Abstract
Recurrent gain-of-function mutations in the transcription factors STAT5A and much more in STAT5B were found in hematopoietic malignancies with the highest proportion in mature T- and natural killer-cell neoplasms (peripheral T-cell lymphoma, PTCL). No targeted therapy exists for these heterogeneous and often aggressive diseases. Given the shortage of models for PTCL, we mimicked graded STAT5A or STAT5B activity by expressing hyperactive Stat5a or STAT5B variants at low or high levels in the hematopoietic system of transgenic mice. Only mice with high activity levels developed a lethal disease resembling human PTCL. Neoplasia displayed massive expansion of CD8+ T cells and destructive organ infiltration. T cells were cytokine-hypersensitive with activated memory CD8+ T-lymphocyte characteristics. Histopathology and mRNA expression profiles revealed close correlation with distinct subtypes of PTCL. Pronounced STAT5 expression and activity in samples from patients with different subsets underline the relevance of JAK/STAT as a therapeutic target. JAK inhibitors or a selective STAT5 SH2 domain inhibitor induced cell death and ruxolitinib blocked T-cell neoplasia in vivo. We conclude that enhanced STAT5A or STAT5B action both drive PTCL development, defining both STAT5 molecules as targets for therapeutic intervention.
Introduction
Peripheral (mature) T-cell lymphomas (PTCL) are heterogeneous neoplasms often accompanied by aggressive courses and extranodal organ infiltration. PTCL have variable histology, immunophenotype, and molecular features.1,2 The World Health Organization (WHO) classification of lymphoid neoplasms distinguishes more than 30 mature T- and natural killer (NK)-cell neoplasms. The most common subtype is PTCL, not otherwise specified (NOS), which collects together cases not attributable to other, better defined, entities. PTCL, NOS is a highly dynamic category with respect to consensus features, proposed cell of origin, and prognostic subsets based on molecular signatures.3 PTCL, NOS with a follicular T-helper (TFH) cell phenotype relates to angioimmunoblastic T-cell lymphoma (AITL), with which it is co-categorized in a provisional group.4 Another distinguishable PTCL, NOS subset is constituted by cases with cytotoxic features. High-throughput methodologies helped in such histogenetic assignments and in the prognostically relevant separation of non-TFH type PTCL, NOS from AITL and anaplastic large cell lymphoma.2,3,5–7 Eminent problems, particularly in PTCL, NOS, are the inefficacy of the poly-chemotherapies historically designed for aggressive B-cell lymphomas and the lack of high-fidelity mouse models8–10 in which to investigate biological principles and to address preclinical questions.
The molecular landscape of PTCL, NOS reveals that altered T-cell receptor signaling, epigenetic modifiers, and immune evasion mechanisms are common.3,5–7,11–17 Activating mutations in the JAK-STAT pathway affecting mostly the interleukin-2 receptor (IL2R), JAK1, JAK3, STAT3, and STAT5B were found in many mature T- and NK-cell neoplasms.18,19 The entities with the highest incidence of STAT5B and STAT3 mutations are anaplastic large cell lymphoma, cutaneous T-cell lymphoma (CTCL; comprising mycosis fungoides and Sézary syndrome), enteropathy-associated T-cell lymphoma, hepatosplenic T-cell lymphoma, NK/T-cell lymphoma, T-cell prolymphocytic leukemia, and the auto-aggressive CD8+ T-large granular lymphocyte leukemia.15,20–22 Furthermore, mutations in chromatin remodelers, GTPases, DNA repair machinery or co-repressors have been associated with JAK/STAT hyperactivation.19
STAT5BN642H is the most frequent recurrent gain-of-function mutation in the closely related genes encoding for the transcription factors STAT5A and STAT5B. It is associated with unfavorable disease progression in patients15,23–31 and leads to an aggressive CD8+ T-cell neoplasia in mice.32 JAK-STAT signaling is a central cancer pathway driving survival and cell cycle progression, but it also promotes differentiation and senescence as safety pathways. STAT5A and STAT5B play important roles in immune cells33 and absence of lymphoid STAT5 results in loss of CD8+, gδ, and regulatory T cells (Treg)34 Differentiation of CD8+ T cells is regulated by STAT5 in a dose-dependent manner35 and enhances effector and memory CD8+ T-cell survival and proliferation. High levels of tyrosine phos-phorylated STAT5 (pYSTAT5) are associated with a negative prognosis in many myeloid neoplasms.36
Aggressive CD8+ T-cell neoplasia resulted in early death upon STAT5BN642H expression.32 Enhanced pYSTAT5 can also be mimicked by the hyperactive Stat5aS710F variant (cS5F).37 We generated and compared graded STAT5 activity mouse models within the hematopoietic system. Low activity models displayed only a modest CD8+ T-cell expansion, whereas those with high STAT5 activity developed aggressive CD8+ PTCL-like disease reminiscent of human PTCL, NOS with cytotoxic features. Although STAT5A- and STAT5B-induced changes largely overlap, STAT5B hyperactivation was more aggressive than STAT5A hyperactivation. Comparative analyses revealed that STAT5A and STAT5B overexpression is common in human mature T-cell lymphomas. The clinical JAK1/2/3 inhibitors ruxolitinib and tofacitinib38 as well as a selective STAT5 inhibitor39 specifically reduced viability of PTCL cells. Ruxolitinib blocked PTCL disease in vivo. We conclude that STAT5 activation drives PTCL and that patients with PTCL can benefit from JAK/STAT inhibitors.
Methods
Animals and generation of transgenic mice
Mice were maintained on a C57BL/6N background, housed in a specific-pathogen-free facility under standardized conditions and monitored daily for signs of disease. All animal experiments were carried out according to the animal license protocols (BMWF-66.009/0281-I/3b/2012, BMWFW-68.205/0166-WF/V/3b/2015, BMWFW-68.205/0117-WF/V/3b/2016 and BMWFW-68.205/0103-WF/V/3b/2015) approved by the institutional Ethics Committee and the Austrian Ministry BMWF authorities. All transgenic mice were hemizygous. Non-transgenic littermates served as controls. We used the vav-hematopoietic vector vav-hCD4 (HS21/45)40 to generate transgenic mice expressing cS5F in the hematopoietic system at different levels (called cS5Alo [B6N-Tg(Vav-cS5F)564Biat] and cS5Ahi [B6N-Tg(Vav-cS5F)565Biat]), as described in the Online Supplementary Methods. Details of hSTAT5B and hSTAT5BN642H mice have been published.32 All primers used are listed in Online Supplementary Table S1.
Patients’ samples
Retrospective immunohistological analysis, approved by the ethics committee of the Medical University of Vienna (1437/2016), was done on formalin-fixed, paraffin-embedded patients’ specimens of 35 PTCL, NOS, 14 AITL, 6 mycosis fungoides, and 29 CTCL (from 23 patients) cases and 5 non-diseased lymph nodes, kindly provided by the Medical University of Vienna, Austria, the Karl Landsteiner University of Health Sciences, St. Poelten, Austria, Wilheminenspital (Wiener Krankenanstaltenverbund), Vienna, Austria, and the University Hospital Brno, Czech Republic. Samples were included in this study after patients had given informed consent in accordance with the Declaration of Helsinki. Diagnoses of samples were made according to the 2008 WHO criteria by experienced hematopathologists or expert dermatopathologists. Patients with CTCL were diagnosed according to the WHO-EORTC classification for cutaneous lymphomas as follows: mycosis fungoides (stage IA: n=10, stage IB: n=1, stage IIA: n=1, stage IIB: n=6), Sézary syndrome (n=2), and lymphomatoid papulosis (n=3).
Histological analysis of murine and human sections
Formalin-fixed, paraffin-embedded 3 mm consecutive mouse organ sections were stained with hematoxylin (Merck, Darmstadt, Germany) and eosin G (Carl Roth, Karlsruhe, Germany). Immunohistochemistry was performed using antibodies against CD3, Ki67, pYSTAT5, STAT5A and STAT5B (Online Supplementary Table S4, Online Supplementary Methods). For immunohistochemical analysis of STAT5A and STAT5B expression in human samples, tissue microarrays were built including 14 AITL, 35 PTCL, NOS, 7 CTCL, and 6 mycosis fungoides cases each represented by duplicate or triplicate core biopsies. In addition, paraffin-preserved sections of 29 CTCL samples (from 23 patients) were analyzed. Fivc non-neoplastic lymph nodes served as controls. Quantification of the staining is described in the Online Supplementary Methods. Images were taken with a Zeiss Imager Z1 microscope (Carl Zeiss, Oberkochen, Germany).
