Abstract
Staphylococcus epidermidis is a Gram-positive saprophytic bacterium found in the microaerobic/anaerobic layers of the skin that becomes a health hazard when it is carried across the skin through punctures or wounds. Pathogenicity is enhanced by the ability of S. epidermidis to associate into biofilms, where it avoids attacks by the host and antibiotics. To test the effect of oxygen on metabolism and biofilm generation, cells were cultured at different oxygen concentrations ([O2]). As [O2] decreased, S. epidermidis metabolism went from respiratory to fermentative. Remarkably, the rate of growth decreased at low [O2] while a high concentration of ATP ([ATP]) was kept. Under hypoxic conditions bacteria associated into biofilms. Aerobic activity sensitized the cell to hydrogen peroxide-mediated damage. In the presence of metabolic inhibitors, biofilm formation decreased. It is suggested that at low [O2] S. epidermidis limits its growth and develops the ability to form biofilms.
Keywords: Staphylococcus epidermidis, Oxygen concentration, Metabolism, Biofilms, Rate of oxygen consumption, Fermentation
Introduction
Saprophytic microorganisms control pathogenic bacteria, digest nutrients and synthesize coenzymes, prosthetic groups and amino acids (Foster et al. 2005; Berg 1996; Sender et al. 2016). In the skin, Staphylococcus epidermidis inhibits colonization by Staphylococcus aureus or Streptococcus pyogenes secreting antimicrobial compounds and proteases (Cogen et al. 2010; Iwase et al. 2010). In the skin, S. epidermidis inhabits the epidermis, dermis and the nearly anoxic sebaceous glands (Grice and Segre 2011).
Staphylococcus epidermidis is frequently introduced through wounds and surgical procedures. A recent study reported the presence of antibiotic-resistant S. epidermidis strains in 46% of hospital secondary infections (Chabi and Momtaz 2019). Many of these strains were resistant to at least three antibiotics (Chabi and Momtaz 2019). Indeed, many antibiotics have to be tested in order to treat S. epidermidis nosocomial infections (Roujansky et al. 2020). S. epidermidis is also found frequently in implanted devices such as valves and catheters. There is an active search for materials to coat implant surfaces which may prevent biofilm formation (Rabin et al. 2015). Among these, zirconium nitride has shown promise in orthopaedic implants (Pilz et al. 2019), while sphingosine coating is being used with success on implant titanium-surfaces (Beck et al. 2019). Inside the body, this bacterium has to face attack from the immune system, high [O2] (Fang et al. 2016) and antibiotics (Leid 2009), most likely triggering a stress response. Within the organism, S. epidermidis may find areas with low [O2], similar to its natural habitat; it is likely that the bacterium will make an effort to remain in the hypoxic area, adhering to the surface and organizing into biofilms (Lewis 2007; Uribe-Alvarez et al. 2016). In regard to hypoxic environments within the host, these are often found at or near artificial devices such as catheters or prosthetic valves, where biofilms may force removal of implanted devices (Fey and Olson 2010; Büttner et al. 2015).
Understanding the S. epidermidis response to different [O2] would help optimize treatments (Cotter et al. 2009). We have reported that growing S. epidermidis at different [O2] modifies expression of respiratory chain enzymes and the ability to form biofilms (Uribe-Alvarez et al. 2016). At high [O2], cytochrome oxidases and NADH dehydrogenases are abundant and biofilms are minimal. In contrast, [O2] depletion increases nitrate reductase expression and association into biofilms (Uribe-Alvarez et al. 2016).
Here, the effect of [O2] on both, the aerobic and anaerobic metabolism of S. epidermidis was evaluated, together with [ATP]. In addition, the sensitivity of S. epidermidis to the toxic effects of hydrogen peroxide was tested. In each case, the biofilm-forming activity of cells was measured (Lewis 2007). When ATP synthesis was inhibited to different degrees by inhibitors of respiration (cyanide) (Uribe-Alvarez et al. 2016) or glycolysis (1,4-bisphosphobutane) (Hartman and Barker 1965; Rosas-Lemus et al. 2016a), biofilm formation also decreased. It is suggested that S. epidermidis associates into biofilms as a strategy to avoid high [O2].
Materials and methods
Bacterial strain and growth media
Staphylococcus epidermidis strain ATCC 12228 was a kind donation from Dr. Juan Carlos Cancino Díaz (Instituto Politécnico Nacional, México). A loophole from the bacterium was suspended in 5 mL of 3% tryptic soy broth (Fluka, Sigma) and incubated at 37 °C for 24 h. Pre-cultures were added to 1 L LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) plus 2% glucose and incubated 24 h at 30 °C under aerobic (shaking 150 rpm), microaerobic (5% CO2, no agitation) or anaerobic (static in oxygen-depleted sealed acrylic chamber) conditions. Then the cells were washed three times at 5000×g for 10 min with distilled water and resuspended in 10 mM HEPES pH 7.4 (Uribe-Alvarez et al. 2016).
Cytoplasmic extracts
All procedures were conducted at 4 °C. Cells (grown under aerobic, microaerobic or anaerobic conditions) were centrifuged at 5000×g for 10 min, washed three times with distilled water and resuspended in 50 mL 10 mM HEPES, pH 7.4, supplemented with one tablet of protease-inhibitor cocktail (Complete) and 1 mM PMSF. Cells were disrupted by sonication using a Sonics VibraCell sonicator (Sonics & materials, Inc., Newtown, CT) 7 × 20 s with 20 s intervals. To remove unbroken cells the suspension was centrifuged at 10,000×g for 10 min and the supernatant was recovered.
