Abstract
Proposals to update the harmonised monitoring and reporting of antimicrobial resistance (AMR) from a public health perspective in Salmonella, Campylobacter coli, Campylobacter jejuni, Escherichia coli, Enterococcus faecalis, Enterococcus faecium and methicillin‐resistant Staphylococcus aureus (MRSA) from food‐producing animals and derived meat in the EU are presented in this report, accounting for recent trends in AMR, data collection needs and new scientific developments. Phenotypic monitoring of AMR in bacterial isolates, using microdilution methods for testing susceptibility and interpreting resistance using epidemiological cut‐off values is reinforced, including further characterisation of those isolates of E. coli and Salmonella showing resistance to extended‐spectrum cephalosporins and carbapenems, as well as the specific monitoring of ESBL/AmpC/carbapenemase‐producing E. coli. Combinations of bacterial species, food‐producing animals and meat, as well as antimicrobial panels have been reviewed and adapted, where deemed necessary. Considering differing sample sizes, numerical simulations have been performed to evaluate the related statistical power available for assessing occurrence and temporal trends in resistance, with a predetermined accuracy, to support the choice of harmonised sample size. Randomised sampling procedures, based on a generic proportionate stratified sampling process, have been reviewed and reinforced. Proposals to improve the harmonisation of monitoring of prevalence, genetic diversity and AMR in MRSA are presented. It is suggested to complement routine monitoring with specific cross‐sectional surveys on MRSA in pigs and on AMR in bacteria from seafood and the environment. Whole genome sequencing (WGS) of isolates obtained from the specific monitoring of ESBL/AmpC/carbapenemase‐producing E. coli is strongly advocated to be implemented, on a voluntary basis, over the validity period of the next legislation, with possible mandatory implementation by the end of the period; the gene sequences encoding for ESBL/AmpC/carbapenemases being reported to EFSA. Harmonised protocols for WGS analysis/interpretation and external quality assurance programmes are planned to be provided by the EU‐Reference Laboratory on AMR.
Keywords: antimicrobial resistance monitoring, Salmonella, Campylobacter, E. coli, MRSA, food‐producing animals, food
Summary
Provisions for the monitoring of antimicrobial resistance (AMR) in zoonotic and indicator bacteria in food‐producing animals and derived meat are laid down in Directive 2003/99/EC. Also foreseen is the possibility of broadening the scope of the AMR monitoring to other zoonotic agents in so far as they may present a threat to public health. Commission Implementing Decision 2013/652/EC, implementing Directive 2003/99/EC, lays down detailed and harmonised rules for the monitoring and reporting of AMR, which are applicable from 2014 until the end of 2020. This legislative framework has been partially derived from technical specification documents issued by the European Food Safety Authority (EFSA) in 2012, providing guidance on the harmonised monitoring of AMR in Salmonella, Campylobacter, indicator Escherichia coli and enterococci, as well as on the monitoring of prevalence, genetic diversity and AMR of methicillin‐resistant Staphylococcus aureus (MRSA) in several food‐producing animal categories and derived meat. In 2014, the EFSA also issued detailed functional specifications on randomised sampling for harmonised monitoring of AMR. The implementation of the legislation/these specifications by the European Union (EU) Member States (MSs) has led to greater harmonisation and better comparability of data on AMR; however, it seems inevitable that further enhancements and specific adaptations will be required on an ongoing basis to respond effectively to the constantly evolving threat of AMR.
The EFSA received a mandate from the European Commission to review and update the technical specifications issued in 2012 and 2014 and notably, specifically address in these updates the possible use of molecular typing methods, in the light of the latest scientific opinions on AMR, technological developments, recent trends in AMR and relevance for public health, as well as the audits assessing the implementation of the Decision performed by the European Commission in a number of MSs. This report includes proposals for implementing updated guidelines for further harmonised monitoring of AMR in food‐producing animals and food and for ensuring continuity in following up further trends in AMR (a factor which underpins the revision of existing guidelines).
The evidence from the European Union Summary Reports on AMR and the audit reports has shown that legislation has mostly been implemented by the MSs and has increasingly resulted in the production of comparable and reliable phenotypic AMR data over time. This is particularly true for the monitoring of trends and occurrence of resistance in indicator E. coli, which has become of particular relevance, as Salmonella prevalence has become increasingly low, thanks to the success of the control measures in place in the EU MSs.
In reviewing combinations of bacteria/animal/food as candidates for mandatory monitoring, it is proposed to reinforce the approach of prioritising potential consumers’ exposure by targeting zoonotic Salmonella spp. and Campylobacter jejuni and Campylobacter coli, as well as indicator commensal E. coli from the major domestically produced animal populations. These populations include laying hens, broilers, turkeys, fattening pigs and bovine animals under 1 year of age, as well as meat derived from these sources. One of the major aims is the collection of AMR data that can be investigated in combination with data on exposure to antimicrobials, such as data on domestic consumption of antimicrobials in food‐producing animal populations. Although monitoring performed on a yearly basis would allow earlier detection of trends in AMR, in any direction, than monitoring at greater intervals, it is proposed to retain and reinforce the current monitoring performed on a rotating basis, targeting fattening pigs and bovine animals of less than one year, and poultry, separately, every second year. Thus, the potential benefits of an increased frequency of monitoring were reviewed considering competing priorities, as well as the need to get a balanced output from each of the most important sectors.
In addition to the routine monitoring performed on a biennial basis, the undertaking of complementary baseline cross‐sectional surveys in order to assess specifically the situation on certain AMR issues, such as MRSA, AMR in bacteria from sea food and AMR in bacteria from the environment, over the period of validity of the upcoming Commission Implementing Decision in 2021 onwards is suggested. It is envisaged that the detailed harmonised protocols of those specific baseline surveys would be designed at a later stage, considering the most recent data, once a clear agreement to carry out such studies had been reached.
Within the context of the implementation of the EU and MSs’ action plans against the threat of AMR, a further decrease in use of antimicrobials in food‐producing animals is expected to occur in conjunction with implementing complementary mitigation measures in the coming years, resulting in a decrease in the selective pressure on the emergence and/or occurrence of AMR. The approach and the results of the sample size analyses and calculation in the previous EFSA technical specifications were reviewed. The minimum target number of organisms of each bacterial species which should be examined is currently 170 from each type of domestic animal production type. The sample size should be adapted in the case of low Salmonella prevalence and very small production sectors. In order to ensure sufficient statistical power so that even slight decreases in AMR can be detected, it is also recommended that sample size is reviewed by each MS taking into account their own situation and objectives of reduction of AMR in the light of the simulations presented in this report, which also account for the assessment of the occurrence of resistance with sufficient accuracy. It is acknowledged that this approach may lead to an increase in the number of samples to be collected, and that this may require additional resource from the MSs. In designing a sampling scheme, therefore, special efforts have been made to, where possible, exploit samples already collected under existing AMR monitoring programmes, such as caecal samples gathered at the slaughterhouse.
A key principle considered in relation to sampling design is to reinforce harmonised functional procedures for randomised sampling of animal and meat samples at different stages of the food chain, yielding representative and comparable data. Both collection strategies, a prospective and a retrospective sampling plan, of samples and isolates, respectively, are retained. The former involves collecting sufficient numbers of representative animal and chilled meat samples from which recovered isolates are tested for susceptibility; the latter involves selecting randomly Salmonella isolates from collections constituted within the framework of the national control programmes of Salmonella in poultry flocks. A generic proportionate stratified sampling process is proposed for the different sampling plans and numerical illustrations of proportional allocation are also recalled.
Stratified sampling of Salmonella isolates recovered from broiler, laying hen and fattening turkey primary production, and available in the collection of the laboratories involved in the Salmonella national control programmes, with proportional allocation to the size of the collection of isolates recovered from the production, is proposed. An alternative approach is to perform a simple random sampling within the sampling frame of flocks positive for Salmonella in those MS where a database records flocks tested positive for Salmonella. One Salmonella isolate per serovar and epidemiological unit should be retained for susceptibility testing. If more than 170 Salmonella isolates fitting the epidemiological criteria are available, 170 Salmonella isolates selected at random should be tested for antimicrobial susceptibility; if less than 170 Salmonella isolates are available, all available isolates should be tested for antimicrobial susceptibility (without any proportional allocation).
Stratified sampling of caecal content samples (single or pooled) in the slaughterhouses, accounting for at least 60% of the domestic production of the food‐producing animal populations monitored, with proportionate allocation to the slaughterhouse production, allows for the collection of representative isolates of Salmonella, Campylobacter, indicator E. coli and the assessment of the prevalence of ESBL‐/AmpC‐/carbapenemase‐producing E. coli from the populations of broilers, fattening turkeys, fattening pigs and bovine animals of less than 1 year of age, domestically produced. Definitions of ‘domestically produced’ animals are proposed for the sake of harmonisation.
Sampling of different chilled fresh meat categories is targeted at retail outlets serving the final consumer, with proportional allocation of the number of samples to the population of the geographical region (NUTS‐3 area) accounting for at least 80% of the national population, to test for the presence of ESBL‐/AmpC‐/carbapenemase‐producing E. coli.
The epidemiological units are flocks of poultry, slaughter batches of fattening pigs and bovine animals under 1 year, and lot of meat.
As regards the laboratory methodologies, it is confirmed that broth microdilution is the preferred method and that European Committee on Antimicrobial Susceptibility Testing (EUCAST, http://www.eucast.org/) epidemiological cut‐off values should be used as interpretative criteria to define microbiological resistance. The concentration ranges to be used should ensure that both the epidemiological cut‐off value and the clinical breakpoint are included so that comparability of results with human data is made possible. As regards the harmonised set of antimicrobial substances to be used for phenotypic susceptibility testing, it is noted that the panels currently included in the legislation have been used across the MSs. The limited revisions and/or additions that have been proposed should enable to both account for recent trends in AMR and continue following up further temporal trends for the sake of continuity. Also, although in principle, the optimal concentration range should be tested for each substance, for some substances this has been reduced to a minimum range so that wells can be freed on the harmonised plate to allow for inclusion of novel substances.
In particular, it is proposed to complement the current (first) harmonised panel of antimicrobials for Salmonella and E. coli with amikacin to improve the detection of 16S rRNA methyltransferase enzymes that confer resistance to all aminoglycosides except streptomycin. These methyltransferases have been increasingly found in association with carbapenemases, AmpC or ESBL enzymes and fluoroquinolone resistance in Enterobacteriaceae, especially outside Europe. In order to accommodate the additional substance, it is suggested to slightly modify the harmonised panel by reducing some of the dilution ranges, in particular those for ampicillin, nalidixic acid, tetracycline, gentamicin, trimethoprim, sulfamethoxazole and chloramphenicol in the upper end of the scales, as the susceptible end is considered more important to define.
No changes were deemed necessary to the recommended antimicrobial panel currently used to test further those Salmonella and E. coli isolates that exhibit resistance to a third‐generation cephalosporins and/or carbapenems so as to perform the monitoring of extended‐spectrum β‐lactamase‐/AmpC β‐lactamase‐/carbapenemase‐producing bacteria, whether deriving from either the routine monitoring of AMR in Salmonella and indicator E. coli or the specific monitoring of ESBL/AmpC/carbapenemase producers.
As regards Campylobacter, it is proposed to slightly alter the harmonised panel with the removal of nalidixic acid, streptomycin and the lowest concentration of gentamicin so as to allow for the inclusion of additional higher concentrations of erythromycin (for better detection of isolates with high‐level erythromycin resistance which may be presumptively considered to harbour the erm(B) gene) as well as higher concentrations of ciprofloxacin and a phenicol molecule, thereby allowing for the presumptive deduction of genotypes with altered sequence of the CmeABC efflux pump and its regulating region. It is also proposed to include a carbapenem antimicrobial. In addition, in order to improve the comparability of Campylobacter prevalence and AMR data between MSs, it is highly desirable that the EU monitoring be based on harmonised methods for both isolation and antimicrobial susceptibility testing. A harmonised protocol based on the European standard EN ISO 10272‐1, detection procedure C, should be provided for the purpose. It is proposed that the EURL‐Campylobacter perform in 2019–2020 the requested pilots to finalise the setting up of the harmonised protocol. A better knowledge of the prevalence of Campylobacter spp., C. jejuni and C. coli in animal production and food of animal origin in the different MSs and of the prevalence of resistance in these two species will help understanding the epidemiology and especially the potential sources of human C. jejuni and C. coli infections, and the differences in proportions of C. jejuni and C. coli in human cases between European countries. It is also proposed, based on recent source attribution studies, to target cattle in the Campylobacter AMR monitoring. This is also likely to aid development of improved measures for control of this zoonotic agent.
Considering the advantages inherent in the whole genome sequencing (WGS) technology but also its current limitations, as well as the expected evolution of the present situation, it is proposed to follow a gradual, phased approach to integration of WGS within the harmonised AMR monitoring. The integration process could be initiated by complementing the harmonised phenotypic monitoring with WGS on a voluntary basis in the early phase of the period 2021–2026 and at the end of the period envisage the replacement of the standard routine phenotypic antimicrobial susceptibility testing with the systematic use of WGS. The period 2021–2026 should therefore be seen as a transitory period for the implementation of WGS, expected to be a reasonable transition period for the MSs to gain experience and acquire WGS technology.
The proposed flexible approach corresponds to allowing MSs/National Reference Laboratories (NRLs) to use WGS on a voluntary basis for detection of ESBL/AmpC/carbapenemase‐producing E. coli replacing panels 1 and 2 of the phenotypic antimicrobial susceptibility testing method in the monitoring for these organisms. The voluntary replacement of the phenotypic antimicrobial susceptibility testing method for detection of ESBL‐/AmpC/carbapenemase‐producing E. coli is proposed to begin in 2021.
The switch to WGS for commensal E. coli, Campylobacter spp., Salmonella spp. and enterococci should optimally take place in a co‐ordinated way for all MSs at one predetermined point in time and when the necessary developments are in place to allow comparison of the historical (phenotypic) and genotypic results. The phased introduction of WGS for the specific monitoring of ESBL/AmpC/carbapenemase‐producing E. coli would mean that in the course of the upcoming Commission Implementing Decision's validity period, for example by 2025, all MSs should have WGS in place. A road map should be set up to establish methods for comparison of outputs, whether simple (e.g. occurrence of resistance) or complex (e.g. complete susceptibility), from phenotypic data (including historical data) and WGS data that will need to have been developed and implemented at the point of the switch from phenotypic to genotypic monitoring, as well as harmonised procedures for handling and reporting WGS data.
To achieve the goal of implementing WGS across the food and veterinary sectors of the NRLs during the upcoming Commission Implementing Decision's validity period, as well as the generalised use of WGS in the specific monitoring by the end of the period. The performance by the MSs of the WGS part of the ‘Confirmatory Testing’ should become mandatory by the end of the period. Again WGS can be used on a voluntary basis at the start of the period, with the support of the EURL AR.
To implement the use of WGS, it is pivotal for reasons of comparability that all data are processed using the same/harmonised approach. It is proposed that the EURL‐AR continue the effort to provide and organise training in DNA extraction, library preparation and sequencing. Moreover, the EURL‐AR should recommend harmonised standard operating procedure/protocols/guidelines including quality criteria and external quality assessment procedures, overseen by the EURL‐AR, should be developed in 2019–2020. The aim should be to use the same version and curated reference database based on benchmarking efforts, and similar parameters/tools for all the steps of the analysis (trimming, quality, assembly, AMR characterisation). It is also proposed that, by the implementation of WGS, the EURL‐AR develops and establishes a genomic‐based proficiency test to assess the quality of the genomes produced by the NRLs and the reliability of the detection of AMR determinants/genes. MSs will be encouraged to submit voluntarily their sequences, if not already publicly available, to the planned Joint ECDC‐EFSA database for WGS data in order to perform joint analysis across sectors (human, animal and food).
It is proposed to reinforce the monitoring of MRSA in food‐producing animals and food. In particular, the concept of a baseline survey on MRSA in pigs to be performed either on the farm or at the slaughterhouse over the period of validity of the next Decision is put forward. The harmonised panel for testing the antimicrobial susceptibility of MRSA has been reviewed. Some of the antimicrobials included in the panel are critically important in human medicine for the treatment of MRSA and other Gram‐positive bacterial infections in humans, including vancomycin and linezolid. Resistance to vancomycin or linezolid should lead to investigation of the mechanism of resistance, including WGS, to determine whether any of those genes known to confer resistance are present. A number of alterations to the concentration ranges are proposed for gentamicin, trimethoprim, kanamycin, fusidic acid, penicillin, vancomycin and rifampicin; the antimicrobials included in the panel remain unchanged. Characterisation of MRSA isolates is also recommended by genotypic analysis (WGS) to determine strains and lineages as well as to investigate the presence of important virulence and host‐adaptation factors and those specific genetic markers (e.g. phages) associated with certain animal hosts.
It is also suggested that routine monitoring is complemented with specific baseline (cross‐sectional) studies (in addition to that on MRSA in pigs) on AMR in bacteria from seafood and the environment, to be carried out over the validity period of the next legislation. The intention is to propose detailed protocols for those baseline studies later on, once a clear agreement on the conduct of those baseline studies has been reached.
As regards the best format for the reporting of the data, the recommendations are the same as those from the technical specifications relating to the collection and reporting of data at isolate‐based level which have been previously published.
The genes encoding ESBL/AmpC/carbapenemases detected by WGS should be reported to EFSA. Protocols and interpretation of WGS analysis need to be harmonised and supported by external quality assurance programmes.
In addition, it is emphasised that if complementary AMR monitoring in additional animal populations and food categories is carried out, specific representative sampling plans should be devised and results should be reported separately.
Finally, it is proposed that the technical specifications be re‐assessed and updated regularly in the light of the results of the first monitoring campaigns, the most recent literature and the constantly evolving situation.
1. Introduction
The Commission adopted in June 2017 a new European One Health action plan against Antimicrobial Resistance (AMR)1 which provides a framework for continued, more extensive action to reduce the emergence and spread of AMR and to increase the development and availability of new effective antimicrobials inside and outside the EU. Under this new One Health action plan, in order to strengthen One Health surveillance and reporting of AMR and antimicrobial use, the Commission is committed to review EU implementing legislation on monitoring AMR in zoonotic and commensal bacteria in farm animals and food, to take into account new scientific developments and data collection needs.
In accordance with Article 31 of regulation (EC) No 178/2002, the Commission has requested scientific and technical assistance to the European Food Safety Authority (EFSA) in view of reviewing Decision 2013/652/EU. The European Union reference laboratory for antimicrobial resistance (EURL‐AR) in Copenhagen and the European Centre for Disease Prevention and Control (ECDC) have been consulted on this matter as appropriate.
1.1. Background and Terms of Reference as provided by the EC
1.1.1. Background
Combating AMR is a priority for the European Commission (EC). Surveillance of AMR and antimicrobial consumption is essential to have comprehensive and reliable information on the development and spread of drug‐resistant bacteria, to measure the impact of measures taken to reduce AMR and to monitor progress. Such data provide insights to inform decision‐making and facilitate the development of appropriate strategies and actions to manage AMR at European, national and regional levels.
As part of the new European One Health action plan against Antimicrobial Resistance (AMR)2 adopted in June 2017, the Commission is committed to review EU implementing legislation on the harmonised monitoring of AMR in zoonotic and commensal bacteria in food‐producing animals and food, to take into account new scientific developments, including the recent evolutions of epidemiological situations in the Member States, and data collection needs.
Under Directive 2003/99/EC3 on the monitoring of zoonoses and zoonotic agents, Member States must provide comparable monitoring data on the occurrence of AMR in zoonotic agents and, in so far as they present a threat to public health, other agents.
Between 2008 and 2011 EFSA adopted several scientific opinions4, a joint scientific opinion together with the ECDC, the EMA and the Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR)5 and a technical report6 which concluded that in view of the increasing public health concern regarding AMR, the use of harmonised methods and epidemiological cut‐off values is necessary to ensure the comparability of data over time at Member State level, and also to facilitate the comparison of the occurrence of AMR between Member States.
In 2012 the EFSA published two additional scientific reports on the technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator commensal Escherichia coli and Enterococcus spp. bacteria transmitted through food7 and the technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus (MRSA) in food‐producing animals and food.8
Having regard to the findings of these scientific reports, the European Commission adopted in 2013 Decision 2013/652/EU9 on the monitoring and reporting on antimicrobial resistance in zoonotic and commensal bacteria as part of the 2011–2016 EC action plan against the rising threats from AMR.10 This Decision, which applies from 2014 to 2020, lays down detailed rules for the harmonised monitoring and reporting of AMR to be carried out by Member States in accordance with Article 7(3) and 9(1) of Directive 2003/99/EC and Annex II (B) and Annex IV thereto.
Since its publication, the EFSA issued a scientific report on detailed functional specifications on randomised sampling for harmonised monitoring of antimicrobial resistance in zoonotic and commensal bacteria.11 The European Commission has also conducted a series of audits on the implementation of Decision 2013/652/EU. An interim overview report on these series was published in July 2017.12 It highlights the main key implementation barriers faced by Member States, in particular achieving the minimum required number of samples/isolates:
In the case of Salmonella, isolates are obtained either from the Salmonella National Control Programs (SNCPs) or from the application of Regulation (EC) No 2073/2005 on microbiological criteria for foodstuffs. In case of low prevalence all Salmonella isolates available shall be tested. This includes those isolates obtained by food business operators (FBOs) but competent authorities rarely manage to avail these isolates.
In the case of Campylobacter jejuni, in some Member States two phenomena have been observed since Decision 2013/652/EU entered into force, namely, the low prevalence of C. jejuni and the higher prevalence of Campylobacter coli in certain poultry sectors.
In certain circumstances, and due to the structural particularities of some production sectors, the definition of the epidemiological unit, in particular for fattening pigs and bovines under 1 year of age, has also been a limiting factor to collect isolates in certain Member States.
Due to a combination of these factors Member States with a low production, in particular small Member States, have communicated problems to achieve the minimum number of isolates.
1.1.2. Terms of Reference as provided by the EC
In accordance with Article 31 of Regulation (EC) No 178/2002, the Commission therefore requests scientific and technical assistance from the European Food Safety Authority (EFSA) in view of updating:
the 2012 EFSA technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator commensal Escherichia coli and Enterococcus spp. bacteria transmitted through food. In doing so, the EFSA should also provide advice on how to scientifically address the problems identified during the audits in Member States as specified in the background.
the 2012 EFSA technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus (MRSA) in food‐producing animals and food.
the 2014 EFSA technical specifications on randomised sampling for harmonised monitoring of antimicrobial resistance in zoonotic and commensal bacteria.
The EFSA should address in these updates the possible use of molecular typing methods (e.g. Whole Genome Sequencing) to complement and/or replace the phenotypic methods currently used to assess antimicrobial resistance, described in Decision 2013/652/EU, taking into account the necessity to ensure the comparability of the results with the non‐molecular techniques and the possibility to integrate the molecular data with past and future phenotypical data. If molecular techniques are proposed as alternative techniques to the phenotypic methods, EFSA should provide technical specifications on how to ensure comparability of techniques.
The scientific and technical assistance should take into account recent scientific opinions on AMR, technological developments, recent trends in AMR and relevance for public health. It should consider the comparable AMR monitoring data reported by the Member States to EFSA in the period following adoption of Decision 2013/652/EU, as well as the outcomes of audits assessing implementation of the Decision which provide an overview of the main key implementation barriers faced by Member States. It should also take into account the Joint scientific opinion on a list of outcome indicators as regards surveillance of antimicrobial resistance and antimicrobial consumption in humans and food‐producing animals,13 as it establishes a set of indicators for Member States to assess their progress in reducing the consumption of antimicrobials and antimicrobial resistance in both humans and food‐producing animals. Finally, it should also ensure that proposed developments, where possible, enhance the Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) performed by EFSA, EMA and ECDC.
2. Brief outline of the current harmonised monitoring system of AMR
Directive 2003/99/EC14 on the monitoring of zoonoses and zoonotic agents set out generic requirements for the monitoring and reporting of AMR in isolates of zoonotic Salmonella spp. and Campylobacter spp., as well as in selected other bacteria – in so far as they present a threat to public health – from food‐producing animals and food in the EU Member States (MSs). Within the framework of AMR monitoring in food‐producing animals and food, the occurrence of AMR is typically defined as the proportion of bacterial isolates tested for a given antimicrobial and found to present any degree of acquired reduced phenotypic susceptibility when compared to the fully susceptible wild‐type population, i.e. to display ‘microbiological resistance’. Epidemiological cut‐off values (ECOFFs)15 are used as interpretative criteria of microbiological resistance.16
2.1. Description of the data collected by the system
The AMR monitoring in food‐producing animals and food was revised by Commission Decision 2013/652/EU implementing Directive 2003/99/EC, which set out monitoring priorities from a public health perspective and described those combinations of bacterial species, antimicrobial substances, food‐producing animal populations and food products which should be monitored from 2014 onwards, including the frequency with which monitoring should be performed.
2.1.1. Bacterial species, food‐producing animals and food products
Since the implementation of the Commission Decision, the monitoring of AMR in zoonotic Salmonella spp. and C. jejuni, as well as in indicator E. coli from the major food‐producing animal populations domestically produced has become mandatory. Indicator E. coli and Campylobacter spp. isolates derive from active monitoring programmes, based on representative random sampling of carcasses of healthy animals sampled at the slaughterhouse to collect caecal samples. For Salmonella spp. from broilers, laying hens and fattening turkeys, isolates are included which originate from Salmonella national control plans, as well as isolates from carcases of broilers and fattening turkeys, sampled as part of process hygiene criteria. For Salmonella spp. from fattening pigs and bovine animals under 1 year of age, isolates are included originating from carcases of these animals, sampled as part of process hygiene criteria. The target number of organisms of each bacterial species which should be examined is 170 from each type of domestic animal production type (this is reduced to 85 organisms from poultry and pigs, if production is less than 100,000 tonnes per annum). From 2014 onwards, poultry/poultry meat will be monitored in 2014, 2016, 2018 and 2020, and pigs and bovines under 1 year, pork and beef in 2015, 2017 and 2019. Within each MS, the various types of livestock and meat from those livestock should be monitored when production exceeds 10,000 tonnes slaughtered per year (Table 1).
Table 1.
Organisms included in AMR monitoring in 2014 and subsequent years, as set out in Commission Implementing Decision 2013/652/EU
| Animal populations/ Meat | Salmonella spp. | C. jejuni | C. coli | Indicator/ ESBL‐producing E. coli | CP‐producing E. coli | E. faecalis/ E. faecium |
|---|---|---|---|---|---|---|
| Broilers | M: NCP, PHC | M: CSS | V | M: CSS | V | V |
| Laying hens | M: NCP | – | – | – | – | – |
| Fattening turkeys | M: NCP, PHC | M: CSS | V | M: CSS | V | V |
| Bovines, < 1 year old | M: PHC | – | – | M: CSS | V | V |
| Fattening pigs | M: PHC | – | – | M: CSS | V | V |
| Broiler meat | – | – | – | M: R | V | – |
| Pig meat | – | – | – | M: R | V | – |
| Bovine meat | – | – | – | M: R | V | – |
CP: carbapenemase; CSS: caecal samples from healthy animals at slaughter; M: mandatory monitoring; NCP: Salmonella national control plans; PHC: surveillance of process hygiene criteria; R: at retail; V: voluntary monitoring.
2.1.2. Panels of antimicrobial substances
The antimicrobial substances included in the monitoring from 2014 onwards are shown in Table 2. The panel of antimicrobials tested includes those of particular public health relevance as well as those of epidemiological relevance; ECOFFs were used as the interpretative criteria of resistance (Kahlmeter et al., 2003). The harmonised panel of antimicrobials used, particularly for Salmonella spp. and E. coli, was broadened with the inclusion of substances, such as colistin and ceftazidime, that are either important for human health or provide clearer insight into the probable mechanisms of resistance to extended‐spectrum cephalosporins.
Table 2.
Harmonised set of antimicrobial substances used for the monitoring of resistance in zoonotic Salmonella spp. and Campylobacter spp., and indicator Escherichia coli and enterococci isolates from food‐producing animals and food over the period 2014–2020a
| Substances | Salmonella | C. coli/C. jejuni | Indicator E. coli | Enterococci |
|---|---|---|---|---|
| Ampicillin | ● | ● | ● | |
| Azithromycin | ● | ● | ||
| Cefepime | x | x | ||
| Cefotaxime | ● | ● | ||
| Cefotaxime + clavulanic acid | x | x | ||
| Ceftazidime | ● | ● | ||
| Ceftazidime + clavulanic acid | x | x | ||
| Chloramphenicol | ● | ● | ● | |
| Ciprofloxacin | ● | ● | ● | |
| Colistin | ● | ● | ||
| Daptomycin | ● | |||
| Ertapenem | x | x | ||
| Erythromycin | ● | ● | ||
| Gentamicin | ● | ● | ● | ● |
| Imipenem | x | x | ||
| Linezolid | ● | |||
| Meropenem | ● | ● | ||
| Nalidixic acid | ● | ● | ● | |
| Quinupristin/Dalfopristin | ● | |||
| Streptomycin | ● | ● | ||
| Sulfonamides | ● | ● | ||
| Teicoplanin | ● | |||
| Temocillin | x | x | ||
| Tetracycline | ● | ● | ● | ● |
| Tigecycline | ● | ● | ● | |
| Trimethoprim | ● | ● | ||
| Vancomycin | ● |
●: all isolates; x: only for isolates resistant to cefotaxime, ceftazidime and/or meropenem.
Commission Decision 2013/652/EU17.
2.1.3. Specific monitoring of ESBL‐/AmpC‐ and/or carbapenemase‐producing E. coli
Commission Implementing Decision 2013/652/EU stipulates caecal samples from broilers, fattening turkeys, fattening pigs and bovines under 1 year of age, as well as from broiler meat, pork and beef collected at retail should be examined for E. coli using selective media incorporating the third‐generation cephalosporin cefotaxime. This medium is selective for E. coli resistant to third‐generation cephalosporins and is expected to allow the growth of extended‐spectrum β‐lactamases (ESBL), AmpC β‐lactamases (AmpC) or carbapenemase enzyme producers, which are resistant to cefotaxime at the microbiological cut‐off. Three hundred caecal samples should be examined from each type of livestock and meat.
In practice, all presumptive ESBL‐ or AmpC‐ or carbapenemase‐producing E. coli isolates identified through the selective plating, as well as all those randomly selected isolates of Salmonella spp. and E. coli, recovered from non‐selective media, that are resistant to cefotaxime or ceftazidime or meropenem, are further tested with a second panel of antimicrobial substances (Table 2). This second panel of antimicrobials includes cefotaxime and ceftazidime with and without clavulanic acid (to investigate whether synergy is observed with clavulanate), as well as the antimicrobials cefoxitin, cefepime, temocillin, ertapenem, imipenem and meropenem. The second panel of antimicrobials is designed to enable phenotypic characterisation of ESBL‐, AmpC‐ and carbapenemase producers.
Moreover, Commission Implementing Decision 2013/652/EU also foresees the monitoring of carbapenemase‐producing microorganisms using selective medium with a carbapenem, on a voluntary basis. A number of the MSs has performed this specific monitoring focusing on the detection of carbapenemase‐producing E. coli.
2.2. Strength of the system
The monitoring of AMR in food‐producing animals under Commission Implementing Decision 2013/652/EU covers the main food‐producing animal species and where appropriate, includes different production sectors (for example, broilers and laying hens). Randomised, representative sampling is no longer stratified at the level of the different animal species (e.g. Gallus gallus, cattle, pigs) but rather is performed at the level of the major food‐producing animal production categories, such as broilers, laying hens, fattening pigs, fattening turkeys and bovines under 1 year of age.
The effects of consumption of antimicrobials in a given country and animal species, on the occurrence of resistance, can be studied more easily in indicator organisms than in food‐borne pathogens, such as Salmonella spp., because all food‐producing animals generally carry these indicator bacteria. Monitoring resistance in indicator commensal E. coli has become mandatory. Sampling for the collection of indicator bacteria should be representative of the domestically produced population studied, in accordance with the provisions of the Decision and the corresponding detailed technical specifications issued by EFSA (2012b, 2014). The isolates subjected to susceptibility testing have typically been derived from active monitoring programmes in healthy animals, ensuring representativeness of resistance data, especially in the case of indicator bacteria and Campylobacter spp. AMR data from susceptibility testing of Salmonella spp. have remained more dependent on the prevalence and the serovar distribution of the bacteria in the different animal populations.
Commission Implementing Decision 2013/652/EU also ensures that all reporting countries submit data for a common core set of antimicrobials and bacteria set out in Table 2; data for these combinations should therefore be comprehensive. The collection and reporting of data are now performed at the isolate level, which enables in‐depth analyses to be conducted, in particular on the occurrence of multidrug resistance. The analysis of the results at individual isolate level also allows investigation of possible associations between the occurrence of isolates which are fully susceptible to the panel of antimicrobials tested and the consumption of antimicrobials.
An external quality assurance system, based on regular training and yearly proficiency tests, is included in the AMR monitoring programmes. This will detect potential differences between the laboratories performing susceptibility tests relating to methods or interpretative criteria and is coordinated by the National Reference Laboratories on AMR (NRL‐AR) within each reporting country and the EU Reference Laboratory on Antimicrobial Resistance (EURL‐AR).
2.3. Impediments of the system
The European Commission has carried out audits in, up to now, 14 MSs to check the actual implementation of the AMR monitoring programmes laid down by the current EU legislation. The system ensures that there is harmonisation of resistance monitoring in food‐producing animals and comparability of the AMR data recorded in the respective EU MSs.
MSs came across some challenges when implementing Commission Implementing Decision 2013/652/EU in some aspects of the monitoring including planning and sampling, sample processing, laboratory testing and reporting to EFSA. It is worth recalling that those issues have been improved since the audits were conducted. Those challenges were taken into account when drafting the current technical report.
3. Recent developments and the evolution of the AMR situation
The introduction of Commission Implementing Decision 2013/652/EU planning the implementation of revised panels of antimicrobials to be tested and specific monitoring has enabled to enlarge the scope of the AMR monitoring, and to enhance the reliability of the results. Nevertheless, the continually evolving threat from emerging resistance underlines the need to review the data collected, interpret the findings and assess trends in a constant manner, taking also into account recent developments in the methodology. A number of AMR issues have notably emerged since 2013, as summarised below and detailed further in Appendix A. Similarly, the epidemiological situation regarding Salmonella and Campylobacter has evolved over the last years and the revision of the harmonised monitoring of AMR should also account for such evolution. Alert points (e.g. emerging resistances, mechanisms, clones, etc.) might be defined triggering further investigation of individual isolates by molecular methods.
3.1. Salmonella enterica subsp. enterica
Over the last decade, prevalence in targeted serovars of Salmonella in poultry has substantially decreased in most of the MSs. In certain MSs, the decrease may have exacerbated further the lack of a sufficient number of Salmonella isolates from poultry (i.e. broilers, laying hens, fattening turkeys) to be tested for AMR. As AMR in Salmonella continues to develop and new resistance issues have emerged in the population, continuous monitoring of AMR in Salmonella is advisable, although the limited number of isolates does not allow a full statistical trend analysis of the level of resistance. In contrast, antimicrobial susceptibility testing (AST) in Salmonella should predominantly seek to detect and follow new emerging issues including sporadic occurrence of clones, such as multi‐drug resistant (MDR) S. Kentucky with a high resistance to fluoroquinolones and S. Infantis isolates exhibiting combined resistance to highest priority critically important antimicrobials (HPCIAs), such as extended‐spectrum cephalosporins, fluoroquinolones and colistin (Appendix A). To this end, Salmonella obtained from slaughter animals have proved to be of major importance for the assessment of strains entering the food chain.
3.2. Campylobacter spp.
The high level of harmonisation of the method used for susceptibility testing, whether considering the sets of antimicrobials and dilution ranges to be tested, as well as the interpretative criteria of resistance (EUCAST ECOFFs) and the representative sampling design, enables comparison between levels of resistance and resistance profiles observed in bacteria among the MSs. External quality assurance for susceptibility testing is also provided to the NRLs by the EURL‐AR. Still, in the context of the rather recent introduction of the European standard EN ISO 10272‐1, slight differences between the Campylobacter isolation methods used in the MSs may occur and affect the recovery of Campylobacter spp. from samples, the proportions of C. jejuni or C. coli obtained and even the diversity of the isolates recovered, including the antimicrobial susceptibility profiles.
3.3. Indicator commensal E. coli
Under the current version of the implementing legislation, monitoring of AMR in indicator commensal E. coli is the data backbone of the EU‐wide monitoring. This is well justified as commensal E. coli have been shown to mirror exposure of population to antimicrobial selection pressure. Its prevalence in the intestines of production animals is usually > 90%, and target numbers of 170 (or 85) isolates were easily achieved. Therefore, this bacterial population can provide continuous evidence on trends, including possible reduction of AMR when antimicrobial consumption (AMC) has decreased over the last years, as observed in certain MSs.
3.4. Enterococci
In the Commission Implementing Decision of 12 November 2013 (2013/652/EU) on the monitoring and reporting of AMR in zoonotic and commensal bacteria, Enterococcus faecium and Enterococcus faecalis have been included since 2014 in the panel of bacterial species under survey on a voluntary basis. Annually, only a limited number of MSs submit AMR data to EFSA relating to E. faecium and E. faecalis. Furthermore, the AMR data submitted are sparse, sporadic and not well geographically represented, thus, the data is difficult to act and conclude upon. The current data available show, however, also that the AMR trend curve seems quite consistent with minor changes over years (DANMAP 2017). In addition, the level of AMR in enterococci is less predictive of the level of AMR in other Gram‐positive bacteria relevant for public health like staphylococci, where clonal spread of particular strains can be significant. For antimicrobials, whose spectrum of activity mainly includes Gram‐positive organisms (e.g. the macrolides), then mandatory monitoring of the susceptibility of E. faecalis and E. faecium at 4 year rotational basis, will allow investigation of associations between AMC and AMR to be undertaken. The enterococci fulfil a useful and unique function among the organisms which are monitored by representing a common or frequent Gram‐positive indicator organism which is not subject to the pressures from targeted control measures in the way that Campylobacter might be (a Gram‐negative zoonotic organism, though susceptible to macrolides). Monitoring AMR in enterococci as indicator bacteria representing Gram‐positive organisms will complement the data from E. coli is for Gram‐negative bacteria (Enterobacteriaceae) though data might be collected on a less frequent basis, reflecting the lower priority.
3.5. Specific monitorings of ESBL‐/AmpC‐ and/or carbapenemase‐ producing E. coli
Selective isolation of ESBL/AmpC and/or carbapenemase‐producing E. coli has demonstrated that it provides interesting complementary information to the monitoring of indicator commensal E. coli. The prevalence of ESBL/AmpC‐producing E. coli would be largely underestimated if monitored only by the testing of random isolates of indicator E. coli alone. Harmonised methods for the detection of these bacteria have proven to be robust and regular PT trials performed by the EURL‐AR have shown that the NRL‐ARs are capable of detecting this kind of bacteria with sufficient accuracy. The wealth of identified strains of ESBL/AmpC‐producing E. coli have supported further analyses in research projects that helped understand potential spill over from animal populations and food to humans, especially to exposed people working in animal husbandry (Dorado‐Garcıa et al., 2018). Moreover, within this monitoring, a few strains of E. coli co‐producing carbapenemases have been also identified (EFSA and ECDC, 2017a, 2018a, 2019). The voluntary detection of carbapenemase‐producing microorganism from food‐producing animals and/or foods using specific selection with carbapenems, has also allowed the detection of a few isolates belonging to different Enterobacteriaceae (E. coli and Enterobacter spp.). The upcoming results of the ongoing IMPART research project‐topic 218 relating to selective isolation and detection of carbapenemase‐producing Enterobacteriaceae will be taken into account by the EURL‐AR while reviewing the current harmonised method recommended.
3.6. MRSA
Monitoring of MRSA prevalence in animals and food is currently performed on a voluntary basis by a number of MSs, many of whom report results to EFSA for inclusion in the yearly EU Summary Report on AMR. Some MSs also perform molecular typing and susceptibility testing of MRSA isolates, again on a voluntary basis (EFSA, 2012b). The EURL‐AR has published an updated protocol for the isolation of MRSA from food‐producing animals and the farm environment (http://www.eurl.eu).
A range of types of MRSA have been detected in animals (including the farm environment) and food, including healthcare‐associated MRSA, community‐associated MRSA, livestock‐associated MRSA and mecC MRSA, providing a strong rationale for further characterisation and typing of MRSA isolates from animal sources. Most isolates from food‐producing animals are livestock‐associated MRSA.
Those MSs performing AST and providing the results to EFSA have generally used a broth microdilution susceptibility testing method and have also tested very similar or identical panels of antimicrobials, as proposed by EFSA in 2012 (EFSA, 2012b).
The EURL‐AR organises external quality assurance assays for susceptibility testing of MRSA. Since monitoring of MRSA is carried out on a voluntary basis, there has been some minor variation in some of the procedures adopted by the MSs; sampling programmes in particular are subject to national variation in accordance with national priorities.
3.7. Klebsiella pneumoniae
Klebsiella pneumoniae is often multidrug resistant and appears to develop resistance easily (Wyres and Holt, 2018). It occurs in the environment, for example in surface waters, and on the mucous membranes and in the intestinal contents of humans and animals (Podschun and Ullman, 1998). It is therefore of potential interest as an AMR indicator and has high Public Health relevance since MDR clones may cause clinical infections and nosocomial outbreaks. Nevertheless, K. pneumoniae prevalence in samples is variable, and very high number of samples might need to be collected to obtain enough isolates. In the best case, K. pneumoniae could add as extra species for AMR monitoring of a (potential) pathogenic bacteria with an unknown zoonotic potential (if any). It is believed that, ideally, the prevalence/variability of prevalence of K. pneumoniae in food‐producing animals could be monitored. Despite this, as this bacterial species might not offer more as bacterial indicator of AMR than indicator E. coli does already, adding K. pneumoniae will not really improve the harmonised monitoring system of healthy animals at slaughter and may only provide fragmented information which may prove to be hard to interpret. Instead, at this stage, the monitoring of K. pneumoniae should be preferentially considered as part of a targeted monitoring of AMR in bacterial animal pathogens.
3.8. Epidemiological context
Within the context of the implementation of the EU and MSs’ action plans against the threat of AMR, a further decrease in use of antimicrobials in food‐producing animals is expected to occur in conjunction with implementing complementary mitigation measures in the coming years, resulting in a decrease in the selective pressure on the emergence and/or occurrence of AMR. Sampling design, and in particular sample size, should be therefore reviewed accounting for this expected epidemiological context in order to ensure that sufficient statistical power is available so that even slight decreases in AMR can be detected.
4. Views and feedback from NRL‐AR and EFSA Networks
To address further certain specific aspects of the AMR monitoring in the EU MSs/reporting countries, a digital Specific Questionnaire on Harmonised AMR Monitoring in 2017 and/or 2018 was drafted by the EFSA WG and the EURL‐AR using the tool ‘EU‐survey’ (Appendix B). In particular, the questionnaire was designed to better assess the vision of the MSs and to gauge the potential for further harmonisation of procedures and the degree of support from MSs for further monitoring. The online survey was performed in June 2018 and targeted the network of the NRL‐ARs and the EFSA Network on zoonoses/AMR monitoring. The questionnaire covered five aspects, specifically:
I: Methodologies applied to isolate Campylobacter spp. for antimicrobial susceptibility testing,
II: Monitoring of colistin resistance using selective media,
III: Characterisation of the ESBL/AmpC/carbapenemase producers and related gene identification,
IV: Monitoring of MRSA in animals and food, and
V: First and second antimicrobial panels for susceptibility testing of Salmonella and E. coli.
In total, 27 MSs and 4 non‐MSs answered the questionnaire. The main findings of the Campylobacter survey (I) and the MRSA survey (IV) are summarised below, whereas the outcomes of the survey regarding the other aspects are presented in Appendices Appendix E – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: Monitoring of specific colistin resistance, Appendix F – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: Further characterisation of ESBL/AmpC/carbapenemase producers and corresponding genes identification, Appendix G – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: First and second panels for susceptibility testing of Salmonella and E. coli.
4.1. Campylobacter survey
To explore the potential for further harmonisation of procedures relating to the isolation process of Campylobacter isolates used for susceptibility testing among the MSs, a specific questionnaire survey addressed, among others, aspects related to the isolation methods used in laboratories providing Campylobacter isolates to the NRL‐ARs. The procedural aspects revealed by the survey as desirable candidates for potential further harmonisation are shortly presented in Table 3. More detailed information is also provided in Appendix C.
Table 3.
Main findings of the specific questionnaire survey relating to AMR monitoring in Campylobacter spp., EU MSs, June 2018
| Specific Topics | Collected Information |
|---|---|
|
Sampling procedure |
Variability in the number of samples collected and pooled (from 1 to 10) per slaughter batch was reported. For instance, a caecal sample from only one broiler was sampled in 8 MSs, whereas, in 15 MSs, a pool of caeca from ten broilers was collected. |
| Isolation method of Campylobacter |
Although from 1 up to 25 different laboratories were involved in the isolation of Campylobacter spp. for AMR monitoring in the reporting MSs, a single method was reported to be used for isolating Campylobacter strains for AMR monitoring in 23 MSs. Direct plating, without enrichment, of caecal contents, as described in the detection procedure C of the standard EN ISO 10272‐1 for samples containing high numbers of Campylobacter spp. was carried out in 23 MSs. Inoculation of two different agar media was reported in 16 MSs, from which 15 MSs used mCCDA. Either Karmali (this was considered not optimal, as both media contain the same inhibitor (cefoperazone)), Preston, blood agar, CFA, CASA or Skirrow media were used as second plate. |
| Over‐week‐end culturing | There were also differences between laboratories in the procedures used for cultures over week‐end periods, resulting for certain laboratories in cultures set up only on certain week days, in order to avoid any week‐end working. |
| Species identification |
Most countries reported using MALDI‐TOF mass spectrometry or PCR in accordance with the method of Denis et al. (1999). Microscopical examination, and/or oxidase, catalase, hippurate and indoxyl acetate tests were also performed by several countries. Regarding the number of colonies selected for identification, variability between countries occurred, with one to five colonies identified per sample. This may impact the assessment of the prevalence of Campylobacter spp. and the proportion of C. jejuni/C. coli in a sample. |
| Use of ISO Standarda | 21 MSs reported its use in various contexts, and 19 MSs being accredited for this method. |
AMR: antimicrobial resistance; MS: Member State; mCCDA: modified charcoal‐cefoperazone‐deoxycholate agar; CFA: campyfood agar plate; CASA: Campylobacter selective chromogenic medium; MALDI‐TOF: Matrix Assisted Laser Desorption/Ionization – Time of Flight; PCR: polymerase chain reaction.
European standard EN ISO 10272‐1.
The specific questionnaire survey therefore revealed as desirable and reasonable to propose and implement a harmonised protocol based on the European standard EN ISO 10272‐1, detection procedure C, including precise recommendations (e.g. number of carcases sampled, maximum elapsed time before analysis, number and nature of selective media, scheme and number of colonies to be checked for species identification) to the MSs, in order to enhance further the harmonisation of AMR monitoring in Campylobacter.
4.2. MRSA survey
To assess the potential for harmonisation of procedures relating to MRSA and to gauge the degree of support from MSs for further monitoring, the specific questionnaire survey also addressed questions relating to MRSA. The main findings are summarised in Tables 4 and 5, with more detailed information provided in Appendix D.
Table 4.
Main findings of the specific questionnaire survey relating to MRSA monitoring, EU MSs, June 2018
| Specific Topics | Collected Information |
|---|---|
| Sample types | Considering the matrix from which MRSA isolation was attempted, 7/27 MSs tested meat, 10/27 MSs tested nasal and/or skin swabs, 9/27 tested environmental samples (dust or boot swabs) and 5/27 tested other samples such as milk. |
| Animal populations |
Considering poultry, 3 MSs monitored healthy broilers on farms for MRSA; 1 MS also monitored healthy turkeys on farms. The frequency of testing was 3 years (2 MS) or 3–5 years (1 MS). Three MSs monitored healthy cattle on farms (2 MSs) or both at farm and abattoir level (1 MS) at intervals of 3 years (1 MS) or 3–5 years (1 MS) with monitoring set to begin in the Netherlands. Germany also monitored veal calves and milk/bulk milk from dairy cows. Five MSs monitored healthy pigs for MRSA either at abattoirs (2 MSs) or on farms (3 MSs) at a frequency of 2 years (1 MS), 3 years (1 MS), 3–5 years (1 MS) or irregularly (1 MS). |
| MRSA isolation method | 9/27 MSs reported using a two‐step (2S) method for MRSA isolation, while 5/27 used a one‐step (1S) method.a One MS employed a slight variation of the 2S method. The 1S and 2S methods are discussed further elsewhere in this report. |
| Susceptibility testing | The majority of responding MSs determined the susceptibility of MRSA isolates by broth microdilution.b All MSs performing broth microdilution used EUCAST thresholds to interpret resistance results; of the four MSs performing disc diffusion, three used CLSI breakpoints, while one used EUCAST disc diffusion breakpoints. Considering those MSs performing broth microdilution susceptibility testing, all MSs tested a common core panelc (EFSA, 2012b). |
| Samples collected | The preferred sampling site in live pigs or pigs after slaughter was nasal or skin swabs, while in poultry (broilers and turkeys), cloacal, oral and skin (including underwing) swabs were favoured. Nasal swabs were the only reported option for sampling veal calves, while milk or bulk milk was collected from lactating bovine animals. 12/27 MSs reported collecting and testing samples from the farm environment, with 11/27 testing dust and four of these also testing boot swabs. One MS tested only boot swabs. |
| MRSA characterisation | 13/27 MSs reported the use of spa‐typing to characterise MRSA isolates. Four MSs used WGS, SCCmec and multilocus sequence typing to characterise isolates. |
MRSA: methicillin‐resistant Staphylococcus aureus; MS: Member State; EUCAST: European Committee on Antimicrobial Susceptibility Testing; CLSI: Clinical and Laboratory Standards Institute.
An abbreviated protocol described in Larsen et al. (2017).
Susceptibility testing of MRSA isolates was performed by 13/27 MSs using broth microdilution and 4/27 using disc diffusion; 11/27 MSs either did not test or did not respond to this question.
Of the following antimicrobials: clindamycin, tetracycline, rifampicin, streptomycin, fusidic acid, penicillin, chloramphenicol, kanamycin, erythromycin, ciprofloxacin, cefoxitin, tiamulin, linezolid, quinupristin‐dalfopristin, mupirocin, vancomycin, gentamicin, trimethoprim and sulfamethoxazole. One MS tested additionally enrofloxacin, while France tested lincomycin and spiramycin.
Table 5.
National monitoring programmes in place for meat and milk in the MSs, as reported in the specific questionnaire survey, EU MSs, June 2018
| Meat or meat products | MSs | At abattoirs/At retail | Frequency of monitoring |
|---|---|---|---|
| From Broilers |
AT, DE |
At retail (AT), Both (DE) |
Every 2nd year (AT), Every 3–5 years (DE) |
| From Turkeys | DE | Both (DE) | Every 3–5 years (DE) |
| Beef | DE | Both (DE) | Every 3–5 years (DE) |
| Pork |
AT, FI, DE |
At retail (AT, FI), Both (DE) |
Every 2nd year (AT), Every 3–5 years (DE), Not regularly (FI) |
| From Veal Calves, Milk | DE | – | – |
Regarding the possible further EU‐wide surveillance of MRSA, 19/27 MSs (70%) considered that would be useful in food‐producing animals, a view also expressed by two responding non‐MSs. Only one MS disagreed and 7/27 did not respond to this question. The main animal species considered the highest priority for further EU‐wide monitoring were pigs (18/27 MSs), broilers (15/27 MSs), veal calves (10/27 MSs) and turkeys (7/27 MSs). Those species mentioned by MSs infrequently in the responses received included beef and dairy cattle and companion animals. Of the 27 MSs, 7 favoured farm‐based surveys of pig and/or poultry farms to assess herd or flock level prevalence and diversity, while 9/27 MSs favoured abattoir‐based surveillance to assess the diversity of MRSA in pigs and/or poultry, recognising that cross‐contamination between animals may occur at slaughter. Fewer MSs (11/27, 40%) favoured further EU‐wide monitoring of meat, with 30% expressing the contrary view and 30% not expressing a view. The types of meat considered of highest priority were the same as those reported for food‐producing animals (pork and chicken then with less support, meat from turkeys and veal calves).
5. Objectives of monitoring AMR from a public health perspective
The monitoring of AMR in food‐producing animals and food is essential to provide comprehensive, comparable and reliable information on the development and spread of drug‐resistant bacteria, to measure the impact of measures taken to reduce AMR and to monitor progress. The continually evolving threat from emerging resistance has underlined the need to strengthen further AMR monitoring and to constantly review the data collected, interpret the findings and assess trends to inform, update and consolidate national action plans against AMR based on a ‘One Health’ approach.
The monitoring of AMR in food‐producing animals and food is intended to be performed in close relationship with the surveillance of AMC in food‐producing animals, as well as with the AMR and AMC monitoring programmes in humans. Such AMR and AMC data provide valuable insights to inform decision‐making and facilitate the development of appropriate strategies and actions to manage AMR at European, national and regional levels. The first EU‐wide integrated analyses of data on the consumption of antimicrobial agents and the occurrence of AMR in bacteria from humans, food‐producing animals and food, were jointly performed by ECDC, EFSA and EMA (JIACRA I and JIACRA II), and showed positive correlation between AMC and AMR. Such analyses are intended to be carried out on a regular basis.
The present technical specifications aim at improving the comparability and the reliability of the AMR data collected by the MSs and enlarging the scope of the monitoring. Recent developments, including the increasing use of molecular monitoring through whole genome sequencing, are acknowledged and have been incorporated into the monitoring programme. The detailed characterisation of isolates at the molecular level facilitates comparison of AMR in humans and animals at several levels, including the occurrence and types of resistance genes, resistance plasmids and the host bacterial organism. The technical specifications set out proposals whereby molecular monitoring in selected bacteria can be progressively implemented across all MSs, while retaining comparability with the phenotypic monitoring which is currently in place.
Comparability with the monitoring performed in previous years is an important over‐arching consideration, which has been taken into account at all levels when revising the technical specifications. The data collected in previous years provide an important resource against which future trends can be evaluated. AMR in certain bacterial organisms occurring in food or food‐producing animals has been monitored under Commission Implementing Decision 2013/652/EU for several years; the revised technical specifications propose the expansion of the monitoring to include other selected bacterial species and sample types in mandatory monitoring or ‘baseline’ surveys, provide a rationale for their inclusion, and propose recommendations on the frequency of monitoring.
The requirement to provide information on the current AMR situation in food‐producing animals and food, has been considered against the background of EU and national initiatives focussing on AMR and AMC and the reductions in AMC which have been achieved in a number of MSs. The different sizes of the various livestock sectors in the different MSs have been considered and proposals developed to ensure comparability of monitoring while at the same time ensuring their cost‐effectiveness.
6. Rationale for revising the current AMR monitoring system
6.1. To adapt the monitoring to emerging AMR and current priorities
6.1.1. Inclusion of AMR monitoring in C. coli
6.1.1.1.
Campylobacter coli is a human pathogen
The ECDC/EFSA EU Summary Report on zoonoses reported 246,158 confirmed cases of human campylobacteriosis in the EU in 2017. C. jejuni and C. coli were associated with, respectively, 84.4% and 9.2% of those confirmed cases for which Campylobacter species information was reported (EFSA and ECDC, 2018b), representing more than 20,000 human cases of C. coli infections. Certain countries recorded even higher proportions of confirmed C. coli infections, such as France, where 15% of human cases were linked to C. coli (CNR‐Campylobacter, online), whereas lower proportions of confirmed clinical C. coli infections were also registered in certain EU MSs, for example, at 5–6% in Poland (Sadkowska‐Todys and Kucharczyk, 2016). In the USA, approximately 10% of human cases were associated with C. coli (Bolinger and Kathariou, 2017). It is unknown whether those disparities reflect either true differences in epidemiological conditions (varying sources of infection) or discrepancies in surveillance and reporting systems between countries (mandatory/voluntary notification of cases, surveillance coverage, analytical methods).
C. coli is more often resistant than C. jejuni to several antimicrobials, and particularly to erythromycin, a resistance that is usually rare in C. jejuni
Considering Campylobacter isolates from human cases in Europe in 2016, the levels of resistance to erythromycin and gentamicin in C. coli (at 11.0% and 1.7%, respectively) were markedly higher than those observed in C. jejuni (at 2.1% and 0.4%, respectively) (EFSA and ECDC, 2018a). The combined resistance to ciprofloxacin and erythromycin was low in C. jejuni (0.6%) but moderate in C. coli (8.0%). A higher level of resistance in C. coli compared with C. jejuni was also reported at the country level. In France, C. coli was more often resistant to macrolides, fluoroquinolones and tetracyclines (CNR‐Campylobacter, online), whereas, in the USA and Spain, resistance to macrolides was more frequently reported in C. coli than in C. jejuni (Bolinger and Kathariou, 2017).
Considering animal sources, C. coli was found more often resistant to ciprofloxacin than C. jejuni in poultry in the USA, even after the ban of enrofloxacin (Bolinger and Kathariou, 2017). In Europe, in 2016, C. coli in poultry were significantly more often resistant to ciprofloxacin than C. jejuni. The comparison is however limited to a few countries testing susceptibility in both species, as C. coli was not tested in many countries (EFSA and ECDC, 2018a). In the USA, in retail meat (mainly poultry), higher rates of AMR (except for tetracycline) were described in C. coli than in C. jejuni (Zhao et al., 2010). Similar observations were also made in other animal productions. In France, C. coli from calves was more frequently resistant to tetracyclines, ciprofloxacin and erythromycin (Chatre et al., 2010), whereas, in Poland, C. coli from pig and cattle carcasses were significantly more often resistant to streptomycin and tetracycline than C. jejuni (Wieczorek and Osek, 2013). In the USA, high levels of erythromycin resistance were also reported in C. coli from chickens, turkeys, cattle and market swine in comparison with C. jejuni (Bolinger and Kathariou, 2017).
