Abstract
Microorganisms acquire energy and nutrients from dynamic environments, where substrates vary in both type and abundance. The regulatory system responsible for prioritizing preferred substrates is known as carbon catabolite repression (CCR). Two broad classes of CCR have been documented in the literature. The best described CCR strategy, referred to here as classic CCR (cCCR), has been experimentally and theoretically studied using model organisms such as Escherichia coli. cCCR phenotypes are often used to generalize universal strategies for fitness, sometimes incorrectly. For instance, extremely competitive microorganisms, such as Pseudomonads, which arguably have broader global distributions than E. coli, have achieved their success using metabolic strategies that are nearly opposite of cCCR. These organisms utilize a CCR strategy termed ‘reverse CCR’ (rCCR), because the order of preferred substrates is nearly reverse that of cCCR. rCCR phenotypes prefer organic acids over glucose, may or may not select preferred substrates to optimize growth rates, and do not allocate intracellular resources in a manner that produces an overflow metabolism. cCCR and rCCR have traditionally been interpreted from the perspective of monocultures, even though most microorganisms live in consortia. Here, we review the basic tenets of the two CCR strategies and consider these phenotypes from the perspective of resource acquisition in consortia, a scenario that surely influenced the evolution of cCCR and rCCR. For instance, cCCR and rCCR metabolism are near mirror images of each other; when considered from a consortium basis, the complementary properties of the two strategies can mitigate direct competition for energy and nutrients and instead establish cooperative division of labor.
Keywords: Diauxie, Carbon catabolic repression, Reverse carbon catabolic repression, Division of labor, Overflow metabolism
Introduction
Most natural environments are physically, chemically, and temporally complex, with the resident microorganisms exposed to multifactorial selection pressures. It is not possible to optimize all cellular functions simultaneously due to constraints on cellular resources such as anabolic nitrogen or cytoplasmic volume; a concept illustrated by the ‘Darwinian Demon’ thought exercise [1, 2]. Therefore, microorganisms must dynamically change their phenotypes with environmental fluctuations to maintain fitness [3]. Metabolic regulation of substrate-consumption order is paramount to fitness. There are a few broad strategies that can be utilized when an organism is presented with two potential substrates: (1) utilize the ‘optimal’ substrate, (2) utilize the less ‘optimal’ substrate, or (3) co-catabolize both substrates simultaneously (see Box 1 for discussion of ‘optimal’ substrates). It can be more resource effective to catabolize a single substrate at a time as opposed to expressing multiple catabolic pathways simultaneously [4, 5]; although, exceptions exist [6]. Carbon catabolite repression (CCR) is a global regulation system that controls the sequential catabolism of preferred substrates from a milieu (Box 2). CCR is associated with generalist microorganisms that can utilize multiple substrates and exist in environments, where these substrates are often available at varying abundances  .
.
The cCCR regulation scheme, known under specific conditions as diauxie, has been the focus of much experimental and theoretical research [7–12]. The historical basis of cCCR goes back more than 70 years to Jacques Monod and his observations of Escherichia coli growing in the presence of glucose and other sugars [13]. Monod recorded E. coli cultures growing exponentially on glucose until depleted, followed by a lag phase, then a second exponential growth phase on the other sugar. Diauxic growth is a trade-off between specializing for a single nutrient and being able to switch between nutrients as conditions change [14]. Committing to one metabolism at a time, necessitates a metabolic shift when the preferred substrate is exhausted, with a resulting temporal delay.  .
.
The traditional interpretation of diauxie as being carried out by a single population of organisms, synchronized in function, that sequentially catabolizes preferred and non-preferred substrates has been challenged [15, 16]. Recent research has posited an alternative interpretation; rather than a homogenous population, the observed lag phase can be explained by dynamic shifts in subpopulations possessing different fitness. For instance, during growth on glucose, multiple subpopulations catabolize, the sugar but upon glucose depletion only some subpopulations, in an ecological bet-hedging strategy, are phenotypically prepared to consume the less desirable substrate [17]. The lag in growth between the two sugars corresponds to a shift in subpopulation distributions as fit subpopulations outgrow other subpopulations [17]. Control of these subpopulations has been associated with the regulation of bistable genetic switches [18, 19].
cCCR is not used by all glucose-utilizing microorganisms. Pseudomonads and Acinetobacter [20] utilize substrates in an order almost reverse of cCCR. Therefore, this metabolic regulatory strategy was termed reverse carbon catabolite repression (rCCR) or sometimes reverse diauxie. rCCR utilizing microorganisms are very competitive as evidenced by their broad global distributions and are arguably at least as competitive as the better studied members of the Enterobacter and Firmicutes used to define cCCR. The ecological rationale of the reverse diauxie strategy is still an open question and is discussed in later sections.
The study of cCCR and rCCR has traditionally focused on monocultures for the sake of simplicity, possibly limiting the interpretation of regulatory systems, as most microorganisms have existed on evolutionary timeframes within consortia [21–24]. The organisms have interacted with other species for millennia, and these interactions have influenced access to energy and nutrients [25]. Many genes regulated by the CCR systems such as quorum sensing, biofilm formation, and antibiotic or reactive oxygen species (ROS) tolerance have been studied from the perspective of medical challenges and monocultures, even though these behaviors modulate inter-microbial interactions and strategies for resource acquisition in consortia (see sections below).
This review summarizes (1) cCCR and rCCR molecular mechanisms; (2) theory used to interpret and model CCR phenotypes; and (3) the nexus of CCR, metabolism, and cellular interactions. Collectively, the properties of cCCR and rCCR phenotypes can enhance growth and persistence by enabling cooperation via division of labor, as observed in some medical environments such as chronic wounds, where cCCR and rCCR microorganisms commonly cooccur (Fig. 1).
Fig. 1.
Illustration of classic carbon catabolite repression (cCCR) phenotype with overflow metabolism and reverse carbon catabolite repression (rCCR) phenotype as well as an illustration of the complementary relationship between the two global metabolism regulators. cCCR microorganisms prefer glucose to other substrates, while rCCR microorganisms prefer organic acids to glucose
Molecular mechanisms of CCR
Most CCR studies have been performed in model organisms such as E. coli and Bacillus subtilis, which prefer glucose over other substrates. The regulatory outcomes of CCR in E. coli and B. subtilis are similar, though the mechanisms differ (Fig. 2). This review summarizes molecular mechanisms of CCR for context and to highlight similarities and differences between cCCR and rCCR (Table 1). This summary is not exhaustive; there are numerous excellent reviews which discuss finer aspects of CCR molecular biology [7–12].
Fig. 2.
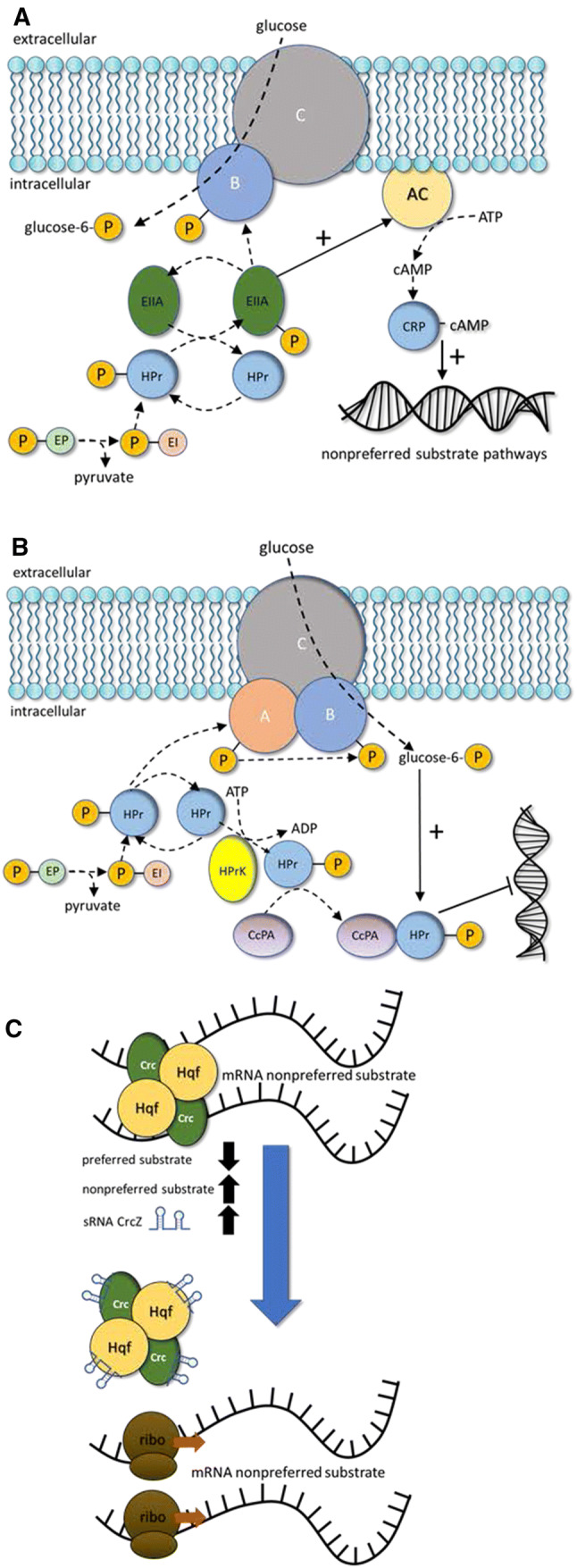
Molecular components of carbon catabolite repression (CCR). a Major molecular components of classic carbon catabolite repression (cCCR) in Gram-negative E. coli. b Major molecular components of cCCR in Gram-positive B. subtilis. c Major molecular components of reverse carbon catabolite repression (rCCR) found in Pseudomonads. See main text for abbreviations and details
Table 1.