RNA sequencing
mRNA was isolated from CD8+ T cells harvested from lymph nodes [wildtype (wt) - 2-3 mice pooled per sample, cS5Alo, cS5Ahi, hSTAT5BN642H n=5, hSTAT5B n=4]. RNA sequencing was performed with Illumina HiSeq-2500 (Illumina, San Diego, CA, USA) at the VBCF next-generation sequencing unit (www.vbcf.ac.at). Details of the analysis are provided in the Online Supplementary Methods.
RNA-sequencing data can be found in the GEO database with the accession identities GSE124102 and GSE93847.
Statistical analysis
Data are reported as mean values ± standard error of mean and were analyzed by GraphPad Prism® 5 (San Diego, CA, USA) or RStudio Version 1.0.153 (Boston, MA, USA). In vitro data, western blots, quantitative reverse transcriptase polymerase chain reactions (qRT-PCR) and viability assays were repeated at least three times (unless indicated otherwise). The numbers of animals or patients are stated in each figure or figure legend. Applied statistical tests are mentioned in the respective figure legend. P values <0.05 were accepted as statistically significant and denoted as follows: *P<0.05; **P<0.01; ***P<0.001, and ****P<0.0001.
Results
STAT5A or STAT5B activation leads to a mature CD8+ T-cell disease in mice
To test whether hyperactive STAT5 signaling alone is sufficient to drive PTCL, we generated transgenic mice with graded expression of wildtype (wt) STAT5B or gain-of-function Stat5a or STAT5B. We used the well-characterized hyperactive Stat5aS710F (cS5F) variant37 expressed at low or high level, human (h)STAT5B served as a negative control and hSTAT5BN642H served as a positive T-cell neoplastic model.32 All transgenes contain a C-terminal FLAG-tag driven under control of the vav-promoter enabling expression from the hematopoietic stem cell stage onwards throughout all blood lineages40 (hereafter referred to as cS5AF, cS5Alo, cS5Ahi, hSTAT5B and hSTAT5BN642H) (Figure 1A, Online Supplementary Figure S1A-D). We confirmed significantly elevated pYSTAT5 levels in hematopoietic organs of cS5Ahi and hSTAT5BN642H mice whereas the cS5Alo animals showed a modest increase in pYSTAT5 (Figure 1B, Online Supplementary Figure S1E). Kidney, colon and liver did not display significant transgene expression (Online Supplementary Figure S1E). Transgene expression was verified by qRT-PCR in FACS-sorted CD4+, CD8+, CD19+, CD11b+ and NK cells of 8-week old cS5Alo and cS5Ahi mice, with CD8+ T cells showing highest transgene mRNA expression (Online Supplementary Figure S1F).
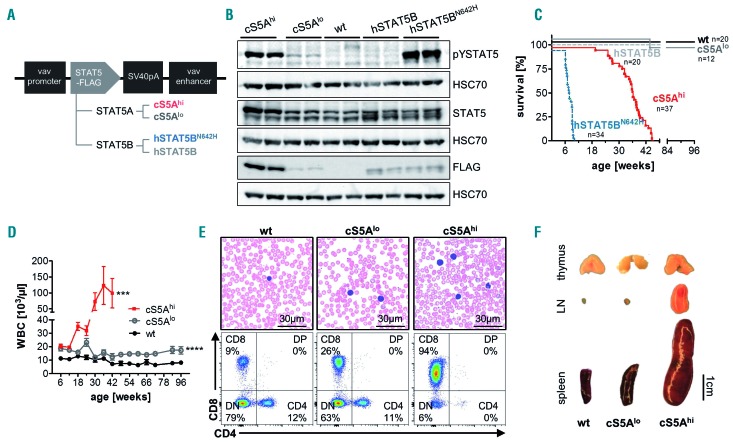
Figure 1.
Expression of a gain-of-function Stat5a or STAT5B variant leads to a polyclonal CD8+ T-cell disease. (A) Schematic representation of the FLAG-tagged STAT5 constructs for generation of transgenic mouse lines expressing hyperactive Stat5a (cS5Alo and cS5Ahi) or human STAT5B (hSTAT5B and hSTAT5BN642H). (B) Immunoblot on lymph node lysates from cS5Ahi, cS5Alo, wildtype (wt), hSTAT5B, and hSTAT5BN642H mice (n=2/genotype) using antibodies to FLAG, phosphotyrosine(Y694)-STAT5 (pYSTAT5) and STAT5. HSC70 was used as a loading control. Representative blot of four experiments. (C) Kaplan-Meier disease-free survival plot of wt (n=20), cS5Alo (n=12), cS5Ahi (n=37), hSTAT5B (n=20) and hSTAT5BN642H (n=34) mice; P≤0.0001 with the log-rank (Mantel-Cox) test. (D) White blood cell (WBC) count measured at 6-week intervals from wt (n≥6), cS5Alo (n≥8) and cS5Ahi (n≥10) mice for 66 weeks (cS5Ahi until 42 weeks, unpaired t-test wt vs. cS5Alo P≤0.0001, wt vs. cS5Ahi P=0.003). (E) Representative blood smears of 32-week old mice, scale bar 30 mm, and representative flow cytometry dot plots of CD4 and CD8 cells in peripheral blood of 32-week old wt, cS5Alo and cS5Ahi mice. (F) Macroscopic appearance of thymi, lymph nodes (LN), and spleens of representative age-matched wt, cS5Alo and diseased cS5Ahi mice.
Enhanced STAT5 activation in the hematopoietic system led to development of neoplasia resulting in death between 25 to 45 weeks of age irrespective of gender in cS5Ahi mice, which was significantly later than in hSTAT5BN642H mice which develop a lethal disease within 10 weeks.32 cS5Alo were as long-lived as wt mice; hSTAT5B mice were also followed for more than 1 year without showing signs of disease (Figure 1C). The strong lymphoma phenotype of hSTAT5BN642H mice suggests a role of STAT5A as a possible balancer due to STAT5A/B heterodimerization, but only high pYSTAT5 levels drive the disease.
White blood cell counts rose steadily in cS5Ahi mice to a level comparable to that in diseased hSTAT5BN642H mice, whereas cS5Alo exerted only a subtle effect, resulting in doubling of the white cell count compared to that in wt mice (Figure 1D, Online Supplementary Figure S1H), also seen in the blood smears (Figure 1E). Like hSTAT5BN642H mice, adult cS5Ahi massively expanded CD8+ T cells and the phenotype in cS5Alo mice was intermediate (Figure 1E, Online Supplementary Figure S1G, H). At 32 weeks of age, cS5Ahi mice had massively enlarged lymph nodes at all lymphatic sites and severe splenomegaly, but cS5Alo mice did not show any pathological abnormalities. Interestingly, macroscopically, STAT5A transgenic thymi showed no obvious differences from those of age-matched controls (Figure 1F). Our further analysis focused on the comparison of cS5Ahi mice and their wt littermates.
Expanded CD8+ T cells exert a mature cytotoxic T-lymphocyte phenotype
At the age of 25-45 weeks all cS5Ahi mice developed terminal disease. CD8+ T cells were dominant in spleen (Figure 2A, Online Supplementary Figure S2A), peripheral blood, lymph nodes and bone marrow of cS5Ahi mice (Online Supplementary Figure S2B). While relative levels of CD4+ T, CD19+ B and CD11b+Gr1hi myeloid cells were decreased (Online Supplementary Figure S2A-C), absolute numbers of these cell types were elevated, although to a lesser extent than CD8+ T cells (Figure 2A). In contrast, both the relative and absolute numbers of NK cells were decreased (Figure 2A, Online Supplementary Figure S2A). Western blot analysis of lymphocyte subpopulations confirmed the highest levels of cS5AF expression in CD8+ T cells (Online Supplementary Figure S2D). The cS5Alo mice did not display a phenotype despite increased CD8+ T-cell numbers and pronounced pYSTAT5 levels (Online Supplementary Figure S2E-I).
