Abstract
The emergence of large gene expression datasets has revealed the need for improved tools to identify enriched gene categories and visualize enrichment patterns. While gene ontogeny (GO) provides a valuable tool for gene set enrichment analysis, it has several limitations. First, it is difficult to graph multiple GO analyses for comparison. Second, genes from some model systems are not well represented. For example, ∼30% of Caenorhabditis elegans genes are missing from the analysis in commonly used databases. To allow categorization and visualization of enriched C. elegans gene sets in different types of genome-scale data, we developed WormCat, a web-based tool that uses a near-complete annotation of the C. elegans genome to identify coexpressed gene sets and scaled heat map for enrichment visualization. We tested the performance of WormCat using a variety of published transcriptomic datasets, and show that it reproduces major categories identified by GO. Importantly, we also found previously unidentified categories that are informative for interpreting phenotypes or predicting biological function. For example, we analyzed published RNA-seq data from C. elegans treated with combinations of lifespan-extending drugs, where one combination paradoxically shortened lifespan. Using WormCat, we identified sterol metabolism as a category that was not enriched in the single or double combinations, but emerged in a triple combination along with the lifespan shortening. Thus, WormCat identified a gene set with potential. phenotypic relevance not found with previous GO analysis. In conclusion, WormCat provides a powerful tool for the analysis and visualization of gene set enrichment in different types of C. elegans datasets.
Keywords: C. elegans, gene set enrichment analysis, RNA sequencing visualization
RNA-SEQ is an indispensable tool for understanding how gene expression changes during development or upon environmental perturbations. As this technology has become less expensive and more robust, it has become more common to generate data from multiple conditions, enabling comparisons of gene expression profiles across biological contexts. The most commonly used method to derive information on the biological function of coexpressed genes is gene ontology (GO) (The Gene Ontology Consortium 2019) (Ashburner et al. 2000), where annotation for each gene follows three major classifications: Biological Process, Molecular Function, or Cellular Component. For example, the Biological Process class refers to genes included in a process that an organism is programmed to execute, and that occurs through specific regulated molecular events. Molecular Function denotes protein activities, and Cellular Component maps the location of activity. Within each of these classifications, functions are broken down in parent–child relationships with increasing functional specificity (Figure 1A). However, child classes can be linked to different parent classes, making statistical analysis not straightforward. For example, the child class phospholipid biosynthetic process can be linked to both of the parent groupings metabolic process and cellular process. Thus, GO provides multiple descriptors per gene. Although GO was developed to compare gene function across newly sequenced genomes, it became apparent that it could also be used to identify shared functional classifications within large-scale gene expression data (Eisen et al. 1998; Spellman et al. 1998). Currently, multiple web-based servers that use different statistical tests can be used to determine the enrichment of GO terms for a gene set of interest. For example, PANTHER (www.pantherdb.org) provides enriched GO terms determined by Fisher’s Exact Test with a Benjamini-Hochberg false discovery rate (FDR) correction for 131 species (Mi et al. 2019). Because the multiplicity of GO term parent–child relationships can produce complex data structures, specialized ontologies such as GO-Slim use a restricted set of terms, searching biological processes as default (Mi et al. 2019). P-values provide relevance for enriched GO terms. Visualization of gene set enrichment data are important for identifying critical elements and communication of information. PANTHER provides pie or bar charts of individual searches (Mi et al. 2019). The GOrilla platform generates tables of P-values (Eden et al. 2009) and links to another service, REVIGO, that uses semantic graphs to visualize GO terms data (Supek et al. 2011). Thus, the GO databases provide a widely used platform for classifying, comparing, and visualizing functional genomic data. However, as outlined below, GO is of limited use for the analysis of Caenorhabditis elegans data and visualization of multiplexed datasets.
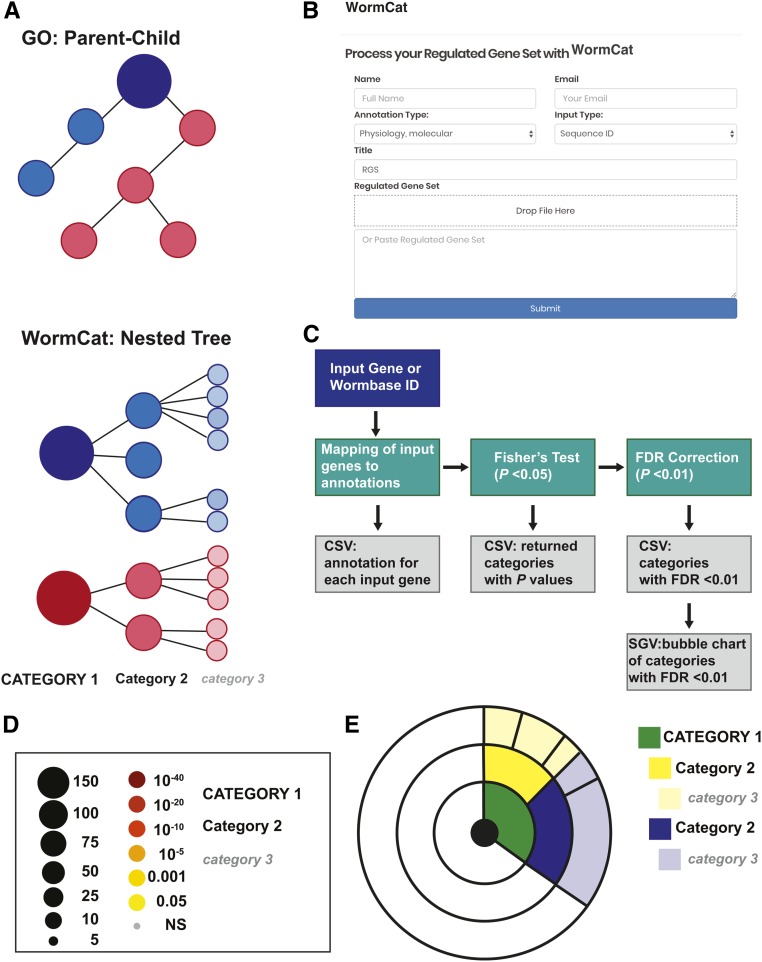
Figure 1.
WormCat annotates and visualizes C. elegans gene enrichment from genome-scale data. (A) Diagram comparing the parent–child methods for linking GO terms with the nested tree strategy used for annotating C. elegans genes in WormCat. (B) Screenshot of the WormCat web page showing the data entry form. (C) Flow chart diagraming steps and outputs from the WormCat program. Data outputs are in tabular comma-separated values (CSV) and scalable vector graphics (SVG) formats. (D) Legend for scaled bubble charts showing the number of genes referenced to size and P-value referenced to color. In graphs, Category 1, 2, and 3 are differentiated by capitalization, size, and italics. (E) Legend for sunburst plots showing concentric rings visualizing Category 1, 2, and 3 data.
The nematode C. elegans has been at the forefront of genomics research. It was the first metazoan organism with a completely sequenced genome (Caenorhabditis elegans Sequencing Consortium 1998). After the discovery of RNA interference (RNAi) (Fire et al. 1998), multiple RNAi libraries were developed for performing genome-wide knockdown screens (Kamath et al. 2003; Rual et al. 2004). Gene expression profiling studies using microarrays or RNA-seq have compared gene expression in sex-specific, developmental/aging-related, specific gene deletion, tissue-specific, and dietary or stress-related animal conditions (Reinke et al. 2000; Hillier et al. 2005; Baugh et al. 2009; Oliveira et al. 2009; Deng et al. 2011; Schwarz et al. 2012; Bulcha et al. 2019). While GO has been used extensively to analyze C. elegans gene expression profiling data, it has several limitations. First, ∼30% of C. elegans genes are not annotated in GO databases (Ding et al. 2018), excluding these genes from the analysis. Thus, these genes are arbitrarily excluded from enrichment statistics. Second, the visualization of enrichment data from comparative RNA-seq datasets is difficult, and this is true not only for C. elegans datasets but for gene expression profile comparisons in any organism. Most users display the output data as lists with P-values (MacNeil et al. 2013) or as pie or bar charts (Ding et al. 2015), which are challenging to multiplex for comparison of multiple datasets. Finally, it can be challenging to determine which input genes are associated with a given GO classification, which is critical for interpreting the accuracy and biological importance of enriched gene sets.
We constructed a web-based gene set enrichment analysis tool we named WormCat (WormCatalog) that works independently from GO to identify potentially coexpressed or cofunctioning genes in genome-wide expression studies or functional screens. WormCat (www.wormcat.com), uses a concise list of nested categories where each gene is first assigned to a category based on physiological function, and then to a molecular function or cellular location. WormCat provides a scaled bubble chart that allows the visualization and direct comparison of complex datasets. The tool also provides csv files containing input gene annotations, P-values from Fisher’s exact tests, and Bonferroni multiple hypothesis testing corrections. We used WormCat to identify functional gene sets in published gene expression data and large-scale RNAi screens. WormCat reproducibly identified prior GO classifications, and provided an easy way to interpret visualization that enables the facile and intuitive comparison of multiple published datasets. We also identified new groups of enriched categories with potentially important biological significance, showing that WormCat provides enrichment information not revealed by GO. Taken together, WormCat offers an alternative and complementary tool for categorizing and visualizing data for genome-wide C. elegans studies, and may provide a platform for similar annotations in other model organisms and humans.
Materials and Methods
Annotations
WormBase version WS270 was used to provide WormBase descriptions and provide phenotype information.
Scripts
The processed data were analyzed using R version 3.4.4 (2018-03-15), and depends on the following R packages: datasets, graphics, grDevices, methods, stats, utils, ggplot2, plot flow, scales, ggthemes, pander, data.table, plyr, gdtools, svglite, and FSA.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The code and annotation lists are available under MIT Open Source License, and can be downloaded from the GitHub repository https://github.com/dphiggs01/wormcat along with version-control information. Alternatively, WormCat can be installed directly as an R package using the devtools library. Supplemental material has been deposited at figshare and includes 12 supplemental figures and 14 supplemental tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10312070.
