Abstract
Tumor necrosis factor receptor 1 (TNFR1) is a central mediator of the inflammatory pathway and is associated with several autoimmune diseases such as rheumatoid arthritis. A revision to the canonical model of TNFR1 activation suggests that activation involves conformational rearrangements of preassembled receptor dimers. Here, we identified small-molecule allosteric inhibitors of TNFR1 activation and probed receptor dimerization and function. Specifically, we used a fluorescence lifetime–based high-throughput screen and biochemical, biophysical, and cellular assays to identify small molecules that noncompetitively inhibited the receptor without reducing ligand affinity or disrupting receptor dimerization. We also found that residues in the ligand-binding loop that are critical to the dynamic coupling between the extracellular and the transmembrane domains played a key gatekeeper role in the conformational dynamics associated with signal propagation. Last, using a simple structure-activity relationship analysis, we demonstrated that these newly found molecules could be further optimized for improved potency and specificity. Together, these data solidify and deepen the new model for TNFR1 activation.
INTRODUCTION
Tumor necrosis factor receptor 1 (TNFR1) plays a key role in the transduction of inflammatory signals (1). Binding of its native ligands, TNFα and lymphotoxin-α (LTα), to TNFR1 stimulates inhibitor of nuclear factor κBα (IκBα) degradation and nuclear factor κB (NF-κB) activation, which has been associated with several autoimmune diseases such as rheumatoid arthritis (1–3). Therapeutic targeting of TNFR1 signaling is a billion-dollar industry (4). Unfortunately and despite the availability of crystal structures for more than two decades (5, 6), currently available anti-TNF therapeutics do not directly or specifically target the receptors and, as a consequence, induce dangerous side effects (7–10). Thus, there is an urgent need to develop a new approach to inhibition of TNFRs. The most promising approach will be to take advantage of the available structures to deepen our understanding of the structure-function relationship of TNFR1, with the goal of rationally maximizing the efficiency of inhibitors.
Ligand binding induces TNFR1 trimerization, which promotes the trimerization of cytosolic death domains and concomitant recruitment of downstream signaling machinery (5, 10). This model is primarily based on the original crystal structure of a ligand-bound, trimeric receptor complex, in which there are no direct receptor-receptor interactions (5). However, this model is confounded by the preassembly of TNFR1 as high-affinity receptor dimers in the plasma membrane (6, 11, 12). On the basis of the crystal structure and on subsequent mutagenesis studies (6, 11–13), preligand dimerization is driven by well-defined monomer-monomer interactions across the preligand assembly domains (PLADs), which are located within the N-terminal cysteine-rich domain 1 (CRD1) and far from the ligand- binding loop. Critically, there is no evidence to suggest that these dimer structures dissociate on ligand binding despite the lack of receptor-receptor interactions in the trimeric structure. Thus, reconciling this apparent inconsistency in the dimer and trimer structures is a long- standing goal within the field (14).
Using TNFR1 and death receptor 5 (DR5), another member of the TNFR superfamily, we have provided evidence for a revision to the accepted model of TNFR activation that reconciles both structural states (13, 15–19). We and others speculate that TNF receptor dimers may form the nexus for larger-scale networks of ligand-bound TNFR trimers (16, 20–22), and we show this to be the case for one of two alternatively spliced isoforms of DR5 (16). On the basis of the crystal structures of TNFR1 (5, 6), we have built a structural model of this oligomeric network, which, in its minimal, active state, consists of a dimer of ligand-bound trimers (Fig. 1A) (15). This minimal size has been supported by superresolution imaging (23). In this model, the preassembled dimer remains intact upon ligand binding and is predicted to undergo a conformational change that propagates from the extracellular domain (ECD) to the cytosol without dimer dissociation. Our focus here was the further exploration of conformational dynamics as central to TNFR1 inhibition and as a potential target for inhibitory small molecules. Although we focused on the idea that dimer itself is the functional signaling unit, the receptor trimer complex (noninteracting receptor monomers stabilized by ligand binding) may still be functional, although perhaps less so. Regardless of this open controversy, the idea that activation is accompanied by conformational changes, be they in dimeric or trimeric form, now has solid support (15, 24). In particular, fluorescence resonance energy transfer (FRET) data suggest, without delineating a clear mechanism, that ligand binding causes a conformational change in the preassembled TNFR1 complex (24, 25). In addition, we have shown by FRET imaging that a constitutively active, disease-related mutant (R92Q) is, like wild type (WT), preassembled as a dimer (15). Moreover, the FRET measurements show that this active R92Q dimer is conformationally distinct from WT, the first direct evidence of a correlation between the backbone structure of the preassembled TNFR1 dimer and receptor activity. As further support, our computational normal mode analysis of an elastic network model of the TNFR1 crystal structures suggests a long-range, anticorrelated motion near the ligand-binding loop that propagates a conformational change through the backbone of the ECD (15).
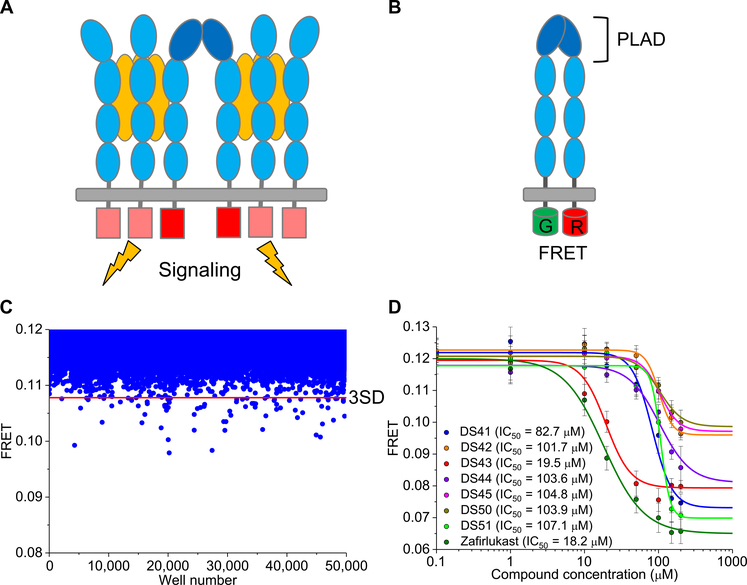
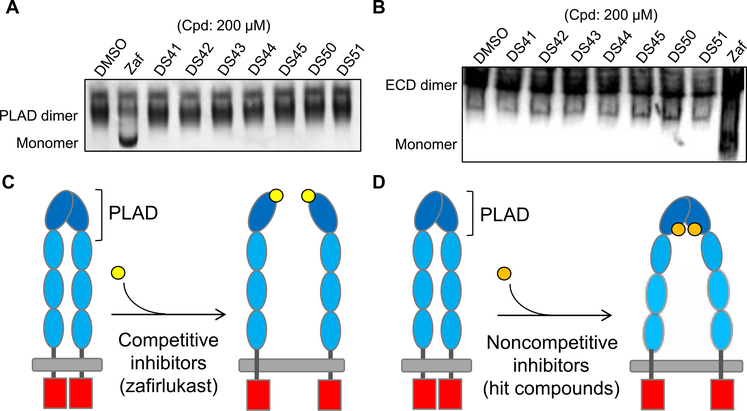
Fig. 1. Discovery of small molecules that perturb conformational states of preassembled TNFR1 dimer.
(A) Schematic of ligand-induced oligomerization of TNFR1 trimers held together by the preligand assembly domain (PLAD). (B) Schematic of the TNFR1ΔCD-FRET biosensor engineered by fusing the green or red fluorescent proteins (GFPs or RFPs) to the C terminus of TNFR1 with truncated cytosolic domain. Ligand-independent association of the fluorophore-tagged receptors through PLAD-PLAD interactions results in fluorescence resonance energy transfer (FRET). The FRET biosensor can detect changes in the cytosolic spacing between receptor monomers. (C) High-throughput screening of ChemBridge DIVERSet 50,000-compound library using the TNFR1 FRET biosensor expressed in HEK293 cells. Compounds that reduced the FRET efficiency below 3SD (red line) were selected for further characterization. Data are representative of one experiment. (D) Secondary FRET analysis of the dose response of the seven hit compounds and zafirlukast (known TNFR1 inhibitor). Data are means ± SD from three independent experiments.
Collectively, these findings suggest that TNFR1 activation is accompanied by backbone conformational changes. However, whether the backbone conformational state of TNFR1 is critical to its activation and whether altering the native conformation of the preassembled dimer is a viable strategy for inhibition remain open and provocative questions. To address these questions, we used small-molecule discovery driven by time-resolved FRET (TR-FRET) measurements of intermonomeric spacing to probe previously undetected conformational states of the preassembled TNFR1 dimer. We found a class of small molecules that perturbed the conformation of TNFR1 and, critically, did so without altering either ligand binding affinity or receptor preassembly. These results provide experimental proof of long-range allosteric coupling in the ECD of TNFR1, which could be exploited to inhibit activation. In addition to providing further evidence for a new model of TNFR1 activation, we showed that these new molecules could be optimized for binding affinity, potency, and receptor specificity, for development as potential therapeutics to directly target TNFR1.
RESULTS
Small-molecule hit compounds modulate conformational states of preassembled dimeric TNFR1
We performed high-throughput screening of the 50,000-compound ChemBridge DIVERSet library to identify small molecules that modulated TNFR1 conformational states. To detect changes in the cytosolic spacing between receptor monomers, we used our previously engineered stable cell lines expressing a TNFR1ΔCD-FRET pair construct, in which either a green or red fluorescent protein (GFP or RFP) is fused to the C terminus of TNFR1 with a truncated cytosolic domain (ΔCD) (TNFR1ΔCD-GFP as donor and TNFR1ΔCD-RFP as acceptor) (Fig. 1B). We have shown that these truncated receptor constructs are properly trafficked to the plasma membrane where they maintain their preassembled dimeric assembly (15), and we validated the constructs for high-throughput screening with a smaller library in the absence of ligand (26). As will be clarified below, changes in FRET could represent either (i) dissociation of the TNFR1 dimer or, (ii) as is our focus in this study, a change in the backbone conformation of the receptor that results in small changes in intermonomeric spacing without inhibiting dimerization. The screen was enabled by a high-throughput fluorescence lifetime plate reader technology with increased precision in TR-FRET–based screening by a factor of 30 (27), which allows for reliable detection of subtle protein structural changes and can monitor allosteric regulation of receptors by small molecules.
