Abstract
Objective
To examine causal associations between risky behavior phenotypes and Parkinson disease using a mendelian randomization approach.
Methods
We used 2-sample mendelian randomization to generate unconfounded estimates using summary statistics from 2 independent, large meta-analyses of genome-wide association studies on risk-taking behaviors (n = 370,771–939,908) and Parkinson disease (cases n = 9,581, controls n = 33,245). We used the inverse variance weighted method as the main method for judging causality.
Results
Our results support a strong protective association between the tendency to smoke and Parkinson disease (odds ratio [OR] 0.714 per log odds of ever smoking, 95% confidence interval [CI] 0.568–0.897, p = 0.0041, Cochran Q test p = 0.238; I2 index 6.3%). Furthermore, we observed risk association trends between automobile speed propensity and the number of sexual partners and Parkinson disease after removal of overlapping loci with other risky traits (OR 1.986 for each 1-SD increase in normalized automobile speed propensity, 95% CI 1.215–3.243, p = 0.0066; OR 1.635 for each 1-SD increase in number of sexual partners, 95% CI 1.165–2.293, p = 0.0049).
Conclusion
These findings provide support for a causal relationship between general risk tolerance and Parkinson disease and may provide new insights into the pathogenic mechanisms leading to the development of Parkinson disease.
Parkinson disease (PD) is the second most prevalent neurodegenerative disorder characterized pathologically by progressive loss of dopaminergic neurons in the substantia nigra.1 The currently available treatment options for PD are symptomatic only. The lack of disease-modifying or protective treatments is due at least in part to the fact that the exact disease mechanisms are currently only partly understood.
The vast majority of cases of PD are caused by the combined action and likely interaction of genetic variants and environmental and lifestyle exposures.2–5 Several common habitual agents such as smoking, coffee intake, and alcohol drinking have shown protective associations with PD in large-scale meta-analyses of observational studies.6 Several recent studies have further shown beneficial effects of cannabidiol, a nonpsychotomimetic compound derived from cannabis, on nonmotor symptoms in patients with PD.7 It is noteworthy that several impulse control disorders such as gambling, hypersexuality, and compulsive eating are observed more frequently in patients with PD, with some studies reporting up to 40% prevalence in patients with PD.8 However, it is believed that these symptoms may be the result of dopamine agonist therapy prescribed to patients with PD.9 The imminent challenge in this context is to decipher whether these PD-associated environmental/lifestyle/behavioral variables contribute to or are an effect of the disease.
The recent development of the mendelian randomization (MR) approach allows investigators to judge causality on the basis of genetic data generated in observational studies. Specifically, this relies on the use of genetic variants as proxy markers of risk factors and takes care of confounding by exploiting random allocation of genetic variants at birth (for more details, see e-methods available from Dryad, doi.org/10.5061/dryad.c84c221).
We have also seen a surge of MR studies in the field of PD specifically exploring the causal role of several circulating biomarkers.10–15 For example, a recent MR study reported a significant causal association with a lifelong PD risk reduction of 3% per 10-μg/dL increase in serum iron levels.12 Most recently, another study further reported a risk reduction of 18% with a lifetime exposure of 5-kg/m2 higher body mass index.13
To date, the majority of studies in the field of PD have focused on modifiable environmental factors only, and MR studies exploring the role of behavioral phenotypes are lacking. Henceforth, our primary aim was to investigate the willingness to take risk as a causal factor in the development of PD. For this, we applied a 2-sample MR to investigate whether people with risk-taking tendency have an altered risk for PD.16 A recent genome-wide association study (GWAS) identified 611 independent loci with several measures of risky behaviors, including general risk tolerance, adventurousness, and risky behaviors in the driving, drinking, smoking, and sexual domains.17 We used all reported loci to mimic the random allocation of loci among cases with PD and controls in available data from a large, recent GWAS on PD.4,18 As secondary analyses, we further considered the wider literature to support inferences drawn from our primary analysis using previously reported GWAS on similar habitual behaviors such as smoking phenotypes, coffee consumption, alcoholism, cannabis dependence, and gambling.19–24
Methods
A brief explanation of MR methods in general is given in e-methods available from Dryad (doi.org/10.5061/dryad.54t59r8).
Study design and identification of datasets
We conducted a 2-sample MR using summary-level estimates to explore the causal role of several risky behaviors on PD.25 We identified genetic instruments that influence risky behaviors using a recently published meta-analysis of GWAS datasets on risky behaviors.17 The study reported statistically significant associations of 611 independent loci (p < 5 × 10−8) in a discovery cohort in up to 939,908 individuals of European ancestry with 6 highly correlated risky behavior phenotypes, including general risk tolerance, adventurousness, automobile speeding propensity, drinks per week, ever vs never smoking, and number of sexual partners. The study further defined general risk tolerance as the willingness to take risks, adventurousness as the self-reported tendency to be adventurous vs cautious, automobile speeding propensity as the tendency to drive faster than the speed limit, drinks per week as the average number of alcoholic drinks consumed per week, ever smoker (tendency to smoke) as whether one has ever been a smoker, and lastly number of sexual partners as the lifetime number of sexual partners.
We further extracted summary estimates of the identified genetic variants from the discovery cohort of a recent meta-analysis of GWAS on 9,581 cases with PD and 33, 245 controls of European ancestry.4 For this, we used data available on the PDGene database.18 Genetic instruments were identified for smoking (cigarettes per day), smoking initiation, smoking cessation, cannabis dependence, pathologic gambling, and alcohol and coffee consumption from independent GWAS as a part of our secondary analyses.19–24
Prioritization of genetic variants and power analysis
We systematically screened all the identified loci for a possible direct involvement in PD. For this, we used data available via PDGene to extract the list of loci shown to be significantly associated with PD (p < 5 × 10−8).18 We further checked overlapping loci for a relevant role in the pathogenesis of PD using a literature search. If substantial evidence was found, the respective loci were excluded from the list of genetic variants (single nucleotide polymorphisms [SNPs], genetic instrument) of the respective behavioral traits.