Summary of carbon catabolite repression (CCR) mechanisms and key molecular components in representative Gram-positive and Gram-negative microorganisms
| Microorganism | Gram staining | Governing mechanism | Important components | References |
|---|---|---|---|---|
| E. coli | − | Positive induction of DNA transcription | EIIA-P, cAMP, CAP | [34, 35] |
| Bacillus subtilis | + | Negative regulation of DNA transcription | CcpA, HPr, HPrK | [36–38] |
| Lactobacillus brevis | + | Negative regulation of DNA transcription | CcpA, HPr, HPrK | [39] |
| Lactococcus lactis | + | Negative regulation of DNA transcription | CcpA, HPr, HPrK | [40] |
| Clostridium cellulyticum | + | Negative regulation of DNA transcription | CcpA, HPr, HPrK | [41] |
| Corynebacterium glutamicum | + | Negative regulation of DNA transcription | Ram A, RamB | [42, 43] |
| Acinetobacter baylyi | − | Repression of mRNA translation | Crc, Hqf, CrcZ | [7–9, 44] |
| Pseudomonas putida | − | Repression of mRNA translation | Crc, Hqf, CrcZ | [7–9, 44] |
| Pseudomonas aeruginosa | − | Repression of mRNA translation | Crc, Hqf, CrcZ | [7–9, 44] |
| Streptomyces coeiicoior | + | Biochemical state of glucose kinase | Glk | [45, 46] |
Molecular mechanisms of cCCR
Gram-negative E. coli utilizes two major cCCR regulation mechanisms: (1) inducer exclusion [9, 44, 47, 48] and (2) cyclic AMP (cAMP)-catabolite repressor protein (CRP) regulation of gene protomer activity; although the condition-specific importance of each mechanism is still an open research question [49, 50]. Glucose can enter the cell using the multisubunit phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS). Regulation of cCCR begins with the phosphorylation state of the glucose-specific subunit of the PTS transporter, EIIAGlc. EIIAGlc is phosphorylated by PEP via a chain of phosphorylation reactions. In the presence of high glucose flux, the phosphate from EIIAGlc is transferred to the hexose producing glucose-6-phosphate, precluding the phosphate group from activating PTS EIIA components specific for less preferred substrates. This process is known as inducer exclusion, because the excluded, non-preferred substrates are inducers for their own catabolic operons [51]. When glucose flux is low, phosphorylated EIIAGlc accumulates and activates EIIA subunits for less preferred substrates and activates the enzyme adenylate cyclase [35, 52] which catalyzes the production of cAMP [53]. cAMP binds to the catabolite gene activator protein (CAP), forming the cAMP–CAP complex which binds to DNA, activating the promoters of ~ 200 genes for a range of cellular processes [49, 50, 54–57]. The classic example of the CCR inducer exclusion and cAMP-CRP regulation occurs during glucose–lactose diauxic growth [58–61].
CCR is complex, with many levels of modulation in addition to inducer exclusion and cAMP-CRP regulation. Small regulatory RNAs (sRNA) also contribute to post-transcriptional regulation of CCR [10, 62, 63]. sRNA Spot 42 is described as a third pillar of CCR in E. coli [10, 64]. Spot 42 represses target genes as opposed to the role of cAMP–CRP, which generally activates gene targets. The Spot 42 regulon has 29 documented genes [65]. Central metabolism intermediates such as a-ketoglutarate, oxaloacetate, PEP, and pyruvate also play important roles in CCR by affecting the levels of cAMP, adding additional complexity to CCR [66, 67].
E. coli cCCR regulation ‘glitches’ can result in substrate selection that does not enable the fastest growth [68]. E. coli cultures growing on a mixture of glucose and lactose and a poor nitrogen source (e.g., arginine, glutamate, and proline) preferentially consume glucose over lactose, but growth on glucose is slower than on lactose due to altered cAMP concentrations. Therefore, this diauxic growth pattern has relatively slow growth during the first phase and faster growth during the second phase. The effect is attributed to laboratory conditions not reflective of habitats that influenced E. coli selection [68].
Gram-positive B. subtilis also prefers glucose and utilizes the PTS system for sugar transport, although the molecular components of CCR are different than in E. coli. cCCR in B. subtilis is mediated largely by negative regulation through a repressor protein expressed in the presence of glucose. The key cCCR components include catabolite control protein A (CcpA), phosphocarrier protein (HPr), HPr kinase (HPrK), and glycolysis intermediates fructose-1,6-bisphosphate and glucose-6-phosphate [36–38]. HPr plays two important roles. First, HPr participates in the phosphorylation cascade by transferring the phosphate group from PEP to glucose, forming glucose-6-P. Second, HPr is a regulator molecule which responds to glycolysis indicators fructose-1,6-bisphosphate or glucose-6-phosphate to form a complex with CcpA. The CcpA–HPr complex, stabilized with glycolysis intermediates, can bind DNA, inhibiting expression of genes associated with non-preferred substrates [69–75]. Approximately 400 B. subtilus genes are influenced by CCR [76, 77].
Molecular mechanisms of rCCR
cCCR-mediated preference for glucose is not a universal strategy for generalist, glucose-catabolizing heterotrophs [9, 78, 79]. Pseudomonas aeruginosa and Pseudomonas putida consume preferred carbon substrates in nearly reverse order from E. coli and B. subtilis. They preferentially catabolize organic acids and amino acids before catabolizing glucose [8, 78, 80–82]. Study of rCCR lags the study of cCCR, highlighting a research area of opportunity.
Known rCCR mechanisms in Pseudomonads operate at the mRNA level; this is contrary to most cCCR microorganisms, which are governed by DNA-binding transcription regulators [83, 84]. RNA chaperone protein Hfq represses expression of catabolic targets for non-preferred substrates by binding directly to the 5′ end of the mRNA. Catabolite repression control (Crc) protein interacts with Hfq, creating a stabilized Hfq/Crc/mRNA complex, enhancing regulation. Hfq and Crc regulons have significant overlap [85–87]. Computational analysis of Crc-binding sites predicted 143 candidate genes in P. putida and 215 candidate genes in P. aeruginosa, while experimental hfq gene deletion studies identified 212 target genes in P. fluorescens [88–90]. Small regulatory RNAs (sRNA) act as antagonists to Hfq, binding the protein and relieving repression of the non-preferred catabolic pathways. P. aeruginosa has 1 characterized sRNA (CrcZ), while P. fluorescens and P. putida have two (CrcY, CrcZ), as does P. syringae (CrcZ, CrcX) [84, 91, 92].
Theoretical aspects of CCR
CCR is the collective result of many interacting molecular components. The complexity necessitates in silico representations for quantitative predictions. Decades of in silico CCR models exist, most focusing on E. coli [67, 93]. The earliest in silico models were primarily mechanistic, comprised of mass balances on enzymes, metabolites, and metabolic regulators, and were represented by ordinary differential equations. As a result, they were limited by the availability of experimentally measured enzyme parameters and metabolite concentrations [67, 93]. These models were not well-suited to address the ecological rationale of CCR strategies. One exception is cybernetic modeling, which examines implicit aspects of resource investment into metabolic strategies and can be used to study diauxic growth [95]. The modeling technique requires a priori calibration data to identify cybernetic parameters which are then used to interpret pathway resource requirements; the cybernetic parameters do not necessary map explicitly to molecular components [96].
The omics revolution has generated huge data bases which have been leveraged by a metabolic modeling family known as stoichiometric modeling; two common types of stoichiometric modeling are flux balance analysis (FBA) and elementary flux mode analysis (EFMA) [97–100]. Stoichiometric models mathematically define the phenotypic capabilities of metabolic systems by accounting for the net transformation of all enzyme-catalyzed reactions as well as relevant abiotic reactions. The mass balances for all system components can be written as a set of linear equations when appropriate time scales are assumed; this negates the requirement for difficult to measure enzyme parameters. The modeling approach does require a minimal set of parameters such as biomass composition and cellular maintenance energy values [101].
A remarkable number of stoichiometric modeling studies have examined aspects of cCCR, including maximum growth rates, overflow metabolisms, and sequential use of substrates. Stoichiometric models define a mathematical solution space of all possible cellular phenotypes based on the linear mass balance equations; context specific flux distributions are identified using optimization criteria. The most commonly used optimization criterion identifies phenotypes that maximize growth rate, consistent with the basic tenets of cCCR [102–104]. All microbial systems are resource constrained and metabolic strategies that maximize fitness must competitively allocate limiting resources. Early applications of stoichiometric models examined limited accessibility of a catabolic resource, O2, and identified metabolisms that maximize energy extraction from glucose while minimizing consumption of O2 [99, 100, 105, 106]. Resource allocation into cCCR-relevant strategies has been a topic of many stoichiometric studies and has resulted in predictions of overflow metabolisms that secrete partially oxidized byproducts such as acetate even in the presence of electron acceptors such as O2. Overflow metabolisms were for decades described as a wasteful metabolism that did not extract all the available energy from a substrate, but recently this view has changed [31]. Representation of resource scarcity in stoichiometric models has expanded to consider anabolic resources, and the simulations have provided ecological explanations for overflow metabolisms occurring at both fast growth rates during batch cultivation as well as slow growth rates during chemostat cultivation [107]. Numerous proxies for anabolic cellular resources have been described in E. coli, with each generating similar outcomes; metabolic fluxes are directed to maximize energy production from the limiting resource, which leads to partial oxidation of substrate and an overflow metabolism. Basically, when glucose is the preferred substrate, glycolysis is an essential pathway, while auxiliary metabolic routes are optional and thus, down regulated when an anabolic resource becomes scarce. Anabolic resource considerations have included (1) accounting of the moles of a resource (e.g., nitrogen, carbon, amino acid) required to synthesize a metabolic pathway relative to the catabolic efficiency of the pathway [31, 32], (2) the volume of the cytosol occupied by enzymes relative to the catabolic efficiency of the flux distribution [103, 104], and (3) the membrane surface area occupied by membrane complexes required for a cellular flux distribution [108, 109] as well as other representations of optimal proteome investment into metabolic fluxes [5, 110]. In silico metabolism representations have also expanded to account for not only metabolic pathways but also the enzymatic machinery required to synthesize RNA and protein [111–113]. The converging of numerous resource allocation problems onto similar phenotypes highlights common relationships between the different proxies regardless of the level of molecular details considered. Resource allocation into enzymes has also been used to justify the sequential catabolizing of glucose and lactose in a mixture. For example, glucose catabolism requires fewer enzymes, permitting larger concentrations of the enzymes and driving a faster growth rate than lactose catabolism [94, 114].
