Abstract
Ets1 is emerging as a key transcription factor that is required to prevent autoimmunity in mice and humans. Ets1 is expressed in both B and T cells, and mice lacking Ets1 are characterized by excess B and T cell activation, leading to enhanced formation of Ab-secreting cells and high titers of autoantibodies. In humans, genome-wide association studies have detected associations of single nucleotide polymorphisms in the human ETS1 gene with autoimmune diseases, including lupus. An increased fraction of CD4+ T cells from Ets1−/− mice have an activated effector-memory phenotype, and there are aberrations in differentiation that contribute to the autoimmune phenotype. In vitro studies of B cells suggest that Ets1 may have B cell–intrinsic effects as well. To confirm B cell–intrinsic roles for Ets1, we crossed CD19-Cre mice to mice with a floxed allele of Ets1. Mice with a B cell–specific deletion of Ets1 show increases in B cell activation, numbers of Ab-secreting cells, and levels of autoantibodies, despite the fact that T cells are normal. However, when compared with conventional Ets1 knockout mice, mice with B cell–specific loss of Ets1 have a significantly milder phenotype. These results demonstrate that Ets1 is required in B cells to prevent autoimmune responses but that loss of Ets1 activity in other cell types is required for maximal autoimmune phenotypes.
INTRODUCTION
Autoimmune diseases such as systemic lupus erythematosus result from immune system recognition of and activation by self-antigens. In aggregate, these diseases are thought to affect 5–10% of the population (1, 2). The causes of autoimmune disease are complex and depend on a variety of genetic and environmental factors. One gene implicated in the development and progression of autoimmune diseases is Ets1 (3), which encodes a transcription factor highly expressed in B and T lymphocytes. Ets1 knockout mice develop a lupus-like autoimmune disease, accompanied by aberrant B and T cell differentiation (4–10). The human ETS1 gene has also been identified as a susceptibility locus for development of lupus and multiple other autoimmune diseases (11–18).
B cells from Ets1−/− mice show a variety of defects including loss of the marginal zone B cell population, increased levels of activation markers in follicular B cells, increased isotype switching to IgG1 and IgE, reduced switching to IgG2a, and increased numbers of Ab-secreting cells (ASCs) (4–6, 19). The increase in ASCs in Ets1−/− mice is accompanied by increases in serum IgM and IgG1 levels and the secretion of autoantibodies that deposit as immune complexes in the kidney glomeruli (4, 5, 8, 19). Transfer of purified Ets1−/− B cells into wild-type hosts results in downregulation of several activation markers including CD23, CD80, and CD86 (4), indicating that part of the B cell phenotype in Ets1−/− mice is B cell extrinsic. T cells from Ets1−/− mice have severe aberrations, including increased differentiation to an effector/memory phenotype and altered differentiation of Th subsets (8–10, 20–22). Freshly isolated CD4+ T cells from Ets1−/− mice express high levels of mRNA for IL-4, IL-5, IL-10, and IL-13 but reduced levels of mRNA for IFN-γ (8). Ets1−/− CD4 T cells cultured in vitro under appropriate skewing conditions show similarly reduced IFN-γ production but reduced rather than enhanced production of Th2 cytokines (IL-4, IL-5, and IL-13) (10). Ets1−/− CD4 T cells give rise to increased percentages of IL-17–secreting cells (21). In addition, there are reduced numbers of Foxp3+ regulatory T cells (Tregs) in Ets1−/− mice, and the Tregs that do develop express low levels of Foxp3 and are poor suppressors of effector T cell responses (8). Transfer of wild-type Tregs into chimeras containing Ets1−/− T cells results in the restoration of Ets1−/− marginal zone B cells and reduced production of IgG1 and IgE Abs (8). Therefore, impaired Treg function was suggested to be the cause of several B cell defects resulting from the absence of Ets1. However, a more recent study in which Ets1 was specifically deleted in the T cell population shows that the major T cell aberration underlying the autoimmune phenotype of Ets1−/− mice is excess T cell differentiation to T follicular helper cells that secrete IL-4 (T follicular helper type 2 [Tfh2] cells) (20).
Together, the results described above indicate that abnormalities in the CD4+ T cell population in Ets1 knockout mice may drive the aberrations in the B cell compartment. However, this does not preclude an important role of Ets1 in B cells and certain data substantiate a function of Ets1 within B cells themselves. For instance, in vitro culture of purified Ets1−/− B cells in the presence of TLR ligands results in hyperproduction of ASCs (4, 23). In addition, retrovirally driven expression of Ets1 in differentiating wild-type B cells prevents their differentiation to ASCs (23–25). B cell–intrinsic functions of Ets1 in regulating ASC generation include the ability of Ets1 to bind to and inhibit the function of the ASC transcription factor Blimp1 (24, 25) and direct regulation of genes important for B cell differentiation (26). Purified Ets1-deficient B cells cultured with LPS and IFN-γ show a defect in switching to IgG2a (5). Furthermore, mixed bone marrow chimeras generated with congenically labeled wild-type and Ets1−/−donor cells showed that Ets1−/− B cells give rise to more ASCs than wild-type B cells when they are found together in the same microenvironment (23). In the current study, analysis of mice with a B cell–specific deletion of Ets1 shows that there is a cell-intrinsic role for Ets1 in regulating B cell activation, development of ASCs, and production of autoantibodies. These B cell–intrinsic functions of Ets1 likely cooperate with T cell–intrinsic functions of Ets1 to modulate immune responses.
MATERIALS AND METHODS
Mice
Ets1 floxed mice have been reported previously (27) and have been bred to the C57BL/6 genetic background for >12 generations. CD19-Cre mice were obtained from Jackson Laboratory and are on a C57BL/6 background. Conventional Ets1 knockout mice and littermate wild-type controls were bred in our colony and are maintained on a mixed genetic background (C57BL/6 × 129Sv), because of perinatal lethality on a pure C57BL/6 background. Animal experiments were performed under the approval and guidance of the Institutional Animal Care and Use Committee of Roswell Park Cancer Institute.
Western blotting
B cells were purified from spleens of mice using magnetic beads to select CD43-negative cells, and lysates were prepared. Western blotting was performed using monoclonal anti-Ets1 Ab (Clone D808A; Cell Signaling Technology). Blots were reprobed with monoclonal anti-GAPDH Ab (Clone 6C5; MilliporeSigma) as a loading control.
Flow cytometry
Single-cell suspensions of spleen and lymph nodes were prepared using standard techniques and then stained with Ghost Dye Violet 510 to label dead cells. Cells were subsequently stained with various combinations of Abs against the following Ags to assess B cell differentiation and activation status: B220, CD21, CD23, CD73, CD80, CD86, CD138, Fas, IgM, IgD, IgG1, and PD-L2 as well as peanut agglutinin (PNA). T cell differentiation was assessed by staining with Abs against CD4, CD8, CD62L, CD44, CD86, and intracellular Foxp3. Flow cytometry data were collected on an LSR II flow cytometer. Flow cytometry data were analyzed using FlowJo software to gate on live (Ghost Dye Violet 510 negative) cells and subsequently gated on singlets based on side scatter area versus width and on the lymphocyte gate based on forward scatter area versus side scatter area. B and T lymphocytes were further gated based on positivity for B220 (B cells) or CD4 (T cells), and expression of relevant markers in these populations was then examined.
