Abstract
Background & Aims
Liver inflammation has been recognized as a hallmark of hepatocarcinogenesis. Although Forkhead Box M1 (FoxM1) is a well-defined oncogenic transcription factor that is overexpressed in hepatocellular carcinoma (HCC), its role in liver inflammation has never been explored.
Methods
We generated hepatocyte-specific FoxM1 conditional transgenic (TG) mice by using the Cre-loxP and Tetracycline (Tet)-on systems to induce FoxM1 expression in a hepatocyte-specific and time-dependent manner.
Results
After treatment of Tet-derivatives doxycycline (DOX) to induce FoxM1, TG mice exhibited spontaneous development of hepatocyte death with elevated serum alanine aminotransferase levels and hepatic infiltration of macrophages. The removal of DOX in TG mice completely removed this effect, suggesting that spontaneous inflammation in TG mice occurs in a hepatocyte FoxM1-dependent manner. In addition, liver inflammation in TG mice was associated with increased levels of hepatic and serum chemokine (C-C motif) ligand 2 (CCL2). In vitro transcriptional analysis confirmed that CCL2 is a direct target of FoxM1 in murine hepatocytes. After receiving FoxM1 induction since birth, all TG mice exhibited spontaneous HCC with liver fibrosis at 12 months of age. Hepatic expression of FoxM1 was significantly increased in liver injury models. Finally, pharmacologic inhibition of FoxM1 reduced liver inflammation in models of liver injury.
Conclusions
Hepatocyte FoxM1 acts as a crucial regulator to orchestrate liver inflammation linking to hepatocarcinogenesis. Thus, hepatocyte FoxM1 may be a potential target not only for the treatment of liver injury but also for the prevention toward HCC.
Keywords: FoxM1, Liver Inflammation, CCL2
Abbreviations used in this paper: ALT, alanine aminotransferase; AS, antisense; bp, base pair; CCL2, chemokine (C-C motif) ligand 2; CCl4, carbon tetrachloride; DOX, doxycycline; FoxM1, Forkhead Box M1 transcription factor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GalNAc, N-acetylgalactosamine; HCC, hepatocellular carcinoma; HFHC, high-fat/high-cholesterol; NPC, nonparenchymal cell; PBS, phosphate-buffered saline; RT-PCR, reverse transcription polymerase chain reaction; S, sense; Tet, tetracycline; TG, transgenic; TUNEL, terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling; WT, wild-type
Graphical abstract
Summary.
Hepatocyte-specific FoxM1 transgenic (TG) mice develop spontaneous liver inflammation, fibrosis, and carcinogenesis. Spontaneous liver inflammation in TG mice was associated with increased expression of chemokine CCL2. FoxM1-specific inhibitor effectively reduces liver inflammation and CCL2 expression in models of liver injury.
Hepatocellular carcinoma (HCC) is one of the most common types of malignant neoplasms among primary liver cancers and a major cause of cancer-related deaths worldwide.1 It is well-known that HCC occurs in patients with chronic liver diseases, including hepatitis virus infections, alcoholic or nonalcoholic fatty liver diseases, and other liver diseases.2 Irrespective of the etiology, chronic liver inflammation promotes subsequent hepatic wound-healing responses, eventually leading to the development of liver fibrosis and HCC.3,4 Thus, the molecular mechanisms driving liver inflammation to HCC need to be better understood to identify a therapeutic strategy to prevent HCC in patients with chronic liver diseases.
The Forkhead box M1 (FoxM1) transcription factor is a member of the Fox family of proteins that share a highly conserved structure in the winged helix DNA-binding domain.5, 6, 7 Several studies have shown that FoxM1 acts as an important regulator for cell proliferation and is frequently expressed at a higher level than normal in a variety of human cancers.7, 8, 9, 10, 11, 12 Previous animal studies have shown that FoxM1 is critical for cancer cell proliferation of HCC.13,14 We previously reported that a high FoxM1 expression defines poor prognosis in patients with HCC after surgical resection,15 and that FoxM1 is regulated via the mevalonate pathway of lipid metabolism in hepatoma cell lines.16 Consistent with our observations, FoxM1 was shown to be overexpressed in tumor tissues of hepatitis B virus–related HCC compared with the surrounding nontumor tissues.17 In addition, FoxM1 expression has been also shown to be increased in the livers of chronic hepatitis or cirrhosis patients with hepatitis B virus infection compared with normal liver samples,17 suggesting that FoxM1 may be involved in the initial step of hepatocarcinogenesis during the development of liver injury.
In adult human organs, FoxM1 is abundantly expressed in testis, thymus, intestine, and colon.18 By contrast, no FoxM1 expression is present in adult murine livers under normal conditions18; however, it is induced in the livers during liver regeneration after partial hepatectomy19 and during toxin-induced liver injury.20 These results suggest that FoxM1 may be involved in not only hepatocyte proliferation but also liver inflammation. Consistent with these results, a previous study demonstrated that FoxM1 is required for allergen-mediated goblet cell proliferation and pulmonary inflammation through the induction of inflammatory cytokines or chemokines in a murine model of chronic lung disorder.21 However, to date the role of hepatocyte FoxM1 in liver inflammation during the development of liver injury has remained unclear.
In this study, we hypothesized that hepatocyte FoxM1 affects the inflammatory microenvironment of the liver. To clarify this issue, we developed a new murine model in which ectopic overexpression of FoxM1 in hepatocytes is controlled in a time-dependent manner.
Results
Overexpression of Forkhead Box M1 Transcription Factor in Murine Livers Causes Spontaneous Liver Injury and Fibrosis
To test whether increased FoxM1 expression is sufficient to drive liver injury, we established hepatocyte-specific FoxM1 conditional transgenic (TG) (TetO7-FoxM1tg/tg/Rosa26-LSL-rtTA/Alb-Cretg/-) mice using the tetracycline (Tet)-on and Cre-loxP systems (Figure 1A). To induce FoxM1 expression, TG and wild-type (WT) (TetO7-FoxM1tg/tg/Rosa26-LSL-rtTA/Alb-Cre-/-) mice were treated with doxycycline (DOX). Western blot and immunohistochemical analyses using antibody against FoxM1 confirmed that ectopic overexpression of FoxM1 was induced in a liver-specific and DOX-specific manner (Figure 1B–D). At 13 weeks of age, TG mice showed spontaneous elevations of serum alanine aminotransferase (ALT) levels and hepatocyte death, confirmed by an increased number of terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL)–positive hepatocytes (Figure 1E–G). Furthermore, TG mice showed spontaneous liver fibrosis, confirmed by collagen-specific picrosirius red staining or immunostaining for α-smooth muscle actin, a marker of activated hepatic stellate cells (Figure 1H and I). This spontaneous liver fibrosis in TG mice was further assessed by the increased expression of fibrosis-related genes in the livers (Figure 1J). Taken together, overexpression of FoxM1 in the livers induced spontaneous liver injury and fibrosis.
Figure 1.
Spontaneous liver injury and fibrosis in hepatocyte-specific FoxM1 transgenic mice. (A) Schematic illustration of strategy for generating hepatocyte-specific FoxM1 TG mice. (B) Western blot analysis showing FoxM1 protein expression in liver, kidney, and heart of WT and TG mice after 3 days of DOX treatment at 8 weeks. FoxM1 protein expression was liver-specific and DOX-specific in TG mice. (C) Representative images of FoxM1 staining in liver sections of WT and TG mice after 3 days of DOX treatment at 8 weeks. Scale bar: 100 μm (original magnification, ×200). (D) Quantification of FoxM1-positive area in liver sections of WT and TG mice after 3 days of DOX treatment at 8 weeks (WT, n = 8; TG, n = 8). (E) Serum ALT levels in WT and TG mice after 13 weeks of DOX treatment since birth (WT, n = 8; TG, n = 8). (F) Representative images of H&E (top) and TUNEL (bottom) staining in liver sections of WT and TG mice after 13 weeks of DOX treatment since birth. Scale bar: 200 μm (original magnification, ×100). (G) Quantification of number of TUNEL-positive hepatocytes in liver sections of WT and TG mice after 13 weeks of DOX treatment since birth (WT, n = 8; TG, n = 8). (H) Representative views of picrosirius red (top) and α-smooth muscle actin (bottom) staining in liver sections of WT and TG mice after 13 weeks of DOX treatment since birth. Scale bar: 200 μm (original magnification, ×100). (I) Quantification of picrosirius red–positive area in livers of WT and TG mice after 13 weeks of DOX treatment since birth (WT, n = 8; TG, n = 8). (J) Gene expression of ColIα1 and ColIα2 in livers of WT and TG mice after 13 weeks of DOX treatment since birth (WT, n = 8; TG, n = 8). Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using unpaired Student t test.
Short-Term Overexpression of Forkhead Box M1 Transcription Factor Induces Reversible Liver Inflammation With Macrophage Recruitment
To investigate whether FoxM1 itself has a direct effect on spontaneous liver injury in TG mice, we introduced transient overexpression of FoxM1 at 8 weeks of age for 3 days and repressed its expression by removing DOX (Figure 2A and B). After 3 days of DOX treatment, TG mice exhibited elevated serum ALT levels and hepatocyte death, confirmed by TUNEL staining; this spontaneous liver injury was completely reversible by removing DOX (Figure 2C–E). Furthermore, the increased liver injury in TG mice was associated with hepatic infiltration of inflammatory cells, consisting of F4/80-positive macrophages; this spontaneous liver inflammation was also completely reversed by removing DOX (Figure 2F). In addition, the fluorescence-activated cell sorting analysis confirmed that the number of CD11b+F4/80+ macrophages was increased in the livers of TG mice in a DOX-dependent manner (Figure 2G). These data indicate that short-term overexpression of FoxM1 is sufficient to induce liver injury and inflammation with macrophage recruitment.