Protein concentration
Protein concentrations from intact S. epidermidis cells were determined by the biuret method (Gornall et al. 1949). Absorbance (540 nm) was measured in a Beckman-Coulter DU50 spectrophotometer. For cytoplasmic extracts, protein concentration was measured by Bradford at 595 nm, using 1 or 2 µL aliquots of the sample in a PolarStar Omega (BMG labtech, Ortenberg, Germany) (Bradford 1976).
Rate of oxygen consumption
The rate of oxygen consumption was measured in 10 mM HEPES pH 7.4 plus the indicated respiratory substrate. Bacteria, 0.5 mg prot mL−1 were added to a water-jacketed 1 mL chamber at 37 °C equipped with a Clark type electrode connected to a Strathkelvin model 782 oxymeter. Data were analyzed using the 782 Oxygen System Software (Warner/Strathkelvin Instruments) (Uribe-Alvarez et al. 2016).
Ethanol production
Fermentation by cell cytoplasmic extracts (0.5 mg prot. mL−1) was measured in 0.1 M MES-TEA, pH 7.0, 1.8 mM NAD plus either glucose or glycerol and incubated at 30 °C for 0, 2.5, 5 or 10 min. The reaction was stopped with 30% TCA, 0.1 mL and neutralized with NaOH. Ethanol was measured adding a 10 µL aliquot (0.005 mg) of the supernatant to 0.2 mL 114 mM K2HPO4, pH 7.6. After 1 min, 30 μg ADH mL−1 was added, the sample was incubated for 30 min and O.D. was determined at 340 nm in a POLARstar Omega. Ethanol is reported as μmol ethanol (mg prot)−1 (Araiza-Olivera et al. 2013).
ATP concentration
ATP was measured in cytoplasm extracts resuspended to 0.025 mg protein in 0.15 mL reaction buffer (20 mM KH2PO4, 40 mM Na2HPO4, 80 mM NaCl, 1 mM MgSO4). An ATP calibration curve was prepared freshly each day using lyophilized luciferase (Sigma-Aldrich). Luciferase was prepared following instructions by the provider and 0.02 mL was added to each sample in a 96-well microplate. Bioluminescence was detected in a POLARstar Omega luminometer (BGM LABTECH, Offenburg, Germany). [ATP] was reported as µmol (mg prot)−1 (Palikaras and Tavernarakis 2016; Mendoza-Hoffmann et al. 2018).
Susceptibility to hydrogen peroxide-mediated damage
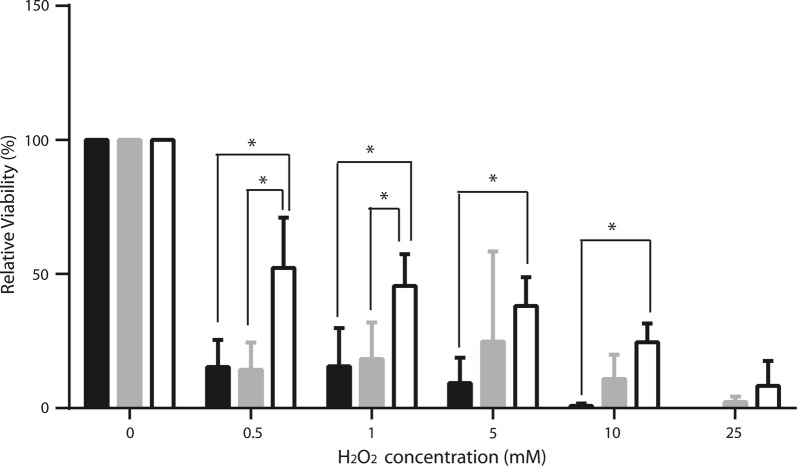
The effect of [H2O2] on the viability of S. epidermidis was determined as previously reported (Macvanin and Hughes 2010). Briefly, cells were adjusted to an O.D. = 0.1 (600 nm) and then H2O2 (0 to 25 mM as indicated) was added to the reaction mixture. After 30 min, serial dilution of the cultures was performed in 0.9% NaCl and 10 µL of the 1:1000 diluted sample was plated in LB, 2% glucose agar plates and incubated 24 h at 37 °C. Colony forming units (CFU) mL−1 were counted. The sample taken before H2O2 addition was assigned as 100%. The average of three experiments is shown with SD. ANOVA test and Tukey’s multiple comparison-test were used. Significance was *P < 0.0001.
Biofilm formation and detection
Biofilm generation was measured in sterile Costar 96-well polystyrene plates as previously reported (Calà et al. 2015; Uribe-Alvarez et al. 2016). Briefly, in each well, 0.4% crystal violet in 33% glacial acetic acid was mixed with the indicated, inhibitors sodium cyanide (NaCN) (100 µM), butane-1,4-bisphosphate (B1,4BP) (1 mM) or, carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (0.1, 0.5, or 1 µM, as indicated). Then bacteria were added to O.D. 0.02. Final volume 200 μL. The plate was incubated 24 h at 37 °C with 5% CO2. After incubation, wells were washed twice with 200 µL phosphate-buffered saline (PBS) to remove non-adherent bacteria. Plates were dried for 1 h at 60 °C, stained with 0.4% crystal violet for 10 min and washed under running tap water to remove excess stain. Absorbance (492 nm) was measured using a microplate reader (Polar Star Omega, BMG Labtech). Each sample was tested in three independent triplicate experiments and compared against the non-treated control using one-way variance analysis (ANOVA) plus Dunnett’s post hoc test.