GO searches:
Genes lists were entered as test sets into GOrilla (http://cbl-gorilla.cs.technion.ac.il/) (Eden et al. 2009) with the WormCat annotation list used as background so that the same background set was used when comparing WormCat and GOrilla. “All” was selected for ontogeny choices, and the P-value thresholds were set to 10−3. Output selections were Microsoft Excel and REVIGO (Supek et al. 2011).
Results
C. elegans gene annotation
The C. elegans genome encodes ∼19,800 protein-coding genes, ∼260 microRNAs, and numerous other noncoding RNAs (WormBase version WS270). We annotated all C. elegans genes first based on physiological functions, and, when these functions were unknown or pleiotropic, according to molecular function or subcellular location (see Supplemental Material, Table S1 for annotations, Table S2 for Category definitions). Our annotations are structured as nested categories, enabling classification into broad (Category 1; Cat1), or more specific categories (Category 2 or 3; Cat2 or Cat3). This annotation has the advantage of including information from multiple sources in addition to GO. For example, we used phenotype information available in WormBase (Lee et al. 2018) for Cat1 assignments. Importantly, the phenotypic data present in WormBase (Lee et al. 2018) was used only if phenotypes were: (1) derived from wild type animals, (2) examined in detail in peer-reviewed publications, and (3) represented in two independent screens. If a gene was ascribed a clear physiological function with these criteria, we assigned it to a physiological category, examples of which include Stress response, Development, and Neuronal function. If gene products have multiple functions within the cell, act in multiple cells type, or different developmental times, we prioritized assignment to molecular categories. Molecular categories harbor both genes whose products comprise molecular machines, as well as the chaperones or regulatory factors that are necessary for the function of such machines. We used information on the molecular function of human orthologs to classify C. elegans genes that had not been molecularly defined in nematodes, and were highly similar in BLAST scores. For example, we classified the C. elegans gene W03D8.8 in Metabolism: lipid: beta-oxidation based on a BLAST score of e = 7 × 10−37 and similarity over 92% of its length to human ACOT4 (acyl-CoA thioesterase 4). For genes with weaker homology to human genes, we further refined assignments using BLAST (Altschul et al. 1990) and the NCBI Conserved Domain server (Marchler-Bauer et al. 2017). We used these tools to determine if there was significant homology or shared domains between C. elegans and human proteins, then used information in UniProt (www.uniprot.org) for the human proteins to determine molecular classification. For example, we placed the C. elegans gene T26E4.3 in Protein modification: carbohydrate-based on a BLAST core of e = 4 × 10−7 over 95% of its length to human alpha fucosyltransferase 1, and identification of a Fut1_Fut2-like domain by the NCBI conserved domain server with an e score of 6.16 × 10−36. However, while the gene BE10.3 is referred to in the WormBase description as an ortholog of human FUT9 (fucosyltransferase (9) (Table S1), we found no homology to human genes by NCBI BLAST or domain conservation across all organisms with the NCBI Conserved Domain server. Therefore, we classified BE10.3 in Unknown. Finally, if no biological or molecular function could be assigned, protein subcellular localization was used for annotation. For example, a protein with a predicted membrane-spanning region that lacks characterization as a receptor would be placed in Transmembrane protein. Genes with no functional information were classified as Unknown (Cat1). A total of 8160 genes lacked sufficient information for classification in physiological, molecular, or subcellular localization categories, and were classified in Unknown. Many of these genes are C. elegans- or nematode-specific; however, some have homology to human genes of unknown function. WormBase also aggregates microarray and RNA-seq information, and annotates genes that respond to pharmacological treatments (Lee et al. 2018). We also used this information to differentiate genes within Unknown: regulated by multiple stresses that respond to at least two commonly used stressors. This classification does not imply that these genes have a function in the stress response. It does allow identification of genes with otherwise unknown functions that are common responders to stress. This classification may be useful to distinguish RNA-seq datasets that respond similarly to pharmacological stressors or can serve as a source to identify specific genes of interest for additional study. We also included pseudogenes and noncoding RNAs in our annotation list. These genes commonly appear in RNA-seq data; including them in the annotation list allows them to be labeled within the user’s input dataset. In this way, we were able to leverage multiple data sources to categorize C. elegans genes into potentially functional biological groups.
WormCat.com allows web-based searches of input genes and generates scaled bubble charts and gene lists
WormCat.com maps annotations to input genes then determine category enrichment for Cat1, Cat2, and Cat3 (Figure 1B). Determination of category enrichment in a gene set of interest compared to the entire genome can rely on several commonly used statistics such as the Fisher’s exact test and the Mann-Whitney test (Mi et al. 2019). We used Fisher’s exact test to determine if categories were over-represented because it is accurate down to small sample sizes, which may occur in high-resolution classifications (McDonald 2014). In addition, we included the Bonferroni FDR correction (McDonald 2014). To determine the number of false positives after Fisher’s test or the FDR correction, we tested randomized gene lists of 100, 500, 1000, or 1500 genes and found that small numbers of genes were returned using a P-value cut-off of 0.05 (for, example 5 genes were returned on the 1000 gene random set). Few genes were returned from any of the randomized sets using an FDR cutoff of 0.01 (Table S3). Because an FDR <0.01 is relatively stringent, Fisher’s exact test P-values will also be provided, allowing users to make independent evaluations on the statistical cut-offs.
The WormCat website (www.wormcat.com) provides gene enrichment outputs in multiple formats (Figure 1C). First, all input genes are listed with mapped annotations (rgs_and_categories.csv). Genes that matched at least one Cat1, Cat2, and Cat3 classification are returned with Fisher’s exact test P-values (Cat1.csv, Cat2.csv, or Cat3.csv). Next, Cat1, Cat2, and Cat3 matches with an FDR correction of <0.01 are returned as CSV files named Cat1.apv, Cat2.apv, and Cat3.apv (appropriate P-value). Finally, the Cat.apv files are used to generate two types of graphical output. First, it constructs scaled heat map bubble charts (Cat1., Cat2., Cat3.sgv) where color signifies P-value, and size specifies the number of genes in the category (Figure 1D). The scaling for these graphs is fixed so that multiple datasets can be graphed together. Second, a sunburst graph is built with concentric rings of Cat1, Cat2, and Cat3 values (Figure 1E). In these graphs, rings sections correspond to categories, with section sizes proportional to numbers of genes in the category. On the website, each ring section is clickable to generate a subgraph-based division within a section. For example, clicking a single Cat1 section would generate a subgraph with all the Cat2 and Cat3 subdivisions located within. This graphical output is likely to be most useful for visualization of a single RNA-seq dataset, or genetic screening data. Thus, WormCat provides multiple outputs to allow inspection of individual input genes, generation of gene tables, and P-values, and graphical visualization of enrichments.
Comparison of GO and WormCat analysis of sams-1(RNAi) enrichment data
To determine the utility of the WormCat annotations, we first analyzed microarray data we had previously generated to compare gene expression changes after knockdown of sams-1, with and without dietary supplementation of choline (Ding et al. 2015). sams-1 encodes an S-adenosylmethionine (SAM) synthase, which is an enzyme that produces nearly all of the methyl groups used in methylation of histones and nucleic acids, in addition to the production of the membrane phospholipid phosphatidylcholine (PC) (Mato and Lu 2007). sams-1 RNAi or loss-of-function (lof) animals have extended lifespan (Hansen et al. 2005), increased lipid stores (Walker et al. 2011), and activated innate immune signatures (Ding et al. 2015). sams-1 animals have low PC (Walker et al. 2011), but those levels are restored with supplementation of choline (Ding et al. 2015), which supports SAM-independent phosphatidylcholine synthesis (Vance 2014) (Figure 2A). Gene expression changes in sams-1(RNAi) animals could result from a perturbation in different SAM-dependent pathways. To determine which transcriptional changes occurred downstream of alterations in PC synthesis, we performed microarrays with RNA from sams-1(RNAi) and sams-1(RNAi) animals supplemented with choline; 90% of genes that changed in expression in sams-1(RNAi) animals returned to wild-type levels after choline supplementation. Therefore, the expression of the remaining 10% of genes was altered by sams-1 RNAi independently of phosphatidylcholine levels (Ding et al. 2015).
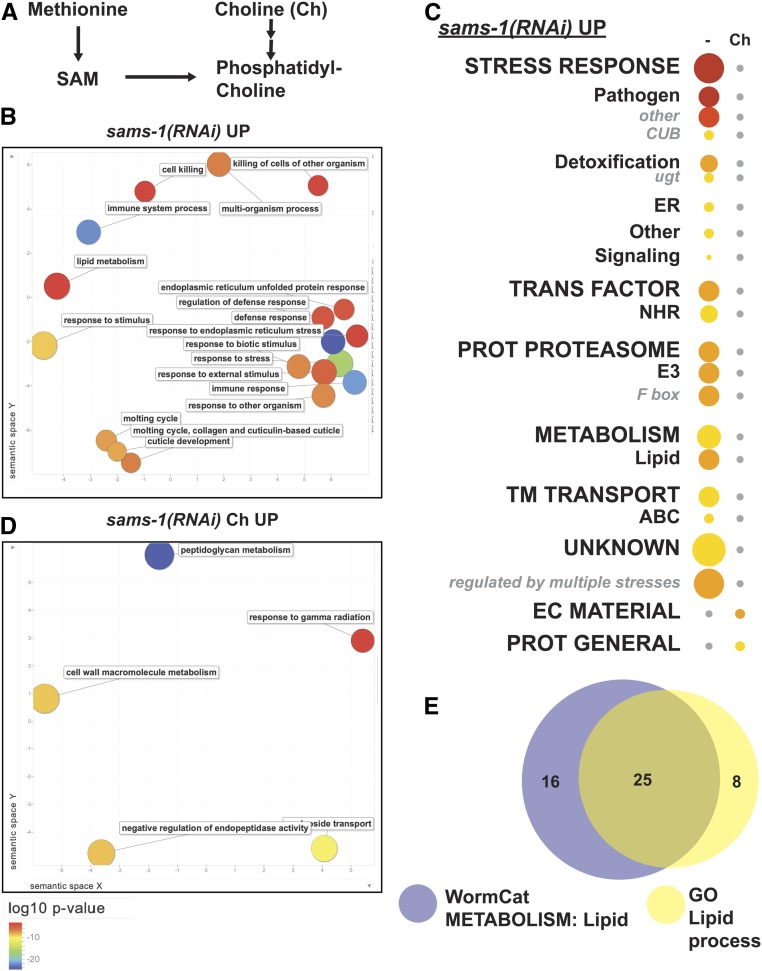
Figure 2.