Figure 2.
Reversible liver inflammation with macrophage recruitment induced by short-term hepatocyte-specific overexpression of FoxM1. (A) Schematic illustration of short-term hepatic overexpression of FoxM1. Hepatic overexpression of FoxM1 was induced by 3 days of DOX treatment (DOX on) and repressed by 3 subsequent days of DOX removal (DOX off). Pretreatment is denoted as DOX (-). (B) Western blot analysis showing FoxM1 protein expression in livers of WT and TG mice at indicated time points. (C) Serum ALT levels of WT and TG mice at indicated time points (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (D) Representative images of TUNEL staining in liver sections of WT and TG mice. Top, DOX(-); middle, DOX on; bottom, DOX off. Scale bar: 200 μm (original magnification, ×100). (E) Quantification of number of TUNEL-positive hepatocytes in liver sections of WT and TG mice at indicated time points (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (F) Representative images of H&E (left) and F4/80 (right) staining in liver sections of WT and TG mice. Top, DOX (-); second, DOX on; third, DOX on (enlarged view of boxed region of second panel); bottom, DOX off. Scale bar: 100 μm (original magnification, ×200). (G) Quantification of number of CD11b+F4/80+ macrophages in livers of WT and TG mice at indicated time points (WT, n = 3 at 8 wk; TG, n = 3 at 8 wk). (H) Quantification of hepatic gene expression of F4/80, Cd68, Nos2, Tnfα, Ccl2, Ccr2, Ccl5, Arg1, and Cd206 in WT and TG mice after 3 days of DOX treatment (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (I) Quantification of serum CCL2 levels in WT and TG mice at indicated time points (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (J) Western blot analysis showing FoxM1 protein expression in livers of WT and TG mice after 1 day of DOX treatment. (K) Serum ALT levels of WT and TG mice after 1 day of DOX treatment (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (L) Gene expression of Ccl2 (left) and serum CCL2 (right) of WT and TG mice after 1 day of DOX treatment (WT, n = 8 at 8 wk; TG, n = 8 at 8 wk). (M) Representative images of FoxM1 (top) and TUNEL (bottom) staining in liver sections of TG mice. Left, DOX (-); middle, DOX low dose; right, DOX on. (N) Quantification of FoxM1-positive area in liver sections of TG mice. DOX (-), n = 8; DOX low dose, n = 6; DOX on, n = 8 at 8 wk. (O) Quantification of number of TUNEL-positive hepatocytes in liver sections of TG mice. DOX (-), n = 8; DOX low dose, n = 6; DOX on, n = 8 at 8 wk. (P) Serum ALT levels of TG mice. DOX (-), n = 8; DOX low dose, n = 6; DOX on, n = 8 at 8 wk. (Q) Ccl2 gene expression in livers of TG mice. DOX (-), n = 8; DOX low dose, n = 6; DOX on, n = 8 at 8 wk. Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using one-way analysis of variance test (C, E, G, I, N, O, P, Q) or Student t test (H, K, L).
To investigate the molecular mechanisms underlying the spontaneous recruitment of hepatic macrophages in TG mice, we examined the gene expression of macrophage-related markers in the livers of TG mice. TG mice showed a significant increase in the hepatic gene expression of macrophage-related markers, including F4/80, Cd68, Nos2, Tnfα, Ccl2, Ccr2, Ccl5, Arg1, and Cd206 (Figure 2H). Among these markers, Ccl2 showed a 145-fold higher hepatic expression in TG mice than in WT mice (Figure 2H). Consistently, 3-day DOX treatment resulted in increased serum chemokine (C-C motif) ligand 2 (CCL2) levels in TG mice, and this increase was completely reversed by removing DOX (Figure 2I).
To rule out the possibility that CCL2 induction results from increased liver injury in TG mice, we overexpressed FoxM1 for 1 day with DOX (Figure 2J). There was no apparent increase in serum ALT levels and no difference in serum ALT levels in WT and TG mice after 1 day of DOX treatment (Figure 2K). However, there was a significant increase in hepatic gene expression of Ccl2 compared with WT mice at this earlier 1-day time point (Figure 2L), suggesting that the increased expression of hepatic Ccl2 gene in TG mice might occur before induction of liver injury.
Next, we titrated the levels of DOX in the drinking water and developed TG mice with lower FoxM1 expression by using low dose of DOX (DOX low dose, 0.01 mg/mL) (Figure 2M). Even DOX low dose induced the moderate expression of FoxM1 (27.4%; Figure 2N) and an increase in TUNEL-positive hepatocytes (Figure 2O) and serum ALT levels (Figure 2P) as was observed with the higher dose of DOX (DOX on, 0.2 mg/mL). Furthermore, DOX low dose increased Ccl2 gene expression in livers of TG mice as observed with DOX on (Figure 2Q).
Chemokine (C-C Motif) Ligand 2 as a Direct Target of Forkhead Box M1 Transcription Factor in Hepatocytes
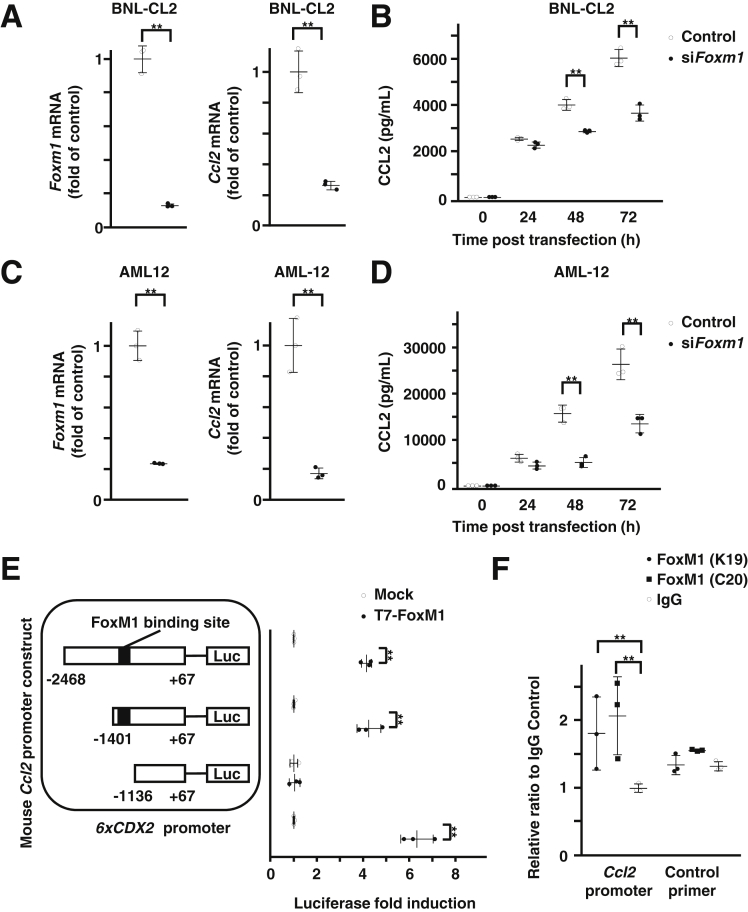
To investigate whether hepatocyte FoxM1 regulates the expression of CCL2, we next performed in vitro experiments using murine hepatocyte cell lines. Small interfering RNA–mediated knockdown of FoxM1 resulted in a significant decrease in the gene expression of Ccl2 and protein expression of CCL2 in murine hepatocyte cell lines BNL-CL2 (Figure 3A and B) and AML12 (Figure 3C and D).
Figure 3.
Direct regulation of CCL2 by FoxM1 in hepatocytes. (A) Quantification of gene expression of FoxM1 (left) and Ccl2 (right) in FoxM1 siRNA-transfected BNL-CL2 cells (n = 3 per group). (B) Quantification of CCL2 levels in supernatant of FoxM1 siRNA-transfected BNL-CL2 cells at indicated time points after transfection (n = 3 per group). (C) Quantification of gene expression of FoxM1 (left) and Ccl2 (right) in FoxM1 siRNA-transfected AML12 cells (n = 3 per group). (D) Quantification of CCL2 levels in supernatant of FoxM1 siRNA-transfected AML12 cells at indicated time points after transfection (n = 3 per group). (E) Schematic illustration of luciferase (Luc) reporter constructs containing −2468/+67 bp murine Ccl2 promoter and its deletion mutants (−1401/+67 bp and −1136/+67 bp) and quantification of transcriptional activities induced by cotransfection of T7-FoxM1 expression vector (T7-FoxM1) compared with CMV-empty vector (Mock) (n = 3 per group). A 6xCDX2 promoter LUC construct was used as a positive control. (F) Quantification of chromatin immunoprecipitation assay to show direct binding of FoxM1 protein to murine Ccl2 promoter DNA using 2 independent antibodies against FoxM1 (K19 and C20) compared with immunoglobulin G control (n = 3 per group). Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated using unpaired Student t test.
