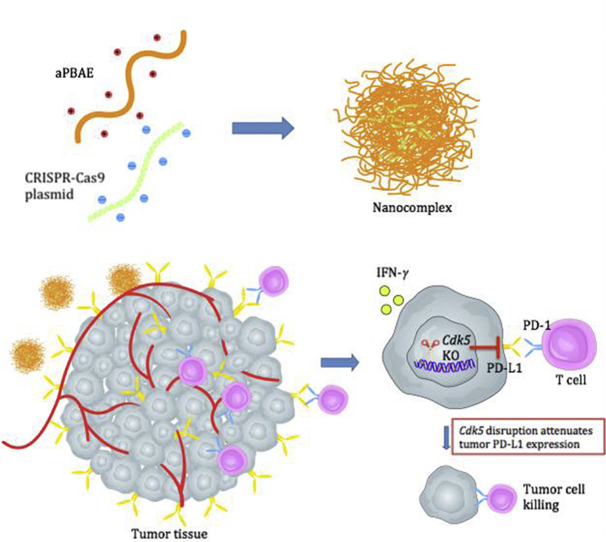
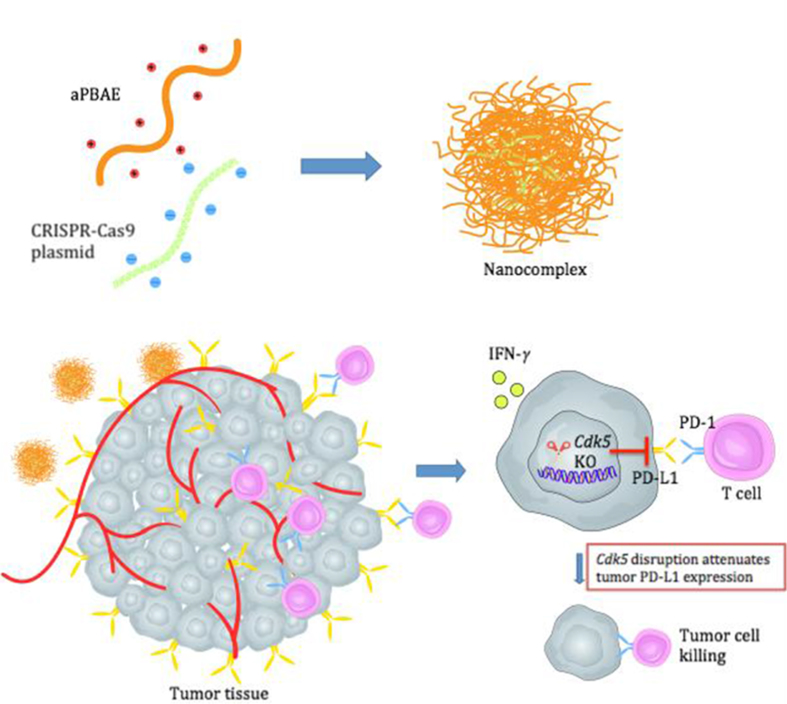
Abstract
Blocking the programmed death-ligand 1 (PD-L1) on tumor cells with monoclonal antibody therapy has emerged as powerful weapon in cancer immunotherapy. However, only a minority of patients presented immune responses in clinical trials. To develop an alternative treatment method based on immune checkpoint blockade, we designed a novel and efficient CRISPR-Cas9 genome editing system delivered by cationic copolymer aPBAE to downregulate PD-L1 expression on tumor cells via specifically knocking out Cyclin-dependent kinase 5 (Cdk5) gene in vivo. The expression of PD-L1 on tumor cells was significantly attenuated by knocking out Cdk5, leading to effective tumor growth inhibition in murine melanoma and lung metastasis suppression in triple-negative breast cancer. Importantly, we demonstrated that aPBAE/Cas9-Cdk5 treatment elicited strong T cell-mediated immune responses in tumor microenvironment that the population of CD8+ T cells was significantly increased while regulatory T cells (Tregs) was decreased. It may be the first case to exhibit direct in vivo PD-L1 downregulation via CRISPR-Cas9 genome editing technology for cancer therapy. It will provide promising strategy for preclinical antitumor treatment through the combination of nanotechnology and genome engineering.
KEY WORDS: CRISPR-Cas9 genome editing system, Cyclin-dependent kinase 5 (Cdk5), Programmed death-ligand 1 (PD-L1), Antitumor immunity, Nanoparticles
Graphical abstract
A novel CRISPR-Cas9 genome editing system delivered by cationic copolymer aPBAE to downregulate PD-L1 expression on tumor cells via specifically knocking out Cdk5 gene in vivo, which provided an alternative treatment method based on immune checkpoint blockade.
1. Introduction
Immune checkpoints are essential for maintaining immune homeostasis and preventing autoimmunity as they are regulators of immune activation. However, in many cancers, the intrinsic immune checkpoint mechanisms are often overactivated resulting in suppression on antitumor immune responses. Engagement of programmed cell death 1 (PD-1) and its ligand PD-L1 is a dominant immune checkpoint pathway that mainly exerts in the tumor microenvironment1. PD-1 is expressed on the surface of activated T cells, B cells, macrophages and many tumor-infiltrating lymphocytes (TILs)2, while PD-L1 is predominantly expressed in a broad spectrum of solid tumors and leukemia3, 4. Due to the high expression of PD-L1, tumor cells could evade immune attack and form immunosuppressive tumor microenvironment. Immune checkpoint therapies could release the inhibitory mechanisms of T-cell mediated immunity and promote cytotoxic T lymphocytes (CTLs) responses, providing a potent strategy for antitumor treatment5. Nowadays, the blocking monoclonal antibodies of PD-1 or PD-L1, approved by the U.S. Food and Drug Administration (FDA), have exhibited notable clinical successes and emerged as powerful weapon in cancer immunotherapy6, 7. However, only a minority of patients presented immune responses and severe side effects also exhibited during the immune checkpoint inhibitors therapy8, 9. The combination of immune checkpoint blockade and nanotechnology facilitated delivery of anti-PD antibodies and enhanced antitumor efficacy10, 11, 12. Except these, increasing researches that focus on the intrinsic immune modulatory pathways are being investigated to explore alternative therapies. For example, gene therapy is speculated to interfere with immune modulatory pathways that could elicit specific and persistent antitumor immune responses to overcome the limits of current antibody therapy.
As we know, the expression of PD-L1 can be mainly downregulated by means of small interfering RNA (siRNA) combined with nanoparticles13, 14, 15. Even though it is evidenced to inhibit tumor growth to some extent, the siRNA treatment has its own limits to overcome. Recently, a newly emerged CRISPR-Cas9 genome editing system provides a tremendous promising strategy for versatile and high specifically targeting genome editing that has been widely applied in treating various genetic diseases16, 17. The CRISPR-Cas9 system was initially found in prokaryote as an immune system resistant to foreign genetic elements18 and now has been used for genome engineering in mammalian cells. Cas9 is an endonuclease that can be guided to the target sites with the aid of two RNAs, which are crRNA and transactivating crRNA (tracrRNA). crRNA defines the genomic target by encoding the guide RNAs which contain 20 nucleotide target sequence. The tracrRNA functionalizes to link the crRNA to Cas9 and facilitates the processing of crRNA into discrete units19. Therefore, the genome of cells can be precisely edited at the desired location by delivering Cas9/gRNA complex, including knocking out the existing genes and knocking in functional genes.
Cyclin-dependent kinase 5 (Cdk5), a serine/threonine kinase whose activity depends on p35 and/or p39, was initially considered as playing a crucial role in center nervous system development20. Cdk5 has also been elucidated to participate in apoptosis, angiogenesis and senescence of various tumor cells21, 22, 23. Strikingly, a novel function of Cdk5 in immunity has been now identified to regulate the PD-1/PD-L1 pathway. The cell line in which Cdk5 was previously knocked out expressed much less PD-L1 due to the interference with interferon regulatory factor-2 (IRF2) and interferon regulatory factor 2-binding protein 2 (IRF2BP2). As a result, such cells failed to grow up to large-sized tumor in mice model owing to enhanced T cells infiltration and improved immune surveillance compared to wide type cells24. Although this research elucidated that the inhibition of Cdk5 might provide a new option for antitumor immunity by downregulation of PD-L1, further work is still needed to realize direct attenuation of PD-L1 in vivo in order to provide promising strategy for preclinical antitumor treatment.
To effectively attenuate the expression of PD-L1 in tumor cells, we first designed CRISPR-Cas9 plasmid to target Cdk5 gene. To further overcome the drawback of CRISPR-Cas9 plasmid with relatively low delivery efficiency in vivo, a biodegradable cationic polymer, poly(β-amino esters) (PBAEs) was applied to facilitate the gene delivery25, 26, 27. It could interact with CRISPR-Cas9 plasmid and condense into nano-sized complex particle. Then the nanoparticles were delivered into tumors to evidence the role of Cdk5 in the modulation of immune checkpoint pathway (Scheme 1). Two kinds of tumor model, melanoma and triple negative breast cancer, which are identified to exhibit co-occurrence of elevated Cdk5 and PD-L1 mRNA expression24, were chosen to investigate the efficacy of Cdk5 genetic intervention. With the combination of nanoparticle delivery system, obvious PD-L1 downregulation was observed with modulated immunosuppressive microenvironment and enhanced antitumor immunity. This research might firstly investigate the in vivo efficacy of PD-L1 downregulation mediated by CRISPR-Cas9 genome editing for cancer therapy. It is speculated to provide powerful evidence for preclinical antitumor treatment and promote the development for translational medicine in future.
Scheme 1.
Preparation of aPBAE/CRISPR-Cas9 nanoparticles and schematic illustration of PD-L1 attenuation therapy.