C. coli may contain and transfer important and emerging antimicrobial resistance genes to C. jejuni
The erm(B) gene encodes an rRNA methylase responsible of resistance to macrolides, lincosamides, and streptogramin B antibiotics and is frequently encountered in various Gram‐positive and Gram‐negative bacteria19 in animals (Roberts, 2008). It was recently described in Campylobacter in China, where it was more frequently observed in C. coli than in C. jejuni, and was shown to confer high‐level resistance to macrolides (Liu et al., 2017). In Europe, up to now, erm(B) has been detected only in a few C. coli isolates from broilers and turkeys in Spain (Florez‐Cuadrado et al., 2017) and from a broiler isolate in Belgium (Elhadidy et al., 2019). In Campylobacter, this gene is present either on plasmids or, more frequently, on multidrug resistance islands (MDRI), also containing other resistance genes, such as tet(O) coding for tetracycline resistance and aminoglycosides resistance genes, such as aad9, aadE, aph(2”)‐IIIa and aacA‐aphD (Florez‐Cuadrado et al., 2017). The MDRI can be transferred to a macrolide susceptible C. jejuni by natural transformation (Wang et al., 2014).
C. coli may contain the cfr(C) gene conferring resistance to phenicols, lincosamides, pleuromutilins and oxazolidinones. It was first detected in C. coli isolates of cattle origin in various states of the USA. The gene is located on a conjugative plasmid and can transfer various traits of resistance to a C. jejuni recipient strain (Tang et al., 2017). The cat gene conferring resistance to phenicols (Li et al., 2017) was found to be more frequent in C. coli than in C. jejuni from broilers in live bird markets in China (Li et al., 2017).
The presence in Campylobacter of these resistance genes, plasmids and MDRI is worrisome, as they not only enable their bacterial host to resist to one or several important therapeutic options, but also because they can lead to co‐selection of multidrug‐resistant Campylobacter isolates, by use of other antimicrobials. As these resistances seem more frequent in C. coli but can be transferred to C. jejuni, it is important to detect their presence in the C. coli population at an early stage.
C. coli is more prevalent than C. jejuni in poultry in several countries
Although C. jejuni has for long been considered as the prominent Campylobacter species in poultry, it has been recently reported that C. coli may be more often detected, for example in France, Hungary and Spain (EFSA and ECDC, 2014).20 The actual prevalence of C. coli and C. jejuni in poultry production likely depends on various factors, including the production type, AMC (Avrain et al., 2003) and the age of the birds sampled (El‐Shibiny et al., 2005). Those factors may vary between countries and likely yield differences. The observed proportions of Campylobacter species among isolates however also depend on the isolation methods, as certain media may favour one Campylobacter species over another (Reperant et al., 2016). The identification of a low number of colonies at the species level in a sample will, in most cases, result in the detection of the dominant species. The detection of the different species present in a sample, particularly when the numbers of cells of each species differ substantially, may necessitate the analysis of many colonies or the use of different species‐selective or species‐favouring media/protocols. It is therefore desirable to ensure a greater degree of harmonisation of the isolation protocols to ensure a relevant comparison between countries in terms of prevalence of C. jejuni and C. coli.
6.1.1.1.1.
Proposal: To include C. coli from poultry, fattening pigs and bovines of less than 1 year of age in the harmonised monitoring of AMR, because of its importance as a food‐borne zoonotic pathogen and a potential reservoir of antimicrobial resistance genes (e.g. transferable macrolide resistance genes). Where samples are already collected and tested for Campylobacter spp., susceptibility testing on isolates of both C. jejuni and C. coli, where relevant, will only induce a limited additional cost compared with susceptibility testing on isolates of C. jejuni only.
6.1.2. Inclusion of monitoring of MRSA
The EFSA scientific report on technical specifications on the harmonised monitoring and reporting of AMR in methicillin‐resistant S. aureus in food‐producing animals and food (EFSA, 2012b) provided the rationale for monitoring of MRSA. An EU‐wide baseline survey was performed in 2008 to obtain comparable preliminary data on the occurrence and diversity of MRSA in primary pig production in all MSs through a harmonised sampling scheme (EFSA, 2009). Pooled dust samples collected from pig holdings were tested for MRSA and all isolates were subjected to spa‐typing and determination of their MRSA status. The survey results indicated that MRSA was common in breeding pig holdings in some MSs, while, in other MSs, the prevalence was low (EFSA, 2010). MRSA ST398 was by far the most predominant MRSA lineage identified. Further investigation of the diversity of MRSA spa‐types also showed that the distribution of spa‐types differed significantly between countries. These revised technical specifications (as in the current report) take account of recent developments and propose a harmonised methodology to be used in the monitoring of MRSA in the most relevant production animals and foodstuffs throughout the EU, updating the previous EFSA technical specifications (EFSA, 2012b) where appropriate.
Relatively few MSs have reported regular monitoring for MRSA in food‐producing or other animals or meat derived from those animals to EFSA (EFSA and ECDC, 2019). Monitoring in recent years has, although performed on a voluntary basis, provided significant results, including data regarding the occurrence of different strains or lineages of MRSA in animals, the occurrence of virulence or host adaptation factors in MRSA detected in animals and the occurrence of resistance to antimicrobials other than methicillin in those MRSA. The features detected in the monitoring performed in recent years by the MSs provide strong evidence of the relevance and value of ongoing monitoring. Considering those MSs reporting the results of monitoring programmes for MRSA in animals or food to EFSA, some have reported data only on the occurrence of MRSA, while others have reported additional molecular typing and antimicrobial susceptibility data. The low numbers of MSs voluntarily reporting MRSA data as well as the lack of complete harmonisation of monitoring programmes, provide only a limited assessment of the occurrence and characteristics of MRSA currently occurring in animals at the EU level. These issues were also discussed in the previous recommendations (EFSA, 2012b) and are summarised below.
The rationale for the stage of the food chain to be monitored for MRSA was set out in the previous EFSA technical specifications (EFSA, 2012b), and the two stages primarily considered were the farm or the slaughterhouse. Monitoring at farm level could facilitate investigation of risk factors and the influence of farm management practices on the occurrence of MRSA, though costs of sampling on farms were likely to be higher than sampling at slaughterhouses; the latter being considered to be more cost‐effective (EFSA, 2012b). A degree of cross‐contamination between batches of animals is considered likely to occur in abattoir‐based monitoring and changes in MRSA status in finished pigs can occur during transport to abattoirs and also while held in the lairage prior to slaughter (Broens et al., 2011). Changes in the MRSA status of pigs have usually been assessed by monitoring through the collection of nasal and/or ear skin swabs and it is possible that animals remain carriers at other sites, such as the tonsil. Longitudinal studies also indicate that carriage of MRSA declines on entry to the fattening period (Bangerter et al., 2016). Nevertheless, studies carried out in Italy in 2008 have shown that the outputs obtained through population studies on MRSA in production pigs on the farm (survey performed on accordance with Decision 2008/55/EC21) or at the abattoir (Battisti et al., 2010) provide a broadly similar picture, with non‐significantly different holding‐level prevalence assessed at around 35% in both approaches and similar distributions of spa‐types and sequence types (STs). There are advantages and disadvantages to farm‐based or abattoir‐based monitoring and comprehensive information is not available covering all relevant technical aspects. The decision as to whether farm‐based or abattoir‐based surveillance should be performed will also be influenced by practical considerations.
6.1.2.1.
Occurrence of MRSA in food‐producing animals
EFSA (EFSA and ECDC, 2019) describes the occurrence of MRSA in animals and meat. Where molecular typing data are available, the MRSA isolates detected are categorised in the EU Summary Report on AMR (where possible) as livestock‐associated (LA‐), healthcare‐associated (HA‐), community‐associated (CA‐) or mecC MRSA. The prevalence and typing data provide useful characterisation of those strains occurring in animals, although the lack of harmonisation of monitoring procedures mean that results relating to prevalence are not in general comparable between MSs. Certain MSs have applied a consistent methodology to monitor the occurrence of MRSA in animals over several years and monitor changes in prevalence, but again, approaches in different MSs are not harmonised and different sampling procedures have been adopted, meaning that data are unlikely to be directly comparable between the different MSs.
Occurrence of virulence/host adaptation factors in MRSA in food‐producing animals
EFSA (EFSA and ECDC, 2018a) also describes the recognised virulence and host‐adaptation genes where appropriate and when these have been reported by MSs in MRSA. These virulence and host‐adaptation genes include, for example, the Panton–Valentin leukocidin (PVL) frequently associated with CA‐MRSA strains and considered a virulence factor for human infection, as well as other leukocidin genes which have been associated with S. aureus in particular animal species (for example lukM in ruminants (Schlotter et al., 2012)). In addition, certain bacterial phages can be associated with particular lineages of S. aureus/MRSA and can carry genes which may confer adaptations facilitating the colonisation of (or virulence in) certain animal hosts (Van Der Mee‐Marquet et al., 2013). The immune evasion cluster which carries (amongst other genes) the immune evasion genes sak (encoding staphylokinase), chp (encoding chemotaxis inhibitory protein) and scn (encoding staphylococcal complement inhibitory protein) has been frequently detected in S. aureus recovered from humans but is rare in isolates recovered from animals (Cuny et al., 2015).
Occurrence of antimicrobial resistance in MRSA in food‐producing animals
Only a few MSs have reported details of the antimicrobial susceptibility of MRSA isolates recovered from animals or meat to EFSA in recent years (EFSA and ECDC, 2019). The antimicrobial susceptibility of MRSA isolates can provide useful information regarding the lineage/clade of MRSA which has been detected; this is particularly relevant for LA‐MRSA CC398 where livestock‐associated strains are typically resistant to tetracyclines, providing one of the markers which assists in distinguishing between the livestock and human clades of CC398 (Kinross et al., 2017).
Resistance to linezolid, a critically important last resort antimicrobial in human medicine, has been detected in livestock in Europe (EFSA and ECDC, 2018a). Resistance to linezolid may be conferred by mutational resistance or by acquisition of resistance genes, some of which also confer resistance to antimicrobials frequently used in veterinary medicine, with the consequence that use of certain veterinary medicines may co‐select for linezolid resistance. Resistance to vancomycin (another critically important antimicrobial in human medicine) has not been detected in MRSA from livestock but is included in the voluntary monitoring performed by those MSs which undertake it. The monitoring programme should also assess the degree of resistance to those antimicrobials which are particularly important in the treatment of humans.
The monitoring of antimicrobial susceptibility patterns can be used, in conjunction with strain typing data to provide useful information on the evolution and dissemination of strains of MRSA.
6.1.2.1.1.
Proposal:
To undertake monitoring for MRSA in food‐producing animals in the EU MSs. The priority species for investigation should be growing/fattening pigs, monitored mandatorily through a harmonised EU‐wide baseline/cross‐sectional survey, performed either on the farm or at the slaughterhouse, for example on 4‐year rotating basis. Monitoring in other food‐producing animal species (e.g. veal calves, turkeys and broilers) and in meat should be done according to national priorities.
To harmonise monitoring approaches in order to improve comparability between monitoring in MSs and facilitate comparison of different approaches to control at national level.
To implement strain typing to allow detailed characterisation of MRSA strains and lineages, virulence and host‐adaptation factors, and other genetic markers (e.g. phages) associated with certain animal hosts.
To include susceptibility testing of MRSA in the monitoring programme.
6.2. To improve comparability of AMR data obtained across the EU MSs
6.2.1. Harmonisation of isolation methods of Campylobacter spp.
The use of different isolation methods, in particular regarding possibly varying inoculum size (Hsieh et al., 2018), enrichment (Carrillo et al., 2014; Reperant et al., 2016), temperature, media (Hsieh et al., 2018), and number of isolates identified at the species level (Carrillo et al., 2014; Reperant et al., 2016), may strongly influence the recovery of Campylobacter spp. from samples, the proportions of C. jejuni or C. coli or even the diversity of the isolates, including their profiles of antimicrobial susceptibility. Variability in Campylobacter detection methods has been evidenced through the specific questionnaire survey (see Section 4.1 and Appendix C). A review assessing the impact of using different transport conditions, culture media, temperature and atmosphere and methods also advocated the need of comparable and consistent methods (Huang et al., 2015). A better knowledge of the prevalence of Campylobacter spp., C. jejuni and C. coli, in animal production and food of animal origin in the MSs and of the prevalence of resistance in these two Campylobacter spp. will help understand the epidemiology and especially, the sources of human C. jejuni and C. coli infections, and the differences of proportions of C. jejuni and C. coli in human cases between European countries. A better understanding of the sources in the different MSs could help design measures for a better control of this zoonotic agent.
6.2.1.1.
Proposal: To improve the comparability of Campylobacter prevalence and antimicrobial resistance data between MS, EU monitoring should be based on harmonised methods for both isolation and antimicrobial susceptibility testing. A harmonised protocol based on the European standard EN ISO 10272‐1, detection procedure C, should be provided in liaison with the EURL‐Campylobacter (see Appendix N).
6.2.2. Harmonisation of isolation methods of MRSA
The EURL‐AR has recently updated the protocol for isolation of MRSA from animals and the environment, to reflect recent findings on the relative sensitivity of different selective culture methods (Larsen et al., 2017). The previous technical specifications (EFSA, 2012b) were based on the data available at that time and recommended a two‐stage isolation method, rather than that currently recommended by the EURL‐AR, which reflects recent findings and adopts a one‐stage protocol (Larsen et al., 2017). The one‐stage method has been shown to have increased relative sensitivity when applied to pigs (Larsen et al., 2017), though differences between the methods were not apparent in studies on cattle or poultry (Nemeghaire et al., 2013, 2014).
It is unlikely that the proposed further monitoring will be directly comparable with the baseline survey for MRSA which was performed in 2008 (EFSA, 2010) and which relied on sampling dust and focused on breeding pigs. Further monitoring provides the opportunity to evaluate further the different isolation methods involving one or two selective stages. This could be particularly useful if the one stage method provides increased sensitivity when applied to dust samples, because of the ease of sampling dust vs collecting samples from animals.
The relative sensitivity of sampling different sites in pigs has been assessed using the two‐stage isolation method, but not using the one‐stage method (Figure 1).
Figure 1.
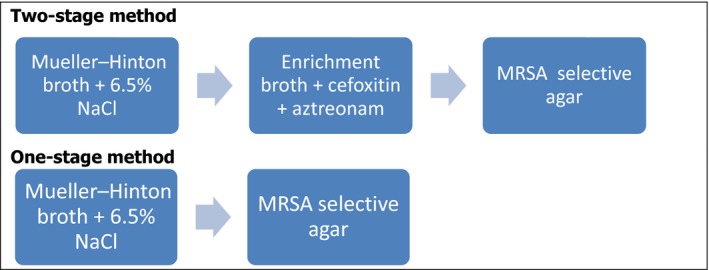
Schematic representation of one‐stage and two‐stage selective isolation methods for MRSA
Sampling nares and ear skin of pigs (60 per farm) on three pig farms in Belgium was found to have a relative sensitivity of 98.2%, higher than sampling ear skin alone (91.4%), perineum (76.5%) or nares (75.3%) or other combinations of these three sites (Pletinckx et al., 2012). Relative sensitivity was also investigated in Denmark (Agersø et al., 2013) who reported relative sensitivity of sampling air filters (78%), dust (43%), nasal swabs (78%) and ear skin swabs (90%).
6.2.2.1.
Proposal: To plan a preliminary evaluation of the relative sensitivity of one‐stage and two‐stage isolation methods applied to different sample matrices in pigs in a separate dedicated essay, to be coordinated by the EURL‐AR and the EFSA, before implementing further monitoring of MRSA, and in particular before drafting the protocol of the proposed baseline survey on MRSA in pigs.
6.2.3. Harmonisation of sampling matrices for MRSA
Considering the available data on the relative sensitivity of different sampling matrices, the following scheme is proposed for the mandatory sampling of food‐producing animals and their immediate environment (Table 6). Meat and other products derived from animals (e.g. milk) should be sampled on a voluntary basis as minced meat or composite milk samples.
Table 6.
Recommended samples for monitoring MRSA in certain food‐producing animals
| Sample type/Animal population | Pigs | Veal Calves | Turkeys | Broilers |
|---|---|---|---|---|
| Dust | ● | ● | ● | ● |
| Nasal swabs | ● | ● | ||
| Oral swabs | ● | ● | ||
| Ear skin swabs | ● | |||
| Under wing swabs | ● | ● |
6.3. To enlarge the scope of the monitoring AMR of food origin
6.3.1. Targeting other animal species when monitoring AMR in Campylobacter
Campylobacter spp. has a broad host range, and many wild and domestic mammal and bird species are recognised as healthy carriers (Dearlove et al., 2016). Food animals are considered as a major source of human campylobacteriosis through contamination of food products. The 2008 EU‐wide baseline study assessed the prevalence of Campylobacter‐colonised broiler batches and Campylobacter‐contaminated broiler carcasses at the EU level as 71.2% and 75.8%, respectively. More recently, in 2016, the proportions of Campylobacter‐positive samples of fresh meat of broilers, turkeys, pigs and bovines in the reporting EU MSs were estimated at 36.7%, 11.0%, 2.9% and 1.0%, respectively (EFSA and ECDC, 2017b). Higher levels of prevalence were also recorded. In Poland, a prevalence of 14.9% was observed on bovine carcasses at slaughterhouse, with mainly C. jejuni, and 29.9% on pig carcasses, with mainly C. coli (Wieczorek and Osek, 2013). In Austria, 30% of dairy calf farms tested positive for Campylobacter, and 14.4% of the calves tested positive for C. jejuni (Klein‐Jöbstl et al., 2016). In 2015, 64.2% of the caecal content samples of bovine animals, of less than 1 year of age, collected at slaughter in Germany, tested positive for Campylobacter spp. (EFSA and ECDC, 2017a). A C. jejuni prevalence of 24.7% was also observed in veal calves in Italy (NRL‐AR, Italy, personal communication) and a very high prevalence (98.5%) of C. jejuni was recently reported in French calves (Thépault et al., 2018).
In 2016, proportions of milk and cheeses found contaminated with Campylobacter were below 1.5% in nine and five European MSs, respectively (EFSA and ECDC, 2017b), whereas in the USA Campylobacter spp. was detected in one‐fourth (25%) of operations in either bulk tank milk or milk filters. Therefore, consumption of unpasteurised or raw milk may be a source of human infection (Del Collo et al., 2017). Although several authors did not detect Campylobacter in mussels (Ripabelli et al., 1999; Lévesque et al., 2010) or fish (Salmo trutta) (Raeisi et al., 2017), antimicrobial‐resistant Campylobacter were isolated from edible bivalve molluscs purchased from markets in Thailand (Soonthornchaikul and Garelick, 2009). This broad host range requires the ranking of the sources of human infections, and several studies have attempted to investigate the main sources of human campylobacteriosis. The more recent studies are principally based on genome sequences.
The first methods used nucleotide sequences from a few housekeeping genes. It was observed that certain multilocus sequence types (MLST) could be strongly associated with specific hosts such as clonal complexes ST‐257 with chickens and ST‐61 with ruminants (Dearlove et al., 2016). Thus, Mossong et al. (2016) reported that studies based on MLST have attributed chicken as the main reservoir of human infections (50–80%) followed by cattle (20–30%). The authors conducted a source attribution study of human campylobacteriosis in Luxembourg and, according to an asymmetric island model, it was concluded that human cases could be attributed to the following sources: poultry (61.2%), ruminants (33.3%), environmental water (4.9%) and swine (0.6%) (Mossong et al., 2016). Nohra et al. (2016) conducted a molecular study on C. coli strains isolated from different sources in New Zealand between 2005 and 2014. Molecular subtyping revealed 27 different STs, of which 18 belonged to clonal complex ST‐828. Modelling indicated that ruminants and poultry were the main sources of C. coli human infection, but the authors did not include pork because they had only one C. coli from this source. In contrast, in a combined case–control and molecular source attribution study of human Campylobacter infections in Germany, Rosner et al. (2017) attributed more than 90% of human campylobacteriosis cases to chicken, and none of the isolates were related to cattle.
Some common strains, notably isolates belonging to the clonal complexes ST‐21 and ST‐45 in C. jejuni and ST‐828 in C. coli, have however broad host ranges, which hampers the use of MLST for performing source attribution (Dearlove et al., 2016). For those human cases related to these three clonal complexes, the whole genome sequences enabled an attribution of 89% of cases to a chicken source; 10% to cattle; and 1% to pigs. In addition, a recent source attribution study, based on fifteen loci, found to be host‐segregating markers, was used to investigate the source of 42 French and 281 UK clinical C. jejuni isolates (Thépault et al., 2017). While 56.8% of the British campylobacteriosis were linked to chicken, French clinical isolates were equally associated with chicken and ruminant reservoirs, which is consistent with a very high prevalence of C. jejuni in French calves (98.5%), and the presence of clusters common to human and cattle isolates (Thépault et al., 2018).
A recent review (Whitehouse et al., 2018) indicated that the part played by swine in human campylobacteriosis remains unclear as, although C. coli is frequently isolated from pigs, different studies showed a low contribution from pigs compared with poultry, cattle or dogs. Pigs are typically rarely colonised by C. jejuni. The same authors indicated that seafood was shown to be responsible of human outbreaks of campylobacteriosis.
6.3.1.1.
Proposal: To target, in addition to poultry and pigs, veal calves in the C. jejuni and C. coli antimicrobial resistance monitoring. The prevalence of Campylobacter is high in bovine animals, and several source attribution studies concluded that ruminants play an important role in human Campylobacter spp. cases (second rank after poultry, or even as frequent as poultry). As, in bovine populations, antimicrobial consumption is mainly related to calves, monitoring should first consider calves, especially since caecal samples have already been collected from this production every second year within the framework of the current monitoring programme.
6.3.2. Carrying out complementary baseline surveys
It is proposed to perform specifically dedicated EU‐wide baseline surveys with changing specific animal/food/environmental categories. This instrument may be used to explore the prevalence of AMR in specific animal/food/bacteria combinations that according to current opinion do not require annual testing but need to be analysed at the European level to obtain an overview over AMR in those animal/food/environmental categories. Ideally, the timing of these more intensive programmes should be harmonised between MSs to optimise comparability of results. Such programmes may be repeated at regular intervals, where necessary.
6.3.2.1.
Possible extension of AMR monitoring in aquaculture and seafood
Seafood production for consumption, including capture fisheries and aquaculture, is growing globally on a yearly basis (https://ourworldindata.org/). Conversely to major meat production systems, no widely adopted standardised AMR monitoring in aquaculture and seafood has been agreed internationally, despite several studies having shown that this commodity can be contaminated by various antimicrobial‐resistant bacteria, including zoonotic pathogens. A large proportion of the seafood consumed in the EU is imported from non‐EU countries (http://www.eumofa.eu/) and it has also been repeatedly shown that seafood imported from countries outside the EU can harbour genes conferring resistance to antimicrobials, including last resort antimicrobials, such as carbapenems (Rubin et al., 2014; Morrison and Rubin, 2015; Janecko et al., 2016; Mangat et al., 2016; Roschanski et al., 2017; Brouwer et al., 2018; Lee et al., 2018), thus exposing EU consumers to AMR genes that would otherwise mainly be restricted to clinical environments or which might be extremely rare or undetected within the EU. Different regulatory framework on AMC and, overall, a possibly higher AMR occurrence in food production in several non‐EU countries compared with the EU MSs, should be taken into account while designing a sampling scheme for AMR monitoring in seafood, depending on the objective of the monitoring.
A programme for AMR monitoring in seafood may be relevant for different purposes, including:
-
To estimate the risk of exposure of consumers to antimicrobial‐resistant bacteria and their resistance genes
via imported seafood from different global sources, and
via seafood originating from the European environment (i.e. in filter feeding molluscs or other seafood originating from European production waters).
To monitor the effect of AMC in aquaculture.
To indirectly assess through filter feeding molluscs produced within the EU the degree of environmental contamination with resistant bacteria in European production waters.
To indirectly monitor the effect of environmental interventions aimed at reducing discharge of resistant bacteria into surface waters and the environment.
6.3.2.1.1.
Proposal: To perform complementary cross‐sectional/baseline surveys on AMR in bacteria from aquaculture and/or (imported) seafood, over the period of validity of the upcoming Commission Implementing Decision in 2021 onwards. For more detailed information, see also 9.2.
Monitoring AMR in bacteria from the environment
Monitoring of the environment for bacteria resistant to antimicrobials has an important role in providing additional targeted data to complement the monitoring performed in humans, animals and food. The occurrence of antimicrobial‐resistant bacteria in humans, animals and food may both influence the occurrence of resistant bacteria in the environment (e.g., through sewage discharges, farm slurry disposal, food waste disposal), as well as itself being influenced by the presence of resistant bacteria in the environment, which may contaminate food or animal feed or otherwise enter and spread (or disseminate their resistance genes) within the animal or human population. The environment provides a route for transfer and dissemination of resistance between humans within the human population, between animals within the animal population, as well as providing opportunities for exposure of animals to human bacteria and humans to animal bacteria. In some cases, environmental pathways for the spread of resistance may be influenced by wildlife vectors, such as gulls; the environment may also act as a reservoir of resistant bacteria.
Monitoring the occurrence of AMR in bacteria in the environment can be performed for a number of different reasons and the underlying rationale or objectives will influence the type of monitoring which is selected. A recent FAO/WHO expert meeting on food‐borne AMR considered the role of the environment, crops and biocides (FAO/WHO, 2018) and particularly the transmission of resistant bacteria or resistance genes from environmental sources to foods and feeds of plant and aquatic animal origin.
Contamination of crops such as fruit, vegetables or grain with antimicrobial‐resistant bacteria can occur; reducing contamination reduces human or animal exposure from crops. Soil, manure, and irrigation water are important sources of microbial contamination.
Antimicrobials may be used for therapeutic purposes in aquaculture; the water component of aquaculture systems may contain resistant bacteria, including bacteria derived from human or terrestrial animal waste. Substances co‐selecting for resistance can also exert an effect in the aqueous environment.
Some antimicrobials and substances co‐selecting for AMR are used in horticulture.
Vegetables, fruit and salads form an important source of exposure to consumers as they are frequently consumed without prior heat treatment. The potential role of irrigation water for the contamination of such commodities has recently been addressed in an EFSA technical report on water reuse (EFSA, 2017). Several recent studies covering vegetables, fruit and salads have however shown that the occurrence of bacteria on these commodities is low and therefore, probably not efficiently addressed by active monitoring with a limited number of samples. To cover AMR bacteria from those commodities, it may be more advisable to continuously test bacteria found during routine testing of these for commodities for AMR in a passive monitoring approach using test methods in line with the methods recommended in this document.
Aquaculture has been dealt with under the previous chapter. Testing domestically produced shellfish may still offer the opportunity to cover food safety aspects along with environmental aspects, as shellfish are exposed to coastal waters that may reflect environmental contamination of these waters by wastewater.
Use of antimicrobials in horticulture is a comparatively rare event and its effect on AMR of products is more adequately addressed by specific studies than by EU‐wide surveys.
6.3.2.1.1.
Proposal: To carry out a baseline survey on AMR in bacteria from domestically produced shellfish that may address simultaneously consumer exposure via shellfish and environmental exposure of shellfish to resistant bacteria from, e.g. wastewater. It is envisaged that the detailed harmonised protocol of that specific baseline survey would be designed at a later stage, considering the most recent data, once a clear agreement to carry out such surveys had been reached.
It is considered that additional interesting data on AMR in the environment might be available from testing bacteria gathered from the monitoring programs on bathing water quality within the framework of the EU ‘Bathing Water’ Directive 2006/7/EC,22 although these are not considered within the realm of this document.
6.4. To perform genetic characterisation of AMR determinants
Over the last few years, high throughput sequencing (HTS) technologies, including the whole genome DNA sequencing technology (WGS), have rapidly evolved and technological progress has led to cheaper, faster, smaller and more efficient sequencing platforms, as well as decreased costs in high‐throughput sequencing. WGS enables to determine the complete DNA sequence of a bacterium, which, once analysed further through bioinformatics tools and pipelines, can potentially provide a wealth of additional information on various traits of the microorganism in question, and possibly even in a single assay, depending on the tool or pipeline used. This may include, for example, AMR and virulence traits, which could not easily be uncovered previously, as well as, still depending on the tool or pipeline used, additional information, such as MLST, predicted serotypes, core‐genome MLST (cgMLST), plasmid replicons, etc.
For the detection of AMR determinants, WGS allows confirmation of the presence of an extended set of AMR determinants and enables differentiation between AMR mediated by chromosomal point mutations and resistance linked to acquired resistance genes, which are more prone to dissemination by horizontal gene transfer. In particular, the location of AMR genes in the genome of the organism can be determined by WGS, distinguishing between localisations on the chromosome and on mobile genetic elements. It is acknowledged, however, that long‐read sequencing can be required to facilitate characterisation of mobile genetic elements, such as plasmids and this is more costly than short‐read sequencing which is cheaper and consequently more frequently performed.
WGS has recently proven to be a powerful tool for epidemiological surveillance of AMR along the food chain. In the ‘One Health’ context, the additional information provided from the genotype (resistance determinants, host bacterial strain type, genetic background, etc.) can be of great significance in facilitating source attribution and informing on the possible contribution of food‐producing animals and derived food to the burden of AMR in humans. In addition, WGS can also be used to perform, for example, source attribution and virulence investigation within the framework of food‐borne outbreak investigation, which makes it a multipurpose tool and increases its cost‐effectiveness.
WGS also confers a practical advantage compared with conventional molecular methods, such as pulse field gel electrophoresis (PFGE), polymerase chain reaction (PCR) or quantitative PCR (qPCR), since, once generated, WGS data can be stored and maintained in databases and therefore, remain easily available for future investigations, regardless of the maintenance of strain collections. This offers the unique opportunity to re‐analyse previously collected data, searching for novel genes, e.g. newly discovered emerging AMR determinants, as recently occurred with mcr‐1 colistin resistance gene (Hasman et al., 2015). In contrast, a current limitation of using WGS for detection of AMR determinants relates to the detection of unknown/novel AMR genes or mutations, as these will not be present in the available reference databases prior to the discovery, making complementation by phenotypic test still indispensable at the current time when searching for new AMR determinants.
In general, high concordance between WGS‐AMR and phenotypic AMR has been described in some studies (see Section 11). Discordant results between WGS‐AMR and phenotypic antimicrobial susceptibility may still occur depending on bacterial species and AMR traits considered. The limited level of discordance is mostly related to AMR mediated by chromosomal point‐mutations and much less to those related to acquired genes. Moreover, the discordance could in very limited cases be observed in the expression of the AMR often related to some aminoglycosides or due to truncated contigs. The occurrence of constitutive and inducible resistance provides another potential source of discrepancy between phenotype and genotype. Therefore, depending on the monitoring objective, the organisms and/or the AMR, complementation with phenotypic data should still be envisaged at least in a transitional period. The technology is still evolving by utilising machine‐learning/deep learning approaches, which might potentially be used for detecting novel AMR genes and network associations linked to AMR.
In the USA, the National AMR Monitoring System (NARMS) already implemented the use of WGS combined with bioinformatics analysis, as part of AMR monitoring for all Salmonella spp. and Campylobacter spp. and some AMR strains of E. coli and Enterococcus spp. NARMS performs in parallel routine conventional phenotypic AST for monitoring. WGS is carried out at the respective NARMS laboratories, and sequences are analysed for the resistome locally. NARMS has begun reporting on a weekly basis all AMR genotypes of Salmonella isolated from retail meats, food‐producing animals, and humans via a public open access web portal. NARMS takes also advantages of the added value of WGS to extract information about bacterial speciation, serotyping, and to improve the understanding of emerging AMR phenomena and the passage of AMR bacteria through the food chain (NARMS).
6.4.1.
Proposal: Considering not only the advantages inherent in the WGS analysis but also its current limitations, as well as the expected evolvement of the present situation, it is proposed to follow a gradual, phased approach to integration of WGS within the harmonised AMR monitoring, over the period of validity of the upcoming Commission Implementing Decision in 2021 onwards.
7. Recommendations on bacterial species, food‐producing animals and food for monitoring AMR
7.1. Bacterial species to be considered
The harmonised monitoring of AMR in animals and food should cover both zoonotic agents, in the first instance Salmonella and C. jejuni and C. coli, and indicator E. coli of the commensal flora. Such monitoring should supplement AMR testing in isolates from humans.
Indicator E. coli organisms of commensal intestinal flora are commonly isolated from animal intestinal content and faeces and can be used as indicators of the Gram‐negative commensal intestinal flora. Commensal bacteria that contaminate food may be also considered a potential AMR hazard, as they can harbour transferable resistance genes. During the passage through the intestine, these bacteria may transfer their resistance genes to host‐adapted bacteria or to pathogens. Exchange of resistance genes between bacteria from different sources can also occur in the environment, including in the kitchen (EFSA, 2008). The effects of use patterns of antimicrobials in a given country and animal population as well as trends in the occurrence of resistance can be studied more accurately in indicator bacteria than in food‐borne pathogens, because all animals generally carry such indicator bacteria. With the current implementation of national control programmes for Salmonella in poultry, the number of Salmonella isolates available has diminished in most MSs, but may still vary markedly between MSs. This decrease makes the monitoring of AMR in indicator E. coli even more relevant.
It is therefore proposed to strongly reinforce the previous recommendation for the monitoring of AMR in commensal indicator E. coli in domestic animals (EFSA, 2012a), as resulting AMR data are more representative and comparable and to consider this monitoring as a priority. Complementary testing of commensal E. coli from retail meat is also relevant as such testing facilitates exposure assessment for consumers, as the prevalence of zoonotic bacteria, such as Salmonella, may be or become low or extremely low.
The rationale for the inclusion on MRSA in future monitoring programmes has been presented in Section 6.1.2 together with the views of MSs on further monitoring of MRSA (Section 4.2 and Appendix D). Collection of meat samples allows testing for multiple pathogens from a single meat sample including MRSA, whereas caecal contents (used for many other target bacteria mentioned in this document) would not be considered optimal for MRSA – which would need to be collected in targeted, specifically designed monitoring studies.
7.2. Combinations of bacterial species/food‐animal populations or food categories
The structuring of harmonised AMR monitoring and reporting in Salmonella spp., C. jejuni and C. coli and indicator E. coli systematically according to production types/animal populations to which the consumer will most likely be exposed through food thereof, in particular meat products, is a recommendation strongly reinforced. These animal populations correspond to various production types of the main food‐producing animal species, such as broilers, laying hens, fattening turkeys, fattening pigs, veal calves, and the related meat and food products. In the EU, beef is more frequently consumed than veal, and raw beef may be also usually consumed. The recommendation on the combinations of bacterial species and animal populations/food categories to be regarded as a priority in a routine AMR monitoring is presented in Table 7. It is reinforced that the harmonised AMR monitoring should primarily be focused on domestic production, so that the relationships between AMR and antimicrobial usage can be analysed further.
Table 7.
Combinations of bacterial species–food‐producing animal populations/meat to be tested for antimicrobial susceptibility within the harmonised AMR monitoring
| Animal populationa/Meatb | Salmonella spp. (at the serovar level) | C. jejuni/ C. coli c | Indicator commensal E. coli | ESBL/AmpC/CP‐producing E. coli | CP‐producing E. coli | E. faecalis/ E. faecium |
|---|---|---|---|---|---|---|
| Broilers | M: NCP, CSS | M: CSS | M: CSS | M: CSS | M/Vd: CSS | M/Vd: CSS |
| Laying hens | M: NCP | – | – | – | – | – |
| Fattening turkeys | M: NCP, CSS | M: CSS | M: CSS | M: CSS | M/Vd: CSS | M/Vd: CSS |
| Bovines, < 1 year old | M: CSS | M: CSS | M: CSS | M: CSS | M/Vd: CSS | M/Vd: CSS |
| Fattening pigs | M: CSS | M: CSS | M: CSS | M: CSS | M/Vd: CSS | M/Vd: CSS |
| Broiler meat | – | – | V: R | M: R | M/Vd: R | – |
| Turkey meat | – | – | V: R | M: R | M/Vd: R | – |
| Pig meat | – | – | V: R | M: R | M/Vd: R | – |
| Bovine meat | – | – | V: R | M: R | M/Vd: R | – |
CSS: caecal samples from healthy animals at slaughter; M: mandatory monitoring; NCP: Salmonella national control plans; R: at retail; V: voluntary monitoring. CP: carbapenemase‐producers.
Domestically produced.
Including imported and domestically produced products.
For each animal species, and for each MS, the target is the more prevalent Campylobacter species; all isolates of the other Campylobacter species that are identified, considering the specification of one isolate per species and epidemiological unit, are to be included. However, for fattening pigs, only C. coli is considered.
Mandatory on a 4‐year rotational basis, voluntary intervening years.
In addition, it would be also desirable to specifically monitor AMR in indicator commensal E. coli from meat imported from third countries, for example poultry meat, at the EU level.
8. Recommendations on sampling design for monitoring AMR
8.1. General considerations on a representative and random sampling
Isolates which are tested for antimicrobial susceptibility should derive from active monitoring programmes so that the determination of bacterial prevalence in the studied animal populations, whether Salmonella, Campylobacter or indicator bacteria, as well as the prevalence of resistant bacteria can be ensured. Bacterial isolates tested for antimicrobial susceptibility should originate from healthy animals sampled from randomly selected epidemiological units, whether poultry flocks or slaughter batches randomly selected within the slaughterhouses. Randomised sampling strategies should be emphasised, allowing for proper statistical data analysis and reducing the effect of sampling bias. A random sample in each animal population targeted ensures the representativeness of the entire population and reflects variability in managerial and hygienic practices in holdings and different country regions. An approximately equal distribution of the collected samples over the year enables the different seasons to be covered.
If diseased animals are sampled, these susceptibility results should be reported separately.
8.2. Sample size calculation
The sample size (i.e. number of isolates to be tested for susceptibility at each sampling time) should allow, within a predetermined accuracy, the calculation of the proportion of resistance to a particular antimicrobial for a given combination of bacterial species/animal populations or food categories and to detect changes in this proportion over time. The approach and the results of the sample size analyses and calculations in the EFSA technical specifications (EFSA, 2007, 2008, 2012a) were reviewed and revised using additional sets of scenarios (Appendix H).
It is proposed that the minimum target number of organisms of each bacterial species which should be examined is 170 from each type of domestic animal production type (this may be reduced to 85 organisms from poultry and pigs, if production is less than 100,000 tonnes per annum). The sample size should be adapted in the case of low Salmonella prevalence and very small production sectors. The number of biological samples to be collected is determined in order to achieve 170 isolates by accounting for the prevalence of the bacteria species monitored. For the MSs with very small production sectors, it is also proposed to perform a pragmatic application of the finite population correction factor as applied to the estimation of a proportion (see Appendix I).
The number of Salmonella isolates deriving from the NCPs to be tested per poultry population is 170. In the case of higher number of Salmonella isolates available, a random selection of 170 isolates should be performed from the collection of yearly available isolates in the MS. In the case of low prevalence, all the Salmonella isolates available – clinical isolates excluded – should be tested for susceptibility.
Within the context of the implementation of the EU and MSs’ action plans against the threat of AMR, a further decrease in use of antimicrobials in food‐producing animals is expected to occur in conjunction with implementing complementary mitigation measures in the coming years, resulting in a decrease in the selective pressure on the emergence and/or occurrence of AMR. In order to ensure sufficient statistical power so that even slight decreases in AMR can be detected, it is therefore recommended that sample size is reviewed by each MS taking into account their own situation and objectives of reduction of AMR in the light of the simulations presented in this report, which also account for the assessment of the occurrence of resistance with sufficient accuracy. It is acknowledged that this approach may lead to an increase in the number of samples to be collected, and that this may require additional resources from the MSs.
8.3. Stratified sampling approach
8.3.1. Sampling plan of Salmonella in certain poultry populations
It is proposed that the AMR monitoring in Salmonella spp. isolates from poultry populations of various production (i.e. laying hens, broilers, fattening turkeys) targeted by national control (and monitoring) programmes (NCPs) of Salmonella, are based on Salmonella isolates collected in the framework of these control programmes. For the national control and monitoring programmes of Salmonella in such poultry populations, minimum requirements on the collection of type of material and where the sampling is to take place are already fixed by the EU legislation. An unbiased estimate of the proportion of resistance may be obtained through a sampling frame covering all epidemiological units (flocks) of the national production. This is most readily achieved if Salmonella isolates originate from the NCPs. The epidemiological unit for the various poultry populations’ concerned (broilers, laying hens and fattening turkeys) is the flock, because most holdings practise all‐in–all‐out production. It is assumed that Salmonella isolates of the same serovar from the same epidemiological unit (poultry flock) show a similar pattern of resistance. To ensure representativeness, susceptibility testing should be done for no more than one isolate per Salmonella serovar from the same epidemiological unit/year.
8.3.2. Sampling plan of bacterial species in food‐producing animal populations and meat at retail
For all the other combinations of bacterial species/animal populations, it is proposed that AMR monitoring is based on the representative/random collection of caecal samples at the slaughterhouse. Sampling performed at the slaughterhouse is emphasised, as in many of the MSs it will be most cost‐effective way to collect the samples. It is recommended that at least 60% of the domestic animal population in a MS are included in the sampling frame, meaning that slaughterhouses processing at least 60% of the domestic animals of the relevant animal category (starting with the slaughterhouses of largest throughput) are eligible for sampling. An active monitoring programme should be based on random sampling of healthy animal carcasses. The sampling plan should be typically stratified per slaughterhouse by allocating the number of samples collected per slaughterhouse proportionally to the annual throughput of the slaughterhouse. An approximately equal distribution of the collected samples over the year enables the different seasons to be covered.
The epidemiological unit for the various poultry populations concerned (broilers and fattening turkeys) is the flock. The epidemiological unit for fattening pigs and bovine animals of less than 1 year of age is the slaughter batch, defined as a group of animals of the same age raised together under the same conditions and exposed to the same risk factors regarding AMR and sent together to the slaughterhouse at the same moment. Only one representative sample of caecal content per epidemiological unit (e.g. flock, batch of animals sent to the slaughterhouse), derived from 1 carcase, is gathered to account for clustering. Regarding broilers, the representative sample of caecal content collected from a given epidemiological unit should derive from 10 animals, in order to collect enough material to test for the presence of Salmonella, indicator commensal E. coli, ESBL‐/AmpC‐producing E. coli, C. jejuni, C. coli and (where applicable) Enterococcus spp.
For the sake of continuity, samples of fresh meat can be collected at retail level. Sampling at retail, without differentiating between domestic and imported products, provides a better estimate of consumers’ exposure to resistant bacteria.
8.3.3. Generic stratified sampling approach and sampling plans
The simple and robust randomised sampling procedure currently in place, mostly relying on a stratified sampling approach23 with proportional allocation of the sample numbers per strata, has been reviewed and is proposed to be reinforced. The general characteristics of the proportional stratified sampling approach, applied to the prospective sampling plans of samples and the retrospective sampling plans of isolates proposed, are briefly presented in Table 8. It illustrates stratified sampling concepts, such as strata, proportional allocation, epidemiological unit, to the sampling plans proposed. Detailed functional indications about every specific sampling plan are provided in Appendix Appendix J – Generic proportionate stratified sampling approach, Appendix K – Sampling plans of Salmonella isolates from primary production of poultry, Appendix L – Sampling plans of representative caecal isolates at slaughter, Appendix M – Sampling designs of representative isolates from meat at retail.
Table 8.
General characteristics of the stratified sampling approach applied to prospective sampling of samples and retrospective sampling of isolates
| Sampling concept | Prospective sampling of samples | Retrospective sampling of isolates | |
|---|---|---|---|
| Sampling of caecal samples at slaughter | Sampling of meat samples at retail | Sampling of Salmonella from primary production of poultryd | |
|
Target populations |
Domestically produced – Broilersa – Fattening Turkeysa – Fattening Pigsa – Bovines < 1 yeara |
– Broiler meat – Turkey meat – Pig meat – Bovine meat |
– Broiler flocks – Laying hen flocks – Fattening turkey flocks |
| Strata (1st stage) | Slaughterhousesb | NUTS‐3 areac | Laboratories involved in NCPs |
| Proportional allocation | Sample size proportionate to the slaughterhouse throughput | Sample size proportionate to the NUTS‐3 area population | Isolate sample size proportionate to the size of the relevante isolate collection in the laboratoryf |
| 2nd stage | Batches/lots of carcases originating from the sample flock/herd | Retailers | NA |
| Epidemiological Unit |
Flock of poultry or slaughter batches of pigs/bovines |
One lot of meat | Flock of poultry |
| Sample/Isolate | 1 sample of caecal content per epidemiological unit g |
1 meat sample per lot |
1 isolate per flock of poultry |
NA: not applicable.
The source population of broilers/fattening turkeys/fattening pigs/bovines of less than 1 year of age covers that domestically produced and slaughtered in the slaughterhouses representing at least 60 % of the all broilers/fattening turkeys/fattening pigs/bovines of less than 1 year of age slaughtered in the Member State.
Those slaughterhouses that accounted for at least 60 % of all broilers/fattening turkeys/fattening pigs/bovines of less than 1 year of age domestically produced in the previous years, according to the most recent statistics.
Those NUTS‐3 areas that accounted for at least 80 % of the national population according to the most recent statistics, as a logistic compromise.
Stratified random sampling (SRS) performed in the sampling frame of the poultry flocks tested positive for Salmonella is also alternatively proposed in the MS where a (central) database continuously records newly detected positive flocks.
The size of the relevant isolate collection is the number of isolates originating from the examined animal population in the study period.
Those laboratories accounting for 80 % of the total number of Salmonella isolates in the poultry production in question within the MS in the previous year, as a logistic compromise, where the Salmonella prevalence is important.
The sample of caecal content may derive from one carcase (single sample) for all animal species, except broilers for which the sample of caecal content derive from 10 carcases, or a number of carcases (pooled sample) per batch/lot of carcases originating from the same epidemiological unit, whether a flock of poultry or a herd of pigs/bovines.
8.4. Sampling frequency and targeted routine monitoring
Annual testing should be the basic rule for the monitoring of AMR, as it may allow earlier detection of evolution in temporal trends. Nevertheless, the biennial monitoring has been acknowledged as a good compromise between scientific needs and MS capacities. For the sake of continuity, it is proposed that for the combinations of interest, the sampling is performed consistently on a biennial basis. In addition, in those MSs where the epidemiological situation requests a more sensitive detection of emerging AMR and trends in the combinations of interest, a switch towards a yearly monitoring may be required.
9. Proposals of complementary baseline surveys on AMR
9.1. Methicillin‐resistant Staphylococcus aureus
The objectives of sampling (Section 6.1.2), appropriate isolation methods (Section 6.2.2) and appropriate sample matrices (Section 6.2.3) and feedback from NRL‐AR and EFSA networks (Section 4.2) have already been considered. It has been shown that pigs in an MRSA positive pig herd are probably repeatedly, intermittently and transiently colonised (Bangerter et al., 2016). Thus individual pigs may change status several times, with the implication that determination of herd MRSA status, based on the investigation of samples from batches of pigs is the optimal parameter to measure (Bangerter et al., 2016) rather than determination of individual pig status or within‐herd prevalence. The inclusion of monitoring of MRSA, the option of sampling on farms or at abattoirs, occurrence of virulence factors and AMR have also already been considered (Section 6.1.2). It is proposed that baseline surveys for MRSA should be performed.
Six batches comprising pooled samples from 10 animals to be tested with each sample type (nasal swabs and ear skin swabs in the case of pigs, as these samples provided the highest relative sensitivity (Section 6.2.2)), with each batch of swabs to be cultured separately. Swabs should be collected at these sites from pigs in the case of both on farm‐ and abattoir‐based sampling. Five dust samples to be collected and tested from each farm, when on farm sampling is performed.
Considering the optimal age of pigs for investigation to determine the MRSA status, data from longitudinal studies suggest that the maximum colonisation of pigs lies during the period between 2 weeks and the point of entry to the fattening period at about 11 weeks of age (Bangerter et al., 2016). The increase in prevalence in MRSA in growing pigs is also supported by data from other longitudinal studies, such as those investigating the effect of zinc oxide supplementation on MRSA in pigs (Slifierz et al., 2015) which found the highest prevalence in groups of pigs (supplemented and un‐supplemented with zinc oxide) was between 21 and 42 days of age.
9.1.1.
Proposal: To sample, in the case of on farm sampling, growing pigs within the age range 3–11 weeks for MRSA; in other species, the available age (or age range) present on the farm on the date of sampling should be sampled.
The baseline survey of 2008 (EFSA, 2010) focused on breeding pigs. Diverse types of herds or flocks may be encountered, including outdoor and extensive production. Studies in Denmark (DANMAP, 2017) have shown that pigs from positive herds moving to outdoor herds may not prove MRSA positive when sampled at the ‘standard’ culture sites (nares, ear skin, etc.) which are generally utilised in such studies. To optimally detect MRSA, it is suggested that herds on which growing pigs at the age of 3–11 weeks are present should be sampled; conventional (housed) herds should be sampled as there is limited data available relating to the optimal sampling methods which should be applied to sampling other types of pig herds.
9.1.2.
Proposal: To sample conventional, housed herds, flocks or groups of animals or if sampling at slaughter, to sample animals originating from conventional, housed herds or flocks. In the case of on farm sampling of pigs, all such herds with pigs present on farm in the age range of 3–11 weeks should be eligible for inclusion in the monitoring.
Relevant epidemiological data should be collected as part of the survey, whether performed on farm or at the abattoir. To maximise participation the monitoring should be performed on an anonymised basis with overall results reported at national level.
9.2. Specific AMR monitoring in aquaculture and seafood products
A number of different bacteria might be considered for inclusion in an AMR monitoring programme of aquaculture and seafood products. Many bacterial species have been recovered from aquaculture and seafood products and it has also been shown that microbiota changes considerably both between and within seafood species according to diet/trophic levels and season, and also based on wild or captive status (Egerton et al., 2018). The development of a ‘universal’ indicator for AMR monitoring in such products therefore may not be possible and different bacterial species may need to be selected based both on the objective of the AMR monitoring and on the sample matrix (wild vs. farmed seafood, geographic area of production, etc.). Furthermore, it is evident that consumption patterns of these products differ widely across EU MSs and thereby consumers are exposed to different aquaculture and seafood production types with their own specific microbiota, implying a need to select different bacterial indicators and sample matrices across EU MSs to fulfil the same objective of the AMR monitoring.
Bacterial species that, when taken individually, may be relevant to fulfil at least one of the objectives listed above include E. coli, Salmonella spp. and other Enterobacteriaceae in particular K. pneumoniae (which survives well in surface waters, as well as in the intestinal tract of animals), Vibrio spp. (including V. parahaemolyticus, V. vulnificus, nonO1–nonO39 V. cholerae), Campylobacter spp. (including C. jejuni, C. lari, C. peloridis), Arcobacter spp., Aeromonas hydrophila, Plesiomonas shigelloides, Shigella spp., Shewanella spp., Clostridium perfringens, Enterococcus spp., Pseudomonas spp. and Listeria spp. among others. The utility of some of these bacterial species as indicators could be however limited by the low prevalence and relative infrequent detection in seafood (for example for Salmonella spp., Plesiomonas shigelloides, Shigella spp., Vibrio spp.).
The development of a programme for AMR monitoring in seafood potentially requires a standardised method for isolation, AST and/or interpretation of AST results for several bacterial species, such as Aeromonas sp., Arcobacter sp., and Vibrio sp. among others. In some species (for example Aeromonas spp.), intrinsic resistance is common as a result of the possession of chromosomal beta‐lactamases. Recent genomics and metagenomics approaches for AMR monitoring would overcome the lack of standardised classical microbiology procedures for susceptibility testing and potentially detect all AMR genes in an isolate or, in the case of metagenomics, in a sample, however such approaches have not been comprehensively applied to seafood yet, mainly due to limited knowledge of AMR genes in some of the potential bacterial targets listed above. There is some development work required to provide validated protocols for AMR monitoring in seafood. Interest to fill in the current knowledge gaps has however grown rapidly as the need for AMR monitoring has become evident along with the expansion of seafood production. It is therefore expected that in a relatively short time further information will be available to allow development of standard, harmonised methods for AMR monitoring in seafood.
9.2.1.
Proposal: To design the detailed harmonised protocols of EU‐wide cross‐sectional/baseline surveys on AMR in bacteria from aquaculture and/or (imported) seafood, once a clear agreement to carry out such surveys had been reached. Nevertheless, it may be necessary to develop preliminary approaches based on current knowledge and then adapt and refine techniques as further information becomes available, in accordance with future priorities. For domestically produced seafood, a preliminary proposal may for example address E. coli recovered from shellfish under Regulation EC/2073/200524 and subjected to susceptibility testing as described in 9.3
9.3. Specific AMR monitoring in the environment
The relative importance of the processes contributing to resistant bacteria occurring in the environment will differ between different countries. As pointed out in a previous chapter, a number of previous studies monitoring of crops and vegetables in Europe and North America have shown that contamination with resistant bacteria (for example ESBL‐producing E. coli) may occur rather infrequently (Reuland et al., 2014; van Hoek et al., 2015; Randall et al., 2017). There are also challenges in developing a cost‐effective, harmonised monitoring system which provides outputs which are representative of the overall or national situation. Some questions relating to the occurrence of antimicrobial‐resistant bacteria in the environment are likely to be best resolved through targeted research studies rather than by ongoing monitoring. Moreover, integration of work carried out in the framework of the EU‐bathing water directive with AMR testing of bacteria may offer additional opportunities to integrate activities under a one health umbrella. However, this is not within the scope of this technical specification.
9.3.1.
Proposal: To carry out a baseline survey on domestic shellfish products covering at the same time exposure of consumers to bacteria from these commodities and the environmental exposure of the shellfish to resistant environmental bacteria (e.g. from wastewater).
10. Recommendations on methodology for testing the antimicrobial susceptibility of bacterial isolates
10.1. Analytical methods in routine monitoring and quality control
10.1.1. Antimicrobial susceptibility testing
Standardised dilution methods give a semi‐quantitative measurement of the susceptibility as an antimicrobial concentration (expressed in mg/L) that is reproducible between different laboratories with a biological variation (one dilution step). As the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website (http://www.eucast.org/) gives access to aggregated distributions of minimum inhibitory concentration (MIC) for these bacterial species, as well as defining ECOFFs (Kahlmeter et al., 2003) and clinical breakpoints (CBPs) in human medicine, data obtained by making use of dilution methods can be interpreted for both epidemiological and clinical purposes, provided that the dilution range used frames both thresholds. It is proposed to reinforce the previous recommendation for the use of standardised dilution methods for AST of bacterial strains targeted by the harmonised monitoring (EFSA, 2007, 2008, 2012a). Standardised dilution methods should be used to test the susceptibility to, at least, a specified concise list of antimicrobials, with predefined appropriate dilution ranges and ECOFFs. There is a need to follow up possible evolutions of ECOFFs, as suggested by EUCAST, and account for them while analysing historical data and temporal trends. The existing quality assurance system should be also reinforced. At the EU level, proficiency tests of the EURL‐AR for susceptibility testing of Campylobacter, Salmonella, E. coli, enterococci and staphylococci, which are performed annually for the NRL‐AMR, support the harmonisation process. In the national AMR monitoring programme, if several laboratories are involved in the isolation and/or testing of the relevant bacteria, proficiency tests should be applied to confirm the capability of those laboratories to detect and where applicable, to test the respective bacterial species in the respective sampling materials.
10.1.2. Isolation methods of Campylobacter spp.
Under the framework of Commission Decision 2013/652/EU, the high level of harmonisation of susceptibility testing enables comparison between the percentages of resistance and resistance profiles reported for the different bacterial targets by the EU MSs. The results of the specific questionnaire survey performed in June 2018, although encouraging, confirmed that certain aspects need to be harmonised further, as they affect the chance of detecting C. jejuni and C. coli and thus, the assessment of the prevalence of resistance. A proposal of harmonised method derived from the ISO Standard, for isolation and identification of C. jejuni and C. coli to be used within the framework of the future AMR monitoring has been put forward in Appendix N. In order to fine tune the harmonised method, the EURL‐Campylobacter has planned to perform the necessary additional assays in 2019–2020.
10.2. Panel of antimicrobial substances for Salmonella and E. coli
10.2.1. Overall proposal
To provide continuity of monitoring data and allow epidemiological tracing of isolates with particular patterns of resistance (particularly in relation to certain Salmonella serovars), it is recommended that those antimicrobials listed in previous recommendations should remain in future testing requirements. The rationale for inclusion of the antimicrobials recommended for use in current monitoring programmes has been previously described elsewhere (EFSA, 2007, 2008, 2012a), in particular regarding the phenotypic monitoring of the presumptive ESBL‐producing and AmpC β‐lactamase‐producing bacteria in animals and food, and the inclusion of last‐resort antimicrobials in the treatment of certain infections with highly resistant Gram‐negative bacteria in humans, such as the carbapenems and colistin. It is reinforced that isolates are tested for susceptibility and MICs interpreted using the epidemiological cut‐off values and concentration ranges shown in Table 9 to determine the susceptibility of Salmonella spp., and indicator commensal E. coli.
Table 9.
Panel of antimicrobial substances, ECOFFs, CBPs and concentration ranges (in mg/L) for Salmonella and E. coli (first panel)