WormCat verifies known category enrichments sams-1(RNAi) upregulated genes. (A) Schematic showing metabolic pathways linking methionine, SAM, choline, and phosphatidylcholine production. Gene expression microarray data for (B–D) were obtained from Ding et al. (2015). (B) Semantic plot of GO enriched classifications generated by REVIGO (Supek et al. 2011) from sams-1(RNAi) Up genes. (C) WormCat visualization of categories enriched in genes upregulated in sams-1(RNAi) animals with and without choline supplementation in order of Cat1 strongest enrichment. Categories 2 and 3 are listed under each Category 1, with Category 2 or 3 sets that appeared independently of a Category 1 listed last. Bubble heat plot key is the same as Figure 1D. (D) sams-1(RNAi) Up plus choline (Ch) genes visualized by REVIGO. (E) Venn diagram showing overlap between WormCat Metabolism: lipid and GO Lipid process gene annotations. ABC, ATP-Binding Cassette; Ch, Choline; CUB, Complement C1r/C1s, Uegf, Bmp1 domain; EC Material, Extracellular Material; NHR, Nuclear Hormone Receptor; Prot General, Proteolysis General; Prot Proteasome, Proteolysis Proteasome; SAM, S-adenosylmethionine; TM Transport, Transmembrane Transport; ugt, UDP-glycosyltransferase
In order to identify GO terms enrichment with WormCat, we submitted genes up- or downregulated twofold or more in sams-1(RNAi) animals to both WormCat and GOrilla (Eden et al. 2009). We used REVIGO (Supek et al. 2011) to visualize GO output. Both GOrilla/REVIGO (Figure 2B, Figure S2, A and B, and Table S4) and WormCat (Figure 2C and Table S5) identified categories of stress-response and metabolism linked to lipid accumulation in the genes that are upregulated upon sams-1 RNAi, which is in agreement with our previous analysis (Ding et al. 2015). Interestingly, the relative importance of lipid metabolism is different in the two analyses. In the WormCat analysis, Metabolism: lipid was the third most enriched Cat2 category with a P-value of 1.2 × 10−9 (Table S5). In the GO analysis, however, lipid metabolic process was found with a modest enrichment of FDR corrected P-value = 5 × 10−2 (Table S4). WormCat identified 41 genes in the Metabolism: lipid category, whereas GOrilla’s GO term search identified 33 genes in lipid metabolic process (Figure 2E and Table S4). Further inspection showed that six of the genes identified by solely by GOrilla were phospholipid lipases or phosphatases, one was an undefined hydrolase with no domain similarity to genes with known lipid functions, and one was a transmembrane protein. Each of these genes may be better classified in other categories (see Table S4 for GO lipid genes annotated by WormCat, tab 5 “GO_lipid_sams_up”). For example, lipases that hydrolyze phospholipids are the endpoints of metabolic pathways but produce second messengers acting in signaling pathways. One of these genes, Y69A2AL.2, has significant similarity to the human phospholipase A2 gene, PLA2G1B (BLAST e score of 2 × 10−11). This class of phospholipases cleave 3-sn-phosphoglycerides to produce the signaling molecule arachidonic acid (Xu et al. 2009); therefore, a classification of Signaling is likely more reflective of its biological function than Metabolism: lipid. Taken together, WormCat identifies more genes that are directly relevant to the increased lipid storage phenotype observed with sams-1(RNAi) or (lof) animals (Walker et al. 2011; Smulan et al. 2016).
Next, we compared WormCat analysis of sams-1(RNAi) upregulated genes to the Gene Set Enrichment Analysis (GSEA) tool located in the WormBase suite (Angeles-Albores et al. 2016). GSEA, a GO-based tool, identified similar categories as GOrilla, with a concurrently high score for the lipid catabolic process (Figure S1). Our test set included 773 genes (Table S5, tab4); however, 286 of these genes were excluded from the GSEA analysis (Table S6), similar to the percentage excluded in a GOrilla analysis (Ding et al. 2018). Unlike GOrilla, GSEA provides the user with gene IDs of excluded genes (Table S6). Therefore, we asked if these genes were excluded because their functions were undefined, or if they were instead capable of classification. We found that 118 of the 286 excluded genes were classified as Unknown by WormCat (Table S6). However, 92 of the 476 genes GSEA included were also Unknown in WormCat analysis (Table S5, tab 4). Thus, genes within this set that are classified as Unknown by WormCat only partially overlap with genes excluded from GO analysis. Furthermore, WormCat classified 117 genes within the 286 genes excluded from GSEA, with 16 in noncoding categories and the remaining 101 in protein-coding categories such as Cytoskeleton, Metabolism, and Proteolysis: proteasome (Table S6). Thus, analysis of genes excluded from GO shows that an important fraction can be annotated and that Unknown WormCat categories are represented in both genes included and excluded from GO analysis.
Next, we used WormCat to analyze genes downregulated in sams-1(RNAi) animals. We noted enrichment in Development: germline and mRNA function categories in sams-1(RNAi) animals, and that this enrichment is lost with choline treatment (Figure S2D and Table S5). This is consistent with the reduction in embryo production after sams-1(RNAi), and the rescue of fertility when choline supplementation restores PC levels (Walker et al. 2011; Ding et al. 2015). Stress response categories, however, are enriched in downregulated genes from both sams-1(RNAi) and sams-1(RNAi) choline-treated animals (Figure S2C and Table S5). This appears to contrast with the complete loss of enrichment after choline treatment in the upregulated stress-response genes (Figure 2C and Table S5). However, an inspection of the annotated gene lists returned by WormCat shows that the individual genes within the downregulated Stress response category are different (Figure S2E and Table S5). Thus, on a gene by gene level, this data shows that the effects of choline supplementation are distinct for the up- and downregulated genes in the Stress response category. In addition, this demonstrates that, by providing both gene set enrichment and annotation of individual genes, WormCat provides a level of analysis that is difficult to achieve by traditional GO methods.
Tau-tubulin kinases family are enriched in spermatogenic germlines
C. elegans is a robust model system for studying development and differentiation. The study of hermaphrodite germline development has been of particular interest, as it first produces sperm, after which it switches to oocyte production (Hubbard and Greenstein 2005). This concurs with distinct gene expression programs for both processes (Greenstein 2005; L’hernault 2006). Recently, the Kimble laboratory performed RNA-seq on dissected germlines from genetically female [fog-2(q71)] and genetically male [fem-3(q96)] animals (Ortiz et al. 2014) (Figure 3A). Genes expressed in both germlines were called gender-neutral (GN), in contrast to genes that are specific to female (Oo, oogenic) or male (Sp, spermatogenic) germlines (Ortiz et al. 2014). We used WormCat to determine enrichment categories in each dataset. We found that GN genes are strongly enriched for growth, DNA, transcription, and mRNA functions (Figure 3B and Table S7), which is expected because the germline is undergoing extensive mitotic and meiotic divisions. We further found that Chromosome dynamics and Meiotic functions were enriched in the GN dataset (Figure 3C and Table S7), as were mRNA functions of Processing and Binding (Figure 3D and Table S7). Oo genes were enriched for mRNA binding proteins, especially the zinc finger (ZF) class (Figure 3D and Table S7). These include such as maternally deposited oma-1, pie-1, pos-1, and mex-1, mex-5, and mex-6 mRNAs, which are known to function in oocytes (Lee and Schedl 2006) (Table S7). ZF proteins with unknown nucleic acid binding specificity were also enriched in the Oo dataset (Figure 3D and Table S7), suggesting that many of these may also be produced in the maternal germline. In an independent dataset comparing RNA from germline-less [glp-4(bn2)], oocyte [fem-3(gof)] and sperm-producing [fem-1(lof)] animals by microarray analysis (Reinke et al. 2000), we also observed enrichment in categories for mRNA functions, transcription, development, and cell cycle control (Figure S3, A–D and Table S8).
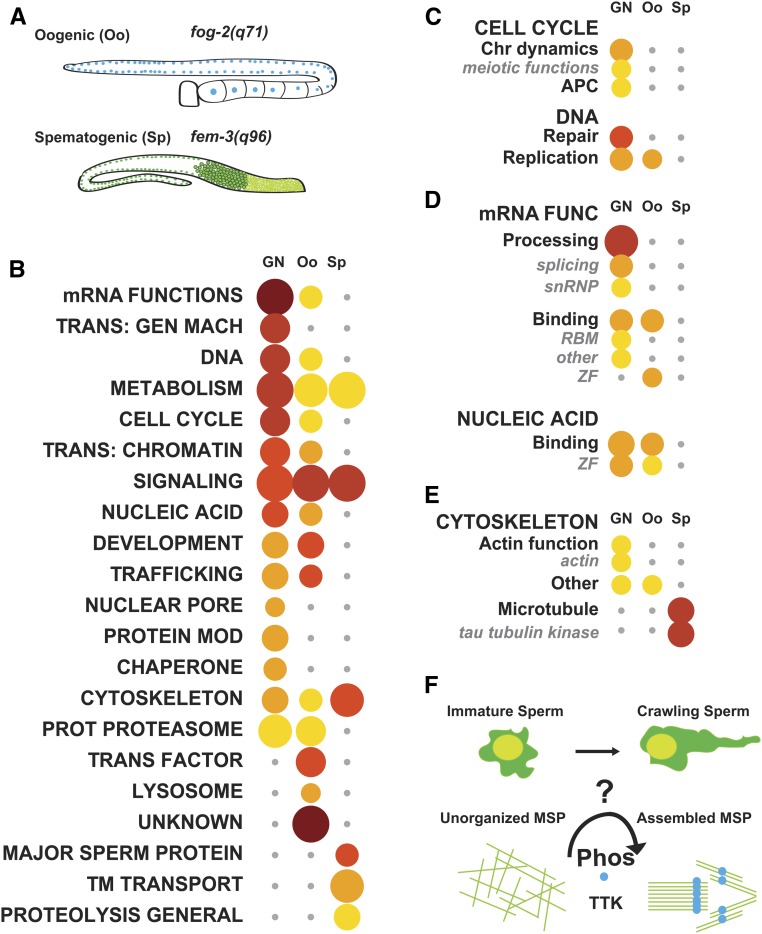
Figure 3.