2. Materials and methods
2.1. Materials and reagents
pX330 plasmid was obtained from Addgene (#42230). pMax-GFP plasmid was acquired from Amaxa (Lonza, Basel, Switzerland). Antibodies used for Western blot were from as the following, Cdk5 (Santa Cruz Biotechnology, SC-6247, Delaware Ave, USA), p35 (Santa Cruz Biotechnology, SC293184), β-actin (Cell Signaling, 4967S, Danvers, USA). Cdk5 (Abcam, ab40773, Cambridge, UK) and PD-L1 (Abcam, ab205921) antibodies were obtainedfor immunostaining. The in vivo anti-mouse PD-L1 (B7-H1) antibody was purchased from BioXcell, USA (BP0101). The antibodies for flow cytometry were purchased from BD Pharmingen™, including PE Rat anti-mouse CD274 (558091), PE-Cy5-conjugated anti-CD3e (553065), PE-conjugated anti-CD4 (553730) and FITC-conjugated anti-CD8a (553030). Treg kit was purchased from Novusbio, USA (MAGM208). (1,4-butanediol) BDD, (5-amino-1-pentanol) AP and (1-(3-aminopropyl)-4-methylpiperazine) AMP were obtained from TCI, Shanghai, China. All the solvents used were of analytical grade and produced by Sinopharm, Beijing, China. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine (Cre), blood urea nitrogen (BUN) and lactate dehydrogenase (LDH) kits were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
2.2. Cell culture and animals
Murine melanoma cell line B16F10 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Hyclone) containing 10% fetal bovine serum, 100 IU/mL of penicillin and 100 μg/mL of streptomycin sulfate. Murine triple negative breast carcinoma cell line 4T1 cells were culture in RPMI 1640 (Hyclone) containing 10% fetal bovine serum, 100 μg/mL of penicillin and 100 μg/mL of streptomycin sulfate. Both cell lines were cultured in a humidified atmosphere incubator with 5% CO2 at 37 °C.
Female C57BL/6 and Balb/C mice (6–8 weeks old) were purchased from Hubei Provincial Center for Disease Control and Prevention, Wuhan, China. All mice were maintained under specific pathogen-free (SPF) condition in the Animal Center of Huazhong University of Science and Technology, Wuhan, China. The animals were maintained under standard condition. All the animals were treated according to the regulations of Chinese law and the local Ethical Committee Quantita.
2.3. Construct sgRNAs expressing vectors
The coding sequence of sgRNA (sgRNA1, sgRNA2, sgRNA3 or sgRNA4) against Cdk5 was cloned into BbsI-pre-digested pX330. The target sequence of sgRNA was 20 bp and two complementary oligonucleotides were annealed to generate dsDNA which had 4 bp overhangs on both ends.
2.4. Synthesis of poly(β-amino ester) (PBAE) copolymer
PBAE was synthesized via Michael-type polymerization similar with our previous work28 (Supporting Information Fig. S2A). In brief, BDD and AP (1.05:1, molar ratio) were mixed and reacted at 90 °C for 24 h. The resultant polymers were precipitated in cold diethyl ether and further washed for three times to remove the unreacted monomers. After vacuum drying, the polymers were dissolved in DMF and reacted with AMP at room temperature (RT) for 24 h to get the final AMP-modified PBAE polymer (aPBAE), which was purified by cold diethyl ether precipitating and washing. The chemical structure of the product was characterized by 1H NMR spectra (Bruker AVANCE III 400 MHz NMR spectrometer, solvent: CDCl3).
2.5. Preparation and characterization of aPBAE/pDNA
The aPBAE/pDNA nanoparticles were formed by 2 μg pDNA with various amount of aPBAE solution (15 mg/mL) at different weight ratios (10:1, 20:1 40:1, 60:1, 80:1. 100:1 and 120:1) in 25 mmol/L sodium acetate solution and incubated at RT for 30 min. The hydrodynamic size and zeta potential were then measured by a dynamic light scattering (DLS, Brookhaven Instruments, Brookhaven, USA). The gel retardation assays of aPBAE/pDNA at different weight ratios were performed to identify the condensation ability. The prepared nanoparticles were loaded into 1% (w/v) agarose gel in Tris-acetate-EDTA buffer and ran at 120 V for 40 min. A UV illuminator with a Gel Doc System (Bio-Rad, 170-8170, California, USA) was used to visualize the retardation results.
2.6. Invitro transfection efficiency
Both cell lines were transfected with pMax-GFP which contained GFP gene sequence to evaluate the transfection efficiency. The cells were seeded at the amount of 2.0 × 105 cells per well 24 h before transfection in 6-well plates. When the cells reached 70%–80% confluent, the nanoparticles contained 2 μg pMax-GFP were added to each well and incubated for additional 48 h and then the cells were observed by an inverted fluorescence microscope (Olympus, Japan). PEI 25K (Sigma) and X-tremeGENE HP DNA transfection reagent (Roche) were used as positive controls at their optimal conditions. The transfection efficiency was further quantified by flow cytometry (Calibur, BD, New Jersey, USA).
2.7. T7EI cleavage assay and indel sequencing
Genomic DNA was extracted from B16F10 cells after transfection for 72 h using QuickExtract DNA Extraction solution (Epicentre). The polymerase chain reaction (PCR) was performed with 100 ng genomic DNA, 1 μL high fidelity phusion polymerase (New England Biolabs) and the primers (details in Supporting Information Table S1) at an annealing temperature of 60 °C. 200 ng purified PCR products were hybridized according to the following condition, heating to 95 °C and ramping down to 25 °C with specific ramp rate. T7EI enzyme (NEB) was used to digest the reannealed PCR products. The cleavage products were loaded into 2% agarose gel and visualized with a Gel Doc gel imaging system. To further analyze the indel mutation sequence, TA cloning was performed with the PCR products and colonies were picked up randomly for sequencing.
2.8. Western blot analysis
Primary antibodies to Cdk5, p35, β-actin and HPR conjugated secondary antibodies were used for analysis. The protein lysate was dissolved in 1 × Laemmli buffer and boiled at 95 °C for 5–10 min. The samples were loaded into 10% SDS-PAGE gel, running at 80 V in resolving gel and 120 V in stacking gel. Afterwards, the proteins on the gel were transferred into a nitrocellulose membrane. The membrane was blocked with 5% non-fat milk in phosphate buffered saline with tween-20 (PBST) for about 1 h and then incubated with primary antibodies at their optimal concentration overnight at 4 °C, respectively. The membrane was washed with PBST and incubated in HPR secondary antibodies for 2 h at RT. The results were visualized by using a chemiluminescence detection system.
2.9. Quantitative RT-PCR (qRT-PCR)
Total RNAs were extracted from fresh tumor tissues of melanoma and triple negative breast cancer with TRIzol Reagent and further purified using the RNeasy Mini Kit (Qiagen, Dusseldorf, Germany). The total RNAs were reverse transcribed into cDNA with iScript cDNA Synthesis Kit (Bio-Rad). The cDNA was quantified by SYBR gene expression assays on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Primers were listed in Supporting Information Table S2.
2.10. Invitro cytotoxicity analysis
B16F10 cells were seeded in 96-well plates at the amount of 5 × 103/well in 100 μL cell culture medium and incubated for 24 h before experiment. The aPBAE/pDNA nanoparticles were added to the wells with different weight ratios. Afterwards, the cells were incubated at 37 °C for additional 24 h. 10 μL MTT (5 mg/mL) was added to each well and incubated for another 2–4 h. Subsequently, the MTT solution was aspirated and 150 μL DMSO was added to dissolve formazan. The absorbance was measured at 490 nm on a microplate reader (Multiskan MK3, Thermo, MA, USA).
2.11. Antitumor evaluation against melanoma
B16F10 cells (5 × 104 cells/per mouse) were inoculated at the right flank of female C57BL/6 mice. The mice were randomly divided into four groups (n = 6). Treatment began ten days after inoculation and was repeated for 4 times with an interval of 3 days. The mice were treated with phosphate buffered saline (PBS), naked pDNA, aPBAE/Cas9-null or aPBAE/Cas9-Cdk5 intratumorally. 5 μg plasmid was used for each injection per mouse. 72 h after the last injection, the tumors were dissected and separately stored in 4% paraformaldehyde at −80 °C for further analysis. Survival assay was also analyzed in four groups (n = 10) with the same tumor model and treatment method. The groups were PBS, naked pDNA, aPBAE/Cas9-Cdk5 and anti-PD-L1 antibody. The dosage of anti-PD-L1 antibody was 25 μg per mouse for each intratumoral injection.
2.12. Invivo evaluation on antitumor immunity of PD-L1 downregulation
The tumors were digested to harvest the infiltrated lymphocytes by using collagenase A-Hanks buffer. The digested tissues were incubated at 37 °C shaking for 70–80 min. After eliminating RBCs, the cells were collected by filtering and followed by centrifugation at 1000×g for 5 min. The lymphocytes were subsequently purified by the mouse lymphocyte isolating solution. To prevent nonspecific binding, the purified lymphocytes were firstly blocked with anti-CD16/CD32 antibody (BD Pharmingen™) for 10 min at 4 °C. Afterwards, the cells were labeled with anti-CD3e, anti-CD4 and anti-CD8a antibodies to evaluate the infiltration of effector T cells. Moreover, the cells were also labeled with anti-CD4, anti-CD25 and anti-Foxp3 antibodies according to the instruction of the kit to analyze the percentage of Tregs.
2.13. Lung metastasis inhibition of triple negative breast cancer
To establish pulmonary metastasis model of triple negative breast cancer, 4T1 cells (1 × 106 cells/per mouse) were inoculated at the right fat pad of female Balb/C mice. Seven days after inoculation, the mice were randomly divided into four groups (n = 6). PBS, naked pDNA, aPBAE/Cas9-Cdk5 or anti-PD-L1 antibody was intratumorally injected into the mice for four times with an interval of 3 days. The dosage was same as above. 72 h after the last injection, the tumors and lungs were collected and fixed in 4% paraformaldehyde for evaluation of antitumor response and lung metastasis inhibition.
2.14. Invivo gene transfection
B16F10 cells (5 × 104) were inoculated at the right flank of C57BL/6 mice. aPBAE/pMax-GFP nanoparticles were injected into the tumors when the tumor volume reached about 200 mm3. For each mouse, 5 μg of pMax-GFP was injected intratumorally. The transfected tumors were harvested 48 h later, and the tissues were embedded in OCT glue for frozen sections. The in vivo transfection results were analyzed by fluorescence microscope.
2.15. Cytotoxicity assay of CTL
The evaluation of antitumor efficacy of primed splenocytes was performed according to previously reported29. 2 × 104 B16F10 cells (with or without aPBAE/Cas9-Cdk5 pretreated for 72 h) were seeded into 96-well plate in triplicate and served as target cells. The lymphocytes isolated and purified from primed mice were added to target cells with different E/T (effector/target) cell ratios (5:1, 20:1 or 50:1) and served as effector cells. After 24-h co-culture, MTT assay was performed to calculate percentage of specific lysis.