Results
Oxygen is among the most important factors driving evolution (Lane 2002). Its partial reduction products, the reactive oxygen species (ROS) destroy nucleic acids, proteins and membranes (Ezraty et al. 2017). Thus, to profit from its remarkable electron acceptor properties, organisms have to deal carefully with the dangerous oxygen molecule (Lane 2002; Rosas-Lemus et al. 2016b). S. epidermidis lives in hypoxic/anoxic environments, although it can adapt to high [O2]. In order to follow the metabolic adaptation of S. epidermidis it was cultivated at different [O2]. After 24 h under aerobic conditions biomass yield was 8.58 g/L, three times higher than under microaerobiosis, 2.11 g/L or anaerobiosis, 1.75 g/L.
In order to further explore the basis for biomass yield variations at different [O2], the activity of the respiratory chain from S. epidermidis grown at different [O2] was measured (Fig. 1). As expected from previous respiratory chain protein expression results (Uribe-Alvarez et al. 2016), the ability of cells to consume oxygen was proportional to [O2] in the growth medium. In aerobic conditions and in the presence of lactate the rate of oxygen consumption was 70 natgO (mg prot. min)−1, at least five times higher than in microaerobic media, where the rate was 5 natgO (mg prot. min)−1 or in those grown under anaerobic conditions, where it was negligible (Fig. 1). Under normoxia the best respiratory fuel was lactate, which was oxidized around three times as fast as glucose or ethanol (Fig. 1).
Fig. 1.
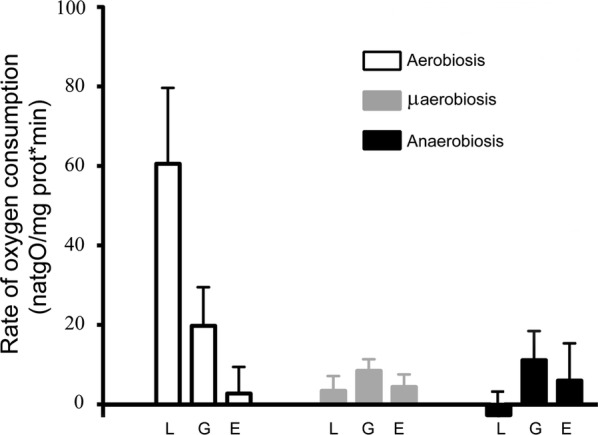
Rate of oxygen consumption by S. epidermidis in the presence of different respiratory substrates. Experimental conditions: 10 mM HEPES (pH 7.4). As indicated, substrates were: L: 10 mM lactate; G: 40 mM glucose or E: 33 mM ethanol. Cells were grown at different [O2] as follows: aerobic (empty bars), microaerobic (gray bars) and anaerobic (black bars)
In S. epidermidis respiratory chain activities correlated with growth rates. However, it was reasoned that in hypoxia glycolysis may constitute an important source of energy (Somerville and Proctor 2009). Furthermore, as S. epidermidis, normally lives at low [O2], fermentation may be the preferred energy-yielding pathway in this bacterium. To test this, S. epidermidis was grown at different [O2] and ethanol production from either glucose (Fig. 2a) or glycerol (Fig. 2b) was measured at 2.5, 5 and 10 min of incubation. Both substrates were equally efficient. However, at different [O2] large variations in the rate of fermentation were observed: bacteria from anaerobic media were the most active, (Fig. 2), suggesting that fermentation increases as [O2] decreases.
Fig. 2.
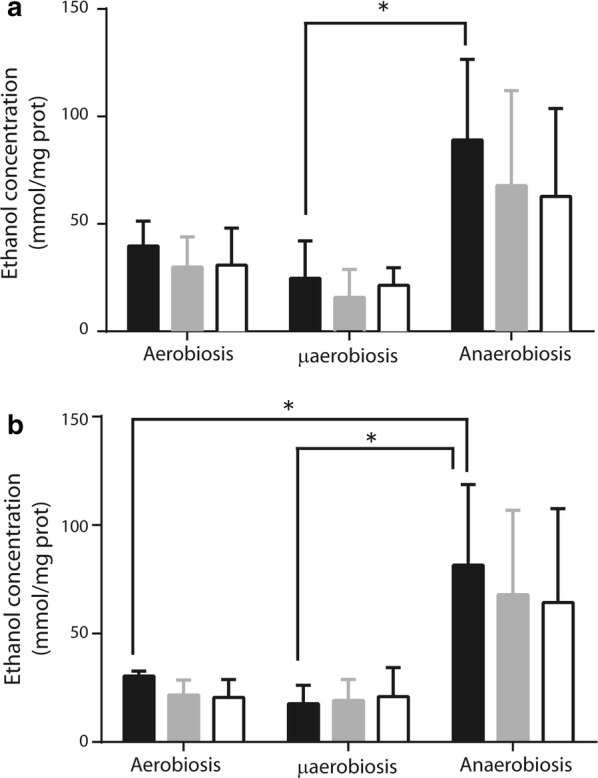
Fermentation by S. epidermidis grown at different [O2]. Cytoplasmic extracts were obtained from S. epidermidis grown under aerobic, microaerobic or anaerobic conditions. Fermentation by cell cytoplasmic extracts (0.5 mg prot. mL−1) was measured using a 20 mM glucose or b 20 mM glycerol. Samples were incubated at 30 °C for: 2.5 min (black columns), 5 min (gray columns) or 10 min (white columns). Results are reported as μmol ethanol per mg protein. Tukey’s comparison test was used to determine significant differences (*P < 0.05)
In S. epidermidis, increasing [O2] increased the rate of oxygen consumption while fermentation was inhibited. To determine which of these pathways produced more energy, the concentration of ATP ([ATP]) was measured in S. epidermidis grown under normoxia, hypoxia or anoxia (Fig. 3). Contrary to what we expected from the low growth rate and the slow respiratory activity observed, in hypoxia- and anoxia-grown cells, [ATP] was higher than in normoxia as aerobiosis, [ATP] increased roughly five times in hypoxia and three times in anoxia as compared to normoxia (Fig. 3).