ELISA
Solutions of 10 μg/ml anti-mouse IgM, anti-mouse IgG, or various potential autoantigens were used to coat MaxiSorp 96-well ELISA plates overnight. Autoantigens included mixed calf thymus histones (Sigma-Aldrich), calf thymus dsDNA (ThermoFisher Scientific), mouse purified myelin basic protein (MBP) (Sigma-Aldrich), and Smith Ag (SMA-3000 containing SmB, SmB’, and SmD Ags; ImmunoVision). Dilutions of serum from mice of various genotypes were added to plates, and ELISA was performed using standard techniques and TMB substrate. Absorbances were read at 450 nm. For total serum IgM and IgG, absorbance values were compared with a standard curve of purified mouse IgM or IgG to calculate exact concentrations. For autoantibodies, data are reported as the absorbance values.
ELISPOT
ELISPOT plates (MultiScreen 96-well plates with Immobilon-P membranes; MilliporeSigma) were coated overnight with anti-IgM (BioLegend) or anti-IgG (SouthernBiotech) Abs. Single-cell suspensions of spleen and lymph node were plated at various dilutions, and ELISPOT assay was performed as previously described (24).
Staining of kidney sections for immune complex deposition
Kidneys were harvested from mice, frozen in Tissue-Tek OCT medium, and sectioned with a cryostat. Sections were fixed in paraformaldehyde and stained with FITC-conjugated anti-mouse IgM or anti-mouse IgG. Images were captured with a Nikon 80i fluorescence microscope and analyzed using Fiji software.
Statistics
All data are shown as mean ± SEM. Means between Ets1+/+ and Ets1−/− and between CD19-Cre Ets1+/+ and CD19-Cre Ets1fl/fl mice were compared using unpaired Student t test or Mann–Whitney U test depending on the distribution of data. A p value ≤0.05 was considered significant.
RESULTS
Generation of mice lacking Ets1 specifically in B cells
To test B cell–intrinsic versus B cell–extrinsic roles for Ets1, mice specifically lacking Ets1 in B cells were generated by crossing the well-characterized B cell–specific CD19-Cre line with mice carrying a floxed allele of Ets1 to generate CD19-Cre Ets1fl/fl mice (Fig. 1A). The floxed allele of Ets1 carries loxP sites flanking the last two exons of the gene (exons 7 and 8), which encode the DNA-binding domain of Ets1. Western blotting showed that Ets1 was efficiently deleted in B cells from CD19-Cre Ets1fl/fl mice, yielding levels similar to that found in conventional Ets1 knockout mice (Ets1−/−) (Fig. 1B). The spleens of CD19-Cre Ets1fl/fl mice were normal in size, whereas spleens of conventional Ets1 knockout mice were enlarged (Fig. 1C). In keeping with the normal spleen size, the total numbers of splenocytes, splenic B cells, and splenic T cells of CD19-Cre Ets1fl/fl mice were similar to those found in wild-type (Ets1+/+) and CD19-Cre only (CD19-Cre Ets1+/+) control mice (Fig. 1D–F).
FIGURE 1. Generation and initial characterization of mice lacking Ets1 in B cells.
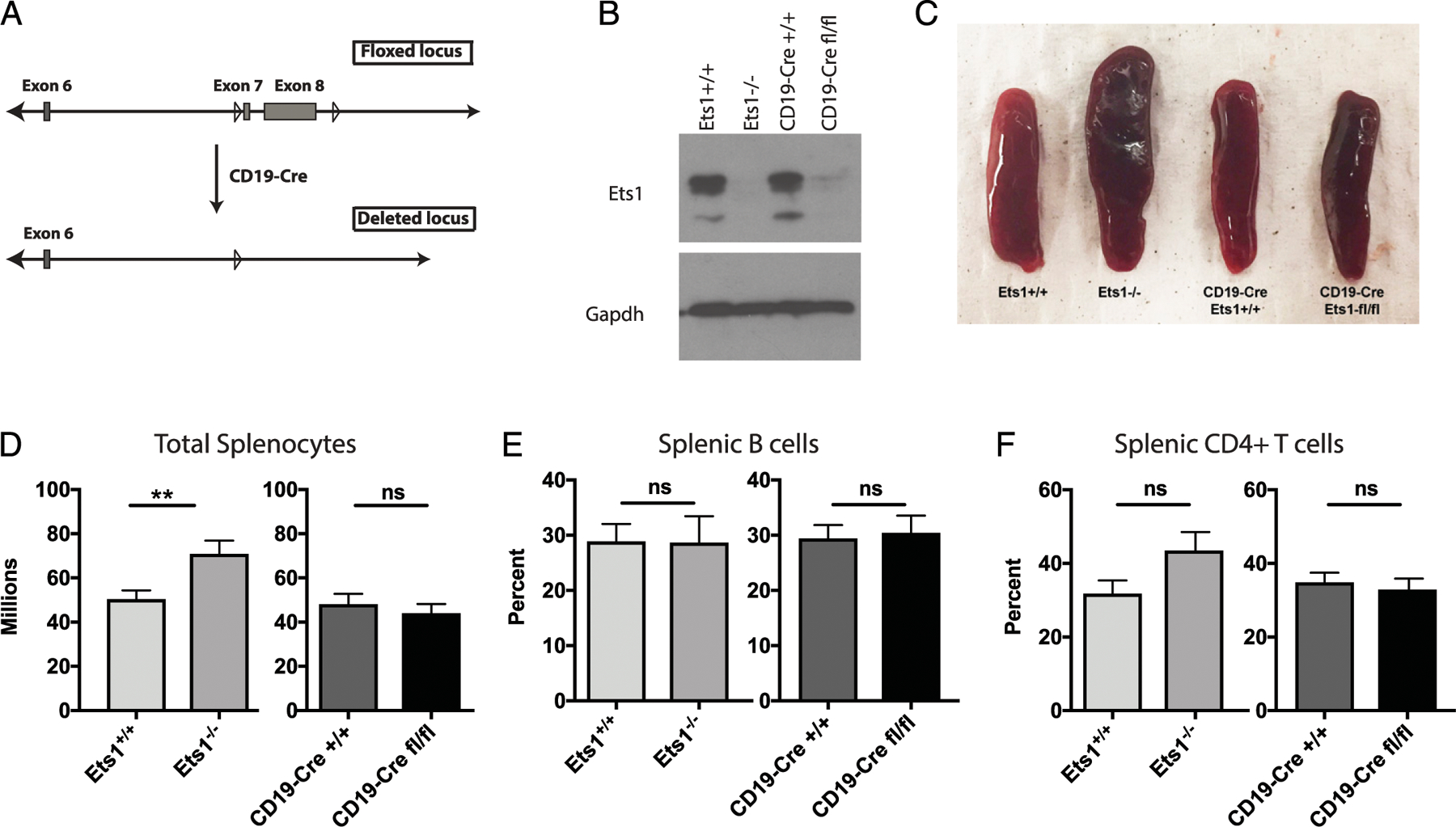
(A) Generation of B cell–specific Ets1 knockout mice. Diagram of the floxed allele (top) in which loxP sites flank the final two exons of the Ets1 gene (exons 7 and 8) and the deleted allele (bottom). (B) Western blot for Ets1 protein (top) using lysates from purified splenic B cells. GAPDH was used as a loading control (bottom). (C) Spleens from mice of the indicated genotypes showing that Ets1−/−, but not CD19-Cre Ets1fl/fl, mice have enlarged spleens. (D–F) Quantification of the number of total splenocytes (n = 20 Ets1+/+ and Ets1−/− and 12 CD19-Cre Ets1+/+ and CD19-Cre Ets1fl/fl mice), splenic B cells (n = 8 Ets1+/+, 6 Ets1−/−, 11 CD19-Cre Ets1+/+, and 12 CD19-Cre Ets1fl/fl mice), and splenic CD4+ T cells (n = 7 Ets1+/+, 5 Ets1−/−, 10 CD19-Cre Ets1+/+, and 8 CD19-Cre Ets1fl/fl mice) in mice of the indicated genotypes. **p < 0.01. ns, not significant.