The ecological basis of rCCR is not well studied. Common hypotheses invoke the availability of substrates during the organisms’ evolutionary histories. In many environments, such as soils, sediments, and plant surfaces, organic acids are plentiful byproducts of biomass degradation or plant secretions [7, 8]. However, these environments also contain sugars [115]. Organisms such as P. aeruginosa and P. putida are competent glucose catabolizers, but their regulation schemes do not prioritize the substrate. A ‘gluttony’ hypothesis has been proposed which postulates that Pseudomonads utilize a strategy of consuming substrates as fast as possible to deny the resources to competitors [90]. The hypothesis states the rCCR regulation is necessary to balance metabolic intermediates which could accumulate to inhibitory concentrations. The theory postulates the outcome of rCCR is not to select ‘preferred substrates’ but rather to repress substrates in a manner that balances metabolism. The theory does not explicitly address the sequential use of substrates or why glucose is not co-metabolized.
Few, if any, modeling studies explicitly address the basic tenets of rCCR such as the preference for organic acids over glucose, the lack of almost any overflow metabolism, and a propensity to not always maximize growth rate in monoculture [9, 30, 116–118]. Resource availability limits bacterial productivity in most environments, so widely distributed bacteria such as Pseudomonads have surely been influenced by resource scarcity. The role of resource allocation to enzymes in rCCR organisms is unknown, but it generally contradicts theories postulated for E. coli. rCCR organism do not optimize investment into glycolysis, at the expense of respiration, resulting in an overflow metabolism; in fact, rCCR organisms catabolizing glucose do not typically exhibit an overflow metabolism and instead completely oxidize the sugar to CO2 [118–120]. All Pseudomonads catabolize glucose using the ED pathway [118, 121]. The ED pathway minimizes investment into enzymes as compared to the higher cellular-energy yielding EMP pathway [2, 31, 32, 122], (Fig. 3) providing hints of potential resource allocation strategies in Pseudomonads. In silico representations of P. aeruginosa and P. putida metabolism using stoichiometric models used the maximization of growth optimization criterion to identify flux distributions [123–126]. rCCR organisms do not always select preferred substrates based on maximizing growth rates [30].
Fig. 3.
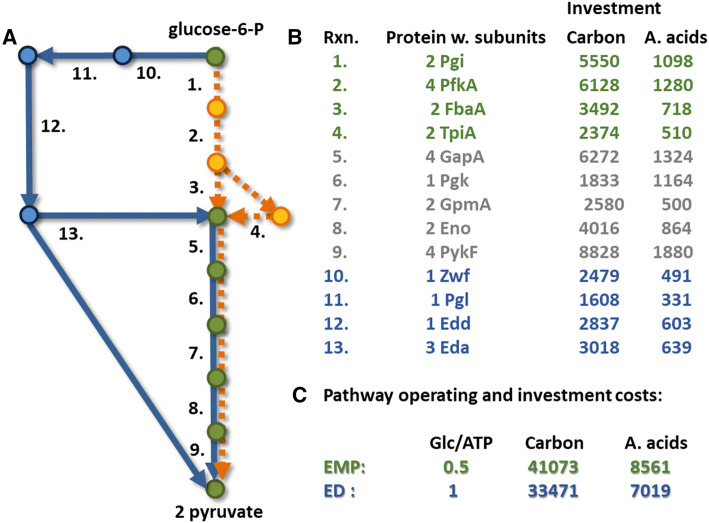
Trade-off between resource investment into enzymes and energetic efficiency of the Embden–Meyerhof–Parnas (EMP) (orange lines) or Entner–Doudoroff (ED) (blue lines) glycolysis pathways. Shared components are colored green. Pseudomonads use the ED pathway to catabolize glucose; the ED pathway requires fewer resources to assemble but also extracts less cellular energy from glucose than the EMP pathway. Reaction numbers map to enzymes, protein subunit compositions, and the resources required to assemble functional enzyme based on E. coli protein sequences. Data from Carlson, 2007 [31], figure modified from Carlson and Taffs, 2010 [2]
CCR and social interactions
Microbial consortia are ubiquitous in nature, and competition for resources often predicts the outcome of microbial interactions [127–130]. Therefore, it is logical that global regulation strategies such as CCR are linked to social behaviors such as quorum sensing, biofilm formation, and virulence mechanisms including antibiotic or ROS sensitivity [131, 132]. Examples of the different social interactions for cCCR and rCCR organisms are summarized in Table 2.
Table 2.
Summary of social behaviors regulated by carbon catabolite repression (CCR) mechanisms in both classic (c) and reverse (r) CCR utilizing microorganisms
| Microorganism (gram stain) | CCR | Social behavior linkage to CCR | ||
|---|---|---|---|---|
| Biofilm formation | E. coli (-) | c | cAMP-CRP regulates csgD and biofilm formation | [155, 156] |
| P. aeruginosa (-) | r | Crc is required for aerobic biofilm formation | [157] | |
| P. aeruginosa (-) | r | CrcZ cross-regulation controls anaerobic biofilm formation | [158] | |
| P. aeruginosa (-) | r | cbrB is linked to biofilm formation and dispersal | [159] | |
| S. aureus (+) | c | CcpA regulates biofilm formation | [160] | |
| S. epidermidis (+) | c | CcpA regulates biofilm formation | [161] | |
| Quotum sensing | E. coli (-) | c | Cocrystal structures of LsrK and HPr establishes link between CCR and QS | [162] |
| E. coli (-) | c | cAMP-CRP influence AI2 synthesis and uptake | [163, 164] | |
| P. aeruginosa (-) | r | Lon and CIpXP proteases link CCR and QS | [136] | |
| S. aureus (+) | c | Ccpa upregulates Agr which is an autoinducing QS peptide | [165] | |
| Stress tolerance | P. aeruginosa (-) | r | Hfq-dependent antibiotic susceptibility | [148] |
| P. aeruginosa (-) | r | Oxidative stress tolerance orchestrated by Crc | [149] | |
| E. coli (-) | c | Acetate CCR regulates biofilm formation and virulence | [150] | |
| S. aureus (+) | c | CCpE control virulence factors including alpha toxin | [166] | |
| P. syringae (-) | r | Crc influences virulence, oxidative tolerance, and biofilm formation | [167] |
Quorum sensing is an intercellular communication mechanism which enables coordination of cellular phenotypes across populations and space [133]. Quorum sensing involves the synthesis and secretion of small soluble molecules that accumulate in the environment. At critical concentrations, the soluble molecules are imported into the cells and initiate gene regulation. Quorum sensing is regulated by CCR [134–136] (Table 2). CCR-influenced quorum sensing is also required for other social behaviors including the development of mature biofilms [137, 138].
Biofilms are microbial communities encapsulated in self-produced polymer and are often associated with interfaces [139, 140]. Biofilms are social structures with high densities of cells (~ 300 g/L), up to two orders of magnitude higher than typical in vitro culture and many orders of magnitude higher than common environmental populations [141–143]. Metabolite transport in biofilms is primarily mediated by diffusion, and high cell density and high cellular activity create mass transfer limitation and nutritional gradients [82]. Biofilms are argued to promote the cellular interaction strategy of altruism [131, 144]. Recent research demonstrated a remarkable substrate ‘flux time-sharing’ and metabolic oscillations used by altruistic B. subtilis cultures to partition resources between cells on the periphery of a biofilm and cells deep within a biofilm [145, 146]. The cells on the interior and exterior of the biofilm oscillate their activity in a coordinated manner, taking turns metabolizing the limiting substrate, which resolves conflict for the substrate and ultimately enhances community growth.
Virulence mechanisms refer to a broad range of cellular strategies that, in a host, enhance resource acquisition, growth, and persistence and are often associated with CCR [7, 147]. Common examples of virulence mechanisms include antibiotic tolerance, production of enzymes such as catalase, superoxide dismutase, and other enzymes for either producing or mitigating ROS, secretion of resource acquisition molecules such as siderophores, proteases, lipases, elastinases, or haemolysins, as well as the quorum sensing and biofilm phenotype mentioned previously (Table 2). Virulence mechanisms, such as antibiotic tolerance, are typically studied from the perspective of monocultures and medical challenges [148–150]. Antibiotics in microbial environments precede human use; these agents are part of natural microorganism interactions used to influence the distribution of species in consortia; secretion and degradation of antibiotics can stabilize consortia [151, 152]. Antibiotic and ROS susceptibility is well documented as a function of CCR [153, 154].