Fig. 3.
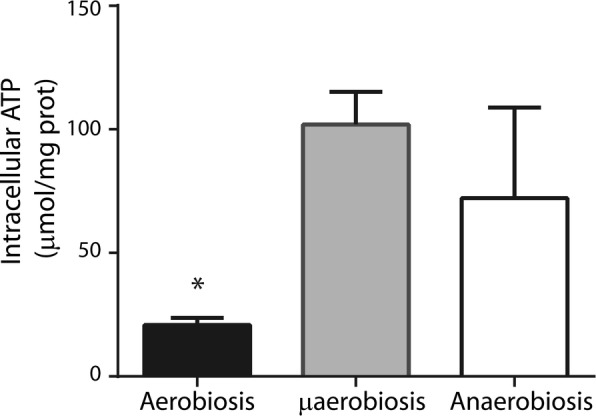
Intracellular ATP concentrations in S. epidermidis grown at different [O2]. Cells were grown at different [O2] in LB plus glucose. Cytoplasmic extracts were obtained from each of these cultures and used to measure intracellular ATP. ATP concentration was estimated using luciferase and interpolating into a standard curve (see “Materials and methods”). The average of three experiments is shown with SD. * indicates significant difference P < 0.05
In S. aureus a deficient respiratory chain confers resistance to H2O2 toxicity (Painter et al. 2017), suggesting that anaerobiosis-adapted cells resist oxidative stress better. Thus, we decided to test S. epidermidis grown at different [O2] for its sensitivity to H2O2 (Fig. 4) (Lobritz et al. 2015). Even at the lowest concentrations of H2O2 we used (0.5 mM), viability decreased in all cells. Aerobic-grown cells exhibited the poorest survival rates, while cells grown under anaerobiosis survived best, such that even at the highest H2O2 concentration tested (25 mM H2O2) a small amount of viable cells was detected (Fig. 4). The increase in sensitivity to ROS observed in aerobically grown S. epidermidis was probably due to increased expression of the redox enzymes in the respiratory chain (Uribe-Alvarez et al. 2016). These redox enzymes contain different coenzymes and prosthetic groups, which normally become free radicals during their catalytic cycle (Quinlan et al. 2013; Rosas-Lemus et al. 2016b). Thus, as reported for S. aureus (Painter et al. 2017) at high [O2] S. epidermidis expressed an active respiratory chain and its sensitivity to H2O2 increased.
Fig. 4.
H2O2 effect on cellular viability. S. epidermidis susceptibility to hydrogen peroxide was determined using 0, 0.5, 1, 5, 10 or 25 mM H2O2 in each group: aerobiosis (black bar), microaerobiosis (gray bar) or anaerobiosis (white bar). After 30 min of incubation with H2O2, the samples were diluted 1:1000, 10 µL were taken and plated in LB plus 2% glucose-agar. CFU/mL were counted. Samples without treatment were assigned as 100% viable cells. The average of three experiments is shown with SD. Significance *P < 0.0001
The highest [ATP] was detected in cells grown at low [O2], which exhibited a slow growth rate. This seemingly contradictory situation may be explained by proposing that when S. epidermidis finds a low [O2], which resembles that found in its normal niche, it makes an effort to attach itself to a surface, redirecting its energy use from growth to produce polysaccharides and proteins for biofilm generation (Beenken et al. 2004; Lewis 2007). To analyze whether biofilm was dependent on [ATP], S. epidermidis was grown under hypoxia and in the presence and absence of different metabolic inhibitors. In hypoxic grown-cells both oxidative phosphorylation and fermentation are active. It was observed that cells incubated in the presence of the respiratory chain inhibitor cyanide or the glycolytic inhibitor 1,4-bisphosphobutane, formed smaller biofilms than the control and that addition of both inhibitors led to even less biofilms (Fig. 5a). This would suggest that biofilm formation activity is proportional to [ATP]. In addition, the uncoupler CCCP was used at concentrations below those where it killed cells (Result not-shown), observing that biofilm generation decreased further as uncoupler concentration increased (Fig. 5b). These results suggest that, regardless of its source, in S. epidermidis high [ATP] is needed to form biofilms.
Fig. 5.