The FRET efficiency for all of the compounds, after removing the potential false negatives, was plotted (Fig. 1C), and the distribution of efficiencies was fitted to a Gaussian distribution to obtain a mean and SD. The top 40 compounds, each of which decreased the average FRET by more than 3 SDs (3SD) greater than the mean of control cells, were selected and purchased from ChemBridge Corporation. For simplicity, we limited our search to compounds that reduce FRET, although others that increase FRET may also be of interest in future studies. The compounds were then tested for dose-dependent FRET changes to confirm their specific interactions with the biosensor and to measure the potency and the extent of receptor perturbation by the compounds. Seven of the 40 hit compounds (fig. S1A) had a dose-dependent decrease in FRET efficiency (Fig. 1D) with relative half-maximal effects (relative EC50) ranging between 20 to 110 μM, with DS43 having the lowest EC50. No FRET change was observed with a negative control compound (fig. S1B). We note that from our previous study, the EC50 of the dimer-breaking compound zafirlukast was at the lower end of this spectrum (18 μM) (Fig. 1D).
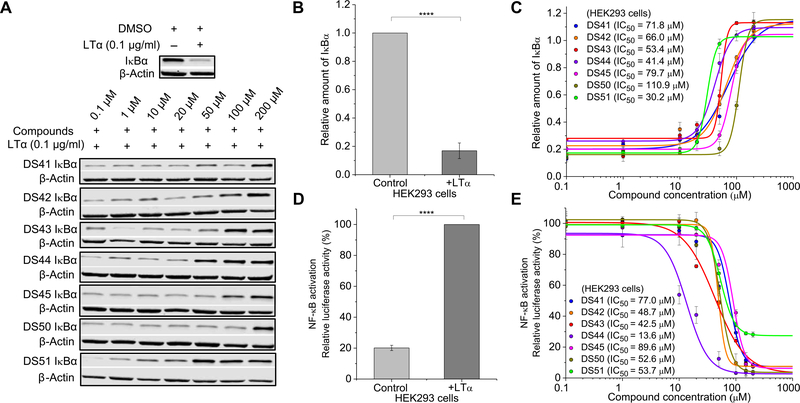
Hit compounds inhibit TNFR1-stimulated IκBα degradation and NF-κB activation
The dose-dependent effect of hit compounds on ligand-induced IκBα degradation was determined by immunoblotting (Fig. 2A). After LTα treatment, IκBα was degraded to 20% of the basal levels in human embryonic kidney (HEK) 293 cells (Fig. 2B). IκBα degradation was inhibited in a dose-dependent manner in cells treated with any of the seven hit compounds, and potency measurements showed relative half-maximal inhibitory concentrations (relative IC50) between 30 and 111 μM (Fig. 2C). To test the inhibition of NF-κB activation, we performed a luciferase assay in the presence of LTα (Fig. 2D). Hit compounds again inhibited ligand-induced NF-κB activation in a dose-dependent manner, with relative IC50 of the compounds between 14 and 90 μM (Fig. 2E). We note that both functional assays produced IC50 values that roughly correspond to the FRET results, although important differences exist that reflect differences in the specificity of the individual compounds for the receptor. This important nuance will be addressed below with studies using a TNFR1 knockout (KO) cell line. In addition, we note that HEK293 cells showed basal NF-κB activation (20% relative luciferase activity) in the absence of exogenous ligands. This has been suggested to be due to constitutive signaling of cytokine receptors, including TNFR1, in the ligand-free state (28, 29). Specific inhibition of TNFR1 such as through up-regulation of silencer of death domain can reduce this constitutive signaling (29).
Fig. 2. Hit compounds inhibit TNFR1-stimulated IκBα degradation and NF-κB activation.
(A to C) Western blot analysis of IκBα abundance in lysates of HEK293 cells treated with LTα and the hit compounds at the indicated doses. Western blots (A) are representative of three independent experiments. Quantified band intensity values (B and C) are means ± SD from all experiments. ****P < 0.0001 compared to control by two-tailed unpaired t test. (D and E) Luciferase assay of NF-κB activation in HEK293 cells transfected with reporter plasmids and treated with LTα and DMSO control (D) or LTα and increasing concentrations of hit compounds (E). Data are means ± SD of three independent experiments. ****P < 0.0001 compared to control by two-tailed unpaired t test.
A subset of hit compounds are receptor specific in blocking TNFR1-induced NF-κB activation
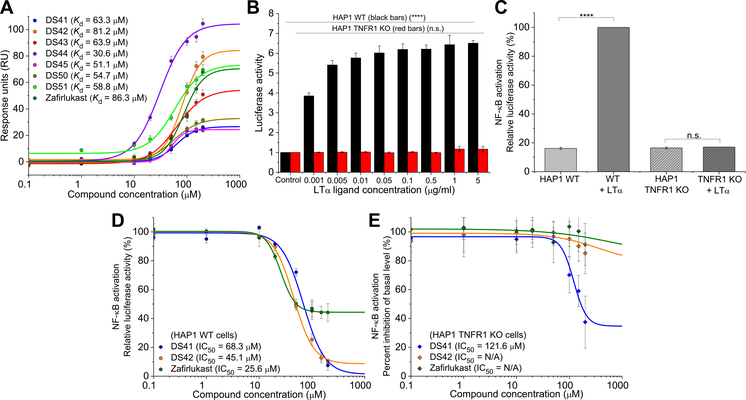
To determine whether any of the seven hit compounds acted directly on TNFR1, we first performed measurements with surface plasmon resonance (SPR). Purified TNFR1 ECD was immobilized onto the SPR chip, which was followed by flowing the compounds through the chip to allow for binding. All hit compounds showed dose-dependent binding to the TNFR1 ECD with binding affinities [Kd (dissociation constant)] between 31 and 81 μM, as compared to 86 μM for zafirlukast (Fig. 3A and fig. S2, A to H).
Fig. 3. Hit compounds bind TNFR1 and require the receptor for their effects.
(A) Direct binding of the hit compounds to the TNFR1 extracellular domain (ECD) was characterized by surface plasmon resonance (SPR). Data are means ± SD from three independent experiments. (B) Dose-dependent ligand-induced NF-κB activation in both wild-type (WT) and TNFR1 knockout (KO) HAP1 cells. Data are means ± SD of three independent experiments. ****P < 0.0001 compared to control by two-tailed unpaired t test, and n.s. indicates not significant. (C) NF-κB activation in WT and TNFR1 KO HAP1 cells with the optimized LTα concentration of 0.1 μg/ml. Data are means ± SD of three independent experiments. ****P < 0.0001 compared to control by two-tailed unpaired t test, and n.s. indicates not significant. (D and E) NF-κB activation in WT HAP1 cells (D) and TNFR1 KO HAP1 cells (E) treated with LTα and increasing concentration of compounds (DS41, DS42, and zafirlukast) to test the specificity of the compounds to TNFR1. Data are means ± SD of three independent experiments. N/A, not applicable.
Next, we aimed to establish whether the functional effects of the hit compounds (Fig. 2E) were specifically due to binding the receptor and alteration of the receptor conformation rather than through the inhibition of proteins in alternate signaling pathways. We first used a TNFR1 KO near-haploid HAP1 cell line established by CRISPR. As expected, WT HAP1 cells showed a dose-dependent increase in luciferase activity in response to LTα, which was not observed in the TNFR1 KO cells, confirming that the TNFR1 KO cells were not functionally sensitive to TNFR1 stimulation (Fig. 3B). A ligand concentration of 0.1 μg/ml was chosen for treatment of both WT and TNFR1 KO HAP1 cells because maximal NF-κB activation as measured by luciferase activity was obtained at this concentration in WT cells but not in TNFR1 KO cells (Fig. 3C).
As with HEK293 cells (Fig. 2D), KO cells showed basal activation of the NF-κB pathway (about 20% relative luciferase activity). This basal activity was unchanged in the presence of ligand. This feature made these cell lines useful as a first control to ensure that the functional effects we observed in the HEK293 cells were due to direct interactions with TNFR1 and not due to the compounds indirectly acting on other proteins in alternate NF-κB pathways. As expected, all compounds reduced ligand-induced NF-κB activation in the WT HAP1 cells to very a similar extent as was observed in the HEK293 cells (Fig. 3D and fig. S3A). Then, the TNFR1 specificity of the compounds was determined by monitoring their effect on the basal level of NF-κB activation in TNFR1 KO cells. Our expectation was that the truly specific compounds should not affect this basal activity. To show the effectiveness of this approach, DS44 and DS50 reduced the basal level of NF-κB activation in the TNFR1 KO cells at similar IC50 to the WT cells (fig. S3B), demonstrating a nonspecific interaction with other proteins in the cells and ruling out these two compounds as specific inhibitors of TNFR1. We noted that DS44 was the compound with the highest binding affinity for TNFR1, as measured by SPR, highlighting the importance of this specificity test. DS41, DS43, DS45, and DS51 were all specific at the range near their IC50, but each of these compounds showed some nonspecificity at higher concentrations (Fig. 3E and fig. S3B). DS42, on the other hand, together with zafirlukast, emerged as the most specific of the newly found inhibitors, showing almost no effect on basal activity in TNFR1 KO cells even at high concentrations (Fig. 3E).
As a second control to further determine the specificity of the compounds, we tested their effect on TNFR1-associated death domain (TRADD)–induced NF-κB activation, which is independent of ligand activation of TNFR1. Because TRADD is the first cytosolic protein in the TNFR1 cascade, testing the effect of the compounds after stimulating TRADD, without involving TFNR1, represents the most complete control for specificity. Overexpression of TRADD significantly increased NF-κB activation in HEK293 cells (fig. S4A), as shown previously (30). Both DS42 (fig. S4B) and zafirlukast (fig. S4C) had little effect on TRADD-induced NF-κB activation, even at relatively high concentrations; DS42 did show partial inhibition at 200 μM, although this was well above its IC50. In contrast, DS41, which showed some nonspecificity in inhibiting basal NF-κB activity in TNFR1 KO HAP1 cells, also inhibited TRADD-induced NF-κB activation at a similar IC50 to its inhibition of the TNFR1-induced NF-κB activation (fig. S4D). These results further confirmed the high specificity of DS42 and zafirlukast in targeting TNFR1.
