Abstract
Drug resistance is the major obstacle of gemcitabine-based chemotherapy for the treatment of pancreatic ductal adenocarcinoma (PDAC). Many long non-coding RNAs (lncRNAs) are reported to play vital roles in cancer initiation and progression. Here, we report that lncRNA SLC7A11-AS1 is involved in gemcitabine resistance of PDAC. SLC7A11-AS1 is overexpressed in PDAC tissues and gemcitabine-resistant cell lines. Knockdown of SLC7A11-AS1 weakens the PDAC stemness and potentiates the sensitivity of resistant PDAC cells toward gemcitabine in vitro and in vivo. SLC7A11-AS1 promotes chemoresistance through reducing intracellular reactive oxygen species (ROS) by stabilizing nuclear factor erythroid-2-related factor 2 (NRF2), the key regulator in antioxidant defense. Mechanically, SLC7A11-AS1 is co-localized with β-TRCP1 in the nucleus. The exon 3 of SLC7A11-AS1 interacts with the F-box motif of β-TRCP1, the critical domain that recruits β-TRCP1 to the SCFβ-TRCP E3 complex. This interaction prevents the consequent ubiquitination and proteasomal degradation of NRF2 in the nucleus. Our results demonstrate that the overexpression of SLC7A11-AS1 in gemcitabine-resistant PDAC cells can scavenge ROS by blocking SCFβ-TRCP-mediated ubiquitination and degradation of NRF2, leading to a low level of intracellular ROS, which is required for the maintenance of cancer stemness. These findings suggest SLC7A11-AS1 as a therapeutic target to overcome gemcitabine resistance for PDAC treatment.
Keywords: SLC7A11-AS1, β-TRCP1, NRF2, gemcitabine, pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive malignancy, characterized by late diagnosis, low 5-year survival rate (9% for all stages), and resistance to chemotherapy.1,2 Gemcitabine (2′, 2′-difluoro-2′-deoxycytidine [dFdC]) has been widely used for the treatment of locally advanced and metastatic PDAC; however, innate and acquired drug resistances have been major barriers in gemcitabine-based therapy.3,4 Emerging evidence demonstrates that a subset of cancer cells, called cancer stem cells (CSCs), are highly resistant to chemotherapy and are responsible for drug resistance and cancer recurrence. Gemcitabine eliminates the bulk cancer cells, leading to enrichment of the CSCs in PDAC.5
A low level of intracellular reactive oxygen species (ROS) is required for the maintenance of cancer stemness.6 Chemotherapeutic agents such as gemcitabine stimulate ROS generation to induce cell death.4 To survive under oxidative stress, chemoresistant cancer cells develop an effective antioxidant system to limit the excessive accumulation of ROS.7 Nuclear factor erythroid-2-related factor 2 (NRF2), the key regulator of redox hemostasis, is frequently observed to be overexpressed in CSCs in contrast with the bulk cancer cells.8 In addition, NRF2 is overexpressed in PDAC tissues and confers their chemoresistance.9,10 NRF2 is tightly regulated by the ubiquitin-proteasome system.11 Two major E3 ligases are involved in NRF2 proteasomal degradation: the KEAP1-Cul3-Rbx1 and the SKP1-Cul1-Rbx1 (SCFβ-TRCP) E3 complex, which mediate NRF2 ubiquitination and subsequent degradation in the cytosol and nucleus, respectively.12,13 Intriguingly, NRF2 protein level is not negatively correlated to the KEAP1 expression in PDAC.9 In addition, the sustained activation of NRF2 in human PDAC cannot be explained by somatic mutations in NRF2 or KEAP1.14 Furthermore, high levels of β-TRCP1 are also detected in the specimens of PDAC.15 Thus, it is necessary to find the underlying mechanism responsible for elevated levels of NRF2 in PDAC, which might be of potential therapeutic value in overcoming gemcitabine resistance.
Long non-coding RNAs (lncRNAs) provide a new perspective in understanding complicated signal transduction. They are transcribed by polymerase II with greater than 200 nt. Increasing evidence demonstrates the crucial roles of lncRNAs in the tumorigenesis and chemoresistance of PDAC.16, 17, 18, 19, 20 For example, Neat1 has been reported to suppress pancreatic cancer initiation in a p53-dependent manner.18 lncRNA MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1) was found to promote tumorigenicity and reduce chemosensitivity of PDAC cells.19 Our previous work indicates that linc-DYNC2H1-4 promotes PDAC stemness by acting as a sponge of miR-145.20
In this study, we found that lncRNA SLC7A11-AS1 was overexpressed in gemcitabine-resistant PDAC cells and was involved in drug resistance. It interacts with the F-box motif of β-TRCP1, preventing NRF2 ubiquitination and subsequent proteasomal degradation in the nucleus, which in turn reduces intracellular ROS for the maintenance of PDAC stemness and chemoresistance.
Results
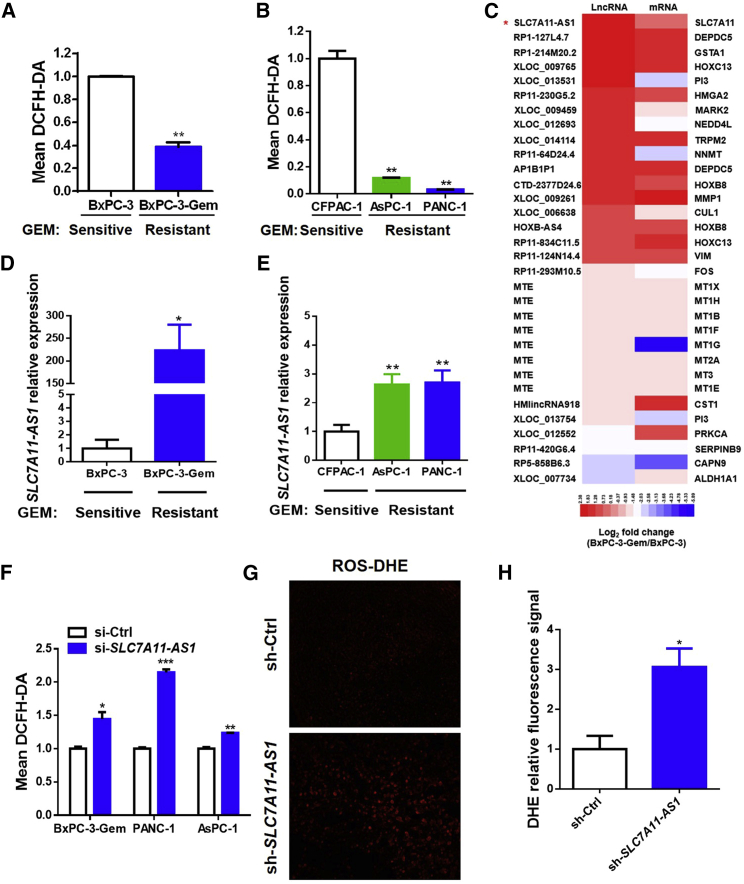
Overexpression of SLC7A11-AS1 in Gemcitabine-Resistant PDAC Cells Reduces the Intracellular Level of ROS
Previously, we established a gemcitabine-resistant subline BxPC-3-Gem (Figure S1A) from the parental sensitive pancreatic cancer BxPC-3 cells and found that gemcitabine-resistant PDAC cells had high cancer stemness properties.20 Because a low level of ROS is required for the maintenance of stemness,6 we determined the ROS level in this paired cell line and found that it was lower in gemcitabine-resistant BxPC-3-Gem than that in the sensitive cells (Figure 1A). Low levels of intracellular ROS were also observed in the other two gemcitabine-resistant cell lines, PANC-1 and AsPC-1 (Figure S1B), in comparison with the sensitive CFPAC-1 cells (Figure 1B). To see lncRNA involvement in ROS regulation, we performed array analysis and enriched lncRNAs with potential involvement in ROS regulation according to the nearby associated genes. Among them, lncRNA SLC7A11-AS1 was the gene that was in highest expression in BxPC-3-Gem compared with BxPC-3 cells (Figure 1C). qRT-PCR confirmed that SLC7A11-AS1 was overexpressed in BxPC-3-Gem compared with BxPC-3 cells (Figure 1D). Different expression levels of SLC7A11-AS1 were also observed in gemcitabine-resistant PANC-1 and AsPC-1 in comparison with the sensitive CFPAC-1 cells (Figure 1E). Next, we knocked down SLC7A11-AS1 in gemcitabine-resistant cell lines to determine its effect in ROS regulation. Three small interfering RNAs (siRNAs) were designed, and siRNA#1 could knock down SLC7A11-AS1 expression by 50% (Figure S1C); thus, it was used in further study. Knockdown of SLC7A11-AS1 in gemcitabine-resistant BxPC-3-Gem, PANC-1, and AsPC-1 cells (Figure S1D) led to significant elevation of intracellular ROS levels (Figure 1F), indicating that SLC7A11-AS1 acts as an antioxidant. To see the in vivo effect, PANC-1 cells with SLC7A11-AS1-knockdown or control shRNA (Figure S1E) were subcutaneously inoculated into BALB/c nude mice. PANC-1 xenograft tissues with SLC7A11-AS1 knockdown showed near 3-fold increase of ROS levels in comparison with control (Figures 1G and 1H). These results indicate that overexpression of SLC7A11-AS1 in gemcitabine-resistant PDAC cells can reduce the intracellular level of ROS.