2.16. Safety evaluation
The blood was collected for biochemistry analysis (ALT, AST, BUN, Cre and LDH). The major organs were dissected for H&E staining to analyze histological changes (heart, liver, spleen, lung and kidney).
2.17. Statistical analysis
Two-tailed Student's t-test with equal variance was performed to compare two groups. One-way ANOVA analysis was performed to evaluate the difference between multiple groups. All the statistical analyses were performed by the Prism software package (PRISM 6.0, GraphPad Software, USA). The statistical significance was set as a threshold of P < 0.05. All the data were expressed as mean ± SD.
3. Results
3.1. Construction of Cas9-Cdk5 system
The pX330 vector is a human codon-optimized SpCas9 and chimeric sgRNA expression plasmid. Four sgRNAs that target different exons of Cdk5 gene were designed and successfully cloned into the pX330 vectors to generate the Cas9-Cdk5 plasmid (Supporting Information Table S1 and Fig. S1).
3.2. Synthesis and characterization of aPBAE nanoparticles
The synthesis of aPBAE copolymer was shown in Fig. S2A. The chemical structure of PBAE and aPBAE was confirmed by 1H NMR spectra. As shown in Fig. S2B, all the signals of 5-amino-1-pentanol (AP) and 1,4-butanediol-diacrylate (BDD) units were clearly represented. Typically, the peak at 4.09 ppm, belonging to –COOCH2‒ protons of BDD units, and the peaks at 1.57, 1.43 and 1.33 ppm, being attributed to –CH2‒ in AP units, could be all found. The peak area ratio was 2:1:1:1, which was consistent with the proton number in the polymer structure. After reacting with 1-(3-aminopropyl)-4-methylpiperazine (AMP), the signal of end diacrylate groups (6.5–5.5 ppm) in PBAE disappeared and some small peaks at about 1.6 and 2.6 ppm occurred, which confirmed the chemical structure of aPBAE with successful AMP modification.
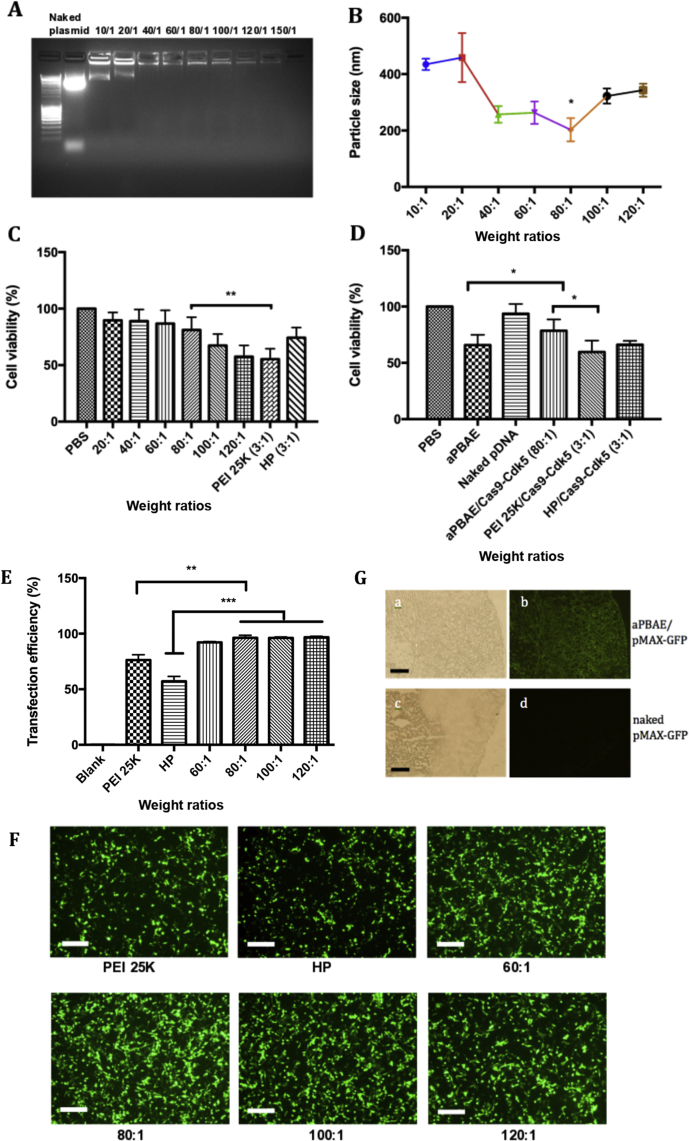
It is spontaneous for aPBAE polymer and CRISPR-Cas9 plasmid to formulate nano-sized electrostatic complex due to the opposite charge. The weight ratios of aPBAE and pDNA (Cas9-Cdk5) from 10 to 120 were analyzed by the gel retardation assay. It demonstrated that the Cas9-Cdk5 can be well condensed by aPBAE when the weight ratio was larger than 40 (Fig. 1A). The hydrodynamic diameters of different weight ratios of aPBAE/Cas9-Cdk5 were also measured by dynamic light scattering (DLS), and the results showed the diameters ranging from 202.9 to 458.8 nm (Supporting Information Table S5). The diameter was relatively smaller when the weight ratio of aPBAE to Cas9-Cdk5 equaled to 80 (Fig. 1B). The optimized nanoparticles were with the diameter of 246.3 ± 30.1 nm and ζ-potential of 23.8 ± 2.0 mV when the weight ratio was 80. The nanoparticles also exhibited a relatively high stability and the diameters increased slightly after 24 h incubation (Supporting Information Table S3).
Figure 1.
Characterization of aPBAE copolymer nanoparticles and evaluation of transfection efficiency. (A) Gel retardation assay of aPBAE/Cas9-Cdk5 nanoparticles with different weight ratios. Lane 1, DNA ladder; Lane 2, naked Cas9-Cdk5 plasmid; Lane 3–10, aPBAE/Cas9-Cdk5 at weight ratios of 10:1, 20:1, 40:1, 60:1, 80:1, 100:1, 120:1 and 150:1. (B) Hydrodynamic particle size of aPBAE/Cas9-Cdk5 with different weight ratios. (C) Viability of B16F10 cells after 24 h incubation with aPBAE/Cas9-Cdk5 of different weight ratios. PEI 25K and HP regent were used as controls. (D) Viability of B16F10 cells after 24 h incubation with different treatments. (E) Quantitative analysis of transfection efficiency in B16F10 cells that transfected with aPBAE/pMax-GFP nanoparticles. (F) Fluorescence images of B16F10 cells that transfected with aPBAE/pMax-GFP nanoparticles. The scale bar is 200 μm. (G) Fluorescence images of frozen sections from B16F10 tumor bearing mice treated with aPBAE/pMax-GFP (80:1) in vivo. The scale bar is 200 μm. Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.005.
The cytotoxicity of aPBAE/Cas9-Cdk5 was evaluated in B16F10 cells. Cells were treated with different weight ratios of aPBAE/Cas9-Cdk5 nanoparticles with PEI 25K and commercial liposome [HP transfection reagents, (20100100, Roche, Basel, Switzerland)] as controls. The cytotoxicity of nanoparticles increased in accordance with the weight ratios. It exhibited similar tendency as it was analyzed in Kwak's research30. Importantly, the cytotoxicity was lower than PEI 25K even when the weight ratio reached 120, approaching to commercial liposome when the weight ratio was 80. Moreover, the aPBAE/Cas9-Cdk5 nanoparticles exhibited lower toxicity than aPBAE copolymer due to charge neutralization. Plasmids were all endotoxin free extracted, therefore, the cell viability was close to that of PBS (Fig. 1C and D).
3.3. Invitro transfection efficiency
Transfection efficiency was assessed both in B16F10 and 4T1 cell lines. pMax-GFP was chosen as model gene that had a strong CMV promoter for GFP gene sequences. The aPBAE/pMax-GFP was evaluated in cell culture medium containing 10% serum, which was consistent with polyplexes PEI 25K and lipoplexes HP transfection reagent. The transfection evaluation for each group was repeated for 3 times. In B16F10 cells the aPBAE/pMax-GFP exhibited ultra-high transfection efficiency of more than 95% when the weight ratio was higher than 80. On contrast, PEI 25K/pMax-GFP and HP/pMax-GFP both with weight ratio of 3:1 only induced about 75% and 60% transfection efficiency respectively (Fig. 1E and F, Supporting Information Fig. S3). The superiority of aPBAE copolymer in transfection was further demonstrated in 4T1 cells. As shown in graphs, the transfection efficiency was performed by flow cytometry. The HP/pMax-GFP remained 60% transfection efficiency while PEI 25K/pMax-GFP decreased to ∼30% transfection efficiency. aPBAE/pMax-GFP still performed high transfection efficiency of 70% for weight ratios higher than 80 (Supporting Information Figs. S4 and S5). Therefore, aPBAE/pMax-GFP was identified to exhibit relatively high transfection efficiency even in the cell culture medium containing 10% serum compared with PEI 25K and HP reagent, indicating the outstanding serum tolerance of aPBAE copolymer. Moreover, the high transfection efficiency of aPBAE/pMax-GFP was stable in various cell lines, providing a versatile transfection reagent for gene delivery.
Combining with transfection results and characterization of the nanoparticles, the weight ratio of 80 for aPBAE/Cas9-Cdk5 was chosen as the most suitable ratio for the following in vitro and in vivo investigations.
In addition, in vivo transfection efficiency was monitored before gene disruption evaluation in the xenograft melanoma model. The tumors were treated with aPBAE/pMax-GFP nanoparticles and harvested 48 h after transfection. As shown in Fig. 1G, ultra-high expression of GFP protein can be visualized in the tumor tissues near the injection site of the frozen sections treated with aPBAE/pMax-GFP. No obvious GFP signal can be detected when the naked pMax-GFP was injected alone. It provided strong basis for in vivo animal experiments afterwards.