Small-molecule inhibitors do not change the binding affinity of ligand for TNFR1
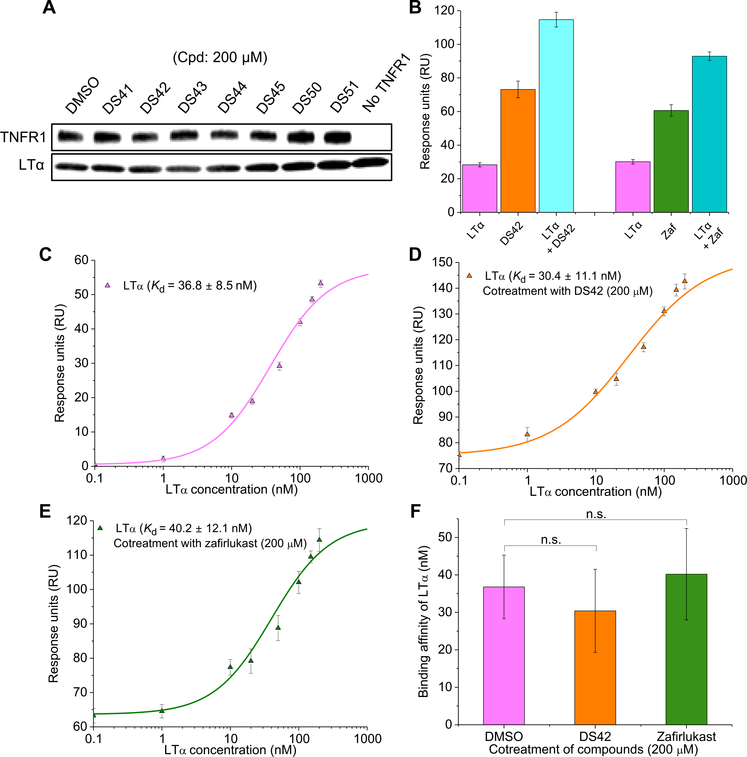
After determining the specificity of each of the compounds, we then aimed to parse whether the mechanisms of action of these compounds is to perturb ligand-receptor interactions, receptor-receptor interactions such as dimerization, or a new mechanism: alteration of conformational states of the receptor. We note that DS42 had only a small, although significant, effect on FRET. If we could show that the compound did not prevent ligand binding or dimerization, then this would highlight the possibility that subtle changes in conformation of the preassembled dimer may be enough to inhibit the receptor in the ligand-bound, fully assembled state.
First, to test whether the compounds act by blocking ligand binding, we monitored ligand-receptor binding in the absence and presence of the compounds using coimmunoprecipitation and SPR. Coimmunoprecipitation experiments qualitatively confirmed that none of the hit compounds eliminated ligand-receptor interactions (Fig. 4A). To quantitatively confirm the noncompetitive nature of the compounds, we performed SPR measurements in the presence of both the ligand and the compounds. Both LTα (50 nM) and the compounds (DS42 and zafirlukast at 200 μM) were passed through the TNFR1 ECD immobilized surface for SPR measurements with individual treatment of ligand only or compound only as controls. For both DS42 and zafirlukast, the response from cotreatment of ligand and compounds was equal to the sum of the individual response from ligand-only or compound-only binding, indicating simultaneous binding of the ligand and the small-molecule inhibitors to the receptor (Fig. 4B). We then investigated the dose dependence of ligand binding to TNFR1 in the presence of the compounds to determine whether either affected ligand binding affinity (Kd). We found that in the absence of compounds, the Kd of LTα was 37 nM (Fig. 4C and fig. S5A), as expected (31). In the presence of saturating concentrations of the compounds, DS42 (200 μM) or zafirlukast (200 μM), LTα binding affinity was essentially unchanged (within error of the measurements) (Fig. 4, D to F, and fig. S5, B and C).
Fig. 4. Small-molecule inhibitors do not block ligand-receptor interactions.
(A) Coimmunoprecipitation between TNFR1 and ligand LTα with treatment of hit compounds (Cpd) at saturation dose of 200 μM. Equal amount of LTα is shown as pull-down controls. Western blots are representative of three independent experiments. (B) Noncompetitive binding assay of LTα (50 nM) and compounds [DS42 or zafirlukast (Zaf) at 200 μM] to TNFR1 ECD was performed by SPR. Data are means ± SD of three independent experiments. (C) Dose-dependent binding of LTα to TNFR1 ECD with increasing concentration of ligand. Data are means ± SD of three independent experiments. (D and E) Dose-dependent binding of LTα in the presence of compounds, DS42 (D) or zafirlukast (E), at saturated compound concentration of 200 μM. Data are means ± SD of three independent experiments. (F) Comparison of the binding affinity of LTα to TNFR1 in the absence and presence of compounds (DS42 or zafirlukast). Data are means ± SD of three independent experiments, and n.s. indicates not significant by two-tailed unpaired t test.
Small-molecule inhibitors also do not change the dimerization of the TNFR1 PLAD or ECD
Having shown that the new compounds do not act by disrupting ligand binding, we next investigated whether, like zafirlukast, they acted by disrupting the dimerization of TNFR1 ECDs through the PLAD. We have shown that purified soluble TNFR1 PLAD exists as dimers under native conditions, which can be disrupted to form a monomer by small molecules that disrupt receptor-receptor interaction such as zafirlukast (26). Using this biochemical approach (26), we first tested the ability of each of the compounds to disrupt soluble PLAD dimers under native conditions. The PLAD dimers were disrupted by zafirlukast but not by any of the new hit compounds (Fig. 5A). We used immunoblotting to show that the higher molecular weight bands in native gels were aggregates of PLAD protein, which were not altered by DS42 (fig. S6A). To test whether the compounds bind elsewhere on the ECD of the receptor and cause PLAD dissociation by inducing long-range allosteric coupling, we tested their ability to disrupt dimers of purified, full-length TNFR1 ECD proteins under native conditions. The results again showed that none of the compounds other than zafirlukast disrupted the TNFR1 ECD dimer (Fig. 5B). In addition, we confirmed that none of the hit compounds affected ligand-ligand interactions (fig. S6B).
Fig. 5. Small-molecule inhibitors do not disrupt receptor-receptor interactions.
(A) Native gel characterization of soluble PLAD of TNFR1 with treatment of DMSO control, zafirlukast, and hit compounds (200 μM). Gels are representative of three independent experiments. (B) Native gel characterization of soluble ECD of TNFR1 with treatment of DMSO control, zafirlukast, and hit compounds (200 μM). Gels are representative of three independent experiments. (C) Schematics illustrating the mechanism of competitive inhibition by zafirlukast in disrupting receptor-receptor interactions. (D) Schematics illustrating the mechanism of noncompetitive inhibition by the new hit compounds in stabilizing the nonfunctional conformational states of TNFR1 without disrupting receptor-receptor interactions.
Collectively, these findings distinguish the new compounds found here from zafirlukast and from previous small molecules that target TNFR1 by blocking the ligand binding (32–34). However, zafirlukast is not competitive in the most traditional sense of preventing ligand binding. Instead, zafirlukast competes with monomer-monomer binding without disrupting ligand binding and diminishes function by reducing dimerization (Fig. 5C). We showed here that in contrast, the new compounds act noncompetitively without affecting ligand binding or TNFR1 monomer-monomer interactions. Instead, the TNFR1-specific compounds exert their inhibitory effect by altering the conformational state of the preassembled dimer (as observed from the FRET changes) and somehow do so in a way that does not prevent ligand binding or dimerization (Fig. 5D). From the perspective of understanding the mechanism of TNFR1 activation, this demonstrates that TNFR1 can be inactive, although the ligand is bound and the receptor dimer is intact.
New noncompetitive inhibitors are more efficient than the competitive inhibitor zafirlukast
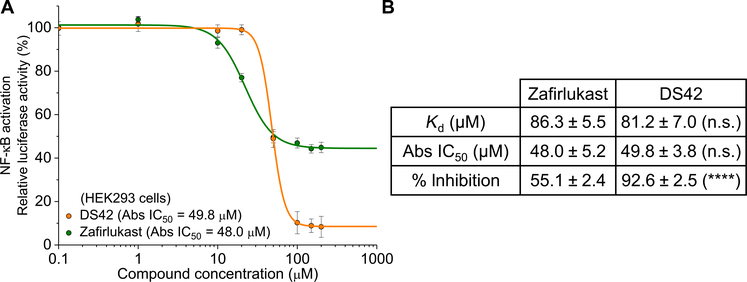
We next compared the efficiency of inhibition by DS42, a noncompetitive inhibitor, with that of the competitive inhibitor zafirlukast. In theory, given equal affinity for its binding site on the receptor, a noncompetitive compound should be more efficient than a competitive one. We observed that DS42 inhibited NF-κB to a greater extent (93%) compared to zafirlukast (55%) (Fig. 6A). By comparing the binding and inhibitory profiles of DS42 and zafirlukast, we found that both compounds had similar binding affinity (Kd) and absolute IC50 values (Fig. 6B). This result indicates that the increase in inhibition by DS42 is due to more efficient allosteric inhibition unencumbered by competition and not due to increased binding. The inability to obtain maximum inhibition by zafirlukast could be attributed to its competition with native receptor monomers, which are in dynamic equilibrium with the dimeric form, making it difficult for zafirlukast to achieve maximum inhibition due to its low binding affinity.
Fig. 6. Noncompetitive inhibitors are more efficient than competitive inhibitor.
(A) NF-κB activation in HEK293 cells treated with LTα and increasing concentration of compounds (DS42 and zafirlukast) to compare the inhibition efficiency between noncompetitive and competitive inhibitors. Data are means ± SD of three independent experiments. (B) Comparison of the binding affinity, the absolute (Abs) IC50, and the percent inhibition of NF-κB activation between DS42 and zafirlukast. Data are means ± SD of three independent experiments. ****P < 0.0001 for DS42 compared to zafirlukast by two-tailed unpaired t test, and n.s. indicates not significant.