Figure 1.
Overexpression of SLC7A11-AS1 in Gemcitabine-Resistant PDAC Cells Reduces Intracellular ROS Level
(A and B) PDAC cells with acquired (A, BxPC-3-Gem vs BxPC-3) or innate (B, PANC-1 and AsPC-1 vs CFPAC-1) gemcitabine resistance were incubated with probe DCFH-DA (10 μM) for 30 min. ROS levels were detected by flow cytometry. (C) The log2 fold changes of lncRNAs and their nearby coding genes that are associated with ROS regulation were presented by heatmap. (D and E) Expression of SLC7A11-AS1 in PDAC cell lines with aquired (D) or innate drug resistance (E) was detected by qRT-PCR and normalized to GAPDH (n = 3). (F) ROS levels were determined in gemcitabine-resistant PDAC cells with SLC7A11-AS1 knockdown or control as described in (A) and (B). (G and H) SLC7A11-AS1-knockdown and control PANC-1 cells (1.5 × 106) were inoculated in BALB/c nude mice (n = 5). Slides from tumor tissues were incubated with DHE for in situ detection of ROS. (G and H) Fluorescence was detected by fluorescent microscope (G) and quantified by using ImageJ (H) (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
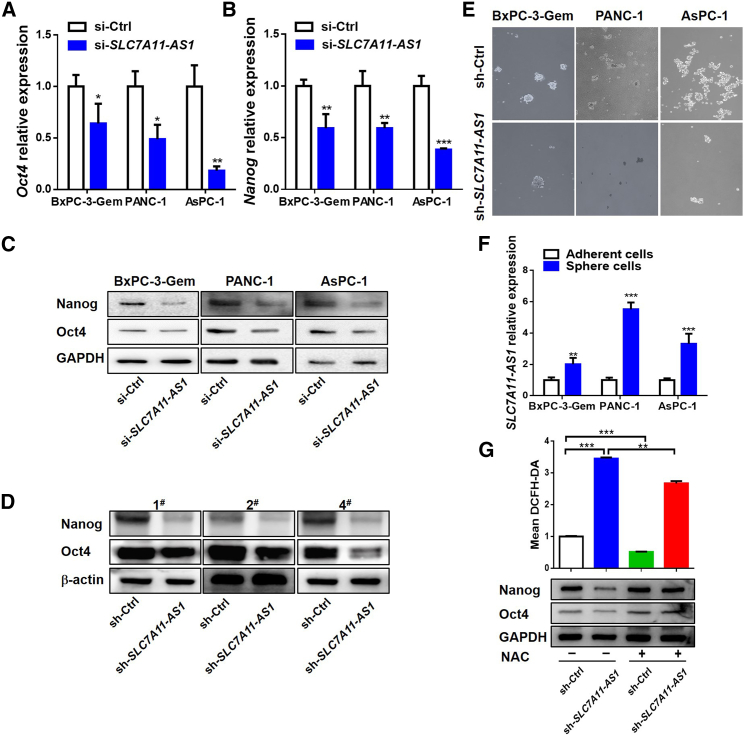
SLC7A11-AS1 Scavenges ROS to Promote Cancer Stemness
Because a low level of ROS is required for the maintenance of cancer stemness,6 we further investigated whether SLC7A11-AS1 could affect cancer stemness. Oct4 and Nanog, the two markers of stemness, were significantly decreased in SLC7A11-AS1-knockdown BxPC-3-Gem, PANC-1, and AsPC-1 cells (Figure S1D) in comparison with control at both mRNA and protein levels (Figures 2A–2C). In vivo, the tumors derived from the SLC7A11-AS1 knockdown group exhibited lower protein levels of Nanog and Oct4 than the control group (Figure 2D). Knockdown of SLC7A11-AS1 in the three gemcitabine-resistant cell lines by shRNA (Figure S1E) significantly suppressed their sphere-forming abilities, a representative trait of CSCs21 (Figure 2E). In addition, compared with the adherent cells, the spheroids exhibited higher expression level of SLC7A11-AS1 (Figure 2F). These results indicate that SLC7A11-AS1 promotes cancer stemness in PDAC cells.
Figure 2.
SLC7A11-AS1 Scavenges ROS to Promote Cancer Stemness
(A–C) Effects of SLC7A11-AS1 knockdown on expressions of Oct4 and Nanog were determined by qRT-PCR (A and B) and western blot (C) in gemcitabine-resistant PDAC cells (n = 3). (D) Expressions of Oct4 and Nanog in tumor tissues (as described in Figure 1G) were determined by western blot (D). (E) Oncosphere-formation assay in SLC7A11-AS1-knockdown and control gemcitabine-resistant PDAC cells. Original magnification ×10. (F) qRT-PCR analysis of SLC7A11-AS1 expression in spheroids and paired attached PDAC cells (n = 3). (G) Flow cytometry analysis of ROS using probe DCFH-DA (upper) and western blot analysis of Oct4 and Nanog (bottom) in SLC7A11-AS1-knockdown and control PANC-1 cells treated with or without NAC (10 mM) for 48 h (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
Then we further determined whether SLC7A11-AS1 promotes cancer stemness through reducing ROS level. We introduced N-acetylcysteine (NAC), a scavenger of ROS. The use of NAC decreased SLC7A11-AS1 silencing-induced ROS (Figure 2G, upper), which also reduced its impact on stemness. SLC7A11-AS1 silencing-induced protein decreases of Nanog and Oct4 were rescued to control level by addition of NAC (Figure 2G, bottom), indicating that SLC7A11-AS1 regulates cancer stemness via regulating ROS level in gemcitabine-resistant PDAC cells.
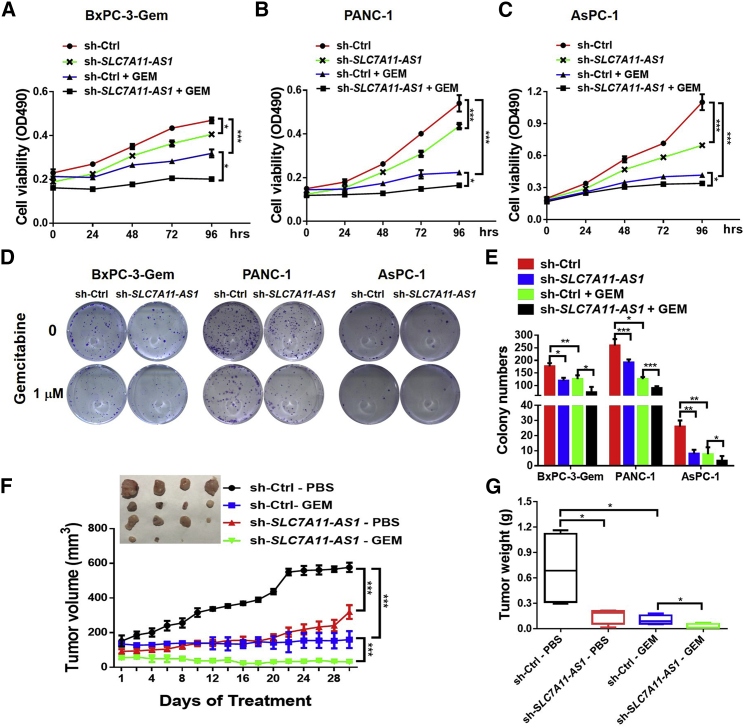
SLC7A11-AS1 Promotes Chemoresistance in PDAC
Cancer stemness confers chemoresistance.22 Next, we investigated SLC7A11-AS1 involvement in chemoresistance. Knockdown of SLC7A11-AS1 caused growth inhibition and further potentiated gemcitabine efficacy in resistant BxPC-3-Gem, PANC-1, and AsPC-1 cells (Figures 3A–3C). Moreover, colony formation showed an almost 2-fold decrease of colony numbers after gemcitabine treatment in SLC7A11-AS1-knockdown BxPC-3-Gem, PANC-1, and AsPC-1 cells compared with control (Figures 3D and 3E). To see SLC7A11-AS1 effects in vivo, PANC-1 xenografts with SLC7A11-AS1 knockdown or control were established subcutaneously in BALB/c mice, followed by gemcitabine treatment every 4 days when control tumor reached 150 mm3. SLC7A11-AS1 knockdown significantly potentiated gemcitabine efficacy as shown by the approximately 80% decrease of tumor growth and 77% decrease of tumor weight in the SLC7A11-AS1-knockdown plus gemcitabine group in comparison with gemcitabine alone group (Figures 3F and 3G). These results indicate that SLC7A11-AS1 can potentiate cancer cell sensitivity toward gemcitabine, suggesting it might be of potential therapeutic value to overcome gemcitabine resistance in PDAC.
