Abstract
Ehrlichia spp. are emerging tick-borne obligatory intracellular bacteria that cause febrile and sometimes fatal diseases with abnormal blood cell counts and signs of hepatitis. Ehrlichia HF strain provides an excellent mouse disease model of fatal human ehrlichiosis. We recently obtained and established stable culture of Ehrlichia HF strain in DH82 canine macrophage cell line, and obtained its whole genome sequence and annotation. To identify genes required for in vivo virulence of Ehrlichia, we constructed random insertional HF strain mutants by using Himar1 transposon-based mutagenesis procedure. Of total 158 insertional mutants isolated via antibiotic selection in DH82 cells, 74 insertions were in the coding regions of 55 distinct protein-coding genes, including TRP120 and multi-copy genes, such as p28/omp-1, virB2, and virB6. Among 84 insertions mapped within the non-coding regions, seven are located in the putative promoter region since they were within 50 bp upstream of the seven distinct genes. Using limited dilution methods, nine stable clonal mutants that had no apparent defect for multiplication in DH82 cells, were obtained. Mouse virulence of seven mutant clones was similar to that of wild-type HF strain, whereas two mutant clones showed significantly retarded growth in blood, livers, and spleens, and the mice inoculated with them lived longer than mice inoculated with wild-type. The two clones contained mutations in genes encoding a conserved hypothetical protein and a staphylococcal superantigen-like domain protein, respectively, and both genes are conserved among Ehrlichia spp., but lack homology to other bacterial genes. Inflammatory cytokine mRNA levels in the liver of mice infected with the two mutants were significantly diminished than those infected with HF strain wild-type, except IL-1β and IL-12 p40 in one clone. Thus, we identified two Ehrlichia virulence genes responsible for in vivo infection, but not for infection and growth in macrophages.
Keywords: Ehrlichia HF strain, Himar1 transposon mutagenesis, mouse virulence, inflammatory cytokines, staphylococcal superantigen-like domain, obligatory intracellular bacteria
Importance
Ehrlichiosis, a sometimes deadly febrile disease in man and animal, is caused by infection with Gram-negative obligatory intracellular bacteria, Ehrlichia. Ehrlichia species lack typical pathogen-associate molecular patterns (PAMPs), such as lipopolysaccharide, peptidoglycan, pili, and flagella, yet they induce acute and/or chronic inflammatory cytokines in infected animals. Studies to identify virulence factors and PAMPs of Ehrlichia species are hampered by the limitation to applicable genetic tools and small laboratory animal models. Mouse infection with Ehrlichia HF strain provides an excellent model for fatal human ehrlichiosis, as the HF strain is highly virulent in laboratory mice. However, due to inability to culture the HF stain, the use of this model has been limited. As we have succeeded in culturing and whole genome sequencing Ehrlichia HF strain, here, we applied Himar1 transposon random mutagenesis system to this bacterium, and analyzed virulence of the mutant Ehrlichia strains in the immunocompetent laboratory mice. The Ehrlichia HF strain mutagenesis and mouse model provide insights toward in vivo virulence factors and PAMPs of Ehrlichia pathogens.
Introduction
Ehrlichiae are obligate intracellular, gram-negative cocci that infect wild and domestic animals, and humans, and cause emerging infectious diseases called ehrlichiosis. Ehrlichiosis is a tick-borne zoonosis, and not directly transmitted between humans and/or animals. Four different species of Ehrlichia are known to infect humans: E. chaffeensis, E. ewingii, Venezuelan Human Ehrlichiosis (VHE) of E. canis, and E. muris subsp. eauclairensis subsp. nov. (Anderson et al., 1991; Perez et al., 1996; Buller et al., 1999; Pritt et al., 2011, 2017). E. chaffeensis, E. canis, and E. muris infect monocytes and macrophages, whereas E. ewingii infects granulocytes, and they cause subclinical or serious sometimes fatal infectious diseases (Buller et al., 1999; Paddock and Childs, 2003; Pritt et al., 2011). Human ehrlichiosis is characterized by fever, headache, myalgia, thrombocytopenia, leucopenia, and elevated liver enzyme levels (Perez et al., 1996, 2006; Buller et al., 1999; Paddock and Childs, 2003; Martinez et al., 2008; Pritt et al., 2011). Complications, such as pulmonary insufficiency, renal failure, encephalopathy, and disseminated intravascular coagulation can occur in fatal human cases of E. chaffeensis infection (Paddock et al., 1997). Human ehrlichiosis have been reported from the United States, Europe, Asia, Africa, and South America, although most of the case reports have originated in the United States. Since many human ehrlichiosis cases are neither reported nor diagnosed, the number of reported cases so far likely significantly underestimates the true incidence of the disease. Despite this, the number of human ehrlichiosis cases reported to CDC in the year 2018 was around 1,525, which showed a 10-fold increase in the incidence of the disease over a 10-year period, with the highest incidence among individuals aged more than 60 years (Centers for Disease Control and Prevention, 2018). Although the infection can be treated with broad-spectrum antibiotic doxycycline, the disease is of particular threat in the immuno-compromised and the elderly, and can result in severe morbidity and mortality if left untreated (Walker and Dumler, 1996; Paddock and Childs, 2003).
No known pathogen-associated molecular patterns (PAMPs) including endotoxin, peptidoglycan, flagella, common pili, or exotoxin has been detected in Ehrlichia spp., yet they induce acute and/or chronic inflammatory cytokines production in MyD88-dependent, but Toll-like receptors (TLR)-independent manner (Koh et al., 2010; Miura et al., 2011; Rikihisa, 2015). Several virulence factors of E. chaffeensis, which are required for bacterial entry, survival and proliferation of macrophages, have been demonstrated, including ehrlichial invasin that binds host cell receptor and triggers its entry into host cells, three type IV secretion system effectors, three pairs of two-component regulatory system, and outer membrane porins for nutrient acquisition (Kumagai et al., 2006, 2008; Mohan Kumar et al., 2013; Rikihisa, 2015, 2017; Lin et al., 2016; Sharma et al., 2017; Teymournejad et al., 2017; Yan et al., 2018). However, whether Ehrlichia spp. have in vivo virulence factors which are not related to bacterial entry and growth in macrophages is unknown.
E. chaffeensis naturally infects dogs and deer with mild to no clinical signs (Dawson and Ewing, 1992; Davidson et al., 2001; Unver et al., 2002). However, the difficulty, expense, and lack of inbred or gene knockout in these animal models, makes screening in vivo virulence factors prohibitive. On the other hand, although E. chaffeensis only transiently infects immunocompetent laboratory mice (Ohashi et al., 1998a), Ehrlichia sp. HF strain (also called Ixodes ovatus Ehrlichia, IOE), which was isolated in Japan from Ixodes ovatus ticks, is highly virulent in laboratory mice (Fujita and Watanabe, 1994; Shibata et al., 2000; Okada et al., 2001, 2003). Mice inoculated with HF strain bacteria develop severe clinical signs within 7 days and die within 10 days (Fujita and Watanabe, 1994; Shibata et al., 2000). Thus, mouse infection with HF strain can serve as a model for fatal human ehrlichiosis. The HF strain causes a toxic shock-like syndrome in mice, involving many inflammatory factors mediated by CD8+ and CD4+ lymphocytes, NKT cells, and neutrophils (Ismail et al., 2004; Stevenson et al., 2008; Yang et al., 2013). The HF strain was not previously culturable, thus all previous studies were performed using the mouse spleen homogenate containing the HF strain (Ismail et al., 2004; Stevenson et al., 2008; Yang et al., 2013). To facilitate studies using the HF strain, we recently stably cultured the HF strain in canine macrophage DH82 cells, and obtained the complete whole genome sequence of the HF strain [GenBank accession NZ_CP007474 (Lin et al., 2013)]. Comparative genome sequence analysis revealed the HF strain is most closely related to E. muris subsp. eauclairensis subsp. nov., the most recent human ehrlichiosis agent (Pritt et al., 2011, 2017) followed by E. chaffeensis.
Obstacles to functional genome analysis of obligate intracellular pathogen are DNA delivery while retaining viability of extracellular bacteria, efficient reintroduction of the transformed bacterial population into host cells, limited selection markers, and the limited efficiency of homologous recombination and transposition systems. Development of the mariner transposase Himar1, which can function in many organisms (Rubin et al., 1999; Pelicic et al., 2000; Ashour and Hondalus, 2003) and the development of hyperactive Himar1 mutants (Lampe et al., 1999) have effectively diminished this last obstacle. Himar transposition provides an ideal system for global genome functional analysis of the AT-rich organism like Ehrlichia species (Munderloh et al., 2012; Cheng et al., 2013; McClure et al., 2017), since it recognizes AT sites and inserts at a single site per genome (Lampe et al., 1999). In the present study, we constructed HF strain Himar1 transposon mutant library, and cloned selected mutants and characterized cloned mutant pathogenicity in laboratory mice.
Results
Ehrlichia sp. HF Strain Himar1 Insertional Mutant Library
Host cell-free Ehrlichia sp. HF strain was transformed with the plasmid, pCis-mCherry-SS Himar A7 (Munderloh et al., 2012; Cheng et al., 2013). This plasmid carries a Himar1 mariner transposase, and genes encoding mCherry and downstream spectinomycin/streptomycin antibiotic resistance (aad) that are flanked by the nine base pair inverted repeats recognizable by the transposase with 1,833 base pairs to be inserted into the target genome. Both genes are controlled by the Anaplasma marginale transcriptional regulator 1 (Am-Tr1) promoter, which provides constitutive expression of downstream genes in Ehrlichia organisms for transposition and antibiotic selection. The transformed bacteria were cultured in DH82 cells in the presence of spectinomycin and streptomycin antibiotics to recover mutant bacteria containing Himar insertions. These preparations were observed using Hema3 staining for ehrlichial growth and DeltaVision deconvolution fluorescence microscope for expression of mCherry fluorescence protein in mutant bacteria. Genomic loci of the insertions were identified by semi-random, two-step PCR (ST-PCR) (Chun et al., 1997) using primers listed in Table S1, and mapped to Ehrlichia sp. HF genome by SeqBuilder program in DNAStar Lasergene12 software package. For 23 successful transformation experiments, 158 genomic insertion sites were mapped: among 866 protein-coding genes 74 insertions were present within the coding regions of 55 genes, and the remaining 84 insertions were present within the non-coding regions of the genome (Figure 1, Table 1 and, Table S2). One intergenic insertion site each was mapped to <50 bp upstream of seven distinct genes, and may modify the transcription of these genes (Table S2). In E. chaffeensis Himar1 mutagenesis studies, frequency of mutants is ~5 distinct clones per electroporation (Cheng et al., 2013). In our study, we obtained 1–39 distinct insertional mutations per electroporation. Since the mutants were obtained via antibiotic selection in DH82 cells, mutants in the genes that are essential for survival in the macrophages could not be recovered. All 55 genes that had intragenic insertions have homologs in two human monocytic Ehrlichiosis agents: E. chaffeensis and E. muris subsp. eauclairensis (Table 1 and Table S2, only 34 genes with annotated functions in Ehrlichia sp. HF genome are shown in Table 1, and the remaining 21 genes encode hypothetical proteins without known functions are also included in Table S2). For all 55 genes, E. muris subsp. eauclairensis has the highest % identity among all Ehrlichia species.