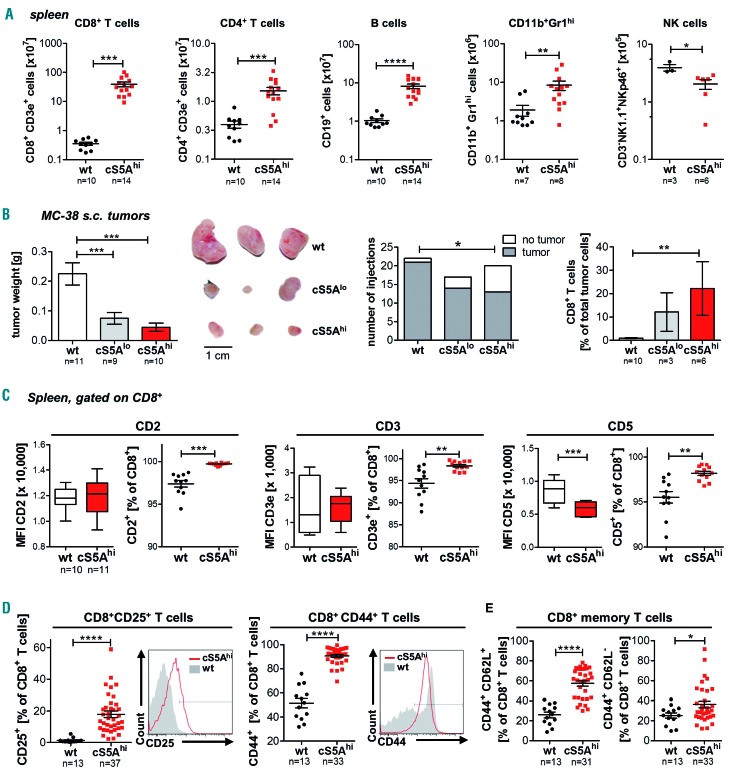
Figure 2.
High STAT5A activation promotes CD8+ T-cell neoplasia. (A) Flow cytometric analysis of splenocytes of diseased cS5Ahi mice and wildtype (wt) littermates showing absolute CD8+ T-cell (P=0.0001), CD4+ T-cell (P=0.0002), B-cell (P≤0.0001, all unpaired t-test with the Welch correction), CD11b+Gr1hi (P=0.0014) and natural killer (NK)-cell numbers (P=0.0237, both unpaired t-test) (B) Left: tumor weight of subcutaneous (s.c.) MC-38 tumors 18 days after injection of 1x106 cells in both flanks of 10-week old wt (n=11), cS5Alo (n=9) and cS5Ahi (n=10) mice (one-way analysis of variance with the Tukey multiple comparison test). Middle: macroscopic view of isolated MC-38 tumors, scale bar represents 1 cm. Right: tumor incidence per injection of MC-38 cells (logistic regression, P=0.031) and percentage of CD8+ T-cell tumor infiltrating cells (Kruskal-Wallis test with the Dunn multiple comparison test). (C) Flow cytometric analysis of CD2, CD3 and CD5 expression on CD8+ wt (n=10) and cS5Ahi (n=11) splenocytes. Mean fluorescent intensity (MFI) unpaired t-test (CD2 P=0.85, CD3 P=0.91, CD5 P=0.0002), relative expression unpaired t-test with the Welch correction (CD2 P=0.0001, CD3 P=0.0044, CD5 P=0.0021). (D) Flow cytometric characterization of splenic wt (n=13) and cS5Ahi (n≥31) CD8+ T cells: CD25 (left, P<0.0001, unpaired t-test) and CD44 expression (right, P<0.0001, unpaired t-test with the Welch correction) with representative histograms and (E) CD44+CD62L+ (left, P<0.0001, unpaired t-test with the Welch correction) and CD44+CD62L- expression (right, P=0.0492, unpaired t-test).
To assess the functionality of the cytotoxic T cells (CTL), we injected the C57BL/6 isogenic CTL-responsive lymphoma cell line E.G7 or the colon carcinoma cell line MC-38 into flanks of 10-week old wt, cS5Alo or cS5Ahi mice. In comparison to tumors in wt hosts, tumors in cS5Alo and cS5Ahi mice were significantly smaller or absent and infiltrated more by CD8+ T cells, indicating that the expanded CD8+ T cells retained functionality (Figure 2B, Online Supplementary Figure S2J).
Next, we sought to further characterize the disease-causing cells. cS5Ahi CD8+ T cells retained CD2, CD3 and CD5 expression (Figure 2C, Online Supplementary Figure S2K). CD25+ and CD44+ CD8+ T cells were expanded in peripheral blood and hematopoietic organs of cS5Ahi animals (Figure 2D, Online Supplementary Figure S2L, M), indicative of an activated/memory-like T-cell phenotype.41 CD8+ T cells can be divided into effector (TEM) and central memory (TCM) subsets, either representing a rapid effector cell or exerting lymph node-homing properties with potent proliferative potential.42,43 cS5Ahi mice displayed elevated levels of CD44+CD62L+CD8+ (TCM) and CD44+CD62L−CD8+ (TEM) T cells (Figure 2E, Online Supplementary Figure S2N, O). Gene set enrichment analysis on differentially expressed genes in CD8+ T cells of cS5Ahi compared to cS5Alo mice also correlated to a memory/effector signature (Online Supplementary Figure S2P).44 The homing and activation marker CCR7 was also expressed on a subpopulation; CD8+CD62L−CD27−CCR7+ cells in particular were more frequent in cS5Ahi than in wt cases (Online Supplementary Figure S2Q, R). Furthermore, hyperactive STAT5A signaling led to more Treg and gδ T cells (Online Supplementary Figure S2S, T).
Together, these data indicate that hyperactivation of STAT5A in cS5Ahi transgenic mice induces mature T-cell neoplasia with an activated cytotoxic CD8+ memory phenotype.
STAT5-driven CD8+ T cells infiltrate organs
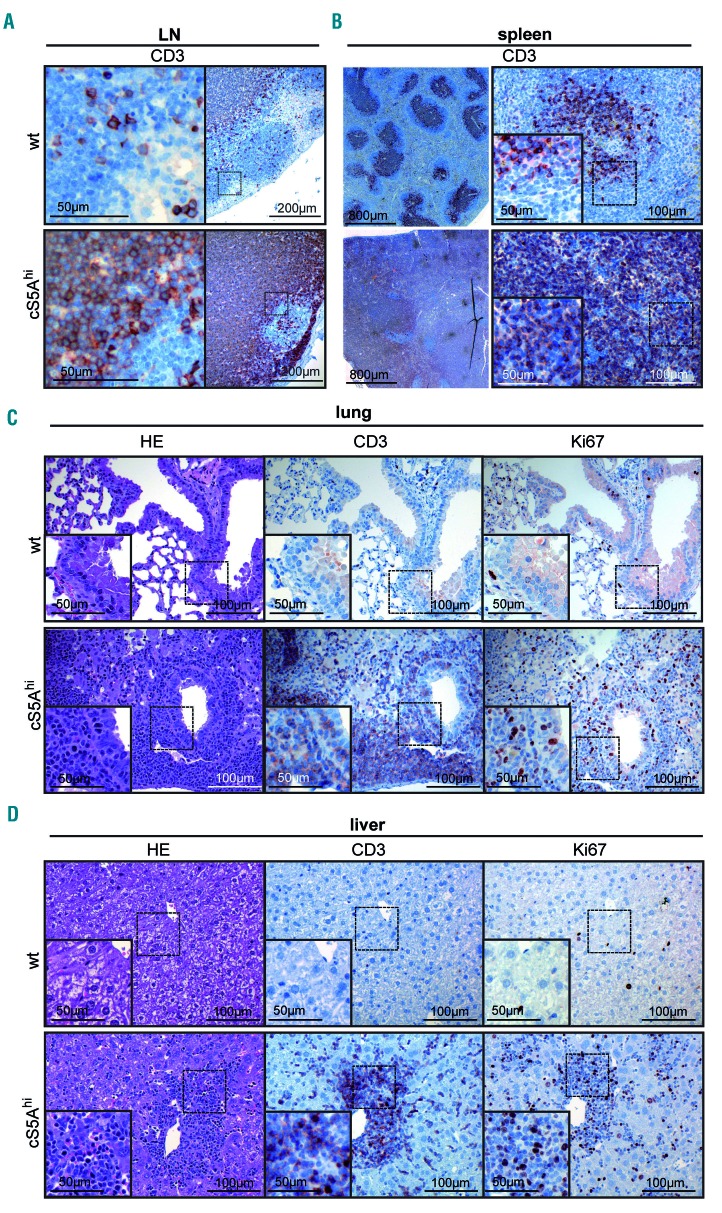
Enlarged lymph nodes, splenomegaly and infiltration of T cells into organs such as the skin, liver, lung and bone marrow are hallmarks in human PTCL. Histological analysis of tissues from cS5Ahi mice revealed disrupted lymph nodes and spleen architecture with dense infiltration of CD3+ T cells and increased proliferation (Ki67+) within lymphomas (Figure 3A, B, Online Supplementary Figure S3A). With regards to skin pathology, diseased mice displayed a thickened dermis with diffuse infiltration of CD3+ T cells (Online Supplementary Figure S3B, C). Moreover, peribronchial and interstitial T-cell infiltrations were detected in lungs of cS5Ahi mice (Figure 3C). Hepatic T-cell infiltration in portal tracts and sinusoids as well as diffuse perivascular and interstitial renal infiltrates were prominent (Figure 3D, Online Supplementary Figure S3D). Higher numbers of Ki67+ cells indicated active proliferation of infiltrating T cells (Figure 3C, D, Online Supplementary Figure S3B, D, quantification summarized in Online Supplementary Figure S3E).