We then performed in vitro transcriptional analysis to investigate whether CCL2 is a direct target of FoxM1. One potential FoxM1 binding site was identified in the −2468/+67 base pair (bp) promoter region of the murine Ccl2 gene at −1343/−1338 bp (Figure 3E). Cotransfection of FoxM1 expression vector resulted in significant increased activity of the −1401/+67 bp Ccl2 luciferase reporter, and the deletion of the FoxM1 binding site in the Ccl2 promoter region −1136/+67 bp abolished the capacity of FoxM1 to stimulate this activity, indicating that −1343/−1338 bp in the Ccl2 promoter region functions as a FoxM1 binding site (Figure 3E). To further confirm the binding of FoxM1 to the promoter region of the Ccl2 gene, the chromatin immunoprecipitation assay was performed in murine hepatocyte BNL-CL2 cells using 2 antibodies against FoxM1. This assay showed the specific binding of FoxM1 protein to the Ccl2 promoter DNA (Figure 3F). Collectively, the data indicate that CCL2 is a direct target of FoxM1 in hepatocytes and suggest the involvement of hepatic CCL2 induction in the spontaneous macrophage recruitment of TG livers.
Hepatocytes Are the Major Source of Chemokine (C-C Motif) Ligand 2 Production in Transgenic Mice
To investigate which cell population produces CCL2 in TG mice, we examined the gene expression of Ccl2 and the protein expression of CCL2 in hepatocytes and nonparenchymal cells (NPCs) isolated from livers of WT and TG mice after 3 days of DOX treatment. Hepatocytes of TG mice showed a significant increase in Ccl2 gene expression compared with those of WT mice, whereas that in NPCs was comparable between the 2 groups (Figure 4A). Western blot analysis showed the increased expression of CCL2 protein in hepatocytes of TG mice compared with those of WT mice (Figure 4B). These data suggest that the major source of CCL2 is hepatocytes rather than NPCs in TG mice.
Figure 4.
Hepatocytes are the major source of CCL2 production in TG mice. (A) Quantification of Ccl2 gene expression in hepatocytes (left) and NPCs (right) from WT and TG mice after 3 days of DOX treatment (n = 3, each group). (B) Western blot analysis of FoxM1 protein expression in hepatocytes and NPCs of WT and TG mice.
Forkhead Box M1 Transcription Factor Induction Has a Modest Effect on Cell Death in Cultured Hepatocytes In Vitro
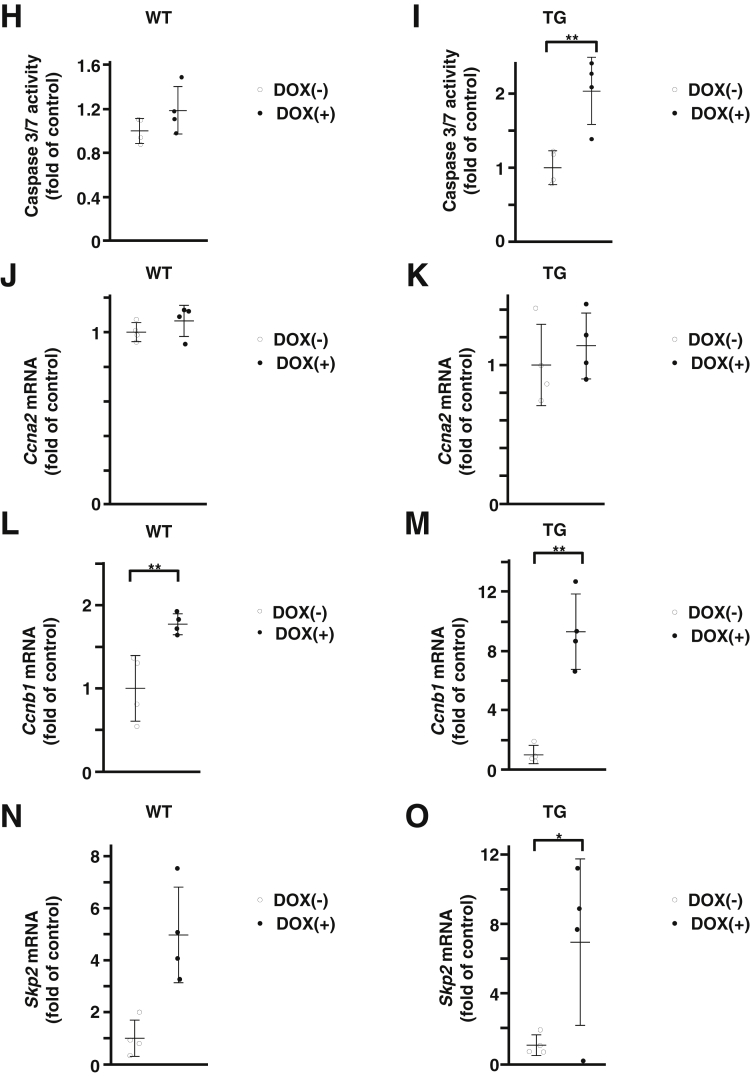
We investigated the effect of FoxM1 induction on hepatocyte cell viability and death in vitro by using primary cultured hepatocytes isolated from WT and TG mice. WT and TG hepatocytes were cultured in vitro and were treated with 100 ng/mL DOX for 24 hours. Western blot analysis confirmed that FoxM1 protein was induced in TG hepatocytes treated with DOX (Figure 5A). Consistent with in vivo data, in vitro treatment with DOX increased in Ccl2 gene expression and CCL2 protein expression in the supernatant of TG hepatocytes but not in the supernatants of WT hepatocytes (Figure 5B–E). The WST assay showed that DOX treatment resulted in a modest reduction (0.95-fold) in cell viability of TG hepatocytes (Figure 5F and G), and the caspase 3/7 activity assay showed a modest increase (2.03-fold) in cell death of TG hepatocytes (Figure 5H and I). Among cell cycle–related genes, gene expression of Ccnb1 and Skp2, known FoxM1-target genes, increased in TG hepatocytes treated with DOX (Figure 5J–O). These in vitro data suggest that FoxM1 induction has a modest effect on hepatocyte death.
Figure 5.
FoxM1 induction has modest effect on cell death in cultured hepatocytes in vitro. (A) Western blot analysis showing FoxM1 protein expression in isolated primary hepatocytes of TG mice treated with 100 ng/mL DOX. (B and C) Quantification of gene expression of Ccl2 in primary hepatocytes from WT (B) and TG (C) mice treated with 100 ng/mL DOX (n = 4 per group). (D and E) Quantification of CCL2 levels in supernatant of primary hepatocytes from WT (D) and TG (E) mice treated with 100 ng/mL DOX (n = 4 per group). (F and G) WST assay of primary hepatocytes from WT (F) and TG (G) mice treated with 100 ng/mL DOX (n = 4 per group). (H and I) Caspase 3/7 activity in primary hepatocytes from WT (H) and TG (I) mice treated with 100 ng/mL DOX (n = 4 per group). (J and K) Quantification of gene expression of Ccna2 in primary hepatocytes from WT (J) and TG (K) mice treated with 100 ng/mL DOX (n = 4 per group). (L and M) Quantification of gene expression of Ccnb1 in primary hepatocytes from WT (L) and TG (M) mice treated with 100 ng/mL DOX (n = 4 per group). (N and O) Quantification of gene expression of Skp2 in primary hepatocytes of WT (N) and TG (O) mice treated with 100 ng/mL DOX (n = 4 per group). Data are expressed as individual values and mean ± standard deviation; *P < .05, **P < .01. Significance was calculated by using unpaired Student t test.
Hepatocyte-Specific Chemokine (C-C Motif) Ligand 2 Inhibition Reduces Liver Injury in Transgenic Mice
To investigate whether hepatocyte-derived CCL2 plays a role in the development of spontaneous liver injury in TG mice, we inhibited CCL2 expression by using hepatocyte-specific N-acetylgalactosamine (GalNAc)-siRNA system.22 Subcutaneous administration of GalNAc-conjugated siRNA against murine Ccl2 (GalNAc-siCcl2) reduced gene expression of hepatic Ccl2 and serum levels of CCL2 compared with administration of GalNAc-siControl (control) in TG mice after 3 days of DOX treatment, confirming that hepatocytes are the major source of CCL2 production in TG mice (Figure 6A). Subcutaneous administration of GalNAc-siCcl2 also reduced liver injury (Figure 6B) as shown by reduced serum ALT levels and reduced number of TUNEL-positive hepatocytes compared with the control in TG mice after 3 days of DOX treatment (Figure 6C–E). Furthermore, subcutaneous administration of GalNAc-siCcl2 reduced infiltration of F4/80-positive macrophages compared with control in TG mice after 3 days of DOX treatment (Figure 6F). Although TG mice did not show apparent liver fibrosis assessed by picrosirius red staining after 3 days of DOX treatment (Figure 6G and H), subcutaneous administration of GalNAc-siCcl2 resulted in significant reduction of gene expression of hepatic Col1α2 compared with control in TG mice after 3 days of DOX treatment (Figure 6I). Collectively, these data suggest that hepatocyte-derived CCL2 contributes to liver injury in TG mice.
Figure 6.