Loss of Ets1 specifically in B cells results in aberrations in B cell differentiation
Conventional Ets1−/− mice have a loss of marginal zone type B cells and a higher than normal expression of CD23 on follicular B cells (Fig. 2A–C). CD19-Cre Ets1fl/fl mice also lack marginal zone B cells (Fig. 2A, 2B), whereas the levels of CD23 on follicular B cells from CD19-Cre Ets1fl/fl mice were not elevated (Fig. 2A, 2B). Like Ets1−/− mice, CD19-Cre Ets1fl/fl mice have an increase in B220lowCD138+ ASCs in the spleen, although the overall percentages and numbers of ASC were not as high in CD19-Cre Ets1fl/fl mice as in Ets1−/− mice. Furthermore, CD19-Cre Ets1fl/fl mice did not have an elevation of ASC in their lymph nodes (Fig. 2C, 2D), despite the fact that Ets1−/− mice do. Because not all ASCs express CD138 (28), ELISPOT was performed to enumerate both IgM- and IgG-secreting cells in the spleens and lymph nodes of CD19-Cre Ets1fl/fl and control mice. The number of IgM-secreting ASCs was elevated in the spleens and lymph nodes of CD19-Cre Ets1fl/fl mice, whereas IgG-secreting cells were only significantly elevated in the lymph nodes of CD19-Cre Ets1fl/fl mice (Supplemental Fig. 1).
FIGURE 2. Mice with a B cell–specific loss of Ets1 have reduced marginal zone B cells and increased ASCs.
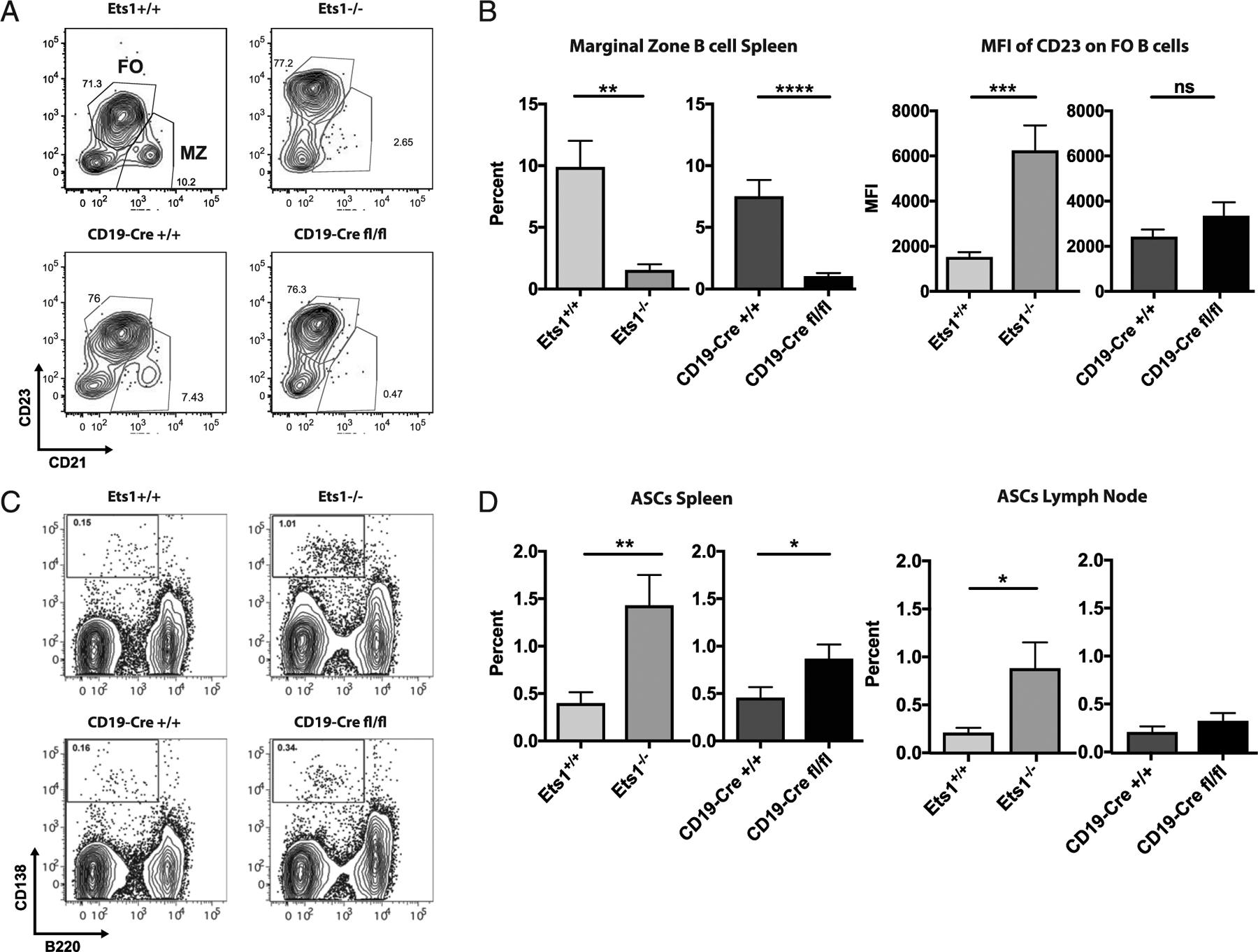
(A) Flow cytometry analysis of CD21 versus CD23 in gated live, B220+ B cells showing that Ets1−/− and CD19-Cre Ets1fl/fl mice lack CD21hiCD23lo marginal zone B cells. Gate positions were adjusted to enclose the major cell populations. (B) Quantification of the percent of marginal zone B cells and the mean fluorescence intensity (MFI) of CD23 on follicular B cells (n = 8 Ets1+/+, 6 Ets1−/−, 11 CD19-Cre Ets1+/+, and 8 CD19-Cre Ets1fl/fl mice). (C) Flow cytometry analysis of B220 versus CD138 in gated live cells showing that Ets1−/− and CD19-Cre Ets1fl/fl mice have increased percentages of B220loCD138+ Ab-secreting plasma cells. (D) Quantification of the percent of ASCs in the spleen and lymph nodes of the various strains of mice (n = 7 Ets1+/+, 6 Ets1−/−, 9 CD19-Cre Ets1+/+, and 11 CD19-Cre Ets1fl/fl mice). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
Follicular B cells in Ets1−/− mice have an activated phenotype with upregulation of CD80 and CD86. A significant fraction of the CD80+ B cells in Ets1−/− mice coexpress PDL2 and CD73 (Fig. 3A and data not shown) and hence have a memory B cell phenotype, as defined previously (29). CD19-Cre Ets1fl/fl mice also have an increase in B cells with a memory phenotype (CD80+PDL2+) in the spleen but not lymph nodes (Fig. 3A, 3B). In contrast, there was no increase of CD86 staining on B cells from CD19-Cre Ets1fl/fl mice (Supplemental Fig. 2A, 2B).
FIGURE 3. Mice with a B cell–specific deletion of Ets1 have increased memory phenotype B cells but no increase in switching to IgG1.