The SNPs constituting each genetic instrument were checked for strong linkage disequilibrium (LD). We used the rAggr database to look for correlated variants in individuals with European descent from the 1,000 Genomes phase 3 data26 and excluded one of the variants for pairs with R2 > 0.25. Finally, if SNPs were not available in the PD GWAS dataset, we identified proxy SNPs using an R2 cutoff of 0.9 based on the rAggr database as above.
The strength of the prioritized genetic instrument was judged with F statistics as explained earlier.12 We computed the variance in exposures explained by prioritized genetic instruments (R2) of the genetic instruments using effect estimates and the standard error of individual SNPs as described elsewhere.27 Lastly, power calculations were done with the method described a previously described method.28,29
Estimation of effect using MR
In cases when the genetic instruments consisted of a single SNP, we used the Wald ratio estimate along with the delta method to obtain the related estimate of the variance. In cases when the genetic instruments consisted of multiple SNPs, we used the inverse variance weighted (IVW) fixed-effect method as the main method to estimate the effect of genetically predicted behavioral phenotypes on PD by combining the genetic loci-specific Wald ratio estimates. The IVW MR analysis using summary-level data has previously been shown to yield effect estimates similar to those generated by a 2-stage least-squares regression method using individual-level data.19 We specifically used the IVW method incorporating second-order weights because casual estimates generated through this method are expected to provide a more accurate reflection of the variance of the Wald ratio estimate.30
However, in the absence of reliable information on functional pathways and proportion and direction of pleiotropic genetic variants, additional MR methods, including MR-Egger, weighted median, and weighted mode-based method, were used to check the consistency of the direction of effect estimates.16,31–33 Unlike IVW, which assumes no intercept term in the model, the MR-Egger method provides less biased effect estimates in the presence of directional pleiotropy and considerable heterogeneity assuming absence of measurement error.16 However, the MR-Egger method is more sensitive to unobserved associations of genetic variants with confounders of the exposure-outcome association and requires a greater sample size for the same underlying variance in exposure.32 Both the IVW and MR-Egger methods further assume that the pleiotropic effects of genetic variants are independent of their associations with the exposure known as the instrument strength independent of direct effect assumption. In the case of violation, the weighted median method may provide consistent effect estimates even if up to 50% of genetic variants do not conform to the instrument strength independent of direct effect assumption. In addition, the weighted mode–based method may provide consistent effect estimates, in particular, even when the no measurement error assumption was not met, but assuming that the most frequent value of the bias of the Wald ratio estimates is zero.34
We used latest functions and packages in R for a variety of statistical and graphical procedures implemented in the current article. Within every MR method, we computed casual estimates as odds ratio (OR) for PD per unit log of odds of the categorical behavioral phenotypes or OR per unit SD of the continuous behavioral phenotypes. Lastly, to address the issue of multiple testing, results were considered statistically significant at the 5% level after a conservative Bonferroni correction of the significance level, thus if p < 8.3 × 10−3 (0.05/6 independent primary MR hypotheses).
Assessment of pleiotropy
We used the Cochran Q statistic and I2 for the IVW method using second-order weights as main methods to identify pleiotropic variants.35 Furthermore, results from the less powerful MR-Egger test were also used to explore heterogeneity, including the test for deviation of the intercept from the null for the MR-Egger model using the χ2 test for independence.30 We further used the Rucker Q′ statistic to describe heterogeneity around the MR-Egger fit.36 The appropriate use of the main MR method for interpretation of effect estimate in the present study was judged by calculating the ratio between the Rucker Q′ and Cochran Q statistics.34 As a rule of thumb, the IVW method is recommended as the main method for judging the effect estimate if the ratio approaches 1.
To evaluate heterogeneity graphically, funnel plots were constructed that plot the spread of the inverse of the standard error of the respective Wald ratio estimates of each individual SNP around the MR estimates. In addition, scatterplots of effect estimates of individual SNPs with outcome vs effect estimates of individual SNPs with exposure are provided as a comparative visual assessment of the effect estimates generated from different MR methods. We further constructed radial MR plots that have been recently suggested as a more suitable approach for visual detection of outliers compared to traditional scatterplots, specifically when the difference between the IVW and MR-Egger estimates is large.37
Sensitivity analyses
We conducted a leave-one-out sensitivity analysis to check for a disproportionate influence of individual SNPs on overall effect estimates using the IVW method. We used forest plots to visually assess the results of the analysis and to further identify the outliers.
Because all the behavioral traits are highly correlated and are expected to exhibit shared genetic influence, we conducted a sensitivity analysis by including only genetic loci specific to each individual behavioral trait. We used a value of R2 > 0.8 to consider loci to be overlapping with other loci in an independent genetic instrument. Such an approach may help us to judge the reliability of independent associations of observed phenotypic traits. We further adopted a conservative approach by using loci unique to each phenotypic trait (R2 < 0.01) at the cost of reduced power.
We used the Phenoscanner database to identify potential pleiotropic variants by checking significant associations of loci prioritized in the present study with phenotypes from previously published GWAS.38,39 We further checked GWAS listed in the GWAS Catalog to search for any missed hits. The identified variants were then grouped into categories depending on their association with potential confounder phenotypes and were checked for an influence on the effect estimate using the leave-one-out approach.
We further evaluated the biological relevance of different brain regions in contribution to the overall effect estimate through analysis of gene expression data for the available loci from our different genetic instruments. Gene expression data were extracted from the Genotype-Tissue Expression Project comprising data on a total of 12 different brain regions.40,41 The identified loci were then grouped into categories as per their expression in specific brain regions and checked for an influence on the causal estimate after their exclusion using a leave-one-out approach.
Data availability
Our study is based on publicly available data only.
Results
Prioritization of genetic instruments and power analysis
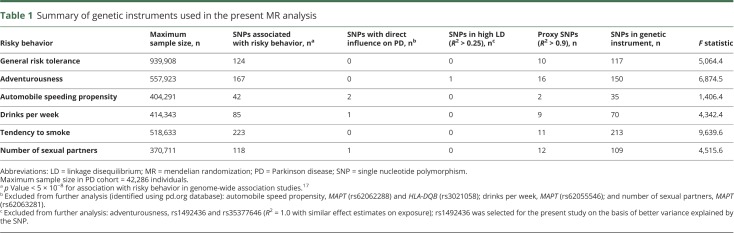
The descriptive statistics of the genetic instruments selected for the MR analyses are presented in table 1. The data used for the analyses are available from Dryad (supplementary table 1, doi.org/10.5061/dryad.54t59r8).