Figure 3.
Expanded CD8+ T cells infiltrate peripheral organs. (A) CD3 staining of a representative enlarged lymph node (LN) of a diseased cS5Ahi mouse and age-matched wildtype (wt) mouse LN. (B) CD3 staining on consecutive cuts of representative spleens of wt controls and diseased cS5Ahi mice. (C, D) Hematoxylin & eosin (HE) (left), CD3 (middle) and Ki67 (right) staining of consecutive sections of lung (C) and liver (D) of diseased cS5Ahi and age-matched wt mice. (A-D) Scale bars indicate 50, 100, 200, or 800 mm.
Transferred cS5Ahi CD8+ T cells induce peripheral T-cell lymphoma in non-irradiated recipients
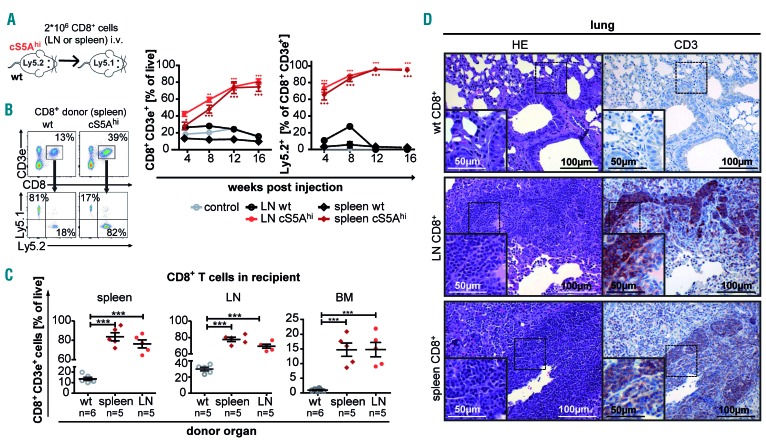
To establish the disease-initiating potency of the mature and differentiated cS5Ahi-CD8+ T cells, we transferred CD8+ Ly5.2+/CD45.2+ T cells from lymph nodes or spleens of diseased cS5Ahi transgenic mice into non-irradiated immunocompetent Ly5.1+/CD45.1+ recipient mice (Figure 4A). Recipients of cS5Ahi CD8+ T cells showed an increase in white blood cell count starting from 8 weeks after transplantation due to an expansion of the Ly5.2+ cS5Ahi-CD8+ T cells in contrast to wt-derived CD8+ T cells (Figure 4B, Online Supplementary Figure S4A). Recipients of cS5Ahi-CD8+ T cells had splenomegaly (Online Supplementary Figure S4B) and high levels of donor-derived cS5Ahi-CD8+ T cells in organs (Figure 4C, D, Online Supplementary Figure S4C, D). We conclude that cS5Ahi-CD8+ T cells can expand and rapidly infiltrate multiple organs in wt recipients. This indicates that high pYSTAT5 in CD8+ T cells induces a transplantable disease.
Figure 4.
cS5Ahi CD8+ T-cell transfer recapitulates an aggressive T-cell lymphoma/leukemia phenotype. (A) Scheme of CD8+ T-cell transfer. LN: lymph node; wt: wildtype; i.v.: intravenous. (B) Representative flow cytometry dot plots of recipient’s peripheral blood 12 weeks after injection showing gating on CD3e+ CD8+ T cells and further Ly5.1/2 gating (left). CD8+ T cells and %Ly5.2+ donor-derived CD8+ T cells (right) in recipient’s peripheral blood measured after injection at 4-week intervals for 16 weeks (n=6/organ source, untreated control n=2, wt LN n=1, P<0.0001). (C) Endpoint analysis: percentage of CD8+ T cells in spleen (left), LN (middle) and bone marrow (BM) (right) of control (wt-CD8+ recipients), spleen- and LN-derived cS5Ahi-CD8+ T-cell recipients (P<0.0001). Two-way analysis of variance (ANOVA) with the Bonferroni post test (B) and one-way ANOVA with the Tukey multiple comparison test. (D) Hematoxylin & eosin (HE) and anti-CD3 staining of representative consecutive sections from lungs of wt and cS5Ahi-CD8+ T-cell recipients. Scale bars indicate 50 or 100 mm.
cS5Ahi- or hSTAT5BN642H-dependent gene expression profiles are highly correlated to human peripheral T-cell lymphoma
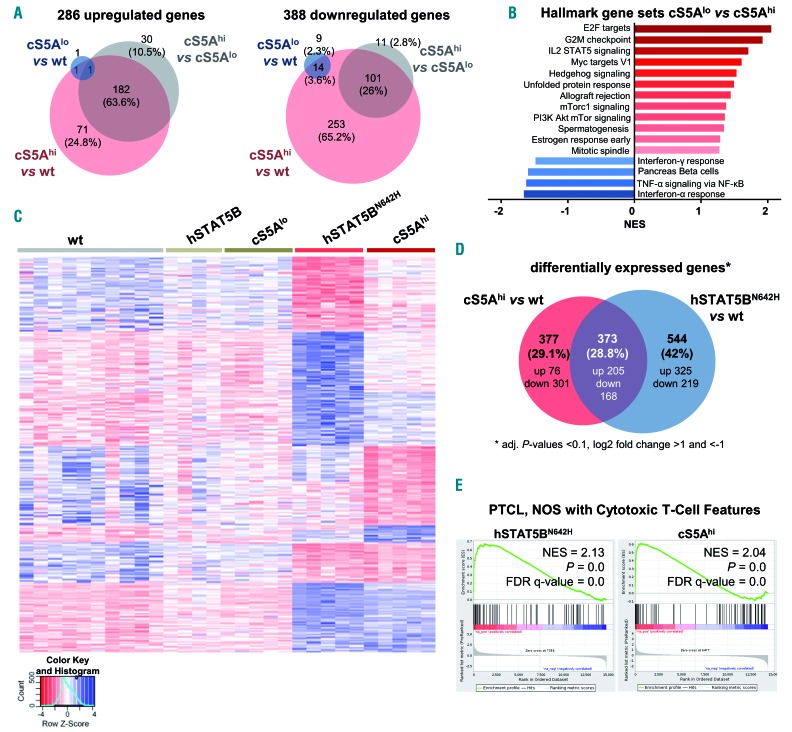
Next, we performed RNA-sequencing and subsequent gene set enrichment analysis with CD8+ T cells from 15-week old wt, cS5Alo and cS5Ahi mice to identify cS5AF- dependent changes in global gene expression patterns. Only very few genes were deregulated in cS5Alo CD8+ T cells, reflecting mild changes. Comparing up- or down-regulated genes in cS5Ahi CD8+ T cells to wt and cS5Alo, there were 182 commonly up- (63.6%) and 101 (26%) commonly down-regulated genes. In addition, 71 (24.8%) or 30 genes (10.5%) were specifically upregulated in cS5Ahi vs. wt or cS5Ahi vs. cS5Alo cells, respectively. Most downregulated genes (253 genes, 65.2%) were seen in cS5Ahi compared to wt mice, whereas only 11 genes (2.8%) showed lowered expression in cS5Ahi compared to cS5Alo animals (Figure 5A, Online Supplementary Figure S5A). We confirmed differential expression of well-described STAT5-target genes Pim1, Bcl2, Bcl6 and Cish by qRT-PCR (Online Supplementary Figure S5B). Gene set enrichment analysis on genes significantly up- or down-regulated in wt vs. cS5Ahi or cS5Alo vs. cS5Ahi mice confirmed the IL-2-STAT5 signaling axis and revealed enrichment of E2F and Myc targets and G2M checkpoint genes as well as a lowered interferon (IFN) response in STAT5 hyperactive mice (Figure 5B, Online Supplementary Figure S5C). This matches the described STAT5-IFN axis in transformation.45 Stat5a and Stat5b share very similar roles in T cells.46 However, sequencing efforts attribute an important role to the activating STAT5BN642H variant.28,32 To compare the phenotypically largely overlapping, though much more aggressive, disease of hSTAT5BN642H and cS5Ahi mice, we contrasted gene expression patterns of wt, cS5Alo, cS5Ahi, hSTAT5B and hSTAT5BN642H CD8+ T cells (Figure 5C, Online Supplementary Figure S5D, RNA-sequencing of hSTAT5B and hSTAT5BN642H as published32). The hSTAT5B and cS5Alo expression profiles cluster with that of wt T cells (Online Supplementary Figure S5D), whereby hyperactive STAT5A and STAT5B signaling share 373 (28.8%) commonly deregulated genes (Figure 5D). Importantly, both CD8+ T-cell neoplasia models are enriched for genes reported to be altered in PTCL, NOS with cytotoxic T-cell features6,7 (Figure 5E and Online Supplementary Table S5), the closest match being to deregulated cS5Ahi genes in the tested T-cell lymphoma gene sets (Online Supplementary Figure S5E). Enrichr pathway analysis47 of the shared cS5Ahi and hSTAT5BN642H gene expression signature revealed an upregulation of cytokine-cytokine receptor interaction and JAK/STAT signaling. The exclusive hSTAT5BN642H genes can be attributed to enhanced cell cycle and division, which may explain its sensitivity to Aurora Kinase inhibition32 (Online Supplementary Figure S5F, Online Supplementary Table S6).
