Abstract
Recently, increasing evidence has revealed that long noncoding RNAs (lncRNAs) play important roles in the pathogenesis of multiple cancers. Although the oncogenic effects of lncRNA nuclear-enriched abundant transcript 1 (NEAT1) in some cancers have been reported, the functional significance and molecular mechanism of NEAT1 in pancreatic cancer (PC) progression remains elusive. In this study, our findings showed that NEAT1 expression was upregulated in PC tissues and cell lines; high NEAT1 expression was associated with tumor size, TNM stage, lymph node and distant metastasis, and also predicted poor prognosis. Functional experiments demonstrated that NEAT1 could promote PC cell proliferation and metastasis both in vitro and in vivo. Mechanistically, NEAT1 could associate with E74 like ETS transcription factor 3 (ELF3) mRNA and enhance the combination of Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) and ELF3 mRNA, subsequently suppressing the degradation of ELF3 mRNA. Overall, our research indicates that NEAT1 might be a potential therapeutic target for patients with PC.
Keywords: mRNA stability, IGF2BP1, invasion, progression
Introduction
Pancreatic cancer (PC) is one of the most fatal malignancy worldwide, which ranks fourth in cancer-related death [1]. Most PC patients are usually clinically silent at the early stage, and often diagnosed in advanced stages accompanied by tumor invasion and metastasis. Radical resection is not suitable for these patients. Moreover, PC is resistance to radiotherapy and chemotherapy [2]. Even though some advancement has been obtained in the development of combination therapies, the therapeutic options for PC patients remain limited [3]. Thus, further investigation of the molecular mechanisms underlying PC initiation and progression is urgently needed and will be helpful to discover novel therapeutic strategies for treating PC patients and improving prognosis.
The high-throughput sequencing technology has identified thousands of long noncoding RNAs (lncRNAs), which are larger than 200 nt in length and do not appear to have protein-coding potential. LncRNAs have emerged as critical regulators in biological processes via acting as scaffolds or guides to regulate the interaction of protein with DNA, as decoys to bind proteins or microRNAs, or modulating the posttranslational modification of their associating proteins [4,5]. Dysregulation of lncRNAs has been found in human cancers. LncRNAs serve as oncogenes or tumor suppressors to regulate cancer cell proliferation, migration, invasion, differentiation and autophagy [6,7]. LncRNA nuclear-enriched abundant transcript 1 (NEAT1) is a newly discovered essential component of nuclear paraspeckles [8]. NEAT1 acts as an oncogenic lncRNA promoting tumorignenesis, metastasis and chemoresisitance via a competing endogenous RNA (ceRNA) mechanism in several types of human cancers, including colon cancer, melanoma, hepatocellular carcinoma, breast cancer and PC [9-13]. Additionally, NEAT1 binds to EZH2 and increases the H3K27me3 level in the promoter region of ICAT, GSK3B, and Axin2, which is critical for glioma progression [14]. NEAT1 is capable to suppresses E-cadherin transcription through association with G9a-DNMT1-Snail complex in osteosarcoma [15]. Recently, many lncRNAs have been revealed to interact with mRNAs and increase the stability of mRNAs, subsequently affecting the tumor growth and metastasis [16-18]. However, whether NEAT1 associates mRNAs to contribute to the PC progression remains unclear.
The present study showed that overexpression of NEAT1 served as a prognostic marker for the PC patients. Then, the functional role of NEAT1 in the malignant phenotypes of PC cells both in vitro and in vivo was explored. Further mechanistic investigation revealed that NEAT1 could associate with E74 like ETS transcription factor 3 (ELF3) mRNA and increase the stability of ELF3 mRNA. Taken together, our study indicates that NEAT1 may be used as a promising therapeutic target for PC.
Materials and methods
Tissues samples and cell culture
60 pairs of PC and adjacent normal pancreatic tissues were collected from PC patients who underwent radical resection at The First People’s Hospital of Shangqiu. The fresh tissues were immediately frozen in liquid nitrogen and stored at -80°C until used. The diagnosis of all specimens was histopathologically confirmed by two pathologists. None of the patients received any preoperative treatment. All patients provided written informed consent. Our research was approved by the Ethics Committee of the First People’s Hospital of Shangqiu in accordance with the Declaration of Helsinki. A normal human pancreatic cell line HPDE6-C7 and the PC cell lines PANC-1, BxPC-1, BxPC-3, AsPC-1, PaCa-2 and SW1990 were purchased from the American Type Culture Collection (ATCC). All cells were cultured in RPMI-1640 (Gibco) medium supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere at 37°C with 5% CO2.
RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using Trizol regent (Invitrogen) according to the standard protocol. 1 μg RNA was reverse transcribed to cDNA by using the Prime Script RT Reagent Kit (Takara). qRT-PCR was performed on an ABI StepOne Plus PCR system (Applied Biosystems, USA) using the SYBR Premix Ex Taq (Takara). GAPDH was used as internal reference. The value of relative gene expression was calculated by using 2-ΔΔCt method and normalized to control group. The primer sequences were shown as follow: NEAT1-F: CCCTTCTTCCTCCCTTTAACTTATC, NEAT1-R: GCCTCTCTTTCTCCACCATTAC; ELF3-F: CTGAGAAAGAATGAGGGAAGGAG, ELF3-R: CGCACGACCTCCTAAATCTAAA; GAPDH-F: CTCCTCACAGTTGCCATGTA, GAPDH-R: GTTGAGCACAGGGTACTTTATTG; IGF2BP1-F: CCAGTATGTGGGTGCCATTAT, IGF2BP1-R: TCCTTCCTATGCACGTCTATCT.