Table 1.
Summary of genetic instruments used in the present MR analysis
Two SNPs from the MAPT (rs62062288; p with PD = 3.1 × 10−21) and HLA-DQB (rs3021058; p with PD = 2.7 × 10−3) genes were excluded from the genetic instrument for automobile speeding propensity phenotype on the basis of a potential direct involvement in PD. Two additional MAPT SNPs were present in the respective genetic instrument for the phenotypes drinks per week (rs62055546; p with PD = 5.2 × 10−21) and number of sexual partners (rs62063281; p with PD = 1.73 × 10−21) and were not carried forward to further analyses. One SNP was observed to be in complete LD with another SNP for the adventurousness phenotype and was excluded. The final number of available SNP data varied from 35 (of 42) for automobile speeding propensity to 213 (of 223) for smoking tendency with F statistics of the pooled genetic instrument ranging from 1,406.4 (for automobile speeding propensity) to 9,639.6 (for ever vs never smoking).
Our power analyses suggest that our study has ≈80% power to detect a true OR of 1.349 or 0.698 for PD per SD of the continuous phenotype assuming that the proportion of the continuous phenotype explained by the genetic instrument is ≥1% at a type 1 error rate of 0.05.
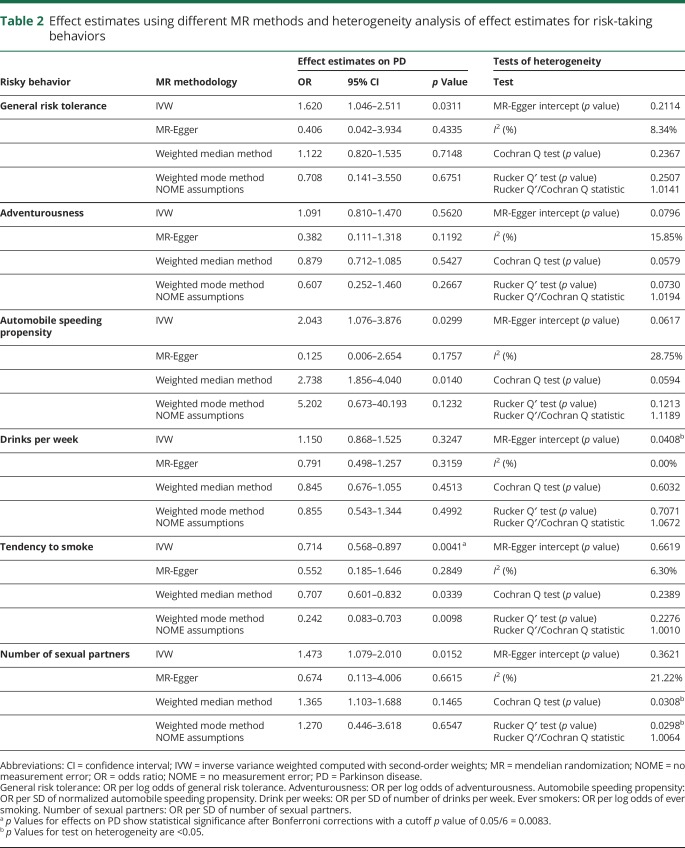
Estimation of effect estimates using MR and assessment of pleiotropy
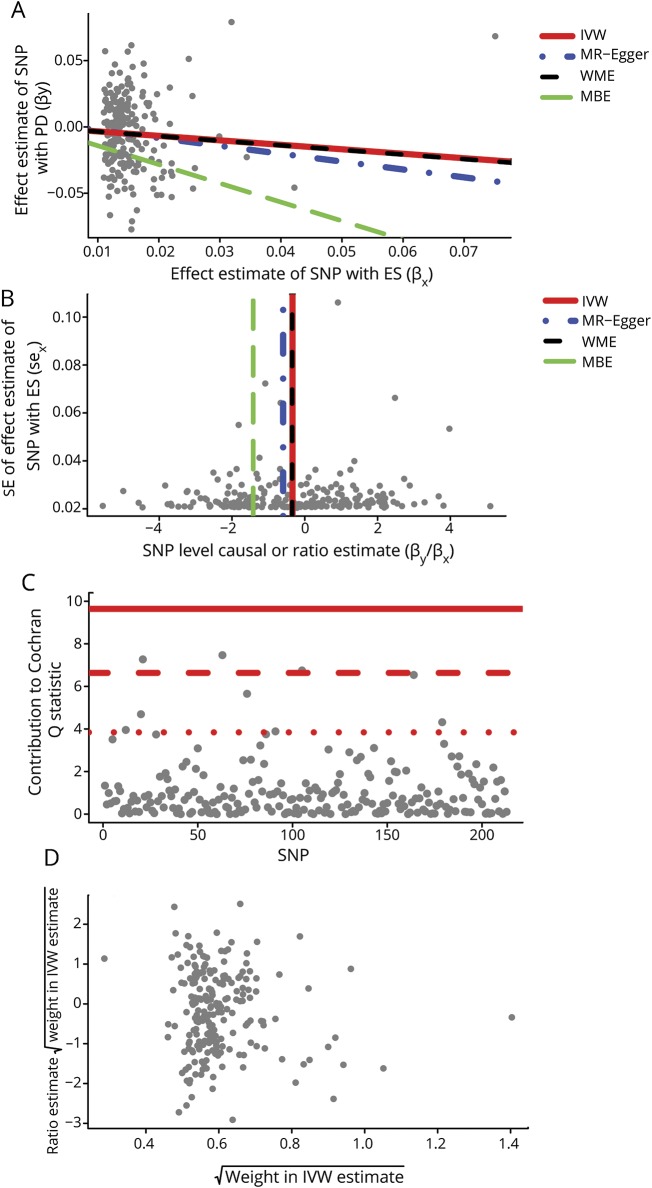
The effect estimates using different MR methods are provided in table 2. With the IVW method, a genetically increased risk of tendency to smoke was associated with a reduced risk of PD per unit increase in log odds of ever smoking (OR 0.714 per log odds of ever smoking, 95% confidence interval [CI] 0.568–0.897, p = 0.0041). Results from the weighted median MR analysis showed similar results (OR 0.707 per log odds of ever smoking, 95% CI 0.601–0.832). There was minimal evidence of heterogeneity of effect estimates between individual variants (I2 = 6.30%, Cochran Q test p = 0.2389), which was confirmed with the MR-Egger intercept test (p = 0.6619). Corresponding plots used for the assessment of pleiotropy are shown in figure 1. We did not detect any potential outlier or pleiotropic variant in the association analysis of the tendency to smoke phenotype and PD.