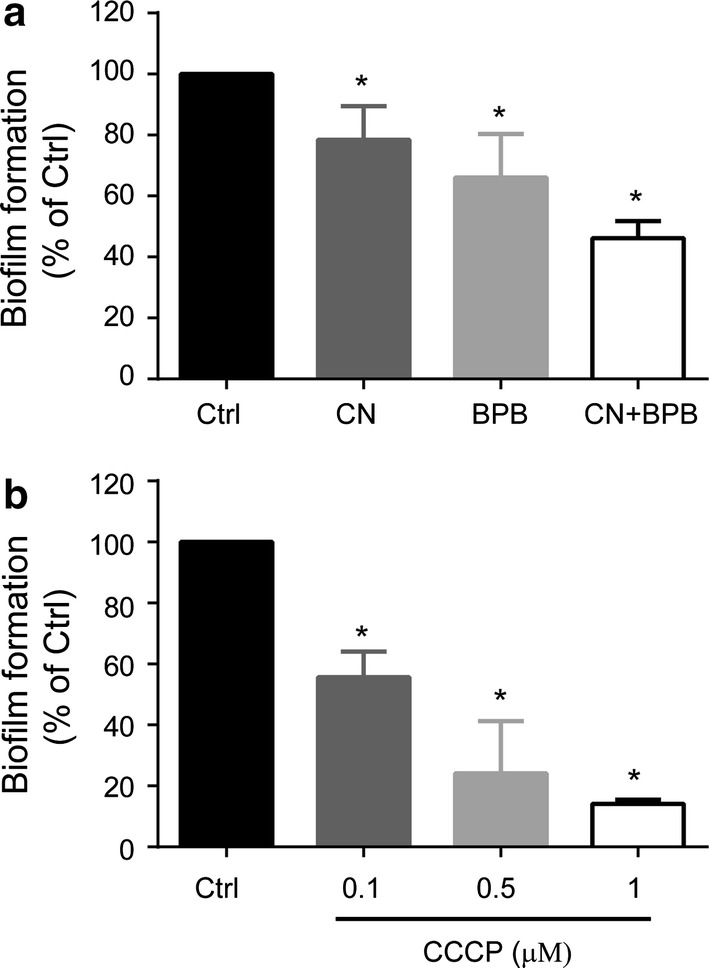
In vitro biofilm inhibition assay. S. epidermidis was grown under microaerobic conditions. a Different metabolic inhibitors were added as indicated: 100 µM NaCN, 1 mM B1,4BP or both inhibitors. b Different concentrations of the uncoupler CCCP (0.5, 1.0 and 1.5 µM) were added to deplete ATP. After 24 h of incubation biofilm generation was evaluated by measuring the absorbance at 492 nm with a microplate reader. Each sample was compared with the control (without additions). Statistics were applied using ANOVA and Dunnett’s post hoc test. Significance *P < 0.0001
Discussion
Antibiotic-resistant strains of S. epidermidis are increasingly found in nosocomial infections (Chabi and Momtaz 2019). Implant removal due to S. epidermidis biofilm colonization is also quite frequent (Gristina 1987; Raad et al. 1998). S. epidermidis is frequently found in coagulase-negative staphylococci-caused prosthetic valve infective endocarditis cases (Mack et al. 2013), in 30–43% implant infections (Zimmerli et al. 2004) and in 50–70% catheter-related infections (von Eiff et al. 2002). Understanding the physiology of the bacterium is a must in order to design new treatment and prevention methods (Uribe-Alvarez et al. 2016). In biofilms, S. epidermidis cells are protected from the host. Thus, it is most important to analyse the association and specialization processes of the cells involved in the genesis of biofilms.
Diverse facultative bacteria adapt to wide [O2], differentially expressing redox enzymes in its respiratory chain. S. epidermidis does express different enzymes at varying [O2] (Uribe-Alvarez et al. 2016). Aerobic metabolism enabled cells to grow more (Baez and Shiloach 2014). Still, enhanced growth resulted in higher sensitivity to H2O2, suggesting that high contents of redox enzymes make cells vulnerable to ROS. Indeed, when grown at high [O2], sensitivity to ROS is enhanced in S. aureus and Enterococcus faecalis, while their mutant counterparts, lacking an efficient respiratory chain resist ROS better (Painter et al. 2017).
When exposing S. epidermidis grown in different [O2] to oxygen peroxide, we observed a similar phenomenon: cells grown in hypoxic or anoxic environments, which exhibited low respiratory rates were more resistant to oxygen peroxide (Fig. 4). Thus, as in S. aureus, the lack of an efficient respiratory chain in S. epidermidis enabled cells to survive ROS. This is probably useful when bacteria detached from a biofilm reach other tissues where they may be confronted with the oxidative burst generated by the immune system (Jensen et al. 1992).
The rate of oxygen consumption in aerobic grown cells was highest when lactate was the substrate. This is probably due to the direct donation of electrons to the menaquinone pool by lactate dehydrogenase (Götz and Mayer 2013; Kane et al. 2016). The slower rates observed for alcohol, may be due to an additional step as alcohol dehydrogenase electrons are first donated to Ndi2 (Artzatbanov and Petrov 1990). The rate of respiration was also slow for glucose, probably for the same reason, as intermediaries have to undergo many reactions before releasing electrons to the respiratory chain (Ferreira et al. 2013). In contrast, under anaerobiosis, lactate-dependent oxygen consumption disappeared completely while a small rate of glucose-dependent oxygen consumption was still present. In contrast, in S. aureus increased lactate dehydrogenase expression anaerobiosis has been reported (Fuchs et al. 2007).