Analysis of germline-specific RNA-seq data identifies the tau tubulin kinase family as a male-specific category. (A) Schematic showing germlines used for female (top) or male (bottom)-specific RNA-seq analysis from Ortiz et al. (2014) and the mutant alleles to cause these phenotypes. (B) WormCat Category 1 analysis of Germline neutral (GN), Oogenic (Oo), or Spermatogenic (Sp) datasets ordered by most enriched in GN data. (C–E) Breakdown of WormCat enrichment from the Category 1 level for Cell Cycle (C), mRNA Functions and Nucleic Acid (D), and Cytoskeleton (E). Bubble heat plot key is the same as Figure 1D. (F) Schematic showing predicted phosphorylation and organization of MSPs during C. elegans sperm maturation based on WormCat findings. APC, Anaphase Promoting Complex; Chr Dynamics, Chromosome Dynamics; mRNA Func., mRNA Function; MSP, Major Sperm Protein; Phos, Phosphorylation; Protein Mod, Protein Modification; Prot Proteasome, Proteolysis Proteasome; RBM, RNA Binding Motif; TTK, Tau Tubulin Kinase; TM Transport, Transmembrane Transport; Trans: Gen Mach, Trans: Chromatin, Transcription: Chromatin; Transcription: General Machinery; Trans Factor, Transcription Factor; ZF, Zinc Finger
As expected, Sp genes are enriched for Major Sperm Proteins (MSPs), which are necessary for sperm crawling (Figure 3B and Table S7). Interestingly, a class of potential cytoskeletal regulators, tau-tubulin kinases (TTKs), were also enriched in Sp genes (64 of 71, P-value of 8.8 × 10−34) (Figure 3E and Table S7). One TTK, spe-6, was previously isolated in a screen for spermatogenesis defects, and is thought to be involved in phosphorylation of MSPs to allow the sperm to crawl (Varkey et al. 1993). Underscoring the potential importance of the TTKs in the male germline, WormCat also produced an enrichment in tau tubulin kinases in the Reinke et al. (2000) spermatogenic gene sets (Figure S3E and Table S8). Thus, WormCat has identified a class of kinases that may be important for sperm-specific functions (Figure 3F).
To directly compare gene set enrichment from WormCat and GO, we analyzed each of these germline-enriched datasets with GOrilla and used REVIGO (Supek et al. 2011) for visualization (Figure S4, A–C, Figure S5, A and B, Table S7, and Table S8). For the GN genes, the top 5 of the 544 significantly enriched categories were nucleic acid metabolic process (GO:0090304), nucleobase-containing compound metabolic process (GO:0006139), heterocycle metabolic process (GO:0046483), cellular aromatic compound metabolic process (GO:0006725), and organic cyclic compound metabolic process (GO:1901360) (Figure S4A and Table S7, see tabs 7, 8). These GO categories are highly overlapping and linked to multiple general processes involving nucleic acids. One gene GO:0006139, gut-2, an LSM RNA binding protein, was present in 23 different GO categories (Table S7). A comparison of these GO categories found that each contains genes placed in distinct WormCat categories. For example, gut-2 was placed in mRNA Functions in WormCat, ama-1, the RNA Pol II large subunit, placed in Transcription: General Machinery, brc-1, the BRCA1 ortholog, placed in DNA and nsun-5, a mitochondrial RNA methyltransferase placed in Metabolism: mitochondria. These WormCat categories are the top five identified in the GN dataset (Figure 3B and Table S7). Thus, while WormCat and GO are both identify nucleic acid-related processed as among the most highly enriched in the GN dataset, the WormCat data are more concise and easily aligned with the molecular processes.
Within the spermatogenic datasets from Ortiz et al. (2014) and Reinke et al. (2000), WormCat identified a class of kinases, tau tubulin kinases (TTKs), that have the potential to function in sperm motility. General categories of phosphorus metabolic process (GO:0006793), phosphate-containing compound metabolic process (GO:0006796), and peptidyl-threonine phosphorylation (GO:0018107) were among the top five most enriched categories by GO from the Spermatogenic dataset; however, the TTKs as a group were not selectively identified from these very broad signaling categories in either spermatogenic data set (Table S7 and Table S8). Thus, WormCat provided advantages over GO in the germline data sets by providing less redundant, and more easily interpreted, data, and, most importantly, by identifying novel categories with potential links to biological function.
Identification of postembryonic tissue-specific gene expression categories
Improved technologies for cell-type-specific marker expression, nematode disruption, and deep sequencing of small RNA quantities have allowed construction of gene expression datasets from larval (Spencer et al. 2011) and adult (Kaletsky et al. 2018) somatic tissues. To generate data from larval cell types, the Miller laboratory used cell-type-specific tagged green fluorescent proteins to label a wide variety of larval tissues, and examined mRNA expression in tiling microarrays (Spencer et al. 2011). RNA from each cell type would include tissue-specific, broadly expressed, and ubiquitously expressed genes. To define cell-type specific transcripts, Spencer et al. (2011) designated selectively enriched genes as expressed more than twofold vs. the whole animal and as present in a few cell types (Spencer et al. 2011). First, we performed WormCat analysis on the selectively enriched gene sets, and found distinct gene set enrichments for each tissue type (Figure 4A and Table S9). For instance, body wall muscle (BWM) was enriched for Muscle Function and Cytoskeleton (Figure 4B and Table S9). The category Metabolism was enriched in both intestine (Int) and hypodermis (Hyp), whereas Stress responses appeared more specific for the intestine, and Extracellular material for the hypodermis (Figure 4, B and C and Table S9). This likely reflects the role of the intestine in mediating contact with the bacterial diet, and the importance of the hypodermis for cuticle formation. While metabolic genes are expected to be required across multiple cell types, some cell types have specialized metabolic requirements. Lipid metabolism gene enrichment appeared at the Cat2 level in both intestine and hypodermis. However, Cat3 analysis shows that sterol and sphingolipid genes drive this enrichment in the intestine, while hypodermal lipid enrichment involves more broad categories, with minor enrichments in Metabolism: lipid: binding and Metabolism: lipid: lipase (P-values of 4.51 × 10−04 and 2.86 × 10−04, which did not satisfy the FDR cutoff) (Figure 4D and Table S9). The Cat1 level analysis showed strong enrichment of transmembrane (TM) transporters in all tissues, including the intestine, excretory cells, and in neurons; however, the Cat2 level shows enrichment of distinct classes of transporters (Figure 4B and Table S9) aligning with functions such as nutrient uptake, waste processing, and channel activity in each of these cell types.
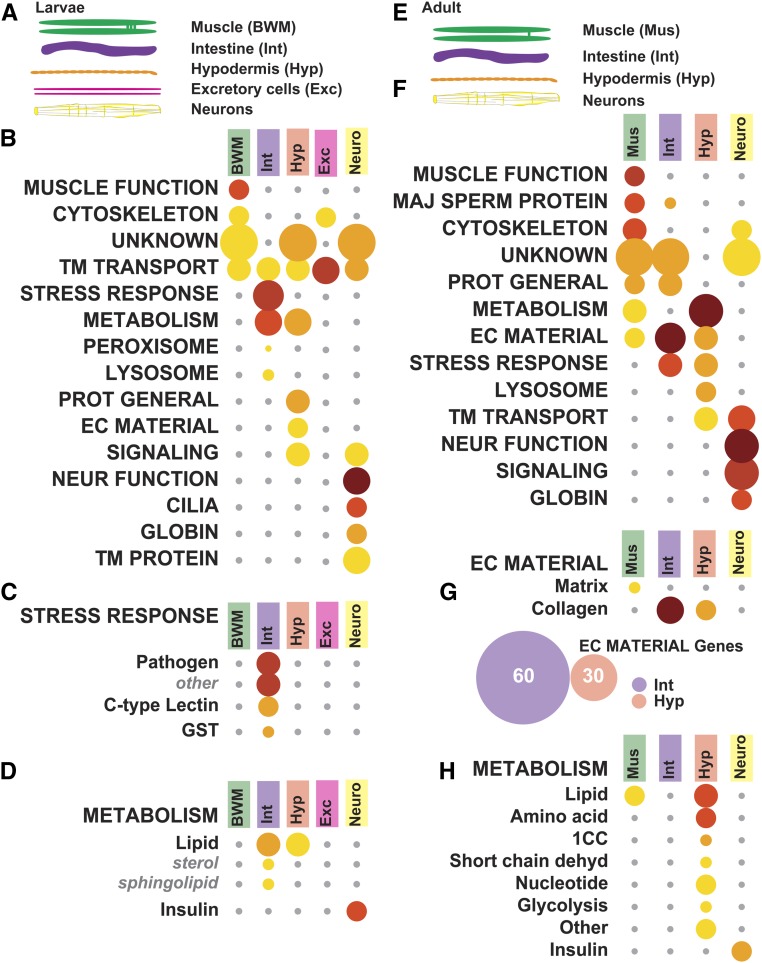
Figure 4.