Figure 3.
SLC7A11-AS1 Knockdown Potentiates Resistant PDAC Cell Response to Gemcitabine
(A–C) SLC7A11-AS1-knockdown and control (A) BxPC-3-Gem, (B) PANC-1, and (C) AsPC-1 cells were treated with gemcitabine (1 μM) for indicated times, and cell viability was analyzed by MTT assay (n = 3). (D and E) Colony formation assays of SLC7A11-AS1-knockdown and control cells with gemcitabine treatment (1 μM) for 2 weeks (n = 3). Representative images (D) and average number of colonies (E) were shown. (F and G) Subcutaneous xenograft analysis of SLC7A11-AS1-knockdown and control PANC-1 cells (1.5 × 106) in nude mice treated with gemcitabine (50 mg/kg body weight) or PBS by intraperitoneal (i.p.) injection every 4 days (n = 4). Tumor volume (F) and weight (G) were shown. *p < 0.05, **p < 0.01, ***p < 0.001.
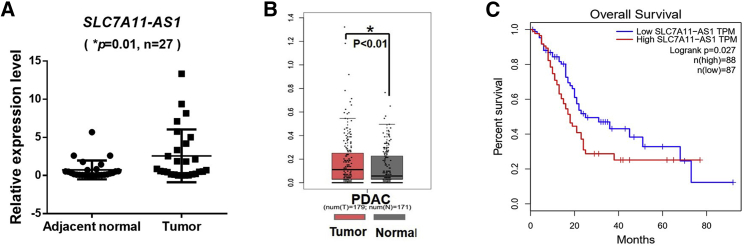
SLC7A11-AS1 Correlates with Poor Prognosis of PDAC Patients
To see the clinical significance of SLC7A11-AS1, we determined SLC7A11-AS1 expressions in PDAC patients and found that it was higher in tumor tissues than that in the adjacent normal tissues (n = 27) (Figure 4A). A large-scale dataset analysis by using Gene Expression Profiling Integrative Analysis (GEPIA; http://gepia.cancer-pku.cn)23 confirmed that SLC7A11-AS1 was overexpressed in PDAC tissues (n = 179) compared with normal tissues (n = 171) (p < 0.01) (Figure 4B). Moreover, Kaplan-Meier survival analysis indicated that high expression of SLC7A11-AS1 in PDAC tissues was associated with a shorter overall lifespan of PDAC patients (p = 0.027) (Figure 4C), indicating that high level of SLC7A11-AS1 correlates with poor prognosis of PDAC patients.
Figure 4.
SLC7A11-AS1 Expression Levels Are Correlated with PDAC Patient’s Survival
(A) Expression of SLC7A11-AS1 in tumor and adjacent normal tissues of PDAC patients was determined by qRT-PCR and normalized to 18S rRNA (n = 27). (B) The expression level of SLC7A11-AS1 in PDAC and normal tissues was analyzed by using GEPIA. (C) Kaplan-Meier curve of overall survival of PDAC patients analyzed by using GEPIA.
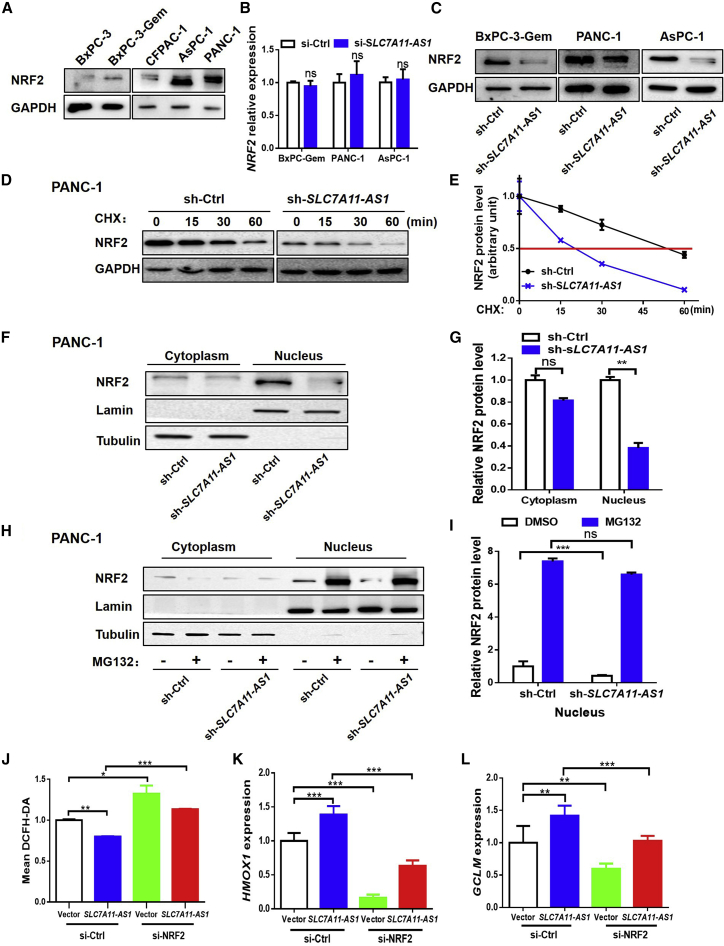
SLC7A11-AS1 Prevents NRF2 Proteasomal Degradation to Defend ROS
Next, we were interested to know how SLC7A11-AS1 affected ROS level in PDAC cells. NRF2 is the key regulator of redox hemostasis.24 We noticed that NRF2 protein level was in line with the endogenous expression level of SLC7A11-AS1, showing that PDAC cells (BxPC-3-Gem, PANC-1, and AsPC-1) with highly expressed SLC7A11-AS1 had high protein levels of NRF2, and conversely, cells with lowly expressed SLC7A11-AS1 had low protein levels of NRF2 (Figure 5A versus Figures 1D and 1E). These results suggest that SLC7A11-AS1 might regulate NRF2 in PDAC cells. Knockdown of SLC7A11-AS1 in SLC7A11-AS1-overexpressed BxPC-3-Gem, PANC-1, and AsPC-1 cells had no effect on NRF2 mRNA levels (Figure 5B). However, it led to reduction of NRF2 protein in the three gemcitabine-resistant PDAC cells (Figure 5C). Then we determined whether SLC7A11-AS1 might affect the stability of NRF2 protein. We treated the cells with protein synthesis inhibitor cycloheximide (CHX) and found that knockdown of SLC7A11-AS1 reduced the stability of NRF2 protein (Figures 5D and 5E). The half-life of NRF2 protein was shortened by about 3-fold by SLC7A11-AS1 knockdown in PANC-1 cells (Figure 5E). Subcellular fractionation revealed that the nuclear portion of NRF2 protein was significantly decreased upon SLC7A11-AS1 knockdown, but not the cytosolic portion (Figures 5F and 5G). MG132, a specific proteasome inhibitor, could completely rescue the loss of nuclear NRF2 protein caused by SLC7A11-AS1 knockdown (Figures 5H and 5I), indicating that SLC7A11-AS1 stabilizes nuclear NRF2 by preventing its proteasomal degradation.