| Antimicrobial | Bacterial species | ECOFFa R: > (mg/L) | CBPa R: > (mg/L) | Current concentration range (mg/L) (No. of wells) | Suggested new concentration range (mg/L) (No. of wells) |
|---|---|---|---|---|---|
| Ampicillin | Salmonella | 8 | 8 | 1–64 (7) | 1–32 (6) |
| E. coli | 8 | 8 | |||
| Cefotaxime | Salmonella | 0.5 | 2 | 0.25–4 (5) | 0.25–4 (5) |
| E. coli | 0.25 | 2 | |||
| Ceftazidime | Salmonella | 2 | 4 | 0.25–8 (6) | 0.25–8 (6) |
| E. coli | 0.5 | 4 | |||
| Meropenem | Salmonella | 0.125 | 8 | 0.03–16 (10) | 0.03–16 (10) |
| E. coli | 0.125 | 8 | |||
| Nalidixic acid | Salmonella | 16 | NA | 4–128 (6) | 4–64 (5) |
| E. coli | 16 | NA | |||
| Ciprofloxacin | Salmonella | 0.06 | 0.06 | 0.015–8 (10) | 0.015–8 (10) |
| E. coli | 0.06 | 0.5 | |||
| Tetracycline | Salmonella | 8 | NA | 2–64 (6) | 2–32 (5) |
| E. coli | 8 | NA | |||
| Colistin | Salmonella | 2 (EFSA) | 2 | 1–16 (5) | 1–16 (5) |
| E. coli | 2 | 2 | |||
| Gentamicin | Salmonella | 2 | 4 | 0.5–32 (7) | 0.5–16 (6) |
| E. coli | 2 | 4 | |||
| Trimethoprim | Salmonella | 2 | 4 | 0.25–32 (8) | 0.25–16 (7) |
| E. coli | 2 | 4 | |||
| Sulfamethoxazole | Salmonella | 256 (EFSA) | NA | 8–1024 (8) | 8–512 (7) |
| E. coli | 64 | NA | |||
| Chloramphenicol | Salmonella | 16 | 8 | 8–128 (5) | 8–64 (4) |
| E. coli | 16 | 8 | |||
| Azithromycin | Salmonella | 16 (EFSA) | NA | 2–64 (6) | 2–64 (6) |
| E. coli | 16 (EFSA) | NA | |||
| Tigecycline | Salmonella | 1 (EFSA) | NA | 0.25–8 (6) | 0.25–8 (6) |
| E. coli | 1 (EFSA) | 2 | |||
| Amikacin | Salmonella | NA | 16 | – | 4–256 (7) |
| E. coli | 8 | 16 | – |
NA: not available. ECOFF: epidemiological cut‐off value; CBP: clinical breakpoint;
EUCAST ECOFFs and CBPs as available on the EUCAST website, last accessed on 6.3.2019.
All E. coli isolates deriving from the specific monitoring of ESBL‐/AmpC‐/carbapenemase producing E. coli, as well as those randomly selected isolates of Salmonella spp. and E. coli deriving from the routine monitoring that, after testing with the first panel of antimicrobials are found to be resistant to cefotaxime, ceftazidime or meropenem, should be further tested with a second panel of antimicrobial substances as shown in Table 10. This panel notably includes cefoxitin, cefepime and clavulanate in combination with cefotaxime and ceftazidime for the detection of presumptive ESBL and AmpC producers, as well as imipenem, meropenem and ertapenem to phenotypically identify presumptive carbapenemase producers.
Table 10.
Panel of antimicrobial substances, EUCAST ECOFFs and concentration ranges used for testing Salmonella spp. and E. coli isolates, resistant to cefotaxime, ceftazidime or meropenem (second panel)
| Antimicrobial | Salmonella EUCAST ECOFFa | E. coli EUCAST ECOFFa | Concentration range, mg/L (no. of wells) |
|---|---|---|---|
| Cefoxitin | > 8 | > 8 | 0.5–64 (8) |
| Cefepime | NAb | > 0.125 | 0.06–32 (10) |
| Cefotaxime + clavulanic acid | NA | NA | 0.06–64 (11) |
| Ceftazidime + clavulanic acid | NA | NA | 0.125–128 (11) |
| Meropenem | > 0.125 | > 0.125 | 0.03–16 (10) |
| Temocillin | NAc | NAc | 0.5–64 (8) |
| Imipenem | > 1 | > 0.5 | 0.12–16 (8) |
| Ertapenem | NAd | NAd | 0.015–2 (8) |
| Cefotaxime | > 0.5 | > 0.25 | 0.25–64 (9) |
| Ceftazidime | > 2 | > 0.5 | 0.25–128 (10) |
ECOFFs: epidemiological cut‐off values; EUCAST: European Committee on Antimicrobial Susceptibility Testing (last accessed on 6.3.2019); NA: not available.
EUCAST epidemiological cut‐off values available as the Decision 2013/652/EU was drafted (2013). For some antimicrobials, these values have been updated. To allow comparison with the data collected in previous years, the ECOFFs laid down in the legislation have been considered.
For cefepime the cut‐off value used in the analysis for Salmonella spp. was > 0.125 mg/L.
For temocillin the cut‐off value used in the analysis was > 32 mg/L.
For ertapenem, the cuff‐off value used in the analysis was > 0.06 mg/L.
10.2.2. Suggested alterations
10.2.2.1.
Panel for routine monitoring
It is proposed to complement the harmonised panel of substances with amikacin. Amikacin is one of the most commonly used aminoglycosides in hospitals for the treatment of infections by Gram‐negative bacteria in a number of MSs. Main indications are urinary tract infections, bacteraemia and intra‐abdominal infections. There are large differences in use across the EU, with very high use in some MSs with high levels of resistance in Gram‐negatives, like in Italy and Greece, while elsewhere it is not used at all.25 Amikacin is not exactly a last resort antibiotic but neither is it ‘first line’. There is partial cross‐resistance with other aminoglycosides, like gentamicin and tobramycin why it could serve as a marker with gentamicin, being more active for the presumptive detection of the 16S rRNA methyltransferases. Adding amikacin to the first panel will therefore improve the detection of 16S rRNA methyltransferases. These enzymes, conferring resistance to all aminoglycosides except streptomycin, seem to increasingly be associated with carbapenemases, AmpC or ESBLs and FQ resistance in Gram‐negative Enterobacteriaceae – especially outside Europe,26 sensitive detection of this type of resistance is considered to be important for monitoring purposes. In drafting this proposal, the suggestions of inclusion of additional substances collected through the specific questionnaire survey were also carefully addressed and considered of less significance for the first panel. To allow for inclusion of amikacin, it is proposed to slightly reduce some of the dilution ranges in the upper end of the scales, in particular those for ampicillin, nalidixic acid, tetracycline, gentamicin, trimethoprim, sulfamethoxazole and chloramphenicol.
Panel for detection and specific monitoring of presumptive ESBL, AmpC and carbapenemase producers
It is proposed to keep the second panel (Table 10) for the sake of continuity (Appendix O).
10.3. Panel of antimicrobial substances for C. jejuni and C. coli
10.3.1. Overall proposal
Commission Implementing Decision 2013/652/EU followed the ranges recommended by EFSA for erythromycin (1–128 mg/L), ciprofloxacin (0.12–16 mg/L), tetracycline (0.5–64 mg/L) and gentamicin (0.12–16 mg/L) but not for streptomycin (0.25–16 mg/L instead of 1–128 mg/L), the latter being tested on a voluntary basis. In contrast to the EFSA technical specifications (EFSA, 2012a), Commission Implementing Decision 2013/652/EU also includes the testing of nalidixic acid (1–64 mg/L).
Recent findings related to emerging mechanisms of resistance in Campylobacter spp. need to be addressed. The presence of the erm(B) gene in Campylobacter spp. has been reported, and its detection is critical, as this gene is present on mobile genetic elements and is usually responsible for a very high level of macrolide resistance (> 128 mg/L). As the erm(B) gene is usually present on MDRI or plasmids bearing other resistance genes. It may be selected by use of various antimicrobials and the presence of such genetic elements in a strain is typically associated with MDR. Increasing the tested concentrations of erythromycin beyond the actual range (up to 512 mg/L instead of 128 mg/L) should enable the (phenotypic) screening of isolates carrying presumably this resistance gene or MDRI. Such isolates should subsequently be analysed by molecular methods, if possible by WGS.
Efflux pumps encoded by bacteria can protect them from natural substances produced by the host, such as bile, hormones and host‐defence molecules, and they are also able to confer different level of resistance against structurally diverse antimicrobials. In C. jejuni, the expression of the chromosomal efflux pump CmeABC is negatively regulated by CmeR which binds to an inverted repeat (IR) on the cmeR‐cmeA intergenic region. Two recent publications (Yang et al., 2017; Zhang et al., 2017) point out the impact of substitutions, insertions or deletions in the IR: strains with such polymorphisms, sometimes in addition to the gyrA C257T mutation, are more often resistant to tetracycline, doxycycline, florfenicol, chloramphenicol and gentamicin and their ciprofloxacin MICs are higher (4–128 mg/L) than the ones of strains without modifications of the IR (4–8 mg/L (Zhang et al., 2017). Besides these modifications in the expression of the CmeABC pump, Yao et al. (2016) identified in China, isolates with a ‘super’ efflux pump variant of CmeABC (named RE‐CmeABC). The ciprofloxacin MICs of the isolates bearing the RE‐CmeABC and the C257T mutation in gyrA were excessively high (256–512 mg/L). These isolates were also resistant to tetracycline and florfenicol, whereas erythromycin and chloramphenicol MICs varied, respectively, from 4 to 256 mg/L (erythromycin ECOFF: 4 mg/L for C. jejuni) and from 16 to 128 mg/L (chloramphenicol ECOFF: 16 mg/L for C. jejuni). The RE‐CmeABC coding region could be transferred between Campylobacter isolates by natural transformation and the MICs of florfenicol, chloramphenicol, ciprofloxacin, erythromycin and tetracycline were increased in the transformants. The acquisition of such ‘super’ efflux pumps may be problematic, as it provides isolates with simultaneous resistance or decreased susceptibility to diverse classes of antimicrobials, including molecules with a therapeutic interest in humans.
The current harmonised panel does not allow detection of the precise ciprofloxacin MIC of isolates not inhibited by 16 mg/L and chloramphenicol or florfenicol are not included in the list of molecules to test. Testing a larger range of ciprofloxacin concentrations (up to 32 mg/L instead of up to 16 mg/L) and the addition of florfenicol or chloramphenicol to the harmonised set of antimicrobial substances seems a valuable option in order to identify isolates in which modifications of the sequence of the CmeABC pump, and its regulating region, should be further investigated via molecular methods.
The emergence and spread of Campylobacter strains resistant to the current antimicrobials used in human medicine, has already been reported. In the USA, a C. jejuni strain was isolated from a patient suffering from gastroenteritis 3 weeks after returning from Malaysia. The strain was resistant to macrolides (azithromycin, clindamycin, erythromycin and telithromycin), fluoroquinolones, gentamicin and tetracycline. Whole genome sequencing revealed mutations in gyrA and the presence of aadE, aph(2’’)‐If, aph (3’)‐III, tet(O), bla OXA‐184 and erm(B) genes (Chen et al., 2018). In China, 58 isolates (57 C. coli and 1 C. jejuni) from pigs, chickens, ducks and human patients were found to be resistant to erythromycin, clindamycin, ciprofloxacin and tetracycline and 67% of these isolates were also resistant to gentamicin. Characterisation of these isolates showed the erm(B) gene located in transferable MDRI and suggested zoonotic transmission (Wang et al., 2014). In South Korea, a C. jejuni isolated from duck meat was found resistant to all eight molecules tested (azithromycin, clindamycin, ciprofloxacin, nalidixic acid, gentamicin, erythromycin and tetracycline) (Wei et al., 2016). In Europe, in 2016, several MSs (France, Spain, Denmark, Italy and Malta) reported C. jejuni isolates of human origin resistant to the four priority antimicrobials (erythromycin, ciprofloxacin, tetracycline and gentamicin). Similar findings were described for C. coli isolates (EFSA and ECDC, 2018a). Resistance to the same four molecules was reported in two C. jejuni isolates from broilers in Switzerland (EFSA and ECDC, 2018a), in 19 C. coli from pigs and in two C. jejuni from calves in different MSs in 2017.
Because Campylobacter isolates resistant to the four priority antimicrobials are already present in animals and patients, it seems now necessary to consider last‐resort options. As carbapenems are sometimes used for serious and/or systemic infections (Ge et al., 2013; Gaudreau et al., 2016; Kim et al., 2017), their inclusion in the panel was deemed valuable to evaluate and monitor the resistance to this class of antimicrobials, for which few data concerning Campylobacter susceptibility are yet available.
Campylobacter strains from animals or their products analysed by disk diffusion were found intermediate to imipenem (Karikari et al., 2017). The E‐test was recently used to test the susceptibility of human isolates, and all were found susceptible to meropenem (Post et al., 2017) or imipenem (Gaudreau et al., 2014). MICs for imipenem of 7 C. coli and 8 C. jejuni isolates ranged from 0.015 to 0.25 mg/L (Gaudreau et al., 2014). Ohishi et al. (2017) determined the imipenem MICs of 106 human and 79 chicken C. jejuni isolates from Japan by broth microdilution. All isolates were found susceptible, with MICs ≤ 0.06 to 0.12 mg/L. Lehtopolku et al. (2010) studied the susceptibility of clinical erythromycin‐susceptible and erythromycin‐resistant C. jejuni or C. coli isolates by agar plate dilution; the authors reported MIC90 of imipenem as 0.5 and 0.25 mg/L for 19 erythromycin‐resistant and 118 erythromycin‐susceptible isolates, respectively. For meropenem, the MIC90 were, respectively, 0.25 and 0.125 mg/L. All isolates were susceptible to both carbapenems. The French National Reference Center for Campylobacter and Helicobacter obtained ertapenem MICs values of 0.01 to 0.5 mg/L for 319 out of 328 (97.25%) Campylobacter isolates of different species (Lehours, personal communication submitted by e‐mail, January 2019).
Carbapenem‐non‐susceptible Campylobacter strains have already been reported (Hagiya et al., 2018). In Japan, C. coli isolates obtained successively from a patient after long‐term treatment against recurrent bacteremia had an elevated MIC for meropenem (1 mg/L, then 4 mg/L). Lehours et al. (2018) detected C. jejuni isolates with ertapenem MIC of 1 to ≥ 32 mg/L.
10.3.2. Suggested alterations
As in the advised technical specifications of 2012 (EFSA, 2012a), we suggest to remove nalidixic acid from the list of tested molecules. Frequently, resistance to ciprofloxacin parallels resistance to nalidixic acid, and percentages of resistance are equal. In a few cases, isolates are found resistant to ciprofloxacin but not to nalidixic acid. From a public health point of view, the ciprofloxacin is, however, the antibiotic which is used in human medicine and only ciprofloxacin and not nalidixic acid is reported for human isolates in the EU summary report on AMR. It is also proposed to remove the lower concentration of gentamicin (0.12 mg/L) to gain one well for the modifications detailed below. Removal of streptomycin should also be considered, as this antimicrobial is not tested and is not used for human campylobacteriosis and was not mandatory for animal isolates.
The previous EFSA specifications (EFSA, 2012a) mentioned the fact that carbapenems should be included in the surveillance of animals and food, if evidence of carbapenems‐resistance in Campylobacter emerges. As carbapenem‐non‐susceptible or ‐resistant Campylobacter isolates have now been detected in human isolates, the inclusion of this class of antimicrobials (six dilutions) seems justified and needs to be considered.
For a better detection of isolates with erm(B) gene, it is also proposed to increase the upper range of erythromycin concentrations.
Better detection of isolates with modifications of the sequence of the CmeABC pump and its regulating region would be facilitated by increasing the range of concentrations of ciprofloxacin (two more concentrations) and including a phenicol molecule (six dilutions). The removal of nalidixic acid (seven wells) and of streptomycin (seven wells) and the lower concentration of gentamicin (one well) would enable the testing of wider ranges for erythromycin (10 instead on 6 dilutions) and ciprofloxacin (nine instead of eight dilutions), and include a phenicol (six dilutions) and a carbapenem molecule (six dilutions).
The carbapenem to be tested will be chosen among imipenem, meropenem or ertapenem, according to data available in 2019 (QC ranges for the reference strain C. jejuni ATCC 33560, ECOFF values or CBPs. Up to now, no ECOFF and no CBPs have been defined by EUCAST for imipenem, meropenem and ertapenem for Campylobacter. CLSI recommends using the breakpoint of 16 mg/L for imipenem and meropenem which was defined for other non‐Enterobacteriaceae by CLSI M100‐S26. A breakpoint of 1 mg/L is proposed by CA‐SFM 2018 for ertapenem, which, according to recent data (Lehours et al., 2018), seems to better detect carbapenems‐resistant isolates, but QC ranges for the reference strain need to be determined. The suggested alterations are given in Table 11. As 46 antimicrobial‐supplemented wells and two positive control wells are used for each strain, it is still possible to test two strains in a 96‐well plate. A harmonised plate is proposed in Appendix P.
Table 11.
Panel of antimicrobial substances, ECOFFs, CBPs and concentration ranges (in mg/L) for Campylobacter jejuni and C. coli
| Antimicrobial | ECOFF 2013 | CBP 2013 | Advised 2012 | Decision 2013 | ECOFF 2019 | CBP 2019 | Suggested range, mg/L (no. of wells) | |
|---|---|---|---|---|---|---|---|---|
| Erythromycin | C. jejuni | 4 | 4 | 1–128 (8) | 1–128 (8) | 4 | 4 | 1–512 (10) |
| C. coli | 8 | 8 | 8 | 8 | ||||
| Ciprofloxacin | C. jejuni | 0.5 | 0.5 | 0.12–16 (8) | 0.12–16 (8) | 0.5 | 0.5 | 0.12–32 (9) |
| C. coli | 0.5 | 0.5 | 0.5 | 0.5 | ||||
| Tetracycline | C. jejuni | 1 | 2 | 0.5–64 (8) | 0.5–64 (8) | 1 | 1 | 0.5–64 (8) |
| C. coli | 2 | 2 | 2 | 2 | ||||
| Streptomycin | C. jejuni | 4 | NA | 1–128 (8) | 0.25–16 (7) | 4 | 4 | |
| C. coli | 4 | NA | 4 | 4 | ||||
| Gentamicin | C. jejuni | 2 | NA | 0.12–16 (8) | 0.12–16 (8) | 2 | 2 | 0.25–16 (7) |
| C. coli | 2 | NA | 2 | 2 | ||||
| Nalidixic acid | C. jejuni | 16 | NA | – | 1–64 (7) | 16 | 16 | – |
| C. coli | 16 | NA | 16 | 16 | ||||
| Chloramphenicol | C. jejuni | – | – | – | – | 16 | 16 | 2–64 (6) |
| C. coli | – | – | 16 | 16 | ||||
| Penemsa | C. jejuni | – | – | – | – | – |
Ertapenem: 1 (CA‐SFM 2018) Imipenem: 16 (CLSI)b Meropenem: 16 (CLSI)b |
Ertapenem: 0.125–4 (6) |
| C. coli | – | – | – | – | – |
Ertapenem: 1 (CA‐SFM 2018) Imipenem: 16 (CLSI)b Meropenem: 16 (CLSI)b |
– |
ECOFF: epidemiological cut‐off value; CBP: clinical breakpoint; CLSI: Clinical and Laboratory Standards Institute.
2012: According to EFSA (2012a).
2013: According to Commission Implementing Decision 2013/652/EU.
2019: According to http://mic.eucast.org/Eucast2 (last accessed on 6.3.2019)
CA‐SFM 2018: http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM%20V1_0%20FEVRIER%202018.pdf
to be chosen among imipenem, meropenem or ertapenem, according to data available in 2019 (QC ranges for the reference strain C. jejuni ATCC 33560, ECOFF values or clinical breakpoints).
CLSI breakpoint of 16 mg/L recommended for imipenem and meropenem for other non Enterobacteriaceae by CLSI M100‐S26.
10.4. Panel of antimicrobial substances for MRSA
10.4.1. Overall proposal
Commission Implementing Decision 2013/652 does not stipulate concentration ranges for testing the susceptibility of MRSA by broth microdilution. The 2012 EFSA technical specifications (EFSA, 2012b), described a recommended and an optional set of antimicrobials for inclusion in the susceptibility testing panel and suggested concentration ranges which should be tested. Subsequently, a commercial microtitre plate has become available which broadly covers the EFSA recommendation. The current EUCAST CBPs and ECOFFs for S. aureus were reviewed. There have been some changes in the availability of CBPs and ECOFFs from EUCAST since the publication of the 2012 EFSA technical specifications. The proposed panel of antimicrobials is in accordance with the previous EFSA technical specifications (EFSA, 2012b).
Ceftobiprole is an important compound in human medicine for treatment of MRSA infections and is one of several compounds (ceftaroline, daptomycin, tigecycline) available for the treatment of MRSA in humans, which are not used in veterinary medicine. Resistance to these compounds is currently rare in MRSA and inclusion in the proposed panel was considered against the expected loss of epidemiological information obtained from replacement of antimicrobials in the existing panel of compounds.
All MRSA isolates might be expected to demonstrate resistance to penicillin which is included in the current panel. Penicillin could therefore be removed from the panel; however, MSs commented during the preparation of these recommendations that this panel of antimicrobials was also useful for testing other organisms (S. aureus). In that situation, the inclusion of penicillin was useful.
The proposed panel includes two antimicrobial compounds used in human medicine which may be last resort compounds for the treatment of MRSA (linezolid and vancomycin). The other antimicrobials have been included based on their therapeutic applications or use in veterinary medicine, as well as because they can provide useful epidemiological information.
Resistance to certain combinations of antimicrobials can suggest the presence of particular resistance genes, which are noteworthy, because the resistance conferred includes resistance to linezolid or the broad spectrum oxazolidinone, tedizolid. These patterns include those conferred by the genes recently described (Shen et al., 2013; Fan et al., 2016; Antonelli et al., 2018) presented in Table 12. Resistance to linezolid should trigger further genotypic characterisation, specifically including investigation for the presence of these linezolid resistance genes.
Table 12.
List of particular resistance genes whose presence in S. aureus can be suggested by resistance to certain combinations of antimicrobials
| Genes | cfr | optrA a | poxtA c |
|---|---|---|---|
|
Resistance conferred to |
|
|
|
primarily detected in enterococci.
expanded‐spectrum oxazolidinone.
related to optrA (32% protein identity).
10.4.2. Suggested alterations
The gentamicin range of 1–16 mg/L was expanded to 0.5–16 mg/L because the CBP is currently > 1 mg/L. The trimethoprim range of 2–32 mg/L was revised to 1–16 mg/L because the ECOFF is currently > 2 mg/L. The kanamycin range of 4–64 mg/L was shortened to 4–32 mg/L. The fusidic acid range of 0.5–4 mg/L was expanded to 0.25–4 mg/L because the ECOFF is currently > 0.5 mg/L. The penicillin range of 0.12–2 mg/L was revised to 0.06–1 mg/L because the ECOFF and CBP are currently > 0.12 mg/L. The vancomycin range of 1–16 mg/L was shortened to 1–8 mg/L; any resistance to vancomycin detected above the ECOFF or CBP should trigger comprehensive further investigation. The rifampicin range of 0.016–0.5 mg/L was not amended and covers the ECOFF but not the CBP.
The existing MIC ranges in the panel and proposed revisions are described in Tables 13 and 14.
Table 13.
Core panel of antimicrobial substances, ECOFFs, CBPs and concentration ranges (in mg/L) for S. aureus
| Antimicrobial | ECOFF[Link] 2012 | CBP[Link] 2012 | Advised 2012 | Decision 2013 | ECOFF 2019 | CBP 2019 | Suggested range, mg/L | Currently available plate format |
|---|---|---|---|---|---|---|---|---|
| Cefoxitin | > 4 | > 4 | > 4 | –a | > 4 | > 4b | 0.5–16 | 0.5–16 |
| Chloramphenicol | > 16 | > 8 | > 16 | – | > 16 | > 8 | 4–64 | 4–64 |
| Ciprofloxacin | > 1 | > 1 | > 1 | – | > 1 | > 1 | 0.25–8 | 0.25–8 |
| Clindamycin | > 0.25 | > 0.5 | > 0.25 | – | > 0.25 | > 0.5 | 0.12–4 | 0.12–4 |
| Erythromycin | > 1 | > 2 | > 1 | – | > 1 | > 2 | 0.25–8 | 0.25–8 |
| Gentamicin | > 2 | – | > 2 | – | > 4 | > 1 | 0.5–16 | 1–16 |
| Linezolid | > 4 | > 4 | > 4 | – | > 4 | > 4 | 1–8 | 1–8 |
| Mupirocin | > 1 | > 256 | > 1 | – | > 1 | NA | 0.5–2256 | 0.5–2256 |
|
Quinupristin/ Dalfopristin |
> 1 | > 2 | > 1 | – | > 1 | > 2 | 0.5–4 | 0.5–4 |
| Sulfamethoxazole | > 128 | > 1024c | > 128 | – | > 128 | NA | 64–512 | 64–512 |
| Tetracycline | > 1 | > 2 | > 1 | – | > 1 | > 2 | 0.5–16 | 0.5–16 |
| Tiamulin | > 2 | – | > 2 | – | > 2 | NA | 0.5–4 | 0.5–4 |
| Trimethoprim | > 2 | > 4 | > 2 | – | > 2 | > 4 | 1–16 | 2–32 |
|
Trimethoprim/ sulfamethoxazoled |
> 0.5 | > 4 | > 0.5 | – | > 0.5 | > 4 | – e | – e |
| Vancomycin | > 2 | > 2 | > 2 | – | > 2 | > 2 | 1–8 | 1–16 |
NA: not available; ECOFF: epidemiological cut‐off value; CBP: clinical breakpoint; MIC: minimum inhibitory concentration.
Not included as combination.
Not given as a clinical breakpoint by EUCAST, but rather stated that S. aureus with cefoxitin MIC values > 4 are methicillin resistant.
Clinical and Laboratory Standards Institute (CLSI).
Breakpoint expressed as trimethoprim concentration; trimethoprim:sulfamethoxazole in the ratio 1:19.
Not included.
EUCAST ECOFFs and CBPs last accessed on 6.3.2019.
Table 14.
Optional additional antimicrobial substances, ECOFFs, CBPs and concentration ranges (in mg/L) for S. aureus
| Antimicrobial | ECOFFb 2012 | CBPb 2012 | Advised 2012 | Decision 2013 | ECOFF 2019 | CBP 2019 | Suggested ranges ‐ mg/L | Currently available plate format |
|---|---|---|---|---|---|---|---|---|
| Ceftobiprole | NA | NA | None | –a | >1 | >2 | ||
| Kanamycin | > 8 | – | > 8 | – | > 8 | NA | 4–32 | 4–64 |
| Tigecycline | > 0.5 | > 0.5 | > 0.5 | – | NA | > 0.5 | ||
| Fusidic acid | > 0.5 | > 1 | > 0.5 | – | > 0.5 | > 1 | 0.25–4 | 0.5–4 |
| Daptomycin | > 1 | > 1 | > 1 | – | > 1 | > 1 | ||
| Rifampicin | NS | NS | NS | – | > 0.032 | > 0.5a | 0.016–0.5 | 0.016–0.5 |
| Penicillin | NS | NS | NS | – | > 0.125 | > 0.125 | 0.06–1 | 0.12–2 |
| Streptomycin | NS | NS | NS | – | > 16 | NA | 4–32 | 4–32 |
NA: Not available; NS: not specified; ECOFF: epidemiological cut‐off value; CBP: clinical breakpoint.
Not included.
EUCAST ECOFFs and CBPs last accessed on 6.3.2019.
10.5. Panel of antimicrobial resistance for E. faecalis and E. faecium
To provide continuity of monitoring data, it is proposed that those antimicrobials listed in the current legislation should remain unchanged in future testing requirements. ECOFFs and CBPs have been updated where necessary in accordance with the recommendations of EUCAST.
11. Genetic characterisation and complementary molecular analyses
11.1. Complementing the phenotypic AMR monitoring
11.1.1. State of play
Because of the strong added value expected to be brought by implementing WGS, the number of laboratories in various sectors (public health, food safety, veterinary medicine) having implemented WGS methods to analyse microorganisms for different purposes (surveillance, outbreak investigations, research) has started to continuously increase in the EU over the recent years. This development has, however, occurred at different paces among EU/EFTA MSs, as illustrated by the recently published EC/EFSA survey on the availability of the use of WGS methods for the main food‐borne pathogens in animals, food, feed and their related environment in the EU/EFTA countries (EFSA, 2018). By the end of 2016, all European Union Reference Laboratories (EURLs) (7, 100%), almost half of the reporting NRLs (31/71 respondents, 44%) and only a few official laboratories (OLs) (5/76 respondents, 7%) reported conducting WGS of isolates from animals, food or feed. The answering laboratories involved in the survey reporting performing WGS originated from 17 out of the 30 countries (27 MSs, Iceland, Norway and Switzerland) (EFSA, 2018).
Nevertheless, for a number of practical reasons, notably relating to limited (1) harmonisation of existing methodologies, (2) provision or access to WGS platforms by NRLs, (3) bioinformatics analytic pipelines, (4) curation of AMR gene libraries and (5) availability of national bioinformatic expertise, as well as for financial reasons, the complete replacement of the harmonised phenotypic monitoring of AMR in food‐producing animals and food with a fully implemented WGS‐based approach seems premature in 2021 but more realistic, following an incremental approach, towards the second part of the validity period of the upcoming Commission Implementing Decision, a priori around 2025–2026. There are also potential sensitivities in relation to the public availability of national WGS data relating to AMR in zoonotic food‐borne bacteria. Reporting WGS data in a closed environment or reporting partial data, such as AMR genes, will facilitate future developments by taking account of such initial sensitivities.
Currently, the genotypic determination of acquired AMR genes is performed by WGS, without confirming the clinical relevance of the genes. Resulting data are not directly comparable to phenotypic data obtained by broth microdilution, with MIC values interpreted by using CBPs as a comparator. The surveillance of AMR in food‐borne pathogens potentially causing human infections needs to take into account the clinical significance of (clinical) resistance and susceptibility (microbiological resistance). The key requirement for AMR surveillance in human clinical medicine is a need to guarantee that bacteria causing infections are fully treatable with selected defined antibiotics. It is therefore highly likely that AST of food‐borne pathogens among human clinical infections is going to continue to rely on phenotypic monitoring in medium term perspective. Those data are also typically collated to perform epidemiological monitoring of AMR in food‐borne pathogens from human infections. As those monitoring data are also used in joint initiatives which utilise the data collected across the human and veterinary sectors, maintaining comparability capability in outputs is, for this purpose, paramount when measuring the level and frequency of AMR. The ECDC has still recently launched tender for provision of proficiency testing in this area, which also includes expertise in and explore further capabilities of in silico prediction of the resistome.
Consequently, in reviewing and updating the current technical specifications for a harmonised phenotypic monitoring, the EFSA WG has taken into account a constantly evolving situation and likely future developments in the area of WGS.
The AMR monitoring performed under Commission Implementing Decision 2013/652/EU is reported in the EU Summary Report on AMR. Those AMR data are also utilised in a number of other ways, for example, to derive summary indicators of AMR (ECDC, EFSA BIOHAZ Panel and CVMP, 2017) and to investigate possible associations between AMC and AMR (ECDC, EFSA and EMA, 2017). The necessity for comparability of results across the EU is therefore a key requirement of the harmonised AMR monitoring, so that comparable data may continue to be utilised in these further reports and studies. Considering a future situation where WGS is provided by some MSs and phenotypic data by other MSs then issues of comparability arise not only at the level of concordance of the genotypic and phenotypic result for the individual antimicrobial, but also in terms of the comparability of the overall output; this is considered in greater detail in the paragraphs below. Much work has been done to determine whether a genotypic resistance result can be considered equivalent to a resistance phenotype and this has been discussed above; concordance between the methods is therefore one aspect for consideration.
The EU Summary Report on AMR produces a range of outputs from the phenotypic monitoring, such as complete susceptibility, as defined within and detected by the phenotypic monitoring, which equates to no microbiological resistance detected to any substance of the harmonised panel tested. Considering the outputs from WGS, complete susceptibility in this case will mean the detection of no resistance genes to any antimicrobial following the analysis of the genome sequence. The phenotypic and genotypic outputs are, in this case, clearly not equivalent. Similar issues might arise with some of the other patterns of resistance which are noted in the EU Summary Report and how they can be identified from phenotypic or genotypic data. While it could be possible to devise a scheme to ‘filter’ the WGS outputs and exclude resistance genes which would not be detected through phenotypic monitoring in order to harmonise the genotypic and phenotypic results, this would be cumbersome to develop and would result in difficulties in nomenclature. The optimal solution to surmount this issue of comparability is for all MSs to switch from phenotypic to genotypic monitoring at a single convenient point, which will be determined at a future date.
11.1.1.1.
Proposal: To set up a road map to establish methods for comparison outputs, whether simple (e.g. occurrence of resistance) or complex (e.g. complete susceptibility) between phenotypic data (including historical data) and WGS data that will need to have been developed and implemented at the point of the switch from phenotypic to genotypic monitoring, as well as harmonised procedures for handling and reporting WGS data.
In the case of the specific monitoring for ESBL/AmpC/carbapenemase‐producing E. coli, the priority is given to the detection and identification of the genes encoding ESBL/AmpC beta‐lactamases and carbapenemases. The specific monitoring therefore allows flexibility for MSs to change from phenotypic to genotypic monitoring at a convenient point between 2021 and 2026. It will also be possible for MSs to change at a time which is most convenient for them according to their national situation. At 2025, it is suggested that it should become mandatory for all MSs to use WGS‐based monitoring (i.e. WGS) for the specific monitoring the ESBL/AmpC/carbapenemase‐producing E. coli. This provides another immediate advantage in that strain typing of the host E. coli in this situation (MLST for example), provides an additional output extremely useful for epidemiological purposes in conjunction with the identification of genes encoding beta‐lactamases/carbapenemases.
11.1.2. Proposed approach to integration of WGS
Considering the advantages inherent in the WGS technology but also its current limitations, as well as the expected evolvement of the present situation, it is proposed to follow a gradual approach to the integration of WGS within the harmonised AMR monitoring. The integration process could be initiated by complementing the harmonised phenotypic monitoring with WGS in the early phase of the period 2021–2026 for the specific monitoring and subsequently, at a gradual pace over the years, to plan for the possible replacement of the remaining standard phenotypic AST with the systematic use of WGS. The period 2021–2026 should therefore be seen as a transitory period for the implementation of WGS, expected to be a reasonable pace for the MSs to gain experience and acquire WGS technology (Table 15). A rendez‐vous clause providing for review of the situation regarding the implementation of WGS in the MSs in 2023 could be also set.
Table 15.
Proposed panel of antimicrobial substances, EUCAST ECOFFs and CBPs, and concentration ranges to be tested in E. faecalis and E. faecium
| Antimicrobial | Species | ECOFF 2012a | CBP 2012a | Advised 2012b | Decision 2013b | ECOFF 2019c | CBP 2019c | Advised 2019b |
|---|---|---|---|---|---|---|---|---|
| Streptomycin | E. faecalis | > 512 | NA | 16–2048 (8) | – | > 512 | NA | – |
| E. faecium | > 128 | NA | > 128 | NA | ||||
| Gentamicin | E. faecalis | > 32 | NA | 8–1024 (8) | 8–1024 (8) | > 32 | NA | 8–1024 (8) |
| E. faecium | > 32 | NA | > 32 | NA | ||||
| Chloramphenicol | E. faecalis | > 32 | NA | 4–128 (6) | 4–128 (6) | > 32 | NA | 4–128 (6) |
| E. faecium | > 32 | NA | > 32 | NA | ||||
| Ampicillin | E. faecalis | > 4 | > 8 | 0.5–64 (8) | 0.5–64 (8) | > 4 | > 8 | 0.5–64 (8) |
| E. faecium | > 4 | > 8 | > 4 | > 8 | ||||
| Vancomycin | E. faecalis | > 4 | > 4 | 1–128 (8) | 1–128 (8) | > 4 | > 4 | 1–128 (8) |
| E. faecium | > 4 | > 4 | > 4 | > 4 | ||||
| Teicoplanin | E. faecalis | > 2 | > 2 | 0.5–64 (8) | 0.5–64 (8) | > 2 | > 2 | 0.5–64 (8) |
| E. faecium | > 2 | > 2 | > 2 | > 2 | ||||
| Erythromycin | E. faecalis | > 4 | NA | 1–128 (8) | 1–128 (8) | > 4 | NA | 1–128 (8) |
| E. faecium | > 4 | NA | > 4 | NA | ||||
| Q/D | E. faecalis | > 16 | NA | 0.5–64 (8) | 0.5–64 (8) | NA | NA | 0.5–64 (8) |
| E. faecium | > 1 | > 4 | NA | > 4 | ||||
| Tetracycline | E. faecalis | > 4 | NA | 1–128 (8) | 1–128 (8) | > 4 | NA | 1–128 (8) |
| E. faecium | > 4 | NA | > 4 | NA | ||||
| Tigecycline | E. faecalis | > 0.25 | > 0.5 | 0.03–4 (8) | 0.03–4 (8) | > 0.5 | > 0.25 | 0.03–4 (8) |
| E. faecium | > 0.25 | > 0.5 | > 0.25 | > 0.25 | ||||
| Linezolid | E. faecalis | > 4 | > 4 | 0.5–64 (8) | 0.5–64 (8) | > 4 | > 4 | 0.5–64 (8) |
| E. faecium | > 4 | > 4 | > 4 | > 4 | ||||
| Daptomycin | E. faecalis | > 4 | NA | 0.25–32 (8) | 0.25–32 (8) | > 4 | NA | 0.25–32 (8) |
| E. faecium | > 4 | NA | > 8 | NA | ||||
| Ciprofloxacin | E. faecalis | > 4 | NA | – | 0.12–16 (8) | > 4 | NA | 0.12–16 (8) |
| E. faecium | > 4 | NA | > 4 | NA |
NA: not available; Q/D: Quinupristin/dalfopristin; ECOFF: epidemiological cut‐off value; CBP: clinical breakpoint.
EUCAST values (mg/L).
Range of concentrations (mg/L) and no of wells in brackets.
EUCAST values (mg/L). March 2019.
11.1.2.1.
Proposal: To allow flexibility to voluntary MSs/NRLs to use WGS for characterisation of putative ESBL‐/AmpC‐/carbapenemase‐producing E. coli (replacing panels 1 and 2 of the phenotypic susceptibility testing method) within the mandatory specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli in 2021 onwards.
The switch to WGS for indicator commensal E. coli, Campylobacter spp., Salmonella spp. and enterococci should optimally take place in a co‐ordinated way for all MSs and when the necessary developments are in place to allow comparison of the historical (phenotypic) and genotypic results. The phased introduction of WGS for the specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli would mean that in the course of the upcoming Commission Implementing Decision's validity period, for example by 2025, all MSs should have WGS in place.
11.1.2.2.
Proposal: To achieve the goal of implementing WGS across the food and veterinary sectors of the NRLs during the upcoming Commission Implementing Decision's validity period, as well as use in the specific monitoring, it is also proposed that the participation to the ‘Confirmatory Testing’ exercise, possibly using WGS on a voluntary basis, with the support of the EURL‐AR, becomes mandatory.
In practice, the approach can be understood as a step of formalisation of the voluntary EURL/EFSA confirmatory testing. Currently, the confirmatory testing is performed by the EURL‐AR with the support of the MSs providing the relevant bacterial strains on a voluntary basis. The use of WGS and bioinformatics tools within the framework of the EURL/EFSA confirmatory testing has already proved valuable by complementing the already generated phenotypic data. It showed different clonalities among S. Infantis from several European countries. It identified new mcr‐variants and also co‐located carbapenemase and colistin determinants on the same mobile element. This genotypic monitoring under the planned approach would be triggered by certain phenotypic traits of resistance or combinations of resistance traits (phenotypic profiles) obtained from the harmonised phenotypic monitoring. Either all or a representative subset of isolates meeting the inclusion criteria of phenotypic profiles could be offered to the complementary molecular monitoring. The list of inclusion criteria should be reviewed on a regular basis to account for the discovery of new genes and the constantly evolving situation.
Table 16.
Approach to integration of WGS by MSs within harmonised monitoring of AMR over the 2021–2026 period
| Year | WGS applied to | ||||
|---|---|---|---|---|---|
| Specific monitoring of ESBL/AmpC/carbapenemase‐producing E. coli | MSs WGS Confirmatory testing | Indicator E. coli a | Salmonella | Campylobacter | |
| 2021 | Voluntary | Voluntary | NA (Phenotypic) | Phenotypic | Phenotypic |
| 2022 | Voluntary | Voluntary | NA (Phenotypic) | NA (Phenotypic) | NA (Phenotypic) |
| 2023 | Voluntary | Voluntary | NA (Phenotypic) | NA (Phenotypic) | NA (Phenotypic) |
| 2024 | Voluntary | Voluntary | NA (Phenotypic) | NA (Phenotypic) | NA (Phenotypic) |
| 2025 | Mandatory | Voluntary | NA (Phenotypic) | NA (Phenotypic) | NA (Phenotypic) |
| 2026 | Mandatory | Voluntary | TBD | TBD | TBD |
TBD: to be determined based on progress achieved; NA: not applicable; WGS: whole genome sequencing.
In the case of commensal E. coli and Salmonella that are resistant to cefotaxime or ceftazidime further characterisation may be performed by WGS (as under the specific monitoring) on a voluntary basis to 2024, and then on a mandatory basis.
WGS of MRSA recovered from animals or meat may be performed as an alternative to phenotypic testing throughout the period of validity of the upcoming implementing Decision.
11.2. Harmonised protocol and quality criteria
Using WGS, several bioinformatic tools/pipelines/sequencing platforms are available to detect and characterise AMR traits. Some examples are shortly presented in Appendix Q. Various strategies can be used to perform genotypic determination of AMR genes and to predict AMR profiles, such as k‐mer approach, sequence comparisons of the DNA raw reads or of the assembled DNA to reference databases; they may thus vary between the different existing tools. Insufficient quality/harmonisation of WGS processes could lead to inconsistent/incompatible results/data. Quality control parameters are therefore needed to be implemented to improve the reliability and comparability of the results obtained, similarly to the importance of using quality control strains for phenotypic AST. Short‐read WGS has difficulties for managing direct repeats and plasmid analysis and can be misleading in investigating plasmid related AMR genes. This is believed to be improved by development in long read technologies. To implement the use of WGS, it is pivotal that all data are processed using the same/a harmonised approach for comparability reasons. The attainment of a suitable level of proficiency and demonstration of competence in performing the techniques will be an essential preliminary part of the implementation process, in exactly the same way that the robustness of phenotypic AMR testing is currently assessed.
In addition, there are various reference AMR gene databases, which require to be constantly curated as soon as new/novel AMR genes/variants and other resistant determinants are described. The curation of AMR databases is a pivotal prerequisite for the detection of AMR genes. In general, any of these tools will be able to detect only known AMR genes/mechanisms. The use of an inadequate curated database may lead to false negative results by failing to detect AMR gene(s) present or even false positive results, if entries in the database are erroneous. It is also important to take into account that the detection of a gene does not always imply phenotypic resistance since, for this, the gene needs to be expressed; the gene could also comprise an intrinsic resistance gene in the organism tested (for example mcr‐1 in some organisms in the genus Moraxella).
The use of different databases/bioinformatic tools and nomenclature can hamper the comparability of the WGS results obtained. As several comprehensive catalogues of chromosomal mutations and genes mediating AMR have been developed, a designated catalogue needs to be agreed on. Likewise, a harmonised computational approach to predict AMR from WGS data needs to be developed. Also, clear criteria to define a gene as ‘novel’ (i.e. % of identity with existing genes) need to be established, based on published literature stating how to name new genes based on homology to existing variants. The proposed implementation of WGS‐based AMR monitoring therefore requires a strong effort of harmonisation at the main steps of the initial DNA preparation (e.g. DNA extraction, library preparation, sequencing, etc.) followed by the bioinformatic analysis, visualisation and interpretation of results.
11.2.1.
Proposal: The EURL‐AR will continue the effort to provide and organise training in DNA extraction, library preparation, and sequencing. Moreover, the EURL‐AR will recommend harmonised standard operating procedures, protocols and guidelines including quality criteria to be developed in 2019–2020, in liaison with EFSA. The aim is to use the same version of a reference database and pipelines based on benchmarking efforts, and similar parameters/tools for all the steps of the analysis (trimming, quality, assembly, AMR characterisation). It is also proposed that, by the implementation of WGS, the EURL‐AR develops and establishes a genomic‐based proficiency test to assess the quality of the genomes produced by the NRLs and of the AMR determinants/genes detection.
11.3. Reporting of AMR determinants/genes to EFSA
It is proposed that identified AMR genes (as interpreted by the MS) are subsequently submitted to EFSA. Along with the collection of phenotypic antimicrobial susceptibility data, EFSA will collect data on AMR genes reported by the MSs according to a well‐defined AMR gene catalogue to be established and maintained by the EURL‐AR in liaison with EFSA. The data requires knowledge and critical review of the submitted results; a validation step of the submitted results by EFSA and/or the EURL‐AR should be foreseen. The current EFSA/reporting system data model allows submission of data regarding ESBL‐/AmpC‐/carbapenemase encoding genes by the MSs (see Figure 2 and Section 12). The list of current gene data was curated and updated by the EURL‐AR in 2018. Inclusion of additional AMR genes for further antimicrobials will require the update of the data catalogues with the support of the EURL‐AR.
Figure 2.
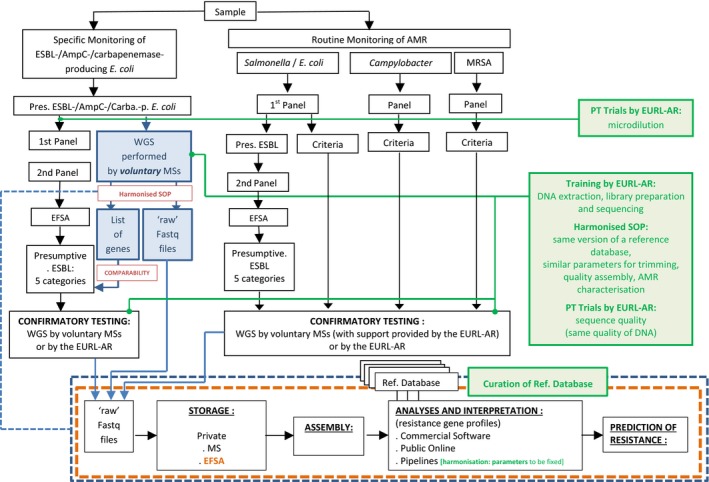
Flow chart presenting a generic process of use of WGS by the MSs on a voluntary basis for the specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli within the framework of the harmonised monitoring of AMR, with possibility to report either encoding genes (by now) or sequences (in future) to EFSA, and the necessary quality steps
Finally, it is proposed that WGS AMR data should be presented in the EU‐Summary Report and/or in interactive tools. The comparability with the results for previous years data is an issue which will need to be considered and any disparities investigated; for some antimicrobials (the aminoglycosides, further mentioned below), a large number of different resistance genes may confer similar phenotypes of resistance and comparability between all results may not be straightforward or easily achieved.
There are potential sensitivities in relation to the availability of national WGS data relating to AMR in zoonotic/indicator food‐borne bacteria and reporting in a closed environment will facilitate future development by taking account of such sensitivities (see Section 11.4).
11.4. Possible further developments in the near future
WGS is considered as a powerful tool for epidemiological surveillance of AMR along the food chain. In addition to AST, WGS can also be used to perform, for example, source attribution, outbreak and virulence investigation, which makes it a multipurpose tool and increases its cost‐effectiveness.
Recent literature has already demonstrated a high concordance27 between the results of WGS‐based genotyping of strains for AMR and the phenotypic antimicrobial susceptibility tests. The concordance depends on the bacterial species, the antimicrobial substances and the data collection considered. Main disagreements in some of the studies notably concerned the combinations Salmonella/streptomycin (Neuert et al., 2018) and Salmonella/E. coli/spectinomycin (Zankari et al., 2013) and E. coli/β‐lactam (DTU, IZSLT, BfR, NIPH, NVRI, PHE, APHA and IZSV, 2018) and failure to detect chromosomal point mutations which upregulate AMR by some of the tools in use.
The rapid technological progress expected to occur before 2026 is acknowledged. As technological developments are constantly and rapidly evolving, it is believed that, in the future, WGS results will allow predicting AMR phenotypes and even MIC values. Although WGS‐based AMR surveillance programmes are being increasingly implemented all over the world, only the NARMS WGS‐based AMR monitoring system appears to be harmonised. WGS combined with the use of bioinformatics tools for AMR analysis needs to be based on common quality criteria and harmonised procedures, thus, has the potential to open perspectives for a EU‐wide WGS data collection, leading to a harmonised AMR monitoring system.
WGS‐based AMR surveillance may provide results for many antimicrobials which are satisfactorily concordant with the equivalent phenotypic test results. Where results are concordant, comparability with historical data and between MSs using WGS or phenotypic monitoring methods is likely to be rapidly achieved. The software necessary to collate the WGS data outputs will need to be developed. The proposed key indicator of complete susceptibility refers to full susceptibility to a defined panel of antimicrobials; an equivalent measure or a suitably revised measure needs to be adopted for WGS data. There are a number of similar examples where an approach needs to be developed to utilise the WGS data in an equivalent or enhanced comparable way to the phenotypic data. Approaches to report these data also need development, as well as procedures for outbreak investigation, source attribution and investigation of virulence factors in relation to the AMR data.
Developments may prove faster than expected and setting priorities for development is likely to result in rapid, incremental improvement in the data obtained from surveillance programmes. There is also likely to be strong synergy in this area with other related initiatives such, as the database under development by ECDC and EFSA for the investigation of multicountry food borne outbreaks of disease. ECDC and EFSA have jointly addressed the EC mandate ‘Request for technical support to collect and analyse whole genome sequencing (WGS) data in the joint ECDC‐EFSA molecular typing database’ (EFSA, 2019). Once the planned EFSA‐ECDC WGS Joint database will be available, WGS sequences can be submitted to this Joint ECDC‐EFSA database by the MSs on a voluntary basis (if not already publicly available), so that joint analysis across sectors (human, animal and food) can be performed (see Figure 2). This system is planned to offer a close environment to host sequences and enable to upload/download sequences from public repositories (as wished by the data providers), and to use of a harmonised pipeline for the analysis.
Consideration may still need to be given to maintaining a degree of limited phenotypic monitoring, to ensure that emerging resistance is not overlooked.
12. Collection and reporting of molecular typing data/phenotypic data
The EFSA has developed a harmonised and streamlined reporting system of AMR data to be transmitted by the EU MSs and other reporting countries under the framework of Directive 2003/99/EC and Commission Implementing Decision 2013/652/EU in food‐producing animals and foodstuffs derived thereof. The system notably aims to ensure that the data collected are relevant and easy to analyse at the EU level, by applying a number of validity checks to the reported data. The reporting of AMR data and text forms relies on the transmission of eXtensible Markup Language (XML) files, through the Data Collection Framework (DCF) of the EFSA, in accordance with detailed guidelines.
A reporting manual is also drafted by EFSA on a yearly basis to provide detailed guidance on reporting AMR data. The manual typically applies to the bacterial agents, antimicrobial substances, animal populations and food categories to be reported. Instructions are given on the description of the sampling and monitoring schemes, as well as on the analyses of the results in the national reports. The manual specifically covers Salmonella, C. coli, C. jejuni, indicator commensal E. coli, indicator enterococci and MRSA, as included in the current data collection. These instructions are applicable to reporting national data on AMR in bacterial agents and national overview information on the general evaluation of the AMR situation and the process of monitoring AMR in relative text forms, through the DCF. Specific guidance is also included for reporting on the prevalence, genetic diversity and AMR of MRSA from food‐producing animals and food derived thereof. The manual also incorporates specific guidance for the mandatory reporting of data on Salmonella spp. and commensal indicator E. coli producers of ESBLs/AmpC/carbapenemases obtained from the harmonised routine monitoring, and data on ESBL‐/AmpC‐/carbapenemase‐producing E. coli derived from the specific monitoring as well as the voluntary reporting of data on the specific monitoring of carbapenemase‐producing E. coli.
Additional technical information about AMR data reporting can be also found in Data dictionaries and guidelines for reporting data on zoonoses, AMR and food‐borne outbreaks, issued on a yearly basis.
12.1.
Proposal: EFSA will continue using the same data collection system of phenotypic antimicrobial susceptibility data, using the same data model and collecting the same data/information. The system already in place enables the collection of ESBL/AmpC/carbapenemase‐encoding genes that could be reported by the MSs voluntarily implementing the specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli by using WGS. The genes will need to be reported according to a well‐defined AMR gene catalogue to be established and maintained by the EURL‐AR and EFSA.
13. Review of the routine monitoring of AMR
It is recommended that these technical specifications be re‐assessed and updated regularly in the light of the results of the first monitoring campaigns, the technological developments and the trends in the constantly emerging and evolving area of AMR.
Abbreviations
- AMC
antimicrobial consumption
- AMEG
Antimicrobial Advice ad hoc Expert Group
- AmpC
AmpC β‐lactamases
- AMR
antimicrobial resistance
- AST
antimicrobial susceptibility testing
- CA
community‐associated
- CASA
Campylobacter selective chromogenic medium
- CBP
clinical breakpoints
- CFA
campyfood agar plate
- cgMLST
core‐genome multilocus sequence type
- CHL
chloramphenicol
- CIA
critically important antimicrobial
- CIP
ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- CSS
caecal samples from healthy animals at slaughter
- DCF
Data Collection Framework
- ECDC
European Centre for Disease Prevention and Control
- ECOFF
Epidemiological cut‐off value
- EMA
European Medicines Agency
- ERY
erythromycin
- ESC
extended‐spectrum cephalosporins
- ESBL
extended‐spectrum β‐lactamases
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- EURL‐AR
European Union reference laboratory for antimicrobial resistance
- FBO
food business operator
- GEN
gentamicin
- HA
healthcare‐associated
- HPCIA
highest priority critically important antimicrobials
- HTS
high throughput sequencing
- IR
inverted repeat
- JIACRA
Joint Interagency Antimicrobial Consumption and Resistance Analysis
- LA
livestock‐associated
- LA‐MRSA
livestock‐associated methicillin‐resistant Staphylococcus aureus
- M
mandatory monitoring
- MALDI‐TOF
Matrix Assisted Laser Desorption/Ionization ‐ Time of Flight
- mCCDA
modified charcoal‐cefoperazone‐deoxycholate agar
- MDR
multi‐drug resistance/multi‐drug resistant
- MDRI
multidrug resistance islands
- MIC
minimum inhibitory concentration
- MLST
multilocus sequence type
- MRSA
methicillin‐resistant Staphylococcus aureus
- MS
Member State
- NRL
National Reference Laboratories
- OL
Official laboratory
- PCR
polymerase chain reaction
- PCU
population correction unit
- PFGE
pulse field gel electrophoresis
- PHC
process hygiene criteria
- POS
positive control
- PT
proficiency testing
- PVL
Panton–Valentin leukocidin
- SCENIHR
Scientific Committee on Emerging and Newly Identified Health Risks
- SNCP
Salmonella National Control Program
- SRS
Simple random sampling
- ST
sequence type
- STR
streptomycin
- TET
tetracycline
- V
voluntary monitoring
- WGS
Whole genome sequencing
- XML
eXtensible Markup Language
Appendix A – Emerging antimicrobial resistance issues since 2013
A.1. Plasmid‐mediated resistance to colistin
In November 2015, transferable resistance to the polymyxin antimicrobial colistin, mediated by the mcr‐1 gene, was reported in E. coli from pigs in China and some other countries in the Far East and Europe (Liu et al., 2016). Subsequently mcr‐1 has been identified in retrospective studies of colistin‐resistant E. coli and Salmonella from many countries world‐wide (Skov and Monnet, 2016). More recently, a novel mcr gene, mcr‐2, has been identified in passive screening of porcine and bovine colistin‐resistant E. coli isolates made in Belgium between 2011 and 2012 that did not show presence of mcr‐1. Of particular note is that this mcr gene was associated with highly transmissible IncX4 plasmids (Xavier et al., 2016). ECDC have published a rapid risk assessment of the implications for human medicine of plasmid‐mediated colistin resistance (ECDC, 2016). As time progresses, more and more different mcr genes have been described; currently, the latest mcr‐gene has been named mcr‐8 (Wang et al., 2018).
In 2016, The EU Antimicrobial Advice ad hoc Expert Group (AMEG) noted that there are wide variations in the use of colistin in EU MSs adjusted for the biomass under exposure (kg livestock, expressed in population correction unit (PCU)) between countries (EMA/AMEG, 2016). Countries with intensive livestock production can have a level of use below 1 mg/PCU (e.g. Denmark and the UK) or much higher, up to 20–25 mg/PCU (Italy and Spain). The AMEG have therefore recommended that for the ‘high and moderate consumers’ the target and desirable levels are set at 5 mg/PCU and 1 or below 1 mg/PCU, respectively, based on the observations on the level of use in other countries. The AMEG further recommended that more information should be gathered to determine the minimum level of colistin use that can be achieved, while maintaining animal welfare and preventing the increased use of other critically important antimicrobials (CIAs).
More recent studies have demonstrated the presence of the mcr‐1 gene mediated by an IncI2 plasmid in isolates of E. coli and Klebsiella pneumoniae from sewage from different waste water plants in Spain (Ovejero et al., 2017), in colistin‐resistant clinical isolates of K. pneumoniae isolated in Laos and France (Rolain et al., 2016) and mcr‐1.2, a new mcr variant carried on a transferable plasmid from a colistin‐resistant KPC carbapenemase‐producing K. pneumoniae strain of human origin in Italy (Di Pilato et al., 2016). These studies clearly demonstrate the ability of mcr‐carrying plasmids to become disseminated in potentially highly pathogenic strains in humans and the environment in Europe.
A.2. The emergence and spread of certain multidrug‐resistant Salmonella serovars
Salmonella Infantis is one of the most commonly isolated serovars and MDR strains have recently been emerging worldwide. Comparative analyses between pre‐emergent and the clonal emergent S. Infantis populations demonstrated the fixation of adaptive mutations in the DNA gyrase (gyrA) and nitroreductase (nfsA) genes, conferring resistance to quinolones and nitrofurans, respectively, and the carriage of an emergent‐specific plasmid, designated pESI. This self‐transferred episome is a mosaic megaplasmid (~ 280 kb), which increases bacterial tolerance to environmental mercury (mer operon) and oxidative stress, and provides further resistance to tetracyclines, sulfamethoxazole and trimethoprim, most likely due to the presence of tetRA, sulI and dfrA genes, respectively. Moreover, pESI codes for the yersiniabactin siderophore system and two novel chaperone‐usher fimbriae. In vitro studies established that pESI conjugation into a plasmidless S. Infantis strain results in superior biofilm formation, adhesion and invasion into avian and mammalian host cells. In vivo mouse infections demonstrated higher pathogenicity and increased intestinal inflammation caused by an S. Infantis strain harbouring pESI compared with the plasmidless parental strain (Aviv et al., 2014).
MDR Salmonella Kentucky strains such as ST198 (see Appendix A above) have been increasingly isolated from across Europe since 2009. Multiple mutations within chromosomal genes gyrA and parC are responsible for high‐level ciprofloxacin resistance. One of the isolates from Poland was extended‐spectrum β‐lactamase‐ (ESBL) positive: the strain 1643/2010 carried a conjugative 167,779 bp plasmid of IncA/C family. The sequence analysis revealed that it carried a bla CTX‐M‐25 gene and an integron with another β‐lactamase‐encoding gene – bla OXA‐21. This is the first known report of a CTX‐M‐25 encoding gene both in Poland and in S. Kentucky world‐wide, as well as in the IncA/C plasmid. Analysis of the integron showed a novel arrangement of gene cassettes – aacA4, aacC‐A1 and bla OXA‐21 where the latter might result from an intergeneric gene transfer. The study confirmed that the S. Kentucky population isolated in Poland belongs to global epidemics of high level fluoroquinolone‐resistant clone ST198 that can carry rare β‐lactamase genes (Wasyl et al., 2015).
The identification in the Netherlands in 2016 of clonal clusters shared by ESC‐resistant S. Heidelberg strains in food‐producing animals and poultry meat that can cause human infections (Liakopoulos et al., 2016) underscores the risk for potential zoonotic or food‐borne transmission of these strains to humans. Although no human infections linked to these contaminated products have been yet documented in the Netherlands, the risk of potential zoonotic or food‐borne transmission of ESC‐resistant S. Heidelberg strains further highlights the necessity for active surveillance and intervention strategies by public health organisations.
The recent identification in Africa of highly drug‐resistant epidemic lineages of S. Enteritidis capable of bloodstream invasion (Feasey et al., 2016) is a major cause of international concern. Such strains, which exhibit resistance to almost all commonly used antimicrobials encoded by a plasmid which also carries a repertoire of virulence genes, are not primarily zoonotic and are transmitted by person‐to‐person spread in the community and hospitals. Nevertheless, it is important to realise that isolates of these organisms have been isolated from travellers returning to EU countries, and as such can potentially influence findings on multiple resistance to antimicrobials such as tetracyclines and other commonly used antimicrobials in cases of S. Enteritidis in EU countries.
A.3. The ongoing spread of LA‐MRSA in certain high‐risk groups of workers in direct contact with live animals and of MRSA in pigs and other species
A zoonotic reservoir in food production animals involving a specific clone, MRSA ST398, which spreads extensively in animals and is found in retail meat, has been increasingly noted in several EU countries. As such this poses a potential threat to public health, as people in contact with food production animals are at higher risk of colonisation. The most probable transmission route seems to be by contact (Huang et al., 2014), and dust in intensive animal housing is often contaminated with MRSA ST398 (Cole et al., 2000; Broens et al., 2008).
Although the clonal relationship between MRSA strains of CC398 is clear in livestock and people, this is less obvious in horses. Small companion animals typically share MRSA strains that seem to originate from and exchange with a human reservoir (Catry et al., 2010), as well as harbouring other resistant species of staphylococcus (Cohn and Middleton, 2010). Most isolates from clinically infected animals carry numerous genetic elements related to AMR and virulence genes, and a phi3 prophage encoding immune‐modulating proteins that is associated with animal‐to‐human transmission. Recent findings suggest clonal expansion and dissemination of a new subpopulation of CC398 isolates, responsible for invasive infections in various animals, with a considerable potential to colonise and infect humans; probably greater than that of the original pig/human‐adapted CC398 isolates (Van der Mee‐Marquet et al., 2014). Recently, specific clones of MRSA have been reported as circulating between pigs and dairy cattle in Italy, raising the possibility of still wider dissemination of such organisms between species (Feltrin et al., 2016).
Livestock‐associated MRSA (LA‐MRSA) was first reported in pigs in 2005 (Voss et al., 2005). To date, LA‐MRSA is only prevalent in certain high‐risk groups of workers in direct contact with live animals and the spatial distribution of MRSA genotypes suggests interspecies transmission and colonisation of different populations, communities, animals and their products, although a relevant association between contamination of food products and consumers has not been confirmed. The carrier rate of MRSA is high in food handlers in restaurants in some countries and this can lead to contamination of food (Alsamarai et al., 2015). Nevertheless, the proportion of human MRSA infections that are due to LA‐MRSA strains remains low overall in the EU and in most EU MSs.
A.4. Transferable resistance to macrolides in Campylobacter spp. from food‐producing animals
Resistance to macrolides in Campylobacter spp. has generally been the result of mutations in ribosomal RNA or ribosomal proteins and these mutations are thought to have incurred fitness costs, accounting for the low occurrence of erythromycin resistance in many countries (Wang et al., 2014). Ribosomal mutations can confer high‐level erythromycin resistance (Gibreel and Taylor, 2006). Transferable resistance to the macrolide erythromycin was first described in campylobacter isolates from food‐producing animals (including pigs, chickens and ducks) from China in 2014 (Qin et al., 2014; Wang et al., 2014) and frequently resulted in high level resistance to erythromycin, with MICs recorded at > 512 mg/L. Resistance was conferred by the rRNA methylase gene erm(B), which can be associated with either chromosomal multidrug resistance islands or transferable plasmids. More recently the transferable erm(B) gene has been identified in a C. coli isolate from broilers and turkeys in Spain (Florez‐Cuadrado et al., 2016) and from a broiler isolate in Belgium (Elhadidy et al., 2019), and this may provide a means whereby macrolide resistance can spread rapidly in Campylobacter spp. in animals in the EU. The situation may be compared with tetracycline resistance, which is frequently plasmid‐mediated in Campylobacter spp., and is frequently detected in many EU MSs at high levels.
A.5. Carbapenemase‐producing Enterobacteriaceae from animals and food
The emergence and spread of bacteria with acquired resistance to carbapenems is of public health concern, and thus their presence and/or emergence in bacteria from food‐producing animals and foods deserves attention, as these bacteria could constitute a reservoir of such bacteria for humans. There are a few scientific reports of carbapenemase‐producing bacteria isolated from farm animals and food derived thereof as well as from companion animals from different parts of the world (Fernández et al., 2018; Köck et al., 2018). In Europe, findings of carbapenemase‐producing Enterobacteriaceae in farm animals and food derived thereof described in the literature include E. coli and S. Infantis with VIM‐1 from pigs and poultry from Germany (Fischer et al., 2017; Irrgang et al., 2017), E. coli with OXA‐181 from pigs and E. coli with VIM‐1 from seafood from Italy as well as Enterobacter cloacae with IMI‐1 from imported frozen seafood sampled in the Netherlands (Brouwer et al., 2018; Köck et al., 2018). One of the studies from Germany also describes long term persistence of carbapenemase‐producing E. coli carrying VIM‐1 at a pig farm (Irrgang et al., 2017). Furthermore, Pseudomonas fluorescens with VIM‐1 and IMP‐1 have been isolated from ready to eat salads in Italy (Iseppi et al., 2018). Salad was also the suspected source of Citrobacter freundii with VIM‐1 causing an outbreak at a hospital in Germany (Pletz et al., 2018).
Since the implementation of the harmonised monitoring, some of these isolates were reported to EFSA: In 2015, Germany reported the detection of a VIM‐1‐producing E. coli isolated from a fattening pig sample and Belgium reported the detection of a VIM‐1‐producing E. coli isolated from a pig meat sample. In 2016, Romania reported three isolates of OXA‐162‐producing E. coli isolated from broilers (n=2) and broiler meat (n=1) (EFSA and ECDC, 2017a, 2018a). In 2017, Germany reported once more the detection of a VIM‐1‐producing E. coli belonging to a different genetic type than the isolates detected in previous years, suggesting the spread of the bla‐VIM‐1 plasmid (Irrgang et al., 2017).
Appendix B – Questionnaire of the Specific Survey on harmonised AMR monitoring
1.
To better assess the vision of the MSs and to gauge the potential for further harmonisation of procedures and the degree of support from MSs for further AMR monitoring, an online survey was performed in June 2018 and targeted the network of the NRL‐ARs and the EFSA Network on zoonoses/AMR monitoring, using a digital Specific Questionnaire on Harmonised AMR Monitoring in 2017 and/or 2018 was drafted by the EFSA WG and the EURL‐AR using the tool ‘EU‐survey’.
Appendix B can be found in the online version of this output, under the section ‘Supporting information’, at: http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2019.5709/full
Appendix C – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: Isolation of Campylobacter spp. for antimicrobial susceptibility testing in 2017/2018
1.
Q1: How many laboratories are involved in your MS/country?