Division of labor and resource partitioning in microbial consortia
Interactions between microorganisms with similar catabolic preferences are often antagonistic [24]. Resource partitioning through mechanisms such as division of labor or reciprocal prioritization of substrates can mitigate direct competition [22, 23, 128, 168–170]. cCCR and rCCR utilizing organisms are often found to coexist in medical environments, including chronic wounds and cystic fibrosis lungs [171]. rCCR organism P. aeruginosa has been isolated from 15 to 80% of chronic wounds, while cCCR organism S. aureus has been found in more than 90% of chronic leg ulcers [172–178]. S. aureus and P. aeruginosa are excellent biofilm formers and often isolated together [179, 180]. Synergistic interactions in polymicrobial infections can lead to greater virulence likely through enhanced resource acquisition and greater efficiency of converting resources into new biomass. For instance, wounds colonized by polymicrobial consortia have more negative patient outcomes than wounds colonized by monocultures [173, 177, 181, 182].
CCR regulates a wide range of social behaviors and likely modulates division of labor or reciprocal substrate prioritization (Table 2). cCCR and rCCR consortia represent an intuitive division of labor, whereby the rCCR organism consumes byproducts of the cCCR metabolism removing metabolites that could be a thermodynamic constraint on metabolism as well as inhibitory [183] (Fig. 1). This positive feedback mechanism would enable consortia to increase biomass productivity by more completely depleting available substrates than a monoculture [184, 185] (Fig. 3).
rCCR microorganisms prefer non-fermentable substrates such as organic acids necessitating the presence of a terminal electron acceptor, such as O2 [116]. The availability of O2 is a fitness challenge especially in diffusion-limited biofilms, where other organisms could consume the catabolic substrate. P. aeruginosa is a very competitive microorganism with mechanisms to acquire limited resources such as O2. First, P. aeruginosa utilizes quorum sensing and other community surveillance tactics to assess and regulate the type of competitive strategies employed [21, 176, 186–188]. These strategies involve the secretion of phenazine moieties such as pyocyanin, quinolones, and cyanide. The phenazine- and quinolone-based molecules have numerous social and resource acquisition activities including antibacterial properties, biofilm formation, iron chelation, cellular communication, extracellular electron transport and the production of ROS [189–193]. These molecules, along with cyanide which is also secreted by P. aeruginosa, can inhibit respiration in neighboring cells [188, 194]. Collectively, these mechanisms would modulate the phenotypes of nearby cells, forcing them into a fermentative metabolism which partitions O2 for P. aeruginosa respiration, while at the same time producing organic acids, a preferred carbon source. Long-term exposure to quinolones selects for small colony variants (SCVs) of S. aureus which have impaired respiratory activity and utilize primarily fermentative metabolisms [180, 188, 195].
Direct measurement of biomass productivity in natural consortia with cooccurring cCCR and rCCR microorganisms is difficult; artificial, in vitro culturing conditions often result in imbalanced resource partitioning, competition for resources and antagonism [181, 196]. Quantitative biomass productivity results are available for both evolved and synthetic systems that behave analogously to a cCCR + rCCR consortium. These systems have defined division of labor consisting of a primary, glucose-catabolizing population and a secondary, byproduct catabolizing population. The primary population prefers glucose, maximizes growth rate, and utilizes an overflow metabolism with the secretion of acetate and other byproducts. The byproduct population catabolizes the overflow byproducts and increases system productivity via two mechanisms. First, it modifies the local environment by removing acetate, which is a potent growth inhibitor that also lowers biomass yields on substrate, and secondly, the population utilizes non-preferred substrates that would otherwise be unutilized by the cCCR microorganism. The system design is simple but has a remarkable effect on system productivity. The in vitro consortium with evolved division of labor has 15% improvement in biomass productivity relative to the original monoculture during planktonic growth [197, 198]. The synthetic consortium with engineered division of labor has a 20% improvement in biomass productivity during planktonic growth and a 50% increase in biomass productivity during biofilm growth [199]. Both the evolved and engineered systems utilize substrate more effectively, driving glucose to lower concentrations [197–199]. The increases in biomass productivity can be explained by an enhanced return on carbon investment derived from the kinetic effect of division of labor, the removal of inhibitory byproducts which promotes more complete depletion of substrate, and the catabolism of non-preferred substrates that would otherwise be wasted. The section below provides a quantitative analysis of the effects.
Theoretical analysis of division of labor enhancing biomass productivity
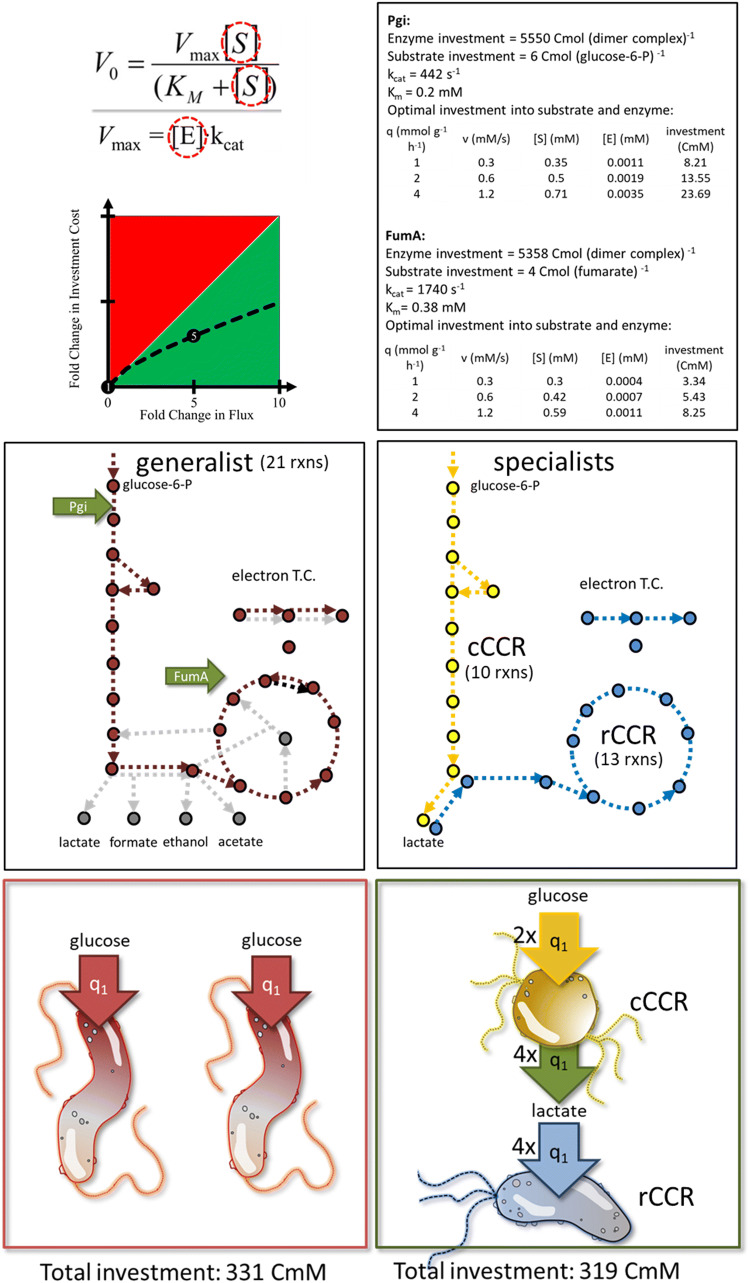
Enzyme flux is realized by investment into both enzymes and substrate pools. There is an optimal relationship between enzyme and substrate concentrations that minimizes the total investment necessary to drive a flux [107, 200–203]. Cross feeding consortia utilizing division of labor can have a better functional return on investment based on the properties of Michaelis–Menten kinetics. An example is presented which demonstrates a reduced total requirement for carbon with cross feeding between two specialist consortium members as compared to two generalist microorganisms. The aggregate specialist consortium and generalist system perform the same net transformation of glucose to CO2 and both systems consume glucose at the same total rate. For the quantitative example, enzymes from glycolysis are represented by properties of the Pgi enzyme and enzymes from the tricarboxylic acid (TCA) cycle are represented by the properties of the FumA enzyme. Irreversible Michaelis–Menten kinetics are assumed for the single substrate reactions; enzyme amino acid sequence and subunit composition for E. coli are used to calculate carbon investment into enzymes and substrates. Figure 4 details the enzyme parameters, overall biochemical pathways, and the cross feeding scheme based on a cCCR and rCCR metabolism.
Fig. 4.