Hepatocyte-specific CCL2 inhibition reduces liver injury in TG mice. (A) Quantification of hepatic gene expression of Ccl2 (left) and serum CCL2 levels (right) in TG mice treated with GalNAc-siControl (control) or GalNAc-siCcl2 (control, n = 4 at 8 wk; GalNAc-siCcl2, n = 4 at 8 wk). (B) Representative images of H&E staining of TG mice treated with control or GalNAc-siCcl2. Scale bar: 200 μm (original magnification, ×100). (C) Serum ALT levels of TG mice treated with control or GalNAc-siCcl2 (control, n = 4 at 8 wk; GalNAc-siCcl2, n = 4 at 8 wk). (D) Representative images of TUNEL staining of TG mice treated with control or GalNAc-siCcl2. Scale bar: 200 μm (original magnification, ×100). (E) Quantification of number of TUNEL-positive hepatocytes in liver sections from control and GalNAc-siCcl2 group mice (control, n = 4 at 8 wk; GalNAc-siCcl2, n = 4 at 8 wk). (F) Representative images of F4/80 staining of TG mice treated with control or GalNAc-siCcl2 (bottom, enlarged views of boxed regions in top image). Scale bar: 100 μm (original magnification, ×200). (G) Representative images of picrosirius red staining in liver sections from TG mice treated with control or GalNAc-siCcl2. Scale bar: 200 μm (original magnification, ×100). (H) Quantification of picrosirius red–positive area (%) in livers of control and GalNAc-siCcl2 group mice (control, n = 4 at 8 wk; GalNAc-siCcl2, n = 4 at 8 wk). (I) Gene expression of ColIα1 (left) and ColIα2 (right) in livers of control and GalNAc-siCcl2 group mice (control, n = 4 at 8 wk; GalNAc-siCcl2, n = 4 at 8 wk). Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using unpaired Student t test.
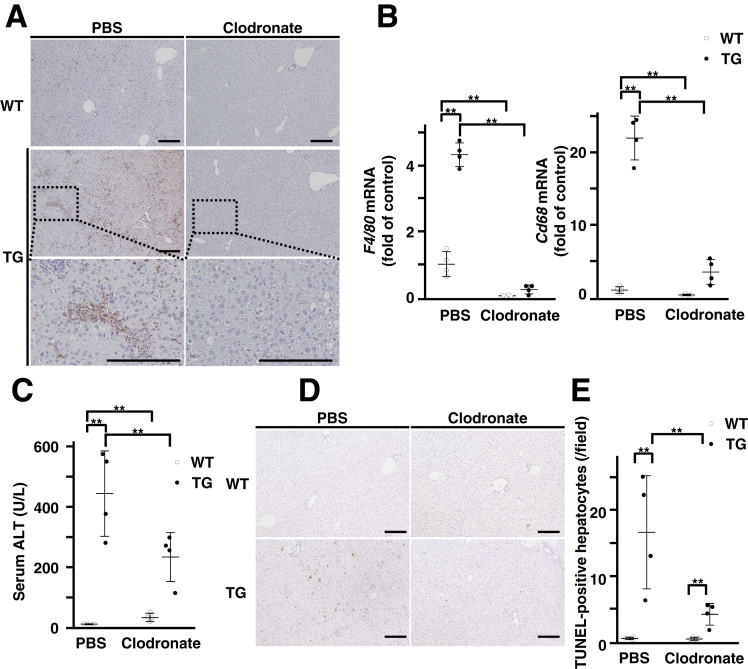
Spontaneous Liver Injury in Transgenic Mice Occurs Through Hepatic Macrophage Recruitment Induced by Forkhead Box M1 Transcription Factor Expression
To investigate the consequences of hepatic macrophage recruitment in liver injury of TG mice, we depleted the macrophages of these mice. Immunohistochemical or real-time reverse transcription polymerase chain reaction (RT-PCR) analysis showed that the administration of clodronate liposomes resulted in the successful depletion of F4/80-positive macrophages in the livers of WT and TG mice (Figure 7A and B). Macrophage-depleted TG mice exhibited lower serum ALT levels and TUNEL-positive hepatocytes than phosphate-buffered saline (PBS) liposomes–treated TG mice (Figure 7C–E). These data suggest that spontaneous hepatocyte death in TG mice is associated with hepatic macrophage recruitment induced by FoxM1 expression.
Figure 7.
Macrophage recruitment is required for spontaneous liver inflammation in TG mice. (A) Representative images of F4/80 staining in liver sections of 3-day DOX-treated WT and TG mice injected with PBS or clodronate liposomes. Top, WT mice; middle, TG mice; bottom, enlarged views of boxed region of middle panels. Scale bar: 200 μm (original magnification, ×100). (B) Quantification of hepatic gene expression of F4/80 (left) and Cd68 (right) in 3-day DOX-treated WT and TG mice injected with PBS or clodronate liposomes. (PBS-treated WT mice, n = 4 at 8 wk; PBS-treated TG mice, n = 4 at 8 wk; clodronate liposome–treated WT mice, n = 4 at 8 wk; clodronate liposome–treated TG mice, n = 4 at 8 wk). (C) Quantification of serum ALT levels in 3-day DOX-treated WT and TG mice injected with PBS or clodronate liposomes (PBS-treated WT mice, n = 4 at 8 wk; PBS-treated TG mice, n = 4 at 8 wk; clodronate liposome–treated WT mice, n = 4 at 8 wk; clodronate liposome–treated TG mice, n = 4 at 8 wk). (D) Representative images of TUNEL staining in liver sections of 3-day DOX-treated WT and TG mice injected with PBS or clodronate liposomes. Scale bar: 200 μm (original magnification, ×100). (E) Quantification of number of TUNEL-positive hepatocytes in liver sections of 3-day DOX-treated WT and TG mice injected with PBS or clodronate liposomes (PBS-treated WT mice, n = 4 at 8 wk; PBS-treated TG mice, n = 4 at 8 wk; clodronate liposome–treated WT mice, n = 4 at 8 wk; clodronate liposome–treated TG mice, n = 4 at 8 wk). Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using one-way analysis of variance test.
Forkhead Box M1 Transcription Factor–Dependent Liver Inflammation Leads to Hepatocarcinogenesis
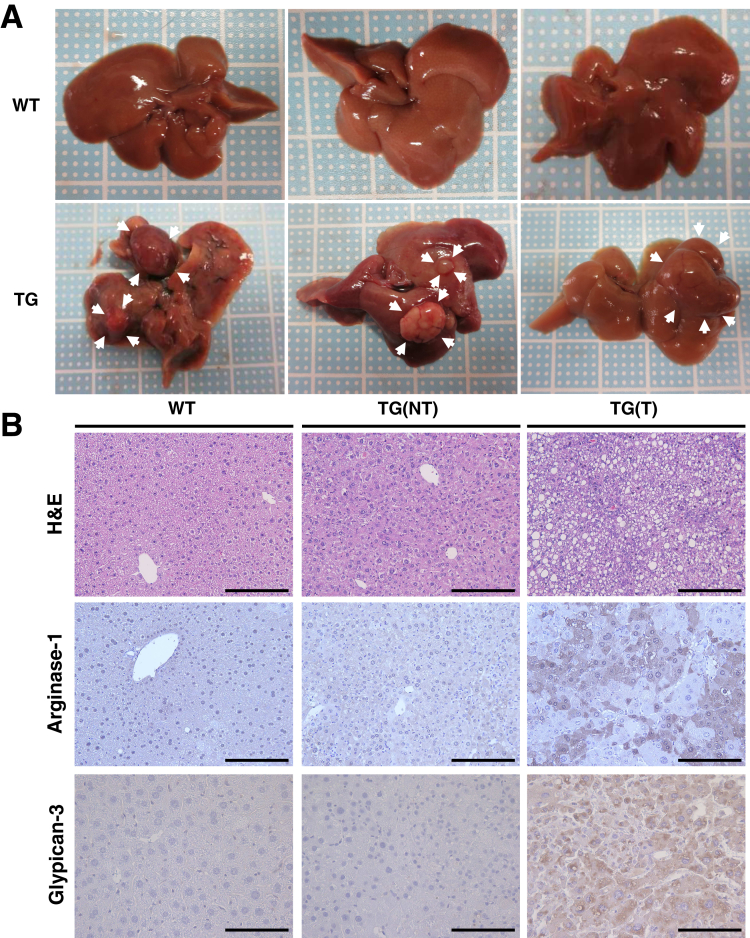
In patients with chronic liver diseases, continuous liver inflammation and fibrosis are known to lead to tumorigenesis. Therefore, we investigated whether long-term overexpression of FoxM1 affects hepatocarcinogenesis. At 48 weeks of age, although all TG mice treated with DOX after birth were found to develop liver tumors, WT mice did not develop any tumors (Figure 8A–C). Histologic analysis showed that liver tumors in TG mice were HCC, because tumor cells were large with hyperchromatic nuclei in compact growth pattern and did not form any tubular structure (Figure 8B). In addition, liver tumors in TG mice showed increased expression of arginase-1 and glypican-3, known makers of HCC,23,24 confirming that these tumors are HCC (Figure 8B). The Ki-67 immunohistochemistry confirmed that liver tumors in TG mice showed higher cell proliferation than the nontumor regions of TG mice or the livers of WT mice (Figure 8D and E). Furthermore, picrosirius red staining and increased hepatic expression of fibrosis-related genes showed that the nontumor regions of TG mice developed liver fibrosis (Figure 8F–H). Our data indicate that overexpression of FoxM1 in hepatocytes results in liver injury and subsequent liver inflammation and fibrosis, leading to hepatocarcinogenesis.
Figure 8.