Albania and Switzerland have one single laboratory each, while Norway has 2 and Iceland has 4.
C.1. Isolation method of Campylobacter spp. from caecal content samples
Q2: Regarding the method of isolation of Campylobacter from caecal content samples, was there a single method used in the ‘field’ laboratory(ies) in charge of isolating Campylobacter strains for AMR monitoring?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes (if there is a single ‘field’ laboratory in charge, or there are several laboratories using the same method, please, answer ‘Yes’) |

|
23 | 85.2% |
| No, different methods exist |

|
4 | 14.8% |
| No Answer | – | 0 | 0% |
Q3: Was an enrichment step performed?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
4 | 14.8% |
| No |

|
23 | 85.2% |
| No Answer | 0 | 0% |
In 23 out of the 27 MSs, a single method is used for isolating Campylobacter strains for AMR monitoring in either one or several laboratories. An enrichment step, consisting in incubating in either Preston or Bolton broth, at 41.5+/−1°C for 40/48 h, is performed in four countries only. It is worth noting that the enrichment step is not recommended by the NF EN ISO 10272‐1:2017 Standard for samples harbouring high numbers of Campylobacters.
Iceland is the only non‐MS in which different methods are used. In none of the non‐MS is performed an enrichment step.
Q4: Which bacterial agar media plates for isolating Campylobacter spp. were used in 2017 and/or 2018?
Twenty‐five MSs and all non‐MSs use mCCDA medium, which is the media recommended by the NF EN ISO 10272‐1:2017 Standard, whereas only 2 MSs used Karmali agar. Oxoid provides the media to 19 of the MSs and three of the non‐MSs.
A second plate is used in 16 MSs, whether Preston, Karmali, blood agar, CFA, CASA, or Skirrow.
All countries incubate the plates at 41.5+/−1°C, except Slovakia, which reports doing so at 37°C. Twenty‐six out of the 27 MSs (and also three non‐MSs) incubate the plates for 40–48 h, while the other two countries incubated for 72–120 h.
Q4.1. Which method(s) of inoculation was/were used?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Cotton swab |