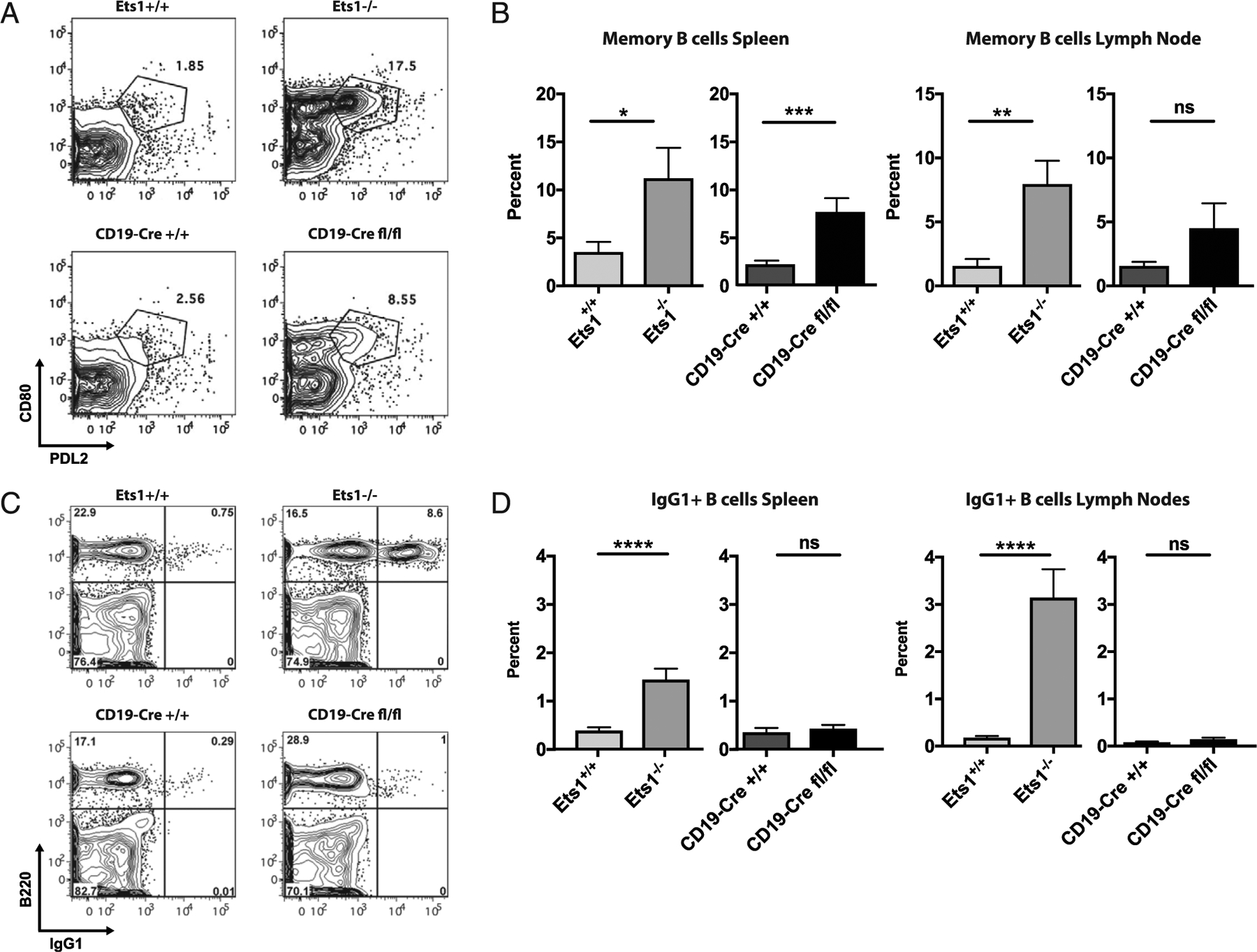
(A) Flow cytometry analysis of PDL2 versus CD80 in gated live B220+ B cells showing that Ets1−/− and CD19-Cre Ets1fl/fl mice have increased percentages of B cells with a memory phenotype (PDL2+CD80+). (B) Quantification of the percentages of memory B cells in spleen and lymph nodes of the various strains of mice (n = 6 Ets1+/+, 6 Ets1−/−, 9 CD19-Cre Ets1+/+, and 6 CD19-Cre Ets1fl/fl mice). (C) Flow cytometry analysis of B220 versus IgG1 in gated live splenocytes showing that Ets1−/− mice, but not CD19-Cre Ets1fl/fl mice, have increased percentages of IgG1+ B cells. (D) Quantification of the percentages of IgG1+ B cells in the spleen and lymph nodes of the various strains of mice (n = 21 Ets1+/+, 17 Ets1−/−, 11 CD19-Cre Ets1+/+, and 12 CD19-Cre Ets1fl/fl mice). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
A significant fraction of B cells in spleens and lymph nodes of conventional Ets1 knockout mice express surface IgG1 (Fig. 3C, 3D). However, very few B cells in CD19-Cre Ets1fl/fl mice switch to IgG1. Class-switching typically occurs in the germinal center, and conventional Ets1 knockout mice have increased percentages of B cells with a germinal center phenotype (B220+PNA+FAS+) in their lymph nodes but not spleens. CD19-Cre Ets1fl/fl mice do not show an increase in the numbers of GC B cells (Supplemental Fig. 2C, 2D). In summary, specific deletion of Ets1 in B cells alone results in the loss of the marginal zone B cell population and an increase in ASCs and memory B cells in the spleen but not lymph nodes. However, B cell–specific deletion of Ets1 is not sufficient to induce class-switchingtoIgG1ortheformation of germinal center B cells.
T cells do not show an activated phenotype in CD19-Cre Ets1fl/fl mice
Both CD4+ and CD8+ T cells from conventional Ets1−/− mice have an activated phenotype (9). B cells can act as APCs to T cells, resulting in T cell activation (30–34). Therefore, it is possible that spontaneous activation of B cells in CD19-Cre Ets1fl/fl mice could result in the secondary activation of the T cells. To test this possibility, markers of T cell activation were examined on cells isolated from CD19-Cre Ets1fl/fl mice. In CD19-Cre Ets1fl/fl mice, the majority of CD4+ T cells are of the naive phenotype, whereas CD4+ T cells are skewed toward an activated effector/memory phenotype in Ets1−/− mice (Fig. 4A, 4B). T cells from conventional Ets1−/− mice upregulate expression of CD86 on their surfaces (Fig. 4C, 4D) (9). CD86 expression is found on memory T cells after their interaction with dendritic cells (35–37), suggesting that the CD86+ T cells in Ets1−/− mice may represent memory T cells. CD19-Cre Ets1fl/fl mice do not upregulate CD86 on T cells, suggesting that they do not have an increase in memory phenotype T cells (Fig. 4C, 4D).
FIGURE 4. Mice with a B cell–specific deletion of Ets1 do not have increased CD4+ T cell activation.