The normal habitat for S. epidermidis is the microaerobic environment found in different epidermic and dermic layers (Peyssonnaux et al. 2008). One strategy S. epidermidis uses when confronted with high [O2] is the differential expression of a diverse number of redox enzymes in the respiratory chain. Reports indicate that when microaerophilic or anaerophilic bacteria find a suitable environment, they react manufacturing proteins and polysaccharides that enable them to form biofilms and attach to surfaces at low [O2]. Avoiding high [O2] involves both, anchoring in low oxygen environments and building biofilms as barriers against penetration of ROS or toxic substances (Palikaras and Tavernarakis 2016). Metabolic adaptation has also been reported for Neisseria gonorrhoeae, when it is stimulated to form biofilms. A proteomic analysis of N. gonorrhoeae biofilms evidenced up-regulation of proteins involved in anaerobic metabolism such as glycolysis and TCA cycle plus increased expression of those proteins involved in biofilm generation like pilus-associated proteins (Phillips et al. 2012). In addition, some oxidative stress genes are required for normal biofilm formation in N. gonorrhoeae (Falsetta et al. 2011).
The increase in ATP prior to biofilm formation has been reported in others bacterium. Bacillus brevis and Escherichia coli react to substrate depletion by adhering to glass surfaces and at the same time increase [ATP] two to fivefold as compared to planktonic cells (Hong and Brown 2009). So, the conditions where bacteria need to make biofilms promote saving ATP even at the expense of the growth rate. ATP is most likely needed to synthesize the extracellular proteins and the polysaccharide fibers that anchor cells to surfaces and to each other. Inhibiting ATP production in micro- or anaerobic conditions by adding cyanide or 1,4-bisphosphobutane resulted in a reduced biofilm formation (Fig. 5). This phenomenon is also observed when treating S. epidermidis with the nitrate reductase inhibitor methylamine in anaerobic conditions (Uribe-Alvarez et al. 2016). In contrast, in aerobiosis cyanide promotes biofilm formation (Uribe-Alvarez et al. 2016).
Even when facultative bacteria such as S. epidermidis survive at high [O2], their habitat in the skin is hypoxic to anoxic. While they survive in aerobic environments their susceptibility to ROS-mediated damage and possibly to attack by macrophages increases. They thus present an oxygen avoidance behavior, anchoring and associating in hypoxic environments (Fig. 6). Learning how avoidance works in S. epidermidis and other bacteria would impact both the physiologic and therapeutic field.
Fig. 6.
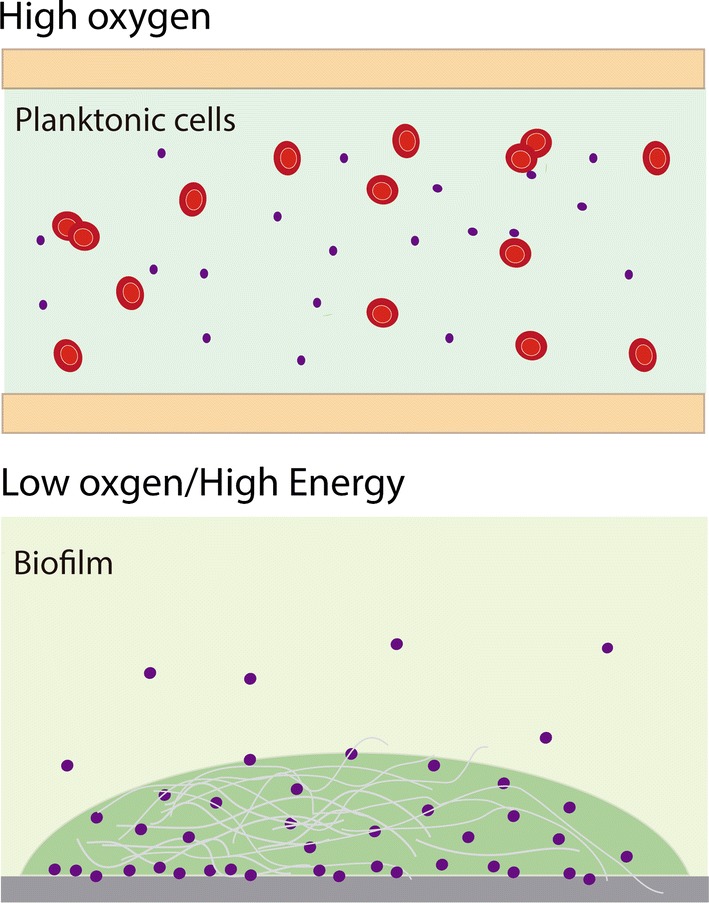
Cartoon depicting the shift that Staphylococcus epidermidis makes when [O2] decreases in the growth medium. When high oxygen concentrations are found in the medium, S. epidermidis are planktonic cells and flow with the blood (top). In contrast, under micro- or anaerobic conditions cells shift to a fermentative metabolism and accumulate ATP adhering to a suitable surface (e.g. epithelia, catheters, artificial valves) and eventually forming a biofilm. In this state the cells exhibit more resistance to H2O2 mediated damage. Excess ATP is probably used to produce adhesion proteins and poly-N-acetylglucosamine (gray fibers in the illustration) (bottom)
Aiming to understand such rise in ATP, we found that other bacteria, e.g. Bacillus brevis and Escherichia coli, react to substrate depletion by adhering to glass surfaces and at the same time increase [ATP] two- to fivefold in comparison to planktonic cells (Hong and Brown 2009). In this regard, it has been reported that hypoxic stimuli induce biofilm formation in S. epidermidis (Uribe-Alvarez et al. 2016).