Western blot
Cells were lysed by RIPA buffer (Beyotime, China) containing protease inhibitor cocktail (Roche). Equivalent amounts of protein samples were separated by SDS-PAGE and then transferred onto PVDF membrane (Millipore). The membranes were incubated with indicated primary antibodies at 4°C overnight, and then incubated with corresponding secondary antibodies at room temperature for 1 hour. Finally, the membranes were detected by Immobilon Western Chemiluminescent HRP Substrate (Millipore). The antibodies used in this research were shown as follow: ELF3 (Abcam), GAPDH (Proteintech) and IGFBP1 (Abcam).
Construction of stable cells and transfection
The lentiviral particles expressing negative control (shNC), NEAT1 or ELF3 shRNAs were purchased from GenPharma (Shanghai, China) and used to repress NEAT1 or ELF3 expression. The lentiviral particles expressing negative control (NC), exogenous NEAT1 or ELF3 were purchased from GenPharma (Shanghai, China) and used to overexpress NEAT1 or ELF3. Cells were infected with indicated lentiviruses in the presence of polybrene. Stable cells were selected using puromycin. The siRNA used to inhibit IGFBP1 expression was purchased from Ribobio (Guangzhou, China). Transfection was performed using the Lipofectamine 2000 kit (Invitrogen, USA) according to the manufacturer’s instructions.
CCK-8 assay
Cell proliferation was measured using Cell Counting Kit-8 (CCK-8, Dojindo). In brief, 2 × 103 cells per well were seeded in 96-well plates. At the indicated time points, 10 μl CCK-8 reagent (Dojindo) was added to the medium, and then incubated for 1 hour at 37°C. The OD value was quantified using an ELx800 Absorbance Microplate Reader (BioTek) at 450 nm.
Transwell assay
Cell migration and invasion was determined by transwell assay. Transwell assay was carried out as previously described [19].
RNA immunoprecipitation (RIP) and MS2-RIP assay
RIP experiments were performed with the Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore) according to the manufacturer’s instructions. 5 μg IGF2BP1 (Abcam) antibody was used for RIP assays to co-precipitate endogenous NEAT1. Normal rabbit IgG antibody was taken as a negative control. The amount of NEAT1 pulled down by IGF2BP1 was then detected by qRT-PCR. For MS2-RIP, cells were co-transfected with pCMV-MS2, pCMV-NEAT1-MS2, or pCMV-NEAT1-MUT-MS2 and pMS2-GFP (Addgene). 48 hours later, cells were used to conduct RIP experiments using a GFP antibody (Cell Signaling). The amount of ELF3 mRNA enriched by NEAT1 was measured by qRT-PCR or RNA-sequencing.
RNA pull-down assay
RNA pull-down assay was conducted using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo) according to the manufacturer’s instructions. In brief, NBAT1 or its antisense RNA was in vitro transcribed and then biotin-labeled. Biotinylated RNA was incubated with streptavidin agarose beads and mixed with cell lysate. The retrieved proteins from RNA-binding protein complexes were subjected to western blot assay or mass spectrometry analysis. The retrieved RNA from RNA-binding protein complexes were subjected to qRT-PCR detection. Antisense NBAT1 was taken as negative control.
In vivo tumorigenic and metastasis assay
4-weeks-old male athymic BALB/c nude mice were maintained under specific pathogen-free conditions. 1 × 107 cells were injected subcutaneously into the right flank of nude mice (n = 5 per group). The tumor growth was measured by the formula (length × width2)/2 every week. For the metastasis model, 2 × 106 cells were injected into the tail vein of nude mice (n = 5 per group). After two months, the mice were sacrificed. Lung tissues were removed and fixed for H&E staining as previously described [20]. The numbers of pulmonary metastases were calculated under a microscope. Animal experiments were approved by the ethical committee of the First People’s Hospital of Shangqiu according to national guidelines.
Statistical analysis
Statistical analysis was assessed by GraphPad Prism software. Each experiment was performed at least three times. Data are presented as mean ± standard deviation. The statistical significance of different groups was determined using Student’s t test or a one-way analysis of variance (ANOVA). The Chi-square test was used to determine relationship between NEAT1 expression and clinicopathological features of PC patients. Kaplan-Meier method and log-rank test was used to calculate survival. Statistical significance was set at *P < 0.05, **P < 0.01 and ***P < 0.001. P < 0.05 was considered statistically significant.
Results
NEAT1 is overexpressed in PC tissues and cell lines and associated with poor prognosis
We first carried out qRT-PCR assays to measure the expression levels of NEAT1 in PC and matched neighboring normal pancreatic tissues from 60 patients with PC. As shown in Figure 1A, NEAT1 expression was overexpressed in PC tissues compared to normal tissues. We further detected the NEAT1 expression in a normal human pancreatic cell line HPDE6-C7 and six PC cell lines PANC-1, BxPC-1, BxPC-3, AsPC-1, PaCa-2 and SW1990. It was found that NEAT1 was also upregulated in PC cell lines compared with HPDE6-C7 cells (Figure 1B).
Figure 1.