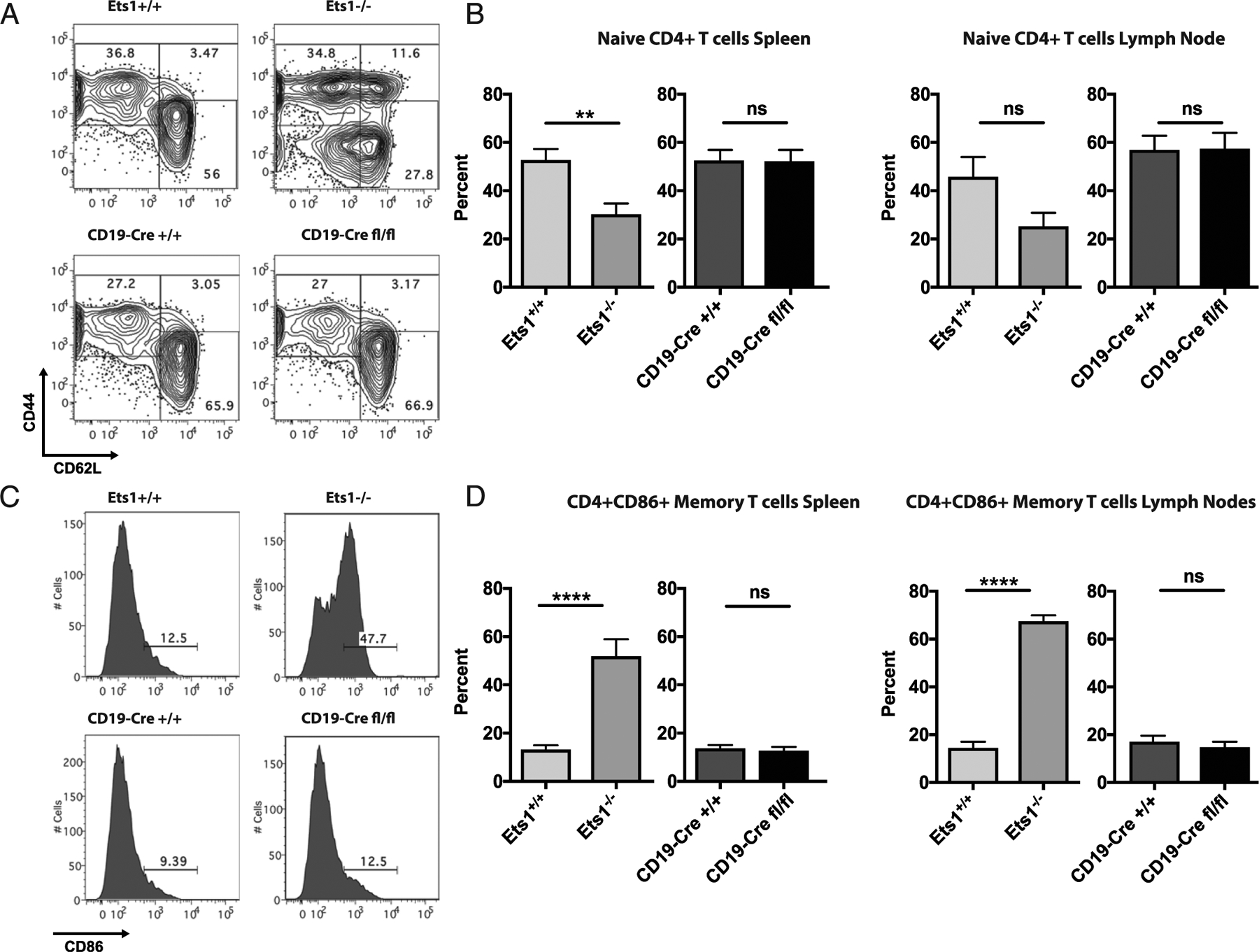
(A) Flow cytometry analysis of CD62L versus CD44 in gated live CD4+ T cells in spleen of the indicated mice. (B) Quantification of the percentages of naive phenotype CD4 T cells (CD62LhiCD44lo) in spleen and lymph nodes of the various strains of mice (n = 7 Ets1+/+, 5 Ets1−/−, 10 CD19-Cre Ets1+/+, and 8 CD19-Cre Ets1fl/fl mice). (C) Flow cytometry analysis of CD86 staining on CD4+ T cells of mice. (D) Quantification of the percent of CD4+ CD86+ T cells in gated live spleen and lymph node cells of the indicated mice (n = 7 Ets1+/+, 5 Ets1−/−, 10 CD19-Cre Ets1+/+, and 8 CD19-Cre Ets1fl/fl mice). **p < 0.01, ****p < 0.0001. ns, not significant.
Impaired Treg activity has also been implicated in driving the changes in B cells lacking Ets1 (8). A normal percentage of CD4+ T cells in spleens and lymph nodes of Ets1−/− mice expressed Foxp3 (Supplemental Fig. 3A–C), unlike what has previously been reported (8). In fact, the percentage of Foxp3+ cells within the CD4+ gate was slightly increased in Ets1−/− mice. However, the overall intensity of Foxp3 staining was reduced in Tregs from the lymph nodes of Ets1−/− mice (Supplemental Fig. 3D). Mice with a B cell–specific knockout of Ets1 showed similar percentages of Foxp3+ CD4+ T cells as wild-type mice and no decrease in the intensity of Foxp3 staining. Therefore, the B cell activation found in CD19-Cre Ets1fl/fl mice is not the result of nor does it lead to secondary activation of the T cell population or changes in the Treg population.
Loss of Ets1 specifically in B cells leads to formation of autoantibodies
As described above, there is increased B cell differentiation to ASCs in CD19-Cre Ets1fl/fl mice. This finding was correlated with a trend toward increased levels of serum IgM in CD19-Cre Ets1fl/fl mice, although this was not statistically significant (Table I). Serum IgG was not elevated in CD19-Cre Ets1fl/fl mice (Table I). IgM and IgG autoantibodies were also examined. As shown in Fig. 5A–D, Ets1−/− mice showed elevated levels of both IgM and IgG autoantibodies for dsDNA, histone proteins, Smith Ag, and MBP. In contrast, CD19-Cre Ets1fl/fl mice have elevated IgM and IgG autoantibodies against histones and MBP and elevated IgM autoantibodies against DNA. However, unlike Ets1−/− mice, CD19-Cre Ets1fl/fl mice lack autoantibodies against the RNA-associated Smith Ag.
TABLE I.
Serum IgM and IgG levels
| Ets1+/+ | Ets1−/− | CD19-Cre Ets1+/+ | CD19-Cre Ets1fl/fl | |
|---|---|---|---|---|
| Total serum IgM (μg/ml) | 347.5 ± 123 | 1947 ± 307*** | 356 ± 204 | 579 ± 102 |
| Total serum IgG (mg/ml) | 6.21 ± 1.60 | 10.19 ± 2.19 | 6.29 ± 2.24 | 4.21 ± 0.94 |
p < 0.001 (comparing Ets1−/− to Ets1+/+ for serum IgM; all other differences are not statistically significant).
FIGURE 5. Mice with a B cell–specific deletion of Ets1 produce IgM and IgG autoantibodies.