Figure 5.
Transcriptional profiling reveals close correlation to human peripheral T-cell lymphoma. (A) CD8+ T-cell RNA-sequencing (performed with Illumina HiSeq2500) analysis showing the number of significantly (left) up- and (right) down-regulated genes in wildtype (wt), cS5Alo and cS5Ahi CD8+ T cells obtained from lymph nodes (n=5/genotype; adjusted P-values <0.1, log2 fold change >1 and <-1). (B) Summary of gene set enrichment analysis (GSEA) of hallmark gene sets enriched in cS5Ahi vs. cS5Alo CD8+ T cells [false discovery rate (FDR) ≤0.25, adjusted P-value ≤0.05)]. (C) RNA isolated from CD8+ T cells of wt (n=10), hSTAT5B (n=4), hSTAT5BN642H, cS5Alo and cS5Ahi (all n=5) mice were subjected to RNA-sequencing. Heatmap of genes deregulated in a comparison of all genotypes to wt controls (n=1,055) clustered for up- and down-regulated genes specific to individual conditions, as well as for genes shared between hSTAT5BN642H and cS5Ahi. Scaled, rlog transformed normalized counts from DESeq2 were used for the analysis. (D) Venn diagram of differentially expressed genes in cS5Ahi vs. wt and hSTAT5BN642H vs. wt CD8+ T cells (adjusted P-values <0.1, log2 fold change >1 and <-1). (E) GSEA of STAT5BN642H and cS5Ahi expression data shows a correlation to peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS) with cytotoxic T-cell features. NES: normalized enrichment score. The gene set was compiled from literature.6,7
We conclude that the immunophenotype, pathology and gene signatures of the cS5Ahi-induced PTCL-like disease overlap with those of human PTCL, NOS with cytotoxic T-cell features, which is associated with a particularly poor prognosis. hSTAT5BN642H correlates similarly but is more aggressive. This implies that a significant threshold of STAT5 activity is not only required to induce, but is also sufficient to promote PTCL development.
Elevated STAT5 activation in human mature T-cell lymphomas
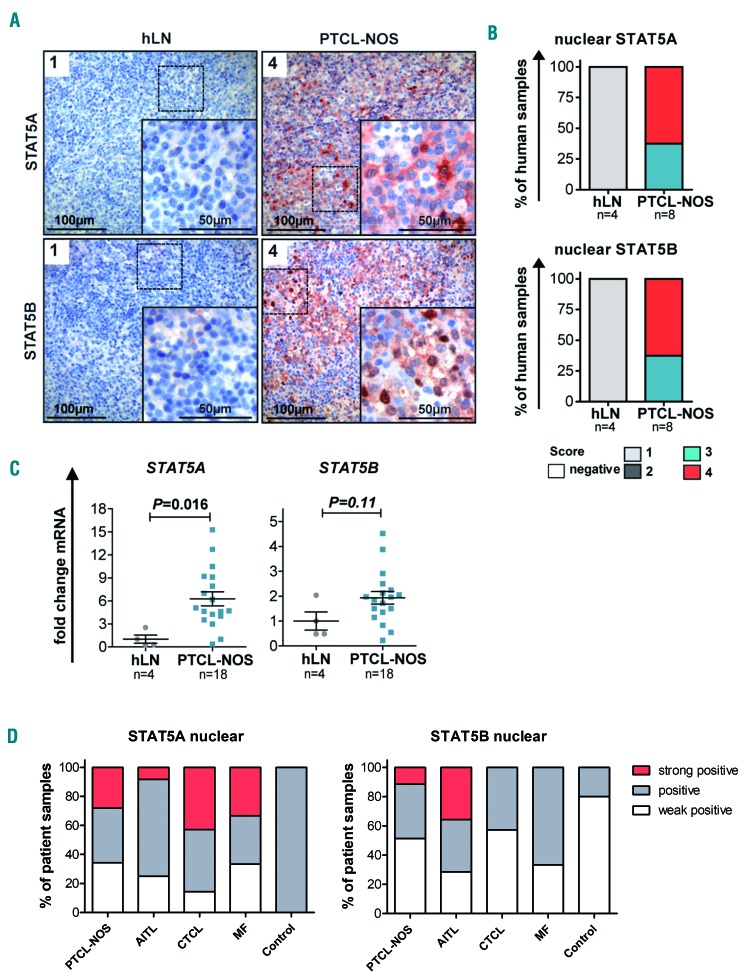
Subsequently, we investigated STAT5A or STAT5B expression and cellular localization in PTCL entities by specific immunohistochemical STAT5A or STAT5B staining. Quantification of nuclear STAT5 staining intensity revealed that PTCL, NOS cases had higher expression and activation of STAT5A and STAT5B (score 3 or 4) than non-reactive lymph node samples (score 1 Figure 6A, B), in line with nuclear staining indicative of enhanced STAT5-mediated transcriptional activity. Comparable results were obtained when samples of AITL and various CTCL cases were analyzed (Online Supplementary Figure S6A-C), in line with previous reports.48–51 pYSTAT5 staining on PTCL and AITL cases confirmed that nuclear STAT5A/B staining corresponds to elevated pYSTAT5 levels (Online Supplementary Figure S6D). Analysis of STAT5A and STAT5B mRNA expression levels in 18 PTCL, NOS samples compared to non-diseased human lymph nodes (n=4) showed six-fold and two-fold upregulation of STAT5A and STAT5B expression, respectively (Figure 6C, Online Supplementary Figure S6E). Similar results were obtained when we compared seven AITL cases to control tissue (Online Supplementary Figure S6F). Enhanced STAT5A expression was strongly correlated with elevated STAT5B levels (Online Supplementary Figure S6E-F). To increase the numbers of patients and disease entities, tissue microarrays were quantified for STAT5A and STAT5B expression – with highly positive STAT5A and STAT5B nuclear staining intensities across PTCL entities (Figure 6D, Online Supplementary Figure S6G).
Figure 6.
Enhanced STAT5 expression and activity in peripheral T-cell lymphoma, not otherwise specified. (A) STAT5A (top) and STAT5B (bottom) staining of representative non-diseased human lymph nodes (hLN, n=4) and peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS) cases (n=8) with nuclear STAT5A/B staining intensity scoring. Scale bars indicate 100 or 50 mm. (B) Summary of scoring of nuclear STAT5A (top) and STAT5B (bottom) staining intensity ranging from 1 (low) to 4 (high). (C) STAT5A (left) and STAT5B (middle) mRNA levels of non-diseased hLN (n=4) vs. PTCL, NOS lymphoma tissue (n=18, STAT5A P=0.016, STAT5B P=0.11, unpaired t-test). Mean STAT5A or STAT5B expression in hLN was normalized to 1. (D) Statistical summary of nuclear STAT5A (left) and STAT5B (right) staining intensity, classified as weakly positive, positive and strongly positive, of 35 PTCL, NOS, 14 angioimmunoblastic T-cell lymphoma (AITL), 7 cutaneous T-cell lymphoma (CTCL), 6 mycosis fungoides (MF), and 5 control samples spotted on a tissue microarray.