3.4. Invitro gene disruption analysis
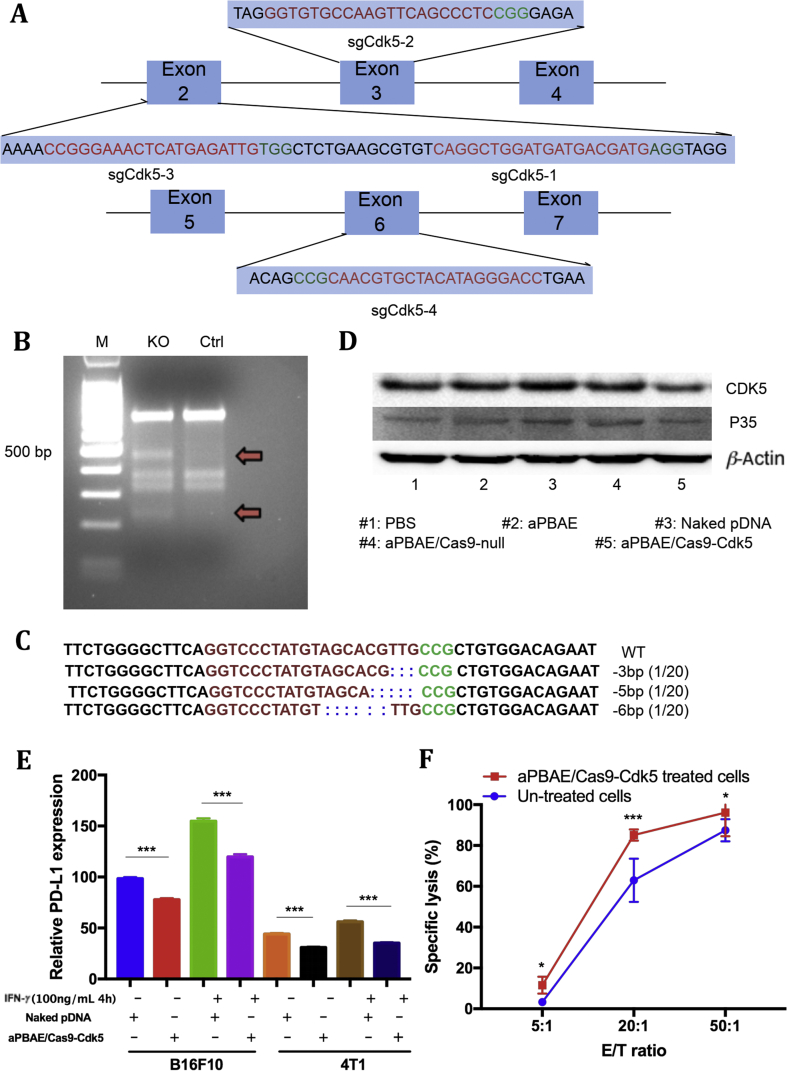
Double-strand breaks (DSBs) at specific genomic locus can be induced in 48–72 h after the CRISPR-Cas9 system delivered into cells31. Each sgRNA containing CRISPR-Cas9 plasmid was used to transfect into B16F10 cells for 72 h separately and different primers were designed for each target site (Fig. 2A, Supporting Information Table S1). To evaluate the targeting efficiency, T7EI endonuclease which can recognize and cleave the mismatched DNA was used to quantify the mutation frequency of target locus. The CRISPR/sgRNA4 system was identified to knockout the target gene most efficiently. Two cleavage bands which were about 400 bps and 200 bps could be clearly visualized by gel electrophoresis, and the cleavage efficiency was about 25%–30% (Fig. 2B)32, 33. Here, insertion/deletion (indel) information was presented to confirm the gene sequence of mutant alleles. The purified PCR products containing indels were subcloned into TA vector, and 20 colonies were randomly picked up for sequencing. Some representative indel sequences were shown in Fig. 2C and Supporting Information Fig. S6. Furthermore, we detected the expression of CDK5 protein and its co-activator P35 protein can be reduced after aPBAE/Cas9-Cdk5 transfection. Naked Cas9-Cdk5 plasmid, aPBAE copolymer alone and aPBAE/Cas9-null which did not contain Cdk5 targeting sequence were used as controls, exhibiting negligible effect (Fig. 2D). Therefore, it was identified that aPBAE/Cas9-Cdk5 could successfully knockout the target gene by disrupting DNA sequences, resulting in inhibition of CDK5 and P35 proteins expression.
Figure 2.
In vitro gene disruption analysis. (A) Schematic diagram of sgRNAs targeting Cdk5 gene. sgRNA targeting sequences are highlighted in red and PAM sequences are highlighted in green. (B) aPBAE/Cas9-Cdk5 mediated cleavage of Cdk5 gene in B16F10 cells detected by T7EI cleavage assay. M, DNA marker. (C) Representative indel mutation sequences of B16F10 cells after aPBAE/Cas9-Cdk5 treatment. (D) Expression of CDK5 and P35 protein of B16F10 cells detected by Western blotting assay after different treatments. (E) PD-L1 expression of B16F10 and 4T1 cells treated with IFN-γ, naked pDNA and aPBAE/Cas9-Cdk5 (n = 4). (F) The cytotoxicity assay of T lymphocytes from spleen against B16F10 cells with or without aPBAE/Cas9-Cdk5 treated (n = 3). Data were expressed as mean ± SD. ∗P < 0.05 and ∗∗∗P < 0.005.
Additionally, PD-L1 is highly expressed in a variety of tumor cells due to the chronic inflammation of tumor microenvironment. The expression of PD-L1 is mainly induced by specific inflammatory signals, in particularly IFN-γ. Thus, we further analyzed PD-L1 expression of both B16F10 and 4T1 cells after aPBAE/Cas9-Cdk5 nanoparticles treatment with or without IFN-γ stimulation. As shown in Fig. 2E, IFN-γ stimulation (100 ng/mL, 4 h) could obviously induce PD-L1 expression in both cell lines. Importantly, aPBAE/Cas9-Cdk5 nanoparticles could downregulate about 20% of PD-L1 in B16F10 cells without IFN-γ stimulation and 30% with IFN-γ stimulation. Although the expression of PD-L1 was lower in 4T1 cells compared to B16F10 cells, aPBAE/Cas9-Cdk5 nanoparticles could still downregulate about 15% of PD-L1 without IFN-γ stimulation and 25% with IFN-γ stimulation.
Moreover, the cytotoxicity assay was performed by isolating T lymphocytes from spleen of C57BL/6 mouse against B16F10 cells with or without aPBAE/Cas9-Cdk5 treated. As shown in Fig. 2F, T lymphocytes exhibited more cytotoxic to aPBAE/Cas9-Cdk5-treated cells compared to untreated cells especially when the E/T ratio was 20:1, which indicated enhanced CTL responses induced by PD-L1 downregulation.
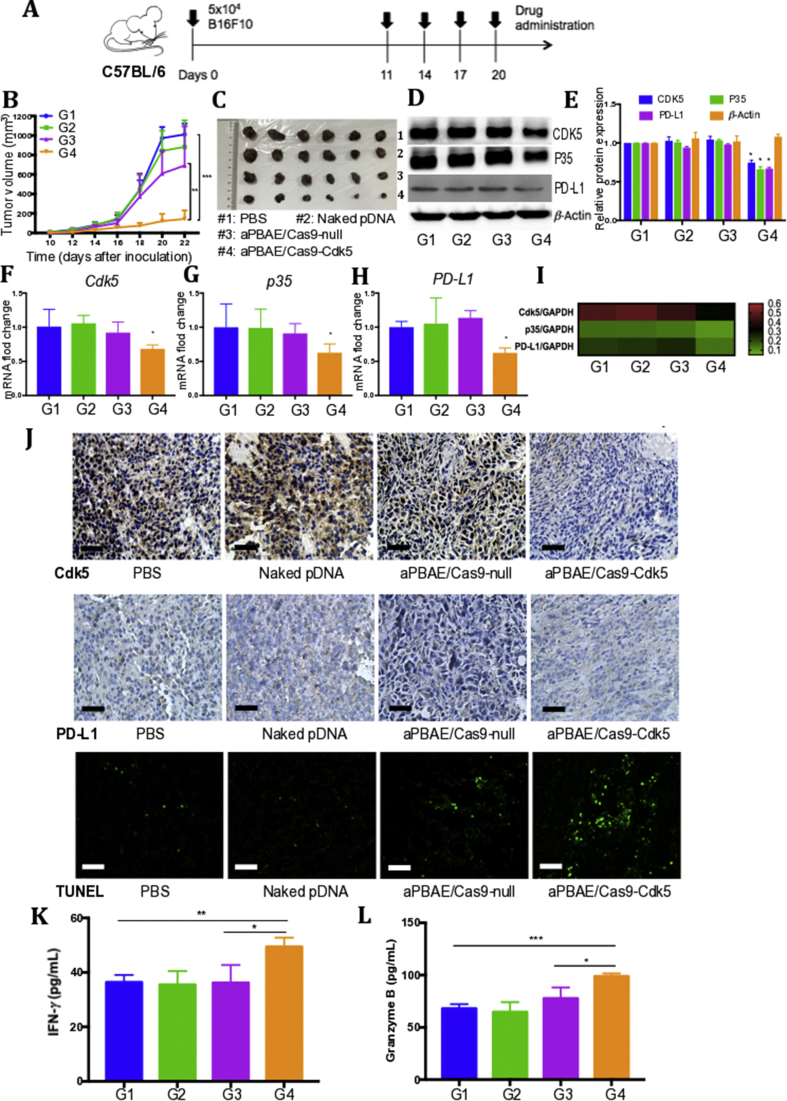
3.5. CRISPR-Cas9 mediated PD-L1 attenuation suppressed B16F10 tumor growth
Inspired by the results of Cdk5 knockout in B16F10 cells in vitro, we further analyzed in vivo antitumor potential of aPBAE/Cas9-Cdk5 nanoparticles. The Cdk5 gene disruption efficacy in melanoma was assessed based on the tumor bearing mouse model. These mice were treated with PBS, naked pDNA (Cas9-Cdk5), aPBAE/Cas9-null and aPBAE/Cas9-Cdk5 nanoparticles, respectively. Cas9-null which is the plasmid without target sequence was used as negative control to exclude the impact of Cdk5 disruption. The mice were treated as shown in Fig. 3A. No obvious fluctuation of the mice body weight was observed during the experiment for all treatment groups, implying no significant toxicity existence of the delivery systems (Supporting Information Fig. S12A). As shown in Fig. 3B, aPBAE/Cas9-Cdk5 treatment could significantly suppress tumor growth compared to aPBAE/Cas9-null treatment, suggesting that knocking out Cdk5 in vivo could enhance antitumor effect. Moreover, it can be evidenced that the Cas9-Cdk5 plasmid could be efficiently delivered in vivo after condensation with aPBAE copolymer as no obvious antitumor response was observed in naked pDNA group. Importantly, the inhibitory rate of tumor growth for aPBAE/Cas9-Cdk5 was 79.2%. Although the aPBAE/Cas9-null-treated mice exhibited smaller tumor volume than PBS group due to the unavoidable slight cytotoxicity of cationic aPBAE copolymer which consistent with MTT results in vitro, it still showed significant antitumor efficacy of aPBAE/Cas9-Cdk5 treatment compared to aPBAE/Cas9-null treatment. The results illustrated the novel function of Cdk5 target gene.