Acknowledgements
Partially funded by UNAM/DGAPA/PAPIIT grant IN203018 to SUC and CONACYT 241670 grant to AMA. UPD (MsSc) and EES (PhD) are graduate students in the Biochemistry Program at UNAM. LMG is in the Biomedical PhD program at UNAM. OMR is a graduate student at CIAD. UPD, LMG and OMR are CONACYT fellows. CUA present address: Fox-Chase Cancer Center, Philadelphia, PA. Technical help from Ramón Méndez-Franco is acknowledged.
Authors’ contributions
UPD, participated in all experiments and in discussions helping to write and edit the manuscript. CUA contributed to the original idea, she was the first author of the previous paper, participated in oxymetry experiments and in discussions helping to write and edit the manuscript. LMG participated in oxygen-consumption experiments and discussions on the manuscript. EES, participated in fermentation experiments and in discussions. OMR participated in the experiments performed both at CIAD and at UNAM and in discussions. AMA contributed with early ideas and designed some protocols, she provided reagents and facilities at CIAD and edited the manuscript. NCF taught graduate students the techniques involved in each experiment, supervising experimental work and protocols. SUC helped develop the original idea, designed the project, helped in technique application and wrote the manuscript. He provided facilities and found funding. Participated in lab discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Araiza-Olivera D, Chiquete-Felix N, Rosas-Lemus M, Sampedro JG, Peña A, Mujica A, Uribe-Carvajal S. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013;280:3887–3905. doi: 10.1111/febs.12387. [DOI] [PubMed] [Google Scholar]
- Artzatbanov VYu, Petrov VV. Branched respiratory chain in aerobically grown Staphylococcus aureus—oxidation of ethanol by cells and protoplasts. Arch Microbiol. 1990;153:580–584. doi: 10.1007/BF00245268. [DOI] [PubMed] [Google Scholar]
- Baez A, Shiloach J. Effect of elevated oxygen concentration on bacteria, yeasts, and cells propagated for production of biological compounds. Microb Cell Fact. 2014;13:181. doi: 10.1186/s12934-014-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Sehl C, Voortmann S, Verhasselt HL, Edwards MJ, Buer J, Hasenberg M, Gulbins E, Becker KA. Sphingosine is able to prevent and eliminate Staphylococcus epidermidis biofilm formation on different orthopedic implant materials in vitro. J Mol Med. 2019 doi: 10.1007/s00109-019-01858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842X(96)10057-3. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Büttner H, Mack D, Rohde H. Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front Cell Infect Microbiol. 2015;5:14. doi: 10.3389/fcimb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calà C, Amodio E, Di Carlo E, Virruso R, Fasciana T, Giammanco A. Biofilm production in Staphylococcus epidermidis strains, isolated from the skin of hospitalized patients: genetic and phenotypic characteristics. N Microbiol. 2015;38:521–529. [PubMed] [Google Scholar]
- Chabi R, Momtaz H. Virulence factors and antibiotic resistance properties of the Staphylococcus epidermidis strains isolated from hospital infections in Ahvaz, Iran. Trop Med Health. 2019;47:56. doi: 10.1186/s41182-019-0180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, Torpey JW, Otto M, Nizet V, Kim JE, Gallo RL. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130(1):192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter JJ, O’Gara JP, Mack D, Casey E. Oxygen-mediated regulation of biofilm development is controlled by the alternative sigma factor sigma(B) in Staphylococcus epidermidis. Appl Environ Microbiol. 2009;75:261–264. doi: 10.1128/AEM.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty B, Gennaris A, Barras F, Collet J-F. The gain of single electrons by oxygen (O2) generates partially reduced reactive oxygen species (ROS), including superoxide anions (O2·−. Nat Publ Gr. 2017 doi: 10.1038/nrmicro.2017.26. [DOI] [Google Scholar]
- Falsetta ML, Steichen CT, McEwan AG, Cho C, Ketterer M, Shao J, Hunt J, Jennings MP, Apicella MA. The composition and metabolic phenotype of Neisseria gonorrhoeae biofilms. Front Microbiol. 2011;2:75. doi: 10.3389/fmicb.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Frawley ER, Tapscott T, Vázquez-Torres A. Bacterial stress responses during host infection. Cell Host Microbe. 2016;20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MT, Manso AS, Gaspar P, Pinho MG, Neves AR. Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE. 2013;8:e58277. doi: 10.1371/journal.pone.0058277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornall AG, Bardawill CJ, David MM. Determination of serum proteins by means of the biuret reaction. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- Götz F, Mayer S. Both terminal oxidases contribute to fitness and virulence during organ-specific Staphylococcus aureus colonization. MBio. 2013;4:e00976-13. doi: 10.1128/mBio.00976-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristina AG. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- Hartman FC, Barker R. An exploration of the active site of aldolase using structural analogs of fructose diphosphate*. Biochemistry. 1965;4:1068–1075. doi: 10.1021/bi00882a014. [DOI] [PubMed] [Google Scholar]