|
5 | 18.5% |
| Sterile loop of 1 microliter |

|
3 | 11.1% |
| Sterile loop of 10 microliters |

|
18 | 66.7% |
| Other method |

|
3 | 11.1% |
| No Answer |

|
1 | 3.7% |
Other methods include a glass stick, spatula or diluting the caecal contents in PBS and then inoculating and spreading a sample of the suspension on the surface of the plate.
AL uses the three main methods. NO and CH use a sterile loop (of either 1 or 10 microliters), whereas IS uses the suspension method described above.
Q5: Which method(s) was(were) performed for the identification of Campylobacter isolates?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Microscope exam (Gram stain, morphology, motility) |

|
13 | 48.2% |
| PCR |

|
13 | 48.2% |
| MALDI‐TOF Mass Spectrometry |

|
14 | 51.9% |
| Biochemical tests |

|
12 | 44.4% |
| Other approaches |

|
2 | 7.4% |
| No Answer | 0 | 0% |
Seven MSs out the 13 countries carrying out PCR use the protocol described by Denis et al. in 1999.28 The most common biochemical tests performed were oxydase test (9), catalase (10), hippurate (7) and indoxyl29 (6). Those MSs who replied ‘Other approaches’ tested the ability to grow aerobically at 25°C or in the presence of nalidixic acid and cephalotin.
Three non‐MSs used microscope exam, while two used MALDI‐TOF and other two used PCR.
Q6: How many characteristic colonies were selected from primary culture plates before considering the caecal sample as negative for Campylobacter spp., C. jejuni or C. coli ?
The mean number of colonies selected is 4.1 for Campylobacter genera, whereas it is 3.7 for C. jejuni and 3.3 for C. coli. In non‐MSs, this number is 4 for Campylobacter genera and 3.5 for both species.
Q7: Under which storage conditions were Campylobacter spp. isolates provided/sent to the NRL‐AR?
| Bars | Answers | Ratio | |
|---|---|---|---|
| Broth |

|
5 | 18.5% |
| Beads |

|
3 | 11.1% |
| Transport swabs |

|
9 | 33.3% |
| No Answer |

|
10 | 37.0% |
The temperatures of storage and transport vary among the countries that provided responses. Non‐MSs did not provide information about the storage conditions.
| Caecal content samples | # MSs | |
|---|---|---|
| Storage temperature | Transport temperature | |
| −80°C to ‐45°C | −80°C to +20°C | 10 MSs |
| +3°C to +8°C | 4°C to 25°C | 8 MSs |
| 22°C to 25°C | 22°C to 25°C | 4 MSs |
C.2. Isolation method of Campylobacter spp. from meat samples
Q8: Was Campylobacter spp. isolated from meat for the purpose of antimicrobial susceptibility testing in 2017 and/or 2018?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
11 | 40.7% |
| No |

|
16 | 59.2% |
| No Answer | 0 | 0% |
Q9: Regarding the method of isolation of Campylobacter from meat samples, was there a single method used in the ‘field’ laboratories in charge of isolating Campylobacter strains for AMR monitoring?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yesa |

|
10 | 37.0% |
| No, different methods exist |

|
1 | 3.7% |
| No Answer |

|
16 | 59.3% |
i.e. if there is a single ‘field’ laboratory in charge, or there are several laboratories using the same method.
Q10: Was an enrichment step performed?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
8 | 29.63% |
| No |

|
3 | 11.11% |
| No Answer |

|
16 | 59.26% |
The enrichment method generally consisted in incubating either Bolton or Preston media at 41.5+/−1°C for 40/48 h, which is what is recommended by the NF EN ISO 10272‐1:2017 Standard for samples with low numbers of campylobacters.
Switzerland is the only non‐MS that provided information about the isolation of Campylobacter from meat samples, using a single method, with an enrichment step.
Q11: Which bacterial agar media plates for isolating Campylobacter spp. were used in 2017 and/or 2018?
In 9 of the 10 MSs that answered, as well as in Switzerland, mCCDA agar is used, which is the recommendation of the NF EN ISO 10272‐1:2017 Standard. Oxoid is typically the main provider. All countries, one excepted, incubate at 41.5+/−1 °C for 40–48 h.
A second plate, either CFA, Karmali/Skirrow, modified Preston, is also used in six countries.
Q11.1: Which method(s) of inoculation was/were used?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Cotton swab |

|
1 | 3.7% |
| Sterile loop of 1 microliter |

|
1 | 3.7% |
| Sterile loop of 10 microliters |

|
6 | 22.2% |
| Other methodsa |

|
3 | 11.1% |
| No Answer |

|
17 | 63.0% |
The other methods used consisted of using the ISO 10272‐1 method for inoculation, or in preparing a suspension and then spreading it in the plate.
Q12: Which method(s) was/were performed for the identification of Campylobacter spp. isolates?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Microscope exam (Gram stain, morphology, motility) |

|
5 | 18.5% |
| PCR |

|
5 | 18.5% |
| MALDI‐TOF Mass Spectrometry |

|
7 | 25.9% |
| Biochemical tests |

|
5 | 18.5% |
| Other approachesa |

|
2 | 7.4% |
| No Answer |

|
16 | 59.3% |
For other approaches, countries used the VIDAS technique of BioMerieux or tested the absence of aerobic growth.
Q13: How many characteristic colonies were selected from primary culture plates before considering the meat sample as negative for Campylobacter spp., C. jejuni or C. coli ?
Nine countries (plus Switzerland) tested 5 colonies of Campylobacter spp., while two countries tested only 1. Seven countries (plus Switzerland) tested 5 colonies of the Campylobacter species.
Q14: Under which storage conditions were Campylobacter spp. isolates provided/sent to the NRL‐AR?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Broth | 0 | 0% | |
| Beads |

|
1 | 3.7% |
| Transport swabs |

|
8 | 29.6% |
| No Answer |

|
19 | 70.4% |
C.3. Standard used in the ‘field’ laboratories for isolating Campylobacter spp. for AMR monitoring
Q15: Is the European standard EN ISO 10272‐1 used in the ‘field’ laboratory(ies) for any purpose?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
21 | 77.8% |
| No |

|
3 | 11.1% |
| No Answer |

|
3 | 11.1% |
AL, IS and NO also use the standard ISO for any purpose, while CH did not provide information for these questions.
Q16: Has the European standard EN ISO 10272‐1 been used in the ‘field’ laboratory(ies) within the framework of the harmonised AMR monitoring in 2017 and/or 2018?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
16 | 59.3% |
| No |

|
8 | 29.6% |
| No Answer |

|
3 | 11.1% |
NO has used the standard ISO, while AL and IS have not.
Q17: Is(Are) the ‘field’ laboratory(ies) accredited for the European standard EN ISO 10272‐1?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
19 | 70.4% |
| No |

|
5 | 18.5% |
| No Answer |

|
3 | 11.1% |
In NO, laboratories are accredited.
C.4. Pooling of sample types for isolating Campylobacter spp. within the framework of the harmonised monitoring of AMR in Campylobacter spp.
Q18: For a given flock of broilers (epidemiological unit), from how many carcases does the sample of caecal content derive?
the caecal content may therefore derive from 1 or 2 caeca of the carcase.
when more than one broiler carcases were used, the number of sampled carcases was ten for 15 MS
Q19: For a given flock of fattening turkeys (epidemiological unit), from how many carcases does the sample of caecal content derive?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| AMR monitoring in Campylobacter spp. from turkeys is not performed |

|
14 | 51.9% |
| 1 carcasea |

|
4 | 14.8% |
| More than 1 carcaseb |

|
7 | 25.9% |
| No Answer |

|
2 | 7.4% |
The caecal content may therefore derive from 1 or 2 caeca of the carcase.
When more than one turkey carcases were used, the number of sampled carcases was ten for 5 MS
Q20: For a given batch of slaughter pigs (originating from the same farm ‐ epidemiological unit), from how many carcases does the sample of caecal content derive?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| AMR monitoring in Campylobacter spp. from pigs is not performed |

|
17 | 63.0% |
| 1 carcase |

|
6 | 22.2% |
| More than 1 carcase |

|
3 | 11.1% |
| No Answer |

|
1 | 3.7% |
Q21: For a given batch of calves of less than 1 year of age (originating from the same farm ‐ epidemiological unit), from how many carcases does the sample of caecal content derive?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| AMR monitoring in Campylobacter spp. from calves is not performed |

|
21 | 77.8% |
| 1 carcase |

|
4 | 14.8% |
| More than 1 carcase |

|
1 | 3.7% |
| No Answer |

|
1 | 3.7% |
Overview graph of sample pooling:
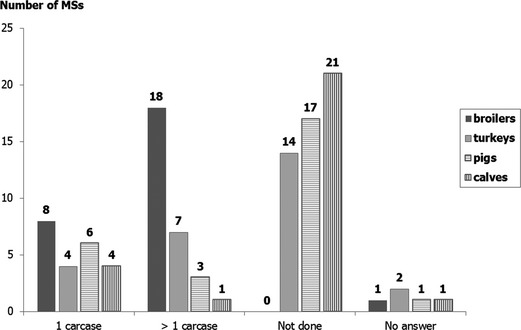
In the case of the four non‐MSs, samples derived from more than 1 carcase (between 4 and 10) for broilers. For the rest of the matrices, only NO sampled in turkeys (10 carcases) and slaughter pigs (1 carcase).
C.5. Procedures used for primary culture of C. jejuni and C. coli over week‐end periods
Q22: Does culture work continue over the week‐end period?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes, as a general rule (To Q23–26) |

|
11 | 40.7% |
| Yes, sometimes (To Q23–26) |

|
12 | 44.4% |
| No, never (To Q27) |

|
3 | 11.1% |
| No Answer |

|
1 | 3.7% |
Q23: May new cultures commence over the week‐end period?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes, as a general rule |

|
2 | 7.4% |
| Yes, sometimes |

|
7 | 25.9% |
| No, never |

|
14 | 51.9% |
| No Answer |

|
4 | 14.8% |
Q24: Are existing cultures further sub‐cultured/read over the week‐end period?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes, as a general rule |

|
7 | 25.9% |
| Yes, sometimes |

|
8 | 29.6% |
| No, never |

|
8 | 29.6% |
| No Answer |

|
4 | 14.8% |
Q25: Are cultures set up only on certain week days in order to avoid any week‐end working?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes, as a general rule cultures are set up to avoid week‐end working |

|
5 | 18.5% |
| Yes, sometimes cultures are set up to avoid week‐end working |

|
4 | 14.8% |
| No, cultures are not set up depending on the week‐end, they are set up any day of the week |

|
14 | 51.9% |
| No Answer |

|
4 | 14.8% |
Q25.1: If cultures are set up to avoid week‐end working, on which week days may culture start?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Monday |

|
7 | 25.9% |
| Tuesday |

|
5 | 18.5% |
| Wednesday |

|
7 | 25.9% |
| Thursday |

|
1 | 3.7% |
| Friday |

|
2 | 7.4% |
| No Answer |

|
19 | 70.4% |
Q26: If culture cannot continue during the week‐end, what is it done with existing cultures?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Extending incubation times over the week‐end period |

|
6 | 22.2% |
| Using incubators which can refrigerate plates after an interval over the week‐end period |

|
8 | 29.6% |
| Other means |

|
2 | 7.4% |
| There is always work continuity during the week‐ends |

|
7 | 25.9% |
| No Answer |

|
5 | 18.5% |
Q27: Are cultures set up only on certain week days, in order to avoid any week‐end working?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes, as a general rule cultures are set up to avoid week‐end working |

|
2 | 7.4% |
| Yes, sometimes cultures are set up to avoid week‐end working | 0 | 0% | |
| No, cultures are not set up depending on the week‐end, they are set up any day of the week |

|
1 | 3.7% |
| No Answer |

|
24 | 88.9% |
Q27.1. If cultures are set up to avoid week‐end working, on which weekdays may culture start?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Monday |

|
2 | 7.41% |
| Tuesday |

|
2 | 7.41% |
| Wednesday |

|
2 | 7.41% |
| Thursday |

|
2 | 7.41% |
| Friday |

|
2 | 7.41% |
| No Answer |

|
25 | 92.59% |
Q27.2. If culture does not continue during the week‐end, what is it done with existing cultures?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Extending incubation times over the week‐end period |

|
1 | 3.7% |
| Using incubators which can refrigerate plates after an interval over the week‐end period |

|
1 | 3.7% |
| Other means | 0 | 0% | |
| No Answer |

|
26 | 96.3% |
Overview of the Section A.5 :
An overview of the procedures used for primary culture of C. jejuni and C. coli over week‐end periods
| Items | Cultures continue over the week‐end |
Cultures Commence over the week‐end |
Subculture/read over the week‐end |
|---|---|---|---|
| Yes (as a rule) | 11a , b | 2 | 7 |
| Yes (sometimes) | 12a | 7 | 8 |
| No, never | 3c | 14 | 8 |
When asked if cultures are set up only on certain week days in order to avoid any week‐end working, nine countries confirmed, as a general rule or sometimes, to set up cultures on Monday (7), Tuesday (5), Wednesday (7), Thursday (1) or Friday (2).
AL, IS, NO also continue work over the week‐end as a general rule and they do not set up cultures depending on the week‐end.
Also CH. Two MSs do not set up cultures depending on the week‐end, while two other countries do.
C.6. Ongoing Campylobacter national studies not part of AMR monitoring
Q29: Do you perform recurrent monitoring for the prevalence of C. coli or C. jejuni in animal species or environmental samples annually or regularly?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
9 | 33.3% |
| No |

|
17 | 63.0% |
| No Answer |

|
1 | 3.7% |
Q29.1: Which Campylobacter species and animal species are monitored?
| Combinations monitored | # of MSs |
|---|---|
| C. coli in broilers | 9 (in 8 abattoirs) |
| C. coli in turkeys | 2 (in 2 abattoirs) |
| C. coli in pigs | 7 (in 6 abattoirs) |
| C. jejuni in veal calves | 4 (in 3 abattoirs) |
| C. coli in veal calves | 4 (in 3 abattoirs) |
Q29.2. Are other combinations of Campylobacter species and animal populations monitored?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
5 | 18.5% |
| No |

|
4 | 14.8% |
| No Answer |

|
18 | 66.7% |
IS performs recurrent monitoring for Campylobacter spp. in broilers (faeces). NO performs recurrent monitoring for Campylobacter coli in broilers and in pigs (annually, at abattoirs and on farms).
Q29.3: Do you use the isolation methodology described above?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
7 | 25.9% |
| No |

|
2 | 7.4% |
| No Answer |

|
18 | 66.7% |
IS and NO use the methodology described above.
Q29.4: Do you use the microdilution method for antimicrobial susceptibility testing of these isolates?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
7 | 25.9% |
| No |

|
2 | 7.4% |
| No Answer |

|
18 | 66.7% |
Q29.5: Is the AMR monitoring harmonised panel of antimicrobials used for these isolates?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
| Yes |

|
7 | 25.9% |
| No |

|
2 | 7.4% |
| No Answer |

|
18 | 66.7% |
IS does not perform the antimicrobial susceptibility testing of isolates. NO does, using disk diffusion methodology and the AMR monitoring harmonised panel of antimicrobials.
Appendix D – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: MRSA
D.1. Questions on isolation of MRSA
Q1: In which matrices do you use selective methods for the culture of MRSA?
| Matrices | Bars | Answers | Ratioa |
|---|---|---|---|
|

|
7 | 25.9% |
|

|
10 | 37.0% |
|

|
6 | 22.2% |
|

|
3 | 11.1% |
|

|
5 | 18.5% |
|

|
11 | 40.7% |
|

|
2 | 7.4% |
The ratio is calculated out of 27 responding MSs.
In the case of non‐MSs, IS uses nasal swabs from pig carcasses. NO uses nasal or skin swabs from animals, environmental dust samples and environmental boot swabs samples, while CH only uses meat and nasal or skin swabs from animals. AL reported that they do not use selective methods for MRSA.
Q2: Which protocols do you currently routinely use for the culture of MRSA?
| Protocol | Bars | Answers | Ratioa |
|---|---|---|---|
|

|
9 | 33.3% |
|

|
5 | 18.5% |
|

|
2 | 7.4% |
|

|
12 | 44.4% |
The ratio is calculated out of 27 responding MSs.
2 S method.
described by Larsen et al. (Eurosurveillance) (1 S method).
From the 12 with ‘No answer’, 11 are those who answered ‘Not performed’ in the previous question.
DE and LU reported to use ‘Another method’. DE employs a variation of the EURL method, using 6.0% NaCl for enrichment and 50 mg/L Aztreonam for selection; ChromID MRSA selective agar is used. LU used MRSA Select II (BioRad) on previously obtained coagulase positive colonies on BP‐RPF.
In the case of non‐MSs, IS uses the 2S method, while NO uses the 1S method. CH uses a variation similar to DE, but selecting with 3.5 mg/L Cefoxitin & 75 mg/L Aztreonam.
D.2. Questions on susceptibility testing of MRSA
Q3: Do you perform susceptibility testing of MRSA isolates?
| Susceptibility testing | Bars | Answer | Ratioa |
|---|---|---|---|
|

|
6 | 22.2% |
|

|
13 | 48.1% |
|

|
4 | 14.8% |
|
0 | 0% | |
|

|
5 | 18.5% |
The ratio is calculated out of 27 responding MSs.
NO and CH use broth microdilution for susceptibility testing of MRSA isolates. AL and IS do not perform it.
Q3.1: If susceptibility testing is performed, please, specify method and interpretation criteria used (e.g. EUCAST, CLSI):
Among those MSs performing the susceptibility testing of MRSA isolates, the interpretative criteria of resistance used are mainly those recommended by EUCAST, as shown in the table below.
| Susceptibility testing | Interpretative criteria of resistance | ||
|---|---|---|---|
| EUCAST | CLSI | Total | |
|
13 | 0 | 13 |
|
3a | 1 | 4 |
| Total | 16 | 1 | 17 |
One respondent also uses CA‐SFM.
NO and CH use EUCAST interpretative criteria as well.
Q3.3: If susceptibility testing is performed, please specify the panel of antimicrobials tested:
In the case of NL and UK, it was not defined or it included only a few of these compounds. NO and CH also uses the EUST panel.
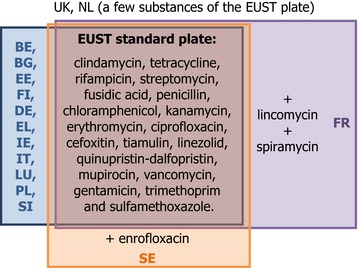
D.3. Questions on National Monitoring Programme of MRSA
Q4: Do you have National Monitoring Programmes in place for MRSA in animals?
| Animal species | MSs | At abattoirs/On farms |
Healthy/Diseased animals |
Frequency of monitoring |
|---|---|---|---|---|
| Broilers |
BE, DE, NL |
On farms | Healthya |
Every 3rd year (BE), Every 3–5 years (DE), starting in 2018 (NL) |
| Turkeys | DE | On farms | Healthya | Every 3–5 years |
| Cattle |
BE, DEb, NL |
On farms (BE, NL), Both (DE) |
Healthya |
Every 3rd year (BE), Every 3–5 years (DE), starting in 2019? (NL) |
| Pigs |
BE, FI, DE, NL, ES |
At abattoirs (FI, ES), On farms (BE, DE, NL) |
Healthya |
Every 2nd year (ES), Every 3rd year (BE), 3–5 years (DE), starting in 2019 (NL), Not regularly (FI) |
In DE, some animals might have suffered disease, but those were not targeted.
In DE, also milk and veal calves.
CH has National Monitoring Programmes in place for calves and pigs (both performed at abattoirs on healthy animals, every second year). NO has a National Monitoring Programme in place for pigs (performed on farm, both for healthy and diseased animals, annually).
Q5: Do you have National Monitoring Programmes in place for MRSA in meat?
|
Meat or meat products |
MSs | At abattoirs/At retail |
Frequency of monitoring |
|---|---|---|---|
| From Broilers |
AT, DE |
At retail (AT), Both (DE) |
Every 2nd years (AT), Every 3–5 years (DE) |
| From Turkeys | DE | Both (DE) | Every 3–5 years (DE) |
| Beef | DE | Both (DE) | Every 3–5 years (DE) |
| Pork |
AT, FI, DE |
At retail (AT, FI), Both (DE) |
Every 2nd years (AT), Every 3–5 years (DE), Not regularly (FI) |
| From Veal Calves | DEa |
DE also reported monitoring MRSA in milk on a regular basis.
CH has a National Monitoring Programme for MRSA in beef, broiler, and pork meat or derived products thereof, performed all at retail every second year.
Q6: How do you routinely perform the characterisation of MRSA isolates?
| Method of characterisation | Bars | Answers | Ratioa |
|---|---|---|---|
|

|
13 | 48.1% |
|

|
4 | 14.8% |
|

|
4 | 14.8% |
|

|
4 | 14.8% |
|

|
1 | 3.7% |
|

|
12 | 44.4% |
The ratio is calculated out of 27 responding MSs.
NO routinely uses the four main methods, while CH uses spa‐typing and other methods (PCR).
Q7: What are your preferred, optimal sampling/culture sites for isolation of MRSA in the different animal species when considering live animals or animals immediately after slaughter?
| MSs | Optimal sampling/culture sites for isolation of MRSA in | ||||
|---|---|---|---|---|---|
| Pigs | Broilers | Turkeys | Veal calves | Other Animals | |
| AT |
• Nasal swabs • Skin swabs a |
||||
| BE | • Nasal swabs | • Nasal swabs | • Nasal swabs |
• Dairy cows/cattle: Nasal swabs at farm (pools of 20/10) |
|
| FI |
• Nasal swabs • Skin swabs |
||||
| FR | • Skin swabs | ||||
| DE | • Skin swabs |
• Dairy cows: milk samples |
|||
| IE |
• Neck swabs • Skin swabs |
||||
| IT |
• Nasal swabs, • Skin swabsa |
• Neck swabs • Under wing swabs |
• Neck swabs, • Under wing swabs |
• Nasal swabs | |
| LV | • Nasal swabs | • Nasal swabs | |||
| LU | • Nasal swabs | ||||
| NL | • Nasal swabs | • Throat swabs | • Nasal swabs | ||
| SI |
• Nasal swabs, • Skin swabsa |
||||
| ES | |||||
| SE | • Skin swabsa | • Nasal swabs |
• Dairy cows: milk samples |
||
| UK |
• Nasal swabs • Skin swabsa |
• Cloacal swabs • Oral swabs • Under wing swabs |
• Cloacal swabs • Oral swabs • Under wing swabs |
• Nasal swabs | |
|
Nasal swabs: 9 Skin swabs: 7 |
Nasal swabs: 6 | ||||
behind the ears.
It is of note that certain MSs, as well as non‐MSs, provided generic indication, such as ‘Live animals’ or ‘After slaughter’.
Q8: What are your preferred, optimal sampling/culture sites for isolation of MRSA in the animal and farm environment?
| Sampling/Culture sites | Bars | Answers | Ratioa |
|---|---|---|---|
|

|
11 | 40.7% |
|

|
5 | 18.5% |
|

|
0 | 0% |
|

|
14 | 51.9% |
The ratio is calculated out of 27 responding MSs.
Overview table:
| Dust | Boot swabs | Both | Other samples |
|---|---|---|---|
| 7 | 1 | 4 | 0 |
| FR, IT, LT, LU, NL, SI, SE | IE | HK, FI, DE, UK | – |
NO used dust and boot swabs, while CH used boot swabs.
D.4. Questions on further EU‐wide monitoring of MRSA
Q9: Do you consider that further EU‐wide surveillance of MRSA would be useful?
Q9.1: In animals?
The ratio is calculated out of 27 responding MSs.
EL.
AL, NO and CH responded Yes.
Q9.1.1: If Yes, please, specify in which animal species:
| Animal species | Bars | Answers | Ratioa |
|---|---|---|---|
|

|
18 | 66.7% |
|

|
15 | 55.6% |
|

|
7 | 25.9% |
|

|
10 | 37.0% |
|

|
5 | 18.5% |
|

|
8b | 29.6% |
The ratio is calculated out of 27 responding MSs.
Includes those MSs that did not answer + EL.
Overview table:
| MSs | Pigs | Broilers | Turkeys | Veal calves | Others |
|---|---|---|---|---|---|
| AT | X | X | X | X | |
| BE | X | X | X | Dairy cows | |
| BG | X | X | |||
| HR | X | X | |||
| EE | X | ||||
| FI | X | Young beef | |||
| FR | X | X | X | ||
| DE | X | X | X | X | Dairy cows |
| IE | X | X | |||
| IT | X | X | X | ||
| LV | X | X | X | ||
| LT | X | Dogs, cats | |||
| LU | X | X | |||
| NL | X | X | X | ||
| PT | X | X | X | X | |
| RO | X | X | X | ||
| SI | X | ||||
| SE | X | X | X | X | All cattle |
| UK | X | X | X | X | |
| Total | 18 | 15 | 7 | 10 | 5 |
AL: in Broilers; NO: in Pigs, Broilers and Others (horses); CH: in Pigs, Broilers and Veal calves.
Q9.2: In meat?
| Bars | Answers | Ratioa | |
|---|---|---|---|
|

|
11 | 40.7% |
|

|
8 | 29.6% |
|

|
8 | 29.6% |
The ratio is calculated out of 27 responding MSs.
AL and CH replied ‘Yes’, whereas NO replied ‘No’.
Q9.2.1: If Yes, please specify in meat derived from which animal species:
| Bars | Answers | Ratio(a) | |
|---|---|---|---|
|

|
11 | 40.7% |
|

|
11 | 40.7% |
|

|
5 | 18.5% |
|

|
4 | 14.8% |
|

|
1 | 3.7% |
|

|
16 | 59.3% |
Overview table:
| MS | Pigs | Broilers | Turkeys | Veal calves | Others |
|---|---|---|---|---|---|
| AT | X | X | X | X | |
| BG | X | X | |||
| DE | X | X | X | X | Beef |
| EL | X | X | |||
| IT | X | X | |||
| LV | X | X | |||
| LU | X | X | |||
| NL | X | X | X | ||
| PT | X | X | X | X | |
| RO | X | X | X | ||
| UK | X | X | X | ||
| Total | 11 | 11 | 5 | 4 | 1 |
AL: in Broilers; CH: in Pigs, Broilers, and Veal calves.
Q10: If a baseline survey was performed on MRSA in pigs and poultry, what type of survey would you consider to be most valuable?
| Items | Bars | Answers | Ratio |
|---|---|---|---|
|

|
7 | 25.9% |
|

|
9 | 33.3% |
|

|
4 | 14.8% |
|

|
7 | 25.9% |
| Type of survey | More detailed description of surveys | Answers |
|---|---|---|
| On the farm | A farm‐based survey, assessing the prevalence and diversity of MRSA on pig and poultry farms | 7 |
| Variation | 3a | |
| At the abattoir | A survey at the slaughterhouse level to assess the diversity of MRSA harboured by the slaughtered animals, recognising that cross‐contamination between animals or carcases may occur at slaughter | 9 |
| Variation | 3b |
On pig farm only (1); On pig, poultry and turkey farms (1); On veal calve farm (1).
At the abattoir for broilers only (2), On meat from broiler and pork (1).
AL and NO considered the farm survey more valuable, while IS and CH consider the slaughterhouse more valuable.
Appendix E – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: Monitoring of specific colistin resistance
1.
Q1: How do you detect phenotypic colistin resistance in indicator commensal E. coli / Salmonella spp.?
| Answers | Ratio | ||
|---|---|---|---|
| Broth dilution MIC determination using colistin |

|
25 | 92.59% |
| Agar dilution MIC determination using colistin | 0 | 0% | |
| Gradient strip MIC determination using colistin | 0 | 0% | |
| Conventional disk diffusion methods using colistin |

|
1 | 3.7% |
| Pre‐diffusion disk diffusion methods using colistin |

|
2 | 7.41% |
| Other approachesa |

|
1 | 3.7% |
| No Answerb |

|
2 | 7.41% |
MacConkey + 1 mg/L colistin.
MT, HR.
Q2. What is the preferred phenotypic method?
All countries (including IS, NO and CH) use Broth dilution MIC determination. In addition, four MSs use a second method. For all countries Broth dilution is their preferred phenotypic method, except in the case of the MS using also pre‐diffusion disk diffusion method, where both methods are used.
Q3. Is a method for selective isolation of colistin‐resistant E. coli used?
| Answers | Ratio | ||
|---|---|---|---|
| Yes (AT, NL, PL, PT, SI, UK) |

|
6 | 22.22% |
| No |

|
20 | 74.07% |
| No Answer (MT) |

|
1 | 3.7% |
Q3.1. If Yes, please describe the method:
6 countries use a method for selective isolation of colistin‐resistant E. coli. Five of the countries use a variation of the EURL‐AR protocol, based on culturing in MacConkey + Colistin. One country uses CHROMID colistin R media, only for research purposes. NO uses SuperPolymyxin agar.
Q3.2. To which matrices is the method applied?:
| Answers | Ratio | ||
|---|---|---|---|
| Meat |

|
3 | 11.11% |
| Caecal contents |

|
6 | 22.22% |
| Other matrices (environmental samples) |

|
1 | 3.7% |
| No Answer (20+1 from before) |

|
21 | 77.78% |
All countries tested caecal contents, also NO.
Q4. Which method(s) is/are used to identify mcr ‐genes?
| Answers | Ratio | ||
|---|---|---|---|
| PCR |

|
6 | 22.22% |
| EURL‐AR PCR method |

|
17 | 62.96% |
| WGS |

|
10 | 37.04% |
| Other methods | 0 | 0% | |
| No Answer |

|
6 | 22.22% |
Q5. Is additional characterisation performed in order to further inform about?
| Answers | Ratio | ||
|---|---|---|---|
| Genes/IS‐elements |

|
10 | 37.04% |
| Plasmids |

|
10 | 37.04% |
| Strains |

|
12 | 44.44% |
| Other (WGS done when PCR for mcr is negative) |

|
1 | 3.7% |
| No Answer |

|
13 | 48.15% |
Q6. Additional information/comments about the monitoring of specific colistin resistance:
Some countries perform characterisation of colistin resistance, but only for research purposes.
Appendix F – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: Further characterisation of ESBL/AmpC/carbapenemase producers and corresponding genes identification
1.
Q1. Have you occasionally or regularly characterised the ESBL/AmpC/carbapenemase E. coli population (by performing enumeration of this E. coli population) in meat samples?
| Answers | Ratio | ||
|---|---|---|---|
| No, never |

|
22 | 81.48% |
| Yes (IT, MT, NL, ES, UK) |

|
5 | 18.52% |
| No Answer | 0 | 0% |
Q1.1. If Yes, please, specify in which kind of meat:
| Answers | Ratio | ||
|---|---|---|---|
| Meat from pigs |

|
5 | 18.52% |
| Meat from broilers |

|
4 | 14.81% |
| Meat from turkeys |

|
2 | 7.41% |
| Meat from cattle |

|
4 | 14.81% |
| Other meat | 0 | 0% | |
| No Answer |

|
22 | 81.48% |
IT, MT, NL, ES and UK were the only countries that reported having occasionally or regularly characterised the ESBL/AmpC/carbapenemase E. coli population, doing it all of them for pigs. From the non‐MSs, only NO responded positively, for both meat from broilers and from turkeys.
Q2. Do you regularly perform the identification of the genes encoding for these enzymes?
| Answers | Ratio | ||
|---|---|---|---|
| Yes |

|
8 | 29.63% |
| No |

|
19 | 70.37% |
| No Answer | 0 | 0% |
Q3. At which level do you regularly perform the identification of the genes encoding for these enzymes?
| Answers | Ratio | ||
|---|---|---|---|
| At group level (e.g. CTX‐M) |

|
1 | 3.70% |
| At variant level (e.g. CTX‐M‐15) |

|
7 | 25.93% |
| No Answer |

|
19 | 70.37% |
IE, IT, LU, NL, PT, SW and UK perform the identification of the genes at variant level, while DE performs it at group level.
Q4. What method(s) do you use to perform this characterisation?
Q4.1. Identification of enzyme group/class/family:
| Answers | Ratio | ||
|---|---|---|---|
| PCR |

|
7 | 25.93% |
| WGS |

|
4 | 14.81% |
| Other (Sanger sequencing) |

|
1 | 3.7% |
| No Answer |

|
19 | 70.37% |
Q4.2. Identification of enzyme variant:
| Answers | Ratio | ||
|---|---|---|---|
| PCR/Sanger sequencing |

|
6 | 22.22% |
| WGS |

|
6 | 22.22% |
| Other | 0 | 0% | |
| No Answer |

|
19 | 70.37% |
Q4.3. If you perform WGS, which bioinformatic tool(s)/pipeline(s) do you work with?
| Answers | Ratio | ||
|---|---|---|---|
| DTU Resfinder |

|
6 | 22.22% |
| Bionumerics |

|
1 | 3.7% |
| CARD |

|
1 | 3.7% |
| Other (Enterobase, SeqFinder) |

|
2 | 7.41% |
| No Answer |

|
21 | 77.78% |
DTU Resfinder was clearly the most widely tool used for the characterisation of the ESBL/AmpC/carbapenemase enzymes. NO was the only non‐MS who regularly performs the identification of the genes encoding for ESBL/AmpC/carbapenemases. They also use PCR/Sanger sequencing and WGS (working with DTU Resfinder and CARD).
Q5. Do you report the results of the genes’ characterisation to EFSA?
| Answers | Ratio | ||
|---|---|---|---|
| Yes (AT, EE, IT, NL, SE) |

|
5 | 18.52% |
| No |

|
18 | 66.67% |
| No Answer (CY, EL, HU, MT) |

|
4 | 14.81% |
Q5.1. If you do not, why?
| Answers | Ratio | ||
|---|---|---|---|
| The reporting deadlines are too short |

|
5 | 18.52% |
| There is lack of resources for reporting |

|
11 | 40.74% |
| Other reason |

|
6 | 22.22% |
| No Answer (previous 4 + 5 Yes) |

|
9 | 33.33% |
Only 5 MSs report the results of the genes characterisation to EFSA. Those who do not is mainly due to a lack of resources. The countries choosing ‘Other reason’ indicated that they were not actually performing the characterisation, either due to a lack of resources or because it is not mandatory to do so.
IS and NO report their results, while AL and CH as they lack resources to perform the analyses.
Q6. For ESBL/AmpC/carbapenemase producing E. coli , do you collect other molecular typing data?
| Answers | Ratio | ||
|---|---|---|---|
| Multilocus sequence type of the host E. coli |

|
5 | 18.52% |
| WGS of the enzyme‐carrying strain |

|
6 | 22.22% |
| Sequencing of the responsible gene carrying plasmid after conjugation or transformation |

|
5 | 18.52% |
| Plasmid typing (inc typing) data for the plasmid carrying the responsible gene |

|
3 | 11.11% |
| Plasmid multilocus sequence typing data for the plasmid carrying the responsible gene |

|
4 | 14.81% |
| Other (2 of the replies indicated ‘none’) |

|
2 | 14.81% |
| No Answer |

|
16 | 59.26% |
NO also collects other molecular typing data.
Q7. Would you be in favour of a collection and analysis of typing data related to ESBL/AmpC/carbapenemase encoding genes, at the EU level?
| Answers | Ratio | ||
|---|---|---|---|
| Yes |

|
19 | 70.37% |
| No (HR, CY, LT, LU) |

|
4 | 14.81% |
| No Answer (CZ, EE, HU, MT) |

|
4 | 14.81% |
Most countries were in favour of a collection and analysis of typing data at EU level. Six countries (4 MSs, plus AL and NO) were not.
Q8. Given the complexity of the typing and analysis, would you be in favour of a timescale for testing, collecting and reporting this data longer than 1 year (after the end of the year in which isolates were collected)?
| Answers | Ratio | ||
|---|---|---|---|
| Yes |