WormCat analysis of tissue-specific gene sets reveals the importance of the intestine in stress-responsive categories. (A) Diagram showing larval tissues isolated in tiling array data used in figures B–D from Spencer et al. (2011) (B) WormCat Category 1 enrichment for larval tissue-specific selective enriched gene sets shows differentiation of Body wall muscle (BWM), Intestine (Int), Hypodermis (Hyp), Excretory cells (Exe), and Neurons (Neuro). (C–D) Category 2 and 3 breakdown of Stress Response (C) and Metabolism (D). (E) Schematic showing adult tissues isolated for RNA-seq used in figures F–I from Kaletsky et al. (2018) (F) Category 1 analysis of enriched genes shows the differentiation of muscle and neuronal functions. (G–H) Category 2 and 3 breakdown of Extracellular Material gene enrichment, including a Venn diagram showing relationships between collagen genes in intestine and hypodermis (G), and Metabolism (H). Bubble heat plot key is the same as Figure 1D. 1CC, 1-Carbon Cycle; EC Material, Extracellular Material; GST, Glutathione-S-transferase; Maj Sperm Protein, Major Sperm Protein; Neur Function, Neuronal Function; Prot General, Proteolysis General; Short Chain Dehyd, Short Chain Dehydrogenase; TM Transport, Transmembrane Transport
Next, we examined the data from Kaletsky et al. (2018), who performed RNA-seq from adult C. elegans sorted for muscle (Mus), intestinal (Int), hypodermal (Hyp), and neurons (Figure 4E and Table S10). They computationally separated genes to distinguish expression specificity, demarking “enriched,” “unique,” and “ubiquitously” expressed categories. We used the “enriched” gene sets in WormCat analysis, and found that WormCat correctly mapped muscle or neuronal genes to those cell types (Figure 4F and Table S10). At the Cat1 level, Extracellular material was enriched in muscle, hypodermis, and intestine (Figure 4F and Table S10). At the Cat2 levels, Extracellular material diverged with matrix showing enrichment in muscle and collagen, showing enrichment in intestine and hypodermis (Figure 4G and Table S10). However, the collagen genes enriched in intestine and hypodermis were distinct (Figure 4G and Table S10), perhaps reflecting differing roles for these collagens in the cuticle vs. in basement membranes. Distinguishing individual genes for this comparison is very cumbersome in commonly used GO servers, and, therefore, represents an advantage of using WormCat. Previous studies found that two intestinal basement membrane collagens were produced in nonhypodermal tissues (Graham et al. 1997); however, this data suggests that the intestine others could produce others locally. Kaletsky et al. (2018) also noted enrichment of metabolic function in adult hypodermis with GO analysis. Metabolic gene enrichment was also detected by WormCat analysis of their data (Figure 4H and Table S10), as well as in the larval data from Spencer et al. (2011) (Figure 4D and Table S9).
In our annotation strategy, we chose to restrict genes in categories such as Neuronal function to those that are specific to that tissue, and that have a described physiological function. Genes that functioned in neurons, as well as other tissues, were placed in more general molecular function-based categories. With this approach, we hoped to reduce false-positive identification of neuronal categories outside the nervous system, yet permit the identification of related, yet functionally less-specific groups. For example, while the WormCat analysis of the neuronal tissues in the Spencer et al. (2011) and Kaletsky et al. (2018) datasets showed strong enrichment of neuronal-specific categories, it also included categories of genes likely to function in both neurons and other tissues, or that contained genes that had not yet been classified in vivo. These categories include Metabolism: insulin (Figure 4, D and H and Table S10), Transmembrane (TM) transport, Signaling (Figure 4, B and F and Table S10), and Transmembrane protein (Figure 4B and Table S10). This is in line with the analysis by both Kaletsky et al. (2018) and Ritter et al. (2013).
In order to distinguish the utility of WormCat from GO for the tissue-specific Spencer et al. (2011) and Kaletsky et al. (2018) datasets, we used GOrilla (Eden et al. 2009) to generate GO analysis, and visualized the data with REVIGO (Supek et al. 2011) (Figure S6, Figure S7, Figure S8, Table S9, and Table S10). There were many similarities between the categories. For example, categories linked to the Cytoskeleton are highly enriched in the muscle datasets from Kaletsky et al. (2018) by GOrilla and WormCat (Figure 4F, Figure S7A, and Table S10). In another example, Stress response categories were highly enriched by both WormCat and GO in the larval (Spencer et al. 2011) and adult (Murphy et al. 2003) intestine (Figure 4F, Figure S6B, Figure S7B, and Table S10). However, as shown above, WormCat identified the insulin gene family as strongly enriched in both larval (Figure 4D) and adult (Figure 4H) neuronal tissue. Insulins were not identified as a class by our GO analysis. Instead, they were distributed among less specific categories such as biological regulation (GO:0065007), regulation of biological process (GO:0050789), and regulation of cellular process (GO:0050794) (Figure S5, Figure S6, Table S9, and Table S10). Thus, WormCat finds the major categories shown by GOrilla in the tissue-specific data, and also identifies additional enriched groups.
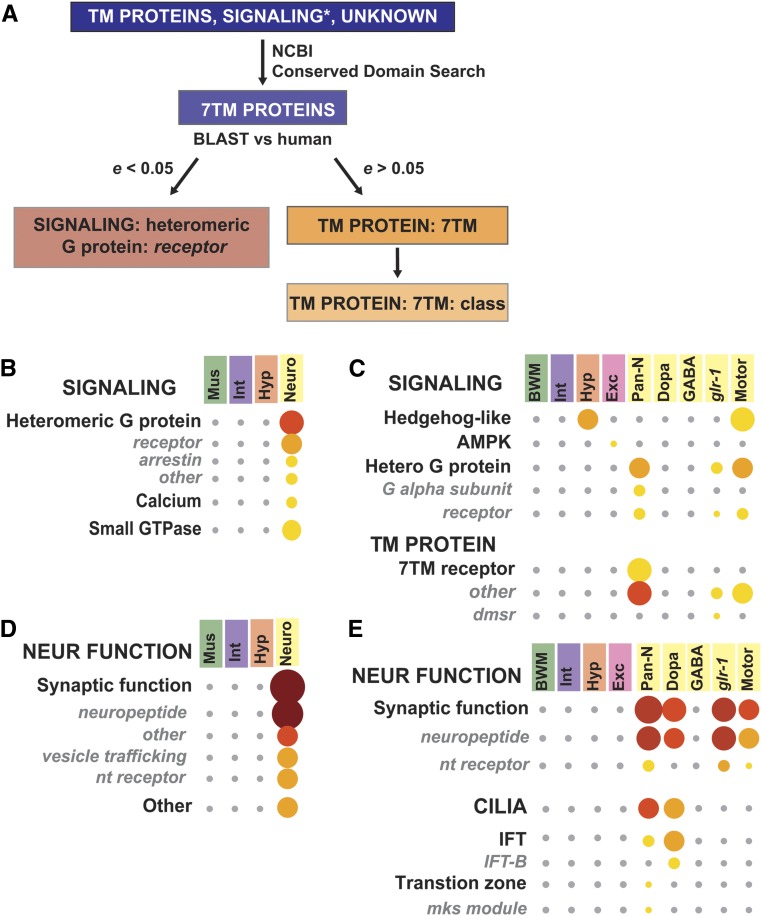
The seven transmembrane (7TM) protein family in C. elegans presented an annotation challenge. This class comprises ∼8% of all protein-coding genes that seem likely to function in neurons, yet whose functions are undescribed (Robertson and Thomas 2006). Some have significant homology to mammalian G protein-coupled receptors (GPCRs), while others are nematode or C. elegans specific (Robertson and Thomas 2006). In order to identify and classify these proteins as accurately as possible, GPCRs with strong evidence for neuron-specific activity were placed in Neuronal function, while all other potential GPCRs were classified by protein domain and homology. For developing a list of potential GPCRs, we selected genes identified in WormBase as containing a transmembrane domain as well as those we initially annotated as GPCRs in the Signaling category. To recover any genes missed by these approaches, we added all Unknown proteins from our annotation list. We submitted the protein sequences for these genes to the NCBI Conserved Domain search tool (Marchler-Bauer et al. 2017), and selected all the genes in these groups that contained a 7TM domain (Figure 5A). Next, we used BLASTP to determine the degree of homology to human GPCRs, which would reflect the conservation of function. Genes that had BLASTP scores of e < 0.05 on the NCBI server were classified in Signaling: heteromeric G protein: receptor. Those with e scores >0.05 were classified as TM protein: 7TM, with class designated by WormBase in Cat3. Thus, genes classified within Neuronal function or Signaling have a strong likelihood of GPCR function, whereas those in TM protein: 7TM have not been sufficiently defined. Signaling: G protein categories are enriched in neuronal genes sets from both Kaletsky et al. (2018) and Spencer et al. (2011) (Figure 5, B and C, Table S9, and Table S10), and 7TM proteins show enrichment in the larval pan-neuronal, glr-1-expressing neurons, and motor neurons (Figure 5C, Table S9, and Table S10). Thus, our annotation strategy allows separation of GPCRs with a high likelihood of neuronal function, yet still permits enrichment of the larger class of 7TM proteins in neuronal tissues.
Figure 5.