Figure 1.
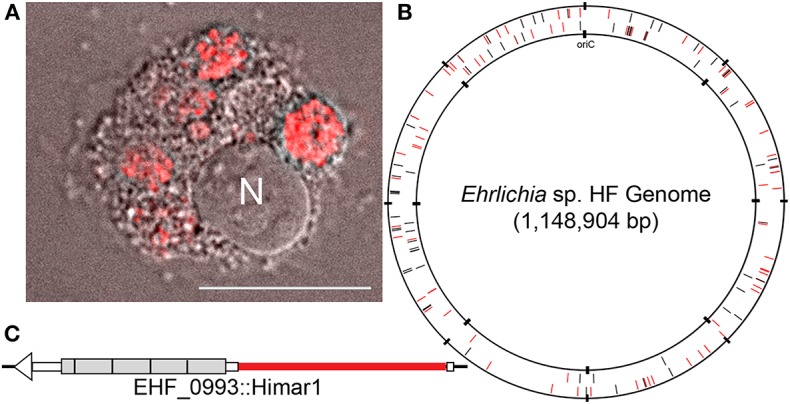
HF mutant and genomic map of the HF strain, showing transposon insertion sites. (A) HF mutant H53 expressing mCherry fluorescence in DH82 cells. N, nucleus. mCherry fluorescence/Differential Interference Microscopy. DeltaVision deconvolution microscopy, Scale bar, 10 μm. (B) Ehrlichia sp. HF chromosomal map showing transposon insertion sites. All mutant bacteria containing the Himar insertions were recovered from DH82 cells by the selection of spectinomycin and streptomycin antibiotics. Black lines represent intragenic insertions and red lines represent intergenic insertions. (C) HF mutant H60E (EHF_0993::Himar1). Insertion site is near 5′-end of the gene encoding TRP120 at upstream of the 4 1/4 of the 300-bp tandem repeats (shown in gray boxes) in TRP120, likely disrupting this protein's expression or function.
Table 1.
Himar1 insertion sites within genes of assigned functions and % identity of the homologous genes in human pathogens E. muris subsp. eauclairensis and E. chaffeensis.
| Locus ID | Genomic insertion site | Isolate ID | Gene product | % aa Identity | |
|---|---|---|---|---|---|
| E. muris | E. chaffeensis | ||||
| EHF_0031 | 28140 | H74C | Chromosome partitioning ATPase ParA | 98 | 95 |
| EHF_0045 | 43288 | H77A-4 | Major outer membrane protein OMP1-19 | 92 | 84 |
| 43813 | H77B-1 | ||||
| EHF_0048 | 45484 | H82B-2 | Major outer membrane protein OMP1-17 | 93 | 85 |
| 45586 | H75B-1 | ||||
| 45987 | H69F-2 | ||||
| 46141 | H77B-3 | ||||
| EHF_0066 | 61587 | H67C | Major outer membrane protein OMP1-2 | 90 | 65 |
| EHF_0069 | 64253 | H79A-3 | Nucleoside diphosphate kinase | 96 | 94 |
| EHF_0075 | 72763 | H73A-1 | Dethiobiotin synthase | 91 | 79 |
| EHF_0115 | 112044 | H80B-3 | Gamma carbonic anhydrase family protein | 92 | 96 |
| EHF_0125 | 122477 | H81C-2 | Peptidase M16 family | 93 | 89 |
| EHF_0135 | 133819 | H43F | Type IV secretion protein VirB2 homolog (VirB2-4) | 84 | 78 |
| EHF_0167 | 179134 | H76F | Aspartate kinase | 95 | 93 |
| EHF_0237 | 248268 | H72E | DNA-3-methyladenine glycosylase | 93a | 87 |
| EHF_0287 | 308227 | H75C | RDD family protein | 90 | 81 |
| EHF_0324 | 353657 | H56B | Lipoate-protein ligase B (LipB) | 95 | 86 |
| EHF_0333 | 366003 | H74B-1 | Dihydropteroate synthase | 94 | 56 |
| EHF_0346 | 382057 | H80C-3 | Cell division ZapA family protein | 96 | 88 |
| EHF_0382 | 406316 | H55 | comEC/Rec2-related domain protein | 93 | 80 |
| EHF_0383 | 407501 | H79E-2 | Sodium:alanine symporter family protein | 95 | 90 |
| EHF_0445 | 490405 | H73E-1 | Type IV secretion system protein, VirB6 family (VirB6-4) | 73b | 82b |
| EHF_0446 | 491800 | H82C | DUF2460 domain-containing protein | 95 | 86 |
| EHF_0513 | 566454 | H73D-2 | Membrane protein, TerC family | 90 | 83 |
| EHF_0515 | 573935 | H81D | Chromosome segregation protein SMC (structural maintenance of chromosomes), archaeal type | 55 | 44 |
| EHF_0522 | 579965 | H69F-1 | DNA recombination protein RmuC family protein | 94 | 89 |
| 580467 | H34 | ||||
| EHF_0702 | 792823 | H77A-1 | Smr domain-containing protein | 87 | 76 |
| 792889 | H67F | ||||
| EHF_0703 | 794748 | H53D-2 | Thiamine biosynthesis protein ThiC | 95 | 92 |
| EHF_0717 | 810577 | H80D-3 | Major Facilitator Superfamily (MFS) transporter | 96 | 83 |
| 810865 | H69E | ||||
| EHF_0718 | 813074 | H79A-2 | Major Facilitator Superfamily (MFS) transporter | 94 | 75 |
| EHF_0723 | 820620 | H80B-4 | DNA mismatch repair protein MutS | 97 | 93 |
| EHF_0732 | 831375 | H75F-4 | Phage conserved hypothetical BR0599 family protein | 90 | 83 |
| EHF_0743 | 847362 | H76A-1 | Glutathione S-transferase family protein | 97 | 94 |
| EHF_0772 | 892485 | H82D-2 | DNA mismatch repair protein MutL | 93 | 81 |
| EHF_0775 | 897331 | H81A-2 | Putative membrane protein | 65 | 51 |
| EHF_0810 | 936950 | H77F | bolA family protein | 83 | 72 |
| EHF_0877 | 1016610 | H73E-2 | Transcriptional repressor NrdR | 99 | 94 |
| EHF_0919 | 1059627 | H72B | Amidophosphoribosyltransferase ComF | 93 | 82 |
| EHF_0956 | 1098537 | H82A-3 | Octaprenyl-diphosphate synthase | 98 | 88 |
| EHF_0993 | 1142873 | H60E | 120 kDa immunodominant surface protein (TRP120) | 38 | 30 |
Homologous match in E. muris subsp. eauclairensis EMUCRT_RS02460 is a frameshifted pseudogene.
Although E. chaffeensis showed higher % identity compared to E. muris subsp. eauclairensis, it only align with the N-terminal 971 aa of E. chaffeensis VirB6-4 (2,940 aa in total). However, HF strain VirB6-4 was aligned with the full length of E. muris VirB6-4 (1,937 aa), and had higher MAX score (alignment score) (2,763) vs. that of E. chaffeensis (1,789).
All Ehrlichia spp. sequenced so far have multiplicated genes encoding immunodominant outer membrane proteins P28/OMP-1 (Ohashi et al., 1998b, 2001; Rikihisa, 2015), some of which were demonstrated to have porin activity in E. chaffeensis (Kumagai et al., 2008), and genes encoding type IV secretion apparatus proteins VirB/D (Rikihisa, 2010, 2017; Gillespie et al., 2016). Of 23 paralogs of p28/omp-1 of the HF strain, insertion was detected in four p28/omp-1s: EHF_0066, EHF_0067, EHF_0045 (two different insertions), and EHF_0048 (four different insertions), showing not all P28/OMP-1s are required for growth in macrophages. There are five virB2 paralogs, and one of which, virB2-4 had insertion. Insertion was also detected in one of four tandem virB6 paralogs, virB6-4. One of two copies of bolA encoding a transcription factor BolA related to stress resistance (Santos et al., 1999; Cheng et al., 2011) had an insertion. In addition to these genes with multiple provisions, a single copy gene trp120, homologous to E. chaffeensis TRP120, a type I secretion system effector, which has been extensively studied (Yu et al., 2000; Luo et al., 2011; Zhu et al., 2011; Dunphy et al., 2013, 2014), had the insertion near 5′-end (Figure 1C), implying this gene is not required for Ehrlichia HF infection of macrophages.
Himar1 intragenic insertion mutants were also identified in genes for vitamin synthesis [thiamine biosynthesis protein ThiC (Lawhorn et al., 2004) and dethiobiotin synthase (Otsuka et al., 1988)], mutation repair [RmuC (Slupska et al., 2000), DNA mismatch repair proteins MutS and MutL (Mansour et al., 2001)], transcriptional regulation [DNA-3-methyladenine glycosylase (Wyatt et al., 1999), transcriptional regulator NrdR (Torrents et al., 2007)], nutrient transport [sodium:alanine symporter family protein (Khani et al., 2018), and two MFS transporters (Quistgaard et al., 2016)], bacterial division and chromosomal segregation [chromosome partitioning ATPase ParA (Lutkenhaus, 2012), cell division ZapA family protein (Small et al., 2007)], and others [amidophosphoribosyltransferase ComF (Bhagavan and Ha, 2015), tellurium resistance protein TerC (Turkovicova et al., 2016), RDD family protein (Shao et al., 2018), glutathione S-transferase (Nebert and Vasiliou, 2004), aspartate kinase (Min et al., 2015), gamma carbonic anhydrase family protein (Hewett-Emmett and Tashian, 1996), octaprenyl-diphosphate synthase (Ashby and Edwards, 1990)] (Table 1). These genes are apparently not required for HF strain infection of macrophages. Six insertions were found within 5% the length of the ORF from the C-terminus of EHF_0075, EHF_0332, EHF_0513, EHF_0717, EHF_0733, EHF_0768; these protein functions may not be disrupted by the Himar1 insert.