|
20 | 74.07% |
| No (HR), and also AL |

|
1 | 3.7% |
| No Answer (CZ, EE, HU, LU, MT, ES) |

|
6 | 22.22% |
Appendix G – Outcome of the Specific Questionnaire Survey, EU MSs, June 2018: First and second panels for susceptibility testing of Salmonella and E. coli
1.
Q1. Do you have any suggestions for one or two additional antimicrobials to be included in the panel for testing susceptibility of Salmonella and indicator commensal E. coli (first panel), which would be of relevance for public health? (in order of priority)
| First antimicrobial suggested | Second antimicrobial suggested | Antimicrobial to remove |
|---|---|---|
| Cefiderocola | Levofloxacina | Ceftazidimea, nalidixic acida |
| Fosfomycinb | Second‐generation cephalosporinb | Reduce meropenem MIC range on the 1st panel. Reduce the MIC range of other agentsb |
| Amikacin (3) | No answer (3) | Reduce the MIC range of other agents (2) |
| Imipenem | No answer | No suggestions/answer (22) |
| No suggestions/answer (21) | No suggestions/answer (21) | Meropenem range could be shortened (1) |
All replies from the same country.
All replies from the same country.
The number of respondents that gave a given answer is shown between brackets.
Non‐MSs did not provide any suggestions.
Q2. Do you have any suggestions for one or two additional antimicrobials to be included in the panel for testing Salmonella and indicator commensal E. coli resistant to cefotaxime or ceftazidime or meropenem (second panel) which would be of value for phenotypic characterisation of these bacteria? (in order of priority)
| Antimicrobial suggested | Antimicrobial to remove |
|---|---|
| Doripenema | Ertapenema |
| Faropenemb | Reduce the MIC range of some agentsb |
| Fosfomycin | No suggestions/answer |
| No suggestions/answer (24) | No suggestions/answer (24) |
All replies from the same country.
All replies from the same country.
The number of respondents that gave a given answer is shown between brackets.
Non‐MSs did not provide any suggestions.
Appendix H – Approach for reviewing the sample size calculation
1.
The approach and the results of the sample size analyses and calculation in the EFSA technical specifications (EFSA, 2007, 2008, 2012a,b) were reviewed and revised using additional sets of scenarios.
H.1. The additional sets of scenarios addressed
Table 17.
Scenarios set 1: Occurrence of resistance (proportion of isolates detected as resistant)
| Point estimatesa | Direction of trendb | Magnitude of trendb |
|---|---|---|
| 80% | Decreasing | − 10% over 3 successive samplings |
| 40% | Decreasing | − 10% over 3 successive samplings |
| 10% | Decreasing | − 2% over 3 successive samplings |
| 5% | Decreasing | − 1% over 3 successive samplings |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
Table 18.
Scenarios set 2: Rate of complete susceptibility (proportion of isolates categorised as susceptible to each of substances of the harmonised panel)
| Point estimatea | Direction of trendb | Magnitude of trendb |
|---|---|---|
| 80% | Increasing | + 10% over 3 successive sampling |
| 40% | Increasing | + 10% over 3 successive sampling |
| 10% | Increasing | + 10% over 3 successive sampling |
| 5% | Increasing | + 10% over 3 successive sampling |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
Table 19.
Scenarios set 3: Emergence of (occurrence of) resistance (probability to detect at least one isolate as resistant) [at different rate of emergence]
| Point estimatea | Direction of trendb | Magnitude of trendb |
|---|---|---|
| 0% | Increasing | + 0.01% over 3 successive sampling |
| 0% | Increasing | + 0.5% over 3 successive sampling |
| 0% | Increasing | + 1% over 3 successive sampling |
| 0% | Increasing | + 5% over 3 successive sampling |
| 0% | Increasing | + 10% over 3 successive sampling |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
H.2. Statistical approach to address the sets of scenarios
Scenario set 1 and 2
The first set of scenarios focuses on particular decreasing trends, starting with an existing assumed proportion of isolates detected as resistant at time 0 (say at year 2019) and reaching a particular lower proportion after three successive sampling times (say at 2021, 2023, 2025). Likewise, scenario set 2 defines particular increasing trends over 3 successive sampling times. In what follows, the ‘proportion’ refers in a generic way to the proportion of isolates detected as resistant, or to the proportion of isolates categorised as susceptible to each of substances of the harmonised panel.
All these particular scenarios were investigated in the generic tables (Tables H.1 and H.2), covering many more scenarios, for sample sizes 100, 150, 170, 200, 250 and 300 at each sampling time. It is of interest to get insights in, for each scenario:
the accuracy of the estimation of the ‘proportion’ based on the sample of a particular size at each of the successive sampling times;
the power to detect the trend as defined by a scenario at the end of the 3 successive sampling times.
Table H.1.
Sample sizes needed for estimating proportions of resistance and detecting decreasing trends in resistance with a given accuracy
| Trend as percentage | Sample Size | Percentage Accurate 2021 | Average Accuracy 2021 | Percentage Accurate 2023 | Average Accuracy 2023 | Percentage Accurate 2025 | Average Accuracy 2025 | Power Trend 2019–2023 | Power Trend 2019–2025 |
|---|---|---|---|---|---|---|---|---|---|
| 95% ↘□ 90% | At (2021, 2023, 2025) estimation of 93.68%, 92.03%, 90% with accuracy 0.0454, 0.0475, 0.05, respectively | ||||||||
| 300 | 100 | 0.028 | 100 | 0.031 | 100 | 0.034 | 31.8 | 75.5 | |
| 250 | 100 | 0.030 | 100 | 0.034 | 100 | 0.037 | 27.9 | 71.7 | |
| 200 | 99.5 | 0.034 | 99.6 | 0.037 | 99.3 | 0.041 | 24.3 | 64.3 | |
| 170 | 97.1 | 0.037 | 92.2 | 0.041 | 87.6 | 0.045 | 22.2 | 55.5 | |
| 150 | 87.6 | 0.039 | 76.3 | 0.044 | 65.2 | 0.048 | 21.6 | 55.6 | |
| 100 | 33.7 | 0.049 | 20.1 | 0.054 | 13.2 | 0.058 | 18.6 | 45.2 | |
| 90% ↘□ 80% | At (2021, 2023, 2025) estimation of 87.29%, 83.98%, 80% with accuracy 0.0534, 0.0575, 0.0625, respectively | ||||||||
| 300 | 100 | 0.038 | 100 | 0.041 | 100 | 0.045 | 64.3 | 97.8 | |
| 250 | 100 | 0.041 | 100 | 0.045 | 100 | 0.049 | 59.2 | 95.9 | |
| 200 | 98.8 | 0.046 | 99.3 | 0.051 | 99.8 | 0.055 | 53.0 | 93.8 | |
| 170 | 80.1 | 0.050 | 77.6 | 0.054 | 81.8 | 0.060 | 47.7 | 91.4 | |
| 150 | 52.0 | 0.053 | 40.1 | 0.058 | 37.6 | 0.063 | 47.0 | 90.0 | |
| 100 | 4.9 | 0.065 | 2.1 | 0.071 | 0.6 | 0.077 | 38.6 | 83.0 | |
| 80% ↘□ 70% | At (2021, 2023, 2025) estimation of 76.97%, 73.63%, 70% with accuracy 0.0663, 0.0705, 0.075, respectively | ||||||||
| 300 | 100.0 | 0.047 | 100.0 | 0.050 | 100.0 | 0.051 | 47.9 | 89.0 | |
| 250 | 100.0 | 0.052 | 100.0 | 0.054 | 100.0 | 0.056 | 44.4 | 84.2 | |
| 200 | 100.0 | 0.058 | 100.0 | 0.060 | 100.0 | 0.063 | 38.3 | 81.8 | |
| 170 | 89.7 | 0.062 | 97.3 | 0.065 | 100.0 | 0.068 | 35.1 | 80.5 | |
| 150 | 43.9 | 0.067 | 59.6 | 0.070 | 82.8 | 0.072 | 33.0 | 76.4 | |
| 100 | 0.5 | 0.081 | 0.2 | 0.085 | 0.3 | 0.088 | 27.5 | 62.5 | |
| 70% ↘□ 60% | At (2021, 2023, 2025) estimation of 66.82%, 63.48%, 60% with accuracy 0.0790, 0.0832, 0.0875, respectively | ||||||||
| 300 | 100 | 0.053 | 100 | 0.054 | 100 | 0.055 | 38.7 | 82.5 | |
| 250 | 100 | 0.058 | 100 | 0.059 | 100 | 0.060 | 34.8 | 78.0 | |
| 200 | 100 | 0.064 | 100 | 0.066 | 100 | 0.067 | 29.8 | 72.1 | |
| 170 | 100 | 0.070 | 100 | 0.071 | 100 | 0.073 | 30.8 | 68.7 | |
| 150 | 99.1 | 0.074 | 100 | 0.076 | 100 | 0.077 | 25.3 | 66.4 | |
| 100 | 0.5 | 0.09 | 0.8 | 0.092 | 1.0 | 0.094 | 24.4 | 56.1 | |
| 60% ↘□ 50% | At (2021, 2023, 2025) estimation of 56.72%, 53.37%, 50% with accuracy 0.0916, 0.0958, 0.10, respectively | ||||||||
| 300 | 100.0 | 0.056 | 100.0 | 0.056 | 100.0 | 0.056 | 35.7 | 79.6 | |
| 250 | 100.0 | 0.061 | 100.0 | 0.061 | 100.0 | 0.061 | 32.7 | 76.6 | |
| 200 | 100.0 | 0.068 | 100.0 | 0.068 | 100.0 | 0.068 | 30.8 | 70.4 | |
| 170 | 100.0 | 0.073 | 100.0 | 0.074 | 100.0 | 0.074 | 28.5 | 66.7 | |
| 150 | 100.0 | 0.078 | 100.0 | 0.079 | 100.0 | 0.079 | 26.3 | 61.1 | |
| 100 | 4.3 | 0.095 | 51.0 | 0.096 | 100.0 | 0.096 | 22.4 | 54.0 | |
| 50% ↘ 40% | At (2021, 2023, 2025) estimation of 46.63%, 43.28%, 40% with accuracy 0.0958, 0.0916, 0.0875, respectively | ||||||||
| 300 | 100 | 0.056 | 100 | 0.056 | 100 | 0.055 | 40.1 | 79.3 | |
| 250 | 100 | 0.061 | 100 | 0.061 | 100 | 0.060 | 32.2 | 75.1 | |
| 200 | 100 | 0.068 | 100 | 0.068 | 100 | 0.067 | 30.8 | 68.0 | |
| 170 | 100 | 0.074 | 100 | 0.073 | 100 | 0.073 | 29.0 | 67.0 | |
| 150 | 100 | 0.079 | 100 | 0.078 | 100 | 0.077 | 25.3 | 61.5 | |
| 100 | 50.7 | 0.096 | 5.2 | 0.095 | 0.9 | 0.094 | 21.8 | 52.4 | |
| 40% ↘□ 30% | At (2021, 2023, 2025) estimation of 36.52%, 33.18%, 30% with accuracy 0.0832, 0.0790, 0.075, respectively | ||||||||
| 300 | 100 | 0.054 | 100 | 0.053 | 100 | 0.052 | 41.5 | 82.4 | |
| 250 | 100 | 0.059 | 100 | 0.058 | 100 | 0.056 | 39.7 | 80.8 | |
| 200 | 100 | 0.066 | 100 | 0.064 | 100 | 0.063 | 35.2 | 75.4 | |
| 170 | 100 | 0.071 | 100 | 0.070 | 99.9 | 0.068 | 32.0 | 72.2 | |
| 150 | 100 | 0.076 | 99.1 | 0.074 | 82.7 | 0.072 | 32.1 | 69.9 | |
| 100 | 0.7 | 0.092 | 0.7 | 0.090 | 0.4 | 0.088 | 25.2 | 58.9 | |
| 30% ↘□ 20% | At (2021, 2023, 2025) estimation of 26.37%, 23.03%, 20% with accuracy 0.0705, 0.0663, 0.0625, respectively | ||||||||
| 300 | 100 | 0.050 | 100 | 0.047 | 100 | 0.045 | 50.1 | 90.0 | |
| 250 | 100 | 0.054 | 100 | 0.052 | 100 | 0.049 | 49.9 | 87.2 | |
| 200 | 100 | 0.060 | 100 | 0.058 | 99.6 | 0.055 | 42.5 | 82.4 | |
| 170 | 97.6 | 0.065 | 89.1 | 0.063 | 82.0 | 0.059 | 41.2 | 82.6 | |
| 150 | 63.2 | 0.069 | 46.0 | 0.066 | 39.5 | 0.063 | 39.8 | 78.5 | |
| 100 | 0.4 | 0.085 | 1.2 | 0.081 | 0.9 | 0.077 | 34.9 | 69.4 | |
| 20% ↘□ 10% | At (2021, 2023, 2025) estimation of 16.02%, 12.71%, 10% with accuracy 0.0575, 0.0534, 0.05, respectively | ||||||||
| 300 | 100 | 0.041 | 100 | 0.038 | 100 | 0.034 | 73.5 | 97.3 | |
| 250 | 100 | 0.045 | 100 | 0.041 | 100 | 0.037 | 68.7 | 97.3 | |
| 200 | 98.9 | 0.051 | 99.1 | 0.046 | 99.2 | 0.042 | 66.0 | 96.7 | |
| 170 | 75.8 | 0.055 | 78.7 | 0.050 | 87.1 | 0.045 | 58.3 | 94.1 | |
| 150 | 43.2 | 0.058 | 52.8 | 0.053 | 64.7 | 0.048 | 59.9 | 92.3 | |
| 100 | 2.7 | 0.071 | 6.4 | 0.065 | 12.4 | 0.059 | 48.7 | 85.9 | |
| 10% ↘□ 5% | At (2021, 2023, 2025) estimation of 7.97%, 6.32%, 5% with accuracy 0.0475, 0.0454, 0.0438, respectively | ||||||||
| 300 | 100 | 0.031 | 100 | 0.028 | 100 | 0.025 | 36.6 | 72.7 | |
| 250 | 100 | 0.034 | 100 | 0.030 | 100 | 0.027 | 36.2 | 65.5 | |
| 200 | 99.7 | 0.038 | 99.8 | 0.034 | 100 | 0.030 | 31.3 | 64.2 | |
| 170 | 92.8 | 0.041 | 96.8 | 0.037 | 99.1 | 0.033 | 28.0 | 60.3 | |
| 150 | 76.7 | 0.043 | 85.9 | 0.039 | 93.6 | 0.036 | 29.2 | 55.3 | |
| 100 | 22.2 | 0.053 | 36.0 | 0.049 | 44.3 | 0.044 | 24.6 | 49.2 | |
| 10% ↘□ 8% | At (2021, 2023, 2025) estimation of 9.29%, 8.62%, 8% with accuracy 0.0491, 0.0484, 0.0475, respectively | ||||||||
| 300 | 100 | 0.033 | 100 | 0.032 | 100 | 0.031 | 6.1 | 12.6 | |
| 5% ↘□ 4% | At (2021, 2023, 2025) estimation of 4.64%, 4.31%, 4% with accuracy 0.0433, 0.0429, 0.0425, respectively | ||||||||
| 300 | 100 | 0.024 | 100 | 0.023 | 100 | 0.023 | 2.6 | 7.1 | |
Table H.2.
Sample sizes needed for estimating proportions of resistance and detecting increasing trends in resistance with a given accuracy
| Trend | Sample Size | Percentage Accurate 2021 | Average Accuracy 2021 | Percentage Accurate 2023 | Average Accuracy 2023 | Percentage Accurate 2025 | Average Accuracy 2025 | Power Trend 2019–2023 | Power Trend 2019–2025 |
|---|---|---|---|---|---|---|---|---|---|
| 5% ↗□ 15% | At (2021, 2023, 2025) estimation of 0.073%, 0.105%, 0.15% with accuracy 0.0466, 0.0507, 0.0563, respectively | ||||||||
| 170 | 94.1 | 0.039 | 84.6 | 0.046 | 77.5 | 0.053 | 62.1 | 97.4 | |
| 100 | 27.1 | 0.051 | 9.3 | 0.06 | 2.9 | 0.069 | 48.9 | 93.4 | |
| 5% ↗□ 10% | At (2021, 2023, 2025) estimation of 6.32%, 7.97%, 10% with accuracy 0.0454, 0.0475, 0.05, respectively | ||||||||
| 300 | 100 | 0.028 | 100 | 0.031 | 100 | 0.034 | 31.7 | 75.1 | |
| 250 | 100 | 0.030 | 100 | 0.034 | 100 | 0.037 | 26.0 | 69.1 | |
| 200 | 99.9 | 0.034 | 99.7 | 0.038 | 99.0 | 0.042 | 23.4 | 66.7 | |
| 170 | 97.5 | 0.037 | 93.1 | 0.041 | 86.7 | 0.045 | 23.8 | 60.0 | |
| 150 | 86.9 | 0.039 | 77.7 | 0.043 | 65.6 | 0.048 | 20.8 | 54.8 | |
| 100 | 36.5 | 0.048 | 21.3 | 0.053 | 12.2 | 0.058 | 15.5 | 43.3 | |
| 10% ↗□ 20% | At (2021, 2023, 2025) estimation of 12.71%, 16.02%, 20% with accuracy 0.0534, 0.0575, 0.0625, respectively | ||||||||
| 300 | 100 | 0.038 | 100 | 0.041 | 100 | 0.045 | 62.9 | 97.4 | |
| 250 | 100 | 0.041 | 100 | 0.045 | 100 | 0.049 | 57.7 | 95.6 | |
| 200 | 97.9 | 0.046 | 98.5 | 0.051 | 99.8 | 0.055 | 53.3 | 93.8 | |
| 170 | 81.5 | 0.050 | 76.2 | 0.055 | 79.7 | 0.060 | 46.1 | 94.1 | |
| 150 | 52.1 | 0.053 | 40.1 | 0.058 | 39.3 | 0.063 | 47.4 | 90.6 | |
| 100 | 5.9 | 0.065 | 3.2 | 0.071 | 1.0 | 0.077 | 35.4 | 82.5 | |
| 20% ↗□ 30% | At (2021, 2023, 2025) estimation of 23.03%, 26.37%, 30% with accuracy 0.0663, 0.0705, 0.075, respectively | ||||||||
| 300 | 100 | 0.047 | 100 | 0.050 | 100 | 0.051 | 48.7 | 87.6 | |
| 250 | 100 | 0.052 | 100 | 0.054 | 100 | 0.056 | 42.3 | 83.8 | |
| 200 | 100 | 0.058 | 100 | 0.061 | 100 | 0.063 | 41.5 | 82.1 | |
| 170 | 90.1 | 0.063 | 97.9 | 0.065 | 100 | 0.068 | 36.1 | 76.3 | |
| 150 | 45.0 | 0.067 | 58.4 | 0.070 | 83.7 | 0.072 | 33.7 | 74.4 | |
| 100 | 0.8 | 0.081 | 0.5 | 0.084 | 0.5 | 0.088 | 24.1 | 63.8 | |
| 30% ↗□ 40% | At (2021, 2023, 2025) estimation of 33.18%, 36.52%, 40% with accuracy 0.0790, 0.0832, 0.0875, respectively | ||||||||
| 300 | 100 | 0.053 | 100 | 0.054 | 100 | 0.055 | 39.2 | 82.9 | |
| 250 | 100 | 0.058 | 100 | 0.059 | 100 | 0.060 | 36.0 | 77.8 | |
| 200 | 100 | 0.065 | 100 | 0.066 | 100 | 0.067 | 32.9 | 73.0 | |
| 170 | 100 | 0.070 | 100 | 0.071 | 100 | 0.073 | 27.5 | 68.5 | |
| 150 | 98.3 | 0.074 | 100 | 0.076 | 100 | 0.077 | 28.7 | 67.9 | |
| 100 | 0.4 | 0.090 | 0.5 | 0.092 | 1.8 | 0.094 | 22.2 | 52.8 | |
| 40% ↗□ 50% | At (2021, 2023, 2025) estimation of 43.28%, 46.63%, 50% with accuracy 0.0916, 0.0958, 0.1, respectively | ||||||||
| 300 | 100 | 0.056 | 100 | 0.056 | 100 | 0.056 | 36.4 | 79.3 | |
| 250 | 100 | 0.061 | 100 | 0.061 | 100 | 0.061 | 33.9 | 75.9 | |
| 200 | 100 | 0.068 | 100 | 0.068 | 100 | 0.069 | 29.9 | 70.4 | |
| 170 | 100 | 0.073 | 100 | 0.074 | 100 | 0.074 | 28.9 | 67.7 | |
| 150 | 100 | 0.078 | 100 | 0.079 | 100 | 0.079 | 26.6 | 63.0 | |
| 100 | 4.5 | 0.095 | 51.7 | 0.096 | 100 | 0.096 | 22.2 | 52.4 | |
| 50% ↗□ 60% | At (2021, 2023, 2025) estimation of 53.37%, 56.72%, 60% with accuracy 0.0958, 0.0916, 0.0875, respectively | ||||||||
| 300 | 100 | 0.056 | 100 | 0.056 | 100 | 0.055 | 38.9 | 81.2 | |
| 250 | 100 | 0.061 | 100 | 0.061 | 100 | 0.060 | 35.8 | 76.9 | |
| 200 | 100 | 0.068 | 100 | 0.068 | 100 | 0.067 | 28.9 | 68.9 | |
| 170 | 100 | 0.074 | 100 | 0.074 | 100 | 0.073 | 27.8 | 64.6 | |
| 150 | 100 | 0.079 | 100 | 0.078 | 100 | 0.077 | 28.4 | 66.4 | |
| 100 | 52.9 | 0.096 | 5.3 | 0.095 | 1.2 | 0.094 | 25.0 | 54.6 | |
| 60% ↗□ 70% | At (2021, 2023, 2025) estimation of 63.48%, 66.82%, 70% with accuracy 0.0832, 0.0790, 0.075, respectively | ||||||||
| 300 | 100 | 0.054 | 100 | 0.053 | 100 | 0.051 | 41.1 | 82.9 | |
| 250 | 100 | 0.059 | 100 | 0.058 | 100 | 0.056 | 37.0 | 79.0 | |
| 200 | 100 | 0.066 | 100 | 0.064 | 100 | 0.063 | 34.2 | 73.3 | |
| 170 | 100 | 0.072 | 100 | 0.070 | 100 | 0.068 | 32.4 | 73.3 | |
| 150 | 100 | 0.076 | 98.7 | 0.075 | 83.0 | 0.072 | 29.9 | 67.0 | |
| 100 | 0.6 | 0.092 | 0.3 | 0.090 | 0.4 | 0.088 | 26.3 | 56.3 | |
| 70% ↗□ 80% | At (2021, 2023, 2025) estimation of 73,63%, 76.97%, 80% with accuracy 0.0705, 0.0663, 0.0625, respectively | ||||||||
| 300 | 100 | 0.050 | 100.0 | 0.047 | 100 | 0.045 | 50.9 | 88.7 | |
| 250 | 100 | 0.054 | 100.0 | 0.052 | 100 | 0.049 | 48.9 | 87.2 | |
| 200 | 100 | 0.060 | 100.0 | 0.058 | 99.8 | 0.055 | 41.6 | 82.8 | |
| 170 | 97.3 | 0.066 | 88.8 | 0.063 | 78.2 | 0.060 | 38.8 | 79.9 | |
| 150 | 61.6 | 0.069 | 45.8 | 0.067 | 40.6 | 0.063 | 39.5 | 75.9 | |
| 100 | 0.2 | 0.085 | 0.6 | 0.081 | 1.2 | 0.077 | 30.6 | 68.1 | |
| 80% ↗□ 90% | At (2021, 2023, 2025) estimation of 83,98%, 87.29%, 90% with accuracy 0.0575, 0.0534, 0.05, respectively | ||||||||
| 300 | 100 | 0.041 | 100 | 0.037 | 100 | 0.034 | 76.4 | 97.4 | |
| 250 | 100 | 0.045 | 100 | 0.041 | 100 | 0.037 | 69.5 | 96.1 | |
| 200 | 98.6 | 0.051 | 98.5 | 0.046 | 99.0 | 0.042 | 65.9 | 96.4 | |
| 170 | 78.6 | 0.055 | 80.8 | 0.050 | 86.5 | 0.045 | 63.1 | 94.6 | |
| 150 | 40.4 | 0.058 | 50.8 | 0.053 | 64.8 | 0.048 | 57.8 | 93.6 | |
| 100 | 2.2 | 0.071 | 4.9 | 0.065 | 11.0 | 0.059 | 46.4 | 87.3 | |
| 90% ↗□ 95% | At (2021, 2023, 2025) estimation of 92,03%, 93.68%, 95% with accuracy 0.0475, 0.0454, 0.0438, respectively | ||||||||
| 300 | 100 | 0.031 | 100 | 0.028 | 100 | 0.025 | 37.6 | 72.6 | |
| 250 | 100 | 0.034 | 100 | 0.030 | 100 | 0.027 | 37.3 | 67.3 | |
| 200 | 99.9 | 0.037 | 99.9 | 0.034 | 100 | 0.031 | 30.3 | 64.3 | |
| 170 | 92.4 | 0.041 | 97.2 | 0.037 | 98.9 | 0.033 | 29.4 | 59.1 | |
| 150 | 74.2 | 0.044 | 86.0 | 0.039 | 94.0 | 0.035 | 30.6 | 57.2 | |
| 100 | 19.9 | 0.053 | 35.0 | 0.049 | 41.1 | 0.044 | 25.2 | 46.0 | |
For this purpose, 1,000 samples of a particular size are simulated according to a particular scenario. For each of the 1,000 samples, Bayesian credible intervals are computed for the ‘proportion’ at each of the three successive sampling times and for the slope parameter defining the particular positive or negative linear trend of the scenario at hand, on the logit scale, at the last sampling time, and additionally also the second sampling time.
Regarding the first objective, the accuracy of the estimation, two measures are included in the generic Tables H.1 and H.2:
the percentage of the 1,000 credible intervals (Bayesian confidence intervals) reaching a predefined accuracy; the predefined accuracy is mentioned in Tables H.1 and H.2 in the top line in bold;
the average accuracy of the 1,000 credible intervals.
For the second objective, the power to detect the trend, the percentage of the 1,000 one‐sided credible intervals not containing the 0 slope of no trend (equivalent to rejecting the null hypothesis of no trend against the one‐sided alternative of a positive or negative trend), is determined at the second and third sampling time.
The criteria to select a sample size are as follows: the minimal number guaranteeing that
at least 80% of the credible intervals reach the predefined accuracy;
a power to detect the trend (defined by the scenario) of at least 80%.
Bayesian methodology has been applied for determining the credible intervals. Main motivation to turn to the Bayesian paradigm was the inclusion of the assumed value of the ‘proportion’ at time 0 for determine the power to detect a given trend with a given sample size. The core component of the Bayesian trend model is the basic logistic model:
with denotes the ‘proportion’ at time t = 0 (year 2019) and the three successive sampling times 2, 4 and 6 (the years 2021, 2023 and 2025). Consider as an example the first scenario of set 2: an increasing trend from to . The proportion is represented in the above logistic model by the intercept, more precisely , and thus the logistic equation can be rewritten as:
with β the only remaining unknown parameter representing the in(de)creasing trend. The value β = 0 corresponds to no trend, and power refers to the probability to reject β = 0 (the probability to detect the trend, having truly a trend from to in each of the 1,000 generated samples). However, in reality is not known exactly and the value 0.8 is considered to be a point estimate based on a historical sample of size 170 taken at time 0 (the recommended required sample size in the previous technical specifications). Hence, in the above logistic regression model, is not taking to be equal to 0.8, but is given an informative beta prior corresponding exactly to the uncertainty of a 95% confidence interval based on a historical sample of size 170 with point estimate 0.8. The slope parameter β gets an uninformative prior, such that its estimation is essentially only based on the ‘future’ data of the three successive sampling times t = 2,4,6.
H.3. Outcomes of the simulations
For the first two scenarios of set 1, Table H.1 (grey shaded lines) shows that for the historically applied target number 170 of bacterial isolates to be susceptibility tested per study animal population, per country and per year (EFSA, 2007, 2008, 2012a,b) at each sampling time t = 2,4,6, is performing reasonably well, with 90 to 100% required accuracy and a power of 80.5 for the first and 72.2 for the second scenario. For the last two scenarios of set 1, even the number of 300, while guaranteeing an accurate estimation of the proportion of isolates detected as resistant at each sampling time, does only reach a power of about 13%, far too low.
These findings are summarised in the table below, leading to the general conclusion that a sample size of 170–250 meets the criteria for decreasing trends of 10% in the occurrence of resistance, and that no practically feasible sample size does so for trends of 1–2%.
Table 20.
Scenarios set 1: Occurrence of resistance (proportion of isolates detected as resistant)
| Point estimatesa | Direction of trendb | Magnitude of trendb | Required sample size |
|---|---|---|---|
| 80% | Decreasing | − 10% over 3 successive sampling | 170 |
| 40% | Decreasing | − 10% over 3 successive sampling | 170–250 |
| 10% | Decreasing | − 2% over 3 successive sampling | >> 300 |
| 5% | Decreasing | − 1% over 3 successive sampling | >> 300 |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
The next table summarises the findings for set 2 of scenarios, from the simulations results shown in Table H.2 (grey shaded lines). The number 170 performs sufficiently well for two scenarios. For the second scenario, starting at 40% and increasing to 50%, a higher number up to 300 is needed.
Table 21.
Scenarios set 2: Rate of complete susceptibility (proportion of isolates categorised as susceptible to each of substances of the harmonised panel)
| Point estimatea | Direction of trendb | Magnitude of trendb | Required sample size |
|---|---|---|---|
| 80% | Increasing | + 10% over 3 successive sampling | 170 |
| 40% | Increasing | + 10% over 3 successive sampling | 200–300 |
| 10% | Increasing | + 10% over 3 successive sampling | 170 |
| 5% | Increasing | + 10% over 3 successive sampling |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
Scenario set 3
A different approach was taken for the emergence scenarios. Instead of determining the sample size in order to reach sufficiently accurate estimation of the proportion of isolates detected as resistant or to reach a sufficiently high power to detect a particular upward or downward trend in that proportion, focus is on detecting at least one isolate as resistant, given a particular trend, increasing from 0.01% up to 10% over 3 successive samplings. The probability to detect at least one isolate as resistant within a sample of a particular size can be calculated analytically and no simulations are needed. The probability to detect at least one isolate as resistant as a function of the sample size is (i) based on only the data at the first sampling time, (ii) based on the data of the first together with second sampling time (accumulated probability), (iii) based on all data across all three successive sampling times. The table below indicates the minimum number required at each sampling time to reach a probability of at least 0.8 based on all data across all three sampling times. The number 170 guarantees to detect at least one isolate as resistant with probability more than 80% after three sampling times for a trend from 0% at time 0 (year 2019) to 1% or higher (6 years later, year 2025).
Table 22.
Scenarios set 3: Emergence of (occurrence of) resistance (probability to detect at least one isolate as resistant) [at different rate of emergence]
| Point estimatea | Direction of trendb | Magnitude of trendb | Required sample size |
|---|---|---|---|
| 0% | Increasing | + 0.01% over 3 successive sampling | >>> 300 |
| 0% | Increasing | + 0.5% over 3 successive sampling | 318 |
| 0% | Increasing | + 1% over 3 successive sampling | 159 |
| 0% | Increasing | + 5% over 3 successive sampling | 32 |
| 0% | Increasing | + 10% over 3 successive sampling | 16 |
of occurrence of resistance
As antimicrobial susceptibility testing is performed ‘every second year’, a trend is obtained over 3 successive sampling, i.e. over 6 years. Linear trend on the logistic scale is considered.
H.4. Accounting for the prevalence of the bacterial species monitored
The number of biological samples to be collected from each animal population in order to achieve for example 170 isolates depends on the prevalence of the bacterial species monitored. In the particular case of very low bacterial prevalence, whenever a large number of samples has to be collected to achieve a sufficient number of isolates, a passive surveillance scheme can be implemented using isolates deriving from oriented or systematic sampling. Nevertheless, such samples must not be used for isolation of the indicator organisms E. coli or enterococci because the samples will not be representative of the total population but would represent the targeted population which was sampled. E. coli and enterococci are to be representative of the total population because a future objective will be to link the amount of antimicrobial usage in the animal population to the occurrence of resistance in that population and, therefore, it is not appropriate to study only a targeted subset of the population when considering this requirement.
Appendix I – Required sample size for small countries equivalent to the required sample size of 170 for large countries (small or large in terms of total number of epidemiological units, referred to as the population)
1.
A pragmatic application of the finite population correction factor as applied to the estimation of a proportion
The finite population correction factor is defined as
where f is the sampling fraction
with n the sample size and N the population size.
This factor FPC corrects the variance of the sample proportion (as an estimator for the population proportion), leading to a smaller standard error (higher precision). Note that the correction factor is close to 1 and becomes negligible in case the sample is relatively small as compared to the population (rule of thumb, if f < 0.05, then FPC > 0.95, being close enough to 1 for being ignored).
Suppose n∞ reflects the required sample size for a so‐called (hypothetical) infinite population (meaning very large so that the sample is always expected to be less than 5% of the population). As a correction with FPC reduces the variance, a smaller sample is required to obtain the same accuracy as n∞ for that infinite population. For the determining the corresponding finite population sample size, the correction factor is
and that corrected required sample size equals SSCF·n∞.
For instance, if n∞ = 170 and N = 500, SSCF = 500/670 = 0.746 and consequently the ‘equivalent’ finite population sample size n = 0.746 · 170 = 126.8, or 127 observations are needed instead of 170.
Suppose N=50,000, then SSCF = 50,000/50,170 = 0.997 and hence the reduction is negligible.
Applying this correction to the population sizes N of the following table … :
| Population size | CY | EE | LU | LV | IS |
|---|---|---|---|---|---|
| No. of laying hen flocks in production/year | 134 | 94 | 29 | 50 | 55 |
| No. of broiler flocks slaughtered/year | 1163 | 600 | 5+2 | 700 | 936 |
| No. of pig holdings | 78 | 68 | < 100 | 80 | 13 |
| No. of slaughter batches of pigsa | – | – | – | – | 688 |
a batch/lot of carcases slaughtered the same day and originating from the same herd of pigs.
… leads to the corrected sample size (‘equivalent’ to size of 170):
| Corrected sample size | CY | EE | LU | LV | IS |
|---|---|---|---|---|---|
| No. of laying hen flocks in production/year | 75 | 61 | 25 | 39 | 42 |
| No. of broiler flocks slaughtered/year | 149 | 133 | 7 | 137 | 144 |
| No. of pig holdings | 54 | 49 | 62 | 55 | 12 |
| No. of slaughter batches of pigsa | – | – | – | – | 137 |
a batch/lot of carcases slaughtered the same day and originating from the same herd of pigs.
Appendix J – Generic proportionate stratified sampling approach
J.1. Generic stratified sampling process
The process of stratified sampling for a particular sampling plan in a given MS is depicted in Figure J.1.
Figure J.1.
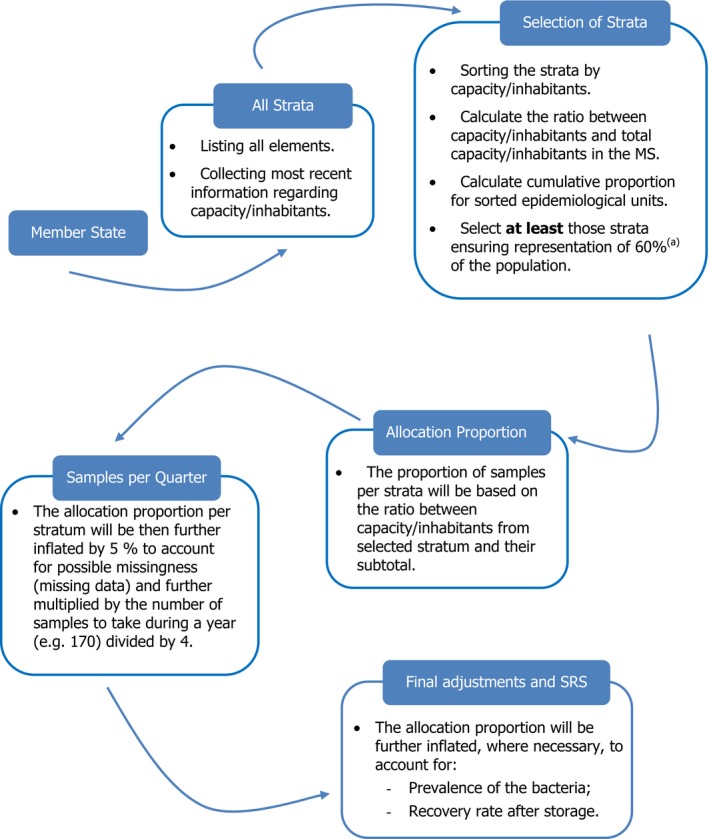
Flow chart presenting the generic process of stratified sampling
- (a): The strata for which cumulative proportion is larger than 0.6 but smaller than 0.7 are selected, respectively.
J.1.1. Identification and selection of strata
Once the targeted population and the elements of the sampling procedure, such as strata and epidemiological units (EpiU), have been identified, the number of strata from which to sample (K) is subsequently selected according to the ‘capacity’ of the stratum. Depending on the sampling plan considered, the ‘capacity’ of the stratum may be (1) the annual throughput of the slaughterhouse in the previous year, (2) the number of inhabitants in the NUTS‐3 region, (3) number of relevant isolates obtained in the previous year and stored in the isolate collection of the laboratory in a MS representing 60%/80% of the total throughput, inhabitants or isolates collected in the MS once sorted by their capacity for the case of slaughterhouse or laboratories or inhabitants in the different regions.
J.1.2. Proportional allocation of sample size per strata and per quarter
Next to the selection of the strata from which to sample, the number of isolates/samples to be taken for each stratum will be proportional to the throughput, inhabitants, or previous isolates collected in the laboratories, considering the total number of isolates/samples to be collected in a year divided into four quarters, further assuming 5% of potential missingness (missing data), the required number of isolates/samples per stratum should be inflated 5% in order to achieve the appropriate number of isolates/samples and power. In the case where the number of isolates/samples to be sampled is N, then the number of isolates/samples to be sampled by quarter for each stratum will be calculated as N divided by 4, further multiplied by 1.05 and then multiplied by the allocation proportion previously described ((XN.Epi.K).
To account for issues that may be encountered in a number of special cases, scenarios have been developed and are illustrated numerically to guide the process of proportional allocation as shown below, always considering simple random sampling within each of the strata. These numerical illustrations of proportional allocation are performed considering that:
the total number of samples/isolates to be collected in a year equals 170 and thus, the number of samples to be collected per quarter (without considering any potential missingness (missing data) should therefore equal at least 43;
the sampling plan focus on those strata ensuring a representation of at least 60%30 of the population (cumulative proportion is larger than 0.6 but smaller than 0.8), as it is foreseen by the legislation for the sampling of caecal samples at the slaughterhouse.
Nevertheless, while calculating the number of caecal samples to be taken at slaughter, it is advisable to account for the prevalence of the zoonotic bacteria and any potential missingness {missing data}.
J.1.2.1. Scenario I
Under Scenario I, it is considered that the number of isolates/samples available/collected per quarter in each of the selected strata ((YN.Epi.K) is larger than the allocated number of isolates/samples to be sampled (XN.Epi.K) (Table J.1).
Table J.1.
| MS | Strata | Stratum capacity | Proportion | Cumulative proportion | Allocation proportion (πN.Epi.K) | Samples per quarter (XN.Epi.K) | Sample unit available (YN.Epi.K) | Samples takenc |
|---|---|---|---|---|---|---|---|---|
| Country X | P | 66,000,000 | 0.1346 | 0.1346 | 0.2075 | 10 | 12 | 12 |
| Country X | G | 65,000,000 | 0.1326 | 0.2672 | 0.2044 | 10 | 8 | 8 |
| Country X | M | 54,000,000 | 0.1101 | 0.3773 | 0.1698 | 8 | 7 | 7 |
| Country X | Q | 46,000,000 | 0.0938 | 0.4711 | 0.1447 | 7 | 9 | 9 |
| Country X | O | 46,000,000 | 0.0938 | 0.5649 | 0.1447 | 7 | 7 | 7 |
| Country X | D | 41,000,000 | 0.0836 | 0.6485 | 0.1289 | 6 | 5 | 5 |
| Country X | K | 39,000,000 | 0.0795 | 0.7280 | – | – | – | – |
| Country X | C | 27,000,000 | 0.0551 | 0.7831 | – | – | – | – |
| Country X | E | 24,000,000 | 0.0489 | 0.8320 | – | – | – | – |
| Country X | A | 22,000,000 | 0.0449 | 0.8769 | – | – | – | – |
| Country X | F | 19,000,000 | 0.0387 | 0.9156 | – | – | – | – |
| Country X | L | 16,000,000 | 0.0326 | 0.9482 | – | – | – | – |
| Country X | J | 13,000,000 | 0.0265 | 0.9747 | – | – | – | – |
| Country X | H | 12,000,000 | 0.0245 | 0.9992 | – | – | – | – |
| Country X | B | 259,000 | 0.0005 | 0.9997 | – | – | – | – |
| Country X | R | 30,000 | 0.0001 | 0.9998 | – | – | – | – |
| Country X | N | 28,000 | 0.0001 | 0.9999 | – | – | – | – |
| Country X | I | 25,000 | 0.0001 | 1 | – | – | – | – |
| Total: | 18 | 490,342,000 | 1 | – | 1 | 48 | 48 | 48 |
The calculation is based on the conventions that the Total number of samples/isolates to be sampled in a year equals 170 and the Number of samples/isolates to be sampled per quarter equals 43, without accounting for any bacteria prevalence nor considering potential missingness (missing data).
Colour legend:
Green Cells: Selected Stratum representing 60% of the total throughput, inhabitants or isolates collected in the Member State.
Yellow Cells: Stratum for which the available samples/isolates is smaller than the number of samples/isolates that should be sampled.
Orange Cells: Stratum for which the available samples/isolates is larger than the number of samples/isolates that should be sampled.
Final adjustments accounting for the prevalence of the bacteria and rate of recovery after storage are not addressed in this illustration.
J.1.2.2. Scenario II
Under Scenario II, it is considered that the number of isolates/samples available/collected in the quarter in some (at least one) each of the selected strata ((YN.Epi.K) is lower than the allocated number of isolates/samples to be sampled (XN.Epi.K), but the total number of available isolates/samples for the selected strata is equal to the required number of isolates/samples to be sampled, thus all available isolates/samples are taken (Table J.2).
Table J.2.
| MS | Strata | Stratum capacity | Proportion | Cumulative proportion | Allocation proportion (πN.Epi.K) | Samples per quarter (XN.Epi.K) | Sample unit available (YN.Epi.K) | Samples takenc |
|---|---|---|---|---|---|---|---|---|
| Country X | P | 66,000,000 | 0.1346 | 0.1346 | 0.2075 | 10 | 12 | 12 |
| Country X | G | 65,000,000 | 0.1326 | 0.2672 | 0.2044 | 10 | 8 | 8 |
| Country X | M | 54,000,000 | 0.1101 | 0.3773 | 0.1698 | 8 | 7 | 7 |
| Country X | Q | 46,000,000 | 0.0938 | 0.4711 | 0.1447 | 7 | 9 | 9 |
| Country X | O | 46,000,000 | 0.0938 | 0.5649 | 0.1447 | 7 | 7 | 7 |
| Country X | D | 41,000,000 | 0.0836 | 0.6485 | 0.1289 | 6 | 5 | 5 |
| Country X | K | 39,000,000 | 0.0795 | 0.7280 | ||||
| Country X | C | 27,000,000 | 0.0551 | 0.7831 | ||||
| Country X | E | 24,000,000 | 0.0489 | 0.8320 | ||||
| Country X | A | 22,000,000 | 0.0449 | 0.8769 | ||||
| Country X | F | 19,000,000 | 0.0387 | 0.9156 | ||||
| Country X | L | 16,000,000 | 0.0326 | 0.9482 | ||||
| Country X | J | 13,000,000 | 0.0265 | 0.9747 | ||||
| Country X | H | 12,000,000 | 0.0245 | 0.9992 | ||||
| Country X | B | 259,000 | 0.0005 | 0.9997 | ||||
| Country X | R | 30,000 | 0.0001 | 0.9998 | ||||
| Country X | N | 28,000 | 0.0001 | 0.9999 | ||||
| Country X | I | 25,000 | 0.0001 | 1 | ||||
| Total: | 18 | 490,342,000 | 1 | 1 | 48 | 48 | 48 |
The calculation is based on the conventions that the Total number of samples/isolates to be sampled in a year equals 170 and the Number of samples/isolates to be sampled per quarter equals 43, without accounting for any bacteria prevalence nor considering potential missingness (missing data).
Colour legend:
Green Cells: Selected Stratum representing 60% of the total throughput, inhabitants or isolates collected in the Member State.
Yellow Cells: Stratum for which the available samples/isolates is smaller than the number of samples/isolates that should be sampled.
Orange Cells: Stratum for which the available samples/isolates is larger than the number of samples/isolates that should be sampled.
Final adjustments accounting for the prevalence of the bacteria and rate of recovery after storage are not addressed in this illustration.
J.1.2.3. Scenario III
Under Scenario III, the number of isolates/samples available/collected in the quarter in some (at least one) of the selected strata (YN.Epi.K) is not larger than the allocated number of isolates/samples to be sampled (XN.Epi.K), but the total number of available isolates/samples is larger than the required number of isolates/samples to be sampled (Table J.3). Next, from those strata (K’) where (XN.Epi.K’ ≥ YN.Epi.K’) all YN.Epi.K’ will be taken, and the rest of the samples to be collected () will be redistributed among the remainder of the strata (Ko). The distribution among the remainder strata will be the minimum value between the samples available () and the sr multiplied by the allocation proportion for each EpiU not yet sampled (K o) divided by the sum of all allocation proportions of the units not yet sampled, the mathematical expression would be:
Table J.3.
| MS | Strata | Capacity | Capacity (%) | Cumulative proportion | Allocation proportion (πN.Epi.K) | Samples per quarter (X N.Epi.K) | Sample unit available (Y N.Epi.K) | Samples takenc |
|---|---|---|---|---|---|---|---|---|
| Country X | P | 66,000,000 | 0.1346 | 0.1346 | 0.2075 | 10 | 15 | 15 |
| Country X | G | 65,000,000 | 0.1326 | 0.2672 | 0.2044 | 10 | 6 | 6 |
| Country X | M | 54,000,000 | 0.1101 | 0.3773 | 0.1698 | 8 | 7 | 7 |
| Country X | Q | 46,000,000 | 0.0938 | 0.4711 | 0.1447 | 7 | 17 | 12 |
| Country X | O | 46,000,000 | 0.0938 | 0.5649 | 0.1447 | 7 | 3 | 3 |
| Country X | D | 41,000,000 | 0.0836 | 0.6485 | 0.1289 | 6 | 5 | 5 |
| Country X | K | 39,000,000 | 0.0795 | 0.7280 | ||||
| Country X | C | 27,000,000 | 0.0551 | 0.7831 | ||||
| Country X | E | 24,000,000 | 0.0489 | 0.8320 | ||||
| Country X | A | 22,000,000 | 0.0449 | 0.8769 | ||||
| Country X | F | 19,000,000 | 0.0387 | 0.9156 | ||||
| Country X | L | 16,000,000 | 0.0326 | 0.9482 | ||||
| Country X | J | 13,000,000 | 0.0265 | 0.9747 | ||||
| Country X | H | 12,000,000 | 0.0245 | 0.9992 | ||||
| Country X | B | 25,9000 | 0.0005 | 0.9997 | ||||
| Country X | R | 30,000 | 0.0001 | 0.9998 | ||||
| Country X | N | 28,000 | 0.0001 | 0.9999 | ||||
| Country X | I | 25,000 | 0.0001 | 1 | ||||
| Total | 18 | 490,342,000 | 1 | 1 | 48 | 53 | 48 |
The calculation is based on the conventions that the Total number of samples/isolates to be sampled in a year equals 170 and the Number of samples/isolates to be sampled per quarter equals 43, without accounting for any bacteria prevalence nor considering potential missingness (missing data).
Colour legend:
Green Cells: Selected Stratum representing 60% of the total throughput, inhabitants or isolates collected in the Member State.
Yellow Cells: Stratum for which the available samples/isolates is smaller than the number of samples/isolates that should be sampled.
Orange Cells: Stratum for which the available samples/isolates is larger than the number of samples/isolates that should be sampled.
Final adjustments accounting for the prevalence of the bacteria and rate of recoverage after storage are not addressed in this illustration.
J.1.2.4. Scenario IV
Under Scenario IV, it is considered that the number of isolates/samples available/collected quarterly in some (at least one) each of the selected strata (YN.Epi.K) is lower than the allocated number of isolates/samples to be sampled (XN.Epi.K), and the total number of available isolates/samples is lower than the required number of isolates/samples to be sampled; thus all available isolates/samples from the selected strata are selected and the number of isolates/samples that need to be still sampled should come from the remainder of the strata (the 40% initially excluded), considering the original ranking of strata performed on the basis of the capacity of stratum (i.e. throughput, inhabitants or isolates collected) (Table J.4).
Table J.4.
| MS | Strata | Stratum capacity | Proportion | Cumulative proportion | Allocation proportion (πN.Epi.K) | Samples per quarter (XN.Epi.K) | Sample unit available (YN.Epi.K) | Samples takenc |
|---|---|---|---|---|---|---|---|---|
| Country X | P | 66,000,000 | 0.1346 | 0.1346 | 0.2075 | 10 | 13 | 13 |
| Country X | G | 65,000,000 | 0.1326 | 0.2672 | 0.2044 | 10 | 6 | 6 |
| Country X | M | 54,000,000 | 0.1101 | 0.3773 | 0.1698 | 8 | 7 | 7 |
| Country X | Q | 46,000,000 | 0.0938 | 0.4711 | 0.1447 | 7 | 11 | 11 |
| Country X | O | 46,000,000 | 0.0938 | 0.5649 | 0.1447 | 7 | 3 | 3 |
| Country X | D | 41,000,000 | 0.0836 | 0.6485 | 0.1289 | 6 | 5 | 5 |
| Country X | K | 39,000,000 | 0.0795 | 0.7280 | 2 | 2 | ||
| Country X | C | 27,000,000 | 0.0551 | 0.7831 | 6 | 1 | ||
| Country X | E | 24,000,000 | 0.0489 | 0.8320 | 3 | 0 | ||
| Country X | A | 22,000,000 | 0.0449 | 0.8769 | 4 | 0 | ||
| Country X | F | 19,000,000 | 0.0387 | 0.9156 | 2 | 0 | ||
| Country X | L | 16000000 | 0.0326 | 0.9482 | 9 | 0 | ||
| Country X | J | 13,000,000 | 0.0265 | 0.9747 | 5 | 0 | ||
| Country X | H | 12,000,000 | 0.0245 | 0.9992 | 2 | 0 | ||
| Country X | B | 259,000 | 0.0005 | 0.9997 | 3 | 0 | ||
| Country X | R | 30,000 | 0.0001 | 0.9998 | 4 | 0 | ||
| Country X | N | 28,000 | 0.0001 | 0.9999 | 8 | 0 | ||
| Country X | I | 25,000 | 0.0001 | 1 | 7 | 0 | ||
| Total | 18 | 490,342,000 | 1 | – | 1 | 48 | 45 | 48 |
The calculation is based on the conventions that the Total number of samples/isolates to be sampled in a year equals 170 and the Number of samples/isolates to be sampled per quarter equals 43, without accounting for any bacteria prevalence nor considering potential missingness (missing data).
Colour legend:
Green Cells: Selected Stratum representing 60% of the total throughput, inhabitants or isolates collected in the MS.
Yellow Cells: Stratum for which the available samples/isolates is smaller than the number of samples/isolates that should be sampled.
Orange Cells: Stratum for which the available samples/isolates is larger than the number of samples/isolates that should be sampled.
Final adjustments accounting for the prevalence of the bacteria and rate of recovery after storage are not addressed in this illustration.
J.1.2.5. Scenario V
Under Scenario V, it is considered that the number of isolates/samples available/collected in the quarter in some of the selected strata (YN.Epi.K) is lower than the allocated total number of isolates/samples to be sampled (XN.Epi.K) and the rest of the strata cannot be sampled (for economical or practical reasons), or the number of samples in the remainder of the strata (the 40% initially excluded) is less than the number of isolates/samples that still need to be sampled, then the next quarter(s) the number of samples to be taken should compensate for the number of samples that were not collected during the previous quarter(s).
Take all isolates/samples from the selected strata (Nq = 45) and the number of isolates/samples to be taken in the next quarter will be equal to the original amount plus the number of samples/isolates that were not taken in the previous quarter e.g. in Table J.5 two possible solutions would be:
To compensate for the number of isolates/samples that were not collected the previous quarter directly in the next quarter, for instance the number of isolates/samples to be collected in the next quarter would be .
The number of samples/isolates to be collected in the following quarters (let nquarter, represent the number of quarter still to be sampled, assuming that nquarter = 3) would be adjusted as
Table J.5.
| MS | Strata | Stratum capacity | Proportion | Cumulative proportion | Allocation proportion (πN.Epi.K) | Samples per quarter (XN.Epi.K) | Sample unit available (YN.Epi.K) | Samples takenc |
|---|---|---|---|---|---|---|---|---|
| Country X | P | 66,000,000 | 0.1346 | 0.1346 | 0.2075 | 10 | 13 | 13 |
| Country X | G | 65,000,000 | 0.1326 | 0.2672 | 0.2044 | 10 | 6 | 6 |
| Country X | M | 54,000,000 | 0.1101 | 0.3773 | 0.1698 | 8 | 7 | 7 |
| Country X | Q | 46,000,000 | 0.0938 | 0.4711 | 0.1447 | 7 | 11 | 11 |
| Country X | O | 46,000,000 | 0.0938 | 0.5649 | 0.1447 | 7 | 3 | 3 |
| Country X | D | 41,000,000 | 0.0836 | 0.6485 | 0.1289 | 6 | 5 | 5 |
| Country X | K | 39,000,000 | 0.0795 | 0.7280 | 0 | 0 | ||
| Country X | C | 27,000,000 | 0.0551 | 0.7831 | 0 | 0 | ||
| Country X | E | 24,000,000 | 0.0489 | 0.8320 | 0 | 0 | ||
| Country X | A | 22,000,000 | 0.0449 | 0.8769 | 0 | 0 | ||
| Country X | F | 19,000,000 | 0.0387 | 0.9156 | 0 | 0 | ||
| Country X | L | 16,000,000 | 0.0326 | 0.9482 | 0 | 0 | ||
| Country X | J | 13,000,000 | 0.0265 | 0.9747 | 0 | 0 | ||
| Country X | H | 12,000,000 | 0.0245 | 0.9992 | 0 | 0 | ||
| Country X | B | 259,000 | 0.0005 | 0.9997 | 0 | 0 | ||
| Country X | R | 30,000 | 0.0001 | 0.9998 | 0 | 0 | ||
| Country X | N | 28,000 | 0.0001 | 0.9999 | 0 | 0 | ||
| Country X | I | 25,000 | 0.0001 | 1 | 0 | 0 | ||
| Total | 18 | 490,342,000 | 1 | 1 | 48 | 45 | 45 |
The calculation is based on the conventions that the Total number of samples/isolates to be sampled in a year equals 170 and the Number of samples/isolates to be sampled per quarter equals 43, without accounting for any bacteria prevalence nor considering potential missingness (missing data).
Colour legend:
Green Cells: Selected Stratum representing 60% of the total throughput, inhabitants or isolates collected in the MS.
Yellow Cells: Stratum for which the available samples/isolates is smaller than the number of samples/isolates to be sampled.
Orange Cells: Stratum for which the available samples/isolates is larger than the number of samples/isolates to be sampled.
Final adjustments accounting for the prevalence of the bacteria and rate of recovery after storage are not addressed in this illustration.
J.1.3. Accounting for the prevalence and recovery rate of the bacteria
The number of samples to be collected from each animal population in order to achieve the number of isolates required depends on the prevalence of the bacteria considered (C. jejuni, C. coli, E. coli, E. faecium and E. faecalis). The CA should estimate the number of samples to be collected in order to obtain the required number of bacterial isolates, given the prevalence of those particular bacteria in caecal samples of the specific animal populations (e.g. the predominant Campylobacter species in broilers, fattening turkeys, fattening pigs and calves). Assuming that the sample is selected randomly and that the prevalence in the animals slaughtered in the selected slaughterhouses is similar to the national prevalence, the number of samples to be collected can be calculated as the inverse of the prevalence. For example, if the prevalence was 70%, the number of samples to be collected in order to ensure the collection of 170 isolates would be equal to 242 (170/0.7 = 242).
Regarding Campylobacter, in those MS where a particularly marked seasonality in the prevalence occurs, it may be desirable, for cost‐effectiveness reasons, to focus on the quarters {seasons} where the prevalence is not low to very low and the chance of isolating Campylobacter higher.
For indicator E. coli, which is highly prevalent, the number of samples to be collected would be slightly higher than the number of isolates required, as it can be assumed that these bacteria are almost constantly recovered from animal caecal samples (prevalence rates/isolation rates ranging between 95 and 100%).
For indicator Enterococcus spp., the monitoring of AMR may be performed on a voluntary basis. The expected prevalence of enterococci is typically much lower, for example around 50% in caecal samples from broilers, and this should be accounted for. The number of samples could be reduced, if one isolate of each of the species E. faecium and E. faecalis, from the same sample, is taken. For this purpose, the samples collected could be tested for the Presence of Enterococcus spp. and more than one isolated strain of Enterococcus spp. per sample (typically between 1 and 5) stored to be subsequently speciated by PCR. Among the E. faecalis and E. faecium isolates available, only one per species and per sample should be finally tested for susceptibility and the results for E. faecium and E. faecalis reported separately. Low numbers of E. faecium and E. faecalis isolates per animal population would mean that for these animal populations there is a decreased precision of the estimate of the proportion of resistance and it would be more difficult to detect trends over time.