In brief, patient-derived PTCL samples displayed STAT5 upregulation and enhanced intensity of STAT5A/B nuclear staining, pointing to an important role of STAT5 in various PTCL subsets. These findings establish elevated expression of STAT5A/B across human PTCL entities, which we finally set out to target pharmacologically.
Proliferation of peripheral T-cell lymphoma cells is highly sensitive to targeted JAK/STAT pathway therapy
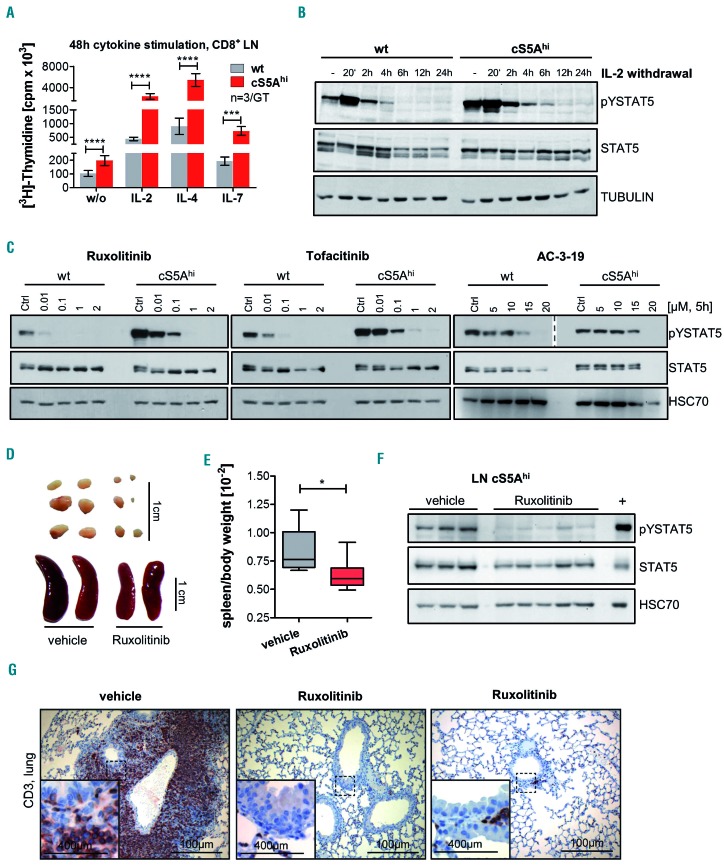
Primary cultures of cS5Ahi CTL were cytokine-dependent and hypersensitive to IL-2, IL-4 and IL-7. This indicates higher cytokine-induced proliferation of cS5Ahi compared to wt cells (Figure 7A, Online Supplementary Figure S7A), also reflected by the longer pYSTAT5 persistence after IL-2 withdrawal (Figure 7B, Online Supplementary Figure S7B). Results from our mouse models32 and recent literature30,52 indicate that inhibition of STAT5 signaling may represent a therapeutic option in PTCL patients. We investigated the effects of the Food and Drug Administration-approved JAK1/2 inhibitor ruxolitinib and JAK1/2/3 inhibitor tofacitinib38, as cS5AF depends on upstream cytokine signaling, as well as the STAT5 inhibitor AC-3-19, which was described to inhibit pYSTAT5 activation and downstream target genes.39,53 In the presence of IL-2, cS5Ahi mice-derived CTL cells were treated with increasing concentrations of these JAK and STAT5 inhibitors, which led to decreased STAT5 activation and cell viability (Figure 7C, Online Supplementary Figure S7C). All three inhibitors reduced the viability of cS5Ahi-derived CTL cells already at low concentrations, with the half maximal inhibitory concentration (IC50) being ~4.1 mM for AC-3-19, ~0.005 mM for ruxolitinib and ~0.03 mM for tofacitinib (Online Supplementary Figure S7C). Sensitivity to STAT5 inhibition could be confirmed in human PTCL cell lines. AC-3-19 treatment reduced the viability of Mac2A, FePD, SU-DHL-1, Mac1 and SR786 (IC50 5-10 mM) (Online Supplementary Figure S7D). When we used ruxolitinib we found that Mac1- and Mac2A cells that harbor a JAK2 translocation were sensitive.54 Control cell lines were only affected at significantly higher concentrations (AC-3-19: <20 mM) (Online Supplementary Figure S7D, summary on mutations in human cells in Online Supplementary Table S7). In addition, ruxolitinib blocked neoplastic cell growth in cS5Ahi mice, as exemplified by reductions of splenomegaly and lymphadenopathy (Figure 7D-E). The specificity of the treatment was validated by reduced pYSTAT5 in lymph nodes and spleen (Figure 7F, Online Supplementary Figure S7E). Most impressively, the T-cell infiltration in peripheral organs was drastically reduced in treated mice (Figure 7G, Online Supplementary Figure S7F, G). Collectively, these data verify that malignant cS5Ahi-CTL and human PTCL cells critically depend on STAT5 signaling.
Figure 7.
Dependence of cS5Ahi on upstream cytokine signaling can be used for targeted pharmacological therapy. (A) [3H]Thymidine incorporation in CD8+ T cells isolated from lymph nodes (LN) of 35-week old wildtype (wt) and cS5Ahi mice after stimulation with interleukin (IL)-2 (100 U/mL, P≤0.0001), IL-4 (100 ng/mL, P≤0.0001), IL-7 (10 ng/mL, P=0.0002) or without (w/o) cytokines (P≤0.0001, n=3/genotype in triplicate, all unpaired t-test). (B) Immunoblot for pYSTAT5 and STAT5 on IL-2-cultured wt and cS5Ahi LN cells sampled at specific time points after IL-2 withdrawal. α-TUBULIN was used as a loading control. Representative blot of three experiments. (C) In vitro treatment of wt and cS5Ahi LN-derived T cells with increasing concentrations of ruxolitinib (left), tofacitinib (middle) or AC-3-19 (right) for 5 h blotted for pYSTAT5 (AC-3-19 – two different exposures are shown indicated by the dashed line) and STAT5. An equal amount of dimethylsulfoxide was used as a control. HSC70 served as a loading control. Representative blot of three experiments. (D) In vivo treatment of cS5Ahi mice with 45 mg/kg ruxolitinib (n=6) or vehicle (n=6) for 30 days. Macroscopic appearance of LN (top) and spleen (bottom) and (E) spleen/body weight ratio after 30 days of treatment (Mann Whitney test, P=0.026). (F) Immunoblot (representative blot of two) on LN lysates of vehicle- or ruxolitinib-treated cS5Ahi mice for pYSTAT5 and STAT5, HSC70 was used as a loading control and diseased cS5Ahi spleen lysate was used as a positive control. (G) CD3 staining of lung sections of vehicle- and ruxolitinib-treated cS5Ahi mice, scale bars represent 400 or 100 mm.
Discussion
PTCL patients face an unfavorable prognosis as chemotherapy results in a poor 5-year overall survival rate.2,55 Targeted treatment options only exist for ALK+ anaplastic large cell lymphoma and are urgently needed for the other 30 PTCL entities. The cohort of patients we investigated revealed increased expression and activity of both STAT5A and STAT5B proteins in mature T-cell lymphomas. Using cS5Ahi transgenic mice we unambiguously associated STAT5A activity with PTCL phenotype. Importantly, mRNA expression changes of cS5Ahi mice closely matched gene expression profiles of human T-cell neoplasia and overlapped with the profile of the more aggressive hSTAT5BN642H mouse model.32 Importantly, clinical JAK1/2/3 inhibitors ruxolitinib and tofacitinib38 and selective STAT5 inhibitors39 reduced neoplastic PTCL cell growth.