Long-range perturbation of TNFR1 conformational dynamics by noncompetitive inhibitors is mediated by residues in the ligand-binding loop
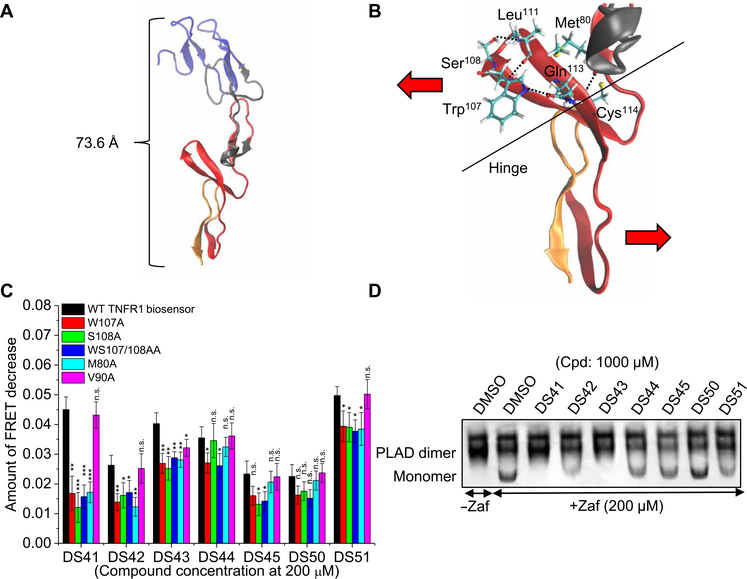
Next, we asked whether we could use the small molecules to elucidate changes in the conformational states in the preassembled dimer. We have shown that the constitutively active, dimeric R92Q mutant adopts an altered, active conformation compared to WT (15). We have also used computational normal mode analysis of the available TNFR1 crystal structures and predict the implicit capacity of the receptor (in both the preligand and ligand-bound states) to undergo an anticorrelated motion that couples conformational changes across the ECD (15). In particular, even in the absence of ligand, we highlight conformational coupling between residues in the ligand-binding loop (residues 77 to 81 and 107 to 113) and the membrane proximal domain. We have predicted the existence of a hinge that connects the domains and, hence, serves a critical role in propagating activating conformational changes. Here, by further examining the residues in the ligand-binding loop in the crystal structure of the preligand dimer [Protein Data Bank (PDB): 1NCF], we found that the key ligand binding residues Trp107, Ser108, and Met80 form four hydrogen pairs (Trp107 with Gln113 and with Leu111, Ser108 also with Leu111, and Met80 with Cys114) in this hinge region (Fig. 7, A and B). This led us to ask whether these critical residues and this same coupling might also be important in propagating the long-range conformational changes induced by our new small-molecule inhibitors, although the inhibited conformation state is, by definition, distinct from the active state.
Fig. 7. Long-range perturbation of TNFR1 conformational dynamics by noncompetitive inhibitors is mediated by residues in the ligand-binding loop.
(A) Crystal structure of the TNFR1 ECD (PDB: 1NCF). The distance between the membrane distal domain, including PLAD, and the membrane proximal domain is estimated to be 73.6 Å, which suggests a potential long-range signal propagation between noncompetitive inhibitors binding at the PLAD or the ECD and the perturbation of the membrane proximal domain as shown by FRET change. Four different cysteine-rich domains (CRD1 to CRD4) are colored in blue, gray, red, and orange, respectively. (B) Surface representation showing the coupling motions between residues in the ligand-binding loop and the membrane proximal domain (PDB: 1NCF). The key ligand binding residues Trp107, Ser108, and Met80 form four hydrogen bonds with Leu111, Gln113, and Cys114, which stabilize the conformation of the region to behave like a hinge in aiding the opening of the receptor. Abolishing the hydrogen bonds may decouple the domains and prevent conformational change acting through the hinge. (C) The amount of FRET decrease in HEK293 cells expressing WT and mutant TNFR1 FRET biosensors (W107A, S108A, WS107/108AA, M80A, and V90A) treated with noncompetitive inhibitors (200 μM) in the absence of ligand. Values were normalized to the DMSO-only control, and data are means ± SD of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 for WT compared to mutant biosensors by two-tailed unpaired t test, and n.s. indicates not significant. (D) Native gel characterization of soluble PLAD in cells cotreated with DMSO control or hit compounds (1000 μM) and zafirlukast (200 μM) to test the competition between the hit compounds and zafirlukast in interacting with PLAD. Gels are representative of three independent experiments.
To test this hypothesis experimentally, we made single and double mutants of these three critical residues (W107A, S108A, WS107/108AA, and M80A) in the FRET biosensor as well as a control mutant (V90A) located far from the ligand-binding loop. Although these residues are essential for ligand binding (15, 35, 36), here, we interrogated the preligand conformations of the receptor to understand the mechanism of inhibition by the small molecules, testing whether the compounds perturb the mutant FRET biosensors in the absence of ligand. If these residues form a critical region in the conformational network within the ECD, then by breaking these hydrogen bonds, we hypothesized that we would decouple the domains and reduce the ability of the small molecules to cause the inhibitory conformational change. To start, the mutations did not affect the basal FRET observed from preligand receptor dimers (fig. S7A). Although the compounds did still reduce FRET in the mutants (fig S7B), the magnitude of the effect was significantly diminished with each of the compounds (except DS50), thus supporting the hypothesis of a loss in conformational coupling between the loop and the remainder of the ECD (Fig. 7C). We note that the biggest effect of the mutations was on DS41, which was a compound that showed one of the largest FRET changes for the WT receptor (Fig. 1D). As a negative control, the extent of FRET change induced by all compounds in the control mutant (V90A) was similar to that of the WT FRET biosensor (Fig. 7C).
We also asked whether we could use zafirlukast to probe the potential binding regions for the small-molecule inhibitors at low resolution. In particular, because the hit compounds bound the TNFR1 ECD with higher affinity than zafirlukast (Fig. 3A) but did not disrupt the PLAD-PLAD interaction (Fig. 5A), we hypothesized that compounds that compete with zafirlukast and prevent its disruption of PLAD-PLAD interaction might bind to the PLAD. Cotreatment of soluble dimeric PLAD with the hit compounds and zafirlukast revealed that some compounds—in particular, DS41, DS42, and DS43—competed with zafirlukast in binding to PLAD and prevented the ability of zafirlukast to disrupt PLAD dimerization (Fig. 7D and fig. S8, A and B). Compounds that have little or no effect in competing with zafirlukast may bind either to the part of CRD2 that is close to the PLAD and is not involved in ligand binding or to CRD4. In addition, we quantitatively showed that cotreatment of DS42 and zafirlukast reduced the binding affinity of DS42 to the TNFR1 ECD by 1.5-fold (fig. S8C) and decreased its potency in inhibiting TNFR1-induced NF-κB activation by 2.7-fold (fig. S8D), as compared to DS42-only treatment. However, whether these compounds directly compete with zafirlukast or are allosterically changing the conformation of the receptor such that it cannot bind zafirlukast remains to be investigated. The PLAD contains both the dimer binding interface and a nonbinding interface, and our results do not distinguish between these two surfaces; however, our results suggest that some compounds interact directly with the PLAD.
To further probe the specific mechanisms of DS42 and zafirlukast, we tested the effects of these small molecules in the presence of H398 human TNFR1-specific antibody. The binding epitope of the H398 antibody is in the regions of the CRD1 and CRD2 of TNFR1, and this antibody inhibits receptor function by blocking ligand binding (37). We hypothesized that DS42 and zafirlukast, which did not ablate ligand binding, would not prevent H398 binding and function. SPR binding indicated that DS42 and zafirlukast (200 μM) did not compete with H398 [at its Kd of 1.0 nM (37)] for binding TNFR1 (fig. S9A). Furthermore, H398 antibody inhibited TNFR1-induced NF-κB activation with an IC50 of 1.2 nM (fig. S9B), similar to a previous report (37). Consistent with our hypothesis, the IC50 values for inhibition of NF-κB by DS42 and zafirlukast were unchanged by the presence of H398, illustrating that neither compound competed with H398 in inhibiting receptor function (fig. S9, C and D).
A representative hit compound (DS41) is optimizable for binding affinity, potency, and specificity
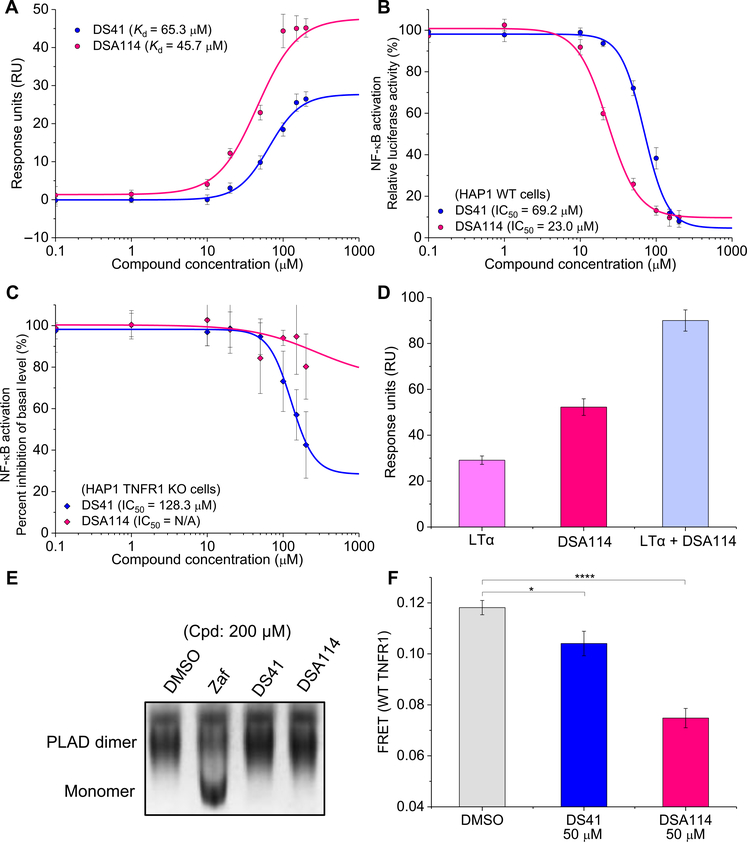
The best compounds identified from screening the 50,000-compound DIVERSet library have absolute IC50 values (as assessed by inhibition of NF-κB activation) around 50 μM, similar to the value we obtain for zafirlukast (50 μM) (26). To test whether we could improve the IC50 values for our hit compounds, we performed a preliminary structure-activity relationship (SAR) analysis of DS41. We focused on DS41 for several reasons. First, DS41 showed the largest amount of FRET decrease in the mutant TNFR1 biosensors (Fig. 7C), suggesting that the chemical aspects of this compound, if harnessed, may be most efficient in targeting the allosteric mechanism of action described in this study. Second, DS41 had nonspecific effects as shown by its inhibition of basal NF-κB activation (Fig. 3E and fig. S4D). Thus, we attempted to optimize DS41 by simultaneously eliminating this nonspecificity and increasing binding affinity, thereby improving the potency such that the compound was specific at its effective concentration (IC50). We obtained 19 analogs from ChemBridge (table S1) to determine SAR. One analog, DSA114, showed increased binding affinity to the TNFR1 ECD (Fig. 8A and fig. S10A), strong inhibition of IκBα degradation (fig. S10, B and C), and a threefold increase in the potency of inhibiting NF-κB activation as compared to the lead compound, DS41 (Fig. 8B and fig. S10D). DSA114 was substantially more specific than DS41 (Fig. 8C), especially at its IC50 value (fig. S10, D and E). Specifically, it did not block ligand-receptor interactions (Fig. 8D and fig. S10, F to H), did not disrupt PLAD-PLAD interactions (Fig. 8E), and prevented zafirlukast from disrupting the PLAD-PLAD interactions (fig. S10I). In addition, DSA114 decreased FRET in the WT TNFR1 biosensor to a greater extent than DS41 at the same concentration (Fig. 8F).