Table 2.
Effect estimates using different MR methods and heterogeneity analysis of effect estimates for risk-taking behaviors
Figure 1. Causal association analysis and assessment of pleiotropy for the ever smoking phenotype with PD.
(A) Scatterplot showing causal effect estimates computed with various mendelian randomization (MR) methods. (B) Funnel plot showing the extent of heterogeneity among the individual Wald ratio estimates. (C) Plot of Cochran Q estimates for individual single nucleotide polymorphisms (SNPs) constituting the genetic instrument for ever smoker (ES) phenotype using inverse variance weighted (IVW) method using second-order weights. (D) Radial MR plot showing the distribution of weights contributed by individual SNPs in the causal effect estimation by IVW method using second-order weights. MBE = mode-based estimate; PD = Parkinson disease; SE = standard error; WME = weighted median estimate.
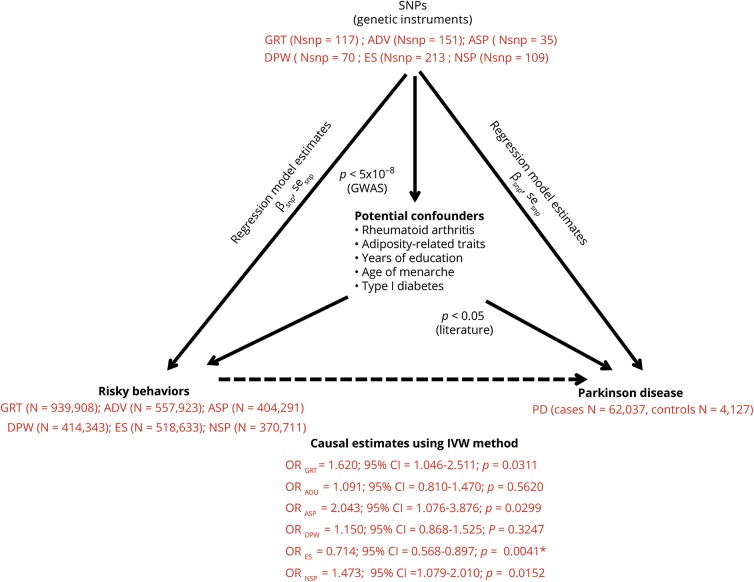
For other risk-taking behaviors, including general risk tolerance, automobile speeding propensity, and the number of sexual partners, we observed a trend toward positive associations (general risk tolerance: OR 1.620 per log odds of general risk tolerance, 95% CI 1.046–2.511, p = 0.0311; automobile speeding propensity: OR 2.043 for each 1-SD increase in normalized automobile speed propensity, 95% CI 1.076–3.876, p = 0.0299; number of sexual partners: OR 1.473 for each 1-SD increase in the number of sexual partners, 95% CI 1.079–2.010, p = 0.0152). A triangulation flowchart summarizing the findings of the study in the context of the MR workflow is given in figure 2.
Figure 2. Triangular representation of results from the present MR study.
ADV = adventurousness; ASP = automobile speeding propensity; βsnp = regression coefficient corresponding to each specific SNP for respective arm of the triangle corresponding to each relationship; CI = confidence interval; DPW = drinks per week; ES = ever vs never smoking; GRT = general risk tolerance; GWAS = genome-wide association studies; Nsnp = number of SNPs in the genetic instrument for each respective risky behavior; NSP = number of sexual partners; OR = odds ratio per log odds of risky behavior; PD = Parkinson disease; sesnp = standard error estimates corresponding to each specific SNP for the respective arm of the triangle corresponding to each relationship; SNP = single nucleotide polymorphism.
Sensitivity analyses
Leave-one-out analysis
In the forest plots, we observed outlier SNPs for the phenotypes general risk tolerance (rs993137), adventurousness (rs10433500), and drinks per week (rs1229984) (data not shown). Leaving out outlier SNPs for each of the respective phenotypes did not alter the results substantially (general risk tolerance: OR 1.763 per log odds of general risk tolerance, 95% CI 1.133–2.744, p = 0.0125; adventurousness: OR 1.164 per log odds of adventurousness, 95% CI 0.858–1.580, p = 0.3270; drinks per week: OR 1.368 for each 1-SD increase in the number of drinks per week, 95% CI 0.978–1.915, p = 0.0669).