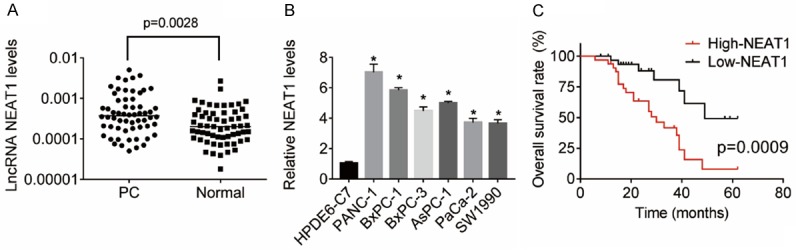
NEAT1 is overexpressed in PC tissues and cell lines and associated with poor prognosis. A. The NEAT1 expression levels in 60 pairs of PC and matched neighboring normal pancreatic tissues were determined by qRT-PCR. B. The NEAT1 expression in a normal human pancreatic cell line HPDE6-C7 and six PC cell lines PANC-1, BxPC-1, BxPC-3, AsPC-1, PaCa-2 and SW1990 was assessed by qRT-PCR. C. PC patients were divided into a high-level group (n = 30) and a low-level group (n = 30) based on the median value of NEAT1 expression in PC tissues. The Kaplan-Meier method and log-rank test was used to evaluate the relationship between NEAT1 expression and overall survival time of patients with PC.
To evaluate the relationship between NEAT1 expression and clinicopathological features, PC patients were divided into a high-level group (n = 30) and a low-level group (n = 30) based on the median value of NEAT1 expression in PC tissues. We found that NEAT1 expression was closely associated with tumor size, TNM stage, lymph node and distant metastasis (Table 1). Additionally, Kaplan-Meier analysis revealed that high-level NEAT1 expression in PC tissues markedly correlated with shorter overall survival time of patients (Figure 1C).
Table 1.
Correlation analysis between NEAT1 expression and clinicopathological features of PC patients
| Parameters | Number of cases | NEAT1 expression | P Value | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Gender | ||||
| Male | 34 | 18 | 16 | 0.602 |
| Female | 26 | 12 | 14 | |
| Age | ||||
| < 60 | 34 | 16 | 18 | 0.602 |
| ≥ 60 | 26 | 14 | 12 | |
| Tumor Size (cm) | ||||
| < 2 | 28 | 8 | 20 | 0.002 |
| ≥ 2 | 32 | 22 | 10 | |
| Differentiation | ||||
| Well/moderate | 32 | 14 | 18 | 0.301 |
| Poor | 28 | 16 | 12 | |
| TNM Stage | ||||
| I-II | 28 | 8 | 20 | 0.002 |
| III-IV | 32 | 22 | 10 | |
| Lymph node metastasis | ||||
| Positive | 27 | 19 | 8 | 0.004 |
| Negative | 33 | 11 | 22 | |
| Vascular infiltration | ||||
| Positive | 28 | 13 | 15 | 0.605 |
| Negative | 32 | 17 | 15 | |
| Distant metastasis | ||||
| Positive | 26 | 18 | 8 | 0.009 |
| Negative | 34 | 12 | 22 | |
The medium of NEAT1 expression in PC tissues was taken as cutoff.
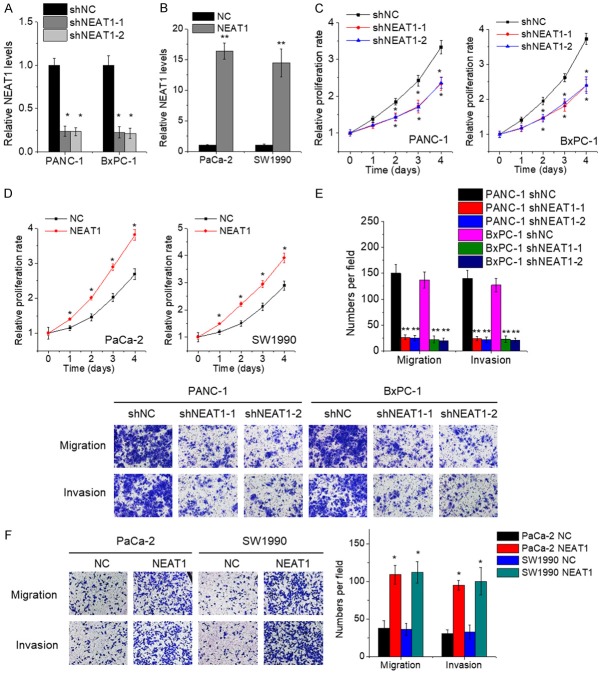
NEAT1 facilitates PC cell proliferation, migration and invasion in vitro
The effect of NEAT1 on the in vitro malignant behavior of PC cells was determined by gain- and loss-of-function approaches. PANC-1 and BxPC-1 cells endogenously expressed higher levels of NEAT1, while PaCa-2 and SW1990 cells expressed lower levels of NEAT1 (Figure 1C). Hence, we developed PANC-1 and BxPC-1 cells with NEAT1 knockdown (Figure 2A), and PaCa-2 and SW1990 cells with NEAT1 overexpression (Figure 2B). The results of CCK-8 assays showed that depletion of endogenous NEAT1 expression attenuated the proliferative capacity of PANC-1 and BxPC-1 cells compared to that of control cells (Figure 2C). Conversely, the cell proliferation was enhanced after overexpressing NEAT1 in PaCa-2 and SW1990 cells (Figure 2D). Moreover, cell migration and invasion was measured by transwell assays. The results revealed that the repression of NEAT1 inhibited the migration and invasion ability of PANC-1 and BxPC-1 cells (Figure 2E), whereas ectopic expression of NEAT1 resulted in opposite phenotypes in PaCa-2 and SW1990 cells (Figure 2F).