Figure 3.
CRISPR-Cas9-mediated PD-L1 attenuation suppressed B16F10 tumor growth. (A) Treatment scheme. (B) Tumor growth curves of PBS, naked pDNA, aPBAE/Cas9-null and aPBAE/Cas9-Cdk5 (n = 6). (C) Photographs of tumors dissected from C57BL/6 mice after different treatments (n = 6). (D) Representative protein expression and (E) light intensities analysis of Western blot for the tumor tissues dissected from mice treated with PBS, naked pDNA, aPBAE/Cas9-null or aPBAE/Cas9-Cdk5 (n = 3). (F) mRNA fold change of Cdk5, (G) p35 and (H) PD-L1 for the tumor tissues dissected from mice treated with PBS, naked pDNA, aPBAE/Cas9-null or aPBAE/Cas9-Cdk5 (n = 3). (I) Heat map of the corresponding mRNA levels compared to GAPDH analyzed by qRT-PCR. (J) Immunohistochemistry analysis of the expression of Cdk5, PD-L1 and TUNEL in tumor sections after different treatments. The scale bars are 50, 50 and 100 μm, respectively. The levels of (K) IFN-γ and (L) granzyme B in tumor microenvironment were analyzed (n = 3). PBS (G1), naked pDNA (G2), aPBAE/Cas9-null (G3) and aPBAE/Cas9-Cdk5 (G4). Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.005.
In addition, to further compare with anti-PD-L1 antibody, the overall survival was performed by analyzing the tumor bearing mice with different treatments, including PBS, naked pDNA, aPBAE/Cas9-Cdk5 and anti-PD-L1 antibody. As shown in Supporting Information Fig. S8, aPBAE/Cas9-Cdk5 can significantly prolong survival of B16F10 bearing mice with the overall survival time to 40 days, compared with 32 days for anti-PD-L1 antibody and 26 days for PBS treatment. The results indicated that more stable antitumor efficacy of aPBAE/Cas9-Cdk5 compared to anti-PD-L1 antibody in melanoma. And the tumor volume and tumor weight were significant reduced in the mice treated with aPBAE/Cas9-Cdk5 compared to PBS and anti-PD-L1 antibody group (Supporting Information Fig. S9). As demonstrated in clinical trials, only a minority of patients exhibited antitumor response to anti-PD therapy due to tumor heterogeneity34, 35. Thus, our delivery system based on Cdk5 knockout to downregulate PD-L1 expression showed distinct advantages compared to the anti-PD-L1 antibody, and it can be incorporated into various antitumor therapies in future.
To further investigate the intrinsic mechanism of retarded tumor growth by aPBAE/Cas9-Cdk5 nanoparticles, protein and mRNA expression levels of relative genes were analyzed. The results showed that the protein expression of CDK5 and its activator P35 in aPBAE/Cas9-Cdk5-treated group were relatively less than those of PBS, naked pDNA and aPBAE/Cas9-null-treated groups (Fig. 3D). As mentioned above, Cdk5 gene disruption could attenuate the expression of PD-L1 of tumor cells with the chronic existence of IFN-γ in tumor microenvironment24. Therefore, the Western blotting results confirmed the downregulation of PD-L1 expression by aPBAE/Cas9-Cdk5 nanoparticles as expected (Fig. 3D). The relative protein expression was further quantified. In aPBAE/Cas9-Cdk5-treated mice, the protein expressions were about 74.7% for CDK5, 65.9% for P35 and 66.9% for PD-L1 compared to PBS group (Fig. 3E). In addition, the upregulation of IRF2 and IRF2BP2 protein expression after aPBAE/Cas9-Cdk5 treatment was also verified to confirm the pathway (Supporting Information Fig. S15A). Furthermore, the relative mRNA expression levels were assessed by qRT-PCR with the GAPDH house keeping gene as the normalization control. After aPBAE/Cas9-Cdk5 treatment, the mRNA levels of Cdk5 (68.2%), p35 (62.6%) and PD-L1 (62.4%) were significantly decreased compared to PBS group (Fig. 3F–H). The heat map showed the tendency of the corresponding mRNA levels mediated by different treatment formulations (Fig. 3I). The protein expression showed a similar trend with the mRNA regulation, and the results illustrated that there existed positive correlation between Cdk5 and PD-L1 which was consistent with our speculation. Besides, we performed T7EI assay and demonstrated the in vivo gene disruption in melanoma tumor model (Supporting Information Fig. S7).
Immunohistochemistry (IHC) analysis and terminal deoxynucleotidyl transferase-mediated dUTP-biotinnick end labeling (TUNEL) assay were also performed to evidence the results. As shown in Fig. 3J, the expression of CDK5 protein was obviously reduced after aPBAE/Cas9-Cdk5 treatment, indicating efficient knocking out of Cdk5 target gene in tumor tissues. And the IHC analysis also confirmed our speculation on the downregulation of PD-L1. The apoptosis of tumor cells was also investigated after aPBAE/Cas9-Cdk5 treatment by TUNEL assay. Engagement of PD-L1 on tumor cells with its receptor PD-1 on immune cells can transmit an inhibitory signal to induce immune evasion. The downregulation of PD-L1 expression could release the “brake” on effector T cells to some extent. Therefore, B16F10 cells were susceptible to be killed by T lymphocytes. Consistently, the results showed that the population of apoptotic tumor cells in tumors received aPBAE/Cas9-Cdk5 was greater than that of PBS, naked pDNA and aPBAE/Cas9-null treatment. The H&E staining sections of tumors further confirmed the superiority of aPBAE/Cas9-Cdk5 delivery system (Supporting Information Fig. S10).
Moreover, we further performed ELISAs of the tumor tissues to evaluate T cell activity in vivo. IFN-γ and granzyme B are important mark cytokines of activated TILs and play critical role in T cell mediated tumor cell killing. As shown in Fig. 3K and L, the level of IFN-γ and granzyme B were both significantly increased in aPBAE/Cas9-Cdk5-treated mice compared to other groups, which indicated that enhanced CTL responses in tumor microenvironment.
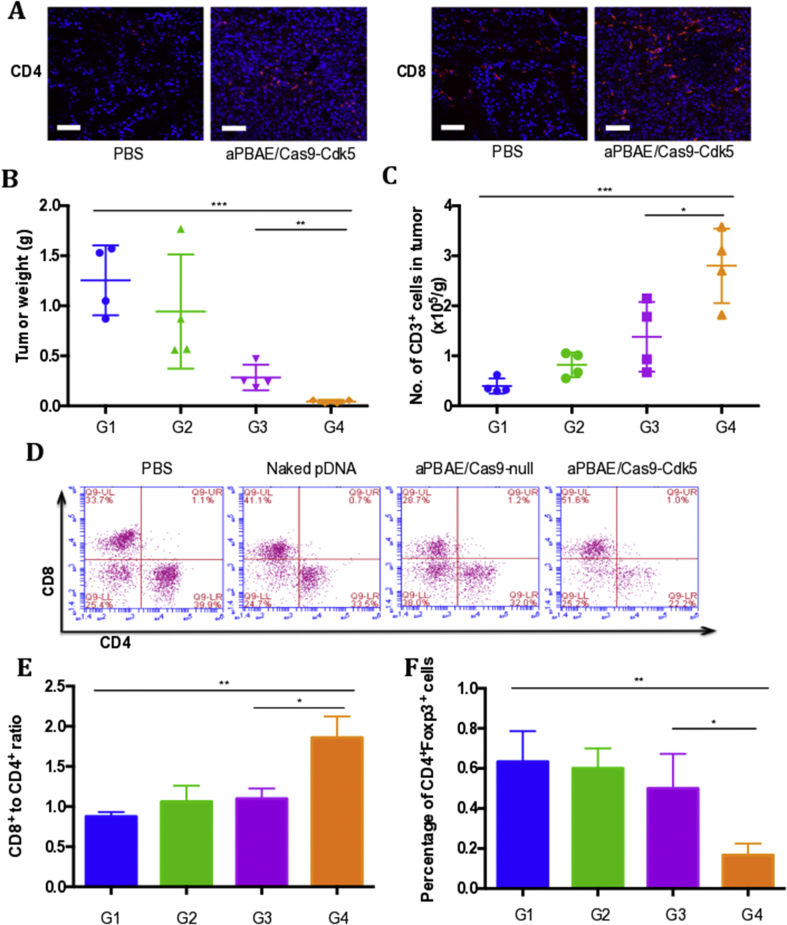
3.6. CRISPR-Cas9 mediated PD-L1 attenuation triggered T cell-mediated antitumor immune response
As shown above, knocking out Cdk5 in melanoma could attenuate PD-L1 expression. Here, we further investigated the alternation of TILs in tumor microenvironment after aPBAE/Cas9-Cdk5 treatment. The immunofluorescence staining results showed that higher infiltration of CD8+ and CD4+ T cells could be detected in the mice treated with aPBAE/Cas9-Cdk5 than PBS. Only a small amount of T cells was infiltrated in the control group (Fig. 4A). Tumor tissues were also digested to harvest the TILs for flow cytometer analysis. As shown in Fig. 4B, the tumor weight of aPBAE/Cas9-Cdk5 group was significantly lower than the other three groups. In accordance with this result, the number of CD3+ T cells was the highest after aPBAE/Cas9-Cdk5 treatment, which was nearly 10-fold compared to PBS, implying elevated T cell infiltration in tumor microenvironment by PD-L1 downregulation (Fig. 4C).
Figure 4.
CRISPR-Cas9-mediated PD-L1 attenuation triggered T cell-mediated antitumor immune response in murine melanoma model. (A) Immunofluorescence images of tumor sections showed CD4+ T cells and CD8+ T cells infiltration. The scale bar is 100 μm. (B) Tumor weight of mice treated with PBS, naked pDNA, aPBAE/Cas9-null or aPBAE/Cas9-Cdk5 (n = 4). (C) Number of CD3+ cells per gram tumor after different treatments (n = 4). (D) Representative scatter plots of CD4+ T cells and CD8+ T cells in tumors (n = 4). (E) Ratios of CD8+ T cells and CD4+ T cells in tumors (n = 4). (F) Percentage of CD4+Foxp3+ cells in tumors (n = 4). PBS (G1), naked pDNA (G2), aPBAE/Cas9-null (G3) and aPBAE/Cas9-Cdk5 (G4). Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.005.