Figure 5.
SLC7A11-AS1 Prevents NRF2 Proteasomal Degradation to Defend ROS
(A) Western blot analysis of NRF2 in PDAC cell lines. (B and C) Effects of SLC7A11-AS1 knockdown on NRF2 expression were determined by qRT-PCR (B) and western blot (C) in SLC7A11-AS1-overexpressed PDAC cells (n = 3). (D) Effect of SLC7A11-AS1 knockdown on half-life of NRF2 was determined by western blot in PANC-1 cells treated with CHX (100 μg/mL). (E) Quantification of NRF2 was normalized to the loading control and expressed relative to 0 h. (F) NRF2 protein levels in the nucleus and cytosol in PANC-1 cells with SLC7A11-AS1 knockdown or sh-Ctrl were determined by western blot. (G) Quantifications of nuclear and cytoplasmic NRF2 were normalized to the loading control and expressed relative to sh-Ctrl. (H) Western blot analysis of nuclear and cytoplasmic fractions from SLC7A11-AS1-knockdown and control PANC-1 cells treated with or without MG132 (1 μM) for 24 h. (I) Quantification of nuclear NRF2 as in (H) (n = 3). (J–L) PANC-1 cells with or without siRNA-mediated NRF2 knockdown were transfected with SLC7A11-AS1 or empty vector for 48 h, followed by ROS detection using flow cytometry (J), qRT-PCR analysis of NRF2 target genes, HMOX1 (K), and GCLM (L). *p < 0.05, **p < 0.01, ***p < 0.001. ns, not significant.
Then, we determined whether SLC7A11-AS1 exhibited antioxidant ability through NRF2 regulation. Flow cytometry (FCM) analysis showed that ectopic expression of SLC7A11-AS1 caused ROS reduction, whereas it lost antioxidant function when NRF2 was silenced by siRNA (Figure 5J). NRF2 activates the expressions of target antioxidant genes such as GCLM and HMOX1.25,26 qRT-PCR showed that ectopic expression of SLC7A11-AS1 upregulated the expressions of GCLM and HMOX1, whereas the upregulations were decreased to or even under basal level when NRF2 was knocked down (Figures 5K and 5L). These results indicate that SLC7A11-AS1 prevents the proteasomal degradation of nuclear NRF2, leading to decreased ROS.
SLC7A11-AS1 Interacts with β-TRCP1 to Block NRF2 Ubiquitination in the Nucleus
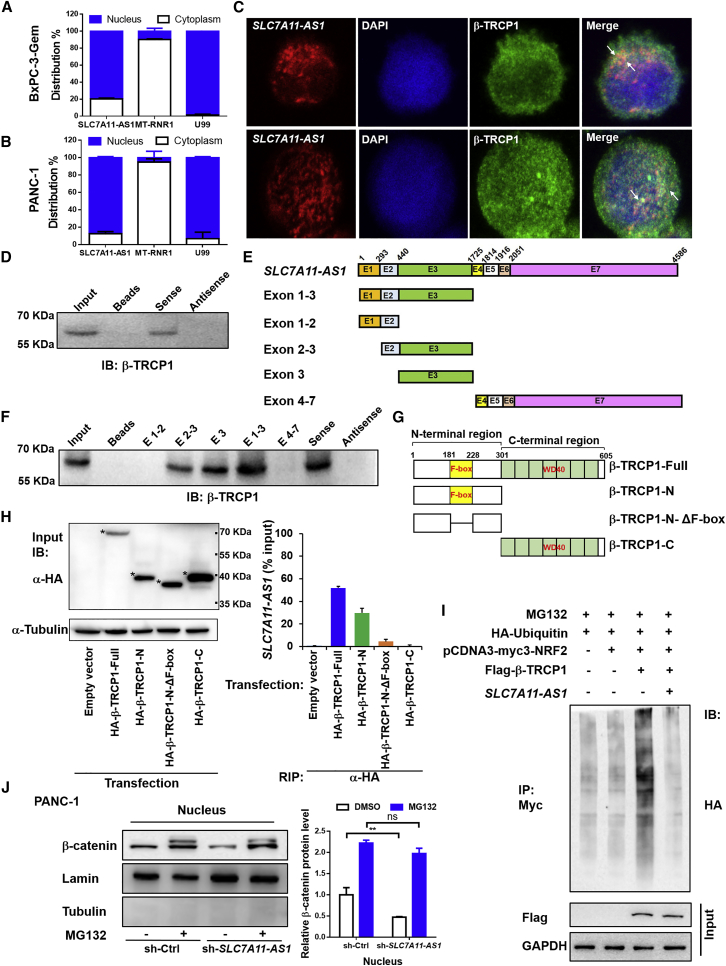
To see the underlying mechanism that SLC7A11-AS1 prevents NRF2 proteasomal degradation in the nucleus, we determined the intracellular location of SLC7A11-AS1 in BxPC-3-Gem and PANC-1 cells. qRT-PCR showed that SLC7A11-AS1 was mainly located in the nucleus, and it was marginally detected in the cytosol (Figures 6A and 6B). Considering that SLC7A11-AS1 regulates NRF2 posttranscription and lncRNAs within the nucleus function through protein interaction,27,28 we hypothesized that SLC7A11-AS1 might be interfering with a nuclear E3 ligase to affect the stability of NRF2. β-TRCP1 is reported to be responsible for NRF2 ubiquitination and proteasomal degradation in the nucleus.13 Therefore, we performed nuclear co-localization of β-TRCP1 with SLC7A11-AS1. Immunofluorescence showed that β-TRCP1 and SLC7A11-AS1 were co-localized in the nucleus of BxPC-3-Gem and PANC-1 cells (Figure 6C).
Figure 6.
SLC7A11-AS1 Prevents β-TRCP1-Mediated Ubiquitination and Degradation of NRF2
(A and B) Subcellular distribution of SLC7A11-AS1 was detected by qRT-PCR in (A) BxPC-3-Gem and (B) PANC-1 cells. U99 and MT-RNR1 were as nuclear and cytoplasmic marker, respectively (n = 3). (C) The immunofluorescence staining of SLC7A11-AS1 (red), β-TRCP1 (green), and DAPI (blue) in BxPC-3-Gem and PANC-1 cells. (D) Nuclear extracts from BxPC-3-Gem cells were incubated with biotinylated sense and antisense SLC7A11-AS1 generated in vitro, and proteins were precipitated with streptavidin beads and subjected to immunoblotting (IB) analysis with anti-TRCP1 antibody. (E) A schematic diagram of full-length SLC7A11-AS1 and its series of truncates. (F) Nuclear extracts from BxPC-3-Gem cells were incubated with biotinylated SLC7A11-AS1 truncates and antisense SLC7A11-AS1 generated in vitro, and proteins were precipitated with streptavidin beads and subjected to IB analysis with anti-TRCP1 antibody. (G) A schematic diagram of β-TRCP1 and its truncates. (H) RIP assay analysis of the interaction of β-TRCP1 and its truncates (β-TRCP1-N, β-TRCP1-N-ΔF-box, and β-TRCP1-C) with SLC7A11-AS1 in PANC-1 cells. Whole-cell expression (input) of proteins was detected by IB with indicated antibodies (left). The asterisks indicate β-TRCP1 and its truncates bands. The immunoprecipitated SLC7A11-AS1 by using anti-HA antibody was measured by qRT-PCR and represented as a fraction of input RNA (% input) prior to immunoprecipitation (right) (n = 3). (I) Effect of SLC7A11-AS1 on β-TRCP1-mediated ubiquitination of NRF2. 293T cells co-transfected with indicated plasmids were treated with MG132 (1 μM) for 12 h and subjected to immunoprecipitation (IP) with the anti-Myc antibody, followed by IB with anti-HA antibody. Whole-cell expression (input) of proteins was detected by IB with anti-FLAG or anti-GAPDH antibodies. (J) SLC7A11-AS1 prevents β-catenin proteasomal degradation. Western blot (left) analysis of nuclear β-catenin in SLC7A11-AS1-knockdown and control PANC-1 cells treated with or without MG132 (5 μM) for 24 h. Quantification of β-catenin protein (right) was normalized to the loading control (Lamin) and expressed relative to sh-Ctrl without MG132 (n = 3). **p < 0.01. ns, not significant.
