. (2006). Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 47: 1158–1168. [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22: 53–78. [DOI] [PubMed] [Google Scholar]
- Steiner A., Müller L., Rybak K., Vodermaier V., Facher E., Thellmann M., Ravikumar R., Wanner G., Hauser M.T., Assaad F.F. (2016). The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis. Mol. Plant 9: 528–540. [DOI] [PubMed] [Google Scholar]
- Strasser R. (2016). Plant protein glycosylation. Glycobiology 26: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjanowicz P.M., McKinnon I., Taylor N.G., Gardiner J., Jarvis M.C., Turner S.R. (2004). The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 37: 730–740. [DOI] [PubMed] [Google Scholar]
- Takahashi J., et al. (2009). KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol. 50: 1099–1115. [DOI] [PubMed] [Google Scholar]
- Turner S., Kumar M. (2018). Cellulose synthase complex organization and cellulose microfibril structure. Philos. Trans. A Math. Physiol. Eng. Sci. 376: 20170048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain T., Crowell E.F., Timpano H., Biot E., Desprez T., Mansoori N., Trindade L.M., Pagant S., Robert S., Höfte H., Gonneau M., Vernhettes S. (2014). The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D., Gadeyne A., Vanstraelen M., Inzé D., Van Montagu M.C., De Jaeger G., Russinova E., Geelen D. (2011). Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. USA 108: 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J.E., Thole J.M., Goedhart J., Nielsen E., Munnik T., Gadella T.W. Jr. (2009). Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 57: 356–372. [DOI] [PubMed] [Google Scholar]
- von Schaewen A., Rips S., Jeong I.S., Koiwa H. (2015). Arabidopsis thaliana KORRIGAN1 protein: N-Glycan modification, localization, and function in cellulose biosynthesis and osmotic stress responses. Plant Signal. Behav. 10: e1024397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li J., Yuan M. (2007). Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 48: 1534–1547. [DOI] [PubMed] [Google Scholar]
- Wang S., Kurepa J., Hashimoto T., Smalle J.A. (2011). Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 23: 3412–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cai Y., Wang H., Zeng Y., Zhuang X., Li B., Jiang L. (2014). Trans-Golgi network-located AP1 gamma adaptins mediate dileucine motif-directed vacuolar targeting in Arabidopsis. Plant Cell 26: 4102–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G., Inzé D., Beemster G.T.S. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135: 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Imaizumi N., Kaneko S., Kawagoe Y., Tagiri A., Tanaka H., Nishitani K., Komae K. (2006). Carbohydrate-binding module of a rice endo-beta-1,4-glycanase, OsCel9A, expressed in auxin-induced lateral root primordia, is post-translationally truncated. Plant Cell Physiol. 47: 1555–1571. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Komae K. (2006). A rice family 9 glycoside hydrolase isozyme with broad substrate specificity for hemicelluloses in type II cell walls. Plant Cell Physiol. 47: 1541–1554. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2016). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7: 11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Persson S., Hirst J., Robinson M.S., van Damme D., Sánchez-Rodríguez C. (2015). Change your TPLATE, change your fate: Plant CME and beyond. Trends Plant Sci. 20: 41–48. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000a). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Nishizawa N., Wu Y., Kost B., Chua N.H. (2000b). KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]