Previous transgenic models addressing leukemogenesis downstream of STAT5 employed either wt or STAT5 gain-of-function variants in a lymphoid-restricted manner resulting in expansion of mature T cells, lymphoblastic or B-cell lymphoma.56–59 Broad hematopoietic expression resulted in enhanced granulopoiesis.60 Here, we engineered expression of the well-characterized hyperactive STAT5A variant cS5F using the vav-promoter starting expression from the hematopoietic stem cell stage.40 cS5Ahi mice developed effector/memory- CD8+ T-cell malignancies featuring pronounced organ infiltration. Transgene expression was observed in all major blood lineages tested, but neoplastic expansion of CD8+ T cells was most prominent. This argues for the susceptibly of the T-cell lineage to STAT5 hyperactivation and it emphasizes the role of JAK1/3/STAT3/5 signaling in the outgrowth of T-cell lymphomas. We suggest that CD8+ T cells outcompete cS5AF-mediated effects on other lineages due to faster cell cycle progression, higher survival, cytokine sensitivity and cell fate-specific expression differences. We speculate that STAT5-mediated effects are more negatively controlled in myeloid over lymphoid cell types and that low pYSTAT5 levels, as mimicked in cS5Alo mice, do not promote neoplasia. Gene-dosage and graded STAT5 activity levels in our study showed a clear positive correlation between STAT5 activation status and CD8+ T-cell numbers.34,35 Senescence, diminished oligomer formation or differentiation capacity might contribute to the differences between cS5Alo and cS5Ahi. Although the level of pYSTAT5 in cS5Alo mice was significant, it was nevertheless lower than in cS5Ahi or hSTAT5BN642H mice, and malignant transformation did not occur in the cS5Alo animals. Thus, the real driver of PTCL disease might be the amount of pYSTAT5 resulting in enhanced and prolonged transcriptional activity.
In PTCL several factors account for STAT5 activation – such as mutations in STAT5 or in upstream signaling components (e.g. IL-2R, JAK1, JAK3). Surprisingly, recurrent mutations in epigenetic and chromatin remodeling factors parallel JAK/STAT activation and the epigenetic landscape is also shaped by STAT3/5 and its versatile interaction partners.52,61 However, until now a detailed understanding of these factors in PTCL is lacking. Increased pYSTAT5 levels have also been reported upon autocrine PDGFα signaling in PTCL, NOS.62 In CTCL, overexpression of oncogenic miR-155,49 downregulation of tumor-suppressive miR-22,50 enhanced progression by lymphotoxin-α-dependent lymphangiogenesis,63 STAT5-dependent CD80 expression,51 resistance to vorinostat48 and risk of disease progression64 were attributed to enhanced STAT5 signaling. The identification of recurrent and mutually exclusive gain-of-function mutations in STAT3 suggests that these proteins share redundant functions in PTCL.18,23,28 Thus, dual STAT3/5 inhibition is needed in future therapy, which could be approached by SH2-domain blockers. Certainly, mutations or translocations in the upstream JAK proteins boost STAT5 signaling,25,29,30,65 further emphasizing the potential benefit of JAK and STAT inhibitors.
The cS5Ahi model recapitulates clinical features of PTCL with high STAT5 activation. Among the few high-fidelity mouse models for other PTCL subsets,9,66–69 the cS5Ahi model closely phenocopies pathological hallmarks of cytotoxic CD8+ T-cell PTCL. Importantly, cS5Ahi-expressing cells are hypersensitive to the Food and Drug Administration-approved JAK1/2 inhibitor ruxolitinib, which is currently being studied in phase II clinical trials for the treatment of relapsed PTCL and adult T-cell leukemia/lymphoma (NCT01431209, NCT02974647, NCT01712659). In line with this, JAK-inhibitor treatment of Sézary syndrome,29 T-cell prolymphocytic leukemia,54,70 and adult T-cell leukemia/lymphoma71 revealed reduced proliferation and immunomodulatory effects on the PTCL tumor microenvironment.3 Moreover, a small molecule inhibitor of STAT5 SH2 domain-phosphopeptide interactions39 resulted in strongly reduced viability in PTCL lines with high STAT5 activity and its successor compound showed significant effects in acute myeloid leukemia.53 Although these inhibitors are currently considered lead compounds, specific STAT5 inhibitors are also expected to enter clinical trials and combinations with kinase inhibitors could be tested.
In conclusion, our cS5Ahi transgenic mouse model indicates that STAT5 is a driver and STAT5 itself or its interaction partners are potential drug targets in PTCL. The STAT5-dependent CD8+ PTCL mouse model described here represents a tool to investigate molecular mechanisms of PTCL development. Our model serves for identification and pre-clinical testing of novel interventional strategies to target STAT5-dependent PTCL. We conclude that both STAT5A and STAT5B are oncogenes in PTCL, and STAT5B is more transforming. Overall, mutation induced-cytokine sensitivity drives PTCL due to enhanced STAT3/STAT5 signaling.
Supplementary Material
Acknowledgments
Jerry Adams (WEHI, Australia) kindly provided us with the vav-hCD4 (HS21/45) plasmid and Dagmar Stoiber (LBI-CR, Vienna, Austria) with the E.G7 cell line. We thank Safia Zahma (LBI-CR, Vienna, Austria), Elisabeth Gurnhofer, Sigurd Krieger (both at the Department of Clinical Pathology, Medical University Vienna, Austria), Sabine Fajmann (Institute of Pharmacology and Toxicology, University of Veterinary Medicine, Vienna, Austria), and Boris Kovacic (Institute of Medical Genetics, Medical University of Vienna, Vienna, Austria) for technical support. We thank also our mouse facility team and Jisung Park (Department of Chemistry, University of Toronto Mississauga, Canada) for synthesizing the inhibitor AC-3-19 used in this study.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/2/435
References
- 1.Foss FM, Zinzani PL, Vose JM, Gascoyne RD, Rosen ST, Tobinai K. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756–6767. [DOI] [PubMed] [Google Scholar]
- 2.Armitage JO. The aggressive peripheral T-cell lymphomas: 2017. Am J Hematol. 2017;92(7):706–715. [DOI] [PubMed] [Google Scholar]
- 3.Wang T, Feldman AL, Wada DA, et al. GATA-3 expression identifies a high-risk subset of PTCL, NOS with distinct molecular and clinical features. Blood. 2014;123(19):3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siaghani PJ, Song JY. Updates of peripheral T cell lymphomas based on the 2017 WHO classification. Curr Hematol Malig Rep. 2018;13(1):25–36. [DOI] [PubMed] [Google Scholar]
- 5.Laginestra MA, Piccaluga PP, Fuligni F, et al. Pathogenetic and diagnostic significance of microRNA deregulation in peripheral T-cell lymphoma not otherwise specified. Blood Cancer J. 2014;4(11):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iqbal J, Wright G, Wang C, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123(19):2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal J, Weisenburger DD, Greiner TC, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115(5):1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinrich T, Rengstl B, Muik A, et al. Mature T-cell lymphomagenesis induced by retroviral insertional activation of Janus kinase 1. Mol Ther. 2013;21(6):1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner K, Crispatzu G, Al-Ghaili N, et al. Models for mature T-cell lymphomas—a critical appraisal of experimental systems and their contribution to current T-cell tumorigenic concepts. Crit Rev Oncol Hematol. 2013;88(3):680–695. [DOI] [PubMed] [Google Scholar]
- 10.Spinner S, Crispatzu G, Yi JH, et al. Re-activation of mitochondrial apoptosis inhibits T-cell lymphoma survival and treatment resistance. Leukemia. 2016;30(7):1520–1530. [DOI] [PubMed] [Google Scholar]
- 11.Piccaluga P, Tabanelli V, Pileri S. Molecular genetics of peripheral T-cell lymphomas. Int J Hematol. 2014;99(3):219–226. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, McKeithan TW, Gong Q, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilcox RA. A three-signal model of T-cell lymphoma pathogenesis. Am J Hematol. 2016;91(1):113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–1315 [DOI] [PubMed] [Google Scholar]
- 15.Schrader A, Crispatzu G, Oberbeck S, et al. Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat Commun. 2018;9(1):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litvinov IV, Tetzlaff MT, Thibault P, et al. Gene expression analysis in cutaneous T-cell lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology. 2017;6(5):e1306618–e1306618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warner K, Weit N, Crispatzu G, Admirand J, Jones D, Herling M. T-cell receptor signaling in peripheral T-cell lymphoma – a review of patterns of alterations in a central growth regulatory pathway. Curr Hematol Malig Rep. 2013;8(3):163–172. [DOI] [PubMed] [Google Scholar]
- 18.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Arnam JS, Lim MS, Elenitoba-Johnson KSJ. Novel insights into the pathogenesis of T-cell lymphomas. Blood. 2018;131(21):2320–2330. [DOI] [PubMed] [Google Scholar]
- 20.Lone W, Alkhiniji A, Manikkam Umakanthan J, Iqbal J. Molecular insights into pathogenesis of peripheral T cell lymphoma: a review. Curr Hematol Malig Rep. 2018;13(4):318–328. [DOI] [PubMed] [Google Scholar]