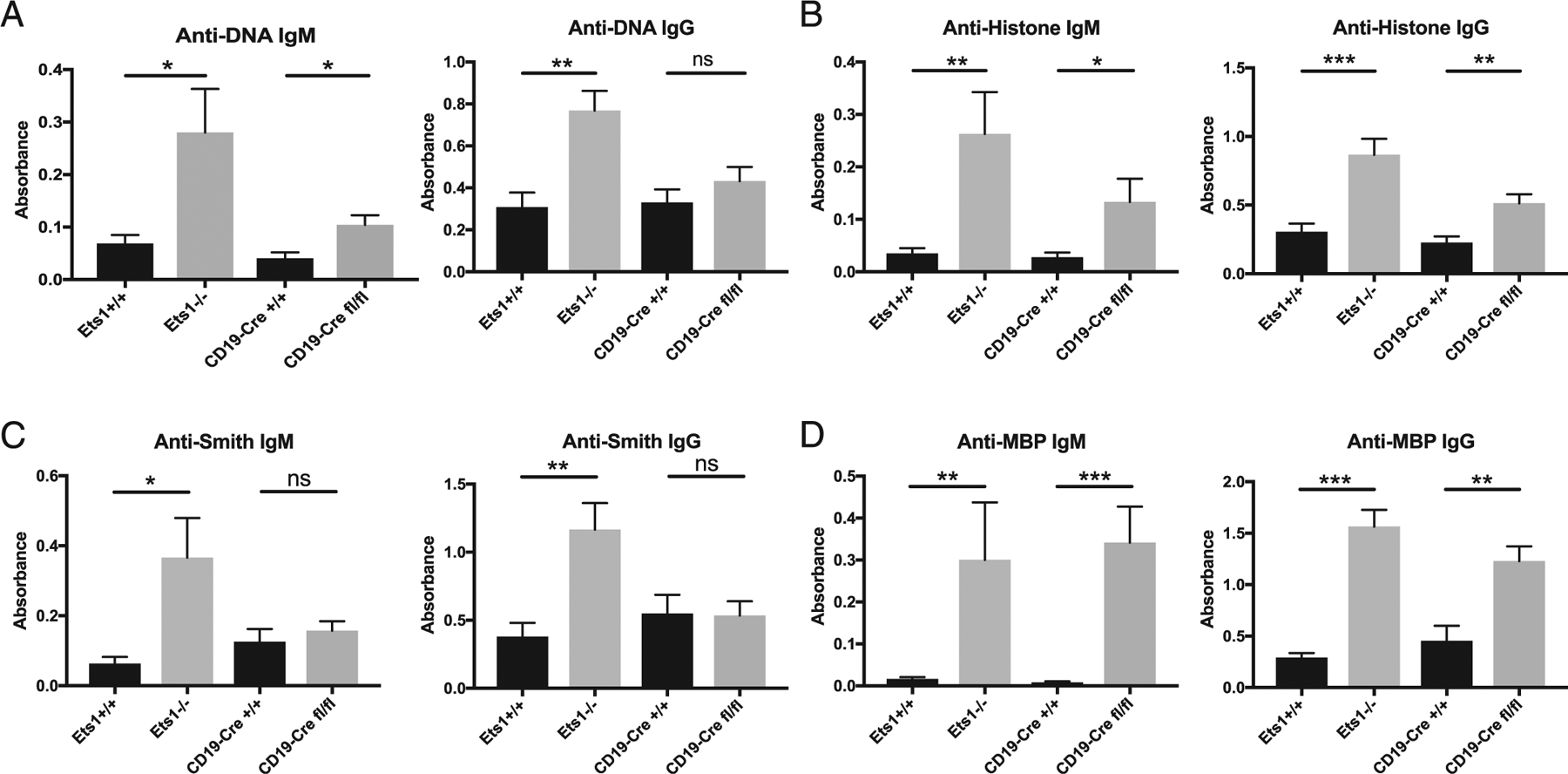
ELISA quantification of IgM and IgG autoantibodies against (A) dsDNA (n = 7 Ets1+/+, 7 Ets1−/−, 7 CD19-Cre Ets1+/+, and 9 CD19-Cre Ets1fl/fl mice), (B) mixed histone proteins (H2A, H2B, H3, and H4) (n = 8 Ets1+/+, 8 Ets1−/−, 7 CD19-Cre Ets1+/+, and 8 CD19-Cre Ets1fl/fl mice), (C) Smith Ag (n = 6 Ets1+/+, 6 Ets1−/−, 6 CD19-Cre Ets1+/+, and 6 CD19-Cre Ets1fl/fl mice), or (D) MBP (n = 7 Ets1+/+, 8 Ets1−/−, 7 CD19-Cre Ets1+/+, and 10 CD19-Cre Ets1fl/fl mice). *p < 0.05, **p < 0.01, ***p < 0.001. ns, not significant.
To further confirm the production of autoantibodies, immune complex deposition in the kidneys of mice was examined. As shown in Fig. 6A and 6B, kidneys fromEts1−/−mice have both IgM and IgG autoantibody deposits. In contrast, CD19-Cre Ets1fl/fl mice showed similar deposition of IgM immune complexes, whereas deposition of IgG immune complexes was comparatively weak.
FIGURE 6. CD19-Cre Ets1fl/fl mice have IgM immune complexes that deposit in the kidneys.
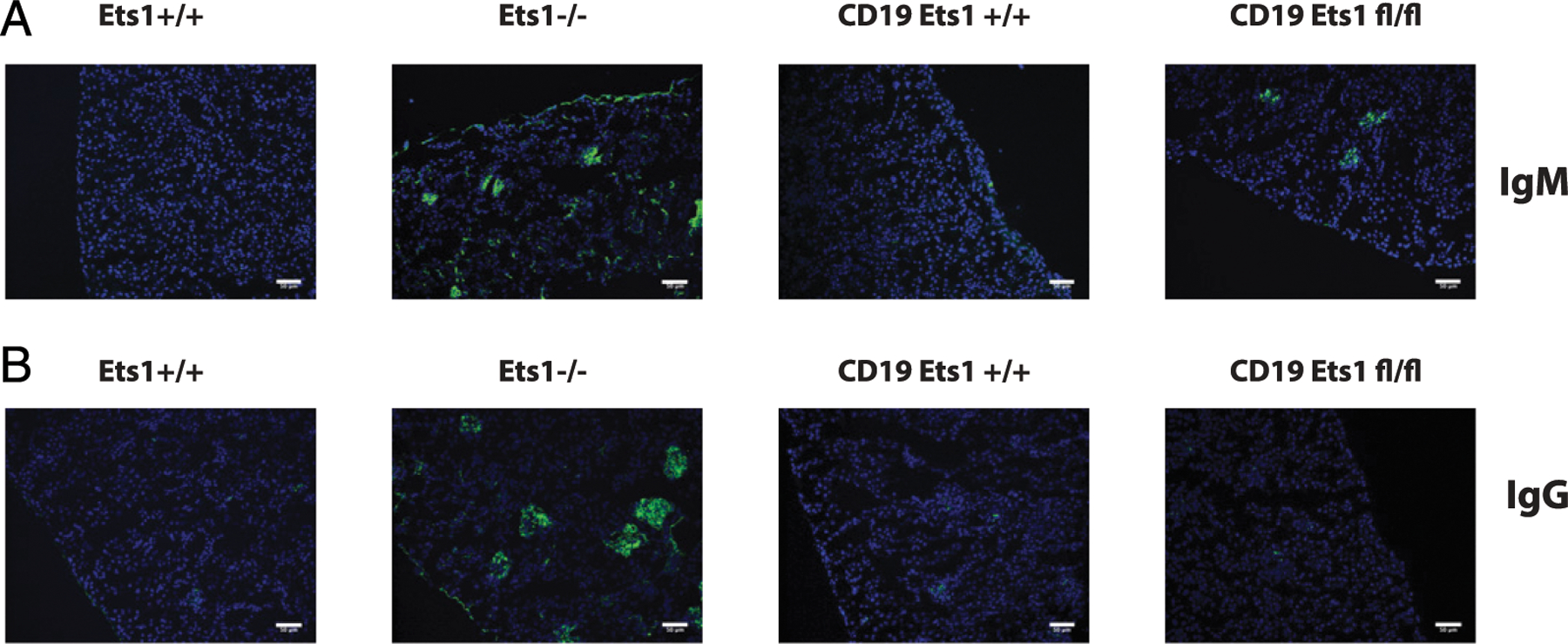
Immunostaining of kidney sections with FITC-labeled (green) anti-mouse IgM (A) or anti-mouse IgG (B). Nuclei are counterstained with DAPI (blue) (n = 4 mice of each genotype for each Ab isotype). All scale bars are 50 μm.