Cloning HF Strain Mutants
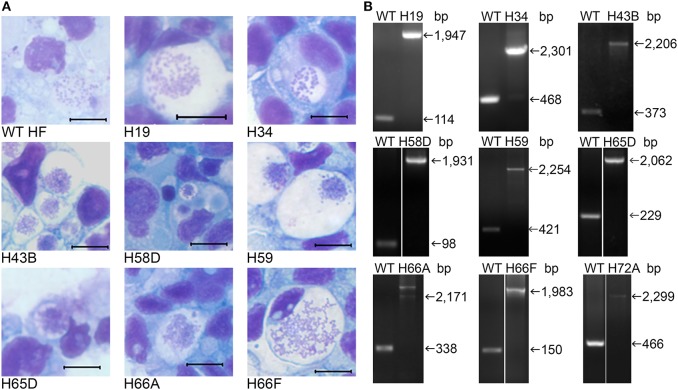
Himar1 transposon insertions occur mostly once or rarely twice per genome (Le Breton et al., 2006; Liu et al., 2007; Cartman and Minton, 2010; Cheng et al., 2013). As previous studies with Himar1 transposon mutagenesis of Anaplasma and Ehrlichia (Felsheim et al., 2006; Cheng et al., 2013), in many rounds of transformation, we obtained a mixture of several insertional mutants, as shown by ST-PCR results from the same cultures. To confirm the presence of Himar1 insertion mutants, insert-specific PCR experiments were performed with primers designed to anneal near identified insertion sites paired with primers annealing to mCherry or aad genes in the inserted transposon (Figure 2A and Table S1). To isolate individual clones from the current Himar1 insertion library with 158 genomic insertion sites, limiting dilution of infected DH82 cells was used. To verify the clonality, specific PCR experiments that amplify the genome sequences flanking the insertion site were developed for each mutant clone (Table S1 and Figure 2B). For the flank PCR, primers flanking both sides of the insertion were designed and used to generate PCR products up to 2,500 bp (Figure 2B and Table S1). Wild-type (WT) or other Himar1 insertional mutants, if present, would show the smaller PCR product as they lack the Himar1 insert at this genomic locus. Multiple samples from frozen-thawed cells of 10–20 culture passages (~2 passages/week), were examined by the insertion-specific flank PCR. Of nine clones identified, four were obtained in DH82 cell culture after limiting dilution, and five were clones from original transformation and confirmed by flank PCR (Table 2). Except one clone where the insertion site is at the upstream of type I secretion outer membrane/TolC family protein, the remaining eight clones contain intragenic insertions. All nine clones could infect ISE6 tick cells in culture as shown by HEMA3 staining of infected cells and verified by the flank PCR (Figure 3 and Table 2).
Figure 2.
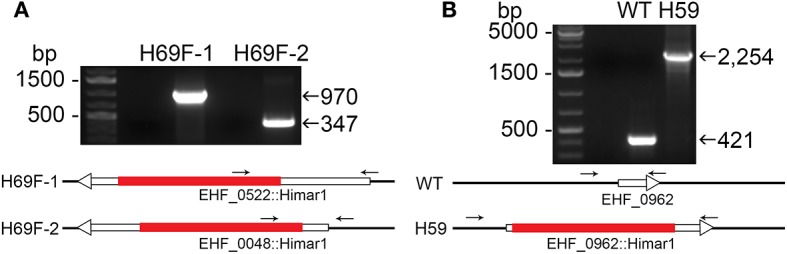
Insertion site-specific PCR to verify insertion site and flank PCR to verify clonality. (A) Specific insert PCR showing insertion sites at specific genomic loci of EHF_0522::Himar1 and EHF_0048::Himar1. (B) Flank PCR demonstrating clonality of EHF_0962::Himar1, arrows—PCR primers, red bar—Himar1 insert containing aad/mCherry, white boxes with arrow—ORFs. The lengths of ORFs and Himar1 insert were drawn to scale except primers that are enlarged.
Table 2.
Characteristics of Cloned HF Himar1 mutants.
| Isolate ID | Locus ID | Gene product | AA | Infects ISE6? | Infects mice? |
E. chaffeensis ortholog (proteomics coverage)a |
|---|---|---|---|---|---|---|
| H19b | EHF_0231 | Hypothetical protein | 222 | Yes | Yes | n/a |
| H34b | EHF_0522 | rmuC family protein | 433 | Yes | Yes | ECH_0577 (0. 584383) |
| H43Bb | EHF_ RS04100 |
Conserved hypothetical protein with SSL domain | 651 | Yes | Delayed mortality, decreased infection | ECH_0150 (0.383929) |
| H58D | Intergenic, 184 bp up EHF_0151 |
Type I secretion outer membrane/TolC family protein | n/a | Yes | Yes | n/a |
| H59b | EHF_0962 | Conserved hypothetical protein | 119 | Yes | Delayed mortality, same infection | ECH_0079 (0.597015) |
| H65D | EHF_0880 | Conserved hypothetical protein | 101 | Yes | Yes | ECH_0181 (0.252427) |
| H66A | EHF_0933 | Conserved hypothetical protein | 84 | Yes | Yes | ECH_0130 (0. 357143) |
| H66Fb | EHF_0332 | Conserved hypothetical protein | 353 | Yes | Yes | ECH_0379 (0. 45584) |
| H72A-1 | EHF_0150 | Conserved hypothetical protein | 244 | Yes | Yes | ECH_1021 (0.383562) |
E. chaffeensis Arkansas ortholog protein expression in HL-60 cells (Lin et al., 2011).
Originally clone.
Figure 3.
Infectivity of cloned HF Himar1 mutants in ISE6 tick cells. Cloned HF Himar1 mutants and WT HF cultured in ISE6 tick cells. (A) HEMA3 staining. Scale bar, 10 μm. (B) Insertion site-specific flank PCR confirming clonality of each mutant in tick cells.
Determination of Mouse LD50 of HF Strain Cultured in DH82 Cells
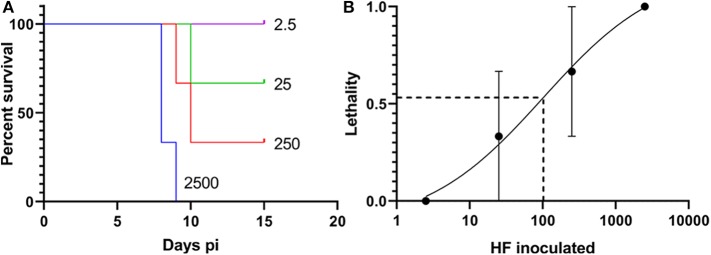
The mouse virulence of the HF strain cultured in DH82 cells has not been determined. Therefore, four 10-fold serially diluted WT HF strain (2,500, 250, 25, and 2.5 bacteria) were intraperitoneally inoculated into three female ICR (CD-1) mice each to obtain Kaplan-Meier survival curve. Based on the result, LD50 of HF strain in mice determined by Questgraph LD50 calculator (AAT Bioquest, 2020), was 100 bacteria (Figure 4).
Figure 4.
LD50 of the Wild-type (WT) HF strain. (A) Kaplan-Meier survival curves for WT HF bacteria, number of inoculated bacteria/mouse is shown for each curve, N = 3, Log-rank, Gehan-Breslow-Wilcoxon test among each group: P < 0.01. (B) Sigmoidal curve showing the calculation of WT HF culture. LD50 is based on survival with 10-fold dilutions, calculated from AAT Bioquest LD50 calculator, with means and standard error (N = 3).
Mouse Virulence of Cloned HF Mutants
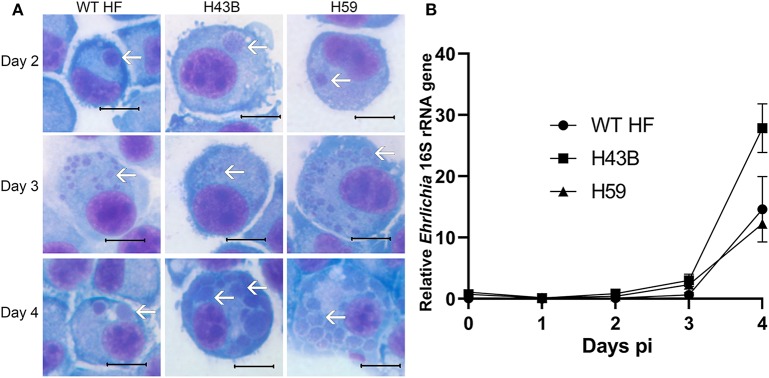
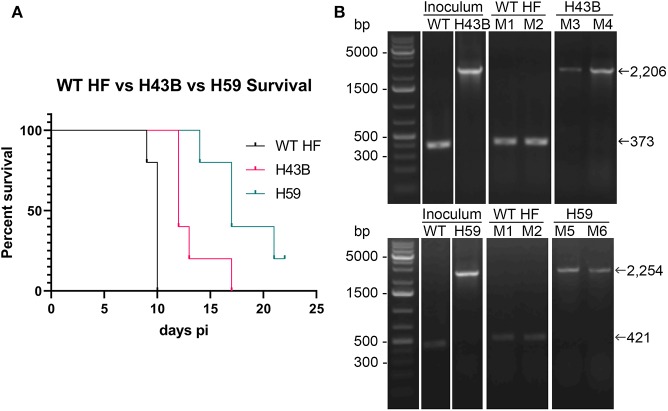
We investigated whether Himar1 insertions affected the HF strain's mouse infectivity and pathogenesis. Immunocompetent mice were inoculated intraperitoneally with nine cloned mutants and WT HF strain at ~5–15 × 106 bacteria/mouse. With this high dosage, most mice became severely moribund and lost >20% body weight and were euthanized, or died at 4–12 days post-inoculation (pi) (Figure 5). However, mice inoculated with H59 and H43B mutants survived significantly longer at 8–12 days pi (Figure 5), hence, these two mutants were selected for further study. H59 has insertion in EHF_0962 (conserved hypothetical protein, 119 aa) and H43B has insertion in EHF_RS04100 (conserved hypothetical protein, 651 aa) (Table 2). Although no known motif was detected in EHF_0962, staphylococcal superantigen-like (SSL) domain (Fraser and Proft, 2008) (aa 429–537, E-value 2.47 × e−04) was detected in EHF_RS04100. Compared with WT, H59, and H43B infection and growth in DH82 macrophage were similar by quantitative PCR (qPCR) (Figure 6). When ~4 × 104 WT HF were inoculated per mouse, all five mice were severely moribund necessitating euthanasia or died on day 9–10 pi, however mice inoculated with H43B were euthanized or died from day 12 to 17 pi. Mice inoculated with H59 were euthanized or died from day 13 to 21 pi, with one mouse apparently recovering from illness on day 22, when the experiment was terminated (Figure 7A). Clonality of H43B and H59 in inoculated mice were verified by insertion site-specific flank PCR at euthanasia in the liver samples of inoculated mice (Figure 7B). Thus, H43B and particularly H59 mutants have partially reduced virulence in mice.