Illustration of enhanced functional return on carbon investment in a cross feeding consortium relative to a generalist system. Enzyme flux requires investment into both enzyme and metabolite pools which can be optimized to reduce total cellular investment. Operating enzymes at higher fluxes represents a better functional return on resource investment (e.g., aggregate carbon atoms in enzyme and substrate). Analysis considers Michaelis–Menten-type kinetics, 2 cells using a generalist strategy, or 2 specialist cells cross feeding. The overall glucose flux and glucose transformation are the same for both scenarios. Glycolysis reactions are represented by the enzyme Pgi, while tricarboxylic acid (TCA) cycle enzymes are represented by the enzyme FumA. Enzyme values are from E. coli and obtained from Brenda and EcoCyc. The specialist consortium requires a smaller investment of carbon to attain the same flux and transformation as the generalist system. Specific glucose uptake rate (q1) was set to 1 mmol glucose g cdw−1 h−1. Portion of figure modified from Beck et al. 2016 [200]
The generalist system is comprised of two cells each operating a complete oxidation of glucose and each consuming glucose at a specific rate of 1 mmol glucose g cdw−1 h−1. The generalist cells have 21 considered enzymatic steps. The specialist consortium is also comprised of two cells with the first cell consuming glucose at a rate of 2 mmol glucose g cdw−1 h−1 and secreting lactate at 4 mmol lactate g cdw−1 h−1. The first cell has 10 considered enzymatic reactions. The second cell consumes the lactate at the same rate it is secreted. The second cell has 13 considered enzymatic reactions. When carbon investment into both enzymes and substrate pools is considered, the cross feeding system requires ~ 4% smaller investment to operate the same net transformation even though it contains two more enzyme-catalyzed steps than the generalist system. This system is analogous to cross feeding between S. aureus and P. aeruginosa. The reduced resource requirement is available for enhanced growth, providing an explanation for the enhanced biomass productivity.
The catabolism of overflow byproducts removes inhibitors, which improves overall consortium productivity, as mentioned above. This effect has been quantified in a recent stoichiometric modeling study that examined biofilm growth of an engineered, division of labor consortium which cross feeds acetate [204]. The model incorporated the genome-encoded metabolic potential of the two microorganisms, the diffusion-based transport of metabolites in the biofilm, and considered the inhibitory properties of the exchanged acetate (Fig. 5). The simulations captured the enhanced biomass productivity, approximately 50% improvement, observed in the corresponding experimental study based on the removal of the inhibitory acetate [199]. This modeling study did not account for resource investment into intracellular enzymes and metabolites highlighting how each mechanism in isolation can enhance productivity. Integrating both mechanisms into a computational representation is anticipated to further capture in situ mechanisms of productivity.
Fig. 5.
Role of byproduct inhibition in cross feeding consortia growing as biofilms. a Schematic of metabolic interactions between cCCR- and rCCR-cell types. b Example of the inhibitory properties of an organic acid as a function of concentration. cIn vitro data for an engineered, cross feeding consortium growing as a biofilm. d In silico prediction of biomass productivity for an engineered consortium growing as a biofilm. e Micrograph of a cryosectioned biofilm comprised of an engineered consortium, rCCR-cell type expressing rfp, cCCR-cell type expressing gfp. fIn silico predictions of cell-type spatial distributions within a biofilm. Portions of the figure are modified from Patel et al. 2019 [204] and Bernstein et al. 2012 [199]
Conclusions
Carbon catabolite repression is the result of natural selection within dynamic environments. Two common forms of carbon catabolite repression, classic carbon catabolite repression and reverse carbon catabolite repression have broad global distributions in natural microbial populations and have near mirror-image preferences for substrates. These metabolic strategies are typically studied from the perspective of monocultures. Here we present a consortia-based argument for the two metabolic strategies based on reciprocal social interactions including quorum sensing, biofilm formation, and virulence mechanisms. Appropriate ecological interpretation of global regulation schemes can open many new avenues of research leading to a greater understanding of the microbial world and its uses in agriculture, industry, and medicine. Box 3 highlights interesting, unresolved questions regarding rCCR  .
.
Acknowledgements
HP was supported by the Army Research Office award W911NF-16-1-0463, SLM was supported by the National Institutes of Health award U01EB019416, ADA was supported by the National Science Foundation award OIA 1736255, and RPC was supported by National Science Foundation award 1361240.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Law R. Optimal life histories under age-specific predation. Am Nat. 1979;114(3):399–417. doi: 10.1086/283488. [DOI] [Google Scholar]
- 2.Carlson RP, Taffs RL. Molecular-level tradeoffs and metabolic adaptation to simultaneous stressors. Curr Opin Biotechnol. 2010;21(5):670–676. doi: 10.1016/j.copbio.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoval AO, Sheftel H, Shinar G, Hart Y, Ramote O, Mayo A, Dekel E, Alon U, Science S, Series N, June N. Evolutionary trade-offs, pareto optimality, and the geometry of phenotype space. Science. 2012;336(6085):1157–1160. doi: 10.1126/science.1217405. [DOI] [PubMed] [Google Scholar]
- 4.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl Environ Microbiol. 2017;83(13):e00410–e00417. doi: 10.1128/AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Xia K, Yang X, Tang C. Growth strategy of microbes on mixed carbon sources. Nat Commun. 2019;10(1):1279. doi: 10.1038/s41467-019-09261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol. 2008;67(2):305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6(8):613. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 8.Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev. 2010;34(5):658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 9.Collier DN, Hager PW, Phibbs PV. Catabolite repression control in the Pseudomonads. Res Microbiol. 1996;147(6):551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 10.Durica-Mitic S, Gopel Y, Gorke B. Carbohydrate utilization in bacteria: making the most out of sugars with the help of small regulatory RNAs. Microbiol Spectr. 2018;6(2):1–19. doi: 10.1128/microbiolspec.rwr-0013-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinier A, Deutscher J. Sophisticated regulation of transcriptional factors by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. J Mol Biol. 2017;429(6):773–789. doi: 10.1016/j.jmb.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Vinuselvi P, Kim MK, Lee SK, Ghim CM. Rewiring carbon catabolite repression for microbial cell factory. BMB Rep. 2012;45(2):59–70. doi: 10.5483/bmbrep.2012.45.2.59. [DOI] [PubMed] [Google Scholar]
- 13.Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3(1):371–394. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 14.Chu D. Limited by sensing– A minimal stochastic model of the lag-phase during diauxic growth. J Theor Biol. 2017;414:137–146. doi: 10.1093/bjc/azq038. [DOI] [PubMed] [Google Scholar]
- 15.Chu DF. In silico evolution of diauxic growth. BMC Evol Biol. 2015;15(1):211. doi: 10.1186/s12862-015-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermsen R, Okano H, You C, Werner N, Hwa T. A growth-rate composition formula for the growth of E coli on co-utilized carbon substrates. Mol Syst Biol. 2015;11(4):801. doi: 10.15252/msb.20145537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci. 2014 doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotte O, Volkmer B, Radzikowski JL, Heinemann M. Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol Syst Biol. 2014;10:736. doi: 10.15252/msb.20135022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Succurro A, Segre D, Ebenhoh O. Emergent subpopulation behavior uncovered with a community dynamic metabolic model of Escherichia coli diauxic growth. mSystems. 2019 doi: 10.1128/msystems.00230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann T, Sorg T, Siehler SY, Gerischer U. Role of Acinetobacter baylyi Crc in catabolite repression of enzymes for aromatic compound catabolism. J Bacteriol. 2009;191(8):2834–2842. doi: 10.1128/JB.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Roder HL, Madsen JS, Bjarnsholt T, Sorensen SJ, Burmolle M. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol. 2016;7:1366. doi: 10.3389/fmicb.2016.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren D, Madsen JS, Sorensen SJ, Burmolle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9(1):81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russel J, Roder HL, Madsen JS, Burmolle M, Sorensen SJ. Antagonism correlates with metabolic similarity in diverse bacteria. Proc Natl Acad Sci USA. 2017;114(40):10684–10688. doi: 10.1073/pnas.1706016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein HC, Carlson RP. Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput Struct Biotechnol J. 2012;3(4):e201210017. doi: 10.5936/csbj.201210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scitable by Nature EDUCATION (2005) Nature Publishing Group. www.nature.com/scitable/definition/catabolite-repression-113/. Accessed 3 Sep 2018
- 27.Zhang YH, Lynd LR. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci USA. 2005;102(20):7321–7325. doi: 10.1073/pnas.0408734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parche S, Jacobs D, Arigoni F, Titgemeyer F, Jankovic I. Lactose-over-glucose preference in bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. Society. 2006;188(4):1260–1265. doi: 10.1128/JB.188.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng TK, Zeikus JG. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982;150(3):1391–1399. doi: 10.1128/JB.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ Microbiol. 2010;12(6):1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- 31.Carlson RP. Metabolic systems cost-benefit analysis for interpreting network structure and regulation. Bioinformatics. 2007;23(10):1258–1264. doi: 10.1093/bioinformatics/btm082. [DOI] [PubMed] [Google Scholar]