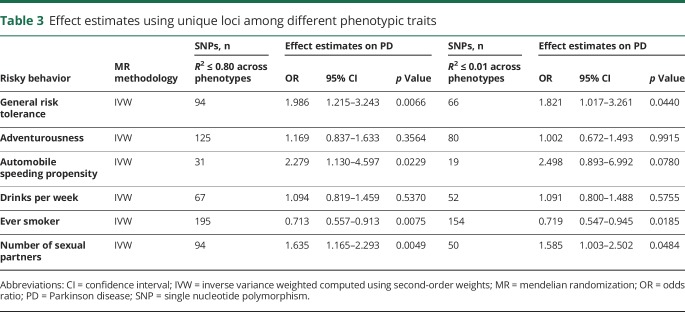
Genetic overlap between risky behaviors
We identified a reduction in the number of unique SNPs in the genetic instruments for each of the phenotypes using 2 different LD cutoffs (R2 ≤ 0.8 and R2 ≤ 0.01) (table 3). However, regardless of the cutoff criteria, there was no change in the protective association of the tendency to smoke phenotype with PD (number of SNPs in the genetic instrument with R2 ≤ 0.8: 195, OR 0.713 per log odds of ever smoking, 95% CI 0.557–0.913, p-value = 0.0037; number of SNPs in the genetic instrument with R2 ≤ 0.01: OR 0.719 per log odds of ever smoking, 95% CI 0.547–0.945, p = 0.0185). Furthermore, consistent with the risk-increasing trend observed for general risk tolerance and the number of sexual partners, we observed a stronger suggestion for causal association with PD for both phenotypes after reducing the number of SNPs from 117 to 94 (R2 ≤ 0.8) for general risk tolerance and from 109 to 94 (R2 ≤ 0.8) for the number of sexual partners (OR 1.986 per log odds of general risk tolerance, 95% CI 1.215–3.243, p = 0.0066; OR 1.635 for each 1-SD increase in number of sexual partners, 95% CI 1.165–2.293, p = 0.0049). The associations persisted when a stringent lower R2 cutoff of 0.01 was used (p = 0.0440 and 0.0484).
Table 3.
Effect estimates using unique loci among different phenotypic traits
Genetic variants associated with potential confounders
We did a comprehensive screening of the Phenoscanner database for potential associations of genetic loci used in the current study and reported to be associated with other phenotypes. The identified associated phenotypes were then investigated for association with PD through a thorough literature search. Using this strategy, we identified rheumatoid arthritis, years of educational attainment, adiposity-related traits, age at menarche, and type 1 diabetes mellitus as potential confounders45–49 (figure 2). We identified 8 genetic variants or loci from our genetic instrument for the tendency to smoke phenotype associated with different confounding traits. SNP rs12042017 has been previously reported to be associated with years of educational attainment (p = 4.48 × 10−10). SNPs rs13396935 and rs6265 were observed to be associated with several adiposity-related measures. The proxy variant rs1514174 of rs4650277 (R2 = 0.99) was further associated with body mass index (p = 2.99 × 10−27). Rheumatoid arthritis, age at menarche, and type 1 diabetes mellitus were further identified as potential confounders associated with rs2734971, rs4650277, and rs1701704 (proxy for rs772921 with complete LD). Our sensitivity analysis excluding each of the SNPs or combinations of SNPs based on their common associated trait showed no overall influence on effect estimate for the tendency to smoke phenotype (data not shown).
Genetic variants involved in brain expression
Using brain-specific expression quantitative trait loci retrieved from the Genotype-Tissue Expression Project, we identified 27 different SNPs from the genetic instrument for the tendency to smoke phenotype with varied influence in different brain regions (data not shown). Surprisingly, the corresponding candidate genes were least represented in the substantia nigra, while as many as 10 genetic variants were observed to significantly influence gene expression in cerebellar hemisphere and cerebellum. Our sensitivity analysis showed that excluding genetic variants mapping to genes overexpressed in the cerebellum had maximum influence on the overall causal effect estimation (OR 0.761, 95% CI 0.606–0.957, p = 0.0197). A similar influence was observed after the exclusion of all the genetic variants mapping to genes expressed in brain (OR 0.735, 95% CI 0.581–0.930, p = 0.0106). Our sensitivity analysis thereby suggested an important role of the cerebellum in the smoking tendency phenotype. Our literature search for the excluded genetic variants in the tendency to smoke genetic instrument influencing expression in cerebellum for potential influence on other biological pathways rules out pleiotropic effects of these variants.
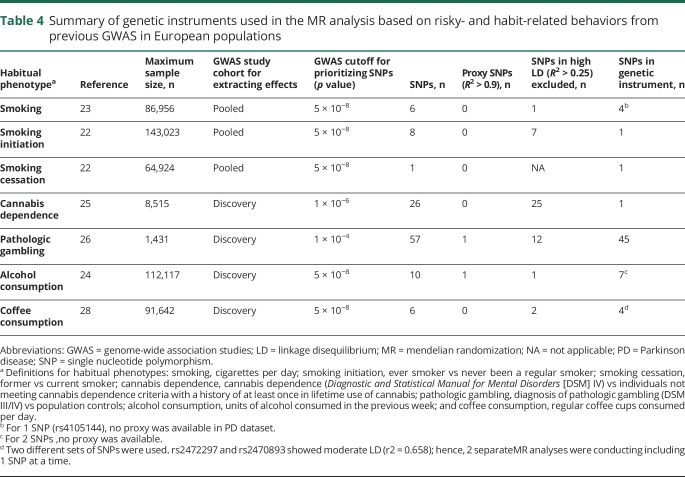
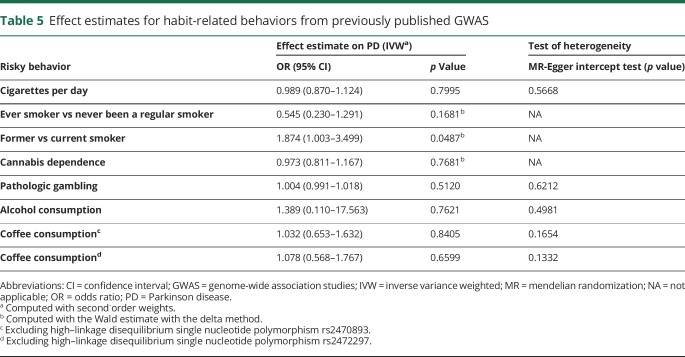
Secondary MR analysis
The descriptive statistics of the genetic instruments selected for the secondary MR analyses are presented in table 4. The effect estimates generated with MR are shown in table 5. We first used genetic instruments for different traits representative of the smoking phenotype (ever smoker vs never been a regular smoker, former vs current smoker, and cigarettes per day).42–46 A previous meta-analysis of GWAS on the ever smoker phenotype in 143,023 individuals of European ancestry identified genetic variants from the BDNF gene to be associated with the ever smoker phenotype.22 Because all the variants were in high LD with each other, we used rs6265, a nonsynonymous variant with a functional effect on gene expression, as a proxy for all other variants for the MR analysis. We failed to observe any association (OR 0.545, 95% CI 0.230–1.291, p = 0.1681). Interestingly, a genetic instrument (rs3025343) for former smokers vs current smokers showed a trend toward risk for predisposition to PD (OR 1.874, 95% CI 1.003–3.499, p = 0.0487). We further extracted a genetic instrument made up of 4 uncorrelated genetic variants for a related phenotype of cigarettes per day from a meta-analysis of GWAS on 86,956 individuals, again failing to observe any trend (OR 0.989, 95% CI 0.870–1.124, p = 0.7995).23
Table 4.