Figure 5.
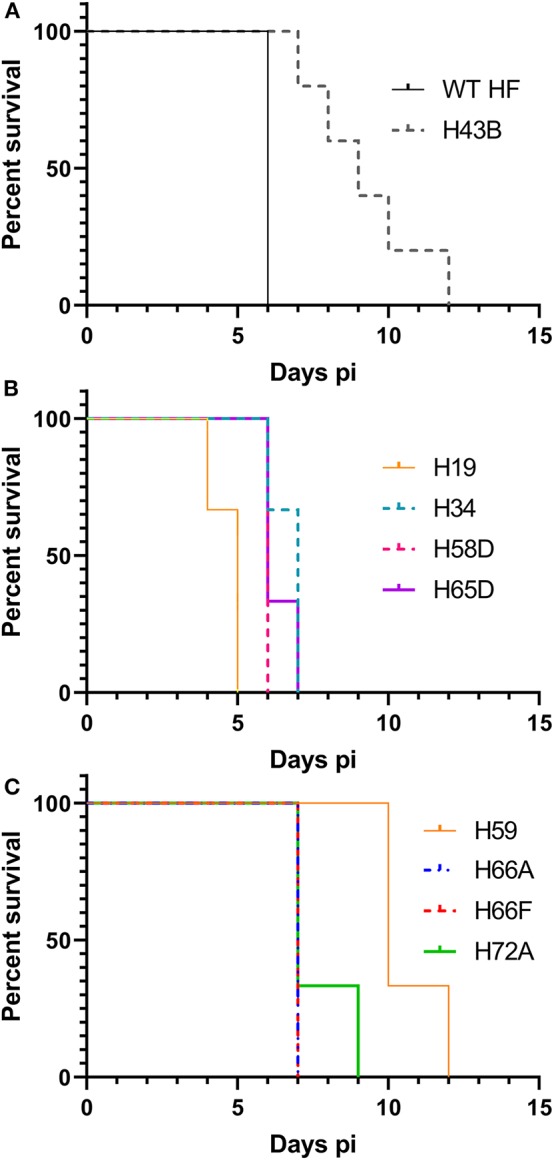
Kaplan-Meier survival curves for mice inoculated with nine cloned mutants. (A) Wild type and H43B. Compared with WT, H43B survived significantly longer, P < 0.05 by the Log-rank test (N = 5). (B,C) Compared to H59, each clone P < 0.05 by the Log-rank test (N = 3). (B,C) Were separated for figure clarity.
Figure 6.
Growth curves of H43B and H59 clones. (A) DH82 cells were infected with WT HF, H43B, and H59 clones. Representative HEME3 staining images were shown at day 2, 3, or 4 post-infection. White arrows, ehrlichial microcolonies (morulae). Scale bar, 10 μm. (B) Growth curve showing WT HF, H43B, and H59 in DH82 cells as determined by qPCR. Infection and growth of H59 and H43B mutants in DH82 cells were similar by repeated-measures ANOVA, P > 0.05 (N = 3).
Figure 7.
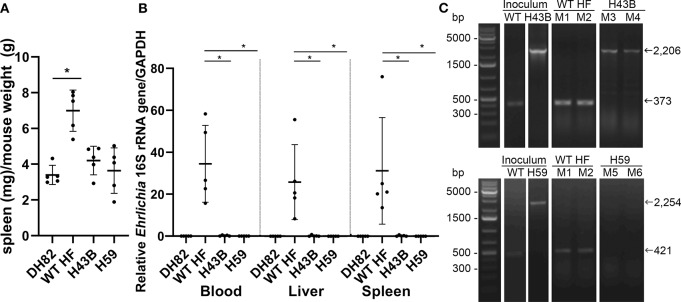
Kaplan-Meier survival curves and flank PCR to detect the clonality of H34B and H59. (A) Kaplan-Meier survival curves for two Himar1 mutant clones, P < 0.001 by Log-rank, Gehan-Breslow-Wilcoxon test (N = 5). (B) Flank PCR showing that WT-, H43B-, and H59-infected DH82 cells (inoculum), and liver tissues of Mouse 1 (M1) and Mouse 2 (M2) inoculated with WT and died at day 10 (WT HF), Mouse 3 (M3) and Mouse 4 (M4) died at day 12 and 17, respectively (H43B, top panels), and Mouse 5 (M5) and Mouse 6 (M6) died at day 14 and 21, respectively (H59, bottom panels). For H43B, EHF_RS04100 primers (Table S1) flanking H43B insertion site were used, showing 373 and 2,206 bp without and with Himar1 insert, respectively. For H59, EHF_0962 primers (Table S1) flanking H59 insertion site were used, showing 421 and 2,254 bp without and with Himar1 insert, respectively.
Bacterial Load and Cytokine Gene Expression in Mice Inoculated With Cloned Mutants
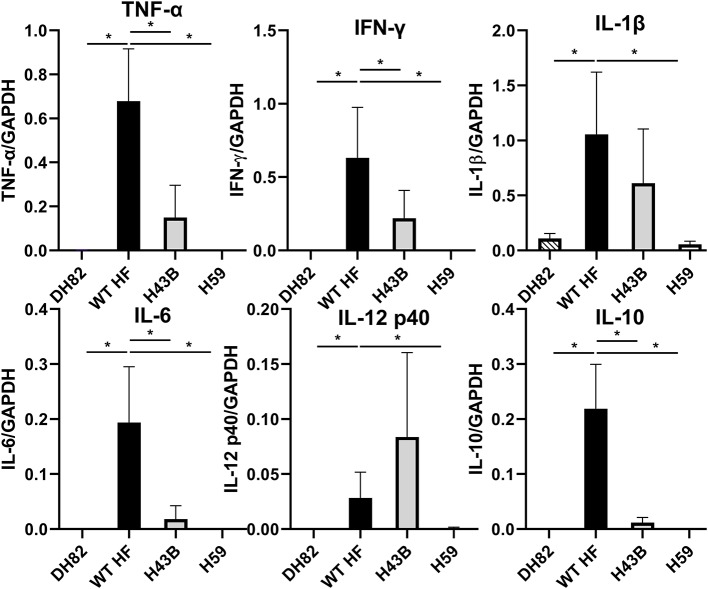
To compare mouse infection and inflammation, mice were inoculated with 600 ~ 3,760 WT, H43B, and H59 bacteria and euthanized at day 7 pi. At day 7 pi, slight splenomegaly was detected with WT, but not with H43B or H59, compared to control mice inoculated with DH82 cells (Figure 8A). In the blood, spleen, and liver, similar levels of bacteria were detected with WT, but dramatically low levels of H43B mutant, and no H59 mutant was detected by qPCR (Figure 8B). Flank PCR showed the clonality of H43B and H59 mutants in DH82 cells prior to mouse inoculation, and the clonality of H43B was confirmed in liver tissues at day 7 pi (Figure 8C). Quantitative reverse transcription PCR (qRT-PCR) results showed that WT HF upregulated proinflammatory cytokines (TNF-α, IL-1β, IFN-γ, IL-6) and immunosuppressive cytokines IL-10 in the liver at day 7 pi (Figure 9). These cytokine mRNAs were undetectable or negligible in the liver of H59-inoculated mice. H43B induced significantly reduced levels of TNF-α, IFN-γ, IL-6, and IL-10 mRNA in the liver compared with WT HF, although IL-1β and IL-12 p40 mRNA levels were similar (Figure 9). H43B mouse inoculation experiment was repeated with ~30-fold more bacteria, but obtained similar results (data not shown).
Figure 8.
Bacterial load in mice inoculated with H59, H43B, and WT HF strains. (A) Spleen index at day 7 pi, *P < 0.05 with ANOVA and Tukey's test (N = 5). (B) Bacterial load in mouse blood, liver, spleen day 7 pi. 16S rRNA gene was normalized to GAPDH, *P < 0.05 using ANOVA (N = 5). (C) Flank PCR using primers described in Figure 7B, showing the clonality of H43B- and H59-infected DH82 cells, and in liver tissues of Mouse 3 (M3) and Mouse 4 (M4) for H34B, and Mouse 5 (M5) and Mouse 6 (M6) for H59, respectively at day 7 pi. Mouse 1 (M1) and mouse 2 (M2) were inoculated with WT HF strain, and liver tissues at day 7 pi were used as controls.
Figure 9.
Cytokine gene expression in livers of mice inoculated with H59, H43B, and WT HF strains. qRT-PCR analysis of differentially expressed cytokine genes in livers of mice inoculated with WT and mutant HF strains at day 7 pi. The data shown are means and standard deviations, with mRNA levels normalized by GAPDH, *P < 0.05 using ANOVA followed by Tukey (N = 5).