- 21.Coppe A, Andersson EI, Binatti A, et al. Genomic landscape characterization of large granular lymphocyte leukemia with a systems genetics approach. Leukemia. 2017;31(5):1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koskela HLM, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajala HLM, Eldfors S, Kuusanmäki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandapalli OR, Schuessele S, Kunz JB, et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica. 2014;99(10):e188–e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiel MJ, Velusamy T, Rolland D, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124(9):1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontro M, Kuusanmaki H, Eldfors S, et al. Novel activating STAT5B mutations as putative drivers of T-cell acute lymphoblastic leukemia. Leukemia. 2014;28(8):1738–1742. [DOI] [PubMed] [Google Scholar]
- 27.Nicolae A, Xi L, Pittaluga S, et al. Frequent STAT5B mutations in γδ hepatosplenic T-cell lymphomas. Leukemia. 2014;28(11):2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Küçük C, Jiang B, Hu X, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells. Nat Commun. 2015;6:6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiel MJ, Sahasrabuddhe AA, Rolland DCM, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015;6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dufva O, Kankainen M, Kelkka T, et al. Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat Commun. 2018;9(1):1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cross NCP, Hoade Y, Tapper WJ, et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia. 2018;33(2):415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5B(N642H) is a driver mutation for T cell neoplasia. J Clin Invest. 2018;128(1):387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heltemes-Harris LM, Farrar MA. The role of STAT5 in lymphocyte development and transformation. Curr Opin Immunol. 2012;24(2):146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoelbl A, Kovacic B, Kerenyi MA, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107(12):4898–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ermakova O, Piszczek L, Luciani L, et al. Sensitized phenotypic screening identifies gene dosage sensitive region on chromosome 11 that predisposes to disease in mice. EMBO Mol Med. 2011;3(1):50–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriggl R, Sexl V, Kenner L, et al. Stat5 tetramer formation is associated with leuke-mogenesis. Cancer Cell. 2005;7(1):87–99. [DOI] [PubMed] [Google Scholar]
- 38.Kontzias A, Kotlyar A, Laurence A, Changelian P, O’Shea JJ. Jakinibs: a new class of kinase inhibitors in cancer and autoimmune disease. Curr Opin Pharmacol. 2012; 12(4):464–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cumaraswamy AA, Lewis AM, Geletu M, et al. Nanomolar-potency small molecule inhibitor of STAT5 protein. ACS Med Chem Lett. 2014;5(11):1202–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogilvy S, Metcalf D, Gibson L, Bath ML, Harris AW, Adams JM. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94(6):1855–1863. [PubMed] [Google Scholar]
- 41.Berard M, Tough DF. Qualitative differences between naïve and memory T cells. Immunology. 2002;106(2):127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan L, Gurnani K, Dicaire C, et al. Rapid clonal expansion and prolonged maintenance of memory CD8+ T cells of the effector (CD44highCD62Llow) and central (CD44highCD62Lhigh) phenotype by an archaeosome adjuvant independent of TLR2. J Immunol. 2007;178(4):2396–2406. [DOI] [PubMed] [Google Scholar]
- 43.Grange M, Buferne M, Verdeil G, Leserman L, Schmitt-Verhulst A, Auphan-Anezin N. Activated STAT5 promotes long-lived cytotoxic CD8+ T cells that induce regression of autochthonous melanoma. Cancer Res. 2012;72(1):76–87. [DOI] [PubMed] [Google Scholar]
- 44.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103(9):3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kollmann S, Grundschober E, Maurer B, et al. Twins with different personalities: STAT5B – but not STAT5A – has a key role in BCR/ABL-induced leukemia. Leukemia. 2019. January 24 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villarino A, Laurence A, Robinson GW, et al. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. eLife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fantin VR, Loboda A, Paweletz CP, et al. Constitutive activation of signal transducers and activators of transcription predicts vorinostat resistance in cutaneous T-cell lymphoma. Cancer Res. 2008;68(10): 3785–3794. [DOI] [PubMed] [Google Scholar]
- 49.Kopp KL, Ralfkiaer U, Gjerdrum LMR, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12(12):1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sibbesen NA, Kopp KL, Litvinov IV, et al. Jak3, STAT3, and STAT5 inhibit expression of miR-22, a novel tumor suppressor microRNA, in cutaneous T-cell lymphoma. Oncotarget. 2015;6(24):20555–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Wang HY, Wei F, et al. Cutaneous T cell lymphoma expresses immunosup-pressive CD80 (B7-1) cell surface protein in a STAT5-dependent manner. J Immunol. 2014;192(6):2913–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orlova A, Wingelhofer B, Neubauer HA, et al. Emerging therapeutic targets in myeloproliferative neoplasms and peripheral T-cell leukemia and lymphomas. Expert Opin Ther Targets. 2018;22(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wingelhofer B, Maurer B, Heyes EC, et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia. 2018;32(5): 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng SY, Yoshida N, Christie AL, et al. Targetable vulnerabilities in T- and NK-cell lymphomas identified through preclinical models. Nat Commun. 2018;9(1):2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy NM, Evens AM. Chemotherapeutic advancements in peripheral T-cell lymphoma. Semin Hematol. 2014;51(1):17–24. [DOI] [PubMed] [Google Scholar]
- 56.Kelly JA, Spolski R, Kovanen PE, et al. Stat5 Synergizes with T cell receptor/antigen stimulation in the development of lymphoblastic lymphoma. J Exp Med. 2003;198(1):79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen B, Yi B, Mao R, et al. Enhanced T cell lymphoma in NOD.Stat5b transgenic mice is caused by hyperactivation of Stat5b in CD8(+) thymocytes. PLoS One. 2013;8(2): e56600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burchill MA, Goetz CA, Prlic M, et al. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol. 2003;171(11):5853–5864. [DOI] [PubMed] [Google Scholar]
- 59.Joliot V, Cormier F, Medyouf H, Alcalde H, Ghysdael J. Constitutive STAT5 activation specifically cooperates with the loss of p53 function in B-cell lymphomagenesis. Oncogene. 2006;25(33):4573–4584. [DOI] [PubMed] [Google Scholar]
- 60.Lin W-C, Schmidt JW, Creamer BA, Triplett AA, Wagner K-U. Gain-of-function of Stat5 leads to excessive granulopoiesis and lethal extravasation of granulocytes to the lung. PLoS One. 2013;8(4):e60902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wingelhofer B, Neubauer HA, Valent P, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32(8):1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piccaluga PP, Rossi M, Agostinelli C, et al. Platelet-derived growth factor alpha medi ates the proliferation of peripheral T-cell lymphoma cells via an autocrine regulatory pathway. Leukemia. 2014;28(8):1687–1697. [DOI] [PubMed] [Google Scholar]
- 63.Lauenborg B, Christensen L, Ralfkiaer U, et al. Malignant T cells express lymphotoxin α and drive endothelial activation in cutaneous T cell lymphoma. Oncotarget. 2015;6(17):15235–15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Litvinov IV, Netchiporouk E, Cordeiro B, et al. The use of transcriptional profiling to improve personalized diagnosis and management of cutaneous T-cell lymphoma (CTCL). Clin Cancer Res. 2015;21(12):2820–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bergmann AK, Schneppenheim S, Seifert M, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2014;53(4):309–316. [DOI] [PubMed] [Google Scholar]
- 66.Beachy SH, Onozawa M, Chung YJ, et al. Enforced expression of Lin28b leads to impaired T-cell development, release of inflammatory cytokines, and peripheral T-cell lymphoma. Blood. 2012;120(5):1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muto H, Sakata-Yanagimoto M, Nagae G, et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J. 2014;4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pechloff K, Holch J, Ferch U, et al. The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma. J Exp Med. 2010;207(5): 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tezuka K, Xun R, Tei M, et al. An animal model of adult T-cell leukemia: humanized mice with HTLV-1–specific immunity. Blood. 2014;123(3):346–355. [DOI] [PubMed] [Google Scholar]
- 70.Andersson EI, Pützer S, Yadav B, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2017;32(3):774–787. [DOI] [PubMed] [Google Scholar]
- 71.Zhang M, Mathews Griner LA, Ju W, et al. Selective targeting of JAK/STAT signaling is potentiated by Bcl-xL blockade in IL-2-dependent adult T-cell leukemia. Proc Natl Acad Sci U S A. 2015;112(40):12480–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.