Figure 2.
NEAT1 facilitates PC cell proliferation, migration and invasion in vitro. A. The PANC-1 and BxPC-1 cells were transfected with lentiviral vector containing NEAT1 shRNA (shNEAT1) or negative control shRNA (shNC), and the expression of NEAT1 was detected by qRT-PCR. B. The PaCa-2 and SW1990 cells were transfected with lentiviral vector containing NEAT1 or negative control vector (NC), and the expression of NEAT1 was detected by qRT-PCR. C. The proliferation of control and NEAT1-depleted PANC-1 and BxPC-1 cells was detected by CCK-8 assay. D. The proliferation of control and NEAT1-overexpressed PaCa-2 and SW1990 cells was detected by CCK-8 assay. E. The cell migration and invasion of control and NEAT1-depleted PANC-1 and BxPC-1 cells was detected by transwell assay. F. The cell migration and invasion of control and NEAT1-overexpressed PaCa-2 and SW1990 cells was detected by transwell assay.
Depletion of NEAT1 expression inhibits PC growth and metastasis in vivo
To confirm the pro-proliferative effect of NEAT1 in vivo, control and NEAT1-depleted PANC-1 cells were subcutaneously injected into the right flank of nude mice. Tumor growth curve was measured. We observed that xenograft tumors grown from NEAT1-depleted PANC-1 cells formed more slowly, and had smaller volumes and weights than tumors formed from control cells (Figure 3A and 3B).
Figure 3.
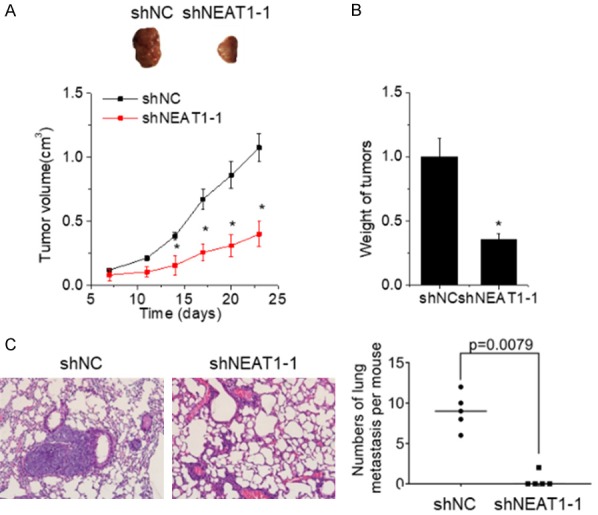
Depletion of NEAT1 expression inhibits PC growth and metastasis in vivo. (A) Xenograft tumors derived from PANC-1 cells with NEAT1 silence. Volumes of xenograft tumors derived from control and NEAT1-depleted PANC-1 cells were measured at the indicated times. (B) Tumor weight from (A) was measured. (C) Representative image of lung metastases in mice injected with control and NEAT1-depleted PANC-1 cells (left). The numbers of lung metastasis per mouse was calculated (right).
To validate the pro-metastatic effect of NEAT1 in vivo, control and NEAT1-depleted PANC-1 cells were injected into the tail vein of nude mice. Two months later, the mice were sacrificed, and the pulmonary metastasis was detected. All mice in the control group had lung metastatic nodules, but only 20% (1/5) of mice in the NEAT1-depleted group had lung metastatic nodules. The number of metastatic foci in the NEAT1-depleted group was obviously decreased compared with the control group (Figure 3C). Consistent with the in vitro results, these data suggest that NEAT1 facilitates PC growth and metastasis in vivo.
NEAT1 interacts with ELF3 mRNA and increases its stability
We then investigated whether NEAT1-mediated the stability of its interacting mRNA involved in the PC progression. An RIP assay with MS2-binding protein (MS2bp) which specifically binds RNA containing MS2-binding sequences (MS2bs) was conducted. The pCMV plasmid containing NEAT1 combined with MS2bs elements was constructed and then cotransfected into PaCa-2 cells with a plasmid expressing MS2bp-GFP. The RIP assay was then carried out using anti-GFP antibody. The mRNA pulled down by NEAT1 was then subjected to RNA sequencing analysis. Notably, E74 like ETS transcription factor 3 (ELF3) mRNA was one of the most enriched transcripts (Supplementary Figure 1). For confirmation of the interaction between NEAT1 and ELF3 mRNA, MS2-RIP was performed and the results showed that ELF3 transcripts were significantly enriched by NEAT1 compared to the empty vector MS2 or IgG (Figure 4A). The RNA pull-down assay further validated this interaction (Figure 4B).
Figure 4.