Discussion
We have demonstrated that use of the transposon insertional mutagenesis method identified a broad range of 55 ehrlichial genes, which might be unnecessary for this bacterial infection and growth in macrophages. Conservation of these genes among sequenced Ehrlichia species suggest that these genes have important roles in the Ehrlichia life cycle, other than infecting, surviving, and growing in mammalian leukocytes. Nearly 40% of these genes are predicted to encode hypothetical proteins that lack homology to other bacterial genes, suggesting these genes have evolved to serve unique functions of Ehrlichia spp. Among sequenced Ehrlichia spp. these genes had the highest homology with genes from E. muris subsp. eauclairensis, the newest member of human monocytic ehrlichiosis agent discovered in the US. As little is known for pathogenesis of E. muris subsp. eauclairensis, studies on HF strain will help illuminate this bacterium and its pathogenesis.
Despite reductive genome evolution resulting in a budget genome, P44/Msp2/OMP-1/P28/P30 genes were expanded by duplication in Ehrlichia and Anaplasma species (Rikihisa, 2010), and genes encoding VirB/D type IV secretion apparatus are expanded among the order Rickettsiales (Rikihisa, 2017), suggesting these duplications are important for the bacterial life cycle. But not all of these genes are required for in vitro growth in mammalian cell cultures in the absence of in vivo immune pressure. Indeed, our current study and previous publications showed virB6-4, the largest and most downstream gene in the virB6 operon (EHF_0445, APH_0377, MC1_00820) is consistently disrupted by Himar1 transposon without affecting Ehrlichia, Anaplasma, or Rickettsia growth in mammalian host cells (Felsheim et al., 2006; Lamason et al., 2018). Despite extensive arrays of in vitro functions proposed for TRP120 (former gp120) for E. chaffeensis infection of macrophages (Yu et al., 2000; Luo et al., 2011; Zhu et al., 2011; Dunphy et al., 2013, 2014), the gene encoding HF strain TRP120 homolog had the insertion near 5′ end, implicating this gene is dispensable for Ehrlichia HF infection of macrophages. TRP120 appears to be an abundantly produced protein in E. chaffeensis (Yu et al., 2000), TRP mutant of HF obtained in our study would help elucidate in vivo or other functions of TRP120 in the future.
The present study found Himar1 insertions in 55 distinct genes. Felsheim et al. (2006) reported isolation of five intragenic insertions APH_0928, APH_0798/0797, APH_0546, APH_0377, and APH_0584 in Anaplasma phagocytophilum cultured in HL-60 cells. APH_0584 encodes O-methyltransferase and the mutant is unable to grow in ISE6 tick cells (Oliva Chavez et al., 2015). In the present study, all nine cloned mutants, which were obtained through limited dilutions or antibiotic selection in DH82 cell cultures, could infect ISE6 tick cells. Cheng et al. (2013) identified ECH_0379, ECH_0601, and ECH_0660 intragenic insertions. The mutant of ECH_0660 encoding a phage head-tail connector protein was cloned, which was unable to infect deer, and this mutant was used to vaccinate dogs and protect dogs from E. chaffeensis infection by syringe and tick inoculation of E. chaffeensis (Nair et al., 2015; McGill et al., 2016). In the present study, ECH_0379 (encoding a conserved hypothetical protein) homolog, EHF_0332 had an intragenic insertion, corroborating that function of this gene is not required for Ehrlichia spp. infection of macrophages. Lamason et al. (2018) reported Himar1 insertion in 75 distinct genes of Rickettsia parkeri str. Portsmouth (GenBank # NC_017044.1). In the present study, in addition to the VirB6-4 (MC1_00820) ortholog described above, M16 family peptidase (MC1_01650) homolog, EHF_0125, and DNA mismatch repair protein MutS (MC1_02260) homolog, EHF_0723 had insertions, suggesting functions of these genes are universally dispensable for mammalian cell infection by members of the order Rickettsiales. All other mutants were newly detected in the present study. As members of the order Rickettsiales evolutionary share many genes and lifestyle, growing numbers of mutants and their functional characterization, undoubtedly advance our understanding of obligatory intracellular bacteria, their pathogenesis, and adaptation to invertebrate vectors.
There are many virulence factors that were not detected by this method because mutants cannot infect or grow in DH82 cells, or grow too slowly. Of 866 predicted protein-coding open reading frames (ORFs) in the HF genome, over 60% (533) ORFs are house-keeping genes, and knockout mutants of these genes cannot survive in DH82 cell culture systems. Of remaining ORFs, ~60 ORFs of the HF strain are likely required for macrophage infection, as orthologs of E. chaffeensis are essential for infection of macrophages including type IV secretion system apparatus (23 ORFs) and effector proteins (3), two-component regulatory system (6), and outer membrane proteins (porin, lipoproteins, invasin; ~30 ORFs) etc. (Kumagai et al., 2006, 2008; Huang et al., 2008; Mohan Kumar et al., 2013; Rikihisa, 2015, 2017; Lin et al., 2016; Sharma et al., 2017; Teymournejad et al., 2017; Yan et al., 2018). Of the remaining ~330 genes, most encode hypothetical proteins or proteins with unknown functions. Our previous RNASeq data (NCBI GEO accession number GPL18497) (Lin et al., 2013) revealed that over 60 ORFs of HF strain were differentially expressed (>2-fold differences, ~70% of which were hypothetical proteins) between ISE6 and canine DH82 cells, suggesting differential requirement of these gene products in two types of host cells. Of 55 ORFs of Ehrlichia HF strain with Himar1 insertions that can infect macrophages, six ORFs (EHF_0470, EHF_0051, EHF_0962, EHF_0775, EHF_0242, EHF_0623) were upregulated 2- to 5-fold in the HF strain cultured in ISE6 cells than the HF strain cultured in DH82 cells based on the RNASeq data. Thus, these HF strain genes may function in ticks rather in mammals. Based on these information, we expect maximum 330 knockout intragenic insertional mutants can be recovered by using both tick and mammalian cell lines in the future.
This study identifies two mutants, H43B and H59, which have insertions in a hypothetical protein (EHF_RS04100) and a protein containing staphylococcal superantigen-like domain (EHF_0962), respectively, are required for rapid mouse infection and death. Unlike spotted fever group Rickettsia sp. (Lamason et al., 2018), Ehrlichia sp. does not produce distinct plaques (host cell lysis), therefore, the limited dilution procedure is required to obtain the clonal population of Ehrlichia sp. We established a procedure to verify clonality at every step of cloning and in vitro and in vivo experiments in this study. We could not, however, deny the possibility that reduced growth of H43B and H59 in mice is due to secondary mutation as complementation experiments are not feasible at this time. However, studies with recombinant proteins, and/or anti-sense protein nucleic acid (Sharma et al., 2017; Yan et al., 2018) will potentially verify these results in the future.
Previously fatal murine ehrlichiosis due to HF strain (derived from infected mouse spleen homogenate) infection is associated with T cell-mediated tissue damage, high levels of serum TNF-α and IL-10, and CD4-Th1 hyporesponsiveness (Stevenson et al., 2008), which are associated with liver pathology and failure to clear HF strain in mice (Ismail et al., 2004). In the present study, we also found cultured WT HF induces strong TNF-α and IL-10 mRNA as well as other proinflammatory cytokines in the liver, which perhaps killed mice; however, reduced levels of these cytokines helped mice inoculated with H43B or H59 lived longer. Although IFN-γ is critical in reducing bacterial growth in macrophages (Barnewall and Rikihisa, 1994), reduced infection of these two mutants in mice did not correlate with IFN-γ mRNA levels. Furthermore, mRNA levels of IL-12 p40 which stimulates NK cells and T helper cells to produce IFN-γ (Hodge et al., 2002) did not correlate with slow growth of mutants in the mice. Interplay of bacteria and host immune system is complex, but liver cytokine response of WT and mutant HF strain suggests hepatitis in ehrlichiosis is related to bacterial load in the liver. Despite of delayed growth of H34B and H59 in mice, they eventually proliferated and killed mice at above certain dosages. Ehrlichia HF is, therefore, expected to have additional virulence factors that eventually overcome the mouse immune defense.
Hence, more Ehrlichia mouse virulence-related genes are waiting to be discovered. The immunocompetent animal model lends itself to the discovery of genes for in vivo virulence but not for infection or growth in macrophage in cell culture, and could be important for differentiating between liver and non-liver virulence mechanisms, which is of major benefit in the absence of accurate in vitro measures of the liver and hematopoietic disease caused by Ehrlichia. Finally, there are virulence factors that we could not have detected in an animal model. These include factors important for survival in ticks, multiplication within ticks and subsequent spread to humans.
Materials and Methods
Ethics Statement
All animal experiments were performed in accordance with the Ohio State University Institutional Animal Care and Use Committee guidelines and approved protocol. The university program has full continued accreditation by the Association for Assessment and Accreditation of Laboratory Animal Care International under 000028, dated 9 June 2000, and has Public Health Services assurance renewal A3261-01, dated 6 February 2019–28 February 2023. The program is licensed by the USDA, number 31-R-014, and is in full compliance with Animal Welfare Regulations.
Ehrlichia sp. HF Culture and Purification
Canine histiocytic leukemia DH82 cells were cultured in DMEM (Dulbecco minimal essential medium; Mediatech, Manassas, VA) supplemented with 5% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 2 mM l-glutamine (l-Gln; GIBCO, Waltham, MA) at 37°C under 5% CO2 in a humidified atmosphere as described previously (Rikihisa et al., 1991). Ehrlichia sp. HF (Shibata et al., 2000) was cultured in DH82 cells with the addition of 0.1 μg/mL cycloheximide (Millipore Sigma, Burlington, MA) at the same condition. The ISE6 cell line, derived from the Ixodes scapularis tick embryo, was cultured in L15C300 medium at 34°C as described previously (Munderloh et al., 1999). To assess the degree of bacterial infection in host cells, a drop of infected cells was centrifuged onto a slide in a Shandon Cytospin 4 cytocentrifuge (Thermo Fisher, Waltham, MA), then fixed and stained with HEMA 3 staining solutions (Thermo Fisher).