Detailed analysis of neuronal tissue-specific gene sets reveals specific enrichment for cilia gene expression on dopaminergic neurons. (A) Flow chart showing the process for annotating seven transmembrane (7TM) proteins. e value is the statistical score provided by the NCBI BLAST server. Asterisk on Signaling notes that only predicted GPCRs within this category were submitted to the NCBI conserved domain server. (B–E) Breakdown of Neuronal Function to Category 2 and 3 from larval data in Kaletsky et al. (2018) (B and D) or adult data in Spencer et al. (2011) (C and E). 7TM receptor, Seven Transmembrane Receptor; BWM, Body Wall Muscle; dmsr, DroMyoSuppressin Receptor Related; Dopa, Dopaminergic Neurons; Exe, Excretory Cells; GABA, Gamma-Aminobutyric Acid-Specific Neurons; glr-1, Glutamate Receptor-Specific Neurons; Hetero G protein, Heterotrimeric G Protein; Hyp, Hypodermis; IFT, Intraflagellar Transport; Int, Intestine; mks module, Meckel-Gruber syndrome Module; Motor, Motor Neurons; nt Receptor, Neurotransmitter Receptor; Neuro, Neurons; Pan-N, Pan-Neuronal
In order to directly compare WormCat and GO on the larval neuronal data sets, we examined category enrichment of Spencer et al. (2011) pan-neuronal and motor neuron genes in GO by GOrilla (Eden et al. 2009), using REVIGO (Supek et al. 2011) for visualization (Figure S6, Figure S8, and Table S9). The most enriched category in the pan-neuronal or motor neuron datasets was G protein-coupled receptor signaling (GO:0007186). Next, we used WormCat to determine how we had annotated genes within GO:0007186, and found that this GO category included genes we had classified in Signaling: Heteromeric G protein (G-alpha subunits and receptors), Neuronal Function: Synaptic function (neuropeptides and neurotransmitter receptors), and TM protein: 7TM receptor (Figure 5C and Table S9). While inclusion of the G protein signaling apparatus and neuropeptide ligands is appropriate for the broad category of G protein signaling, the GO categories do not differentiate between GPCRs with a high likelihood of function from the poorly classified 7TM proteins. In addition, many of the nlp genes listed in GO:0007186 are functionally uncharacterized, and, thus, it is not clear if they are bona fide GPCR ligands or could interact with other receptors outside of GPCR signaling (Li and Kim 2008). Therefore, WormCat improves on GO analysis for these datasets by providing more nuanced information on the function of these genes in GPCR pathways.
Neuronal genes from adult (Kaletsky et al. 2018) and larval (Spencer et al. 2011) gene sets also showed strong enrichment in Cat2 and Cat3 classifications within Neuronal function, such as Synaptic function, neuropeptide, and neurotransmitter (nt) receptor (Figure 5, D and E, Table S9, and Table S10). Cilia gene enrichment was also apparent in the pan-neuronal and dopaminergic larval gene sets (Figure 5D and Table S9). Neurons are the only ciliated cells in C. elegans, and cilia occur on multiple neuronal subtypes (Inglis et al. 2007). However, all dopaminergic neurons are ciliated (Inglis et al. 2007), and, are, therefore, more likely to show enrichment. Taken together, our WormCat analysis of these large tissue-specific gene sets provides a detailed view of gene classes specific to muscle, hypodermis, intestine, and neurons in larvae and adults. We have identified differential enrichment in lipid metabolism genes, and collagens from intestine and hypodermis defined a classification system for GPCRs and 7TMs, and identified Cilia as a major enriched category in dopaminergic neurons. Much of this information goes beyond what GO analysis reveals, and provides predictions that can be useful to design future studies. Identification of these types of nuanced tissue-specific patterns is an important step to understanding how specific cell types function.
Drug interactions limiting lifespan induce changes in sterol metabolism
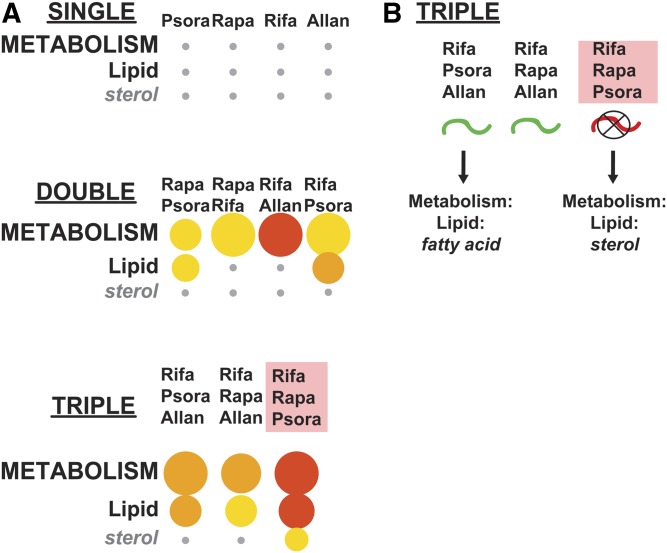
C. elegans is particularly suited to studies determining gene expression changes in response to a panel of treatments in a whole animal, and to correlate these changes to physiological function. For example, Admasu et al. (2018) generated a complex gene expression dataset by performing parallel RNA-seq on animals treated with five lifespan-increasing drugs that affect distinct pathways (Allantoin, Rapamycin, Metformin, Psora-5, and Rifampicin). They used five pairwise combinations and three triple-drug combinations to determine if any combination lead to further lifespan extension, and to identify gene expression profiles associated with increased longevity (Admasu et al. 2018). They found that one triple-drug combination (Rifa/Psora/Allan) activated lipogenic metabolism through the transcription factor SBP-1/SREBP-1, and determined that the drug-induced longevity was dependent on SBP-1 function (Admasu et al. 2018). The authors also made the striking observation that a distinct triple-drug combination (Rifa/Rapa/Psora) reduced lifespan, even though each single drug or drug pairs increased longevity (Admasu et al. 2018). To determine if any gene expression categories might explain this effect, we used WormCat to analyze category enrichment for the up and downregulated genes for each single drug, pairwise, or triple-drug combination (Figure 6A, Figure S9, Figure S10, Table S11, and Table S12). Similar to the author’s KEGG analysis (Admasu et al. 2018), we observed Metabolism: lipid enrichment in long-lived Rifa/Rapa/Psora-treated animals (Figure 6A and Table S11); however, we also noted that Metabolism: lipid was enriched in all three combinations with WormCat. Next, we examined the up and downregulated genes to determine if any categories correlated with the failure to survive in the Rifa/Rapa/Psora treated animals. We did not find category signatures in the downregulated genes that appeared to correlate with the decrease in longevity (Figure S10 and Table S12). However, upregulated genes from the short-lived Rifa/Rapa/Psora treated animals were enriched in another specific class of lipid metabolic genes: sterol metabolism (Figure 6A and Figure S9). Closer examination of the single and pairwise combinations showed that the enrichment of sterol metabolic genes only appeared in the triple combination with poor survival (Figure 6B). C. elegans does not use cholesterol as a membrane component (Ashrafi 2007). Thus, this category does not include cholesterol synthesis genes, but does include genes involved in modification of sterols, for example, in steroid hormone production (Watts and Ristow 2017). Examination of individual genes (Table S11, Tab 18 Sterol Genes) showed that 5 of the 19 had lifespan phenotypes, and 4 had lethality related phenotypes in WormBase, consistent with their effects on survival in Admasu et al. (2018). Furthermore, Murphy et al. (2003) showed that 3 of the 19 sterol genes are upregulated in another long-lived model, daf-2(mu150), and two of these, stdh-1 and stdh-3 are required for lifespan extension in daf-2(mu150) animals (Murphy et al. 2003). Thus, the category enrichments captured by WormCat for this drug study have identified sterol metabolism genes as potential players in the paradoxical lifespan shortening effects of the Rifa/Rapa/Psora combination.
Figure 6.
WormCat analysis of RNA-seq data from C. elegans treated with combinations of lifespan-lengthening drugs reveals the emergence of sterol metabolism in drug combinations, limiting survival. (A) Comparison of Metabolism: lipid: sterol enrichment in single, double, and triple-drug combinations shows sterol emergence in the Rifa/Rapa/Psora gene set (Admasu et al. 2018). (B) Diagram showing a summary of data from lifespan changes after triple-drug treatment from Admasu et al. (2018). Pink box denotes drug combination that causes premature death. Bubble heat plot key is the same as Figure 1D. Allan, Allantoin; Psora, Psora-4; Rapa, Rapamycin; Rifa, Rifampicin
In order to compare gene set enrichment of the triple-drug combinations from WormCat with GO, we analyzed upregulated genes from the Rifa/Psora/Allan-, Rifa/Rapa/Allan-, and Rifa/Rapa/Psora-treated animals in GOrilla (Eden et al. 2009), and visualized the data with REVIGO (Supek et al. 2011) (Figure S11 and Table S11). WormCat and GO showed multiple similarities. For example, WormCat and GO identified extracellular matrix-linked categories in all three triple combinations (WormCat: EC MATERIAL; GOrilla: GO:0030198: extracellular matrix organization) (Figure S9 and Table S11). However, WormCat identified Metabolism: lipid in all three combinations, whereas GO analysis by GOrilla only identified categories linked to lipid metabolism (GO:0006629: lipid metabolic process (q = 5.63 × 10−03), GO:0044255 cellular lipid metabolic process (q = 1.49 × 10−02) and GO:0006631 fatty acid metabolic process (q = 2.16 × 10−02) in the Rifa/Rapa/Psora dataset (Table S11). WormCat also showed a much higher enrichment score for Metabolism: lipid, P = 2.00 × 10−14) (Table S11). Thus, as in the sams-1 microarray data discussed previously, WormCat provides an improved tool for determining the enrichment of metabolic genes.
WormCat also found an enrichment of transcription factors in each of the triple combinations, with specific enrichments in nuclear hormone receptors and homeodomain genes in the Rifa/Psora/Allan-upregulated set (Figure S9) Enrichments of nuclear hormone receptors in C. elegans is potentially of interest as they may regulate multiple metabolic regulatory networks (Arda et al. 2010). However, GOrilla only identified categories linked to transcription factors (GO:0006355: regulation of transcription, DNA-templated, GO:0051252: regulation of RNA metabolic process, GO:2001141: regulation of RNA biosynthetic process, GO:1903506 regulation of nucleic acid-templated transcription, and GO:0019219 regulation of nucleobase-containing compound metabolic process) in the Rifa/Psora/Allan dataset. No individual class of transcription factors showed enrichment in any of the triple combinations by GO (Table S11); thus, WormCat offers a clear advantage over GO by providing increased coverage across diverse categories of gene function.