Summary of genetic instruments used in the MR analysis based on risky- and habit-related behaviors from previous GWAS in European populations
Table 5.
Effect estimates for habit-related behaviors from previously published GWAS
We further used a recently published GWAS on alcohol consumption using data from 112,117 individuals from the UK Biobank.24 Using 7 genetic variants as a genetic instrument, we were able to replicate our finding of suggestive absence of causal association of alcohol consumption with PD (OR 1.389, 95% CI 0.110–17.563, p = 0.7621). We further failed to observe any association of number of regular coffee cups per day with PD (OR 1.032, 95% CI 0.653–1.632, p = 0.8405).28
We additionally investigated a protective causal role of cannabis dependence in PD by exploiting a recent GWAS study on cannabis dependence in 2080 cannabis-dependent cases and 6,435 cannabis-exposed controls of European descent.25 The study reported a potential role of a cluster of highly linked 26 SNPs spanning a region on chromosome 10. The study further identified a putative functional SNP rs1409568 among this cluster responsible for the observed phenotypic association. Our investigation of a potential causal role of rs1409568 did not show evidence for a role of cannabis dependence in PD predisposition (OR 0.973, 95% CI 0.811–1.167, p = 0.7681). A recent GWAS in 1,531 whites further reported an absence of any significant SNPs with pathologic gambling.27 We generated a genetic instrument based on the top hits from the study with a cutoff of p < 10−4 using 45 uncorrelated genetic variants. We observed no association of PD with pathologic gambling (OR 1.004, 95% CI 0.991–1.018, p = 0.5120).
Discussion
This comprehensive study exploring the role of risky behaviors as potential causal factors for PD using an MR approach suggests that the tendency-to-smoke trait is possibly causally related to PD, with individuals who started smoking being protected against PD. Our sensitivity analysis further demonstrated robustness of the reported association in the absence of any detectable pleiotropic effect. Furthermore, our secondary MR analysis did not show any association of other habitual traits, including other smoking phenotypes such as number of cigarettes per day, cannabis dependence, pathologic gambling, and alcohol and coffee consumption with PD.
Numerous observational studies have previously shown an inverse association of smoking with PD. A meta-analysis merging smoking status trait from 33 different populations demonstrated a risk reduction by 36% for ever vs never smokers with consistent results in both case-control and cohort studies.2 Other epidemiologic studies suggested significant gene-by-smoking interaction effects in PD.47,48 In our study, we observed a PD risk reduction of 31% for ever smokers vs never smokers. Although risk reduction effects demonstrated in an observational study and in an MR study may not be comparable, a consistency in the direction of protective associations by both the approaches is an important finding.
To validate our results, we performed secondary MR analyses using other habit-related behaviors from other GWAS. The lack of association with a previously reported genetic instrument for the ever smoker instrument and former smoker vs current smoker may be explained by lower power of the GWAS, with only 1 significant variant contributing to the instrument for both MR analyses. We also did not observe an association of PD risk and the number of cigarettes per day. One explanation would be that this continuous phenotype mainly reflects just the tobacco and nicotine exposure, whereas the ever vs never smoking might rather be a sign of risk-taking behavior. Our results thereby clearly imply the need for careful dissection of different smoking phenotypes. This will improve our understanding the causal role of the tendency to smoke on PD and provide further insight into the development of the disease.
As outlined in the Results section, our MR results on coffee consumption (cups per week) and alcohol consumption (drinks per week) also did not show significant associations. However, we cannot exclude that the analysis of coffee and alcohol consumption as quantitative traits may have the same limitations as the analysis of cigarettes smoked per day. Furthermore, we could have missed the association with coffee consumption, possibly owing to the low variance explained by the genetic instruments, eventually resulting in low statistical power. Evidently, the lack of a role of cannabis dependence observed in the present study also needs to be evaluated with stronger genetic instruments.
The absence of association with gambling in our analysis, however, could be attributed to the winner's curse because SNP exposure estimates used to calculate effect estimates may be overestimated due to limited power of the study on gambling phenotype. There has also been growing debate in recent years on the diagnosis of gambling in the fifth edition of the DSM with changes in criteria expected to improve classification accuracy.49 Because the diagnostic criteria adopted for pathologic gambling classification in the source GWAS of our current MR study were based on DSM III and IV, the possibility of potential selection bias on account of an incorrect classification of pathologic gambling in a subset of individuals cannot be ruled out. Instead, pathologic gambling could be the result of dopamine agonist therapy prescribed to patients with PD and may not lead to PD itself. In summary, we hope that in the future, with the availability of large-scale GWAS datasets on secondary analysis conducted in the current study, some light could be shed on the causality of these important habit-forming substances.
Another important finding of our comprehensive MR analyses was the absence of any potential causal role of drinks per week with PD. Using data from the UK Biobank, we were able to replicate our finding of absence of association of alcohol consumption with risk of PD. However, our study suggests a potential causal association of the number of sexual partners and PD risk. To the best of our knowledge, no epidemiologic population-based study has yet examined the role of promiscuity on PD risk. Therefore, our MR results need to be interpreted cautiously, and independent lines of validation of this association are required to confirm these results.