For transformation, confluent 90% Ehrlichia-infected DH82 cells were harvested from three T25 flasks. Host cell–free bacteria were purified by sonication on ice for 8 s twice at output setting 3 using a fine tip on a W380 Sonicator (Heat Systems, Newtown, CT). Lysed cells were centrifuged at 700 × g (Sorvall 6000D, Thermo Fisher) to remove unbroken cells and nuclei, filtered sequentially through 5.0- and 2.7-μm nylon syringe filters (Millipore, Billerica, MA), and centrifuged in 40 mL polycarbonate high speed centrifuge tubes (Nalgene Nunc International Corporation, Rochester, NY) at 10,000 × g (Sorvall RC 5C Plus using SS-34 rotor) to pellet host cell–free bacteria as described previously (Lin et al., 2016).
pCis-mCherry-SS-Himar A7 Plasmid Purification and Transformation of Ehrlichia sp. HF
pCis-mCherry-SS-Himar A7 plasmid (Cheng et al., 2013) was used for transformation of Ehrlichia sp. HF. The plasmids were purified from overnight transformed dam−/dcm− E. coli cultures (200 ml) using EndoFree plasmid purification Maxi kit according to the manufacturer's instructions (Qiagen, Germantown, MD). Freshly purified host cell-free bacteria were washed twice with 1 ml ice-cold 0.3 M sucrose and resuspended in 85 μl of 0.3 M sucrose. Bacteria were mixed with 6–8 μg plasmid, transferred to a 2-mm gap electroporation cuvette (Bio-Rad, Hercules, CA), and incubated on ice for 15 min (Yan et al., 2018). Bacteria were electroporated at 2,500 V, 25 μF, and 400 Ω using a Gene Pulser Xcell™ Electroporation System (Bio-Rad). Transformed HF were cultured in 6-well plate with a confluent monolayer of DH82 cells in 1 mL DMEM supplemented with 30% FBS, 4 mM l-Gln, and 10 mM sodium pyruvate (Corning, Corning, NY), incubated at 30°C for 16–20 h to enhance aad expression, and then transferred to a 37°C incubator. After 2 days, transformed HF expressing aad were selected in the presence of 100 μg/ml spectinomycin, 100 μg/ml streptomycin, and 0.1 μg/ml cycloheximide, and the culture medium containing antibiotics was replaced twice a week until % infected cells reached above 10% (~2 weeks).
Image Acquisition and Analysis by DeltaVision Microscopy
DH82 cells infected with Ehrlichia sp. HF transformed with pCis mCherry-SS Himar A7 plasmids were cytocentrifuged onto a glass slide and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) for 20 min at room temperature. After washing the cells with PBS, glass coverslips were mounted on the slide with 70% glycerol mounting medium (Thermo Fisher), and sealed by a nail polish. Fluorescence images with overlay differential interference contrast (DIC) images were captured using a DeltaVision PersonalDV Deconvolution microscope system using TRITC filter sets (GE Healthcare, Marlborough, MA).
Determine Genomic Insertion Loci Using Semi-random Nested PCR, and Bioinformatics Analysis
The genomic loci of the Himar1 insertion sites were determined by semi-random nested PCR as previously described (Cheng et al., 2013), using primers listed in Table S1. Briefly, DNA was purified from Ehrlichia sp. HF mutants using QIAamp DNA blood mini kit according to the manufacturer's instructions (Qiagen). The first-step PCR used transposon-specific outer primers P1 or P5 (Table S1) and a semi-conserved degenerate primer P2, which has a defined adapter sequence at the 5′-end that pairs with the universal primer P4 and a random sequence near the 3′-end to allow for random annealing in Ehrlichia chromosome. The first PCR reaction yielded a longer product that served as the template in the second-step PCR reaction using transposon-specific inner primers (P3 or P6) and a second universal primer (P4) that anneals to the 5′-end of primer P2, allowing for specific amplification of the longer PCR product. Residual primers, dNTPs, and enzymes were removed from the first-step PCR products using GeneJET DNA purification kit (Thermo Fisher). The final PCR products were resolved by 1% agarose gel electrophoresis, and the specific DNA bands were purified from the gel using a QIAquick Gel Extraction Kit (Qiagen), and sequenced using transposon-specific primers P3 or P6. The resulting sequences were compared with Ehrlichia sp. HF genome sequence (NCBI GenBank accession number: NZ_CP007474.1) by BLASTN (Altschul et al., 1990) to map the transposon insertion sites on the genome. These insertion loci were mapped to the circular Ehrlichia sp. HF genome using GenomeViz software (Ghai et al., 2004).
NCBI conserved domain search was used to identify potential function of targeted proteins, and NCBI BLASTP was used to identify homology within the family Anaplasmataceae and other related bacteria. SignalP was used to predict signal peptides and membrane localization (Bendtsen et al., 2004).
Cloning Mutants and Clonality Confirmation
DH82 cells suspended at 104 cells in 50 μL DMEM culture medium (supplemented with 5% FBS, 2 mM l-Gln, and 0.1 μg/mL cycloheximide) were seeded into each wells of a 96 well flat-bottomed plate (Greiner, Monroe, NC). Approximately 3 Ehrlichia sp. HF mutant-infected DH82 cells suspended in 50 μL culture medium were inoculated into each well. After overnight incubation, 100 μL of additional culture medium was added to each well and cells were allowed to grow to confluency. Then monolayers were scraped with sterile pipette tips and ~104 cells in 20 μL were transferred to a new 96-well flat, clear-bottom black plate (Tecan, Morrisville, NC). After cells reached confluency at ~4 days, mCherry fluorescence was detected using a Tecan Infinite M Nano+ microplate reader (Tecan). Positive wells were expanded and checked for single insert clones using PCR analysis with primers flanking specifically to the target insertion sites (Table S1). Primers were designed using the NCBI primer-BLAST (Ye et al., 2012).
DNA Purification and Quantification of Ehrlichia Bacterial Numbers
DNA was purified from uninfected and Ehrlichia sp. HF-infected DH82 cells and mouse tissues with a QIAamp DNA Mini Kit (Qiagen). To quantify Ehrlichia, an absolute quantification method was used by creating a standard curve of the Ehrlichia 16S rDNA cloned into a pUC19 plasmid as a standard template (Teymournejad et al., 2017), and bacterial numbers were determined by 16S rDNA copy numbers by qPCR analysis according to the manufacturer's protocol (Stratagene, Waltham, MA) (Liu et al., 2012). The qPCR mixture (20 μl) included 0.25 μM each primer, and 10 μl of SYBR green qPCR master mix (Thermo Fisher). Primer sequences for the Ehrlichia 16S rRNA gene are shown in Table S1. PCR was performed in the Mx3000P instrument (Stratagene). The copy number of the targeted gene in DNA samples was calculated by comparing the Ct value with the standard curve. The value was normalized against mouse GAPDH levels using specific primers (Table S1).
qRT-PCR Analysis
WT and mutant-infected mouse liver were preserved in 0.5 cm thickness in RNAlater reagent (Thermo Fisher). Total cellular RNA was extracted from 30 mg tissue per sample using RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA concentrations and quality were determined by NanoDrop (Thermo Fisher). Total RNA (0.5 μg) was reverse transcribed using a Maxima H Minus First Strand cDNA Synthesis Kit and oligo dT primers (Thermo Fisher). The qPCR reaction mixture (20 μl) included 5 μl cDNA (corresponding to 0.1 μg of total RNA), 0.25 μM each primer, and 10 μl SYBR Green qPCR master mix (Thermo Fisher). PCR was performed in an Mx3000P instrument. Primer sequences are described in Table S1.
In vitro Growth Curve
To generate growth curves of WT, H43B, and H59 mutants of Ehrlichia sp. HF in DH82 cell cultures, bacteria were isolated by sonication without filtration as described above, and centrifuged at 10,000 × g for 5 min at 4°C. Supernatant was discarded, and host cell-free bacteria were resuspended in DMEM supplemented with 5% FBS, 2 mM l-Gln, and 0.1 μg/mL cycloheximide, and inoculated on DH82 monolayers in 5 wells/group of 12-well plates. Monolayers were washed three times with media at 2 h pi to remove unbound bacteria. To assess bacterial growth, monolayers were washed three times with PBS, and cells were scraped off in 1 ml PBS. DNA was extracted at each day pi for 4 days, and bacteria numbers were determined by qPCR and normalized by total DNA amount, which corresponds to total cell numbers. One sample of WT and mutant cultures were evaluated by Hema3 staining each day (Oliva Chavez et al., 2015).
Mouse Infection
In each experiment, infected DH82 cells were resuspended in DMEM medium without FBS and antibiotics, and 0.5 mL per mouse were intraperitoneally inoculated using a 1 mL TB syringe with a 26-gauge needle (BD, Franklin Lakes, NJ). Mice were monitored daily for weight loss and moribund condition indicated by hunched posture, squinted eyes, slow movement, reduced response to stimulation, and loss of 20% body weight according to IACUC. Moribund mice were euthanized by CO2 inhalation and cervical dislocation. As bacteria need to be inoculated into mice immediately after harvesting, bacterial numbers were first estimated by counting percent infected cells and approximate numbers of bacteria per cell, and verified by qPCR later as described above. Four sets of experiments were carried out. (1) For LD50 determination, female ICR (CD-1) at 4 weeks old (Envigo, Indianapolis, IN) were intraperitoneally inoculated in groups of three with serially diluted 100, 10, 1, and 0.1 DH82 cells containing ~2,500, 250, 25, and 2.5 bacteria, respectively. (2) For screening mutants that lost virulence, male and female C57BL/6 mice at 8–20 weeks old (Envigo) in groups of three to five mice each were intraperitoneally inoculated with 5–15 × 106 Ehrlichia sp. HF mutant bacteria from infected DH82 cells. (3) For analysis of mortality of mice infected with H43B and H59, female ICR mice at 10 weeks old (Envigo) in groups of five mice each were intraperitoneally inoculated with 200 infected DH82 cells containing 42,500 WT, 28,400 H43B, or 42,700 H59 bacteria, respectively. Tissue samples were frozen for use in DNA extraction. (4) For H43B and H59 bacterial burden and cytokine analysis, female ICR mice at 6 weeks old (Envigo) in groups of five mice each were intraperitoneally inoculated with 200 infected cells containing 1,330 WT, 600 H43B, and 3,760 H59 bacteria. Mice were euthanized on day 7 pi. Cardiac puncture blood samples were collected and tissue samples were stored in RNALater (Qiagen) for RNA or DNA extraction.