Identification of gene set enrichments in RNAi screening data
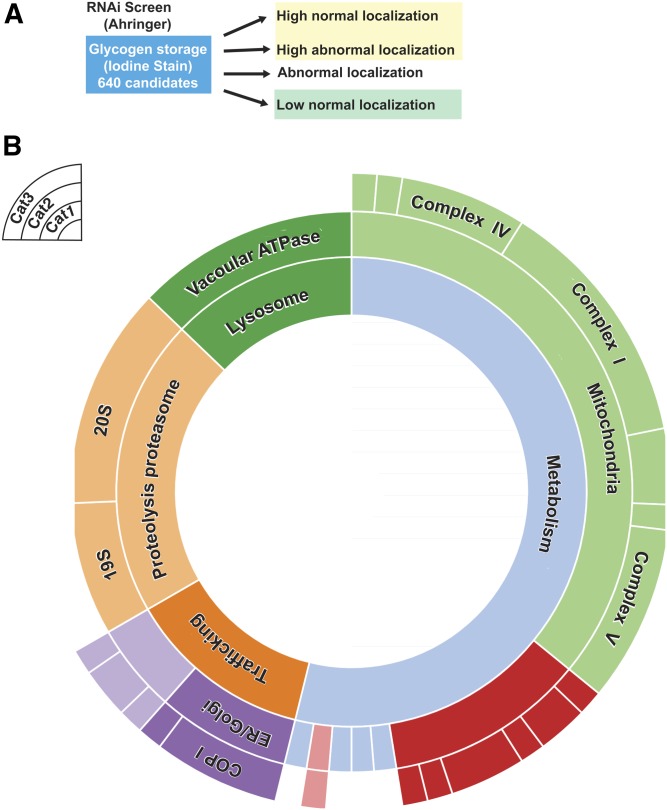
In order to use WormCat to analyze genome-scale RNAi screening data, we mapped WormCat annotations to the list of genes in the Ahringer library (Kamath et al. 2003) (Table S13). To test this approach, we used data from the Roth laboratory, who screened the Ahringer library for changes in glycogen storage in C. elegans and identified >600 genes, scored as glycogen high, glycogen low, and abnormal localization (LaMacchia et al. 2015) (Figure 7A and Table S14). The authors functionally classified all hits from the screen with an inhouse annotation list, graphed the percentage within each group, and noted high percentages of genes with roles in metabolism (electron transport chain), signaling, protein synthesis or stability, and trafficking (LaMacchia et al. 2015); however, they were unable to assign statistical significance to any of the groups. WormCat identified similar groups as the LaMacchia et al. (2015) functional classification for the “glycogen low” candidates. For example, we identified Metabolism: mitochondria, complex I, III, IV, and V and found statistical enrichment in these categories (Figure 7B and Table S14). However, signaling had no enrichment (Table S14). Thus, WormCat can identify statistically relevant pathways in genome-scale RNAi screen data.
Figure 7.
WormCat analysis of a genome-scale RNAi screen quantitates categories of candidate genes. (A) Schematic of the RNAi screen from LaMacchia et al. (2015) identifying candidate genes that altered glycogen staining. (B) Sunburst diagram from low glycogen candidates showing significantly enriched categories.
To provide a direct comparison between WormCat and GO with this dataset, we determined the GO term associated with the “glycogen low” data by GOrilla (Eden et al. 2009), and visualized the data with REVIGO (Supek et al. 2011) (Figure S12 and Table S14). A total of 185 separate GO terms were identified in this data set compared to the 4 Cat1 level terms identified by WormCat (Metabolism, Lysosome, Proteolysis Proteasome, and Trafficking) (Figure 7B and Table S14). WormCat also finds a limited number of Cat2 groupings within these sets, including Metabolism: mitochondria, Lysosome: vacuolar ATPase, Proteolysis Proteasome:19S, 20S, and Trafficking: ER/Golgi) (Figure 7B and Table S14). This large difference in the number of significantly enriched categories stems from the multiple, overlapping categories present in the GO analysis. For example, the mitochondrial gene cyc-1 (cytochrome c oxidase) is represented in 87 of the GO terms, whereas the annotation in WormCat is METABOLISM: mitochondria (Table S14, tab 8).
Similarly, the vacuolar ATPase vha-6 appears in 39 of GO terms returned, the proteasomal component pbs-7 is present in 23, and the ER/Golgi COP I component Y71F9AL.17 is in 21 (see Table S14, tabs 9–11). This GO term redundancy provides the user with a complex, hard to interpret, list. In addition, GO terms that are repeated fewer times (such as those containing the trafficking gene Y71F9AL.17) become marginalized in a complex list. Thus, with this dataset, WormCat provides easily distinguished categories with clear links to biological or molecular functions. The GO terms show the same genes repeated in a large fraction of the categories and obscure categories with less gene redundancy.
Discussion
WormCat provides new insights into comparative RNA-seq data
Current technology allows for the routine use of genome-scale experiments for the generation of gene expression data. The goal of these experiments is often to identify classes of genes that add insight to biological functions, as well as to highlight selected genes for individual analysis. GO analysis, while widely used, is difficult to apply to datasets with multiple combinations of treatments or genetic perturbations. Further, for C. elegans, current GO analysis is often inaccurate, and misses useful physiological and molecular information. Here, we have shown that WormCat can annotate gene categories, provide enrichment statistics, and display user-friendly graphics for gene sets identified from C. elegans gene expression studies. Furthermore, our visualization strategy allows comparison across multiple datasets, facilitating the identification of categories with shared biological functions.
Our initial, script-based, smaller-scale version of WormCat highlighted changes in metabolic gene expression in C. elegans with changes in levels of the methyl donor SAM or methyltransferases modifying H3K4me3 (Ding et al. 2018). In this study, we have expanded the annotation list, developed a web-based server, and added a new graphical output. We used WormCat to successfully analyze data from metabolic, tissue-specific, and drug-induced expression changes. This analysis provides not only validation and use-case examples, but also additional insights into the known gene expression patterns. For example, our examination of germline gene expression datasets from the Kimble and Kim laboratories (Reinke et al. 2000; Ortiz et al. 2014) identified a large class of microtubule kinases (TTK) as enriched in spermatogenic gene sets, and as a coenriched gene set with MSPs. One TTK, spe-6, has been previously identified in a screen for mutants with defects in sperm development (Varkey et al. 1993). Our results suggest that many genes in this family could have important functions in spermatogenesis, and that the appearance of MSPs and TTKs in a dataset could also serve as a marker for maleness. Finally, we used WormCat to analyze a dataset consisting of RNA-seq from C. elegans treated with multiple lifespan-changing drugs, alone or in combination, plus one mutation animal strain that extends lifespan (Admasu et al. 2018). The classification and graphical output allowed us to identify the upregulation of sterol metabolism genes in a triple-drug combination that was not present in the single or double drug treatments. Thus, WormCat identified a gene set that may be important for the effects of the lifespan-altering drugs in this assay.
Strengths and weaknesses of WormCat
We developed WormCat to overcome some of the limitations of GO analysis when analyzing C. elegans gene expression data, and to utilize specific phenotype data available in WormBase. In addition, we specifically engineered WormCat to classify data for the identification of coexpressed or cofunctioning gene sets. Finally, we developed two graphical outputs: a scaled heat map/bubble plot and a sunburst plot. The modular nature of the bubble plot allows multiple datasets to be grouped and compared, while the sunburst plot gives a concise view of single datasets, as may be obtained with screening data. Our validation with random gene testing and analysis of C. elegans gene expression data from metabolic, tissue-specific, and drug-treated animals shows that WormCat is a robust tool that provides biologically relevant gene enrichment information. There are three main areas that WormCat provides an advantage over using GO that are apparent in our case studies. First, as discussed above, we found that, in some of our test cases, WormCat identified broader sets of genes within categories or categories that were not identified by GO. Second, the WormCat output is much easier to interpret; the bubble charts provide intuitive visualization, and the tables provide clear access to the enrichment statistics and annotation of the input genes. Third, the availability of the annotations for each input gene enables comparisons between genes in categories. For example, we found that while Extracellular material: collagen was enriched in both intestine and hypoderm in the Kaletsky et al. (2018) data set, the genes were nonoverlapping, suggesting tissue-specific expression of collagen genes. This comparison would be difficult to make with GO, as many common GO servers do not supply the genes with each category in an easily accessible manner. Directly comparing the genes within WormCat and GO categories from our previously published dataset of gene expression after sams-1 knockdown, we found that WormCat identified a broader set of lipid metabolic genes than GO analysis from GOrilla, and that the genes identified only by GO analysis might be better classified in different categories to reflect their biological functions. Thus, WormCat provides an alternative to GO with advantages in output that improve data interpretation and access to gene annotations that allow deeper comparisons among categories. In some cases, WormCat also identifies categories that are not found by GO.
However, there are several limitations to WormCat. First, while multiple researchers with varied expertise curated our annotation list, some genes may be misannotated, or some Cat2 or Cat3 groups may fit better in other Cat1 classifications. We will update the WormCat annotation list at periodic intervals while providing access to the previous annotation lists. Second, each C. elegans gene received a single, nested, annotation, rather than a group of annotations as in GO. We chose to prioritize the visualization of enriched gene sets in this instance, using a single annotation per gene to permit graphing in scaled heat maps. Access to the program and annotation lists for the local application also allows users to customize the annotation lists according to their preferences.