Furthermore, researches demonstrated that continuous overexpression of PD-L1 could lead to T cell anergy and exhaustion at late stage of diseases. The generation of Tregs are also elevated36. In our work, the absolute population of CD8+ T cells treated with aPBAE/Cas9-Cdk5 (49.65%) increased by almost 1.5-fold over the PBS (37.6%), naked pDNA (40.45%) and aPBAE/Cas9-null group (38.5%). The population of CD4+ T cells exhibited no obvious fluctuation. Moreover, the population of CD8+ T cells was nearly 2-fold versus CD4+ T cells in the tumors of aPBAE/Cas9-Cdk5 treatment compared with control groups, indicating enhanced CTLs infiltration as expected (Fig. 4D and E, Supporting Information Table S4).
In addition, for the control groups of PBS, naked pDNA and aPBAE/Cas9-null, the populations of CD4+Foxp3+ cells were 0.63%, 0.6% and 0.5%, respectively. However, it was significantly decreased in aPBAE/Cas9-Cdk5 group to 0.15% (Fig. 4F, Supporting Information Fig. S13 and Table S4). The population ratios of both CD8+ T cells to CD4+Foxp3+ T cells and CD4+Foxp3- T cells to CD4+Foxp3+ T cells in B16F10 tumors were significantly increased in the aPBAE/Cas9-Cdk5 treated mice (Supporting Information Fig. S14). The results suggested that the aPBAE/Cas9-Cdk5 nanoparticles could be effectively delivered in vivo, resulting in efficiently Cdk5 target gene disruption and consequently inhibition of PD-L1 expression. Most importantly, aPBAE/Cas9-Cdk5 nanoparticles could trigger robust T cell-mediated antitumor immune responses and reverse the immunosuppressive tumor microenvironment.
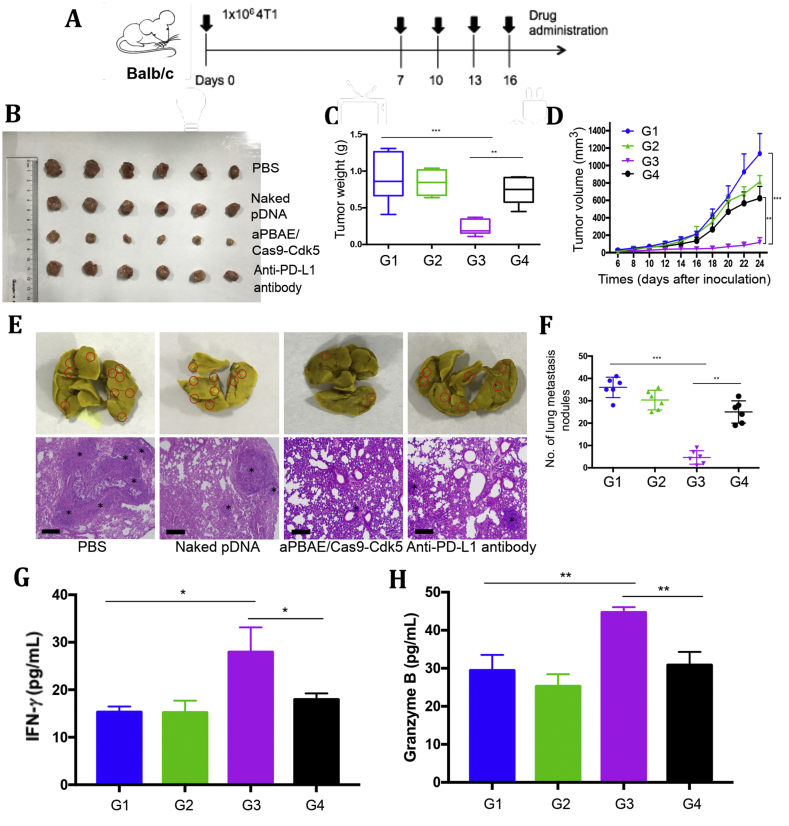
3.7. CRISPR-Cas9 mediated PD-L1 attenuation suppressed 4T1 tumor growth and inhibited lung metastasis
4T1 cells, the triple negative breast cancer cell line, were inoculated into the fat pads of female Balb/C mice to establish the tumor model. This animal model can closely mimic the stage IV human breast cancer to investigate tumor proliferation and spontaneous lung metastasis. As we have demonstrated the novel function of Cdk5 in melanoma with comparing aPBAE/Cas9-Cdk5 and aPBAE/Cas9-null to emphasis the role of target gene, in the following antitumor experiment, we did not demonstrate and compare these two groups anymore, but mainly focused on comparing antitumor efficacy of aPBAE/Cas9-Cdk5 and anti-PD-L1 antibody.
To observe the antitumor effect and lung metastasis inhibition by Cdk5 gene disruption, PBS, naked pDNA, aPBAE/Cas9-Cdk5 or anti-PD-L1 antibody was intratumorally injected into 4T1 tumor bearing mice, respectively. The treatment scheme was presented in Fig. 5A. As shown in Fig. 5B, the tumor volume of naked pDNA showed no obvious difference compared with PBS. Anti-PD-L1 antibody exhibited weak antitumor effect with inhibitory rate of 45.1%. Remarkably, aPBAE/Cas9-Cdk5 nanoparticles suppressed tumor growth with 89.6% reduction of tumor volume and 75.7% reduction of tumor weight, indicating that downregulation of PD-L1 expression by knocking out Cdk5 was more effective to inhibit tumor growth than anti-PD-L1 antibody alone (Fig. 5C and D). Moreover, body weight of mice was monitored during whole course of experiments which showed no significant change, demonstrating the safety of these delivery systems (Supporting Information Fig. S12B). Furthermore, to assess the lung metastasis inhibition efficacy, tumor nodules on the surface of lungs were counted and sections of lung tissues were stained with H&E. Mice treated with anti-PD-L1 antibody showed weak effect on lung metastasis inhibition, while aPBAE/Cas9-Cdk5 can effectively reduce the number of tumor nodules which was consistent with the antitumor results. The average number of tumor nodules was 36 ± 4 and 25 ± 5 for PBS and anti-PD-L1 antibody, respectively. Notably, the mice treated with aPBAE/Cas9-Cdk5 exhibited only a few tumor nodules with an average number of 5 ± 3 (Fig. 5E). The results were further demonstrated by representing the metastasis area in H&E staining (Fig. 5F). The H&E staining sections of tumors further confirmed the superiority of aPBAE/Cas9-Cdk5 delivery system (Supporting Information Fig. S11). Besides, the production of IFN-γ and granzyme B in aPBAE/Cas9-Cdk5 treated mice were both obviously elevated which indicated enhanced CTL responses in tumor microenvironment (Fig. 5G and H).
Figure 5.
CRISPR-Cas9-mediated PD-L1 attenuation suppressed 4T1 tumor growth and inhibited lung metastasis. (A) Treatment scheme. (B) Photographs of tumor dissected from Balb/C mice treated with PBS, naked pDNA, aPBAE/Cas9-Cdk5 or anti-PD-L1 antibody (n = 6). (C) Tumor weight of the mice (n = 6). (D) Tumor growth curve of the mice after different treatments (n = 6). (E) Representative photographs and H&E staining sections of lungs. The black stars mark the metastatic area. The scale bar is 200 μm. (F) Number of lung metastasis nodules (n = 6). The levels of (G) IFN-γ and (H) granzyme B in tumor microenvironment were analyzed (n = 3). PBS (G1), naked pDNA (G2), aPBAE/Cas9-Cdk5 (G3) and anti-PD-L1 antibody (G4). Data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.005.
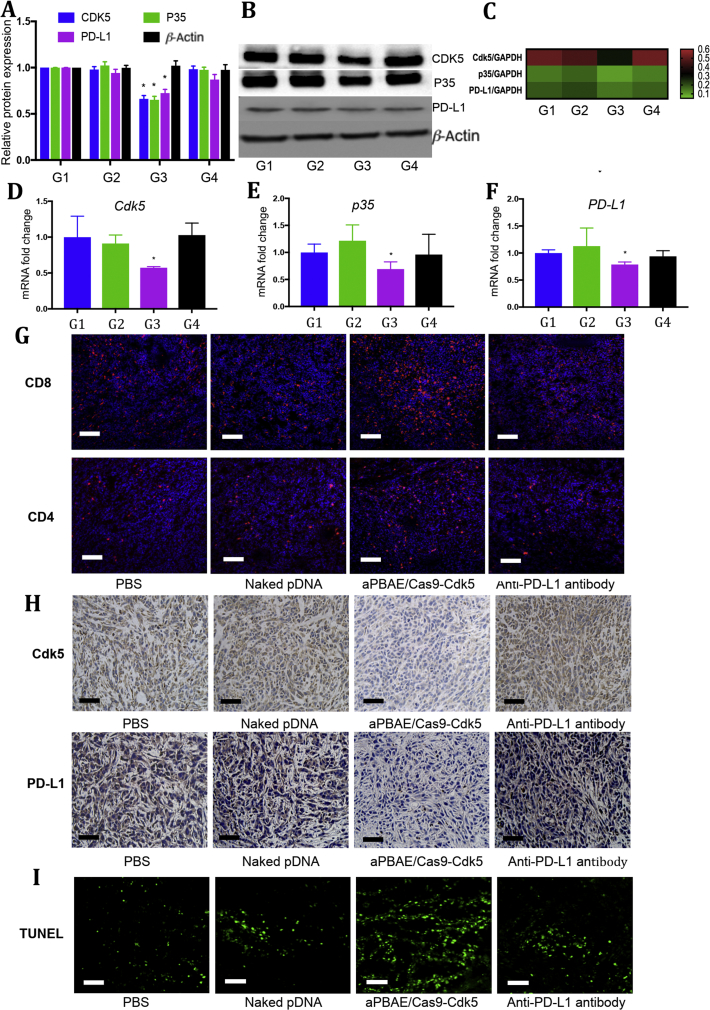
Moreover, the intrinsic mechanism was also investigated to evidence our speculation. The protein expression of CDK5 and its activator P35 in aPBAE/Cas9-Cdk5-treated group were significantly less than those of PBS, naked pDNA and anti-PD-L1 antibody-treated groups, and the PD-L1 expression was obviously downregulated compared to controls. The relative protein expression was further quantified. In aPBAE/Cas9-Cdk5, the protein expressions were about 66.2% for CDK5, 65.2% for P35 and 72.3% for PD-L1 compared to PBS group (Fig. 6A and B). The expression of IRF2 and IRF2BP2 protein after aPBAE/Cas9-Cdk5 treatment were upregulated as demonstrated in pathway (Supporting Information Fig. S15B). The relative mRNA expression level was also quantified which showed well correlation with protein expression. After aPBAE/Cas9-Cdk5 treatment, the mRNA levels of Cdk5 (57.2%), p35 (69.2%) and PD-L1 (78.7%) were significantly decreased compared to PBS group. The heat map also showed the tendency of the corresponding mRNA levels mediated by different treatment formulations (Fig. 6C‒F).