Fig. 8. Lead compounds are optimizable for binding affinity, potency and specificity.
(A) SPR characterization of the binding affinity of the hit compound DS41 or its analog DSA114 to TNFR1 ECD. Data are means ± SD of three independent experiments. (B and C) NF-κB activation in WT HAP1 cells (B) and TNFR1 KO HAP1 cells (C) treated with LTα and increasing concentration of DS41 or DSA114 to test the improvements in the potency and specificity of the analog. Data are means ± SD of three independent experiments. N/A, not applicable. (D) Noncompetitive binding test of LTα (50 nM) and DSA114 (200 μM) to TNFR1 ECD was performed by SPR. Data are means ± SD of three independent experiments. (E) Native gel characterization of soluble PLAD with treatment of DMSO control, zafirlukast, DS41, or DS114 (200 μM) to test the disruption of PLAD dimerization by the compounds. Gels are representative of three independent experiments. (F) FRET measurements in HEK293 cells expressing WT TNFR1 FRET biosensor treated with DMSO control, DS41 (50 μM), and DSA114 (50 μM) in the absence of ligand to compare the extent of receptor perturbation by the hit compound and its analog. Data are means ± SD of three independent experiments. *P < 0.05 and ****P < 0.0001 compared to control by two-tailed unpaired t test.
Structurally, DS41 contains a 4-piperidinyl-1H-pyrazol-5-yl core with 2-methyl-3-phenyl-2-propen-1-yl (R1) attached to the nitrogen of the piperidine and cyclopentanecarboxamide (R2) to the pyrazole (table S1, compound 1). Our preliminary SAR analysis of DS41 illustrated that both its R1 and R2 groups played important roles in determining compound binding affinity and potency. In particular, the R1 group was preferred to maintain potent activity (as assessed by IC50 and percent inhibition of NF-κB activation) because deviation from the current 2-methyl-3-phenyl-2-propen-1-yl group resulted in a substantial decreased percent inhibition (table S1, compound 2 to 14). However, modification of R1 modulated the potency in terms of IC50, and hydrophobic groups were better tolerated than substituents containing polar moieties. Simplification of the 2-methyl-3-phenyl-2-propen-1-yl substituent by reduction of the olefin and removal of the phenyl ring substantially enhanced potency at the expense of percent inhibition (table S1, compound 8). Our data suggest that the 2-methyl and 3-phenyl groups on the 2-propene of the R1 of DS41 may be important for increasing the percent inhibition, and a flexible carbon chain (changing propene to propane) may favor binding and increase potency (table S1, compound 2 versus 5). On the other hand, the R2 group was more tolerant to modification, and five of the six derivatives containing benzyl, substituted phenyl, or 3-phenylpropyl groups maintained or exceeded the percent inhibition of the initial hit while exhibiting IC50 values ranging from 26 to 115 μM (table S1, compound 15 to 20). This limited series suggests that this position offers more opportunities for modulating potency and drug disposition properties while maintaining high percent inhibition. A comparison between the structures of DS41 and DSA114 and other hit compounds such as DS42 indicates moderate structural similarity because all contain a substituted piperidine core (fig. S1A). This preliminary SAR analysis, together with the comparison with other hit compounds, suggests that some of these structurally similar compounds may act through the same inhibitory mechanism.
DISCUSSION
There are currently five U.S. Food and Drug Administration–approved biologic agents including monoclonal antibodies (infliximab, adalimumab, certolizumab pegol, and golimumab) and soluble TNF receptor (etanercept) that target TNFα for treatment of TNFR1-related inflammatory diseases (10, 38–40). Despite high potency, these therapeutics result in global TNFα blockade, which has several negative consequences: low rates of disease remission, the development of fatal side effects such as lupus-like symptoms and lymphomas, and the generation of antibodies against biologic TNFα inhibitors (40, 41). In addition, compared to small molecules, antibodies are expensive and often fail to cross the blood-brain barrier and can also lead to injection site reactions or infusion reactions (10, 42, 43).
To overcome these limitations, the therapeutic paradigm in this field has shifted from targeting TNFα to developing inhibitors that directly target TNFR1. To a large extent, TNFR1 receptor-specific inhibitors are antibodies (37, 44–46) or small molecules that competitively block ligand-receptor interactions (32, 34). Because the ligand binding affinity of TNFα to TNFR1 is very high (Kd = 0.38 nM) (33), small molecules that work by competitively eliminating ligand binding have a very steep hill to climb. In addition, it has been suggested that TNFR1 antagonists that block ligand binding reduce the TNFα neutralizing capacity of soluble TNFR1 because the circulating forms of the receptor may function as decoys for the ligand and their concentrations may reflect long-term exposure to this proinflammatory cytokine (47–49). At low concentrations, soluble TNFR1 enhances the actions of TNFα, but at higher concentrations, the effects of TNFα are abrogated (50). Furthermore, cell-autonomous interaction between TNFα and TNFR1 is critical for cell survival, maintenance, and function and is neuroprotective (51, 52).
Thus, approaches that do not involve eliminating ligand binding are highly attractive, although have proved elusive. One approach has been to block the preassembly of TNFR1 dimers by targeting the PLAD. Competitively eliminating the PLAD-PLAD interaction is an attractive alternative to blocking ligand binding because the monomer-monomer interaction (low micromolar) is weaker than the ligand-receptor affinity (12, 53). In a seminal study, it was suggested that soluble TNFR1 PLAD was able to disrupt TNFR1 receptor- receptor interaction and to inhibit TNFα-induced inflammatory signaling in vitro as well as to ameliorate arthritis in a mouse model (54). However, in that study, the glutathione S-transferase–tagged PLAD protein ablated ligand binding, making it unclear whether the effect of inhibition was due to disruption of the receptor dimer (54). In our previous high-throughput screening study, we showed that a small molecule (zafirlukast) can disrupt the dimeric PLAD-PLAD interaction without blocking ligand binding, inhibiting ligand-induced NF-κB activation with a potency of 50 μM (absolute IC50) (26). However, zafirlukast has relatively low affinity and only partially inhibits activation. In addition to the higher homotypic affinity of the PLAD-PLAD interaction compared to the zafirlukast-PLAD affinity, we speculate that the inability of zafirlukast to completely inhibit TNFR1 could be due to preservation of a functionally inefficient ligand-bound trimeric structure, despite elimination of the dimer (22).
The relatively low affinity and inhibition efficiency of zafirlukast and related compounds suggest a need for a noncompetitive targeting strategy for more effective inhibition of TNF receptors. To make progress in this way has required that the field exploit developments in understanding the structure and dynamics of the receptors (14–16, 19, 23, 55). We are aware of one important study that used computational design to find a small molecule (called F002) that binds to a cavity distal to the ligand-binding loop and inhibits TNFR1 allosterically (33). Although the binding cavity of the small molecule is located away from the ligand interaction site and the binding of the small molecule may not directly prevent ligand binding to the receptor, evidence is lacking to rule out the possibility of reduced or eliminated ligand binding. Moreover, the functional efficacy of F002 is 60-fold weaker than its binding affinity, suggesting that this small molecule competes with some other process, perhaps ligand binding.
In theory, small molecules that act at allosteric sites but cause conformational changes that reduce ligand binding suffer the same competitive disadvantage of orthosteric modulators that directly eliminate ligand binding (56). Allosteric small-molecule inhibitors that do not influence ligand binding or require outcompeting receptor-receptor interactions may be more efficient because they are unencumbered by competition (either ligand-receptor or receptor- receptor) (57–59). However, by what mechanism could such a small molecule operate? Here, we have demonstrated one approach to receptor inhibition by small molecules acting noncompetitively on the receptor: stabilization of nonfunctional conformational states of the receptor that is independent of ligand binding or receptor dimerization. These noncompetitive inhibitors are more efficient at inhibiting TNFR1 signaling than our previously found competitive inhibitor. However, we note that most of the compounds (with the exception of DS42), by acting through this allosteric mechanism, may also exert some effects downstream of TRADD or on other signaling proteins. Moreover, small modifications to the chemical structure of a lead compound allowed us to easily improve its potency and specificity. These scaffolds could be optimized even further by medicinal chemistry for potential therapeutic developments.