An important limitation of our current study is that we could not directly assess associations of individual genetic variants with potential confounders of association between risk behavior and PD due to the lack of knowledge of potential confounders and the unavailability of individual-level data. Nevertheless, our sensitivity analysis demonstrated that exclusion of loci being associated with PD-associated phenotypes from the MR analyses had no effect on the overall association. We could not further provide data on the degree of sample overlap among GWAS datasets on exposure and outcome in our 2-sample MR design. A considerable overlap could bias the results toward the estimates generated through observational studies. However, this potential limitation could not have any impact on our results because the IVW method using second-order weights used in the current study is known to address this bias. MR methodology relies on exposure data collected in previously conducted observational studies, the majority of them being case-control studies. Although the methodology provides an additional line of evidence to what has been frequently observed in the observational study, it may not be able to exclude recall bias involved at the time of collection of self-reported data.50 Several studies have shown that retrospective assessment of alcohol intake or smoking profoundly influences the association with PD, because the recall of past behaviors can be affected by current exposures.51 In addition, prospective studies often showed nonsignificant associations with PD.52 Nevertheless, exposure data used for MR studies are less likely to have recall bias because MR studies use exposure data collected from the general population compared to case-control studies involving patients with PD who are more likely to be biased on the basis of being a case (reporting bias) and to have recall bias due to age-related cognitive decline.
A recent study showed a disproportionate focus on European populations in GWAS, with 75% of studies in the National Human Genome Research Institute GWAS Catalog using European or mostly European populations.53 Moreover, the majority of remaining studies are usually underpowered to detect associations, thereby limiting their use for inferring conclusions on causality using the MR approach. Furthermore, genetic variants show a remarkable ethnicity-specific heterogeneity not only in their distribution but also in their functional activity. For instance, the same genetic variant could exhibit different pleiotropic effects in different populations. Henceforth, our findings in a European population cannot readily be generalized to populations with different ethnic and racial backgrounds.
It may be noted that we may have missed some of the potential cases of PD among controls in our PD cohort. Because the controls represent individuals taken from the general European population and assuming a prevalence of 108 to 257 per 100,000 in the European population, we assume that the number of potential cases of PD among controls could be 50 to 100 in our cohort of 33,235 controls.54 This small number of possibly undetected cases of PD among controls is unlikely to have a relevant influence on the overall results.
Despite these limitations, to the best of our knowledge, our study represents the most comprehensive MR study to date on risky behavior phenotypes and PD. An extensive sensitivity analysis including use of genetic instruments specific to individual phenotypic traits, use of previous studies, a literature search for potential pleiotropic variants, and brain expression analysis collectively demonstrates a potentially strong causal protective role of smoking tendency on PD. The present study also demonstrates that careful interpretation of pleiotropic signals and sensitivity analysis based on biological function could lead to fine filtering of GWAS signals. Such an approach may assist in differentiating between mediators and exposures, thereby helping us to construct the causal pathways leading to PD.55 We further stress the need for interpreting the evidence of causality from our study cautiously in light of existing biological knowledge and further evidence from other epidemiologic study designs.
Acknowledgment
The authors acknowledge the investigators of the original study4 and the PDGene database team18 for sharing the PD GWAS data used in this study.
Glossary
- CI
confidence interval
- DSM
Diagnostic and Statistical Manual for Mental Disorders
- GWAS
genome-wide association study
- IVW
inverse variance weighted
- LD
linkage disequilibrium
- MR
mendelian randomization
- OR
odds ratio
- PD
Parkinson disease
- SNP
single nucleotide polymorphism
Appendix. Authors
Footnotes
Editorial, page 653
Study funding
This study was supported by grants from the German Research Foundation (Research Unit ProtectMove, FOR 2488).
Disclosure
S. Grover, C. Lill, and M. Kasten report no disclosures to the manuscript. C. Klein reports personal fees from Centogene and Biogen. F. Del Greco M and I. König report no disclosures to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.de Lau LM, Breteler MM. Epidemiology of Parkinson's disease. Lancet Neurol 2006;5:525–535. [DOI] [PubMed] [Google Scholar]
- 2.Noyce AJ, Bestwick JP, Silveira-Moriyama L, et al. Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann Neurol 2012;72:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lill CM. Genetics of Parkinson's disease. Mol Cel Probes 2016;30:386–396. [DOI] [PubMed] [Google Scholar]
- 4.Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 2014;46:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls MA, Blauwendraat C, Vallerga CL, et al. Parkinson's disease genetics: identifying novel risk loci, providing causal insights and improving estimates of heritable risk. bioRxiv 2018:388165. [Google Scholar]
- 6.Noyce AJ, Nalls MA. Mendelian randomization: the key to understanding aspects of Parkinson's disease causation? Mov Disord 2016;31:478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peres FF, Lima AC, Hallak JEC, Crippa JA, Silva RH, Abilio VC. Cannabidiol as a promising strategy to treat and prevent movement disorders? Front Pharmacol 2018;9:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossu G, Rinaldi R, Colosimo C. The rise and fall of impulse control behavior disorders. Parkinson Relat Disord 2018;46(suppl 1):S24–S29. [DOI] [PubMed] [Google Scholar]
- 9.Moore TJ, Glenmullen J, Mattison DR. Reports of pathological gambling, hypersexuality, and compulsive shopping associated with dopamine receptor agonist drugs. JAMA Intern Med 2014;174:1930–1933. [DOI] [PubMed] [Google Scholar]