To see whether SLC7A11-AS1 could interact with β-TRCP1, nuclear extracts from BxPC-3-Gem cells were incubated with biotinylated sense or antisense SLC7A11-AS1 RNA generated in vitro. Proteins precipitated with streptavidin beads were resolved by SDS-PAGE. The association of β-TRCP1 with SLC7A11-AS1 sense RNA, but not antisense or the beads, was confirmed by immunoblotting analysis with anti-β-TRCP1 antibody (Figure 6D). To identify the regions of SLC7A11-AS1 that are responsible for binding with β-TRCP1, we generated five fragments of SLC7A11-AS1 (Figure 6E). The results from RNA pull-down and immunoblotting showed that SLC7A11-AS1 exon 3 (440–1,725 nt of SLC7A11-AS1) was essential for the interaction with β-TRCP1 protein (Figure 6F). Reciprocally, RNA immunoprecipitation (RIP) assay was performed to confirm the interaction between SLC7A11-AS1 and β-TRCP1. β-TRCP1 has two major domains: the F-box domain, which is responsible for recruiting β-TRCP1 to the SKP1-Cul1-Rbx1 (SCFβ-TRCP) E3 ligase complex through interaction with adaptor protein SKP1; and the WD domain responsible for interaction with the substrates.29 To see which domain is responsible for interacting with SLC7A11-AS1, β-TRCP1 fragments with deletions of either one (β-TRCP1-N without WD domain and β-TRCP1-C without F-box motif) or both domains (β-TRCP1-N-ΔF-box) were constructed (Figure 6G). Western blot using input lysates from PANC-1 cells transfected with hemagglutinin (HA)-tagged β-TRCP1 and its truncations confirmed their expressions (Figure 6H, left). The immunoprecipitates revealed that SLC7A11-AS1 was enriched by β-TRCP1 antibody. Almost 50% of input SLC7A11-AS1 could be pulled down by full-length β-TRCP1 (Figure 6H, right). However, β-TRCP1 fragment without F-box domain (β-TRCP1-C, containing WD) completely lost its interaction with SLC7A11-AS1, showing a SLC7A11-AS1 level similar to empty vector. β-TRCP1 fragment without WD domain (β-TRCP1-N, containing F-box motif) could interact with SLC7A11-AS1, but its binding ability dropped nearly 40% compared with the full-length β-TRCP1 (Figure 6H, right). Further deletion showed that β-TRCP1 fragment without both F-box and WD domains (β-TRCP1-N-ΔF-box) was not able to interact with SLC7A11-AS1; the binding activity dropped ∼90% (Figure 6H, right). These results demonstrate that the F-box domain of β-TRCP1 is responsible for its direct interaction with SLC7A11-AS1, but it needs the WD domain to enhance the binding ability.
Next, we determined whether the interaction between SLC7A11-AS1 and β-TRCP1 was functional. As shown in Figure 6I, β-TRCP1 led to NRF2 ubiquitination, but it was blocked in the presence of SLC7A11-AS1 in 293T cells (Figure 6I), indicating that the interaction between SLC7A11-AS1 and β-TRCP1 blocks NRF2 ubiquitination. The F-box domain is responsible for β-TRCP1 interacting with SKP1 of the SCFβ-TRCP1 E3 complex. If this domain was blocked by SLC7A11-AS1, the degradation of other substrates of SCFβ-TRCP1 E3 would be affected as well. Indeed, we found that the proteasomal degradation of nuclear β-catenin, another substrate of SCFβ-TRCP1,30 was blocked by SLC7A11-AS1 (Figure 6J), further confirming that SLC7A11-AS1 interacts with the F-box of β-TRCP1.
Discussion
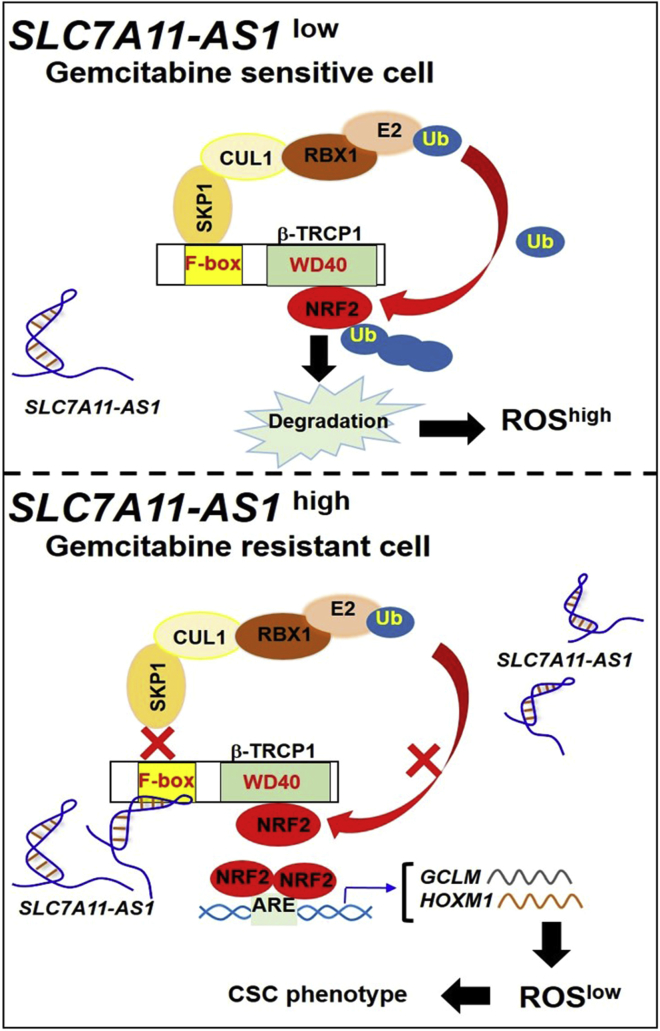
lncRNAs play important roles in governing cell response to chemotherapeutics.31,32 In the current study, we found that SLC7A11-AS1 promotes cancer stemness and chemoresistance by scavenging ROS. SLC7A11-AS1 stabilizes nuclear NRF2 protein by blocking SCFβ-TRCP-mediated ubiquitination and subsequent proteasomal degradation, leading to a low level of intracellular ROS, which is required for the maintenance of PDAC stemness and chemoresistance (Figure 7).
Figure 7.
A Proposed Working Diagram of SLC7A11-AS1
SLC7A11-AS1 stabilizes NRF2 by interacting with β-TRCP1 to lower ROS for the maintenance of PDAC stemness.
SLC7A11-AS1 is the anti-sense transcript of SLC7A11 that encodes xCT, a transporter of glutamine and cysteine.33 It has been reported as a tumor suppressor in gastric and epithelial ovarian cancer, which is downregulated in these tumor tissues compared with the adjacent non-tumorous counterparts.33,34 Our data indicate that SLC7A11-AS1 is an oncogene in PDAC that contributes to sphero-formation and the upregulation of CSC markers. Furthermore, overexpression of SLC7A11-AS1 confers to gemcitabine resistance of PDAC cells both in vitro and in vivo. High expressions of SLC7A11-AS1 are detected in human PDAC specimens and associate with a shorter overall lifespan of PDAC patients, further supporting the oncogenic function of SLC7A11-AS1 in PDAC. The discrepancy of SLC7A11-AS1 function in gastric, epithelial ovary cancer and PDAC might be because of different cancer types.
Accumulating evidence has demonstrated that CSCs are responsible for tumorigenesis, chemoresistance, metastasis, and progression,35 and a low level of ROS is required for the maintenance of cancer stemness.6 PDAC cells resistant to gemcitabine exert enhanced property of cancer stemness.20,36 Here, we found that the resistant PDAC cells also showed a decreased level of ROS in comparison with the sensitive cells. SLC7A11-AS1 is overexpressed in gemcitabine-resistant PDAC cells. Knockdown of SLC7A11-AS1 results in elevation of the intracellular ROS, leading to the loss of stemness. SLC7A11-AS1 silencing-induced reduction of stemness can be rescued by NAC, a scavenger of ROS. These results indicate that SLC7A11-AS1 plays a role in the maintenance of cancer stemness by suppressing ROS. Furthermore, our results demonstrate that SLC7A11-AS1 exerts antioxidant activity through NRF2. It lost antioxidant function when NRF2 was silenced.