MATERIALS AND METHODS
Molecular biology
The DNAs (TNFR1ΔCD-GFP and TNFR1ΔCD-RFP) used to engineer the TNFR1 FRET biosensor were generated in our previous work (26). Briefly, complementary DNAs encoding truncated TNFR1ΔCD (amino acids 1 to 242) were fused to the N terminus of the enhanced GFP and TagRFP vectors using standard cloning techniques. The mutations of the key ligand binding residues (Q107A, S108A, QS107/108AA, and M80A) and the control mutant (V90A) in the TNFR1ΔCD-GFP and TNFR1ΔCD-RFP plasmids were introduced by QuikChange mutagenesis (Agilent Technologies). To prevent constitutive fluorophore clustering, all vectors contain the monomeric mutation A206K in the fluorescent proteins (60).
Cell culture and generation of stable cell lines
HEK293 cells (American Type Culture Collection) were cultured in phenol red–free Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 2 mM L-glutamine (Invitrogen), heat-inactivated 10% fetal bovine serum (FBS HI; Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco). HAP1 WT and TNFR1 KO cells (Horizon) were cultured in Iscove’s modified Dulbecco’s medium (Gibco) supplemented with 10% FBS (Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco). Cell cultures were maintained in an incubator with 5% CO2 (Forma Series II Water Jacket CO2 Incubator; Thermo Fisher Scientific) at 37°C. The HAP1 TNFR1 KO cell line was edited by CRISPR-Cas to contain a 70–base pair insertion in a coding exon of TNFRSF1A. The TNFR1ΔCD-FRET pair stable cell line was generated as described in our previous work and has been monitored continuously for more than 3 years with expression above 95%, as characterized by flow cytometry (26). The high expression of the TNFR1ΔCD-FRET pair in the stable cell line indicates that they are suitable for high-throughput screening. The mutant forms of the TNFR1 FRET biosensor (Q107A, S108A, QS107/108AA, M80A, and V90A) were generated by transiently transfecting HEK293 cells using Lipofectamine 3000 (Invitrogen) with TNFR1ΔCD-GFP and TNFR1ΔCD-RFP DNAs containing the respective mutations.
Large-scale high-throughput screening with ChemBridge DIVERSet library
The DIVERSet library containing 50,000 compounds was purchased from ChemBridge Corporation, formatted into 96-well mother plates using an FX liquid dispenser, and subsequently formatted across 157 plates of the 384-well plate at 50 nl (10 μM final concentration per well) using an Echo liquid dispenser. Dimethyl sulfoxide (DMSO) was loaded in matching %v/v as in-plate no-compound controls as well as in columns 1, 2, 23, and 24 as negative controls. The 384-well flat, black-bottom polypropylene plates (PN 781209, Greiner Bio-One) were used because of their low autofluorescence and low interwell cross-talk. The plates were sealed and stored at −20°C until use. The screening was carried out across 5 days with ~10,000 compounds (~32-compound plates) screened each day. A week before screening, a fresh vial of TNFR1ΔCD-GFP/RFP (TNFR1ΔCD-FRET pair) cells was thawed, plated in a 225-cm2 flask (Corning), and checked for expression. The cells were then expanded into six 225-cm2 flasks for 3 days and further expanded into 36 flasks for another 3 days. Before each day of screening, the stable cells expressing the TNFR1 FRET biosensor were harvested to check for expression and response variation in fluorescent intensity. Stable cells were then dispensed into the drug plates (50,000 cells per well) and incubated with the compounds or DMSO as a negative control, followed by fluorescence lifetime measurements. The fluorescence waveforms were acquired by a prototype fluorescence lifetime plate reader (Fluorescence Innovations Inc.) as described (26).
High-throughput screening data analysis
Time-resolved fluorescence waveforms obtained for each well were fitted to single-exponential decays using least-squares minimization global analysis software (Fluorescence Innovations Inc.) to give donor-acceptor lifetime (τDA) from the TNFR1ΔCD-GFP/RFP (FRET pair) cell line and donor lifetime (τD) from a TNFR1ΔCD-GFP donor-only control cell line. FRET efficiency (E) was then calculated on the basis of Eq. 1
| (1) |
The Z-factor, a high-throughput screening assay quality indicator, was determined with zafirlukast as a positive control and DMSO as a negative control and calculated on the basis of Eq. 2 (61)
| (2) |
In Eq. 2, σp and σn are the SDs of the observed τDA values, and μp and μn are the mean τDA values of the positive and negative controls, respectively. To make this metric less sensitive to strong outliers, we used the normalized median absolute deviation (1.4826 × MAD) and median in place of the SD and mean, respectively (62). The Z-factor obtained was 0.76 ± 0.02, indicating excellent assay quality.
Fluorescent compounds were flagged as potential false positives due to interference from compound fluorescence by a set of stringent fluorescent compound filters based on analysis of the spectral waveforms of each well from the DIVERSet screen (63). After removal of fluorescent compounds, a histogram of the FRET distribution from all compounds in the screen was plotted and fitted to a Gaussian curve to obtain a mean and SD. A hit was defined as a compound that decreased the FRET efficiency by more than 3SD relative to the mean.
FRET dose-response assay
The hit compounds—DS41 (ID 19298144), DS42 (ID 43812755), DS43 (ID 23420063), DS44 (ID 95020298), DS45 (ID 45055796), DS50 (ID 33619467), and DS51 (ID 74188632)—were purchased from ChemBridge Corporation. A negative control compound (ID 65311687) was also purchased. These drug compounds were dissolved in DMSO to make 10 mM stock solution, which were then subsequently serially diluted in 96-well mother plates to obtain eight doses at 50× concentrations. Hits were screened at eight different concentrations (0.1 to 200 μM). Compounds (1 μl) were transferred from the mother plates into assay plates using a Mosquito HV liquid handler (TTP Labtech Ltd.). The cell preparation of the WT TNFR1 FRET biosensor in the FRET dose-response assays was carried out through the same way as the high-throughput screening.
IκBα degradation and NF-κB activation assays
IκBα degradation assay and NF-κB activation assay with HEK293 cells were carried out as described (26). Densitometry of the Western blots was performed using Image Studio (LI-COR Biosciences), and data were normalized to the β-actin loading control and the amount of IκBα in the control cells in the absence of ligand. For TRADD-induced NF-κB activation in HEK293 cells, cells (1 × 106) in a six-well plate were transfected with 1 μg of NF-κB firefly luciferase reporter genes, 0.1 μg of Renilla luciferase reporter genes, 1 μg of TRADD plasmid, and 0.4 μg of control plasmid for a total of 2.5 μg of DNA. In the control cells, the TRADD plasmid was replaced with control plasmid. After 3 hours of transfection, cells were harvested and plated (30,000 cells per well; total volume, 50 μl) into 96-well white solid-bottom assay plates (Greiner Bio-One North America). Drug treatments (0.1 to 200 μM) or DMSO treatment (negative control) was performed 5 hours after cell plating. Luciferase activities were determined 24 hours after treatments. Briefly, 50 μl of Dual-Glo Luciferase Reagent (Promega) was added and incubated at room temperature for 15 min and measured firefly luminescence using a Cytation 3 Cell Imaging Multi-Mode Reader luminometer (BioTek). Next, 50 μl of Dual-Glo Stop & Glo Reagent (Promega) was added and incubated at room temperature for 15 min and measured Renilla luminescence using luminometer. The luciferase activities were normalized on the basis of Renilla expression levels.
For NF-κB activation luciferase assay with HAP1 WT and TNFR1 KO cells, the cells were transfected with the NF-κB–luciferase reporter genes (10 μg of firefly luciferase genes and 1 μg of Renilla luciferase genes) in a 100-mm plate with Lipofectamine 3000 (Invitrogen). On the following day, cells were lifted with TrypLE and resuspended in phenol red–free DMEM (Gibco). These cells were dispensed into 96-well assay plates (30,000 cells per well; total volume, 50 μl) and incubated with drugs (0.1 to 200 μM) or DMSO (negative control) in the presence (0.1 μg/ml) and absence of LTα for 18 hours at 37°C. Readings for luciferase activities were acquired as described above. All data from NF-κB activation assays were normalized to luciferase activity of cells in the presence of ligand or TRADD overexpression.
Coimmunoprecipitation and native gel characterization
Coimmunoprecipitation between endogenous TNFR1 and ligand LTα in the presence and absence of hit compounds was performed as described previously (26). Briefly, the anti-FLAG magnetic beads (M8823, MilliporeSigma) were incubated with soluble FLAG-tagged LTα. After washing to remove unbound LTα, magnetic beads with LTα were then incubated with HEK293 cell lysates in the presence of DMSO or hit compounds. Immunoprecipitate samples were then resolved using 4 to 15% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel with staining of FLAG antibody (2044S, Cell Signaling Technology) and TNFR1 antibody (Ab19139, Abcam) to probe for the amount of FLAG-tagged LTα and TNFR1, respectively. No TNFR1 control was performed with magnetic beads with LTα incubated with respective lysis buffer only without the cell lysates. The overexpression and purification of the N-terminal FLAG-tagged LTα and FLAG-tagged TNFR1 PLAD (residues 30 to 82) were carried out as described previously, and the soluble PLAD protein exists as dimers under native conditions (26). The recombinant human TNFR1 ECD was purchased from Abcam. The purity of the proteins was assessed by 4 to 15% SDS-PAGE gels (Bio-Rad) under reducing conditions, followed by Coomassie staining. Protein concentrations were measured using the bicinchoninic acid assay (Thermo Fisher Scientific). To test the disruption of receptor-receptor or ligand- ligand interactions, purified soluble TNFR1 PLAD or ECD and LTα (5 μg) were assessed using Native PAGE gels (Bio-Rad) in the absence and presence of hit compounds (200 μM) under nonreducing conditions, followed by Coomassie staining. Native PAGE gel of soluble PLAD was also characterized by Western blot with TNFR1-specific antibody (H-5) (sc-8436, Santa Cruz Biotechnology) that targets the ECD of the receptor. Competition assay between the hit compounds and zafirlukast was performed by the cotreatment of both the hit compounds (200 or 1000 μM) and zafirlukast (200 μM) at the same time to soluble PLAD and observed the extent of disruption of PLAD-PLAD interactions.