Figure 6.
Regulation of intrinsic mechanisms in 4T1 tumor after aPBAE/Cas9-Cdk5 treatment. (A) Light intensities analysis and (B) representative protein expression of Western blot for the breast carcinoma tissues dissected from mice treated with PBS, naked pDNA, aPBAE/Cas9-Cdk5 or anti-PD-L1 antibody (n = 3). (C) Heat map of the corresponding mRNA levels compared to GAPDH analyzed by qRT-PCR. (D) mRNA fold change of Cdk5, (E) p35 and (F) PD-L1 for the tumor tissues dissected from mice treated with PBS, naked pDNA, aPBAE/Cas9-Cdk5 or anti-PD-L1 antibody (n = 3). PBS (G1), naked pDNA (G2), aPBAE/Cas9-Cdk5 (G3) and anti-PD-L1 antibody (G4). (G) Immunofluorescence images of tumor sections showed CD8+ T cells and CD4+ T cells infiltration. The scale bar is 200 μm. (H) Representative IHC analysis of the expression of Cdk5 and PD-L1 after treatment. The scale bar is 50 μm. (I) TUNEL analysis in tumor sections after different treatments. The scale bar is 100 μm. Data were expressed as mean ± SD. ∗P < 0.05.
In addition, as shown in Fig. 6G, the infiltration of CD8+ T cells was much higher in the mice treated with aPBAE/Cas9-Cdk5 compared to PBS, naked pDNA and anti-PD-L1 antibody groups, while no obvious elevation of the infiltrated CD4+ T cells could be observed. Moreover, the expression of Cdk5 was reduced after aPBAE/Cas9-Cdk5 treatment and consequently the PD-L1 expression level was also inhibited, indicating the successfully Cdk5 gene knockout in vivo (Fig. 6H). Besides, as shown in Fig. 6I, the TUNEL staining of tumor tissues also indicated enhanced apoptosis of tumor cells after aPBAE/Cas9-Cdk5 treatment.
It is worth noting that our results are in accordance with most researches that anti-PD-L1 antibody alone has limited antitumor responses in 4T1 tumor37. Two critical reasons were speculated in the phenomena. One is the restricted targeting delivery efficiency of anti-PD-L1 antibody in vivo, and the other is that 4T1 is not a highly PD-L1 expressed cell line. It is reported that conjugating anti-PD-L1 antibody to the surface of platelets could facilitate efficiently targeting to the residual tumor sites after surgery compared to anti-PD-L1 antibody alone38. Moreover, combination of anti-PD-L1 antibody with other therapies exhibited enhanced antitumor efficacy39, 40.
3.8. Safety evaluation
The toxicity of these delivery systems was evaluated by analyzing the serum chemistry indexes and histology of major organs. The levels of the biomarkers for liver health, heart failure and kidney injury, were all in normal range for each treatment group (Supporting Information Fig. S16). No significant evidence of histopathologic changes was observed in H&E staining sections of major organs after treatment, including heart, liver, spleen, lung and kidney (Supporting Information Fig. S17).
4. Discussion
The immune checkpoint inhibitors have got durable responses with long-term survival in various cancers in clinical trials, such as melanoma, breast cancer, non-small cell lung cancer (NSCLC), kidney cancer, bladder cancer, Hodgkin's lymphoma and many other tumor types. Increasing human monoclonal antibodies approved by FDA that targeting the PD-1/PD-L1 pathway have shown notable efficacy in antitumor treatment. For example, Nivolumab is demonstrated to have strong therapeutic efficacy on chemotherapy-refractory squamous NSCLC41. It also exhibited higher performance in advanced melanoma compared to standard chemotherapy42. However, the immune checkpoint inhibitors still face several challenges. First, due to the individual heterogeneity, only a minority of patients can respond to therapy. It is reported that the immune genes and pathways, host micro-biome, and the nature of tumor itself can be the critical reasons that limit the clinical outcomes of immune checkpoint therapy35. Second, the therapeutic antibody can also target normal organs during treatment and elicit severe immune-related adverse events, including inflammation of lung, endocrine organs and many other organs43. Therefore, some patients have to receive lifelong hormone therapy. Third, although these immune checkpoint inhibitors can release the “brake” on cancer immunotherapy resulting in enhanced tumor killing, they cannot reverse the immunosuppressive tumor microenvironment fundamentally. Thus, a minority of patients exhibits drug resistance and tumor recurrence in long-term treatment34, 44. Considering these severe factors, the alternative therapies that could improve the applicability of these immune checkpoint inhibitors to benefit majority of patients are the central theme for cancer immunotherapy in future.
Genetic interventions are explored in recent researches to achieve high-target specificity and limit side effects of immune checkpoint antibodies. A polymeric carrier consisting of disulfide-crosslinked polyethyleneimine and dermatan sulfate is formulated for siPD-L1 delivery. By delivering the siPD-L1 in vivo, it cannot only influence the PD-1/PD-L1 pathway but also the mTOR intrinsic pathways for tumorigenesis. Significantly suppression of tumor growth was observed both in the immunocompetent and non-immunocompetent melanoma mouse models30. It provides an alternative strategy of incorporating nanoparticle delivery system into genetic interventions for improving cancer immunotherapy.
However, there are some limitations by using siRNA for genetic engineering. First, siRNA is used for knocking down the target gene expression, but not completely eliminating it, and thus the incomplete inhibition of target gene limits its therapeutic efficiency in clinic. Second, the duration of siRNA inhibitory effects could last only for a short time. Normally the therapeutic effect reaches the peak two days after transfection and is then faded45. Third, siRNA is easily to be degraded and hard to be operated. Therefore, novel genome engineering techniques are in urgent demand. The emergence of CRISPR-Cas9 genome editing system greatly expands the potential for treating genetic diseases. This genetic engineering tool shows the advantages of simplicity, highly targeting efficiency and completely knocking out genetic codes. Thus, we applied the CRISPR-Cas9 system in cancer immunotherapy to target the PD-1/PD-L1 pathway. Utilizing this newly emerging technology, target genes involved in immune modulatory pathways can be specifically and persistently knocked out, hence boosting antitumor immune responses and modulating the immunosuppressive tumor microenvironment.
Normally, the CRISPR-Cas9 system can be delivered in three forms, the CRISPR-Cas9 plasmid, a complex with Cas9 protein and sgRNA, and a mixture of Cas9 mRNA and sgRNA. We chose CRISPR-Cas9 plasmid with Cdk5-targeting sequence as it is more stable and versatile. Cdk5 was chosen as the target gene in our work as it was identified when knocked out could consequently attenuate PD-L1 expression. In addition, considering the systemic safety we did not target PD-L1 directly as it was demonstrated that PD-L1-deficient mice exhibited hyperactive immune responses leading to autoimmune diseases46. aPBAE copolymer was identified as versatile gene delivery system that can well condense plasmid. We chose PBAE based on the concerns of transfection efficiency and safety. PEI 25K is the golden standard for gene transfection. However, the toxicity is the major concern. Some other biodegradable cationic polymers, e.g., polylysine (PLL) and chitosan were ineffective for transfection unless their molecular weights were high enough. For cationic dendrimers, such as PAMAM, polypropylenimine (PPI) and PLL dendrimers, high generation is also required for gene transfection but also accompanied with high toxicity47. PBAE is a kind of biodegradable cationic polymers which can be easily synthesized with various chemical structures. The transfection efficiency of aPBAE/pMax-GFP was ultra-high in both B16F10 and 4T1 cells, which was more than 95% and 70%, respectively (Fig. 1E and F, Supporting Information Figs. S3‒S5). Moreover, in melanoma and breast cancer models, aPBAE/Cas9-Cdk5 suppressed tumor growth and prevented lung metastasis (Figure 3, Figure 5). Strikingly, aPBAE/Cas9-Cdk5 played an essential role in remodeling the tumor microenvironment. The population of CD8+ T cells and the ratio of CD8+ T cells to CD4+ T cells were significantly increased in melanoma after aPBAE/Cas9-Cdk5 treatment. At the meantime, the population of Tregs was decreased (Figure 4, Figure 6G, and Fig. S13). Systemic delivery of Cas9-Cdk5 will be further investigated to treat various tumor types in the future.
Moreover, our approach showed better therapeutic efficacy compared with anti-PD-L1 antibody. First, CRISPR/Cas9 system can permanently knockout target gene which is fundamentally different from other gene therapy, such as siRNA. Therefore, PD-L1 can be permanently downregulated in the Cdk5-knocked out tumor cells. Importantly, both of the membrane and cytosolic protein of PD-L1 can be downregulated by re-editing the endogenous Cdk5 gene, which is different from antibody because the latter neutralize membrane protein only. Second, as PD-L1 is also expressed on healthy tissues and cells, immune-related adverse effects can be observed during the clinical treatment by PD-1/PD-L1 antibodies among a minority of patients, especially the irreversible toxic effects in endocrine systems. The aPBAE/Cas9-Cdk5 nanoparticles exhibited higher transfection efficiency to tumor cells with higher proliferation ability compared to normal cells. Thus, it can avoid the “on-target but off-tumor” side effect to some extent. Third, for some tumor types, single anti-PD-L1 antibody therapy is not enough to inhibit tumor growth, and our system can serve as a robust platform for combination immunotherapy by delivering a CRISPR/Cas9 pool targeting multiple immune evasion mechanism.