Long-term hepatocyte-specific overexpression of FoxM1 causes hepatocarcinogenesis with significant fibrosis in the background liver. (A) Gross appearance of livers in WT and TG mice after 12 months of DOX treatment since birth. Arrowheads indicate liver tumors in TG mice. (B) Representative images of H&E (top), arginase-1 (middle), and glypican-3 (bottom) staining of WT and TG mice after 12 months of DOX treatment since birth. Nontumor (NT) lesions in WT mice (left), nontumor lesions in TG mice, denoted as TG(NT) (middle), and tumor lesions in TG mice, which are denoted as TG(T) (right). Scale bar: 100 μm (original magnification, ×400). (C) Quantification of tumor numbers (left), maximal size (mm, middle), and tumor incidence (%, right) in WT and TG mice after 12 months of DOX treatment since birth (WT mice, n = 10; TG mice, n = 10). (D) Representative images of Ki-67 staining of WT and TG mice after 12 months of DOX treatment since birth. Scale bar: 200 μm (original magnification, ×100). (E) Quantification of number of Ki-67 positive hepatocytes in liver sections of NT lesions in WT mice, NT lesions in TG mice, denoted as TG(NT), and tumor lesions in TG mice, which are denoted as TG(T). (WT mice, n = 10; TG mice, n = 10). (F) Representative images of picrosirius red staining of WT and TG mice after 12 months of DOX treatment since birth. Scale bar: 200 μm (original magnification, ×100). (G) Quantification of picrosirius red–positive area (%) in liver sections of WT and TG mice (WT mice, n = 10; TG mice, n = 10). (H) Quantification of ColIα1 and ColIα2 gene expression in livers of WT and TG mice (WT mice, n = 10; TG mice, n = 10). Data are expressed as individual values and mean ± standard deviation; NT, nontumor; T, tumor. *P < .05, **P < .01. Significance was calculated by using one-way analysis of variance test (E) or Student t test (C, G, H).
Hepatic Forkhead Box M1 Transcription Factor Is Induced in Humans and Mice With Chronic Liver Injury
To confirm the significance of FoxM1 in liver injury, we examined the expression of FoxM1 in the liver tissues of patients with chronic liver diseases such as chronic hepatitis B, chronic hepatitis C, nonalcoholic steatohepatitis, and cirrhosis. As we reported previously,15,16 an immunohistochemical analysis confirmed increased FoxM1expression in HCC tissues (Figure 9A). This analysis also showed higher FoxM1 expression in liver tissues of chronic liver diseases compared with normal liver tissues (Figure 9A). To confirm an increase in hepatic FoxM1 expression during the development of chronic liver injury, we next used 2 murine models of chronic liver injury induced by a high-fat/high-cholesterol (HFHC) diet or chronic administration of carbon tetrachloride (CCl4). The gene expression of Foxm1 was increased as serum ALT levels were elevated in both models (Figure 9B). Immunohistochemical analysis also confirmed a higher hepatic expression of FoxM1 in chronic liver disease tissues than in normal liver tissues (Figure 9C). Collectively, these data suggest a role of FoxM1 in liver injury.
Figure 9.
Increased expression of FoxM1 in liver sections of patients with chronic liver diseases and murine model of liver injury. (A) Representative images of FoxM1 staining in liver sections of patients with chronic liver diseases. Scale bar: 100 μm (original magnification, ×400). Quantification of FoxM1-positive area was indicated on lower left. CHB, chronic hepatitis B; CHC, chronic hepatitis C; LC, liver cirrhosis; NASH, nonalcoholic steatohepatitis; NL, normal liver. (B) Quantification of hepatic FoxM1 expression (left) and serum ALT levels (right) in C57BL/6 mice fed a HFHC diet for 8 weeks or treated with CCl4 for 4 weeks. Mice fed a normal diet (ND) were used as the control (ND, n = 6 at 14 wk; HFHC, n = 6 at 14 wk; CCl4, n = 4 at 14 wk). (C) Representative images of FoxM1 staining (left) and quantification of FoxM1-positive area (right) in liver sections of C57BL/6 mice fed a ND or a HFHC diet or treated with CCl4. ND-fed mice were used as the control. Scale bar: 100 μm (original magnification, ×200). Quantification positive rate was indicated on the lower left. Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using unpaired Student t test.
Forkhead Box M1 Transcription Factor Inhibitor Reduces Liver Inflammation in a Liver Injury Model
We finally investigated whether the pharmacologic inhibition of FoxM1 reduces liver inflammation during the development of liver injury. For this purpose, we used a murine model of liver injury fed a HFHC diet, which induced hepatic FoxM1 expression (Figure 9B and C). Mice fed a HFHC diet were treated with a known FoxM1 inhibitor, thiostrepton.25,26 The thiostrepton-treated mice showed significantly lower serum ALT levels and fewer TUNEL-positive hepatocytes than the control mice (Figure 10A–D). The thiostrepton-treated mice also showed significantly fewer F4/80-positive macrophages in the livers than the control mice (Figure 10E). Furthermore, thiostrepton treatment resulted in a decrease of hepatic Ccl2 expression or serum CCL2 levels in mice with HFHC diet (Figure 10F and G). On the other hand, thiostrepton treatment did not affect hepatocyte proliferation in HFHC diet–fed mice (Figure 10H and I). The anti-inflammatory effect of thiostrepton was also confirmed in TG mice, showing that thiostrepton treatment effectively reduced spontaneous liver inflammation without significant antiproliferation effect on hepatocytes (Figure 11). These findings suggest that FoxM1 inhibition may represent a therapeutic approach against liver injury.
Figure 10.
FoxM1 inhibitor reduces liver inflammation in mice fed a HFHC diet. (A) Serum ALT levels of C57BL/6 mice fed a HFHC diet for 8 weeks and treated with vehicle (control) or thiostrepton for 2 weeks (control, n = 8 at 14 wk; thiostrepton, n = 8 at 14 wk). (B) Representative images of H&E staining of control and thiostrepton groups. Scale bar: 200 μm (original magnification, ×100). (C) Representative images of TUNEL staining of control and thiostrepton groups. Scale bar: 200 μm (original magnification, ×100). (D) Quantification of number of TUNEL-positive hepatocytes in liver sections of control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). (E) Representative images of F4/80 staining of C57BL/6 mice treated with vehicle or thiostrepton (bottom, enlarged views of top boxed regions). Scale bar: 100 μm (original magnification, ×200). (F) Quantification of F4/80 and Ccl2 gene expression in livers of control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). (G) Serum CCL2 levels of control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). (H) Representative images of Ki-67 staining in control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). Scale bar: 100 μm (original magnification, ×200). (I) Quantification of number of Ki-67 positive hepatocytes in liver sections of control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). Data are expressed as individual values and mean ± standard deviation; **P < .01. Significance was calculated by using unpaired Student t test.
Figure 11.
FoxM1 inhibitor reduces liver inflammation in TG mice. (A) Serum ALT levels of TG mice treated with vehicle (control) or thiostrepton every 3 days from 3 days before the experiment (control, n = 8 at 8 wk; thiostrepton, n = 8 at 8 wk). Hepatic overexpression of FoxM1 was induced by 3 days of DOX treatment. (B) Representative images of H&E staining of TG mice treated with vehicle or thiostrepton. Scale bar: 100 μm (original magnification, ×200). (C) Representative images of TUNEL staining of TG mice treated with vehicle or thiostrepton. Scale bar: 200 μm (original magnification, ×100). (D) Quantification of number of TUNEL-positive hepatocytes in liver sections of control and thiostrepton groups (control, n = 8 at 8 wk; thiostrepton, n = 8 at 8 wk). (E) Representative images of F4/80 staining of TG mice treated with vehicle or thiostrepton (bottom, enlarged views of top boxed regions). Scale bar: 100 μm (original magnification, ×200). (F) Quantification of hepatic gene expression of F4/80 and Ccl2 in TG mice treated with vehicle or thiostrepton (control, n = 8 at 8 wk; thiostrepton, n = 8 at 8 wk). (G) Serum CCL2 levels in TG mice treated with vehicle or thiostrepton (control, n = 8 at 8 wk; thiostrepton, n = 8 at 8 wk). (H) Representative images of Ki-67 staining in control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). Scale bar: 100 μm (original magnification, ×200). (I) Quantification of number of Ki-67 positive hepatocytes in liver sections of control and thiostrepton groups (control, n = 8; thiostrepton, n = 8). Data are expressed as individual values and mean ± standard deviation; *P < .05, **P < .01. Significance was calculated by using unpaired Student t test.
Discussion
Targeting liver inflammation is crucial for the development of new treatments against chronic liver diseases.27,28 Here we identified hepatocyte FoxM1 as a key driver to trigger chronic liver inflammation. Similar to the clinical course of patients with chronic liver disease, hepatocyte-specific FoxM1 transgenic mice developed spontaneous liver inflammation and fibrosis, leading to hepatocarcinogenesis. This FoxM1-dependent liver inflammation with hepatic macrophage recruitment was associated with hepatocyte-derived chemokine CCL2 that was identified as a novel direct target of FoxM1. Finally, we showed that inhibiting FoxM1 with its specific inhibitor effectively reduced liver inflammation in a model of liver injury induced by HFHC diet. Thus, our current study raised a possibility that targeting FoxM1 could be a novel therapeutic approach for liver injury toward HCC.
FoxM1 has a cell-autonomous role in forcing the cell proliferation of a variety of cancers including HCC.29 Several lines of evidence demonstrated that FoxM1 is overexpressed in cancer tissues compared with surrounding noncancer tissues.7, 8, 9, 10, 11, 12 However, little is known whether chronic tissue damage might affect FoxM1 expression in the damaged tissues. According to previous reports, FoxM1 is more expressed in the damaged lung tissues of patients with idiopathic pulmonary fibrosis than those of normal patients,30 and FoxM1 expression is also increased in human gastritis tissues with Helicobacter pylori infection compared with normal gastric tissues.31 In this study, we showed that FoxM1 expression was induced in the damaged livers of murine models and was implicated in human specimens of chronic liver diseases. These data indicate that FoxM1 might be involved in the initial process of liver damage and inflammation before HCC development.