- 32.Carlson RP. Decomposition of complex microbial behaviors into resource-based stress responses. Bioinformatics. 2009;25(1):90–97. doi: 10.1093/bioinformatics/btn589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton WA, Dawes EA. A diauxic effect with Pseudomonas aeruginosa. Biochem J. 1959;71:25P–26P. [Google Scholar]
- 34.Harwood JP, Gazdar C, Prasad C, Peterkofsky A. A involvement of the glucose enzymes II of the sugar phosphotransferase system in the regulation of adenylate cyclase by glucose in Escherichia coli. J Biol Chem. 1976;25(8):2462–2468. [PubMed] [Google Scholar]
- 35.Feucht BU, Saier MH. Fine control of adenylate cyclase by the phosphoenolpyruvate:sugar phosphotransferase systems in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;141(2):603–610. doi: 10.1128/JB.141.2.603-610.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Titgemeyer F, Hillen W. Global control of sugar metabolism: a Gram-positive solution. Antonie Van Leeuwenhoek. 2002;82(1):59–71. doi: 10.1023/A:1020628909429. [DOI] [PubMed] [Google Scholar]
- 37.Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67(4):475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of α amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5(3):575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 39.Djordjevic GM, Tchieu JH, Saier MH. Genes involved in control of galactose uptake inlactobacillus brevis and reconstitution of the regulatory system in Bacillus subtilis. J Bacteriol. 2001;183(10):3224–3236. doi: 10.1128/JB.183.10.3224-3236.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol. 2007;189(4):1366–1381. doi: 10.1128/jb.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdou L, Boileau C, Philip Pd, Pagès S, Fiérobe H-P, Tardif C. Transcriptional regulation of the clostridium cellulolyticum cip-cel Operon: a complex mechanism involving a catabolite-responsive element. J Bacteriol. 2008;190(5):1499–1506. doi: 10.1128/JB.01160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arndt A, Eikmanns BJ. The alcohol dehydrogenase gene adhA in corynebacterium glutamicum is subject to carbon catabolite repression. J Bacteriol. 2007;189(20):7408–7416. doi: 10.1128/JB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J Bacteriol. 2004;186(9):2798–2809. doi: 10.1128/JB.186.9.2798-2809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178(7):2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwakman JH, Postma PW. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J Bacteriol. 1994;176(9):2694–2698. doi: 10.1128/jb.176.9.2694-2698.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Commichau FM, Stülke J. Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol Microbiol. 2008;67(4):692–702. doi: 10.1111/j.1365-2958.2007.06071.x. [DOI] [PubMed] [Google Scholar]
- 47.Chu D, Barnes DJ. The lag-phase during diauxic growth is a trade-off between fast adaptation and high growth rate. Sci Rep. 2016;6:25191. doi: 10.1038/srep25191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wezel GPV, König M, Mahr K, Nothaft H, Thomae AW, Bibb M, Titgemeyer F. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in streptomyces coelicolor A3(2) MMB. 2007;12(1–2):67–74. doi: 10.1159/000096461. [DOI] [PubMed] [Google Scholar]
- 49.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293(2):199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 50.Malan TP, Kolb A, Buc H, McClure WR. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- 51.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11(2):87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Harwood JP, Gazdar C, Prasad C, Peterkofsky A, Curtis SJ, Epstein W. Involvement of the glucose enzymes II of the sugar phosphotransferase system in the regulation of adenylate cyclase by glucose in Escherichia coli. J Biol Chem. 1976;251(8):2462–2468. [PubMed] [Google Scholar]
- 53.Park Y-H, Lee BR, Seok Y-J, Peterkofsky A. In vitro reconstitution of catabolite repression in Escherichia coli. J Biol Chem. 2006;281(10):6448–6454. doi: 10.1074/jbc.M512672200. [DOI] [PubMed] [Google Scholar]
- 54.Tagami H, Aiba H. A common role of CRP in transcription activation: CRP acts transiently to stimulate events leading to open complex formation at a diverse set of promoters. EMBO J. 1998;17(6):1759–1767. doi: 10.1093/emboj/17.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cases I, Velázquez F, de Lorenzo V. The ancestral role of the phosphoenolpyruvate–carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res Microbiol. 2007;158(8):666–670. doi: 10.1016/j.resmic.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70(4):939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Mol Biol Rev. 1993;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hogema BM, Arents JC, Bader R, Postma PW. Autoregulation of lactose uptake through the LacYpermease by enzyme IIAGlc of the PTS in Escherichia coli K-12. Mol Microbiol. 1999;31(6):1825–1833. doi: 10.1046/j.1365-2958.1999.01319.x. [DOI] [PubMed] [Google Scholar]
- 59.Inada T, Kimata K, Aiba H. Mechanism responsible for glucose–lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells. 1996;1(3):293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 60.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. cAMP receptor protein–cAMP plays a crucial role in glucose–lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc Natl Acad Sci. 1997;94(24):12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beckwith JR. Regulation of the Lac operon. Science. 1967;156(3775):597–604. doi: 10.1126/science.156.3775.597. [DOI] [PubMed] [Google Scholar]
- 62.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43(6):880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31(8):1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beisel CL, Storz G. Discriminating tastes: physiological contributions of the Hfq-binding small RNA Spot 42 to catabolite repression. RNA Biol. 2011;8(5):766–770. doi: 10.4161/rna.8.5.16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41(3):286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500(7462):301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bettenbrock K, Fischer S, Kremling A, Jahreis K, Sauter T, Gilles ED. A quantitative approach to catabolite repression in Escherichia coli. J Biol Chem. 2006;281(5):2578–2584. doi: 10.1074/jbc.M508090200. [DOI] [PubMed] [Google Scholar]
- 68.Bren A, Park JO, Towbin BD, Dekel E, Rabinowitz JD, Alon U. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci Rep. 2016;6:24834. doi: 10.1038/srep24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15(6):1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 70.Jones BE, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit RE. Binding of the catabolite repressor protein CcpA to Its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272(42):26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 71.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci. 1998;95(4):1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jault J-M, Fieulaine S, Nessler S, Gonzalo P, Pietro AD, Deutscher J, Galinier A. The HPr kinase from Bacillus subtilis is a homo-oligomeric enzyme which exhibits strong positive cooperativity for nucleotide and fructose 1,6-bisphosphate binding. J Biol Chem. 2000;275(3):1773–1780. doi: 10.1074/jbc.275.3.1773. [DOI] [PubMed] [Google Scholar]
- 73.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27(6):1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 74.Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118(6):731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 75.Seidel G, Diel M, Fuchsbauer N, Hillen W. Quantitative interdependence of coeffectors, CcpA and cre in carbon catabolite regulation of Bacillus subtilis. FEBS J. 2005;272(10):2566–2577. doi: 10.1111/j.1742-4658.2005.04682.x. [DOI] [PubMed] [Google Scholar]
- 76.Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2004;39(5):1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida K, Kobayashi K, Miwa Y, Kang CM, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29(3):683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim SU, Kim DC, Dhurjati P. Mathematical modeling for mixed culture growth of two bacterial populations with opposite substrate preferences. Biotechnol Bioeng. 1988;31(2):144–159. doi: 10.1002/bit.260310208. [DOI] [PubMed] [Google Scholar]
- 79.Ramkrishna D, Kompala DS, Tsao GT. Are microbes optimal strategists? Biotechnol Progr. 1987;3(3):121–126. doi: 10.1002/btpr.5420030302. [DOI] [Google Scholar]
- 80.Kim SU, Dhurjati P. Analysis of two interacting bacterial populations with opposite substrate preferences. Biotechnol Bioeng. 1987;29(8):1015–1023. doi: 10.1002/bit.260290813. [DOI] [PubMed] [Google Scholar]
- 81.Wolff JA, MacGregor CH, Eisenberg RC, Phibbs PV. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol. 1991;173(15):4700–4706. doi: 10.1128/JB.173.15.4700-4706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart PS. Diffusion in Biofilms. J Bacteriol. 2003;185(5):1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sonnleitner E, Blasi U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 2014;10(6):e1004440. doi: 10.1371/journal.pgen.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci. 2009;106(51):21866–21871. doi: 10.1073/pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sonnleitner E, Wulf A, Campagne S, Pei XY, Wolfinger MT, Forlani G, Prindl K, Abdou L, Resch A, Allain FH, Luisi BF, Urlaub H, Blasi U. Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res. 2018;46(3):1470–1485. doi: 10.1093/nar/gkx1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moreno R, Hernandez-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol. 2015;17(1):105–118. doi: 10.1111/1462-2920.12499. [DOI] [PubMed] [Google Scholar]
- 87.Kambara TK, Ramsey KM, Dove SL. Pervasive targeting of nascent transcripts by Hfq. Cell Rep. 2018;23(5):1543–1552. doi: 10.1016/j.celrep.2018.03.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grenga L, Chandra G, Saalbach G, Galmozzi CV, Kramer G, Malone JG. Analyzing the complex regulatory landscape of Hfq—an integrative, multi-omics approach. Front Microbiol. 2017;8:1784. doi: 10.3389/fmicb.2017.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moreno R, Fonseca P, Rojo F. Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol. 2012;83(1):24–40. doi: 10.1111/j.1365-2958.2011.07912.x. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Gokhale CS, Rainey PB, Zhang XX. Unravelling the complexity and redundancy of carbon catabolic repression in Pseudomonas fluorescens SBW25. Mol Microbiol. 2017;105(4):589–605. doi: 10.1111/mmi.13720. [DOI] [PubMed] [Google Scholar]