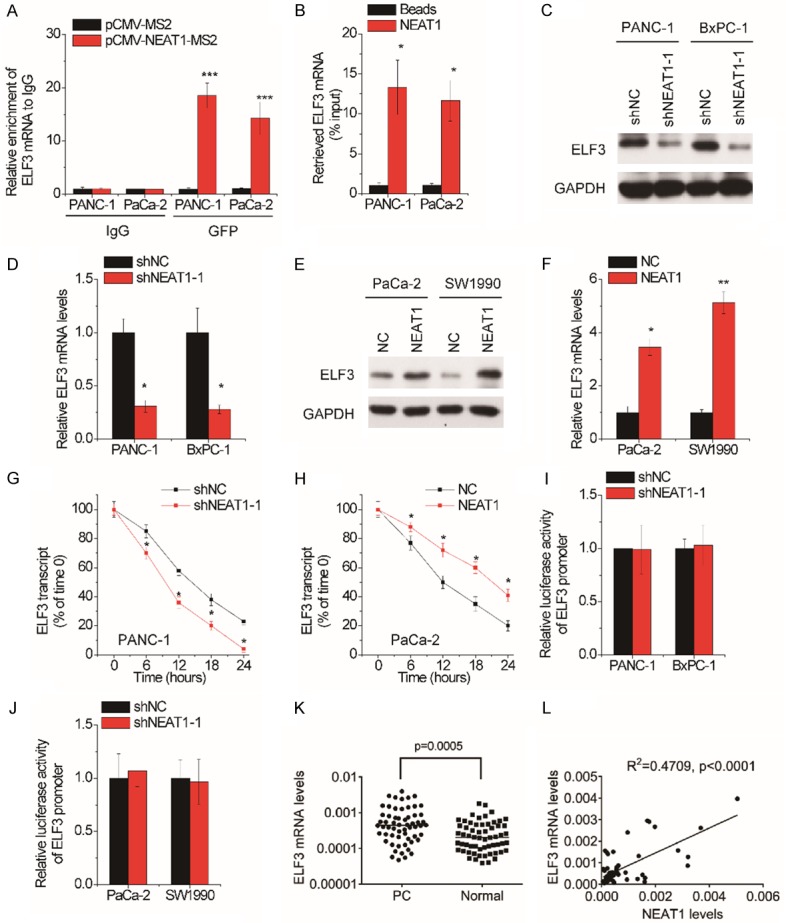
NEAT1 interacts with ELF3 mRNA and increases its stability. A. MS2-RIP assay was performed to detect the amount of ELF3 mRNA pulled down by NEAT1 in PANC-1 and PaCa-2 cells. IgG was taken as negative control. B. PANC-1 and PaCa-2 cell lysates were incubated with biotin-labeled NEAT1; after pull-down, ELF3 mRNA was determined by qRT-PCR. C. Western blot analysis of the ELF3 protein levels in control and NEAT1-depleted PANC-1 and BxPC-1 cells. D. qRT-PCR analysis of the ELF3 mRNA levels in control and NEAT1-depleted PANC-1 and BxPC-1 cells. E. Western blot analysis of the ELF3 protein levels in control and NEAT1-overexpressed PaCa-2 and SW1990 cells. F. qRT-PCR analysis of the ELF3 mRNA levels in control and NEAT1-overexpressed PaCa-2 and SW1990 cells. G. Control and NEAT1-depleted PANC-1 cells were treated with α-amanitin (50 mM) to block new RNA synthesis. The degradation of ELF3 mRNA was measured. H. Control and NEAT1-overexpressed PaCa-2 cells were treated with α-amanitin (50 mM) to block new RNA synthesis. The degradation of ELF3 mRNA was measured. I. The luciferase activity analysis of ELF3 promoter in control and NEAT1-depleted PANC-1 and BxPC-1 cells. J. The luciferase activity analysis of ELF3 promoter in control and NEAT1-overexpressed PaCa-2 and SW1990 cells. K. The ELF3 mRNA expression levels in 60 pairs of PC and matched neighboring normal pancreatic tissues were determined by qRT-PCR. L. The Pearson correlation analysis between NEAT1 and ELF3 mRNA levels in PC tissues.
In the next step, we explored the possible biological roles of the interaction between NEAT1 and ELF3 mRNA. We examined whether NEAT1 affected ELF3 expression using qRT-PCR and western blot experiments. Our findings demonstrated that knockdown of NEAT1 significantly decreased both mRNA and protein levels of ELF3 in PANC-1 and BxPC-1 cells (Figure 4C and 4D), while exogenous expression of NEAT1 increased the ELF3 expression in PaCa-2 and SW1990 cells (Figure 4E and 4F). To examine whether NEAT1 stabilized ELF3 mRNA, PC cells were treated with α-amanitin to block RNA synthesis, and the loss of ELF3 mRNA was tested by qRT-PCR. The half-life of ELF3 mRNA was shortened by NEAT1 silence in PANC-1 cells (Figure 4G). In contrast, ectopic expression of NEAT1 elongated the half-life of ELF3 mRNA in PaCa-2 cells (Figure 4H). Additionally, neither overexpression nor knockdown of NEAT1 affected the luciferase activity of ELF3 promoter (Figure 4I and 4J), supporting that NEAT1 upregulated ELF3 expression through inhibiting its degradation. The pathological correlation between NEAT1 and ELF3 was also tested. We found that ELF3 mRNA expression was upregulated (Figure 4K) and positively correlated with NEAT1 expression in PC tissues (Figure 4L).