SPR binding assay
Binding affinity between TNFR1 ECD and compounds or ligand was determined by SPR analysis using Biacore S200. Recombinant human TNFR1 ECD (Abcam) was immobilized on the CM5 sensor chip (Biacore, GE Healthcare) by amine coupling. Briefly, the dextran surface was activated with a 1:1 mixture of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.1 M N-hydroxysuccinimide. TNFR1 ECD (20 μg/ml) in 10 mM sodium acetate at pH 5 was flowed past a working surface before blocking the remaining activated carboxymethyl groups with 1 M ethanolamine at pH 8.5 to achieve a level of 2500 response units suitable for binding analysis. The reference surface was activated and reacted with only ethanolamine.
For direct binding assays between the receptor and the small molecules, the hit compounds and the analog at eight different concentrations (0.1 to 200 μM) as well as DMSO-only controls were prepared in HBS-EP buffer (GE Healthcare) containing a total of 2% DMSO. For competition assays between ligand and small molecules, ligand (LTα) at eight different concentrations (0.1 to 200 nM) was prepared in HBS-EP buffer in the presence of saturated dose of compounds (200 μM) or DMSO containing a total of 2% DMSO. For other competition assays, samples containing small molecules and/or antibody were prepared in HBS-EP buffer. The samples were injected over both the reference and ECD immobilized surfaces at 10 μl/min for 90 s and dissociated in glycine-HCl (pH 2.5). Samples and blanks from buffer and DMSO-only controls were measured on a 96-well microplate (Biacore, GE Healthcare) at 25°C. Reflectivity response data points were extracted from response curves at 5 s before the end of the injection to determine steady-state binding. All the data were double referenced with blanks using standard procedures with Biacore S200 evaluation software v1.0.
Computational analysis of crystal structure
For analysis of the TNFR1 crystal structure, visual molecular dynamics (VMD) was used. The crystal structure of the preligand dimer of TNFR1 ECD was downloaded from the PDB (PDB: 1NCF) (6), and the structure preparation was performed in CHARMM-GUI. The distance measurement of the TNFR1 ECD was performed in VMD. The hydrogen bonds between the residues in the ligand-binding loop were determined by VMD with a distance cutoff of <3.2 Å and an angle cutoff of <60°.
SAR analysis
The analogs of the lead compound (DS41) were identified through a search of the company’s database (ChemBridge) that shares more than 90% similarity (based on the chemical functionality and scaffolding as determined by the company’s similarity search engine) to the lead compound. A total of 19 analogs of DS41 (compound IDs in table S1) were purchased from ChemBridge Corporation. The analogs were dissolved in DMSO to make 10 mM stock solution, which were serially diluted in 96-well mother plates at eight different doses (0.1 to 200 μM) at 50× concentration. FRET measurements were performed using the WT TNFR1 FRET biosensor at a compound concentration of 50 μM. The ability of the analog to inhibit IκBα degradation was tested at a compound concentration of 200 μM, and the inhibition of NF-κB activation was determined in a dose-dependent manner to obtain the IC50 values and percent inhibition listed in table S1. SPR binding assays, specificity tests, and mechanistic assays were conducted as described above.
Statistical analysis
Data are presented as means ± SD unless stated otherwise. To determine statistical significance for all experiments, data analysis was performed using a two-tailed unpaired t test (Student’s t test) with P values determined using GraphPad software. Values of P < 0.05 were considered statistically significant. GraphPad style in using asterisks to denote P values in figures was used (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Supplementary Material
Fig. S1. Chemical structures of the novel small-molecule inhibitors of TNFR1 and a negative control compound.
Fig. S2. The seven hit compounds and zafirlukast bind the TNFR1 ECD as characterized by SPR measurements.
Fig. S3. Some hit compounds nonspecifically inhibit TNFR1-stimulated NF-κB activation.
Fig. S4. DS41, but not DS42 and zafirlukast, inhibits TRADD-induced NF-κB activation in HEK293 cells.
Fig. S5. Small-molecule inhibitors do not disrupt ligand-receptor interactions as characterized by SPR measurements.
Fig. S6. Hit compounds do not disrupt either the TNFR1 PLAD dimer or the LTα trimer.
Fig. S7. Hit compounds reduce FRET mediated by the TNFR1 mutant biosensors.
Fig. S8. Hit compounds compete with zafirlukast in binding to the TNFR1 PLAD and in inhibiting NF-κB activation.
Fig. S9. Hit compounds do not compete with the H398 antibody in modulating TNFR1 signaling.
Fig. S10. DSA114, an analog of DS41, shows improved potency and specificity by acting through the same noncompetitive inhibition mechanism.
Table S1. Functional characterization of DS41 and its analogs.
Acknowledgments
We thank N. Vunnam, B. Brummel, and Z. Ding from the Sachs group; J. Li and S. Yuen from the Thomas group; W. Fiers from the Aldrich group; and G. D. Gillispie and K. C. Peterson from Fluorescence Innovations Inc. for technical discussions. We also thank J. Connett for confirming our methods in statistical analysis. Flow cytometry was performed at the University of Minnesota (UMN) Lillehei Heart Institute, compound dispensing and SPR [S10 Shared Instrumentation Grant 1S10OD021539-01 funded by the Office of Research Infrastructure Programs (ORIP), NIH] were performed at the UMN Institute of Therapeutic Drug Discovery and Development (ITDD) High-Throughput Screening Laboratory, and spectroscopy were performed at the UMN Biophysical Technology Center. This work was carried out, in part, using computing resources at the UMN Supercomputing Institute.
Funding: This study was supported by U.S. NIH grants to J.N.S. (R01 GM107175 and R35 GM131814) and D.D.T. (R01 GM27906; AG26160; R42 DA037622, subcontract from grant to Fluorescence Innovations, Gillispie PI). C.H.L. was supported by the Doctoral Dissertation Fellowship from the UMN.
Footnotes
Competing interests: D.D.T. holds equity in and serves as executive officer for Photonic Pharma LLC, a company that owns intellectual property related to technology used in parts of this project. These relationships have been reviewed and managed by the UMN in accordance with its conflict-of-interest polices. The other authors declare that they have no competing interests.
Data and materials availability: All data required to evaluate the conclusions of the paper are present in the main text or the Supplementary Materials of the paper.
SUPPLEMENTARY MATERIALS
http://stke.sciencemag.org/cgi/content/full/12/592/eaav5637/DC1
REFERENCES AND NOTES
- 1.Ashkenazi A, Dixit VM, Death receptors: Signaling and modulation. Science 281, 1305–1308 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Brenner D, Blaser H, Mak TW, Regulation of tumour necrosis factor signalling: Live or let die. Nat. Rev. Immunol. 15, 362–374 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Wajant H, Scheurich P, TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 278, 862–876 (2011). [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health, The Autoimmune Diseases Coordinating Committee, Progress in Autoimmune Diseases Research (National Institutes of Health, 2005), foreword and pp. i, 1, 2, 16, 17, 28–30, 32, 52. [Google Scholar]
- 5.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W, Crystal structure of the soluble human 55 kd TNF receptor-human TNFβ complex: Implications for TNF receptor activation. Cell 73, 431–445 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Naismith JH, Devine TQ, Brandhuber BJ, Sprang SR, Crystallographic evidence for dimerization of unliganded tumor necrosis factor receptor. J. Biol. Chem. 270, 13303–13307 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Brown SL, Greene MH, Gershon SK, Edwards ET, Braun MM, Tumor necrosis factor antagonist therapy and lymphoma development: Twenty-six cases reported to the Food and Drug Administration. Arthritis Rheum. 46, 3151–3158 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Shakoor N, Michalska M, Harris CA, Block JA, Drug-induced systemic lupus erythematosus associated with etanercept therapy. Lancet 359, 579–580 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Sicotte NL, Voskuhl RR, Onset of multiple sclerosis associated with anti-TNF therapy. Neurology 57, 1885–1888 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Sedger LM, McDermott MF, TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants—Past, present and future. Cytokine Growth Factor Rev. 25, 453–472 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Chan FK-M, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ, A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288, 2351–2354 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Lee H-W, Lee S-H, Lee H-W, Ryu Y-W, Kwon M-H, Kim Y-S, Homomeric and heteromeric interactions of the extracellular domains of death receptors and death decoy receptors. Biochem. Biophys. Res. Commun. 330, 1205–1212 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Vunnam N, Lo CH, Grant BD, Thomas DD, Sachs JN, Soluble extracellular domain of death receptor 5 inhibits TRAIL-induced apoptosis by disrupting receptor–receptor interactions. J. Mol. Biol. 429, 2943–2953 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Valley CC, Lewis AK, Sachs JN, Piecing it together: Unraveling the elusive structure-function relationship in single-pass membrane receptors. Biochim. Biophys. Acta Biomembr. 1859, 1398–1416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis AK, Valley CC, Sachs JN, TNFR1 signaling is associated with backbone conformational changes of receptor dimers consistent with overactivation in the R92Q TRAPS mutant. Biochemistry 51, 6545–6555 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Valley CC, Lewis AK, Mudaliar DJ, Perlmutter JD, Braun AR, Karim CB, Thomas DD, Brody JR, Sachs JN, Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces death receptor 5 networks that are highly organized. J. Biol. Chem. 287, 21265–21278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vunnam N, Campbell-Bezat CK, Lewis AK, Sachs JN, Death receptor 5 activation is energetically coupled to opening of the transmembrane domain dimer. Biophys. J. 113, 381–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis AK, Valley CC, Peery SL, Brummel B, Braun AR, Karim CB, Sachs JN, Death receptor 5 networks require membrane cholesterol for proper structure and function. J. Mol. Biol. 428, 4843–4855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis AK, James ZM, McCaffrey JE, Braun AR, Karim CB, Thomas DD, Sachs JN, Open and closed conformations of the isolated transmembrane domain of death receptor 5 support a new model of activation. Biophys. J. 106, L21–L24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reis CR, van Assen AHG, Quax WJ, Cool RH, Unraveling the binding mechanism of trivalent tumor necrosis factor ligands and their receptors. Mol. Cell. Proteomics 10, M110.002808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukai Y, Nakamura T, Yoshikawa M, Yoshioka Y, Tsunoda S.