As previously indicated, in order to also take into account a 5% missingness (missing data), the number of yearly samples to be collected should be further inflated by 5%, e.g. 179 samples (170 × 1.05 = 178.5) or 254 samples (242 × 1.05 = 254) in the examples given above. As the strain recovery rate after storage may be less than 100%, the number of yearly samples to be collected should be further inflated to offset this, based on the experience of the laboratories, for example by an additional 2%.
J.1.4. Simple random sampling of samples/isolates within the stratum
Simple random sampling (SRS) is the simplest form of drawing elements from a targeted population. It involves drawing elements successively such that each population member has equal and a non‐zero probability of being selected.
A SRS procedure is applied in order to select an adequate number of isolates/samples to be taken by quarter. If, in practice, it is more convenient for the central and local CAs, the SRS may also be planned and performed by month. For certain sampling plans, it may be necessary to perform a series of SRS processes, for example, regarding the sampling of caecal samples at slaughter, working days should be selected first, then batch(es)/lot(s) of carcases originating from the same epidemiological unit (flock or slaughter batch) on the selected days, and finally one carcase (or a number of carcases in the case of a grouped sample) from the selected batch(es).
Considering a list with N elements (e.g. isolates in isolate collection or days/working days in a month), a sample of n elements should be drawn. Each list element is assigned a number, e.g. 1,…N, and the selection can proceed the following way: (i) randomly select a number between 1 and N (this can be done with excel using the = RANDBETWEEN(1, N) function and many other statistical software). The element corresponding to the selected number is included in the sample and it cannot be selected again, this is referred to as sampling without replacement.
Appendix K – Sampling plans of Salmonella isolates from primary production of poultry
K.1. Objective
The objective is to collect and test for antimicrobial susceptibility at least 170 representative Salmonella spp. isolates obtained, respectively, from the populations of laying hen flocks, broiler flocks and fattening turkey flocks in the MS on a yearly basis (whether the sampling was performed by the CA or under its supervision, by the FBO). In any case, operators should carefully check that the Salmonella strains randomly selected and submitted for susceptibility testing originate from different (positive) flocks and, optimally, from different farms.
K.2. The delineation of animal populations
Flocks of broilers, laying hens and fattening turkeys (in the MS where more than 10,000 tonnes are slaughtered on a yearly basis) in production between 1 January to 31 December and covered by the Salmonella National Control Programme (NCP) are eligible for AMR Monitoring in Salmonella spp. isolates.
K.3. Samples and the sampling designs
Two approaches are advisable, according to the Salmonella prevalence in the populations of laying hen flocks, broiler flocks, fattening turkey flocks and the availability of a central database recording the positive flocks in real time.
K.3.1. Approach 1: proportionate stratified sampling of isolates
This approach follows a stratified sampling strategy with proportional allocation within a sampling frame of Salmonella spp. strains deriving from the isolate collections available from the official laboratories and/or other laboratories designated by the CA to carry out testing under the NCP requirements31 involved in the Salmonella NCP. For this purpose, the following parameters are considered when adapting the generic stratified sampling approach to this specific sampling:
Strata : the laboratories involved in the Salmonella NCP32
Stratum capacity : the size of the collection of Salmonella strains originating from the animal population examined and isolated within the framework of the NCP in the previous year, available in the laboratory.
Epidemiological unit (EpiU) : For the purpose of this sampling, epidemiological units are represented by flocks of broilers/laying hens/fattening turkeys.
The Salmonella spp. strains, respectively, isolated from broiler flocks, laying hen flocks and fattening turkey flocks, covered by the Salmonella NCP of the MS and in production from 1 January to 31 December are eligible. It is of note that S. Gallinarum Pullorum strains as well as the Salmonella strains isolated within the context of clinical investigations are excluded for the purpose of the AMR monitoring. Each MS shall list all official laboratories and/or other laboratories designated by the CA involved in the testing of samples within the framework of the NCP and rank them by decreasing order by number of eligible Salmonella spp. strains isolated from laying hen/broiler/fattening turkey flocks between 1 January to 31 December in the previous year. A sampling plan is designed by the CA at the beginning of the year in order to plan the activity and adapted as necessary in the course of the year so that the required number of representative bacterial isolates can be collected and eventually submitted for susceptibility testing.
The list of participating laboratories accounting for (at least 80% of) the total number of Salmonella isolates is then compiled starting with the laboratories with largest isolate collection (to be considered separately per animal population of origin). The isolate sample size for each laboratory in the sampling period is allocated proportionally to the laboratory isolate collection size of the previous year. Every quarter, simple random sampling is performed within the sampling frame of strains (see approach presented), unique per positive flock, up to the selection of the quarter of yearly isolate sample size. Either the sampling frame of Salmonella strains in the collections of the laboratories may include only strains unique per positive flock or a check for duplicates per positive flock is carried out after the random selection.
In the particular case where the number of available isolates is lower than the quarter of the yearly isolate sample size, the scenario III, IV and V should apply in order to complement the isolate sample size up to (at least) 170 isolates.
In the case where less than 170 Salmonella isolates have been isolated in each of these poultry populations in a given year, all available isolates should be included in AMR monitoring, provided that there is no more than one isolate per Salmonella serovar from the same flock (epidemiological unit) per year.
K.3.2. Approach 2: simple random sampling of positive flocks
An alternative approach is to perform a simple random sampling (SRS) within the sampling frame of flocks involved in the NCP and which have tested positive for Salmonella. In the MS where a national database records the flocks of laying hens/broilers/fattening turkeys tested positive for Salmonella spp. (all serovars) in real time, it is advisable to devise the sampling design as a quarterly SRS of the flocks tested positive for Salmonella. Every quarter, the number of positive flocks to be randomly selected among the flocks tested positive over the quarter equals 43 flocks. The SRS is performed according to the procedure developed.
A unique strain of Salmonella spp. (in the case all the isolates recovered belong to the same serovar) or a unique strain per Salmonella serovar (in the case the isolate recovered belong to different serovars) recovered from each positive flock randomly selected is submitted for susceptibility testing. If more than one strain has been isolated from the flock, a random selection of a strain among those isolated is performed from the competent laboratory collection and included in the AMR monitoring programme.
If less than 43 flocks tested positive in a given quarter, the total number of strains available is included in the AMR monitoring programme. In this case, it is advisable that the next quarter(s) the number of isolates to be taken should compensate for the number of samples that were not collected during the previous quarter(s), if possible.
Appendix L – Sampling plans of representative caecal isolates at slaughter
L.1. Objective
The objective is to collect and test for antimicrobial susceptibility representative caecal isolates of:
E. coli from broilers, fattening turkeys, fattening pigs and bovines under 1 year of age;
C. jejuni/C. coli from broilers, fattening turkeys and bovines (under 1 year of age);
C. coli from fattening pigs;
Extended‐spectrum beta‐lactamase (ESBL)‐/AmpC‐/carbapenemase‐producing E. coli from broilers, fattening turkeys, fattening pigs and bovines under 1 year of age; and on a voluntary basis:
E. faecium and E. faecalis from broilers, fattening turkeys, fattening pigs and bovines under 1 year of age;
MSs assess which specific combinations of animal populations and bacteria should be sampled, what number of isolates/samples should be collected and tested, and in which years. Table L.1 can be used as a means to identify the isolates/samples to be collected.
Table L.1.
Animal populations to be sampled for caecal samples at slaughter and number of samples/isolates foreseen per bacteria and per year
| Animal population | Bacteria | 2021 | 2022 | 2023 | 2024 | 2025 | 2026 |
|---|---|---|---|---|---|---|---|
| Broilers | C. jejuni/C. coli f | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc |
| Salmonella | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | |
| E. coli | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | |
| E. faecalis, E. faecium b | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | |
| enzyme‐producing E. coli e | – | 300 samplesd | – | 300 samplesd | – | 300 samplesd | |
| Fattening turkeysa | C. jejuni/C. coli f | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc |
| Salmonella | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | ||
| E. coli | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | |
| E. faecalis, E. faecium b | – | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | |
| enzyme‐producing E. coli e | – | 300 samplesd | – | 300 samplesd | – | 300 samplesd | |
| Fattening pigs | C. coli f | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – |
| Salmonella | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| E. coli | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| E. faecalis, E. faecium b | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| enzyme‐producing E. coli e | 300 samplesd | – | 300 samplesd | – | 300 samplesd | – | |
| Bovines < 1 year of agea | C. jejuni/C. coli f | 170 isolatesc | 170 isolatesc | 170 isolatesc | |||
| Salmonella | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| E. coli | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| E. faecalis, E. faecium b | 170 isolatesc | – | 170 isolatesc | – | 170 isolatesc | – | |
| enzyme‐producing E. coli e | 300 samplesd | – | 300 samplesd | – | 300 samplesd | – |
to be sampled only if production (turkey meat, meat of bovines under 1 year of age) is more than 10,000 t per year.
to be sampled on a voluntary basis.
85 isolates if production is less than 100,000 t per year.
150 samples if production (poultry meat, pig meat) is less than 100,000 t per year or if production (bovine meat) is less than 50 000 t per year.
enzyme‐producing E. coli states for those isolates deriving from specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli and specific monitoring of CP‐producing E. coli
For each animal species, and for each MS, the target of 170 isolates applies to the more prevalent Campylobacter species; all isolates of the other Campylobacter species that are identified, considering the specification of one isolate per species and epidemiological unit, are to be included.
L.2. The delineation of the animal populations
The eligible populations of broilers/fattening turkeys/fattening pigs/bovines of less than 1 year of age cover those domestically produced and slaughtered in the slaughterhouses covering at least 60% of the all broilers/fattening turkeys/fattening pigs/bovines of less than 1 year of age slaughtered in the MS.
It is proposed that domestically produced broilers, fattening turkeys, fattening pigs and calves of less than 1 year of age may be, respectively, defined:
Regarding broilers and fattening turkeys, as birds hatched and raised in the MS as well as those imported/traded as day‐old chicks in the MS (i.e. hatched abroad).
Regarding fattening pigs and calves of less than 1 year of age, as those animals produced and slaughtered in the MS and excluding those imported and intended for direct slaughter after importation. Therefore, pigs imported/traded in the MS as post‐weaners, growers and fatteners are considered as domestically produced animals. As a general guideline, domestically produced animals should spend at least about 50% of their lifespan in the MS.
L.3. Samples and sampling design
A sampling plan is to be designed by the CA at the beginning of the year (at the latest) in order to ensure that the required number of representative bacterial isolates is collected and eventually submitted for susceptibility testing. For this purpose, the following parameters are considered when adapting the generic stratified sampling approach to this specific sampling:
Strata (1st stage) : the slaughterhouses where eligible animals are slaughtered within the MS.
Stratum capacity : the annual throughput of domestically produced meat of the slaughterhouse during the previous year or according to the latest data available.33
Strata (2nd stage) : For the purpose of this sampling at slaughter, batches/lots of carcases originating from the same epidemiological unit (flock of broilers/fattening turkeys, slaughter batch of fattening pigs and slaughter batch of bovines under 1 year of age) are used for a second stage selection. As indicated below, no more than one caecal content sample (single or pooled)34 per batch/lot of carcases of the same epidemiological unit should be collected.
Accounting for the bacteria prevalence, possible missingness and possible loss during storage, for the definition of the number of samples to be collected in each quarter of the year from each selected slaughterhouse should ensure that the isolates required by the monitoring programme are collected. If after the first quarter the CA realises that the number of samples is not sufficient to obtain the expected number of isolates, the number of samples to be collected in the following quarter(s) may be adapted and recalculated (see Scenario V).
L.3.1. Proportional allocation
Each MS will rank all slaughterhouses by throughput of eligible animals between 1 January to 31 December, in the previous year or according to the latest data available. Starting with the slaughterhouses of largest throughput, sufficient slaughterhouses should be enrolled to cover at least 60% of the national throughput of animals domestically produced. A list of participating slaughterhouses is then compiled, and the predetermined number of carcases to be sampled is distributed according to the proportional throughput from 1 January to 31 December in the previous year.
L.3.2. Simple random sampling of days, batch(es) of carcases and carcases
For each month, sampling sessions should be allocated randomly to the days of the month, based on operating days for that particular slaughterhouse. Simple random sampling techniques, as previously described, should be used. Preferably, a maximum of one caecal sample per slaughterhouse is sampled per day to ensure that there is no correlation between positive results that may derive from direct or indirect contact between sampled animals before slaughter. For this purpose, a number of sampling days (equal to the sample size) is randomly selected from the operating days of the slaughterhouse.
For each slaughterhouse each month, a number between 1 and 31 shall be selected at random. If the randomly selected number is a slaughtering day, for that month, then that day is selected for sampling. If not, then a new number is selected randomly. This process is preferentially performed once a month and repeated so many times as there are samples to be collected at the slaughterhouse. In addition, a number of practical issues may need to be taken into account when defining sampling days and samples to be taken at slaughterhouse. Sampling days may need to be restricted based on laboratory and courier limitations, e.g. in some situations it may be necessary to exclude Fridays. For each sampling day, if more batches are slaughtered and more than one sample a day is collected, batches to be sampled should be selected randomly. From each selected batch/lot of carcases originating from the same epidemiological unit, one caecal content sample (single or pooled) from healthy35 animal(s) should be randomly selected and collected. This would ensure that only one isolate per epidemiological unit of origin per year is finally submitted to susceptibility testing. Where it is not possible to sample only one caecal sample per day per slaughterhouse due to logistical reasons, it may be proposed that up to five caecal samples per day can be sampled.36
A stratification by slaughterhouse does not rule out the possibility of taking account of production types (e.g. for broilers: organic, free‐range, standard), notably where relevant statistics/information are available.
Appendix M – Sampling designs of representative isolates from meat at retail
M.1. Objective
The objective is to collect 300 representative random samples of fresh meat of broilers, pig meat and bovine meat, respectively, and to test them for the presence of ESBL‐/AmpC‐/carbapenemase‐producing isolates of E. coli. However, in MS with a production of less than 100,000 tonnes of poultry meat slaughtered per year, less than 100,000 tonnes of pig meat slaughtered per year and less than 50,000 tonnes bovine meat slaughtered per year37 the MS shall analyse 150 samples instead of 300 samples for each corresponding specific combination.
M.2. The delineation of the meat categories
The eligible categories of fresh meat of broilers, pig meat and bovine meat cover those marketed at the retail stage in the NUTS‐3 area representing at least 80% of the population in the MS.
Within the framework of this sampling plan, retail is understood as outlets selling directly to the final consumer for subsequent domestic consumption, i.e. outlets such as supermarkets, specialist shops, markets and excluding catering activities, restaurants, wholesalers and similar outlets.
Within the framework of this sampling plan, fresh meat is understood as chilled meat (meaning that frozen meat is excluded), including meat that is wrapped, vacuum‐wrapped or wrapped in a controlled atmosphere. As there is a specific interest as regards the consumer being exposed further up the food chain, then carcases or meat portions with skin on from broilers should be sampled as a measure of the likely heaviest degree of contamination from the animals. Selecting a standard type of meat that is readily available in all MS is important for harmonised monitoring. Un‐skinned carcases/meat portions (diced or breast) for broilers and samples of meat of pigs, and meat of cattle (both displayed without skin, typically) (sliced or diced) should be sampled.
M.2.1. Sampling design
A proportionate stratified sampling scheme is used at the MS level whereby the samples are allocated proportionally to the size of the human population in the regions (NUTS‐3 area) accounting for at least 80% of the national population. Next, at the second level, are the retail outlets to be sampled. At the third level, samples within the different meat categories to be sampled are selected. The 300/150 samples (of each meat category) are to be allocated in proportion to the size of the human population in the NUTS‐3 area. The assumption underpinning the allocation of sample numbers to the regions, within MS, according to the size of their human populations, is that the human population sizes are fairly proportional to the volume sizes of the selected food categories on the market.
For the purpose of the proportional allocation of sample numbers, the following parameters are considered when adapting the generic stratified sampling approach to this specific sampling plan:
Strata: the NUTS‐3 area accounting for at least 80% of the national population38;
Stratum capacity: the number of inhabitants according to the most recent data available;
Epidemiological unit (EpiU): lot of chilled fresh meat. No more than one sample per epidemiological unit per year should be collected.
Ideally, the central CA should draw up a sampling plan following the rules described below and based on the best marketing data available.
Selection of the retail outlet categories to be targeted
The CAs are responsible for choosing the retail outlets to be included. Typical types of retail outlets that could be included for sampling are: supermarkets and small shops/speciality delicatessens (e.g. traditional craft butcher's businesses). Stratification on the major types of retail outlets allows for accounting for potential differences in supply chains (imported vs. domestic) and meat production types (e.g. for broiler meat: farmer's, organic, free‐range, standard, etc.). The following rule shall be used to choose the types of retail outlets to be sampled and needs to be followed for each of meat categories:
If the biggest category of outlets (for example supermarkets) supply at least 80% of the market of a meat category then samples only need to be taken from those outlets. Where that is not the case, the second largest outlet category should be added and so on until at least 80% of the market is covered.
The number of samples that should be taken from each retail outlet category included in the sampling plan should be proportionate to the market share of that outlet category within the targeted outlet categories.
If no major differences in supply chains and meat production types are expected between the categories of outlets, sampling may be limited to the biggest category of outlets in the MS to reduce logistical constraints.
With respect to the definition of sampling days and selection of the samples to be taken within each epidemiological unit, simple random sampling techniques should be used. In addition, a number of practical issues may need to be taken into account when defining sampling days and samples to be taken at retail. For each month sampling sessions should be allocated randomly to the days of the month, based on working days. Sampling days may need to be restricted based on laboratory and courier limitations, e.g. in some situations it may be necessary to exclude Fridays.
At the same visit to a retail outlet, up to five different meat batches/lot per meat category can be sampled without preselecting samples based on the origin of the food. It is essential that cross contamination is avoided during the collection of fresh meat samples. Precautions must therefore be taken at all stages to ensure that the equipment used during sampling, transport and storage is not contaminated.
M.2.2. Complementary AMR monitoring in additional animal populations and food categories
To complement AMR monitoring previously described, MS may be interested in monitoring AMR in other animal species/populations, food categories and points of the food chain other than those foreseen by Decision 2013/652/EC. This may include may include monitoring of imported (fresh or frozen) meat from third countries at the border inspection posts, non‐domestically produced meat (e.g. animals slaughter in the country but born and raised abroad) or frozen meat at retail, and other animal species (e.g. rabbit, sheep).
For imported meat, a stratified sampling design per MS with proportional allocations of the number of isolates to the quantities imported from third countries of origin, appears to be the best and the most cost‐effective option. For example, the monitoring of AMR in poultry meat imported from third‐countries may be a specific target of this kind of complementary monitoring regarding imported meat. The statistics on broiler meat importations issued by EUROSTAT could be used to construct the sampling plan. In any case, AMR data deriving from the monitoring of imported meat from third countries should be reported separately.
Appendix N – Harmonised method for isolation, identification and storage of Campylobacter jejuni and/or C. coli from broilers, fattening turkeys, fattening pigs and calves
N.1. Scope of the method
The protocol describes the detection and identification of Campylobacter jejuni and C. coli by direct plating from caecal content of broilers, turkeys, and bovine of less than 1 year of age and C. coli by direct plating from caecal content of fattening pigs. The aim is to obtain, as far as possible, one isolate of C. jejuni and one isolate of C. coli from each epidemiological unit of poultry and calves and one isolate of C. coli from each epidemiological unit of pigs.
The first step consists in determining, by direct plating, the presence or absence of thermo‐tolerant Campylobacter species in the sample, the second step being the identification of C. jejuni and C. coli.
N.2. References
This protocol is based on the EN ISO 10272‐1: ‘Microbiology of the food chain – horizontal method for detection and enumeration of Campylobacter spp.’
N.3. Detection of the presence of thermo‐tolerant Campylobacter
N.3.1. Transport of samples and storage before analysis
Samples should be maintained at a temperature of 5+/−3°C. Analysis should begin as soon as possible, preferably within 96 hours* after collecting the samples. Samples should not be left to dry.
*The EURL‐Campylobacter will validate in 2019 the delay of 96 hours for samples from pig. This delay has already been validated for samples from poultry and bovine animals.
N.3.2. Inoculation
Using a sterile loop of 10 microliters, the well mixed samples are plated directly onto the first half of selective mCCD agar (and possibly on to a second media)*. A second loop is used to streak out on the second half of the plate.
*A study to evaluate the benefit of using a second isolation media in addition to mCCDA is planned to be conducted in 2019 by the EURL‐Campylobacter and laboratories from four other MSs. mCCDA, Preston and Butzler media will be used in the participating countries to analyse chicken and pig caecum samples according to the present protocol, and isolation rates of C. jejuni and C. coli will be compared. The present protocol is written as in the case two media are used. Texts to be amended in the case only mCCDA is used are indicated with an asterisk.
A quality control procedure to validate the productivity and selectivity of the agar media should be carried out as described in Annex B of the EN ISO 10272‐1: ‘Microbiology of the food chain – horizontal method for detection and enumeration of Campylobacter spp.’, using reference strains of C. jejuni, C. coli, E. coli and S. aureus.
N.3.3. Incubation
The (two)* plates are incubated at 41.5°C +/−1°C in a microaerobic atmosphere and examined after 44 h+/−4h to detect the presence of suspect Campylobacter colonies.
N.3.4. Purification before confirmation and storage
Based on colony morphology (greyish, flat and moist colonies, sometimes with metallic sheen and tendency to spread, on mCCD agar (and typical appearance on second media)*, five typical or suspect colonies are selected for confirmation and identification.
Tests should be carried out without delay to avoid culturability loss of Campylobacter isolates in air.
Re‐streak each selected colony to purify on a non‐selective blood agar plate (e.g. Columbia blood agar), in order to obtain well isolated colonies.
Incubate at – 41.5°C for 24–48 h in jars under microaerophilic environment.
From each plate of Columbia blood agar medium, select one well isolated colony to streak again onto a plate of blood agar medium in order to obtain a heavy growth of a pure culture for identification and storage. Incubate at 41.5°C for 24–48 h in jars under microaerophilic environment.
N.3.5. Identification
Identify selected subcultures until one isolate of C. jejuni and one isolate of C. coli are detected. However, for samples from pigs, only one C. coli is to be identified. Identification can be performed using either matrix‐assisted laser desorption/ionisation time of flight mass spectrometer (MALDI‐TOF) according to the manufacturer's instructions or PCR according to the protocol recommended by the Antimicrobial resistance reference laboratory (https://www.eurl-ar.eu/protocols.aspx), eventually after one or several of the tests described in EN ISO 10272‐1 (examination of morphology and motility, aerobic growth at 25°C, oxidase activity, catalase activity, hippurate hydrolysis and indoxyl acetate hydrolysis). Other methods may be used provided the suitability of the method is verified (ISO 7218).
Positive (C. jejuni and C. coli) and negative (E. coli) reference strains must be used for controls.
N.3.6. Expression of results
For each sample of caeca, according to the identification of the typical or suspect colonies, the result is indicated as:
-
–
Presence of C. jejuni detected or not detected (for poultry and calves samples)
-
–
Presence of C. coli detected or not detected (for poultry, pigs and calves samples).
(The media from which each isolate was obtained is planned to be recorded for further transmission to EFSA).
N.3.7. Selection of isolates for MIC testing
Determination of MICs should be performed on a maximum of one C. jejuni and one C. coli per batch of animals. In case more than two isolates are found to belong to the same species, one is randomly chosen for MIC testing. (Data concerning the media from which this isolate was obtained are registered for further transmission)*.
The AST can be performed either directly after identification or after appropriate storage.
Storage
Store at –70°C in glycerol peptone water or beads
-
–
The randomly selected C. jejuni isolate and the C. coli isolate, or
-
–
The five presumptive Campylobacter from the last blood agar plate inoculated, if identification is performed on thawed cultures, after storage at −70°C and/or transport. In this case, measures to ensure viability of the cultures during storage and transport must be taken.
Appendix O – Harmonised parameters for the specific monitoring of ESBL‐/carbapenemase‐producing E. coli
O.1. Isolation of ESBL‐/AmpC‐/carbapenemase‐producing E. coli
For the sake of continuity, it is proposed that for the specific monitoring of ESBL‐/AmpC‐/carbapenemase‐producing E. coli, the isolation method starts with a non‐selective pre‐enrichment step, followed by inoculation on MacConkey agar containing a third‐generation cephalosporin in a selective concentration (cefotaxime 1 mg/L), in accordance with the most recent version of the detailed protocol for standardisation of the EU Reference Laboratory for Antimicrobial Resistance (EURL‐AR).39 Using this protocol, also carbapenemase‐producing isolates can also be recovered.
If available, one presumptive ESBL‐/AmpC‐/carbapenemase‐producing E. coli isolate obtained from each positive caecal sample and meat sample is tested for its antimicrobial susceptibility to the first panel of antimicrobials to confirm the microbiological resistance to cefotaxime (expected as the antimicrobial is present in the isolation medium at a concentration higher than the ECOFF), and identify possible resistance to ceftazidime and/or ceftazidime and/or meropenem. In a second step, the isolate should be tested using the second panel of antimicrobials to infer the presumptive ESBL‐/AmpC‐/carbapenemase‐producing phenotype according to the β‐lactam resistance phenotype obtained.
O.2. Identification of presumptive ESBL, AmpC and/or carbapenemase producers
Definition of ESBL, AmpC, ESBL + AmpC, CP‐phenotypes
The categorisation of isolates resistant to third‐generation cephalosporins and/or carbapenems in presumptive ESBL, AmpC or carbapenemase producers was carried out based on the EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance (EUCAST, 2013). In these expert guidelines, and based on other EUCAST and CLSI guidelines to detect ESBL/AmpC producers, a screening breakpoint of > 1 mg/L is recommended for cefotaxime and ceftazidime. This screening breakpoint is higher than the ECOFFs applied for antimicrobial susceptibility of both antimicrobials for E. coli, and to cefotaxime for Salmonella. For this report, a first condition for classifying isolates as presumptive ESBL/AmpC producers related to their MIC for either cefotaxime or ceftazidime, was to apply this screening breakpoint of MICs > 1 mg/L. Only isolates which presented MIC values accomplishing with this requisite (as expected for most of the ESBL/AmpC producers) were further considered. In total, for the third generation cephalosporin‐ and/or carbapenem‐resistant isolates, five main categorisations are made: 1. ESBL phenotype; 2. AmpC phenotype; 3. ESBL + AmpC phenotype; 4. CP‐phenotype; and 5. Other phenotypes (see Figure O.1).
Figure O.1.
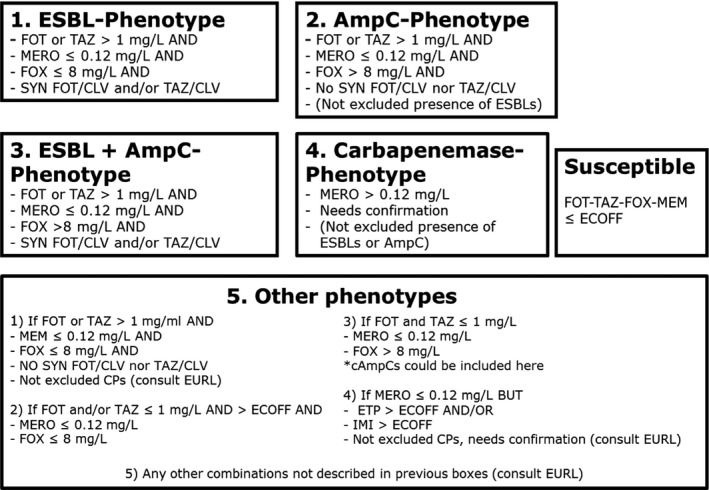
Phenotypes inferred based on the resistance to the β‐lactams included in Panel 2
- Presumptive ESBL‐producers include isolates exhibiting Phenotype 1 or 3.
- Presumptive AmpC producers include isolates exhibiting Phenotype 2 or 3.
To detect the production of ESBLs, a synergy test for cefotaxime and ceftazidime, in combination with clavulanic acid was performed. An eight‐fold reduction in the MIC for the cephalosporin combined with clavulanic acid compared with that obtained for the cephalosporin alone was interpreted as a positive synergy test. In all other cases, the synergy test was considered negative. For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and a synergy test positive for any of these antimicrobials, together with susceptibility to cefoxitin (≤ 8 mg/L) and meropenem (MEM ≤ 0.125 mg/L see CP phenotype) were classified as ESBL phenotype.
For the AmpC phenotype, the combination MIC > 8 mg/L (ECOFF) for cefoxitin together with MICs > 1 mg/L for cefotaxime and/or ceftazidime was used as phenotypic criteria to investigate the presence of AmpC production in E. coli. It should be also underlined that there are a few AmpC enzymes that do not confer resistance to cefoxitin (i.e. ACC‐1), and that there are other mechanisms (porin loss, presence of carbapenemases, a few ESBLs like cefotaxime (CTX‐M)‐5 that could generate similar MIC values for the different antimicrobials (EFSA, 2012a; EUCAST, 2013). Phenotypic AmpC confirmation tests (i.e. cloxacillin synergy) were not required for the present monitoring. For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime and cefoxitin MIC > 8 mg/L, together with negative synergy test for both cefotaxime and ceftazidime/clavulanic acid, together with susceptibility to meropenem (MEM ≤ 0.125 mg/L) were classified in the AmpC phenotype category. No distinction between acquired AmpC and natural AmpC was made.
For the present report, isolates with MICs > 1 mg/L for cefotaxime and/or ceftazidime, positive synergy tests for any of these antimicrobials with clavulanic acid and cefoxitin MIC > 8 mg/L, together with susceptibility to meropenem (MEM ≤ 0.12 mg/L) were classified under the ESBL + AmpC phenotype category. In some isolates, several mechanisms can be present at the same time, making it very difficult to differentiate the phenotypes. Also the high‐level expression of AmpC β‐lactamases can mask the presence of ESBLs. AmpC can also be present in isolates with positive ESBL tests (clavulanic acid synergy). In this case, the cefepime/clavulanic acid synergy test should be used to overturn/confirm the presence of ESBLs in these isolates (EUCAST, 2013) but, unfortunately, the combination cefepime/clavulanic acid was not included among the substances tested for monitoring. The inclusion of resistance to cefepime with a MIC value ≥ 4 mg/L as an additional criterion proposed elsewhere (EFSA, 2012a), could be useful to ascertain the presence of an ESBL‐producer.
For the classification of isolates into the putative carbapenem producers (CPs), a meropenem screening cut‐off of > 0.125 mg/L (which coincides with the harmonised ECOFF) was chosen. It is known that other mechanisms (i.e. hyperproduction or combination of ESBLs and/or AmpC and porin loss) can also affect to the MIC values generated for the different carbapenems, especially for ertapenem. The confirmation of the carbapenemase production recommended by the EUCAST guidelines cannot be inferred from the carbapenem susceptibility testing data reported, but needs further phenotypic or molecular testing. Those MSs that reported data suggesting the presence of putative CPs were recommended to validate the results by performing further confirmatory testing, and the EURL‐AR offered to apply WGS of the isolates. For the present report, isolates with MIC > 0.125 mg/L for meropenem would be considered as presumptive CP producers and were classified under the CP phenotype. The presence of other resistance mechanisms (ESBLs, AmpC, etc.) within the isolates placed in this group cannot be ruled out.
In this group, phenotypes not included in the categorisations defined above were included: isolates with a MIC > 0.125 mg/L for ertapenem and/or MIC > 1 mg/L for imipenem (EUCAST screening cut‐offs, one dilution step higher than the currently defined ECOFFs) but no resistance to meropenem (MIC < 125 mg/L) were classified under the category ‘other phenotype’. Finally, isolates with MICs ≤ 1 mg/L for cefotaxime and ceftazidime would be considered as not ESBL and/or AmpC producers. This implied that some isolates considered as microbiologically resistant (MICs over the ECOFFs) would not be further classified, as probably other mechanisms or technical issues in the MIC testing (i.e. MIC value close to the ECOFF) would be responsible for the MIC values obtained. For the present report, cefotaxime‐ and ceftazidime‐resistant isolates with MICs ≤ 1 mg/L for both antimicrobials were considered as putative non‐ESBL/AmpC producers and were classified under the category ‘other phenotype’.
We are aware that without a further molecular characterisation of the isolates, it will not be possible to know exactly which resistance mechanisms are present. For epidemiological purposes and based on the EUCAST guidelines, the classification of ‘presumptive’ producers for the different mechanism conferring resistance to third‐generation cephalosporins and/or carbapenems was considered. Molecular characterisation of these mechanisms is recommended.
Appendix P – Harmonised plates for Campylobacter spp.
P.1. Previous plate (in mg/L)
Suggestions for removal are indicated in red font below.
| ERY 128 | CIP 16 | TET 64 | GEN 16 | NAL64 | STR 16 |
| ERY 64 | CIP 8 | TET 32 | GEN 8 | NAL32 | STR 8 |
| ERY 32 | CIP 4 | TET 16 | GEN 4 | NAL16 | STR 4 |
| ERY 16 | CIP 2 | TET 8 | GEN 2 | NAL8 | STR 2 |
| ERY 8 | CIP 1 | TET 4 | GEN 1 | NAL4 | STR 1 |
| ERY 4 | CIP 0,5 | TET 2 | GEN 0,5 | NAL2 | STR 0,5 |
| ERY 2 | CIP 0,25 | TET 1 | GEN 0,25 | NAL1 | STR 0,25 |
| ERY 1 | CIP 0,12 | TET 0,5 | GEN 0,12 | POS | POS |
P.2. Proposal
New concentrations (mg/L) are indicated in grey shading in Figure P.1 below. Cut‐offs for C. jejuni (A) and C. coli (B) are indicated with horizontal black line.
Figure P.1.
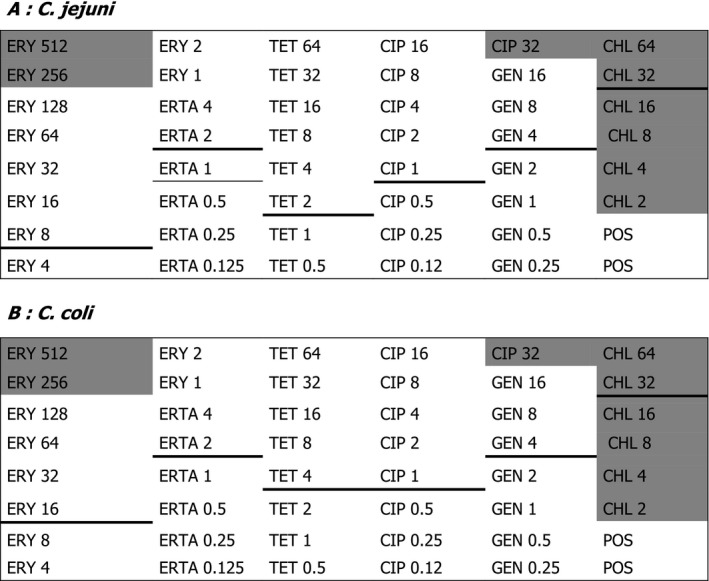
Design of previous plate and suggestions of alterations
- ERY: erythromycin; CIP: ciprofloxacin; TET: tetracycline; STR: streptomycin; GEN: gentamicin; CHL: chloramphenicol; ERTA: ertapenem; POS: positive control.
Appendix Q – Examples of reference databases/Tools/Pipelines/software
| Reference Databases (examples of) a | Tools (examples of) a | Pipelines/platforms/software (examples of) a |
|---|---|---|
|
Resfinder (V.3.1): The ResFinder databases, acquired genes and chromosomal mutations, are manually curated. Versions V.2 and previous versions of ResFinder did not include chromosomal mutations. (DTU, http://www.genomicepidemiology.org/) CARD reference database: The reference database used by the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/) is a manually curated resource containing high quality reference data on the molecular basis of AMR, with an emphasis on the genes, proteins and mutations involved in AMR as well as their associated phenotypes able to detect genes as well and mutations conferring AMR to fluoroquinolones, and cephalosporins. ARG‐ANNOT |
Resfinder 3.1: ResFinder consists of two programs, ResFinder which identifies acquired genes, and PointFinder identifing chromosomal mutations ResFinder is an open source tool. Allows choosing between searching for AMR genes or for chromosomal point mutations. For the first option, it is possible to select some or all the families of antibiotics for which genes should be found, as well as choosing the threshold for %ID and the minimum length. For the chromosomal point mutations, the species queried should be selected and whether if all mutations should be reported or only those known. Versions V.2 and previous versions of ResFinder did not detect chromosomal mutations. (DTU, http://www.genomicepidemiology.org/) CARD Resistance Gene Identifier (RGI): Uses CARD's curated AMR detection models to predict complete resistome from genome sequences. The RGI provides a preliminary annotation of the submitted DNA sequence(s) or protein sequences based upon the data available in the CARD. ABBRICATE: Combines the results of mapping different databases: CARD, Resfinder V.2, ARG‐ANNOT. ‘Genefinder’ algorithm (used at PHE): Maps the sequencing reads to a set of reference sequences using Bowtie 2 followed by generation of an mpileup file using Samtools (Langmead and Salzberg, 2012). The reference database used included acquired genes and mutations for beta‐lactamases (Day et al., 2017; Sadouki et al., 2017) as well as reference genes from CARD and Resfinder (Neuert et al., 2018). ABRES Finder: The Antibiotic Resistance gene Finder henceforth abbreviated ABRES Finder is a consortium of antibiotic resistance genes where one can get all the relevant information regarding antibiotic resistance at single stop. Currently, ABRES Finder is focussed about the prevalence of antibiotic resistance in India. ABRES is centralised on specific genes in microorganism which causes the phenomenon of antibiotic resistance. The aim is to provide a complete set of information about resistance genes, subfamily of the genes, mechanism of antibiotic resistance and organism. Periodic addition of new resistance genes and sub‐types will be done in the database. http://scbt.sastra.edu/ABRES/ ARGminer: Antibiotic Resistance Gene miner database – A new web‐based curation system, ARG‐miner, which supports annotation of ARGs at multiple levels, including: gene name, antibiotic category, resistance mechanism, and evidence for mobility and occurrence in clinically important bacterial strains. To overcome limitations of manual curation, we employ crowdsourcing as a novel strategy for expanding curation capacity towards achieving a truly comprehensive, up‐to‐date database. We develop and validate the approach by comparing performance of multiple cohorts of curators with varying levels of expertise, demonstrating that ARG‐miner is more cost effective and less time‐consuming relative to traditional expert curation. https://bench.cs.vt.edu/argminer/#/classify;gene_id=A0A0P9K4B7 ARGO: Antibiotic Resistance Genes Online – A database containing gene sequences conferring resistance to these two classes of antibiotics, vancomycin and β−lactam resistance genes. It is designed as a resource to enhance research on the prevalence and spread of antibiotic resistance genes. ARGO is the first attempt to compile the resistance gene sequence data with state specific information. http://www.argodb.org/ BLDB: The aim of the Beta‐Lactamase DataBase (BLDB) is to compile sequence information as well as biochemical and structural information on all the currently known β‐lactamases. BLDB offers in addition tools to analyse β‐lactamases and provides important links to the related web resources (NCBI, PDB, etc.). This comprehensive web‐based database, which is updated on a weekly basis, may provide at a glance useful insights in the structure‐function relationships of β‐lactamases, and thus allowing a better understanding of substrate specificities, determine key residues involved in substrate recognition and hydrolysis, and to foresee the impact of mutations in the hydrolysis profile. http://bldb.eu/ BacMet: A database of biocide and metal resistance genes with highly reliable content. In BacMet version 1.0, there were 470 resistance genes in the experimentally confirmed database, whereas the predicted database had 25,477 resistance genes. In BacMet version 1.1, the experimentally confirmed database contained 704 resistance genes, whereas the predicted database contained 40,556 resistance genes. Currently, in BacMet version 2, the experimentally confirmed database contains 753 resistance genes, while the predicted database contains 155,512 genes. The much bigger increase in the predicted database is largely a consequence of the rapidly increasing number of reported bacterial genes in public databases. http://bacmet.biomedicine.gu.se/about_BacMet.html Lahey: Tables of β‐Lactamase Classification and Amino Acid Sequences for TEM, SHV and OXA Extended‐Spectrum and Inhibitor Resistant Enzymes. As of July 1 2018, the Lahey site is no longer assigning beta‐lactamase allele numbers. Please contact https://www.ncbi.nlm.nih.gov/pathogens/submit-beta-lactamase for further details https://www.lahey.org/Studies/ CBMAR: A database, which facilitates comprehensive molecular annotation and discovery of novel β‐lactamases. As against the limited scope of other existing similar databases, CBMAR provides information useful for molecular and biochemical characterisation of each family of β‐lactamase. The basic architecture of CBMAR is based on Ambler classification, which divides β‐lactamases as serine (Classes A, C and D) and metallo‐β‐lactamases (Class B). Each class is further divided into several families on the basis of their hydrolytic character. In CBMAR, each family is annotated with (i) sequence variability, (ii) antibiotic resistance profile, (iii) inhibitor susceptibility, (iv) active site, (v) family fingerprints, (vi) mutational profile, (vii) variants, (viii) gene location, (ix) phylogenetic tree and several other features. Each entry also has external links to the relevant protein/nucleotide sequence and structure databases. The database also supports sequence similarity searches using BLAST and assigns a new β‐lactamase protein to its respective family on the basis of family‐specific fingerprint. http://14.139.227.92/mkumar/lactamasedb FARME DB: FARMEDB is a database of DNA and protein sequences derived exclusively from environment sequences showing AR in laboratory experiments. The Functional Antibiotic Resistant Metagenomic Element (FARME) database is a compilation of publicly available DNA sequences, predicted protein sequences conferring antibiotic resistance and additional regulatory and mobile genetic elements and predicted proteins flanking the antibiotic‐resistant genes. FARME is the first database to focus on functional metagenomic AR gene elements and provides a resource to better understand antibiotic resistance in the 99% of bacteria which cannot be cultured as well as their relationship to known antibiotic‐resistant genes derived from cultured isolates. http://staff.washington.edu/jwallace/farme/index.html Galileo AMR: A unique system (Attacca) that has allowed creation an archive of relevant elements and identification of these elements in bacterial DNA sequences. The aim of this project is to make our repository of antibiotic resistance genes available to the wider research community, to allow researchers to annotate sequences containing resistance genes and associated mobile elements using our system and to contribute new entries to the repository as these are found. https://galileoamr.arcbio.com/mara/ MEGARes: AmrPlusPlus is a Galaxy‐based metagenomics pipeline that is intuitive and easy to use. The pipeline takes advantage of current and new tools to help identify and characterise resistance genes within metagenomic sequence data. http://megares.meglab.org/amrplusplus/latest/html/ NDARO: NCBI National Database of Antibiotic Resistant Organisms, a collaborative, cross‐agency, centralised hub for researchers to access AMR data to facilitate real‐time surveillance of pathogenic organisms. https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/ PATRIC: Bacterial bioinformatics resource center, the Pathosystems Resource Integration Center, provides integrated data and analysis tools to support biomedical research on bacterial infectious diseases. https://patricbrc.org/ Resfams: Resfams is a curated database of protein families and associated profile hidden Markov models (HMMs), confirmed for antibiotic resistance function and organised by ontology. http://www.dantaslab.org/ ARGs‐OAP: An online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG‐database. https://smile.hku.hk/SARGs |