NRF2 is the key regulator of antioxidant defense.8 Overactivation of NRF2 promotes cancer cell survival and enhances cancer stemness, as well as chemoresistance and/or radioresistance.37 Dysregulation on the ubiquitin-proteasomal degradation of NRF plays an important role in the sustained activation of NRF2 in cancers. For example, KEAP1 methylation leads to reduced KEAP1 level, which correlates with increased level of nuclear NRF2 in colon cancer.38 Somatic mutation of NRF2 or KEAP1 that disrupts NRF2/KEAP1 interaction confers to NRF2 activation in renal cell carcinoma and other types of solid tumors.39,40 Recently, β-TRCP has been identified as an E3 ligase that mediates NRF2 degradation in the nucleus.41 β-TRCP belongs to the F-box protein families, also known as F-box/WD repeat-containing protein 1A (FBXW1A).29 It consists of two major functional domains: the carboxy-terminal WD domain that binds to specific substrates; and the F-box motif, which recruits F-box protein to the SKP1-Cul1-Rbx1 (SCFβ-TRCP) E3 ligase complex through interaction with adaptor protein SKP1.29 β-TRCP recognizes and binds to the DSGIS and DSAPGS motifs in the Neh6 domain of NRF2 and ubiquitylates NRF2 for proteasome degradation in the nucleus.13,41 NRF2 protein in normal pancreatic ductal epithelial cells is defined in the cytosol at weak level and without nuclear expression. It shows increased cytoplasmic level and positive staining in the nucleus in PDAC tissues.9 However, NRF2 protein level is not negatively correlated to the KEAP1 expression in PDAC tissues.9 In addition, the sustained activation of NRF2 in human PDAC cannot be explained by somatic mutations of NRF2 or KEAP1.14 Furthermore, high levels of β-TRCP1 are also detected in the specimens of PDAC.15 Hence it is necessary to find an additional degradation mechanism to understand sustained activation of NRF2 in PDAC. In the present study, our results reveal that the overexpression of SLC7A11-AS1 in gemcitabine-resistant PDAC cells can block β-TRCP1-mediated ubiquitination and subsequent degradation of NRF2. SLC7A11-AS1 is co-localized with β-TRCP1 in the nucleus of PDAC cells. Pull-down assays show that β-TRCP1 binds to exon 3 (440–1725 nt) of SLC7A11-AS1. RIP assays reveal that SLC7A11-AS1 interacts with the F-box domain of β-TRCP1. Moreover, SLC7A11-AS1 blocks β-TRCP1-mediated ubiquitination and subsequent proteasomal degradation of NRF2 in the nucleus of PDAC cells. There is another mechanism reported to affect the stability of nuclear NRF2. NRF2 traffics to promyelocytic leukemia-nuclear bodies (PMLNBs), where RNF4 polyubiquitylates polysumoylated NRF2 in PML-NBs, subsequently leading to its degradation.42 In this study, we cannot rule out whether SLC7A11-AS1 might affect RNF4-mediated nuclear NRF2 degradation. However, our results clearly demonstrate that SLC7A11-AS1 is involved in SCFβ-TRCP-mediated NRF2 degradation in the nucleus.
β-TRCP plays critical roles in many key processes, such as cell cycle, apoptosis, and migration.43 A number of important proteins, including β-catenin, cell division cycle 25 (Cdc25), vascular endothelial growth factor receptor 2 (VEGFR2), inhibitor of nuclear factor-κB (IκB), and mouse double minute 2 (Mdm2), are ubiquitinated by β-TRCPs.29 Given that SLC7A11-AS1 binds to the F-box domain of β-TRCP1, which is responsible for recruiting β-TRCP1 to the SCFβ-TRCP E3 complex, it raises a possibility that SLC7A11-AS1 may inhibit ubiquitination and degradation of other substrates of β-TRCP1. Indeed, we found that knockdown of SLC7A11-AS1 led to enhanced degradation of nuclear β-catenin, further supporting that SLC7A11-AS1 interacts with the F-box domain of β-TRCP1, blocking the formation of the SKP1-Cul1-Rbx1 E3 ligase complex. Hence SLC7A11-AS1 is a very important lncRNA that can affect the degradation of extensive substrates of β-TRCP1. Several lncRNAs have been reported to interfere with E3 ligase, such as OCC-144 and UPAT.45 These lncRNAs interact with E3 ligase binding sites to substrates, thus affecting specific substrate protein degradation. To date, SLC7A11-AS1 is, to the best to our knowledge, the first lncRNA reported that can disrupt SCFβ-TRCP E3 complex formation, thus affecting the degradation of an extensive number of proteins that are important for cancer progression. Our work highlights the important role of SLC7A11-AS1 in regulating β-TRCP1 function.
In summary, our work demonstrates that SLC7A11-AS1 promotes cancer stemness and chemoresistance by blocking the nuclear NRF2 protein degradation through interaction with β-TRCP1. Our study suggests that SLC7A11-AS1 may be of potential value as a novel therapeutic target for PDAC treatment.
Materials and Methods
Cell Culture
Human PDAC cell lines BxPC-3, PANC-1, and AsPC-1 were cultured in RPMI-1640 medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin/streptomycin. CFPAC-1 and 293T cells were cultured in Iscove’s modified Dulbecco’s medium (GIBCO) and Dulbecco’s modified Eagle’s medium (GIBCO), respectively, with the same supplement as shown above. The gemcitabine-resistant subline BxPC-3-Gem was maintained in medium containing 50 nM gemcitabine (LC Laboratories, Woburn, MA, USA).20
PDAC Patient Samples
PDAC samples and adjacent non-tumor tissues were obtained from patients diagnosed with PDAC at the Affiliated Tumor Hospital of Harbin Medical University (Harbin, China). All samples were collected immediately from resection, snap frozen in liquid nitrogen, and stored at −80°C. Written consent was obtained from all patients. Ethical consent was approved by the Committees for Ethical Review of Research involving Human Subjects of Harbin Medical University.
RNA Isolation and qRT-PCR
Total RNA was isolated using TRIzol (Invitrogen, Grand Island, NY, USA) and reverse transcribed into cDNA using a reverse transcription kit (TOYOBO, Japan). qRT-PCR was performed using GoTaq qPCR Master Mix (Promega, Madison, WI, USA) with gene-specific primers (Comate Bioscience, China) listed in Table S1. The results were calculated using the 2−ΔΔCt method and normalized against internal control.
Transfection of Plasmid and Small siRNA
Transfection experiments were conducted by using Lipofectamine 3000 (Invitrogen) and blended within Opti-MEM I Reduced Serum Media (GIBCO). siRNA targeting SLC7A11-AS1 (RiboBio, China) or NRF2 (GenePharma, China) was transfected at the final concentration of 50 nmol/L. PCDNA3-myc3-NRF2 and HA-ubiquitin were provided by Profs. Ying Hu and Yu Li, respectively (Harbin Institute of Technology). Primers for plasmid construction and siRNA sequences were listed in Table S1.
Lentivirus Production and Infection
To establish stable knockdown cell lines, we cloned a short hairpin RNA (shRNA) sequence that specifically targets SLC7A11-AS1 into pLKO.1-Puro vector (Addgene, Cambridge, MA, USA). The sequence of shRNA was listed in Table S1. Lentivirus was produced in 293T cells co-transfected with pCMV-VSV-G (vesicular stomatitis virus G protein) (1 μg), pGag/Pol/PRE (9 μg), and pLKO.1-puro empty or pLKO.1-puro-sh-SLC7A11-AS1 plasmid (10 μg). The medium containing lentivirus was collected every 24 h and filtered (0.45 μm). Gemcitabine-resistant BxPC-3-Gem, PANC-1, and AsPC-1 cells were incubated with the medium for 48 h, and stable lines were selected using 10 μg/mL of puromycin (Sigma). The knockdown efficiency was evaluated by qRT-PCR.
Sphere-Formation Assay
Cells (500 cells/well) were seeded into six-well low-attachment plates (Corning, Tewksbury, MA, USA) and cultured in Dulbecco’s modified Eagle’s medium-F12 medium (GIBCO), containing 2% B27 (GIBCO), 10 ng/mL of epidermal growth factor (EGF; GIBCO), and 10 ng/mL of basic fibroblast growth factor (FGF; GIBCO) for 7 days. Spheroids were photographed.