Chronic inflammation is a key characteristic of chronic liver disease.27,28 The interaction between hepatocytes and NPCs via a variety of mediators such as cytokines or chemokines32 has been shown to be crucial for driving liver inflammation; however, the signaling pathways in hepatocytes are not yet fully understood. In this study, we showed that increased FoxM1 in murine livers resulted in enhanced liver inflammation. Among inflammatory mediators that were increased in TG livers, CCL2 was found to have a putative FoxM1 binding site in its promoter region, and in vitro experiments confirmed that FoxM1 is able to regulate CCL2 directly in hepatocytes. CCL2 is a well-defined chemokine that recruits CCR2-positive monocytes/macrophages to trigger liver inflammation and promote hepatocarcinogenesis.33,34 Spontaneous liver inflammation in TG mice was consistently associated with increased macrophage recruitment in the injured livers, suggesting a novel cell-nonautonomous role of hepatocyte FoxM1 to trigger chronic liver inflammation via direct regulation of inflammatory mediators and recruitment of inflammatory cells.
Therapeutic strategies to target FoxM1 in liver diseases, especially in HCC, have been investigated with in vivo and in vitro experiments so far.14,16 It has been shown that the inhibition of FoxM1 activity effectively reduces liver tumors in a murine model of HCC.14 We recently demonstrated that statins, well-known cholesterol-lowering drugs, also reduce FoxM1 expression and thereby induce cell death in human HCC cells.16 It has been reported that hepatocyte-specific FoxM1 knockout mice exhibit impaired liver regeneration after partial hepatectomy,19 and macrophage-specific FoxM1 knockout mice show delayed wound-healing response and worsened liver function in a liver injury model induced by CCl4,35 implying that FoxM1 inhibition may worsen liver damage and inflammation. However, in this study we showed that thiostrepton, a well-established FoxM1 inhibitor,25,26 effectively improved liver damage and reduced liver inflammation with macrophage recruitment and CCL2 expression in a murine liver injury model induced by HFHC diet. Consistent with our data, a recent publication demonstrated that a FoxM1 inhibitor effectively reduces lung inflammation in response to house dust mite allergens in mice.36 These findings suggest that FoxM1 inhibitors target the cell-nonautonomous function of FoxM1 rather than its cell-autonomous function in vivo. Further studies will be needed to elucidate this issue.
In conclusion, we have demonstrated that hepatocyte FoxM1 drives inflammatory response linking liver fibrosis and carcinogenesis via induction of inflammatory mediators. Because HCC arises in a background of liver inflammation and fibrosis, targeting hepatocyte FoxM1 would be a potential application not only for the treatment of liver injury but also for early intervention or prevention toward HCC.
Materials and Methods
All authors had access to the study data and had reviewed and approved the final manuscript.
Mice
Albumin promoter-driven Cre recombinase (Alb-Cre) transgenic C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were crossed with TetO-GFP-FoxM1-ΔN mice (TetO7-FoxM1)37,38 and Rosa26-loxP-STOP-loxP-reverse tetracycline transcriptional activator (rtTA) mice (Rosa26-LSL-rtTA) (Figure 1A). Transgenic (TetO7-FoxM1tg/tg/Rosa26-LSL-rtTA/Alb-Cretg/-) and WT (TetO7-FoxM1tg/tg/Rosa26-LSL-rtTA/Alb-Cre-/-) mice were treated with low dose DOX (0.01 mg/dL) or regular dose DOX on (0.2 mg/dL) (catalog D9891; Sigma-Aldrich, St Louis, MO) dissolved in 5% sucrose (catalog 30403-55; Nacalai Tesque, Kyoto, Japan) and supplied as drinking water to induce hepatocyte-specific FoxM1 expression. The Alb-Cre transgene was detected by using the following sense (S) and antisense (AS) primers: 5′-GCGGCATGGTGCAAGTTGAAT-3′ (S) and 5′-CGTTCACCGGCATCAACGTTT-3′ (AS). PCR analysis was performed to detect for the TetO-GFP-FoxM1-ΔN transgene with the following primers: 5′-CGACAAGCAGAAGAACGGCATC-3′ (S) and 5′-AGTAGGGAAAGTGGTCCTCAATCC-3′ (AS). The primers for detection of the ROSA26-LSL-rtTA transgene were as follows: 5′-GAGTTCTCTGCTGCCTCCTG-3′ (S) and 5′-AAGACCGCGAAGAGTTTGTC-3′ (AS). The WT ROSA26 allele was detected by using the following primers: 5′-GAGTTCTCTGCTGCCTCCTG-3′ (S) and 5′-CGAGGCGGATACAAGCAATA-3′ (AS). All mice were maintained under specific pathogen-free conditions in automated watered and ventilated cages on a 12-hour light/dark cycle. All animal experiments of this study were performed with humane care under approval from the Animal Care and Use Committee of Osaka University Medical School.
Real-Time Reverse Transcription Polymerase Chain Reaction
Total RNA was prepared by using the QIAshredder and the RNeasy Mini kit (catalog 79656 and 74106; Qiagen, Hilden, Germany), and quantitative real-time RT-PCR was performed by using the ReverTra Ace qPCR RT Master Mix (catalog FSQ-201; Toyobo, Osaka, Japan) according to the manufacturer’s protocol. Quantitative real-time RT-PCR reaction was performed with a Thunderbird SYBR qPCR Mix (catalog QPS-201; Toyobo) on a Quant Studio 6 Flex Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA). The following primers used in this study were purchased from Qiagen: murine Foxm1 (QT00120498), Gapdh (QT00199388), ColIα1 (QT00162204), ColIα2 (QT01055572), F4/80 (QT00099617), Cd68 (QT00254051), Nos2 (QT00100275), Tnfα (QT00104006), Ccl2 (QT00167832), Ccl5 (QT01747165), Arg1 (QT00134288), Ccna2 (QT00102151), Ccnb1 (QT01757007), and Skp2 (QT00118573). The following primers were synthesized by Sigma-Genosys: 5′-TCTGGGCTCACTATGCTGCA-3′ (S) and 5′-CCAAAGGTAACTGTCCTGGC-3′ (AS) for murine Ccr2; 5′-CAAAAACTGACTGGGCTTCC-3′ (S) and 5′-GCCCTTGATTCCAAAGAGTG-3′ (AS) for Cd206. The expression values of the murine genes were normalized to the glyceraldehyde 3-phosphate dehydrogenase (Gapdh) level.
Serum Analysis
Blood samples were harvested from the inferior vena cava and centrifuged at 700g for 10 minutes. Serum ALT levels were measured by using a colorimetric assay kit (catalog 431-30901; Wako, Kyoto, Japan) according to the manufacturer’s protocol.
Immunohistochemical Analysis
Liver tissues were fixed overnight with 10% formaldehyde neutral buffer solution (catalog 37152-51; Nacalai Tesque), embedded in paraffin, and sectioned (4-μm thick). Tissue sections were subjected to Mayer’s hematoxylin-eosin and immunohistochemical staining. We used the following antibodies: rabbit anti-mouse FoxM1 (A-11, 1:500, catalog sc-271746; Santa Cruz Biotechnology, Santa Cruz, CA), rat anti-mouse F4/80 (1:100, catalog MCA497GA; Serotec, Oxford, UK), rabbit anti-mouse α-smooth muscle actin (1:200, catalog ab5694; Abcam, Cambridge, MA), rabbit anti-mouse Ki-67 (D3B5) (1:400, catalog 12202; Cell Signaling Technology, Danvers, MA), goat anti-mouse arginase-1 (M-20,1:100, catalog sc-18355; Santa Cruz Biotechnology), or mouse glypican-3 (F-3, 1:100, catalog sc-390587; Santa Cruz Biotechnology). Overexpression of FoxM1 in hepatocytes of TG mice was assessed by immunohistochemical staining with FoxM1 (D12D5) (1:200, catalog 5436; Cell Signaling Technology). Liver fibrosis was quantified by measuring the picrosirius red (catalog 24901-500; Polysciences Inc, Warrington, PA) stained area using ImageJ (version 1.80; US National Institute of Health, Bethesda, MD). FoxM1 expression was quantified by measuring the area stained with anti-FoxM1 antibodies using ImageJ software (version 1.80). Cells with nuclear DNA fragmentation were detected by using a TUNEL staining kit (catalog S7100; Millipore, Molsheim, France) according to the manufacturer’s protocol. The numbers of TUNEL or Ki-67 positive hepatocytes per view field were counted at ×100 magnification.
Western Blot Analysis
Murine liver samples were homogenized in RIPA buffer containing 50 mmol/L Tris (pH 7.5), 0.15 mol/L NaCl, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 1 mmol/L EDTA, 1% Nonidet P-40, phosphatase inhibitor cocktail (catalog 0574-61; Nacalai Tesque), and protease inhibitor cocktail (catalog 25955-11; Nacalai Tesque). For Western blot analysis, the primary antibodies anti-human FoxM1 (D12D5) (1:1000, catalog 5436; Cell Signaling Technology) and GAPDH (1:1000, catalog 5174; Cell Signaling Technology) were used. The donkey anti-rabbit immunoglobulin G–horseradish peroxidase (1:3000, catalog sc-2357; Santa Cruz Biotechnology) was used as a secondary antibody.
Preparation of Mononuclear Cells
The livers of WT or TG mice were passed through a 70-μm nylon mesh to create single cell suspensions. The cell suspension was collected, and parenchymal cells were isolated from mononuclear cells by centrifugation at 50g for 5 minutes. The mononuclear cells were washed in PBS and suspended in 40% Percol (catalog 17089101; GE Healthcare Biosciences AB, Uppsala, Sweden). The suspended cells were gently overlaid onto 70% Percol (GE Healthcare Biosciences AB) and centrifuged at 750g for 20 minutes. The mononuclear cells were collected from the interface, washed twice in PBS, and used for flow cytometry.