- 91.Filiatrault MJ, Stodghill PV, Wilson J, Butcher BG, Chen H, Myers CR, Cartinhour SW. CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000. RNA Biol. 2013;10(2):245–255. doi: 10.4161/rna.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia-Maurino SM, Perez-Martinez I, Amador CI, Canosa I, Santero E. Transcriptional activation of the CrcZ and CrcY regulatory RNAs by the CbrB response regulator in Pseudomonas putida. Mol Microbiol. 2013;89(1):189–205. doi: 10.1111/mmi.12270. [DOI] [PubMed] [Google Scholar]
- 93.Kremling A, Geiselmann J, Ropers D, de Jong H. Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbiol. 2015;23(2):99–109. doi: 10.1016/j.tim.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 94.Kremling A, Geiselmann J, Ropers D, de Jong H. An ensemble of mathematical models showing diauxic growth behaviour. BMC Syst Biol. 2018;12(1):1–16. doi: 10.1186/s12918-018-0604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kompala DS, Ramkrishna D, Jansen NB, Tsao GT. Investigation of bacterial growth on mixed substrates: experimental evaluation of cybernetic models. Biotechnol Bioeng. 1986;28(7):1044–1055. doi: 10.1002/bit.260280715. [DOI] [PubMed] [Google Scholar]
- 96.DeVilbiss F, Mandli A, Ramkrishna D. Consistency of cybernetic variables with gene expression profiles: a more rigorous test. Biotechnol Prog. 2018;34(4):858–867. doi: 10.1002/btpr.2654. [DOI] [PubMed] [Google Scholar]
- 97.Schuster R, Schuster S. Refined algorithm and computer program for calculating all non–negative fluxes admissible in steady states of biochemical reaction systems with or without some flux rates fixed. Bioinformatics. 1993;9(1):79–85. doi: 10.1093/bioinformatics/9.1.79. [DOI] [PubMed] [Google Scholar]
- 98.Schuster S, Dandekar T, Fell DA. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999;17(2):53–60. doi: 10.1016/S0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- 99.Varma A, Boesch BW, Palsson BO. Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates. Appl Environ Microbiol. 1993;59(8):2465–2473. doi: 10.1128/AEM.59.8.2465-2473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia-Coli W3110. Appl Environ Microb. 1994;60(10):3724–3731. doi: 10.1128/AEM.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beck AE, Hunt KA, Carlson RP. Measuring cellular biomass composition for computational biology applications. Processes. 2018;6(5):38. doi: 10.3390/pr6050038. [DOI] [Google Scholar]
- 102.Schuster S, Pfeiffer T, Fell DA. Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol. 2008;252(3):497–504. doi: 10.1016/j.jtbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 103.Beg QK, Vazquez A, Ernst J, de Menezes MA, Bar-Joseph Z, Barabasi AL, Oltvai ZN. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci USA. 2007;104(31):12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vazquez A, Oltvai ZN. Macromolecular crowding explains overflow metabolism in cells. Sci Rep. 2016;6:31007. doi: 10.1038/srep31007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carlson R, Srienc F. Fundamental Escherichia coli biochemical pathways for biomass and energy production: identification of reactions. Biotechnol Bioeng. 2004;85(1):1–19. doi: 10.1002/bit.10812. [DOI] [PubMed] [Google Scholar]
- 106.Carlson R, Srienc F. Fundamental Escherichia coli biochemical pathways for biomass and energy production: creation of overall flux states. Biotechnol Bioeng. 2004;86(2):149–162. doi: 10.1002/bit.20044. [DOI] [PubMed] [Google Scholar]
- 107.Carlson RP, Beck AE, Phalak P, Fields MW, Gedeon T, Hanley L, Harcombe WR, Henson MA, Heys JJ. Competitive resource allocation to metabolic pathways contributes to overflow metabolisms and emergent properties in cross-feeding microbial consortia. Biochem Soc Trans. 2018;46(2):269–284. doi: 10.1042/BST20170242. [DOI] [PubMed] [Google Scholar]
- 108.Zhuang K, Vemuri GN, Mahadevan R. Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol. 2011;7:500. doi: 10.1038/msb.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Szenk M, Dill KA, de Graff AMR. Why do fast-growing bacteria enter overflow metabolism? Testing the membrane real estate hypothesis. Cell Syst. 2017;5(2):95–104. doi: 10.1016/j.cels.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 110.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528(7580):99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goelzer A. Fromion V (2011) Bacterial growth rate reflects a bottleneck in resource allocation. Biochim Biophys Acta. 1810;10:978–988. doi: 10.1016/j.bbagen.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 112.Goelzer A, Fromion V. Resource allocation in living organisms. Biochem Soc Trans. 2017;45(4):945–952. doi: 10.1042/BST20160436. [DOI] [PubMed] [Google Scholar]
- 113.Yang L, Ma D, Ebrahim A, Lloyd CJ, Saunders MA, Palsson BO. solveME: fast and reliable solution of nonlinear ME models. BMC Bioinformatics. 2016;17(1):391. doi: 10.1186/s12859-016-1240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mahadevan R, Edwards JS, Doyle FJ. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83(3):1331–1340. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rentz JA, Alvarez PJ, Schnoor JL. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ Microbiol. 2004;6(6):574–583. doi: 10.1111/j.1462-2920.2004.00589.x. [DOI] [PubMed] [Google Scholar]
- 116.Tiwari N, Campbell J. Enzymatic control of the metabolic activity of Pseudomonas aeruginosa grown in glucose or succinate media. Biochim Biophys Acta (BBA) Gen Subj. 1969;192(3):395–401. doi: 10.1016/0304-4165(69)90388-2. [DOI] [PubMed] [Google Scholar]
- 117.Trautwein K, Grundmann O, Wohlbrand L, Eberlein C, Boll M, Rabus R. Benzoate mediates repression of C(4)-dicarboxylate utilization in “Aromatoleum aromaticum” EbN1. J Bacteriol. 2012;194(2):518–528. doi: 10.1128/JB.05072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berger A, Dohnt K, Tielen P, Jahn D, Becker J, Wittmann C. Robustness and plasticity of metabolic pathway flux among uropathogenic isolates of Pseudomonas aeruginosa. PLoS One. 2014;9(4):e88368. doi: 10.1371/journal.pone.0088368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hintermayer SB, Weuster-Botz D. Experimental validation of in silico estimated biomass yields of Pseudomonas putida KT2440. Biotechnol J. 2017;12(6):1600720. doi: 10.1002/biot.201600720. [DOI] [PubMed] [Google Scholar]
- 120.La Rosa R, Behrends V, Williams HD, Bundy JG, Rojo F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ Microbiol. 2016;18(3):807–818. doi: 10.1111/1462-2920.13126. [DOI] [PubMed] [Google Scholar]
- 121.Nikel PI, Chavarria M, Fuhrer T, Sauer U, de Lorenzo V. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the entner-doudoroff, embden-meyerhof-parnas, and pentose phosphate pathways. J Biol Chem. 2015;290(43):25920–25932. doi: 10.1074/jbc.M115.687749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci USA. 2013;110(24):10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Oberhardt MA, Goldberg JB, Hogardt M, Papin JA. Metabolic network analysis of Pseudomonas aeruginosa during chronic cystic fibrosis lung infection. J Bacteriol. 2010;192(20):5534–5548. doi: 10.1128/JB.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu Z, Fang X, Wood TK, Huang ZJ. A systems-level approach for investigating Pseudomonas aeruginosa biofilm formation. PLoS One. 2013;8(2):e57050. doi: 10.1371/journal.pone.0057050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nogales J, Palsson BO, Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Borgos SE, Bordel S, Sletta H, Ertesvag H, Jakobsen O, Bruheim P, Ellingsen TE, Nielsen J, Valla S. Mapping global effects of the anti-sigma factor MucA in Pseudomonas fluorescens SBW25 through genome-scale metabolic modeling. BMC Syst Biol. 2013;7:19. doi: 10.1186/1752-0509-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goyal A, Dubinkina V, Maslov S. Multiple stable states in microbial communities explained by the stable marriage problem. ISME J. 2018;12(12):2823–2834. doi: 10.1038/s41396-018-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ. Engineering microbial consortia by division of labor. Microb Cell Fact. 2019;18(1):35. doi: 10.1186/s12934-019-1083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shuler ML, Kargi F, DeLisa M. Bioprocess engineering: basic concepts. 3. Boston: Prentice Hall; 2017. [Google Scholar]
- 131.Kreft JU. Biofilms promote altruism. Microbiology. 2004;150(Pt 8):2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- 132.Boyle KE, Monaco HT, Deforet M, Yan J, Wang Z, Rhee K, Xavier JB. Metabolism and the evolution of social behavior. Mol Biol Evol. 2017;34(9):2367–2379. doi: 10.1093/molbev/msx174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol. 2006;59(5):1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 135.Zhang L, Gao Q, Chen W, Qin H, Hengzhuang W, Chen Y, Yang L, Zhang G. Regulation of pqs quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiology. 2013;159(Pt 9):1931–1936. doi: 10.1099/mic.0.066266-0. [DOI] [PubMed] [Google Scholar]
- 136.Yang N, Lan L. Pseudomonas aeruginosa Lon and ClpXP proteases: roles in linking carbon catabolite repression system with quorum-sensing system. Curr Genet. 2016;62(1):1–6. doi: 10.1007/s00294-015-0499-5. [DOI] [PubMed] [Google Scholar]
- 137.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13(1):27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 138.Coenye T. Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing. Future Microbiol. 2010;5(7):1087–1099. doi: 10.2217/fmb.10.68. [DOI] [PubMed] [Google Scholar]
- 139.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 140.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 141.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104(3):876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14(9):589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 143.Stewart PS. A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnol Bioeng. 1998;59(3):261–272. doi: 10.1002/(SICI)1097-0290(19980805)59:3<261::AID-BIT1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 144.Kreft JU, Bonhoeffer S. The evolution of groups of cooperating bacteria and the growth rate versus yield trade-off. Microbiology. 2005;151(Pt 3):637–641. doi: 10.1099/mic.0.27415-0. [DOI] [PubMed] [Google Scholar]
- 145.Liu J, Prindle A, Humphries J, Gabalda-Sagarra M, Asally M, Lee DY, Ly S, Garcia-Ojalvo J, Suel GM. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature. 2015;523(7562):550–554. doi: 10.1038/nature14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu J, Martinez-Corral R, Prindle A, Lee DD, Larkin J, Gabalda-Sagarra M, Garcia-Ojalvo J, Suel GM. Coupling between distant biofilms and emergence of nutrient time-sharing. Science. 2017;356(6338):638–642. doi: 10.1126/science.aah4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johnson Douglas I. Bacterial Pathogens and Their Virulence Factors. Cham: Springer International Publishing; 2017. Pseudomonas spp; pp. 325–338. [Google Scholar]