MTT Assay
3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) was performed as described previously.20 In brief, PDAC cells were seeded in 96-well plates (5 × 103 cells/well) and treated with various doses of gemcitabine for 72 h. MTT (5 mg/mL; Sigma) was added into each well after treatment, and the formed MTT products were dissolved in DMSO (Sigma). The optical density was measured at a wavelength of 490 nm using an iMark Microplate Absorbance Reader (Bio-Rad, USA). The half-maximal inhibitory concentration (IC50) value was determined by using GraphPad 6 software (Prism). All of the experiments were performed in triplicate.
Colony Formation Assay
Cells seeded in six-well plates (500 cells/well) were treated with gemcitabine (1 μM) and continuously cultured for 14 days without disturbance. Culture medium was replaced every 5 days. Colonies were fixed in formaldehyde for 30 min, stained with 0.1% crystal violet for 30 min, and photographed.
ROS Detection
Cellular ROS level was measured by incubating cells with 2′, 7’ dichlorodihydrofluorescein diacetate (DCFH-DA; 10 μM; Beyotime, China) for 30 min at 37°C after treatments. The reduced DCFH-DA can be oxidized and converted into fluorescent 2′, 7’-dichlorofluorescein (DCF) by intracellular ROS. Fluorescence intensity of cell suspension was determined by FCM (BD FACSCalibur; BD Biosciences). In total, 10,000 cells were analyzed per sample.
For ROS detection in tumor tissue, frozen tumors were sectioned into 10 μm, and the slides were incubated with dihydroethidium (DHE; 5 μM; Vigorous, China) in a humidified condition at 37°C for 30 min.46 The fluorescent signals were detected by using a fluorescence microscope. The resulting DHE-mediated fluorescence was quantified by ImageJ.
Subcellular Fractionation
The nuclear and cytoplasmic fractions were prepared as described previously.47 In brief, PANC-1 and BxPC-3-Gem cells were washed with cold PBS twice and resuspended in pre-chilled cell disruption buffer (1.5 mM MgCl2, 10 nM KCl, 20 mM Tris-HCl, 1 mM DTT). Cell suspensions were incubated on ice for 10 min, followed by homogenization and addition of 0.1% Triton X-100. The homogenates were visually inspected under a microscope to ensure that the nuclei remained intact while the membranes were broken in over 90% of cells. The nuclei were separated from the cytosol by centrifuging at 1,500 × g for 5 min.
Western Blot Analysis
Whole-cell lysates were prepared using RIPA lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM sodium chloride, 1.0% Nonidet P-40 [NP-40], 0.1% SDS) with 1% protease inhibitor cocktail. Western blot was performed as described previously20 with primary antibodies against NRF2, Nanog, Oct4, β-Catenin, GAPDH, α-Tubulin, FLAG tag (Proteintech, China), and Lamin (Santa Cruz, Dallas, TX, USA).
RNA Pull-Down Assay
For RNA in vitro transcription, full-length SLC7A11-AS1 and its truncates were amplified and inserted into Peasy-T1 (TransGen, China). Linearized fragments were used as a template for in vitro transcription of biotin-labeled RNAs by using TranscriptAid T7 High Yield Transcription Kit (Thermos Scientific, Pittsburgh, PA, USA) and Label IT Nucleic Acid Labeling Kit (Mirus, Madison, WI, USA). After denaturing for 10 min at 65°C, the labeled transcripts (5 μg) were incubated with nuclear extracts (2 mg) with 100 U/mL RNaseOUT (Invitrogen) at 4°C with rotation. Unlabeled RNA with nuclear protein was used as control. After 2-h incubation, streptavidin beads (MedChemExpress [MCE]; Monmouth Junction, NJ, USA) were added, and the enriched components were analyzed by western blot.
RNA In Situ Hybridization and Immunofluorescence Staining
Biotin-labeled SLC7A11-AS1 probe (1,420 nt in exon 3) was generated as described above. Cells were fixed with 4% paraformaldehyde for 15 min, followed by permeabilization with 0.1% Triton X-100 for 5 min and hybridization with biotin-labeled SLC7A11-AS1 probe in 2× saline sodium citrate (SSC; Sigma) at 65°C overnight in a moist chamber.
For co-localization study, cells were co-stained with rabbit anti-β-TRCP1 antibody (1:100 dilution; ABclonal, China) and streptavidin, Alexa Flour 555 conjugate (1:200 dilution; Thermo Fisher) at 37°C for 1 h, then washed with PBS three times, and incubated with Chicken anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:200 dilution, Thermo Fisher) for 2 h at room temperature. The nuclei were counterstained with DAPI (Sigma). The fluorescence images were taken by confocal microscope.
RIP Assay
Full-length β-TRCP1 and its truncations (β-TRCP1-N, β-TRCP1-C) were amplified and sub-cloned into pIRM-3xHA vector using appropriate restriction enzyme digestion. β-TRCP1-N-ΔF-box was generated using two-stepwise PCR. HA-empty vector, HA-β-TRCP1, and its truncations were transfected into PANC-1 cells in the presence of SLC7A11-AS1 expression vector PCDNA3.1-SLC7A11-AS1 for 48 h. Cells were lysed with 1 mL buffer (150 mM NaCl, 1 mM EDTA, 20 mM HEPES, 1% NP-40, 1 mM PMSF, 1 mM DTT, 100 U/mL RNaseOUT). 100 μL of supernatant was saved as input for qRT-PCR analysis, and another 100 μL of supernatant as input for western blot analysis. Cell lysates were immunoprecipitated with anti-HA-tag antibody (ABclonal) overnight. The precipitated RNAs were eluted with TRIzol and analyzed by qRT-PCR using SLC7A11-AS1-specific primers (Table S1). The amount of immunoprecipitated RNAs is represented as the percentile of the amount of input RNA (% input).
Ubiquitination Assay
293T cells were transfected with HA-ubiquitin along with PCDNA3-myc3-NRF2 and pCXN2-FLAG-β-TRCP1 in the presence or absence of PCDNA3.1-SLC7A11-AS1 for 24 h, followed by treatment with MG132 (1 μM, MCE) for an additional 12 h. Collected cells were lysed in 100 μL of dilution buffer containing 10 mM Tris-HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl and 1% Triton X-100, and an equal amount of cell lysis buffer (2% SDS, 150 mM NaCl, 10 mM Tris-HCl [pH 8.0]). The diluted samples were centrifuged at 20,000 × g for 30 min, and the supernatant (1.5 mg) was used for immunoprecipitation with anti-Myc antibody (Cell Signaling Technology, Danvers, MA, USA), followed by immunoblotting with anti-HA antibody.
Protein Half-Life Assay
Cells were treated with CHX (100 μg/mL; Sigma) for up to 60 min and lysed with RIPA buffer. Whole-cell lysates (20 μg) were separated by SDS-PAGE, and protein levels were measured by immunoblot.
Mice Xenograft Study
All of the animal care and experimental procedures were approved by the Animal Care and Use Committee of Harbin Institute of Technology. Female athymic BALB/c nude mice (4–5 weeks old) were obtained from Beijing HFK Bioscience. For in vivo ROS detection, sh-SLC7A11-AS1 or sh-Ctrl cells (1.5 × 106) were injected subcutaneously into the left or right posterior flank of nude mice, respectively (n = 5). For chemoresistance study, cells were injected into the right side of the mice (n = 4). Tumor volumes were calculated using the equation: V = a × b2/2, where a is the largest diameter, and b is the perpendicular diameter to a.
Statistical Analysis
Statistical analysis was carried out using GraphPad software, v.6. Student’s t test (two-tailed) was used when comparing only two groups. Differences between more than two groups were analyzed by using two-way ANOVA. Data are presented as the mean ± standard deviation (SD) and repeated from at least three independent experiments. p < 0.05 was considered to be statistically significant.
Author Contributions
H.Y. and Q.Y. conceived the idea and designed the experiments. H.Y., Q.Y., and K.L. wrote the manuscript. Q.Y. and K.L. performed the experiments and analyzed the data. C.Z. generated PCDNA3.1-SLC7A11-AS1. Some of the experiments were performed with help from X.H., Y.M., X.L., and J.L.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by National Natural Science Foundation of China (grants 81872439 and 31700780) and China Postdoctoral Science Foundation Funded Project (grant 2018T110281). We thank Drs. Y. Li and H. Nie for providing clinical samples.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.035.