Statistical Analysis
Statistical analysis was performed with analysis of variance (ANOVA) followed by Tukey multiple comparison, and with repeated measures analysis of variance (rANOVA). P < 0.05 was considered statistically significant. Kaplan-Meier survival curves were analyzed by Log-rank, Gehan-Breslow-Wilcoxon test or the Log-rank test. All statistical analyses were performed using Prism 8 software (GraphPad, La Jolla, CA).
Data Availability Statement
Genome sequences for Ehrlichia sp. HF (GenBank accession NZ_CP007474) and transcriptome data (GEO accession number GPL18497) were deposited at NCBI.
Ethics Statement
The animal study was reviewed and approved by the Ohio State University Institutional Animal Care and Use Committee.
Author Contributions
YR and ML contributed the conception and design of the study. HB, ML, and OT performed the experiments and analysis. YR, ML, and HB wrote the manuscript. All authors read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ulrike Munderloh at the University of Minnesota for providing plasmid pCis-mCherry-SS-Himar A7, and ISE6 tick cells.
Footnotes
Funding. This project was partially funded by R01 AI121124 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services. The whole genome sequencing was partially funded from NIH under contract number HHSN272200900009C. HB was partially supported by C. Glenn Barber Fund from College of Veterinary Medicine, The Ohio State University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00002/full#supplementary-material
References
- AAT Bioquest I. (2020). Quest Graph™ LD50 Calculator. Available online at: https://www.aatbio.com/tools/ld50-calculator (accessed May 31, 2019).
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anderson B. E., Dawson J. E., Jones D. C., Wilson K. H. (1991). Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29, 2838–2842. 10.1128/JCM.29.12.2838-2842.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby M. N., Edwards P. A. (1990). Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 265, 13157–13164. [PubMed] [Google Scholar]
- Ashour J., Hondalus M. K. (2003). Phenotypic mutants of the intracellular actinomycete Rhodococcus equi created by in vivo Himar1 transposon mutagenesis. J. Bacteriol. 185, 2644–2652. 10.1128/JB.185.8.2644-2652.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnewall R. E., Rikihisa Y. (1994). Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect. Immun. 62, 4804–4810. 10.1128/IAI.62.11.4804-4810.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., Von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Bhagavan N. V., Ha C.-E. (2015). Nucleotide metabolism, in Essentials of Medical Biochemistry, 2nd Edn., eds Bhagavan N. V., Ha C.-E. (Cambridge, MA: Academic Press; ), 465–487. 10.1016/B978-0-12-416687-5.00025-7 [DOI] [Google Scholar]
- Buller R. S., Arens M., Hmiel S. P., Paddock C. D., Sumner J. W., Rikhisa Y., et al. (1999). Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N. Engl. J. Med. 341, 148–155. 10.1056/NEJM199907153410303 [DOI] [PubMed] [Google Scholar]
- Cartman S. T., Minton N. P. (2010). A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76, 1103–1109. 10.1128/AEM.02525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2018). RE: National Notifiable Diseases Surveillance System, 2017 Annual Tables of Infectious Disease Data. Atlanta, GA: CDC Division of Health Informatics and Surveillance. [Google Scholar]
- Cheng C., Nair A. D., Indukuri V. V., Gong S., Felsheim R. F., Jaworski D., et al. (2013). Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 9:e1003171. 10.1371/annotation/322c8bba-8dae-4196-b85d-5de315e44d76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Miura K., Popov V. L., Kumagai Y., Rikihisa Y. (2011). Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol. Microbiol. 82, 1217–1234. 10.1111/j.1365-2958.2011.07885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun K. T., Edenberg H. J., Kelley M. R., Goebl M. G. (1997). Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13, 233–240. [DOI] [PubMed] [Google Scholar]
- Davidson W. R., Lockhart J. M., Stallknecht D. E., Howerth E. W., Dawson J. E., Rechav Y. (2001). Persistent Ehrlichia chaffeensis infection in white-tailed deer. J. Wildl. Dis. 37, 538–546. 10.7589/0090-3558-37.3.538 [DOI] [PubMed] [Google Scholar]
- Dawson J. E., Ewing S. A. (1992). Susceptibility of dogs to infection with Ehrlichia chaffeensis, causative agent of human ehrlichiosis. Am. J. Vet. Res. 53, 1322–1327. [PubMed] [Google Scholar]
- Dunphy P. S., Luo T., Mcbride J. W. (2013). Ehrlichia moonlighting effectors and interkingdom interactions with the mononuclear phagocyte. Microbes Infect. 15, 1005–1016. 10.1016/j.micinf.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy P. S., Luo T., Mcbride J. W. (2014). Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect. Immun. 82, 4154–4168. 10.1128/IAI.01984-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim R. F., Herron M. J., Nelson C. M., Burkhardt N. Y., Barbet A. F., Kurtti T. J., et al. (2006). Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 6:42. 10.1186/1472-6750-6-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. D., Proft T. (2008). The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225, 226–243. 10.1111/j.1600-065X.2008.00681.x [DOI] [PubMed] [Google Scholar]
- Fujita H., Watanabe Y. (1994). Ehrlichial organisms isolated from Ixodes ovatus ticks and field rodents in Japan. Ann. Rep. Ohara Hosp. 37, 13–17. [Google Scholar]
- Ghai R., Hain T., Chakraborty T. (2004). GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. 10.1186/1471-2105-5-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. J., Phan I. Q., Driscoll T. P., Guillotte M. L., Lehman S. S., Rennoll-Bankert K. E., et al. (2016). The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog. Dis. 74:ftw058. 10.1093/femspd/ftw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett-Emmett D., Tashian R. E. (1996). Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Mol. Phylogenet. Evol. 5, 50–77. 10.1006/mpev.1996.0006 [DOI] [PubMed] [Google Scholar]
- Hodge D. L., Martinez A., Julias J. G., Taylor L. S., Young H. A. (2002). Regulation of nuclear gamma interferon gene expression by interleukin 12 (IL-12) and IL-2 represents a novel form of posttranscriptional control. Mol. Cell. Biol. 22, 1742–1753. 10.1128/MCB.22.6.1742-1753.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Lin M., Wang X., Kikuchi T., Mottaz H., Norbeck A., et al. (2008). Proteomic analysis of and immune responses to Ehrlichia chaffeensis lipoproteins. Infect. Immun. 76, 3405–3414. 10.1128/IAI.00056-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N., Soong L., Mcbride J. W., Valbuena G., Olano J. P., Feng H. M., et al. (2004). Overproduction of TNF-alpha by CD8+ type 1 cells and down-regulation of IFN-gamma production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J. Immunol. 172, 1786–1800. 10.4049/jimmunol.172.3.1786 [DOI] [PubMed] [Google Scholar]
- Khani A., Popp N., Kreikemeyer B., Patenge N. (2018). A glycine riboswitch in streptococcus pyogenes controls expression of a sodium:alanine symporter family protein gene. Front. Microbiol. 9:200. 10.3389/fmicb.2018.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y. S., Koo J. E., Biswas A., Kobayashi K. S. (2010). MyD88-dependent signaling contributes to host defense against ehrlichial infection. PLoS ONE 5:e11758. 10.1371/journal.pone.0011758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Cheng Z., Lin M., Rikihisa Y. (2006). Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74, 5014–5022. 10.1128/IAI.00735-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y., Huang H., Rikihisa Y. (2008). Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J. Bacteriol. 190, 3597–3605. 10.1128/JB.02017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R. L., Kafai N. M., Welch M. D. (2018). A streamlined method for transposon mutagenesis of Rickettsia parkeri yields numerous mutations that impact infection. PLoS ONE 13:e0197012. 10.1371/journal.pone.0197012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe D. J., Akerley B. J., Rubin E. J., Mekalanos J. J., Robertson H. M. (1999). Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U.S.A. 96, 11428–11433. 10.1073/pnas.96.20.11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhorn B. G., Gerdes S. Y., Begley T. P. (2004). A genetic screen for the identification of thiamin metabolic genes. J. Biol. Chem. 279, 43555–43559. 10.1074/jbc.M404284200 [DOI] [PubMed] [Google Scholar]
- Le Breton Y., Mohapatra N. P., Haldenwang W. G. (2006). In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72, 327–333. 10.1128/AEM.72.1.327-333.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Daugherty S. C., Parankush S., Kumar N., Cheng Z., Xiong Q., et al. (2013). Sequencing and comparison of genomes of Ehrlichia strains and transcriptome profiles in mammalian host and tick cells, in ASM 113th General Meeting (Denver, CO: American Society for Microbiology; ). [Google Scholar]
- Lin M., Kikuchi T., Brewer H. M., Norbeck A. D., Rikihisa Y. (2011). Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front. Microbiol. 2:24. 10.3389/fmicb.2011.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Liu H., Xiong Q., Niu H., Cheng Z., Yamamoto A., et al. (2016). Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12, 2145–2166. 10.1080/15548627.2016.1217369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Bao W., Lin M., Niu H., Rikihisa Y. (2012). Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell. Microbiol. 14, 1037–1050. 10.1111/j.1462-5822.2012.01775.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. M., Tucker A. M., Driskell L. O., Wood D. O. (2007). Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 73, 6644–6649. 10.1128/AEM.01727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T., Kuriakose J. A., Zhu B., Wakeel A., Mcbride J. W. (2011). Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect. Immun. 79, 4382–4391. 10.1128/IAI.05608-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. (2012). The ParA/MinD family puts things in their place. Trends Microbiol. 20, 411–418. 10.1016/j.tim.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour C. A., Doiron K. M., Cupples C. G. (2001). Characterization of functional interactions among the Escherichia coli mismatch repair proteins using a bacterial two-hybrid assay. Mutat. Res. 485, 331–338. 10.1016/S0921-8777(01)00071-4 [DOI] [PubMed] [Google Scholar]