-i, Nakagawa S, Yamagata Y, Tsutsumi Y, Solution of the structure of the TNF-TNFR2 complex. Sci. Signal. 3, ra83 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Vanamee ÉS, Faustman DL, Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal. 11, eaao4910 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Fricke F, Malkusch S, Wangorsch G, Greiner JF, Kaltschmidt B, Kaltschmidt C, Widera D, Dandekar T, Heilemann M, Quantitative single-molecule localization microscopy combined with rule-based modeling reveals ligand-induced TNF-R1 reorganization toward higher-order oligomers. Histochem. Cell Biol. 142, 91–101 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Chan FK-M, Siegel RM, Zacharias D, Swofford R, Holmes KL, Tsien RY, Lenardo MJ, Fluorescence resonance energy transfer analysis of cell surface receptor interactions and signaling using spectral variants of the green fluorescent protein. Cytometry 44, 361–368 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Chan FK-M, Three is better than one: Pre-ligand receptor assembly in the regulation of TNF receptor signaling. Cytokine 37, 101–107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo CH, Vunnam N, Lewis AK, Chiu T-L, Brummel BE, Schaaf TM, Grant BD, Bawaskar P, Thomas DD, Sachs JN, An innovative high-throughput screening approach for discovery of small molecules that inhibit TNF receptors. SLAS Discov. 22, 950–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruber SJ, Cornea RL, Li J, Peterson KC, Schaaf TM, Gillispie GD, Dahl R, Zsebo KM, Robia SL, Thomas DD, Discovery of enzyme modulators via high-throughput time-resolved FRET in living cells. J. Biomol. Screen. 19, 215–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamińska A, Bartosik A, Banach-Orłowska M, Pilecka I, Jastrzębski K, Zdżalik-Bielecka D, Castanon I, Poulain M, Neyen C, Wolińska-Nizioł L, Toruń A, Szymańska E, Kowalczyk A, Piwocka K, Simonsen A, Stenmark H, Fürthauer M, González-Gaitán M, Miaczynska M, ESCRT proteins restrict constitutive NF-κB signaling by trafficking cytokine receptors. Sci. Signal. 9, ra8 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y, Woronicz JD, Liu W, Goeddel DV, Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 283, 543–546 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Hsu H, Xiong J, Goeddel DV, The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 81, 495–504 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Day ES, Cote SM, Whitty A, Binding efficiency of protein–protein complexes. Biochemistry 51, 9124–9136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter PH, Scherle PA, Muckelbauer JK, Voss ME, Liu RQ, Thompson LA, Tebben AJ, Solomon KA, Lo YC, Li Z, Strzemienski P, Yang G, Falahatpisheh N, Xu M, Wu Z, Farrow NA, Ramnarayan K, Wang J, Rideout D, Yalamoori V, Domaille P, Underwood DJ, Trzaskos JM, Friedman SM, Newton RC, Decicco CP, Photochemically enhanced binding of small molecules to the tumor necrosis factor receptor-1 inhibits the binding of TNF-alpha. Proc. Natl. Acad. Sci. U.S.A. 98, 11879–11884 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murali R, Cheng X, Berezov A, Du X, Schön A, Freire E, Xu X, Chen YH, Greene MI, Disabling TNF receptor signaling by induced conformational perturbation of tryptophan-107. Proc. Natl. Acad. Sci. U.S.A. 102, 10970–10975 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Feng Z, Wang Y, Ma S, Hu Z, Yang P, Chai Y, Xie X, Discovery of novel ligands for TNF-α and TNF receptor-1 through structure-based virtual screening and biological assay. J. Chem. Inf. Model. 57, 1101–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takasaki W, Kajino Y, Kajino K, Murali R, Greene MI, Structure-based design and characterization of exocyclic peptidomimetics that inhibit TNFα binding to its receptor. Nat. Biotechnol. 15, 1266–1270 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN, The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 287, 34979–34991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zettlitz KA, Lorenz V, Landauer K, Münkel S, Herrmann A, Scheurich P, Pfizenmaier K, Kontermann RE, ATROSAB, a humanized antagonistic anti-tumor necrosis factor receptor one-specific antibody. MAbs 2, 639–647 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Zheng Y, Chen X, Drugs for autoimmune inflammatory diseases: From small molecule compounds to anti-TNF biologics. Front. Pharmacol. 8, 460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lis K, Kuzawińska O, Bałkowiec-Iskra E, Tumor necrosis factor inhibitors—State of knowledge. Arch. Med. Sci. 10, 1175–1185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalliolias GD, Ivashkiv LB, TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marc F, Maini RN, Perspectives from masters in rheumatology and autoimmunity: Can we get closer to a cure for rheumatoid arthritis? Arthritis Rheumatol. 67, 2283–2291 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Schabert VF, Watson C, Joseph GJ, Iversen P, Burudpakdee C, Harrison DJ, Costs of tumor necrosis factor blockers per treated patient using real-world drug data in a managed care population. J. Manag. Care Pharm. 19, 621–630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaltsonoudis E, Voulgari PV, Konitsiotis S, Drosos AA, Demyelination and other neurological adverse events after anti-TNF therapy. Autoimmun. Rev. 13, 54–58 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Shibata H, Yoshioka Y, Ohkawa A, Abe Y, Nomura T, Mukai Y, Nakagawa S, Taniai M, Ohta T, Mayumi T, Kamada H, S.-i. Tsunoda, Y. Tsutsumi, The therapeutic effect of TNFR1-selective antagonistic mutant TNF-α in murine hepatitis models. Cytokine 44, 229–233 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Steeland S, Puimège L, Vandenbroucke RE, Van Hauwermeiren F, Haustraete J, Devoogdt N, Hulpiau P, Leroux-Roels G, Laukens D, Meuleman P, De Vos M, Libert C, Generation and characterization of small single domain antibodies inhibiting human tumor necrosis factor receptor 1. J. Biol. Chem. 290, 4022–4037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kontermann RE, Munkel S, Neumeyer J, Müller D, Branschädel M, Scheurich P, Pfizenmaier K, A humanized tumor necrosis factor receptor 1 (TNFR1)-specific antagonistic antibody for selective inhibition of tumor necrosis factor (TNF) action. J. Immunother. 31, 225–234 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Fischer R, Kontermann RE, Maier O, Targeting sTNF/TNFR1 signaling as a new therapeutic strategy. Antibodies 4, 48–70 (2015). [Google Scholar]
- 48.Gohda T, Niewczas MA, Ficociello LH, Walker WH, Skupien J, Rosetti F, Cullere X, Johnson AC, Crabtree G, Smiles AM, Mayadas TN, Warram JH, Krolewski AS, Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J. Am. Soc. Nephrol. 23, 516–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng M, Loughran PA, Zhang L, Scott MJ, Billiar TR, Shedding of the tumor necrosis factor (TNF) receptor from the surface of hepatocytes during sepsis limits inflammation through cGMP signaling. Sci. Signal. 8, ra11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D, Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J. Exp. Med. 175, 323–329 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf Y, Shemer A, Polonsky M, Gross M, Mildner A, Yona S, David E, Kim K-W, Goldmann T, Amit I, Heikenwalder M, Nedospasov S, Prinz M, Friedman N, Jung S, Autonomous TNF is critical for in vivo monocyte survival in steady state and inflammation. J. Exp. Med. 214, 905–917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brambilla L, Guidotti G, Martorana F, Iyer AM, Aronica E, Valori CF, Rossi D, Disruption of the astrocytic TNFR1-GDNF axis accelerates motor neuron degeneration and disease progression in amyotrophic lateral sclerosis. Hum. Mol. Genet. 25, 3080–3095 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Cao J, Meng F, Gao X, Dong H, Yao W, Expression and purification of a natural N-terminal pre-ligand assembly domain of tumor necrosis factor receptor 1 (TNFR1 PLAD) and preliminary activity determination. Protein J. 30, 281 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Deng G-M, Zheng L, Ka-Ming Chan F, Lenardo M, Amelioration of inflammatory arthritis by targeting the pre-ligand assembly domain of tumor necrosis factor receptors. Nat. Med. 11, 1066–1072 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Bugge K, Kragelund BB, Lindorff-Larsen K, Role of protein dynamics in transmembrane receptor signalling. Curr. Opin. Struct. Biol. 48, 74–82 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Wootten D, Christopoulos A, Sexton PM, Emerging paradigms in GPCR allostery: Implications for drug discovery. Nat. Rev. Drug Discov. 12, 630–644 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Wells JA, McClendon CL, Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Schön A, Lam SY, Freire E, Thermodynamics-based drug design: Strategies for inhibiting protein-protein interactions. Future Med. Chem. 3, 1129–1137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schön A, Madani N, Smith AB, Lalonde JM, Freire E, Some binding-related drug properties are dependent on thermodynamic signature. Chem. Biol. Drug Des. 77, 161–165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zacharias DA, Violin JD, Newton AC, Tsien RY, Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Zhang J-H, Chung TD, Oldenburg KR, A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE, Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6, 569–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaaf TM, Peterson KC, Grant BD, Bawaskar P, Yuen S, Li J, Muretta JM, Gillispie GD, Thomas DD, High-throughput spectral and lifetime-based FRET screening in living cells to identify small-molecule effectors of SERCA. SLAS Discov. 22, 262–273 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Chemical structures of the novel small-molecule inhibitors of TNFR1 and a negative control compound.
Fig. S2. The seven hit compounds and zafirlukast bind the TNFR1 ECD as characterized by SPR measurements.
Fig. S3. Some hit compounds nonspecifically inhibit TNFR1-stimulated NF-κB activation.
Fig. S4. DS41, but not DS42 and zafirlukast, inhibits TRADD-induced NF-κB activation in HEK293 cells.
Fig. S5. Small-molecule inhibitors do not disrupt ligand-receptor interactions as characterized by SPR measurements.
Fig. S6. Hit compounds do not disrupt either the TNFR1 PLAD dimer or the LTα trimer.
Fig. S7. Hit compounds reduce FRET mediated by the TNFR1 mutant biosensors.
Fig. S8. Hit compounds compete with zafirlukast in binding to the TNFR1 PLAD and in inhibiting NF-κB activation.
Fig. S9. Hit compounds do not compete with the H398 antibody in modulating TNFR1 signaling.
Fig. S10. DSA114, an analog of DS41, shows improved potency and specificity by acting through the same noncompetitive inhibition mechanism.
Table S1. Functional characterization of DS41 and its analogs.