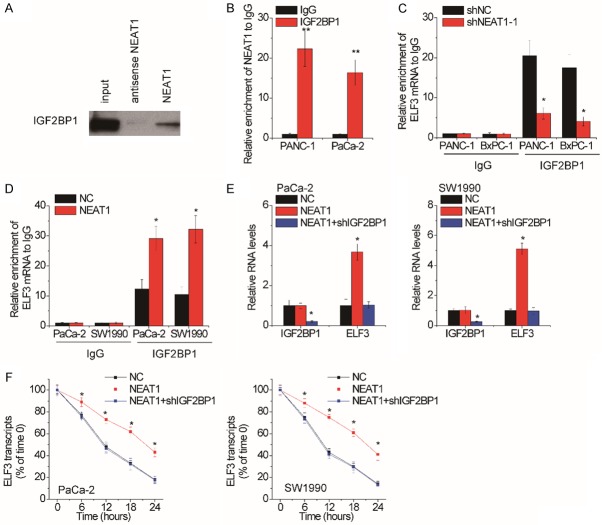
NEAT1 suppresses the degradation of ELF3 mRNA via association with IGF2BP1
Many studies have reported that RNA-binding proteins play critical roles in lncRNA-mediated mRNA stability [21-23]. To discover which RNA-binding protein involved in NEAT1-mediated ELF3 mRNA stability, we conducted RNA pull-down assay followed by mass spectrometry analysis to identify proteins interacting with NEAT1. Interestingly, insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) was identified to potentially associate with NEAT1 (Supplementary Figure 2). To validate the association between NEAT1 and IGF2BP1, the NEAT1-pull-down proteins were subjected to western blot using IGF2BP1 antibody. NEAT1 instead of its antisense RNA could pull down IGF2BP1 (Figure 5A). For further confirmation, an RIP assay using IGF2BP1 antibody was performed and the results showed that the IGF2BP1 antibody could significantly enrich NEAT1 transcripts (Figure 5B).
Figure 5.
NEAT1 suppresses the degradation of ELF3 mRNA via association with IGF2BP1. A. The biotinylated NEAT1 or antisense RNA was incubated with PANC-1 cell extracts, and associated IGF2BP1 were detected by western blot. B. RIP assay using IgG or IGF2BP1 antibody was performed in PANC-1 and PaCa-2 cells, and then the pull-down NEAT1 was detected by qRT-PCR. C. RIP assay using IgG or IGF2BP1 antibody was performed in NEAT1-depleted PANC-1 and BxPC-1 cells, and then the pull-down ELF3 mRNA was detected by qRT-PCR. D. RIP assay using IgG or IGF2BP1 antibody was performed in NEAT1-overexpressed PaCa-2 and SW1990 cells, and then the pull-down ELF3 mRNA was detected by qRT-PCR. E. The IGF2BP1 shRNA was transfected into NEAT1-overexpressed PaCa-2 and SW1990 cells, and then the ELF3 mRNA levels were assessed by qRT-PCR. F. The IGF2BP1 shRNA was transfected into NEAT1-overexpressed PaCa-2 and SW1990 cells, and then the ELF3 stability was measured.
We speculated that NEAT1 might recruit IGF2BP1 to ELF3 mRNA. To confirm this, we performed RIP assays and found that knockdown of NEAT1 attenuated the binding level of IGF2BP1 on ELF3 mRNA (Figure 5C), while overexpression of NEAT1 exerted the opposite effect (Figure 5D). Moreover, IGF2BP1 shRNA was transfected into NEAT1-overexpressed PaCa-2 and SW1990 cells. It was demonstrated that deletion of IGF2BP2 expression could abolish the NEAT1-mediated ELF3 upregulation and stability (Figure 5E and 5F). Together, these data indicate that NEAT1 stabilizes ELF3 mRNA via association with IGF2BP1.
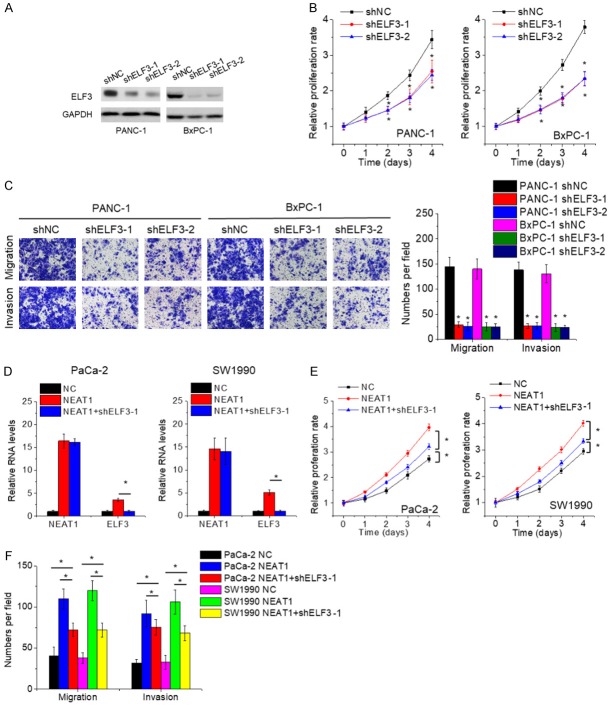
NEAT1 exerts oncogenic effects partially via ELF3
Since ELF3 could be regulated by NEAT1, we explored the effects of ELF3 on PC cells. We developed PANC-1 and BxPC-1 cells with ELF3 silence (Figure 6A). Results of CCK-8 and transwell assays demonstrated that inhibition of ELF3 suppressed cell proliferation, migration and invasion (Figure 6B and 6C), which was similar to the phenotypes induced by NEAT1 knockdown. Similarly, suppression of IGF2BP1 resulted in the same phenomenon (Supplementary Figure 3A and 3B).
Figure 6.