- Martinez M. C., Gutierrez C. N., Monger F., Ruiz J., Watts A., Mijares V. M., et al. (2008). Ehrlichia chaffeensis in child, Venezuela. Emerg. Infect. Dis. 14, 519–520. 10.3201/eid1403.061304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure E. E., Chavez A. S. O., Shaw D. K., Carlyon J. A., Ganta R. R., Noh S. M., et al. (2017). Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat. Rev. Microbiol. 15, 544–558. 10.1038/nrmicro.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill J. L., Nair A. D., Cheng C., Rusk R. A., Jaworski D. C., Ganta R. R. (2016). Vaccination with an attenuated mutant of Ehrlichia chaffeensis induces pathogen-specific CD4+ T cell immunity and protection from tick-transmitted wild-type challenge in the canine host. PLoS ONE 11:e0148229. 10.1371/journal.pone.0148229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W., Li H., Li H., Liu C., Liu J. (2015). Characterization of aspartate kinase from Corynebacterium pekinense and the critical site of Arg169. Int. J. Mol. Sci. 16, 28270–28284. 10.3390/ijms161226098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Matsuo J., Rahman M. A., Kumagai Y., Li X., Rikihisa Y. (2011). Ehrlichia chaffeensis induces monocyte inflammatory responses through MyD88, ERK, and NF-kappaB but Not through TRIF, interleukin-1 receptor 1 (IL-1R1)/IL-18R1, or toll-like receptors. Infect. Immun. 79, 4947–4956. 10.1128/IAI.05640-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan Kumar D., Yamaguchi M., Miura K., Lin M., Los M., Coy J. F., et al. (2013). Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog. 9:e1003666. 10.1371/journal.ppat.1003666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh U. G F. R, Burkhardt N., Herron M., Oliva Chavez A., Nelson C., Kurtti T. (2012). Intracellular pathogens II: Rickettsiales, in The Way Forward: Improving Genetic Systems, ed Palmer Gh A. A. A. (New York, NY: ASM Press; ), 271–272. 10.1128/9781555817336.ch14 [DOI] [Google Scholar]
- Munderloh U. G., Jauron S. D., Fingerle V., Leitritz L., Hayes S. F., Hautman J. M., et al. (1999). Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37, 2518–2524. 10.1128/JCM.37.8.2518-2524.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A. D., Cheng C., Jaworski D. C., Ganta S., Sanderson M. W., Ganta R. R. (2015). Attenuated mutants of Ehrlichia chaffeensis induce protection against wild-type infection challenge in the reservoir host and in an incidental host. Infect. Immun. 83, 2827–2835. 10.1128/IAI.00487-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W., Vasiliou V. (2004). Analysis of the glutathione S-transferase (GST) gene family. Hum. Genomics 1, 460–464. 10.1186/1479-7364-1-6-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N., Rikihisa Y., Unver A. (2001). Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69, 2083–2091. 10.1128/IAI.69.4.2083-2091.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N., Unver A., Zhi N., Rikihisa Y. (1998a). Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J. Clin. Microbiol. 36, 2671–2680. 10.1128/JCM.36.9.2671-2680.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N., Zhi N., Zhang Y., Rikihisa Y. (1998b). Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66, 132–139. 10.1128/IAI.66.1.132-139.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Tajima T., Kawahara M., Rikihisa Y. (2001). Ehrlichial proliferation and acute hepatocellular necrosis in immunocompetent mice experimentally infected with the HF strain of Ehrlichia, closely related to Ehrlichia chaffeensis. J. Comp. Pathol. 124, 165–171. 10.1053/jcpa.2000.0447 [DOI] [PubMed] [Google Scholar]
- Okada H., Usuda H., Tajima T., Kawahara M., Yoshino T., Rikihisa Y. (2003). Distribution of ehrlichiae in tissues as determined by in-situ hybridization. J. Comp. Pathol. 128, 182–187. 10.1053/jcpa.2002.0624 [DOI] [PubMed] [Google Scholar]
- Oliva Chavez A. S., Fairman J. W., Felsheim R. F., Nelson C. M., Herron M. J., Higgins L., et al. (2015). An O-methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum. PLoS Pathog. 11:e1005248. 10.1371/journal.ppat.1005248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A. J., Buoncristiani M. R., Howard P. K., Flamm J., Johnson C., Yamamoto R., et al. (1988). The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide-sequence of the bio operon. J. Biol. Chem. 263, 19577–19585. [PubMed] [Google Scholar]
- Paddock C. D., Childs J. E. (2003). Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64. 10.1128/CMR.16.1.37-64.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock C. D., Sumner J. W., Shore G. M., Bartley D. C., Elie R. C., Mcquade J. G., et al. (1997). Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J. Clin. Microbiol. 35, 2496–2502. 10.1128/JCM.35.10.2496-2502.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicic V., Morelle S., Lampe D., Nassif X. (2000). Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182, 5391–5398. 10.1128/JB.182.19.5391-5398.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M., Bodor M., Zhang C., Xiong Q., Rikihisa Y. (2006). Human infection with Ehrlichia canis accompanied by clinical signs in Venezuela. Ann. N. Y. Acad. Sci. 1078, 110–117. 10.1196/annals.1374.016 [DOI] [PubMed] [Google Scholar]
- Perez M., Rikihisa Y., Wen B. (1996). Ehrlichia canis-like agent isolated from a man in Venezuela: antigenic and genetic characterization. J. Clin. Microbiol. 34, 2133–2139. 10.1128/JCM.34.9.2133-2139.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B. S., Allerdice M. E. J., Sloan L. M., Paddock C. D., Munderloh U. G., Rikihisa Y., et al. (2017). Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int. J. Syst. Evol. Microbiol. 67, 2121–2126. 10.1099/ijsem.0.001896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B. S., Sloan L. M., Johnson D. K., Munderloh U. G., Paskewitz S. M., Mcelroy K. M., et al. (2011). Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N. Engl. J. Med. 365, 422–429. 10.1056/NEJMoa1010493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistgaard E. M., Low C., Guettou F., Nordlund P. (2016). Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol. 17, 123–132. 10.1038/nrm.2015.25 [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. (2010). Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8, 328–339. 10.1038/nrmicro2318 [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. (2015). Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu. Rev. Microbiol. 69, 283–304. 10.1146/annurev-micro-091014-104411 [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. (2017). Role and function of the type IV secretion system in Anaplasma and Ehrlichia species. Curr. Top. Microbiol. Immunol. 413, 297–321. 10.1007/978-3-319-75241-9_12 [DOI] [PubMed] [Google Scholar]
- Rikihisa Y., Stills H., Zimmerman G. (1991). Isolation and continuous culture of Neorickettsia helminthoeca in a macrophage cell line. J. Clin. Microbiol. 29, 1928–1933. 10.1128/JCM.29.9.1928-1933.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E. J., Akerley B. J., Novik V. N., Lampe D. J., Husson R. N., Mekalanos J. J. (1999). In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 96, 1645–1650. 10.1073/pnas.96.4.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. M., Freire P., Vicente M., Arraiano C. M. (1999). The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 32, 789–798. 10.1046/j.1365-2958.1999.01397.x [DOI] [PubMed] [Google Scholar]
- Shao L., Abdel-Motaal H., Chen J., Chen H., Xu T., Meng L., et al. (2018). Characterization of a functionally unknown arginine-aspartate-aspartate family protein from Halobacillus andaensis and functional analysis of its conserved arginine/aspartate residues. Front. Microbiol. 9:807. 10.3389/fmicb.2018.00807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Teymournejad O., Rikihisa Y. (2017). Peptide nucleic acid knockdown and intra-host cell complementation of Ehrlichia type IV secretion system effector. Front. Cell. Infect. Microbiol. 7:228. 10.3389/fcimb.2017.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S., Kawahara M., Rikihisa Y., Fujita H., Watanabe Y., Suto C., et al. (2000). New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J. Clin. Microbiol. 38, 1331–1338. 10.1128/JCM.38.4.1331-1338.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska M. M., Chiang J. H., Luther W. M., Stewart J. L., Amii L., Conrad A., et al. (2000). Genes involved in the determination of the rate of inversions at short inverted repeats. Genes Cells 5, 425–437. 10.1046/j.1365-2443.2000.00341.x [DOI] [PubMed] [Google Scholar]
- Small E., Marrington R., Rodger A., Scott D. J., Sloan K., Roper D., et al. (2007). FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J. Mol. Biol. 369, 210–221. 10.1016/j.jmb.2007.03.025 [DOI] [PubMed] [Google Scholar]
- Stevenson H. L., Crossley E. C., Thirumalapura N., Walker D. H., Ismail N. (2008). Regulatory roles of CD1d-restricted NKT cells in the induction of toxic shock-like syndrome in an animal model of fatal ehrlichiosis. Infect. Immun. 76, 1434–1444. 10.1128/IAI.01242-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teymournejad O., Lin M., Rikihisa Y. (2017). Ehrlichia chaffeensis and its invasin EtpE block reactive oxygen species generation by macrophages in a DNase X-dependent manner. MBio 8:e01551-17. 10.1128/mBio.01551-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents E., Grinberg I., Gorovitz-Harris B., Lundstrom H., Borovok I., Aharonowitz Y., et al. (2007). NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 189, 5012–5021. 10.1128/JB.00440-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkovicova L., Smidak R., Jung G., Turna J., Lubec G., Aradska J. (2016). Proteomic analysis of the TerC interactome: novel links to tellurite resistance and pathogenicity. J. Proteomics 136, 167–173. 10.1016/j.jprot.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Unver A., Rikihisa Y., Stich R. W., Ohashi N., Felek S. (2002). The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts. Infect. Immun. 70, 4701–4704. 10.1128/IAI.70.8.4701-4704.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. H., Dumler J. S. (1996). Emergence of the ehrlichioses as human health problems. Emerg. Infect. Dis. 2, 18–29. 10.3201/eid0201.960102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt M. D., Allan J. M., Lau A. Y., Ellenberger T. E., Samson L. D. (1999). 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays 21, 668–676. [DOI] [PubMed] [Google Scholar]
- Yan Q., Lin M., Huang W., Teymournejad O., Johnson J. M., Hays F. A., et al. (2018). Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation. Proc. Natl. Acad. Sci. U.S.A. 115, E8977–E8986. 10.1073/pnas.1806904115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Ghose P., Ismail N. (2013). Neutrophils mediate immunopathology and negatively regulate protective immune responses during fatal bacterial infection-induced toxic shock. Infect. Immun. 81, 1751–1763. 10.1128/IAI.01409-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. J., Mcbride J. W., Diaz C. M., Walker D. H. (2000). Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Kuriakose J. A., Luo T., Ballesteros E., Gupta S., Fofanov Y., et al. (2011). Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect. Immun. 79, 4370–4381. 10.1128/IAI.05422-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences for Ehrlichia sp. HF (GenBank accession NZ_CP007474) and transcriptome data (GEO accession number GPL18497) were deposited at NCBI.