DISCUSSION
The Ets1 gene is a crucial regulator of immune cell functions and a susceptibility locus for numerous autoimmune and inflammatory conditions. Ets1 is expressed at high levels in both B and T lymphocytes and at lower levels in other cell types. Mice with a conventional deletion of the Ets1 gene, where Ets1 is absent in all cell types, develop an autoimmune syndrome similar to lupus (4, 8, 20), with increased activation of both B and T cells. A recent study has shown that deletion of Ets1 specifically in T cells results in severe autoimmune symptoms that mimic complete deletion of the gene (20). Ets1 was shown to suppress development of Tfh2 cells that secrete IL-4. The increases in Tfh2 cells drive B cell activation with development of germinal centers and increased isotype switching, ASC generation, and secretion of autoantibodies. In the same study, a B cell-specific knockout of Ets1 using CD19-Cre was also reported, although not extensively characterized (20). In fact, the only phenotype reported for the B cell deletion of Ets1 was increased serum IgM (20). In this study, deletion of Ets1 in B cells was shown to lead to numerous additional cell-intrinsic changes in the B cell compartment, including decreased percentages of marginal zone B cells, increased numbers of Ab-secreting plasma cells, increased numbers of B cells with a memory phenotype, and increased titers of autoantibodies. These changes are achieved without any apparent hyperactivation of the T cell compartment. Mice with a conventional deletion of the Ets1 gene, in which Ets1 is lacking in all tissues, show an increase in germinal center B cells and switching to IgG1. However, this phenotype is not found in mice with a B cell–specific deletion of Ets1. B cells from conventional Ets1 knockout mice also show upregulation of CD23 and CD86, which are not upregulated on B cells from CD19-Cre Ets1fl/fl mice. These phenotypes are likely driven by Ets1-deficient autoreactive T cells. Some parameters controlled by Ets1, such as the development of ASCs and memory B cells and the production of autoantibodies, have a B cell–intrinsic component but show a more significant deviation from normal in conventional Ets1 knockout mice than in B cell–specific Ets1 knockout mice. This suggests that Ets1 plays roles in both B and T cells to control these differentiation steps.
Ets1−/− mice produce IgM and IgG autoantibodies against a wide range of Ags including DNA, histones, Smith Ag, and MBP. CD19-Cre Ets1fl/fl mice produce only a subset of these autoantibodies. For instance, both IgM and IgG autoantibodies were detected against histone proteins and MBP, whereas only IgM autoantibodies were found that recognized dsDNA. In contrast, neither IgM nor IgG autoantibodies against the RNA-associated Smith Ag were found in CD19-Cre Ets1fl/fl mice. These observations hint that B cells may be activated in different ways in conventional Ets1−/− mice versus CD19-Cre Ets1fl/fl mice. One possibility is that autoantibodies that develop in CD19-Cre Ets1fl/fl mice may be derived from T cell–independent activation of B cells, whereas T cell–dependent responses may predominate in Ets1−/− mice. Costimulation of B cells via nucleic acid–specific TLRs (e.g., TLR9) may stimulate autoantibody production against DNA or histone proteins. However, if TLR signaling is involved in autoantibody production in CD19-Cre Ets1fl/fl mice, it seems not to be sufficient to costimulate autoantibody production against the RNA-associated Smith Ag, despite the fact that this autoantigen signals via TLR7 (38, 39). An alternate T cell–independent pathway might involve T-independent type II activation of B cells. This may be the case for Ags present in repetitive structures, such as the polymerized nucleotides in the DNA, the histone subunits in nucleosomes of chromatin, or the network of repetitive subunits on the cytoplasmic face of the myelin membrane formed by MBP. Death of cells may lead to exposure of these repetitive elements, which could potentially extensively cross-link the BCR of Ag-specific B cells and activate them in the absence of T cell help.
It is also possible that T cells could provide help to B cells from CD19-Cre Ets1fl/fl mice, leading to their activation and ASC differentiation. Although the bulk population of T cells did not show an activated phenotype and Tregs were present in normal numbers, it remains possible that a small population of Ag-specific T cells become activated and coordinate B cell responses. If so, they most likely drive extrafollicular B cell responses, given that few germinal center B cells were found in CD19-Cre Ets1fl/fl mice. The lack of germinal center B cells in CD19-Cre Ets1fl/fl mice is somewhat at odds with increased formation of memory B cells (CD80+PDL2+), which are typically thought to be derived from germinal centers. However, T cell–independent extrafollicular B cell responses can also promote memory B cell formation (40, 41).
Previously, Ets1−/− mice were reported to harbor reduced numbers of Foxp3+ Tregs (8). In this study, no reduction in the numbers of Foxp3+ Tregs in spleen or lymph nodes of either conventional Ets1 knockout mice or B cell–specific Ets1 knockout mice was observed. However, CD4+ T cells from the lymph nodes of conventional Ets1 knockout mice expressed low levels of Foxp3. This is consistent with previous studies, which demonstrate that Ets1 can directly regulate the expression of the Foxp3 gene itself (8, 42). The lower than normal levels of Foxp3 in Tregs of conventional Ets1 knockout mice may contribute to the immune aberrations in those animals. However, because Foxp3 expression is normal in CD19-Cre Ets1fl/fl mice, this cannot explain B cell phenotypes in these mice.
In humans, single nucleotide polymorphisms in the ETS1 gene locus have been associated with multiple autoimmune and inflammatory diseases including lupus, rheumatoid arthritis, psoriasis, and atopic dermatitis (43). A number of studies have also shown low levels of Ets1 mRNA in PBMCs of patients with autoimmune disease (12, 13, 16, 44). Given the B cell–intrinsic roles for Ets1 described in this article, low expression of Ets1 in human B cells could result in increased differentiation and Ab secretion. Low Ets1 levels have also been described in purified T cells from autoimmune patients (45, 46) and could also promote disease pathogenesis. Therefore, genetic changes that reduce Ets1 expression in B cells or T cells or both may contribute to the development of an autoimmune response.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Satrajit Sinha at the University of Buffalo for helpful discussions and Kirsten Smalley for help with maintaining the mouse colony.
This work was supported by grants from the Lupus Research Alliance and the National Institute of Allergy and Infectious Diseases (R01 AI122720) as well as a National Cancer Institute Core Center grant to Roswell Park Cancer Institute (P30CA016056).