Annotation lists of genome-scale data are likely to contain errors. We have defined several sources of error, and have taken corrective steps. In some cases, a gene may be simply misannotated. For example, a component of the General transcription machinery was placed in Signaling by the annotator. In others, the classification system may be incorrect. An example of this would be classifying enzymes that modify small molecules as protein modification. To estimate the misclassification error rate, we generated a list of 3000 random WormBase IDs. We mapped each ID to our annotation list and rechecked the annotations. We found 29/2294 genes (1.3%) whose annotations were incorrect by our criteria (13 of these were Unknown genes that could be classified in other categories). This suggests ∼300 genes in the entire dataset may be misannotated by our criteria, many representing Unknown genes that could acquire classification. We will periodically update the WormCat annotation lists to accommodate new gene information and correct errors.
It is important to note that some gene classifications depend on criteria that are open to interpretation. For example, transcription factors regulating genes within a pathway are grouped within a linked category to allow identification of cofunctioning genes. For instance, efl-1, a master regulator of cell cycle genes, is annotated as Cell cycle: transcriptional regulator, instead of with the more broadly acting trans-regulatory factors in Transcription factor: E2F. To allow for different interpretations of the annotation strategy, we have set up a GitHub site (https://github.com/dphiggs01/wormcat), where the annotation list and scripts for executing WormCat can be downloaded and customized by the user to accommodate differences in annotation preference.
The value of gene set enrichment is also highly dependent on the criteria used to specify the regulated genes. In the present study, we used the same criteria as the respective authors, except that we separated up and downregulated genes where necessary. For example, in the Kaletsky et al. (2018) tissue-specific data, the authors provided data for all genes expressed in each tissue, enriched genes (expressed at FDR >0.05, and log2 fold change >2 relative to other tissues), or unique genes (log2 RPKM >5) significantly differentially expressed in comparison to the expression of each of the three other tissues (FDR >0.05, log2 fold change >2 for each comparison) (Kaletsky et al. 2018). We found the best resolution of WormCat categories between the tissues occurred with the enriched datasets, rather than with all genes or unique gene sets. This suggests that gene lists with all expressed genes may require more stringent statistical cutoffs, but also that WormCat may not be as suited to highly filtered data.
Application to other organisms
By developing WormCat specifically for analyzing C. elegans gene sets, we were able to take advantage of available data on WormBase, but this limited the applicability of our annotation list with other organisms. Although researchers in mammalian fields can access pathway analysis pipelines such as Ingenuity Pathway Analysis (Qiagen; Krämer et al. 2014) that identify functionally linked genes, these programs do not necessarily provide a simple graphical output for comparative analysis. WormCat analysis generating the scaled heat/bubble charts can be adapted for use with other organisms by running the program locally with altered annotation lists. Replacing gene IDs and the Cat1, Cat2, and Cat3 values with any annotation allows customization of the pipeline to any other organism. Thus, the modular nature of WormCat allows adaptation to multiple annotation strategies within C. elegans or to other organisms, allowing a streamlined visualization for examining genome-scale expression or screen data.
Acknowledgments
We wish to thank members of the Walker and Walhout laboratories for helpful discussion. Funding to A.K.W. National Institutes of Health (NIH) National Institute on Aging (NIA) 1R01AG053355 and A.J.M.W. grants NIH grants DK068429 and GM122502.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.10312070.
Communicating editor: V. Reinke
Literature Cited
- Admasu T. D., Chaithanya Batchu K., Barardo D., Ng L. F., Lam V. Y. M. et al. , 2018. Drug synergy slows aging and improves healthspan through IGF and SREBP lipid signaling. Dev. Cell 47: 67–79.e5. 10.1016/j.devcel.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Angeles-Albores D., N Lee R. Y., Chan J., and Sternberg P. W., 2016. Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics 17: 366 10.1186/s12859-016-1229-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda H. E., Taubert S., MacNeil L. T., Conine C. C., Tsuda B. et al. , 2010. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol. 6: 367 10.1038/msb.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., 2007. Obesity and the regulation of fat metabolism (March 9, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.130.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Demodena J., and Sternberg P. W., 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94. 10.1126/science.1169628 [DOI] [PubMed] [Google Scholar]
- Bulcha J. T., Giese G. E., Ali M. Z., Lee Y. U., Walker M. D. et al. , 2019. A persistence detector for metabolic network rewiring in an animal. Cell Rep. 26: 460–468.e4. 10.1016/j.celrep.2018.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [corrigenda: Science 283: 35 (1999)]; [corrigenda: Science 283: 2103 (1999)]; [corrigenda: Science 285: 1493 (1999)]. [DOI] [PubMed]
- Deng X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D. et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans, and Drosophila melanogaster. Nat. Genet. 43: 1179–1185. 10.1038/ng.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., Smulan L. J., Hou N. S., Taubert S., Watts J. L. et al. , 2015. s-adenosylmethionine levels govern innate immunity through distinct methylation-dependent pathways. Cell Metab. 22: 633–645. 10.1016/j.cmet.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., Higgins D. P., Yadav D. K., Godbole A. A., Pukkila-Worley R. et al. , 2018. Stress-responsive and metabolic gene regulation are altered in low S-adenosylmethionine. PLoS Genet. 14: e1007812 10.1371/journal.pgen.1007812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., and Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., and Botstein D., 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Graham P. L., Johnson J. J., Wang S., Sibley M. H., Gupta M. C. et al. , 1997. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol. 137: 1171–1183. 10.1083/jcb.137.5.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., 2005. Control of oocyte meiotic maturation and fertilization (December 28, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.53.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A. L., Dillin A., and Kenyon C., 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1: 119–128. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Coulson A., Murray J. I., Bao Z., Sulston J. E. et al. , 2005. Genomics in C. elegans: so many genes, such a little worm. Genome Res. 15: 1651–1660. 10.1101/gr.3729105 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., and Greenstein D., 2005. Introduction to the germ line (September 1, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.18.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis P. N., Ou G., Leroux M. R., and Scholey J. M., 2007. The sensory cilia of Caenorhabditis elegans (March 8, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.126.2, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R., Yao V., Williams A., Runnels A. M., Tadych A. et al. , 2018. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14: e1007559 10.1371/journal.pgen.1007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R. et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J. Jr., and Tugendreich S., 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30: 523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMacchia J. C., Frazier H. N. III, and Roth M. B., 2015. Glycogen fuels survival during hyposmotic-anoxic stress in Caenorhabditis elegans. Genetics 201: 65–74. 10.1534/genetics.115.179416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., and Schedl T., 2006. RNA-binding proteins (April 18, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.79.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y. N., Howe K. L., Harris T. W., Arnaboldi V., Cain S. et al. , 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46: D869–D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault, S. W., 2006 Spermatogenesis (February 20, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.85.1, http://www.wormbook.org.
- Li, C., and K. Kim, 2008 Neuropeptides (September 25, 2008), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.142.1, http://www.wormbook.org.
- MacNeil L. T., Watson E., Arda H. E., Zhu L. J., and Walhout A. J. M., 2013. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153: 240–252. 10.1016/j.cell.2013.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J. et al. , 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45: D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato J. M., and Lu S. C., 2007. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology 45: 1306–1312. 10.1002/hep.21650 [DOI] [PubMed] [Google Scholar]
- McDonald J. H., 2014. Handbook of Biological Statistics. Sparky House Publishing, Baltimore. [Google Scholar]
- Mi H., Muruganujan A., Huang X., Ebert D., Mills C. et al. , 2019. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14: 703–721. 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S. et al. , 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Oliveira R. P., Porter Abate J., Dilks K., Landis J., Ashraf J. et al. , 2009. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8: 524–541. 10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., and Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765–1772. 10.1534/g3.114.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Smith H. E., Nance J., Wang J., Van Doren C. et al. , 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605–616. 10.1016/S1097-2765(00)00059-9 [DOI] [PubMed] [Google Scholar]
- Ritter A. D., Shen Y., Fuxman Bass J., Jeyaraj S., Deplancke B. et al. , 2013. Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. 23: 954–965. 10.1101/gr.150466.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., and Thomas J. H., 2006. The putative chemoreceptor families of C. elegans (January 06, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.66.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S. et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E. M., Kato M., and Sternberg P. W., 2012. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 109: 16246–16251. 10.1073/pnas.1203045109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulan L. J., Ding W., Freinkman E., Gujja S., Edwards Y. J. et al. , 2016. Cholesterol-independent SREBP-1 maturation is linked to ARF1 inactivation. Cell Rep. 16: 9–18. 10.1016/j.celrep.2016.05.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K. et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. 10.1091/mbc.9.12.3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L. et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. 10.1101/gr.114595.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., and Smuc T., 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2019. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47: D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance D. E., 2014. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim. Biophys. Acta 1838: 1477–1487. 10.1016/j.bbamem.2013.10.018 [DOI] [PubMed] [Google Scholar]
- Varkey J. P., Jansma P. L., Minniti A. N., and Ward S., 1993. The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics 133: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K. et al. , 2011. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147: 840–852. 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., and Ristow M., 2017. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 207: 413–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Yi L., Feng Y., Chen L., and Liu J., 2009. Structural insight into the activation mechanism of human pancreatic prophospholipase A2. J. Biol. Chem. 284: 16659–16666. 10.1074/jbc.M808029200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The code and annotation lists are available under MIT Open Source License, and can be downloaded from the GitHub repository https://github.com/dphiggs01/wormcat along with version-control information. Alternatively, WormCat can be installed directly as an R package using the devtools library. Supplemental material has been deposited at figshare and includes 12 supplemental figures and 14 supplemental tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10312070.
GO searches:
Genes lists were entered as test sets into GOrilla (http://cbl-gorilla.cs.technion.ac.il/) (Eden et al. 2009) with the WormCat annotation list used as background so that the same background set was used when comparing WormCat and GOrilla. “All” was selected for ontogeny choices, and the P-value thresholds were set to 10−3. Output selections were Microsoft Excel and REVIGO (Supek et al. 2011).