Flow Cytometric Analysis
Prepared liver mononuclear cells were suspended in a solution of PBS, 0.3% w/v bovine serum albumin, and 0.1% w/v sodium azide. To avoid nonspecific bindings of antibodies to Fc receptors, the cells were pretreated with anti-CD16/32 antibody (clone 2.4G2, catalog 553141; BD Biosciences, Franklin Lakes, NJ) for 15 minutes. Then the cells were stained for 20 minutes at 4°C with the following antibodies: allophycocyanin-conjugated anti-mouse CD11b (clone M1/70, catalog 553312; BD Biosciences) and phycoerythrin-conjugated anti-mouse F4/80 (clone T45-2342, catalog 565410; BD Biosciences) for macrophages. To label dead cells, 7-amino-actinomycin D (catalog 559925; BD Biosciences) was used. These samples were subjected to flow cytometric analysis using a fluorescence-activated cell sorting Canto II system (BD Biosciences). Data were analyzed using FlowJo software (TreeStar, Ashland, OR). The number of CD11b+F4/80+ macrophages in a cell subset was determined by using the following calculation: total liver mononuclear cell number × corresponding cell subset proportion to the total cells.
Enzyme-Linked Immunosorbent Assay
The levels of serum CCL2 and secreted CCL2 protein from cell lines were measured by using MCP-1/CCL2 mouse uncoated enzyme-linked immunosorbent assay kit (catalog 88-7391-86; Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Cell Culture
Murine primary hepatocytes and NPCs were isolated from WT and TG mice by 2-step collagenase-pronase perfusion as previously described.39 Briefly, the murine liver was perfused with Liver Perfusion Medium (catalog 17701038; Thermo Fisher Scientific) for 5 minutes to remove blood. For digestion of murine liver, the tissue was perfused with 53% pronase solution and 0.27% collagenase solution for 1 and 5 minutes. All of the solutions were warmed to 37°C before use and perfused at a flow rate of 4 mL/min. The perfused murine liver was transferred into Dulbecco modified Eagle medium containing 10% fetal calf serum and minced to release the hepatocytes. The suspension was filtered through a cell strainer with a pore size of 70 μm (catalog 352350; Corning, NY) and was washed 3 times with centrifugation at 50g for 1 minute at 4°C. After collecting the supernatant, the cell pellet was resuspended in William’s medium E with 10% fetal calf serum. In a select experiment, the NPCs in the supernatant were pelleted at 400g for 5 minutes and subjected to Western blot and real-time RT-PCR analysis. Hepatocytes (25,000 cells/cm2) were seeded on type I collagen-coated microplate (catalog 4820-010; Iwaki Glass, Shizuoka, Japan) in William’s medium E with 10% fetal calf serum, 2 mmol/L L-glutamine, 10–7 mol/L insulin, 10–7 mol/L dexamethasone, 100 U/mL penicillin, and 100 U/mL streptomycin. Isolated hepatocytes with >90% viability, determined by trypan blue exclusion, were cultured overnight. To induce FoxM1 protein expression, primary hepatocytes isolated from WT and TG mice were treated with DOX for 24 hours. After treating with DOX for 24 hours, WST assay and caspase-3/7 activity were measured in primary hepatocytes of WT and TG mice by using Cell count Reagent SF (catalog 07553-44; Nacalai Tesque) and a luminescent substrate assay (Caspase-Glo assay, catalog G8093; Promega, Madison, WI) according to the manufacturer’s instructions. The murine nontransformed hepatocyte cell lines BNL-CL2 and AML12 were obtained from the American Type Culture Collection (Manassas, VA). For the small RNA interference assay, cells were transfected with either 20 nmol/L murine Foxm1 siRNA (catalog MSS204338; Thermo Fisher Scientific) or control siRNA (catalog 12935112; Thermo Fisher Scientific) by using Lipofectamine RNAiMAX (catalog 13778150; Invitrogen Corporation, Carlsbad, CA) according to the manufacturer’s instructions.
Dual Luciferase Assay
The promoter sequences for murine Ccl2 genes were obtained from the National Center for Biotechnology Information database. Murine Ccl2 promoter fragments were amplified by PCR of murine genomic DNA using the following S and AS primers: 5′-TCCGGCCCATGAGAGAACTGCTT-3′ (S1, –2468/–2445), 5′-ACTATGCCTGGCTCCTGGTA-3′ (S2, –1401/–1381), 5′-GAAGACTCCGCTCAGCCAC-3′ (S3, –1136/–1117), and 5′-TGGCTTCAGTGAGAGTTGGCTGGT-3′ (AS, +67/+44). The Ccl2 promoter nucleotide sequence was confirmed by DNA sequencing and cloned into a pGL3 basic luciferase vector (Promega) to generate a reporter plasmid. We transfected BNL-CL2 with either cytomegalovirus promoter-T7–tagged FoxM1 plasmid (CMV-T7-FoxM1) or cytomegalovirus-empty expression plasmid, as well as with luciferase reporters driven by the murine Ccl2 promoter lesions. After seeding cells and culturing them overnight, the plasmid was transfected with FuGENE HD transfection reagent (catalog E2311; Promega) according to the manufacturer’s instructions. We used the 6×CDX2-Luc plasmid, a known FoxM1 reporter, as a positive control for CMV-T7-FoxM1 transcriptional activity. The CMV-Renilla luciferase plasmid was used as internal controls to normalize transfection efficiency. A dual luciferase assay (Promega) was performed 48 hours after transfection, as described previously.38
Chromatin Immunoprecipitation Assay
BNL-CL2 cells were prepared by using the SimpleChIP Plus Enzymatic Chromatin IP kit (catalog 9005; Cell Signaling Technology). Nuclear extracts from BNL-CL2 cells were cross-linked by the addition of formaldehyde, sonicated, and used for immunoprecipitation with FoxM1 rabbit polyclonal antibodies (K-19 and C-20) (catalog sc-500, sc-502; Santa Cruz Biotechnology). Reverse cross-linked chromatin immunoprecipitation DNA samples were subjected to real-time PCR using the oligonucleotides specific to promoter regions of the murine Ccl2 gene: 5′-TCAGGTCCAGGGAAGCATTCTG-3′ (S) and 5′-TGTGTTTATTCCATGGCAAGTGGTC-3′ (AS).
Synthesis of the Triantennary N-Acetylgalactosamine–siRNA
GalNAc-siRNA conjugates were synthesized by Nihon Gene Research Laboratories Inc, as described previously.22 The target sequences for GalNAc-siCcl2 and GalNAc-siControl (control) were 5′-TGAATGAGTAGCAGCAGGTGAGTGG-3′ and 5′-GGTCTGATCACTGCTCGGT-3′.40 The designed triantennary GalNAc-siCcl2 and control are shown in Table 1. TG mice were injected subcutaneously with 3 mg/kg GalNAc-siCcl2 or control 24 hours before DOX treatment, as described previously.41
Table 1.
Designs of the GalNAc-siRNA Conjugates
| Compound | Strand | Sequence (5' to 3') |
|---|---|---|
| GalNAc-siCcl2 | S | UGAAUGAGUAGCAGCAGGUGAGUGG-(GalNAc) |
| AS | CCACUCACCUGCUGCUACUCAUUCATT | |
| GalNAc-siControl | S | GGUCUGAUCACUGCUCGGU-(GalNAc) |
| AS | ACCGAGCAGUGAUCAGACCTT |
Depletion of Macrophages
Clodronate liposomes were prepared as previously described.42 To deplete macrophages, the mice were injected intravenously with either 50 mg/kg of body weight clodronate liposomes or PBS liposomes from 3 days before the experiment. Macrophage depletion in the livers was verified by using F4/80 immunostaining 3 days after intravenous injection.
Induction of Chronic Liver Injury
To induce chronic liver injury, C57BL/6 mice were subjected to a HFHC diet (catalog D09100308; Research Diet, New Brunswick, NJ) for 8 weeks or intraperitoneal administration of CCl4 (0.5 mL/kg body weight) 2 times a week for 4 weeks. The mice were killed at 48 hours after final injection.
Inhibition of Forkhead Box M1 Transcription Factor Activity
To inhibit FoxM1 activity, we used thiostrepton (catalog T8902; Sigma-Aldrich), a FoxM1 inhibitor.25,26 In a chronic liver injury model, the mice were injected intraperitoneally with either 30 mg/kg body weight thiostrepton dissolved in 10% dimethyl sulfoxide or vehicle as a control for a total of 7 injections over 2 weeks, as described previously.43 TG mice were injected intraperitoneally with thiostrepton or vehicle for 2 times from 3 days before DOX treatment.
Clinical Samples
Tumor and nontumor liver tissues were obtained from patients who underwent surgical liver resection from primary liver tumors. This study was performed according to the ethical guidelines of the Declaration of Helsinki. Approval to use resected samples was obtained from the Institutional Review Board Committee at Osaka University Hospital (No. 13556), and written informed consent was obtained from all patients.
Statistical Analysis
Statistical analysis was performed by using JMP Pro 13.0 software (SAS Institute, Cary, NC). Individual values (symbols) and means (bar) ± standard deviations (error bars) from at least 3 independent experiments are shown. The Student t test was used to analyze the significant difference between the 2 groups, and one-way analysis of variance test was performed to analyze the difference among groups. P values less than .05 were considered to be statistically significant.