- 10.Prins BP, Abbasi A, Wong A, et al. Investigating the causal relationship of C-reactive protein with 32 complex somatic and psychiatric outcomes: a large-scale cross-consortium mendelian randomization study. PLoS Med 2016;13:e1001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benn M, Nordestgaard BG, Frikke-Schmidt R, Tybjaerg-Hansen A. Low LDL cholesterol, PCSK9 and HMGCR genetic variation, and risk of Alzheimer's disease and Parkinson's disease: mendelian randomisation study. BMJ 2017;357:j1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichler I, Del Greco MF, Gogele M, et al. Serum iron levels and the risk of Parkinson disease: a mendelian randomization study. PLoS Med 2013;10:e1001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyce AJ, Kia DA, Hemani G, et al. Estimating the causal influence of body mass index on risk of Parkinson disease: a mendelian randomisation study. PLoS Med 2017;14:e1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson SC, Singleton AB, Nalls MA, Richards JB; International Parkinson's Disease Genomics Consortium (IPDGC). No clear support for a role for vitamin D in Parkinson's disease: a mendelian randomization study. Mov Disord 2017;32:1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kia DA, Noyce AJ, White J, et al. Mendelian randomization study shows no causal relationship between circulating urate levels and Parkinson's disease. Ann Neurol 2018;84:178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S, Bowden J. Integrating summarized data from multiple genetic variants in mendelian randomization: bias and coverage properties of inverse-variance weighted methods. [Preprint]. 2015:arXiv:151204486 2015. Available from arxiv.org/abs/1512.04486. [Google Scholar]
- 17.Karlsson Linnér R, Biroli P, Kong E, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 2019;51:245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lill CM, Roehr JT, McQueen MB, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: the PDGene database. PLoS Genet 2012;8:e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobacco Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010;42:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010;42:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke TK, Adams MJ, Davies G, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112,117). bioRxiv 2017;22:1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal A, Chou YL, Carey CE, et al. Genome-wide association study identifies a novel locus for cannabis dependence. Mol Psychiatry 2018;23:1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang M, Lemenager T, Streit F, et al. Genome-wide association study of pathological gambling. Eur Psychiatry 2016;36:38–46. [DOI] [PubMed] [Google Scholar]
- 24.Coffee and Caffeine Genetics Consortium, Cornelis MC, Byrne EM, Esko T, et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry 2015;20:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grover S, Del Greco MF, Stein CM, Ziegler A. Mendelian randomization. Methods Mol Biol 2017;1666:581–628. [DOI] [PubMed] [Google Scholar]
- 26.Edlund CK, Conti DV, Van Den Berg, DJ.. rAggr 2017. Available at http://raggr.usc.edu/. Accessed June 22, 2018. [Google Scholar]
- 27.Li B, Martin E. An approximation to the F-distribution using the chi-square distribution. Computat Stat Data Anal 2002;40:6. [Google Scholar]
- 28.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in mendelian randomization studies. Int J Epidemiol 2013;42:1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.mRnd. Available at cnsgenomics.com/shiny/mRnd. Accessed August 22, 2019.
- 30.Bowden J, Del Greco MF, Minelli C, et al. Improving the accuracy of two-sample summary data mendelian randomization: moving beyond the nome assumption. Int J Epidemiol Epub 2018 Dec 18. [DOI] [PMC free article] [PubMed]
- 31.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017;46:1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J. A framework for the investigation of pleiotropy in two-sample summary data mendelian randomization. Stat Med 2017;36:1783–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in mendelian randomisation studies with summary data and a continuous outcome. Stat Med 2015;34:2926–2940. [DOI] [PubMed] [Google Scholar]
- 36.Rucker G, Schwarzer G, Carpenter JR, Binder H, Schumacher M. Treatment-effect estimates adjusted for small-study effects via a limit meta-analysis. Biostatistics 2011;12:122–142. [DOI] [PubMed] [Google Scholar]
- 37.Bowden J, Spiller W, Del Greco MF, et al. Improving the visualization, interpretation and analysis of two-sample summary data mendelian randomization via the radial plot and radial regression. Int J Epidemiol 2018;47:1264–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32:3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamat MA, Blackshaw JA, Young R, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019; btz469: doi.org/10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed]
- 40.GTEx Consortium. Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GTEx Portal. 2017. Available at: gtexportal.org. Accessed June 22, 2018. [Google Scholar]
- 42.Rugbjerg K, Friis S, Ritz B, Schernhammer ES, Korbo L, Olsen JH. Autoimmune disease and risk for Parkinson disease: a population-based case-control study. Neurology 2009;73:1462–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotagal V, Bohnen NI, Muller ML, et al. Educational attainment and motor burden in Parkinson's disease. Mov Disord 2015;30:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park K, Oeda T, Kohsaka M, Tomita S, Umemura A, Sawada H. Low body mass index and life prognosis in Parkinson's disease. Parkinson Relat Disord 2018;55:81–85. [DOI] [PubMed] [Google Scholar]
- 45.Cereda E, Barichella M, Cassani E, Caccialanza R, Pezzoli G. Reproductive factors and clinical features of Parkinson's disease. Parkinson Relat Disord 2013;19:1094–1099. [DOI] [PubMed] [Google Scholar]
- 46.De Pablo-Fernandez E, Goldacre R, Pakpoor J, et al. Association between diabetes and subsequent Parkinson disease: a record-linkage cohort study. Neurology 2018;91:e139–e142. [DOI] [PubMed] [Google Scholar]
- 47.Chuang YH, Lee PC, Vlaar T, et al. Pooled analysis of the HLA-DRB1 by smoking interaction in Parkinson disease. Ann Neurol 2017;82:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee PC, Ahmed I, Loriot MA, et al. Smoking and Parkinson disease: evidence for gene-by-smoking interactions. Neurology 2018;90:e583–e592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petry NM, Blanco C, Stinchfield R, Volberg R. An empirical evaluation of proposed changes for gambling diagnosis in the DSM-5. Addiction 2013;108:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coughlin SS. Recall bias in epidemiologic studies. J Clin Epidemiol 1990;43:87–91. [DOI] [PubMed] [Google Scholar]
- 51.Bettiol SS, Rose TC, Hughes CJ, Smith LA. Alcohol consumption and Parkinson's disease risk: a review of recent findings. J Parkinsons Dis 2015;5:425–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palacios N, Gao X, O'Reilly E, et al. Alcohol and risk of Parkinson's disease in a large, prospective cohort of men and women. Mov Disord 2012;27:980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales J, Welter D, Bowler EH, et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol 2018;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Campenhausen S, Bornschein B, Wick R, et al. Prevalence and incidence of Parkinson's disease in Europe. Eur Neuropsychopharmacol 2005;15:473–473. [DOI] [PubMed] [Google Scholar]
- 55.Grover S, Del Greco MF, König I. Evaluating the current state of mendelian randomization studies: a protocol for a systematic review on methodological and clinical aspects using neurodegenerative disorder as a model outcome. Syst Rev 2018;7:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Our study is based on publicly available data only.