CGE Analysis pipeline: Batch upload (DTU, http://www.genomicepidemiology.org/), currently being updated and uses Resfinder. V.2); INNUENDO: uses ABRICATE. Bionumerics: a commercial software developed by Applied Maths (BioMérieux) (http://www.applied-maths.com/bionumerics). It uses the Resfinder database (V.2, further updated in November 2018.) |
Information presented to the best knowledge of the WG experts, based on their own expertise and publicly available information. Status as of 30 September 2018. Further developments to the different tools since that date are not presented here.
Supporting information
Questionnaire of the Specific Survey on harmonised AMR monitoring
Suggested citation: EFSA (European Food Safety Authority) , Aerts M, Battisti A, Hendriksen R, Kempf I, Teale C, Tenhagen B‐A, Veldman K, Wasyl D, Guerra B, Liébana E, Thomas‐López D and Belœil P‐A, 2019. Scientific report on the technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food‐producing animals and food. EFSA Journal 2019;17(6):5709, 122 pp. 10.2903/j.efsa.2019.5709
Requestor: European Commission
Question number: EFSA‐Q‐2018‐00051
Acknowledgements: EFSA wishes to thank the following hearing experts for the support provided to this scientific output: Patrick McDermott, Valeria Bortolaia and Francesca Leoni.
Approved: 30 April 2019
Notes
COM (2017) 0339
COM (2017) 0339 ‐ https://ec.europa.eu/health/amr/sites/amr/files/amr_action_plan_2017_en.pdf
OJ L 325, 12.12.2003 p. 31.
Scientific Opinion on Foodborne antimicrobial resistance as a biological hazard (EFSA Journal (2008) 765, 1‐87); Scientific opinion on the assessment of the public health significance of meticillin resistant Staphylococcus aureus (MRSA) (EFSA Journal (2009) 993, 1‐73); Scientific opinion on the public health risks of bacterial strains producing extended‐spectrum β‐lactamases (ESBL) and/or AmpC β‐lactamases (AmpC) in food and food‐producing animals (EFSA Journal (2011; 9(8):2322).
Joint scientific opinion on antimicrobial resistance focused on infections transmitted to humans from animals and food (zoonoses) (EFSA Journal 2009; 7(11):1372).
Technical report on EFSA approaches to risk assessment in the area of antimicrobial resistance, with an emphasis on commensal microorganisms (EFSA Journal 2011; 9(10):196).
EFSA Journal 2012; 10(6):2742.
EFSA Journal 2012; 10(10):2897.
OJ L 309, 14.11.2013, p. 26.
COM (2011) 748.
EFSA Journal 2014;12(5):3686.
Directive 2003/99/EC of the European Parliament and of the Council of 17 November 2003 on the monitoring of zoonoses and zoonotic agents, amending Council Decision 90/424/EEC and repealing Council Directive 92/117/EEC. OJ L 325, 12.12.2003, p. 31–40.
ECOFFs separate the naive, susceptible wild‐type bacterial populations from isolates that have developed reduced susceptibility to a given antimicrobial agent (Kahlmeter et al., 2003). The ECOFFs may differ from breakpoints used for clinical purposes, which are defined against a background of clinically relevant data, including therapeutic indication, clinical response data, dosing schedules, pharmacokinetics and pharmacodynamics.
For various combinations of bacteria/antimicrobial substance, the ECOFFs equal the clinical breakpoints set for humans by EUCAST (for example: colistin/Salmonella, fluoroquinolones/Campylobacter jejuni, macrolides/Campylobacter jejuni).
Commission Implementing Decision 2013/652/EU of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria. OJ L 303, 14.11.2013, p. 26–39
Acinetobacter, Aerococcus, Arcanobacterium, Bacillus, Bacteroides, Citrobacter, Corynebacterium, Enterobacter, Enterococcus, Eubacterium, Fusobacterium, Haemophilus Lactobacillus, Micrococcus, Pantoeae Peptostreptococcus, Porphyromonas, Proteus, Pseudomonas, Ruminococcus, Streptococcus, Treponema.
This was also mentioned in the interim overview report (http://ec.europa.eu/food/audits-analysis/overview_reports/details.cfm?rep_id=110).
Commission Decision 2008/55/EC of 20 December 2007 concerning a financial contribution from the Community towards a survey on the prevalence of Salmonella spp. and Methicillin‐resistant Staphylococcus aureus in herds of breeding pigs to be carried out in the Member States. OJ L 14, 17.1.2008, p. 10–25.
Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC. OJ L 64, 4.3.2006, p. 37–51.
Stratified sampling is a method of sampling from a population which involves, in the first step, the division of a population into smaller groups known as strata. The strata are formed based on members’ shared attributes or characteristics. In the second step, a random sample from each stratum is taken in a number proportional to the stratum's size when compared to the population. These subsets of the strata are then pooled to form a random sample. In practice, stratified random sampling is typically employed in large‐scale surveys/routine monitoring to reduce some of the logistical costs associated with collecting information from a sample.
Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. OJ L 338, 22.12.2005, p. 1–26.
Several studies have shown a high concordance (> 96%) between the presence of known acquired resistance genes or mutations and MICs at or above the clinical breakpoint for resistance. High levels of sensitivity (> 87%) and specificity (> 98%) have been also observed, depending of the species analysed (Rene Hendriksen, EURL‐AR, personal communication).
Available on the EURL‐AR resistance website (https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/280_protocol-for-campylobacter-november-2013.pdf).
As described by the NF EN ISO 10272‐1:2017 Standard.
In the case where the plan should ensure a representation of at least 80% of the population, the numerical calculation should be adapted accordingly.
Designated under Regulation 2160/2003 article 12(1)(a).
As a logistic compromise, it may be acceptable that the sampling plan focus on those laboratories accounting for 80% of the total number of Salmonella isolates in the poultry production in question within the MS in the previous year. If the Salmonella prevalence in the poultry productions is important and the laboratories involved in the NCPs numerous, it may be envisaged to account for organisational constraints to further limit the strata (laboratories) involved in the sampling plans, provided that a good geographical representation is assured and any particular subpopulations are not systematically included/excluded.
In the case statistics on the annual throughput of domestically produced meat of slaughterhouses are not available, statistics on the annual throughput of meat (whatever is the origin of the animals slaughtered) are used.
A single caecal sample derives from the caecal content of both caecal diverticula from one carcase of broiler, while a pooled caecal sample derives from the caecal content of 10 carcases of broilers. For turkeys, pigs and calves, some caecal content collected from one carcase should provide enough material to perform the requested testing. In the case of low Campylobacter prevalence, pooled caecal samples may be envisaged.
Condemned or underweight carcases and animals undergoing emergency slaughter should be notably excluded.
In exceptional cases, when it appears for particular economical and organisational reasons that it would be more cost‐effective, a greater number of samples in a day could be collected, e.g. more than five caecal samples may be sampled per day per slaughterhouse, provided that a potentially introduced clustering effect is accounted for by increasing the sample size of the monitoring.
According to the most recent data available at EUROSTAT (http://epp.eurostat.ec.europa.eu).
A sampling frame representing at least 80% of the target population is proposed as a logistic compromise, since the legislation does not foresee any restriction of the study population. However, in certain MSs it might be necessary to further limit the NUTS‐3 areas involved in the sampling frame to the regions with high population density to account for logistical and organisational constraints, in particular in those geographically large MSs with sparsely populated regions.
Available online: http://www.eurl-ar.eu
References
- Agersø Y, Vigre H, Cavaco L and Josefsen M, 2013. Comparison of air samples, nasal swabs, ear‐skin swabs and environmental dust samples for detection of methicillin‐resistant Staphylococcus aureus (MRSA) in pig herds. Epidemiology and Infection., 142, 1–10. 10.1017/S095026881300280X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsamarai AM, Abbas HM and Atia QM, 2015. Nasal carriage of methicillin resistant Staph aureus in food provider in restaurant at Samara city. World Journal of Pharmacy and Pharmaceutical Sciences, 4, 50–58. [Google Scholar]
- Antonelli A, Maria D'Andrea M, Brenciani A, Galeotti CL, Morroni G, Pollini S, Emanuele Varaldo P and Rossolini MG, 2018. Characterisation of poxtA, a novel phenicol‐oxazolidinone‐tetracycline resistance gene from an MRSA of clinical origin. Journal of Antimicrobial Chemotherapy, 73, 1763–1769. [DOI] [PubMed] [Google Scholar]
- Aviv G, Tsyba K, Steck N, Salmon‐Divon M, Cornelius A, Rahav G, Grassl GA and Gal‐Mor O, 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environmental Microbiology, 16, 977–994. 10.1111/1462-2920.12351 [DOI] [PubMed] [Google Scholar]
- Avrain L, Humbert F, L'Hospitalier R, Sanders P, Vernozy‐Rozand C and Kempf I, 2003. Antimicrobial resistance in Campylobacter from broilers: Association with production type and antimicrobial use. Veterinary Microbiology, 96, 267–276. [DOI] [PubMed] [Google Scholar]
- Bangerter PD, Sidler X, Perreten V and Overesch G, 2016. Longitudinal study on the colonisation and transmission of methicillin‐resistant Staphylococcus aureus in pig farms. Veterinary Microbiology, 183, 125–134. [DOI] [PubMed] [Google Scholar]
- Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R, Feltrin F, Zini M and Aarestrup FM, 2010. Heterogeneity among methicillin‐resistant Staphylococcus aureus from Italian pig finishing holdings. Veterinary Microbiology, 142, 361–366. [DOI] [PubMed] [Google Scholar]
- Bolinger H and Kathariou S, 2017. The Current State of Macrolide Resistance in Campylobacter spp.: Trends and Impacts of Resistance Mechanisms. Applied and Environmental Microbiology, 83, pii, e00416–e00417. 10.1128/AEM.00416-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broens EM, van Cleef BAGL, Graat EAM and Kluytmans AJW, 2008. Transmission of methicillin‐resistant Staphylococcus aureus from food production animals to humans: a review. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources, 3, 1–12. 10.1079/PAVSNNR20083095 [DOI] [Google Scholar]
- Broens EM, Graat EA, Van der Wolf PJ, Van de Giessen AW and De Jong MC, 2011. Transmission of methicillin resistant Staphylococcus aureus among pigs during transportation from farm to abattoir. Veterinary Journal, 189, 302–305. [DOI] [PubMed] [Google Scholar]
- Brouwer MSM, Rapallini M, Geurts Y, Harders F, Bossers A, Mevius DJ, Wit B and Veldman KT, 2018. Enterobacter cloacae complex isolated from shrimps from Vietnam carrying blaIMI‐1 resistant to carbapenems but not cephalosporins. Antimicrobial Agents Chemotherapy, 62, e00398–18. 10.1128/AAC.00398-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo CD, Plante D, Iugovaz I, Kenwell R, Belanger G, Boucher F, Poulin N and Trottier YL, 2014. Method‐dependent variability in determination of prevalence of Campylobacter jejuni and Campylobacter coli in Canadian retail poultry. Journal of Food Protection, 77, 1682–1688. [DOI] [PubMed] [Google Scholar]
- Catry B, Van Duijkeren E, Pomba MC, Greko C, Moreno MA, Pyorala S, Ruzauskas M, Sanders P, Threlfall EJ, Ungemach F, Torneke K, Munoz‐Madero C and Torren‐Edo J, 2010. Reflection paper on MRSA in food‐producing and companion animals: epidemiology and control options for human and animal health. Epidemiology and Infection, 138, 626–644. [DOI] [PubMed] [Google Scholar]
- Chatre P, Haenni M, Meunier D, Botree MA, Calavas D and Madec JY, 2010. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from cattle between 2002 and 2006 in France. Journal of Food Protection, 73, 825–831. [DOI] [PubMed] [Google Scholar]
- Chen JC, Tagg KA, Joung YJ, Bennett C, Watkins LF, Eikmeier D and Folster JP, 2018. Report on erm(B)+ Campylobacter jejuni in the United States. Antimicrobial Agents and Chemotherapy, 62, e02615–17. 10.1128/AAC.02615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNR‐Campylobacter , online. Bilan de la surveillance des infections à Campylobacter en France en 2015. Van Cauteren D, Lehours P, Bessède E, De Valk H and Mégraud F. Available online: http://invs.santepubliquefrance.fr/content/download/126529/449965/version/1/file/Bilan_Campylobacter_2015.pdf [Accessed: 22.03.2019].
- Cohn LA and Middleton JR, 2010. A veterinary perspective on methicillin‐resistant staphylococci. Journal of veterinary emergency and critical care. Journal of Veterinary Emergency and Critical Care, 20, 31–45. 10.1111/j.1476-4431.2009.00497.x [DOI] [PubMed] [Google Scholar]
- Cole D, Todd L and Wing S, 2000. Concentrated swine feeding operations and public health: a review of occupational and community health effects. Environmental Health Perspectives, 108, 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C, Abdelbary M, Layer F, Werner G and Witte W, 2015. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Veterinary Microbiology, 177, 219–223. [DOI] [PubMed] [Google Scholar]
- DANMAP , 2017. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria From Food Animals, Food and Humans in Denmark., ISSN, 1600–2032. [Google Scholar]
- Day M, Doumith M, Jenkins C, Dallman TJ, Hopkins KL, Elson R, Godbole G and Woodford N, 2017. Antimicrobial resistance in Shiga toxin‐producing Escherichia coli serogroups O157 and O26 isolated from human cases of diarrhoeal disease in England, 2015. Journal of Antimicrobial Chemotherapy, 72, 145–152. 10.1093/jac/dkw371 [DOI] [PubMed] [Google Scholar]
- Dearlove BL, Cody AJ, Pascoe B, Meric G, Wilson DJ and Sheppard SK, 2016. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. The ISME Journal, 10, 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Collo LP, Karns JS, Biswas D, Lombard JE, Haley BJ, Kristensen RC, Kopral CA, Fossler CP and Van Kessel JAS, 2017. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter spp. in bulk tank milk and milk filters from US dairies. Journal of Dairy Science, 100, 3470–3479. [DOI] [PubMed] [Google Scholar]
- Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G and Colin P, 1999. Development of a m‐PCR assay for simultaneous identification of Campylobacter jejuni and C. coli . Letters in Applied Microbiology, 29, 406–410. [DOI] [PubMed] [Google Scholar]
- Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, Giani T, Menichetti F and Rossolini GM, 2016. mcr‐1.2, a new mcr variant carried on a transferable plasmid from a colistin‐resistant KPC carbapenemase‐producing Klebsiella pneumoniae strain of sequence type 512. Antimicrobial Agents and Chemotherapy, 60, 5612–5615. 10.1128/AAC.01075-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorado‐Garcıa A, Smid JH, van Pelt W, Bonten MJM, Fluit AC, van den Bunt G, Wagenaar JA, Hordijk J, Dierikx CM, Veldman KT, de Koeijer A, Dohmen W, Schmitt H, Liakopoulos A, Pacholewicz E, Lam TJGM, Velthuis AG, Heuvelink A, Gonggrijp MA, van Duijkeren E, van Hoek AHAM, de Roda Husman AM, Blaak H, Havelaar AH, Mevius DJ and Heederik DJJ, 2018. Molecular relatedness of ESBL/AmpC‐producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. Journal of Antimicrobial Chemotherapy, 73, 339–347. 10.1093/jac/dkx397 [DOI] [PubMed] [Google Scholar]
- DTU, IZSLT, BfR, NIPH, NVRI, PHE, APHA and IZSV (Technical University of Denmark ‐ National Food Institute, Istituto Zooprofilattico Sperimentale del Lazio e della Toscana, Federal Institute for Risk Assessment, National Institute of Public Health – National Institute of Hygiene, National Veterinary Research Institute, Public Health England, Animal and Plant Health Agency and Istituto Zooprofilattico Sperimentale delle Venezie), 2018. Final report of ENGAGE ‐ Establishing Next Generation sequencing Ability for Genomic analysis in Europe. EFSA supporting publication 2018:15(6):EN‐1431. 252 pp. 10.2903/sp.efsa.2018.EN-1431 [DOI]
- ECDC (European Centre for Disease Prevention and Control), 2016. Plasmid‐mediated colistin resistance in Enterobacteriaceae. Available online: http://ecdc.europa.eu/en/publications/Publications/%3fenterobacteriaceae-risk-assessment-diseases-caused-by-antimicrobial-resistant-microorganisms-europe-june-2016.pdf
- ECDC and EFSA (European Centre for Disease Prevention and Control and European Food Safety Authority), Van Walle I, Guerra B, Borges V, Carriço JA, Cochrane G, Dallman T, Franz E, Karpíšková R, Litrup E, Mistou M‐Y, Morabito S, Mossong J, Alm E, Barrucci F, Bianchi C, Costa G, Kotila S, Mangone I, Palm D, Pasinato L, Revez J, Struelens M, Thomas‐López D and Rizzi V, 2019. EFSA and ECDC technical report on the collection and analysis of whole genome sequencing data from food‐borne pathogens and other relevant microorganisms isolated from human, animal, food, feed and food/feed environmental samples in the joint ECDC–EFSA molecular typing database. EFSA supporting publication 2019:EN‐1337. 92 pp. 10.2903/sp.efsa.2019.en-1336 [DOI]
- ECDC, EFSA BIOHAZ Panel and CVMP (European Centre for Disease Prevention and Control, European Food Safety Authority Panel on Biological Hazards and EMA Committee for Medicinal Products for Veterinary Use), 2017. ECDC, EFSA and EMA Joint Scientific Opinion on a list of outcome indicators as regards surveillance of antimicrobial resistance and antimicrobial consumption in humans and food‐producing animals. EFSA Journal 2017;15(10):5017, 70 pp. 10.2903/j.efsa.2017.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC, EFSA and EMA (European Centre for Disease Prevention and Control, European Food Safety Authority and European Medicines Agency), 2017. ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food‐producing animals – Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA Journal 2017;15(7):4872, 135 pp. 10.2903/j.efsa.2017.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2007. Report of the Task Force of Zoonoses Data Collection including a proposal for a harmonized monitoring scheme of antimicrobial resistance in Salmonella in fowl (Gallus gallus), turkeys, and pigs and Campylobacter jejuni and C. coli in broilers. EFSA Journal 2007;5(2):96, 46 pp. 10.2903/j.efsa.2007.96r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2008. Report from the Task Force on Zoonoses Data Collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. from food animals. EFSA Journal 2007;6(4):141, 44 pp. 10.2903/j.efsa.2008.141r [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2009. Analysis of the baseline survey on the prevalence of methicillin‐resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. Part A: MRSA prevalence estimates; on request from the European Commission. EFSA Journal 2009;7(11):1376, 82 pp. 10.2903/j.efsa.2009.1376 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2010. Analysis of the baseline survey on the prevalence of methicillin‐resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008 ‐ Part B: factors associated with MRSA contamination of holdings. EFSA Journal 2010;8(6):1597, 67 pp. 10.2903/j.efsa.2010.1597 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012a. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator Escherichia coli and Enterococcus spp. bacteria transmitted through food. EFSA Journal 2012;10(6):2742, 64 pp. 10.2903/j.efsa.2012.2742 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012b. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus in food‐producing animals and food. EFSA Journal 2012;10(10):2897, 56 pp. 10.2903/j.efsa.2012.2897 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2014. Technical specifications on randomised sampling for harmonised monitoring of antimicrobial resistance in zoonotic and commensal bacteria. EFSA Journal 2014;12(5):3686, 33 pp. 10.2903/j.efsa.2014.3686 [DOI] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2014. The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2012. EFSA Journal 2014;12(3):3590, 336 pp. 10.2903/j.efsa.2014.3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2017a. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA Journal 2017;15(2):4694, 212 pp. 10.2903/j.efsa.2017.4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2017b. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2016. EFSA Journal 2017;15(12):5077, 228 pp. 10.2903/j.efsa.2017.5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2018a. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA Journal 2018;16(2):5182, 270 pp. 10.2903/j.efsa.2018.5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2018b. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2017. EFSA Journal 2018;16(12):5500, 262 pp. 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2019. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA Journal 2019;17(2):5598, 278 pp. 10.2903/j.efsa.2019.5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), Allende A, Barceló Culleres D, Gironés Llop R, Laval A, Robertson L, da Silva Felício MT, Gervelmeyer A, Ramos Bordajandi L and Liebana E, 2017. Request for scientific and technical assistance on proposed EU minimum quality requirements for water reuse in agricultural irrigation and aquifer recharge. EFSA supporting publication 2017;15(7):EN‐1247, 19 pp. 10.2903/sp.efsa.2017.en-1247 [DOI]
- EFSA (European Food Safety Authority), García Fierro R, Thomas‐Lopez D, Deserio D, Liebana E, Rizzi V and Guerra B, 2018. Outcome of EC/EFSA questionnaire (2016) on use of Whole Genome Sequencing (WGS) for food‐ and waterborne pathogens isolated from animals, food, feed and related environmental samples in EU/EFTA countries. EFSA supporting publication 2018:15(6):EN‐1432. 49 pp. 10.2903/sp.efsa.2018.en-1432 [DOI]
- Egerton S, Culloty S, Whooley J, Stanton C and Ross RP, 2018. The Gut Microbiota of Marine Fish. Frontiers in Microbiology, 9, 873 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhadidy M, Miller WG, Arguello H, Álvarez‐Ordóñez A, Dierick K and Botteldoorn N, 2019. Molecular epidemiology and antimicrobial resistance mechanisms of Campylobacter coli from diarrhoeal patients and broiler carcasses in Belgium. Transboundary and Emerging Diseases, 66, 463–475. [DOI] [PubMed] [Google Scholar]
- El‐Shibiny A, Connerton PL and Connerton IF, 2005. Enumeration and diversity of campylobacters and bacteriophages isolated during the rearing cycles of free‐range and organic chickens. Applied and Environmental Microbiology, 71, 1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA/AMEG (European Medicines Agency/Antimicrobial Advice ad hoc Expert Group), 2016. Updated advice on the use of colistin products in animals within the European Union: development of resistance and possible impact on human and animal health (EMA/CVMP/CHMP/231573/2016). Available online: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/%3f2016/05/WC500207233.pdf
- EN ISO 10272‐1 : 2017. Microbiology of the food chain – horizontal method for detection and enumeration of Campylobacter spp. ‐ Part 1: Detection method. 24 pp. Available online: https://www.iso.org/standard/63225.html
- EUCAST (European Committee for Antimicrobial Susceptibility Testing), 2013. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 1.0 December 2013. Available on: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf
- Fan R, Li D, Wang Y, He T, Feßler AT, Schwarz S and Wu C, 2016. Presence of the optrA gene in methicillin‐resistant Staphylococcus sciuri of porcine origin. Antimicrobial Agents and Chemotherapy, 60, 7200–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO/WHO (Food and Agriculture Organization of the United Nations/World Health Organization), 2018. FAO/WHO expert meeting on foodborne antimicrobial resistance: Role of environment, crops and biocides. Rome, 11‐15 June 2018. Available online: http://www.fao.org/3/CA0963EN/ca0963en.pdf
- Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, Wigley P, Barquist L, Langridge GC, Feltwell T, Harris SR, Mather AE, Fookes M, Aslett M, Msefula C, Kariuki S, Maclennan CA, Onsare RS, Weill FX, Le Hello S, Smith AM, McClelland M, Desai P, Parry CM, Cheesbrough J, French N, Campos J, Chabalgoity JA, Betancor L, Hopkins KL, Nair S, Humphrey TJ, Lunguya O, Cogan TA, Tapia MD, Sow SO, Tennant SM, Bornstein K, Levine MM, Lacharme‐Lora L, Everett DB, Kingsley RA, Parkhill J, Heyderman RS, Dougan G, Gordon MA and Thomson NR, 2016. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high‐income settings and invasive disease in low‐income settings. Nature Genetics, 48, 1211–1217. 10.1038/ng.3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin F, Alba P, Kraushaar B, Ianzano A, Argudín MA, Di Matteo P, Porrero MC, Aarestrup FM, Butaye P, Franco A and Battisti A, 2016. A livestock‐associated, multidrug‐resistant, methicillin‐resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Applied and Environmental Microbiology, 82, 816–821. 10.1128/AEM.02854-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J, Guerra B and Rodicio MR, 2018. Resistance to carbapenems in non‐typhoidal Salmonella enterica serovars from humans, animals and food. Veterinary Science, 5 pii: E40 10.3390/vetsci5020040. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, San José M, Roschanski N, Schmoger S, Baumann B, Irrgang A, Friese A, Roesler U, Helmuth R and Guerra B, 2017. Spread and persistence of VIM‐1 Carbapenemase‐producing Enterobacteriaceae in three German swine farms in 2011 and 2012. Veterinary Microbiology, 200, 118–123. [DOI] [PubMed] [Google Scholar]
- Florez‐Cuadrado D, Ugarte‐Ruiz M, Quesada A, Palomo G, Domínguez L and Porrero MC, 2016. Description of an erm (B)‐carrying Campylobacter coli isolate in Europe. Journal of Antimicrobial Chemotherapy, 71, 841–843. 10.1093/jac/dkv383 [DOI] [PubMed] [Google Scholar]
- Florez‐Cuadrado D, Ugarte‐Ruiz M, Meric G, Quesada A, Porrero MC, Pascoe B, Saez‐Llorente JL, Orozco GL, Dominguez L and Sheppard SK, 2017. Genome comparison of erythromycin resistant Campylobacter from turkeys identifies hosts and pathways for horizontal spread of erm(B) genes. Frontiers in Microbiology, 8, 2240 10.3389/fmicb.2017.02240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau C, Boucher F and Bekalb S, 2014. Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli isolates obtained in Montreal, Quebec, Canada, from 2002 to 2013. Journal of Clinical Microbiology, 52, 2644–2646. 10.1128/JCM.00362-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau C, Pilon PA, Sylvestre JL, Boucher F and Bekal S, 2016. Multidrug‐resistant Campylobacter coli in men who have sex with men, Quebec, Canada, 2015. Emerging Infectious Diseases, 22, 1661–1663. 10.3201/eid2209.151695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Wang F, Sjölund‐Karlsson M and McDermott PF, 2013. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. Journal of Microbiological Methods, 95, 57–67. 10.1016/j.mimet.2013.06.021 [DOI] [PubMed] [Google Scholar]
- Gibreel A and Taylor DE, 2006. Macrolide resistance in Campylobacter jejuni and Campylobacter coli . Journal of Antimicrobial Chemotherapy, 58, 243–255. [DOI] [PubMed] [Google Scholar]
- Hagiya H, Kimura K, Nishi I, Yoshida H, Yamamoto N, Akeda Y and Tomono K, 2018. Emergence of Carbapenem Non‐susceptible Campylobacter coli after Long‐term Treatment against Recurrent Bacteremia in a Patient with X‐linked Agammaglobulinemia. Internal Medicine, 57, 2077–2080. 10.2169/internalmedicine.0312-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM and Skov RL, 2015. Detection of mcr‐1 encoding plasmid‐mediated colistin‐resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eurosurveillance, 20(49). Erratum in: Eurosurveillance 2015;20(50). [DOI] [PubMed] [Google Scholar]
- van Hoek AH, Veenman C, van Overbeek WM, Lynch G, de Roda Husman AM and Blaak H, 2015. Prevalence and characterization of ESBL‐ and AmpC‐producing Enterobacteriaceae on retail vegetables. International Journal of Food Microbiology, 204, 1–8. 10.1016/j.ijfoodmicro.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Hsieh YH, Simpson S, Kerdahi K and Sulaiman IM, 2018. A comparative evaluation study of growth conditions for culturing the isolates of Campylobacter spp. Current Microbiology, 75, 71–78. [DOI] [PubMed] [Google Scholar]
- Huang E, Gurzau AE, Hanson BM, Kates AE, Smith TC, Pettigrew MM, Spinu M and Rabinowitz PM, 2014. Detection of livestock‐associated methicillin‐resistant Staphylococcus aureus among swine workers in Romania. Journal of Infection and Public Health, 7, 323–332. 10.1016/j.jiph.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Huang H, Brooks BW, Lowman R and Carrillo CD, 2015. Campylobacter species in animal, food, and environmental sources, and relevant testing programs in Canada. Canadian Journal of Microbiology, 61, 701–721. [DOI] [PubMed] [Google Scholar]
- Irrgang A, Fischer J, Grobbel M, Schmoger S, Skladnikiewicz‐Ziemer T, Thomas K, Hensel A, Tenhagen BA and Käsbohrer A, 2017. Recurrent detection of VIM‐1‐producing Escherichia coli clone in German pig production. Journal of Antimicrobial Chemotherapy., 72, 944–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseppi R, de Niederhäusern S, Bondi M, Messi P and Sabia C, 2018. Extended‐Spectrum β‐Lactamase, AmpC, and MBL‐Producing Gram‐Negative Bacteria on Fresh Vegetables and Ready‐to‐Eat Salads Sold in Local Markets. Microbial Drug Resistance, 24, 1156–1164. 10.1089/mdr.2017.0198 [DOI] [PubMed] [Google Scholar]
- Janecko N, Martz SL, Avery BP, Daignault D, Desruisseau A, Boyd D, Irwin RJ, Mulvey MR and Reid‐Smith RJ, 2016. Carbapenem‐resistant Enterobacter spp. in retail seafood imported from Southeast Asia to Canada. Emerging Infectious Diseases, 22, 1675–1677. 10.3201/eid2209.160305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlmeter G, Brown DF, Goldstein FW, MacGowan AP, Mouton JW, Osterlund A, Rodloff A, Steinbakk M, Urbaskova P and Vatopoulos A, 2003. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. Journal of Antimicrobial Chemotherapy, 52, 145–148. 10.1093/jac/dkg312 [DOI] [PubMed] [Google Scholar]
- Karikari AB, Obiri‐Danso K, Frimpong EH and Krogfelt KA, 2017. Antibiotic Resistance of Campylobacter Recovered from Faeces and Carcasses of Healthy Livestock. BioMed Research International, 10.1155/2017/4091856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shin JA, Han SB, Cho B, Jeong DC and Kang JH, 2017. Recurrent Campylobacter jejuni bacteremia in a patient with hypogammaglobulinemia: A case report. Medicine (Baltimore), 96, e7238 10.1097/MD.0000000000007238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross P, Petersen A, Skov R, Van Hauwermeiren E, Pantosti A, Laurent F, Voss A, Kluytmans J, Struelens MJ, Heuer O and Monnet DL and the European human LA‐MRSA study group , 2017. Livestock‐associated meticillin‐resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Eurosurveillance, 22, pii=16‐0. 10.2807/1560-7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein‐Jöbstl D, Sofka D, Iwersen M, Drillich M and Hilbert F, 2016. Multilocus sequence typing and antimicrobial resistance of Campylobacter jejuni isolated from dairy calves in Austria. Frontiers in Microbiology, 7, 72 10.3389/fmicb.2016.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck R, Daniels‐Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S and Jurke A, 2018. Carbapenem‐resistant Enterobacteriaceae in wildlife, food‐producing, and companion animals: a systematic review. Clinical Microbiology and Infection, 24, 1241–1250. 10.1016/j.cmi.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Langmead B and Salzberg S, 2012. Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Sunde M, Islam MZ, Urdahl AM, Barstad AS, Larsen AR, Grøntvedt CA and Angen Ø, 2017. Evaluation of a widely used culture‐based method for detection of livestock‐associated meticillin‐resistant Staphylococcus aureus (MRSA), Denmark and Norway, 2014 to 2016. Eurosurveillance, 22, pii=30573. 10.2807/1560-7917.es.2017.22.28.30573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L‐H, Ab Mutalib N‐S, Law JW‐F, Wong SH and Letchumanan V, 2018. Discovery on Antibiotic Resistance Patterns of Vibrio parahaemolyticus in Selangor Reveals Carbapenemase Producing Vibrio parahaemolyticus in Marine and Freshwater Fish. Frontiers in Microbiology, 9, 2513 10.3389/fmicb.2018.02513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehours P, Oleastro M, Nunes A, Liassine N, Lowe DM and Godbole G, 2018. Recurrent Campylobacter jejuni infections with in vivo selection of resistance to macrolides and carbapenems: molecular characterization of resistance determinants. ECCMID 28th, European Congress of Clinical Microbiology and Infectious Diseases, MADRID 21‐24 April 2018. [DOI] [PMC free article] [PubMed]
- Lehtopolku M, Nakari UM, Kotilainen P, Huovinen P, Siitonen A and Hakanen AJ, 2010. Antimicrobial susceptibilities of multidrug‐resistant Campylobacter jejuni and C. coli strains: in vitro activities of 20 antimicrobial agents. Antimicrobial Agents and Chemotherapy, 54, 1232–1236. 10.1128/AAC.00898-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque B, Barthe C, Dixon BR, Parrington LJ, Martin D, Doidge B, Proulx JF and Murphy D, 2010. Microbiological quality of blue mussels (Mytilus edulis) in Nunavik, Quebec: A pilot study. Canadian Journal of Microbiology, 56, 968–977. [DOI] [PubMed] [Google Scholar]
- Li B, Ma L, Li Y, Jia H, Wei J, Shao D, Liu K, Shi Y, Qiu Y and Ma Z, 2017. Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathogens and Disease, 14, 96–102. [DOI] [PubMed] [Google Scholar]
- Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MS, Kant A, Wit B, Heymans R, van Pelt W and Mevius DJ, 2016. Extended‐spectrum cephalosporin‐resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerging Infectious Diseases, 22, 1257–1261. 10.3201/eid2207.151377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH and Shen J, 2016. Emergence of plasmid‐mediated colistin resistance mechanism MCR‐1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases, 16, 161‐168 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Liu D, Deng F, Gao Y, Yao H, Shen Z, Wu C, Wang Y and Shen J, 2017. Dissemination of erm(B) and its associated multidrug‐resistance genomic islands in Campylobacter from 2013 to 2015. Veterinary Microbiology, 204, 20–24. [DOI] [PubMed] [Google Scholar]
- Mangat CS, Boyd D, Janecko N, Martz SL, Desruisseau A, Carpenter M, Reid‐Smith RJ and Mulvey MR, 2016. Characterization of VCC‐1, a Novel Ambler Class A Carbapenemase from Vibrio cholerae Isolated from Imported Retail Shrimp Sold in Canada. Antimicrobial Agents and Chemotherapy, 60, 1819–1825. 10.1128/AAC.02812-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BJ and Rubin JE, 2015. Carbapenemase producing bacteria in the food supply escaping detection. PLoS ONE, 10, e0126717 10.1371/journal.pone.0126717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossong J, Mughini‐Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie HM, van Pelt W and Ragimbeau C, 2016. Human Campylobacteriosis in Luxembourg, 2010‐2013: A case‐control study combined with multilocus sequence typing for source attribution and risk factor analysis. Scientific Reports, 6, 20939 10.1038/srep20939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeghaire S, Roelandt S, Argudín MA, Haesebrouck F and Butaye P, 2013. Characterization of methicillin‐resistant Staphylococcus aureus from healthy carrier chickens. Avian Pathology, 42, 342–346. [DOI] [PubMed] [Google Scholar]
- Nemeghaire S, Argudín MA, Haesebrouck F and Butaye P, 2014. Epidemiology and molecular characterization of methicillin resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Veterinary Research, 10, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuert S, Nair S, Day MR, Doumith M, Ashton PM, Mellor KC, Jenkins C, Hopkins KL, Woodford N, de Pinna E, Godbole G and Dallman TJ, 2018. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non‐typhoidal Salmonella enterica . Frontiers in Microbiology, 9, 592 10.3389/fmicb.2018.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohra A, Grinberg A, Midwinter AC, Marshall JC, Collins‐Emerson JM and French NP, 2016. Molecular epidemiology of Campylobacter coli strains isolated from different sources in New Zealand between 2005 and 2014. Applied and Environmental Microbiology, 82, 4363–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi T, Aoki K, Ishii Y, Usui M, Tamura Y, Kawanishi M, Ohnishi K and Tateda K, 2017. Molecular epidemiological analysis of human‐ and chicken‐derived isolates of Campylobacter jejuni in Japan using next‐generation sequencing. Journal of Infection and Chemotherapy, 23, 165–172. 10.1016/j.jiac.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Ovejero CM, Delgado‐Blas JF, Calero‐Caceres W, Muniesa M and Gonzalez‐Zorn B, 2017. Spread of mcr‐1‐carrying Enterobacteriaceae in sewage water from Spain. Journal of Antimicrobial Chemotherapy, 72, 1050–1053. 10.1093/jac/dkw533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletinckx LJ, De Bleecker Y, Dewulf J, Rasschaert G, Goddeeris BM and De Man I, 2012. Evaluation of salt concentrations, chromogenic media and anatomical sampling sites for detection of methicillin‐resistant Staphylococcus aureus in pigs. Veterinary Microbiology, 154, 363–368. [DOI] [PubMed] [Google Scholar]
- Pletz MW, Wollny A, Dobermann UH, Rödel J, Neubauer S, Stein C, Brandt C, Hartung A, Mellmann A, Trommer S, Edel B, Patchev V, Makarewicz O and Maschmann J, 2018. A Nosocomial Foodborne Outbreak of a VIM Carbapenemase‐Expressing Citrobacter freundii . Clinical Infectious Disease, 67, 58–64. 10.1093/cid/ciy034 [DOI] [PubMed] [Google Scholar]
- Podschun R and Ullman U, 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical Microbiology Reviews, 11, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post A, Martiny D, van Waterschoot N, Hallin M, Maniewski U, Bottieau E, Van Esbroeck M, Vlieghe E, Ombelet S, Vandenberg O and Jacobs J, 2017. Antibiotic susceptibility profiles among Campylobacter isolates obtained from international travelers between 2007 and 2014. European Journal of Clinical Microbiology and Infectious Diseases, 36, 2101–2107. 10.1007/s10096-017-3032-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, Wu C, Wang S, Zhang J and Shen J, 2014. Report of ribosomal RNA methylase gene erm(B) in multidrug‐resistant Campylobacter coli . Journal of Antimicrobial Chemotherapy, 69, 964–968. 10.1093/jac/dkt492 [DOI] [PubMed] [Google Scholar]
- Raeisi M, Khoshbakht R, Ghaemi EA, Bayani M, Hashemi M, Seyedghasemi NS and Shirzad‐Aski H, 2017. Antimicrobial resistance and virulence‐associated genes of Campylobacter spp. isolated from raw milk, fish, poultry, and red meat. Microbial Drug Resistance, 23, 925–933. [DOI] [PubMed] [Google Scholar]
- Randall LP, Lodge MP, Elviss NC, Lemma FL, Hopkins KL, Teale CJ and Woodford N, 2017. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended‐spectrum beta‐lactamase (ESBL)‐producing and carbapenem‐resistant Escherichia coli . International Journal of Food Microbiology, 241, 283–290. [DOI] [PubMed] [Google Scholar]
- Reperant E, Laisney MJ, Nagard B, Quesne S, Rouxel S, Le Gall F, Chemaly M and Denis M, 2016. Influence of enrichment and isolation media on the detection of Campylobacter spp. in naturally contaminated chicken samples. Journal of Microbiological Methods, 128, 42–47. [DOI] [PubMed] [Google Scholar]
- Reuland EA, Al Naiemi N, Raadsen SA, Savelkoul PHM, Kluytmans JAJW and Vandenbroucke‐Grauls CMJE, 2014. Prevalence of ESBL‐producing Enterobacteriaceae in raw vegetables. European Journal of Clinical Microbiology and Infectious Diseases, 33, 1843–1846. 10.1007/s10096-014-2142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripabelli G, Sammarco ML, Grasso GM, Fanelli I, Caprioli A and Luzzi I, 1999. Occurrence of Vibrio and other pathogenic bacteria in Mytilus galloprovincialis (mussels) harvested from Adriatic Sea, Italy. International Journal of Food Microbiology, 49, 43–48. [DOI] [PubMed] [Google Scholar]
- Roberts MC, 2008. Update on macrolide‐lincosamide‐streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiology Letters, 282, 147–159. [DOI] [PubMed] [Google Scholar]
- Rolain JM, Kempf M, Leangapichart T, Chabou S, Olaitan AO, Le Page S, Morand S and Raoult D, 2016. Plasmid‐mediated mcr‐1 gene in colistin‐resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrobial Agents and Chemotherapy, 60, 6994–6995. 10.1128/AAC.00960-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschanski N, Guenther S, Vu TTT, Fischer J, Semmler T, Huehn S, Alter T and Roesler U, 2017. VIM‐1 carbapenemase‐producing Escherichia coli isolated from retail seafood, Germany 2016. Eurosurveillance, 10.2807/1560-7917.ES.2017.22.43.17-00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner BM, Schielke A, Didelot X, Kops F, Breidenbach J, Willrich N, Gölz G, Alter T, Stingl K and Josenhans C, 2017. A combined case‐control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Scientific Reports, 7, 5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin JE, Ekanayake S and Fernando C, 2014. Carbapenemase‐producing organism in food, 2014. Emerging Infectious Diseases, 20, 1264–1265. 10.3201/eid2007.140534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadkowska‐Todys M and Kucharczyk B, 2016. Campylobacteriosis in Poland in 2013 and 2014. Przeglad Epidemiologiczny, 70, 209–215. [PubMed] [Google Scholar]
- Sadouki Z, Day MR, Doumith M, Chattaway MA, Dallman TJ, Hopkins KL, Elson R, Woodford N, Godbole G and Jenkins C, 2017. Comparison of phenotypic and WGS‐derived antimicrobial resistance profile of Shigella sonnei isolated from cases of diarrhoeal disease in England and Wales, 2015. Journal of Antimicrobial Chemotherapy, 72, 2496–2502. 10.1093/jac/dkx170 [DOI] [PubMed] [Google Scholar]
- Schlotter K, Ehricht R, Hotzel H, Monecke S, Pfeffer M and Donat K, 2012. Leukocidin genes lukF‐P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia. Germany. Veterinary Research, 43, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Wang Y and Schwarz S, 2013. Presence and dissemination of the multiresistance gene cfr in Gram‐positive and Gram‐negative bacteria. Journal of Antimicrobial Chemotherapy, 68, 1697–1706. [DOI] [PubMed] [Google Scholar]
- Skov RL and Monnet DL, 2016. Plasmid‐mediated colistin resistance (mcr‐1 gene): three months later, the story unfolds. Eurosurveillance, 21, 30155 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- Slifierz MJ, Friendship RM and Weese JS, 2015. Methicillin‐resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Applied and Environmental Microbiology, 81, 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonthornchaikul N and Garelick H, 2009. Antimicrobial resistance of campylobacter species isolated from edible bivalve molluscs purchased from Bangkok markets, Thailand. Foodborne Pathogens and Disease, 6, 947–951. [DOI] [PubMed] [Google Scholar]
- Tang Y, Dai L, Sahin O, Wu Z, Liu M and Zhang Q, 2017. Emergence of a plasmid‐borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter . Journal of Antimicrobial Chemotherapy, 72, 1581–1588. [DOI] [PubMed] [Google Scholar]
- Thépault A, Meric G, Rivoal K, Pascoe B, Mageiros L, Touzain F, Rose V, Beven V, Chemaly M and Sheppard SK, 2017. Genome‐wide identification of host‐segregating epidemiological markers for source attribution in Campylobacter jejuni . Applied and Environmental Microbiology, 83, pii, e03085–16. 10.1128/AEM.03085-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépault A, Poezevara T, Quesne S, Rose V, Chemaly M and Rivoal K, 2018. Prevalence of thermophilic Campylobacter in cattle production at slaughterhouse level in France and link between C. jejuni bovine strains and campylobacteriosis. Frontiers in Microbiology, 9, 471 10.3389/fmicb.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Mee‐Marquet N, Corvaglia A, Valentin A, Hernandez D, Bertrand X, Girard M, Kluytmans J, Donnio P, Quentin R and François P, 2013. Analysis of prophages harbored by the human‐adapted subpopulation of Staphylococcus aureus CC398. Infection, Genetics and Evolution, 18, 299–308. [DOI] [PubMed] [Google Scholar]
- Van der Mee‐Marquet NL, Corvaglia A, Haenni M, Bertrand X, Franck JB, Kluytmans J, Girard M, Quentin R and François P, 2014. Emergence of a novel subpopulation of CC398 Staphylococcus aureus infecting animals is a serious hazard for humans. Frontiers in Microbiology, 5, 652 10.3389/fmicb.2014.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss A, Loeffen F, Bakker J, Klaassen and Wulf M, 2005. Methicillin‐resistant Staphylococcus aureus in pig farming. Emerging Infectious Diseases, 11, 1965–1966. 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, Zhang Q and Shen J, 2014. Emergence of multidrug‐resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrobial Agents and Chemotherapy, 58, 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z and Wang Y, 2018. Emergence of a novel mobile colistin resistance gene, mcr‐8, in NDM‐producing Klebsiella pneumoniae . Emerging Microbes and Infections, 7, 122 10.1038/s41426-018-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasyl D, Kern‐Zdanowicz I, Domańska‐Blicharz K, Zając M and Hoszowski A, 2015. High‐level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying bla CTX‐M‐25 gene. Veterinary Microbiology, 175, 85–91. 10.1016/j.vetmic.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Wei B, Cha S‐Y, Yoon R‐H, Kang M, Roh J‐H, Seo H‐S, Lee J‐A and Jang H‐K, 2016. Prevalence and antimicrobial resistance of Campylobacter spp. isolated from retail chicken and duck meat in South Korea. Food Control, 62, 63–68. [Google Scholar]
- Whitehouse CA, Zhao S and Tate H, 2018. Antimicrobial resistance in Campylobacter species: mechanisms and genomic epidemiology. Advances in Applied Microbiology, 10.1016/bs.aambs.2018.01.001 [DOI] [PubMed] [Google Scholar]
- Wieczorek K and Osek J, 2013. Characteristics and antimicrobial resistance of Campylobacter isolated from pig and cattle carcasses in Poland. Polish Journal of Veterinary Sciences, 16, 501–508. [DOI] [PubMed] [Google Scholar]
- Wyres KL and Holt KE, 2018. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Current Opinions in Microbiology, 45, 131–139. 10.1016/j.mib.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Xavier BB, Lammens C, Ruhal R, Kumar‐Singh S, Butaye P, Goossens H and Malhotra‐Kumar S, 2016. Identification of a novel plasmid‐mediated colistin‐resistance gene, mcr‐2, in Escherichia coli, Belgium, June 2016. Eurosurveillance, 21 10.2807/1560-7917.es.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yang W, Zhang M, Zhou J, Pang L, Wang G and Hou F, 2017. The molecular mechanisms of ciprofloxacin resistance in clinical Campylobacter jejuni and their genotyping characteristics in Beijing, China. Foodborne Pathogens and Disease, 14, 386–392. [DOI] [PubMed] [Google Scholar]
- Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, Wu C, Yu EW, Zhang Q and Shen J, 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. MBio, 7:e01543–16. 10.1128/mbio.01543-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV and Aarestrup FM, 2013. Genotyping using whole‐genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. Journal of Antimicrobial Chemotherapy, 68, 771–777. 10.1093/jac/dks496 [DOI] [PubMed] [Google Scholar]
- Zhang T, Cheng Y, Luo Q, Lu Q, Dong J, Zhang R, Wen G, Wang H, Luo L, Wang H, Liu G and Shao H, 2017. Correlation between gyrA and CmeR box polymorphism and fluoroquinolone resistance in Campylobacter jejuni isolates in China. Antimicrobial Agents and Chemotherapy, 61 pii: e00422‐17. 10.1128/aac.00422-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL and McDermott PF, 2010. Antimicrobial resistance of campylobacter isolates from retail meat in the United States between 2002 and 2007. Applied and Environmental Microbiology, 76, 7949–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Questionnaire of the Specific Survey on harmonised AMR monitoring