- 148.Pusic P, Sonnleitner E, Krennmayr B, Heitzinger DA, Wolfinger MT, Resch A, Blasi U. Harnessing metabolic regulation to increase Hfq-dependent antibiotic susceptibility in Pseudomonas aeruginosa. Front Microbiol. 2018;9:2709. doi: 10.3389/fmicb.2018.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Corona F, Martinez JL, Nikel PI. The global regulator Crc orchestrates the metabolic robustness underlying oxidative stress resistance in Pseudomonas aeruginosa. Environ Microbiol. 2019;21(3):898–912. doi: 10.1111/1462-2920.14471. [DOI] [PubMed] [Google Scholar]
- 150.Bernal V, Castano-Cerezo S, Canovas M. Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl Microbiol Biotechnol. 2016;100(21):8985–9001. doi: 10.1007/s00253-016-7832-x. [DOI] [PubMed] [Google Scholar]
- 151.Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521(7553):516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24(2):195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yeung AT, Bains M, Hancock RE. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol. 2011;193(4):918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hufnagel DA, Evans ML, Greene SE, Pinkner JS, Hultgren SJ, Chapman MR. The catabolite repressor protein-cyclic AMP complex regulates csgD and biofilm formation in uropathogenic Escherichia coli. J Bacteriol. 2016;198(24):3329–3334. doi: 10.1128/JB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jackson DW, Simecka JW, Romeo T. Catabolite repression of Escherichia coli biofilm formation. J Bacteriol. 2002;184(12):3406–3410. doi: 10.1128/jb.184.12.3406-3410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.O’Toole GA, Gibbs KA, Hager PW, Phibbs PV, Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2000;182(2):425–431. doi: 10.1128/jb.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pusic P, Tata M, Wolfinger MT, Sonnleitner E, Haussler S, Blasi U. Cross-regulation by CrcZ RNA controls anoxic biofilm formation in Pseudomonas aeruginosa. Sci Rep. 2016;6:39621. doi: 10.1038/srep39621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Amador CI, Lopez-Sanchez A, Govantes F, Santero E, Canosa I. A Pseudomonas putida cbrB transposon insertion mutant displays a biofilm hyperproducing phenotype that is resistant to dispersal. Environ Microbiol Rep. 2016;8(5):622–629. doi: 10.1111/1758-2229.12414. [DOI] [PubMed] [Google Scholar]
- 160.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun. 2008;76(5):2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sadykov MR, Hartmann T, Mattes TA, Hiatt M, Jann NJ, Zhu Y, Ledala N, Landmann R, Herrmann M, Rohde H, Bischoff M, Somerville GA. CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology. 2011;157(Pt 12):3458–3468. doi: 10.1099/mic.0.051243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ha JH, Hauk P, Cho K, Eo Y, Ma X, Stephens K, Cha S, Jeong M, Suh JY, Sintim HO, Bentley WE, Ryu KS. Evidence of link between quorum sensing and sugar metabolism in Escherichia coli revealed via cocrystal structures of LsrK and HPr. Sci Adv. 2018;4(6):eaar7063. doi: 10.1126/sciadv.aar7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mitra A, Herren CD, Patel IR, Coleman A, Mukhopadhyay S. Integration of AI-2 based cell-cell signaling with metabolic cues in Escherichia coli. PLoS One. 2016;11(6):e0157532. doi: 10.1371/journal.pone.0157532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J Bacteriol. 2005;187(6):2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 166.Hartmann T, Baronian G, Nippe N, Voss M, Schulthess B, Wolz C, Eisenbeis J, Schmidt-Hohagen K, Gaupp R, Sunderkotter C, Beisswenger C, Bals R, Somerville GA, Herrmann M, Molle V, Bischoff M. The catabolite control protein E (CcpE) affects virulence determinant production and pathogenesis of Staphylococcus aureus. J Biol Chem. 2014;289(43):29701–29711. doi: 10.1074/jbc.M114.584979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chakravarthy S, Butcher BG, Liu Y, D’Amico K, Coster M, Filiatrault MJ. Virulence of Pseudomonas syringae pv. tomato DC3000 is influenced by the catabolite repression control protein Crc. Mol Plant Microbe Interact. 2017;30(4):283–294. doi: 10.1094/mpmi-09-16-0196-r. [DOI] [PubMed] [Google Scholar]
- 168.Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR. Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. MBio. 2017;8(5):e01068-17. doi: 10.1128/mbio.01068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Riviere A, Selak M, Geirnaert A, Van den Abbeele P, De Vuyst L. Complementary mechanisms for degradation of inulin-type fructans and arabinoxylan oligosaccharides among bifidobacterial strains suggest bacterial cooperation. Appl Environ Microbiol. 2018 doi: 10.1128/AEM.02893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 171.Orazi G, O’Toole GA (2017) Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio 8(4). 10.1128/mbio.00873-17 [DOI] [PMC free article] [PubMed]
- 172.Rhoads DD, Wolcott RD, Sun Y, Dowd SE. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci. 2012;13(3):2535–2550. doi: 10.3390/ijms13032535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One. 2011;6(11):e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J. 2006;3(3):225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, Moser C, Tolker-Nielsen T, Hoiby N, Givskov M, Bjarnsholt T. Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol. 2008;46(8):2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA. 2013;110(3):1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52(3):188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Siddiqui AR, Bernstein JM. Chronic wound infection: facts and controversies. Clin Dermatol. 2010;28(5):519–526. doi: 10.1016/j.clindermatol.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 179.Sanchez CJ, Jr, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, Murray CK. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Noto MJ, Burns WJ, Beavers WN, Skaar EP. Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J Bacteriol. 2017;199(17):e00221-17. doi: 10.1128/jb.00221-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82(11):4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25(1):193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Brileya KA, Camilleri LB, Zane GM, Wall JD, Fields MW. Biofilm growth mode promotes maximum carrying capacity and community stability during product inhibition syntrophy. Front Microbiol. 2014;5:693. doi: 10.3389/fmicb.2014.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107(5):2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.de Mazancourt C, Schwartz MW. A resource ratio theory of cooperation. Ecol Lett. 2010;13(3):349–359. doi: 10.1111/j.1461-0248.2009.01431.x. [DOI] [PubMed] [Google Scholar]
- 186.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci USA. 2015;112(7):2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Allegretta G, Maurer CK, Eberhard J, Maura D, Hartmann RW, Rahme L, Empting M. In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front Microbiol. 2017;8(MAY):1–12. doi: 10.3389/fmicb.2017.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O’Toole GA. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015;197(14):2252–2264. doi: 10.1128/jb.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Das T, Kutty SK, Tavallaie R, Ibugo AI, Panchompoo J, Sehar S, Aldous L, Yeung AW, Thomas SR, Kumar N, Gooding JJ, Manefield M. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep. 2015;5:8398. doi: 10.1038/srep08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol. 2014;92(2):399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101(5):1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Meirelles LA, Newman DK. Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol Microbiol. 2018;110(6):995–1010. doi: 10.1111/mmi.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hall Susan, McDermott Catherine, Anoopkumar-Dukie Shailendra, McFarland Amelia, Forbes Amanda, Perkins Anthony, Davey Andrew, Chess-Williams Russ, Kiefel Milton, Arora Devinder, Grant Gary. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins. 2016;8(8):236. doi: 10.3390/toxins8080236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2(2):71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 195.Hoffman LR, Deziel E, D’Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci. 2006;103(52):19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187(2):554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kinnersley M, Wenger J, Kroll E, Adams J, Sherlock G, Rosenzweig F. Ex uno plures: clonal reinforcement drives evolution of a simple microbial community. PLoS Genet. 2014;10(6):e1004430. doi: 10.1371/journal.pgen.1004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kinnersley MA, Holben WE, Rosenzweig F. E Unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 2009;5(11):e1000713. doi: 10.1371/journal.pgen.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bernstein HC, Paulson SD, Carlson RP. Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity. J Biotechnol. 2012;157(1):159–166. doi: 10.1016/j.jbiotec.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Beck AE, Hunt K, Bernstein HC, Carlson RP. Interpreting and designing microbial communities for bioprocess applications, from components to interactions to emergent properties. In: Eckert CA, Trinh CT, editors. Biotechnology for biofuel production and optimization. Amsterdam: Elsevier; 2016. pp. 407–432. [Google Scholar]
- 201.Carlson RP, Oshota OJ, Taffs RL. Systems analysis of microbial adaptations to simultaneous stresses. In: Wang X, Chen J, Quinn P, editors. Reprogramming microbial metabolic pathways. Dordrecht: Springer; 2012. [DOI] [PubMed] [Google Scholar]
- 202.Schepens D, Beck AE, Heys JJ, Gedeon T, Carlson RP (2017) The Benefits of resource partitioning and division of labor in microbial consortia. In: P Amar FK, V Norris (ed) Advances in Systems and Synthetic Biology EDP Sciences Publishing, pp 137–148
- 203.Schepens D, Carlson RP, Heys J, Beck AE, Gedeon T. Role of resource allocation and transport in emergence of cross-feeding in microbial consortia. J Theor Biol. 2019;467:150–163. doi: 10.1016/j.jtbi.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 204.Patel A, Carlson RP, Henson MA. In silico metabolic design of two-strain biofilm systems predicts enhanced biomass production and biochemical synthesis. Biotechnol J. 2019;14(7):e1800511. doi: 10.1002/biot.201800511. [DOI] [PubMed] [Google Scholar]