Abbreviations used in this article:
- ASC
Ab-secreting cell
- MBP
myelin basic protein
- Tfh2
T follicular helper type 2
- Treg
regulatory T cell
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
REFERENCES
- 1.Hayter SM, and Cook MC. 2012. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun. Rev 11: 754–765. [DOI] [PubMed] [Google Scholar]
- 2.Cooper GS, Bynum ML, and Somers EC. 2009. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun 33: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrett-Sinha LA 2013. Review of Ets1 structure, function, and roles in immunity. Cell. Mol. Life Sci. 70: 3375–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, John SA, Clements JL, Percy DH, Barton KP, and Garrett-Sinha LA. 2005. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int. Immunol 17: 1179–1191. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen HV, Mouly E, Chemin K, Luinaud R, Despres R, Fermand JP, Arnulf B, and Bories JC. 2012. The Ets-1 transcription factor is required for Stat1-mediated T-bet expression and IgG2a class switching in mouse B cells. Blood 119: 4174–4181. [DOI] [PubMed] [Google Scholar]
- 6.Eyquem S, Chemin K, Fasseu M, and Bories JC. 2004. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA 101: 15712–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyquem S, Chemin K, Fasseu M, Chopin M, Sigaux F, Cumano A, and Bories JC. 2004. The development of early and mature B cells is impaired in mice deficient for the Ets-1 transcription factor. Eur. J. Immunol 34: 3187–3196. [DOI] [PubMed] [Google Scholar]
- 8.Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-defranoux O, Bandeira A, and Bories JC. 2010. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J. Exp. Med 207: 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements JL, John SA, and Garrett-Sinha LA. 2006. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J. Immunol 177: 905–912. [DOI] [PubMed] [Google Scholar]
- 10.Grenningloh R, Kang BY, and Ho IC. 2005. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med 201: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, et al. 2009. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet 41: 1234–1237. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, et al. ; Asian Lupus Genetics Consortium. 2010. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 6: e1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan S, Dang J, Li J, Yang Z, Zhao H, Xin Q, Ma X, Liu Y, Bian X, Gong Y, and Liu Q. 2014. ETS1 variants confer susceptibility to ankylosing spondylitis in Han Chinese. Arthritis Res. Ther 16: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudenberg J, Lee HS, Han BG, Shin HD, Kang YM, Sung YK, Shim SC, Choi CB, Lee AT, Gregersen PK, and Bae SC. 2011. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 63: 884–893. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki A, Yamashita K, Hirano F, Sada KE, Tsukui D, Kondo Y, Kimura Y, Asako K, Kobayashi S, Yamada H, et al. 2018. Association of ETS1 polymorphism with granulomatosis with polyangiitis and proteinase 3-anti-neutrophil cytoplasmic antibody positive vasculitis in a Japanese population. J. Hum. Genet 63: 55–62. [DOI] [PubMed] [Google Scholar]
- 16.Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, Xiang Q, Zhou Y, Kijlstra A, and Yang P. 2014. MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS One 9: e91199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al. ; Wellcome Trust Case Control Consortium 2. 2012. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet 44: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lill CM, Luessi F, Alcina A, Sokolova EA, Ugidos N, de la Hera B, Guillot-Noël L, Malhotra S, Reinthaler E, Schjeide BM, et al. 2015. Genome-wide significant association with seven novel multiple sclerosis risk loci. J. Med. Genet 52: 848–855. [DOI] [PubMed] [Google Scholar]
- 19.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, and Leiden JM. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity 9: 555–563. [DOI] [PubMed] [Google Scholar]
- 20.Kim CJ, Lee CG, Jung JY, Ghosh A, Hasan SN, Hwang SM, Kang H, Lee C, Kim GC, Rudra D, et al. 2018. The transcription factor Ets1 suppresses T follicular helper type 2 cell differentiation to halt the onset of systemic lupus erythematosus. [Published erratum appears in 2019 Immunity 50: 272.] Immunity 49: 1034–1048.e8. [DOI] [PubMed] [Google Scholar]
- 21.Moisan J, Grenningloh R, Bettelli E, Oukka M, and Ho IC. 2007. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med 204: 2825–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muthusamy N, Barton K, and Leiden JM. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377: 639–642. [DOI] [PubMed] [Google Scholar]
- 23.Luo W, Mayeux J, Gutierrez T, Russell L, Getahun A, Müller J, Tedder T, Parnes J, Rickert R, Nitschke L, et al. 2014. A balance between B cell receptor and inhibitory receptor signaling controls plasma cell differentiation by maintaining optimal Ets1 levels. J. Immunol 193: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John S, Russell L, Chin SS, Luo W, Oshima R, and Garrett-Sinha LA. 2014. Transcription factor Ets1, but not the closely related factor Ets2, inhibits antibody-secreting cell differentiation. Mol. Cell. Biol 34: 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.John SA, Clements JL, Russell LM, and Garrett-Sinha LA. 2008. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J. Biol. Chem 283: 951–962. [DOI] [PubMed] [Google Scholar]
- 26.Saelee P, Kearly A, Nutt SL, and Garrett-Sinha LA. 2017. Genome-wide identification of target genes for the key B cell transcription factor Ets1. Front. Immunol 8: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zook EC, Ramirez K, Guo X, van der Voort G, Sigvardsson M, Svensson EC, Fu YX, and Kee BL. 2016. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J. Exp. Med 213: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, and Nutt SL. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med 200: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, and Shlomchik MJ. 2014. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol 15: 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan OT, Hannum LG, Haberman AM, Madaio MP, and Shlomchik MJ. 1999. A novel mouse with B cells but lacking serum antibody reveals an antibody-independent role for B cells in murine lupus. J. Exp. Med 189: 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Neill SK, Shlomchik MJ, Glant TT, Cao Y, Doodes PD, and Finnegan A. 2005. Antigen-specific B cells are required as APCs and autoantibody-producing cells for induction of severe autoimmune arthritis. J. Immunol 174: 3781–3788. [DOI] [PubMed] [Google Scholar]
- 32.Pierson ER, Stromnes IM, and Goverman JM. 2014. B cells promote induction of experimental autoimmune encephalomyelitis by facilitating reactivation of T cells in the central nervous system. J. Immunol 192: 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serreze DV, Fleming SA, Chapman HD, Richard SD, Leiter EH, and Tisch RM. 1998. B lymphocytes are critical antigen-presenting cells for the initiation of T cell-mediated autoimmune diabetes in nonobese diabetic mice. J. Immunol 161: 3912–3918. [PubMed] [Google Scholar]
- 34.Wong FS, Wen L, Tang M, Ramanathan M, Visintin I, Daugherty J, Hannum LG, Janeway CA Jr., and Shlomchik MJ. 2004. Investigation of the role of B-cells in type 1 diabetes in the NOD mouse. Diabetes 53: 2581–2587. [DOI] [PubMed] [Google Scholar]
- 35.Ferlazzo G, Semino C, Meta M, Procopio F, Morandi B, and Melioli G. 2002. T lymphocytes express B7 family molecules following interaction with dendritic cells and acquire bystander costimulatory properties. Eur. J. Immunol 32: 3092–3101. [DOI] [PubMed] [Google Scholar]
- 36.Hakamada-Taguchi R, Kato T, Ushijima H, Murakami M, Uede T, and Nariuchi H. 1998. Expression and co-stimulatory function of B7–2 on murine CD4+ T cells. Eur. J. Immunol 28: 865–873. [DOI] [PubMed] [Google Scholar]
- 37.Jeannin P, Herbault N, Delneste Y, Magistrelli G, Lecoanet-Henchoz S, Caron G, Aubry JP, and Bonnefoy JY. 1999. Human effector memory T cells express CD86: a functional role in naive T cell priming. J. Immunol 162: 2044–2048. [PubMed] [Google Scholar]
- 38.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, and Shlomchik MJ. 2006. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25: 417–428. [DOI] [PubMed] [Google Scholar]
- 39.Pawar RD, Ramanjaneyulu A, Kulkarni OP, Lech M, Segerer S, and Anders HJ. 2007. Inhibition of toll-like receptor-7 (TLR-7) or TLR-7 plus TLR-9 attenuates glomerulonephritis and lung injury in experimental lupus. J. Am. Soc. Nephrol 18: 1721–1731. [DOI] [PubMed] [Google Scholar]
- 40.Toyama H, Okada S, Hatano M, Takahashi Y, Takeda N, Ichii H, Takemori T, Kuroda Y, and Tokuhisa T. 2002. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity 17: 329–339. [DOI] [PubMed] [Google Scholar]
- 41.Taylor JJ, Pape KA, and Jenkins MK. 2012. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med 209: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polansky JK, Schreiber L, Thelemann C, Ludwig L, Krüger M, Baumgrass R, Cording S, Floess S, Hamann A, and Huehn J. 2010. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J. Mol. Med. (Berl.) 88: 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrett-Sinha LA, Kearly A, and Satterthwaite AB. 2016. The role of the transcription factor Ets1 in lupus and other autoimmune diseases. Crit. Rev. Immunol 36: 485–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Sun LD, Lu WS, Hu WL, Gao JP, Cheng YL, Yu ZY, Yao S, He CF, Liu JL, et al. 2010. Expression analysis of ETS1 gene in peripheral blood mononuclear cells with systemic lupus erythematosus by real-time reverse transcription PCR. Chin. Med. J. (Engl.) 123: 2287–2288. [PubMed] [Google Scholar]
- 45.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, and Pei G. 2009. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol 10: 1252–1259. [DOI] [PubMed] [Google Scholar]
- 46.Xiang N, Li XP, Li XM, Wang GS, Tao JH, Pan HF, Fang X, Ma Q, and Yu N. 2014. Expression of Ets-1 and FOXP3 mRNA in CD4(+)CD25 (+) T regulatory cells from patients with systemic lupus erythematosus. Clin. Exp. Med 14: 375–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