Supplemental Information
References
- 1.Adamska A., Domenichini A., Falasca M. Pancreatic ductal adenocarcinoma: current and evolving therapies. Int. J. Mol. Sci. 2017;18:1338–1380. doi: 10.3390/ijms18071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hung S.W., Mody H.R., Govindarajan R. Overcoming nucleoside analog chemoresistance of pancreatic cancer: a therapeutic challenge. Cancer Lett. 2012;320:138–149. doi: 10.1016/j.canlet.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju H.Q., Gocho T., Aguilar M., Wu M., Zhuang Z.N., Fu J., Yanaga K., Huang P., Chiao P.J. Mechanisms of overcoming intrinsic resistance to gemcitabine in pancreatic ductal adenocarcinoma through the redox modulation. Mol. Cancer Ther. 2015;14:788–798. doi: 10.1158/1535-7163.MCT-14-0420. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Duan Q., Zhao H., Liu T., Wu H., Shen Q., Wang C., Yin T. Gemcitabine treatment promotes pancreatic cancer stemness through the Nox/ROS/NF-κB/STAT3 signaling cascade. Cancer Lett. 2016;382:53–63. doi: 10.1016/j.canlet.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X., Zhang Y., Zheng J., Pan J. Reactive oxygen species in cancer stem cells. Antioxid. Redox Signal. 2012;16:1215–1228. doi: 10.1089/ars.2012.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryoo I.G., Lee S.H., Kwak M.K. Redox modulating NRF2: a potential mediator of cancer stem cell resistance. Oxid. Med. Cell. Longev. 2016;2016:2428153–2428166. doi: 10.1155/2016/2428153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong Y.B., Kang H.J., Kwon S.Y., Kim H.J., Kwon K.Y., Cho C.H., Lee J.M., Kallakury B.V., Bae I. Nuclear factor (erythroid-derived 2)-like 2 regulates drug resistance in pancreatic cancer cells. Pancreas. 2010;39:463–472. doi: 10.1097/MPA.0b013e3181c31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lister A., Nedjadi T., Kitteringham N.R., Campbell F., Costello E., Lloyd B., Copple I.M., Williams S., Owen A., Neoptolemos J.P. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol. Cancer. 2011;10:37–49. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder B., Jiang T., Wu T., Tao S., Rojo de la Vega M., Tian W., Chapman E., Zhang D.D. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem. Soc. Trans. 2015;43:680–686. doi: 10.1042/BST20150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müerköster S., Arlt A., Sipos B., Witt M., Grossmann M., Klöppel G., Kalthoff H., Fölsch U.R., Schäfer H. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–1324. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 16.Li Z., Zhao X., Zhou Y., Liu Y., Zhou Q., Ye H., Wang Y., Zeng J., Song Y., Gao W. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J. Transl. Med. 2015;13:84–99. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan H.X., Wang Y., Li C., Xu J.W., Zhou B., Zhu J.K., Han H.F., Wang L., Wang Y.S., Hu S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374:261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Mello S.S., Sinow C., Raj N., Mazur P.K., Bieging-Rolett K., Broz D.K., Imam J.F.C., Vogel H., Wood L.D., Sage J. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao F., Hu H., Han T., Yuan C., Wang L., Jin Z., Guo Z., Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y., Zhang Z., Li K., Gong L., Yang Q., Huang X., Hong C., Ding M., Yang H. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis. 2017;8:e2924. doi: 10.1038/cddis.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastrana E., Silva-Vargas V., Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S., Lee T.K., Zheng B.J., Chan K.W., Guan X.Y. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 23.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menegon S., Columbano A., Giordano S. The dual roles of NRF2 in cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H.R., Kim S., Kim E.J., Park J.H., Yang S.H., Jeong E.T., Park C., Youn M.J., So H.S., Park R. Suppression of Nrf2-driven heme oxygenase-1 enhances the chemosensitivity of lung cancer A549 cells toward cisplatin. Lung Cancer. 2008;60:47–56. doi: 10.1016/j.lungcan.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Chen L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Sun Q., Hao Q., Prasanth K.V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat. Rev. Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ougolkov A., Zhang B., Yamashita K., Bilim V., Mai M., Fuchs S.Y., Minamoto T. Associations among β-TrCP, an E3 ubiquitin ligase receptor, β-catenin, and NF-kappaB in colorectal cancer. J. Natl. Cancer Inst. 2004;96:1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 31.Cai Q., Wang S., Jin L., Weng M., Zhou D., Wang J., Tang Z., Quan Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer. 2019;18:82–97. doi: 10.1186/s12943-019-1016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu Z., Schmöllerl J., Cuiffo B.G., Karnoub A.E. Microenvironmental regulation of long noncoding RNA LINC01133 promotes cancer stem cell‐like phenotypic traits in triple‐negative breast cancers. Stem Cells. 2019;37:1281–1292. doi: 10.1002/stem.3055. [DOI] [PubMed] [Google Scholar]
- 33.Yuan J., Liu Z., Song R. Antisense lncRNA As-SLC7A11 suppresses epithelial ovarian cancer progression mainly by targeting SLC7A11. Pharmazie. 2017;72:402–407. doi: 10.1691/ph.2017.7449. [DOI] [PubMed] [Google Scholar]
- 34.Luo Y., Wang C., Yong P., Ye P., Liu Z., Fu Z., Lu F., Xiang W., Tan W., Xiao J. Decreased expression of the long non-coding RNA SLC7A11-AS1 predicts poor prognosis and promotes tumor growth in gastric cancer. Oncotarget. 2017;8:112530–112549. doi: 10.18632/oncotarget.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peitzsch C., Tyutyunnykova A., Pantel K., Dubrovska A. Cancer stem cells: The root of tumor recurrence and metastases. Semin. Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Niess H., Camaj P., Renner A., Ischenko I., Zhao Y., Krebs S., Mysliwietz J., Jäckel C., Nelson P.J., Blum H. Side population cells of pancreatic cancer show characteristics of cancer stem cells responsible for resistance and metastasis. Target. Oncol. 2015;10:215–227. doi: 10.1007/s11523-014-0323-z. [DOI] [PubMed] [Google Scholar]
- 37.Wu S., Lu H., Bai Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 2019;8:2252–2267. doi: 10.1002/cam4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanada N., Takahata T., Zhou Q., Ye X., Sun R., Itoh J., Ishiguro A., Kijima H., Mimura J., Itoh K. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12:66–76. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo N.J., Kim H.R., Kim Y.R., An C.H., Lee S.H. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology. 2012;60:943–952. doi: 10.1111/j.1365-2559.2012.04178.x. [DOI] [PubMed] [Google Scholar]
- 40.Ooi A., Dykema K., Ansari A., Petillo D., Snider J., Kahnoski R., Anema J., Craig D., Carpten J., Teh B.T., Furge K.A. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013;73:2044–2051. doi: 10.1158/0008-5472.CAN-12-3227. [DOI] [PubMed] [Google Scholar]
- 41.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malloy M.T., McIntosh D.J., Walters T.S., Flores A., Goodwin J.S., Arinze I.J. Trafficking of the transcription factor Nrf2 to promyelocytic leukemia-nuclear bodies: implications for degradation of NRF2 in the nucleus. J. Biol. Chem. 2013;288:14569–14583. doi: 10.1074/jbc.M112.437392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J., Zhou W., Yang F., Chen G., Li H., Zhao Y., Liu P., Li H., Tan M., Xiong X., Sun Y. The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat. Commun. 2017;8:14002–14017. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan Y., Xiao X., He Z., Luo Y., Wu C., Li L., Song X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taniue K., Kurimoto A., Sugimasa H., Nasu E., Takeda Y., Iwasaki K., Nagashima T., Okada-Hatakeyama M., Oyama M., Kozuka-Hata H. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. USA. 2016;113:1273–1278. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schluterman M.K., Chapman S.L., Korpanty G., Ozumi K., Fukai T., Yanagisawa H., Brekken R.A. Loss of fibulin-5 binding to β1 integrins inhibits tumor growth by increasing the level of ROS. Dis. Model. Mech. 2010;3:333–342. doi: 10.1242/dmm.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rio D.C., Ares M., Jr., Hannon G.J., Nilsen T.W. Preparation of Cytoplasmic and Nuclear RNA from Tissue Culture Cells. Cold Spring Harb. Protoc. 2010. 2010 doi: 10.1101/pdb.prot5441. pdb.prot5441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.