NEAT1 exerts oncogenic effects partially via ELF3. A. The PANC-1 and BxPC-1 cells were transfected with lentiviral vector containing ELF3 shRNA (shELF3) or negative control shRNA (shNC), and the expression of ELF3 was detected by qRT-PCR. B. The proliferation of control and ELF3-depleted PANC-1 and BxPC-1 cells was detected by CCK-8 assay. C. The cell migration and invasion of control and ELF3-depleted PANC-1 and BxPC-1 cells was detected by transwell assay. D. ELF3 shRNA was transfected into NEAT1-overexpressed PaCa-2 and SW1990 cells, and then the ELF3 mRNA levels were assessed by qRT-PCR. E. ELF3 knockdown partially abolished the NEAT1-mediated promotion of PaCa-2 and SW1990 cell proliferation. F. ELF3 knockdown partially abolished the NEAT1-mediated promotion of PaCa-2 and SW1990 cell migration and invasion.
Finally, to assess the role of ELF3 in the oncogenic effects of NEAT1, rescue experiments were conducted. We employed the shRNA to abolish ELF3 elevation in NEAT1-overexpressed PaCa-2 and SW1990 (Figure 6D). The proliferation, migration and invasion ability enhanced by NEAT1 was repressed by ELF3 downregulation in part (Figure 6E and 6F). Moreover, depletion of ELF3 expression strengthened the proliferation, migration and invasion inhibition mediated by NEAT1 knockdown (Supplementary Figure 3C and 3D). Together, our data suggest that NEAT1 exerts oncogenic function at least in part via regulating ELF3 expression.
Discussion
The molecular mechanism of the protein-coding genes in cancer was well documented before the discovery of nonconding RNAs. Recently, abnormal expression of lncRNAs has been found in PC tissues and cells, such as GSTM3TV2, PXN-AS1, MTA2TR and PVT1 [24-27]. Here, our research validated a significant increase of NEAT1 expression in PC tissue samples and cell lines. Moreover, overexpression of NEAT1 was significantly associated with larger tumor size, advanced TNM stage, lymph node and distant metastasis, as well as predicted poorer prognosis of PC patients. These findings indicated that NEAT1 may serve as a new diagnostic and unfavourable prognostic marker for patients with PC.
Quite a few lncRNAs have been identified to affect gene expression through transcription control, chromatin modification, or acting as ceRNA to sponge microRNAs, implying that the dysregulated lncRNAs exert crucial regulatory functions in tumor progression [7]. However, to date, little is known about the involvement of NEAT1 in PC tumorigenesis and progression. Cao et al. reported that NEAT1 upregulates c-met by competitively binding miR-335-5p in PC cells [28]. Another group also demonstrated that NEAT1 acts a ceRNA for miR-506-3p, thus facilitating PC proliferation [13]. Here, our study revealed a novel mechanism that NEAT1 stabilized its interacting mRNA to regulate PC progression. The results of MS2-RIP and RNA pull-down assays identified a direct interaction between NEAT1 and ELF3 mRNA. NEAT1 could not affect the ELF3 transcription, but suppressed the degradation of ELF3 mRNA. Mechanistically, NEAT1 associated with RNA-binding protein IGF2BP1 and promoted the binding of IGF2BP1 to ELF3 mRNA, thus stabilizing ELF3 mRNA. IGF2BP1 is a conserved RNA-binding protein which play an important role in post-transcriptional regulation of its target mRNA. IGF2BP1 functions as an oncogene via associating with the mRNA of classical oncogenes and suppressing their degradation, such as GLI1, c-Myc, MDR1, and KRAS [29,30]. Recently, some lncRNAs have been revealed to participate in the regulation of IGF2BP1. For example, LINC01093 directly binds IGF2BP1, which attenuates the interaction between IGF2BP1 and GLI1 mRNA and subsequently induces the degradation of GLI1 mRNA [21]. Moreover, GHET1 associates with IGF2BP1 and then enhances its interaction with c-Myc mRNA, consequently delaying the degradation of c-Myc mRNA [22]. Combined with these studies, our research highlighted the functional significance of lncRNAs in the mRNA stability regulated by IGF2BP1. Notably, IGF2BP1 recognizes the consensus N6-methyladenosine (m6A) sequence of mRNAs and recruits co-factor ELAVL1 to protect m6A-containing mRNAs from degradation [31]. Hence, we speculated that NEAT1 might also affect the m6A modification of ELF3 mRNA, which needs further exploration.
ELF3 is a member of the epithelial-specific subfamily of ETS transcription factors. The function of ELF3 is paradoxical in different types of cancer. In hepatocellular carcinoma, ELF3 promotes epithelial-mesenchymal transition (EMT) and metastasis via inhibiting miR-141-3p transcription [32]. However, ELF3 acts a tumor suppressor inhibiting proliferation and EMT in ovarian cancer cells [33]. Here, we found that ELF3 expression was upregulated in PC tissues. Knockdown of ELF3 significantly repressed PC cell proliferation, migration and invasion, suggesting that ELF3 functions as an oncogene in PC.
Conclusion
Collectively, this research reveals that NEAT1 as an oncogenic lncRNA to facilitate PC growth and metastasis in an IGF2BP1/ELF3-dependent manner. These findings imply that NEAT1 may be a promising effective target for patients with PC.
Disclosure of conflict of interest
None.