- Hong Y, Brown DG. Variation in bacterial ATP level and proton motive force due to adhesion to a solid surface. Appl Environ Microbiol. 2009;75:2346–2353. doi: 10.1128/AEM.02671-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- Jensen ET, Kharazmi A, Høiby N, Costerton JW. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS. 1992;100:727–733. doi: 10.1111/j.1699-0463.1992.tb03991.x. [DOI] [PubMed] [Google Scholar]
- Kane AL, Brutinel ED, Joo H, Maysonet R, VanDrisse CM, Kotloski NJ, Gralnick JA. Formate metabolism in Shewanella oneidensis generates proton motive force and prevents growth without an electron acceptor. J Bacteriol. 2016;198:1337–1346. doi: 10.1128/JB.00927-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. Oxygen: the molecule that made the world. Oxford: Oxford University Press; 2002. [Google Scholar]
- Leid JG. Bacterial biofilms resist key host defenses. Microbe. 2009;4:66–70. [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Lobritz MA, Belenky P, Porter CBM, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Davies AP, Harris LG, Jeeves R, Pascoe B, Knobloch JK-M, Rohde H, Wilkinson TS. Biomaterials associated infection. New York: Springer; 2013. Staphylococcus epidermidis in biomaterial-associated infections; pp. 25–56. [Google Scholar]
- Macvanin M, Hughes D. Methods in molecular biology. Clifton: Humana Press; 2010. Assays of sensitivity of antibiotic-resistant bacteria to hydrogen peroxide and measurement of catalase activity; pp. 95–103. [DOI] [PubMed] [Google Scholar]
- Mendoza-Hoffmann F, Pérez-Oseguera Á, Cevallos MÁ, Zarco-Zavala M, Ortega R, Peña-Segura C, Espinoza-Simón E, Uribe-Carvajal S, García-Trejo JJ. The biological role of the ζ subunit as unidirectional inhibitor of the F1FO-ATPase of Paracoccus denitrificans. Cell Rep. 2018;22:1067–1078. doi: 10.1016/j.celrep.2017.12.106. [DOI] [PubMed] [Google Scholar]
- Painter KL, Hall A, Ha KP, Edwards AM. The electron transport chain sensitizes Staphylococcus aureus and Enterococcus faecalis to the oxidative burst. Infect Immun. 2017 doi: 10.1128/IAI.00659-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Tavernarakis N. Intracellular assessment of ATP levels in Caenorhabditis elegans. Bio-Protocol. 2016 doi: 10.21769/BioProtoc.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C, Boutin AT, Zinkernagel AS, Datta V, Nizet V, Johnson RS. Critical role of HIF-1α in keratinocyte defense against bacterial infection. J Invest Dermatol. 2008;128:1964–1968. doi: 10.1038/JID.2008.27. [DOI] [PubMed] [Google Scholar]
- Phillips NJ, Steichen CT, Schilling B, Post DMB, Niles RK, Bair TB, Falsetta ML, Apicella MA, Gibson BW. Proteomic analysis of Neisseria gonorrhoeae biofilms shows shift to anaerobic respiration and changes in nutrient transport and outermembrane proteins. PLoS ONE. 2012;7:e38303. doi: 10.1371/journal.pone.0038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz M, Staats K, Tobudic S, Assadian O, Presterl E, Windhager R, Holinka J. Zirconium nitride coating reduced Staphylococcus epidermidis biofilm formation on orthopaedic implant surfaces: an in vitro study. Clin Orthop Relat Res. 2019;477:461–466. doi: 10.1097/CORR.0000000000000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, Orr AL, Brand MD. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol. 2013;1:304–312. doi: 10.1016/j.redox.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raad I, Alrahwan A, Rolston K. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin Infect Dis. 1998;26:1182–1187. doi: 10.1086/520285. [DOI] [PubMed] [Google Scholar]
- Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Fut Med Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- Rosas-Lemus M, Chiquete-Félix N, Ruíz-Pérez K, Rigoulet M, Devin A, Hernández-Rodríguez M, Uribe-Carvajal S. Sensitivity of the mitochondrial unspecific channel of Saccharomyces cerevisiae to butane-1,4-bisphosphate, a competitive inhibitor of fructose-1,6-bisphosphate-aldolase. ChemistrySelect. 2016;1:2930–2934. doi: 10.1002/slct.201600303. [DOI] [Google Scholar]
- Rosas-Lemus M, Uribe-Alvarez C, Contreras-Zentella M, Luévano-Martínez LA, Chiquete-Félix N, Morales-García NL, Simón EE, Muhlia-Almazán A, Escamilla-Marván E, Uribe-Carvajal S. Free radicals and diseases. InTech: Rijeka; 2016. Oxygen: from toxic waste to optimal (toxic) fuel of life. [Google Scholar]
- Roujansky A, Martin M, Gomart C, Hulin A, Mounier R. Multidrug-resistant Staphylococcus epidermidis ventriculostomy-related infection successfully treated by intravenous ceftaroline after failure of daptomycin treatment. World Neurosurg. 2020 doi: 10.1016/j.wneu.2020.01.013. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Alvarez C, Chiquete-Félix N, Contreras-Zentella M, Guerrero-Castillo S, Peña A, Uribe-Carvajal S. Staphylococcus epidermidis: metabolic adaptation and biofilm formation in response to different oxygen concentrations. Pathog Dis. 2016;74:ftv111. doi: 10.1093/femspd/ftv111. [DOI] [PubMed] [Google Scholar]
- von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2:677–685. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med. 2004;351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]