In addition, the stromal cells in tumor microenvironment include macrophages, fibroblast, vascular endothelial cells and so on. Although the transfection efficiency of Cas9-Cdk5 nanoparticles in these cells is much lower than tumor cells, PD-L1 expression can also somehow be downregulated in these cells. As these cells are important factors that contribute to tumor microenvironment, PD-L1 downregulation of these cells could help to change the immunosuppressive tumor microenvironment into immunosupportive, hence making the tumors more sensitive to immune therapy48, 49.
5. Conclusion
In summary, we developed a gene delivery system that can deliver the CRISPR-Cas9 genome editing system with high efficiency both in vitro and in vivo. The CRISPR-Cas9 plasmid can be well condensed into cationic copolymer to form nanoparticles which showed superior transfection efficiency in B16F10 and 4T1 cells compared to PEI 25K and HP transfection reagent. Moreover, the Cdk5 target gene can be efficiently knocked out in vitro utilizing the nanoparticles. The aPBAE/Cas9-Cdk5 nanoparticles could significantly disrupt Cdk5 target gene leading to PD-L1 expression downregulation in vivo, and subsequently inhibition of tumor growth and prevention of lung metastasis. Furthermore, PD-L1 expression was attenuated by permanently knocking out Cdk5, which can release the immune checkpoint blockade, restore the CTLs mediated immune responses and reverse the immunosuppressive tumor microenvironment. Compared to the anti-PD-L1 antibody, our research inspires new treatment for cancer immunotherapy that can maintain relatively long-term therapeutic effects. It provides an alternative strategy for blocking the immune checkpoint pathways. Most importantly, it provides powerful strategy for preclinical treatment by combining immune checkpoint genetic intervention and nanotechnology.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81872810, 81673374 and 81871473), Wuhan University of Science and Technology Plan for Applied Fundamental Research (2017060201010146, China), the Fundamental Research Funds for the Central Universities (2018KFYYXJJ019, 2019KFYRCPY049 and 2016YXMS138, China). We thank Weijun Feng, Ph.D, Professor, Fudan University, for professional advice.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2019.07.004.
Appendix A. Supporting information
The following is the Supporting information to this article:
References
- 1.Dong H., Strome S.E., Salomao D.R., Tamura H., Hirano F., Files D.B. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 2.Keir M.E., Butte M.J., Freeman G.J., Sharp A.H. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taube J.M., Anders R.A., Young G.D., Xu H., Sharma R., McMiller T.L. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spranger S., Spaapen R.M., Zha Y., Williams J., Meng Y., Ha T.T. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selby M.J., Engelhardt J.J., Quigley M., Henning K.A., Chen T., Srinivasan M. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Canc Immunol Res. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 6.Alsaab H.O., Sau S., Alzhrani R., Tatiparti K., Bhise K., Kashaw S.K. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol. 2017;8:561. doi: 10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao M.X., Guo W.J., Wu Y.Y., Yang C.X., Zhong L., Deng G.L. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm Sin B. 2019;2:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tompson R.H., Gillett M.D., Cheville J.C., Lohse C.M., Dong H., Webster W.S. Costimulatory molecule B7-H1 in primary and metastatic clear cell renal cell carcinoma. Cancer. 2005;104:2084–2091. doi: 10.1002/cncr.21470. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein M.B., Garnett C.T., Zhang H., Velcich A., Wattenberg M.M., Gameiro S.R. Radiation-induced modulation of costimulatory and coinhibitory T-cell signaling molecules on human prostate carcinoma cells promotes productive antitumor immune interactions. Cancer Biother Radiopharm. 2014;29:153–161. doi: 10.1089/cbr.2013.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C., Duan X., Guo N., Chan C., Poon C., Weichselbaum R.R. Core-shell nanoscale coordination polymers combine chemotherapy and photodynamic therapy to potentiate checkpoint blockade cancer immunotherapy. Nat Commun. 2016;7:12499. doi: 10.1038/ncomms12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., Wang J., Zhang X., Yu S., Wen D., Hu Q. In situ formed reactive oxygen species-responsive scaffold with gemcitabine and checkpoint inhibitor for combination therapy. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aan3682. [DOI] [PubMed] [Google Scholar]
- 12.Kuai R., Yuan W., Son S., Nam J., Xu Y., Fan Y. Elimination of established tumors with nanodisc-based combination chemoimmunotherapy. Sci Adv. 2018;4 doi: 10.1126/sciadv.aao1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D., Wang T., Liu J., Yu H., Jiao S., Feng B. Acid-activatable versatile micelleplexes for PD-L1 blockade-enhanced cancer photodynamic immunotherapy. Nano Lett. 2016;16:5503–5513. doi: 10.1021/acs.nanolett.6b01994. [DOI] [PubMed] [Google Scholar]
- 14.Dai L., Li K., Li M., Zhao X., Luo Z., Lu L. Size/Charge changeable acidity-responsive micelleplex for photodynamic-improved PD-L1 immunotherapy with enhanced tumor penetration. Adv Funct Mater. 2018;28:1707249. [Google Scholar]
- 15.Teo P.Y., Yang C., Whilding L.M., Parente-Pereira A.C., Maher J., George A.J. Ovarian cancer immunotherapy using PD-L1 siRNA targeted delivery from folic acid-functionalized polyethylenimine: strategies to enhance T cell killing. Adv Healthc Mater. 2015;4:1180–1189. doi: 10.1002/adhm.201500089. [DOI] [PubMed] [Google Scholar]
- 16.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 17.Ledford H. CRISPR, the disruptor. Nature. 2015;522:20–24. doi: 10.1038/522020a. [DOI] [PubMed] [Google Scholar]
- 18.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 19.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhavan R., Tsai L.H. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 21.Contreras-Vallejos E., Ultreras E., Gonzalez-Billault C. Going out of the brain: non-nervous system physiological and pathological functions of Cdk5. Cell Signal. 2012;24:44–52. doi: 10.1016/j.cellsig.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 22.Arif A. Extraneuronal activities and regulatory mechanisms of the atypical cyclin-dependent kinase Cdk5. Biochem Pharmacol. 2012;84:985–993. doi: 10.1016/j.bcp.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Pareek T.K., Lam E., Zheng X., Askew D., Kulkarni A.B., Chance M.R. Cyclin-dependent kinase 5 activity is required for T cell activation and induction of experimental autoimmune encephalomyelitis. J Exp Med. 2010;207:2507–2519. doi: 10.1084/jem.20100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorand R.D., Nthale J., Myers JT., Barkauskas D.S., Avril S., Chirieleison S.M. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn D.M., Langer R. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 26.Akinc A., Lynn D.M., Anderson D.G., Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 27.Cheng W., Wu D., Liu Y. Michael addition polymerization of trifunctional amine and acrylic monomer: a versatile platform for development of biomaterials. Biomacromolecules. 2016;17:3115–3126. doi: 10.1021/acs.biomac.6b01043. [DOI] [PubMed] [Google Scholar]
- 28.Yin M., Bao Y., Gao X., Wu Y., Sun Y., Zhao X. Redox/pH dual-sensitive hybrid micelles for targeting delivery and overcoming multidrug resistance of cancer. J Mater Chem B. 2017;5:2964–2978. doi: 10.1039/c6tb03282f. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang X., Wu T., Zhao Y., Hu X., Bao Y., Guo Y. Lipid-enveloped zinc phosphate hybrid nanoparticles for codelivery of H-2Kb and H-2Db-restricted antigenic peptides and monophosphoryl lipid A to induce antitumor immunity against melanoma. J Control Release. 2016;228:26–37. doi: 10.1016/j.jconrel.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 30.Kwak G., Kim D., Nam G.H., Wang S.Y., Kim I.S., Kim S.H. Programmed cell death protein ligand-1 silencing with polyethylenimine-Dermatan sulfate complex for dual inhibition of melanoma growth. ACS Nano. 2017;11:10135–10146. doi: 10.1021/acsnano.7b04717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L., Song L., Liu X., Yang X., Li X., He T. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 33.Kim S.M., Yang Y., Oh S.J., Hong Y., Seo M., Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Robert C., Schachter J., Long G.V., Arance A., Grob J.J., Mortier L. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 35.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 36.He J., Hu Y., Hu M., Li B. Development of PD-1/PD-L1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. Sci Rep. 2015;5:13110. doi: 10.1038/srep13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan X., Chan C., Guo N., Han W., Weichselbaum R.R., Lin W. Photodynamic therapy mediated by nontoxic core-shell nanoparticles synergizes with immune checkpoint blockade to elicit antitumor immunity and antimetastatic effect on breast cancer. J Am Chem Soc. 2016;138:16686–16695. doi: 10.1021/jacs.6b09538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C., Sun W., Ye Y., Hu Q., Bomba H.N., Gu Z. In situ activation of platelets with checkpoint inhibitors for post-surgical cancer immunotherapy. Nat Biomed Eng. 2017;1 [Google Scholar]
- 39.Sagiv-Barfi I., Kohrt H.E.K., Czerwinski D.K., Ng P.P., Chang B.Y., Levy R. Therapeutic antitumor immunity by checkpoint blockade is enhanced by ibrutinib, an inhibitor of both BTK and ITK. Proc Natl Acad Sci U S A. 2015;112:E966–E972. doi: 10.1073/pnas.1500712112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfirschke C., Engblom C., Rickelt S., Cortez-Retamozo V., Garris C., Pucci F. Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity. 2016;44:343–354. doi: 10.1016/j.immuni.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powles T., Eder J.P., Fine G.D., Braiteh F.S., Loriot Y., Cruz C. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 42.Robert C., Long G.V., Brady B., Dutriaux C., Maio M., Mortier L. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 43.Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenberg J.E., Hoffman-Censits J., Powles T., van der Heijden M.S., Balar A.V., Necchi A. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uprichard S.L. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura H., Nose M., Hiai H., Minato N., Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 47.Astruc D., Boisseller E., Omelas C. Dendrimers designed for functions: from physical, photophysical, and supramolecular properties to applications in sensing, catalysis, molecular electronics, photonics, and nanomedicine. Chem Rev. 2010;110:1857–1959. doi: 10.1021/cr900327d. [DOI] [PubMed] [Google Scholar]
- 48.Ros X.R., Vermeulen L. Turning cold tumors hot by blocking TGF-β. Trends Canc. 2018;4:335–337. doi: 10.1016/j.trecan.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Galluzzi L., Chan T.A., Kroemer G., Wolchok J.D., López-Soto A. The hallmarks of successful anticancer immunotherapy. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aat7807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.