Abbreviations
- NEAT1
Nuclear-enriched abundant transcript 1
- ELF3
E74 like ETS transcription factor 3
- IGF2BP1
Insulin-like growth factor 2 mRNA-binding protein 1
- ceRNA
Competing endogenous RNA
- RIP
RNA immunoprecipitation
- MS2bp
MS2-binding protein
Supporting Information
References
- 1.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aier I, Semwal R, Sharma A, Varadwaj PK. A systematic assessment of statistics, risk factors, and underlying features involved in pancreatic cancer. Cancer Epidemiol. 2019;58:104–110. doi: 10.1016/j.canep.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Melisi D, Calvetti L, Frizziero M, Tortora G. Pancreatic cancer: systemic combination therapies for a heterogeneous disease. Curr Pharm Des. 2014;20:6660–6669. doi: 10.2174/1381612820666140826154327. [DOI] [PubMed] [Google Scholar]
- 4.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34:153–180. doi: 10.1080/02648725.2018.1471566. [DOI] [PubMed] [Google Scholar]
- 6.Barangi S, Hayes AW, Reiter R, Karimi G. The therapeutic role of long non-coding RNAs in human diseases: a focus on the recent insights into autophagy. Pharmacol Res. 2019;142:22–29. doi: 10.1016/j.phrs.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8 doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW, Wang X, Jin H, Kwong A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10:270. doi: 10.1038/s41419-019-1513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Z, Dang J, Song A, Cui X, Ma Z, Zhang Z. NEAT1 promotes colon cancer progression through sponging miR-495-3p and activating CDK6 in vitro and in vivo. J Cell Physiol. 2019;234:19582–19591. doi: 10.1002/jcp.28557. [DOI] [PubMed] [Google Scholar]
- 11.Xia Y, Zhou Y, Han H, Li P, Wei W, Lin N. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J Cell Physiol. 2019;234:19592–19601. doi: 10.1002/jcp.28559. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Xia X. Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. J Cell Physiol. 2019 doi: 10.1002/jcp.27567. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Liu C, Wu Q, Zhang J, Min Q, Sheng T, Wang X, Zou Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482:828–834. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, Zhou J, Kang C, Li M, Jiang C. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am J Cancer Res. 2018;8:81–90. [PMC free article] [PubMed] [Google Scholar]
- 16.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, Byrom D, Riera A, Rossell D, Mangues R, Massague J, Sancho E, Batlle E. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Bian C, Yuan L, Gai H. A long non-coding RNA LINC01288 facilitates non-small cell lung cancer progression through stabilizing IL-6 mRNA. Biochem Biophys Res Commun. 2019;514:443–449. doi: 10.1016/j.bbrc.2019.04.132. [DOI] [PubMed] [Google Scholar]
- 19.Xie CR, Wang F, Zhang S, Wang FQ, Zheng S, Li Z, Lv J, Qi HQ, Fang QL, Wang XM, Yin ZY. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol Ther Nucleic Acids. 2017;9:440–451. doi: 10.1016/j.omtn.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y, Zhang F, Lu Y, Zheng L, Zhang W, Li X, Li X. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376:62–73. doi: 10.1016/j.canlet.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 21.He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z, Deng X, Yang C, Ruan H, Yu C, Zhao F, Yao M, Fang J, Gu J, Zhou J, Fan J, Qin W, Yang XR, Wang H. A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019;450:98–109. doi: 10.1016/j.canlet.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 23.Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, Liu B, Sun S, Yang F, Wang L, Qu L. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10:154. doi: 10.1038/s41419-019-1331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong G, Liu C, Yang G, Feng M, Xu J, Zhao F, You L, Zhou L, Zheng L, Hu Y, Wang X, Zhang T, Zhao Y. Long noncoding RNA GSTM3TV2 upregulates LAT2 and OLR1 by competitively sponging let-7 to promote gemcitabine resistance in pancreatic cancer. J Hematol Oncol. 2019;12:97. doi: 10.1186/s13045-019-0777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J, Jia Y, Chen H, Chen W, Zhou X. Long non-coding RNA PXN-AS1 suppresses pancreatic cancer progression by acting as a competing endogenous RNA of miR-3064 to upregulate PIP4K2B expression. J Exp Clin Cancer Res. 2019;38:390. doi: 10.1186/s13046-019-1379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, Zhong JX, Deng SJ, Liu ML, Huang K, Li Q, Li W, Hu YH, Wang F, Wang CY, Zhao G. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1alpha. Theranostics. 2019;9:5298–5314. doi: 10.7150/thno.34559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M, Wei F, Wu J, Jiang J, Jia Y, Mo F, Zhang S, Liang X, Mou X, Tang J. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0381-y. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao J, Zhang Y, Yang J, He S, Li M, Yan S, Chen Y, Qu C, Xu L. NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335-5p/c-met axis. Am J Cancer Res. 2016;6:2361–2374. [PMC free article] [PubMed] [Google Scholar]
- 29.Noubissi FK, Goswami S, Sanek NA, Kawakami K, Minamoto T, Moser A, Grinblat Y, Spiegelman VS. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 2009;69:8572–8578. doi: 10.1158/0008-5472.CAN-09-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, Huttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Huttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang Y, Zhou S, Cen D, Ji L, Han W, Cai X. ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 2018;9:387. doi: 10.1038/s41419-018-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeung TL, Leung CS, Wong KK, Gutierrez-Hartmann A, Kwong J, Gershenson DM, Mok SC. ELF3 is a negative regulator of epithelial-mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:16951–16963. doi: 10.18632/oncotarget.15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.