Acknowledgments
The authors thank Dr Pradip Raychaudhuri (University of Illinois at Chicago) for providing CMV-T7-FoxM1 and 6×CDX2-Luc plasmids.
Footnotes
Author contributions Study concept and design: T. Kurahashi, Y. Yoshida, S. Ogura, and T. Takehara. Acquisition of data: T. Kurahashi, S. Ogura, M. Egawa, and K. Furuta. Analysis and interpretation of data: T. Kurahashi, Y. Yoshida, S. Ogura, M. Egawa, K. Furuta, H. Hikita, T. Kodama, R. Sakamori, S. Kiso, Y. Kamada, I. C. Wang, H. Eguchi, E. Morii, Y. Doki, M. Mori, V. V. Kalinichenko, T. Tatsumi, and T. Takehara. Writing, review, or revision of the manuscript: T. Kurahashi, Y. Yoshida, and T. Takehara.
Conflicts of interest The authors disclose no conflicts.
Funding Supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Contributor Information
Yuichi Yoshida, Email: yyoshida@gh.med.osaka-u.ac.jp.
Tetsuo Takehara, Email: takehara@gh.med.osaka-u.ac.jp.
References
- 1.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273 e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He G., Karin M. NF-kappaB and STAT3: key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi K., Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 5.Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 6.Costa R.H. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7:108–110. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 7.Kalin T.V., Ustiyan V., Kalinichenko V.V. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilarsky C., Wenzig M., Specht T., Saeger H.D., Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6:744–750. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim I.M., Ackerson T., Ramakrishna S., Tretiakova M., Wang I.C., Kalin T.V., Major M.L., Gusarova G.A., Yoder H.M., Costa R.H., Kalinichenko V.V. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 10.Kalin T.V., Wang I.C., Ackerson T.J., Major M.L., Detrisac C.J., Kalinichenko V.V., Lyubimov A., Costa R.H. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida Y., Wang I.C., Yoder H.M., Davidson N.O., Costa R.H. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Park H.J., Carr J.R., Chen Y.J., Zheng Y., Li J., Tyner A.L., Costa R.H., Bagchi S., Raychaudhuri P. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 2011;71:4292–4302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalinichenko V.V., Major M.L., Wang X., Petrovic V., Kuechle J., Yoder H.M., Dennewitz M.B., Shin B., Datta A., Raychaudhuri P., Costa R.H. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gusarova G.A., Wang I.C., Major M.L., Kalinichenko V.V., Ackerson T., Petrovic V., Costa R.H. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egawa M., Yoshida Y., Ogura S., Kurahashi T., Kizu T., Furuta K., Kamada Y., Chatani N., Hamano M., Kiso S., Hikita H., Tatsumi T., Eguchi H., Nagano H., Doki Y., Mori M., Takehara T. Increased expression of Forkhead box M1 transcription factor is associated with clinicopathological features and confers a poor prognosis in human hepatocellular carcinoma. Hepatology Research. 2017;47:1196–1205. doi: 10.1111/hepr.12854. [DOI] [PubMed] [Google Scholar]
- 16.Ogura S., Yoshida Y., Kurahashi T., Egawa M., Furuta K., Kiso S., Kamada Y., Hikita H., Eguchi H., Ogita H., Doki Y., Mori M., Tatsumi T., Takehara T. Targeting the mevalonate pathway is a novel therapeutic approach to inhibit oncogenic FoxM1 transcription factor in human hepatocellular carcinoma. Oncotarget. 2018;9:21022–21035. doi: 10.18632/oncotarget.24781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia L., Huang W., Tian D., Zhu H., Zhang Y., Hu H., Fan D., Nie Y., Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Ye H., Kelly T.F., Samadani U., Lim L., Rubio S., Overdier D.G., Roebuck K.A., Costa R.H. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Kiyokawa H., Dennewitz M.B., Costa R.H. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Hung N.J., Costa R.H. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33:1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 21.Ren X., Shah T.A., Ustiyan V., Zhang Y., Shinn J., Chen G., Whitsett J.A., Kalin T.V., Kalinichenko V.V. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol Cell Biol. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel'in A.V., Milstein S., Taneja N., O'Shea J., Shaikh S., Zhang L., van der Sluis R.J., Jung M.E., Akinc A., Hutabarat R., Kuchimanchi S., Fitzgerald K., Zimmermann T., van Berkel T.J., Maier M.A., Rajeev K.G., Manoharan M. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 23.Capurro M., Wanless I.R., Sherman M., Deboer G., Shi W., Miyoshi E., Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 24.Timek D.T., Shi J., Liu H., Lin F. Arginase-1, HepPar-1, and Glypican-3 are the most effective panel of markers in distinguishing hepatocellular carcinoma from metastatic tumor on fine-needle aspiration specimens. Am J Clin Pathol. 2012;138:203–210. doi: 10.1309/AJCPK1ZC9WNHCCMU. [DOI] [PubMed] [Google Scholar]
- 25.Bhat U.G., Halasi M., Gartel A.L. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PloS One. 2009;4 doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde N.S., Sanders D.A., Rodriguez R., Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nature Chemistry. 2011;3:725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 27.Elsharkawy A.M., Mann D.A. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 28.Luedde T., Schwabe R.F. NF-kappaB in the liver: linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raychaudhuri P., Park H.J. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penke L.R., Speth J.M., Dommeti V.L., White E.S., Bergin I.L., Peters-Golden M. FOXM1 is a critical driver of lung fibroblast activation and fibrogenesis. J Clin Invest. 2018;128:2389–2405. doi: 10.1172/JCI87631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y., Wang L., Zeng J., Shen L., Liang X., Yu H., Liu S., Liu Z., Sun Y., Li W., Chen C., Jia J. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol Cancer Res. 2013;11:834–844. doi: 10.1158/1541-7786.MCR-13-0007. [DOI] [PubMed] [Google Scholar]
- 32.Marra F., Tacke F. Roles for chemokines in liver disease. Gastroenterology. 2014;147:577–594.e1. doi: 10.1053/j.gastro.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 33.Li X., Yao W., Yuan Y., Chen P., Li B., Li J., Chu R., Song H., Xie D., Jiang X., Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 34.Teng K.Y., Han J., Zhang X., Hsu S.H., He S., Wani N.A., Barajas J.M., Snyder L.A., Frankel W.L., Caligiuri M.A., Jacob S.T., Yu J., Ghoshal K. Blocking the CCL2-CCR2 axis using CCL2-neutralizing antibody is an effective therapy for hepatocellular cancer in a mouse model. Mol Cancer Ther. 2017;16:312–322. doi: 10.1158/1535-7163.MCT-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X., Zhang Y., Snyder J., Cross E.R., Shah T.A., Kalin T.V., Kalinichenko V.V. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L., Ren X., Wang I.C., Pradhan A., Zhang Y., Flood H.M., Han B., Whitsett J.A., Kalin T.V., Kalinichenko V.V. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Science Signaling. 2017;10(475) doi: 10.1126/scisignal.aai8583. pii: eaai8583. [DOI] [PubMed] [Google Scholar]
- 37.Wang I.C., Zhang Y., Snyder J., Sutherland M.J., Burhans M.S., Shannon J.M., Park H.J., Whitsett J.A., Kalinichenko V.V. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev Biol. 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balli D., Ustiyan V., Zhang Y., Wang I.C., Masino A.J., Ren X., Whitsett J.A., Kalinichenko V.V., Kalin T.V. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32:231–244. doi: 10.1038/emboj.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furuta K., Yoshida Y., Ogura S., Kurahashi T., Kizu T., Maeda S., Egawa M., Chatani N., Nishida K., Nakaoka Y., Kiso S., Kamada Y., Takehara T. Gab1 adaptor protein acts as a gatekeeper to balance hepatocyte death and proliferation during acetaminophen-induced liver injury in mice. Hepatology. 2016;63:1340–1355. doi: 10.1002/hep.28410. [DOI] [PubMed] [Google Scholar]
- 40.Taniuchi K., Cerny R.L., Tanouchi A., Kohno K., Kotani N., Honke K., Saibara T., Hollingsworth M.A. Overexpression of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell growth. Oncogene. 2011;30:4843–4854. doi: 10.1038/onc.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willoughby J.L.S., Chan A., Sehgal A., Butler J.S., Nair J.K., Racie T., Shulga-Morskaya S., Nguyen T., Qian K., Yucius K., Charisse K., van Berkel T.J.C., Manoharan M., Rajeev K.G., Maier M.A., Jadhav V., Zimmermann T.S. Evaluation of GalNAc-siRNA conjugate activity in pre-clinical animal models with reduced asialoglycoprotein receptor expression. Mol Ther. 2018;26:105–114. doi: 10.1016/j.ymthe.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura K., Yang L., van Rooijen N., Ohnishi H., Seki E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1310–G1321. doi: 10.1152/ajpgi.00365.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiler S.M.E., Pinna F., Wolf T., Lutz T., Geldiyev A., Sticht C., Knaub M., Thomann S., Bissinger M., Wan S., Rossler S., Becker D., Gretz N., Lang H., Bergmann F., Ustiyan V., Kalin T.V., Singer S., Lee J.S., Marquardt J.U., Schirmacher P., Kalinichenko V.V., Breuhahn K. Induction of chromosome instability by activation of yes-associated protein and Forkhead Box M1 in liver Cancer. Gastroenterology. 2017;152:2037–2051.e22. doi: 10.1053/j.gastro.2017.02.018. [DOI] [PubMed] [Google Scholar]